- 1Department of Functional Ecology, Institute of Botany of the Czech Academy of Sciences, Třeboň, Czechia
- 2Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
- 3Department of Geography, Institute of Ecology and Earth Sciences, University of Tartu, Tartu, Estonia
- 4Faculty of Science, Department of Botany, University of South Bohemia, České Budějovice, Czechia
- 5Swiss Federal Institute for Forest, Snow and Landscape Research WSL, Birmensdorf, Switzerland
Modern analytical tools are essential for advancing research and facilitating interdisciplinary collaboration. The R software serves as a comprehensive solution for statistical computing and graphics in all scientific disciplines, including dendrochronology. Beyond managing traditional tasks like data processing, analysis, and results visualization, R is pivotal in integrating innovative techniques, such as multi-proxy datasets, artificial intelligence or machine learning, to address emerging challenges in tree-ring research. However, a comprehensive overview of R’s functionalities in dendrochronology is lacking, despite its growing importance and increasing role in interdisciplinary research. Here we present an overview of 38 R packages relevant to tree-ring research, categorized by functionality. For each R package, concise descriptions and examples of usage are provided to facilitate the identification and selection of suitable tools for researchers, academicians, and students within and outside the field. We further discuss the transformative potential of R in building a centralized, open-access ecosystem, emphasizing its role in standardizing workflows, enhancing reproducibility, and expanding dendrochronology’s integration with other scientific disciplines in a digital era. We propose that these advancements not only streamline dendrochronological workflows but also provide valuable insights for addressing global environmental and ecological challenges.
1 Introduction
The rapid evolution of computer hardware (serving computational power) and open-source and free computer-supported programming languages (e.g., Python, R, Java) has been revolutionizing scientific research by democratizing data analysis. This not only enabled researchers of all disciplines to efficiently perform complex data measurements, statistical computations, and data visualizations but also, importantly, to contribute to building up flexible and customizable applications at the service of the whole scientific community. Such programming languages provide easy accessibility to a vast community of developers to actively and continuously improve and enhance languages and libraries while increasing trust and confidence thanks to its transparency, eventually thriving community collaboration. Hence, the utilization of freely available, broadly applicable, and continuously improvable applications has played a pivotal role in accelerating scientific progress.
Currently, R is an open-source programming language and environment that offers an extensive array of statistical and graphical techniques (R Core Team, 2022), has emerged as one of the most powerful and versatile platforms within the broad scientific community (Westgate, 2019), and has become a dominant software in academia and across scientific fields (Amezquita et al., 2020; Joo et al., 2020; Sousa et al., 2020). Its utilization has offered a remarkable opportunity for knowledge exchange and fostered interdisciplinary collaboration (Atkins et al., 2022). With R’s popularity among researchers across diverse scientific domains, it has become easier to share methodologies, exchange insights, and build upon existing knowledge (Raasch et al., 2013). This collaborative environment has stimulated innovation and enabled a rapid dissemination of findings. Hence, such collective efforts not only enhance the quality of research but also contribute to the development and broad application of standardized best practices and promote greater reproducibility and transparency within the scientific community (Muenchow et al., 2019; Powers and Hampton, 2019). This collaborative nature empowered researchers to customize analytical tools according to specific research objectives, ensuring that the software remained up-to-date and responsive to the evolving demands within and among individual research disciplines.
The field of dendrochronology, which uses the radial growth patterns of trees to reveal valuable environmental and historical information, is no exception from the above-described trends. Currently, there is a diverse ecosystem of R packages focusing on tree-ring analysis. These packages have facilitated researchers in performing many of their tasks, from parameter measurement, through data preparation and analysis, to data and results visualization. Hence, some fields of dendrochronology are relatively well covered by functionalities of currently available R package(s). Nevertheless, there is still room for new packages or development of those currently available to fill in the full range of tree-ring research-related tasks which are not currently addressed. Arguably, there are also other open-source programming languages and environments, with Python (Van Rossum and Drake, 2009) being used in ecology and dendrochronology (see https://opendendro.org/). Currently, however, there is a limited number of Python packages in dendrochronology (see, e.g., Lozhkin et al., 2024), and, thus, we focus here on R, which currently is the most popular software among ecologists (Lai et al., 2019).
Despite the availability of search engines, discovering and accessing desired R functions or collections remains challenging due to the diverse modalities used to promote them. Before applying them, users must first be aware of their existence and functionalities. Often these packages are deposited on platforms like CRAN (a repository for R packages and documentation, https://cran.r-project.org/) and might also be described in scientific publications. However, some packages are exclusively found on specific software development platforms (e.g., GitHub, https://github.com/), or personal or institutional websites. Despite the growing number of tree-ring-specific R packages, a comprehensive overview has currently been missing (but see Atkins et al. (2022) for an example of forest research). It is, thus, challenging for users to navigate and select the most suitable set of R functions/packages for their needs.
Creating an inventory of comprehensive tree-ring-related R packages and Shiny applications (a web-based interactive application built using the Shiny framework in the R programming language), including examples of their applications, would serve as a valuable tool for students, teachers, and researchers across different disciplines. Further, enhancing awareness of a consistent R ecosystem and, consequently, its use, would lead to reducing the variability in data measurement introduced by (i) the use of numerous platforms (Lara et al., 2015; Maes et al., 2017) or (ii) personal subjectivity (Maxwell et al., 2011). Further, it would also reduce the space for errors introduced during data processing and analysis when developing and utilizing new code for tasks already covered in R, which may be more prone to mistakes than tools approved by peer review and consequently used and tested by the community. Consequently, this will eliminate discrepancies in comparative, synthesis, and review studies within dendrochronology and in interdisciplinary research contexts.
Here, we aim to present a comprehensive and organized overview of the currently available R packages, including Shiny applications, developed for dendrochronological analyses. This will guide readers through the specific focus of each package, making it easier to pinpoint the most appropriate package or combination for their research inquiries. We also discuss the prospects and challenges for dendrochronological research to stay competitive and consolidate its role in interdisciplinary research in the era of digitalization.
2 Methods
The inventory of tree-ring-related R packages was performed by applying a systematic search (at the beginning of March 2025) in Google Scholar (https://scholar.google.com/), CRAN (https://cran.r-project.org/), and GitHub (https://github.com/) using a list of defined keywords. As keywords, we used “R package dendrochronology”, and “R package tree rings” for Google Scholar, and “dendrochronology”, “dendroclimatology”, “dendroecology”, “dendroarchaeology”, “dendrogeomorphology”, “wood anatomy”, “wood formation”, “dendrometer”, “dendrochemistry”, “tree-ring”, “tree ring”, “tree-growth”, “tree growth”, for CRAN and GitHub. The systematic search was complemented by the screening of dendrochronology-related literature and web pages. It is important to note that we focused only on R packages and, thus, separate R function(s) were not considered. From the full list of identified R packages, we only considered those being (i) publicly available (i.e., not upon request), (ii) developed for broader use (i.e., not limited for a case-specific purpose), and (iii) with sufficient documentation describing at least the main functions (via scientific article, vignettes, README.md, or video). The identified R packages were categorized into (i) those geared towards measuring tree-ring characteristics and (ii) those centered on data processing, analysis, and visualization. Within the latter category, further distinctions were made between packages of general applicability in dendrochronology and those tailored to specific subdisciplines within the field.
3 Structured presentation of tree-ring-specific R packages
Altogether, we identified 38 tree-ring-related R packages (Figure 1). From all these packages, 28 are available in CRAN and 10 on GitHub only (Figure 1; Supplementary Table S1). Overall, 32 R packages were associated with at least one scientific publication focused solely on the presentation of the package and its functionalities (Supplementary Table S1). Further, we mention example(s) of the studies using individual packages (wherever possible), reflecting the package (i) complexity, (ii) usage (see Figure 1) as well as (iii) long-term relevance (in the developing fields, such as dendroanatomy). The purpose is to provide a short resume of the focus of each package, and readers are referred to the original publication or other sources for more information (see Supplementary Table S1). Below, the packages are listed in alphabetical order within individual sections.
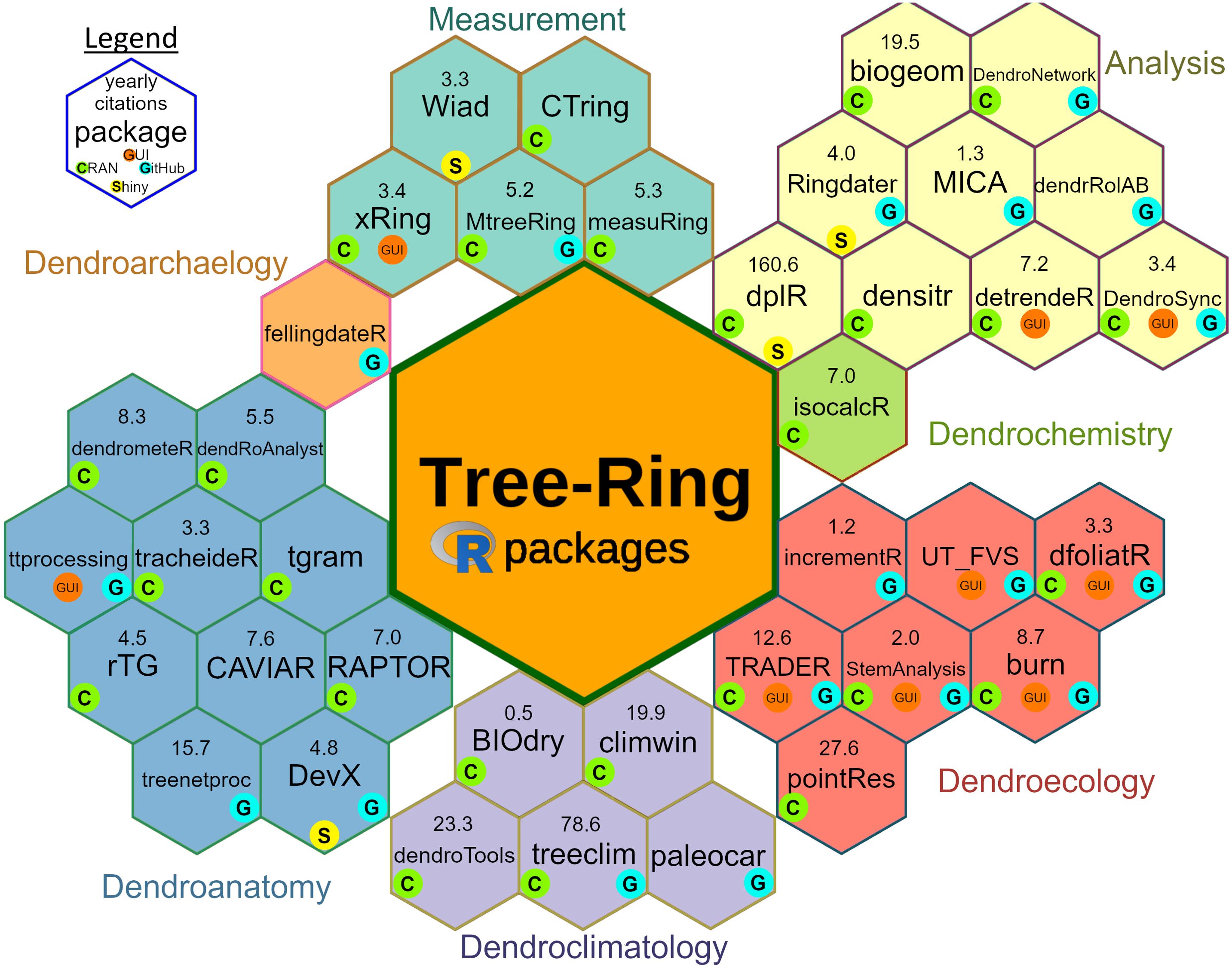
Figure 1. Structured overview of tree-ring-related R packages. Each hexagonal icon represents the package name, accompanied by the number of yearly citations (November 2024) from an associated introductory publication (if available, see Supplementary Table S1). The smaller icons denote the package’s presence on CRAN (C) and GitHub (G) and the availability of a related Shiny application (S) or a graphical user interface (GUI) (as in March 2025). The indication of subdiscipline does not exclude that the individual packages can also be used in another category.
3.1 R packages facilitating the measurement of tree-ring parameters
Thanks to the accessibility of flatbed scanners and digital cameras, it is now feasible to extract measurements of tree-ring width and wood density from cross-sections of pre-prepared wood surfaces using image analysis. Consequently, five R packages have been specifically developed to automate the processes of ring detection and measurement directly on the images (Figure 1; Table 1).
