- 1Department of Pulmonology, Shanghai Children’s Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 2Department of Pulmonology, Tianjin Children’s Hospital (Children’s Hospital, Tianjin University), Machang Compus, Tianjin, China
- 3Department of Pediatric Pulmonology and Immunology, West China Second University Hospital, Sichuan University, Chengdu, China
- 4Department of Pediatric Respiratory Medicine, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, China
- 5Department of Pediatric Pulmonology, Yaan People’s Hospital, Yaan, China
Objective: To investigate the prevalence and clinical significance of respiratory pathogen co-detection in children diagnosed with Mycoplasma pneumoniae pneumonia (MPP).
Methods: A prospective observational multicenter study was conducted, collecting clinical data from pediatric patients diagnosed with MPP in four hospitals across China from December 1 to December 31, 2023. Targeted next-generation sequencing (tNGS) results and clinical characteristics were analyzed. Participants were divided into mild and severe groups according to disease severity. Severe cases were further subdivided into an MP alone group and a multi-pathogen co-detection group. Receiver operating characteristic (ROC) curve analysis was performed to assess the predictive performance of inflammatory biomarkers for multi-pathogen co-detection.
Results: A total of 266 children were enrolled. Severe cases had significantly higher C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), lactate dehydrogenase (LDH), D-dimer, interleukin-6 (IL-6), IL-10, and interferon-γ (IFN-γ) levels, as well as longer hospital stays (all P < 0.05). Multi-pathogen co-detection was found in 49.62% of MPP patients, and was more frequent in severe cases than in mild cases (54.05% vs. 39.51%, P < 0.05). The most common co-detected pathogens were rhinovirus, adenovirus, influenza A virus, Haemophilus influenzae, and Streptococcus pneumoniae. Among severe cases, the white blood cell (WBC) count and LDH, IL-6, and IL-10 levels were significantly higher in the multi-pathogen co-detection group compared to the MP alone group (P < 0.05).ROC analysis revealed that IL-6 and IL-10, especially in combination, effectively predicted multi-pathogen co-detection.
Conclusions: Multi-pathogen co-infections substantially influence the severity of pediatric MPP. The findings highlight the diagnostic value of tNGS for identifying co-pathogens and underscore the predictive potential of inflammatory biomarkers (especially IL-6 and IL-10). The integration of tNGS and these biomarkers may facilitate early detection and targeted therapeutic interventions, thereby improving prevention and treatment outcomes in pediatric MPP.
1 Introduction
Mycoplasma pneumoniae (MP) is one of the principal pathogens responsible for community-acquired pneumonia (CAP) in children (1). MP spreads predominantly through respiratory droplets and exhibits cyclical outbreaks every 3–7 years, particularly in densely populated regions (2). While Mycoplasma pneumoniae pneumonia (MPP) is generally a self-limiting condition, severe cases can lead to significant complications, both within and outside the pulmonary system. These complications include necrotizing pneumonia, bronchiolitis obliterans, and acute respiratory distress syndrome, which can be life-threatening in severe cases (3, 4).
Co-infection with other pathogens in children with MPP is common, with reported co-infection rates ranging from 30% to 60% (5, 6). However, most previous studies were single-center or limited to certain pathogen types, providing insufficient comprehensive evidence regarding the clinical impact of co-infections on pediatric MPP severity.
In this prospective observational multicenter study, we utilized targeted next-generation sequencing (tNGS) to comprehensively detect respiratory co-pathogens among children hospitalized with MPP across different regions of China. By correlating pathogen profiles with clinical severity and inflammatory responses, we aimed to clarify how co-infections affect pediatric MPP outcomes. Additionally, receiver operating characteristic (ROC) curve analysis was conducted to determine whether inflammatory biomarkers could accurately predict multi-pathogen co-detection, thereby supporting early identification and optimized management strategies for Severe Mycoplasma pneumoniae pneumonia.
2 Method
2.1 Study patients
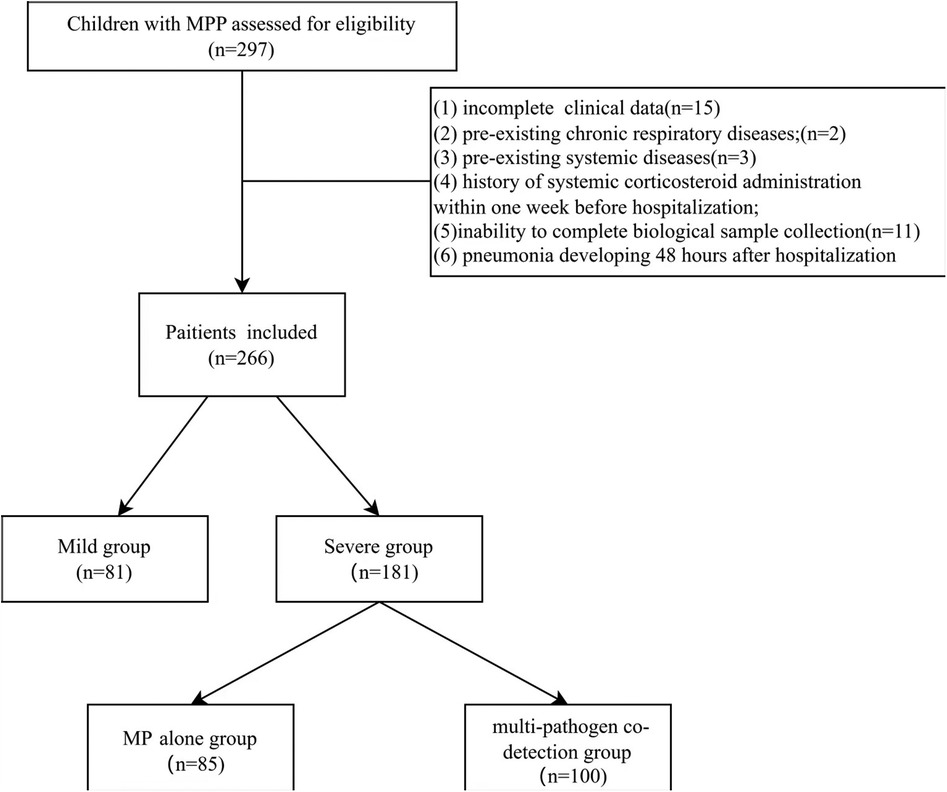
Clinical and laboratory data were prospectively collected from children with MPP who were hospitalized between December 1 and 31, 2023, at four centers in China: West China Second University Hospital (Chengdu), Tianjin Children's Hospital (Tianjin), Shanghai Children's Hospital (Shanghai), and Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University (Wenzhou). Initially, 297 patients were assessed for eligibility. After excluding 31 patients based on the exclusion criteria, a total of 266 patients were finally enrolled in the study (Figure 1).
