- Department of Horticultural Science, University of Minnesota, Saint Paul, MN, United States
Legumes are of primary importance for agroecosystems because they provide protein-rich foods and enhance soil fertility through fixed atmospheric nitrogen. The legume-rhizobia symbiosis that makes this possible has been extensively studied, from basic research on biochemical signaling to practical applications in cropping systems. While rhizobia are the most-studied group of associated microorganisms, the functional benefit they confer to their legume hosts by fixing nitrogen is not performed in isolation. Indeed, non-rhizobia members of the rhizosphere and nodule microbiome are now understood to contribute in multiple ways to nodule formation, legume fitness, and other agroecosystem services. In this review, we summarize advances contributing to our understanding of the diversity and composition of bacterial members of the belowground legume microbiome. We also highlight applied work in legume food and forage crops that link microbial community composition with plant functional benefits. Ultimately, further research will assist in the development of multi-species microbial inoculants and cropping systems that maximize plant nutrient benefits, while reducing sources of agricultural pollution.
Introduction
Legumes are at the heart of sustainable agricultural strategies to alleviate global hunger, reduce the carbon footprint of farming, and improve soil fertility (Peoples et al., 2009; Smith et al., 2016). As one of the main sources of nitrogen (N) in terrestrial soils, biological nitrogen fixation (BNF) performed via interkingdom legume-rhizobia symbiosis is critical for ecosystem functioning. It is estimated that agriculturally important legumes fix between 40 and 90 Tg N yr−1 (Galloway et al., 2008; Taylor et al., 2020). Although humans have studied BNF for nearly 200 years, our understanding of how legumes impact agroecosystem function is still evolving. While scientific advances have propelled cereal crop yields to historic levels in the past 50 years, advances in grain legume production lag far behind with annual yield increases stalling at 0–2% (Foyer et al., 2016).
In the following sections, we highlight work that has expanded our understanding of the legume microbiome beyond rhizobia-associated benefits to soil fertility from BNF. Plant microbiomes are defined as complex networks of bacteria, fungi, viruses, and other microfauna that interact with the plant host on a cellular level or may impact plant fitness by modifying the immediate soil environment (Quiza et al., 2015; Leach et al., 2017). In this mini-review, we limit our focus to legume interactions with bacterial communities within the nodule and rhizosphere.
Legume-Associated Rhizobacterial Communities
Mounting evidence pointing to the breadth of microbial taxa that influence and are influenced by legumes suggests that this is an area scientists should critically examine to maximize potential benefits from legume crops. Importantly, we do not yet understand the specific mechanisms involved in rhizobacterial recruitment, their functional implications at the field scale, or the specificity of the interaction. This is partly due to difficulties in parsing out effects of rhizodeposition from other dynamic soil processes in the rhizosphere (Tsiknia et al., 2021). Below, we describe work that examines legume-mediated impacts on rhizobacterial communities with potential consequences for broader agroecosystem functioning.
Legume-Microbial Signaling in the Rhizosphere
Rhizospheric legume-microbe interactions and bacterial community composition are largely mediated by root exudates (Tian et al., 2020), the synthesis of which can account for 10–20% of net photosynthetic carbon (Tsiknia et al., 2021). Intracellular symbiosis with rhizobia and capacity for BNF result in some specific differences in how legumes modify rhizobacterial communities compared to non-legumes.
Legumes initiate microbial interactions in the rhizosphere through communication pathways involving root-exudates known as flavonoids which trigger the production of nodulation (Nod) factors by compatible rhizobia symbionts (Peck et al., 2006; Wang et al., 2018). Flavonoid production is further stimulated by legume root perception of Nod factors, creating a positive feedback between legume and rhizosphere signaling that is reinforced by rhizobia presence (Liu and Murray, 2016). Studies involving mutant legumes unable to nodulate or form mycorrhizal symbioses revealed significant effects of symbiont absence on bacteriome structure in the rhizosphere (Uroz et al., 2019). Nodulation induces unique metabolic changes within the legume host that may explain this effect, including the production of cysteine-rich peptides in some legumes that control bacteroid differentiation but also act as antimicrobials against non-symbionts (Maróti et al., 2015). However, the role of these peptides in modulating microbial community structure has not yet been thoroughly tested.
Flavonoids not only initiate symbiosis with rhizobia but also play a central role in shaping legume rhizosphere community structure. In vitro experiments using RNAi soybean roots deficient in flavonoid production showed that flavonoids significantly affect rhizosphere bacterial community composition (White et al., 2015). One explanation might be that the common symbiosis pathway in legumes is shared beyond rhizobia and arbuscular mycorrhizal fungi (AMF) (Skiada et al., 2020). For example, Medicago truncatula mutants unable to establish AMF symbiosis grown in field soil under greenhouse conditions had reduced microbial abundance in the rhizosphere, suggesting a role for conserved symbiotic pathways in rhizosphere enrichment processes (Wang et al., 2021). Although flavonoids are present across the plant kingdom (e.g., Lapcik et al., 2006; Tian et al., 2021), accumulated evidence suggests they are particularly prevalent among legumes (Veitch, 2007; Jasiński et al., 2009) and their importance in multiple initiation and regulatory functions pertaining to intracellular symbiosis cannot be understated. While there is limited research on differences in flavonoid profiles among legumes and non-legumes, Weston and Mathesius (2013) note that species-specific differences in root flavonoid exudation could be responsible for various root-microbe interactions. Extensive metabolic and proteomic root exudate profiling under field conditions is needed to clarify the role of legume-derived flavonoids in rhizobacterial selection and enrichment.
