- Core Technology Laboratories, Asahi Quality & Innovations, Ltd., Moriya-Shi, Japan
Reduction of greenhouse gas emissions is important to limit climate change. Because ruminant animals emit greenhouse gases, the worldwide plant-based alternative market is an emerging trend for eating less meat and dairy products. To produce plant-based dairy products such as yogurt alternatives, certain lactic acid bacterial species, which are used for cow's milk fermentation, are often used. Substrate changes from cow's milk to plant-based milk caused nutritional changes, and unsaturated fatty acids are more enriched in plant-based milk alternatives than in cow's milk. Unsaturated fatty acids can lead to the formation of aldehydes, some of which are off-flavors; therefore, substrate changes have the potential to alter the suitable lactic acid bacterial species used for fermentation to control flavor formation, such as aldehyde compounds. However, differences in the effect of the fermentation processes on aldehyde compounds have not been evaluated among lactic acid bacterial species. In this study, we comprehensively evaluated the effect of lactic acid bacterial fermentation on aldehyde compounds in synthetic medium and plant-based milk alternatives using 20 species of lactic acid bacteria. Heterofermentative lactic acid bacteria such as strains belonging to Limosilactobacillus had a strong aldehyde-reducing ability, likely from differences in alcohol dehydrogenase function. Because the odor detection threshold of ethanol compounds was lower than that of their equivalent aldehyde compounds, our findings are valuable for the fermentation of plant-based milk alternatives with lactic acid bacteria with the goal of decreasing off-flavors derived from aldehyde compounds.
Introduction
Climate change is one of the greatest concerns to humankind. To limit climate change, global action is needed to reduce greenhouse gas (GHG) emissions in many sectors, including the food sector. The role of the food sector involves reducing GHG emissions in agriculture and across the food chain and dietary changes within the population toward environmentally sustainable diets. One of the main dietary changes to reduce GHG emissions is to eat less meat and dairy products from ruminant animals, which emit GHGs (Macdiarmid, 2022). Under these circumstances, the worldwide plant-based alternative market is continuously growing, while one of the food trends in the dairy industry is the demand for alternative plant-based products. Plant-based dairy alternatives are not an entirely new product category, as they have been part of many food cultures historically, such as soy milk in China and tiger nut milk in Spain (Vaikma et al., 2021). A previous study defined plant-based milk alternatives as fluids that result from the size reduction of plant material extracted in water and further homogenization of such fluids, resulting in a particle size distribution in the range of 5–20 μm, which imitates cow's milk in appearance and consistency (Sethi et al., 2016). The plant-based milk alternatives are generally classified into five categories: cereal-based (e.g., oat milk, rice milk, corn milk, spelt milk), legume-based (e.g., soy milk, peanut milk, lupin milk, cowpea milk), nut-based (e.g., almond milk, coconut milk, hazelnut milk, pistachio milk, walnut milk), seed-based (e.g., sesame milk, flax milk, hemp milk, sunflower milk) and pseudocereal-based (e.g., quinoa milk, teff milk, amaranth milk).
In addition, plant-based yogurt alternatives, which are produced from plant-based milk alternatives, have been sold in many different countries, and several papers have discussed plant-based yogurt alternatives (Wu et al., 2021; Gupta et al., 2022). Boeck et al. (2021) reviewed the plant-based yogurt alternatives and indicated the lactic acid bacteria used for their fermentation. To produce plant-based yogurt alternatives, Streptococcus thermophilus (S. thermophilus), Lactobacillus delbrueckii subsp. bulgaricus (L. delbrueckii subsp. bulgaricus) and Lactobacillus acidophilus (L. acidophilus), have often been used for fermentation. These species are also used for fermentation of cow's milk because cow's yogurt has been traditionally manufactured using S. thermophilus and L. delbrueckii subsp. bulgaricus (Hamann and Marth, 1984), and acidophilus milk has been manufactured using L. acidophilus (Young and Nelson, 1978). There are many nutritional changes due to substrate changes from cow's milk to plant-based milk alternatives (Verduci et al., 2019). For example, the fatty acid profile differs from cow's milk to plant-based milk alternatives. While saturated fatty acids are dominant in cow's milk, unsaturated fatty acids such as oleic acid (C18:1) and linoleic acid (C18:2, ω-6) are dominant in plant-based milk alternatives (Martínez-Padilla et al., 2020). It is known that unsaturated fatty acids can lead to the formation of aldehydes as secondary oxidation products, and some of these aldehydes are known to be compounds that cause off-flavors in cow's milk (Huang et al., 2022). Therefore, plant-based milk alternatives rich in unsaturated fatty acids need to require even more attention to off-flavor formation than the cow's milk. The nutritional changes due to substrate changes from cow's milk to plant-based milk may alter the suitable lactic acid bacterial species used for fermentation for flavor formation, such as aldehydes. Previous report indicated that aldehydes such as hexanal, trans-2-nonenal and trans, trans-2,4-nonadienal were reduced by a few lactic acid bacterial strains (Vermeulena et al., 2007), however, there are few reports that have comprehensively evaluated lactic acid bacteria fermentation properties and their effect on aldehydes with using many lactic acid bacterial species. In this study, we evaluated the effect of aldehyde compounds, some of which are known as off-flavors in milk, on lactic acid bacterial fermentation. A total of 20 species, including 6 genera of lactic acid bacteria, all of which were classified into the genus Lactobacillus in the past (Zheng et al., 2020), were used for this evaluation. Here, we used rice, which is the most important crop in Asia. In recent report about plant-based materials for milk alternatives (Pua et al., 2022), nutritional compositions of carbohydrate, fiber, total protein, and total fat contents in the rice were showed as 80 %, 1.3 %, 7.1 % and 0.7 %, respectively, and the rice had a higher ratio of carbohydrate than the other plant-based materials. In additions, the rice is known as low allergenic food material than the other plant-based material (Pantoa et al., 2020), and is gluten-free food material (Rai et al., 2018), which is beneficial for people with celiac disease.
