- 1Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
- 2Innovative Genomics Institute, University of California, Berkeley, Berkeley, CA, United States
- 3Plant Gene Expression Center, US Department of Agriculture-Agricultural Research Service, University of California, Berkeley, Albany, CA, United States
- 4Horticultural Sciences Department, University of Florida, Gainesville, FL, United States
- 5Department of Global Development, Cornell University, Ithaca, NY, United States
Climate change imposes a severe threat to agricultural systems, food security, and human nutrition. Meanwhile, efforts in crop and livestock gene editing have been undertaken to improve performance across a range of traits. Many of the targeted phenotypes include attributes that could be beneficial for climate change adaptation. Here, we present examples of emerging gene editing applications and research initiatives that are aimed at the improvement of crops and livestock in response to climate change, and discuss technical limitations and opportunities therein. While only few applications of gene editing have been translated to agricultural production thus far, numerous studies in research settings have demonstrated the potential for potent applications to address climate change in the near future.
Introduction
Climate change poses a severe threat to the future of the environment as it pertains to agriculture, biodiversity, human society, and nearly every facet of our world. The primary cause of climate change is the anthropogenic addition of greenhouse gases to the atmosphere. Due to these human emissions, the average temperature of the planet has risen by nearly 1°C since 1850 (IPCC, 2018; Nunez et al., 2019). Even if warming were to be halted at 1.5°C, which would require drastic and immediate global action, long-term effects of past emissions would linger for centuries or millennia (IPCC, 2018). The magnitude of the effects depends on the amount of emissions; in general, more frequent heatwaves, droughts, floods, and persistent sea level rise and global temperature increases are expected (IPCC, 2018). Indeed, many of these effects are already being observed (IPCC, 2018; Nunez et al., 2019; Shukla et al., in press).
In both natural ecosystems and agricultural settings, plants and animals are being forced to contend with novel conditions that change more quickly than their pace of adaptation. Rising temperatures and shifting precipitation regimes will drastically alter the biological landscape, resulting in species migration, invasion, and extinction (Urban, 2015; Nunez et al., 2019). One meta-review of more than 130 studies has estimated that one in six species may go extinct due to the changing climate (Urban, 2015). Simultaneously, global food supplies are declining as droughts and floods impact agricultural output. Under a range of warming scenarios, agricultural output is expected to decline globally. Productivity of major commodity crops will be affected, especially in lower latitudes where the effects of climate change on yield will be more severe (Shukla et al., in press).
In response to these challenges, the use of gene editing, also referred to as genome editing or genome engineering, has emerged as a method to either aid in the adaptation of organisms to climate change or help mitigate the effects of climate change on agriculture.
Gene editing is a method to generate DNA modifications at precise genomic locations. These modifications can result in knockout or knockdown of one or multiple genes without the permanent insertion of any foreign DNA. Alternatively, genes from within the organism's genepool or from other organisms can be inserted into precise locations within the genome to knock-in a new trait. Transcription activator-like effector nucleases (TALENs), Zinc Finger Nucleases (ZFNs), and CRISPR/Cas systems have all been utilized to achieve precise gene edits (Gaj et al., 2016; Khalil, 2020). The precision and efficiency of generating edits has been tremendously improved by the introduction of CRISPR/Cas systems, although there is certainly still a role for other gene editing technologies. The application of gene editing techniques has generated great potential for developing crops and livestock that can better manage the impositions of climate change.
We seek here to illuminate the ways in which gene editing may help combat the deleterious effects of climate change by highlighting current efforts to apply these techniques in crops and livestock. We will summarize the efforts undertaken thus far and describe the limitations and opportunities that exist with gene editing technologies. Tables 1–4, provided at the end of the review, summarizes the breadth of applications of gene editing in crops and livestock.
Climate Change Will Inhibit Agricultural Productivity
The effects of climate change have already started to emerge and will undoubtedly worsen. Currently, crops in lower latitude regions have begun to experience yield declines, while higher-latitude regions have experienced an increase in yield (Iizumi et al., 2018; Shukla et al., in press). However, global declines in yield and crop suitability are projected over the course of the century as a direct result of climate change. According to the Intergovernmental Panel on Climate Change (IPCC), extreme weather events will disrupt and decrease global food supply and drive higher food prices (Shukla et al., in press). In dry areas of the planet especially, climate change and desertification are likely to reduce agricultural productivity. Areas closer to the equator will be most vulnerable to declines in crop yield as temperature increases, with the continents of Asia and Africa having the largest populations vulnerable to increased desertification. Indeed, desertification has already started to reduce agricultural productivity and biodiversity, compounded by unsustainable land management and increased population pressure. While it is unclear to what extent aridity will increase on a global scale, it is likely that the area at risk of salinization will increase. Climate change will also contribute to current land degradation with increased droughts, floods, rising seas, and more intense tropical storms (Shukla et al., in press).
Effects of Climate Change on Crops
The major contributing greenhouse gas to climate change is carbon dioxide (CO2) (IPCC, 2018; Shukla et al., in press), which generally has a positive effect on plant growth. As CO2 concentration increases, so too does the rate of photosynthesis and carbon assimilation (an effect known as CO2 fertilization) (Wang et al., 2020). Simultaneously, however, the nutritional quality of food decreases in response to heightened CO2 (Shukla et al., in press). Furthermore, the increased growth associated with higher CO2 may be offset by other environmental factors; there has been an observed decline in this CO2 fertilization effect in the past 30 years, likely due to shifting nutrient concentrations and lower availability of water (Wang et al., 2020). Given the aforementioned increase in extreme temperature and precipitation events, combined with the shifting prevalence and range of diseases across the globe (Fisher et al., 2012; Bett et al., 2017), the overall effect of climate change on crops will be detrimental (Iizumi et al., 2018; Shukla et al., in press). Already, global yields of maize, wheat, and soybeans have slightly decreased from 1981 to 2010 relative to the pre-industrial climate (Iizumi et al., 2018).
Effects of Climate Change on Livestock
Livestock will similarly be negatively affected by climate change. Increasing temperature and shifting precipitation directly impact livestock themselves, the crops grown for their feed, and diseases that infect them (Rojas-Downing et al., 2017; Shukla et al., in press). Increasing temperatures will have perhaps the most profound effects on livestock: heat stress impacts feed intake and can reduce weight gain, decreases reproductive efficiency, has multiple negative health effects, and increases mortality in many livestock species (Rojas-Downing et al., 2017).
Climate Change Impacts Biodiversity and Food Systems
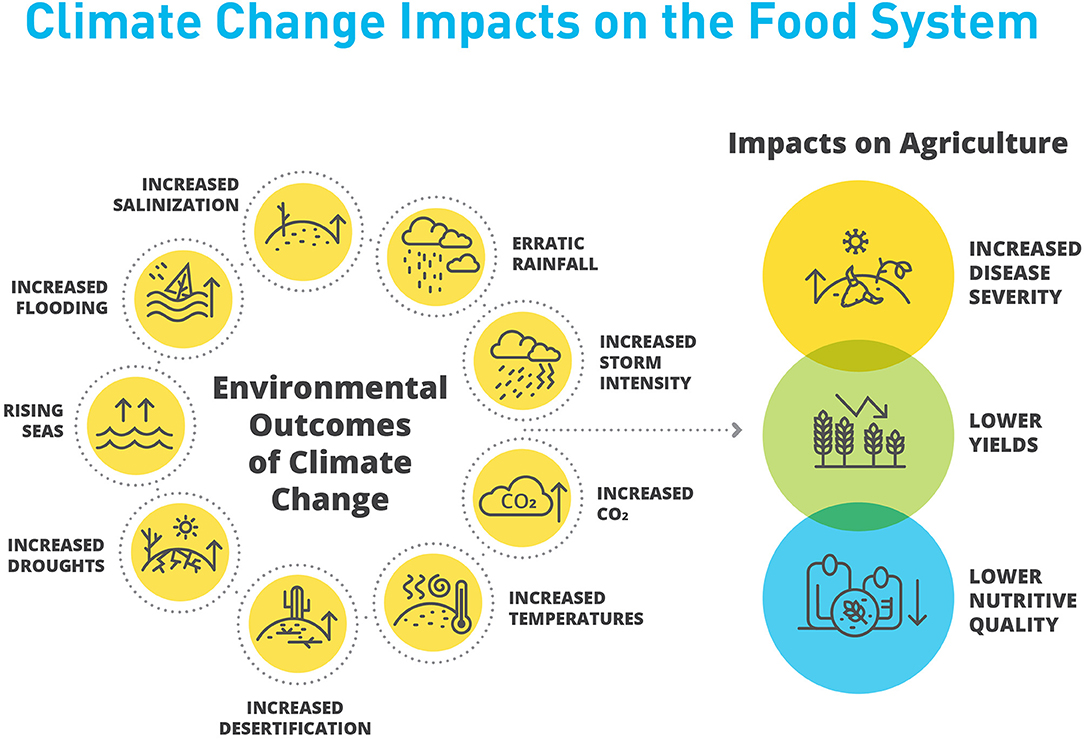
Beyond agriculture, the effects of climate change on biodiversity are no less severe. A recent meta-review of 97 studies found that even with only moderate increases of global temperature, biodiversity will suffer significant declines (Nunez et al., 2019). The pressures of climate change on biodiversity, in combination with increased agricultural demand, have also served to exacerbate the oftentimes antagonistic relationship between agricultural and natural landscapes. The impact of individual climate change effects and their intersections are complex. Broadly, the direct and indirect consequences of climate change will be deleterious to plant and animal performance in cultivated systems (Figure 1).
Mitigating the deleterious impacts of climate change on biodiversity is paramount. However, most applications of gene editing have converged on agricultural commodities: there are very few instances of gene editing for climate change in non-commodity organisms. This review therefore focuses on how gene editing solutions can address the broad effects of climate change on agriculture while maintaining the importance of applying these transformative technologies to the totality of biodiversity threatened by climate change.
Applications of Gene Editing in Agriculture
Here we present an extensive exploration of gene editing-based solutions in response to the daunting limitations to agricultural productivity imposed by climate change. We note that these examples are mostly from public institutions and represent proof-of-concept experiments rather than commercialized technologies.
Increasing Abiotic Stress Tolerance
Abiotic stresses, including but not limited to drought, salinity, and flooding, pose some of the most severe threats to agricultural productivity in the face of climate change. Abiotic stress is anticipated to become more severe in agricultural systems as a result of climate change. Current research efforts demonstrate that gene editing is an effective tool in broadening resistance of crop tolerance as described in the following examples (Table 1, Figure 2).
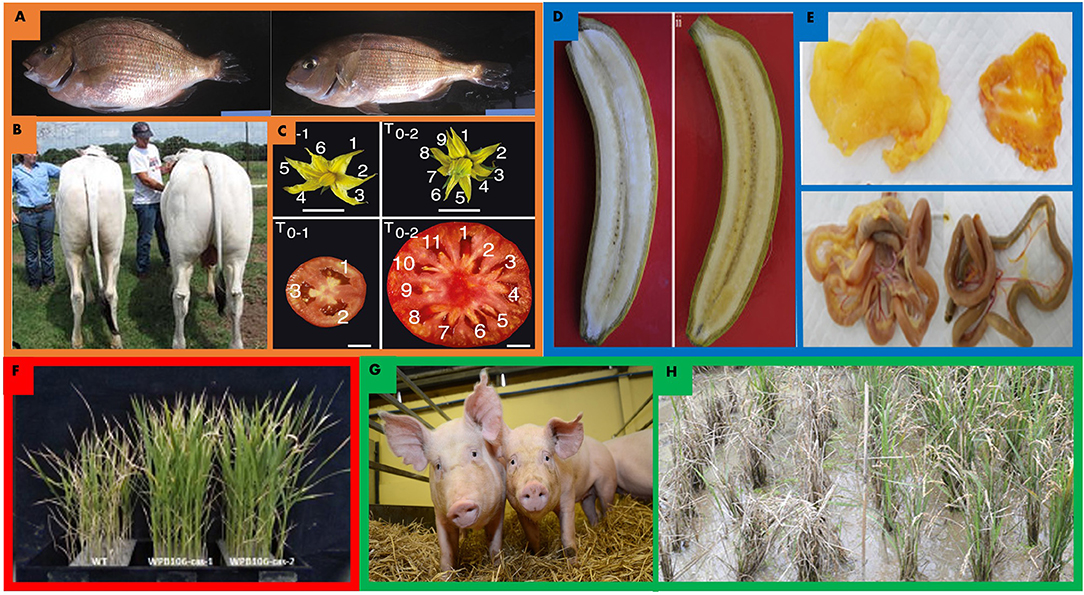
Figure 2. Gene editing improvements for land sparing in orange, nutrition in blue, abiotic stress tolerance in red, and disease in green. (A,B) MSTN gene edits in livestock enhance muscle yields in a variety of organisms. (B) CRISPR/Cas9 mediated MSTN edited red sea bream (left) and wild type (right) (Kishimoto et al., 2018). Edited red sea bream exhibited 16% skeletal muscle mass increases on average. (C) TALEN enabled MSTN edited cow (right) and wild type (left) exhibit increased overall mass and greater muscle mass (Proudfoot et al., 2015). (C) CRISPR/Cas9 promoter editing of tomato CLV3, S, SP facilitated novel variation and enhancements to fruit size, floral architecture and overall architecture in tomato. Edited tomato (right) exhibits enlarged fruit size and increased locule number (Rodríguez-Leal et al., 2017). (D) CRISPR/Cas9 edited LCYε enhanced beta-carotene accumulation in edited banana (right) relative to wild type (left) by nearly six fold in some lines (Kaur et al., 2020). (E) CRISPR/Cas9 mediated editing of G0S2 in chicken (right) accumulates less abdominal and gastrointestinal fat (Park et al., 2019). (F) Improvements to saline tolerance in rice. Knockout of OsRR22 enhances yield in high saline environments. Wild type (left) and edited rice (right) grown in 0.75% saline solution. Wild type plants are 13% shorter than edited plants in saline conditions (Zhang A. et al., 2019). (G) CD163 locus edited by CRISPR/Cas9 yielded gene edited pigs that are entirely resistant to porcine reproductive and respiratory virus when challenged with the virus (Whitworth et al., 2014, 2016). (H) bsr-k1 edited rice in field (right) greatly outperforms wild type (left) after being challenged with rice blast. Edited lines performed 50% better than wild type in field after inoculation (Zhou X. et al., 2018).
