- 1Medical Clinic III, Synergy and Experimental Medicine Research Group, University Hospital Bonn (UKB), Bonn, Germany
- 2Institute of Forestry and Nature Management, Volga State University of Technology, Yoshkar-Ola, Russia
- 3Pharmacognosy, Department of Pharmaceutical Biosciences, Uppsala University, Uppsala, Sweden
- 4Medicinal Plants Innovation Center, Mae Fah Luang University, Chiang Rai, Thailand
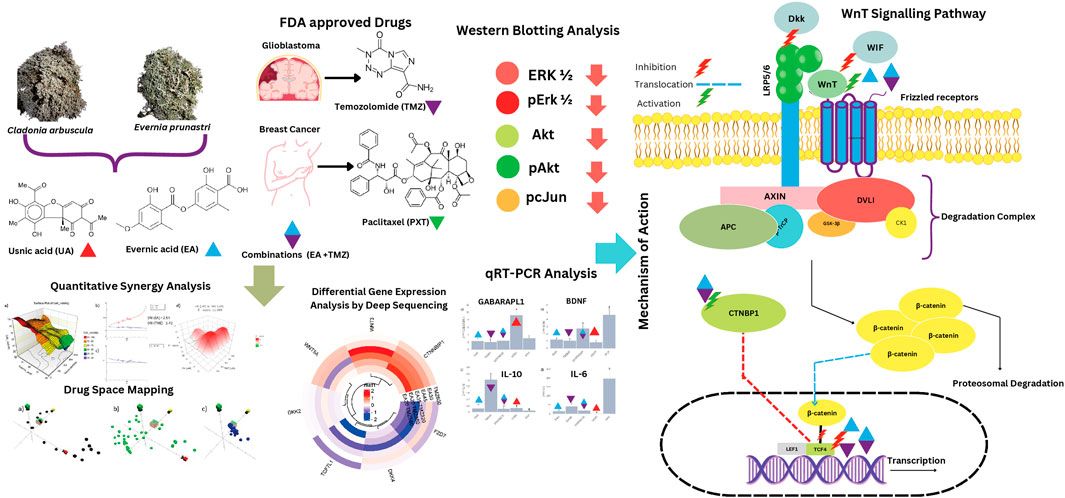
Introduction: Temozolomide (TMZ) and Paclitaxel (PXT), crucial anti-cancer drugs for glioblastoma (GBM) and primary breast cancer (BC), respectively, face drug resistance. Therefore, we investigated the adjuvant potential of characterized extracts of the lichens Evernia prunastri (L.) Ach. (Epr), Cladonia arbuscula (Wallr.) Flot (Car) and their metabolites, evernic acid (EA) and usnic acid (UA) alone or in combination with TMZ and PTX for their immunomodulatory and chemosensitivity increasing potential.
Methods: TMZ-resistant U-87 cells, MCF7 BC-cells, and normal human skin fibroblasts (HSKF) were treated with hexane (Hex), dichloromethane (DCM), and acetonitrile (ACN) extracts of Epr (EprDCM, EprACN), Car (CarHex, CarACN), and with EA and UA to measure cell metabolic activity. Molecular mechanisms were predicted using ChemGPS-NP and validated by Western blot, RNA sequencing, quantitative RT-PCR, and Wnt inhibitory factor 1 (WIF1) protein expression. Combinatory effects were calculated by Combination Index (CI) and Zero Interaction Potency methods (ZIP).
Results: Extracts and selected metabolites reduced concentration-dependent cellular metabolic activity in U-87 and MCF7 cells. EprACN and EA (U-87 cells: IC50 30 μg/mL), safe to HSKF, regulated key proteins in MAP kinases pathways, supporting predictions made by ChemGPS-NP. The combination EA-TMZ showed additive effects (TMZ-reduction: 3.4 fold), reduced transcription of Wnt pathway members, and increased in U-87 cells protein releases of WiF1, the central inhibitor of Wnt-signaling. Further gene expression data (GE) suggest involvement of IL-17 receptor and BDNF.
Discussion: The combination EA-TMZ interacts with the Wnt pathway regulation associated with sensitizing U-87 cells, without increasing GEs of pro-inflammatory cytokines. EA deserves further investigation as an adjuvant.
1 Introduction
Breast cancer (BC) is the most common cancer worldwide and ranks fifth among cancer-related deaths (Zhang et al., 2023). Projections indicate a significant increase in the global burden, with cases expected to rise by nearly 40% and deaths by 68% by 2050 if current trends continue (Liao, 2025). Brain and central nervous system cancer ranks approximately 12th in incidence and 8th in mortality in Europe based on age-standardized rates (DeCordova et al., 2020). Glioblastoma (GBM), classified as a WHO Grade 4 CNS tumor according to the 2021 WHO Classification of Tumors of the Central Nervous System, represents the most aggressive form of brain cancer (Louis et al., 2021). The 5- and 10-year survival rates still remain at 5% and 2.6%, respectively (Ma et al., 2022; Alcantara Llaguno et al., 2009). The current standard of care for GBM follows the Stupp protocol, established in 2005, which includes maximal safe surgical resection, concurrent chemoradiotherapy, and adjuvant temozolomide (TMZ) (Jezierzański et al., 2024).
The poor treatment prognosis, notably GBM, has been linked to its diverse molecular profiles, resulting in distinct phenotypes also associated with TMZ-resistance (DeCordova et al., 2020). Several fundamental mechanisms contribute to GBM’s treatment resistance and incurability. The blood-brain barrier represents a critical obstacle, preventing most systemic chemotherapeutic agents from reaching therapeutic concentrations in brain tissue (Zuccarini et al., 2018). Glioblastoma stem cells (GSCs) constitute another major resistance mechanism, exhibiting enhanced DNA repair capacity, resistance to apoptosis, metabolic reprogramming, and self-renewal capacity that maintains the tumor cell population (Nowak et al., 2021; Guan et al., 2020). Methylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter plays a critical role in TMZ-resistance. Tumors with an unmethylated MGMT promoter are significantly more resistant to TMZ, and prolonged treatment may induce loss of MGMT methylation, further contributing to acquired resistance (Li et al., 2021).
Recently, Weighted Gene Co-Expression Network Analysis (WGCNA) was used to capture the molecular heterogeneity of GBM patients on the molecular level. Authors constructed an immune-related prognostic model to predict patient sensitivity to checkpoint inhibitor blockade therapy (Ma et al., 2022). The high-risk group (non-survival) was associated with epithelial-mesenchymal transition (EMT), high immune cell infiltration, immune activation, a low mutation number, and high methylation, while the low-risk group had an adverse status (Ma et al., 2022).
BC is also extremely variable in morphology and at the molecular level, necessitating combinatorial therapy modalities depending on the molecular subtype, which is defined by hormone receptor (HR) status and human epidermal growth factor receptor-2 (HER2) expression (Li et al., 2017; Kashyap et al., 2022). These include HR-positive/HER2-negative (HR+/HER2-), HR+/HER2+, HR-/HER2+, and triple-negative breast cancer (TNBC), which lacks estrogen receptor, progesterone receptor, and HER2 expression (Li et al., 2017). TNBC is particularly aggressive, accounting for approximately 10%–15% of all breast cancers but contributing to a disproportionately high percentage of breast cancer-related deaths globally, estimated at around 40% (Rosińska et al., 2024). The 5-year relative survival rate for TNBC, combining all stages, is approximately 77%, with significantly lower rates for distant metastatic disease (Baranova et al., 2022).
The current standard of care in breast cancer varies by subtype and stage. Early-stage breast cancer typically involves surgical resection, adjuvant or neoadjuvant chemotherapy, radiation therapy, hormone therapy for hormone receptor-positive tumors, and targeted therapy for HER2-positive tumors (Bravo et al., 2023; Grant et al., 2024).
The Wnt pathway is implicated in both BC and GBM, significantly contributing to treatment resistance (Zhong and Virshup, 2020), even though precise regulatory mechanisms remain unclear (Kashyap et al., 2022). Crosstalk between Wnt signaling and other pathways contributes to cancer development and spread, including resistance to pathway inhibitors (Zhong and Virshup, 2020). This understanding drives the development of novel combination therapies to minimize toxicity and resistance (Zhong and Virshup, 2020). Therefore, exploring unexamined plant extracts or phytocompounds contributes to discovering safe, effective, and novel combination therapies (Ulrich-Merzenich et al., 2017).
TMZ and paclitaxel (PTX), key chemotherapeutics for GBM and BC, respectively, are under investigation for combination therapies (Tan et al., 2020). TMZ, a DNA alkylating agent, induces cell death by causing base mismatches and DNA strand breaks (Tan et al., 2020). Ongoing 583 clinical trials worldwide explore treatments for GBM (https://clinicaltrials.gov/), including immune checkpoint inhibitors paired with CNS-penetrant or potent inhibitors (Gueble et al., 2022). Despite multimodal treatments, GBM patients face a low median survival of 12.1–16.6 months (Neff et al., 2022; Minea et al., 2024; Fekete et al., 2023), emphasizing the urgency for novel treatment strategies (Tan et al., 2020).
Taxanes, like PTX, used in BC treatment, were derived from the Pacific yew’s bark (Wani et al., 1971). PTX induces cell death by stabilizing microtubules, causing G2/M arrest and initiating apoptosis (Manthey et al., 1992). Resistance to PTX and other anti-cancer drugs (Das et al., 2021; Bukowsk et al., 2020) is common for BC, just as resistance in GBM to TMZ (Lee, 2016). The resistance of BC to PTX is thought to be a consequence of a disequilibrium in various signaling pathways, mutations in certain genes, and epigenetic deregulations (Abu et al., 2019). In particular, the genes of the ATP-binding cassette (ABC) superfamily of drug efflux, including P-glycoprotein (P-gp), are involved in the resistance to PTX by leading to an overexpression of P-gp in BC-cells (Abu et al., 2019). In MCF7 cells, aberrantly regulated expression of FOXM1 and KIF20A was associated with PTX-resistance (Abu et al., 2019; Khongkow et al., 2016). Further, the overexpression of miR-200c-3p contributed to the resistance of BC cells to PTX by an aberrant regulation of SOX2 (Abu et al., 2019).
This diversity of resistance mechanisms promotes a search for natural compounds as adjuvants (Persano et al., 2022). Therapies triggering multiple pathways (and specifically addressing crucial survival pathways) may be more promising (Sestito et al., 2018).
