- 1Institute of Food and Agricultural Sciences, Department of Microbiology and Cell Science, University of Florida, Gainesville, FL, United States
- 2Department of Biology, The College of Wooster, Wooster, OH, United States
- 3Department of Agricultural Education and Communication, University of Florida, Gainesville, FL, United States
- 4Department of Natural and Applied Sciences, University of Dubuque, Dubuque, IA, United States
- 5Center for New Designs in Learning and Scholarship, Georgetown University, Washington, DC, United States
- 6Formerly of the Division of Undergraduate Education, Directorate for Education and Human Resources, National Science Foundation, Alexandria, VA, United States
- 7DNA Learning Center, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, United States
- 8Department of Biology, Eastern Connecticut State University, Willimantic, CT, United States
- 9Department of Biology, Georgetown University, Washington, DC, United States
Introduction: Bioinformatics is an interdisciplinary field at the intersection of computational and biological sciences that focuses on the analysis and interpretation of large biological data sets. Although recognized as essential in the life sciences, bioinformatics is not commonly integrated in undergraduate life science education programs. Based on a national survey in 2016, the Network for Integrating Bioinformatics into Life Sciences Education (NIBLSE) published a community-sourced set of core competencies in bioinformatics education. The survey also identified barriers that prevent incorporation of these competencies into the curriculum. In the current study, the NIBLSE group reports the findings of a new survey to 509 life science educators across the US in 2022 to identify current barriers of bioinformatics integration and to determine if the landscape of bioinformatics education has changed since the 2016 survey.
Results: Similar to previous results, a majority of respondents who currently teach bioinformatics or plan to teach bioinformatics report barriers. The top two barriers reported are students lacking prerequisite skills/knowledge and instructors lacking time to restructure course content. As in 2016, women reported experiencing barriers to bioinformatics teaching significantly more often than men; faculty from underrepresented minority backgrounds reported barriers more often than non-URM faculty; and educators at minority-serving institutions (MSIs) reported barriers more frequently than colleagues at non-MSIs. For additional insight into the barriers facing these educators, we conducted focus groups which provided qualitative data that supported the survey findings and revealed common themes including faculty perceptions of the relevance of bioinformatics in the curriculum. Despite the perceived value of bioinformatics education, many focus group members cited lack of student preparation and interest, and technological access as barriers. Participants also discussed how professional development and community support would enhance and sustain bioinformatics teaching.
Discussion: Taken all together, this study indicates that challenges remain, which vary among faculty types and settings, but that more educators are attempting to integrate bioinformatics into life sciences education. In summary, our results suggest that redoubled efforts to provide training and community support to life sciences faculty is necessary.
Introduction
Bioinformatics is a rapidly expanding interdisciplinary field that encompasses computer science, mathematics, statistics, and biological data (Gauthier et al., 2019). Bioinformatics has greatly influenced the types of questions addressed by contemporary researchers, and given the sheer scale of datasets, how biological research is done. Typically, bioinformatics involves retrieving, processing, filtering, visualizing, and analyzing omics-focused datasets. The rapid advances in inexpensive high-throughput technology driving the sustained torrent of “big data” makes it critical for today’s life science trainees to learn the skills necessary to analyze biological data and make appropriate inferences. Furthermore, many of these disciplinary data science skills may be applied to datasets beyond the scope of biology. Consequently, providing early skill development in bioinformatics helps emerging scientists advance their research potential and primes them to seek out opportunities in higher education and the technical workforce (Porter and Smith, 2019). The field of bioinformatics has grown from early studies on protein sequences in the 1950’s. With the ability to sequence DNA, it has become easier to gather large numbers of sequences; at the same time, computer technology (both hardware and software) have advanced in order to analyze these sequences. It has been suggested that the term “bioinformatics” may soon become completely synonymous with biology (Gauthier et al., 2019).
Currently, training in the use of bioinformatics tools is a major bottleneck in this fast-moving field. In response to this bottleneck, there have been numerous calls for professional development in this area (Carvalho and Rustici, 2013; Attwood et al., 2019) and integration into educational frameworks (Altman, 1998; Machluf and Yarden, 2013; Wilson Sayres et al., 2018; Attwood et al., 2019; Niepielko and Shumskaya, 2021; Rocha et al., 2022; Isik et al., 2023). Integration of bioinformatics learning objectives across undergraduate life science degree programs can theoretically address the issue of bioinformatics literacy (Dinsdale et al., 2015). Further, bioinformatics-driven undergraduate curricula, when carefully designed, also have the potential to address traditional STEM access and diversity barriers (Elgin et al., 2021; Handelsman et al., 2022; The Genomic Data Science Community Network, 2022).
The Network for Integrating Bioinformatics into Life Sciences Education (NIBLSE) began in 2014 as an NSF-funded research coordination network focused on fostering community efforts to promote the integration of bioinformatics within undergraduate biological sciences (Dinsdale et al., 2015; Toby et al., 2022).1 The network’s overarching goals are to identify best practices for (1) preparation of students for bioinformatics instruction; (2) integration of bioinformatics into life science curriculum at all levels; (3) assessment of outcomes; and (4) preparation of faculty trained in life sciences to deliver bioinformatics-based curricula.
One of NIBLSE’s primary accomplishments was to develop a community-driven set of competencies for undergraduate bioinformatics instruction (Wilson Sayres et al., 2018). These competencies were based in part on a national survey of life sciences undergraduate faculty conducted in 2016, which gathered information about the nature of bioinformatics education in the US. The survey also provided the first deep investigation of the barriers that undergraduate life sciences faculty face in efforts to integrate bioinformatics (Williams et al., 2019). The barriers questions were open-response items that were then investigated by keyword analysis to quantify thematic responses. We grouped the reported barriers into several “super-categories”: faculty issues (e.g., lack of training, competing time commitments), student issues (e.g., lack of adequate math preparation, lack of adequate coding experience, lack of student interest), curriculum issues (e.g., lack of time for course development, lack of time for inclusion in an overfilled curriculum, speed at which materials and tools change) and less frequently, facilities, resource, and institutional issues (Williams et al., 2019).
The results from the 2016 survey included several key findings (Williams et al., 2019). Overall, lack of expertise and training at the faculty level was identified as the most common barrier among respondents followed by a lack of prerequisite skills and knowledge at the student level. Furthermore, the reported prevalence of barriers varied by demographic groups. In particular, women and faculty identifying as under-represented minorities (URM) reported barriers at higher rates than their comparison groups, men and non-URM faculty.
In the current study, we investigated whether the landscape of barriers for implementing bioinformatics in undergraduate curricula has changed since 2016 by conducting a new national survey followed by focus group discussions. Results from the 2016 survey were used to design Likert-scale items to explore faculty perceptions of specific barriers. For example, in the keyword analysis approach in 2016, many respondents included the word “time” as a barrier but upon reading the responses, time meant different things (time for course development, time/space in the curriculum, time for training), so we used more specific language to explore these issues further in the current survey. One impetus for the study was to further investigate the extent to which barriers to integration of bioinformatics were perceived by faculty who are members of under-represented minority groups and/or faculty at minority-serving institutions (MSIs). Because the number of survey respondents from these faculty was limited, we augmented the information collected in the new survey with a series of focus groups to understand these faculty experiences in more detail. In summary, many of the same barriers still exist although a greater proportion of faculty are in fact attempting to incorporate bioinformatics. Their efforts can be supported by faculty training and up-to-date curricular materials (Ryder et al., 2020; Drew et al., 2021; Kleinschmit et al., 2021, 2023), efforts in which NIBLSE continues to engage.
Materials and methods
Solicitation of survey participants
A mailing list of approximately 13,000 life sciences faculty at 2- and 4-year US institutions was purchased from ExactData.com, a targeted, direct-marketing company that maintains lists of U.S. college and university faculty. As we were particularly interested in responses from faculty from historically underrepresented minority (URM) groups in STEM (e.g., African Americans, Hispanics, Native Americans and Native Alaskans, and Pacific Islanders), we included all available URM biology faculty records from ExactData, approximately 3,000 individual entries. The balance of the records was non-URM faculty. Among other information, each record contained the name, e-mail address, ethnicity, gender, and current institution of the individual.
