- 1Plant Genomics and Bioinformatics Laboratory, National Center for Biotechnology, Astana, Kazakhstan
- 2Laboratory of Flora and Plant Resources, Astana Botanical Garden, Orynbor, Astana, Kazakhstan
- 3Faculty of Biology and Biotechnology, Al-Farabi Kazakh National University, Almaty, Kazakhstan
Medicinal plants are highly vulnerable to overexploitation and environmental pressures, leading to a risk of extinction. Local ecological heterogeneity influences phenotypic variability and adaptive responses to stress. Species survival under specific conditions depends on genome plasticity, which can be altered by the activation of retrotransposons that are sensitive to environmental changes. Genome profiling using the inter-primer binding site (iPBS) polymerase chain reaction (PCR) method enables the assessment of genetic polymorphisms within and between populations over small geographical ranges, thereby providing crucial insights for conservation efforts. Paeonia anomala L. (P. anomala) is a valuable medicinal plant that has experienced a population decline in Kazakhstan due to extensive harvesting for medicinal use. This study investigated and analyzed the morphometric traits, genetic diversity, and environmental conditions of five P. anomala populations in the Kazakh Altai. Phenotypic variability analysis revealed significant interpopulation differences in traits such as plant diameter, plant height, raw biomass, and leaf blade length and width. We found that spatial orientation, slope angle, and human activity had a significant effect on the phenotypic variability of P. anomala plants in the studied populations. Genetic analysis using iPBS genome profiling identified 1,176 PCR fragments, of which 860 were polymorphic, with polymorphism levels ranging from 46% to 64%. The IVA population exhibited the highest genetic variability (He = 0.212; I = 0.315), whereas the ASU population exhibited the lowest genetic diversity (He = 0.163; I = 0.244). Our study provides a better understanding of P. anomala population differentiation under local environmental conditions and supports the development of effective conservation strategies.
Introduction
The study and preservation of plant biodiversity has recently emerged as a global conservation priority owing to the severe threats posed by biodiversity loss to human well-being (Niinemets, 2010; Walters and Pence, 2020). Overexploitation of plant resources has rendered species with medicinal or ornamental properties particularly vulnerable, with some approaching extinction. Among these species is P. anomala L., an anomalous peony also known as Maryin root. The distribution of this species in the Kazakh Altai region is highly restricted and continues to decline due to relentless harvesting during the flowering period. Populations of P. anomala are dwindling, as local communities use them as raw medicinal materials (Baitulin and Kotukhov, 2011).
Peony (P. anomala L.) is a valuable medicinal plant used in both traditional and conventional medicine. It is listed in the Red Book of the Republic of Kazakhstan as a rare or endangered species due to the unsustainable exploitation of natural resources. The roots and rhizomes of P. anomala contain iridoids, tannins, flavonoids, and essential oils (Lu et al., 2019; Jiang et al., 2020). It is also an important source of medicinal preparations in traditional Chinese medicine, particularly for treating disorders of the central nervous system (Qiu et al., 2018; Wang et al., 2021). The plant’s therapeutic effects are associated with monoterpene glycoside paeoniflorin, whose derivatives have been investigated for the treatment of senile dementia (Lu et al., 2019). Paeoniflorin also exhibits antitumor activity against various human cancers. Another notable compound found in P. anomala is gnetin, a derivative of resveratrol. Gnetin reduces the expression of tumor necrosis factor-α and interleukin-4, rendering P. anomala extracts suitable for cancer therapy (Espinoza and Inaoka, 2017; Wang, 2020).
P. anomala is an important resource, and studying its biodiversity is crucial for developing strategies to conserve and manage its bioresource potential and to prevent population extinction. The predominant natural populations of P. anomala in Kazakhstan are primarily located in the forests and meadows of the Kazakh Altai, a region known for its high biological diversity and presence of plant species that are rare or absent elsewhere in the country (Marina et al., 2010; Paltsyn, 2015). The Kazakh Altai region exhibits unique climatic and geomorphological features and considerable ecosystem diversity. Plant life cycles in this area are influenced by extreme daily temperature fluctuations, intense solar radiation, summer snowfall, winter soil exposure, and deep soil freezing—reaching depths of 120–140 cm in certain years. Local environmental conditions (microclimate), including elevation, slope, and human activity, also have a significant effect on plant distribution and survival strategies (Opedal et al., 2014; Süle et al., 2020). Additionally, ecological factors, such as soil pH, humus content, nutrient availability, and heavy metal concentrations, further influence plant development (Aung et al., 2022).
Adapting to local environmental conditions influences the functioning and genetic biodiversity of a population. To survive in changing natural conditions, plants undergo various morphological and physiological adjustments to cope with environmental stress (Engelhardt et al., 2014). This adaptability is particularly important for perennial herbaceous plants whose reproductive capacity can decline under stressful conditions. Plants respond to stress at multiple organizational levels through stress factors that trigger physiological and genomic changes. Stress-induced genomic responses often involve the activation of mobile genetic elements, particularly retrotransposons, which contain cis-regulatory promoter elements resembling transcriptional protective factors (Lanciano and Mirouze, 2018). Retrotransposons comprise a substantial portion of eukaryotic genomes—ranging from 10% to 80% in various plant species—and are actively transcribed, modulating the expression of nearby genes in response to stress (Kumar, 2018; Xia et al., 2020; Lovell et al., 2021).
The displacement of certain retrotransposons near genes sensitive to environmental change has been hypothesized to serve as an evolutionary mechanism for stress adaptation (Baduel and Colot, 2021). Bursts of retrotransposon activity triggered by stress may induce structural genomic changes that facilitate rapid adaptation to external conditions—particularly in wild populations with low genetic diversity (Madlung and Comai, 2004; Casacuberta and Gonzalez, 2013). These bursts result in new insertions at different genomic loci and increase mutation and recombination rates, both of which are crucial for plants, especially perennials, which cannot avoid stress (Madlung and Comai, 2004; Casacuberta and Gonzalez, 2013; Schrader and Schmitz, 2019; Ashapkin et al., 2020)through dispersal.
Continuous selection under specific environmental conditions can lead to genomic structural changes associated with retrotransposon activation, thereby enhancing genetic variability through the formation of adaptive alleles (Belyayev, 2014). Several retrotransposon-based molecular marker systems have been developed to detect genetic polymorphisms based on the heterogeneous distribution of identifiable repeat sequences. These systems vary in the types of primers used for amplification. In species with unknown genomes, identifying polymorphisms requires knowledge of retrotransposon long terminal repeat (LTR) sequences, which is difficult without cloning and sequencing technologies (Abdollahi Mandoulakani et al., 2015; Milovanov et al., 2019; Arvas et al., 2023; Kalendar and Kairov, 2024; Kalendar et al., 2024b).
The inter-primer binding site (iPBS) method developed by Kalendar et al. is highly versatile and applicable to all eukaryotic organisms without requiring genomic sequence data. This is due to the universal and highly conserved PBS domain shared by all LTR retrotransposons. PBS sequences—typically up to 18 nucleotides long—are highly repetitive and characteristic of higher eukaryotes. Therefore, the iPBS method provides an efficient marker system for assessing genetic diversity within and among populations, species, and varieties, as well as for detecting somaclonal variation in eukaryotic organisms (Kalendar et al., 2010; Doungous et al., 2015, 2019; Milovanov et al., 2019).
The biodiversity of the genus Paeonia has been examined using multiple marker systems, including iPBS markers (Yuan et al., 2012; Lim et al., 2013; Duan et al., 2015; Xu et al., 2016; Fan et al., 2019; Wang, 2020; Xue et al., 2021). However, investigations into natural populations of P. anomala remain limited, and their geographic range is often inadequately covered. A few studies have employed microsatellite markers (Ji et al., 2014; Tanhuanpää et al., 2021) and chloroplast and nuclear genome markers. A single study has applied retrotransposon markers to analyze natural populations of P. anomala, and it revealed minimal genetic diversity (Pan et al., 2007; Boronnikova and Kalendar, 2010) (Boronnikova and Kalendar, 2010). Previously, we assessed the diversity of P. anomala populations with a focus on floristic, anatomical–morphological, and ecological–phytogenetic characteristics of their habitats, as well as their genetic relationships (Kubentayev et al., 2023).
In this study, we aimed to address two key objectives: (1) to evaluate the influence of local ecological factors on the variability of P. anomala populations in Kazakhstan and (2) to assess the utility of retrotransposons as markers for detecting genetic diversity associated with environmental variation. Studies on the genetic diversity of natural P. anomala populations in East Kazakhstan are crucial for assessing their current status and for developing informed strategies for the conservation of this valuable medicinal species.
Materials and methods
Research objects
This study examined samples from five Kazakh populations of P. anomala collected from natural habitats in the Altai Mountains of Kazakhstan (Western, Southern, and Kalbin Altai). All populations were located at similar elevations (898–1,195 m above sea level). Field studies were conducted in the foothills of the Sarymsakty Range and Bukhtarma River Valley (Southern Altai), the western foothills of the Kalbin Ridge (Kalbin Altai), and the Seriy Lug region (Ivanovsky Ridge, Western Altai). The population distributions and habitat conditions are listed in Table 1.
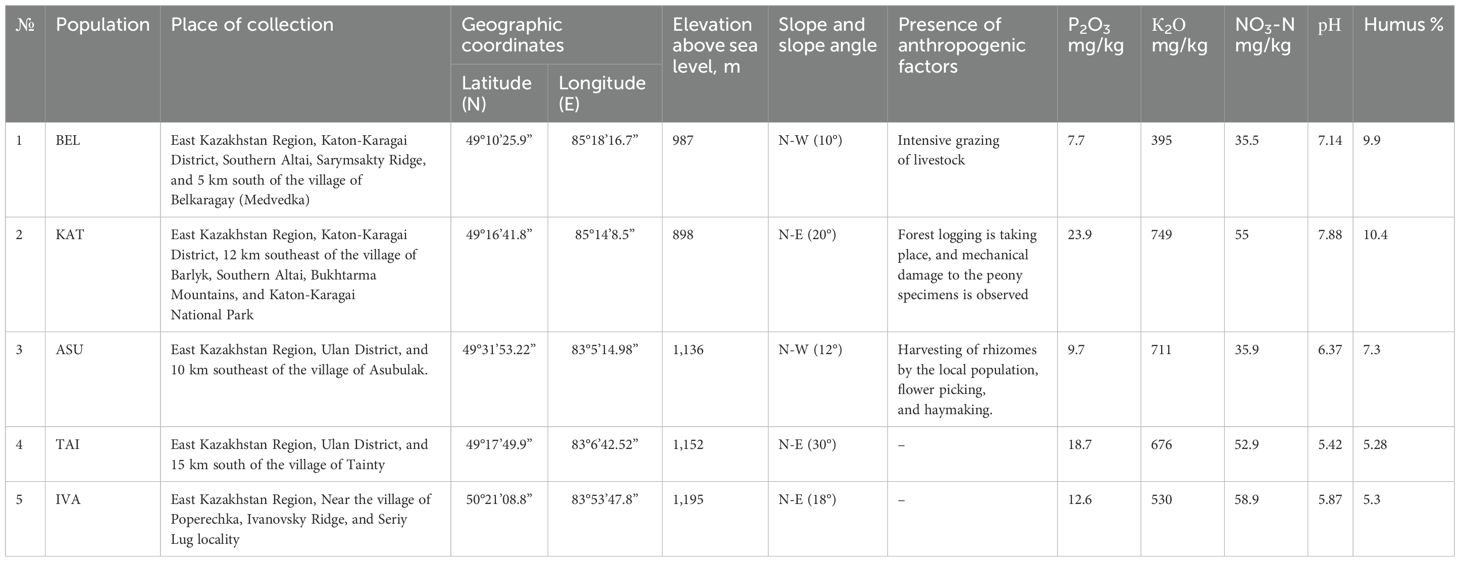
Table 1. Characteristics of the habitat conditions of P. anomala populations in the Kazakhstan Altai region.
