- 1Shaanxi Provincial Key Laboratory of Infection and Immune Diseases, Shaanxi Provincial People’s Hospital, Xi’an, Shaanxi, China
- 2Shaanxi Engineering Research Center of Cell Immunology, Shaanxi Provincial People’s Hospital, Xi’an, Shaanxi, China
- 3Department of Pediatrics, Shaanxi Provincial People’s Hospital, Xi’an, Shaanxi, China
- 4Department of Graduate School, Yan’an University, Yan’an, Shaanxi, China
Background: Kawasaki disease is the leading cause of acquired heart disease in children, yet timely diagnosis remains difficult due to overlapping symptoms with other febrile illnesses.
Methods: In a retrospective case–control study of 38 children with Kawasaki disease and 44 febrile controls, we measured hematological parameters and C-reactive protein (CRP) using standardized analyzers and profiled seven serum microRNAs by qRT-PCR. Biomarkers showing significant differences were used to build logistic regression models with a 70/30 train–test split, and diagnostic accuracy was assessed by receiver operating characteristic analysis. Functional enrichment of miRNA targets was explored using network analysis.
Results: CRP and three microRNAs (miR-223-3p, miR-19a-3p, miR-18a-5p) were significantly elevated in Kawasaki disease. Individually, these markers achieved strong discrimination (AUC: 0.846–0.986), while their combination yielded an AUC of 0.990, sensitivity 1.000, and specificity 0.923. The three microRNAs were positively correlated and enriched for pathways including p53 signaling and cell cycle regulation, with KCNQ1OT1 identified as a shared lncRNA interactor.
Conclusion: Integrating CRP with a concise serum miRNA panel demonstrates promising discriminatory potential for Kawasaki disease vs. other febrile illnesses and suggests mechanistic involvement of p53-associated pathways, supporting future validation in larger, independent cohorts.
Introduction
Kawasaki Disease (KD), clinically referred to as mucocutaneous lymph node syndrome, is an acute febrile illness primarily affecting children under the age of 5 rears, with etiology yet to be fully understood (1). It is the leading cause of acquired heart disease in children, with approximately 25% of untreated cases developing coronary artery lesions (CAL), potentially leading to myocardial infarction or sudden death (2). KD affects numerous ethnic groups globally, with East Asian countries having the highest incidence, notably China (3). Currently, KD diagnosis relies heavily on clinical features, with no confirmatory laboratory test available to date (4). However, resembling clinical features of KD are also presenting in many other childhood infectious febrile illnesses (5, 6), such as viral infections and scarlet fever (7). Consequently, discriminating KD from these conditions can pose a significant diagnostic challenge. Misdiagnosis can result in treatment delays, substantially elevating the risk of developing CAL and associated morbidity and mortality. Early intravenous immunoglobulin (IVIG) administration can reduce CAL risk to 5% (8). Therefore, there is an urgent demand for developing a more specific, sensitive, accurate, reproducible, and rapid biochemical test to facilitate a definitive diagnosis and timely initiation of appropriate therapeutic interventions.
MicroRNAs (miRNAs) are small non-coding RNA molecules, usually 18–25 nucleotides in length that play a critical role in regulating cellular and immune functions through post-transcriptional modifications (9). MiRNAs are implicated in various pathological abnormalities, ranging from cancer (10) to adult and pediatric cardiovascular diseases (11–13). Specific to KD, miRNAs have emerged as potential diagnostic biomarkers and critical players in disease pathogenesis. Initial studies by Shimizu et al. highlighted the differential expression of miRNAs in acute KD, notably miR-145, which influences TGF-β signaling in coronary arteries (14). Subsequent research has expanded our understanding of miRNAs in KD diagnosis, pathogenesis, and treatment (15). For instance, Yun et al. found that miR-200c and miR-371-5p upregulation could be implicated in KD pathogenesis by modulating the inflammatory response (16). John et al. reported a distinctive circulating miRNA profile in KD, including miR-210-3p, miR-184, and miR-19a-3p, which are positively correlated. Moreover, Jia et al. characterized a set of four serum exosomal miRNAs, miR1246, miR-4436b-5p, miR-197-3p, and miR-671-5p, that are capable of differentiating KD patients from healthy individuals (17). Furthermore, bioinformatic analysis has led to specific microRNAs such as mir-126-3p, mir-375, and mir-146a-5p being identified as potential biomarkers for KD (18). Recent investigations have identified ten miRNAs holding substantial promise as valuable biomarkers for KD and may participate in KD pathogenesis via regulation of the TGF-β signaling pathway (19). Despite significant progress, a universally accepted miRNA-based diagnostic approach for KD has yet to be established in clinical practice, reflecting the ongoing need for research in this area.
In this study, we aimed to assess the efficacy of a novel multi-biomarker test in distinguishing KD from non-KD febrile controls. Our focus was on a combination of conventional and novel biomarkers: two widely recognized inflammatory markers, C-reactive protein (CRP) and platelet count (PLT), along with seven miRNAs (miR-223-3p, miR-19a-3p, miR-18a-5p, miR-21-5p, miR-155-5p, miR-145, and miR-146a-5p) identified for their involvement in KD. CRP and PLT are established nonspecific indicators of inflammation and infection (20, 21), with documented elevations in acute KD cases (22, 23). However, while CRP demonstrates high sensitivity, its limited disease specificity and frequent elevation in various viral or bacterial infections reduce its standalone diagnostic utility. PLT reflects systemic inflammation and thrombocytosis, commonly observed in KD after day 5, but also lacks disease specificity. In contrast, circulating miRNAs provide molecular-level insight into disease-specific immunopathological responses and may complement conventional markers by capturing regulatory signatures unique to KD. Therefore, we hypothesized that integrating CRP and PLT with selected miRNAs could enhance diagnostic accuracy by leveraging both systemic inflammatory markers and post-transcriptional regulatory signals. This approach aims to improve diagnostic precision in real-world cases where clinical presentations often overlap between KD and other febrile illnesses. Among the seven selected candidate miRNAs, miR-223-3p (24), miR-19a-3p (25), miR-18a-5p (26), miR-145 exhibit an elevated pattern in KD, while miR-21-5p and miR-155-5p were shown to be downregulated (27). MiR-146a-5p has been pinpointed as a direct target of differentially expressed genes (DEGs) in KD through network analysis, and miR-146a gene polymorphisms have been associated with KD susceptibility (28).
We conducted a retrospective case–control analysis involving 38 blood samples from patients diagnosed with KD and 44 samples from febrile control children to evaluate the expression levels of CRP, PLT, and selected serum miRNAs. Beyond confirming the individual discriminatory value of each biomarker, we demonstrated that their combined application significantly enhances diagnostic performance. Additionally, we investigated the interrelationships among these biomarkers and performed functional enrichment analyses to explore their involvement in molecular pathways relevant to KD pathophysiology, aiming to provide further insight into their clinical utility and mechanistic significance.
Materials and methods
Patients and clinical data
We retrospectively analyzed the medical records of children admitted to Shaanxi Provincial People's Hospital (China) between June 2022 and June 2024 with a diagnosis of Kawasaki disease (KD) or other febrile illnesses. A total of 42 children with KD and 46 with febrile illnesses were initially considered. However, 4 KD patients and 2 febrile patients were excluded due to withdrawal from the study. The final cohort included 38 KD patients (16 with complete KD and 22 with incomplete KD) and 44 febrile controls. All patients in the KD group met the diagnostic criteria established by the American Heart Association for complete or incomplete KD.
