- 1Centre for Respiratory Diseases and Meningitis, National Institute for Communicable Diseases of the National Health Laboratory Service, Johannesburg, South Africa
- 2Department of Clinical Microbiology and Infectious Diseases, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
- 3Department of Medical Virology, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
- 4Department of Paediatrics, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
- 5School of Public Health, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
- 6MassGenics, Duluth, GA, United States
Introduction: The burden of morbidity and mortality of severe respiratory illness (SRI) remains disproportionately high among young children, and in low-and middle-income countries. We used a multi-pathogen respiratory PCR assay to detect pathogens in children aged <5 years hospitalized with SRI.
Methods: Prospective syndromic surveillance for SRI was performed at two sentinel hospitals in South Africa between January and December 2017. Nasopharyngeal aspirates and sputa were collected and tested using a real-time polymerase chain reaction based TaqMan Array Card (TAC) for the detection of 21 respiratory pathogens. Pathogen detection was compared by age group using the chi-squared test and seasonal frequency analysed.
Results: From January through December 2017, 361 children were enrolled and of these, 198 cases with sufficient specimen volume were included in this study. Overall, 189/198 (95%) of the children tested positive for at least one pathogen. Common viruses identified included rhinovirus (65/198; 33%), respiratory syncytial virus (RSV) (54/198; 27%), adenovirus (34/198; 17%), and enterovirus (28/198; 14%). Common bacteria detected included Haemophilus influenzae (121/198; 61%), Streptococcus pneumoniae (114/198; 58%), Klebsiella pneumoniae (61/198; 31%), Staphylococcus aureus (52/198; 26%), and Acinetobacter baumannii (27/198; 14%).
Discussion: Bacterial detections were high in our study driven by the high detection of S. pneumoniae and H. influenzae. Co-detections of pathogens were common and require clinical evaluation to determine their relevance in clinical management. Further, given the high prevalence of RSV amongst children hospitalized with SRI, there is an urgent need for continued efforts towards access to maternal RSV vaccines and therapeutic interventions such as monoclonal antibodies particularly in low- and middle-income countries which experience the highest burden of RSV-associated disease.
1 Introduction
The burden of morbidity and mortality of severe respiratory illness (SRI) remains disproportionately high among children aged <5 years globally and is highest in the first year of life (1, 2). In this age group, respiratory viruses, such as respiratory syncytial virus (RSV) and influenza are commonly responsible for the majority of infections however, bacterial infections, such as with Streptococcus pneumoniae and non-typeable Haemophilus influenzae may occur following an acute viral infection due to mucosal invasion or aspiration of nasopharyngeal bacteria and are associated with more severe outcomes of SRI (3–7).
The South African Thoracic Society's SRI treatment guidelines recommend that all children with signs of severe pneumonia be given empiric antibiotic therapy for at least five days, the escalation, de-escalation or discontinuation of which should be advised by the patient's response to therapy and/or microbiology results (8). Polymerase chain reaction (PCR) is standard for the detection of respiratory viruses and atypical pneumonia-causing bacteria, however, in South Africa, PCR is not routinely performed as part of the diagnostic workflow in SRI as treatment is usually empiric. Accurate diagnosis of the causative pathogen is critical for optimizing effective patient management decisions, such as the need for antibiotic or antiviral prescriptions or to inform empiric therapy.
We used a multi-pathogen real-time PCR-based respiratory panel to detect viruses and bacteria in children aged <5 years hospitalized with SRI at two sentinel hospitals in South Africa.
2 Materials and methods
2.1 Study population
Prospective hospital-based syndromic surveillance for SRI in all ages was initiated in 2009 in South Africa to describe the aetiology of and risk factors for community-acquired pneumonia. Children aged <5 years hospitalized with SRI and enrolled at two sentinel hospitals in the North-West and Mpumalanga Provinces between January and December 2017 were included in the current study. In-hospital outcome (discharge or death) was recorded.
2.2 Case definition
A case of SRI was defined as any hospitalized child, regardless of symptom duration, in age-defined categories as follows: any child aged 2 days to <3 months with diagnosis of suspected sepsis or physician diagnosed lower respiratory tract infection (LRTI) irrespective of signs and symptoms and, any child ≥3 months to <5 years with physician-diagnosed acute LRTI including bronchiolitis, pneumonia, bronchitis and pleural effusion (9). Surveillance officers administered a questionnaire with demographic information and obtained clinical information from medical records.
2.3 Specimen collection
Nasopharyngeal aspirates (NPA) were collected from all children and induced sputum specimens were collected from individuals aged ≥3 months. If induced sputum collection was contraindicated or was not advised by the attending clinician and a patient was able to expectorate, expectorated sputum was collected. Specimens were transported to the National Institute for Communicable Diseases, Johannesburg, South Africa, for testing and thereafter stored at −80°C.
2.4 Laboratory testing
From January through December 2017, 361 children were enrolled and of these, 198 cases with sufficient specimen volume (200 µl) were included in this study. Total nucleic acids (TNA) were extracted from 200 µl of specimen using the MagNA Pure Compact instrument (Roche Diagnostics, Mannheim, Germany) with Total Nucleic Acid Isolation Kit I according to the manufacturer's instructions and thereafter stored at −20°C.
