- 1Immunology, Inflammation and Metabolism, Division of Translational Medicine, SIDRA Medical and Research Center, Doha, Qatar
- 2Division of Neonatology, SIDRA Medical and Research Center, Doha, Qatar
- 3Weill Cornell Medical College, New York, NY, United States
- 4Pediatric Nephrology and Hypertension, SIDRA Medical and Research Center, Doha, Qatar
- 5Medical University of South Carolina, Charleston, SC, United States
The surfaces of the human body are heavily populated by a highly diverse microbial ecosystem termed the microbiota. The largest and richest among these highly heterogeneous populations of microbes is the gut microbiota. The collection of microbes and their genes, called the microbiome, has been studied intensely through the past few years using novel metagenomics, metatranscriptomics, and metabolomics approaches. This has enhanced our understanding of how the microbiome affects our metabolic, immunologic, neurologic, and endocrine homeostasis. Hypertension is a leading cause of cardiovascular disease worldwide; it contributes to stroke, heart disease, kidney failure, premature death, and disability. Recently, studies in humans and animals have shown that alterations in microbiota and its metabolites are associated with hypertension and atherosclerosis. In this review, we compile the recent findings and hypotheses describing the interplay between the microbiome and blood pressure, and we highlight some prospects by which utilization of microbiome-related techniques may be incorporated to better understand the pathophysiology and treatment of hypertension.
Background
The human microbiota is a mixed community of microorganisms composed of bacteria, viruses, archaea, and eukaryotic microbes that coinhabit the human body surfaces (1). The collection of those microbes and their genes is named the human microbiome (2).
Multiple studies have shown that each body site is characterized by unique ecological communities of microbial species (1) and each person has a unique microbiome (3). Those interpersonal variabilities are likely related to differences in our genetic background, origin, geographical location, age, life style, diet, and early exposure to various microbes, as well as exposure to antibiotics or probiotics (3). The microbiome composition is also affected by early life events; including delivery mode, gestational age, hospitalization, and the method of feeding (4).
Hypertension is a global public health problem and contributes to the burden of heart disease, stroke, kidney failure, premature death, and disability. It is considered the most prevalent modifiable cardiovascular disease (CVD) risk factor. High blood pressure affects 1.13 billion people worldwide. In the US, 75 million American adults (29%) suffer from high blood pressure, which is around one out of three adults (5). It is also estimated that more than 3% of children suffer from hypertension; this number increases in obese children, since the prevalence of primary hypertension rises progressively with increases in BMI percentile from less than the fifth percentile (2%) to more than the 95th percentile (11%) (6). Globally, 42 million preschool children were overweight in 2013. The prevalence of obesity in the US is about 17%; it affects about 12.7 million children and adolescents (7, 8). It is important to mention that while obesity is associated with increased prevalence of hypertension, not all obese children are hypertensive.
Recently, multiple animal and human studies have examined the relationship between the gut and the oral microbiome and blood pressure (9–11). These studies aimed to explore different hypotheses linking the microbiota and its metabolites to blood pressure. Here, we provide an overview of the literature and discuss the proposed mechanisms. We also discuss potential microbiota-altering therapies and lifestyle modifications and their effect on blood pressure.
The Microbiome in Health and Disease
Our microbiota is highly dynamic and continuously changing. This is in part a reflection of age-related changes like growth and development, where the highest variation takes place during childhood but later decreases with age (12). Newborns’ and infants’ microbiota differentiates and becomes distinct and site-specific as they grow older (13). The microbiota in infants less than 6 months old is different from that of older infants, where by the age of 3 years, a child’s microbiome is highly similar to that of an adult and is considered relatively stable (14, 15). However, the microbiome composition remains subject to changes related to any disease, change of diet, use of antibiotics, and in response to major life events like pregnancy and puberty (16–18).
Microbiota composition is also determined by the physical characteristics and chemical properties of the site that is being colonized (19). Therefore, the primary determinant of community composition is the anatomical location: within the same habitat, interpersonal variation is significant (20) and is more complex than the temporal variability observed in multiple sites within the same individual (21). Those site-specific signatures of the microbiome help elucidate the many changes associated with health and disease.
Tools Used to Study the Microbiome
Recent breakthroughs in high-throughput Next Generation Sequencing techniques, summarized in Figure 1, have leveraged our understanding of the composition and the function of the microbiome and have substantially advanced our knowledge on the crucial role of the microbiome in maintaining host physiology and homeostasis (22). Those techniques range from sequencing of the 16S rRNA-encoding genes, used to characterize the microbial phylogenetic composition of a sample collected from a specific body site, to the shotgun metagenomic approaches, used to identify all the genomes of microbes coexisting in the same site along with their biological functions (23–25). In addition to genomic sequencing-based analysis, other methods have been developed to study the microbial transcriptome, proteome, and metabolome, as they provide additional information at successive levels of microbial physiology (26). Metabolomics aim to study the metabolic functions by which the microbiota contributes to the human physiology; those functions include energy harvest, bile acid transformations, choline transformation, and the production of short-chain fatty acids (SCFAs), vitamins, and amino acids (27).
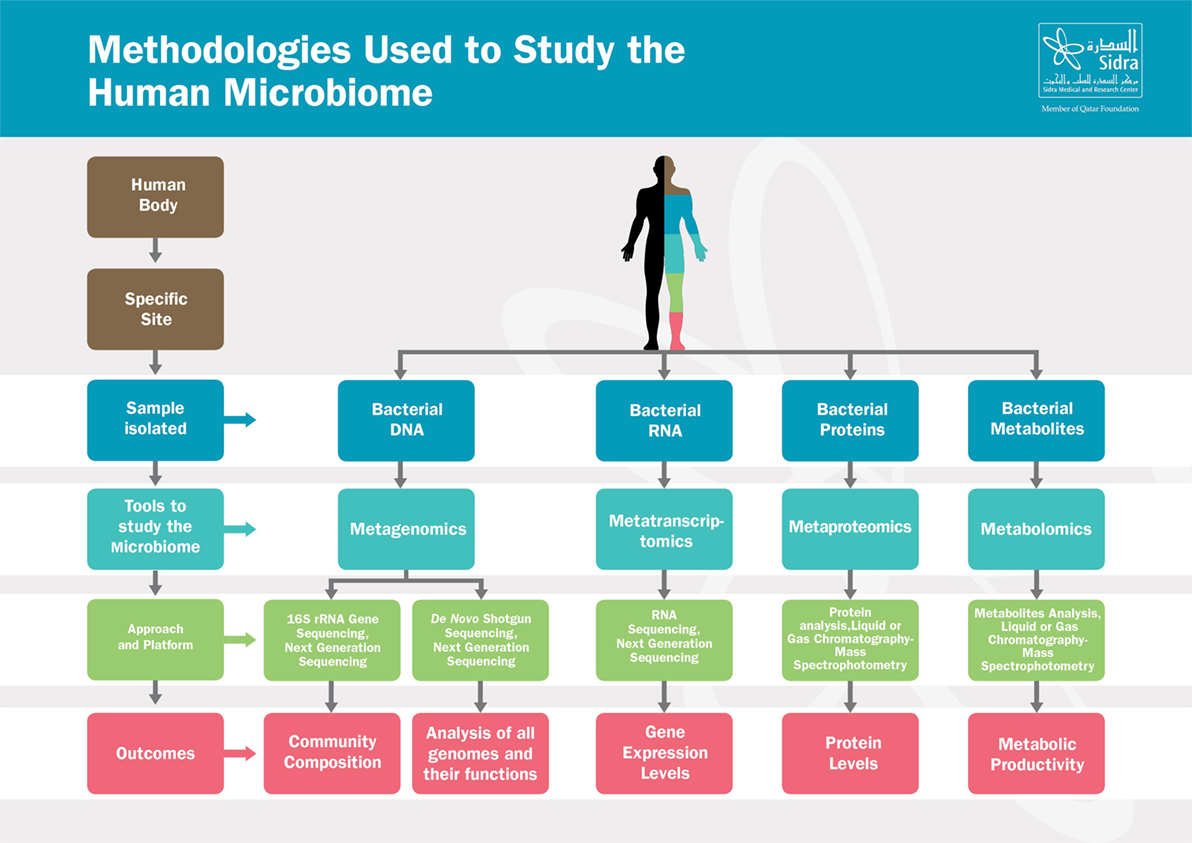
Figure 1. Methodologies used to study the human microbiome. The human body harbors large amounts of microbes that vary from one site to another. From each specific site, microbes can be studied in various ways depending on the samples isolated. Those tools range from metagenomics, metatranscriptomics to metaproteomics and metabolomics. Based on the approach and platform used, results include composition of the microbial communities, microbial genes function, gene expression or proteins levels, and metabolites activities.
