- 1The Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital, Qingyuan, China
- 2Department of Human Nutrition, Food and Animal Sciences, College of Tropical Agriculture and Human Resources, University of Hawaii at Manoa, Honolulu, HI, United States
Lipid metabolism is an essential biological process involved in nutrient adjustment, hormone regulation, and lipid homeostasis. An irregular lifestyle and long-term nutrient overload can cause lipid-related diseases, including atherosclerosis, myocardial infarction (MI), obesity, and fatty liver diseases. Thus, novel tools for efficient diagnosis and treatment of dysfunctional lipid metabolism are urgently required. Furthermore, it is known that lncRNAs based regulation like sponging microRNAs (miRNAs) or serving as a reservoir for microRNAs play an essential role in the progression of lipid-related diseases. Accordingly, a better understanding of the regulatory roles of lncRNAs in lipid-related diseases would provide the basis for identifying potential biomarkers and therapeutic targets for lipid-related diseases. This review highlighted the latest advances on the potential biomarkers of lncRNAs in lipid-related diseases and summarised current knowledge on dysregulated lncRNAs and their potential molecular mechanisms. We have also provided novel insights into the underlying mechanisms of lncRNAs which might serve as potential biomarkers and therapeutic targets for lipid-related diseases. The information presented here may be useful for designing future studies and advancing investigations of lncRNAs as biomarkers for diagnosis, prognosis, and therapy of lipid-related diseases.
Introduction
Lipid metabolism is an intricate and complex physiological process that is involved in the progression of lipid-related diseases (Li et al., 2017). Importantly, since modern society is associated with irregular lifestyle patterns and long-term nutrient overload, severe lipid metabolism disorders and lipid accumulation have become commonplace (Liu and Ding, 2017; Dłubek et al., 2021). Abnormal lipid metabolism is the primary feature of several refractory chronic diseases (Yang et al., 2016), such as atherosclerotic disease (Michos et al., 2019), obesity (Wang et al., 2014), fatty liver disease (Vernon et al., 2011), and diabetes mellitus (Garde et al., 2019). Thus, developing novel tools and strategies for maintaining cholesterol homeostasis is urgently required to prevent and treat these diseases.
Long non-coding RNAs (lncRNAs) are a class of RNA that do not encode proteins (Kim et al., 2009). Instead, they are involved in complex biological processes and pathophysiological conditions, including lipid metabolism disorders (Zeng et al., 2018; Simion et al., 2019). Recently, numerous clinical studies have shown that lncRNAs impair cholesterol homeostasis and play a critical role in the progression of lipid-related diseases (Han et al., 2019; Ou et al., 2020). For example, a primate-specific lncRNA (CHROME) was found to be elevated in the plasma and atherosclerotic plaques of patients with coronary heart disease (CHD) (Hennessy et al., 2019). Similarly, highly up-regulated in liver cancer (HULC) lncRNA was discovered to modulate the deregulation of lipid metabolism in hepatoma cells and result in malignant development (Cui et al., 2015). These findings suggest that lncRNAs regulate lipid metabolism and promote the development of lipid-related diseases. LncRNAs might also function as the miRNAs sponges and affect lipid metabolism and related diseases (Lan et al., 2019). Importantly, lncRNAs also play an essential in the progression of some other diseases, such as cancer (Hahne and Valeri, 2018). Much research has been conducted on the specific functions of lncRNAs in these diseases.
The emerging role of lncRNAs as potential biomarkers and therapeutic targets for lipid-related diseases has not explicitly been summarised, and the present review aims to fill this gap in the literature. LncRNAs have been increasingly recognized as potential biomarkers for various human diseases, including atherosclerosis (Simion et al., 2020), MI (Spiroski et al., 2021), liver disease (Yang et al., 2021), and cancer (Xing et al., 2021). Here, we mainly reviewed the recent investigations of the role of lncRNAs as potential biomarkers and therapeutic targets in lipid-related diseases. Findings from this review would summarize the mechanisms by which lncRNAs act as biomarkers and therapeutic targets for lipid-related diseases.
LncRNAs Mechanisms of Action
Recent studies have illustrated that lncRNAs can bind to the proteins, RNA, DNA, or a combination of them to exert their functions (Fasolo et al., 2019; Hu Y. et al., 2019). As regulators of gene expression, lncRNAs involve in various biological processes (Fernandes et al., 2019; Mumtaz and Online, 2017), acting as miRNA sponge, decoys, scaffolds, guides, and post-translation regulation (Rinn and Chang, 2012) (Figure 1). For instance, many lncRNAs act as a miRNA sponge to regulate miRNAs and their targets. For example, small nucleolar RNA host gene 16 (SNHG16) facilitated the development and progression of neuroblastoma by upregulating homeobox A7 (HOXA7) expression via sponging miR-128–3p (Bao et al., 2020). Decoying lncRNAs mediated transcriptional repression by guiding chromatin modifiers such as m6A formation and recognition to genomic targets, such as XIST (Patil et al., 2016), HOTAIR (Loewen et al., 2014), and GAS5 (Sun et al., 2017). LncRNAs can be used as scaffolds to form enhancer loops or as structural components of ribonucleoprotein complexes (Stackhouse et al., 2020). Nuclear paraspeckle assembly transcript 1 (NEAT1) scaffolds broadly interacts with NONO/PSF and other RNA-binding proteins (RBPs) and that globally enhance pri-miRNA processing (Jiang et al., 2017). Additionally, many lncRNAs exert their functions by sequestering regulatory factors in the nucleus or cytoplasm: for example, colon cancer-associated transcript-2 (CCAT2) can block miR-145 maturation by inhibiting pre-miR-145 export to cytoplasm (Yu Y. et al., 2017); whereas cytoplasmic lncRNAs, such as lincRNA-p21, interact with RNA-binding protein HuR to recruit let-7/Ago2 to inhibit their repression of lincRNA-p21 stability (Yoon et al., 2012). Finally, lncRNAs can act as enhancers or co-activators of target gene activation, such as H19 and GAS5. LncRNA may have more than one function, varying by subcellular localization, stimuli, and/or cell types. With the continuous increase of lncRNA-mediated functions, it has become clear that they are important regulators of multiple biological and cellular processes and can be used as candidate diagnostic and prognostic biomarkers for human diseases.
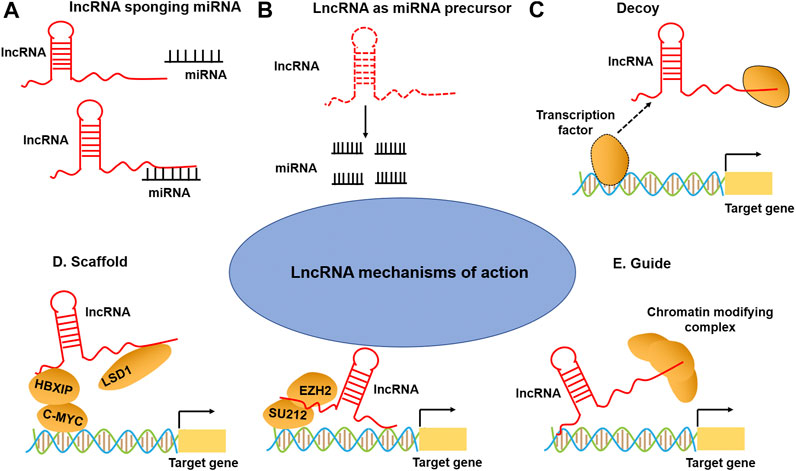
FIGURE 1. LncRNA mechanisms of action. (A) LncRNAs can act as a sponge to titrate miRNAs away from their mRNA targets. (B) The lncRNAs can act as miRNA precursors. (C) LncRNA can bind to transcription factors or other proteins as a decoy and sequester them away from chromatin (lower-right). (D) LncRNA can also serve as a scaffold to promote the assembling of chromatin remodeling complexes. (E) LncRNA can guide transcription factors to specific genomic locations for regulating gene expression.
LncRNAs Participate in the Development of Lipid-Related Diseases
Lipid metabolism is the biosynthesis and biodegradation of lipids in cells (Santos and Schulze, 2012). It involves the breakdown and storage of fats for energy and the synthesis of structural and functional lipids (de Carvalho and Caramujo, 2018). Lipid biosynthesis is a part of metabolic abnormalities in cells, which require large quantities of lipids to synthesize cytomembranes, organelles, and signaling molecules during cell proliferation (Xu et al., 2020). Importantly, fatty acid oxidation (FAO) can provide abundant ATP for cells (Jeon et al., 2012), and fatty acids are a major source of ATP molecules (Fhu and Ali, 2020). In addition, lncRNAs affect gene expression that is involved in lipid metabolism (Table 1). Numerous studies have shown that lncRNAs participate in lipid metabolism by influencing the expression of key genes, networks, and pathways involved in lipid biosynthesis, cholesterol transport, lipid uptake, and cholesterol efflux (Figure 2).