Real-time PCR was carried out for the detection of 21 viruses and bacteria using a custom-made TaqMan Array Card (TAC) developed from previously published assays (10–12). Pathogens tested, in duplicate, included Mycobacterium tuberculosis, Staphylococcus aureus, S. pneumoniae, H. influenzae, Group B streptococci (GBS), Mycoplasma pneumoniae, Bordetella spp., Legionella spp., Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, adenovirus, human metapneumovirus (hMPV), enterovirus, influenza types A and B, parainfluenza virus types 1–3, RSV, and rhinovirus. TACs included two controls; the internal positive control (IPC) for monitoring of the real-time PCR reaction and a human RNAseP gene control for monitoring of specimen quality.
TAC assays were performed using 50 µl qScript XLT 1-step RT-qPCR Toughmix (Quantabio, Beverly, Massachusetts, USA) and 50 µl of TNA extract. TAC PCR was carried out on the Applied Biosystems ViiA7 and QuantStudio 7 Real Time PCR systems (Life Technologies, New York, USA) using the following cycling conditions: 45°C for 10 min, 94°C for 10 min, 45 cycles of 94°C for 30 s and 60°C for 1 min. A no template control (NTC) and positive control consisting of combined RNA transcripts generated as previously described by Kodani et al. were included on each TAC (13). A positive result was recorded if amplification occurred in at least one of the duplicate reactions with cycle threshold (Ct) <40.
2.5 Statistical analysis
Stata 14 (Stata Corporation, College Station, TX) was used for statistical analysis. McNemar's χ2 test or Fischer's exact test were used, where appropriate, to compare categorical variables (p-value <0.05 was considered statistically significant). Pathogen prevalence was stratified by season in which December—February were classified as summer, March—May as autumn, June—August as winter, and September—November as spring (14).
3 Results
3.1 Study population
From January through December 2017, 361 patients aged <5 years were enrolled in the SRI surveillance program, of which 228 (63%) were diagnosed with LRTI at discharge, 36 (10%) with bronchiolitis while 141 (39%) were diagnosed with other illnesses including bronchitis, tuberculosis, and sepsis. Of the 361 cases enrolled, 198 (55%) were included in this study if at least one respiratory specimen (NPA and/or sputum) were available for testing (Table 1).The median age of patients tested on TAC was 10 months [interquartile range (IQR) 4–21 months], 57% (113/198) were aged <1 year and 89% (177/195) were HIV-uninfected. Antibiotic therapy was initiated in 96% (191/196) of the patients at admission and the in-hospital case-fatality proportion was 1% (2/192).
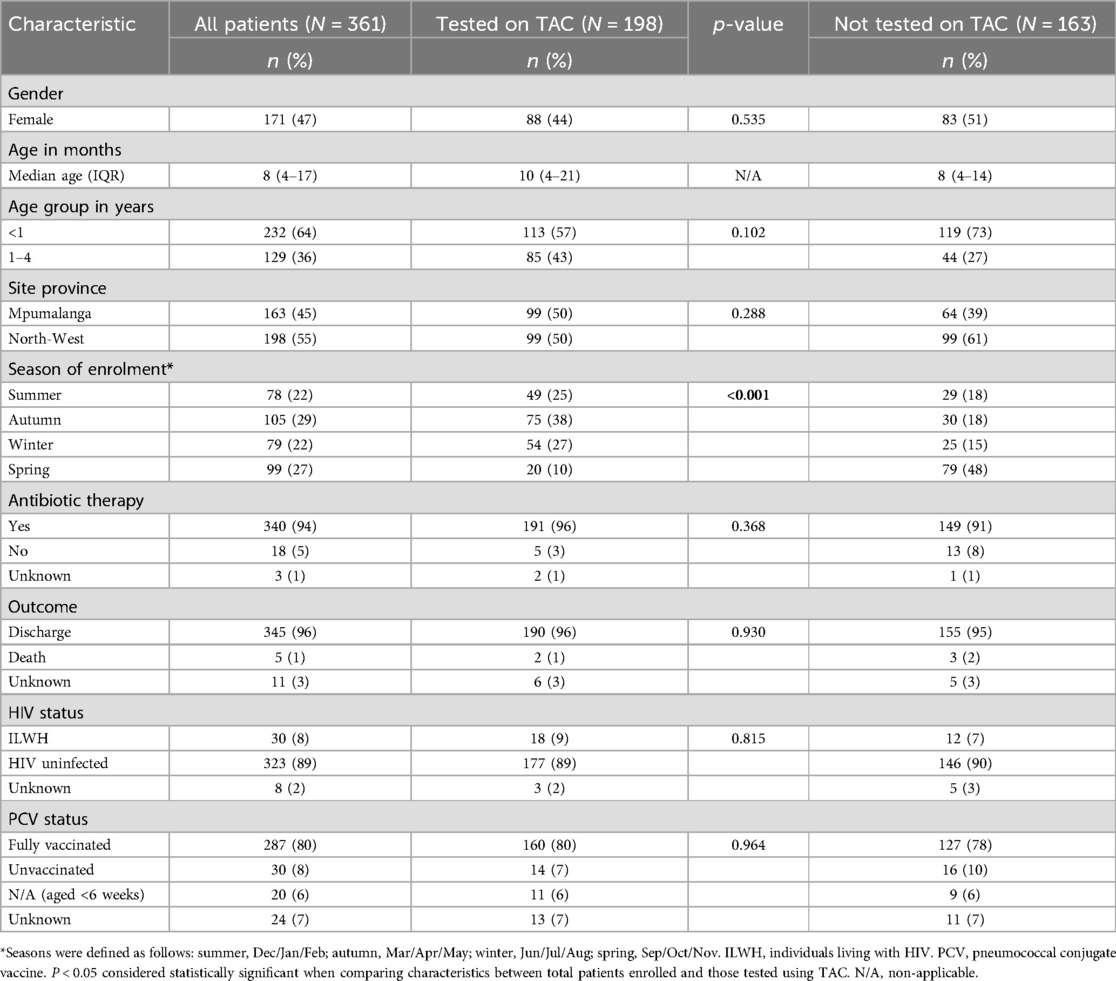
Table 1. Demographic and clinical characteristics of children aged <5 years hospitalized with severe respiratory illness (SRI) at two sentinel sites in South Africa, January–December 2017.
