- 1Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hong Kong, China
- 2Food Safety and Technology Research Center, Hong Kong PolyU Shen Zhen Research Institute, Shenzhen, China
- 3State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention (ICDC), Chinese Center for Disease Control and Prevention, Beijing, China
Vibrio parahaemolyticus is a clinically important foodborne pathogen that causes acute gastroenteritis worldwide. It has been shown that horizontal gene transfer (HGT) contributes significantly to virulence development of V. parahaemolyticus. In this study, we identified a novel znuA homolog (vpa1307) that belongs to a novel subfamily of ZnuA, a bacterial zinc transporter. The vpa1307 gene is located upstream of the V. parahaemolyticus pathogenicity island (Vp-PAIs) in both tdh-positive and trh-positive V. parahaemolyticus strains. Phylogenetic analysis revealed the exogenous origin of vpa1307 with 40% of V. parahaemolyticus clinical isolates possessing this gene. The expression of vpa1307 gene in V. parahaemolyticus clinical strain VP3218 is induced under zinc limitation condition. Gene deletion and complementation assays confirmed that vpa1307 contributes to the growth of VP3218 under zinc depletion condition and that conserved histidine residues of Vpa1307 contribute to its activity. Importantly, vpa1307 contributes to the cytotoxicity of VP3218 in HeLa cells and a certain degree of virulence in murine model. These results suggest that the horizontally acquired znuA subfamily gene, vpa1307, contributes to the fitness and virulence of Vibrio species.
Introduction
Vibrio parahaemolyticus is a motile and facultative anaerobe that frequently inhabits in estuarine and marine environments. It is a well-known pathogen that leads to acute gastroenteritis worldwide, especially in areas with high level consumption of seafood such as raw oysters and shrimps (Blake et al., 1980).
The pathogenicity of this pathogen is highly associated with the thermostable direct hemolysin (TDH), TDH-related hemolysin (TRH), and two type III secretion systems (T3SSs), T3SS1 and T3SS2 (Shirai et al., 1990; Hiyoshi et al., 2010). TDH is a pore-forming toxin that leads to the lysis of human erythrocytes, activation of calcium influx, disruption of cytoskeleton and epithelial barrier, while TRH has only been linked to hemolytic activity. T3SS1 is prevalent in all V. parahaemolyticus strains, whereas T3SS2 is more popular among clinical isolates (Meador et al., 2007). Comparative genomic analysis of pre-pandemic and pandemic V. parahaemolyticus strains, as well as molecular profiling studies, revealed that the organization of mobile gene cassettes and pathogenicity islands were divergent in these V. parahaemolyticus strains and that the emergence of pandemic strain could be associated with the recombination events and novel gene acquisition (Hurley et al., 2006; Gonzalez-Escalona et al., 2008; Han et al., 2008; Caburlotto et al., 2011; Chen et al., 2011; Yan et al., 2011; Garcia et al., 2012; Gennari et al., 2012; Gavilan et al., 2013; Ottaviani et al., 2013; Theethakaew et al., 2013; Turner et al., 2013). The genetic divergence of V. parahaemolyticus strongly correlates with their diverse virulence potentials (Vongxay et al., 2008; Caburlotto et al., 2010).
T3SS2 is located within the pathogenicity island (Vp-PAI), implying the acquisition of T3SS2 via horizontal gene transfer (HGT) (Okada et al., 2009). Recent works also support that T3SS2 is not only transferable among V. parahaemolyticus strains but also among other Vibrio spp. (Dziejman et al., 2005; Caburlotto et al., 2009; Okada et al., 2010). Functional analysis of genes encoded in the T3SS2 gene cluster revealed that T3SS2 in V. parahaemolyticus contributes to its colonization and competition to protists in aquatic environment (Matz et al., 2011; Ritchie et al., 2012). Apart from the genes of T3SS2, other horizontally acquired genes can also contribute to the virulence of V. parahaemolyticus. VpaH in V. parahaemolyticus TH3996, an acquired gene through HGT, can significantly enhance its motility, biofilm formation and adherence (Park et al., 2005). This observation has prompted the need to characterize novel horizontally acquired virulence factors in V. parahaemolyticus.
Zinc is an important component for many bacterial metalloenzymes. Intracellular pathogens require zinc for invasion, survival, and replication in the host. However, zinc concentration in the host is very low, therefore, it is essential for pathogenic bacteria to take up zinc effectively in the host (Hantke, 2005). Most pathogenic bacteria take up zinc using single or multiple zinc transportation systems. ZnuACB is the most important high-affinity zinc acquisition system in many bacteria, where ZnuA is responsible for zinc binding, ZnuB is for transportation of zinc across the inner membrane, and ZnuC works as an ATPase providing energy for zinc intake process (Hantke, 2005). ZnuACB was shown to be essential for virulence of Brucella abortus, Campylobacter jejuni, Moraxella catarrhalis, Salmonella enterica, and Haemophilus ducreyi (Lewis et al., 1999; Yang et al., 2006; Ammendola et al., 2007; Davis et al., 2009; Murphy et al., 2013), but not for uropathogenic Escherichia coli, Proteus mirabilis, or Yersinia pestis (Sabri et al., 2009; Desrosiers et al., 2010; Nielubowicz et al., 2010). This is due to the possessing of other zinc uptake genes, such as zupT, encoding a low-affinity zinc acquisition protein in E. coli (Sabri et al., 2009). In V. parahaemolyticus, the mechanisms of zinc transportation and its contribution to the pathogenesis of V. parahaemolyticus are not well-defined.
In this study, we identified a unique znuA homolog (vpa1307) that represents a novel subfamily of ZnuA in V. parahaemolyticus. Vpa1307 is localized upstream of Vp-PAIs in both tdh-positive V. parahaemolyticus RIMD2210633 and trh-positive V. parahaemolyticus TH3996 strains. Phylogenetic analysis suggested that vpa1307 is acquired by V. parahaemolyticus through HGT. The role of Vpa1307 as ZnuA was confirmed and its contribution to the pathogenesis of V. parahaemolyticus was determined.
