- 1The First Affiliated Hospital of University of Science and Technology of China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Heifei, China
- 2Institute of Immunology and the Chinese Academy of Sciences (CAS) Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medicine and Medical Center, University of Science and Technology of China, Hefei, China
SIRT6 is a member of the Sir2-like family in mammals. Recent structural and biochemical studies have characterized SIRT6 as having deacetylation, defatty-acylation, and mono-ADP-ribosylation activities, which determine its important regulatory roles during physiological and pathological processes. This review focuses mainly on the regulatory functions of SIRT6 in aging, cancer, and, especially, immunity. Particular attention is paid to studies illustrating the critical role of SIRT6 in the regulation of immune cells from the viewpoints of immunesenescence, immunometabolism, and tumor immunology. Owing to its role in regulating the function of the immune system, SIRT6 can be considered to be a potential therapeutic target for the treatment of diseases.
Introduction
SIRT6 is a multifunctional protein with several enzymatic activities. First, SIRT6 is categorized as a class III histone deacetylase with deacetylation activity (1). However, its substrates are not limited to the acetyl groups of histone H3 and H4 lysine residues but can also include other proteins with acetyl groups on their lysine residues (2), allowing SIRT6 to regulate gene expression or protein activity through post-translational modifications (PTMs). Second, SIRT6 has defatty-acylation activity, which enables it to regulate the secretion of proteins including tumor necrosis factor alpha (TNF-α) (3). Finally, SIRT6 is a mono-ADP-ribosylation enzyme; it can activate poly(ADP-ribose) polymerase-1 (PARP-1), thereby promoting repair of DNA damage (4). These three enzymatic characteristics form the foundation of the ability of SIRT6 to regulate various physiological and pathological processes.
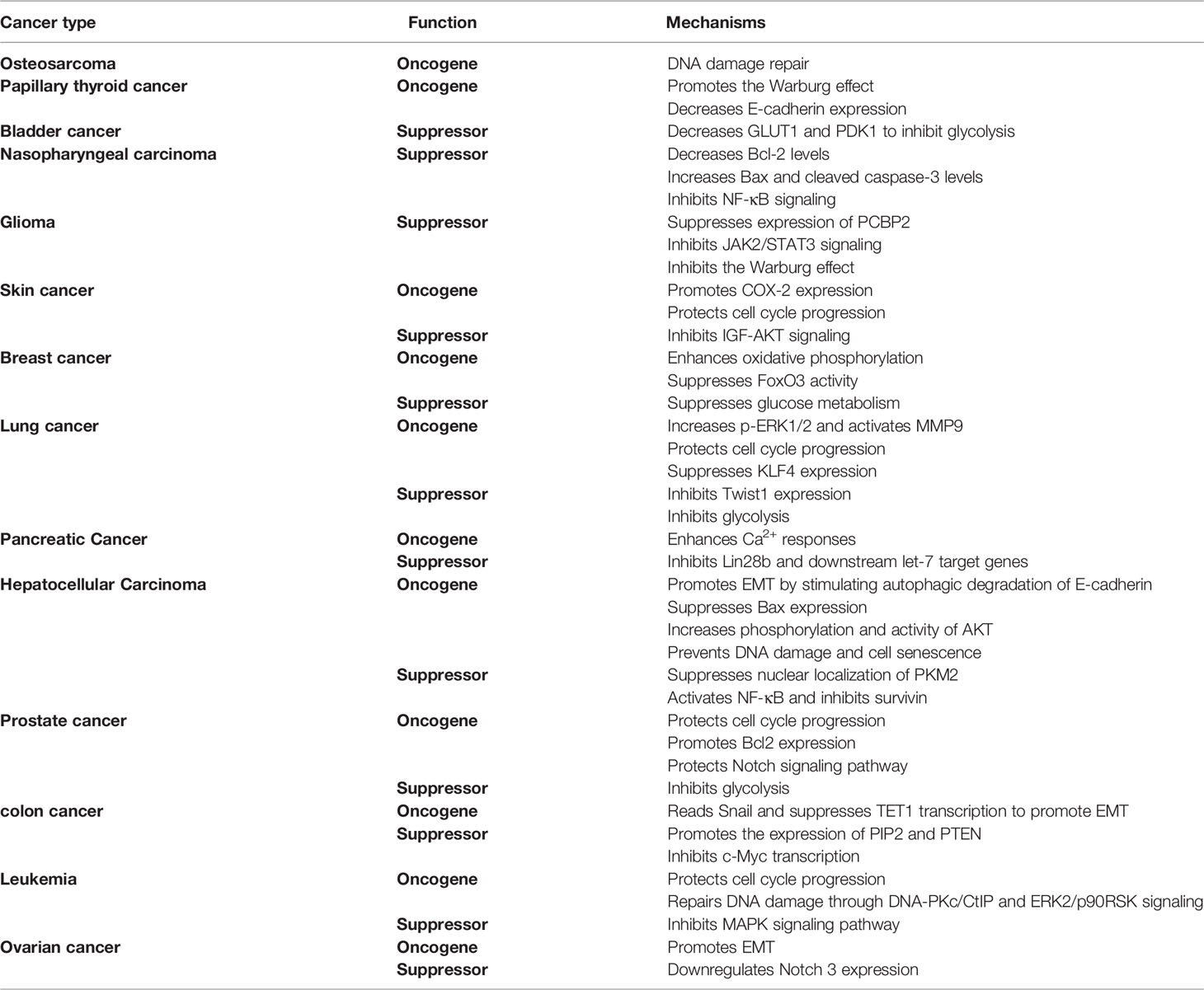
SIRT6 is a longevity protein that can inhibit the aging of cells, tissues, organs and the body by promoting DNA damage repair (5–7), maintaining normal chromosome structure (8–10), and regulating energy metabolism (11, 12) and the senescence-associated secretory phenotype (SASP) (13, 14). Immunosenescence, an aspect of aging, is also inhibited by SIRT6 (15). In addition, SIRT6 can regulate the development of inflammation. At present, most evidence reflects the anti-inflammatory effects of SIRT6 via inhibition of the production of inflammatory cytokines and promotion of polarization of immune cells to an immunosuppressive phenotype. For example, SIRT6 has been shown to promote the M2 polarization of macrophages (16–19). However, a few studies have demonstrated pro-inflammatory activity of SIRT6, manifested in the promotion of infiltration and survival of inflammatory immune cells and inflammatory cytokine production (3, 14, 20). SIRT6 also regulates the development process of cancer, an aging-related disease. In melanoma, breast, lung, pancreatic, liver, prostate, colon, ovarian, and blood cancers, SIRT6 has contradictory roles as either oncogene or tumor suppresser at different stages of the cancer or in different tumor cell lines. It shows only tumorigenic effects in osteosarcoma (21) and papillary thyroid cancer (22, 23) and only antitumorigenic effects in bladder cancer (24), nasopharyngeal carcinoma (25), and glioma (26–28). Here, we briefly summarize the enzymatic characteristics of SIRT6; describe in detail the roles of these biochemical and molecular characteristics in aging, immunity, and cancer; and explore the role of SIRT6 in epigenetic immunity and tumor immunology.
Enzymatic Characteristics of SIRT6
SIRT6 is a member of the Sir2-like protein family. Mammalian sirtuins can be divided into four classes according to their core domain sequences. SIRT6 and SIRT7 are class IV sirtuins (29). SIRT6 has three major enzymatic activities: deacetylation, defatty-acylation, and mono-ADP-ribosylation. These form an important basis for its participation in the regulation of physiological and pathological processes in mammals, including aging, immunity, and cancer occurrence and development.
Deacetylation Activity
As SIRT6 is located in the nucleus and only diffuses into the cytoplasm (30), it is mainly involved in the regulation of acetylation of lysines on proteins in the nucleus. SIRT6 is a NAD+-dependent deacetylase (31). The deacetylation activity of SIRT6 is dependent on nucleosomes, as it adopts its active structure through binding to nucleosomes. When histone H3 and H4 are packaged as nucleosomes instead of free histones, SIRT6 binds to substrates and catalyzes deacetylation (32). Lysine acetylations of multiple sites of histones H3 and H4 are substrates of SIRT6. For example, SIRT6 dynamically combines with chromatin to deacetylate acetylated H3K9 (H3K9ac), thereby regulating telomeric chromatin and inhibiting end-to-end chromosomal fusion and premature aging of cells (31). SIRT6 deacetylates H3K18ac on pericentric heterochromatin to prevent mitotic errors and cell senescence (33). SIRT6 deacetylates H3K56ac. In SIRT6-knockdown cells, hyperacetylation of H3K56 at telomeres was found to influence telomeric chromatin structure, leading to telomere dysfunction and genomic instability (34). SIRT6 can be recruited to the proximal promoter region of the Pcsk9 gene by transcription factor forkhead box O3 (FOXO3), deacetylating H3K9ac and H3K56ac, and thereby inhibiting gene expression (35). Knockdown of SIRT6 in mouse oocytes induces hyperacetylation of H4K16, which significantly increases the incidence of aneuploidy and severely impairs kinetochore-microtubule interactions (36). The deacetylation efficiency of SIRT6 on different substrates is different. SIRT6 can effectively remove the acetyl groups from H3K9, H3K18, and H3K27, whereas its deacetylation activity is weak on H3K4ac, H3K14ac, H3K23ac, H3K36ac, H3K56ac, and H3K79ac (37). In addition to histones, the substrates of SIRT6 include acetylated lysine residues of other proteins, including GCN5 K549ac, PKM2 K433ac, Ku70 K542ac, NAMPT K53ac/369ac, XBP1s K257/297ac, SOD2 K68/122ac, and p53 K382ac (2). The catalytic mechanism of SIRT6-mediated enzymatic function has been well reviewed by Fiorentino et al. (38). Similar to many enzymatic reactions, the catalytic activity of SIRT6 is adjustable. SIRT6 shows poor deacetylase activity in vitro (32); however, free fatty acids could increase SIRT6 deacetylation activity 35-fold at physiological concentrations (39).
Defatty-Acylation Activity
In addition to its deacetylation activity, SIRT6 has defatty-acylation activity, which has emerged as a mechanism regulating the secretion of many proteins. Lysine fatty acylation of TNF-α promotes its lysosomal targeting and degradation (40). A large hydrophobic pocket was identified in the crystal structure of SIRT6 that could accommodate long-chain fatty acyl groups. SIRT6 promotes TNF-α secretion by removing the fatty acyl modification on TNF-α K19 and K20 (3). Furthermore, many ribosomal proteins are secreted by the exosomes of SIRT6-knockout MEFs; this was shown to increase the proliferation of NIH 3T3 mouse embryonic fibroblasts (MEFs) (41). In the same cell line, a SIRT6-knockout mutant showed upregulation of R-Ras2 lysine fatty acylation, which facilitated the localization of R-Ras2 to the plasma membrane and promoted its interaction with phosphatidylinositol 3-kinase (PI3K), thereby activating the Akt signaling pathway and increasing cell proliferation (42).
Mono-ADP-Ribosylation Activity
SIRT6 is a mono-ADP-ribosyltransferase. Mouse SIRT6 (mSIRT6) relies on NAD+ for intramolecular single ADP-riboglycosylation and can be recognized by an antibody specific to mono-ADP-ribose (30). Purified recombinant mSIRT6 has been shown to catalyze radiolabel transfer of [32P] NAD. The transfer of NAD+ to mSIRT6 occurs via an intra-molecular mechanism, indicating that SIRT6 is an auto-ADP-ribosyltransferase (30). SIRT6 mono-ADP-ribosylates PARP1 K521 to enhance DSB repair (4). It can also mono-ADP-ribosylates BAF170 K312 to promote NRF2 target gene transcription (43). Pan et al. solved the structure of the human SIRT6-ADP-ribose complex (44).
