- 1Department of Molecular Cell Biology, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
- 2Institute of Biotechnology, Vietnam Academy of Science and Technology, Hanoi, Vietnam
- 3Department of Environment and Health, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
- 4Mientrung Institute for Scientific Research, Vietnam Academy of Science and Technology, Hanoi, Vietnam
- 5Department of Preventive Dentistry, Academic Centre for Dentistry Amsterdam (ACTA), University of Amsterdam and Vrije Universiteit Amsterdam, Amsterdam, Netherlands
- 6Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
- 7BioDetection Systems, Amsterdam, Netherlands
- 8Department of Ecological Science, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
We determined the degradation rates of the herbicides 2,4-D and 2,4,5-T by two different bacterial communities. One of these originated from soil heavily contaminated with herbicides from Bien Hoa airbase, the other from the same soil but amended with additional carbon and Gibbs energy sources. The community from the contaminated, but untreated, soil degraded both 2,4-D and 2,4,5-T within 5 days of cultivation. The one from the amended soil, however, hardly showed any degradation of the compounds throughout 23 days of cultivation. After refreshment of the medium and prolonged culturing, however, this community degraded both herbicides within 25 days with progressively increasing rates. nMDS analysis revealed a highly significant differentiation pattern of the two communities. Cultures inoculated with amended soil showed a significant increase of Bacillus and Paenibacillus upon prolonged exposure to the herbicides. The succession in the culture from untreated soil, on the other hand, was dominated by species from the Proteobacteria. We were able to isolate two of them and they were shown to be related to Bordetella petrii and Sphingomonas histidinilytica, successively. Subsequent PCR analyses of their DNA revealed the presence of key genes involved in the degradation of the herbicides. This study provides a more fundamental understanding of the biodegradation of 2,4-D and 2,4,5-T by displaying part of the bacterial community succession during their breakdown allowing a comprehensive view on potential key degraders.
Introduction
In the period 1961–1971 during the Vietnam War, the United States military army sprayed ~20 million gallons of herbicides over large areas in Vietnam, Cambodia, and Laos to defoliate trees and to destroy crops. The most widely used herbicide mixture consisted of the n-butyl esters of two phenoxyacetic acids termed 2,4-D and 2,4,5-T (Westing, 1984; Stellman et al., 2003). During the chemical production of these herbicides, trace amounts of dioxins were formed. Dioxins are persistent and have a great impact on human health even at very low concentrations (Van den Berg et al., 2006; Squadrone et al., 2015). Bien Hoa airbase in South Vietnam was an important airbase during the United States military operation called Operation Ranch Hand. This operation was part of the herbicidal warfare and Bien Hoa airbase served as a storage site for barrels containing the herbicides. Soils and sediments in the vicinity of Bien Hoa airbase were polluted with high concentrations of herbicides and dioxins due to large spills (Dwernychuk et al., 2002; Mai et al., 2007). The toxicity equivalent (TEQ) concentration of polychlorinated dibenzo-p-dioxins and polychlorinated dibenzofurans (PCDD/Fs) in soil and sediment varied from 7.6 to 962,000 and 17 to 4,860 ng/kg dry weight, respectively (Thuong et al., 2015). Since the end of Operation Ranch Hand in 1971, there has been an increasing concern about the human and environmental health effects associated with these spills. Therefore, actions have been taken to control the situation and to prevent further damage to human health and the environment.
One of the options to remove toxic substances from soils is via bioremediation using microorganisms, during which these compounds are degraded by dedicated microorganisms. The pollution of soil and water by herbicides is a worldwide problem, and bioremediation by microorganisms has always been a popular research topic because it is a clean, cheap, and relatively efficient process (Chatterjee et al., 1982; Golovleva et al., 1990; Oh and Tuovinen, 1990; Daubaras et al., 1996; Maltseva et al., 1996; Macur et al., 2007; Cycoń et al., 2011; Stibal et al., 2012; Han et al., 2015; Hayashi et al., 2016). Indeed, many reports indicated that bacterial strains isolated from different environments, such as Cupriavidus pinatubonensis JMP 134, Sphingomonas sp. TFD44, Achromobacter xylosoxidans subsp. denitrificans EST4002, Bradyrhizobium sp. HW13, Pseudomonas aeruginosa PAOlc, and Halomonas sp. EF43 are able to degrade 2,4-D aerobically (Filer and Harker, 1997; Laemmli et al., 2000; Kleinsteuber et al., 2001; Kitagawa et al., 2002; Vedler et al., 2004; Thiel et al., 2005). They have been mainly isolated from soils polluted with herbicides (Ka et al., 1994; Tonso et al., 1995). The characteristic feature of most of these bacteria is that they have a set of 6 so-called tfd genes which encode the enzymes for the sequential breakdown of 2,4-D into maleylacetate (Serbent et al., 2019; Pimviriyakul et al., 2020).
In contrast to the diversity of aerobic 2,4-D-degrading bacteria, only a few aerobic 2,4,5-T degraders have been reported. The best known are members of the genera Burkholderia (Kellogg et al., 1981; Daubaras et al., 1996; Huong et al., 2007), Nocardioides (Golovleva et al., 1990), Sphingomonas (Huong et al., 2007), and Bradyrhizobium (Rice et al., 2005; Huong et al., 2007; Hayashi et al., 2016), all of which were capable of utilizing 2,4,5-T as the major carbon and energy source. A characteristic enzyme involved in the corresponding degradation pathway is a Tft-type ring hydroxylating dioxygenase responsible for the initial removal of the acetate side chain to convert 2,4,5-T into 2,4,5 trichlorophenol (2,4,5-TCP) (Danganan et al., 1994). Its subunits, TftA and TftB, are key in the kyoto encyclopedia of genes and genomes (KEGG) pathway “chlorocyclohexane and chlorobenzene degradation” including the pathways for 2,4-D and 2,4,5-T degradation (Kanehisa and Goto, 2000). Well-studied homologs of TftA/TftB are HadA/HadX, which catalyze dehalogenation of halogenated phenol derivatives (Pimviriyakul and Chaiyen, 2018), and TcpA/Fre, which dehalogenate dihalophenols (Fang et al., 2020). They belong to a larger family of monooxygenases including PcpA/PcpB, CphC/CphB, and DcmB1/DcmB2 (Pimviriyakul et al., 2020).
Importantly, the degradation pathways via TftA and TftB work only under aerobic conditions as they involve mono- and dioxygenases. Anaerobic degradation of 2,4-D and 2,4,5-T and other chlorinated aromatic compounds involves many types of reductive dehalogenases such as 2,5-dichlorohydroquinone reductive dehalogenase (Miyauchi et al., 1998), and chlorohydroquinone dehalogenase (Arora and Jain, 2012; Arora et al., 2014), but knowledge about the processes is scarce (Ghattas et al., 2017; Al-Fathi et al., 2019). Yet, one has to be aware of the fact that major portions of environmental pollutants flow to anaerobic zones. There, the degradation of the herbicides may well be less efficient resulting in more persistency of them under anoxic conditions (Albrechtsen et al., 2001; Shareef et al., 2014; Al-Fathi et al., 2019).
As part of the cleaning approach at Bien Hoa airbase in Vietnam, an active landfill contaminated with herbicides and dioxins was biotreated in four bioremediation cells for 40 months (Nguyen and Dang, 2012). The details of the biotreatments are described in the supplement of a related study (Lechner et al., 2018). In short, the landfill consisted of four cells containing 3,384 m3 of herbicide and dioxin contaminated soil. The soil contaminated with polychlorinated dibenzo-p-dioxins and herbicides were well mixed with different agents such as rice straw, hulls, biosurfactants, nutrients, electron acceptors, electron donors and vitamins and was periodically supplied with water (moisture range from 16 to 24%). The 2 m thick soil layer was sealed above and below with four layers of bentonite and geological protection material and 50 cm of non-contaminated soil as the top layer, which was planted with grass. The result of that bioremediation study showed that the average total toxicity equivalence from all four cells decreased from around 10,000 to 14 ng TEQ/kg dry weight (Dang, 2013). Lechner et al. further demonstrated that enrichment cultures obtained after cultivation of soil from these bioremediation cells in medium with 2,4,5-T and pyruvate and lactate degraded 2,4,5-T via 2,4,5-trichlorophenol (TCP) to 3,4-dichlorophenol in the presence of pyruvate as carbon and Gibbs energy donor (Lechner et al., 2018). A follow-up study aimed at displaying the effect of different amendments to mesocosms filled with soil contaminated with herbicides collected from Bien Hoa airbase on the degradation rate of 2,4-D and 2,4,5-T. The average 2,4-D concentration at the start of that study was about 1,500 mg/kg soil Preliminary data indicated that soils amended with additional carbon and energy sources performed worse with respect to the degradation of the herbicides as compared to soils left untouched. More specifically, microbial communities from T1 soil degraded 97% of 2,4-D and 95% of 2,4,5-T during 38 months of incubation, while only 20% of 2,4-D and 17% of 2,4,5-T was degraded by the microbial communities from T4A soil (Thi Lan Anh Nguyen, unpublished data).
In this study, we aimed at getting a more fundamental understanding of the processes involved in the degradation of 2,4-D and 2,4,5-T, and at identifying the key microorganisms responsible for the breakdown of them. The specific approach was to enrich bacteria from soils from two of the mesocosms mentioned above under defined conditions with 2,4-D and 2,4,5-T as the major carbon and energy sources, and to monitor the degradation of these herbicides over time. We performed two independent experiments to correlate the resulting degradation kinetics to changes in community structures. In the first experiment, the herbicides were added to enrichment cultures in powder form. In the second experiment, the herbicides were added either in powder form or dissolved in acetone to study if dissolving the herbicides prior to the addition would stimulate their breakdown. Altogether, we obtained a more fundamental view of 2,4-D and 2,4,5-T degradation and the role of key species in that process. Most importantly, we show that initial starvation of bacterial communities in soils not amended with additional carbon and energy sources on the one hand and enrichment of these communities via pre-exposure to the herbicides, on the other hand, are key to a better performance of cultures degrading herbicides in particular and perhaps of xenobiotics in general.