Grazing intensity was assessed through comparative analysis with background (control) sites that were not subject to anthropogenic pressure. Key ecological and vegetation parameters, including vegetation height, canopy density, and species composition, were compared. This approach allowed us to estimate the impact of grazing on the studied populations.
Fieldwork was conducted after snowmelt, depending on local weather conditions and temperature. The geographic coordinates, aspect, and elevation above sea level for each population were determined using a global positioning system (GPS) navigator. Terrain slope was assessed according to the scale established by (Leont′ev, 1988).
Experimental design
For each population, fifteen–10 m × 10 m plots were randomly distributed throughout the study area. Soil samples were collected from each plot along two diagonals from the 0–30 cm soil layer and subsequently combined into a composite sample for laboratory analysis.
Soil sample analysis
Soil samples were manually collected from a depth of 0–20 cm using a soil auger. Each sample was sieved through a 2-mm mesh to remove coarse particles and plant residues. The samples were air-dried at room temperature for seven days until a constant weight was reached, then stored in paper bags in a dry room. Humus content, soil pH, and concentrations of key mineral elements (N, P, K) were analyzed at the Analytical Center of the A.I. Barayev Research and Production Center. Nitrate nitrogen (NO3-N) and phosphorus (P) contents were determined using the CINAO method with a Cary 50 spectrophotometer (Varian, Australia). Mobile potassium (K) forms were analyzed using the Machigin method, as modified by CINAO, with a Jenway PFP-7 flame photometer (UK) (GOST, 1985, 1991a). Humus content was determined using the Tyurin method, as modified by CINAO, with a DR-3900 spectrophotometer (Hach-Lange, Germany) (GOST, 1991b).
Morphometric analysis of P. anomala plants
Morphological analysis was conducted on both vegetative and generative parts of the plants during the full flowering stage. A minimum of 10 mature, representative P. anomala plants uniformly distributed across each population were selected for quantitative trait analysis. Morphometric parameters were measured using a standard millimeter ruler with 1-mm accuracy. To ensure analytical precision, each parameter was measured in 30 replicates. The mean value for each parameter was then calculated based on these measurements, and sampling was carried out in the morning hours (from 8:00 to 11:00) under dry weather conditions. The same individuals were also used for DNA extraction. Voucher specimens were deposited in the Astana Botanical Garden Herbarium (NUR). Sample collection was performed from May to July 2022.
DNA extraction and iPBS genotyping
Plant material was placed in polyethylene bags, transported on ice from the field, and preserved at −20°C in the laboratory.
DNA was extracted from individual P. anomala leaves using either a high-salt gel electroelution trap or an acidic CTAB solution (2% CTAB, 2 M NaCl, 10 mM Na3EDTA, and 50-mM HEPES, pH 5.3) (Kalendar et al., 2021, 2023, 2024). DNA integrity was assessed via electrophoresis on a 1% agarose gel in 1 × buffer (20-mM Tris-HEPES, pH 8.06), followed by imaging with the iBright™ CL1500 Imaging System (Invitrogen™, Thermo Fisher Scientific, USA) gel documentation system.
Genetic diversity assessment
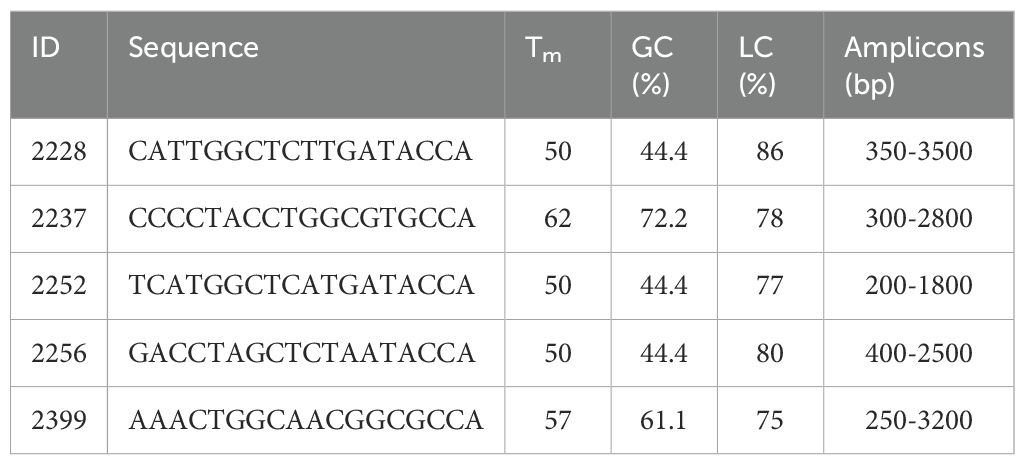
DNA samples from 10 mature plants per population were used for genetic analysis. Universal PBS primers were employed to assess the genetic diversity of the P. anomala populations (Kalendar et al., 2010, 2017). The sequences of primers complementary to different retrotransposon regions are listed in Table 2.
The efficiency of each primer was evaluated based on the uniformity of amplification patterns and the degree of polymorphism detected. A preliminary primer screening was conducted to test amplification quality. Primers producing low numbers of PCR products or predominantly monomorphic bands were excluded from further analysis. The five most effective primers—2228, 2237, 2252, 2256, and 2399—were selected for the final study based on their ability to consistently generate clear, reproducible, and polymorphic banding patterns.
PCR reaction and data analysis
The PCR reaction was conducted in a 20-µL reaction volume, consisting of 10 ng DNA, 1 × Phire Reaction Buffer with MgCl2, 1 µM PBS primer, 0.2 mM dNTP mixture, and 0.5 U Phire Hot Start II DNA Polymerase. The amplification protocol was as follows: preliminary denaturation at 98°C for 2 min; 30 cycles at 98°C for 10 s, 55–60°C for 30 s, and 72°C for 1 min; and a final elongation step at 72°C for 2 min. Amplification was performed using a SimpliAmp thermal cycler (Thermo Fisher Scientific, Inc., MA, USA). PCR products were separated on a 1.5% agarose gel stained with ethidium bromide. Fragment sizes were determined by comparison with a DNA marker (Thermo Fisher Scientific GeneRuler DNA Ladder Mix, 100–10,000 bp). The fragment lengths were analyzed using the iBright™ CL1500 Imaging System (Invitrogen) gel documentation system.
Data analysis
Only distinct and scorable amplicons were included in the genetic variability analysis. The level of polymorphism for each primer was calculated as the ratio of polymorphic amplicons to the total number of amplified fragments, assuming that each unique band size represented a distinct locus. A binary matrix was constructed in which each reproducible fragment was scored as present (1) or absent (0). This matrix was used to compute the total number of alleles, Shannon’s information index (I), genetic differentiation index (PhiPT) among populations, and the number of private alleles per population using GenAlEx 6.5 software (Peakall and Smouse, 2012). Analysis of molecular variance (AMOVA) was performed to assess variation within and among populations. Differences between populations were tested using the Mann–Whitney U test in R version 4.1.2. The significance level was set at p < 0.05.
Results
Ecological characteristics of P. anomala habitats
P. anomala is located in the Kazakh Altai region at elevations ranging from 1,030 m to 1,800 m above sea level. The populations examined in this study were situated in various regions of Western and Southern Altai, as well as in the foothills of the Kalbin Ridge, at elevations between 987 and 1,195 m above sea level (Figure 1).
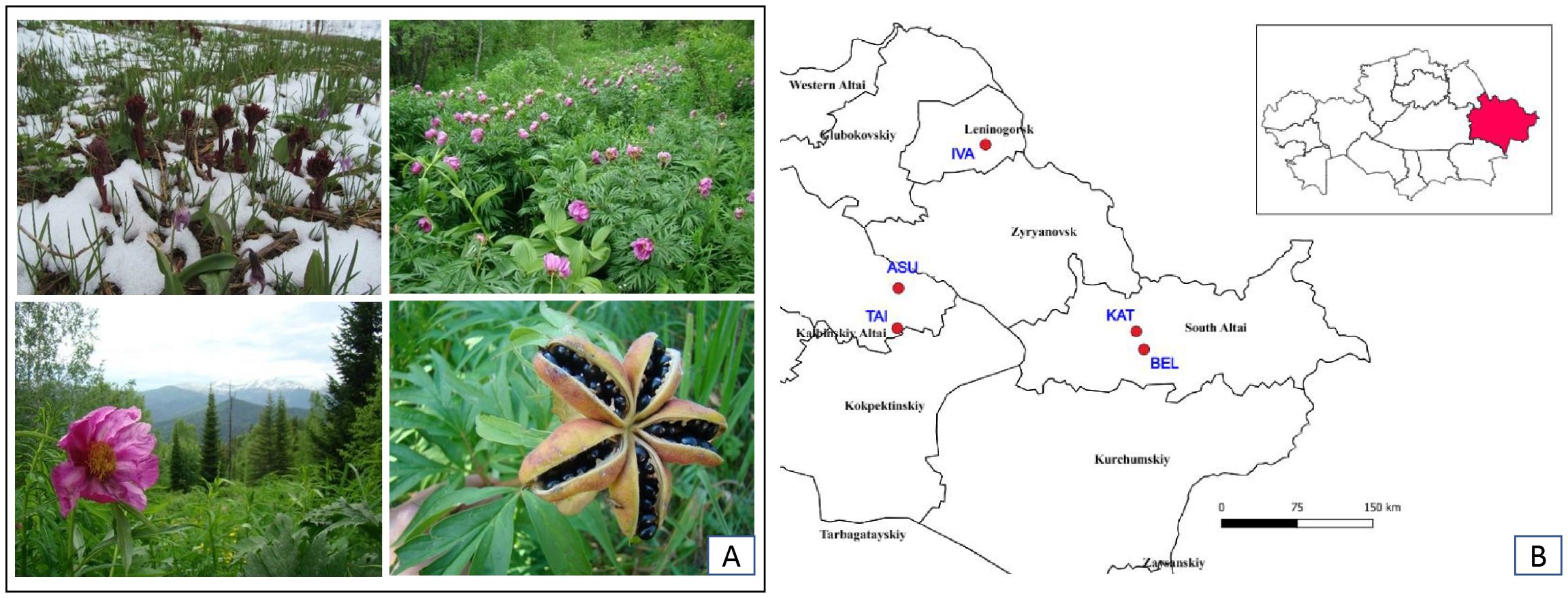
Figure 1. Seasonal development and distribution of P.anomala in the Kazakh Altai. (A) P. anomala plants at the start of the growing, flowering, and fruiting seasons in the Kazakh Altai (photo by N. Premina). (B) Distribution of P. anomala populations in the Kazakh Altai.
