- Unit of Gynecology and Obstetrics, Fondazione Poliambulanza Hospital, Brescia, Italy
Human endogenous retroviruses (HERVs) are transcribed in many cancer types, including head and neck cancer. Because of accumulating mutations at proviral loci over evolutionary time, HERVs are functionally defective and cannot complete their viral life cycle. Despite that, HERV transcripts, including full-length viral RNAs and viral RNAs spliced as expected at the conventional viral splice sites, can be detected in particular conditions, such as cancer. Interestingly, non-viral–related transcription, including aberrant, non-conventionally spliced RNAs, has been reported as well. The role of HERV transcription in cancer and its contribution to oncogenesis or progression are still debated. Nonetheless, HERVs may constitute a suitable cancer biomarker or a target for therapy. Thus, ongoing research aims both to clarify the basic mechanisms underlying HERV transcription in cancer and to exploit its potential toward clinical application. In this mini-review, we summarize the current knowledge, the most recent findings, and the future perspectives of research on HERV transcription and splicing, with particular focus on head and neck cancer.
Introduction
Human endogenous retroviruses (HERVs) are remnants of ancient retroviral infections integrated into the human genome. Because of recombination events over evolutionary time, most HERV loci are reduced to solitary Long Terminal Repeats (LTRs) (soloLTRs) or viral genomic fragments of various lengths. There are many different families of HERVs, and altogether, they comprise 8% of the human genome. Among these, the HERV-H family is the largest group, consisting of full-length elements (about 100 copies), fragmented elements (800–900 copies), and soloLTRs (about 1,000 copies) (1, 2). None of the full-length proviruses can complete their viral life cycle, due to mutations that accumulated at the provirus loci over evolutionary time, not even HERV-K, although it is the most recent retrovirus that infected the human germline and thus the most intact among all the HERVs. There are approximately 30 full-length HERV-Ks in the human genome out of the several hundreds of HERV-K loci detected, most of which contain fragments or soloLTRs (3). Although none of such full-length HERVs is any longer fully active as virus, transcription has been detected in particular conditions, such as cancer. RNAs from members of multiple HERV families, including HERV-H, HERV-K, HERV-W, HERV-E, HERV-P, HERV-T, HERV-F, HERV-R, and HERV-S, have been identified in cancer (4–13). HERVs have been found active as well as in other conditions such as autoimmune diseases (14, 15), particularly in Systemic Lupus Erythematosus (16, 17), in neurodegenerative diseases (18, 19), particularly in multiple sclerosis (18, 20, 21), and have implications with immunity and other viral infections (22), particularly with HIV (23, 24). Transcription at HERV loci may start from the viral promoter within the 5′LTR and from an upstream promoter. In fact, transcription has been detected also at heavily rearranged and mutated HERV genomic fragments, even in the absence of a functional viral promoter (25–27). The role of such transcripts, often part of long non-coding RNAs (lncRNAs), and their function as RNAs or potential for translating into proteins are largely unknown. However, some viral fragments have been highly conserved through evolution and the derived translated protein exploited by the host for its own purposes. The most notable example is Syncytin-1 that is a protein encoded by ERVW-1 gene, a remnant of an ancient HERV-W env gene and that is a fundamental protein for placenta formation in humans. On the other hand, translation of canonical viral proteins (Gag, Pro, Pol, and Env) from full-length HERV-K loci has been described. It is relevant to point out that some HERV-K proteins (Env and accessory proteins Rec and Np9) are translated from spliced viral transcript. In an effort to better characterize full-length HERV-K transcription at specific proviral loci, in our previous work (28), we detected transcripts derived from aberrant alternative splicing events, across different cancer types, including head and neck cancer cell lines.
Human endogenous retroviruses are, in essence, retroviruses
Endogenous retroviruses (ERVs), including HERVs, follow the same replicative life cycle of all retroviruses, with the specific characteristic of infecting the genome of germ cells and thus integrating into the genome of the host and being passed to the progeny.
The provirus, which is the integrated DNA form of the retrovirus, is composed of an internal genome segment containing the viral protein coding genes and an LTR at each end of the viral genome. Enhancers and promoters and initiation start site for viral transcription are located in the LTR.
The retroviral genome consists of at least four basic genes: gag, pro, pol, and env. These genes encode structural and enzymatic proteins that are essential for viral replication, including the viral capsid components (gag); protease (pro); reverse transcriptase (RT), ribonuclease H, and integrase (pol); and the envelope protein (env). Many, but not all, retroviruses have additional genes that encode accessory proteins, such as rec in HERV-Ks. For almost all retroviruses, some of these genes (gag, pro, and pol) are translated from the primary mRNA transcript. For the other viral genes including env, one or more splicing events must occur.
Death of the virus: HERVs are now viral remnants
ERVs, once stably integrated in the host genome in the form of DNA proviruses, have to face several challenges to survive. Mutations and other recombination events inevitably accumulate at these loci over evolutionary time, just as they do at any host locus. Because, per se, there is no selection pressure on the host to maintain the ERV proviruses in an intact form, with certain exceptions at least for parts of certain proviral genomes, these will eventually include lethal mutations that prevent each ERV from encoding new, fully infectious viral particles and thus the genetic death of that element.
These mutagenic events include nucleotide substitutions, deletions, insertions, and translocations. One particularly important and common type of event for ERVs is the recombination between two homologous LTRs in the same provirus, leading to a soloLTR, with one LTR and the internal viral genome being excised. Even point mutations can irremediably inactivate a provirus if they compromise the function of essential viral proteins.
Thus, to date, no individual HERV-K provirus present in the human genome has been found to retain a full capability of encoding an infectious genome capable of completing the viral life cycle.
Life in death: HERVs are transcribed, nonetheless
HERV expression is regulated at the level of the LTRs, which function as promoter and contain the transcription starting site and numerous transcription factor binding sites (29, 30). Because HERVs infect the germline, it is reasonable that specific transcription factors that are active during oogenesis, spermiogenesis, or early embryo development are exploited for viral activation. Such factors have not been clearly characterized yet (31, 32). In addition to the viral life cycle, HERVs have been detected in several physiologic and pathologic conditions (33–37). Thus, the LTR is likely to be able to bind also with transcription factors other than those specifically active in germline (30).
The implications of transcription of heavily mutated or rearranged HERV genes are mostly unknown. HERVs may translate for viral proteins, which are mostly non-functional as explained above, truncated viral proteins, and non-viral proteins resulting from frameshift mutations, indels, or various genetic rearrangements. In addition, transcripts from these loci may lack obvious Open Reading Frame (ORF).
However, most HERV loci are soloLTRs. In such cases, the virus no longer exists at the specific genomic locus, and the soloLTR constitutes a relic of the previous infection (38). Nonetheless, sequences within the LTR may still be usable by the host transcription factors. In fact, LTRs can serve as promoters in both sense and antisense orientations (39). Thus, they can influence the expression of nearby host genes (27, 40, 41). Moreover, soloLTR may promote transcription of long intergenic non-coding RNAs (lincRNAs) (42). The SAMMSON lncRNA is promoted by a soloLTR (LTR1A2) and was recently reported as involved in oncogenesis in melanoma (43). Other examples of LTR-promoted oncogenic lncRNAs include HULC in hepatocarcinoma (44), UCA1 in urotelial carcinoma (45), and LCT13, which is another lncRNA that is promoted by an antisense soloLTR (L1PA2), in colorectal cancer (46).
To avoid potentially deleterious transcription, both from soloLTRs and full-length proviruses of fragments, it is commonly assumed that HERVs are mostly silenced by epigenetic regulation (47, 48). Multiple strategies are used to regulate HERV transcription such as localization of soloLTRs and proviruses to heterochromatin and CpG methylation and histone deacetylation (49).
HERVs in cancer and other conditions: Friend or foe?
