- 1College of Animal Science and Technology, Inner Mongolia Minzu University, Tongliao, China
- 2Tongliao City Animal Quarantine Technical Service Centre, Tongliao, China
- 3Tongliao Institute of Agriculture and Animal Husbandry Sciences, Tongliao, China
Babesiosis, a zoonotic parasitic disease caused by Babesia protozoa, poses significant infection risks across mammalian species. Clinical manifestations in vertebrate hosts range from spontaneous abortion to fatal outcomes, with immunocompromised individuals potentially transmitting the pathogen through blood products or transplanted organs, thereby amplifying epidemiological risks. Effective disease management carries substantial public health implications for livestock production, companion animal welfare, and food safety in endemic regions. In global endemic zones, conventional diagnostic approaches combine morphological identification of Babesia spp. with complementary serological assays. Contemporary molecular diagnostics, particularly nucleic acid amplification techniques, have emerged as valuable adjunctive tools. A critical challenge in veterinary practice involves persistent subclinical carriers among treated livestock populations, necessitating precise parasite speciation for effective transmission control. This review synthesizes recent advancements in babesiosis detection methodologies, with particular emphasis on their implementation in clinical microbiology laboratories. This article introduces the latest progress in Babesiosis detection technology and its application in clinical microbiology laboratories, to provide a theoretical and practical basis for the comprehensive prevention and control of Babesiosis.
1 Introduction
Babesiosis, caused by apicomplexan parasites of the genus Babesia, represents the second most prevalent blood-borne parasitic disease in vertebrates after trypanosomiasis (Penzhorn, 2006; Cook and Puri, 2024). Since Victor Babes’ seminal description of bovine babesiosis in Romania (1888), over 100 Babesia spp.have been taxonomically identified. The primary transmission vector remains ticks, with secondary routes including erythrocyte transfusion, transplacental transmission, and organ transplantation (Wudhikarn et al., 2011; Obeta et al., 2020; Handel et al., 2021; Kumar et al., 2021). Babesiosis is most prevalent in rodents, carnivores, and cattle, and it is considered relatively rare for the general public to be infected with babesiosis (Dvoraková and Dvorácková, 2007; Jerzak et al., 2023). Clinically significant species are categorized by erythrocytic trophozoite dimensions: Small-form Babesia (1.0-2.5 μm): Includes pathogenic species Babesia gibsoni (B. gibsoni) and Babesia microti (B.microti); Large-form Babesia (2.5-5.0 μm): Encompasses Babesia bovis (B.bovis), Babesia caballi (B.caballi), Babesia canis(B. canis) and Babesia bigemina (B.bigemina) (Panti-May and Rodríguez-Vivas, 2020). Epidemiologically critical zoonotic strains include B. microti, B. duncani, and B. divergens. Regional distribution patterns reveal: Pastoral areas: B.bovis, B. bigemina, and B. caballi; Companion animals: B. canis and B.gibsoni; Wild animals: B. microti. As obligate vertebrate pathogens, Babesia infections pose substantial challenges to both public health systems and veterinary management practices.
2 Epidemiologically significant babesiosis
Six Babesia spp. with confirmed zoonotic transmission include B.microti, B. duncani, B. divergens, B. motasi (KO-1 strain), B. crassa-like agent, and B. venatorum, alongside two genetically distinct pathogenic subtypes: B. divergens-like MO1 and B. microti-like protozoa (Kumar et al., 2021). Human babesiosis exhibits distinct geographic pathogen profiles: B. microti (asymptomatic to severe cases) and B. duncani (high-fatality infections) predominate in the Americas (Scott and Scott, 2018; Menis et al., 2021); B. divergens causes acute disease in splenectomized patients across Europe (Hunfeld et al., 2008); B. crassa-like and B.venatorum are hyperendemic in northeastern Asia (Jiang et al., 2015; Vannier and Krause, 2015; Jia et al., 2018; Chen et al., 2019); while Africa and Australia report indigenous strains (e.g., Egyptian variants) and imported pathogens (e.g., South African lineages) (Fang et al., 2015; Saleh et al., 2015; Kumar et al., 2021). Babesia spp. has a wide distribution and a complex life cycle. These apicomplexans maintain an obligate two-host lifecycle involving ixodid ticks (definitive hosts) and vertebrate reservoirs, with transmission dynamics diverging between morphological groups. Small Babesia spp. (B.microti clade) propagate through a rodent-tick-rodent cycle: larval ticks acquire parasites from infected rodents, transmit them during nymphal feeding, and reinfect new rodent hosts. Large Babesia spp. (B.divergens group) follow a ruminant-tick-ruminant cycle, where adult female ticks transmit parasites to grazing hosts, perpetuating through subsequent tick generations (Karbowiak et al., 2018). Sexual reproduction occurs exclusively within tick vectors, complemented by asexual replication in vertebrate hosts, as illustrated in Figure 1.
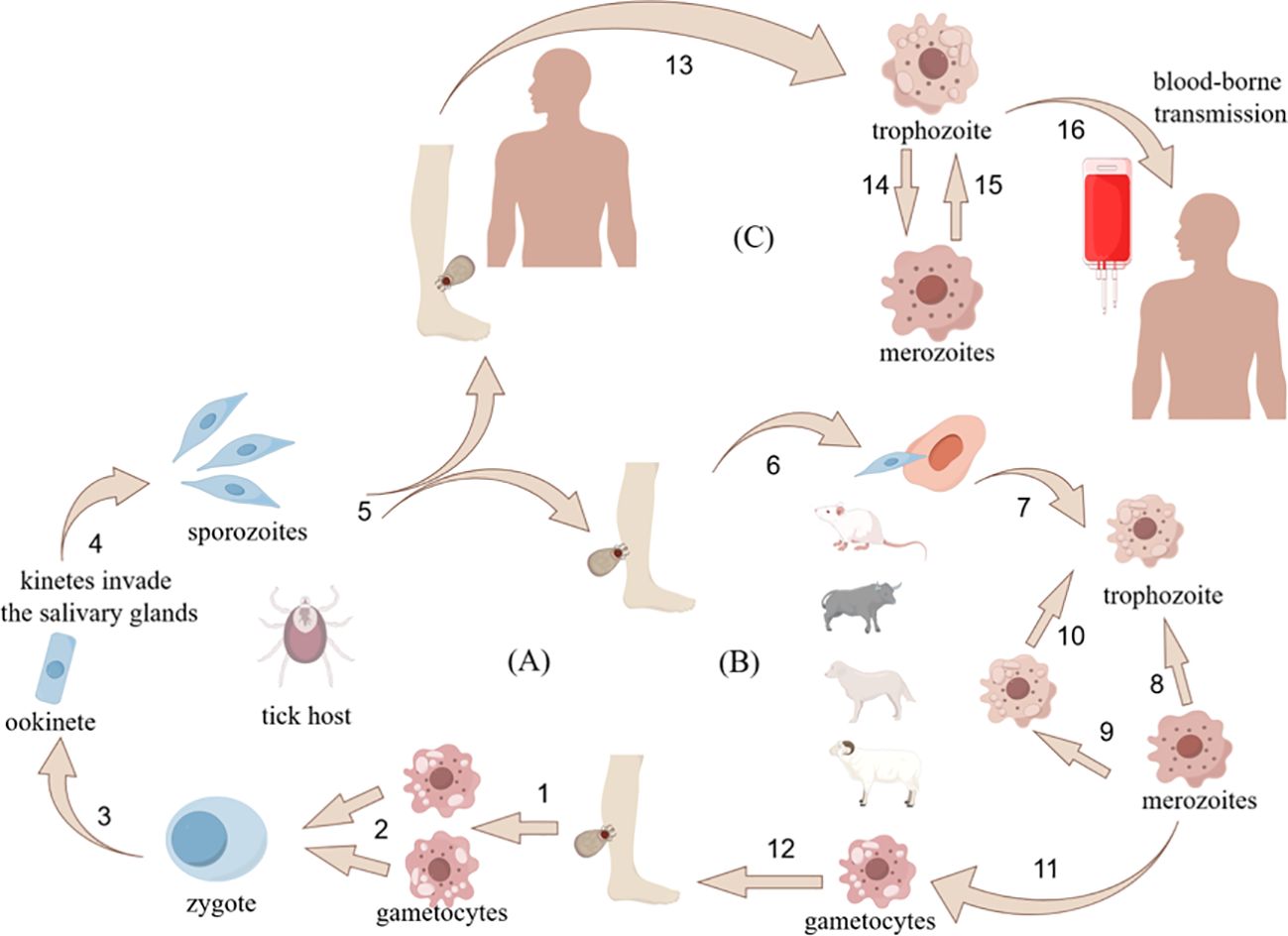
Figure 1. Detailed information on sexual and asexual reproduction of Babesia spp. Image created from the platform - Researchers' House license code exported ID:IWUAS50a2a.
3 Clinical manifestations and pathogenic mechanisms
After the invasion of host erythrocytes by Babesia, multiple systemic pathological reactions are triggered by destroying the morphology of erythrocytes and inhibiting the oxygen-carrying function and hematopoietic ability. Clinical manifestations range from characteristic presentations—including hemolytic anemia, icterus, pyrexia, and respiratory distress—to potentially fatal complications such as splenic rupture and infarction. Notably, emerging evidence documents atypical immunopathological sequelae encompassing acute phase responses and secondary hemophagocytic lymphohistiocytosis (sHLH) (Milanović et al., 2020; Mojtahed et al., 2020; Santos et al., 2020; Jacobs et al., 2025). Babesia infection elicits host-specific pathophysiological responses across mammalian species. Rodent models demonstrate B. microti-induced hematological alterations characterized by accelerated coagulation kinetics and thrombotic predisposition, with gestational infections provoking placental vascular pathology through collagen deposition and erythrocyte-endothelial adhesion (Jasik et al., 2024). Bovine infections by B. bovis or B. bigemina clinically manifest as cerebral hyperemia, inappetence, lactation suppression, and hemoglobinuria crisis (Sivakumar et al., 2018). Canine cases present complex systemic involvement beyond febrile episodes and anorexia, progressing to renal impairment, pancreatic inflammation, and acute respiratory failure (“pulmonary shock”), with persistent subclinical carriage observed post-treatment (Muench et al., 2023; Antognoni et al., 2025). immunocompromised hosts, particularly splenectomized individuals, exhibit exacerbated disease trajectories confirming immune status-dependent pathogenesis (Mojtahed et al., 2020; Torianyk, 2021b; Kakoullis et al., 2025). Various species of Babesia have been reported to further exacerbate the risk of multi-organ dysfunction by activating the endogenous coagulation system and inducing immunopathologic responses (Welzl et al., 2001; Okła et al., 2014; Preena et al., 2021).
4 Detection of important babesiosis
In addition to the internationally recognized gold standard microscopic examination, detection techniques for babesiosis also rely on molecular diagnosis, genotyping, and serological methods. While molecular techniques, particularly nucleic acid amplification testing (NAAT), enable precise mapping of Babesia spp. distribution, population genetics, and co-infection epidemiology, their field utility remains constrained by instrumentation dependency and technical complexity. Conversely, seroepidemiological profiling captures immune response patterns across endemic regions, offering critical insights into antigenic variation mechanisms, albeit challenged by cross-reactivity with related apicomplexans, false-negative results during seroconversion windows, and prolonged antibody persistence (>12 months post-clearance). According to the diagnostic recommendations summarized in the analysis of the IDSA Guidelines for the Diagnosis and Management of Babesiosis, confirmation of a diagnosis of babesiosis requires a combination of peripheral blood smear microscopy (the gold standard) and PCR testing, and antibody testing alone is not recommended because antibodies may persist for more than 1 year after clearance of the infection (Krause et al., 2021).
