- 1College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 2Guangzhou National Laboratory, Guangzhou, China
- 3State Key Laboratory of Oral & Maxillofacial Reconstruction and Regeneration, National Clinical Research Center for Oral Diseases, Shaanxi Key Laboratory of Stomatology, Department of Prosthodontics, School of Stomatology, The Fourth Military Medical University, Xi’an, China
Head and Neck Squamous Cell Carcinoma (HNSCC), ranking among the six most prevalent malignancies worldwide, is characterized by significant heterogeneity. Conventional monotherapeutic approaches, including surgical intervention, radiotherapy, and chemotherapy, often fail to achieve complete tumor cell elimination, consequently leading to disease recurrence and metastatic progression. In this context, personalized immunotherapeutic strategies, particularly cancer vaccines and immune checkpoint inhibitors, have emerged as promising therapeutic modalities for patients with recurrent/metastatic (R/M) HNSCC. Neoantigens, which exhibit selective expression in tumor tissues while remaining absent in normal tissues, have garnered considerable attention as novel targets for HNSCC personalized immunotherapy. However, the marked heterogeneity of HNSCC, coupled with patient-specific HLA variations, necessitates precise technical identification and evaluation of neoantigens at the individual level-a significant contemporary challenge. This comprehensive review systematically explores the landscape of neoantigen-based immunotherapy in HNSCC, including neoantigen sources, screening strategies, identification methods, and their clinical applications. Additionally, it evaluates the therapeutic potential of combining neoantigen-based approaches with other immunotherapeutic modalities, particularly immune checkpoint inhibitors, providing valuable insights for future clinical practice and research directions in HNSCC treatment.
1 Introduction
According to the data released by the World Health Organization (WHO) in 2022, head and neck cancer ranks as the sixth most common malignant tumor globally (1). It exhibits a relatively high incidence and mortality rate in Asia, and both rates being notably higher in men (Figures 1A, C) compared to women (Figures 1B, D) (source: https://gco.iarc.fr/today). Head and neck cancer encompasses a group of malignant tumors that arise in the epithelial tissues of the paranasal sinuses, lips, oral cavity, nasal cavity, pharynx, and larynx (2). The most common histological subtype is squamous cell carcinoma, which accounts for approximately 90% of cases (3). The incidence of HNSCC continues to rise and is projected to increase by 30% by 2030, leading to an estimated 1.08 million new cases annually (4).
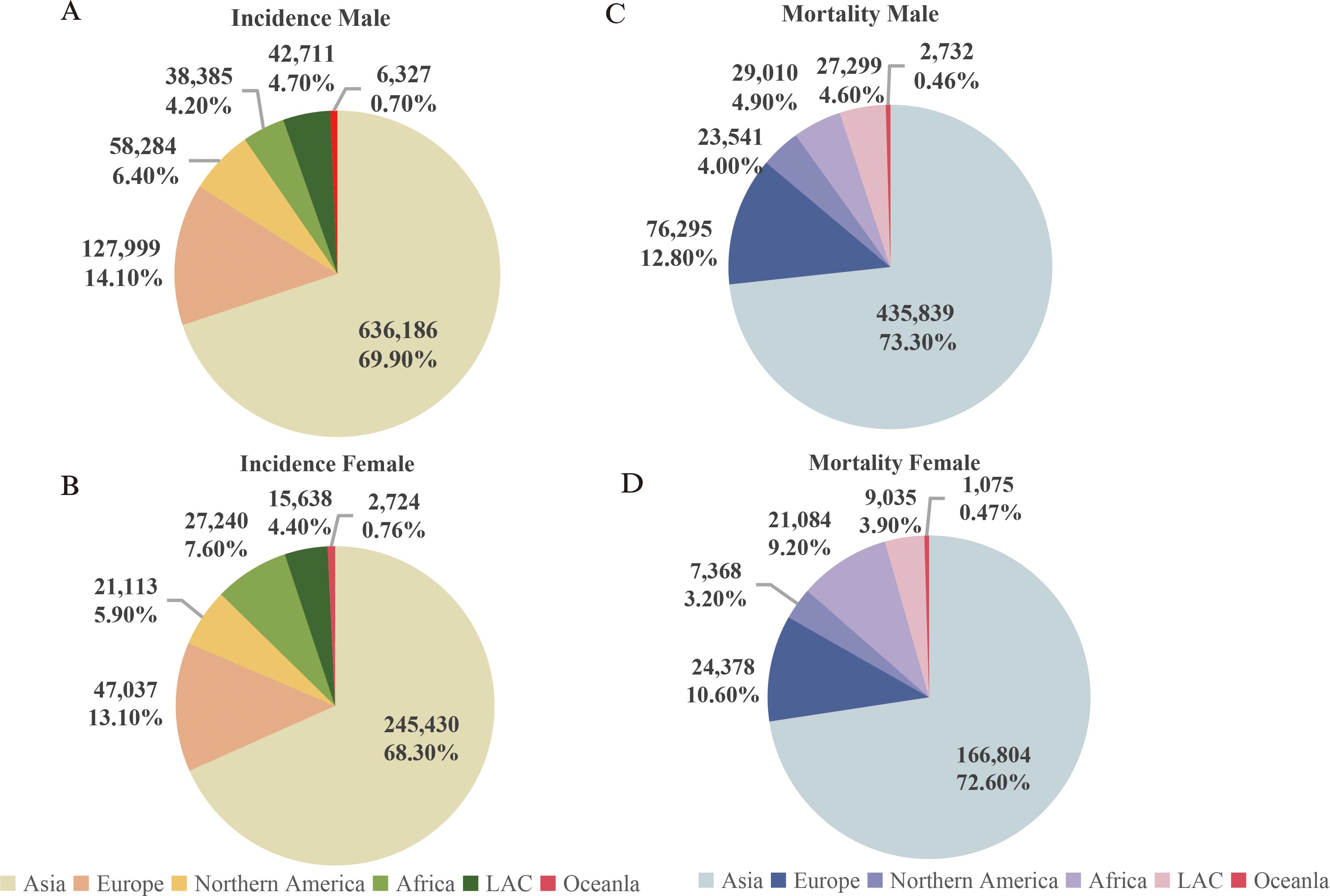
Figure 1. Incidence and mortality rates of head and neck cancer (Updated 2022). (A) Incidence rate of head and neck cancer in males; (B) Incidence rate of head and neck cancer in females; (C) Mortality rate of head and neck cancer in males; (D) Mortality rate of head and neck cancer in females. Note: Source: https://gco.iarc.fr/today.
Head and Neck Squamous Cell Carcinoma (HNSCC) can be induced by various factors, including chronic excessive alcohol consumption, poor oral hygiene, viral infections, betel nut chewing, and smoking (5). The complex anatomical and physiological structures of the head and neck contribute to highly heterogeneous nature of HNSCC (6). The majority of patients are diagnosed at the locally advanced stages of the disease (7). Conventional treatment modalities for HNSCC include surgery, chemotherapy, and radiotherapy (8). However, these treatments are often associated with severe late-stage adverse effects, such as renal impairment, hearing loss, myelosuppression, and aspiration pneumonia (9). A significant shift in treatment strategies involves the combination of immune checkpoint inhibitors, such as anti-PD-1 monoclonal antibodies, with chemotherapy. This approach has led to improvements in the survival rates of patients with recurrent or metastatic (R/M) HNSCC (10). Despite these advances, the four-year survival rate remains disappointingly low, ranging from only 15% to 19% (11).
HNSCC demonstrates significantly heterogeneity in terms of molecular characteristics, cellular phenotypes, and the composition of the tumor microenvironment (TME) (12, 13). For instance, the distribution of cancer-associated fibroblasts (CAFs) and the expression of various CAF markers exhibit notable variations among individual HNSCC patients (14). Additionally, there are differences in the mutational burden between human papillomavirus-positive (HPV+) and human papillomavirus-negative (HPV−) tumors (12). These factors collectively influence the outcome of conventional clinical treatments. Therefore, the development of more effective targeted therapies is essential to improving the prognosis for HNSCC patients.
Personalized immunotherapy is rapidly reshaping the treatment landscape of HNSCC. Neoantigens play a central role in tumor vaccines and immune checkpoint blockade (ICB) strategies (15). Tumor antigens can be categorized into two types. The first category, known as tumor-associated antigens (TAAs), are highly expressed in tumor tissues while exhibiting low expression in normal tissues. The second category, tumor neoantigens (also referred to as tumor-specific antigens, or TSAs), consists of unique peptide segments generated by genomic aberrations, transcriptomic irregularities, abnormal post-translational modifications, and other factors in tumor cells (16, 17). These neoantigens can be recognized by the major histocompatibility complex (MHC) on antigen-presenting cells (APCs), and the resulting tumor-specific peptide-HLA complexes can be identified by T cells, thereby triggering an anti-tumor immune response (18). Immunogenomic techniques have been employed to predict a large number of neoantigens arising from mutations based on cancer genomic data. However, proteomic studies examining peptides binding to HLA have shown that most of the predicted neoantigens were not detected (19). The discrepancy between predicted and observed highlights limitations in the current prediction methods and technologies. To improve the identification of clinically relevant neo-epitopes, epitope prediction should incorporate multiple methods, alongside advanced quality assessment metrics for neo-epitopes.
2 Challenges in the treatment of HNSCC
HNSCC exhibits a high degree of both inter-tumor and intra-tumor heterogeneity (20). HPV-positive and HPV-negative HNSCC not only originate from distinct anatomical sites but also display divergent mutation spectra, molecular characteristics, immune landscapes, and clinical prognoses (21). Additionally, HNSCC is a notable immunosuppressive malignancy. For instance, regulatory T cells (Tregs), tumor-associated macrophages, and myeloid-derived suppressor cells (MDSCs) have been shown to enhance immune evasion of HNSCC (22). Human leukocyte antigens (HLAs), which play a central role in the immune response, exhibit considerable genetic polymorphism and vary across HNSCC patients. For example, Tang et al. identified that the genetic susceptibility to nasopharyngeal carcinomas is linked to the HLA haplotype A*0206, a subtype associated with a heightened risk of the disease in East and Southeast Asia (23). Similarly, Makni et al. found that HLA-A31, A33, A19, B16, B53, and the alleles DRB103, DRB113, and DQB1*02 are potential susceptibility genes for nasopharyngeal carcinoma, while HLA-B14, HLA-B35, and DRB1*01, DQB1*05 may offer a protective effects (24). Understanding the HLA genotypes of different HNSCC patients can help elucidate the pathogenesis of the disease and facilitate the development of more precise, targeted therapies.
The primary goal in treating patients with HNSCC are to achieve complete tumor resection or effective tumor control, extend survival, and minimize treatment-related adverse effects (25). For patients with locally advanced or recurrent/metastatic (R/M) HNSCC, surgery is typically the first-line curative approach. In cases where surgery is not feasible, a combination of radiotherapy and cisplatin is commonly used (26). For patients who cannot tolerate cisplatin or are elderly (over 70 years of age), radiotherapy alone is an alternative (27). The U.S. Food and Drug Administration (FDA) has approved the use of nivolumab and pembrolizumab for treating patients HNSCC patients who have developed resistance to platinum-based therapies (28–30). Results from the phase II KEYNOTE-055 study showed that pembrolizumab monotherapy had some efficacy in platinum-resistant HNSCC. The study reported an objective response rate (ORR) of 16%, a duration of response (DOR) of 8 months, progression-free survival (PFS) of 2.1 months, and an overall survival (OS) of 8 months (31). Despite these findings, patients still face challenges such as a relatively low response rate and a high risk of recurrence during treatment.
Patients with HNSCC often present with malnutrition or are complicated by other comorbidities. Traditional treatment methods, while effective in some cases, can cause significant damage to the immune system and trigger severe side effects (32). Additionally, HNSCC is a highly heterogeneous malignancy, and variations in human leukocyte antigen (HLA) genotypes among patients contribute to substantial differences in prognosis (33). Given these challenges, there is an urgent need for continued research to explore alternative strategies for HNSCC (11). This review focuses on the development of personalized treatment regimens based on neoantigens. Such approaches offer promising application potential and substantial clinical value, contributing to more efficient treatment strategies for patients with HNSCC.
