- 1College of Life and Health Sciences, Northeastern University, Shenyang, Liaoning, China
- 2Department of Emergency, Xinqiao Hospital, Army Medical University, Chongqing, China
Fungal infections are becoming more prevalent globally, particularly affecting immunocompromised populations, such as people living with HIV, organ transplant recipients and those on immunomodulatory therapy. Globally, approximately 6.55 million people are affected by invasive fungal infections annually, leading to serious health consequences and death. Mitochondria are membrane-bound organelles found in almost all eukaryotic cells and play an important role in cellular metabolism and energy production, including pathogenic fungi. These organelles possess their own genome, the mitochondrial genome, which is usually circular and encodes proteins essential for energy production. Variation and evolutionary adaptation within and between species’ mitochondrial genomes can affect mitochondrial function, and consequently cellular energy production and metabolic activity, which may contribute to pathogenicity and drug resistance in certain fungal species. This review explores the link between the mitochondrial genome and mechanisms of fungal pathogenicity and drug resistance, with a particular focus on Cryptococcus neoformans and Candida albicans. These insights deepen our understanding of fungal biology and may provide new avenues for developing innovative therapeutic strategies.
Introduction
The rising incidence of fungal infections, with morbidity and mortality rates rising globally, poses a significant public health challenge (Tu et al., 2021; Fisher et al., 2022; Roe, 2023; Denning, 2024). These infections are not only causing substantial health losses but also imposing a considerable financial burden. In the United States alone, direct medical costs are estimated to reach billions of dollars (Koffi et al., 2021; Iliev et al., 2024). The widespread use of antifungal agents has led to the emergence of drug resistance, which has now become a critical public health concern. This resistance not only limits treatment options for patients but also increases the risk of recurrent infections and mortality (Kontoyiannis and Lewis, 2002; Lockhart et al., 2023; Vitiello et al., 2023; Zhang et al., 2023). The World Health Organization (WHO) has released its first Fungal Priority Pathogen List (FPPL), focusing on the most threatening global fungal threats. This has been a major concern for the scientific community, calling on the global research community to strengthen research on fungal infections and drug resistance (Fisher and Denning, 2023; Casalini et al., 2024).
Mitochondria are important organelles in eukaryotic cells, with the role of ATP production and participation in various cellular processes (Singh, 2021; Ziegler et al., 2021; Borcherding and Brestoff, 2023; Ryu et al., 2024). Mitochondria possess their own genomes, termed the mitochondrial genomes, which enable them to synthesize a subset of proteins independently of the nucleus (Sandor et al., 2018; Fu et al., 2020). These proteins play an essential role in cellular energy metabolism, the respiratory chain, and various essential biochemical processes (Wallace, 2016; Kim et al., 2022; Liu et al., 2022; Vercellino and Sazanov, 2022). As eukaryotes evolved, the mitochondrial genome appeared to vary considerably among the major eukaryotic taxa (Zardoya, 2020). In animals, the mitochondrial genomes are typically closed-loop DNA molecules encoding a small number of genes involved in energy production (Boore, 1999). Animal mitochondrial genomes generally lack intergenic regions and introns, rendering them relatively simple (Miyazawa et al., 2021; Pimentel et al., 2024). However, exceptions have been reported in some metazoans with regenerative abilities, such as placozoans and bryozoans (Jenkins et al., 2022). In contrast, the mitochondrial genome of plants is primarily in the form of closed circular DNA molecules containing intergenic regions and varying numbers of introns (Kan et al., 2020; Grosser et al., 2023). Fungal mitochondrial genomes are more similar to plant mitochondria than to animal mitochondria in terms of structure and composition (Sandor et al., 2018). For example, Saccharomyces cerevisiae is the first fungus reported to have a cyclic mitochondrial genome and has been extensively studied (Lipinski et al., 2010). Research related to the role of the mitochondrial genome in human fungal pathogens is also on the rise. In pathogenic fungi, the functionality of mitochondria directly influences their physiological status and pathogenic potential (Shingu-Vazquez and Traven, 2011; Gao et al., 2022; Alves and Gourlay, 2024). Mitochondrial proteins are critical for establishing infection through energy metabolism and efficient oxidative stress responses (Bambach et al., 2009; Verma et al., 2018; Soggiu et al., 2019; Zhai et al., 2021). There is also a link between mitochondria and the resistance to antibiotic drugs in pathogenic fungi (Shingu-Vazquez and Traven, 2011; Li et al., 2020; Song et al., 2020; Zhang et al., 2021). The introns of the mitochondrial genome, the stability of the genome, and the generation of reactive oxygen species (ROS) as byproducts can all influence the ability of pathogenic fungi to infect hosts and resist antibiotic drugs (Yildiz and Ozkilinc, 2020; Liu and Pyle, 2021, 2024). In this review, we will focus on outlining the composition and diversity of the mitochondrial genome of pathogenic fungi, the relationship between fungal mitochondrial genome and pathogenicity, and the impact of fungal mitochondrial genome on their drug resistance, with a particular focus on the human opportunistic pathogens Cryptococcus neoformans and Candida albicans. This review provides valuable information for future studies of fungal mitochondrial systems as well as innovative new therapeutic strategies.
Mitochondrial genome structure and diversity in pathogenic fungi
Fungal mitochondrial genomes typically exhibit a single circularly mapped chromosome, near-standard bacterial-like tRNA and rRNA structures, and a minimal genetic code (Fonseca et al., 2021; Tang et al., 2024). These mitochondrial genomes encode essential genes required for oxidative phosphorylation and other mitochondrial functions (Lavin et al., 2008; Sandor et al., 2018). Variations in core gene count and organization, along with intergenic region sequences, considerably affect the size of fungal mitochondrial genomes (Aguileta et al., 2014; Franco et al., 2017; Korovesi et al., 2018; de Almeida et al., 2021; Tang et al., 2024). Animal mitochondrial genomes are relatively small, ranging from 14 kb to 20 kb, while plant mitochondrial genomes are larger, ranging from 180 kb to 600 kb (Boore, 1999; Morley and Nielsen, 2017). Fungal mitochondrial genomes fall between these two size ranges, varying from 11,223 bp in Hanseniaspora pseudoguilliermondii to 332,165 bp in Golovinomyces cichoracearum (Zaccaron and Stergiopoulos, 2021; Christinaki et al., 2022). Most proteins encoded by fungal mitochondrial genomes are key components of oxidative phosphorylation. Nevertheless, there are obvious differences in the amount of these proteins at the species level. For example, seven proteins are encoded in the mitochondrial genome of the model yeast S. cerevisiae (Foury et al., 1998; De Chiara et al., 2020) or the related human pathogen Candida glabrata (Koszul et al., 2003). However, the mitochondrial genomes of the pathogens Cryptococcus deneoformans and Cryptococcus deuterogattii encode 13 proteins (Ma and May, 2010), and the mitochondrial genomes of Aspergillus and Penicillium encode 14 proteins (Joardar et al., 2012). Introns are the main drivers of size variations in fungal mitochondrial genomes. They are widely present in these genomes and are generally classified into Group I and Group II based on RNA secondary structure (Saldanha et al., 1993; Gomes et al., 2023). Variation in intron content is observed even within the same fungal genus. The number of introns, including the COX1, COX2, COX3, NAD1 and NAD5 genes, varies considerably between species and occasionally within a single species. Notably, the COX1 gene is an important reservoir for introns (Ye et al., 2020; Tan et al., 2022), known as a key place where introns are stored. The heterogeneity of mitochondrial introns is closely linked to species evolution. In genera like Aspergillus and Penicillium, intron addition and deletion occur in a cyclic manner (Nielsen et al., 2004; Joardar et al., 2012). In the Cordyceps militaris strain, each intron in the mitochondrial genome may have experienced one to four gain and/or loss events (Zhang et al., 2015). Research indicates that, although the large subunit of ribosomal RNA (LSU rRNA) genes in the majority of fungal species contain highly conserved introns, certain Candida species do not possess these introns. This observation implies a correlation between the presence of introns and evolutionary development, emphasizing important variability both among different species and within individual species (Miletti and Leibowitz, 2000; Krol et al., 2019). A recent study used an RNA structure-based bioinformatics approach to identify Group I introns in key mitochondrial genes of pathogenic fungi, including all high-priority pathogens identified by the World Health Organization. The study also assessed the prevalence of these introns in different fungal phylogenies and their fixation within a few genetic hotspots (Liu and Pyle, 2024). It was also found that in C. albicans and Candida auris, the splicing catalysis of these introns demonstrated greater efficiency compared to previously identified group I introns. Also, these introns exhibited rapid catalytic turnover under ambient temperatures and physiological concentrations of magnesium ions (Liu and Pyle, 2024). This finding implies a potential close relationship between introns and the environmental adaptability of pathogenic fungi.