Although HERVs’ role in health and disease has been explored for many years, it remains poorly understood (33–37).
An oncogenic role for HERV-K has been proposed. In fact, HERV-K transcripts and proteins have been detected in several cancer types, including teratocarcinoma (50–55), trophoblastic tumors and germ cell tumors (56, 57), seminomas (58–60), breast cancer (61–67), prostate cancer (68–71), leukemias (72–77), renal cancer (78), ovarian cancer (79–81), cervical cancer (28, 82), melanoma (83–87), soft tissue sarcoma (88), osteosarcoma (89), Kaposi’s sarcoma (90), glioblastoma (91), astrocytic tumors (92), hepatoblastoma (93, 94), and hepatocellular carcinoma (95).
The correlation between HERV-K expression and cancer is not sufficient in itself to clarify whether HERV-K has tumorigenic activity. In fact, overexpression of Rec or Np9 in transgenic mice has been shown to cause tumors (96).
HERV-K expression has also been detected in normal tissues and physiological conditions. In the human placenta, in addition to the abovementioned HERV-W and HERV-FRD, HERV-K has been detected as well (73, 97), but its role in physiology or disease, if any, has not been elucidated yet. In fact, HERV-K has been speculated to have a role in placental dysfunctions such as preclampsia (98). HERV-K viral particles can be detected by electron microscopy in normal human placenta (36).
As abovementioned, the well-conserved env genes from HERV-W1 and HERV- FRD transcribe for Syncytin-1 and Synicitin-2, respectively, and mediate the fusion of villous cytotrophoblasts during placentation (99, 100). In fact, Env protein has fusogenic properties and has been proposed to have a role in cancer, including in epithelial-to-mesenchymal transition (101, 102).
Moreover, env genes have been proposed to exhibit an immunosuppressive role that is important for preventing maternal rejection of the semi-allogenic fetus during pregnancy (103). Such property has been speculated to be exploited by cancer cells as well to facilitate tolerance.
HERV-H has been detected in human normal tissue, specifically in placenta, and cancer, including breast cancer (104), pancreatic cancer (105), liver cancer (106, 107), prostate cancer (108), ovarian cancer (109), lung cancer (110), colon cancer (111, 112), cervical cancer (104), and others (2, 107, 113).
An interesting regulatory role for pluripotency in human embryonic stem cells and induced pluripotent stem cells has been described for HERV-H, including ncRNAs, enhancers, and alternative promoters, and markers of topologically associating domain (TAD) boundaries (114).
Little is known regarding HERV-K and HERV-H transcription in normal tissues other than placenta.
Bioinformatic search for HERV-conserved ORFs led to the identification of a small but interesting number of env-related genes with a full-length coding sequence, such as syncytin-1 and syncytin-2 (115, 116).
Recent research led to the identification of a peculiar HERV locus transcribing for a unique Env protein, called HEMO, which is released in the human blood circulation. HEMO has also been detected in placenta and various cancers as well, including ovarian cancer and endometrial cancer, and in germline, liver, lung, or breast tumors (117). The HEMO retroviral env gene belongs to the MER34 family, which comprise only highly degenerated and rearranged elements.
Little or none is known about the vast world of HERV-containing lncRNAs. Several examples have been described but functions and significance is mostly unknown (118–120).
One example is lncMER52A, a liver cancer–specific oncogenic lncRNA transcribed by MER52A LTR retrotransposon of the ERV1 class (121).
Another example is the overexpression in breast cancer of the TROJAN lncRNA, which contains a complete LTR70 sequence of several mosaic LTRs flanked by MER67C and LTR56 (122).
The good, the bad, and the ugly: Canonical, alternative, and aberrant splicing of HERV transcripts
Intact full-length HERVs produce a single full-length RNA transcript, from 5′LTR to 3′LTR. Then, a single splicing event must occur to produce the 1X-spliced env transcript. An additional splicing event is needed to produce the 2X-spliced np9 and rec transcripts in HERV-Ks.
In the previous paragraph, we list the many examples of detection of Env, Rec, and Np9 proteins in cancer. All of them must have derived from canonical splicing events of the HERV full-length transcript. Splicing occurs when specific splicing signal sequences, namely, splice donor (SD) and splice acceptor (SA), are recognized by the spliceosome machinery, and intronic sequence is excised. Typically, only a fraction of HERV full-length transcript undergoes splicing, because the full-length transcript constitutes the genome for the forming virions.
It has been speculated that the highly mutated HERV genome may disrupt canonical SD/SA signals and allow cryptic splicing sites to emerge. In fact, many of such sites have been identified throughout the HERV genome (123, 124).
Previous work from Lindeskong and Blomberg (113) has shown alternative splicing for HERV-H env transcripts from normal and leukemia lymphocytes (Figure 1). These alternative splicing sites use canonical consensus signals for major spliceosome. Although the function and significance of such findings is unknown, it can be speculated that these constitute cryptic SD/SA sequences. The shift to these cryptic signals was not due to mutations at the canonical splice sites, as the functional canonical splice sites could be sequenced. In fact, multiple SD and SA signals have been predicted in HERV-H by bioinformatic analysis (125). Moreover, normal and leukemia lymphocytes showed different levels of amplification of the spliced env transcripts, thus indicating that the cellular type could determine which alternative splicing event to favor. Thus, the molecular mechanisms for the SD/SA choice were not identified. It is relevant to point out that some of these transcripts have ORFs that could possibly be translated into novel proteins, not per se related to the original viral proteins.
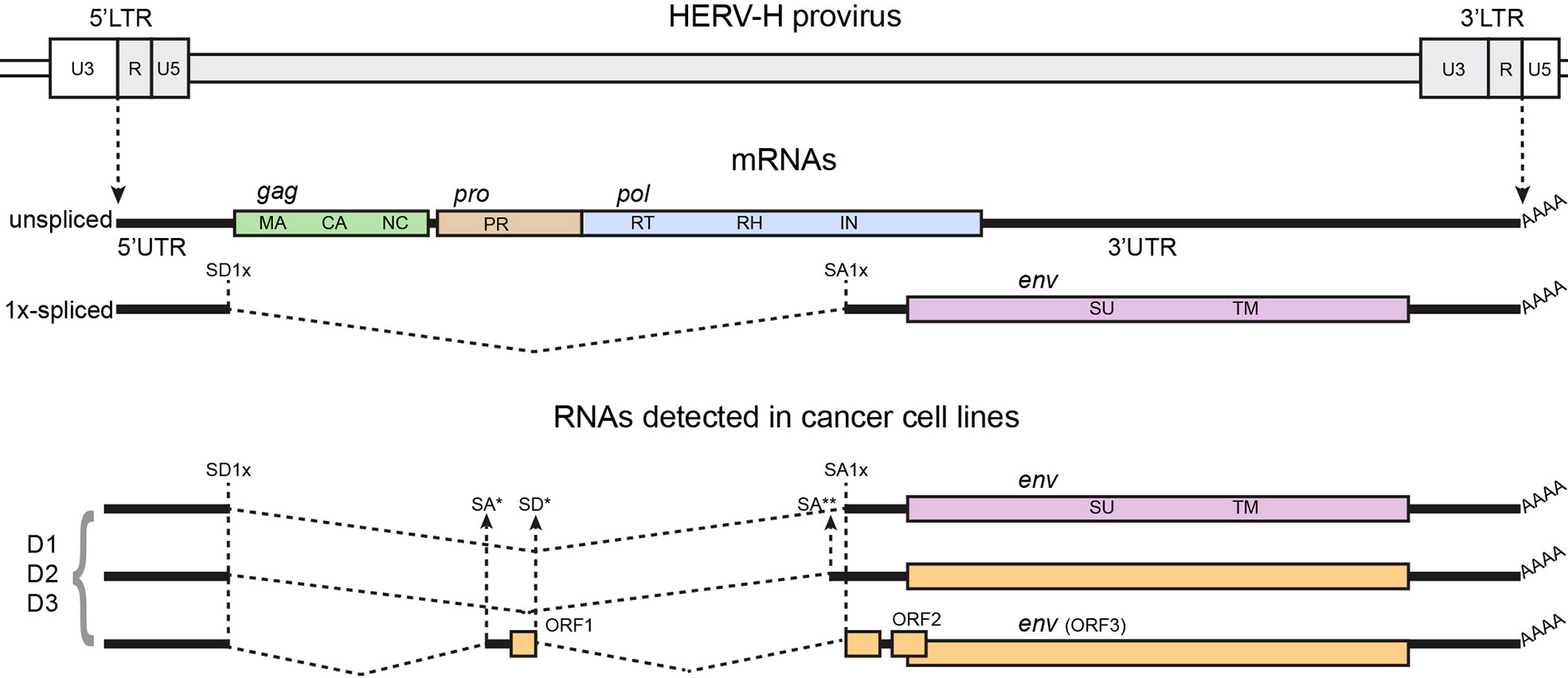
Figure 1 Structure of the HERV-H genome, canonical spliced mRNAs, and spliced RNAs detected in (113), from which the figure was adapted. A genetic map of a HERV-H provirus (gray) inserted into flanking host genome sequences is shown. The unspliced primary viral transcript and singly spliced env mRNA are shown below the viral genome. Poly (A) tails are indicated (AAAA). The dashed, angled line shows the excised intronic sequences. Colored boxes indicated the different genes, indicated in the figure by name and ORF. Splice donor (SD) and splice acceptor (SA) sites are indicated on map, including canonical sites for 1x-spliced mRNA (SD1x and SA1x) and canonical sites for 2x-spliced mRNAs (SD2x’ for type I proviruses, SD2x” for type II proviruses, and SA2x), and individual alternative SD/SA are identified by stars (*, **). Black vertical dashed lines identify canonical SD/SA sites across the panel; black arrows show alternative SD/SA sites. D1, D2, and D3 are fictitious names of individual HERV-H viruses detected in the study.
