- 1Department of Laboratory Medicine, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, China
- 2Department of Dermatology, Sichuan Provincial People’s Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, China
- 3Department of Critical Care Medicine, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, China
- 4Department of Radiology, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, China
Mucormycosis is a lethal fungal infection disease with high mortality rate. However, investigations assessing the value of metagenomic next-generation sequencing (mNGS) for distinguishing Mucorales infection from colonization are currently insufficient. A retrospective analysis of clinical date from 71 patients at Sichuan Provincial People’s Hospital from September 2021 to September 2024 was conducted. The performance of mNGS in distinguishing Mucorales infection from colonization, along with the differences in patients’ characteristics, imaging characteristics, antimicrobial adjustment, and microbiota, were examined. Among the 71 patients, 51 were identified as Mucorales infection group (3 proven and 48 probable cases), and 20 were colonization group (possible cases). Receiver operating characteristic (ROC) curve for mNGS indicated an area under the curve of 0.7662 (95%CI: 0.6564-0.8759), with an optimal threshold value of 51 for discriminating Mucorales infection from colonization. The infection group exhibited a higher proportion of antimicrobial adjustments compared to the colonization group (64.71% vs. 35.00%, P < 0.05), with antifungal agent changed being more dominant (43.14% vs. 10.00%, P < 0.01). Mucorales RPTM value, length of hospital stays, hsCRP, immunocompromised, malignant blood tumor, and antifungal changed were significantly positively correlated with Mucorales infection. Rhizomucor pusillus showed significant differences between the two groups. The abundance of Torque teno virus significantly increased in the infection group, whereas the colonization group exhibited higher abundance of Rhizomucor delemar. mNGS is a valuable tool for differentiating colonization from infection of Mucorales. Malignant blood tumor, immunocompromised, length of hospital stays and hsCRP were significant different indicators between patients with Mucorales infection from colonization.
1 Introduction
Mucormycosis, a lethal and opportunistic infection disease caused by fungi of the order Mucorales, aggressively invades human blood, organs, and tissues (Donnelly et al., 2020; Panda et al., 2024; Pappas et al., 2021). The Mucorales order comprises 55 genera and 261 species, with 38 recognized as pathogenic to humans. Rhizopus arrhizus is the most prevalent pathogenic genus globally, followed by Mucor and Rhizomucor, while Apophysomyces and Cunninghamella are less frequently implicated. These fungi are ubiquitous in the environment and exhibit a high propensity for colonizing the human respiratory tract (Liang et al., 2024; Roden et al., 2005). Although Mucorales colonization does not immediately provoke disease, it serves as a prerequisite for chronic and allergic mycoses, as well as localized airway infections in invasive fungal diseases. The diagnosis relies on histopathological analysis, conventional microbiological testing (CMT), and imaging, with histopathology or culture considered as the “gold standard” for diagnosis of mucormycosis (Donnelly et al., 2020; Pappas et al., 2021). Histopathological analysis of sterile specimens was critical for confirmation, but there exist difficulties in sampling (Hammer et al., 2018). For CMT, including culture and direct microscopic examination of specimens, also faces limitations in timely diagnosing mucormycosis (Skiada et al., 2018; Wang et al., 2024). While other microbiological testing (OMT) like the galactomannan (GM) antigen testing and (1–3)-β-D-glucan (G) testing have difficulties with accuracy (Lass-Flörl et al., 2021; Lmoth et al., 2021). On account of nonspecific symptoms and signs, early mucormycosis identification is still a challenge in clinic. Definitive diagnosis of mucormycosis, particularly distinguishing between colonization and active infection, remains a significant clinical hurdle (Sipsas et al., 2018; Skiada et al., 2018). However, there remains a paucity of studies focused on differentiating Mucorales infection from colonization.
mNGS is a unbiased sequencing of all nucleic acids (DNA/RNA) in clinical samples (blood, cerebrospinal fluid, respiratory secretions, etc.), and the identification of pathogens (bacteria, viruses, fungi, parasites) through bioinformatics comparison. It does not require pre-assumption of pathogens and is suitable for detecting unknown infections, mixed infections, or rare pathogens (Gu et al., 2019). While, nucleic acid testing by PCR for single agents to multiplexed PCR testing using syndromic panels generally include the most common pathogens associated with a defined clinical syndrome (Chiu and Miller, 2019). The application of metagenomic next-generation sequencing (mNGS) has gained prominence in the clinical diagnosis of infectious diseases, particularly when empirical anti-infective therapies prove ineffective or when CMT fails to identify the etiology. Compared to CMT, mNGS demonstrates superior diagnostic performance for invasive fungal infections, and multiple studies highlight its ability to detect fungal pathogens undiagnosable by traditional methods (Jia et al., 2023; Jiang et al., 2024; Liu et al., 2021; Wang et al., 2024; Zhang et al., 2024, 2024).
Furthermore, the utility of mNGS in differentiating fungal colonization from infection have been explored, primarily by establishing thresholds for pathogen-specific read counts. For instance, Liu et al. demonstrated that bronchoalveolar lavage fluid (BALF) mNGS could distinguish Pneumocystis jirovecii colonization from infection with an area under the curve (AUC) of 0.973, identifying an optimal threshold of 14 reads (Liu et al., 2021). Jia et al. reported a species-specific read number (SSRN) cut-off of 2.5 for diagnosing invasive pulmonary aspergillosis (IPA) versus non-IPA, with distinct thresholds of 1 and 4.5 for immunocompromised and diabetic IPA patients, respectively (Jia et al., 2023). Similarly, the study of Jiang et al. discovered an optimal mNGS RPTM (reads per ten million) cut-off value of 23 for discriminating between Aspergillus infection and colonization (Jiang et al., 2024). Despite these advancements, critical gaps persist in understanding the clinical characteristics of patients and the microbial compositional differences between those with Mucorales colonization and infection.
In this study, we evaluated the efficacy of mNGS, culture and OMT in distinguishing Mucorales infection from colonization. Furthermore, we delineated variations in antimicrobial management strategies, clinical indicators, and shifts in pulmonary microbial composition between these patient groups.
2 Materials and methods
2.1 Study design and participants
This retrospective study included 71 patients with mucormycosis hospitalized at the Sichuan Provincial People’s Hospital from September 2021 to September 2024. The corresponding medical records were reviewed, and the clinical data analyzed including demographic characteristics, type of underlying disease, diagnosis, clinical course, treatment, and outcome.
BALF, blood, SCF and tissue were used for pathogen identification through CMT, including culture for bacteria (blood agar plates, Chocolate, and MacConkey) and fungi (Sabouraud agar plates), and OMT methods, including 1-3-β-D-glucan (G) test (Fungi (1,3)-β-D-glucan assay kit, Gold Mountainriver Tech Development Co.,LTD, Beijing, China), galactomannan (GM) test (Galactomannan test kit, Dana Biotechnology Co.,LTD, Tianjing, China) and smear microscopy for fungi (KOH or Phenol cotton orchid stain), aiming to provide a methodological assessment.
2.2 Criteria for Mucorales infection diagnosis
In this study, the diagnoses of invasive Mucorales infection were classified into proven, probable and possible cases based on the guidelines performed by the European Organization for Research and Treatment of Cancer/Mycoses Study Group Education and Research Consortium (EORTC/MSGERC) (Donnelly et al., 2020). Proven cases required adhere to host factors, clinical signs or symptoms, and positive results from microbiological and/or histopathological examination. The microbiological criteria include microscopic examination and Mucorales recovered by culture from specimens obtained through aseptic procedures from normally sterile, clinically, or radiologically abnormal sites consistent with an infectious disease process. For histopathology, needle aspiration or biopsy revealed hyphae, and accompanied by evidence of associated tissue damage. Probable Mucorales infection is definite as the presence of at least one host factor, a clinical feature and mycologic evidence. Alternatively, a joint diagnosis by imaging experts and clinical doctors of the hospital was needed in case of mycological evidence has not been found or detection of the same Mucorales pathogen through mNGS on more than two occasions. Possible cases meet the criteria of with a host factor and a clinical feature of Mucorales infection, but not mycologic criteria. Proven and probable cases were classified into Mucorales infection group, and possible cases were classified into Mucorales colonization group (Donnelly et al., 2020; Feys et al., 2022; Jiang et al., 2024). Two experienced physicians made clinical diagnoses; when they gave different results, another senior physician made a judgement. Therefore, patients with host factors, obvious clinical signs or symptoms but without positive mycological results were classified as Mucorales infection, as well the cases were considered as colonization when Mucorales was identified but without a final diagnoses of Mucorales infection (Donnelly et al., 2020; Feys et al., 2022; Jiang et al., 2024).
