- 1Department of Land Resources and Environmental Sciences, Montana State University, Bozeman, MT, United States
- 2Central Agricultural Research Center, Montana State University, Moccasin, MT, United States
- 3School of Food and Agriculture, University of Maine, Orono, ME, United States
Industrialized agriculture results in simplified landscapes where many of the regulatory ecosystem functions driven by soil biological and physicochemical characteristics have been hampered or replaced with intensive, synthetic inputs. To restore long-term agricultural sustainability and soil health, soil should function as both a resource and a complex ecosystem. In this study, we examined how cropping systems impact soil bacterial community diversity and composition, important indicators of soil ecosystem health. Soils from a representative cropping system in the semi-arid Northern Great Plains were collected in June and August of 2017 from the final phase of a 5-year crop rotation managed either with chemical inputs and no-tillage, as a USDA-certified organic tillage system, or as a USDA-certified organic sheep grazing system with reduced tillage intensity. DNA was extracted and sequenced for bacteria community analysis via 16S rRNA gene sequencing. Bacterial richness and diversity decreased in all farming systems from June to August and was lowest in the chemical no-tillage system, while evenness increased over the sampling period. Crop species identity did not affect bacterial richness, diversity, or evenness. Conventional no-till, organic tilled, and organic grazed management systems resulted in dissimilar microbial communities. Overall, cropping systems and seasonal changes had a greater effect on microbial community structure and diversity than crop identity. Future research should assess how the rhizobiome responds to the specific phases of a crop rotation, as differences in bulk soil microbial communities by crop identity were not detectable.
Introduction
Modern industrial agriculture results in overly simplified landscapes where soil mediated regulatory ecosystem functions have been hampered or replaced with intensive off-farm inputs aimed principally at securing high yields. The overreliance on synthetic chemical and mechanical inputs, however, comes with a high potential for soil degradation and soil loss (Landis, 2017; Vanwalleghem et al., 2017). Worldwide, agricultural erosion has diminished soil capacity to function as a resource and a living ecosystem on 40% of all agricultural land, underscoring the need to restore and maintain soil health (Doran and Zeiss, 2000). In particular, tillage is used in many conventional and organic agricultural systems to cultivate fields, remove crop and weed residues, and incorporate manure inputs. Intensive conventional tillage is known to facilitate soil erosion and a loss of nutrients (Jat et al., 2019; Schneekloth et al., 2020), particularly in dry regions (Clay et al., 2014). Moreover, this practice can dramatically reduce bacterial (de Quadros et al., 2012; García-Orenes et al., 2016; Ishaq et al., 2020a) and fungal (Drijber et al., 2000; Castillo et al., 2006) diversity in soil, potentially impairing the functionality of these systems. Alternatives to conventional tillage are being sought for organic systems which maintain crop productivity but sustain soil or biodiversity loss.
The soil microbiome—the array of bacteria, fungi, archaea, protozoa, viruses, and their collective genomes which interact with each other and with macroorganisms, is an essential component of the soil ecosystem underpinning numerous ecosystem functions (Lal, 2016). Soil microorganisms influence soil physical structure, drive nutrient cycling through decomposition of organic matter and mineralization of nitrogen, and suppress disease in plants (Miller and Jastrow, 2000; Brussaard et al., 2007; Martínez-García et al., 2018). Soil bacteria can produce hormones that directly influence plant growth or modify the production of plant growth hormones (Patten and Glick, 2002; Mohite, 2013), and mutualistic soil biota aid in plant water and nutrient uptake, increasing aboveground productivity (Bender and van der Heijden, 2015; van der Heijden et al., 2015). Understanding how these interactions between aboveground and belowground communities condition productivity is essential to enhance the sustainability of farming, especially as intense agricultural management is known to reduce micro- and macro-biological diversity with associated negative consequences to ecosystem functions.
Organic farming is one approach to improve agricultural sustainability through increasing reliance on ecological processes rather than synthetic inputs. Compared to chemically managed systems, organic systems place a greater emphasis on soil regeneration and aboveground biodiversity. Previous literature has examined the differential impact of organic and conventional management systems on soil microbiota, which can lead to distinct belowground communities (Li et al., 2012; Hartmann et al., 2015; Harkes et al., 2019). For example, soil microbiota is reported to increase in phylogenetic richness and community variability in organic systems (Lupatini et al., 2017), but this may be linked to the greater bioavailability of nutrient inputs in a more neutral soil pH (Lauber et al., 2009; Zhalina et al., 2015). Nutrient input, in particular, may be the driving force of these observations (Hartmann et al., 2015; Zhang et al., 2017; Semenov et al., 2020). There is also a knowledge gap of how soil microbial communities vary among contrasting organic systems.
In the semiarid regions of the Northern Great Plains, fallow periods where fields are left bare for a growing season to aid in the preservation of soil moisture do not contribute as many benefits to the soil as more diverse cropping systems (Rosenzweig et al., 2018). Also, the highly simplified landscapes that dominate conventional agriculture of the region are characterized by low plant diversity and a high dependence on synthetic inputs (Adhikari et al., 2019) which negatively impact soil biota and result in less diverse belowground communities (Chaudhry et al., 2012; Ishaq et al., 2017). In contrast, organic cropping systems are characterized by more complex crop rotations and diverse associated biodiversity, including weed communities, than those observed in conventional wheat-summer fallow rotations across the region (Adhikari and Menalled, 2018).
Diversified crop rotations that include for-profit or for-health (i.e., green manure or bioremediation) plant species reduce or replace fallow periods, benefiting soil ecosystems and increasing soil biodiversity (Maarastawi et al., 2018; Peralta et al., 2018). Carbon and nitrogen from microbial biomass can increase by roughly 20 and 27%, respectively, in diversified crop rotations compared to a simple monoculture (McDaniel et al., 2014). Functional and metabolic diversity in heterotrophic soil bacterial communities also increase under more diverse crop rotations (D'Acunto et al., 2018). It is unclear whether the soil physicochemical changes, increases in crop detritus, host-specific promotion, or inclusion of various functional groups associated with diversifying crop rotations have the greatest impact on soil microbial communities (Venter et al., 2016). While there is not a specific sequence of crops that will achieve these benefits, cover crop mixtures are often included in these rotations toward the same end.
Cover crops can recruit specific microorganisms via plant inputs (Ishaq et al., 2017), and while there is not a comprehensive understanding of what microbial communities are recruited by specific plant species, there is some evidence that certain plants act more selectively than others (Massenssini et al., 2015; Trognitz et al., 2016; Aguilera et al., 2017). It is also known that cover crops can alter soil microbial community dynamics because microbiota with fast growth rates and the capacity to utilize temporally limited nutrients can take advantage of organic carbon from cover crop roots, root turnover, and crop residue (Wortman et al., 2013) and may favor bacterial communities while those that become low quality residue favor fungi (Frasier et al., 2016). These inputs of biomass and root exudates shape soil microbial activity and diversity, even after cover crops are terminated for the season (Wortman et al., 2013; Calderón et al., 2016).
In semi-arid agroecosystems, cover crops must be terminated early enough in the growing season to preserve soil moisture for the commercial crop. Cropping systems employ various methods of cover crop termination that have their own effects on soil microbial communities. For example, conservation, or reduced tillage lessens disruption to soil organic carbon pools and no tillage leaves cover crop residue on the soil surface. This can result in increased soil carbon, soil nitrogen, and microbial biomass at the surface of the soil (Helgason et al., 2010; Sapkota et al., 2012; Nivelle et al., 2016) but has been criticized for its heavy reliance on herbicides and the associated selection of herbicide-resistant weed biotypes (Menalled et al., 2016). In recent years, consumer demands and market opportunities have driven an expansion of organic agricultural systems (USDA ERS, 2020) which use tillage to terminate cover crops and incorporate residue into the soils. However, tillage disturbs the soil ecosystem by accelerating soil organic matter oxidation and labile carbon cycling (McLauchlan, 2006), factors that have driven a growing interest to reduce soil disturbance practices in organic systems (Carr, 2017). The integration of crop and livestock practices has been explored as an approach to reduce tillage intensity in organic cropping systems (Miller et al., 2015, McKenzie et al., 2016) while increasing soil nitrogen, soil carbon, and microbial biomass (Ishaq et al., 2017). However, little is known about specific effects of grazing on soil microbial communities.
Semiarid, dryland agriculture makes up a significant portion of global crop production, and understanding how soil communities interact with crops under different farming practices is crucial for global agricultural security (Delgado-Baquerizo et al., 2017). In this study, we assessed soil bacterial community responses to contrasting cropping systems in semiarid regions of the Northern Great Plains. To do so, we took advantage of a field experiment that began in 2012 at the Montana State University Fort Ellis Research and Teaching Center in Bozeman, MT to test a 5-year crop rotation under three different management systems: chemical inputs with no-tillage, a USDA-certified organic system with tillage, and a USDA-certified organic system with targeted sheep grazing for cover crop and weed termination (Ishaq et al., 2020a). We hypothesized that (1) organically managed systems support higher soil bacterial diversity than chemically managed systems, (2) the two organic systems result in dissimilar soil bacterial communities, and (3) diversity is highest in the cover crop phases. We did this by comparing bacterial communities from winter wheat, safflower/sweet clover, and sweet clover under different management systems in fields where the crop rotation has been established for several years.