Lichens have been utilized in traditional medicine for ages (Crawford and Ranković, 2015). They are symbiotic organisms consisting of a fungus (mycobiont) and either algae or cyanobacteria (photobiont) (Schwendener, 2011). They are a promising source for novel organic small molecules and synergistic therapeutic strategies. Evernia prunastri L. and Cladonia arbuscula (Wallr.) Fot. were selected based on their published bioactivity. Evernic acid (EA), the main metabolite of E. prunastri L., has shown antimicrobial, cytotoxic, neuroprotective, and anti-inflammatory properties in prior studies (Fernández-Moriano et al., 2017; Lee et al., 2021; Gökalsın and Sesal, 2016; Shcherbakova et al., 2019). However, its role in cancer therapy remains largely unexplored. Usnic acid (UA), a prominent secondary metabolite isolated from C. arbuscula (Wallr.) Fot., has shown potent antiproliferative effects in several cancer types. Notably, UA exhibited promising cytotoxicity against T-47D breast cancer cells, Capan-2 pancreatic cancer cells (Einarsdottir et al., 2010), and MCF7 breast cancer cells as well (Bačkorová et al., 2011; Galanty et al., 2017; Kiliç et al., 2019; Brisdelli et al., 2013).
We investigated extracts from E. prunastri L. (Parmeliaceae) and C. arbuscula (Wallr.) Fot. (Cladoniaceae) along with their major metabolites, evernic acid (EA) and usnic acid (UA), for their potential to reduce metabolic activity in U-87 glioma and MCF7 breast cancer cells.
Both cell lines were selected as representative in vitro models for glioblastoma and breast cancer, respectively, due to their widespread use, well-characterized molecular profiles, and relevance to the mechanisms under investigation. U-87 cells are among the most commonly used GBM models and exhibit key features of primary glioblastoma, such as rapid proliferation, resistance to temozolomide (TMZ) (Xie et al., 2015). Similarly, MCF7 cells are commonly used for BC, characterized by estrogen receptor positivity and moderate sensitivity to chemotherapeutics like paclitaxel (PTX), making them a standard model for studying HR+ BC and mechanisms of taxane resistance (Neve et al., 2006).
U-87 and MCF7 remain widely accepted platforms for early-stage anticancer research. In this study, they were used to evaluate not only the ability of the lichen extracts and their metabolites to influence cancer cell metabolic activity, but also their potential to synergize with standard chemotherapeutics (TMZ or PTX). Extracts and combinations showing efficacy were further assessed for possible mechanisms of action. This included chemographic prediction tools, cytokine response profiling, and evaluation of their potential to modulate cellular pathways involved in drug sensitivity, forming a foundation for future translational investigations.
2 Materials and methods
2.1 Chemicals, media, and assays
Usnic acid (UA) (purity 98%), paclitaxel, resazurin tox kit, insulin (Sigma Aldrich, Germany); evernic acid (EA) (purity 98%), temozolomide (Cayman Chemical, United States); (phospho-AKT rabbit polyclonal antibody (1:2000), ß-actin rabbit polyclonal antibody (1:2000), phospho-p44/42 (Thr202/Tyr204) rabbit polyclonal antibody (1:2000), p44/42 mouse clonal antibody (1:2000), goat anti-rabbit IgG-HRP conjugated to horseradish peroxidase (1:2000) (Santa Cruz Biotechnology, United States); phospho-c-Jun (Ser73) rabbit polyclonal antibody (1:2000) (Cell Signaling Technology, United States); Minimum Essential Medium (MEM), Roswell Park Memorial Institute (RPMI) 1,640 Medium, Dulbecco’s Modified Eagle’s Medium (DMEM, fetal bovine serum (FBS), non-essential amino acids (MEM NEAA), sodium pyruvate, penicillin/streptomycin (Gibco™, United States); Human WIF-1 DuoSet ELISA (R&D Systems, United States), RNeasy MiniPlus kit (QIAGEN, Netherlands).
2.2 Lichens material
Samples of E. prunastri (L.) Ach. and C. arbuscula (L.) Hoffm. were collected in the Mari El Republic of Russia on the campus of the Volga State University of Technology. The lichens were identified by lichenologist G.A. Bogdanov at the Bolshaya Kokshaga Natural Reserve. The voucher specimens of the lichens were deposited at the Institute of Forestry and Nature Management, Volga State University of Technology, Yoshkar-Ola, Russia, with the references Epr_06.2012 (E. prunastri) and Car_06.2012 (C. arbuscula).
2.3 Extraction and characterisation
Extracts were obtained and characterized as described earlier (Shcherbakova et al., 2021). Air-dried powdered thalli of the lichens were extracted by sequential maceration with hexane, dichloromethane (DCM), or with 60% acetonitrile in water (ACN) at room temperature (RT) for 24 h with each solvent. The extracts were filtered and then concentrated under reduced pressure in a rotary evaporator Rotavapor R (Buchi Labortechnik AG, Flawil, Switzerland). The dry extracts were stored at RT until usage.
2.4 Chemographic prediction of the mode of action
ChemGPS-NP (http://chemgps.bmc.uu.se) provides a multidimensional (8D) map of physicochemical property space. On this map, molecules are positioned based on their estimated physico-chemical properties. Compounds with similar structures and hence properties are positioned on the map in mutual proximity. Thus positions and distances between compounds can be used to predict their biological activities. This method has been specifically validated for anti-cancer modes of action as well as for a broad range of other experimentally demonstrated activities (Buonfiglio et al., 2015). It was used to predict possible modes of action of EA.
2.5 Cell culture and cytotoxicity assay
2.5.1 Cell lines and culture
Human primary glioblastoma (U-87), human breast adenocarcinoma (MCF7), and human skin fibroblast (HSKF) cell lines were purchased via Cell Line Services (CLS) from the German Collection of Microorganisms and Cell Cultures (DSMZ) and Promocell and grown as described earlier (Ammar and Ulrich-Merzenich, 2017). HSKF was included as a non-malignant cell line to assess the general cytotoxicity and selectivity of the tested extracts, metabolites, and compounds, distinguishing between broad cellular effects and specific anti-cancer activity.
2.5.2 Metabolic activity assay as measure for cell viability
Cells (5 × 103 cells/well) were seeded into 96-well plates and treated as described (Ulrich-Merzenich et al., 2017). For the treatment with the CarHex, CarACN, EprDCM, and EprACN extracts, a concentration range of 6.25–100 μg/mL was used. For EA and UA, the tested concentration range was 4.15–66.46 μg/mL and 4.30–68.86 μg/mL, respectively (corresponding to 12.5–200 µM). TMZ was tested in a range of 9.71–155.32 μg/mL (50–800 µM), and PTX in a range of 21.35–341.56 μg/mL (25–400 µM). All treatments were performed for 24 h. Metabolic activity of the cells, as an indirect measure of cell viability, was measured by resazurin fluorometric assay (Sigma) as described (Ulrich-Merzenich et al., 2017). The concentration range was selected based on the published data (Einarsdottir et al., 2010; Kiliç et al., 2019; Brisdelli et al., 2013; Emsen et al., 2018; Bézivin et al., 2004). The 24-h time point was selected for all viability assays to ensure consistency and comparability across all experimental conditions. This time frame is widely used for resazurin-based cytotoxicity assays and is sufficient to capture early drug-induced effects on cell viability, as well as to minimize secondary effects such as nutrient depletion or over-confluence in culture.
2.6 Western blot analysis
Western blot analysis was performed to investigate the expression and phosphorylation levels of Akt, Erk1/2, and c-Jun in U-87 cells treated with EprACN or EA as described (Ulrich-Merzenich et al., 2007) and in Supplementary Material S1. Antibody (Ab) details (pAKT, pErk1/2, Erk 1/2, p-cJun, ß-actin, secondary Abs) are provided in Supplementary Material Table S1.
2.7 Synergy screening
Cells (as described under Section 2.5.2) were treated with 7 different combinations of metabolites/compounds. Concentrations for the combinations were chosen based on the results with the single extracts/metabolites (Chou, 2010) (see also Section 3.1 and Supplementary Material S2). A total of five concentrations were chosen with the following concentration ranges for the different metabolites/drugs: 1) TMZ (9.75–155.32 μg/mL = 50–800 µM) and EprACN (6.25–100 μg/mL); 2) TMZ (9.75–155.32 μg/mL = 50–800 µM) and EA (4.15–66.46 μg/mL = 12.5–200 µM); 3) TMZ (9.75–155.32 μg/mL = 50–800 µM) and UA (4.30–68.86 μg/mL = 12.5–200 µM); 4) PTX (5.34–85.39 μg/mL = 6.25–100 µM) and EprACN (6.25–100 μg/mL); 5) PTX (5.34–85.39 μg/mL = 6.25–100 µM) and EA (4.15–66.46 μg/mL = 12.5–200 µM); 6) PTX (5.34–85.39 μg/mL = 6.25–100 µM) and UA (4.30–68.86 μg/mL = 12.5–200 µM); 7) TMZ (62.13–124.26 μg/mL = 320–640 µM) and EA (6.65–13.29 μg/mL = 20–40 µM). For details, see Supplementary Material S3. Drug combinations were tested in comparison to their respective controls.
2.7.1 Synergy calculation
Synergism was calculated using the CompuSyn software (https://compusyn.software.informer.com/) based on the Chou and Talalay Combination Index (CI) method (Chou, 2010) and by the web application “SynergyFinder” (v.1) employing the Zero Interaction Potency (ZIP) model (Ianevski et al., 2017). For the CI and the Dose Reduction Index (DRI) calculation, the following ratios of the combined drugs were used: 1) TMZ: EP – 1.5:1 (c:c); 2) TMZ: EA – 8:1 (c:c); 3) TMZ: UA – 8:1 (c:c); 4) PTX: EP – 1:1.2 (c:c); 5) 4) PTX: EA – 1:2 (c:c); 6) 4) PTX: UA – 1:2 (c:c); 7) TMZ: EA – 16:1 (c:c).
2.8 RNA deep sequencing of primary glioblastoma cells (U-87)
2.8.1 Cell culture
U-87 cells (106 cells/well) were seeded in 6-well plates and stimulated for 24 h with the following treatments: 1) EA (9.97 μg/mL = 30 µM); 2) EA (14.95 μg/mL = 45 µM); 3) TMZ (116.49 μg/mL = 600 µM); 4) EA (6.65 μg/mL = 20 µM) + TMZ (73.78 μg/mL = 380 µM); 5) EA (6.65 μg/mL = 20 µM) + TMZ (112.61 μg/mL = 580 µM); 6) EA (11.63 μg/mL = 35 µM) + TMZ (62.13 μg/mL = 320 µM); 7) untreated controls.
2.8.2 RNA isolation and sequencing
RNA was extracted as described (Ulrich-Merzenich et al., 2017) 100 ng/μL was used for RNA sequencing. The RNA sequencing was performed by the NGSCore Facility of the University Hospital Bonn. RNASeq data were deposited into the Gene Expression Omnibus database under accession number GSE245919 (URL: 154 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245919).
2.8.3 Data evaluation
Data evaluation was performed according to the guidelines provided on the Galaxy Training website (https://training.galaxyproject.org/) and with Ingenuity Pathway Analysis (QIAGEN IPA). GeneMANIA (Warde-Farley et al., 2010) was used to construct the network based on data on co-expression, genetic interaction, and pathways (https://genemania.org/).
2.9 Quantitative RT-PCR (qRT-PCR)
The same RNA samples analyzed by RNASeq were used for quantitative reverse transcription (qRT-PCR) as described earlier (Abdel-Aziz et al., 2015). See also Supplementary Material S3, S4.