2.3 Sample collation and mNGS detection
Clinical samples, including blood, BALF, CSF, pus, pleural fluids, and tissue, were collected using aseptic techniques when clinicians suspects a pathogenic microorganism infection but has not yet found etiological evidence. And chemical DNA or RNA stabilizers were used to minimize the possibility of nucleic acid degradation at the time of sample collection. The detailed methods regarding the wet lab and bioinformatics had been described previously (Zhou et al., 2022). Briefly, nucleic acids were extracted using the TIANamp Micro DNA Kit (DP316, TIANGEN BIOTECH, Beijing, China). The extracted DNA underwent fragmentation, end repair, adapter ligation and sequencing. Quality assessment was performed using the Agilent 2100 system and sequencing was conducted on the MGISEQ-2000 platform (BGI Genomics Co.,Ltd., Shenzhen, China).
2.4 ROC curve construction
The ROC curve is constructed based on the Mucorales RPTM values detected by mNGS. The RPTM value reflects the load of Mucorales in the sample and is a core indicator for distinguishing infection from colonization. According to guidelines performed by the EORTC/MSGERC, patients were divided into infection group and colonization group. By calculating the sensitivity and specificity at different RPTM thresholds, ROC curves were plotted, and the Youden index (sensitivity+specificity -1) was used to determine the optimal cut-off value.
2.5 Statistical analysis
The data were analyzed by descriptive statistics. The chi-square test was applied to the categorical variables. A student t-test was used for continuous variables. P-value less than 0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism (Version 8.0.2, GraphPad Software Inc) and SPSS (Version 25, IBM Corp). The diagnostic performance of mNGS was evaluated using the area under the curve of receiver operating characteristic (ROC), where the best cut-off value was obtained. The sensitivity and specificity of the detection method were analyzed as reference (Blauwkamp et al., 2019). The correlation analysis was conducted in R by the corrplot package. The alpha diversity index was calculated based on Shannon and Simpson indexes. Beta-diversity was visualized using principal coordinate analysis (PCoA), and an ANOSIM test was performed in R with the Vegan package. The stacked bar plot of the community composition was visualized in R using the ggplot2 package. Linear discriminant analysis (LDA) effect size (LEfSe) was utilized by R with microeco package to identify significantly different species among the groups, with thresholds of log10 LDA Score ≥ 2 and P value ≤ 0.05.
3 Results
3.1 Baseline characteristics and sample classification
Totally, 71 patients were included and diagnosed as proven (n = 3), probable (n = 48) and possible (n = 20) mucormycosis. Among them, 51 were identified as Mucorales infection, and 20 were colonization group.
According to Table 1, the median age at diagnosis was 57 years old (ranged from 9 to 103), and most were males (70.42%, n = 50). The significant differences in Mucorales infection and colonization groups were observed including malignant blood tumor (n =15 vs. n = 1, P = 0.0294), longer length of hospital stays (LOHS) (29.57 vs. 19.45 days, P = 0.0494), immunocompromised (n = 26 vs. n = 4, P = 0.0311), and hsCRP level (127.25 vs. 54.16 ug/mL, P = 0.0014).
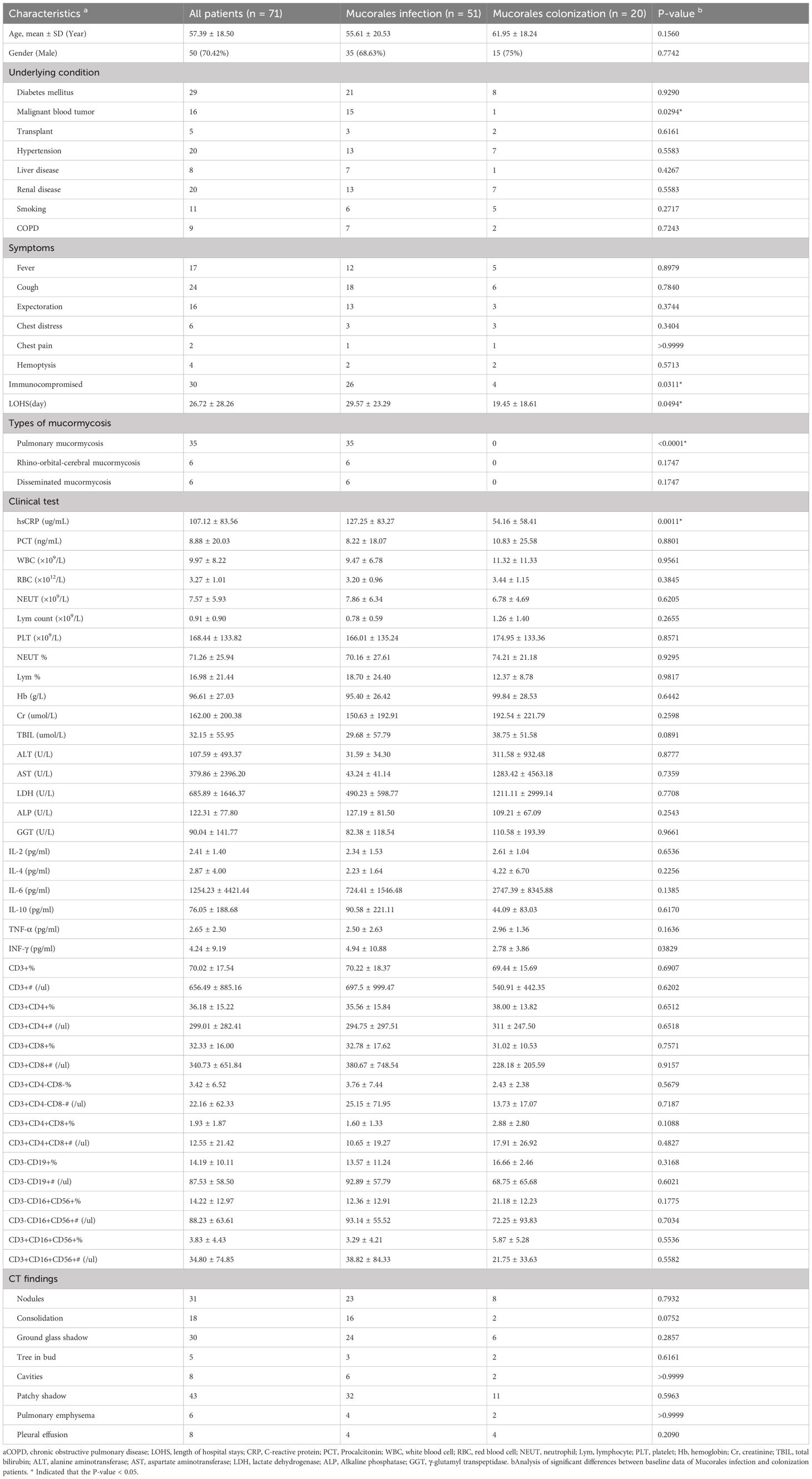
Table 1. General demographic and clinical characteristics of the patients with Mucorales infection and colonization.
