- Simon Hippenmeyer Lab, Institute of Science and Technology, Klosterneuburg, Austria
Scientific research is to date largely restricted to wealthy laboratories in developed nations due to the necessity of complex and expensive equipment. This inequality limits the capacity of science to be used as a diplomatic channel. Maker movements use open-source technologies including additive manufacturing (3D printing) and laser cutting, together with low-cost computers for developing novel products. This movement is setting the groundwork for a revolution, allowing scientific equipment to be sourced at a fraction of the cost and has the potential to increase the availability of equipment for scientists around the world. Science education is increasingly recognized as another channel for science diplomacy. In this perspective, we introduce the idea that the Maker movement and open-source technologies have the potential to revolutionize science, technology, engineering and mathematics (STEM) education worldwide. We present an open-source STEM didactic tool called SCOPES (Sparking Curiosity through Open-source Platforms in Education and Science). SCOPES is self-contained, independent of local resources, and cost-effective. SCOPES can be adapted to communicate complex subjects from genetics to neurobiology, perform real-world biological experiments and explore digitized scientific samples. We envision such platforms will enhance science diplomacy by providing a means for scientists to share their findings with classrooms and for educators to incorporate didactic concepts into STEM lessons. By providing students the opportunity to design, perform, and share scientific experiments, students also experience firsthand the benefits of a multinational scientific community. We provide instructions on how to build and use SCOPES on our webpage: http://scopeseducation.org.
Introduction
Scientific advances and the technologies leading to these discoveries are the principal drivers for sustained wealth (Giovannini et al., 2015; Coccia, 2018)1. Science transcends borders, governments and languages, a notion that Louis Pasteur elegantly pointed out: “Science knows no country, because knowledge belongs to humanity, and is the torch which illuminates the world,” (Dubos, 1951). The rise of large global scientific consortia in the 20th century saw a fundamental shift in how scientific progress was achieved (Aboukhalil, 2014). This cooperative approach of scientists working together for a common goal has spanned national and political differences and formed the foundation for what is now termed science diplomacy (Gluckman et al., 2017). In many cases, science itself exists beyond the realm of politics, putting scientists in a unique position to form international bonds between countries that may not occur through foreign policy alone (Lord and Turekian, 2007). Countries with an active academic community like the United States, use science diplomacy extensively to shape governmental policies and improve their public perception abroad (Lord and Turekian, 2007; Hormats, 2012). However, for science diplomacy to be successful, active investment of resources at the national level must occur. The mobility of students, researchers, and technical workers is also an important factor, with more open and internationally engaged nations achieving a higher scientific impact (Wagner et al., 2018).
The United States, China, and other developed countries are dominant when it comes to natural-science research output (Csomós, 2018)2. One source for this inequality in output stems from the enormous financial investments required for upgrading and maintaining state-of-the-art scientific equipment. Some of these costs can be mitigated by incorporating freely available (open-source) technologies including additive manufacturing (3D printing), laser cutting and computer numerical control (CNC) milling to manufacture scientific equipment locally at a fraction of the cost. It is therefore not surprising that in particular, 3D printing has become a central component of many modern research laboratories (Maia Chagas, 2018). Applications range from creating custom apparatuses and equipment for experiments to finding simple solutions for everyday items such as racks for test tubes. This “open-source revolution” gives labs with limited budgets the opportunity to perform experiments that may have once been prohibitively expensive (Pearce, 2012; Baden et al., 2015; Barbera et al., 2016; Maia Chagas et al., 2017; Maia Chagas, 2018; Scott et al., 2018; Chen et al., 2019; Collins et al., 2019; Gilbert et al., 2019; Gopalakrishnan et al., 2019; Sule et al., 2019; Tomlinson et al., 2019). Recent advances such as the AttoBright microscope to detect biomarkers for Parkinson’s disease with single-molecule resolution demonstrates that 3D printed equipment can even be used for diagnosis (Brown et al., 2019). If this trend of open-source iterative design in laboratory hardware continues, the opportunities for more global research participation will be within reach.
Obtaining scientific equipment is not enough as most requires skilled and educated professionals to operate. Therefore, educational systems need to help facilitate a scientifically-literate society that supports scientific endeavors. Political strategies increasingly recognize the importance of education for effective science diplomacy (Fedoroff, 2009; Zewail, 2010). However, the strength of scientific collaborations originates from their pragmatic roots which extend beyond the stability of political relations between nations (Taubes, 1995; Stone, 2017). This is made possible by multi-faceted, discipline-specific communication channels and scientific networks3. For education to work similarly, dedicated networks must be built to empower educators to make global connections that aid their working goal. This need has already been partially recognized and a plethora of online platforms exist that allow classrooms to connect globally4.
There is an increasing demand for modernization of science, technology, engineering, and mathematics (STEM) education curriculums (Leshner and Scherer, 2019; Long et al., 2019). While many developed nations have been quick to adopt digital communication platforms in the classroom, without dedicated didactic concepts it has been demonstrated these methods can negatively influence learning outcomes (OECD, 2015). One source of inspiration for making dedicated didactic tools in education might come from “maker communities.” Here people connect both locally and globally to share data and ideas to solve problems in their communities or produce new and innovative products. This development has benefited heavily from the establishment of Makerspaces where the costs for open-source manufacturing technologies are shared by the local maker community (Niaros et al., 2017). Some classrooms have already embraced this philosophy and incorporate Maker Education as a core component of their STEM curriculum (Lecorchick et al., 2019).