2.10 Protein Wnt-inhibitory factor 1 (WIF1) release
Cells were treated as described under 2.5.2. Treatment was either with EA (6.65, 13.29, and 19.94 μg/mL = 20, 40, and 60 µM) or TMZ (58.25, 87.37, and 116.49 μg/mL = 300, 450, and 600 µM) alone or in combinations. The WIF1 release was determined by Human WIF-1 DuoSet ELISA (R&D Systems).
2.11 Statistical analysis
All values are expressed as mean ± SEM of three independent experiments. Experiments were performed with at least 3 replicates for each condition, if not otherwise mentioned. Statistical analyses were performed with SigmaStat (v.4.0) (http://www.systat.de/SigmaStat4_PR.html) and Origin 2018 software (https://www.originlab.com/origin) packages.
3 Results
3.1 Effects of lichen extracts and metabolites on cancer cells and fibroblasts
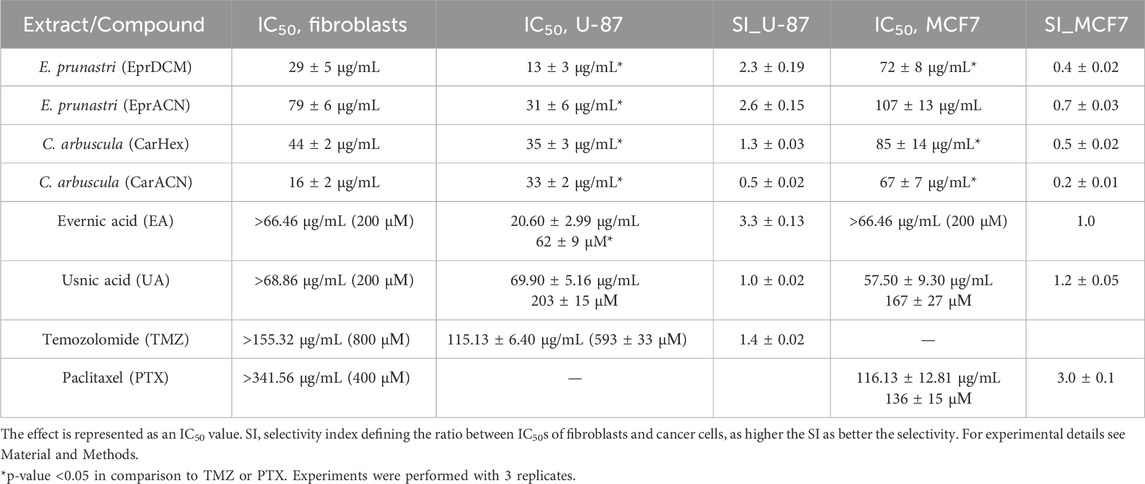
The ability of the lichen extracts, metabolites, and reference drugs to reduce the metabolic activity of TMZ-resistant U-87 and MCF7 cell lines and normal human skin fibroblasts (HSKF) is displayed in Table 1 and Supplementary Material S2.
The IC50 of TMZ against U-87 cells was high, confirming resistance to TMZ (Lee, 2016). All lichen extracts and metabolites significantly reduced the metabolic activity of U-87 cells. EprDCM had the highest potency, however, it also affected HSKF at a similar concentration, showing undesirable off-target action. EprACN, CarHex, and CarACN showed comparable reduction in U-87 cell metabolic activity. CarACN showed a high effect on HSKF, while EprACN demonstrated favorable specificity with a high selectivity index (SI) for U-87.
EA and UA did not significantly affect the metabolic activity of HSKF at concentrations up to 66.46 μg/mL and 68.86 μg/mL (equivalent to 200 µM), respectively. In U-87 cells, EA demonstrated greater activity and more favorable response comparing to UA. The results indicated that while the compounds were effective against cancer cell lines, they exhibited reduced activity toward HSKF cells, suggesting a degree of selectivity toward malignant cells.
The lichen extracts did not reduce the metabolic activity of MCF7 with the SI value below 1, indicating a lack of selectivity. Both EA and UA showed activity against MCF7 cells, but their SI was comparatively lower than that of PTX, leading to the discontinuation of further experiments with MCF7 cells. In the next step, combinations were investigated.
3.2 Combinatorial effects of lichen metabolites with TMZ or PTX
Figure 1a demonstrates the effects of combining TMZ or PTX with EprACN, EA, or UA on the metabolic activity of U-87 and MCF7 cells. The combination Index (CI) and the Dose Reduction Index (DRI) are shown in Figure 1b and the δ-score in Figure 1c.

Figure 1. Synergy assessments (a) Surface plots illustrating the cell viability expressed as a percentage of the control (Z-axes) depending on the treatment with the combination of TMZ or PTX (X-axes) and EprACN or EA or UA (Y-axes) at different concentrations. (b) Plots indicating the relation of CI and DRI to Fa. The CI < 1 denotes synergy, CI = 1 addition, and CI > 1 antagonism. (c) Surface plots illustrating the δ-score (Z-axes) indicating synergy (red), additive effect (white), or antagonism (green) for the combinations of TMZ or PTX (Y-axes) with EprACN, EA, or UA (X-axes) at various concentrations. Experiments were performed with 3 replicates.
The combination of EprACN with TMZ exhibited antagonism (CI = 3.07) and with PTX additive effects (CI = 1.04) (Figure 1b). Despite the additive effect of EprACN-PTX, metabolic activity remained above 50%. Consequently, combinations involving EprACN were not pursued. The UA-TMZ combination also displayed limited modulation of the metabolic activity (Figure 1a), and was therefore excluded from further investigations.
Although the EA-TMZ combination demonstrated antagonism, EA increased the sensitivity of U-87 cells to TMZ by 3.4 times (DRI = 3.4). Synergistic effects were observed at Fa < 0.5 (Figure 1b) with a CI of 0.97 for the combination of EA (8.31 μg/mL = 25 µM) and TMZ (77.66 μg/mL = 400 µM) at a 1:4 ratio (Figure 1c), resulting in approximately 50% reduction in metabolic activity (Figure 1a).
Effects of the EA-PTX and UA-PTX combinations were similar, with enhanced effectiveness leading to up to 60% in metabolic activity reduction (Figure 1a). These combinations had a primarily additive effect (Figure 1b, CI ≈ 1), transitioning to synergistic effects at concentrations of EA (4.15–33.23 μg/mL = 12.5–100 µM) – PTX (5.34–85.39 μg/mL = 6.25–100 µM) and UA (4.30–34.43 μg/mL = 12.5–100 µM) – PTX (5.34–85.39 μg/mL = 6.25–100 µM) (Figure 1c, δ-score > 0).
Since synergistic effects are influenced not only by drug concentrations but also by their ratio (Ammar and Ulrich-Merzenich, 2017), further research focused on the ratio (1:16) that demonstrated synergy (Figure 2).

Figure 2. U-87 cell viability and synergy calculations for the combinations of TMZ with EA at a ratio of 1:16. (a) Surface plot illustrating the cell viability as a percentage of the control (Z-axes) with the combination of TMZ (X-axes) and EA (Y-axes) at different concentrations. (b) The plot illustrates the relation of DRI to Fa. (c) The plot illustrates the CI according to Fa. (d) The surface plot illustrates the δ-score (Z-axes) indicating synergy for TMZ (Y-axes) with EA (X-axes) combination at different concentrations. Experiments were performed with 3 replicates.
At a ratio of 1:16, the TMZ-EA combination reached the maximum reduction of metabolic activity (75%). The IC50 of the drug combination was lower than the IC50 values of the single drugs (Figure 2a). Both metabolites showed a dose reduction (DRI > 1), and the CI value indicated additive effects (Figures 2 b,c). The δ-score demonstrated synergistic effects (Figure 2d). These promising results led to the investigation of the potential mechanisms underlying EA’s action (Figure 3).
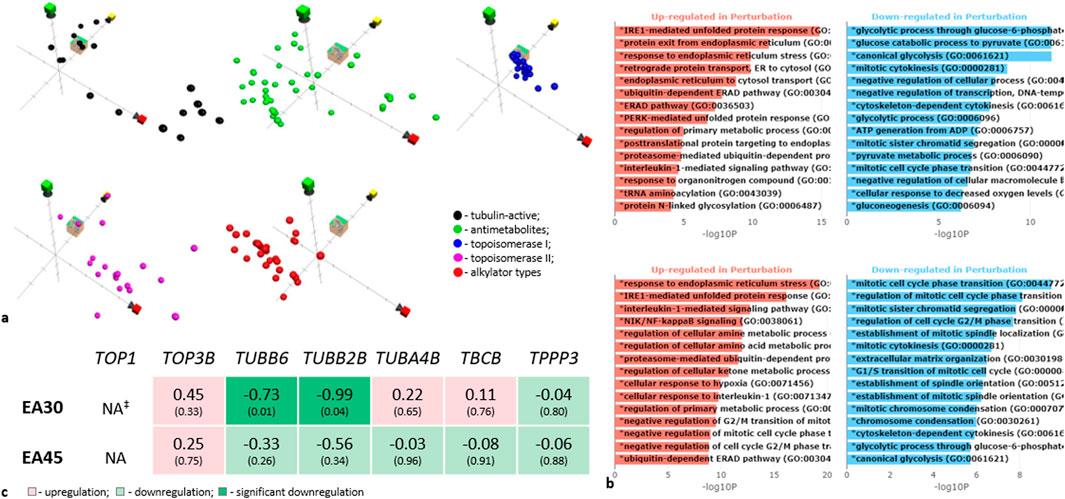
Figure 3. Investigation of the mechanism of action of Evernia prunastri ingredients. (a) Position in chemical property space of EA (cube), compared to reference sets of compounds in ChemGPS. The axis markers: red–size-related properties, yellow–conjugation and aromaticity-related properties, and green–lipophilicity and solubility-related properties. (b) Gene ontology enrichment analysis bar plot based on RNA deep sequencing data. Y-axis indicates significantly enriched GO biological processes, X-axis shows -log10 (p-values). Upper panel -EA 30 µM (9.97 μg/mL), lower panel -EA 45 µM (14.95 μg/ml). For details see Supplementary Material S5. (c) Gene sequencing data for tubulin-encoding genes. ‡NA: the gene is “not available” or the gene was excluded from analysis because it contained an extreme count outlier–large counts (during the gene expression analysis in Galaxy). Experiments were performed with 3 replicates.
3.3 Mechanistic insights into EA’s mode of action
In a first step, we used ChemGPs for a chemographic prediction of EA’s mode of action. The Euclidean distance calculation over all eight dimensions of the ChemGPS-NP chemical property space suggests that EA’s most probable mode of action is either topoisomerase I inhibition or disturbance of tubulin activity (Figure 3a). EA shares chemical properties with topoisomerase I (TOP1) inhibitors. Few TOP-1 inhibitors are semi-synthetic derivatives of the plant alkaloid camptothecin, stabilizing TOP1-DNA cleavable complexes (Top1cc) (Thomas and Pommier, 2019/06). In a second step, we compared the prediction with the RNAseq data analyses of the experiments. Here, the level of TOP1 was insignificant (Figure 3c). Thus, EA may interact with TOP1 either by blocking the DNA-helix or by disrupting, e.g., electrostatic interactions on the surface of TOP1 by binding to the active site.