Materials and Methods
Soil Sampling and Culturing Conditions
This study describes two experiments to deepen our insight in the biodegradation of 2,4-D and 2,4,5-T. In the first experiment, we used two different herbicide-contaminated soils from the Bien Hoa Airbase, one that had been amended with organic materials and the other left unamended. In the second experiment, we used the same unamended soil but with herbicides added in two different forms, in powder or dissolved in acetone. The soil amendments were carried out in a previous study. In that previous study, herbicide and dioxin-contaminated soil was collected from Bien Hoa airbase (10°58′14.3″ N 106°48′19.3″ E), Dong Nai Province, Vietnam (Thi Cam Ha Dang, unpublished data). For that, five holes were dug 2.5–3 m deep from the surface by an excavator. Samples were taken at about 20–25 different positions of the piles of soil from each hole and well mixed on-site, transferred to Hanoi, and then divided over 6 containers, each with a volume of 200 L and filled with 140 kg of soil. The containers were periodically supplied with water to keep the moisture about 12–20%. The soils had an average dioxin toxicity of 21.605 ngTEQ/kg and 2,4-D and 2,4,5-T concentrations of 1,500 and 1,200 mg/kg soil, respectively. These soils were used for a bioremediation experiment during 38 months. One container, called T1, was left untreated for 38 months. The other five were biotreated for the same period each with a different design. The soil of one of these, called T4A, was amended with an inoculum of soil from Bien Hoa airbase after in situ bioremediation, 20% of agricultural waste (rice straw and rice husk) and compost from cattle manure and corn stalks, 17% of rice bran and soybean powder, along with 46% of a mixture of inorganic salts (NH4NO3, NH4Cl, K2HPO4, KH2PO4, Ca3(PO4)2 and MgSO4) and trace elements, 5% of plant surfactants and a mixture of vitamins (B12, B1, C), and 12% of organic acids and salts. The above components were supplied at the start and after 6, 21, 30 and 38 months of bioremediation (Thi Cam Ha Dang, unpublished data). In the study described here, we used the 38 months old soil from containers T1 and T4A to culture and enrich bacterial communities in order to correlate the potential of the communities to degrade the herbicides to their community compositions. The carbon and nitrogen concentrations in T1 and T4A soil were measured using a FlashEA 1112 (Thermo Scientific), with EAGER300 operating software and a thermoconductive detector (Aerts et al., 2019). T1 soil contained 0.43% carbon and 0.05% nitrogen, T4A soil contained 2.14% carbon and 0.31% nitrogen, which values are higher than in T1 soil as a result of the amendments. For all enrichment studies, we inoculated soil samples of 10 grams in 250 ml Erlenmeyers filled with basal salt medium (BS, 40 ml per culture) containing KH2PO4 0.5 g/l, (NH4)2SO4 0.25 g/l, MgSO4 0.2 g/l, CaCl2 0.5 g/l and NaNO3 0.4 g/l, pH 7.0 (van der Zaan et al., 2012). Compounds 2,4-D (>95% purity) and 2,4,5-T (>95% purity) were purchased from Sigma. In the first experiment, they were provided in powder form at final concentrations of 100 mg/l 2,4-D and 100 mg/l 2,4,5-T in cultures with soil from either the T1 or T4A containers that were incubated in the dark on a rotary shaker at 30°C and 200 rpm (1st enrichment). After a given cultivation period, 10 ml of these cultures were then used to inoculate 90 ml fresh BS medium with final concentrations of 200 mg/l 2,4-D and 100 mg/l 2,4,5-T and further incubated by shaking at 200 rpm and 30°C (2nd enrichment). In a second experiment, the herbicides were added either in powder form or dissolved in acetone at final concentrations of 100 mg/l 2,4-D and 100 mg/l 2,4,5-T in cultures with soil from the T1 container that were incubated in the dark on a rotary shaker at 30°C and 200 rpm (1st enrichment). Obtaining the 2nd, 3rd and 2nd spike enrichments was done essentially as described in the first experiment but again with the herbicides added either in powder form or dissolved in acetone at final concentrations of 200 mg/l 2,4-D and 100 mg/l 2,4,5-T. The number of cultivation days for each specific experiment is mentioned in the results section. From the initial carbon concentrations in T1 and T4A soils, we calculated that the residual carbon concentrations in the 1st, 2nd, and 3rd enrichments of T1 soil were 1075, 107.5, and 10.75 mg/L, respectively, and those of T4A soil in the 1st and 2nd enrichments 5,350 and 535 mg/L, respectively. All experiments were performed in triplicate. The control flask for non-biological degradation of the herbicides contained T1 soil that was sterilized by heating it at 121°C during 15 mins. A diagram with the set up of the mesocosms and the two enrichment experiments described in this study is shown in Supplementary Figure S1.
Rationale for Random Sampling vs. Destructive Sampling
We aimed to study the succession over time of bacterial cultures growing on 2,4-D and 2,4,5-T as the major carbon and Gibbs energy sources and to correlate community structures and their changes to their corresponding degradation capacities. We rationalized that it should be best if samples were taken over time from the same bottle rather than via destructive sampling of parallel cultures in time. Therefore, we investigated if the herbicides would be homogenously distributed during random sampling. For that, 2 ml of homogenate from flasks containing 10 ml of BS medium and 200 mg/l each of 2,4-D and 2,4,5-T were taken for herbicide extraction. For destructive sampling, three vials that had 2 ml of BS medium and 200 mg/l of each 2,4-D and 2,4,5-T were extracted using the QuEChERS method. The results showed no significant differences between either of the two sampling methods (Supplementary Figure S2). Therefore, we used random sampling from the same cultures to study the biological succession of their bacterial communities.
Determination of 2,4-D and 2,4,5-T Concentrations by Tandem Liquid Chromatography-Mass Spectrometry (LC-MS/MS)
To track the decline of herbicide concentrations over time, herbicides were extracted using the Quick, Easy, Cheap, Effective, Rugged, and Safe (QuEChERS) method (Rejczak and Tuzimski, 2015). At various time points, aliquots of the cultures (2 ml) were transferred to 15-ml Falcon tubes. Next, 2 ml of acetonitrile with 1% formic acid was added and vortexed for 15 min. A mixture of salts (2 g of MgSO4, 0.5 g of NaCl, 0.5 g of C6H5NaO7·2H2O, and 0.25 g of C6H6Na2O7·1.5 H2O) was added to the homogenate and mixed for 5 min, followed by centrifugation for 5 min at 12,000 × g. Then, 2 ml of the supernatant was collected for the LC-MS/MS analysis. The amount of residual 2,4-D and 2,4,5-T in the culture medium at different time points was determined by LC-MS/MS using an Elute ultra-high-pressure liquid chromatography (UHPLC) system coupled to an EVOQ triple quadrupole mass selective detector (TQ-MS; both Bruker, Bremen, Germany). The target compounds were separated on a 50 × 2 mm, 3 μm Luna C18 column (Phenomenex, Utrecht, The Netherlands) at a temperature of 40°C, applying a gradient of 0.1% formic acid and methanol (both Biosolve, Valkenswaard, The Netherlands). The gradient started with 5% methanol, increased in 2 mins to 30% followed by an elevation to 100% in 4 mins with a constant flow of 400 μl/min. The TQ-MS was set at electrospray ionization in a negative ion mode. The transitions for 2,4-D were 221 > 163 and 219 > 161 (quantifier/qualifier). For 2,4,5-T the transitions were 253 > 195 and 255 > 197 (quantifier/qualifier). All the transitions were scanned with a rate of 300 ms. Before analysis, samples were diluted in de-ionized water and stored at 4°C until injection. Data acquisition and analyses were performed with MS Data review software. Data analyses of the biodegradation tests were performed using Prism version 8.2.1.
Detection of 2,4-D and 2,4,5-T-Degrading Genes by PCR
Primers for the PCR amplification of tfdA, tfdB, tfdC, and tftA genes are listed in Table 1. The PCR amplification conditions were as follows: 3 min at 94°C, 30 cycles with 60 s at 94°C, 60 s at 53°C, 60 s at 72°C, and final elongation for 6 min at 72°C. All PCR products were separated on 1.5% agarose gel, stained with Roti®-Gelstain, visualized under UV light, and photographed. DNA of the PCR fragments was sequenced by Macrogen Europe.
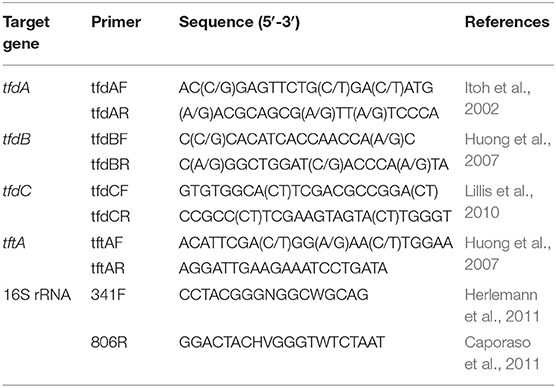
Table 1. List of primers used for detection of genes involving in break-down of 2,4-D and 2,4,5-T and the V3-V4 region of bacterial 16S rRNA genes.
Community Profiling Using Illumina Sequencing of the 16S rRNA Gene Amplicon
DNA of enrichment samples was extracted using the MoBio PowerSoil® DNA Isolation kit (Carlsbad, CA, USA). The quality and quantity of the extracted DNA were checked by agarose gel electrophoresis and Qubit dsDNA HS Assay kit (ThermoFischer Scientific, cat. no. Q32851).
The primers used to target the V3–V4 region of bacterial 16S rRNA genes are listed in Table 1. These amplicon primers contained Illumina adapters and an 8-nucleotide index barcode sequence (Kozich et al., 2013). The PCR system and amplification conditions were similar to those described by Mao et al. (2015). The composition of a PCR reaction was as follows: 5 μM primers, 5x Phusion HF Green Buffer, 10 mM dNTP mix, 2 U/μL Phusion II HS HF Polymerase, and 1.5 ng DNA template. The PCR program with Veriti Thermal Cycler (Thermo Fisher Scientific) consisted of the following steps: initial denaturation at 98°C for 30 s, 25 cycles at 98°C for 30 s, 55°C for 30 s and 72°C for 30 s, and a final extension at 72°C for 5 min. The PCR products were purified with Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) on a BILATEST magnetic separator plate (Sigma Aldrich, cat. no.: Z662429-1EA). Subsequently, purified PCR products were visualized using gel electrophoresis and quantified with a Qubit dsDNA HS Assay Kit on a Qubit 2.0 fluorometer (Thermo Fisher Scientific). The libraries were sequenced on the Illumina MiSeq platform (Illumina Inc; San Diego, CA, USA) with 150 cycles for forward and reverse reads.
