Abstract
Staphylococci are a versatile genus of bacteria that are capable of causing acute and chronic infections in diverse host species. The success of staphylococci as pathogens is due in part to their ability to mitigate endogenous and exogenous oxidative and nitrosative stress. Endogenous oxidative stress is a consequence of life in an aerobic environment; whereas, exogenous oxidative and nitrosative stress are often due to the bacteria's interaction with host immune systems. To overcome the deleterious effects of oxidative and nitrosative stress, staphylococci have evolved protection, detoxification, and repair mechanisms that are controlled by a network of regulators. In this review, we summarize the cellular targets of oxidative stress, the mechanisms by which staphylococci sense oxidative stress and damage, oxidative stress protection and repair mechanisms, and regulation of the oxidative stress response. When possible, special attention is given to how the oxidative stress defense mechanisms help staphylococci control oxidative stress in the host.
Introduction
The genus staphylococcus represents a broad family of related species and subspecies that are widely distributed in nature and that colonize skin, skin glands, and mucous membranes of humans and other warm-blooded animals (Götz et al., 2006). Several staphylococcal species have commercial value and are routinely used as starter cultures in fermentation processes (Schleifer and Fischer, 1982; Geisen et al., 1992; Tanasupawat et al., 1992; Hammes et al., 1995; Montel et al., 1996; Probst et al., 1998; Place et al., 2003), whereas others have achieved importance as animal and human pathogens. On this latter point, some staphylococci, such as Staphylococcus epidermidis, are pathogens primarily due to their ability to colonize indwelling medical devices and to form biofilms (von Eiff et al., 2002; Vuong and Otto, 2002; McCann et al., 2008; Rogers et al., 2009). In contrast, Staphylococcus aureus is capable of causing a variety of diseases ranging from soft tissue infections to life-threatening septicemia. The ability of S. aureus to cause this wide array of infections is due to its diverse range of virulence factors and its resistance to numerous antibiotics. In addition, S. aureus is a prevalent cause of infections due to the fact that 20–30% of humans carry S. aureus in their anterior nares (Kluytmans et al., 1997; von Eiff et al., 2001; Wertheim et al., 2005). Lastly, the prominence of S. aureus as a pathogen is also due to its ability to evade or defend itself from the host immune system (Voyich et al., 2005; Palazzolo-Ballance et al., 2008; Foster, 2009). For these reasons, S. aureus is the most prominent staphylococcal pathogen of nosocomial and community-acquired infections and a leading cause of human infections worldwide (Lowy, 1998; Diekema et al., 2001; Stevens, 2003; Grundmann et al., 2006; Chambers and DeLeo, 2009; Rosenthal et al., 2010; Johnson, 2011). As oxidative and nitrosative killing mechanisms are important for the host immune response, this review will focus on the ability of staphylococci to resist oxidative stress with an emphasis on S. aureus, for which the greatest amount of information is available.
Endogenous and exogenous oxidative and nitrosative stress
Endogenous oxidative stress can be caused by many things, including aerobic respiration, autooxidation reactions, intracellular redox reactions, and antibiotics (Pomposiello and Demple, 2002; Imlay, 2003; Kohanski et al., 2007; Yeom et al., 2010). During aerobic respiration, oxygen functions as a final electron acceptor in the electron transport chain where its complete reduction results in the formation of H2O. Occasionally, oxygen undergoes incomplete reduction on interaction with flavoproteins (e.g., oxidases and monooxygenases) and can generate reactive oxygen species (ROS) (Messner and Imlay, 1999). Upon interaction with the reduced FAD cofactor of flavoenzymes, one or two electrons are transferred to molecular oxygen (Müller, 1987), leading to the generation of endogenous superoxide anions (O−2) and hydrogen peroxide (H2O2) (Massey et al., 1969; Korshunov and Imlay, 2010). In addition to flavoenzyme catalyzed reactions generating ROS, other reactions can also produce reactive oxygen intermediates (Imlay et al., 1988); specifically, Fenton chemistry can produce the highly-reactive hydroxyl radicals (HO·). This chemistry occurs when iron reacts with H2O2 and generates HO· (Figures 1 and 2). Fenton chemistry has been hypothesized to be possible with other metals such as Cu and Cd; however, questions remain about the physiological significance of non-ferrous metals catalyzing this chemistry (Macomber and Imlay, 2009).
Figure 1
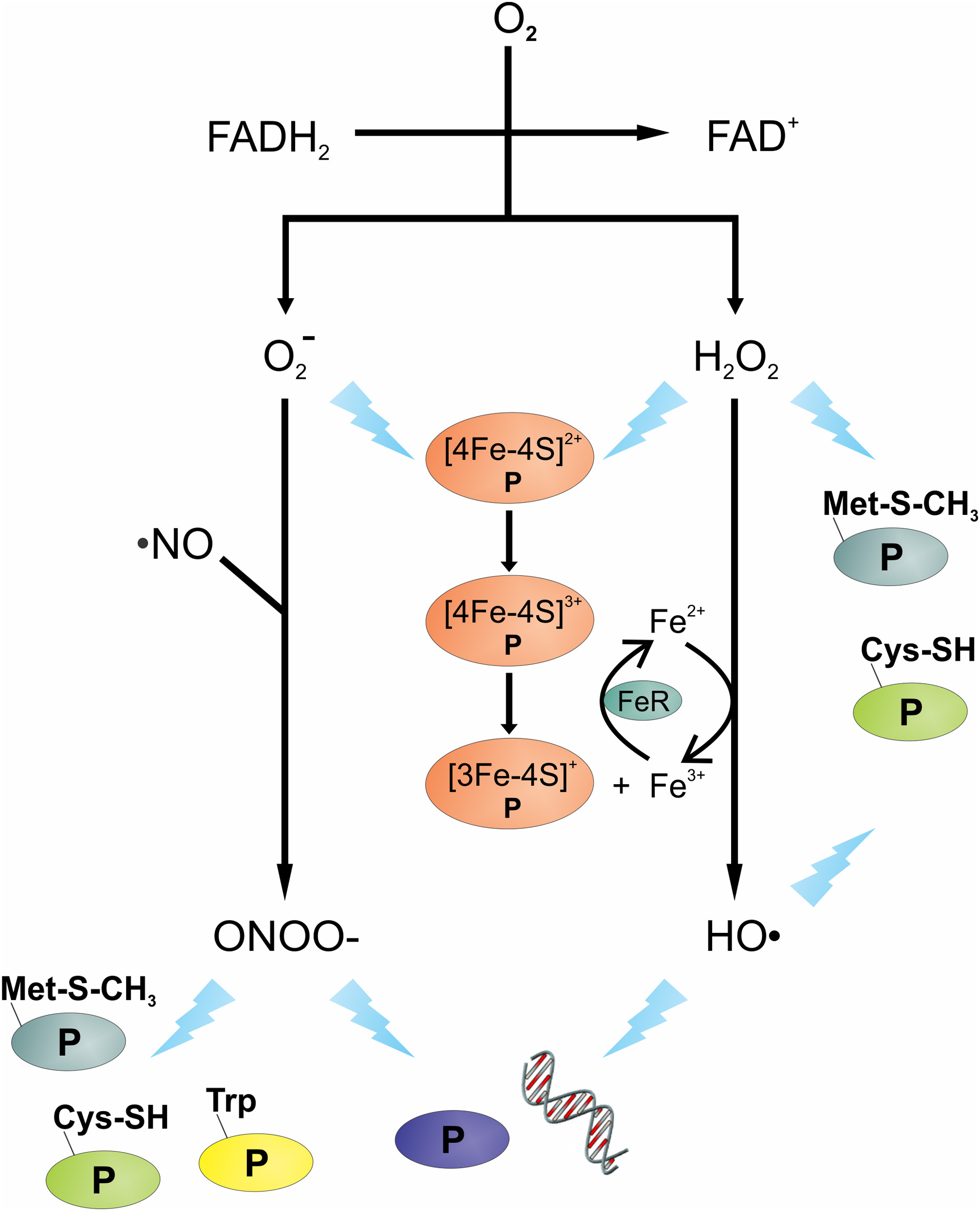
Overview of oxidative and nitrosative stressors and their potential targets. The transfer of electron(s) from the reduced FAD of flavoenzymes to oxygen (O2) can produce superoxide anions (O−2) and/or hydrogen peroxide (H2O2). Reaction of O−2 with nitric oxide (·NO) can lead to the formation of peroxynitrite (OONO−). Intracellular ferric (Fe3+) reduction is catalyzed by ferric reductase (FeR), the Fe2+ can react with H2O2 to generate hydroxyl radicals (HO·). Damage to DNA and protein(s) is shown as a lightning bolt. Proteins are presented using letter “P”.
Figure 2
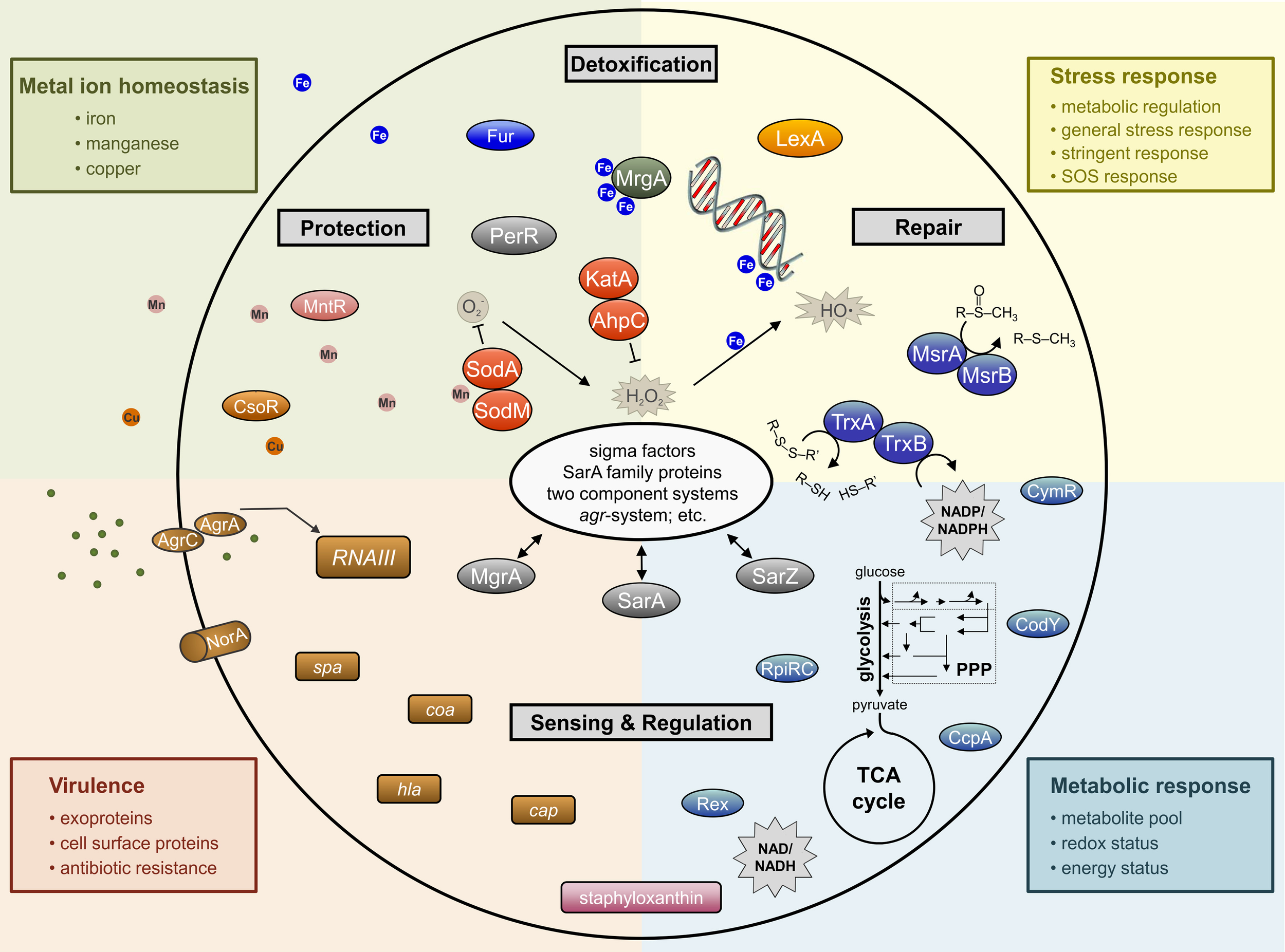
Simplified schematic overview of important determinants involved in staphylococcal response to oxidative stress affecting whole cell physiology.
In addition to endogenous oxidative stress, exogenous oxidative stress is a common challenge that bacteria must overcome in order to survive. Host innate immune cells such as macrophages, monocytes, and neutrophils have NADPH oxidase (NOX) that is responsible for the generation of the O−2 during an oxidative burst. Superoxide is generated when the catalytic subunit of NOX transfers electrons from NADPH to oxygen; a process requiring FAD and heme (Nauseef, 2004). Once formed, O−2 can undergo dismutation to H2O2. This H2O2 that is formed after dismutation of O−2 can be used by the myeloperoxidase (MPO) complex to produce the bactericidal compound hypochlorite (OCl−).
MPO, a 150 KDa heme-containing protein complex, is released into the phagosome when azurophilic granules fuse with the phagosome. In the phagosome, MPO binds to bacteria and catalyzes the H2O2-dependent conversion of Cl− to OCl− (Harrison and Schultz, 1976; Klebanoff, 1999). In addition to the generation of OCl−, MPO is involved in the oxidation of L-tyrosine, such as that found in enkephalins, to the potent cytotoxic tyrosyl radical (o,o′-dityrosine) (Heinecke et al., 1993). That being said, the susceptibility of bacteria to MPO-mediated killing varies; hence, MPO is not considered essential for the innate immune response to bacteria (Lehrer et al., 1969; Lanza, 1998; Allen and Stephens, 2011).
Nitric oxide (·NO) is produced by all immune cells and it is important in the control of pathogens; however, like MPO, it is not equally effective against all pathogens (Bogdan et al., 2000). As an example, in mice, the function of ·NO in S. aureus control is limited, whereas for Salmonella enterica serovar Typhimurium it is critical (Nathan and Shiloh, 2000; Vazquez-Torres et al., 2000). Like ROS, NO is a reactive oxidant with potent cytotoxic properties against bacteria. In human macrophage, nitric oxide synthase (iNOS or NOS2) is induced on encountering a pathogen or by activation via cytokines. Once induced, iNOS or NOS2 catalyzes the conversion of L-arginine to L-citrulline and ·NO, a reaction that also reduces oxygen and oxidizes NADPH. While ·NO is toxic to bacteria by itself, ·NO has a synergistic effect with H2O2 to facilitate bacterial killing (Brunelli et al., 1995; Woodmansee and Imlay, 2003; Han et al., 2009). In addition, ·NO and O−2 can form the bactericidal compound peroxynitrite (OONO−) (Figure 1), a highly reactive nitrogen intermediate (Huie and Padmaja, 1993).
Bacterial targets of oxidative damage
The toxicity of ROS is due to its ability to damage any oxidizable moiety in a biological molecule. In E. coli, the importance of ROS damage has been demonstrated in mutants that lack components of the oxidative stress response system (i.e., superoxide dismutase, catalase, and peroxidase) (Carlioz and Touati, 1986; Park et al., 2005b; Jang and Imlay, 2007). In mutants lacking multiple genes of the oxidative stress response, the bacteria were highly sensitive to oxidants such as paraquat and H2O2. In this section we will attempt to address some of the consequences of oxidative stress in bacteria.