3.2 Prevalence of respiratory pathogens
Among the 198 patients tested, 183 (92%) had a NPA tested and 44 (22%) had sputum tested. Among these, 154 (78%) had a NPA only tested, 15 (8%) sputum only and 29 (15%) had both sputum and NPA tested. Overall, 189/198 (95%) of children tested positive for at least one respiratory pathogen: 89% (177/198) for ≥1 bacterial pathogen and 79% (156/198) for ≥1 viral pathogen. Common viruses identified included rhinovirus (65/198; 33%), RSV (54/198; 27%), adenovirus (34/198; 17%), and enterovirus (28/198; 14%). Common bacteria detected included H. influenzae (121/198; 61%), S. pneumoniae (114/198; 58%), K. pneumoniae (61/198; 31%), S. aureus (52/198; 26%), and A. baumannii (27/198; 14%) (Table 2). Of the patients in which both sputum and NPA specimens were available for testing (n = 29), there was a high congruence in the detection of H. influenzae (91%) and S. pneumoniae (82%) between specimen types (Supplementary Table S1). On the other hand, congruence was moderate to low for the detection of K. pneumoniae (57%), S. aureus (40%), and A. baumannii (20%), which were more commonly detected in NPA compared to sputum.
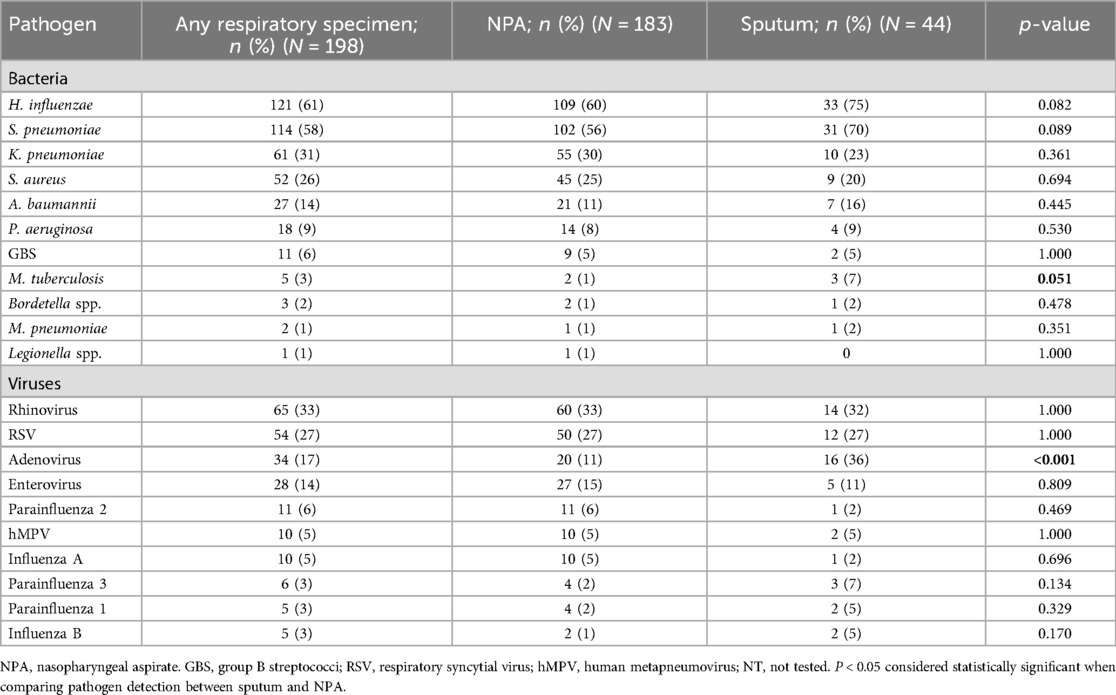
Table 2. Pathogens detected in children aged <5 years hospitalized at two sentinel sites with severe respiratory illness by specimen type, South Africa, January–December 2017.