Sequence Processing and Analyses
The sequencing data reads were processed as described previously (Persoon et al., 2017). In short, the reads were merged (Edgar and Flyvbjerg, 2015), filtered, and clustered into operational taxonomic units (OTUs) in line with the UPARSE method using USEARCH version 8.0.1623 (Edgar, 2013). Sequences to be mapped to the cluster centroids were filtered on a maximum error rate of 0.005, while no ambiguous bases were allowed. QIIME version 1.8.0 (Caporaso et al., 2010) was used to select the most abundant sequence of each OTU, the taxonomy of which was assigned using the RDP classifier (Wang et al., 2007) with a minimum confidence of 0.8 and the 97% representative sequence set (V3–V4 region only, made non-redundant [cf. Koopman et al., 2016] based on the SILVA rRNA database v132 (Quast et al., 2013). For the taxonomic summaries of the bacterial communities at phylum and genus levels, the counts in the OTU table were converted to relative abundances. Before the alpha- and beta-diversity analyses (including nMDS), the OTU table was standardized using the decostand function (method = “hellinger”) (Steinert et al., 2017, 2019; Helber et al., 2019) of the R package vegan v.2.5-6 (Oksanen et al., 2016) in R v3.3.1 (R Core Team, 2016). The alpha diversity indices, species richness (S, number of OTUs/sample), and Shannon index (H) were calculated in R using vegan. The difference in alpha diversity indices between two groups (i.e., spikes, soils, and enrichments of soils) was analyzed with the Wilcoxon rank-sum test using function wilcox.test, whereas the difference in alpha diversity indices among enrichments of each spike was analyzed with the Kruskal–Wallis rank-sum test using the function kruskal.test followed by Dunn's Kruskal-Wallis multiple comparisons test using the function dunnTest within the package FSA v.0.8.30 (Ogle, 2017) with the Benjamini-Hochberg (BH) p value correction for multiple testing (Benjamini and Hochberg, 1995). The non-metric multidimensional scaling (nMDS) plot was created via the function metaMDS (Bray–Curtis distances). Confidence ellipses in nMDS plots were shown at a 0.95 level using the function ordiellipse (kind “sd”) of the vegan package. Multivariate analysis based on Bray–Curtis dissimilarities (1,000 permutations) of bacterial communities based on soils, spikes, enrichments, followed by pairwise comparisons of enrichments were performed using the functions betadisper, permutest (dispersion), adonis (PERMANOVA) from vegan, and the function pairwise.perm.manova of the package RVAideMemoire v.0.9-77 (Hervé, 2020) with the BH p-value correction for multiple testing.
The OTU differential abundance (log2-fold change in abundance of each OTU) between two groups (i.e., between two culture periods of enrichment was calculated using the R package phyloseq v.1.32.0 (McMurdie and Holmes, 2013) and “DESeq2” v1.28.1 (Love et al., 2014). For this analysis, the original OTU table was normalized internally with DESeq2. The p values were adjusted with the BH correction method and an OTU was considered as differentially abundant if its mean proportion was significantly different between sample classes (p < 0.05). Only OTUs with a relative abundance of more than 2% in total were selected for these analyses.
Data Accessibility
The raw Illumina sequencing data were deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession number PRJNA596903. The link to this data, initially for review only, is https://submit.ncbi.nlm.nih.gov/subs/sra/SUB6464612/overview. After acceptance of the manuscript, the data will become publicly available along with an assigned DOI number. The 6 sequenced gene fragments have GenBank accession numbers from MT149894 to MT149899.
Results
Biodegradation of 2,4-D and 2,4,5-T by Bacterial Soil Communities Originating From Bien Hoa Airbase
Bacterial communities enriched from untreated soil were more potent in the degradation of 2,4-D and 2,4,5-T than those enriched from biotreated soil. We show that in the first set of experiments, during which we studied the degradation of the herbicides over time in cultures inoculated with soil from either T1 (untreated control) or T4A (biotreated) mesocosms. The results of that degradation experiment are shown in Figure 1. It should be noted that the initial concentrations of the herbicides in the T4A culture were higher than those of the control and the T1 culture due to residual 2,4-D and 2,4,5-T in the T4A soil. The flask inoculated with T1 soil degraded both herbicides within 5 days of cultivation. This is in contrast to the culture with T4A soil that hardly showed degradation throughout 23 days of cultivation (Figures 1A,B). When these cultures were refreshed with new medium and herbicides (see “Soil Sampling and Culturing Conditions” in Materials and Methods section), we noticed that the T1 cultures were still able to degrade the herbicides within 5 days. Now, also the T4A cultures showed degradation of both herbicides in this 2nd enrichment after 25 days of cultivation (Figures 1C,D). The data further show an apparent increase in the levels of the herbicides in T4A cultures (Figures 1C,D). We hypothesize that 2,4,5-T is slowly released from soil particles resulting in enhanced concentrations in solution. The levels of the herbicides stayed constant in the sterile controls. We also determined the number of colony forming units (CFUs) over time in both enrichments to get an impression of the biomass over time (Supplementary Figure S3).
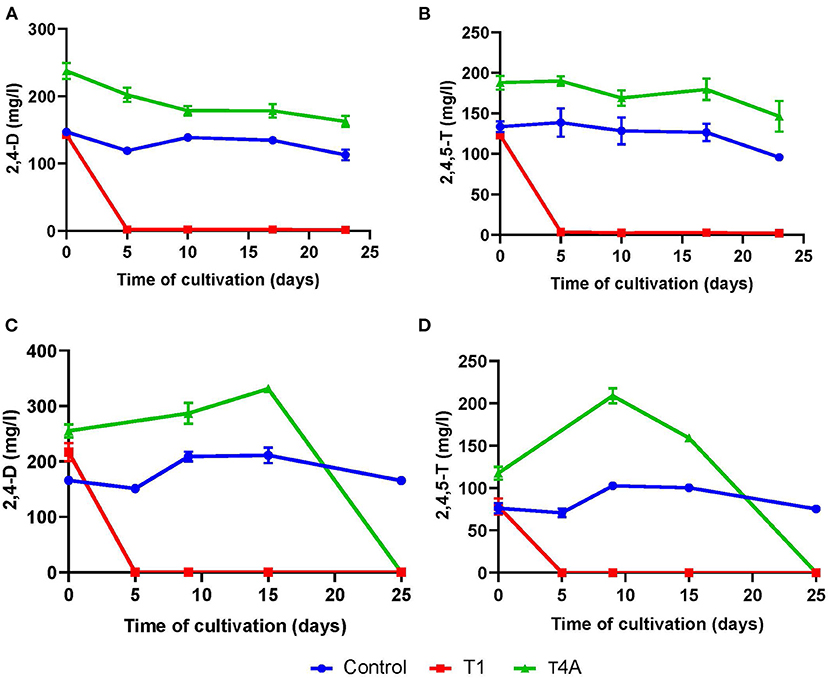
Figure 1. Degradation of 2,4-D (A,C) and 2,4,5-T (B,D) in cultures inoculated with T1 (red curves) and T4A (green curves) soil of the 1st enrichment and the 2nd enrichment (C,D) in time. The blue curves are data from the sterilized control flask. Each data point is the average of three independent measurements, and the error bars are their corresponding standard deviations.
Community Dynamics of Herbicide Degrading Cultures Enriched From T1 and T4A Soil
This paragraph shows that the taxonomic diversity and succession of the bacterial communities in the enrichment cultures of T1 soil and T4A soil described above were significantly different. To obtain the taxonomic data of these bacterial communities, samples were taken at different time points for DNA extraction and subsequent community profiling via the Illumina MiSeq platform. Taxonomic analysis of the normalized (relative abundance) OTU tables yielded a total of eleven classifiable phyla (Supplementary Figure S4A). Three of these were dominant in all samples of communities enriched from T1 soil, namely Proteobacteria (63–74%), Actinobacteria (6.4–28%), and Bacteroidetes (3.0–19%). Bacterial consortia developed from T4A soil on the other hand contained a high percentage of Firmicutes (57–99.9%). Species from three genera, Sphingomonas, Glutamicibacter, and Pseudomonas, became dominant after 5 days of cultivation with relative abundances of 31, 27, and 7%, respectively. The relative abundance of species from some other genera, Castellaniella, Proteiniphilum, and from the family Microscillaceae, on the other hand, decreased in that period (Supplementary Figure S4B). Enrichments from T4A soil showed a significant increase of species from the genera Bacillus, Symbiobacterium, and Paenibacillus upon prolonged exposure to the herbicides. Representatives of the Atopostipes, on the other hand, were significantly decreased in their relative abundance (Supplementary Figure S4B). In the 2nd enrichment of all cultures enriched from T1 soil, members of three phyla, Proteobacteria (87–93%), Actinobacteria (1.3–14%), and Bacteroidetes (2–12.8%), were dominant (Supplementary Figure S5A). These included 9 relatively abundant genera amongst which Sphingomonas (17.7–59%), Luteimonas (4.4–22.5%), Proteiniphilum (0.7–6.4%), Rhodanobacter (0.6–14.6%), Bordetella (1–4%), Glutamicibacter (0.7–11%), Sphingopyxis (2.6–10%), Acinetobacter (0.2–2.6%) and Achromobacter (1.3–11.1%). Members of the genus Sphingopyxis were present at relatively high abundances at all time points of cultivation. The relative abundances of some other genera, however, changed over time. Those of the genera Sphingomonas and Pseudomonas increased during 25 days of cultivation. In contrast, the relative abundances of members of the genera Luteimonas (4.4–22.5%), Proteiniphilum and Acinetobacter decreased over time (Supplementary Figure S5B). The genera that were dominant in the enrichments of T1 soil were hardly present in those of T4A soil. In these cultures, members of the phylum Firmicutes were present at relatively high abundances (>99%). The most dominant genera after 25 days of cultivation were Paenibacillus (49.7%), Symbiobacterium (17.0%). The relative abundance of genera belonging to the Clostridiales, on the other hand, decreased in all cultures during 25 days of cultivation (Supplementary Figure S5B).
The OTU tables were further analyzed for alpha diversity indices (Supplementary Figure S6), both the OTU richness index (left panels of each set) and the Shannon index (right panels of each set), in order to determine the richness and diversity of species within these cultures. The results show that the alpha diversity indices of T1 soil were significantly higher than those of the T4A bacterial communities (Supplementary Figure S6A).
OTU richness and Shannon indices of the soil T1 were higher significant than those of the soil T4A. The bacterial diversity was statistically different as judged by the OTU richness index in the 1st and 2nd enrichment of T1 soil communities. In contrast, the results of the Shannon index did not reach the level of significance in either of the enrichment communities (Supplementary Figure S6B). We also compared the microbial species richness of the 1st and 2nd enrichment originating from T4A soil, as estimated by the OTU richness index and Shannon index, and the results show that they are not significantly different (Supplementary Figure S6C).