In addition to flavonoids and other compounds required for the symbiosis pathway, legumes differ from non-legumes by N rhizodeposition. Legumes release N-rich compounds, principally driven by the decomposition of senescing nodules and exudation of soluble nitrogenous compounds (Fustec et al., 2010). Due to the difficulty of measuring root-derived N under field conditions, estimates of legume N rhizodeposition as a percentage of total plant N vary widely from 4 to 71% (Fustec et al., 2010). Legume-derived N transforms the structure and function of rhizobacterial communities in ways that are distinct from non-legumes. Legume root zones are associated with greater microbial abundance and diversity, potentially by favoring certain metabolic profiles (e.g., N-cyclers) or fast growing r-strategists (Hamel et al., 2018). For example, a potted greenhouse experiment demonstrated that intercropping the legume Medicago sativa with Dactylis glomerata significantly increased root-associated N-cycling bacteria relative to the single-species D. glomerata (cat grass) treatment (Zhao et al., 2017). In another greenhouse study, legumes increased soil bacterial diversity and the abundance of certain groups (Verrumicrobia, Actinobacteria, and Nitrospirae) compared to grass treatments (Zhou et al., 2017).
Legumes and non-legumes alike selectively modify the composition of microbial communities through root exudates in ways that promote their own growth and fitness (Zhalnina et al., 2018). Mucilage, lysed cells, carbohydrates, volatile organic compounds (VOCs), and other secondary metabolites or signaling molecules (e.g., strigolactones) serve important functions in recruiting plant growth promoting bacteria (PGPBs) to alleviate biotic and abiotic stress in legumes and non-legumes (Vives-Peris et al., 2020; Figure 1). Compared to grasses, some legumes secrete a greater quantity of sugars and amino acids, potentially because of positive feedbacks with symbionts and PGPBs in the rhizosphere (Cesco et al., 2010). Weisskopf et al. (2008) found that greater organic acid exudation by lupin roots compared to wheat in a microcosm explained 15% of the observed variance in rhizobacterial abundance between the two species. More evidence is needed to clarify unique features of legume root exudate profiles and functional impacts on rhizobacteria across a broader species range.
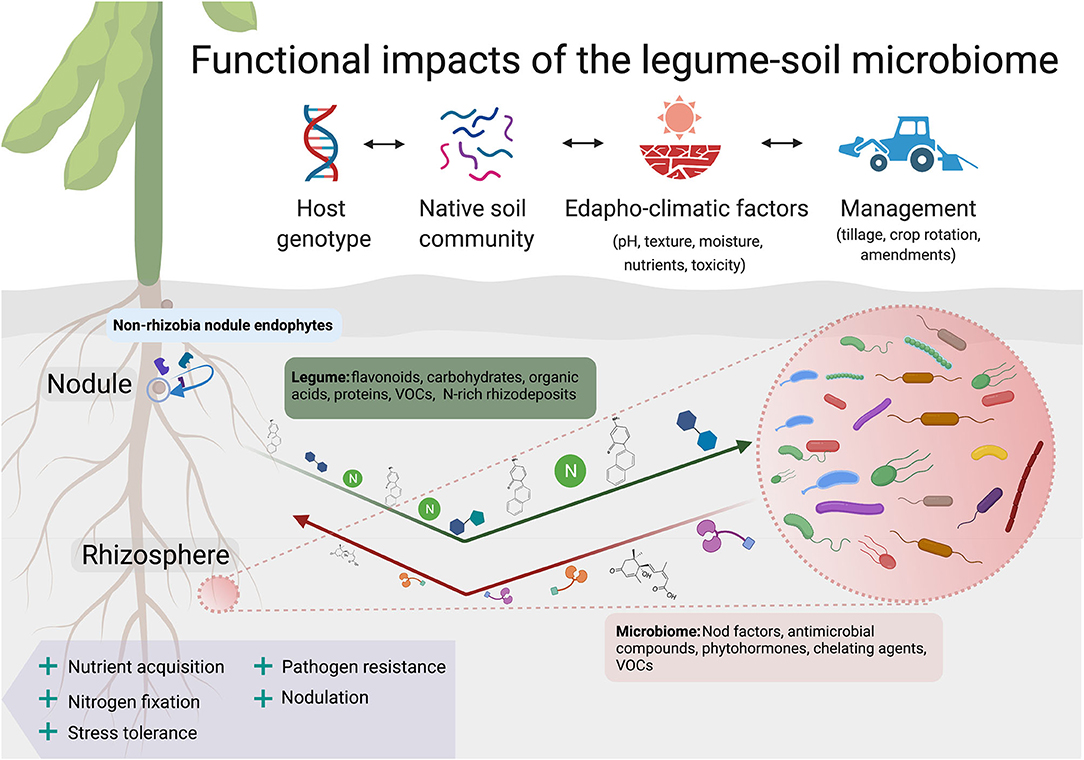
Figure 1. Legumes form interspecific associations with endophytes and enrich the growth of other microbial communities within the rhizosphere. Plant-derived factors in the rhizosphere such as flavonoids, allelopathic phytochemicals, and nitrogenous compounds preferentially favor the growth of certain groups over others. This “filtering effect” (Xiao et al., 2017) ultimately leads to a decrease in diversity with increasing proximity to the legume root surface. The soil microbiome in turn influences legume performance through rhizospheric signaling that includes Nod factors, phytohormones, volatile organic compounds (VOCs), and antimicrobial compounds. Following legume root and nodule senescence, rhizobia, and other endophytes are released back into the rhizosphere. Multiple seasons of legume cropping may cause shifts in the abundance and activity of symbionts, PGPBs, and other rhizosphere-associated groups over time (Thies et al., 1995; Ai et al., 2015). Legume impacts on the structure and function of the soil microbiome are mediated by numerous environmental factors and management practices, such as soil pH, tillage, and residue management. Created with BioRender.com.