Using data from the Carnegie Classification of Institutions of Higher Education,2 information about an individual’s current institution was added to each record, including basic Carnegie classification, size and setting, “control” (whether the institution is public or private), and whether the institution is an HBCU, Tribal, Hispanic-serving, Minority-serving, or women-only institution. For ease of data analysis, the 34 basic Carnegie classifications were collapsed into nine: (1) Not classified; (2) Associate’s Colleges; (3) Baccalaureate/Associate’s Colleges: Associate’s Dominant; (4) Doctoral/Professional Universities; (5) Master’s Colleges and Universities; (6) Baccalaureate Colleges; (7) Baccalaureate/Associate’s Colleges: Mixed Baccalaureate/Associate’s; (8) Special Focus Four-Year; and (9) Tribal Colleges. Similarly, the 19 Carnegie size and setting classifications were binned into five: (1) Not classified; (2) Small or Very Small; (3) Medium; (4) Large or Very Large; and (5) Exclusively graduate/professional.
The 2022 barriers survey
A 24-item survey instrument, informed by the previous 2016 NIBLSE survey (Williams et al., 2019), was developed to probe the barriers faculty face in integrating bioinformatics into their teaching (See Supplementary material 1). The survey began with a consent statement, then contained several types of items (multiple-choice, free-response write-ins, and Likert-scale) organized into three blocks: (1) questions that asked how much bioinformatics the respondent currently included in their teaching, (2) the barriers they encountered when doing so, and (3) self-reported demographics including degree(s), current institution(s) and academic role, and extent of bioinformatics training. To streamline the survey for respondents, branching logic was used when appropriate to filter questions (for example, respondents who indicated no barriers to integrating bioinformatics did not see follow up questions about specific barriers).
The survey was administered using Qualtrics (Qualtrics, Provo, UT, 2020), after approval from the University of Florida’s Institutional Review Board (UF IRB#202102370). Following recommendations from Dillman et al. (2014), a link to the survey was distributed by email to the 13,398 individuals in the mailing list on November 4, 2021 and remained open until January 10, 2022 with three reminders sent throughout that period. There were 563 responses to the survey. All potentially identifying information was removed from the data before analysis.
Survey analysis methods
Data from respondents who did not teach undergraduate courses were removed from the data set, as was data from individuals who did not provide consent, even if they completed other questions in the survey. As in Williams et al. (2019), the barriers reported by respondents were analyzed with respect to several demographic criteria including sex, race/ethnicity, year of highest degree, level of bioinformatics training, extent of current bioinformatics teaching, institutional Carnegie Classification, and MSI status. For a given demographic, respondents who did not answer were not included in that demographic category.
Data analysis was performed in R version 4.1.2 (R Core Team, 2021) (Refer to our Supplementary material at https://github.com/niblse/barriers_survey_2022 for all R scripts used in this analysis). Data preparation, statistical summation, and graph preparation were performed using tidyverse (Wickham et al., 2019). Statistical tests (chi-squared, difference in proportions) were performed using infer R package (Couch et al., 2021). The false discovery rate method of Benjamini and Hochberg (1995) was used to adjust p-values for multiple hypothesis testing when appropriate.
Focus groups
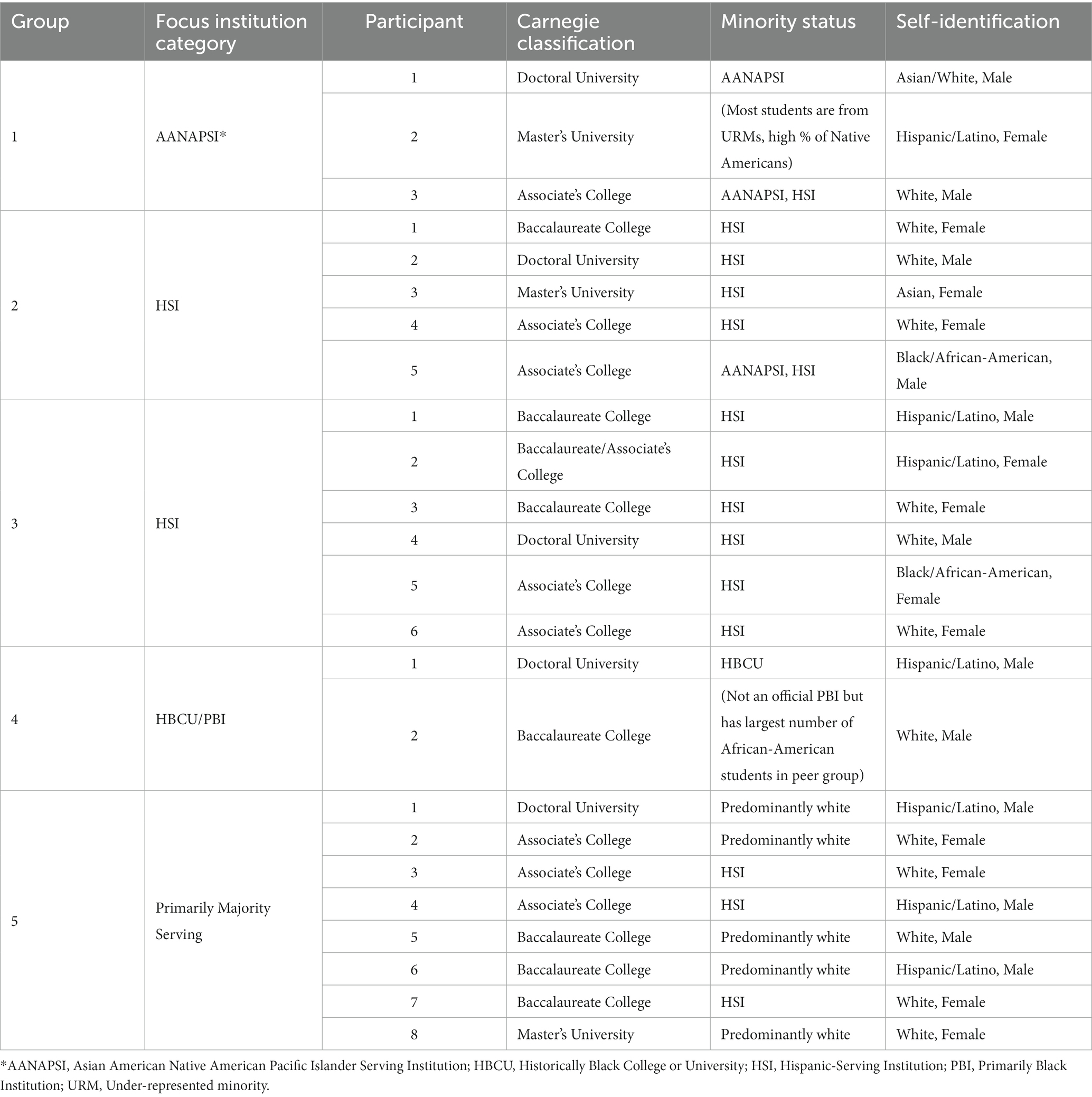
Five focus groups, each 60–90 min long (Saldana and Omasta, 2022), were conducted with life sciences faculty about the perceived barriers to integrating bioinformatics into their courses. IRB approval for the focus groups was sought from Georgetown University’s Institutional Review Board (GU IRB# STUDY00005685), and the study was deemed exempt. In order to volunteer, participants were asked to complete a secure electronic application form (Supplementary material 2), which explicitly served as consent. A link to the form was sent to individuals on the mailing list. In addition, a flier containing a QR code that linked to the application form was disseminated by social media (specifically, Twitter) and at the 2022 Society for Advancement of Chicanos/Hispanics & Native Americans in Science meeting to elicit additional volunteers. All individuals who completed the form were encouraged to send the form to others who might also be interested (“snow-ball” sampling). A total of 118 individuals responded to the form. Of these, 35 individuals were invited to participate based on their availability and the type of institution at which they taught (as listed in Table 1) as we aimed for faculty at a diverse set of institutions. As an incentive for participation, those who participated were sent a NIBLSE-branded beverage set.
Ultimately, 23 faculty participated in the focus groups, the majority of whom were from Minority-Serving Institutions (Table 1). One focus group was comprised primarily of faculty at Asian-American and Native Pacific Islander serving institutions (AANPI), two groups were comprised of faculty at Hispanic-Serving Institutions (HSIs), one group was comprised of mostly faculty at Historically Black Colleges and Universities (HBCUs) or Primarily Black Institutions (PBIs). The final focus group was composed of faculty from majority-serving institutions, several of whom identified as either African-American or Hispanic (Table 1). The focus group participants also taught at a variety of institution types, including community colleges (Table 1).