The nMDS analysis revealed different profiles of bacterial communities between cultures enriched from T1 and T4A soils, which was significant as judged by permutational multivariate analysis of variance (PERMANOVA p < 0.001) (Supplementary Figure S7A). However, it should be noted that the multivariate homogeneity of group dispersions also indicated a significant difference between the T1 and T4A cultures (dispersion p < 0.01). Such dispersion of bacterial communities may well be explained by the fact that the bacterial communities were sampled and analyzed at different time points after the start of the experiment. The number of samples taken and analyzed at each time point were only two, which is not enough for statistics. Therefore, we do not display them in Supplementary Figure S7 and do not mention the effects of the sampling time on the bacterial communities structures. T1 cultures displayed significantly different bacterial community profiles between the 1st and 2nd enrichments, whereas they were homogeneous within each of the enrichments (dispersion p > 0.05 and PERMANOVA p < 0.001) (Supplementary Figure S7B). In the case of the T4A cultures, we noticed significant differences of bacterial community profiles between 1st and 2nd enrichments by both permutational multivariate analysis of variance (PERMANOVA p < 0.01) and the multivariate homogeneity of groups dispersions (dispersion p < 0.01) (Supplementary Figure S7C).
The different taxa whose relative abundances significantly changed over time during the cultivation are shown in Figure 2. The 1st enrichment of T1 soil showed 3 OTUs (genera Castellaniella, Proteiniphilum, and a member of the Microscillaceae) with decreased and 6 OTUs (genera Sphingomonas, Sphingobium, Achromobacter, Pseudomonas, Glutamicibacter, and Novosphingobium) with increased relative abundances over the time of cultivation. The latter six OTUs had a low relative abundance in the original inoculum, but they became more dominant during growth in the 1st enrichment cultures with 2,4-D and 2,4,5-T as the major carbon and energy sources. In the 2nd enrichment, 6 OTUs that belong to genera Sphingomonas, Pseudomonas, Bordetella, Achromobacter, Sphingobium, and Novosphingobium were significantly enriched after 5 days of cultivation (Figure 2C).
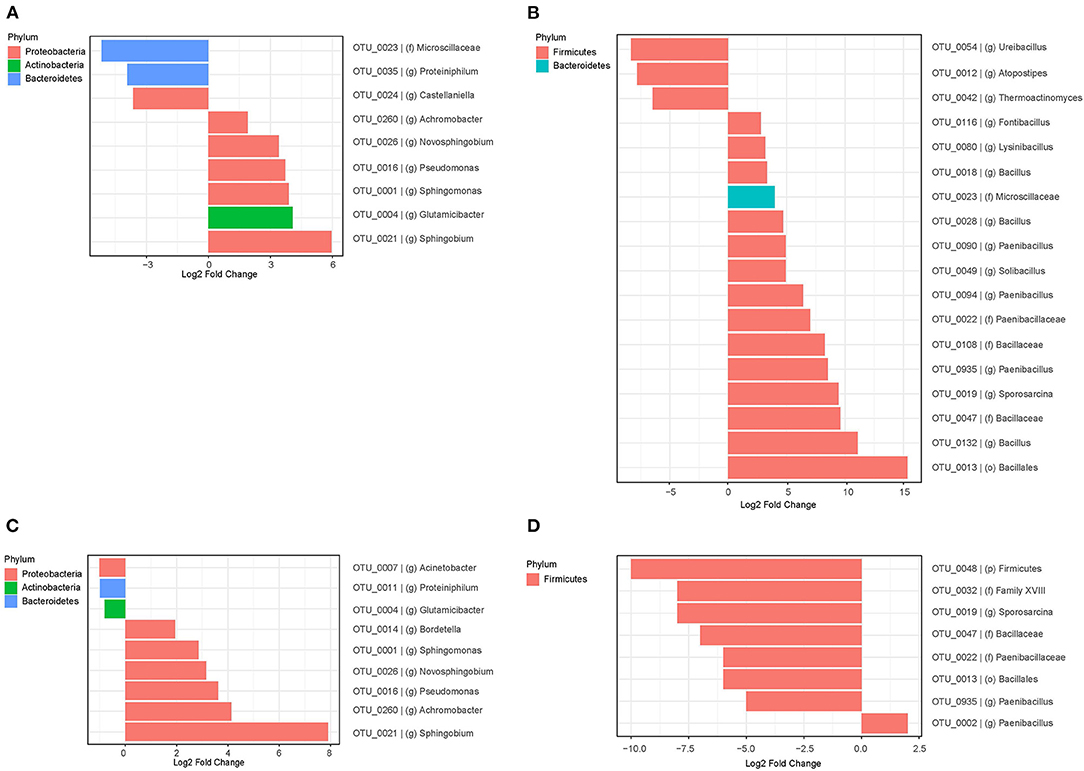
Figure 2. Differential analysis of OTUs whose relative abundance changed significantly after 5 days of cultivation as compared to the start of the experiment. (A,B) Show the results from bacterial communities in the 1st enrichments of T1 and T4A soil, respectively, and (C,D) those from bacterial communities in the 2nd enrichments of T1 and T4A soil, respectively. The phyla of the respective OTUs are indicated at the upper left of each figure with the corresponding colors.
The 1st 5 days of enrichment of T4A soil resulted in decreased relative abundances of species from the genera Atopostipes, Ureibacillus, and Thermoactinomyces all 3 of which are from the Firmicutes (Figure 2B). Other genera such as Bacillus, Paenibacillus, Solibacillus, Lysinibacillus and Fontibacillus from the Firmicutes and Bacteroidetes became relatively more abundant.
Relation Between Bioavailability and Degradation Kinetics of 2,4-D and 2,4,5-T by T1 Communities
From the second set of experiments, we concluded that the breakdown of 2,4-D and 2,4,5-T improved when they were dissolved in acetone prior to addition to the enrichment cultures. To show this, we supplied the herbicides either in powder form (P) or dissolved in acetone (S), with the assumption that in the latter forms the herbicides should be more bioavailable. We decided to use the 38 months old T1 soil as inoculum as this soil showed the highest capacity to degrade the herbicides in the initial experiments. The resulting cultures degraded both herbicides within 25 days of cultivation during the 1st enrichment (Figures 3A,B), and possibly faster than that, but here we were interested in the enrichment of degrading species.
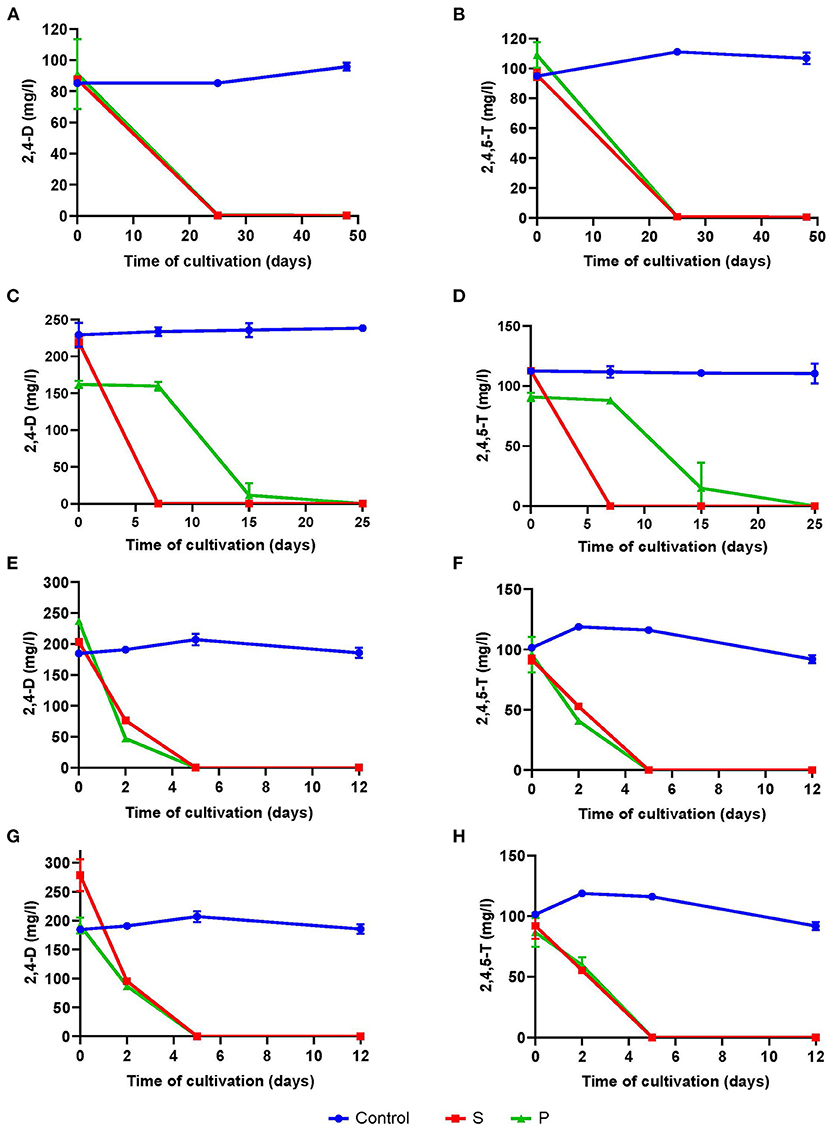
Figure 3. Degradation of 2,4-D (A,C,E,G) and 2,4,5-T (B,D,F,H) in mineral salts medium inoculated with T1 soil in the 1st enrichment (A,B), degradation profiles of the 2nd enrichment (C,D), of the 3rd enrichment (E,F), and of the 2nd enrichment after addition of an extra spike of the herbicides (G,H) during a 12-day cultivation period. Each point and shape represent the average of three independent measurements, and the error bars are their standard deviations. Blue circles, control with sterilized soil (see “Soil Sampling and Culturing Conditions” in Materials and methods section); green triangles, herbicides in powder form; red squares, herbicides dissolved in acetone.