This species prefers shaded slopes and intermountain valleys, inhabiting forested and shrub-covered slopes facing the northwest and northeast. Populations are represented by individual, well-developed bushes, or clusters consisting of several individuals, depending on the dominant type of reproduction (sexual or vegetative. Vegetative reproduction occurs via rhizome fragmentation in aging individuals, forming dense clonal nests.
The timing of the growing season depends on meteorological conditions, specifically average daily temperature and snow cover depth. In years with minimal snowfall, characterized by significant soil freezing, the growing season commenced in late April and ends in early May, when the average daily temperature reaches 6°C. In high-snowfall years, vegetative growth begins even earlier—during the first 10 days of April—when snowmelt occurs and average daily temperature reaches 2.9°C (Figure 1).
P. anomala experienced intensive growth in May and ceased completely by the time of flowering in June. Each flower blooms for 5–7 days; however, late frosts and rainy weather may hinder flowering and fruit set, thereby extending the ripening period. Seeds mature in the first half of August and fall near the parent plant. Vegetation ceases in September when severe frosts kill the aboveground parts of the plant.
Field expeditions were conducted during the peak flowering period in the last 10 days of June. Although all P. anomala populations were located at similar elevations (Figure 1), other environmental variables contributed to differences in habitat conditions (Table 1).
Soil nutrient content in the 0–30 cm layer varied among the populations. Available potassium content ranged from 395 mg/100 g in the BEL population to 749 mg/100 g in the KAT population. Soil pH ranged from weakly acidic (pH 5.42–6.37) in the ASU, TAI, and IVA populations to weakly alkaline (pH 7.14–7.88) in the BEL and KAT populations. NO3-N content ranged from 35.5 mg/kg (BEL) to 58.9 mg/kg (IVA), while humus content ranged from 5.3% (IVA) to 10.4% (KAT).
Considering the locations of the populations relative to cardinal direction and slope steepness, the BEL and ASU populations were located on gentle northwestern slopes, whereas the other populations were situated on steep northeastern slopes. The analysis of various environmental factors revealed that anthropogenic factors (intense livestock grazing) significantly affected the BEL population.
These plants grow in soils with low mineral content and minimal levels of available forms of NPK. Conversely, plants in the KAT population grew in soils with high fertility levels and faced similar anthropogenic pressures. Other populations exhibited various combinations of environmental factors that determine their adaptability and survival in specific habitats.
Variability of morphometric traits in P. anomala plants in Kazakhstan populations
The variability of morphometric traits in plants from five P. anomala populations situated in distinct geomorphological regions of Kazakhstan, Altai was heterogeneous due to differences in habitat conditions. Figure 2 presents a comparative analysis of the morphometric characteristics of P. anomala plants from the BEL, KAT, ASU, TAI, and IVA populations. Each population was characterized by the number of generative and vegetative shoots, density of adult individuals per unit area, wet biomass of one plant, plant and stem diameter, leaf blade length and width, petiole length, and plant height. Each column represents the mean value along with the standard error and standard deviation for a specific trait in a given population.
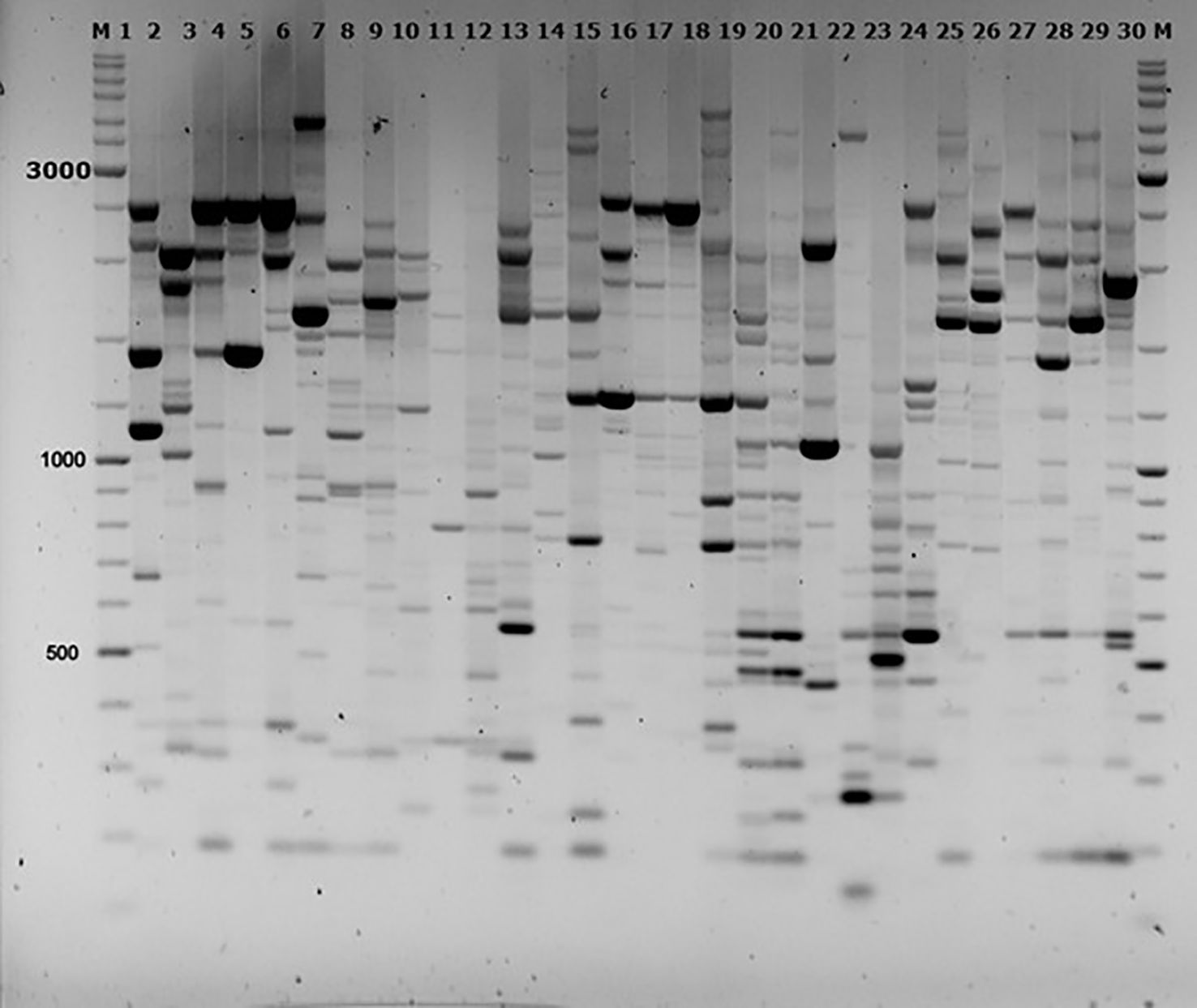
Figure 2. Morphometric characteristics of P. anomala plants from different populations in the Kazakh Altai.
The results showed that the TAI population was the most productive in terms of the number of adult and generative shoots. However, the highest average values for most morphological traits were observed in the IVA population. Maximum average values were recorded for traits such as plant diameter, plant height, wet biomass, and leaf blade length and width. Conversely, the lowest values for most of the studied traits were observed in the BEL population.
Plants from the KAT population exhibited the fewest generative shoots per adult bush, which affected the overall plant mass. In general, taller and more sprawling bushes were characteristic of the adults in the TAI and IVA populations.
No statistically significant differences were observed between populations for several traits, such as the number of leaves on one generative shoot, stem diameter, or number of fruit sections. Statistically significant differences (p < 0.05) were found between populations in traits related to leaf size. Other traits also showed statistically significant interpopulation differences with a high level of confidence (p < 0.05).
We analyzed the relationships between phenotypic diversity and external environmental factors. The P. anomala populations were clustered into groups based on habitat conditions, soil nutrient content, and presence of anthropogenic factors (Figure 2; Table 3). This classification enabled us to identify relationships between environmental conditions and the morphological traits of P. anomala.
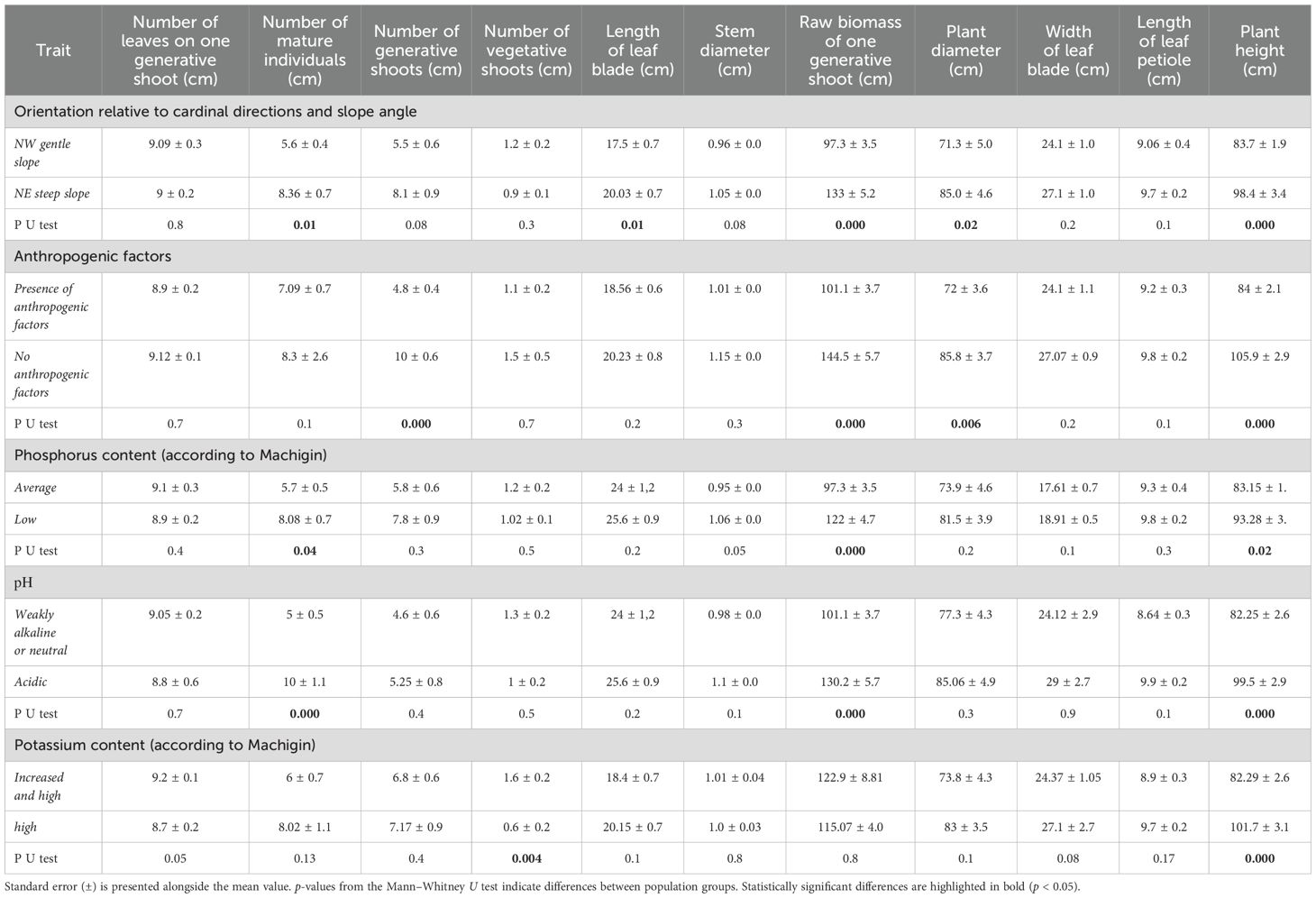
Table 3. Comparison of local environmental factors on the morphological variability of 10 mature P. anomala.
