Abstract
Background:
DNA is generally considered the ultimate target of cisplatin, so DNA repair has become the hallmark for cisplatin chemoresistance that is attributed to the poor overall survival (50%) among patients with head and neck cancer (HNC). As the efficacy of cisplatin is dose-dependent, we conducted the first study in an Asian population to characterize the DNA repair genes ACTL6A and ERCC1 based on the dosing of cisplatin-based chemoradiotherapy (CRT).
Methods:
Locally advanced HNC (LAHNC) patients who were planning to undergo cisplatin-based CRT were enrolled in a prospective study to quantify the dose-dependent expressions of ACTL6A and ERCC1 from peripheral blood mononuclear cells via quantitative polymerase chain reaction; these results were integrated with computational analysis and systematic review/meta-analysis to formulate evidence-based translation decisions. The Friedman test and Wilcoxon’s test were used to compare the expressions of the two genes before and after CRT, and Spearman’s rank correlation was used to find the correlation between ACTL6A and ERCC1 expressions. All statistical analyses were performed using SPSS version 29.
Results:
A total of 77 LAHNC patients were enrolled in this study, of which 96.1% were men and 3.9% were women with a mean age of 52.88 ± 9.68 years. The median expressions of ERCC1 were significantly increased (p < 0.001) after 50% (0.19) and 100% CRT (0.23) compared to the baseline value (0.14), whereas ACTL6A expression decreased from 4.77 to 3.87 after 50% CRT (p < 0.05) and increased to 5.43 after 100% CRT. From the computational analysis, ACTL6A and ERCC1 were found to be overexpressed among HNC patients and observed to regulate 10 repair pathways. Overexpressions of ERCC1 and ACTL6A were predicted to infiltrate the tumors with CD4+ cells, macrophages, dendritic cells, and B cells. The hazard ratios for overall survival were found to be 1.67 among the ACTL6A overexpressed and 1.82 among the ERCC1 overexpressed HNC patients via computational analysis and meta-analysis, respectively. Furthermore, FDA-approved drugs like gemcitabine and panobinostat were found to be the best candidates for downregulating ERCC1 and ACTL6A expressions based on binding affinities of −3.707 and −4.198 kcal/mol, respectively.
Conclusion:
The increased expressions of ACTL6A and ERCC1 during/after cisplatin-based CRT are expected to mediate DNA repair leading to chemoresistance, which could result in poor overall survival in HNC patients. Thus, FDA-approved drugs like panobinostat and gemcitabine can be repurposed to target the chemoresistance genes ACTL6A and ERCC1, respectively.

Highlights
• Of the 77 LAHNC patients in the study cohort, men outnumbered women and had a mean age of 52.88 ± 9.68 years.
• ACTL6A expression increased after CRT (5.43) compared to the baseline value (4.77).
• ERCC1 expression significantly increased with CRT, indicating high nucleotide excision repair capacity.
• ERCC1/ACTL6A overexpressions were linked to poor overall survival (hazard ratio: 1.82/1.67).
• Gemcitabine and panobinostat can downregulate ERCC1 and ACTL6A, respectively.
1 Introduction
Head and neck cancer (HNC) refers to a group of heterogenous malignancies generally originating from the mucosal epithelial regions of the head and neck, such as the oral cavity, pharynx, larynx, oro/hypo/naso-pharynx, and salivary glands (Chaudhary et al., 2023). According to GLOBOCAN 2022, HNC has collectively secured the top spot among Indian patients in terms of incidence (17.53% or 247,924 new cases), 5-year prevalence (18.94% per 100,000 out of 613,841), and mortality (15.05% or 137,925) (Global Cancer Observatory, 2024: India Fact Sheet). Alcohol consumption, tobacco use (smoke/smokeless), poor oral hygiene, viral infections (human papilloma virus/Epstein–Barr virus), altered expressions of tumor suppressors, and oncogenes are the predominant etiopathophysiological factors associated with the development of HNC (Chaudhary et al., 2023). Localized/early-stage (stages I/II) HNCs are generally managed through surgery or radiation therapy, whereas locally advanced HNC (LAHNC) is generally managed using concurrent chemoradiotherapy (CCRT) with/without surgery (Chaudhary et al., 2023). Approximately 66.6% of the Indian population of HNC patients are diagnosed at the locally advanced stage, which makes CCRT with/without surgery as the popular choice of treatment among clinicians (Chaudhary et al., 2023; Mathur et al., 2020). Thus, chemotherapy serves as the cornerstone of the treatment strategy for managing HNC.
Cisplatin is the most widely preferred and broad-spectrum frontline dose-dependent antineoplastic drug in HNC that exerts its anticancer effects via the formation of interstrand and intrastrand cross-linking with nuclear/mitochondrial DNA at the N7 positions of adenine and guanine, thereby arresting the cell cycle at the G2 phase and causing apoptosis to interfere with DNA repair (Chaudhary et al., 2023; Ranasinghe et al., 2022). In addition, aqueous cisplatin is known to enhance the mitochondrial outer membrane permeabilization, which further induces the caspases and causes apoptosis of tumor cells through the release of protein cytochrome c into the cytoplasm (Kanno et al., 2021). Generally, once-weekly intravenous administration of cisplatin at 30–40 mg/m2 for 6–7 weeks has been proven to be the best alternative to 3-weekly intravenous administration of cisplatin at 100 mg/m2 as the former is associated with minimal toxicity (Chaudhary et al., 2023; NCCN Guidelines, 2024). Furthermore, cisplatin is often combined with other anticancer agents, such as paclitaxel, docetaxel, 5-fluorouracil (TPF regimen), hydroxyurea, etoposide, pembrolizumab, nivolumab, and cetuximab, to manage locally LAHNC and recurrent/metastatic HNC (R/MHNC) (Chaudhary et al., 2023; NCCN Guidelines, 2024). However, it is disheartening that almost 65% of LAHNC patients do not reap any benefits from such therapy, which is attributable to the recurrence, metastasis, and poor survival among LAHNC patients (Chaudhary et al., 2023; Mathur et al., 2020). Furthermore, approximately 70%–90% of R/MHNC patients do not respond to immunotherapy (Chaudhary et al., 2023). Collectively, these hurdles in the management of HNC have resulted in poor 5-year overall survival rates (50%) (Gormley et al., 2022). According to the Surveillance, Epidemiology, and End Results (SEER) registry, there has been a modest increase in the 5-year relative survival rate among HNC patients to approximately 65.25% between 2014 and 2020 (i.e., 5-year relative survival rates of oral cavity and pharynx cancer is 69% and larynx cancer is 61.5%) (SEER Cancer Stat Facts, 2024). The mortality rate of Indian patients accounts for approximately 71% of all HNC-related deaths in southeast Asia and 28% globally (Chauhan et al., 2018). The disease burden of HNC and its ineffective response to cisplatin have necessitated investigations into the causes behind the limited benefits of cisplatin therapy, which are achieved by exploring the possible biological markers involved in the molecular mechanisms of chemoresistance (Kanno et al., 2021).
Chemoresistance is a multifaceted condition that is often associated with increased DNA repair, deregulated influx/efflux pump, enzymatic inactivation of drugs, aberrant autophagy and apoptosis, regulation of EGFR/FAK/NF-kB pathways, cancer stem cells, and metabolic reprogramming (Ranasinghe et al., 2022; Kanno et al., 2021; Hu et al., 2023). As DNA is the ultimate target of cisplatin therapy, the pathways associated with repair of damaged DNA are crucially linked to cisplatin resistance. Nucleotide excision repair (NER) is a crucial DNA repair pathway that is responsible for clearing cisplatin-DNA adducts compared to other repair pathways, such as double-strand break repair, mismatch repair, and base excision repair (Kanno et al., 2021). NER is further subdivided into two important pathways, namely, the transcription-coupled repair (TCR-NER) and global genome repair (GGR-NER) pathways (KEGG Pathway, 2024: map03420). The current study explores the roles of the excision repair cross-complementation group1 (ERCC1) and actin-like protein 6A (ACTL6A) genes as attractive biological markers associated with DNA repair in HNC.
ERCC1 and ACTL6A are the core proteins of the NER pathway and switch/sucrose non-fermentable (SWI/SNF) complex, respectively (Kanno et al., 2021; Xiao et al., 2021). ERCC1 is a catalytically inactive protein that is capable of initiating DNA/protein–protein interaction (PPI) binds that can cause XPF activation and form the ERCC1-XPF1 heterodimer. Collectively, the ERCC1-XPF1 endonuclease protein complex is responsible for detection and repair of DNA damage. ERCC1 is a high-capacity gene of the NER pathway that mediates cisplatin resistance in HNC patients (Prochnow et al., 2019). However, there exist controversies regarding its expression and clinical significance. Recently, the novel oncogene ACTL6A (a subunit of the SWI/SNF complex) has garnered attention for its DNA repair capacity (Xiao et al., 2021). Biologically, ACTL6A has been reported to be involved in chromatin remodeling and transcription regulation. ACTL6A encodes for the actin-related proteins comprising actin folds that are responsible for the binding and hydrolysis of adenosine triphosphate to remodel chromatin and promote gene expression by enhancing DNA accessibility (Xiao et al., 2021; Dang et al., 2020). Thus, ACTL6A mediates DNA repair via utilization of the SWI/SNF complex that might also promote such repair via NER (Dang et al., 2020). However, this mechanism remains unresolved.
The formation of DNA–cisplatin adducts as well as the anticancer efficacy of cisplatin are attributed to the therapeutic dose administered (Ranasinghe et al., 2022). Thus, it is crucial to determine the biomarkers for cisplatin resistance in relation to the therapeutic dose. Furthermore, investigating the expressions of the chemoresistance genes from blood samples before and after therapy is less invasive, inexpensive, easy, less time-consuming, and ethically safe compared to using tissue samples that are difficult to obtain after therapy as this procedure may disturb the healing process or trigger recurrence, causing harm to the patient. Till date, there are no reported studies on detecting ACTL6A expression and very few studies on detecting ERCC1 expression from blood samples. Furthermore, there are no available studies on characterizing the dose-dependent expressions of ACTL6A and ERCC1 in HNC patients. Thus, to the best of our knowledge, this is the first study on Asian subjects to demonstrate the dose-dependent expressions of ERCC1/ACTL6A (zero cisplatin dose (zero cycle: 0 mg/m2), after administration of 50% dose (3-cycle cisplatin: 90 mg/m2), and after last cycle of cisplatin (4- or 5- or 6-cycle cisplatin: 120 or 150 or 180 mg/m2)) and their correlations at these three phases. Additionally, computational analysis and meta-analysis were performed to investigate regulation of the repair pathways through ACTL6A and ERCC1 interactions with the platinum resistance genes to understand their expression patterns and impacts on overall survival to establish ACTL6A and ERCC1 as the chemoresistance genes. The detailed workflow of the present study is depicted in Figure 1.
FIGURE 1
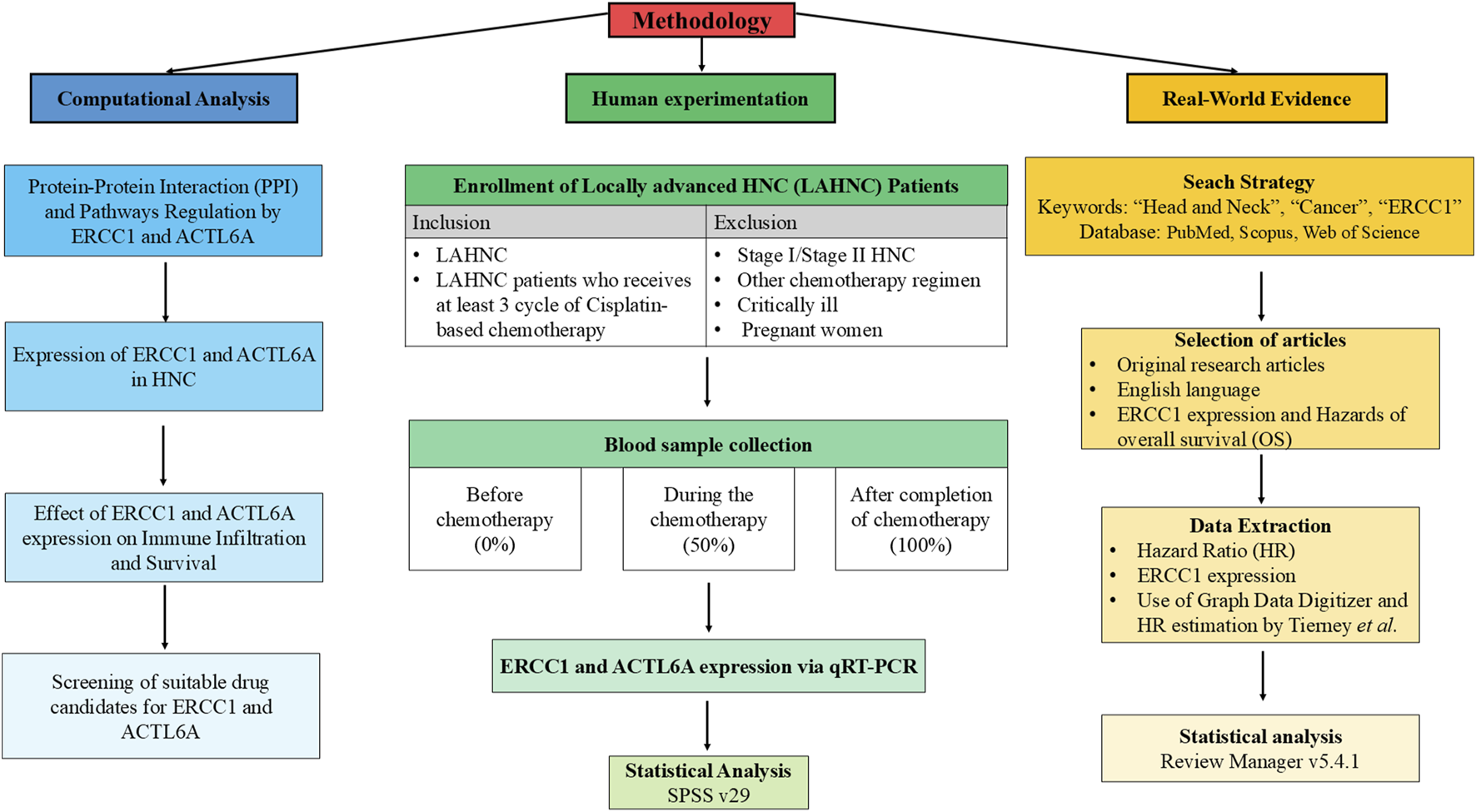
Detailed workflow showing integration of human experimentation with computational analysis and meta-analysis.