The semi-structured focus groups were conducted by a facilitator from Georgetown’s Center for New Designs in Learning and Scholarship. An interview guide [written with reference to Saldana and Omasta (2022)] was used by the facilitator (Supplementary material 3) but spontaneous conversations among the participants were encouraged. At least one member of the NIBLSE Leadership Team sat in on each focus group and, as disciplinary experts, occasionally interjected with questions. The focus groups were conducted on Zoom and recorded. The Zoom-derived transcripts of the recordings were analyzed using simple thematic analysis (Harding, 2013) to identify quotes that illustrated themes uncovered by the survey and the focus groups themselves. The quotes were edited to remove stammers, repetitions, and grammatical errors.
Results
The survey
A broad spectrum of educators participated in the 2022 NIBLSE survey
Altogether, 556 individuals responded to the current NIBLSE survey on barriers to integrating bioinformatics in undergraduate life science education. The 509 educators who agreed to participate in the study and taught undergraduate students were from diverse demographic groups and institution types, most often with some training in bioinformatics (Figure 1). The composite survey respondent is a woman or man of European descent with a PhD earned in 2000–2009, with some bioinformatics experience, working at a non-minority-serving, doctoral-granting, large institution.
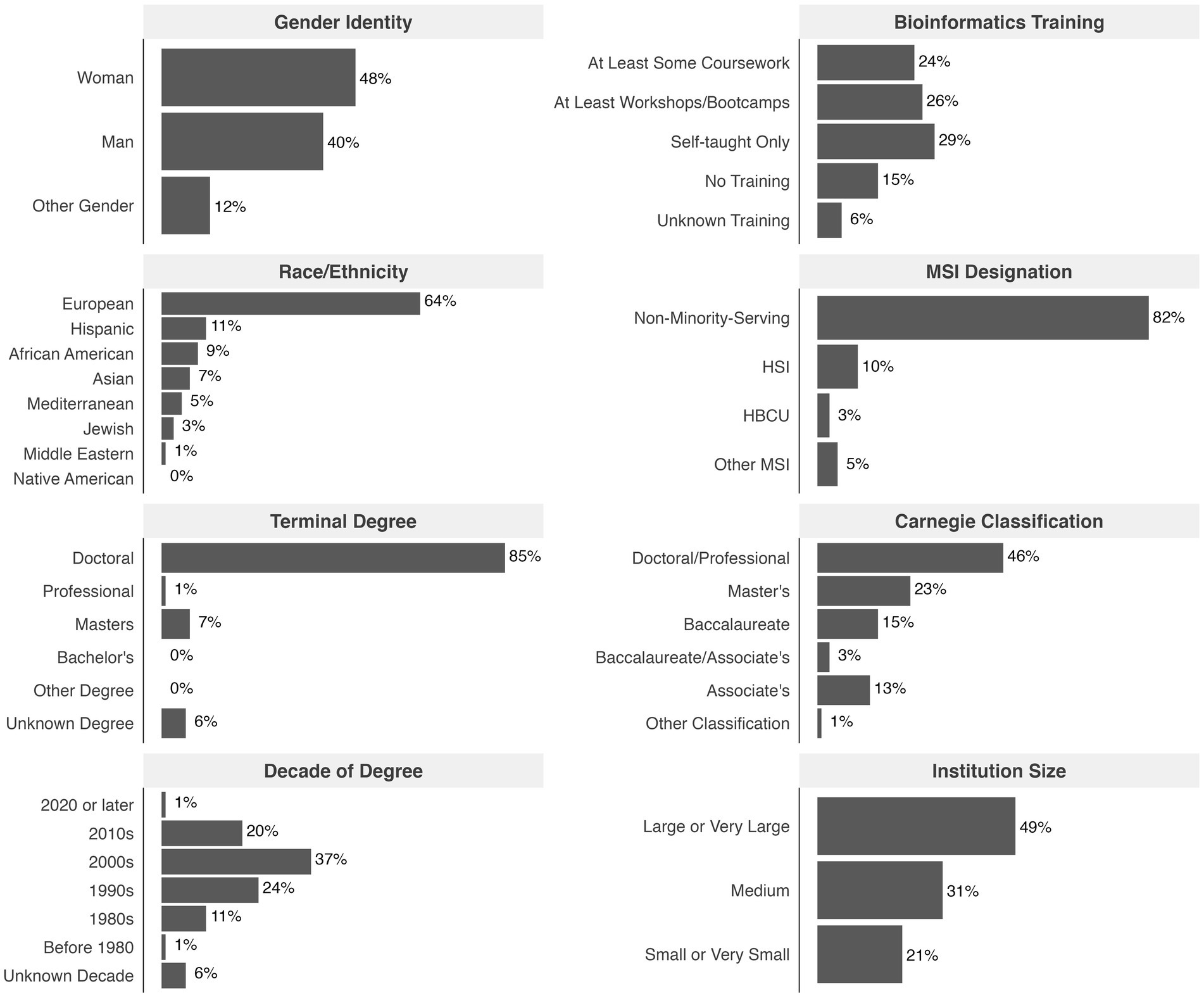
Figure 1. Summary of respondent demographics. Summary demographics shown as percentages of participants (n = 509, the total number of respondents teaching undergraduates who agreed to participate). Participants self-identified gender and race/ethnicity; European included Western European, Eastern European, and Scandinavian; Asian included East Asian, South Asian, and Southeast Asian. Participants could select multiple items for bioinformatics training and were categorized based on most formal training (e.g., if coursework and workshops/bootcamps marked, then indicated as “At Least Some Coursework”).
More undergraduate bioinformatics education is needed, but instructors face barriers
While 90% of those who answered (435 of 484 respondents) agreed that more bioinformatics content is needed in undergraduate courses at their institutions, over 20% (n = 119) of the 509 survey participants did not plan to integrate bioinformatics into their courses. The most frequently cited reasons were because teaching bioinformatics was not their responsibility or appropriate for their course (n = 51), but many others said there were too many barriers (n = 31). On the other hand, over 60% (n = 315) of the survey participants currently teach some bioinformatics in a life science course (twice the rate seen in the 2016 survey), and 30% of those (n = 100) within a classroom-based undergraduate research experience (CURE) or a summer undergraduate research experience (SURE).
Nevertheless, integrating bioinformatics into undergraduate life science courses continues to be a challenge. Over 70% (259 of 362 respondents) of the instructors teaching bioinformatics or who plan to do so reported facing barriers. In particular, instructors identified students lacking prerequisite skills and lack of time to restructure course content as minor to severe challenges more frequently than the other barriers (adj. value of p < 0.01; Figure 2). In contrast, instructors cited not having the autonomy to add course content and lack of student interest less frequently as challenges (adj. value of p < 0.01).
