- 1College of Pharmacy, Binzhou Medical University, Binzhou, Shandong, China
- 2Non-Clinical Research Department, RemeGen Co., Ltd, Yantai, Shandong, China
Introduction: Age-related macular degeneration (AMD) is a leading cause of vision loss in older adults, with limited effective treatments available. This study aimed to investigate the pharmacological effects of dihydromyricetin (DHM) on AMD and to identify its putative pharmacological targets through network analysis and molecular docking approaches.
Methods: In vitro experiments established an AMD model using sodium iodate (SI)-induced ARPE-19 cells, with CCK-8 assays determining 15 mM SI as the optimal modeling concentration and 100 μM DHM as the optimal treatment concentration. For in vivo validation, an AMD model was generated in C57 mice via tail vein injection of SI (30 mg/kg). Subsequent oral gavage with DHM (50 or 100 mg/kg) was administered. Integrated network analysis, molecular docking, and RT- qPCR validation were employed.
Results: RT-qPCR analysis revealed that DHM reversed SI-induced aberrant expression of AMD-associated biomarkers (ICAM-1, APOE, HTRA1, ABCA4). Light microscopy and flow cytometry demonstrated DHM's significant mitigation of SI-triggered cellular morphological alterations and apoptosis (35% reduction). Western blot analysis further confirmed DHM-mediated suppression of apoptosis through regulation of p53, Bax, cleaved caspase-3, and Bcl-2 expression. High-dose DHM significantly attenuated retinal thinning (10.7% reduction), decreased pigment loss, and ameliorated structural disorganization in the outer nuclear layer (ONL). These analyses predicted seven putative targets implicated in functional categories including neurodegeneration, apoptosis, and DNA modification. Subsequent PPI network construction and GO/KEGG enrichment analyses revealed these targets' involvement in biological processes such as angiogenesis and extracellular matrix organization.
Conclusion: In conclusion, the present study demonstrates that DHM can mitigate AMD-related damage in both in vitro and in vivo models, while predicting putative targets and signaling pathways through which DHM may exert its effects against AMD. These findings offer promising directions for the development of AMD therapies and lay the groundwork for further investigation into DHM as a candidate drug for treating and preventing AMD.
1 Introduction
Age-related macular degeneration (AMD) is a complex, multifactorial eye disease primarily affecting adults over 55 years old. It is a leading cause of vision loss in older adults, significantly impairing daily activities that require high-acuity central vision, such as reading and driving (Guymer and Campbell, 2023; Schmidt-Erfurth et al., 2014). With a global prevalence affecting approximately 196 million people worldwide, the prevalence of AMD is projected to rise to 288 million by 2040, a major public health concern (Fleckenstein et al., 2024). The disease is characterized by progressive degeneration of the retinal pigment epithelium (RPE), photoreceptor cells, and, in advanced stages, the choroidal vasculature (Blasiak, 2020).
AMD development is influenced by multiple risk factors, including age, genetic predisposition, and environmental triggers (Ambati and Fowler, 2012; Heesterbeek et al., 2020). Genetic studies have identified key pathogenic factors, such as variants in the ARMS2/HTRA1 (Age-Related Maculopathy Susceptibility 2/HtrA Serine Peptidase 1) gene locus, which are strongly associated with an increased risk of neovascular AMD (Fritsche et al., 2016; DeWan et al., 1979). Additionally, complement-mediated activation and chronic inflammation have been linked to disease progression through ICAM-1(Intercellular Adhesion Molecule 1) secretion from choroidal endothelial cells, while drusen deposits beneath the RPE play a central role in initiating inflammatory responses (Skei et al., 2010). These extracellular deposits, composed of proteins, lipids, and cellular debris, can activate the complement system, recruiting immune cells that exacerbate RPE damage and retinal degeneration (Crabb et al., 2002; Wang et al., 2010; Mullins et al., 2000). Despite extensive research, the pathogenesis of AMD remains incompletely understood.
Oxidative stress and inflammation are recognized as critical factors driving disease progression (Datta et al., 2017; Heloterä and Kaarniranta, 2022). Oxidative stress, resulting from an imbalance between the production of reactive oxygen species (ROS) and cellular antioxidant defenses, is a key contributor to AMD. Persistent oxidative stress leads to mitochondrial dysfunction, lipid peroxidation, and protein/DNA damage, ultimately causing RPE apoptosis (Chan et al., 2019; Kaarniranta et al., 2019). The loss of RPE cells disrupts retinal structural integrity and function, accelerating the progression of AMD (Tang et al., 2021). Inflammation also plays a crucial role in AMD pathogenesis: drusen accumulation—extracellular deposits containing lipids, proteins, and cellular debris—beneath the RPE induces a chronic inflammatory response (Sakurada et al., 2023). This response involves complement system activation and immune cell recruitment, which further damage the RPE and surrounding structures (Crabb, 2014). Consequently, chronic inflammation and oxidative damage create a vicious cycle that promotes retinal degeneration and disease progression. Given the roles of oxidative stress and inflammation, targeting these pathways may slow the onset and progression of AMD.
Sodium iodate (SI) is a stable oxidizing agent widely used to induce retinal degeneration characterized by localized RPE loss and retinal toxicity in mice (NILSSON et al., 1977; Hariri et al., 2013), rats (Shen et al., 2010) and rabbits (Obata et al., 2005). Consequently, it serves as a well-established method for generating in vitro and in vivo AMD models.
Current AMD treatments primarily target the advanced neovascular form of the disease (Heloterä and Kaarniranta, 2022; Fabre et al., 2022). Anti-vascular endothelial growth factor (anti-VEGF) therapies inhibit pathological retinal angiogenesis, slowing vision loss progression (Girgis and Lee, 2023). However, these treatments require frequent intravitreal injections—an invasive approach associated with patient discomfort—and fail to address the underlying oxidative stress and inflammatory drivers of AMD. Critically, no effective therapies exist for dry AMD, which constitutes most cases and is defined by geographic atrophy of the RPE and photoreceptors (Stahl, 2020). These limitations underscore the urgent need for novel therapeutic strategies targeting upstream pathogenic mechanisms, particularly oxidative stress and inflammation.
Dihydromyricetin (DHM), a flavonoid metabolite derived from Nekemias grossedentata (Hand.-Mazz.) J. Wen and Z.L.Nie (Zeng et al., 2023), exhibits potent antioxidant, anti-inflammatory, and neuroprotective properties across multiple disease models. These include antitumour (Sun et al., 2019; Liu et al., 2014),antithrombotic (Jing and Li, 2020; Weng et al., 2017), and glucose/lipid regulatory effects (Liu et al., 2017; He et al., 2019), with particularly significant pharmacological effects in liver diseases (Chen et al., 2021; Chen et al., 2020). Studies demonstrate that DHM ameliorates dextran sulfate sodium-induced colitis in mice by modulating gut microbiota bile acid metabolism (Dong et al., 2021). Furthermore, DHM alleviates high glucose-induced oxidative stress and apoptosis in human retinal pigment epithelial cells via miR-34a downregulation, confirming its protective role against AMD in vitro (Li and Xiao, 2021). Collectively, these findings suggest DHM may mitigate AMD-associated oxidative stress and inflammation. By reducing Reactive Oxygen Species (ROS) production and regulating inflammatory pathways, DHM could preserve retinal structure and function, suggesting therapeutic potential for AMD management.
Network analysis combines systems biology principles to identify multi-target drug molecules by targeting key signaling nodes (Nogales et al., 2022; Zhao et al., 2023). When combined with molecular docking—which predicts optimal binding conformations between interacting molecules—this integrated approach effectively discovers putative targets for complex diseases (Li et al., 2022). Numerous studies have applied this strategy to uncover new drug targets (Zhou et al., 2022; Sun et al., 2023; Zhang et al., 2021), including isobavachin’s anti-hyperuricemic effect and fenugreek-derived multi-target therapies for diabetes (Luo J. et al., 2023; Luo W. et al., 2023).
This study aims to investigate the protective effects of DHM against sodium iodate-induced AMD both in vitro and in vivo, while predicting putative targets through network analysis and molecular docking techniques. This research approach investigated the pharmacological effects of DHM in AMD treatment and predicted putative targets using network analysis and molecular docking methodologies, thereby providing a foundation for developing novel therapeutic strategies targeting oxidative stress and inflammation in ophthalmic diseases with DHM.
2 Materials and methods
2.1 Cell culture and drugs treatment
The adult retinal pigment epithelial cell line ARPE-19 (ATCC) was routinely cultured in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) (ATCC, Manassas, VA, United States, 30–2006) containing 10% fetal bovine serum (BI) and 100 U/mL penicillin/0.1 mg/mL streptomycin at 37°C in an incubator with 5% CO2. For model induction, SI was added to DMEM/F12 medium to a final concentration of 15 mM (Sigma-Aldrich, Saint Louis, MO, United States, S4007), and ARPE-19 cells were treated for 24 h to induce the AMD cell model. For drug treatment, DHM (0.1 mM) (MeilunBio, Dalian, LN, China, MB5834) and NaIO3 (15 mM) were added to DMEM/F12 medium and ARPE-19 cells were treated for 24 h.