2 Materials and methods
2.1 Computational analysis to investigate ACTL6A and ERCC1 in cisplatin resistance
2.1.1 PPIs and pathway regulations of ERCC1 and ACTL6A
In a previous study, we identified 21 genes that were regulated in the platinum drug resistance pathway (ERCC1, MAPK1, MLH1, MDM2, PIK3CA, TP53, ERBB2, BAX, GSTM1, FAS, CASP8, FASLG, ABCC2, XIAP, BCL2, GSTP1, CDKN1A, TOP2A, CDKN2A, BRCA1, and BIRC2) and five hub genes in cisplatin resistance (CCND1, AXL, CDKN2A, TERT, and EZH2), among which ERCC1 was the only NER gene that was regulated for cisplatin resistance (Chaudhary et al., 2023). Furthermore, it has been reported that ACTL6A may contribute to DNA repair via the NER pathway, but its exact mechanism remains a mystery (Xiao et al., 2021). Thus, we investigated the interactions of ACTL6A with these 21 genes and the NER genes to clearly map the contributions of ERCC1/ACTL6A in DNA repair using STRING version 12.0 (https://string-db.org/). Furthermore, an unsupervised analysis was performed via K-means clustering to obtain similar protein clusters (https://string-db.org/) (Szklarczyk et al., 2023).
2.1.2 mRNA and tissue expressions of ERCC1 and ACTL6A in HNC
Overexpression of the DNA repair genes could contribute to the development of cisplatin resistance. Thus, ERCC1 and ACTL6A were investigated for their mRNA- and tissue-level expressions using the UALCAN database (https://ualcan.path.uab.edu/analysis.html) (Chandrashekar et al., 2022, 2017) and Human Protein Atlas (https://www.proteinatlas.org/; The Human Protein Atlas, 2024), respectively. Furthermore, the ERCC1 and ACTL6A genes were queried as target inputs in the muTarget platform to identify the top-5 genes undergoing somatic mutations with prevalence rates of at least 1% among the HNC patients while significantly overexpressing ERCC1 and ACTL6A (https://www.mutarget.com/; Nagy and Győrffy, 2021). Here, muTarget is a platform that links gene expressions with the mutation statuses of the provided genes in solid cancers.
2.1.3 Effects of ERCC1 and ACTL6A expressions on immune infiltration and survival
The infiltration of cancer cells is often linked to compromised tumor responses to anticancer agents, leading to poor clinical outcomes. Thus, the impacts of ERCC1 and ACTL6A expressions on immune cell infiltration in HNC were predicted using TIMER 2.0 (http://timer.cistrome.org/; Li et al., 2020); further, their effects on the overall survival of LAHNC patients (stages III and IV) were investigated using the Kaplan–Meier plotter (https://kmplot.com/analysis/; Győrffy, 2024). The survival analysis was independent of therapy as the Kaplan–Meier plotter database does not have the option to restrict analysis based on treatment time framework, i.e., pre- and post-therapy.
2.1.4 Screening and binding of suitable drug candidates for ERCC1 and ACTL6A
The DNA repair genes ERCC1 and ACTL6A were screened for possible interactions with suitable FDA-approved or non-approved drug candidates using DGIbd (https://www.dgidb.org/; Cannon et al., 2024). Furthermore, the drug candidates were docked against their corresponding targets (ERCC1 and ACTL6A) using Schrodinger version 2022-1 (https://www.schrodinger.com/) to investigate potential molecules other than platinum drugs that could downregulate ERCC1 and ACTL6A. The protein structures of ERCC1 (PDBID: 2A1I) and ACTL6A (PDBID: 9C4B) were obtained from protein data bank (https://www.rcsb.org/; RCSB PDB, 2024), and the structures of their corresponding drug candidates were obtained from PubChem database (https://pubchem.ncbi.nlm.nih.gov/; PubChem, 2024).
2.2 Human study for ERCC1 and ACTL6A expressions among HNC patients
A prospective observational study was carried out at the Department of Oncology at a tertiary care hospital, where LAHNC patients above 18 years of age who were planning to undergo cisplatin-based CRT were enrolled after obtaining written informed consent. However, HNC patients with localized tumors (stage I/II) or those who were scheduled for other treatment modalities, critically ill patients, and pregnant women were excluded from the study. The study was initiated after obtaining approval from the Central Ethics Committee of the university (Ref. no. NU/CEC/2022/307 dated 21 September 2022 and revised on 31 January 2024 with Ref. no. NU/CEC/2024/526) and was also registered as a clinical trial in India (CTRI/2022/10/046142).
The sample size was calculated using the following formula for HNC prevalence of 30% (Dandekar et al., 2017; Prabhash et al., 2020) (P = 0.3) and marginal error of 9% (d = 0.09) at the 95% confidence interval (CI; Zα/2 = 1.96). Thus, the total number of HNC patients was calculated to be 99.59 (rounded to 100). However, only 66.6% of the people in this population 100 belong to the LAHNC group (Mathur et al., 2020). Hence, the minimum sample size required for the study was 67 (N). The final sample size to be enrolled was estimated to be 77 after adjusting the study population for a 15% dropout rate.
2.2.1 Blood sampling and clinical data collection
All LAHNC patients who were enrolled in the study had been scheduled to receive CCRT, i.e., six cycles of cisplatin at 30 mg/m2 weekly along with radiation of 60–70 Gy. After obtaining the informed consent and enrolling the participants, approximately 2 mL of peripheral blood sample was collected from each LAHNC patient in EDTA vacutainers and stored at −80°C. The patient blood was sampled at three different phases, i.e., zero cisplatin (before initiation of cisplatin-based CRT), after 50% of the planned cisplatin was administered (after third cycle of cisplatin therapy), and after completion or last cycle of cisplatin therapy. For patients who received only three cycles of cisplatin therapy, the third phase of blood sampling was conducted after completion of radiation therapy. Furthermore, we collected the demographic details and clinical characteristics of the patients.
2.2.2 Primer selection, verification, and confirmation for ERCC1 and ACTL6A genes
The primers for the quantitative real-time polymerase chain reaction (qRT-PCR) were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/; PrimerBank-MGH-PGA, 2023) and verified with the protein-coding regions of the cDNA sequences for selected transcripts of ERCC1 and ACTL6A from the Ensembl database (https://www.ensembl.org/; Ensembl genome browser, 2023). The amplicon size of the primer selected for ERCC1 was 175 base pairs (forward: TTTGGCGACGTAATTCCCGAC; reverse: CCTGCTGGGGATCTTTCACA) and that for ACTL6A was 83 base pairs (forward: GACAGCATTTGCTAATGGTCGT; reverse: CATCGTGGACTGGAATTGCAG); further, the predesigned primer for β-actin (ACTB) had 249 base pairs (forward: CATGTACGTTGCTATCCAGGC; reverse: CTCCTTAATGTCACGCACGAT). The primers for ERCC1 and ACTL6A along with the predesigned ACTB were confirmed experimentally via conventional PCR followed by DNA gel electrophoresis.
2.2.3 ERCC1 and ACTL6A expressions via PCR
2.2.3.1 Total RNA extraction
The blood samples were centrifuged at 3,000 rpm to separate the plasma, followed by treatment of the blood cells with 1× RBC lysis solution. The mixture of blood cells and RBC lysis solution was left for 15–20 min to ensure RBC lysis and then centrifuged at 3,000 rpm to obtain Peripheral blood mononuclear cells (PBMC). The PBMC were used to extract the total RNA via the TRIzol reagent method using the RNAiso Plus kit (Takara, cat. no. 9109_v201904Da). The purity and concentration of the extracted RNA were confirmed via the nanodrop method.
2.2.3.2 cDNA synthesis
cDNA was synthesized from the total RNA using the PrimeScript™ RT reagent kit (Takara, cat. no. RR037A_v202008Da) in a thermal cycler (Prima 96, HiMedia, India), i.e., reverse transcription was performed at 55°C for 60 min, followed by inactivation of reverse transcriptase at 87°C for 5 s and 4°C thereafter. The synthesized cDNA was stored at −20°C.
2.2.4 qRT-PCR
The expressions of ERCC1 and ACTL6A were obtained using TB Green Premix EX Taq (Tli RNase H Plus, Takara, cat. no. RR820A_ v201903Da) and quantified with the Applied Biosystems™ QuantStudio™ 6 RT-PCR System. The cDNA templates of the targets (ERCC1 and ACTL6A) and reference (ACTB) were amplified using the QuantStudio™ system and SYBR Green PCR Master Mix fluorochrome dye. The qRT-PCR involved three stages: initial denaturation at 95°C for 3 min; PCR-based quantification at 95°C for 15 s followed by 60°C for 30 s and 72°C for 40 s (40 cycles); melting curve at 95°C for 15 s and 60°C for 1 min followed by 95°C for 15 s. The baseline and follow-up samples from a particular patient were processed together to avoid technical errors with the gene expressions. In addition, all samples were processed in duplicate along with RNAase-free water as the negative control.
3 Relative gene expressions and statistical analysis
The relative expressions of ERCC1 and ACTL6A at baseline, after the third cycle of cisplatin therapy, and after the last cycle of cisplatin therapy were estimated by comparing the cycle threshold (Ct) value of a given sample for a particular gene of interest (GOI) (ERCC1 and ACTL6A) with the Ct value of a given sample for the reference gene (ACTB). The relative expressions of the GOIs compared to the reference were calculated using the 2−△CT formula [△CT = Ct (GOI) – Ct (ACTB)]. The patient data were analyzed using descriptive statistics (mean, standard deviation, frequency, percentage, and interquartile range) and were checked for normal distribution using the one-sample Kolmogorov–Smirnov test (p < 0.05). The ERCC1 and ACTL6A expressions between the baseline and follow-ups were compared using the Friedman test. Furthermore, paired comparisons were performed via Wilcoxon’s test. Correlations between ERCC1 and ACTL6A expressions were examined using Spearman’s rank test. All statistical analyses were performed using SPSS version 29 and figure was constructed using GraphPad Prism 8.0.2.
4 Real-world evidence for ERCC1/ACTL6A expressions and survival in HNC via meta-analysis
4.1 Research question and registration
Previous published meta-analyses by Xuelei et al. (2015) and Bišof et al. (2016) revealed that overexpression of ERCC1 is the root cause of unfavorable overall survival outcomes (hazard ratio (HR): 2.14 and 1.95) among HNC patients, with the Asian population being the most affected victims (HR: 2.97 and 3.13, respectively). Thus, we conducted a further meta-analysis to update the predictive value of ERCC1 on overall survival. This meta-analysis was registered prospectively with the International Prospective Register of Systematic Review (PROSPERO, 2024) under the title “Impact of ERCC1 expression on overall survival rate in head and neck cancer” with the registration ID CRD42024542859. However, data on the expression of ACTL6A and its impact on the survival of HNC patients are unavailable; thus, we could not conduct a meta-analysis for ACTL6A.
4.2 Search strategy
An electronic search was conducted for articles in the PubMed, Scopus, and Web of Science databases. The search strategy involved a combination of the following keywords: “Head and Neck,” “ERCC1,” “Cancer.” Further, “Head and Neck,” “ACTL6A,” and “Cancer” were used as the keywords to retrieve articles related to the ACTL6A gene in HNC (see also the Supplementary File). In addition, the reference citations in these articles were manually checked for additional studies. Rayyan Software was used to import and manage the articles.
4.3 Eligibility criteria and selection process
The studies included in this meta-analysis/review were original research articles published in English language that evaluated the relationships between overall survival rate and expression of ERCC1 or ACTL6A in HNC. After removing duplicate articles, the title and abstract were screened for eligibility by two independent authors. Any disagreements between these authors were resolved by a third author after a consensus discussion. Later, the selected studies were assessed for eligibility by two different authors based on the full text, and any disagreements were resolved by a third author. Articles without full text and ineligible articles were excluded from the study.
4.4 Data extraction
Data were extracted from eligible articles using the predesigned proforma containing the following information: author details, country, year of publication, sample size, gender, age, study design, disease details, TNCM/clinical staging, molecular technique used, ERCC1 expression, and outcomes of the study. The data were extracted by two independent authors, and any disagreements were resolved by a third author. If the survival data were represented using the Kaplan–Meier curve, and then the relevant information was interpreted from the graph using Graph Data Digitizer 2.4. If the HR was not reported by the authors, then it was estimated using the method proposed by Tierney et al. (2007).
4.5 Statistical analysis
Review Manager software v5.4.1 (Cochrane Collaboration, Copenhagen, Denmark) was used to generate the forest plots, and the inverse variance method was used for pooled estimates. The outcome variables of all the included studies were represented in terms of the HR and 95% CI. The analyses were performed using RevMan calculator by incorporating the log(HR) with standard error (SE), and the results were presented as HR with 95% CI. All results were presented graphically and numerically in the forest plot along with the weights imparted by the individual studies. The Higgins I2 statistic and visual inspection were used to assess heterogenicity, and the percentage with p-value was used to represent interstudy variability. Both random and fixed effects were used; the fixed-effects model was used when the percentage of heterogenicity was I2 ≤ 40%, whereas the random effects model was used when I2 > 40%. Furthermore, publication bias was assessed using funnel plots and Egger’s regression test.
5 Results
5.1 Computational analysis to investigate cisplatin resistance in HNC via ERCC1/ACTL6A
5.1.1 PPIs associated with ERCC1/ACTL6A and enrichment analysis
A total of 35 genes were investigated for PPIs, including 21 genes for platinum drug resistance, ACTL6A, and eight genes for GGR-NER, with medium confidence (0.400); this revealed interactions between 30 nodes, resulting in a total of 260 edges at an average node degree of 14.9 and average local clustering coefficient of 0.737 (Figure 2A). The PPIs significantly enriched (<1.0e−16) 177 biological process (BP), 17 cellular component (CC), and 15 molecular function (MF) terms of gene ontology along with four KEGG pathways that were modulated by ERCC1 and/or ACTL6A. The ACTL6A gene was found to significantly modulate BPs such as DNA repair, regulation of DNA repair, positive regulation of DNA repair, and regulation of NER, whereas the ERCC1 gene was found to modulate BPs like DNA repair, NER, UV damage excision repair, mismatch repair, pyrimidine dimer repair, pyrimidine dimer repair by NER, and double-strand break repair (Figure 2C). All these repair BPs were modulated via the PPIs of 14 identical genes belonging to the same cluster (red color), namely, ACTL6A, ERCC1, PCNA, MLH1, BRCA1, TP53, XPC, CETN2, CUL4A, DDB1, DDB2, RAD23A, RAD23B, and RBX1 (Figure 2B). Additionally, the DNA repair and NER complexes were the CCs modulated by ERCC1. This shows that both ERCC1 and ACTL6A are involved in DNA repair processes, particularly via the NER pathway.