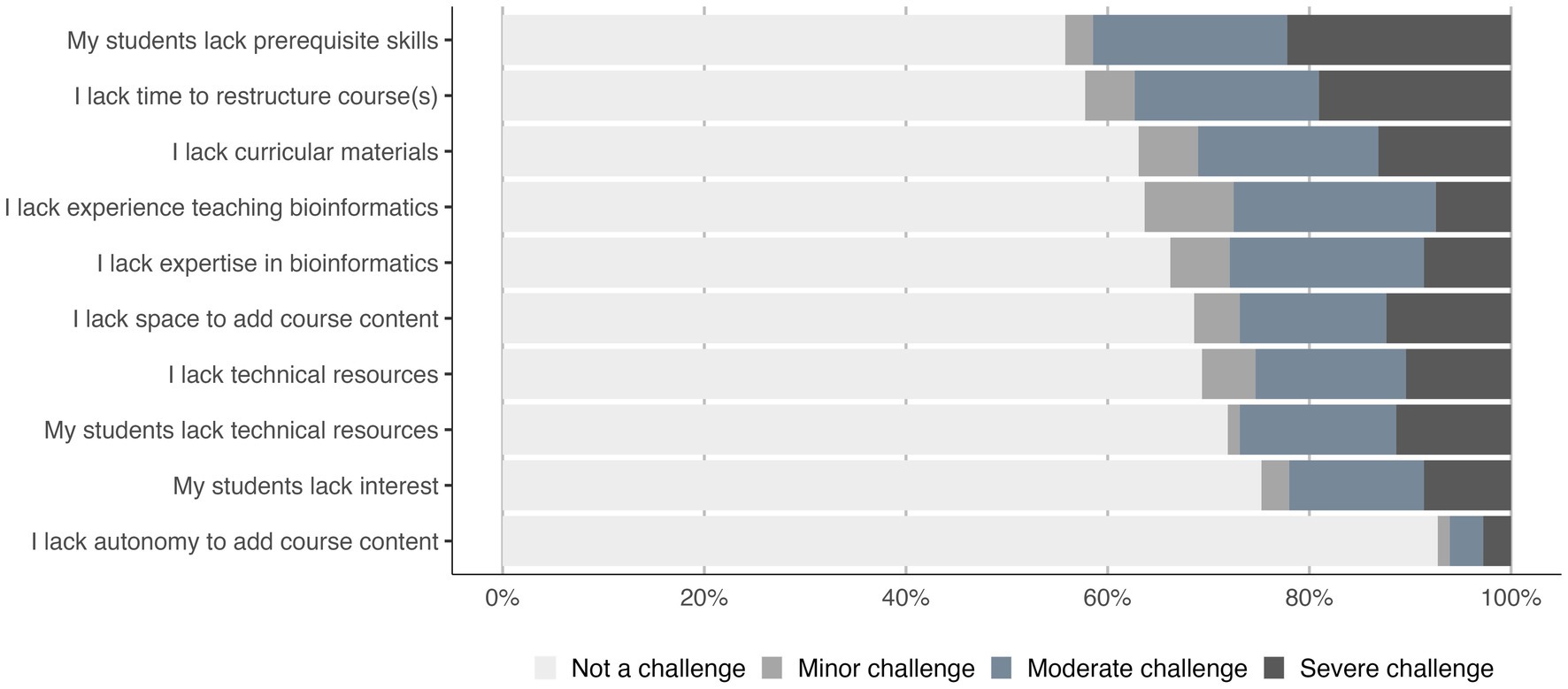
Figure 2. Some barriers were more challenging than others. The frequency of each challenge category is shown for the 10 queried barriers (n = 509).
Women experience more barriers than men
We previously reported in 2016 that women engaged in undergraduate life science education face barriers more frequently than men (Williams et al., 2019). We saw a similar pattern in this 2022 survey with women (and other gender identities) more likely to report barriers to integrating bioinformatics into their teaching than men (Men = 61%, Women = 78%, Others = 77%; value of p < 0.01). For the 10 queried barriers, women and other gender identities more often identified lack of bioinformatics expertise and lack of curricular materials as challenges than men did (adj. value of p < 0.01; Figure 3).
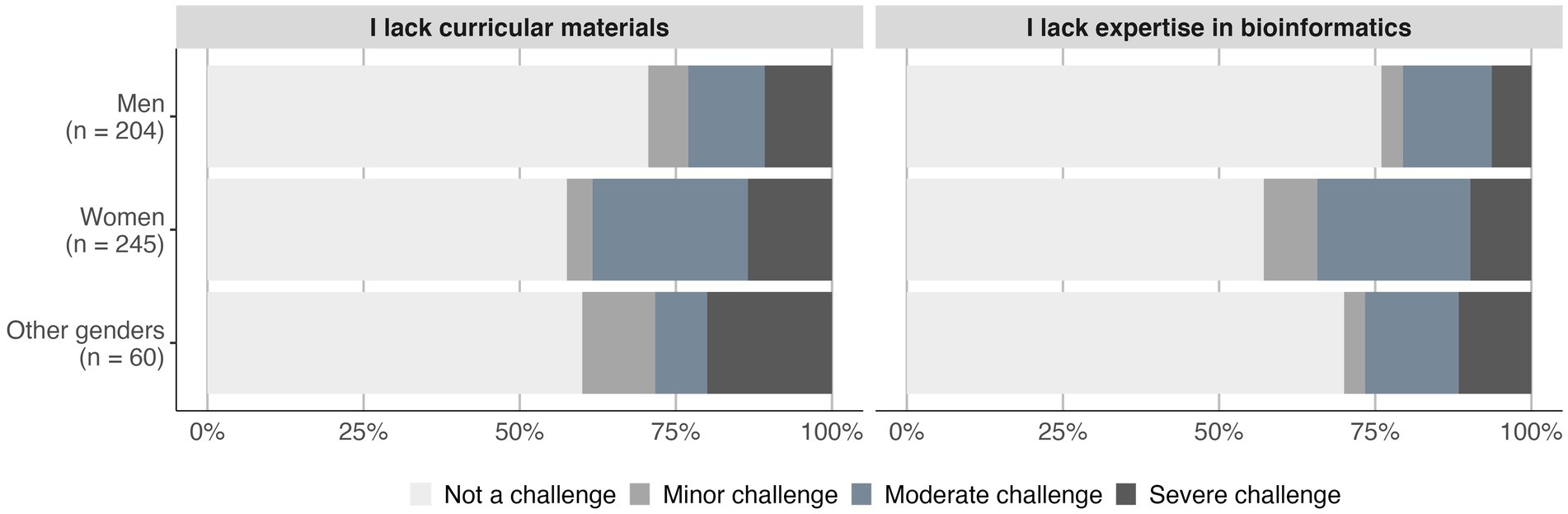
Figure 3. Two barriers were significantly more challenging for women and other genders than men. The frequency of each challenge category is shown for the barriers that differed significantly among gender identities (chi-squared test; adj. value of p < 0.01).
The challenge of integrating bioinformatics varies with Carnegie classification
We reported in 2016 that the frequency of identified barriers diverged between instructors at different institution types (Williams et al., 2019). Similarly, in the 2022 survey we found a significant association between Carnegie classification and encountering barriers (value of p = 0.036). Faculty at baccalaureate colleges reported facing barriers more frequently than the overall faculty average (82.3% vs. 71.4%), while faculty at doctoral institutions reported facing barriers less frequently (64.5%). In particular, the frequency that a lack of expertise in bioinformatics was identified as a challenge varied between faculty at different institution types (adj. value of p < 0.01; Figure 4). No significant difference between institution types was observed for the other queried barriers.
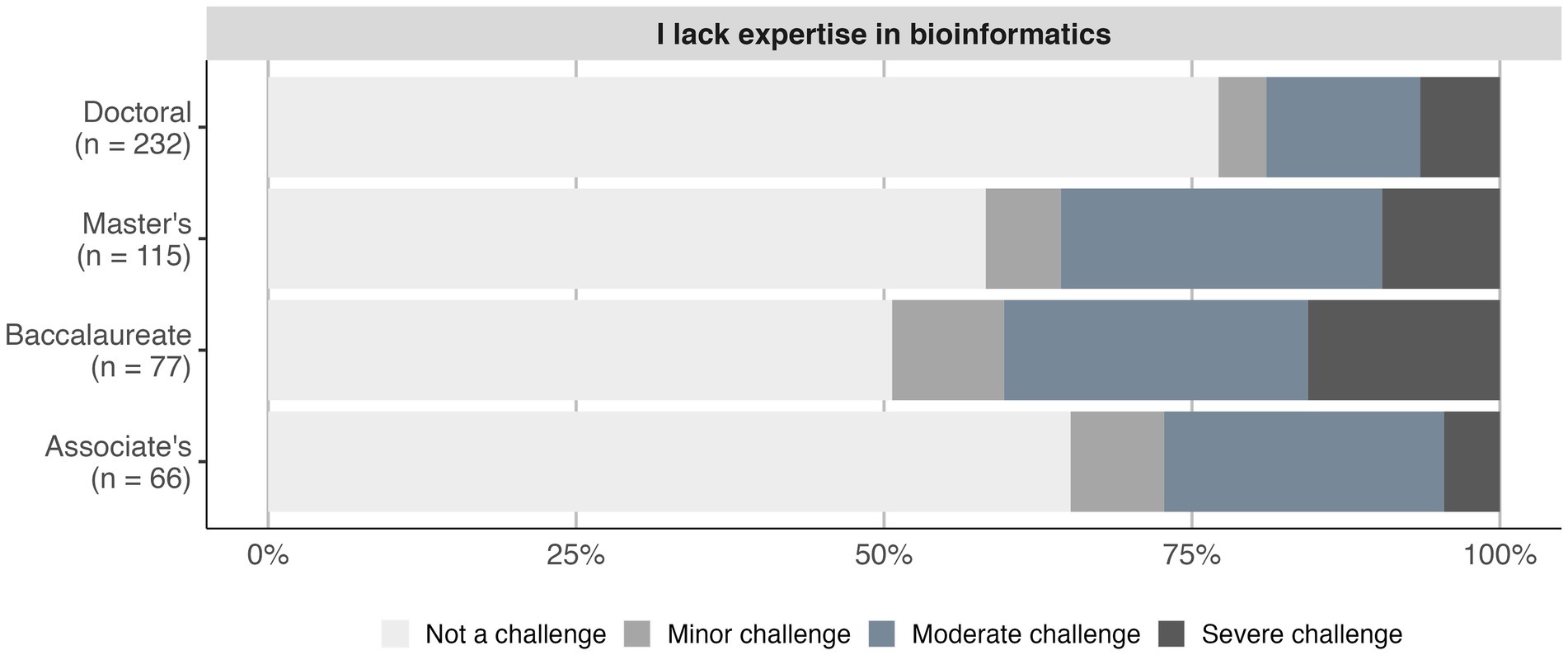
Figure 4. Faculty at baccalaureate colleges more frequently cited a lack of bioinformatics expertise as a challenge. The frequency of each challenge category (none to severe) is shown for the barriers that differed significantly among Carnegie classifications with at least 50 respondents (chi-squared test; adj. value of p < 0.01).