4.1 Blood smear microscopy and emerging binding techniques
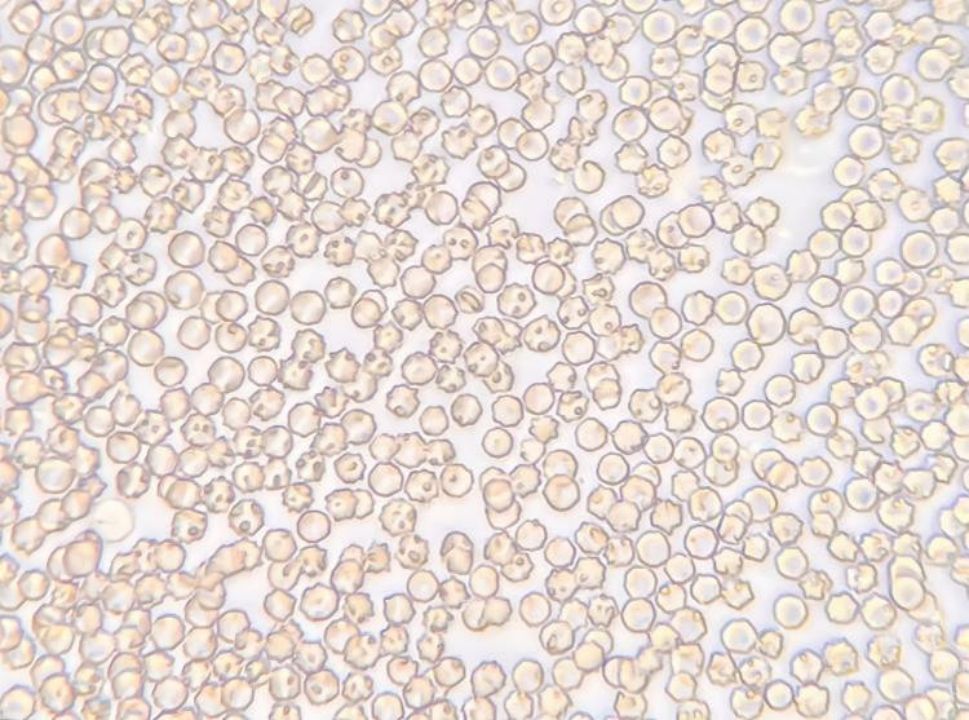
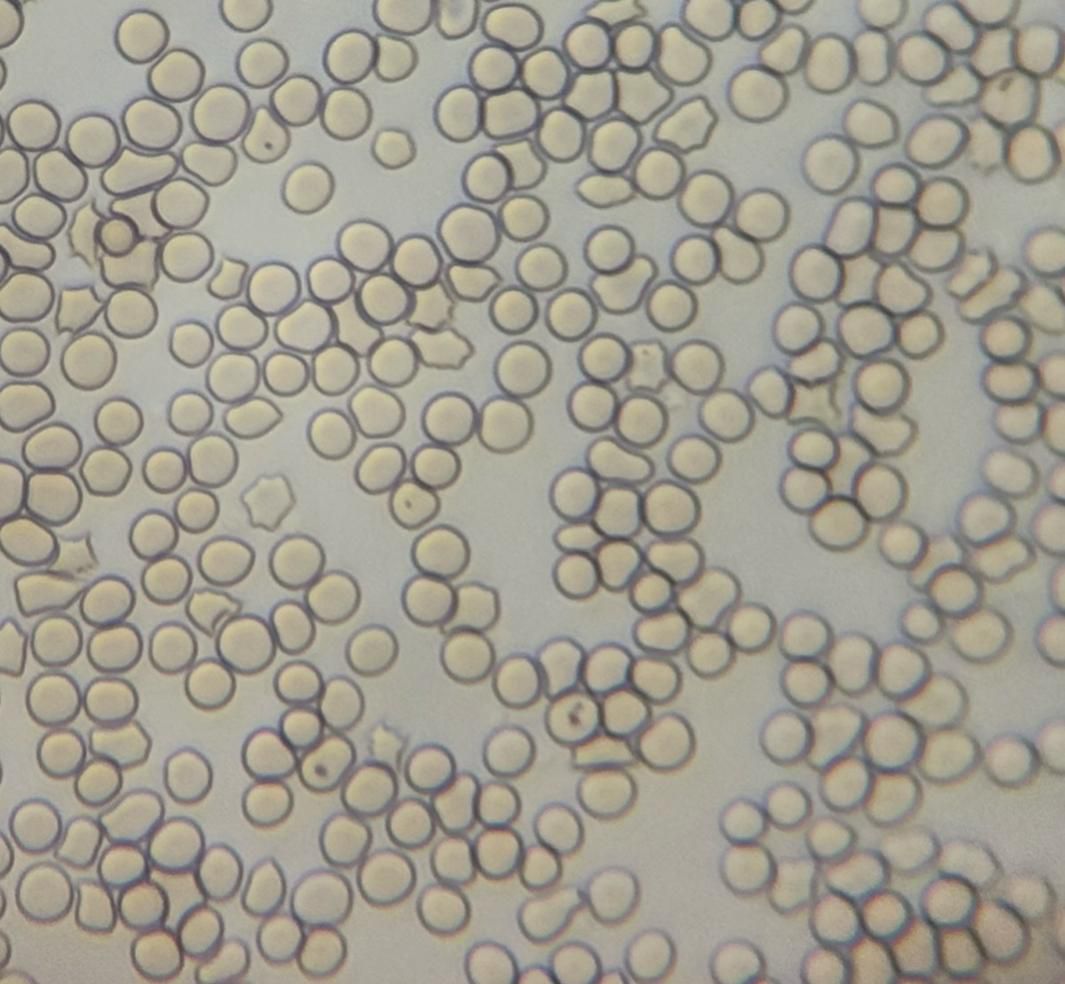
A common method of detecting Babesia is to collect blood from terminal capillaries such as ear tips and tail tips to prepare blood smears, which are observed using a light microscope and the morphology of the parasite’s body (length of cleistothecia, etc.) is measured (Figures 2, 3). In the acute infection phase, when the erythrocyte staining rate is high, only thin smears are required, conversely, thick smears prove essential during subclinical stages or post-acute phases when low parasitemia (<0.1%) challenges detection sensitivity (Alvarez et al., 2019; Dos Santos et al., 2021). While this method offers rapid field applicability, diagnostic accuracy suffers from morphological ambiguities between Babesia subspecies and Plasmodium spp., particularly in early infections lacking overt clinical manifestations. Notably, asymptomatic carriers often evade microscopic diagnosis despite harboring latent infections, with necropsy studies revealing striking tissue-specific parasitemia disparities—cerebral capillaries in B. bovis-infected hosts demonstrate 90-fold higher parasite burdens compared to peripheral blood (Everitt et al., 1986). One study successfully used cerebellar samples obtained from the foramen magnum of the occipital bone to achieve microscopic detection of persistently infected cattle (Hadani et al., 1982).
In recent years, recent advancements in diagnostic hematology integrate digital microscopy with artificial intelligence (AI) to revolutionize Babesia detection, exemplified by predictive models utilizing hematology analyzer parameters (ADVIA) for Babesia canis (B. canis) identification (Pijnacker et al., 2022). One study used a dual-model comparison strategy, i.e., constructing a conventional statistical model (CS) and a machine learning model (ML) based on data from infected cases diagnosed by blood smear or PCR, respectively. The results showed platelet count (PLT), mean platelet volume (MPV), and percentage of unstained cells (LUC%) as key predictors. The CS model achieved 84.6% sensitivity and 97.7% specificity (LR+ 36.78), while the ML model optimized via tree-based algorithms demonstrated perfect sensitivity (100%) with 95.7% specificity (LR+ 23.2). Algorithm-triggered alerts increase microscopic detection probability 37-fold versus routine screening, demonstrating scalable laboratory implementation. Complementing these developments, AI-augmented digital microscopy (IAAI) enables automated parasite speciation and parasitemia quantification, significantly reducing false-negative rates through computational erythrocyte analysis. This synergistic diagnostic ecosystem combines ML-driven prescreening with IAAI confirmation, leveraging routine hematological data for early infection alerts—particularly valuable during low parasitemia phases or microscopy oversights. While ML models enhance sensitivity through nonlinear multiparameter analysis, CS frameworks provide high-specificity decision support, collectively optimizing diagnostic workflows from initial suspicion to pathogen characterization (Durant et al., 2021).
4.2 Experimental inoculation and parasite culture
Experimental inoculation and heterologous diagnostic approaches have historically served as fundamental methodologies in Babesia research, tracing back to Victor Babes’ seminal application of Koch’s postulates through disease recapitulation in inoculated rabbits. Contemporary models employ species-specific vertebrate systems to mirror natural infection dynamics: B. microti pathogenesis is frequently studied in vole (Microtus) reservoirs, while B. bovis and B. bigemina isolation protocols typically utilize splenectomized bovine hosts via infected blood transfusion. The B. divergens-gerbil (Meriones) model has proven particularly valuable for investigating human-relevant pathogenesis. These controlled inoculation systems not only fulfill classical etiological criteria but also enable precise dissection of parasite-host interactions across zoonotic spectra (Krause et al., 1996; Gorenflot et al., 1998; Zintl et al., 2003; Holman et al., 2005). Experimental inoculation is an alternative diagnostic option for cases of hypoparasitemia, which occupied an important place in clinical practice until the advent of molecular biology tests (Krause et al., 1994). The Mongolian gerbil (Meriones unguiculatus) has emerged as a pivotal experimental model for Babesia divergens research, exhibiting acute and frequently lethal symptomatology distinct from bovine-hosted infections (Dkhil et al., 2014). Torianyk, Inna I et al. (Torianyk, 2021a) recent methodological innovations employ dual-host systems—combining Syrian hamsters (Mesocricetus auratus), gerbils, and murine models—to establish in vivo cultivation platforms through immune modulation. Host susceptibility stratification enables targeted propagation: hamsters maintain B. microti reservoirs while gerbils and mice sustain B. divergens cultures. Immunocompromised animal models coupled with multimodal diagnostics (clinical metrics: weight loss, motor dysfunction; laboratory confirmation: fluorescence microscopy/PCR) enhance Babesia detection sensitivity through standardized protocols, achieving parasitological confirmation at submicroscopic thresholds. This integrated approach not only facilitates pathogen isolation but also elucidates complication mechanisms, particularly erythrocyte adhesion-mediated vasculopathies and cytokine dysregulation. Transfecting Babesiosis into animals helps understand the immune mechanisms that clear or worsen the infection and the pathological processes leading to complications. While constrained by prolonged incubation periods (>7 days) and molecular confirmation requirements, these models provide unparalleled resolution for studying hypoparasitemic states and immune evasion strategies, establishing a critical methodological framework for both basic parasitology and translational therapeutic development.
The SCID murine model exhibits unique diagnostic utility in Babesia research through engineered erythrocyte replacement protocols. Immunocompromised mice receiving bovine red blood cell (BoRBC) transfusions alongside anti-murine erythrocyte monoclonal antibody treatment achieve rapid circulatory RBC substitution, creating a humanized hematological niche. Splenectomized SCID hosts inoculated with grazing calf-derived Babesia spp. develop marked parasitemia accompanied by pathognomonic manifestations including hemoglobinuria, hemolytic crisis, and neurological sequelae, enabling reliable pathogen isolation (Tsuji et al., 1995).
4.3 Immunological testing
Immunodiagnostic approaches constitute a critical pillar in comprehensive babesiosis management, enabling precise infection staging through serological antibody profiling, pathogen antigen detection, and immune complex characterization. Serological surveillance proves most effective during convalescent phases with established antibody responses, whereas antigenic assays are indispensable for early infection diagnosis before seroconversion or chronic carrier state identification. These immunological tools synergistically enhance blood product safety through donor screening protocols, significantly mitigating transfusion-mediated transmission risks. In clinical practice, integrated diagnostic algorithms combining antigen testing with PCR amplification demonstrate particular utility for confirming subclinical infections and hypoparasitemic cases, with subsequent microscopic verification ensuring diagnostic rigor. This multimodal strategy effectively bridges the diagnostic window period while addressing the inherent limitations of individual methodologies, thereby optimizing both epidemiological surveillance accuracy and therapeutic intervention timelines.
Pathogen-specific antigens serve as critical diagnostic biomarkers across infectious diseases, yet systematic antigen detection frameworks remain underdeveloped for babesiosis despite identification of multiple candidate targets. Take B. microti for example: Pioneering work by Lodes et al. first characterized B. microti’s immunodominant antigens through serological screening, establishing foundational targets for assay development (Lodes et al., 2000); Subsequent investigations by Homer’s team comprehensively validated the immunogenicity of novel antigenic candidates, advancing diagnostic reagent prototypes (Homer et al., 2003); Breakthrough research from Cornillot’s group identified the secreted antigen BmBAHCS1, demonstrating early IgM seroconversion at 4 days post-infection followed by IgG responses by day 8—a temporal profile enabling acute-phase detection (Cornillot et al., 2016). These cumulative discoveries highlight the evolving antigenic landscape while underscoring the need for standardized multi-antigen panels to address diagnostic window limitations and interspecies variability in Babesia infections.