Superoxide and H2O2 can facilitate the release of iron from Fe-S cluster containing proteins, such as aconitase or serine dehydratase. Both O−2 and H2O2 can oxidize the [4Fe-4S]2+ to [4Fe-4S]3+, which can be further oxidized, leading to the release of iron and the inactivation of the enzyme (Kuo et al., 1987; Flint et al., 1993; Jang and Imlay, 2007). In the presence of H2O2, the iron liberated from Fe-S clusters creates an intracellular environment permissive to Fenton chemistry, which generates the highly reactive HO·. Hydroxyl radicals will react with virtually the first molecule that it encounters; hence, to induce damage it must be in close proximity to its cellular target. DNA is a charged molecule that attracts positively charged molecules, like Fe2+; hence, charge-charge interaction brings iron in close proximity to the DNA phosphodiester backbone. The close proximity of Fe2+ to DNA means that HO· generated by Fenton chemistry will likely react with DNA, inducing lethal or non-lethal mutations (Keyer and Imlay, 1996).
In addition to DNA being a target of ROS, amino acids, and proteins can be oxidized and/or modified by ROS. H2O2 can react with the Fe2+ of an iron-containing protein, presumably through Fenton chemistry, to cause irreversible protein carbonylation and the formation of protein aggregates (Dukan et al., 1999; Davies, 2005). Cysteine, methionine, and tryptophan can be oxidized by H2O2, HO·, and ONOO−, which can lead to reversible or irreversible enzymatic inactivation. As an example, oxidation of cysteine residues can lead to reversible modifications (i.e., sulfenic acid or S-thiolation) or irreversible modifications (i.e., sulfinic acid, sulfonic acid) (Chouchani et al., 2011). Similarly, oxidation of methionine can lead to the formation of methionine sulfoxides, which are reversible through the action of the methionine sulfoxide reductase (discussed in Section “Methionine sulfoxide reductase”). The formation of some important ROS and the potential damage they cause are summarized in Figure 1.
Oxidative stress resistance mechanisms
Pigmentation
As a general rule, most S. aureus strains isolated from human infections will form yellowish-orange or golden colonies due to the presence of carotenoid pigments. These pigments become more pronounced after 24 h of growth and when held at room temperature (Willis and Turner, 1962; Jacobs and Willis, 1964). An exception to this rule are the small colony variant (SCV) S. aureus, which are non-pigmented and may have auxotrophies for hemin, menadione, thiamine, or thymidine. SCVs are often associated with persistent and recurrent infections and are characterized by numerous phenotypic changes (Proctor et al., 2006). The main pigment of S. aureus is the membrane-bound orange-red C30 triterpenoid staphyloxanthin, which is synthesized from the enzymes coded within the crtOPQMN operon (Marshall and Wilmoth, 1981a,b; Pelz et al., 2005). The synthesis of staphyloxanthin involves the head-to-head condensation of two C15 isoprenoid molecules of farnesyl diphosphate to form dehydrosqualene, a reaction catalysed by dehydrosqualene synthase (CrtM). Dehydrosqualene is converted into 4,4′-diaponeurosporene by dehydrosqualene desaturase (CrtN), which is further oxidized, glycosylated, and esterified to yield staphyloxanthin (Wieland et al., 1994; Pelz et al., 2005). The crtOPQMN operon is under positive transcriptional control from the rsbUVW-σB system (Kullik et al., 1998; Giachino et al., 2001; Palma and Cheung, 2001; Bischoff et al., 2004) and under negative regulation by the small RNA, SsrA RNA (Liu et al., 2010).
Carotenoid pigments protect S. aureus against desiccation and photosensitization, and are known to quench toxic singlet oxygen. On this latter point, carotenoids are potent antioxidants due to their numerous conjugated double bonds, which make them an important survival factor for detoxifying ROS (Grinsted and Lacey, 1973; Mathews-Roth et al., 1974; Dahl et al., 1989; Krinsky, 1993; El-Agamey et al., 2004). The importance of staphyloxanthin in protecting S. aureus against ROS is seen in non-pigmented crt mutants, that grow normally, but have increased sensitivity toward ROS, OONO−, and HOCl (Liu et al., 2005; Clauditz et al., 2006). One consequence of this increased sensitivity to oxidants is that S. aureus strains deficient in carotenoid biosynthesis are more readily cleared by the innate immune response (Salamah, 1992; Liu et al., 2005, 2008; Clauditz et al., 2006; Olivier et al., 2009). In a mouse subcutaneous abscess model as well as a systemic S. aureus infection model, non-pigmented S. aureus have reduced virulence and survival relative to the pigmented wild-type strain (Liu et al., 2005, 2008). The resistance to phagocytic killing that is conferred by pigments is primarily attributed to the antioxidant properties of carotenoids, since the effect is not seen in NOX deficient mice or when the oxidative burst is inhibited (Liu et al., 2005). In addition to the antioxidant properties of pigments, staphyloxanthin can also alter membrane rigidity, which is important in protecting against non-oxidative host defences mediated by cationic peptides (Mishra et al., 2011). Because of the contribution of carotenoid pigments to staphylococcal fitness, inhibition of carotenoid biosynthesis is viewed as a potential therapeutic target in treating S. aureus infections (Daum, 2008; Haebich and von Nussbaum, 2008; Liu et al., 2008; Walsh and Fischbach, 2008; Song et al., 2009a,b; Oldfield, 2010).
Detoxifying enzymes
In addition to pigments, most staphylococci possess several enzymes that are used in the detoxification of reactive oxygen and nitrogen intermediates; particularly, superoxide dismutases, catalases, glutathione peroxidases, globins, and peroxiredoxins (Figure 2).
Superoxide dismutase (SOD)
Superoxide dismutases are metalloenzymes that catalyse the dismutation of O−2 to oxygen and H2O2, which can be further reduced to water and oxygen by catalase or alkyl hydroperoxide reductase. By preventing the accumulation of O−2, SODs not only protect the bacterium from damage caused by O−2, but also against products that are derived from reactions requiring O−2, such as OONO− (Figure 1). SODs are classified into one of four types based on the metal ion cofactor; specifically, these are the copper-zinc type (Cu/Zn-SOD), the manganese type (Mn-SOD), the iron type (Fe-SOD), and the nickel type (Ni-SOD) (Fridovich, 1995; Kim et al., 1996, 1998). S. aureus possess two monocistronic superoxide dismutase genes, sodA and sodM (Poyart et al., 1995; Clements et al., 1999; Valderas and Hart, 2001); whereas, coagulase-negative staphylococci lack the sodM gene (Barrière et al., 2001a,b; Valderas et al., 2002).
The S. aureussodA gene was identified during a screen for amino acid starvation survival mutants (Watson et al., 1998) and during characterization of that mutant, it was observed in a zymogram analysis that S. aureus had three zones of SOD activity (Clements et al., 1999). The three zones of activity were attributed to the presence of two homodimers and a heterodimer, which confirmed the presence of a previously identified second SOD (SodM) (Poyart et al., 1995; Valderas and Hart, 2001). During in vitro aerobic growth of S. aureus, the transcription and activity of both SODs increase in the post-exponential growth phase and remain high during the stationary phase, with SodA being responsible for the majority of SOD activity (Clements et al., 1999; Valderas and Hart, 2001; Karavolos et al., 2003). The transcription and activity of both SODs can also be increased by the addition of oxidants; specifically, sodA is induced by internal stressors and sodM by exogenous O−2 stress. That being said, both SODs function in maintaining cell viability during exogenous O−2 stress (Clements et al., 1999; Valderas and Hart, 2001; Karavolos et al., 2003). In vivo, the importance of superoxide dismutase in S. aureus infections is unclear, as some reports suggest that SOD is important (Kanafani and Martin, 1985; Karavolos et al., 2003; Das et al., 2008; Das and Bishayi, 2009), while others suggest that SOD has only a minimal effect on virulence (Mandell, 1975; Clements et al., 1999; Schneider et al., 2002). The difficulty in determining the function of SOD in virulence may be due to the observation that calprotectin sequesters Mn and Zn, decreasing SodA and SodM activity, and rendering the bacteria more susceptible to neutrophil-dependent killing (Kehl-Fie et al., 2011).
Both SODs in S. aureus are transcibed from σA-type promoters. The transcription of sodA initiates from one of two σA-type promoters; however, the first promoter is negatively influenced by σB. While the effect of σB on sodA transcription and activity remains unclear, transcription of sodM and activity are elevated in σB-deficient strains (Karavolos et al., 2003; Bischoff et al., 2004). Additionally, the staphylococcal accessory regulator (SarA) functions as a repressor of sodM transcription independent of σB. Other proteins of the SarA family have only slight effects (i.e., SarR) or no effect on sodM transcription. A less pronounced regulatory effect of SarA on sodA transcription has also been observed (Ballal and Manna, 2009).
Catalase and peroxiredoxin AhpC
As mentioned above, the detoxification of H2O2 is accomplished by catalases that catalyze the degradation of H2O2 to water and oxygen. The catalase family of proteins is divided into monofunctional or typical catalases, bifunctional catalase-peroxidases, and manganese-containing catalases (Chelikani et al., 2004). In the Staphylococcus genus, all species are catalase-positive with the exception of S. saccharolyticus and S. aureus subspecies anaerobius (Götz et al., 2006). S. aureus has a single monofunctional heme-containing tetrameric catalase encoded by the monocistronic katA gene (Sanz et al., 2000; Horsburgh et al., 2001a). In contrast, S. xylosus, S. equorum, and S. saprophyticus each have two catalase genes (Blaiotta et al., 2010). In addition to catalase, staphylococci have several peroxiredoxins that are induced upon treatment of S. aureus with H2O2 (i.e., Tpx, Ohr-like protein, and AhpC) (Wolf et al., 2008; Chen et al., 2009). Peroxiredoxins detoxify alkyl hydroperoxides by converting them to their corresponding alcohols using NADH or NADPH as the reducing equivalents. This reducing activity is dependent on redox-active cysteines in the active site. The alkyl hydroperoxide reductase (ahpC) gene forms an operon with ahpF, which codes for a homodimeric flavoenzyme that acts as a dedicated disulfide reductase to facilitate the reducing equivalent-dependent reduction and regeneration of AhpC (Poole, 2005). In E. coli, AhpC detoxifies low levels of H2O2, whereas KatA is the primary scavenger of H2O2 at high levels (Seaver and Imlay, 2001). Similar to E. coli, the S. aureus catalase is the major determinant in resistance toward H2O2 (Martin and Chaven, 1987; Horsburgh et al., 2001a), while AhpC confers resistance to a broader spectrum of ROS (Cosgrove et al., 2007). In S. aureus mutants lacking both catalase and AhpC, their ability to scavenge exogenous and endogenous H2O2 is inhibited, leading to the accumulation of H2O2 (Cosgrove et al., 2007).
In S. aureus, the katA gene and the ahpCF operon are negatively regulated by PerR (discussed in Section “PerR”), and putative PerR boxes are found in the promoter regions of both genes (Horsburgh et al., 2001a). In addition, the transcription of katA is positively affected by the ferric uptake regulator [Fur, discussed in Section “Ferric uptake regulator (Fur)”] (Horsburgh et al., 2001b). As Fur normally functions as a repressor, it is likely that the positive regulation of katA is due to the repression of a positive regulator of katA, such as a small regulatory RNA. Together, PerR and Fur regulate transcription of the katA gene in response to peroxide and the availability of manganese and iron. As expected, maximal transcription of katA and catalase activity occur under aerobic conditions in the post-exponential growth phase, when tricarboxylic acid (TCA) cycle activity and electron transport are also maximal (Martin and Chaven, 1987; Horsburgh et al., 2001b; Cosgrove et al., 2007). The function of catalase in S. aureus virulence is not completely understood (Mandell, 1975; Kanafani and Martin, 1985; Horsburgh et al., 2001a; Messina et al., 2002; Cosgrove et al., 2007; Das et al., 2008; Das and Bishayi, 2009; Martínez-Pulgarín et al., 2009; Sen et al., 2009); however, both KatA and AhpC are important for nasal colonization (Cosgrove et al., 2007).
Flavohemoglobin (Hmp)
The discovery of a hemoglobin-like protein in E. coli (Vasudevan et al., 1991), and subsequent genome sequencing projects, led to realization that globins are widely distributed in nature. In S. aureus, the hemoglobin-like protein is a flavohemoglobin (Hmp), which has a N-terminal heme-containing globin domain and C-terminal NAD- and FAD-binding domains that together form a ferredoxin-NADP+ oxidoreductase-like domain (Ermler et al., 1995). Hmp family members commonly demonstrate three enzymatic activities: NO-reductase, NO-dioxygenase, and alkylhydroperoxide reductase (Bonamore and Boffi, 2008). During aerobic growth the Hmp from E. coli utilizes NAD(P)H and O2 to convert ·NO to nitrate; however, under anaerobic conditions it converts ·NO to N2O, albeit less efficiently (Gardner et al., 1998; Kim et al., 1999). In vitro experiments using E. coli Hmp have demonstrated an NADH-dependent alkyhydroperoxide reductase activity; however, the in vivo significance of this activity remains to be elucidated (Bonamore et al., 2003). Like E. coli, S. aureus Hmp activity is greatest during microaerobic/anaerobic growth or during nitrosative stress conditions (Gonçalves et al., 2006; Richardson et al., 2006). Regulation of Hmp activity in response to reduced oxygen tension is mediated in part by the SrrAB two-component system; presumably, at the transcriptional level. That being said, the regulators of hmp transcription remain unknown in S. aureus, although, it has been postulated to involve the NO−2-sensing transcription repressor, NsrR (Richardson et al., 2006).
Metal homeostasis
Transition metal ions (i.e., Fe, Cu, Mn, and Zn) give structure to proteins, act as cofactors to enzymes, and are essential for electron transfer; hence, they are required by all forms of life (Lippard and Berg, 1994). The ability of transition metals to transfer electrons is beneficial under some circumstances; however, it is this same ability that facilitates the generation of ROS through Fenton chemistry (Gutteridge et al., 1982; Imlay et al., 1988). For this reason, the transport of metal ions is very tightly regulated to maintain an appropriate intracellular concentration and to avoid the accumulation of metals to toxic levels (Figure 2). To maintain metal ion homeostasis, bacteria have evolved active transporters, efflux systems, and metallochaperones (Finney and O'Halloran, 2003; Maier et al., 2007; Bagai et al., 2008).
Iron
Iron is an important cofactor for numerous enzymes; hence, it is essential for bacterial viability (Griffiths, 1999). Despite iron being one of the most abundant elements in nature, it primarily exists in the insoluble Fe3+ form, which is difficult for bacteria to acquire (Ratledge and Dover, 2000). In the host, the low solubility of iron and the presence of heme, ferritin, and lactoferrin, create an environment in which free iron is essentially non-existent (Brown and Holden, 2002). This iron-limited environment coupled with the bacterial need for iron, creates a major challenge for bacteria in the host (Weinberg, 1978). To counter this challenge, S. aureus have adapted to extract iron from heme, which is carried out in part using the proteins encoded by the iron-regulated surface determinant (Isd) genes, isdA, isdB, isdCDEF, isdG, isdH, and isdI (Skaar et al., 2004; Skaar and Schneewind, 2004; Torres et al., 2006). In addition, S. aureus can extract iron from transferrin using the siderophores staphyloferrin A and B (encoded by sfaABCD and sbnABCDEFGHI, respectively) (Dale et al., 2004; Park et al., 2005a; Cheung et al., 2009; Cotton et al., 2009). When complexed with iron, these siderophores are transported into the bacterial cytoplasm through the HtsABC and SirABC transport systems using the energy from FhuC catalyzed ATP hydrolysis to drive the importation (Speziali et al., 2006; Beasley et al., 2011). In addition, S. aureus can acquire iron from hydroxymate siderophores produced by other bacteria and from catecholamine (Morrissey et al., 2000; Sebulsky et al., 2003, 2004; Beasley et al., 2011). In S. aureus, these iron acquisition and transport systems are regulated in part by the Fur [(Xiong et al., 2000); discussed in Section “Ferric uptake regulator (Fur)”].