Another driver for the establishment of maker communities were technical developments, especially the manufacturing of extremely cost-efficient single-board microcontrollers and computers. Examples include the Arduino (D’Ausilio, 2012) and Raspberry Pi (Severance, 2013)5, that can be used in a wide range of electronic products. The philosophy for designing low-cost computing options was to empower more students to acquire skills in programming and electronics (Severance, 2013) and their importance for students’ success in STEM education is becoming increasingly clear6. Widespread adoption of Raspberry Pis in elementary schools is occurring in developed countries (Quinlan and Baloro, 2018). They have been incorporated into numerous projects to deliver didactic concepts in programming7 and robotics8. In many cases, Raspberry Pis can now serve as low-cost alternatives to traditional desktop computers, making their use in the classroom more feasible (Nuñez-Unda et al., 2018).
One caveat is that many of the current projects designed for Raspberry Pis are reliant on instructions from teachers (Quinlan and Baloro, 2018). Open-ended projects where the students have more freedom to acquire knowledge at their own pace with minimal guidance, while still fitting within the constraints of lessons and teaching goals remains a challenge. Empirical data shows that these flipped learning or flipped classroom approaches generally have a positive influence on learning outcomes in STEM education. However, increasing workloads of educators and demands to incorporate ever-changing technology have limited the application of such approaches (Huber and Werner, 2016).
Inspired by a recent call for modern technology-driven devices to aid didactic learning concepts (OECD, 2015), we present here a STEM didactic tool that we termed Sparking Curiosity through Open-source Platforms in Education and Science (SCOPES). SCOPES is built on five key principles that allow it to be integrated into modern global educational frameworks. (1) To facilitate a flipped classroom approach, SCOPES motivates student engagement and promotes guided discussions by using real-world items to interact with digital content. (2) The core concepts do not rely on written text but rather universal visual content that can be shared globally, by scientists and educators, and used locally with minimal adaptation. To aid worldwide adoption, SCOPES is (3) largely independent of the availability of local resources and (4) has minimal implementation and maintenance costs. (5) Finally, SCOPES is built on open-source technologies to be further developed by the community. In a broader context SCOPES bridges the divide between science labs and classrooms by allowing students to explore real-world experiments and data. We present SCOPES as a proof-of-concept with three activities that can be adapted or integrated into already existing didactic concepts.
SCOPES Proof-Of-Concept
We developed the SCOPES kit as a cost-effective platform to bring life science labs to the classroom. SCOPES is designed as a modular platform that can be easily adapted for the specific needs of the educator, even without any computer or programming skills. During the design of SCOPES, we considered resource limitations educators might face. While dedicated computer classrooms in schools are becoming standard in developed countries, implementing new hardware is both time consuming and costly. Access to a stable source of electricity might also not be available in some areas9. Therefore we chose the Raspberry Pi Zero W (referred to as “Pi” in the following text)10, the most power-efficient member of the Raspberry Pi family, as the core processor for SCOPES. The Pi is powered via a 5 V USB port that is commonly used for charging smartphones and tablets and can be run remotely from a battery or small solar panel11. Another resource limitation are the displays used to show visual output from SCOPES (such as monitors). Mobile access through smartphones and tablets are largely responsible for the increased adoption of social media worldwide12. Therefore we chose to make use of mobile devices to display the visual output of SCOPES since it can be concluded that these devices are abundantly present throughout most countries of the world13,14. The Pi acts as a wireless access point that mobile devices can connect to and display information provided by the Pi. Several devices can connect to SCOPES in parallel, allowing for multiple students to interact with an individual SCOPES. Interactions with SCOPES is done via a web-based interface, running locally on the Pi, allowing set-up and control of SCOPES with no prior programming knowledge. Thus, SCOPES is self-contained and can be operated in the complete absence of any infrastructure with just a mobile device and a battery. We provide tutorials and further details of SCOPES on our website.
SCOPES Modules
The minimal SCOPES module is comprised of a Pi, a Raspberry Pi camera, a small LED strip and an SD card (Figure 1A). Additional access to a personal computer (PC) is necessary to prepare the SD card for use. We provide instructions on how to set-up the SD-card on our website. Once this is prepared, SCOPES is immediately usable. To use SCOPES efficiently, we designed the SCOPEScase. With increasing demands for the use of renewable materials in manufacturing (Mohanty et al., 2018), we constructed the SCOPEScase from laser-cut plywood. The assembly of the SCOPEScase can be achieved within one hour and the only necessary tool is a screwdriver. SCOPEScase conveniently holds and positions the Pi, camera, and LED strip (Figure 1A). The LEDs provide visual feedback for the user and serve as a light source during different activities. The SCOPEScase is not required for the operation of SCOPES, however, we are convinced it provides improved didactic user experience and increases consistency of experiments.