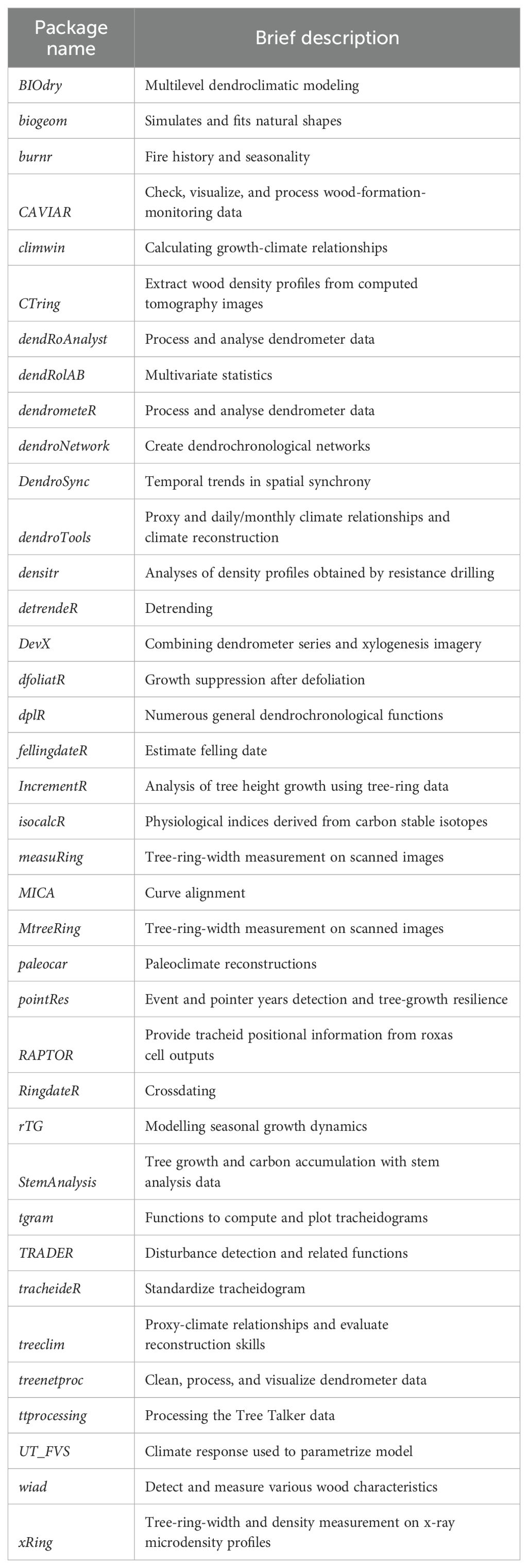
Table 1. List of identified 38 tree-ring-related R packages with a short description of their focus (packages in alphabetical order).
CTring (Mahatara et al., 2024) enables accurate determination of pith positions and generation of tree-ring density profiles from computed tomography images of tree cross sections. It converts grey values into density, combines the adapted Hough Transform method and a one-dimensional edge detector for precise pith detection, and allows users to inspect and adjust tree-ring boundaries via a visual, user-friendly interface. The package’s functions are flexible and adaptable for further applications, such as deriving ring-width time series and exploring low-quality images and ring-porous species.
measuRing (Lara et al., 2015) is used to detect tree-ring borders and measure tree-ring widths from scanned images of wood samples. Tree-ring borders can be automatically identified using a linear detection algorithm or visually assigned by the user. The measurement of tree-ring widths using measuRing can be applied to both coniferous and broadleaved tree species (Caselli et al., 2021).
MtreeRing (Shi et al., 2019) enables the detection of tree-ring boundaries and measuring tree-ring widths from scanned images of wood samples (see, e.g., Zhao et al., 2022; Diao et al., 2023). In addition to its linear detection algorithm, it incorporates alternative methods such as watershed-based segmentation and the Canny edge detector. The choice of automatic detection technique depends on the specific anatomical features of the wood species being analyzed. Tree-ring borders can also be manually assigned by the user. Further, the functionality of MtreeRing was used for the development of ρ-MtreeRing (García-Hidalgo et al., 2021), which provides a graphical user interface for x-ray microdensity analysis.
wiad (Rademacher et al., 2021b) is a versatile measurement-oriented R package. In addition to its capabilities in detecting and measuring annual ring widths from wood images, it goes further by offering tools for identifying specific intra-annual features like earlywood and latewood, density fluctuations, and fire scars. Additionally, wiad is designed to assist in the archival of images, data, and metadata, providing a comprehensive solution for tree-ring analysis. wiad has been used to measure ring widths from scanned images of microsections from broadleaved trees (Rademacher et al., 2022) as well as conifers (Rademacher et al., 2021a). Additionally, it has been applied to measure the radial position of intra-annual density fluctuations within tree rings (Miller et al., 2022).
xRing (Campelo et al., 2019) automatically derives tree-ring widths and density measurements from X-ray micro-density profiles. Specifically, xRing offers functions to identify tree-ring borders and earlywood-latewood transition, allowing for precise measurements of earlywood, latewood, and total ring widths and densities. The package has been used to measure ring width and density on density profiles from both coniferous (Hevia et al., 2020) and broadleaved species (Cahuana et al., 2023).
3.2 R packages facilitating data processing, analysis, and visualization
Dendrochronology encompasses various subdisciplines, each employing distinct statistical methods for data processing and analysis (Speer, 2010). To facilitate navigation through the wide array of available tools, this section is organized based on the packages’ intended purposes. We differentiate between packages with broad applicability across multiple dendrochronological disciplines (Section 3.2.1), as well as packages tailored towards specific subdisciplines (Section 3.2.2).
3.2.1 R packages with broad applicability in dendrochronology
The packages listed and described here have broader applicability across multiple subdisciplines of dendrochronology without being limited to a single area of study. These packages are designed for general data preparation and offer a range of commonly used tree-ring statistical functions. Altogether, we identified eight different R packages (Figure 1; Table 1).
biogeom (Shi et al., 2022) simulates and quantifies natural geometries, such as leaves, eggs, seeds, and tree rings. It allows users to fit empirical datasets to parametric equations using the fitGE function, which models tree-ring boundaries and estimates parameters that describe their shape. This package aids in exploring morphological variation in tree rings, revealing insights into environmental conditions, growth patterns, and ontogeny. By modelling growth trajectories, biogeom helps to predict maximum growth rates and provides a versatile tool for the geometric analysis of natural shapes, including tree rings. It was employed to show that the superellipse equation describes the tree-ring boundaries and estimates the basal area increment of six coniferous species (Huang et al., 2024).
dendRolAB (Buras et al., 2016, 2022) analyzes tree-ring data with a focus on multivariate statistics, like Principal Component Gradient Analysis (PCGA) and the Standardized Growth Change (SGC) method (see, e.g., Mašek et al., 2023). It helps users to: (1) detect significant growth changes, such as droughts or disturbances; (2) quantify the impact and recovery of extreme events; (3) estimate event durations; and (4) improve growth analysis accuracy with robust results. Its key innovation, the Bias-Adjusted SGC (BSGC), refines event detection for more precise insights (see, e.g., Netsvetov et al., 2023). The package also aids in visualizing growth dynamics and environmental responses.
DendroSync (Alday et al., 2018) contains functions for the estimation and visualization of synchronicity in tree-ring networks and for assessing temporal changes in those patterns. This package has been devised to work with traits derived from tree rings (e.g., ring-width). It employs unique variance-covariance mixed models to quantify a shared temporal signal among chronologies over a fixed period. Moreover, it identifies temporal trends in spatial synchrony using a moving window algorithm. Alternative R packages suitable for the evaluation of synchrony visualization spatial trends by correlograms are “synchrony” and “ncf”. The package is often used for analyses of growth sensitivity to climate (Kasper et al., 2023) or climate change-induced stress (Popa et al., 2024).
densitr (Krajnc et al., 2021) contains various tools to analyze density profiles obtained by the resistance drilling of trees. It includes functions to remove the trend from measurements using various methods, plot the profiles, or detect tree rings automatically. The package densitr has been used to measure resistance drilling density and perform subsequent detrending (Arnič et al., 2022; Krajnc et al., 2022).
detrendeR (Campelo et al., 2012) is a package specifically designed for the detrending and standardization of time series with the graphical control of the process. The package also allows chronology building and testing temporal changes of the common signal using standard chronology statistics. detrendeR includes four classical detrending techniques, and the package was frequently used in dendroclimatological studies (see, e.g., Altman et al., 2017; Tumajer et al., 2017; Rozas et al., 2024).
dplR (Bunn, 2008), which stands for “Dendrochronology Program Library in R,” is arguably the most widely used R package within the dendrochronological community and a cornerstone of the open-source dendrochronological framework openDendro (https://opendendro.org/). This comprehensive package was the first to incorporate the R functionality of DOS dendrochronological programs [e.g., COFECHA (Holmes, 1983) or ARSTAN (Cook, 1985)] and offers a sophisticated and diverse array of functions for conducting various standard dendrochronological analyses. These analyses include crossdating (Bunn, 2010), standardization, chronology construction, computation of standard descriptive statistics, data visualization, and much more. dplR is a dynamically evolving package continually incorporating new functions to its repertoire.
Hence, dplR is broadly used for various purposes, starting with functions for importing tree-ring-specific formats to the R environment, statistical validation of dendrochronological datasets, or plotting the data. Further, it was used for crossdating (Pretzsch et al., 2017), detrending (Ols et al., 2023), chronology building (Thakur et al., 2024), or general data treatment as calculating basal area increment (Rai et al., 2023), fitting various filters and power transformation (Wu et al., 2024). It is also employed for various analyses, such as power spectrum analysis (Arroyo-Morales et al., 2023) or superposed epoch analysis (Altman et al., 2021).
MICA (Stangler et al., 2016; Mann et al., 2018), a Multiple Interval-based Curve Alignment, is designed to analyze tree growth by addressing spatial variation in wood density profiles. It identifies key points in these profiles and aligns them to create representative average profiles, reducing noise and providing clearer insights into how environmental factors influence wood density and growth. MICA uses a heuristic approach to synchronize discrete data curves (see, e.g., Raden et al., 2020). This technique is especially useful for analyzing intra-annual data like wood density profiles or wood anatomy, improving accuracy in studying tree responses to environmental variations.
RingdateR (Reynolds et al., 2021) is an integrated tool for dendrochronological research, improving crossdating accuracy and efficiency. It offers two primary modes: Pairwise Analysis Mode, which compares multiple time series to find matches, and Chronology Analysis Mode, which allows users to compare individual measurements to an existing chronology. The package employs statistical correlation analysis, various detrending methods, and visualization tools to support crossdating efforts. It allows the performance of numerous pairwise comparisons among individual records. It also supports graphical analysis to identify false or missing rings and assess adjustments on chronology statistics. Though efficient, crossdating results should be manually verified for accuracy. RingdateR was used to facilitate the crossdating of tree-ring data (Greer et al., 2023) and the annual growth of marine bivalves (Edge et al., 2021).
dendroNetwork (Visser, 2024) is a package for creating dendrochronological (provenance) networks based on the statistical relations between individual tree-ring series or chronologies. After creating the dendrochronological network in R, it is recommended to visualize the network using Cytoscape (https://cytoscape.org/). The development of dendrochronological networks is suitable for the estimation of wood provenance (see, e.g., Visser, 2021) or wood use patterns (Visser and Vorst, 2022).
3.2.2 R packages tailored to specific subdisciplines
Below we introduce the packages grouped by main subdisciplines of dendrochronology (Speer, 2010), i.e. dendroclimatology, dendroecology, dendroanatomy, dendroarchaeology, and dendrochemistry, for which we identified at least one R package (Figure 1; Table 1). The assignment to a given subdiscipline, however, does not exclude that the package can also be used in others.
3.2.2.1 Dendroclimatology
Exploring the connection between tree growth and climate represents a fundamental aspect of tree-ring research. As a result, a multitude of software tools have emerged for conducting dendroclimatological analyses. Given that a significant portion of tree-ring researchers specialize in dendroclimatology, several R packages have been created to address various specific analytical needs within this field. Altogether, we identified five R packages focused on dendroclimatology (Figure 1; Table 1).