The human pathogenic fungus C. neoformans possesses a mitochondrial genome that ranges in size from 24,740 to 31,327 base pairs (Wang and Xu, 2020). The mitochondrial genome contains 17 genes, exhibiting a conserved gene order across closely related species, such as C. deneoformans and Cryptococcus gattii (Litter et al., 2005; Ma and May, 2010). This genome encompasses genes responsible for proteins involved in the electron transport chain, ATP synthesis, and genes encoding the mitochondrial ribosomal protein S3 (Rps3) and the small and large subunits of mitochondrial ribosomal RNA (Wang and Xu, 2020). Comparative genomics has revealed that a 40 kb region containing 14 genes was transferred from var. grubii to var. neoformans (Kavanaugh et al., 2006). These two varieties diverged approximately 18 million years ago (MYA) and share 85-90% nucleotide identity at the genomic level. This nearly identical region is prevalent in most clinical and environmental var. neoformans strains globally and the result of a non-reciprocal transfer event from var. grubii to var. neoformans about 2 MYA. This genetic exchange likely occurred through a hybrid intermediate, potentially formed by incomplete sexual cycling between the variants (Kavanaugh et al., 2006). A study by Yue Wang et al. analyzed 184 sequenced mitochondrial genomes from C. neoformans isolates and revealed variations in size, intron distribution, and single nucleotide polymorphisms in the mitochondrial genomes of Cryptococcus complex (Wang and Xu, 2020). This genetic diversity is likely associated with the fungal geographic distribution, ecological niche, mating type, and genetic lineage. These variability traits of mitochondrial genomes provide important sights into the genetic diversity and evolution of Cryptococcus (Wang and Xu, 2020). The mitochondrial genome of C. neoformans shows evidence of frequent intron gains and losses throughout its evolutionary history, as indicated by the distribution and phylogenies of introns and their corresponding exons. The occurrence of self-scissoring introns within mitochondrial genes differs across various complexes and genotypes, presumably as a result of recurrent intron loss and gain events during the homing mechanism (Gomes et al., 2023). In the mitochondrial genomes of C. neoformans and C. gattii, Group I introns are primarily distributed in the COB and COX1 genes, with varying frequencies among different genotypes and species. For example, the VNIII genotype, a cross between C. neoformans and C. deneoformans, exhibits the highest intron frequencies in four loci of mtLSU, two loci of COB, and five loci of COX1. In contrast, lower intron frequencies were observed in the more virulent and frequently isolated VNI genotype in clinical samples worldwide (Gomes et al., 2023). Phylogenetic analyses of COB and COX1 introns reveal that introns occupying the same insertion site form well-defined monophyletic groups. Introns located at the first insertion site in COB exhibit greater similarities to one another, whereas those at the second insertion site reveal a similar pattern (Gomes et al., 2023). A survey of Group I introns in the mitochondrial LSU rRNA genes of 77 C. neoformans and C. gattii isolates identified two new introns in the LSU rRNA genes. These introns form a monophyletic group closely related to the COX1 introns of certain ascomycete and basidiomycete genera. The structures and sequences of these introns differ from known introns, and their presence is highly associated with pathogenicity and antifungal resistance, suggesting a unique evolutionary history (Gomes et al., 2018).
C. albicans is a common commensal and opportunistic pathogenic fungus, and its mitochondrial DNA molecule is 41 kb in size (Jones et al., 2004). This dimorphic fungus is petite-negative, meaning that the absence of mitochondrial DNA renders it non-viable (Bulder, 1964; Chen and Clark-Walker, 2000; Joers et al., 2007). Its mitochondrial genome contains genes encoding complex I subunits that have been lost in closely related fungi, such as in S. cerevisiae and Saccharomyces pombe (Nosek and Fukuhara, 1994; Anderson et al., 2001). The mitochondrial genome of C. albicans has a GC content of approximately 32%, a characteristic shared by many other yeasts (Kolondra et al., 2015), suggesting that it has been conserved to some extent during evolution. Within this mitochondrial genome, GC-enriched fragments are typically dispersed as clusters of short palindromic sequences that may adopt hairpin structures, serving as mobile genetic elements and regulatory signals (Kolondra et al., 2015). The mitochondrial genome of C. albicans primarily exists as multiple head-to-tail tandems and includes 14 genes for subunits of the oxidative phosphorylation pathway, two ribosomal RNA genes, and 24 tRNA genes (Gerhold et al., 2010; Kolondra et al., 2015). The protein-coding genes encompass subunits of NADH dehydrogenase (Complex I), cytochrome c oxidase (Complex IV), and ATP synthase (Complex V). The mitochondrial genome has a limited number of introns, specifically two in the RNL gene, two in the COB gene, and four in the COX1 gene (Kolondra et al., 2015). The genome consists of two main coding regions: the short coding region (SCR) and the long coding region (LCR), along with two inverted repeat regions (IRa and IRb). These non-coding regions exhibit limited transcriptional activity in RNA sequencing, confined to a few short segments (Kolondra et al., 2015). In C. albicans, mitochondrial intergenic regions are useful for examining microvariation (Bartelli et al., 2013). A study conducted the sequencing of mitochondrial genomes from two clinical isolates of C. albicans, revealing 372 polymorphic sites. Of these, 230 were located within coding regions and 142 within non-coding regions, indicating a notable presence of neutral substitutions. The high variability and size differences in these non-coding regions, with up to a 56 bp size difference, indicate that they may be subject to neutral evolution (Bartelli et al., 2013). Another study sequenced seven regions of the mitochondrial genomes from 36 C. albicans strains and identified 66 polymorphic sites, resulting in the construction of 19 distinct haplotypes. Notably, strains sharing the same mitochondrial haplotypes were found across various geographic regions. The absence of a strong correlation between mitochondrial haplotypes and the origin of strains, whether geographic or anatomical, indicates considerable mobility among various populations (Jacobsen et al., 2008). These findings suggest that the distribution of mitochondrial haplotypes in C. albicans is probably independent of geographic distribution or host specificity. Therefore, caution is advised when employing mitochondrial haplotypes in phylogenetic studies of C. albicans. A study of mitochondrial genomes from sequenced C. albicans isolates representing different sites of infection and countries of origin showed that, in addition to the nuclear genome, the mitochondrial genome also undergoes genetic recombination. This indicates that the mitochondrial genome of C. albicans exhibits variability, influenced by sexual or parasexual mating processes (Wang et al., 2018).
Mitochondrial genome and pathogenic mechanisms
Human fungal pathogens are responsible for severe, life-threatening diseases and predominantly exhibit aerobic characteristics. Therefore, the morphology, genetics, and metabolism of their mitochondria are essential for their survival in environmental contexts and during infections in hosts (Alspaugh, 2015; Chang and Doering, 2018; Song et al., 2020; Rokas, 2022; Thambugala et al., 2024). Mitochondrial dynamics, including their fusion and fission, can impact fungal virulence (Chang and Doering, 2018; Liang et al., 2018). For instance, in C. albicans, loss of FZO1, which is involved in mitochondrial fusion, leads to increased susceptibility to peroxide stress and altered drug susceptibility (Thomas et al., 2013). Similarly, in Aspergillus fumigatus, a conditional mutant strain of the essential gene Mgm1, encoding another GTPase required for mitochondrial fusion, was completely devoid of virulence (Neubauer et al., 2015). Thus, understanding the functional diversity of fungal mitochondria can provide valuable insights into their biology and pathogenic mechanisms, potentially leading to more effective treatments for fungal infections.
The mitochondrial function of C. neoformans is directly related to the pathogenicity (Figure 1). Specifically, the disruption of mitochondrial fusion attenuates the resistance to oxidative and nitrosative stresses and the virulence of C. neoformans (Chang and Doering, 2018). The correlation between the mitochondrial genomes of pathogenic fungi and their virulence is complex. Abnormal expression of mitochondrial genome genes can affect the pathogenicity of Cryptococcus. The increased atypical expression of the mitochondrial genome-encoded NADH dehydrogenase gene, caused by a mutation of the NADH dehydrogenase subunit 1 promoter, leads to heightened ATP production, accelerated metabolic activity, and enhanced virulence in Cryptococcus (Merryman et al., 2020). Besides, the recombination of mitochondrial genomes may affect the virulence of fungal populations. In Cryptococcus species, there is a recombination event of mitochondrial genome in every VNIa-5 isolate, which is associated with disease in HIV-uninfected patients, but not in every non-VNIa-5 isolate, indicating mitochondrial genome recombination may contribute to the emergence of more virulent strains (Ashton et al., 2019). Recent findings indicate that homing endonuclease genes contribute to variations in intron sizes within the mitochondrial LSU rRNA gene of Cryptococcus species. These differences may play a role in mitochondrial functionality, potentially affecting virulence (Gomes et al., 2018). The study by Gomes et al. identified two novel introns in the mitochondrial LSU rRNA gene of C. neoformans. Notably, the presence of these introns was statistically associated with genotypes reported to be less pathogenic, indicating a potential link between intron presence and reduced virulence (Gomes et al., 2018). Another study revealed evidence of frequent gains and losses of mitochondrial introns during the evolution of C. neoformans (Gomes et al., 2023). This indicates a dynamics evolutionary process that could be associated with the fungal adaptation to various host environments and its potential for pathogenicity. Introns within mitochondrial genes can influence the expression of proteins essential for energy production and the response to oxidative stress, both of which are essential for the survival of C. neoformans in hostile environments (Fox, 2012; Chang and Doering, 2018; Gomes et al., 2023). The mitochondrial genome is involved in the production of ROS and the ability to manage excessive ROS is also critical for C. neoformans to survive the hostile environment (Meng and Ding, 2023). The overexpression of the protein encoded by the mitochondrial genome notably enhanced resistance to increased levels of ROS induced by heat stress, enabling C. neoformans to endure high temperatures during host infection (Gao et al., 2022). Nevertheless, the complex relationship between the mitochondrial genome and the virulence of Cryptococcus remains largely known and requires further investigation.