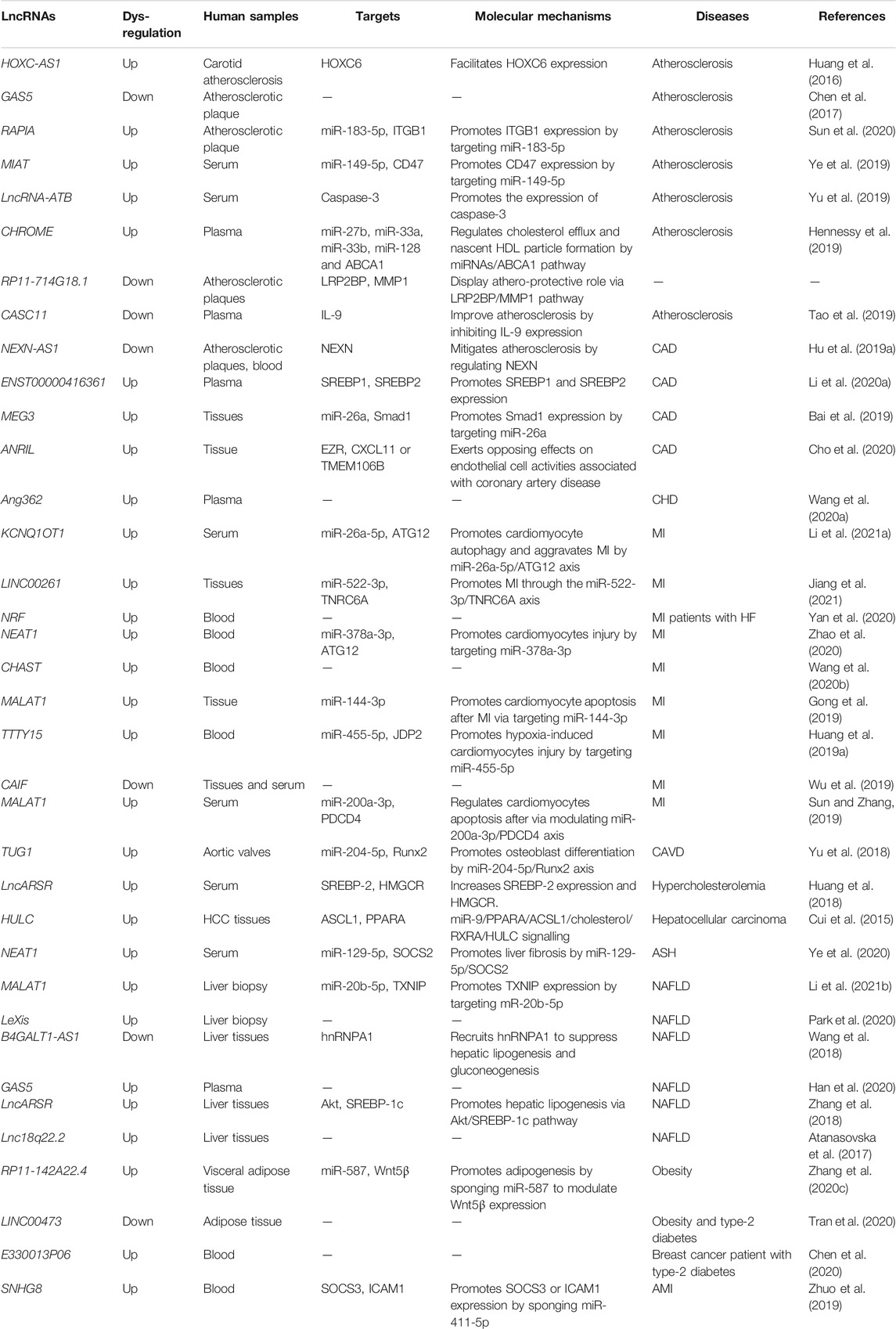
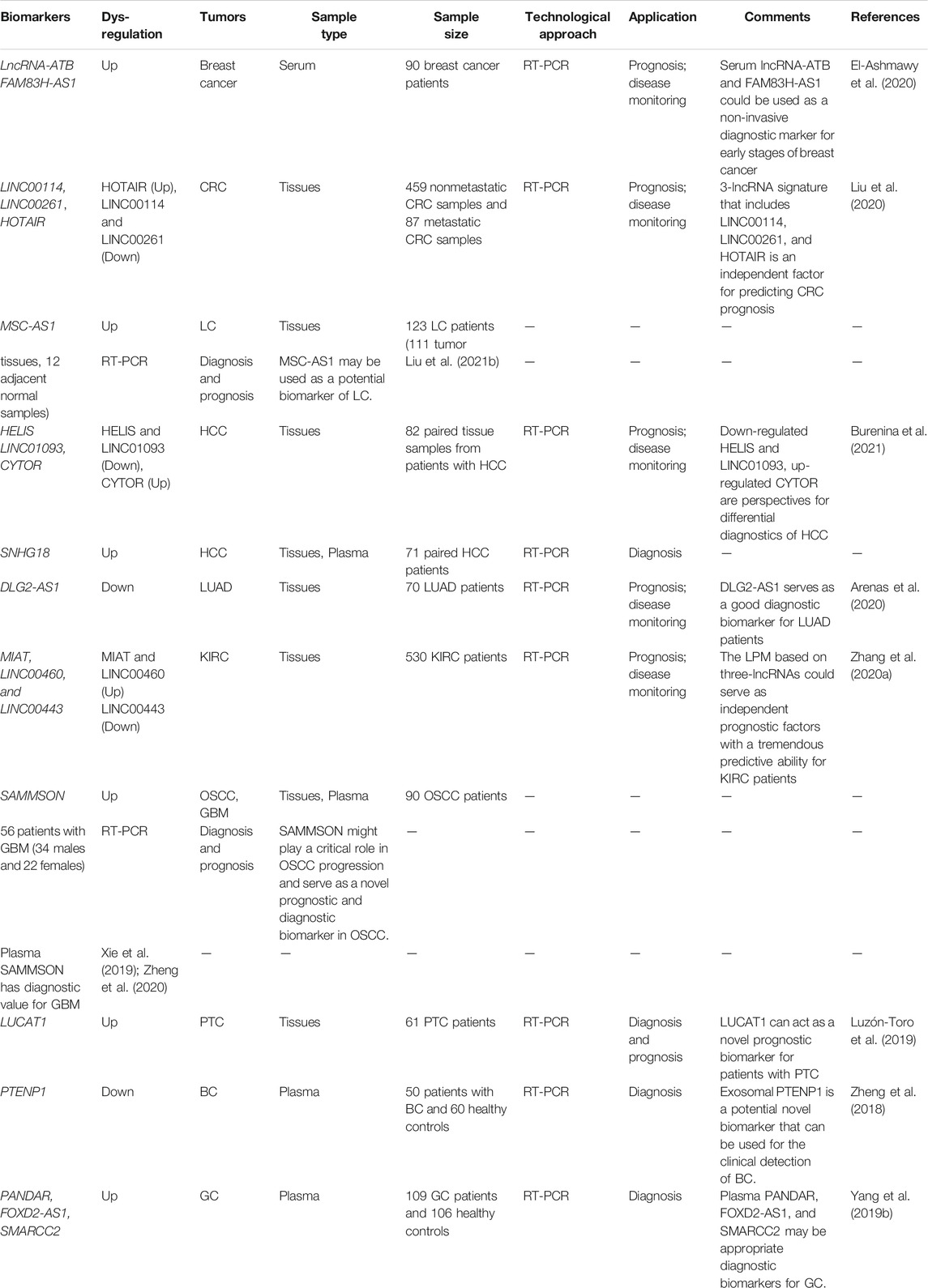
TABLE 1. Summary of the act of lncRNAs as therapeutic targets and potential biomarkers for lipid-related diseases.
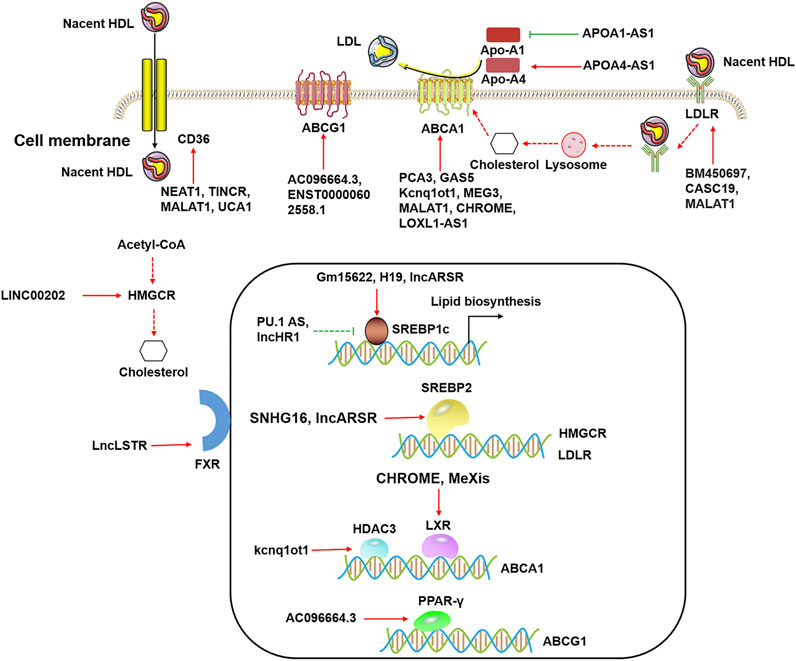
FIGURE 2. The functional roles of lncRNAs in lipid metabolism. LncRNAs modulate cholesterol efflux by ABCA1, ABCG1, CD36, and LDLR in the cytoplasm. LncRNAs regulate ABCA1 expression by HDAC3 and LXRs, and lncRNAs regulate ABCG1 expression by PPAR-γ in the nucleus. LncRNAs influence lipid biosynthesis by SREBP1c and SREBP2 in the nucleus.
Recent studies have reported that lncRNAs participate in the regulation of various genes expression in lipid metabolism that was induced by hormones (Fu et al., 2020), environmental stress (Wen et al., 2020), lipid/cholesterol (Ma et al., 2018), and obesity/type 2 diabetes (Hu et al., 2020). A single lncRNA often targets multiple mRNAs, and these mRNAs are linked to the different metabolic pathways (Huang, 2018). It is important to note that each mRNA is typically targeted by several lncRNAs, enabling coordinated gene expression. Many molecules are involved in lipid metabolism, including nuclear transcription factors such as LXR, FXR, SREBP, and the scavenger receptor CD36 (Figure 2) (Shimano and Sato, 2017; Yan et al., 2018; Piccinin et al., 2021). These regulatory molecules, along with lncRNAs, are implicated in the regulation of lipid metabolism.
Given the fact that lipid metabolism is distributed different cellular organelles also transport of the intermediates between the different organelles is an important point in lipid metabolism (Khor et al., 2013; Xu and Taubert, 2021). Fox example, lipid metabolism is located in the endoplasmic reticulum (ER) for lipid biosynthesis (Jacquemyn et al., 2017), mitochondria and peroxisomes for β-oxidation (Zhou et al., 2018), lipid droplets (LDs) for storage and transport (Freyre et al., 2019), and lysosomes for lipid hydrolysis and recycling (Go et al., 2012). Lipid metabolism includes processes such as lipid uptake, biosynthesis, catabolism, and secretion. LncRNAs can affect biological functions in many ways, such as the miRNA sponge, guide or decoy, scaffold, and chromatin remodeling. Currently, numerous lncRNAs have been identified to be involved in the regulation of lipid metabolism. However, many lncRNAs with lipid metabolism functions do not directly target genes involved in lipid metabolism pathways (He et al., 2019; Lan et al., 2019), such as triglyceride and cholesterol biosynthesis and fatty acid oxidation. Instead, they target the lncRNA-miRNA-mRNA and lncRNA-mRNA axes. For example, the lncRNA HULC has been shown to regulate abnormal lipid metabolism by decreasing miR-9 expression, leading to the upregulation of RXRA expression (Cui et al., 2015). RXRA, a member of the RXR family that can be activated by sterol (Costet et al., 2000), modulates the lipid metabolism disorders by activating acyl-CoA synthetase long-chain family member 1 (ACSL1) (Cui et al., 2015). Similarly, lncRNA PU.1 AS regulates lipid metabolism via the sterol regulatory element-binding protein-1c (SREBP-1c) pathway, resulting in reduced triglyceride synthesis (Dong et al., 2019). Transcription factors of the SREBP family, including SREBP-1a, SREBP-1c, and SREBP-2, are central to transcriptional control of genes related to lipid and fatty acid metabolism (Brown and Goldstein, 1999). Interestingly, overexpression of SREBP-1c is known to facilitate fatty acid and triglyceride synthesis and lead to lipid accumulation in the liver (Yan et al., 2016). On the other hand, the inhibition of SREBP-1c is shown to alleviate lipid accumulation and lipotoxicity (Jin et al., 2020). The involvement of a lncRNA derived from hepatocytes (lnc-HC) in lipid metabolism has been extensively reported. For example, lnc-HC was found to regulate PPARγ-mediated lipid metabolism and triglyceride (TG) concentration via miR-130b-3p, where lnc-HC expression was positively correlated with the miR-130b-3p expression (Lan et al., 2019). Furthermore, it has been illustrated that lnc-HC forms a complex with hnRNPA2B1 and negatively regulates Cyp7a1 and Abca1 expressions; both are implicated in hepatocytic cholesterol metabolism (Lan et al., 2016). Another lncRNA and hnRNP complex has also been identified with LeXis and RALY hnRNP, which are involved in lipid metabolism and influence metabolic gene expression (Sallam et al., 2016).
Diseases Associated With lncRNA-Related Lipid Dysregulation
Several diseases, including atherosclerosis, MI, liver disease, and hypercholesterolemia, are caused by or associated with lipid dysregulation (Butt et al., 2017; Gluchowski et al., 2017; Michos et al., 2019). Importantly, studies focused on these diseases were performed using patient specimens, animal models (ApoE−/− and LDL−/−), and atherosclerosis model cell lines, such as human umbilical vein endothelial cells (HUVECs) (Chen L. et al., 2019), human peripheral blood monocytes (THP-1) (Choi et al., 2021), human vascular smooth muscle cells (HVSMCs) (Li X. et al., 2021). Therefore, we only summarised several representative studies that mainly focused on lncRNA functions in lipid-related disease processes.