Some MSI faculty report a higher frequency of barriers than non-MSI faculty
Similar to findings in the 2016 survey, we found in the 2022 survey that while faculty at historically-black colleges and universities (HBCUs) or Hispanic-serving institutions (HSIs) more frequently reported barriers to integrating bioinformatics into their teaching than faculty elsewhere (HBCU = 82%, n = 17; HSI = 81%, n = 37; other-HSI = 65%, n = 20; non-MSI = 73%, n = 306), the differences were not statistically significant.
Faculty with more recent degrees have more bioinformatics training but are equally likely to integrate bioinformatics in their courses
We reported in 2016 that faculty receiving their terminal degree more recently were more likely to have had formal bioinformatics training, but less likely to integrate bioinformatics than faculty who obtained their degrees a decade or more earlier (Williams et al., 2019). The 2022 survey also found that faculty with more recent terminal degrees were more likely than more experienced faculty to have some training, including one or more courses in bioinformatics (value of p < 0.001; Figure 5A). Despite having more extensive formal training in bioinformatics, faculty with more recent terminal degrees somewhat more often (but not significantly) reported facing challenges to integrating bioinformatics (Figure 5B). Nevertheless, in the 2022 survey we did not see an association between the decade of receiving the terminal degree and integrating at least some bioinformatics instruction in life science classes (Figure 5C). Overall, however, faculty with some bioinformatics training were much more likely to integrate bioinformatics instruction (72%, n = 403) than those with no training (12%, n = 76, value of p <0.001).
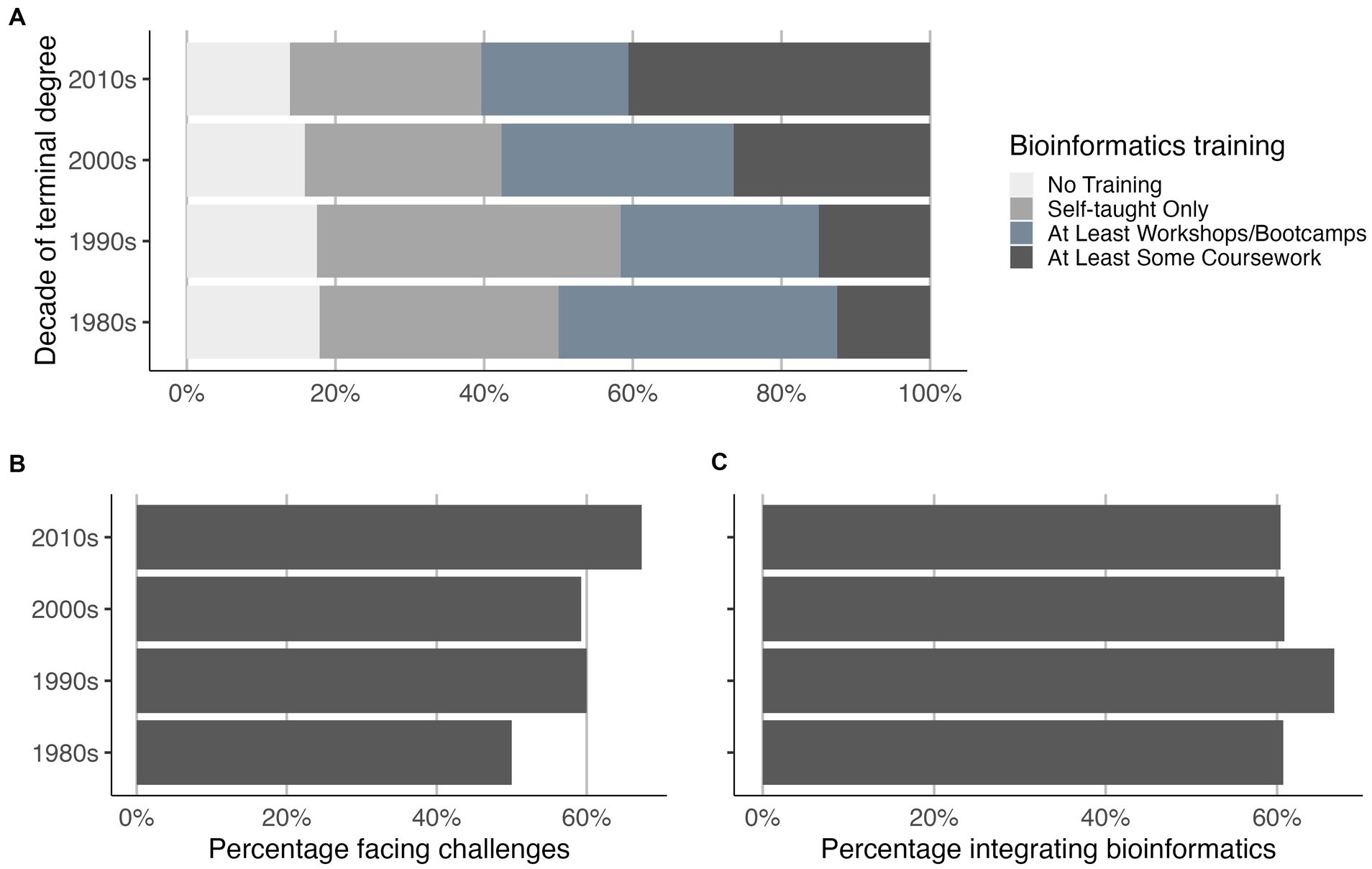
Figure 5. Relationship of decade of terminal degree with bioinformatics training, barriers, and teaching bioinformatics. Faculty who received their terminal degrees more recently (A) more frequently had some course work in bioinformatics but were (B) more likely to report barriers to integrating bioinformatics. Nevertheless, respondents integrated bioinformatics at nearly the same frequency regardless of decade of terminal degree (C). The small number of instructors who obtained their terminal degree before 1980 or after 2019 were excluded from this analysis. Participants could select multiple items for bioinformatics training and were categorized based on most formal training (e.g., if coursework and workshops/bootcamps marked, then indicated as “At Least Some Coursework”).
URM and some MSI faculty report a higher frequency of barriers
The 2016 survey suggested that faculty who are members of underrepresented minority (URM) groups and/or at minority-serving institutions (MSIs) experienced barriers at a higher rate compared to other participants (Williams et al., 2019). While URM faculty more frequently reported barriers to integrating bioinformatics into their teaching than non-URM faculty (URM = 81%, n = 77; non-URM = 72%, n = 317), the 2022 survey differences were not statistically significant. Similarly, while faculty at historically-black colleges and universities (HBCUs) or Hispanic-serving institutions (HSIs) more frequently reported barriers to integrating bioinformatics into their teaching than faculty elsewhere (HBCU = 82%, n = 17; HSI = 81%, n = 37; other-HSI = 65%, n = 20; non-MSI = 73%, n = 306), the differences were also not statistically significant. We explored these trends further in the focus groups (see below).