SIRT6 and Aging
SIRT6 is an important protein with anti-aging effects on cells, tissue, organs, and the body. It inhibits aging via four main pathways: promotion of DNA damage repair, maintenance of the normal telomere structure of chromosomes, regulation of glucose and NAD+ metabolic balance, and regulation of SASP.
Physiology and Pathology of SIRT6 in Aging
SIRT6 is a longevity protein that delays the aging process and participates in the maintenance of telomere and genome stability. A study at the organism level showed that SIRT6-deficient mice had small body size, with loss of subcutaneous fat, profound lymphopenia, lordokyphosis, and severe metabolic defects 2–3 weeks after birth, and eventually died at approximately 4 weeks of age (5). SIRT6 deficiency leads to hyperacetylation of histones at the imprinting control region of developmental repressor H19, which results in severe prenatal developmental delay and death several hours after birth in SIRT6-deficient monkeys (45). By contrast, transgenic mice overexpressing SIRT6 have a longer life span than wild-type mice (11, 46). A study at the organ and tissue levels also demonstrated the anti-aging activity of SIRT6 and proposed SIRT6 as a potential marker of ovarian aging. Its expression was positively correlated with the number of primordial follicles; both SIRT6 protein expression and ovarian reserves decreased with increasing age (47). SIRT6-deficient mice showed symptoms of myocardial hypertrophy and heart failure, and the expression of SIRT6 in failing human hearts was reduced (48).
Cell-level studies have also shown that SIRT6 inhibits cell senescence. SIRT6 could reduce cardiac hypertrophy and cardiomyocyte senescence (49). The addition of SIRT6 increased the resistance of elderly human dermal fibroblasts to classic Yamanaka factor-induced reprogramming (50). SIRT6-deficient human mesenchymal stem cells exhibited accelerated functional decline, which was mainly characterized by redox metabolism disorders and increased sensitivity to oxidative stress and was different from typical cell senescence (51). By contrast, overexpression of SIRT6 inhibited the replicative senescence of chondrocytes (52).
Mechanism of SIRT6 in Anti-Aging
Cell and molecular biology can explain the anti-aging effect of SIRT6. Firstly, SIRT6 is a nucleolar chromatin-related protein involved in various DNA damage repair processes. SIRT6 is related to base excision repair (BER), which has been shown to promote resistance to DNA damage in mouse cells, suppress genomic instability, and promote normal DNA recombination (5). Mechanistically, SIRT6 activates PARP1 through mono-ADP-ribosylation activity and interacts with two BER enzymes (hMYH and hAPE1) to promote BER (6, 53). SIRT6 was recruited to ultraviolet-induced DNA damage sites and deacetylated damaged DNA binding protein 2 (DDB2) at K35 and K77, promoting segregation of DDB2 from chromatin and thereby facilitating nucleotide excision repair (NER) (7). In mammalian cells, oxidative stress activates the protein kinase c-Jun N-terminal kinase to phosphorylate SIRT6 at serine 10; SIRT6 is then recruited to DNA double-strand break (DSB) sites. SIRT6 mono-ADP-ribosylates the K521 lysine of PARP1; this activates PARP1 and enhances DSB repair under oxidative stress (4, 54). SIRT6 recruits chromatin recombinant SNF2H to the DNA cleavage site and deacetylates histone H3K56ac, preventing genomic instability through chromatin remodeling and facilitating the repair of damaged sites (55).
Secondly, SIRT6 can maintain the normal chromosome structure. The stability of chromosomes and telomeres is particularly important for cell anti-aging (56). SIRT6-deficient cells exhibit abnormal telomere structures, including increased chromosomal fragmentation, detached centromeres, and chromosomal gaps, similar to cell defects observed in Werner syndrome (a premature aging disorder) (5, 31). The deacetylation activity of SIRT6 plays an important part in maintaining genome stability. SIRT6 deacetylates H3K9ac, stabilizes the binding of WRN to telomeric chromatin to resist replication-related telomere defects, and prevents end-to-end chromosomal fusion and premature senescence of cells (31). SIRT6 interacts with the nuclear factor-κB (NF-κB) RELA subunit and deacetylates H3K9ac at the promoter of the NF-κB target gene to suppress cell senescence (8). SIRT6 promotes H3K18ac deacetylation, silences pericentric heterochromatin at centromeres, and prevents aberrant accumulation of pericentric transcripts (33). SIRT6 is necessary for maintaining the silencing of the telomere position effect in human cells and plays a key part in maintaining the structure of silent telomeric chromatin (9). As a powerful repressor of retrotransposon L1, SIRT6 mono-ADP-ribosylates KRAB-associated protein 1 (KAP1) and promotes the interaction between KAPI and heterochromatin protein 1 alpha (HP1α), thereby contributing to the packaging of L1 gene elements into heterochromatin to reduce their expression (10).
Thirdly, in addition to maintaining genomic stability as mentioned above, SIRT6 slows the process of aging by regulating glucose homeostasis and the NAD+ metabolic balance (11, 12). Overexpression of SIRT6 is conducive to a “young state” of blood glucose and gluconeogenesis in aged mice. SIRT6 promotes hepatic gluconeogenesis by increasing lipolysis, and increases the levels of precursors of the gluconeogenesis and tricarboxylic acid (TCA) cycles, thereby maintaining the young state of these two cycles (11). In addition, SIRT6 is helps to maintain NAD+ levels by increasing the expression of de novo NAD+ synthesis genes (11). However, SIRT6 is a NAD+-dependent enzyme and also consumes NAD+ during the processes of deacetylation (38), defatty-acylation (42), and mono-ADP-ribosylation (30). NAD+ can delay aging by inhibiting P53 activity. Disruption of the NAD+/NADH ratio in cell solute could promote aging through mitochondrial dysfunction (12). The above conclusions are described in detail in Roichman's research (11) and Wiley’s review (12).
Finally, some studies have shown that SIRT6 affects the SASP to prevent aging. Aging is closely related to the immune system, and immunosenescence is a part of aging. Immune cell components, functions, and intercellular interactions in the innate and adaptive immune systems tend to develop immunotolerance in the process of immunosenescence (57, 58). SIRT6 levels in the articular chondrocytes of osteoarthritis patients are significantly reduced; therefore, overexpression of SIRT6 could reduce the senescence of chondrocytes and prevent the development of osteoarthritis (52). Studies have shown that SIRT6 is directly involved in the regulation of immunosenescence. Dendritic cell (DC) dysfunction is at the core of various common chronic diseases and contributes to the reduction in immunocompetence that occurs during aging. SIRT6-knockout (SIRT6KO) mice showed a lower frequency of bone marrow conventional DC (cDC) precursors and lower numbers of bone marrow-derived cDCs. SIRT6KO mouse cDCs expressed low levels of MHCII, chemokine receptor CCR7, and costimulatory molecules and had lower immunostimulatory activity than wild-type cDCs. The ability of SIRT6KO cDCs to produce IL-12 was generally reduced. SIRT6 deficiency prevented the maturation of BMDCs (bone marrow-derived DCs) generated in vitro in a partial TNF-α-dependent manner. SIRT6 helps BMDCs respond to Toll-like receptor (TLR) ligands. The proliferation of allogeneic lymphocytes in a mixed leukocyte reaction (MLR) stimulated by cells cultured in the presence of SIRT6 inhibitors was also significantly reduced. Therefore, SIRT6 plays a crucial part in the differentiation and function of cDCs, and loss of function of SIRT6 may promote immunosenescence (15). Inflamm-aging (a long-term low levels of inflammatory mediators in aging individuals) (57) will follow. This is often accompanied by the production of a SASP (senescent cells secrete pro-inflammatory cytokines, chemokines, and proteases) (13). SIRT6 increases TNF secretion in BMDCs and THP-1 through post-transcriptional steps (3, 14). Overexpression of SIRT6 proved sufficient to delay the replicative senescence of diploid fibroblast WI38 by attenuating NF-κB signaling. Knockdown of SIRT6 leads to accelerated cell senescence and overactivation of NF-κB (59). Therefore, SIRT6 affects the NK-κB pathway, which regulates cytokine production (discussed in more detail later), and then regulates SASP. In short, SIRT6 may also participate in inflamm-aging via affects on the SASP through regulating the synthesis and release of inflammatory factors.
SIRT6 and Immunity
SITR6 regulates inflammatory development, and plays a complex role. It inhibits inflammatory by promoting M2 macrophage polarization, decreasing number of lymphocytes, inhibiting T cell differentiation, and inhibiting innate immunity response. However it also promotes inflammatory by promoting neutrophils and dendritic cells migration, and promoting TNF-α secretion. Besides, SIRT6 could regulate immunometabolism. All of these will be discussed in detail below.
SIRT6 Regulates Inflammatory Development
Some studies have shown that SIRT6 can inhibit inflammation. In myeloid cells, most studies have focused on the role of SIRT6 in regulating macrophage polarization. Sirt6f/f:Fabp4-Cre mice (with SIRT6 deficiency in preadipocytes and mature adipocytes) exhibited increased expression of inflammatory genes including F4/80, Tnfα, Mcp-1, and Il6 in both brown and white adipose tissues (16). Adipocyte-specific S6KO mice showed increased infiltration of macrophages in epididymal white adipose tissue, many M1 macrophage genes (Il1b, Ccl2, Tnfα, Il6, Nos2, and Ccr2) were upregulated at the mRNA level, and inflammation was increased. However, the mRNA levels of M2 macrophage genes (Mrc1, Mgl1, Arg1, and Il10) were downregulated (17). In the same mouse model, macrophages infiltrated into adipose tissue. Moreover, the ratio of M1/M2 macrophages was significantly increased, and the mRNA expression of inflammatory genes including CD11b+, Cxcl2, CD68, Tnfα, Mcp-1, and Il6 was also increased when mice were fed a high-fat diet (18). By contrast, SIRT6 regulated the expression of IL-4 by adipocytes through an autocrine route, thereby promoting the polarization of macrophages to the M2 type and reducing inflammation (17). In a full-thickness excisional lesion model of dorsal skin, the infiltration of M1 macrophages in myeloid-cell-specific S6KO (mS6KO) mice increased, whereas numbers of M2 macrophages decreased. Therefore, inflammation at the wound site increased, and wound healing was impaired. By contrast, under the condition of M2 polarization stimulated by IL-4, transducing mS6KO bone marrow macrophages (BMMs) with adenovirus expressing Sirt6 promoted the polarization of M2 macrophages by protecting the PI3K-Akt pathway (19). Clinical analysis has shown that levels of SIRT6 in particular chondrocytes of osteoarthritis patients are significantly reduced. Expression levels of SIRT6 in peripheral blood mononuclear cells, monocytes, and macrophages are lower in patients with rheumatoid arthritis compared with those with osteoarthritis. The activity of SIRT6 is negatively correlated with the severity of the disease. Overexpression of SIRT6 can reduce the inflammatory response by reducing the expression of NF-κB-dependent genes, thereby preventing the development of arthritis (52, 60). The arthritis of mS6KO mice is more serious than that of wild-type mice. Lack of SIRT6 leads to upregulation of acetylated-FoxO1 protein levels and CCR3 expression in macrophages, and the migration of macrophages to synoviocyte-derived chemoattractants is enhanced so that more macrophages gather in the synovium (60). In collagen-induced arthritis (CIA), overexpression of SIRT6 inhibits the differentiation of osteoclasts in BMDCs induced by macrophage colony-stimulating factor. The severity of arthritis is reduced, and levels of local and systemic pro-inflammatory cytokines are also downregulated (61). In neuro-inflammation, SIRT6 activation inhibits lipopolysaccharide (LPS)-stimulated inflammatory responses of RAW264.7 macrophages and primary mouse microglia (62). In vitro experiments have also confirmed the anti-inflammatory effect of SIRT6. SIRT6-deficient macrophages promote the activation of NF-κB and the production of IL-6, which results in signal transducer and activator of transcription 3 (STAT3) activation and a positive feedback loop for NF-κB stimulation, and finally accelerates the polarization of pro-inflammatory M1 macrophages (63). By contrast, icariin (ICA) upregulates the expression and increases the activity of SIRT6. ICA treatment inhibits the NF-κB inflammatory signaling pathway and reduces mRNA levels of the NF-κB downstream target genes: Tnfα, Il2, ICAM-1, and Il6, thereby inhibiting inflammatory development (64). BMMs of Sirt6-null mice show high expression of pro-inflammatory genes encoding MCP-1, TNF-α, and IL-6 and hypersensitivity to LPS stimulation due to the hyperacetylation of H3K9 and the increase in c-JUN occupancy in the promoters of these genes (65). By contrast, macrophages overexpressing SIRT6 are transformed into the M2 phenotype, which can avoid the damage induced by high glucose levels (66). S6KO mice have abnormal immune systems and metabolism. There are inextricable links between immune cells and immune cells, and between metabolism and immune cells. It is thus difficult to accurately study a particular type of immune cell under these conditions of multifactorial changes.
