- 1Central Laboratory, Cancer Hospital of Dalian University of Technology, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning, China
- 2China Medical University, Shenyang, Shenyang, Liaoning, China
- 3Graduate School, Dalian Medical University, Dalian, Liaoning, China
- 4Department of Medicine, Kingbio Medical Co., Ltd., Chongqing, China
- 5Department of Pharmacy, Liaoning Vocational College of Medicine, Shenyang, Liaoning, China
- 6Department of Gastroenterological Surgery, Peking University International Hospital, Beijing, China
- 7Department of Hepatobiliary and Pancreatic Surgery, Cancer Hospital of Dalian University of Technology, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning, China
Since the early 20th century, there has been extensive discussion on the intricate relationship between pathogenic infection and tumors. However, most studies on host-pathogen interactions are performed based on the in-vitro culture, immortalized cell lines or animal experiments. A significant challenge lies in accurately establishing a coculture model between tumors and pathogens under the three-dimensional (3D) context. Recently, the hybrid model system that incorporates 3D tumor organoids and two-dimensional cell lines have been gradually used to analyze the intricate relationship between pathogens and tumors, and several coculture techniques for tumor organoids and pathogens have also been developed. Therefore, this study systematically reviewed the preparation and identification of tumor organoids, coculture techniques with pathogens, and their clinical applications, aiming to further understand and simulate the interaction mechanism between the hosts and pathogens.
1 Introduction
Organoids serve as a transitional model between in vitro cancer cell lines and xenografts, offering a unique approach to study cancer biology. Differing from traditional cell culture, the organoid model can preserve cell-cell and cell-matrix interactions by cultivating cancer cells under the three-dimensional (3D) context (Veninga and Voest, 2021; Xu et al., 2022), more closely resembling the characteristics of the original tumor (McCauley and Wells, 2017; Ouchi et al., 2019; Kim et al., 2020). Organoids are classified based on the cellular source, including pluripotent stem cells (PSCs), adult stem cells (ASCs), and patient-derived tumor organoids (PDTOs) (Driehuis et al., 2020; Tindle et al., 2021; Li et al., 2023) (Table 1), among which the PDTOs are small tissue spheroids and are generated following tumor resection (Walsh et al., 2017; Rosenbluth et al., 2020). The advent of PDTOs has enabled the implementation of patient-specific drug screening, personalized treatment, and identification of prognostic biomarkers and mechanisms of drug resistance (Neal and Kuo, 2016).
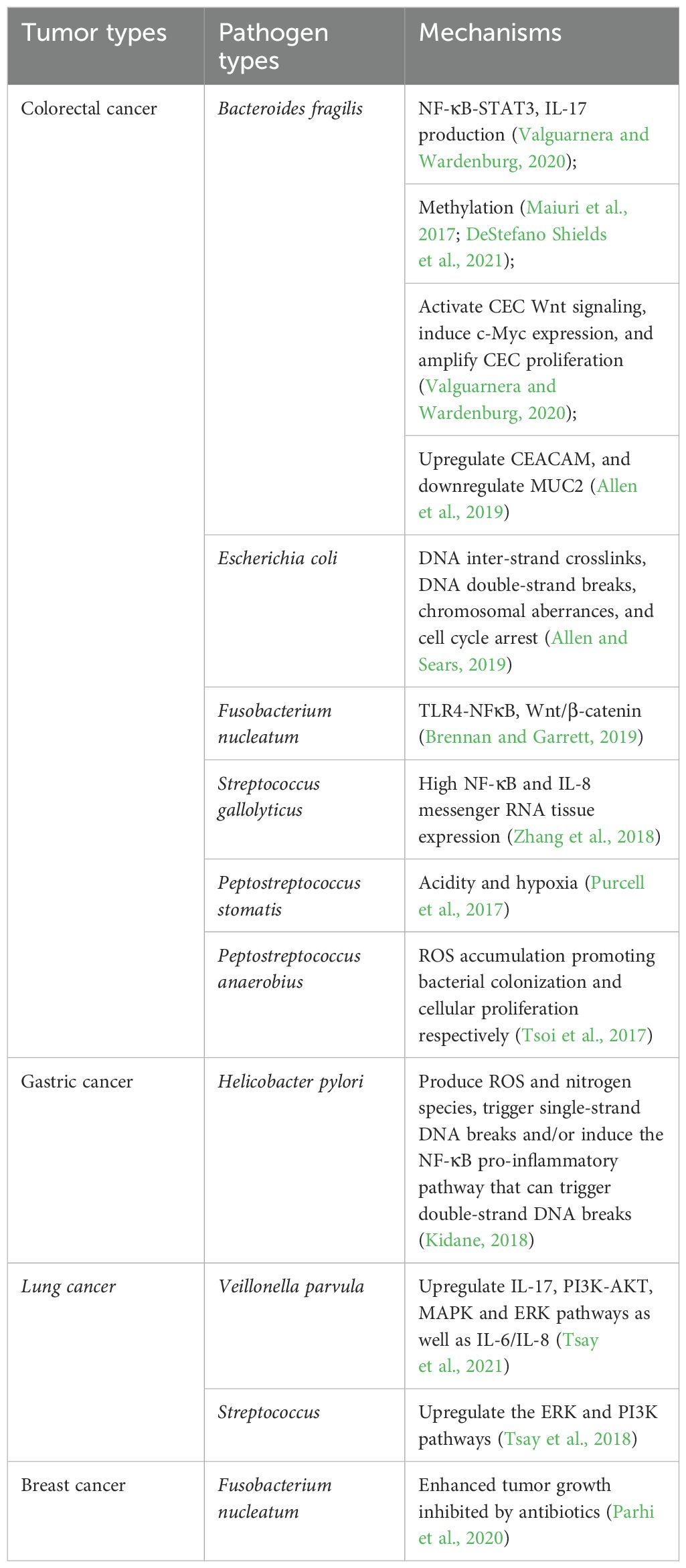
The intricate relationship between infectious diseases and cancers has been extensively studied since the early 20th century. In 2012, approximately 2.2 million new cancer cases were attributed to infections, among which helicobacter pylori (H. pylori), human papillomavirus (HPV), hepatitis B virus (HBV), hepatitis C virus (HCV), and Epstein-Barr virus (EBV) play important roles (Qu et al., 2021). Previous studies have confirmed that tumorigenesis is closely associated with a variety of pathogenic microorganisms comprising a heterogeneous assemblage of bacteria, fungi, protozoa, viruses, and phages (Maiuri et al., 2017; Purcell et al., 2017; Tsoi et al., 2017; Kidane, 2018; Tsay et al., 2018; Zhang et al., 2018; Allen and Sears, 2019; Allen et al., 2019; Brennan and Garrett, 2019; Parhi et al., 2020; Valguarnera and Wardenburg, 2020; DeStefano Shields et al., 2021; Tsay et al., 2021) (Table 2). Nejman et al. undertook an exhaustive examination of the microbiomes in 1,526 tumors (breast, lung, ovarian, pancreatic, melanoma, bone, and brain tumors) and their corresponding normal tissues across 7 distinct cancer types. The findings revealed that each tumor category exhibited a distinct microbiome profile, with breast cancer demonstrating a notably abundant and varied microbiome (Plummer et al., 2016; Nejman et al., 2020). This indicates that coculturing pathogenic microorganisms with tumor organoids offers a new approach for diagnosis, prognostic prediction, and treatment decision in cancer. Although bacterial therapy has shown a greater promise in cancer treatment over the last decade due to its ability to lyse the tumor cells and deliver therapeutic products, the potential cytotoxicity of bacteria for healthy tissues and their inability to entirely lyse cancerous cells poses challenges for cancer treatment (Sepich-Poore et al., 2021; Soleimani and Javadi, 2022). Hence, the investigation into pathogenic microorganisms is crucial for understanding the mechanisms of tumorigenesis and promoting the development of innovative vaccine technologies.
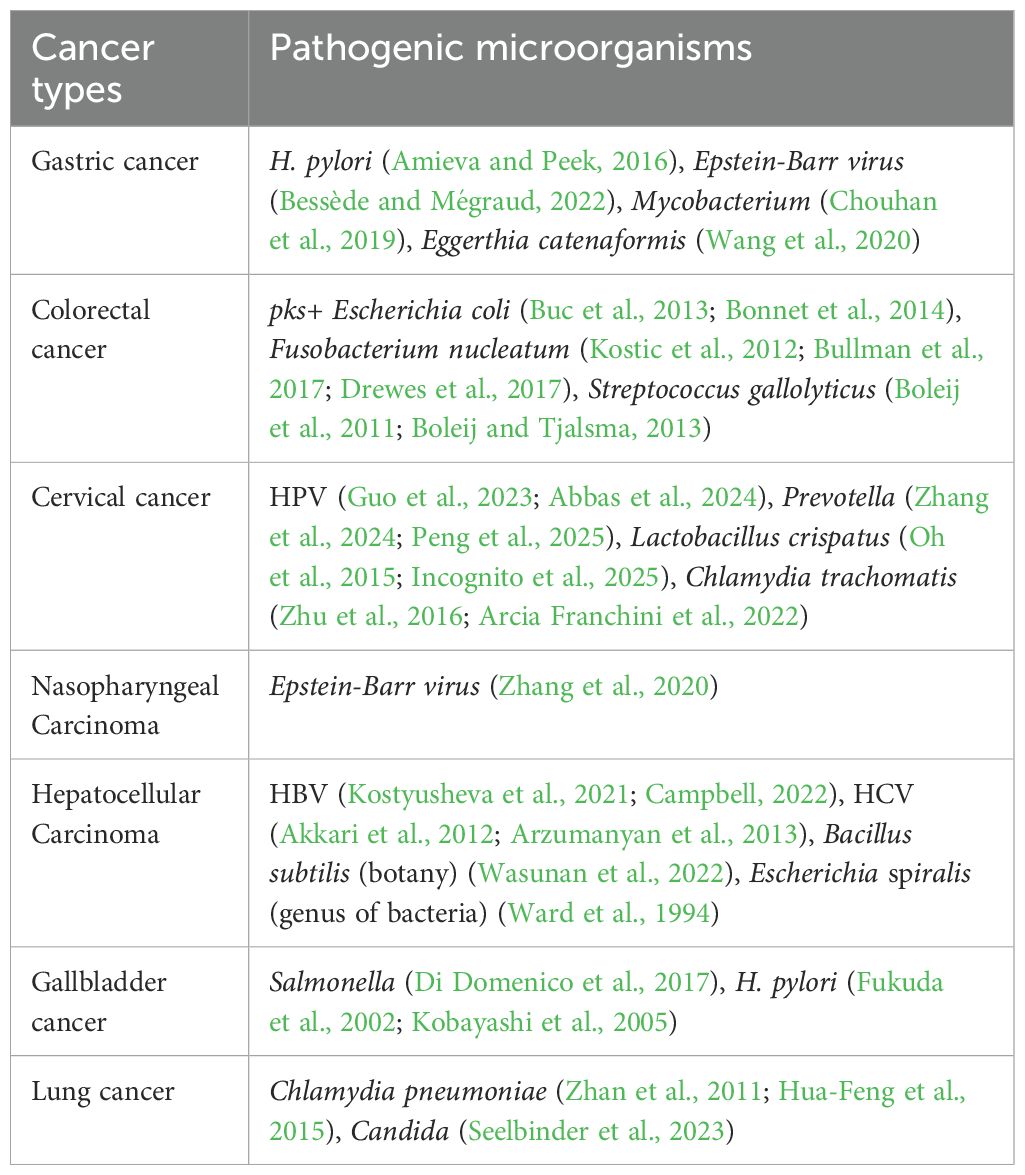
Certain bacteria can induce cancers in diverse organs and tissues, including lung, liver, colorectum, kidney, cervix, brain, gastrointestinal tract, etc (Ward et al., 1994; Fukuda et al., 2002; Kobayashi et al., 2005; Boleij et al., 2011; Zhan et al., 2011; Akkari et al., 2012; Kostic et al., 2012; Arzumanyan et al., 2013; Boleij and Tjalsma, 2013; Buc et al., 2013; Bonnet et al., 2014; Hua-Feng et al., 2015; Oh et al., 2015; Amieva and Peek, 2016; Zhu et al., 2016; Bullman et al., 2017; Di Domenico et al., 2017; Drewes et al., 2017; Chouhan et al., 2019; Wang et al., 2020; Zhang et al., 2020; Kostyusheva et al., 2021; Arcia Franchini et al., 2022; Bessède and Mégraud, 2022; Campbell, 2022; Wasunan et al., 2022; Guo et al., 2023; Seelbinder et al., 2023; Abbas et al., 2024; Zhang et al., 2024; Incognito et al., 2025; Peng et al., 2025) (Table 3). These bacteria contribute to tumorigenesis or malignant progression through various mechanisms (Wong and Yu, 2023; Kwon et al., 2024). Here, we systematically reviewed the preparation and identification of tumor organoids, coculture techniques of tumor organoids and pathogenic microorganisms, and their clinical application.