Plant Growth Promotion
We are at an emergent stage of understanding the functional relevance of recruited rhizobacteria among major agricultural grain legumes, although early studies have suggested a role in adaptation to abiotic stress. In a greenhouse experiment using field collected soils, soybean rhizosphere isolates (Bacillus cereus) alleviated the inhibitory effects of salt stress on nodulation when co-inoculated with Sinorhizobia (Han et al., 2020).
Recent studies have documented rhizobacterial-mediated stress adaptation under field conditions that is strongly influenced by legumes and rhizobia rather than soil factors. Kamutando et al. (2017) showed that the invasive legume Acacia dealbata associates with a core set of microbial rhizosphere taxa across eight geographically diverse locations in South Africa. Subsequent metagenomic sequence analysis revealed higher abundance of plant growth promoting traits in the rhizosphere compared to bulk soil, with the majority involved in nutrient metabolism and membrane transport systems (Kamutando et al., 2019). Amplicon sequence analysis of Kersting's groundnut rhizobacterial communities from a field trial in Ghana similarly identified the enrichment of a core group of taxa that could be involved in plant growth promoting functions such as indole-3-acetic acid (IAA) production (Jaiswal et al., 2019). These results suggest the potential to exploit legume rhizospheric interactions to improve agroecosystem productivity under a changing climate.
Studies focused on agroecosystem-wide benefits suggest legume-mediated PGPB enrichment in the rhizosphere also improves non-legume companion crop performance. Benefits accrued to non-legume crops in rotational systems with legumes could be due to soil “legacy effects,” which are residual soil-transferred effects imparted to the microbiome following legume removal. We have known about legume-derived benefits to succeeding crops for many years: Stevenson and van Kessel (1996) found that field pea rotations benefited wheat growth, with 91% of the yield benefit attributed to reduced disease pressure. While there was no connection made to plant-mediated microbiome shifts at the time, subsequent work made this link more apparent. For instance, Latz et al. (2012) found that legumes from a long-term field experiment promoted antifungal gene abundance among soil bacteria. Similarly, long term soybean cropping systems are known to reduce the diversity, abundance, and pathogenicity of the root rot-causing fungal pathogen Fusarium within the rhizosphere (Wei et al., 2014).
Duchene et al. (2017) recently proposed a “complementarity and facilitation” framework, wherein legume-mediated changes to rhizobacterial communities promote non-legume crop growth via niche differentiation and the enrichment of PGPBs. This means that legume residues or root metabolites favor the persistence of microbial groups such as symbionts (e.g., Burkholderia, Frankia, and Bradyrhizobia), N cyclers (e.g., Nitrospira and Nitrosomonas), or microbes involved in soil organic matter conversions (Fox et al., 2020). Hannula et al. (2020) found strong evidence for plant species-specific effects on soil microbes in a potted greenhouse experiment. The most abundant phyla in legume-conditioned soils were groups (Actinobacteria, Planctomycetes, and Alphaproteobacteria) that are known to form associations with rhizobia (Hartman et al., 2017; Hannula et al., 2020). Similarly, pea and chickpea–conditioned soils benefited wheat growth in a greenhouse study, potentially by increasing the abundance of PGPB including Proteobacteria (Yang et al., 2013).
The degree to which legacy effects from legumes persist over time is still unclear, especially in field conditions. Furthermore, many of the putative growth-promoting functions of enriched rhizosphere taxa have been inferred from sequence analysis that may not reflect their actual performance in vivo. Nevertheless, studying enriched PGPBs within the legume rhizosphere can help researchers identify taxa that can be used to benefit crop growth and inform future field experiments (Hartman et al., 2017).
The Legume Nodule Microbiome
In addition to the previously discussed co-occurrence of rhizobia with certain bacterial taxa in the rhizosphere, non-rhizobia endophytes (NREs) co-inhabit legume root nodules with rhizobia (Table 1). There is growing evidence that rhizobia are not the sole, nor always the most abundant (Muresu et al., 2008) residents of legume root nodules. This discovery can be attributed to advances in sequencing technology, as previous culture-based studies excluded colonies that did not conform to known rhizobia morphology or were not culturable at all (Howieson and Dilworth, 2016). For a comprehensive review on NREs, we refer readers to Martínez-Hidalgo and Hirsch (2017). Below, we focus on recent advances in this area.
Contribution of NREs to Legume Growth and Performance
Domestication and adaptation of legumes to the highly managed conditions of agricultural soils may have reduced nodule-associated microbial diversity, which could provide clues as to the functional benefits of NREs (Cardoso et al., 2018). Divergent NRE diversity between domestic crops and closely related wild relatives may be due to differences in root exudates and other evolutionary adaptations to high-nutrient environments (Pérez-Jaramillo et al., 2017; Fitzpatrick et al., 2018). Domesticated legume varieties often have larger seeds, a result of adaptation to high-input environments where soil N uptake is less costly than the symbiotic relationship required for BNF (Liu et al., 2020; Martínez-Romero et al., 2020). This could suggest an energetic benefit for wild legumes in diversifying nodule occupancy in N-limited soils.