The focus groups
In addition to the survey, we conducted 5 different focus groups with faculty who are attempting to integrate bioinformatics into their teaching. Using a semi-structured format, which included a common script (Supplementary material 3) but allowed the conversations to follow the flow of thoughts, we gained insights into the perceived barriers for faculty and their students. Although we initially arranged the focus groups based on the type of institution (minority-serving or not), common themes emerged across all 5 focus groups, and participants in a given focus group were generally in agreement when discussing the various questions.
Is bioinformatics instruction important for students?
Each session started by asking the participants why they felt it was important to teach bioinformatics to their students. There were a range of responses, but most felt it was important for students to have the data analysis skills necessary for future work or advanced training. Several participants acknowledged they had no hard data regarding the extent to which students were more attractive as candidates for work or higher education, but their impression was that students with some data analysis skills had more opportunities available to them (Table 2). Several faculty members talked about the changes in professional health fields specifically as an impetus for including bioinformatics in their teaching.
What barriers do the students exhibit?
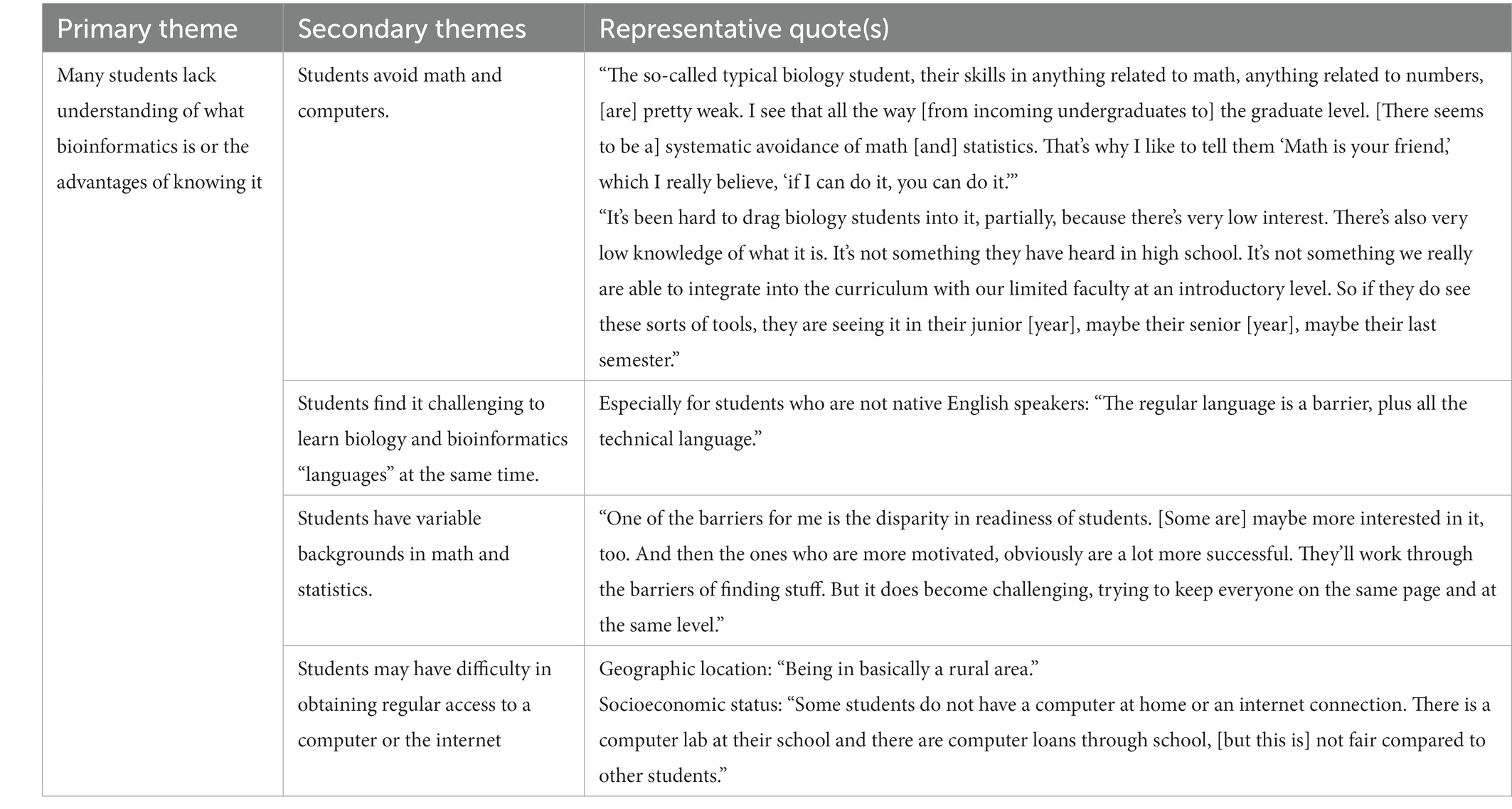
We then asked participants what barriers they encountered with respect to their students. Many said most students do not really understand what bioinformatics is or appreciate the advantages of knowing something about bioinformatics or data science (Table 3). However, several individuals also said that once students had some exposure, they understood better and “got into” it. On the other hand, one participant in Focus Group 3 suggested that some of the savvier students are aware of bioinformatics and data science and are pushing for access to courses.
[There is] a lot of interest from around the university, but my classes are [currently] over full. [Some of] my students have that drive to really push their computer skills and have already recognized the economic need. But there is a huge proportion of our students who are graduating without ever having taken a formal computer-focused course. And so the phrase that I keep bringing up in meetings with administrators is, do you want our graduates to be able to visualize data in spreadsheets and have installed programs on a computer before? [There are] some essential basic skills that are easily integrated into a more formalized education in my field, biology, and how [they] interact with computer systems.
Thus, some biology students are aware of the advantages of learning about bioinformatics, but such students appear to be in the minority, as this statement was not echoed by other members of this group or members of the other groups.
Some participants talked about technical and logistical barriers that students face, such as the difficulty students at their institutions have with regular access to the internet or computers. Reasons for this difficulty included geographic location and socioeconomic status of the students.
What prevents you from teaching bioinformatics?
In the course of listening to the focus group participants, we learned that many faculty, in fact, have some experience teaching bioinformatics but they still encountered a number of difficulties (Table 4).

Table 4. Responses to the question, “What barriers do you have in attempts to incorporate bioinformatics into your teaching?”
First, many faculty are alone in trying to teach bioinformatics at their institution. On the other hand, some participants are working to develop programs at their institutions and are involved in training others. Second, although a number of people said they incorporated some bioinformatics, virtually everyone said that they found it difficult to keep up with this fast-moving field. Instructors have a range of abilities to teach bioinformatics; for example, some have programming expertise and can convey these skills to their students, while others incorporate only web-based tools that require no coding skills. In short, professional development is still important for many. Third, many expressed the thought that there are too many things to cover in the curriculum already. Finally, several participants mentioned the need to get administrative support in efforts to implement bioinformatics more broadly as well as the need for technical support, including computers with operating systems capable of running the necessary software.
Discussion
We conducted a new survey to understand the barriers undergraduate life sciences faculty face in efforts to bring bioinformatics to the classroom and to determine to what extent the barriers observed in our 2016 study (Williams et al., 2019) are still present. We augmented the findings in the 2022 survey with focus groups composed of faculty from minority- and majority-serving institutions of different sizes and Carnegie classifications. The qualitative information gleaned from the focus groups corroborated the survey data.
As before, the vast majority of respondents agreed that bioinformatics is needed in life sciences curricula. We had more than 500 respondents in the current (2022) survey, about 40% of the number of respondents in the 2016 survey. Importantly, in the 2016 survey, only about 30% of participants indicated they were incorporating bioinformatics, while in this survey more than 60% of participants were doing so. This is a significant increase (chi-squared; p < 0.01) in the ensuing 6 years. Nevertheless, barriers are still apparent. The majority of respondents reported facing barriers to integrating bioinformatics, although only a handful of barriers were identified as presenting moderate or severe challenges (Figure 2). In particular, instructors more frequently cited lack of experience teaching bioinformatics, time to restructure course content, and lack of student prerequisite skills. These sentiments were echoed in the focus group conversations (Tables 3, 4).