2.2 Cell viability assay
The survival rate of ARPE-19 cells was determined using a Cell Counting Kit-8 (Beyotime, Shanghai, China, C0037). ARPE-19 cells were inoculated into 96-well culture plates (8 × 104 cells/well) and treated with DHM or NaIO3 for 24 h, followed by incubation with CCK-8 reagent mixed 1:10 with fresh medium for 4 h, and absorbance was measured with an enzyme marker at 570 nm.
2.3 RNA extraction and RT-qPCR analysis
Total RNA was extracted from cultivated cells using Quick cell RNA extraction kit (SparkJade, Jinan, SD, China, AC0205-A). The cDNAs were synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Kyoto, Japan, 3735A). Quantitative RT-qPCR was performed with the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, United States). Ct values were calculated as: ∆Ct = Ct (target gene) – Ct (reference gene); ∆∆Ct = ∆Ct (treatment group) − ∆Ct (control group). The relative expression of the target genes was calculated using 2−ΔΔCT. All the primer sequences used in the experiment can be found in Supplementary Material 1.
2.4 Detection of cell death by flow cytometry
ARPE-19 cells were stained with Annexin V-FITC/PI using a commercial Annexin V-FITC/PI Apoptosis Detection Kit (Biolegend, Santiago, CA, United States, 640,914) according to the manufacturer’s instructions. ARPE-19 cells were treated with or without drugs for 24 h and then subjected to centrifugation to collect the cells, with 5 × 105 cells per group. The cells were then incubated with Annexin V-FITC and PI, after which they were transferred to flow tubes and analyzed by flow cytometry (SONY, Kyoto, Japan, SA3800). Data analysis was performed using SA3800 software.
2.5 Immunoblotting
The cell pellet for protein expression detection was lysed by RIPA buffer (Beyotime, Shanghai, China, P0013B) containing a cocktail protease inhibitor (Beyotime, Shanghai, China, P1006). Protein concentration was quantified using the BCA method (Beyotime, Shanghai, China, P0009). According to the quantitative results, all the groups were unified at the same concentration. The lysate was then boiled at 95°C for 10 min. For SDS-PAGE, gradient gels (GenScript, Nanjing, JS, China, M00657) with a concentration range of 4%–20% were used to separate protein samples. The separated proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 5% milk in PBST at room temperature for 1 h. The corresponding primary antibody was used to incubate the blocked membrane overnight at 4 °C. After combining with secondary antibodies from the same species, protein bands were visualized by chemiluminescent substrates (Thermo Fisher Scientific, Waltham, MA, United States, 34,095). All the antibodies used in the experiment can be found in Supplementary Material 2.
2.6 In vivo animal model
This protocol of the animal experiment was approved by the Institutional Animal Care and Use Committee of Binzhou Medical University. Six-week-old male C57BL/6 J mice were purchased from Pengyue (Jinan, SD, China). The experimental mice were divided into four groups, each comprising eight mice, and their body weights were measured at designated time points throughout the dosing cycle. Mouse AMD model was prepared and each mouse received tail vein injection of sodium iodate (Sigma-Aldrich, Saint Louis, MA, United States, S4007) (30 mg/kg) (Wang et al., 2014). DHM was dissolved in 0.9% NaCl to prepare low-dose (50 mg/kg) and high-dose (100 mg/kg) solutions for post-modeling treatment (Dong et al., 2021; Xie et al., 2022; Yan et al., 2024). DHM was administered by gavage starting 3 days before SI injection, continuing for 8 days post-injection, with a total administration duration of 11 days.
2.7 Spectral domain optical coherence tomography (SD-OCT) and funduscopic examination
Mice were anesthetized by intraperitoneal injection of 1.25% tribromoethanol (AibeiBio, Nanjing, JS, China, M2910), followed by bilateral administration of 5 μL compound tropicamide ophthalmic solution (Novartis, Basel, Switzerland, S01FA51) to induce pupillary dilation; after achieving maximal mydriasis (>1.5 mm diameter), residual drops were removed and corneas protected with Carbomer 940 gel (Siccanove, Nice, France, CBM-940-OG) to prevent dehydration-induced opacity. Animals were immobilized in a stereotaxic platform with optimized ocular alignment for sequential imaging. Fundus photography was performed using a Micron III system (Phoenix Research Laboratories, Pleasanton, CA, United States), with pigment deposit area quantified using ImageJ (1.54i, NIH, Montgomery, MD, United States). High-resolution SD-OCT scans were acquired using radial B-scan patterns (Bioptigen, Morrisville, NC, United States). Retinal thickness (ILM-to-RPE distance) was quantified from optic disc-centered images via automated segmentation in InVivoVue Diver software (v3.1.7, Bioptigen).
2.8 Histological analysis of the retina
Enucleated mouse eyes were immediately fixed in 10% neutral buffered formalin (Sigma-Aldrich, Saint Louis, MO, United States, HT501640) at 4°C for 24 h to preserve retinal cytoarchitecture. Post-fixation, specimens were dehydrated through a graded ethanol series (Sinopharm, Shanghai, China, 64–17-5), cleared in xylene (Sinopharm, Shanghai, China, 1,330–20-7), and embedded in paraffin (Sinopharm, Shanghai, China, 8,002–74-2) per standard histological protocols. Serial sagittal sections containing optic nerve heads were cut at 3 μm thickness using a rotary microtome (Huasu Technology, Jinhua, ZJ, China, HS-3315), mounted on poly-L-lysine-coated slides (Epredia, Portsmouth, NH, United States, J2800AMNZ), and stained with hematoxylin and eosin (H&E) under controlled conditions: hematoxylin (Sigma-Aldrich, St. Louis, MO, United States, GHS316; 5 min), differentiation in 1% acid alcohol (30 s), eosin counterstain (Sigma-Aldrich, St. Louis, MO, United States, HT110316; 2 min). Bright-field images were acquired at × 200 magnification using an AxioScope 5 microscope (Carl Zeiss, Oberkochen, Germany, AxioScope5).
2.9 Target acquisition
DHM targets were retrieved from Swiss Target Prediction (http://www.swisstargetprediction.ch/) (Daina et al., 2019). AMD-related targets were collected from the OMIM database (http://omim.org/) (Hamosh et al., 2000) and GeneCards Database (http://www.genecards.org/). Then, the false-positive and duplicate targets were deleted and integrated. Finally, the targets of DHM and the targets of AMD were intersected to obtain the putative targets of DHM treating AMD.
2.10 Construction of the protein–protein interaction (PPI) network and functional enrichment analysis of genes in networks
Importing putative targets of DHM for AMD into GeneMANIA Database (https://genemania.org/) (Warde-Farley et al., 2010), and constructing protein-protein interaction networks. A cluster of genes highly associated with putative targets was obtained by constructing a network. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets were downloaded from the official GSEA website (https://www.gsea-msigdb.org/gsea/downloads.jsp) (Subramanian et al., 2005). Utilizing this as a foundation, all network genes were mapped to the background set and enriched through the R package clusterProfiler (version 3.14.3) to obtain gene set enrichment results.
2.11 Molecular docking of DHM with putative AMD targets
The 3D structure of DHM was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The PDB files of the target protein structures were obtained from the RCSB PDB database (http://www.rcsb.org/) (Subramanian et al., 2005). Proteins were prepared using PyMOL 2.3.0 (https://www.pymol.org/). Docking was accomplished using AutoDock Vina 1.1.2 software (Trott and Olson, 2010). Subsequently, visual analysis was performed using PyMOL 2.3.0. The protein structures were prepared by removing water molecules and adding polar hydrogen atoms. The grid box was centered on the ligand-binding site with dimensions of 20 × 20 × 20 Å. A total of 10 conformational poses were generated for each ligand-target complex. The pose with the lowest binding energy (ΔG, kcal/mol) was selected for further analysis.
2.12 Statistical analyses
Statistical analyses and graphs were generated using GraphPad Prism 8.0.1 (San Diego, CA, United States). Normality was tested for all datasets. Data are presented as mean ± standard deviation (SD). For normally distributed data, differences among multiple groups were analyzed by one-way ANOVA; for non-normally distributed data, the Kruskal–Wallis test was used, supplemented by homogeneity of variance testing. All data met homogeneity of variance requirements. Thus, pairwise comparisons for normally distributed data used Tukey’s test, while Dunn’s test was applied to non-normally distributed data. Statistical significance between groups was determined by these post-hoc tests (*p < 0.05, **p < 0.01, ***p < 0.001).
3 Results
3.1 In Vitro verification of DHM’s inhibition of AMD progression
SI-induced RPE cell injury is a well-established in vitro AMD model. Reported SI concentrations vary across studies due to methodological differences.