In the Mucorales infection and colonization groups, 13 and 7 Mucorales species were identified by mNGS, respectively (Supplementary Table 1). In Mucorales infection group, Rhizopus microsporus (33.33%, 17/51) was the most common species, followed by Rhizopus arrhizus (23.53%, 12/51) and Rhizomucor pusillus (17.65%, 9/51). Three patients were found to be co-infected with Rhizopus and Mucor, including two patients co-infected with Rhizopus microsporus and Mucor, and one patient co-infected with Rhizopus microsporus and Mucor racemosus. In Mucorales colonization group, Rhizopus delemar (30%, 6/20), Rhizopus arrhizus (25%, 5/20) and Rhizomucor pusillus (15%, 3/20) were the top three of the Mucorales species detected (Figure 1A). The most frequent sample type observed was BALF, followed by blood (Figure 1B; Supplementary Table 1). The Mucorales load was significantly higher in the infection group compared with colonization group, with a median mNGS read number of 1.82 ± 0.98 vs. 1.12 ± 0.53 (P = 0.004) (Figure 1C). Besides, over 68% of patients in the infection group had an RPTM value larger than 20, while the percentage of colonization group less than 20 was 60%. (Figure 1D).
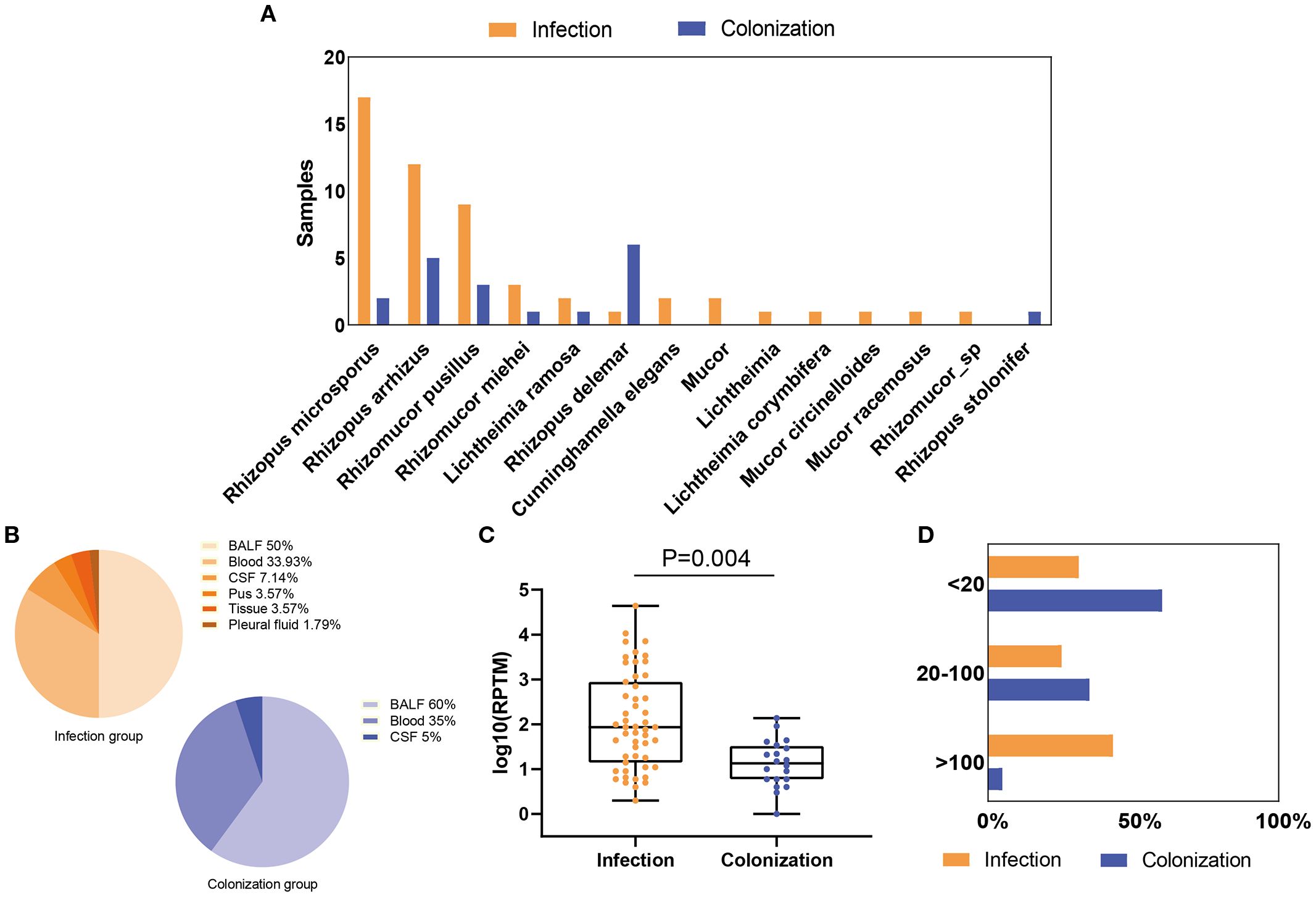
Figure 1. Distribution and abundance of Mucorales species in patients with Mucorales infection and colonization. (A) Comparison of Mucorales species in patients with Mucorales infection and colonization. (B) Distribution of sample types in patients with Mucorales infection and colonization. (C) Differences in mNGS RPTM for Mucorales in patients with Mucorales infection and colonization. (D) Proportion of patients with different mNGS Mucorales reads in the infection and colonization groups.
3.2 Diagnostic efficacy of mNGS for Mucorales infection and colonization
To calculate the cut-off that best discriminated between patients with Mucorales infection from colonization, we created a ROC curve using the Mucorales RPTM of mNGS from the patients. The calculated area under curve was 0.7662 (95% CI: 0.6564-0.8759), with the optimal cut-off value was determined to be 51 (Figure 2A).
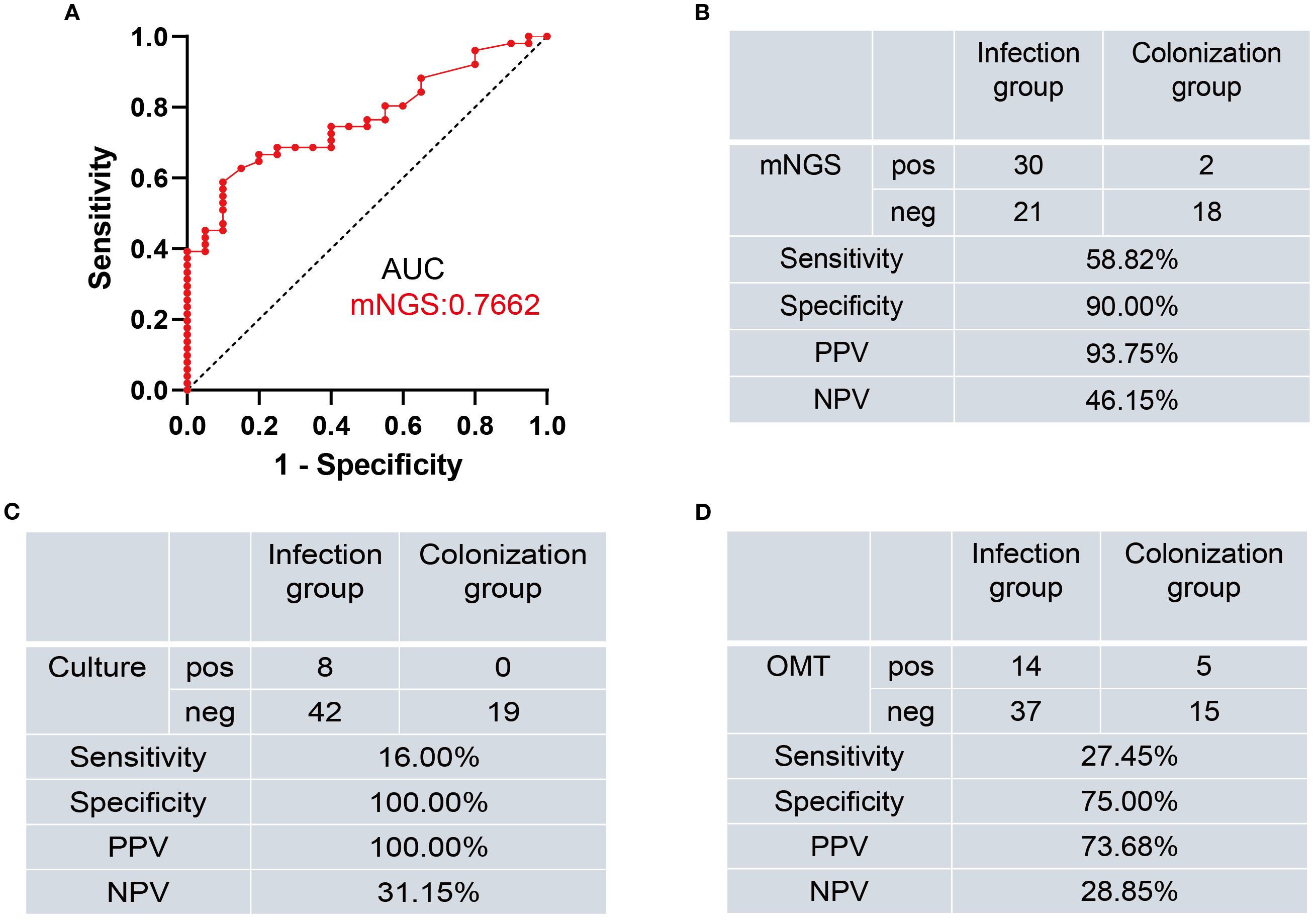
Figure 2. Diagnostic performance of mNGS, Culture, and OMT methods for distinguishing Mucorales infection from colonization. (A) ROC curve of mNGS for discrimination between Mucorales infection and colonization. (B–D) Diagnostic performance of mNGS (B), Culture (C), and OMT (D) methods for differentiating between Mucorales infection and colonization. AUC, area under curve; PPV, positive predictive value; NPV, negative predictive value; pos, positive; neg, negative.