BIOdry (Lara et al., 2018) is a toolkit for analyzing climate-growth relationships. It models allometric relationships in tree growth components like tree-ring width diameter, basal area, and biomass. Key functions include modelFrame, which evaluates allometric parameters, formats TRW units, and incorporates random effects to account for hierarchical variability. The package addresses autocorrelation and manages serial correlations with standard correlation structures. It also models annual aridity indices, allowing custom functions for drought indicators. In addition, it can compare dendroclimatic fluctuations across multilevel data and categorical variables.
climwin (Bailey and van de Pol, 2016; Rubio-Cuadrado et al., 2022) offers a collection of functions designed to compute associations between climate and tree-ring parameters. This package enables calculating various models that account for multiple time windows, establishing connections between a response variable and environmental factors at diverse time resolutions. It identifies influential periods that impact tree proxies and selects parsimonious models. While the climwin package has been widely used in animal ecology, for example to study the impacts of climate change on bird populations (Bailey et al., 2022; McLean et al., 2022), it has also been successfully applied in dendrochronology to calculate climate-growth with optimal climate window (Camarero et al., 2024; Rubio-Cuadrado et al., 2024).
dendroTools (Jevšenak and Levanič, 2018) determines the statistical relationships between tree-ring parameters and daily environmental data by a progressive sliding of moving window of specified length through daily environmental data. It also allows the calculation of bootstrapped and partial correlation coefficients and may be also applied to monthly data (Jevšenak, 2020). Next to the traditionally used linear regression, it provides the opportunity to employ robust nonlinear machine learning functions to investigate climate-growth relationships, including nonlinear artificial neural networks with the Bayesian regularization training algorithm. Further, dendroTools enables the comparison of various regression algorithms for climate reconstruction, and the packages can be used directly to calibrate models for climate reconstruction (Jevšenak et al., 2018b). Due to the broad functionality of dendroTools, including high-quality graphical outputs, it is frequently used in dendroclimatological studies (see, e.g., Lopez-Saez et al., 2023; Tumajer et al., 2023; Rai et al., 2024).
paleocar (Bocinsky et al., 2016) contains functions to carry out tree-ring-based spatiotemporal paleoclimate reconstruction over large geographic areas. It uses the CAR (Correlation-Adjusted (marginal) coRelation) approach as implemented in the “care” package (Zuber and Strimmer, 2011), with speed and memory use optimization. The package has been used to perform paleoclimate reconstructions of precipitation over the Southwestern United States (Bocinsky et al., 2016; Strawhacker et al., 2020).
treeclim (Zang and Biondi, 2015) calibrates proxy-climate relationships in tree-ring chronologies. It offers tools for bootstrapped response and correlation functions, seasonal correlation analysis, and reconstruction skill assessment. The package features a stationary bootstrap method to handle temporal autocorrelation, ensuring robust statistical analysis. It supports flexible variable selection, various bootstrapping schemes, and moving correlation functions. Users can input monthly climate data to calculate response and correlation functions and evaluate the temporal stability of dendroclimatic relationships. This package represents an enhanced version of the no longer available bootres (Zang and Biondi, 2013) and it is broadly used in dendroclimatology (see e.g., Altman et al., 2020; Grover et al., 2023; Arco Molina et al., 2024).
3.2.2.2 Dendroecology
Dendroecological studies, here including also the fields of dendrogeomorphology and dendropyrology, focus on the imprint of ecological and environmental processes in tree rings. These studies mostly focus on detecting disturbances, environmental pollution, specific forest dynamic patterns or other discrete events using tree rings. We have identified seven R packages tailored for such analyses (Figure 1; Table 1).
burnr (Malevich et al., 2018) was developed for tree-ring fire-scar analysis, however, it applies to other event analyses. It reads, writes, and analyses tree-ring fire history (FHX) files. burnr produces fire demography charts, calculates fire frequency and seasonality statistics, and runs superposed epoch analysis. A unique benefit of the package is that it enables automatic analysis and visualization (that is useful for large datasets), but it also facilitates the integration of other R packages (e.g., dplR). Studies using burnr often focus on fire regime (Badeau et al., 2024) and fire history related to climate and human-driven changes (Şahan et al., 2023).
dfoliatR (Guiterman et al., 2020) identifies, quantifies, analyses, and visualizes growth suppression events in tree rings that are often produced by insect defoliation. It infers defoliation events in individual trees based on user-specified thresholds with functions for summary statistics and graphics of tree- and site-level series. dfoliatR is based on OUTBREAK program and improves it in many ways (e.g., identification of events, user control, computation capacity). Moreover, it can remove climatic signals in the series by an indexing procedure. The package is often used for outbreak detection (Jiang et al., 2024) or dendrochronological reconstruction of outbreaks (Fraver et al., 2024).
IncrementR (Kašpar et al., 2019) contains a set of functions for the analysis of height growth along the stem of trees and shrubs (Rita et al., 2020). The main computed parameters provided by the package include height growth along the stem, changes in stem eccentricity and taper. It also contains functions utilizing standard procedures for the estimation of the number of missing tree rings near the pith (Kašpar et al., 2020). The package is useful in ecological and dendrogeomorphological studies as well as serial sectioning.
pointRes (van der Maaten-Theunissen et al., 2015) identifies event and pointer years in tree growth patterns and assesses resilience components. It offers two primary methods: normalizing tree-ring data within a moving window or comparing growth to average growth in preceding years. Users can define thresholds to tailor analyses. The package calculates resilience metrics such as resistance, recovery, and relative resilience, offering insights into tree responses to environmental stressors (Charlet de Sauvage et al., 2023; Camarero et al., 2024). It also provides various plotting functions for visualizing results. An update, pointRes 2.0 (van der Maaten-Theunissen et al., 2021), introduced new indices characterizing tree-growth resilience and new functions for calculating pointer years (see, e.g., Jetschke et al., 2023).
StemAnalysis (Wu et al., 2023) provides a set of procedures for the reconstruction and exploration of tree-growth patterns and estimating tree biomass and carbon storage by volume and allometric models. It is designed to determine age class, stem growth profiles (height, outside-bark diameter, and volume), construct height–diameter relationships, and, consequently, estimate tree biomass and carbon for an individual tree. The package contains several allometric relationships, such as height–diameter (e.g., Richards, Logistic, Weibull, and Gompertz curves), outside-bark DBH, outside-bark stem volume, and carbon accumulation with tree age.
TRADER (Altman et al., 2014) contains four methods commonly used for the detection of disturbance events, (i) radial-growth averaging criteria developed by (Nowacki and Abrams, 1997), (ii) the boundary-line method (Black and Abrams, 2003), (iii) the absolute-increase method (Fraver and White, 2005), and (iv) the combination of radial-growth averaging and boundary-line techniques (Splechtna et al., 2005). Moreover, TRADER enables the detection of past disturbance events using tree-ring data using 24 published methods by setting the various parameters (see also Altman, 2020). The function of comparison of results between different methods is also implemented. Lastly, TRADER contains functions for the detection of tree recruitment and growth trends. Since its introduction, TRADER has been used, e.g., to detect growth releases (Carter et al., 2021; González de Andrés et al., 2024), growth suppression (Camarero and Valeriano, 2023), and growth pattern determination (Janda et al., 2021) to understand forest dynamics.
UT_FVS (Giebink et al., 2022) combines tree-ring and forest inventory data to create a species-specific tree-growth model. By combining forest monitoring data with climate response recorded in tree rings, it parameterizes a widely used forest management tool – Forest Vegetation Simulator (i.e., Forestry growth and yield models, FVS) for several tree species. It allows the parsing of the multiple drivers of tree growth and forest stand development, such as climate, competition, and site characteristics. Further, it will provide more accurate projections of carbon uptake, allowing foresters to anticipate stand vulnerability to climate change and adaptively manage to increase stand resilience.
3.2.2.3 Dendroanatomy and wood formation
Dendroanatomy, which encompasses here xylogenesis observations, dendrometer measurements, and quantitative wood anatomy, is a relatively new and rapidly evolving area within dendrochronology. Hence, in contrast to other tree-ring disciplines, many of the R packages listed in this section lack alternatives in commercial software. This underscores the demand for open-source tools in this research domain, which is evident from the considerable number of R packages available. We identified ten R packages dedicated to dendroanatomy and wood formation (Figure 1; Table 1).
CAVIAR (Rathgeber et al., 2011, 2018) is a package dedicated to the verification, visualization, and manipulation of wood-formation-monitoring data for conifers growing in temperate and cold environments. It contains algorithms to check, display, and process wood-formation-monitoring data, including the computation of dates of wood-formation phenology. This package is currently used as a standard for detecting outliers in the observational data (e.g., Rathgeber et al., 2016), to assess the onset, duration, and end of wood formation processes such as cell enlargement, wall thickening, and maturation (e.g., Wang et al., 2023); and to fit Gompertz function to wood formation dynamics data (e.g., Debel et al., 2024).
dendRoAnalyst (Aryal et al., 2020) is designed for managing and cleaning dendrometer data, separating radial growth from daily cyclic shrinkage and expansion, calculating daily statistics, and linking it to daily meteorological data. The cleaning process includes identifying and erasing sudden jumps in dendrometer data, identifying time gaps, and changing temporal resolutions. Similar to dendrometeR (van der Maaten et al., 2016) and treenetproc (Knüsel et al., 2021), this package is used to extract intra-annual growth rates to link them to co-occurring environmental data (e.g., Zhou et al., 2023; Zhang et al., 2024).
dendrometeR (van der Maaten et al., 2016) facilitates the analysis of dendrometer data using both daily and stem-cycle approaches. The package contains customizable functions to prepare, verify, process, and plot dendrometer series, as well as functions that facilitate the analysis of dendrometer data (i.e. daily statistics or extracted phases) in relation to environmental data. For example, it was used for processing raw dendrometer data to obtain the daily maximum radius series (Sanmiguel-Vallelado et al., 2021).
DevX (Cruz-García et al., 2019) enables the presentation of dendrometer curves and the overlay of growth model fits (Gompertz and Weibull) derived from xylogenesis observations. Wood phenology estimates, such as growth onset, cessation, and duration, are calculated using Gompertz fits (see, e.g., Yin et al., 2024).
RAPTOR (Peters et al., 2018) is designed to transform spatially-resolved tracheid anatomical data – produced by software such as ROXAS, WinCELL, and ImageJ – into radial row positional information for tracheidograms. Using advanced local search and alignment algorithms, RAPTOR assigns tracheids to radial files and determines their positions within them. The outputs from RAPTOR consolidate various intra-ring cell measurements into representative radial files, which are invaluable for linking cell properties – such as size (Cuny et al., 2019; Peters et al., 2021), biomass (Martínez-Sancho et al., 2022), chemical composition (Martínez-Sancho et al., 2023), and functional structure (Sviderskaya et al., 2021) – to the timing of cell formation and corresponding environmental conditions. This linkage is usually achieved through the integration of observed data, such as xylogenesis (e.g., Pérez-de-Lis et al., 2022), modelled data (e.g., Vaganov et al., 2006; Popkova et al., 2018), or climate-growth derived data (e.g., Castagneri et al., 2017).
rTG (Jevšenak et al., 2022) is designed for analyzing tree-ring data and studying tree-growth dynamics, focusing on modelling the temporal patterns of secondary growth in trees, particularly xylem and phloem formation. It offers three key modelling approaches: the Gompertz equation, general additive models, and artificial neural networks with Bayesian regularization, allowing users to compare methods for optimal modelling (see, e.g., Mousavisangdehi et al., 2024). The package is valuable for studying the impacts of environmental factors and climate change on tree growth over time, providing tools for modelling intra-annual growth dynamics and comparing growth across individuals, sites, and seasons. It helps advance understanding of tree responses to ecological and climatic influences. It was used to simulate intra-annual growth dynamics of Fagus orientalis (Mousavisangdehi et al., 2024).
tgram (DeSoto et al., 2011) computes and plots tracheidograms of tracheid anatomical data obtained from line profiles performed along radial files.
tracheideR (Campelo et al., 2016) provides functions for identifying and measuring the tracheid’s radial lumen and wall thickness from line profile data performed on radial files and calculates standardized tracheidograms. It was used for standardization of data based on the relative position of each tracheid inside the tree ring (Vieira et al., 2020).
treenetproc (Knüsel et al., 2021) cleans, processes, and visualizes highly resolved time series of dendrometer data. In two steps, raw dendrometer data is aligned to regular time intervals and cleaned. Further, the package offers functions to extract the day of the start and end of the growing season as well as several characteristics of shrinkage and expansion phases. For instance, it allows users to calculate daily and seasonal growth increments, assess water deficit responses through stem shrinkage patterns, and analyze diurnal cycles to infer tree water status and address ecological and physiological questions. Treenetproc has been employed to determine the climatic drivers of radial growth (Plavcová et al., 2024), the borders and the effective days with growth during the growing season and its variability across different species (Etzold et al., 2022), to reveal how drought conditions alter tree water storage and elastic shrinkage (Peters et al., 2023), and to assess tree growth responses to heatwaves or late frosts (e.g., Salomón et al., 2022).
ttprocessing (Kabala et al., 2024) processes the data produced by the modular monitoring system, the Tree Talker. Most relevantly for dendrochronology, one of the sensors integrated into the Tree Talker system is an infrared dendrometer, which allows the tracking of the growth patterns of trees over time. The package allows the user to download the data, convert raw data to physical quantities, or estimate sap flow from recorded data. Further, it facilitates (i) data treatment, such as removing defective values or removing incomplete records, (ii) visual inspection of the data, and (iii) technical management of the monitoring system. For example, it was employed for obtaining the hourly sap flux values (Kabala et al., 2025).