Methods
Site Description
A field experiment to evaluate approaches to minimize soil disturbances in organic cropping systems was established in 2012 at the Montana State University Fort Ellis Research and Teaching Center in Bozeman, MT (45.653 N, −110.972 W). The underlying soil is a Blackmore silt loam (University of California, Davis, 2019) (fine-silty, mixed superactive, frigid Typic Argiustolls) with 0–4% slopes, composed of about 22% clay, 10% sand, and 68% silt down to 25 cm (Natural Resources Conservation Service Soil Survey Staff, 1999). Fort Ellis usually receives 465 mm of precipitation annually, with monthly mean air temperatures between −5.7 and 18.9°C (PRISM Climate Group, 2020). Prior to 2004, the study site was planted with perennial grasses [Bromus inermis L., Thinopyrum intermedium (Host) Barkworth and D.R. Dewey, and Poa compressa L.]. Between 2004 and 2009, the experimental site followed either continuous spring wheat (Triticum aestivum L.), spring wheat-fallow, or winter wheat-fallow crop rotation. From 2009 to 2012, the study site followed either a continuous alfalfa (Medicago sativa L.) or a 3-year crop rotation consisting of spring wheat in the first year followed by pea (Pisum sativum L.), and hay barley (Hordeum vulgare L.) in the second and third years, respectively. In the spring of 2012, the entire experimental site was planted with glyphosate tolerant rapeseed (Brassica napus L.) and treated with herbicide. The rapeseed was tilled to a depth of 15 cm in July 2012 and planted in September 2012 following the experimental design described below. Additional site information can be found in Miller et al. (2015) and Barsotti et al. (2013).
Study Design
This study followed a split-plot design with three replications where cropping systems were the main plot level and crop phases the split-plots (Supplementary Figure 1). The cropping systems included a chemical no-till system where synthetic off-farm inputs were utilized to manage weeds, pathogens, and soil nutrient levels (hereafter, chemical no-till). This system, often referred to as conventional in the context of industrial-age farming, was considered the basis of comparison for other treatments. The other cropping system treatments included a USDA-certified organic system with tillage used for cover crop termination and weed management (hereafter, organic tilled), and a USDA-certified organic system that employed reduced tillage and targeted sheep grazing (Ovis aries L.) for cover crop termination and weed management with the overall goal of reducing tillage intensity (hereafter, organic grazed) (Ishaq et al., 2020a). The organic tilled treatment represents current organic practices which are effective yet not ecologically sustainable, and the organic grazed treatment provided an experimental approach to reduce tillage intensity. Each cropping system was randomly assigned to a 75 × 90 m plot with three entire field replications and further divided into five 90 × 13 m split-plots separated by a 1 m fallow track, and randomly assigned to one phase of a 5-year crop rotation: Year 1, safflower (Carthamus tinctorius L.) under-sown with yellow sweet clover (Melilotus officinalis (L.) Lam); Year 2, yellow sweet clover; Year 3, winter wheat (Triticum aestivum L.); Year 4, lentils (Lens culinaris Medik); and Year 5, winter wheat. Austrian winter pea (Pisum sativum subsp. arvense.) was planted in fall 2012 for the first year (2013) of the experiment because the biennial nature of yellow sweet clover would have required a previous year of seeding. In 2017, sweet clover was terminated in early July (with allowance for regrowth) and winter wheat was harvested in the last week of July. Safflower, a late spring crop, was performing negligible soil chemistry by the time samples were collected in August and was harvested in September.
A no-till double-disk seeder was used on all cropping systems to minimize soil disturbance. Chemical inputs in the conventional system mimicked standard practices in the Northern Great Plains and included 2,4-D, bromoxynil, dicamba, fluroxypyr, glyphosate, MCPA, pinoxaden, and urea to manage weeds and nutrient availability (Adhikari and Menalled, 2020). Crops from both organic treatments were USDA certified by 2015, after completing the transition to organic that began in 2012. A chisel plow, tandem disk, and field cultivator were utilized in the organic tilled treatment as needed to terminate cover crops, prepare the seedbed, and incorporate cover crop residue into the soil (Ishaq et al., 2020a). Targeted sheep grazing at a stocking density of 50 sheep/ha for 30 days terminated cover crops and managed weeds in the organic grazed treatment (Menalled et al., 2020). The agronomic management details are provided in more detail elsewhere [see Adhikari and Menalled (2020)].
Soil Collection
Soil samples for microbial DNA extraction and sequencing were collected from the safflower/sweet clover, sweet clover, and year 3 winter wheat split-plots, from each of the three cropping systems, in each of the three cropping system replicates, in June and in August of 2017 (i.e., 3 systems × 3 replications × 3 rotations × 2 time points). Due to logistic problems, two safflower/clover split-plots (one chemical no-till and the other organic grazed) in the second field replication were not sampled in June, but all were sampled in mid-August following residue removal in the respective treatments for a total of 52 soil samples. Each split-plot was divided into quartiles to account for spatial variation in the soil environment and allow for more representative microbial community samples. One bulk soil sample from each of these quartiles was taken with a 2 cm diameter core sampler to a depth of 15 cm after clearing the ground of debris and crop residue. The four cores were manually homogenized to one sample per subplot and kept at −20°C until analysis. Extremely dry soil conditions in August 2017 necessitated the use of a pickaxe to break the ground before sampling. The soil sampler was sterilized with 70% isopropyl alcohol and air dried between samples to prevent cross contamination of microbial communities.
Laboratory Methods
Metagenomic soil DNA was extracted from 0.25 g of the 20–30 g homogenized samples using a Qiagen DNeasy PowerSoil kit (Qiagen, Hilden, Germany). Amplicon libraries of the V4 region of the bacterial 16S rRNA gene were prepared following procedures outlined by Kozich et al. (2013), using dual indexed primers (515F and 806R) to target soil bacteria active at the time of sampling. This method amplifies both bacteria and archaea, but the resulting libraries are primarily bacterial as the latter are not amplified well. Amplicons were normalized using SequalPrep plate normalization kit (Invitrogen) and the resulting product was pooled to equimolar concentrations. These pooled amplicons were cleaned with AmpureXP magnetic beads at a concentration of 0.8 (vol/vol) beads to pool ratio. Paired-end sequencing was performed at Michigan State University's RTSF Genomics Core using an Illumina MiSeq with a maximum read length of 250 base pairs on MiSeq v2 reagent cartridge (Illumina Inc.). Illumina Real Time Analysis v1.18.54 was used to perform base calling, and the resulting output was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.19.1. Raw sequences and metadata are available from NCBI under BioProject Accession PRJNA672991.
Bioinformatics
The DADA2 pipeline run in the software environment R 3.6.1 was used to filter paired end reads, remove chimeric sequences, and conduct taxonomic assessment (Callahan et al., 2016; R Development Core Team, 2020). Sequences were trimmed by 10 bases at the start and end positions during filtering, with no ambiguous bases allowed and a maximum expected error of two. The error rates were learned on 2 × 106 randomly selected dereplicated reads and then used to identify Amplicon Sequence Variants (SVs), which are analogous to individuals in that sequences have been grouped down to single-nucleotide polymorphism/single base differences by assessing overall error rate and probability of base error vs. polymorphism occurrence.
Then, using DADA2, two-parent chimeras (bimeras) were removed. The Silva NR version 138 database was used to assign taxonomy to sequence variants (Yilmaz et al., 2014). Data were first rarefied to the size of the smallest sample library (38,776 sequenced reads), which allowed all 52 samples to pass this quality control step. One sample with only 220 sequence variants made it through as a result. This low outlier was from an organic tilled safflower/clover plot sampled in August 2017. Data were rarified to the next lowest number of reads per sample (65,165 sequenced reads). The outlier was subsequently removed along with 42 absolute sequence variants and the 6,932 sequenced reads unique to them.
Statistical Procedures
All data analysis was performed in R 3.6.1–4.0.2 (R Development Core Team, 2020). The phyloseq and vegan packages were used for statistical analysis (McMurdie and Holmes, 2013; Okansen et al., 2019), while ggplot2 was used for data visualization (Wickham, 2016). Alpha or within-community diversity was assessed via observed sequence variants (bacterial community richness), Shannon's Diversity, and Shannon's Evenness. Linear regression with mixed effect models explored the relationships between treatments and alpha diversity. The model for bacterial richness and diversity included cropping system and sampling month as fixed effects and plot as a random effect, while the model for evenness used only month as a fixed effect and plot as a random effect. Crop phase (i.e., plant species identity) was tested as a predictor for richness, diversity, and evenness, but was not a significant predictor for any of them. Field replication was also considered as a random effect because of variation at the main plot level, but split-plot explained more variation in the model for bacterial richness, Shannon's Diversity, and Shannon's Evenness. Nesting split-plot in plot also did not increase the conditional R2 of these models enough to be considered an improvement. Soil data on percent carbon, percent nitrogen, C:N ratio, and pH were available along with crop and weed biomass but could not be used to model alpha diversity because all except C:N ratio and weed biomass (Supplementary Figures 3, 4) varied by cropping system, split-plot, or were highly correlated with one another (r = 0.90). There were no interactions between treatments in any of the models for alpha diversity. Type III One-Way Analysis of Variance determined which variables significantly affected alpha diversity while Tukey's post-hoc comparisons evaluated differences in means among treatment levels.