Studies have also shown that SIRT6 inhibits inflammation via effects on lymphocyte differentiation and function. In S6KO mice, lordokyphosis, colitis caused by erosion of the superficial colonic epithelium, acute loss of subcutaneous fat, and severe lymphopenia relevant to increased lymphocyte apoptosis were observed. Flow cytometry analysis showed that in the thymus, the number of CD4+-CD8 + double-positive cells decreased 50-fold, and in bone marrow, the number of splenic lymphocytes and progenitor B cells decreased 10-fold (5). In CIA rats, a low percentage of regulatory T cells (Tregs) was observed following treatment with C3G (an inhibitor of CD38) and the Sirt6 inhibitor OSS_128167 (67). SIRT6 interacts with and deacetylates GATA3, inhibits the Th2 immune response, and reduces the expression of IL-4, IL-5, and IL-13, thereby weakening airway allergic inflammation induced by ovalbumin or house dust mites (68). S6KO mice developed chronic liver inflammation at approximately 2 months old, and the absence of SIRT6 in T cells was sufficient to induce liver fibrosis and inflammation (65).
Furthermore, SIRT6 negatively regulates the innate immune response during dengue virus (DENV) infection. SIRT6 silencing enhances the production of pro-inflammatory cytokines and chemokines. Overexpression of SIRT6 inhibits NF-κB activation mediated by RIG-I-like receptor and TLR3. The sirtuin core domain of SIRT6 is important for inhibiting NF-κBp65 function. SIRT6 interacts with the p65 DNA-binding domain and competes with p65 to bind the IL-6 promoter and reduce the expression of IL-6 during DENV infection (69).
However, some studies have confirmed that SIRT6 promotes inflammatory development and has pro-inflammatory potential. Human SIRT6 promotes TNF-α secretion by removing the fatty acyl modifications on TNF-α K19 and K20 (3). After CpG stimulation of BMDCs from SIRT6-deficient mice, the amounts of TNF-α synthesized by the cells decreased, which confirmed the pro-inflammatory effect of SIRT6 (14). In autoimmune diseases, SIRT6 inhibitors effectively delay the onset of experimental autoimmune encephalomyelitis (EAE) via the following mechanism: inhibition of SIRT6 reduces the expression of CD40 on lymph node DCs, decreases encephalitogenic T cell infiltration, and decreases the ability of CXCR4+ DCs to migrate into the lymph nodes of EAE mice. Levels of IFN-γ, TNF-α, and IL-12 were also decreased, but the expression of IL-10 was increased with an anti-inflammatory effect (20). SIRT6 also increased the levels of cAMP/Ca2+-dependent transcription factors and nuclear factor of activated T cells through its deacetylation activity, thereby enhancing the expression of TNF- α and chemokine IL-8 (70).
SIRT6 Regulates Immunometabolism
Many studies have confirmed that SIRT6 is involved in cell metabolism regulation (71). Metabolism is an important factor affecting the development and function of immune cells (72, 73). For example, Shun et al. reported that hypoxia may stimulate cell glycolysis and autophagy, and that autophagy promotes the formation of DNA-containing immune complexes and trafficking of TLR9 to the signaling compartment, leading to hyper-responses of immune cells, which are related to the formation of nasal polyps (74–76). The expression of SIRT6 is inhibited in a chronic inflammatory state. If SIRT6 expression is increased, autophagy can be inhibited by inhibiting anaerobic glycolysis, which is conducive to disease treatment (77).Cyclosporine A inhibits neutrophil migration and apoptosis by inhibiting SIRT6, promoting the upregulation of HIF-1α expression and enhancing glycolysis and the TCA cycle, which is conducive to the remission of acute severe ulcerative colitis (78). During the transformation from early inflammation to late inflammation, SIRT6 deacetylates H3K9ac and H3K56ac on several glycolysis gene promoters, inhibits HIF-1α transcriptional activity, and reduces glycolysis activity to promote M2 polarization of macrophages (79).
SIRT6 might regulate T cell differentiation by metabolism. SIRT6 deacetylates and activates FoxO1 to regulate lipid metabolism in brown adipocytes (80). The absence of FoxO1 seriously inhibits the development of Foxp3+ Tregs (81). SIRT6 inhibits HIF-1α activation to inhibit glycolysis, and HIF-1α activates RORγt to enhance Th17 cell polarization (82). HIF-1α promotes IL-9 expression to induce Th9 cell polarization (83). It is well known that after challenge by pathogens, some T cells will continue to exist as longevity memory T cells, which maintain self-renewal capacity, allowing them to proliferate many times over a long duration to prevent rechallenge by the same pathogen. They are long-lived immune cells (84, 85). Unlike naive CD8+ T cells, the percentages of effector memory CD8+ T cells and central memory CD8+ T cells increased with age (86). Subsequently, researchers found that the central memory cells of older individuals shifted toward a chromatin-opening pattern and determined that the gene regulation driven by NRF1 and BATF was a potential target for delaying CD8+ T cell aging (87). Other studies have shown that NRF-1 itself is regulated by PTMs (acetylation, methylation, and phosphorylation) that enhance binding to its target genes (88). The above results suggest that SIRT6 may be involved in the accumulation of memory CD8+ T cells in the aging population because of its capacity to deacetylate.
Although few studies have shown SIRT6 to affect immune cell function through directly regulating immunometabolism, SIRT6 has been found to regulate immune cell activity via effects on PI3K/Akt, NF-κB, and HIF-1α as discussed above. The above three signaling pathways could be involved in regulation of the TCA cycle, glycolysis, pentose phosphate pathway, and Warburg effect, thereby affecting the energy metabolism of immune cells in the resting state and activated state (72, 89–94). SIRT6 also regulates intracellular levels of NAD+, which is an electron acceptor with a key role in cell metabolism. Therefore, SIRT6 is likely to regulate immune cell proliferation, growth, and function by affecting immunometabolism. Pillai’s review discusses this point in detail and proposes that SIRT6 participates in short-term regulation of immune cells through PTMs and long-term regulation through transcriptional regulation of metabolism-related genes (89). Therefore, how SIRT6 regulates immunometabolism and how this regulation affects cell function will be an interesting research direction.
SIRT6 and Cancer
On the one hand, frequent DNA damage and mutation will lead to canceration (95, 96), which provides important support for the development of tumors; on the other hand, chronic inflammation caused by aging will drive tumor initiation, growth, progression, and metastasis (97). Moreover, immunosenescence is conducive to escape of cancer cells from immune system attacks and their eventual development into cancer. As mentioned earlier, SIRT6 plays a positive role in maintaining genomic stability and preventing aging. So SIRT6 is involved in cancer regulation is obvious.In addition, large amounts of data indicate that SIRT6 is directly involved in the occurrence and development of cancer.
SIRT6 Promotes/Inhibits Cancer Development
In certain cancers, SIRT6 promotes cancer development. In osteosarcoma, inhibition of SIRT6 enhances the antitumor effect of doxorubicin by inhibiting the DNA damage repair pathway (21). In papillary thyroid cancer (PTC), SIRT6 increases generation of reactive oxygen species to promote the Warburg effect in PTC cells, and high levels of SIRT6 reduce expression of E-cadherin, thereby promoting the invasion and migration of PTC cells and promoting cancer development (22, 23).
However, in other cancers, SIRT6 exhibits tumor suppressor activity. When bladder cancer develops from T2 to T4, the expression of SIRT6 is significantly decreased. Low SIRT6 expression increases the acetylation of H3K9 and levels of Glut1 and PDK1, enhances glycolysis, and increases the proliferation ability of tumor cells (24). In nasopharyngeal carcinoma, SIRT6 overexpression reduces levels of anti-apoptotic protein Bcl-2 but increases levels of cleaved caspase-3 and pro-apoptotic protein Bax. High levels of SIRT6 inhibit NF-κB signaling and promote apoptosis of nasopharyngeal carcinoma cells (25). SIRT6 is downregulated in human glioma tissues and deacetylates H3K9ac on the promoter of PCBP2 to downregulate PCBP2 expression and inhibit glioma cell growth (26). Elevated SIRT6 expression leads to tumor cell apoptosis by upregulating the expression of Bax and cleaved caspase-8, and downregulating Bcl-2, and inhibiting the Janus kinase 2 (JAK2)-STAT3 pathway (27). FOXO3a transcriptionally activates SIRT6 to inhibit the Warburg effect in glioblastoma cells, thereby inhibiting the development of glioblastoma (28).
The Dual Role of SIRT6 in Cancer Development
In melanoma, CRISPR/Cas9 or lentivirus short hairpin RNA-mediated knockout or knockdown of the SIRT6 gene in A375 melanoma cells, leding to significantly reduced growth, vitality, and clonogenic survival rates of cancer cells, induced cell cycle arrest in G1 phase, and increased senescence-associated β-galactosidase staining (98, 99), reflecting the oncogenic activity of SIRT6. However, in BRAFV600E melanoma cells, SIRT6 haploinsufficiency induced resistance of melanoma cells to mitogen-activated protein kinase (MAPK) inhibitors by activating IGF signaling (100), suggesting an anti-tumor effect of SIRT6. The expression of SIRT6 is decreased in primary melanoma compared with melanocytic nevus. An increase in SIRT6 induces inhibition of cell proliferation, cell cycle arrest, and apoptosis. However, in the metastatic stage of melanoma, the expression of SIRT6 increases (possibly induced by FOXO3a) and promotes the development of melanoma in an autophagy-dependent manner by inhibiting IGF-AKT signaling (101, 102).