FIGURE 2
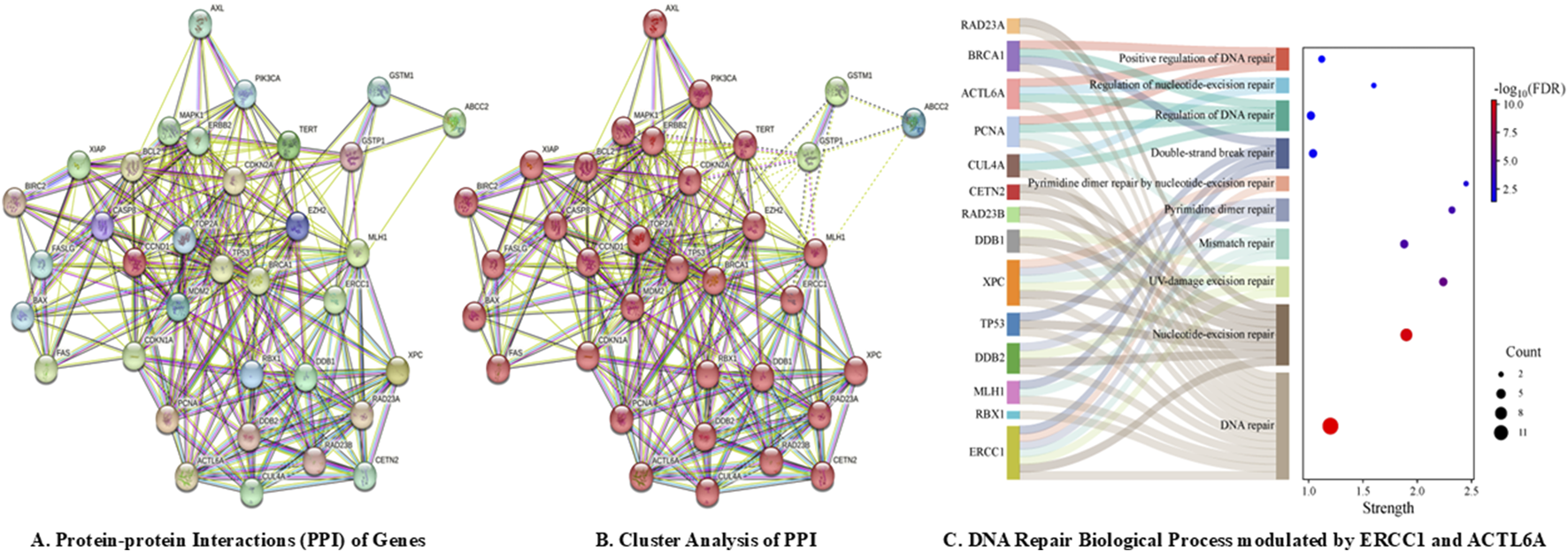
DNA repair pathways modulated by ACTL6A and ERCC1 via computational analysis: (A) protein–protein interactions; (B) cluster analysis; (C) biological process modulations by ERCC1 and ACTL6A.
5.1.2 mRNA and tissue expressions of ERCC1 and ACTL6A in HNC
The median transcripts per million of ERCC1 and ACTL6A were found to be significantly higher in primary tumors (64.801 and 71.98, respectively) than normal samples (46.256 and 30.826, respectively) (Figure 3A). Both ERCC1 and ACTL6A were found to be overexpressed in advanced stages except stage III HNC patients (stage IV > stage II > stage III > stage I) (Figure 3B), particularly among African-American and Caucasian people compared to Asian patients. Furthermore, ERCC1 was found to be overexpressed greatly among persons aged 61–80 years, followed by those in the age groups of 41–60 years, 81–100 years, and 21–40 years; however, ACTL6A was found to be overexpressed greatly among persons aged 41–60 years, followed by those in the age groups of 61–80 years, 21–40 years, and 81–100 years. Additionally, ERCC1 is inconsistently expressed with advancing tumor grade (grade 3 > grade 2 > grade 4 > grade 1), whereas ACTL6A shows significant overexpression with the grade of tumor progression (grade 4 > grade 3 > grade 2 > grade 1) (see Supplementary Table S1 and Supplementary Figure S1). Thus, we can predict that both ERCC1 and ACTL6A are upregulated in HNC patients, particularly in individuals with advanced stages and tumor grades and mostly in persons aged 40–80 years.
FIGURE 3
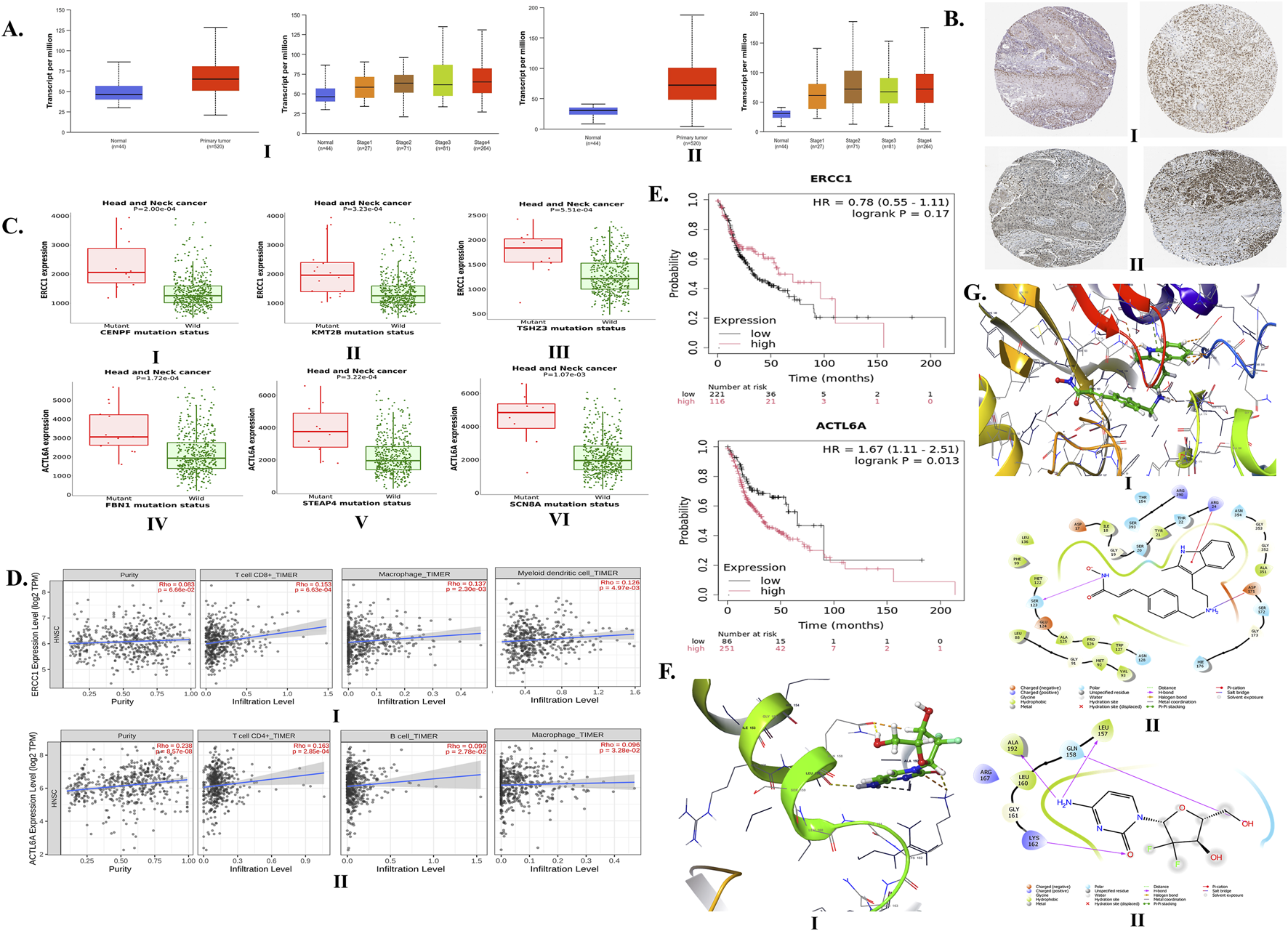
Expressions of ERCC1 and ACTL6A in head and neck cancer (HNC) and their impacts on tumor infiltration and overall survival via computational analysis. (A) mRNA expressions of (I)ERCC1 and (II)ACTL6A in tumor vs. normal samples based on stage of HNC. (B) Expressions of (I)ERCC1 and (II)ACTL6A in tumor tissues via immunohistochemistry. (C) Gene mutations upregulating (I–III)ERCC1 and (IV–VI)ACTL6A expressions. (D) Infiltration of (I) CD8+ cells, macrophages, and myeloid dendritic cells by ERCC1 expression and (II) CD4+ cells, B cells, and macrophages by ACTL6A expression. (E) Impacts of ERCC1 and ACTL6A expressions on overall survival. (F) (I) 3D and (II) 2D interactions of gemcitabine with ERCC1. (G) (I) 3D and (II) 2D interactions of panobinostat with ACTL6A.
Furthermore, ERCC1 and ACTL6A were found to show moderate and moderate-to-strong expressions, respectively. The immunohistochemistry (IHC) report of HNC tissue enclosed in HPA shows moderate expression of ERCC1 at nuclear-level staining using antibodies, such as HPA029773, CAB004390, CAB072859, and CAB072860 (Figure 3B.I and Supplementary Figure S2). Furthermore, ACTL6A shows moderate-to-strong expression at the nuclear- and cytoplasmic/membranous-nuclear-level staining using the CAB012188 antibody (Figure 3B.II, Supplementary Figure S2). These findings confirm the expressions of ERCC1 and ACTL6A in various types of HNC tumor tissues. Moreover, there are certain mutations observed in HNC patients that can alter the expressions of ERCC1 and ACTL6A. The top-5 genes undergoing somatic mutations with at least 1% prevalence rates contribute to the overexpressions of ERCC1 (namely, CASK, CENPF, KMT2B, TSHZ3, and DVL1) and ACTL6A (namely, FBN1, STEAP4, SCN8A, OR8H2, and CASZ1) (Figure 3C, Supplementary Tables S2, S3, Supplementary Figures S3, S4).
5.1.3 Impacts of ERCC1 and ACTL6A expressions on tumor cell infiltration and survival in HNC
The overexpressions of ERCC1 and ACTL6A were found to significantly enhance the infiltration of CD8+ T-cells, macrophages, dendritic cells, CD4+ T-cells, and B-cells into HNC tumor cells (Figure 3D, Supplementary Figure S5). Overexpression of ERCC1 was found to be linked with 22% less risk of death compared to reduced expression in HNC patients (HR: 0.78, p = 0.17), whereas overexpression of ACTL6A was found to be linked with 67% more risk of death among HNC patients (HR: 1.67, p = 0.013) (Figure 3E). Thus, computational analysis reveals that ACTL6A is a significant gene responsible for the poor survival of HNC patients.
5.1.4 Potential drug candidates and their binding affinities with ERCC1 and ACTL6A
A total of 12 drug candidates, i.e., eight FDA-approved drugs (cyclosporine, carboplatin, cisplatin, 5-fluorouracil, doxorubicin hydrochloride, gemcitabine, paclitaxel, and thalidomide) and four drugs not approved by the FDA (staurosporine, herbimycin A, platinum, and platinum compound), were found to interact with the ERCC1 protein. Similarly, a total of four drug candidates (two FDA-approved drugs: panobinostat and cisplatin; two unapproved drugs: sphingosine-1-phosphate and sphingosylphosphorylcholine) were found to interact with the ACTL6A protein (Supplementary Table S4). In the case of cisplatin resistance, the predicted FDA-approved drug candidates other than platinum may be repurposed to downregulate both ERCC1 and ACTL6A genes. Thus, gemcitabine (interaction score: 0.047) and paclitaxel (interaction score: 0.040) were found to have high interactions with ERCC1, whereas panobinostat (interaction score: 0.398) was found to interact with ACTL6A. From the molecular docking studies, we found that gemcitabine and panobinostat possessed higher binding affinities toward ERCC1 and ACTL6A with binding energies of −3.707 kcal/mol (Figure 3F) and −4.198 kcal/mol (Figure 3G), respectively.
5.2 Human study for ERCC1 and ACTL6A expressions among HNC patients
5.2.1 Demographic details and clinical characteristics of the HNC patients
A total of 77 LAHNC patients were enrolled in the study, of which 96.1% patients were men and 3.9% were women with a mean age of 52.88 ± 9.68 years. The majority of patients were in the age group of 41–60 years (61.04%), followed by 61–80 years (29.87%) and 21–40 years (9.09%). Nearly half of the enrolled patients (49.35%) were from the upper part of the lower socioeconomic class and had abnormal body mass index (BMI) values, i.e., they were underweight (42.86%) or overweight (6.49%). Most of the LAHNC patients (89.61%) had a history of social habits, such as drinking alcohol (58.44%), smoking tobacco (46.75%), or chewing tobacco (38.96%) or betel leaf (33.77%) or areca nut (33.77%). Approximately 16.88% of the patients reported both alcohol consumption and smoking. Furthermore, we observed that approximately 24.68% of the HNC patients had comorbidities, where hypertension (15.58%) and diabetes (11.69%) were the most prevalent types followed by cerebrovascular accidents (5.19%), respiratory diseases (2.60%), and ischemic heart disease (1.30%). We found that approximately 29.87% of patients had a history of cancer in their family. Surprisingly, HNC was the most commonly reported type of cancer (15.58%) in the family histories, which was attributed to prevailing social habits in their families (Table 1).