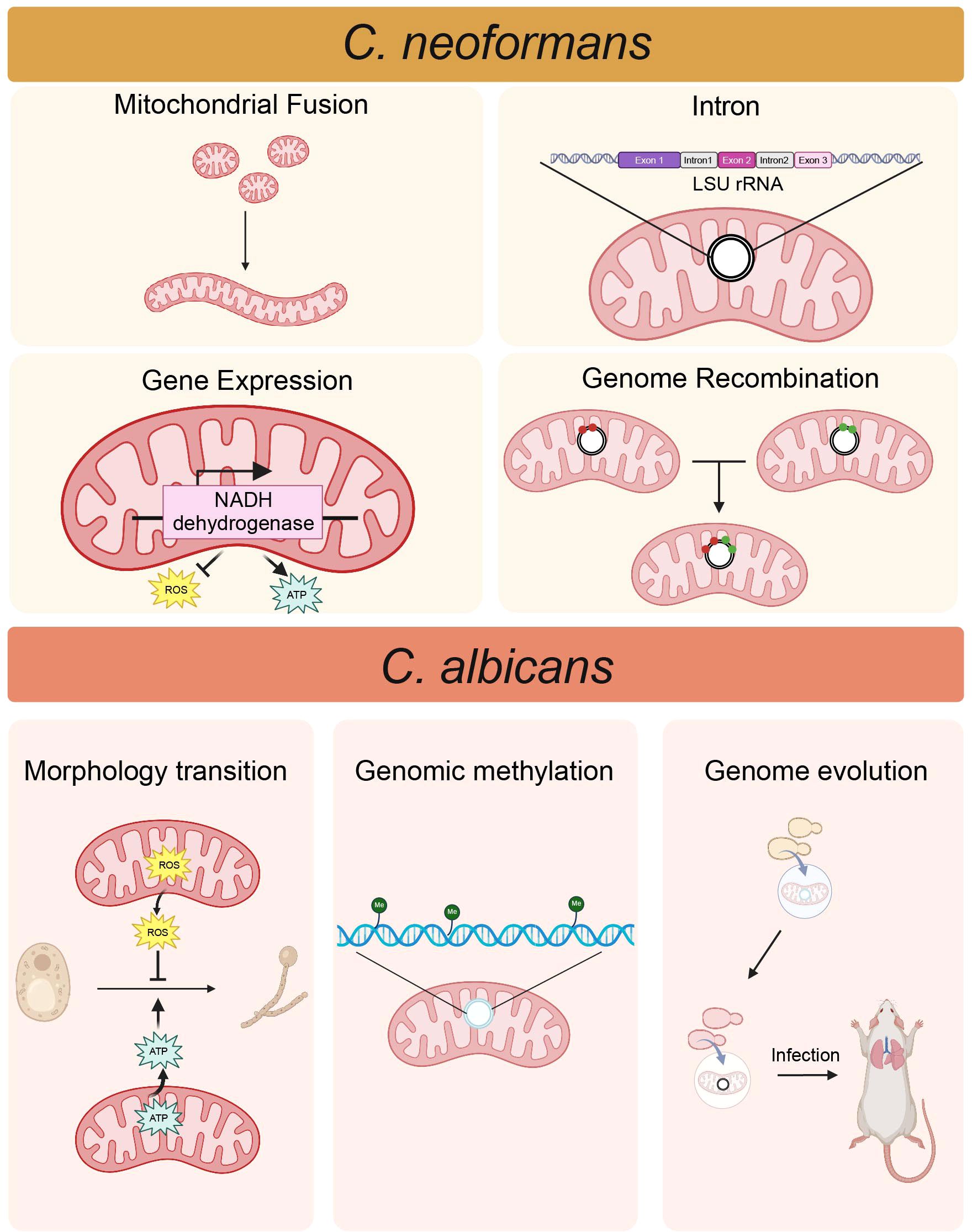
Figure 1. The correlation between mitochondria and the pathogenicity of fungi. In C. neoformans, the process of mitochondrial fusion is associated with pathogenicity. Elevated expression of the mitochondrial genome-encoded NADH dehydrogenase gene increases ATP levels while reducing ROS level, thereby enhancing virulence. Mitochondrial genome recombination may facilitate the emergence of more virulent strains. The presence of two novel introns in the mitochondrial LSU rRNA gene correlates with attenuated pathogenicity. In C. albicans, the yeast-to-hypha transition necessitates high ATP level, and ROS can inhibit this process. Mitochondrial genome evolution may contribute to the important consequences for host adaption. Mitochondrial genome methylation, recognized as a new epigenetic mechanism, can drive adaptive changes in the mitochondrial genome. (Created with BioRender.com).
In C. albicans, genetic diversity within the mitochondrial genomes can have a substantial impact on its pathogenicity by altering the efficiency of energy metabolism, morphological transitions, and responses to oxidative stress (Noble et al., 2010; Bonhomme et al., 2011; Brown et al., 2014; Dantas Ada et al., 2015; Grahl et al., 2015) (Figure 1). Energy metabolism plays a critical role during the transition from yeast to hyphae, a process that requires substantial ATP levels and is essential for tissue invasion and damage within the host (Alonso-Monge et al., 2024). It has been reported that reduced respiratory capacity due to mutations in the mitochondrial complex of C. albicans attenuates its fitness and virulence (Sun et al., 2019). Moreover, mitochondrial mutants of C. albicans exhibited reduced virulence in a mouse model of disseminated disease, while the virulence of all respiratory mutants was similarly diminished in the Galleria mellonella infection model (Bambach et al., 2009; Becker et al., 2010; She et al., 2015; Sun et al., 2019). Biofilm is the key virulence factor for C. albicans and linked to increased resistance to oxidative stress due to the production of ROS, with mitochondria being the primary source of ROS in eukaryotic cells (Seneviratne et al., 2008; Gulati and Nobile, 2016; Pemmaraju et al., 2016), which indicates a link between mitochondrion and biofilm formation. In addition, mitochondria function as the antioxidant defense system, safeguarding the cell from oxidative damage due to immune system in C. albicans. In C. albicans strains exhibiting compromised mitochondrial function, there is a decrease in the copy number of the mitochondrial genome, accompanied by an increase in ROS levels. This abnormal ROS level compromises hyphal formation and alters interaction with macrophages, thereby reducing the virulence of the infection (Liang et al., 2018). Genome evolution in C. albicans, including changes in the mitochondrial genome such as the accumulation of point mutations, loss of heterozygosity (LOH) events, large-scale chromosomal rearrangements, and even ploidy change, may enhance its ability to survive and reproduce in different host environments (Selmecki et al., 2006; Wu et al., 2007; Ford et al., 2015; Bartelli et al., 2018; Ene et al., 2019). Mitochondrial genome methylation in C. albicans has been identified as a novel epigenetic mechanism for the adaptive changes in the mitochondrial genome (Bartelli et al., 2018). Researchers have observed that environmental conditions, such as continuous exposure to hypoxia and 37°C, can decrease mitochondrial genome methylation in strains SC5314 and L757 (Bartelli et al., 2018). This process is closely related to the pathogenicity of Candida, as strains that are able to grow and reproduce rapidly are more likely to cause infections. This decreased methylation varies across strains at specific genome locations (Bartelli et al., 2018), suggesting that this response may be lineage-specific and related to the adaptation and different pathogenicity of these strains. However, more direct evidence is needed to further validate whether the altered methylation levels of these loci can directly affect the pathogenicity and environmental adaptability of C. albicans.
Mitochondrial genome and drug resistance
Fungal drug resistance presents a serious and growing challenge in the medical field (Fisher et al., 2022). This resistance is a complex phenomenon influenced by multiple mechanisms that enable fungi to endure and flourish in the presence of antifungal agents. Acquired resistance often involves mutations in target protein binding sites, alterations in drug targets, and the activation of drug efflux mechanisms (Lockhart et al., 2023; Zhang et al., 2023). The mitochondrial function is closely related to the drug resistance in fungi. In S. cerevisiae, the lack of the mitochondrial genome results in an obvious upregulation of PDR5, a gene that encodes an ATP-binding cassette (ABC) transporter essential for the efflux of antifungal agents, finally enhancing drug resistance (Moye-Rowley, 2005; Rahman et al., 2018; Harris et al., 2021). For pathogenic fungi, such as C. glabrata, the loss of the mitochondrial genome can result in increased drug resistance (Sanglard et al., 2001; Batova et al., 2008; Ferrari et al., 2011). In A. fumigatus, mitochondrion fission mutant strains exhibit increased resistance to azole drugs (Valero et al., 2020). The loss of mitochondrial genome can also disrupt lipid biosynthesis and subsequently alter membrane permeability, influencing the fungal response to antifungal agents (Calderone et al., 2015; Rella et al., 2016). For C. neoformans and C. albicans, the loss of mitochondrial function due to tetracycline treatment can increase the susceptibility of C. neoformans and C. albicans to amphotericin B, a powerful antifungal agent widely used in clinical settings to treat severe fungal infections (Oliver et al., 2008), suggesting the relationship between mitochondrial function and drug resistance varies among different pathogenic fungi.
C. neoformans, like other pathogenic fungi, exhibits drug resistance that is closely associated with mitochondrial functions (Figure 2). Research has shown that azoles kill C. neoformans by increasing intracellular level of ROS, which are mainly produced by mitochondria (Peng et al., 2018), implying a link between mitochondrion and drug resistance. Impaired mitochondrial fusion in C. neoformans results in severe growth defects under hydrogen peroxide stress and is associated with reduced resistance to antifungal agents (Chang and Doering, 2018). Also, a study has pointed out that Cryptococcal mitochondria play a unique role in drug resistance. Age-dependent increases in mitochondrial ROS resulted in regulatory changes in membrane transport proteins and ergosterol synthesis, which enhanced fluconazole tolerance in old Cryptococcus cells (Yoo et al., 2024). Thus, ROS have a complex regulatory role in antifungal drug tolerance in C. neoformans, with drug-induced ROS inhibiting fungal growth and endogenous ROS in old C. neoformans cells enhancing resistance to antifungals. Therefore, further studies are needed to determine whether the differential effects of ROS on drug resistance are due to differences in the types of ROS produced under various conditions. Additionally, dysfunctional mitochondria in C. neoformans have been reported to result in reduced mitochondrial membrane potential, increased susceptibility to oxidative stress, while enhanced tolerance to fluconazole (Telzrow et al., 2023), indicating a complex relationship between mitochondrial function and drug resistance. The mitochondrial genome exhibits a profound relationship with antifungal resistance in C. neoformans. The investigation focused on group I introns in the mitochondrial LSU rRNA gene and their association with drug susceptibility in 77 Cryptococcus isolates, revealing that two novel introns in the LSU rRNA gene may influence the minimum inhibitory concentration of amphotericin B and 5-flucytosine (Gomes et al., 2018). This indicates that group I introns in the mitochondrial genome of Cryptococcus may represent valuable molecular markers and therapeutic targets considering antifungal resistance (Gomes et al., 2018). Also, in the process of asexual reproduction, C. neoformans passes its mitochondrial genome from parent cells to their progeny. This mechanism can lead to the emergence of new mutations within the mitochondrial genome, especially when subjected to selective pressures like antifungal treatments (Dong et al., 2019; Hua et al., 2019; Wang and Xu, 2020).