There is no conclusive evidence that NRE themselves fix nitrogen from within the nodule, although N-fixing genes including nifH and narG have been successfully amplified from multiple NREs isolated from crimson clover nodules in the field (Moore et al., 2020). Importantly, the presence of these genes implies a genetic capability to reduce atmospheric nitrogen to ammonia although we don't have evidence that NREs transcribe these genes in planta. Not all NREs house N fixation genes: Bacillus, accounting for 17.9% of nodule endophytes among a survey of wild legumes in Belgium, was not found to contain N fixation genes (De Meyer et al., 2015).
Besides a putative role in N fixation, NREs may provide other benefits such as phosphate (P) solubilization (Hakim et al., 2020) that serve to increase rhizobia and host fitness. This may be particularly important in weathered and nutrient-deficient tropical soils, where many NREs have been recovered from wild legumes (Muresu et al., 2019). The mechanism and ecological relevance of NRE P solubilization is unknown, as it is not a function that has been confirmed outside of culture-based studies. Interestingly, P plasticity increases in nodules and surrounding root cells under P-limiting conditions (Vardien et al., 2016). Under such conditions, intracellular phosphohydrolases improve P availability for N fixation by “recycling” other cellular proteins. The link between NRE P solubilization phenotypes in vitro and cellular P cycling in planta requires further clarification.
NREs may also suppress plant pathogens. Tokgöz et al. (2020) found that NREs isolated from soybean nodules have antifungal and antibacterial properties that extend beyond the nodule and interact with non-legumes. The authors found that an isolate of the genus Proteus was effective in inhibiting the growth of several bacterial and fungal pathogens in vitro. Growth chamber inoculation experiments with the same strain suppressed bacterial speck disease in tomato seedlings. To our knowledge, this is the only study that has demonstrated that NREs provide pathogen protection in non-legume hosts. If legume nodules are indeed active sites of antimicrobial production (Hansen et al., 2020), it is possible that these benefits could accrue in adjacent or succeeding plants in cropping systems as nodules ultimately senesce (Müller et al., 2001) and the antibiotic producing NREs are released into the soil.
Nodule Colonization by NREs
Understanding NRE nodule entry mechanisms is critical to designing climate-adaptive microbial inocula to improve legume productivity. Legume selection of NREs may occur via host resource sanctioning within the nodule as with rhizobia (Kiers et al., 2003), or during nodule formation. While immunofluorescence microscopy has confirmed the presence of non-rhizobia in nodules in vitro (Muresu et al., 2008), the stage (i.e., root hair curling, infection thread) of nodule formation at which these NREs enter the symbiosome is not yet well-understood (Martínez-Hidalgo and Hirsch, 2017).
There is limited research on the molecular signaling involved in NRE legume colonization, although some studies have reported an absence of nodC among cultured NRE isolates (Ibáñez et al., 2009; Zgadzaj et al., 2015). Importantly, nodC encodes the N-acetyl glucosamine backbone of Nod factors required for root hair curling and infection (Perret et al., 2000). The absence of nod genes thus implies that NREs opportunistically gain entry to the legume host via rhizobia-initiated infection threads rather than initiating the symbiosis themselves (Ibáñez et al., 2009). Alternatively, NREs could also enter through root cracks that may not require Nod factor signaling, as is the case with some photosynthetic Bradyrhizobia (Giraud et al., 2007), although this has not been thoroughly tested. Several NREs are known to produce IAA which stimulates lateral root growth and therefore increases the number of suitable infection sites for root colonization by “crack entry” (Ibáñez et al., 2009; Hakim et al., 2020).
Evidence suggests that NREs may “eavesdrop” on molecules involved in the symbiosis pathway to opportunistically gain entry to the nodule via the infection thread. For example, Zgadzaj et al. (2016) found that rhizobacterial community assembly and root colonization by NREs in Lotus japonicus requires a functional symbiosis pathway. Involvement of legume-derived root signals in NRE enrichment was further supported by evidence that both WT and mutant L. japonicus enriched significantly different root microbiota from both Arabidopsis and three Brassicaceae species (Zgadzaj et al., 2016). More recent work has also found evidence for legume-specific changes to rhizobacterial communities in both field (Hamel et al., 2018) and greenhouse (Zhou et al., 2017) settings. However, no studies have yet been successful in identifying which signals are required for successful NRE nodule colonization or if these signals are legume-specific. For instance, both cytokinin and abscisic acid responses are crucial for nodulation and different in nodulating legumes compared to other plant species; this may present another avenue for future research in this area (Liang and Harris, 2005; Gauthier-Coles et al., 2019).
The specificity of legume-NRE interactions and whether the host is able to select for beneficial NREs (as some legumes select for efficient N-fixing rhizobia) is not yet clear. Evaluation of NREs obtained from diverse wild legume species demonstrated that the majority belonged to the genera Flavobacterium and Pseudomonas with little evidence of host-symbiont specificity (Cardoso et al., 2018). Similarly, low abundance and inconsistent occurrence of NRE strains among nodules collected from the same soybean host plant led to the recent conclusion that legumes exert little selective pressure (Mayhood and Mirza, 2021). Nevertheless, recent work has shown that host genetic control exerts a stronger influence on nodule community composition than edaphic factors (Brown et al., 2020; Shah et al., 2021), suggesting at least some degree of specificity. Legume selection of NREs across diverse environments merit further study to understand how we can leverage plant-soil-management drivers to address critical priorities in agriculture.
Discussion
While there are a growing number of studies (e.g., McDaniel et al., 2016; Yang et al., 2016; Zhou et al., 2017; Wang et al., 2020) that link soil microbial community composition and function to legumes, the practical implications of these changes are difficult to predict. Moving from controlled laboratory and greenhouse settings to testing legume-specific microbiome shifts under field conditions is a critical step toward providing practical applications of this line of research.