Inclusion criteria: (1) presence of fever and respiratory symptoms; (2) pneumonia confirmed by physical examination and chest imaging; (3) laboratory-confirmed Mycoplasma pneumoniae infection.
Exclusion criteria: (1) incomplete clinical data; (2) pre-existing chronic respiratory diseases (e.g., bronchopulmonary dysplasia, primary ciliary dyskinesia, cystic fibrosis); (3) pre-existing systemic diseases (e.g., cardiovascular, hepatic, renal, hematologic, neurologic, or immunologic diseases); (4) history of systemic corticosteroid administration within one week before hospitalization; (5) inability to complete biological sample collection; (6) pneumonia developing 48 h after hospitalization.
2.2 Definitions
Mycoplasma pneumoniae pneumonia (MPP): Pneumonia accompanied by fever and respiratory symptoms, confirmed by physical examination and chest imaging, with laboratory-confirmed Mycoplasma pneumoniae infection.
Severe Mycoplasma pneumoniae pneumonia (SMPP) (7): Defined by the presence of one or more of the following criteria: (1) poor general condition; (2) altered consciousness; (3) cyanosis, dyspnea, pulse oxygen saturation ≤92%,or significant increase in breathing rate (>70 breaths/min in infants, >50 breaths/min in older children); (4) ultra-high fever or persistent high fever lasting more than 5 days; (5) refusal of food or signs of dehydration; (6) extensive pulmonary involvement (≥2/3 of a lobe or multilobar), pleural effusion, pneumothorax, atelectasis, pulmonary necrosis, or lung abscess; (7) extrapulmonary complications.
Based on disease severity, patients were classified into a mild group (uncomplicated MPP) and a severe group (meeting SMPP criteria). Severe cases were further subdivided according to pathogen detection results into an MP alone group (MP detected as the sole pathogen) and a multi-pathogen co-detection group (MP plus ≥1 additional respiratory pathogen detected).
2.3 Data collection
Clinical and microbiological data for hospitalized patients were obtained from the hospital clinical information system. Data included complete blood counts, CRP, ESR, liver function tests, other biochemical parameters, D-dimer, inflammatory cytokines, and length of hospital stay. For each patient, laboratory values at admission (within 24 h of hospitalization) were used as baseline values for comparison between groups.
2.4 Sample collection
Pharyngeal swabs and blood samples were collected within 24 h of hospital admission. In the mild group, trained physicians collected pharyngeal swabs by swabbing the back of the pharynx at least three times. In the severe group, bronchoalveolar lavage fluid (BALF) samples were collected during bronchoscopy within 7 days after admission. The pharyngeal swabs and BALF samples were stored at −80°C and transported to a designated laboratory for targeted next-generation sequencing (tNGS) analysis at ≤−20°C.
2.5 Targeted next-generation sequencing
Pharyngeal swabs (mild group) and BALF samples (severe group) were transported to a central laboratory for targeted next-generation sequencing (tNGS) analysis (Respiratory 100 panel, KingMed Diagnostics, Guangzhou, China). According to the manufacturer's instructions, the tNGS method enables the early diagnosis of respiratory infections by detecting 198 pathogens. After nucleic acid extraction, specific primers targeting bacterial, mycobacterial, viral, fungal, and selected antimicrobial resistance sequences were used to enrich multiplex targets, followed by library preparation for NGS sequencing (8, 9). The reads of sequence (normalized sequence number in 100,000 primary sequences detected) shown in the report were calculated based on internal control. When estimating the concentration of the target pathogen, exogenous plasmids of known concentrations were used as the internal control. The number of normalized sequence reads of the target pathogen could be calculated using the amplification efficiency ratio of the internal control.
To ensure accurate clinical interpretation, all tNGS results were independently reviewed by two experienced clinicians, who carefully assessed each detected pathogen in the context of the patient’s medical history, symptoms, imaging findings, and laboratory results to distinguish true infections from colonization. Given that pharyngeal swabs may detect upper airway commensals, independent interpretation by two clinicians helped ensure accurate pathogen identification. BALF samples from severe cases, representing direct lower respiratory specimens, further reduced concerns about colonization and enhanced diagnostic accuracy.
2.6 Statistical analysis
Data analyses were performed using SPSS software (version 22.0). Normally distributed continuous data were presented as mean ± standard deviation (SD) and compared using independent-sample t-tests. Skewed distribution data were reported as medians with interquartile ranges (IQR) and analyzed using Mann–Whitney U tests. Categorical variables were presented as frequencies and percentages, with comparisons between groups performed using the chi-square test or Fisher's exact test. ROC curve analysis was performed to evaluate the predictive value of inflammatory biomarkers for identifying multi-pathogen co-detection. A Sankey diagram was constructed to visualize the distribution of respiratory pathogens co-detected with MP. The difference was considered statistically significant at P < 0.05.