After iron has been acquired by S. aureus, it is utilized or it is bound by ferritin, bacterioferritin comigratory protein (Bcp), or the Dps homolog MrgA (Metallo regulated gene A), which function as iron chelator/storage proteins. Ferritin, a polypeptide with a ferroxidase center, is encoded by ftnA in S. aureus and sefA in S. epidermidis (Horsburgh et al., 2001a; Morrissey et al., 2004) and both function primarily as iron-storage proteins (Andrews, 1998). In contrast, MrgA has an iron-chelating function and protects DNA from oxidative damage (discussed in Section “MrgA”). Bcp is homologous to peroxiredoxin and as such is likely involved in the thiol-dependent reduction of peroxides. In S. aureus, transcription of bcp, ftnA, and mrgA is regulated by PerR, highlighting the importance of sequestering iron during periods of oxidative stress (Horsburgh et al., 2001a; Chang et al., 2006; Wolf et al., 2008).
Manganese
Like iron, manganese is an essential cofactor in bacteria that is involved in diverse cellular functions such as, sugar metabolism, signal transduction, the stringent response, and oxidative stress resistance (Kehres and Maguire, 2003; Papp-Wallace and Maguire, 2006). In contrast to iron, Mn2+ is soluble in a physiological pH range and it has a higher reduction potential than Fe2+, meaning that it is less likely to facilitate deleterious redox reactions. For these reasons, Mn2+ is important for the detoxification of ROS in bacteria. As examples, the activity of SodA (discussed in Section “Superoxide dismutase”) requires Mn2+ and SodM is also believed to require Mn2+ (Clements et al., 1999; Valderas and Hart, 2001). To supply the S. aureus Mn2+ requirement, there are two transporters for manganese, encoded by mntABC (MntABC-type) and mntH (Nramp-type) and in S. epidermidis, sitABC codes for the MntABC-type transporter (Cockayne et al., 1998; Horsburgh et al., 2002).
The importance of Mn2+ to bacteria can be inferred from the observation that host phagocytic cells transport Mn2+ out of the phagosome upon engulfing a bacterium. To do this, phagocytic cells recruit the efflux protein Nramp1 to the phagosome; thus, reducing the availability of Mn2+ to the bacterium (Jabado et al., 2000). In addition, the heterodimeric host protein calprotectin decreases Mn2+ availability by chelating it (Corbin et al., 2008). Because bacteria need Mn2+ and host cells attempt to deny the bacteria Mn2+, it is not surprising that S. aureus virulence is attenuated in mutant strains lacking both mntA and mntH relative to the isogenic wild-type bacteria. Similarly, mutation of mntA, mntH, or mntR reduces S. aureus survival in human endothelial cells (Horsburgh et al., 2001a, 2002; Ando et al., 2003).
Regulation of Mn2+ transport is critical for maintaining metal ion homeostasis. In S. epidermidis, the DtxR homolog SirR binds to a Sir box consensus sequence in the promoter region of sitABC, in a Mn/Fe-dependent manner, to repress transcription (Cockayne et al., 1998; Hill et al., 1998). Similarly, in S. aureus, the DtxR homolog MntR represses mntABC, but not mntH, in a Mn2+-dependent manner by binding to a MntR box in the promoter region. In addition to MntR, PerR also regulates mntABC transcription and a putative PerR box is located in the promoter region of this operon. This communal regulation of mntABC by MntR and PerR is also seen with other members of the PerR regulon (Horsburgh et al., 2002).
Zinc
Zinc is an essential nutrient that is required as a cofactor for a few enzymes; however, it's more important function is in protein stability (Lippard and Berg, 1994). While Zn may be essential for bacterial viability, an over abundance of Zn is toxic due to the fact that it competes with other metals for binding to the active centers of enzymes (Beard et al., 1997; Xiong and Jayaswal, 1998). The essential nature of Zn and its potential to be toxic make obvious the importance of Zn ion homeostasis, and demonstrate why bacteria have evolved zinc transporters (Blencowe and Morby, 2003). In S. aureus, Zn homeostasis is maintained by the plasmid encoded CadCA (cadCA) transporter and/or the chromosomally encoded ZntRA (zntRA or czrAB) transporter (Endo and Silver, 1995; Xiong and Jayaswal, 1998; Kuroda et al., 1999). Both transport systems code for ArsR/SmtB family transcriptional regulators (CadC and ZntR) (Busenlehner et al., 2003) and metal-exporting membrane proteins CadA and ZntA (Guffanti et al., 2002). In the presence of excess zinc, CadC and ZntR bind Zn, decreasing their affinity for their cognate promoters and this de-represses transcription of cadA and zntA (Nucifora et al., 1989; Singh et al., 1999; Ye et al., 2005).
In B. subtilis, Zn transport is primarily mediated by the proteins encoded within the znuABC operon, and zosA. Transcription of znuABC is regulated by the Zn-responsive Fur homolog known as Zur, while the transcription of zosA is regulated by PerR [discussed in Section “PerR” (Lee and Helmann, 2007)]. In complex with Zn, the Zur homodimer functions as a transcriptional repressor by binding to a Zur-box (AAATCGTAATNATTACGATTT) present in the promoter/operator region of znuABC (Gaballa et al., 2002; Ma et al., 2011). When the availability of Zn is low, Zur releases from the DNA, de-repressing transcription of the ATP binding cassette Zn transporter coded by znuABC. The P-type metal-transporting ATPase coded by zosA is induced in response to H2O2, consistent with its proposed function in oxidative stress resistance (Gaballa and Helmann, 2002; Gaballa et al., 2002). Although zosA has not been identified in S. aureus, it does have genes homologous to znuA and znuB; designated as mreA and mreB, respectively. As stated, Zn has important physiological functions; however, the role of Zn transport in S. aureus pathogenesis remains to be elucidated (Lindsay and Foster, 2001).
Copper
Although limited in number, copper-requiring enzymes have critical roles in bacterial respiration, biosynthesis, and oxidative stress resistance; hence, Cu is considered an essential trace element (Halliwell and Gutteridge, 1984; Puig and Thiele, 2002). The enzymatic utility of Cu is due in part to its ability to act as an electron donor or acceptor by cycling between the Cu2+ to Cu1+ oxidation states. Like iron, the properties of Cu that make it a useful redox cofactor, also allow it to facilitate the generation of ROS (Baker et al., 2010). For this reason, bacteria have evolved mechanisms to protect themselves from the toxic effects of Cu, while maintaining the intracellular concentration of Cu at the minimum necessary for growth (Solioz and Stoyanov, 2003; Liu et al., 2007; Wolschendorf et al., 2011). In S. aureus, copper homeostasis is primarily maintained by the P1-type ATPase CopA and the copper chaperone CopZ; CopZ sequesters intracellular Cu1+ and delivers it to the Cu exporter CopA (Sitthisak et al., 2007). While CopA and CopZ are highly conserved in S. aureus, some strains also have a second P1-type ATPase copper transporter, CopB, and a Cu oxidizing enzyme, multicopper oxidase (mco) (Sitthisak et al., 2005). In contrast to the chromosomally encoded genes copA and copZ, both copB and mco are carried on a plasmid or on a plasmid integrated into the chromosome (Holden et al., 2004; Baker et al., 2011). Importantly, the plasmid carrying copB and mco can be transferred between S. aureus strains (Baker et al., 2011). Regulation of copA, copZ, copB, and mco is dependent upon the Cu1+ responsive copper-sensitive operon repressor (CsoR) (Baker et al., 2011; Grossoehme et al., 2011). CsoR binds to DNA in the absence of Cu1+ and represses transcription; however, as the intracellular concentration of copper increases, CsoR complexes with Cu1+, releases from the DNA, and de-represses transcription of copper resistance genes. The in vivo importance of Cu resistance for S. aureus is likely due to the fact that macrophage increase the Cu concentration in phagosomes via the ATP7A Cu transporter, which enhances the bactericidal activity of the phagosome (Wagner et al., 2005; White et al., 2009).
DNA protection and repair
MrgA
The genomic DNA of bacteria is organized in a nucleoid that requires DNA supercoiling, molecular crowding, and several architectural proteins (e.g., Hu, H-NS, Fis, Dps) (Dame, 2005; Luijsterburg et al., 2006). Dps (DNA-binding protein from starved cells) is a member of the ferritin super family of proteins and a non-specific DNA binding protein that functions as a major determinant for protecting DNA by nucleoid condensation (Martinez and Kolter, 1997; Wolf et al., 1999; Nair and Finkel, 2004). During the stationary phase of growth in E. coli, the conformation of DNA changes from a relaxed state to a compacted state, a process requiring Dps (Kim et al., 2004; Ohniwa et al., 2006). This complex DNA condensation process is regulated during the exponential growth phase by Fis and H-NS binding to the dps promoter and repressing transcription. (Ohniwa et al., 2006; Grainger et al., 2008). Unlike E. coli, S. aureus appears to lack both Fis and H-NS homologs (Ohniwa et al., 2011); hence, transcription of the Dps homolog MrgA is likely linked to growth via other means. One possible linkage between growth and regulation of mrgA is PerR (discussed in Section “PerR”).
Transcription of mrgA is induced by H2O2 and iron (Horsburgh et al., 2001a; Morrissey et al., 2004; Chang et al., 2006; Morikawa et al., 2006; Wolf et al., 2008), suggesting that PerR and Fur regulate mrgA transcription. This suggestion is partially true as PerR is a repressor of mrgA transcription; whereas, mrgA transcription is independent of Fur. Consistent with PerR regulation of mrgA, inactivation of perR results in a compacted nucleoid in the absence of oxidative stress, which is similar to that found in a strain over-expressing mrgA (Morikawa et al., 2006, 2007). The activating effect of iron on mrgA transcription, while not completely understood, is related to the fact that the MrgA/Dps protein is a ferritin-like Fe2+ binding and storage protein (Grant et al., 1998; Zhao et al., 2002; Su et al., 2005). The compact nature of the nucleoid and the susceptibility of DNA to oxidative damage (discussed in Section “Bacterial targets of oxidative damage”) likely led to the evolution of a bi-functional protein involved in DNA condensation and protection from Fe2+ generated HO· (Figure 2).
Excision repair
Oxidative damage to DNA can occur at the bases or sugars, producing lesions such as strand breakage or base alterations (Demple and Harrison, 1994; Lu et al., 2001). Due to the obvious importance of DNA in species propagation, DNA repair mechanisms have evolved to maintain genetic integrity. These DNA repair mechanisms can be divided into two broad catagories; excision repair [i.e., base excision repair (BER), mismatch repair (MMR), and nucleotide excision repair (NER)] and recombinational repair. As with many aspects of bacterial physiology, much of our knowledge has been derived from the study of model bacteria; therefore, we will use this knowledge to draw inferences into staphylococcal DNA repair mechanisms. In time, these inferences will likely be proven correct as S. aureus has homologs of many of the DNA repair enzymes discussed below; specifically, for BER/MMR S. aureus has Nfo, MutM, MutY, MutT, MutS, MutL, RecJ, and Nth (O'Neill and Chopra, 2002; Prunier and Leclercq, 2005; Ambur et al., 2009).
Multistep BER pathways rely on damage-specific DNA glycosylases that scan DNA, recognize base lesions, and initiate removal by cleaving the base–deoxyribose glycosyl bond, forming apurinic/apyrimidimic sites (AP site). The next step in the repair process is restoration of the correct DNA sequence via short-patch (1-nucleotide patch size) or long-patch (multiple nucleotide patch size) pathways. Depending on the enzymatic properties of the initiating DNA glycoslyase, the activities of AP endonucleases and/or DNA deoxyribosephosphodiesterase (drPase) are required to break the DNA during this process. In E. coli, AP endonuclease activity is primarily due to exonuclease III (ExoIII or Xth) and endonuclease IV (EndoIV or Nfo), while drPase activity is due to RecJ and exonuclease I (ExoI) (Ljungquist, 1977; Lindahl, 1979; Rogers and Weiss, 1980; Franklin and Lindahl, 1988; Sandigursky and Franklin, 1992; Mol et al., 2000).
During periods of oxidative stress, a common lesion found in DNA is the oxidized base 7,8-dihydro-8-oxoguanine (8-oxoG or GO lesion), which can mispair with adenine (Shibutani et al., 1991). In E. coli, the formamidopyrimidine DNA glycosylase (Fpg or MutM) acts on oxidized and ring-opened purines, while the adenine DNA glycosylase MutY prevents mutagenic transversions by removing already misincorporated adenine (Michaels et al., 1992a, b; Michaels and Miller, 1992). In addition to repairing DNA, oxidized guanine is removed from the nucleotide pool by the pyrophosphohydrolase activity of MutT (Maki and Sekiguchi, 1992).
The 5,6-double bond of pyrimidines is susceptible to reaction with HO· creating a number of oxidatively damaged products, such as thymine glycol. In E. coli, pyrimidine lesions are recognized by endonuclease III (EndoIII or Nth) and endonuclease VIII (EndoVIII or Nei) (Radman, 1976; Demple and Linn, 1980; Katcher and Wallace, 1983; Breimer and Lindahl, 1984; Wallace, 1988). The removal of misincorporated bases occurs by the MMR system, which requires MutSL for recognition and initiation of excision repair (Modrich, 1991; Marti et al., 2002). Genetic defects in MMR and/or GO systems are associated with mutator phenotypes and as such, these damage repair systems are important in adaptive mutagenesis and the generation of genetic diversity, which was also demonstrated for S. aureus (O'Neill and Chopra, 2002; Chopra et al., 2003; Prunier and Leclercq, 2005; Vidales et al., 2009).
In contrast to BER, NER lesions are repaired in 12–13 nucleotide-long segments, followed by synthesis of a repair patch using the intact strand as a template and ligation of the repaired ends. Briefly, damaged DNA is detected by a complex of UvrA and UvrB. After the DNA damage is detected, UvrB binds the damaged DNA, displacing UvrA, and allowing UvrC to complex with UvrB. It is this UvrBC complex that cleaves the phosphodiester backbone to create a 12 base excision. UvrD facilitates dissociation of base pairing, causing the release of the nucleotide segment. At this point, the deletion can be filled by DNA polymerase I and then the newly synthesized DNA is ligated to the existing DNA. In addition to UvrAB, the transcription-repair coupling factor Mfd can recruit the DNA excision-repair machinery to damaged DNA (Sancar, 1996). This process is likely similar in S. aureus as it has the uvrABC genes and mfd (Ambur et al., 2009).
Recombinational repair
The predominant consequence of oxidative damage to the sugar in DNA is strand breakage, which can be repaired by mechanisms used in recombination. In E. coli, repair of DNA strand breaks initiates when RecBCD binds to the blunt end of a double stranded DNA break and the helicase activity of RecB and RecD unwinds the DNA. Due to severe consequences of strand breakage (i.e., death), there is redundancy in the initiation of recombinational repair systems (i.e. RecF and SbcCD pathways). While the repair of most double strand DNA breaks in E. coli are initiated by the RecBCD exonuclease/helicase complex, low-GC content Gram-positive bacteria, including staphylococci, lack this pathway. Gram-positive bacteria initiate double strand break repair using the AddAB nuclease/helicase complex, functional homologs of RecBCD, or homologs of the RecF and SbcCD pathways (Alonso et al., 1993; Eisen and Hanawalt, 1999; Ambur et al., 2009; Yeeles and Dillingham, 2010). After initiating recombinational repair and as the DNA is unwound, RecA binds to the single stranded DNA and pairs with the homologous DNA sequence and initiates strand invasion. Following strand invasion, RuvAB drive branch migration and then in concert with RuvC, cuts the DNA to resolve the Holliday junction. The process of branch migration and resolution can be catalyzed by RecG as well. These additional components of double strand break repair are conserved in S. aureus (i.e., RecA, RuvAB, and RecG) (Niga et al., 1997; Ambur et al., 2009).