2 Preparation and characterization of tumor organoids
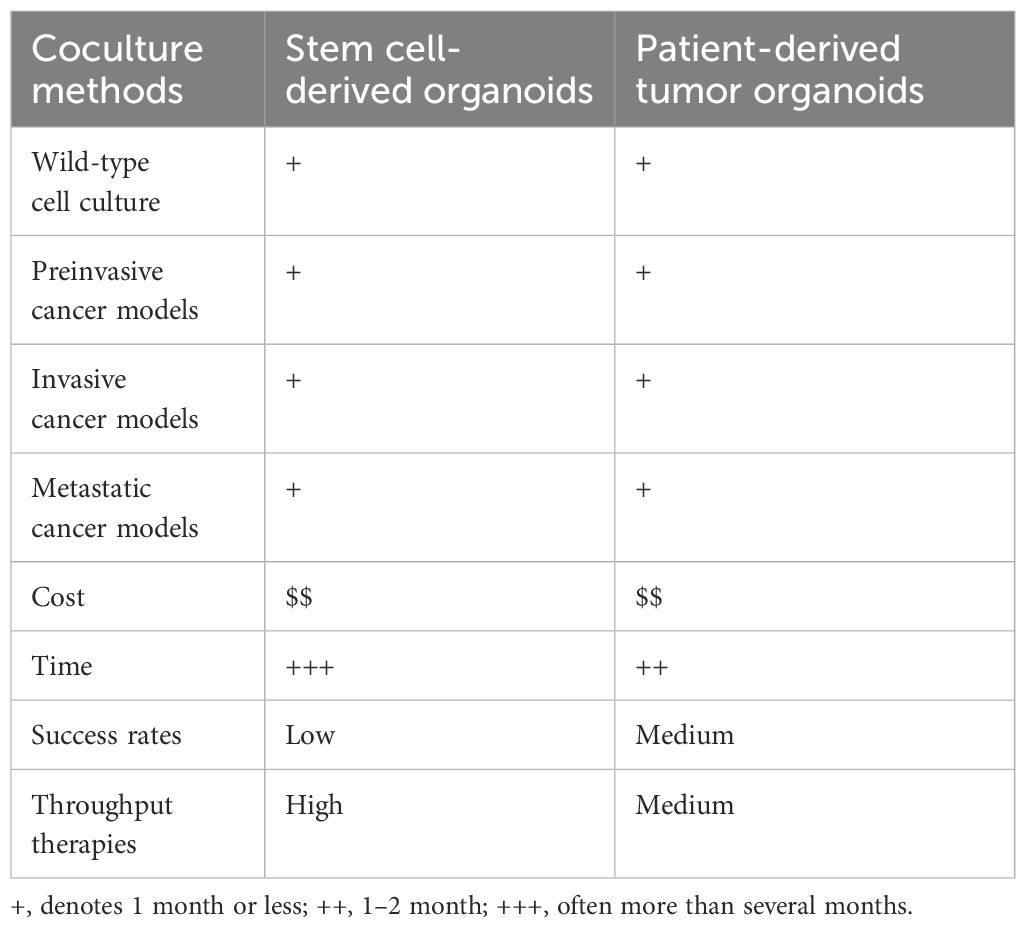
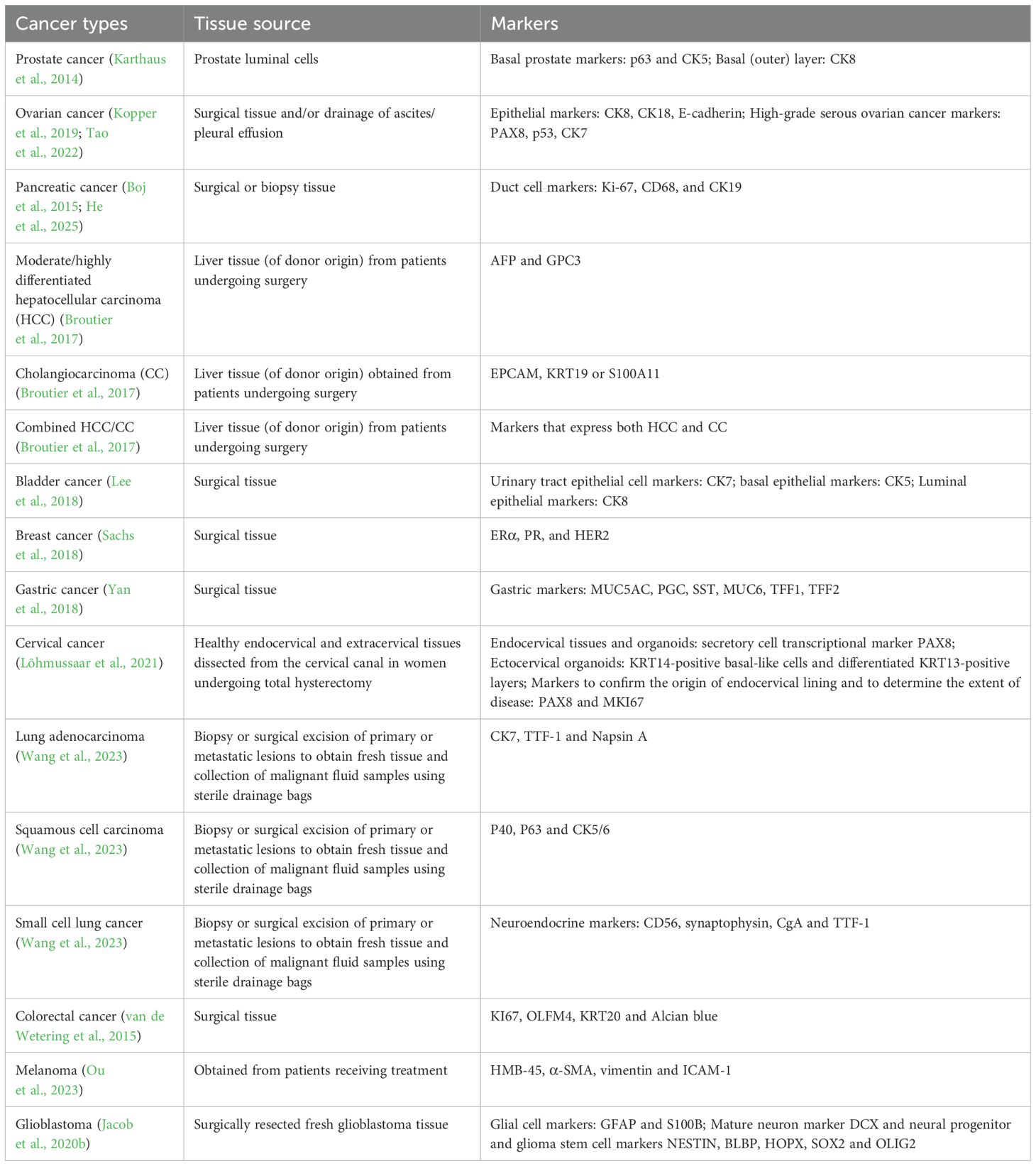
In recent years, PDTOs have been widely used to study various cancer types, including pancreatic cancer (Boj et al., 2015), prostate cancer (Gao et al., 2014), liver cancer (Broutier et al., 2017), bladder cancer (Lee et al., 2018), breast cancer (Sachs et al., 2018), ovarian cancer (Kopper et al., 2019) and gastric cancer (Yan et al., 2018). Cancer is an extremely complex disease, and its heterogeneity is manifested by the fact that the same cancer subtype may vary significantly among the patients, such as the cell shape, size, and gene expression (Figure 1). The quality control of different tumor organoids, especially the stable expression of markers, plays a very important role in identifying successful establishment. The morphology and culture conditions of tumor organoids have been reported in several studies (Karthaus et al., 2014; Yoshida, 2020; Jeong et al., 2023). To provide a basis for standardized quality control of tumor organoids, we summarized the markers applied in the identification of tumor organoids (Karthaus et al., 2014; Boj et al., 2015; van de Wetering et al., 2015; Broutier et al., 2017; Lee et al., 2018; Sachs et al., 2018; Yan et al., 2018; Kopper et al., 2019; Jacob et al., 2020b; Lõhmussaar et al., 2021; Tao et al., 2022; Ou et al., 2023; Wang et al., 2023; He et al., 2025) (Table 4).