Materials and methods
Bacterial strains and culture conditions
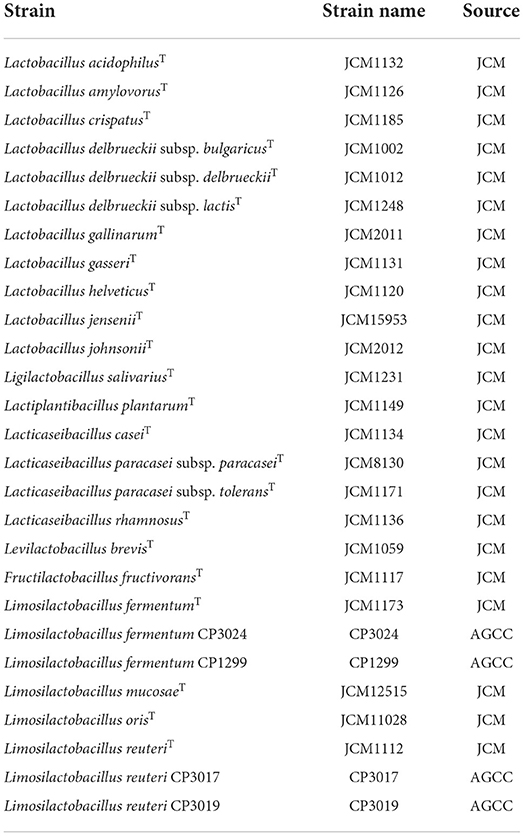
Lactic acid bacterial strains were obtained from the JCM (Japan Collection of Microorganisms, Ibaraki, Japan) and AGCC (Asahi Group Culture Collection, Ibaraki, Japan) (Table 1). Bacterial cultures were subcultured in de Man, Rogosa and Sharpe broth (Becton, Dickinson and Company, Sparks, MD) containing 0.05% L-cysteine hydrochloride (Nacalai Tesque, Inc, Kyoto, Japan) (MRSc broth) and incubated anaerobically at 37 °C for 16 h.
Chemical reagents and materials
The chemical reagents trans, trans-2,4-dinitrophenyl-hydrazine hydrochloride (DNPH), trans, trans-2,4-decadienal butylated hydroxytoluene (BHT), 2-methylpropanal, benzaldehyde, 3-methylbutanal, 2-methylbutanal, hexanal, 2,4-nonadienal, trans, and trans-2,4-decadienal were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Hexanal-d12 was purchased from Sigma–Aldrich (MO, USA). Acetaldehyde, ethanol, acetic acid, hexane and LC–MS grade acetonitrile were obtained from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan).
Sample collection using synthetic medium
Each aldehyde compound, which is acetaldehyde, hexanal or benzaldehyde, was used as a test compound in an experiment using synthetic medium. For experimental evaluation, cultivation method for anaerobic condition was performed as previously described (Prasad et al., 1998) with minor modifications. Briefly, the precultures of MRSc broth were inoculated with 5% v/v into MRSc broths containing test compounds, and the culture broths were incubated anaerobically at 37 °C for 16 h. MRSc broths containing test compounds without bacterial inoculation were also incubated as a negative control. After incubations, the optical density of the broth at 600 nm (OD 600) was measured by a microplate reader (Synergy HTX, Agilent Technologies, Santa Clara, CA). Then, the culture broths were centrifuged at 8,000 × g for 5 min, and the supernatants were collected. The supernatants were frozen at −20 °C until use.
Sample collection using a saccharified liquid of rice
A saccharified liquid of brown rice was obtained from Kikkoman Co., Ltd. (Chiba, Japan.) as a food product. The liquid of brown rice was diluted 1.67 times with water and sterilized at 95 °C for 15 min. For assuming industrial incubation, we used static condition, which was used in previous study (Sugahara et al., 2021). The precultures of MRSc broth were inoculated with 5% v/v into a saccharified liquid of brown rice. The liquid without bacterial inoculation was also incubated as a negative control. All of the liquid was incubated statically at 37 °C for 16 h. After incubation, the cultures were frozen at −20 °C until use.
For the study using the saccharified liquid of white rice, white rice powder (Oryza sativa subsp. japonica) and water were mixed by weight at a ratio of 3:7, and 0.03% v/v amylase (BAN 480 L, Novozymes Japan Ltd., Chiba, Japan) was added. After incubation at 60 °C for 6 h, the liquid was sterilized at 95 °C for 15 min. The precultures of MRSc broth were inoculated with 5% v/v into a saccharified liquid of white rice. The liquid without bacterial inoculation was also incubated as a negative control. All of the liquid was incubated statically at 37 °C for 16 h. After incubation, the cultures were frozen at −20°C until use.
Quantification of acetaldehyde, ethanol and acetic acid
Acetaldehyde, ethanol and acetic acid concentrations in the samples were determined by enzymatic analysis using an F-kit (Roche Diagnostics, Mannheim, Germany) as previously described (Ishiguro et al., 2014).
Sample preparation for aldehyde quantification using the LC–MS/MS method
Sample preparation was performed as previously described (Douny et al., 2016) with some modifications. For preparing standards, 440 μl of standard solutions dissolved in 50% ethanol, 220 μl of water and 220 μl of ethanol were mixed. For preparing samples, 440 μl of sample solution and 440 μl of ethanol were mixed. Then, 80 μl of BHT solution (1 mg/ml in ethanol) and 40 μl of hexanal-d12 solution (25 μg/ml in 50% ethanol) as an internal standard were added to each solution. After vortexing for 15 s, the solutions were centrifuged at 3,700 × g for 10 min. Then, 325 μl of the supernatant was mixed with 325 μl of DNPH solution [0.05 M in acetonitrile/acetic acid/water 8:1:1 (v/v)], and the reaction was carried out for 2 h at 60°C. After derivatization with DNPH, 1 ml of water was added to the reacted solution, and the solution was vortexed. The DNPH derivatives formed in the solution were extracted with 4 x 1 ml hexane. The solvent containing the DNPH derivatives was then evaporated to dryness in a centrifugal evaporator, and 1000 μl of 0.03% acetic acid solution/acetonitrile 60/40 (v/v) was added. The solution was vortexed and transferred to an injection vial.