We evaluated the influence of various factors—including exposure, slope steepness, presence or absence of anthropogenic impacts, mobile forms of phosphorus and potassium, and soil pH—on the primary phenotypic characteristics of adult P. anomala plants. According to the classifications of Kravkov (for nitrogen content) and Tyurin (for humus content), all soil samples were characterized by consistently high levels of these elements. As a result, humus and nitrogen were excluded from the analysis as they showed insufficient variability to influence the morphological variation in P. anomala.
The biometric indicators of P. anomala varied significantly among the studied regions, as assessed using the Mann–Whitney U test (p < 0.05) (Table 3).
This analysis revealed varying degrees of influence on phenotypic variability. The traits most strongly affected by environmental factors were the biomass of generative shoots and plant height, both of which exhibited statistically significant variation across nearly all environmental parameters. The only exception was exchangeable potassium, which had a minimal effect on generative shoot biomass but significantly affected the number of vegetative shoots per plant.
For example, plants in the KAT and TAI populations—growing in soils with low phosphorus content (≤15 mg/kg)—exhibited significantly higher petiole length, higher numbers of leaves on generative shoots, and corresponding raw biomass of generative shoots compared to plants from the BEL, ASU, and IVA populations, which grew in soils with moderate phosphorus content. All differences were statistically significant (p < 0.05).
Significant factors influencing trait variability in P. anomala included slope orientation and steepness. These factors resulted in statistically significant differences in most examined traits, with plants on steep northeastern slopes exhibiting superior biometric characteristics compared to those on gentle northwestern slopes.
Anthropogenic factors reduced the overall plant habitat (height and bush diameter) and raw biomass of the wild P. anomala populations. Plants in the TAI and IVA populations, which were not subject to anthropogenic pressure, exhibited significantly larger size and a higher number of adult individuals compared to those in the BEL, ASU, and KAT populations, where anthropogenic effects were evident. Specifically, average plant height was 84 ± 2.1 cm versus 105.9 ± 2.9 cm, plant diameter was 72 ± 3.6 cm versus 85.8 ± 3.7 cm, and the raw biomass of a single generative shoot was 101.1 ± 3.7 g versus 144.5 ± 5.7 g for populations with anthropogenic pressure versus those without, respectively.
Soil pH also significantly affected the phenotypic characteristics. A decrease in pH correlated with reduced plant height, leaf count on generative shoots, and raw weight. These values were significantly higher in plants from the BEL and KAT populations. Plant height at pH ≥ 7 was 99.5 ± 2.9 versus 82.25 ± 2.6 and 130.2 ± 5.7 versus 101.1 ± 3.7 for acidic versus weakly alkaline or neutral soils, respectively.
iPBS analysis of genetic polymorphism in P. anomala
The inter-primer binding site (iPBS) genome profiling PCR method was used to identify genetic polymorphisms and examine the population structure of P. anomala based on the amplification of highly repetitive PBS sequences. Single primers complementary to PBS regions of retrotransposons were used to assess genetic polymorphism in natural populations of P. anomala (Table 2).
Electrophoretic separation of the amplification products revealed bands ranging from 300 to 6,000 base pairs. The number of amplified fragments and their distribution patterns varied depending on the specific population and the primers used. A total of 1,176 fragments were obtained, of which 860 were polymorphic (Figures 3–5).
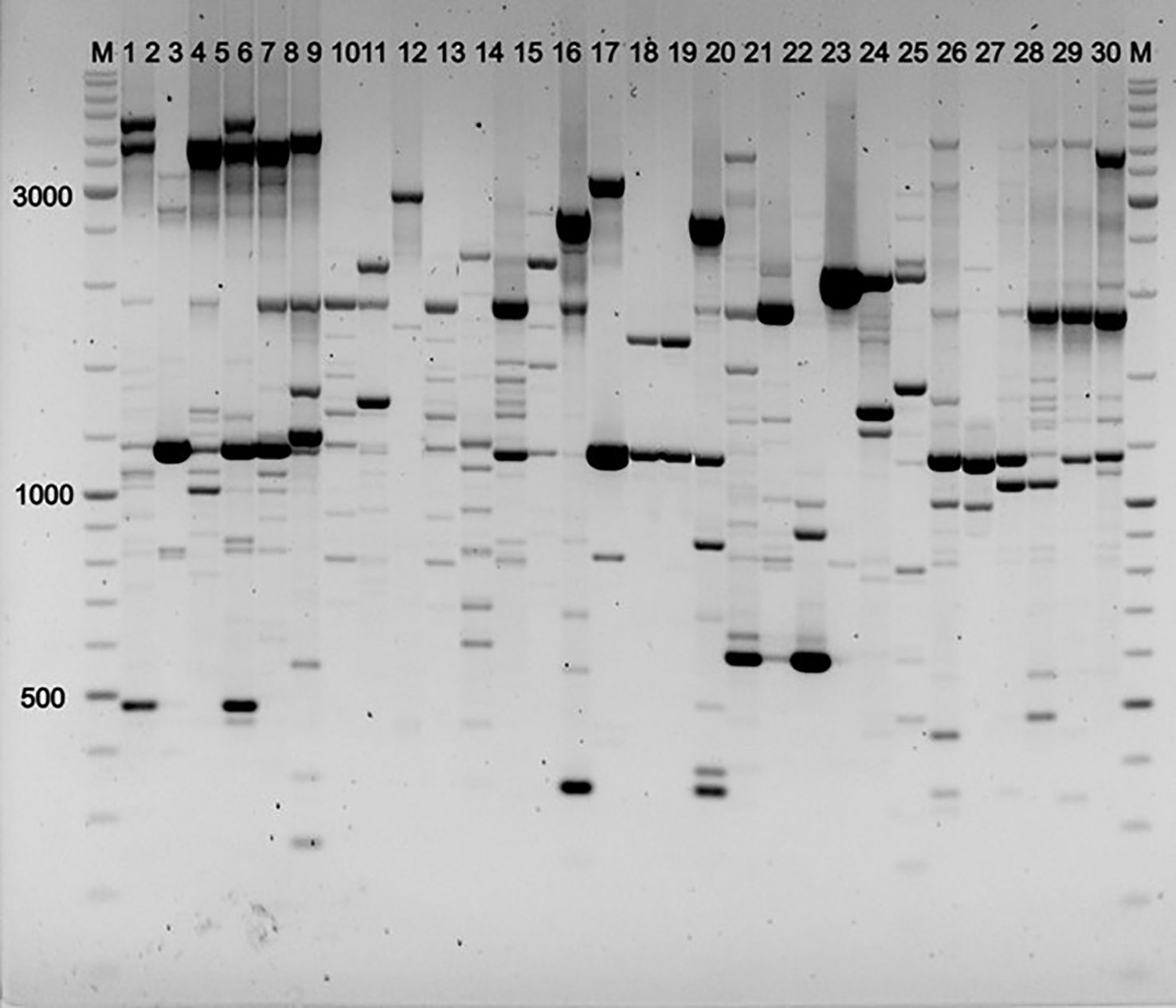
Figure 3. Electrophoretic analysis of iPBS profiling of individual DNA samples from P. anomala populations using primer 2228. Samples from populations: P1 (1–6), P2 (7–12), P3 (13–18), P4 (19–24), P5 (25–30); M = Thermo Scientific GeneRuler DNA Ladder Mix (100–10,000 bp).
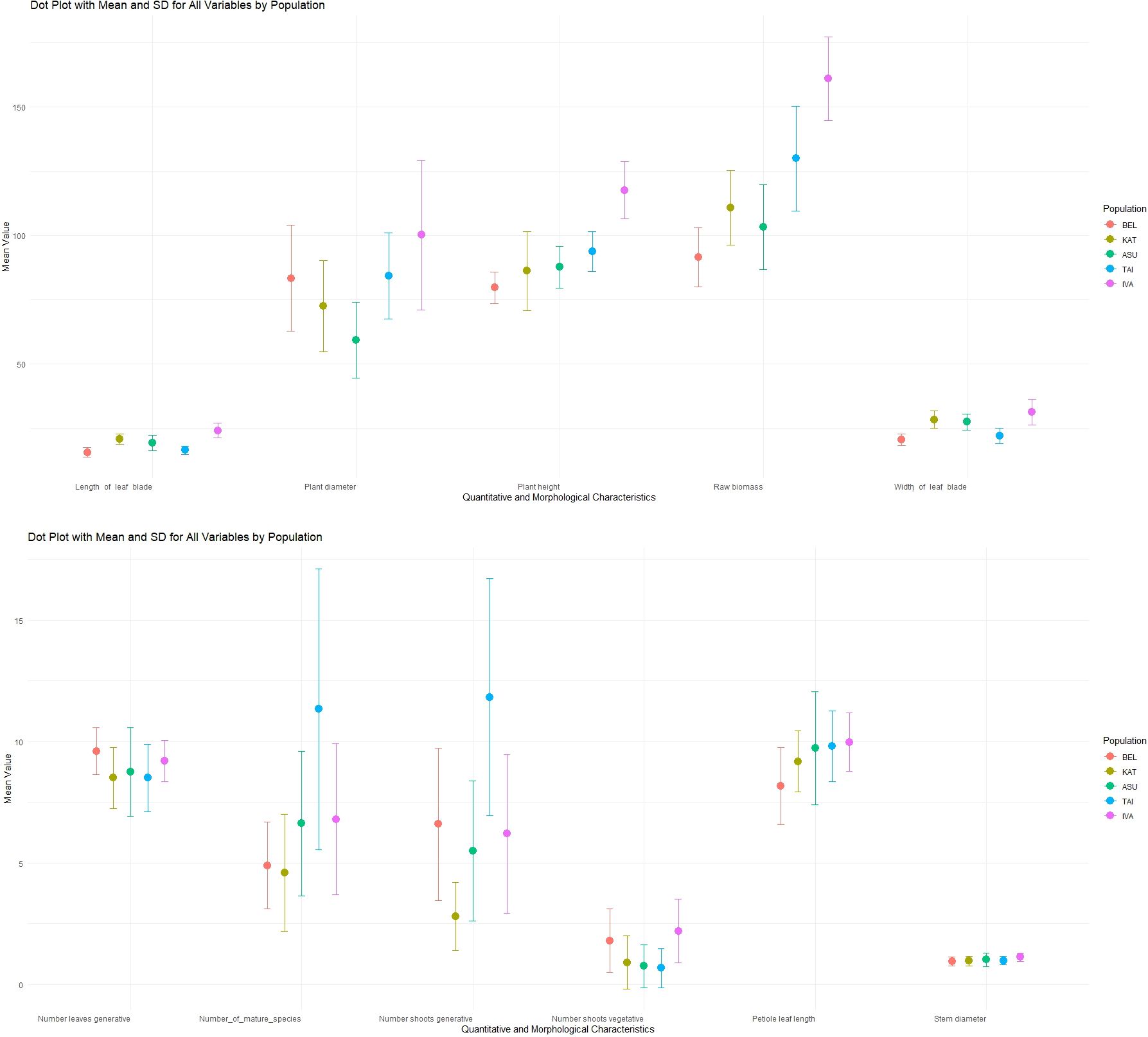
Figure 4. Electrophoretic analysis of iPBS profiling of individual DNA samples from P. anomala populations using primer 2256. Samples from populations: P1 (1–6), P2 (7–12), P3 (13–18), P4 (19–24), P5 (25–30); M = Thermo Scientific GeneRuler DNA Ladder Mix (100–10,000 bp).