Tubulin-active compounds bind to the tubulin microtubules, affecting their dynamics (Janke and Magiera, 2020). Although we did not examine the polymerization of microtubules and their dynamics, we evaluated the transcription of tubulins. EA decreased transcription of the β-tubulins TUBB6 and TUBB2B (Figure 3c). Since microtubules consist of α-β tubulin heterodimers (Janke and Magiera, 2020), a downregulation of β-isotypes may change the microtubule formations and prevent cell division. The Gene Ontology analysis of genes regulated by EA demonstrated an involvement of mitotic cell cycle phase transition, mitotic sister chromatid segregation, the establishment of mitotic spindle localization, mitotic cytokinesis, and mitotic chromosome condensation in such an activity (Figure 3b). This supports the prediction for EA to be a tubulin-active compound (see also Supplementary Material S5 for results of the Expression of genes coding tubulin-related proteins and Supplementary Material S6 for results of the Gene Ontology analysis).
Western blot analyses revealed that EA, along with EprACN, also modulates key signaling pathways. Figures 4a,b depict the modulation of Erk1/2, c-Jun, and Akt by EprACN and EA over 6, 12, and 24 h. EprACN and EA initially stimulated ERK1/2 formation at 6 h, followed by a downregulation after 12 h and 24 h compared to the control. Higher concentrations of EprACN and EA decreased the ERK phosphorylation at all times. They also downregulated c-Jun phosphorylation after 24 h, with a transient increase observed at 6 h. EprACN downregulated Akt phosphorylation after 6 h, whereas EA insignificantly upregulated it. Significant downregulation was observed after 12 h with EprACN (30 μg/mL), with no further change at 24 h, and with EA (13.29 μg/mL = 40 µM) after 24 h. The RNAseq data of the MAPK and PI3K pathways members’ expression, shown in Figure 4c, demonstrated no significant effect on their transcription.

Figure 4. Western blot for key proteins of the MAP Kinase and mTOR pathways expressed in U-87 cells treated with (a) EprACN (0, 20, 30, 40 μg/mL) and (b) EA (0, 40, 50, 60 µM) over a time period of 6, 12, and 24 h. (c) Gene expression of MAPK and PI3K pathways at 24 h #p-value <0.05: time-dependent comparison to control, *p-value <0.05: concentration-dependent protein expression over time. Experiments were performed with 3 replicates.
3.4 Multitarget mechanisms of EA-TMZ synergy
Expanding the RNA Seq analyses, we compared significantly regulated genes from the GE-analyses for all treatments using Venn diagrams (Figure 5a) to identify unique genes for the combinations. A network built using these genes revealed the canonical Wnt pathway as a pathway regulated by these genes (Figure 5b). The canonical Wnt signaling pathway significantly contributes to the development of resistance to chemo- and radiotherapy in GBM (Zhong and Virshup, 2020). Figure 5c illustrates the expressed genes involved in Wnt signaling upon treatment.
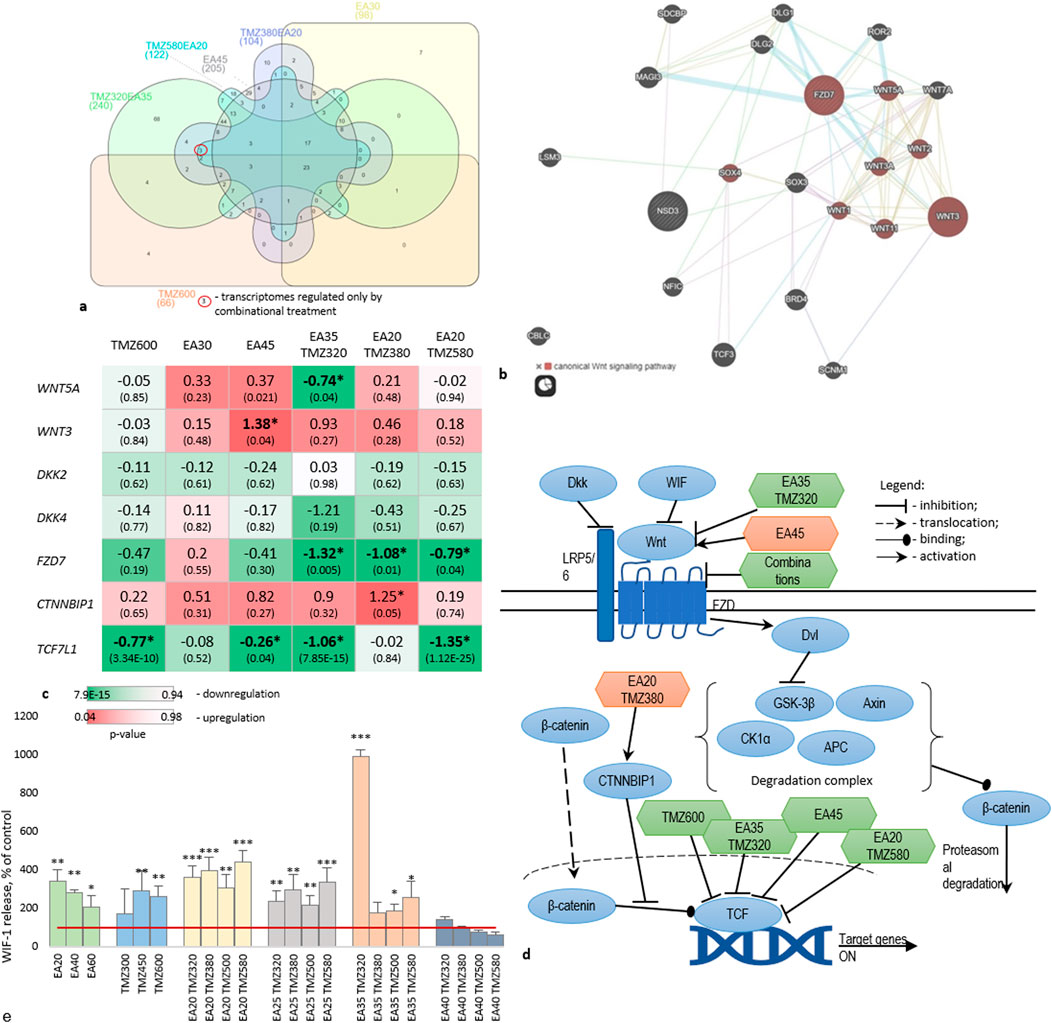
Figure 5. Regulation of Wnt signaling during treatment of U-87 cells with TMZ, EA, and their combinations. (a) Venn-diagram (https://www.interactivenn.net/) on significantly regulated genes obtained from the transcriptome analyses. (b) Network depicting a possible pathway uniquely regulated by the combinations, obtained from GeneMania (https://genemania.org/). (c) Gene expression of Wnt signaling members. Data are presented as logFC. Cell colors represent p-values. (d) Schematic representation of the proposed pathway regulated by combinations. (e) Level of WIF1 protein release into the culture medium. *p < 0.05, **p < 0.01, ***: p < 0.001. The red line indicates the control. Experiments were performed with 3 replicates.
Particularly, EA35TMZ320 significantly downregulated the upstream member WNT5A, whereas other treatments did not show a significant effect. Conversely, WNT3, a ligand of LRP5/6 (Low-density lipoprotein receptor-related protein 5/6), exhibited significant undesirable upregulation by EA45, whereas DKK2 and DKK4, which are inhibitors of LRP5/6, remained unaffected by any treatment.
All combinations significantly downregulated FZD7 transcription, which encodes a transmembrane receptor crucial for Wnt downstream. Activation of this receptor by Wnts inhibits the β-catenin degradation complex. Additionally, CTNNBIP1, an intracellular member known to prevent β-catenin activity, showed significant upregulation of gene expression by EA20TMZ380.
TCF7L1, an intranuclear member of the pathway involved in cell cycle regulation and proliferation, was significantly downregulated by TMZ600, EA45, EA35TMZ320, and EA20TMZ580.
Figure 5d summarizes the interaction of affected targets within the Wnt pathway. The combinations regulated the transcription of more targets compared to single metabolites.
After analyzing the GE of members of the Wnt-signaling pathway under treatment, we wanted to know whether Wnt-signaling is affected on the protein level. Therefore, we measured the Wnt inhibitory factor 1 (WIF1). WIF1 binds to Wnt-proteins, thereby inhibiting the activation of the Wnt-signaling (see also Supplementary Material S7).
Figure 5e shows the release of WIF1 protein by U-87 cells treated with TMZ and EA alone or in combination. Both single metabolites and combinations significantly increased the WIF1 release compared to the control. Combinations demonstrated a dose-dependent increase in WIF1 release, with the highest activity for EA35TMZ320. The effect of TMZ and EA on the WIF1 release showed a concentration-dependent inverse pattern: higher TMZ concentration increases the effect, while higher EA concentration reduces it. Significant up-regulations were observed by the following combinations: EA20TMZ500, EA20TMZ580, EA25TMZ580, EA35TMZ320, EA35TMZ380, and EA30TMZ500. However, EA35TMZ320 showed the highest influence on the WIF1 release.
The Wnt pathway is a regulatory system interacting directly or indirectly with other signaling pathways, including the NF-kB pathway, as a central pathway in inflammation (Guo et al., 2024). This pathway is essential for connecting inflammation and cancer, as well as for tumor growth and resistance (Guo et al., 2024). For BC, it was shown that NF-kB signaling boosts the growth potential of BC cells and facilitates the spread of tumors (Guo et al., 2024). Therefore, we were interested in the influence of our metabolites and their combinations with TMZ on inflammation. At the same time, we were interested in EA’s reported neuroprotective and anti-inflammatory properties (Lee et al., 2021). Figure 6 shows the influence of the single metabolites and the combinations on the GE of GABRs, BDNF, and major cytokines regulating inflammatory processes (IL6, IL10, and TNFA) based on RNA deep sequencing (Figure 6a) and RT-PCR (Figure 6b).