Aldehyde compound quantification using LC–MS/MS
LC–MS/MS analysis was performed as previously described (Douny et al., 2016) with some modifications. We used the Nextera HPLC System (communication bus module: CBM-20A; pump: LC-30AD; autosampler: SIL-30AC; degasser: DGU-20A5R; column oven: CTO-20AC, Shimadzu, Kyoto, Japan) connected to a Sciex Triple quad 6500+ mass spectrometer (AB SCIEX, Concord, Canada). The Sciex Triple quad 6500+ (temperature: 500 °C; curtain gas: 30 psi; collision activated dissociation gas: 8 psi; ion source gas 1: 30 psi; ion source gas 2: 70; entrance potential:−10 V) was used with the parameters described in Table 2.
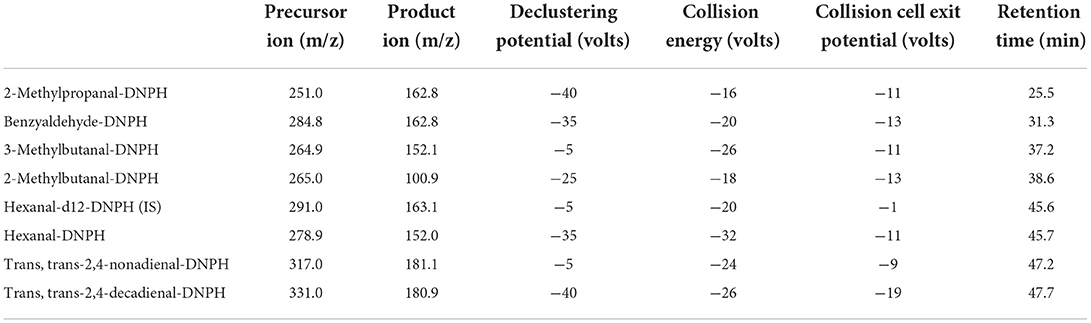
Table 2. Optimized multiple reaction monitoring transition and parameters for seven aldehydes and the internal standard (IS).
Separation was achieved on an Atlantis T3 C18 column (3 μm, 2.1 x 150 mm) with an Atlantis guard column T3 C18 (3 μm, 2.1 x 10 mm), both from Waters Corporation (MA, USA). The solvent flow was 0.25 ml/min, the column temperature was set at 40 °C, and the injection volume was 1 μl. The mobile phase was 0.03% acetic acid solution (solvent A) and acetonitrile (solvent B). The gradient elution conditions included a change from 40 to 48% solvent B in 2 min, and the conditions were held for 37 min. Then, the conditions were a change from 48 to 100% in 4 min, with a hold for 8 min. After that, the condition of solvent B was decreased to 40% in 4 min and maintained for another 6 min reconditioning. The analysis with the mass spectrometer was performed in MS/MS mode with negative ionization.
Results
Acetaldehyde metabolism in lactic acid bacterial strains
The growth ability of lactic acid bacterial strains in MRSc broth with or without 0.05% acetaldehyde was measured as OD 600 values to evaluate the acetaldehyde metabolizing ability associated with bacterial growth (Figure 1A). The OD 600 levels in some strains (L. acidophilusT, Lactobacillus crispatusT, Lactobacillus delbrueckii subsp. delbrueckiiT, Lactobacillus delbrueckii subsp. lactisT, Lactiplantibacillus plantarumT, Lacticaseibacillus caseiT, Lacticaseibacillus paracasei subsp. paracaseiT) showed significant differences between the MRSc broth with and without 0.05% acetaldehyde. Acetaldehyde, ethanol and acetic acid concentrations in the supernatants of the 23 type strains after cultivation in MRSc broth with 0.05% acetaldehyde are shown in Figures 1B–D. The acetaldehyde levels in the supernatants of all strains were significantly lower than the level in the negative control. The levels in the 6 strains (Levilactobacillus brevisT, Fructilactobacillus fructivoransT, Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT) were close to zero. The ethanol levels in the supernatants of 7 strains (Ligilactobacillus salivariusT, Levilactobacillus brevisT, Fructilactobacillus fructivoransT, Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT) showed significantly higher levels compared to the negative control. The levels of the supernatant of the 6 strains (Levilactobacillus brevisT, Fructilactobacillus fructivoransT, Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT) were notably higher than those of the other supernatants. The acetic acid levels in the supernatants of all strains did not show significantly higher levels compared to the negative control.