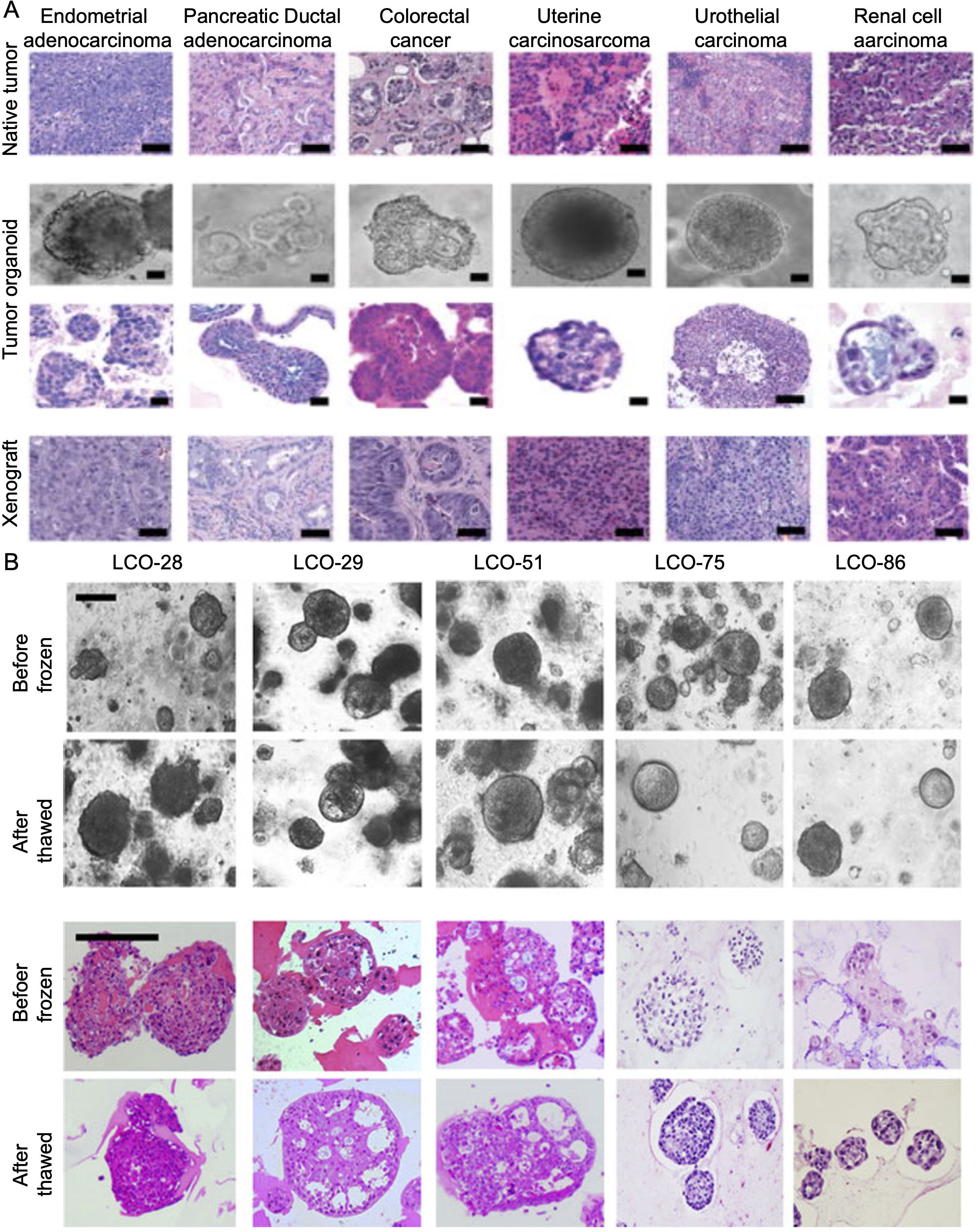
Figure 1. (A) Natural tumor specimens and their derived tumor organoids and xenografts (detailed procedure can be found in reference (Pauli et al., 2017). (B) Bright field microscopy images and H&E-stained images of LCOs before freezing and after thawing. After the thawing test on cryopreserved organoids, the morphology of organoids and the histologic features of original tissues were reconstituted. Scale bar, 200 μm. Information about the LCOs in these images: LCO-28, squamous cell carcinoma; LCO-29, large cell carcinoma; LCO-51, adenocarcinoma; LCO-75, small cell carcinoma; LCO-86, adenosquamous carcinoma (detailed procedure can be found in reference (Kim et al., 2019). LCO, Lung cancer organoids.
3 Development of organoid coculture techniques with pathogens
Coculture techniques play pivotal roles in the examination of host-pathogen interactions and the simplification of in vivo systems. The predictive capacity of cell culture-based assays is constrained by their inability to replicate the intricate organ complexity and inter-tissue communication present in vivo (LeSavage et al., 2022). The advent of microphysiological systems, exemplified by organoid cocultures, has achieved great progress in the fields of stem cell biology, disease modeling, and host-pathogen interactions. Nevertheless, there still exist intricate microbe-disease relationships. Hence, it is very necessary to develop simplified and meaningful approaches to model host-microbe interactions, and to visualize and analyze the mechanisms of bacterial adhesion and internalization at the microscopic level.
There are several methods for cocultures, such as direct coculture of viruses with organoids and injection of microorganisms into the organoid lumen. In the study of Nie et al., the HBV-containing supernatant of HepG2.2.15.7 cells, a HepG2.2.15 clone producing a higher level of HBV, was utilized to coculture with human induced pluripotent stem cell (hiPSC)-liver organoids, hiPSCs-hepatic-like cells, HepG2-tet-Na+-taurocholate cotransporting polypeptide organoids, and primary human hepatocytes in 24-well plates at a specific ratio (Figure 2A) (Nie et al., 2018). The harvested cells were then subjected to HBV covalent closed circular DNA (cccDNA) assay after infection for 10–20 days. This study successfully developed a stable HBV infection model through direct coculture of pathogens and PSCs-induced organoids. However, the coculture period is long, and organoids for passage and clonal growth following exposure to pathogens were limited after long-term culture.
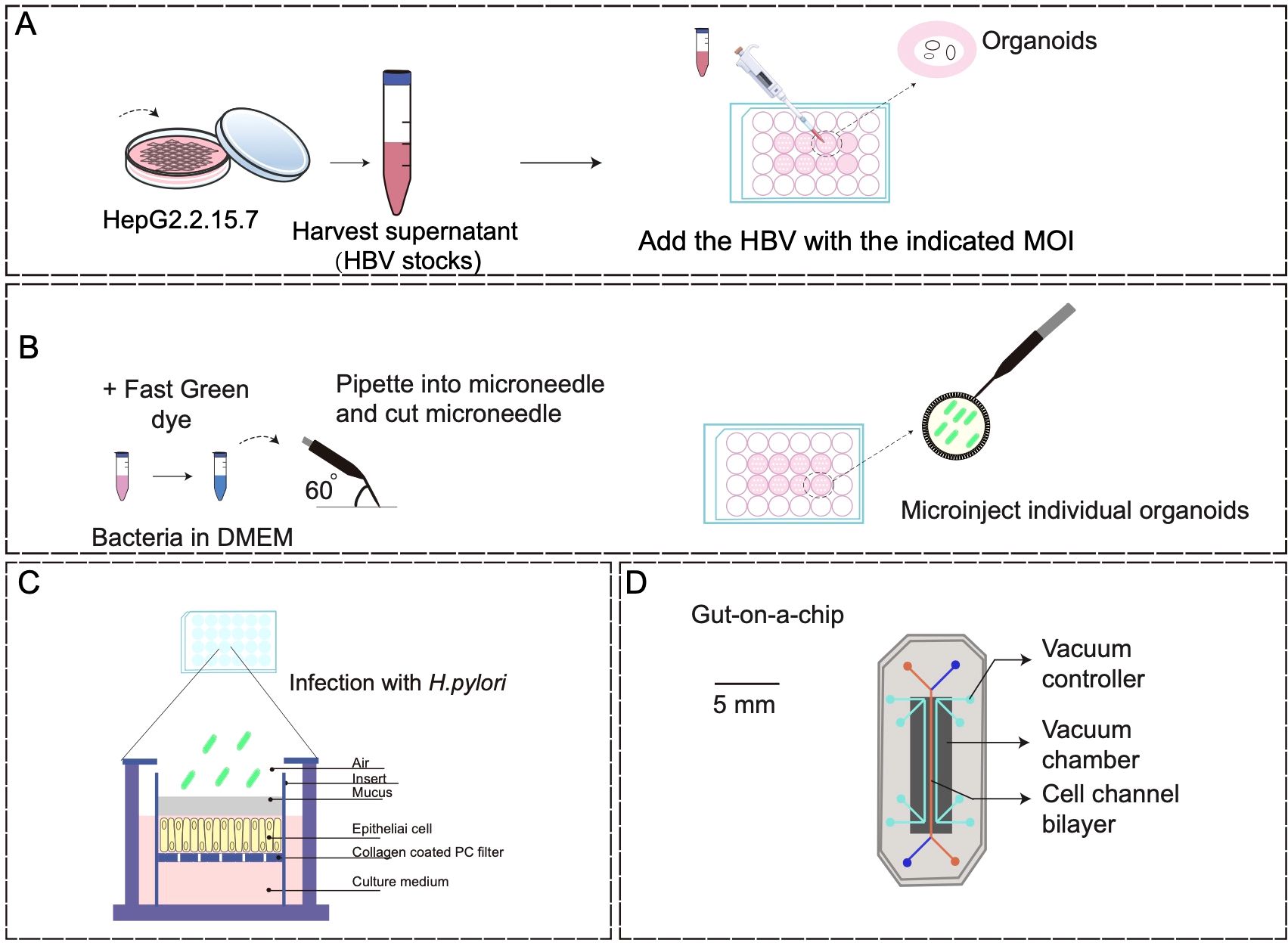
Figure 2. Coculture models of organoids with pathogenic microorganisms. (A) A schematic diagram of direct infection of HepG2.2.15.7 cells with HBV stock solution (detailed procedure can be found in reference (Nie et al., 2018). (B) Organoid microinjections (detailed procedure can be found in reference (Puschhof et al., 2021). (C) Human gastric mucosal columnar epithelium was regenerated using the air-liquid interface culture method (detailed procedure can be found in reference (Boccellato et al., 2019). (D) Human gut-on-a-chip inhabited by microbial flora (detailed procedure can be found in reference (Kim et al., 2012).
With the development of modern biotechnology, the microinjection of microorganisms into the organoid lumen has further enhanced the efficacy of coculture techniques (Figure 2B). The utility of microinjection lies in its capacity to accurately regulate the specific physiological localization of bacteria, although it is not conducive to conducting extensive infection studies. Furthermore, the adoption of transwell-based cell culture methods for investigating bacterial interactions with physiological tissue barriers is steadily increasing (Figure 2C). This method offers the benefit of ensuring consistent exposure of individual cells to microorganisms, but the absence of spatial and environmental protection in bacterial compartments results in reduced viability of specialized anaerobes or unregulated proliferation of other bacterial strains (Boccellato et al., 2019).
Microfluidic organoids-on-a-chip, derived from host tissue cells, offers a valuable tool for in vitro organ mimicry. This allows researchers to manipulate various cellular, molecular, chemical, and biophysical parameters in a controlled manner, either individually or in combination, to study their impact on the development and progression of human cancers, as well as the efficacy of therapeutic interventions (Kim et al., 2012; Bhatia and Ingber, 2014; Benam et al., 2016; Kasendra et al., 2018) (Figure 2D). Organoids-on-a-chip is a microfluidic cell culture device made of materials, such as optically transparent plastics, glass, or flexible polymers like polydimethylsiloxane, which contains perfused hollow microchannels filled with living cells (Kim et al., 2012; Kasendra et al., 2018). For instance, humans exhibit a significant vulnerability to enterohemorrhagic Escherichia coli (EHEC) infection, whereas mice display a relatively low susceptibility to this pathogen (Tovaglieri et al., 2019). Through the utilization of human colon microarray microfluidic culture technology, researchers simulated EHEC infection-induced epithelial damage in the human colon and found that exposure to metabolites originating from the human intestinal microbiomes resulted in more pronounced epithelial damage compared to mice (Tovaglieri et al., 2019). This study employed a multi-omics approach to identify 4 human microbiome metabolites as the mediators of this effect, including 4-methylbenzoic acid, 3,4-dimethylbenzoic acid, hexanoic acid, and heptanoic acid. Previous research on human host-microbiome-pathogen interactions primarily relied on the relevant genomic or macrogenomic studies, posing great challenges to establish causality in human pathogenesis (Surana and Kasper, 2017). The in vitro system described in this study demonstrates species-specificity and highlights the advantages of coculture systems based on the organoids-on-a-chip compared with organoid cultures alone. Additionally, Sun et al. presented an oncolytic virus (OV) evaluation system using microfluidic organ-on-a-chip systems and patient-derived hypopharyngeal and breast cancer organoids, and found that AD4-GHPE, a novel OV, had three antitumor mechanisms: tumor-specific cytotoxicity, a reduction in PD-L1 expression in tumor cells to increase CD8+ T-cell activity, and granulocyte-macrophage colony-stimulating factor secretion (Sun et al., 2025). This evaluation system based on tumor organoids is efficient and reliable, offering a personalized OV treatment recommendation for patients and providing industrialized and standardized research ideas for OV development.