TABLE 1
| Parameters | Frequency | Percentage (%) |
|---|---|---|
| Gender | ||
| Male | 74 | 96.1 |
| Female | 3 | 3.90 |
| Age Groups (52.88 ± 9.68 years) | ||
| 21–40 years | 7 | 9.09 |
| 41–60 years | 47 | 61.04 |
| 61–80 years | 23 | 29.87 |
| BMI Categories (19.39 ± 3.72) | ||
| Overweight | 5 | 6.49 |
| Normal BMI | 39 | 50.65 |
| Underweight | 33 | 42.86 |
| Kuppuswamy Socioeconomic Class | ||
| Lower Middle | 30 | 38.96 |
| Upper Lower | 38 | 49.35 |
| Upper Middle | 9 | 11.69 |
| Social Habits | ||
| No habits | 7 | 9.09 |
| Alcohol consumption | 45 | 58.44 |
| Tobacco smoking | 36 | 46.75 |
| Tobacco chewing | 30 | 38.96 |
| Betel leaf or paan chewing | 26 | 33.77 |
| Areca nut or gutka chewing | 26 | 33.77 |
| Sharp Teeth Associated Injury | ||
| Tongue bite | 7 | 9.09 |
| Cheek bite | 5 | 6.49 |
| Comorbidities | ||
| No comorbidities | 58 | 75.32 |
| Hypertension | 12 | 15.58 |
| Diabetes mellitus | 9 | 11.69 |
| Cerebrovascular accident | 4 | 5.19 |
| Respiratory diseases: Asthma, COPD, and Old TB | 2 | 2.60 |
| Ischemic heart disease | 1 | 1.30 |
| Family History of Cancer | ||
| No familial history of cancer | 54 | 70.13 |
| Patients with familial history of cancer | 23 | 29.87 |
| Breast | 1 | 1.30 |
| Breast and Brain | 1 | 1.30 |
| Hematological | 3 | 3.90 |
| HNC | 13 | 16.88 |
| HNC and Breast | 1 | 1.30 |
| Thyroid | 1 | 1.30 |
| Uterus | 2 | 2.60 |
| Brain | 1 | 1.30 |
| Histopathology | ||
| Well differentiated (Grade 1) | 27 | 35.06 |
| Moderately differentiated (Grade 2) | 43 | 55.84 |
| Poorly differentiated (Grade 3) | 7 | 9.09 |
| 8th Edition of the American Joint Committee on Cancer (AJCC) staging | ||
| Stage III | 16 | 20.78 |
| Stage IV | 61 | 79.22 |
| Stage IVA | 47 | 61.04 |
| Stage IVB | 14 | 18.18 |
| Types of HNC | ||
| Oral Cavity Cancer | 40 | 51.95 |
| Buccal mucosa cancer | 13 | 16.88 |
| Tongue cancer | 18 | 23.38 |
| Floor of the mouth | 3 | 3.90 |
| Gingivobuccal sulcus | 1 | 1.30 |
| Hard palate | 1 | 1.30 |
| Retromolar trigone | 4 | 5.19 |
| Laryngeal cancer | 13 | 16.88 |
| Supraglottic cancer | 5 | 6.49 |
| Aryepiglottic cancer | 2 | 2.60 |
| Epiglottic cancer | 1 | 1.30 |
| Vocal cord cancer | 5 | 6.49 |
| Hypopharynx cancer | 11 | 14.29 |
| Cricopharynx | 2 | 2.60 |
| Pyriform fossa | 9 | 11.69 |
| Oropharynx cancer | 8 | 10.39 |
| Base of tongue cancer | 5 | 6.49 |
| Soft palate | 3 | 3.90 |
| Cancer of unknown primary cause | 3 | 3.90 |
| Lymph node | 2 | 2.60 |
| Brachial cleft cyst | 1 | 1.30 |
| Nasopharynx cancer (Nasal cavity) | 2 | 2.60 |
| Treatment Modalities | ||
| Total concurrent CRT (CCRT) | 55 | 71.43 |
| Concurrent CRT (CCRT) | 46 | 59.74 |
| CCRT with adjuvant chemotherapy | 9 | 11.69 |
| Surgery with adjuvant CCRT | 22 | 28.57 |
| Number of Cycles of Cisplatin Chemotherapy | ||
| 3 | 8 | 10.39 |
| 4 | 6 | 7.79 |
| 5 | 44 | 57.14 |
| 6 | 19 | 24.68 |
| Radiation Dose | ||
| 60 Gy | 17 | 22.08 |
| 66 Gy | 43 | 55.84 |
| 70 Gy | 17 | 22.08 |
| Radiation Fraction | ||
| 30 | 23 | 29.87 |
| 33 | 37 | 48.05 |
| 35 | 17 | 22.08 |
Demographic details and clinical characteristics of the HNC patients in this study.
Clinically, the majority of the enrolled LAHNC patients belonged to grade 2 (55.84%) followed by grades 1 and 3 and were diagnosed at stage IV (79.22%) followed by stage III (20.78%). Approximately half of the HNC patients were diagnosed with carcinoma of the oral cavity (51.95%), followed by laryngeal cancer (16.88%), hypopharyngeal cancer (14.29%), oropharyngeal cancer (10.39%), cancer of unknown primary cause (3.90%), and nasopharyngeal cancer (2.60%). CCRT was the most popular choice of treatment (55.84%), followed by surgery with adjuvant CRT (32.47%) and CCRT with adjuvant chemotherapy (11.69%). All patients in the study cohort were scheduled to undergo six cycles of cisplatin therapy. However, approximately half of the patients received five cycles of cisplatin (57.14%), followed by six cycles (24.68%), three cycles (10.39%), and four cycles (7.79%). The dosage for radiation therapy ranged from 60 to 70 Gy and was administered in 30–35 fractions. The demographics and clinical characteristics of the LAHNC patients are depicted in Table 1.
5.2.2 ERCC1 and ACTL6A expressions from peripheral blood samples via qPCR
Considering the expression of the reference gene as 1 (with >1 being high expression and <1 being low expression), ERCC1 was highly expressed among 14.29% patients out of the total of 77 HNC patients while 85.71% of patients showed low expressions compared to the baseline. Furthermore, 9.09% and 20.78% of patients were observed to have higher expressions of ERCC1 after 50% CCRT and 100% CCRT, respectively. Similarly, ACTL6A was highly expressed in 88.31% of the patients while 11.69% of the patients had low expressions compared to the baseline. After administration of 50% and 100% CCRT dosing, ACTL6A expressions were found to be highly expressed among 75.32% and 84.42% of the patients, respectively (Supplementary Table S5). This shows that cisplatin-based CCRT initially decreases the expressions of ERCC1 (14.29%–9.09%) and ACTL6A (88.31%–75.32%) among HNC patients via initial response to therapy, whereas the expressions of ERCC1 (9.09%–20.78%) and ACTL6A (75.32%–84. 42%) increase later to confer possible resistance to cisplatin therapy.
Comparative analyses on the impacts of cisplatin-based CCRT on the gene expressions showed that the overall median expression of ERCC1 significantly increased (p < 0.001) by 1.64-fold compared to the baseline (from 0.14 to 0.19 and 0.23), signifying that ERCC1 could potentially be involved in DNA repair (Table 2 and Figure 4.I). Similarly, the median expression of ACTL6A significantly decreased by 0.81-fold (from 4.77 to 3.87) after the initial three cycles of CCRT but later increased by 1.14-fold (from 3.87 to 5.43), showing the ability of ACTL6A to bounce back and mediate DNA repair (Table 2 and Figure 4.II). Furthermore, the subgroup analysis of variables showed that patients with advanced ages (40–80 years), advanced stages (stage IV), highly differentiated tumors (grades 1 and 2), low BMIs (underweight/normal), social habits (tobacco smoking, alcohol consumption, betel leaf chewing), oral cavity cancers, and hypopharyngeal cancer who received CCRT alone or five cycles of cisplatin are at high risk of developing ERCC1-mediated cisplatin resistance as ERCC1 was found to be significantly increased in these patients. In contrast, patients with no history of tobacco use or betel leaf chewing also showed significant increases in ERCC1 expressions. Interestingly, we observed that ACTL6A expressions were significantly lower in patients with no history of tobacco smoking, alcohol consumption, or tobacco/betel leaf/areca nut/gutka chewing (Table 2). This indicates that patients with a history of social habits may be at a greater risk of developing chemoresistance to CCRT than patients without such history. Additionally, correlation analysis did not indicate any correlation in the baseline expressions of ERCC1 and ACTL6A (ρ = 0.201, p = 0.08). However, the expressions of these genes were significantly (ρ = 0.331, p = 0.003) and marginally (ρ = 0.215, p = 0.060) correlated after receiving 50% and 100% cisplatin-based CCRT, indicating that ACTL6A could indirectly influence DNA repair via the NER pathways.
TABLE 2
| Parameters | Median expressions of genes | ||||||
|---|---|---|---|---|---|---|---|
| Genes | 0% CRT | 50% CRT | 100% CRT | p-value (0% vs. 50%) | p-value (50% vs. 100%) | p-value (0% vs. 100%) | Overall p-value |
| ERCC1 | 0.14 (0.05, 0.41) | 0.19 (0.06, 0.44) | 0.23 (0.08, 0.68) | 0.301 | 0.001** | 0.011* | p < 0.001*** |
| ACTL6A | 4.77 (1.92, 12.065) | 3.87 (1.00, 8.81) | 5.43 (1.535, 9.26) | 0.028* | 0.459 | 0.362 | 0.729 |
| Sociodemographic/Clinical | Median Expression of ERCC1 and ACTL6A | ||||||
|---|---|---|---|---|---|---|---|
| Age Groups | |||||||
| 21–40 years ERCC1 | 0.12 (0.04, 0.36) | 0.19 (0.08, 0.23) | 0.12 (0.07, 0.32) | 0.397 | 0.672 | 0.499 | 0.651 |
| 21–40 years ACTL6A | 11.89 (1.12, 21.88) | 6.39 (0.48, 9.82) | 4.71 (1.42, 14.57) | 0.128 | 0.612 | 0.499 | 0.565 |
| 41–60 years ERCC1 | 0.22 (0.05, 0.6) | 0.17 (0.06, 0.48) | 0.34 (0.13, 0.1.05) | 0.782 | 0.005* | 0.077 | 0.005* |
| 41–60 years ACTL6A | 4.49 (2.21, 11.73) | 5.93 (2.61, 9.07) | 6.78 (2.53, 9.95) | 0.800 | 0.582 | 0.691 | 0.587 |
| 61–80 years ERCC1 | 0.01 (0.05, 0.32) | 0.19 (0.04, 0.35) | 0.21 (0.08, 0.37) | 0.217 | 0.079 | 0.068 | 0.009* |
| 61–80 years ACTL6A | 4.76 (1.42, 11.98) | 1.14 (0.77, 6.05) | 2.06 (0.85, 7.16) | 0.007 | 0.783 | 0.066 | 0.199 |
| BMI | |||||||
| Underweight ERCC1 | 0.11 (0.05, 0.385) | 0.12 (0.06, 0.25) | 0.2 (0.075, 0.56) | 0.543 | 0.031* | 0.136 | 0.029* |
| Underweight ACTL6A | 5.19 (1.23, 11.55) | 3.15 (0.81, 7.15) | 4.37 (1.54, 8.90) | 0.183 | 0.432 | 0.357 | 0.754 |
| Normal ERCC1 | 0.15 (0.05, 0.6) | 0.20 (0.06, 0.44) | 0.26 (0.12, 0.88) | 0.619 | 0.033* | 0.121 | 0.021* |
| Normal ACTL6A | 4.05 (2.14, 11.89) | 4.25 (1.05, 8.84) | 5.84 (1.17, 9.95) | 0.264 | 0.596 | 0.967 | 0.975 |
| Overweight ERCC1 | 0.27 (0.04, 0.345) | 0.19 (0.085, 1.32) | 0.51 (0.21, 2.975) | 0.225 | 0.08 | 0.068 | 0.076 |
| Overweight ACTL6A | 21.59 (7.02, 25.62) | 9.04 (5.89, 12.75) | 6.78 (4.89, 12.89) | 0.080 | 0.686 | 0.225 | 0.549 |
| Social Habits | |||||||
| No habits ERCC1 | 0.25 (0.09, 0.40) | 0.10 (0.04, 0.14) | 0.10 (0.05, 0.31) | 0.236 | 0.610 | 0.917 | 0.772 |
| Yes habits ERCC1 | 0.13 (0.05, 0.41) | 0.19 (0.06, 0.45) | 0.26 (0.097, 0.89) | 0.153 | 0.001** | 0.007* | p < 0.001*** |