In this mini-review, we have discussed a broad range of legume-bacteria interactions to illustrate that future research beyond N fixation by rhizobia has the capacity to broaden legume agroecosystem services. As our understanding of legume-microbe interactions grows, we will be better equipped to design sustainable cropping systems and inoculants that not only boost crop production, but also improve ecological diversity, retain plant-available N, and facilitate the long-term enrichment of PGPBs to reduce our reliance on inorganic inputs.
Author Contributions
MS and GH conducted literature research and developed the table and figure. JG provided editorial comments and assisted with scope definition. All authors contributed to the article and approved the submitted version.
Funding
This material was based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. (00074041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, C., Liang, G., Sun, J., Wang, X., He, P., Zhou, W., et al. (2015). Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biol. Biochem. 80, 70–78. doi: 10.1016/j.soilbio.2014.09.028
Brígido, C., Menéndez, E., Paço, A., Glick, B. R., Belo, A., Félix, M. R., et al. (2019). Mediterranean native leguminous plants: a reservoir of endophytic bacteria with potential to enhance chickpea growth under stress conditions. Microorganisms 7:392. doi: 10.3390/microorganisms7100392
Brown, S. P., Grillo, M. A., Podowski, J. C., and Heath, K. D. (2020). Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 8:139. doi: 10.1186/s40168-020-00915-9
Cardoso, P., Alves, A., Silveira, P., Sá, C., Fidalgo, C., Freitas, R., et al. (2018). Bacteria from nodules of wild legume species: phylogenetic diversity, plant growth promotion abilities and osmotolerance. Sci. Total Environ. 645, 1094–1102. doi: 10.1016/j.scitotenv
Cesco, S., Neumann, G., Tomasi, N., Pinton, R., and Weisskopf, L. (2010). Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329, 1–25. doi: 10.1007/s11104-009-0266-9
Chibeba, A. M., Guimarães, M. D. F., Rodrigues Brito, O., Nogueira, M. A., Silva Araujo, R., and Hungria, M. (2015). Co-inoculation of soybean with Bradyrhizobium and Azospirillum promotes early nodulation. Am. J. Plant Sci. 6, 1641–1649. doi: 10.4236/ajps.2015.610164
Chibeba, A. M., Pereira, C. S., Antunes, J. E. L., Ribeiro, R. A., de Almeida Lopes, A. C., Gomes, R. L. F., et al. (2020). Polyphasic characterization of nitrogen-fixing and co-resident bacteria in nodules of Phaseolus lunatus inoculated with soils from Piauí State, Northeast Brazil. Symbiosis 80, 279–292. doi: 10.1007/s13199-020-00672-1
de Almeida Lopes, K. B., Carpentieri-Pipolo, V., Fira, D., Balatti, P. A., López, S. M. Y., Oro, T. H., et al. (2018). Screening of bacterial endophytes as potential biocontrol agents against soybean diseases. J. Appl. Microbiol. 125, 1466–1481. doi: 10.1111/jam.14041
De Meyer, S. E., De Beuf, K., Vekeman, B., and Willems, A. (2015). A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 83, 1–11. doi: 10.1016/j.soilbio.2015.01.002
Duchene, O., Vian, J. F., and Celette, F. (2017). Intercropping with legume for agroecological cropping systems: complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 240, 148–161. doi: 10.1016/j.agee.2017.02.019
Dutta, S., Morang, P., Nishanth Kumar, S., and Dileep Kumar, B. S. (2014). Fusarial wilt control and growth promotion of pigeon pea through bioactive metabolites produced by two plant growth promoting rhizobacteria. World J. Microbiol. Biotechnol. 30, 1111–1121. doi: 10.1007/s11274-013-1532-9
Egamberdieva, D., Wirth, S. J., Shurigin, V. V., Hashem, A., and Abd Allah, E. F. (2017). Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under salt stress. Front. Microbiol. 8:1887. doi: 10.3389/fmicb.2017.01887
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Fox, A., Lusher, A., and Widmer, F. (2020). Plant speices identity drives soil microbial community structures that persist under a following crop. Ecol. Evol. 10, 8652–8668. doi: 10.1002/ece3.6560
Foyer, C. H., Lam, H.-M., Nguyen, H. T., Siddique, K. H. M., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2:16112. doi: 10.1038/nplants.2016.112
Fustec, J., Lesuffleur, F., Mahieu, S., and Cliquet, J. B. (2010). Nitrogen rhizodeposition of legumes. A review. Agron. Sustain. Dev. 30, 57–66. doi: 10.1051/agro/2009003
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Gauthier-Coles, C., White, R. G., and Mathesius, U. (2019). Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Front. Plant Sci. 9:1901. doi: 10.3389/FPLS.2018.01901
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J.-C., et al. (2007). Legumes symbioses: absence of Nod genes in photosynthetic Bradyrhizobia. Am. Assoc. Adv. Sci. 316, 1307–1312. doi: 10.1126/science.1139548
Hakim, S., Mirza, B. S., Imran, A., Zaheer, A., Yasmin, S., Mubeen, F., et al. (2020). Illumina sequencing of 16S rRNA tag shows disparity in rhizobial and non-rhizobial diversity associated with root nodules of mung bean (Vigna radiata L.) growing in different habitats in Pakistan. Microbiol. Res. 231, 126356–126356. doi: 10.1016/j.micres.2019.126356
Hamel, C., Gan, Y., Sokolski, S., and Bainard, L. D. (2018). High frequency cropping of pulses modifies soil nitrogen level and the rhizosphere bacterial microbiome in four-year rotation systems of the semiarid prairie. Appl. Soil Ecol. 126, 47–56. doi: 10.1016/j.apsoil.2018.01.003
Han, Q., Ma, Q., Chen, Y., Tian, B., Xu, L., Bai, Y., et al. (2020). Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 14, 1915–1928. doi: 10.1038/s41396-020-0648-9
Hannula, E., Ma, H., Pérez-Jaramillo, J. E., Pineda, A., and Bezemer, T. (2020). Structure and ecological function of the soil microbiome affecting plant-soil feedbacks in the presence of a soil-borne pathogen. Environ. Microbiol. 22, 660–676. doi: 10.1111/1462-2920.14882
Hansen, B. L., Pessotti, R. D. C., Fischer, M. S., Collins, A., El-Hifnawi, L., Liu, M. D., et al. (2020). Cooperation, competition, and specialized metabolism in a simplified root nodule microbiome. MBio 11:e01917-20. doi: 10.1128/mBio.01917-20
Hartman, K., van der Heijden, M. G. A., Roussely-Provent, V., Walser, J.-C., and Schlaeppi, K. (2017). Deciphering composition and function of the root microbiome of a legume plant. Microbiome 5:2. doi: 10.1186/s40168-016-0220-z
Howieson, J. G., and Dilworth, M. J. (2016). Working with Rhizobia. Australian Centre for International Agricultural Research. Available online at: http://aciar.gov.au/files/aciar_mn_173_web-updated_31_may_2016.pdf (accessed March 27, 2020).