As in the 2016 survey (Williams et al., 2019), in the 2022 survey, women experienced more barriers than men (Figure 3). These findings suggest that women in particular would benefit from targeted professional development opportunities. In cooperation with QUBES,3 NIBLSE will continue to offer bioinformatics-focused Faculty Mentoring Networks (FMNs) (Ryder et al., 2020; Kleinschmit et al., 2023). Facilitated by faculty with expertise in the topic under consideration, FMNs support groups of educators working over the course of a semester on a defined task. For example, NIBLSE-led FMNs have assisted faculty with integrating and adapting an existing bioinformatics exercise to their own classroom learning goals.
The 2016 survey suggested that URM faculty experience barriers at higher rates than non-URM faculty; however, the number of URM respondents was too low to detect statistical significance. Therefore, for this survey (as well as the focus groups), we targeted URM faculty (who composed approximately 23% of the names on the mailing list). In the 2022 study, we had about the same absolute number of URM respondents as in the 2016 study, but they were a larger percentage of the total. While we observed the same trend in the current survey, suggesting that URM faculty might experience barriers at higher rates, the results were not statistically significant again this time due to insufficient sample size. In addition, in the 2016 survey (Williams et al., 2019), faculty at MSIs showed a slight trend toward fewer barriers than faculty at majority institutions. When we examined this issue with the current survey data, there was a slight trend in the opposite direction, that MSI faculty experienced barriers at a slightly higher rate than faculty at majority institutions, although again, the results were not statistically significant. Nevertheless, making opportunities for professional development and curricular support available to these faculty would also be advantageous. For example, it was previously reported that faculty at MSIs are disadvantaged in efforts to seek federal funding (Chavela Guerra and Wilson, 2021), yet a recent report from the National Academies of Science, Engineering, and Medicine documents that MSIs are an underutilized resource for strengthening STEM fields in the US (National Academies of Sciences, Engineering, and Medicine, 2019).
When comparing faculty at different institution types, faculty at baccalaureate institutions experienced the most barriers, followed by those at master’s institutions (Figure 4). Based on the focus groups, such faculty are likely to be alone in their efforts to incorporate bioinformatics into their teaching and would benefit from participating in networks outside of their home institution. One opportunity for network participation by undergraduate instructors is through FMNs (see above); others are cross-institutional course-based undergraduate research experience (CURE) and summer undergraduate research experience (SURE) initiatives incorporating bioinformatics components, such as the Genomics Education Partnership (GEP) (Elgin et al., 2017), Genome Solver (Mathur et al., 2019), Prevalence of Antibiotic-Resistance in the Environment (PARE) (Fuhrmeister et al., 2021; Bliss et al., 2023), SEA-PHAGES (Hanauer et al., 2017), Small World Initiative (SWI,4) and Tiny Earth (Hurley et al., 2021). These programs provide centralized support for faculty, including training and professional development, as well as access to computational resources, all of which address barriers identified in the survey and articulated in the focus groups, and may be of particular help to faculty at smaller institutions (Muth and McEntee, 2014). In addition, several of these projects promote development of bioinformatics skills in students as early as freshman year, allowing for growth of skills over the ensuing undergraduate years. While the finding that one-third of faculty integrating bioinformatics do so through some type of undergraduate research experience indicates that many faculty already perceive the value of this approach, expansion of such programs would further promote efforts to support bioinformatics education. Somewhat surprisingly, faculty at associate’s colleges said they experienced fewer barriers than faculty at baccalaureate or master’s institutions. The reasons for this are presently unclear but might reflect differences in content delivered, clientele served, and/or emphasis on practical skills for job seekers.
We found in the 2016 survey (Williams et al., 2019) that faculty who had received their terminal degree more recently had more formal training in bioinformatics than faculty who received their degrees in previous decades. At the same time, more recently awarded degree recipients experienced more barriers in attempts to implement bioinformatics into their teaching. These findings were replicated in the current survey (Figure 5A) and may reflect the more limited teaching experience of newer professors. Alternatively, these results might suggest that there are structural barriers, not explicitly investigated in this study, that hinder progress. It will be necessary to gather information from administrators and other stake-holders to examine such barriers in more detail, but this echoed comments from the focus group conversations that stated administrative support was necessary for expansion of bioinformatics offerings to students.
Arguably, the most difficult barrier to address and overcome for the integration of bioinformatics is lacking the autonomy to add course content (Figure 2). Addressing this issue is typically beyond the reach of individual instructors as changing departmental culture and acquiring administrative approval is necessary. Interestingly, however, respondents cited this potential barrier least frequently. The other barriers identified were those over which individual faculty have at least some autonomy whether it be seeking external assistance with curricula and/or professional development or reprioritizing individual commitments. Information from the focus groups suggested that students often believe they are not interested in bioinformatics upon initial introduction, but after they are exposed to the power of these concepts to address biological questions, as well as the appropriate tools and techniques, they tend to be very interested in further exploration. Likewise, although prerequisite student skills are perceived to be a barrier, incremental exposure to introductory quantitative techniques through a bioinformatics lens by individual instructors could be an effective mechanism to build needed skills.
In summary, in the ensuing interval since our first survey in 2016, notable progress has been made. About twice as many faculty are incorporating at least some bioinformatics learning into their teaching. However, as with our previous results, undergraduate faculty nearly uniformly signaled the need for training in bioinformatics and help in developing curricular resources to teach bioinformatics as revealed in both the survey data and the conversations in the focus groups. NIBLSE has been working in both arenas to address these barriers (Ryder et al., 2020; Kleinschmit et al., 2023). As mentioned previously, other groups have also made efforts to address these barriers, including GEP, SEA-PHAGES, SWI, Tiny Earth, PARE, etc. These groups also have curriculum available for use. The combination of these ongoing initiatives provides a robust framework for future efforts to ensure that tomorrow’s researchers possess the necessary skills and attitudes for conducting research in the 21st century.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by the University of Florida IRB and the Georgetown University IRB. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JD: Conceptualization, Data curation, Methodology, Writing – original draft, Writing – review & editing. WM: Data curation, Formal analysis, Funding acquisition, Methodology, Visualization, Writing – original draft, Writing – review & editing. SG: Conceptualization, Methodology, Writing – review & editing. AK: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. MM: Investigation, Writing – review & editing. MP: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. ET: Conceptualization, Funding acquisition, Writing – review & editing. JW: Conceptualization, Formal analysis, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. BM: Writing – review & editing. AR: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Previous NIBLSE network activities were funded by the National Science Foundation, grant number 1539900.
Acknowledgments
The authors acknowledge the contributions of past members of the NIBLSE Leadership Team, Elizabeth Dinsdale and William Tapprich. We also thank Sam Donovan, Liz Ryder, and Jessica Siltberg-Liberales for their comments and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2023.1317191/full#supplementary-material
Footnotes
References
Altman, R. B. (1998). A curriculum for bioinformatics: the time is ripe. Bioinformatics 14, 549–550. doi: 10.1093/bioinformatics/14.7.549
Attwood, T. K., Blackford, S., Brazas, M. D., Davies, A., and Schneider, M. V. (2019). A global perspective on evolving bioinformatics and data science training needs. Brief. Bioinform. 20, 398–404. doi: 10.1093/bib/bbx100
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Bliss, S. S., Abraha, E. A., Fuhrmeister, E. R., Pickering, A. J., and Bascom-Slack, C. A. (2023). Learning and stem identity gains from an online module on sequencing-based surveillance of antimicrobial resistance in the environment: an analysis of the Pare-Seq curriculum. PLoS One 18:E0282412. doi: 10.1371/journal.pone.0282412
Carvalho, B. S., and Rustici, G. (2013). The challenges of delivering bioinformatics training in the analysis of high-throughput data. Brief. Bioinform. 14, 538–547. doi: 10.1093/bib/bbt018
Chavela Guerra, R. C., and Wilson, C. (2021). From lack of time to stigma: barriers facing faculty at minority-serving institutions pursuing federally funded research. ASEE Virtual Ann. Conf. Available at: https://par.nsf.gov/biblio/10304434
Couch, S. P., Bray, A. P., Ismay, C., Chasnovski, E., Baumer, B. S., and Çetinkaya-Rundel, M. (2021). Infer: an R package for tidyverse-friendly statistical inference. J. Open Source Softw. 6:3661. doi: 10.21105/joss.03661
Dillman, D. A., Smyth, J. D., and Christian, L. M. (2014). Internet, Phone, Mail, and Mixed Mode Surveys: the Tailored Design Method. Hoboken, NJ: John Wiley & Sons, Inc.