| No habits ACTL6A | 11.73 (2.78, 21.59) | 3.10 (1.14, 9.04) | 8.37 (4.46, 10.63) | 0.063 | 0.176 | 1.00 | 0.565 |
| Yes habits ACTL6A | 4.62 (1.67, 11.95) | 3.88 (.937, 8.79) | 5.41 (1.38, 9.14) | 0.089 | 0.652 | 0.366 | 0.876 |
| No smoking ERCC1 | 0.26 (0.085, 0.96) | 0.17 (0.06, 0.44) | 0.21 (0.095, 1.09) | 0.559 | 0.003* | 0.340 | 0.032 |
| Smoking ERCC1 | 0.095 (0.05, 0.25) | 0.195 (0.07, 0.44) | 0.265 (0.08, 0.62) | 0.016* | 0.087 | 0.003* | 0.001** |
| No Smoking ACTL6A | 6.36 (2.66, 14.13) | 4.87 (0.95, 9.06) | 7.04 (3.19, 10.60) | 0.013* | 0.390 | 0.496 | 0.552 |
| Smoking ACTL6A | 3.68 (1.33, 9.71 | 3.315 (1.16, 7.62) | 4.695 (1.09, 7.93) | 0.530 | 0.888 | 0.599 | 1.000 |
| No Alcohol consumption ERCC1 | 0.24 (0.09, 0.64) | 0.11 (0.04, 0.38) | 0.21 (0.07, 0.67) | 0.206 | 0.014* | 0.742 | 0.103 |
| Alcohol consumption ERCC1 | 0.11 (0.04, 0.30) | 0.19 (0.09, 0.30) | 0.32 (0.125, 0.78) | 0.011* | 0.024* | 0.002* | p < 0.001*** |
| No alcohol consumption ACTL6A | 4.27 (1.49, 11.52) | 2.75 (0.84, 7.30) | 5.26 (1.17, 9.82) | 0.166 | 0.176 | 0.601 | 0.680 |
| Alcohol consumption ACTL6A | 6.27 (2.44, 13.72) | 5.93 (1.48, 9.27) | 5.59 (1.95, 8.17) | 0.079 | 0.906 | 0.09 | 0.766 |
| No tobacco chewing ERCC1 | 0.11 (0.04, 0.26) | 0.19 (0.06, 0.29) | 0.25 (0.08, 0.61) | 0.137 | 0.002* | 0.006* | p < 0.001*** |
| Tobacco chewing ERCC1 | 0.28 (0.067, 0.89) | 0.19 (0.06, 0.78) | 0.22 (0.13, 1.17) | 0.982 | 0.094 | 0.447 | 0.126 |
| No tobacco chewing ACTL6A | 4.05 (1.57, 11.98) | 3.10 (1.05, 7.49) | 5.15 (1.17, 8.78) | 0.030* | 0.174 | 0.608 | 0.722 |
| Tobacco chewing ACTL6A | 6.01 (2.54, 12.54) | 6.74 (0.82, 11.45) | 6.97 (1.89, 11.38) | 0.441 | 0.586 | 0.417 | 0.905 |
| No betel leaf chewing ERCC1 | 0.14 (0.05, 0.41) | 0.19 (0.06, 0.29) | 0.22 (0.08, 0.51) | 0.927 | 0.007* | 0.176 | 0.009* |
| Betel leaf chewing ERCC1 | 0.13 (0.037, .0397) | 0.205 (0.06, 0.72) | 0.44 (0.115, 1.32) | 0.092 | 0.029* | 0.014* | 0.002* |
| No Betel leaf chewing ACTL6A | 4.77 (1.41, 11.73) | 3.77 (1.14, 8.42) | 4.46 (1.17, 8.26) | 0.033* | 0.940 | 0.275 | 0.662 |
| Betel leaf chewing ACTL6A | 4.8 (3.21, 13.54) | 6.1 (0.86, 9.04) | 6.91 (3.53, 14.87) | 0.439 | 0.382 | 0.929 | 0.832 |
| No areca nut or gutka chewing ERCC1 | 0.14 (0.05, 0.40) | 0.17 (0.06, 0.44) | 0.31 (0.14, 0.88) | 0.234 | 0.010* | 0.008* | p < 0.001*** |
| Areca nut or gutka chewing ERCC1 | 0.12 (0.047, 0.537) | 0.20 (0.045, 0.44) | 0.195 (0.07, 0.61) | 0.957 | 0.020* | 0.493 | 0.137 |
| No areca nut or gutka chewing ACTL6A | 7.37 (2.68, 13.73) | 5.50 (1.14, 9.47) | 5.43 (1.29, 9.02) | 0.034* | 0.593 | 0.058 | 0.494 |
| Areca nut or gutka chewing ACTL6A | 3.26 (1.22, 6.61) | 2.99 (0.85, 7.49) | 5.49 (1.59, 11.56) | 0.657 | 0.035* | 0.209 | 0.173 |
| Comorbidity | |||||||
| No comorbidity ERCC1 | 0.13 (0.05, 0.465) | 0.19 (0.077, 0.48) | 0.225 (0.09, 0.97) | 0.227 | 0.006* | 0.038* | 0.003* |
| No comorbidity ACTL6A | 3.83 (1.42, 12.49) | 3.82 (1.06, 8.93) | 5.72 (1.59, 9.61) | 0.277 | 0.619 | 0.642 | 0.852 |
| Presence of comorbidity ERCC1 | 0.15 (0.03, 0.32) | 0.12 (0.03, 0.23) | 0.31 (0.05, 0.51) | 0.948 | 0.036* | 0.121 | 0.011* |
| Presence of comorbidity ACTL6A | 5.19 (3.76, 11.73) | 3.89 (0.96, 7.50) | 5.33 (1.29, 8.27) | 0.027* | 0.601 | 0.398 | 0.229 |
| Stages | |||||||
| Stage III ERCC1 | 0.245 (0.08, 1.27) | 0.21 (0.045, 0.43) | 0.19 (0.08, 0.55) | 0.535 | 0.211 | 0.623 | 0.867 |
| Stage III ACTL6A | 2.71 (0.90, 11.91) | 3.83 (1.33, 8.54) | 5.62 (3.47, 14.31) | 0.756 | 0.148 | 0.469 | 0.269 |
| Stage IV ERCC1 | 0.12 (0.05, 0.395) | 0.19 (0.06, 0.435) | 0.27 (0.095, 0.78) | 0.153 | 0.002* | 0.001** | p < 0.001*** |
| Stage IV ACTL6A | 5.11 (2.44, 12.82) | 3.87 (.915, 8.91) | 5.43 (1.35, 8.90) | 0.027* | 0.947 | 0.156 | 0.452 |
| Histopathology | |||||||
| Poorly differentiated ERCC1 | 0.29 (0.09, 1.15) | 0.26 (0.14, 0.84) | 0.25 (0.10, 1.57) | 0.672 | 0.446 | 0.866 | 0.651 |
| Poorly differentiated ACTL6A | 2.68 (1.41, 7.73) | 4.25 (2.89, 8.89) | 3.85 (1.15, 8.78) | 0.612 | 0.866 | 0.398 | 0.368 |
| Moderately differentiated ERCC1 | 0.12 (0.05, 0.36) | 0.15 (0.06, 0.40) | 0.21 (0.09, 0.95) | 0.974 | 0.001** | 0.053 | 0.001** |
| Moderately differentiated ACTL6A | 6.27 (2.93, 13.49) | 3.15 (0.94, 8.78) | 5.89 (1.93, 9.02) | 0.001** | 0.358 | 0.098 | 0.108 |
| Well differentiated ERCC1 | 0.14 (0.04, 0.42) | 0.20 (0.06, 0.43) | 0.27 (0.08, 0.68) | 0.082 | 0.237 | 0.038* | 0.030* |
| Well differentiated ACTL6A | 4.05 (1.25, 13.7) | 6.05 (.75, 8.84) | 5.18 (1.17, 13.90) | 0.829 | 1.00 | 0.848 | 0.772 |
| Type of HNC | |||||||
| Oral cavity cancer ERCC1 | 0.13 (0.05, 0.69) | 0.185 (0.06, 0.44) | 0.29 (0.12, 1.03) | 0.632 | 0.017* | 0.050* | 0.007* |
| Oral cavity cancer ACTL6A | 7.57 (2.48, 13.72) | 6.43 (1.97, 11.03) | 7.33 (1.45, 14.40) | 0.162 | 0.707 | 0.364 | 0.928 |
| Laryngeal cancer ERCC1 | 0.22 (0.04, 0.36) | 0.15 (0.05, 0.34) | 0.22 (0.05, 0.43) | 0.506 | 0.208 | 0.649 | 0.146 |
| Laryngeal cancer ERCC1 | 4.77 (1.32, 9.55) | 2.84 (0.94, 7.59) | 5.33 (2.39, 6.94) | 0.249 | 0.753 | 0.701 | 0.584 |
| Hypopharyngeal cancer ERCC1 | 0.09 (0.04, 0.36) | 0.10 (0.06, 0.70) | 0.15 (0.06, 1.14) | 0.046* | 0.333 | 0.139 | 0.027* |
| Hypopharyngeal cancer ERCC1 | 3.38 (0.66, 7.36) | 5.50 (0.80, 7.50) | 4.46 (0.80, 8.78) | 0.859 | 0.721 | 0.929 | 0.368 |
| Oropharyngeal cancer ERCC1 | 0.085 (0.027, 0.24) | 0.24 (0.19, 0.28) | 0.20 (0.11, 0.76) | 0.123 | 0.345 | 0.161 | 0.079 |
| Oropharyngeal cancer ACTL6A | 5.02 (2.33, 10.92) | 1.49 (0.74, 2.64) | 5.49 (1.75, 7.97) | 0.123 | 0.017* | 0.674 | 0.223 |
| CUP ERCC1 | 0.39 (0.23, 0.42) | 0.35 (0.02, 0.39) | 0.62 (0.27, 0.68) | 0.593 | 0.285 | 0.285 | 0.717 |
| CUP ACTL6A | 4.05 (3.03, 6.36) | 1.75 (0.5, 8.84) | 3.69 (1.09, 20.88) | 0.593 | 0.285 | 1.00 | 0.717 |
| Nasopharyngeal cancer ERCC1 | 0.91 (0.20, 1.16) | 0.44 (0.03, 0.63) | 0.81 (0.37, 1.17) | 0.655 | 0.180 | 0.655 | 0.607 |
| Nasopharyngeal cancer ERCC1 | 7.07 (2.085, 8.51) | 6.88 (3.65, 6.67) | 9.46 (.86, 13.33) | 0.655 | 0.655 | 0.655 | 1.000 |
| Therapy | |||||||
| CCRT ERCC1 | 0.12 (0.05, 0.39) | 0.19 (0.06, 0.29) | 0.21 (0.09, 0.51) | 0.418 | 0.010* | 0.071 | 0.003* |
| CCRT ACTL6A | 4.77 (1.71, 11.98) | 3.25 (0.84, 7.66) | 5.33 (1.42, 8.37) | 0.053* | 0.639 | 0.135 | 0.608 |
| Surgery plus adjuvant CCRT ERCC1 | 0.165 (0.05, 0.465) | 0.195 (0.057, 0.52) | 0.345 (0.080, 1.34) | 0.490 | 0.032* | 0.044* | 0.027* |
| Surgery plus adjuvant CCRT ACTL6A | 4.90 (1.92, 13.59) | 5.03 (2.59, 9.26) | 7.40 (1.74, 15.84) | 0.306 | 0.661 | 0.465 | 0.580 |
| Cycles of chemotherapy | |||||||
| 3 cycles ERCC1 | 0.185 (0.06, 0.38) | 0.15 (0.075, 0.87) | 0.29 (0.22, 1.88) | 0.779 | 0.025* | 0.233 | 0.093 |
| 3 cycles ACTL6A | 3.89 (0.94, 8.59) | 2.57 (0.80, 6.44) | 8.75 (1.91, 13.9) | 0.401 | 0.069 | 0.674 | 0.417 |
| 4 cycles ERCC1 | 0.105 (0.07, 0.28) | 0.145 (0.05, 2.83) | 0.21 (0.082, 1.85) | 0.345 | 0.6 | 0.463 | 0.607 |
| 4 cycles ACTL6A | 1.31 (1.16,12.60) | 4.96 (1.59, 16.10) | 8.18 (5.86, 9.23) | 0.917 | 0.463 | 0.345 | 0.607 |
| 5 cycles ERCC1 | 0.14 (0.05, 0.55) | 0.19 (0.053, 0.42) | 0.29 (0.097, 0.67) | 0.741 | 0.005* | 0.083 | 0.007* |
| 5 cycles ACTL6A | 4.94 (1.82, 13.2) | 4.38 (1.23, 9.36) | 5.27 (1.38, 7.66) | 0.327 | 0.462 | 0.143 | 0.853 |
| 6 cycles ERCC1 | 0.22 (0.03, 0.40) | 0.17 (0.06, 0.44) | 0.13 (0.07, 1.14) | 0.354 | 0.176 | 0.212 | 0.055 |
| 6 cycles ACTL6A | 5.75 (3.38,13.49) | 4.25 (0.94,7.49) | 5.18 (1.10,15.78) | 0.022* | 0.711 | 0.778 | 0.698 |
Comparison of ERCC1 and ACTL6A expressions across chemoradiotherapy.
FIGURE 4
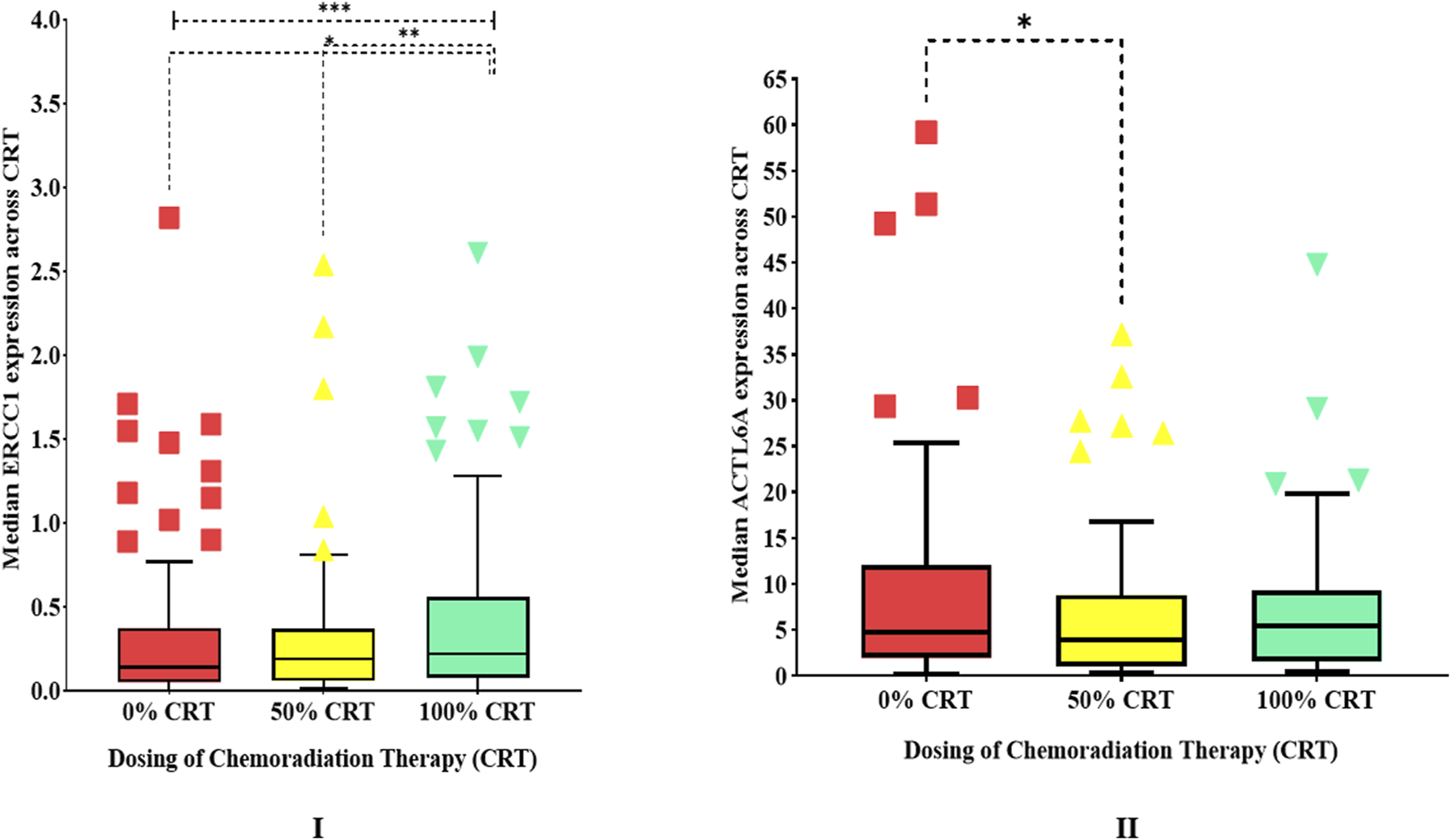
Human experimentation results showing box plots of (I)ERCC1 expressions (outliers with median expressions >4 have been removed) and (II)ACTL6A expressions over the duration of chemoradiotherapy.
5.3 Real-world evidence for ERCC1/ACTL6A expressions and survival in HNC via meta-analysis
A total of 266 articles related to ERCC1 and HNC were obtained by searching the three databases, of which only 12 articles met the criteria for meta-analysis (Supplementary Figure S6). Out of these 12 studies, only four were conducted prospectively while the remaining eight were conducted retrospectively. The aggregate sample size from all included studies was 2,041, of which 1,810 samples (high ERCC1 expression: 911 patients, low ERCC1 expression: 899 patients) were in our analysis (Table 3). Based on the random effects analysis of the pooled data of the 1,810 samples, we found that ERCC1 expression was linked to poor overall survival among HNC patients, i.e., overexpression of the ERCC1 gene significantly increased the risk of mortality among HNC patients by 82% (HR: 1.82, 95% CI: 1.26–2.63, p = 0.0001) compared to patients who had low expressions of ERCC1. However, the analysis showed moderate heterogenicity (X2: 26.77, I2: 56%, p = 0.0005) (Figure 5A). Subgroup analysis of the pooled data also showed that high ERCC1 expression was significantly linked to poor survival rate among Asians (HR: 1.73, 95% CI: 1.16–2.59, p = 0.007) (Figure 5B). Additionally, the funnel plot of the pooled data showed symmetricity with an Egger regression coefficient of −0.152 (p = 0.603, 95% CI: −0.783 to 0.479), suggesting no publication bias. A total of 13 articles were identified from the three databases for ACTL6A and its association with HNC, of which only four articles were found to have the necessary information; however, none of these articles contained information on ACTL6A expression and its impact on survival. Thus, we were unable to conduct a meta-analysis for the ACTL6A gene.