The children in the non-KD febrile control cohort have been diagnosed with a range of illnesses, such as infantile exanthem (6 cases), urticaria (7 cases), acute pharyngitis (5 cases), acute purulent tonsillitis (4 cases), conjunctivitis (5 cases), sepsis (3 cases), various viral (including enterovirus, 6 cases) and bacterial infections (8 cases). During this time, all of the individuals' age, sex, days of fever duration, Hemoglobin, Red blood cell count (RBC), White blood cell count (WBC), Neutrophil count, PLT and CRP were gathered.
The Shaanxi Provincial People's Hospital Ethics Committee evaluated and approved this study (No. 2022114). Prior to sample collection, the patient's parents or legal guardians provided written informed consent. Every experiment was carried out in accordance with the rules and regulations that were in effect at the time.
The following criteria were established for the KD cohort's inclusion:
I. The patient meets the diagnostic criteria for KD, as defined by the American Heart Association (28). This includes having a fever that is higher than 38.0°C and lasts for more than five days. Additionally, the patient must exhibit at least two of the following clinical features: polymorphous rash, bilateral conjunctivitis, cervical lymphadenopathy, diffuse oral mucosal congestion, and edema or erythema of the hands or feet;
II. First-time incidence of the disease;
III. Age under 14;
IV. Full clinical data;
V. Children's guardians have informed consent to this study.
The following were established as the KD cohort's exclusion criteria:
I. Pre-admission treatment with immunoglobulin, aspirin, or other medications;
II. Pre-existing conditions such as congenital heart disease, myocarditis, arrhythmias, and systemic diseases affecting organs like the liver, kidneys, lungs, and malignancies, hematological disorders, and genetic metabolic diseases;
III. Accompanied with autoimmune diseases;
IV. Subjects who withdrew midway.
Sample collection
Peripheral venous blood (PVB) samples were collected from KD and febrile control patients within 24 h of inpatient admission (i.e., on illness days 5–7), and prior to treatment with intravenous immunoglobulin, aspirin, or other disease-specific interventions. PLT, hemoglobin, RBC, WBC and neutrophils were measured by the Sysmex XN-2000 Hematology Analyzer using the impedance flow cytometry method (Sysmex et al., Japan). CRP levels were assessed using an automatic specific protein analyzer using the nephelometric method (PA-990; Lifotronic, Shenzhen, China). All procedures followed the operating protocols and Standard Operating Procedures (SOP) documentation of the reagent and instrument manufacturers.
miRNA isolation and qRT-PCR
Total RNA from serum samples was extracted with the RNeasy plus mini kit (Qiagen), and the RNA concentration was quantified using a Nanodrop 3000 Spectrophotometer (Thermo et al., USA). MiRNAs were reverse transcribed to cDNA with the PrimeScript RT reagent kit (TaKaRa, Dalian, China), and then detected using specific primers synthesized by Sangon Biotech (Shanghai, China) in an ABI 7500 Real-Time PCR System 3 (Applied Biosystems, Foster City, CA), with U6 being the internal control. The expression of miRNAs was calculated and represented using the 2−ΔΔCT method. Quantitative RT-PCR primer sequences were in Table 1.
Construction of miRNA interaction network and functional analysis
miRNet 2.0 tool was used to map the documented interactions between miRNAs and target genes or lncRNAs, and to perform functional enrichment analysis (29). miRNet 2.0 is an interactive platform integrating major databases to elucidate and construct networks between and among experimentally validated target genes, proteins, lncRNAs, circRNAs, and sncRNAs. In miRNet 2.0, the human peripheral blood-specific miRNA-gene interaction data was extracted from latest miRNA annotation databases including miR Base (30), miTarBase (31), DIANA-TarBase (32), TSmiR (33) and IMOTA (34). The miRNA-lncRNA interactions were collected from starBase (35). Functional enrichment analysis was performed using GO and KEGG pathways, implementing hypergeometric tests and empirical sampling enrichment algorithms. The visual analytics was enhanced by methods including the prize-colleting Steiner Forest (PCSF) algorithm. The backbone layout was used in visual representation to effectively reveal hidden patterns in large networks and emphasize the most embedded edges (36). It was manually built by Cytoscape.
Statistical analysis
Data collected for demographic and clinical characteristics was assessed for Gaussian distribution with the Shapiro–Wilk test. Variations in age, PLT and all miRNAs among KD patients and other febrile control cohorts were presented as median values and interquartile ranges (IQR), and the intergroup comparisons were performed using Mann–Whitney U test. Variations in days of fever duration, hemoglobin (g/L), RBC count, WBC count (×109/L), neutrophil count (×109/L) and CRP were presented as mean ± SD, and the intergroup comparisons were performed using unpaired t-test. Variations in sex were presented as frequency (n) and percentage (%), and the intergroup comparisons were performed using Fisher's exact test. To account for multiple comparisons and control the family-wise error rate, the Holm-Šídák correction was applied to all p-values obtained from the statistical tests. This method was chosen due to its balance between controlling Type I error and maintaining statistical power, especially in the context of multiple hypothesis testing involving the evaluation of several biomarkers. P-values were ranked, and adjusted significance levels were calculated accordingly for each test. Adjusted p-values are reported throughout the manuscript, and updated figures reflect these adjustments. Outliers were winsorized at the values of three times the median absolute deviation or greater than the upper limit of quantification. Only variables that demonstrated statistical significance were recruited in the following assessments.
Multiple logistic regression with backward selection was used to assess the predictive value of the candidate biomarkers (individual and combined) for the differentiation of KD vs. other febrile diseases. Data were randomly split into training and test sets to prevent overfitting and to validate the diagnostic models. The training set comprised 70% of the total dataset, while the remaining 30% constituted the test set. This split was performed to ensure that the diagnostic models were trained and tested on independent samples. The model was trained using the training dataset, optimizing for maximum Youden's J statistic to identify the optimal cut-off points for each biomarker. The trained logistic regression models were then validated using the independent test set. Receiver operating characteristic (ROC) curves were generated for each biomarker and the combined model using the test set to evaluate their diagnostic performance. The area under the ROC curve (AUC) was calculated to assess the overall ability of the biomarkers to differentiate between KD and controls. The optimal cut-off values, sensitivity, specificity, and maximum Youden's J statistics were also calculated to determine the best threshold for KD diagnosis. Confidence intervals for AUCs were computed using DeLong's method to assess the statistical significance of the diagnostic performance.
Pearson's correlation coefficient was applied to determine the relationships between candidate biomarkers. A p < 0.05 was considered to be statistically significant.
GraphPad Prism version 10.0 (GraphPad Software, San Diego, California, USA) and RStudio Team (RStudio: Integrated Development for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/, 2020) was used for all statistical analysis. The pROC package was used for ROC analysis.
Results
Patient characteristics
Demographic and clinical characteristics were recorded and analyzed for the KD patients and febrile controls enrolled in this study. The baseline characteristics, containing Age, Sex, days of fever duration, Hemoglobin, RBC, WBC, Neutrophil count, and CRP were not significantly different between the KD and non-KD febrile cohorts, but a significant difference was shown in CRP between the two cohorts, with KD patients demonstrating a higher level of CRP (Table 2).
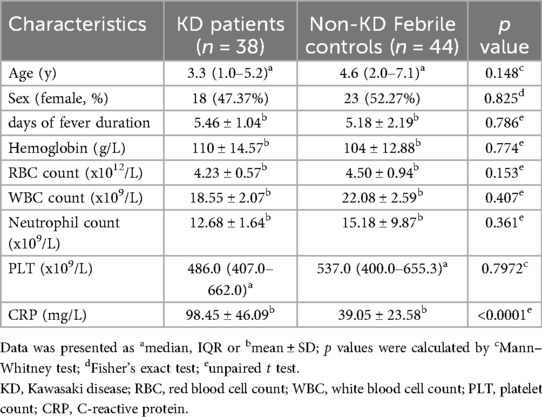
Table 2. Demographic and clinical characteristics of KD patients and non-KD febrile controls cohorts.