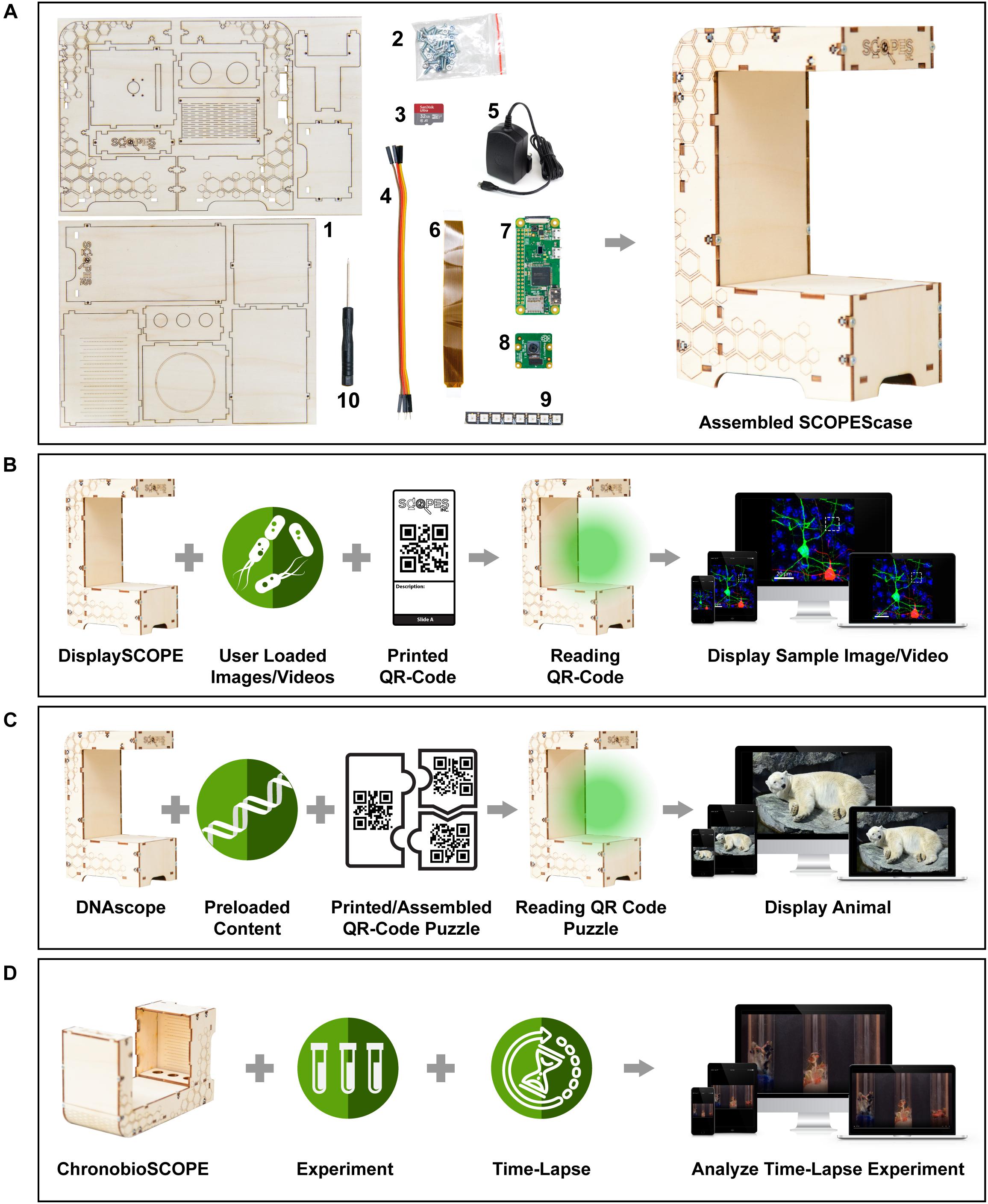
Figure 1. Overview of SCOPES Education Platform. (A) Components required to assemble a functioning SCOPEScase. Parts include (1) Laser-cut wood case, (2) M3 Screws and nuts, (3) SD Card, (4) Male to female jumper cables, (5) 5V power adapter/power supply, (6) Pi Zero camera cable, (7) Raspberry Pi Zero W, (8) Raspberry Pi Camera v1 or v2, (9) Pimorini Blinkt LED, and a (10) Screwdriver. Right panel depicts a fully assembled SCOPEScase. The assembled SCOPEScase is not strictly necessary for all activities in (B–D), but greatly improves the user experience and ease-of-use. (B) Operation in DisplaySCOPE mode. Requires user to load an image and/or video content and link to QR-code through the web interface as well as to print QR-codes. When the QR-code slide is scanned by the DisplaySCOPE, the sample is displayed on the connected device. (C) Operation in DNAScope mode. Content is provided preloaded on the Raspberry Pi disk image (https://scopeseducation.org/downloads/). Requires users to print provided QR-code puzzle pieces. When a complete puzzle is assembled and scanned by the DNAscope the corresponding animal/plant is displayed on the connected device. (D) Operation in ChronobioSCOPE mode. Depending on the samples, users can perform experiments with SCOPEScase in a vertical or horizontal position. Time-lapse settings are defined in the web interface and analysis of the experiment can be viewed on the connected device. For complete assembly and operation instructions please see https://scopeseducation.org.