TABLE 3
| S. No. | Author Year | Country Continent | Sample size 2041–231 = 1810 | Study Design | Cancer site | Stages | Assay | High vs. low ERCC1 expression | HR | 95% CI | p-value | Data extraction model |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Liang et al. (2015) | China Asia | 76 (M:59, F:17) | P | Nasopharynx | III, IVA | IHC | 32 (42.1%) vs. 44 (57.89%) | 1.43 | 0.49–4.16 | - | KM survival curve |
| 2 | Ciaparrone et al. (2015) | Italy Europe | 48 (M:39, F:9) | R | Head/Neck | III–IVB | IHC | 36 (75%) vs. 12 (25%) | 9.53 | 1.27–71.35 | 0.028 | Multivariate |
| 3 | Lu et al. (2017) | China Asia | 334 (M:244, F:90) | R | Nasopharynx | I–IVB | IHC | 118 (35.32%) vs. 216 (64.7%) | 2.65 | 1.16–6.05 | - | KM survival curve |
| 4 | Xu et al. (2017) | China Asia | 201 (M:132, F:69) | P | Nasopharynx | III–IV | IHC | 136 (56.6%) vs. 65 (76.9%) | 5.582 | 1.23–25.27 | 0.026 | Multivariate KM survival curve |
| 5 | An et al. (2017) | Korea Asia | 204 (M:173, F:31) | R | Head/Neck | I–IV | IHC | 136 (66.66%) vs. 68 (33.33%) | 1 | 0.52–1.93 | 0.99 | Multivariate KM survival curve |
| 6 | Prochnow et al. (2019) | Germany Europe | 453 (159 patients excluded) (M:335, F:118) | R | Head/Neck | I–III | IHC | 135 (45.92%) vs. 159 (54.08%) | 1.85 | 1.03–3.35 | - | KM survival curve |
| 7 | Gong et al. (2019) | China Asia | 156 (67 patients excluded) (M:87, F:69) | R | Oral cavity | III, IVA | IHC | 41 (22.4%) vs. 48 (84.7%) | 5.61 | 2.51–12.53 | - | KM survival curve |
| 8 | Raturi et al. (2020) | India Asia | 98 (M:98) | P | Larynx | III–IVB | RT-PCR | 49 (50%) vs. 49 (50%) | 1.26 | 0.73–2.20 | - | KM survival curve |
| 9 | Aksoy et al. (2019) | Turkey Asia | 33 (5 patients excluded) (M:24, F:9) | R | Nasopharynx | II–IVB | IHC | 15 (53.57%) vs. 13 (46.43%) | 1.63 | 0.40–6.68 | - | KM survival curve |
| 10 | Chitapanarux et al. (2020) | Thailand Asia | 262 (M:183, F:79) | R | Nasopharynx | I–IV | IHC | 135 (51.52%) vs. 127 (48.48%) | 1.08 | 0.79–1.47 | 0.647 | Multivariate KM survival curve |
| 11 | Wang et al. (2021) | Taiwan Asia | 98 (M:92, F:6) | R | Oral cavity | I–IV | IHC | 58 (59.18%) vs. 40 (40.82%) | 1.06 | 0.45–2.50 | 0.9 | Multivariate KM survival curve |
| 12 | Hua et al. (2022) | China Asia | 78 (M:59, F:19) | P | Nasopharynx | II | RT-PCR | 20 (25.6%) vs. 58 (74.4%) | 4.59 | 0.65–32.60 | - | KM survival curve |
Characteristics of all the studies included in the meta-analysis.
Note: M: male, F: female, P: prospective, R: retrospective, IHC: immunohistochemistry, RT-PCR: real-time polymerase chain reaction, HR: hazard ratio, KM: Kaplan–Meier. A total of 231 patients were excluded from the analysis because Prochnow et al. and Aksoy et al. did not perform ERCC1 expression analyses for 159 and 5 patients, respectively, whereas Gong et al. compared ERCC1 low vs. high for only 89 patients.
FIGURE 5
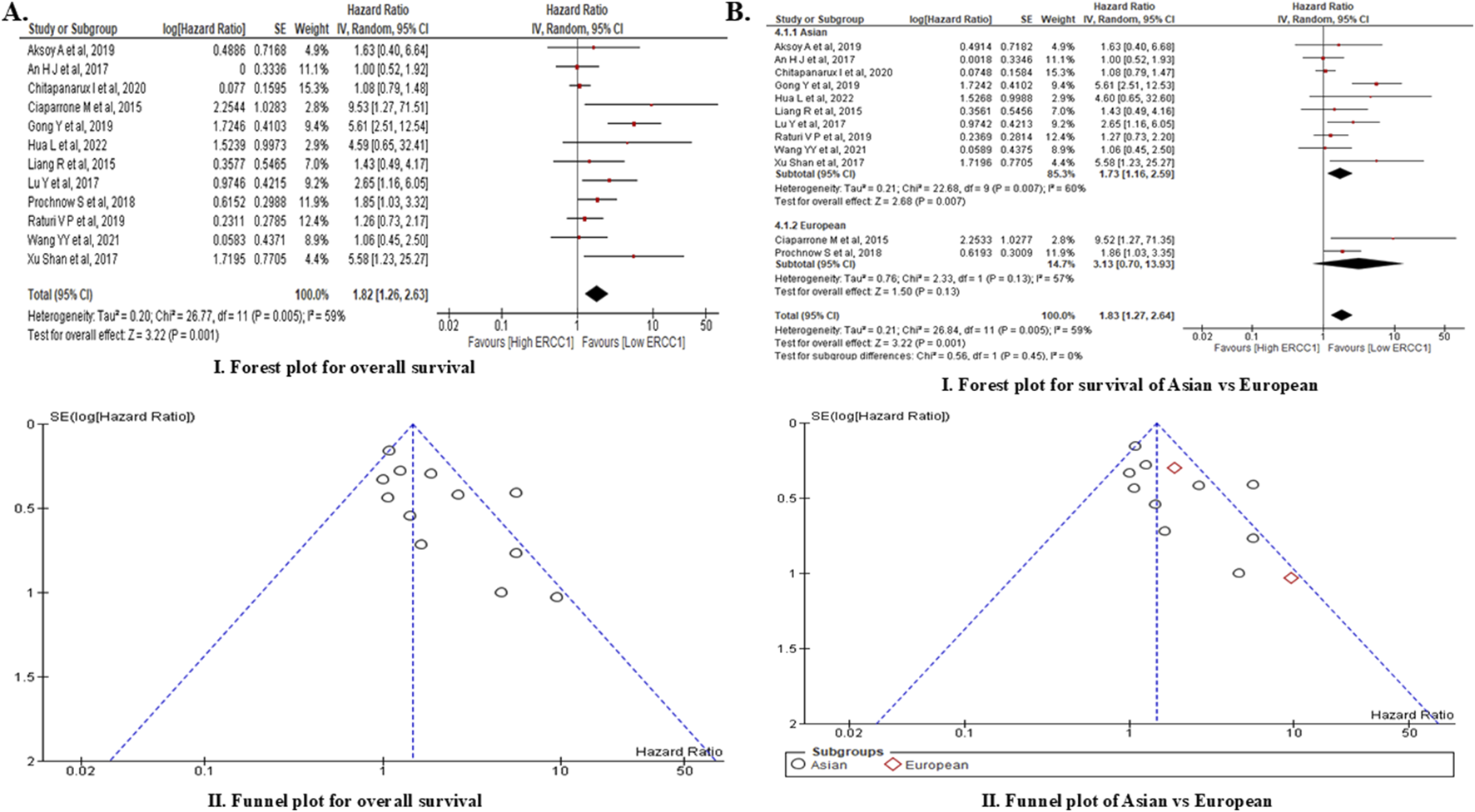
Meta-analysis of ERCC1 expression and overall survival showing forest plot and funnel plot for (A) overall survival of HNC patients and (B) comparison of overall survival of Asian vs. European subjects.
6 Discussion
The increases in the median expressions of ERCC1 and ACTL6A before and after CCRT as well as their associations with the poor overall survival outcomes in HNC patients (revealed by integrating computational analysis with meta-analysis) in the present study predict the chemoresistance of genotoxic regimens like cisplatin-based CCRT as these genes are reported to mediate DNA repair via the NER and/or SWI/SNF pathways (Figure 6). Sociodemographically, our findings are consistent with recent epidemiological studies from north India by Badola et al. (2023) and Chauhan et al. (2022), who reported that HNC is more prevalent in men than women, i.e., 87% vs. 13% and 89.4% vs. 10.6%, respectively. Furthermore, Chauhan et al. (2022) and a study on south Indians by SathiyaPriya et al. (2024) observed that nearly half of the study population (48% and 51%, respectively) was aged 40–60 years; in contrast to our study, Badola et al. (2023) and Bagal et al. (2023) found that most of the HNC patients were above 60 years of age followed by those aged 40–60 years. Furthermore, the socioeconomic classes and social habits of the patients in our study resemble those reported by SathiyaPriya et al. (2024), where most of the HNC patients were from the lower middle (62.3%) or lower (37.7%) socioeconomic class and were most commonly associated with tobacco smoking (47.6%) and alcohol consumption (42.4%) followed by tobacco chewing (30.6%) with betel leaf (27.3%) or areca nut (3.3%). Sharp teeth and teeth-mediated injuries to the oral mucosa or tongue have been infrequently linked to cancer of the oral cavity. Lateral tongue carcinoma (odds ratio (OR): 9.1) has been reported as a teeth-mediated injury (Singhvi et al., 2017), while another study reported that lesions due to trauma (OR: 4.5) were observed to be higher among oral cancer patients than lesions in the control group (Piemonte et al., 2018).
FIGURE 6
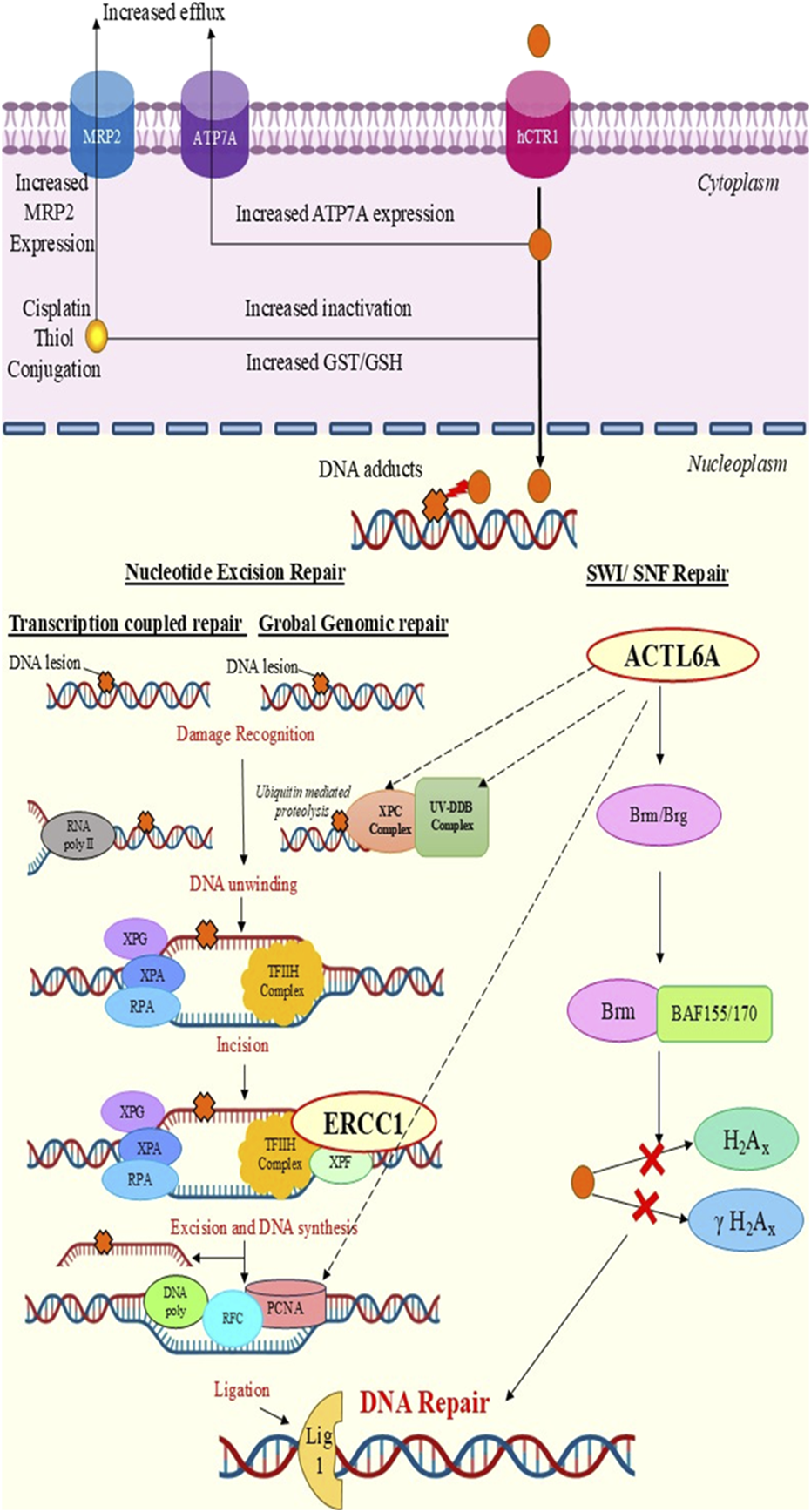
Chemoresistance mechanisms of ERCC1 and ACTL6A. DNA repair is promoted by ERCC1 via the nucleotide excision repair pathway and by ACTL6A through the SWI/SNF complex.