3 The source of neoantigens
Tumor neoantigens are defined by their exclusive expression in tumor tissues, with no detectable expression in healthy tissues. Additionally, they evade the central tolerance mechanism of T-cells, making them highly immunogenic (34). Due to those properties, neoantigens can elicit a robust anti-tumor immune response and are considered highly promising targets in personalized immunotherapy (35). Neoantigens primarily arise from genomic alterations (Figure 2A), including single-nucleotide variants (SNVs), small insertions or deletions (INDELs), and gene fusions. Beyond genetic mutations, additional mechanisms contribute to neoantigen formation, including alternative splicing, aberrant RNA editing (36), non-coding RNA expression (37), and abnormal post-translational modifications (PTMs) of proteins (38) (Figure 2A). In HNSCC, viral oncogenes can integrate into the host genome, providing an additional source of neoantigens and further diversifying the tumor antigen landscape (39) (Figure 2B).
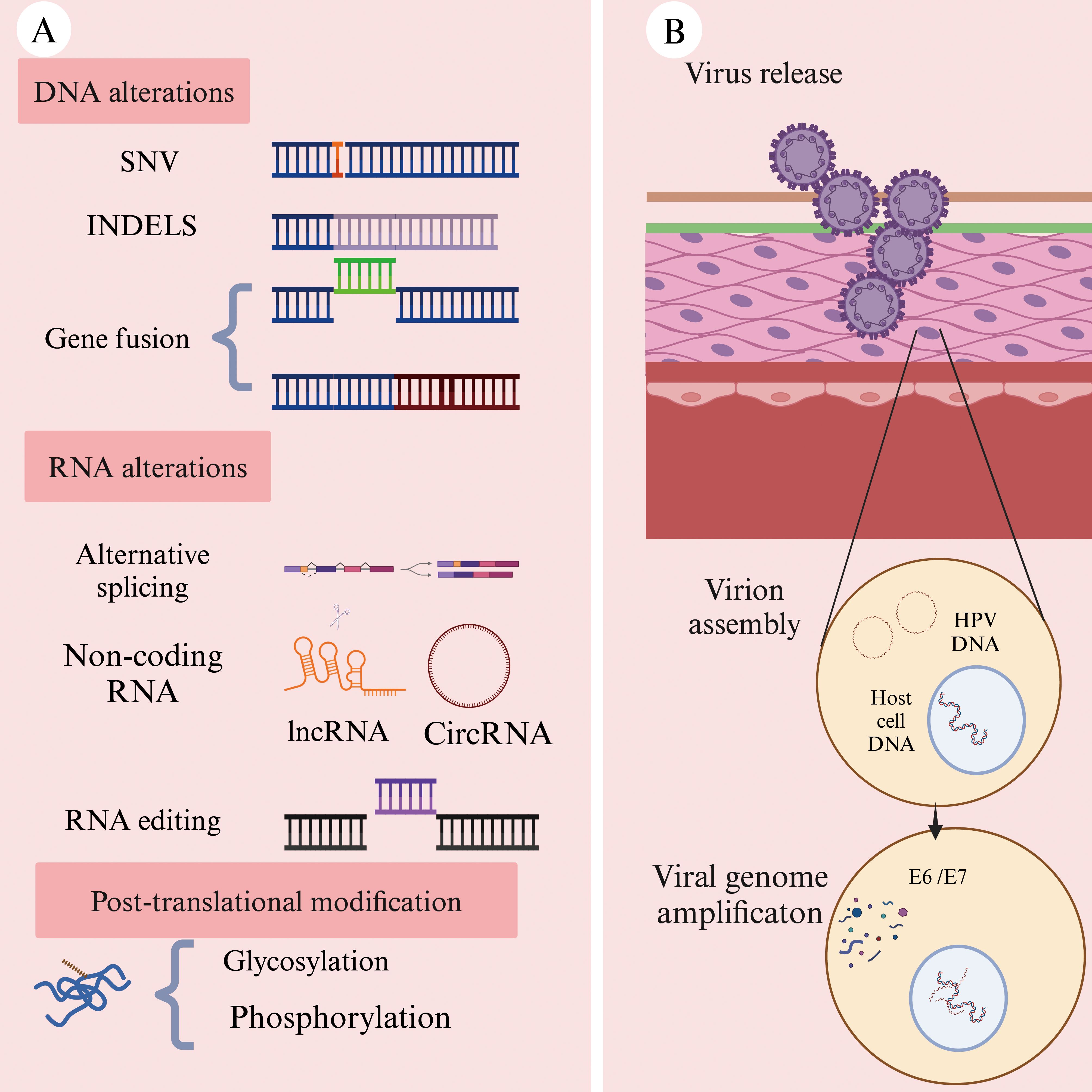
Figure 2. Common sources of neoantigens in HNSCC (Created with BioRender.com). (A) Neoantigens can arise from multiple molecular levels: at the genomic level through single-nucleotide variants (SNVs), insertions or deletions (INDELs), and gene fusions; at the transcriptomic level through alternative splicing, polyadenylation (pA), RNA editing, and non-coding regions; and at the proteomic level through dysregulated translation and post-translational modifications (PTMs). (B) Oncogenic viruses, such as human papillomavirus (HPV), can integrate their genetic material into the host cell genome. The expression of these viral genes within hose cells may result in the production of tumor-specific neoantigens.
3.1 Non-viral-derived neoantigens
Single-nucleotide variants (SNV) are the most common type of genetic mutation in tumor cells. When a single-nucleotide alteration occurs in DNA, it can affect the coding sequence of the gene, leading to the generation of neoantigens (40). Small insertion or deletions (INDELs) refer to the addition or removal of one or more bases at specific loci in the DNA sequence compared to the reference genome (41). If an INDEL occurs within a gene’s coding region or splice site, it can alter protein structure and function, thereby triggering the production of neoantigens (42).
Compared to SNVs, INDELs can result in the formation of novel open reading frames (ORFs), leading to the synthesis of a large number of non-self peptides (34). Notably, neoantigens generated by INDELs exhibit significantly higher immunogenicity than those derived from SNVs (43). Although INDELs are less frequent than SNVs, the results of a large-scale analysis of 5,777 solid tumors across 19 cancer types in The Cancer Genome Atlas (TCGA) demonstrated that INDEL-derived neoantigens are more immunogenic than those produced by SNVs (44).
Gene fusions represent another important class of mutations. When a gene fusion occurs, it can create open reading frame(ORF) that encodes an entirely new protein (45). For instance, in sinus cancer and nasopharyngeal carcinoma, the DEK-AFF2 fusion gene has been identified. A peptide (DKESEEEVS) derived from DEK-AFF2 has been shown to stimulate the activation of autologous peripheral blood mononuclear cells (PBMCs) in a major histocompatibility complex (MHC)-dependent manner (46). The discovery of fusion gene-derived neoantigens has expanded the repertoire of potential targets for cancer immunotherapy, positioning them as a key driving force in the development of novel therapeutic strategies (47).
RNA alternative splicing involves mutations in cis-acting elements and alterations in trans-acting regulatory factors. Neoantigens derived from this process often exhibit enhanced immunogenicity (48). Cis-acting mutations primarily include intron retention and exon skipping, while trans-acting alterations can induce neoantigen generation across the entire genome (49). Pan-cancer analysis using data from The Cancer Genome Atlas (TCGA) has revealed that somatic mutations in key splicing factors, such as SF3B1 and U2AF1, can lead to the formation of splicing-derived neoantigens across various solid tumors (37). In a study of uveal melanoma patients with SF3B1 mutations, tumor and normal tissue samples were collected to predict splicing variants induced by these mutations. The results demonstrated that the resulting neoantigens exhibited strong immunogenicity (50). Wang et al. applied two computational tools, ScanExitron and ScanNeo, to identify and analyze neoantigens generated through exitron skipping. In this process, exitron are defined as cryptic intronic sequences located within annotated protein-coding exons. Their findings indicated that exitron skipping could produce highly immunogenic neoantigens (51).
Although circular RNAs (circRNAs) do not directly encode proteins, their regulatory functions can influence neoantigen generation. Recent advancements in high-throughput circRNA reporter gene screening and mass spectrometry-based peptidomics workflows have facilitated the identification of circRNAs as a novel source of neoantigens. Zheng et al. successfully demonstrated that circRNAs could induce tumor-specific T cell responses, leading to selective tumor cell elimination (52). This discovery provides new insights and potential therapeutic targets for tumor immunotherapy.
A Phase I clinical trial (NCT06530082) utilized mass spectrometry to analyze human leukocyte antigen class I (HLA-I) in breast cancer samples. This analysis combined with ribosome profiling, successfully identified cryptic antigen peptides generated through atypical translation of circFAM53B, which were capable of binding to HLA-I. The study further evaluated a dendritic cell (DC) vaccine based on CircFAM53B-219aa, administered in combination with camrelizumab for the treatment of HER2-negative advanced breast cancer (53). The results demonstrated that the DC vaccine, designed using circRNA-derivedneoantigens, effectively stimulate T cell activation and induced cytotoxic responses against tumor cells.
Abnormal post-translational modifications (PTM) of proteins can also contribute to the generations of neoantigens. Key modification types include glycosylation, O-linked β-N-acetylglucosamine (O-GlcNAc) modification, and phosphorylation, among others (36). These modifications play a critical role in the shaping neoantigen formation and influencing their immunogenic properties (54).
3.2 Virus-derived neoantigens
Viral proteins represent a distinct class of neoantigens in virus-associated tumors, capable of eliciting high-affinity T cell receptor (TCR) responses (55). Certain solid tumors are directly linked to viral infections, such as nasopharyngeal carcinoma, which is associated with caused by Epstein-Barr virus (EBV) infection (56). When viral genes integrate into the host genome, the expression of these foreign genetic elements may lead to the formation of neoantigens, as observed in oropharyngeal cancer caused by human papillomavirus (HPV) infection (57).
High-risk HPV infection accounts for approximately 40% to 70% of head and neck cancers with HPV16 and HPV18 being the most prevalent subtypes (58). The oncoproteins E6 and E7, expressed by high-risk HPV strains, play a pivotal role in disrupting genomic stability and driving tumor progression (59). MEDI0457, a DNA-based immunotherapy, utilizes plasmids encoding interleukin-12 (IL-12) to target the HPV16/18-derived E6 and E7 proteins, demonstrating the potential to elicit a durable anti-tumor immune response (60).
4 Identification and prediction of neoantigens
The identification of neoantigens with high immunogenicity is a critical step in developing effective personalized immunotherapies (61). Next-generation sequencing (NGS), also referred to as high-throughput sequencing, is widely used to detect tumor-specific genetic alterations (62). Whole-exome sequencing (WES) and RNA sequencing (RNA-seq) are fundamental tools in genomics research, facilitating the identification of mutations that may give rise to neoantigens (63).
NGS plays a pivotal role in proteogenomics, a field that integrates genomic data, transcriptomic, and proteomic data to enhance cancer research (64, 65). Tretter et al. developed an innovative approach by combining RNA sequencing, proteomic profiling, and whole-exome sequencing to identify neoantigens at the protein level (66). Mass spectrometry (MS) has also advanced the validation of computationally predicted neoantigens, providing a more comprehensive understanding of their immunogenic potential (67).
Genomic approaches primarily predict neoantigens based on DNA and RNA mutation data, making them suitable for the preliminary identification and screening of potential targets (68); In contrast, MS focuses on directly detecting and characterizing MHC-bound polypeptides, offering a reliable method for confirming neoantigens at the proteomic level (69). The integration of these methodologies enhances the accuracy of neoantigen identification, ultimately facilitating the development of targeted cancer immunotherapies.