3 Results
3.1 Demographic and clinical characteristics of children with MPP
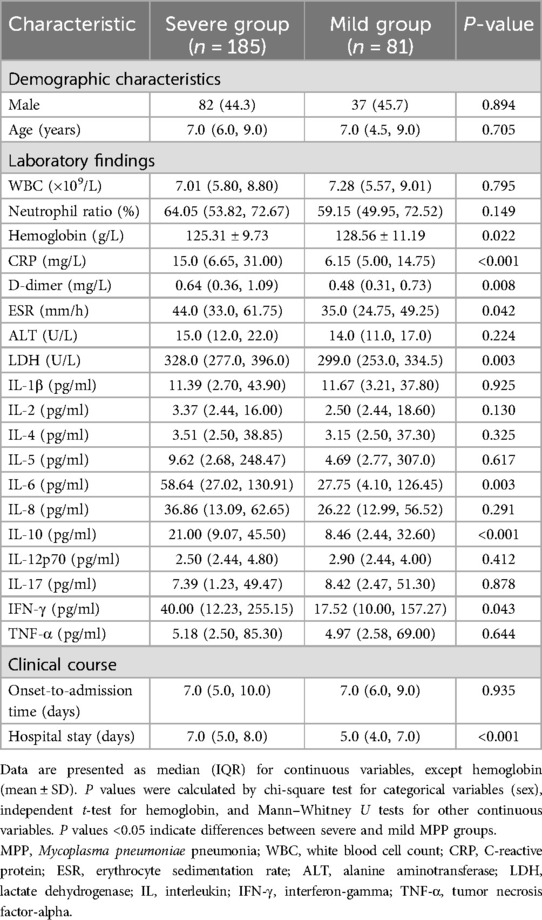
A total of 266 children with MPP were enrolled in this study (119 males and 147 females), including 185 severe cases and 81 mild cases. The median age at onset was 7.0 years in both groups, with no significant differences in age or gender distribution between severe and mild cases (P > 0.05 for both). However, patients in the severe group had significantly longer hospital stays (P < 0.001; Table 1).
Compared to the mild group, patients in the severe group exhibited significantly higher levels of inflammatory biomarkers (CRP, ESR, LDH, D-dimer) and serum cytokines (IL-6, IL-10, IFN-γ) (all P < 0.05; Table 1).
3.2 Detection and regional distribution of respiratory pathogens in children with MPP
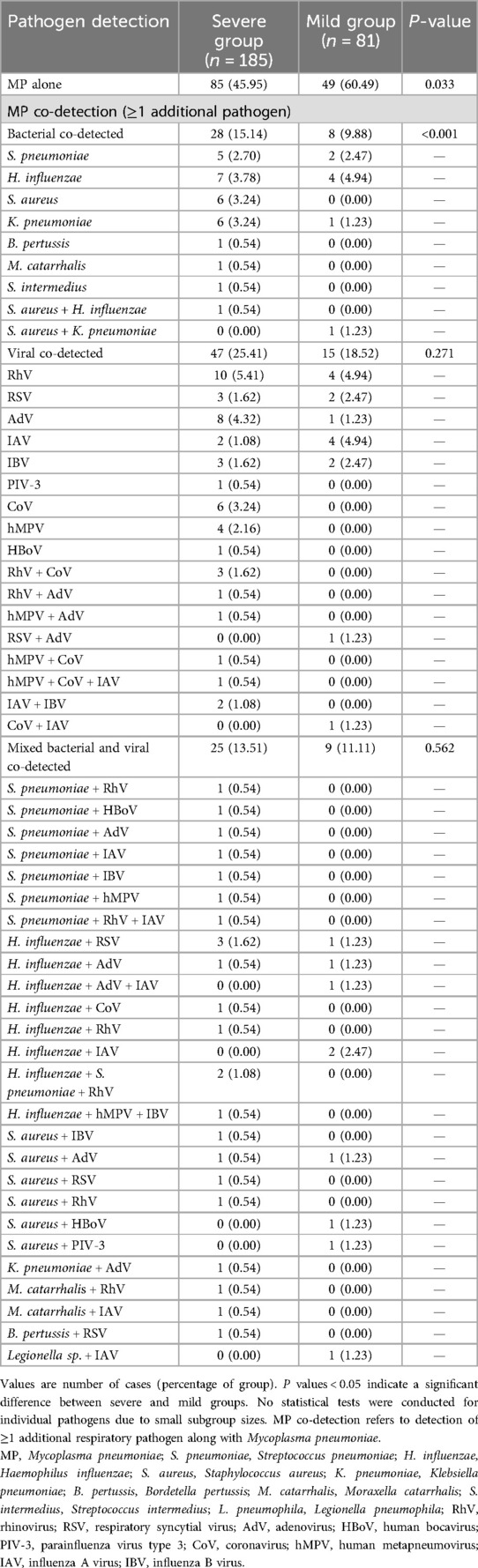
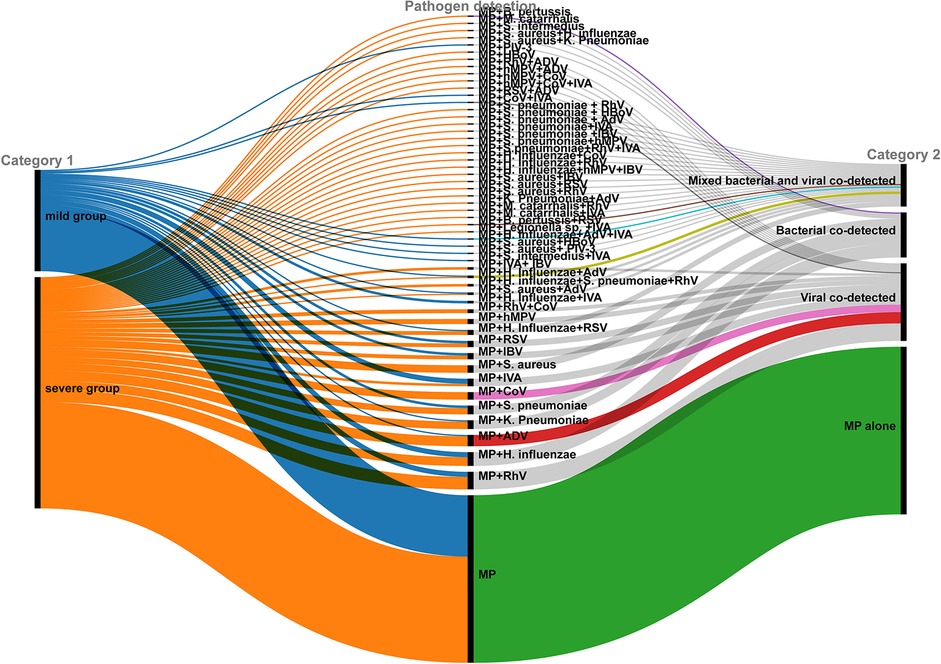
Respiratory pathogens other than MP were identified in 132 out of 266 children (49.62%). Multi-pathogen co-detection rate was significantly higher in the severe group compared to the mild group (54.05% vs. 39.51%, χ2 = 4.769, P = 0.033), especially bacterial co-detection rate (P < 0.001; Table 2). A Sankey diagram further illustrated clear differences in respiratory pathogen profiles between severe and mild groups (Figure 2).