CRP and miRNAs differentiate patients with KD from those with non-KD febrile diseases
First, we aimed to determine the differences between these selected candidate biomarkers in KD patients and non-KD febrile control cohorts. Seven candidate miRNAs were evaluated for their relative expression using the qRT-PCR method. A total of nine candidate biomarkers (including PLT and CRP) were analyzed to test statistical significance between the two cohorts (Figure 1). For all analyses involving multiple comparisons, the p-values were adjusted using the Holm-Šídák correction to control for multiple testing. The revised p-values are reported in the corresponding sub-figures. Among these biomarkers, 4 out of 9 demonstrate significant differences, including one conventional protein biomarker (CRP) (Figure 1A) and three miRNAs (miR-223-3p, miR-19a-3p and miR-18a-5p) (Figures 1C–E), of which an elevated pattern was demonstrated in KD patients compared to patients with other febrile diseases. These four potential biomarkers were then recruited in the following assessments. No statistical difference was found in PLT levels (Figure 1B) or relative expression of miR-21-5p, miR-155-5p, miR-145, or miR-146a-5p (Figures 1F–I).
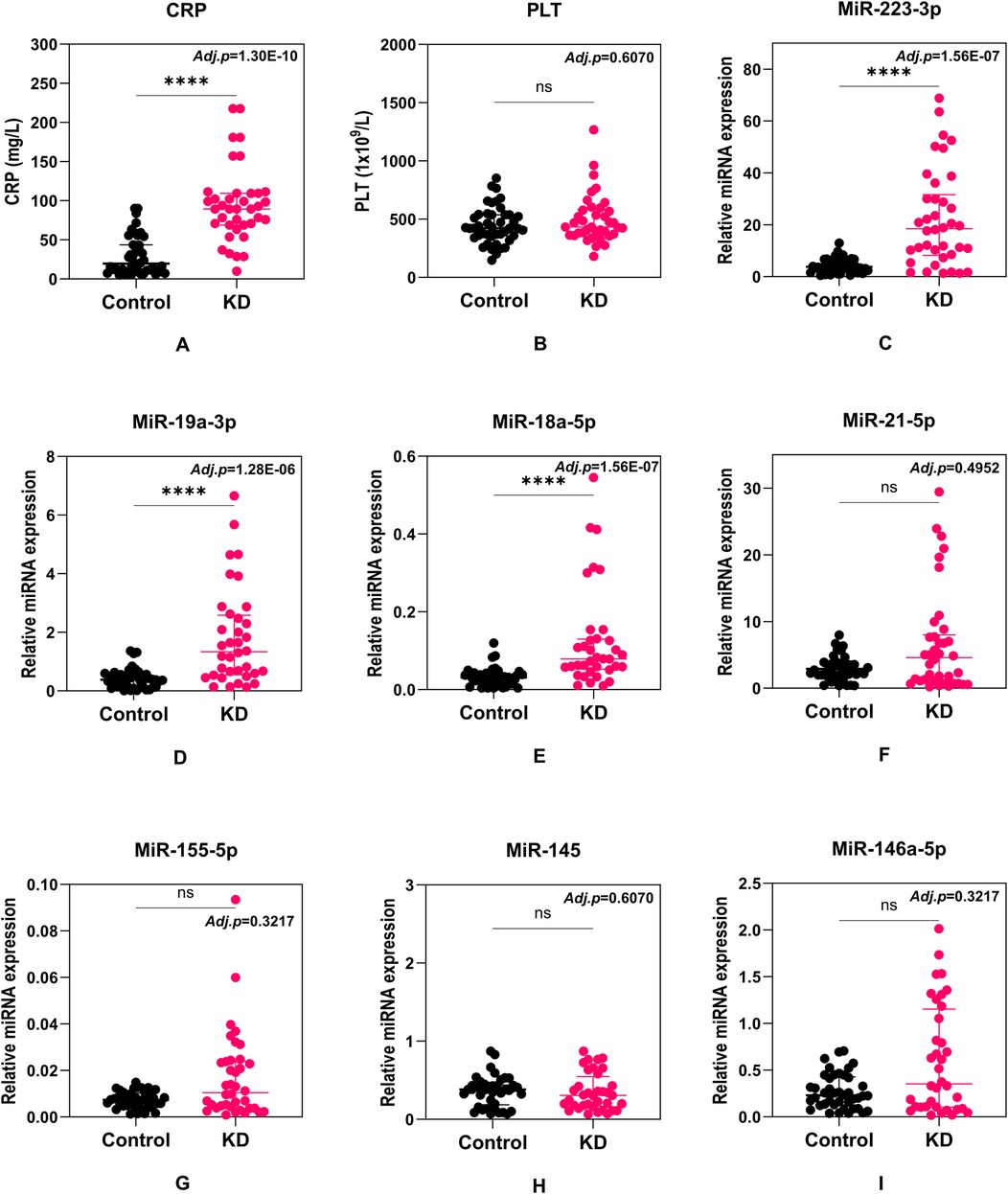
Figure 1. Comparison of C-reactive protein (CRP) (A), platelet count (PLT) (B), miR-223-3p (C), miR-19a-3p (D), miR-18a-5p (E), miR-21-5p (F), miR-155-5p (G), miR-145 (H), and miR-146a-5p (I) levels between KD and non-KD febrile cohorts. Each dot represents a unique patient. Median values and interquartile ranges are shown in the figures. Statistical differences were calculated with the Mann–Whitney U test. Adjusted p values were calculated using the Holm-Šídák test and shown on the figures.
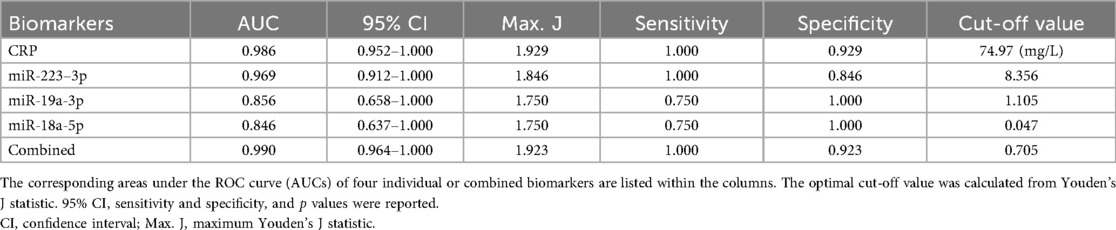
We then evaluated the performance of these four selected potential biomarkers in discriminating patients with KD from those with non-KD febrile diseases. Receiver operator characteristic (ROC) curves were plotted for each biomarker (Figure 2). By training the diagnostic models on a separate dataset and validating them with an independent test set, we ensured robust evaluation of the biomarkers' diagnostic capabilities, mitigating the risk of overfitting and providing more realistic estimates of model performance. The ROC analysis demonstrated that all biomarkers, including CRP, miR-223-3p, miR-19a-3p, miR-18a-5p, and their combined model, had significant diagnostic ability in differentiating KD from non-KD febrile controls (Table 3). CRP exhibited the highest individual diagnostic performance with an AUC of 0.986 (95% CI: 0.952–1.000), a sensitivity of 1.000, a specificity of 0.929, and an optimal cut-off value of 74.97 mg/L. MiR-223-3p followed with an AUC of 0.969 (95% CI: 0.912–1.000), a sensitivity of 1.000, and a specificity of 0.846. The optimal cut-off value for miR-223-3p was 8.356. MiR-19a-3p and miR-18a-5p displayed moderate diagnostic capabilities with AUCs of 0.856 (95% CI: 0.658–1.000) and 0.846 (95% CI: 0.637–1.000), respectively. Both had sensitivities of 0.750 and specificities of 1.000 at cut-off values of 1.105 and 0.047, respectively. The combined model of all biomarkers showed the highest overall diagnostic performance with an AUC of 0.990 (95% CI: 0.964–1.000), sensitivity of 1.000, specificity of 0.923, and an optimal cut-off value of 0.705. This exploratory diagnostic approach, integrating conventional biomarkers with miRNA profiling, demonstrates promising discriminatory potential in differentiating KD from other febrile illnesses. While the model's performance was high in internal validation, its application remains preliminary due to the limited sample size and warrants further evaluation in larger, independent cohorts.