Those tools have tremendously contributed to our current knowledge about the microbiota and their metabolites (22, 27). In recent years, the microbiome was shown to constitute a unique “fingerprint marking” that may play a role in interindividual phenotypic variation in disease presentation, prognosis, progression, and even response to treatment (15, 20).
The Gut Microbiome
Colonization of the human gut with a wide variety of microbes takes place just before birth as evident from the diverse microbial composition of the meconium (28). Maternal microbiota contributes to the formation of the first microbial inoculum, and then soon after birth, the microbial diversity increases and converges toward an adult-like microbiota by the end of the first 3–5 years of life (29). The ecosystem of an adult human gut contains a complex array of microbes with more than 100 trillion microbial cells and more than 1,000 bacterial species (30). The composition of the gut microbiota is highly variable between subjects, as each subject harbors a unique set of microbial species, which is in general highly stable over time in healthy individuals (31). A healthy human adult gut is dominated by the Gram-positive Firmicutes and the Gram-negative Bacteroidetes (31). Most nutrients consumed through a diet are processed by an array of various human enzymes before being absorbed by the small intestine; however, the gut microbiota contributes to the metabolism of dietary fibers that are not usually digested by those enzymes (32). In the large intestine, a group of microbes including clostridial clusters IV, XIVa, Lactobacillus, and Actinobacteria (Bifidobacterium spp.) contribute to the fermentation of dietary plant polysaccharides or fibers, indigestible oligosaccharides, non-digested proteins, and intestinal mucin in order to produce SCFAs (acetate, propionate, and butyrate) (33). In addition to its role in food digestion, the gut microbiota plays a vital role in inhibiting pathogen-invasion by creating colonization resistance, it also contributes to the education and stimulation of the immune system, maintenance of the intestinal epithelial homeostasis and integrity, synthesis of vitamin B and vitamin K, as well as the enhancement of the motility and function of the gastrointestinal tract (GIT) (31).
The Oral Microbiome
The oral cavity is considered one of the most highly dynamic ecosystems in the human body (34–36). 16S rRNA gene sequencing estimated 50–100 billion bacteria in the mouth, comprised of nearly 700 identified bacterial species (36–38). Up to 80% of oral bacteria are dominated by about 200 species of Firmicutes and Proteobacteria, along with Bacteroidetes, Actinobacteria, and Fusobacteria totaling upwards of 95% of all identified oral microbiota (36–38). Further diversity exists between the varied niches of the mouth (34), such as the tooth surface, gingiva, hard and soft palate, and even regionally on the tongue (34). Within this complexity, an ever-growing number of specific microbial taxa have been associated with both oral and systemic diseases (39). For instance, saccharolytic (sugar metabolizing) bacteria, such as Streptococcus and Lactobacillus, have been associated with dental caries, while proteolytic (protein metabolizing) bacteria, such as Prevotella and Porphyromonas, have been associated with periodontitis and halitosis (40). Presence of Porphyromonas gingivalis is shown to be associated with atherosclerosis (41), smoking (42), and several cancers (43).
Microbial Dysbiosis and Disease
Dysbiosis is defined as the change in the composition and structure of the human microbiota of a given site, a change that may help explain why some individuals are more likely to develop certain diseases or develop a more severe form of the illness (44).
Although the relationship between microbial composition and stability to disease predisposition is not a cause-and-effect relationship, the microbiome is a contributor in many disease states, a link that has been previously overlooked. In fact, changes in the microbiome are increasingly linked to the development of several non-communicable diseases, including diabetes (45, 46), obesity (47), CVD (48, 49), cancer (50, 51), inflammatory bowel disease (IBD) (52, 53), asthma (54), and others. Recently, researchers have examined the relationship between kidney disease and the human microbiome, summarized in Ref. (55). Studies have shown a bidirectional relationship between chronic kidney disease and the gut microbiome; where microbiota-derived metabolites contribute to the progression of CKD and the state of chronic kidney disease and inflammation contributes to changes in the diversity and richness of the microbiota. Other studies showed differences in IgA disease progression and the gut microbiota composition, SCFA (derived from the microbiota) to modulate renal dysfunction in states of acute kidney injury via their anti-inflammatory properties, and kidney transplantation and immunosuppressive medications to significantly impact the gut microbiota composition.
It is, therefore, essential to understand the interface between the microbes and the host in any disease condition, as this may help uncover possible disease etiologies and pathogenesis. It may also be possible to use any novel microbial factors or host-related inflammatory markers as diagnostic and therapeutic targets for prediction, prevention, and treatment of some common diseases. Deciphering the possible interindividual variations in the microbial composition in various body sites and identifying the changes in site-specific microbiota composition during onset or progression of various diseases will help to pave the way toward developing microbiota-driven personalized medicine.
While large-scale studies addressed the role of the microbiome in some conditions including cancer, IBD, obesity, and diabetes, only fewer studies describing the role of the microbiome in regulating blood pressure have been published (10, 11, 56). Those studies suggested that hypertension is directly and/or indirectly linked to microbial dysbiosis, and that some microbial metabolites contribute to the regulation of blood pressure (10, 11, 56). Therefore, understanding the nature of hypertension-related microbial aberrations in various body sites may enable us to better understand the pathophysiology of high blood pressure and possibly develop personalized microbiome-based diagnostics for individuals at risk.
In this review, we discuss potential mechanisms by which the microbiota contributes to our blood pressure regulation and describe the link between dysbiosis and hypertension.
Hypertension and the Microbiome
Blood pressure regulation is complex. Multiple physiological systems interact, influenced by the environment and genes, to maintain blood pressure (57). These include but are not limited to: the renin–angiotensin–aldosterone system, the sympathetic nervous system (SNS), the nitrate–nitrite–nitric oxide signaling pathway (NO), uric acid, endothelin, the vasopressin system, natriuretic peptides, vasodilator peptides, the tissue kallikrein–kinin system, the immune system, the adipose tissue, and adipokines (58).
Recently, multiple animal and human studies have examined the relationship between the oral and gut microbiome and blood pressure; they demonstrated a significant decrease in microbial richness and diversity in the presence of hypertension. In addition, studies have demonstrated an altered microbial composition and modified metabolite profiles, suggesting a role for microbial dysbiosis and microbial metabolites in hypertension (11). In a rat model of hypertension, the number of cecal “good bacteria” from the phylum Bacteroidetes is reduced, which is accompanied by a proportional increase in the number of “bad bacteria” from the phylum Firmicutes (11). Studies have also shown that transplant of cecal microbial content from donor hypertensive animals can reproduce hypertension in previously normotensive recipient animals (56). A third set of studies demonstrated a beneficial effect for microbial mass reduction using antibiotics on blood pressure (11). Furthermore, absence of gut microbiota was found to protect mice from angiotensin II (AngII)-induced arterial hypertension, vascular dysfunction, and hypertension-induced end-organ damage (59).
The relationship between the microbiota and blood pressure is a complex one. Researchers have identified multiple possible hypotheses to link dysbiosis and hypertension. Many of which are indirect links that contribute to the metabolic syndrome and the overall increased cardiovascular risk. It is also important to point out that most of the studies were conducted in animal models, and many examined newly proposed hypotheses which yielded results that have yet to be reproduced. Some hypotheses focused on the association between microbial species in the microbiota and their relationship to blood pressure, whereas other hypotheses examined the role of dysbiosis in the pathogenesis (e.g., increased SNS activity), sustenance (e.g., inflammation), and worsening/progression of hypertension (e.g., endothelial dysfunction and vascular remodeling). Here, we provide an overview of the proposed hypotheses linking the microbiota to blood pressure, Figure 2.