The T cell receptor (TCR) exhibits remarkable diversity and is capable of recognizing and specifically binding to neoantigen peptides presented by MHC molecules (70). This recognition primarily occurs through specific interactions with complementarity-determining region 1 (CDR1) and 2 (CDR2) (71). Traditional methods for studying TCRs are often based on bulk cell population analysis, which limits their ability to accurately reflect the TCR expression profile of individual T cells.
High-throughput single-cell RNA sequencing (scRNA-seq) has emerged as a powerful technique for studying gene expression at the individual level within a heterogeneous population (72). By applying scRNA-seq to T cells in tumor samples, researchers can identify multiple neoantigen-specific TCRs with high specificity and affinity, capable of precisely recognizing and binding to neoantigens (73). For instance, scRNA-seq libraries were constructed from nasopharyngeal carcinoma (NPC) patient samples and subsequently sequenced using a high-throughput sequencing platform. In-depth analysis of the sequencing data revealed significant clonal expansion of T cells in EBV-positive tumor samples (74). This finding suggests that the sequence characteristics of these TCRs are closely associated with the EBV-derived neoantigens. Despite its advantages, scRNA-seq has certain limitations. One major challenge is maintaining cell integrity and viability throughout the process, as these factors are critical for ensuring accurate single-cell analysis (75). Addressing these challenges will be essential for further advancing the application of scRNA-seq in neoantigen research and personalized immunotherapy.
The human leukocyte antigen (HLA) system, located on the short arm of chromosome 6, is equivalent to the MHC in humans (76). Humans possess more than 24,000 different alleles of HLA class I (HLA-I, including HLA-A, HLA-B, and HLA-C) and HLA class II (HLA-II, including HLA-DR, HLA-DQ, and HLA-DP), with their combined effects leading to significant polymorphism (77). Wichmann et al. have conducted a study on patients with HNSCC presenting distinct HLA characteristics, observing that disease progression varied among these patients based on their HLA profiles (78). Additionally, previous studies have indicated associations between specific HNSCC subtypes and HLA genotypes (79). As a result, determining the HLA genotype of HNSCC patients is an essential step in the neoantigen prediction process (80). Predicting the binding affinity of HLA class II (HLA-II) is more challenging compared to HLA class I (HLA-I). HLA-I molecules present shorter peptide sequences (8–11 amino acids), while HLA-II molecules present longer sequences (11–20 amino acids or even longer) due to their open peptide-binding grooves (81). Consequently, most studies primarily focus on tumor vaccines that utilize antigenic peptides presented by HLA-IA molecules (82). However, the expression of HLA-IA is often downregulated in tumors, which can promote immune evasion by the tumor (83). In contrast, the expression level of HLA-IB is associated with prognosis and immune infiltration (84). When HLA-IA expression is downregulated, peptides that bind to HLA-IB can emerge as highly promising therapeutic targets. Machine learning and deep learning techniques are increasingly used to predict peptide-HLA binding affinities by analyzing the binding sites of neoantigen peptide sequences and HLA molecules (85). A high binding affinity indicates that the mutated peptide is more likely to bind to HLA and be recognized by T cells (86). Several tools have been developed to identify HLA-I alleles, including OptiType (87) and HLA-HD (88), and tools for HLA-II alleles, such as SOAPHLA (89) and PHLAT (90) (Figure 3). These tools demonstrate prediction accuracies as high as 99% when compared to HLA-specific typing methods used in clinical practice (91). Among them (Figure 3), OptiType currently reports the highest accuracy.
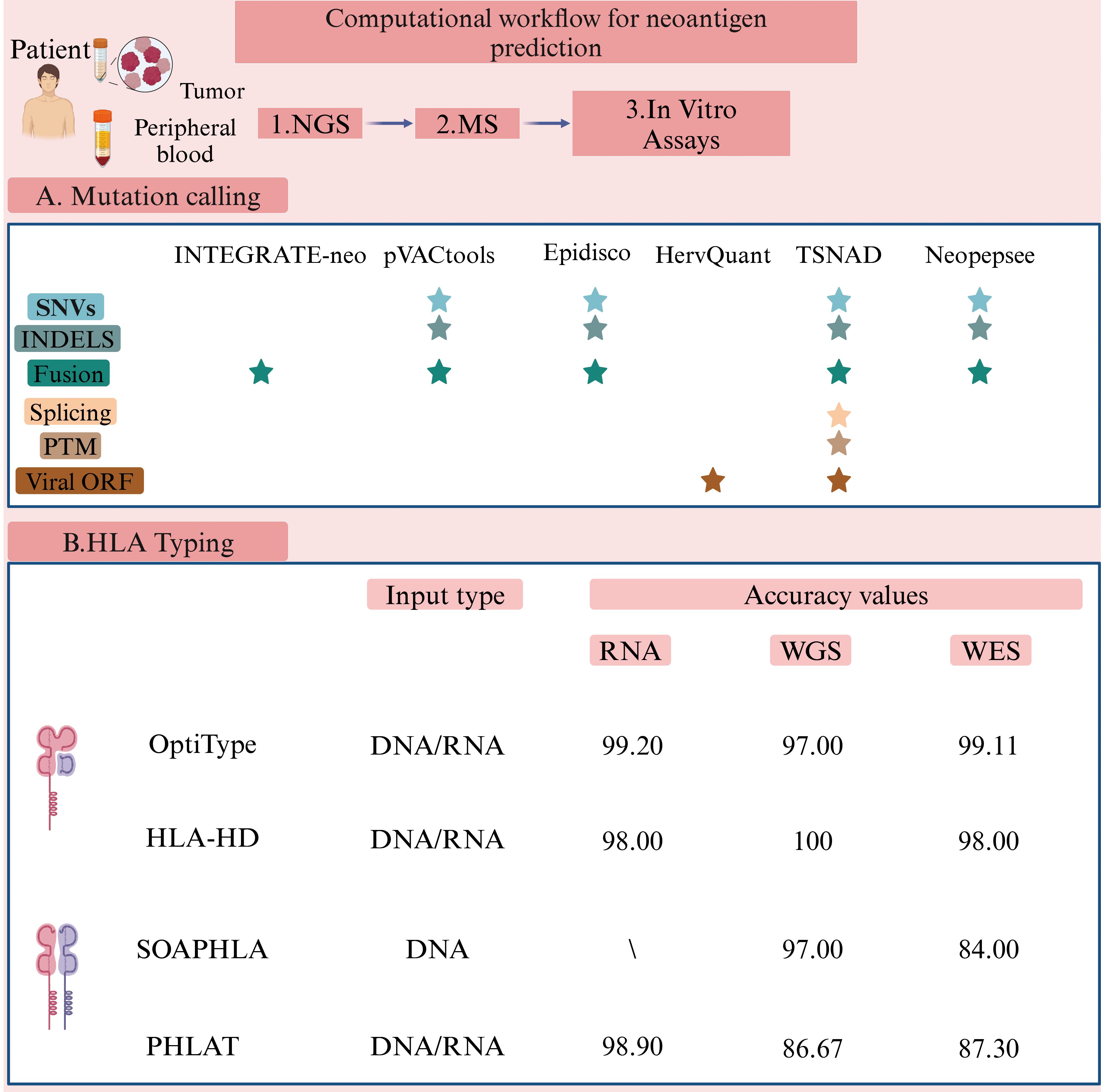
Figure 3. Computational workflow for neoantigen prediction (Created with BioRender.com). Various software packages are utilized to identify sequence variations between tumor and normal cells. These tools predict and prioritize antigen immunogenicity, thus facilitating the selection of optimal tumor neoantigens for therapeutic application. The pVACtools suite consists of multiple functional modules, including pVACseq (personalized Variant Antigens by Cancer Sequencing) for predicting neoantigens derived from somatic mutations, and pVAC-fuse, which identifies neoantigens originating from gene fusions (151) (http://www.pvactools.org). INTEGRATE-Neo is an open-source pipeline designed to utilize NGS data for identifying neoantigens arising from gene fusions, thereby expanding the scope of neoantigen discovery (152) (https://github.com/ChrisMaherLab/INTEGRATE-Neo). Epidisco serves primarily as a workflow orchestration tool, coordinating the parallel execution of analytical processes such as variant detection and neoantigen prediction. It ensures efficient utilization of computational resources, thus accelerating vaccine development (153). HervQuant is specifically designed to analyze and quantify human endogenous retroviruses (hERVs) expression, potentially identifying novel targets for tumor vaccines or immunotherapy (154). TSNAD (Tumor-Specific Neoantigen Detector) follows Genome Analysis Toolkit (GATK) best practice for detecting somatic mutations in cancers and predicting potential neoantigens (155) (https://github.com/jiujiezz/tsnad). Neopepsee integrates sequence features and amino acid immunogenicity profiles through machine learning algorithms to enhance the accuracy and specificity of neoantigen prediction (156) (http://sourceforge.net/projects/neopepsee/).
However, HLA-II typing algorithms still require further development to improve their predictive accuracy compared to HLA-I typing algorithms.
Relying solely on algorithmic predictions to screen candidate neoantigens may lead to false-positive results and fail to accurately predict the binding affinities of all possible HLA alleles to peptides (62). Therefore, functional verification is a essential to ensure the efficacy of candidate neoantigens. This process typically involves testing whether candidate peptides can stimulate T-cell proliferation or induce immune responses such as cytokine production in vitro, as well as evaluating whether the peptides can provoke effective anti-tumor immune responses in vivo (92). One of the key techniques for assessing the immunogenicity of neoantigens is the enzyme-linked immunospot (ELISPOT) assay. This method detects changes in cytokine secretion by T cells after stimulation by neoantigens, using specific antibodies (93). In this assay, patient-derived T cells are co-cultured with dendritic cells (DCs) loaded with candidate neoantigen peptides (94). By measuring the release of cytokines (such as IFN-γ) or the upregulation of activation markers (e.g., CD25), researchers can assess whether T cells are effectively activated (95). Cytotoxicity assays provide another critical approach to evaluate the immunogenicity of neoantigens. In these assays, tumor cell lines expressing patient-specific neoantigens serve as target cells. Patient-derived T cells are co-cultured with the target cells, and subsequent death of the target cells is monitored. This directly reflects the cytotoxic capacity of T cells against tumor cells in vitro and is serve as a valuable measure of neoantigens immunogenicity (96).
5 Neoantigen-based treatment for HNSCC
Identifying the optimal combination of various immunotherapies is crucial for the effective treatment of recurrent and metastatic (R/M) HNSCC (6). A search using the keywords “Head and neck cancer” and “neoantigen” to search on ClinicalTrials.gov has revealed several registered clinical trials targeting neoantigens for treating head and neck cancer. These trials explore the use of neoantigen vaccines, both in combination with and without immune checkpoint inhibitors (ICIs). A summary of these clinical trials is provided in Table 1 (Source: https://clinicaltrials.gov/, update on February 20th, 2025).