Ibáñez, F., Angelini, J., Taurian, T., Tonelli, M. L., and Fabra, A. (2009). Endophytic occupation of peanut root nodules by opportunistic Gammaproteobacteria. Syst. Appl. Microbiol. 32, 49–55. doi: 10.1016/j.syapm.2008.10.001
Jaiswal, S. K., Mohammed, M., and Dakora, F. D. (2019). Microbial community structure in the rhizosphere of the orphan legume Kersting's groundnut (Macrotyloma geocarpum). Mol. Biol. Rep. 46, 4471–4481. doi: 10.1007/s11033-019-04902-8
Jasiński, M., Kachlicki, P., Rodziewicz, P., Figlerowicz, M., and Stobiecki, M. (2009). Changes in the profile of flavonoid accumulation in Medicago truncatula leaves during infection with fungal pathogen Phoma medicaginis. Plant Physiol. Biochem. 47, 847–853. doi: 10.1016/J.PLAPHY.2009.05.004
Kamutando, C. N., Vikram, S., Kamgan-Nkuekam, G., Makhalanyane, T. P., Greve, M., Le Roux, J., et al. (2017). Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 7:6472. doi: 10.1038/s41598-017-07018-w
Kamutando, C. N., Vikram, S., Kamgan-Nkuekam, G., Makhalanyane, T. P., Greve, M., Le Roux, J. J., et al. (2019). The functional potential of the rhizospheric microbiome of an invasive tree species, Acacia dealbata. Microb. Ecol. 77, 191–200. doi: 10.1007/s00248-018-1214-0
Kiers, E. T., West, S. A., Denison, R. F., and Rousseau, R. A. (2003). Host sanctions and the legume-rhizobium mutualism. Nature 425, 78–81. doi: 10.1038/nature01931
Korir, H., Mungai, N. W., Thuita, M., Hamba, Y., and Masso, C. (2017). Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 8:141. doi: 10.3389/fpls.2017.00141
Lapcik, O., Honys, D., Koblovska, R., Mackova, Z., Vitkova, M., and Klejdus, B. (2006). Isoflavonoids are present in Arabidopsis thaliana despite the absence of any homologue to known isoflavonoid synthases. Plant Physiol. Biochem. 44, 106–114. doi: 10.1016/J.PLAPHY.2005.11.006
Latz, E., Eisenhauer, N., Rall, B. C., Allan, E., Roscher, C., Scheu, S., et al. (2012). Plant diversity improves protection against soil-borne pathogens by fostering antagonistic bacterial communities. J. Ecol. 100, 597–604. doi: 10.1111/j.1365-2745.2011.01940.x
Leach, J. E., Triplett, L. R., Argueso, C. T., and Trivedi, P. (2017). Communication in the phytobiome. Cell 169, 587–596. doi: 10.1016/j.cell.2017.04.025
Leite, J., Fischer, D., Rouws, L. F. M., Fernandes-Júnior, P. I., Hofmann, A., Kublik, S., et al. (2017). Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front. Plant Sci. 7:2064. doi: 10.3389/fpls.2016.02064
Liang, Y., and Harris, J. M. (2005). Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am. J. Bot. 92, 1675–1683. doi: 10.3732/AJB.92.10.1675
Liu, C. W., and Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants 5:33. doi: 10.3390/plants5030033
Liu, J., Yu, X., Qin, Q., Dinkins, R. D., and Zhu, H. (2020). The impacts of domestication and breeding on nitrogen fixation symbiosis in legumes. Front. Genet. 11:973. doi: 10.3389/fgene.2020.00973
Maróti, G., Downie, J. A., and Kondorosi, É. (2015). Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr. Opin. Plant Biol. 26, 57–63. doi: 10.1016/J.PBI.2015.05.031
Martínez-Hidalgo, P., and Hirsch, A. M. (2017). The nodule microbiome: N2 fixing rhizobia do not live alone. Phytobiomes J. 1, 70–82. doi: 10.1094/PBIOMES-12-16-0019-RVW
Martínez-Romero, E., Aguirre-Noyola, J. L., Taco-Taype, N., Martínez-Romero, J., and Zuñiga-Dávila, D. (2020). Plant microbiota modified by plant domestication. Syst. Appl. Microbiol. 43, 126106–126106. doi: 10.1016/j.syapm.2020.126106
Mayhood, P., and Mirza, B. S. (2021). Soybean root nodule and rhizosphere microbiome: Distribution of rhizobial and non rhizobial endophytes. Appl. Environ. Microbiol. 87:e02884-20. doi: 10.1128/AEM.02884-20