Salinity Tolerance in Rice
Rice, a staple food for more than half of the world's population, is of primary importance for global food security (Chauhan et al., 2017). Two major abiotic stresses that affect rice are drought and salinity, necessitating research explorations into the potential of leveraging gene editing for developing tolerant varieties. One such exploration was the use of CRISPR/Cas9 to knockout OsRR22, a gene associated with salt susceptibility in rice (Zhang A. et al., 2019). Rice plant performance in high salinity environments (0.75% NaCl) was improved with no concomitant decreases in grain yield, plant biomass, or grain quality. Edited lines were on average 19% shorter in saline solution whereas wild type plants were 32% shorter. Edited plants also had much less severe biomass reductions due to salt exposure compared to unedited plants, and showed no significant differences to unedited plants in the absence of saline. Saline studies were conducted in the greenhouse and overall agronomic performance was evaluated in the field. Researchers found that edited plants had much less severe biomass reductions due to salt exposure compared to wild type plants (Zhang A. et al., 2019).
Drought Tolerance in Rice
Rice has also been engineered to improve drought and high temperature tolerance by targeting stomatal development. Stomata, which are anatomical features on the surface of all crop plant tissues, serve as the major sites of water loss. In this study researchers targeted a positive regulator of stomatal density in rice (Yin et al., 2017); while they did not explicitly test the effects of this editing on water use efficiency, other research has shown that stomatal reductions in rice have clear, positive implications for water use efficiency (Caine et al., 2019). Rice lines with reductions in stomatal densities had better yield in severe drought and were able to maintain lower temperatures despite no differences in yield. Stomatal density reductions achieved by a cisgenic approach in this case mirrored stomatal density reductions achieved by a knockout-based, gene-editing approach. Thus, reducing stomatal densities by gene-editing or cisgenic approaches could enable plants to resist water deficits and could also increase heat tolerance.
Enabling Northern Production of Rice
An additional editing effort in rice produced early maturing rice that is more amenable to production in northern latitudes (Li et al., 2017). Northern latitudes experience longer day lengths and cooler temperatures. Applications of CRISPR/Cas9 to the flowering related genes Hd2,4 and 5, generated rice plants that flowered significantly earlier than their wild-type counterparts, making them more fit for northern production. These varieties may be well-suited for use in future conditions where temperatures and other climatic conditions near equatorial regions render farmlands less fertile. Early flowering plants may also be a useful adaptation against water deficit: by shortening the life span of crops through early flowering, less cumulative water may be required.
Semi-dwarf Banana Varieties
In banana, gene editing using CRISPR/Cas9 to generate knockouts of genes for the biosynthesis of gibberellins has facilitated the development of a semi-dwarfed variety. This variety may be more resistant to lodging as a result of intense winds, typhoons, and storms, anticipated to increase in severity as a result of climate change (Shao et al., 2020). Semi-dwarfed varieties have historically been an important trait in crop improvement, as was the case with rice and wheat that enabled the Green Revolution (Khush, 1999).
Promoter Editing for Drought Tolerance in Maize
Beyond generating knockouts, gene editing tools can also facilitate knock-ins. Researchers have used CRISPR/Cas9 to insert a promoter at a specific maize locus to increase drought tolerance (Shi et al., 2017). Specifically, an alternate maize promoter was inserted before ARGOS8, a gene associated with drought tolerance. This precise insertion enabled greater grain yield during flowering water stress, while maintaining yields in normal growth conditions. This approach represents an intragenic technique facilitated by gene-editng in which a native maize genetic sequence was introduced at a new locus to increase plant adaptation to an abiotic stressor.
Enhancing Thermotolerance of Cattle
In animals, gene editing has also been applied to mitigate abiotic stress imposed by climate change. Acceligen, a subsidiary of Recombinetics Inc., has undertaken an initiative to improve the thermotolerance of cattle, with support from the Foundation for Food and Agriculture Research (FFAR) and Semex. Researchers are focused on replicating the SLICK phenotype originally identified in Senepol cattle through gene editing. Variations of this phenotype in cattle contribute to thermotolerance (Dikmen et al., 2014; Porto-Neto et al., 2018). Conventionally bred cattle possessing SLICK genetics are more thermotolerant, as exhibited by lower vaginal temperatures, lower rectal temperatures, lower respiration rates, and more sweating, thus leading to increased milk production during summer months (Dikmen et al., 2014). Using gene editing approaches, Acceligen seeks to replicate SLICK genetics, to improve thermotolerance of important cattle breeds (Bellini, 2018).
Managing Disease
A majority of diseases affecting plants and animals are anticipated to become more widespread with climate change (Madgwick et al., 2011; Fisher et al., 2012; Bebber, 2015; Bett et al., 2017; Tang X. et al., 2017; Yan et al., 2017). Fortunately, many current gene editing efforts have shown promise in conferring disease resistance. This work will become even more necessary in coming years as climate change increases disease severity and incidence. Increased range of vectors, rising temperatures fostering reproduction of pathogens, and host organisms becoming more susceptible to pathogens are some of many climate change driven causes of worsening disease. Gene editing can provide a solution to managing these current and emerging global threats to agricultural productivity precipitated by climate change (Table 2, Figure 2).
Plant Gene Editing for Disease Resistance
In plants, genes have been identified that increase disease resistance when knocked out. Altering genetic elements involved in susceptibility has thus far been the primary form of disease mitigation through gene editing. While there are few such genes available for increasing disease resistance, researchers have already successfully leveraged many of these loci for heightened resistance. The following examples demonstrate the effectiveness of knocking-out susceptibility loci for enhanced resistance.
Increased Rice Resistance to a Range of Disease
Climate change may have varying effects on diseases across geographies and temporal scales, as was indicated by modeling two prevalent rice diseases in Tanzania: leaf blast and bacterial leaf blight (Duku et al., 2016). It should be noted that these diseases are two of the most devastating rice diseases globally. The model indicated that bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae (PthXo2) will likely become more severe due to climate change in Tanzania, whereas rice infection by leaf blast caused by Magnaporthe oryzae will decrease due to changing climate. Modeling two diseases in the same geography revealed that climate change will not always result in increased disease severity, thereby necessitating careful considerations of diseases that will indeed become worse due to climate change.
Gene editing for a range of rice diseases has proven remarkably effective. CRISPR/Cas9 was used to produce knockouts of OsSWEET13. SWEET family genes encode sucrose transporters that can be exploited by pathogens (Jiang et al., 2013). Mutating this gene increased disease resistance dramatically (Zhou et al., 2015). A similar approach to address bacterial leaf blight used CRISPR/Cas9 to target the promoter region of multiple OsSWEET genes in two studies (Oliva et al., 2019; Xu et al., 2019). Lines that had been edited were broadly resistant to many Xanthomonas pathogens. The knockout of an alternative gene, Os8N3, similarly resulted in plants that were also resistant to this pathogen. Os8N3 was selected as a target based on naturally occurring resistance alleles in this gene despite the mechanisms of resistance not being well-defined (Kim et al., 2019). In the case of leaf blight, gene editing effectively reduced rice susceptibility to a pathogen that is anticipated to become more damaging in some regions of the world due to climate change.
CRISPR/Cas9 mediated knockout of OsERF922, an ethylene response gene previously implicated in blast resistance, had significantly reduced lesion sizes in response to being infected with leaf blast with no concomitant alterations to agronomic traits (Wang et al., 2016).
Editing a eukaryotic elongation factor, eIF4G, in rice, using CRISPR/Cas9 yielded plants entirely resistant to rice tungro virus. Edited plants infected with virus had no detectable viral proteins and exhibited higher yields relative to wild type (Macovei et al., 2018). Additional efforts to engineer broad-scale resistance to multiple rice pathogens simultaneously are described below.
Engineering Broad-Scale Resistance in Rice, Barley, and Tomato
Some crops have been edited to establish resistance to numerous diseases simultaneously. Engineering broad-scale resistance to disease in staple crops could provide a single approach to addressing many simultaneously worsening diseases (Xu et al., 2019). An example of this approach is the editing of bsr-k1, a rice gene identified to be important in disease resistance. Bsr-k1 binds to defense related genes and promotes their turnover (Zhou X. et al., 2018). Editing this gene using CRISPR/Cas9 yielded rice plants that were simultaneously resistant to leaf blast and bacterial leaf blight by stabilizing important defense genes. Field trials of edited lines yielded 50% greater when challenged with rice leaf blast in the field. Other agronomic properties of the edited rice plants were not affected (Zhou X. et al., 2018).
In barley, CRISPR/Cas9-mediated editing of MORC1, a defense related gene previously identified in Arabidopsis thaliana, simultaneously increased resistance to Blumeria graminis f. sp. Hordei the causative agent of barley powdery mildew and Fusarium graminearum. Edited barley plants contained less fungal DNA and exhibited fewer lesions (Kumar et al., 2018).
Likewise, editing of a single locus in tomato conferred broad-spectrum resistance. Mutations to SlDMR6-1. Loss of function mutants in Arabidopsis thaliana maintain higher levels of salicylic acid. CRISPR/Cas9 edited tomato lines were more resistant to P. syringae, P. capsici, and Xanthomonas spp., indicated by markedly less severe disease symptoms and lowered pathogen presence (de Toledo Thomazella et al., 2016).
Managing Cassava Brown Streak Virus
In cassava, gene editing was used to address brown streak virus which can cause yield reductions of 70% in severe cases. Similar to host eukaryotic translation initiation factors in rice (eiF), eIF4E isoforms encoded by the cassava genome are required by Potyviridae viruses for infection. Simultaneous targeting of ncbp1 and ncbp2, two such eiF4E genes, by CRISPR/Cas9 enhanced plant resistance: root disease severity and viral titer were lowered significantly in edited cassava lines (Gomez et al., 2019).
Engineering Cucumber Viral Resistance
Likewise in cucumber, CRISPR/Cas9 was used to generate deletions in the eIF4e gene to inhibit viral infections. Lines with homozygous mutations were resistant to cucumber vein yellowing virus, zucchini yellow mosaic virus, and papaya ringspot virus-W as demonstrated by reduced symptoms and viral accumulation (Chandrasekaran et al., 2016).
Wheat Powdery Mildew Mitigation
Wheat powdery mildew is also anticipated to become increasingly severe on winter wheat in China with the changing climate (Tang X. et al., 2017). In anticipation of this increased disease pressure, researchers in China have undertaken a gene editing effort to address wheat susceptibility to this disease. Employing TALENs and CRISPR/Cas9, researchers successfully edited the mildew resistance locus (MLO) in the wheat genome (Wang et al., 2014). As a result, the percentage of viable powdery mildew-causing pathogens was effectively 0% in edited lines and nearly 20% in wild type. Edited plants exhibited marked improvements in powdery mildew resistance relative to wild type.
An additional effort targeting EDR1, as an alternate mechanism for achieving powdery mildew resistance was undertaken using CRISPR/Cas9: edited wheat plants like the MLO edited lines were resistant to powdery mildew indicated by reductions in fungal structures and microcolonies (Zhang et al., 2017).
Powdery Mildew Mitigation in Tomato and Grape
Gene editing for disease resistance in non-grain crops has also been a successful endeavor. For example, in tomato, the MLO locus was also edited using CRISPR/Cas9, leading to the development of elite tomato varieties that are resistant to the powdery mildew disease as indicated by heightened hydrogen peroxide accumulation after infection with powdery mildew (Nekrasov et al., 2017). Targeting of MLO homologs in grapevine similarly increased powdery mildew resistance; in grapevine there was about a 2-fold reduction in powdery mildew sporulation in an edited line (Wan et al., 2020). It is noteworthy that homologous loci in alternate crop species gave rise to powdery mildew resistance when edited, providing an example of transferable applications of gene editing among distantly related species.