With the purpose of screening for viral splicing transcripts in cancer cell lines, in our previous work (28), we undertook RT-PCR across splicing sites of full-length HERV-K in various cancer cell lines, including breast cancer, cervical cancer, prostate cancer, and head and neck cancer, which are expected to find 1X-spliced transcript for env and 2X-spliced transcript for np9 or rec, respectively, for type I and type II HERV-K-HML2 viruses (25). Indeed, the spliced transcripts were detected in almost all cancer cell lines across the tested panel (Figure 2). Sequencing of the RT-PCR products was performed to identify the specific loci of origin of the transcripts. These analyses identified a total of seven different individual HERV-K loci among the 12 cell lines tested: HERV-K102, HERV-K108, K(I), HERV-K106, HERV-K107, HERV-K111, and HERV-K117.
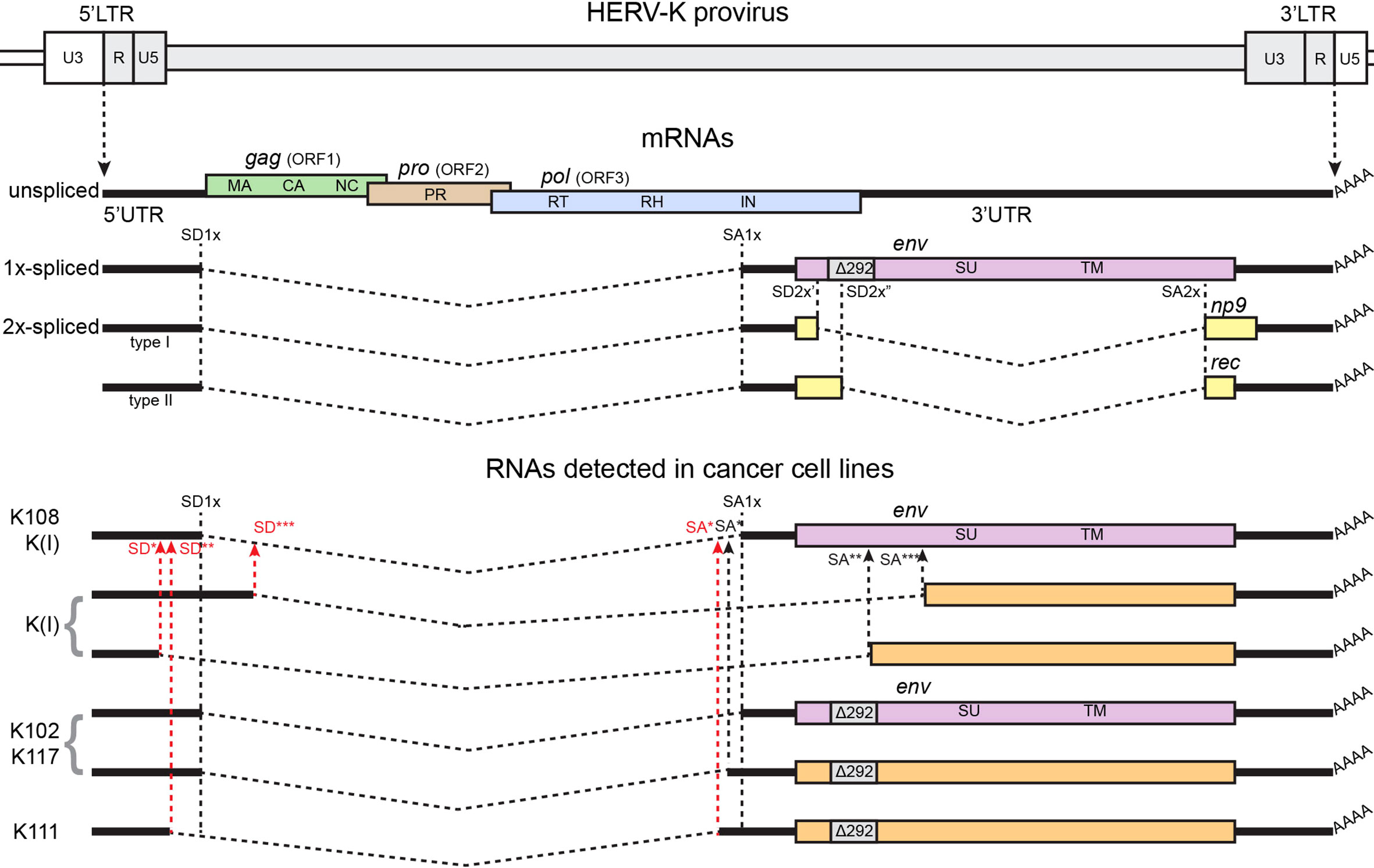
Figure 2 Structure of the HERV-K genome, canonical spliced mRNAs, and spliced RNAs detected in (28), from which the figure was adapted. A genetic map of a HERV-K provirus (gray) inserted into flanking host genome sequences is shown. The unspliced primary viral transcript, singly spliced env mRNA, and doubly spliced rec and np9 mRNAs are shown below the viral genome. Poly (A) tails are indicated (AAAA). The 292 nucleotide deletion of type 1 HERV-K proviruses spanning the pol-env junction is indicated (Δ292). The dashed, angled line shows the excised intronic sequences. Colored boxes indicated the different genes, indicated in the figure by name and ORF. Splice donor (SD) and splice acceptor (SA) sites are indicated on map, including canonical sites for 1x-spliced mRNA (SD1x and SA1x) and canonical sites for 2x-spliced mRNAs (SD2x’ for type I proviruses, SD2x” for type II proviruses, and SA2x), and individual alternative SD/SA are identified by stars (*, **, and ***) and are written in red in case of aberrant sites, without canonical sequences for spliceosomes. Black vertical dashed lines identify canonical SD/SA sites across the panel, black arrows show alternative SD/SA sites, and red arrows show SD/SA aberrant sites. HERV-K108, HERV-K(I), HERV-K102, HERV-K117, and HERV-K111 are the names of individual HERV-Ks detected in the study.