McDaniel, M. D., Grandy, A. S., Tiemann, L. K., and Weintraub, M. N. (2016). Eleven years of crop diversification alters decomposition dynamics of litter mixtures incubated with soil. Ecosphere 7:e01426. doi: 10.1002/ecs2.1426
Moore, V., Davis, B., Poskaitis, M., Maul, J. E., Kucek, L. K., and Mirsky, S. (2020). Phenotypic and nodule microbial diversity among crimson clover (Trifolium incarnatum L.) accessions. Agronomy 10:1434. doi: 10.3390/AGRONOMY10091434
Müller, J., Wiemken, A., and Boller, T. (2001). Redifferentiation of bacteria isolated from Lotus japonicus root nodules colonized by Rhizobium sp. NGR234. J. Exp. Bot. 52, 2181–2186. doi: 10.1093/jexbot/52.364.2181
Muresu, R., Polone, E., Sulas, L., Baldan, B., Tondello, A., Delogu, G., et al. (2008). Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 63, 383–400. doi: 10.1111/j.1574-6941.2007.00424.x
Muresu, R., Porceddu, A., Sulas, L., and Squartini, A. (2019). Nodule-associated microbiome diversity in wild populations of Sulla coronaria reveals clues on the relative importance of culturable rhizobial symbionts and co-infecting endophytes. Microbiol. Res. 221, 10–14. doi: 10.1016/j.micres.2019.01.004
Peck, M. C., Fisher, R. F., and Long, S. R. (2006). Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J. Bacteriol. 188, 5417–5427. doi: 10.1128/JB.00376-06
Peoples, M. B., Brockwell, J., Herridge, D. F., Rochester, I. J., Alves, B. J. R., Urquiaga, S., et al. (2009). The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48, 1–17. doi: 10.1007/BF03179980
Pérez-Jaramillo, J. E., Carrion, V., Bosse, M., Ferrão, L. F. V., de Hollander, M., Garcia, A. A. F., et al. (2017). Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 11, 2244–2257. doi: 10.1038/ismej.2017.85
Perret, X., Staehelin, C., and Broughton, W. J. (2000). Molecular basis of symbiotic promiscuity. Am. Soc. Microbiol. 64, 180–201. doi: 10.1128/MMBR.64.1.180-201.2000
Quiza, L., St-Arnaud, M., and Yergeau, E. (2015). Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front. Plant Sci. 6:507. doi: 10.3389/fpls.2015.00507
Rajendran, G., Sing, F., Desai, A. J., and Archana, G. (2008). Enhanced growth and nodulation of pigeon pea by co-inoculation of Bacillus strains with Rhizobium spp. Bioresour. Technol. 99, 4544–4550. doi: 10.1016/j.biortech.2007.06.057
Schlatter, D., Fubuh, A., Xiao, K., Hernandez, D., Hobbie, S., and Kinkel, L. (2009). Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol. 57, 413–420. doi: 10.1007/s00248-008-9433-4
Shah, A. S., Wakelin, S. A., Moot, D. J., Blond, C., Laugraud, A., and Ridgway, H. J. (2021). Trifolium repens and T. subterraneum modify their nodule microbiome in response to soil pH. J. Appl. Microbiol. 131, 1858–1869. doi: 10.1111/jam.15050
Sharaf, H., Rodrigues, R. R., Moon, J., Zhang, B., Mills, K., and Williams, M. A. (2019). Unprecedented bacterial community richness in soybean nodules vary with cultivar and water status. Microbiome 7:63. doi: 10.1186/s40168-019-0676-8
Skiada, V., Avramidou, M., Bonfante, P., Genre, A., and Papadopoulou, K. K. (2020). An endophytic Fusarium–legume association is partially dependent on the common symbiotic signalling pathway. New Phytol. 226, 1429–1444. doi: 10.1111/nph.16457
Smith, A., Snapp, S., Dimes, J., Gwenambira, C., and Chikowo, R. (2016). Doubled-up legume rotations improve soil fertility and maintain productivity under variable conditions in maize-based cropping systems in Malawi. Agric. Syst. 145, 139–149. doi: 10.1016/j.agsy.2016.03.008
Stevenson, F. C., and van Kessel, C. (1996). The nitrogen and non-nitrogen rotation benefits of pea to succeeding crops. Can. J. Plant Sci. 76, 735–745.