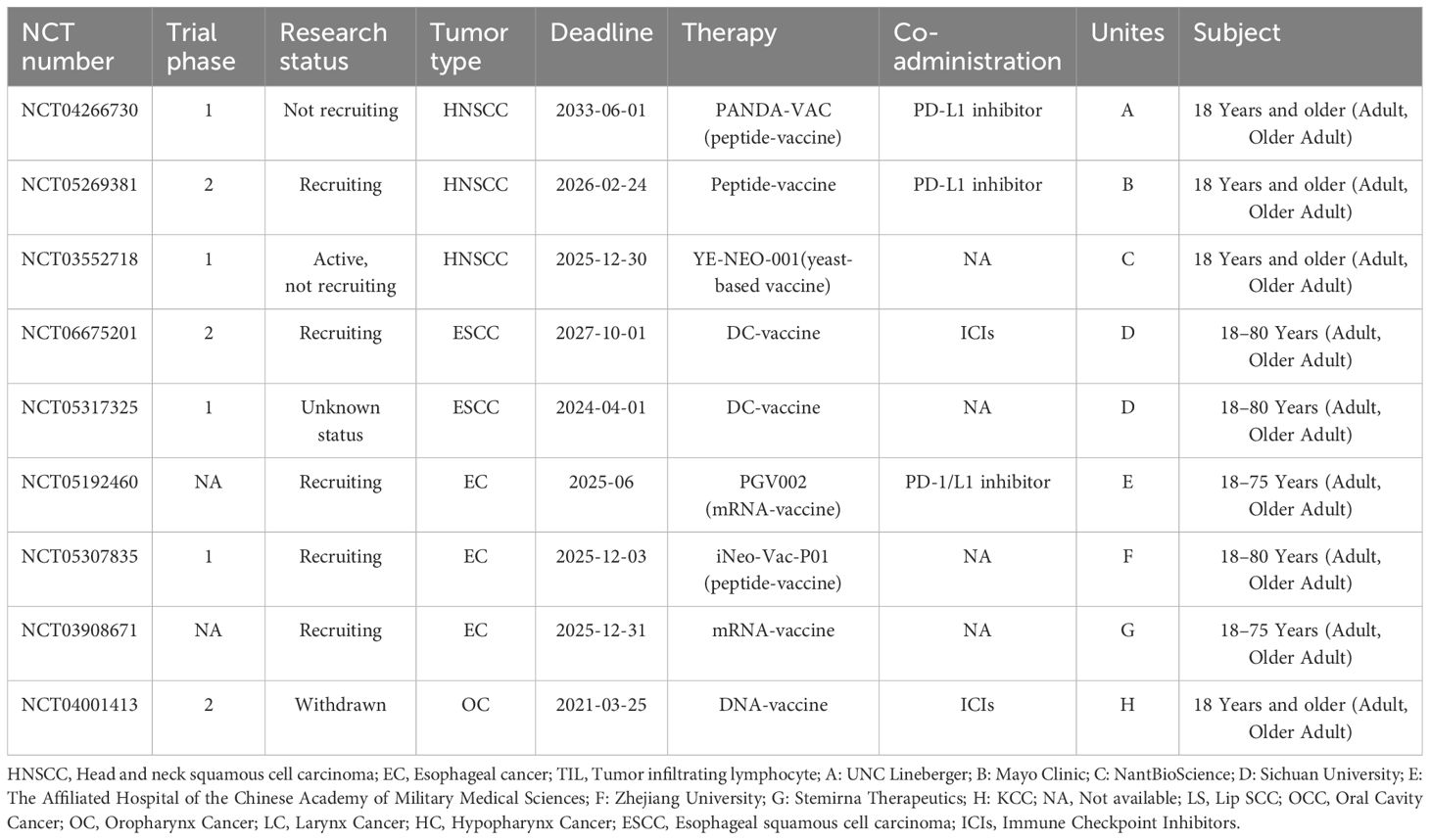
Table 1. Neoantigen-based clinical trials for “head and neck cancer” registered on ClinicalTrials.gov.
5.1 Tumor vaccines targeting neoantigens
Given the high heterogeneity of HNSCC, patients with this malignance may benefit from personalized neoantigen-based vaccines tailored to their specific tumor mutations (97). Although these vaccines are in the early stage of development, they have demonstrated significant potential in cancer immunotherapy. Neoantigen-based tumor vaccines can be broadly categorized into nucleic acid vaccines, peptide vaccines, and dendritic cell (DC) vaccines (Figure 4) (98). Each vaccine type utilizes distinct antigen-presenting cell (APC) processing pathways. For DNA vaccines, the genetic material is internalized into the cytoplasm, where it undergoes transcription and translation before being processed into antigenic peptides. For RNA vaccines, translation occurs directly on cytoplasm ribosomes, followed by proteasomal degradation of the resulting neoantigen proteins (99). The antigen peptides generated through these processes follow two primary pathways: 1) intracellular antigens: Proteins degraded by the proteasome are further processed in the endoplasmic reticulum and loaded onto MHC class I molecules. These peptides are subsequently presented to cytotoxic T lymphocytes (CTLs), triggering a targeted immune response against tumor cells (100); 2) extracellular antigens: Proteins degraded within lysosomes are loaded onto MHC class II molecules and presented to CD4+ T cells, which play a crucial role in orchestrating the immune response (101). Additionally, activated immune cells secrete cytokines such as granzyme, perforin, TNFα, and IFNγ, which contribute to tumor cell lysis and immune-mediated tumor control.
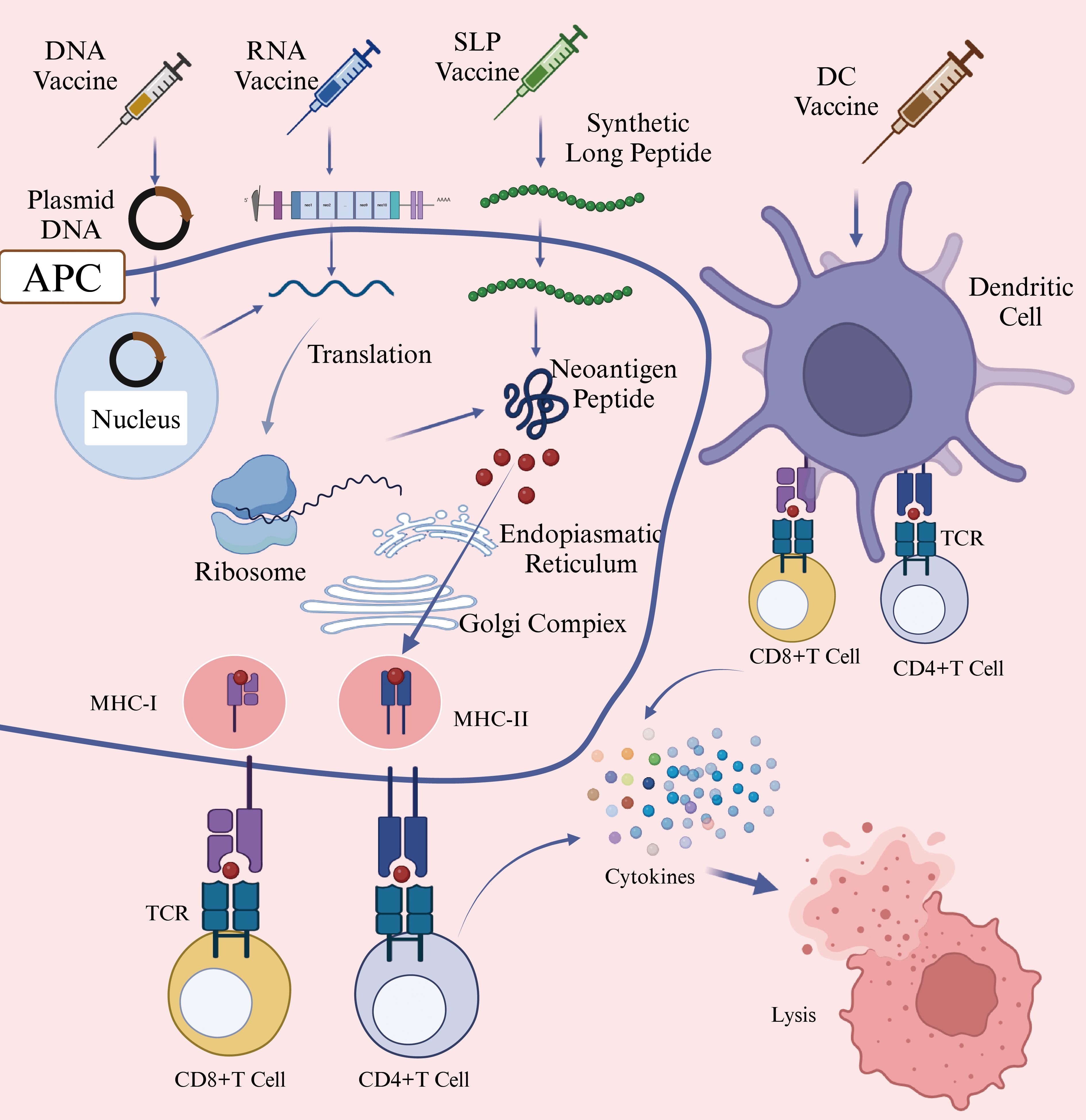
Figure 4. Mechanism of Action of DNA/RNA/SLP/DC Vaccines (Created with BioRender.com). This figure illustrated the mechanisms through which various neoantigen vaccines stimulate immune responses against tumor cells. DNA vaccines introduce genes encoding neoantigens into host cells, leading to neoantigen protein synthesis. RNA vaccines deliver neoantigen-encoding mRNA into cytoplasm, where cellular machinery translates the mRNA directly into neoantigen proteins. Synthetic long peptide (SLP) vaccines consist of multiple neoantigen peptide sequences and are directly internalized by antigen-presenting cells (APCs). Dendritic cell (DC) vaccines involve the ex vivo loading of neoantigens onto DC cells, followed by their reinfusion into patients. Subsequently, APCs process these neoantigens and present peotide fragments via major histocompatibility complex (MHC) molecules to T cells. Activated T cells recognize these presented neoantigens, resulting in targeted immune responses and the elimination of tumor cells. Abbreviations: APC, antigen-presenting cell; DC, Dendritic Cell; MHC-I, major histocompatibility complex class I; MHC-II, major histocompatibility complex class II; TCR, T-Cell receptor; SLP, synthetic long peptide.
Nucleic acid vaccines are primarily classified into DNA vaccines and RNA vaccines. These vaccines can deliver multiple tumor neoantigens in a single administration, triggering both cellular and humoral anti-tumor immune responses (102). A clinical trial utilizing linear DNA amplicons-small DNA fragments encoding neoantigens, demonstrated a significant enhancement in antigen-specific cytotoxic T lymphocyte (CTL) responses and tumor regression (103). However, DNA neoantigen vaccines face several challenges. For instance, DNA sequence introduced into the body is susceptible to degradation by nucleases, which can reduce vaccine efficacy (104).
In contrast, mRNA vaccines offer advantages such as good tolerability, lower cost, and rapid production (105). Once inside host cells, mRNA neoantigen vaccines direct ribosomes to translate the encoded genetic information they carry into neoantigen proteins (106). Compared to DNA vaccines, mRNA vaccines eliminate the risk of insertional mutations and transcriptional abnormalities, avoiding potential side effects associated with DNA integration (107). Additionally, since neoantigen proteins are synthesized through the cell’s natural translation mechanism, they may elicit a more robust immune response than peptide vaccines, which deliver pre-processed peptide segments (108). One notable example is mRNA-4157, a vaccine co-developed by Merck and Moderna, capable of encoding up to 30 different neoantigens. When administered in combination with the PD-1 inhibitor pembrolizumab, it reduced the risk of recurrence or death by 49% in patients with surgically resected high-risk melanoma (stage III/IV) and decreased the risk of distant metastasis or death by 62% (109). Personalized mRNA vaccines combined with anti-PD-1 monoclonal antibodies have also demonstrated clinical efficacy in treatmenting HNSCC. A clinical trial (NCT03468244) has reported a case in which a patient with advanced esophageal squamous cell carcinoma (ESCC)-characterized by microsatellite stability and a low likelihood of benefiting from immune checkpoint inhibitors (ICIs), achieved a partial response (PR) after treatment with mRNA personalized cancer vaccine and PD-1 monoclonal antibodies. The patient exhibited progression-free survival (PFS) of 457 days, overall survival (OS) of 457 days, and a duration of response (DOR) of 377 days (110). These findings underscore the potential of mRNA vaccines to express multiple tumor-specific neoantigens and enhance immune response when used in combination with other therapies (62). Despite these advantages, mRNA has a linear structure that is prone to degradation by RNases, resulting in relatively poor stability (111). To address this, modifications are necessary to enhance mRNA stability and further research is needed to determine optimal delivery methods and dosing strategies. Circular RNA (circRNA), which has a covalently closed-loop structure and lacks a 5’ cap and a 3’ tail, exhibits high resistance to RNases degradation and a longer intracellular half-life (112). This unique stability makes circRNA a promising candidate for next-generation vaccines. A study by Wang et al. used the permuted intron-exon (PIE) strategy to generate circRNA molecules containing internal ribosome entry site (IRES) elements and coding sequences for hepatocellular carcinoma (HCC) neoantigens. The expressed neoantigens were captured and internalized by DCs, which then presented them to CD8+ and CD4+ T cells, thereby initiating an adaptive immune response (113). DCs are considered the most potent professional antigen-presenting cells (APCs) in the human immune system (114).