In breast cancer, SIRT6 can enhance the expression and activity of pyruvate dehydrogenase (PDH), thereby enhancing oxidative phosphorylation in breast cancer cells and promoting the occurrence of breast tumors in mice (103). High nuclear levels of SIRT6 promot cancer development and is significantly associated with poor overall survival (104). Low levels of SIRT6 increase acetylated FOXO3, thereby inhibiting tumor development (105). However, another study showed that ectopic expression of SIRT6 reduced pAkt, hexokinase-2, and PDH kinase-1 protein levels, thereby inhibiting metabolic pathways in breast cancer (106).
In lung cancer, SIRT6 is overexpressed in non-small-cell lung cancer (NSCLC) cell lines (107–109). SIRT6 increases extracellular signal-regulated kinase (p-ERK) 1/2 phosphorylation and activates matrix metalloproteinase 9 (MMP9) to facilitate tumor cell migration and invasion (109). Silencing of SIRT6 impaired the proliferation and differentiation of NSCLC cell lines, arresting cells in the S and G0/G1 phases (107). miR-34 inhibited the proliferation of A549 cells by inhibiting SIRT6 expression (108). A lack of SIRT6 leads to upregulation of Kruppel-like factor 4 (KLF4) in NSCLC cells to reduce their invasiveness (110). However, studies have shown that patients with low nuclear expression of SIRT6 have cancer that is more aggressive and shorter survival (111). SIRT6 inhibits cell proliferation by inhibiting the expression of Twist1 in NSCLC (112). In the A549 lung cancer cell line, α-hederin was shown to inhibit c-Myc and HIF-1α by increasing the expression of SIRT6 to inhibit glycolysis and further inhibit the proliferation of A549 cells (113).
In pancreatic cancer, the SIRT6 inhibitor quinazolinedione synergistically kills pancreatic cancer cells with gemcitabine (114). SIRT6 enhances Ca2+ responses, which promotes the migration ability of pancreatic cancer cells (70). However, another study found that SIRT6 was an important tumor suppressor in pancreatic ductal adenocarcinoma. The absence of SIRT6 leads to hyperacetylation of the Lin28b promoter, Myc recruitment, and significant induction of Lin28b and its downstream let-7 target genes IGF2BP1, IGF2BP3, and HMGA2, thereby promoting the development of cancer (115).
In liver cancer, upregulation of SIRT6 is very common in liver cancer tissues and is highly correlated with poor overall survival rate, disease-free survival, hepatocellular carcinoma (HCC) cell migration, tumor size, tumor grade, and vascular invasion (116, 117). The suppression of SIRT6 in various liver cancer cell lines can inhibit cell growth and induce apoptosis in vitro. In vivo experiments also confirmed that the suppression of SIRT6 inhibits tumor growth (117). SIRT6 promotes the migration, invasion, and epithelial–mesenchymal transition (EMT) of HCC cells. Mechanistically, SIRT6 overexpression induces E-cadherin degradation to improve cancer invasion and migration ability. SIRT6 deacetylates the promoter of Bax (the main determinant of apoptosis of cancer cells) at H3K9 and suppresses its promoter activity to prevent cancer cell apoptosis (117). SIRT6 reduces the acetylation of AKT, resulting in increased phosphorylation of AKT and promoting its activity. Activated AKT promotes phosphorylation of anti-apoptotic protein X-linked inhibitor of apoptosis protein to prevent cancer cell apoptosis (118). SIRT6 silencing inhibits the growth of HCC cell lines by inducing p53/p21- and p16/Rb-independent cell senescence (119). However, some studies have found that SIRT6 inhibits the development of liver cancer. The level of SIRT6 decreases with increasing liver cancer grade, and increasing the level of SIRT6 at the initiation stage could significantly impair the development of cancer (120, 121). Mechanistically, the decrease in SIRT6 levels increases the acetylation level of the lysine residue at position 433 of nuclear pyruvate kinase M2 (PKM2) and promotes the oncogenic functions of PKM2, which is conducive to cell proliferation, migration, and invasion (120). Increasing SIRT6 levels represses survivin and inhibits cancer progression by reducing histone H3K9ac and NF-κB activation (121).
In prostate cancer, SIRT6 is overexpressed in prostate tumors. Knockdown of SIRT6 in prostate cancer cells results in cell cycle arrest at sub-G1 phase, increased apoptosis, increased DNA damage, and decreased BCL2 expression, thereby reducing cancer cell viability and enhancing chemotherapeutic sensitivity (122). The absence of SIRT6 significantly inhibits the activation of prostate cancer-related signaling pathways such as the Notch pathway, thereby inhibiting the proliferation and metastasis of prostate cancer cell lines (123). However, studies have shown that E2F promotes tumor growth by suppressing SIRT6 transcription to enhance glycolysis (124).
In colon cancer, SIRT6 deacetylates H3K9ac to promote the EMT process by reading snail and inhibiting TET1 transcription, further promoting tumorigenesis. Knockdown of SIRT6 in HCT116 cells leads to reduced colony formation (125). However, studies have shown that the expression of SIRT6 protein in colon cancer tissues is downregulated, and patients with higher SIRT6 expression show better prognosis (126). Upregulation of SIRT6 promoted the expression of PIP2 and PTEN and improved the stability of PTEN. The apoptosis levels of SW620 colon cancer cells overexpressing SIRT6 increased, and their proliferation ability was weakened (127). USP10 protects SIRT6 from proteasome-mediated degradation. SIRT6 inhibits c-Myc transcriptional activity, thereby inhibiting cell cycle progression, cancer cell growth, and tumor initiation in the colon cancer cell line HCT116 (128).
In ovarian cancer, SIRT6 knockdown in OVCAR3 and OVCAR5 ovarian cancer cells significantly inhibited cell migration and invasion (129). However, the expression of SIRT6 in human ovarian cancer tissues was significantly decreased, and the expression of Notch3 was increased, which further promoted the development of cancer (130).
In addition to its role in solid tumors, SIRT6 regulates blood cancer in a similarly complex manner. SIRT6 is overexpressed in CD34+ hematopoietic progenitors and multiple myeloma in patients with acute myeloid leukemia, and high SIRT6 levels are associated with poor prognosis (131, 132). SIRT6 deacetylates DNA-Pkc and CtIP and inactivates ERK2/p90RSK signaling to increase DNA repair, conferring DNA damage resistance (132). In diffuse large B-cell lymphoma cells, knockdown of SIRT6 increased sensitivity to chemotherapy, apoptosis rates, dysfunctional cell proliferation, and cell cycle arrest between the G2 and M phases, reflecting the tumor-promoting activity of SIRT6 (133). However, some studies have shown that SIRT6 deacetylates H3K9ac at the promoter of transcription factor ELK1 and ERK signal-related genes, thereby downregulating the signal transduction of the MAPK pathway and decreasing proliferation (132).
In summary, SIRT6 has shown contradictory results, promoting or suppressing cancer among different cancers, and even at different stages of development or different cell lines of the same cancer (Table 1). If SIRT6 is to be used to regulate the development of cancer via effects on the metabolism, proliferation, and apoptosis of cancer cells, it will be necessary to conduct a very comprehensive study of its role in the occurrence and development of various cancers. Effective activators and inhibitors of SIRT6 are also needed to suit the remedy to the case. As SIRT6 also has important roles in aging and immune regulation, how to reduce drug side effects will be another urgent problem to be solved in the future. At present, few studies have shown whether SIRT6 could enhance the anti-tumor ability by regulating the activity of immune cells; this may become a new research direction in the future.
Conclusion and Future Perspectives
SIRT6 has a range of PTM capabilities including deacetylation, defatty-acylation, and mono-ADP-ribosylation activities. This multifunctional PTM protein is widely involved in aging, immunity, and cancer regulation. The substrates of SIRT6 during aging, immunity, and cancer regulation are summarized in Table 2.
SIRT6 is a longevity protein that prevents cells, tissues, organs, and the body from aging. Although the mechanisms underlying these effects are diverse, they all involve resistance of aging by promoting of DNA damage repair, maintaining of the normal telomere structure of chromosomes, regulating of glucose and NAD+ metabolic balance, and by regulating of SASP (Figure 1). SIRT6 can also affect the differentiation and function of immune cells by regulating PTM affecting cells or the immunometabolism. However, the role of SIRT6 in immune regulation is complex. Although most studies have shown it to have anti-inflammatory activity, there is no lack of evidence regarding its pro-inflammatory potential (Figure 2). There has been insufficient research on how SIRT6 affects inflammation by regulating immune cells; SIRT6 has rarely been studied in many immune cells including granulocytes, monocytes, B cells, natural killer (NK) cells, and NKT cells. However, according to the recent research, the SIRT6-PTM or immunometabolism axes represent new directions with research potential. Further studies are required to clarify the role of SIRT6 in the regulation of inflammation, for example, its impact on different immune cells in different diseases or at different stages of aging, as well as on the differentiation, maturation, and function of immune cells.
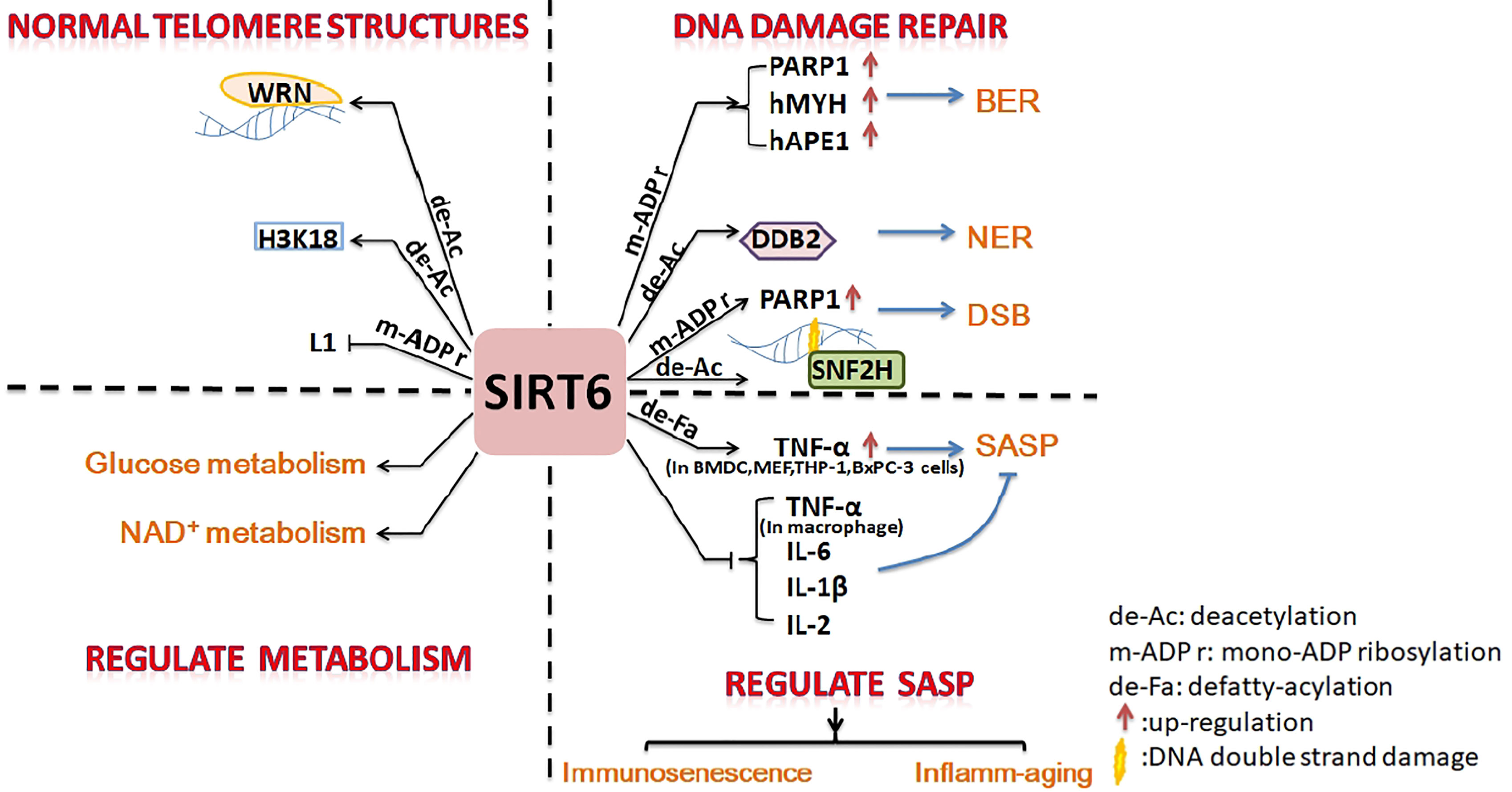
Figure 1 Summary of inhibition of cell senescence by SIRT6. SIRT6 promotes DNA damage repair, maintains the normal structure of telomere chromatin, regulates energy metabolism, and inhibits SASP to prevent cell senescence.