For the 2nd enrichment culture, we used a 10% volume from these 25 days-old cultures to inoculate fresh media with the herbicides and monitored the kinetics of their degradation. Both herbicides were fully removed within 7 days of culturing when they had been dissolved in acetone, but it took around 15 days when they were added in powder form (Figures 3C,D). When these 2nd enrichments had again been refreshed with new medium and herbicides, the rates of 2,4-D and 2,4,5-T degradation in this 3rd enrichment were similar regardless of whether they were supplied in powder form or dissolved in acetone. For both compounds, more than 60% was consumed within the first 2 days of culturing and complete removal was apparent within 5 days (Figures 3E,F). We also determined the degradation kinetics in the 2nd enrichment after the herbicides had been consumed and spiked with additional herbicides (2nd spike). The results are comparable with those from the 3rd enrichment (Figures 3G,H). Again, we determined the number of CFUs over time in all enrichments and the spike experiment to get an impression of biomass formation (Supplementary Figure S8).
Community Structures and Dynamics of Enrichment Cultures From T1 Soil
The nMDS analysis revealed that the addition of the herbicides in powder form or dissolved in acetone to the cultures resulted in somewhat different bacterial communities in the second enrichments (Figure 4A). Furthermore, we also observed significant changes in the bacterial consortia during successive enrichments of the cultures (Figures 4B,C). Pairwise comparisons between the enrichments after the spike P (herbicides in powder form) showed that the bacterial consortia of all pairwise comparisons of the enrichments were significantly different (Supplementary Table S1). Pairwise comparisons between the enrichments after the spike S (herbicides dissolved in acetone) showed that the bacterial consortia of almost pairwise comparisons of the enrichments were significantly different except for the pairwise comparison of the 2nd spike (2nd Sp) and 3rd enrichments, which had similar bacterial consortia (Supplementary Table S1).
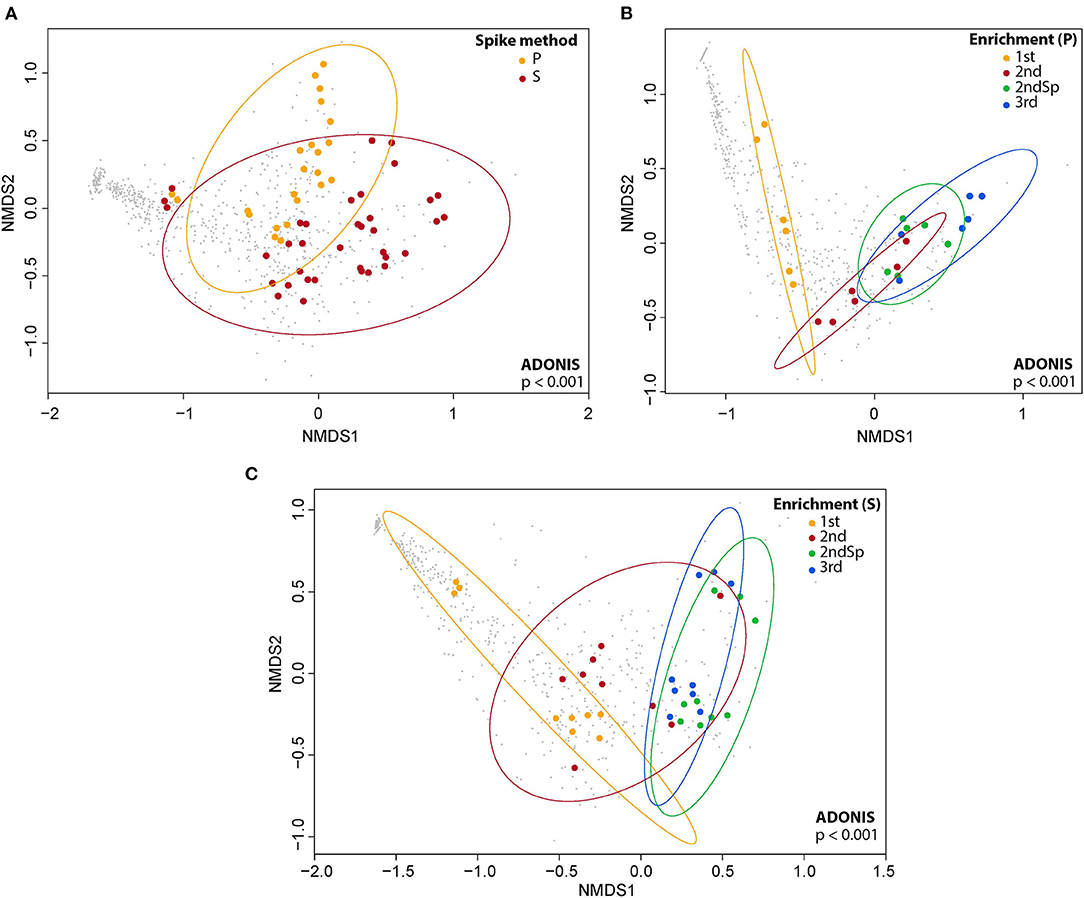
Figure 4. nMDS ordination of the Bray-Curtis dissimilarity between soil bacterial communities of cultures where the herbicides were added in powder form or dissolved in acetone (A), between soil bacterial communities of different enrichments where both 2,4-D and 2,4,5-T were added in powder form (B) and between soil bacterial communities of different enrichments where 2,4-D and 2,4,5-T were dissolved in acetone (C). P, herbicides in powder form; S, herbicides dissolved in acetone. Taxonomic variables (OTUs) have been added as gray dots to the background of each ordination using function argument “points,” display = “species” of the function “metaMDS” in R.
The alpha diversity indices, Richness indices (left panels of each set in Supplementary Figure S9), and Shannon indices (right panels of each set in Supplementary Figure S9) were determined during the enrichments. The result shows that these indices do not differ significantly over time depending on whether the herbicides had been added in powder form or dissolved in acetone (Supplementary Figure S9A). We also compared the bacterial species richness of the 1st, 2nd, 2nd spike, and 3rd enrichment cultures with herbicides supplied in either powder form or dissolved in acetone, as estimated by the Richness index and the Shannon index. These results indicated that the diversity quickly decreased over time in each enrichment regardless of the way the herbicides were added (Supplementary Figures S9B,C).
The bacterial genera from the enrichments of T1 soil grouped into 13 phyla, the most dominant of which are Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes (Supplementary Figures S10–S13). The profiles of the abundant phyla in the 1st enrichment were comparable regardless of whether the herbicides were added in powder form or dissolved in acetone (Supplementary Figure S10A), but they diverged in the 2nd, 3rd, and 2nd spike enrichments (Supplementary Figures S11A, S12A, S13A). The succession of the communities in the 3rd enrichment and in the 2nd enrichment with the additional spike was relatively comparable. Cultures, where the herbicides were dissolved in acetone, showed an increase in members of the Bacteroidetes and a decrease of those of the Proteobacteria. This is in complete contrast to the subsequent enrichments where the herbicides had been supplied in powder form. Here, we observed an increase in the Proteobacteria and a decrease in Bacteroidetes (Supplementary Figures S11A, S12A, S13A).
The 5 most abundant genera in the 1st enrichment where 2,4-D and 2,4,5-T had been spiked in powder form were Sphingomonas, Proteiniphilum, Luteimonas, Sphingobium, and Sphingopyxis. Most of the corresponding OTUs had a low relative abundance in the original inoculum, but they became increasingly abundant after 25 days of cultivation (Figure 5A). All of these had a significantly differential relative abundance increase over the time of cultivation. The relative abundance of members of the genera Sphingomonas and Pseudomonas further increased over time in the next enrichments (Supplementary Figure S14A).
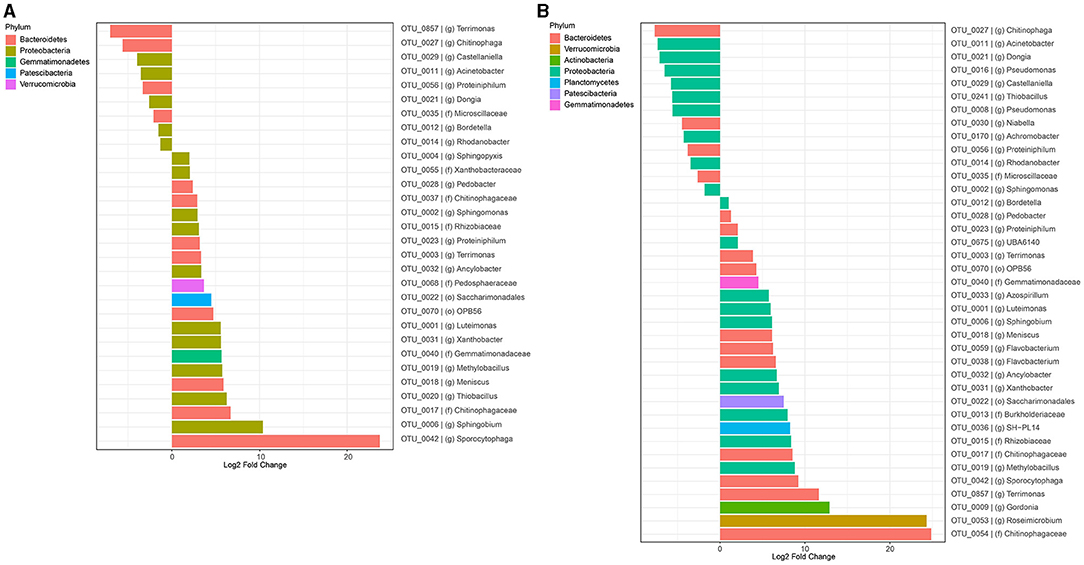
Figure 5. Differential analysis representing each OTU whose relative abundance changed significantly (p < 0.01) over time from the original inoculum (EPA), in 1st enrichment of T1 soil communities with 2,4-D and 2,4,5-T in powder form (A) and 2,4-D and 2,4,5-T dissolved in acetone (B). Colors represent the phylum of the presented OTUs.
Cultures spiked with 2,4-D and 2,4,5-T dissolved in acetone, showed differential increases of 26 OTUs, the most dominant of which belong to the genera Sphingomonas, Luteimonas, Gordonia, and families Burkholderiaceae, and Rhizobiaceae in the 1st enrichment after 25 days of cultivation (Figure 5B; Supplementary Figure S10B). The 2nd enrichment further boosted the growth of species from the genera Bordetella, Sphingomonas, Achromobacter, and Sphingobium after 7 days of cultivation (Supplementary Figures S11B, S15A). Interestingly, members of the genera Sphingomonas and Achromobacter became even more dominant in the 3rd enrichments and after the 2nd spike in the 2nd enrichment (Supplementary Figures S12B, S13B). This is also apparent from the differential abundance analyses (Supplementary Figures S15B,C).