Materials and Methods
Bioinformatics Analysis
Multiple sequence alignments were performed by the use of Clustal W2. Three-dimensional (3D) structure was predicated and modeled using Swiss-model. Structural alignment was generated using TM-align servers from Zhang's lab at University of Michigan. Phylogenetic analysis was performed using MEGA version 5 after multiple alignment of the data via CLUSTAL_X. Distances were obtained using options according to Kimura's two-parameter model and clustering was performed by using the neighbor-joining method. The topology of the neighbor-joining phylogenetic tree was evaluated by using bootstrap resampling with 1000 replications.
Bacterial Strains, Plasmid, and Growth Conditions
Plasmids pDM4 and pMMB207 were used for gene deletion and complementation experiments, respectively. E. coli SY327 λpir was used for conjugation (Miller and Mekalanos, 1988). Clinical V. parahaemolyticus strains were obtained from hospitals in Hong Kong. Other strains and plasmids used in this study were listed in Table 1. V. parahaemolyticus was cultured in LB medium supplemented with 2.5% sodium chloride (NaCl) at 37°C. Chloramphenicol (25 μg/ml for E. coli and 5 μg/ml for V. parahaemolyticus), kanamycin (50 μg/ml for E. coli), and 1 mM Isopropyl β-D-1-Thiogalactopyranoside (IPTG) were supplied if necessary. Zinc depletion was carried out using specific zinc chelator, N,N,N′,N′-Tetrakis (2-pyridylmethyl) ethylenediamine (TPEN, Sigma) dissolved in ethanol.
Construction of Deletion and Complementary Strains
The vpa1307 gene was deleted from V. parahaemolyticus strain VP3218 by homologous recombination using the methods described previously (Liverman et al., 2007; Zhou et al., 2010). Briefly, the upstream and downstream sequences of vpa1307 gene were amplified using primers vpa1307-1F/vpa1307-1R and vpa1307-2F/vpa1307-2R, respectively (Table 2). These two fragments were used as templates for the second round of PCR using primers vpa1307-1F/vpa1307-2R. The purified overlapping PCR product was digested and cloned into the same digested suicide vector, pDM4. E. coli SY327 λ pir carrying the recombinant plasmid, the helper plasmid pPK2013, and V. parahaemolyticus strain VP3218 were mixed (5:5:1, v/v/v), spun down and resuspended in 100 μ l LB broth, poured onto a filter on LB agar plate, and incubated overnight. The bacteria on the filter were resuspended, spread on Thiosulfate-citrate-bile salts-sucrose agar (TCBS) containing 5 μg/ml chloramphenicol to select transconjugants. Randomly selected transconjugants were cultured on LB agar in the presence of 5% sucrose and subjected to repeated serial passages. The knockout mutant, Δ vpa1307 was obtained.
To construct the complementary strain, the vpa1307 gene with additional ribosome-binding site was amplified using primers vpa1307com-F and vpa1307com-R (Zhou et al., 2010) (Table 2). PCR product was digested and cloned into the same digested pMMB207 to create pMMB207:vpa1307. This recombinant plasmid was transformed into E. coli SY327 λ pir and then conjugated into Δvpa1307 with the presence of helper plasmid pPK2013 carrying E. coli SY327 λ pir. Transconjugants were selected on TCBS containing 5 μg/ml chloramphenicol and the strain Δ vpa1307::pvpa1307 was obtained.
Site-directed mutagenesis was generated using GENEART® Site-Directed Mutagenesis kit (Invitrogen Co., NY, USA) with primer pairs H69A-F/H69A-R and H148A-F/H148A-R, (Table 2). Plasmid pMMB207:vpa1307 was used as template. Successful mutations were confirmed by sequencing. Δ vpa1307::pvpa1307 (H69A, H148A) was obtained by the use of the method described above.
RT-PCR, PCR, and Growth Assay
Thirty-five micrometers TPEN was added to wild type (WT) log-phase V. parahaemolyticus culture. After induction for 30 min, 1 ml culture was collected and used to extract RNA using Trizol (Invitrogen) following the manufacturer's instructions. DNA was removed from the sample with DNase (Turbo DNase, Ambion) according to the manufacturer's instructions. 0.5 μg RNA was used as template using Superscript one-step RT-PCR system (Invitrogen). No TPEN culture was used as negative control. Primers rtvpa1307-F/rtvpa1307-R and rt16S-F/rt16S-R were used, respectively (Table 2). Primers vpa1307-F and vpa1307-R (Table 2) were used for screening the distribution of vpa1307 in V. parahaemolyticus clinical isolates by PCR approach. The tdh and trh genes were also screened by PCR.
For growth assay, overnight V. parahaemolyticus culture was diluted in LB broth and grown to the exponential growth phase (OD600 ≈ 0.6–0.7). The cells were diluted 1:100 into fresh LB broth with or without 35 μ M TPEN, respectively and grown at 37°C with shaking (250 rpm). OD600 was monitored at specific time points. A similar procedure was used in relative growth assay, except that OD600 was only monitored at 6 h. 1 mM IPTG plus 5μg/ml chloramphenicol was added when culturing the complementary strains. Relative growth rate was calculated as culture grown with TPEN to that grown without 35 μ M TPEN.