Protein damage repair
Thioredoxin
In bacteria, the cytoplasm is in a reduced state; hence, protein thiols can be maintained in their reduced form (Pollitt and Zalkin, 1983; Derman and Beckwith, 1991). This reduced state of the bacterial cytoplasm depends heavily on the thioredoxin and glutaredoxin systems and the low-molecular-weight thiol reductants coenzyme A (CoASH) and bacillithiol (BSH) (Brown, 1959; Derman et al., 1993; Prinz et al., 1997; Di Simplicio et al., 2003). While many bacteria have both the thioredoxin and glutaredoxin systems, most Gram-positive bacteria, including S. aureus, lack the glutaredoxin system (Vido et al., 2005; Diep et al., 2006); thus, these bacteria rely heavily on the thioredoxin system to maintain a reduced cytoplasm (Scharf et al., 1998; Uziel et al., 2004). The importance of thioredoxin cannot be overstated as it is essential for a large variety of cellular processes, including acting as a hydrogen donor to ribonucleotide reductase and methionine sulfoxide reductases (Russel and Model, 1986; Aberg et al., 1989). As important, thioredoxins are major contributors to oxidative stress resistance by facilitating the reduction of H2O2, scavenging HO·, and donating reducing equivalents to peroxiredoxins and peroxidase (Arnér and Holmgren, 2000) (Figure 2).
The thioredoxin system is comprised of thioredoxin (trxA) and the thioredoxin reductase (trxB). Thioredoxins are small disulfide reductase proteins, while thioredoxin reductase uses the electrons from NADPH to maintain thioredoxin in a reduced state (Holmgren, 1985). In B. subtilis, trxA and trxB transcription is maintained at a basal level during growth but it is increased in response to diamide, H2O2, heat, salt, or ethanol stress (Scharf et al., 1998; Leichert et al., 2003; Mostertz et al., 2004). These increases in trxA and trxB transcription are primarily mediated by σA, σB, and Spx (Scharf et al., 1998), an RNA polymerase-dependent transcriptional activator that responds to diamide stress (Nakano et al., 2003). Similar to B. subtilis, transcription of trxA and trxB in S. aureus is maintained at a basal level under aerobic and anaerobic growth conditions, with Spx being required for transcription under all growth conditions (Horsburgh et al., 2001a; Pamp et al., 2006; Ballal and Manna, 2010). Also similar to B. subtilis, stressors such as copper, diamide, menadione, and tert-butyl hydroperoxide induce transcription of trxA and trxB (Uziel et al., 2004; Wolf et al., 2008; Baker et al., 2010).
As stated above, bacteria rely on the cysteine-containing small proteins thioredoxin and glutaredoxin to carry out the thiol-disulfide redox cycling reactions and maintain a reduced cytoplasm (Holmgren, 1989); therefore, cysteine biosynthesis is critical for sustaining the reducing environment of the cytoplasm. This can be seen by the fact that under diamide or H2O2-induced oxidative stress, S. aureus increases cysteine biosynthesis and uptake (Chang et al., 2006; Wolf et al., 2008). In B. subtilis and S. aureus, cysteine biosynthesis and transport are negatively regulated by CymR in complex with CysK [o-acetyl serine (OAS) thiol-lyase] (Even et al., 2006; Soutourina et al., 2009). Although cysteine is needed to maintain the reducing environment of the cytoplasm via thioredoxin, the intracellular concentration of cysteine is kept low due the ability of free cysteine to reduce Fe3+ to Fe2+, which can facilitate Fenton chemistry (Park and Imlay, 2003). Thus, the intracellular concentration of cysteine must be finely balanced, or the reducing environment of the cytoplasm will be compromised. This can be seen in cymR mutants where cysteine accumulates in the cytoplasm and susceptibility to H2O2 also increases (Soutourina et al., 2009, 2010).
CoA reductase
Coenzyme A functions as a substrate for biosynthesis and the oxidation of pyruvate and fatty acids. These functions rely on the ability of CoA to form high-energy thioester bonds, such as that found in acetyl-CoA (Magnuson et al., 1993; del Cardayre et al., 1998). In addition to the metabolic importance of CoA, staphylococci use CoA and, possibly, bacillithiol as low molecular weight free thiols to help maintain the reducing environment of the cytoplasm (Newton et al., 1993; del Cardayre et al., 1998; Fahey, 2001; Pöther et al., 2009). To fill the need for reduced CoA (CoASH), coenzyme A disulfide reductase catalyzes the NADPH-dependent reduction of CoASSCoA to CoASH and protein-SH (Coulter et al., 1998). The importance of coenzyme A disulfide reductase is reflected in the observation that inactivation of the S. aureus coenzyme A disulfide reductase results in the attenuation of virulence for mice relative to the isogenic strain (Coulter et al., 1998; Schneider et al., 2002). Because of the metabolic demand for CoA and the use of CoA as a free thiol, it is difficult to predict which function is more responsible for this attenuation.
Methionine sulfoxide reductase
Methionine is highly susceptible to oxidation (Dean et al., 1997; Grimaud et al., 2001), which can lead to structural changes that alter or inhibit enzymatic functions. When oxidized, methionine forms diastereomeric S and R forms of methionine sulfoxide (Brot et al., 1981; Moskovitz et al., 1996; Sharov et al., 1999). To repair this change, most life forms rely on methionine sulfoxide reductases (Moskovitz et al., 1996). The oxidized S form of methionine is reduced by the enzyme MsrA, while the R form is reduced by MsrB. The reduction of methionine sulfoxide to methionine is dependent on the thioredoxin thiol-disulfide redox system or other reducing agents that can donate electrons (Russel and Model, 1986). The importance of methionine sulfoxide reductase can be seen in E. coli where GroEL, a chaperone involved in the folding of polypeptides, is highly susceptible to inactivation by host immune cell generated ONOO− and HOCl (Khor et al., 2004; Sasindran et al., 2007).
In most bacteria genes encoding for the enzymes, MsrA and MsrB, are commonly present as a single copy for each one (Ezraty et al., 2005). In S. aureus, there are three paralogs of msrA (i.e., msrA1, msrA2, msrA3), and a single msrB gene. msrA1 and msrB are present in an operon with a PTS permease and a fourth gene of unknown function; whereas, msrA2 and msrA3 are unlinked with msrA1 and msrB (Singh et al., 2001; Singh and Moskovitz, 2003). As expected, msrA1 deletion makes S. aureus more sensitive to H2O2; however, the other msrA paralogs do not complement for the msrA1 deletion. In the case of msrA2, this may be due poor transcription rather than inability to function as a methionine sulfoxide reductase. Interestingly, msrA1 transcription can be induced by oxacillin and other cell wall antibiotics, but not by peroxide (Singh et al., 2001; Singh and Moskovitz, 2003). While some work has been performed on methionine sulfoxide reductase in S. aureus, the fitness benefit of having three msrA paralogs remains to be determined.
Fe-S cluster repair
Iron-sulfur clusters are ubiquitous prosthetic groups that are involved in diverse cellular processes such as electron transfer, enzyme activity, environmental sensing, and gene regulation. The biogenesis of these Fe-S clusters requires the presence of assembly systems (e.g., isc, suf, and csd operons/systems), containing cysteine desulfurases, Fe-S scaffold proteins, and other accessory proteins (Johnson et al., 2005; Fontecave and Ollagnier-de-Choudens, 2008). As mentioned in Section “Bacterial targets of oxidative damage”, Fe-S clusters are very susceptible to oxidative inactivation; hence, to survive in an aerobic environment, bacteria have evolved Fe-S cluster repair mechanisms. In some bacteria, the suf system appears to maintain Fe-S cluster assembly under oxidative stress conditions, while IscS is thought to repair Fe-S clusters (Nachin et al., 2003; Djaman et al., 2004). In addition to IscS, the recovery of aconitase and fumarase activity after oxidative damage is facilitated by the Fe-S cluster repair proteins YtfE in E. coli and ScdA in S. aureus (Justino et al., 2006, 2007; Overton et al., 2008; Vine et al., 2010). ScdA is a di-iron protein that is widely distributed among bacteria and is believed to be a new protein family that repairs Fe-S clusters (Overton et al., 2008). In S. aureus, the monocistronic scdA gene was first identified as being involved in autolysis and cell division (Brunskill et al., 1997). Transcription of scdA is de-repressed in the post-exponential growth phase when TCA cycle activity and electron transport are maximal and the bacterial demand for iron is at its greatest (Brunskill et al., 1997; Somerville et al., 2003), a process mediated by MgrA [(Ingavale et al., 2003); discussed in Section “MgrA”]. Consistent with the MgrA-dependent repression of scdA, transcription of scdA can be induced by exposure to H2O2 (Chang et al., 2006) and scdA inactivation leads to increased sensitivity to H2O2 (Overton et al., 2008). The importance of scdA to staphylococcal pathogenesis remains to be determined.
Sensing and regulation
As oxidative stress affects all aspect of bacterial physiology, it is understandable that regulation of cellular processes that respond to oxidative stress is complex (Figure 2). In fact, the complexity is significantly increased when one considers that the staphylococcal response to oxidative stress depends on the chemical nature of the oxidant. This was demonstrated when S. aureus was treated with three different oxidative stress inducing compounds (i.e., paraquat, H2O2, diamide) and the protein profiles for each stress had only limited overlap (Wolf et al., 2008). Because many of the regulators that affect the oxidative stress response are discussed in other areas of this Frontiers Research Topic, we have limited our discussion to only a few of the sensing and regulatory systems in staphylococci.
Environmental sensing—the metabolic response
Despite the large percentage of the staphylococcal genomes dedicated to metabolism and physiology, staphylococci only require 13 biosynthetic intermediates to synthesize all macromolecules in the cell. These 13 biosynthetic intermediates are derived from the three metabolic pathways of central metabolism: glycolysis, the pentose phosphate pathway (PPP), and the TCA cycle. Because of the importance of these 13 intermediates, staphylococci have evolved metabolite responsive regulators (e.g., CcpA, CodY, RpiR) to “sense” the availability of these intermediates or compounds derived from them (Somerville and Proctor, 2009). As stated above, oxidative stress leads to the rapid inactivation of Fe-S cluster containing enzymes and the reversible and irreversible oxidation of some cysteine and methionine-containing proteins. Hence, oxidative stress alters enzymatic activity, resulting in changes in metabolite concentrations as well as the redox poise. These changes in the bacterial metabolic status create signals that alter the activity of redox-responsive and metabolite-responsive regulators (e.g., Rex, CcpA, CodY, RpiR) (Egeter and Brückner, 1996; Seidl et al., 2008a, b; Pagels et al., 2010). It is for this reason that metabolite-responsive regulators, such as CodY (Majerczyk et al., 2010) and RpiRC (Zhu et al., 2011) are involved in regulating components of the oxidative stress response (Figure 2).
Ferric uptake regulator (Fur)
The ferric uptake regulator (Fur) is a transcriptional regulator that is partially responsible for maintenance of iron homeostasis in many bacteria, including S. aureus and S. epidermidis (Ernst et al., 1978; Hantke, 1981; Heidrich et al., 1996; Xiong et al., 2000). Fur is a homodimeric metalloprotein with an N terminal DNA binding domain and C terminal dimerization domain that may be occupied by structural zinc (Jacquamet et al., 1998; Gonzalez de Peredo et al., 1999; Sheikh and Taylor, 2009). Fur when complexed with iron can regulate the transcription of genes by binding to a 19 bp inverted repeat sequence known as the Fur box (GATAATTGATAATCATTATC) in the promoter region (Ochsner et al., 1995; Escolar et al., 1999; Xiong et al., 2000; Baichoo and Helmann, 2002). Fur primarily functions as a repressor; therefore, changes in gene expression during growth in iron-limited medium are similar to those changes observed in a fur mutant (Johnson et al., 2011). Interestingly, in vitro studies of fur mutants have demonstrated that the number of genes regulated by iron far outweighs the number of genes identified as being directly regulated by Fur. It is hypothesized that Fur-independent transcription of genes that are regulated by the availability of iron may be regulated by agr, rot, and sae, as the transcription of these regulators is influenced by Fur during iron-limited growth. As stated, Fur is primarily a transcriptional repressor; however, Fur is a positive effector of katA transcription and consistent with this positive effect, fur mutants have reduced catalase activity and increased sensitivity to peroxide stress (Horsburgh et al., 2001b). The more likely explanation for the positive effect of Fur on katA transcription is that Fur represses transcription of a positive regulator of katA transcription; specifically, small RNAs have been implicated in regulating S. aureus transcription and translation (Allard et al., 2006; Felden et al., 2011). In S. aureus, Fur is speculated to regulate the accumulation of immunomodulatory proteins, cytolytic proteins, and to protect against neutrophil-mediated killing (Torres et al., 2010). Despite this speculation, fur inactivation has a slight effect on S. aureus virulence in one type of experimental model (Horsburgh et al., 2001b).
PerR
PerR is a second member of the Fur family of regulators in staphylococci, and it has been identified as a peroxide sensing protein (Horsburgh et al., 2001a; Lee and Helmann, 2006a, 2007). The PerR regulon includes many genes involved in the oxidative stress response and iron storage, including katA, ahpCF, mrgA, bcp, and trxA genes. The fact that PerR regulates part of the oxidative stress response would suggest it is important for surviving the host immune response during an infection; however, in a mouse model of infection, perR mutants are only slightly attenuated in virulence relative to the parental strain (Horsburgh et al., 2001a). As a member of the Fur family of regulators, the activity of PerR is dependent upon metal ions. In B. subtilis, PerR carries structural zinc and its DNA binding activity is enhanced when complexed with either Fe or Mn (Lee and Helmann, 2006a). PerR containing Fe or Mn will function as a transcriptional repressor by binding to a consensus DNA sequence, termed the PerR box (AAGTATTATTTATTATTATTA) (Chen et al., 1995; Horsburgh et al., 2001a). In the presence of H2O2, the iron in PerR leads to formation of HO·, which oxidizes the iron-coordinating histidines, causing the loss of iron and DNA binding activity (Lee and Helmann, 2006b). When PerR is complexed with Mn, it is less likely to be inactivated by H2O2 because Mn is a poor mediator of Fenton chemistry; thus, in the absence of HO· there is little oxidation of the metal coordinating histidines and PerR retains its DNA binding properties. Based on this mechanism of activity regulation, it is understandable that in the presence of high Mn2+ and low Fe2+ PerR regulon members remain repressed in the presence of H2O2 (Horsburgh et al., 2001a, 2002; Fuangthong et al., 2002).
MgrA
MgrA (multiple gene regulator) is a member of MarR family of regulators that positively affects capsule biosynthesis and nuclease accumulation, represses α-toxin, coagulase, and protein A synthesis, and represses autolysis (Ingavale et al., 2003; Luong et al., 2003, 2006). In addition, MgrA regulates transcription of several multidrug efflux pumps (i.e., NorA, NorB, NorC, and Tet38) (Figure 2); thus, MgrA functions in staphylococcal resistance to different antibiotics including fluoroquinolones, tetracycline, vancomycin or penicillin (Truong-Bolduc et al., 2003, 2006; Cui et al., 2005; Kaatz et al., 2005; Truong-Bolduc et al., 2005; Chen et al., 2006; Truong-Bolduc et al., 2006). In total, transcriptional profiling has revealed that MgrA affects the transcription of as many as 350 genes (Luong et al., 2006). This global reach of MgrA is achieved by binding to target genes as well as by indirect regulation through its affects on other regulators (i.e., SarS, SarV, SigB, LytRS, and ArlRS) (Ingavale et al., 2003, 2005; Truong-Bolduc et al., 2003; Manna et al., 2004; Luong et al., 2006). The global nature of MgrA is also reflected in the fact that it is required for the establishment and progression of S. aureus infections in murine abscess, septic arthritis, and sepsis models (Chen et al., 2006; Jonsson et al., 2008; Sun et al., 2011). S. aureus MgrA is structurally similar to the MarR of E. coli in that it contains a DNA binding, helix-turn-helix domain and a dimerization domain (Chen et al., 2006). In addition, MgrA contains a single cysteine (Cys12) in the dimerization domain that is accessible to oxidizing agents. This dimer interface domain is similar to that of the B. subtilis peroxide-sensing regulator OhrR (Fuangthong et al., 2001; Fuangthong and Helmann, 2002; Lee et al., 2007; Soonsanga et al., 2007), which when the cysteines of the two monomers are oxidized leads to dissociation of MgrA from the DNA (Chen et al., 2006). In addition, to regulating activity via Cys oxidation, the activity of MgrA is modulated by the eukaryotic-like serine/threonine kinase (Stk1 or PknB) (Truong-Bolduc et al., 2008).