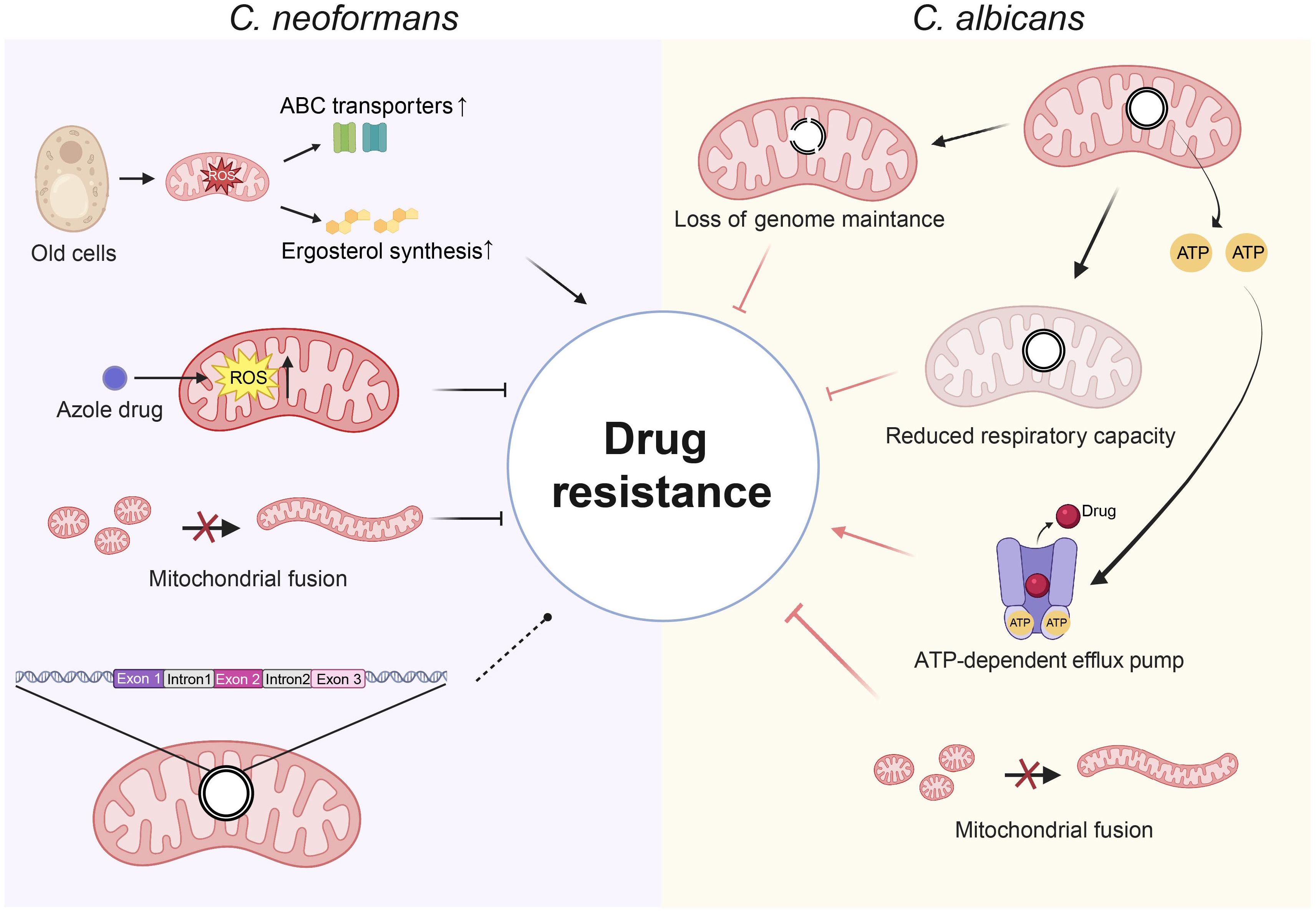
Figure 2. The correlation between mitochondria and antifungal drug resistance. In C. neoformans, azoles exert their antifungal effects by increasing intracellular levels of ROS. In old cells, elevated endogenous mitochondrial ROS drive resistance to drugs by increasing ergosterol synthesis and upregulating ABC transporter. The impaired mitochondrial fusion results in reduced resistance to antifungal drugs. Introns in the mitochondrial LSU rRNA gene has an impact on drug susceptibility for amphotericin B and 5-flucytosine. In C. albicans, defects in mitochondrial fusion and losses in mitochondrial genome maintenance are associated with diminished resistance to azole drugs. The reduced respiratory capacity diminishes the ability to withstand azole drugs. ATP supply enhances the efficacy of drug efflux pumps, increasing azole efflux and promoting drug resistance. Solid arrows indicate activation, Arrows with a perpendicular bar indicate suppression. Dashed arrows with a circular dot indicate dual effects. (Created with BioRender.com).
In C. albicans, deficiencies in mitochondrial fusion and a substantial decline in mitochondrial genome maintenance are linked to diminished resistance to azole antifungals, indicating a connection between the integrity of the mitochondrial genome and drug resistance (Ror and Panwar, 2019), suggesting a relationship between the integrity of the mitochondrial genome and drug resistance (Figure 2). The mitochondrial genome is important for respiration and ROS production, both key to developing drug resistance. Mutations in the mitochondrial complex of C. albicans lead to reduced respiratory capacity, thereby decreasing its ability to resist azole drugs (Shingu-Vazquez and Traven, 2011; Sun et al., 2013; Thomas et al., 2013; Sun et al., 2019). The function of efflux pumps for antifungal medications requires energy, indicating a possible relationship between the mitochondrial genome and the regulation of drug efflux pump activity (Guo et al., 2014, 2017). In azole-tolerant C. albicans, elevated mitochondrial aerobic respiration is associated with an increase in ATP levels, indicating a greater transfer of ATP from the mitochondria to the cytoplasm (Guo et al., 2017). This ensures that drug efflux pumps have a greater supply of available ATP, thereby enhancing intracellular azole efflux. A study has indicated that photodynamic treatment (PDT) presents a promising therapeutic option for C. albicans, particularly in light of its multi-drug resistance profile. Notably, the respiratory-deficient strain of C. albicans demonstrated an increased susceptibility to PDT compared to its parental strain (Chabrier-Rosello et al., 2008). This finding suggests that the integrity of the mitochondrial genome may play an essential role in the varying responses of C. albicans to different therapeutic approaches.
Although few antifungals currently target the mitochondrial genome directly, several mitochondrion-targeting drugs have shown potential as antifungals (Qin et al., 2023) (Figure 3). These agents target unique proteins in the electron transport chain, which are considered promising antifungal targets. For example, T-2307, which inhibits complexes CIII and CIV of the electron transport chain, is in phase II clinical trials, indicating its potential to overcome drug resistance (Shibata et al., 2012; Gerlach et al., 2021). Preclinical agents such as ML316, which targets the mitochondrial phosphate carrier Mir1, exhibit potential in enhancing the effectiveness of current antifungal treatments (McLellan et al., 2018). Licicolin H is a highly effective, broad-range antifungal agent that specifically targets cytochrome bc1 reductase (mitochondrial respiratory complex III). It has shown major inhibitory effects against various species, including Candida, Cryptococcus, and Aspergillus (Singh et al., 2012, 2013). Similarly, indazole compounds Inz-1 and Inz-5 inhibit cytochrome bc1, reducing the mitochondrial respiration of C. albicans (Vincent et al., 2016). However, the connection between the mitochondrial genome and drug resistance in pathogenic fungi is not yet fully understood. Therapeutic strategies targeting mitochondrial genome of pathogenic fungi and their encoded proteins will largely have the potential to improve fungal disease treatment, because their genomes and products have characteristics unique to different species.