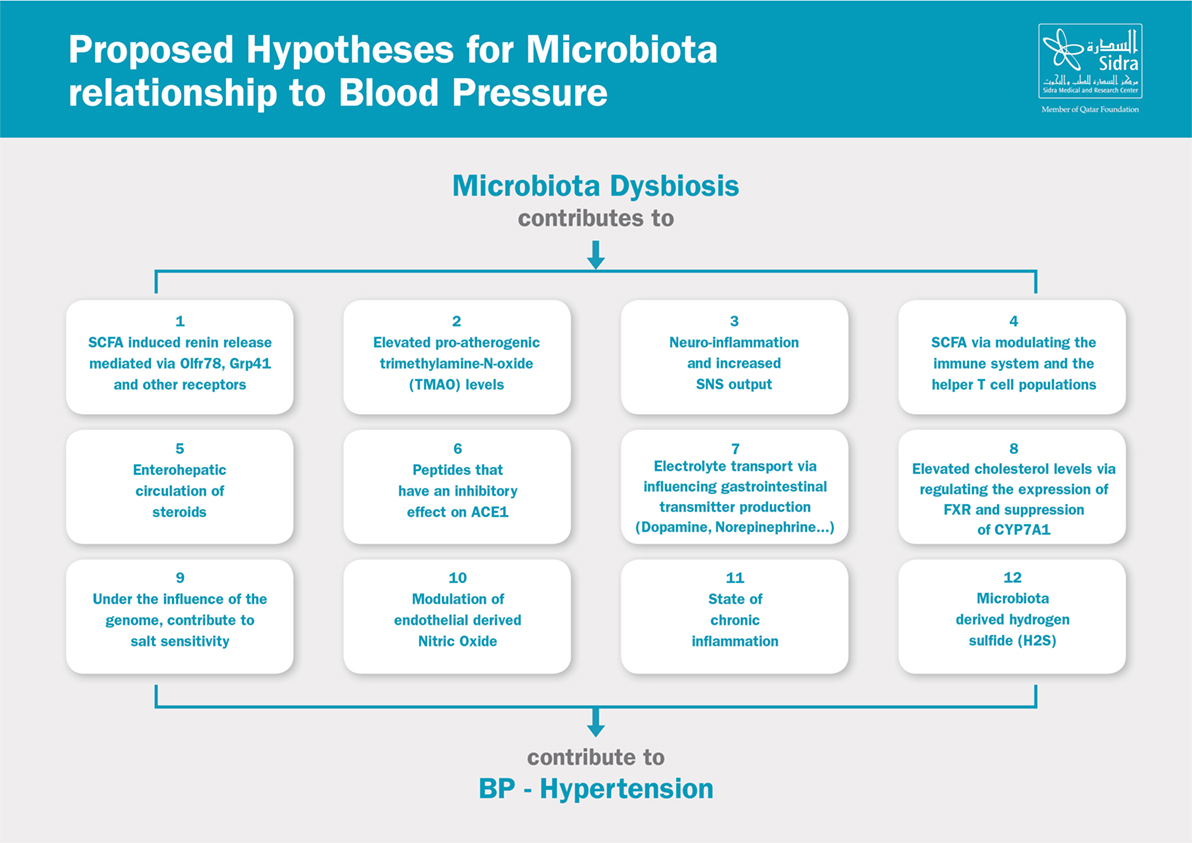
Figure 2. Proposed hypotheses for microbiota relationship to blood pressure. SCFA, short-chain fatty acids; SNS, sympathetic nervous system; ACE, angiotensin-converting enzyme; FXR, farnesoid X receptor.
Microbial SCFA Metabolites Regulate Blood Pressure via Olfactory Receptors
Researchers have described a less rich and diverse microbiota in hypertensive compared to control subjects (11). SCFAs are products resulting from fermentation of various nutrients by the gut microbiota and are later absorbed into the blood stream (60). Members of the olfactory signaling pathway are expressed in the human kidneys. Olfr78 is an olfactory receptor that mediates renin secretion after stimulation by SCFAs (61). Other SCFA receptors including Gpr41 and Gpr43 (also called free fatty acid receptor 3 or FFAR3 and FFAR2, respectively) are also expressed in the renal vasculature. Propionate administration to Gpr41-deficient mice induced blood pressure elevation, suggesting that Gpr41 is needed to counterbalance the pressor response to SCFA (10).
Increased Risk of Atherosclerosis from Microbiota-Generated Trimethylamine-N-Oxide (TMAO)
Another indirect connection between the gut microbiome composition and hypertension derives from the role of the gut microbes in the metabolism of choline and phosphatidylcholine to trimethylamine (TMA), which is further metabolized to the pro-atherogenic species, TMAO (62). Koeth and colleagues showed that the metabolism of dietary l-carnitine, a TMA abundant in red meat, by the intestinal microbiota results in the production of TMAO and accelerates the development and progression of atherosclerosis in mice (62). Recently, a study in a healthy human volunteer showed that microbiota in the small intestine generated the phosphatidylcholine breakdown product TMA (63). The resulting TMAO was suppressed by topical-acting antibiotics (63). It is important to point out that this link is with atherosclerosis and not directly to hypertension.
Microbiota-Induced Neuroinflammation and Increased Sympathetic Activity
Vagal nerve stimulation and blocking sympathetic derive prevent breakdown of the intestinal lumen–blood barrier and enhance epithelia cell barrier function (64, 65). Santisteban and colleagues hypothesized that hypertensive stimuli (such as Ang II, salt, and stress) trigger autonomic neural pathways resulting in increases in sympathetic and dampening of the parasympathetic activities which in turn contribute to the overall increase in blood pressure (66). Increased sympathetic nervous activity to the gut could result in increased gut permeability, gut inflammation, and dysbiosis, leading to an imbalance in the microbial-derived metabolites in the plasma, possibly contributing to chronic inflammation and sustained hypertension.
Modulation of Blood Pressure via the Effects of the Microbiota on the Immune System
Researchers have proposed a brain–gut–bone marrow axis in which sympathetic activity to the bone marrow induces mobilization of hematopoietic stem cells (66). In this hypothesis, hematopoietic stem cells may migrate to the brain or to the gut and contribute to local inflammation and immune responses. This may further increase the sympathetic activity and contribute to blood pressure elevation. On the other hand, SCFAs, such as acetate and butyrate, have been shown to have anti-inflammatory effects on myeloid and intestinal epithelial cells (67). Recently, Kim and colleagues reported marked decreases in microbial richness and diversity in hypertensive patients and also observed marked differences in circulating inflammatory cells in hypertensive individuals compared to controls (68); T-helper 17 cells were particularly relevant because activation of these cells is regulated by gut-intrinsic mechanisms, and their increase may be a result of dysbiosis in hypertension (68). In germ-free mice, the absence of gut microbiota seems to protect the animals from AngII-induced arterial hypertension, vascular dysfunction, and hypertension-induced end-organ damage. This protection appears to be mediated by inhibiting the accumulation of the inflammatory myelomonocytic cells in the vasculature and altered cytokine signaling (59).
Role of the Microbiota in the Enterohepatic Circulation of Steroids
Another intriguing hypothesis involves the role of the gut microbiota in the enterohepatic circulation of steroids. Using antibiotic therapy, the bacterial flora of rats was modified to interrupt the enterohepatic circulation of steroids excreted in bile. Antibiotic and corticosterone were administered simultaneously to these animals. After 5 days, rats receiving both steroids and antibiotics had an average elevation of 9.2 mmHg in their blood pressure compared with 24.6 mmHg in rats given steroid alone. These findings are consistent with the possibility that metabolites of steroids, when reabsorbed in the enterohepatic circulation, contribute to the physiological response to exogenous steroids (69). Further studies are needed to examine the role of gut-derived steroids.