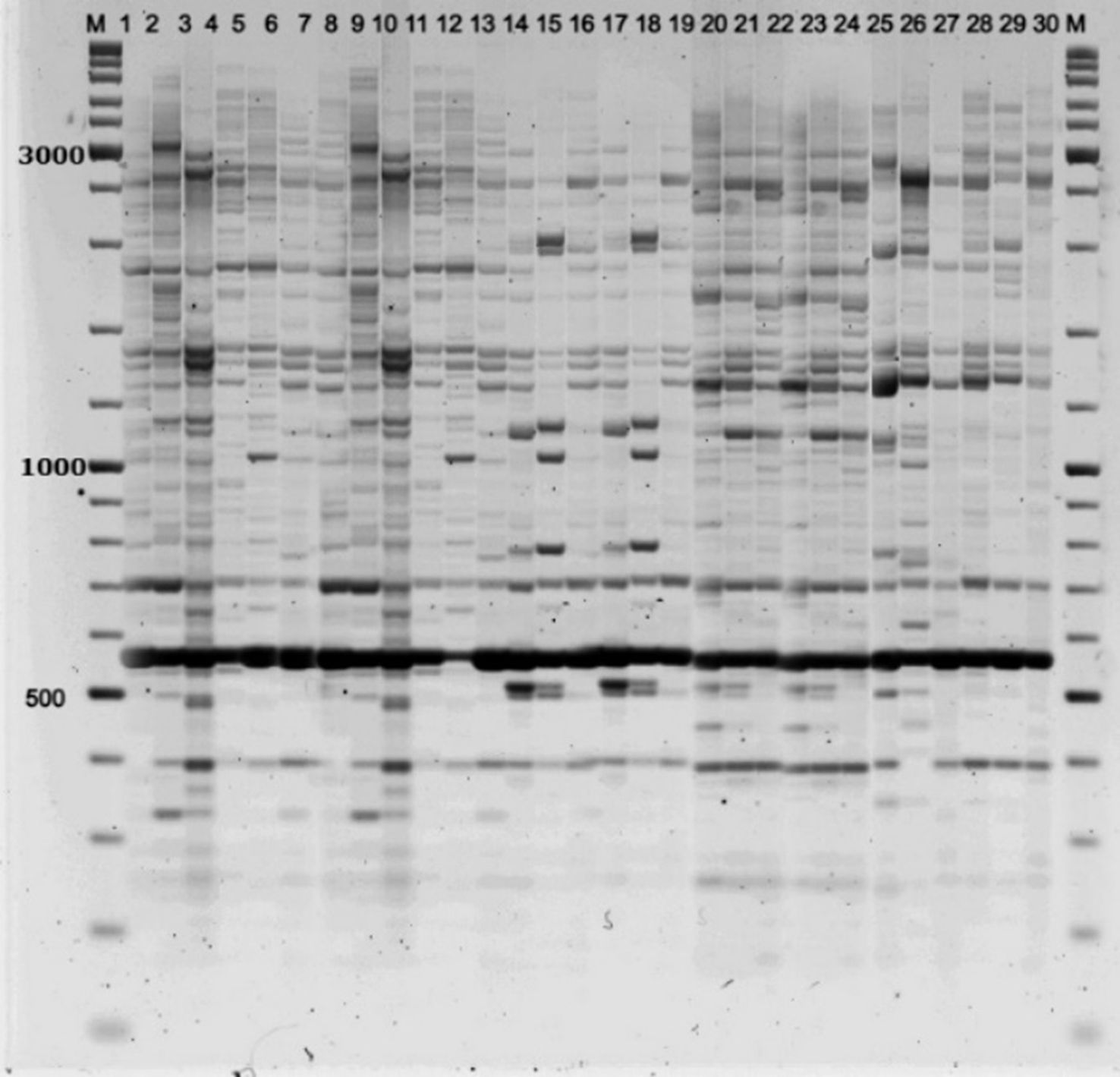
Figure 5. Electrophoretic analysis of iPBS profiling of individual DNA samples from P. anomala populations using primer 2339 Samples from populations: P1 (1–6), P2 (7–12), P3 (13–18), P4 (19–24), P5 (25–30); M = Thermo Scientific GeneRuler DNA Ladder Mix (100–10,000 bp).
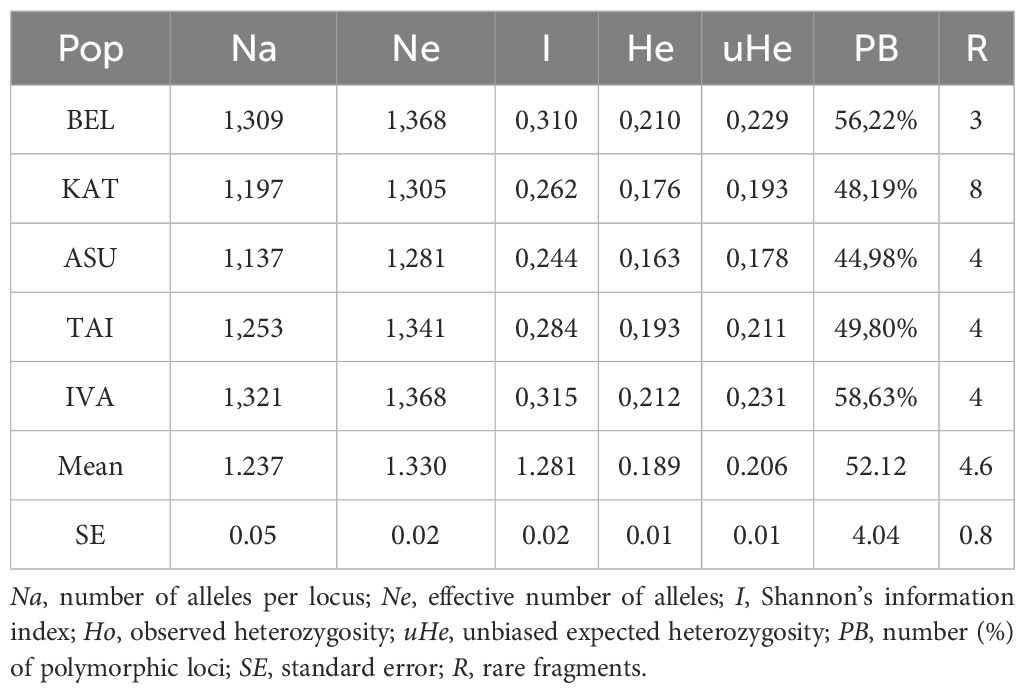
The levels of detectable polymorphism and saturation of the amplification profiles ranged from 46% (primer 2230) to 64% (primer 2399). Genetic diversity parameters—including the number of effective alleles, Shannon’s information index (I), and expected heterozygosity (He)—were calculated for each population (Table 4). These metrics provide a quantitative assessment of genetic variability within populations.
In assessing indicators of intrapopulation diversity in P. anomala, the IVA population exhibited the highest polymorphism level (58%), along with high values for the effective number of alleles (Ne = 1.368), Shannon’s information index (I = 0.315), and expected heterozygosity (He = 0.231) (Table 4). High genetic variability was also observed in the BEL population, which had the highest average number of alleles and the lowest proportion of rare alleles. Rare alleles were defined as DNA fragments occurring at a frequency of less than 5%. A total of 23 rare alleles were identified in the P. anomala samples. The maximum number of rare alleles (eight) was recorded in the KAT population, whereas the other populations exhibited a range of 3-4.
The ASU population showed the lowest levels of genetic variability, with values for Ne (1.281), I (0.244), He (0.163), and uHe (0.178). Low diversity values may reflect historical genetic bottlenecks or ongoing genetic drift in these populations.
The KAT and TAI populations displayed intermediate levels of genetic diversity, with the KAT population exhibiting the highest number of rare and unique alleles (R).
Analysis of genetic diversity in P. anomala populations revealed that the main diversity parameters remained largely consistent regardless of the presence or absence of environmental factors. However, the ratio of within-to between-population variability was altered. The smallest differences across P. anomala populations were observed in relation to the soil availability of mobile phosphorus and potassium, which accounted for 6% and 8% of the variation, respectively (Table 4).
Genetic polymorphism in the Kazakh P. anomala populations, as revealed by the iPBS profiling method, was predominantly shaped by within-population variability, which accounted for 92–94% of the total genetic variation. Environmental parameters such as mobile phosphorus content, soil pH, slope angle, and spatial orientation contributed to increased interpopulation differences, reaching 21–23%, with a ∮∮PT value <0.001 (Table 5).