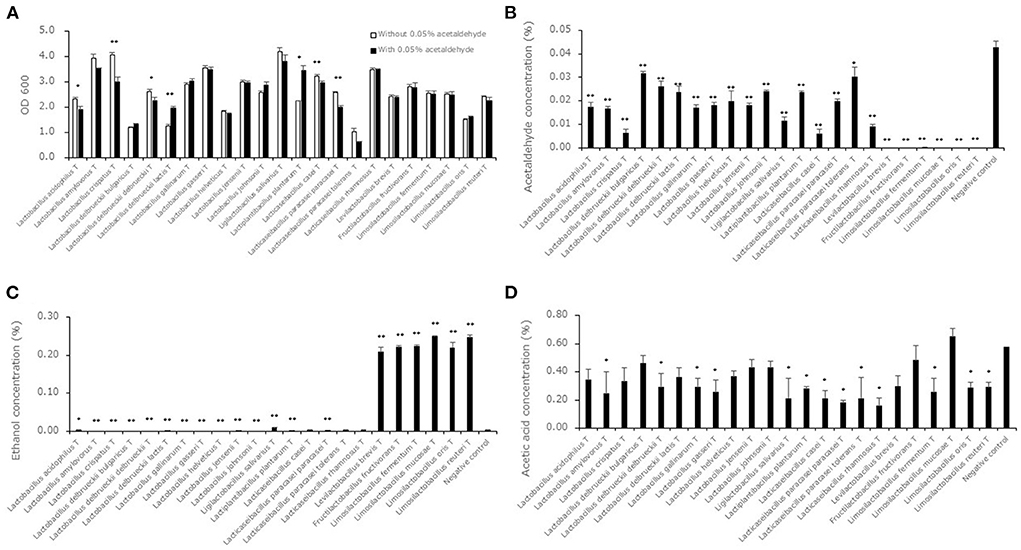
Figure 1. Acetaldehyde metabolism in lactic acid bacterial strains. Data are shown as the mean ± SD (n = 3). (A) White bars indicate the OD 600 values after cultivation in MRSc broth. Black bars indicate OD 600 values after cultivation in MRSc broth with 0.05% acetaldehyde. P values were calculated using Welch's t test between the levels in the MRSc broth and the MRSc broth with 0.05% acetaldehyde. *p < 0.05, **p < 0.01. (B) Black bars indicate acetaldehyde concentrations after cultivation in MRSc broth with 0.05% acetaldehyde. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01. (C) Black bars indicate ethanol concentrations after cultivation in MRSc broth with 0.05% acetaldehyde. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01. (D) Black bars indicate ethanol concentrations after cultivation in MRSc broth with 0.05% acetaldehyde. P values compared with the negative control were calculated using Welch's t test. *p < 0.05.
Ability of lactic acid bacterial strains to decrease hexanal and benzaldehyde
Hexanal and benzaldehyde concentrations in the supernatants of the 23 type strains after cultivation in MRSc broth with 0.05% hexanal or benzaldehyde are shown in Figures 2A,B. Although the acetaldehyde level of the supernatants of all strains was significantly lower than the level in the negative control, the hexanal level in the supernatant of the 7 strains (L. acidophilusT, L. delbrueckii subsp. bulgaricusT, Lactobacillus gallinarumT, Lactobacillus gasseriT, Lactobacillus helveticusT, Lacticaseibacillus paracasei subsp. paracaseiT, Lacticaseibacillus paracasei subsp. toleransT) showed no significant difference compared to that in the negative control. The benzaldehyde level in the supernatant of the 8 strains (L. acidophilusT, L. delbrueckii subsp. bulgaricusT, Lactobacillus gallinarumT, Lactobacillus gasseriT, Lactobacillus helveticusT, Ligilactobacillus salivarius T, Lacticaseibacillus paracasei subsp. paracaseiT, Lacticaseibacillus paracasei subsp. toleransT) showed no significant difference compared to that in the negative control. To evaluate the decrease in hexanal and benzaldehyde in high-concentration environments, we selected 6 strains (Levilactobacillus brevisT, Fructilactobacillus fructivoransT, Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT) as high-decreasing strains and Lactobacillus gasseriT as a low-decreasing strain. Hexanal and benzaldehyde concentrations in the supernatants of the seven selected strains after cultivation in MRSc broth with 0.2% hexanal or benzaldehyde are shown in Figures 2C,D. The hexanal and benzaldehyde levels in the supernatant of the four strains (Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT) were significantly lower than those in the supernatant of Levilactobacillus brevisT.
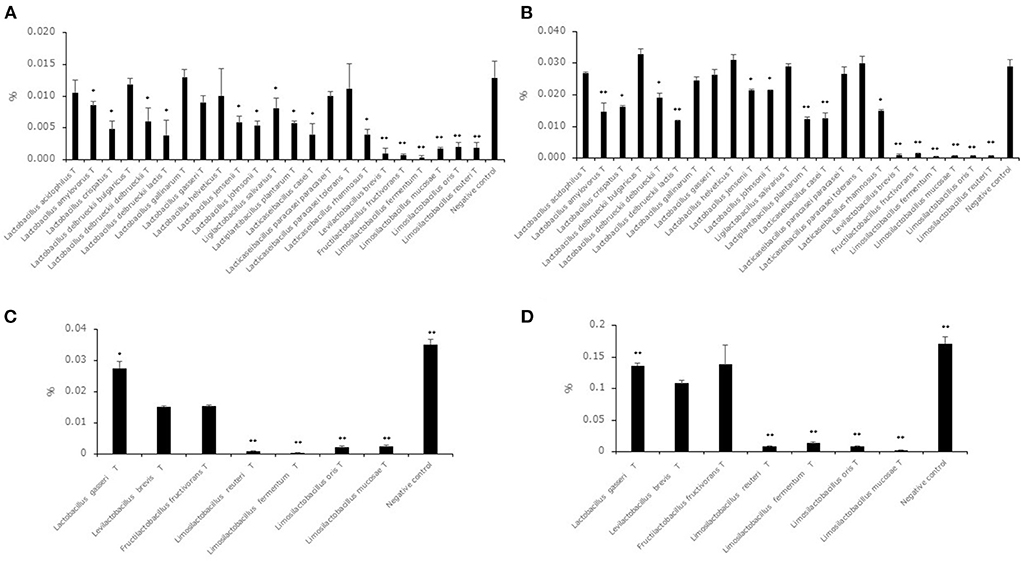
Figure 2. Hexanal and benzaldehyde decreasing ability in lactic acid bacterial strains. Data are shown as the mean ± SD (n = 3). (A) Bars indicate hexanal concentrations after cultivation in MRSc broth with 0.05% hexanal. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01. (B) Bars indicate benzaldehyde concentrations after cultivation in MRSc broth with 0.05% benzaldehyde. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01. (C) Bars indicate hexanal concentrations after cultivation in MRSc broth with 0.2% hexanal. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01. (D) Bars indicate benzaldehyde concentrations after cultivation in MRSc broth with 0.2% benzaldehyde. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01.