Unexpectedly, while most RT-PCR products showed splicing at the expected positions within the HERV-K genome, the transcripts spliced at the additional previously unidentified sites were also detected (Figure 2). The conventional 1X-env splicing sites were detected for HERV-K108, HERV-K109, HERV-K(I), HERV-K102, and HERV-K117. Four loci—HERV-K102, HERV-K(I), HERV-K117, and HERV-K111—showed unusual 1X-env splicing variants formed from the use of alternative SD/SA sites. The splice sites that were detected in some instances matched the consensus signals for the major or minor spliceosomes, whereas, in other instances, they did not. For HERV-K102, the same splice sites were detected in six different cell lines, and for HERV-K117, the same sites were detected in two. Such aberrant spliced transcripts were detected in the majority of the cell lines, and in some instances, both the conventional and aberrant splice sites were detected for particular proviruses [HERV-K102, HERV-K(I), and HERV-K117].
This finding was unexpected, and the possible biological significance and the underlying molecular mechanism are unknown.
What about HERVs in head and neck cancer?
Although HERV expression has been detected in many cancer types, in head and neck cancers, it has been rarely tested.
The most relevant example comes from Kolbe et al. (126) who analyzed 43 paired tumor and adjacent normal tissue samples from The Cancer Genome Atlas program. Transcripts were detected from over 3,000 specific HERV loci, in tumor and adjacent normal tissue. Approximately one-third of them were differentially expressed between the two tissue types. Most differentially expressed HERVs showed higher levels in tumor tissue. Differentially expressed HERVs were enriched in members of the HERV-H family. A hierarchical clustering based on HERV expression was performed, and the two resulting distinct clusters showed significant difference in survival.
Although this study was performed by looking at the single loci level, a clustering algorithm was used to generate two prognostic distinct clusters. Thus, the contribution of specific HERV loci to survival was not investigated—not the details at the molecular level regarding which portion of the viral genome and genes were transcribed and whether any splicing event occurred.
Cuffel et al. (127) studied the expression of cancer-testis and other tumor-associated antigens in head and neck squamous cell carcinoma (HNSCC). Samples from 57 HNSCC patients were analyzed by RT/PCR, Immunohistochemistry (IHC), and correlated with survival. Among the results, it is relevant to highlight that a HERV-K–related antigen, HERV-K-MEL, was among the most frequently expressed genes as it was detected in 42% of the patients. However, in their analysis, HERV-K-MEL expression did not impact survival. HERV-K-MEL is an HERV-K env-related antigen that was first identified as recognized by cytolytic T lymphocytes in melanoma (84).
A recent study by Zapatka et al. (128) has shown ERV1 expression in HNSCC. The authors, as part of the Pan-Cancer Analysis of Whole Genomes Consortium, performed a whole-transcriptome sequencing data from over 2,000 cancer samples across 38 tumor types. Among these, HERV expression, particularly ERV1, was detected in HNSCC, although no correlation with survival was evident.
Michna et al. (129) reported the expression of ERVMER34-1 and ERV3-1 (HERV-R) in CAL-33 HNSCC cell line. Interestingly, ERV3-1 was upregulated in response to irradiation. The authors speculated that these findings may have implications in radiosensitivity in cancer.
Landriscina et al. (130) have shown that RT is active in FRO, WRO, and ARO human thyroid carcinoma cell lines. They showed that nevirapine and efavirenz, two RT inhibitors that are usually employed in HIV treatment, reversibly inhibit cell proliferation in the undifferentiated thyroid carcinoma ARO and FRO cells. However, they did not characterize RT origin. In fact, in addition to HERVs, most retroelements, including LINE elements, have an RT coding gene (131).
Indeed, HERV-K is expressed in head and neck cancer cell lines, and splicing events are detected.
In our abovementioned study (28), we tested the head and neck cancer cell lines FaDu, UPCI-SCC-90, and UM-SCC-47. 1X-env and 2X-rec transcripts were detected in all three cell lines, whereas 2X-np9 was only detected in UPCI-SCC-90. Specifically, HERV-K102, HERV-K108, K(I), and HERV-K106 were detected. In all three cell lines, aberrant 1X-env transcripts were detected: HERV-K(I) in UPCI-SCC-90, and HERV-K102 in FaDu and UM-SCC-47.
To the author’s knowledge, no other examples of HERV detection in head and neck cancer appear available in the scientific literature.
Discussion
HERV activation is a common feature in cancer. However, its role, if any, has not been fully elucidated yet.
Although HERVs are highly mutated, they have been shown to retain, at least in some cases, enough viral functions to translate to the canonical viral proteins, including those that require splicing events. Moreover, alternative and aberrant splicing variants have been detected.
Pre-mRNA splicing is a common post-transcriptional event for both the host cell and retroviruses, including HERVs. There are several different types of alternative splicing, among which the most common include exon skipping, selection of splice donor, and selection of splice acceptor. Alternative splicing allows to expand the variety of encoded proteins, but it may also be involved in regulation of translation, for example, by including an early stop codon. The splicing process involves the spliceosome machinery, which is formed by five small nuclear ribonuleoproteins and several proteins. The cell has two different types of spliceosome: the U2-type “major” spliceosome and the U12-type “minor” spliceosome. They recognize different specific intronic sequences for the splice sites. Most splicing occurs through the major spliceosome. Alternative and aberrant splicings have been involved in cancer initiation and progression (132).
Transcription of the HERV provirus, by RNA polymerase II, generates a primary viral RNA that is both 5′ capped and 3′ polyadenylated, as any other mRNA of the host cell (124). Similarly to the host RNA, the primary RNA can be regulated by internally m6A and m5C methylations (133).
The primary transcript translates the gag-pro-pol genes into capside (MA, CA, and NC), protease (Pro), RT, and integrase (IN) proteins and constitutes the genome for the new virions. Env requires a splicing event to occur, and the accessory proteins Rec and Np9 require a second splicing event. Both alternative splicings require simple intron excision. Thus, the primary RNA must escape splicing to reach packaging into the forming virions. How this selection between splicing and packaging is made is unclear. It has been speculated that this may depend on splice site efficiency. In fact, the spliceosome machinery would intercept every single primary RNA transcript displaying splice sites with high efficiency, thus entirely preventing full-length RNA packaging. Conversely, the presence of low-efficiency splice sites would allow both RNA forms to co-exist. This ensures the correct life cycle of the retrovirus, but it may also facilitate alternative, cryptic, splice sites to emerge and be utilized. This may explain the findings of alternative splicing of HERV-H (113) and HERV-K (28) of the splice site selection type. Similar mechanisms have been observed for HIV (134) and other retroviruses (124). However, some splice site sequences in HERV-K alternative splicing do not match with signal sequences for either U2- or U12-type spliceosomes. This observation is unexpected and unexplained.
The alternative and aberrant splice sites detected highlight the possibility of new research in the field of spliceosome functions. In fact, the sequences of most of the alternative sites that were detected shared only partial overlap with those of the conventional splicing signals. What determinants within the viral genome affect the utilization of alternative splice sites is currently unknown, and further studies may shed light on the basic mechanisms of splicing.
The use of non-canonical SA/SD sequences has been shown to some extent in trans-splicing (135), which is an unusual form of splicing between two individual pre-RNA transcripts. Most trans-splicing is expected to follow the canonical spliceosome-mediated splicing process. However, a transfer RNA (tRNA)-mediated splicing, which does not require the canonical consensus sequences for splice sites nor the spliceosome machinery, has been described, at least for trans-splicing (136).