Nucleic acid vaccines require the host cell’s transcription or translation machinery to synthesize antigenic proteins in vivo, after which antigen presentation can occur. In contrast, neoantigen peptide vaccines directly provide antigenic peptide fragments. Neoantigen peptide vaccines are the most widely used form of neoantigen-based cancer vaccines. They feature well-defined sequences, straightforward preparation and storage processes, and the ability to directly bind to MHC molecules, effectively triggering strong CD8+ T cell responses in tumors with both high and low mutation burdens (115). A clinical trial investigating a personalized neoantigen peptide vaccine for HNSCC (NCT04183166) demonstrated that in resected HPV-negative HNSCC patients, these vaccines could stimulate tumor-specific immune responses and reduce the risk of recurrence (116). The immunogenicity of neoantigen peptide vaccines can be further enhanced through the use of immunostimulatory adjuvants (117). For instance, a neoantigen peptide vaccine (NCT02897765) formulated with the polyinosinic-polycytidylic acid (poly-ICLC) as an adjuvant successfully activated CD8+ and CD4+ T cells in patients with advanced melanoma, non-small cell lung cancer (NSCLC), or bladder cancer, all of which exhibit high mutation burdens. In high-risk melanoma patients, this vaccine prevent recurrence for up to 25 months following treatment (118). Several studies have reported that neoantigen peptide vaccines are generally tolerated, with only a few cases of serious adverse events (AEs) (119). Importantly, these AEs cannot be solely attributed to the vaccines themselves but are primarily linked to cancer progression. Given their favorable safety profile and patient tolerability, personalized neoantigen peptide vaccines are regarded as a promising therapeutic approach for cancer immunology. The efficacy of neoantigen peptide vaccines is significantly influenced by peptide length, making it a critical factor in vaccine design. Short peptides typically refer to minimal peptide epitopes with optimal binding sequences. For CD8+ T cells, these epitopes consist of 8–11 amino acids, which fit within the MHC-I antigen-binding groove (120). In contrast, for CD4+ T cells, minimal peptide epitopes range from 13–18 amino acids (121). However, short peptide vaccines have a short half-life and limited immunogenicity, making it challenging to generate sustained T cell response. To overcome these limitations, many studies choose for long peptide vaccines, as they can bind to multiple HLA alleles and are more likely to induce a robust and long-lasting anti-tumor immune response (122).
As previously discussed, neoantigen vaccines based on DNA, RNA, or peptide segments typically require endogenous APCs, such as dendritic cells (DCs), to internalize the antigens and present peptide-major histocompatibility complex (pMHC) complexes to T cells (123). In this section, we focus on a distinct therapeutic approach -personalized neoantigen peptide-pulsed autologous dendritic cells vaccines. Unlike conventional vaccines, this strategy involves isolating autologous DCs from the patient, loading them in vitro with individualized neoantigen peptides, and subsequently reinfusing the mature, peptide-loaded DCs to stimulate a targeted T-cell response. The Neo-DCVac, a personalized neoantigen dendritic cell vaccine, is specifically designed to deliver neoantigen-loaded DCs into patients to prime the adaptive immune system. Neoantigen-pulsed DC vaccines have shown encouraging anti-tumor activity in patients with advanced or relapsed malignancies (124). In a 2024 clinical study (NCT05023928) conducted by Chen et al., the safety and feasibility of Neo-DCVac as a postoperative adjuvant treatment for esophageal squamous cell carcinoma (ESCC) were evaluated. Twelve patients were enrolled in the study, which report one- and two-year overall survival (OS) rates of 100% and 91.7%, respectively, and disease-free survival (DFS) rates of 88.3% and 66.7% (125). Despite its promising efficacy, the widespread clinical application of Neo-DCVac remains constrained by high costs, complex manufacturing protocols, and procedural risks related to leukapheresis, including vascular injury and electrolyte imbalances (126).
5.2 ICI combined with neoantigen-based treatment for HNSCC
Immune checkpoint inhibitors (ICIs) have demonstrated significant survival benefits in R/M HNSCC patients while maintaining a favorable safety profile. The five-year OS rate has increased from 5.0% to 15.4%-23.9% with ICI treatment (127). Before administering ICIs, PD-L1 CPS scoring, tumor mutational burden (TMB) assessment, and clinical symptom evaluation should be conducted to guide personalized treatment strategies (128, 129). The CPS (combined positive score) is reported as an integer between 0 and 100 (130). A higher CPS score correlates with an improved objective response rate (ORR) and survival benefit (31). For R/M HNSCC patients with PD-L1 CPS ≥ 1, the combined first-line treatment includes either pembrolizumab plus platinum-based chemotherapy and 5-FU or pembrolizumab monotherapy (131). For patients with unknown PD-L1 status or PD-L1 CPS < 1, the preferred first-line regimen is pembrolizumab combined with platinum-based chemotherapy and 5-FU (132). TMB has been established as a predictive biomarker for immunotherapy efficacy across multiple tumors types and serve as an indirect indicator of neoantigen generation (133). Both the 2022 ASCO guidelines and the 2023 National Comprehensive Cancer Network (NCCN) guidelines recommend pembrolizumab for first-line or later-line treatment in R/M HNSCC patients with TMB-high (≥ 10 mut/Mb) (134).
In HNSCC, combination therapy involving anti-PD-L1 and anti-CTLA-4 inhibitors demonstrates superior efficacy compared to anti-PD-L1 monotherapy, as it promotes recruitment of CD4+ T cells to tumor-draining lymph nodes (TDLN), where they differentiate into effector T cells capable of targeting and eliminating tumor cells (135). However, immune checkpoint inhibitors primarily target one or two stages of the anti-cancer immune pathway, and only a small subset of patients develop a robust anti-tumor response with single-agent therapy (67). Therefore, the combination of ICIs with personalized neoantigen vaccines has been shown to significantly enhance tumor regression compared to monotherapy (136).
Immune escape is a key mechanism driving R/M HNSCC, leading to T cell anergy and CD8+ T cell exhaustion. These exhausted T cells typically exhibit high expression of PD-1 and CD39, which suppresses the body’s ability to eliminate tumor cells through the immune system (137). Neoantigen vaccines stimulate the patient’s immune system, particularly by enhancing the response of tumor-specific CD8+ T cells (138). However, interferon-γ (IFNγ) produced by CD8+ T cells and Th1 CD4+ cells can regulate PD-L1 expression (139), which may ultimately impair the effectiveness of the vaccine by reinforcing immune suppression. To counteract this, immune checkpoint inhibitors (ICIs), including anti-CTLA-4 antibodies, anti-PD-1 antibodies, and anti-PD-L1 antibodies, bind to immune checkpoint proteins on T cells, effectively reversing tumor-mediated immune suppression and restoring T cell function (140). A study by Ott et al. demonstrated the potential of synthetic neoantigen peptide vaccines in enhancing immune responses. In this study, six melanoma patients who had undergone surgical resection were treated with neoantigen vaccines, followed by PD-1 antibody therapy. Among them, two patients achieved complete tumor regression, highlighting the synergistic effect of neoantigen vaccines and ICIs (118). Several ongoing clinical trials are investigating personalized neoantigen vaccines in combination with immune checkpoint inhibitors for HNSCC treatment. Examples include PANDA-VAC (NCT04266730), PGV002 (NCT05192460), and NeoDC-Vac (NCT06675201), as detailed in Table 1.
6 Conclusion
This review provides an overview of HNSCC treatment strategies targeting tumor neoantigens and presets theoretical evidence supporting the clinical relevance of neoantigen-based immunotherapies. However, personalized neoantigen vaccines for HNSCC remain limited in both market availability and clinical development. A primary challenge lies in the screening and identification of highly immunogenic neoantigens, which heavily rely on high-throughput sequencing and bioinformatics techniques. Technical inaccuracies in these processes may result in false positives, leading to the selection of ineffective neoantigens that fail to elicit robust immune responses (141). Neoantigen identification can be enhanced by employing computational algorithms to construct virtual peptidomes from NGS data, combined with mass spectrometry(MS)-based analysis of peptides bound to MHC molecules. Integrating genomic and transcriptomic sequencing data with HLA-associated peptideome analysis has further improved the sensitivity and specificity of neoantigen identification (142). For clinical applications, an efficient computational workflow is essential for precise neoantigen selection. Despite rapid advances in sequencing technologies, neoantigen identification and validation remain time-consuming and costly. The process of developing neoantigen vaccines from patient tumor samples typically requires three to five months (108), significantly limiting their clinical feasibility.
By targeting PD-L1 overexpression, CD58 genetic alterations, and the immunosuppressive microenvironments, more effective combination treatment strategies can be designed, potentially improving the prognosis of patients with HNSCC. In diffuse large B-cell lymphoma (DLBCL), translocation of the PD-L1 gene locus with the IGH gene frequently results in PD-L1 overexpression (143). Similar overexpression of PD-L1 has been observed in HNSCC. Future studies investigating the genetic basis for PD-L1 upregulation in HNSCC could benefit from methodologies previously employed in DLBCL research. Elevated PD-L1 expression is associated with tumor aggressiveness and poor clinical outcomes; accordingly, combined blockade of PD-L1 and CD73 has demonstrated significant inhibition of tumor growth and metastasis (144). While PD-L1 inhibitors have shown clinical efficacy in treating HNSCC, their combination with CD73 inhibitors or other immune checkpoint inhibitors may further enhance antitumor immune responses. Furthermore, CD58 has been shown to suppress PDL-1 and IDO expression by inhibiting the JAK2/STAT1 pathway through activation of the LYN/CD22/SH2 domain-containing phosphatase 1 (SHP1) axis (145). Therefore, combining PD-L1 inhibitors with approaches designed to enhance CD58 signaling may offer a promising strategy to overcome PD-L1-mediated immune suppression (146).
However, neoantigen vaccines alone are unlikely to achieve complete tumor eradication (147). First, tumors with low TMB might be unsuitable for existing neoantigen vaccine strategies. For example, nasopharyngeal carcinoma exhibits a lower mutation rate of only 1 mutation/Mb (148). This relatively low rate complicates the identification of immunogenic neoantigens. Second, immune escape mechanisms in tumors remain a significant obstacle to the efficacy of tumor vaccines (149). Combining neoantigen vaccines and ICIs offers a promising strategy to overcome tumor immune evasion (150). However, research on neoantigen vaccine-ICI combination therapy is still in the clinical trial phase (102), requiring further investigation and clinical validation to establish its safety and efficacy. With continued scientific advancements and technological progress, the widespread clinical adoption of neoantigen-based immunotherapy-particularly in HNSCC is expected. Future developments may lead to enhanced treatment efficacy, reduced preparation time, and greater accessibility, ultimately improving patient outcomes.