Comparing pathogen prevalence between the infants aged <1 year and the children aged 1–4 years, RSV [34% [38/113] vs. 19% [16/85]; p = 0.031], A. baumannii [19% [22/113] vs. 6% [5/85]; p = 0.011], and P. aeruginosa [13% [15/113] vs. 4% [3/85]; p = 0.035] were more commonly detected among infants (Figure 1; Table 3). Further, PIV3 (6/113; 5%) and Bordetella spp. (3/113; 3%) were only detected in infants. Adenovirus was more prevalent among the older children [25% [21/85] vs. 12% [13/113]; p = 0.025] and was also more commonly detected in sputum compared to NPA [36% [16/44] vs. 11% [20/183]; p < 0.001] (Tables 2, 3).
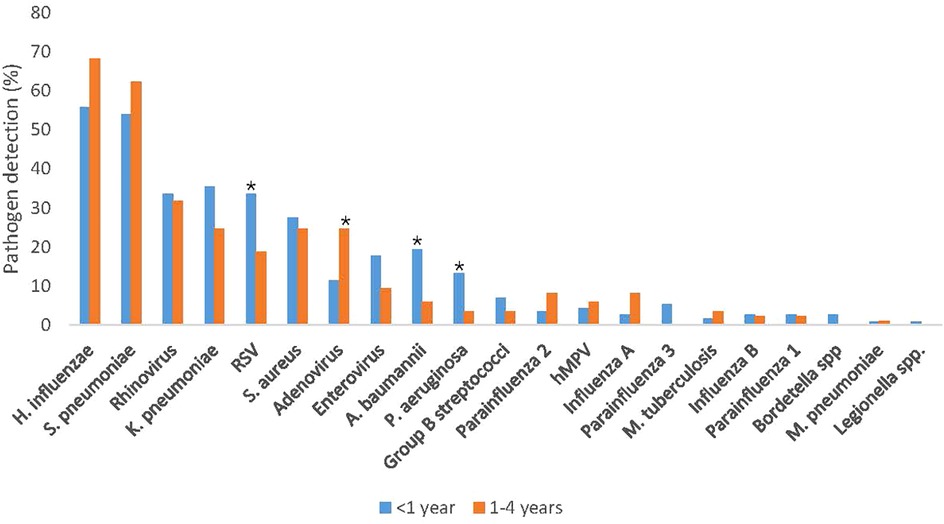
Figure 1. Pathogen detection rate stratified by age, among children aged <5 years hospitalized with severe respiratory illness, January–December 2017 (N = 198). RSV, respiratory syncytial virus. *P < 0.05 considered statistically significant.
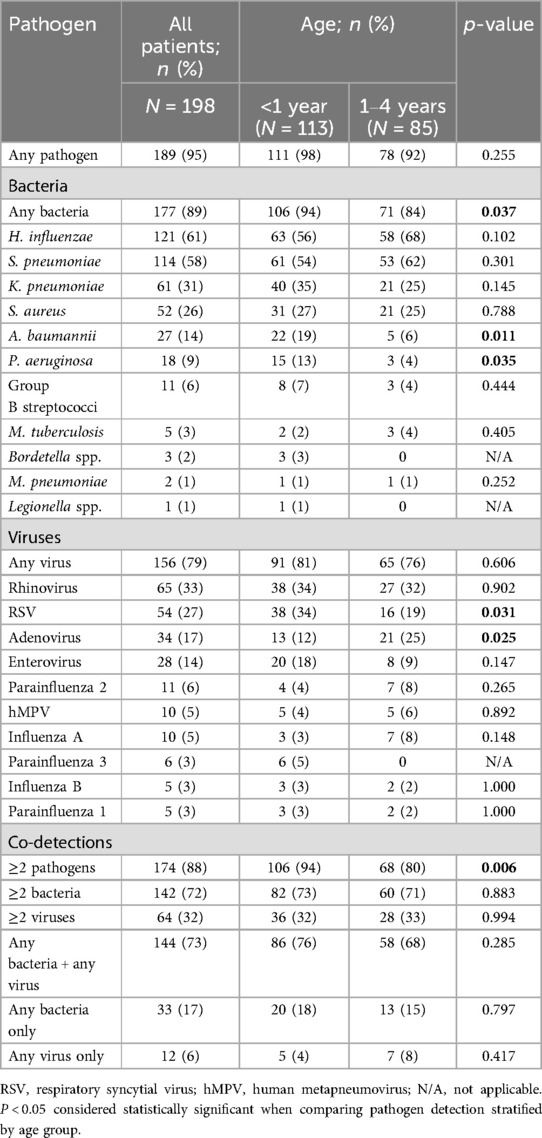
Table 3. Prevalence of bacterial and viral pathogens, stratified by age group, among young children aged <5 years hospitalized with severe respiratory illness, January–December 2017.