Enhancing Resistance of Tomato to Alternate Diseases
Also in tomato, gene editing approaches have improved resistance to bacterial speck and tomato yellow leaf curl virus (Tashkandi et al., 2018; Ortigosa et al., 2019). Editing of JAZ2 in tomato using CRISPR/Cas9 reduced infection by Pseudomonas syringae pv. tomato, the causal agent of bacterial speck, by reducing pathogen mediated stomatal opening. Edited plants maintained significantly reduced levels of Pseudomonas syringae pv. tomato relative to wild type. CRISPR/Cas9 mediated resistance in tomato has also been effectively applied to manage yellow leaf curl virus. Tomato plants were engineered to contain guides targeting multiple sequences in the TYCV genome. Viral accumulation was markedly decreased in engineered tomatoes and this resistance was heritable over many generations.
Securing Banana Resistance to BSV
In bananas, banana streak virus (BSV) presents a major barrier to breeding and distribution of banana cultivars (Musa spp.) in various parts of the world (Tripathi J. N. et al., 2019). This virus integrates into the B subgenome of Musa species and remains latent until plants encounter stress such as drought. Many important agronomic species of banana such as plantains are affected by this virus, and breeding programs are restricted in their ability to use Musa balbisiana as source material due to the presence of the latent virus. Knockouts of the endogenous virus produced lines in which 75% of edited plants remained asymptomatic after being exposed to water stress. This is the first study to show the efficacy of targeting an integrated plant virus in a plant genome and provides a promising mechanism to address a significant barrier in banana production and breeding (Tripathi J. N. et al., 2019).
Transient Resistance in Cacao
In cacao, knockout-based improvement of resistance to Phytopthora tropicalis was achieved in a transient assay. Cacao plants transiently expressing CRISPR/Cas9 targeting the TcNPR3 gene, a suppressor of disease response, exhibited smaller lesions after infection with Phytopthora tropicalis. This work represents the first application of gene editing in cacao, and paves the way for future stably heritable edited lines of cacao (Fister et al., 2018).
Grapefruit Resistance to Citrus Canker
Citrus canker is a devastating disease in most citrus fruits especially as most commercial varieties remain susceptible to infection. Application of CRISPR/Cas9 to grapefruit has successfully increased resistance of edited fruit to infection. The causative agent of citrus canker, Xanthomonas citri subsp. citri (Xcc), is able to increase expression of CsLOB1 thus generating cankers (Jia et al., 2016). CRISPR/Cas9 mediated editing of CsLOB1 promoter binding sites (Jia et al., 2016) and the CsLOB1 coding region (Jia et al., 2017) were both effective in generating resistant grapefruit lines indicated by tremendously reduced symptoms.
Increased Virus and Abiotic Stress Resistance in Potato
CRISPR/Cas9 mediated editing of a gene encoding coilin in potato plants has facilitated increased resistance to potato virus Y. Coilin is a major structural component of the subnuclear Cajal bodies previously implicated in virus interactions in planta. Modification of the potato coilin gene C-terminal domain significantly increased resistance of potato to Potato virus Y and also increased salt and drought tolerance (Makhotenko et al., 2019).
Curing Cotton Cancer
Cotton verticillium or “cotton cancer” is a severe disease of cotton caused by Verticillium dahliae (Zhang Z. et al., 2018) with little resistant germplasm available in natural populations. 14-3-3 proteins c and d had been previously identified as negative regulators of disease response. Knockouts of 14-3-3 c and 14-3-3 d simultaneously yielded cotton plants that were more resistant to cotton verticillium indicated by fewer disease symptoms and lowered pathogen presence.
Animal Gene Editing for Disease Resistance
Diseases affecting animal hosts are projected to be affected by climate change as well. The intersection of multiple climate and human variables makes it difficult to determine how climate change will affect animal pathogens (Samy and Peterson, 2016; Bett et al., 2017). It is likely that currently temperate locations will become more favorable to tropical vector-borne diseases, exposing new host populations with no previous immunity and potentially creating new disease transmission modes and patterns (De et al., 2008). Overall, there is high confidence that diseases and disease vectors will worsen due to climate change (Shukla et al., in press).
Gene editing has been used to target several animal diseases; several such studies are described below to demonstrate the state of the field. These examples are meant to demonstrate a substantial basis for the capacity of gene editing to ameliorate diseases that may potentially worsen due to climate change. The vast majority of editing for animal disease resistance centers on the creation of disease-resistant livestock, with the exception of CRISPR-based gene drives in mosquitoes and other vector and reservoir species (Esvelt et al., 2014; Hammond et al., 2016; Esvelt and Gemmell, 2017; Champer et al., 2018; Scudellari, 2019).
Viral Resistance in Chicken
In chickens, avian leukosis viral subgroup J is a disease that can infect meat and laying chickens, resulting in relatively high mortality rates. Researchers used CRISPR/Cas9 and homologous recombination to create an amino acid deletion in the extracellular portion of the gene chNHE1 (chicken Na+/H+ exchanger type 1), which encodes the virus receptor in chickens that allows the virus to infect cells. The deletion was completed in chicken primordial germ cells, which through transplantation and subsequent breeding resulted in chickens resistant to infection by the virus (Koslov et al., 2020).
Tuberculosis and Mastitis Resistance in Cattle
Tuberculosis resistance in cattle has been addressed in two studies. In the first study, researchers focused on the mouse gene SP110 (SP110 Nuclear Body Protein), which controls Mycobacterium tuberculosis (MTB) infections and induces apoptosis in infected cells (Wu et al., 2015). The authors used TALENickases to insert the gene into a specific location in the bovine genome via homologous recombination; the knock-in of this resistance gene improved tuberculosis resistance (Wu et al., 2015). A second study using CRISPR/Cas9 was able to knock-in the NRAMP1 gene (natural resistance-associated macrophage protein-1, a gene associated with innate immunity) from bovine via homologous recombination. The resulting cattle likewise exhibited increased tuberculosis resistance (Gao et al., 2017).
Similarly, gene editing has been utilized to prevent mastitis, the most significant disease of dairy cows. In two studies, homologous recombination in cattle facilitated by ZFNickases enabled the insertion of two genes that confer resistance to infection from S. aureus, a causative agent of mastitis: the gene encoding lysostaphin from Staphylococcus simulans (Liu et al., 2013) and the human lysozyme (hLYZ) gene (Liu et al., 2014) into an intron of the β-casein locus of dairy cattle. Because casein is a protein found in dairy milk, the genes inserted into this locus would mimic expression of β-casein and the exogenous proteins would be present in milk produced from the edited cows (Liu et al., 2013). Both of these studies yielded dairy cows with milk that was able to prevent S. aureus infection of the lactating cow.
M. haemolytica is a causative agent of pneumonia in cattle. ZFNs were used to make a precise edit in CD18, a gene encoding a surface protein on cattle leukocytes. Leukocytes extracted from the edited cow exhibited very low levels of cell toxicity in the presence of M. haemolytica leukotoxin relative to wild type cattle. Thus, a gene edit markedly improved the tolerance of gene edited cattle to M. haemolytica, a pervasive agent of disease (Shanthalingam et al., 2016).
Disease Resistance in Pigs
Progress has also been made in developing disease resistance in pigs using gene editing. In a 2014 study, researchers were able to knock out two genes, CD163 and CD1D (Whitworth et al., 2014). The former is required for infection by porcine reproductive and respiratory syndrome virus (PRRS virus), and the latter is involved in innate immunity. In a follow-up to this study, the researchers assessed the Cd163 knockout pigs for resistance to PRRS, finding that they displayed no symptoms when infected. In comparison, wild-type offspring developed severe symptoms necessitating their euthanization (Whitworth et al., 2016). Similar results were also obtained by a later study, also using CRISPR/Cas9 editing to knock out CD163 to produce pigs fully resistant to PRRSV (Burkard et al., 2017, 2018).
Further work in pigs was able to demonstrate the utility of CRISPR/Cas9 to knock-in resistance to classical swine fever virus (CSFV) at the Rosa26 locus (Xie et al., 2018). This locus is a preferred site for transgene insertion due to its ubiquitous and strong expression, coupled with a lack of gene-silencing effects (Irion et al., 2007; Xie et al., 2018). The edited pigs were resistant to CSFV, whereas all wild type pigs exhibited 100% mortality. Researchers were also able to knock-in the C. elegans fat-1 gene into the Rosa26 locus in pigs (Li M. et al., 2018). As fat-1 is implicated in both disease resistance and nutritive quality of meat, this study served as a proof-of-concept to demonstrate the possibility of simultaneously improving the nutritional value of pork and increasing general disease resistance in pigs. Finally, a recent study was able to use CRISPR/Cas9 editing to knock out the ANPEP (aminopeptidase N) gene, conferring resistance to infections caused by coronaviruses (Whitworth et al., 2019).
Viral Resistance in Aquatic Species
Use of gene editing to combat disease in aquatic species is more limited; the first use of a CRISPR system to test enhanced disease resistance was in 2018 (Ma et al., 2018). Here, researchers were able to use CRISPR/Cas9 in grass carp cell lines to knock out gcJAM-A (grass carp Junctional Adhesion Molecule-A), a gene involved in grass carp reovirus (GCRV) infection. When challenged with GCRV, the edited cells were shown to suppress viral replication (Ma et al., 2018).
In crops and livestock, genes have been identified that increase disease resistance when knocked out. Altering genetic elements involved in susceptibility has thus far been the primary form of disease mitigation through gene editing for crops. Whereas, in livestock, gene editing has facilitated the knockout and knock-in of genes to improve disease resistance. Successful gene editing studies in crops and animals provide a reasonable foundation for further use of this technology to address a range of diseases, some of which may be exacerbated by climate change.
Increasing Yields
Global climate change will continue to broadly reduce crop and livestock yields (Lobell and Gourdji, 2012). Some landscapes will experience yield improvements, but largely climate change stands to lower productivity (Shukla et al., in press). Coupled with population increases, these coinciding phenomena will necessitate the expansion of agricultural lands into currently non-cultivated geographical areas. This expansion imposes a severe threat to biodiversity and the associated ecological services of non-agricultural lands. Land sparing through the augmentation of yield can mitigate the deleterious effects of agricultural expansion, with gene editing as a potential tool toward this solution (Table 3, Figure 2).
Crop Yield Improvement
Improving Rice Yields
In crop plants, a variety of gene edits have been produced to increase yields. In rice, for example, targeting different combinations of genes associated with yield restrictions has produced lines with 11–68% increased yields (Huang et al., 2018; Miao et al., 2018; Zhou et al., 2019). DEP1 and Gn1a are yield-associated genes that have been previously characterized for their involvement in yield attributes. One novel allele of Gn1A and three novel alleles of DEP1 generated by CRISPR/Cas9 produced significantly higher yields relative to wild type alleles.Gn1A novel mutants yielded >24% which was slightly better than the 21% yield advantage conferred by natural mutant Gn1A. Novel mutants in DEP1 yielded up to 51% greater than WT which is 11% greater than the naturally occurring mutants (Huang et al., 2018).
In another case simultaneous and individual knockouts of three yielded related genes, GS3, GW2, and Gn1a, identified to negatively regulate grain size, width, and number, respectively, increased yield in rice in three different cultivars. Simultaneous KOs generated greatest yield increases, with gains of up to 68% in one cultivar. Multiple knockouts can thus be additive in yield improvements (Zhou et al., 2019).
CRISPR-Cas9 induced mutations in class I PYL genes were also able to increase yield. Abscisic acid (ABA) is a. PYL/RCARs are genes that encode receptors for ABA, a phytohormone essential in drought responses and in seed dormancy. Simultaneous mutations of class I PYL genes resulted in rice plants that were higher yielding in paddy conditions. The stomata in PYL mutants were much less responsive to the addition of ABA, maintaining larger apertures despite the presence of the drought signal. Thus, the mutant plants lost water more readily. The resultant plants had larger panicles, greater panicle branches, more tillers, and overall higher yield when tested in field conditions. This approach could be appropriate for rice grown in paddy conditions but would be deleterious in conditions where water is limited. Triple knockout of PYLs 1,4,6 afforded a 30% increase in yield in well-watered conditions (Miao et al., 2018).
Knockout-Based Wheat Yield Improvement
In wheat, simultaneous knockouts of GW2, LPX-1, and MLO were meant to enhance yield and disease resistance. Genes selected were previously shown in separate studies to increase yield and wheat pathogen resistance. Homozygous, simultaneous mutations in these three loci yielded wheat plants that had significantly elevated grain weights and size, while disease resistance was not evaluated (Wang et al., 2018).
Improving Yields of Waxy Maize
In maize, CRISPR/Cas9 was used to generate high amylopectin varieties from elite cultivars by knocking out the waxy gene. Gene edited varieties yielded 5.5 bushels more per acre relative to high amylopectin varieties generated by conventional breeding and were produced in less time, highlighting the throughput and utility of gene editing relative to conventional breeding in certain specific applications (Gao et al., 2020).
Engineering Tomato Architecture and Domestication for Yield
Engineering of tomato architecture using CRISPR/Cas9 has enabled improvements such as drastically increased fruit size and altered plant morphology better suited for urban environments (Rodríguez-Leal et al., 2017; Lemmon et al., 2018). Promoter editing of SlCLV3, a mobile peptide important in floral stem cell regulation, S, and inflorescence architecture gene, and SP, an overall architecture gene, were able to generate novel variation and enhancement in fruit size, floral architecture and overall architecture in tomato. This study also provided major breakthroughs in the use of gene editing cis-regulatory elements for crop improvements.