Changes in relative bacterial abundance from June to August among cropping systems were represented with heatmaps that displayed the fifty most abundant sequence variants labeled by genera. Permutational random forest analysis was used with the rfpermute package to determine the taxa which were significantly discriminatory between treatment states, highlighting the ones that were unique or a defining feature of the three cropping systems. For each comparison, 500 trees were made with 100 permutations each. Relative abundance of important taxa was then visualized by sampling month within each cropping system. Unspecified genera in both the heatmaps and random forest analysis were identified at the family level.
Bray-Curtis dissimilarities (based on species' presence/absence and relative abundance) were calculated to assess between-community diversity and visualized using non-metric multidimensional scaling ordination. Mean dissimilarities of soil bacterial communities grouped by treatment were compared using analysis of variance and Tukey's post-hoc tests. A permutational analysis of variance (PERMANOVA) compared differences among bacterial communities using the adonis function with 9,999 permutations. Data were stratified by split-plot to account for repeated measures. Homogeneity of dispersions—an assumption for using a PERMANOVA—was evaluated using the betadisper function.
Results
Alpha Diversity
There were 11,193 unique sequence variants in the 51 soil samples collected in 2017. Bacterial richness decreased in every crop across all three cropping systems between June and August but varied the most in the chemical no-till cropping system and the least in the organic systems (Figure 1A). Safflower/clover and winter wheat in the chemical no-till systems had bacterial communities with a wider range of Shannon's diversity compared to those under organic management (Figure 1B). Bacterial evenness changed the least in organic tilled winter wheat, organic tilled safflower/clover, and chemical no-till safflower/clover (Figure 1C).
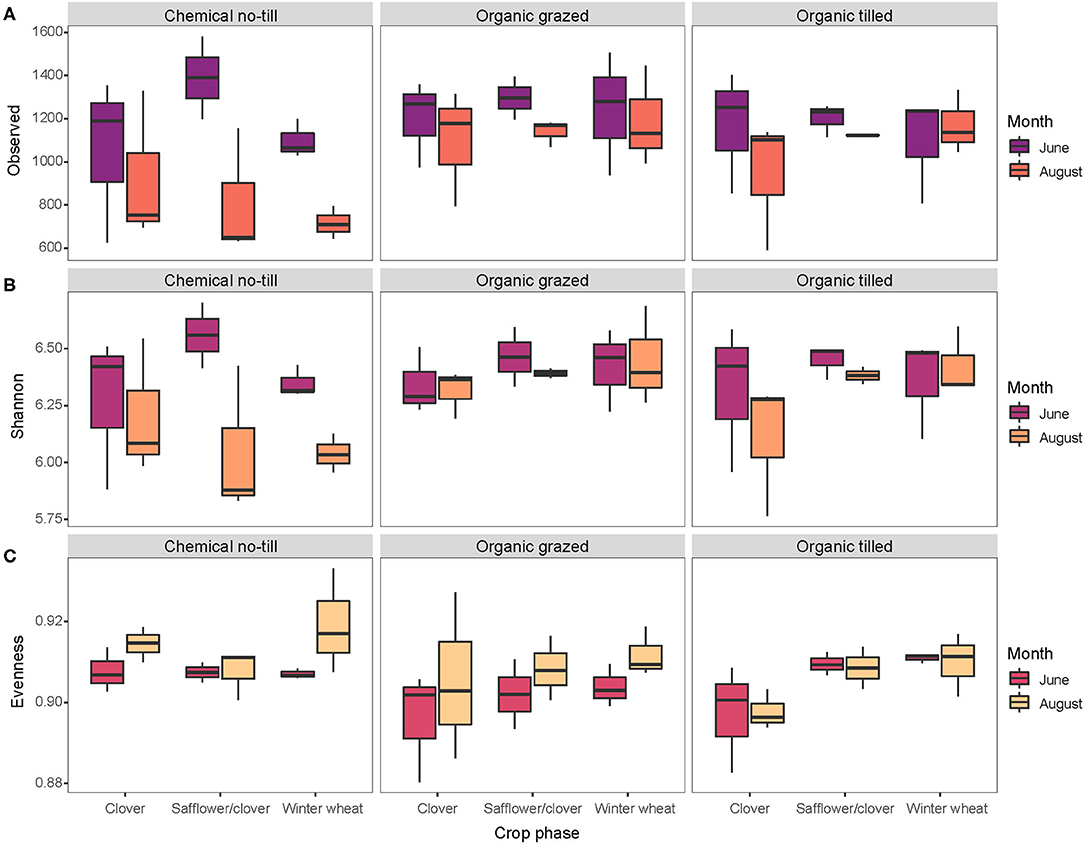
Figure 1. Alpha diversity metrics for soil bacterial communities associated with three crops in three contrasting dryland cropping systems in June and August. Diversity metrics include (A) richness, (B) Shannon's Diversity, and (C) Shannon's Evenness.
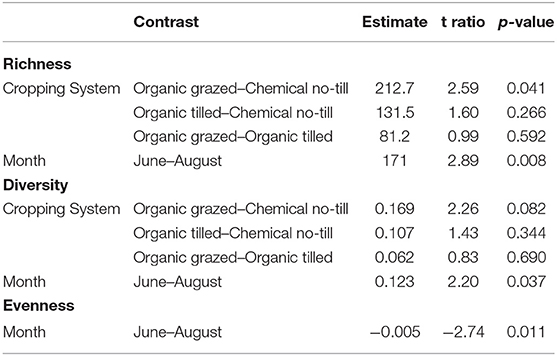
All measures of alpha diversity changed with respect to sampling month (Table 1). Bacterial richness varied a function of cropping system and month (p = 0.049, equivocal, 0.008, respectively, Table 1). Richness was higher in the organic grazed system than in the chemical no-till system, an estimated difference in means of 212 observed sequence variants when averaged across months (p = 0.041, Table 2). Bacterial richness was significantly higher in June than in August with an estimated difference in means of 171 observed sequence variants when averaged across cropping systems (p = 0.008, Table 2). Bacterial richness did not differ between the grazed and tilled organic systems. Cropping system as a predictor of Shannon's diversity was trending toward significance (p = 0.09, Table 1). The chemical no-till system was trending toward less diversity than the organic grazed system, with an estimated difference in mean Shannon's diversity measure of 0.169 when averaged across months (p = 0.08, Table 2). Soil bacterial diversity and evenness also changed significantly by month (p = 0.037 and p = 0.011, respectively, Table 1). Shannon's diversity was higher in June than in August, with an estimated difference in means of 0.123 (p = 0.011, Table 2). Mean Shannon's evenness, however, increased by an estimate of 0.005 by the end of the summer (p = 0.011, Table 2).
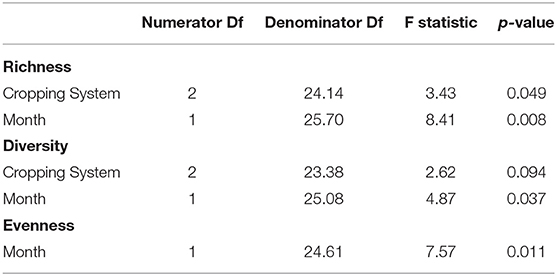
Table 1. Statistics from Type III Analysis of Variance for mixed effect linear models of bacterial richness, Shannon's Diversity, and Shannon's Evenness.
Overall Community Composition: Abundance
Taxa from the phyla Actinobacteria, Proteobacteria, Verrucomicrobia, and Firmicutes consistently had the highest relative abundance across all three cropping systems (Supplementary Figure 2), though there were some shifts in community composition. Sequence variants from Bradyrhizobium (Proteobacteria) and Candidatus Udaeobacter (Verrucomicrobia) were more abundant in June while those from Candidatus Nitrososphaera (Thaumarchaeota, Archaea) and Blastococcus (Actinobacteria) were more abundant in August (Figure 2). Pseudarthobacter (Actinobacteria) remained highly abundant throughout the summer in almost every treatment plot (Figure 2). Some changes in bacterial abundance occurred within particular crop phases or cropping systems. For example, one sample in an organic tilled clover plot had a high abundance of Candidatus Udaeobacter in June but not in August, while organic tilled plots had higher abundance of Bacillaceae in August than in June. Other sequence variants were abundant throughout the summer, such as Nitrososphaeraceae, Xanthobacteraceae (Proteobacteria), Sphingomonas (Proteobacteria), and a number of taxa not identified at the genus level (Figure 2).