Cytotoxicity Assay
HeLa cells were washed five times with PBS to completely wash the serum-off before bacteria was added and incubated in DMEM (without serum and antibiotics). Overnight V. parahaemolyticus strains were diluted 100-fold using fresh LB broth and grown at 37°C for 4 h. Cultures were then collected, washed, resuspended in DMEM (without serum) and used to infect HeLa cells at a multiplicity of infection (MOI) of ~50 cfu per cell. Supernatants were collected at specific time points and the amounts of LDH released were determined using CytoTox 96 Non-Radioactive Cytotoxicity kit (Promega) following the manufacturer's instructions. Percentage of cytotoxicity was calculated using formula: (test LDH release—spontaneous release)/maximal release. Test LDH release represents the LDH release after infection with different V. parahaemolyticus strains; spontaneous release represents the baseline cell LDH release without infecting with any bacteria, whereas maximal release represents the release of LDH when cells were lysed using lysis solution from the kit.
Murine Infection Assay
V. parahaemolyticus strains (108 CFU) were intraperitoneally injected into 6- to 10-week-old C57BL/6 mice (n = 10) as previously described (Hiyoshi et al., 2010; Pineyro et al., 2010; Whitaker et al., 2012) and mice that were alive were measured at the indicated time points. Three independent replicated experiments were performed. The animal experiments were conducted in the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention (CDC) following the guidelines and policies approved by the Chinese CDC.
Results
Bioinformatics Analysis of the vpa1307 Gene and its Distribution in V. parahaemolyticus
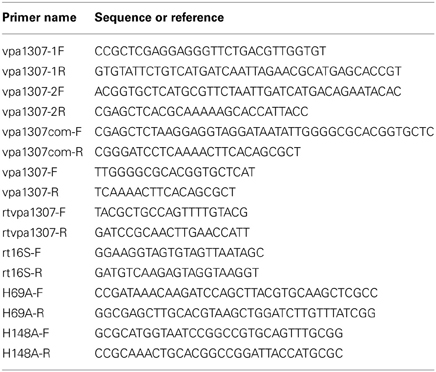
Since PAI is important for the virulence, we focused on genes related to Vp-PAI (Dobrindt et al., 2004). After a close examination of the Vp-PAI region from tdh-positive V. parahaemolyticus RIMD2210633, we found a hypothetical gene, vpa1307 that is localized upstream of the Vp-PAI. A similar vpa1307 was also identified upstream of Vp-PAI of a trh-positive V. parahaemolyticus TH3996 (Figure 1).
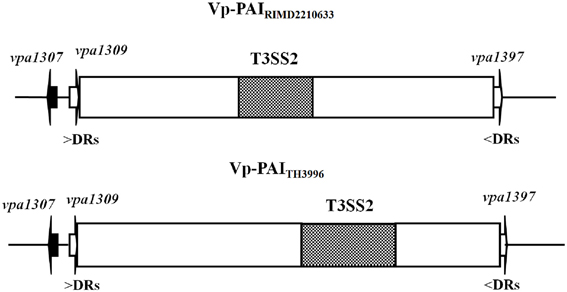
Figure 1. Schematic of vpa1307 gene location in V. parahaemolyticus RIMD2210633 and TH3996. Vp-PAI (open box) is flanked by direct repeats (DRs, 5′-AACTC-3′). The white arrows (vpa1309 and vpa1397) indicate the first and last genes outside the VP-PAI. The vpa1307 gene and T3SS2 gene cluster are also indicated.
BLAST analysis showed that Vpa1307 shares 23% amino acid sequence identity to the zinc binding protein from V. cholerae O1 biovar EI Tor strain N16961. In addition, Vpa1307 possesses three conserved histidine residues, H69, H148, and H202 that are the hallmark of ZnuA family of proteins (Figure 2). It was shown that residues H69, H148, and H202 are critical for zinc binding and activity (Banerjee et al., 2003; Li and Jogl, 2007; Loisel et al., 2008; Yatsunyk et al., 2008; Ilari et al., 2011). The 3D structure of Vpa1307 was modeled and aligned with the crystal structure of ZnuA from Synechocystis sp. PCC 6803, even through the similarity between these two proteins is only 24%. The TM score, an algorithm to calculate the structural similarity of two protein models, is 0.97, which strongly suggests that Vpa1307 is likely to be a member of ZnuA family (Figure 3).

Figure 2. Multiple sequence alignment of Vpa1307 and ZnuA proteins. The amino acid sequences (GenBank accession No.CAB72627, P73085, AAC74927, Q8Z5W7, AAC45725, and Vpa1307 from C. jejuni, Synechocystis sp., E. coli, S. enterica, Treponema pallidum, and V. parahaemolyticus, respectively) were aligned using the CLUSTAL W2. Three conserved histidine residues were indicated by black arrows.
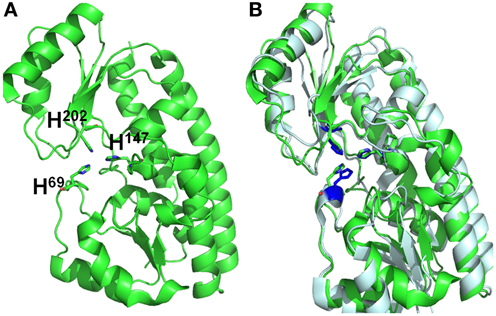
Figure 3. Structural alignments between VPA1307 and ZnuA from Synechocystis sp. (A) VPA1307 modeled structure. Structure of VPA1307 was modeled using SWISS-MODEL program and three conserved histidine residues were labeled. (B) Structural comparison of VPA1307 (green) and the crystal structure of ZnuA from Synechocystis sp. (PDB accession number 1PQ4) template (gray). A TM score of 0.97 was obtained over 252 aligned residues.
To explore the evolution history of Vpa1307, a phylogenetic tree was constructed (Figure 4). The neighbor-joining phylogenetic tree showed that Vpa1307 together with its four homologs from other Vibrio spp. fell within the lineage of ZnuA family and formed a distinct cluster within members of ZnuA from other genera. Intriguingly, the phylogenetic analysis also showed that Vpa1307 was excluded from the Vibrionaceae clade of ZnuA, suggesting an exogenous origin of Vpa1307 and representing a novel subfamily of ZnuA. The data suggested that vpa1307 is very likely acquired by V. parahaemolyticus through HGT.