Efforts have been made to increase yield as well as other agronomic properties of tomatoes using CRISPR/Cas9 to domesticate a wild tomato variety. By identifying and editing six genes associated with key domestication traits, researchers were able to domesticate a wild tomato relative, increasing fruit size 3-fold and fruit number 10-fold. and yield while also improving nutrition, abiotic stress tolerance, and disease tolerance. This study also provides the basis for future gene editing mediated wild relative domestications (Zsögön et al., 2018).
Livestock Yield Improvement
Enhancing Livestock Yield Through MSTN Knockouts
In animal gene editing, many efforts have converged on targeting the MSTN gene in species such as pig, cattle, sheep, goat, rabbit, and several aquatic animals including carp, catfish, and red sea bream (Dong et al., 2011; Han et al., 2014; Luo et al., 2014; Ni et al., 2014; Crispo et al., 2015; Proudfoot et al., 2015; Qian et al., 2015; Wang K. et al., 2015; Wang X. et al., 2015; Bi et al., 2016; Guo et al., 2016; Li et al., 2016; Rao et al., 2016; Tanihara et al., 2016; Yu et al., 2016, 2019; Zhong et al., 2016; Khalil et al., 2017; Wang et al., 2017; He et al., 2018; Kishimoto et al., 2018; Su et al., 2018a; Zhang J. et al., 2018; Zhang Y. et al., 2019; Sun et al., 2020). The MSTN gene (also known as GDF8) encodes the gene for myostatin, a growth differentiation factor that inhibits muscle growth. In natural cattle populations MSTN mutations underlie the double-muscled phenotype (McPherron and Lee, 1997). Successful MSTN knockout animals exhibit significantly higher muscle mass than those with the functional MSTN gene. Assessment of MSTN knockouts tend to vary, with some studies comparing birth weight, body weight-to-muscle mass, muscle fiber number, muscle weight, and muscle size between edited and unedited animals. Combined with several bottlenecks in efficiency of editing, comparing the outcomes of each study to another can be challenging. ZFN, TALENs, and CRISPR/Cas9 have all been utilized to achieve MSTN-edited animals.
Studies that assessed the phenotypes of edited MSTN pigs reported increased birth weight (Wang K. et al., 2015), body weight-to-muscle mass ratio equaling 170% that of unedited pigs (Rao et al., 2016), and upwards of 100% increased muscle mass (Qian et al., 2015). Other studies reported an obvious double-muscled phenotype (Tanihara et al., 2016; Wang et al., 2017), or significantly larger muscles (Bi et al., 2016), but not all studies reported an assessment of the edited pig phenotype (Su et al., 2018a). Comparatively fewer studies on MSTN knockouts have been conducted in cattle, although two performed in 2014 resulted in edited animals with an obvious double-muscled phenotype (Luo et al., 2014; Proudfoot et al., 2015).
Faster growth (Crispo et al., 2015) and increased body weight of up to 60% (Li et al., 2016) were found in sheep, with similar results in goats (Ni et al., 2014; Yu et al., 2016; He et al., 2018). Studies performed as early as 2014 reported successful editing in sheep (Proudfoot et al., 2015), but phenotypic assessment in all cases is still lacking (Han et al., 2014), or limited to microscopy of muscle tissue (Zhang Y. et al., 2019). One study edited both goats and rabbits; while both exhibited increased weight ratios of biceps and quadriceps upwards of 50%, the rabbits tended to have very large tongues and low viability (Guo et al., 2016). Other studies in goats successfully targeted another gene, FGF5, in addition to MSTN (Wang X. et al., 2015), or inserted an additional gene, fat-1, into the MSTN locus (Zhang J. et al., 2018).
MSTN has also been targeted in several aquatic species, with the first heritable MSTN knockout in an aquaculture species being performed by ZFN in 2011 (Dong et al., 2011). TALENs and CRISPR/Cas9 were later used to edit carp, a tetraploid species, although severe bone defects were present in addition to enhanced muscle mass (Zhong et al., 2016). Successful editing followed in several aquaculture species such as catfish, which resulted in a 29.7% increase in catfish fry body weight (Khalil et al., 2017), red sea bream, which resulted in a 16% increase in muscle mass (Kishimoto et al., 2018), and blunt snout bream, which resulted in a 7% increase in body weight (Sun et al., 2020). Outside of fish there has also been one successful MSTN knockout in pacific oyster, a major aquaculture bivalve (Yu et al., 2019).
While MSTN editing appears quite promising for improving animal yields, the drawbacks of this gene target must also be considered. For example, increased birth weight of edited animals can result in birthing challenges, and viability has been an issue in several studies (Wang X. et al., 2015; Rao et al., 2016). Fine-tuning MSTN mutations beyond complete knockouts could serve to optimize the use of this gene as a land sparing tool. Additional targets for increasing biomass beyond MSTN should also be considered. For instance, of MSTN mutations, other studies in pigs have targeted knockouts for increased muscle mass (Xiang et al., 2018; Zou Y. et al., 2018; Liu et al., 2019).
Enhancing Livestock Yields Through Alteration of Sex Ratios
Livestock gene editing has also been employed to alter sex ratios of offpsring. In many production schemes, only a single sex is required (i.e., female chickens in laying operations). Increasing ratios of the preferred sex in offspring stands to lower inputs and space typically allocated to rearing animals which are unfit for the desired production. An effort in chicken editing used CRISPR/Cas9 to insert a fluorescent protein into the male sex chromosomes thus facilitating high-throughput sex determination during embryogenesis (Lee et al., 2019). A system to produce exclusively female offspring in mice by targeting exclusively male genes has been developed with potential for transferral to alternate mammalian species (Yosef et al., 2019). Ongoing works seeks to improve sex-determining and sex-biasing technologies to facilitate land and resource sparing in livestock operations (Kurtz and Petersen, 2019; Douglas, 2020; CSIRO, 2021), and avenues for increasing litter size in general are being explored: one study succeeded in mutating the GDF9 gene to increase litter size in goats (Niu et al., 2018).
Land sparing is not the only method by which the ecological consequences of agriculture can be buffered; one alternative is land sharing (Phalan et al., 2011; Soga et al., 2014). Most of the efforts undertaken by researchers to date are intended to be compatible with a land sparing approach however. Gene editing for increased yields of plants and animals has been considerably effective in research studies and could act to prevent the sprawl of agricultural production.
Enhancing Nutrition
At the end of 2019, 690 million people worldwide were suffering from undernourishment, or the insufficient consumption of calories (Von Grebmer et al., 2020). This pandemic stands to worsen as climate change exacerbates yield deficits and pest pressure. Much more pervasive however is malnutrition, which encompasses undernourishment as well as micronutrient deficiencies and overconsumption of calories. As of 2014, ~2 billion individuals suffered from micronutrient deficiencies (Von Grebmer et al., 2014).
Climate change is currently contributing to malnourishment in several ways and is predicted to worsen. More extreme climatic conditions will disrupt food chains and increase food prices, with tropical and subtropical regions experiencing the worst effects of crop yield decline (Shukla et al., in press). Prolonged droughts, which are projected to increase, reduce root acquisition of water soluble nutrients such as nitrate, sulfate, calcium, magnesium, and silicon. Additionally as erratic rainfall episodes worsen, more nitrate is expected to leach from soils (St.Clair and Lynch, 2010).
The increased level of CO2 will also be detrimental to nutritional quality of many crops (Soares et al., 2019; Shukla et al., in press). Wheat grown at projected mid-to-late 21st century CO2 levels has been found to have reduced protein, zinc, and iron (Shukla et al., in press), and similar nutrient decreases have been observed in rice, legumes, and several vegetables (Soares et al., 2019). A modeling study predicted that climate change has placed many fruits and vegetable crops at a high risk of going extinct (Springmann et al., 2016). In addition, lack of reduced intake of fruits and vegetables caused by limited access could double the number of deaths caused by malnutrition by 2050 (Springmann et al., 2016). Projections indicate that at the current rate of CO2 emissions, an additional 1.9% of the global population will become deficient in zinc, 1.3% will be protein deficient, and 57% of children and childbearing-aged women will live in geographies at high-risk of iron deficiencies, by 2050 (Smith and Myers, 2018).
Gene editing may play a role in ameliorating the current and future states of the malnourishment pandemic beyond providing increased yields. Recent work applied to plants and livestock has indicated that gene editing may effectively increase desirable nutritional metabolites, reduce anti-nutrients, and alter macronutrients in ways that can be advantageous for human health (Table 4, Figure 2).
Increasing Beneficial Metabolites
The color pigments of crop plants such as anthocyanins, lycopene, carotenoids are known for possessing high antioxidant vitamins, necessary for improving nutrition and fighting disease. Anthocyanins help in reducing inflammation and preventing oxidative damage to cells while beta-carotene (a precursor to vitamin A) is essential for vision and other immune functions (Tanaka and Ohmiya, 2008). In rice, CRISPR/Cas9 editing facilitated the insertion of carotenoid biosynthesis genes in a precise genomic location to increase carotenoid accumulation (Tanaka and Ohmiya, 2008) that could further advance earlier efforts to engineer “Golden Rice.” These fortified lines stand to benefit the poorest women and children in developing countries with rice-dominant diets, such as Bangladesh. Likewise, proanthocyanidins, and anthocyanins augmentation has been pursued in rice (Zafar et al., 2020) and other species. In tomato, CRISPR/Cas9 editing has been used to target a series of genes, resulting in yellow, pink and purple colored tomatoes respectively (Cermák et al., 2015; Filler Hayut et al., 2017; Deng et al., 2018). The insertion of a strong promoter via CRISPR/Cas9 upstream of ANT1, which codes for a Myb transcription factor, resulted in anthocyanin accumulation manifested by an intense purple color in tomatoes (Cermák et al., 2015). The Phytoene synthase gene (Psy1) and other carotenoid biosynthesis genes have also been targets of genome editing that significantly increase lycopene content in tomatoes (Li X. et al., 2018). In addition, domestication of a wild tomato using gene editing produced tomatoes with 500% increased accumulation of lycopene (Zsögön et al., 2018). Dramatic enhancements of beta-carotene in banana fruit were facilitated by a CRISPR/Cas9 mediated knockout (Kaur et al., 2020). This color variation holds a great potential for improving consumers' appeal for variety, while broadening intake of healthy pigments.
Gene editing approaches have been most commonly applied to commodity crops. However, great potential of these tools lies in improving nutrition of non-staple, regionally relevant fruits and vegetables. For example, the tools of gene editing are being applied to further domesticate and improve new “super foods” from tomato's lesser known relatives in the Physalis genus such as goldenberry and groundcherry (Lemmon et al., 2018). Many of these species are rich in minerals, macro- and micronutrients and bioactive compounds such as antioxidants, vitamins A, B, and C, and have been long-used as folk remedies for diseases still relevant today (Shenstone et al., 2020). Early results suggest that CRISPR/Cas9-mediated deletions of previously identified agronomic genes may improve the cultivability of nutritional berries such as the ground cherry (Lemmon et al., 2018), rendering it more economically viable for commercial production.
The Fatty Acid Desaturase 2 (FAD2) gene determines the levels of monounsaturated fats in most oil producing crops; gene editing tools have been used to induce mutations in this gene, leading to a significant change in their level. In the emerging oil seed crop, Camelina sativa, targeted CRISPR/Cas9 editing, increased total seed oleic oil composition by more than 50% in some cases while reducing linoleic and linolenic acid levels (Jiang et al., 2017; Morineau et al., 2017). Similarly, oleic acid content has been improved in Brassica napus (Okuzaki et al., 2018). Shifts in seed oil composition are thought to be advantageous nutritionally and to extend shelf-life of oil extracts (Jiang et al., 2017; Morineau et al., 2017). Applications of TALEN-mediated editing to soybean facilitated the development of a high oleic and low linolenic acid soybeans (HOLL). Oil extracted from these modified soybeans could bypass the need for hydrogenation, a process which generates unhealthy trans-fats (Demorest et al., 2016). HOLL soybeans are being developed for commercialization by Calyxt Inc. with hopes of release by 2022 (Calyxt, 2019).
Similar efforts are underway using TALEN-mediated mutagenesis of FAD2 gene to increase oleic acid content of peanut (Wen et al., 2018).
Modifying Macronutrients
In addition to augmenting levels of beneficial metabolites, gene editing has made improvements to macronutrients possible. In rice, knockouts of the SBEIIb gene associated with amylopectin biosynthesis decreased levels of amylopectin in favor of amylose in the grain endosperm (Sun et al., 2017). Conversely, other food crops such as cassava have been targeted for reduced starchy content through editing two genes involved in amylose biosynthesis (Bull et al., 2018). A knockout of the GBSS gene in potato using CRISPR-Cas9 also led to an altered starch content (Andersson et al., 2017). In strawberry, gene editing has been used to develop a continuum of altered sugar content (Xing et al., 2020). As demonstrated, gene editing can be used to fundamentally alter the composition of macronutrients for nutritional improvements.