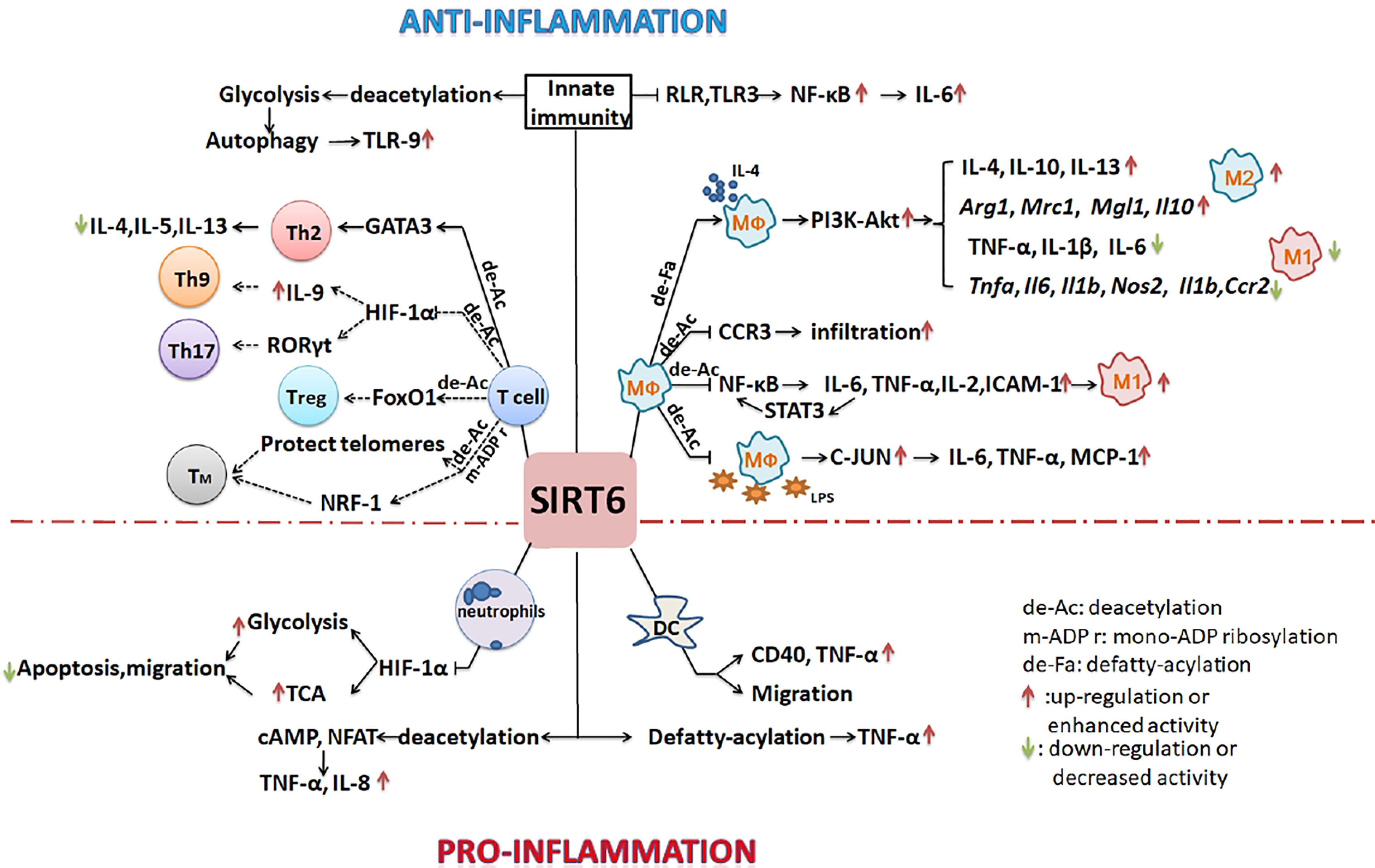
Figure 2 SIRT6 regulates the development of inflammation by regulating of the differentiation and function of immune cells. SIRT6 mainly inhibits the polarization of M1 macrophages, promotes the polarization of M2 macrophages, inhibits the overreaction of T cells, and suppresses innate immunity to inhibit the inflammatory process. By promoting the migration of neutrophils and DCs, it may also enhance inflammation. (Solid lines indicate that there is data support; dotted lines indicate a possible situation).
The role of SIRT6 in cancer development is complex. SIRT6 shows differential expression in cancer tissues compared with normal tissues; its expression levels may also vary among different cancers, at different stages of the same cancer, and in different cell lines of the same tumor type. It also has both positive and negative effects on the regulation of cancer. Possible reasons for this include the following. 1. SIRT6 participates in the NAD+ decomposition pathway and regulates the activity of many proteins related to cancer developmental pathways by controlling levels of NAD+. Cancers that occur at different ages or in different stages of cancer development show different intracellular NAD+ concentrations; thus, the role of SIRT6 will be different. 2. As SIRT1 and SIRT7 are also located in the nucleus, they may compete with SIRT6 for NAD+ consumption and also, importantly, regulate PTMs, thereby affecting cancer-related signaling pathways. The dynamic changes in SIRT1, SIRT6, and SIRT7 levels in different cancers, as well as their PTMs on histones and non-histone substrates, increase the complexity of the role of SIRT6 in pathological processes. To elucidate the complex influence of SIRT6 on cancer, it will be necessary to carry out more studies focusing on specific patient ages and tumor stages.
Few studies have analyzed whether SIRT6 could achieve anti-cancer effects via regulation of immune cell function. This could represent a new direction for future research. For example, it may be possible to adjust the polarization of macrophages through SIRT6 to affect tumor progression. In the healthy state, higher SIRT6 levels promote the polarization of M2 macrophages and maintain a low level of inflammation, which can prevent chronic inflammation and cancer development. On the other hand, in the initial stage of cancer, reduced SIRT6 levels promote the polarization of M1 macrophages. This in turn increases the pro-inflammatory ability of macrophages, leading to more immune cells being recruited to the cancer tissue to eliminate cancer cells by forming a strong immune protective barrier. In the stage of primary tumor formation and the development of aggressive tumors, reducing SIRT6 levels can increase the M1/M2 ratio, preventing the formation of an immunosuppressive tumor microenvironment and thereby inhibiting tumor development and invasion. Such tumor immunotherapy needs to be adjusted according to the progression of cancer and the different types of immune cells. More research is needed to further understand the regulatory role of SIRT6 in the immune system and in cancer.
As SIRT6 can regulate immune cell function, it could also promote or inhibit cancer development by influencing cancer cell metabolism, survival, proliferation, apoptosis, migration, and other pathways. Therefore, when designing SIRT6 activators or inhibitors to treat cancer, comprehensive consideration is necessary of the differential impact on cancer cells and immune cells to avoid conflicting drug effects. Precise administration using cell-targeted drugs is a potential approach.
Taken together, these findings indicate that SIRT6 will serve as an important target candidate for regulating immunosenescence and immune cell function. Drugs designed to target SIRT6 will also make an important contribution to the fight against chronic inflammation and cancer. SIRT6, as an important regulator throughout immunosenescence, inflamm-aging, and cancer, is a potential target for the regulation of the immune system.