SarZ
In addition to MgrA, a second MarR family regulator, SarZ, is involved in sensing oxidative stress. Like MgrA, SarZ has a DNA-binding helix–turn–helix motif, a single cysteine (Cys13), and a dimerization domain. The oxidation of Cys13 to sulfenic acid by peroxides is insufficient to disrupt SarZ DNA-binding properties; however, generation of a mixed disulfide or further oxidation to sulfinic acid or sulfonic acid leads to a de-repression of transcription. Thus, in S. aureus, SarZ and MgrA function as thiol switches, similar to the B. subtilis OhrR (Chen et al., 2006, 2011; Poor et al., 2009). Among the SarZ affected genes, many code for proteins involved in intermediary, amino acid, fatty acid, nucleotide, and sugar metabolism, including regulators of pyrimidine synthesis (pyrR) and gluconate catabolism (gntR, GntR-like protein). In addition, SarZ regulates transcription of the H2O2-inducible Ohr-like peroxiredoxin (Chen et al., 2009). Interestingly, there is little regulatory overlap between SarZ and MgrA affected genes (Luong et al., 2006; Chen et al., 2009). Since the Cys oxidation mechanisms for regulating the activity of MgrA and SarZ are similar, this absence of overlap may only partially explain why different oxidants create different proteome profiles (Wolf et al., 2008).
SarA
The SarA (Cheung et al., 1992) is a promiscuous DNA binding protein containing a single cysteine (Cys9) at the dimerization interface that may be involved in oxidative stress sensing; however, SarA is more sensitive to alkylation than it is to oxidation (Chen et al., 2011). The DNA binding activity of SarA may be influenced by the redox poise or the oxidative status of the cytoplasm (Chan and Foster, 1998; Lindsay and Foster, 1999; Fujimoto et al., 2009). This may explain why SarA is a negative effector of superoxide dismutase and thioredoxin reductase transcription (Ballal and Manna, 2009, 2010). In addition to regulating some aspects of the oxidative stress response, SarA affects transcription of genes involved in many cellular processes, including virulence related genes and amino acid, nucleotide, and cell wall metabolism genes (Dunman et al., 2001).
SOS response
If the ROS burden is high and the general stress response systems (e.g., σB-system) are overwhelmed, then the SOS response can become activated. As mentioned above, oxidative stress frequently induces DNA damage. For this reason, it is not surprising that exposure of S. aureus to H2O2 can induce the LexA regulated SOS response (Chang et al., 2006; Wolf et al., 2008). The SOS response is a highly conserved global DNA damage repair system that can be triggered by numerous DNA damaging agents, including fluoroquinolone or β-lactam antibiotics (Anderson et al., 2006; Cirz et al., 2007; Erill et al., 2007). During an SOS response, the sensor protein RecA becomes activated by non-specific binding to single-stranded DNA that is derived from recombinational repair or stalled replication. Activated RecA stimulates the autocatalytic cleavage of the SOS transcriptional repressor LexA in the C-terminal dimerization domain and in the N-terminal DNA binding domain, leading to the de-repression of SOS genes. When RecA no longer encounters ssDNA, the concentration of non-cleaved LexA increases and the SOS repair system is deactivated. While the SOS system is important for staphylococcal survival, it has also been linked to virulence, antibiotic resistance, and the dissemination of mobile genetic elements (Úbeda et al., 2005, 2007; Goerke et al., 2006; Kelley, 2006; Maiques et al., 2006). As an example, the gene encoding fibronectin binding protein B (fnbpB) is part of the LexA regulon in S. aureus (Bisognano et al., 2004).
Conclusion
Staphylococci face the near constant challenge of surviving in the presence of exogenous and endogenous oxidants. To meet this challenge, staphylococci have evolved a multitude of oxidative defense strategies that require a coordinated regulatory response (Figure 2). This regulatory response relies on molecular sentinels to detect oxidative stress or the damage caused by oxidative stress and to transduce these signals to regulators that enhance or repress transcription of the defence genes in proportion to the challenge. Once activated, the defence machinery must repair or degrade and replace damaged DNA and proteins. Disruptions in the ability of staphylococci to sense, respond, or repair oxidative stress, and the damage caused by oxidative stress, results in a fitness cost that makes the bacterium more sensitive to oxidative damage.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Aberg A. Hahne S. Karlsson M. Larsson A. Ormö M. Ahgren A. Sjöberg B. M. (1989). Evidence for two different classes of redox-active cysteines in ribonucleotide reductase of Escherichia coli. J. Biol. Chem. 264, 12249–12252.
2
Allard M. Moisan H. Brouillette E. Gervais A. L. Jacques M. Lacasse P. Diarra M. S. Malouin F. (2006). Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 8, 1679–1690.
3
Allen R. C. Stephens J. T. Jr. (2011). Myeloperoxidase selectively binds and selectively kills microbes. Infect. Immun. 79, 474–485. 10.1128/IAI.00910-09
4
Alonso J. C. Stiege A. C. Lüder G. (1993). Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 239, 129–136.
5
Ambur O. H. Davidsen T. Frye S. A. Balasingham S. V. Lagesen K. Rognes T. Tønjum T. (2009). Genome dynamics in major bacterial pathogens. FEMS Microbiol. Rev. 33, 453–470. 10.1111/j.1574-6976.2009.00173.x
6
Anderson K. L. Roberts C. Disz T. Vonstein V. Hwang K. Overbeek R. Olson P. D. Projan S. J. Dunman P. M. (2006). Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188, 6739–6756. 10.1128/JB.00609-06
7
Ando M. Manabe Y. C. Converse P. J. Miyazaki E. Harrison R. Murphy J. R. Bishai W. R. (2003). Characterization of the role of the divalent metal ion-dependent transcriptional repressor MntR in the virulence of Staphylococcus aureus. Infect. Immun. 71, 2584–2590. 10.1128/IAI.71.5.2584-2590.2003
8
Andrews S. C. (1998). Iron storage in bacteria. Adv. Microb. Physiol. 40, 281–351.
9
Arnér E. S. Holmgren A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. 10.1046/j.1432-1327.2000.01701.x
10
Bagai I. Rensing C. Blackburn N. J. McEvoy M. M. (2008). Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry47, 11408–11414. 10.1021/bi801638m
11
Baichoo N. Helmann J. D. (2002). Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184, 5826–5832. 10.1128/JB.184.21.5826-5832.2002
12
Baker J. Sengupta M. Jayaswal R. K. Morrissey J. A. (2011). The Staphylococcus aureus CsoR regulates both chromosomal and plasmid-encoded copper resistance mechanisms. Environ. Microbiol. 13, 2495–2507. 10.1111/j.1462-2920.2011.02522.x
13
Baker J. Sitthisak S. Sengupta M. Johnson M. Jayaswal R. K. Morrissey J. A. (2010). Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl. Environ. Microbiol. 76, 150–160. 10.1128/AEM.02268-09
14
Ballal A. Manna A. C. (2009). Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J. Bacteriol. 191, 3301–3310. 10.1128/JB.01496-08
15
Ballal A. Manna A. C. (2010). Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol. 192, 336–345. 10.1128/JB.01202-09
16
Barrière C. Brückner R. Talon R. (2001a). Characterization of the single superoxide dismutase of Staphylococcus xylosus. Appl. Environ. Microbiol. 67, 4096–4104. 10.1128/AEM.67.9.4096-4104.2001
17
Barrière C. Leroy-Sétrin S. Talon R. (2001b). Characterization of catalase and superoxide dismutase in Staphylococcus carnosus 833 strain. J. Appl. Microbiol. 91, 514–519. 10.1046/j.1365-2672.2001.01411.x
18
Beard S. J. Hashim R. Membrillo-Hernández J. Hughes M. N. Poole R. K. (1997). Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25, 883–891. 10.1111/j.1365-2958.1997.mmi518.x
19
Beasley F. C. Marolda C. L. Cheung J. Buac S. Heinrichs D. E. (2011). Staphylococcus aureus transporters Hts, Sir, and Sst capture iron liberated from human transferrin by Staphyloferrin A, Staphyloferrin B, and catecholamine stress hormones, respectively, and contribute to virulence. Infect. Immun. 79, 2345–2355. 10.1128/IAI.00117-11
20
Bischoff M. Dunman P. Kormanec J. Macapagal D. Murphy E. Mounts W. Berger-Bächi B. Projan S. (2004). Microarray-based analysis of the Staphylococcus aureussigmaB regulon. J. Bacteriol. 186, 4085–4099. 10.1128/JB.186.13.4085-4099.2004
21
Bisognano C. Kelley W. L. Estoppey T. Francois P. Schrenzel J. Li D. Lew D. P. Hooper D. C. Cheung A. L. Vaudaux P. (2004). A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279, 9064–9071. 10.1074/jbc.M309836200
22
Blaiotta G. Fusco V. Ercolini D. Pepe O. Coppola S. (2010). Diversity of Staphylococcus species strains based on partial kat (catalase) gene sequences and design of a PCR-restriction fragment length polymorphism assay for identification and differentiation of coagulase-positive species (S. aureus, S. delphini, S. hyicus, S. intermedius, S. pseudintermedius, and S. schleiferi subsp. coagulans). J. Clin. Microbiol. 48, 192–201. 10.1128/JCM.00542-09
23
Blencowe D. K. Morby A. P. (2003). Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27, 291–311.
24
Bogdan C. Röllinghoff M. Diefenbach A. (2000). Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12, 64–76. 10.1016/S0952-7915(99)00052-7
25
Bonamore A. Boffi A. (2008). Flavohemoglobin: structure and reactivity. IUBMB Life60, 19–28. 10.1002/iub.9
26
Bonamore A. Gentili P. Ilari A. Schininà M. E. Boffi A. (2003). Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase. J. Biol. Chem. 278, 22272–22277. 10.1074/jbc.M301285200
27
Breimer L. H. Lindahl T. (1984). DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J. Biol. Chem. 259, 5543–5548.
28
Brot N. Weissbach L. Werth J. Weissbach H. (1981). Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. U.S.A. 78, 2155–2158.
29
Brown G. M. (1959). The metabolism of pantothenic acid. J. Biol. Chem. 234, 370–378.
30
Brown J. S. Holden D. W. (2002). Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4, 1149–1156. 10.1016/S1286-4579(02)01640-4
31
Brunelli L. Crow J. P. Beckman J. S. (1995). The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch. Biochem. Biophys. 316, 327–334. 10.1006/abbi.1995.1044
32
Brunskill E. W. de Jonge B. L. Bayles K. W. (1997). The Staphylococcus aureusscdA gene: a novel locus that affects cell division and morphogenesis. Microbiology143(Pt 9), 2877–2882. 10.1099/00221287-143-9-2877
33
Busenlehner L. S. Pennella M. A. Giedroc D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143.
34
Carlioz A. Touati D. (1986). Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life?EMBO J. 5, 623–630.
35
Chambers H. F. DeLeo F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. 10.1038/nrmicro2200
36
Chan P. F. Foster S. J. (1998). Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180, 6232–6241.
37
Chang W. Small D. A. Toghrol F. Bentley W. E. (2006). Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188, 1648–1659. 10.1128/JB.188.4.1648-1659.2006
38
Chelikani P. Fita I. Loewen P. C. (2004). Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61, 192–208. 10.1007/s00018-003-3206-5
39
Chen L. Keramati L. Helmann J. D. (1995). Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U.S.A. 92, 8190–8194.
40
Chen P. R. Bae T. Williams W. A. Duguid E. M. Rice P. A. Schneewind O. He C. (2006). An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2, 591–595. 10.1038/nchembio820
41
Chen P. R. Brugarolas P. He C. (2011). Redox signaling in human pathogens. Antioxid. Redox Signal. 14, 1107–1118. 10.1089/ars.2010.3374
42
Chen P. R. Nishida S. Poor C. B. Cheng A. Bae T. Kuechenmeister L. Dunman P. M. Missiakas D. He C. (2009). A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 71, 198–211. 10.1111/j.1365-2958.2008.06518.x
43
Cheung A. L. Koomey J. M. Butler C. A. Projan S. J. Fischetti V. A. (1992). Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U.S.A. 89, 6462–6466.
44
Cheung J. Beasley F. C. Liu S. Lajoie G. A. Heinrichs D. E. (2009). Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74, 594–608. 10.1111/j.1365-2958.2009.06880.x
45
Chopra I. O'Neill A. J. Miller K. (2003). The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updat. 6, 137–145.
46
Chouchani E. T. James A. M. Fearnley I. M. Lilley K. S. Murphy M. P. (2011). Proteomic approaches to the characterization of protein thiol modification. Curr. Opin. Chem. Biol. 15, 120–128. 10.1016/j.cbpa.2010.11.003
47
Cirz R. T. Jones M. B. Gingles N. A. Minogue T. D. Jarrahi B. Peterson S. N. Romesberg F. E. (2007). Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189, 531–539. 10.1128/JB.01464-06
48
Clauditz A. Resch A. Wieland K. P. Peschel A. Götz F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953. 10.1128/IAI.00204-06
49
Clements M. O. Watson S. P. Foster S. J. (1999). Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181, 3898–3903.
50
Cockayne A. Hill P. J. Powell N. B. Bishop K. Sims C. Williams P. (1998). Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66, 3767–3774.
51
Corbin B. D. Seeley E. H. Raab A. Feldmann J. Miller M. R. Torres V. J. Anderson K. L. Dattilo B. M. Dunman P. M. Gerads R. Caprioli R. M. Nacken W. Chazin W. J. Skaar E. P. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science319, 962–965. 10.1126/science.1152449
52
Cosgrove K. Coutts G. Jonsson I. M. Tarkowski A. Kokai-Kun J. F. Mond J. J. Foster S. J. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. 10.1128/JB.01524-06
53
Cotton J. L. Tao J. Balibar C. J. (2009). Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry48, 1025–1035. 10.1021/bi801844c
54
Coulter S. N. Schwan W. R. Ng E. Y. Langhorne M. H. Ritchie H. D. Westbrock-Wadman S. Hufnagle W. O. Folger K. R. Bayer A. S. Stover C. K. (1998). Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30, 393–404. 10.1046/j.1365-2958.1998.01075.x
55
Cui L. Lian J. Q. Neoh H. M. Reyes E. Hiramatsu K. (2005). DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49, 3404–3413. 10.1128/AAC.49.8.3404-3413.2005
56
Dahl T. A. Midden W. R. Hartman P. E. (1989). Comparison of killing of gram-negative and gram-positive bacteria by pure singlet oxygen. J. Bacteriol. 171, 2188–2194.
57
Dale S. E. Sebulsky M. T. Heinrichs D. E. (2004). Involvement of SirABC in ironsiderophore import in Staphylococcus aureus. J. Bacteriol. 186, 8356–8362. 10.1128/JB.186.24.8356-8362.2004
58
Dame R. T. (2005). The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56, 858–870. 10.1111/j.1365-2958.2005.04598.x
59
Das D. Bishayi B. (2009). Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb. Pathog. 47, 57–67. 10.1016/j.micpath.2009.04.012
60
Das D. Saha S. S. Bishayi B. (2008). Intracellular survival of Staphylococcus aureus: correlating production of catalase and superoxide dismutase with levels of inflammatory cytokines. Inflamm. Res. 57, 340–349. 10.1007/s00011-007-7206-z
61
Daum R. S. (2008). Removing the golden coat of Staphylococcus aureus. N. Engl. J. Med. 359, 85–87. 10.1056/NEJMcibr0803278
62
Davies M. J. (2005). The oxidative environment and protein damage. Biochim. Biophys. Acta1703, 93–109. 10.1016/j.bbapap.2004.08.007
63
Dean R. T. Fu S. Stocker R. Davies M. J. (1997). Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 324(Pt 1), 1–18.
64
del Cardayre S. B. Stock K. P. Newton G. L. Fahey R. C. Davies J. E. (1998). Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 273, 5744–5751. 10.1074/jbc.273.10.5744
65
Demple B. Harrison L. (1994). Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63, 915–948. 10.1146/annurev.bi.63.070194.004411
66
Demple B. Linn S. (1980). DNA N-glycosylases and UV repair. Nature287, 203–208.