Aldehyde compound levels in the saccharified liquid of brown rice after cultivation
To evaluate the decreasing aldehyde compounds in Limosilactobacillus, which had a high ability to reduce aldehyde compounds in this study, we used nine strains belonging to Limosilactobacillus, and Lactobacillus gasseriT was used as a low-ability strain. Seven aldehyde compound (trans, trans-2,4-decadienal, trans, trans-2,4-nonadienal, 2-methylbutanal, 2-methylpropanal, 3-methylbutanal, benzaldehyde, hexanal) levels in the saccharified liquid of brown rice after cultivation were quantified (Figures 3A–H). Except for trans, trans-2,4-nonadienal levels, all aldehyde compound levels in the culture of 8 strains belonging to Limosilactobacillus were significantly lower than in the negative control. In addition, the total levels of seven aldehyde compounds in the culture of 8 strains belonging to Limosilactobacillus also showed significant differences compared to the negative control. The 3-methylbutanal level in the culture of Lactobacillus gasseriT was significantly higher than that in the negative control.
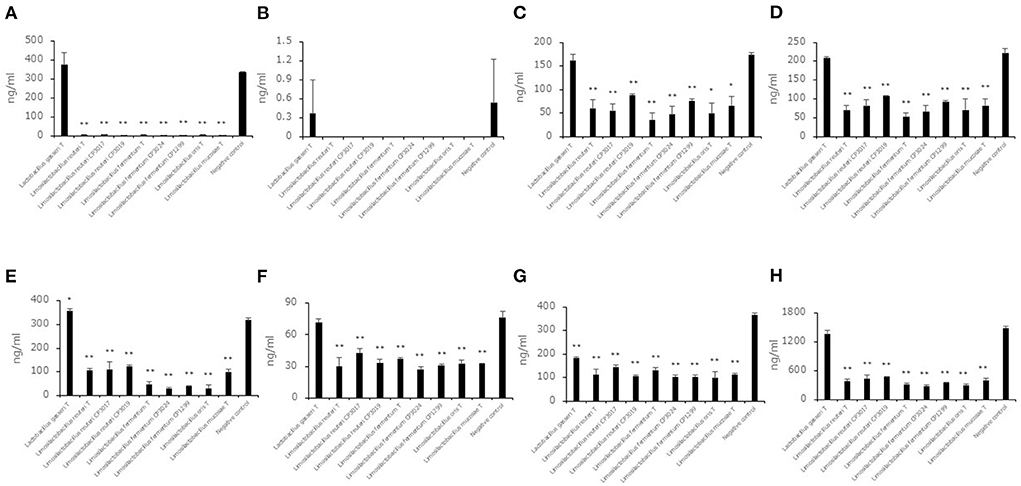
Figure 3. Aldehyde compound levels in the saccharified liquid of brown rice after cultivation. Data are shown as the mean ± SD (n = 3). Bars indicate aldehyde compound levels [(A) trans, trans-2,4-decadienal, (B) trans, trans-2,4-nonadienal, (C) 2-methylbutanal, (D) 2-methylpropanal, (E) 3-methylbutanal, (F) benzaldehyde, (G) hexanal, (H) total of seven aldehyde compounds] in the saccharified liquid of brown rice after cultivation. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01.
Aldehyde compound levels in the saccharified liquid of white rice after fermentation
Each of seven aldehyde compound levels in the saccharified liquid of white rice after fermentation was also measured (Figures 4A–H). Unlike the saccharified liquid of brown rice, the aldehyde compound levels, trans, trans-2,4-decadienal, trans, trans-2,4-nonadienal, 2-methylbutanal, 2-methylpropanal, 3-methylbutanal and benzaldehyde did not show a significant difference in the culture of some strains belonging to Limosilactobacillus. The hexanal level and total levels of seven aldehyde compounds in the culture of eight strains belonging to Limosilactobacillus showed significantly lower levels compared to the negative control. The 2-methylbutanal, 2-methylpropanal and 3-methylbutanal levels in the cultures of Lactobacillus gasseriT and Limosilactobacillus reuteriT were significantly higher than those in the negative control. The benzaldehyde level and total levels of seven aldehyde compounds in the culture of Lactobacillus gasseriT were significantly higher than those in the negative control.
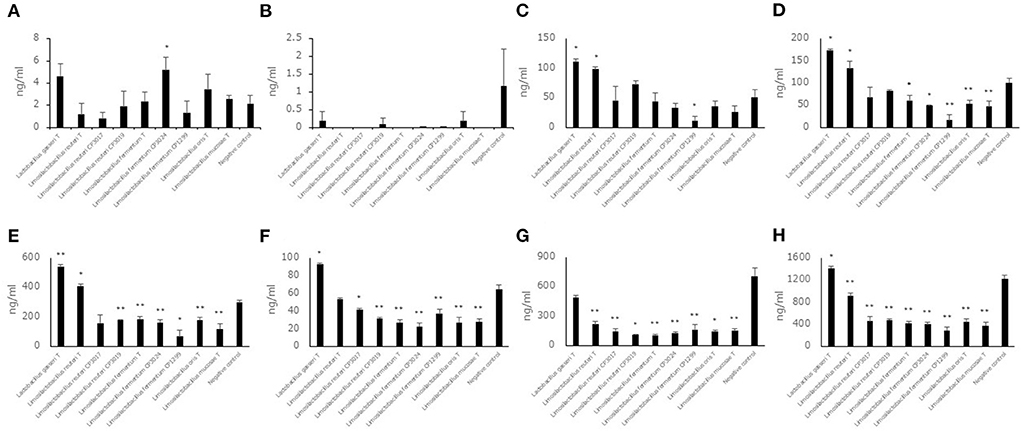
Figure 4. Aldehyde compound levels in the saccharified liquid of white rice after cultivation. Data are shown as the mean ± SD (n = 3). Bars indicate aldehyde compound levels [(A) trans, trans-2,4-decadienal, (B) trans, trans-2,4-nonadienal, (C) 2-methylbutanal, (D) 2-methylpropanal, (E) 3-methylbutanal, (F) benzaldehyde, (G) hexanal, (H) total of seven aldehyde compounds] in saccharified liquid of white rice after cultivation. P values compared with the negative control were calculated using Welch's t test. *p < 0.05, **p < 0.01.