Subsequently, we evaluated the diagnostic efficacy of mNGS, culture and OMT in distinguishing infection from colonization (Supplementary Table 1). When using RPTM ≥ 51 as the threshold criterion for Mucorales infection and colonization, the sensitivity of mNGS was 58.82%, which was significantly higher than culture (16.00%, P < 0.001) and OMT (27.45%, P = 0.0025). For specificity, there was no significant difference between mNGS and culture (90.00% vs. 100.00%, P = 0.4872), nor between mNGS and OMT (90.00% vs. 75.00%, P = 0.4075). While the specificity of culture was significantly higher than that of OMT (P = 0.0471) (Figures 2B–D).
3.3 Diagnostic value of imaging for Mucorales infection and colonization
To evaluate the value of imaging in diagnosing Mucorales infection, we reviewed the imaging results of all cases. As shown in Figure 3, the Brain MRI of patient No.9 showed abnormal lesions, but it can’t indicate which pathogen caused it. Patient No.10 displayed a mixed infection, but cannot be distinguished. Patient No.61 presented no abnormalities. The remaining patients of No. 11, No. 26, No. 42, No. 44, No.65 are all not that obvious for Mucorales infection diagnosis. Altogether, it is difficult to determine whether the detected abnormalities are caused by Mucorales. Therefore, it is of great significance to combine other laboratory tests for the diagnosis of Mucorales infection.
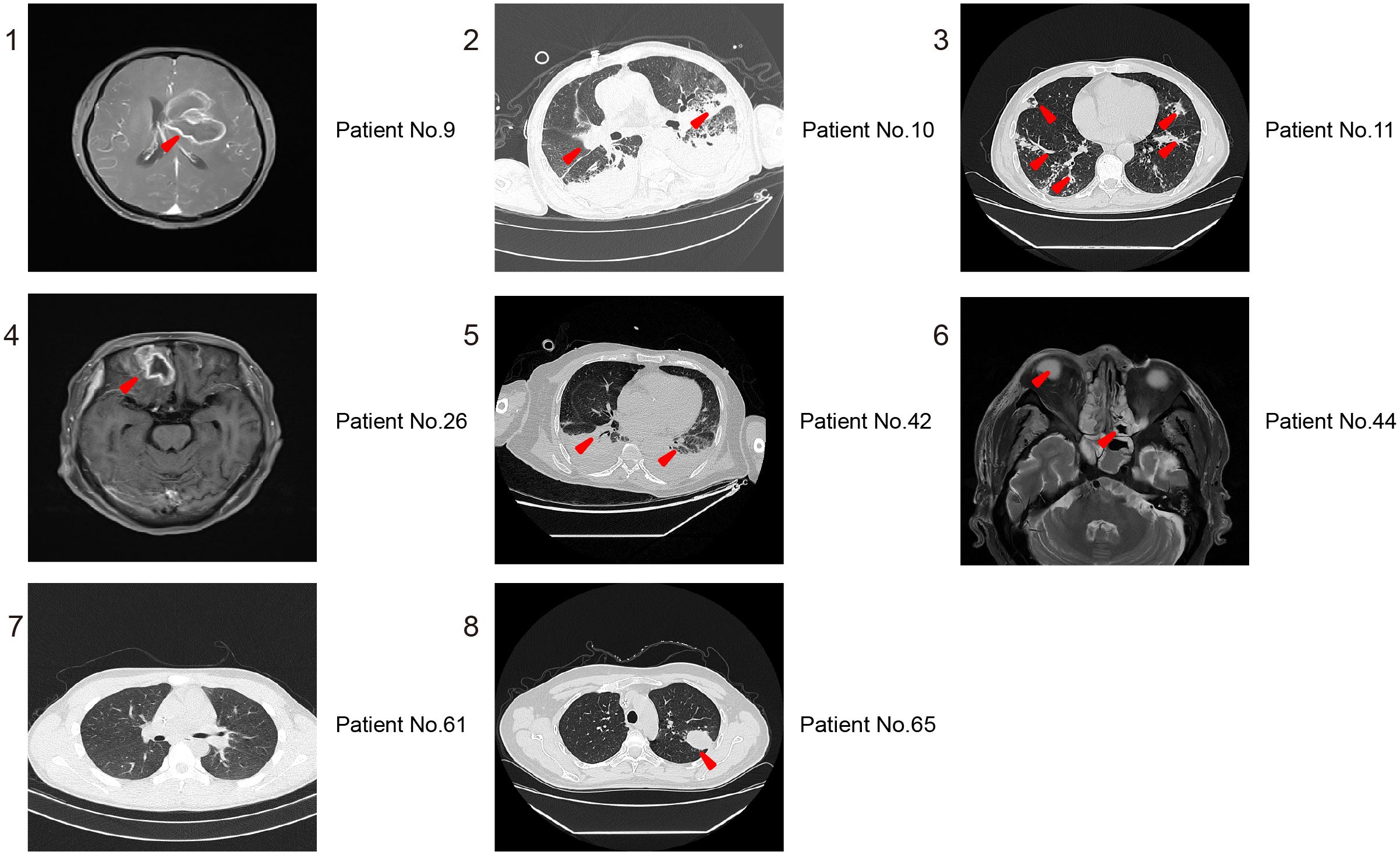
Figure 3. Representative imaging results for distinguishing Mucorales infection from colonization. 1,4, and 6 are MRI of Brain; 2, 3, 5, 7, and 8 are CT of lung. The red arrowheads showed the abnormal lesions.
3.4 Impacts of mNGS on antimicrobial usage of Mucorales infection patients
The incidence of bacterial and fungal co-infection was higher in both infection group (56.86%, 29/51) and colonization group (25.00%, 5/20) (Figures 4A, B). To explore the influence of mNGS results on antimicrobial usage, we analyzed variations in antimicrobial regimens of antibacterial and antifungal agent before and after mNGS detection. As results in Figure 4C, the antimicrobial regimens were adjusted in 33 out of 51 (64.71%) samples from patients with Mucorales infection, which was significantly higher than that in Mucorales colonization (35.00%, P < 0.05). Among the 33 samples, 22 samples had their antifungal agent changed, 10 cases had both antibacterial and antifungal agents adjusted, while one case had their antibacterial changed. The percentage of patients requiring antifungal agent adjusted was significantly higher in Mucorales infection group compared to colonization group (43.14% vs. 10.00%, P < 0.01). Moreover, among 22 patients of infection group who received antifungal treatment, 15 (68.18%) showed improvements, 2 (9.09%) died, and 5 (22.73%) were discharged voluntarily. And, among 10 patients who received both antibacterial and antifungal treatment, 7 (70.00%) have improved, 1 (10.00%) died, and 2 (20.00%) were discharged voluntarily.