Inhibition of Angiotensin 1-Converting Enzyme
Nakamura and colleagues (70) reported a significant decrease in the systolic blood pressure in rats fed sour milk (contains the two tripeptides Val–Pro–Pro and Ile–Pro–Pro). Research on the health effects of pasteurized sour milk, which is fermented by a starter culture containing Lactobacillus helveticus as the predominant microorganism, indicated that antihypertensive effects and angiotensin-I converting enzyme inhibitory peptides are present in sour milk (71, 72). On the other hand, researchers have shown ACE2 has a RAS-independent function, regulating intestinal amino acid homeostasis, expression of antimicrobial peptides, and the ecology of the gut microbiome (73).
Electrolyte Transport via Influencing Gastrointestinal Transmitter Production
The gut microbiota can influence the ability of the gastric and intestinal enterochromaffin cells to produce serotonin, dopamine, and norepinephrine. These transmitters have been found to influence Na–K ATPase and electrolyte transport in the intestine (74). It is still unclear whether enterochromaffin cell-derived transmitter effects are clinically significant in blood pressure regulation. Further studies are needed to examine the role of gastrointestinal transmitter production on blood pressure regulation.
Elevated Cholesterol Levels via Regulating the Expression of RXR and Suppression of CYP7A1
Cholesterol is the precursor to bile acids synthesis in the liver. Bile acids are further metabolized by the gut microbiota into secondary bile acids. In the ileum and liver, nuclear farnesoid X receptor (FXR) plays a key role in the bile acid synthesis; when activated, it exerts negative feedback to control bile acid synthesis (75). Gut microbiota may indirectly play a role in the increased risk of atherosclerotic disease by increasing cholesterol levels; this may be attributed to its role in reducing the levels of tauro-beta-muricholic acid, a FXR antagonist, as well as by suppressing the rate-limiting enzyme CYP7A1 in bile acid synthesis from cholesterol (75). It is important to point out that this link is with atherosclerosis and increased cholesterol levels and not directly to hypertension.
Under the Influence of the Host Genome, Microbiota May Contribute to Salt Sensitivity
Mell and colleagues hypothesized that the interaction between the host and the gut microbiota influences the development of salt-sensitive hypertension (76). They reported differences in the gut (cecal) microbiota composition between the salt-sensitive (S) and the salt-resistant (R) Dahl rats. After a single bolus of R rat cecal content to S rats, they showed exacerbated hypertension in high salt-fed S rats, with systolic blood pressure to be consistently and significantly elevated during the rest of the recipient rat life, which also had a shorter lifespan. They speculated that this effect may be mediated via SCFAs, as both acetate and hetanoate were higher in the R to S transfer group (76).
Modulation of Endothelial-Derived Nitric Oxide (NO)
It is also worth mentioning that the nitrate–nitrite–nitric oxide signaling pathway involved in the pathogenesis of hypertension is highly affected by microbial diversity through the formation of nitrite, NO, and other bioactive nitrogen oxides (77). NO is an endogenously produced, lipophilic, and diffusible molecule that exerts a diverse array of critical autocrine and paracrine signaling activities. NO acts directly on smooth muscle cells to promote relaxation and inhibits both platelet function and vascular smooth muscle cell proliferation and migration (78). Formation of nitrite and propagation of its downstream NO-signaling effects depend on the oral bacterial reduction of inorganic nitrate by a set of bacterial nitrate reductase enzymes that are largely absent from the human genome (77, 79, 80). Studies in Sprague-Dawley rats (77) and normal human volunteers (81) showed that depletion of oral bacterial nitrate reductases by chlorhexidine mouthwash correlated with a 90% decrease in oral nitrite levels in humans, along with a 25% decrease in plasma levels (p = 0.001), and 2–3.5 mmHg increase in blood pressure (81). Several studies in hypertensive, overweight, and other patient populations have been performed, all revealing predictable acute or chronic blood pressure reduction with varying types of nitrate supplementation (82), this beneficial effect is abolished both acutely and chronically by antimicrobial mouthwash use [reviewed in Ref. (83, 84)].
State of Chronic Inflammation
Dysbiosis, gut wall inflammation, and increased gut wall permeability have been shown to contribute to the state of chronic systemic inflammation. Endotoxemia has been linked to the development of low-grade systemic inflammation and vascular inflammation via toll-like receptor-dependent mechanisms (85). In obese individuals, intestinal microbiota composition was associated with local and systemic inflammation (elevated C-reactive protein) (86).
Microbiota-Derived Hydrogen Sulfide (H2S)
Microbiota, like many mammalian cells and tissues, also produce H2S (87). Microbes exploit this gaseous molecule as an antioxidant defense mechanism, for energy production, and for cell cycle regulation. It is estimated that 50% of fecal H2S is derived from bacteria, thus the total plasma H2S pool varies depending on the individual’s microbiota milieu in the gut. H2S plays a crucial role in a variety of physiological functions, including smooth muscle relaxation, oxidant regulation, inflammation, and angiogenesis (88). Hydrogen sulfide is synthesized primarily from the amino acids cysteine and homocysteine. H2S biosynthesis deregulation, particularly in the renal vasculature, may play a role in hypertension or possibly contribute to existing high blood pressure (89). Although theoretically plausible, it is unknown to what extent does microbiota-derived H2S contribute to blood pressure regulation in humans? This area is still in need of further research.
Hypertension Effects on the Microbiota Composition and Function
While the focus of most studies was to examine how the microbiota at different body sites can modulate blood pressure, a few studies looked at the other direction of this relationship, that is, can hypertension affect our microbiota composition and cause dysbiosis? Is dysbiosis a target organ injury due to hypertension? And does dysbiosis precede, accompany or result from hypertension? It is important to point out that the majority of published studies were cross-sectional and were designed to examine associations and not to determine cause-and-effect relationships. Hypertensive animals and humans were found to have decreased microbial richness, diversity, and composition (11). Santisteban and colleagues tested the hypothesis that increased sympathetic drive to the gut in hypertensive animals is associated with increased gut wall permeability, increased inflammatory status, and microbial dysbiosis (90). Changes in gut pathology were present and were associated with alterations in microbial communities relevant to blood pressure control. However, whether gut permeability and dysbiosis played a role in the pathogenesis of hypertension or were a consequence of hypertension is still not clear. It is very well possible that the relationship between dysbiosis and hypertension is bidirectional or an amplifying one. Further studies are needed to decipher this relationship.
Antihypertensive Medications: Gut Microbiota-Mediated Drug Interactions
The hepatic enzyme system is the key player when it comes to drug metabolism; however, the gut bacteria also exert a variety of metabolic changes to orally ingested drugs, including reductive and hydrolytic reactions. Researchers have reported gut microbiota-mediated drug interactions between multiple medications and antibiotics (91). Those interactions were mediated by alterations in the gut microbiota. The drug amlodipine’s plasma concentration area under curve was increased by up to 133% in ampicillin-treated rats. This increase in its bioavailability was attributed to the reduction of gut microbiota that usually contributes to amlodipine metabolism (92). The authors went on to caution clinicians regarding the use of antibiotics in patients treated with amlodipine. On the other hand, antibiotic treatment alone using minocycline was able to “rebalance” the microbiota and was associated with blood pressure reduction (11). Another aspect to the interaction between antihypertensive medications and the microbiota was described by Santisteban and colleagues (90). In their study, they found increased permeability and stiffness of the gut barrier, decreased levels of tight junction proteins, increased gut fibrosis, thickening of the gut muscularis layer, decreased villi length, and goblet cell loss in spontaneously hypertensive rats and in rats with AngII-induced hypertension. Treatment of SHR with captopril reduced gut permeability and completely restored fibrosis levels and thickness of the muscularis layer and only partially restored villi length (90). Some studies are underway (clinical trial NCT02188381), and more are needed to further examine the microbiota interaction and its role in the metabolism of different antihypertensive medications, as well as the effect of concomitant antibiotic treatment.