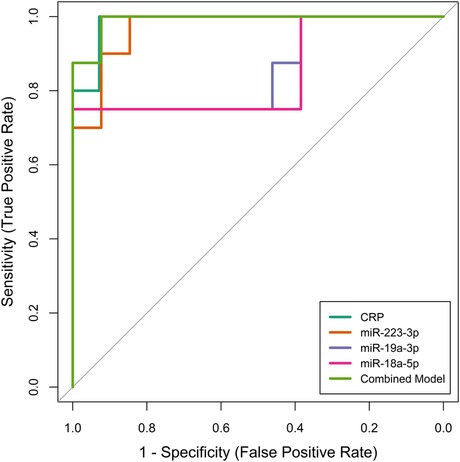
Figure 2. Receiver operating characteristic (ROC) curves illustrating the diagnostic performance of individual biomarkers (CRP, miR-223-3p, miR-19a-3p, miR-18a-5p) and their combined model in differentiating patients with kawasaki disease (KD) from those with non-KD febrile diseases. The ROC curves were generated using an independent test set to validate the diagnostic accuracy of each biomarker and the combined model.
We then examined the potential relationships between these potential biomarkers using Pearson's coefficient analysis. Notably, the relative expression of miR-223-3p, miR-19a-3p, and miR-18a-5p were highly consistent and positively correlated (all p < 0.05) (Figure 3).
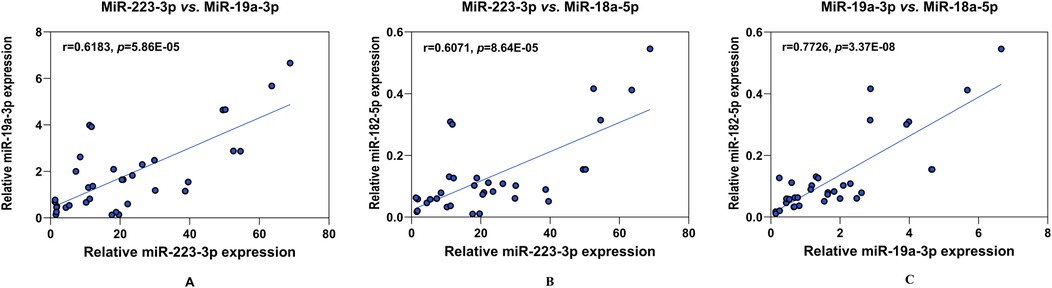
Figure 3. Analysis of the correlation between the four potential biomarkers of KD. The positive paired correlation of the relative expression is shown between miR-223-3p and miR-19a-3p (A); miR-223-3p and miR-18a-5p (B); and miR-19a-3p and miR-18a-5p (C), respectively, using Pearson's coefficient analysis (all, p < 0.05). CRP is not corelated with any miRNAs (data not shown). r, Spearman's rank correlation coefficient.
MiRNA interaction network and functional analysis
To investigate the functions of the identified candidate miRNAs (miR-223-3p, miR-19a-3p and miR-18a-5p) shown to be discriminative of KD and other febrile diseases and how these functions are implicated in the development and progression of KD, we first utilized the miRnet 2.0 tool to obtain downstream gene targets of these miRNAs. The human peripheral blood-specific miRNA-gene interaction data was extracted from central databases, including miTarBase v8.0, DIANA-TarBase v8.0, TSmiR, and IMOTA.
A total of 2,714 gene targets was identified to be interacting with all three miRNAs (Supplementary Table S1), among which 14 were commonly shared, being F3, TWF1, TP53, ZNF460, GFPT1, MSMO1, WASL, SECISBP2l, TMEM64, STMN1, LIF, MDM2, XPO1 and POU2F1 (Figure 4A). These gene targets were validated from multiple methods such as CLASH, next-generation sequencing, PAR-CLIP, luciferase reporter assay and qRT-PCR.
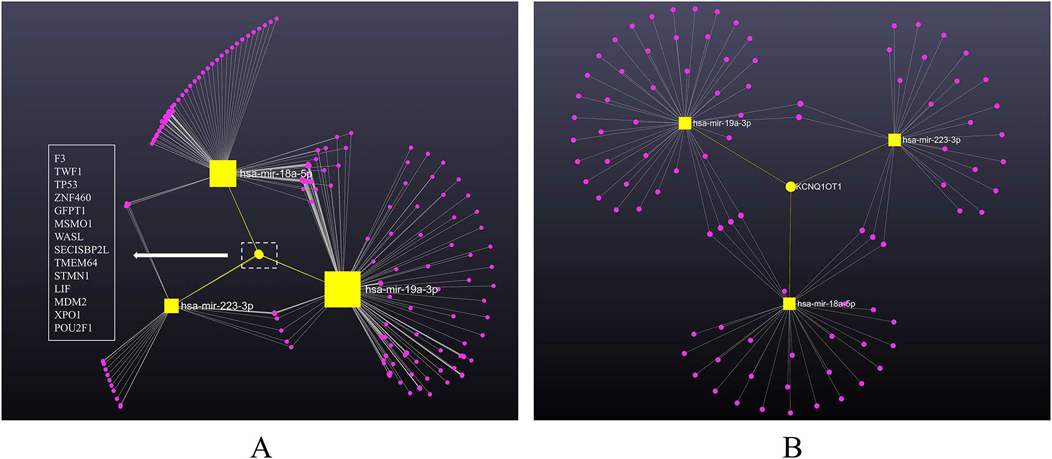
Figure 4. MiRNA-gene/lncRNA interaction network showing the genes associated with three miRNAs, constructed using cytospace. MiRNAs are labeled and shown in yellow squares sized by the number of interactions. (A) MiRNA-gene network. The target genes are shown in pink dots. Edges between nodes indicate miRNA-gene interaction. The yellow dot represents the identified 14 commonly shared gene targets of the miRNAs, listed in the left panel. (B) MiRNA-lncRNA network. The target lncRNAs are shown in pink dots. Edges between nodes indicate miRNA-lncRNA interaction. The yellow dot represents the identified commonly shared lncRNA target (LncRNA KCNQ1OT1) of the miRNAs.
To expand the network further, we integrated starBase to include lncRNAs. According to miRnet 2.0, 94 lncRNAs was identified as directly linked to the three miRNAs (Supplementary Table S2). Intriguingly, lncRNA KCNQ1OT1 was the only lncRNA highlighted to be commonly shared by all three miRNAs (Figure 4B), indicating a potential signature in KD worth exploring. The interactions between miRNAs and target genes/lncRNAs were plotted in the backbone layout using Cytospace software.
To further explore the roles of the identified miRNA-gene network in KD pathophysiology and better understand how this crosstalk contributes to altering specific pathways, we performed functional enrichment analysis in miRnet 2.0 on all the miRNAs and associated genes using KEGG pathways. A total of 100 pathways were identified to be implicated (Supplementary Table S3), among which 32 pathways encompass all three miRNAs (Supplementary Table S4), including “bladder cancer,” “glioma,” “p53 signaling pathway”, “melanoma,” “chronic myeloid leukemia,” “prostate cancer,” “Epstein–Barr virus infection,” “cell cycle” and “steroid biosynthesis.” Considering the functions of interest for KD, a p-value < 0.05 and total hits in the gene targets, the “p53 signaling pathway” (p = 0.00701) and “cell cycle” (p = 0.0222) demonstrated the highest potential to be associated with all three candidate miRNAs and hence in this study, the discrimination between KD and other febrile diseases.