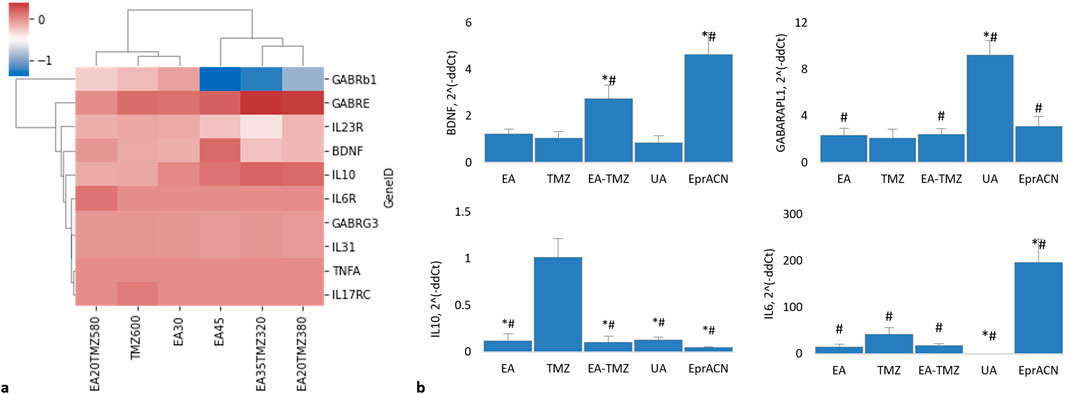
Figure 6. Expression of genes encoding brain-related proteins and cytokines. (a) Clustered HeatMap of RNAseq data showing the expression levels of various genes. None of the genes were found to be significantly regulated (p > 0.05). Genes include GABRB1: gamma-aminobutyric acid receptor (GABR) subunit beta 1; GABRE: GABR subunit epsilon; GABRG3: GABR subunit gamma 3; BDNF: brain-derived neurotrophic factor; IL10: interleukin 10; IL6R: interleukin 6 receptor; TNFa: tumor necrosis factor a; IL23R: interleukin 23 receptor; IL31: interleukin 31; IL17RC: interleukin 17 receptor. (b) Histograms showing mRNA levels based on qRT-PCR data. Concentrations: EA – 45 μM, TMZ – 600 μM, EA-TMZ – 35–320 μM, UA – 200 μM, and EprACN – 30 μg/mL. GABARAPL1: GABAA receptor-associated protein-like 1. *p < 0.05 comparing to TMZ, #p < 0.05 comparing to untreated control. Experiments were performed with 3 replicates.
The pro-inflammatory cytokine TNFA was neither detected by RNA deep sequencing nor by qRT-PCR. The anti-inflammatory IL10, detected by deep sequencing, was not significantly influenced. The RNAseq data showed no regulation of IL6. However, qRT-PCR data showed a significantly amplified expression of IL6 for the single treatment with EprACN or with TMZ, whereas UA downregulated the GE.
RNAseq analysis showed no significant differences in the expression of GABA receptor subunits. However, GABRB1 was downregulated by EA45 (p = 0.07), while GABARAPL1 was desirably upregulated for all treatments, with the highest expression seen with UA.
RNAseq analysis further revealed insignificant influences of all treatments on BDNF transcription. However, in qRT-PCR investigations, BDNF was upregulated by EA-TMZ and EprACN and downregulated by UA.
IL10 gene expression was significantly downregulated by all treatments except TMZ, while IL6 gene expression was significantly increased by EprACN and decreased by UA.
4 Discussion
The effectiveness of standard chemotherapeutics such as temozolomide (TMZ) in glioblastoma (GBM) and paclitaxel (PTX) in breast cancer (BC) is often limited by developing resistance and eventual treatment failure (Das et al., 2021; Lee, 2016). Therefore, we investigated the adjuvant potential of characterized extracts of E. prunastri and C. arbuscula and their metabolites evernic acid (EA) and usnic acid (UA) alone or in combination with TMZ and PTX for their immunomodulatory and chemosensitivity-increasing potential. HSKF served to evaluate general cytotoxicity and selectivity, even though these cells do not fully replicate the physiological environment or cellular characteristics of normal brain or mammary tissue. U-87 and MCF7 cells have their limitation, too. U-87 cells differ genetically from primary GBM tumors and do not fully recapitulate the intratumoral heterogeneity, stem cell populations, or invasive behavior observed in patient-derived GBM (Xie et al., 2015; Allen et al., 2016). Likewise, MCF7 cells, while representative of HR+ BC, do not model the full spectrum of BC subtypes, particularly TNBC or HER2+ disease, and lack the tumor microenvironment components that influence drug response in vivo (Neve et al., 2006; Holliday and Speirs, 2011). Both models are grown in two-dimensional monolayers, which do not capture the complexity of tumor-stroma interactions, hypoxia, or immune modulation present in human tumors. Despite these limitations, the use of U-87 and MCF7 in this study provides a well-established platform for assessing the antiproliferative and resistance-modulating effects of lichen-derived metabolites with findings that can inform subsequent validation in more complex models such as patient-derived cells, organoids, or in vivo systems.
The initial experiments revealed a reduction in cellular metabolic activity in U-87 and MCF7 cells after treated with the lichen extracts and their metabolites. However, they were less effective against MCF7 cells compared to PTX. Therefore, experiments with MCF7 cells were discontinued, even though UA had exhibited activity against MCF7 cells in earlier studies (Bačkorová et al., 2011; Hawrył et al., 2020).
For extracts of E. prunastri, IC50 values of 90.1 μg/mL and 81.8 μg/mL were reported (Bézivin et al., 2004), which is in the range of our results. Effects of EA against MCF7 and U-87 have not been reported earlier. However, previous reports suggest its activity against A-172 and T98G glioblastoma cell lines (Studzińska-Sroka et al., 2021), supporting our findings.
The extracts of C. arbuscula were not further investigated due to their non-selective effect. Further experiments focused on the TMZ-resistant U-87 cells (IC50 > 500 µM) (Lee, 2016) and the combination of TMZ and EA, since this combination yielded the most promising reduction in cellular metabolic activity, while showing minimal effect on HSKF.
To substantiate the antiproliferative effects, we performed Western blot (WB) analyses of members of the central MAPK families. They play a central role by regulating the cell cycle engine and other proliferation-related proteins (Bézivin et al., 2004; Studzińska-Sroka et al., 2021; Zhang and Liu, 2002). In addition, we measured AKT. In the context of cancer, Akt signaling promotes tumor cell survival, proliferation, growth, and metabolism by activating its downstream effectors. Both the PI3K/Akt and the MEK/ERK pathways cooperate in tumor growth and are involved in the development of therapeutic resistance in GBM cells (Singh et al., 2023).
WB analyses revealed that EprACN and EA reduced the expression and phosphorylation of ERK1/2 over time, explaining the observed decrease in the metabolic activity in U-87 cells and revealing an antiproliferative activity, as in most cells, sustained ERK activation is required to induce cell cycle entry. In Glioma, an aberrant activity of the RAS/MAPK/ERK pathway appears to play a crucial role in the development of gliomas (Stachyra and Grzybowska-Szatkowska, 2025). Even a large proportion of resistance mechanisms are associated with reactivation of the RTK/Raf/Ras/MEK/ERK pathway. Co-treatment with inhibitors targeting these pathways is meanwhile regarded as a compelling strategy to overcome resistance mechanisms in GBM (Yakubov et al., 2025).
Another member of the MAPK-pathway, the so-called proto-oncogene c-Jun, is also central to cancer-altered signalling: an upregulated c-Jun was described for variable tumor cells, specifically in brain tumors, contributing to its malignancy (Blau et al., 2012). Blau et al. demonstrated that the accumulation of c-Jun in tumors is regulated more translationally than transcriptionally (Blau et al., 2012) corresponding to our data with the regulation of c-Jun at the protein but not at the mRNA level.
EprACN and EA downregulated pAkt in a time- and dose-dependent manner corresponding to the reduction in metabolic activity in U-87 cells. This is particularly relevant, given that overexpression and high phosphorylation of Akt correlate with a poor prognosis in glioblastoma patients (Shahcheraghi et al., 2020). Increased pAkt activates transcription factors by phosphorylating GSK3β, leading to its inactivation and subsequent translocation of β-catenin into the nucleus. Both pathways, when activated independently, may contribute to resistance (Manoranjan et al., 2020). Conversely, β-catenin also induces the expression of Akt1 and Akt2 and the phosphorylation of Akt (Zhong and Virshup, 2020). Transcription of WNT3 was contradictorily upregulated by EA, indicating a limitation as a single treatment at higher concentrations. Nevertheless, the downregulation of Akt by EprACN and EA may contribute to the enhanced sensitivity of U-87 cells to TMZ.
In our combinatory study, GE-profiling of U-87 cells revealed the modulation of multiple components within the Wnt/β-catenin pathway. Combination EA35TMZ320 reduced the transcription of WNT5A, an upstream intracellular member (Yu et al., 2007; Chen et al., 2021) known to play a pro-tumor role in glioma (Zuccarini et al., 2018; Chen et al., 2021) to induce the migration of GBM cells (Lee, 2016) to increase cell proliferation (Yu et al., 2007; Chen et al., 2021) and to correlate with higher WHO histological glioma classification grades (Alkailani et al., 2022). A Wnt5a knockdown inhibited the activity of the GSK3β/β-catenin pathway related to glioma-derived endothelial cell angiogenesis (Chen et al., 2021).
Frizzleds (FZDs) are transmembrane receptors (Alkailani et al., 2022) inhibiting the β-catenin degradation complex (Shahcheraghi et al., 2020). In glioma, FZD7 is upregulated, correlating with poor patient outcomes (Zuccarini et al., 2018). The significant downregulation of FZD7 GE by all combinations is promising. Alike EA20TMZ380 significantly upregulated the GE of CTNNBIP1, which prevents the interaction of β-catenin and TCF (transcription factors) family members. Negative regulation of CTNNBIP1 correlates with higher grades of glioma (Tong et al., 2015).
To support the transcriptional findings, we measured the release of Wnt inhibitory factor (WIF1) protein via ELISA. WIF1 binds to Wnt-proteins, thereby inhibiting Wnt pathway signalling. Both single metabolites and combinations increased the release of WIF1 protein. In contrast, the combination of high concentrations of EA (40 µM) and TMZ (≥380 µM) reduced WIF1 releases, supporting the dose-dependent effects observed in the GE analyses, where higher concentrations induced an upregulation of WNT3. This inverse dose-dependent WIF1 regulation underscores the importance of optimized dosing to balance pathway modulation.
Cross-talks between signaling pathways are known to play a role in resistance development f. e. Wnt/ß-catenin signaling activates NF-kB in the cytoplasm, whereas Dvl inhibits NF-kB signaling in the nucleus (Guo et al., 2024). Even more in breast cancer, NF-kB has been confirmed to be a crucial link between resistance signaling pathways (Zhao et al., 2021). Activated NF-kB promotes the production of Wnt, ß-catenin, and ß-TrCP, which can lead to cytokine storms up to death (Guo et al., 2024; Jang et al., 2021). We did not measure NF-kB, but the gene expression of IL6, a product of NF-kB, which can activate, via STAT3, cell survival, proliferation, and inflammation (Guo et al., 2024). The resolution of inflammation is regarded as a novel host-focused option to complement existing therapies for glioma (Bazan et al., 2021). IL6 is frequently upregulated in GBM, where it activates JAK/STAT3 signaling to promote tumor cell survival, proliferation, and therapy resistance, and contributes to an immunosuppressive milieu (West et al., 2018). IL10 primarily exerts immunosuppressive effects in the GBM microenvironment by inhibiting effective anti-tumor immune responses and, in some contexts, directly enhancing glioma proliferation via JAK–STAT3 activation (Widodo et al., 2021). EprACN and TMZ increased the transcription of IL6 (RT-PCR), and IL10 transcripts were detectable. However, we previously observed that plant ingredients and acetylsalicylic acid stimulated the GE of IL6 and IL10 under non-stress conditions, which turned into an anti-inflammatory response under inflammatory conditions. Such cytokine regulations may keep the immune-regulatory system active and influence the cytokine release dynamics (Ulrich-Merzenich et al., 2017). This hypothesis aligns with Ahmad et al.‘s proposition that the simultaneous expression of IL6 and IL10 in tumor tissues improves the survival of breast cancer patients, although underlying mechanisms remain unclear (Ahmad et al., 2018).