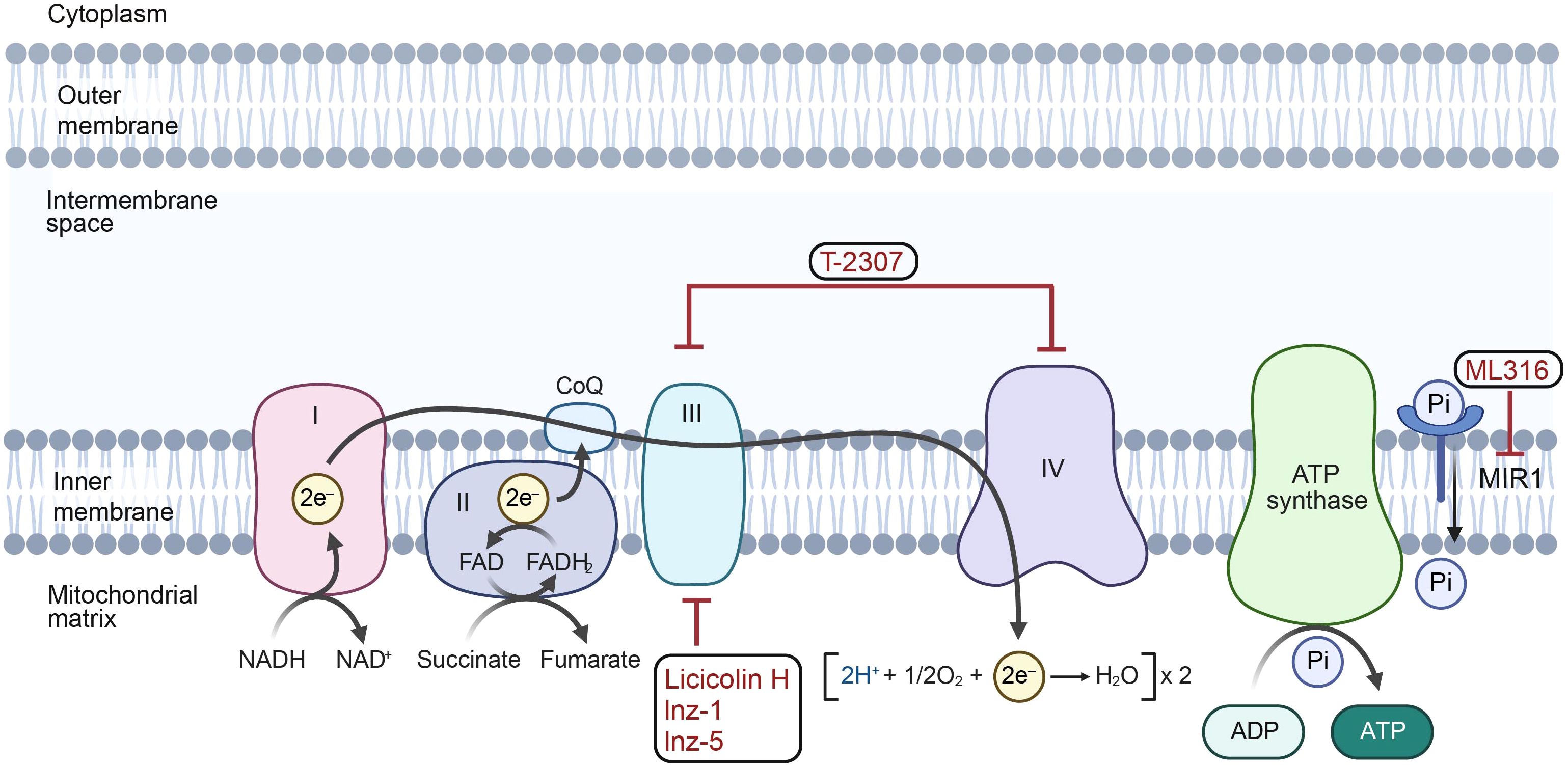
Figure 3. The mechanisms of antifungal agents on mitochondria. T-2307 inhibits the activity of mitochondrial complexes III and IV in the electron transport chain. ML316 suppresses the mitochondrial phosphate carrier Mir1, thus reducing the generation of ATP. Licicolin H, lnz-1 and lnz-5 target mitochondrial respiratory complex III. (Created with BioRender.com).
Conclusions
Mitochondrial genome structure and diversity
The mitochondrial genomes of pathogenic fungi display considerable structural and size variability. Typically circular in form, these genomes encode important proteins involved in energy metabolism. Variations in fungal mitochondrial size primarily arise from differences in introns and intergenic regions, along with distinctions in core protein-coding genes. The presence of introns is notable, as they play an important role in contributing to these size differences, thus contributing to genetic diversity within fungal mitochondrial genomes. The dynamics characteristics of these introns, manifested through their acquisition and loss, are intricately linked to the evolutionary processes of species, emphasizing the adaptability of fungal mitochondria.
Mitochondrial genomes and pathogenicity
The connection between the mitochondrial genome and pathogenicity in fungi is complex. In C. neoformans, variations in mitochondrial genome size, mainly due to the presence of introns, are associated with levels of virulence. Strains exhibiting defective tubular mitochondria tend to display reduced virulence characteristics. Besides, the mitochondrial genome plays a critical role in the generation and regulation of ROS, which is essential for the survival of C. neoformans within the host. In the case of C. albicans, the genetic diversity observed within mitochondrial genomes considerably impacts its pathogenic potential. This impact arises from its influence on the efficiency of energy metabolism, morphological transitions, and responses to oxidative stress. These findings emphasize the essential role that the mitochondrial genome has in fungal pathogenicity.
Mitochondrial function and drug resistance
The mitochondrial function in pathogenic fungi is key for understanding drug resistance. In C. neoformans, the dysfunction of mitochondrion and its byproducts ROS has a complex impact on the resistance to azole drug. In addition, variations in drug susceptibility are linked to introns found in the mitochondrial LSU rRNA gene, indicating that these introns could be used as molecular markers for antifungal resistance. In C. albicans, mutations affecting mitochondrial morphology result in reduced resistance against azole drugs. Importantly, the mitochondrial genome plays a role in respiration and ROS production, which is related to drug resistance. However, the direct connection between antifungal drug resistance and the mitochondrial genome remains largely unexplored. More studies are needed to determine whether the mitochondrial genome could be targeted to create better therapeutic strategies for managing fungal infections.
Author contributions
YN: Writing – original draft, Writing – review & editing. XG: Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funds for this research were supported by National Natural Science Foundation of China (82302549 to X.G.), Young Doctoral Talent Incubation Program of Xinqiao Hospital of Army Medical University (2023YQB017 to X.G.) and the Municipal Natural Science Foundation of Chongqing (CSTB2024NSCQ-MSX1230 to XG).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguileta, G., de Vienne, D. M., Ross, O. N., Hood, M. E., Giraud, T., Petit, E., et al. (2014). High variability of mitochondrial gene order among fungi. Genome Biol. Evol. 6, 451–465. doi: 10.1093/gbe/evu028
Alonso-Monge, R., Cortes-Prieto, I., Roman, E., Pla, J. (2024). Morphogenetic transitions in the adaptation of Candida albicans to the mammalian gut. Microbes Infect. 26, 105253. doi: 10.1016/j.micinf.2023.105253
Alspaugh, J. A. (2015). Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 78, 55–58. doi: 10.1016/j.fgb.2014.09.004
Alves, R., Gourlay, C. W. (2024). Editorial: Mitochondrial function and dysfunction in pathogenic fungi. Front. Physiol. 15. doi: 10.3389/fphys.2024.1506684
Anderson, J. B., Wickens, C., Khan, M., Cowen, L. E., Federspiel, N., Jones, T., et al. (2001). Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J. Bacteriol 183, 865–872. doi: 10.1128/JB.183.3.865-872.2001
Ashton, P. M., Thanh, L. T., Trieu, P. H., Van Anh, D., Trinh, N. M., Beardsley, J., et al. (2019). Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat. Commun. 10, 2035. doi: 10.1038/s41467-019-10092-5
Bambach, A., Fernandes, M. P., Ghosh, A., Kruppa, M., Alex, D., Li, D., et al. (2009). Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell 8, 1706–1720. doi: 10.1128/EC.00066-09
Bartelli, T. F., Bruno, D. C. F., Briones, M. R. S. (2018). Evidence for mitochondrial genome methylation in the yeast Candida albicans: A potential novel epigenetic mechanism affecting adaptation and pathogenicity? Front. Genet. 9. doi: 10.3389/fgene.2018.00166
Bartelli, T. F., Ferreira, R. C., Colombo, A. L., Briones, M. R. (2013). Intraspecific comparative genomics of Candida albicans mitochondria reveals non-coding regions under neutral evolution. Infect. Genet. Evol. 14, 302–312. doi: 10.1016/j.meegid.2012.12.012
Batova, M., Borecka-Melkusova, S., Simockova, M., Dzugasova, V., Goffa, E., Subik, J. (2008). Functional characterization of the CgPGS1 gene reveals a link between mitochondrial phospholipid homeostasis and drug resistance in Candida glabrata. Curr. Genet. 53, 313–322. doi: 10.1007/s00294-008-0187-9
Becker, J. M., Kauffman, S. J., Hauser, M., Huang, L., Lin, M., Sillaots, S., et al. (2010). Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. U.S.A. 107, 22044–22049. doi: 10.1073/pnas.1009845107
Bonhomme, J., Chauvel, M., Goyard, S., Roux, P., Rossignol, T., d’Enfert, C. (2011). Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol. Microbiol. 80, 995–1013. doi: 10.1111/j.1365-2958.2011.07626.x
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Borcherding, N., Brestoff, J. R. (2023). The power and potential of mitochondria transfer. Nature 623, 283–291. doi: 10.1038/s41586-023-06537-z
Brown, A. J., Brown, G. D., Netea, M. G., Gow, N. A. (2014). Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 22, 614–622. doi: 10.1016/j.tim.2014.07.001
Bulder, C. J. (1964). Induction of petite mutation and inhibition of synthesis of respiratory enzymes in various yeasts. Antonie Van Leeuwenhoek 30, 1–9. doi: 10.1007/BF02046695
Calderone, R., Li, D., Traven, A. (2015). System-level impact of mitochondria on fungal virulence: to metabolism and beyond. FEMS Yeast Res. 15, fov027. doi: 10.1093/femsyr/fov027
Casalini, G., Giacomelli, A., Antinori, S. (2024). The WHO fungal priority pathogens list: a crucial reappraisal to review the prioritisation. Lancet Microbe 5, 717–724. doi: 10.1016/S2666-5247(24)00042-9
Chabrier-Rosello, Y., Foster, T. H., Mitra, S., Haidaris, C. G. (2008). Respiratory deficiency enhances the sensitivity of the pathogenic fungus Candida to photodynamic treatment. Photochem. Photobiol. 84, 1141–1148. doi: 10.1111/j.1751-1097.2007.00280.x
Chang, A. L., Doering, T. L. (2018). Maintenance of mitochondrial morphology in Cryptococcus neoformans is critical for stress resistance and virulence. mBio 9 (6), e01375-18. doi: 10.1128/mBio.01375-18
Chen, X. J., Clark-Walker, G. D. (2000). The petite mutation in yeasts: 50 years on. Int. Rev. Cytol 194, 197–238. doi: 10.1016/s0074-7696(08)62397-9
Christinaki, A. C., Kanellopoulos, S. G., Kortsinoglou, A. M., Andrikopoulos, M., Theelen, B., Boekhout, T., et al. (2022). Mitogenomics and mitochondrial gene phylogeny decipher the evolution of Saccharomycotina yeasts. Genome Biol. Evol. 14 (5), evac073. doi: 10.1093/gbe/evac073
Dantas Ada, S., Day, A., Ikeh, M., Kos, I., Achan, B., Quinn, J. (2015). Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5, 142–165. doi: 10.3390/biom5010142