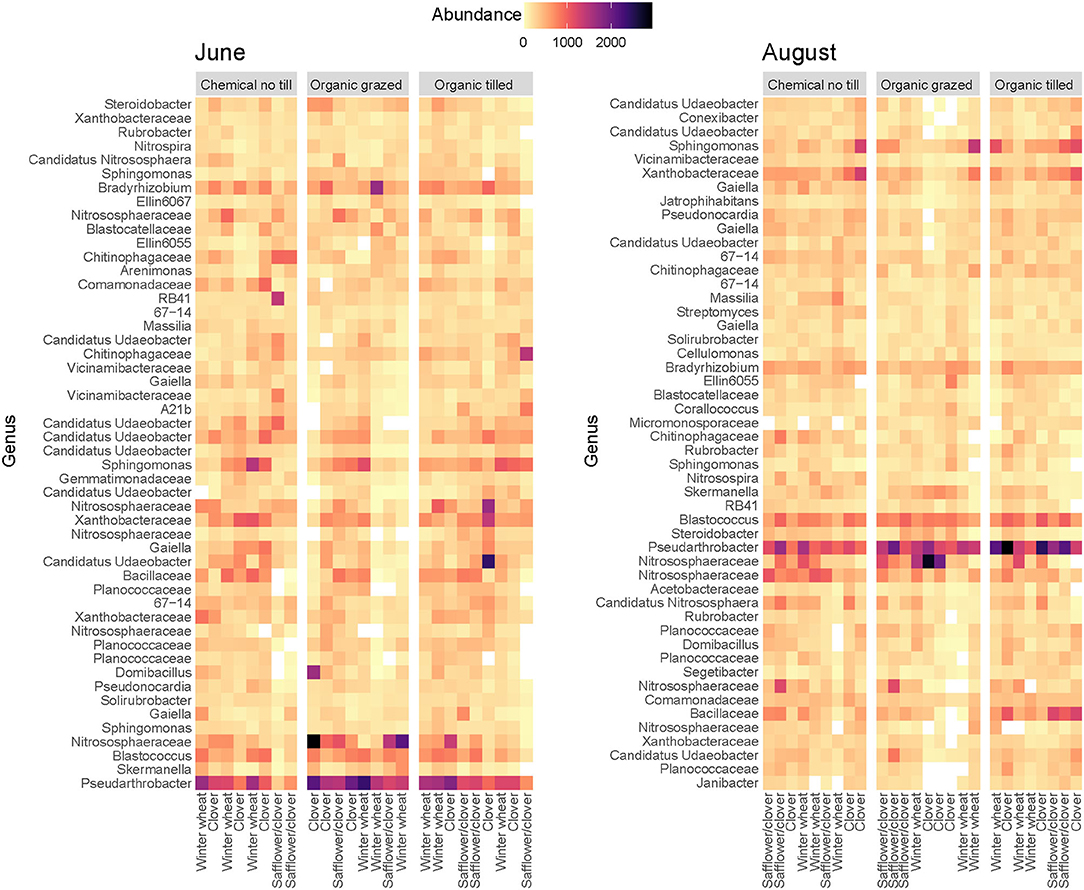
Figure 2. Abundance of the most abundant sequence variants identified at the genus level in three contrasting dryland cropping systems. Sequence variants from unspecified genera are identified at the family level.
A random forest analysis identified the top 50 sequence variants, by calculated importance (p < 0.05), that responded to cropping systems in each month (Figure 3). The relative abundances of these taxa faceted by month indicated which predictor taxa were important to different cropping systems in June and August (Figure 3). Pseudarthobacter was the most abundant, followed by Nitrososphaeraceae and Candidatus Udaeobacter. Log relative abundance of Pseudarthobacter, increased in all three cropping systems from June to August. Nitrososphaeraceae increased in the chemical no-till and organic tilled systems and decreased in the organic grazed system while Candidatus Udaeobacter did the opposite (Figure 3). The random forest analysis had an out-of-bag error estimate of 19.61% as a prediction error for bootstrapped samples that did not contain elements of the original dataset.
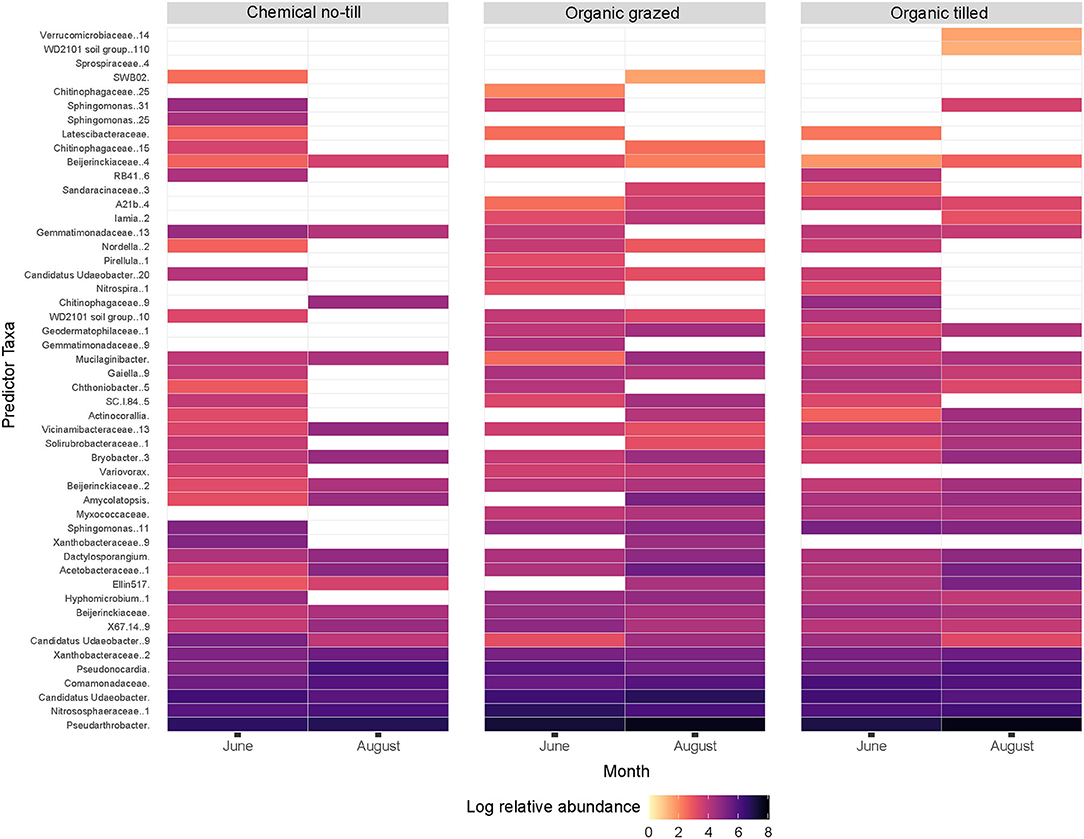
Figure 3. Log relative abundance of the top 50 predictor sequence variants in all cropping systems by month represented by a random forest analysis. White spaces indicate taxa that are absent or at very low abundance. Sequence variants from unspecified genera are identified at the family level.
Beta (Between-Community) Diversity
The PERMANOVA indicated that cropping system, crop, month, and the interaction between cropping systems and crop identity impacted bacterial communities (Table 3). Of the three treatments, cropping system explained the most variation among communities (10.1%) and the interaction between cropping system and crops explained second (8.3%). The organic grazed system had different soil bacterial communities from the organic tilled and chemical no-till systems (Figure 4). Organic grazed communities and chemical no-till communities had a mean dissimilarity of 55.06%, followed by 54.94% between organic grazed and organic tilled communities. Chemical no-till and organic tilled systems had less dissimilar communities in comparison with a mean dissimilarity of 52.26%. Dissimilarity decreased in the chemical no-till system, increased in the organic grazed system, and changed little in the organic tilled system as the summer progressed (Figure 4). Communities in organic grazed systems had the highest within-group mean dissimilarity of 53.25% on a weighted Bray-Curtis scale. Communities in the organic tilled and chemical no-till systems had within-group dissimilarities of 50.65 and 51.04%, respectively.
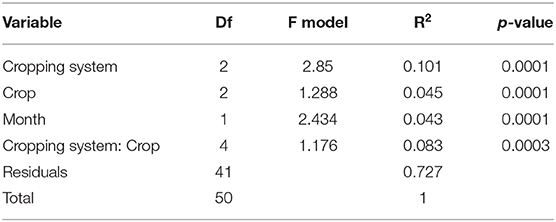
Table 3. PERMANOVA of the effects of cropping system, crop, month, and the interaction between cropping system and crop on soil bacterial communities.