The potential for translating into protein may vary. In fact, the assumption that a viral transcript will translate into a viral protein must be verified, especially for HERVs that carry numerous mutations in their sequence and that not only may affect viral protein function but also may prevent correct polypeptide three-dimensional conformation. In fact, HERV transcripts may be part of lncRNAs, by polymerase read-through transcription of proviral loci (137). Such transcripts may bear regulatory functions, particularly on gene expression (138). lncRNAs may interact with DNA, RNA, or proteins: lncRNAs may promote or repress transcription, by working as signals or decoys, respectively, or may function as epigenetic regulators or even as scaffolds by interacting with various protein partners (139–141). The inclusion of HERV components into lncRNAs has been reported (142).
HERVs’ proteins and transcripts constitute an attractive target for therapy. Many authors have been exploiting such knowledge to refine various strategies against HERVs to selectively target cancer cells (84, 103, 143–145).
The need for locus-specific analyses is evident. However, the high degree of repetitive and highly similar sequences in HERV elements has made locus-specific characterization of HERVs a significant challenge.
Although HERVs have been detected in head and neck cancer, the research in this cancer type is still at a very early stage and warrants further study.
Author contributions
LA has conceived, performed literature research for, and written the article. The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mager DL, Hunter DG, Schertzer M, Freeman JD. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3). Genomics (1999) 59(3):255–63. doi: 10.1006/geno.1999.5877
2. Yi JM, Kim HM, Kim HS. Human endogenous retrovirus HERV-h family in human tissues and cancer cells: expression, identification, and phylogeny. Cancer Lett (2006) 231(2):228–39. doi: 10.1016/j.canlet.2005.02.001
3. Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology (2011) 8(1):90. doi: 10.1186/1742-4690-8-90
4. Yi JM, Kim HS. Expression analysis of endogenous retroviral elements belonging to the HERV-f family from human tissues and cancer cells. Cancer Lett (2004) 211(1):89–96. doi: 10.1016/j.canlet.2004.01.026
5. Yi JM, Kim TH, Huh JW, Park KS, Jang SB, Kim HM, et al. Human endogenous retroviral elements belonging to the HERV-s family from human tissues, cancer cells, and primates: expression, structure, phylogeny and evolution. Gene (2004) 342(2):283–92. doi: 10.1016/j.gene.2004.08.007
6. Yi JM, Kim HM, Kim HS. Expression of the human endogenous retrovirus HERV-W family in various human tissues and cancer cells. J Gen Virol (2004) 85(5):1203–10. doi: 10.1099/vir.0.79791-0
7. Yi JM, Kim HS. Expression and phylogenetic analyses of human endogenous retrovirus HC2 belonging to the HERV-T family in human tissues and cancer cells. J Hum Genet (2007) 52(4):285–96. doi: 10.1007/s10038-007-0115-8
8. Yi JM, Schuebel K, Kim HS. Molecular genetic analyses of human endogenous retroviral elements belonging to the HERV-p family in primates, human tissues, and cancer cells. Genomics (2007) 89(1):1–9. doi: 10.1016/j.ygeno.2006.08.010
9. Yi JM, Kim HS. Molecular phylogenetic analysis of the human endogenous retrovirus e (HERV-e) family in human tissues and human cancers. Genes Genet Syst (2007) 82(1):89–98. doi: 10.1266/ggs.82.89
10. Salavatiha Z, Soleimani-Jelodar R, Jalilvand S. The role of endogenous retroviruses-K in human cancer. Rev Med Virol (2020) 30(6):1–13. doi: 10.1002/rmv.2142
11. Kim HS, Ahn K, Kim DS. Quantitative expression of the HERV-W env gene in human tissues. Arch Virol (2008) 153(8):1587. doi: 10.1007/s00705-008-0159-x
12. Ahn K, Kim HS. Structural and quantitative expression analyses of HERV gene family in human tissues. Mol Cells (2009) 28(2):99–103. doi: 10.1007/s10059-009-0107-y
13. Jansz N, Faulkner GJ. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol (2021) 22(1):147. doi: 10.1186/s13059-021-02357-4
14. Posso-Osorio I, Tobón GJ, Cañas CA. Human endogenous retroviruses (HERV) and non-HERV viruses incorporated into the human genome and their role in the development of autoimmune diseases. J Transl Autoimmun (2021) 4:100137. doi: 10.1016/j.jtauto.2021.100137
15. Grandi N, Tramontano E. HERV envelope proteins: Physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol (2018) 9:462. doi: 10.3389/fmicb.2018.00462
16. Quaglia M, Merlotti G, Andrea MD, Borgogna C, Cantaluppi V. Viral infections and systemic lupus erythematosus: New players in an old story. Viruses (2021) 13(2):277. doi: 10.3390/v13020277
17. Illescas-Montes R, Corona-Castro CC, Melguizo-Rodríguez L, Ruiz C, Costela-Ruiz VJ. Infectious processes and systemic lupus erythematosus. Immunology (2019) 158(3):153–60. doi: 10.1111/imm.13103
18. Dolei A, Perron H. The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: a biological interface between virology, genetics, and immunology in human physiology and disease. J neurovirol (2009) 15(1):4–13. doi: 10.1080/13550280802448451
19. Christensen T. HERVs in neuropathogenesis. J Neuroimmune Pharm (2010) 5(3):326–35. doi: 10.1007/s11481-010-9214-y
20. Mentis A --A, Dardiotis E, Grigoriadis N, Petinaki E, Hadjigeorgiou GM. Viruses and endogenous retroviruses in multiple sclerosis: From correlation to causation. Acta Neurol Scand (2017) 136(6):606–16. doi: 10.1111/ane.12775
21. Tarlinton RE, Martynova E, Rizvanov AA, Khaiboullina S, Verma S. Role of viruses in the pathogenesis of multiple sclerosis. Viruses (2020) 12(6):643. doi: 10.3390/v12060643
22. Alcazer V, Bonaventura P, Depil S. Human endogenous retroviruses (HERVs): Shaping the innate immune response in cancers. Cancers (2020) 12(3):610. doi: 10.3390/cancers12030610
23. Liu CH, Grandi N, Palanivelu L, Tramontano E, Lin LT. Contribution of human retroviruses to disease development–a focus on the HIV– and HERV–cancer relationships and treatment strategies. Viruses (2020) 12(8):852. doi: 10.3390/v12080852
24. van der Kuyl AC. HIV Infection and HERV expression: a review. Retrovirology (2012) 9(1):6. doi: 10.1186/1742-4690-9-6
25. Vargiu L, Rodriguez-Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology (2016) 13(1):7. doi: 10.1186/s12977-015-0232-y
26. Kovalskaya E, Buzdin A, Gogvadze E, Vinogradova T, Sverdlov E. Functional human endogenous retroviral LTR transcription start sites are located between the r and U5 regions. Virology (2006) 346(2):373–8. doi: 10.1016/j.virol.2005.11.007
27. Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene (2009) 448(2):105–14. doi: 10.1016/j.gene.2009.06.020
28. Agoni L, Lenz J, Guha C. Variant splicing and influence of ionizing radiation on human endogenous retrovirus K (HERV-K) transcripts in cancer cell lines. PloS One (2013) 8(10):e76472. doi: 10.1371/journal.pone.0076472
29. Akopov SB, Nikolaev LG, Khil PP, Lebedev YB, Sverdlov ED. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett (1998) 421(3):229–33. doi: 10.1016/S0014-5793(97)01569-X
30. Ruda VM, Akopov SB, Trubetskoy DO, Manuylov NL, Vetchinova AS, Zavalova LL, et al. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res (2004) 104(1):11–6. doi: 10.1016/j.virusres.2004.02.036