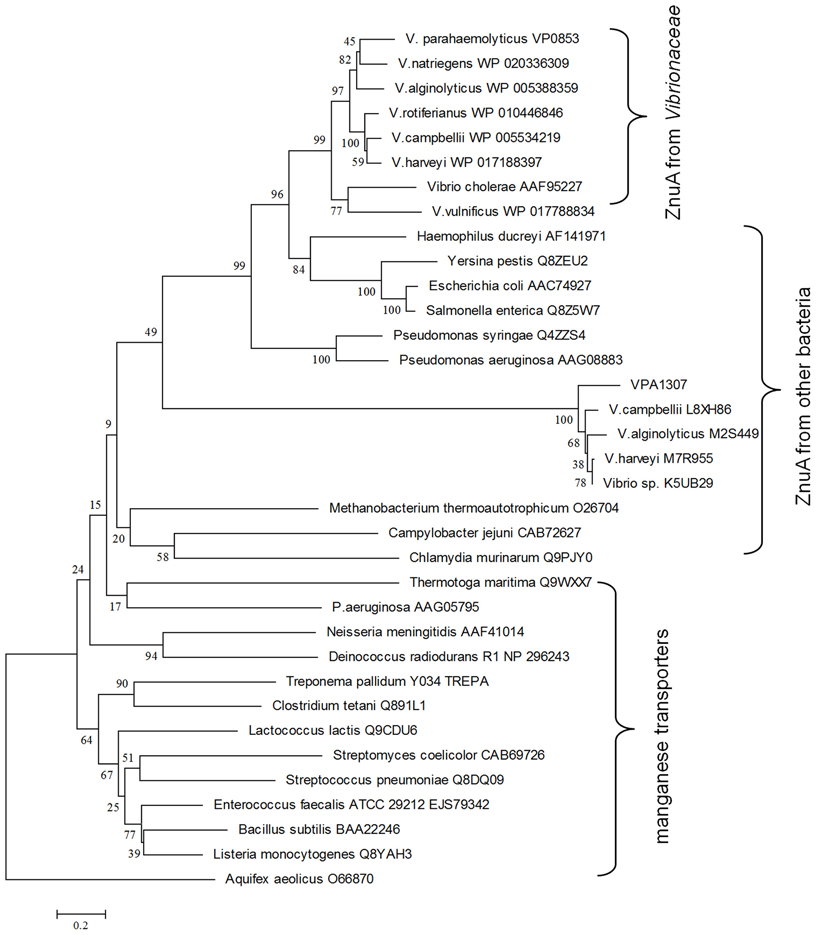
Figure 4. Neighbor-joining tree of VPA1307 and related genes. The protein sequences were obtained from NCBI except that L8XH86, M2S449, M7R955, and K5UB29 were obtained from EBI. ZnuA homologs were represented in red (except that of Aquifex aeolicus) and manganese transporters were represented in black. Bootstrap values (>50%) are shown at branch nodes. ZnuA homolog protein sequence from Aquifex aeolicus was used as outgroup. Bar, 0.2 difference at the amino acid level.
Prevalence of vpa1307 in V. parahaemolyticus Clinical Isolates
Given that vpa1307 group genes were exogenously acquired by Vibrio spp., the prevalence of this gene among V. parahaemolyticus strains was evaluated. Our data showed that the vpa1307 gene was detectable in 8 out of 20 (40%) of the tdh-positive strains but not in tdh- and trh-negative strains, suggesting the exogenous origin of vpa1307.
Role of Vpa1307 on V. parahaemolyticus Growth
To test the contribution of Vpa1307 to V. parahaemolyticus growth, both WT V. parahaemolyticus VP3218 clinical strains and the vpa1307 deletion mutant, Δ vpa1307, were grown in normal and zinc depletion conditions. All test strains showed similar growth rate in normal medium, while the growth of Δvpa1307 was inhibited by ~70% in the medium containing 35 μ M TPEN, a zinc chelating agent, compared to growth in normal medium. However, the growth of WT was only slightly inhibited in the medium containing 35 μ M TPEN (Figure 5). This indicated that vpa1307 contributes to the growth of V. parahaemolyticus under the zinc limitation condition. The data prompt us to examine the expression status of vpa1307 in V. parahaemolyticus. It was shown that vpa1307 only expressed under zinc depletion condition (35 μ M TPEN added) (Figure 6), but not in normal medium. The expression regulation feature of vpa1307 was consistent with the contribution of vpa1307 to V. parahaemolyticus growth under zinc depletion condition.
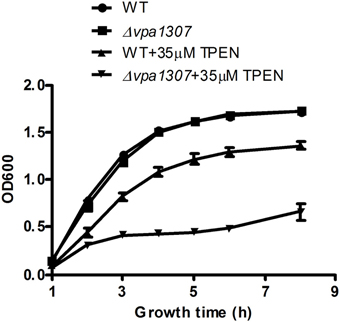
Figure 5. Growth rates of V. parahaemolyticus strains. V. parahaemolyticus strains were cultured in LB or LB supplemented with 35 μ M TPEN. V. parahaemolyticus growth (OD600) was monitored. The data represents three independent experiments ± the SD.
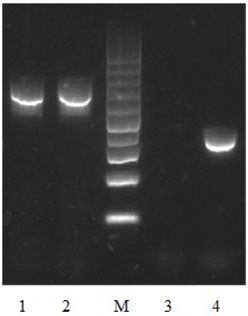
Figure 6. Detection of vpa1307 transcript by RT-PCR in V. parahaemolyticus. V. parahaemolyticus was grown in LB and RNA was extracted after incubation with/without 35 μ M TPEN. 16S RNA was used as a loading control. Lane 1, 16S RNA (with TPEN); lane 2, 16S RNA (without TPEN); lane 3, vpa1307 (without TPEN); lane 4, vpa1307 (with TPEN); lane M, 100-bp maker (Thermo Scientific).