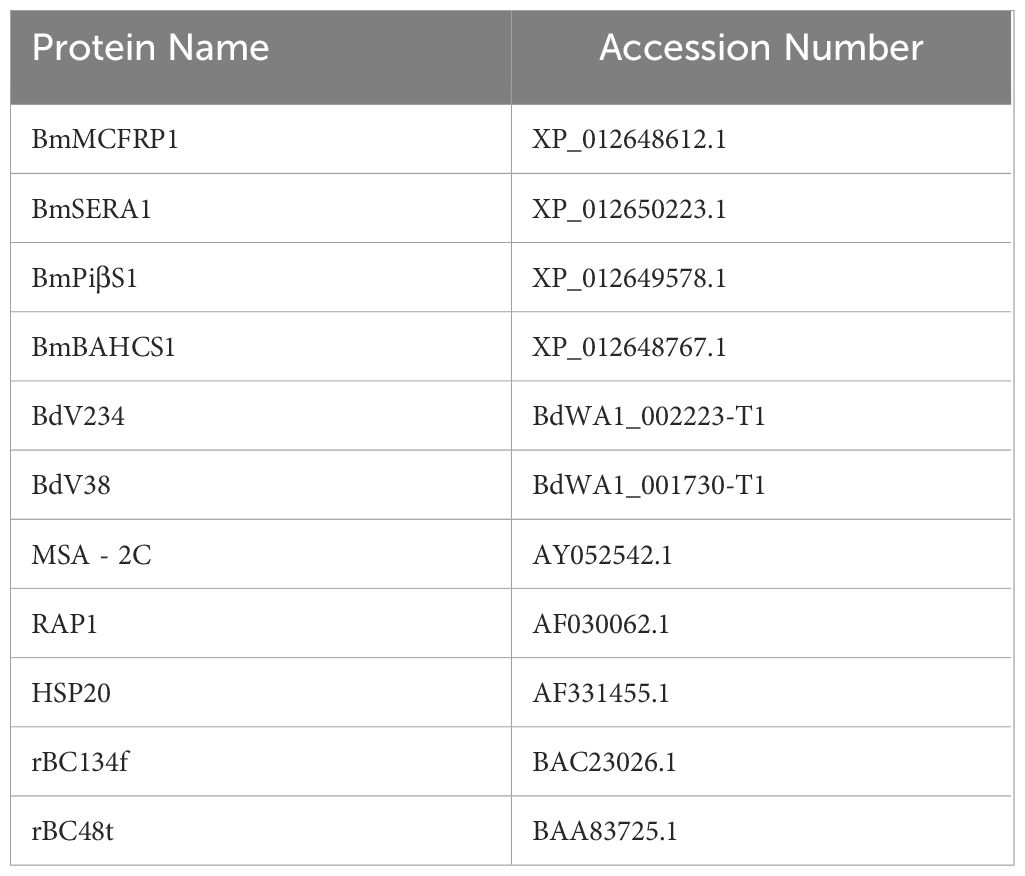
Recent advancements integrating genomic screening with transcriptomic profiling have revolutionized Babesia antigen discovery, enabling systematic identification of immunodominant targets spanning secreted and membrane-anchored proteins. While serological antibodies remain pivotal biomarkers for exposure history, their diagnostic utility is constrained during early infection windows prior to seroconversion, complicating differentiation between active and resolved infections. In the early 1970s, experiments were conducted to establish indirect immunofluorescence assays (IFA) for antibody detection in animal models (Cox and Turner, 1970; Goldman et al., 1972; Leeflang and Perié, 1972), although antibodies can be effectively detected, there are limitations such as lack of early sensitivity, today only indirect fluorescent antibody assays (IFAs) are commercially available to detect antibodies against Babesia divergens in humans (Jian et al., 2010; Cheng et al., 2018; Wang et al., 2020a; Tijani et al., 2024). To contemporary enzyme-linked immunosorbent assay (ELISA) platforms. Early ELISA iterations utilized crude Babesia spp.-infected host lysates, progressively transitioning to recombinant antigen systems (Meeusen et al., 1985; Houghton et al., 2002; Loa et al., 2004; Luo et al., 2011). Recently, breakthrough innovations now achieve 100% diagnostic sensitivity through multiplex antigen panels combining BmMCFRP1, BmSERA1, BmPiβS, and BmBAHCS1 (Verma et al., 2020). Antigen-capture ELISA (e.g., Babesia duncani antigen capture assays (BdACAs)) further enhances specificity by targeting secretory antigens (BdV234, BdV38), achieving detection thresholds of 115 infected erythrocytes/μL while circumventing PCR false positives from residual DNA (Chand et al., 2024). These integrated approaches bridge critical diagnostic gaps, enabling precise infection staging from acute parasitemia to chronic carrier states.
Serodiagnostic innovations continue to evolve species-specific strategies for Babesia detection across host systems. In bovine babesiosis, a recombinant chimeric antigen (rMABbo) ELISA integrating B. bovis MSA-2c, RAP-1, and HSP20 epitopes achieves 95.9% sensitivity and 94.3% specificity, outperforming conventional single-antigen assays (Jaramillo Ortiz et al., 2018). Equine diagnostics benefit from multiplex antigen cocktails combining B. caballi rBC134f and rBC48t, advancing detection windows to 4 days post-infection through stage-specific epitope targeting (El-Sayed et al., 2023). Human diagnostics leverage peptide array technology to screen B. microti MSP2 and SA-1 antigenic motifs, demonstrating conserved immunoreactivity that minimizes false negatives from genetic variability, even in Anaplasma phagocytophilum co-infection scenarios (Tagliafierro et al., 2022). Despite these advances, serological limitations persist: IFA’s subjective interpretation and unstandardized reagents yield elevated false positives for B. divergens (Pokhil et al., 2020); while RAP-1 conservation between B. bovis and B. bigemina induces cross-reactivity (Jaramillo Ortiz et al., 2018). Notably, B. microti IFA exhibits superior specificity, with minimal cross-reactivity to other Babesia spp. except at low antibody titers (Chisholm et al., 1978). These findings underscore the necessity for standardized antigen panels and automated interpretation systems to harmonize diagnostic accuracy across zoonotic spectra.
Immunodiagnostic approaches for babesiosis demonstrate variable utility across infection stages and host systems. Complement fixation tests (CFT) targeting Babesia bovis-specific IgM antibodies show diagnostic value in acute infections but fail to detect chronic carriers due to transient complement-binding immunoglobulin responses (Mahoney, 1967; Ogunremi et al., 2007, 2008); Comparatively, indirect fluorescent antibody tests (IFAT) exhibit superior practicality through earlier seroconversion detection (3–5 days post-infection), reduced operational complexity (complement-independent), and lower costs (≈33% of CFT expenses), while maintaining >92% accuracy for B. bovis and B. bigemina identification (Kumar et al., 2003); Emerging rapid diagnostics like immunochromatographic tests (ICT) and automated fluorescence immunoassays (AFIA) enable high-throughput screening, though require geographic validation across diverse epidemiologic contexts (Moritz et al., 2017; Stuart Tayebwa et al., 2020; Andrade et al., 2024; Jongejan et al., 2024); The monoclonal antibody capture assay (mGPAC) offers precise diagnostic capabilities by targeting parasite-secreted antigens such as BmGPI12, specifically distinguishing active Babesia microti infections from prior exposures. This method demonstrates absolute specificity for B. microti without cross-reactivity to related species like B. divergens or B. duncani, making it particularly valuable for immunocompromised and elderly patient populations (Gagnon et al., 2022), With a detection limit of 7.3 pg/µl - 20 pg/µl, mGPAC surpasses both real-time PCR and microscopy in sensitivity and specificity for identifying active B. microti infections (Thekkiniath et al., 2018; Wilhelmsson et al., 2020; Montero et al., 2023), Its unique ability to differentiate current parasitemia from historical immune responses addresses critical diagnostic challenges in vulnerable cohorts, while maintaining rigorous species discrimination crucial for accurate clinical management; While traditional methods such as rapid conglutination tests (RCT) show strong concordance with ELISA/IFAT (κ=0.89) and favorable performance metrics (90.9% sensitivity, 97.6% specificity), their exclusion from World Organisation for Animal Health (OIE)-certified protocols stems from unstandardized procedures and inadequate quality control frameworks (Kappmeyer et al., 1999). Despite technological advancements, most serologic assays remain excluded from international diagnostic guidelines due to insufficient standardization. IFA persists as the serological gold standard, particularly valuable for transfusion-transmission investigations (Herwaldt et al., 2011; Moritz et al., 2016). Crucially, immunodiagnostics should complement—not replace—direct detection methods (microscopy/PCR), given their limitations in early infection windows (<5 days post-exposure) and immunocompromised hosts, coupled with prolonged antibody persistence post-cure (Vannier and Krause, 2012; Sanchez et al., 2016). This diagnostic synergy ensures accurate infection confirmation while addressing the temporal and immunological constraints inherent to antibody-based detection.
4.4 Molecular biology testing
Recent advances in molecular biology and immunological methods have enabled the development of nucleic acid probe assays for Babesia spp. identification. These assays employ primers and probes targeting conserved variable regions of parasite genes, allowing qualitative detection of parasite DNA in host blood samples. Complementing this approach, fluorescence in situ hybridization (FISH) technology specifically targets the 18S ribosomal RNA (rRNA) of Babesia spp. using labeled DNA probes, with positive hybridization signals visualized as green fluorescence under microscopy (Shah et al., 2020). The streamlined FISH protocol involves blood sample preparation with smear fixation reagents, followed by a 30-minute hybridization at 37°C and fluorescence detection, completing the entire diagnostic process within two hours (Shah and Ramasamy, 2022). This cost-effective platform requires minimal equipment, as conventional light microscopes can be adapted for fluorescence detection by installing LED light sources instead of specialized fluorescence microscopes. Unlike DNA-based PCR methods, FISH demonstrates superior performance by distinguishing viable from non-viable parasites while avoiding interference from PCR inhibitors in blood samples. The current assay panel covers zoonotic species (B. microti, B. duncani, B. divergens) and veterinary pathogens (B. bovis, B. bigemina in cattle, B. caballi in horses), with a detection limit of 57–58 parasites/μL. Notably, the technology maintains specificity even during co-infections with Plasmodium species or Lyme disease spirochetes. When combined with indirect fluorescent antibody (IFA) testing, this rRNA-targeted FISH approach enhances screening efficiency in endemic regions and shows particular promise for resource-limited settings due to its minimal infrastructure requirements and elimination of nucleic acid amplification steps.
Reverse line hybridization (RLB) has emerged as a robust platform for detecting tick-borne Babesia spp., exemplified by integrated systems like the Tekenscanner that enable rapid confirmation of host infections through species-specific probes (Stoltsz et al., 2020; Byaruhanga et al., 2023; Hoxha et al., 2025). Parallel advancements include Tick - Borne Disease Capture Sequencing (TBDCapSeq), a high-throughput probe technology capable of simultaneous genomic detection for 11 tick-borne pathogens across 50 samples per run. This method demonstrates exceptional sensitivity (1–10 genome copies), outperforming ultra-high-throughput sequencing (UHTS) by 25 to >10,000-fold and surpassing qPCR detection limits, making it particularly effective for large-scale surveillance. While its dual barcoding system reduces per-sample costs, TBDCapSeq requires optimized protocols for low-parasitemia specimens (e.g., increased blood volumes or sequencing depth) and faces limitations in detecting novel pathogens due to probe dependency on known sequences. Cross-reactivity risks from conserved rRNA gene regions further necessitate complementary genetic analyses for species confirmation (Jain et al., 2021).
These technological advancements position FISH as a rapid, cost-effective clinical/veterinary tool for detecting viable parasites, while TBDCapSeq and RLB provide high-throughput surveillance solutions. Future development should focus on probe specificity enhancements and integrated diagnostic workflows combining multiple techniques to address current limitations in pathogen coverage and analytical specificity.
Polymerase chain reaction (PCR) has established itself as a cornerstone technology for detecting Babesia infections and cryptic carrier states, driven by its exceptional specificity and sensitivity. Continuous methodological refinements have expanded its utility across diverse strains, including B. caballi, B. microti, B. bovis, and B. bigemina. Early conventional PCR systems (1990s) targeted conserved regions such as the 18S rRNA gene’s V4 domain in B. bovis, achieving detection limits of 10–100 organisms/mL (Fahrimal et al., 1992; Xu et al., 2018). The advent of reverse transcription PCR (RT-PCR) in the 2000s enabled transcriptional-level analyses, including B. bovis MSA-2c antigenic gene expression and B. bigemina rap-1 family transcriptional networks, while boosting sensitivity tenfold to 10 organisms/mL for low-parasitemia detection (Suarez et al., 2003; Wilkowsky et al., 2003; Colasante et al., 2024). Modern advancements center on real-time quantitative PCR (qPCR), exemplified by multiplex assays targeting the sbp4 gene for B. caballi genotyping (Type A/B/C) with 5 copies/μL sensitivity (Giglioti et al., 2018); and extraction-free protocols enabling direct Anaplasma/Babesia/Ehrlichia triplex screening from diluted blood (43 copies/mL detection limit, 98% agreement with microscopy) (Patiño et al., 2024), Optimized qPCR achieves remarkable sensitivity for B. bigemina (1.5 infected RBCs/μL), surpassing microscopy by 100-fold (Stoltsz et al., 2020). Optimized nested PCR (nPCR) (Nicolaiewsky et al., 2001; Tian et al., 2015; Faria et al., 2017; Kumar et al., 2022) while nested PCR (nPCR) enhances B. bovis apocytochrome b gene detection to 2 parasites/0.5 mL blood – a 1,000-fold improvement over blood smears (Bhat et al., 2017; Radzijevskaja et al., 2018; Hong et al., 2019; Swei et al., 2019; Cai et al., 2024). Despite these advances, nPCR carries risks of underdetection in low-load samples and cross-species reactivity. Emerging approaches, such as genotype-specific qPCR (100% detection probability, 95% CI) (Venter et al., 2024). and semi-nested PCR coupled with SacII digestion (sensitivity to 0.00000012% parasitemia for B. orientalis) (Liu et al., 2007). Iterative optimizations in multiplex design, probe engineering, and workflow portability have collectively elevated PCR’s diagnostic precision for hypoparasitemia, mixed infections, and strain differentiation, solidifying its role in modern parasitology.