Key Genes of 2,4-D- and 2,4,5-T-Degrading Bacteria
We also isolated two bacterial species and characterized them for key genes involved in degradation of the herbicides. For that, we plated serial dilutions of the 2nd enrichments on nutrient agar plates and isolated bacterial cells from two of the most abundant colonies that appeared, for an initial characterization of their genetic potential to degrade 2,4-D and/or 2,4,5-T. Sequencing of their 16S rRNA genes revealed that they were related to Bordetella petrii and Sphingomonas histidinilytica (Nguyen et al., submitted). We have tentatively designated these two species B. petrii BT1 9.2 and S. histidinilytica BT1 5.2, respectively. Targeted PCR on their DNA revealed that B. petrii BT1 9.2 has tfdA, B, and C genes (Table 2). It did not show a PCR product amplified from a homologous tftA gene. S. histidinilytica BT1 5.2 on the other hand did show the latter product, along with PCR fragments of the tfdB and tfdC genes. The tfdA gene was apparently absent from this species as we did not obtain the appropriate PCR fragment.
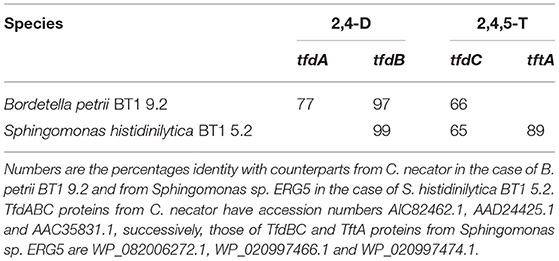
Table 2. Summary of species and genes involved in break-down of 2,4-D (columns 2,4-D) and 2,4,5-T (column 2,4,5-T).
Discussion
We here report on the bacterial community structure, activity, and dynamics of cultures originating from Vietnamese soil heavily contaminated with herbicides. Cultures from both the untreated control soil (T1) and biotreated soil (T4A; amended with various ingredients amongst which different carbon sources such as rice straw and compost. See the Materials and Methods section for details) were incubated with the herbicides 2,4-D and 2,4,5-T as the major sources of carbon and energy. Other sources were those still present in the original soil and amounted to about 10–50 mg/L in the 3rd enrichments, which is at about the same order of magnitude as the total concentrations of the herbicides, i.e., 200–300 mg/L. Both T1 and T4A soil communities showed degradation of the herbicides coupled to cellular growth, but the T1 culture did this significantly faster than the one from T4A. The U.S. Environmental Protection Agency estimated the half-life of 2,4-D in natural environments at around 10 days in aerobic soils and 41–333 days in anaerobic aquatic environments (EPA, 2005). Our T1 enrichment showed a half-life of 2,4-D of 1–2 days, which is even faster than described in that document. The initial T1 community was dominated by Alphaproteobacteria as opposed to some genera of the phylum Firmicutes, like Paenibacillus, Symbiobacterium, Bacillus, and Ureibacillus in the T4A community. Indeed, many species from the Alphaproteobacteria are known for their ability to degrade herbicides (Huong et al., 2007; Cycoń et al., 2011; Giri et al., 2016). We hypothesize that the community from untreated soil is relatively poor in carbon and Gibbs energy sources and as such offers niches for specialized strains that can use the herbicides as alternative sources much more efficiently than those from cultures raised from richer soil. In the latter case, the specialists may well have been outcompeted by those growing on the amendments as a result of competition for oxygen, for vitamins or for essential nutrients. Indeed, many reports indicate that indigenous microbial populations are good candidates for the breakdown of aromatic compounds that pollute the environment that they live in (Ren et al., 2015; Liu et al., 2019). More detailed studies also showed a clear correlation between expression of tfdA and cad genes involved in phenoxyacetic acid degradation and mineralisation rate of these herbicides along with the evolution of the soil microbial communities (Bælum et al., 2008; Ditterich et al., 2013).
The approach to expose the communities to the herbicides as the major sources of carbon and energy turned out to be successful. Firstly, the T4A community started degrading the herbicides, albeit only after a prolonged duration of the enrichment. Many studies have also reported the adaptation of microbial communities to persistent organic pollutants in laboratory systems (Macedo et al., 2006; Poursat et al., 2019a,b). Secondly, it has led to the enrichment of species that suggest their importance in their breakdown. Indeed, we isolated two species from the enrichments, termed them B. petrii BT1 9.2 and S. histidinilytica BT1 5.2 after their closest relatives, and showed that they contain genes relevant for the breakdown of 2,4-D and/or 2,4,5-T. Sphingomonas species are versatile organisms with a high potential to degrade (chlorinated) aromatics as well as 2,4-D (Smejkal et al., 2001; Fukuda et al., 2002; Xia et al., 2005; Sørensen et al., 2013). However, to our knowledge, this is the first study showing that a B. petrii species has that potential as well. Only one report describes genes involved in the degradation of 2,4-D, but that genetic potential has never been verified by physiological experiments (Gross et al., 2008).
In addition to these 2 species, several other ones that showed increased contributions upon successive enrichments were detected in the Illumina profiles. Sphingomonas, Pseudomonas, and Achromobacter are dominant genera that were enriched from T1 soil and these may play a crucial role in the degradation of 2,4-D and 2,4,5-T. Members of the genus Sphingomonas are becoming important in environmental microbiology because they have been reported to degrade 2,4-D as judged from many studies (Smejkal et al., 2001; Thiel et al., 2005; Shimojo et al., 2009; Cycoń et al., 2011). In addition to Sphingomonas, also species from the genera Pseudomonas and Achromobacter are known for their potential to degrade 2,4-D (Amjad, 2003; Vedler et al., 2004; Xia et al., 2017).
We further rationalized that bioavailability of 2,4-D and 2,4,5-T would be improved when we supplied them dissolved in acetone, rather than in powder form. Adding 2,4-D and 2,4,5-T dissolved in acetone to the incubations keeps these chemicals largely in the dissolved form and thereby avoids the dependence of their biodegradation rates on the rates of dissolution of the crystalline substrates. In soil, their rates of biodegradation will depend on their partitioning to soil particles and, in particular, on their rates of desorption. There is evidence that these chemicals form bound residues in soil with low bioavailability, which will therefore resist biodegradation (Davies et al., 1999; Botero et al., 2017). Indeed, we observed a fast breakdown of both herbicides dissolved in acetone at the first 7 days of the 2nd enrichment (see Supplementary Figure S1), while it took some 7 days before the start of degradation of the herbicides when they were added in powder form. When both cultures were refreshed with a new medium, the rates of the breakdown of the herbicides were comparable regardless of the way the herbicides were supplied. Apparently, both communities became optimally adapted to their specific culturing conditions. Not only were the kinetic patterns of herbicide degradation different, but also the community structures after the subsequent enrichments were different. The dominance of Sphingomonas was apparent in both types of culture, but the one with herbicides added as powder additionally showed relatively high abundances of Achromobacter and Pseudomonas, whereas the one with herbicides dissolved in acetone showed relatively high abundances of Luteimonas and Terrimonas. Perhaps this difference in community structure reflects the preference of certain bacterial groups concerning adhesion and uptake of the herbicides. Likewise, some bacterial species from these genera may feed on necromass as part of a more extended food web in these enrichment cultures.
Taken together, our study contributes to a more fundamental understanding of the succession of bacterial communities concerning structure, function, and dynamics upon exposure to herbicides and demonstrates the appearance of key species potentially speeding up the rate of herbicide degradation. Here, we assume that dominance of such key species reflects the potential to use the herbicides 2,4-D and 2,4,5-T as the major sources of carbon and energy, but this still requires verification. Most importantly, the rate of breakdown of the herbicides not only depends on the history of the indigenous community and a successful enrichment by supplying the herbicides as carbon and energy sources but also on the bioavailability of these herbicides on the one hand and a certain degree of starvation concerning alternative carbon and energy sources on the other hand. These conditions may sum up to a more general principle for biodegradation of other types of xenobiotics as well. More specifically, our study results, under laboratory conditions, suggest that keeping a carbon starvation condition during the onsite treatment may probably result in an efficient, low-cost and environmentally friendly remediation process for herbicide-contaminated soils.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/ repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA596903; https://www.ncbi.nlm.nih.gov/genbank/, MT149894; https://www.ncbi.nlm.nih.gov/genbank/, MT149895; https://www.ncbi.nlm.nih.gov/genbank/, MT149896; https://www.ncbi.nlm.nih.gov/genbank/, MT149897; https://www.ncbi.nlm.nih.gov/genbank/, MT149898; https://www.ncbi.nlm.nih.gov/genbank/, MT149899.
Author Contributions
TLAN designed and performed the research, analyzed the data, prepared figures and tables, authored or reviewed drafts of the paper, and approved the final draft. HTCD delivered soil material, reviewed drafts of the paper, and approved the final draft. JK determined the concentration of the herbicides and approved the final draft. MB assisted in the laboratory experiments, reviewed the draft, and approved the final draft. TTHD analyzed the Illumina sequencing, prepared figures, and tables, reviewed the draft, and approved the final draft. BB analyzed the Illumina sequencing, reviewed the draft, and approved the final draft. JP gave suggestions, reviewed the draft, and approved the final draft. AB reviewed drafts of the paper and approved the final draft. RS designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by a BE Basic Foundation-FES grant (Grant No: FES0905) from the Dutch Ministry of Economic Affairs and by a grant from the Ministry of Science and Technology of Vietnam (MOST, project code 826/QD-BKHCN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Brett Olivier and Chrats Melkonian for their assistance in data management.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frsc.2021.692012/full#supplementary-material
References
Aerts, J. W., van Spanning, R. J. M., Flahaut, J., Molenaar, D., Bland, P. A., Genge, M. J., et al. (2019). Microbial communities in sediments from four mildly acidic ephemeral salt lakes in the Yilgarn Craton (Australia)—terrestrial analogs to ancient Mars. Front. Microbiol. 10:779. doi: 10.3389/fmicb.2019.00779
Albrechtsen, H. J., Mills, M. S., Aamand, J., and Bjerg, P. L. (2001). Degradation of herbicides in shallow Danish aquifers: an integrated laboratory and field study. Pest Manag. Sci. 57, 341–350. doi: 10.1002/ps.305
Al-Fathi, H., Koch, M., Lorenz, W. G., and Lechner, U. (2019). Anaerobic degradation of 2,4,5-trichlorophenoxyacetic acid by enrichment cultures from freshwater sediments. Environ. Sci. Pollut. Res. Int. 26, 34459–34467. doi: 10.1007/s11356-019-06584-y
Amjad, B. K. (2003). Isolation and characterization of 2,4-Dichlorophenoxyacetic acid degrading organisms from soil in Jordan Valley. Biotechnology 2, 73–85. doi: 10.3923/biotech.2003.73.85
Arora, P. K., and Jain, R. K. (2012). Metabolism of 2-chloro-4-nitrophenol in a gram negative bacterium, Burkholderia sp. RKJ 800. PLoS ONE 7:e38676. doi: 10.1371/journal.pone.0038676
Arora, P. K., Srivastava, A., and Singh, V. (2014). Novel degradation pathway of 2-chloro-4-aminophenol in arthrobacter sp. SPG: PeerJ Preprints 2:e194v1. doi: 10.7287/peerj.preprints.194v1
Bælum, J., Nicolaisen, M. H., Holben, W. E., Strobel, B. W., Sørensen, J., and Jacobsen, C. S. (2008). Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2, 677–687. doi: 10.1038/ismej.2008.21
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300.