Conserved Histidine Residues of vpa1307 Contribute to its Activity
To further confirm that Vpa1307 is a homolog of ZnuA, the contribution of three conserved histidine residues to the activity of Vpa1307 was tested. An elegant design helps us to achieve this goal. First, a complementary construct was made by incorporating a vpa1307 into plasmid pMMB207 and designated as pvpa1307. Second, two histidine residues, H69 and H148 that may be essential for Vpa1307 function, were mutated to alanine to obtain pvpa1307 (H69A, H148A). Mutations of two of the three histidines are enough to inactivate a ZnuA function (Li and Jogl, 2007; Loisel et al., 2008; Ilari et al., 2011). These two constructs were then used to complement the loss of function by vpa1307 deletion mutant, Δvpa1307. As showed in Figure 7, compared to WT V. parahaemolyticus VP3218, vpa1307 deletion mutant, Δ vpa1307, showed about 25% of growth rate. When complemented with vap1307, Δ vpa1307::pvpa1307, the growth rate of V. parahaemolyticus increased to ~75% compared to the WT strain. However, when complemented with the vpa1307 double histidine mutant Δ vpa1307::pvpa1307 (H69A, H148A), the growth rate of V. parahaemolyticus remained at the same level as Δ vpa1307 (25%) suggesting that the mutations, H69A, H148A, completely abolished the activity of Vpa1307. Furthermore, RT-PCR assay has confirmed that the expression level of vpa1307 in WT V. parahemolyticus VP3218 strain, Δ vpa1307::pvpa1307, and Δ vpa1307::pvpa1307 (H69A, H148A) were similar suggesting that the loss of function of vpa1307 (H69A, H148A) was due to the mutation of conserved histidine residues (data not shown). These data further confirmed that Vpa1307 is a member of ZnuA.
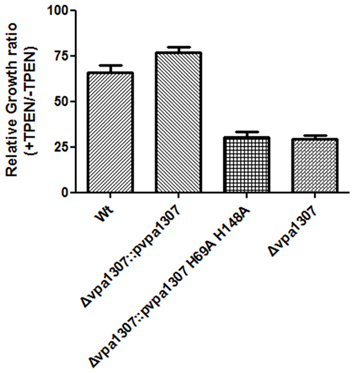
Figure 7. Relative growth rate of different V. parahaemolyticus strains under normal and zinc depletion conditions. Different V. parahaemolyticus strains as indicated were cultured in LB or LB supplemented with 35 μ M TPEN. Their growth (OD600) was monitored at 6 h and relative growth rates were calculated as culture grown with TPEN to that of grown without TPEN. The data represents three independent experiments ± the SD.
Vpa1307 Contributes to Cytotoxicity of v. parahaemolyticus in Hela Cells
Since ZnuA contributed to host cell infection in B. abortus, M. catarrhalis, and S. enterica (Yang et al., 2006; Ammendola et al., 2007; Murphy et al., 2013), we further tested whether vpa1307 gene contributed to the virulence of V. parahaemolyticus. HeLa cells that were maintained in serum-free DMEM were infected with Δ vpa1307, Δ vpa1307::pvpa1307 and WT strains, respectively. Similar to no infection cell control, strain Δ vpa1307 did not cause notable cell rounding and detachment, while strain Δ vpa1307::pvpa1307 showed more cell rounding than infected with WT strain of V. parahaemolyticus and all cells were detached after longer incubation (Data not shown). The cytotoxicity of V. parahaemolyticus was also determined by measuring the amount of LDH released from damaged cells. WT V. parahaemolyticus caused about 70% of LDH release, whereas Δ vpa1307 strain caused ~20% of LDH release after 4 h infection (Figure 8). The complementation of vpa1307, Δ vpa1307::pvpa1307, regained its toxicity back to 60% of LDH release, a level of toxicity similar to that of WT strain. These data indicated that Vpa1307 contributes to the cytotoxicity of V. parahaemolyticus strain VP3218 in HeLa cells. It also suggested that acquisition of zinc from cells is required for the successful infection of V. parahaemolyticus.
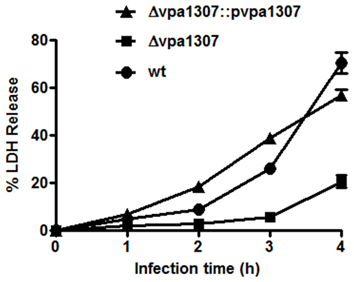
Figure 8. Cytotoxic effect of vpa1037 on HeLa cells. HeLa cells were infected with V. parahaemolyticus at MOI of ~50 cfu per cell. Supernatants were collected at specific time points and the amounts of LDH released were determined using CytoTox 96 Non-Radioactive Cytotoxicity kit (Promega) following the manufacturer's instructions. Percentage of cytotoxicity was calculated using formula: (test LDH release—spontaneous release)/maximal release. Test LDH release represents the LDH release after infection with different V. parahaemolyticus strains; spontaneous release represents the baseline cell LDH release without infecting with any bacteria, whereas maximal release represents the release of LDH when cells were lysed using lysis solution from the kit. The data represents three independent experiments ± the sem.