67
Derman A. I. Beckwith J. (1991). Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 173, 7719–7722.
68
Derman A. I. Prinz W. A. Belin D. Beckwith J. (1993). Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science262, 1744–1747. 10.1126/science.8259521
69
Di Simplicio P. Franconi F. Frosalí S. Di Giuseppe D. (2003). Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids25, 323–339. 10.1007/s00726-003-0020-1
70
Diekema D. J. Pfaller M. A. Schmitz F. J. Smayevsky J. Bell J. Jones R. N. Beach M. (2001). Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 32(Suppl. 2), S114–S132. 10.1086/320184
71
Diep B. A. Gill S. R. Chang R. F. Phan T. H. Chen J. H. Davidson M. G. Lin F. Lin J. Carleton H. A. Mongodin E. F. Sensabaugh G. F. Perdreau-Remington F. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet367, 731–739. 10.1016/S0140-6736(06)68231-7
72
Djaman O. Outten F. W. Imlay J. A. (2004). Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 279, 44590–44599. 10.1074/jbc.M406487200
73
Dukan S. Belkin S. Touati D. (1999). Reactive oxygen species are partially involved in the bacteriocidal action of hypochlorous acid. Arch. Biochem. Biophys. 367, 311–316. 10.1006/abbi.1999.1265
74
Dunman P. M. Murphy E. Haney S. Palacios D. Tucker-Kellogg G. Wu S. Brown E. L. Zagursky R. J. Shlaes D. Projan S. J. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353. 10.1128/JB.183.24.7341-7353.2001
75
Egeter O. Brückner R. (1996). Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol. Microbiol. 21, 739–749. 10.1046/j.1365-2958.1996.301398.x
76
Eisen J. A. Hanawalt P. C. (1999). A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435, 171–213. 10.1016/S0921-8777(99)00050-6
77
El-Agamey A. Lowe G. M. McGarvey D. J. Mortensen A. Phillip D. M. Truscott T. G. Young A. J. (2004). Carotenoid radical chemistry and antioxidant/pro-oxidant properties. Arch. Biochem. Biophys. 430, 37–48. 10.1016/j.abb.2004.03.007
78
Endo G. Silver S. (1995). CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 177, 4437–4441.
79
Erill I. Campoy S. Barbé J. (2007). Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656. 10.1111/j.1574-6976.2007.00082.x
80
Ermler U. Siddiqui R. A. Cramm R. Friedrich B. (1995). Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution. EMBO J. 14, 6067–6077.
81
Ernst J. F. Bennett R. L. Rothfield L. I. (1978). Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135, 928–934.
82
Escolar L. Pérez-Martín J. de Lorenzo V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229.
83
Even S. Burguière P. Auger S. Soutourina O. Danchin A. Martin-Verstraete I. (2006). Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 188, 2184–2197. 10.1128/JB.188.6.2184-2197.2006
84
Ezraty B. Aussel L. Barras F. (2005). Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta1703, 221–229. 10.1016/j.bbapap.2004.08.017
85
Fahey R. C. (2001). Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55, 333–356. 10.1146/annurev.micro.55.1.333
86
Felden B. Vandenesch F. Bouloc P. Romby P. (2011). The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 7:e1002006. 10.1371/journal.ppat.1002006
87
Finney L. A. O'Halloran T. V. (2003). Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science300, 931–936. 10.1126/science.1085049
88
Flint D. H. Tuminello J. F. Emptage M. H. (1993). The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268, 22369–22376.
89
Fontecave M. Ollagnier-de-Choudens S. (2008). Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474, 226–237. 10.1016/j.abb.2007.12.014
90
Foster T. J. (2009). Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet. Dermatol. 20, 456–470. 10.1111/j.1365-3164.2009.00825.x
91
Franklin W. A. Lindahl T. (1988). DNA deoxyribophosphodiesterase. EMBO J. 7, 3617–3622.
92
Fridovich I. (1995). Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64, 97–112. 10.1146/annurev.bi.64.070195.000525
93
Fuangthong M. Atichartpongkul S. Mongkolsuk S. Helmann J. D. (2001). OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183, 4134–4141. 10.1128/JB.183.14.4134-4141.2001
94
Fuangthong M. Helmann J. D. (2002). The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U.S.A. 99, 6690–6695. 10.1073/pnas.102483199
95
Fuangthong M. Herbig A. F. Bsat N. Helmann J. D. (2002). Regulation of the Bacillus subtilisfur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184, 3276–3286. 10.1128/JB.184.12.3276-3286.2002
96
Fujimoto D. F. Higginbotham R. H. Sterba K. M. Maleki S. J. Segall A. M. Smeltzer M. S. Hurlburt B. K. (2009). Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol. Microbiol. 74, 1445–1458. 10.1111/j.1365-2958.2009.06942.x
97
Gaballa A. Helmann J. D. (2002). A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45, 997–1005. 10.1046/j.1365-2958.2002.03068.x
98
Gaballa A. Wang T. Ye R. W. Helmann J. D. (2002). Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184, 6508–6514. 10.1128/JB.184.23.6508-6514.2002
99
Gardner P. R. Gardner A. M. Martin L. A. Salzman A. L. (1998). Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U.S.A. 95, 10378–10383.
100
Geisen R. Lucke F. K. Kröckel L. (1992). Starter and protective cultures for meat and meat-products. Fleischwirtschaft72, 894–898.
101
Giachino P. Engelmann S. Bischoff M. (2001). Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183, 1843–1852. 10.1128/JB.183.6.1843-1852.2001
102
Goerke C. Köller J. Wolz C. (2006). Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 171–177. 10.1128/AAC.50.1.171-177.2006
103
Gonçalves V. L. Nobre L. S. Vicente J. B. Teixeira M. Saraiva L. M. (2006). Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 580, 1817–1821. 10.1016/j.febslet.2006.02.039
104
Gonzalez de Peredo A. Saint-Pierre C. Adrait A. Jacquamet L. Latour J. M. Michaud-Soret I. Forest E. (1999). Identification of the two zinc-bound cysteines in the ferric uptake regulation protein from Escherichia coli: chemical modification and mass spectrometry analysis. Biochemistry38, 8582–8589. 10.1021/bi9902283
105
Götz F. Bannerman T. Schleifer K. H. (2006). “The Genera Staphylococcus and Macrococcus,” in The Procaryotes, eds DworkinM.FalkowS.RosenbergE.SchleiferK. H.StackebrandtE. (New York, NY: Springer), 5–75.
106
Grainger D. C. Goldberg M. D. Lee D. J. Busby S. J. (2008). Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68, 1366–1377. 10.1111/j.1365-2958.2008.06253.x
107
Grant R. A. Filman D. J. Finkel S. E. Kolter R. Hogle J. M. (1998). The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5, 294–303.
108
Griffiths E. (ed.). (1999). “Iron in biological systems.” in Iron and Infection. (New York, NY: Wiley). 1–26
109
Grimaud R. Ezraty B. Mitchell J. K. Lafitte D. Briand C. Derrick P. J. Barras F. (2001). Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48915–48920. 10.1074/jbc.M105509200
110
Grinsted J. Lacey R. W. (1973). Ecological and genetic implications of pigmentation in Staphylococcus aureus. J. Gen. Microbiol. 75, 259–267.
111
Grossoehme N. Kehl-Fie T. E. Ma Z. Adams K. W. Cowart D. M. Scott R. A. Skaar E. P. Giedroc D. P. (2011). Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531. 10.1074/jbc.M111.220012
112
Grundmann H. Aires-de-Sousa M. Boyce J. Tiemersma E. (2006). Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet368, 874–885. 10.1016/S0140-6736(06)68853-3
113
Guffanti A. A. Wei Y. Rood S. V. Krulwich T. A. (2002). An antiport mechanism for a member of the cation diffusion facilitator family: divalent cations efflux in exchange for K+ and H+. Mol. Microbiol. 45, 145–153. 10.1046/j.1365-2958.2002.02998.x
114
Gutteridge J. M. Rowley D. A. Halliwell B. (1982). Superoxide-dependent formation of hydroxyl radicals and lipid peroxidation in the presence of iron salts. Detection of ‘catalytic’ iron and anti-oxidant activity in extracellular fluids. Biochem. J. 206, 605–609.
115
Haebich D. von Nussbaum F. (2008). “Superbugs bunny” outsmarts our immune defense. ChemMedChem3, 1173–1177. 10.1002/cmdc.200800171
116
Halliwell B. Gutteridge J. M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1–14.
117
Hammes W. P. Bosch I. Wolf G. (1995). Contribution of Staphylococcus carnosus and Staphylococcus piscifermentas to the fermentation of protein foods. J. Appl. Bacteriol. Symp. Suppl. 79, 76–83.
118
Han G. Martinez L. R. Mihu M. R. Friedman A. J. Friedman J. M. Nosanchuk J. D. (2009). Nitric oxide releasing nanoparticles are therapeutic for Staphylococcus aureus abscesses in a murine model of infection. PLoS One4:e7804. 10.1371/journal.pone.0007804
119
Hantke K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182, 288–292.
120
Harrison J. E. Schultz J. (1976). Studies on the chlorinating activity of myeloperoxidase. J. Biol. Chem. 251, 1371–1374.
121
Heidrich C. Hantke K. Bierbaum G. Sahl H. G. (1996). Identification and analysis of a gene encoding a Fur-like protein of Staphylococcus epidermidis. FEMS Microbiol. Lett. 140, 253–259.
122
Heinecke J. W. Li W. Francis G. A. Goldstein J. A. (1993). Tyrosyl radical generated by myeloperoxidase catalyzes the oxidative cross-linking of proteins. J. Clin. Invest91, 2866–2872. 10.1172/JCI116531
123
Hill P. J. Cockayne A. Landers P. Morrissey J. A. Sims C. M. Williams P. (1998). SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66, 4123–4129.
124
Holden M. T. Feil E. J. Lindsay J. A. Peacock S. J. Day N. P. Enright M. C. Foster T. J. Moore C. E. Hurst L. Atkin R. Barron A. Bason N. Bentley S. D. Chillingworth C. Chillingworth T. Churcher C. Clark L. Corton C. Cronin A. Doggett J. Dowd L. Feltwell T. Hance Z. Harris B. Hauser H. Holroyd S. Jagels K. James K. D. Lennard N. Line A. Mayes R. Moule S. Mungall K. Ormond D. Quail M. A. Rabbinowitsch E. Rutherford K. Sanders M. Sharp S. Simmonds M. Stevens K. Whitehead S. Barrell B. G. Spratt B. G. Parkhill J. (2004). Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U.S.A. 101, 9786–9791. 10.1073/pnas.0402521101
125
Holmgren A. (1985). Thioredoxin. Annu. Rev. Biochem. 54, 237–271. 10.1146/annurev.bi.54.070185.001321
126
Holmgren A. (1989). Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264, 13963–13966.
127
Horsburgh M. J. Clements M. O. Crossley H. Ingham E. Foster S. J. (2001a). PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69, 3744–3754. 10.1128/IAI.69.6.3744-3754.2001
128
Horsburgh M. J. Ingham E. Foster S. J. (2001b). In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. 10.1128/JB.183.2.468-475.2001
129
Horsburgh M. J. Wharton S. J. Cox A. G. Ingham E. Peacock S. Foster S. J. (2002). MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286. 10.1046/j.1365-2958.2002.02944.x
130
Huie R. E. Padmaja S. (1993). The reaction of NO with superoxide. Free Radic. Res. Commun. 18, 195–199.
131
Imlay J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. 10.1146/annurev.micro.57.030502.090938
132
Imlay J. A. Chin S. M. Linn S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science240, 640–642. 10.1126/science.2834821
133
Ingavale S. van Wamel W. Luong T. T. Lee C. Y. Cheung A. L. (2005). Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73, 1423–1431. 10.1128/IAI.73.3.1423-1431.2005
134
Ingavale S. S. van Wamel W. Cheung A. L. (2003). Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48, 1451–1466.
135
Jabado N. Jankowski A. Dougaparsad S. Picard V. Grinstein S. Gros P. (2000). Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192, 1237–1248.
136
Jacobs S. I. Willis A. T. (1964). Some physiological characteristics of neomycin and kanamycin-resistant strains of Staphylococcus aureus. J. Clin. Pathol. 17, 612–616.
137
Jacquamet L. Aberdam D. Adrait A. Hazemann J. L. Latour J. M. Michaud-Soret I. (1998). X-ray absorption spectroscopy of a new zinc site in the Fur protein from Escherichia coli. Biochemistry37, 2564–2571. 10.1021/bi9721344
138
Jang S. Imlay J. A. (2007). Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282, 929–937. 10.1074/jbc.M607646200
139
Johnson A. P. (2011). Methicillin-resistant Staphylococcus aureus: the European landscape. J. Antimicrob. Chemother. 66(Suppl. 4), iv43–iv48. 10.1093/jac/dkr076
140
Johnson D. C. Dean D. R. Smith A. D. Johnson M. K. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. 10.1146/annurev.biochem.74.082803.133518
141
Johnson M. Sengupta M. Purves J. Tarrant E. Williams P. H. Cockayne A. Muthaiyan A. Stephenson R. Ledala N. Wilkinson B. J. Jayaswal R. K. Morrissey J. A. (2011). Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int. J. Med. Microbiol. 301, 44–52. 10.1016/j.ijmm.2010.05.003
142
Jonsson I. M. Lindholm C. Luong T. T. Lee C. Y. Tarkowski A. (2008). mgrA regulates staphylococcal virulence important for induction and progression of septic arthritis and sepsis. Microbes Infect. 10, 1229–1235. 10.1016/j.micinf.2008.07.026
143
Justino M. C. Almeida C. C. Gonçalves V. L. Teixeira M. Saraiva L. M. (2006). Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257, 278–284. 10.1111/j.1574-6968.2006.00179.x
144
Justino M. C. Almeida C. C. Teixeira M. Saraiva L. M. (2007). Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 282, 10352–10359. 10.1074/jbc.M610656200
145
Kaatz G. W. Thyagarajan R. V. Seo S. M. (2005). Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob. Agents Chemother. 49, 161–169. 10.1128/AAC.49.1.161-169.2005
146
Kanafani H. Martin S. E. (1985). Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J. Clin. Microbiol. 21, 607–610.
147
Karavolos M. H. Horsburgh M. J. Ingham E. Foster S. J. (2003). Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology149, 2749–2758. 10.1099/mic.0.26353-0
148
Katcher H. L. Wallace S. S. (1983). Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry22, 4071–4081.
149
Kehl-Fie T. E. Chitayat S. Hood M. I. Damo S. Restrepo N. Garcia C. Munro K. A. Chazin W. J. Skaar E. P. (2011). Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe10, 158–164. 10.1016/j.chom.2011.07.004
150
Kehres D. G. Maguire M. E. (2003). Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27, 263–290.
151
Kelley W. L. (2006). Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 62, 1228–1238. 10.1111/j.1365-2958.2006.05444.x
152
Keyer K. Imlay J. A. (1996). Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U.S.A. 93, 13635–13640.
153
Khor H. K. Fisher M. T. Schöneich C. (2004). Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO−). J. Biol. Chem. 279, 19486–19493. 10.1074/jbc.M310045200
154
Kim E. J. Chung H. J. Suh B. Hah Y. C. Roe J. H. (1998). Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol. Microbiol. 27, 187–195. 10.1046/j.1365-2958.1998.00674.x
155
Kim F. J. Kim H. P. Hah Y. C. Roe J. H. (1996). Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur. J. Biochem. 241, 178–185. 10.1111/j.1432-1033.1996.0178t.x
156
Kim J. Yoshimura S. H. Hizume K. Ohniwa R. L. Ishihama A. Takeyasu K. (2004). Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 32, 1982–1992. 10.1093/nar/gkh512
157
Kim S. O. Orii Y. Lloyd D. Hughes M. N. Poole R. K. (1999). Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445, 389–394.
158
Klebanoff S. J. (1999). Myeloperoxidase. Proc. Assoc. Am. Physicians111, 383–389.
159
Kluytmans J. van Belkum A. Verbrugh H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520.