Co-detections of ≥2 pathogens were common (174/198; 88%) and were more prevalent among infants compared to older children [94% [106/113] vs. 80% [68/85]; p = 0.006] (Table 3). Most co-detections were bacterial-viral (144/198, 73%) while single viral infections were rare (12/198, 6%). Common bacterial pathogens that were detected as a co-pathogen with a virus were H. influenzae (102/144, 71%), S. pneumoniae (100/144, 69%), K. pneumoniae (43/144, 30%), S. aureus (41/144, 28%), and A. baumannii (20/144, 14%) as was noted in one of the two cases that died. We detected A. baumannii, S. pneumoniae, K. pneumoniae, H. influenzae, P. aeruginosa, S. aureus and human parainfluenza type 3 in a 6-month-old decedent, and only bacteria (S. pneumoniae, K. pneumoniae, P. aeruginosa, and S. aureus) in the second deceased patient (aged 11 months).
The majority (129/198; 65%) of the specimens were collected in the autumn (75/198; 38%) and winter months (54/198; 27%) (Tables 1; Supplementary Table S2). However, overall pathogen detection was the same throughout the seasons (Supplementary Table S2). At individual pathogen level, S. pneumoniae and H. influenzae were more prevalent in winter months, each at 70% (38/54), compared to the other seasons, albeit not statistically significant (p = 0.136 and 0.263, respectively). RSV, on the other hand, was more common in the summer (19/49; 39%) and autumn months (22/75; 29%) compared to the other seasons (p < 001).
4 Discussion
This study reports on the detection of respiratory pathogens, using a multi-pathogen PCR assay, among children aged <5 years hospitalized with SRI in South Africa in 2017. A potential respiratory pathogen was detected in 95% of cases: leading viruses included rhinovirus, RSV, adenovirus, and enteroviruses while the leading bacteria, in both NPA and induced sputum, included H. influenzae, S. pneumoniae and K. pneumoniae as per previous reports (4, 15–19). RSV, A. baumannii, and P. aeruginosa were more prevalent among infants compared to children aged 1–4 years. Bordetella spp. and PIV3 were also only detected in infants but the detection rates were low owing to our small sample size and thus hindering the ability to accurately identify any differences between the age groups.
The high prevalence of RSV (27%) among children aged <5 years hospitalized with SRI is well documented (18, 20). Pretorius et al. previously reported a 25% prevalence of RSV among children aged <5 years hospitalized with severe acute respiratory illness in South Africa between 2012 and 2015 (18). Similar findings for RSV in this age group were also reported in Niger in 2015 (23%) (16), and in the Pneumonia Etiology Research for Child Health (PERCH) study (20%–40%) in The Gambia, Mali, Kenya, Zambia, South Arica, Bangladesh and Thailand between 2011 and 2014 (15). Further, our findings that the prevalence of RSV infection was higher among infants than older children is supported by findings from the PERCH study which reported a RSV prevalence of 40% in this age group compared to 20% in children aged 1–4 years (15). Moyes et al. estimated a mean annual total of 96,220 (95% CI 66,470–132,844) cases of RSV-associated SRI among young children aged <5 years in South Africa between 2011 and 2016. During this period, the burden of RSV disease was the highest among the <1-month age group with RSV-associated deaths highest in the first and second month of life (20). Recently, the FDA has licensed two products which prevent RSV disease in infants, a maternal vaccine and a long-acting monoclonal antibody prophylactic (21). Baral et al. estimated that RSV maternal immunisation has the potential to prevent about 28% to 31% of RSV cases, and 40% to 44% of RSV hospitalizations (22). While a number of these RSV maternal immunisation trials are ongoing, Kampmann et al. reported on the high efficacy of the bivalent RSV prefusion F protein–based (RSVpreF) vaccine administered through maternal immunisation in the 24–36 weeks gestation period; the vaccine was efficacious against severe medically-attended RSV-associated lower respiratory tract illness in the first 3 months of life (23). Further, Mahtab et al. recently identified RSV among the leading pathogens in deaths due to pneumonia in sub-Saharan African and South Asia between 2016 and 2022, particularly in the first 6 months of life (24). These and our findings highlight the urgency for continued efforts towards preventative public health interventions to reduce the burden of RSV-associated SRI, particularly in the first year of life and in low- and middle-income countries where the burden is highest.
In our study, a bacterial pathogen was detected in 89% of the cases and were more prevalent amongst infants compared to children aged 1–4 years. Streptococcus pneumoniae and H. influenzae were the most commonly detected at 58% and 61% prevalence, respectively. This is similar to a previous report from South Africa by Zar et al. (6). In their study, however, they reported similar detection rates between cases and healthy controls thus attributing the high detection rates to carriage.
Pathogen co-detections were common with bacterial and viral co-infections occurring the most frequently, as reported elsewhere (6, 25). Approximately 74% and 70% of cases testing positive for A. baumannii and K. pneumoniae, respectively, were also positive for a virus. Similarly, S. pneumoniae and H. influenzae were co-detected with a virus in 88% and 84% of the cases, respectively. Viral and bacterial co-infections increase the risk of severe disease (26, 27); single infections with a virus were rare (6%) among the SRI cases in our study.