Discussion
In the present study, it was considered important to evaluate the lactic acid bacterial fermentation properties of the effect on aldehyde compounds in plant-based milk alternatives because aldehyde compounds are known to cause off-flavors in cow milk. Therefore, we evaluated the ability of lactic acid bacterial strains to decrease aldehydes by using synthetic medium with aldehyde compounds, and we found that heterofermentative lactic acid bacterial strains had a strong ability to decrease aldehyde compounds. The strains belonging to the genus Limosilactobacillus, which is known as heterofermentative lactic acid bacterium, also showed a high ability to decrease aldehyde compounds in plant-based milk alternatives.
It is known that lactic acid bacteria have alcohol dehydrogenases (Halloum et al., 2015), and a previous study indicated that some lactic acid bacterial strains produce acetic acid through acetaldehyde metabolism (Nosova et al., 2000). Therefore, we analyzed acetaldehyde, ethanol and acetic acid levels in the supernatants of the synthetic medium after fermentation (Figure 1). The bacterial growth in some strains was influenced by the addition of 0.05% acetaldehyde to MRSc broth (Figure 1A). Because 0.5 of the OD600 level was approximately considered to be 500 million cells (Dimov et al., 2007), it was assumed that all bacterial strains were capable of growing in the broth. The acetaldehyde levels were decreased by lactic acid bacterial fermentation with bacterial growth (Figure 1B). The results for belonging to the genera Levilactobacillus, Fructilactobacillus and Limosilactobacillus, which showed acetaldehyde levels close to zero, indicated an obvious increase in ethanol levels (Figure 1C). Because all strains showed constant or decreased acetic acid levels (Figure 1D), it was suggested that acetaldehyde decreased in the genera Levilactobacillus, Fructilactobacillus and Limosilactobacillus because of alcohol dehydrogenase. A previous study, which showed core and pangenomes of 174 type strains of lactic acid bacteria, indicated that alcohol dehydrogenase was present in most heterofermentative lactobacilli but absent in most homofermentative lactobacilli (Zheng et al., 2015). Because the genera Levilactobacillus, Fructilactobacillus and Limosilactobacillus were defined as heterofermentative organisms, our results are consistent with the core and pangenomic analysis. In additions, previous report indicated that Limosilactobacillus reuteri and Fructilactobacillus sanfranciscensis, which are heterofermentative lactic acid bacteria, promptly reduced aldehydes, but Latilactobacillus sakei, which is homofermentative lactic acid bacterium, did not promptly (Vermeulena et al., 2007). The phenotypic result supports our results.
Although all strains decreased the acetaldehyde level in the synthetic medium with acetaldehyde, 7 strains and 8 strains did not decrease the level in the synthetic medium with hexanal and benzaldehyde, respectively (Figures 2A,B). This result suggested that middle-chain aldehydes and aromatic aldehydes were more difficult to reduce than short-chain aldehydes. Furthermore, benzaldehyde might be more difficult to reduce than middle-chain aldehydes from the result of Ligilactobacillus salivariusT. A previous study indicated that alcohol dehydrogenase derived from lactic acid bacteria had the ability to convert aldehyde compounds to alcohol compounds (Halloum et al., 2015). There is a possibility that alcohol dehydrogenase has different effects on compound structure, and it was thought that the high decrease in hexanal and benzaldehyde levels in the genera Levilactobacillus, Fructilactobacillus and Limosilactobacillus was derived from alcohol dehydrogenase. Previous studies indicated that the odor detection thresholds of hexanal, hexanol, benzaldehyde and benzyl alcohol were 0.019, 4.865, 0.35 and 10 mg/kg, respectively (Eriksson et al., 1976; Buttery et al., 1987). In addition, the value of the odor detection threshold in aldehyde compounds such as 2-methylpropanal, 2-methylbutanal and 3-methylbutanal showed higher values than in their equivalent ethanol compounds such as 2-methyl-1-propanol, 2-methyl-1-butanol and 3-methyl-1-butanol (Abraham et al., 2012). From these insights, a strong decreasing ability in the genera Levilactobacillus, Fructilactobacillus and Limosilactobacillus was thought to be useful for reducing off-flavors derived from aldehyde compounds in plant-based milk alternatives when aldehyde compounds were considered off flavors. In addition, we evaluated which strains used in this study had the highest decrease in aldehyde compounds among the Levilactobacillus brevisT, Fructilactobacillus fructivoransT, Limosilactobacillus fermentumT, Limosilactobacillus mucosaeT, Limosilactobacillus orisT and Limosilactobacillus reuteriT by using synthetic medium with 0.2% hexanal or benzaldehyde (Figures 2C,D). The results suggested that strains belonging to genus Limosilactobacillus were suitable for reducing aldehyde compounds in the lactic acid bacterial strains evaluated in this study.