Disruption of lipid metabolism has been confirmed as a significant factor in the pathogenesis of atherosclerosis (Sukhorukov et al., 2020). The progression of atherosclerosis is known to be regulated by disturbances of lipid metabolism (Lovren et al., 2015), which impairs endothelial cells’ function. Recent studies have identified H19 as a well-known lncRNA associated with atherosclerosis (Huang Y. et al., 2019). H19 expression has been reported to be up-regulated in patients with atherosclerosis and may be a potential therapeutic target for atherosclerosis (Yang Y. et al., 2019). Knockdown of H19 inhibits hyperlipidemia and alleviates atherosclerotic lesions in HFD-treated ApoE−/− mice (Pan and sciences, 2017; Shi et al., 2020), while lentivirus-mediated H19-forced expression increase the plaque area size (Huang Y. et al., 2019). Technically, H19 acts as a molecular sponge for miR-148b-3p and activates its expression of ELF5 (E74 like ETS transcription factor 5), resulting in the restoration of ELF5 that inhibit the cell migration in ox-LDL-stimulated HUVECs (Liu S. et al., 2021). Additionally, lncARSR, a lncRNA regulator of Akt signaling associated with HCC and RCC, has recently been studied as a potential therapeutic target for cholesterol disorder, and its downstream target SREBP-2 was identified. SREBP-2 has been found to bind to HMG-CoA reductase (HMGCR) to promote hepatic cholesterol biosynthesis, resulting in aberrant regulation of cholesterol metabolism (Huang et al., 2018). Collectively, lncARSR-SREBP-2-HMGCR plays a pivotal role in regulating lipid metabolism and the development of atherosclerosis (Xiao and Song, 2013).
Dysregulated lipid metabolism is a hallmark of non-alcoholic steatohepatitis (NASH), a very common liver disorder (Musso et al., 2013). Recently, growing evidence has suggested that dysregulated lncRNA expression is associated with inflammation and fibrosis in NASH (Leti et al., 2017). Whole transcriptome analysis and identified differentially expressed lncRNAs (RP11-128N14.5 and TGFB2-OT1) in patients with non-alcoholic fatty liver disease (NAFLD) (Di Mauro et al., 2019). Several lncRNAs, including hepatocellular carcinoma up-regulated lncRNA, NEAT1, and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), were highly expressed in liver biopsies from NAFLD patients (Leti et al., 2017). Furthermore, expression of MALAT1 was upregulated in livers of ob/ob mice and hepatocytes exposed to palmitate (Yan et al., 2016). Another lncRNA, Alu-mediated p21 transcriptional regulator (APTR), was discovered to be significantly increased in human cirrhosis and activate hepatic stellate cells (Yu et al., 2015). Hepatic LeXis expression is a mediator of cholesterol biosynthesis (Sallam et al., 2016). Thus, raising or lowering LeXis levels influence the expression of genes involved in cholesterol biosynthesis and alter liver and plasma cholesterol levels (Sallam et al., 2016). Brown fat-enriched lncRNA 1 (Blnc1) was strongly elevated in obesity and NAFLD in mice (Zhao et al., 2018). Hepatic Blnc1 deficiency is suggested to abrogate high-fat diet-induced hepatic steatosis and insulin resistance and ameliorate NASH pathogenesis (Zhao et al., 2018). These findings provide a further rationale for analyzing global changes in lncRNA expression in NAFLD and NASH.
Recent bioinformatics and high-throughput sequencing studies have revealed that lncRNAs are differentially expressed in patients with hypoalphalipoproteinemia and MI caused by abnormal lipid metabolism (Wang et al., 2019). Differently expressed lncRNAs and mRNAs in atherosclerosis by analyzing dataset GSE28829 (Wang et al., 2019). A total of 654 lncRNAs and 5,784 mRNAs were significantly dysregulated in the progression of atherosclerosis (Wang et al., 2019). Moreover, six lncRNAs, ZFAS1 (ZNFX1 antisense RNA 1), LOC100506730, LOC100506691, DOCK9-AS2, RP11-6I2.3, and LOC100130219, were confirmed as potential novel therapeutic and prognostic targets for atherosclerosis (Wang et al., 2019). LncRNA ENST00000416361 was higher in the plasma of 50 patients with coronary artery disease (CAD) than the 50 healthy volunteers (Li P. et al., 2020). SREBP1 and SREBP2 were also up-regulated in CAD patients and showed positive correlations with ENST00000416361 (Li P. et al., 2020). Single nucleotide polymorphisms (SNPs) on the cyclin-dependent kinase inhibitor 2B antisense RNA (ANRIL) and MALAT1, two lncRNAs, affect the prognosis of MI (Li Y. et al., 2020). ANRIL rs9632884 and MALAT1 rs3200401 were significantly associated with the lipid levels of both controls and MI patients (Li Y. et al., 2020). KCNQ1 overlapping transcript 1 (KCNQ1OT1) was found to be increased in the serum of myocardial infarction (MI) patients, ischemia/reperfusion (I/R) mouse and hypoxia/reoxygenation (H/R)-induced cell model (Li J. et al., 2021). Moreover, several SNPs interacted with sex and age and modified the total cholesterol (rs9632884), LDL-C (rs1537373), and creatinine levels, affecting the risk of MI (Li Y. et al., 2020). These studies using clinical specimens and in vitro disease models have suggested that lncRNAs are involved in lipid-related diseases. However, the results should be further validated via in vitro and in vivo systems. Further research is required to analyze potential biomarkers and therapeutic targets in various lipid-related diseases (see Figure 3). This review provides a comprehensive insight into the current knowledge regarding the involvement of lncRNAs in regulating lipid metabolism, which may unveil the potential biomarkers and therapeutic targets for treating lipid-related diseases (Table 1).
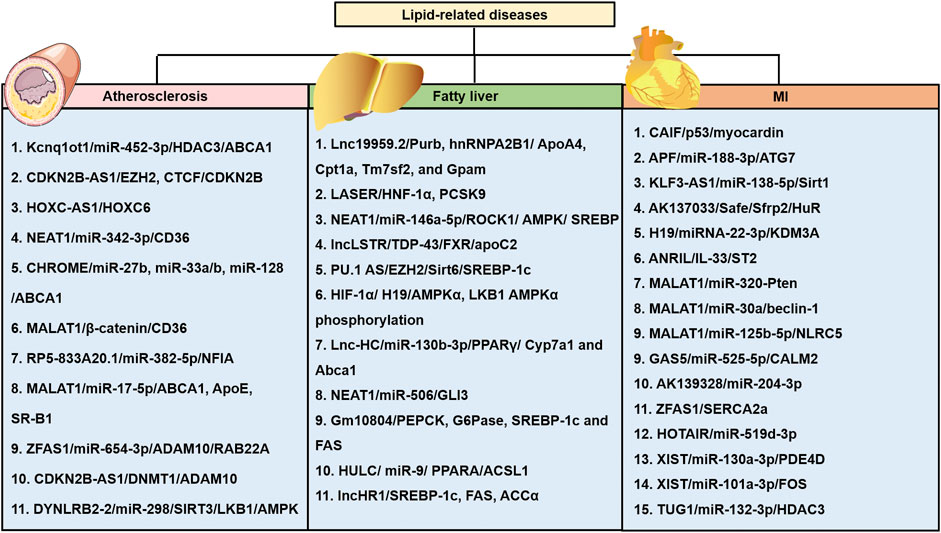
FIGURE 3. LncRNAs are involved in the three major diseases, including atherosclerosis, NAFLD, and myocardial infarction (MI) caused by abnormal cholesterol levels and various lipid fractions. Various lncRNAs and their mechanisms are illustrated. APF: autophagy promoting factor; CAIF: cardiac autophagy inhibitory factor; CALM2: calmodulin 2; GSA5: growth arrest-specific transcript 5; lncRNA XIST: long non-coding RNA X-inactive specific transcript; NLRC5: nucleotide-binding and oligomerization domain-like receptor C5; Sfrp2: secreted frizzled-related protein 2.
LncRNAs are Ideal Diagnostic Biomarkers and Therapeutic Targets
Diagnosis of several lipid-related diseases and their associated disease risks are mainly accomplished by analyzing the concentrations of lipid components such as total cholesterol, HDL, LDL, and triglycerides in the blood (Gotto, 2011; Paredes et al., 2019). This method only obtains accurate results when patients are fasted for at least 9–12 h. However, it provides limited information on cholesterol levels. Thus, it is necessary to search for better diagnostics and novel biomarkers for lipid-related diseases to overcome these disadvantages. LncRNAs are present in body fluids and are as stable as mRNA. Due to their tissue-specific properties, lncRNAs can be used as clinical indicators for diagnosis and are expected to become a new target for disease treatment (Table 1). Therefore, the application of lncRNAs as diagnostic biomarkers can result in a timely collection of more accurate and detailed disease information and risk factor data.
Previous attempts to use lncRNAs as biomarkers for disease diagnosis have been demonstrated in several cancer studies (Ratti et al., 2020) (Table 2). They revealed the functional roles of lncRNAs during cancer progression, including tumorigenesis, metastasis, and resistance to cancer treatment (Shen et al., 2015; Bin et al., 2018). Interestingly, some lipid-related lncRNAs mentioned in this review have also been emphasized in some cancer studies and proposed as potential diagnostic biomarkers (Peng et al., 2020). For example, the CHROME, which is mainly involved in cholesterol efflux and HDL biogenesis, was elevated in the plasma and atherosclerotic plaques of individuals and identified as a novel biomarker for the progression of CAD (Hennessy et al., 2019). On the other hand, plasma LeXis, which participates in cholesterol metabolism and the development of hepatic steatosis, was found to act as a non-invasive diagnostic biomarker for NASH (Park et al., 2020). NEAT1 and ANRIL, which are associated with cholesterol synthesis and MI, respectively, were suggested to be biomarkers that identify non-small cell lung carcinoma (NSCLC) (Yu X. et al., 2017; Osielska and Jagodziński, 2018). Furthermore, elevated plasma levels of HULC, which is involved in cholesterol synthesis, were identified as a biomarker for liver cancer (Xie et al., 2013). Additionally, the correlation between MALAT1, known to participate in cholesterol efflux, and lung cancer has been suggested as a diagnostic indicator (Lin et al., 2018). Moreover, the role of TUG1, an atherosclerosis-associated lncRNA, in various cancers has been previously studied (Niu et al., 2017; Guo et al., 2019). TUG1 was found to recruit specific RNA-binding proteins to facilitate cancer progression (Duan et al., 2019). These results suggest that lncRNAs play multiple functional roles in various disease processes and, as has frequently been reported in recent studies, cholesterol homeostasis is closely related to cancer occurrence. Collectively, these reports on lncRNAs in cancer indicate that the development of lncRNA biomarkers for diagnosing lipid-related diseases is very promising.
Importantly, from a therapeutic perspective, the best approach to prevent and treat lipid-related diseases is to make certain lifestyle modifications, such as exercising more and consuming a healthy diet (Mannu et al., 2013). However, if high lipid levels persist, medication must be taken to lower them. As mentioned earlier, the diagnosis criteria for lipid-related diseases are based on detecting cholesterol levels present in plasma (Płaczkowska et al., 2014). Thus, the primary purpose of treatment is to reduce cholesterol to appropriate levels. However, it is essential to note that the relationship between cholesterol and lipid-related diseases is ever-changing, which means that treatments also vary depending on the type and condition of the related disease. For instance, statin-based drugs, bile acid sequestrants, and cholesterol absorption inhibitors (Ezetimibe) are used clinically for different conditions. Specifically, statins decrease substances required for liver cholesterol production, bile oxides or bile acid sequestrants facilitate bile acid production from cholesterol, and cholesterol absorption inhibitors reduce cholesterol and limit cholesterol absorption from the small intestine (Taoufiq et al., 2011). In addition, drugs that only increase the absorption of LDL cholesterol have also been increasingly used recently (Lee et al., 2020). Due to their specific actions and side effects, these drugs are commonly used in combination in clinical and surgical treatments.
Importantly, lncRNAs involved in lipid metabolism can also be used as potential therapeutic targets to maintain cholesterol levels in the normal range. In general, RNA interference (RNAi), using shRNA, siRNA, or anti-sense oligonucleotide (ASO), is the most promising approach to target lncRNA silencing (Chi et al., 2017). This approach has been proven effective at the whole animal and cellular levels through various research (Liu et al., 2017; Zhang L. et al., 2020). For instance, the lentiviral shRNA targeting of lncRNA myocardial infarction associated transcript (MIAT) significantly attenuates atherosclerosis progression and increases plaque stability in vivo (Ye et al., 2019). Thus, a novel method for achieving safe and efficient RNAi delivery should be investigated and developed by further research. Furthermore, ASO-based methods are also studied for more stable and less off-target occurrence in addition to RNA interference technology (Maruyama and Yokota, 2020). For example, MALAT1 targeted ASO has been developed, and its inhibitory effect has been identified using animal models of malignancy (Amodio et al., 2018). Moreover, besides the method that targets lncRNA itself, controlling lncRNA function by inhibiting its interaction with the RNA-binding proteins has also been attempted (Kung et al., 2013; Bhat et al., 2016). However, note that RNA interference therapeutics have recently been progressed through preclinical development into clinical trials (Bobbin and Rossi, 2016). Thus, applying these as ideal clinical therapeutics requires the development of safe and effective delivery systems.