Author contributions
MJ: Writing – review & editing, Writing – original draft. JL: Conceptualization, Methodology, Writing – review & editing. JW: Methodology, Writing – review & editing, Conceptualization. XY: Writing – original draft, Writing – review & editing. WW: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Key R&D Program Project in Shaanxi Province (Grant number: 2023-YBSF-230).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wang CW, Biswas PK, Islam A, Chen MK, and Chueh PJ. The use of immune regulation in treating head and neck squamous cell carcinoma (HNSCC). Cells. (2024) 13(5):413. doi: 10.3390/cells13050413
2. El-Bayoumy K, Chen KM, Zhang SM, Sun YW, Amin S, Stoner G, et al. Carcinogenesis of the oral cavity: environmental causes and potential prevention by black raspberry. Chem Res Toxicol. (2017) 30:126–44. doi: 10.1021/acs.chemrestox.6b00306
3. Barsouk A, Aluru JS, Rawla P, Saginala K, and Barsouk A. Epidemiology, risk factors, and prevention of head and neck squamous cell carcinoma. Med Sci (Basel). (2023) 11(2):42. doi: 10.3390/medsci11020042
4. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
5. Tsou HH, Tsai HC, Chu CT, Cheng HW, Liu CJ, Lee CH, et al. Cigarette smoke containing acrolein upregulates EGFR signaling contributing to oral tumorigenesis in vitro and in vivo. Cancers (Basel). (2021) 13(14):3544. doi: 10.3390/cancers13143544
6. Hashim D, Genden E, Posner M, Hashibe M, and Boffetta P. Head and neck cancer prevention: from primary prevention to impact of clinicians on reducing burden. Ann Oncol. (2019) 30:744–56. doi: 10.1093/annonc/mdz084
7. Wang H, Zheng Z, Zhang Y, Bian C, Bao J, Xin Y, et al. Locally advanced head and neck squamous cell carcinoma treatment efficacy and safety: a systematic review and network meta-analysis. Front Pharmacol. (2023) 14:1269863. doi: 10.3389/fphar.2023.1269863
8. Martinez-Useros J and Garcia-Foncillas J. The challenge of blocking a wider family members of EGFR against head and neck squamous cell carcinomas. Oral Oncol. (2015) 51:423–30. doi: 10.1016/j.oraloncology.2015.02.092
9. Wang Y, Yang T, Gan C, Wang K, Sun B, Wang M, et al. Temporal and spatial patterns of recurrence in oral squamous cell carcinoma, a single-center retrospective cohort study in China. BMC Oral Health. (2023) 23:679. doi: 10.1186/s12903-023-03204-7
10. Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. (2019) 394:1915–28. doi: 10.1016/s0140-6736(19)32591-7
11. Harrington KJ, Burtness B, Greil R, Soulières D, Tahara M, de Castro G, et al. Pembrolizumab with or without chemotherapy in recurrent or metastatic head and neck squamous cell carcinoma: updated results of the phase III KEYNOTE-048 study. J Clin Oncol. (2023) 41:790–802. doi: 10.1200/jco.21.02508
12. Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. (2015) 21:870–81. doi: 10.1158/1078-0432.Ccr-14-2481
13. Hanna GJ, Liu H, Jones RE, Bacay AF, Lizotte PH, Ivanova EV, et al. Defining an inflamed tumor immunophenotype in recurrent, metastatic squamous cell carcinoma of the head and neck. Oral Oncol. (2017) 67:61–69. doi: 10.1016/j.oraloncology.2017.02.005
14. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. (2017) 171:1611–1624.e1624. doi: 10.1016/j.cell.2017.10.044
15. Zhang Z, Lu M, Qin Y, Gao W, Tao L, Su W, et al. Neoantigen: A new breakthrough in tumor immunotherapy. Front Immunol. (2021) 12:672356. doi: 10.3389/fimmu.2021.672356
16. Xie N, Shen G, Gao W, Huang Z, Huang C, and Fu L. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther. (2023) 8:9. doi: 10.1038/s41392-022-01270-x
17. Apavaloaei A, Hardy MP, Thibault P, and Perreault C. The origin and immune recognition of tumor-specific antigens. Cancers (Basel). (2020) 12(9):2607. doi: 10.3390/cancers12092607
18. Minati R, Perreault C, and Thibault P. A roadmap toward the definition of actionable tumor-specific antigens. Front Immunol. (2020) 11:583287. doi: 10.3389/fimmu.2020.583287
19. Wu J, Zhao W, Zhou B, Su Z, Gu X, Zhou Z, et al. TSNAdb: A database for tumor-specific neoantigens from immunogenomics data analysis. Genomics Proteomics Bioinf. (2018) 16:276–82. doi: 10.1016/j.gpb.2018.06.003
20. Razzaghi H, Saraiya M, Thompson TD, Henley SJ, Viens L, and Wilson R. Five-year relative survival for human papillomavirus-associated cancer sites. Cancer. (2018) 124:203–11. doi: 10.1002/cncr.30947
21. Leemans CR, Snijders PJF, and Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. (2018) 18:269–82. doi: 10.1038/nrc.2018.11
22. Patini R, Cordaro M, Marchesini D, Scilla F, Gioco G, Rupe C, et al. Is systemic immunosuppression a risk factor for oral cancer? A systematic review and meta-analysis. Cancers (Basel). (2023) 15(12):3077. doi: 10.3390/cancers15123077
23. Tang M, Zeng Y, Poisson A, Marti D, Guan L, and Zheng Y. Haplotype-dependent HLA susceptibility to nasopharyngeal carcinoma in a Southern Chinese population. Genes Immun. (2010) 11:334–42. doi: 10.1038/gene.2009.109
24. Makni H, Daoud J, Ben Salah H, Mahfoudh N, Haddar O, Karray H, et al. HLA association with nasopharyngeal carcinoma in southern Tunisia. Mol Biol Rep. (2010) 37:2533–9. doi: 10.1007/s11033-009-9769-y
25. Patel SG, Amit M, Yen TC, Liao CT, Chaturvedi P, Agarwal JP, et al. Lymph node density in oral cavity cancer: results of the International Consortium for Outcomes Research. Br J Cancer. (2013) 109:2087–95. doi: 10.1038/bjc.2013.570
26. Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, et al. Molecular mechanisms of cisplatin resistance. Oncogene. (2012) 31:1869–83. doi: 10.1038/onc.2011.384
27. Lacas B, Carmel A, Landais C, Wong SJ, Licitra L, Tobias JS, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): An update on 107 randomized trials and 19,805 patients, on behalf of MACH-NC Group. Radiother Oncol. (2021) 156:281–93. doi: 10.1016/j.radonc.2021.01.013
28. Wijdeven RH, Pang B, Assaraf YG, and Neefjes J. Old drugs, novel ways out: Drug resistance toward cytotoxic chemotherapeutics. Drug Resist Updat. (2016) 28:65–81. doi: 10.1016/j.drup.2016.07.001
29. Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. (2016) 17:956–65. doi: 10.1016/s1470-2045(16)30066-3
30. Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. (2016) 375:1856–67. doi: 10.1056/NEJMoa1602252
31. Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV, Gilbert J, et al. Pembrolizumab for platinum- and cetuximab-refractory head and neck cancer: results from a single-arm, phase II study. J Clin Oncol. (2017) 35:1542–9. doi: 10.1200/jco.2016.70.1524
32. Agne G, Kohler H, Chulam T, Pinto C, and Kowalski L. Oncologic outcomes of microscopic tumor cut-through in locally advanced oral squamous cell carcinoma. Arch Head Neck Surg. (2022) 51:e20220013. doi: 10.4322/ahns.2022.0013
33. Dyckhoff G, Herold-Mende C, Scherer S, Plinkert PK, and Warta R. Human leucocyte antigens as prognostic markers in head and neck squamous cell carcinoma. Cancers (Basel). (2022) 14(15):3828. doi: 10.3390/cancers14153828
34. Shang S, Zhao Y, Qian K, Qin Y, Zhang X, Li T, et al. The role of neoantigens in tumor immunotherapy. BioMed Pharmacother. (2022) 15:1113118. doi: 10.1016/j.biopha.2022.113118
35. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, and Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. (2017) 17:209–22. doi: 10.1038/nrc.2016.154
36. Capietto AH, Hoshyar R, and Delamarre L. Sources of cancer neoantigens beyond single-nucleotide variants. Int J Mol Sci. (2022) 23(17):10131. doi: 10.3390/ijms231710131
37. Wang E and Aifantis I. RNA splicing and cancer. Trends Cancer. (2020) 6:631–44. doi: 10.1016/j.trecan.2020.04.011
38. Pounraj S, Chen S, Ma L, Mazzieri R, Dolcetti R, and Rehm BHA. Targeting tumor heterogeneity with neoantigen-based cancer vaccines. Cancer Res. (2024) 84:353–63. doi: 10.1158/0008-5472.Can-23-2042
39. Welters MJ, van der Sluis TC, van Meir H, Loof NM, van Ham VJ, van Duikeren S, et al. Vaccination during myeloid cell depletion by cancer chemotherapy fosters robust T cell responses. Sci Transl Med. (2016) 8:334ra352. doi: 10.1126/scitranslmed.aad8307
40. Wang L, Shamardani K, Babikir H, Catalan F, Nejo T, Chang S, et al. The evolution of alternative splicing in glioblastoma under therapy. Genome Biol. (2021) 22:48. doi: 10.1186/s13059-021-02259-5
41. Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, and Serody JS. Alternative tumour-specific antigens. Nat Rev Cancer. (2019) 19:465–78. doi: 10.1038/s41568-019-0162-4
42. Juhari WKW, Ahmad Amin Noordin KB, Zakaria AD, Rahman W, Mokhter W, Hassan MRA, et al. Whole-genome profiles of malay colorectal cancer patients with intact MMR proteins. Genes (Basel). (2021) 12(9):1448. doi: 10.3390/genes12091448
43. Abbott CW, Boyle SM, Pyke RM, McDaniel LD, Levy E, Navarro FCP, et al. Prediction of immunotherapy response in melanoma through combined modeling of neoantigen burden and immune-related resistance mechanisms. Clin Cancer Res. (2021) 27:4265–76. doi: 10.1158/1078-0432.Ccr-20-4314
44. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. (2017) 18:1009–21. doi: 10.1016/s1470-2045(17)30516-8
45. TaNiue K and Akimitsu N. Fusion genes and RNAs in cancer development. Noncoding RNA. (2021) 7(1):10. doi: 10.3390/ncrna7010010
46. Yang J, Annala M, Ji P, Wang G, Zheng H, Codgell D, et al. Recurrent LRP1-SNRNP25 and KCNMB4-CCND3 fusion genes promote tumor cell motility in human osteosarcoma. J Hematol Oncol. (2014) 7:76. doi: 10.1186/s13045-014-0076-2
47. Wang Y, Shi T, Song X, Liu B, and Wei J. Gene fusion neoantigens: Emerging targets for cancer immunotherapy. Cancer Lett. (2021) 506:45–54. doi: 10.1016/j.canlet.2021.02.023
48. Rosenberg-Mogilevsky A, Siegfried Z, and Karni R. Generation of tumor neoantigens by RNA splicing perturbation. Trends Cancer. (2025) 11:12–24. doi: 10.1016/j.trecan.2024.10.008
49. Venkataramany AS, Schieffer KM, Lee K, Cottrell CE, Wang PY, Mardis ER, et al. Alternative RNA splicing defects in pediatric cancers: new insights in tumorigenesis and potential therapeutic vulnerabilities. Ann Oncol. (2022) 33:578–92. doi: 10.1016/j.annonc.2022.03.011
50. Bigot J, Lalanne AI, Lucibello F, Gueguen P, Houy A, Dayot S, et al. Splicing patterns in SF3B1-mutated uveal melanoma generate shared immunogenic tumor-specific neoepitopes. Cancer Discov. (2021) 11:1938–51. doi: 10.1158/2159-8290.Cd-20-0555
51. Wang TY and Yang R. Integrated protocol for exitron and exitron-derived neoantigen identification using human RNA-seq data with ScanExitron and ScanNeo. STAR Protoc. (2021) 2:100788. doi: 10.1016/j.xpro.2021.100788