3.2.2.4 Dendroarchaeology
Dendroarchaeology is one of the most traditional disciplines within tree-ring research and it is mostly dependent on absolute dating of dead wood samples. Hence, the packages introduced in part “3.2.1. General dendrochronological R packages” cover a significant portion of calculations used within dendroarchaeology. Further, we identified one R package developed specifically for the use of dendroarchaeology (Figure 1; Table 1).
fellingdateR (Haneca, 2024) estimates the felling date from the dated tree-ring series of historical timbers considering the presence of partially preserved sapwood. It is especially useful for wood samples where the sapwood is incomplete, and the last formed tree ring is missing. The current version of fellingdater package is designed for analyzing tree-ring data from European oak (Quercus sp.) However, it also allows users to input custom sapwood datasets, enabling the analysis of tree-ring data from other regions and species. Further, the package also facilitates crossdating procedures by computing commonly used correlation values between dated tree-ring series and reference chronologies. The package has been used to estimate felling dates of oak timbers in medieval constructions in Belgium, linking building activity to economic, social, and demographic dynamics (Haneca et al., 2020).
3.2.2.5 Dendrochemistry
Dendrochemistry uses various techniques, mostly elements, radiocarbon, and stable isotopes, to trace environmental changes. We identified one R package relevant exclusively to dendrochemistry (Figure 1; Table 1).
isocalcR (Mathias and Hudiburg, 2022) was developed to standardize the calculations of leaf and tree physiological indices derived from carbon stable isotopes. The isocalcR package offers a set of functions to calculate leaf carbon isotope discrimination, leaf intercellular CO2 concentration, the ratio of intercellular to atmospheric CO2 concentration, the difference between atmospheric and intercellular CO2 concentration, and intrinsic water-use efficiency. These calculations are based on carbon isotope signatures in leaf or wood tissue and require minimal input from the user. It also implements data products of atmospheric [CO2] (ppm) and atmospheric δ13CO2 (‰) composition for the period 0–2021 C.E. The package has been used to perform carbon isotope calculations (Treml et al., 2022; Mathias et al., 2023; Santini et al., 2024).
4 Prospects and future challenges
4.1 An evolving dendrochronology
Since its foundation as a scientific discipline, tree-ring research has been a cornerstone for dating and environmental reconstruction, serving as a transdisciplinary tool bridging diverse fields of study (Speer, 2010; Schweingruber, 2012). Traditional methodologies primarily relied on ring width measurements, using well-established techniques such as crossdating and standardization to interpret environmental changes with annual precision and accuracy (Fritts, 1976). For decades, widely used statistical and graphical tools, such as COFECHA (Holmes, 1983) for cross-dating or ARSTAN for detrending and chronology building (Cook, 1985), along with other DOS-based programs (Holmes, 1994), were the principal analytical tools for tree-ring studies.
In recent decades, however, tree-ring research has experienced a transformative shift driven by the digital revolution (Hey et al., 2009), shaping its role in understanding tree growth and environmental interactions. These changes have broadened the scope of the field, integrating disciplines such as ecophysiology (Arco Molina et al., 2024), wood anatomy (von Arx et al., 2021), dendroprovenancing (Hellmann et al., 2017; Housset et al., 2018), or growth phenology (Cuny et al., 2015; Gao et al., 2022). This expansion has provided deeper insights into tree growth mechanisms and propelled tree-ring research into new frontiers (Altman, 2020; Domínguez-Delmás, 2020; Pearl et al., 2020).
This transformation is evident at multiple levels. Advances in computational power and the inclusion of innovative techniques reduce data processing costs and time, leading to the reshaping of traditional approaches. Methods such as X-ray densitometry (Björklund et al., 2019), blue light reflectance (Rydval et al., 2024), and digital microscopy for quantitative wood anatomy (Fonti et al., 2010) enable precise evaluation of sub-annual resolution. Additionally, cutting-edge instrumentation like MICADAS radiocarbon analysis (Wacker et al., 2014) and laser ablation for stable isotope measurements (Saurer et al., 2023) enhance accuracy and detail in isotopic analyses.
Simultaneously, tools for monitoring tree physiology (Steppe et al., 2015) and environmental conditions (Correa-Díaz et al., 2019) offer unprecedented spatial and temporal resolution, spanning scales from cellular to ecosystem levels (Puchi et al., 2020). Remote sensing and advanced monitoring systems further enhance the link between high-resolution environmental datasets and tree-ring observations, enabling the contextualization of growth responses and facilitating nuanced ecosystem analyses. Powerful data analysis tools, including artificial intelligence and machine learning, amplify these technological advancements. These methods streamline the recognition of anatomical patterns (Resente et al., 2021; Katzenmaier et al., 2023) and facilitate the integration of increasingly large and complex datasets (Babst et al., 2018).
However, the need for integrating R with other programming languages and platforms is growing. Python and C++, for example, are becoming essential in handling more complex tasks, such as automated image analysis for tree-ring measurement (e.g., deep learning and machine learning techniques) and high-resolution CT imaging for wood anatomy. These languages excel at processing large image datasets and running intensive computational analyses, complementing R’s strengths in statistical modeling, data visualization, and ecosystem modeling. Furthermore, cloud-based computing platforms and distributed processing are poised to support large-scale data workflows, enabling seamless integration with R for processing and analyzing data in parallel.
Looking ahead, combining R’s strengths with other tools and languages, as well as leveraging emerging technologies such as machine learning and cloud computing, is expected to create even more efficient and scalable workflows in dendrochronology. By streamlining the integration of different approaches, researchers will be better equipped to handle the increasing complexity of tree-ring datasets, unlocking new insights into forest dynamics, climate variability, and long-term ecological change.
4.2 R’s role in advancing and standardizing dendrochronology
In addition to these technological advances, the integration of diverse tools and programming languages is becoming increasingly relevant in modern dendrochronology. R, for example, continues to play a central role in data analysis, with various packages developed for tree-ring measurement and statistical analysis. These packages, such as CTring, MtreeRing, and dplR, offer flexibility for tree-ring measurement workflows, from earlywood–latewood delineation to chronology building and statistical analysis.
In this context, R, with its established use within the research community, represents a significant opportunity to drive dendrochronology’s digital transformation. This powerful, open-source, and accessible platform can further support versatile, standardized, and transparent approaches to data handling and analysis (see https://opendendro.org as a first existing example), which is essential for advancing the field (e.g., Bunn, 2008) and fostering integration with related disciplines (e.g., Jevšenak et al., 2018a). Already widely adopted by the community, R offers an extensive range of R packages for dendrochronology for data manipulation, statistical analysis, and visualization (see Section 3 for an overview). Thus, R-related functionalities can serve as a central hub for modernizing and streamlining workflows within the discipline.
However, several key areas could be further developed to fully capitalize on R’s potential in transforming dendrochronology. First, while R currently supports a range of standard tasks in dendrochronology, the integration of emerging data types and structures from both tree-ring analysis and related fields remains a challenge. These include tree-ring-based multi-proxy datasets, which combine various records of tree characteristics beyond the standard single-value-per-ring approach, such as wood density, cell structure and size, stable isotopes, and chemical compositions. Additionally, the integration of diverse proxy records, such as ice-core, shells, corals, fish otoliths, speleothems, and DNA, remains underdeveloped. Moreover, as dendrochronology increasingly intersects with other disciplines, such as remote sensing, genetics, and climate science, R packages that facilitate interdisciplinary integration will become more critical. For example, integrating remote sensing data with tree-ring chronologies could deepen our understanding of how vegetation responds to climate change (Correa-Díaz et al., 2019; Tumajer et al., 2023), while linking genetic markers with dendrochronological data might offer new insights into tree adaptability and genetic diversity (Housset et al., 2018; Isaac-Renton et al., 2018; Martínez-Sancho et al., 2021). Furthermore, the inclusion of sub-annual resolved datasets, which provide finer temporal resolution of tree growth and environmental responses (Lopez-Saez et al., 2023), is still a challenge, as are differences in temporal and spatial scales (Babst et al., 2018; Altmanová et al., 2025). These challenges are particularly pronounced when combining tree-ring data with other data types at different scales, such as ecosystem-level or pixel-level data. The integration of such disparate datasets requires careful alignment of both temporal and spatial dimensions to ensure meaningful analyses.
Moreover, R’s existing statistical capabilities are vast, but the integration of cutting-edge techniques–such as machine learning and artificial intelligence in dendrochronological tools—remains underdeveloped (but see, e.g., Jevšenak et al., 2018a; Resente et al., 2021; Katzenmaier et al., 2023; Poláček et al., 2023; García-Hidalgo et al., 2024; Keret et al., 2024). These methods hold great promise for automating image-based, labor-intensive tasks, such as tree-ring measurement or cross-dating, thereby reducing subjectivity, increasing repeatability, and significantly enhancing efficiency. However, as these techniques are further integrated into workflows, there is a growing need to ensure that the “magic box” of AI and machine learning models remains transparent (Cheong, 2024). This transparency is crucial to ensure that the underlying processes are understandable, reproducible, and scientifically valid, which helps improve interpretation and build trust in the results. Additionally, the effectiveness of AI models relies heavily on the quality and diversity of their training datasets. Ensuring that these datasets are publicly available, comprehensive, representative, and free from bias is essential for developing robust and reliable models that can be confidently applied in dendrochronology.
Furthermore, R’s capabilities extend beyond analysis. By integrating with other open-access tools (e.g., ImageJ, QuPath, PostgreSQL, Shiny), R can provide a foundation for building workflows that not only standardize analyses but also centralize, archive, and make both raw data and metadata interactively visible, along with their corresponding analyzed digital images (e.g., Rademacher et al., 2021b). Advanced visualization techniques will play a crucial role in interpreting complex, multi-proxy datasets that combine tree-ring data with other environmental or ecological variables. Future R packages could include tools for creating interactive visualizations of temporal structures in intra-annual data, which would enhance the interpretation of high-resolution datasets and reveal subtle environmental signals. Additionally, sophisticated statistical approaches, such as spatiotemporal, hierarchical, and mechanistic modeling, offer transformative potential for analyzing complex patterns across diverse environmental conditions and geographic regions. Mechanistic models, in particular, could simulate the processes driving tree growth and environmental responses, offering deeper insights into how various factors interact over time and space (Babst et al., 2018). Together, these approaches would enhance data processing efficiency, comply with the FAIR data principles, and offer significant opportunities to strengthen and modernize the invaluable International Tree-Ring Data Bank (ITRDB, see Zhao et al. (2019) for a description of its content). As the volume and diversity of data structure continue to grow, developing R packages incorporating these advanced workflows will facilitate the systematic organization and long-term storage of data, enabling broader temporal and spatial coverage. This would not only push the boundaries of dendrochronology but also expand the ITRDB’s capacity for future research and collaborations (Guiterman et al., 2023).
Lastly, the growing emphasis on reproducibility and transparency in research highlights the critical need for well-integrated metadata management. While initiatives such as TRiDaS (Jansma et al., 2010), Tellervo (Brewer, 2016), and LiPD (McKay and Emile-Geay, 2016) have significantly improved metadata accessibility, R packages still require further development to standardize metadata handling. Such enhancements would ensure better interoperability across datasets and studies, making it easier to share and compare data. Addressing these gaps will not only improve the accessibility of data but also promote greater collaboration, enabling more impactful interdisciplinary research.
4.3 Building an R-centered dendrochronological ecosystem
To fully capitalize on the potential of R in dendrochronology, it is essential to develop a centralized and coordinated ecosystem for managing and distributing R packages and related datasets. This ecosystem requires contributions from all stakeholders.
Package developers could play a pivotal role by prioritizing the creation of well-documented, actively maintained, and user-friendly tools. Hosting these packages on structured repositories, preferably CRAN, ensures standardized documentation with detailed descriptions of functions, arguments, and working examples, facilitating broader adoption. Meanwhile, GitHub offers flexibility for collaborative development during the early stages, providing an invaluable space for innovation. Publishing research articles alongside R packages would further amplify their visibility and impact by providing scientific context, detailed use cases, and peer-reviewed workflows, thereby fostering trust and alignment with research standards.
Users, as key contributors, could complement these efforts by reporting bugs, suggesting improvements, and supporting updates, especially for newly developed or evolving packages. Such collaboration strengthens tools, builds professional networks, and enhances the ecosystem, ultimately benefiting the broader research community.
Leading organizations such as the Association for Tree-Ring Research (ATR) and the Tree-Ring Society (TRS) could support the ecosystem by (i) maintaining an up-to-date field-specific list of R packages as well as other software’s/tools related to dendrochronology, (ii) promoting best practices for package usage and development, and (iii) curating structured repositories of tree-ring data and associated metadata. A centralized platform hosted by such well-established organizations could serve as a reliable and long-term hub for collaboration, reproducibility, and open science, ensuring that researchers have easy access to the most relevant tools and documentation.