de Almeida, J. R., Riano Pachon, D. M., Franceschini, L. M., Dos Santos, I. B., Ferrarezi, J. A., de Andrade, P. A. M., et al. (2021). Revealing the high variability on nonconserved core and mobile elements of Austropuccinia psidii and other rust mitochondrial genomes. PLoS One 16, e0248054. doi: 10.1371/journal.pone.0248054
De Chiara, M., Friedrich, A., Barre, B., Breitenbach, M., Schacherer, J., Liti, G. (2020). Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC Biol. 18, 49. doi: 10.1186/s12915-020-00786-4
Denning, D. W. (2024). Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438. doi: 10.1016/S1473-3099(23)00692-8
Dong, K., You, M., Xu, J. (2019). Genetic changes in experimental populations of a hybrid in the Cryptococcus neoformans species complex. Pathogens 9 (1), 3. doi: 10.3390/pathogens9010003
Ene, I. V., Bennett, R. J., Anderson, M. Z. (2019). Mechanisms of genome evolution in Candida albicans. Curr. Opin. Microbiol. 52, 47–54. doi: 10.1016/j.mib.2019.05.001
Ferrari, S., Sanguinetti, M., De Bernardis, F., Torelli, R., Posteraro, B., Vandeputte, P., et al. (2011). Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55, 1852–1860. doi: 10.1128/AAC.01271-10
Fisher, M. C., Alastruey-Izquierdo, A., Berman, J., Bicanic, T., Bignell, E. M., Bowyer, P., et al. (2022). Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 20, 557–571. doi: 10.1038/s41579-022-00720-1
Fisher, M. C., Denning, D. W. (2023). The WHO fungal priority pathogens list as a game-changer. Nat. Rev. Microbiol. 21, 211–212. doi: 10.1038/s41579-023-00861-x
Fonseca, P. L. C., De-Paula, R. B., Araujo, D. S., Tome, L. M. R., Mendes-Pereira, T., Rodrigues, W. F. C., et al. (2021). Global characterization of fungal mitogenomes: new insights on genomic diversity and dynamism of coding genes and accessory elements. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.787283
Ford, C. B., Funt, J. M., Abbey, D., Issi, L., Guiducci, C., Martinez, D. A., et al. (2015). The evolution of drug resistance in clinical isolates of Candida albicans. Elife 4, e00662. doi: 10.7554/eLife.00662
Foury, F., Roganti, T., Lecrenier, N., Purnelle, B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440, 325–331. doi: 10.1016/s0014-5793(98)01467-7
Fox, T. D. (2012). Mitochondrial protein synthesis, import, and assembly. Genetics 192, 1203–1234. doi: 10.1534/genetics.112.141267
Franco, M. E. E., Lopez, S. M. Y., Medina, R., Lucentini, C. G., Troncozo, M. I., Pastorino, G. N., et al. (2017). The mitochondrial genome of the plant-pathogenic fungus Stemphylium lycopersici uncovers a dynamic structure due to repetitive and mobile elements. PLoS One 12, e0185545. doi: 10.1371/journal.pone.0185545
Fu, Y., Tigano, M., Sfeir, A. (2020). Safeguarding mitochondrial genomes in higher eukaryotes. Nat. Struct. Mol. Biol. 27, 687–695. doi: 10.1038/s41594-020-0474-9
Gao, X., Fu, Y., Sun, S., Gu, T., Li, Y., Sun, T., et al. (2022). Cryptococcal Hsf3 controls intramitochondrial ROS homeostasis by regulating the respiratory process. Nat. Commun. 13, 5407. doi: 10.1038/s41467-022-33168-1
Gerhold, J. M., Aun, A., Sedman, T., Joers, P., Sedman, J. (2010). Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 39, 851–861. doi: 10.1016/j.molcel.2010.09.002
Gerlach, E. S., Altamirano, S., Yoder, J. M., Luggya, T. S., Akampurira, A., Meya, D. B., et al. (2021). ATI-2307 exhibits equivalent antifungal activity in Cryptococcus neoformans clinical isolates with high and low fluconazole IC(50). Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.695240
Gomes, F., Arantes, T. D., Fernandes, J. A. L., Ferreira, L. C., Romero, H., Bosco, S. M. G., et al. (2018). Polymorphism in Mitochondrial Group I Introns among Cryptococcus neoformans and Cryptococcus gattii Genotypes and Its Association with Drug Susceptibility. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00086
Gomes, R., da Silva, K. J. G., Ferreira, L. C., Arantes, T. D., Theodoro, R. C. (2023). Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii. J. Fungi (Basel) 9 (6), 629. doi: 10.3390/jof9060629
Grahl, N., Demers, E. G., Lindsay, A. K., Harty, C. E., Willger, S. D., Piispanen, A. E., et al. (2015). Mitochondrial activity and cyr1 are key regulators of Ras1 activation of C. albicans virulence pathways. PLoS Pathog. 11, e1005133. doi: 10.1371/journal.ppat.1005133
Grosser, M. R., Sites, S. K., Murata, M. M., Lopez, Y., Chamusco, K. C., Love Harriage, K., et al. (2023). Plant mitochondrial introns as genetic markers - conservation and variation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1116851
Gulati, M., Nobile, C. J. (2016). Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 18, 310–321. doi: 10.1016/j.micinf.2016.01.002
Guo, H., Xie, S. M., Li, S. X., Song, Y. J., Lv, X. L., Zhang, H. (2014). Synergistic mechanism for tetrandrine on fluconazole against Candida albicans through the mitochondrial aerobic respiratory metabolism pathway. J. Med. Microbiol. 63, 988–996. doi: 10.1099/jmm.0.073890-0
Guo, H., Xie, S. M., Li, S. X., Song, Y. J., Zhong, X. Y., Zhang, H. (2017). Involvement of mitochondrial aerobic respiratory activity in efflux-mediated resistance of C. albicans to fluconazole. J. Mycol Med. 27, 339–344. doi: 10.1016/j.mycmed.2017.04.004
Harris, A., Wagner, M., Du, D., Raschka, S., Nentwig, L. M., Gohlke, H., et al. (2021). Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat. Commun. 12, 5254. doi: 10.1038/s41467-021-25574-8
Hua, W., Vogan, A., Xu, J. (2019). Genotypic and Phenotypic Analyses of Two “Isogenic” Strains of the Human Fungal Pathogen Cryptococcus neoformans var. neoformans. Mycopathologia 184, 195–212. doi: 10.1007/s11046-019-00328-9
Iliev, I. D., Brown, G. D., Bacher, P., Gaffen, S. L., Heitman, J., Klein, B. S., et al. (2024). Focus on fungi. Cell 187, 5121–5127. doi: 10.1016/j.cell.2024.08.016
Jacobsen, M. D., Rattray, A. M., Gow, N. A., Odds, F. C., Shaw, D. J. (2008). Mitochondrial haplotypes and recombination in Candida albicans. Med. Mycol 46, 647–654. doi: 10.1080/13693780801986631
Jenkins, H. L., Graham, R., Porter, J. S., Vieira, L. M., de Almeida, A. C. S., Hall, A., et al. (2022). Unprecedented frequency of mitochondrial introns in colonial bilaterians. Sci. Rep. 12, 10889. doi: 10.1038/s41598-022-14477-3
Joardar, V., Abrams, N. F., Hostetler, J., Paukstelis, P. J., Pakala, S., Pakala, S. B., et al. (2012). Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics 13, 698. doi: 10.1186/1471-2164-13-698
Joers, P., Gerhold, J. M., Sedman, T., Kuusk, S., Sedman, J. (2007). The helicase CaHmi1p is required for wild-type mitochondrial DNA organization in Candida albicans. FEMS Yeast Res. 7, 118–130. doi: 10.1111/j.1567-1364.2006.00132.x
Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., et al. (2004). The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101, 7329–7334. doi: 10.1073/pnas.0401648101
Kan, S. L., Shen, T. T., Gong, P., Ran, J. H., Wang, X. Q. (2020). The complete mitochondrial genome of Taxus cuspidata (Taxaceae): eight protein-coding genes have transferred to the nuclear genome. BMC Evol. Biol. 20, 10. doi: 10.1186/s12862-020-1582-1
Kavanaugh, L. A., Fraser, J. A., Dietrich, F. S. (2006). Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol. Biol. Evol. 23, 1879–1890. doi: 10.1093/molbev/msl070
Kim, M., Mahmood, M., Reznik, E., Gammage, P. A. (2022). Mitochondrial DNA is a major source of driver mutations in cancer. Trends Cancer 8, 1046–1059. doi: 10.1016/j.trecan.2022.08.001
Koffi, D., Bonouman, I. V., Toure, A. O., Kouadjo, F., N’Gou, M. R. E., Sylla, K., et al. (2021). Estimates of serious fungal infection burden in Cote d’Ivoire and country health profile. J. Mycol Med. 31, 101086. doi: 10.1016/j.mycmed.2020.101086
Kolondra, A., Labedzka-Dmoch, K., Wenda, J. M., Drzewicka, K., Golik, P. (2015). The transcriptome of Candida albicans mitochondria and the evolution of organellar transcription units in yeasts. BMC Genomics 16, 827. doi: 10.1186/s12864-015-2078-z
Kontoyiannis, D. P., Lewis, R. E. (2002). Antifungal drug resistance of pathogenic fungi. Lancet 359, 1135–1144. doi: 10.1016/S0140-6736(02)08162-X