We did not observe changes in the GE of IL4 and IL8. However, both exert their most critical actions via crosstalk with immune and endothelial cells. In a tumor-cell-only model, the roles of IL4 and IL8 would likely appear less central compared to their significance in the complex in vivo GBM microenvironment (Brat et al., 2005; Losur et al., 2024).
Furthermore, the IL-17B/ IL-17RB pathway has been implicated in tumorigenesis and resistance to anticancer therapies (Briukhovetska et al., 2021). The IL-17A/F, binding to the IL17RC receptor, demonstrated pro-tumoral effects (Briukhovetska et al., 2021). Although its precise role in tumor resistance remains unclear. Our RNAseq data revealed exclusive expression of RNA encoding IL17RC in resistant U-87 cells treated with high TMZ concentrations, suggesting that the IL-17 pathway may contribute to the development of resistance in U-87 cells, a finding warranting further study.
Brain-derived neurotrophic factor (BDNF), an endogenous signaling molecule, is involved in the carcinogenesis of glioma (Zheng and Chen, 2020), especially in tumor growth and metastasis in neuroblastoma (Chen et al., 2016), whereas a precursor of BDNF (proBDNF) plays a role in the modulation of cell apoptosis (Xiong et al., 2013). In our study, EA35TMZ320 and EprACN30 induced significant over-expression of BDNF without activating GE of the PI3K and MAPK pathway members. We hypothesize that such an effect does not induce cell growth or counteract the reduction in metabolic activity seen in U-87 cells upon different treatments.
GABAA forms a heterotetrameric complex (Huang et al., 2022). The expression of subunits α, β, and γ-subfamilies correlates with the malignancy grade of gliomas (Smits et al., 2012). Patients with high GABAA receptor-associated protein (GABARAPL1) expression levels were reported to have a lower risk for metastasis (Le Grand et al., 2011). In MCF7 cells, GABARAPL was demonstrated to inhibit Dvl2 (disheveled segment polarity protein 2), an inhibitor of the β-catenin degradation complex (Boyer-Guittaut et al., 2014). While TMZ, EA, and TMZ-EA did not affect the GE of Dvl2, TMZ and EA upregulated the GE of GABARAPL1. Thus, we suggest that the downregulation of the Wnt signaling cascade observed in our study is likely a direct effect on Wnt signaling members rather than through GABARAPL1.
5 Summary and conclusion
This study demonstrates the potential of lichen-derived metabolites, particularly evernic acid (EA), to modulate key pathways associated with TMZ resistance in U-87 cells, suggesting a promising multitarget mechanism for future investigation. EA reduced the metabolic activity of TMZ-resistant U-87 cells, synergizing with TMZ to reduce viability by 75% at optimized ratios. The prediction of ChemGPS that EA acts on tubulin activity was supported by deep sequencing. Mechanistically, EA suppressed GE of oncogenic Wnt/β-catenin signaling while upregulating the protein expression of WIF1 as a central inhibitor of Wnt-signaling. Combinatorial EA-TMZ treatment further modulated MAPK/PI3K pathways, inhibiting ERK1/2, c-Jun, and Akt phosphorylation, which are critical for glioblastoma survival and resistance.
The discovery of IL17RC overexpression in resistant cells underscores a novel pathway implicated in TMZ resistance, warranting further exploration.
Even though present studies lack an in vivo validation, findings form a base for subsequent validation in more complex models. Future studies should clarify EA’s direct role in tubulin dynamics, its influence on the IL-17 pathway, on established mechanisms of drug resistance, such as MGMT promoter methylation status, DNA repair pathways, or efflux transporter activity in primary patient-derived cells, as well as in vivo, for example, in genetically engineered glioma models (GEGMs) or orthotopic animal models including pharmacodynamic and pharmacokinetic evaluations to explore clinical translation of EA-TMZ combinations. Integrating computational tools like ChemGPS-NP with multi-omics approaches will accelerate the development of natural product-based therapies to address refractory cancers. This work advances the paradigm of combinatorial, mechanism-driven strategies to disrupt resistance-associated pathways and enhance chemosensitivity in oncology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because humans were not involved in the study. Only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because animals were not involved in the study. Only commercially available established cell lines were used.
Author contributions
AS: Conceptualization, Software, Investigation, Formal Analysis, Writing – review and editing, Resources, Visualization, Writing – original draft. LN: Formal Analysis, Writing – review and editing, Investigation, Visualization, Validation. AK: Writing – review and editing, Methodology, Conceptualization. AB: Conceptualization, Methodology, Writing – review and editing, Software. SB: Methodology, Conceptualization, Writing – review and editing, Software. ER: Writing – review and editing, Resources. GU-M: Supervision, Conceptualization, Writing – review and editing, Software, Methodology, Funding acquisition, Data curation, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. A. Shcherbakova was supported by the German Academic Exchange Service (DAAD) with a long-term scholarship at Bonn University (Medical Clinic III, University Hospital Bonn, Germany) in 2014/15, a short-term scholarship in 2019, by a grant from the President of the Russian Federation for Young Scientists [number MK5290.2012.4] and a grant for international mobility from the Volga State University of Technology at Uppsala University in 2013. Institutional funding was provided for the experimental research at UKB. This publication was supported by the Open Access Publication Fund of the University of Bonn.
Acknowledgments
Part of the work on extract preparation was undertaken in the laboratory of Ulf Göransson, Uppsala University (Shcherbakova et al., 2021). The authors thank Bernd Merzenich for his skillful language editing of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1633978/full#supplementary-material
Abbreviations
TMZ, Temozolomide; PXT, Paclitaxel; GBM, glioblastoma; BC, breast cancer; EprDCM, dichloromethane extract from E. prunastri; EprACN–acetonitrile extract from E. prunastri; CarHex, hexane extract from C. arbuscula; CarACN–acetonitrile extract from C. arbuscula; EA, evernic acid; UA, usnic acid; TMZ320EA35, combination of TMZ (320 µM = 62.13 μg/mL) with EA (35 µM = 11.63 μg/mL); TMZ380EA20, TMZ (380 µM = 73.78 μg/mL) with EA (20 µM = 6.65 μg/mL); TMZ580EA20, TMZ (580 µM = 112.61 μg/mL) with EA (20 µM = 6.65 μg/mL).
References
Abdel-Aziz, H., Schneider, M., Neuhuber, W., Kassem, A. M., Khailah, S., Müller, J., et al. (2015). GPR84 and TREM-1 signaling contribute to the pathogenesis of reflux esophagitis. Mol. Med. 21, 1011–1024. doi:10.2119/molmed.2015.00098
Abu, S. T. M., Samec, M., Liskova, A., Kubatka, P., and Büsselberg, D. (2019). Paclitaxel’s mechanistic and clinical effects on breast cancer. Biomolecules 9, 789. doi:10.3390/biom9120789
Ahmad, N., Ammar, A., Storr, S. J., Green, A. R., Rakha, E., Ellis, I. O., et al. (2018). IL-6 and IL-10 are associated with good prognosis in early stage invasive breast cancer patients. Cancer Immunol. Immunother. 67 (4), 537–549. doi:10.1007/s00262-017-2106-8
Alcantara Llaguno, S. R., Chen, J., and Parada, L. F. (2009). Signaling in malignant astrocytomas: role of neural stem cells and its therapeutic implications. Clin. Cancer Res. 15 (23), 7124–7129. doi:10.1158/1078-0432.CCR-09-0433
Alkailani, M. I., Aittaleb, M., and Tissir, F. (2022). WNT signaling at the intersection between neurogenesis and brain tumorigenesis. Front. Mol. Neurosci. 15, 1017568. doi:10.3389/fnmol.2022.1017568
Allen, M., Bjerke, M., Edlund, H., Nelander, S., and Westermark, B. (2016). Origin of the U87MG glioma cell line: good news and bad news. Sci. Transl. Med. 8 (354), 354re3. doi:10.1126/scitranslmed.aaf6853
Ammar, R., and Ulrich-Merzenich, G. (2017). Curcumin synergizes with the endocannabinoid reuptake inhibitor OMDM-2 in human MCF-7 breast cancer and U-87 glioblastoma cells. Synergy. 5, 7–14. doi:10.1016/j.synres.2017.11.001
Bačkorová, M., Bačkor, M., Mikeš, J., Jendželovský, R., Fedoročko, P., Backorova, M., et al. (2011). Variable responses of different human cancer cells to the lichen compounds parietin, atranorin, usnic acid and gyrophoric acid. Toxicol Vitro. 25(1), 37–44. doi:10.1016/j.tiv.2010.09.004
Baranova, A., Krasnoselskyi, M., Starikov, V., Kartashov, S., Zhulkevych, I., Vlasenko, V., et al. (2022). Triple-negative breast cancer: current treatment strategies and factors of negative prognosis. J. Med. Life 15 (2), 153–161. doi:10.25122/jml-2021-0108
Bazan, N. G., Reid, M. M., Flores, V. A. C., Gallo, J. E., Lewis, W., and Belayev, L. (2021). Multiprong control of glioblastoma multiforme invasiveness: blockade of pro-inflammatory signaling, anti-angiogenesis, and homeostasis restoration. Cancer Metastasis Rev. 40 (3), 643–647. doi:10.1007/s10555-021-09987-x
Bézivin, C., Tomasi, S., Rouaud, I., Delcros, J. G. G., Boustie, J., Bezivin, C., et al. (2004). Cytotoxic activity of compounds from the lichen: Cladonia convoluta. Planta Med. 70, 874–877. doi:10.1055/s-2004-827240
Blau, L., Knirsh, R., Ben-Dror, I., Oren, S., Kuphal, S., Hau, P., et al. (2012). Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc. Natl. Acad. Sci. 109 (42), E2875–E2884. doi:10.1073/pnas.1203659109
Boyer-Guittaut, M., Poillet, L., Liang, Q., Bôle-Richard, E., Ouyang, X., Benavides, G. A., et al. (2014). The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells. Autophagy 10 (6), 986–1003. doi:10.4161/auto.28390
Brat, D. J., Bellail, A. C., and Van Meir, E. G. (2005). The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol. 7 (2), 122–133. doi:10.1215/S1152851704001061
Bravo, P., Dois, A., Villarroel, L., González-Agüero, M., Fernández-González, L., Sánchez, C., et al. (2023). Factors influencing the implementation of shared decision-making in breast cancer care: protocol for a mixed-methods study. BMJ Open 13 (7), e074111. doi:10.1136/bmjopen-2023-074111
Brisdelli, F., Perilli, M., Sellitri, D., Piovano, M., Garbarino, J. A., Nicoletti, M., et al. (2013). Cytotoxic activity and antioxidant capacity of purified lichen metabolites: an in vitro study. Phytotherapy Res. 27 (3), 431–437. doi:10.1002/ptr.4739
Briukhovetska, D., Dörr, J., Endres, S., Libby, P., Dinarello, C. A., and Kobold, S. (2021). Interleukins in cancer: from biology to therapy. Nat. Rev. Cancer 21 (8), 481–499. doi:10.1038/s41568-021-00363-z