Vpa1307 Contributes to the Pathogenesis of V. parahaemolyticus
To further evaluate the role of Vpa1307 in the pathogenesis of V. parahaemolyticus, mouse infection model was employed. As shown in Figure 9, during the early infection period (within 6h), both WT V. parahaemolyticus and vpa1307 deletion mutant, Δ vpa1307, showed similar effect on mice and caused about 10% death. However, the mortality of Δ vpa1307 was dramatically delayed and attenuated when compared with WT strain at the later infection period. At post 24 h of infection, mortality rate of WT stain reached 80%, while that of Δ vpa1307 was about 30~50%. This indicated that the Vpa1307 contributes to the pathogenesis of V. parahaemolyticus at certain extent.
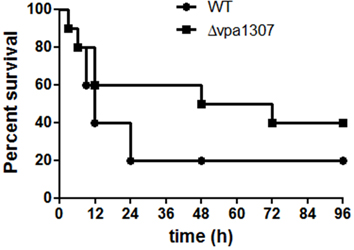
Figure 9. Survival rates of murine model infected with different V. parahaemolyticus strains. C57BL/6 mice (n = 10) were infected intraperitoneally with WT or Δ vpa1307 strains (108 CFU) and mice were monitored for the duration of 96 h. The mortality rate was measured at different time points (0, 3, 6, 9, 12, 24, 48, 72, and 96 h) for three independent experiments. Each data point in the figure represents the average of the data from three experiments. Kaplan–Meier and log rank tests were used to analyze the data.
Discussion
A pathogenic bacterial species is usually a group of diverse strains that inhabit in different environments. These strains usually display different infection abilities, which are highly correlated with their variation in the genomes. The evolutionary forces for the genomic flexibility involve gene gain, gene loss, gene duplication as well as mutations. It is known that HGT greatly affects the virulence of bacterium. Comparative genomic analysis of pre-pandemic and pandemic V. parahaemolyticus strains as well as molecular profiling studies revealed that the organization of mobile gene cassettes and PAIs were divergent in V. parahaemolyticus strains. The genetic divergence of this bacterium suggests that it evolves quickly in response to different pressures in the aquatic environment (Han et al., 2008; Chen et al., 2011; Gavilan et al., 2013), which results in a diverse virulence potential(Caburlotto et al., 2010). The genomic flexibility greatly affects the fitness to the hosts and the virulence potential of the pathogen. In this study, we identified and characterized a novel znuA homolog gene, vpa1307 in V. parahaemolyticus. Vpa1307 is localized upstream of Vp-PAI, and was annotated as an adhesion protein in strain RIMD2210633 (Makino et al., 2003). Our study revealed that vpa1307 is a zinc transporter from a novel group of ZnuA family. Most interestingly, vpa1307 was acquired by V. parahaemolyticus through HGT.
The exogenous origin of vpa1307 was confirmed by studying the prevalence of this gene in clinical strains; our results show that this gene is uniquely present in tdh-positive strains but not in tdh- and trh-negative strains. Given that tdh-positive V. parahaemolyticus strains are commonly associated with clinical infections, the close association of vpa1307 to tdh-positive strains may suggest that vpa1307 could be one of the virulence factors contributing to clinical infection. Similar to what was found for the znuA family genes; the expression of vpa1307 was induced in zinc limitation condition and contributed to V. parahaemolyticus growth under zinc starvation condition. Considering that zinc concentration is low in seawater (Bruland, 1989), the acquisition of vpa1307 gene may facilitate V. parahaemolyticus to persist in the marine environment.
It has been shown that when more than one zinc uptake systems exist in the pathogenic bacterium, such as in uropathogenic E. coli, P. mirabilis, Y. pestis, and Listeria monocytogenes, deletion of one of them did not affect their virulence in vivo (Sabri et al., 2009; Desrosiers et al., 2010; Nielubowicz et al., 2010; Corbett et al., 2012). Instead, the additional zinc acquiring systems contributed to the competitive advantage, such as in uropathogenic E. coli and P. mirabilis (Sabri et al., 2009; Nielubowicz et al., 2010). Given that many V. parahaemolyticus strains harbored more than one znuA homolog genes; it is not surprising to see that Vpa1307 only contributed partially to the pathogenesis of V. parahaemolyticus. It is interesting to see that Vpa1307 contributes to the cytotoxicity in HeLa cells and certain degree of pathogenesis in mice.
This is the first report of a functional exogenous znuA homolog acquired by V. parahaemolyticus via HGT. The gain of this gene might enhance the survival of Vibrio spp. in adverse condition. HGT has been shown to contribute to bacterial fitness in new environment and virulence of a pathogen. The reason that V. parahaemolyticus strains caused variable cytotoxicity is probably due to acquisition of novel virulence genes, such as vpa1307. Considering that chitin is abundant in the aquatic environments and that it has been shown to stimulate the process of natural competence and transformation (Meibom et al., 2005), it could be a great concern that V. parahaemolyticus may easily acquire other genes that can strengthen its pathogenicity or antibiotic resistance in the aquatic environments.
Funding
This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200) and the Research Fund for the Control of Infectious Diseases from the Food and Health Bureau, the Government of Hong Kong SAR (K-ZJG2 to Sheng Chen).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the critical reading of the manuscript by Dr. Edward Chan and useful discussion with members from Sheng's lab. We also thank Drs. Hans Wolf-Watz, Douglas R. Call and Kim Orth for kindly providing E. coli SY327 λpir strain and plasmids pDM4, pMMB207 and pPK2013 for gene knockout experiments.