Restoring the Balance: Current and Potential Interventions
Lifestyle changes and dietary interventions are key modifiable factors in the management of hypertension. Recently, researchers have started to examine changes in blood pressure as they manipulate the microbiome by introducing dietary and lifestyle changes.
Lifestyle Modifications and Their Effect on Hypertension and the Microbiome
Sufficient sleep is vital for maintaining physical and mental health. Chronic sleep deficiency is related to a wide variety of diseases, including CVD and metabolic disease (93). Epidemiologic studies have established the best amount of sleep for adults as approximately 7 h and that this range correlates best with a lower prevalence of CVD and reduced risk of hypertension (94). Lately an intricate, bidirectional relationship between sleep, circadian rhythms, and the composition of the microbiome in mice was described (95, 96). Benedict et al. showed that sleep deprivation induced changes in microbial families of bacterial gut species in humans (97). Furthermore, Durgan et al. established a link between gut dysbiosis and the development of obstructive sleep apnea-induced hypertension (56). On the other hand, Zhang et al. reported that sleep restriction over several consecutive days does not overtly influence the composition of the microbiome of either rats or humans (98). Further studies are needed to examine the triangular relationship between sleep (duration and quality), blood pressure, and the microbiome.
Sedentary lifestyle is linked to poor health, increased cardiovascular, and metabolic disease risk (99). On the other hand, exercise offers a protective effect; it has a positive effect on body composition, immunity, and cardiovascular health (94, 99). Exercise affects the gut microbiome composition (100). Athletes have a more diverse gut microbiota; Clarke and colleagues showed that a positive effect exists between physical activity, increased dietary protein, and the diversity of the gut microbiome (101). Furthermore, Allen and colleagues demonstrated that different exercise modalities (forced and voluntary) can evoke changes in richness and evenness in the microbiome at varying body sites (102). It remains unclear whether the effect of physical activity on the microbiome is independent of any accompanying adjustment of dietary intake (mainly protein) (100).
Diet and Its Effect on Hypertension and the Microbiome
Multiple dietary components have been shown to affect blood pressure (103), and various studies have examined the effect of manipulating those components on the blood pressure (104). The Dietary Approaches to Stop Hypertension (DASH) diet is one of the interventions used to reduce blood pressure (98). This diet is rich in fruits and vegetables, as well as low-fat dairy, and at the same time has a low content of saturated and total fat (105). DASH-sodium trial demonstrated significant dose–response decreases in blood pressure when the DASH diet and sodium restriction were combined (105, 106). This reduction in blood pressure was accompanied by 30 and 20% risk reduction of CVD after a long-term follow-up for 15–20 years, respectively (107). It is possible that components (high fiber, dairy) of such interventions alter the microbiota in favor of a more balanced one and contribute to its blood pressure-lowering effects (70).
Food Supplements and Their Effect on Hypertension and the Microbiome
Prebiotics
Prebiotics are non-digestible food ingredients that escape digestion in the upper part of the GIT, only to be available for breakdown and fermentation by the gut microbiota within the lower parts of the GIT (108). Most prebiotics are derived from plants. Their role in lowering the CVD risk has been attributed to their ability to lower serum lipid and cholesterol levels (109). Population studies indicate that higher dietary fiber intake was significantly associated with a lower risk of obesity and hypertension (110). Another possible mechanism by which prebiotics could regulate blood pressure is through the attenuation of insulin resistance (111). Additionally, prebiotics have also been reported to reduce the risk of hypertension by improving the absorption of minerals such as calcium in the GIT (112).
Probiotics
Probiotics are living microorganisms that confer a health benefit on the host when administered in sufficient amounts (113). Some probiotic strains exhibit antihypertensive effects: for example, consumption of a dairy product mixture, including Enterococcus faecium and two strains of Streptococcus thermophiles, for 8 weeks lowered systolic blood pressure (114). Administration of Lactobacillus plantarum 299v for 6 weeks was also found to reduce systolic blood pressure in heavy smokers (115). Furthermore, consumption of probiotics-fermented potato yogurt could reduce hypertension-induced cardiac myocyte apoptosis in hypertensive rats and, therefore, can promote cardiac protection against hypertension (116). Lactobacillus casei and Streptococcus thermophilus TMC 1543 were also proven to lower systolic blood pressure and risk factors that caused ischemic heart disease (117).
Synbiotics
Synbiotics are nutritional supplements containing both probiotics and prebiotics in a form of synergy (118). Synbiotics improved survival and distribution of microbial supplements within the GIT by facilitating selective stimulation and activation of growth and metabolism of probiotics (118). Just like prebiotics and probiotics, synbiotics can modulate the gut metabolic activities without modifying the overall structure. Predominant strains of probiotics used in synbiotic preparations include Lacbobacilli, Bifidobacteria species, Saccharomyces boulardii, and Bacillus coagulans, whereas Oligosaccharides, inulin, and other dietary fibers from natural sources form the basis of the prebiotic component. It is worth mentioning that an animal study examining the role of the synbiotic dietary supplement of Lactobacillus plantarum HEAL19 together with fermented blueberry was not effective in lowering blood pressure in hypertensive rats (119). To our knowledge, there are no human trials evaluating the effects of synbiotics on hypertension, such trials are warranted.
Xenobiotics
Xenobiotics are chemicals or substances that are foreign to an organism or biological system. They are not nutrients and enter the body through ingestion, inhalation, or dermal exposure (120). Xenobiotics have the potential to induce gut dysbiosis and influence disease states. Previously published reviews elegantly shed the light on potential mechanisms that link the human gut microbiome to the efficacy and toxicity of xenobiotics (drugs, dietary compounds, and environmental toxins), even after short periods of exposure (121, 122). However, more research is needed to understand the interactions between xenobiotics, blood pressure, and the gut microbiome.
Fecal Microbiota Transplants (FMT)
In FMT, the fecal matter is collected from a tested donor, then blended with saline or other solutions, filtered, and drained and later administered to the recipient via colonoscopy, endoscopy, sigmoidoscopy, or enema.
As the use of FMT in the management of severe or recurrent Clostridium difficile infection is becoming well established (123, 124) and its use in the treatment of IBD, especially ulcerative colitis, is being intensely studied (125), FMT is being increasingly evaluated for use in other areas. Vrieze and colleagues reported improved insulin sensitivity by transfer of microbiota from lean donors to individuals with metabolic syndrome (126). This result gives hope that FMT may become part of treatment regimens for metabolic syndrome and resistant hypertension in the future. Studies have also shown that transplant of cecal microbial content from donor hypertensive animals can reproduce hypertension in previously normotensive recipient animals (56); whether this can be reproduced in humans and whether reverse transplantation will achieve blood pressure reduction in hypertensive subjects remain to be tested.
More work is required to establish the effect of FMT in resistant hypertension. This will include careful evaluation, screening and donor selection, transplant composition, as well as mode of delivery of the transplant (127, 128). Also, it is not known whether the microbiota manipulation can be sustained without continuous application or the need for a concurrent change in dietary or lifestyle habits.
Conclusion
Studies have described how dysbiosis may modulate blood pressure and contribute to CVD. While most studies were performed using animal models, a few studies were conducted in adult hypertensive subjects and none were conducted in children. Given the differences in the gut microbiota composition between children and adults, there is a pressing need for more studies in the pediatric population; it is necessary to characterize the microbiome profile in hypertensive and obese hypertensive children compared to their siblings and their healthy counterparts.
Understanding the nature of hypertension-related microbial aberrations in various body sites, may enable future development of personalized microbiome-based diagnostics and therapies for individuals at risk. Identifying specific microbial signatures associated with the high-risk population may potentially serve as a biomarker to develop non-invasive diagnostics tools. Multiple promising interventions have been described to restore a more balanced microbiome; such treatments need to be further examined in a systematic way to evaluate their potential in lowering blood pressure through modulation of the microbiota. It is worth mentioning that clinical trial NCT02188381 is currently recruiting participants to study gut microbiota involvement in the neuroinflammation-mediated initiation and establishment of resistant hypertension, as well as the possible beneficial role of minocycline therapy on outcomes in resistant hypertension.