4 Application of tumor organoids cocultured with pathogenic microorganisms
Coculture techniques have been widely utilized in the field of biology to investigate interactions between various cell populations, or cells and pathogenic microorganisms (Goers et al., 2014). Under this context, we focus on the cocultivation of pathogenic microorganisms and tumor cells. Traditional coculture systems using the cancer cell lines, such as direct coculture, indirect coculture, and co-immobilized mixed culture, are complex and lack versatility, and are unable to accurately replicate the host environment. In contrast, tumor organoid coculture models offer a more effective means of simulating the intricate interactions that occur within tumor tissues.
4.1 Brain tumor organoids and viral infections
Gliomas are the most common and lethal primary malignant adult brain tumors, in which glioblastomas are the most common (Zavala-Vega et al., 2019). EBV, a member of the herpesviridae family, was the first oncolytic virus to be described. Since then, several viruses associated with cancer have been identified (Cobbs, 2013; Lisyany et al., 2019).
In 2013, Lancaster and Knoblich developed a methodology for culturing brain organoids comprising multiple brain regions. As the organoid develops, cerebrospinal fluid similar to that in the lateral ventricles is found within the neuroepithelial buds. Concurrently, the neuroepithelial cells undergo additional differentiation and migration towards the outer layers, culminating in the formation of brain organoid cultures with various brain regions, including the forebrain, choroid plexus, hippocampal region, and prefrontal lobe (Lancaster et al., 2013). In 2014, Lancaster et al. developed a human PSC-derived 3D organoid culture system, known as brain organoids, which can generate various distinct and interconnected brain regions (Lancaster et al., 2013). Importantly, these brain organoids have been effectively utilized in modeling the pathogenesis of primary microcephaly through lentiviral shRNA targeting of CDK5/RAP2-dependent pathways.
The human brain is frequently susceptible to viral infections, and numerous viral families contain neurotropic viruses (Ruiz-Guillen et al., 2017; Tavcar et al., 2021). Neurological infections can cause central nervous system disorders, consequently leading to fatality or long-term consequences (Hopkins et al., 2021). Human cytomegalovirus (HCMV) infection is linked to human glioblastoma, but the precise mechanisms of infection remain incompletely elucidated. Dong et al. utilized the tissues from the glioblastoma margin to establish glioblastoma organoids (GBOs), and then cocultured the GBOs with HCMV after treatment with a 2,5-dimethylpyrrolizidine benzoic acid derivative, an EphA2 antagonist (Dong et al., 2023). The results revealed that EphA2 might serve as a potential therapeutic target for inhibiting HCMV infection in glioblastoma cells. The use of brain organoids offers a versatile human cellular platform for investigating cellular susceptibility, disease mechanisms, and therapeutic interventions (Jacob et al., 2020a). With the development of organoids cocultured with pathogenic microorganisms, the potential mechanism of brain tumors may be further illuminated.
4.2 Lung organoids and viruses
Lung cancer stands as the leading cause of cancer-related death worldwide (Hirsch et al., 2017). Although organoids established from human lung cancer resections and metastatic biopsies can preserve tumor histopathological and molecular features (Kim et al., 2019), there are rare studies regarding the association between lung cancer and infection based on the organoid platform. In view of this, we mainly investigated the relationship between organoids and respiratory viruses.
Recently, lung organoids have shown their suitability as the models for studying respiratory viruses. In a previous study, respiratory syncytial virus (RSV) and human parainfluenza virus (HPIV) were found to successfully infect human airway organoids (Porotto et al., 2019), which might serve as a versatile model for studying hereditary, malignant, and infectious pulmonary diseases (Sachs et al., 2019). There are also studies that use differentiated airway organoids to predict the infectivity of emerging respiratory viruses, including human and avian influenza viruses and zoonotic coronaviruses (Hui et al., 2018; Zhou et al., 2018; Han et al., 2021; Lamers et al., 2021). Importantly, the lung organoid platform can be used to screen therapeutic drugs and anti-microbial drugs (Sachs et al., 2019).
Regarding respiratory infectious diseases, virologists are trying to use organoid models as platforms to understand the mechanisms of viral infection, cell deregulation and drug screening, but there is still much to do in bacterial and parasitic infections (Fonseca et al., 2017). Heo et al. utilized organoids to illustrate the interaction of a human protozoan parasite, Cryptosporidium, with intestinal and lung epithelia that were considered as the two major sites of infection (Heo et al., 2018). After injection of Cryptosporidium oocysts into the organoid lumen, the parasite propagated within the organoids and completed its life cycle. Additionally, this study also highlighted the importance of interferon-I signaling in response to Cryptosporidium infection through transcriptomic analysis (Heo et al., 2018). In the future, we believe that cocultures of pathogens with lung organoids will be better established to understand and predict human infectious diseases.
4.3 Nasopharyngeal carcinoma organoids and EBV
Nasopharyngeal carcinoma (NPC) is a highly aggressive malignant tumor. Its etiology is multifactorial, in which EBV infection may be a major pathogenic factor (Chen et al., 2019). In 2022, Wang et al. successfully cultured NPCOs from a total of 77 samples, including 34 primary samples, 28 recurrent samples, and 15 samples of normal mucosa. The corresponding success rates of NPCOs were 47.06%, 81.25%, and 86.5%, respectively (Wang et al., 2022). All non-keratinizing NPCO samples exhibited positive for EBV-encoded small RNA (EBER) and negative for CK7. The recurrent NPCOs demonstrated increased expression of stem cell markers, including BMI-1, CD44, and CD133. Furthermore, the recurrent NPCOs could be successfully cultured up to the 4th generation and underwent multiple freeze-thaw cycles, unlike primary NPCOs which proved challenging to culture. Through histological staining, immunohistochemistry, and EBER in situ hybridization (ISH) assays, it was observed that NPCOs could retain the pathological characteristics of the original tumors and EBV infection status to a significant extent.
4.4 Gastric cancer organoids and H. pylori
H. pylori is an organism related to ulcer disease and gastric cancer, and its oncogenic actions fully reflect the intricate interplay between human cells, microorganisms, and the environment (Wroblewski et al., 2010). H. pylori infection can cause chronic inflammation of the gastric mucosa, resulting in gastric mucosal cell changes and atrophy to promote development of precancerous lesions and cancer.
Over a decade ago, human gastric organoids (hGOs) were successfully established utilizing gastric cancer tissue, cancerous site tissue, and induced PSCs (Kalabis et al., 2012; McCracken et al., 2014; Broda et al., 2019; Holokai et al., 2019). In 2014, McCracken et al. successfully developed a 3D hGO in vitro through directed differentiation of human PSCs (McCracken et al., 2014). The formation of these organoids depends on the regulation of various signaling pathways, including FGF, Wnt, BMP, retinoic acid, and EGF. The development of hGOs follows similar molecular and morphogenetic stages as observed in the mouse gastric development. In 2019, Holokai et al. demonstrated that H. pylori can induce the expression of the immune checkpoint molecule PD-L1 (CD274) via the Shh signaling pathway in a human organ culture model (Holokai et al., 2019). This study employed a coculture system involving patient-derived organoids infected with H. pylori and autologous immune cells to develop the therapy of H. pylori and PD-1 inhibitors and explore the protective role of PD-L1 against bacterial infection. In 2023, Wuputra et al. developed an organoid model of H. pylori infection by constructing a cytotoxin-associated gene A-GFP-tagged strain of H. pylori and infecting gastric organoids through microinjection (Wuputra et al., 2023). This resulted in the successful creation of a gastric organoid model capable of simulating H. pylori infection in vivo. To elucidate the functions of HDGF and TNFα secreted by H. pylori-infected tumor organoids, this study prepared recombinant HDGF and TNFα, and assessed the cytotoxicity and invasiveness of gastric cancer organoids. The findings suggest that HDGF and TNFα act as independent signaling molecules in the progression of gastric cancer infected by H. pylori.
The timeline from H. pylori infection to gastric atrophy, intestinal metaplasia and intraepithelial neoplasia may be months to years long (Piazuelo et al., 2021). During this period, the loss of acid-secreting parietal cells makes the stomach in a relatively hypochlorous environment, promoting changes in the composition of the gastric microbiota (Li and Perez Perez, 2018; Lahner et al., 2020; Barra et al., 2021). In humans with chronic gastritis, Prevotella, Streptococcus, Pseudomonas, Sphingobacterium, Bacillus, and Fusobacterium have also been found in normal mucosa adjacent to tumors. However, H. pylori remains an organism consistently identified at different stages of progression (Barra et al., 2021). The utilization of organoid models that are more sophisticated than the conventional models, such as cell lines, would enhance research on gastric epithelial repair, the function of gastric hormones, and the mechanisms of vaccine-induced protection.
4.5 Hepatocellular cancer organoids and HBV
HBV infection is the primary etiological factor for chronic cirrhosis and HCC (Di Bisceglie, 2009; MacLachlan and Cowie, 2015; An et al., 2018). The infection and replication of HBV are characterized by high specificity in host species and organs, which is believed to govern the intricate interplay between the immune response and virus-specific factors to culminate in the development of HCC. Epidemiological investigations have predominantly elucidated the molecular pathways involved in HBV-induced HCC, and genome-wide analyses of viral and host features are conducted (Fattovich et al., 2008; El-Serag, 2012; Fujimoto et al., 2012; Huang et al., 2012; Jiang et al., 2012; Ji et al., 2014; Shibata and Aburatani, 2014; Nantasanti et al., 2016; Cancer Genome Atlas Research Network, 2017; Sartorius et al., 2019; Sagnelli et al., 2020). Moreover, HCC cell lines are utilized in vitro studies (Zhang et al., 2014; Thomas and Liang, 2016). Nevertheless, the lack of appropriate animal or in vitro model systems for studying HBV infection poses a significant challenge due to the virus-specific host and cell type preferences. Chimpanzees are currently the sole animal model capable of supporting the entire HBV replication process, as they exhibit distinctly different gene expression profiles compared with primary cells (Protzer, 2017).
In 2021, a research team successfully cultured a liver organoid-derived primary in vitro HBV infection model from a healthy donor (De Crignis et al., 2021). These organoids were demonstrated to generate HBV cccDNA and HBeAg, and express intracellular HBV RNA and proteins, consequently producing infectious HBV. HBV-infected hepatocyte organoid platforms hold promise for drug screening to assess anti-HBV efficacy and drug-induced toxicity. Additionally, this study also utilized lentivirus to create transgenic organoid lines with integrated copies of HBV, contributing to viral production and HBV transcriptional research. Due to the diverse nature and immunosuppressive conditions, a significant majority (80-90%) of HCC patients do not exhibit objective responses to immunotherapy. Zou et al. developed chimeric antigen receptor T cells targeting HBV surface proteins (HBV-car-T cells) and personalized tumor-reactive CD8+ T cells (Zou et al., 2021). Subsequently, a coculture system involving autologous HBV+ HCC organoids and T cells was employed to assess their anti-tumor efficacy and mechanisms. Based on the microfluidic chip, a liver organoid system containing CD8+ T cells and ASCs was developed (Natarajan et al., 2022). This microfluidic coculture system supported the capability of targeted killing liver organoids with HCV non-structural protein 3-specific peptides under the circumstance of patient-derived KLVALGINAV CD8+ T cells. Furthermore, this study further underscored the innovative utility of the co-culture system for investigating the molecular mechanisms underlying the adaptive immune response to HCV in an in vitro model employing primary human cells.