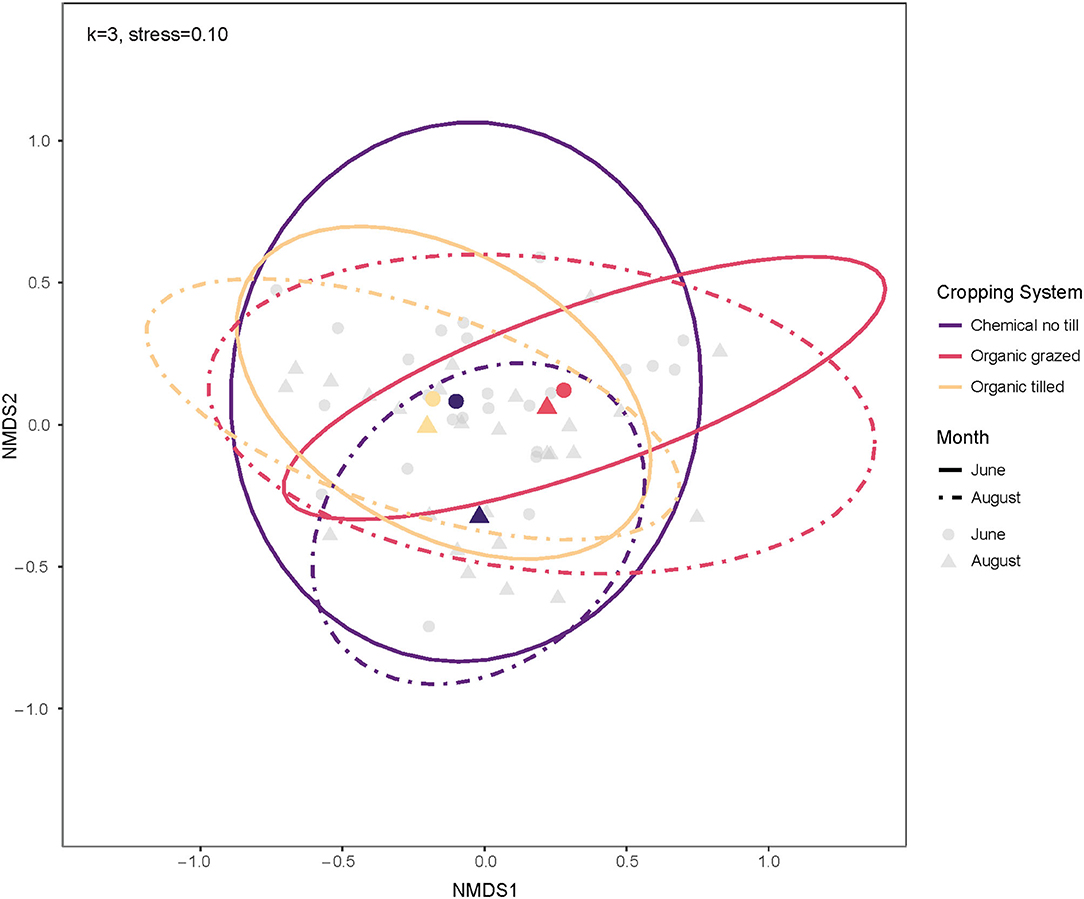
Figure 4. Ordination of soil bacterial communities by cropping system for June and August using non-metric multidimensional scaling on weighted Bray-Curtis dissimilarities. The centroids and multivariate t-distribution ellipses for bacterial communities in the three cropping systems are designated by color for both June (circles, solid line) and August (triangles, dashed line). For the NMDS calculation, k = 3, and stress = 0.10.
Soil microbial communities become more dissimilar throughout the summer, and this was driven more by cropping system than the crop phase (i.e., plant species identity) (Figure 4). June communities were 53.95% dissimilar from August communities on average. An analysis of dissimilarities between just the two organic systems was performed to determine if the chemical no-till system masked differences between organic grazed and organic tilled microbial communities. However, between-group and within-group mean dissimilarities for bacterial communities did not significantly change for cropping system, crop phase, or month.
Within crop phases, safflower/clover split-plots and clover split-plots had soil microbial communities with the highest mean dissimilarity at 54.65%, followed by clover and winter wheat split-plots at 53.60%. Bacterial communities in safflower/clover and winter wheat split-plots were 51.73% dissimilar, on average. Winter wheat bacterial communities were less dissimilar across cropping systems and had the lowest within-group mean dissimilarity of 51.09%. Organic grazed clover split-plots had dissimilar soil bacterial communities from the clover split-plots of the organic tilled and chemical no-till systems, and communities under organic tilled safflower/clover split plots were more dissimilar from the organic grazed and chemical no-till safflower/clover (Figure 5). Clover communities had the highest mean within-group dissimilarity in all cropping systems at 56.05%. Bray-Curtis dissimilarities did not differ among crop phases in the cropping systems, but there was an interaction between cropping system and month (p = 0.033). However, there were no significant post-hoc comparisons to indicate which systems changed in beta diversity as a function of month.
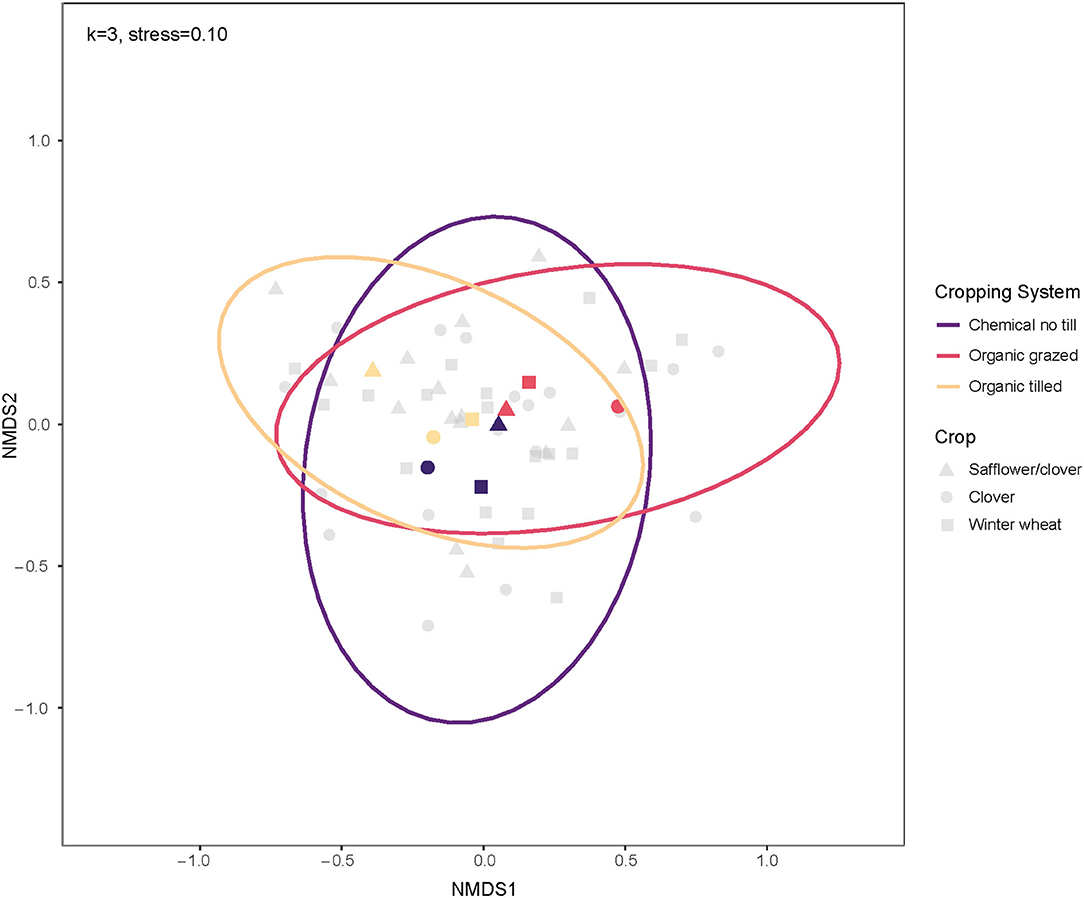
Figure 5. Ordination of soil bacterial communities by crop for three contrasting dryland cropping systems using non-metric multidimensional scaling on weighted Bray-Curtis dissimilarities. The centroids and multivariate t-distribution ellipses for bacterial communities in the three cropping systems are designated by color for cropping system and shape for soil growing clover (circles), safflower/clover (triangles), or winter wheat (squares). For the NMDS calculation, k = 3, and stress = 0.10.
Discussion
The sustainable productivity of arable land is dependent on preventing or remediating soil degradation, and alternatives to common agricultural practices need to be thoroughly studied to allow researchers to make accurate recommendations for producers. With that goal in mind, this study was part of a field experiment comparing the effects of cropping systems on crop production, soil bacterial communities, and other ecosystem dynamics. In this study, we confirmed that organically grazed systems support higher soil bacterial richness and Shannon's diversity than chemically managed systems but did not find evidence suggesting that soil bacterial diversity differed between organic systems or was higher in cover crops compared to winter wheat, when accounting for cropping systems and sampling month.