We present here three activities that showcase the capabilities of SCOPES and should serve as inspiration for the development of new projects using SCOPES in the classroom. Firstly, we incorporated the ability for digitized samples to be viewed and explored, referred to as the DisplaySCOPE. This provides an avenue for scientists around the world to share their discoveries directly with classrooms and educators. It also lowers maintenance costs as fresh samples can be constantly downloaded and the burden of sourcing such samples is not on the educator. Secondly, DNAscope provides a fun and explorative way for students to learn about DNA. Thirdly, the ChronobioSCOPE allows students to observe lengthy biological processes, by running time-lapse experiments that can span days to weeks. Below we elaborate on each of these activities in more detail.
DisplaySCOPE: Explore Digitized Samples
Modern biology is heavily reliant on interpreting visual data obtained through imaging. We propose the use of DisplaySCOPE in a flipped classroom approach where students perform a hypothetical experiment while exploring digitized visual content (in this case microscope images) by themselves with minimal guidance. The learning materials required include 3D printable or paper microscope slides with quick response (QR) codes. Once the individual QR-codes are placed under the camera, the code is read and the corresponding microscope image can then be explored by the student (Figure 1B). SCOPES provides an easy-to-use, browser-based interface to upload digital content (images or movies) and to link these files to one of the provided QR-codes. SCOPES also allows several students to interact with one SCOPE simultaneously through their own devices (i.e., tablet, smartphone, computer) and explore the content at their own pace.
DNASCOPE: Learn About DNA
DNAscope is an activity that allows students to assemble the sequence of DNA fragments and identify the species of animal or plant that this DNA belongs to (Figure 1C). In total 27 different genetic signatures (animals/plants) are currently supported. The activity is performed by having students assemble three puzzle pieces into a complete puzzle, representing the gene sequence of an animal. Each puzzle piece contains a short DNA sequence on one side and a unique QR-code on the back. When a complete puzzle is placed under the DNAscope it displays the image of an animal specific to that QR-code combination. DNAscope is delivered with SCOPES and therefore needs no additional activity from the educator to work. This activity was developed to complement an outreach activity, described below, that is aimed to improve students’ understanding that DNA is the molecule that contains the instructions for life.
ChronobioSCOPE: Perform Real Experiments
Many intriguing biological processes are painstakingly slow. For example, plant growth can take several days before any observable changes occur. Therefore, typical biological experiments in a classroom are started and then explored at a defined endpoint, which might not capture the interest of students. To make these experiments more informative, we designed ChronobioSCOPE in a way that allows students to prepare a time-lapse video of long biological processes (Figure 1D). In a vertical position, the SCOPEScase is designed to monitor experiments done in a flat reaction chamber (i.e., petri dish). When placed horizontally the SCOPEScase is optimized to act as a stand for reaction tubes. The progress of the experiment is monitored by taking consecutive pictures of the transparent reaction chamber or tubes. Finally, all images can be downloaded and processed on a computer, transforming the lengthy experiment into a brief movie sequence. SCOPES streamlines the process of filming the time-lapse, monitoring its status and downloading files. On our webpage, we provide examples of experiments we have performed using the ChonobioSCOPE.
SCOPES in Science Education
DisplaySCOPE
To demonstrate SCOPES in science education, we provide a brief report of using the mosaic analysis with double marker (MADM) Brain Explorer (DisplaySCOPE predecessor) during the IST Austria Open Campus Day 2018 and the Austrian Long Night of Research 2018, and the DisplaySCOPE at Maker Faire Vienna 2019. SCOPES was used to explain MADM technology at both events (Zong et al., 2005; Hippenmeyer et al., 2010). Mosaic analysis with double marker is a powerful, visually informative genetic technology used in modern labs and as a detailed explanation on its purpose and capabilities is beyond the scope of this perspective, we refer the interested reader to previous publications (Hippenmeyer et al., 2010, 2013; Hippenmeyer, 2013; Beattie et al., 2017; Laukoter et al., 2020)15. Simply put, MADM allows the sparse labeling of cells in any mouse tissue with fluorescent markers that can be visualized under a microscope. When MADM labeling occurs in a dividing stem cell or progenitor, all daughter cells are permanently labeled with either green fluorescent protein (GFP) or red fluorescent protein (tdTomato; Hippenmeyer, 2013). In the brain, MADM has been used to label neural stem cells that have the potential to produce two classes of cells, neurons and glia, with dramatically different function and morphology. To aid the didactic experience, visitors selected a physical representation of a microscope slide (laser-cut acrylic or paper) and placed it on the SCOPEScase. The LED flashed to mimic scanning of the slide, then displayed an interactive, high-resolution microscopic image of a MADM brain16. We had more than 2000 visitors at both events and we found that the combination of physically interacting with the SCOPEScase, the visual LED feedback and the exploration of digitized samples, provided a powerful didactic experience. Most visitors could immediately identify neurons and glia as two distinct cell types in the brain based on morphology. We observed that by allowing visitors to explore the samples on their own, they became more intellectually engaged. Not only did it spark questions about brain development, but also confocal microscopy, which was the main piece of equipment used to acquire the digital images. In previous outreach events, similar interest was mostly absent when conveying the same message using static images on a screen. The overwhelmingly positive feedback we received from the MADM Brain Explorer was the underlying motivation for us to develop this didactic concept into the now more universal DisplaySCOPE.