Small molecules have been extensively used for the therapeutic targeting of various diseases. These compounds have greater cellular uptake and fewer administrative challenges than antisense oligonucleotides and viral vectors for RNAi delivery. Small molecule inhibitors target lncRNAs by preventing them from binding to their RNA-binding proteins (RBPs). After analysing the lncRNA expression profiles from lncRNA modulator atlas in pan-cancer (LncMAP) database by bioinformatics analysis, the lncRNA network consists of 1,206 nodes and 4,770 drug-lncRNA associations to examine the global relationship between small molecule drugs and their affected lncRNAs (He et al., 2019). In addition, small molecules were screened to modulate the lncRNA HOX transcript antisense RNA (HOTAIR)-enhancer of zeste homolog2 (EZH2) interaction using alphaScreen technology (Pedram Fatemi et al., 2015). The interaction was inhibited with HOTAIR-polycomb repressive complex 2 (PRC2) binding through small-molecule intervention resulting in reduced metastatic phenotypes in many cancers, including breast (Gupta et al., 2010), colorectal (Kogo et al., 2011), and hepatocellular carcinomas (El-Khazragy et al., 2020). However, it is necessary to investigate the lncRNA-protein interaction and pharmacological trends further to develop more effective small molecule drugs (Figure 3).
Conclusion and Future Perspectives
Recent studies have shown that lncRNAs are involved in various lipid-related diseases (Table 1), thereby opening up a new research field and providing insight for lncRNAs as important eukaryotic transcripts. Concerning the correlation between lncRNAs regulation and lipid-related diseases, atherosclerosis is the most frequently studied disease (Ye et al., 2021). The occurrence of lipid-related diseases is due to the inactivation of suppressor genes and the activation of pathogenic genes. Thus, screening and identifying candidate biomarkers for prognosis, monitoring, and evaluating patients’ responses to therapies is required to develop novel strategies for lipid-related disease therapies. Also, ncRNAs (miRNAs and lncRNAs), DNA methylation, and histone modifications can epigenetically regulate gene expression. LncRNAs have recently served as important regulators of lipid-related diseases via various biological processes, including lipid metabolism, lipid accumulation, lipid synthesis, and cholesterol efflux (Sallam et al., 2018; Chen X. et al., 2019; Wang Z. et al., 2020; Zuo et al., 2020). Thus, there is a considerable thrill in using lncRNAs as a critical therapeutic target in treating lipid-related diseases.
Recent studies have demonstrated that lncRNAs could be detected in the blood plasma, tumor tissue, and urine, making them serve as promising biomarkers for development as disease, including atherosclerosis, MI, and cancer diseases (Dastmalchi et al., 2020; Fattahi et al., 2020). Genome-wide sequencing techniques have emerged as an important technology and reported a large number of newly dysregulated lncRNAs, implying promising results about the broad application prospects of lncRNAs in the prognosis and diagnosis of lipid-related diseases. Deregulation of many lncRNAs, such as H19 (Pan, 2017), TUG1 (Li et al., 2018), GAS5 (Chen et al., 2017), RAPIA (Sun et al., 2020), MIAT (Ye et al., 2019), CASC11 (Tao et al., 2019), NEXN-AS1 (Hu Y.-W. et al., 2019), and lnc00113 (Yao et al., 2018), has been detected in patients with atherosclerosis. LncRNAs including H19, TUG1, MIAT, and CASC11 could be detected in serum samples as a potential diagnostic marker in patients with atherosclerosis. In addition to establishing the functional role of lncRNAs in diagnosis, some lncRNAs such as AL117190.1, COL4A2-AS1, LINC00184, MEG3 and MIR22HG could function as crucial prognostic markers for patients (Yao et al., 2019). Besides, as diagnostic and prognostic markers, lncRNAs such as H19 (Yörüker et al., 2018), MEG3 (Wan and Zhao, 2020), PVT1 (Pan et al., 2019), FAM83H antisense RNA 1 (FAM83H-AS1) (El-Ashmawy et al., 2020), SNHG1 (Xiao et al., 2018), and LUCAT1 (Xing et al., 2021) are involved in the process of various cancer progression. Thus, we speculate that dysregulated lncRNAs may be used as biomarkers to provide diagnosis and prognostic of lipid-related diseases but also are useful in therapeutic applications.
Although it is well established that high concentrations of serum cholesterol levels facilitate the development of atherosclerosis (Johnston et al., 2017), the association of LDL-C or other lipids with atherosclerosis remains controversial. To date, a large number of lncRNAs associated with lipid metabolism and lipid-related diseases have been identified through RNA-seq and bioinformatics analyses. The functions of these lncRNAs may have important clinical implications in lipid metabolism and lipid-related diseases since they provide a myriad of possibilities for the diagnostics and treatment of these diseases. Furthermore, lncRNAs have been described as high tissue- and cell type-specific expression patterns (Kopp and Mendell, 2018; Antonov et al., 2019), which could be classified as different subclasses of lipid-related diseases or even predict responses to treatments. However, our current knowledge of the effect of lncRNAs on lipid-related diseases is possibly only the tip of the iceberg. Thus, more comprehensive investigations should be conducted to better understand how lncRNAs affect lipid-related diseases and develop new therapies.
The study of lncRNAs involved in controlling the cholesterol levels, specifically lncRNAs that directly interact with target genes or epigenetic proteins at the transcriptional level, may contribute to developing novel drugs to treat lipid-related diseases. Importantly, the latest next-generation sequencing-based big data research has identified numerous lncRNAs associated with various lipid-related diseases (Ye et al., 2021). However, further molecular biological research is needed to deepen the understanding of the association between various lncRNAs discovered and actual genetic mechanisms.
This review summarised various lipid-related lncRNAs and their target genes that play essential roles in lipid metabolism and lipid-related diseases. The involvement of lncRNAs was abnormally expressed in certain disease conditions, including atherosclerosis (Gao and Guo, 2021), myocardial infarction (Li J. et al., 2021), non-alcoholic fatty liver disease (Li J.-z. et al., 2021), and hypercholesterolemia (Tontonoz et al., 2017). Furthermore, a large number of lncRNAs identified from various studies were found to be associated with a diverse range of diseases. As lncRNAs are structurally and functionally conserved, further research is required to develop more effective diagnostics and therapeutics in this field or reveal the mechanism of certain diseases (see Figure 4 and Table 1). Altogether, advancing the knowledge of these lncRNAs and their functions is crucial for developing novel detection and modification methods.
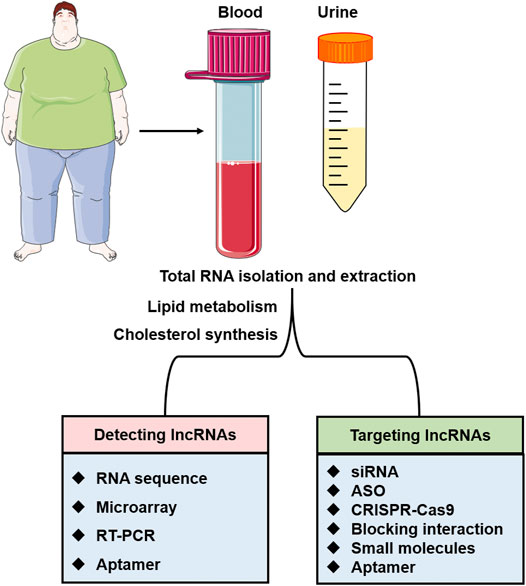
FIGURE 4. The application of lncRNAs as therapeutic targets and diagnostic biomarkers. LncRNAs in urine or blood specimens can be detected by various methods such as RNA sequence, microarray, RT-PCR, and aptamer. The interactions of lncRNAs with target proteins and lncRNAs involved in lipid metabolism and cholesterol synthesis will be the potential therapeutic targets for lipid-related diseases.
Author Contributions
W-CY and X-FP conceived and outlined the article. W-CY, S-FH, CH, and LJ surveyed the literature and wrote the article. W-CY and S-FH researched the literature and provided suggestions. W-CY and X-FP conceived ideas and the initial design. All the authors have approved the manuscript for submission.