Initiatives and platforms such as (i) Dendro-Hub (https://www.dendrohub.com, the online community for dendrochronologists), (ii) OpenDendro (https://opendendro.org, the home of an open-source framework for the essential analytic software used in dendrochronology), (iii) Dendro-Elevator (https://dendro.elevator.umn.edu/, a cyberinfrastructure model for online archive curation, visualization, and analysis with images of tree-rings and related environmental proxy datatypes), or (iv) Dendro-R (https://ronaldvisser.github.io/Dendro_R, a list of R-packages for dendrochronology), together with (v) relevant review papers, provide an excellent starting point for centralizing knowledge and building a collaborative, open-access infrastructure. By enabling the sharing of datasets, methods, images, and analytical scripts, they promote transparency, reproducibility, and standardized workflows, allowing data to be more easily accessed, compared, and integrated across studies. Moreover, such platforms can foster cross-disciplinary research by enabling seamless integration between dendrochronology and related environmental sciences. As these tools mature and their adoption broadens, they hold the potential to advance the accessibility, interoperability, and impact of dendrochronological research.
Ultimately, a unified R-centred ecosystem could overcome the inconsistencies that arise from using disparate software tools. Fostering a more standardized and consistent approach would support robust interdisciplinary collaboration, positioning dendrochronology as a critical contributor to environmental and climate science. This ecosystem would provide a powerful foundation for sharing and visualizing tree-ring data, incorporating AI and machine learning tools, and developing innovative approaches for analyzing complex, multi-proxy datasets.
We are convinced that the R environment provides the conditions for the above-suggested transition to be successful. We thus encourage the dendro-community to persist in the development of R packages to further enhance the quality of tree-ring research by equipping researchers with state-of-the-art tools. To facilitate this evolution, scientific publications introducing new methods should be required to include publicly available working codes upon submission. Without this requirement, incorporating novel methods becomes challenging, increasing the risk of errors and reducing reproducibility. Journals must adopt clear guidelines requiring working code with examples for new methods to ensure their accessibility and effective integration into research workflow.
5 Conclusion and call to action
The success of this R-centered dendrochronological ecosystem will hinge on community engagement, the sharing of resources, and the alignment of efforts to tackle the critical challenges facing modern dendrochronology. We urge researchers and organizations to invest in the development of R-based platforms and contribute to the ecosystem’s sustainability. By adopting open science principles and ensuring the sustainability of the ecosystem through regular updates and active collaboration, the field will be well-positioned to address future environmental challenges.
In conclusion, the future of dendrochronology hinges on our ability to adapt to the complexities of modern data and computational methods. By developing and maintaining specialized R packages and promoting standardized workflows, we can streamline data analysis, enhance reproducibility, and support interdisciplinary collaboration. For example, integrating tree-ring data with remote sensing or genomic information could provide unprecedented insights into how forests respond to climate change. The continued evolution of the R ecosystem for dendrochronology will not only unlock new opportunities to address pressing environmental challenges but also empower researchers to advance our understanding of tree growth processes and refine reconstructions of past and present climate dynamics. A collective commitment to innovation and collaboration within the dendrochronological community will ensure these tools reach their full potential in shaping the future of environmental science, ultimately advancing our understanding of tree growth and past and present climate dynamics.
Author contributions
JA: Writing – review & editing, Supervision, Conceptualization, Writing – original draft. NA: Writing – original draft, Conceptualization, Writing – review & editing. PFi: Writing – original draft, Conceptualization, Writing – review & editing. KK: Conceptualization, Writing – original draft, Writing – review & editing. PFo: Supervision, Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Research Grant 25-15727S of the Czech Science Foundation; long-term research development project No. RVO 67985939 of the Czech Academy of Sciences; Estonian Research Council grant PSG1044; and the ORD program of the ETH Domain with the project “Towards an Open and Fair International Tree-Ring database for intra-annual tree-ring data”. Open access funding by Swiss Federal Institute for Forest, Snow and Landscape Research (WSL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2025.1593675/full#supplementary-material
References
Alday J. G., Shestakova T. A., Resco de Dios V., and Voltas J. (2018). DendroSync: An R package to unravel synchrony patterns in tree-ring networks. Dendrochronologia 47, 17–22. doi: 10.1016/j.dendro.2017.12.003
Altman J. (2020). Tree-ring-based disturbance reconstruction in interdisciplinary research: Current state and future directions. Dendrochronologia 63, 125733. doi: 10.1016/j.dendro.2020.125733
Altman J., Fibich P., Dolezal J., and Aakala T. (2014). TRADER: A package for tree ring analysis of disturbance events in R. Dendrochronologia 32, 107–112. doi: 10.1016/j.dendro.2014.01.004
Altman J., Fibich P., Santruckova H., Dolezal J., Stepanek P., Kopacek J., et al. (2017). Environmental factors exert strong control over the climate-growth relationships of Picea abies in Central Europe. Sci. Total Environ. 609, 506–516. doi: 10.1016/j.scitotenv.2017.07.134
Altman J., Saurer M., Dolezal J., Maredova N., Song J.-S., Ho C.-H., et al. (2021). Large volcanic eruptions reduce landfalling tropical cyclone activity: Evidence from tree rings. Sci. Total Environ. 775, 145899. doi: 10.1016/j.scitotenv.2021.145899
Altman J., Treydte K., Pejcha V., Cerny T., Petrik P., Srutek M., et al. (2020). Tree growth response to recent warming of two endemic species in Northeast Asia. Clim. Change 162, 1345–1364. doi: 10.1007/s10584-020-02718-1
Altmanová N., Fibich P., Doležal J., Bažant V., Černý T., Molina J. G. A., et al. (2025). Spatial heterogeneity of tree-growth responses to climate across temperate forests in Northeast Asia. Agric. For. Meteorol. 362, 110355. doi: 10.1016/j.agrformet.2024.110355
Amezquita R. A., Lun A. T. L., Becht E., Carey V. J., Carpp L. N., Geistlinger L., et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145. doi: 10.1038/s41592-019-0654-x
Arco Molina J. G., Saurer M., Altmanova N., Treydte K., Dolezal J., Song J. S., et al. (2024). Recent warming and increasing CO2 stimulate growth of dominant trees under no water limitation in South Korea. Tree Physiol. 44, tpae103. doi: 10.1093/treephys/tpae103
Arnič D., Krajnc L., Gričar J., and Prislan P. (2022). Relationships between wood-anatomical features and resistance drilling density in Norway spruce and European beech. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.872950
Arroyo-Morales S., Astudillo-Sánchez C. C., Aguirre-Calderón O. A., Villanueva-Díaz J., Soria-Díaz L., and Martínez-Sifuentes A. R. (2023). A precipitation reconstruction based on pinyon pine tree rings from the northeastern Mexican subtropic. Theor. Appl. Climatol. 151, 635–649. doi: 10.1007/s00704-022-04303-1
Aryal S., Häusser M., Grießinger J., Fan Z., and Bräuning A. (2020). dendRoAnalyst”: A tool for processing and analysing dendrometer data. Dendrochronologia 64, 125772. doi: 10.1016/j.dendro.2020.125772
Atkins J. W., Stovall A. E. L., and Alberto Silva C. (2022). Open-Source tools in R for forestry and forest ecology. For. Ecol. Manage. 503, 119813. doi: 10.1016/j.foreco.2021.119813
Babst F., Bodesheim P., Charney N., Friend A. D., Girardin M. P., Klesse S., et al. (2018). When tree rings go global: Challenges and opportunities for retro- and prospective insight. Quaternary Sci. Rev. 197, 1–20. doi: 10.1016/j.quascirev.2018.07.009
Badeau J., Guibal F., Fulé P. Z., Chauchard S., Moneglia P., and Carcaillet C. (2024). 202 years of changes in Mediterranean fire regime in Pinus nigra forest, Corsica. For. Ecol. Manage. 554, 121658. doi: 10.1016/j.foreco.2023.121658
Bailey L. D. and van de Pol M. (2016). climwin: an R toolbox for climate window analysis. PloS One 11, e0167980. doi: 10.1371/journal.pone.0167980
Bailey L. D., van de Pol M., Adriaensen F., Arct A., Barba E., Bellamy P. E., et al. (2022). Bird populations most exposed to climate change are less sensitive to climatic variation. Nat. Commun. 13, 2112. doi: 10.1038/s41467-022-29635-4
Björklund J., von Arx G., Nievergelt D., Wilson R., Van den Bulcke J., Günther B., et al. (2019). Scientific merits and analytical challenges of tree-ring densitometry. Rev. Geophys. 57, 1224–1264. doi: 10.1029/2019RG000642
Black B. A. and Abrams M. D. (2003). Use of boundary-line growth patterns as a basis for dendroecological release criteria. Ecol. Appl. 13, 1733–1749. doi: 10.1890/02-5122
Bocinsky R. K., Rush J., Kintigh K. W., and Kohler T. A. (2016). Exploration and exploitation in the macrohistory of the pre-Hispanic Pueblo Southwest. Sci. Adv. 2, e1501532. doi: 10.1126/sciadv.1501532
Brewer P. W. (2016). Data management in dendroarchaeology using tellervo. Radiocarbon 56, S79–S83. doi: 10.2458/azu_rc.56.18320
Bunn A. G. (2008). A dendrochronology program library in R (dplR). Dendrochronologia 26, 115–124. doi: 10.1016/j.dendro.2008.01.002
Bunn A. G. (2010). Statistical and visual crossdating in R using the dplR library. Dendrochronologia 28, 251–258. doi: 10.1016/j.dendro.2009.12.001
Buras A., Ovenden T., Rammig A., and Zang C. S. (2022). Refining the standardized growth change method for pointer year detection: Accounting for statistical bias and estimating the deflection period. Dendrochronologia 74, 125964. doi: 10.1016/j.dendro.2022.125964
Buras A., van der Maaten-Theunissen M., van der Maaten E., Ahlgrimm S., Hermann P., Simard S., et al. (2016). Tuning the voices of a choir: Detecting ecological gradients in time-series populations. PloS One 11, e0158346. doi: 10.1371/journal.pone.0158346
Cahuana L. A. P., Piña E. A. G., Tuesta G. P., and Tomazello-Filho M. (2023). Radial variation of wood density and fiber morphology of two commercial species in a tropical humid forest in Southeastern Peru. Cerne 29, e103143. doi: 10.1590/01047760202329013143
Camarero J. J., Salinas-Bonillo M. J., Valeriano C., Rubio-Cuadrado Á., Fernández-Cortés Á., Tamudo E., et al. (2024). Watering the trees for the forest: Drought alleviation in oaks and pines by ancestral ditches. Sci. Total Environ. 950, 175353. doi: 10.1016/j.scitotenv.2024.175353
Camarero J. J. and Valeriano C. (2023). Responses of ancient pollarded and pruned oaks to climate and drought: Chronicles from threatened cultural woodlands. Sci. Total Environ. 883, 163680. doi: 10.1016/j.scitotenv.2023.163680
Campelo F., Garcia-Gonzalez I., and Nabais C. (2012). detrendeR - A Graphical User Interface to process and visualize tree-ring data using R. Dendrochronologia 30, 57–60. doi: 10.1016/j.dendro.2011.01.010
Campelo F., Mayer K., and Grabner M. (2019). xRing—An R package to identify and measure tree-ring features using X-ray microdensity profiles. Dendrochronologia 53, 17–21. doi: 10.1016/j.dendro.2018.11.002
Campelo F., Nabais C., Carvalho A., and Vieira J. (2016). tracheideR—An R package to standardize tracheidograms. Dendrochronologia 37, 64–68. doi: 10.1016/j.dendro.2015.12.006
Carter D. R., Bialecki M. B., Windmuller-Campione M., Seymour R. S., Weiskittel A., and Altman J. (2021). Detecting growth releases of mature retention trees in response to small-scale gap disturbances of known dates in natural-disturbance-based silvicultural systems in Maine. For. Ecol. Manage. 502, 119721. doi: 10.1016/j.foreco.2021.119721
Caselli M., Loguercio G.Á., Urretavizcaya M. F., and Defossé G. E. (2021). Stand level volume increment in relation to leaf area index of Austrocedrus Chilensis and Nothofagus dombeyi mixed forests of Patagonia, Argentina. For. Ecol. Manage. 494, 119337. doi: 10.1016/j.foreco.2021.119337
Castagneri D., Fonti P., von Arx G., and Carrer M. (2017). How does climate influence xylem morphogenesis over the growing season? Insights from long-term intra-ring anatomy in Picea abies. Ann. Bot. 119, 1011–1020. doi: 10.1093/aob/mcw274
Charlet de Sauvage J., Bugmann H., Bigler C., and Lévesque M. (2023). Species diversity and competition have minor effects on the growth response of silver fir, European larch and Douglas fir to drought. Agric. For. Meteorol. 341, 109664. doi: 10.1016/j.agrformet.2023.109664
Cheong B. C. (2024). Transparency and accountability in AI systems: safeguarding wellbeing in the age of algorithmic decision-making. Front. Hum. Dyn. 6. doi: 10.3389/fhumd.2024.1421273
Cook E. R. (1985). A time series analysis approach to tree ring standardization. PhD Thesis, Univ. Arizona, Tucson.