First, we determined SI cytotoxicity in ARPE-19 cells under our experimental conditions. The half-maximal inhibitory concentration (IC50) of SI was 12.86 mM (CCK-8 assay; Figure 1A). For model consistency, 15 mM SI was selected for subsequent experiments. Six DHM concentrations (0, 12.5, 25, 50, 100, 200 μM) were tested based on prior studies (Dong et al., 2021; Li and Xiao, 2021). DHM’s protective effect against SI-induced injury was assessed via CCK-8 assay across six concentrations. Under the treatment of 100 μM DHM, the cell viability was about 86%, and with a 120% improvement compared to the SI-only group. Thus, 100 μM DHM was selected as the optimal concentration (Figure 1B).
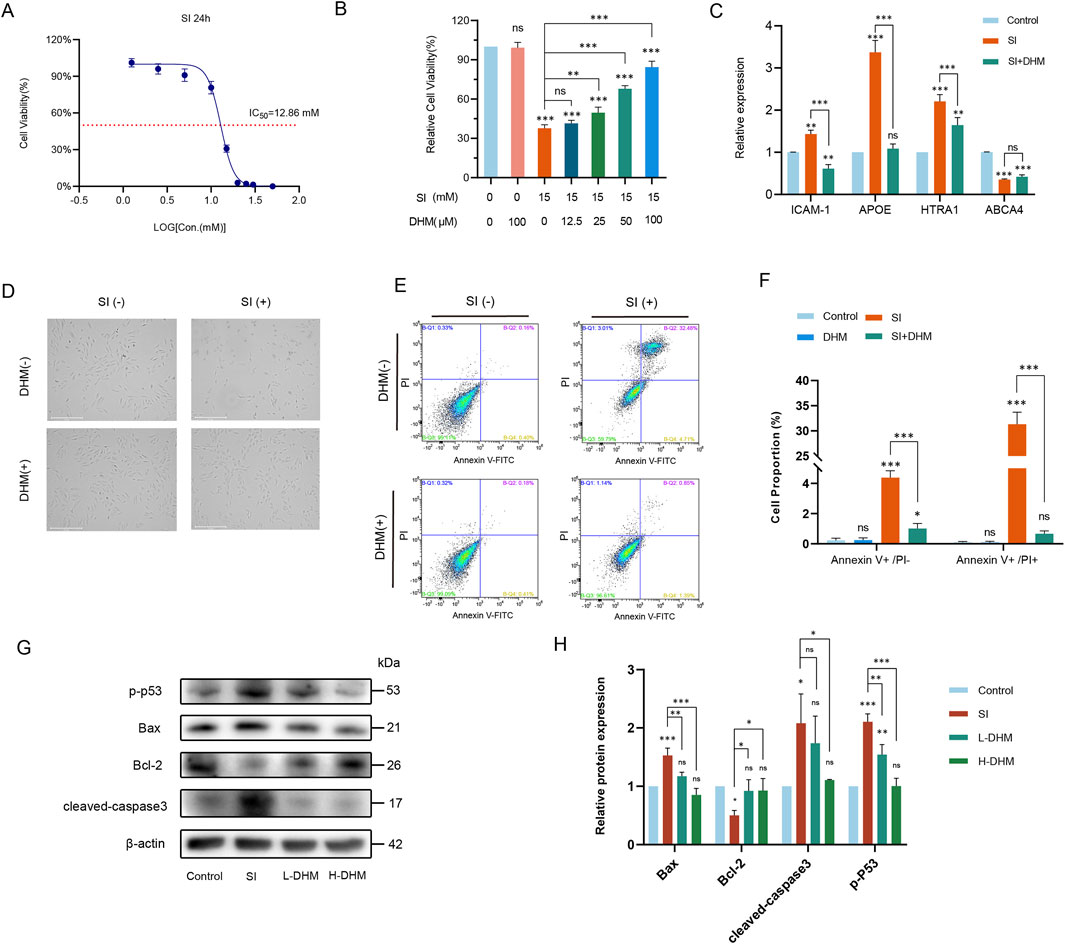
Figure 1. DHM attenuates sodium iodate-induced AMD pathology in vitro. (A) IC50 determination of SI in ARPE-19 cells via CCK-8 assay. (B) Cell viability after 24 h co-treatment with 15 mM SI and DHM (CCK-8 assay). (C) mRNA expression of AMD-associated markers (ICAM-1, APOE, HTRA1, ABCA4) by RT-qPCR. (D) Morphological changes in ARPE-19 cells post 24 h treatment. (E) Apoptosis analysis by Annexin V-FITC/PI flow cytometry. (F) Quantification of flow cytometry data. (G) Western blot analysis of apoptosis-related proteins (p-p53, Bax, cleaved caspase-3, Bcl-2). (H) Densitometric quantification of protein expression. Data were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test and are presented as mean ± standard deviation (n = 3 per group). ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001. Significance symbols: Above bars: vs. untreated control; Between bars: vs. SI model group.
The mRNA expression of four AMD-associated markers—ICAM-1, APOE, HTRA1, and ABCA4—was analyzed by RT-qPCR (Yang et al., 2016; Vessey et al., 2022; Pan et al., 2023; Farnoodian et al., 2022). Compared to untreated controls, modeling with 15 mM SI significantly upregulated ICAM-1, APOE, and HTRA1 transcripts while downregulating ABCA4. Notably, 100 μM DHM treatment reversed these alterations, restoring marker expression toward baseline levels relative to the model group (Figure 1C).
Light microscopy revealed comparable cellular morphology between the DHM-only (100 μM) and untreated control groups. Following 15 mM SI treatment, ARPE-19 cells exhibited pronounced morphological alterations and extensive cell death. However, DHM co-treatment markedly reduced SI-induced cell death and restored normal morphology (Figure 1D). Flow cytometric analysis demonstrated that 100 μM DHM alone induced no significant cytotoxicity relative to negative controls. After sodium iodate treatment, approximately 5% of the cells were FITC Annexin V positive and PI negative (early apoptosis), and approximately 30% of the cells were FITC Annexin V and PI positive (terminal apoptosis or necrosis). DHM co-treatment significantly reduced both apoptotic populations and normalized cellular viability (Figures 1E,F).
To elucidate DHM’s role in SI-induced apoptosis, Western blot analysis revealed that SI upregulated phosphorylated p53 (p-p53), Bax, and cleaved caspase-3 while downregulating the anti-apoptotic protein Bcl-2 relative to controls. Critically, both high and low DHM concentrations attenuated these alterations: suppressing p-p53, Bax, and cleaved caspase-3 overexpression, and restoring Bcl-2 levels. Thus, DHM inhibited SI-triggered apoptosis in ARPE-19 cells (Figures 1G,H).
3.2 In Vivo pharmacological effects of DHM in AMD mouse models
To validate DHM responses in murine AMD, we established an AMD model in C57 mice by injecting SI (30 mg/kg) into the tail vein. Mice received oral DHM (50 or 100 mg/kg) for 3 days before SI injection, and treatment was continued for 7 days post-injection (Figure 2A). Body weights were measured every 3 days throughout the experiment. No significant differences in body weight were observed among the four groups, and no adverse effects (e.g., piloerection, convulsions, motor disorders, or abnormal somnolence) occurred (Figure 2B). Retinal thickness (from the ILM to the ROST) was measured by fundus photography and SD-OCT before DHM administration and at the end of the treatment cycle. The SI model group showed a significant reduction in retinal thickness (approximately 39.4 μm). Both DHM doses attenuated this reduction, but only the high-dose group (100 mg/kg) showed statistically significant effects, with an average improvement of 10.7 μm (27% improvement rate) (Figures 2C,D). Fundus examination revealed that DHM reduced retinal pigment loss by approximately 5% (low-dose) and 6.5% (high-dose) (Figures 2E,F). Furthermore, HE staining showed pronounced disorganization in the outer nuclear layer (ONL) of SI-treated mice versus controls, while DHM treatment reduced ONL disorganization (Figure 2G). These findings indicate that oral DHM markedly attenuates SI-induced retinal injury.