Bukowski, K., Kciuk, M., and Kontek, R. (2020). Mechanisms of multidrug resistance in cancer chemotherapy. Int. J. Mol. Sci. 21 (9), 3233. doi:10.3390/ijms21093233
Buonfiglio, R., Engkvist, O., Várkonyi, P., Henz, A., Vikeved, E., Backlund, A., et al. (2015). Investigating pharmacological similarity by charting chemical space. J. Chem. Inf. Model 55 (11), 2375–2390. doi:10.1021/acs.jcim.5b00375
Chen, B., Liang, Y., He, Z., An, Y., Zhao, W., and Wu, J. (2016). Autocrine activity of BDNF induced by the STAT3 signaling pathway causes prolonged TrkB activation and promotes human non-small-cell lung cancer proliferation. Sci. Rep. 6 (1), 30404. doi:10.1038/srep30404
Chen, Z. F., Liu, J., Huang, Z., Zheng, Y., Deng, S., et al. (2021). Dual role of WNT5A in promoting endothelial differentiation of glioma stem cells and angiogenesis of glioma derived endothelial cells. Oncogene 40 (32), 5081–5094. doi:10.1038/s41388-021-01922-2
Chou, T. C. (2010). Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70 (2), 440–446. doi:10.1158/0008-5472.CAN-09-1947
Crawford, S. D. (2015). “Lichens used in traditional medicine,” in Lichen secondary metabolites: bioactive properties and pharmaceutical potential. Editor B. Ranković (Cham, Switzerland: Springer International Publishing), 27–80. doi:10.1007/978-3-319-13374-4_2
Das, T., Anand, U., Pandey, S. K., Ashby, C. R., Assaraf, Y. G., Chen, Z. S., et al. (2021). Therapeutic strategies to overcome taxane resistance in cancer. Drug Resist. Updat. 55, 100754. doi:10.1016/j.drup.2021.100754
DeCordova, S., Shastri, A., Tsolaki, A. G., Yasmin, H., Klein, L., Singh, S. K., et al. (2020). Molecular heterogeneity and immunosuppressive microenvironment in glioblastoma. Front. Immunol. 11, 1402. doi:10.3389/fimmu.2020.01402
Einarsdottir, E., Groeneweg, J., Bjornsdottir, G. G., Harethardottir, G., Omarsdottir, S., Ingolfsdottir, K., et al. (2010). Cellular mechanisms of the anticancer effects of the lichen compound usnic acid. Planta Med. 76 (10), 969–974. doi:10.1055/s-0029-1240851
Emsen, B., Aslan, A., Türkez, H., Taghizadehghalehjoughi, A., and Kaya, A. (2018). The anti-cancer efficacies of diffractaic, lobaric, and usnic acid: in vitro inhibition of glioma. J. Cancer Res. Ther. 14, 941–951. doi:10.4103/0973-1482.177218
Fekete, B., Werlenius, K., Tisell, M., Pivodic, A., Smits, A., Jakola, A. S., et al. (2023). What predicts survival in glioblastoma? A population-based study of changes in clinical management and outcome. Front. Surg. 10, 1249366. doi:10.3389/fsurg.2023.1249366
Fernández-Moriano, C., Divakar, P. K., Crespo, A., and Gómez-Serranillos, M. P. (2017). Protective effects of lichen metabolites evernic and usnic acids against redox impairment-mediated cytotoxicity in central nervous system-like cells. Food Chem. Toxicol. 105, 262–277. doi:10.1016/j.fct.2017.04.030
Galanty, A., Koczurkiewicz, P., Wnuk, D., Paw, M., Karnas, E., Podolak, I., et al. (2017). Usnic acid and atranorin exert selective cytostatic and anti-invasive effects on human prostate and melanoma cancer cells. Toxicol Vitro 40, 161–169. doi:10.1016/j.tiv.2017.01.008
Gökalsın, B., and Sesal, N. C. (2016). Lichen secondary metabolite evernic acid as potential quorum sensing inhibitor against Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 32 (9), 150. doi:10.1007/s11274-016-2105-5
Grant, S. J., Kay, S., Lacey, J., Kumar, S., Kerin-Ayres, K., Stehn, J., et al. (2024). Feasibility study of a multimodal prehabilitation programme in women receiving neoadjuvant therapy for breast cancer in a major cancer hospital: a protocol. BMJ Open 14 (3), e080239. doi:10.1136/bmjopen-2023-080239
Guan, R., Zhang, X., and Guo, M. (2020). Glioblastoma stem cells and Wnt signaling pathway: molecular mechanisms and therapeutic targets. Chin. Neurosurg. J. 6 (1), 25. doi:10.1186/s41016-020-00207-z
Gueble, S. E., Vasquez, J. C., and Bindra, R. S. (2022). The role of PARP inhibitors in patients with primary malignant central nervous system tumors. Curr. Treat. Options Oncol. 23 (11), 1566–1589. doi:10.1007/s11864-022-01024-5
Guo, Q., Jin, Y., Chen, X., Ye, X., Shen, X., Lin, M., et al. (2024). NF-κB in biology and targeted therapy: new insights and translational implications. Signal Transduct. Target Ther. 9 (1), 53. doi:10.1038/s41392-024-01757-9
Hawrył, A., Hawrył, M., Hajnos-Stolarz, A., Abramek, J., Bogucka-Kocka, A., and Komsta, Ł. (2020). HPLC fingerprint analysis with the antioxidant and cytotoxic activities of selected lichens combined with the chemometric calculations. Molecules 25 (18), 4301. doi:10.3390/molecules25184301
Holliday, D. L., and Speirs, V. (2011). Choosing the right cell line for breast cancer research. Breast Cancer Res. 13 (4), 215. doi:10.1186/bcr2889
Huang, Q., Chen, L., Liang, J., Huang, Q., and Sun, H. (2022). Neurotransmitters: potential targets in glioblastoma. Cancers (Basel) 14 (16), 3970. doi:10.3390/cancers14163970
Ianevski, A., He, L., Aittokallio, T., and Tang, J. (2017). SynergyFinder: a web application for analyzing drug combination dose – response matrix data. Bioinformatics 33 (15), 2413–2415. doi:10.1093/bioinformatics/btx162
Jang, J., Song, J., Sim, I., Kwon, Y. V., and Yoon, Y. (2021). Wnt-signaling inhibitor Wnt-C59 suppresses the cytokine upregulation in multiple organs of lipopolysaccharide-induced endotoxemic mice via reducing the interaction between β-catenin and NF-κB. Int. J. Mol. Sci. 22 (12), 6249. doi:10.3390/ijms22126249
Janke, C., and Magiera, M. M. (2020). “The tubulin code and its role in controlling microtubule properties and functions. Vol. 21,” in Nature reviews molecular cell biology. Berlin, Germany: Nature Research, 307–326.
Jezierzański, M., Nafalska, N., Stopyra, M., Furgoł, T., Miciak, M., Kabut, J., et al. (2024). Temozolomide (TMZ) in the treatment of glioblastoma multiforme—a literature review and clinical outcomes. Curr. Oncol. 31 (7), 3994–4002. doi:10.3390/curroncol31070296
Kashyap, D., Pal, D., Sharma, R., Garg, V. K., Goel, N., Koundal, D., et al. (2022). Global increase in breast cancer incidence: risk factors and preventive measures. Biomed. Res. Int. 2022, 9605439–16. doi:10.1155/2022/9605439
Khongkow, P., Gomes, A. R., Gong, C., Man, E. P. S., Tsang, J. W. H., Zhao, F., et al. (2016). Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene 35 (8), 990–1002. doi:10.1038/onc.2015.152
Kiliç, N., Islakoğlu, Y. Ö., Büyük, İ., Gür-Dedeoğlu, B., and Cansaran-Duman, D. (2019). Determination of usnic acid responsive miRNAs in breast cancer cell lines. Anticancer Agents Med. Chem. 19 (12), 1463–1472. doi:10.2174/1871520618666181112120142
Le Grand, J., Chakrama, F., Seguin-Py, S., Fraichard, A., Delage-Mourroux, R., Jouvenot, M., et al. (2011). GABARAPL1 (GEC1): original or copycat? Autophagy 7, 1098–1107. doi:10.4161/auto.7.10.15904
Lee, S. Y. (2016). Temozolomide resistance in glioblastoma multiforme. Genes Dis. 3 (3), 198–210. doi:10.1016/j.gendis.2016.04.007
Lee, S., Suh, Y. J., Yang, S., Hong, D. G., Ishigami, A., Kim, H., et al. (2021). Neuroprotective and anti-inflammatory effects of evernic acid in an MPTP-induced Parkinson’s disease model. Int. J. Mol. Sci. 22 (4), 2098. doi:10.3390/ijms22042098
Li, J., Xia, Y., Wu, Q., Zhu, S., Chen, C., Yang, W., et al. (2017). Outcomes of patients with inflammatory breast cancer by hormone receptor- and HER2-defined molecular subtypes: a population-based study from the SEER program. Oncotarget 8 (30), 49370–49379. doi:10.18632/oncotarget.17217
Li, H., Liu, S., Jin, R., Xu, H., Li, Y., Chen, Y., et al. (2021). Pyrvinium pamoate regulates MGMT expression through suppressing the Wnt/β-catenin signaling pathway to enhance the glioblastoma sensitivity to temozolomide. Cell Death Discov. 7 (1), 288. doi:10.1038/s41420-021-00654-2
Liao, L. (2025). Inequality in breast cancer: global statistics from 2022 to 2050. Breast 79, 103851. doi:10.1016/j.breast.2024.103851
Losurdo, A., Di Muzio, A., Cianciotti, B. C., Dipasquale, A., Persico, P., Barigazzi, C., et al. (2024). T cell features in glioblastoma may guide therapeutic strategies to overcome microenvironment immunosuppression. Cancers (Basel) 16 (3), 603. doi:10.3390/cancers16030603
Louis, D. N., Perry, A., Wesseling, P., Brat, D. J., Cree, I. A., Figarella-Branger, D., et al. (2021). The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 23 (8), 1231–1251. doi:10.1093/neuonc/noab106
Ma, S., Wang, F., Wang, N., Jin, J., Ba, Y., Ji, H., et al. (2022). Multiomics data analysis and identification of immune-related prognostic signatures with potential implications in prognosis and immune checkpoint blockade therapy of glioblastoma. Front. Neurol. 20, 886913. doi:10.3389/fneur.2022.886913
Manoranjan, B., Chokshi, C., Venugopal, C., Subapanditha, M., Savage, N., Tatari, N., et al. (2020). A CD133-AKT-Wnt signaling axis drives glioblastoma brain tumor-initiating cells. Oncogene 39 (7), 1590–1599. doi:10.1038/s41388-019-1086-x
Manthey, C. L., Brandes, M. E., Perera, P. Y., and Vogel, S. N. (1992). Taxol increases steady-state levels of lipopolysaccharide-inducible genes and protein-tyrosine phosphorylation in murine macrophages. J. Immunol. 149 (7), 2459–2465. doi:10.4049/jimmunol.149.7.2459
Minea, R. O., Thein, T. Z., Yang, Z., Campan, M., Ward, P. M., Schönthal, A. H., et al. (2024). NEO212, temozolomide conjugated to NEO100, exerts superior therapeutic activity over temozolomide in preclinical chemoradiation models of glioblastoma. Neurooncol Adv. 6 (1), vdae095. doi:10.1093/noajnl/vdae095
Neff, C., Chavez, G., Proescholdt, C., Kruchko, C., Cioffi, G., Waite, K., et al. (2022). EPID-06. improvements in survival for glioblastoma in the post-stupp protocol era. Neuro Oncol. 24 (Suppl. ment_7), vii110. doi:10.1093/neuonc/noac209.416
Neve, R. M., Chin, K., Fridlyand, J., Yeh, J., Baehner, F. L., Fevr, T., et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10 (6), 515–527. doi:10.1016/j.ccr.2006.10.008
Nowak, B., Rogujski, P., Janowski, M., Lukomska, B., and Andrzejewska, A. (2021). Mesenchymal stem cells in glioblastoma therapy and progression: how one cell does it all. Biochimica Biophysica Acta (BBA) - Rev. Cancer 1876 (1), 188582. doi:10.1016/j.bbcan.2021.188582
Persano, F., Gigli, G., and Leporatti, S. (2022). Natural compounds as promising adjuvant agents in the treatment of gliomas. Int. J. Mol. Sci. 23 (6), 3360. doi:10.3390/ijms23063360
Rosińska, M., Dubiański, R., Konieczna, A., Poleszczuk, J., Pawlik, H., Nowecki, Z. I., et al. (2024). Retrospective observational study to determine the epidemiology and treatment patterns of patients with triple-negative breast cancer. Cancers (Basel) 16 (6), 1087. doi:10.3390/cancers16061087
Schwendener, S. (2011). Untersuchungen über den Flechtenthallus. In: Beiträge zur wissenschaftlichen Botanik. Ann Arbor, MI: University of Michigan. p. 127–195.