Author Contributions
YW, JJ, and YL figured out the idea of writing this review. YL summarized the published results and drafted the manuscript. YW and JJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported in partial by the National Natural Science Foundation of China, No. 21977121 (YW) and in partial by important Direction Project Cultivation Fund, Institute of Immunology and the CAS Key Laboratory of Innate Immunity and Chronic Disease (2020) & University of Science and Technology of China (2021) (YW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Seto E, Yoshida M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb Perspect Biol (2014) 6(4):A018713. doi: 10.1101/cshperspect.a018713
2. Liu G, Chen H, Liu H, Zhang W, Zhou J. Emerging Roles of SIRT6 in Human Diseases and its Modulators. Med Res Rev (2021) 41(2):1089–137. doi: 10.1002/med.21753
3. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 Regulates TNF-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature (2013) 496(7443):110–3. doi: 10.1038/nature12038
4. Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 Promotes DNA Repair Under Stress by Activating Parp1. Science (2011) 332(6036):1443–6. doi: 10.1126/science.1202723
5. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic Instability and Aging-Like Phenotype in the Absence of Mammalian Sirt6. Cell (2006) 124(2):315–29. doi: 10.1016/j.cell.2005.11.044
6. Xu Z, Zhang L, Zhang W, Meng D, Zhang H, Jiang Y, et al. SIRT6 Rescues the Age Related Decline in Base Excision Repair in a PARP1-Dependent Manner. Cell Cycle (2015) 14(2):269–76. doi: 10.4161/15384101.2014.980641
7. Geng A, Tang H, Huang J, Qian Z, Qin N, Yao Y, et al. The Deacetylase SIRT6 Promotes the Repair of UV-Induced DNA Damage by Targeting Ddb2. Nucleic Acids Res (2020) 48(16):9181–94. doi: 10.1093/nar/gkaa661
8. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappaB-Dependent Gene Expression and Organismal Life Span. Cell (2009) 136(1):62–74. doi: 10.1016/j.cell.2008.10.052
9. Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is Required for Maintenance of Telomere Position Effect in Human Cells. Nat Commun (2011) 2:433. doi: 10.1038/ncomms1443
10. Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. SIRT6 Represses LINE1 Retrotransposons by Ribosylating KAP1 But This Repression Fails With Stress and Age. Nat Commun (2014) 5:5011. doi: 10.1038/ncomms6011
11. Roichman A, Elhanati S, Aon MA, Abramovich I, Di Francesco A, Shahar Y, et al. Restoration of Energy Homeostasis by SIRT6 Extends Healthy Lifespan. Nat Commun (2021) 12(1):3208. doi: 10.1038/s41467-021-23545-7
12. Wiley CD, Campisi J. The Metabolic Roots of Senescence: Mechanisms and Opportunities for Intervention. Nat Metab (2021) 3(10):1290–301. doi: 10.1038/s42255-021-00483-8
13. Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. Cellular Senescence and the Senescent Secretory Phenotype: Therapeutic Opportunities. J Clin Invest (2013) 123(3):966–72. doi: 10.1172/JCI64098
14. Van Gool F, Galli M, Gueydan C, Kruys V, Prevot PP, Bedalov A, et al. Intracellular NAD Levels Regulate Tumor Necrosis Factor Protein Synthesis in a Sirtuin-Dependent Manner. Nat Med (2009) 15(2):206–10. doi: 10.1038/nm.1906
15. Lasiglie D, Boero S, Bauer I, Morando S, Damonte P, Cea M, et al. Sirt6 Regulates Dendritic Cell Differentiation, Maturation, and Function. Aging (Albany NY) (2016) 8(1):34–49. doi: 10.18632/aging.100870
16. Xiong X, Zhang C, Zhang Y, Fan R, Qian X, Dong XC. Fabp4-Cre-Mediated Sirt6 Deletion Impairs Adipose Tissue Function and Metabolic Homeostasis in Mice. J Endocrinol (2017) 233(3):307–14. doi: 10.1530/JOE-17-0033
17. Song MY, Kim SH, Ryoo GH, Kim MK, Cha HN, Park SY, et al. Adipose Sirtuin 6 Drives Macrophage Polarization Toward M2 Through IL-4 Production and Maintains Systemic Insulin Sensitivity in Mice and Humans. Exp Mol Med (2019) 51(5):1–10. doi: 10.1038/s12276-019-0256-9
18. Kuang J, Zhang Y, Liu Q, Shen J, Pu S, Cheng S, et al. Fat-Specific Sirt6 Ablation Sensitizes Mice to High-Fat Diet-Induced Obesity and Insulin Resistance by Inhibiting Lipolysis. Diabetes (2017) 66(5):1159–71. doi: 10.2337/db16-1225
19. Koo JH, Jang HY, Lee Y, Moon YJ, Bae EJ, Yun SK, et al. Myeloid Cell-Specific Sirtuin 6 Deficiency Delays Wound Healing in Mice by Modulating Inflammation and Macrophage Phenotypes. Exp Mol Med (2019) 51(4):1–10. doi: 10.1038/s12276-019-0248-9
20. Ferrara G, Benzi A, Sturla L, Marubbi D, Frumento D, Spinelli S, et al. Sirt6 Inhibition Delays the Onset of Experimental Autoimmune Encephalomyelitis by Reducing Dendritic Cell Migration. J Neuroinflamm (2020) 17(1):228. doi: 10.1186/s12974-020-01906-1
21. Zhang Z, Ha SH, Moon YJ, Hussein UK, Song Y, Kim KM, et al. Inhibition of SIRT6 Potentiates the Anti-Tumor Effect of Doxorubicin Through Suppression of the DNA Damage Repair Pathway in Osteosarcoma. J Exp Clin Cancer Res (2020) 39(1):247. doi: 10.1186/s13046-020-01759-9
22. Yu W, Yang Z, Huang R, Min Z, Ye M. SIRT6 Promotes the Warburg Effect of Papillary Thyroid Cancer Cell BCPAP Through Reactive Oxygen Species. Onco Targets Ther (2019) 12:2861–8. doi: 10.2147/OTT.S194256
23. Yang Z, Yu W, Huang R, Ye M, Min Z. SIRT6/HIF-1alpha Axis Promotes Papillary Thyroid Cancer Progression by Inducing Epithelial-Mesenchymal Transition. Cancer Cell Int (2019) 19:17. doi: 10.1186/s12935-019-0730-4
24. Wu M, Dickinson SI, Wang X, Zhang J. Expression and Function of SIRT6 in Muscle Invasive Urothelial Carcinoma of the Bladder. Int J Clin Exp Pathol (2014) 7(10):6504–13.
25. Ouyang L, Yi L, Li J, Yi S, Li S, Liu P, et al. SIRT6 Overexpression Induces Apoptosis of Nasopharyngeal Carcinoma by Inhibiting NF-kappaB Signaling. Onco Targets Ther (2018) 11:7613–24. doi: 10.2147/OTT.S179866
26. Chen X, Hao B, Liu Y, Dai D, Han G, Li Y, et al. The Histone Deacetylase SIRT6 Suppresses the Expression of the RNA-Binding Protein PCBP2 in Glioma. Biochem Biophys Res Commun (2014) 446(1):364–9. doi: 10.1016/j.bbrc.2014.02.116
27. Chang M, Qiao L, Li B, Wang J, Zhang G, Shi W, et al. Suppression of SIRT6 by miR-33a Facilitates Tumor Growth of Glioma Through Apoptosis and Oxidative Stress Resistance. Oncol Rep (2017) 38(2):1251–8. doi: 10.3892/or.2017.5780
28. Dong Z, Zhong X, Lei Q, Chen F, Cui H. Transcriptional Activation of SIRT6 via FKHRL1/FOXO3a Inhibits the Warburg Effect in Glioblastoma Cells. Cell Signal (2019) 60:100–13. doi: 10.1016/j.cellsig.2019.04.009
29. Frye RA. Phylogenetic Classification of Prokaryotic and Eukaryotic Sir2-Like Proteins. Biochem Biophys Res Commun (2000) 273(2):793–8. doi: 10.1006/bbrc.2000.3000
30. Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 Homolog SIRT6 is a Nuclear ADP-Ribosyltransferase. J Biol Chem (2005) 280(22):21313–20. doi: 10.1074/jbc.M413296200
31. Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, et al. SIRT6 is a Histone H3 Lysine 9 Deacetylase That Modulates Telomeric Chromatin. Nature (2008) 452(7186):492–6. doi: 10.1038/nature06736
32. Gil R, Barth S, Kanfi Y, Cohen HY. SIRT6 Exhibits Nucleosome-Dependent Deacetylase Activity. Nucleic Acids Res (2013) 41(18):8537–45. doi: 10.1093/nar/gkt642
33. Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, et al. SIRT6 Deacetylates H3K18ac at Pericentric Chromatin to Prevent Mitotic Errors and Cellular Senescence. Nat Struct Mol Biol (2016) 23(5):434–40. doi: 10.1038/nsmb.3202
34. Michishita E, McCord R, Boxer L, Barber M, Tao H, Gozani O, et al. Cell Cycle-Dependent Deacetylation of Telomeric Histone H3 Lysine K56 by Human SIRT6. Cell Cycle (2009), 26648(16):. doi: 10.4161/cc.8.16.9367
35. Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. FoxO3 Transcription Factor and Sirt6 Deacetylase Regulate Low Density Lipoprotein (LDL)-Cholesterol Homeostasis Via Control of the Proprotein Convertase Subtilisin/Kexin Type 9 (Pcsk9) Gene Expression. J Biol Chem (2013) 288(41):29252–9. doi: 10.1074/jbc.M113.481473
36. Han L, Ge J, Zhang L, Ma R, Hou X, Li B, et al. Sirt6 Depletion Causes Spindle Defects and Chromosome Misalignment During Meiosis of Mouse Oocyte. Sci Rep (2015) 5:15366. doi: 10.1038/srep15366
37. Wang WW, Zeng Y, Wu B, Deiters A, Liu WR. A Chemical Biology Approach to Reveal Sirt6-Targeted Histone H3 Sites in Nucleosomes. ACS Chem Biol (2016) 11(7):1973–81. doi: 10.1021/acschembio.6b00243
38. Fiorentino F, Mai A, Rotili D. Emerging Therapeutic Potential of SIRT6 Modulators. J Med Chem (2021) 64(14):9732–58. doi: 10.1021/acs.jmedchem.1c00601
39. Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-Chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J Biol Chem (2013) 288(43):31350–6. doi: 10.1074/jbc.C113.511261
40. Jiang H, Zhang X, Lin H. Lysine Fatty Acylation Promotes Lysosomal Targeting of TNF-Alpha. Sci Rep (2016) 6:24371. doi: 10.1038/srep24371
41. Zhang X, Khan S, Jiang H, Antonyak MA, Chen X, Spiegelman NA, et al. Identifying the Functional Contribution of the Defatty-Acylase Activity of SIRT6. Nat Chem Biol (2016) 12(8):614–20. doi: 10.1038/nchembio.2106
42. Zhang X, Spiegelman NA, Nelson OD, Jing H, Lin H. SIRT6 Regulates Ras-Related Protein R-Ras2 by Lysine Defatty-Acylation. Elife (2017) 6:e25158. doi: 10.7554/eLife.25158
43. Rezazadeh S, Yang D, Tombline G, Simon M, Regan SP, Seluanov A, et al. SIRT6 Promotes Transcription of a Subset of NRF2 Targets by Mono-ADP-Ribosylating Baf170. Nucleic Acids Res (2019) 47(15):7914–28. doi: 10.1093/nar/gkz528
44. Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and Biochemical Functions of SIRT6. J Biol Chem (2011) 286(16):14575–87. doi: 10.1074/jbc.M111.218990
45. Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, et al. SIRT6 Deficiency Results in Developmental Retardation in Cynomolgus Monkeys. Nature (2018) 560(7720):661–5. doi: 10.1038/s41586-018-0437-z
46. Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The Sirtuin SIRT6 Regulates Lifespan in Male Mice. Nature (2012) 483(7388):218–21. doi: 10.1038/nature10815
47. Zhang J, Fang L, Lu Z, Xiong J, Wu M, Shi L, et al. Are Sirtuins Markers of Ovarian Aging? Gene (2016) 575(2 Pt 3):680–6. doi: 10.1016/j.gene.2015.09.043
48. Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, et al. The Sirtuin SIRT6 Blocks IGF-Akt Signaling and Development of Cardiac Hypertrophy by Targeting C-Jun. Nat Med (2012) 18(11):1643–50. doi: 10.1038/nm.2961
49. Pillai VB, Samant S, Hund S, Gupta M, Gupta MP. The Nuclear Sirtuin SIRT6 Protects the Heart From Developing Aging-Associated Myocyte Senescence and Cardiac Hypertrophy. Aging (Albany NY) (2021) 13(9):12334–58. doi: 10.18632/aging.203027
50. Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, et al. The Role of SIRT6 Protein in Aging and Reprogramming of Human Induced Pluripotent Stem Cells. J Biol Chem (2013) 288(25):18439–47. doi: 10.1074/jbc.M112.405928
51. Pan H, Guan D, Liu X, Li J, Wang L, Wu J, et al. SIRT6 Safeguards Human Mesenchymal Stem Cells From Oxidative Stress by Coactivating Nrf2. Cell Res (2016) 26(2):190–205. doi: 10.1038/cr.2016.4
52. Wu Y, Chen L, Wang Y, Li W, Lin Y, Yu D, et al. Overexpression of Sirtuin 6 Suppresses Cellular Senescence and NF-kappaB Mediated Inflammatory Responses in Osteoarthritis Development. Sci Rep (2015) 5:17602. doi: 10.1038/srep17602
53. Hwang BJ, Jin J, Gao Y, Shi G, Madabushi A, Yan A, et al. SIRT6 Protein Deacetylase Interacts With MYH DNA Glycosylase, APE1 Endonuclease, and Rad9-Rad1-Hus1 Checkpoint Clamp. BMC Mol Biol (2015) 16:12. doi: 10.1186/s12867-015-0041-9
54. Van Meter M, Simon M, Tombline G, May A, Morello TD, Hubbard BP, et al. JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks. Cell Rep (2016) 16(10):2641–50. doi: 10.1016/j.celrep.2016.08.006
55. Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, et al. SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability Through Chromatin Remodeling. Mol Cell (2013) 51(4):454–68. doi: 10.1016/j.molcel.2013.06.018
56. Arsenis NC, You T, Ogawa EF, Tinsley GM, Zuo L. Physical Activity and Telomere Length: Impact of Aging and Potential Mechanisms of Action. Oncotarget (2017) 8(27):45008–19. doi: 10.18632/oncotarget.16726
57. Fulop T, Dupuis G, Baehl S, Le Page A, Bourgade K, Frost E, et al. From Inflamm-Aging to Immune-Paralysis: A Slippery Slope During Aging for Immune-Adaptation. Biogerontology (2016) 17(1):147–57. doi: 10.1007/s10522-015-9615-7
58. Fulop T, Larbi A, Dupuis G, Le Page A, Frost EH, Cohen AA, et al. Immunosenescence and Inflamm-Aging As Two Sides of the Same Coin: Friends or Foes? Front Immunol (2017) 8:1960. doi: 10.3389/fimmu.2017.01960
59. Zhang N, Li Z, Mu W, Li L, Liang Y, Lu M, et al. Calorie Restriction-Induced SIRT6 Activation Delays Aging by Suppressing NF-kappaB Signaling. Cell Cycle (2016) 15(7):1009–18. doi: 10.1080/15384101.2016.1152427
60. Woo SJ, Noh HS, Lee NY, Cheon YH, Yi SM, Jeon HM, et al. Myeloid Sirtuin 6 Deficiency Accelerates Experimental Rheumatoid Arthritis by Enhancing Macrophage Activation and Infiltration Into Synovium. EBioMedicine (2018) 38:228–37. doi: 10.1016/j.ebiom.2018.11.005
61. Lee HS, Ka SO, Lee SM, Lee SI, Park JW, Park BH. Overexpression of Sirtuin 6 Suppresses Inflammatory Responses and Bone Destruction in Mice With Collagen-Induced Arthritis. Arthritis Rheum (2013) 65(7):1776–85. doi: 10.1002/art.37963
62. He T, Shang J, Gao C, Guan X, Chen Y, Zhu L, et al. A Novel SIRT6 Activator Ameliorates Neuroinflammation and Ischemic Brain Injury Via EZH2/FOXC1 Axis. Acta Pharm Sin B (2021) 11(3):708–26. doi: 10.1016/j.apsb.2020.11.002
63. Lee Y, Ka SO, Cha HN, Chae YN, Kim MK, Park SY, et al. Myeloid Sirtuin 6 Deficiency Causes Insulin Resistance in High-Fat Diet-Fed Mice by Eliciting Macrophage Polarization Toward an M1 Phenotype. Diabetes (2017) 66(10):2659–68. doi: 10.2337/db16-1446
64. Chen Y, Sun T, Wu J, Kalionis B, Zhang C, Yuan D, et al. Icariin Intervenes in Cardiac Inflammaging Through Upregulation of SIRT6 Enzyme Activity and Inhibition of the NF-Kappa B Pathway. BioMed Res Int (2015) 2015:895976. doi: 10.1155/2015/895976
65. Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, et al. Progression of Chronic Liver Inflammation and Fibrosis Driven by Activation of C-JUN Signaling in Sirt6 Mutant Mice. J Biol Chem (2012) 287(50):41903–13. doi: 10.1074/jbc.M112.415182
66. Ji L, Chen Y, Wang H, Zhang W, He L, Wu J, et al. Overexpression of Sirt6 Promotes M2 Macrophage Transformation, Alleviating Renal Injury in Diabetic Nephropathy. Int J Oncol (2019) 55(1):103–15. doi: 10.3892/ijo.2019.4800
67. Wang H, Li S, Zhang G, Wu H, Chang X. Potential Therapeutic Effects of Cyanidin-3-O-Glucoside on Rheumatoid Arthritis by Relieving Inhibition of CD38+ NK Cells on Treg Cell Differentiation. Arthritis Res Ther (2019) 21(1):220. doi: 10.1186/s13075-019-2001-0
68. Jang HY, Gu S, Lee SM, Park BH. Overexpression of Sirtuin 6 Suppresses Allergic Airway Inflammation Through Deacetylation of GATA3. J Allergy Clin Immunol (2016) 138(5):1452–5 e13. doi: 10.1016/j.jaci.2016.05.019
69. Li P, Jin Y, Qi F, Wu F, Luo S, Cheng Y, et al. SIRT6 Acts as a Negative Regulator in Dengue Virus-Induced Inflammatory Response by Targeting the DNA Binding Domain of NF-kappaB P65. Front Cell Infect Microbiol (2018) 8:113. doi: 10.3389/fcimb.2018.00113
70. Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, et al. The NAD+-Dependent Histone Deacetylase SIRT6 Promotes Cytokine Production and Migration in Pancreatic Cancer Cells by Regulating Ca2+ Responses. J Biol Chem (2012) 287(49):40924–37. doi: 10.1074/jbc.M112.405837
71. Tasselli L, Zheng W, Chua KF. SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol Metab (2017) 28(3):168–85. doi: 10.1016/j.tem.2016.10.002
72. Pearce EL, Pearce EJ. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity (2013) 38(4):633–43. doi: 10.1016/j.immuni.2013.04.005
73. Loftus RM, Finlay DK. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J Biol Chem (2016) 291(1):1–10. doi: 10.1074/jbc.R115.693903
74. Ramanathan M Jr., Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. Sinonasal Epithelial Cell Expression of Toll-Like Receptor 9 is Decreased in Chronic Rhinosinusitis With Polyps. Am J Rhinol (2007) 21(1):110–6. doi: 10.2500/ajr.2007.21.2997
75. Chaturvedi A, Dorward D, Pierce SK. The B Cell Receptor Governs the Subcellular Location of Toll-Like Receptor 9 Leading to Hyperresponses to DNA-Containing Antigens. Immunity (2008) 28(6):799–809. doi: 10.1016/j.immuni.2008.03.019
76. Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical Autophagy is Required for Type I Interferon Secretion in Response to DNA-Immune Complexes. Immunity (2012) 37(6):986–97. doi: 10.1016/j.immuni.2012.09.014
77. Shun CT, Lin SK, Hong CY, Lin CF, Liu CM. Sirtuin 6 Modulates Hypoxia-Induced Autophagy in Nasal Polyp Fibroblasts Via Inhibition of Glycolysis. Am J Rhinol Allergy (2016) 30(3):179–85. doi: 10.2500/ajra.2016.30.4282
78. Lu H, Lin J, Xu C, Sun M, Zuo K, Zhang X, et al. Cyclosporine Modulates Neutrophil Functions Via the SIRT6-HIF-1alpha-Glycolysis Axis to Alleviate Severe Ulcerative Colitis. Clin Transl Med (2021) 11(2):e334. doi: 10.1002/ctm2.334
79. Fortuny L, Sebastian C. Sirtuins as Metabolic Regulators of Immune Cells Phenotype and Function. Genes (Basel) (2021) 12(11):1698. doi: 10.3390/genes12111698
80. Jung SM, Hung CM, Hildebrand SR, Sanchez-Gurmaches J, Martinez-Pastor B, Gengatharan JM, et al. Non-Canonical Mtorc2 Signaling Regulates Brown Adipocyte Lipid Catabolism Through SIRT6-Foxo1. Mol Cell (2019) 75(4):807–22 e8. doi: 10.1016/j.molcel.2019.07.023
81. Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch'en IL, Stockmann C, et al. Foxo Transcription Factors Control Regulatory T Cell Development and Function. Immunity (2010) 33(6):890–904. doi: 10.1016/j.immuni.2010.12.002
82. Liu X, Hu Z, Zhou H. N-Acetylcysteine Improves Inflammatory Response in COPD Patients by Regulating Th17/Treg Balance Through Hypoxia Inducible Factor-1alpha Pathway. BioMed Res Int (2021) 2021:6372128. doi: 10.1155/2021/6372128
83. Wang Y, Bi Y, Chen X, Li C, Li Y, Zhang Z, et al. Histone Deacetylase SIRT1 Negatively Regulates the Differentiation of Interleukin-9-Producing CD4(+) T Cells. Immunity (2016) 44(6):1337–49. doi: 10.1016/j.immuni.2016.05.009
84. Kumar BV, Connors TJ, Farber DL. Human T Cell Development, Localization, and Function Throughout Life. Immunity (2018) 48(2):202–13. doi: 10.1016/j.immuni.2018.01.007
85. Bellon M, Nicot C. Telomere Dynamics in Immune Senescence and Exhaustion Triggered by Chronic Viral Infection. Viruses (2017) 9(10):289. doi: 10.3390/v9100289
86. Wertheimer AM, Bennett MS, Park B, Uhrlaub JL, Martinez C, Pulko V, et al. Aging and Cytomegalovirus Infection Differentially and Jointly Affect Distinct Circulating T Cell Subsets in Humans. J Immunol (2014) 192(5):2143–55. doi: 10.4049/jimmunol.1301721
87. Moskowitz DM, Zhang DW, Hu B, Le Saux S, Yanes RE, Ye Z, et al. Epigenomics of Human CD8 T Cell Differentiation and Aging. Sci Immunol (2017) 2(8):eaag0192. doi: 10.1126/sciimmunol.aag0192
88. Joseph JS, Ayeleso AO, Mukwevho E. Exercise Increases Hyper-Acetylation of Histones on the Cis-Element of NRF-1 Binding to the Mef2a Promoter: Implications on Type 2 Diabetes. Biochem Biophys Res Commun (2017) 486(1):83–7. doi: 10.1016/j.bbrc.2017.03.002
89. Pillai VB, Gupta MP. Is Nuclear Sirtuin SIRT6 a Master Regulator of Immune Function? Am J Physiol Endocrinol Metab (2021) 320(3):E399–414. doi: 10.1152/ajpendo.00483.2020