31. Mao J, Zhang Q, Cong YS. Human endogenous retroviruses in development and disease. Comput Struct Biotechnol J (2021) 19:5978–86. doi: 10.1016/j.csbj.2021.10.037
32. Mastora E, Christodoulaki A, Papageorgiou K, Zikopoulos A, Georgiou I. Expression of retroelements in mammalian gametes and embryos. In Vivo (2021) 35(4):1921–7. doi: 10.21873/invivo.12458
33. Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV transcriptome project. BMC Genomics (2008) 9(1):354. doi: 10.1186/1471-2164-9-354
34. Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, et al. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol (2005) 79(1):341–52. doi: 10.1128/JVI.79.1.341-352.2005
35. Colmegna I, Garry RF. Role of endogenous retroviruses in autoimmune diseases. Infect Dis Clin N Am (2006) 20(4):913–29. doi: 10.1016/j.idc.2006.09.008
36. Sugimoto J, Schust DJ. Human endogenous retroviruses and the placenta. Reprod Sci (2009) 16(11):1023–33. doi: 10.1177/1933719109336620
37. Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci (2008) 65(21):3366–82. doi: 10.1007/s00018-008-8496-1
38. Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, et al. Rate of recombinational deletion among human endogenous retroviruses. J Virol (2007) 81(17):9437–42. doi: 10.1128/JVI.02216-06
39. Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mobile Dna-uk. (2016) 7(1):24. doi: 10.1186/s13100-016-0080-x
40. Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene (2006) 366(2):335–42. doi: 10.1016/j.gene.2005.09.003
41. Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. At Least 50% of human-specific HERV-K (HML-2) long terminal repeats serve In vivo as active promoters for host nonrepetitive DNA transcription. J Virol (2006) 80(21):10752–62. doi: : 10.1128/JVI.00871-06
42. Laurent GS, Shtokalo D, Dong B, Tackett MR, Fan X, Lazorthes S, et al. VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer. Genome Biol (2013) 14(7):R73. doi: 10.1186/gb-2013-14-7-r73
43. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature (2016) 531(7595):518–22. doi : 10.1038/nature17161
44. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology (2007) 132(1):330–42. doi: 10.1053/j.gastro.2006.08.026
45. Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J Cancer Res Clin (2016) 142(7):1407–19. doi: : 10.1007/s00432-015-2042-y
46. Cruickshanks HA, Vafadar-Isfahani N, Dunican DS, Lee A, Sproul D, Lund JN, et al. Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res (2013) 41(14):6857–69. doi: 10.1093/nar/gkt438
47. Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene (2009) 448(2):151–67. doi: 10.1016/j.gene.2009.08.006
48. Hurst TP, Magiorkinis G. Epigenetic control of human endogenous retrovirus expression: Focus on regulation of long-terminal repeats (LTRs). Viruses (2017) 9(6):130. doi: 10.3390/v9060130
49. Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci (2012) 37(4):127–33. doi: 10.1016/j.tibs.2011.11.006
50. Chan SM, Sapir T, Park SS, Rual JF, Contreras-Galindo R, Reiner O, et al. The HERV-K accessory protein Np9 controls viability and migration of teratocarcinoma cells. PloS One (2019) 14(2):e0212970. doi: 10.1371/journal.pone.0212970
51. Morozov VA, Morozov AV. A comprehensive analysis of human endogenous retroviruses HERV-K (HML.2) from teratocarcinoma cell lines and detection of viral cargo in microvesicles. Int J Mol Sci (2021) 22(22):12398. doi: 10.3390/ijms222212398
52. Bhardwaj N, Montesion M, Roy F, Coffin JM. Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line tera-1. Viruses (2015) 7(3):939–68. doi: 10.3390/v7030939
53. Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology (1993) 196(1):349–53. doi: 10.1006/viro.1993.1487
54. Li MD, Bronson DL, Lemke TD, Faras AJ. Restricted expression of new HERV-K members in human teratocarcinoma cells. Virology (1995) 208(2):733–41. doi: 10.1006/viro.1995.1205
55. Löwer R, Löwer J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-h and HERV-K in teratocarcinoma cells. Virology (1993) 192(2):501–11. doi: 10.1006/viro.1993.1066
56. Herbst H, Sauter M, Mueller-Lantzsch N. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am J Pathol (1996) 149(5):1727–35.
57. Herbst H, Sauter M, Kühler-Obbarius C, Löning T, Mueller-Lantzsch N. Human endogenous retrovirus (HERV)-K transcripts in germ cell and trophoblastic tumours. Apmis (1998) 106(1-6):216–20. doi: 10.1111/j.1699-0463.1998.tb01338.x
58. Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, et al. Human endogenous retrovirus K10: expression of gag protein and detection of antibodies in patients with seminomas. J Virol (1995) 69(1):414–21. doi: 10.1128/jvi.69.1.414-421.1995
59. Vinogradova T, Leppik L, Kalinina E, Zhulidov P, Grzeschik KH, Sverdlov E. Selective differential display of RNAs containing interspersed repeats: analysis of changes in the transcription of HERV-K LTRs in germ cell tumors. Mol Genet Genomics (2002) 266(5):796–805. doi: 10.1007/s00438-001-0596-7
60. Rakoff-Nahoum S, Kuebler PJ, Heymann JJ, Sheehy ME, Ortiz GM, Ogg GS, et al. Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res Hum Retrov. (2006) 22(1):52–6. doi: 10.1089/aid.2006.22.52
61. Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol (1987) 61(6):2059–62. doi: 10.1128/jvi.61.6.2059-2062.1987
62. Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2001) 7(6):1553–60.
63. Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res (2008) 68(14):5869–77. doi: 10.1158/0008-5472.CAN-07-6838
64. Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, et al. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J Virol (2008) 82(4):1808–18. doi: 10.1128/JVI.02115-07
65. Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia (2008) 10(6):521–IN2. doi: 10.1593/neo.07986
66. Tavakolian S, Goudarzi H, Faghihloo E. Evaluating the expression level of HERV-K env, np9, rec and gag in breast tissue. Infect Agents Cancer (2019) 14(1):42. doi: 10.1186/s13027-019-0260-7
67. Wei Y, Wei H, Wei Y, Tan A, Chen X, Liao X, et al. Screening and identification of human endogenous retrovirus-K mRNAs for breast cancer through integrative analysis of multiple datasets. Front Oncol (2022) 12:820883. doi: 10.3389/fonc.2022.820883
68. Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun (2007) 8:15.
69. Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol (2013) 3:180. doi: 10.3389/fonc.2013.00180
70. Rezaei SD, Hayward JA, Norden S, Pedersen J, Mills J, Hearps AC, et al. HERV-K gag RNA and protein levels are elevated in malignant regions of the prostate in males with prostate cancer. Viruses (2021) 13(3):449. doi: 10.3390/v13030449
71. Goering W, Schmitt K, Dostert M, Schaal H, Deenen R, Mayer J, et al. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate (2015) 75(16):1958–71. doi: 10.1002/pros.23095
72. Brodsky I, Foley B, Gillespie D. Expression of human endogenous retrovirus (HERV-K) in chronic myeloid leukemia. Leuk Lymphoma (2009) 11(sup1):119–23. doi: 10.3109/10428199309047874
73. Simon M, Haltmeier M, Papakonstantinou G, Werner T, Hehlmann R, Leib-Mösch C. Transcription of HERV-k-related LTRs in human placenta and leukemic cells. Leukemia (1994) 8 Suppl 1:S12–7.