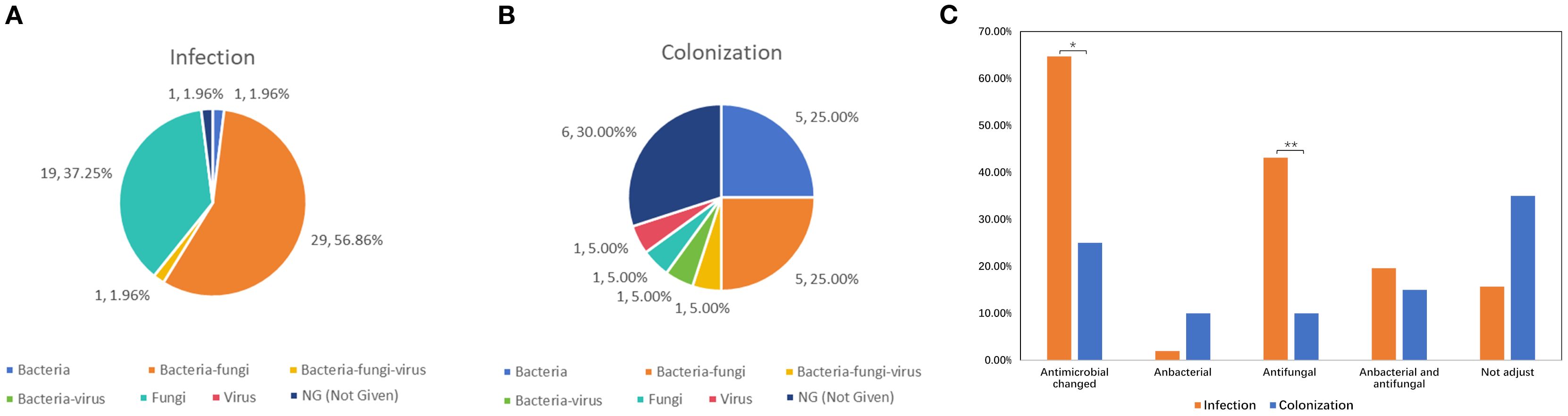
Figure 4. Impacts of mNGS on antimicrobial adjustment in patients with Mucorales infection and colonization. (A, B) The infection types of patients with Mucorales infection (A) and colonization (B). (C) Variations in antimicrobial regimens of antibacterial and antifungal agent before and after mNGS detection. *P < 0.05, **P < 0.01.
3.5 Correlations between the characteristics and Mucorales infection
We conducted Spearman correlation analyses to examine the relationship between various characteristics and Mucorales infection. The results showed significant positive correlations between Mucorales infection and the following variables: Mucorales RPTM value, LOHS, hsCRP, immunocompromised, malignant blood tumor, and antifungal changed. Significant negative correlations between Mucorales infection and not adjust drug level were observed. Additionally, significant positive correlations were observed between Mucorales RPTM value and the following variables: hsCRP, PCT, immunocompromised, malignant blood tumor, and liver disease. Notably, OMT Mucorales positivity was positively correlated with age, and CD3+ index; and negatively correlated with lymphocyte ratio and Alanine Aminotransferase (ALT). Furthermore, positive correlations were found between LOHS and the following variants: B cells, NK cells and NKT cells. Pleural effusion was significantly positive with IL-1, IL-2, IL-4, IL-6, IL-8, IL-17, and TNF-α (Figure 5).
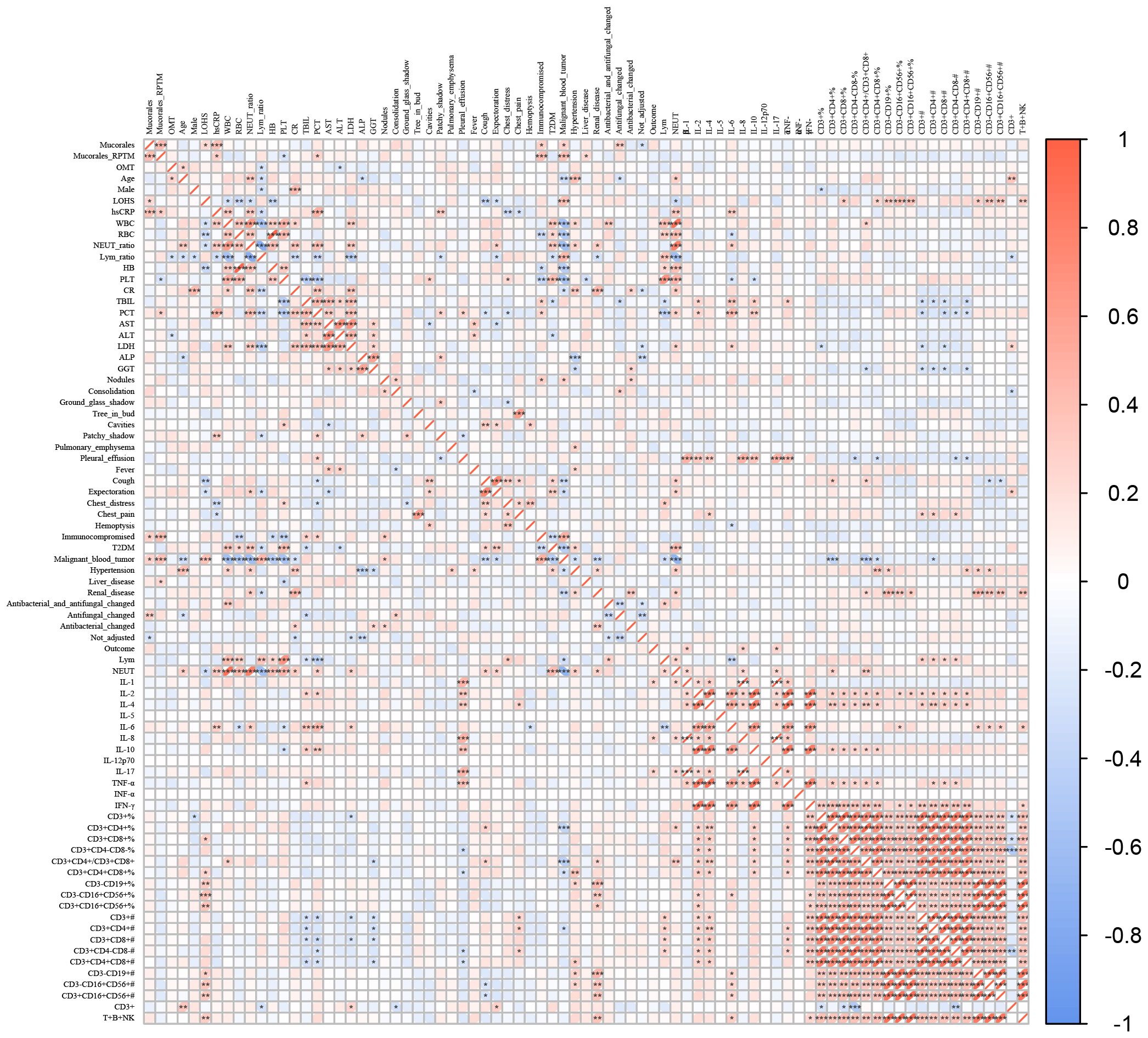
Figure 5. Correlations between the characteristics and Mucorales infection. Spearman correlations analysis between Mucorales infection and characteristics of patients. *P < 0.05, **P < 0.01, ***P < 0.001.