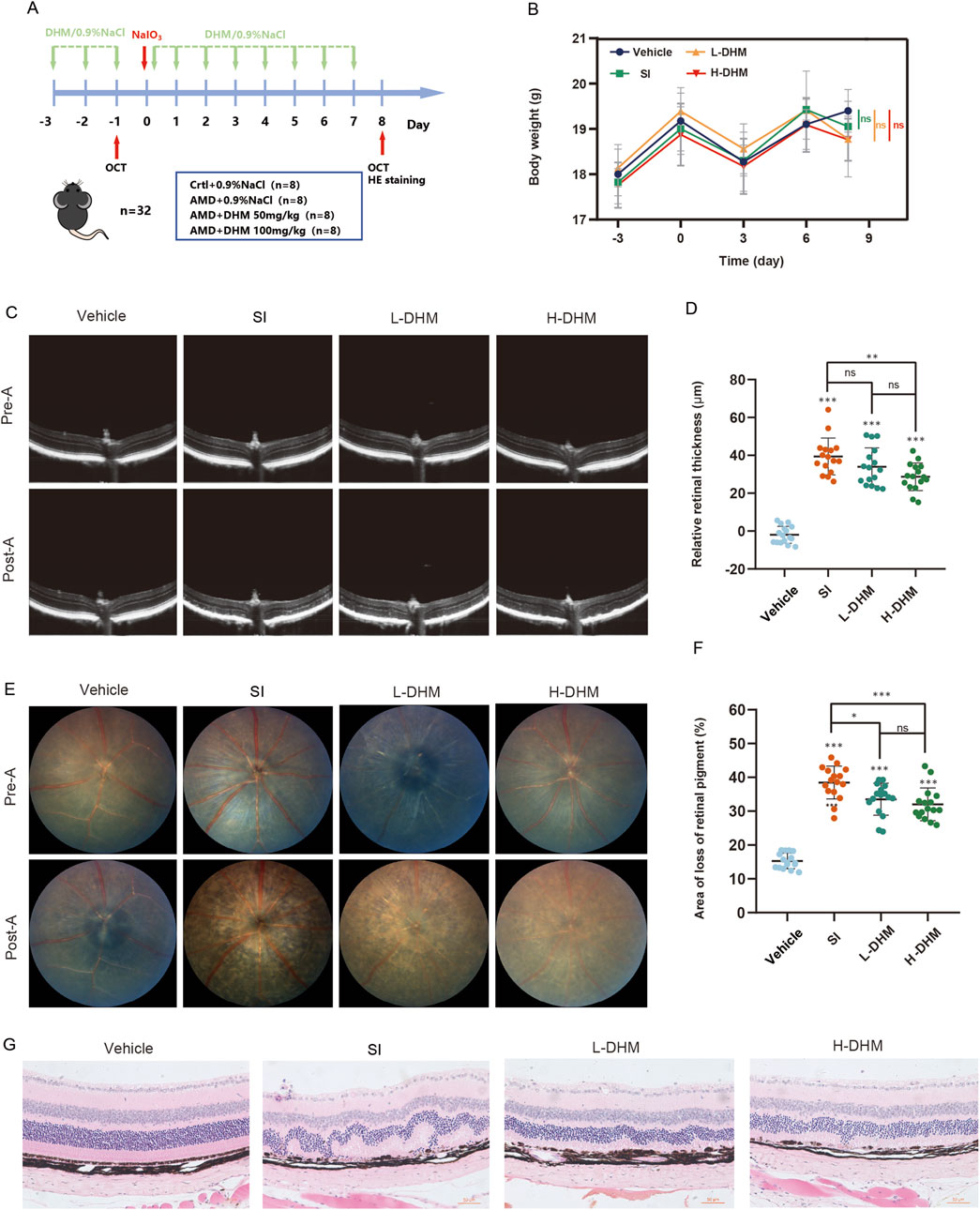
Figure 2. DHM alleviates AMD pathology in vivo. (A) Experimental timeline of DHM and SI administration in C57 mice. (B) Body weight changes across treatment groups during the study. (C) Retinal thickness measurements by SD-OCT pre- and post-treatment. (D) Quantitative analysis of retinal thickness changes. (E) Representative fundus images showing retinal pigment status. (F) Quantification of retinal pigment loss improvement. (G) H&E staining of retinal sections (The scale bar = 50 μm, and the magnification is ×200). Data were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test and are presented as mean ± SD (n = 16 per group). ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001. Significance symbols: Above bars: vs. untreated control; Between bars: vs. SI model group.
3.3 Prediction of putative DHM targets against AMD
AMD-related targets were collected from OMIM (gene map) and GeneCards (relevance score >10) and a total of 636 AMD-related target genes were obtained after deletion of duplicates. The DHM drug target genes were collated from the Swiss Target Prediction (Probability*>0). Resulting in a total of 70. AMD disease target genes and DHM drug target genes were taken and intersection was taken to obtain putative targets of DHM against AMD. Eventually, a target gene set of 12 targets was obtained (Figure 3A). These targets can be broadly classified into seven categories according to their functions, including neurodegenerative proteins (APP, MAPT), apoptosis-related proteins (BCL2), Enzymes of DNA Modification and Maintenance Category (DNMT1, TERT), Transcriptional regulatory factors (HIF1A), Growth factors and cognate receptors (KDR, PGF, VEGFA), protein modifying enzymes (MMP2, MMP9) and nuclear receptors (PPARG) (Figure 3B).
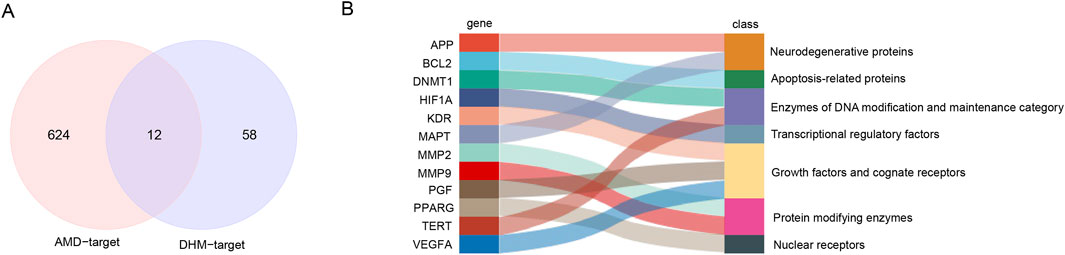
Figure 3. Predicting putative targets of DHM for alleviating AMD. (A) Venn diagram of candidate targets identified through multi-database screening. (B) Functional enrichment analysis of the 12 putative targets.
3.4 PPI network construction and Functional Enrichment analysis of target gene sets
A protein-protein interaction (PPI) network comprising 12 core targets and 20 functionally related genes was constructed using GeneMANIA, visually presenting interactions among target genes (Figure 4A). The full set of 32 genes underwent GO and KEGG enrichment analyses. GO biological process analysis revealed top 10 pathways enriched in angiogenesis regulation, including vascular endothelial growth factor signaling, vasculature development, and endothelial cell proliferation, indicating core targets’ roles in vascular morphogenesis and proliferation control (Yan et al., 2024; Gui et al., 2023) (Figure 4B). GO cellular component analysis showed enrichment in subcellular structures: platelet α granule lumen, telomerase holoenzyme complex, collagen-containing extracellular matrix, secretory vesicles, and axons, suggesting involvement in vesicle transport, extracellular matrix organization, and neuronal maintenance (Figure 4C). Notably, VEGF from platelet α granules may promote angiogenesis via vesicular transport (Manne et al., 2017). GO molecular function terms included VEGF receptor binding, growth factor activity, and chemotactic factor activity, implying regulation of angiogenesis and immune cell recruitment (Figure 4D) (Sharma et al., 2012; Rosenfeld, 2011). DHM may inhibit angiogenesis by blocking VEGF receptor binding or chemokine activity, aligning with anti-angiogenic therapies (e.g., bevacizumab). KEGG pathway enrichment highlighted Pathways in Cancer, MAPK signaling, and PI3K-Akt signaling, linking targets to oncogenic signaling and metabolic disease mechanisms (Figure 4E) (Wu et al., 2025; Muraleva and Kolosova, 2023). Complete GO/KEGG results are in Supplementary Material 3.