To evaluate the reduction ability of aldehyde compounds in plant-based milk alternatives, we used the saccharified liquid of brown and white rice as brown and white rice milk (Figure 3, 4). In brown rice milk, all aldehyde compound levels except for those of trans, trans-2,4-nonadienal were decreased by fermentation with Limosilactobacillus (Figures 3A–H). In white rice milk, the total 7 aldehyde levels were decreased by fermentation with Limosilactobacillus, and some aldehyde compounds, such as 2-methylbutanal, 2-methylpropanal, 3-methylbutanal and benzaldehyde, were increased by fermentation with Lactobacillus gasseriT. A previous study indicated that 2-methylbutanal, 2-methylpropanal, 3-methylbutanal and benzaldehyde were produced from the lactic acid bacterial fermentation of branched-chain amino acids and phenylalanine (Yvon and Rijnen, 2001). Because some aldehyde compounds, such as 2-methylbutanal, 2-methylpropanal, 3-methylbutanal and benzaldehyde, were produced by lactic acid bacterial fermentation, the concentrations of these aldehyde compounds were increased by lactic acid bacterial fermentation in some cases. However, strains belonging to Limosilactobacillus had a strong aldehyde-reducing ability, and it is thought that this ability was the reason that aldehyde levels were maintained at a low level. Previous study indicated that an alcohol dehydrogenase derived from Levilactobacillus brevis LB19 reacted different rate with different substrate such as propionaldehyde, butyraldehyde and valeraldehyde (Halloum et al., 2015), therefore, there is possibility that the difference between production rates from the amino acids and the reduction rate by the alcohol dehydrogenase may affect the aldehydes concentrations after fermentation. In addition, the levels of aldehyde compounds in brown rice milk were the same between the negative control and that fermented by Lactobacillus gasseriT, and the levels of aldehyde compounds in white rice milk were increased by fermentation with Lactobacillus gasseriT. The results suggested that the different fermentation substrates showed increased levels of off-flavors. In this study, the ability to decrease aldehyde compounds in plant-based milk alternatives was not evaluated in the other heterofermentative lactic acid bacterial strains such as strains belonging to Levilactobacillus or Fructilactobacillus, at least strains belonging to genus Limosilactobacillus were found to strongly reduce aldehydes in plant-based milk alternatives compared with homofermentative lactic acid bacteria.
Conclusion
In conclusion, we evaluated the ability of lactic acid bacteria to decrease aldehyde compounds in synthetic medium and plant-based milk alternatives, and we found that heterofermentative lactic acid bacteria such as Limosilactobacillus had a strong aldehyde-reducing ability, and the ability was highly likely to be derived from differences in the function of alcohol dehydrogenase. We believe that our findings will be valuable for off-flavor suppression in the fermentation of plant-based milk alternatives with lactic acid bacteria, and that heterofermentative lactic acid bacteria would make a small but beneficial contribution to reducing greenhouse gas emissions by improving sensory properties of the fermented plant-based milk alternatives.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Author contributions
HS conceived and designed the experiments. HS, SK, and KN performed the experiments. HS and SK analyzed the data. HS, KS, and YN wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Asahi Quality & Innovations, Ltd.
Conflict of interest
Authors HS, SK, KN, KS, and YN were employed by company Asahi Quality & Innovations, Ltd.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abraham, M. H., Sánchez-Moreno, R., Cometto-Muñiz, J. E., and Cain, W. S. (2012). An algorithm for 353 odor detection thresholds in humans. Chem. Senses. 37, 207–218. doi: 10.1093/chemse/bjr094
Boeck, T., Sahin, A. W., Zannini, E., and Arendt, E. K. (2021). Nutritional properties and health aspects of pulses and their use in plant-based yogurt alternatives. Compr. Rev. Food Sci. Food Saf. 20, 3858–3880. doi: 10.1111/1541-4337.12778
Buttery, R. G., Ling, L. C., and Light, D. M. (1987). Tomato leaf volatile aroma components. J. Agric. Food Chem. 35, 1039–1042. doi: 10.1021/jf00078a043
Dimov, S., Kirilov, N., Peykov, S., and Ivanova, I. (2007). Growth characteristics of twenty Lactobacillus delbrueckii strains isolated from bulgarian home made yoghurts. Biotechnol. and Biotechnol. Eq. 21, 172–176. doi: 10.1080/13102818.2007.10817439
Douny, C., Bayram, P., Brose, F., Degand, G., and Scippo, M. L. (2016). Development of an LC-MS/MS analytical method for the simultaneous measurement of aldehydes from polyunsaturated fatty acids degradation in animal feed. Drug Test. Anal. 8, 458–464. doi: 10.1002/dta.2013
Eriksson, C. E., Lundgren, B., and Vallentin, K. (1976). Odor detectability of aldehydes and alchols originating from lipid oxidation. Chem. Senses. 2, 3–15. doi: 10.1093/chemse/2.1.3
Gupta, M. K., Torrico, D. D., Ong, L., Gras, S. L., Dunshea, F. R., and Cottrell, J. J. (2022). Plant and dairy-based yogurts: a comparison of consumer sensory acceptability linked to textural analysis. Foods. 11, 463. doi: 10.3390/foods11030463