Discussion
To date, a rapid and effective test for accurate KD diagnosis is still unavailable. This gap complicates the differentiation of KD from other febrile diseases or infections, often leading to treatment delays and the risk of developing coronary artery lesions (CALs). Over the years, different approaches have been employed in the search for potential biomarkers other than routinely used clinical parameters, including but not limited to DNA methylation profiling, particularly detecting HAMP promoter hypomethylation (37), quantitative protein array for cytokines and chemokines, and plasma antibody profiling using proteome microarrays. Cross-sectional studies also identify a broad panel of biomarkers, such as myeloid-related protein 8/14 and the CLEC4D, GPR84, and HP genes in peripheral leukocytes (38). However, a definitive, universally accepted biomarker for KD remains unidentified.
In our study, we evaluated both conventional biomarkers (CRP, PLT) and a novel serum miRNA panel (miR-223-3p, miR-19a-3p, miR-18a-5p, miR-21-5p, miR-155-5p, miR-145, and miR-146a-5p) to assess their diagnostic potential for distinguishing KD from other febrile illnesses (Figure 1). Our primary finding suggests that a multimarker approach—integrating CRP with miR-223-3p, miR-19a-3p, and miR-18a-5p—achieves high discriminative accuracy (AUROC = 0.990, sensitivity = 1.000, specificity = 0.923) (Figure 2). While CRP alone shows relatively high diagnostic utility, its limited disease specificity and overlap with other inflammatory febrile conditions—including viral or bacterial infections—reduce its reliability as a standalone diagnostic marker. Meanwhile, miRNAs offer molecular-level signatures that can capture disease-specific immune regulation patterns not reflected by conventional biomarkers. Therefore, integrating CRP with select miRNAs likely captures both general inflammatory burden and disease-specific regulatory signals. This combined approach may improve diagnostic confidence, particularly in clinically ambiguous cases where conventional parameters alone are inconclusive. It is also important to note that, while the model yielded a sensitivity of 1.000 and specificity of 0.923 in our internal validation cohort, these results must be interpreted with caution due to a relatively limited sample size and potential for overfitting. Nonetheless, this analysis serves as an important proof-of-concept, illustrating the feasibility and potential utility of integrating clinical and molecular biomarkers for improving KD diagnostic specificity. Future multicenter studies with larger, prospectively enrolled cohorts are needed to validate and refine this biomarker combination, confirm its clinical generalizability, and assess its performance in early-stage KD or incomplete presentations.
CRP, an acute inflammatory protein synthesized by hepatocytes in response to infection or tissue damage, plays a critical role in the body's innate immune response. It stimulates phagocytosis, the classical complement pathway, and pathogen and necrotic cell elimination by the immune system (39). Moreover, CRP also stimulates platelet aggregation, epithelial cell activation, and vascular permeability, which can cause vasculitis (40). Therefore, CRP's high serum levels suggest inflammation, making it a useful clinical diagnostic for myocardial injury severity and CAL prediction in acute KD. Our study indicated that KD patients had considerably higher CRP levels than non-KD febrile controls, suggesting its usefulness as a diagnostic tool.
MiRNAs, being small, stable molecules easily extractable from blood, urine, and other bodily fluids via cost-effective assays, have gained attention in KD biomarker research due to their high sensitivity, specificity, and stability. Over the last decade, numerous clinical studies have reported that dysregulated miRNAs may serve as potential KD indicators, utilizing a variety of methods, including qRT-PCR, miRNA microarray, next-generation sequencing, and bioinformatic analysis of Gene Expression Omnibus (GEO) database (41). However, no consensus miRNA-based KD diagnosis strategy has been widely applied in clinical practice.
Our findings on the upregulation of specific miRNAs in KD patients, including miR-222-3p, are consistent with previous reports. MiR-222-3p has been found significantly elevated in serum, platelets, and vascular endothelial cells in acute KD cases, playing a key role in vascular injury associated with the disease (42). This miRNA is shown to be predominantly involved in immune-related signalling pathways, notably affecting T cell and B cell receptor pathways, as shown by a recent KEGG pathway analysis. Research indicates miR-223-3p has a protective effect against vascular endothelial damage in KD (43). Studies by Wang et al. and Guo et al. highlight miR-223-3p's capacity to mitigate inflammation and attenuate endothelial cell injury by targeting and suppressing IL6ST, pSTAT3, NF-kB p65, and ICAM-1 in TNF-α treated human coronary artery endothelial cells (HCAECs) (44). Additionally, miR-223-3p influences cell proliferation, apoptosis, and inflammatory cytokine expression in HCAECs, partly by inhibiting FOXP3 expression. Its reduced levels have been linked to severe coronary artery lesions in KD through the modulation of vascular smooth muscle cell (VSMC) differentiation via the suppression of platelet-derived growth factor receptor b (PDGFRb) (45).
While miR-223-3p has been extensively studied, miR-19a-3p and miR-18a-5p have been less investigated in KD. Recent evidence indicates miR-19a-3p is upregulated in KD compared to non-KD febrile controls, however, researchers found it ineffective for predicting CAL or IVIG treatment responses. Subsequently, Liao et al. have identified a potential mechanism by which miR-19a-3p overexpression leads to endothelial dysfunction in KD. They demonstrated that miR-19a-3p suppresses Argonaute 2 (AGO2), disrupting the AGO2/PTEN/VEGF regulatory axis. This finding offers insight into the molecular pathways affected by miR-19a-3p in KD, highlighting its contribution to endothelial dysfunction. MiR-19a serve as a promising biomarker in heart and is essential for maintaining tissue homeostasis. Its increased levels have been associated with enhanced proliferation and reduced apoptosis in cardiomyocytes, contributing to protective effects against ischemia-induced heart failure in animal models and mitigating hypertension-induced cardiac hypertrophy by targeting PDE5A (46). Notably, miR-19a-3p also directly inhibits tumour necrosis factor alpha (TNF-α) (47), a pro-inflammatory cytokine linked to coronary artery abnormalities and is elevated in KD patients. MiR-18a-5p is also shown to be upregulated in KD, and its potential involvement in miRNA-18a-5p/Samd3 signaling pathway regulates endothelial injury, a key feature of KD pathophysiology. Beyond KD, miR-18a-5p has been shown to inhibit endothelial-mesenchymal transition through modulating the Notch2 pathway in cardiac fibrosis. Additionally, its elevated levels have been observed in atherosclerosis, indicating a broader impact on cardiovascular health (48). Furthermore, miR-18a-5p promotes the proliferation of vascular smooth muscle cells (VSMCs) by activating the AKT/ERK signaling pathway, highlighting its significant function in vascular pathology and potential as a biomarker across a spectrum of cardiovascular diseases.