Large, prospective clinical trials to establish a more definitive relationship between dysbiosis and high blood pressure and to identify specific microbial signatures in hypertensive subjects are needed before deploying any therapies targeted at altering the microbiota composition.
Author Contributions
All authors (IS, BR, and SA) have equally contributed to the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Bernice Lo for her assistance in the proofreading of this manuscript.
References
1. Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res (2009) 19(7):1141–52. doi:10.1101/gr.085464.108
3. Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol (2012) 129(5):1204–8. doi:10.1016/j.jaci.2012.03.010
4. Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics (2012) 129(5):950–60. doi:10.1542/peds.2011-2736
5. Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011–2012. NCHS Data Brief (2013) 133:1–8.
6. Sorof J, Daniels S. Obesity hypertension in children: a problem of epidemic proportions. Hypertension (2002) 40(4):441–7. doi:10.1161/01.HYP.0000032940.33466.12
7. Spear BA, Barlow SE, Ervin C, Ludwig DS, Saelens BE, Schetzina KE, et al. Recommendations for treatment of child and adolescent overweight and obesity. Pediatrics (2007) 120(Suppl 4):S254–88. doi:10.1542/peds.2007-2329F
8. Flynn JT, Falkner BE. Obesity hypertension in adolescents: epidemiology, evaluation, and management. J Clin Hypertens (Greenwich) (2011) 13(5):323–31. doi:10.1111/j.1751-7176.2011.00452.x
9. Desvarieux M, Demmer RT, Jacobs DR Jr, Rundek T, Boden-Albala B, Sacco RL, et al. Periodontal bacteria and hypertension: the oral infections and vascular disease epidemiology study (INVEST). J Hypertens (2010) 28(7):1413–21. doi:10.1097/HJH.0b013e328338cd36
10. Pevsner-Fischer M, Blacher E, Tatirovsky E, Ben-Dov IZ, Elinav E. The gut microbiome and hypertension. Curr Opin Nephrol Hypertens (2017) 26(1):1–8. doi:10.1097/MNH.0000000000000293
11. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension (2015) 65(6):1331–40. doi:10.1161/HYPERTENSIONAHA.115.05315
12. Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Aging of the human metaorganism: the microbial counterpart. Age (Dordr) (2012) 34(1):247–67. doi:10.1007/s11357-011-9217-5
13. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107(26):11971–5. doi:10.1073/pnas.1002601107
14. Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol (2014) 87(1):280–90. doi:10.1111/1574-6941.12223
15. Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol (2016) 16:90. doi:10.1186/s12866-016-0708-5
16. Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med (2012) 4(10):77. doi:10.1186/gm378
17. Neuman H, Koren O. The pregnancy microbiome. Nestle Nutr Inst Workshop Ser (2017) 88:1–9. doi:10.1159/000455207
18. Neu J. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med (2016) 21(6):373–9. doi:10.1016/j.siny.2016.05.001
19. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet (2012) 13(4):260–70. doi:10.1038/nrg3182
20. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308(5728):1635–8. doi:10.1126/science.1110591
21. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (2009) 326(5960):1694–7. doi:10.1126/science.1177486
22. Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol (2012) 7:99–122. doi:10.1146/annurev-pathol-011811-132421
23. Ram JL, Karim AS, Sendler ED, Kato I. Strategy for microbiome analysis using 16S rRNA gene sequence analysis on the Illumina sequencing platform. Syst Biol Reprod Med (2011) 57(3):162–70. doi:10.3109/19396368.2011.555598
24. Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol (2014) 16(9):2659–71. doi:10.1111/1462-2920.12250
25. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet (2012) 13(1):47–58. doi:10.1038/nrg3129
26. Boyang Ji JN. New insight into the gut microbiome through metagenomics. Adv Genomics Genet (2015) 5:77–91. doi:10.2147/AGG.S57215
27. Vernocchi P, Del Chierico F, Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol (2016) 7:1144. doi:10.3389/fmicb.2016.01144
28. Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One (2015) 10(7):e0133320. doi:10.1371/journal.pone.0133320
29. Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis (2015) 26:26050. doi:10.3402/mehd.v26.26050
30. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464(7285):59–65. doi:10.1038/nature08821
31. Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology (2011) 140(6):1713–9. doi:10.1053/j.gastro.2011.02.011
32. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes (2012) 3(4):289–306. doi:10.4161/gmic.19897
33. Arora T, Sharma R. Fermentation potential of the gut microbiome: implications for energy homeostasis and weight management. Nutr Rev (2011) 69(2):99–106. doi:10.1111/j.1753-4887.2010.00365.x
34. Xu X, He J, Xue J, Wang Y, Li K, Zhang K, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol (2015) 17(3):699–710. doi:10.1111/1462-2920.12502
35. Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev (2010) 74(3):453–76. doi:10.1128/MMBR.00014-10
36. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol (2010) 192(19):5002–17. doi:10.1128/JB.00542-10
37. Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) (2010) 2010:baq013. doi:10.1093/database/baq013
38. Xue J, Xiao LY, Zhou XD. [The latest progress in studies of human oral microbiome]. Hua Xi Kou Qiang Yi Xue Za Zhi (2010) 28(1):5–8.
39. Chen H, Jiang W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol (2014) 5:508. doi:10.3389/fmicb.2014.00508
40. Takahashi N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J Dent Res (2015) 94(12):1628–37. doi:10.1177/0022034515606045
41. Ford PJ, Gemmell E, Timms P, Chan A, Preston FM, Seymour GJ. Anti-P. gingivalis response correlates with atherosclerosis. J Dent Res (2007) 86(1):35–40. doi:10.1177/154405910708600105
42. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med (2013) 187(10):1067–75. doi:10.1164/rccm.201210-1913OC
43. Atanasova KR, Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Mol Oral Microbiol (2014) 29(2):55–66. doi:10.1111/omi.12047
44. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis (2015) 26:26191. doi:10.3402/mehd.v26.26191
45. Dunne JL, Triplett EW, Gevers D, Xavier R, Insel R, Danska J, et al. The intestinal microbiome in type 1 diabetes. Clin Exp Immunol (2014) 177(1):30–7. doi:10.1111/cei.12321
46. Gross M. Does the gut microbiome hold clues to obesity and diabetes? Curr Biol (2013) 23(9):R359–62. doi:10.1016/j.cub.2013.04.047
47. Bradlow HL. Obesity and the gut microbiome: pathophysiological aspects. Horm Mol Biol Clin Investig (2014) 17(1):53–61. doi:10.1515/hmbci-2013-0063
48. Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One (2014) 9(8):e105221. doi:10.1371/journal.pone.0105221
49. Rajendhran J, Shankar M, Dinakaran V, Rathinavel A, Gunasekaran P. Contrasting circulating microbiome in cardiovascular disease patients and healthy individuals. Int J Cardiol (2013) 168(5):5118–20. doi:10.1016/j.ijcard.2013.07.232
50. Rogers CJ, Prabhu KS, Vijay-Kumar M. The microbiome and obesity-an established risk for certain types of cancer. Cancer J (2014) 20(3):176–80. doi:10.1097/PPO.0000000000000049
51. Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J (2014) 20(3):181–9. doi:10.1097/PPO.0000000000000048
52. Major G, Spiller R. Irritable bowel syndrome, inflammatory bowel disease and the microbiome. Curr Opin Endocrinol Diabetes Obes (2014) 21(1):15–21. doi:10.1097/MED.0000000000000032
53. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology (2014) 146(6):1489–99. doi:10.1053/j.gastro.2014.02.009
54. Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe (2015) 17(5):592–602. doi:10.1016/j.chom.2015.04.007
55. Al Khodor S, Shatat IF. Gut microbiome and kidney disease: a bidirectional relationship. Pediatr Nephrol (2017) 32(6):921–31. doi:10.1007/s00467-016-3392-7
56. Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension (2016) 67(2):469–74. doi:10.1161/HYPERTENSIONAHA.115.06672
57. Kunes J, Zicha J. The interaction of genetic and environmental factors in the etiology of hypertension. Physiol Res (2009) 58(Suppl 2):S33–41.