DNASCOPE
Inspired by projects using different colored LEGO bricks to represent the nucleotides in DNA17,18, we prepared a didactic concept for the IST Austria Summer School 2017, a 5-day event for children age 6–10 years. We introduced the idea that we (Homo sapiens) share 99% of our genetic information with chimpanzees, one of our nearest evolutionary ancestors (Wildman et al., 2003). However, children quickly provided a list of differences between humans and chimps. With the help of trained instructors, we then explained that the instructions for how a cell, and more broadly, a human or any animal is built is encoded by our DNA, which can be envisioned as a long string of four letters19. Some parts of this code are similar or conserved between species and other segments are unique to a single species. Organisms are constantly interacting with their surroundings and leave behind traces of their DNA within feces, mucus, hair, leaves, roots, or shed skin. Collectively, this is referred to as environmental DNA (eDNA) and can be purified from water, soil or even air. The subsequent analysis allows scientists to monitor the different species living in or transiting through an area even when the animal or plant is long gone (Bohmann et al., 2014). In the following activity, we performed an eDNA experiment on the IST Austria campus to convey the concept that a DNA sequence can unambiguously identify a species. In preparation, we hid pre-made DNA sequences made of colored LEGO bricks on the IST Austria campus for the children to find. Back in the lab, the children performed the “sequencing” by identifying the letter code linked to the color sequence using a conversion table. Once this was done, the children had to assemble their sequence with prepared paper pieces that had the same color/letter-code printed on them. Once their sequence was aligned, children turned the paper pieces over to find an image of the organism linked to the DNA sequence. Based on the positive response we received from the children, we considered this concept highly successful in explaining the complex nature of eDNA. We built on these experiences when developing DNAScope to reduce the preparation workload for instructors and to meet the demands of children growing up in the digital age. In its current form, we envision that children would search and assemble re-usable 3D printed or paper puzzle pieces that contain a short DNA sequence on one side and a QR-code on the back. After recording their sequence, students would then scan the QR-codes of their completed puzzle with the DNAscope and discover which plant or animal the DNA sequence came from originally (Figure 1C).
Discussion
Much of modern life science research is deemed too expensive or difficult to be adapted to a classroom setting. This creates a divide between the science students are exposed to, and the current state of science research (Gage, 2019). SCOPES can be adapted to communicate complex subjects from genetics to neurobiology, perform real-world biological experiments and explore digitized scientific samples. We envision SCOPES to enhance interactions between scientists and educators by directly sharing their findings using didactic concepts. By lowering the barrier to entry for educators in costs, manufacturing, and maintenance of SCOPES, we hope to enhance science diplomacy sustainably by building and strengthening global communities. Ideally, SCOPES gives all parties involved the opportunity to connect and experience the rewards of a multinational community.
While SCOPES is designed to be adaptable, it is not without limitations. Firstly, SCOPES currently does not provide an interaction network. We do not see this as a major limitation as several platforms exist where didactic concepts can be shared20. Our vision is to develop our online website as an interactive platform, ultimately bridging scientists with educators. We encourage educators and scientists to keep us updated on novel learning materials via the contact form on the SCOPES homepage so that we can make new SCOPES projects available for the community.
Secondly, SCOPES is not a microscope and will not achieve the resolution required for visualizing individual cells. If educators wish to build such a device, we recommend other excellent open-source projects (Cybulski et al., 2014; Geissmann et al., 2017; Tovar et al., 2018; Amann et al., 2019) including the FlyPi (Maia Chagas et al., 2017), multi-fluorescent (Nuñez et al., 2017), and OpenFlexure microscopes (Collins et al., 2019). While some of these microscopes achieve a high degree of optical resolution, many are purpose-built and some require additional components that may be expensive or difficult to source. We believe it is important for students to learn how real data is acquired in life science labs and recommend complementing lesson plans with open-source microscopes whenever possible. Nevertheless, we argue that the greatest value for younger students comes from the self-exploration of samples imaged with cutting edge technology. We present SCOPES as a tool that allows scientists to share and explain their work with a broad and enthusiastic audience by providing high-quality images with explanations. At the same time, SCOPES allows students to explore samples they would otherwise not have access to. All this comes with the significant benefit that there is minimal effort required by the educator when using these images. In summary, we designed SCOPES as a cost-effective device that inspires students to be inquisitive, ask questions, and discuss actual lab results.
Another limitation of the current SCOPEScase is that it requires access to a laser cutter. These devices have become common in developed countries and are typically available in public Makerspaces. Laser-cut items can even be ordered at relatively low-cost via the internet. However, we realize that technologies such as 3D printing are even more widespread, especially in the classroom, and we plan to incorporate these manufacturing processes into future designs of the SCOPEScase.
Through proof-of-concept, this perspective intends to demonstrate that the Maker movement and use of open-source technology have the potential to revolutionize classroom education. Activities presented here are to highlight the capabilities of SCOPES and not meant to be finalized didactic concepts. SCOPES extends the use of electronic image databases (for example: Williams et al., 2017) by allowing students to use real-world items (SCOPEScase, QR-codes) to interact with digital content. We anticipate that these features will empower instructors to create interactive concepts with relatively little effort. Our mission is to develop SCOPES further and we are adapting SCOPES into the framework of lesson plans with classroom instructors. Updates, improvements and new teaching material will be published on the SCOPES website. We encourage users to communicate requests for new features as well as share their experiences and concepts via the contact form on our homepage.