Additionally, our study revealed a positive correlation among the expressions of miR-223-3p, miR-19a-3p, and miR-18a-5p, suggesting their involvement in shared biological pathways (Figure 3). We further attempted to illustrate the pathophysiologic role of these identified serum miRNAs in KD using bioinformatic tool miRNet 2.0, which serves as a powerful and convenient tool for target identification and visualisation. we identified 14 gene targets common to these miRNAs, including F3, TWF1, TP53, and others (Figure 4A). A further KEGG functional enrichment analysis highlighted their significant enrichment in pathways such as the p53 signalling pathway and cell cycle, among others, with the p53 signalling pathway showing a notable association with KD pathophysiology, considering a p-value <0.05, the functions of interest for KD, and total hits in the gene targets. P53, also known as TP53 or tumour protein, is a well-studied tumour suppressor that plays a crucial role in maintaining cellular and genetic stability and regulating cancer formation (49). Recent research has reported that miRNAs are implicated in the p53/miRNA network, that miRNAs regulated by p53 may mediate cellular processes such as cell cycle progression, epithelial–mesenchymal transition, metabolism, cell survival and angiogenesis (50). For instance, research has demonstrated that miR-223-3p directly targets p53 to suppress cell proliferation and migration in lung squamous cell carcinoma (51). MiR-19a-3p was also implicated in p53 pathway in ameliorating age-related bone loss by altering p53 and p21 expression (52). Moreover, genome-wide miRNA expression analysis has found that miR-18a-5p was regulated by p53 in neuroblastoma to promote apoptosis in neuroblastoma cell lines (53). In KD, the activation of P53 by 1,25-Dihydroxyvitamin D3 suggests its involvement in regulating T lymphocyte proliferation and modulating ERK1/2 signaling pathway (54). Our study suggests a key role for the p53/miRNA network in KD's pathogenesis, revealing an underexplored area that warrants additional study.
In addition to the 14 gene targets, our network analysis revealed a new lncRNA target, KCNQ1OT1 (Figure 4B). Long non-coding RNAs (lncRNAs), which are transcripts longer than 200 nucleotides, play crucial roles in regulating gene expression and signalling pathways, impacting tissue homeostasis and diseases, including those affecting the cardiovascular system relevant to KD complications. LncRNA KCNQ1OT1, an antisense non-coding RNA implicated in cardiac development, has been observed to increase in myocardial infarction patients (55). Its downregulation has been shown to mitigate myocardial ischemia, reperfusion injury, and cardiac hypertrophy post-acute myocardial infarction by influencing the miR-2054/AKT3 and miR-204-5p/LGALS3 pathways (56). Interestingly, the specific involvement of LncRNA KCNQ1OT1 in KD has yet to be extensively explored. Our findings suggest a potential interaction between miRNAs and LncRNA KCNQ1OT1 in KD's pathophysiology, presenting a novel avenue for research. Further experimental research are needed to understand this relationship and its effects on KD.
In recent years, bioinformatics tools have been integrated in search of KD biomarkers and pathogenesis pathways. Biomarkers evaluated by urine peptidome profiling and whole blood cell type-specific gene expression analysis were used to construct the first diagnostic algorithm to distinguish KD from febrile controls in 2011 (57). However, its application was limited by a small sample size and extensive processing time. Recently, machine learning methods have been employed in KD diagnosis. For instance, Tsai et al. has recently introduced a KD prediction model established with XGBoost, which used 5 markers (Pyuria, white blood cell counts in urine, ALT level, CRP level, and eosinophil percentage) originated from 74,641 children with fever, and demonstrated a sensitivity of 93% and a specificity of 97% (58). Furthermore, a dynamic nomogram, developed using LASSO and incorporating 33 biological parameters from routine blood tests, effectively distinguishes KD from sepsis-a condition with similar symptoms with KD but requiring very different treatment (59). Our investigation offers a promising diagnostic approach for KD, suggesting the potential for a robust, clinically applicable algorithm. Future studies with expanded sample sizes are essential to validate our findings and refine this strategy. A more comprehensive and dynamic model, validated through extensive research, could significantly enhance KD diagnosis in clinical settings, contributing to timely and precise treatment interventions.
While this study focused on patients with ≥5 days of fever to align with diagnostic thresholds for KD, assessing the utility of this biomarker combination during the earlier febrile phase (<5 days) may enhance the timeliness of diagnosis and intervention. Future investigations should aim to evaluate the discriminative performance of CRP and miRNAs in this critical early window. Moreover, although febrile controls were used to simulate real-world diagnostic dilemmas, incorporating a healthy pediatric cohort would help establish baseline expression levels for these miRNAs and further clarify disease-specific alterations.
Limitations
This study has several limitations. First, it was designed as a single-center, retrospective case-control study with a predetermined diagnosis. While this design allowed for timely sample collection and focused biomarker analysis, it is inherently different from a cohort study and does not permit longitudinal assessment of disease development or outcomes. Second, the relatively small sample size—particularly the limited number of complete KD case—may reduce the statistical robustness of the diagnostic model and raise concerns about potential overfitting. Although the model demonstrated excellent discrimination in this dataset, the findings should be considered exploratory and interpreted with caution. Validation in larger, independent cohorts is necessary to confirm the model's performance and ensure generalizability. Third, the study population consisted exclusively of Chinese pediatric patients, limiting extrapolation of the results to other ethnic groups. Future studies should aim to include diverse populations to assess the consistency of biomarker expression and model performance across different genetic and environmental backgrounds. Lastly, biomarker assessment was conducted at a single time point before IVIG administration. Evaluating longitudinal changes in CRP and miRNA expression—including in IVIG responders vs. non-responders—would further clarify their diagnostic and prognostic potential.
Conclusions
This study presents a preliminary diagnostic approach combining CRP and serum miRNAs to distinguish KD from other febrile conditions. The integrated model demonstrated strong discriminative potential in this initial dataset and may offer a complementary strategy to enhance diagnostic confidence in clinically ambiguous cases. In addition, the involvement of these miRNAs in pathways such as p53 signaling and lncRNA interaction suggests possible mechanistic links that warrant further exploration. While the findings are promising, future studies with larger, diverse cohorts and prospective validation are essential before clinical implementation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by The Shaanxi Provincial People's Hospital Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants' legal guardians/next of kin.
Author contributions
XH: Conceptualization, Visualization, Writing – original draft, Methodology, Funding acquisition. HL: Writing – original draft, Formal analysis, Data curation. XZ: Resources, Visualization, Methodology, Writing – review & editing. HZ: Writing – review & editing, Resources, Data curation. YL: Resources, Writing – review & editing. QN: Investigation, Funding acquisition, Writing – review & editing. JW: Data curation, Resources, Writing – review & editing. CX: Supervision, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by International Science and Technology Cooperation Program Project of Key Research and Development Plan of Shaanxi Province (2020KWZ-20), Shaanxi Province Innovation Capability Support Plan (2024RS-CXTD-84), Key Research and Development Plan of Shaanxi Province (2023-GHZD-41), Shaanxi Provincial Health and Medical Research Innovation Capacity Enhancement Plan (2024PT-01) and Shaanxi Provincial Health High-Level Talents Cultivation Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2025.1678095/full#supplementary-material
Supplementary Table S1 | Gene targets identified to be interacting with all three candidate miRNAs (miR-223-3p, miR-19a-3p and miR-18a-5p) by miRnet 2.0.
Supplementary Table S2 | LncRNAs identified to be interacting with all three candidate miRNAs (miR-223-3p, miR-19a-3p and miR-18a-5p) by miRnet 2.0.
Supplementary Table S3 | KEGG pathways implicated in candidate miRNAs and associated genes analyzed by miRnet 2.0.
Supplementary Table S4 | KEGG pathways encompass all three miRNAs analyzed by miRnet 2.0.