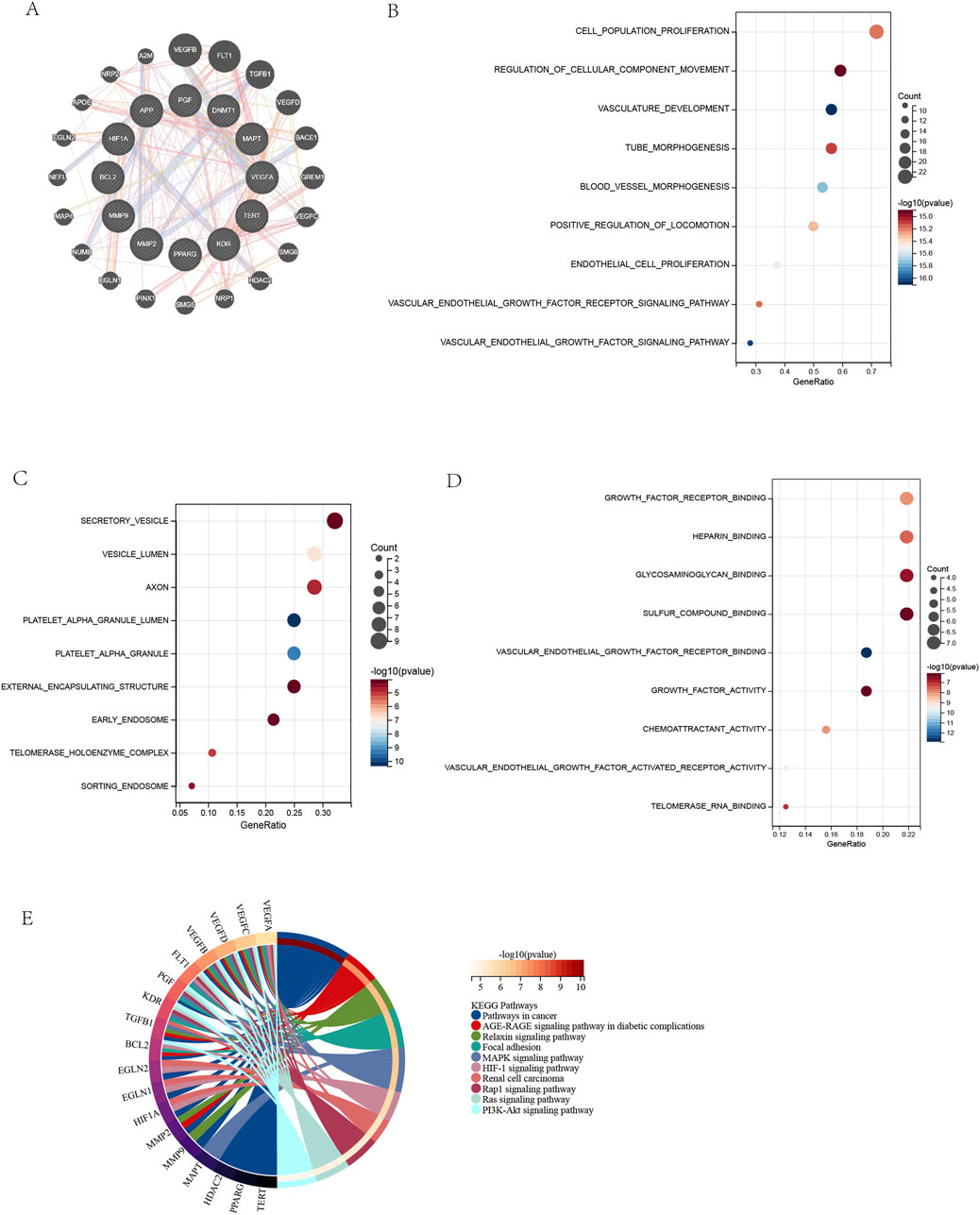
Figure 4. Protein-Protein Interaction Network and Functional Enrichment of Target Genes. (A) PPI network of DHM-AMD targets (12 core +20 related genes). (B) GO: Biological Process enrichment (top 10 terms). (C) GO: Cellular Component enrichment (top 10 terms). (D) GO: Molecular Function enrichment (top 10 terms). (E) KEGG pathway enrichment (top 10 terms). Enrichment criteria: FDR <0.05 (Benjamini and Hochberg correction).
3.5 In Vitro assessment of DHM effects on target genes
RT-qPCR was used to detect the mRNA expression levels of target proteins in NC, SI and DHM treatment groups. PGF, KDR, MMP9, VEGFA and APP were significantly upregulated in the SI group, and significantly downregulated after DHM treatment. MMP2, BCL2 were significantly downregulated in the SI group, and significantly upregulated after DHM treatment. MAPT, HIF1A, DNMT1, PPARG and TERT showed no obvious alleviating effects after DHM treatment compared with the SI treatment group. (Figures 5A–L). Notably, some predicted targets have been experimentally validated as pivotal regulators in AMD pathological processes. For example, MMP2 supplementation has been shown to ameliorate drusen accumulation, while MMP9 and APP have been identified as potential AMD biomarkers (Lynn et al., 2022; Hussain et al., 2011). Co-inhibition of VEGFA and PGF has been demonstrated to restrict retinal neovascularization in mice (Balser et al., 2019).
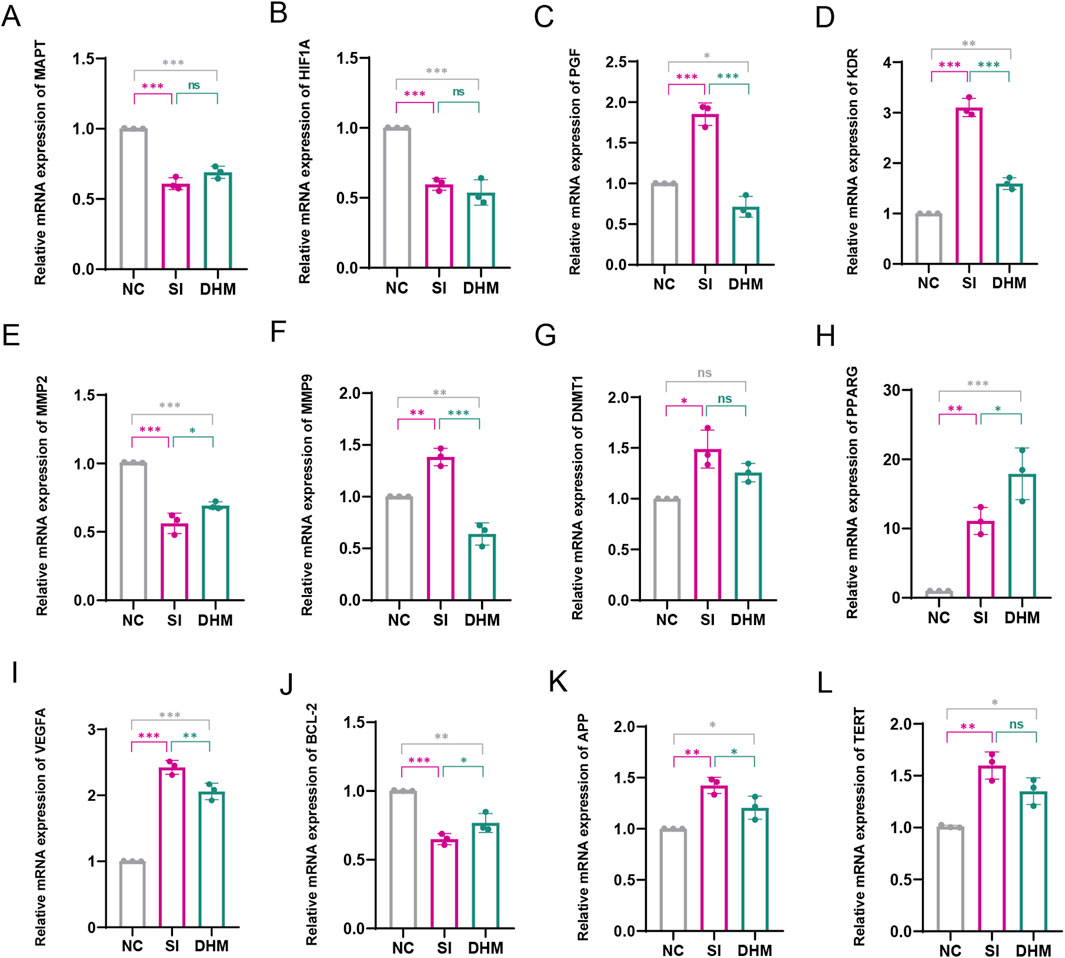
Figure 5. Effect of DHM on target proteins. The mRNA expression levels of MAPT (A), HIF1A (B), PGF (C), KDR (D), MMP2 (E), MMP9 (F), DNMT1 (G), PPARG (H), VEGFA (I), BCL2 (J), APP (K), TERT (L). Data in Figure G (non-normally distributed) were analyzed by Kruskal–Wallis with Dunn’s test; other data (normally distributed) were assessed by one-way ANOVA with Tukey’s test. Values represent mean ± SD (n = 3 per group). ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001. Significance symbols: Above bars: vs. untreated control; Between bars: vs. SI model group.
3.6 Molecular docking analysis of DHM-Target binding
RT-qPCR results confirmed DHM’s inhibition of aberrant mRNA expression in these seven targets. Therefore, molecular docking between DHM and the targets was performed using AutoDockTools-1.5.7 to validate binding affinity. The lowest-energy docking poses were retained for analysis. DHM exhibited the strongest binding to MMP9 (binding energy = −10.2 kcal/mol), followed by KDR, MMP2, VEGFA, APP, BCL2, and PGF (Figure 6H). Molecular interactions at binding sites are visualized in Figures 6A–G.