In South Africa, the administration of empiric antibiotic therapy depends on the child's age, among other factors, and can include amoxicillin-clavulanate, ampicillin and gentamicin (8). In our study, 96% of the children received antibiotics on admission; the low prevalence of virus-only infections and high prevalence of bacterial detections whether as a single or a co-pathogen highlight the importance of investigating the interplay between viruses and colonising bacteria in the pathogenesis of SRI.
The main limitation of our study is the lack of healthy controls which would have been useful to determine the etiological fraction of individual pathogens to disease. Studies have reported high carriage rates of S. pneumoniae, S. aureus, K. pneumoniae, H. influenzae, A. baumannii, and P. aeruginosa in the upper respiratory tract, particularly in healthy young children (6, 15, 28–31). Testing of upper respiratory tract samples only may not, therefore, discriminate between colonizing and pathogenic organisms, making it difficult to attribute etiology. Future studies can apply pathogen-specific density thresholds in upper respiratory tract specimens when establishing SRI etiology as some studies have shown a higher median density for some bacteria in cases compared to healthy controls (32, 33). Also, the lack of data from analysis of invasive clinical specimens also limits our ability to discriminate between colonizing and pathogenic organisms as the clinical utility of testing respiratory specimens is in the detection of viruses and atypical pneumonia pathogens. However, viruses including adenovirus and rhinovirus have been detected amongst healthy controls (15) and therefore their roles in disease could not be established. Some case-control studies have shown association of rhinoviruses and enteroviruses with SRI whereas others, such as adenovirus and parainfluenza viruses type 1–3 have shown weak or no attribution to disease (6, 15, 18). Pathogen co-detections were common with bacterial and viral co-infections occurring the most frequently. Primary infection with a virus predisposes an individual to a secondary bacterial infection however, with our study design, we were not able to determine whether one pathogen preceded the other. We were unable to determine seasonality of individual pathogens over longer periods of time as our study period was only one year, we could however describe the prevalence of each pathogen during each season in the one year of the study. Lastly, we do not have data on the number of SRI cases presenting at the study sites and our small sample size only represents 55% of the cases enrolled at the study sites and may therefore not reflect the true distribution of pathogens in these settings and thus limiting our ability to generalize our findings.
5 Conclusion
In our study we show a disproportionately higher number of SRI cases in infants aged <1 year compared to children aged 1–4 years. The rate of bacterial detections was high particularly among infants compared to the children aged 1–4 years. RSV, A. baumannii, and P. aeruginosa were more commonly identified in infants compared to older children, and the majority of the cases tested positive for more than one pathogen of which bacterial-viral co-detections occurred in 73% of all cases tested. The high rate of co-detections requires clinical evaluation to determine their relevance for individual treatment and management decisions. Given the high prevalence of RSV amongst children hospitalized with SRI, particularly in the first year of life, there is an urgent need for continued efforts towards early-life RSV prevention strategies to help reduce RSV-associated morbidity and mortality. These include access to maternal vaccines and monoclonal antibodies particularly in low- and middle-income countries which experience the highest burden of RSV-associated disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Human Research Ethics Committee (HREC) of the University of the Witwatersrand, Johannesburg, South Africa. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
MM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft, Writing – review & editing. CR: Investigation, Methodology, Writing – review & editing. KN: Investigation, Methodology, Writing – review & editing. MD: Conceptualization, Data curation, Investigation, Methodology, Project administration, Writing – review & editing. OH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. OM: Conceptualization, Formal analysis, Investigation, Project administration, Writing – review & editing. SW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing – review & editing. ST: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Writing – review & editing. CC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing. AvG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing. NW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the US Centers for Disease Control and Prevention (Cooperative Agreement Number: U51/IP000155). The funders had no role in study design, data collection, analysis and interpretation, decision to submit the work for publication, or preparation of the manuscript.
Acknowledgments
The authors would like to thank GERMS-SA for the study design, patient enrolment and data collection; the Centre for Respiratory Diseases and Meningitis of the National Institute for Communicable Diseases, a division of the National Health Laboratory Service for laboratory testing and the U.S. CDC for the development and optimization of the TaqMan Array Card assays.