Botero, L. R., Mougin, C., Peñuela, G., and Barriuso, E. (2017). Formation of 2,4-D bound residues in soils: New insights into microbial metabolism. Sci. Total Environ. 584–585, 715–722. doi: 10.1016/j.scitotenv.2017.01.105
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 108(Supplement 1):4516. doi: 10.1073/pnas.1000080107
Chatterjee, D. K., Kilbane, J. J., and Chakrabarty, A. M. (1982). Biodegradation of 2,4,5-trichlorophenoxyacetic acid in soil by a pure culture of Pseudomonas cepacia. Appl. Environ. Microbiol. 44, 514–516.
Cycoń, M., Zmijowska, A., and Piotrowska-Seget, Z. (2011). Biodegradation kinetics of 2,4-D by bacterial strains isolated from soil. Cent. Eur. J. Biol. 6, 188–198. doi: 10.2478/s11535-011-0005-0
Dang, T. C. H. (2013). Final Project Report: Treatment of Herbicide/Dioxin Contaminated Soil by Bioremediation Technology in Active Landfill. Hanoi: Ministry of Science and Technology Hanoi.
Danganan, C. E., Ye, R. W., Daubaras, D. L., Xun, L., and Chakrabarty, A. M. (1994). Nucleotide sequence and functional analysis of the genes encoding 2,4,5-trichlorophenoxyacetic acid oxygenase in Pseudomonas cepacia AC1100. Appl. Environ. Microbiol. 60, 4100–4106.
Daubaras, D. L., Danganan, C. E., Hübner, A., Ye, R. W., Hendrickson, W., and Chakrabarty, A. M. (1996). Biodegradation of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia strain AC1100: evolutionary insight. Gene 179, 1–8. doi: 10.1016/S0378-1119(96)00326-5
Davies, N. A., Edwards, P. A., Lawrence, M. A. M., Taylor, M. G., and Simkiss, K. (1999). Influence of particle surfaces on the bioavailability to different species of 2,4-dichlorophenol and pentachlorophenol. Environ. Sci. Technol. 33, 2465–2468. doi: 10.1021/es9900499
Ditterich, F., Poll, C., Pagel, H., Babin, D., Smalla, K., Horn, M. A., et al. (2013). Succession of bacterial and fungal 4-chloro-2-methylphenoxyacetic acid degraders at the soil–litter interface. FEMS Microbiol. Ecol. 86, 85–100. doi: 10.1111/1574-6941.12131
Dwernychuk, L. W., Cau, H. D., Hatfield, C. T., Boivin, T. G., Hung, T. M., Dung, P. T., et al. (2002). Dioxin reservoirs in southern Viet Nam—a legacy of agent orange. Chemosphere 47, 117–137. doi: 10.1016/S0045-6535(01)00300-9
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., and Flyvbjerg, H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482. doi: 10.1093/bioinformatics/btv401
EPA (2005). Reregistration Eligibility Decision for 2,4-D. 738-R-05-002. Prevention, Pesticides and Toxic Substances (7508C). Washington, DC: United States Environmental Protection Agency
Fang, L., Qin, H., Shi, T., Wu, X., Li, Q. X., and Hua, R. (2020). Ortho and para oxydehalogenation of dihalophenols catalyzed by the monooxygenase TcpA and NAD(P)H:FAD reductase Fre. J. Hazard. Mater. 388, 121787. doi: 10.1016/j.jhazmat.2019.121787
Filer, K., and Harker, A. R. (1997). Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl. Environ. Microbiol. 63, 317–320.
Fukuda, K., Nagata, S., and Taniguchi, H. (2002). Isolation and characterization of dibenzofuran-degrading bacteria. FEMS Microbiol. Lett. 208, 179–185. doi: 10.1111/j.1574-6968.2002.tb11079.x
Ghattas, A.-K., Fischer, F., Wick, A., and Ternes, T. A. (2017). Anaerobic biodegradation of (emerging) organic contaminants in the aquatic environment. Water Res. 116, 268–295. doi: 10.1016/j.watres.2017.02.001
Giri, K., Pandey, S., Kumar, R., and Rai, J. P. N. (2016). Biodegradation of isoproturon by Pseudoxanthomonas sp. isolated from herbicide-treated wheat fields of Tarai agro-ecosystem, Pantnagar. 3 Biotech 6:190. doi: 10.1007/s13205-016-0505-8
Golovleva, L. A., Pertsova, R. N., Evtushenko, L. I., and Baskunov, B. P. (1990). Degradation of 2,4,5-trichlorophenoxyacetic acid by a Nocardioides simplex culture. Biodegradation 1, 263–271. doi: 10.1007/BF00119763
Gross, R., Guzman, C. A., Sebaihia, M., dos Santos, V. A., Pieper, D. H., Koebnik, R., et al. (2008). The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Genomics 9:449. doi: 10.1186/1471-2164-9-449
Han, L., Zhao, D., and Li, C. (2015). Isolation and 2,4-D-degrading characteristics of Cupriavidus campinensis BJ71. Braz. J. Microbiol. 46, 433–441. doi: 10.1590/S1517-838246220140211
Hayashi, S., Sano, T., Suyama, K., and Itoh, K. (2016). 2,4-Dichlorophenoxyacetic acid (2,4-D)- and 2,4,5-trichlorophenoxyacetic acid (2,4,5-T)-degrading gene cluster in the soybean root-nodulating bacterium Bradyrhizobium elkanii USDA94. Microbiol. Res. 188–189, 62–71. doi: 10.1016/j.micres.2016.04.014
Helber, S. B., Steinert, G., Wu, Y.-C., Rohde, S., Hentschel, U., Muhando, C. A., et al. (2019). Sponges from Zanzibar host diverse prokaryotic communities with potential for natural product synthesis. FEMS Microbiol. Ecol. 95:fiz026. doi: 10.1093/femsec/fiz026
Herlemann, D. P. R., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hervé, M. (2020). RVAideMemoire: Testing and Plotting Procedures for Biostatistics, Version 0.9-77. Available online at: https://cran.r-project.org/web/packages/RVAideMemoire/index.html.
Huong, N. L., Itoh, K., and Suyama, K. (2007). Diversity of 2,4-dichlorophenoxyacetic acid (2,4-D) and 2,4,5-trichlorophenoxyacetic acid (2,4,5-T)-degrading bacteria in Vietnamese soils. Microbes Environ. 22, 243–256. doi: 10.1264/jsme2.22.243
Itoh, K., Kanda, R., Sumita, Y., Kim, H., Kamagata, Y., Suyama, K., et al. (2002). tfdA-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in alpha-Proteobacteria. Appl. Environ. Microbiol. 68, 3449–3454. doi: 10.1128/aem.68.7.3449-3454.2002
Ka, J. O., Holben, W. E., and Tiedje, J. M. (1994). Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60, 1106–1106.
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kellogg, S. T., Chatterjee, D. K., and Chakrabarty, A. M. (1981). Plasmid-assisted molecular breeding: new technique for enhanced biodegradation of persistent toxic chemicals. Science 214, 1133–1135. doi: 10.1126/science.7302584
Kitagawa, W., Takami, S., Miyauchi, K., Masai, E., Kamagata, Y., Tiedje, J. M., et al. (2002). Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184, 509–518. doi: 10.1128/jb.184.2.509-518.2002
Kleinsteuber, S., Müller, R. H., and Babel, W. (2001). Expression of the 2,4-D degradative pathway of pJP4 in an alkaliphilic, moderately halophilic soda lake isolate, Halomonas sp. EF43. Extremophiles 5, 375–384. doi: 10.1007/s007920100202
Koopman, J. E., Buijs, M. J., Brandt, B. W., Keijser, B. J. F., Crielaard, W., and Zaura, E. (2016). Nitrate and the origin of Saliva influence composition and short chain fatty acid production of oral microcosms. Microb. Ecol. 72, 479–492. doi: 10.1007/s00248-016-0775-z
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/aem.01043-13
Laemmli, C. M., Leveau, J. H., Zehnder, A. J., and van der Meer, J. R. (2000). Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182, 4165–4172. doi: 10.1128/jb.182.15.4165-4172.2000
Lechner, U., Türkowsky, D., Dinh, T. T. H., Al-Fathi, H., Schwoch, S., Franke, S., et al. (2018). Desulfitobacterium contributes to the microbial transformation of 2,4,5-T by methanogenic enrichment cultures from a Vietnamese active landfill. Microb. Biotechnol. 11, 1137–1156. doi: 10.1111/1751-7915.13301
Lillis, L., Clipson, N., and Doyle, E. (2010). Quantification of catechol dioxygenase gene expression in soil during degradation of 2,4-dichlorophenol. FEMS Microbiol. Ecol. 73, 363–369. doi: 10.1111/j.1574-6941.2010.00906.x
Liu, X., Chen, K., Chuang, S., Xu, X., and Jiang, J. (2019). Shift in bacterial community structure drives different atrazine-degrading efficiencies. Front. Microbiol. 10:88. doi: 10.3389/fmicb.2019.00088
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Macedo, A. J., Neu, T. R., Kuhlicke, U., and Abraham, W. R. (2006). Adaptation of microbial communities in soil contaminated with polychlorinated biphenyls, leading to the transformation of more highly chlorinated congeners in biofilm communities. Biofilms 3, 37–46. doi: 10.1017/S1479050507002116
Macur, R. E., Wheeler, J. T., Burr, M. D., and Inskeep, W. P. (2007). Impacts of 2,4-D application on soil microbial community structure and on populations associated with 2,4-D degradation. Microbiol. Res. 162, 37–45. doi: 10.1016/j.micres.2006.05.007
Mai, T. A., Doan, T. V., Tarradellas, J., de Alencastro, L. F., and Grandjean, D. (2007). Dioxin contamination in soils of Southern Vietnam. Chemosphere 67, 1802–1807. doi: 10.1016/j.chemosphere.2006.05.086
Maltseva, O., McGowan, C., Fulthorpe, R., and Oriel, P. (1996). Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142 (Pt 5), 1115–1122. doi: 10.1099/13500872-142-5-1115
Mao, S., Zhang, M., Liu, J., and Zhu, W. (2015). Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5, 16116–16116. doi: 10.1038/srep16116
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Miyauchi, K., Suh, S. K., Nagata, Y., and Takagi, M. (1998). Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of gamma-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180, 1354–1359. doi: 10.1128/JB.180.6.1354-1359.1998
Nguyen, B. H., and Dang, T. C. H. (2012). Characterization of bacterial community structure in bioremediation treatment of herbicide/dioxin-contaminated soil at field trials. Tap Chi Sinh Hoc 32, 88–93. doi: 10.15625/0866-7160/v32n1.659
Oh, K. H., and Tuovinen, O. H. (1990). Degradation of 2,4-dichlorophenoxyacetic acid by mixed cultures of bacteria. J. Ind. Microbiol. 6, 275–278. doi: 10.1007/BF01575873
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al (2016). Vegan Community Ecology Package. R package version 2.4.0. Available online at: http://CRAN.Rproject.org/package=vegan (accessed August 21, 2019).