58. Chopra S, Baby C, Jacob JJ. Neuro-endocrine regulation of blood pressure. Indian J Endocrinol Metab (2011) 15(Suppl 4):S281–8. doi:10.4103/2230-8210.86860
59. Karbach SH, Schonfelder T, Brandao I, Wilms E, Hormann N, Jackel S, et al. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J Am Heart Assoc (2016) 5(9):e003698. doi:10.1161/JAHA.116.003698
60. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res (2013) 54(9):2325–40. doi:10.1194/jlr.R036012
61. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A (2013) 110(11):4410–5. doi:10.1073/pnas.1215927110
62. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med (2013) 19(5):576–85. doi:10.1038/nm.3145
63. Stremmel W, Schmidt KV, Schuhmann V, Kratzer F, Garbade SF, Langhans CD, et al. Blood trimethylamine-N-oxide originates from microbiota mediated breakdown of phosphatidylcholine and absorption from small intestine. PLoS One (2017) 12(1):e0170742. doi:10.1371/journal.pone.0170742
64. Schaper J, Wagner A, Enigk F, Brell B, Mousa SA, Habazettl H, et al. Regional sympathetic blockade attenuates activation of intestinal macrophages and reduces gut barrier failure. Anesthesiology (2013) 118(1):134–42. doi:10.1097/ALN.0b013e3182784c93
65. Krzyzaniak M, Peterson C, Loomis W, Hageny AM, Wolf P, Reys L, et al. Postinjury vagal nerve stimulation protects against intestinal epithelial barrier breakdown. J Trauma (2011) 70(5):1168–1175; discussion 1175–1166. doi:10.1097/TA.0b013e318216f754
66. Santisteban MM, Kim S, Pepine CJ, Raizada MK. Brain-gut-bone marrow axis: implications for hypertension and related therapeutics. Circ Res (2016) 118(8):1327–36. doi:10.1161/CIRCRESAHA.116.307709
67. Iraporda C, Errea A, Romanin DE, Cayet D, Pereyra E, Pignataro O, et al. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology (2015) 220(10):1161–9. doi:10.1016/j.imbio.2015.06.004
68. Kim S, Rodriguez V, Santisteban M, Yang T, Qi Y, Raizada M, et al. 6b.07: hypertensive patients exhibit gut microbial dysbiosis and an increase in Th17 cells. J Hypertens (2015) 33(Suppl 1):e77–8. doi:10.1097/01.hjh.0000467562.03337.a5
69. Honour J. The possible involvement of intestinal bacteria in steroidal hypertension. Endocrinology (1982) 110(1):285–7. doi:10.1210/endo-110-1-285
70. Nakamura Y, Yamamoto N, Sakai K, Takano T. Antihypertensive effect of sour milk and peptides isolated from it that are inhibitors to angiotensin I-converting enzyme. J Dairy Sci (1995) 78(6):1253–7. doi:10.3168/jds.S0022-0302(95)76689-9
71. Seppo L, Jauhiainen T, Poussa T, Korpela R. A fermented milk high in bioactive peptides has a blood pressure-lowering effect in hypertensive subjects. Am J Clin Nutr (2003) 77(2):326–30.
72. Takano T. Anti-hypertensive activity of fermented dairy products containing biogenic peptides. Antonie Van Leeuwenhoek (2002) 82(1–4):333–40. doi:10.1023/A:1020600119907
73. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature (2012) 487(7408):477–81. doi:10.1038/nature11228
74. Feng XY, Li Y, Li LS, Li XF, Zheng LF, Zhang XL, et al. Dopamine D1 receptors mediate dopamine-induced duodenal epithelial ion transport in rats. Transl Res (2013) 161(6):486–94. doi:10.1016/j.trsl.2012.12.002
75. Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab (2013) 17(2):225–35. doi:10.1016/j.cmet.2013.01.003
76. Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics (2015) 47(6):187–97. doi:10.1152/physiolgenomics.00136.2014
77. Petersson J, Carlstrom M, Schreiber O, Phillipson M, Christoffersson G, Jagare A, et al. Gastroprotective and blood pressure lowering effects of dietary nitrate are abolished by an antiseptic mouthwash. Free Radic Biol Med (2009) 46(8):1068–75. doi:10.1016/j.freeradbiomed.2009.01.011
78. Lundberg JO, Gladwin MT, Weitzberg E. Strategies to increase nitric oxide signalling in cardiovascular disease. Nat Rev Drug Discov (2015) 14(9):623–41. doi:10.1038/nrd4623
79. Sparacino-Watkins C, Stolz JF, Basu P. Nitrate and periplasmic nitrate reductases. Chem Soc Rev (2014) 43(2):676–706. doi:10.1039/C3CS60249D
80. Lundberg JO, Govoni M. Inorganic nitrate is a possible source for systemic generation of nitric oxide. Free Radic Biol Med (2004) 37(3):395–400. doi:10.1016/j.freeradbiomed.2004.04.027
81. Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate-reducing oral bacteria in blood pressure control. Free Radic Biol Med (2013) 55:93–100. doi:10.1016/j.freeradbiomed.2012.11.013
82. Gee LC, Ahluwalia A. Dietary nitrate lowers blood pressure: epidemiological, pre-clinical experimental and clinical trial evidence. Curr Hypertens Rep (2016) 18(2):17. doi:10.1007/s11906-015-0623-4
83. Hobbs DA, George TW, Lovegrove JA. The effects of dietary nitrate on blood pressure and endothelial function: a review of human intervention studies. Nutr Res Rev (2013) 26(2):210–22. doi:10.1017/S0954422413000188
84. Siervo M, Lara J, Ogbonmwan I, Mathers JC. Inorganic nitrate and beetroot juice supplementation reduces blood pressure in adults: a systematic review and meta-analysis. J Nutr (2013) 143(6):818–26. doi:10.3945/jn.112.170233
85. Smith BJ, Lightfoot SA, Lerner MR, Denson KD, Morgan DL, Hanas JS, et al. Induction of cardiovascular pathology in a novel model of low-grade chronic inflammation. Cardiovasc Pathol (2009) 18(1):1–10. doi:10.1016/j.carpath.2007.07.011
86. Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) (2013) 21(12):E607–15. doi:10.1002/oby.20466
87. Pimentel M, Mathur R, Chang C. Gas and the microbiome. Curr Gastroenterol Rep (2013) 15(12):356. doi:10.1007/s11894-013-0356-y
88. Weber GJ, Pushpakumar S, Tyagi SC, Sen U. Homocysteine and hydrogen sulfide in epigenetic, metabolic and microbiota related renovascular hypertension. Pharmacol Res (2016) 113(Pt A):300–12. doi:10.1016/j.phrs.2016.09.002
89. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A (2010) 107(33):14691–6. doi:10.1073/pnas.1005963107
90. Santisteban MM, Qi Y, Zubcevic J, Kim S, Yang T, Shenoy V, et al. Hypertension-linked pathophysiological alterations in the gut. Circ Res (2017) 120(2):312–23. doi:10.1161/CIRCRESAHA.116.309006
91. Yoo DH, Kim IS, Van Le TK, Jung IH, Yoo HH, Kim DH. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos (2014) 42(9):1508–13. doi:10.1124/dmd.114.058354
92. Yoo HH, Kim IS, Yoo DH, Kim DH. Effects of orally administered antibiotics on the bioavailability of amlodipine: gut microbiota-mediated drug interaction. J Hypertens (2016) 34(1):156–62. doi:10.1097/HJH.0000000000000773
93. Mullington JM, Haack M, Toth M, Serrador JM, Meier-Ewert HK. Cardiovascular, inflammatory, and metabolic consequences of sleep deprivation. Prog Cardiovasc Dis (2009) 51(4):294–302. doi:10.1016/j.pcad.2008.10.003
94. Johnson DA. Enhancing the microbiome through diet, sleep, and exercise. Medscape (2016). Available from: http://www.medscape.com/viewarticle/860179
95. Rosselot AE, Hong CI, Moore SR. Rhythm and bugs: circadian clocks, gut microbiota, and enteric infections. Curr Opin Gastroenterol (2016) 32(1):7–11. doi:10.1097/MOG.0000000000000227
96. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell (2013) 153(4):812–27. doi:10.1016/j.cell.2013.04.020
97. Benedict C, Vogel H, Jonas W, Woting A, Blaut M, Schurmann A, et al. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol Metab (2016) 5(12):1175–86. doi:10.1016/j.molmet.2016.10.003
98. Zhang SL, Bai L, Goel N, Bailey A, Jang CJ, Bushman FD, et al. Human and rat gut microbiome composition is maintained following sleep restriction. Proc Natl Acad Sci U S A (2017) 114(8):E1564–71. doi:10.1073/pnas.1620673114
99. Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. CMAJ (2006) 174(6):801–9. doi:10.1503/cmaj.051351
100. O’Sullivan O, Cronin O, Clarke SF, Murphy EF, Molloy MG, Shanahan F, et al. Exercise and the microbiota. Gut Microbes (2015) 6(2):131–6. doi:10.1080/19490976.2015.1011875
101. Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut (2014) 63(12):1913–20. doi:10.1136/gutjnl-2013-306541
102. Allen JM, Miller MEB, Pence BD, Whitlock K, Nehra V, Gaskins HR, et al. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol (2015) 118(8):1059–66. doi:10.1152/japplphysiol.01077.2014
103. Vasdev S, Stuckless J. Antihypertensive effects of dietary protein and its mechanism. Int J Angiol (2010) 19(1):e7–20. doi:10.1055/s-0031-1278362
104. Nguyen H. A review of nutritional factors in hypertension management. Int J Hypertens (2013) 1–12. doi:10.1155/2013/698940
105. Bray GA. A further subgroup analysis of the effects of the DASH diet and three dietary sodium levels on blood pressure: results of the DASH-sodium trial (vol 94, pg 222, 2004). Am J Cardiol (2010) 105(4):579–579. doi:10.1016/j.amjcard.2009.10.002
106. Sacks FM, Svetkey LP, Vollmer WM, Appel LJ, Bray GA, Harsha D, et al. Effects on blood pressure of reduced dietary sodium and the dietary approaches to stop hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N Engl J Med (2001) 344(1):3–10. doi:10.1056/NEJM200101043440101
107. Cook NR, Cutler JA, Obarzanek E, Buring JE, Rexrode KM, Kumanyika SK, et al. Long term effects of dietary sodium reduction on cardiovascular disease outcomes: observational follow-up of the trials of hypertension prevention (TOHP). Br Med J (2007) 334(7599):885b–8b. doi:10.1136/bmj.39147.604896.55
108. Hutkins RW, Krumbeck JA, Bindels LB, Cani PD, Fahey G Jr, Goh YJ, et al. Prebiotics: why definitions matter. Curr Opin Biotechnol (2016) 37:1–7. doi:10.1016/j.copbio.2015.09.001
109. Ferrier KE, Muhlmann MH, Baguet JP, Cameron JD, Jennings GL, Dart AM, et al. Intensive cholesterol reduction lowers blood pressure and large artery stiffness in isolated systolic hypertension. J Am Coll Cardiol (2002) 39(6):1020–5. doi:10.1016/S0735-1097(02)01717-5
110. Lairon D, Arnault N, Bertrais S, Planells R, Clero E, Hercberg S, et al. Dietary fiber intake and risk factors for cardiovascular disease in French adults. Am J Clin Nutr (2005) 82(6):1185–94.
111. Saad MF, Rewers M, Selby J, Howard G, Jinagouda S, Fahmi S, et al. Insulin resistance and hypertension: the insulin resistance atherosclerosis study. Hypertension (2004) 43(6):1324–31. doi:10.1161/01.HYP.0000128019.19363.f9
112. Streppel MT, Arends LR, van’t Veer P, Grobbee DE, Geleijnse JM. Dietary fiber and blood pressure: a meta-analysis of randomized placebo-controlled trials. Arch Intern Med (2005) 165(2):150–6. doi:10.1001/archinte.165.2.150
113. Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis (2008) 46(Suppl 2):S58–61; discussion S144–151. doi:10.1086/523341
114. Agerholm-Larsen L, Raben A, Haulrik N, Hansen AS, Manders M, Astrup A. Effect of 8 week intake of probiotic milk products on risk factors for cardiovascular diseases. Eur J Clin Nutr (2000) 54(4):288–97. doi:10.1038/sj.ejcn.1600937
115. Naruszewicz M, Johansson ML, Zapolska-Downar D, Bukowska H. Effect of Lactobacillus plantarum 299v on cardiovascular disease risk factors in smokers. Am J Clin Nutr (2002) 76(6):1249–55.
116. Lin PP, Hsieh YM, Kuo WW, Lin YM, Yeh YL, Lin CC, et al. Probiotic-fermented purple sweet potato yogurt activates compensatory IGFIR/PI3K/Akt survival pathways and attenuates cardiac apoptosis in the hearts of spontaneously hypertensive rats. Int J Mol Med (2013) 32(6):1319–28. doi:10.3892/ijmm.2013.1524
117. Kawase M, Hashimoto H, Hosoda M, Morita H, Hosono A. Effect of administration of fermented milk containing whey protein concentrate to rats and healthy men on serum lipids and blood pressure. J Dairy Sci (2000) 83(2):255–63. doi:10.3168/jds.S0022-0302(00)74872-7
118. Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics – a review. J Food Sci Technol (2015) 52(12):7577–87. doi:10.1007/s13197-015-1921-1
119. Xu J, Ahren IL, Prykhodko O, Olsson C, Ahrne S, Molin G. Intake of blueberry fermented by Lactobacillus plantarum affects the gut microbiota of L-NAME treated rats. Evid Based Complement Alternat Med (2013) 2013:809128. doi:10.1155/2013/809128
120. Bojić G, Goločorbin-Kohn S, Stojančević M, Mikov M, Suvajdzić L. Metabolic activity of gut microbiota and xenobiotics. Matica Srpska J Nat Sci Novi Sad (2015) 128:47–55.
121. Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol (2016) 14(5):273–87. doi:10.1038/nrmicro.2016.17
122. Lu K, Mahbub R, Fox JG. Xenobiotics: interaction with the intestinal microflora. ILAR J (2015) 56(2):218–27. doi:10.1093/ilar/ilv018
123. Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol (2011) 9(2):88–96. doi:10.1038/nrgastro.2011.244
124. Choi HH, Cho YS. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc (2016) 49(3):257–65. doi:10.5946/ce.2015.117
125. Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet (2017) 389(10075):1218–28. doi:10.1016/S0140-6736(17)30182-4
126. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143(4):913–6.e7. doi:10.1053/j.gastro.2012.06.031
127. Sha S, Liang J, Chen M, Xu B, Liang C, Wei N, et al. Systematic review: faecal microbiota transplantation therapy for digestive and nondigestive disorders in adults and children. Aliment Pharmacol Ther (2014) 39(10):1003–32. doi:10.1111/apt.12699
Keywords: hypertension, dysbiosis, microbiota, lifestyle, blood pressure, short-chain fatty acid, microbial metabolites
Citation: Al Khodor S, Reichert B and Shatat IF (2017) The Microbiome and Blood Pressure: Can Microbes Regulate Our Blood Pressure? Front. Pediatr. 5:138. doi: 10.3389/fped.2017.00138
Received: 28 March 2017; Accepted: 01 June 2017;
Published: 19 June 2017
Edited by:
Robert P. Woroniecki, State University of New York, United StatesReviewed by:
Litwin Mieczysła˛w, Children’s Memorial Health Institute, PolandChristine B. Sethna, Cohen Children’s Medical Center, United States
Copyright: © 2017 Al Khodor, Reichert and Shatat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibrahim F. Shatat, aXNoYXRhdEBzaWRyYS5vcmc=
 Souhaila Al Khodor
Souhaila Al Khodor Bernd Reichert
Bernd Reichert Ibrahim F. Shatat
Ibrahim F. Shatat