We have designed SCOPES to be built and used with no computer or programming skills. As SCOPES builds on open-source Raspberry Pi hardware, it can be readily adapted for novel projects. This will be necessary as studies have shown that when students can relate the material they learn in the classroom to their own lives, their motivation and performance improves (Hulleman and Harackiewicz, 2009). We also anticipate that through collaborative learning, SCOPES will encourage interdisciplinary experiential learning where programming skills are used to explore biological concepts and thus bridge several STEM disciplines. An example might be that students build the SCOPEScase, follow tutorials on how to program time-lapses on the Raspberry Pi21, and then use this to perform a biological experiment as described for the ChronobioSCOPE. In this way, SCOPES facilitates hands-on interdisciplinary learning and problem-solving opportunities for students. SCOPES can also be expanded using the exquisite capabilities of the Raspberry Pi to interact with a spectrum of sensors to record environmental parameters. We are therefore convinced that SCOPES can be adapted to aid didactic learning in the classroom and encourage students to engage cognitively and develop critical problem-solving skills.
Data Availability Statement
All details for building and operating SCOPES are available at our webpage: http://scopeseducation.org.
Author Contributions
RB and FP conceptualized the study, wrote the original draft, and prepared the figure. All authors wrote, reviewed, and edited the final version of the manuscript.
Funding
RB received support from Austrian Science Fund (FWF) Lise-Meitner program (M 2416). This study was also supported by IST Austria institutional funds to SH, a grant by NÖ Forschung und Bildung n[f + b] (C13-002) to SH; and the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement No. 725780 LinPro) to SH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Hippenmeyer laboratory for discussion. We would also like to thank the IST Austria Events and Communications team for providing multiple venues for us to develop and present our work. Thank you to the Bioimaging Facility, LSF and PCF at IST Austria for technical support. Thank you to Happy Lab Wien (https://www.happylab.at/en_vie/home) for access to equipment necessary for prototyping the SCOPEScase. We would also like to thank Daniel Gütl for his discussions regarding additive manufacturing techniques. Icons for Figure 1 provided by https://pixabay.com/users/mcmurryjulie-2375405/, image of mobile devices was adapted from an image by https://pixabay.com/users/nick_h-1821910/ and image of a polar bear was from https://pixabay.com/photos/polar-bear-zoo-white-bear-animal-1959068/. We apologize to all colleagues whose work we could not cite due to space restrictions.
Footnotes
- ^ https://www.un.org/en/development/desa/policy/untaskteam_undf/thinkpieces/28_thinkpiece_science.pdf
- ^ Nature Index 2019 – https://www.nature.com/articles/d41586-019-01921-0
- ^ http://connectedresearchers.com
- ^ https://www.digitalhumanlibrary.org/teachers/global-connections-for-teachers-and-students/
- ^ https://www.raspberrypi.org/
- ^ https://teachcomputing.org/
- ^ Raspberry Pi based Piper computers: https://www.playpiper.com/
- ^ https://camjam.me/
- ^ https://www.iea.org/reports/sdg7-data-and-projections
- ^ https://www.raspberrypi.org/blog/raspberry-pi-zero-w-joins-family/
- ^ https://learn.pi-supply.com/make/pijuice-naturejuice-project/
- ^ https://www.statista.com/statistics/330695/number-of-smartphone-users-worldwide
- ^ https://newsroom.cisco.com/feature-content?articleId=1872300
- ^ https://www.internetworldstats.com/stats.htm
- ^ SCOPES Education tutorial on MADM Brain Explorer: www.scopeseducation.org/tutorials/MADM
- ^ MADM Brain Explorer at IST Austria Open Campus 2018: https://youtu.be/-xByJ5erNTo
- ^ https://samnicholls.net/2017/03/15/lego-sequencer/
- ^ https://www.earlham.ac.uk/articles/earlham-institute-lego-sequencer
- ^ DNA Structure and Replication: Crash Course Biology: https://www.youtube.com/watch?v=8kK2zwjRV0M
- ^ https://www.digitalhumanlibrary.org/teachers/global-connections-for-teachers-and-students/
- ^ https://projects.raspberrypi.org/en/projects/timelapse-setup
References
Amann, S., Witzleben, M. V., and Breuer, S. (2019). 3D-printable portable open-source platform for low-cost lens-less holographic cellular imaging. Sci. Rep. 9:11260. doi: 10.1038/s41598-019-47689-1
Baden, T., Chagas, A. M., Gage, G., Marzullo, T., Prieto-Godino, L. L., and Euler, T. (2015). Open Labware: 3-D printing your own lab equipment. PLoS Biol, 13:e1002086. doi: 10.1371/journal.pbio.1002086
Barbera, G., Liang, B., Zhang, L., Gerfen, Charles, R., Culurciello, E., et al. (2016). Spatially Compact neural clusters in the dorsal striatum encode locomotion relevant information. Neuron 92, 202–213. doi: 10.1016/j.neuron.2016.08.037
Beattie, R., Postiglione, M. P., Burnett, L. E., Laukoter, S., Streicher, C., Pauler, F. M., et al. (2017). Mosaic analysis with double markers reveals distinct sequential functions of lgl1 in neural stem cells. Neuron 94, 517.e3–533.e3. doi: 10.1016/j.neuron.2017.04.012
Bohmann, K., Evans, A., Gilbert, M. T. P., Carvalho, G. R., Creer, S., Knapp, M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. 29, 358–367. doi: 10.1016/j.tree.2014.04.003
Brown, J. W. P., Bauer, A., Polinkovsky, M. E., Bhumkar, A., Hunter, D. J. B., Gaus, K., et al. (2019). Single-molecule detection on a portable 3D-printed microscope. Nat. Commun. 10:5662. doi: 10.1038/s41467-019-13617-0
Chen, K. L., Ven, T. N., Crane, M. M., Chen, D. E., Feng, Y.-C., Suzuki, N., et al. (2019). An inexpensive microscopy system for microfluidic studies in budding yeast. Transl. Med. Aging 3, 52–56. doi: 10.1016/j.tma.2019.05.001
Coccia, M. (2018). Socioeconomic driving forces of scientific research. arXiv [preprint]. Avaliable online at: https://arxiv.org/abs/1806.05028 (accessed March 15, 2020).