The tNGS analysis identified multi-pathogen co-detection in 132 children (49.62%). The most commonly co-detected organisms were Haemophilus influenzae (H. influenzae, 26 cases, 9.8%), rhinovirus (RhV, 25 cases, 9.4%), adenovirus (AdV, 19 cases, 7.1%), influenza A virus (IAV, 18 cases, 6.8%), Streptococcus pneumoniae (S. pneumoniae, 16 cases, 6.0%), Staphylococcus aureus (S. aureus, 15 cases, 5.6%), and human coronavirus (CoV, 13 cases, 4.9%). RhV, AdV, and IAV were the predominant viral pathogens, while H. influenzae and S. pneumoniae were the primary bacterial pathogens co-detected with MP. Although detection rates of AdV, CoV, RhV, human metapneumovirus (hMPV), influenza B virus (IBV), S. pneumoniae and S. aureus were higher in the severe group than in the mild group, these differences were not statistically significant (Table 3).
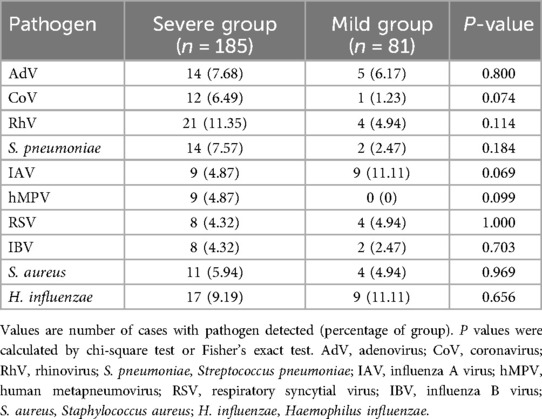
Table 3. Detection rates of specific pathogens co-detected with MP: comparison between severe and mild groups.
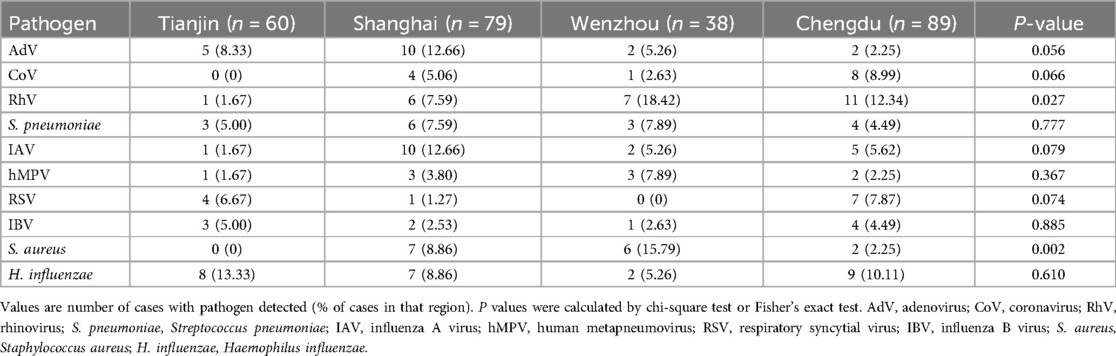
No significant differences were observed in the rates of multi-pathogen co-detection with MP between mild and severe cases across different regions (all P > 0.05; Table 4). However, the predominant pathogens detected varied by region, with H. influenzae being most common in Tianjin (13.33%), AdV and IAV in Shanghai (each 12.65%), and RhV predominating in both Wenzhou (18.42%) and Chengdu (12.36%). The detection rates of RhV and S. aureus differed significantly among the four regions (P < 0.05; Table 5).
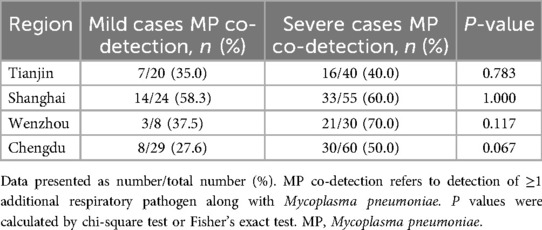
Table 4. Comparison of multi-pathogen co-detection rates between mild and severe cases of MPP across different regions.
3.3 Demographic and clinical characteristics of SMPP patients in the MP alone and multi-pathogen co-detection groups
Among the 185 children with SMPP, 85 had MP detected alone and 100 had co-detection of additional pathogens. No significant differences were observed between the two groups regarding gender distribution, age of onset, time from symptom onset to admission, or length of hospital stay (all P > 0.05; Table 6).