Clinically, a significant proportion of the patients in our study were underweight, so we hypothesize that low BMI may be associated with HNC occurrence; this is also supported by the findings from a Korean study, where the incidence of HNC was observed to be higher among underweight individuals (HR: 1.32) than normal weight and overweight patients (HR: 0.89). Furthermore, it was noted that tobacco smoking (HR: 1.448) and alcohol drinking (HR: 1.448) along with low BMI could impose a significantly higher (p < 0.05) risk of developing HNC (Kim et al., 2022). A study by Eytan et al. (2019) among 10,524 HNC patients in the United States showed that hypertension, hyperlipidemia, chronic obstructive pulmonary disease, and diabetes were the most common comorbidities at the time of diagnosis, which is consistent with the conditions among our population. Although HNC incidence is not believed to depend on a family history of cancer, we observed that approximately 29.87% of our HNC patients presented with such family history; of these, 16.88% reported a family history of HNC, which is a serious concern. A recent study by Pachuau et al. (2022) on north Indians reporting a family history of cancer among first-degree relatives showed that the risk of developing cancer was significantly higher (OR: 1.921, p = 0.037). Furthermore, another study by Li et al. (2021) revealed that the risk of developing HNC among family members increased by 2-fold if the parents/siblings developed HNC. Carcinomas of the oral cavity, larynx, and hypo/oro/nasopharynx were the most predominant types of HNC among our patients, which conform with the sites of HNC development reported from an analysis of 37 Indian cancer registries (Bagal et al., 2023); however, there is a slight disagreement with the findings of Badola et al. (2023) and Chauhan et al. (2022) who reported larynx cancer as the second most-common type after oral cavity cancer. The treatment strategies adopted for our patients (i.e., surgery and CCRT or CCRT alone) comply with the standard treatment guidelines for the management of LAHNC (NCCN Guidelines, 2024; Badola et al., 2023).
Till date, there is only one report of a European study on the dose-dependent expressions of NER genes (Psyrri et al., 2021) among 43 HNC patients, where 35 were responders (81.4%) and 8 were non-responders (18.60%) to cisplatin-based CRT; it was also found that DNA damage, oxidative stress, and NER pathway capacity were significantly higher (p < 0.05) in the cisplatin non-responders than responders owing to diminished apoptosis of the tumor cells among the non-responders. This is in agreement with the findings of our study that ERCC1 expression was significantly increased by 1.64-fold after CCRT compared to the baseline, confirming the increase in NER capacity to clear damaged DNA-cisplatin adducts. Furthermore, approximately 20.78% of the patients in our study showed overexpression of ERCC1 after 100% CCRT, which is nearly equal to that of the non-responder group reported by Psyrri et al. (2021). Although the DNA repair capacity of ERCC1 was found increase with therapy, the overall median expression of ERCC1 was lower than that of the reference gene in our study; this is in agreement with the findings of Psyrri et al. (2021) who observed downregulation of the NER genes, such as ERCC1, ERCC2/XPD, XPA, and XPC, among HNC patients. Even though we predicted no link between overexpression of ERCC1 and overall survival via computational analysis, we found that upregulation of ERCC1 is significantly linked to poor overall survival (HR: 1.82) through the meta-analysis of dose-independent expressions in ERCC1 studies; this is consistent with the previously reported HRs (2.14 and 1.95) among ERCC1 overexpressing HNC patients (Xuelei et al., 2015; Bišof et al., 2016). These findings are attributed to the increased NER capacity via ERCC1, which may be associated with CCRT resistance and poor clinical outcomes among HNC patients. Furthermore, nearly half of the HNC patients (50.33%) among the studies included in the meta-analysis showed high ERCC1 expressions, which is comparatively higher than that observed in our study where 14.29% and 20.78% of the patients had high expressions at baseline and after 100% CCRT, respectively. The details of the studies included in the meta-analysis are outlined in Table 3 (Prochnow et al., 2019; Liang et al., 2015; Ciaparrone et al., 2015; Lu et al., 2017; Xu et al., 2017; An et al., 2017; Gong et al., 2019; Aksoy et al., 2019; Raturi et al., 2020; Chitapanarux et al., 2020; Wang et al., 2021; Hua et al., 2022).
Presently, there are no available studies on evaluating the dose-dependent expression of ACTL6A. However, ACTL6A has been applauded as a novel gene responsible for cisplatin resistance in various cancers, such as ovarian, lung, and esophageal cancers (Xiao et al., 2021). Overexpression of ACTL6A is believed to mediate DNA repair via the SWI/SNF complex by regulating the expression of the Brahma related gene 1 (Brg1) or Brahma (Brm) and promoting its binding to BRAF155/BRAF170 to hinder cisplatin-mediated H2AX or γH2AX activation (Xiao et al., 2021). Out of the four documents that we retrieved through a systematic search, three studies used human tissue samples to explore ACTL6A as a biomarker for cell proliferation, invasion, or metastasis, leading to unfavorable/poor prognosis among HNC patients (Xiao et al., 2021; Liu et al., 2024; Dang et al., 2020; Saladi et al., 2017). A recently published Chinese study by Liu et al. (2024) reported that ACTL6A is significantly overexpressed in oral cancer tissues compared to normal tissues and proposed that tumor factors like E2F7, TP63, and microRNA has-mir-381 regulate ACTL6A expression to promote cell proliferation, migration, and invasion through the WNT and TP53 signaling pathways. It has also been reported that high ACTL6A expression is significantly linked to TP53 mutation rate, which could contribute to chemoresistance to CRT (Xiao et al., 2021). Similarly, studies by Dang et al. (2020) and Saladi et al. (2017) confirmed overexpression of ACTL6A in HNC, anticipating that ACTL6A interacts with P63 and activates the Yes-associated protein (YAP); this could lead to translocation of YAP into the nucleus, which promotes tumorigenesis via the Hippo-YAP signaling pathway (Dang et al., 2020; Saladi et al., 2017). These findings are correlated with those of our study, where we predicted and demonstrated ACTL6A overexpression in HNC via computational analysis and qPCR across the therapy. Furthermore, overexpression of ACTL6A was also predicted to be a significant contributor to poor overall survival. However, none of these studies have demonstrated the involvement of ACTL6A in DNA repair in HNC or its relation to NER. The present study indicates that ACTL6A interacts with the UV-DDB complex, XPC complex of GGR-NER, and PCNA of TCR-NER, thereby contributing to DNA repair. We also found significant and marginally significant correlations between ERCC1 and ACTL6A expressions after 50% (p = 0.003) and 100% (p = 0.06) CCRT, respectively, among the HNC patients, which supports the hypothesis of ACTL6A-mediated NER activation.
Immune cell infiltration of the tumor cells and their interactions with the tumor microenvironment have been proposed to modulate the immune cells, leading to immunosuppression and chemoresistance, thereby resulting in poor clinical outcomes like metastasis and poor survival (Wondergem et al., 2020; Jumaniyazova et al., 2022). However, the inconsistencies in these findings pose conflicts for acceptability in clinical practice. Neutrophil-infiltrating tumor cells undergo polarization to form two phenotypes N1 and N2 that exbibit antitumor and protumor properties, respectively. Here, the N2 phenotype makes the tumor more aggressive by inducing genetic instabilities, angiogenesis, metastasis, and immunosuppression (Wondergem et al., 2020; Jumaniyazova et al., 2022). However, infiltration of the tumor cells by myeloid dendritic cells was reported to exert antitumor and anti-inflammatory effects via increased tumor leucocyte infiltration, whereas plasmacytoid dendritic cell infiltration was reported to be linked with unfavorable outcomes (Wondergem et al., 2020; Jumaniyazova et al., 2022). Similar to neutrophils, macrophages also polarize into M1 and M2 phenotypes, of which the M2 phenotype is linked with protumoral activities, such as tumor migration, invasion, metastasis, and poor survival (Wondergem et al., 2020; Jumaniyazova et al., 2022). To some extent, CD8+ infiltration has been reported to be associated with favorable outcomes, whereas the effects of CD4+ are yet to be clarified (Wondergem et al., 2020; Jumaniyazova et al., 2022). These findings may be important in chemoresistance as both ERCC1 and ACTL6A expressions were found to increase the infiltration of immune cells, such as CD4+ cells, macrophages, myeloid dendritic cells, and B cells.
Nevertheless, knockdown of DNA repair expression could reverse the chemoresistance of or restore sensitivity to the cisplatin or platinum drugs. Among the HNC patients with cisplatin-based CRT resistance or platinum drug resistance, FDA-approved drugs like cyclosporin, 5-fluorouracil, doxorubicin, gemcitabine, paclitaxel, thalidomide, and panobinostat can be repurposed to downregulate ERCC1 and ACTL6A genes. Although paclitaxel and 5-fluorouracil are used for the management of HNC (NCCN Guidelines, 2024), there are no data regarding the use of these anticancer agents against ERCC1 and ACTL6A genes among HNC patients. Thus, we recommend the clinical investigation of these anticancer agents in combination with platinum therapy to mitigate platinum drug resistance or achieve better efficacy of CCRT among HNC patients. Moreover, E-X PPI2, E-X AS7, and panobinostat (a HDAC inhibitor) have been reported to silence ERCC1 and ACTL6A expressions in melanoma and ovarian/lung cancers, respectively, via in vitro and preclinical experiments (Xiao et al., 2021; McNeil et al., 2015). Similarly, siRNA- and shRNA-transfected HNC cell lines have shown promising results for downregulating ACTL6A expressions (Liu et al., 2024; Dang et al., 2020; Saladi et al., 2017); these findings offer hope for tackling chemoresistance in cancer therapy.
7 Limitations and future directions
Although the present study was conducted with a unique methodology to decipher the dose-dependent expressions of chemoresistance genes and has the advantage of a molecularly sensitive technique like qPCR compared to IHC, we were unable to evaluate the tumor burden via the RECIST criteria, which should be addressed in the future to generalize our findings. However, the findings of the current study can also be utilized to conduct a novel clinical trial to investigate the dose-dependent expressions of ERCC1 and ACTL6A among large HNC cohorts along with RECIST mapping of the tumor burden for clinical applicability. Furthermore, ACTL6A (Liu et al., 2024; Dang et al., 2020; Saladi et al., 2017) and ERCC1 (Seetharam et al., 2010; Wang et al., 2017) can be targeted using siRNA and shRNA to silence their expressions to counteract chemoresistance. The present study also offers a hypothesis regarding the associations between chromatin remodeling genes and their DNA repair capacities via the SWI/SNF as well as NER pathways, which could motivate future research in this field.
8 Conclusion
We demonstrate that increased expressions of ERCC1 and ACTL6A during and/or after cisplatin-based CRT can mediate DNA repair, leading to chemoresistance in HNC as well as poor overall survival thereof. ERCC1 and ACTL6A are known to regulate several repair pathways that participate in DNA repair processes. ACTL6A is also known to promote DNA repair activity by interacting with the UV-DDB complex, XPC complex of GGR-NER, and PCNA of TCR-NER. Thus, ERCC1 and ACTL6A are critical evolutionarily conserved core proteins with theranostic potential for cisplatin or cisplatin-based CRT resistance that can be detected via liquid biopsy. Furthermore, repurposing some of the available FDA-approved drugs for targeting ERCC1 and ACTL6A is proposed as a novel approach to counteract chemoresistance in clinical practice.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The study involving humans was approved by the Central Ethics Committee (CEC) of Nitte (Deemed to be University) (Ref. no. NU/CEC/2022/307, revised ref. no. NU/CEC/2024/526) and was registered as a clinical trial in India (CTRI/2022/10/046142). The study was conducted in abeyance with all guidelines of the Declaration of Helsinki. The studies were conducted in accordance with the local legislation and institutional requirements. All participants provided their written informed consent to participate in this study. No potentially identifiable images or data are presented in this work.
Author contributions
RKC: conceptualization, data curation, formal analysis, investigation, methodology, project administration, writing – original draft, and writing – review and editing. PP: formal analysis, methodology, and writing – review and editing. VVS: project administration, resources, supervision, and writing – review and editing. UVM: conceptualization, resources, supervision, validation, and writing – review and editing. PS: formal analysis, resources, validation, and writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors wish to thank the late Dr. Jayarama Shetty. The authors also express their gratitude to the Central Research Laboratory, Nitte (Deemed to be University) along with Mrs. Shweta Shetty K, Mrs. Prajna Bhandary, and Dr. Ananthesh L for their support during the study. The authors are grateful to the Japanese Society of Medical Oncology for providing an opportunity to present the human experimentation part of this research work at the “22nd Annual Meeting of the Japanese Society of Medical Oncology (JSMO2025)” held from 6–8 March 2025 at Kobe, Japan. The abstract (no: 101030) might be included in the supplement of the Annals of Oncology. The graphical abstract was constructed using study images as well as images from free server bioicons.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1541987/full#supplementary-material
Abbreviations
DNA: deoxyribonucleic acid, HNC: head and neck cancer, ERCC1: excision repair cross-complementation group 1, ACTL6A: actin-like protein 6A, CRT: chemoradiotherapy, FDA: Food and Drug Administration, NER: nucleotide excision repair, SWI/SNF: switch/sucrose non-fermenting, LAHNC: locally advanced head and neck cancer, CCRT: concurrent chemoradiotherapy, R/MHNC: recurrent/metastatic head and neck cancer, HR: hazard ratio, TCR: transmission-coupled repair, GGR: global genome repair, PPI: protein–protein interaction, BP: biological process, CC: cellular component, MF: molecular function, IHC: immunohistochemistry, BMI: body mass index, OR: odds ratio.