Collins, J. T., Knapper, J., Stirling, J., Mduda, J., Mkindi, C., Mayagaya, V., et al. (2019). Robotic microscopy for everyone: the OpenFlexure microscope. bioRxiv [preprint]. doi: 10.1101/861856
Csomós, G. (2018). A spatial scientometric analysis of the publication output of cities worldwide. J. Informetrics 12, 547–566.
Cybulski, J. S., Clements, J., and Prakash, M. (2014). Foldscope: origami-based paper microscope. PloS One 9:e98781. doi: 10.1371/journal.pone.0098781
D’Ausilio, A. (2012). Arduino: a low-cost multipurpose lab equipment. Behav. Res. Methods 44, 305–313. doi: 10.3758/s13428-011-0163-z
Gage, G. J. (2019). The case for neuroscience research in the classroom. Neuron 102, 914–917. doi: 10.1016/j.neuron.2019.04.007
Geissmann, Q., Garcia Rodriguez, L., Beckwith, E. J., French, A. S., Jamasb, A. R., and Gilestro, G. F. (2017). Ethoscopes: an open platform for high-throughput ethomics. PLoS Biol. 15:e2003026. doi: 10.1371/journal.pbio.2003026
Gilbert, K. M., Schaeffer, D. J., Gati, J. S., Klassen, L. M., Everling, S., and Menon, R. S. (2019). Open-source hardware designs for MRI of mice, rats, and marmosets: integrated animal holders and radiofrequency coils. J. Neurosci. Methods 312, 65–72. doi: 10.1016/j.jneumeth.2018.11.015
Giovannini, E., Niestroy, I., Nilsson, M., Roure, F., and Spanos, M. (2015). The Role of Science, Technology and Innovation Policies to Foster the Implementation of the Sustainable Development goals. Report of the Expert group “Follow-up to Rio 20. Luxemburg: Publications of European Union.
Gluckman, P. D., Turekian, V. C., Grimes, R. W., and Kishi, T. (2017). Science diplomacy: a pragmatic perspective from the inside. Sci. Dipl. 6, 1–13.
Gopalakrishnan, V., Krishnan, N. P., McClure, E., Pelesko, J., Crozier, D., Williamson, D. F. K., et al. (2019). A low-cost, open source, self-contained bacterial EVolutionary biorEactor (EVE). bioRxiv [preprint]. doi: 10.1101/729434
Hippenmeyer, S. (2013). Dissection of gene function at clonal level using mosaic analysis with double markers. Front. Biol. 8, 557–568. doi: 10.1007/s11515-013-1279-6
Hippenmeyer, S., Johnson, R. L., and Luo, L. (2013). Mosaic analysis with double markers reveals cell-type-specific paternal growth dominance. Cell Rep. 3, 960–967. doi: 10.1016/j.celrep.2013.02.002
Hippenmeyer, S., Youn, Y. H., Moon, H. M., Miyamichi, K., Zong, H., Wynshaw-Boris, A., et al. (2010). Genetic mosaic dissection of lis1 and ndel1 in neuronal migration. Neuron 68, 695–709. doi: 10.1016/j.neuron.2010.09.027
Huber, E., and Werner, A. (2016). “A review of the literature on flipping the STEM classroom: preliminary findings,” in Paper Presented at: 33rd International Conference of Innovation, Practice and Research in the Use of Educational Technologies in Tertiary Education-ASCILITE 2016-Show Me the Learning, Adelaide).
Hulleman, C. S., and Harackiewicz, J. M. (2009). Promoting interest and performance in high school science classes. Science 326:1410. doi: 10.1126/science.1177067
Laukoter, S., Beattie, R., Pauler, F. M., Amberg, N., Nakayama, K. I., and Hippenmeyer, S. (2020). Imprinted Cdkn1c genomic locus cell-autonomously promotes cell survival in cerebral cortex development. Nat. Commun. 11:195. doi: 10.1038/s41467-019-14077-2
Lecorchick, D., Spires, H. A., and Gallo, L. (2019). “Makerspace curriculum development and implementation toward cross-cultural integrative stem learning: a united states and china high school collaboration,” in ASEE Southeastern Section Conference, Raleigh, CA.
Leshner, A. I., and Scherer, L. (2019). Critical steps toward modernizing graduate STEM education. Issues Sci. Technol. 35, 46–49.
Long, L. N., Blanchette, S., Kelley, T. D., and Hohnka, M. (2019). “The crucial need to modernize engineering education,” in IEEE Aerospace Conference, Big Sky, MT, 1–9.