4.6 Cervical cancer organoids and HPV
Over 90% of cervical cancer patients are attributed to high-risk HPV infection, particularly HPV-16 and HPV-18. High-risk HPV is known to cause cervical cancer through the expression of its E6/E7 proto-oncoproteins (Pal and Kundu, 2020). The squamocolumnar junction (SCJ) is the primary site of HPV infection (Rajendra and Sharma, 2019). Nevertheless, the absence of human-derived in vitro models for the SCJ has hindered the research on precancerous lesions and HPV-related cancers.
In 2020, researchers successfully generated organoids derived from the normal SCJ region using stromal gel 3D culture technology. These SCJ organoids primarily consisted of squamous cells in a compact structure, with some mucin-secreting uterine cervical canal cells present alongside the squamous cell population. Transcriptome analysis revealed elevated expression levels of SCJ marker genes in these organoids compared to immortalized cervical cell lines originating from non-SCJ regions (Maru et al., 2020). As a predominant subtype of cervical cancer, squamous cell carcinoma (SqCa) (Sahasrabuddhe et al., 2012) comprises 70% of all cases and typically follows a progression from HPV infection to low-grade squamous intraepithelial lesion (LSIL), then to high-grade squamous intraepithelial lesion (HSIL), with a process that may span over a decade (Gravitt and Winer, 2017). Thus, there is an urgent need for enhanced comprehension of the precancerous status. In 2024, Hu et al. collected HSIL/SqCa tissues from HPV-positive patients undergoing surgical biopsies to create a biobank containing cervical precancerous pathogens and tumor organoids, which retained genomic and transcriptomic profiles, as well as the causative HPV genome. Through coculturing the organoid models with HPV antigenic peptide-stimulated peripheral blood immune cells (Hu et al., 2024), different immune responses were observed in the two organoid models. This study established an experimental platform and biobank for conducting in vitro mechanistic studies on HPV-associated cervical diseases, screening therapeutic vaccines, and developing personalized treatment options.
Small cell carcinoma of the cervix (scCC) is also a rare and highly aggressive cancer associated with HPV. In a previous study, the organoids from a patient with HPV18-positive scCC were generated. Through whole exome sequencing and RNA-seq, therapeutic targets specific to HPV-derived scCC were identified. Additionally, utilizing organoids and organoid-derived mouse xenograft models, drug sensitivity testing was conducted. The findings all suggest the potential of tumor organoids in uncovering targets for rare cancers (Kusakabe et al., 2023).
5 Conclusions and prospects
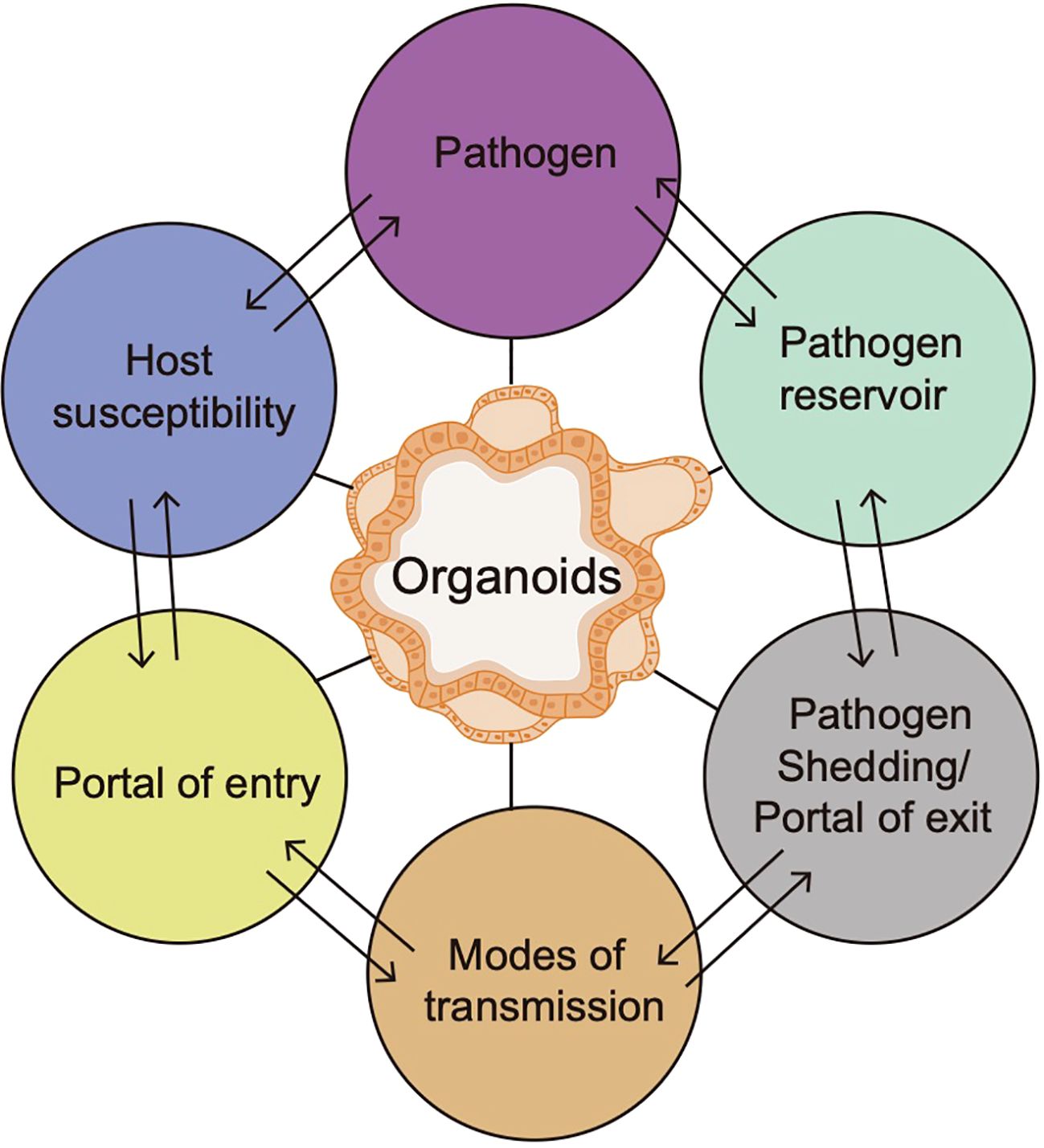
Pathogenic infection may appear in various anatomical locations within the host, which is usually considered to be an inducement for diseases (Gilbert and Lewis, 2019). Due to infection, host-pathogen interactions can result in either host immunity or an aggravated immune response mainly based on 6 factors, including the host susceptibility, portal of entry, modes of transmission, portal of exit, pathogen reservoir and pathogens (Dutta and Clevers, 2017). Organoids, a platform for studying pathogen-induced tumorigenesis, can be used to study diverse links of the chain of infection model and help to develop more efficacious control measures against emerging pathogens, thus promoting the understanding of the host-pathogen interactions (Figure 3). Nevertheless, there are still several challenges that should be considered. First, the full impact of tumor microenvironment on tumor behaviors is difficult to be captured in organoids due to lack of stromal components. Second, the microbial colonization efficiency is variable, and the study of anaerobic bacteria requires specialized techniques, including specific culture methods and manipulation of microbes (Strobel, 2009). Moreover, high-throughput experimental setups are limited by the manual nature of the microinjection procedure (Bartfeld et al., 2015). Although the technique of directly coculturing pathogenic microorganisms with organoids at a specific multiplicity of infection (MOI) has been extensively employed, there remains a lack of standard protocols for MOI and infection timing. Notably, the mutations in organoids are typically subclonal, random, and primarily impact non-coding regions, but refinement and standardization of reagents and protocols for organoid culture are very necessary for their effective utilization in clinical settings, including precancerous study and beyond.
Notwithstanding these challenges, the coculture system of tumor organoids with pathogenic microorganisms is significant in comprehending and simulating the status of human viral infection, in vivo homeostasis, and disease progression. Outside the gastrointestinal tract, the microbiota can affect the immune function by regulating the balance of Treg cells, γδT cells, and cytokine production. The brain interacts with the gastrointestinal system through a vast network described as the gut-brain axis, which may be expanded to include the gut microbiota, thus labeling the gut-microbiota-brain axis (Patterson et al., 2019). The existing preclinical data show that head injury can cause structural and functional damage to the digestive tract, but there is no experimental model that directly reflects this research (Sundman et al., 2017). Despite this gap, the coculture method proposed in this study may be used as a reference.
In the future, efforts will be made to gradually overcome the constraints above. The utilization of tumor organoid-based coculture models holds promise for enhancing patient-derived disease models, drug screening and stem cell research, as well as elucidating the interactions between pathogen-induced infection and tumor mechanisms, which paves the way for translational research and personalized treatment.
Author contributions
XZ: Writing – original draft, Investigation, Conceptualization, Writing – review & editing, Supervision. SS: Supervision, Writing – review & editing, Conceptualization, Investigation, Writing – original draft. SC: Formal analysis, Writing – original draft, Investigation, Data curation, Writing – review & editing. JD: Data curation, Investigation, Formal analysis, Writing – review & editing, Writing – original draft. FD: Resources, Writing – review & editing, Writing – original draft, Methodology, Investigation. JW: Writing – original draft, Resources, Writing – review & editing, Investigation, Methodology. DW: Writing – original draft, Methodology, Resources, Investigation, Writing – review & editing. YY: Supervision, Writing – original draft, Writing – review & editing, Conceptualization. YL: Funding acquisition, Writing – original draft, Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Key Laboratory of Precision Medicine for Malignancies in Liaoning Province (No.: k2229).