Lowering Anti-nutritional Compounds
Gene editing has also been applied to numerous plant species to limit the accumulation of anti-nutritional components such as phytic acid in maize, which is disruptive to the nutrition of monogastric animals (Shukla et al., 2009; Liang et al., 2014). To mitigate the deleterious effects of phytic acid on iron, zinc and calcium absorption, CRISPR/Cas9 and ZFNs were used in separate studies to mutagenize genes in the phytic acid pathway, indicating the potential utility of gene editing for anti-nutritional mitigation (Liang et al., 2014). In sorghum, an important food crop in drought-prone areas, a major class of storage proteins called kafirins leads to a poor protein digestibility. By targeting genes that synthesize kafirins, researchers have successfully reduced kafirin levels and improved protein quality and digestibility (Li et al., 2018). In a similar effort, Tang et al. engineered a rice variety to prevent the accumulation of cadmium in rice grains using CRISPR/Cas9 (Tang L. et al., 2017).
In chickens, gene editing has driven improvements of nutritive properties by reducing fat. Using CRISPR/Cas9 to target the GOS2 gene known to influence lipid catabolism yielded chickens with dramatically reduced abdominal fat deposition, with no observed side-effects (Park et al., 2019). As previously mentioned, the knockin of fat-1 in pigs enhanced the nutritional value of pork by altering accumulation of beneficial fatty acids (Li M. et al., 2018). Gene editing has also been used to reduce the allergenicity of globally important commodities such as milk and wheat (Yu et al., 2011; Su et al., 2018b; Sánchez-León et al., 2018).
In animals and crops, gene editing has been leveraged to improve the nutritional aspects of these foodstuffs. This technology has enabled micronutrient improvements, anti-nutritional component reduction, macronutrient modifications, and removal of allergens. In anticipation of the severe impacts of climate change on nutrition, gene editing could serve as a potent adaptive mechanism across a suite of commodities and nutritional traits.
Limitations and Opportunities
Gene editing has already been applied successfully to plants and animals in research settings to address the effects of climate change Tables 1–4, Figures 2, 3. However, there are still significant limitations to its efficacy in enabling climate change mitigation and adaptation. One of the most prominent limitations of current applications is the narrow scope of potential solutions without the use of intra-, cis-, and transgenic approaches. In most of the examples described, researchers produce mutations of genes whose knockout contributes positively to a trait. However, it is more common that loss-of-function mutation detracts from the organism's performance. Thus, seeking to produce improved plants solely through loss-of-function mutation significantly limits the range of possible improvements. Intragenics, cisgenics, and transgenics have a proven record in research settings of improving plant performance in a range of environments and conditions (Klümper and Qaim, 2014; Laible et al., 2015; Steinwand and Ronald, 2020). These approaches are able to leverage a vast range of genomic sequences for use in plant and animal improvements. Gene editing in conjunction with intra-, cis-, and transgenic approaches is especially beneficial. For example, CRISPR/Cas9 was used to precisely insert a promoter present in the maize genome upstream of a gene that contributes to drought tolerance (Shi et al., 2017). This novel recombination of genetic elements native to the maize genome via CRISPR/Cas9 in an intragenic approach improved drought tolerance. However, genetic elements from sexually incompatible organisms can also provide potent mechanisms for improving performance. In cows for instance, a knock-in of a mouse resistance gene using TALENs enhanced tuberculosis resistance (Wu et al., 2015). Gene editing in conjunction with ICT approaches is especially beneficial. While significant improvements through loss-of-function gene editing and intragenic approaches are demonstrably effective, greater opportunity exists in leveraging the totality of genetic diversity. Thus, regulatory barriers to use of gene editing in conjunction with ICT approaches must be addressed to maximize the potential of agricultural improvements.
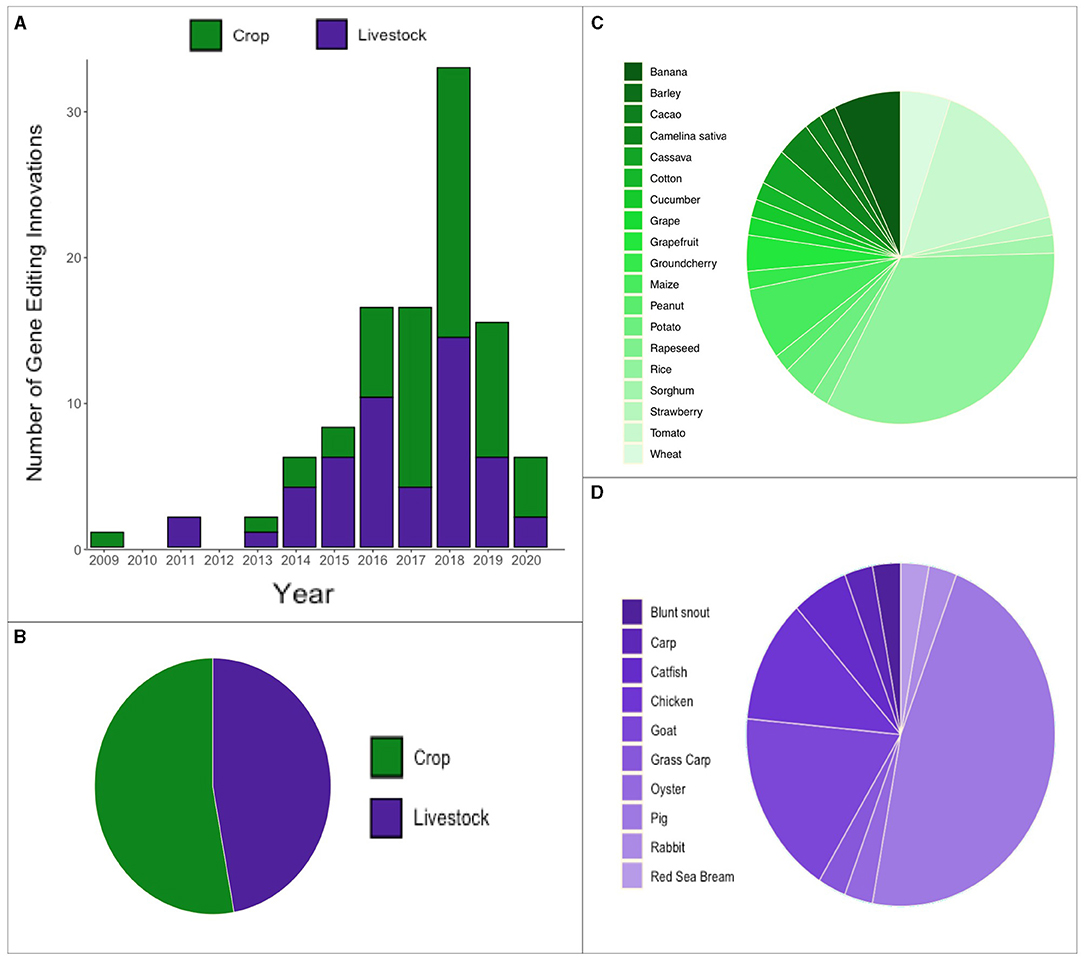
Figure 3. Overview of gene editing innovation emergence over time and species. (A) Annual count of gene editing innovations since 2009. (B) Proportion of gene editing innovations (B) in crops and livestock (C) a diverse array of crops (D) a diverse array of livestock.
Opportunities to address climate change with gene editing continue to expand as new technologies emerge. For example, alternatives to the traditional Cas9 protein for editing are being developed. Base editors that facilitate precise nucleotide modifications, epigenome modifiers that alter DNA confirmation and associated expression levels, and prime editing for precise insertion of short DNA fragments are promising candidates in this regard (Xie et al., 2018; Roca Paixão et al., 2019; Lin et al., 2020). Additionally, new techniques are emerging to improve the rates of homologous recombination, a current major limitation in plants (Huang and Puchta, 2019).
To maximize the utility of these emerging editing tools, bottlenecks in delivery must be surmounted. For livestock, the current largest barrier to editing tends to be the production of homozygous, non-mosaic, gene edited animals. Two methods currently exist to generate edited embryos prior to their transfer to a surrogate mother: one uses somatic cell nuclear transfer (SCNT) from an edited cell line to produce an edited embryo, while the other uses direct editing of a zygote. In the former case, sequencing can quickly confirm homozygous edits prior to embryo implantation, but is burdened by the inefficiency of SCNT. In the latter case, breeders can forgo SCNT but are unable to verify homozygous edits until the animal is born (Bishop and Eenennaam, 2020).
Similarly in plant regeneration, tissue culture is limiting in its species range and efficacy. To enable a broad spectrum of solutions in this space, tissue culture resources for a broader set of species must be developed. Tools for increasing the transformability of recalcitrant varieties and species are currently underway and show great promise for gene editing-based climate change solutions (Altpeter et al., 2016; Maher et al., 2020). These emerging technologies could foster further improvements in climate change performance.
Explorations of limitations beyond technical applications are not within the scope of this review, but are germane nonetheless. Consumer acceptance, policy frameworks, and economic feasibility will all factor into the ultimate success of the applications discussed.
Gene Editing Opportunities for African Crops and Livestock
The African continent is poised to benefit greatly from advances in gene editing research for agriculture. The Green Revolution, which bolstered the agricultural yields in Central America, South America, and Asia, largely overlooked African agriculture (Toenniessen et al., 2008). Despite significant progress made to improve food insecurity in developing nations, diseases and pests still plague most of the region's agriculture. Irregular weather patterns, insect swarms and natural disasters have also impacted agriculture in the region. Climate change will not only affect Africa severely and variably according to the diverse geographies, but Africa's susceptibility to climate change hinges on non-climate related vulnerabilities including but not limited to high prevalence of malnutrition, low literacy rates, and high population growth rates (Nkomo et al., 2006). As such, there is much potential to focus research efforts on the crops and livestock that are dominant and specific to Africa.
Gene Editing for Disease Resistance
African staples including African banana, plantain, cassava, and yam are actively being edited for eventual application in agricultural production. Comparable to crops like wheat, rice, maize, or potatoes in other parts of the world, these staples contribute majorly to the income of small-holder farmers in Africa. Biotic and abiotic factors including diseases and pest infestation, drought, flooding, etc affects the productivity of these crops. Using gene editing, scientists are targeting these crops to withstand these challenges.
For example, researchers are using this breeding approach to improve resistance to cassava brown streak disease (Gomez et al., 2019), a disease that significantly limits the production of the crop. In addition, attempts were made to engineer resistance to African cassava mosaic virus (ACMV). However, resistance to ACMV was not achieved in the glasshouse experiment, providing evidence of the mutagenic nature of this virus and potential for further studies (Mehta et al., 2019). In banana and plantain, a gene editing system has been applied in developing disease resistant varieties. Using Cas9, Cas12, Cas13, and Cas14 enzymes, this system enabled the detection of gene sequences that informed its potential application in plant disease diagnosis (Tripathi J. N. et al., 2019).
Furthermore, a proof of concept on the potential of using gene-editing technology for the development of novel sources of resistance to rice yellow mottle virus; maize lethal necrosis, a severe disease of maize in East Africa; and striga resistance in sorghum through a Low Germination Stimulant 1 (LGS1) gene knock-out are actively being developed by numerous researchers in Africa (Karembu, 2021).
Gene Editing for Yield Increase and Abiotic Stress Resistance
Targeting abiotic stresses, the first successful CRISPR/Cas9-mediated gene knockout system in African sorghum varieties and cowpea have recently been reported (Che et al., 2018). Recently, gene-editing has been used to target a visual marker gene in yam enabling potential rapid improvements by bypassing yam's long breeding-cycle (Syombua et al., 2019) and to develop climate-smart banana varieties with multiple and durable resistances to extreme temperature and drought (Tripathi L. et al., 2019). In maize, researchers have used CRISPR/Cas9 to develop lines tolerant to drought, DNA damage, and oxidative stress by altering the expression of poly(ADP-ribose) polymerase (PARP), which is responsible for maintenance of the energy homeostasis during stress conditions (Njuguna et al., 2017).
Furthermore, researchers have provided an example of how gene editing could be used to target and improve the yield potential of Kabre rice, thus enhancing the domestication of African rice landraces (Lacchini et al., 2020). Similarly, a proof of concept is being developed for the application of genome editing in developing wheat varieties tolerant to drought. Through employing CRISPR-Cas9 to inactivate the Sal1 genes in wheat, the project will determine if wheat sal1 mutant plants will show an improved tolerance to drought stress (Karembu, 2021; USDA ARS, 2021).
Gene Editing for Nutritional Enhancement
Nutritional enhancement is another major focus of some gene editing research in Africa. Scientists have demonstrated how gene editing could be used to improve the nutritional contents and yield of cassava. Similarly, the technology is being used to reduce the level of erucic acid in Ethiopian mustard (Brassica carinata) varieties. While current research shows that the mustard varieties available in market are above the acceptable level of total fatty acid content, gene editing could be used to edit targeted genes for improvement of nutritional content of these mustard varieties (Karembu, 2021). In targeting both improved yield and enhanced beta carotene content in sweet potato, scientists are employing gene editing technology to knock out the genes involved in converting beta carotene into other products (Joseph, 2021).