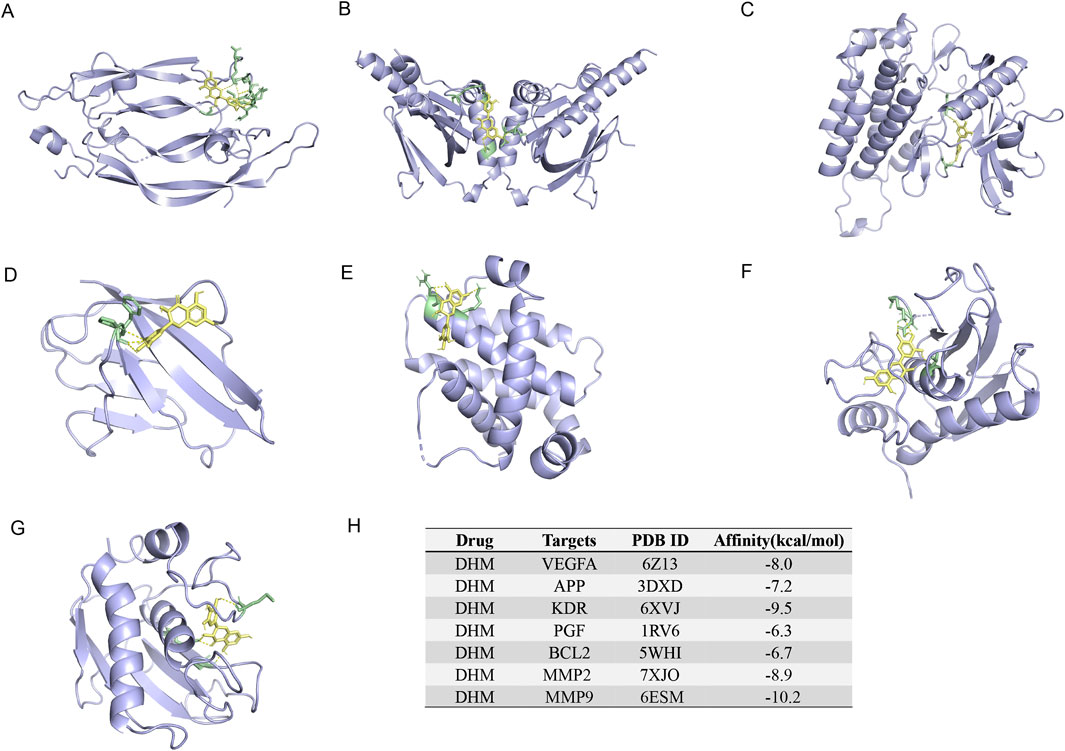
Figure 6. The 3-dimensional map of the binding sites between DHM and target proteins. (A) VEGFA, (B) APP, (C) KDR, (D) PGF, (E) BCL2, (F) MMP2, (G) MMP9. (H) Docking information and binding capacity of individual proteins.
4 Discussion
This study demonstrates that DHM effectively mitigates AMD pathology, as evidenced by protective effects in both in vitro and in vivo models. In vitro, DHM significantly suppressed SI-induced apoptosis in ARPE-19 cells, preserving cellular integrity against oxidative damage. In vivo, DHM alleviated key retinal degeneration markers in SI-induced AMD mice: attenuating retinal thinning, reducing pigment loss, and restoring ONL organization. These dose-dependent pharmacological effects (maximal at high doses) suggest DHM’s potential for AMD prevention and treatment.
The study builds on previous research, such as the work by Li and Xiao (2021), which demonstrated DHM’s protective effects on high glucose-induced oxidative stress and apoptosis in RPE cells. However, Li et al.‘s study was limited to in vitro experiments, leaving DHM’s in vivo effects unexamined. Beyond confirming the protective effect of DHM against RPE cell apoptosis in vitro—mediated through inhibition of the p53-dependent Bcl-2 apoptotic pathway—this study extended these findings to an in vivo setting. We established a sodium iodate-induced AMD murine model, enabling comprehensive evaluation of DHM’s therapeutic potential. This study supports DHM as a prospective AMD pharmacological agent by demonstrating its attenuation of AMD progression in a human-mirroring animal model.
Current AMD therapies (e.g., anti-VEGF injections) primarily slow neovascular AMD progression but face limitations (Rosenfeld, 2011; Markham, 2019). These therapies require frequent intravitreal injections, which can be invasive, uncomfortable, and costly for patients. Furthermore, they do not address the underlying oxidative and inflammatory processes that contribute to both neovascular and dry forms of AMD. As a result, many patients with the dry form of the disease, which accounts for a majority of cases, have no effective treatment options available. DHM, as demonstrated here, offers a promising alternative. By simultaneously modulating oxidative stress and inflammation, DHM addresses core AMD pathogenesis. Its oral bioavailability further enhances clinical potential, potentially reducing treatment burden and costs while providing systemic protection. However, despite the convenience of oral administration, DHM’s bioavailability and long-term toxicity require validation in large animal models. In the future, phase I clinical trials should be conducted to evaluate its safety.
Network analysis predicted 12 putative targets associated with DHM’s counteraction of AMD, all of which represent key pathogenic pathways involved in angiogenesis, apoptosis regulation, and oxidative stress response. Seven targets were preliminarily validated through quantification of mRNA expression in vitro, showing significant modulation upon DHM treatment. Molecular docking analysis demonstrated robust binding affinities between DHM and these targets—including MMP9, VEGFA, and BCL2—suggesting that DHM may exert its pharmacological effects by modulating relevant signaling pathways through interactions with these critical proteins.
Notably, this study has several limitations that should be acknowledged. First, as a flavonoid metabolite, DHM may act as a pan-assay interfering substance (PAINS), exerting non-specific effects via mechanisms such as redox cycling or metal chelation (Bolz et al., 2021; Lagorce et al., 2017). While our in vitro, in vivo, and target-binding data support specific pharmacological effects, further studies (e.g., with structural analogs of DHM lacking PAINS-associated moieties) are needed to confirm target specificity. Second, the sodium iodate tail vein injection mouse model represents a validated and widely used approach for establishing experimental AMD, specifically mimicking the geographic atrophy stage of advanced dry AMD (Wang et al., 2014; Kiuchi et al., 2002). Its primary advantage lies in the precise and reproducible induction of RPE cell oxidative damage and apoptosis, followed by secondary photoreceptor degeneration—processes that recapitulate the core pathological cascade of human AMD (Énzsöly et al., 2014; Yang et al., 2021). This method, characterized by acute and single-factor induction, fails to recapitulate the chronic, age-related, multifactorial-driven complexity of human AMD. In the future, it is necessary to further validate in combination with gene-editing models or laser-induced models. It does not recapitulate early features (e.g., drusen) or induce neovascularization, so findings should be cautiously extrapolated to human AMD. Third, although the putative targets identified via network analysis were validated at the mRNA level in vitro, this validation is limited. The network analysis itself is hypothesis-generating, and false positives cannot be ruled out; thus, we plan to conduct protein-level validation (e.g., Western blotting) and functional studies (e.g., target knockout) for each candidate in future research.
5 Conclusion
This study demonstrates through in vitro and in vivo models that DHM significantly ameliorates SI-induced retinal damage. Its protective effects encompass suppression of RPE apoptosis and preservation of retinal architecture. Network analysis and molecular docking predicted seven putative targets, with enrichment analyses revealing these targets’ modulation of key AMD-associated pathological pathways—including angiogenesis (e.g., VEGFA pathway), apoptosis (e.g., BCL2 pathway), and extracellular matrix metabolism (e.g., MMP2/MMP9 pathway). It must be emphasized that these computational predictions require experimental validation. Our findings provide preliminary evidence supporting DHM as a potential multi-target prophylactic/therapeutic candidate for AMD. Nevertheless, its precise mechanisms of action and clinical translatability warrant further in-depth investigation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Institutional Animal Care and Use Committee of Binzhou Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HZ: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing. JP: Formal Analysis, Methodology, Validation, Writing – review and editing, Investigation. BW: Methodology, Visualization, Writing – review and editing, Validation. YZ: Validation, Writing – review and editing, Investigation. SL: Project administration, Supervision, Writing – review and editing, Resources. JJ: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Writing – review and editing, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
Authors JP, YZ, SL, and JJ were employed by RemeGen Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1588970/full#supplementary-material
References
Ambati, J., and Fowler, B. J. (2012). Mechanisms of age-related macular degeneration. Neuron 75, 26–39. doi:10.1016/j.neuron.2012.06.018
Balser, C., Wolf, A., Herb, M., and Langmann, T. (2019). Co-inhibition of PGF and VEGF blocks their expression in mononuclear phagocytes and limits neovascularization and leakage in the murine retina. J. Neuroinflammation 16, 26. doi:10.1186/s12974-019-1419-2
Blasiak, J. (2020). Senescence in the pathogenesis of age-related macular degeneration. Cell. Mol. Life Sci. 77, 789–805. doi:10.1007/s00018-019-03420-x
Bolz, S. N., Adasme, M. F., and Schroeder, M. (2021). Toward an understanding of pan-assay interference compounds and promiscuity: a structural perspective on binding modes. J. Chem. Inf. Model 61, 2248–2262. doi:10.1021/acs.jcim.0c01227