Persoon, I. F., Buijs, M. J., Özok, A. R., Crielaard, W., Krom, B. P., Zaura, E., et al. (2017). The mycobiome of root canal infections is correlated to the bacteriome. Clin. Oral Investig. 21, 1871–1881. doi: 10.1007/s00784-016-1980-3
Pimviriyakul, P., and Chaiyen, P. (2018). A complete bioconversion cascade for dehalogenation and denitration by bacterial flavin–dependent enzymes. J. Biol. Chem. 293, 18525–18539. doi: 10.1074/jbc.RA118.005538
Pimviriyakul, P., Wongnate, T., Tinikul, R., and Chaiyen, P. (2020). Microbial degradation of halogenated aromatics: molecular mechanisms and enzymatic reactions. Microb. Biotechnol. 13, 67–86. doi: 10.1111/1751-7915.13488
Poursat, B. A. J., van Spanning, R. J. M., Braster, M., Helmus, R., de Voogt, P., and Parsons, J. R. (2019a). Biodegradation of metformin and its transformation product, guanylurea, by natural and exposed microbial communities. Ecotoxicol. Environ. Saf. 182, 109414. doi: 10.1016/j.ecoenv.2019.109414
Poursat, B. A. J., van Spanning, R. J. M., de Voogt, P., and Parsons, J. R. (2019b). Implications of microbial adaptation for the assessment of environmental persistence of chemicals. Crit. Rev. Environ. Sci. Technol. 49, 2220–2255. doi: 10.1080/10643389.2019.1607687
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ (accessed on August 21, 2019).
Rejczak, T., and Tuzimski, T. (2015). A review of recent developments and trends in the QuEChERS sample preparation approach. Open Chem. 13, 980–1010. doi: 10.1515/chem-2015-0109
Ren, G., Ren, W., Teng, Y., and Li, Z. (2015). Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Front. Microbiol. 6:22. doi: 10.3389/fmicb.2015.00022
Rice, J. F., Menn, F. M., Hay, A. G., Sanseverino, J., and Sayler, G. S. (2005). Natural selection for 2,4,5-trichlorophenoxyacetic acid mineralizing bacteria in agent orange contaminated soil. Biodegradation 16, 501–512. doi: 10.1007/s10532-004-6186-8
Serbent, M. P., Rebelo, A. M., Pinheiro, A., Giongo, A., and Tavares, L. B. B. (2019). Biological agents for 2,4-dichlorophenoxyacetic acid herbicide degradation. Appl. Microbiol. Biotechnol. 103, 5065–5078. doi: 10.1007/s00253-019-09838-4
Shareef, A., Page, D., Vanderzalm, J., Williams, M., Gupta, V. V. S. R., Dillon, P., et al. (2014). Biodegradation of simazine and diuron herbicides under aerobic and anoxic conditions relevant to managed aquifer recharge of storm water. Clean 42, 745–752. doi: 10.1002/clen.201300092
Shimojo, M., Kawakami, M., and Amada, K. (2009). Analysis of genes encoding the 2,4-dichlorophenoxyacetic acid-degrading enzyme from Sphingomonas agrestis 58-1. J. Biosci. Bioeng. 108, 56–59. doi: 10.1016/j.jbiosc.2009.02.018
Smejkal, C. W., Vallaeys, T., Burton, S. K., and Lappin-Scott, H. M. (2001). A rapid method to screen degradation ability in chlorophenoxyalkanoic acid herbicide-degrading bacteria. Lett. Appl. Microbiol. 32, 273–277. doi: 10.1046/j.1472-765x.2001.00900.x
Sørensen, S. R., Juhler, R. K., and Aamand, J. (2013). Degradation and mineralisation of diuron by Sphingomonas sp. SRS2 and its potential for remediating at a realistic μg L(-1) diuron concentration. Pest Manag. Sci. 69, 1239–1244. doi: 10.1002/ps.3490
Squadrone, S., Brizio, P., Nespoli, R., Stella, C., and Abete, M. C. (2015). Human dietary exposure and levels of polychlorinated dibenzo-p-dioxins (PCDDs), polychlorinated dibenzofurans (PCDFs), dioxin-like polychlorinated biphenyls (DL-PCBs) and non-dioxin-like polychlorinated biphenyls (NDL-PCBs) in free-range eggs close to a secondary aluminum smelter, Northern Italy. Environ. Pollut. 206, 429–436. doi: 10.1016/j.envpol.2015.07.048
Steinert, G., Rohde, S., Janussen, D., Blaurock, C., and Schupp, P. J. (2017). Host-specific assembly of sponge-associated prokaryotes at high taxonomic ranks. Sci. Rep. 7:2542. doi: 10.1038/s41598-017-02656-6
Steinert, G., Wemheuer, B., Janussen, D., Erpenbeck, D., Daniel, R., Simon, M., et al. (2019). Prokaryotic diversity and community patterns in Antarctic continental shelf sponges. Front. Mar. Sci. 6:297. doi: 10.3389/fmars.2019.00297
Stellman, J. M., Stellman, S. D., Christian, R., Weber, T., and Tomasallo, C. (2003). The extent and patterns of usage of Agent Orange and other herbicides in Vietnam. Nature 422, 681–687. doi: 10.1038/nature01537
Stibal, M., Bælum, J., Holben, W. E., Sørensen, S. R., Jensen, A., and Jacobsen, C. S. (2012). Microbial degradation of 2,4-dichlorophenoxyacetic acid on the greenland ice sheet. Appl. Environ. Microbiol. 78, 5070–5076. doi: 10.1128/AEM.00400-12
Thiel, M., Kaschabek, S. R., Gröning, J., Mau, M., and Schlömann, M. (2005). Two unusual chlorocatechol catabolic gene clusters in Sphingomonas sp. TFD44. Arch. Microbiol. 183, 80–94. doi: 10.1007/s00203-004-0748-3
Thuong, N. V., Hung, N. X., Mo, N. T., Thang, N. M., Huy, P. Q., Van Binh, H., et al. (2015). Transport and bioaccumulation of polychlorinated dibenzo-p-dioxins and dibenzofurans at the Bien Hoa Agent Orange hotspot in Vietnam. Environ. Sci. Pollut. Res. Int. 22, 14431–14441. doi: 10.1007/s11356-014-3946-9
Tonso, N. L., Matheson, V. G., and Holben, W. E. (1995). Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30, 3–24. doi: 10.1007/BF00184510
Van den Berg, M., Birnbaum, L. S., Denison, M., De Vito, M., Farland, W., Feeley, M., et al. (2006). The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 93, 223–241. doi: 10.1093/toxsci/kfl055
van der Zaan, B. M., Saia, F. T., Stams, A. J., Plugge, C. M., de Vos, W. M., Smidt, H., et al. (2012). Anaerobic benzene degradation under denitrifying conditions: Peptococcaceae as dominant benzene degraders and evidence for a syntrophic process. Environ. Microbiol. 14, 1171–1181. doi: 10.1111/j.1462-2920.2012.02697.x
Vedler, E., Vahter, M., and Heinaru, A. (2004). The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186, 7161–7174. doi: 10.1128/jb.186.21.7161-7174.2004
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261. doi: 10.1128/AEM.00062-07
Xia, Y., Min, H., Rao, G., Lv, Z.-M., Liu, J., Ye, Y.-F., et al. (2005). Isolation and characterization of phenanthrene-degrading Sphingomonas paucimobilis strain ZX4. Biodegradation 16, 393–402. doi: 10.1007/s10532-004-2412-7
Keywords: chlorophenoxyacetic acid herbicides, microbial community dynamics, microbial community composition analysis, biodegradation, niche partitioning
Citation: Nguyen TLA, Dang HTC, Koekkoek J, Dat TTH, Braster M, Brandt BW, Parsons JR, Brouwer A and van Spanning RJM (2021) Correlating Biodegradation Kinetics of 2,4-Dichlorophenoxyacetic Acid (2,4-D) and 2,4,5-Trichlorophenoxyacetic Acid (2,4,5-T) to the Dynamics of Microbial Communities Originating From Soil in Vietnam Contaminated With Herbicides. Front. Sustain. Cities 3:692012. doi: 10.3389/frsc.2021.692012
Received: 07 April 2021; Accepted: 14 September 2021;
Published: 20 October 2021.
Edited by:
Brenda B. Casper, University of Pennsylvania, United StatesReviewed by:
Paunita Iuliana Boanca, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaCaroline Michel, Bureau de Recherches Géologiques et Minières, France
Copyright © 2021 Nguyen, Dang, Koekkoek, Dat, Braster, Brandt, Parsons, Brouwer and van Spanning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rob J. M. van Spanning, cm9iLnZhbi5zcGFubmluZ0B2dS5ubA==; Thi Lan Anh Nguyen, dC5sLmEubmd1eWVuQHZ1Lm5s; bGFuYW5oZG44NkBnbWFpbC5jb20=
 Thi Lan Anh Nguyen
Thi Lan Anh Nguyen Ha Thi Cam Dang2
Ha Thi Cam Dang2 Ton That Huu Dat
Ton That Huu Dat Bernd W. Brandt
Bernd W. Brandt Abraham Brouwer
Abraham Brouwer Rob J. M. van Spanning
Rob J. M. van Spanning