Korovesi, A. G., Ntertilis, M., Kouvelis, V. N. (2018). Mt-rps3 is an ancient gene which provides insight into the evolution of fungal mitochondrial genomes. Mol. Phylogenet Evol. 127, 74–86. doi: 10.1016/j.ympev.2018.04.037
Koszul, R., Malpertuy, A., Frangeul, L., Bouchier, C., Wincker, P., Thierry, A., et al. (2003). The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 534, 39–48. doi: 10.1016/s0014-5793(02)03749-3
Krol, J., Nawrot, U., Bartoszewicz, M. (2019). The susceptibility of Candida albicans strains to selected anticancer drugs and flucytosine, relevance of the presence of self-splicing intron in 25S rDNA. J. Mycol Med. 29, 39–43. doi: 10.1016/j.mycmed.2018.11.002
Lavin, J. L., Oguiza, J. A., Ramirez, L., Pisabarro, A. G. (2008). Comparative genomics of the oxidative phosphorylation system in fungi. Fungal Genet. Biol. 45, 1248–1256. doi: 10.1016/j.fgb.2008.06.005
Li, Y., Zhang, Y., Zhang, C., Wang, H., Wei, X., Chen, P., et al. (2020). Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 117, 1711–1721. doi: 10.1073/pnas.1911560116
Liang, C., Zhang, B., Cui, L., Li, J., Yu, Q., Li, M. (2018). Mgm1 is required for maintenance of mitochondrial function and virulence in Candida albicans. Fungal Genet. Biol. 120, 42–52. doi: 10.1016/j.fgb.2018.09.006
Lipinski, K. A., Kaniak-Golik, A., Golik, P. (2010). Maintenance and expression of the S. cerevisiae mitochondrial genome–from genetics to evolution and systems biology. Biochim. Biophys. Acta 1797, 1086–1098. doi: 10.1016/j.bbabio.2009.12.019
Litter, J., Keszthelyi, A., Hamari, Z., Pfeiffer, I., Kucsera, J. (2005). Differences in mitochondrial genome organization of Cryptococcus neoformans strains. Antonie Van Leeuwenhoek 88, 249–255. doi: 10.1007/s10482-005-8544-x
Liu, H., Liu, Y., Wang, H., Zhao, Q., Zhang, T., Xie, S. A., et al. (2022). Geometric constraints regulate energy metabolism and cellular contractility in vascular smooth muscle cells by coordinating mitochondrial DNA methylation. Adv. Sci. (Weinh) 9, e2203995. doi: 10.1002/advs.202203995
Liu, T., Pyle, A. M. (2021). Discovery of highly reactive self-splicing group II introns within the mitochondrial genomes of human pathogenic fungi. Nucleic Acids Res. 49, 12422–12432. doi: 10.1093/nar/gkab1077
Liu, T., Pyle, A. M. (2024). Highly reactive group I introns ubiquitous in pathogenic fungi. J. Mol. Biol. 436, 168513. doi: 10.1016/j.jmb.2024.168513
Lockhart, S. R., Chowdhary, A., Gold, J. A. W. (2023). The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol. 21, 818–832. doi: 10.1038/s41579-023-00960-9
Ma, H., May, R. C. (2010). Mitochondria and the regulation of hypervirulence in the fatal fungal outbreak on Vancouver Island. Virulence 1, 197–201. doi: 10.4161/viru.1.3.11053
McLellan, C. A., Vincent, B. M., Solis, N. V., Lancaster, A. K., Sullivan, L. B., Hartland, C. L., et al. (2018). Inhibiting mitochondrial phosphate transport as an unexploited antifungal strategy. Nat. Chem. Biol. 14, 135–141. doi: 10.1038/nchembio.2534
Meng, Y., Ding, C. (2023). Mitochondria in Cryptococcus: an update of mitochondrial transcriptional regulation in Cryptococcus. Curr. Genet. 69, 1–6. doi: 10.1007/s00294-023-01261-7
Merryman, M., Crigler, J., Seipelt-Thiemann, R., McClelland, E. (2020). A mutation in C. neoformans mitochondrial NADH dehydrogenase results in increased virulence in mice. Virulence 11, 1366–1378. doi: 10.1080/21505594.2020.1831332
Miletti, K. E., Leibowitz, M. J. (2000). Pentamidine inhibition of group I intron splicing in Candida albicans correlates with growth inhibition. Antimicrob. Agents Chemother. 44, 958–966. doi: 10.1128/AAC.44.4.958-966.2000
Miyazawa, H., Osigus, H. J., Rolfes, S., Kamm, K., Schierwater, B., Nakano, H. (2021). Mitochondrial genome evolution of placozoans: gene rearrangements and repeat expansions. Genome Biol. Evol. 13 (1), evaa213. doi: 10.1093/gbe/evaa213
Morley, S. A., Nielsen, B. L. (2017). Plant mitochondrial DNA. Front. Biosci. (Landmark Ed) 22, 1023–1032. doi: 10.2741/4531
Moye-Rowley, W. S. (2005). Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354, 15–21. doi: 10.1016/j.gene.2005.03.019
Neubauer, M., Zhu, Z., Penka, M., Helmschrott, C., Wagener, N., Wagener, J. (2015). Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Mol. Microbiol. 98, 930–945. doi: 10.1111/mmi.13167
Nielsen, C. B., Friedman, B., Birren, B., Burge, C. B., Galagan, J. E. (2004). Patterns of intron gain and loss in fungi. PloS Biol. 2, e422. doi: 10.1371/journal.pbio.0020422
Noble, S. M., French, S., Kohn, L. A., Chen, V., Johnson, A. D. (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598. doi: 10.1038/ng.605
Nosek, J., Fukuhara, H. (1994). NADH dehydrogenase subunit genes in the mitochondrial DNA of yeasts. J. Bacteriol 176, 5622–5630. doi: 10.1128/jb.176.18.5622-5630.1994
Oliver, B. G., Silver, P. M., Marie, C., Hoot, S. J., Leyde, S. E., White, T. C. (2008). Tetracycline alters drug susceptibility in Candida albicans and other pathogenic fungi. Microbiol. (Reading) 154, 960–970. doi: 10.1099/mic.0.2007/013805-0
Pemmaraju, S. C., Padmapriya, K., Pruthi, P. A., Prasad, R., Pruthi, V. (2016). Impact of oxidative and osmotic stresses on Candida albicans biofilm formation. Biofouling 32, 897–909. doi: 10.1080/08927014.2016.1212021
Peng, C. A., Gaertner, A. A. E., Henriquez, S. A., Fang, D., Colon-Reyes, R. J., Brumaghim, J. L., et al. (2018). Fluconazole induces ROS in Cryptococcus neoformans and contributes to DNA damage in vitro. PLoS One 13, e0208471. doi: 10.1371/journal.pone.0208471
Pimentel, L. G. P., Soares, R. A. S., de Assis, P. M., Batista da Silva, I., Rodrigues-Oliveira, I. H., Rocha, R. R., et al. (2024). Mitochondrial genomes of mammals from the Brazilian cerrado and phylogenetic considerations for the orders artiodactyla, carnivora, and chiroptera (Chordata: Mammalia). Life (Basel) 14 (12), 1597. doi: 10.3390/life14121597
Qin, Y., Wang, J., Lv, Q., Han, B. (2023). Recent progress in research on mitochondrion-targeted antifungal drugs: a review. Antimicrob. Agents Chemother. 67, e0000323. doi: 10.1128/aac.00003-23
Rahman, H., Carneglia, J., Lausten, M., Robertello, M., Choy, J., Golin, J. (2018). Robust, pleiotropic drug resistance 5 (Pdr5)-mediated multidrug resistance is vigorously maintained in Saccharomyces cerevisiae cells during glucose and nitrogen limitation. FEMS Yeast Res. 18 (4), foy029. doi: 10.1093/femsyr/foy029
Rella, A., Farnoud, A. M., Del Poeta, M. (2016). Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 61, 63–72. doi: 10.1016/j.plipres.2015.11.003
Roe, K. (2023). Increased fungal infection mortality induced by concurrent viral cellular manipulations. Lung 201, 467–476. doi: 10.1007/s00408-023-00642-6
Rokas, A. (2022). Evolution of the human pathogenic lifestyle in fungi. Nat. Microbiol. 7, 607–619. doi: 10.1038/s41564-022-01112-0
Ror, S., Panwar, S. L. (2019). Sef1-regulated iron regulon responds to mitochondria-dependent iron-sulfur cluster biosynthesis in Candida albicans. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01528
Ryu, K. W., Fung, T. S., Baker, D. C., Saoi, M., Park, J., Febres-Aldana, C. A., et al. (2024). Cellular ATP demand creates metabolically distinct subpopulations of mitochondria. Nature 635, 746–754. doi: 10.1038/s41586-024-08146-w
Saldanha, R., Mohr, G., Belfort, M., Lambowitz, A. M. (1993). Group I and group II introns. FASEB J. 7, 15–24. doi: 10.1096/fasebj.7.1.8422962
Sandor, S., Zhang, Y., Xu, J. (2018). Fungal mitochondrial genomes and genetic polymorphisms. Appl. Microbiol. Biotechnol. 102, 9433–9448. doi: 10.1007/s00253-018-9350-5
Sanglard, D., Ischer, F., Bille, J. (2001). Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45, 1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001
Selmecki, A., Forche, A., Berman, J. (2006). Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370. doi: 10.1126/science.1128242
Seneviratne, C. J., Wang, Y., Jin, L., Abiko, Y., Samaranayake, L. P. (2008). Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8, 2936–2947. doi: 10.1002/pmic.200701097