Funding
The authors gratefully acknowledge the financial support from the National Natural Sciences Foundation of China (Grant No. 82000407), the Guangdong Natural Sciences Foundation (Grant No. 2019A1515110080), the Guangdong Medical Research Foundation (Grant No. B2021153), and the Medical Research Fund project of Qingyuan People’s Hospital (20190219 and 20190226).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amodio, N., Stamato, M. A., Juli, G., Morelli, E., Fulciniti, M., Manzoni, M., et al. (2018). Drugging the lncRNA MALAT1 via LNA gapmeR ASO Inhibits Gene Expression of Proteasome Subunits and Triggers Anti-multiple Myeloma Activity. Leukemia 32, 1948–1957. doi:10.1038/s41375-018-0067-3
Antonov, I. V., Mazurov, E., Borodovsky, M., and Medvedeva, Y. A. (2019). Prediction of lncRNAs and Their Interactions with Nucleic Acids: Benchmarking Bioinformatics Tools. Brief. Bioinformatics 20, 551–564. doi:10.1093/bib/bby032
Arenas, A., Cuadros, M., Andrades, A., García, D., Coira, I., Rodríguez, M., et al. (2020). LncRNA DLG2-AS1 as a Novel Biomarker in Lung Adenocarcinoma. Cancers 12. doi:10.3390/cancers12082080
Atanasovska, B., Rensen, S. S., van der Sijde, M. R., Marsman, G., Kumar, V., Jonkers, I., et al. (2017). A Liver-specific Long Noncoding RNA with a Role in Cell Viability Is Elevated in Human Nonalcoholic Steatohepatitis. Hepatology 66, 794–808. doi:10.1002/hep.29034
Bai, Y., Zhang, Q., Su, Y., Pu, Z., and Li, K. (2019). Modulation of the Proliferation/Apoptosis Balance of Vascular Smooth Muscle Cells in Atherosclerosis by lncRNA-MEG3 via Regulation of miR-26a/Smad1 Axis. Int. Heart J. 60, 444–450. doi:10.1536/ihj.18-195
Bao, J., Zhang, S., Meng, Q., and Qin, T. (2020). SNHG16 Silencing Inhibits Neuroblastoma Progression by Downregulating HOXA7 via Sponging miR-128-3p. Neurochem. Res. 45, 825–836. doi:10.1007/s11064-020-02955-x
Bhat, S. A., Ahmad, S. M., Mumtaz, P. T., Malik, A. A., Dar, M. A., Urwat, U., et al. (2016). Long Non-coding RNAs: Mechanism of Action and Functional Utility. Non-coding RNA Res. 1, 43–50. doi:10.1016/j.ncrna.2016.11.002
Bin, X., Hongjian, Y., Xiping, Z., Bo, C., Shifeng, Y., and Binbin, T. (2018). Research Progresses in Roles of LncRNA and its Relationships with Breast Cancer. Cancer Cel. Int. 18, 179. doi:10.1186/s12935-018-0674-0
Bobbin, M. L., and Rossi, J. J. (2016). RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 56, 103–122. doi:10.1146/annurev-pharmtox-010715-103633
Brown, M. S., and Goldstein, J. L. (1999). A Proteolytic Pathway that Controls the Cholesterol Content of Membranes, Cells, and Blood. Proc. Natl. Acad. Sci. 96, 11041–11048. doi:10.1073/pnas.96.20.11041
Burenina, O. Y., Lazarevich, N. L., Kustova, I. F., Shavochkina, D. A., Moroz, E. A., Kudashkin, N. E., et al. (2021). Panel of Potential lncRNA Biomarkers Can Distinguish Various Types of Liver Malignant and Benign Tumors. J. Cancer Res. Clin. Oncol. 147, 49–59. doi:10.1007/s00432-020-03378-5
Butt, A. A., Yan, P., Chew, K. W., Currier, J., Corey, K., Chung, R. T., et al. (2017). Risk of Acute Myocardial Infarction Among Hepatitis C Virus (HCV)-Positive and HCV-Negative Men at Various Lipid Levels: Results from ERCHIVES, Clin. Infect. Dis., 65. 557–565. doi:10.1093/cid/cix359
Chen, L., Zheng, S. Y., Yang, C. Q., Ma, B. M., and Jiang, D. (2019a). MiR-155-5p Inhibits the Proliferation and Migration of VSMCs and HUVECs in Atherosclerosis by Targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 23, 2223–2233. doi:10.26355/eurrev_201903_17270
Chen, L., Yang, W., Guo, Y., Chen, W., Zheng, P., Zeng, J., et al. (2017). Exosomal lncRNA GAS5 Regulates the Apoptosis of Macrophages and Vascular Endothelial Cells in Atherosclerosis. PloS one 12, e0185406. doi:10.1371/journal.pone.0185406
Chen, R., Shi, P., Zhang, Y., Wu, H., Li, X., Yang, W., et al. (2020). Long Non-coding RNAE330013P06 Promotes Progression of Breast Cancer with Type 2 Diabetes. J. Clin. Lab. Anal. 34, e23172. doi:10.1002/jcla.23172
Chen, X., Tan, X.-R., Li, S.-J., and Zhang, X.-X. (2019b). LncRNA NEAT1 Promotes Hepatic Lipid Accumulation via Regulating miR-146a-5p/ROCK1 in Nonalcoholic Fatty Liver Disease. Life Sci. 235, 116829. doi:10.1016/j.lfs.2019.116829
Chi, X., Gatti, P., and Papoian, T. (2017). Safety of Antisense Oligonucleotide and siRNA-Based Therapeutics. Drug Discov. Today 22, 823–833. doi:10.1016/j.drudis.2017.01.013
Cho, H., Li, Y., Archacki, S., Wang, F., Yu, G., Chakrabarti, S., et al. (2020). Splice Variants of lncRNA RNA ANRIL Exert Opposing Effects on Endothelial Cell Activities Associated with Coronary Artery Disease. RNA Biol. 17, 1391–1401. doi:10.1080/15476286.2020.1771519
Choi, S.-H., Agatisa-Boyle, C., Gonen, A., Kim, A., Kim, J., Alekseeva, E., et al. (2021). Intracellular AIBP (Apolipoprotein A-I Binding Protein) Regulates Oxidized LDL (Low-Density Lipoprotein)-Induced Mitophagy in Macrophages. Atvb 41, e82–e96. doi:10.1161/atvbaha.120.315485
Costet, P., Luo, Y., Wang, N., and Tall, A. R. (2000). Sterol-dependent Transactivation of theABC1 Promoter by the Liver X Receptor/Retinoid X Receptor. J. Biol. Chem. 275, 28240–28245. doi:10.1074/jbc.m003337200
Cui, M., Xiao, Z., Wang, Y., Zheng, M., Song, T., Cai, X., et al. (2015). Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells through an miR-9-Mediated RXRA Signaling Pathway. Cancer Res. 75, 846–857. doi:10.1158/0008-5472.can-14-1192
Dastmalchi, N., Safaralizadeh, R., and Nargesi, M. M. (2020). LncRNAs: Potential Novel Prognostic and Diagnostic Biomarkers in Colorectal Cancer. Cmc 27, 5067–5077. doi:10.2174/0929867326666190227230024
de Carvalho, C. C. C. R., and Caramujo, M. J. (2018). The Various Roles of Fatty Acids, 23. Basel, Switzerland: Molecules. doi:10.3390/molecules23102583
Di Mauro, S., Scamporrino, A., Petta, S., Urbano, F., Filippello, A., Ragusa, M., et al. (2019). Serum Coding and Non‐coding RNAs as Biomarkers of NAFLD and Fibrosis Severity. Liver Int. 39, 1742–1754. doi:10.1111/liv.14167
Dong, Z., Li, C., Yin, C., Xu, M., Liu, S., and Gao, M. (2019). LncRNA PU.1 AS Regulates Arsenic-Induced Lipid Metabolism through EZH2/Sirt6/SREBP-1c Pathway. J. Environ. Sci. 85, 138–146. doi:10.1016/j.jes.2019.05.019
Duan, W., Nian, L., Qiao, J., and Liu, N. N. (2019). LncRNA TUG1 Aggravates the Progression of Cervical Cancer by Binding PUM2. Eur. Rev. Med. Pharmacol. Sci. 23, 8211–8218. doi:10.26355/eurrev_201910_19128
Dłubek, J., Rysz, J., Jabłonowski, Z., Gluba-Brzózka, A., and Franczyk, B. (2021). The Correlation between Lipid Metabolism Disorders and Prostate Cancer. Curr. Med. Chem. 28, 2048–2061. doi:10.2174/0929867327666200806103744
El-Ashmawy, N. E., Hussien, F. Z., El-Feky, O. A., Hamouda, S. M., and Al-Ashmawy, G. M. (2020). Serum LncRNA-ATB and FAM83H-AS1 as Diagnostic/prognostic Non-invasive Biomarkers for Breast Cancer. Life Sci. 259, 118193. doi:10.1016/j.lfs.2020.118193
El-Khazragy, N., Elshimy, A. A., Hassan, S. S., Shaaban, M. H., Bayoumi, A. H., El Magdoub, H. M., et al. (2020). Lnc-HOTAIR Predicts Hepatocellular Carcinoma in Chronic Hepatitis C Genotype 4 Following Direct-Acting Antivirals Therapy. Mol. Carcinog 59, 1382–1391. doi:10.1002/mc.23263
Fasolo, F., Patrucco, L., Volpe, M., Bon, C., Peano, C., Mignone, F., et al. (2019). The RNA-Binding Protein ILF3 Binds to Transposable Element Sequences in SINEUP lncRNAs. FASEB J. 33, 13572–13589. doi:10.1096/fj.201901618rr
Fattahi, S., Kosari‐Monfared, M., Golpour, M., Emami, Z., Ghasemiyan, M., Nouri, M., et al. (2020). LncRNAs as Potential Diagnostic and Prognostic Biomarkers in Gastric Cancer: A Novel Approach to Personalized Medicine. J. Cel Physiol 235, 3189–3206. doi:10.1002/jcp.29260
Fernandes, J. C. R., Acuña, S. M., Aoki, J. I., Floeter-Winter, L. M., and Muxel, S. M. (2019). Long Non-coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 5 (1), 17. doi:10.3390/ncrna5010017
Fhu, C. W., and Ali, A. (2020). Fatty Acid Synthase: An Emerging Target in Cancer, 25. Molecules. 25, 3935. doi:10.3390/molecules25173935
Freyre, C., Rauher, P. C., Ejsing, C. S., and Klemm, R. W. J. M. C. (2019). MIGA2 Links Mitochondria, the ER, and Lipid Droplets and Promotes De Novo Lipogenesis in Adipocytes, Mol. Cel, 76, 811–825. doi:10.1016/j.molcel.2019.09.011
Fu, X., Cong, H., Zhao, S., Li, Y., Liu, T., Sun, Y., et al. (2020). Construction of Glycometabolism- and Hormone-Related lncRNA-Mediated Feedforward Loop Networks Reveals Global Patterns of lncRNAs and Drug Repurposing in Gestational Diabetes. Front. Endocrinol. 11, 93. doi:10.3389/fendo.2020.00093
Gao, H., and Guo, Z. (2021). LncRNA XIST Regulates Atherosclerosis Progression in Ox-LDL-Induced HUVECs. Open Med. (Warsaw, Poland) 16, 117–127. doi:10.1515/med-2021-0200
Garde, S., Akhter, R., Nguyen, M., Chow, C., and Eberhard, J. (2019). Periodontal Therapy for Improving Lipid Profiles in Patients with Type 2 Diabetes Mellitus: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 20. doi:10.3390/ijms20153826
Gluchowski, N. L., Becuwe, M., Walther, T. C., and Farese, R. V. (2017). Lipid Droplets and Liver Disease: from Basic Biology to Clinical Implications. Nat. Rev. Gastroenterol. Hepatol. 14, 343–355. doi:10.1038/nrgastro.2017.32
Go, G. W., and Mani, A. (2012). Low-density Lipoprotein Receptor (LDLR) Family Orchestrates Cholesterol Homeostasis. Yale J. Biol. Med. 85, 19–28.
Gong, X., Zhu, Y., Chang, H., Li, Y., and Ma, F. (2019). Long Noncoding RNA MALAT1 Promotes Cardiomyocyte Apoptosis after Myocardial Infarction via Targeting miR-144-3p. Biosci. Rep. 39. doi:10.1042/bsr20191103
Gotto, A. M. (2011). Jeremiah Metzger Lecture: Cholesterol, Inflammation and Atherosclerotic Cardiovascular Disease: Is it All LDL? Trans. Am. Clin. Climatol Assoc. 122, 256–289.