52. Zheng Y-C and Chen C-K. Abstract B001: CircRNA-derived neoantigen identification in cancer vaccine development. Mol Cancer Ther. (2024) 23:B001–1. doi: 10.1158/1538-8514.Rnadrivers24-b001
53. Su S. Beneath the surface: neoantigens beyond chromosomal DNA mutations. Cancer Discov. (2024) 14:2066–70. doi: 10.1158/2159-8290.Cd-24-0830
54. RodrÍguez E, Schetters STT, and van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol. (2018) 18:204–11. doi: 10.1038/nri.2018.3
55. Krump NA and You J. From merkel cell polyomavirus infection to merkel cell carcinoma oncogenesis. Front Microbiol. (2021) 12:739695. doi: 10.3389/fmicb.2021.739695
56. Zhang WT, Zhu GL, Xu WQ, Zhang W, Wang HZ, Wang YB, et al. Association of PD-1/PD-L1 expression and Epstein–Barr virus infection in patients with invasive breast cancer. Diagn Pathol. (2022) 17:61. doi: 10.1186/s13000-022-01234-3
57. Pal A and Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front Microbiol. (2019) 10:3116. doi: 10.3389/fmicb.2019.03116
58. Sabatini ME and Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer. (2020) 122:306–14. doi: 10.1038/s41416-019-0602-7
59. Spriggs CC and Laimins LA. Human papillomavirus and the DNA damage response: exploiting host repair pathways for viral replication. Viruses. (2017) 9(8):232. doi: 10.3390/v9080232
60. Aggarwal C, Cohen RB, Morrow MP, Kraynyak KA, Sylvester AJ, Knoblock DM, et al. Immunotherapy targeting HPV16/18 generates potent immune responses in HPV-associated head and neck cancer. Clin Cancer Res. (2019) 25:110–24. doi: 10.1158/1078-0432.Ccr-18-1763
61. Katsikis PD, Ishii KJ, and Schliehe C. Challenges in developing personalized neoantigen cancer vaccines. Nat Rev Immunol. (2024) 24:213–27. doi: 10.1038/s41577-023-00937-y
62. Barbier AJ, Jiang AY, Zhang P, Wooster R, and Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. (2022) 40:840–54. doi: 10.1038/s41587-022-01294-2
63. Halima A, Vuong W, and Chan TA. Next-generation sequencing: unraveling genetic mechanisms that shape cancer immunotherapy efficacy. J Clin Invest. (2022) 132(12):e154945. doi: 10.1172/jci154945
64. Ren Y, Yue Y, Li X, Weng S, Xu H, Liu L, et al. Proteogenomics offers a novel avenue in neoantigen identification for cancer immunotherapy. Int Immunopharmacol. (2024) 142:113147. doi: 10.1016/j.intimp.2024.113147
65. Jaffe JD, Berg HC, and Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. (2004) 4:59–77. doi: 10.1002/pmic.200300511
66. Tretter C, de Andrade Krätzig N, Pecoraro M, Lange S, Seifert P, von Frankenberg C, et al. Proteogenomic analysis reveals RNA as a source for tumor-agnostic neoantigen identification. Nat Commun. (2023) 14:4632. doi: 10.1038/s41467-023-39570-7
67. Zhou C, Zhu C, and Liu Q. Toward in silico identification of tumor neoantigens in immunotherapy. Trends Mol Med. (2019) 25:980–92. doi: 10.1016/j.molmed.2019.08.001
68. Laumont CM, Vincent K, Hesnard L, Audemard É, Bonneil É, Laverdure JP, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. (2018) 10(470):eaau5516. doi: 10.1126/scitranslmed.aau5516
69. Zhang X, Qi Y, Zhang Q, and Liu W. Application of mass spectrometry-based MHC immunopeptidome profiling in neoantigen identification for tumor immunotherapy. BioMed Pharmacother. (2019) 120:109542. doi: 10.1016/j.biopha.2019.109542
70. De Mattos-Arruda L, Vazquez M, Finotello F, Lepore R, Porta E, Hundal J, et al. Neoantigen prediction and computational perspectives towards clinical benefit: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. (2020) 31:978–90. doi: 10.1016/j.annonc.2020.05.008
71. Finotello F, Rieder D, Hackl H, and Trajanoski Z. Next-generation computational tools for interrogating cancer immunity. Nat Rev Genet. (2019) 20:724–46. doi: 10.1038/s41576-019-0166-7
72. Pasetto A and Lu YC. Single-cell TCR and transcriptome analysis: an indispensable tool for studying T-cell biology and cancer immunotherapy. Front Immunol. (2021) 12:689091. doi: 10.3389/fimmu.2021.689091
73. Lu YC, Zheng Z, Lowery FJ, Gartner JJ, Prickett TD, Robbins PF, et al. Direct identification of neoantigen-specific TCRs from tumor specimens by high-throughput single-cell sequencing. J Immunother Cancer. (2021) 9(7):e002595. doi: 10.1136/jitc-2021-002595
74. Zhao J, Guo C, Xiong F, Yu J, Ge J, Wang H, et al. Single cell RNA-seq reveals the landscape of tumor and infiltrating immune cells in nasopharyngeal carcinoma. Cancer Lett. (2020) 477:131–43. doi: 10.1016/j.canlet.2020.02.010
75. Erfanian N, Derakhshani A, Nasseri S, Fereidouni M, Baradaran B, Jalili Tabrizi N, et al. Immunotherapy of cancer in single-cell RNA sequencing era: A precision medicine perspective. BioMed Pharmacother. (2022) 146:112558. doi: 10.1016/j.biopha.2021.112558
76. Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE, et al. IPD-IMGT/HLA database. Nucleic Acids Res. (2020) 48:D948–d955. doi: 10.1093/nar/gkz950
77. Okada M, Shimizu K, and Fujii SI. Identification of neoantigens in cancer cells as targets for immunotherapy. Int J Mol Sci. (2022) 23(5):2594. doi: 10.3390/ijms23052594
78. Wichmann G, Herchenhahn C, Boehm A, Mozet C, Hofer M, Fischer M, et al. HLA traits linked to development of head and neck squamous cell carcinoma affect the progression-free survival of patients. Oral Oncol. (2017) 69:115–27. doi: 10.1016/j.oraloncology.2017.04.017
79. Lu CC, Chen JC, Jin YT, Yang HB, Chan SH, and Tsai ST. Genetic susceptibility to nasopharyngeal carcinoma within the HLA-A locus in Taiwanese. Int J Cancer. (2003) 103:745–51. doi: 10.1002/ijc.10861
80. Montesion M, Murugesan K, Jin DX, Sharaf R, Sanchez N, Guria A, et al. Somatic HLA class I loss is a widespread mechanism of immune evasion which refines the use of tumor mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov. (2021) 11:282–92. doi: 10.1158/2159-8290.Cd-20-0672
81. Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. (2015) 520:692–6. doi: 10.1038/nature14426
82. Voogd L, Ruibal P, Ottenhoff THM, and Joosten SA. Antigen presentation by MHC-E: a putative target for vaccination? Trends Immunol. (2022) 43:355–65. doi: 10.1016/j.it.2022.03.002
83. DhatChinamoorthy K, Colbert JD, and Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. (2021) 12:636568. doi: 10.3389/fimmu.2021.636568
84. Wieten L, Mahaweni NM, Voorter CE, Bos GM, and Tilanus MG. Clinical and immunological significance of HLA-E in stem cell transplantation and cancer. Tissue Antigens. (2014) 84:523–35. doi: 10.1111/tan.12478
85. Mei S, Li F, Leier A, Marquez-Lago TT, Giam K, Croft NP, et al. A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction. Brief Bioinform. (2020) 21:1119–35. doi: 10.1093/bib/bbz051
86. Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. (2020) 48:D1057–d1062. doi: 10.1093/nar/gkz874
87. Claeys A, Merseburger P, Staut J, Marchal K, and Van den Eynden J. Benchmark of tools for in silico prediction of MHC class I and class II genotypes from NGS data. BMC Genomics. (2023) 24:247. doi: 10.1186/s12864-023-09351-z
88. Li X, Zhou C, Chen K, Huang B, Liu Q, and Ye H. Benchmarking HLA genotyping and clarifying HLA impact on survival in tumor immunotherapy. Mol Oncol. (2021) 15:1764–82. doi: 10.1002/1878-0261.12895
89. Xie C, Yeo ZX, Wong M, Piper J, Long T, Kirkness EF, et al. Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc Natl Acad Sci U S A. (2017) 114:8059–64. doi: 10.1073/pnas.1707945114
90. Lee H and Kingsford C. Kourami: graph-guided assembly for novel human leukocyte antigen allele discovery. Genome Biol. (2018) 19:16. doi: 10.1186/s13059-018-1388-2
91. Kiyotani K, Mai TH, and Nakamura Y. Comparison of exome-based HLA class I genotyping tools: identification of platform-specific genotyping errors. J Hum Genet. (2017) 62:397–405. doi: 10.1038/jhg.2016.141
92. Johanns TM, Miller CA, Liu CJ, Perrin RJ, Bender D, Kobayashi DK, et al. Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma. Oncoimmunology. (2019) 8:e1561106. doi: 10.1080/2162402x.2018.1561106
93. Mysore V, Cullere X, Settles ML, Ji X, Kattan MW, Desjardins M, et al. Protective heterologous T cell immunity in COVID-19 induced by the trivalent MMR and Tdap vaccine antigens. Med. (2021) 2:1050–71:e1057. doi: 10.1016/j.medj.2021.08.004
94. Cimen Bozkus C, Blazquez AB, Enokida T, and Bhardwaj N. A T-cell-based immunogenicity protocol for evaluating human antigen-specific responses. STAR Protoc. (2021) 2:100758. doi: 10.1016/j.xpro.2021.100758
95. Lowery FJ, Krishna S, Yossef R, Parikh NB, Chatani PD, Zacharakis N, et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science. (2022) 375:877–84. doi: 10.1126/science.abl5447
96. Zhang W, Yin Q, Huang H, Lu J, Qin H, Chen S, et al. Personal neoantigens from patients with NSCLC induce efficient antitumor responses. Front Oncol. (2021) 11628456:628456. doi: 10.3389/fonc.2021.628456
97. Ottensmeier CHH, Delord J-P, Lalanne A, Lantz O, Jamet C, TAVERNARO A, et al. Safety and immunogenicity of TG4050: A personalized cancer vaccine in head and neck carcinoma. J Clin Oncol. (2023) 41:6082–2. doi: 10.1200/JCO.2023.41.16_suppl.6082
98. Morse MA, Gwin WR 3rd, and Mitchell DA. Vaccine therapies for cancer: then and now. Target Oncol. (2021) 16:121–52. doi: 10.1007/s11523-020-00788-w
99. Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong F, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. (2019) 18:128. doi: 10.1186/s12943-019-1055-6
100. Chen S, Pounraj S, Sivakumaran N, Kakkanat A, Sam G, Kabir MT, et al. Precision-engineering of subunit vaccine particles for prevention of infectious diseases. Front Immunol. (2023) 14:1131057. doi: 10.3389/fimmu.2023.1131057
101. Jahanafrooz Z, Baradaran B, Mosafer J, Hashemzaei M, Rezaei T, Mokhtarzadeh A, et al. Comparison of DNA and mRNA vaccines against cancer. Drug Discov Today. (2020) 25:552–60. doi: 10.1016/j.drudis.2019.12.003
102. Fritah H, Rovelli R, Chiang CL, and Kandalaft LE. The current clinical landscape of personalized cancer vaccines. Cancer Treat Rev. (2022) 106:102383. doi: 10.1016/j.ctrv.2022.102383
103. Conforti A, Salvatori E, Lione L, Compagnone M, Pinto E, Shorrock C, et al. Linear DNA amplicons as a novel cancer vaccine strategy. J Exp Clin Cancer Res. (2022) 41:195. doi: 10.1186/s13046-022-02402-5
104. Ledesma-Feliciano C, Chapman R, Hooper JW, Elma K, Zehrung D, Brennan MB, et al. Improved DNA vaccine delivery with needle-free injection systems. Vaccines (Basel). (2023) 11(2):280. doi: 10.3390/vaccines11020280