Sestito, S., Runfola, M., Tonelli, M., Chiellini, G., and Rapposelli, S. (2018). New multitarget approaches in the war against glioblastoma: a mini-perspective. Front. Pharmacol. 9, 874. doi:10.3389/fphar.2018.00874
Shahcheraghi, S. H., Tchokonte-Nana, V., Lotfi, M., Lotfi, M., Ghorbani, A., and Sadeghnia, H. R. (2020). Wnt/beta-catenin and PI3K/Akt/mTOR signaling pathways in glioblastoma: two main targets for drug design: a review. Curr. Pharm. Des. 26 (15), 1729–1741. doi:10.2174/1381612826666200131100630
Shcherbakova, A., Strömstedt, A. A., Göransson, U., Gnezdilov, O., Turanov, A., Boldbaatar, D., et al. (2021). Antimicrobial and antioxidant activity of Evernia prunastri extracts and their isolates. World J. Microbiol. Biotechnol. 37 (8), 129. doi:10.1007/s11274-021-03099-y
Shcherbakova, A., Nyugen, L., and Ulrich-Merzenich, G. (2019). The Lichen compounds evernic acid and usnic acid synergize with Temozolomide in the glioblastoma cellline U-87. Oncol. Res. Treat. 42 (S4), 275–276.
Singh, K., Han, C., Fleming, J. L., Becker, A. P., McElroy, J., Cui, T., et al. (2023). TRIB1 confers therapeutic resistance in GBM cells by activating the ERK and Akt pathways. Sci. Rep. 13 (1), 12424. doi:10.1038/s41598-023-32983-w
Smits, A., Jin, Z., Elsir, T., Pedder, H., Nistér, M., Alafuzoff, I., et al. (2012). GABA-A channel subunit expression in human glioma correlates with tumor histology and clinical outcome. PLoS One 7 (5), e37041. doi:10.1371/journal.pone.0037041
Stachyra, P., and Grzybowska-Szatkowska, L. (2025). Signaling pathways in gliomas. Genes (Basel) 16 (5), 600. doi:10.3390/genes16050600
Studzińska-Sroka, E., Majchrzak-Celińska, A., Zalewski, P., Szwajgier, D., Baranowska-Wójcik, E., Kaproń, B., et al. (2021). Lichen-derived compounds and extracts as biologically active substances with anticancer and neuroprotective properties. Pharmaceuticals 14 (12), 1293. doi:10.3390/ph14121293
Tan, A. C., Ashley, D. M., López, G. Y., Malinzak, M., Friedman, H. S., and Khasraw, M. (2020). Management of glioblastoma: state of the art and future directions. CA Cancer J. Clin. 70 (4), 299–312. doi:10.3322/caac.21613
Thomas, A., and Pommier, Y. (2019/06/21. 2019). Targeting topoisomerase I in the era of precision medicine. Clin. Cancer Res. 25 (22), 6581–6589. doi:10.1158/1078-0432.CCR-19-1089
Tong, Y. Q., Liu, B., Zheng, H. Y., Gu, J., Liu, H., Li, F., et al. (2015). MiR-215, an activator of the CTNNBIP1/β-catenin pathway, is a marker of poor prognosis in human glioma. Oncotarget 6 (28), 25024–25033. doi:10.18632/oncotarget.4622
Ulrich-Merzenich, G., Zeitler, H., Panek, D., Bokemeyer, D., and Vetter, H. (2007). Vitamin C promotes human endothelial cell growth via the ERK-signaling pathway. Eur. J. Nutr. 46 (2), 87–94. doi:10.1007/s00394-006-0636-5
Ulrich-Merzenich, G., Hartbrod, F., Kelber, O., Müller, J., Koptina, A., and Zeitler, H. (2017). Salicylate-based phytopharmaceuticals induce adaptive cytokine and chemokine network responses in human fibroblast cultures. Phytomedicine 34, 202–211. doi:10.1016/j.phymed.2017.08.002
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971). Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93 (9), 2325–2327. doi:10.1021/ja00738a045
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38 (Suppl. l_2), W214–W220. doi:10.1093/nar/gkq537
West, A., Tsui, V., Stylli, S., Nguyen, H., Morokoff, A., Kaye, A., et al. (2018). The role of interleukin-6-STAT3 signalling in glioblastoma. Oncol. Lett. 16, 4095–4104. doi:10.3892/ol.2018.9227
Widodo, S. S., Dinevska, M., Furst, L. M., Stylli, S. S., and Mantamadiotis, T. (2021). IL-10 in glioma. Br. J. Cancer 125 (11), 1466–1476. doi:10.1038/s41416-021-01515-6
Xie, Y., Bergström, T., Jiang, Y., Johansson, P., Marinescu, V. D., Lindberg, N., et al. (2015). The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine 2 (10), 1351–1363. doi:10.1016/j.ebiom.2015.08.026
Xiong, J., Zhou, L., Lim, Y., Yang, M., Zhu, Y. H., Li, Z. W., et al. (2013). Mature BDNF promotes the growth of glioma cells in vitro. Oncol. Rep. 30 (6), 2719–2724. doi:10.3892/or.2013.2746
Yakubov, R., Kaloti, R., Persaud, P., McCracken, A., Zadeh, G., and Bunda, S. (2025). It’s all downstream from here: RTK/Raf/MEK/ERK pathway resistance mechanisms in glioblastoma. J. Neurooncol 172 (2), 327–345. doi:10.1007/s11060-024-04930-w
Yu, J. M., Jun, E. S., Jung, J. S., Suh, S. Y., Han, J. Y., Kim, J. Y., et al. (2007). Role of Wnt5a in the proliferation of human glioblastoma cells. Cancer Lett. 257 (2), 172–181. doi:10.1016/j.canlet.2007.07.011
Zhang, W., and Liu, H. T. (2002). MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12 (1), 9–18. doi:10.1038/sj.cr.7290105
Zhang, M. I., Lucas, E., Rol, M. L., Carvalho, A. L., Basu, P., et al. (2023). CanScreen5, a global repository for breast, cervical and colorectal cancer screening programs. Nat. Med. 29 (5), 1135–1145. doi:10.1038/s41591-023-02315-6
Zhao, H., Wu, L., Yan, G., Chen, Y., Zhou, M., Wu, Y., et al. (2021). Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Target Ther. 6 (1), 263. doi:10.1038/s41392-021-00658-5
Zheng, B., and Chen, T. (2020). MiR-489-3p inhibits cell proliferation, migration, and invasion, and induces apoptosis, by targeting the BDNF-mediated PI3K/AKT pathway in glioblastoma. Open Life Sci. 15 (1), 274–283. doi:10.1515/biol-2020-0024
Zhong, Z., and Virshup, D. M. (2020). Wnt signaling and drug resistance in cancer. Mol. Pharmacol. 97 (2), 72–89. doi:10.1124/mol.119.117978
Keywords: lichens, glioblastoma, synergy, Wnt signaling, prediction, resistance
Citation: Shcherbakova A, Nguyen L, Koptina A, Backlund A, Banerjee S, Romanov E and Ulrich-Merzenich G (2025) Phytochemical combinations of lichen Evernia prunastri (L.) Ach. reduce drug resistance to temozolomide but not to paclitaxel in vitro. Front. Pharmacol. 16:1633978. doi: 10.3389/fphar.2025.1633978
Received: 23 May 2025; Accepted: 12 August 2025;
Published: 15 September 2025.
Edited by:
Xinyu Wang, Philadelphia College of Osteopathic Medicine (PCOM), United StatesReviewed by:
Ali Hafez El-Far, Damanhour University, EgyptMariana Magalhães, University of Coimbra, Portugal
Copyright © 2025 Shcherbakova, Nguyen, Koptina, Backlund, Banerjee, Romanov and Ulrich-Merzenich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: G. Ulrich-Merzenich, R3VkcnVuLnVscmljaC1tZXJ6ZW5pY2hAdWtib25uLmRl
†ORCID: Anna Koptina, orcid.org/0000-0003-2030-1604; Anders Backlund, orcid.org/0000-0003-2043-4183
 A. Shcherbakova
A. Shcherbakova L. Nguyen1,3
L. Nguyen1,3 S. Banerjee
S. Banerjee G. Ulrich-Merzenich
G. Ulrich-Merzenich