Conflict of interest
CC has received grant support from Sanofi, Advanced Vaccine Initiative, and payment of travel costs from Parexel. NW and AvG have received grant support from Sanofi Pasteur and the Bill and Melinda Gates Foundation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2025.1498197/full#supplementary-material
References
1. Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. (2013) 381(9875):1405–16. doi: 10.1016/S0140-6736(13)60222-6
2. McAllister DA, Liu L, Shi T, Chu Y, Reed C, Burrows J, et al. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob Health. (2019) 7(1):e47–57. doi: 10.1016/S2214-109X(18)30408-X
3. Gutiérrez F, Masiá M, Rodríguez JC, Mirete C, Soldán B, Padilla S, et al. Community-acquired pneumonia of mixed etiology: prevalence, clinical characteristics, and outcome. Eur J Clin Microbiol Infect Dis. (2005) 24(6):377–83. doi: 10.1007/s10096-005-1346-2
4. Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. (2015) 372(9):835–45. doi: 10.1056/NEJMoa1405870
5. le Roux DM, Myer L, Nicol MP, Zar HJ. Incidence and severity of childhood pneumonia in the first year of life in a South African birth cohort: the Drakenstein Child Health Study. Lancet Glob Health. (2015) 3(2):e95–e103. doi: 10.1016/S2214-109X(14)70360-2
6. Zar HJ, Barnett W, Stadler A, Gardner-Lubbe S, Myer L, Nicol MP. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: a nested case-control study of the drakenstein child health study. Lancet Respir Med. (2016) 4(6):463–72. doi: 10.1016/S2213-2600(16)00096-5
7. Moore DP, Baillie VL, Mudau A, Wadula J, Adams T, Mangera S, et al. The etiology of pneumonia in HIV-uninfected South African children: findings from the pneumonia etiology research for child health (PERCH) study. Pediatr Infect Dis J. (2021) 40(9S):S59–68. doi: 10.1097/INF.0000000000002650
8. Zar HJ, Moore DP, Andronikou S, Argent AC, Avenant T, Cohen C, et al. Diagnosis and management of community-acquired pneumonia in children: south African thoracic society guidelines. Afr J Thorac Crit Care Med. (2020) 26(3):10.7196/AJTCCM.2020.v26i3.104. doi: 10.7196/AJTCCM.2020.v26i3.104
9. Moleleki M, du Plessis M, Ndlangisa K, Reddy C, Hellferscee O, Mekgoe O, et al. Pathogens detected using a syndromic molecular diagnostic platform in patients hospitalized with severe respiratory illness in South Africa in 2017. Int J Infect Dis. (2022) 122:389–97. doi: 10.1016/j.ijid.2022.06.011
10. Diaz MH, Waller JL, Napoliello RA, Islam MS, Wolff BJ, Burken DJ, et al. Optimization of multiple pathogen detection using the TaqMan array card: application for a population-based study of neonatal infection. PLoS One. (2013) 8(6):e66183. doi: 10.1371/journal.pone.0066183
11. Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. (2011) 49(6):2175–82. doi: 10.1128/JCM.02270-10
12. Velaphi SC, Westercamp M, Moleleki M, Pondo T, Dangor Z, Wolter N, et al. Surveillance for incidence and etiology of early-onset neonatal sepsis in Soweto, South Africa. PLoS One. (2019) 14(4):e0214077. doi: 10.1371/journal.pone.0214077
13. Kodani M, Winchell JM. Engineered combined-positive-control template for real-time reverse transcription-PCR in multiple-pathogen-detection assays. J Clin Microbiol. (2012) 50(3):1057–60. doi: 10.1128/JCM.05987-11
14. van der Walt AJ, Fitchett JM. Statistical classification of South African seasonal divisions on the basis of daily temperature data. S Afr J Sci. (2020) 116:1–15. doi: 10.17159/sajs.2020/7614
15. Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet. (2019) 394(10200):757–79. doi: 10.1016/S0140-6736(19)30721-4
16. Lagare A, Ousmane S, Dano ID, Issaka B, Issa I, Mainassara HB, et al. Molecular detection of respiratory pathogens among children aged younger than 5 years hospitalized with febrile acute respiratory infections: a prospective hospital-based observational study in Niamey, Niger. Health Sci Rep. (2019) 2(11):e137. doi: 10.1002/hsr2.137
17. Cohen C, Walaza S, Moyes J, Groome M, Tempia S, Pretorius M, et al. Epidemiology of viral-associated acute lower respiratory tract infection among children <5 years of age in a high HIV prevalence setting, South Africa, 2009–2012. Pediatr Infect Dis J. (2015) 34(1):66–72. doi: 10.1097/INF.0000000000000478
18. Pretorius MA, Tempia S, Walaza S, Cohen AL, Moyes J, Variava E, et al. The role of influenza, RSV and other common respiratory viruses in severe acute respiratory infections and influenza-like illness in a population with a high HIV sero-prevalence, South Africa 2012–2015. J Clin Virol. (2016) 75:21–6. doi: 10.1016/j.jcv.2015.12.004