90. Burns JS, Manda G. Metabolic Pathways of the Warburg Effect in Health and Disease: Perspectives of Choice, Chain or Chance. Int J Mol Sci (2017) 18(12):2755. doi: 10.3390/ijms18122755
91. Sommermann TG, O'Neill K, Plas DR, Cahir-McFarland E. IKKbeta and NF-kappaB Transcription Govern Lymphoma Cell Survival Through AKT-Induced Plasma Membrane Trafficking of GLUT1. Cancer Res (2011) 71(23):7291–300. doi: 10.1158/0008-5472.CAN-11-1715
92. Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, et al. EGFR-Induced and PKCepsilon Monoubiquitylation-Dependent NF-kappaB Activation Upregulates PKM2 Expression and Promotes Tumorigenesis. Mol Cell (2012) 48(5):771–84. doi: 10.1016/j.molcel.2012.09.028
93. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int J Mol Sci (2019) 20(2):238. doi: 10.3390/ijms20020238
94. Robey RB, Hay N. Is Akt the "Warburg Kinase"?-Akt-Energy Metabolism Interactions and Oncogenesis. Semin Cancer Biol (2009) 19(1):25–31. doi: 10.1016/j.semcancer.2008.11.010
95. Basu AK. DNA Damage, Mutagenesis and Cancer. Int J Mol Sci (2018) 19(4):970. doi: 10.3390/ijms19040970
96. Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell (2017) 168(4):644–56. doi: 10.1016/j.cell.2017.01.002
97. Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity (2019) 51(1):27–41. doi: 10.1016/j.immuni.2019.06.025
98. Garcia-Peterson LM, Ndiaye MA, Chhabra G, Singh CK, Guzman-Perez G, Iczkowski KA, et al. CRISPR/Cas9-Mediated Knockout of SIRT6 Imparts Remarkable Antiproliferative Response in Human Melanoma Cells In Vitro and In Vivo. Photochem Photobiol (2020) 96(6):1314–20. doi: 10.1111/php.13305
99. Garcia-Peterson LM, Ndiaye MA, Singh CK, Chhabra G, Huang W, Ahmad N. SIRT6 Histone Deacetylase Functions as a Potential Oncogene in Human Melanoma. Genes Cancer (2017) 8(9-10):701–12. doi: 10.18632/genesandcancer.153
100. Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, et al. SIRT6 Haploinsufficiency Induces BRAF(V600E) Melanoma Cell Resistance to MAPK Inhibitors Via IGF Signalling. Nat Commun (2018) 9(1):3440. doi: 10.1038/s41467-018-05966-z
101. Wang L, Guo W, Ma J, Dai W, Liu L, Guo S, et al. Aberrant SIRT6 Expression Contributes to Melanoma Growth: Role of the Autophagy Paradox and IGF-AKT Signaling. Autophagy (2018) 14(3):518–33. doi: 10.1080/15548627.2017.1384886
102. Dong Z, Yang J, Li L, Tan L, Shi P, Zhang J, et al. FOXO3aSIRT6 Axis Suppresses Aerobic Glycolysis in Melanoma. Int J Oncol (2020) 56(3):728–42. doi: 10.3892/ijo.2020.4964
103. Becherini P, Caffa I, Piacente F, Damonte P, Vellone VG, Passalacqua M, et al. SIRT6 Enhances Oxidative Phosphorylation in Breast Cancer and Promotes Mammary Tumorigenesis in Mice. Cancer Metab (2021) 9(1):6. doi: 10.1186/s40170-021-00240-1
104. Khongkow M, Olmos Y, Gong C, Gomes AR, Monteiro LJ, Yague E, et al. SIRT6 Modulates Paclitaxel and Epirubicin Resistance and Survival in Breast Cancer. Carcinogenesis (2013) 34(7):1476–86. doi: 10.1093/carcin/bgt098
105. Mahmud Z, Gomes AR, Lee HJ, Aimjongjun S, Jiramongkol Y, Yao S, et al. EP300 and SIRT1/6 Co-Regulate Lapatinib Sensitivity Via Modulating FOXO3-Acetylation and Activity in Breast Cancer. Cancers (Basel) (2019) 11(8):1067. doi: 10.3390/cancers11081067
106. Choe M, Brusgard JL, Chumsri S, Bhandary L, Zhao XF, Lu S, et al. The RUNX2 Transcription Factor Negatively Regulates SIRT6 Expression to Alter Glucose Metabolism in Breast Cancer Cells. J Cell Biochem (2015) 116(10):2210–26. doi: 10.1002/jcb.25171
107. Krishnamoorthy V, Vilwanathan R. Silencing Sirtuin 6 Induces Cell Cycle Arrest and Apoptosis in non-Small Cell Lung Cancer Cell Lines. Genomics (2020) 112(5):3703–12. doi: 10.1016/j.ygeno.2020.04.027
108. Ruan L, Chen J, Ruan L, Tan A, Wang P. miR-34a Inhibits Tumorigenesis of NSCLC Via Targeting SIRT6. Int J Clin Exp Pathol (2018) 11(3):1135–45.
109. Bai L, Lin G, Sun L, Liu Y, Huang X, Cao C, et al. Upregulation of SIRT6 Predicts Poor Prognosis and Promotes Metastasis of non-Small Cell Lung Cancer Via the ERK1/2/MMP9 Pathway. Oncotarget (2016) 7(26):40377–86. doi: 10.18632/oncotarget.9750
110. Li Z, Huang J, Shen S, Ding Z, Luo Q, Chen Z, et al. SIRT6 Drives Epithelial-To-Mesenchymal Transition and Metastasis in non-Small Cell Lung Cancer Via Snail-Dependent Transrepression of KLF4. J Exp Clin Cancer Res (2018) 37(1):323. doi: 10.1186/s13046-018-0984-z
111. Azuma Y, Yokobori T, Mogi A, Altan B, Yajima T, Kosaka T, et al. SIRT6 Expression is Associated With Poor Prognosis and Chemosensitivity in Patients With non-Small Cell Lung Cancer. J Surg Oncol (2015) 112(2):231–7. doi: 10.1002/jso.23975
112. Han Z, Liu L, Liu Y, Li S. Sirtuin SIRT6 Suppresses Cell Proliferation Through Inhibition of Twist1 Expression in non-Small Cell Lung Cancer. Int J Clin Exp Pathol (2014) 7(8):4774–81.
113. Fang C, Liu Y, Chen L, Luo Y, Cui Y, Zhang N, et al. Alpha-Hederin Inhibits the Growth of Lung Cancer A549 Cells In Vitro and In Vivo by Decreasing SIRT6 Dependent Glycolysis. Pharm Biol (2021) 59(1):11–20. doi: 10.1080/13880209.2020.1862250
114. Sociali G, Galeno L, Parenti MD, Grozio A, Bauer I, Passalacqua M, et al. Quinazolinedione SIRT6 Inhibitors Sensitize Cancer Cells to Chemotherapeutics. Eur J Med Chem (2015) 102:530–9. doi: 10.1016/j.ejmech.2015.08.024
115. Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, et al. SIRT6 Suppresses Pancreatic Cancer Through Control of Lin28b. Cell (2016) 165(6):1401–15. doi: 10.1016/j.cell.2016.04.033
116. Han LL, Jia L, Wu F, Huang C. Sirtuin6 (SIRT6) Promotes the EMT of Hepatocellular Carcinoma by Stimulating Autophagic Degradation of E-Cadherin. Mol Cancer Res (2019) 17(11):2267–80. doi: 10.1158/1541-7786.MCR-19-0321
117. Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH, Zhou L, et al. SIRT6 Overexpression Potentiates Apoptosis Evasion in Hepatocellular Carcinoma Via BCL2-Associated X Protein-Dependent Apoptotic Pathway. Clin Cancer Res (2016) 22(13):3372–82. doi: 10.1158/1078-0432.CCR-15-1638
118. Zhou HZ, Zeng HQ, Yuan D, Ren JH, Cheng ST, Yu HB, et al. NQO1 Potentiates Apoptosis Evasion and Upregulates XIAP Via Inhibiting Proteasome-Mediated Degradation SIRT6 in Hepatocellular Carcinoma. Cell Commun Signal (2019) 17(1):168. doi: 10.1186/s12964-019-0491-7
119. Lee N, Ryu HG, Kwon JH, Kim DK, Kim SR, Wang HJ, et al. SIRT6 Depletion Suppresses Tumor Growth by Promoting Cellular Senescence Induced by DNA Damage in HCC. PloS One (2016) 11(11):e0165835. doi: 10.1371/journal.pone.0165835
120. Bhardwaj A, Das S. SIRT6 Deacetylates PKM2 to Suppress its Nuclear Localization and Oncogenic Functions. Proc Natl Acad Sci U S A (2016) 113(5):E538–47. doi: 10.1073/pnas.1520045113
121. Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, et al. Liver Cancer Initiation is Controlled by AP-1 Through SIRT6-Dependent Inhibition of Survivin. Nat Cell Biol (2012) 14(11):1203–11. doi: 10.1038/ncb2590
122. Liu Y, Xie QR, Wang B, Shao J, Zhang T, Liu T, et al. Inhibition of SIRT6 in Prostate Cancer Reduces Cell Viability and Increases Sensitivity to Chemotherapeutics. Protein Cell (2013) 4(9):702–10. doi: 10.1007/s13238-013-3054-5
123. Han Q, Xie QR, Li F, Cheng Y, Wu T, Zhang Y, et al. Targeted Inhibition of SIRT6 Via Engineered Exosomes Impairs Tumorigenesis and Metastasis in Prostate Cancer. Theranostics (2021) 11(13):6526–41. doi: 10.7150/thno.53886
124. Wu M, Seto E, Zhang J. E2F1 Enhances Glycolysis Through Suppressing Sirt6 Transcription in Cancer Cells. Oncotarget (2015) 6(13):11252–63. doi: 10.18632/oncotarget.3594
125. Geng CH, Zhang CL, Zhang JY, Gao P, He M, Li YL. Overexpression of Sirt6 is a Novel Biomarker of Malignant Human Colon Carcinoma. J Cell Biochem (2018) 119(5):3957–67. doi: 10.1002/jcb.26539
126. Zhang Y, Nie L, Xu K, Fu Y, Zhong J, Gu K, et al. SIRT6, a Novel Direct Transcriptional Target of FoxO3a, Mediates Colon Cancer Therapy. Theranostics (2019) 9(8):2380–94. doi: 10.7150/thno.29724
127. Tian J, Yuan L. Sirtuin 6 Inhibits Colon Cancer Progression by Modulating PTEN/AKT Signaling. BioMed Pharmacother (2018) 106:109–16. doi: 10.1016/j.biopha.2018.06.070
128. Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, et al. USP10 Antagonizes C-Myc Transcriptional Activation Through SIRT6 Stabilization to Suppress Tumor Formation. Cell Rep (2013) 5(6):1639–49. doi: 10.1016/j.celrep.2013.11.029
129. Bae JS, Noh SJ, Kim KM, Park SH, Hussein UK, Park HS, et al. SIRT6 Is Involved in the Progression of Ovarian Carcinomas Via Beta-Catenin-Mediated Epithelial to Mesenchymal Transition. Front Oncol (2018) 8:538. doi: 10.3389/fonc.2018.00538
130. Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M, Zhang H, et al. The Histone Deacetylase SIRT6 Inhibits Ovarian Cancer Cell Proliferation Via Down-Regulation of Notch 3 Expression. Eur Rev Med Pharmacol Sci (2015) 19(5):818–24.
131. Cagnetta A, Soncini D, Orecchioni S, Talarico G, Minetto P, Guolo F, et al. Depletion of SIRT6 Enzymatic Activity Increases Acute Myeloid Leukemia Cells' Vulnerability to DNA-Damaging Agents. Haematologica (2018) 103(1):80–90. doi: 10.3324/haematol.2017.176248
132. Cea M, Cagnetta A, Adamia S, Acharya C, Tai YT, Fulciniti M, et al. Evidence for a Role of the Histone Deacetylase SIRT6 in DNA Damage Response of Multiple Myeloma Cells. Blood (2016) 127(9):1138–50. doi: 10.1182/blood-2015-06-649970
Keywords: aging, immunity, inflammation, cancer, SIRT6, epigenetics
Citation: Li Y, Jin J and Wang Y (2022) SIRT6 Widely Regulates Aging, Immunity, and Cancer. Front. Oncol. 12:861334. doi: 10.3389/fonc.2022.861334
Received: 24 January 2022; Accepted: 14 March 2022;
Published: 06 April 2022.
Edited by:
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2022 Li, Jin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wang, d3k4M0B1c3RjLmVkdS5jbg==; Jing Jin, amluamluZzE5QHVzdGMuZWR1LmNu
 Yunjia Li
Yunjia Li Jing Jin
Jing Jin Yi Wang
Yi Wang