Chan, C. M., Huang, D. Y., Sekar, P., Hsu, S. H., and Lin, W. W. (2019). Reactive oxygen species-dependent mitochondrial dynamics and autophagy confer protective effects in retinal pigment epithelial cells against sodium iodate-induced cell death. J. Biomed. Sci. 26, 40. doi:10.1186/s12929-019-0531-z
Chen, L., Yao, M., Fan, X., Lin, X., Arroo, R., Silva, A., et al. (2020). Dihydromyricetin attenuates streptozotocin-induced liver injury and inflammation in rats via regulation of NF-κB and AMPK signaling pathway. eFood 1, 188–195. doi:10.2991/efood.k.200207.001
Chen, J., Wang, X., Xia, T., Bi, Y., Liu, B., Fu, J., et al. (2021). Molecular mechanisms and therapeutic implications of dihydromyricetin in liver disease. Biomed. Pharmacother. 142, 111927. doi:10.1016/j.biopha.2021.111927
Crabb, J. W. (2014). The proteomics of drusen. Cold Spring Harb. Perspect. Med. 4, a017194. doi:10.1101/cshperspect.a017194
Crabb, J. W., Miyagi, M., Gu, X., Shadrach, K., West, K. A., Sakaguchi, H., et al. (2002). Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 99, 14682–14687. doi:10.1073/pnas.222551899
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364. doi:10.1093/nar/gkz382
Datta, S., Cano, M., Ebrahimi, K., Wang, L., and Handa, J. T. (2017). The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 60, 201–218. doi:10.1016/j.preteyeres.2017.03.002
DeWan, A., Liu, M., Hartman, S., Zhang, S. S. M., Liu, D. T. L., Zhao, C., et al. (1979). HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 2006, 314. doi:10.1126/science.1133807
Dong, S., Zhu, M., Wang, K., Zhao, X., Hu, L., Jing, W., et al. (2021). Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Res. 171, 105767. doi:10.1016/j.phrs.2021.105767
Énzsöly, A., Szabó, A., Kántor, O., Dávid, C., Szalay, P., Szabó, K., et al. (2014). Pathologic alterations of the outer retina in streptozotocin-induced diabetes. Invest. Ophthalmol. Vis. Sci. 55, 3686–3699. doi:10.1167/iovs.13-13562
Fabre, M., Mateo, L., Lamaa, D., Baillif, S., Pagès, G., Demange, L., et al. (2022). Recent advances in age-related macular degeneration therapies. Molecules 27, 5089. doi:10.3390/molecules27165089
Farnoodian, M., Bose, D., Khristov, V., Susaimanickam, P. J., Maddileti, S., Mariappan, I., et al. (2022). Cell-autonomous lipid-handling defects in Stargardt iPSC-derived retinal pigment epithelium cells. Stem Cell Rep. 17, 2438–2450. doi:10.1016/j.stemcr.2022.10.001
Fleckenstein, M., Schmitz-Valckenberg, S., and Chakravarthy, U. (2024). Age-related macular degeneration: a review. JAMA 331, 147–157. doi:10.1001/jama.2023.26074
Fritsche, L. G., Igl, W., Bailey, J. N. C., Grassmann, F., Sengupta, S., Bragg-Gresham, J. L., et al. (2016). A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 48, 134–143. doi:10.1038/ng.3448
Girgis, S., and Lee, L. R. (2023). Treatment of dry age-related macular degeneration: a review. Clin. Exp. Ophthalmol. 51, 835–852. doi:10.1111/ceo.14294
Gui, S. Y., Wang, X. C., Huang, Z. H., Li, M. M., Wang, J. H., Gui, S. Y., et al. (2023). Nanoscale coordination polymer Fe-DMY downregulating Poldip2-Nox4-H2O2 pathway and alleviating diabetic retinopathy. J. Pharm. Anal. 13, 1326–1345. doi:10.1016/j.jpha.2023.05.002
Guymer, R. H., and Campbell, T. G. (2023). Age-related macular degeneration. Lancet 401, 1459–1472. doi:10.1016/s0140-6736(22)02609-5
Hamosh, A., Scott, A. F., Amberger, J., Valle, D., and McKusick, V. A. (2000). Online mendelian inheritance in man (OMIM). Hum. Mutat. 15, 57–61. doi:10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G
Hariri, S., Tam, M. C., Lee, D., Hileeto, D., Moayed, A. A., and Bizheva, K. (2013). Noninvasive imaging of the early effect of sodium iodate toxicity in a rat model of outer retina degeneration with spectral domain optical coherence tomography. J. Biomed. Opt. 18, 26017. doi:10.1117/1.JBO.18.2.026017
He, J., Zhang, J., Dong, L., Dang, X., Wang, L., Cheng, L., et al. (2019). Dihydromyricetin attenuates metabolic syndrome and improves insulin sensitivity by upregulating insulin receptor substrate-1 (Y612) tyrosine phosphorylation in db/db mice. Diabetes, Metabolic Syndrome Obes. 12, 2237–2249. doi:10.2147/DMSO.S218487
Heesterbeek, T. J., Lorés-Motta, L., Hoyng, C. B., Lechanteur, Y. T. E., and den Hollander, A. I. (2020). Risk factors for progression of age-related macular degeneration. Ophthalmic Physiological Opt. 40, 140–170. doi:10.1111/opo.12675
Heloterä, H., and Kaarniranta, K. (2022). A linkage between angiogenesis and inflammation in neovascular age-related macular degeneration. Cells 11, 3453. doi:10.3390/cells11213453
Hussain, A. A., Lee, Y., Zhang, J. J., and Marshall, J. (2011). Disturbed matrix metalloproteinase activity of Bruch’s membrane in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 52, 4459–4466. doi:10.1167/iovs.10-6678
Jing, N., and Li, X. (2020). Dihydromyricetin attenuates inflammation through TLR4/NF-kappaB pathway. Open Med. Pol. 14, 719–725. doi:10.1515/med-2019-0083
Kaarniranta, K., Pawlowska, E., Szczepanska, J., Jablkowska, A., and Blasiak, J. (2019). Role of mitochondrial dna damage in ros-mediated pathogenesis of age-related macular degeneration (Amd). Int. J. Mol. Sci. 20, 2374. doi:10.3390/ijms20102374
Kiuchi, K., Yoshizawa, K., Shikata, N., Moriguchi, K., and Tsubura, A. (2002). Morphologic characteristics of retinal degeneration induced by sodium iodate in mice. Curr. Eye Res. 25, 373–379. doi:10.1076/ceyr.25.6.373.14227
Lagorce, D., Oliveira, N., Miteva, M. A., and Villoutreix, B. O. (2017). Pan-assay interference compounds (PAINS) that may not be too painful for chemical biology projects. Drug Discov. Today 22, 1131–1133. doi:10.1016/j.drudis.2017.05.017
Li, W., and Xiao, H. (2021). Dihydromyricetin alleviates high glucose-induced oxidative stress and apoptosis in human retinal pigment epithelial cells by downregulating miR-34a expression. Diabetes, Metabolic Syndrome Obes. 14, 387–397. doi:10.2147/dmso.s290633
Li, T., Guo, R., Zong, Q., and Ling, G. (2022). Application of molecular docking in elaborating molecular mechanisms and interactions of supramolecular cyclodextrin. Carbohydr. Polym. 276, 118644. doi:10.1016/j.carbpol.2021.118644
Liu, J., Shu, Y., Zhang, Q., Liu, B., Xia, J., Qiu, M., et al. (2014). Dihydromyricetin induces apoptosis and inhibits proliferation in hepatocellular carcinoma cells. Oncol. Lett. 8, 1645–1651. doi:10.3892/ol.2014.2330
Liu, T. T., Zeng, Y., Tang, K., Chen, X. M., Zhang, W., and Xu, X. L. (2017). Dihydromyricetin ameliorates atherosclerosis in LDL receptor deficient mice. Atherosclerosis, 262. doi:10.1016/j.atherosclerosis.2017.05.003
Luo, J., Chen, X. H., Liang, P. Y., Zhao, Z., Wu, T., huang, Li Z., et al. (2023a). Mechanism of anti-hyperuricemia of isobavachin based on network pharmacology and molecular docking. Comput. Biol. Med. 155, 106637. doi:10.1016/j.compbiomed.2023.106637
Luo, W., Deng, J., He, J., Yin, L., You, R., Zhang, L., et al. (2023b). Integration of molecular docking, molecular dynamics and network pharmacology to explore the multi-target pharmacology of fenugreek against diabetes. J. Cell Mol. Med. 27, 1959–1974. doi:10.1111/jcmm.17787
Lynn, S. A., Soubigou, F., Dewing, J. M., Smith, A., Ballingall, J., Sass, T., et al. (2022). An exploratory study provides insights into MMP9 and aβ levels in the vitreous and blood across different ages and in a subset of AMD patients. Int. J. Mol. Sci. 23, 14603. doi:10.3390/ijms232314603
Manne, B. K., Xiang, S. C., and Rondina, M. T. (2017). Platelet secretion in inflammatory and infectious diseases. Platelets 28, 155–164. doi:10.1080/09537104.2016.1240766