References
1
AksoyA.ElkiranE. T.HarputluogluH.DagliA. F.IsikdoganA.UrakciZ. (2019). Is excision repair cross-complementation Group1 expression a biological marker in nasopharynx carcinoma?J. Cancer Res. Ther.15 (3), 550–555. 10.4103/0973-1482.206865
2
AnH. J.JoH.JungC. K.KangJ. H.KimM. S.SunD. I.et al (2017). Prognostic implication of ERCC1 protein expression in resected oropharynx and oral cavity cancer. Pathol. Res. Pract.213 (8), 949–955. 10.1016/j.prp.2017.05.006
3
BadolaA.MehtaP.MehraS.SoodS. (2023). Epidemiology and survival analysis of head and neck cancer: results from comprehensive care center in North India. Oral Oncol. Rep.6, 100022–109060. 10.1016/j.oor.2023.100022
4
BagalS.BudukhA.ThakurJ. S.DoraT.QayyumiB.KhannaD.et al (2023). Head and neck cancer burden in India: an analysis from published data of 37 population-based cancer registries. Ecancermedicalscience17, 1603. 10.3332/ecancer.2023.1603
5
BišofV.Zajc PetranovićM.RakušićZ.SamardžićK. R.JuretićA. (2016). The prognostic and predictive value of excision repair cross-complementation group 1 (ERCC1) protein in 1288 patients with head and neck squamous cell carcinoma treated with platinum-based therapy: a meta-analysis. Eur. Arch. Otorhinolaryngol.273 (9), 2305–2317. 10.1007/s00405-015-3710-x
6
CannonM.StevensonJ.StahlK.BasuR.CoffmanA.KiwalaS.et al (2024). DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res.52 (D1), D1227–D1235. 10.1093/nar/gkad1040
7
ChandrashekarD. S.BashelB.BalasubramanyaS. A. H.CreightonC. J.Ponce-RodriguezI.ChakravarthiB. V. S. K.et al (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia19 (8), 649–658. 10.1016/j.neo.2017.05.002
8
ChandrashekarD. S.KarthikeyanS. K.KorlaP. K.PatelH.ShovonA. R.AtharM.et al (2022). UALCAN: an update to the integrated cancer data analysis platform. Neoplasia25, 18–27. 10.1016/j.neo.2022.01.001
9
ChaudharyR. K.KhanalP.MatetiU. V.ShastryC. S.ShettyJ. (2023). Identification of hub genes involved in cisplatin resistance in head and neck cancer. J. Genet. Eng. Biotechnol.21 (1), 9. 10.1186/s43141-023-00468-y
10
ChauhanA. S.PrinjaS.GhoshalS.VermaR.OinamA. S. (2018). Cost of treatment for head and neck cancer in India. PLoS One13 (1), e0191132. 10.1371/journal.pone.0191132
11
ChauhanR.TrivediV.RaniR.SinghU. (2022). A study of head and neck cancer patients with reference to tobacco use, gender, and subsite distribution. South Asian J. Cancer11 (1), 46–51. 10.1055/s-0041-1740601
12
ChitapanaruxI.LekawanvijitS.SripanP.MahanupabP.ChakrabandhuS.OnchanW.et al (2020). The prognostic value of excision repair cross-complementing Group 1 expression in nasopharyngeal cancer patients. J. Res. Med. Sci.25, 34. 10.4103/jrms.JRMS_787_18
13
CiaparroneM.CaspianiO.BiccioloG.SignorelliD.SimonelliI.de CamporaL.et al (2015). Predictive role of ERCC1 expression in head and neck squamous cell carcinoma patients treated with surgery and adjuvant cisplatin-based chemoradiation. Oncology89 (4), 227–234. 10.1159/000430447
14
DandekarM.TuljapurkarV.DharH.PanwarA.DcruzA. K. (2017). Head and neck cancers in India. J. Surg. Oncol.115 (5), 555–563. 10.1002/jso.24545
15
DangY.ZhangL.WangX. (2020). Actin-like 6A enhances the proliferative and invasive capacities of laryngeal squamous cell carcinoma by potentiating the activation of YAP signaling. J. Bioenerg. Biomembr.52 (6), 453–463. 10.1007/s10863-020-09855-3
16
Ensembl genome browser 112 (2023). Available online at: https://www.ensembl.org/index.html (Accessed on March 6, 2023).
17
EytanD. F.BlackfordA. L.EiseleD. W.FakhryC. (2019). Prevalence of comorbidities among older head and neck cancer survivors in the United States. Otolaryngol. Head. Neck Surg.160 (1), 85–92. 10.1177/0194599818796163
18
Global Cancer Observatory (2024). India Fact sheet. Available online at: https://gco.iarc.who.int/media/globocan/factsheets/populations/356-india-fact-sheet.pdf (Accessed on June 1, 2024).
19
GongY.JuH.RenG.WuY. (2019). Cisplatin based induction chemotherapy modified by ERCC1 improved the outcome of young adults with locally advanced oral squamous cell carcinoma. J. Cancer10 (9), 2083–2090. 10.7150/jca.28959
20
GormleyM.CreaneyG.SchacheA.IngarfieldK.ConwayD. I. (2022). Reviewing the epidemiology of head and neck cancer: definitions, trends and risk factors. Br. Dent. J.233 (9), 780–786. 10.1038/s41415-022-5166-x
21
GyőrffyB. (2024). Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innov. (Camb)5 (3), 100625. 10.1016/j.xinn.2024.100625
22
HuH.LiB.WangJ.TanY.XuM.XuW.et al (2023). New advances into cisplatin resistance in head and neck squamous carcinoma: mechanisms and therapeutic aspects. Biomed. Pharmacother.163, 114778. 10.1016/j.biopha.2023.114778
23
HuaL.ChenS.WeiM.ShenY.LongJ.LinZ.et al (2022). Predictive value of ERCC1 mRNA level from receiver-operator characteristic and pretreatment EBV-DNA Virus load in stage II nasopharyngeal carcinoma patients receiving intensity-modulated radiotherapy with concurrent cisplatin. Cancer Biother. Radiopharm.37 (1), 2–10. 10.1089/cbr.2020.4474
24
JumaniyazovaE.LokhoninaA.DzhalilovaD.KosyrevaA.FatkhudinovT. (2022). Immune cells in head-and-neck tumor microenvironments. J. Pers. Med.12 (9), 1521. 10.3390/jpm12091521
25
KannoY.ChenC. Y.LeeH. L.ChiouJ. F.ChenY. J. (2021). Molecular mechanisms of chemotherapy resistance in head and neck cancers. Front. Oncol.11, 640392. 10.3389/fonc.2021.640392
26
KEGG Pathway: (2024). Nucleotide excision repair. Available online at: https://www.kegg.jp/pathway/map03420. Accessed on 1 . Accessed on 1 June 2024
27
KimC. S.ParkJ. O.NamI. C.ParkS. J.LeeD. H.KimH. B.et al (2022). Associations of body mass index and waist circumference with the risk of head and neck cancer: a national population-based study. Cancers (Basel)14 (16), 3880. 10.3390/cancers14163880
28
LiT.FuJ.ZengZ.CohenD.LiJ.ChenQ.et al (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res.48 (W1), W509–W514. 10.1093/nar/gkaa407
29
LiX.KoskinenA. I.HemminkiO.FörstiA.SundquistJ.SundquistK.et al (2021). Family history of head and neck cancers. Cancers (Basel)13 (16), 4115. 10.3390/cancers13164115
30
LiangR.LinY.LiuZ. H.LiaoX. L.YuanC. L.LiaoS. N.et al (2015). Correlation between ERCC1 expression and concurrent chemotherapy and radiotherapy in patients with locally advanced nasopharyngeal cancer. Genet. Mol. Res.14 (2), 5804–5811. 10.4238/2015.May.29.12
31
LiuY.LiuY.LiY.WangT.LiB.KongX.et al (2024). High expression of ACTL6A leads to poor prognosis of oral squamous cell carcinoma patients through promoting malignant progression. Head Neck46 (6), 1450–1467. 10.1002/hed.27742
32
LuY.HuangH.KangM.YiM.YangH.WuS.et al (2017). Combined Ki67 and ERCC1 for prognosis in non-keratinizing nasopharyngeal carcinoma underwent chemoradiotherapy. Oncotarget8 (51), 88552–88562. 10.18632/oncotarget.19158
33
MathurP.SathishkumarK.ChaturvediM.DasP.SudarshanK. L.SanthappanS.et al (2020). Cancer statistics, 2020: report from national cancer registry programme, India. JCO Glob. Oncol.6, 1063–1075. 10.1200/GO.20.00122
34
McNeilE. M.AstellK. R.RitchieA. M.ShaveS.HoustonD. R.BakraniaP.et al (2015). Inhibition of the ERCC1-XPF structure-specific endonuclease to overcome cancer chemoresistance. DNA Repair (Amst)31, 19–28. 10.1016/j.dnarep.2015.04.002
35
NagyÁ.GyőrffyB. (2021). muTarget: a platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer148 (2), 502–511. 10.1002/ijc.33283
36
NCCN Guidelines for Head and neck cancers, version 4. (2024). NCCN clinical practice guidelines in Oncology. Available online at: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf. Accessed 1, June 2024.
37
PachuauL.ZamiZ.NungaT.ZodinglianaR.ZoramthariR.LalnuntluangaR.et al (2022). First-degree family history of cancer can be a potential risk factor among head and neck cancer patients in an isolated Mizo tribal population, northeast India. Clin. Epidemiol. Glob. Health13 (2213-3984), 100954. 10.1016/j.cegh.2021.100954
38
PiemonteE.LazosJ.BelardinelliP.SecchiD.BrunottoM.Lanfranchi-TizeiraH. (2018). Oral cancer associated with chronic mechanical irritation of the oral mucosa. Med. Oral Patol. Oral Cir. Bucal23 (2), e151–e160. 10.4317/medoral.22017
39
PrabhashK.BabuG.ChaturvediP.KuriakoseM.BirurP.AnandA. K.et al (2020). Indian clinical practice consensus guidelines for the management of very advanced disease of squamous cell carcinoma of head and neck. Indian J. Cancer57 (Suppl. ment), S22–S25. 10.4103/0019-509X.278977
40
PrimerBank-MGH-PGA (2023). Available online at: https://pga.mgh.harvard.edu/primerbank/(Accessed on March 6, 2023).
41
ProchnowS.WilczakW.BoschV.ClauditzT. S.MuenscherA. (2019). ERCC1, XPF and XPA-locoregional differences and prognostic value of DNA repair protein expression in patients with head and neck squamous cell carcinoma. Clin. Oral Investig.23 (8), 3319–3329. 10.1007/s00784-018-2751-0
42
PROSPERO (2024). International prospective register of systematic reviews. Available online at: https://www.crd.york.ac.uk/prospero/(Accessed on May 13, 2024).
43
PsyrriA.GkotzamanidouM.PapaxoinisG.KrikoniL.EconomopoulouP.KotsantisI.et al (2021). The DNA damage response network in the treatment of head and neck squamous cell carcinoma. ESMO Open6 (2), 100075. 10.1016/j.esmoop.2021.100075
44
PubChem (2024). Available online at: https://pubchem.ncbi.nlm.nih.gov/(Accessed on June 8, 2024).
45
RanasingheR.MathaiM. L.ZulliA. (2022). Cisplatin for cancer therapy and overcoming chemoresistance. Heliyon8 (9), e10608. 10.1016/j.heliyon.2022.e10608
46
RaturiV. P.WuC. T.MohammadS.HojoH.BeiY.NakamuraM.et al (2020). Could excision repair cross-complementing group-1 mRNA expression from peripheral blood lymphocytes predict locoregional failure with cisplatin chemoradiation for locally advanced laryngeal cancer?Asia Pac. J. Clin. Oncol.16 (2), e19–e26. 10.1111/ajco.13239
47
RCSB PDB (2024). Homepage. Available online at: https://www.rcsb.org/(Accessed on June 8, 2024).
48
SaladiS. V.RossK.KaraayvazM.TataP. R.MouH.RajagopalJ.et al (2017). ACTL6A is Co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell31 (1), 35–49. 10.1016/j.ccell.2016.12.001
49
SathiyaPriyaS.ShaviG. R.SangaR.ShankarS.GunasekaranL.RahilaC.et al (2024). Assessment of the prevalence of types of head and neck cancer in a tertiary cancer center at Madurai, Tamil Nadu: a cross-sectional study. J. Cancer Res. Ther.20 (3), 959–965. 10.4103/jcrt.jcrt_2081_22
50
SEER Cancer Stat Facts (2024). Available online at: https://seer.cancer.gov/statfacts/(Accessed on July 15, 2024).
51
SeetharamR. N.SoodA.Basu-MallickA.AugenlichtL. H.MariadasonJ. M.GoelS. (2010). Oxaliplatin resistance induced by ERCC1 up-regulation is abrogated by siRNA-mediated gene silencing in human colorectal cancer cells. Anticancer Res.30 (7), 2531–2538.
52
SinghviH. R.MalikA.ChaturvediP. (2017). The role of chronic mucosal trauma in oral cancer: a review of literature. Indian J. Med. Paediatr. Oncol.38 (1), 44–50. 10.4103/0971-5851.203510
53
SzklarczykD.KirschR.KoutrouliM.NastouK.MehryaryF.HachilifR.et al (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res.51 (D1), D638–D646. 10.1093/nar/gkac1000
54
The Human Protein Atlas (2024). Available online at: https://www.proteinatlas.org/(Accessed on June 5, 2024).
55
TierneyJ. F.StewartL. A.GhersiD.BurdettS.SydesM. R. (2007). Practical methods for incorporating summary time-to-event data into meta-analysis. Trials8, 16. 10.1186/1745-6215-8-16
56
WangW.ZhangL.LiuL.ZhengY.ZhangY.YangS.et al (2017). Chemosensitizing effect of shRNA-mediated ERCC1 silencing on a Xuanwei lung adenocarcinoma cell line and its clinical significance. Oncol. Rep.37 (4), 1989–1997. 10.3892/or.2017.5443
57
WangY. Y.FangP. T.SuC. W.ChenY. K.HuangJ. J.HuangM. Y.et al (2021). Excision repair cross-complementing group 2 upregulation is a potential predictive biomarker for oral squamous cell carcinoma recurrence. Oncol. Lett.21 (6), 450. 10.3892/ol.2021.12711
58
WondergemN. E.NautaI. H.MuijlwijkT.LeemansC. R.van de VenR. (2020). The immune microenvironment in head and neck squamous cell carcinoma: on subsets and subsites. Curr. Oncol. Rep.22 (8), 81. 10.1007/s11912-020-00938-3
59
XiaoY.LinF. T.LinW. C. (2021). ACTL6A promotes repair of cisplatin-induced DNA damage, a new mechanism of platinum resistance in cancer. Proc. Natl. Acad. Sci. U. S. A.118 (3), e2015808118. 10.1073/pnas.2015808118
60
XuS.YuY.RongJ.HuD.ZhangL.FuS.et al (2017). Expression of BRCA1 and ERCC1 as predictive clinical outcome after radiochemotherapy in patients with locoregionally moderate-advanced nasopharyngeal carcinoma. Oncotarget8 (19), 31355–31367. 10.18632/oncotarget.15565
61
XueleiM.JingwenH.WeiD.HongyuZ.JingZ.ChangleS.et al (2015). ERCC1 plays an important role in predicting survival outcomes and treatment response for patients with HNSCC: a meta-analysis. Oral Oncol.51 (5), 483–492. 10.1016/j.oraloncology.2015.02.094
Summary
Keywords
chemoradiotherapy, chemoresistance, cisplatin, DNA repair, drug repurposing, evidence
Citation
Chaudhary RK, Patil P, Shetty VV, Mateti UV and Shetty P (2025) A multimodal approach for establishing ACTL6A and ERCC1 as chemoresistance genes in locally advanced head and neck cancer. Front. Pharmacol. 16:1541987. doi: 10.3389/fphar.2025.1541987
Received
09 December 2024
Accepted
17 April 2025
Published
29 May 2025
Volume
16 - 2025
Edited by
Luis Abel Quiñones, University of Chile, Chile
Reviewed by
Ashwin Kotnis, All India Institute of Medical Sciences, Bhopal, India
Yabing Nan, Department of Cancer Biology, Dana–Farber Cancer Institute, United States
Updates
Copyright
© 2025 Chaudhary, Patil, Shetty, Mateti and Shetty.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uday Venkat Mateti, udayvenkatmateti@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.