Differences Between Cropping Systems
Management practices used by chemical and organic systems can result in divergent microbial communities, principally due to the effects of tillage, fungicides, fertilizers, herbicides, and divergent plant communities, factors which are known to affect macro- and microbial diversity (Hartmann et al., 2015; Smith et al., 2015; Nettles et al., 2016; Zuber and Villamil, 2016). In this study, the chemical no-till system resulted in the least diverse microbial communities, which were dissimilar to those communities under grazed but not tilled organic management. Both organic systems in this study utilized different levels of tillage for weed management and cover crop termination. Specifically, the organic grazed system did not receive any tillage for the first 3 years of the rotation while soil at the organic tilled system was mechanically disturbed on a regular basis during this study. Tillage can act as a selective pressure leading to a difference in soil microbial community composition under contrasting soil disturbance regimes (Navarro-Noya et al., 2013). Breaking apart soil aggregates exposes previously protected soil organic matter to oxidizing conditions (Six et al., 2000; Van Groenigen et al., 2010) favorable to fast growing copiotrophic bacteria (Srour et al., 2020). However, findings on the effects of tillage on soil microbial communities are not consistent. Studies have reported higher microbial richness and diversity in no-till systems compared to tilled systems while others find no difference between the two (Navarro-Noya et al., 2013).
No Impact of Crop Rotation Phase
Soil microbial community alpha diversity was less affected by plant species in the crop rotation than by the overall cropping system and seasonality, a result previously demonstrated in this field experiment (Ishaq et al., 2020a). The lack of a response to crop phase (i.e., plant species identity) indicates other factors were more important in determining microbial community composition. It is presumed that chemical- vs. organic-based inputs and soil disruptions from tillage (Ishaq et al., 2020a), as well as seasonal temperature and precipitation (Ishaq et al., 2020b), are stronger selective pressures of soil microbial communities than the effects that crop species have on quality of plant residue inputs (Schmatz et al., 2017) or microbial-recruitment by plants (Ishaq et al., 2017). A similar study of crop rotations and management strategies in the Central Great Plains found that conservation tillage and arable weed diversity affected soil microbial communities much more than cover crop diversity (Wortman et al., 2013).
This study identified that soil microbial communities were impacted by an interaction between cropping systems and crop identity. In particular, within-group community dissimilarity was higher in the organic grazed system, regardless of crop. Soil bacteria from clover subplots had the highest within-group dissimilarity in all cropping systems, indicating that local environmental factors created more variation in the membership of bacterial communities. This may be due to the effect of local soil conditions on the soil microbial assembly (Brown et al., 2020), or to genetic variation in plants and their relative interactions with soil microbiota (Pérez-Jaramillo et al., 2019; Brown et al., 2020).
Previous research on the impact of tillage and crop identity on soil microbial communities indicated that tillage selects for faster growing taxa while cover crops select for moderate-growth taxa with more biochemical capacities (Schmidt et al., 2018). Moreover, the crop species used in rotations can alter soil microbial communities both taxonomically and functionally. While clover is known to recruit nitrogen-fixing bacteria in root nodules, clover and grass crops increase soil respiration from the community as a whole and presumably digestion of complex carbohydrates (Martínez-García et al., 2018). The effects of safflower planted with clover on soil microbiota has not been thoroughly evaluated, but previous studies linked safflower fertility to high soil bacterial abundance and low fungal abundance (Lu et al., 2013). This may be due to the phosphate-solubilizing bacteria recruited to the safflower rhizosphere (Zhang et al., 2019) taking the place of mutualistic soil fungi that create bioavailable phosphorus, which form the basis of their nutritional symbiosis with plants in exchange for sugars.
In the present study, given the drought resistance of safflower, the diversity of two plant species in the safflower/clover year, and the bacterial recruitment capacity of clover, it was anticipated that the bacterial diversity in soil would differ among crop phases. It was expected that winter wheat would recruit the lowest bacterial diversity, clover the next highest, and safflower/clover would result in the highest bacterial diversity. Variation in bacterial diversity among crop phases was moderated by cropping system and did not differ based solely on crop. Safflower/clover did not have a higher bacterial richness, but it did exhibit less variability in richness between plots and appeared to lose fewer taxa between June and August. Climate data at the experimental site show a trend of decreasing precipitation and increasing temperature over the course of the growing season (Adhikari and Menalled, 2020), which can lead to a decrease in microbial community diversity (Naylor and Coleman-Derr, 2018).
Differences Between Sampling Months
Low bacterial richness observed in August was presumably related to low soil moisture due to severely dry late summer conditions, an effect which has been observed previously (Fuchslueger et al., 2014; de Vries et al., 2018; Naylor and Coleman-Derr, 2018; Ishaq et al., 2020b). Soil bacteria are in closer contact with their surroundings and are limited by the availability of resources. In addition to the lack of moisture itself, microbial communities are altered by a reduction in plant-soil feedbacks which occur under drought conditions (Fuchslueger et al., 2014; de Vries et al., 2018). The large decrease in alpha diversity observed in the chemical no-till safflower/clover split-plots may have been compounded by crop senescence in these plots, heightening community vulnerability to dry soil conditions.
Dissimilarity in soil communities between June and August reflect a reduction in species richness and an increase in evenness associated with late summer dry soil conditions. In agreement with Ishaq et al. (2020b), we observed that soil bacterial communities become more disparate toward the end of the growing season when plant growth and moisture are no longer selecting bacterial growth and localized differences in fields may determine which bacteria can survive. If these spatially specific effects persist over time, it can lead to legacy effects on the microbial community over several growing seasons, affecting system resiliency (Seipel et al., 2019).
Overall, soil microbial communities in bulk soil respond to overarching management systems but not necessarily crop species. Conditions within the growing season such as precipitation and soil moisture have a more pronounced effect on community richness and composition than crop phase. Looking to future research, this study generated additional hypotheses and considerations. The rhizosphere needs to be sampled rather than bulk soil to assess how microbiota respond to the specific phases of a crop rotation (i.e., plant species identity), as differences in bulk soil microbial communities among the crop rotation plots were not detectable. Additionally, studies need to incorporate multiple time points in the growing season and assess long-term changes in soil microbial communities caused by disturbances. This knowledge, in turn, will allow an improved understanding of how management systems and their associated ecological disturbances create circumstances from which it is more difficult for microbial communities, soil health, and plant productivity to recover.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna672991, PRJNA672991.
Author Contributions
TO performed data analysis, interpretation, and wrote the manuscript. JE performed laboratory protocols and facilitated sequencing. SI contributed to data analysis code development. JE, TS, FM, and SI performed high-level development of the experimental design and analysis, as well as review of the manuscript. FM was principal investigator for USDA funding and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded with a grant from the United States Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA)'s Agriculture and Food Research Initiative (AFRI) Competitive Grant (Menalled, Accession: 1014774). This publication was supported with funds from the University of Maine.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Fort Ellis personnel for sheep maintenance, Pat Hatfield and Perry Miller for farm administration, and to members of the Menalled/Seipel lab for assistance with soil collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.624242/full#supplementary-material
References
Adhikari, S., Adhikari, A., Weaver, D. K., Bekkerman, A., and Menalled, F. D. (2019). Impacts of agricultural management systems on biodiversity and ecosystem services in highly simplified dryland landscapes. Sustainability 11:3223. doi: 10.3390/su11113223
Adhikari, S., and Menalled, F. D. (2018). Impacts of dryland farm management systems on weeds and ground beetles (Carabidae) in the Northern Great Plains. Sustainability 10:2146. doi: 10.3390/su10072146
Adhikari, S., and Menalled, F. D. (2020). Supporting beneficial insects for agricultural sustainability: the role of livestock-integrated organic and cover cropping to enhance ground beetle (Carabidae) communities. Agronomy 10:1210. doi: 10.3390/agronomy10081210