Conflict of interest
Authors FD and JW were employed by the company Kingbio Medical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, K., Yoo, K. J., Prem, K., and Jit, M. (2024). Equity impact of HPV vaccination on lifetime projections of cervical cancer burden among cohorts in 84 countries by global, regional, and income levels, 2010-22: a modelling study. EClinicalMedicine 70, 102524. doi: 10.1016/j.eclinm.2024.102524
Akkari, L., Grégoire, D., Floc'h, N., Moreau, M., Hernandez, C., Simonin, Y., et al. (2012). Hepatitis C viral protein NS5A induces EMT and participates in oncogenic transformation of primary hepatocyte precursors. J. Hepatol. 57, 1021–1028. doi: 10.1016/j.jhep.2012.06.027
Allen, J., Hao, S., Sears, C. L., and Timp, W. (2019). Epigenetic changes induced by bacteroides fragilis toxin. Infect. Immun. 87, e00447–e00418. doi: 10.1128/IAI.00447-18
Allen, J. and Sears, C. L. (2019). Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 11, 11. doi: 10.1186/s13073-019-0621-2
Amieva, M. and Peek, R. M., Jr. (2016). Pathobiology of helicobacter pylori-induced gastric cancer. Gastroenterology 150, 64–78. doi: 10.1053/j.gastro.2015.09.004
An, P., Xu, J., Yu, Y., and Winkler, C. A. (2018). Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front. Genet. 9, 261. doi: 10.3389/fgene.2018.00261
Arcia Franchini, A. P., Iskander, B., Anwer, F., Oliveri, F., Fotios, K., Panday, P., et al. (2022). The role of chlamydia trachomatis in the pathogenesis of cervical cancer. Cureus 14, e21331. doi: 0.7759/cureus.21331
Arzumanyan, A., Reis, H. M., and Feitelson, M. A. (2013). Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 13, 123–135. doi: 10.1038/nrc3449
Barra, W. F., Sarquis, D. P., Khayat, A. S., Khayat, B. C. M., Demachki, S., Anaissi, A. K. M., et al. (2021). Gastric cancer microbiome. Pathobiology 88, 156–169. doi: 10.1159/000512833
Bartfeld, S., Bayram, T., van de Wetering, M., Huch, M., Begthel, H., Kujala, P., et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–36 e6. doi: 10.1053/j.gastro.2014.09.042
Benam, K. H., Villenave, R., Lucchesi, C., Varone, A., Hubeau, C., Lee, H. H., et al. (2016). Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13, 151–157. doi: 10.1038/nmeth.3697
Bessède, E. and Mégraud, F. (2022). Microbiota and gastric cancer. Semin. Cancer Biol. 86, 11–17. doi: 10.1016/j.semcancer.2022.05.001
Bhatia, S. N. and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Boccellato, F., Woelffling, S., Imai-Matsushima, A., Sanchez, G., Goosmann, C., Schmid, M., et al. (2019). Polarised epithelial monolayers of the gastric mucosa reveal insights into mucosal homeostasis and defence against infection. Gut 68, 400–413. doi: 10.1136/gutjnl-2017-314540
Boj, S. F., Hwang, C. I., Baker, L. A., Chio, I. I., Engle, D. D., Corbo, V., et al. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. doi: 10.1016/j.cell.2014.12.021
Boleij, A. and Tjalsma, H. (2013). The itinerary of Streptococcus gallolyticus infection in patients with colonic Malignant disease. Lancet Infect. Dis. 13, 719–724. doi: 10.1016/S1473-3099(13)70107-5
Boleij, A., van Gelder, M. M., Swinkels, D. W., and Tjalsma, H. (2011). Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin. Infect. Dis. 53, 870–878. doi: 10.1093/cid/cir609
Bonnet, M., Buc, E., Sauvanet, P., Darcha, C., Dubois, D., Pereira, B., et al. (2014). Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 20, 859–867. doi: 10.1158/1078-0432.CCR-13-1343
Brennan, C. A. and Garrett, W. S. (2019). Fusobacterium nucleatum-symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Broda, T. R., McCracken, K. W., and Wells, J. M. (2019). Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat. Protoc. 14, 28–50. doi: 10.1038/s41596-018-0080-z
Broutier, L., Mastrogiovanni, G., Verstegen, M. M., Francies, H. E., Gavarró, L. M., Bradshaw, C. R., et al. (2017). Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435. doi: 10.1038/nm.4438
Buc, E., Dubois, D., Sauvanet, P., Raisch, J., Delmas, J., Darfeuille-Michaud, A., et al. (2013). High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PloS One 8, e56964. doi: 10.1371/journal.pone.0056964
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240
Campbell, K. (2022). Hepatitis B and the liver cancer endgame. Nature 603, S64–S65. doi: 10.1038/d41586-022-00821-0
Cancer Genome Atlas Research Network (2017). Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 169, 1327–41 e23. doi: 10.1016/j.cell.2017.05.046
Chen, Y. P., Chan, A. T. C., Le, Q. T., Blanchard, P., Sun, Y., and Ma, J. (2019). Nasopharyngeal carcinoma. Lancet 394, 64–80. doi: 10.1016/S0140-6736(19)30956-0
Chouhan, D., Barani Devi, T., Chattopadhyay, S., Dharmaseelan, S., Nair, G. B., Devadas, K., et al. (2019). Mycobacterium abscessus infection in the stomach of patients with various gastric symptoms. PloS Negl. Trop. Dis. 13, e0007799. doi: 10.1371/journal.pntd.0007799
Cobbs, C. S. (2013). Cytomegalovirus and brain tumor: epidemiology, biology and therapeutic aspects. Curr. Opin. Oncol. 25, 682–688. doi: 10.1097/CCO.0000000000000005
De Crignis, E., Hossain, T., Romal, S., Carofiglio, F., Moulos, P., Khalid, M. M., et al. (2021). Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife 10, e60747. doi: 10.7554/eLife.60747.sa2
DeStefano Shields, C. E., White, J. R., Chung, L., Wenzel, A., Hicks, J. L., Tam, A. J., et al. (2021). Bacterial-driven inflammation and mutant BRAF expression combine to promote murine colon tumorigenesis that is sensitive to immune checkpoint therapy. Cancer Discov. 11, 1792–1807. doi: 10.1158/2159-8290.CD-20-0770
Di Bisceglie, A. M. (2009). Hepatitis B and hepatocellular carcinoma. Hepatology 49, S56–S60. doi: 10.1002/hep.22962
Di Domenico, E. G., Cavallo, I., Pontone, M., Toma, L., and Ensoli, F. (2017). Biofilm producing salmonella typhi: chronic colonization and development of gallbladder cancer. Int. J. Mol. Sci. 18, 1887. doi: 10.3390/ijms18091887
Dong, X. D., Li, Y., Li, Y., Sun, C., Liu, S. X., Duan, H., et al. (2023). EphA2 is a functional entry receptor for HCMV infection of glioblastoma cells. PloS Pathog. 19, e1011304. doi: 10.1371/journal.ppat.1011304
Drewes, J. L., White, J. R., Dejea, C. M., Fathi, P., Iyadorai, T., Vadivelu, J., et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34. doi: 10.1038/s41522-017-0040-3
Driehuis, E., Kretzschmar, K., and Clevers, H. (2020). Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409. doi: 10.1038/s41596-020-0379-4
Dutta, D. and Clevers, H. (2017). Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 48, 15–22. doi: 10.1016/j.coi.2017.07.012
El-Serag, H. B. (2012). Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–73 e1. doi: 10.1053/j.gastro.2011.12.061
Fattovich, G., Bortolotti, F., and Donato, F. (2008). Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J. Hepatol. 48, 335–352. doi: 10.1016/j.jhep.2007.11.011
Fonseca, K. L., Rodrigues, P. N. S., Olsson, I. A. S., and Saraiva, M. (2017). Experimental study of tuberculosis: From animal models to complex cell systems and organoids. PloS Pathog. 13, e1006421. doi: 10.1371/journal.ppat.1006421
Fujimoto, A., Totoki, Y., Abe, T., Boroevich, K. A., Hosoda, F., Nguyen, H. H., et al. (2012). Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764. doi: 10.1038/ng.2291
Fukuda, K., Kuroki, T., Tajima, Y., Tsuneoka, N., Kitajima, T., Matsuzaki, S., et al. (2002). Comparative analysis of Helicobacter DNAs and biliary pathology in patients with and without hepatobiliary cancer. Carcinogenesis 23, 1927–1931. doi: 10.1093/carcin/23.11.1927
Gao, D., Vela, I., Sboner, A., Iaquinta, P. J., Karthaus, W. R., Gopalan, A., et al. (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187. doi: 10.1016/j.cell.2014.08.016
Gilbert, N. M. and Lewis, A. L. (2019). Covert pathogenesis: Transient exposures to microbes as triggers of disease. PloS Pathog. 15, e1007586. doi: 10.1371/journal.ppat.1007586
Goers, L., Freemont, P., and Polizzi, K. M. (2014). Co-culture systems and technologies: taking synthetic biology to the next level. J. R Soc. Interface 11, 20140065. doi: 10.1098/rsif.2014.0065
Gravitt, P. E. and Winer, R. L. (2017). Natural history of HPV infection across the lifespan: role of viral latency. Viruses 9, 267. doi: 10.3390/v9100267
Guo, C., Qu, X., Tang, X., Song, Y., Wang, J., Hua, K., et al. (2023). Spatiotemporally deciphering the mysterious mechanism of persistent HPV-induced Malignant transition and immune remodelling from HPV-infected normal cervix, precancer to cervical cancer: Integrating single-cell RNA-sequencing and spatial transcriptome. Clin. Transl. Med. 13, e1219. doi: 10.1002/ctm2.v13.3
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275. doi: 10.1038/s41586-020-2901-9
He, Y., Zhu, Y., Wang, W., Yi, Y., Wang, Z., Zhao, C., et al. (2025). Clinical efficacy and chemoresistance analysis of precision neoadjuvant chemotherapy for borderline resectable pancreatic cancer: a prospective, single-arm pilot study. Int. J. Surg. 111, 3269–3280. doi: 10.1097/JS9.0000000000002342
Heo, I., Dutta, D., Schaefer, D. A., Iakobachvili, N., Artegiani, B., Sachs, N., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. doi: 10.1038/s41564-018-0177-8
Hirsch, F. R., Scagliotti, G. V., Mulshine, J. L., Kwon, R., Curran, W. J., Jr., Wu, Y. L., et al. (2017). Lung cancer: current therapies and new targeted treatments. Lancet 389, 299–311. doi: 10.1016/S0140-6736(16)30958-8
Holokai, L., Chakrabarti, J., Broda, T., Chang, J., Hawkins, J. A., Sundaram, N., et al. (2019). Increased programmed death-ligand 1 is an early epithelial cell response to helicobacter pylori infection. PloS Pathog. 15, e1007468. doi: 10.1371/journal.ppat.1007468
Hopkins, H. K., Traverse, E. M., and Barr, K. L. (2021). Methodologies for generating brain organoids to model viral pathogenesis in the CNS. Pathogens 10, 1510. doi: 10.3390/pathogens10111510
Hu, B., Wang, R., Wu, D., Long, R., Fan, J., Hu, Z., et al. (2024). A promising new model: establishment of patient-derived organoid models covering HPV-related cervical pre-cancerous lesions and their cancers. Adv. Sci. (Weinh) 11, e2302340. doi: 10.1002/advs.202302340
Hua-Feng, X., Yue-Ming, W., Hong, L., and Junyi, D. (2015). A meta-analysis of the association between Chlamydia pneumoniae infection and lung cancer risk. Indian J. Cancer 52 Suppl 2, e112–e115. doi: 10.4103/0019-509X.172506
Huang, J., Deng, Q., Wang, Q., Li, K. Y., Dai, J. H., Li, N., et al. (2012). Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 44, 1117–1121. doi: 10.1038/ng.2391