105. Cheng R, Xu Z, Luo M, Wang P, Cao H, Jin X, et al. Identification of alternative splicing-derived cancer neoantigens for mRNA vaccine development. Brief Bioinform. (2022) 23(2):bbab553. doi: 10.1093/bib/bbab553
106. Yuan Y, Gao F, Chang Y, Zhao Q, and He X. Advances of mRNA vaccine in tumor: a maze of opportunities and challenges. biomark Res. (2023) 11:6. doi: 10.1186/s40364-023-00449-w
107. Lang F, Schrörs B, Löwer M, Türeci Ö, and Sahin U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov. (2022) 21:261–82. doi: 10.1038/s41573-021-00387-y
108. Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. (2017) 547:222–6. doi: 10.1038/nature23003
109. Khattak A, Weber JS, Meniawy T, Taylor MH, Ansstas G, Kim KB, et al. Distant metastasis-free survival results from the randomized, phase 2 mRNA-4157-P201/KEYNOTE-942 trial. J Clin Oncol. (2023) 41:LBA9503–LBA9503. doi: 10.1200/JCO.2023.41.17_suppl.LBA9503
110. Wang B, Peng X, Li J, Wang Y, Chen L, Wu M, et al. Personalized mRNA vaccine combined with PD-1 inhibitor therapy in a patient with advanced esophageal squamous cell carcinoma. Am J Cancer Res. (2024) 14:3896–904. doi: 10.62347/nvfb3780
111. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. (2019) 76:96–109.e109. doi: 10.1016/j.molcel.2019.07.016
112. Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. (2018) 13:312–21. doi: 10.1016/j.omtn.2018.09.010
113. Wang F, Cai G, Wang Y, Zhuang Q, Cai Z, Li Y, et al. Circular RNA-based neoantigen vaccine for hepatocellular carcinoma immunotherapy. MedComm. (2020) 5(8):e667. doi: 10.1002/mco2.667
114. Chen MY, Zhang F, Goedegebuure SP, and Gillanders WE. Dendritic cell subsets and implications for cancer immunotherapy. Front Immunol. (2024) 15:1393451. doi: 10.3389/fimmu.2024.1393451
115. Blass E and Ott PA. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat Rev Clin Oncol. (2021) 18:215–29. doi: 10.1038/s41571-020-00460-2
116. Delord J-P, Block MS, Ottensmeier C, Colon-Otero G, Le Tourneau C, Lalanne A, et al. Phase 1 studies of personalized neoantigen vaccine TG4050 in ovarian carcinoma (OC) and head and neck squamous cell carcinoma (HNSCC). J Clin Oncol. (2022) 40:2637–7. doi: 10.1200/JCO.2022.40.16_suppl.2637
117. Khong H and Overwijk WW. Adjuvants for peptide-based cancer vaccines. J Immunother Cancer. (2016) 456:56. doi: 10.1186/s40425-016-0160-y
118. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. (2017) 547:217–21. doi: 10.1038/nature22991
119. Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. (2019) 565:240–5. doi: 10.1038/s41586-018-0810-y
120. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, and McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. (2015) 33:169–200. doi: 10.1146/annurev-immunol-032414-112334
121. Waki K, Ozawa M, and Yamada A. Suppression of high mobility group box 1 in B16F10 tumor does not inhibit the induction of neoantigen-specific T cells. Cancer Sci. (2022) 113:4082–91. doi: 10.1111/cas.15563
122. Bijker MS, van den Eeden SJ, Franken KL, Melief CJ, Offringa R, and van der Burg SH. CD8+ CTL priming by exact peptide epitopes in incomplete Freund’s adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J Immunol. (2007) 179:5033–40. doi: 10.4049/jimmunol.179.8.5033
123. Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. (2015) 348:803–8. doi: 10.1126/science.aaa3828
124. Bol KF, Schreibelt G, Gerritsen WR, de Vries IJ, and Figdor CG. Dendritic cell-based immunotherapy: state of the art and beyond. Clin Cancer Res. (2016) 22:1897–906. doi: 10.1158/1078-0432.Ccr-15-1399
125. Chen Y, Shi H, Hu Y, Yuan Y, Yang Y, Xu H, et al. Abstract 4108: Safety and feasibility of personalized neoantigen DC vaccines in postoperative esophagus squamous cell carcinoma: A Phase 1b trial. Cancer Res. (2024) 84:4108–8. doi: 10.1158/1538-7445.Am2024-4108
126. Delamarre L, Mellman I, and Yadav M. Cancer immunotherapy. Neo approaches to cancer vaccines. Science. (2015) 348:760–1. doi: 10.1126/science.aab3465
127. Tahara M, Greil R, Rischin D, Harrington KJ, Burtness B, Castro GD, et al. 659MO Pembrolizumab with or without chemotherapy for first-line treatment of recurrent/metastatic (R/M) head and neck squamous cell carcinoma (HNSCC): 5-year results from KEYNOTE-048. Ann Oncol. (2022) 33:S844. doi: 10.1016/j.annonc.2022.07.783. S844.
128. Keam B, Machiels JP, Kim HR, Licitra L, Golusinski W, Gregoire V, et al. Pan-Asian adaptation of the EHNS-ESMO-ESTRO Clinical Practice Guidelines for the diagnosis, treatment and follow-up of patients with squamous cell carcinoma of the head and neck. ESMO Open. (2021) 6:100309. doi: 10.1016/j.esmoop.2021.100309
129. Wang FH, Zhang XT, Li YF, Tang L, Qu XJ, Ying JE, et al. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021. Cancer Commun (Lond). (2021) 41:747–95. doi: 10.1002/cac2.12193
130. de Ruiter EJ, Mulder FJ, Koomen BM, Speel EJ, van den Hout M, de Roest RH, et al. Comparison of three PD-L1 immunohistochemical assays in head and neck squamous cell carcinoma (HNSCC). Mod Pathol. (2021) 34:1125–32. doi: 10.1038/s41379-020-0644-7
131. Song Y, Zhang B, Xin D, Kou X, Tan Z, Zhang S, et al. First-line serplulimab or placebo plus chemotherapy in PD-L1-positive esophageal squamous cell carcinoma: a randomized, double-blind phase 3 trial. Nat Med. (2023) 29:473–82. doi: 10.1038/s41591-022-02179-2
132. Black CM, Hanna GJ, Wang L, Ramakrishnan K, Goto D, Turzhitsky V, et al. Real-world treatment patterns and outcomes among individuals receiving first-line pembrolizumab therapy for recurrent/metastatic head and neck squamous cell carcinoma. Front Oncol. (2023) 13:1160144. doi: 10.3389/fonc.2023.1160144
133. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. (2017) 9:34. doi: 10.1186/s13073-017-0424-2
134. Yilmaz E, Ismaila N, Bauman JE, Dabney R, Gan G, Jordan R, et al. Immunotherapy and biomarker testing in recurrent and metastatic head and neck cancers: ASCO guideline. J Clin Oncol. (2023) 41:1132–46. doi: 10.1200/jco.22.02328
135. Franken A, Bila M, Mechels A, Kint S, Van Dessel J, Pomella V, et al. CD4(+) T cell activation distinguishes response to anti-PD-L1+anti-CTLA4 therapy from anti-PD-L1 monotherapy. Immunity. (2024) 57:541–558.e547. doi: 10.1016/j.immuni.2024.02.007
136. Mohsin SF. Vaccine a promising immunotherapy option for head and neck cancer patients. Pak J Med Sci. (2024) 40:1578–83. doi: 10.12669/pjms.40.7.8791
137. Zhang H, Dai Z, Wu W, Wang Z, Zhang N, Zhang L, et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res. (2021) 40:184. doi: 10.1186/s13046-021-01987-7
139. Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. (2013) 5:200ra116. doi: 10.1126/scitranslmed.3006504
140. Topalian SL, Hodi SF, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. (2012) 366:2443–54. doi: 10.1056/NEJMoa1200690
141. Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. (2019) 565:234–9. doi: 10.1038/s41586-018-0792-9
142. Sidhom JW, Oliveira G, Ross- MacDonald P, Wind-Rotolo M, Wu CJ, et al. Deep learning reveals predictive sequence concepts within immune repertoires to immunotherapy. Sci Adv. (2022) 8:eabq5089. doi: 10.1126/sciadv.abq5089
143. Georgiou K, Chen L, Berglund M, Ren M, de Miranda NFFC, Lisboa S, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. (2016) 127:3026–34. doi: 10.1182/blood-2015-12-686550
144. Zhang T, Liu H, Jiao L, Zhang Z, He J, Li L, et al. Genetic characteristics involving the PD-1/PD-L1/L2 and CD73/A2aR axes and the immunosuppressive microenvironment in DLBCL. J Immunother Cancer. (2022) 10(4):e004114. doi: 10.1136/jitc-2021-004114
145. Xu X, Zhang Y, Lu Y, Zhang X, Zhao C, Wang J, et al. CD58 alterations govern antitumor immune responses by inducing PDL1 and IDO in diffuse large B-cell lymphoma. Cancer Res. (2024) 84:2123–40. doi: 10.1158/0008-5472.Can-23-2874
146. Ho P, Melms JC, Rogava M, Frangieh CJ, Poźniak J, Shah SB, et al. The CD58-CD2 axis is co-regulated with PD-L1 via CMTM6 and shapes anti-tumor immunity. Cancer Cell. (2023) 41:1207–1221.e1212. doi: 10.1016/j.ccell.2023.05.014
147. Melief CJ, van Hall T, Arens R, Ossendorp F, and van der Burg SH. Therapeutic cancer vaccines. J Clin Invest. (2015) 125:3401–12. doi: 10.1172/jci80009
148. Dai W, Zheng H, Cheung AK, and Lung ML. Genetic and epigenetic landscape of nasopharyngeal carcinoma. Chin Clin Oncol. (2016) 5:16. doi: 10.21037/cco.2016.03.06
149. Ali M, Foldvari Z, Giannakopoulou E, Böschen M-L, Strønen E, Yang W, et al. Induction of neoantigen-reactive T cells from healthy donors. Nat Protoc. (2019) 14:1926–43. doi: 10.1038/s41596-019-0170-6
150. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. (2017) 16:130. doi: 10.1186/s12943-017-0699-3
151. Hundal J, Kiwala S, McMichael J, Miller CA, Xia H, Wollam AT, et al. pVACtools: A computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res. (2020) 8:409–20. doi: 10.1158/2326-6066.Cir-19-0401
152. Zhang J, Mardis ER, and Maher CA. INTEGRATE-neo: a pipeline for personalized gene fusion neoantigen discovery. Bioinformatics. (2017) 33:555–7. doi: 10.1093/bioinformatics/btw674
153. Rubinsteyn A, Kodysh J, Hodes I, Mondet S, Aksoy BA, Finnigan JP, et al. Computational pipeline for the PGV-001 neoantigen vaccine trial. Front Immunol. (2017) 8:1807. doi: 10.3389/fimmu.2017.01807
154. Smith CC, Beckermann KE, Bortone DS, De Cubas AA, Bixby LM, Lee SJ, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. (2018) 128:4804–20. doi: 10.1172/jci121476
155. Zhou Z, Lyu X, Wu J, Yang X, Wu S, Zhou J, et al. TSNAD: an integrated software for cancer somatic mutation and tumour-specific neoantigen detection. R Soc Open Sci. (2017) 4:170050. doi: 10.1098/rsos.170050
Keywords: head and neck squamous cell carcinoma (HNSCC), neoantigen, personalized neoantigen vaccines, immune checkpoint inhibitors (ICIs), combination therapy
Citation: Jiang M, Li J, Wei J, Yang X and Wang W (2025) Advances in neoantigen-based immunotherapy for head and neck squamous cell carcinoma: a comprehensive review. Front. Oncol. 15:1593048. doi: 10.3389/fonc.2025.1593048
Received: 13 March 2025; Accepted: 17 April 2025;
Published: 15 May 2025.
Edited by:
Xianhuo Wang, Tianjin Medical University Cancer Institute and Hospital, ChinaReviewed by:
Shigenori Goto, Seta Clinic Tokyo, JapanSyed Fareed Mohsin, Qassim University, Saudi Arabia
Copyright © 2025 Jiang, Li, Wei, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuerong Yang, eHJ5YW5nQHNkYXUuZWR1LmNu; Weiqi Wang, MTUwMjkwNjM0NzhAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Manzhu Jiang1†
Manzhu Jiang1† Jianhua Wei
Jianhua Wei Xuerong Yang
Xuerong Yang