Guo, B. H., Zhao, Q., and Li, H. Y. (2019). TUG1 Promotes the Development of Prostate Cancer by Regulating RLIM. Eur. Rev. Med. Pharmacol. Sci. 23, 1926–1933. doi:10.26355/eurrev_201903_17230
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long Non-coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 464, 1071–1076. doi:10.1038/nature08975
Hahne, J. C., and Valeri, N. (2018). Non-Coding RNAs and Resistance to Anticancer Drugs in Gastrointestinal Tumors. Front. Oncol. 8, 226. doi:10.3389/fonc.2018.00226
Han, M., Lee, J., Kim, G., Lee, E., Lee, Y., Jang, S., et al. (2020). Expression of the Long Noncoding RNA GAS5 Correlates with Liver Fibrosis in Patients with Nonalcoholic Fatty Liver Disease. Genes 11. doi:10.3390/genes11050545
Han, X., Huang, S., Xue, P., Fu, J., Liu, L., Zhang, C., et al. (2019). PTPRE-AS1LncRNA Modulates M2 Macrophage Activation and Inflammatory Diseases by Epigenetic Promotion of PTPRE. Sci. Adv. 5, eaax9230. doi:10.1126/sciadv.aax9230
He, W., Liang, B., Wang, C., Li, S., Zhao, Y., Huang, Q., et al. (2019). MSC-regulated lncRNA MACC1-AS1 Promotes Stemness and Chemoresistance through Fatty Acid Oxidation in Gastric Cancer. Oncogene 38, 4637–4654. doi:10.1038/s41388-019-0747-0
Hennessy, E. J., van Solingen, C., Scacalossi, K. R., Ouimet, M., Afonso, M. S., Prins, J., et al. (2019). The Long Noncoding RNA CHROME Regulates Cholesterol Homeostasis in Primates. Nat. Metab. 1, 98–110. doi:10.1038/s42255-018-0004-9
Hu, W., Ding, Y., Wang, S., Xu, L., and Yu, H. (2020). The Construction and Analysis of the Aberrant lncRNA-miRNA-mRNA Network in Adipose Tissue from Type 2 Diabetes Individuals with Obesity. J. Diabetes Res. 2020, 3980742. doi:10.1155/2020/3980742
Hu, Y.-W., Guo, F.-X., Xu, Y.-J., Li, P., Lu, Z.-F., McVey, D. G., et al. (2019a). Long Noncoding RNA NEXN-AS1 Mitigates Atherosclerosis by Regulating the Actin-Binding Protein NEXN. J. Clin. Invest. 129, 1115–1128. doi:10.1172/jci98230
Hu, Y., Jin, Y., Wu, X., Yang, Y., Li, Y., Li, H., et al. (2019b). LncRNA-HGBC Stabilized by HuR Promotes Gallbladder Cancer Progression by Regulating miR-502-3p/SET/AKT axis. Mol. Cancer 18, 167. doi:10.1186/s12943-019-1097-9
Huang, C., Hu, Y.-W., Zhao, J.-J., Ma, X., Zhang, Y., Guo, F.-X., et al. (2016). Long Noncoding RNA HOXC-AS1 Suppresses Ox-LDL-Induced Cholesterol Accumulation through Promoting HOXC6 Expression in THP-1 Macrophages. DNA Cel. Biol. 35, 722–729. doi:10.1089/dna.2016.3422
Huang, J., Chen, S., Cai, D., Bian, D., and Wang, F. (2018). Long Noncoding RNA lncARSR Promotes Hepatic Cholesterol Biosynthesis via Modulating Akt/SREBP-2/HMGCR Pathway. Life Sci. 203, 48–53. doi:10.1016/j.lfs.2018.04.028
Huang, S., Tao, W., Guo, Z., Cao, J., and Huang, X. (2019a). Suppression of Long Noncoding RNA TTTY15 Attenuates Hypoxia-Induced Cardiomyocytes Injury by Targeting miR-455-5p. Gene 701, 1–8. doi:10.1016/j.gene.2019.02.098
Huang, Y. (2018). The Novel Regulatory Role of lncRNA-miRNA-mRNA axis in Cardiovascular Diseases. J. Cel Mol Med 22, 5768–5775. doi:10.1111/jcmm.13866
Huang, Y., Wang, L., Mao, Y., and Nan, G. (2019b). Long Noncoding RNA-H19 Contributes to Atherosclerosis and Induces Ischemic Stroke via the Upregulation of Acid Phosphatase 5. Front. Neurol. 10, 32. doi:10.3389/fneur.2019.00032
Jacquemyn, J., Cascalho, A., and Goodchild, R. E. J. E. R. (2017). The Ins and Outs of Endoplasmic Reticulumヽontrolled Lipid Biosynthesis, EMBO Rep., 18, e201643426. doi:10.15252/embr.201643426
Jeon, S.-M., Chandel, N. S., and Hay, N. (2012). AMPK Regulates NADPH Homeostasis to Promote Tumour Cell Survival during Energy Stress. Nature 485, 661–665. doi:10.1038/nature11066
Jiang, C., Zhao, Q., Wang, C., Peng, M., Hao, G., Liu, Z., et al. (2021). Downregulation of Long Noncoding RNA LINC00261 Attenuates Myocardial Infarction through the miR-522-3p/Trinucleotide Repeat-Containing Gene 6a (TNRC6A) Axis. Cardiovasc. Ther. 2021, 6628194. doi:10.1155/2021/6628194
Jiang, L., Shao, C., Wu, Q.-J., Chen, G., Zhou, J., Yang, B., et al. (2017). NEAT1 Scaffolds RNA-Binding Proteins and the Microprocessor to Globally Enhance Pri-miRNA Processing. Nat. Struct. Mol. Biol. 24, 816–824. doi:10.1038/nsmb.3455
Jin, M., Feng, H., Wang, Y., Yan, S., Shen, B., Li, Z., et al. (2020). Gentiopicroside Ameliorates Oxidative Stress and Lipid Accumulation through Nuclear Factor Erythroid 2-Related Factor 2 Activation. Oxidative Med. Cell. longevity 2020, 2940746. doi:10.1155/2020/2940746
Johnston, T. P., Korolenko, T. A., Pirro, M., and Sahebkar, A. (2017). Preventing Cardiovascular Heart Disease: Promising Nutraceutical and Non-nutraceutical Treatments for Cholesterol Management. Pharmacol. Res. 120, 219–225. doi:10.1016/j.phrs.2017.04.008
Khor, V. K., Shen, W.-J., Kraemer, F. B., and Care, M. (2013). Lipid Droplet Metabolism. Curr. Opin. Clin. Nutr. Metab. Care 16, 632–637. doi:10.1097/mco.0b013e3283651106
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cel Biol 10, 126–139. doi:10.1038/nrm2632
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T., et al. (2011). Long Noncoding RNA HOTAIR Regulates Polycomb-dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 71, 6320–6326. doi:10.1158/0008-5472.can-11-1021
Kopp, F., and Mendell, J. T. (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Kung, J. T. Y., Colognori, D., and Lee, J. T. (2013). Long Noncoding RNAs: Past, Present, and Future. Genetics 193, 651–669. doi:10.1534/genetics.112.146704
Lan, X., Wu, L., Wu, N., Chen, Q., Li, Y., Du, X., et al. (2019). Long Noncoding RNA Lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Ther. - Nucleic Acids 18, 954–965. doi:10.1016/j.omtn.2019.10.018
Lan, X., Yan, J., Ren, J., Zhong, B., Li, J., Li, Y., et al. (2016). A Novel Long Noncoding RNA Lnc-HC Binds hnRNPA2B1 to Regulate Expressions of Cyp7a1 and Abca1 in Hepatocytic Cholesterol Metabolism. Hepatology 64, 58–72. doi:10.1002/hep.28391
Lee, K., Hwang, H., and Cho, J. (2020). Long Non-coding RNA Associated with Cholesterol Homeostasis and its Involvement in Metabolic Diseases. Int. J. Mol. Sci. 21. doi:10.3390/ijms21218337
Leti, F., Legendre, C., Still, C. D., Chu, X., Petrick, A., Gerhard, G. S., et al. (2017). Altered Expression of MALAT1 lncRNA in Nonalcoholic Steatohepatitis Fibrosis Regulates CXCL5 in Hepatic Stellate Cells. Translational Res. 190, 25–39. doi:10.1016/j.trsl.2017.09.001
Li, F. P., Lin, D. Q., and Gao, L. Y. (2018). LncRNA TUG1 Promotes Proliferation of Vascular Smooth Muscle Cell and Atherosclerosis through Regulating miRNA-21/PTEN axis. Eur. Rev. Med. Pharmacol. Sci. 22, 7439–7447. doi:10.26355/eurrev_201811_16284
Li, J.-z., Ye, L.-h., Wang, D.-h., Zhang, H.-c., Li, T.-y., Liu, Z.-q., et al. (2021b). The Identify Role and Molecular Mechanism of the MALAT1/hsa-Mir-20b-5p/TXNIP axis in Liver Inflammation Caused by CHB in Patients with Chronic HBV Infection Complicated with NAFLD. Virus. Res. 298, 198405. doi:10.1016/j.virusres.2021.198405
Li, J., Tong, Y., Zhou, Y., Han, Z., Wang, X., Ding, T., et al. (2021a). LncRNA KCNQ1OT1 as a miR-26a-5p Sponge Regulates ATG12-Mediated Cardiomyocyte Autophagy and Aggravates Myocardial Infarction. Int. J. Cardiol.
Li, P., Yan, X., Xu, G., Pang, Z., Weng, J., Yin, J., et al. (2020a). A Novel Plasma lncRNA ENST00000416361 Is Upregulated in Coronary Artery Disease and Is Related to Inflammation and Lipid Metabolism. Mol. Med. Rep. 21, 2375–2384. doi:10.3892/mmr.2020.11067
Li, X., Li, L., Dong, X., Ding, J., Ma, H., and Han, W. (2021c). Circ_GRN Promotes the Proliferation, Migration, and Inflammation of Vascular Smooth Muscle Cells in Atherosclerosis through miR-214-3p/FOXO1 Axis. J. Cardiovasc. Pharmacol. 77, 470–479. doi:10.1097/fjc.0000000000000982
Li, Y., Ma, Z., Jiang, S., Hu, W., Li, T., Di, S., et al. (2017). A Global Perspective on FOXO1 in Lipid Metabolism and Lipid-Related Diseases. Prog. Lipid Res. 66, 42–49. doi:10.1016/j.plipres.2017.04.002
Li, Y., Zhang, D., Zhang, Y., Xu, X., Bi, L., Zhang, M., et al. (2020b). Association of lncRNA Polymorphisms with Triglyceride and Total Cholesterol Levels Among Myocardial Infarction Patients in Chinese Population. Gene 724, 143684. doi:10.1016/j.gene.2019.02.085
Lin, L., Li, H., Zhu, Y., He, S., and Ge, H. (2018). Expression of Metastasis-Associated Lung Adenocarcinoma Transcript 1 Long Non-coding RNA In Vitro and in Patients with Non-small Cell Lung Cancer. Oncol. Lett. 15, 9443–9449. doi:10.3892/ol.2018.8531
Liu, S., Cao, Q., An, G., Yan, B., and Lei, L. (2020). Identification of the 3-lncRNA Signature as a Prognostic Biomarker for Colorectal Cancer. Int. J. Mol. Sci. 21. doi:10.3390/ijms21249359
Liu, S., Horlbeck, M., Cho, S., Birk, H., Malatesta, M., He, D., et al. (2017). CRISPRi-Based Genome-Scale Identification of Functional Long Noncoding RNA Loci in Human Cells. New York, N.Y.: Science, 355.
Liu, S., Xu, D.-s., Li, M., Zhang, Y., Li, Q., Li, T.-t., et al. (2021a). Icariin Attenuates Endothelial-Mesenchymal Transition via H19/miR-148b-3p/ELF5 in Ox-LDL-Stimulated HUVECs. Mol. Ther. - Nucleic Acids 23, 464–475. doi:10.1016/j.omtn.2020.11.021
Liu, Y., and Ding, Z. (2017). Obesity, a Serious Etiologic Factor for Male Subfertility in Modern Society. Reproduction (Cambridge, England) 154, R123–R131. doi:10.1530/rep-17-0161
Liu, Y., Meng, W., Cao, H., and Wang, B. (2021b). Identification of MSC-AS1, a Novel lncRNA for the Diagnosis of Laryngeal Cancer. Eur. Arch. Otorhinolaryngol. 278, 1107–1118. doi:10.1007/s00405-020-06427-4
Loewen, G., Jayawickramarajah, J., Zhuo, Y., and Shan, B. (2014). Functions of lncRNA HOTAIR in Lung Cancer. J. Hematol. Oncol. 7, 90. doi:10.1186/s13045-014-0090-4
Lovren, F., Teoh, H., and Verma, S. (2015). Obesity and Atherosclerosis: Mechanistic Insights. Can. J. Cardiol. 31, 177–183. doi:10.1016/j.cjca.2014.11.031
Luzón-Toro, B., Fernández, R., Martos-Martínez, J., Rubio-Manzanares-Dorado, M., Antiñolo, G., and Borrego, S. (2019). LncRNA LUCAT1 as a Novel Prognostic Biomarker for Patients with Papillary Thyroid Cancer. Scientific Rep. 9, 14374. doi:10.1038/s41598-019-50913-7
Ma, Y., Zhang, J., Wen, L., and Lin, A. (2018). Membrane-lipid Associated lncRNA: A New Regulator in Cancer Signaling. Cancer Lett. 419, 27–29. doi:10.1016/j.canlet.2018.01.008
Mannu, G. S., Zaman, M. J., Gupta, A., Rehman, H. U., and Myint, P. K. (2013). Evidence of Lifestyle Modification in the Management of Hypercholesterolemia. Curr. Cardiol. Rev. 9, 2–14. doi:10.2174/157340313805076313
Maruyama, R., and Yokota, T. (2020). Knocking Down Long Noncoding RNAs Using Antisense Oligonucleotide Gapmers. Methods Mol. Biol. (Clifton, N.J.) 2176, 49–56. doi:10.1007/978-1-0716-0771-8_3
Michos, E. D., McEvoy, J. W., and Blumenthal, R. S. (2019). Lipid Management for the Prevention of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 381, 1557–1567. doi:10.1056/nejmra1806939
Mumtaz, P. T., and Online, S. B. J. B. P. (2017). LncRNAs and Immunity: Watchdogs for Host Pathogen Interactions, 19. doi:10.1186/s12575-017-0052-7
Musso, G., Gambino, R., and Cassader, M. (2013). Cholesterol Metabolism and the Pathogenesis of Non-alcoholic Steatohepatitis. Prog. lipid Res. 52, 175–191. doi:10.1016/j.plipres.2012.11.002
Niu, Y., Ma, F., Huang, W., Fang, S., Li, M., Wei, T., et al. (2017). Long Non-coding RNA TUG1 Is Involved in Cell Growth and Chemoresistance of Small Cell Lung Cancer by Regulating LIMK2b via EZH2. Mol. Cancer 16, 5. doi:10.1186/s12943-016-0575-6
Osielska, M. A., and Jagodziński, P. P. (2018). Long Non-coding RNA as Potential Biomarkers in Non-small-cell Lung Cancer: What Do We Know So Far? Biomed. Pharmacother. 101, 322–333. doi:10.1016/j.biopha.2018.02.099
Ou, M., Li, X., Zhao, S., Cui, S., and Tu, J. (2020). Long Non-coding RNA CDKN2B-AS1 Contributes to Atherosclerotic Plaque Formation by Forming RNA-DNA Triplex in the CDKN2B Promoter. EBioMedicine 55, 102694. doi:10.1016/j.ebiom.2020.102694
Pan, J. X. (2017). LncRNA H19 Promotes Atherosclerosis by Regulating MAPK and NF-kB Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 322–328.