Gene Editing for Livestock Improvement
Ongoing work in the application of gene editing for African livestock holds promise for meeting the agricultural demands of this uniquely poised continent. Private-public partnerships between African livestock research centers, with support and funding from the Bill & Melinda Gates Foundation and other donors seek to improve African livestock. Numerous opportunities exist for editing African breeds of cows and chickens for improved stress and disease tolerance. For example, to combat animal trypanosomiasis, a proof of concept has been demonstrated to introduce the Apolipoprotein L1 (ApoL 1) gene, which has been found to confer resistance to trypanosomiasis in primates (O'Toole et al., 2017), into goats using CRISPR/Cas9 (Karembu, 2021). Additionally, researchers are accelerating the rate of development of the African Swine Fever Virus (ASFV) vaccine using CRISPR/Cas9 and synthetic biology technologies (Abkallo, 2020). Furthermore, gene editing is being used to edit virulence genes of Theileria parva to increase the immune response of cattle when they are vaccinated against East Coast fever (ECF) (Karembu, 2021). To confer protection against the Avian influenza virus (AIV) infection in poultry, researchers are using gene-editing tools to generate variations in ANP32A, a gene that affects AIV replication. This will enable them to predict which of these targeted changes on the ANP32A gene will make the most significant restrictive effect on avian influenza virus replication (Karembu, 2021).
While most gene editing product development is at the proof-of-concept phase, the tools present many opportunities to potentially reduce the amount of time it takes to bring improved crop and animal varieties to the market in sub-Saharan Africa. Interestingly, countries like Kenya and Nigeria have made adequate provisions in their biosafety framework to ensure that these gene edited products will not be treated differently from conventional crops (Komen et al., 2020). These favorable policies will also provide the basis for future editing with broad application in enhancing climate adaptations for African agriculture.
Conclusion
Gene editing is an emerging and increasingly prominent approach in plants and animals applied in response to current, and anticipation of future, climate change. Solutions provided by gene editing have the potential to stand alone or be adopted concomitantly with other climate-smart solutions. The range of gene editing applications for climate change is summarized in Tables 1–4, Figures 2, 3. Many of these applications have converged on traits such as disease resistance, nutritional improvements, abiotic stress tolerance and yield increases, but these are by no means representative of the breadth of opportunities that exist.
The emergence and application of CRISPR/Cas9 based editing in crops and livestock has facilitated a recent increase in the application of gene editing for climate change.
Despite its success in a research context, gene editing for climate change has largely not yet transitioned to real application. Adoption of these technological innovations has been stifled by regulatory barriers, social barriers, and prohibitive policies, among other externalities beyond the technical limitations described. A majority of the advances in gene editing applications for agriculture have occurred recently, which also explains, in part, the relatively low throughput to agricultural production. The ongoing efforts of public and private institutions alike, are rapidly expanding on current technological innovations.
While it should be noted that gene editing is not the sole promising agricultural improvement mechanism, the potency of this technology in providing solutions for climate change in agriculture cannot be overlooked. Gene editing, as demonstrated by the numerous studies summarized herein, stands to provide marked enhancements in agriculture to address climate change.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
NK and WH developed the concept and completed much of the research, and writing. SE and MA supported the writing and editing of numerous sections. All authors contributed to the article and approved the submitted version.
Funding
WH receives funding from UC Berkeley and is supported by NIH grant DP5-OD023072. NK gratefully acknowledges financial support from the Foundation for Food and Agriculture Research (FFAR) Fellows program and the National Science Foundation Graduate Research Fellowships Program. MA currently receives support from the Genetics and Genomics Scholars Program, North Carolina State University. SE currently receives funding from the Bill & Melinda Gates Foundation and USDA. This work was supported by funding for an early draft of the manuscript which was modified for publication was provided by the Keystone Policy Institute.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge the authors whose work was not included due to space limitations for their important contributions to the field. We thank Maci Mueller for her careful reading and feedback on the manuscript.
References
Abkallo, H. (2020). Accelerating African Swine Fever Virus (ASFV) Vaccine Development via CRISPR-Cas9 and Synthetic Biology Technologies. Nairobi.
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Andersson, M., Turesson, H., Nicolia, A., Fält, A.-S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum Tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Bebber, D. P. (2015). Range-expanding pests and pathogens in a warming world. Annu. Rev. Phytopathol. 53, 335–356. doi: 10.1146/annurev-phyto-080614-120207
Bellini, J. (2018). This Gene-Edited Calf Could Transform Brazil's Beef Industry. Available online at: https://www.wsj.com/video/series/moving-upstream/this-gene-edited-calf-could-transform-brazil-beef-industry/D2D93B49-8251-405F-BC35-1E5C33FA08AF (accessed January 27, 2021).
Bett, B., Kiunga, P., Gachohi, J., Sindato, C., Mbotha, D., Robinson, T., et al. (2017). effects of climate change on the occurrence and distribution of livestock diseases. Prev. Vet. Med. 137, 119–129. doi: 10.1016/j.prevetmed.2016.11.019
Bi, Y., Hua, Z., Liu, X., Hua, W., Ren, H., Xiao, H., et al. (2016). Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 6:31729. doi: 10.1038/srep31729
Bishop, T. F., and Eenennaam, A. L. V. (2020). Genome editing approaches to augment livestock breeding programs. J. Exp. Biol. 223 (Suppl 1):jeb207159. doi: 10.1242/jeb.207159
Bull, S. E., Seung, D., Chanez, C., Mehta, D., Kuon, J.-E., Truernit, E., et al. (2018). Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 4:eaat6086. doi: 10.1126/sciadv.aat6086
Burkard, C., Lillico, S. G., Reid, E., Jackson, B., Mileham, A. J., Ait-Ali, T., et al. (2017). Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLOS Pathog. 13:e1006206. doi: 10.1371/journal.ppat.1006206
Burkard, C., Opriessnig, T., Mileham, A. J., Stadejek, T., Ait-Ali, T., Lillico, S. G., et al. (2018). Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J. Virol. 92:e00415–18. doi: 10.1128/JVI.00415-18
Caine, R. S., Yin, X., Sloan, J., Harrison, E. L., Mohammed, U., Fulton, T., et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221, 371–384. doi: 10.1111/nph.15344
Calyxt (2019). Calyxt's High Oleic Low Linolenic Soybean Deemed Non-Regulated by USDA. ≫ Calyxt. Available online at: https://calyxt.com/calyxts-high-oleic-low-linolenic-soybean-deemed-non-regulated-by-usda/ (accessed January 28, 2021).
Cermák, T., Baltes, N. J., Cegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16:232. doi: 10.1186/s13059-015-0796-9
Champer, J., Liu, J., Oh, S. Y., Reeves, R., Luthra, A., Oakes, N., et al. (2018). Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. U.S.A. 115, 5522–5527. doi: 10.1073/pnas.1720354115
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chauhan, B. S., Jabran, K., and Mahajan, G.. (2017). Rice Production Worldwide. Cham: Springer International Publishing.
Che, P., Anand, A., Wu, E., Sander, J. D., Simon, M. K., Zhu, W., et al. (2018). Developing a flexible, high-efficiency agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol. J. 16, 1388–1395. doi: 10.1111/pbi.12879
Crispo, M., Mulet, A. P., Tesson, L., Barrera, N., Cuadro, F., Santos-Neto, P. C., et al. (2015). Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS ONE 10:e0136690. doi: 10.1371/journal.pone.0136690
CSIRO (2021). Sex Determination Techniques for the Egg and Poultry Industries. Available online at: https://www.csiro.au/en/research/production/biotechnology/chicken-sex-selection (accessed March 16, 2021).
de Toledo Thomazella, D. P., Brail, Q., Dahlbeck, D., and Staskawicz, B. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv [Preprint]. doi: 10.1101/064824
De, L. R. S., Jean-Antoine, R., and Jan, S. (2008). Climate change: effects on animal disease systems and implications for surveillance and control. Rev. Sci. Tech. 27, 339–354. doi: 10.20506/rst.27.2.1807
Demorest, Z. L., Coffman, A., Baltes, N. J., Stoddard, T. J., Clasen, B. M., Luo, S., et al. (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 16:225. doi: 10.1186/s12870-016-0906-1
Deng, L., Wang, H., Sun, C., Li, Q., Jiang, H., Du, M., et al. (2018). Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genomics 45, 51–54. doi: 10.1016/j.jgg.2017.10.002
Dikmen, S., Khan, F. A., Huson, H. J., Sonstegard, T. S., Moss, J. I., Dahl, G. E., et al. (2014). The SLICK hair locus derived from senepol cattle confers thermotolerance to intensively managed lactating holstein cows. J. Dairy Sci. 97, 5508–5520. doi: 10.3168/jds.2014-8087
Dong, O. X., Yu, S., Jain, R., Zhang, N., Duong, P. Q., Butler, C., et al. (2020). Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 11:1178. doi: 10.1038/s41467-020-14981-y
Dong, Z., Ge, J., Li, K., Xu, Z., Liang, D., Li, J., et al. (2011). Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus Fulvidraco) using engineered zinc finger nucleases. PLoS ONE 6:e28897. doi: 10.1371/journal.pone.0028897
Douglas, M. (2020). Chicken CRISPR – Teaming for Interdisciplinary Research Pre-Seed Program. Available online at: https://research.uga.edu/team-pre-seeds/projects/chicken-crispr/ (accessed March 16, 2021).
Duku, C., Sparks, A. H., and Zwart, S. J. (2016). Spatial Modelling of rice yield losses in tanzania due to bacterial leaf blight and leaf blast in a changing climate. Clim. Change 135, 569–583. doi: 10.1007/s10584-015-1580-2
Esvelt, K. M., and Gemmell, N. J. (2017). Conservation demands safe gene drive. PLOS Biol. 15:e2003850. doi: 10.1371/journal.pbio.2003850
Esvelt, K. M., Smidler, A. L., Catteruccia, F., and Church, G. M. (2014). Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3:e03401. doi: 10.7554/eLife.03401
Filler Hayut, S., Melamed Bessudo, C., and Levy, A. A. (2017). Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8:15605. doi: 10.1038/ncomms15605
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Fister, A. S., Landherr, L., Maximova, S. N., and Guiltinan, M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in theobroma cacao. Front. Plant Sci. 9:268. doi: 10.3389/fpls.2018.00268
Gaj, T., Sirk, S. J., Shui, S., and Liu, J. (2016). Genome-editing technologies: principles and applications. Cold Spring Harb. Perspect. Biol. 8:a023754. doi: 10.1101/cshperspect.a023754
Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., et al. (2020). Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 38, 579–581. doi: 10.1038/s41587-020-0444-0
Gao, Y., Wu, H., Wang, Y., Liu, X., Chen, L., Li, Q., et al. (2017). Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol. 18:13. doi: 10.1186/s13059-016-1144-4
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava EIF4E isoforms NCBP-1 and NCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Guo, R., Wan, Y., Xu, D., Cui, L., Deng, M., Zhang, G., et al. (2016). Generation and evaluation of myostatin knock-out rabbits and goats using CRISPR/Cas9 system. Sci. Rep. 6:29855. doi: 10.1038/srep29855
Hammond, A., Galizi, R., Kyrou, K., Simoni, A., Siniscalchi, C., Katsanos, D., et al. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector anopheles gambiae. Nat. Biotechnol. 34, 78–83. doi: 10.1038/nbt.3439
Han, H., Yonghe, M. A., Wang, T., Ling, L., Tian, X., Hu, R., et al. (2014). One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agric. Sci. Eng. 1, 2–5. doi: 10.15302/J-FASE-2014007
He, Z., Zhang, T., Jiang, L., Zhou, M., Wu, D., Mei, J., et al. (2018). Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. 38:BSR20180742. doi: 10.1042/BSR20180742
Huang, L., Zhang, R., Huang, G., Li, Y., Melaku, G., Zhang, S., et al. (2018). Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. Crop J. 6, 475–481. doi: 10.1016/j.cj.2018.05.005
Huang, T.-K., and Puchta, H. (2019). CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 38, 443–453. doi: 10.1007/s00299-019-02379-0
Iizumi, T., Shiogama, H., Imada, Y., Hanasaki, N., Takikawa, H., and Nishimori, M. (2018). Crop production losses associated with anthropogenic climate change for 1981–2010 compared with preindustrial levels. Int. J. Climatol. 38, 5405–5417. doi: 10.1002/joc.5818
IPCC (2018). “Summary for policymakers,” in Global Warming of 1.5°C. An IPCC Special Report on the Impacts of Global Warming of 1.5°C Above Pre-industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty, eds V. Masson-Delmotte, P. Zhai, H.-O. Pörtner, D. Roberts, J. Skea, P. R. Shukla, et al. (Geneva: World Meteorological Organization), 32.
Irion, S., Luche, H., Gadue, P., Fehling, H. J., Kennedy, M., and Keller, G. (2007). Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 25, 1477–1482. doi: 10.1038/nbt1362
Jia, H., Orbovic, V., Jones, J. B., and Wang, N. (2016). Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using Cas9/SgRNA to produce transgenic duncan grapefruit alleviating XccΔpthA4:DCsLOB1.3 Infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/SgRNA-mediated targeted gene modification in arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., and Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Joseph, O. G. (2021). Ghana Scientist Turns to Gene Editing to Improve Sweet Potato Crop. Alliance for Science.
Karembu, M. (2021). “Genome editing in Africa's agriculture 2021: an early take-of,” in International Service for the Acquisition of Agri-biotech Applications (ISAAA AfriCenter) (Nairobi).