4.5 Isothermal amplification technology
Loop-mediated isothermal amplification (LAMP) has emerged as a transformative nucleic acid amplification technique, enabling rapid, equipment-independent detection of Babesia spp. under constant temperature conditions. This method excels in identifying early-stage infections (e.g., feline hosts), chronic cases (canines), and low-level or persistent parasitemia in diverse hosts including sheep, goats, and yaks (Guan et al., 2008; He et al., 2009; Mandal et al., 2015). Demonstrating superior analytical performance, LAMP achieves 100-fold greater sensitivity than PCR-agarose gel electrophoresis (PCR-AGE), 10-fold improvement over nested PCR (nPCR), and enhanced reliability compared to modified semi-nested PCR in baseline controls (Yang et al., 2016; Arnuphapprasert et al., 2023). Its detection threshold reaches 50 fg per reaction for B. motasi (Hong et al., 2019); with additional applications in species differentiation, such as distinguishing bovine Babesia from Theileria parasites (Tian et al., 2015). Recent innovations have expanded LAMP’s utility through hybrid platforms: LAMP-lateral flow dipstick (LAMP-LFD) integrates chromatographic strips for visual readouts, achieving 100-fold higher positivity rates than conventional PCR while maintaining absolute specificity for Babesia DNA, Complementary modifications like LAMP-LED and LAMP-UV incorporate portable light sources for result visualization, demonstrating 10-fold greater sensitivity than PCR-AGE for bovine babesiosis detection with 100% specificity (Yang et al., 2016; Schmidt et al., 2017). These advancements position LAMP as a field-deployable alternative to PCR, offering equivalent rapidity and sensitivity while eliminating the need for sophisticated thermocycling equipment.
Isothermal amplification technologies including recombinase polymerase amplification (RPA), recombinase-aid amplification (RAA), and cross-priming amplification (CPA) have demonstrated significant potential for field-deployable diagnostics of Babesia spp (Lei et al., 2020). RPA leverages recombinase-mediated strand invasion to amplify target sequences at low temperatures (37–42°C), achieving rapid detection thresholds of 22.5 copies/μL within 30 minutes when paired with lateral flow strips. Innovations like the rpaBab264-primer-based RPA-LFD system enable B. vogeli detection at 40°C in 10 minutes with comparable accuracy to conventional PCR. For B. orientalis, RPA targeting the mitochondrial COXI gene achieves 0.25 parasites/μL sensitivity in 15 minutes at 37°C—40-fold more sensitive than standard PCR—while maintaining specificity against apicomplexan relatives and host DNA (An et al., 2021; Nie et al., 2021; Onchan et al., 2022). RAA shares mechanistic similarities with RPA but operates at reduced costs, detecting 10 copies/reaction in 20 minutes (Lin et al., 2022). CPA distinguishes itself through cross-priming multiplex amplification, exemplified by a B. duncani-specific assay achieving 0.98 pg/μL sensitivity (~20 parasites/μL) at 59°C within 60 minutes via vertical flow strip visualization (CPA-VF), with 98.7% accuracy and a per-test cost of $0.20. Further optimizations yield CPA-VF sensitivities of 50 fg/reaction for B. motasi (5-fold superior to qPCR) and 320 fg/reaction for B. bovis, demonstrating field performance equivalent to PCR without cross-reactivity to co-circulating bovine pathogens (Wang et al., 2020b; Nian et al., 2024).
These platforms collectively address critical needs for rapid, low-resource Babesia detection, balancing high sensitivity, cost-effectiveness, and operational simplicity. TBDCapSeq demonstrates unparalleled analytical sensitivity (1–10 genome copies), surpassing qPCR, RPA, nested PCR (nPCR), FISH, and conventional PCR in detection limits. Isothermal amplification methods (LAMP, RPA, CPA) collectively outperform conventional PCR in both sensitivity and operational simplicity. Regarding processing speed, RPA achieves results within 10–30 minutes—the fastest among compared techniques—followed by FISH (2 hours), qPCR (~1 hour), and LAMP (~1 hour). Equipment needs vary substantially: FISH requires only a modified light microscope with LED illumination (lowest cost), while LAMP and RPA depend on basic thermostatic devices. qPCR necessitates fluorescence quantification instruments, and TBDCapSeq relies on high-throughput sequencers.
For clinical settings prioritizing rapidity and ease-of-use, FISH, LAMP, and RPA are optimal due to their minimal infrastructure demands and short turnaround times. Large-scale surveillance programs benefit from high-throughput platforms like TBDCapSeq, RLB, and multiplex qPCR. In scenarios requiring ultra-sensitive detection of low-parasitemia samples, nPCR and TBDCapSeq emerge as preferred choices, leveraging their enhanced limits of detection.
4.6 New technical applications for Babesia diagnostics
4.6.1 High-throughput sequencing tool (Haemabiome)
The Haemabiome high-throughput sequencing platform represents a paradigm shift in Babesia diagnostics, employing dual-stage PCR amplification of conserved 16S/18S rDNA regions coupled with barcoding and Illumina MiSeq sequencing. This approach enables comprehensive pathogen identification through alignment with the SILVA ribosomal RNA database project (SILVA) reference database, achieving high-resolution detection of Babesia spp. alongside co-circulating genera such as Theileria. By integrating multi-pathogen detection capability with exceptional sensitivity, the system addresses the critical limitations of traditional single-target assays. Its capacity to resolve mixed infections and uncover genetic diversity within parasite populations positions Haemabiome as a powerful tool for epidemiological surveillance and complex clinical case analysis, particularly in regions endemic to multiple tick-borne pathogens (Yalcindag et al., 2024). Emerging omics technologies are revolutionizing Babesia diagnostics and pathogenesis research through multi-dimensional analytical approaches. Label-based high-throughput quantitative proteomics (HQP) employs tandem mass tagging (TMT) with LC-MS/MS to profile serum/urine proteomes, revealing early renal injury biomarkers like NGAL and L-FABP in canine babesiosis through integrated KEGG/GO pathway analysis (Bilić et al., 2023). Metagenomic next-generation sequencing (mNGS) provides hypothesis-free pathogen detection across diverse specimens (whole blood, plasma, urine), recently proving instrumental in diagnosing Babesia-induced hemolytic anemia from sputum samples—a case initially misattributed to Epstein-Barr virus complications (Lu et al., 2024). Metagenomic next-generation sequencing (mNGS) provides hypothesis-free pathogen detection across diverse specimens (whole blood, plasma, urine), recently proving instrumental in diagnosing Babesia-induced hemolytic anemia from sputum samples—a case initially misattributed to Epstein-Barr virus complications (Gyarmati et al., 2016; Schmidt et al., 2017; Vijayvargiya et al., 2019; Lu et al., 2024). At the cellular level, single-cell RNA sequencing (scRNA-seq) enables unprecedented resolution of Babesia’s asexual replication cycle through pseudotime gene expression mapping (Rezvani et al., 2022). Singh et al. (2023) complementing genomic advances exemplified by B. duncani’s fully sequenced genome with detailed epigenomic/transcriptomic annotations. Comparative multi-omics leveraging Plasmodium models reveal divergent drug sensitivity mechanisms between these apicomplexans, informing targeted therapeutic development (Si et al., 2023). Whole genome sequencing facilitates phylogenetic reconstruction and gene family evolution analysis, distinguishing Babesia lineages while identifying host-specific invasion determinants and virulence factors (Wang et al., 2022). Integrated genomic-proteomic workflows further enable rational vaccine antigen discovery through computational epitope prediction and functional validation (Liu et al., 2023).
4.6.2 Microfluidics and biomedical engineering technology
Microfluidic platforms leveraging insulator-based dielectrophoresis (iDEP) offer a transformative approach for rapid Babesia detection by exploiting dielectric property differences between infected and healthy erythrocytes. Infection-induced structural alterations in red blood cell membranes—such as ridge formation—modify their polarization characteristics, enabling iDEP systems to separate parasitized cells via dielectrophoretic (DEP) forces in non-uniform electric fields. Validated through GFP labeling and Diff-Quik staining in B. bovis models, this technology processes 1 μL samples in <1 minute, concentrating 0.1% parasitemia specimens to 70% purity while achieving >98% healthy cell recovery at optimized voltages. With a sensitivity threshold of 0.1% parasitemia (5,000 parasites/μL), iDEP outperforms conventional microscopy but remains less sensitive than PCR (1–5 parasites/μL) (Adekanmbi et al., 2016). The platform employs a single-shell model to quantify infection-induced erythrocyte heterogeneity (e.g., membrane deformation, cellular swelling) through S-curve fitting, combining operational simplicity with analytical reliability. By eliminating time-consuming microscopy and complex nucleic acid amplification, iDEP significantly enhances transfusion screening efficiency and reduces babesiosis transmission risks. While currently validated for bovine B. bovis, adaptation to human pathogens like B. microti represents a critical next step for clinical translation (Oladokun et al., 2023).
4.6.3 Biomarker tests
Emerging urinary biomarker assays and nanotechnology platforms are advancing early detection of Babesia-associated complications in veterinary medicine. The uNGAL/uKIM-1 test quantifies urinary neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 levels, demonstrating superior sensitivity for identifying acute kidney injury (AKI) in canine babesiosis compared to traditional renal markers. In dogs meeting International Renal Interest Society (IRIS) AKI criteria (serum creatinine elevation >0.3 mg/dL within 48 hours), uNGAL and uKIM-1 levels showed significant elevation (P<0.001) even at IRIS grade I, proving particularly effective for detecting renal damage during non-azotemic stages (Asma Idress et al., 2024).
Complementing these biomarker approaches, nanoparticle-mass spectrometry (NanoCage-MS) employs engineered nanocages to capture B. microti-specific urinary proteins for subsequent mass spectrometric identification, enabling parasite detection through targeted proteomic signatures (Magni et al., 2020). While both technologies show promise for early clinical intervention—with uNGAL/uKIM-1 enhancing AKI management and NanoCage-MS providing direct pathogen detection—their veterinary application requires further validation through large-scale, multi-etiology studies to confirm diagnostic accuracy across diverse infection scenarios.
4.6.4 Genome and gene editing technologies
Next-generation sequencing platforms and CRISPR-based systems are driving transformative advances in Babesia genomics and diagnostics. Oxford Nanopore Technology (ONT) excels in resolving complex genomic architectures, while Illumina short-read sequencing provides high-fidelity assembly—complementary approaches that revealed B. duncani’s compact genome size and distinct phylogenetic lineage from B. microti and Theileria species (Wang et al., 2022). Targeted amplicon deep sequencing (TADS), though limited in parasite studies by host DNA interference and genetic diversity challenges, has been adapted through nested universal parasite detection (nested UPDx) to achieve qPCR-comparable sensitivity for discriminating Babesia from Plasmodium and Trypanosoma in blood samples (Flaherty et al., 2021).
CRISPR-Cas systems have emerged as precision tools for apicomplexan genome engineering, with Hakimi et al. pioneering CRISPR-Cas9 applications in B. bovis for epitope tagging and gene replacement via Cas9/hDHFR plasmid systems (Hakimi et al., 2019). The compact CRISPR-Cas12a platform enables rapid field diagnostics through recombinase polymerase amplification (RPA) integration, detecting Babesia spp. via lateral flow strips in <2 hours with high specificity (Muriuki et al., 2024). Cas12 is a smaller nuclease that is easier to deliver to smaller parasites and has the potential for AAV-mediated delivery, providing an advantage for editing the apicomplexan genome (Webi et al., 2024). Due to the lack of RNAi inducible genes, RNAi has limitations in some apicomplexes, and Cas13, which targets RNA, offers new opportunities to study gene expression and regulation, but there is no Cas13 applied to Babesia.
These synergistic technologies—spanning long-read genomics, enhanced amplicon sequencing, and programmable nucleases—collectively accelerate functional genomics and field-deployable diagnostics for babesiosis management.
4.6.5 Automated testing equipment and mass spectrometry
The Sysmex XN-31m automated hematology analyzer, originally developed for human malaria detection, has been successfully adapted for equine babesiosis diagnosis through fluorescence flow cytometry. This system identifies Babesia-infected red blood cells (iRBCs) by detecting parasite-specific fluorescent signatures, augmented by machine learning algorithms for pattern recognition. Capable of real-time parasitemia quantification, it reduces treatment decision times during acute infections while minimizing manual interpretation errors through automation (Ochi et al., 2024). With a limit of detection (LoD) surpassing conventional microscopy (>100 iRBCs/μL), the platform excels in high-throughput screening scenarios. Integrated multi-parameter analysis simultaneously evaluates routine hematological indices (red cell counts, hemoglobin levels), enhancing diagnostic comprehensiveness.
Complementing this approach, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) enables direct pathogen identification from clinical specimens, recently extended to arthropod vectors (Ixodes, Rhipicephalus, Amblyomma ticks) and parasitic protozoa (Sánchez-Juanes et al., 2022). In canine babesiosis, the technology detects a unique 51–52 kDa serum protein biomarker absent in healthy dogs, demonstrating high specificity for B. canis diagnosis (Dzięgiel et al., 2016). While proven effective in malaria-endemic regions, broader Babesia applications require optimization of species-specific proteomic databases and standardized data protocols. Together, these automated systems bridge clinical and vector surveillance needs, offering scalable solutions for endemic disease management through rapid, operator-independent diagnostics.