Pan, J. X., and sciences, p. (2017). LncRNA H19 Promotes Atherosclerosis by Regulating MAPK and NF-kB Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 322–328.
Pan, X., Cheng, R., Zhu, X., Cai, F., Zheng, G., Li, J., et al. (2019). Prognostic Significance and Diagnostic Value of Overexpressed lncRNA PVT1 in Colorectal Cancer. Clin. Lab. 65. doi:10.7754/clin.lab.2019.190412
Paredes, S., Fonseca, L., Ribeiro, L., Ramos, H., Oliveira, J., and Palma, I. (2019). Novel and Traditional Lipid Profiles in Metabolic Syndrome Reveal a High Atherogenicity. Scientific Rep. 9, 11792. doi:10.1038/s41598-019-48120-5
Park, J., Kim, G., Jang, S., Lee, Y., Lee, E., Lee, H., et al. (2020). LeXisPlasma Long Noncoding RNA Is a Potential Diagnostic Marker for Non-alcoholic Steatohepatitis, 10. Basel, Switzerland: Life. doi:10.3390/life10100230
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537, 369–373. doi:10.1038/nature19342
Pedram Fatemi, R., Salah-Uddin, S., Modarresi, F., Khoury, N., Wahlestedt, C., and Faghihi, M. A. (2015). Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. J. Biomol. Screen. 20, 1132–1141. doi:10.1177/1087057115594187
Peng, L., Jiang, J., Tang, B., Nice, E. C., Zhang, Y.-Y., and Xie, N. (2020). Managing Therapeutic Resistance in Breast Cancer: from the lncRNAs Perspective. Theranostics 10, 10360–10377. doi:10.7150/thno.49922
Piccinin, E., Cariello, M., and Moschetta, A. (2021). Lipid Metabolism in colon Cancer: Role of Liver X Receptor (LXR) and Stearoyl-CoA Desaturase 1 (SCD1). Mol. aspects Med. 78, 100933. doi:10.1016/j.mam.2020.100933
Płaczkowska, S., Pawlik-Sobecka, L., et al. Placzkowska, S., Pawlik-Sobecka, L., Kokot, I., and Piwowar, A. (2014). [Incidence of Complex Metabolic Disorders Among Young People-Ppreliminary Report]. Pol. Merkur Lekarski 37, 269–273.
Ratti, M., Lampis, A., Ghidini, M., Salati, M., Mirchev, M. B., Valeri, N., et al. (2020). MicroRNAs (miRNAs) and Long Non-coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Targ Oncol. 15, 261–278. doi:10.1007/s11523-020-00717-x
Rinn, J. L., and Chang, H. Y. (2012). Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Sallam, T., Jones, M. C., Gilliland, T., Zhang, L., Wu, X., Eskin, A., et al. (2016). Feedback Modulation of Cholesterol Metabolism by the Lipid-Responsive Non-coding RNA LeXis. Nature 534, 124–128. doi:10.1038/nature17674
Sallam, T., Jones, M., Thomas, B. J., Wu, X., Gilliland, T., Qian, K., et al. (2018). Transcriptional Regulation of Macrophage Cholesterol Efflux and Atherogenesis by a Long Noncoding RNA. Nat. Med. 24, 304–312. doi:10.1038/nm.4479
Santos, C. R., and Schulze, A. (2012). Lipid Metabolism in Cancer. FEBS J. 279, 2610–2623. doi:10.1111/j.1742-4658.2012.08644.x
Shen, X.-h., Qi, P., and Du, X. (2015). Long Non-coding RNAs in Cancer Invasion and Metastasis. Mod. Pathol. 28, 4–13. doi:10.1038/modpathol.2014.75
Shi, X., Wei, Y., Li, H., Jiang, T., Zheng, X., Yin, K., et al. (2020). Long Non-coding RNA H19 in Atherosclerosis: what Role? Mol. Med. (Cambridge, Mass 26, 72. doi:10.1186/s10020-020-00196-w
Shimano, H., and Sato, R. (2017). SREBP-regulated Lipid Metabolism: Convergent Physiology - Divergent Pathophysiology. Nat. Rev. Endocrinol. 13, 710–730. doi:10.1038/nrendo.2017.91
Simion, V., Haemmig, S., and Feinberg, M. W. (2019). LncRNAs in Vascular Biology and Disease. Vasc. Pharmacol. 114, 145–156. doi:10.1016/j.vph.2018.01.003
Simion, V., Zhou, H., Haemmig, S., Pierce, J. B., Mendes, S., Tesmenitsky, Y., et al. (2020). A Macrophage-specific lncRNA Regulates Apoptosis and Atherosclerosis by Tethering HuR in the Nucleus. Nat. Commun. 11, 6135. doi:10.1038/s41467-020-19664-2
Spiroski, A.-M., Sanders, R., Meloni, M., McCracken, I. R., Thomson, A., Brittan, M., et al. (2021). The Influence of the LINC00961/SPAAR Locus Loss on Murine Development, Myocardial Dynamics, and Cardiac Response to Myocardial Infarction. Int. J. Mol. Sci. 22. doi:10.3390/ijms22020969
Stackhouse, C., Gillespie, G., and Willey, C. (2020). Exploring the Roles of lncRNAs in GBM Pathophysiology and Their Therapeutic Potential. Cells 9, 236910.3390/cells9112369.
Sukhorukov, V. N., Khotina, V. A., Chegodaev, Y. S., Ivanova, E., Sobenin, I. A., and Orekhov, A. N. (2020). Lipid Metabolism in Macrophages: Focus on Atherosclerosis. Biomedicines 8. doi:10.3390/biomedicines8080262
Sun, C., Fu, Y., Gu, X., Xi, X., Peng, X., Wang, C., et al. (2020). Macrophage-Enriched lncRNA RAPIA. Atvb 40, 1464–1478. doi:10.1161/atvbaha.119.313749
Sun, D., Yu, Z., Fang, X., Liu, M., Pu, Y., Shao, Q., et al. (2017). Lnc RNA GAS 5 Inhibits Microglial M2 Polarization and Exacerbates Demyelination. EMBO Rep. 18, 1801–1816. doi:10.15252/embr.201643668
Sun, R., and Zhang, L. (2019). Long Non-coding RNA MALAT1 Regulates Cardiomyocytes Apoptosis after Hypoxia/reperfusion Injury via Modulating miR-200a-3p/PDCD4 axis. Biomed. Pharmacother. 111, 1036–1045. doi:10.1016/j.biopha.2018.12.122
Tao, K., Hu, Z., Zhang, Y., Jiang, D., and Cheng, H. (2019). LncRNA CASC11 Improves Atherosclerosis by Downregulating IL-9 and Regulating Vascular Smooth Muscle Cell Apoptosis and Proliferation. Biosci. Biotechnol. Biochem. 83, 1284–1288. doi:10.1080/09168451.2019.1597621
Taoufiq, Z., Pino, P., N'Dilimabaka, N., Arrouss, I., Assi, S., Soubrier, F., et al. (2011). Atorvastatin Prevents Plasmodium Falciparum Cytoadherence and Endothelial Damage. Malar. J. 10, 52. doi:10.1186/1475-2875-10-52
Tontonoz, P., Wu, X., Jones, M., Zhang, Z., Salisbury, D., and Sallam, T. (2017). Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation 136, 776–778. doi:10.1161/circulationaha.117.029002
Tran, K.-V., Brown, E. L., DeSouza, T., Jespersen, N. Z., Nandrup-Bus, C., Yang, Q., et al. (2020). Human Thermogenic Adipocyte Regulation by the Long Noncoding RNA LINC00473. Nat. Metab. 2, 397–412. doi:10.1038/s42255-020-0205-x
Vernon, G., Baranova, A., and Younossi, Z. M. (2011). Systematic Review: the Epidemiology and Natural History of Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis in Adults. Aliment. Pharmacol. Ther. 34, 274–285. doi:10.1111/j.1365-2036.2011.04724.x
Wan, S., and Zhao, H. (2020). Analysis of Diagnostic and Prognostic Value of lncRNA MEG3 in Cervical Cancer. Oncol. Lett. 20, 183. doi:10.3892/ol.2020.12044
Wang, C., Shi, H., Chen, L., Li, X., Cao, G., and Hu, X. (2019). Identification of Key lncRNAs Associated with Atherosclerosis Progression Based on Public Datasets. Front. Genet. 10, 123. doi:10.3389/fgene.2019.00123
Wang, H., Gong, H., Liu, Y., and Feng, L. (2020a). Relationship between lncRNA-Ang362 and Prognosis of Patients with Coronary Heart Disease after Percutaneous Coronary Intervention. Biosci. Rep. 40. doi:10.1042/bsr20201524
Wang, J., Yang, W., Chen, Z., Chen, J., Meng, Y., Feng, B., et al. (2018). Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis. Diabetes 67, 581–593. doi:10.2337/db17-0799
Wang, S., Moustaid-Moussa, N., Chen, L., Mo, H., Shastri, A., Su, R., et al. (2014). Novel Insights of Dietary Polyphenols and Obesity. J. Nutr. Biochem. 25, 1–18. doi:10.1016/j.jnutbio.2013.09.001
Wang, X., Wang, L., Ma, Z., Liang, W., Li, J., Li, Y., et al. (2020b). Early Expressed Circulating Long Noncoding RNA CHAST Is Associated with Cardiac Contractile Function in Patients with Acute Myocardial Infarction. Int. J. Cardiol. 302, 15–20. doi:10.1016/j.ijcard.2019.12.058
Wang, Z., Yang, X., Kai, J., Wang, F., Wang, Z., Shao, J., et al. (2020c). HIF-1α-upregulated lncRNA-H19 Regulates Lipid Droplet Metabolism through the AMPKα Pathway in Hepatic Stellate Cells. Life Sci. 255, 117818. doi:10.1016/j.lfs.2020.117818
Wen, X., Ding, Y., Tan, Z., Wang, J., Zhang, D., and Wang, Y. (2020). Identification and Characterization of Cadmium Stress-Related LncRNAs from Betula Platyphylla. Plant Sci. 299, 110601. doi:10.1016/j.plantsci.2020.110601
Wu, D., Zhou, Y., Fan, Y., Zhang, Q., Gu, F., Mao, W., et al. (2019). LncRNA CAIF Was Downregulated in End-Stage Cardiomyopathy and Is a Promising Diagnostic and Prognostic Marker for This Disease. Biomarkers 24, 735–738. doi:10.1080/1354750x.2019.1677778
Xiao, B., Huang, Z., Zhou, R., Zhang, J., and Yu, B. (2018). The Prognostic Value of Expression of the Long Noncoding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) in Patients with Solid Malignant Tumors: A Systematic Review and Meta-Analysis. Med. Sci. Monit. 24, 5462–5472. doi:10.12659/msm.911687
Xiao, X., and Song, B.-L. (2013). SREBP: a Novel Therapeutic Target. Acta Biochim. Biophys. Sinica 45, 2–10. doi:10.1093/abbs/gms112
Xie, H., Ma, H., and Zhou, D. (2013). Plasma HULC as a Promising Novel Biomarker for the Detection of Hepatocellular Carcinoma. Biomed. Research International 2013, 136106. doi:10.1155/2013/136106