References
Ammendola, S., Pasquali, P., Pistoia, C., Petrucci, P., Petrarca, P., Rotilio, G., et al. (2007). High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75, 5867–5876. doi: 10.1128/IAI.00559-07
Banerjee, S., Wei, B., Bhattacharyya-Pakrasi, M., Pakrasi, H. B., and Smith, T. J. (2003). Structural determinants of metal specificity in the zinc transport protein ZnuA from synechocystis (6803). J. Mol. Biol. 333, 1061–1069. doi: 10.1016/j.jmb.2003.09.008
Blake, P. A., Weaver, R. E., and Hollis, D. G. (1980). Diseases of humans (other than cholera) caused by vibrios. Annu. Rev. Microbiol. 34, 341–367. doi: 10.1146/annurev.mi.34.100180.002013
Bruland, W. K. (1989). Complexation of zinc by natural organic-ligands in the central north pacific. Limnol. Oceanogr. 34, 269–285. doi: 10.4319/lo.1989.34.2.0269
Caburlotto, G., Gennari, M., Ghidini, V., Tafi, M., and Lleo, M. M. (2009). Presence of T3SS2 and other virulence-related genes in tdh-negative Vibrio parahaemolyticus environmental strains isolated from marine samples in the area of the Venetian Lagoon, Italy. FEMS Microbiol. Ecol. 70, 506–514. doi: 10.1111/j.1574-6941.2009.00764.x
Caburlotto, G., Lleo, M. M., Gennari, M., Balboa, S., and Romalde, J. L. (2011). The use of multiple typing methods allows a more accurate molecular characterization of Vibrio parahaemolyticus strains isolated from the Italian Adriatic Sea. FEMS Microbiol. Ecol. 77, 611–622. doi: 10.1111/j.1574-6941.2011.01142.x
Caburlotto, G., Lleo, M. M., Hilton, T., Huq, A., Colwell, R. R., and Kaper, J. B. (2010). Effect on human cells of environmental Vibrio parahaemolyticus strains carrying type III secretion system 2. Infect. Immun. 78, 3280–3287. doi: 10.1128/IAI.00050-10
Chen, Y., Stine, O. C., Badger, J. H., Gil, A. I., Nair, G. B., Nishibuchi, M., et al. (2011). Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12:294. doi: 10.1186/1471-2164-12-294
Corbett, D., Wang, J., Schuler, S., Lopez-Castejon, G., Glenn, S., Brough, D., et al. (2012). Two zinc uptake systems contribute to the full virulence of Listeria monocytogenes during growth in vitro and in vivo. Infect. Immun. 80, 14–21. doi: 10.1128/IAI.05904-11
Davis, L. M., Kakuda, T., and DiRita, V. J. (2009). A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640. doi: 10.1128/JB.01394-08
Desrosiers, D. C., Bearden, S. W., Mier, I. Jr., Abney, J., Paulley, J. T., Fetherston, J. D., et al. (2010). Znu is the predominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect. Immun. 78, 5163–5177. doi: 10.1128/IAI.00732-10
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi: 10.1038/nrmicro884
Dziejman, M., Serruto, D., Tam, V. C., Sturtevant, D., Diraphat, P., Faruque, S. M., et al. (2005). Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 102, 3465–3470. doi: 10.1073/pnas.0409918102
Garcia, K., Gavilan, R. G., Hofle, M. G., Martinez-Urtaza, J., and Espejo, R. T. (2012). Microevolution of pandemic Vibrio parahaemolyticus assessed by the number of repeat units in short sequence tandem repeat regions. PLoS ONE 7:e30823. doi: 10.1371/journal.pone.0030823
Gavilan, R. G., Zamudio, M. L., and Martinez-Urtaza, J. (2013). Molecular epidemiology and genetic variation of pathogenic Vibrio parahaemolyticus in Peru. PLoS Negl. Trop. Dis. 7:e2210. doi: 10.1371/journal.pntd.0002210
Gennari, M., Ghidini, V., Caburlotto, G., and Lleo, M. M. (2012). Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol. Ecol. 82, 563–573. doi: 10.1111/j.1574-6941.2012.01427.x
Gonzalez-Escalona, N., Martinez-Urtaza, J., Romero, J., Espejo, R. T., Jaykus, L. A., and DePaola, A. (2008). Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190, 2831–2840. doi: 10.1128/JB.01808-07
Han, H., Wong, H. C., Kan, B., Guo, Z., Zeng, X., Yin, S., et al. (2008). Genome plasticity of Vibrio parahaemolyticus: microevolution of the ‘pandemic group.’ BMC Genomics 9:570. doi: 10.1186/1471-2164-9-570
Hantke, K. (2005). Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8, 196–202. doi: 10.1016/j.mib.2005.02.001
Hiyoshi, H., Kodama, T., Iida, T., and Honda, T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780. doi: 10.1128/IAI.01051-09
Hurley, C. C., Quirke, A., Reen, F. J., and Boyd, E. F. (2006). Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. doi: 10.1186/1471-2164-7-104
Ilari, A., Alaleona, F., Petrarca, P., Battistoni, A., and Chiancone, E. (2011). The X-ray structure of the zinc transporter ZnuA from Salmonella enterica discloses a unique triad of zinc-coordinating histidines. J. Mol. Biol. 409, 630–641. doi: 10.1016/j.jmb.2011.04.036
Lewis, D. A., Klesney-Tait, J., Lumbley, S. R., Ward, C. K., Latimer, J. L., Ison, C. A., et al. (1999). Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 67, 5060–5068.
Li, H., and Jogl, G. (2007). Crystal structure of the zinc-binding transport protein ZnuA from Escherichia coli reveals an unexpected variation in metal coordination. J. Mol. Biol. 368, 1358–1366. doi: 10.1016/j.jmb.2007.02.107
Liverman, A. D., Cheng, H. C., Trosky, J. E., Leung, D. W., Yarbrough, M. L., Burdette, D. L., et al. (2007). Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. U.S.A. 104, 17117–17122. doi: 10.1073/pnas.0703196104
Loisel, E., Jacquamet, L., Serre, L., Bauvois, C., Ferrer, J. L., Vernet, T., et al. (2008). AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis. J. Mol. Biol. 381, 594–606. doi: 10.1016/j.jmb.2008.05.068
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Matz, C., Nouri, B., McCarter, L., and Martinez-Urtaza, J. (2011). Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS ONE 6:e20275. doi: 10.1371/journal.pone.0020275
Meador, C. E., Parsons, M. M., Bopp, C. A., Gerner-Smidt, P., Painter, J. A., and Vora, G. J. (2007). Virulence gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 45, 1133–1139. doi: 10.1128/JCM.00042-07
Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C. Y., and Schoolnik, G. K. (2005). Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827. doi: 10.1126/science.1120096
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583.