Kaur, N., Alok, A., Shivani K.umar, P., Kaur, N., Awasthi, P., Chaturvedi, S., et al. (2020). CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metab. Eng. 59, 76–86. doi: 10.1016/j.ymben.2020.01.008
Khalil, A. M. (2020). The genome editing revolution: review. J. Genet. Eng. Biotechnol. 18:68. doi: 10.1186/s43141-020-00078-y
Khalil, K., Elayat, M., Khalifa, E., Daghash, S., Elaswad, A., Miller, M., et al. (2017). Generation of myostatin gene-edited channel catfish (Ictalurus Punctatus) via zygote injection of CRISPR/Cas9 system. Sci. Rep. 7:7301. doi: 10.1038/s41598-017-07223-7
Khush, G. S. (1999). Green revolution: preparing for the 21st century. Genome 42, 646–655. doi: 10.1139/g99-044
Kim, Y.-A., Moon, H., and Park, C.-J. (2019). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to xanthomonas oryzae pv. Oryzae. Rice 12:67. doi: 10.1186/s12284-019-0325-7
Kishimoto, K., Washio, Y., Yoshiura, Y., Toyoda, A., Ueno, T., Fukuyama, H., et al. (2018). Production of a breed of red sea bream pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture 495, 415–427. doi: 10.1016/j.aquaculture.2018.05.055
Klümper, W., and Qaim, M. (2014). A meta-analysis of the impacts of genetically modified crops. PLoS ONE 9:e111629. doi: 10.1371/journal.pone.0111629
Komen, J., Tripathi, L., Mkoko, B., Ofosu, D. O., Oloka, H., and Wangari, D. (2020). Biosafety regulatory reviews and leeway to operate: case studies from Sub-Sahara Africa. Front. Plant Sci. 11:130. doi: 10.3389/fpls.2020.00130
Koslov,á, A., Trefil, P., Mucksov,á, J., Reinišov,á, M., Plach,ý, J., Kalina, J., et al. (2020). Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J Subgroup of avian leukosis virus. Proc. Natl. Acad. Sci. U.S.A. 117, 2108–2112. doi: 10.1073/pnas.1913827117
Kumar, N., Galli, M., Ordon, J., Stuttmann, J., Kogel, K., and Imani, J. (2018). Further analysis of barley MORC1 using a highly efficient RNA-guided Cas9 Gene-editing system. Plant Biotechnol. J. 16, 1892–1903. doi: 10.1111/pbi.12924
Kurtz, S., and Petersen, B. (2019). Pre-determination of sex in pigs by application of CRISPR/Cas system for genome editing. Theriogenology 137, 67–74. doi: 10.1016/j.theriogenology.2019.05.039
Lacchini, E., Kiegle, E., Castellani, M., Adam, H., Jouannic, S., Gregis, V., et al. (2020). CRISPR-Mediated accelerated domestication of African rice landraces. PLoS ONE 15:e0229782. doi: 10.1371/journal.pone.0229782
Laible, G., Wei, J., and Wagner, S. (2015). Improving livestock for agriculture – technological progress from random transgenesis to precision genome editing heralds a new era. Biotechnol. J. 10, 109–120. doi: 10.1002/biot.201400193
Lee, H. J., Yoon, J. W., Jung, K. M., Kim, Y. M., Park, J. S., Lee, K. Y., et al. (2019). Targeted gene insertion into Z chromosome of chicken primordial germ cells for avian sexing model development. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 33, 8519–8529. doi: 10.1096/fj.201802671R
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438. doi: 10.1104/pp.18.00200
Li, H., Wang, G., Hao, Z., Zhang, G., Qing, Y., Liu, S., et al. (2016). Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Sci. Rep. 6:33675. doi: 10.1038/srep33675
Li, M., Ouyang, H., Yuan, H., Li, J., Xie, Z., Wang, K., et al. (2018). Site-specific Fat-1 knock-in enables significant decrease of n-6PUFAs/n-3PUFAs ratio in pigs. G3 Genes Genomes Genet. 8, 1747–1754. doi: 10.1534/g3.118.200114
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Li, X., Zhou, W., Ren, Y., Tian, X., Lv, T., Wang, Z., et al. (2017). High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genomics 44, 175–178. doi: 10.1016/j.jgg.2017.02.001
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, X., Liu, H., Wang, M., Li, R., Zeng, J., Mo, D., et al. (2019). Disruption of the ZBED6 binding site in Intron 3 of IGF2 by CRISPR/Cas9 leads to enhanced muscle development in liang guang small spotted pigs. Transgenic Res. 28, 141–150. doi: 10.1007/s11248-018-0107-9
Liu, X., Wang, Y., Guo, W., Chang, B., Liu, J., Guo, Z., et al. (2013). Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat. Commun. 4:2565. doi: 10.1038/ncomms3565
Liu, X., Wang, Y., Tian, Y., Yu, Y., Gao, M., Hu, G., et al. (2014). Generation of mastitis resistance in cows by targeting human lysozyme gene to β-Casein locus using zinc-finger nucleases. Proc. Biol. Sci. 281:20133368. doi: 10.1098/rspb.2013.3368
Lobell, D. B., and Gourdji, S. M. (2012). The influence of climate change on global crop productivity. Plant Physiol. 160, 1686–1697. doi: 10.1104/pp.112.208298
Luo, J., Song, Z., Yu, S., Cui, D., Wang, B., Ding, F., et al. (2014). Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PLOS ONE 9:e95225. doi: 10.1371/journal.pone.0095225
Ma, J., Fan, Y., Zhou, Y., Liu, W., Jiang, N., Zhang, J., et al. (2018). Efficient resistance to grass carp reovirus infection in JAM-A knockout cells using CRISPR/Cas9. Fish Shellfish Immunol. 76, 206–215. doi: 10.1016/j.fsi.2018.02.039
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Cermák, T., et al. (2018). Novel alleles of rice EIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Madgwick, J. W., West, J. S., White, R. P., Semenov, M. A., Townsend, J. A., Turner, J. A., et al. (2011). Impacts of climate change on wheat anthesis and fusarium ear blight in the UK. Eur. J. Plant Pathol. 130, 117–131. doi: 10.1007/s10658-010-9739-1
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., Makarov, V. V., Suprunova, T. P., et al. (2019). Functional analysis of coilin in virus resistance and stress tolerance of potato solanum tuberosum using CRISPR-Cas9 editing. Dokl. Biochem. Biophys. 484, 88–91. doi: 10.1134/S1607672919010241
McPherron, A. C., and Lee, S. J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U. S. A. 94, 12457–12461. doi: 10.1073/pnas.94.23.12457
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S. S.-A., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20:80. doi: 10.1186/s13059-019-1678-3
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. U.S.A. 115, 6058–6063. doi: 10.1073/pnas.1804774115
Morineau, C., Bellec, Y., Tellier, F., Gissot, L., Kelemen, Z., Nogué, F., et al. (2017). Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid camelina sativa. Plant Biotechnol. J. 15, 729–739. doi: 10.1111/pbi.12671
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482. doi: 10.1038/s41598-017-00578-x
Ni, W., Qiao, J., Hu, S., Zhao, X., Regouski, M., Yang, M., et al. (2014). Efficient gene knockout in goats using CRISPR/Cas9 system. PLOS ONE 9:e106718. doi: 10.1371/journal.pone.0106718
Niu, Y., Zhao, X., Zhou, J., Li, Y., Huang, Y., Cai, B., et al. (2018). Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9. Reprod. Fertil. Dev. 30, 307–312. doi: 10.1071/RD17068
Njuguna, E., Coussens, G., Aesaert, S., Neyt, P., Anami, S., and Lijsebettens, M. V. (2017). Modulation of energy homeostasis in maize and arabidopsis to develop lines tolerant to drought, genotoxic and oxidative stresses. Afr. Focus 30, 66–76. doi: 10.21825/af.v30i2.8080
Nkomo, J. C., Nyong, A. O., and Kulindwa, K. (2006). “The impacts of climate change in Africa,” in Final Draft Submitted to the Stern Review on the Economics of Climate Change.
Nunez, S., Arets, E., Alkemade, R., Verwer, C., and Leemans, R. (2019). Assessing the impacts of climate change on biodiversity: is below 2 °C enough? Clim. Change 154, 351–365. doi: 10.1007/s10584-019-02420-x
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-Mediated genome editing of the fatty acid desaturase 2 gene in brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
O'Toole, J. F., Bruggeman, L. A., Madhavan, S., and Sedor, J. R. (2017). The cell biology of APOL1. Semin. Nephrol. 37, 538–545. doi: 10.1016/j.semnephrol.2017.07.007
Park, T. S., Park, J., Lee, J. H., Park, J.-W., and Park, B.-C. (2019). Disruption of G0/G1 Switch Gene 2 (G0S2) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 33, 1188–1198. doi: 10.1096/fj.201800784R
Phalan, B., Onial, M., Balmford, A., and Green, R. E. (2011). Reconciling food production and biodiversity conservation: land sharing and land sparing compared. Science 333, 1289–1291. doi: 10.1126/science.1208742
Porto-Neto, L. R., Bickhart, D. M., Landaeta-Hernandez, A. J., Utsunomiya, Y. T., Pagan, M., Jimenez, E., et al. (2018). Convergent evolution of slick coat in cattle through truncation mutations in the prolactin receptor. Front. Genet. 9:57. doi: 10.3389/fgene.2018.00057
Proudfoot, C., Carlson, D. F., Huddart, R., Long, C. R., Pryor, J. H., King, T. J., et al. (2015). Genome edited sheep and cattle. Transgenic Res. 24, 147–153. doi: 10.1007/s11248-014-9832-x
Qian, L., Tang, M., Yang, J., Wang, Q., Cai, C., Jiang, S., et al. (2015). Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in meishan pigs. Sci. Rep. 5:14435. doi: 10.1038/srep14435
Rao, S., Fujimura, T., Matsunari, H., Sakuma, T., Nakano, K., Watanabe, M., et al. (2016). Efficient modification of the myostatin gene in porcine somatic cells and generation of knockout piglets. Mol. Reprod. Dev. 83, 61–70. doi: 10.1002/mrd.22591
Roca Paixão, J. F., Gillet, F.-X., Ribeiro, T. P., Bournaud, C., Lourenço-Tessutti, I. T., Noriega, D. D., et al. (2019). Improved drought stress tolerance in arabidopsis by CRISPR/DCas9 fusion with a histone acetyltransferase. Sci. Rep. 9:8080. doi: 10.1038/s41598-019-44571-y
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8. doi: 10.1016/j.cell.2017.08.030
Rojas-Downing, M. M., Nejadhashemi, A. P., Harrigan, T., and Woznicki, S. A. (2017). Climate change and livestock: impacts, adaptation, and mitigation. Clim. Risk Manag. 16, 145–163. doi: 10.1016/j.crm.2017.02.001
Samy, A. M., and Peterson, A. T. (2016). Climate change influences on the global potential distribution of bluetongue virus. PLOS ONE 11:e0150489. doi: 10.1371/journal.pone.0150489
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Scudellari, M. (2019). Self-destructing mosquitoes and sterilized rodents: the promise of gene drives. Nature 571, 160–162. doi: 10.1038/d41586-019-02087-5
Shanthalingam, S., Tibary, A., Beever, J. E., Kasinathan, P., Brown, W. C., and Srikumaran, S. (2016). Precise gene editing paves the way for derivation of mannheimia haemolytica leukotoxin-resistant cattle. Proc. Natl. Acad. Sci. U. S. A. 113, 13186–13190. doi: 10.1073/pnas.1613428113
Shao, X., Wu, S., Dou, T., Zhu, H., Hu, C., Huo, H., et al. (2020). Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 18, 17–19. doi: 10.1111/pbi.13216
Shenstone, E., Lippman, Z., and Van Eck, J. (2020). A review of nutritional properties and health benefits of physalis species. Plant Foods Hum. Nutr. Dordr. Neth. 75, 316–325. doi: 10.1007/s11130-020-00821-3
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 Variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shukla, P. R., Skea, J., Calvo Buendia, E., Masson-Delmotte, V., Pörtner, H.-O., and Roberts, D. C., (eds.). (in press). “Technical Summary, 2019,” in Climate Change Land: an IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, Greenhouse Gas Fluxes in Terrestrial Ecosystems.