References
1. McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. (2017) 135(17):e927–99. doi: 10.1161/CIR.0000000000000484
2. Rife E, Gedalia A. Kawasaki disease: an update. Curr Rheumatol Rep. (2020) 22(10):75. doi: 10.1007/s11926-020-00941-4
3. Elakabawi K, Lin J, Jiao F, Guo N, Yuan Z. Kawasaki disease: global burden and genetic background. Cardiol Res. (2020) 11(1):9–14. doi: 10.14740/cr993
4. Zandstra J, van de Geer A, Tanck MWT, van Stijn-Bringas Dimitriades D, Aarts CEM, Dietz SM, et al. Biomarkers for the discrimination of acute Kawasaki disease from infections in childhood. Front Pediatr. (2020) 8:355. doi: 10.3389/fped.2020.00355
5. Seki M, Minami T. Kawasaki disease: pathology, risks, and management. Vasc Health Risk Manag. (2022) 18:407–16. doi: 10.2147/VHRM.S291762
6. Huerta BS, Sanchez AA. Biomarkers in Kawasaki disease. Biomark Med. (2022) 16:51–62. doi: 10.2217/bmm-2021-0855
7. Singh S, Jindal AK, Pilania RK. Diagnosis of Kawasaki disease. Int J Rheum Dis. (2018) 21(1):36–44. doi: 10.1111/1756-185X.13224
8. Newburger JW, Takahashi M, Beiser AS, Burns JC, Bastian J, Chung KJ, et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med. (1991) 324(23):1633–9. doi: 10.1056/NEJM199106063242305
9. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). (2018) 9:402. doi: 10.3389/fendo.2018.00402
10. Yuan L, Jiang X, Gong Q, Gao N. Arsenic resistance protein 2 and microRNA biogenesis: biological implications in cancer development. Pharmacol Ther. (2023) 244:108386. doi: 10.1016/j.pharmthera.2023.108386
11. Miyamoto SD, Karimpour-Fard A, Peterson V, Auerbach SR, Stenmark KR, Stauffer BL, et al. Circulating microRNA as a biomarker for recovery in pediatric dilated cardiomyopathy. J Heart Lung Transplant. (2015) 34(5):724–33. doi: 10.1016/j.healun.2015.01.979
12. Wojciechowska A, Braniewska A, Kozar-Kaminska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. (2017) 26(5):865–74. doi: 10.17219/acem/62915
13. Pozniak T, Shcharbin D, Bryszewska M. Circulating microRNAs in medicine. Int J Mol Sci. (2022) 23(7):3996. doi: 10.3390/ijms23073996
14. Shimizu C, Kim J, Stepanowsky P, Trinh C, Lau HD, Akers JC, et al. Differential expression of miR-145 in children with Kawasaki disease. PLoS One. (2013) 8(3):e58159. doi: 10.1371/journal.pone.0058159
15. Xiong Y, Xu J, Zhang D, Wu S, Li Z, Zhang J, et al. Micrornas in Kawasaki disease: an update on diagnosis, therapy and monitoring. Front Immunol. (2022) 13:1016575. doi: 10.3389/fimmu.2022.1016575
16. Zhang W, Wang Y, Zeng Y, Hu L, Zou G. Serum miR-200c and miR-371-5p as the useful diagnostic biomarkers and therapeutic targets in Kawasaki disease. Biomed Res Int. (2017) 2017:8257862. doi: 10.1155/2017/8257862
17. Jia H-L, Liu C-W, Zhang L, Xu W-J, Gao X-J, Bai J, et al. Sets of serum exosomal microRNAs as candidate diagnostic biomarkers for Kawasaki disease. Sci Rep. (2017) 7:44706. doi: 10.1038/srep44706
18. Cai Y, Hu W. Identifying differentially expressed genes and miRNAs in Kawasaki disease by bioinformatics analysis. Sci Rep. (2022) 12(1):21879. doi: 10.1038/s41598-022-26608-x
19. Chen C-C, Chu H-Y, Chang IY-F, Chang Y-S, Weng K-P, Chang L-S, et al. Symptom-correlated miRNA signature as a potential biomarker for Kawasaki disease. Biomed J. (2023) 47:100684. doi: 10.1016/j.bj.2023.100684
20. Sproston NR, Ashworth JJ. Role of C-reactive protein at sites of inflammation and infection. Front Immunol. (2018) 9:754. doi: 10.3389/fimmu.2018.00754
21. Chaudhary H, Nameirakpam J, Kumrah R, Pandiarajan V, Suri D, Rawat A, et al. Biomarkers for Kawasaki disease: clinical utility and the challenges ahead. Front Pediatr. (2019) 7:242. doi: 10.3389/fped.2019.00242
22. Arora K, Guleria S, Jindal AK, Rawat A, Singh S. Platelets in Kawasaki disease: is this only a numbers game or something beyond? Genes Dis. (2020) 7(1):62–6. doi: 10.1016/j.gendis.2019.09.003
23. Nakagama Y, Inuzuka R, Hayashi T, Shindo T, Hirata Y, Shimizu N, et al. Fever pattern and C-reactive protein predict response to rescue therapy in Kawasaki disease. Pediatr Int. (2016) 58(3):180–4. doi: 10.1111/ped.12762
24. Zheng R, Xie J, Li W, Shang J, Shi Z, Zhu S, et al. MiR-223-3p affects the proliferation and apoptosis of HCAECs in Kawasaki disease by regulating the expression of FOXP3. Immun Inflamm Dis. (2023) 11(7):e939. doi: 10.1002/iid3.939
25. Liao J, Guo X, Fan X, Zhang X, Xu M. Upregulation of miR-184 and miR-19a-3p induces endothelial dysfunction by targeting AGO2 in Kawasaki disease. Cardiol Young. (2023) 33(10):1962–6. doi: 10.1017/S1047951122003523
26. Li X, Sun Y, Cai Y, Zhang X, Zhang X. Identification of miRNA profile in the peripheral blood and clinical significance of miR-355 and miR-2911 expression in children with Kawasaki disease. Am J Transl Res. (2022) 14(11):7820–30. Available online at: https://pmc.ncbi.nlm.nih.gov/articles/PMC9730091/36505330
27. Ni F-F, Li C-R, Li Q, Xia Y, Wang G-B, Yang J. Regulatory T cell microRNA expression changes in children with acute Kawasaki disease. Clin Exp Immunol. (2014) 178(2):384–93. doi: 10.1111/cei.12418
28. Zha L, Li S, Liu X, Li Z, Jiang J, Huang L, et al. Association of miR-146a gene polymorphism at loci rs2910164G/C, rs57095329 A/G, and rs6864584 T/C with susceptibility to Kawasaki disease in Chinese children. Pediatr Cardiol. (2019) 40(3):504–12. doi: 10.1007/s00246-018-2002-9
29. Chang L, Zhou G, Soufan O, Xia J. MiRNET 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. (2020) 48(W1):W244–51. doi: 10.1093/nar/gkaa467
30. Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. (2019) 47(D1):D155–62. doi: 10.1093/nar/gky1141
31. Hosen MR, Goody PR, Zietzer A, Xiang X, Niepmann ST, Sedaghat A, et al. Circulating microRNA-122-5p is associated with a lack of improvement in left ventricular function after transcatheter aortic valve replacement and regulates viability of cardiomyocytes through extracellular vesicles. Circulation. (2022) 146(24):1836–54. doi: 10.1161/CIRCULATIONAHA.122.060258
32. Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. (2018) 46(D1):D239–45. doi: 10.1093/nar/gkx1141
33. Guo Z, Maki M, Ding R, Yang Y, zhang B, Xiong L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep. (2014) 4:5150. doi: 10.1038/srep05150