Halloum, I., Thompson, B., Pugh, S., and Nielsen, D. R. (2015). Activity of Lactobacillus brevis alcohol dehydrogenase on primary and secondary alcohol biofuel precursors. Fermentation. 1, 24–37. doi: 10.3390/fermentation1010024
Hamann, W. T., and Marth, E. H. (1984). Survival of Streptococcus thermophilus and Lactobacillus bulgaricus in commercial and experimental yogurts. J. Food Prot. 47, 781–786. doi: 10.4315/0362-028x-47.10.781
Huang, G., Li, N., Liu, K., Yang, J., Zhao, S., Zheng, N., et al. (2022). Effect of flaxseed supplementation in diet of dairy cow on the volatile organic compounds of raw milk by HS-GC-IMS. Front. Nutr. 9, 831178. doi: 10.3389/fnut.2022.831178
Ishiguro, K., Ando, T., Maeda, O., Watanabe, O., and Goto, H. (2014). Suppressive action of acetate on interleukin-8 production via tubulin-α acetylation. Immunol. Cell Biol. 92, 624–630. doi: 10.1038/icb.2014.31
Macdiarmid, J. I. (2022). The food system and climate change: are plant-based diets becoming unhealthy and less environmentally sustainable? Proc. Nutr. Soc. 81, 162–167. doi: 10.1017/S0029665121003712
Martínez-Padilla, E., Li, K., Frandsen, H. B., Joehnke, M. S., Vargas-Bello-Pérez, E., and Petersen, I. L. (2020). In vitro protein digestibility and fatty acid profile of commercial plant-based milk alternatives. Foods. 9, 1784. doi: 10.3390/foods9121784
Nosova, T., Jousimies-Somer, H., Jokelainen, K., Heine, R., and Salaspuro, M. (2000). Acetaldehyde production and metabolism by human indigenous and probiotic Lactobacillus and Bifidobacterium strains. Alcohol Alcohol. 35, 561–568. doi: 10.1093/alcalc/35.6.561
Pantoa, T., Baricevic-Jones, I., Suwannaporn, P., Kadowaki, M., Kubota, M., Roytrakul, S., et al. (2020). Young rice protein as a new source of low allergenic plant-base protein. Journal of Cereal Science. 93, 102970. doi: 10.1016/j.jcs.2020.102970
Prasad, J., Gillab, H., Smarta, J., and Gopala, K. P. (1998). Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 8, 993–1002. doi: 10.1016/S0958-6946(99)00024-2
Pua, A., Tang, V. C. Y., Goh, R. M. V., Sun, J., Lassabliere, B., and Liu, S. Q. (2022). Ingredients, processing, and fermentation: addressing the organoleptic boundaries of plant-based dairy analogues. Foods. 11, 875. doi: 10.3390/foods11060875
Rai, S., Kaur, A., and Chopra, C. S. (2018). Gluten-free products for celiac susceptible people. Front. Nutr. 5, 116. doi: 10.3389/fnut.2018.00116
Sethi, S., Tyagi, S. K., and Anurag, R. K. (2016). Plant-based milk alternatives an emerging segment of functional beverages: a review. J. Food Sci. Technol. 53, 3408–3423. doi: 10.1007/s13197-016-2328-3
Sugahara, H., Nagayama, K., Ikeda, S., Hirota, T., and Nakamura, Y. (2021). D- and L-amino acid concentrations in culture broth of Lactobacillus are highly dependent on the phylogenetic group of Lactobacillus. Biochem. Biophys. Rep. 27, 101073. doi: 10.1016/j.bbrep.2021.101073
Vaikma, H., Kaleda, A., Rosend, J., and Rosenvald, S. (2021). Market mapping of plant-based milk alternatives by using sensory (RATA) and GC analysis. Future Foods. 4, 100049. doi: 10.1016/j.fufo.2021.100049
Verduci, E., D'Elios, S., Cerrato, L., Comberiati, P., Calvani, M., Palazzo, S., et al. (2019). Cow's milk substitutes for children: nutritional aspects of milk from different mammalian species, special formula and plant-based beverages. Nutrients. 11, 1739. doi: 10.3390/nu11081739
Vermeulena, N., Czernyb, M., Gänzle, M. G., Schieberleb, P., and Vogela, R. F. (2007). Reduction of (E)-2-nonenal and (E,E)-2,4-decadienal during sourdough fermentation. J. Cereal Sci. 45, 78–87. doi: 10.1016/j.jcs.2006.07.002
Wu, L., Wen, Y., Chen, W., Yan, T., Tian, X., and Zhou, S. (2021). Simultaneously deleting ADH2 and THI3 genes of Saccharomyces cerevisiae for reducing the yield of acetaldehyde and fusel alcohols. FEMS Microbiol. Lett. 368, fnab094. doi: 10.1093/femsle/fnab094
Young, C. K., and Nelson, F. E. (1978). Survival of Lactobacillus acidophilus in “sweet acidophilus milk” during refrigerated storage. J. Food Prot. 41, 248–250. doi: 10.4315/0362-028x-41.4.248
Yvon, M., and Rijnen, L. (2001). Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11, 185–201. doi: 10.1016/S0958-6946(01)00049-8
Zheng, J., Ruan, L., Sun, M., and Gänzle, M. (2015). A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 81, 7233–7243. doi: 10.1128/aem.02116-15
Zheng, J., Wittouck, S., Salvetti, E., Franz, C., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: lactic acid bacteria, Limosilactobacillus, plant-based milk, aldehyde, off-flavor, heterofermentative
Citation: Sugahara H, Kato S, Nagayama K, Sashihara K and Nagatomi Y (2022) Heterofermentative lactic acid bacteria such as Limosilactobacillus as a strong inhibitor of aldehyde compounds in plant-based milk alternatives. Front. Sustain. Food Syst. 6:965986. doi: 10.3389/fsufs.2022.965986
Received: 10 June 2022; Accepted: 16 August 2022;
Published: 02 September 2022.
Edited by:
B. N. Dar, Islamic University of Science and Technology, IndiaReviewed by:
Michael Gänzle, University of Alberta, CanadaSheikh Rafeh, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, India
Copyright © 2022 Sugahara, Kato, Nagayama, Sashihara and Nagatomi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirosuke Sugahara, aGlyb3N1a2Uuc3VnYWhhcmFAYXNhaGktcWkuY28uanA=
 Hirosuke Sugahara
Hirosuke Sugahara Sayaka Kato
Sayaka Kato