Correa-Díaz A., Silva L., Horwath W., Gómez-Guerrero A., Vargas-Hernández J., Villanueva-Díaz J., et al. (2019). Linking remote sensing and dendrochronology to quantify climate-induced shifts in high-elevation forests over space and time. J. Geophys. Res.: Biogeosci. 124, 166–183. doi: 10.1029/2018JG004687
Cruz-García R., Balzano A., Čufar K., Scharnweber T., Smiljanić M., and Wilmking M. (2019). Combining dendrometer series and xylogenesis imagery—DevX, a simple visualization tool to explore plant secondary growth phenology. Front. Forests Global Change 2. doi: 10.3389/ffgc.2019.00060
Cuny H. E., Fonti P., Rathgeber C. B., von Arx G., Peters R. L., and Frank D. C. (2019). Couplings in cell differentiation kinetics mitigate air temperature influence on conifer wood anatomy. Plant Cell Environ. 42, 1222–1232. doi: 10.1111/pce.13464
Cuny H. E., Rathgeber C. B. K., Frank D., Fonti P., Mäkinen H., Prislan P., et al. (2015). Woody biomass production lags stem-girth increase by over one month in coniferous forests. Nat. Plants 1, 15160. doi: 10.1038/nplants.2015.160
Debel A., Foroozan Z., Häusser M., Raspe S., and Bräuning A. (2024). Assessing intra-annual growth dynamics in climatically contrasting years, sites, and tree species using dendrometers and wood anatomical data. Front. Forests Global Change 7. doi: 10.3389/ffgc.2024.1342413
DeSoto L., Cruz M., and Fonti P. (2011). Intra-annual patterns of tracheid size in the Mediterranean tree Juniperus thurifera as an indicator of seasonal water stress. Can. J. For. Res. 41, 1280–1294. doi: 10.1139/x11-045
Diao H., Wang A., Gharun M., Saurer M., Yuan F., Guan D., et al. (2023). Tree-ring δ13C of Pinus koraiensis is a better tracer of gross primary productivity than tree-ring width index in an old-growth temperate forest. Ecol. Indic. 153, 110418. doi: 10.1016/j.ecolind.2023.110418
Domínguez-Delmás M. (2020). Seeing the forest for the trees: New approaches and challenges for dendroarchaeology in the 21st century. Dendrochronologia 62, 125731. doi: 10.1016/j.dendro.2020.125731
Edge D. C., Reynolds D. J., Wanamaker A. D., Griffin D., Bureau D., Outridge C., et al. (2021). A multicentennial proxy record of northeast Pacific Sea surface temperatures from the annual growth increments of Panopea generosa. Paleoceanogr. Paleoclimatol. 36, e2021PA004291. doi: 10.1029/2021PA004291
Etzold S., Sterck F., Bose A. K., Braun S., Buchmann N., Eugster W., et al. (2022). Number of growth days and not length of the growth period determines radial stem growth of temperate trees. Ecol. Lett. 25, 427–439. doi: 10.1111/ele.13933
Fonti P., von Arx G., García-González I., Eilmann B., Sass-Klaassen U., Gärtner H., et al. (2010). Studying global change through investigation of the plastic responses of xylem anatomy in tree rings. New Phytol. 185, 42–53. doi: 10.1111/j.1469-8137.2009.03030
Fraver S., Bosely-Smith C., Seirup C., Guiterman C. H., Schmeelk T., Teets A., et al. (2024). Dendrochronological reconstruction of arborvitae leafminer (Argyresthia spp.) outbreaks on northern white-cedar (Thuja occidentalis) in Maine, USA. Can. J. For. Res. 54, 479–485. doi: 10.1139/cjfr-2023-0193
Fraver S. and White A. S. (2005). Identifying growth releases in dendrochronological studies of forest disturbance. Can. J. For. Res. 35, 1648–1656. doi: 10.1139/x05-092
Gao S., Liang E., Liu R., Babst F., Camarero J. J., Fu Y. H., et al. (2022). An earlier start of the thermal growing season enhances tree growth in cold humid areas but not in dry areas. Nat. Ecol. Evol. 6, 397–404. doi: 10.1038/s41559-022-01668-4
García-Hidalgo M., García-Pedrero Á.M., Caetano-Sánchez C., Gómez-España M., Lillo-Saavedra M., and Olano J. M. (2021). ρ-MtreeRing: A graphical user interface for X-ray microdensity analysis. Forests 12, 1405. doi: 10.3390/f12101405
García-Hidalgo M., García-Pedrero Á., Rozas V., Sangüesa-Barreda G., García-Cervigón A. I., Resente G., et al. (2024). Tree ring segmentation using UNEt TRansformer neural network on stained microsections for quantitative wood anatomy. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1327163
Giebink C. L., DeRose R. J., Castle M., Shaw J. D., and Evans M. E. K. (2022). Climatic sensitivities derived from tree rings improve predictions of the Forest Vegetation Simulator growth and yield model. For. Ecol. Manage. 517, 120256. doi: 10.1016/j.foreco.2022.120256
González de Andrés E., Gazol A., Camarero J. J., Bonet J. A., Caballol M., Ceausu A., et al. (2024). Forest and soil fungal community dynamics are fuelled by root rot pathogen-induced gaps. J. Ecol. 112, 1952–1966. doi: 10.1111/1365-2745.14362
Greer K., Csank A., Calvert K., Maddison-MacFadyen M., Smith A., Monk K., et al. (2023). Understanding the historic legacies of empire from the timbers left behind: Towards critical dendroprovenancing in the British North Atlantic. Can. Geographies/Géographies canadiennes 67, 124–138. doi: 10.1111/cag.12831
Grover Z. S., Forrester J. A., Keyser T. L., King J. S., and Altman J. (2023). Growth response, climate sensitivity and carbon storage vary with wood porosity in a southern Appalachian mixed hardwood forest. Agric. For. Meteorol. 332, 109358. doi: 10.1016/j.agrformet.2023.109358
Guiterman C. H., Gille E., Shepherd E., McNeill S., Payne C. R., and Morrill C. (2023). The International Tree-Ring Data Bank at Fifty: Status of stewardship for future scientific discovery. Tree-Ring Res. 80, 13–18. doi: 10.3959/2023-2
Guiterman C. H., Lynch A. M., and Axelson J. N. (2020). dfoliatR: An R package for detection and analysis of insect defoliation signals in tree rings. Dendrochronologia 63, 125750. doi: 10.1016/j.dendro.2020.125750
Haneca K. (2024). fellingdater: a toolkit to estimate, report and combine felling dates derived from historical tree-ring series. J. Open Source Softw. 9, 6716. doi: 10.21105/joss.06716
Haneca K., Debonne V., and Hoffsummer P. (2020). The ups and downs of the building trade in a medieval city: Tree-ring data as proxies for economic, social and demographic dynamics in Bruges (c. 1200–1500). Dendrochronologia 64, 125773. doi: 10.1016/j.dendro.2020.125773
Hellmann L., Tegel W., Geyer J., Kirdyanov A. V., Nikolaev A. N., Eggertsson Ó., et al. (2017). Dendro-provenancing of arctic driftwood. Quaternary Sci. Rev. 162, 1–11. doi: 10.1016/j.quascirev.2017.02.025
Hevia A., Campelo F., Chambel R., Vieira J., Alía R., Majada J., et al. (2020). Which matters more for wood traits in Pinus halepensis Mill., provenance or climate? Ann. For. Sci. 77, 55. doi: 10.1007/s13595-020-00956-y
Hey T., Tansley S., and Tolle K. M. (2009). The fourth paradigm: data-intensive scientific discovery (Redmond, WA: Microsoft research). doi: 10.1109/JPROC.2011.2155130
Holmes R. L. (1983). Computer-assisted quality control in tree-ring dating and measurement. Tree-Ring Bull. 43, 69–78.
Holmes R. L. (1994). Dendrochronology program library user’s manual (Tucson: Laboratory of tree-ring research, University of Arizona).
Housset J. M., Nadeau S., Isabel N., Depardieu C., Duchesne I., Lenz P., et al. (2018). Tree rings provide a new class of phenotypes for genetic associations that foster insights into adaptation of conifers to climate change. New Phytol. 218, 630–645. doi: 10.1111/nph.14968
Huang W., Ma K., and Gladish D. K. (2024). Ellipse or superellipse for tree-ring geometries? evidence from six conifer species. Trees 38, 1403–1413. doi: 10.1007/s00468-024-02561-2
Isaac-Renton M., Montwé D., Hamann A., Spiecker H., Cherubini P., and Treydte K. (2018). Northern forest tree populations are physiologically maladapted to drought. Nat. Commun. 9, 5254. doi: 10.1038/s41467-018-07701-0
Janda P., Ukhvatkina O. N., Vozmishcheva A. S., Omelko A. M., Doležal J., Krestov P. V., et al. (2021). Tree canopy accession strategy changes along the latitudinal gradient of temperate Northeast Asia. Global Ecol. Biogeogr. 30, 738–748. doi: 10.1111/geb.13259
Jansma E., Brewer P. W., and Zandhuis I. (2010). TRiDaS 1.1: The tree-ring data standard. Dendrochronologia 28, 99–130. doi: 10.1016/j.dendro.2009.06.009
Jetschke G., van der Maaten E., and van der Maaten-Theunissen M. (2023). Pointer years revisited: Does one method fit all? A clarifying discussion. Dendrochronologia 78, 126064. doi: 10.1016/j.dendro.2023.126064
Jevšenak J. (2020). New features in the dendroTools R package: Bootstrapped and partial correlation coefficients for monthly and daily climate data. Dendrochronologia 63, 125753. doi: 10.1016/j.dendro.2020.125753
Jevšenak J., Džeroski S., Zavadlav S., and Levanič T. (2018a). A machine learning approach to analyzing the relationship between temperatures and multi-proxy tree-ring records. Tree-Ring Res. 74, 210–224. doi: 10.3959/1536-1098-74.2.210
Jevšenak J., Gričar J., Rossi S., and Prislan P. (2022). Modelling seasonal dynamics of secondary growth in R. Ecography 2022, e06030. doi: 10.1111/ecog.06030
Jevšenak J. and Levanič T. (2018). dendroTools: R package for studying linear and nonlinear responses between tree-rings and daily environmental data. Dendrochronologia 48, 32–39. doi: 10.1016/j.dendro.2018.01.005
Jevšenak J., Levanič T., and Džeroski S. (2018b). Comparison of an optimal regression method for climate reconstruction with the compare_methods() function from the dendroTools R package. Dendrochronologia 52, 96–104. doi: 10.1016/j.dendro.2018.10.001
Jiang Y., Wang Z., Zhang Z., Ding X., Jiang S., and Huang J. (2024). Enhancing forest insect outbreak detection by integrating tree-ring and climate variables. J. For. Res. 35, 106. doi: 10.1007/s11676-024-01759-x
Joo R., Boone M. E., Clay T. A., Patrick S. C., Clusella-Trullas S., and Basille M. (2020). Navigating through the r packages for movement. J. Anim. Ecol. 89, 248–267. doi: 10.1111/1365-2656.13116
Kabala J. P., Massari C., Niccoli F., Natali M., Avanzi F., and Battipaglia G. (2025). Reconstruction of the dynamics of sap-flow timeseries of a beech forest using a machine learning approach. Agric. For. Meteorol. 362, 110379. doi: 10.1016/j.agrformet.2024.110379
Kabala J., Niccoli F., and Battipaglia G. (2024). Update to ttprocessing: the R-package to handle the TreeTalker monitoring data. Dendrochronologia 84, 126167. doi: 10.1016/j.dendro.2024.126167
Kašpar J., Šamonil P., Krůček M., and Daněk P. (2020). Changes in the radial growth of trees in relation to biogeomorphic processes in an old-growth forest on flysch, Czechia. Earth Surf. Processes Landforms 45, 2761–2772. doi: 10.1002/esp.4928
Kašpar J., Tumajer J., and Treml V. (2019). IncrementR: Analysing height growth of trees and shrubs in R. Dendrochronologia 53, 48–54. doi: 10.1016/j.dendro.2018.11.001
Kasper J., Leuschner C., Walentowski H., and Weigel R. (2023). Higher growth synchrony and climate change-sensitivity in European beech and silver linden than in temperate oaks. J. Biogeogr. 50, 209–222. doi: 10.1111/jbi.14525
Katzenmaier M., Garnot V. S. F., Björklund J., Schneider L., Wegner J. D., and von Arx G. (2023). Towards ROXAS AI: Deep learning for faster and more accurate conifer cell analysis. Dendrochronologia 81, 126126. doi: 10.1016/j.dendro.2023.126126
Keret R., Schliephack P. M., Stangler D. F., Seifert T., Kahle H.-P., Drew D. M., et al. (2024). An open-source machine-learning approach for obtaining high-quality quantitative wood anatomy data from E. grandis and P. radiata xylem. Plant Sci. 340, 111970. doi: 10.1016/j.plantsci.2023.111970