34. Palmieri V, Backes C, Ludwig N, Fehlmann T, Kern F, Meese E, et al. IMOTA: an interactive multi-omics tissue atlas for the analysis of human miRNA-target interactions. Nucleic Acids Res. (2018) 46(D1):D770–5. doi: 10.1093/nar/gkx701
35. Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. (2014) 42(Database issue):D92–7. doi: 10.1093/nar/gkt1248
36. Bleazard T, Lamb JA, Griffiths-Jones S. Bias in microRNA functional enrichment analysis. Bioinformatics. (2015) 31(10):1592–8. doi: 10.1093/bioinformatics/btv023
37. Huang Y-H, Kuo H-C, Li S-C, Cai X-Y, Liu S-F, Kuo H-C. Hamp promoter hypomethylation and increased hepcidin levels as biomarkers for Kawasaki disease. J Mol Cell Cardiol. (2018) 117:82–7. doi: 10.1016/j.yjmcc.2018.02.017
38. Kuo K-C, Yang Y-L, Lo M-H, Cai X-Y, Kuo H-C, Huang Y-H. The expression of glycoprotein genes in the inflammatory process of Kawasaki disease. Front Pediatr. (2020) 8:592122. doi: 10.3389/fped.2020.592122
39. Puthucheary Z, Tadie JM, Patel JJ. C-reactive protein in immunometabolism: spared from ‘paying the piper’. Intensive Care Med. (2022) 48:103–05. doi: 10.1007/s00134-021-06586-w
40. Xu PC, Lin S, Yang XW, Gu DM, Yan TK, Wei L, et al. C-reactive protein enhances activation of coagulation system and inflammatory response through dissociating into monomeric form in antineutrophil cytoplasmic antibody-associated vasculitis. BMC Immunol. (2015) 16:10. doi: 10.1186/s12865-015-0077-0
41. An HS, Kim GB, Song MK, Lee SY, Kwon HW, Lee JW, et al. The occurrence of coronary artery lesions in Kawasaki disease based on C-reactive protein levels: a retrospective cohort study. Pediatr Rheumatol Online J. (2021) 19(1):78. doi: 10.1186/s12969-021-00566-6
42. Wang B, Wang L-n, Cheng F-f, Lv H-t, Sun L, Wei D-k, et al. miR-222-3p in platelets serves as a distinguishing marker for early recognition of Kawasaki disease. Front Pediatr. (2019) 7:237. doi: 10.3389/fped.2019.00237
43. Parra-Izquierdo I, Mccarty O, Aslan JE. Platelet miR-223 delivery rescues vascular cells in Kawasaki disease. Circ Res. (2020) 127(7):874–6. doi: 10.1161/CIRCRESAHA.120.317796
44. Guo M, Fan S, Chen Q, Jia C, Qiu M, Bu Y, et al. Platelet-derived microRNA-223 attenuates TNF-alpha induced monocytes adhesion to arterial endothelium by targeting ICAM-1 in Kawasaki disease. Front Immunol. (2022) 13:922868. doi: 10.3389/fimmu.2022.922868
45. Zhang Y, Wang Y, Zhang L, Xia L, Zheng M, Zeng Z, et al. Reduced platelet miR-223 induction in Kawasaki disease leads to severe coronary artery pathology through a miR-223/PDGFRbeta vascular smooth muscle cell axis. Circ Res. (2020) 127(7):855–73. doi: 10.1161/CIRCRESAHA.120.316951
46. Liu K, Hao Q, Wei J, Li G-H, Wu Y, Zhao Y-F. MicroRNA-19a/b-3p protect the heart from hypertension-induced pathological cardiac hypertrophy through PDE5a. J Hypertens. (2018) 36(9):1847–57. doi: 10.1097/HJH.0000000000001769
47. Liu M, Wang Z, Yang S, Zhang W, He S, Hu C, et al. TNF-alpha is a novel target of miR-19a. Int J Oncol. (2011) 38(4):1013–22. doi: 10.3892/ijo.2011.924
48. Chang L, Xia J. MicroRNA regulatory network analysis using miRNET 2.0. Methods Mol Biol. (2023) 2594:185–204. doi: 10.1007/978-1-0716-2815-7_14
49. Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ. (2015) 22(8):1239–49. doi: 10.1038/cdd.2015.53
50. Sargolzaei J, Etemadi T, Alyasin A. The P53/microRNA network: a potential tumor suppressor with a role in anticancer therapy. Pharmacol Res. (2020) 160:105179. doi: 10.1016/j.phrs.2020.105179
51. Luo P, Wang Q, Ye Y, Zhang J, Lu D, Cheng L, et al. MiR-223-3p functions as a tumor suppressor in lung squamous cell carcinoma by miR-223-3p-mutant P53 regulatory feedback loop. J Exp Clin Cancer Res. (2019) 38(1):74. doi: 10.1186/s13046-019-1079-1
52. Kaur J, Saul D, Doolittle ML, Farr JN, Khosla S, Monroe DG. MicroRNA-19a-3p decreases with age in mice and humans and inhibits osteoblast senescence. JBMR Plus. (2023) 7(6):e10745. doi: 10.1002/jbm4.10745
53. Rihani A, Van Goethem A, Ongenaert M, De Brouwer S, Volders P-J, Agarwal S, et al. Genome wide expression profiling of P53 regulated miRNAs in neuroblastoma. Sci Rep. (2015) 5:9027. doi: 10.1038/srep09027
54. Qi XL, Chen LL, Sun XG, Li XM, Zhao LH, Kong DJ. 1,25-dihydroxyvitamin D3 regulates T lymphocyte proliferation through activation of p53 and inhibition of ERK1/2 signaling pathway in children with Kawasaki disease. Eur Rev Med Pharmacol Sci. (2017) 21(16):3714–22. Available online at: https://www.europeanreview.org/article/1328928925469
55. Ginckels P, Holvoet P. Oxidative stress and inflammation in cardiovascular diseases and cancer: role of non-coding RNAs. Yale J Biol Med. (2022) 95(1):129–52. Available online at: https://pmc.ncbi.nlm.nih.gov/articles/PMC8961704/35370493
56. Rong J, Pan H, He J, Zhang Y, Hu Y, Wang C, et al. Long non-coding RNA KCNQ1OT1/microRNA-204-5p/LGALS3 axis regulates myocardial ischemia/reperfusion injury in mice. Cell Signal. (2020) 66:109441. doi: 10.1016/j.cellsig.2019.109441
57. Ling XB, Lau K, Kanegaye JT, Pan Z, Peng S, Ji J, et al. A diagnostic algorithm combining clinical and molecular data distinguishes Kawasaki disease from other febrile illnesses. Bmc Med. (2011) 9:130. doi: 10.1186/1741-7015-9-130
58. Tsai C-M, Lin C-HR, Kuo H-C, Cheng F-J, Yu H-R, Hung T-C, et al. Use of machine learning to differentiate children with Kawasaki disease from other febrile children in a pediatric emergency department. JAMA Network Open. (2023) 6(4):e237489. doi: 10.1001/jamanetworkopen.2023.7489
Keywords: Kawasaki disease, biomarker, C-reactive protein, microRNA, diagnostic strategy
Citation: Huang X, Li H, Zhao X, Zhang H, Li Y, Niu Q, Wang J and Xu C (2025) Combination of CRP and miRNA signature as a potential diagnostic strategy for Kawasaki disease. Front. Pediatr. 13:1678095. doi: 10.3389/fped.2025.1678095
Received: 1 August 2025; Accepted: 26 September 2025;
Published: 15 October 2025.
Edited by:
Amar Taksande, Datta Meghe Institute of Medical Sciences, IndiaReviewed by:
Mingming Zhang, Children’s Hospital of Capital Institute of Pediatrics, ChinaXiaoling Zhong, Chengdu Third People’s Hospital, China
Copyright: © 2025 Huang, Li, Zhao, Zhang, Li, Niu, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuixiang Xu, eHVjdWl4aWFuZ0BzcHBoLXN4LmFjLmNu
†These authors have contributed equally to this work
 Xiaoyan Huang1,2,†
Xiaoyan Huang1,2,† Huiting Li
Huiting Li Haixiang Zhang
Haixiang Zhang