74. Tatkiewicz W, Dickie J, Bedford F, Jones A, Atkin M, Kiernan M, et al. Characterising a human endogenous retrovirus(HERV)-derived tumour-associated antigen: enriched RNA-seq analysis of HERV-K(HML-2) in mantle cell lymphoma cell lines. Mobile Dna-uk. (2020) 11(1):9. doi: 10.1186/s13100-020-0204-1
75. Fischer S, Echeverría N, Moratorio G, Landoni AI, Dighiero G, Cristina J, et al. Human endogenous retrovirus np9 gene is over expressed in chronic lymphocytic leukemia patients. Leuk Res Rep (2014) 3(2):70–2. doi: 10.1016/j.lrr.2014.06.005
76. Bergallo M, Montanari P, Mareschi K, Merlino C, Berger M, Bini I, et al. Expression of the pol gene of human endogenous retroviruses HERV-K and -W in leukemia patients. Arch Virol (2017) 162(12):3639–44. doi: 10.1007/s00705-017-3526-7
77. Januszkiewicz-Lewandowska D, Nowicka K, Rembowska J, Fichna M, Żurawek M, Derwich K, et al. Env gene expression of human endogenous retrovirus-K and human endogenous retrovirus-W in childhood acute leukemia cells. Acta Haematol-basel (2013) 129(4):232–7. doi: 10.1159/000345407
78. Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA Methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Brit J Cancer (1999) 80(9):1312–21. doi: 10.1038/sj.bjc.6690524
79. Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer (2007) 120(1):81–90. doi: 10.1002/ijc.22256
80. Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi L, et al. Cytotoxicity of human endogenous retrovirus K–specific T cells toward autologous ovarian cancer cells. Clin Cancer Res (2015) 21(2):471–83. doi: 10.1158/1078-0432.CCR-14-0388
81. Jeon KY, Ko EJ, Oh YL, Kim H, Eo WK, Kim A, et al. Analysis of KAP1 expression patterns and human endogenous retrovirus env proteins in ovarian cancer. Genes Genom (2020) 42(10):1145–50. doi: 10.1007/s13258-020-00979-9
82. Tavakolian S, Goudarzi H, Moridi A, Faghihloo E. Analysing the HERV-K env, np9, rec and gag expression in cervical tissues. New Microbes New Infect (2021) 44:100936. doi: 10.1016/j.nmni.2021.100936
83. Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res (2003) 63(24):8735–41.
84. Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res (2002) 62(19):5510–6.
85. Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res (2005) 65(10):4172–80. doi: 10.1158/0008-5472.CAN-04-2983
86. Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol (2013) 5(2):307–28. doi: 10.1093/gbe/evt010
87. Huang G, Li Z, Wan X, Wang Y, Dong J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J Carcinog (2013) 12(1):5. doi: 10.4103/1477-3163.109032
88. Giebler M, Staege MS, Blauschmidt S, Ohm LI, Kraus M, Würl P, et al. Elevated HERV-K expression in soft tissue sarcoma is associated with worsened relapse-free survival. Front Microbiol (2018) 9:211. doi: 10.3389/fmicb.2018.00211
89. Wang C, Liang C. The insertion and dysregulation of transposable elements in osteosarcoma and their association with patient event-free survival. Sci Rep-uk. (2022) 12(1):377. doi: 10.1038/s41598-021-04208-5
90. Dai L, Valle LD, Miley W, Whitby D, Ochoa AC, Flemington EK, et al. Transactivation of human endogenous retrovirus K (HERV-K) by KSHV promotes kaposi’s sarcoma development. Oncogene (2018) 37(33):4534–45. doi: 10.1038/s41388-018-0282-4
91. Shah AH, Govindarajan V, Doucet-O’Hare TT, Rivas S, Ampie L, DeMarino C, et al. Differential expression of an endogenous retroviral element [HERV-K(HML-6)] is associated with reduced survival in glioblastoma patients. Sci Rep-uk. (2022) 12(1):6902. doi: 10.1038/s41598-022-10914-5
92. Kessler AF, Wiesner M, Denner J, Kämmerer U, Vince GH, Linsenmann T, et al. Expression-analysis of the human endogenous retrovirus HERV-K in human astrocytic tumors. BMC Res Notes (2014) 7(1):159. doi: 10.1186/1756-0500-7-159
93. Grabski DF, Ratan A, Gray LR, Bekiranov S, Rekosh D, Hammarskjold ML, et al. Human endogenous retrovirus-K mRNA expression and genomic alignment data in hepatoblastoma. Data Brief (2020) 31:105895. doi: 10.1016/j.dib.2020.105895
94. Grabski DF, Ratan A, Gray LR, Bekiranov S, Rekosh D, Hammarskjold ML, et al. Upregulation of human endogenous retrovirus-K (HML-2) mRNAs in hepatoblastoma: Identification of potential new immunotherapeutic targets and biomarkers. J Pediatr Surg (2021) 56(2):286–92. doi: 10.1016/j.jpedsurg.2020.05.022
95. Ma W, Hong Z, Liu H, Chen X, Ding L, Liu Z, et al. Human endogenous retroviruses-K (HML-2) expression is correlated with prognosis and progress of hepatocellular carcinoma. BioMed Res Int (2016) 2016:8201642. doi: 10.1155/2016/8201642
96. Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, et al. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res Off J Am Assoc Cancer Res (2002) 8(6):1800–7.
97. Kämmerer U, Germeyer A, Stengel S, Kapp M, Denner J. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol (2011) 91(1–2):1–8. doi: 10.1016/j.jri.2011.06.102
98. Simpson GR, Patience C, Löwer R, RalfR Tönjes, Moore HDM, Weiss RA, et al. Endogenous d-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology (1996) 222(2):451–6. doi: 10.1006/viro.1996.0443
99. Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, et al. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci (2004) 101(6):1731–6. doi: 10.1073/pnas.0305763101
100. Mi S, Lee X, Li XP, Veldman GM, Finnerty H, Racie L, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature (2000) 403(6771):785–9. doi: 10.1038/35001608
101. Lemaître C, Tsang J, Bireau C, Heidmann T, Dewannieux M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PloS Pathog (2017) 13(6):e1006451. doi: 10.1371/journal.ppat.1006451
102. Dittmar T, Weiler J, Luo T, Hass R. Cell-cell fusion mediated by viruses and HERV-derived fusogens in cancer initiation and progression. Cancers (2021) 13(21):5363. doi: 10.3390/cancers13215363
103. Müller MD, Holst PJ, Nielsen KN. A systematic review of expression and immunogenicity of human endogenous retroviral proteins in cancer and discussion of therapeutic approaches. Int J Mol Sci (2022) 23(3):1330. doi: 10.3390/ijms23031330
104. Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer immunity : J Acad Cancer Immunol (2004) 4:2.