4.6.6 Flow Cytometry Combined with Artificial Intelligence
Emerging synergies between fluorescence technologies and artificial intelligence are advancing Babesia diagnostics through automated fluorescence flow cytometry (FLC), which enables direct parasite quantification from venous blood samples (Vanderboom et al., 2024). Current implementations require development of specialized machine learning (ML) algorithms to overcome the absence of dedicated analytical software, facilitating automated parasite recognition and quantification. The integration of fluorescence in situ hybridization (FISH) with flow cytometry presents a promising frontier, combining FISH’s species-specific detection capabilities with the high-throughput automation of flow systems to enhance both speed and analytical precision (Shah and Ramasamy, 2022). This hybrid approach could leverage optical enhancements to improve sensitivity while maintaining specificity, addressing critical gaps in rapid babesiosis screening. Future implementations may incorporate advanced imaging flow cytometry to simultaneously analyze morphological and molecular markers, enabling real-time parasite characterization. Such innovations position FLC-AI systems as transformative tools for clinical laboratories, offering scalable solutions that reduce operator dependency while improving diagnostic accuracy in both acute and surveillance settings.
4.6.7 Other innovative technologies
Vibrational spectroscopy techniques are advancing Babesia diagnostics by capturing molecular fingerprints of host-parasite interactions across multiple scales. At single-cell resolution, atomic force microscopy-infrared spectroscopy (AFM-IR) enables nanoscale chemical imaging of infected erythrocytes, while confocal Raman microscopy maps molecular alterations during intracellular invasion. For population-level analysis, attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) coupled with partial least squares discriminant analysis (PLS-DA) achieves 92.0% sensitivity and 91.7% specificity in <2 minutes post-sample processing. Rüther et al. optimized this approach for B. bovis through hemoglobin-depletion protocols and spectral signature modeling, enabling scalable detection from individual cells to clinical samples (Rüther et al., 2020).
Complementing these biochemical analyses, the TFinder deep learning system revolutionizes field diagnostics through automated Babesia quantification in bovine blood smears. This CNN-based platform, trained on 2,871 infected erythrocyte images, integrates multi-scale feature fusion and adaptive thresholding to achieve 98.0% sensitivity and 96.9% accuracy in qualitative diagnosis, with quantitative analysis reaching 99.7% specificity. By eliminating manual interpretation errors, TFinder detects parasitemia as low as 0.005%—20-fold below microscopy thresholds—while accelerating analysis 5–8-fold compared to conventional methods. Validated against 750 clinical samples, this first AI-driven solution for bovine tick fever meets medical device certification standards, with ongoing development focused on multi-pathogen detection models and cross-border field validation (Trindade et al., 2025). Together, these technologies exemplify the convergence of molecular phenotyping and artificial intelligence in transforming parasitic disease diagnostics.
5 Conclusion
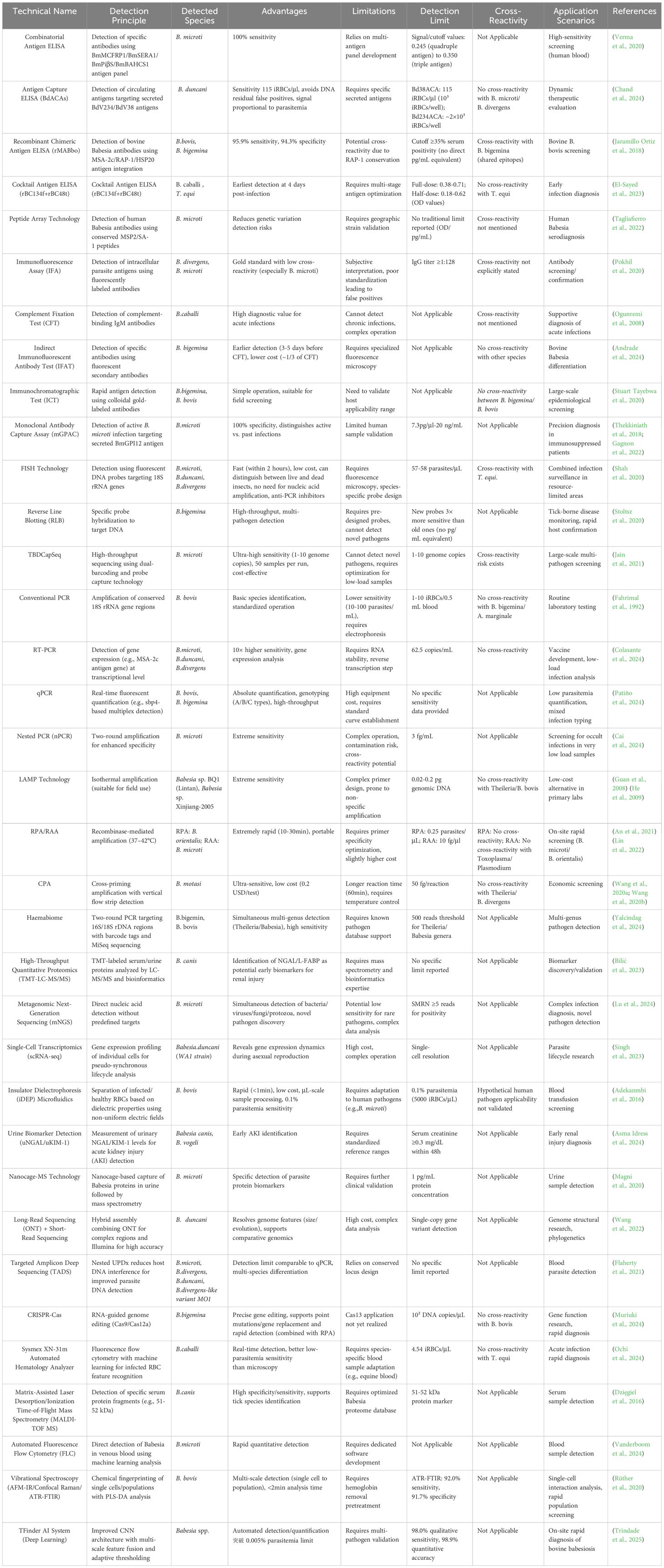
We have compiled a list of important testing methods covered in the text (Table 1 in the Attachments). Accurate species-specific diagnosis is fundamental for effectively preventing and controlling blood-borne parasitic infections. While microscopy remains the most accessible first-line tool due to low cost and rapid results, its clinical utility is constrained by limited sensitivity (particularly in low-parasitemia cases) and operator-dependent interpretation. Immunoassays address throughput limitations of conventional methods but require well-characterized species-specific antigens, with current platforms facing challenges of undefined antigen targets and cross-reactivity (Table 2). Serological approaches detect circulating antibodies yet suffer from persistent antibody retention post-recovery and false negatives in chronic carriers, compounded by reduced specificity when using crude antigen preparations. In contrast, nucleic acid amplification techniques (NAATs) offer superior sensitivity and speciation capacity, enabling discrimination between morphologically similar species like Babesia spp. and Plasmodium spp. Modern molecular advances now permit high-throughput screening and asymptomatic carrier identification – critical capabilities for endemic region surveillance. Current diagnostic integration strategies recommend: Primary screening: ELISA/ICT for clinical samples; Confirmatory testing: In vivo culture with microscopic examination/IFAT; Low-parasitemia cases: LAMP-assisted detection; Comprehensive analysis: PCR-based methods covering all parasitic life stages. In vitro culture systems serve dual purposes: pathogen isolation and supporting vaccine development through antigen characterization and drug susceptibility profiling. To minimize diagnostic errors, parallel testing with complementary assays is strongly advised. Future directions should focus on: Functional genomics of Babesia virulence factors through comparative interspecies transcriptomics; Multi-omics integration (proteomic/metabolomic) to elucidate gene expression networks governing life cycle progression; Next-generation assay development targeting differentially expressed biomarkers.
Author contributions
ZJ: Writing – original draft, Writing – review & editing. YZ: Writing – original draft. DZ: Methodology, Writing – review & editing. HW: Conceptualization, Writing – review & editing. MY: Data curation, Writing – review & editing. ZL: Formal Analysis, Project administration, Writing – review & editing. XZ: Investigation, Writing – review & editing. JC: Supervision, Visualization, Writing – review & editing. XW: Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Inner Mongolia Natural Science Foundation (2021LHMS03008).Multidisciplinary Interdisciplinary Research Project of Basic Scientific Research Operating Expenses for Colleges and Universities directly under Inner Mongolia (GXKY22003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adekanmbi, E. O., Ueti, M. W., Rinaldi, B., Suarez, C. E., and Srivastava, S. K. (2016). Insulator-based dielectrophoretic diagnostic tool for babesiosis. Biomicrofluidics 10, 033108. doi: 10.1063/1.4954196
Alvarez, J. A., Rojas, C., and Figueroa, J. V. (2019). Diagnostic tools for the identification of babesia sp. in persistently infected cattle. Pathogens 8, 3. doi: 10.3390/pathogens8030143
An, X., Zhao, Y., Cui, J., Liu, Q., Yu, L., Zhan, X., et al. (2021). Recombinase polymerase amplification lateral flow dipstick (RPA-LF) detection of Babesia orientalis in water buffalo (Bubalus babalis, Linnaeus 1758). Vet Parasitol. 296, 109479. doi: 10.1016/j.vetpar.2021.109479
Andrade, L. S., de Souza, R. S., Carvalho de Araujo, A., Silva, S. O., Melo, M. N., Melo, F. G., et al. (2024). Hemopathogens in naturally infected bovine fetuses in Brazil. Ticks Tick Borne Dis. 15, 102351. doi: 10.1016/j.ttbdis.2024.102351
Antognoni, M. T., Cremonini, V., Misia, A. L., Gobbo, F., Toniolo, F., and Miglio, A. (2025). Case report: First autochthonous Babesia vulpes infection in a dog from Italy. Front. Vet Sci. 12. doi: 10.3389/fvets.2025.1498721
Arnuphapprasert, A., Nugraheni, Y. R., Aung, A., Asada, M., and Kaewthamasorn, M. (2023). Detection of Babesia bovis using loop-mediated isothermal amplification (LAMP) with improved thermostability, sensitivity and alternative visualization methods. Sci. Rep. 13, 1838. doi: 10.1038/s41598-023-29066-1
Asma Idress, M., Deepa, P. M., Rathish, R. L., Vinodkumar, K., and Pradeep, M. (2024). Diagnostic efficacy of urinary neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 for early detection of acute kidney injury in dogs with leptospirosis or babesiosis. Vet Res. Commun. 48, 2813–2818. doi: 10.1007/s11259-024-10416-x
Bhat, S. A., Singh, N. K., Singh, H., and Rath, S. S. (2017). Molecular prevalence of Babesia bigemina in Rhipicephalus microplus ticks infesting cross-bred cattle of Punjab, India. Parasite Epidemiol. Control 2, 85–90. doi: 10.1016/j.parepi.2017.04.002
Bilić, P., Horvatić, A., Kuleš, J., Gelemanović, A., Beer Ljubić, B., Mũnoz-Prieto, A., et al. (2023). Serum and urine profiling by high-throughput TMT-based proteomics for the investigation of renal dysfunction in canine babesiosis. J. Proteomics 270, 104735. doi: 10.1016/j.jprot.2022.104735
Byaruhanga, C., Makgabo, S. M., Choopa, C. N., Mulandane, F. C., Vorster, I., Troskie, M., et al. (2023). Genetic diversity in Babesia bovis from southern Africa and estimation of B. bovis infection levels in cattle using an optimised quantitative PCR assay. Ticks Tick Borne Dis. 14, 102084. doi: 10.1016/j.ttbdis.2022.102084
Cai, Y., Xu, B., Liu, X., Yang, W., Mo, Z., Zheng, B., et al. (2024). Transmission risk evaluation of transfusion blood containing low-density Babesia microti. Front. Cell Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1334426
Chand, M., Vydyam, P., Pal, A. C., Thekkiniath, J., Darif, D., Li, Z., et al. (2024). A set of diagnostic tests for detection of active Babesia duncani infection. Int. J. Infect. Dis. 147, 107178. doi: 10.1016/j.ijid.2024.107178
Chen, Z., Li, H., Gao, X., Bian, A., Yan, H., Kong, D., et al. (2019). Human Babesiosis in China: a systematic review. Parasitol. Res. 118, 1103–1112. doi: 10.1007/s00436-019-06250-9
Cheng, K., Coller, K. E., Marohnic, C. C., Pfeiffer, Z. A., Fino, J. R., Elsing, R. R., et al. (2018). Performance evaluation of a prototype architect antibody assay for babesia microti. J. Clin. Microbiol. 56, 5. doi: 10.1128/jcm.00460-18
Chisholm, E. S., Ruebush, T. K., 2nd, Sulzer, A. J., and Healy, G. R. (1978). Babesia microti infection in man: evaluation of an indirect immunofluorescent antibody test. Am. J. Trop. Med. Hyg. 27, 14–19. doi: 10.4269/ajtmh.1978.27.14
Colasante, G., Makari, K., Hummel, T. I., and Murphy, C. (2024). Sample-to-answer direct real-time PCR detection of Anaplasma phagocytophilum, Ehrlichia spp., and Babesia spp. infections in whole-blood specimens. Microbiol. Spectr. 12, e0065524. doi: 10.1128/spectrum.00655-24
Cook, M. J. and Puri, B. K. (2024). Babesiosis: analysis of the evidence for infections in the United Kingdom. Int. J. Gen. Med. 17, 4627–4631. doi: 10.2147/ijgm.S485759
Cornillot, E., Dassouli, A., Pachikara, N., Lawres, L., Renard, I., Francois, C., et al. (2016). A targeted immunomic approach identifies diagnostic antigens in the human pathogen Babesia microti. Transfusion 56, 2085–2099. doi: 10.1111/trf.13640
Cox, F. E. and Turner, S. A. (1970). Antigenic relationships between the malaria parasites and piroplasms of mice as determined by the fluorescent-antibody technique. Bull. World Health Organ 43, 337–340.