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). precise genome modification in the crop species zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Smith, M. R., and Myers, S. S. (2018). Impact of anthropogenic CO 2 emissions on global human nutrition. Nat. Clim. Change 8, 834–839. doi: 10.1038/s41558-018-0253-3
Soares, J. C., Santos, C. S., Carvalho, S. M. P., Pintado, M. M., and Vasconcelos, M. W. (2019). Preserving the nutritional quality of crop plants under a changing climate: importance and strategies. Plant Soil 443, 1–26. doi: 10.1007/s11104-019-04229-0
Soga, M., Yamaura, Y., Koike, S., and Gaston, K. J. (2014). Land sharing vs. land sparing: does the compact city reconcile urban development and biodiversity conservation? J. Appl. Ecol. 51, 1378–1386. doi: 10.1111/1365-2664.12280
Springmann, M., Mason-D'Croz, D., Robinson, S., Garnett, T., Godfray, H. C. J., Gollin, D., et al. (2016). Global and regional health effects of future food production under climate change: a modelling study. Lancet 387, 1937–1946. doi: 10.1016/S0140-6736(15)01156-3
St.Clair, S. B., and Lynch, J. P. (2010). The opening of Pandora's box: climate change impacts on soil fertility and crop nutrition in developing countries. Plant Soil 335, 101–115. doi: 10.1007/s11104-010-0328-z
Steinwand, M. A., and Ronald, P. C. (2020). Crop biotechnology and the future of food. Nat. Food 1, 273–283. doi: 10.1038/s43016-020-0072-3
Su, X., Cui, K., Du, S., Li, H., Lu, F., Shi, D., et al. (2018a). Efficient genome editing in cultured cells and embryos of debao pig and swamp buffalo using the CRISPR/Cas9 system. Vitro Cell. Dev. Biol. - Anim. 54, 375–383. doi: 10.1007/s11626-018-0236-8
Su, X., Wang, S., Su, G., Zheng, Z., Zhang, J., Ma, Y., et al. (2018b). Production of microhomologous-mediated site-specific integrated LacS gene cow using TALENs. Theriogenology 119, 282–288. doi: 10.1016/j.theriogenology.2018.07.011
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8:298. doi: 10.3389/fpls.2017.00298
Sun, Y., Zheng, G.-D., Nissa, M., Chen, J., and Zou, S.-M. (2020). Disruption of mstna and mstnb gene through CRISPR/Cas9 leads to elevated muscle mass in blunt snout bream (Megalobrama Amblycephala). Aquaculture 528:735597. doi: 10.1016/j.aquaculture.2020.735597
Syombua, E. D., Zhang, Z., Tripathi, J. N., Ntui, V. O., Kang, M., George, O. O., et al. (2019). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea Spp.). Plant Biotechnol. J. 19, 645–647. doi: 10.1111/pbi.13515
Tanaka, Y., and Ohmiya, A. (2008). Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 19, 190–197. doi: 10.1016/j.copbio.2008.02.015
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7:14438. doi: 10.1038/s41598-017-14832-9
Tang, X., Cao, X., Xu, X., Jiang, Y., Luo, Y., Ma, Z., et al. (2017). Effects of climate change on epidemics of powdery mildew in winter wheat in China. Plant Dis, 101, 1753–1760. doi: 10.1094/PDIS-02-17-0168-RE
Tanihara, F., Takemoto, T., Kitagawa, E., Rao, S., Do, L. T. K., Onishi, A., et al. (2016). Somatic cell reprogramming-free generation of genetically modified pigs. Sci. Adv. 2:e1600803. doi: 10.1126/sciadv.1600803
Tashkandi, M., Ali, Z., Aljedaani, F., Shami, A., and Mahfouz, M. M. (2018). Engineering resistance against tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 13:e1525996. doi: 10.1080/15592324.2018.1525996
Toenniessen, G., Adesina, A., and DeVries, J. (2008). Building an alliance for a green revolution in Africa. Ann. N. Y. Acad. Sci. 1136, 233–242. doi: 10.1196/annals.1425.028
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 Editing of endogenous banana streak virus in the B genome of Musa Spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 1–11. doi: 10.1038/s42003-019-0288-7
Tripathi, L., Ntui, V. O., and Tripathi, J. N. (2019). Application of genetic modification and genome editing for developing climate-smart banana. Food Energy Secur. 8:e00168. doi: 10.1002/fes3.168
Urban, M. C. (2015). Accelerating extinction risk from climate change. Science 348, 571–573. doi: 10.1126/science.aaa4984
USDA ARS (2021). Developing Abiotic Stress Tolerant Wheat Using Gene Editing. Available online at: https://www.ars.usda.gov/research/project?accnNo=434920 (accessed June 29, 2021).
Von Grebmer, K., Bernstein, J., Wiemers, M., Acheampong, K., Hanano, A., Higgins, B., et al. (2020). 2020 Global Hunger Index: One Decade to Zero Hunger - Linking Health and Sustainable Food Systems (Washington, DC), 80.
Von Grebmer, K., Saltzman, A., Birol, E., Wiesmann, D., Prasai, N., Yin, S., et al. (2014). 2014 Global Hunger Index: The Challenge of Hidden Hunger (Washington, DC), 56.
Wan, D.-Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., et al. (2020). CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis Vinifera). Hortic. Res. 7, 1–14. doi: 10.1038/s41438-020-0339-8
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLOS ONE 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, K., Ouyang, H., Xie, Z., Yao, C., Guo, N., Li, M., et al. (2015). Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System. Sci. Rep. 5, 1–11. doi: 10.1038/srep16623
Wang, K., Tang, X., Xie, Z., Zou, X., Li, M., Yuan, H., et al. (2017). CRISPR/Cas9-mediated knockout of myostatin in chinese indigenous erhualian pigs. Transgenic Res. 26, 799–805. doi: 10.1007/s11248-017-0044-z
Wang, S., Zhang, Y., Ju, W., Chen, J. M., Ciais, P., Cescatti, A., et al. (2020). Recent global decline of CO2 fertilization effects on vegetation photosynthesis. Science 370, 1295–1300. doi: 10.1126/science.abb7772
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. doi: 10.1089/crispr.2017.0010
Wang, X., Yu, H., Lei, A., Zhou, J., Zeng, W., Zhu, H., et al. (2015). Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 5:13878. doi: 10.1038/srep13878
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wen, S., Liu, H., Li, X., Chen, X., Hong, Y., Li, H., et al. (2018). TALEN-mediated targeted mutagenesis of fatty acid Desaturase 2 (FAD2) in peanut (Arachis Hypogaea L.) promotes the accumulation of oleic acid. Plant Mol. Biol. 97, 177–185. doi: 10.1007/s11103-018-0731-z
Whitworth, K. M., Lee, K., Benne, J. A., Beaton, B. P., Spate, L. D., Murphy, S. L., et al. (2014). Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 91:78. doi: 10.1095/biolreprod.114.121723
Whitworth, K. M., Rowland, R. R. R., Ewen, C. L., Trible, B. R., Kerrigan, M. A., Cino-Ozuna, A. G., et al. (2016). Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22. doi: 10.1038/nbt.3434
Whitworth, K. M., Rowland, R. R. R., Petrovan, V., Sheahan, M., Cino-Ozuna, A. G., Fang, Y., et al. (2019). Resistance to coronavirus infection in amino peptidase N-Deficient pigs. Transgenic Res. 28, 21–32. doi: 10.1007/s11248-018-0100-3
Wu, H., Wang, Y., Zhang, Y., Yang, M., Lv, J., Liu, J., et al. (2015). TALE nickase-mediated SP110 Knockin endows cattle with increased resistance to tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 112, E1530–E1539. doi: 10.1073/pnas.1421587112
Xiang, G., Ren, J., Hai, T., Fu, R., Yu, D., Wang, J., et al. (2018). Editing porcine IGF2 regulatory element improved meat production in chinese bama pigs. Cell. Mol. Life Sci. 75, 4619–4628. doi: 10.1007/s00018-018-2917-6
Xie, Z., Pang, D., Yuan, H., Jiao, H., Lu, C., Wang, K., et al. (2018). Genetically modified pigs are protected from classical swine fever virus. PLOS Pathog. 14:e1007193. doi: 10.1371/journal.ppat.1007193
Xing, S., Chen, K., Zhu, H., Zhang, R., Zhang, H., Li, B., et al. (2020). Fine-tuning sugar content in strawberry. Genome Biol. 21:230. doi: 10.1186/s13059-020-02146-5
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446. doi: 10.1016/j.molp.2019.08.006
Yan, Y., Wang, Y.-C., Feng, C.-C., Wan, P.-H. M., and Chang, K. T.-T. (2017). Potential distributional changes of invasive crop pest species associated with global climate change. Appl. Geogr. 82, 83–92. doi: 10.1016/j.apgeog.2017.03.011
Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757. doi: 10.1007/s00299-017-2118-z
Yosef, I., Edry-Botzer, L., Globus, R., Shlomovitz, I., Munitz, A., Gerlic, M., et al. (2019). A genetic system for biasing the sex ratio in mice. EMBO Rep. 20:e48269. doi: 10.15252/embr.201948269
Yu, B., Lu, R., Yuan, Y., Zhang, T., Song, S., Qi, Z., et al. (2016). Efficient TALEN-mediated myostatin gene editing in goats. BMC Dev. Biol. 16:26. doi: 10.1186/s12861-016-0126-9
Yu, H., Li, H., Li, Q., Xu, R., Yue, C., and Du, S. (2019). Targeted gene disruption in pacific oyster based on CRISPR/Cas9 ribonucleoprotein complexes. Mar. Biotechnol. 21, 301–309. doi: 10.1007/s10126-019-09885-y
Yu, S., Luo, J., Song, Z., Ding, F., Dai, Y., and Li, N. (2011). Highly efficient modification of Beta-Lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 21, 1638–1640. doi: 10.1038/cr.2011.153
Zafar, K., Sedeek, K. E. M., Rao, G. S., Khan, M. Z., Amin, I., Kamel, R., et al. (2020). Genome editing technologies for rice improvement: progress, prospects, and safety concerns. Front. Genome Ed. 2:5. doi: 10.3389/fgeed.2020.00005
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 Gene. Mol. Breed. 39:47. doi: 10.1007/s11032-019-0954-y
Zhang, J., Cui, M.-L., Nie, Y.-W., Dai, B., Li, F.-R., Liu, D.-J., et al. (2018). CRISPR/Cas9-mediated specific integration of Fat-1 at the goat MSTN locus. FEBS J. 285, 2828–2839. doi: 10.1111/febs.14520
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. Cell Mol. Biol. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, Y., Wang, Y., Yulin, B., Tang, B., Wang, M., Zhang, C., et al. (2019). CRISPR/Cas9-mediated sheep MSTN gene knockout and promote SSMSCs differentiation. J. Cell. Biochem. 120, 1794–1806. doi: 10.1002/jcb.27474
Zhang, Z., Ge, X., Luo, X., Wang, P., Fan, Q., Hu, G., et al. (2018). Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 9:842. doi: 10.3389/fpls.2018.00842
Zhong, Z., Niu, P., Wang, M., Huang, G., Xu, S., Sun, Y., et al. (2016). Targeted disruption of Sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 6:22953. doi: 10.1038/srep22953
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J.-S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhou, J., Xin, X., He, Y., Chen, H., Li, Q., Tang, X., et al. (2019). Multiplex QTL Editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 38, 475–485. doi: 10.1007/s00299-018-2340-3
Zhou, X., Liao, H., Chern, M., Yin, J., Chen, Y., Wang, J., et al. (2018). Loss of function of a rice TPR-Domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 3174–3179. doi: 10.1073/pnas.1705927115
Zou, Y., Li, Z., Zou, Y., Hao, H., Li, N., and Li, Q. (2018). An FBXO40 knockout generated by CRISPR/Cas9 causes muscle hypertrophy in pigs without detectable pathological effects. Biochem. Biophys. Res. Commun. 498, 940–945. doi: 10.1016/j.bbrc.2018.03.085
Zsögön, A., Cermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., et al. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi: 10.1038/nbt.4272
Glossary
Cisgenic: an organism engineered to contain genetic material derived from a sexually compatible species.
Climate adaptation: the process of adjusting to an anticipated climate and its associated conditions, including but not limited to altered temperature and precipitation.
Climate mitigation: the process of limiting the magnitude of climate change, usually by reduction or removal of greenhouse gases.
Homologous recombination: exchange of genetic material between a host organism and a desired template strand of DNA that encodes for the genetic material to be inserted flanked by regions that are complementary to adjacent sites in the host genome.
Intragenic: an organism engineered to contain only native genetic elements in a novel configuration.
Knock-in: a gene-editing approach in which genetic elements of interest are precisely inserted into a pre-defined locus.
Knockdown: a targeted mutation that reduces the expression of a gene.
Knockout: a targeted mutation that yields a non-functional gene product, synonymous with loss of function.
Promoter: the nucleotide sequence preceding a gene that specifies that gene's expression profile.
Transformation: process in which DNA is inserted into the genome of an organism.
Transgenic: an organism engineered to contain genetic material from a sexually incompatible organism.
Wild type: the typical form of a gene or organism in an unedited state.
Keywords: gene edited crops, livestock genetics, climate change, agriculture, food system, food security, livestock genetic resources, crop biotechnology
Citation: Karavolias NG, Horner W, Abugu MN and Evanega SN (2021) Application of Gene Editing for Climate Change in Agriculture. Front. Sustain. Food Syst. 5:685801. doi: 10.3389/fsufs.2021.685801
Received: 17 May 2021; Accepted: 28 July 2021;
Published: 07 September 2021.
Edited by:
Everlon Cid Rigobelo, São Paulo State University, BrazilReviewed by:
Eveline M. Ibeagha-Awemu, Agriculture and Agri-Food Canada (AAFC), CanadaMaura Santos Reis De Andrade Da Silva, Universidade Estadual de São Paulo, Brazil
Copyright © 2021 Karavolias, Horner, Abugu and Evanega. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah N. Evanega, c25kMkBjb3JuZWxsLmVkdQ==
 Nicholas G. Karavolias
Nicholas G. Karavolias Wilson Horner
Wilson Horner Modesta N. Abugu
Modesta N. Abugu Sarah N. Evanega
Sarah N. Evanega