160
Kohanski M. A. Dwyer D. J. Hayete B. Lawrence C. A. Collins J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell130, 797–810. 10.1016/j.cell.2007.06.049
161
Korshunov S. Imlay J. A. (2010). Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 75, 1389–1401. 10.1111/j.1365-2958.2010.07059.x
162
Krinsky N. I. (1993). Actions of carotenoids in biological systems. Annu. Rev. Nutr. 13, 561–587. 10.1146/annurev.nu.13.070193.003021
163
Kullik I. Giachino P. Fuchs T. (1998). Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180, 4814–4820.
164
Kuo C. F. Mashino T. Fridovich I. (1987). alpha, beta-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 262, 4724–4727.
165
Kuroda M. Hayashi H. Ohta T. (1999). Chromosome-determined zinc-responsible operon czr in Staphylococcus aureus strain 912. Microbiol. Immunol. 43, 115–125.
166
Lanza F. (1998). Clinical manifestation of myeloperoxidase deficiency. J. Mol. Med. (Berl.)76, 676–681.
167
Lee J. W. Helmann J. D. (2006a). Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281, 23567–23578. 10.1074/jbc.M603968200
168
Lee J. W. Helmann J. D. (2006b). The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature440, 363–367. 10.1038/nature04537
169
Lee J. W. Helmann J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals20, 485–499. 10.1007/s10534-006-9070-7
170
Lee J. W. Soonsanga S. Helmann J. D. (2007). A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748. 10.1073/pnas.0702081104
171
Lehrer R. I. Hanifin J. Cline M. J. (1969). Defective bactericidal activity in myeloperoxidase-deficient human neutrophils. Nature223, 78–79.
172
Leichert L. I. Scharf C. Hecker M. (2003). Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185, 1967–1975. 10.1128/JB.185.6.1967-1975.2003
173
Lindahl T. (1979). DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol. 22, 135–192.
174
Lindsay J. A. Foster S. J. (1999). Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol. Gen. Genet. 262, 323–331.
175
Lindsay J. A. Foster S. J. (2001). zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology147, 1259–1266.
176
Lippard S. J. Berg J. M. (1994). Principles of bioinorganic chemistry. Mill Valley, CA: University Science Books.
177
Liu C. I. Liu G. Y. Song Y. Yin F. Hensler M. E. Jeng W. Y. Nizet V. Wang A. H. Oldfield E. (2008). A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science319, 1391–1394. 10.1126/science.1153018
178
Liu G. Y. Essex A. Buchanan J. T. Datta V. Hoffman H. M. Bastian J. F. Fierer J. Nizet V. (2005). Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215. 10.1084/jem.20050846
179
Liu T. Ramesh A. Ma Z. Ward S. K. Zhang L. George G. N. Talaat A. M. Sacchettini J. C. Giedroc D. P. (2007). CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68. 10.1038/nchembio844
180
Liu Y. Wu N. Dong J. Gao Y. Zhang X. Shao N. Yang G. (2010). SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. FEBS Lett. 584, 4325–4329. 10.1016/j.febslet.2010.09.024
181
Ljungquist S. (1977). A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem. 252, 2808–2814.
182
Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. 10.1056/NEJM199808203390806
183
Lu A. L. Li X. Gu Y. Wright P. M. Chang D. Y. (2001). Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophys. 35, 141–170. 10.1385/CBB:35:2:141
184
Luijsterburg M. S. Noom M. C. Wuite G. J. Dame R. T. (2006). The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156, 262–272. 10.1016/j.jsb.2006.05.006
185
Luong T. T. Dunman P. M. Murphy E. Projan S. J. Lee C. Y. (2006). Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910. 10.1128/JB.188.5.1899-1910.2006
186
Luong T. T. Newell S. W. Lee C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185, 3703–3710. 10.1128/JB.185.13.3703-3710.2003
187
Ma Z. Gabriel S. E. Helmann J. D. (2011). Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138. 10.1093/nar/gkr625
188
Macomber L. Imlay J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. 10.1073/pnas.0812808106
189
Magnuson K. Jackowski S. Rock C. O. Cronan J. E. Jr. (1993). Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542.
190
Maier R. J. Benoit S. L. Seshadri S. (2007). Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals20, 655–664. 10.1007/s10534-006-9061-8
191
Maiques E. Úbeda C. Campoy S. Salvador N. Lasa I. Novick R. P. Barbé J. Penadés J. R. (2006). beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188, 2726–2729. 10.1128/JB.188.7.2726-2729.2006
192
Majerczyk C. D. Dunman P. M. Luong T. T. Lee C. Y. Sadykov M. R. Somerville G. A. Bodi K. Sonenshein A. L. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. 10.1128/JB.00220-10
193
Maki H. Sekiguchi M. (1992). MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature355, 273–275. 10.1038/355273a0
194
Mandell G. L. (1975). Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal–leukocyte interaction. J. Clin. Invest. 55, 561–566. 10.1172/JCI107963
195
Manna A. C. Ingavale S. S. Maloney M. van Wamel W. Cheung A. L. (2004). Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186, 5267–5280. 10.1128/JB.186.16.5267-5280.2004
196
Marshall J. H. Wilmoth G. J. (1981a). Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147, 900–913.
197
Marshall J. H. Wilmoth G. J. (1981b). Proposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutants. J. Bacteriol. 147, 914–919.
198
Marti T. M. Kunz C. Fleck O. (2002). DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191, 28–41. 10.1002/jcp.10077
199
Martin S. E. Chaven S. (1987). Synthesis of catalase in Staphylococcus aureus MF-31. Appl. Environ. Microbiol. 53, 1207–1209.
200
Martinez A. Kolter R. (1997). Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179, 5188–5194.
201
Martínez-Pulgarín S. Domínguez-Bernal G. Orden J. A. de la Fuente R. (2009). Simultaneous lack of catalase and beta-toxin in Staphylococcus aureus leads to increased intracellular survival in macrophages and epithelial cells and to attenuated virulence in murine and ovine models. Microbiology155, 1505–1515. 10.1099/mic.0.025544-0
202
Massey V. Strickland S. Mayhew S. G. Howell L. G. Engel P. C. Matthews R. G. Schuman M. Sullivan P. A. (1969). The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem. Biophys. Res. Commun. 36, 891–897. 10.1016/0006-291X(69)90287-3
203
Mathews-Roth M. M. Wilson T. Fujimori E. Krinsky N. I. (1974). Carotenoid chromophore length and protection against photosensitization. Photochem. Photobiol. 19, 217–222. 10.1111/j.1751-1097.1974.tb06501.x
204
McCann M. T. Gilmore B. F. Gorman S. P. (2008). Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J. Pharm. Pharmacol. 60, 1551–1571. 10.1211/jpp/60.12.0001
205
Messina C. G. Reeves E. P. Roes J. Segal A. W. (2002). Catalase negative Staphylococcus aureus retain virulence in mouse model of chronic granulomatous disease. FEBS Lett. 518, 107–110.
206
Messner K. R. Imlay J. A. (1999). The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J. Biol. Chem. 274, 10119–10128. 10.1074/jbc.274.15.10119
207
Michaels M. L. Cruz C. Grollman A. P. Miller J. H. (1992a). Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 7022–7025.
208
Michaels M. L. Tchou J. Grollman A. P. Miller J. H. (1992b). A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry31, 10964–10968.
209
Michaels M. L. Miller J. H. (1992). The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325.
210
Mishra N. N. Liu G. Y. Yeaman M. R. Nast C. C. Proctor R. A. McKinnell J. Bayer A. S. (2011). Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55, 526–531. 10.1128/AAC.00680-10
211
Modrich P. (1991). Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25, 229–253. 10.1146/annurev.ge.25.120191.001305
212
Mol C. D. Hosfield D. J. Tainer J. A. (2000). Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 460, 211–229. 10.1016/S0921-8777(00)00028-8
213
Montel M. C. Reitz J. Talon R. Berdagué J. L. Rousset-Akrim S. (1996). Biochemical activities of Micrococcaceae and their effects on the aromatic profiles and odours of a dry sausage model. Food Microbiol. 13, 489–499.
214
Morikawa K. Ohniwa R. L. Kim J. Maruyama A. Ohta T. Takeyasu K. (2006). Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells11, 409–423. 10.1111/j.1365-2443.2006.00949.x
215
Morikawa K. Ohniwa R. L. Kim J. Takeshita S. L. Maruyama A. Inose Y. Takeyasu K. Ohta T. (2007). Biochemical, molecular genetic, and structural analyses of the staphylococcal nucleoid. Microsc. Microanal. 13, 30–35. 10.1017/S1431927607070080
216
Morrissey J. A. Cockayne A. Brummell K. Williams P. (2004). The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 72, 972–979. 10.1128/IAI.72.2.972-979.2004
217
Morrissey J. A. Cockayne A. Hill P. J. Williams P. (2000). Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68, 6281–6288. 10.1128/IAI.68.11.6281-6288.2000
218
Moskovitz J. Weissbach H. Brot N. (1996). Cloning the expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 2095–2099.
219
Mostertz J. Scharf C. Hecker M. Homuth G. (2004). Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology150, 497–512. 10.1099/mic.0.26665-0
220
Müller F. (1987). Flavin radicals: chemistry and biochemistry. Free Radic. Biol. Med. 3, 215–230.
221
Nachin L. Loiseau L. Expert D. Barras F. (2003). SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22, 427–437. 10.1093/emboj/cdg061
222
Nair S. Finkel S. E. (2004). Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186, 4192–4198. 10.1128/JB.186.13.4192-4198.2004
223
Nakano S. Küster-Schöck E. Grossman A. D. Zuber P. (2003). Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 100, 13603–13608. 10.1073/pnas.2235180100
224
Nathan C. Shiloh M. U. (2000). Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 97, 8841–8848. 10.1073/pnas.97.16.8841
225
Nauseef W. M. (2004). Assembly of the phagocyte NADPH oxidase. Histochem. Cell Biol. 122, 277–291. 10.1007/s00418-004-0679-8
226
Newton G. L. Fahey R. C. Cohen G. Aharonowitz Y. (1993). Low-molecular-weight thiols in streptomycetes and their potential role as antioxidants. J. Bacteriol. 175, 2734–2742.
227
Niga T. Yoshida H. Hattori H. Nakamura S. Ito H. (1997). Cloning and sequencing of a novel gene (recG) that affects the quinolone susceptibility of Staphylococcus aureus. Antimicrob. Agents Chemother. 41, 1770–1774.
228
Nucifora G. Chu L. Misra T. K. Silver S. (1989). Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. U.S.A. 86, 3544–3548.
229
O'Neill A. J. Chopra I. (2002). Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutators in clinical isolates is low. J. Antimicrob. Chemother. 50, 161–169. 10.1093/jac/dkf118
230
Ochsner U. A. Vasil A. I. Vasil M. L. (1995). Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177, 7194–7201.
231
Ohniwa R. L. Morikawa K. Kim J. Ohta T. Ishihama A. Wada C. Takeyasu K. (2006). Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J. 25, 5591–5602. 10.1038/sj.emboj.7601414
232
Ohniwa R. L. Ushijima Y. Saito S. Morikawa K. (2011). Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One6:e19172. 10.1371/journal.pone.0019172
233
Oldfield E. (2010). Targeting isoprenoid biosynthesis for drug discovery: bench to bedside. Acc. Chem. Res. 43, 1216–1226. 10.1021/ar100026v
234
Olivier A. C. Lemaire S. van Bambeke F. Tulkens P. M. Oldfield E. (2009). Role of rsbU and staphyloxanthin in phagocytosis and intracellular growth of Staphylococcus aureus in human macrophages and endothelial cells. J. Infect. Dis. 200, 1367–1370. 10.1086/606012
235
Overton T. W. Justino M. C. Li Y. Baptista J. M. Melo A. M. Cole J. A. Saraiva L. M. (2008). Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 190, 2004–2013. 10.1128/JB.01733-07
236
Pagels M. Fuchs S. Pané-Farré J. Kohler C. Menschner L. Hecker M. McNamarra P. J. Bauer M. C. von Wachenfeldt C. Liebeke M. Lalk M. Sander G. von Eiff C. Proctor R. A. Engelmann S. (2010). Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161. 10.1111/j.1365-2958.2010.07105.x
237
Palazzolo-Ballance A. M. Reniere M. L. Braughton K. R. Sturdevant D. E. Otto M. Kreiswirth B. N. Skaar E. P. DeLeo F. R. (2008). Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180, 500–509.
238
Palma M. Cheung A. L. (2001). sigma(B) activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69, 7858–7865. 10.1128/IAI.69.12.7858-7865.2001
239
Pamp S. J. Frees D. Engelmann S. Hecker M. Ingmer H. (2006). Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188, 4861–4870. 10.1128/JB.00194-06
240
Papp-Wallace K. M. Maguire M. E. (2006). Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60, 187–209. 10.1146/annurev.micro.60.080805.142149
241
Park R. Y. Sun H. Y. Choi M. H. Bai Y. H. Shin S. H. (2005a). Staphylococcus aureus siderophore-mediated iron-acquisition system plays a dominant and essential role in the utilization of transferrin-bound iron. J. Microbiol. 43, 183–190.
242
Park S. Imlay J. A. (2003). High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 185, 1942–1950. 10.1128/JB.185.6.1942-1950.2003
243
Park S. You X. Imlay J. A. (2005b). Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 9317–9322. 10.1073/pnas.0502051102
244
Pelz A. Wieland K. P. Putzbach K. Hentschel P. Albert K. Götz F. (2005). Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280, 32493–32498. 10.1074/jbc.M505070200
245
Place R. B. Hiestand D. Gallmann H. R. Teuber M. (2003). Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 26, 30–37.
246
Pollitt S. Zalkin H. (1983). Role of primary structure and disulfide bond formation in beta-lactamase secretion. J. Bacteriol. 153, 27–32.
247
Pomposiello P. J. Demple B. (2002). Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46, 319–341.
248
Poole L. B. (2005). Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433, 240–254. 10.1016/j.abb.2004.09.006
249
Poor C. B. Chen P. R. Duguid E. Rice P. A. He C. (2009). Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 284, 23517–23524. 10.1074/jbc.M109.015826
250
Pöther D. C. Liebeke M. Hochgräfe F. Antelmann H. Becher D. Lalk M. Lindequist U. Borovok I. Cohen G. Aharonowitz Y. Hecker M. (2009). Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 191, 7520–7530. 10.1128/JB.00937-09
251
Poyart C. Berche P. Trieu-Cuot P. (1995). Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol. Lett. 131, 41–45.
252
Prinz W. A. Aslund F. Holmgren A. Beckwith J. (1997). The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272, 15661–15667. 10.1074/jbc.272.25.15661
253
Probst A. J. Hertel C. Richter L. Wassill L. Ludwig W. Hammes W. P. (1998). Staphylococcus condimenti sp. nov., from soy sauce mash, and Staphylococcus carnosus (Schleifer and Fischer 1982) subsp. utilis subsp. nov. Int. J. Syst. Bacteriol. 48(Pt 3), 651–658. 10.1099/00207713-48-3-651
254
Proctor R. A. von Eiff C. Kahl B. C. Becker K. McNamara P. Herrmann M. Peters G. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. 10.1038/nrmicro1384
255
Prunier A. L. Leclercq R. (2005). Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 187, 3455–3464. 10.1128/JB.187.10.3455-3464.2005
256
Puig S. Thiele D. J. (2002). Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6, 171–180. 10.1016/S1367-5931(02)00298-3
257
Radman M. (1976). An endonuclease from Escherichia coli that introduces single polynucleotide chain scissions in ultraviolet-irradiated DNA. J. Biol. Chem. 251, 1438–1445.
258
Ratledge C. Dover L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. 10.1146/annurev.micro.54.1.881
259
Richardson A. R. Dunman P. M. Fang F. C. (2006). The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939. 10.1111/j.1365-2958.2006.05290.x
260
Rogers K. L. Fey P. D. Rupp M. E. (2009). Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23, 73–98. 10.1016/j.idc.2008.10.001
261
Rogers S. G. Weiss B. (1980). Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 65, 201–211.