3.6 Differences in the microbial community structure
The study compared the overall composition and diversity of the microbial signature in patients with Mucorales infection and colonization. Although no significant difference was observed, patients with Mucorales infection showed a higher diversity according to both the Shannon and Simpson indices, indicating a trend towards increased richness and evenness of microbial composition (Figure 6A). PCoA results indicated that the samples from both groups were intermixed. However, the infection group displayed a wider spread of data compared to the colonization group (Figure 6B). Moreover, no significant difference in the microbial community structure between the two groups was observed (Figure 6C).
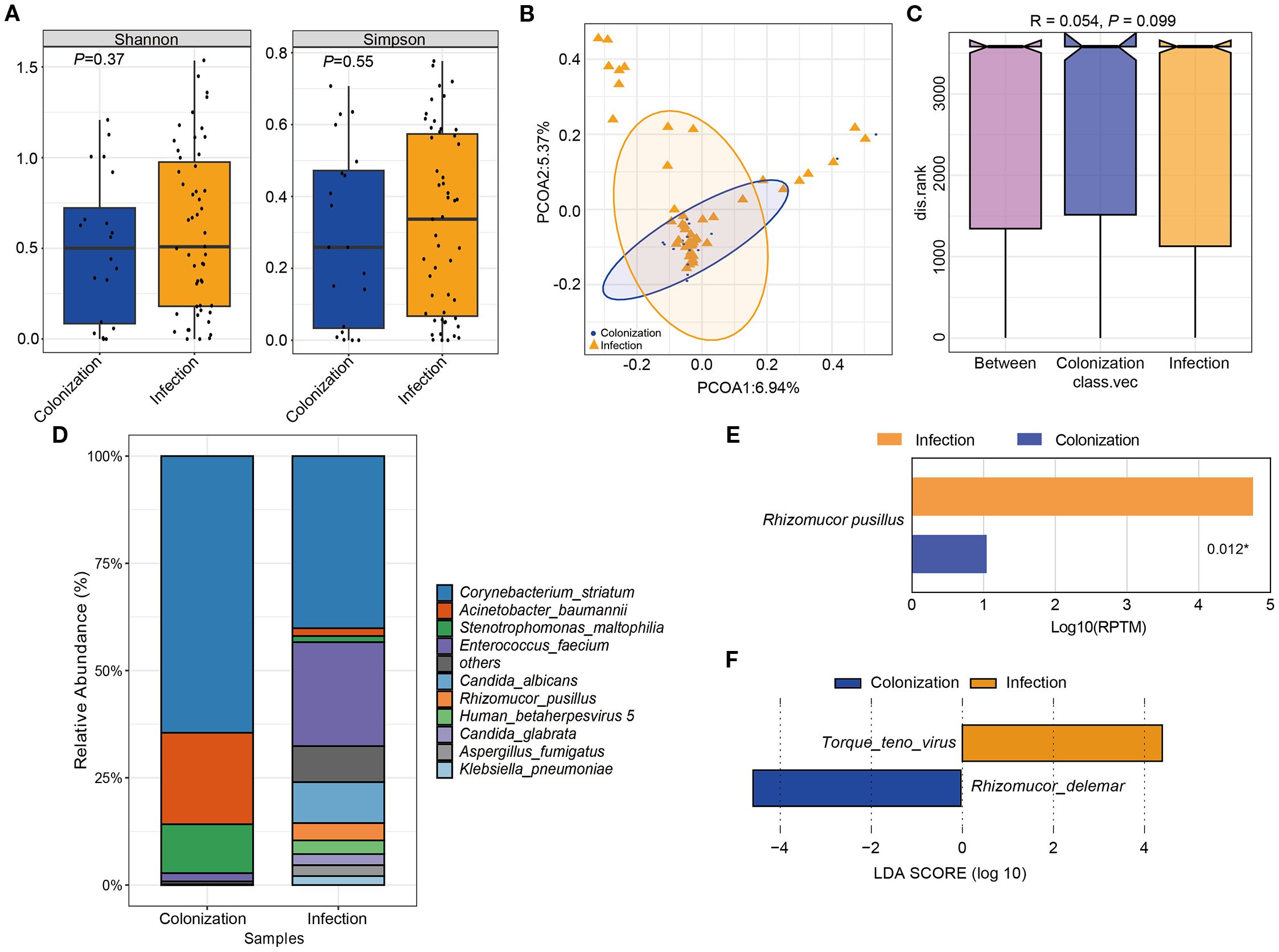
Figure 6. The difference of microbial composition for patients with Mucorales infection and colonization. (A) Alpha diversity was showed by Shannon and Simpson index. (B) PCoA analysis of the microbial composition. (C) ANOSIM for the analysis of microbial community structure. (D) Barplot showed the top 10 species with the highest abundance between two groups. (E) Significant different analysis of the species between two groups with Kruskal-Wallis test. (F) Lefse analysis for enriched species for the two groups. P < 0.05.
The relative abundance of the top 10 species were Corynebacterium striatum, Acinetobacter baumannii, Stenotrophomonas maltophilia, Enterococcus faecium, Candida albicans, Rhizomucor pusillus, Human betaherpesvirus 5, Candida glabrata, Aspergillus fumigatus, and Klebsiella pneumoniae. Among them, only Rhizomucor pusillus showed significant differences between the two groups (Figures 6D, E). Additionally, two species with LDA scores ≥ 2 and P < 0.05 were identified. Torque teno virus (TTV) was significantly more abundant in Mucorales infection group, whereas Rhizomucor delemar was more enriched in Mucorales colonization group (Figure 6F).
4 Discussion
Mucormycosis, a disease with high morbidity and mortality rate, is difficult to diagnose and treat (Cornely et al., 2019). Although Mucorales infection and colonization have clear definitions (Cornely et al., 2014a, 2014b; Donnelly et al., 2020), timely and precise diagnosis of invasive Mucorales infection or colonization is still complicated and difficult in clinic. However, studies focus on distinguishing Mucorales infection from colonization are barely reported. This study was carried out to evaluate the efficacy of mNGS in differentiating Mucorales infection from colonization. Moreover, it also outlined the distribution characteristics of mucormycosis, clinical characteristics, immune changes, outcome, antibiotic adjustment of Mucorales infection and colonization patients, as well as the variations in sample microbiota.
The main reported pathogens in mucormycosis are Rhizopus, Mucor, and Lichtheimia, followed by Rhizomucor, Cunninghamella, Apophysomyces, and Saksenaea (Cornely et al., 2019, 2014a). Consistent with previous researches, our study identified 14 Mucorales species among the patients, with 13 species leading to Mucorales infection. Among them, Rhizopus microsporus, Rhizopus arrhizus, and Rhizomucor pusillus were the most prevalent in patients with Mucorales infection. Rhizopus delemar, while Rhizopus arrhizus, and Rhizomucor pusillus were the most common species in patients with Mucorales colonization (Figure 1). Additionally, from Table 1 and spearson correlation analyses, we found that the Mucorales RPTM value, LOHS, hsCRP, immunocompromised, malignant blood tumor, and antifungal changed accounted for the Mucorales infection, which may be beyond the existed research findings (Cornely et al., 2019).
With the widespread application of mNGS, it offers a hypothesis-free, unbiased approach to pathogen detection, enabling the identification of novel or unexpected organisms, semi-quantitative analysis, and comprehensive genomic coverage. However, its limitations include high host background noise, substantial cost and turnaround time, incomplete reference databases, and susceptibility to environmental contamination (Gu et al., 2019). However, the benefits of using mNGS for pathogen detection have become increasingly apparent, especially for rare and emerging pathogens, such as mucormycosis, hyalohyphomycosis (Fusarium, Paecilomyces, Scedosporium, etc.), and phaeohyphomycosis (Alternaria, Bipolaris, Cladosporium, Rhinocladiella, etc.) (Ling et al., 2024; Xing et al., 2023; Wang et al., 2024; Li et al., 2024). A research has shown that mNGS of infected body fluids by Illumina sequencing has a combined sensitivity and specificity of 79% (95% CI 73.5–85.2%) and 91% (95% CI 87.3–93.8%) for bacteria and 91% (95% CI 84.2–100%) and 89% (95% CI 85.7–92.5%) for fungi, respectively (Gu et al., 2021). The above indicates that mNGS is a highly effective option even before OMT results are available. Early and precise detection of pathogen of severe or rare infectious patients is critical for clinicians to give a timely fast intervention and targeted therapy as quickly as possible. It suggests that the medical related organisms including Candida, Cryptococcus, Mucorales, and Aspergillus has increased in subjects with impaired immune function, and the thick cell wall of fungi is difficult to break to release nucleic acid which lead to false negative mNGS results (Bittinger et al., 2014). While the diagnostic performance of mNGS has improved with optimized extraction methods (Gu et al., 2021). Besides, the positive diagnostic threshold criteria for mNGS should be defined according to different host and pathogen status. Based on these, this study laid the foundation for the establishment of the positive threshold criteria according to different host and pathogen status in some ways.