Dinsdale, E., Elgin, S. C., Grandgenett, N., Morgan, W., Rosenwald, A., Tapprich, W., et al. (2015). NIBLSE: a network for integrating bioinformatics into life sciences education. CBE Life Sci. Educ. 14, 1–4. doi: 10.1187/cbe.15-06-0123
Drew, J. C., Grandgenett, N., Dinsdale, E. A., Vazquez Quinones, L. E., Galindo, S., Morgan, W. R., et al. (2021). There is more than multiple choice: crowd-sourced assessment tips for online, hybrid, and face-to-face environments. J. Microbiol. Biol. Educ. :e00205-21:22. doi: 10.1128/jmbe.00205-21
Elgin, S. C. R., Hauser, C., Holzen, T. M., Jones, C., Kleinschmit, A., Leatherman, J., et al. (2017). The Gep: crowd-sourcing big data analysis with undergraduates. Trends Genet. 33, 81–85. doi: 10.1016/j.tig.2016.11.004
Elgin, S. C., Hays, S., Mingo, V., Shaffer, C. D., and Williams, J. (2021). Building back more equitable stem education: teach science by engaging students in doing science. bioRxiv. doi: 10.1101/2021.06.01.446616
Fuhrmeister, E. R., Larson, J. R., Kleinschmit, A. J., Kirby, J. E., Pickering, A. J., and Bascom-Slack, C. A. (2021). Combating antimicrobial resistance through student-driven research and environmental surveillance. Front. Microbiol. 12:577821. doi: 10.3389/fmicb.2021.577821
Gauthier, J., Vincent, A. T., Charette, S. J., and Derome, N. (2019). A brief history of bioinformatics. Brief. Bioinform. 20, 1981–1996. doi: 10.1093/bib/bby063
Hanauer, D. I., Graham, M. J., Sea, P., Betancur, L., Bobrownicki, A., Cresawn, S. G., et al. (2017). An Inclusive Research Education Community (IREC): impact of the Sea-Phages program on research outcomes and student learning. Proc. Natl. Acad. Sci. U. S. A. 114, 13531–13536. doi: 10.1073/pnas.1718188115
Handelsman, J., Elgin, S., Estrada, M., Hays, S., Johnson, T., Miller, S., et al. (2022). Achieving stem diversity: fix the classrooms. Science 376, 1057–1059. doi: 10.1126/science.abn9515
Harding, J. (2013). Qualitative data analysis from start to finish. Thousand Oaks, CA: Sage Publications.
Hurley, A., Chevrette, M. G., Acharya, D. D., Lozano, G. L., Garavito, M., Heinritz, J., et al. (2021). Tiny earth: a big idea for stem education and antibiotic discovery. mBio 12:e03432-20. doi: 10.1128/mBio.03432-20
Isik, E. B., Brazas, M. D., Schwartz, R., Gaeta, B., Palagi, P. M., Van Gelder, C. W. G., et al. (2023). Grand challenges in bioinformatics education and training. Nat. Biotechnol. 41, 1171–1174. doi: 10.1038/s41587-023-01891-9
Kleinschmit, A. J., Rosenwald, A., Ryder, E. F., Donovan, S., Murdoch, B., Grandgenett, N. F., et al. (2023). Accelerating stem education reform: linked communities of practice promote creation of open educational resources and sustainable professional development. IJ Stem Ed 10:16. doi: 10.1186/s40594-023-00405-y
Kleinschmit, A. J., Ryder, E. F., Kerby, J. L., Murdoch, B., Donovan, S., Grandgenett, N. F., et al. (2021). Community development, implementation, and assessment of a NIBLSE bioinformatics sequence similarity learning resource. PLoS One 16:E0257404. doi: 10.1371/journal.pone.0257404
Machluf, Y., and Yarden, A. (2013). Integrating bioinformatics into senior high school: design principles and implications. Brief. Bioinform. 14, 648–660. doi: 10.1093/bib/bbt030
Mathur, V., Arora, G. S., Mcwilliams, M., Russell, J., and Rosenwald, A. G. (2019). The genome solver project: faculty training and student performance gains in bioinformatics. J. Microbiol. Biol. Educ. 20:20.1.4. doi: 10.1128/jmbe.v20i1.1607
Muth, T. R., and Mcentee, C. M. (2014). Undergraduate urban metagenomics research module. J. Microbiol. Biol. Educ. 15, 38–40. doi: 10.1128/jmbe.v15i1.645
National Academies of Sciences, Engineering, and Medicine. (2019). Minority serving institutions: America’s underutilized resource for strengthening the stem workforce. Washington, DC: The National Academies Press.
Niepielko, M. G., and Shumskaya, M. (2021). Early requirement for bioinformatics in undergraduate biology curricula. Front. Bioinform. 1:656531. doi: 10.3389/fbinf.2021.656531
Porter, S. G., and Smith, T. M. (2019). Bioinformatics for the masses: the need for practical data science in undergraduate biology. OMICS 23, 297–299. doi: 10.1089/omi.2019.0080
R Core Team (2021). R: a language and environment for statistical computing. Vienna, Austria: R Foundation For Statistical Computing
Rocha, M., Massarani, L., Souza, S. J., and Vasconcelos, A. T. R. (2022). The past, present and future of genomics and bioinformatics: a survey of Brazilian scientists. Genet. Mol. Biol. 45:E20210354. doi: 10.1590/1678-4685-gmb-2021-0354
Ryder, E. F., Morgan, W. R., Sierk, M., Donovan, S. S., Robertson, S. D., Orndorf, H. C., et al. (2020). Incubators: building community networks and developing open educational resources to integrate bioinformatics into life science education. Biochem. Mol. Biol. Educ. 48, 381–390. doi: 10.1002/bmb.21387
Saldana, J., and Omasta, M. (2022). Qualitative research: analyzing life. Thousand Oaks, CA: Sage Publications
The Genomic Data Science Community Network (2022). Diversifying the genomic data science research Community. Genome Res. 32, 1231–1241. doi: 10.1101/gr.276496.121
Toby, I., Williams, J. J., Lu, G., Cai, C., Crandall, K. A., Dinsdale, E. A., et al. (2022). Making change sustainable: Network for Integrating Bioinformatics into Life Sciences Education (NIBLSE) meeting review. Coursesource 9. doi: 10.24918/cs.2022.10
Wickham, H., Averick, M., Bryan, J., Chang, W., Mcgowan, L. D., Francois, R., et al. (2019). Welcome to the tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686
Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., et al. (2019). Barriers to integration of bioinformatics into undergraduate life sciences education: a national study of us life sciences faculty uncover significant barriers to integrating bioinformatics into undergraduate instruction. PLoS One 14:E0224288. doi: 10.1371/journal.pone.0224288
Keywords: undergraduate education, stem education, bioinformatics, barriers to integration, education reform
Citation: Drew J, Morgan W, Galindo S, Kleinschmit AJ, McWilliams M, Pauley M, Triplett EW, Williams J, Murdoch B and Rosenwald A (2023) Revisiting barriers to implementation of bioinformatics into life sciences education. Front. Educ. 8:1317191. doi: 10.3389/feduc.2023.1317191
Edited by:
Melissa C. Srougi, North Carolina State University, United StatesReviewed by:
Michael J. Wolyniak, Hampden–Sydney College, United StatesHeather Bennett Miller, High Point University, United States
Copyright © 2023 Drew, Morgan, Galindo, Kleinschmit, McWilliams, Pauley, Triplett, Williams, Murdoch and Rosenwald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne Rosenwald, cm9zZW53YWFAZ2VvcmdldG93bi5lZHU=
†ORCID: Adam J. Kleinschmit, https://orcid.org/0000-0003-3820-524X
 Jennifer Drew
Jennifer Drew William Morgan
William Morgan Sebastian Galindo
Sebastian Galindo Adam J. Kleinschmit
Adam J. Kleinschmit Mindy McWilliams
Mindy McWilliams Mark Pauley6
Mark Pauley6 Eric W. Triplett
Eric W. Triplett Barbara Murdoch
Barbara Murdoch Anne Rosenwald
Anne Rosenwald