Dkhil, M. A., Al-Quraishy, S., and Al-Khalifa, M. S. (2014). The effect of Babesia divergens infection on the spleen of Mongolian gerbils. BioMed. Res. Int. 2014, 483854. doi: 10.1155/2014/483854
Dos Santos, F. B., Gazêta, G. S., Corrêa, L. L., Lobão, L. F., Palmer, J. P. S., Dib, L. V., et al. (2021). Microscopic detection, hematological evaluation and molecular characterization of piroplasms from naturally infected dogs in Rio de Janeiro, Brazil. Acta Parasitol. 66, 1548–1560. doi: 10.1007/s11686-021-00426-z
Durant, T. J. S., Dudgeon, S. N., McPadden, J., Simpson, A., Price, N., Schulz, W. L., et al. (2021). Applications of digital microscopy and densely connected convolutional neural networks for automated quantification of babesia-infected erythrocytes. Clin. Chem. 68, 218–229. doi: 10.1093/clinchem/hvab237
Dvoraková, H. M. and Dvorácková, M. (2007). Babesiosis, a little known zoonosis. Epidemiol. Mikrobiol. Imunol. 56, 176–180.
Dzięgiel, B., Adaszek, Ł., Banach, T., and Winiarczyk, S. (2016). Specificity of mass spectrometry (MALDI-TOF) in the diagnosis of Babesia canis regarding to other canine vector-borne diseases. Ann. Parasitol. 62, 101–105. doi: 10.17420/ap6202.39
El-Sayed, S. A. E., Rizk, M. A., Baghdadi, H. B., Ringo, A. E., Sambuu, G., Nugraha, A. B., et al. (2023). Development of a promising antigenic cocktail for the global detection of Babesia caballi in horse by ELISA. PloS One 18, e0284535. doi: 10.1371/journal.pone.0284535
Everitt, J. I., Shadduck, J. A., Steinkamp, C., and Clabaugh, G. (1986). Experimental Babesia bovis infection in Holstein calves. Vet Pathol. 23, 556–562. doi: 10.1177/030098588602300503
Fahrimal, Y., Goff, W. L., and Jasmer, D. P. (1992). Detection of Babesia bovis carrier cattle by using polymerase chain reaction amplification of parasite DNA. J. Clin. Microbiol. 30, 1374–1379. doi: 10.1128/jcm.30.6.1374-1379.1992
Fang, L. Q., Liu, K., Li, X. L., Liang, S., Yang, Y., Yao, H. W., et al. (2015). Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect. Dis. 15, 1467–1479. doi: 10.1016/s1473-3099(15)00177-2
Faria, A. R., Pires, S. D. F., Reis, A. B., Coura-Vital, W., Silveira, J., Sousa, G. M., et al. (2017). Canine visceral leishmaniasis follow-up: a new anti-IgG serological test more sensitive than ITS-1 conventional PCR. Vet Parasitol. 248, 62–67. doi: 10.1016/j.vetpar.2017.10.020
Flaherty, B. R., Barratt, J., Lane, M., Talundzic, E., and Bradbury, R. S. (2021). Sensitive universal detection of blood parasites by selective pathogen-DNA enrichment and deep amplicon sequencing. Microbiome 9, 1. doi: 10.1186/s40168-020-00939-1
Gagnon, J., Timalsina, S., Choi, J. Y., Chand, M., Singh, P., Lamba, P., et al. (2022). Specific and sensitive diagnosis of babesia microti active infection using monoclonal antibodies to the immunodominant antigen bmGPI12. J. Clin. Microbiol. 60, e0092522. doi: 10.1128/jcm.00925-22
Giglioti, R., de Oliveira, H. N., Okino, C. H., and de Sena Oliveira, M. C. (2018). qPCR estimates of Babesia bovis and Babesia bigemina infection levels in beef cattle and Rhipicephalus microplus larvae. Exp. Appl. Acarol. 75, 235–240. doi: 10.1007/s10493-018-0260-0
Goldman, M., Pipano, E., and Rosenberg, A. S. (1972). Fluorescent antibody tests for Babesia bigemina and B. berbera. Res. Vet Sci. 13, 77–81. doi: 10.1016/S0034-5288(18)34092-X
Gorenflot, A., Moubri, K., Precigout, E., Carcy, B., and Schetters, T. P. (1998). Human babesiosis. Ann. Trop. Med. Parasitol. 92, 489–501. doi: 10.1080/00034989859465
Guan, G., Chauvin, A., Luo, J., Inoue, N., Moreau, E., Liu, Z., et al. (2008). The development and evaluation of a loop-mediated isothermal amplification (LAMP) method for detection of Babesia spp. infective to sheep and goats in China. Exp. Parasitol. 120, 39–44. doi: 10.1016/j.exppara.2008.04.012
Gyarmati, P., Kjellander, C., Aust, C., Song, Y., Öhrmalm, L., and Giske, C. G. (2016). Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci. Rep. 6, 23532. doi: 10.1038/srep23532
Hadani, A., Guglielmone, A. A., Gonzalez de Rios, L., Bermudez, A., Mangold, A., and Barnett, S. F. (1982). Use of cerebellar brain smears in the diagnosis of babesiosis (Babesia bovis) in cattle. Trop. Anim. Health Prod. 14, 242–246. doi: 10.1007/bf02242167
Hakimi, H., Ishizaki, T., Kegawa, Y., Kaneko, O., Kawazu, S. I., and Asada, M. (2019). Genome editing of babesia bovis using the CRISPR/cas9 system. mSphere 4, 8. doi: 10.1128/mSphere.00109-19
Handel, A. S., Krugman, J., Hymes, S., Inkeles, S., and Beneri, C. (2021). A case of relapsed vertically transmitted babesiosis. J. Pediatr. Infect. Dis. Soc. 10, 386–388. doi: 10.1093/jpids/piaa104
He, L., Zhou, Y. Q., Oosthuizen, M. C., and Zhao, J. L. (2009). Loop-mediated isothermal amplification (LAMP) detection of Babesia orientalis in water buffalo (Bubalus babalis, Linnaeus 1758) in China. Vet Parasitol. 165, 36–40. doi: 10.1016/j.vetpar.2009.06.036
Herwaldt, B. L., Linden, J. V., Bosserman, E., Young, C., Olkowska, D., and Wilson, M. (2011). Transfusion-associated babesiosis in the United States: a description of cases. Ann. Intern. Med. 155, 509–519. doi: 10.7326/0003-4819-155-8-201110180-00362
Holman, P. J., Spencer, A. M., Telford, S. R., Goethert, H. K., Allen, A. J., Knowles, D. P., et al. (2005). Comparative infectivity of Babesia divergens and a zoonotic Babesia divergens-like parasite in cattle. Am. J. Trop. Med. Hyg. 73, 865–870. doi: 10.4269/ajtmh.2005.73.865
Homer, M. J., Lodes, M. J., Reynolds, L. D., Zhang, Y., Douglass, J. F., McNeill, P. D., et al. (2003). Identification and characterization of putative secreted antigens from Babesia microti. J. Clin. Microbiol. 41, 723–729. doi: 10.1128/jcm.41.2.723-729.2003
Hong, S. H., Kim, S. Y., Song, B. G., Rho, J. R., Cho, C. R., Kim, C. N., et al. (2019). Detection and characterization of an emerging type of Babesia sp. similar to Babesia motasi for the first case of human babesiosis and ticks in Korea. Emerg. Microb. Infect. 8, 869–878. doi: 10.1080/22221751.2019.1622997
Houghton, R. L., Homer, M. J., Reynolds, L. D., Sleath, P. R., Lodes, M. J., Berardi, V., et al. (2002). Identification of Babesia microti-specific immunodominant epitopes and development of a peptide EIA for detection of antibodies in serum. Transfusion 42, 1488–1496. doi: 10.1046/j.1537-2995.2002.00215.x
Hoxha, I., Xhekaj, B., Halimi, G., Wijnveld, M., Ruivo, M., Çaushi, D., et al. (2025). Zoonotic tick-borne pathogens in ixodes ricinus complex (Acari: ixodidae) from urban and peri-urban areas of Kosovo. Zoon. Public Health 72, 174–183. doi: 10.1111/zph.13197
Hunfeld, K. P., Hildebrandt, A., and Gray, J. S. (2008). Babesiosis: recent insights into an ancient disease. Int. J. Parasitol. 38, 1219–1237. doi: 10.1016/j.ijpara.2008.03.001
Jacobs, M. W., Rocco, J. M., Andersen, L. K., and Robertson, T. E. (2025). Babesiosis with low parasitemia as a cause of secondary hemophagocytic lymphohistiocytosis in a previously healthy adult. IDCases 39, e02172. doi: 10.1016/j.idcr.2025.e02172
Jain, K., Tagliafierro, T., Marques, A., Sanchez-Vicente, S., Gokden, A., Fallon, B., et al. (2021). Development of a capture sequencing assay for enhanced detection and genotyping of tick-borne pathogens. Sci. Rep. 11, 12384. doi: 10.1038/s41598-021-91956-z
Jaramillo Ortiz, J. M., Montenegro, V. N., de la Fournière, S. A. M., Sarmiento, N. F., Farber, M. D., and Wilkowsky, S. E. (2018). Development of an indirect ELISA based on a recombinant chimeric protein for the detection of antibodies against bovine babesiosis. Vet Sci. 5, 5. doi: 10.3390/vetsci5010013
Jasik, K. P., Kleczka, A., and Franielczyk, A. (2024). Histopathological aspects of the influence of babesia microti on the placentas of infected female rats. Vet Sci. 11, 3. doi: 10.3390/vetsci11010018
Jerzak, M., Gandurski, A., Tokaj, M., Stachera, W., Szuba, M., and Dybicz, M. (2023). Advances in babesia vaccine development: an overview. Pathogens 12, 2. doi: 10.3390/pathogens12020300
Jia, N., Zheng, Y. C., Jiang, J. F., Jiang, R. R., Jiang, B. G., Wei, R., et al. (2018). Human babesiosis caused by a babesia crassa-like pathogen: A case series. Clin. Infect. Dis. 67, 1110–1119. doi: 10.1093/cid/ciy212
Jian, Z., Ma, S., Sun, Q., Shen, J., Miao, Z., and Lv, W. (2010). Development of an indirect ELISA assay for the GST-HSP20(exon) fusion protein of bovine double-budded Babesia germanica. Xinjiang Agric. Sci. 47, 980–985.