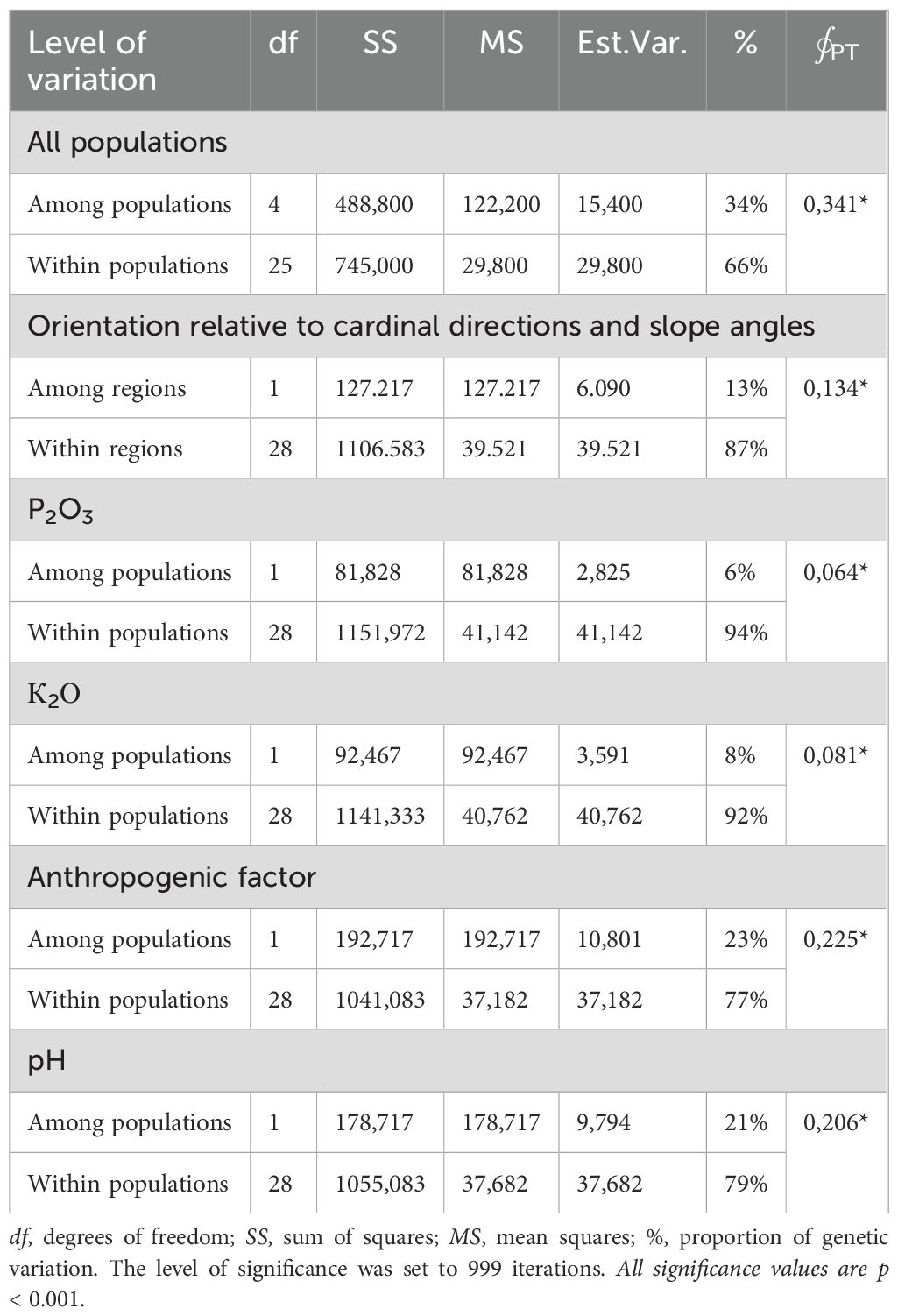
Table 5. Molecular variance within and among populations from the two study regions as calculated by analyses of molecular variance (AMOVA).
df = degrees of freedom; SS = sum of squares; MS = mean squares; % = proportion of genetic variation. Significance was assessed using 999 permutations. All values are significant at p < 0.001.
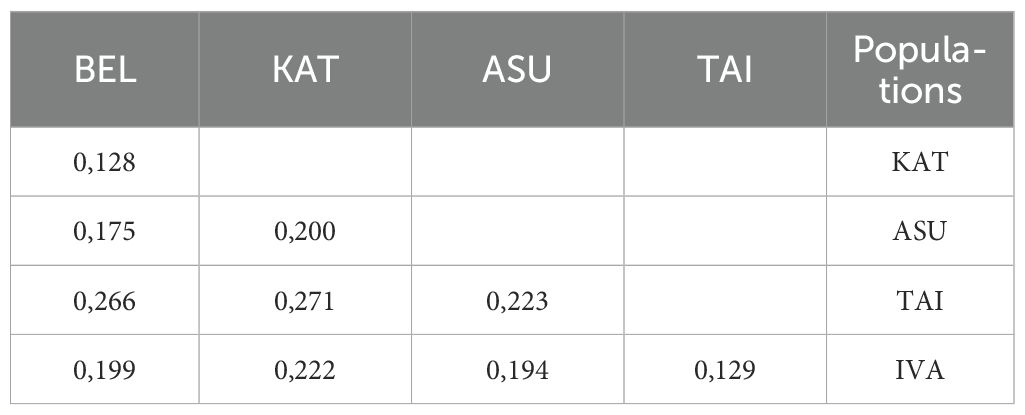
The analysis showed that interpopulation differences contributed minimally to the overall genetic heterogeneity of the species. Population differentiation was largely driven by the individual characteristics of plants growing under specific environmental conditions. The overall ∮PT value across all populations was 0.341, indicating a moderate level of genetic differentiation among populations (Rudmann-Maurer et al., 2007). To assess the genetic similarity among P. anomala populations, we calculated pairwise Nei’s genetic identity values, which are summarized in Table 6.
The Nei genetic distance between P. anomala populations ranged from 0.128 (between BEL and KAT populations) to 0.271 (between BEL and TAI populations) (Table 6). The most extreme environmental conditions were characteristic of the P3 and P4 populations in the Kalbinsky Altai region, which experiences the lowest annual precipitation and the highest radiation balance.
Discussion
Human activities have led to rapid changes in habitat ecology by reshaping the dynamic equilibrium between abiotic and biogenic processes. The overexploitation of natural resources has disrupted the natural cycles within the biosphere (Antonelli et al., 2020; Gallego‐Zamorano et al., 2022). It is currently estimated that two out of every five plant species are at risk of extinction. According to the IUCN Red List of Threatened Species, the primary threats to in situ plant populations are anthropogenic activities, including agriculture, overexploitation, and urbanization. These factors facilitate the transformation of natural ecosystems and contribute to biodiversity decline (Ebert and Engels, 2020). Conserving the biodiversity of valuable resource species is a pressing issue in many countries, including Kazakhstan. Effective conservation strategies for flora of medicinal or ecological importance depend on long-term monitoring studies that gather essential data on the status of wild populations. Population-based studies provide a reliable foundation for assessing the status of endangered plants, identifying the nature and extent of changes under environmental factors, and formulating appropriate conservation strategies to prevent extinction at the local, regional, or global level.
Examining plant population variability with respect to their adaptation to diverse local conditions is a pertinent research direction. Plant genomes possess the unique ability to adapt to various environmental conditions. Over time, plants have evolved metabolic and physiological mechanisms essential for survival under (Reyer et al., 2013; Negi et al., 2016)stress. Ecologically induced variability in morphological traits reflects functional adaptation (Zunzunegui et al., 2009). Individuals of the same species from different populations inhabiting distinct ecological conditions may differ significantly in habitat and other morphological criteria (e.g., leaf size, leaf type, or shoot length—characteristics that reflect various adaptive strategies (Chelli et al., 2019).
The diversity of forms capable of surviving stressful conditions may be partially attributed to polymorphisms in allelic variations in retrotransposons, which regulate gene expression for adaptive responses to changing external conditions. Retrotransposons do not eliminate stress but may enhance adaptation and phenotypic variability by modulating the expression of nearby genes. These elements can form novel cis-regulatory sequences that are evolutionarily significant and enable rapid adaptation to climate change (Belyayev et al., 2010). This supports the view that retrotransposons act as biodiversity inducers, increasing genetic variability in plant populations and potentially providing advantageous traits for stress tolerance (Baduel and Colot, 2021). The activation of LTR retrotransposons—particularly those belonging to the Ty1/Copia and Ty3/Gypsy families—is crucial for species diversity (Papolu et al., 2022). These factors collectively enhance genomic plasticity as part of an adaptive strategy that fosters biodiversity capable of enduring specific environmental constraints (Pimpinelli et al., 2019).
This study examined morphometric variability and genetic structure in five Kazakh populations of the medicinal plant P. anomala L. Although these populations occurred at similar elevations, they inhabited distinct ecological and geographical settings, with variation in precipitation and temperature regimes. Morphological differences in P. anomala populations exhibited considerable variability in bush height and diameter and in the quantitative parameters of vegetative and generative shoots. Moderate variability was observed in certain traits such as leaf count per plant, stem diameter, and fruit count. Similar results were obtained for stem variability in P. peregrina Mill. var. romanica, and leaf variability in peonies (P. delavayi, P. potanini, P. ludlowii, P. lutea, and P. qiui) have been noted in relation to microrelief and plant age (Olteanu et al., 2022).
Key ecological factors considered in this study included anthropogenic impacts, slope orientation, and soil fertility. Anthropogenic stressors—such as grazing, logging, and uncontrolled rhizome harvesting —were evident in three of the five populations and had a substantial effect on morphological traits. The morphometric indicators of P. anomala were significantly lower (p < 0.05) in populations under anthropogenic pressure (Table 2). The most significant interpopulation differences were observed in the number and weight of generative shoots and in plant height. Numerous studies have reported that anthropogenic disturbance reduces genetic diversity in Paeonia species, underscoring the need for state-sponsored biodiversity conservation initiatives (Andrieu et al., 2007; Yang et al., 2022; Chirilă and Vassilev, 2024).
Noteworthy results were observed in relation to slope inclination and orientation. Populations on steep, north-facing slopes (KAT, TAI, and IVA) showed greater plant height and lateral spread compared to those on gentle, northwest-facing slopes. This may be related to variations in heat, light, and precipitation based on the slope exposure. Differences in diurnal soil temperatures between slopes can reach 18°C, with east-facing slopes experiencing significantly higher temperatures. On the eastern slope oriented toward the sun (with a gradient of 30°), the soil temperature may exceed that on the less sunlit western slope by 27°. The west-facing slopes received sunlight after 12 PM, with solar radiation declining after 4 PM (Wang et al., 2024). The lower morphological values observed on northwest-facing slopes were not solely due to reduced sunlight exposure, as several wild peony species (P. veitchii, P. intermedia, P. anomala) are well adapted to low light conditions, often displaying larger leaves and prolonged flowering periods. However, these adaptations did not offset reductions in stem mass and overall plant morphology. Soil pH, P, and K analyses have indicated optimal plant development under conditions of reduced P and excess K; however, other studies have reported contradictory findings (Zhao et al., 2013; Wang et al., 2019; Aung et al., 2022). Anthropogenic stress (e.g., grazing, logging, and hay mowing) is the primary stress factor in the Kazakh population.