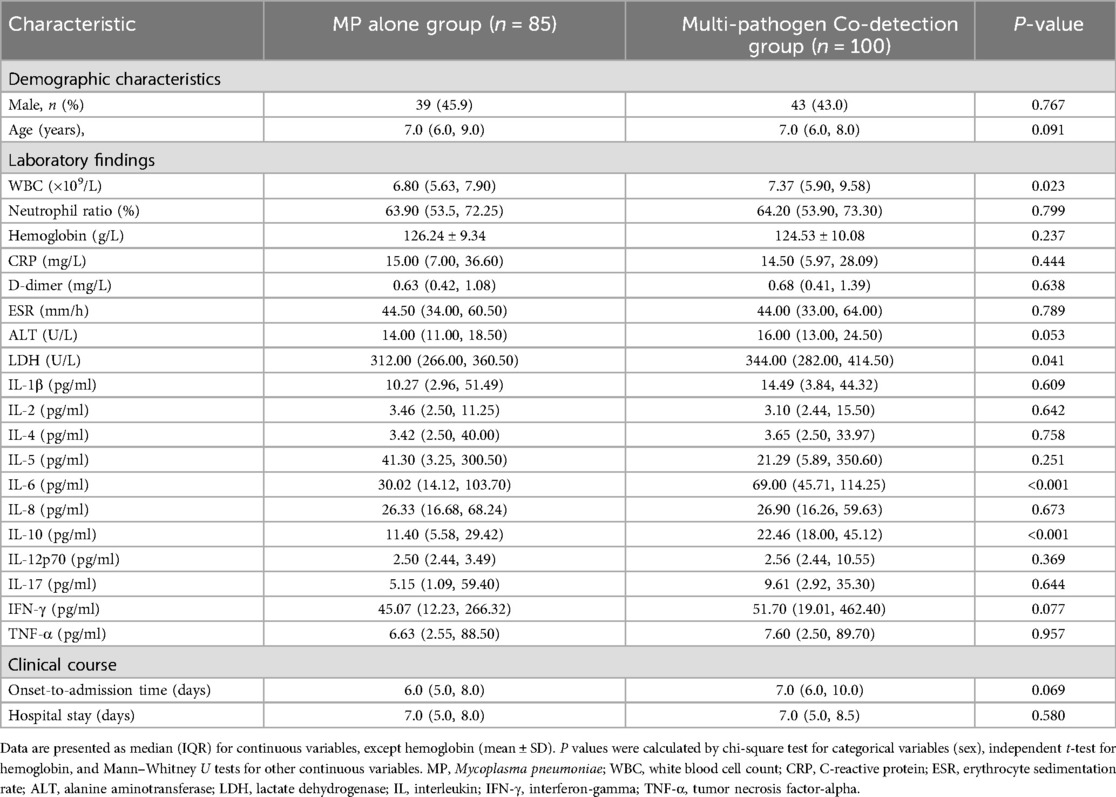
Table 6. Clinical characteristics of children with SMPP: comparison between the MP alone and multi-pathogen co-detection groups.
However, patients in the multi-pathogen co-detection group had significantly higher WBC counts and higher LDH, IL-6, and IL-10 levels compared to those in the MP alone group (all P < 0.05; Table 6).
3.4 ROC analysis of inflammatory markers for predicting co-detection in SMPP
ROC curve analyses were performed to evaluate the predictive value of inflammatory biomarkers (WBC, LDH, IL-6, IL-10, and the combination of IL-6 and IL-10) for identifying multi-pathogen co-detection in children with SMPP. Both WBC and LDH individually showed moderate predictive performance. IL-6 and IL-10 demonstrated better discriminative capability individually, and combining IL-6 and IL-10 further enhanced diagnostic sensitivity, achieving the highest overall diagnostic accuracy (Table 7, Figure 3).
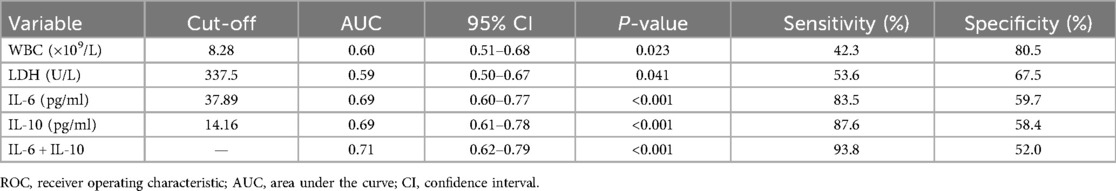
Table 7. ROC curve analysis of inflammatory biomarkers for predicting multi-pathogen co-detection in children with SMPP.
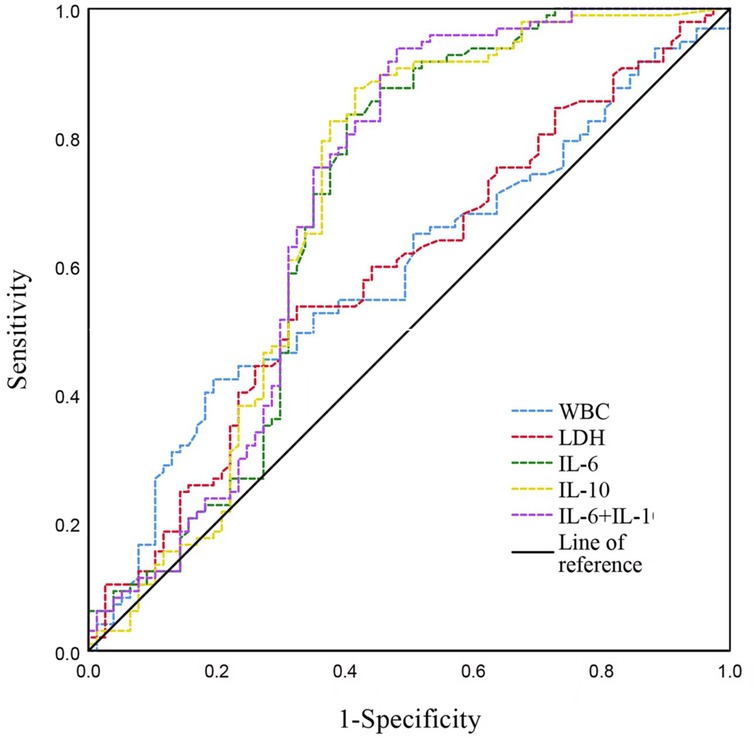
Figure 3. ROC curves illustrating predictive performance of inflammatory biomarkers in children with SMPP. WBC, white blood cell count; ALT, alanine aminotransferase; LDH, lactate dehydrogenase; IL, interleukin.