Numerous studies have investigated the diagnostic ability of mNGS for Mucorales infection, but there remains little research on the distinction of Mucorales colonization and infection (Wang et al., 2024; Zhang et al., 2024). Meaningfully, our study laid the foundation for the establishment of the positive threshold criteria according to different host and pathogen status in some ways. We observed that mNGS displayed superior accuracy in diagnosing Mucorales infection and distinguishing it from colonization when compared to culture and OMT (P < 0.05). The optimal cut-off value of RPTM for mNGS was 51. At this threshold, mNGS achieved a sensitivity of 58.82% and a specificity of 90.00% for the final diagnosis (Figure 2B). Furthermore, multiple (≥10) nodules, pleural effusion and halo sign were reportedly associated with pulmonary mucormycosis (Chamilos et al., 2005; Legouge et al., 2014). However, we found that imaging has limitations in diagnosing mucormycosis in clinical, especially when it comes to co-infection of multiple pathogens. Indeed, this research can serve as a valuable reference for analyzing patients with Mucorales infection and colonization. Notably, even though mNGS serves as a precise pathogen infection test method and has potential diagnosis in clinic, the final diagnosis of the disease counts on clinical experts who integrate the patient’s symptoms, clinical laboratory test results, and etiological findings to make a comprehensive decision. And in the future, it is necessary for us to conduct prospective studies with a large amount of data about distinction of Mucorales infection and colonization.
mNGS had significant impact on treatment regimens, particularly in infectious disease (Zhang et al., 2024). Equally, in this study, 68.18%% and 70.00%% showed improvement among the patients who received only antifungal treatment, and antibacterial combined with antifungal treatment, respectively. This suggested that timely clinical intervention and targeted antifungal therapy for patient prognosis is of great importance. Although Shannon and Simpson indexes were higher in the infection group, no significant differences were observed in species abundance and diversity between the two groups (Figure 6). Even the microbial diversity differences are minimal and not statistically significant, these microbiome findings are as exploratory and mainly hypothesis generating. Incidentally, Rhizomucor pusillus appeared more frequently in Mucorales infection group. Additionally, TTV, and Rhizomucor delemar were significantly more abundant in patients with Mucorales infection and colonization individually. TTV is a member of Anellovirida, which is commonly present in patients with various blood diseases, organ transplants, tumors, periodontitis, and even the healthy population (Maggi and Bendinelli, 2010; Nishizawa et al., 1997). In our study, nine patients were diagnosed with TTV infection, with five patients immunocompromised and three patients suffered from blood disease. However, whether the value of TTV in the infected group indeed existed or was influenced by confounding factors like patients’ immune status, further prospective clinical studies are needed to verify. And further exploration is necessary to deeply understand the potential interaction mechanism between TTV, Rhizomucor delemar and Mucorales infection. The disparity of the different results of microbiome analysis may because of the advanced age of our patients, their relatively lower mortality rate, their immune status, and no restrictions on the type of diseases they exhibited.
In this study, we conducted a comprehensively retrospective study to analyze the clinical characteristics, immune changes, outcome, antibacterial and antifungal adjustment, and microbiota changes in individuals with Mucorales infection and colonization. Furthermore, the efficacy of mNGS was evaluated to distinguish Mucorales infection and colonization. With meticulously designed and analyzed, the study also exists limitations. First, not all patients underwent all clinically laboratory tests, which results in a lack of corresponding comparative diagnostic performance results. The second problem relates to the single-center study. Finally, the sample size is indeed small, and the number of some sample types like CSF, pleural fluid, pus, etc. is little, which may cause a bias in the analysis outcomes.
5 Conclusions
In this investigation, the performance of mNGS in distinguishing Mucorales infection from colonization, with the differences in patients’ clinical characteristics, antibacterial and antifungal adjustment, and microbiota analysis, were analyzed. We found that mNGS has a high diagnostic efficacy for distinguishing Mucorales infection and colonization, which was better than culture and OMT used in this retrospective research. Moreover, mNGS played a more important role on the guidance of medication in patients with Mucorales infection. Malignant blood tumor, immunocompromised, LOHS, and hsCRP were significant different indicators between patients with Mucorales infection from colonization.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Medical Professional Committee of the Sichuan Provincial People’s Hospital (Permit Number: 2022172). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
XZ: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. CY: Data curation, Investigation, Writing – review & editing. XL: Data curation, Investigation, Writing – review & editing. JW: Data curation, Writing – review & editing. YL: Investigation, Writing – review & editing. LP: Investigation, Writing – review & editing. SP: Data curation, Writing – review & editing. HY: Conceptualization, Methodology, Validation, Writing – review & editing. XD: Conceptualization, Methodology, Resources, Writing – review & editing, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Research Fund of Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital (2022QN55 and 2022QN21).
Acknowledgments
We sincerely thank the patients for participating in this original study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1631960/full#supplementary-material
Abbreviations
BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; CMT, conventional microbiological testing; OMT, other microbiological testing; mNGS, metagenomic next-generation sequencing; ROC, receiver operating characteristic; AUC, area under the curve; LOHS, length of hospital stays; RPTM, reads per ten million; G, (1–3)- β-D-glucan; GM, galactomannan; PCoA, principal coordinate analysis; EORTC/MSGERC, European Organization for Research and Treatment of Cancer/Mycoses Study Group Education and Research Consortium.