Conservation strategies for P. anomala should prioritize the maintenance of high-quality soils to support genetic diversity and morphological variability. Future conservation efforts should explore the role of soil amendments, such as organic enrichment, in sustaining populations under less favorable conditions. To conserve this species, we propose the development of a plantation cultivation system to enable its sustainable use in the pharmaceutical industry. Additionally, it is recommended that the species be preserved in living collections of botanical gardens and in vitro collections, as well as in seed banks, to ensure the long-term conservation of their genetic material and support the potential restoration of populations in natural habitats.
Our findings demonstrated that morphological and genetic variability among P. anomala populations is closely linked to ecological and anthropogenic factors. For instance, populations exposed to anthropogenic pressure (BEL, KAT, and ASU) showed significantly lower values for morphological traits such as raw biomass, plant height, and the number of generative shoots compared to populations with minimal human impact (TAI and IVA). Genetically, the BEL and IVA populations demonstrated the highest levels of polymorphism and may possess greater adaptive potential and resilience to environmental changes, making them valuable genetic reservoirs for long-term survival of the species. However, the BEL population experienced significant livestock grazing, unlike the IVA population.
We hypothesized that the high genetic variability in the BEL population, despite grazing pressure, may be associated with adaptive responses to intensive grazing. Grazing may increase light availability by reducing tall vegetation, thereby benefiting P. anomala. In addition, grazing can reduce the thickness of the litter layer, which otherwise inhibits seed germination. This might enhance sexual reproduction and contribute to the maintenance of a broader genetic pool. Similar increases in genetic diversity under grazing pressure have been reported in other studies (Rudmann-Maurer et al., 2007).
Conversely, populations with low genetic diversity, such as ASU, may be at a greater risk of inbreeding depression, reduced fitness, and local extinction (Weeks et al., 2011). Therefore, it is not possible to identify any specific population of P. anomala as a priority for conservation, as the species as a whole is a relatively rare medicinal plant and requires measures for the preservation or restoration of its gene pool.
Genetic polymorphism analysis using iPBS markers and genetic distances between populations indicated the existence of a large P. anomala population in the Kazakh Altai region, which enhances our understanding of the ecological processes that affect the dynamic structure of P. anomala populations and will assist in the formulation of effective conservation strategies. The study of polymorphisms using PBS markers is an alternative method, alongside the use of SSR, ISSR, and SNP markers. iPBS markers are based on conserved PBS (primer binding site) regions of retrotransposons. Their main advantages lie in their universality, cost-effectiveness, and the fact that they do not require prior sequence information from eukaryotic genomes (Kalendar et al., 2010).
In contrast to this method, the use of SSR and SNP markers requires significant investment because of the need for whole-genome sequencing and bioinformatic analysis specific to plant species, as well as the subsequent selection of informative markers capable of detecting a sufficiently large number of loci. However, a limiting factor of PBS profiling is the requirement for high-quality extracted DNA, which is essential for the effective assessment of amplicon polymorphisms using the fingerprinting method (Hosid et al., 2012).
Conclusion
This study highlights the significant influence of ecological and anthropogenic factors on the morphometric variability and genetic diversity of P. anomala populations in the Kazakh Altai. Variability in plant traits was closely associated with environmental conditions such as slope orientation, soil composition, and human disturbance, with pronounced reductions in plant size and reproductive output observed in impacted populations. Taken together, our data suggest that retrotransposons may indirectly influence plant adaptation to environmental stress.
We believe that this type of marker can aid in detecting genetic changes in the genome arising from various stress factors in wild P. anomala populations inhabiting the same region but exposed to different environmental conditions. These findings underscore the importance of preserving diverse habitats and minimizing human interference to maintain genetic diversity and population stability.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Author contributions
AT: Writing – original draft, Visualization, Writing – review & editing. SK: Data curation, Writing – review & editing, Investigation. SM: Data curation, Writing – review & editing, Investigation, Visualization. AS: Investigation, Software, Writing – review & editing. OK: Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan (BR24992881).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdollahi Mandoulakani B., Yaniv E., Kalendar R., Raats D., Bariana H. S., Bihamta M. R., et al. (2015). Development of IRAP- and REMAP-derived SCAR markers for marker-assisted selection of the stripe rust resistance gene Yr15 derived from wild emmer wheat. Theor. Appl. Genet. 128, 211–219. doi: 10.1007/s00122-014-2422-8
Andrieu E., Thompson J. D., and Debussche M. (2007). The impact of forest spread on a marginal population of a protected peony (Paeonia officinalis L.): the importance of conserving the habitat mosaic. Biodiversity Conserv. 16, 643–658. doi: 10.1007/s10531-005-2357-0
Antonelli A. S. R., Fry C., Simmonds M. S., Kersey P. J., Pritchard H., Abbo M., et al. (2020). State of the World’s Plants and Fungi (Kew: Royal Botanic Gardens).
Arvas Y. E., Marakli S., Kaya Y., and Kalendar R. (2023). The power of retrotransposons in high-throughput genotyping and sequencing. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1174339
Ashapkin V. V., Kutueva L. I., Aleksandrushkina N. I., and Vanyushin B. F. (2020). Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21207457
Aung T. T., Shi F., Zhai Y., Xue J., Wang S., Ren X., et al. (2022). Acidic and alkaline conditions affect the growth of tree peony plants via altering photosynthetic characteristics, limiting nutrient assimilation, and impairing ROS balance. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23095094
Baduel P. and Colot V. (2021). The epiallelic potential of transposable elements and its evolutionary significance in plants. Philos. Trans. R Soc. Lond B Biol. Sci. 376, 20200123. doi: 10.1098/rstb.2020.0123
Baitulin I. O. and Kotukhov Y. A. (2011). Flora vascular plants of Kazakhstan Altai Russian: Almaty.
Belyayev A. (2014). Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 27, 2573–2584. doi: 10.1111/jeb.12513
Belyayev A., Kalendar R., Brodsky L., Nevo E., Schulman A. H., and Raskina O. (2010). Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mob DNA 1, 6. doi: 10.1186/1759-8753-1-6
Boronnikova S. V. and Kalendar R. N. (2010). Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants. Russian J. Genet. 46, 36–42. doi: 10.1134/s1022795410010060
Casacuberta E. and Gonzalez J. (2013). The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517. doi: 10.1111/mec.12170
Chelli S., Marignani M., Barni E., Petraglia A., Puglielli G., Wellstein C., et al. (2019). Plant–environment interactions through a functional traits perspective: a review of Italian studies. Plant Biosyst. 153, 853–869. doi: 10.1080/11263504.2018.1559250
Chirilă S. D. and Vassilev K. (2024). Phytocoenology and habitat preference of Paeonia tenuifolia, a vulnerable species in Romania. Biologia 79, 3489–3502. doi: 10.1007/s11756-024-01791-6
Doungous O., Kalendar R., Adiobo A., and Schulman A. H. (2015). Retrotransposon molecular markers resolve cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esculenta) by type and variety. Euphytica 206, 541–554. doi: 10.1007/s10681-015-1537-6
Doungous O., Kalendar R., Filippova N., and Ngane B. K. (2019). Utility of iPBS retrotransposons markers for molecular characterization of African Gnetum species. Plant Biosyst. 154, 587–592. doi: 10.1080/11263504.2019.1651782
Duan Y. B., Guo D. L., Guo L. L., Wei D. F., and Hou X. G. (2015). Genetic diversity analysis of tree peony germplasm using iPBS markers. Genet. Mol. Res. 14, 7556–7566. doi: 10.4238/2015.july.3.31
Ebert A. W. and Engels J. M. M. (2020). Plant biodiversity and genetic resources matter. Plants (Basel) 9. doi: 10.3390/plants9121706
Engelhardt K. A. M., Lloyd M. W., and Neel M. C. (2014). Effects of genetic diversity on conservation and restoration potential at individual, population, and regional scales. Biol. Conserv. 179, 6–16. doi: 10.1016/j.biocon.2014.08.011
Espinoza J. L. and Inaoka P. T. (2017). Gnetin-C and other resveratrol oligomers with cancer chemopreventive potential. Ann. N Y Acad. Sci. 1403, 5–14. doi: 10.1111/nyas.13450
Fan Y., Wang Q., Dong Z., Yin Y., Teixeira da Silva J. A., and Yu X. (2019). Advances in molecular biology of Paeonia L. Planta 251, 23. doi: 10.1007/s00425-019-03299-9
Gallego-Zamorano J., Huijbregts M. A. J., Schipper A. M., and Taylor A. (2022). Changes in plant species richness due to land use and nitrogen deposition across the globe. Diversity Distributions 28, 745–755. doi: 10.1111/ddi.13476
GOST (1985). Soils. Determination of nitrates by CINAO method. Available online at: https://docs.cntd.ru/document/1200023496 (Accessed December 28, 2024) .
GOST (1991a). Soils. Determination of mobile compounds of phosphorus and potassium by Machigin method modified by CINAO. Moscow. Available online at: https://gostexpert.ru/gost/getDoc/38501 (Accessed December 28, 2024).