Knüsel S., Peters R. L., Haeni M., Wilhelm M., and Zweifel R. (2021). Processing and extraction of seasonal tree physiological parameters from stem radius time series. Forests 12, 765. doi: 10.3390/f12060765
Krajnc L., Hafner P., and Gričar J. (2021). The effect of bedrock and species mixture on wood density and radial wood increment in pubescent oak and black pine. For. Ecol. Manage. 481, 118753. doi: 10.1016/j.foreco.2020.118753
Krajnc L., Prislan P., Božič G., Westergren M., Arnič D., Mátyás C., et al. (2022). A comparison of radial increment and wood density from beech provenance trials in Slovenia and Hungary. Eur. J. For. Res. 141, 433–446. doi: 10.1007/s10342-022-01449-5
Lai J., Lortie C. J., Muenchen R. A., Yang J., and Ma K. (2019). Evaluating the popularity of R in ecology. Ecosphere 10, e02567. doi: 10.1002/ecs2.2567
Lara W., Bogino S., and Bravo F. (2018). Multilevel analysis of dendroclimatic series with the R-package BIOdry. PloS One 13, e0196923. doi: 10.1371/journal.pone.0196923
Lara W., Bravo F., and Sierra C. A. (2015). measuRing: An R package to measure tree-ring widths from scanned images. Dendrochronologia 34, 43–50. doi: 10.1016/j.dendro.2015.04.002
Lopez-Saez J., Corona C., von Arx G., Fonti P., Slamova L., and Stoffel M. (2023). Tree-ring anatomy of Pinus cembra trees opens new avenues for climate reconstructions in the European Alps. Sci. Total Environ. 855, 158605. doi: 10.1016/j.scitotenv.2022.158605
Lozhkin G., Dolgova E., and Matskovsky V. (2024). A python package implementing Direct Reconstruction Technique (DIRECT) for dendroclimatological studies. Dendrochronologia 86, 126217. doi: 10.1016/j.dendro.2024.126217
Maes S. L., Vannoppen A., Altman J., Van den Bulcke J., Decocq G., De Mil T., et al. (2017). Evaluating the robustness of three ring-width measurement methods for growth release reconstruction. Dendrochronologia 46, 67–76. doi: 10.1016/j.dendro.2017.10.005
Mahatara D., Campelo F., Houle L., Caron A., Barrette J., Francus P., et al. (2024). CTRing: An R package to extract wood density profiles from computed tomography images of discs and logs. Dendrochronologia 88, 126274. doi: 10.1016/j.dendro.2024.126274
Malevich S. B., Guiterman C. H., and Margolis E. Q. (2018). burnr: Fire history analysis and graphics in R. Dendrochronologia 49, 9–15. doi: 10.1016/j.dendro.2018.02.005
Mann M., Kahle H.-P., Beck M., Bender B. J., Spiecker H., and Backofen R. (2018). MICA: Multiple interval-based curve alignment. SoftwareX 7, 53–58. doi: 10.1016/j.softx.2018.02.003
Martínez-Sancho E., Cernusak L. A., Fonti P., Gregori A., Ullrich B., Pannatier E. G., et al. (2023). Unenriched xylem water contribution during cellulose synthesis influenced by atmospheric demand governs the intra-annual tree-ring δ18O signature. New Phytol. 240, 1743–1757. doi: 10.1111/nph.19278
Martínez-Sancho E., Rellstab C., Guillaume F., Bigler C., Fonti P., Wohlgemuth T., et al. (2021). Post-glacial re-colonization and natural selection have shaped growth responses of silver fir across Europe. Sci. Total Environ. 779, 146393. doi: 10.1016/j.scitotenv.2021.146393
Martínez-Sancho E., Treydte K., Lehmann M. M., Rigling A., and Fonti P. (2022). Drought impacts on tree carbon sequestration and water use–evidence from intra-annual tree-ring characteristics. New Phytol. 236, 58–70. doi: 10.1111/nph.18224
Mašek J., Tumajer J., Lange J., Kaczka R., Fišer P., and Treml V. (2023). Variability in tree-ring width and NDVI responses to climate at a landscape level. Ecosystems 26, 1144–1157. doi: 10.1007/s10021-023-00822-8
Mathias J. M. and Hudiburg T. W. (2022). isocalcR: An R package to streamline and standardize stable isotope calculations in ecological research. Global Change Biol. 28, 7428–7436. doi: 10.1111/gcb.16407
Mathias J. M., Smith K. R., Lantz K. E., Allen K. T., Wright M. J., Sabet A., et al. (2023). Differences in leaf gas exchange strategies explain Quercus rubra and Liriodendron tulipifera intrinsic water use efficiency responses to air pollution and climate change. Global Change Biol. 29, 3449–3462. doi: 10.1111/gcb.16673
Maxwell R. S., Wixom J. A., and Hessl A. E. (2011). A comparison of two techniques for measuring and crossdating tree rings. Dendrochronologia 29, 237–243. doi: 10.1016/j.dendro.2010.12.002
McKay N. P. and Emile-Geay J. (2016). Technical note: The Linked Paleo Data framework – a common tongue for paleoclimatology. Clim. Past 12, 1093–1100. doi: 10.5194/cp-12-1093-2016
McLean N., Kruuk L. E. B., van der Jeugd H. P., Leech D., van Turnhout C. A. M., and van de Pol M. (2022). Warming temperatures drive at least half of the magnitude of long-term trait changes in European birds. Proc. Natl. Acad. Sci. 119, e2105416119. doi: 10.1073/pnas.2105416119
Miller E. W., Rademacher T., Fonti P., Seyednasrollah B., and Richardson A. D. (2022). Assessing intra-annual density fluctuations across and along white pine stems. Botany 100, 583–591. doi: 10.1139/cjb-2021-0218
Mousavisangdehi A., Oladi R., Pourtahmasi K., Etemad V., Koprowski M., and Tumajer J. (2024). Higher temperatures promote intra-annual radial growth of Oriental beech (Fagus orientalis Lipsky) in the humid Hyrcanian forests. Trees 38, 1569–1580. doi: 10.1007/s00468-024-02574-x
Muenchow J., Schäfer S., and Krüger E. (2019). Reviewing qualitative GIS research—Toward a wider usage of open-source GIS and reproducible research practices. Geogr. Compass 13, e12441. doi: 10.1111/gec3.12441
Netsvetov M., Prokopuk Y., Holiaka D., Klisz M., Porté A. J., Puchałka R., et al. (2023). Is there Chornobyl nuclear accident signature in Scots pine radial growth and its climate sensitivity? Sci. Total Environ. 878, 163132. doi: 10.1016/j.scitotenv.2023.163132
Nowacki G. J. and Abrams M. D. (1997). Radial-growth averaging criteria for reconstructing disturbance histories from presettlement-origin oaks. Ecol. Monogr. 67, 225–249. doi: 10.1890/0012-9615(1997)067[0225:rgacfr]2.0.co;2
Ols C., Klesse S., Girardin M. P., Evans M. E. K., DeRose R. J., and Trouet V. (2023). Detrending climate data prior to climate–growth analyses in dendroecology: A common best practice? Dendrochronologia 79, 126094. doi: 10.1016/j.dendro.2023.126094
Pearl J. K., Keck J. R., Tintor W., Siekacz L., Herrick H. M., Meko M. D., et al. (2020). New frontiers in tree-ring research. Holocene 30, 923–941. doi: 10.1177/0959683620902230
Pérez-de-Lis G., Rathgeber C. B., Fernández-de-Uña L., and Ponton S. (2022). Cutting tree rings into time slices: how intra-annual dynamics of wood formation help decipher the space-for-time conversion. New Phytol. 233, 1520–1534. doi: 10.1111/nph.17869
Peters R. L., Balanzategui D., Hurley A. G., von Arx G., Prendin A. L., Cuny H. E., et al. (2018). RAPTOR: Row and position tracheid organizer in R. Dendrochronologia 47, 10–16. doi: 10.1016/j.dendro.2017.10.003
Peters R. L., Steppe K., Cuny H. E., De Pauw D. J., Frank D. C., Schaub M., et al. (2021). Turgor–a limiting factor for radial growth in mature conifers along an elevational gradient. New Phytol. 229, 213–229. doi: 10.1111/nph.16872
Peters R. L., Steppe K., Pappas C., Zweifel R., Babst F., Dietrich L., et al. (2023). Daytime stomatal regulation in mature temperate trees prioritizes stem rehydration at night. New Phytol. 239, 533–546. doi: 10.1111/nph.18964
Plavcová L., Tumajer J., Altman J., Svoboda M., Stegehuis A. I., Pejcha V., et al. (2024). High inter-specific diversity and seasonality of trunk radial growth in trees along an afrotropical elevational gradient. Plant Cell Environ. 48, 2285–2297. doi: 10.1111/pce.15295
Poláček M., Arizpe A., Hüther P., Weidlich L., Steindl S., and Swarts K. (2023). Automation of tree-ring detection and measurements using deep learning. Methods Ecol. Evol. 14, 2233–2242. doi: 10.1111/2041-210X.14183
Popa A., van der Maaten E., Popa I., and van der Maaten-Theunissen M. (2024). Early warning signals indicate climate change-induced stress in Norway spruce in the Eastern Carpathians. Sci. Total Environ. 912, 169167. doi: 10.1016/j.scitotenv.2023.169167
Popkova M. I., Vaganov E. A., Shishov V. V., Babushkina E. A., Rossi S., Fonti M. V., et al. (2018). Modeled tracheidograms disclose drought influence on Pinus sylvestris tree-rings structure from Siberian forest-steppe. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01144
Powers S. M. and Hampton S. E. (2019). Open science, reproducibility, and transparency in ecology. Ecol. Appl. 29, e01822. doi: 10.1002/eap.1822
Pretzsch H., Biber P., Uhl E., Dahlhausen J., Schütze G., Perkins D., et al. (2017). Climate change accelerates growth of urban trees in metropolises worldwide. Sci. Rep. 7, 15403. doi: 10.1038/s41598-017-14831-w
Puchi P. F., Castagneri D., Rossi S., and Carrer M. (2020). Wood anatomical traits in black spruce reveal latent water constraints on the boreal forest. Global Change Biol. 26, 1767–1777. doi: 10.1111/gcb.14906
Raasch C., Lee V., Spaeth S., and Herstatt C. (2013). The rise and fall of interdisciplinary research: The case of open source innovation. Res. Policy 42, 1138–1151. doi: 10.1016/j.respol.2013.01.010
Rademacher T., Fonti P., LeMoine J. M., Fonti M. V., Basler D., Chen Y., et al. (2021a). Manipulating phloem transport affects wood formation but not local nonstructural carbon reserves in an evergreen conifer. Plant Cell Environ. 44, 2506–2521. doi: 10.1111/pce.14117
Rademacher T., Fonti P., LeMoine J. M., Fonti M. V., Bowles F., Chen Y., et al. (2022). Insights into source/sink controls on wood formation and photosynthesis from a stem chilling experiment in mature red maple. New Phytol. 236, 1296–1309. doi: 10.1111/nph.18421
Rademacher T., Seyednasrollah B., Basler D., Cheng J., Mandra T., Miller E., et al. (2021b). The Wood Image Analysis and Dataset (WIAD): Open-access visual analysis tools to advance the ecological data revolution. Methods Ecol. Evol. 12, 2379–2387. doi: 10.1111/2041-210X.13717
Raden M., Mattheis A., Spiecker H., Backofen R., and Kahle H.-P. (2020). The potential of intra-annual density information for crossdating of short tree-ring series. Dendrochronologia 60, 125679. doi: 10.1016/j.dendro.2020.125679
Rai S., Altman J., Kopecký M., Sohar K., Fibich P., Pejcha V., et al. (2023). Contrasting impacts of climate warming on Himalayan Hemlock growth: Seasonal and elevational variations. Dendrochronologia 82, 126144. doi: 10.1016/j.dendro.2023.126144
Rai S., Breme N., Jandova V., Lanta V., Altman J., Ruka A. T., et al. (2024). Growth dynamics and climate sensitivities in alpine cushion plants: insights from Silene acaulis in the Swiss Alps. Alpine Bot. doi: 10.1007/s00035-024-00318-8
Rathgeber C. B. K., Cuny H. E., and Fonti P. (2016). Biological basis of tree-ring formation: A crash course. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00734
Rathgeber C. B. K., Longuetaud F., Mothe F., Cuny H., and Le Moguédec G. (2011). Phenology of wood formation: Data processing, analysis and visualisation using R (package CAVIAR). Dendrochronologia 29, 139–149. doi: 10.1016/j.dendro.2011.01.004
Rathgeber C. B. K., Santenoise P., and Cuny H. E. (2018). CAVIAR: an R package for checking, displaying and processing wood-formation-monitoring data. Tree Physiol. 38, 1246–1260. doi: 10.1093/treephys/tpy054
R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing).