4 Discussion
Previous studies have demonstrated that co-infections with viral and bacterial pathogens frequently occur in pediatric MPP, with reported co-infection rates generally ranging from 30% to 60% (5, 6, 10). Some studies have reported even higher rates, exceeding 88% (11). Clinically, pediatric MPP cases complicated by co-infections typically present with more severe and diverse symptoms, resulting in prolonged hospital stays (6, 10, 12). These findings highlight that co-infection with multiple respiratory pathogens significantly contributes to the severity and clinical complexity of MPP.
In the current prospective observational multicenter study, additional pathogens were detected in 132 of 266 children (49.62%) with MPP, a rate consistent with prior reports (5, 6, 10). Furthermore, the multi-pathogen co-detection rate was significantly higher in the severe group than in the mild group (54.05% vs. 39.51%, χ2 = 4.769; P = 0.033), suggesting that co-infections may be associated with increased disease severity in pediatric MPP.
Interestingly, subgroup analysis by region revealed no significant difference in multi-pathogen co-detection rates between severe and mild cases within each center (Tianjin, Shanghai, Wenzhou, Chengdu; all P > 0.05). This is likely attributable to limited statistical power due to smaller sample sizes at each center, combined with the relatively short duration of the study (one winter month). Additionally, similar respiratory pathogens may have concurrently circulated across these regions during the study period, potentially obscuring regional variations. Future larger-scale studies conducted across multiple seasons are warranted to clarify potential geographic differences in co-infection patterns.
Retrospective analyses of 396 children with MPP identified S. pneumoniae, H. influenzae, and S. aureus as the most common bacterial co-infections, while viral co-infections commonly involved human bocavirus (HBoV), RhV, and respiratory syncytial virus (RSV) (6). In the current study, RhV (9.4%), AdV (7.1%), and IAV (6.8%) were the predominant viral pathogens co-detected with MP, while H. influenzae (9.8%) and S. pneumoniae (6.0%) were the primary bacterial co-pathogens. Some studies identified S. pneumoniae as the most common bacterial co-infection (7); however, our study found H. influenzae to be predominant, possibly due to seasonal differences and limited sample size.
Indeed, regional variations were notable, with H. influenzae (13.33%) being the most common co-pathogen in Tianjin, AdV and IAV (12.65%) dominating in Shanghai, and RhV being prominent in Wenzhou (18.42%) and Chengdu (12.36%). Such differences underscore the importance of considering local epidemiology, environmental factors, and healthcare practices when developing diagnostic and therapeutic strategies.
Given the high frequency and complexity of pathogen co-infections in severe pediatric MPP, there is an urgent need for rapid and cost-effective multiplex diagnostic tools. Recent evidence highlighting neurological dysfunction associated with atypical pathogens such as ammonia-producing microorganisms further underscores the importance of comprehensive pathogen screening in clinical practice (13). Metagenomic next-generation sequencing (mNGS) is of high value in detecting novel, rare, and atypical pathogens (14), but its use in routine practice is limited by its high cost, the influence of human genomic material, and the inability to simultaneously detect both DNA and RNA pathogens in one run (15). Targeted NGS (tNGS), in contrast, uses panels of known pathogen sequences to screen clinical isolates. These panels can target multiple types of pathogens (bacteria, viruses, fungi, atypical agents) with high specificity and sensitivity, and provide faster turnaround times while sequencing directly from clinical specimens (16, 17). In our study, the tNGS panel covered 198 pathogens (over 95% of common respiratory pathogens) and cost roughly one-quarter that of mNGS, making it a practical approach for early and comprehensive pathogen detection in MPP.
MP infection impairs cellular and humoral immunity, leading to an immunosuppressed state that facilitates secondary infections (18). Previous studies have reported elevated levels of inflammatory cytokines, such as IL-1β, IL-6, IL-8, IL-10, and TNF-α, in patients with MPP compared to healthy controls (19, 20). Consistent with these findings, our study showed significantly higher levels of IL-6, IL-10, and IFN-γ levels in severe cases compared to mild cases, with further elevations of IL-6 and IL-10 observed in patients with multi-pathogen co-infections. These results suggest that co-infections intensify inflammatory responses, thereby exacerbating disease severity.
IL-6 and IL-10 play key roles in inflammation and immune regulation, and their marked elevation may reflect more severe disease progression and greater tissue damage. Our further ROC analysis confirmed that IL-6 and IL-10, individually and especially in combination, exhibit high sensitivity (>80%, and approximately 94% combined) for detecting co-infections. Thus, combined IL-6 and IL-10 measurement might be clinically valuable for early identification of SMPP cases at risk of co-infection, facilitating timely intervention and improved patient outcomes.
In addition to cytokines demonstrating significant differences (e.g., IL-6, IL-10, and IFN-γ), several other cytokines, including IL-4, IL-8, and TNF-α, did not differ significantly between the mild and severe groups. This might indicate that these cytokines play a less direct role in the immunopathology of SMPP, or it may reflect sampling timing that did not align with their peak concentrations. Future studies examining the dynamic changes of these cytokines during the course of illness may clarify their roles in MPP progression.