Hosid E., Brodsky L., Kalendar R., Raskina O., and Belyayev A. (2012). Diversity of long terminal repeat retrotransposon genome distribution in natural populations of the wild diploid wheat Aegilops speltoides. Genetics 190, 263–274. doi: 10.1534/genetics.111.134643
Ji L., Teixeira da Silva J. A., Zhang J., Tang Z., and Yu X. (2014). Development and application of 15 novel polymorphic microsatellite markers for sect. Paeonia (Paeonia L.). Biochem. Systematics Ecol. 54, 257–266. doi: 10.1016/j.bse.2014.02.009
Jiang H., Li J., Wang L., Wang S., Nie X., Chen Y., et al. (2020). Total glucosides of paeony: a review of its phytochemistry, role in autoimmune diseases, and mechanisms of action. J. Ethnopharmacology 258, 112913. doi: 10.1016/j.jep.2020.112913
Kalendar R., Antonius K., Smykal P., and Schulman A. H. (2010). iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 121, 1419–1430. doi: 10.1007/s00122-010-1398-2
Kalendar R., Boronnikova S., and Seppanen M. (2021). Isolation and purification of DNA from complicated biological samples. Methods Mol. Biol. 2222, 57–67. doi: 10.1007/978-1-0716-0997-2_3
Kalendar R., Ivanov K. I., Akhmetollayev I., Kairov U., Samuilova O., Burster T., et al. (2024a). An improved method and device for nucleic acid isolation using a high-salt gel electroelution trap. Anal. Chem. 96, 15526–15530. doi: 10.1021/acs.analchem.4c03720
Kalendar R., Ivanov K. I., Samuilova O., Kairov U., and Zamyatnin A. A. Jr. (2023). Isolation of high-molecular-weight DNA for long-read sequencing using a high-salt gel electroelution trap. Anal. Chem. 95, 17818–17825. doi: 10.1021/acs.analchem.3c03894
Kalendar R. and Kairov U. (2024). Genome-Wide Tool for Sensitive de novo Identification and Visualisation of Interspersed and Tandem Repeats. Bioinform. Biol. Insights 18, 11779322241306391. doi: 10.1177/11779322241306391
Kalendar R., Khassenov B., Ramankulov Y., Samuilova O., and Ivanov K. I. (2017). FastPCR: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109, 312–319. doi: 10.1016/j.ygeno.2017.05.005
Kalendar R., Shevtsov A., Otarbay Z., and Ismailova A. (2024b). In silico PCR analysis: a comprehensive bioinformatics tool for enhancing nucleic acid amplification assays. Front. Bioinform. 4. doi: 10.3389/fbinf.2024.1464197
Kubentayev S. A., Khapilina O. N., Ishmuratova M. Y., Sarkytbayeva A. K., Turzhanova A. S., Imanbayeva A. A., et al. (2023). Current state of natural populations of Paeonia anomala (Paeoniaceae) in East Kazakhstan. Diversity 15, 1127. doi: 10.3390/d15111127
Kumar S. (2018). Epigenomics of plant responses to environmental stress. Epigenomes 2. doi: 10.3390/epigenomes2010006
Lanciano S. and Mirouze M. (2018). Transposable elements: all mobile, all different, some stress responsive, some adaptive? Curr. Opin. Genet. Dev. 49, 106–114. doi: 10.1016/j.gde.2018.04.002
Leont′ev A. F. (1988). “Representation of functions in convex domains by generalized exponential series,” in 'American Mathematical Society Translations (Szeged: American Mathematical Society, Washington), 121–130. doi: 10.1090/trans2/140/09
Lim M. Y., Jana S., Sivanesan I., Park H. R., Hwang J. H., Park Y. H., et al. (2013). Analysis of genetic variability using RAPD markers in paeonia spp. Grown in korea. Korean J. Hortic. Sci. Technol. 31, 322–327. doi: 10.7235/hort.2013.12210
Lovell J. T., MacQueen A. H., Mamidi S., Bonnette J., Jenkins J., Napier J. D., et al. (2021). Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature 590, 438–444. doi: 10.1038/s41586-020-03127-1
Lu M., Hu Q., Zhang Y., Zhai Y., Zhou Y., and Jiang J. (2019). Comparative chemical profiling of three TCM drugs in the Paeoniaceae family by UPLC-MS/MS combined with chemometric methods. Biochem. Systematics Ecol. 83, 121–129. doi: 10.1016/j.bse.2019.02.002
Madlung A. and Comai L. (2004). The effect of stress on genome regulation and structure. Ann. Bot. 94, 481–495. doi: 10.1093/aob/mch172
Marina V. O., DaoYuan Z., ShiMing D., LinKe Y. I. N., and BoRong P. A. N. (2010). Rare and endangered plant species of the Chinese Altai Mountains. J. Arid Land 2, 222–230. doi: 10.3724/SP.J.1227.2010.00222
Milovanov A., Zvyagin A., Daniyarov A., Kalendar R., and Troshin L. (2019). Genetic analysis of the grapevine genotypes of the Russian Vitis ampelographic collection using iPBS markers. Genetica 147, 91–101. doi: 10.1007/s10709-019-00055-5
Negi P., Rai A. N., and Suprasanna P. (2016). Moving through the stressed genome: emerging regulatory roles for transposons in plant stress response. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01448
Niinemets Ü. (2010). Responses of forest trees to single and multiple environmental stresses from seedlings to mature plants: Past stress history, stress interactions, tolerance and acclimation. For. Ecol. Manage. 260, 1623–1639. doi: 10.1016/j.foreco.2010.07.054
Olteanu L. S. L., Ciulca S., Ciulca A., Velicevici G., Danci M., and Madoşă E. (2022). Studies on the variability of the population of Romanian peony (Paeonia peregrina Mill. var. romanica) from Fântâna Mare, Tulcea county. J. Horticulture Forestry Biotechnol. 26, 81–87.
Opedal Ø. H., Armbruster W. S., and Graae B. J. (2014). Linking small-scale topography with microclimate, plant species diversity and intra-specific trait variation in an alpine landscape. Plant Ecol. Diversity 8, 305–315. doi: 10.1080/17550874.2014.987330
Paltsyn M. (2015). Protecting the wild nature and biodiversity of the Altai-Sayan Ecoregion (Protecting the Wild: Parks and Wilderness, the Foundation for Conservation). Washington, DC: Island Press/Center for Resource Economics.
Pan J., Zhang D., and Sang T. (2007). Molecular phylogenetic evidence for the origin of a diploid hybrid of Paeonia (Paeoniaceae). Am. J. Bot. 94, 400–408. doi: 10.3732/ajb.94.3.400
Papolu P. K., Ramakrishnan M., Mullasseri S., Kalendar R., Wei Q., Zou L. H., et al. (2022). Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1064847
Peakall R. and Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pimpinelli S., Piacentini L., and Herrel A. (2019). Environmental change and the evolution of genomes: Transposable elements as translators of phenotypic plasticity into genotypic variability. Funct. Ecol. 34, 428–441. doi: 10.1111/1365-2435.13497
Qiu Z. K., He J. L., Liu X., Zeng J., Xiao W., Fan Q. H., et al. (2018). Anxiolytic-like effects of paeoniflorin in an animal model of post traumatic stress disorder. Metab. Brain Dis. 33, 1175–1185. doi: 10.1007/s11011-018-0216-4
Reyer C. P., Leuzinger S., Rammig A., Wolf A., Bartholomeus R. P., Bonfante A., et al. (2013). A plant's perspective of extremes: terrestrial plant responses to changing climatic variability. Glob Chang Biol. 19, 75–89. doi: 10.1111/gcb.12023
Rudmann-Maurer K., Weyand A., Fischer M., and Stocklin J. (2007). Microsatellite diversity of the agriculturally important alpine grass Poa alpina in relation to land use and natural environment. Ann. Bot. 100, 1249–1258. doi: 10.1093/aob/mcm203
Schrader L. and Schmitz J. (2019). The impact of transposable elements in adaptive evolution. Mol. Ecol. 28, 1537–1549. doi: 10.1111/mec.14794
Süle G., Balogh J., Fóti S., Gecse B., and Körmöczi L. (2020). Fine-scale microclimate pattern in forest-steppe habitat. Forests 11, 1078. doi: 10.3390/f11101078
Tanhuanpää P., Juhanoja S., Oskarsson L., Marstein M., and Hartikainen M. (2021). Dear old peonies–for gene banks and gardeners; microsatellite fingerprinting of herbaceous peonies in Fennoscandia. Genet. Resour. Crop Evol. 68, 3413–3426. doi: 10.1007/s10722-021-01201-9
Walters C. and Pence V. C. (2020). The unique role of seed banking and cryobiotechnologies in plant conservation. Plants People Planet 3, 83–91. doi: 10.1002/ppp3.10121
Wang S. Q. (2020). Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant Biol. 20, 510. doi: 10.1186/s12870-020-02682-z
Wang B., Cheng W., Xu H., Wang R., Song K., Bao A., et al. (2024). Vegetation differentiation characteristics and control mechanisms in the Altay region based on topographic gradients. Ecol. Indic. 160. doi: 10.1016/j.ecolind.2024.111838
Wang X., Hao J. C., Shang B., Yang K. L., He X. Z., Wang Z. L., et al. (2021). Paeoniflorin ameliorates oxidase stress in Glutamate-stimulated SY5Y and prenatally stressed female offspring through Nrf2/HO-1 signaling pathway. J. Affect. Disord. 294, 189–199. doi: 10.1016/j.jad.2021.07.054
Wang Q., Zhao R., Chen Q., Teixeira da Silva J. A., Chen L., and Yu X. (2019). Physiological and biochemical responses of two herbaceous peony cultivars to drought stress. HortScience 54, 492–498. doi: 10.21273/HORTSCI13704-18
Weeks A. R., Sgro C. M., Young A. G., Frankham R., Mitchell N. J., Miller K. A., et al. (2011). Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 4, 709–725. doi: 10.1111/j.1752-4571.2011.00192.x
Xia E., Tong W., Hou Y., An Y., Chen L., Wu Q., et al. (2020). The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 13, 1013–1026. doi: 10.1016/j.molp.2020.04.010
Xu X.-X., Cheng F.-Y., Xian H.-L., and Peng L.-P. (2016). Genetic diversity and population structure of endangered endemic Paeonia jishanensis in China and conservation implications. Biochem. Systematics Ecol. 66, 319–325. doi: 10.1016/j.bse.2016.05.003
Xue Y., Liu R., Xue J., Wang S., and Zhang X. (2021). Genetic diversity and relatedness analysis of nine wild species of tree peony based on simple sequence repeats markers. Hortic. Plant J. 7, 579–588. doi: 10.1016/j.hpj.2021.05.004
Yang Y., Wu Y., Chen Q., Liu C., Liu G., Cheng S., et al. (2022). Potential spatiotemporal distribution changes and conservation recommendations of two connected endangered tree peony species (Paeonia decomposita & P. rotundiloba). Flora 294. doi: 10.1016/j.flora.2022.152131
Yuan J. H., Cheng F. Y., and Zhou S. L. (2012). Genetic structure of the tree peony (Paeonia rockii) and the Qinling Mountains as a geographic barrier driving the fragmentation of a large population. PloS One 7, e34955. doi: 10.1371/journal.pone.0034955
Zhao D., Hao Z., Wang J., and Tao J. (2013). Effects of pH in irrigation water on plant growth and flower quality in herbaceous peony (Paeonia lactiflora Pall.). Scientia Hortic. 154, 45–53. doi: 10.1016/j.scienta.2013.02.023
Keywords: Paeonia anomala L., biodiversity, retrotransposons, iPBS markers, morphometric variability, ecological adaptation
Citation: Turzhanova AS, Kubentayev SA, Magzumova SM, Sarkytbayeva AK and Khapilina ON (2025) The impact of local environmental differences on the phenotypic plasticity and genetic variation of Kazakhstani populations of Paeonia anomala. Front. Ecol. Evol. 13:1608776. doi: 10.3389/fevo.2025.1608776
Received: 24 April 2025; Accepted: 27 June 2025;
Published: 22 July 2025.
Edited by:
Neeta Lohani, Donald Danforth Plant Science Center, United StatesReviewed by:
Rania Kouki, High Agronomic Institute of Chott Mariem, TunisiaHossein Zeinalzadeh Tabrizi, Kyrgyz Turkish Manas University, Kyrgyzstan
Copyright © 2025 Turzhanova, Kubentayev, Magzumova, Sarkytbayeva and Khapilina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oxana N. Khapilina, b2tzZnVyQG1haWwucnU=
 Ainur S. Turzhanova
Ainur S. Turzhanova Serik A. Kubentayev
Serik A. Kubentayev Saule M. Magzumova
Saule M. Magzumova Aisulu K. Sarkytbayeva
Aisulu K. Sarkytbayeva Oxana N. Khapilina
Oxana N. Khapilina