Aguilera, A. G., Morey, S., Gammon, M., Jiang, M., Ramos, S., and Kesseli, R. (2017). Effect of plant-soil feedbacks on the growth and competition of Lactuca species. Plant Ecol. 218, 359–372. doi: 10.1007/s11258-016-0697-3
Barsotti, J. L., Sainju, U. M., Lenssen, A. W., Montagne, C., and Hatfield, P. G. (2013). Crop yields and soil organic matter responses to sheep grazing in US northern Great Plains. Soil Tillage Res. 134, 133–141. doi: 10.1016/j.still.2013.07.015
Bender, S. F., and van der Heijden, M. G. A. (2015). Soil biota enhance agricultural sustainability by improving crop yield, nutrient uptake and reducing nitrogen leaching losses. J. Appl. Ecol. 52, 228–239. doi: 10.1111/1365-2664.12351
Brown, S. P., Grillo, M. A., Podowski, J. C., and Heath, K. D. (2020). Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 8:139. doi: 10.1186/s40168-020-00915-9
Brussaard, L., de Ruiter, P. C., and Brown, G. G. (2007). Soil biodiversity for agricultural sustainability. Agric. Ecosyst. Environ. 121, 233–244. doi: 10.1016/j.agee.2006.12.013
Calderón, F. J., Nielsen, D., Acosta-Martínez, V., Vigil, M. F., and Lyon, D. (2016). Cover crop and irrigation effects on soil microbial communities and enzymes in semiarid agroecosystems of the Central Great Plains of North America. Pedosphere 26, 192–205. doi: 10.1016/S1002-0160(15)60034-0
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carr, P. M. (2017). Guest editorial: Conservation tillage for organic farming. Agriculture 7:19. doi: 10.3390/agriculture7030019
Castillo, C. G., Rubio, R., Rouanet, J. L., and Borie, F. (2006). Early effects of tillage and crop rotation on arbuscular mycorrhizal fungal propagules in an Ultisol. Biol. Fertil. Soils 43, 83–92. doi: 10.1007/s00374-005-0067-0
Chaudhry, V., Rehman, A., Mishra, A., Chauhan, P. S., and Nautiyal, C. S. (2012). Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 64, 450–460. doi: 10.1007/s00248-012-0025-y
Clay, D. E., Clay, S. A., Reitsma, K. D., Dunn, B. H., Smart, A. J., Carlson, G. G., et al. (2014). Does the conversion of grasslands to row crop production in semi-arid areas threaten global food supplies? Glob. Food Secur. 3, 22–30. doi: 10.1016/j.gfs.2013.12.002
D'Acunto, L., Andrade, J. F., Poggio, S. L., and Semmartin, M. (2018). Diversifying crop rotation increased metabolic soil diversity and activity of the microbial community. Agric. Ecosyst. Environ. 257, 159–164. doi: 10.1016/j.agee.2018.02.011
de Quadros, P. D., Zhalnina, K., Davis-Richardson, A., Fagen, J. R., Drew, J., Bayer, C., et al. (2012). The effect of tillage system and crop rotation on soil microbial diversity and composition in a subtropical acrisol. Diversity 4, 375–395. doi: 10.3390/d4040375
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3022. doi: 10.1038/s41467-018-05516-7
Delgado-Baquerizo, M., Eldridge, D. J., Ochoa, V., Gozalo, B., Singh, B. K., and Maestre, F. T. (2017). Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 20, 1295–1305. doi: 10.1111/ele.12826
Doran, J. W., and Zeiss, M. R. (2000). Soil health and sustainability: managing the biotic component of soil quality. Appl. Soil Ecol. 15, 3–11. doi: 10.1016/S0929-1393(00)00067-6
Drijber, R. A., Doran, J. W., Parkhurst, A. M., and Lyon, D. J. (2000). Changes in soil microbial community structure with tillage under long-term wheat-fallow management. Soil Biol. Biochem. 32, 1419–1430. doi: 10.1016/S0038-0717(00)00060-2
Frasier, I., Noellemeyer, E., Figuerola, E., Erijman, L., Permingeat, H., and Quiroga, A. (2016). High quality residues from cover crops favor changes in microbial community and enhance C and N sequestration. Glob. Ecol. Conserv. 6, 242–256. doi: 10.1016/j.gecco.2016.03.009
Fuchslueger, L., Bahn, M., Fritz, K., Hasibeder, R., and Richter, A. (2014). Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. New Phytol. 201, 916–927. doi: 10.1111/nph.12569
García-Orenes, F., Morugán-Coronado, A., Zornoza, R., Cerdà, A., and Scow, K. (2016). Correction: Changes in soil microbial community structure influenced by agricultural management practices in a Mediterranean agro-ecosystem. PLoS ONE 11:e0152958. doi: 10.1371/journal.pone.0152958
Harkes, P., Suleiman, A. K. A., van den Elsen, S., de Haan, J., Holterman, M., Kuramae, E. E., et al. (2019). Conventional and organic soil management as divergent drivers of resident and active fractions of major soil food web constituents. Sci. Rep. 9:13521. doi: 10.1038/s41598-019-49854-y
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Helgason, B. L., Walley, F. L., and Germida, J. J. (2010). Long-term no-till management affects microbial biomass but not community composition in Canadian prairie agroecosytems. Soil Biol. Biochem. 42, 2192–2202. doi: 10.1016/j.soilbio.2010.08.015
Ishaq, S. L., Johnson, S. P., Miller, Z. J., Lehnhoff, E. A., Olivo, S. K., Yeoman, C. J., et al. (2017). Impact of cropping systems, soil inoculum, and plant species identity on soil bacterial community structure. Microb. Ecol. 73, 417–434. doi: 10.1007/s00248-016-0861-2
Ishaq, S. L., Seipel, T., Yeoman, C. J., and Menalled, F. D. (2020a). Soil bacterial communities of wheat vary across the growing season and among dryland farming systems. Geoderma 358:113989. doi: 10.1016/j.geoderma.2019.113989
Ishaq, S. L., Seipel, T., Yeoman, C. J., and Menalled, F. D. (2020b). Dryland cropping systems, weed communities, and disease status modulate the effect of climate conditions on wheat soil bacterial communities. mSphere 5, e00340–e00320. doi: 10.1128/mSphere.00340-20
Jat, H. S., Datta, A., Choudhary, M., Yadav, A. K., Choudhary, V., Sharma, P. C., et al. (2019). Effects of tillage, crop establishment and diversification on soil organic carbon, aggregation, aggregate associated carbon and productivity in cereal systems of semi-arid Northwest India. Soil Tillage Res. 190, 128–138. doi: 10.1016/j.still.2019.03.005
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-indexed sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lal, R. (2016). Soil health and carbon management. Food Energy Secur. 5, 212–222. doi: 10.1002/fes3.96
Landis, D. A. (2017). Designing agricultural landscapes for biodiversity-based ecosystem services. Basic Appl. Ecol. 18, 1–12. doi: 10.1016/j.baae.2016.07.005
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Li, R., Khafipour, E., Krause, D. O., Entz, M. H., de Kievit, T. R., and Fernando, W. G. D. (2012). Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PLoS ONE 7:12. doi: 10.1371/journal.pone.0051897
Lu, S., Quan, W., Wang, S. M., Liu, H. L., Tan, Y., Zeng, et al. (2013). Correlation of soil microbes and soil micro-environment under long-term safflower (Carthamus tinctorius L.) plantation in China. J. Environ. Biol. 34, 471–479.
Lupatini, M., Korthals, G. W., de Hollander, M., Janssens, T. K. S., and Kuramae, E. E. (2017). Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 7:2064. doi: 10.3389/fmicb.2016.02064
Maarastawi, S. A., Frindte, K., Linnartz, M., and Knief, C. (2018). Crop rotation and straw application impact microbial communities in Italian and Philippine soils and the rhizosphere of Zea mays. Front. Microbiol. 9:1295. doi: 10.3389/fmicb.2018.01295
Martínez-García, L. B., Korthals, G., Brussaard, L., Jørgensen, H. B., and De Deyn, G. B. (2018). Organic management and cover crop species steer soil microbial community structure and functionality along with soil organic matter properties. Agric. Ecosyst. Environ. 263, 7–17. doi: 10.1016/j.agee.2018.04.018
Massenssini, A. M., Bonduki, V. H. A., Melo, C. A. D., Tótola, M. R., Ferreira, F. A., and Costa, M. D. (2015). Relative importance of soil physicochemical characteristics and plant species identity to the determination of soil microbial community structure. Appl. Soil Ecol. 91, 8–15. doi: 10.1016/j.apsoil.2015.02.009
McDaniel, M. D., Tiemann, L. K., and Grandy, A. S. (2014). Does agricultural crop diversity enhance soil microbial biomass and organic matter dynamics? A meta-analysis. Ecol. Appl. 24, 560–570. doi: 10.1890/13-0616.1
McKenzie, S. C., Goosey, H. B., O'Neill, K. M., and Menalled, F. D. (2016). Integration of sheep grazing for cover crop termination into market gardens: agronomic consequences of an ecologically based management strategy. Renew. Agric. Food Syst. 32, 389–402. doi: 10.1017/S1742170516000326
McLauchlan, K. (2006). The nature and longevity of agricultural impacts on soil carbon and nutrients: a review. Ecosystems 9, 1364–1382. doi: 10.1007/s10021-005-0135-1
McMurdie, P. J., and Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:4. doi: 10.1371/journal.pone.0061217
Menalled, F. D., Peterson, R. K. D., Smith, R. G., Curran, W. S., Páez, D. J., and Maxwell, B. D. (2016). The eco-evolutionary imperative: revisiting weed management in the midst of an herbicide resistance crisis. Sustainability 8:1297. doi: 10.3390/su8121297
Menalled, U. D., Seipel, T., and Menalled, F. D. (2020). Farming system effects on biologically mediated plant–soil feedbacks. Renew. Agric. Food Syst. 36, 1–7. doi: 10.1017/S1742170519000528
Miller, R. M., and Jastrow, J. D. (2000). “Mycorrhizal Fungi Influence Soil Structure” in Arbuscular Mycorrhizas: Physiology and Function, eds Y. Kapulnik and D. Jr. Douds (Dordrecht: Kluwer Academic Publishers), 3–18. doi: 10.1007/978-94-017-0776-3_1
Miller, Z. J., Menalled, F. D., Sainju, U. M., Lenssen, A. W., and Hatfield, P. G. (2015). Integrating sheep grazing into cereal-based crop rotations: spring wheat yields and weed communities. Agronomy 107, 104–112. doi: 10.2134/agronj14.0086
Mohite, B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. Soil Sci. Plant Nutr. 13, 638–649. doi: 10.4067/S0718-95162013005000051
Natural Resources Conservation Service Soil Survey Staff (1999). Soil Taxonomy: A Basic System of Soil Classification for Making and Interpreting Soil Surveys. 2nd Edn. Washington, DC: Natural Resources Conservation Service; U.S. Department of Agriculture Handbook.