Hui, K. P. Y., Ching, R. H. H., Chan, S. K. H., Nicholls, J. M., Sachs, N., Clevers, H., et al. (2018). Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 6, 846–854. doi: 10.1016/S2213-2600(18)30236-4
Incognito, G. G., Ronsini, C., Palmara, V., Romeo, P., Vizzielli, G., Restaino, S., et al. (2025). The interplay between cervicovaginal microbiota diversity, lactobacillus profiles and human papillomavirus in cervical cancer: A systematic review. Healthcare (Basel) 13, 599. doi: 10.37766/inplasy2025.3.0027
Jacob, F., Pather, S. R., Huang, W. K., Zhang, F., Wong, S. Z. H., Zhou, H., et al. (2020a). Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-coV-100. Neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 27, 937–50 e9. doi: 10.1016/j.stem.2020.09.016
Jacob, F., Salinas, R. D., Zhang, D. Y., Nguyen, P. T. T., Schnoll, J. G., Wong, S. Z. H., et al. (2020b). A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204.e22. doi: 10.1016/j.cell.2019.11.036
Jeong, Y. J., Knutsdottir, H., Shojaeian, F., Lerner, M. G., Wissler, M. F., Henriet, E., et al. (2023). Morphology-guided transcriptomic analysis of human pancreatic cancer organoids reveals microenvironmental signals that enhance invasion. J. Clin. Invest 133, e162054. doi: 10.1172/JCI162054
Ji, X., Zhang, Q., Du, Y., Liu, W., Li, Z., Hou, X., et al. (2014). Somatic mutations, viral integration and epigenetic modification in the evolution of hepatitis B virus-induced hepatocellular carcinoma. Curr. Genomics 15, 469–480. doi: 10.2174/1389202915666141114213833
Jiang, Z., Jhunjhunwala, S., Liu, J., Haverty, P. M., Kennemer, M. I., Guan, Y., et al. (2012). The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 22, 593–601. doi: 10.1101/gr.133926.111
Kalabis, J., Wong, G. S., Vega, M. E., Natsuizaka, M., Robertson, E. S., Herlyn, M., et al. (2012). Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat. Protoc. 7, 235–246. doi: 10.1038/nprot.2011.437
Karthaus, W. R., Iaquinta, P. J., Drost, J., Gracanin, A., van Boxtel, R., Wongvipat, J., et al. (2014). Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175. doi: 10.1016/j.cell.2014.08.017
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 8, 2871. doi: 10.1038/s41598-018-21201-7
Kidane, D. (2018). Molecular mechanisms of H. pylori-induced DNA double-strand breaks. Int. J. Mol. Sci. 19, 2891. doi: 10.3390/ijms19102891
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12, 2165–2174. doi: 10.1039/c2lc40074j
Kim, J., Koo, B. K., and Knoblich, J. A. (2020). Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584. doi: 10.1038/s41580-020-0259-3
Kim, M., Mun, H., Sung, C. O., Cho, E. J., Jeon, H. J., Chun, S. M., et al. (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10, 3991. doi: 10.1038/s41467-019-11867-6
Kobayashi, T., Harada, K., Miwa, K., and Nakanuma, Y. (2005). Helicobacter genus DNA fragments are commonly detectable in bile from patients with extrahepatic biliary diseases and associated with their pathogenesis. Dig Dis. Sci. 50, 862–867. doi: 10.1007/s10620-005-2654-1
Kopper, O., de Witte, C. J., Lõhmussaar, K., Valle-Inclan, J. E., Hami, N., Kester, L., et al. (2019). An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 25, 838–849. doi: 10.1038/s41591-019-0422-6
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Kostyusheva, A., Brezgin, S., Glebe, D., Kostyushev, D., and Chulanov, V. (2021). Host-cell interactions in HBV infection and pathogenesis: the emerging role of m6A modification. Emerg. Microbes Infect. 10, 2264–2275. doi: 10.1080/22221751.2021.2006580
Kusakabe, M., Taguchi, A., Tanikawa, M., Hoshi, D., Tsuchimochi, S., Qian, X., et al. (2023). Application of organoid culture from HPV18-positive small cell carcinoma of the uterine cervix for precision medicine. Cancer Med. 12, 8476–8489. doi: 10.1002/cam4.v12.7
Kwon, S. Y., Thi-Thu Ngo, H., Son, J., Hong, Y., and Min, J. J. (2024). Exploiting bacteria for cancer immunotherapy. Nat. Rev. Clin. Oncol. 21, 569–589. doi: 10.1038/s41571-024-00908-9
Lahner, E., Conti, L., Annibale, B., and Corleto, V. D. (2020). Current perspectives in atrophic gastritis. Curr. Gastroenterol. Rep. 22, 38. doi: 10.1007/s11894-020-00775-1
Lamers, M. M., van der Vaart, J., Knoops, K., Riesebosch, S., Breugem, T. I., Mykytyn, A. Z., et al. (2021). An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J. 40, e105912. doi: 10.15252/embj.2020105912
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Lee, S. H., Hu, W., Matulay, J. T., Silva, M. V., Owczarek, T. B., Kim, K., et al. (2018). Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–28.e17. doi: 10.1016/j.cell.2018.03.017
LeSavage, B. L., Suhar, R. A., Broguiere, N., Lutolf, M. P., and Heilshorn, S. C. (2022). Next-generation cancer organoids. Nat. Mater. 21, 143–159. doi: 10.1038/s41563-021-01057-5
Li, Y., Gao, X., Ni, C., Zhao, B., and Cheng, X. (2023). The application of patient-derived organoid in the research of lung cancer. Cell Oncol. (Dordr) 46, 503–519. doi: 10.1007/s13402-023-00771-3
Li, J. and Perez Perez, G. I. (2018). Is there a role for the non-helicobacter pylori bacteria in the risk of developing gastric cancer? Int. J. Mol. Sci. 19, 1353. doi: 10.3390/ijms19051353
Lisyany, N. I., Klyuchnikova, A. A., Belskaya, L. N., Lisyany, A. A., and Gnedkova, I. A. (2019). Cytomegaloviruses and Malignant brain tumors. Exp. Oncol. 41, 300–303. doi: 10.32471/10.32471/exp-oncology.2312-8852.vol-41-no-4
Lõhmussaar, K., Oka, R., Espejo Valle-Inclan, J., Smits, M. H. H., Wardak, H., Korving, J., et al. (2021). Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell. 28, 1380–96.e6. doi: 10.1016/j.stem.2021.03.012
MacLachlan, J. H. and Cowie, B. C. (2015). Hepatitis B virus epidemiology. Cold Spring Harb. Perspect. Med. 5, a021410. doi: 10.1101/cshperspect.a021410
Maiuri, A. R., Peng, M., Podicheti, R., Sriramkumar, S., Kamplain, C. M., Rusch, D. B., et al. (2017). Mismatch repair proteins initiate epigenetic alterations during inflammation-driven tumorigenesis. Cancer Res. 77, 3467–3478. doi: 10.1158/0008-5472.CAN-17-0056
Maru, Y., Kawata, A., Taguchi, A., Ishii, Y., Baba, S., Mori, M., et al. (2020). Establishment and molecular phenotyping of organoids from the squamocolumnar junction region of the uterine cervix. Cancers (Basel) 12, 694. doi: 10.3390/cancers12030694
McCauley, H. A. and Wells, J. M. (2017). Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962. doi: 10.1242/dev.140731
McCracken, K. W., Cata, E. M., Crawford, C. M., Sinagoga, K. L., Schumacher, M., Rockich, B. E., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. doi: 10.1038/nature13863
Nantasanti, S., de Bruin, A., Rothuizen, J., Penning, L. C., and Schotanus, B. A. (2016). Concise review: organoids are a powerful tool for the study of liver disease and personalized treatment design in humans and animals. Stem Cells Transl. Med. 5, 325–330. doi: 10.5966/sctm.2015-0152
Natarajan, V., Simoneau, C. R., Erickson, A. L., Meyers, N. L., Baron, J. L., Cooper, S., et al. (2022). Modelling T-cell immunity against hepatitis C virus with liver organoids in a microfluidic coculture system. Open Biol. 12, 210320. doi: 10.1098/rsob.210320
Neal, J. T. and Kuo, C. J. (2016). Organoids as models for neoplastic transformation. Annu. Rev. Pathol. 11, 199–220. doi: 10.1146/annurev-pathol-012615-044249
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189
Nie, Y. Z., Zheng, Y. W., Miyakawa, K., Murata, S., Zhang, R. R., Sekine, K., et al. (2018). Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 35, 114–123. doi: 10.1016/j.ebiom.2018.08.014
Oh, H. Y., Kim, B. S., Seo, S. S., Kong, J. S., Lee, J. K., Park, S. Y., et al. (2015). The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin. Microbiol. Infect. 21, 674.e1–674.e9. doi: 10.1016/j.cmi.2015.02.026
Ou, L., Liu, S., Wang, H., Guo, Y., Guan, L., Shen, L., et al. (2023). Patient-derived melanoma organoid models facilitate the assessment of immunotherapies. EBioMedicine 92, 104614. doi: 10.1016/j.ebiom.2023.104614
Ouchi, R., Togo, S., Kimura, M., Shinozawa, T., Koido, M., Koike, H., et al. (2019). Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–84 e6. doi: 10.1016/j.cmet.2019.05.007
Pal, A. and Kundu, R. (2020). Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front. Microbiol. 10, 3116. doi: 10.3389/fmicb.2019.03116
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 11, 3259. doi: 10.1038/s41467-020-16967-2
Patterson, T. T., Nicholson, S., Wallace, D., Hawryluk, G. W. J., and Grandhi, R. (2019). Complex feed-forward and feedback mechanisms underlie the relationship between traumatic brain injury and the gut-microbiota-brain axis. Shock 52, 318–325. doi: 10.1097/SHK.0000000000001278
Pauli, C., Hopkins, B. D., Prandi, D., Shaw, R., Fedrizzi, T., Sboner, A., et al. (2017). Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 7, 462–477. doi: 10.1158/2159-8290.CD-16-1154
Peng, Y., Tang, Q., Wu, S., and Zhao, C. (2025). Associations of Atopobium, Garderella, Megasphaera, Prevotella, Sneathia, and Streptococcus with human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: a systematic review and meta-analysis. BMC Infect. Dis. 25, 708. doi: 10.1186/s12879-025-10851-4
Piazuelo, M. B., Bravo, L. E., Mera, R. M., Camargo, M. C., Bravo, J. C., Delgado, A. G., et al. (2021). The Colombian chemoprevention trial: 20-year follow-up of a cohort of patients with gastric precancerous lesions. Gastroenterology 160, 1106–17 e3. doi: 10.1053/j.gastro.2020.11.017
Plummer, M., de Martel, C., Vignat, J., Ferlay, J., Bray, F., and Franceschi, S. (2016). Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 4, e609–e616. doi: 10.1016/S2214-109X(16)30143-7
Porotto, M., Ferren, M., Chen, Y. W., Siu, Y., Makhsous, N., Rima, B., et al. (2019). Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. MBio 10, e00723–e00719. doi: 10.1128/mBio.00723-19
Protzer, U. (2017). Viral hepatitis: The bumpy road to animal models for HBV infection. Nat. Rev. Gastroenterol. Hepatol. 14, 327–328. doi: 10.1038/nrgastro.2017.44
Purcell, R. V., Visnovska, M., Biggs, P. J., Schmeier, S., and Frizelle, F. A. (2017). Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 7, 11590. doi: 10.1038/s41598-017-11237-6
Puschhof, J., Pleguezuelos-Manzano, C., Martinez-Silgado, A., Akkerman, N., Saftien, A., Boot, C., et al. (2021). Intestinal organoid cocultures with microbes. Nat. Protoc. 16, 4633–4649. doi: 10.1038/s41596-021-00589-z
Qu, J., Kalyani, F. S., Liu, L., Cheng, T., and Chen, L. (2021). Tumor organoids: synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun. (Lond) 41, 1331–1353. doi: 10.1002/cac2.v41.12