Resente G., Gillert A., Trouillier M., Anadon-Rosell A., Peters R. L., von Arx G., et al. (2021). Mask, train, repeat! Artificial intelligence for quantitative wood anatomy. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.767400
Reynolds D. J., Edge D. C., and Black B. A. (2021). RingdateR: A statistical and graphical tool for crossdating. Dendrochronologia 65, 125797. doi: 10.1016/j.dendro.2020.125797
Rita A., Pericolo O., Saracino A., and Borghetti M. (2020). Sperry’s packing rule affects the spatial proximity but not clustering of xylem conduits: the case of Fagus sylvatica L. IAWA J. 42, 191–203. doi: 10.1163/22941932-bja10036
Rozas V., Olano J. M., Gazol A., Alonso-Ponce R., Cuende-Arribas S., and Rodríguez-Puerta F. (2024). Elevation and local climate variation control changes in Aleppo pine growth responses to hydroclimate and drought in semi-arid Spain. Regional Environ. Change 24, 87. doi: 10.1007/s10113-024-02256-x
Rubio-Cuadrado Á., Camarero J. J., and Bosela M. (2022). Applying climwin to dendrochronology: A breakthrough in the analyses of tree responses to environmental variability. Dendrochronologia 71, 125916. doi: 10.1016/j.dendro.2021.125916
Rubio-Cuadrado Á., Montes F., Alberdi I., Cañellas I., Aulló-Maestro I., Sánchez-Salguero R., et al. (2024). Analyses from stand to tree level allow disentangling the effects of age, size, origin and competition on tree growth sensitivity to climate in natural and afforested Scots pine forests. Agric. For. Meteorol. 355, 110148. doi: 10.1016/j.agrformet.2024.110148
Rydval M., Björklund J., von Arx G., Begović K., Lexa M., Nogueira J., et al. (2024). Ultra-high-resolution reflected-light imaging for dendrochronology. Dendrochronologia 83, 126160. doi: 10.1016/j.dendro.2023.126160
Şahan E. A., Gürçay B., and Tuncay Güner H. (2023). The history of fire, human and climate in black pine forests of western Anatolia: The Taurus mountains. Dendrochronologia 82, 126149. doi: 10.1016/j.dendro.2023.126149
Salomón R. L., Peters R. L., Zweifel R., Sass-Klaassen U. G. W., Stegehuis A. I., Smiljanic M., et al. (2022). The 2018 European heatwave led to stem dehydration but not to consistent growth reductions in forests. Nat. Commun. 13, 28. doi: 10.1038/s41467-021-27579-9
Sanmiguel-Vallelado A., Camarero J. J., Morán-Tejeda E., Gazol A., Colangelo M., Alonso-González E., et al. (2021). Snow dynamics influence tree growth by controlling soil temperature in mountain pine forests. Agric. For. Meteorol. 296, 108205. doi: 10.1016/j.agrformet.2020.108205
Santini L., Craven D., Rodriguez D. R. O., Quintilhan M. T., Gibson-Carpintero S., Torres C. A., et al. (2024). Extreme drought triggers parallel shifts in wood anatomical and physiological traits in upper treeline of the Mediterranean Andes. Ecol. Processes 13, 10. doi: 10.1186/s13717-024-00486-9
Saurer M., Sahlstedt E., Rinne-Garmston K. T., Lehmann M. M., Oettli M., Gessler A., et al. (2023). Progress in high-resolution isotope-ratio analysis of tree rings using laser ablation. Tree Physiol. 43, 694–705. doi: 10.1093/treephys/tpac141
Schweingruber F. H. (2012). Tree Rings: Basics and Applications of Dendrochronology (Dordrecht: Springer).
Shi P., Gielis J., Quinn B. K., Niklas K. J., Ratkowsky D. A., Schrader J., et al. (2022). ‘biogeom’: An R package for simulating and fitting natural shapes. Ann. New York Acad. Sci. 1516, 123–134. doi: 10.1111/nyas.14862
Shi J., Xiang W., Liu Q., and Shah S. (2019). MtreeRing: An R package with graphical user interface for automatic measurement of tree ring widths using image processing techniques. Dendrochronologia 58, 125644. doi: 10.1016/j.dendro.2019.125644
Sousa D., Rodrigues S., Lima H., and Chagas L. T. (2020). R software packages as a tool for evaluating soil physical and hydraulic properties. Comput. Electron. Agric. 168, 105077. doi: 10.1016/j.compag.2019.105077
Splechtna B. E., Gratzer G., and Black B. A. (2005). Disturbance history of a European old-growth mixed-species forest - A spatial dendro-ecological analysis. J. Veget. Sci. 16, 511–522. doi: 10.1111/j.1654-1103.2005.tb02391.x
Stangler D. F., Mann M., Kahle H.-P., Rosskopf E., Fink S., and Spiecker H. (2016). Spatiotemporal alignment of radial tracheid diameter profiles of submontane Norway spruce. Dendrochronologia 37, 33–45. doi: 10.1016/j.dendro.2015.12.001
Steppe K., Sterck F., and Deslauriers A. (2015). Diel growth dynamics in tree stems: linking anatomy and ecophysiology. Trends Plant Sci. 20, 335–343. doi: 10.1016/j.tplants.2015.03.015
Strawhacker C., Snitker G., Peeples M. A., Kinzig A. P., Kintigh K. W., Bocinsky K., et al. (2020). A landscape perspective on climate-driven risks to food security: exploring the relationship between climate and social transformation in the prehispanic U.S. Southwest. Am. Antiquity 85, 427–451. doi: 10.1017/aaq.2020.35
Sviderskaya I. V., Vaganov E. A., Fonti M. V., and Fonti P. (2021). Isometric scaling to model water transport in conifer tree rings across time and environments. J. Exp. Bot. 72, 2672–2685. doi: 10.1093/jxb/eraa595
Thakur D., Altman J., Jandová V., Fibich P., Münzbergová Z., and Doležal J. (2024). Global warming alters Himalayan alpine shrub growth dynamics and climate sensitivity. Sci. Total Environ. 916, 170252. doi: 10.1016/j.scitotenv.2024.170252
Treml V., Tumajer J., Jandová K., Oulehle F., Rydval M., Čada V., et al. (2022). Increasing water-use efficiency mediates effects of atmospheric carbon, sulfur, and nitrogen on growth variability of central European conifers. Sci. Total Environ. 838, 156483. doi: 10.1016/j.scitotenv.2022.156483
Tumajer J., Altman J., and Lehejček J. (2023). Linkage between growth phenology and climate-growth responses along landscape gradients in boreal forests. Sci. Total Environ. 905, 167153. doi: 10.1016/j.scitotenv.2023.167153
Tumajer J., Altman J., Stepanek P., Treml V., Dolezal J., and Cienciala E. (2017). Increasing moisture limitation of Norway spruce in Central Europe revealed by forward modelling of tree growth in tree-ring network. Agric. For. Meteorol. 247, 56–64. doi: 10.1016/j.agrformet.2017.07.015
Vaganov E. A., Hughes M. K., and Shashkin A. V. (2006). Growth dynamics of conifer tree rings: images of past and future environments (Berlin New York: Springer Science & Business Media).
van der Maaten E., van der Maaten-Theunissen M., Smiljanić M., Rossi S., Simard S., Wilmking M., et al. (2016). dendrometeR: Analyzing the pulse of trees in R. Dendrochronologia 40, 12–16. doi: 10.1016/j.dendro.2016.06.001
van der Maaten-Theunissen M., Trouillier M., Schwarz J., Skiadaresis G., Thurm E. A., and van der Maaten E. (2021). pointRes 2.0: New functions to describe tree resilience. Dendrochronologia 70, 125899. doi: 10.1016/j.dendro.2021.125899
van der Maaten-Theunissen M., van der Maaten E., and Bouriaud O. (2015). pointRes: An R package to analyze pointer years and components of resilience. Dendrochronologia 35, 34–38. doi: 10.1016/j.dendro.2015.05.006
Van Rossum G. and Drake F. L. (2009). Python. Python Software Foundation. Available online at: https://www.python.org/.
Vieira J., Carvalho A., and Campelo F. (2020). Tree growth under climate change: evidence from xylogenesis timings and kinetics. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00090
Visser R. M. (2021). Dendrochronological provenance patterns. Network analysis of tree-ring material reveals spatial and economic relations of roman timber in the continental north-western provinces. J. Comput. Appl. Archaeol. doi: 10.5334/jcaa.79
Visser R. (2024). dendroNetwork: a R-package to create dendrochronological networks (0.5.4) (Zenodo). 4, 230–253. doi: 10.5281/zenodo.11073471
Visser R. M. and Vorst Y. (2022). Connecting ships: using dendrochronological network analysis to determine the wood provenance of Roman-period river barges found in the Lower Rhine region and visualise wood use patterns. Int. J. Wood Culture 3, 123–151. doi: 10.1163/27723194-bja10014
von Arx G., Carrer M., Crivellaro A., De Micco V., Fonti P., Lens F., et al. (2021). Q-NET – a new scholarly network on quantitative wood anatomy. Dendrochronologia 70, 125890. doi: 10.1016/j.dendro.2021.125890
Wacker L., Güttler D., Goll J., Hurni J., Synal H.-A., and Walti N. (2014). Radiocarbon dating to a single year by means of rapid atmospheric 14C changes. Radiocarbon 56, 573–579. doi: 10.2458/56.17634
Wang W., Huang J.-G., Zhang T., Qin L., Jiang S., Zhou P., et al. (2023). Precipitation regulates the responses of xylem phenology of two dominant tree species to temperature in arid and semi-arid forest of the southern Altai Mountains. Sci. Total Environ. 886, 163951. doi: 10.1016/j.scitotenv.2023.163951
Westgate M. J. (2019). revtools: An R package to support article screening for evidence synthesis. Res. Synthesis Methods 10, 606–614. doi: 10.1002/jrsm.1374
Wu H., Fang K., and Li X. (2024). Examining the effect of sample size on the estimation of low-frequency signals in tree-ring chronologies. Dendrochronologia 85, 126213. doi: 10.1016/j.dendro.2024.126213
Wu H., Sun S., Forrester D. I., Shi J., Deng W., Deng X., et al. (2023). StemAnalysis: An R-package for reconstructing tree growth and carbon accumulation with stem analysis data. Comput. Electron. Agric. 210, 107924. doi: 10.1016/j.compag.2023.107924
Yin X.-H., Sass-Klaassen U., Hao G.-Y., and Sterck F. (2024). Ring- and diffuse-porous tree species coexisting in cold and humid temperate forest diverge in stem and leaf phenology. Dendrochronologia 86, 126220. doi: 10.1016/j.dendro.2024.126220
Zang C. and Biondi F. (2013). Dendroclimatic calibration in R: The bootRes package for response and correlation function analysis. Dendrochronologia 31, 68–74. doi: 10.1016/j.dendro.2012.08.001
Zang C. and Biondi F. (2015). treeclim: an R package for the numerical calibration of proxy-climate relationships. Ecography 38, 431–436. doi: 10.1111/ecog.01335
Zhang J., Gou X., Wang Y., Sun Q., Liu J., Wang F., et al. (2024). Impacts of site aridity on intra-annual radial variation of two alpine coniferous species in cold and dry ecosystems. Ecol. Indic. 158, 111420. doi: 10.1016/j.ecolind.2023.111420
Zhao S., Pederson N., D’Orangeville L., HilleRisLambers J., Boose E., Penone C., et al. (2019). The International Tree-Ring Data Bank (ITRDB) revisited: Data availability and global ecological representativity. J. Biogeogr. 46, 355–368. doi: 10.1111/jbi.13488
Zhao H., Wu J., Wang A., Guan D., and Liu Y. (2022). Microtopography mediates the climate–growth relationship and growth resilience to drought of Pinus tabulaeformis plantation in the hilly site. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1060011
Zhou B., Sterck F., Kruijt B., Fan Z.-X., and Zuidema P. A. (2023). Diel and seasonal stem growth responses to climatic variation are consistent across species in a subtropical tree community. New Phytol. 240, 2253–2264. doi: 10.1111/nph.19275
Keywords: forestry, software, time series, paleoecology, paleoclimatology, dendrochronology, data measurement, data analysis
Citation: Altman J, Altmanova N, Fibich P, Korznikov K and Fonti P (2025) Advancing dendrochronology with R: an overview of packages and future perspectives. Front. Ecol. Evol. 13:1593675. doi: 10.3389/fevo.2025.1593675
Received: 14 March 2025; Accepted: 30 April 2025;
Published: 21 May 2025.
Edited by:
Martin De Luis, University of Zaragoza, SpainReviewed by:
Marieke van der Maaten-Theunissen, Technical University Dresden, GermanyErnesto Tejedor, Spanish National Research Council (CSIC), Spain
Copyright © 2025 Altman, Altmanova, Fibich, Korznikov and Fonti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Altman, YWx0bWFuLmphbkBnbWFpbC5jb20=
 Jan Altman
Jan Altman Nela Altmanova1,4
Nela Altmanova1,4 Patrick Fonti
Patrick Fonti