262
Rosenthal V. D. Maki D. G. Jamulitrat S. Medeiros E. A. Todi S. K. Gomez D. Y. Leblebicioglu H. Abu Khader I. Miran da Novales M. G. Berba R. Ramírez Wong F. M. Barkat A. Pino O. R. Dueñas L. Mitrev Z. Bijie H. Gurskis V. Kanj S. S. Mapp T. Hidalgo R. F. Ben Jaballah N. Raka L. Gikas A. Ahmed A. Thu le T. A. Guzmán Siritt M. E. (2010). International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003–2008, issued June 2009. Am. J. Infect. Control. 38, 95–104. e102. 10.1016/j.ajic.2009.12.004
263
Russel M. Model P. (1986). The role of thioredoxin in filamentous phage assembly. Construction, isolation, and characterization of mutant thioredoxins. J. Biol. Chem. 261, 14997–15005.
264
Salamah A. A. (1992). Association of coagulase and/or pigmentation with the virulence of a capsule-lacking Staphylococcus aureus in iron-compromised mice. Microbiologica15, 75–78.
265
Sancar A. (1996). DNA excision repair. Annu. Rev. Biochem. 65, 43–81. 10.1146/annurev.bi.65.070196.000355
266
Sandigursky M. Franklin W. A. (1992). DNA deoxyribophosphodiesterase of Escherichia coli is associated with exonuclease I. Nucleic Acids Res. 20, 4699–4703. 10.1093/nar/20.18.4699
267
Sanz R. Marín I. Ruiz-Santa-Quiteria J. A. Orden J. A. Cid D. Diez R. M. Silhadi K. S. Amils R. de la Fuente R. (2000). Catalase deficiency in Staphylococcus aureus subsp. anaerobius is associated with natural loss-of-function mutations within the structural gene. Microbiology146(Pt 2), 465–475.
268
Sasindran S. J. Saikolappan S. Dhandayuthapani S. (2007). Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2, 619–630. 10.2217/17460913.2.6.619
269
Scharf C. Riethdorf S. Ernst H. Engelmann S. Völker U. Hecker M. (1998). Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180, 1869–1877.
270
Schleifer K. H. Fischer U. (1982). Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32, 153–156.
271
Schneider W. P. Ho S. K. Christine J. Yao M. Marra A. Hromockyj A. E. (2002). Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture. Infect. Immun. 70, 1326–1333. 10.1128/IAI.70.3.1326-1333.2002
272
Seaver L. C. Imlay J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181. 10.1128/JB.183.24.7173-7181.2001
273
Sebulsky M. T. Shilton B. H. Speziali C. D. Heinrichs D. E. (2003). The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J. Biol. Chem. 278, 49890–49900. 10.1074/jbc.M305073200
274
Sebulsky M. T. Speziali C. D. Shilton B. H. Edgell D. R. Heinrichs D. E. (2004). FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: a case of gene duplication and lateral transfer. J. Biol. Chem. 279, 53152–53159. 10.1074/jbc.M409793200
275
Seidl K. Bischoff M. Berger-Bächi B. (2008a). CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect. Immun. 76, 5093–5099. 10.1128/IAI.00724-08
276
Seidl K. Goerke C. Wolz C. Mack D. Berger-Bächi B. Bischoff M. (2008b). Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76, 2044–2050. 10.1128/IAI.00035-08
277
Sen R. Das D. Bishayi B. (2009). Staphylococcal catalase regulates its virulence and induces arthritis in catalase deficient mice. Indian J. Physiol. Pharmacol. 53, 307–317.
278
Sharov V. S. Ferrington D. A. Squier T. C. Schöneich C. (1999). Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 455, 247–250.
279
Sheikh M. A. Taylor G. L. (2009). Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72, 1208–1220. 10.1111/j.1365-2958.2009.06718.x
280
Shibutani S. Takeshita M. Grollman A. P. (1991). Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature349, 431–434. 10.1038/349431a0
281
Singh V. K. Moskovitz J. (2003). Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology149, 2739–2747. 10.1099/mic.0.26442-0
282
Singh V. K. Moskovitz J. Wilkinson B. J. Jayaswal R. K. (2001). Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology147, 3037–3045.
283
Singh V. K. Xiong A. Usgaard T. R. Chakrabarti S. Deora R. Misra T. K. Jayaswal R. K. (1999). ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol. Microbiol. 33, 200–207.
284
Sitthisak S. Howieson K. Amezola C. Jayaswal R. K. (2005). Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl. Environ. Microbiol. 71, 5650–5653. 10.1128/AEM.71.9.5650-5653.2005
285
Sitthisak S. Knutsson L. Webb J. W. Jayaswal R. K. (2007). Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology153, 4274–4283. 10.1099/mic.0.2007/009860-0
286
Skaar E. P. Humayun M. Bae T. DeBord K. L. Schneewind O. (2004). Iron-source preference of Staphylococcus aureus infections. Science305, 1626–1628. 10.1126/science.1099930
287
Skaar E. P. Schneewind O. (2004). Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6, 390–397.
288
Solioz M. Stoyanov J. V. (2003). Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27, 183–195.
289
Somerville G. A. Cockayne A. Dürr M. Peschel A. Otto M. Musser J. M. (2003). Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 185, 6686–6694.
290
Somerville G. A. Proctor R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248. 10.1128/MMBR.00005-09
291
Song Y. Lin F. Y. Yin F. Hensler M. Rodrígues Poveda C. A. Mukkamala D. Cao R. Wang H. Morita C. T. González Pacanowska D. Nizet V. Oldfield E. (2009a). Phosphonosulfonates are potent, selective inhibitors of dehydrosqualene synthase and staphyloxanthin biosynthesis in Staphylococcus aureus. J. Med. Chem. 52, 976–988. 10.1021/jm801023u
292
Song Y. Liu C. I. Lin F. Y. No J. H. Hensler M. Liu Y. L. Jeng W. Y. Low J. Liu G. Y. Nizet V. Wang A. H. Oldfield E. (2009b). Inhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic results. J. Med. Chem. 52, 3869–3880. 10.1021/jm9001764
293
Soonsanga S. Fuangthong M. Helmann J. D. (2007). Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J. Bacteriol. 189, 7069–7076. 10.1128/JB.00879-07
294
Soutourina O. Dubrac S. Poupel O. Msadek T. Martin-Verstraete I. (2010). The pleiotropic CymR regulator of Staphylococcus aureus plays an important role in virulence and stress response. PLoS Pathog6:e1000894. 10.1371/journal.ppat.1000894
295
Soutourina O. Poupel O. Coppée J. Y. Danchin A. Msadek T. Martin-Verstraete I. (2009). CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73, 194–211. 10.1111/j.1365-2958.2009.06760.x
296
Speziali C. D. Dale S. E. Henderson J. A. Vinés E. D. Heinrichs D. E. (2006). Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 188, 2048–2055. 10.1128/JB.188.6.2048-2055.2006
297
Stevens D. L. (2003). Community-acquired Staphylococcus aureus infections: increasing virulence and emerging methicillin resistance in the new millennium. Curr. Opin. Infect. Dis. 16, 189–191. 10.1097/01.qco.0000073765.11390.d5
298
Su M. Cavallo S. Stefanini S. Chiancone E. Chasteen N. D. (2005). The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry44, 5572–5578. 10.1021/bi0472705
299
Sun F. Zhou L. Zhao B. C. Deng X. Cho H. Yi C. Jian X. Song C. X. Luan C. H. Bae T. Li Z. He C. (2011). Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem. Biol. 18, 1032–1041. 10.1016/j.chembiol.2011.05.014
300
Tanasupawat S. Hashimoto Y. Ezaki T. Kozaki M. Komagata K. (1992). Staphylococcuspiscifermentans sp. nov., from fermented fish in Thailand. Int. J. Syst. Bacteriol. 42, 577–581. 10.1099/00207713-42-4-577
301
Torres V. J. Attia A. S. Mason W. J. Hood M. I. Corbin B. D. Beasley F. C. Anderson K. L. Stauff D. L. McDonald W. H. Zimmerman L. J. Friedman D. B. Heinrichs D. E. Dunman P. M. Skaar E. P. (2010). Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78, 1618–1628. 10.1128/IAI.01423-09
302
Torres V. J. Pishchany G. Humayun M. Schneewind O. Skaar E. P. (2006). Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429. 10.1128/JB.01335-06
303
Truong-Bolduc Q. C. Ding Y. Hooper D. C. (2008). Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190, 7375–7381. 10.1128/JB.01068-08
304
Truong-Bolduc Q. C. Dunman P. M. Strahilevitz J. Projan S. J. Hooper D. C. (2005). MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187, 2395–2405. 10.1128/JB.187.7.2395-2405.2005
305
Truong-Bolduc Q. C. Strahilevitz J. Hooper D. C. (2006). NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 1104–1107. 10.1128/AAC.50.3.1104-1107.2006
306
Truong-Bolduc Q. C. Zhang X. Hooper D. C. (2003). Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185, 3127–3138. 10.1128/JB.185.10.3127-3138.2003
307
Úbeda C. Maiques E. Knecht E. Lasa I. Novick R. P. Penadés J. R. (2005). Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56, 836–844. 10.1111/j.1365-2958.2005.04584.x
308
Úbeda C. Maiques E. Tormo M. A. Campoy S. Lasa I. Barbé J. Novick R. P. Penadés J. R. (2007). SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol. Microbiol. 65, 41–50. 10.1111/j.1365-2958.2007.05758.x
309
Uziel O. Borovok I. Schreiber R. Cohen G. Aharonowitz Y. (2004). Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186, 326–334.
310
Valderas M. W. Gatson J. W. Wreyford N. Hart M. E. (2002). The superoxide dismutase gene sodM is unique to Staphylococcus aureus: absence of sodM in coagulase-negative staphylococci. J. Bacteriol. 184, 2465–2472. 10.1128/JB.184.9.2465-2472.2002
311
Valderas M. W. Hart M. E. (2001). Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J. Bacteriol. 183, 3399–3407. 10.1128/JB.183.11.3399-3407.2001
312
Vasudevan S. G. Armarego W. L. Shaw D. C. Lilley P. E. Dixon N. E. Poole R. K. (1991). Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 226, 49–58.
313
Vazquez-Torres A. Jones-Carson J. Mastroeni P. Ischiropoulos H. Fang F. C. (2000). Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192, 227–236. 10.1084/jem.192.2.227
314
Vidales L. E. Cárdenas L. C. Robleto E. Yasbin R. E. Pedraza-Reyes M. (2009). Defects in the error prevention oxidized guanine system potentiate stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 191, 506–513. 10.1128/JB.01210-08
315
Vido K. Diemer H. van Dorsselaer A. Leize E. Juillard V. Gruss A. Gaudu P. (2005). Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J. Bacteriol. 187, 601–610. 10.1128/JB.187.2.601-610.2005
316
Vine C. E. Justino M. C. Saraiva L. M. Cole J. (2010). Detection by whole genome microarrays of a spontaneous 126-gene deletion during construction of a ytfE mutant: confirmation that a ytfE mutation results in loss of repair of iron-sulfur centres in proteins damaged by oxidative or nitrosative stress. J. Microbiol. Methods81, 77–79. 10.1016/j.mimet.2010.01.023
317
von Eiff C. Becker K. Machka K. Stammer H. Peters G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 344, 11–16. 10.1056/NEJM200101043440102
318
von Eiff C. Peters G. Heilmann C. (2002). Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2, 677–685.
319
Voyich J. M. Braughton K. R. Sturdevant D. E. Whitney A. R. Saïd-Salim B. Porcella S. F. Long R. D. Dorward D. W. Gardner D. J. Kreiswirth B. N. Musser J. M. DeLeo F. R. (2005). Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175, 3907–3919.
320
Vuong C. Otto M. (2002). Staphylococcus epidermidis infections. Microbes Infect. 4, 481–489. 10.1016/S1286-4579(02)01563-0
321
Wagner D. Maser J. Lai B. Cai Z. Barry C. E. 3rd Höner Zu Bentrup K. Russell D. G. Bermudez L. E. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174, 1491–1500.
322
Wallace S. S. (1988). AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ. Mol. Mutagen. 12, 431–477.
323
Walsh C. T. Fischbach M. A. (2008). Inhibitors of sterol biosynthesis as Staphylococcus aureus antibiotics. Angew. Chem. Int. Ed. Engl. 47, 5700–5702. 10.1002/anie.200801801
324
Watson S. P. Antonio M. Foster S. J. (1998). Isolation and characterization of Staphylococcus aureus starvation-induc ed, stationary-phase mutants defective in survival or recovery. Microbiology144(Pt 11), 3159–3169. 10.1099/00221287-144-11-3159
325
Weinberg E. D. (1978). Iron and infection. Microbiol. Rev. 42, 45–66.
326
Wertheim H. F. Melles D. C. Vos M. C. van Leeuwen W. van Belkum A. Verbrugh H. A. Nouwen J. L. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762. 10.1016/S1473-3099(05)70295-4
327
White C. Lee J. Kambe T. Fritsche K. Petris M. J. (2009). A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956. 10.1074/jbc.M109.070201
328
Wieland B. Feil C. Gloria-Maercker E. Thumm G. Lechner M. Bravo J. M. Poralla K. Götz F. (1994). Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176, 7719–7726.
329
Willis A. T. Turner G. C. (1962). Staphylococcal lipolysis and pigmentation. J. Pathol. Bacteriol. 84, 337–347.
330
Wolf C. Hochgräfe F. Kusch H. Albrecht D. Hecker M. Engelmann S. (2008). Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics8, 3139–3153. 10.1002/pmic.200701062
331
Wolf S. G. Frenkiel D. Arad T. Finkel S. E. Kolter R. Minsky A. (1999). DNA protection by stress-induced biocrystallization. Nature400, 83–85. 10.1038/21918
332
Wolschendorf F. Ackart D. Shrestha T. B. Hascall-Dove L. Nolan S. Lamichhane G. Wang Y. Bossmann S. H. Basaraba R. J. Niederweis M. (2011). Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 1621–1626. 10.1073/pnas.1009261108
333
Woodmansee A. N. Imlay J. A. (2003). A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol. Microbiol. 49, 11–22. 10.1046/j.1365-2958.2003.03530.x
334
Xiong A. Jayaswal R. K. (1998). Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 180, 4024–4029.
335
Xiong A. Singh V. K. Cabrera G. Jayaswal R. K. (2000). Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology146(Pt 3), 659–668.
336
Ye J. Kandegedara A. Martin P. Rosen B. P. (2005). Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J. Bacteriol. 187, 4214–4221. 10.1128/JB.187.12.4214-4221.2005
337
Yeeles J. T. Dillingham M. S. (2010). The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair (Amst.)9, 276–285. 10.1016/j.dnarep.2009.12.016
338
Yeom J. Imlay J. A. Park W. (2010). Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J. Biol. Chem. 285, 22689–22695. 10.1074/jbc.M110.127456
339
Zhao G. Ceci P. Ilari A. Giangiacomo L. Laue T. M. Chiancone E. Chasteen N. D. (2002). Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277, 27689–27696. 10.1074/jbc.M202094200
340
Zhu Y. Nandakumar R. Sadykov M. R. Madayiputhiya N. Luong T. T. Gaupp R. Lee C. Y. Somerville G. A. (2011). RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 193, 6187–6196. 10.1128/JB.05930-11
Summary
Keywords
Staphylococcus , oxidative stress
Citation
Gaupp R, Ledala N and Somerville GA (2012) Staphylococcal response to oxidative stress. Front. Cell. Inf. Microbio. 2:33. doi: 10.3389/fcimb.2012.00033
Received
05 January 2012
Accepted
29 February 2012
Published
16 March 2012
Volume
2 - 2012
Edited by
David Heinrichs, University of Western Ontario, Canada
Reviewed by
Michael L. Vasil, University of Colorado Medical School, USA; Anthony Richardson, University of North Carolina at Chapel Hill, USA
Copyright
© 2012 Gaupp, Ledala and Somerville.
This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Greg A. Somerville, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, VBS, Fair St. and East Campus Loop, Lincoln, NE, 68583-0905, USA. e-mail: gsomerville3@unl.edu
†These authors contributed equally to this work.
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.