Rajendra, S. and Sharma, P. (2019). Transforming human papillomavirus infection and the esophageal transformation zone: prime time for total excision/ablative therapy? Dis. Esophagus 32, doz008. doi: 10.1093/dote/doz008
Rosenbluth, J. M., Schackmann, R. C. J., Gray, G. K., Selfors, L. M., Li, C. M., Boedicker, M., et al. (2020). Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 11, 1711. doi: 10.1038/s41467-020-15548-7
Ruiz-Guillen, M., Abrescia, N. G., and Smerdou, C. (2017). Neurotropic alphaviruses can propagate without capsid. Oncotarget 8, 8999–9000. doi: 10.18632/oncotarget.13993
Sachs, N., de Ligt, J., Kopper, O., Gogola, E., Bounova, G., Weeber, F., et al. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–86.e10. doi: 10.1016/j.cell.2017.11.010
Sachs, N., Papaspyropoulos, A., Zomer-van Ommen, D. D., Heo, I., Böttinger, L., Klay, D., et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300. doi: 10.15252/embj.2018100300
Sagnelli, E., Macera, M., Russo, A., Coppola, N., and Sagnelli, C. (2020). Epidemiological and etiological variations in hepatocellular carcinoma. Infection 48, 7–17. doi: 10.1007/s15010-019-01345-y
Sahasrabuddhe, V. V., Parham, G. P., Mwanahamuntu, M. H., and Vermund, S. H. (2012). Cervical cancer prevention in low- and middle-income countries: feasible, affordable, essential. Cancer Prev. Res. (Phila) 5, 11–17. doi: 10.1158/1940-6207.CAPR-11-0540
Sartorius, K., Makarova, J., Sartorius, B., An, P., Winkler, C., Chuturgoon, A., et al. (2019). The regulatory role of microRNA in hepatitis-B virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells 8, 1504. doi: 10.3390/cells8121504
Seelbinder, B., Lohinai, Z., Vazquez-Uribe, R., Brunke, S., Chen, X., Mirhakkak, M., et al. (2023). Candida expansion in the gut of lung cancer patients associates with an ecological signature that supports growth under dysbiotic conditions. Nat. Commun. 14, 2673. doi: 10.1038/s41467-023-38058-8
Sepich-Poore, G. D., Zitvogel, L., Straussman, R., Hasty, J., Wargo, J. A., and Knight, R. (2021). The microbiome and human cancer. Science 371, eabc4552. doi: 10.1126/science.abc4552
Shibata, T. and Aburatani, H. (2014). Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 11, 340–349. doi: 10.1038/nrgastro.2014.6
Soleimani, N. and Javadi, M. M. (2022). Future prospects of bacteria-mediated cancer therapies: Affliction or opportunity? Microb. Pathog. 172, 105795. doi: 10.1016/j.micpath.2022.105795
Strobel, H. J. (2009). Basic laboratory culture methods for anaerobic bacteria. Methods Mol. Biol. 581, 247–261. doi: 10.1007/978-1-60761-214-8_16
Sun, Y., Liu, J., Zhu, L., Huang, F., Dong, Y., Liu, S., et al. (2025). Treatment response to oncolytic virus in patient-derived breast cancer and hypopharyngeal cancer organoids: evaluation via a microfluidics organ-on-a-chip system. Bioengineering (Basel) 12, 146. doi: 10.3390/bioengineering12020146
Sundman, M. H., Chen, N. K., Subbian, V., and Chou, Y. H. (2017). The bidirectional gut-brain-microbiota axis as a potential nexus between traumatic brain injury, inflammation, and disease. Brain Behav. Immun. 66, 31–44. doi: 10.1016/j.bbi.2017.05.009
Surana, N. K. and Kasper, D. L. (2017). Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247. doi: 10.1038/nature25019
Tao, M., Sun, F., Wang, J., Wang, Y., Zhu, H., Chen, M., et al. (2022). Developing patient-derived organoids to predict PARP inhibitor response and explore resistance overcoming strategies in ovarian cancer. Pharmacol. Res. 179, 106232. doi: 10.1016/j.phrs.2022.106232
Tavcar, P., Potokar, M., Kolenc, M., Korva, M., Avsic-Zupanc, T., Zorec, R., et al. (2021). Neurotropic viruses, astrocytes, and COVID-19. Front. Cell Neurosci. 15, 662578. doi: 10.3389/fncel.2021.662578
Thomas, E. and Liang, T. J. (2016). Experimental models of hepatitis B and C - new insights and progress. Nat. Rev. Gastroenterol. Hepatol. 13, 362–374. doi: 10.1038/nrgastro.2016.37
Tindle, C., Fuller, M., Fonseca, A., Taheri, S., Ibeawuchi, S. R., Beutler, N., et al. (2021). Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. Elife 10, e66417. doi: 10.7554/eLife.66417
Tovaglieri, A., Sontheimer-Phelps, A., Geirnaert, A., Prantil-Baun, R., Camacho, D. M., Chou, D. B., et al. (2019). Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 7, 43. doi: 10.1186/s40168-019-0650-5
Tsay, J. J., Wu, B. G., Badri, M. H., Clemente, J. C., Shen, N., Meyn, P., et al. (2018). Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 198, 1188–1198. doi: 10.1164/rccm.201710-2118OC
Tsay, J. J., Wu, B. G., Sulaiman, I., Gershner, K., Schluger, R., Li, Y., et al. (2021). Lower airway dysbiosis affects lung cancer progression. Cancer Discov. 11, 293–307. doi: 10.1158/2159-8290.CD-20-0263
Tsoi, H., Chu, E. S. H., Zhang, X., Sheng, J., Nakatsu, G., Ng, S. C., et al. (2017). Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 152, 1419–33.e5. doi: 10.1053/j.gastro.2017.01.009
Valguarnera, E. and Wardenburg, J. B. (2020). Good gone bad: one toxin away from disease for bacteroides fragilis. J. Mol. Biol. 432, 765–785. doi: 10.1016/j.jmb.2019.12.003
van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi: 10.1016/j.cell.2015.03.053
Veninga, V. and Voest, E. E. (2021). Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell. 39, 1190–1201. doi: 10.1016/j.ccell.2021.07.020
Walsh, A. J., Cook, R. S., and Skala, M. C. (2017). Functional optical imaging of primary human tumor organoids: development of a personalized drug screen. J. Nucl. Med. 58, 1367–1372. doi: 10.2967/jnumed.117.192534
Wang, X. W., Xia, T. L., Tang, H. C., Liu, X., Han, R., Zou, X., et al. (2022). Establishment of a patient-derived organoid model and living biobank for nasopharyngeal carcinoma. Ann. Transl. Med. 10, 526. doi: 10.21037/atm-22-1076
Wang, H. M., Zhang, C. Y., Peng, K. C., Chen, Z. X., Su, J. W., Li, Y. F., et al. (2023). Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: A real-world study. Cell Rep. Med. 4, 100911. doi: 10.1016/j.xcrm.2022.100911
Wang, M., Zhang, M., and Xu, H. (2020). First report of bacteremia caused by Eggerthia catenaformis in a patient with gastric Malignancy in China. Anaerobe 64, 102218. doi: 10.1016/j.anaerobe.2020.102218
Ward, J. M., Fox, J. G., Anver, M. R., Haines, D. C., George, C. V., Collins, M. J., Jr, et al. (1994). Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst. 86, 1222–1227. doi: 10.1093/jnci/86.16.1222
Wasunan, P., Maneewong, C., Daengprok, W., and Thirabunyanon, M. (2022). Bioactive Earthworm Peptides Produced by Novel Protease-Producing Bacillus velezensis PM 35 and Its Bioactivities on Liver Cancer Cell Death via Apoptosis, Antioxidant Activity, Protection Against Oxidative Stress, and Immune Cell Activation. Front. Microbiol. 13, 892945. doi: 10.3389/fmicb.2022.892945
Wong, C. C. and Yu, J. (2023). Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20, 429–452. doi: 10.1038/s41571-023-00766-x
Wroblewski, L. E., Peek, R. M., Jr., and Wilson, K. T. (2010). Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin. Microbiol. Rev. 23, 713–739. doi: 10.1128/CMR.00011-10
Wuputra, K., Ku, C. C., Pan, J. B., Liu, C. J., Kato, K., Lin, Y. C., et al. (2023). Independent signaling of hepatoma derived growth factor and tumor necrosis factor-alpha in human gastric cancer organoids infected by helicobacter pylori. Int. J. Mol. Sci. 24, 6567. doi: 10.3390/ijms24076567
Xu, H., Jiao, D., Liu, A., and Wu, K. (2022). Tumor organoids: applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 15, 58. doi: 10.1186/s13045-022-01278-4
Yan, H. H. N., Siu, H. C., Law, S., Ho, S. L., Yue, S. S. K., Tsui, W. Y., et al. (2018). A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 23, 882–97.e11. doi: 10.1016/j.stem.2018.09.016
Yoshida, G. J. (2020). Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 13, 4. doi: 10.1186/s13045-019-0829-z
Zavala-Vega, S., Palma-Lara, I., Ortega-Soto, E., Trejo-Solis, C., de Arellano, I. T., Ucharima-Corona, L. E., et al. (2019). Role of epstein-barr virus in glioblastoma. Crit. Rev. Oncog 24, 307–338. doi: 10.1615/CritRevOncog.2019032655
Zhan, P., Suo, L. J., Qian, Q., Shen, X. K., Qiu, L. X., Yu, L. K., et al. (2011). Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. Eur. J. Cancer 47, 742–747. doi: 10.1016/j.ejca.2010.11.003
Zhang, B., Miao, T., Shen, X., Bao, L., Zhang, C., Yan, C., et al. (2020). EB virus-induced ATR activation accelerates nasopharyngeal carcinoma growth via M2-type macrophages polarization. Cell Death Dis. 11, 742. doi: 10.1038/s41419-020-02925-9
Zhang, X. D., Wang, Y., and Ye, L. H. (2014). Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol. Med. 11, 182–190. doi: 10.7497/j.issn.2095-3941.2014.03.004
Zhang, Y., Weng, Y., Gan, H., Zhao, X., and Zhi, F. (2018). Streptococcus gallolyticus conspires myeloid cells to promote tumorigenesis of inflammatory bowel disease. Biochem. Biophys. Res. Commun. 506, 907–911. doi: 10.1016/j.bbrc.2018.10.136
Zhang, W., Yin, Y., Jiang, Y., Yang, Y., Wang, W., Wang, X., et al. (2024). Relationship between vaginal and oral microbiome in patients of human papillomavirus (HPV) infection and cervical cancer. J. Transl. Med. 22, 396. doi: 10.1186/s12967-024-05124-8
Zhou, J., Li, C., Sachs, N., Chiu, M. C., Wong, B. H., Chu, H., et al. (2018). Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. U S A 115, 6822–6827. doi: 10.1073/pnas.1806308115
Zhu, H., Shen, Z., Luo, H., Zhang, W., and Zhu, X. (2016). Chlamydia trachomatis infection-associated risk of cervical cancer: A meta-analysis. Med. (Baltimore) 95, e3077. doi: 10.1097/MD.0000000000003077
Keywords: tumor organoids, microorganisms, pathogenic infection, coculture, interactions
Citation: Zhang X, Sun S, Cheng S, Dai J, Du F, Wang J, Wei D, Yan Y and Liu Y (2025) Coculture of tumor organoids with pathogenic microorganisms: a novel system to mimic in vivo pathogenic infection. Front. Cell. Infect. Microbiol. 15:1601688. doi: 10.3389/fcimb.2025.1601688
Received: 28 March 2025; Accepted: 11 June 2025;
Published: 30 June 2025.
Edited by:
Tania Wong, Rutgers University, Newark, United StatesReviewed by:
Cristina Giogha, Hudson Institute of Medical Research, AustraliaUpasana Das Adhikari, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2025 Zhang, Sun, Cheng, Dai, Du, Wang, Wei, Yan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yefu Liu, OTc5MDIxNTNAY211LmVkdS5jbg==; Yichao Yan, eWFueWljaGFvQHBrdWloLmVkdS5jbg==
†These authors share first authorship
 Xue Zhang
Xue Zhang Shulan Sun
Shulan Sun Siqi Cheng2
Siqi Cheng2 Yichao Yan
Yichao Yan Yefu Liu
Yefu Liu