Murphy, T. F., Brauer, A. L., Kirkham, C., Johnson, A., Koszelak-Rosenblum, M., and Malkowski, M. G. (2013). Role of the zinc uptake ABC transporter of Moraxella catarrhalis in persistence in the respiratory tract. Infect. Immun. 81, 3406–3413. doi: 10.1128/IAI.00589-13
Nielubowicz, G. R., Smith, S. N., and Mobley, H. L. (2010). Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infect. Immun. 78, 2823–2833. doi: 10.1128/IAI.01220-09
Okada, N., Iida, T., Park, K. S., Goto, N., Yasunaga, T., Hiyoshi, H., et al. (2009). Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77, 904–913. doi: 10.1128/IAI.01184-08
Okada, N., Matsuda, S., Matsuyama, J., Park, K. S., de los Reyes, C., Kogure, K., et al. (2010). Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 10:302. doi: 10.1186/1471-2180-10-302
Ottaviani, D., Leoni, F., Rocchegiani, E., Mioni, R., Costa, A., Virgilio, S., et al. (2013). An extensive investigation into the prevalence and the genetic and serological diversity of toxigenic Vibrio parahaemolyticus in Italian marine coastal waters. Environ. Microbiol. 15, 1377–1386. doi: 10.1111/j.1462-2920.2012.02839.x
Park, K. S., Arita, M., Iida, T., and Honda, T. (2005). vpaH, a gene encoding a novel histone-like nucleoid structure-like protein that was possibly horizontally acquired, regulates the biogenesis of lateral flagella in trh-positive Vibrio parahaemolyticus TH(3996). Infect. Immun. 73, 5754–5761. doi: 10.1128/IAI.73.9.5754-5761.2005
Pineyro, P., Zhou, X., Orfe, L. H., Friel, P. J., Lahmers, K., and Call, D. R. (2010). Development of two animal models to study the function of Vibrio parahaemolyticus type III secretion systems. Infect. Immun. 78, 4551–4559. doi: 10.1128/IAI.00461-10
Ritchie, J. M., Rui, H., Zhou, X., Iida, T., Kodoma, T., Ito, S., et al. (2012). Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. doi: 10.1371/journal.ppat.1002593
Sabri, M., Houle, S., and Dozois, C. M. (2009). Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect. Immun. 77, 1155–1164. doi: 10.1128/IAI.01082-08
Shirai, H., Ito, H., Hirayama, T., Nakamoto, Y., Nakabayashi, N., Kumagai, K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573.
Theethakaew, C., Feil, E. J., Castillo-Ramirez, S., Aanensen, D. M., Suthienkul, O., Neil, D. M., et al. (2013). Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier, and environmental sources in Thailand, determined by multilocus sequence analysis. Appl. Environ. Microbiol. 79, 2358–2370. doi: 10.1128/AEM.03067-12
Turner, J. W., Paranjpye, R. N., Landis, E. D., Biryukov, S. V., Gonzalez-Escalona, N., Nilsson, W. B., et al. (2013). Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS ONE 8:e55726. doi: 10.1371/journal.pone.0055726
Vongxay, K., Wang, S., Zhang, X., Wu, B., Hu, H., Pan, Z., et al. (2008). Pathogenetic characterization of Vibrio parahaemolyticus isolates from clinical and seafood sources. Int. J. Food Microbiol. 126, 71–75. doi: 10.1016/j.ijfoodmicro.2008.04.032
Whitaker, W. B., Parent, M. A., Boyd, A., Richards, G. P., and Boyd, E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80, 1834–1845. doi: 10.1128/IAI.06284-11
Yan, Y., Cui, Y., Han, H., Xiao, X., Wong, H. C., Tan, Y., et al. (2011). Extended MLST-based population genetics and phylogeny of Vibrio parahaemolyticus with high levels of recombination. Int. J. Food Microbiol. 145, 106–112. doi: 10.1016/j.ijfoodmicro.2010.11.038
Yang, X., Becker, T., Walters, N., and Pascual, D. W. (2006). Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect. Immun. 74, 3874–3879. doi: 10.1128/IAI.01957-05
Yatsunyk, L. A., Easton, J. A., Kim, L. R., Sugarbaker, S. A., Bennett, B., Breece, R. M., et al. (2008). Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 13, 271–288. doi: 10.1007/s00775-007-0320-0
Zhou, X., Konkel, M. E., and Call, D. R. (2010). Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 192, 3491–3502. doi: 10.1128/JB.01493-09
Keywords: Vibrio parahaemolyticus, horizontal gene transfer, znuA, fitness, virulence
Citation: Liu M, Yan M, Liu L and Chen S (2013) Characterization of a novel zinc transporter ZnuA acquired by Vibrio parahaemolyticus through horizontal gene transfer. Front. Cell. Infect. Microbiol. 3:61. doi: 10.3389/fcimb.2013.00061
Received: 10 September 2013; Paper pending published: 20 September 2013;
Accepted: 21 September 2013; Published online: 10 October 2013.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Beiyan Nan, University of California, Berkeley, USAQiaobin Xiao, University of Notre Dame, USA
Copyright © 2013 Liu, Yan, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Chen, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom. Kowloon, Hong Kong, China e-mail:c2hlbmcuY2hlbkBwb2x5dS5lZHUuaGs=
 Ming Liu1,2
Ming Liu1,2 Lizhang Liu
Lizhang Liu Sheng Chen
Sheng Chen