Jiang, J. F., Zheng, Y. C., Jiang, R. R., Li, H., Huo, Q. B., Jiang, B. G., et al. (2015). Epidemiological, clinical, and laboratory characteristics of 48 cases of "Babesia venatorum" infection in China: a descriptive study. Lancet Infect. Dis. 15, 196–203. doi: 10.1016/s1473-3099(14)71046-1
Jongejan, F., Du, C., Papadopoulos, E., Blanda, V., Di Bella, S., Cannella, V., et al. (2024). Diagnostic performance of a rapid immunochromatographic test for the simultaneous detection of antibodies to Theileria equi and Babesia caballi in horses and donkeys. Parasit. Vectors 17, 160. doi: 10.1186/s13071-024-06253-1
Kakoullis, L., Alonso, C. D., Burns, R., Aleissa, M. M., Haddad, E. A., Kim, A. J., et al. (2025). Comparative outcomes of babesiosis in immunocompromised and non-immunocompromised hosts: A multicenter cohort study. Clin. Infect. Dis. doi: 10.1093/cid/ciaf034
Kappmeyer, L. S., Perryman, L. E., Hines, S. A., Baszler, T. V., Katz, J. B., Hennager, S. G., et al. (1999). Detection of equine antibodies to babesia caballi by recombinant B. caballi rhoptry-associated protein 1 in a competitive-inhibition enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37, 2285–2290. doi: 10.1128/jcm.37.7.2285-2290.1999
Karbowiak, G., Biernat, B., Stańczak, J., Werszko, J., Szewczyk, T., and Sytykiewicz, H. (2018). The role of particular ticks developmental stages in the circulation of tick-borne pathogens in Central Europe. 6. Babesia. Ann. Parasitol. 64, 265–284. doi: 10.17420/ap6404.162
Krause, P. J., Auwaerter, P. G., Bannuru, R. R., Branda, J. A., Falck-Ytter, Y. T., Lantos, P. M., et al. (2021). Clinical practice guidelines by the infectious diseases society of America (IDSA): 2020 guideline on diagnosis and management of babesiosis. Clin. Infect. Dis. 72, 185–189. doi: 10.1093/cid/ciab050
Krause, P. J., Telford, S. R., Ryan, R., Conrad, P. A., Wilson, M., Thomford, J. W., et al. (1994). Diagnosis of babesiosis: evaluation of a serologic test for the detection of Babesia microti antibody. J. Infect. Dis. 169, 923–926. doi: 10.1093/infdis/169.4.923
Krause, P. J., Telford, S., Spielman, A., Ryan, R., Magera, J., Rajan, T. V., et al. (1996). Comparison of PCR with blood smear and inoculation of small animals for diagnosis of Babesia microti parasitemia. J. Clin. Microbiol. 34, 2791–2794. doi: 10.1128/jcm.34.11.2791-2794.1996
Kumar, S., Kumar, Y., Malhotra, D. V., Dhar, S., and Nichani, A. K. (2003). Standardisation and comparison of serial dilution and single dilution enzyme linked immunosorbent assay (ELISA) using different antigenic preparations of the Babesia (Theileria) equi parasite. Vet Res. 34, 71–83. doi: 10.1051/vetres:2002055
Kumar, B., Maharana, B. R., Thakre, B., Brahmbhatt, N. N., and Joseph, J. P. (2022). 18S rRNA gene-based piroplasmid PCR: an assay for rapid and precise molecular screening of theileria and babesia species in animals. Acta Parasitol. 67, 1697–1707. doi: 10.1007/s11686-022-00625-2
Kumar, A., O'Bryan, J., and Krause, P. J. (2021). The global emergence of human babesiosis. Pathogens 10, 4. doi: 10.3390/pathogens10111447
Leeflang, P. and Perié, N. M. (1972). Comparative immunofluorescent studies on 4 Babesia species of cattle. Res. Vet Sci. 13, 342–346. doi: 10.1016/S0034-5288(18)34012-8
Lei, R., Wang, X., Zhang, D., Liu, Y., Chen, Q., and Jiang, N. (2020). Rapid isothermal duplex real-time recombinase polymerase amplification (RPA) assay for the diagnosis of equine piroplasmosis. Sci. Rep. 10, 4096. doi: 10.1038/s41598-020-60997-1
Lin, H., Zhao, S., Ye, Y., Shao, L., Jiang, N., and Yang, K. (2022). A fluorescent recombinase aided amplification assay for detection of babesia microti. Korean J. Parasitol. 60, 201–205. doi: 10.3347/kjp.2022.60.3.201
Liu, Q., Guan, X. A., Li, D. F., Zheng, Y. X., Wang, S., Xuan, X. N., et al. (2023). Babesia gibsoni whole-genome sequencing, assembling, annotation, and comparative analysis. Microbiol. Spectr. 11, e0072123. doi: 10.1128/spectrum.00721-23
Liu, Q., Zhou, Y. Q., Zhou, D. N., Liu, E. Y., Du, K., Chen, S. G., et al. (2007). Semi-nested PCR detection of Babesia orientalis in its natural hosts Rhipicephalus haemaphysaloides and buffalo. Vet Parasitol. 143, 260–266. doi: 10.1016/j.vetpar.2006.08.016
Loa, C. C., Adelson, M. E., Mordechai, E., Raphaelli, I., and Tilton, R. C. (2004). Serological diagnosis of human babesiosis by IgG enzyme-linked immunosorbent assay. Curr. Microbiol. 49, 385–389. doi: 10.1007/s00284-004-4373-9
Lodes, M. J., Houghton, R. L., Bruinsma, E. S., Mohamath, R., Reynolds, L. D., Benson, D. R., et al. (2000). Serological expression cloning of novel immunoreactive antigens of Babesia microti. Infect. Immun. 68, 2783–2790. doi: 10.1128/iai.68.5.2783-2790.2000
Lu, Y., Zhang, D., Han, D., Yu, F., Ye, X., and Zheng, S. (2024). Case report: diagnosis of hemolytic anemia from babesia and secondary multi-pathogen pneumonia using a metagenomic next-generation sequencing approach. Infect. Drug Resist. 17, 3785–3791. doi: 10.2147/idr.S472861
Luo, Y., Jia, H., Terkawi, M. A., Goo, Y. K., Kawano, S., Ooka, H., et al. (2011). Identification and characterization of a novel secreted antigen 1 of Babesia microti and evaluation of its potential use in enzyme-linked immunosorbent assay and immunochromatographic test. Parasitol. Int. 60, 119–125. doi: 10.1016/j.parint.2010.11.001
Magni, R., Luchini, A., Liotta, L., and Molestina, R. E. (2020). Proteomic analysis reveals pathogen-derived biomarkers of acute babesiosis in erythrocytes, plasma, and urine of infected hamsters. Parasitol. Res. 119, 2227–2235. doi: 10.1007/s00436-020-06712-5
Mahoney, D. F. (1967). Bovine babesiosis: preparation and assessment of complement fixing antigens. Exp. Parasitol. 20, 232–241. doi: 10.1016/0014-4894(67)90043-4
Mandal, M., Banerjee, P. S., Kumar, S., Ram, H., Garg, R., and Pawde, A. M. (2015). Development of loop-mediated isothermal amplification (LAMP) for detection of Babesia gibsoni infection in dogs. Vet Parasitol. 209, 50–55. doi: 10.1016/j.vetpar.2015.02.008
Meeusen, E., Lloyd, S., and Soulsby, E. J. (1985). Antibody levels in adoptively immunized mice after infection with Babesia microti or injection with antigen fractions. Aust. J. Exp. Biol. Med. Sci. 63, 261–272. doi: 10.1038/icb.1985.29
Menis, M., Whitaker, B. I., Wernecke, M., Jiao, Y., Eder, A., Kumar, S., et al. (2021). Babesiosis occurrence among United States medicare beneficiaries, ages 65 and older, during 2006-2017: overall and by state and county of residence. Open Forum Infect. Dis. 8, ofaa608. doi: 10.1093/ofid/ofaa608
Milanović, Z., Beletić, A., Vekić, J., Zeljković, A., Andrić, N., Božović, A. I., et al. (2020). Evidence of acute phase reaction in asymptomatic dogs naturally infected with Babesia canis. Vet Parasitol. 282, 109140. doi: 10.1016/j.vetpar.2020.109140
Mojtahed, A., Bates, D. D. B., and Hahn, P. F. (2020). Splenic findings in patients with acute babesiosis. Abdom. Radiol. (NY) 45, 710–715. doi: 10.1007/s00261-019-02362-z
Montero, E., Folgueras, M., Rodriguez-Pérez, M., Pérez-Ls, L., Díaz-Arias, J., Meana, M., et al. (2023). Retrospective study of the epidemiological risk and serological diagnosis of human babesiosis in Asturias, Northwestern Spain. Parasit. Vectors 16, 195. doi: 10.1186/s13071-023-05817-x
Moritz, E. D., Tonnetti, L., Hewins, M. E., Berardi, V. P., Dodd, R. Y., and Stramer, S. L. (2017). Description of 15 DNA-positive and antibody-negative "window-period" blood donations identified during prospective screening for Babesia microti. Transfusion 57, 1781–1786. doi: 10.1111/trf.14103
Moritz, E. D., Winton, C. S., Tonnetti, L., Townsend, R. L., Berardi, V. P., Hewins, M. E., et al. (2016). Screening for babesia microti in the U.S. Blood supply. N Engl. J. Med. 375, 2236–2245. doi: 10.1056/NEJMoa1600897
Muench, J. R., Jha, P., and Wojtkowski, A. (2023). Babesiosis: an atypical cause of respiratory failure. Cureus 15, e39028. doi: 10.7759/cureus.39028
Muriuki, R., Ndichu, M., Githigia, S., and Svitek, N. (2024). CRISPR-Cas-Based Pen-Side Diagnostic Tests for Anaplasma marginale and Babesia bigemina. Microorganisms 12, 8. doi: 10.3390/microorganisms12122595
Nian, Y., Zhang, S., Wang, J., Li, X., Wang, Y., Liu, J., et al. (2024). A novel and low-cost cross-priming amplification assay for rapid detection of Babesia duncani infection. Exp. Parasitol. 265, 108813. doi: 10.1016/j.exppara.2024.108813
Nicolaiewsky, T. B., Richter, M. F., Lunge, V. R., Cunha, C. W., Delagostin, O., Ikuta, N., et al. (2001). Detection of Babesia equi (Laveran 1901) by nested polymerase chain reaction. Vet Parasitol. 101, 9–21. doi: 10.1016/s0304-4017(01)00471-x
Nie, Z., Zhao, Y., Shu, X., Li, D., Ao, Y., Li, M., et al. (2021). Recombinase polymerase amplification with lateral flow strip for detecting Babesia microti infections. Parasitol. Int. 83, 102351. doi: 10.1016/j.parint.2021.102351
Obeta, S. S., Ibrahim, B., Lawal, I. A., Natala, J. A., Ogo, N. I., and Balogun, E. O. (2020). Prevalence of canine babesiosis and their risk factors among asymptomatic dogs in the federal capital territory, Abuja, Nigeria. Parasite Epidemiol. Control 11, e00186. doi: 10.1016/j.parepi.2020.e00186
Ochi, A., Toya, Y., Sengoku, M., Tsuchiya, S., Kishi, D., and Ueno, T. (2024). In vitro evaluation of the automated hematology analyzer XN-31 for rapid diagnosis of equine piroplasmosis. Microbiol. Spectr. 12, e0058224. doi: 10.1128/spectrum.00582-24
Ogunremi, O., Georgiadis, M. P., Halbert, G., Benjamin, J., Pfister, K., and Lopez-Rebollar, L. (2007). Validation of the indirect fluorescent antibody and the complement fixation tests for the diagnosis of Theileria equi. Vet Parasitol. 148, 102–108. doi: 10.1016/j.vetpar.2007.06.006
Ogunremi, O., Halbert, G., Mainar-Jaime, R., Benjamin, J., Pfister, K., Lopez-Rebollar, L., et al. (2008). Accuracy of an indirect fluorescent-antibody test and of a complement-fixation test for the diagnosis of Babesia caballi in field samples from horses. Prev. Vet Med. 83, 41–51. doi: 10.1016/j.prevetmed.2007.06.009
Okła, H., Jasik, K. P., Słodki, J., Rozwadowska, B., Słodki, A., Jurzak, M., et al. (2014). Hepatic tissue changes in rats due to chronic invasion of Babesia microti. Folia Biol. (Krakow) 62, 353–359. doi: 10.3409/fb62_4.353
Oladokun, R., Adekanmbi, E., Ueti, M., and Srivastava, S. (2023). Dielectric characterization of Babesia bovis using the dielectrophoretic crossover frequency. Electrophoresis 44, 988–1001. doi: 10.1002/elps.202200263
Onchan, W., Ritbamrung, O., Changtor, P., Pradit, W., Chomdej, S., Nganvongpanit, K., et al. (2022). Sensitive and rapid detection of Babesia species in dogs by recombinase polymerase amplification with lateral flow dipstick (RPA-LFD). Sci. Rep. 12, 20560. doi: 10.1038/s41598-022-25165-7
Panti-May, J. A. and Rodríguez-Vivas, R. I. (2020). Canine babesiosis: A literature review of prevalence, distribution, and diagnosis in Latin America and the Caribbean. Vet Parasitol. Reg. Stud. Rep. 21, 100417. doi: 10.1016/j.vprsr.2020.100417
Patiño, L. H., Castañeda, S., Camargo, M., Cao, L. Y., Liggayu, B., Paniz-Mondolfi, A., et al. (2024). Validation of real-time PCR assays for detecting Plasmodium and Babesia DNA species in blood samples. Acta Trop. 258, 107350. doi: 10.1016/j.actatropica.2024.107350
Penzhorn, B. L. (2006). Babesiosis of wild carnivores and ungulates. Vet Parasitol. 138, 11–21. doi: 10.1016/j.vetpar.2006.01.036
Pijnacker, T., Bartels, R., van Leeuwen, M., and Teske, E. (2022). Identification of parameters and formulation of a statistical and machine learning model to identify Babesia canis infections in dogs using available ADVIA hematology analyzer data. Parasit. Vectors 15, 41. doi: 10.1186/s13071-022-05163-4
Pokhil, S. I., Bondarenko, A. V., Bocharova, T. V., Lepilina, K. M., Lytvynenko, M. V., and Gargin, V. V. (2020). Implementation and analysis of Babesia immunoassay testing. Pol. Merkur Lekarski 48, 170–173.