She, X., Khamooshi, K., Gao, Y., Shen, Y., Lv, Y., Calderone, R., et al. (2015). Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse cell functions including cell wall synthesis. Cell Microbiol. 17, 1350–1364. doi: 10.1111/cmi.12438
Shibata, T., Takahashi, T., Yamada, E., Kimura, A., Nishikawa, H., Hayakawa, H., et al. (2012). T-2307 causes collapse of mitochondrial membrane potential in yeast. Antimicrob. Agents Chemother. 56, 5892–5897. doi: 10.1128/AAC.05954-11
Shingu-Vazquez, M., Traven, A. (2011). Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell 10, 1376–1383. doi: 10.1128/EC.05184-11
Singh, H. (2021). Mitochondrial ion channels in cardiac function. Am. J. Physiol. Cell Physiol. 321, C812–C825. doi: 10.1152/ajpcell.00246.2021
Singh, S. B., Liu, W., Li, X., Chen, T., Shafiee, A., Card, D., et al. (2012). Antifungal spectrum, in vivo efficacy, and structure-activity relationship of ilicicolin h. ACS Med. Chem. Lett. 3, 814–817. doi: 10.1021/ml300173e
Singh, S. B., Liu, W., Li, X., Chen, T., Shafiee, A., Dreikorn, S., et al. (2013). Structure-activity relationship of cytochrome bc1 reductase inhibitor broad spectrum antifungal ilicicolin H. Bioorg Med. Chem. Lett. 23, 3018–3022. doi: 10.1016/j.bmcl.2013.03.023
Soggiu, A., Roncada, P., Bonizzi, L., Piras, C. (2019). Role of mitochondria in host-pathogen interaction. Adv. Exp. Med. Biol. 1158, 45–57. doi: 10.1007/978-981-13-8367-0_3
Song, J., Zhou, J., Zhang, L., Li, R. (2020). Mitochondria-mediated azole drug resistance and fungal pathogenicity: opportunities for therapeutic development. Microorganisms 8 (10), 1574. doi: 10.3390/microorganisms8101574
Sun, N., Fonzi, W., Chen, H., She, X., Zhang, L., Zhang, L., et al. (2013). Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob. Agents Chemother. 57, 532–542. doi: 10.1128/AAC.01520-12
Sun, N., Parrish, R. S., Calderone, R. A., Fonzi, W. A. (2019). Unique, diverged, and conserved mitochondrial functions influencing Candida albicans respiration. mBio 10 (3), e00300-19. doi: 10.1128/mBio.00300-19
Tan, H., Yu, Y., Fu, Y., Liu, T., Wang, Y., Peng, W., et al. (2022). Comparative analyses of Flammulina filiformis mitochondrial genomes reveal high length polymorphism in intergenic regions and multiple intron gain/loss in cox1. Int. J. Biol. Macromol 221, 1593–1605. doi: 10.1016/j.ijbiomac.2022.09.110
Tang, J., Zhang, L., Su, J., Ye, Q., Li, Y., Liu, D., et al. (2024). Insights into fungal mitochondrial genomes and inheritance based on current findings from yeast-like fungi. J. Fungi (Basel) 10 (7), 441. doi: 10.3390/jof10070441
Telzrow, C. L., Esher Righi, S., Cathey, J. M., Granek, J. A., Alspaugh, J. A. (2023). Cryptococcus neoformans Mar1 function links mitochondrial metabolism, oxidative stress, and antifungal tolerance. Front. Physiol. 14. doi: 10.3389/fphys.2023.1150272
Thambugala, K. M., Daranagama, D. A., Tennakoon, D. S., Jayatunga, D. P. W., Hongsanan, S., Xie, N. (2024). Humans vs. Fungi: An Overview of Fungal Pathogens against Humans. Pathogens 13 (5), 426. doi: 10.3390/pathogens13050426
Thomas, E., Roman, E., Claypool, S., Manzoor, N., Pla, J., Panwar, S. L. (2013). Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob. Agents Chemother. 57, 5580–5599. doi: 10.1128/AAC.00889-13
Tu, Y., Lineaweaver, W. C., Breland, A., Zhang, F. (2021). Fungal infection in burn patents: A review of 36 case reports. Ann. Plast. Surg. 86, S463–S467. doi: 10.1097/SAP.0000000000002865
Valero, C., Colabardini, A. C., Chiaratto, J., Pardeshi, L., de Castro, P. A., Ferreira Filho, J. A., et al. (2020). Aspergillus fumigatus transcription factors involved in the Caspofungin paradoxical effect. mBio 11 (3), e00816-20. doi: 10.1128/mBio.00816-20
Vercellino, I., Sazanov, L. A. (2022). The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 23, 141–161. doi: 10.1038/s41580-021-00415-0
Verma, S., Shakya, V. P. S., Idnurm, A. (2018). Exploring and exploiting the connection between mitochondria and the virulence of human pathogenic fungi. Virulence 9, 426–446. doi: 10.1080/21505594.2017.1414133
Vincent, B. M., Langlois, J. B., Srinivas, R., Lancaster, A. K., Scherz-Shouval, R., Whitesell, L., et al. (2016). A fungal-selective cytochrome bc(1) inhibitor impairs virulence and prevents the evolution of drug resistance. Cell Chem. Biol. 23, 978–991. doi: 10.1016/j.chembiol.2016.06.016
Vitiello, A., Ferrara, F., Boccellino, M., Ponzo, A., Cimmino, C., Comberiati, E., et al. (2023). Antifungal drug resistance: an emergent health threat. Biomedicines 11 (4), 1063. doi: 10.3390/biomedicines11041063
Wallace, D. C. (2016). Genetics: Mitochondrial DNA in evolution and disease. Nature 535, 498–500. doi: 10.1038/nature18902
Wang, J. M., Bennett, R. J., Anderson, M. Z. (2018). The genome of the human pathogen Candida albicans is shaped by mutation and cryptic sexual recombination. mBio 9 (5), e01205-18. doi: 10.1128/mBio.01205-18
Wang, Y., Xu, J. (2020). Mitochondrial genome polymorphisms in the human pathogenic fungus Cryptococcus neoformans. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00706
Wu, W., Lockhart, S. R., Pujol, C., Srikantha, T., Soll, D. R. (2007). Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol. Microbiol. 64, 1587–1604. doi: 10.1111/j.1365-2958.2007.05759.x
Ye, J., Cheng, J., Ren, Y., Liao, W., Li, Q. (2020). The first mitochondrial genome for geastrales (Sphaerobolus stellatus) reveals intron dynamics and large-scale gene rearrangements of basidiomycota. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01970
Yildiz, G., Ozkilinc, H. (2020). First characterization of the complete mitochondrial genome of fungal plant-pathogen Monilinia laxa which represents the mobile intron rich structure. Sci. Rep. 10, 13644. doi: 10.1038/s41598-020-70611-z
Yoo, K., Bhattacharya, S., Oliveira, N. K., Pereira de Sa, N., Matos, G. S., Del Poeta, M., et al. (2024). With age comes resilience: how mitochondrial modulation drives age-associated fluconazole tolerance in Cryptococcus neoformans. mBio 15, e0184724. doi: 10.1128/mbio.01847-24
Zaccaron, A. Z., Stergiopoulos, I. (2021). Characterization of the mitochondrial genomes of three powdery mildew pathogens reveals remarkable variation in size and nucleotide composition. Microb. Genom 7 (12), 000720. doi: 10.1099/mgen.0.000720
Zardoya, R. (2020). Recent advances in understanding mitochondrial genome diversity. F1000Res 9, F1000 Faculty Rev-270. doi: 10.12688/f1000research.21490.1
Zhai, P., Shi, L., Zhong, G., Jiang, J., Zhou, J., Chen, X., et al. (2021). The oxrA protein of Aspergillus fumigatus is required for the oxidative stress response and fungal pathogenesis. Appl. Environ. Microbiol. 87, e0112021. doi: 10.1128/AEM.01120-21
Zhang, Z., Bills, G. F., An, Z. (2023). Advances in the treatment of invasive fungal disease. PLoS Pathog. 19, e1011322. doi: 10.1371/journal.ppat.1011322
Zhang, H., Xu, G. Y., Mao, C., Xu, Y., Dong, W., Inam, M., et al. (2021). Characterization and complete genome analysis of Bacillus velezensis CB6 revealed ATP synthase subunit alpha against foodborne pathogens. Arch. Microbiol. 203, 1061–1069. doi: 10.1007/s00203-020-02102-8
Zhang, Y., Zhang, S., Zhang, G., Liu, X., Wang, C., Xu, J. (2015). Comparison of mitochondrial genomes provides insights into intron dynamics and evolution in the caterpillar fungus Cordyceps militaris. Fungal Genet. Biol. 77, 95–107. doi: 10.1016/j.fgb.2015.04.009
Keywords: mitochondrial genome, Cryptococcus neoformans, Candida albicans, pathogenicity, drug resistance
Citation: Ni Y and Gao X (2025) Uncovering the role of mitochondrial genome in pathogenicity and drug resistance in pathogenic fungi. Front. Cell. Infect. Microbiol. 15:1576485. doi: 10.3389/fcimb.2025.1576485
Received: 14 February 2025; Accepted: 28 March 2025;
Published: 16 April 2025.
Edited by:
Yuanwei Zhang, Nanjing Normal University, ChinaReviewed by:
Somanon Bhattacharya, Wuxi Advanced Therapeutics, Inc., United StatesAlex Zaccaron, Oregon State University, United States
Copyright © 2025 Ni and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xindi Gao, Z2FveGluZGkxOTkzQHRtbXUuZWR1LmNu
 Yue Ni1
Yue Ni1 Xindi Gao
Xindi Gao