Markham, A. (2019). Brolucizumab: first approval. Drugs 79, 1997–2000. doi:10.1007/s40265-019-01231-9
Mullins, R. F., Russell, S. R., Anderson, D. H., and Hageman, G. S. (2000). Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 14, 835–846. doi:10.1096/fasebj.14.7.835
Muraleva, N. A., and Kolosova, N. G. (2023). P38 MAPK signaling in the retina: effects of aging and age-related macular degeneration. Int. J. Mol. Sci. 24, 11586. doi:10.3390/ijms241411586
Nilsson, S. E. G., Knave, B., and Persson, H. E. (1977). Changes in ultrastructure and function of the sheep pigment epithelium and retina induced by sodium iodate: I. The Ultrastructure of the Normal Pigment Epithelium of the Sheep. Acta Ophthalmol. 55, 994–1006. doi:10.1111/j.1755-3768.1977.tb05681.x
Nogales, C., Mamdouh, Z. M., List, M., Kiel, C., Casas, A. I., and Schmidt, HHHW (2022). Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 43, 136–150. doi:10.1016/j.tips.2021.11.004
Obata, R., Yanagi, Y., Tamaki, Y., Hozumi, K., Mutoh, M., and Tanaka, Y. (2005). Retinal degeneration is delayed by tissue factor pathway inhibitor-2 in RCS rats and a sodium-iodate-induced model in rabbits. Eye 19, 464–468. doi:10.1038/sj.eye.6701531
Pan, Y., Fu, Y., Baird, P. N., Guymer, R. H., Das, T., and Iwata, T. (2023). Exploring the contribution of ARMS2 and HTRA1 genetic risk factors in age-related macular degeneration. Prog. Retin. Eye Res. 97, 101159. doi:10.1016/j.preteyeres.2022.101159
Rosenfeld, P. J. (2011). Bevacizumab versus ranibizumab for AMD. N. Engl. J. Med. 364, 1966–1967. doi:10.1056/NEJMe1103334
Sakurada, Y., Tanaka, K., and Fragiotta, S. (2023). Differentiating drusen and drusenoid deposits subtypes on multimodal imaging and risk of advanced age-related macular degeneration. Jpn. J. Ophthalmol. 67, 1–13. doi:10.1007/s10384-022-00943-y
Schmidt-Erfurth, U., Kaiser, P. K., Korobelnik, J. F., Brown, D. M., Chong, V., Nguyen, Q. D., et al. (2014). Intravitreal aflibercept injection for neovascular age-related macular degeneration: ninety-six-week results of the VIEW studies. Ophthalmology 121, 193–201. doi:10.1016/j.ophtha.2013.08.011
Sharma, N. K., Prabhakar, S., Gupta, A., Singh, R., Gupta, P. K., Gupta, P. K., et al. (2012). New biomarker for neovascular age-related macular degeneration: eotaxin-2. DNA Cell Biol. 31, 1618–1627. doi:10.1089/dna.2012.1786
Shen, Y., Zhuang, P., Lin, B. Q., Zhang, W. Y., and Chiou, G. C. Y. (2010). Effect of Tetramethylpyrazine on RPE degeneration, choroidal blood flow and oxidative stress of RPE cells. Int. J. Ophthalmol. 3, 205–210. doi:10.3980/j.issn.2222-3959.2010.03.06
Skei, J. M., Fingert, J. H., Russell, S. R., Stone, E. M., and Mullins, R. F. (2010). Complement component C5a activates ICAM-1expression on human choroidal endothelial cells. Invest. Ophthalmol. Vis. Sci. 51, 5336. doi:10.1167/iovs.10-5322
Stahl, A. (2020). The diagnosis and treatment of age-related macular degeneration. Dtsch. Arztebl Int. 117, 513–520. doi:10.3238/arztebl.2020.0513
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi:10.1073/pnas.0506580102
Sun, Y., Liu, W., Wang, C., Meng, Q., Liu, Z., Huo, X., et al. (2019). Combination of dihydromyricetin and ondansetron strengthens antiproliferative efficiency of adriamycin in K562/ADR through downregulation of SORCIN: a new strategy of inhibiting P-glycoprotein. J. Cell Physiol. 234, 3685–3696. doi:10.1002/jcp.27141
Sun, Z., Wang, Y., Pang, X., Wang, X., and Zeng, H. (2023). Mechanisms of polydatin against spinal cord ischemia–reperfusion injury based on network pharmacology, molecular docking and molecular dynamics simulation. Bioorg Chem. 140, 106840. doi:10.1016/j.bioorg.2023.106840
Tang, Z., Ju, Y., Dai, X., Ni, N., Liu, Y., Zhang, D., et al. (2021). HO-1-mediated ferroptosis as a target for protection against retinal pigment epithelium degeneration. Redox Biol. 43, 101971. doi:10.1016/j.redox.2021.101971
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Vessey, K. A., Jobling, A. I., Tran, M. X., Wang, A. Y., Greferath, U., and Fletcher, E. L. (2022). Treatments targeting autophagy ameliorate the age-related macular degeneration phenotype in mice lacking APOE (apolipoprotein E). Autophagy 18, 2368–2384. doi:10.1080/15548627.2022.2034131
Wang, L., Clark, M. E., Crossman, D. K., Kojima, K., Messinger, J. D., Mobley, J. A., et al. (2010). Abundant lipid and protein components of drusen. PLoS One 5, e10329. doi:10.1371/journal.pone.0010329
Wang, J., Iacovelli, J., Spencer, C., and Saint-Geniez, M. (2014). Direct effect of sodium iodate on neurosensory retina. Invest. Ophthalmol. Vis. Sci. 55, 1941–1953. doi:10.1167/iovs.13-13075
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38 (Suppl. 2), W214–W220. doi:10.1093/nar/gkq537
Weng, L., Zhang, H., Li, X., Zhan, H., Chen, F., Han, L., et al. (2017). Ampelopsin attenuates lipopolysaccharide-induced inflammatory response through the inhibition of the NF-κB and JAK2/STAT3 signaling pathways in microglia. Int. Immunopharmacol. 44, 1–8. doi:10.1016/j.intimp.2016.12.018
Wu, P., Zhao, L., Du, Y., Lu, J., He, Y., Shu, Q., et al. (2025). Melatonin protects retinal pigment epithelium cells against ferroptosis in AMD via the PI3K/AKT/MDM2/P53 pathway. Front. Pharmacol. 16, 1543575. doi:10.3389/fphar.2025.1543575
Xie, J., Zhang, T., Li, P., Wang, D., Liu, T., and Xu, S. (2022). Dihydromyricetin attenuates cerebral ischemia reperfusion injury by inhibiting SPHK1/mTOR signaling and targeting ferroptosis. Drug Des. Devel Ther. 16, 3071–3085. doi:10.2147/DDDT.S378786
Yan, Q., Li, P., Liu, S., Sun, Y., Chen, C., Long, J., et al. (2024). Dihydromyricetin treats pulmonary hypertension by modulating CKLF1/CCR5 axis-induced pulmonary vascular cell pyroptosis. Biomed. Pharmacother. 180, 117614. doi:10.1016/j.biopha.2024.117614
Yang, P. M., Wu, Z. Z., Zhang, Y. Q., and Wung, B. S. (2016). Lycopene inhibits ICAM-1 expression and NF-κB activation by Nrf2-regulated cell redox state in human retinal pigment epithelial cells. Life Sci. 155, 94–101. doi:10.1016/j.lfs.2016.05.006
Yang, S., Zhou, J., and Li, D. (2021). Functions and diseases of the retinal pigment epithelium. Front. Pharmacol. 12, 727870. doi:10.3389/fphar.2021.727870
Zeng, T., Song, Y., Qi, S., Zhang, R., Xu, L., and Xiao, P. (2023). A comprehensive review of vine tea: origin, research on Materia Medica, phytochemistry and pharmacology. J. Ethnopharmacol. 317, 116788. doi:10.1016/j.jep.2023.116788
Zhang, H., Zhang, Y., Li, Y., Wang, Y., Yan, S., Xu, S., et al. (2021). Bioinformatics and network pharmacology identify the therapeutic role and potential mechanism of melatonin in AD and rosacea. Front. Immunol. 12, 756550. doi:10.3389/fimmu.2021.756550
Zhao, L., Zhang, H., Li, N., Chen, J., Xu, H., Wang, Y., et al. (2023). Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J. Ethnopharmacol. 309, 116306. doi:10.1016/j.jep.2023.116306
Keywords: age-related macular degeneration (AMD), dihydromyricetin (DHM), network analysis, molecular docking, putative targets
Citation: Zhou H, Pang J, Wu B, Zhuang Y, Li S and Jiang J (2025) Dihydromyricetin attenuates age-related macular degeneration: pharmacological effects and exploration of putative targets. Front. Pharmacol. 16:1588970. doi: 10.3389/fphar.2025.1588970
Received: 17 March 2025; Accepted: 29 July 2025;
Published: 21 August 2025.
Edited by:
Javier Echeverria, University of Santiago, ChileReviewed by:
Lihui Zhu, Shanghai Academy of Agricultural Sciences, ChinaSheraz Khoja, University of California, Irvine, United States
Chia-I Tsai, National Chung Hsing University, Taiwan
Copyright © 2025 Zhou, Pang, Wu, Zhuang, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Jiang, al9qaWFuZ0Biem1jLmVkdS5jbg==
 Hongyu Zhou
Hongyu Zhou Jianhui Pang2
Jianhui Pang2