Preena, P., Sarangom, S. B., Ramesh Kumar, K. V., Seeja, S., and Rajalekshmi, S. (2021). Hematological alterations in large Babesia species infection in dogs of Kannur District of Kerala. J. Parasit. Dis. 45, 1090–1095. doi: 10.1007/s12639-021-01404-y
Radzijevskaja, J., Mardosaitė-Busaitienė, D., Aleksandravičienė, A., and Paulauskas, A. (2018). Investigation of Babesia spp. in sympatric populations of Dermacentor reticulatus and Ixodes ricinus ticks in Lithuania and Latvia. Ticks Tick Borne Dis. 9, 270–274. doi: 10.1016/j.ttbdis.2017.09.013
Rezvani, Y., Keroack, C. D., Elsworth, B., Arriojas, A., Gubbels, M. J., Duraisingh, M. T., et al. (2022). Comparative single-cell transcriptional atlases of Babesia species reveal conserved and species-specific expression profiles. PloS Biol. 20, e3001816. doi: 10.1371/journal.pbio.3001816
Rüther, A., Perez-Guaita, D., Poole, W. A., Cooke, B. M., Suarez, C. E., Heraud, P., et al. (2020). Vibrational spectroscopic based approach for diagnosing babesia bovis infection. Anal. Chem. 92, 8784–8792. doi: 10.1021/acs.analchem.0c00150
Saleh, A. M., Adam, S. M., Abdel-Motagaly, A. M., Mohammad, A., Ibrahim, A., and Morsy, T. A. (2015). Human babesiosis: A general review with special reference to Egypt. J. Egypt Soc. Parasitol. 45, 493–510. doi: 10.12816/0017910
Sanchez, E., Vannier, E., Wormser, G. P., and Hu, L. T. (2016). Diagnosis, treatment, and prevention of lyme disease, human granulocytic anaplasmosis, and babesiosis: A review. Jama 315, 1767–1777. doi: 10.1001/jama.2016.2884
Sánchez-Juanes, F., Calvo Sánchez, N., Belhassen García, M., Vieira Lista, C., Román, R. M., Álamo Sanz, R., et al. (2022). Applications of MALDI-TOF mass spectrometry to the identification of parasites and arthropod vectors of human diseases. Microorganisms 10, 9. doi: 10.3390/microorganisms10112300
Santos, M. A., Tierney, L. M., Jr., and Manesh, R. (2020). Babesiosis-associated warm autoimmune hemolytic anemia. J. Gen. Intern. Med. 35, 928–929. doi: 10.1007/s11606-019-05506-5
Schmidt, K., Mwaigwisya, S., Crossman, L. C., Doumith, M., Munroe, D., Pires, C., et al. (2017). Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 72, 104–114. doi: 10.1093/jac/dkw397
Scott, J. D. and Scott, C. M. (2018). Human Babesiosis Caused by Babesia duncani Has Widespread Distribution across Canada. Healthc. (Basel) 6, 2. doi: 10.3390/healthcare6020049
Shah, J. S., Mark, O., Caoili, E., Poruri, A., Horowitz, R. I., Ashbaugh, A. D., et al. (2020). A fluorescence in situ hybridization (FISH) test for diagnosing babesiosis. Diagn. (Basel) 10. doi: 10.3390/diagnostics10060377
Shah, J. S. and Ramasamy, R. (2022). Fluorescence in situ hybridization (FISH) tests for identifying protozoan and bacterial pathogens in infectious diseases. Diagn. (Basel) 12, 6. doi: 10.3390/diagnostics12051286
Si, W., Fang, C., Liu, C., Yin, M., Xu, W., Li, Y., et al. (2023). Why is Babesia not killed by artemisinin like Plasmodium? Parasit. Vectors 16, 193. doi: 10.1186/s13071-023-05783-4
Singh, P., Lonardi, S., Liang, Q., Vydyam, P., Khabirova, E., Fang, T., et al. (2023). Babesia duncani multi-omics identifies virulence factors and drug targets. Nat. Microbiol. 8, 845–859. doi: 10.1038/s41564-023-01360-8
Sivakumar, T., Tuvshintulga, B., Zhyldyz, A., Kothalawala, H., Yapa, P. R., Kanagaratnam, R., et al. (2018). Genetic analysis of babesia isolates from cattle with clinical babesiosis in Sri Lanka. J. Clin. Microbiol. 56, 3. doi: 10.1128/jcm.00895-18
Stoltsz, H., Byaruhanga, C., Troskie, M., Makgabo, M., Oosthuizen, M. C., Collins, N. E., et al. (2020). Improved detection of Babesia bigemina from various geographical areas in Africa using quantitative PCR and reverse line blot hybridisation. Ticks Tick Borne Dis. 11, 101415. doi: 10.1016/j.ttbdis.2020.101415
Stuart Tayebwa, D., Magdy Beshbishy, A., Batiha, G. E., Komugisha, M., Joseph, B., Vudriko, P., et al. (2020). Assessing the immunochromatographic test strip for serological detection of bovine babesiosis in Uganda. Microorganisms 8, 5. doi: 10.3390/microorganisms8081110
Suarez, C. E., Palmer, G. H., Florin-Christensen, M., Hines, S. A., Hötzel, I., and McElwain, T. F. (2003). Organization, transcription, and expression of rhoptry associated protein genes in the Babesia bigemina rap-1 locus. Mol. Biochem. Parasitol. 127, 101–112. doi: 10.1016/s0166-6851(02)00311-0
Swei, A., O'Connor, K. E., Couper, L. I., Thekkiniath, J., Conrad, P. A., Padgett, K. A., et al. (2019). Evidence for transmission of the zoonotic apicomplexan parasite Babesia duncani by the tick Dermacentor albipictus. Int. J. Parasitol. 49, 95–103. doi: 10.1016/j.ijpara.2018.07.002
Tagliafierro, T., Joshi, S., Sameroff, S., Marques, A., Dumler, J. S., Mishra, N., et al. (2022). Detection of antibodies to Anaplasma phagocytophilum and Babesia microti using linear peptides. Ticks Tick Borne Dis. 13, 101999. doi: 10.1016/j.ttbdis.2022.101999
Thekkiniath, J., Mootien, S., Lawres, L., Perrin, B. A., Gewirtz, M., Krause, P. J., et al. (2018). BmGPAC, an antigen capture assay for detection of active babesia microti infection. J. Clin. Microbiol. 56, 5. doi: 10.1128/jcm.00067-18
Tian, Z., Du, J., Yang, J., Liu, A., Liu, X., Liu, G., et al. (2015). A PCR-RFLP Assay targeting RPS8 gene for the discrimination between bovine Babesia and Theileria species in China. Parasit. Vectors 8, 475. doi: 10.1186/s13071-015-1085-x
Tijani, M. K., Svensson, J., Adlerborn, P., Danielsson, L., Teleka, A., Lövmar, M. L., et al. (2024). How to detect antibodies against babesia divergens in human blood samples. Open Forum Infect. Dis. 11, ofae028. doi: 10.1093/ofid/ofae028
Torianyk, I. I. (2021a). Biological method for babesiosis detection: the unified version in vivo. Wiad Lek 74, 268–272. doi: 10.36740/WiadLek
Torianyk, I. I. (2021b). Morphofunctional changes in the spleen of dogs as the components of pathogenetic substantiation of babesiosis. Pol. Merkur Lekarski 49, 416–419.
Trindade, A. S. N., de Moraes, I. M. L., Leal, L., Salvador, V. F., Heller, L. M., Zapa, D. M. B., et al. (2025). TFinder App: Artificial intelligence to diagnose tick fever agents and assess parasitemia/bacteremia in bovine blood smears. Vet Parasitol. 334, 110415. doi: 10.1016/j.vetpar.2025.110415
Tsuji, M., Terada, Y., Arai, S., Okada, H., and Ishihara, C. (1995). Use of the Bo-RBC-SCID mouse model for isolation of a Babesia parasite from grazing calves in Japan. Exp. Parasitol. 81, 512–518. doi: 10.1006/expr.1995.1144
Vanderboom, P. M., Misra, A., Rodino, K. G., Eberly, A. R., Greenwood, J. D., Morris, H. E., et al. (2024). Detection and quantification of Babesia species intraerythrocytic parasites by flow cytometry. Am. J. Clin. Pathol. 161, 451–462. doi: 10.1093/ajcp/aqad168
Vannier, E. and Krause, P. J. (2012). Human babesiosis. N Engl. J. Med. 366, 2397–2407. doi: 10.1056/NEJMra1202018
Vannier, E. and Krause, P. J. (2015). Babesiosis in China, an emerging threat. Lancet Infect. Dis. 15, 137–139. doi: 10.1016/s1473-3099(14)71062-x
Venter, A., Vorster, I., Nkosi, N. F., Sibeko-Matjila, K. P., and Bhoora, R. V. (2024). Molecular genotyping of Babesia caballi. Vet Parasitol. 329, 110214. doi: 10.1016/j.vetpar.2024.110214
Verma, N., Puri, A., Essuman, E., Skelton, R., Anantharaman, V., Zheng, H., et al. (2020). Antigen discovery, bioinformatics and biological characterization of novel immunodominant babesia microti antigens. Sci. Rep. 10, 9598. doi: 10.1038/s41598-020-66273-6
Vijayvargiya, P., Jeraldo, P. R., Thoendel, M. J., Greenwood-Quaintance, K. E., Esquer Garrigos, Z., Sohail, M. R., et al. (2019). Application of metagenomic shotgun sequencing to detect vector-borne pathogens in clinical blood samples. PloS One 14, e0222915. doi: 10.1371/journal.pone.0222915
Wang, J., Chen, K., Yang, J., Zhang, S., Li, Y., Liu, G., et al. (2022). Comparative genomic analysis of Babesia duncani responsible for human babesiosis. BMC Biol. 20, 153. doi: 10.1186/s12915-022-01361-9
Wang, J., Gao, S., Zhang, S., He, X., Liu, J., Liu, A., et al. (2020b). Rapid detection of Babesia motasi responsible for human babesiosis by cross-priming amplification combined with a vertical flow. Parasit. Vectors 13, 377. doi: 10.1186/s13071-020-04246-4
Wang, H., Wang, Y., Huang, J., Xu, B., Chen, J., Dai, J., et al. (2020a). Babesia microti protein bmSP44 is a novel protective antigen in a mouse model of babesiosis. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01437
Webi, E., Abkallo, H. M., Obiero, G., Ndegwa, P., Xie, S., Zhao, S., et al. (2024). Genome editing in apicomplexan parasites: current status, challenges, and future possibilities. Crispr. J. 7, 310–326. doi: 10.1089/crispr.2024.0032
Welzl, C., Leisewitz, A. L., Jacobson, L. S., Vaughan-Scott, T., and Myburgh, E. (2001). Systemic inflammatory response syndrome and multiple-organ damage/dysfunction in complicated canine babesiosis. J. S Afr. Vet Assoc. 72, 158–162. doi: 10.4102/jsava.v72i3.640
Wilhelmsson, P., Lövmar, M., Krogfelt, K. A., Nielsen, H. V., Forsberg, P., and Lindgren, P. E. (2020). Clinical/serological outcome in humans bitten by Babesia species positive Ixodes ricinus ticks in Sweden and on the Åland Islands. Ticks Tick Borne Dis. 11, 101455. doi: 10.1016/j.ttbdis.2020.101455
Wilkowsky, S. E., Farber, M., Echaide, I., Torioni de Echaide, S., Zamorano, P. I., Dominguez, M., et al. (2003). Babesia bovis merozoite surface protein-2c (MSA-2c) contains highly immunogenic, conserved B-cell epitopes that elicit neutralization-sensitive antibodies in cattle. Mol. Biochem. Parasitol. 127, 133–141. doi: 10.1016/s0166-6851(02)00329-8
Wudhikarn, K., Perry, E. H., Kemperman, M., Jensen, K. A., and Kline, S. E. (2011). Transfusion-transmitted babesiosis in an immunocompromised patient: a case report and review. Am. J. Med. 124, 800–805. doi: 10.1016/j.amjmed.2011.03.009
Xu, B., Liu, X. F., Cai, Y. C., Huang, J. L., Zhang, R. X., Chen, J. H., et al. (2018). Screening for biomarkers reflecting the progression of Babesia microti infection. Parasit. Vectors 11, 379. doi: 10.1186/s13071-018-2951-0
Yalcindag, E., Vasoya, D., Hemmink, J. D., Karani, B., Hernandez Castro, L. E., Callaby, R., et al. (2024). Development of a novel Haemabiome tool for the high-throughput analysis of haemopathogen species co-infections in African livestock. Front. Vet Sci. 11. doi: 10.3389/fvets.2024.1491828
Yang, Y., Li, Q., Wang, S., Chen, X., and Du, A. (2016). Rapid and sensitive detection of Babesia bovis and Babesia bigemina by loop-mediated isothermal amplification combined with a lateral flow dipstick. Vet Parasitol. 219, 71–76. doi: 10.1016/j.vetpar.2016.02.004
Keywords: Babesia, diagnostic techniques, serological detection, molecular biology, research advances
Citation: Jia Z, Zhang Y, Zhao D, Wang H, Yu M, Liu Z, Zhang X, Cui J and Wang X (2025) Research progress on diagnostic techniques for different Babesia species in persistent infections. Front. Cell. Infect. Microbiol. 15:1575227. doi: 10.3389/fcimb.2025.1575227
Received: 12 February 2025; Accepted: 24 April 2025;
Published: 16 May 2025.
Edited by:
Sudhir Kumar, Iowa State University, United StatesReviewed by:
Hong-Juan Peng, Southern Medical University, ChinaMansoor Azeem Siddiqui, National Institutes of Health (NIH), United States
Copyright © 2025 Jia, Zhang, Zhao, Wang, Yu, Liu, Zhang, Cui and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueli Wang, d2FuZ3hsOTU3N0BhbGl5dW4uY29t
†These authors share first authorship
 Zelin Jia
Zelin Jia Yuliang Zhang2†
Yuliang Zhang2† Zhilin Liu
Zhilin Liu Xin Zhang
Xin Zhang Jiayu Cui
Jiayu Cui Xueli Wang
Xueli Wang