Markers of infection and inflammation such as CRP, ESR, and LDH were also higher in our severe cases than in mild cases, in line with previous findings (18). MP infection activates exogenous and endogenous coagulation systems, leading to coagulation abnormalities and promoting thrombosis (21). Higher D-dimer levels existed in children with SMPP children compared to mild cases in the current study. This indicates a heightened state of coagulation, suggesting that potential thrombotic complications may contribute to increased disease severity and should be carefully considered in the management of SMPP.
In contrast to previous single-center studies or those limited to detecting specific pathogens, our multicenter approach utilizing tNGS provided comprehensive, accurate, and generalizable identification of respiratory co-pathogens in pediatric MPP. Furthermore, our ROC curve analyses of inflammatory biomarkers—particularly the combined use of IL-6 and IL-10—provided novel and valuable diagnostic insights, significantly improving the predictive accuracy for detecting severe cases complicated by co-infections. The integration of clinical characteristics and biomarker analysis further enhances diagnostic precision and holds potential for improving clinical decision-making and patient outcomes.
Several limitations should be noted. The relatively short duration of our study (one winter month) and moderate sample size (n = 266) may restrict the generalizability of pathogen prevalence across different seasons and geographic regions. Additionally, distinguishing colonization from infection remains challenging with current tNGS technology, particularly for pharyngeal swab samples, as uniform diagnostic standards are lacking. Although we mitigated this concern by employing BALF samples for severe cases and independent clinical assessments by experienced clinicians, differences in sampling methods remain a limitation. Specifically, pathogen concentrations likely varied between mild (pharyngeal swabs) and severe cases (BALF samples), potentially resulting in higher observed co-detection rates among severe patients. Furthermore, logistic regression analysis was not performed, thus limiting our ability to identify co-detected pathogens as independent predictors of SMPP.
Future studies with larger cohorts, extended durations, and spanning multiple seasons are essential for validating and generalizing our preliminary results. Uniform sampling methods or the concurrent collection of paired upper and lower respiratory tract samples should be considered to accurately explore pathogen co-detection rates and their relationship with disease severity. Further studies employing multivariate logistic regression analysis would clarify whether multi-pathogen co-detection independently predicts SMPP.
In conclusion, this prospective observational multicenter study demonstrated a high prevalence of multi-pathogen co-infections among pediatric patients with MPP, particularly in severe cases. The presence of these co-infections was associated with significantly elevated inflammatory markers, including WBC count, LDH, IL-6, and IL-10, indicating more intense inflammatory responses. Understanding the complex interactions between MP and co-detected pathogens, along with regional and seasonal variations, is critical for improving early diagnosis and targeted management strategies, ultimately enhancing clinical outcomes in pediatric MPP.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Institutional Review Board (No. 074-2 of 2022) at the West China Second University Hospital, Sichuan University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants' legal guardians/next of kin. Written informed consent was obtained from the minor(s)' legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author contributions
XD: Data curation, Writing – original draft. RL: Data curation, Writing – original draft. YZ: Writing – review & editing. LC: Writing – review & editing. HZ: Writing – review & editing. FL: Formal analysis, Writing – original draft. WY: Formal analysis, Writing – original draft. YN: Formal analysis, Writing – original draft, Data curation. HW: Data curation, Writing – original draft, Formal analysis. RG: Data curation, Writing – original draft, Formal analysis. XW: Data curation, Writing – original draft, Formal analysis. LLi: Data curation, Writing – original draft, Formal analysis. ZL: Data curation, Writing – original draft, Formal analysis. LuL: Data curation, Writing – original draft, Formal analysis. DL: Data curation, Writing – original draft. QL: Investigation, Supervision, Writing – review & editing. HL: Investigation, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the project “Early Diagnosis Model for Refractory Pneumonia in Children Based on Multimodal Data and Integrated Learning and Computing Peaks” (No. 202340070) and National Natural Science Foundation of China (No. 82370001)., Tianjin Municipal Health Commission Key Discipline Special Fund (No. TJWJ2022XK038) and Major Scientific and Technological Project from Science & Technology Department of Sichuan Province (No. 2022ZDZX0021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tong L, Huang S, Zheng C, Zhang Y, Chen Z. Refractory Mycoplasma pneumoniae pneumonia in children: early recognition and management. J. Clin Med. (2022) 11:2824. doi: 10.3390/jcm11102824
2. Youn YS, Lee KY. Mycoplasma pneumoniae pneumonia in children. Korean J Pediatr. (2012) 55:42–7. doi: 10.3345/kjp.2012.55.2.42