19. Ning G, Wang X, Wu D, Yin Z, Li Y, Wang H, et al. The etiology of community-acquired pneumonia among children under 5 years of age in mainland China, 2001–2015: a systematic review. Hum Vaccin Immunother. (2017) 13(11):2742–50. doi: 10.1080/21645515.2017.1371381
20. Moyes J, Tempia S, Walaza S, McMorrow ML, Treurnicht F, Wolter N, et al. The burden of RSV-associated illness in children aged <5 years, South Africa, 2011 to 2016. BMC Med. (2023) 21(1):139. doi: 10.1186/s12916-023-02853-3
21. Hodgson D, Wilkins N, van Leeuwen E, Watson CH, Crofts J, Flasche S, et al. Protecting infants against RSV disease: an impact and cost-effectiveness comparison of long-acting monoclonal antibodies and maternal vaccination. The Lancet Reg Health Eur. (2024) 38:100829. doi: 10.1016/j.lanepe.2023.100829
22. Baral R, Li X, Willem L, Antillon M, Vilajeliu A, Jit M, et al. The impact of maternal RSV vaccine to protect infants in gavi-supported countries: estimates from two models. Vaccine. (2020) 38(33):5139–47. doi: 10.1016/j.vaccine.2020.06.036
23. Kampmann B, Madhi SA, Munjal I, Simões EAF, Pahud BA, Llapur C, et al. Bivalent prefusion F vaccine in pregnancy to prevent RSV illness in infants. N Engl J Med. (2023) 388(16):1451–64. doi: 10.1056/NEJMoa2216480
24. Mahtab S, Blau DM, Madewell ZJ, Ogbuanu I, Ojulong J, Lako S, et al. Post-mortem investigation of deaths due to pneumonia in children aged 1–59 months in sub-Saharan Africa and South Asia from 2016 to 2022: an observational study. Lancet Child Adolesc Health. (2024) 8(3):201–13. doi: 10.1016/S2352-4642(23)00328-0
25. Bezerra PGM, Britto MCA, Correia JB, Duarte M, Fonceca AM, Rose K, et al. Viral and atypical bacterial detection in acute respiratory infection in children under five years. PLoS One. (2011) 6(4):e18928. doi: 10.1371/journal.pone.0018928
26. Voiriot G, Visseaux B, Cohen J, Nguyen LB, Neuville M, Morbieu C, et al. Viral-bacterial coinfection affects the presentation and alters the prognosis of severe community-acquired pneumonia. Crit Care. (2016) 20(1):375. doi: 10.1186/s13054-016-1517-9
27. Cawcutt KA, Kalil AC. Viral and bacterial co-infection in pneumonia: do we know enough to improve clinical care? Critical Care. (2017) 21(1):19. doi: 10.1186/s13054-016-1592-y
28. Pan H, Cui B, Huang Y, Yang J, Ba-Thein W. Nasal carriage of common bacterial pathogens among healthy kindergarten children in Chaoshan region, Southern China: a cross-sectional study. BMC Pediatr. (2016) 16(1):161. doi: 10.1186/s12887-016-0703-x
29. Adegbola RA, DeAntonio R, Hill PC, Roca A, Usuf E, Hoet B, et al. Carriage of streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS One. (2014) 9(8):e103293. doi: 10.1371/journal.pone.0103293
30. Navne JE, BØRresen ML, Slotved HC, Andersson M, Melbye M, Ladefoged K, et al. Nasopharyngeal bacterial carriage in young children in Greenland: a population at high risk of respiratory infections. Epidemiol Infect. (2016) 144(15):3226–36. doi: 10.1017/S0950268816001461
31. Farida H, Severin JA, Gasem MH, Keuter M, van den Broek P, Hermans PW, et al. Nasopharyngeal carriage of Klebsiella pneumoniae and other gram-negative bacilli in pneumonia-prone age groups in Semarang, Indonesia. J Clin Microbiol. (2013) 51(5):1614–6. doi: 10.1128/JCM.00589-13
32. Claassen-Weitz S, Lim KYL, Mullally C, Zar HJ, Nicol MP. The association between bacteria colonizing the upper respiratory tract and lower respiratory tract infection in young children: a systematic review and meta-analysis. Clin Microbiol Infect. (2021) 27(9):1262–70. doi: 10.1016/j.cmi.2021.05.034
Keywords: severe respiratory illness, community-acquired pneumonia, TaqMan Array Card, childhood pneumonia, South Africa, respiratory pathogens, RSV
Citation: Moleleki M, Reddy C, Ndlangisa K, du Plessis M, Hellferscee O, Mekgoe O, Walaza S, Tempia S, Cohen C, von Gottberg A and Wolter N (2025) Respiratory pathogens detected in children aged <5 years hospitalized with severe respiratory illness, South Africa, 2017. Front. Pediatr. 13:1498197. doi: 10.3389/fped.2025.1498197
Received: 19 September 2024; Accepted: 12 June 2025;
Published: 27 June 2025.
Edited by:
Wei Shi, Capital Medical University, ChinaReviewed by:
Shubhada Bopegamage, Slovak Medical University, SlovakiaBernard E. Ebruke, International Foundation Against Infectious Disease in Nigeria, Nigeria
Copyright: © 2025 Moleleki, Reddy, Ndlangisa, du Plessis, Hellferscee, Mekgoe, Walaza, Tempia, Cohen, von Gottberg and Wolter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole Wolter, bmljb2xld0BuaWNkLmFjLnph
†Deceased
 Malefu Moleleki
Malefu Moleleki Cayla Reddy
Cayla Reddy Kedibone Ndlangisa1,2
Kedibone Ndlangisa1,2 Orienka Hellferscee
Orienka Hellferscee Nicole Wolter
Nicole Wolter