Navarro-Noya, Y. E., Gómez-Acata, S., Montoya-Ciriaco, N., Rojas-Valdez, A., Suárez-Arriaga, M. C., Valenzuela-Encinas, et al. (2013). Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agroecosystem. Soil Biol. Biochem. 65, 86–95. doi: 10.1016/j.soilbio.2013.05.009
Naylor, D., and Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223
Nettles, R., Watkins, J., Ricks, K., Boyer, M., Licht, M., Atwood, L. W., et al. (2016). Influence of pesticide seed treatments on rhizosphere fungal and bacterial communities and leaf fungal endophyte communities in maize and soybean. Appl. Soil Ecol. 102, 61–69. doi: 10.1016/j.apsoil.2016.02.008
Nivelle, E., Verzeaux, J., Habbib, H., Kuzyakov, Y., Decocq, G., Roger, D., et al. (2016). Functional response of soil microbial communities to tillage, cover crops and nitrogen fertilization. Appl. Soil Ecol. 108, 147–155. doi: 10.1016/j.apsoil.2016.08.004
Okansen, J., Blanchet, F. B., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R Package Version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Peralta, A. L., Sun, Y., McDaniel, M. D., and Lennon, J. T. (2018). Crop rotational diversity increases disease suppressive capacity of soil microbiomes. Ecosphere 9:2235. doi: 10.1002/ecs2.2235
Pérez-Jaramillo, J. E., de Hollander, M., Ramírez, C. A., Mendes, R., Raaijmakers, J. M., and Carrión, V. J. (2019). Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 7:114. doi: 10.1186/s40168-019-0727-1
PRISM Climate Group. (2020). Oregon State University. Available online at: http://www.prism.oregonstate.edu/ (accessed May, 5, 2020).
R Development Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://CRAN.R-project.org
Rosenzweig, S. T., Fonte, S. J., and Schipanski, M. E. (2018). Intensifying rotations increases soil carbon, fungi, and aggregation in semi-arid agroecosystems. Agric Ecosyst Environ 258, 14–22. doi: 10.1016/j.agee.2018.01.016
Sapkota, T. B., Mazzoncini, M., Bàrberi, P., Antichi, D., and Silvestri, N. (2012). Fifteen years of no till increase soil organic matter, microbial biomass and arthropod diversity in cover crop-based arable cropping systems. Agron. Sustain. Dev. 32, 853–863. doi: 10.1007/s13593-011-0079-0
Schmatz, R., Recous, S., Aita, C., Tahir, M. M., Schu, A. L., Chaves, B., et al. (2017). Crop residue quality and soil type influence the priming effect but not the fate of crop residue C. Plant Soil 414, 229–245. doi: 10.1007/s11104-016-3120-x
Schmidt, R., Gravuer, K., Bossange, A. V., Mitchell, J., and Scow, K. (2018). Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS ONE 13:2. doi: 10.1371/journal.pone.0192953
Schneekloth, J., Calderón, F. J., Nielsen, D., and Fonte, S. J. (2020). Tillage and residue management effects on irrigated maize performance and water cycling in a semiarid cropping system of Eastern Colorado. Irrig. Sci. 38, 547–557. doi: 10.1007/s00271-020-00702-2
Seipel, T., Ishaq, S. L., and Menalled, F. D. (2019). Agroecosystem resilience is modified by management system via plant–soil feedbacks. Basic Appl. Ecol. 39, 1–9. doi: 10.1016/j.baae.2019.06.006
Semenov, M. V., Krasnov, G. S., Semenov, V. M., and van Bruggen, A. H. C. (2020). Long-term fertilization rather than plant species shapes rhizosphere and bulk soil prokaryotic communities in agroecosystems. Appl. Soil Ecol. 154:103641. doi: 10.1016/j.apsoil.2020.103641
Six, J., Elliott, E. T., and Paustian, K. (2000). Soil macroaggregate turnover and microaggregate formation: a mechanism for C sequestration under no-tillage agriculture. Soil Biol. Biochem. 32, 2099–2103. doi: 10.1016/S0038-0717(00)00179-6
Smith, P., Cotrufo, M. F., Rumpel, C., Paustian, K., Kuikman, P. J., Elliott, J. A., et al. (2015). Biogeochemical cycles and biodiversity as key drivers of ecosystem services provided by soils. Soil 1, 665–685. doi: 10.5194/soil-1-665-2015
Srour, A. Y., Ammar, H. A., Subedi, A., Pimentel, M., Cook, R. L., Bond, J., et al. (2020). Microbial communities associated with long-term tillage and fertility treatments in a corn-soybean cropping system. Front. Microbiol. 11:1363. doi: 10.3389/fmicb.2020.01363
Trognitz, F., Hackl, E., Widhalm, S., and Sessitsch, A. (2016). The role of plant-microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 92:10. doi: 10.1093/femsec/fiw138
University of California, Davis (UC Davis), California Soil Resource Lab; University of California, Division of Agriculture and Natural Resources; Natural Resources Conservation Service. (2019). SoilWeb. University of California; USDA-NRCS. Available online at: https://casoilresource.lawr.ucdavis.edu/gmap/ (accessed October 19, 2020).
USDA ERS. (2020). Organic Market Overview. Economic Research Service. Available online at: https://www.ers.usda.gov/topics/natural-resources-environment/organic-agriculture/organic-market-overview.aspx (accessed January 4, 2021).
van der Heijden, M. G. A., de Bruin, S., Luckerhoff, L., van Logtestijn, R. S. P., and Schlaeppi, K. (2015). A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. doi: 10.1038/ismej.2015.120
Van Groenigen, K. J., Bloem, J., Bååth, E., Boeckx, P., Rousk, J., Bodé, S., et al. (2010). Abundance, production and stabilization of microbial biomass under conventional and reduced tillage. Soil Biol. Biochem. 42, 48–55. doi: 10.1016/j.soilbio.2009.09.023
Vanwalleghem, T., Gómez, J. A., Infante Amate, J., González de Molina, M., Vanderlinden, K., Guzmán, G., et al. (2017). Impact of historical land use and soil management change on soil erosion and agricultural sustainability during the Anthropocene. Anthropocene 17, 13–29. doi: 10.1016/j.ancene.2017.01.002
Venter, Z. S., Jacobs, K., and Hawkins, H. J. (2016). The impact of crop rotation on soil microbial diversity: a meta-analysis. Pedobiologia 59, 215–223. doi: 10.1016/j.pedobi.2016.04.001
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis, 2nd Edn. New York, NY: Springer-Verlag. doi: 10.1007/978-3-319-24277-4
Wortman, S. E., Drijber, R. A., Francis, C. A., and Lindquist, J. L. (2013). Arable weeds, cover crops, and tillage drive soil microbial community composition in organic cropping systems. Appl. Soil Ecol. 72, 232–241. doi: 10.1016/j.apsoil.2013.07.014
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–648. doi: 10.1093/nar/gkt1209
Zhalina, K., Dias, R., de Quadros, P. D., Davis-Richardson, A., Camargo, F. A. O., Clark, I. M., et al. (2015). Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 69, 395–406. doi: 10.1007/s00248-014-0530-2
Zhang, T., Hu, F., and Ma, L. (2019). Phosphate-solubilizing bacteria from safflower rhizosphere and their effect on seedling growth. Open Life Sci. 14, 246–254. doi: 10.1515/biol-2019-0028
Zhang, Y., Shen, H., He, X., Thomas, B. W., Lupwayi, N. Z., Hao, X., et al. (2017). Fertilization shapes bacterial community structure by alteration of soil pH. Front. Microbiol. 8:1325. doi: 10.3389/fmicb.2017.01325
Keywords: 16S rRNA gene, chemical no-tillage, yellow sweetclover [Melilotus officinalis (L.)], Illumina MiSeq®, organic grazed, organic tilled, winter wheat (Triticum aestivum L), safflower (Carthamus tinctorius L.)
Citation: Ouverson T, Eberly J, Seipel T, Menalled FD and Ishaq SL (2021) Temporal Soil Bacterial Community Responses to Cropping Systems and Crop Identity in Dryland Agroecosystems of the Northern Great Plains. Front. Sustain. Food Syst. 5:624242. doi: 10.3389/fsufs.2021.624242
Received: 30 October 2020; Accepted: 23 February 2021;
Published: 26 March 2021.
Edited by:
Saveetha Kandasamy, A&L Canada Laboratories, CanadaReviewed by:
Luciano Gebler, Brazilian Agricultural Research Corporation (EMBRAPA), BrazilNikolaos Vasileios Paranychianakis, Technical University of Crete, Greece
Vivian A. Rincon-Florez, The University of Queensland, Australia
Copyright © 2021 Ouverson, Eberly, Seipel, Menalled and Ishaq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tindall Ouverson, dGluZGFsbC5vdXZlcnNvbkBnbWFpbC5jb20=
 Tindall Ouverson
Tindall Ouverson Jed Eberly
Jed Eberly Tim Seipel
Tim Seipel Fabian D. Menalled
Fabian D. Menalled Suzanne L. Ishaq
Suzanne L. Ishaq