Taylor, B. N., Simms, E. L., and Komatsu, K. J. (2020). More than a functional group: diversity within the legume-rhizobia mutualism and its relationship with ecosystem function. Diversity 12:50. doi: 10.3390/d12020050
Thies, J., Woomer, P., and Singleton, P. (1995). Enrichment of Bradyrhizobium spp. populations in soil due to cropping of the homologous host legume. Soil Biol. Biochem. 27, 633–636. doi: 10.1016/0038-0717(95)98643-3
Tian, B., Pei, Y., Huang, W., Ding, J., and Siemann, E. (2021). Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 15, 1919–1930. doi: 10.1038/s41396-021-00894-1
Tian, T., Reverdy, A., She, Q., Sun, B., and Chai, Y. (2020). The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep. 12, 160–172. doi: 10.1111/1758-2229.12816
Tokgöz, S., Lakshman, D. K., Ghozlan, M. H., Pinar, H., Roberts, D. P., and Mitra, A. (2020). Soybean nodule-associated non-rhizobial bacteria inhibit plant pathogens and induce growth promotion in tomato. Plants 9:1494. doi: 10.3390/plants9111494
Tsiknia, M., Tsikou, D., Papadopoulou, K. K., and Ehaliotis, C. (2021). Multi-species relationships in legume roots: from pairwise legume-symbiont interactions to the plant-microbiome-soil continuum. FEMS Microbiol. Ecol. 97:fiaa222. doi: 10.1093/femsec/fiaa222
Uroz, S., Courty, P., and Oger, P. (2019). Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci. 24, 905–916. doi: 10.1016/J.TPLANTS.2019.06.008
Vardien, W., Steenkamp, E. T., and Valentine, A. J. (2016). Legume nodules from nutrient-poor soils exhibit high plasticity of cellular phosphorus recycling and consersvation during variable phosphorous supply. J. Plant Physiol. 191, 73–81. doi: 10.1016/j.jplph.2015.12.002
Veitch, N. (2007). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 24, 417–464. doi: 10.1039/B511238A
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Wang, C. H., Wu, L., Wang, Z., Alabady, M. S., Parson, D., Molumo, Z., et al. (2020). Characterizing changes in soil microbiome abundance and diversity due to different cover crop techniques. PLoS ONE 15:e0232453. doi: 10.1371/journal.pone.0232453
Wang, J., Andersen, S. U., and Ratet, P. (2018). Editorial: molecular and cellular mechanisms of the legume-rhizobia symbiosis. Front. Plant Sci. 871:1839–1839. doi: 10.3389/fpls.2018.01839
Wang, X., Feng, H., Wang, Y., Wang, M., Xie, X., Chang, H., et al. (2021). Mycorrhizal symbiosis modulates the rhizosphere microbiota to promote rhizobia–legume symbiosis. Mol. Plant 14, 503–516. doi: 10.1016/j.molp.2020.12.002
Wei, W. E. I., Yan-li, X. U., Lin, Z. H. U., and Si-jia, Z. (2014). Impact of long-term continuous cropping on the Fusarium population in soybean rhizosphere. Yingyong Shengtai Xuebao 25, 497–504.
Weisskopf, L., Le Bayon, R. C., Kohler, F., Page, V., Jossi, M., Gobat, J. M., et al. (2008). Spatio-temporal dynamics of bacterial communities associated with two plant species differing in organic acid secretion: a one-year microcosm study on lupin and wheat. Soil Biol. Biochem. 40, 1772–1780. doi: 10.1016/j.soilbio.2008.02.018
Weston, L. A., and Mathesius, U. (2013). Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 39, 283–297. doi: 10.1007/S10886-013-0248-5
White, L. J., Jothibasu, K., Reese, R. N., Brözel, V. S., and Subramanian, S. (2015). Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol. Plant Microbe Interact. 28, 22–29. doi: 10.1094/MPMI-08-14-0247-R
Xiao, X., Chen, W., Zong, L., Yang, J., Jiao, S., Lin, Y., et al. (2017). Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol. Ecol. 26, 1641–1651. doi: 10.1111/mec.14027
Yang, C., Hamel, C., Gan, Y., and Vujanovic, V. (2013). Pyrosequencing reveals how pulses influence rhizobacterial communities with feedback on wheat growth in the semiarid Prairie. Plant Soil 367, 493–505. doi: 10.1007/s11104-012-1485-z
Yang, Z., Yang, W., Li, S., Hao, J., Su, Z., Sun, M., et al. (2016). Variation of bacterial community diversity in rhizosphere soil of sole-cropped versus intercropped wheat field after harvest. PLoS ONE 11:e0150618. doi: 10.1371/journal.pone.0150618
Zgadzaj, R., Garrido-Oter, R., Jensen, D. B., Koprivova, A., Schulze-Lefert, P., and Radutoiu, S. (2016). Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 113, e7996–e8005. doi: 10.1073/pnas.1616564113
Zgadzaj, R., James, E. K., Kelly, S., Kawaharada, Y., De Jonge, N., Jensen, D. B., et al. (2015). A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet. 11:e1005280. doi: 10.1371/journal.pgen.1005280
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhao, M., Jones, C. M., Meijer, J., Lundquist, P. O., Fransson, P., Carlsson, G., et al. (2017). Intercropping affects genetic potential for inorganic nitrogen cycling by root-associated microorganisms in Medicago sativa and Dactylis glomerata. Appl. Soil Ecol. 119, 260–266. doi: 10.1016/j.apsoil.2017.06.040
Keywords: legumes, sustainable agriculture, microbiome, rhizosphere, plant-microbe interactions
Citation: Schaedel M, Hidrobo G and Grossman J (2021) From Microns to Meters: Exploring Advances in Legume Microbiome Diversity for Agroecosystem Benefits. Front. Sustain. Food Syst. 5:668195. doi: 10.3389/fsufs.2021.668195
Received: 15 February 2021; Accepted: 21 September 2021;
Published: 12 October 2021.
Edited by:
Adam C. N. Wong, University of Florida, United StatesReviewed by:
Jan Jansa, Academy of Sciences of the Czech Republic (ASCR), CzechiaUlrike Mathesius, Australian National University, Australia
Copyright © 2021 Schaedel, Hidrobo and Grossman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie Schaedel, c2NoYWU2NTlAdW1uLmVkdQ==
 Marie Schaedel
Marie Schaedel Gabriela Hidrobo
Gabriela Hidrobo Julie Grossman
Julie Grossman