3. Yang B, Zhang W, Gu W, Zhang X, Wang M, Huang L, et al. Differences of clinical features and prognosis between Mycoplasma pneumoniae necrotizing pneumonia and non-Mycoplasma pneumoniae necrotizing pneumonia in children. BMC Infect Dis. (2021) 21:797. doi: 10.1186/s12879-021-06469-x
4. Zheng HQ, Ma YC, Chen YQ, Xu YY, Pang YL, Liu L. Clinical analysis and risk factors of bronchiolitis obliterans after Mycoplasma pneumoniae pneumonia. Infect Drug Resist. (2022) 15:4101–8. doi: 10.2147/idr.S372940
5. Diaz MH, Cross KE, Benitez AJ, Hicks LA, Kutty P, Bramley AM, et al. Identification of bacterial and viral codetections with Mycoplasma pneumoniae using the TaqMan array card in patients hospitalized with community-acquired pneumonia. Open Forum Infect. Dis. (2016) 3:ofw071. doi: 10.1093/ofid/ofw071
6. Zhang X, Chen Z, Gu W, Ji W, Wang Y, Hao C, et al. Viral and bacterial co-infection in hospitalised children with refractory Mycoplasma pneumoniae pneumonia. Epidemiol Infect. (2018) 146:1384–8. doi: 10.1017/s0950268818000778
7. Yang S, Lu S, Guo Y, Luan W, Liu J, Wang L. A comparative study of general and severe mycoplasma pneumoniae pneumonia in children. BMC Infect Dis. (2024) 24:449. doi: 10.1186/s12879-024-09340-x
8. Xing FF, Chiu KH, Deng CW, Ye HY, Sun LL, Su YX, et al. Post-COVID-19 pandemic rebound of macrolide-resistant Mycoplasma pneumoniae infection: a descriptive study. Antibiotics (2024) 13:262. doi: 10.3390/antibiotics13030262
9. Deng Z, Li C, Wang Y, Wu F, Liang C, Deng W, et al. Targeted next-generation sequencing for pulmonary infection diagnosis in patients unsuitable for bronchoalveolar lavage. Front Med (Lausanne) (2023) 10:1321515. doi: 10.3389/fmed.2023.1321515
10. Guo Q, Li L, Wang C, Huang Y, Ma F, Cong S, et al. Comprehensive virome analysis of the viral spectrum in paediatric patients diagnosed with Mycoplasma pneumoniae pneumonia. Virol J. (2022) 19:181. doi: 10.1186/s12985-022-01914-y
11. Shin S, Koo S, Yang YJ, Lim HJ. Characteristics of the Mycoplasma pneumoniae epidemic from 2019 to 2020 in Korea: macrolide resistance and co-infection trends. Antibiotics (2023) 12:1623. doi: 10.3390/antibiotics12111623
12. Gao J, Xu L, Xu B, Xie Z, Shen K. Human adenovirus coinfection aggravates the severity of Mycoplasma pneumoniae pneumonia in children. BMC Infect Dis. (2020) 20:420. doi: 10.1186/s12879-020-05152-x
13. Xu J, Ren X, Huang X, Jin Y, Wang M, Zhong L, et al. Epidemiological and clinical characteristics of ammonia-producing microorganisms in the lungs of patients with severe pneumonia: a multicentre cohort study. J Transl Med. (2024) 22:1148. doi: 10.1186/s12967-024-05974-2
14. Xu J, Zhong L, Shao H, Wang Q, Dai M, Shen P, et al. Incidence and clinical features of HHV-7 detection in lower respiratory tract in patients with severe pneumonia: a multicenter, retrospective study. Crit Care. (2023) 27:248. doi: 10.1186/s13054-023-04530-6
15. Li S, Tong J, Liu Y, Shen W, Hu P. Targeted next generation sequencing is comparable with metagenomic next generation sequencing in adults with pneumonia for pathogenic microorganism detection. J Infect. (2022) 85:e127–9. doi: 10.1016/j.jinf.2022.08.022
16. Gaston DC, Miller HB, Fissel JA, Jacobs E, Gough E, Wu J, et al. Evaluation of metagenomic and targeted next-generation sequencing workflows for detection of respiratory pathogens from bronchoalveolar lavage fluid specimens. J Clin Microbiol. (2022) 60:e0052622. doi: 10.1128/jcm.00526-22
17. Zhong Y, Xu F, Wu J, Schubert J, Li MM. Application of next generation sequencing in laboratory medicine. Ann Lab Med. (2021) 41:25–43. doi: 10.3343/alm.2021.41.1.25
18. He J, Liu M, Ye Z, Tan T, Liu X, You X, et al. Insights into the pathogenesis of Mycoplasma pneumoniae. Mol Med Rep. (2016) 14:4030–6. doi: 10.3892/mmr.2016.5765
19. Fan F, Lv J, Yang Q, Jiang F. Clinical characteristics and serum inflammatory markers of community-acquired mycoplasma pneumonia in children. Clin Respir J. (2023) 17:607–17. doi: 10.1111/crj.13620
20. Chen X, Liu F, Zheng B, Kang X, Wang X, Mou W, et al. Exhausted and apoptotic BALF T cells in proinflammatory airway milieu at acute phase of severe Mycoplasma Pneumoniae pneumonia in children. Front Immunol. (2022) 12:760488. doi: 10.3389/fimmu.2021.760488
Keywords: Mycoplasma pneumoniae, child, pneumonia, pathogen co-detection rate, targeted next-generation sequencing
Citation: Dong X, Li R, Zou Y, Chen L, Zhang H, Lyu F, Yang W, Niu Y, Wang H, Guo R, Wang X, Li L, Lin Z, Luo L, Lu D, Lu Q and Liu H (2025) Co-detection of respiratory pathogens in children with Mycoplasma pneumoniae pneumonia: a multicenter study. Front. Pediatr. 13:1482880. doi: 10.3389/fped.2025.1482880
Received: 18 August 2024; Accepted: 29 April 2025;
Published: 23 May 2025.
Edited by:
Janet R. Hume, University of Minnesota Medical Center, United StatesReviewed by:
Yungang Yang, First Affiliated Hospital of Xiamen University, ChinaLingtong Huang, Zhejiang University, China
Baoying Zheng, The Children's Hospital Affiliated to the Capital Institute of Pediatrics, China
Copyright: © 2025 Dong, Li, Zou, Chen, Zhang, Lyu, Yang, Niu, Wang, Guo, Wang, Li, Lin, Luo, Lu, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Lu, bHVxdWFuLXNoQHZpcC5zaW5hLmNvbQ==; Hanmin Liu, aGFubWluQHNjdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiaoyan Dong
Xiaoyan Dong Rui Li
Rui Li Yingxue Zou
Yingxue Zou Lina Chen3,†
Lina Chen3,† Hailin Zhang
Hailin Zhang Fangfang Lyu
Fangfang Lyu Wenhao Yang
Wenhao Yang Yanhua Niu
Yanhua Niu Quan Lu
Quan Lu