105. Yuan Z, Gardiner JC, Maggi EC, Huang S, Adem A, Bagdasarov S, et al. B7 immune-checkpoints as targets for the treatment of neuroendocrine tumors. Endocr-relat Cancer (2021) 28(2):135–49. doi: 10.1530/ERC-20-0337
106. Pérot P, Mullins CS, Naville M, Bressan C, Hühns M, Gock M, et al. Expression of young HERV-h loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget (2015) 6(37):40095–111. doi: 10.18632/oncotarget.5539
107. Wentzensen N, Coy JF, Knaebel H, Linnebacher M, Wilz B, Gebert J, et al. Expression of an endogenous retroviral sequence from the HERV-h group in gastrointestinal cancers. Int J Cancer (2007) 121(7):1417–23. doi: 10.1002/ijc.22826
108. Manca MA, Solinas T, Simula ER, Noli M, Ruberto S, Madonia M, et al. HERV-K and HERV-h env proteins induce a humoral response in prostate cancer patients. Pathogens (2022) 11(1):95. doi: 10.3390/pathogens11010095
109. Hu L, Hornung D, Kurek R, Ostman H, Helen O, Blomberg J, et al. Expression of human endogenous gammaretroviral sequences in endometriosis and ovarian cancer. AIDS Res Hum Retrov. (2006) 22(6):551–7. doi: 10.1089/aid.2006.22.551
110. Zare M, Mostafaei S, Ahmadi A, Jamalkandi SA, Abedini A, Esfahani-Monfared Z, et al. Human endogenous retrovirus env genes: Potential blood biomarkers in lung cancer. Microb Pathog (2018) 115:189–93. doi: 10.1016/j.micpath.2017.12.040
111. Dolci M, Favero C, Tarantini L, Villani S, Bregni M, Signorini L, et al. Human endogenous retroviruses env gene expression and long terminal repeat methylation in colorectal cancer patients. Med Microbiol Immun (2020) 209(2):189–99. doi: 10.1007/s00430-020-00662-6
112. Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-h elements in colon cancer. PloS One (2012) 7(1):e29950. doi: 10.1371/journal.pone.0029950
113. Lindeskog M, Blomberg J. Spliced human endogenous retroviral HERV-h env transcripts in T-cell leukaemia cell lines and normal leukocytes: alternative splicing pattern of HERV-h transcripts. J Gen Virol (1997) 78(10):2575–85. doi: 10.1099/0022-1317-78-10-2575
114. Sexton CE, Tillett RL, Han MV. The essential but enigmatic regulatory role of HERVH in pluripotency. Trends Genet (2021) 38(1):12–21. doi: 10.1016/j.tig.2021.07.007
115. Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol (2003) 77(19):10414–22. doi: 10.1128/JVI.77.19.10414-10422.2003
116. Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology (2004) 1(1):32. doi: 10.1186/1742-4690-1-32
117. Heidmann O, Béguin A, Paternina J, Berthier R, Deloger M, Bawa O, et al. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci (2017) 114(32):E6642–51. doi: 10.1073/pnas.1702204114
118. Chang TC, Goud S, Torcivia-Rodriguez J, Hu Y, Pan Q, Kahsay R, et al. Investigation of somatic single nucleotide variations in human endogenous retrovirus elements and their potential association with cancer. PloS One (2019) 14(4):e0213770. doi: 10.1371/journal.pone.0213770
119. Babarinde IA, Ma G, Li Y, Deng B, Luo Z, Liu H, et al. Transposable element sequence fragments incorporated into coding and noncoding transcripts modulate the transcriptome of human pluripotent stem cells. Nucleic Acids Res (2021) 49(16):gkab710–. doi: 10.1093/nar/gkab710
120. Petrizzo A, Ragone C, Cavalluzzo B, Mauriello A, Manolio C, Tagliamonte M, et al. Human endogenous retrovirus reactivation: Implications for cancer immunotherapy. Cancers (2021) 13(9):1999. doi: 10.3390/cancers13091999
121. Wu Y, Zhao Y, Huan L, Zhao J, Zhou Y, Xu L, et al. An LTR retrotransposon-derived long noncoding RNA lncMER52A promotes hepatocellular carcinoma progression by binding p120-catenin. Cancer Res (2020) 80(5):976–87. doi: 10.1158/0008-5472.CAN-19-2115
122. Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv (2019) 5(3):eaat9820. doi: 10.1126/sciadv.aat9820
123. Katz RA, Skalka AM. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol (1990) 10(2):696–704. doi: 10.1128/mcb.10.2.696-704.1990
125. Jern P, Sperber GO, Ahlseín G, Blomberg J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus h. J Virol (2005) 79(10):6325–37. doi: 10.1128/JVI.79.10.6325-6337.2005
126. Kolbe AR, Bendall ML, Pearson AT, Paul D, Nixon DF, Pérez-Losada M, et al. Human endogenous retrovirus expression is associated with head and neck cancer and differential survival. Viruses (2020) 12(9):956. doi: 10.3390/v12090956
127. Cuffel C, Rivals J, Zaugg Y, Salvi S, Seelentag W, Speiser DE, et al. Pattern and clinical significance of cancer-testis gene expression in head and neck squamous cell carcinoma. Int J Cancer (2011) 128(11):2625–34. doi: 10.1002/ijc.25607
128. Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M, et al. The landscape of viral associations in human cancers. Nat Genet (2020) 52(3):320–30. doi: 10.1038/s41588-019-0558-9
129. Michna A, Schötz U, Selmansberger M, Zitzelsberger H, Lauber K, Unger K, et al. Transcriptomic analyses of the radiation response in head and neck squamous cell carcinoma subclones with different radiation sensitivity: time-course gene expression profiles and gene association networks. Radiat Oncol (2016) 11(1):94. doi: 10.1186/s13014-016-0672-0
130. Landriscina M, Fabiano A, Altamura S, Bagalà C, Piscazzi A, Cassano A, et al. Reverse transcriptase inhibitors down-regulate cell proliferation in vitro and in vivo and restore thyrotropin signaling and iodine uptake in human thyroid anaplastic carcinoma. J Clin Endocrinol Metab (2005) 90(10):5663–71. doi: 10.1210/jc.2005-0367
131. Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res (2008) 134(1–2):221–34. doi: 10.1016/j.virusres.2007.12.010
132. Di C, Syafrizayanti, Zhang Q, Chen Y, Wang Y, Zhang X, et al. Function, clinical application, and strategies of pre-mRNA splicing in cancer. Cell Death Differ (2019) 26(7):1181–94. doi: 10.1038/s41418-018-0231-3
133. Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, et al. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature (2021) 591(7849):312–6. doi: 10.1038/s41586-020-03135-1
134. Stoltzfus C, Madsen J. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res (2006) 4(1):43–55. doi: 10.2174/157016206775197655
135. Herai RH, Yamagishi MEB. Detection of human interchromosomal trans-splicing in sequence databanks. Brief Bioinform (2010) 11(2):198–209. doi: 10.1093/bib/bbp041
136. Segni GD, Gastaldi S, Tocchini-Valentini GP. Cis- and trans-splicing of mRNAs mediated by tRNA sequences in eukaryotic cells. Proc Natl Acad Sci (2008) 105(19):6864–9. doi: 10.1073/pnas.0800420105
137. Klerk E, Hoen PAC ‘t. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet (2015) 31(3):128–39. doi: 10.1016/j.tig.2015.01.001
138. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
139. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
140. Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell (2013) 154(1):26–46. doi: 10.1016/j.cell.2013.06.020
141. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature (2001) 409(6822):860–921. doi: 10.1038/35057062
142. Podgornaya OI, Ostromyshenskii DI, Enukashvily NI. Who needs this junk, or genomic dark matter. Biochem Mosc (2018) 83(4):450–66. doi: 10.1134/S0006297918040156
143. Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. Jnci J Natl Cancer Inst (2012) 104(3):189–210. doi: 10.1093/jnci/djr540
144. Bronte V, Cingarlini S, Apolloni E, Serafini P, Marigo I, Santo CD, et al. Effective genetic vaccination with a widely shared endogenous retroviral tumor antigen requires CD40 stimulation during tumor rejection phase. J Immunol (2003) 171(12):6396–405. doi: 10.4049/jimmunol.171.12.6396
Keywords: HERV human endogenous retroviruses, endogenous retroviruses (ERV), aberrant splicing, non-canonical splicing, non-conventional splicing, head and neck cancer, cancer
Citation: Agoni L (2022) Alternative and aberrant splicing of human endogenous retroviruses in cancer. What about head and neck? —A mini review. Front. Oncol. 12:1019085. doi: 10.3389/fonc.2022.1019085
Received: 14 August 2022; Accepted: 03 October 2022;
Published: 20 October 2022.
Edited by:
Rong Jia, Wuhan University, ChinaReviewed by:
Elena Cherkasova, National Heart, Lung, and Blood Institute (NIH), United StatesCopyright © 2022 Agoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Agoni, bG9yZW56by5hZ29uaUBnbWFpbC5jb20=
 Lorenzo Agoni
Lorenzo Agoni