References
Blauwkamp, T. A., Thair, S., Rosen, M. J., Blair, L., Lindner, M. S., Vilfan, I. D., et al. (2019). Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674. doi: 10.1038/s41564-018-0349-6
Bittinger, K., Charlson, E. S., Loy, E., Shirley, D. J., Haas, A. R., Laughlin, A., et al. (2019). Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 15, 487. doi: 10.1186/s13059-014-0487-y
Chamilos, G., Marom, E. M., Lewis, R. E., Lionakis, M. S., and Kontoyiannis, D. P. (2005). Predictors of pulmonary zygomycosis versus invasive pulmonary aspergillosis in patients with cancer. Clin. Infect. Dis. 41, 60–66. doi: 10.1086/430710
Chiu, C. Y. and Miller, S. A. (2019). Clinical metagenomics. Nat. Rev. Genet. 20, 341–355. doi: 10.1038/s41576-019-0113-7
Cornely, O. A., Alastruey-Izquierdo, A., Arenz, D., Chen, S. C. A., Dannaoui, E., Hochhegger, B., et al. (2019). Global guideline for the diagnosis and management of mucormycosis: an initiative of the European confederation of medical mycology in cooperation with the mycoses study group education and research consortium. Lancet Infect. Diseases. 19, e405–e421. doi: 10.1016/S1473-3099(19)30312-3
Cornely, O. A., Arikan-Akdagli, S., Dannaoui, E., Groll, A. H., Lagrou, K., Chakrabarti, A., et al. (2014a). ESCMID and ECMM joint clinical guidelines for the diagnosis and management of mucormycosis 2013. Clin. Microbiol. Infect. 20, 5–26. doi: 10.1111/1469-0691.12371
Cornely, O. A., Cuenca-Estrella, M., Meis, J. F., and Ullmann, A. J. (2014b). European society of clinical microbiology and infectious diseases (ESCMID) fungal infection study group (EFISG) and European confederation of medical mycology (ECMM) 2013 joint guidelines on diagnosis and management of rare and emerging fungal diseases. Clin. Microbiol. Infect. 20, 1–4. doi: 10.1111/1469-0691.12569
Donnelly, J. P., Chen, S. C., Kauffman, C. A., Steinbach, W. J., Baddley, J. W., Verweij, P. E., et al. (2020). Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin. Infect. Dis. 71, 1367–1376. doi: 10.1093/cid/ciz1008
Feys, S., Gonçalves, S. M., Khan, M., Choi, S., Boeckx, B., Chatelain, D., et al. (2022). Lung epithelial and myeloid innate immunity in influenza-associated or COVID-19-associated pulmonary aspergillosis: an observational study. Lancet Respir. Med. 10, 1147–1159. doi: 10.1016/S2213-2600(22)00259-4
Gu, W., Deng, X., Lee, M., Sucu, Y. D., Arevalo, S., Stryke, D., et al. (2021). Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124. doi: 10.1038/s41591-020-1105-z
Gu, W., Miller, S., and Chiu, C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Hammer, M. M., Madan, R., and Hatabu, H. (2018). Pulmonary mucormycosis: radiologic features at presentation and over time. Am. J. Roentgenol. 210, 742. doi: 10.2214/AJR.17.18792
Jia, H., Liu, H., Tu, M., Wang, Y., Wang, X., Li, J., et al. (2023). Diagnostic efficacy of metagenomic next generation sequencing in bronchoalveolar lavage fluid for proven invasive pulmonary aspergillosis. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1223576
Jiang, Z., Gai, W., Zhang, X., Zheng, Y., Jin, X., Han, Z., et al. (2024). Clinical performance of metagenomic next-generation sequencing for diagnosis of pulmonary aspergillus infection and colonization. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1345706
Lass-Flörl, C., Samardzic, E., and Knoll, M. (2021). Serology anno 2021-fungal infections: from invasive to chronic. Clin. Microbiol. Infect. 27, 1230–1241. doi: 10.1016/j.cmi.2021.02.005
Legouge, C., Caillot, D., Chrétien, M. L., Lafon, I., Ferrant, E., Audia, S., et al. (2014). The reversed halo sign: pathognomonic pattern of pulmonary mucormycosis in leukemic patients with neutropenia? Clin. Infect. Dis. 58, 672–678. doi: 10.1093/cid/cit929
Li, J., Ge, Y., Xin, C., and Jiang, L. (2024). Rhino-orbital-cerebral mucormycosis caused by Rhizopus arrhizus diagnosis via metagenomics next-generation sequencing: a case report. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1375058
Liang, M., Xu, J., Luo, Y., and Qu, J. (2024). Epidemiology, pathogenesis, clinical characteristics, and treatment of mucormycosis: a review. Ann. Med. 56(1), 2396570. doi: 10.1080/07853890.2024.2396570
Ling, J., Liang, L., Liu, X., Wu, W., Yan, Z., Zhou, W., et al. (2024). Invasive Fusarium solani infection diagnosed by traditional microbial detection methods and metagenomic next-generation sequencing in a pediatric patient: a case report and literature review. Front. Med. 11. doi: 10.3389/fmed.2024.1322700
Liu, L., Yuan, M., Shi, Y., and Su, X. (2021). Clinical performance of BAL metagenomic next-generation sequence and serum (1,3)-β-d-glucan for differential diagnosis of Pneumocystis jirovecii pneumonia and Pneumocystis jirovecii colonisation. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.784236
Lmoth, F., Akan, H., Andes, D., Cruciani, M., Marchetti, O., Ostrosky-Zeichner, L., et al. (2021). Assessment of the role of 1,3-β-d-glucan testing for the diagnosis of invasive fungal infections in adults. Clin. Infect. Dis. 72, S102–S108. doi: 10.1093/cid/ciaa1943
Maggi, F. and Bendinelli, M. (2010). Human anelloviruses and the central nervous system. Rev. Med. Virol. 20, 392–407. doi: 10.1002/rmv.668
Nishizawa, T., Okamoto, H., Konishi, K., Yoshizawa, H., Miyakawa, Y., and Mayumi, M. (1997). A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241, 92–97. doi: 10.1006/bbrc.1997.7765
Panda, S., Sahu, M. C., Turuk, J., and Pati, S. (2024). Mucormycosis: a rare disease to notifiable disease. Braz. J. Microbiol. 55, 1065–1081. doi: 10.1007/s42770-024-01315-z
Pappas, P. G., Chen, S. C., and Donnelly, J. P. (2021). The evidence supporting the revised EORTC/MSGERC definitions for invasive fungal infections. Clin. Infect. Dis. 72, S77–S78. doi: 10.1093/cid/ciaa1765
Roden, M. M., Zaoutis, T. E., Buchanan, W. L., Knudsen, T. A., Sarkisova, T. A., Schaufele, R. L., et al. (2005). Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41, 634–653. doi: 10.1086/432579
Sipsas, N. V., Gamaletsou, M. N., Anastasopoulou, A., and Kontoyiannis, D. P. (2018). Therapy of mucormycosis. J. Fungi. 4, 90. doi: 10.3390/jof4030090
Skiada, A., Lass-Floerl, C., Klimko, N., Ibrahim, A., Roilides, E., and Petrikkos, G. (2018). Challenges in the diagnosis and treatment of mucormycosis. Med. Mycol. 56, S93–S101. doi: 10.1093/mmy/myx101
Wang, Q., Huang, Y., Ma, H., and Fan, G. (2024). A case report: comorbidity of rhinocerebral mucormycosis and pulmonary aspergillosis with challenging diagnosis. Front. Med. 11. doi: 10.3389/fmed.2024.1398714
Wang, W., Yao, Y., Li, X., Zhang, S., Zeng, Z., Zhou, H., et al. (2024). Clinical impact of metagenomic next-generation sequencing of peripheral blood for the diagnosis of invasive mucormycosis: a single-center retrospective study. Microbiol. Spectr. 12, e0355323. doi: 10.1128/spectrum.03553-23
Xing, X., Liu, Q., Yuan, W., Zhu, C., and Yang, J. (2023). Pulmonary scedosporiosis caused by Lomentospora prolificans in a patient who is immunocompetent: a rare case report. Int. J. Infect. Dis. 130, 208–210. doi: 10.1016/j.ijid.2023.03.037
Zhang, X., Zhang, L., Li, Y., Wang, N., and Zhang, Y. (2024). Clinical performance of metagenomic next-generation sequencing for diagnosis of invasive fungal disease after hematopoietic cell transplant. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1210857
Zhang, Y., Wei, E., Niu, J., Yan, K., Zhang, M., Yuan, W., et al. (2024). Clinical features of pediatric mucormycosis: role of metagenomic next generation sequencing in diagnosis. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1368165
Keywords: mucormycosis, Mucorales, diagnosis, metagenomic next-generation sequencing, optimal threshold value
Citation: Zhou X, Yang C, Liu X, Wang J, Li Y, Pan L, Peng S, Yu H and Deng X (2025) Clinical performance of metagenomic next-generation sequencing for distinction and diagnosis of Mucorales infection and colonization. Front. Cell. Infect. Microbiol. 15:1631960. doi: 10.3389/fcimb.2025.1631960
Received: 20 May 2025; Accepted: 01 September 2025;
Published: 18 September 2025.
Edited by:
Beiwen Zheng, Zhejiang University, ChinaReviewed by:
Fabianne Carlesse, University of São Paulo, BrazilRex Jeya Rajkumar Samdavid Thanapaul, Walter Reed Army Institute of Research, United States
Copyright © 2025 Zhou, Yang, Liu, Wang, Li, Pan, Peng, Yu and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiren Deng, eHJkZW5nMTk5MkAxNjMuY29t; Hua Yu, eXZodWEyMDAyQDE2My5jb20=
†These authors have contributed equally to this work
 Xiaoli Zhou
Xiaoli Zhou Chenxi Yang1†
Chenxi Yang1† Yanqiao Li
Yanqiao Li Lingai Pan
Lingai Pan Xiren Deng
Xiren Deng