Lord, K. M., and Turekian, V. C. (2007). Time for a new era of science diplomacy. Science 315, 769–770. doi: 10.1126/science.1139880
Maia Chagas, A. (2018). Haves and have nots must find a better way: the case for open scientific hardware. PLoS Biol. 16:e3000014. doi: 10.1371/journal.pbio.3000014
Maia Chagas, A., Prieto-Godino, L. L., Arrenberg, A. B., and Baden, T. (2017). The €100 lab: a 3D-printable open-source platform for fluorescence microscopy, optogenetics, and accurate temperature control during behaviour of zebrafish, Drosophila, and Caenorhabditis elegans. PLoS Biol. 15:e2002702. doi: 10.1371/journal.pbio.2002702.
Mohanty, A. K., Vivekanandhan, S., Pin, J.-M., and Misra, M. (2018). Composites from renewable and sustainable resources: challenges and innovations. Science 362, 536–542. doi: 10.1126/science.aat9072
Niaros, V., Kostakis, V., and Drechsler, W. (2017). Making (in) the smart city: the emergence of makerspaces. Telematics Informatics 34, 1143–1152.
Nuñez, I., Matute, T., Herrera, R., Keymer, J., Marzullo, T., Rudge, T., et al. (2017). Low cost and open source multi-fluorescence imaging system for teaching and research in biology and bioengineering. PloS One 12:e0187163. 3. doi: 10.1371/journal.pone.0187163
Nuñez-Unda, A., Vera, A., Haz, L., Pinos, V., Zurita, R., and Medina, S. (2018). The raspberry Pi as a computer substitute at elementary schools in developing countries: a pilot experiment in ecuador. MATEC Web Conf. 210, 04023.
Quinlan, O., and Baloro, S. (2018). Raspberry Pi Computers in Schools. Cambridge, UK: Raspberry Pi Foundation Research.
Pearce, J. M. (2012). Building research equipment with Free. Open-source hardware. Science 337:1303. doi: 10.1126/science.1228183
Scott, B. B., Thiberge, S. Y., Guo, C., Tervo, D. G. R., Brody, C. D., Karpova, A. Y., et al. (2018). Imaging cortical dynamics in GCaMP transgenic rats with a head-mounted widefield macroscope. Neuron 100, 1045.e5–1058.e5. doi: 10.1016/j.neuron.2018.09.050
Sule, S. S., Petsiuk, L. A., and Pearce, M. J. (2019). Open source completely 3-D printable centrifuge. Instruments 3:30.
Taubes, G. (1995). Cold war rivals find common ground. Science 268, 488–491. doi: 10.1126/science.268.5210.488
Tomlinson, S., Carrington Yates, H., Oruni, A., Njoroge, H., Weetman, D., Donnelly, M. J., et al. (2019). Open source 3D printable replacement parts for the WHO insecticide susceptibility bioassay system. Parasit.Vectors 12:539. doi: 10.1186/s13071-019-3789-9
Tovar, J. C., Hoyer, J. S., Lin, A., Tielking, A., Callen, S. T., Elizabeth Castillo, S., et al. (2018). Raspberry Pi-powered imaging for plant phenotyping. Appl. Plant Sci. 6:e1031. doi: 10.1002/aps3.1031
Wagner, C. S., Whetsell, T., Baas, J., and Jonkers, K. (2018). Openness and impact of leading scientific countries. Front. Res. Metr. Anal. 3:10.
Wildman, D. E., Uddin, M., Liu, G., Grossman, L. I., and Goodman, M. (2003). Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: enlarging genus Homo. Proc. Nal. Acad. Sci. U.S.A. 100:7181. doi: 10.1073/pnas.1232172100
Williams, E., Moore, J., Li, S. W., Rustici, G., Tarkowska, A., Chessel, A., et al. (2017). Image data resource: a bioimage data integration and publication platform. Nat. Methods 14, 775. doi: 10.1038/s41592-018-0169-x
Keywords: science diplomacy, open science, STEM, science education, mosaic analysis with double markers (MADM), neuroscience
Citation: Beattie R, Hippenmeyer S and Pauler FM (2020) SCOPES: Sparking Curiosity Through Open-Source Platforms in Education and Science. Front. Educ. 5:48. doi: 10.3389/feduc.2020.00048
Received: 31 January 2020; Accepted: 14 April 2020;
Published: 08 May 2020.
Edited by:
Mohammed Andres Mostajo-Radji, University of California, San Francisco, United StatesReviewed by:
Andreas Obers, Peter Doherty Institute for Infection and Immunity, AustraliaDiana Alatriste Gonzalez, Charité Medical University of Berlin, Germany
Copyright © 2020 Beattie, Hippenmeyer and Pauler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert Beattie, cm9iZXJ0LmJlYXR0aWVAaXN0LmFjLmF0; Simon Hippenmeyer, c2ltb24uaGlwcGVubWV5ZXJAaXN0LmFjLmF0; Florian M. Pauler, Zmxvcmlhbi5wYXVsZXJAaXN0LmFjLmF0
†These authors have contributed equally to this work
 Robert Beattie
Robert Beattie Simon Hippenmeyer
Simon Hippenmeyer Florian M. Pauler
Florian M. Pauler