Xie, J., Wang, X., Liu, S., Chen, C., Jiang, F., Mao, K., et al. (2019). LncRNA SAMMSON Overexpression Distinguished Glioblastoma Patients from Patients with Diffuse Neurosarcoidosis. Neuroreport 30, 817–821. doi:10.1097/wnr.0000000000001278
Xing, C., Sun, S.-G., Yue, Z.-Q., and Bai, F. (2021). Role of lncRNA LUCAT1 in Cancer. Biomed. Pharmacother. 134, 111158. doi:10.1016/j.biopha.2020.111158
Xu, D., Wang, Z., Xia, Y., Shao, F., Xia, W., Wei, Y., et al. (2020). The Gluconeogenic Enzyme PCK1 Phosphorylates INSIG1/2 for Lipogenesis. Nature 580, 530–535. doi:10.1038/s41586-020-2183-2
Xu, J., and Taubert, S. (2021). Beyond Proteostasis: Lipid Metabolism as a New Player in ER Homeostasis. Metabolites 11, 52. doi:10.3390/metabo11010052
Yan, C., Chen, J., and Chen, N. (2016). Long Noncoding RNA MALAT1 Promotes Hepatic Steatosis and Insulin Resistance by Increasing Nuclear SREBP-1c Protein Stability. Scientific Rep. 6, 22640. doi:10.1038/srep22640
Yan, C., Zhang, Y., Zhang, X., Aa, J., Wang, G., and Xie, Y. (2018). Curcumin Regulates Endogenous and Exogenous Metabolism via Nrf2-FXR-LXR Pathway in NAFLD Mice. Biomed. Pharmacother. 105, 274–281. doi:10.1016/j.biopha.2018.05.135
Yan, L., Zhang, Y., Zhang, W., Deng, S.-Q., and Ge, Z.-R. (2020). lncRNA-NRF Is a Potential Biomarker of Heart Failure after Acute Myocardial Infarction. J. Cardiovasc. Trans. Res. 13, 1008–1015. doi:10.1007/s12265-020-10029-0
Yang, Y., Li, Y., Ma, Z., Jiang, S., Fan, C., Hu, W., et al. (2016). A Brief Glimpse at CTRP3 and CTRP9 in Lipid Metabolism and Cardiovascular protection. Prog. lipid Res. 64, 170–177. doi:10.1016/j.plipres.2016.10.001
Yang, Y., Tang, F., Wei, F., Yang, L., Kuang, C., Zhang, H., et al. (2019a). Silencing of Long Non-coding RNA H19 Downregulates CTCF to Protect against Atherosclerosis by Upregulating PKD1 Expression in ApoE Knockout Mice. Aging 11, 10016–10030. doi:10.18632/aging.102388
Yang, Z., Sun, Y., Liu, R., Shi, Y., and Ding, S. (2019b). Plasma Long Noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as Potential Novel Diagnostic Biomarkers for Gastric Cancer. Cmar Vol. 11, 6175–6184. doi:10.2147/cmar.s201935
Yang, Z., Zhang, T., Han, S., Kusumanchi, P., Huda, N., Jiang, Y., et al. (2021). Long Noncoding RNA H19 - a New Player in the Pathogenesis of Liver Diseases. Translational Res. 230, 139–150. doi:10.1016/j.trsl.2020.11.010
Yao, X., Yan, C., Zhang, L., Li, Y., and Wan, Q. (2018). LncRNA ENST00113 Promotes Proliferation, Survival, and Migration by Activating PI3K/Akt/mTOR Signaling Pathway in Atherosclerosis. Medicine 97, e0473. doi:10.1097/md.0000000000010473
Yao, Y., Zhang, T., Qi, L., Zhou, C., Wei, J., Feng, F., et al. (2019). Integrated Analysis of Co‐expression and ceRNA Network Identifies Five lncRNAs as Prognostic Markers for Breast Cancer. J. Cel Mol Med 23, 8410–8419. doi:10.1111/jcmm.14721
Ye, J., Lin, Y., Yu, Y., and Sun, D. (2020). LncRNA NEAT1/microRNA-129-5p/SOCS2 axis Regulates Liver Fibrosis in Alcoholic Steatohepatitis. J. translational Med. 18, 445. doi:10.1186/s12967-020-02577-5
Ye, W., Huang, S., Hou, L., Long, H., Yin, K., Hu, C., et al. (2021). Potential Therapeutic Targeting of lncRNAs in Cholesterol Homeostasis. Front. Cardiovasc. Med. 8, 688546. doi:10.3389/fcvm.2021.688546
Ye, Z., Yang, S., Xia, Y., Hu, R., Chen, S., Li, B., et al. (2019). LncRNA MIAT Sponges miR-149-5p to Inhibit Efferocytosis in Advanced Atherosclerosis through CD47 Upregulation. Cel Death Dis. 10, 138. doi:10.1038/s41419-019-1409-4
Yoon, J.-H., Abdelmohsen, K., Srikantan, S., Yang, X., Martindale, J. L., De, S., et al. (2012). LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cel. 47, 648–655. doi:10.1016/j.molcel.2012.06.027
Yörüker, E. E., Keskin, M., Kulle, C. B., Holdenrieder, S., and Gezer, U. (2018). Diagnostic and Prognostic Value of Circulating lncRNA H19 in Gastric Cancer. Biomed. Rep. 9, 181–186. doi:10.3892/br.2018.1116
Yu, C., Li, L., Xie, F., Guo, S., Liu, F., Dong, N., et al. (2018). LncRNA TUG1 Sponges miR-204-5p to Promote Osteoblast Differentiation through Upregulating Runx2 in Aortic Valve Calcification. Cardiovasc. Res. 114, 168–179. doi:10.1093/cvr/cvx180
Yu, F., Zheng, J., Mao, Y., Dong, P., Li, G., Lu, Z., et al. (2015). Long Non-coding RNA APTR Promotes the Activation of Hepatic Stellate Cells and the Progression of Liver Fibrosis. Biochem. biophysical Res. Commun. 463, 679–685. doi:10.1016/j.bbrc.2015.05.124
Yu, H., Ma, S., Sun, L., Gao, J., and Zhao, C. (2019). TGF-β1 U-pregulates the E-xpression of lncRNA-ATB to P-romote A-therosclerosis. Mol. Med. Rep. 19, 4222–4228. doi:10.3892/mmr.2019.10109
Yu, X., Li, Z., Zheng, H., Chan, M., and Wu, W. (2017a). NEAT1: A Novel Cancer-Related Long Non-coding RNA. Cel Prolif. 50. doi:10.1111/cpr.12329
Yu, Y., Nangia-Makker, P., Farhana, L., and Majumdar, A. (2017b). A Novel Mechanism of lncRNA and miRNA Interaction: CCAT2 Regulates miR-145 Expression by Suppressing its Maturation Process in colon Cancer Cells. Mol. Cancer 16, 155. doi:10.1186/s12943-017-0725-5
Zeng, Y., Ren, K., Zhu, X., Zheng, Z., and Yi, G. (2018). Long Noncoding RNAs: Advances in Lipid Metabolism. Adv. Clin. Chem. 87, 1–36. doi:10.1016/bs.acc.2018.07.001
Zhang, D., Zeng, S., and Hu, X. (2020a). Identification of a Three-Long Noncoding RNA Prognostic Model Involved Competitive Endogenous RNA in Kidney Renal clear Cell Carcinoma. Cancer Cel. Int. 20, 319. doi:10.1186/s12935-020-01423-4
Zhang, L., Xu, W., Gao, X., Li, W., Qi, S., Guo, D., et al. (2020b). lncRNA Sensing of a Viral Suppressor of RNAi Activates Non-canonical Innate Immune Signaling in Drosophila. Cell Host & Microbe 27, 115–128. doi:10.1016/j.chom.2019.12.006
Zhang, M., Chi, X., Qu, N., and Wang, C. (2018). Long Noncoding RNA lncARSR Promotes Hepatic Lipogenesis via Akt/SREBP-1c Pathway and Contributes to the Pathogenesis of Nonalcoholic Steatohepatitis. Biochem. biophysical Res. Commun. 499, 66–70. doi:10.1016/j.bbrc.2018.03.127
Zhang, T., Liu, H., Mao, R., Yang, H., Zhang, Y., Zhang, Y., et al. (2020c). The lncRNA RP11-142A22.4 Promotes Adipogenesis by Sponging miR-587 to Modulate Wnt5β Expression. Cel Death Dis. 11, 475. doi:10.1038/s41419-020-2550-9
Zhao, J., Chen, F., Ma, W., and Zhang, P. (2020). Suppression of Long Noncoding RNA NEAT1 Attenuates Hypoxia-Induced Cardiomyocytes Injury by Targeting miR-378a-3p. Gene 731, 144324. doi:10.1016/j.gene.2019.144324
Zhao, X., Xiong, X., Liu, T., Mi, L., Peng, X., Rui, C., et al. (2018). Long Noncoding RNA Licensing of Obesity-Linked Hepatic Lipogenesis and NAFLD Pathogenesis. Nat. Commun. 9, 2986. doi:10.1038/s41467-018-05383-2
Zheng, R., Du, M., Wang, X., Xu, W., Liang, J., Wang, W., et al. (2018). Exosome-transmitted Long Non-coding RNA PTENP1 Suppresses Bladder Cancer Progression. Mol. Cancer 17, 143. doi:10.1186/s12943-018-0880-3
Zheng, X., Tian, X., Zhang, Q., Shi, P., and Li, S. (2020). Long Non-coding RNA SAMMSON as a Novel Potential Diagnostic and Prognostic Biomarker for Oral Squamous Cell Carcinoma. J. dental Sci. 15, 329–335. doi:10.1016/j.jds.2019.11.008
Zhou, L., Yu, M., Arshad, M., Wang, W., Lu, Y., Gong, J., et al. (2018). Coordination Among Lipid Droplets, Peroxisomes, and Mitochondria Regulates Energy Expenditure through the CIDE-ATGL-Pparα Pathway in Adipocytes. Diabetes 67, 1935–1948. doi:10.2337/db17-1452
Zhuo, L., Wen, Y., Wang, Y., Liang, Z., Wu, G., Nong, M., et al. (2019). LncRNA SNHG8 Is Identified as a Key Regulator of Acute Myocardial Infarction by RNA-Seq Analysis. Lipids Health Dis. 18, 201. doi:10.1186/s12944-019-1142-0
Zuo, X., Chen, Z., Gao, W., Zhang, Y., Wang, J., Wang, J., et al. (2020). M6A-mediated Upregulation of LINC00958 Increases Lipogenesis and Acts as a Nanotherapeutic Target in Hepatocellular Carcinoma. J. Hematol. Oncol. 13, 5. doi:10.1186/s13045-019-0839-x
Glossary
AMI Acute myocardial infarction
ASH Alcoholic steatohepatitis
ATG12 autophagy-related 12 homologs
CAIF Cardiac autophagy inhibitory factor
CASIMO1 Cancer-associated small integral membrane open reading frame 1
CAVD calcific aortic valve disease
CHAST cardiac hypertrophy-associated transcript
HF heart failure
HMGCR HMG-CoA reductase
HOXC6 homeobox C6
HOXC-AS1 lncRNA HOXC cluster antisense RNA 1
ITGB1 integrin β1
JDP2 Jun dimerization protein 2
LDLR low-density lipoprotein receptor
LRP2BP low-density lipoprotein related receptor 2 binding protein
MALAT1 metastasis-associated lung adenocarcinoma transcript 1
MIAT myocardial infarction associated transcript
MMP1 matrix metalloproteinase 1
NEXN nexilin F-actin binding protein
NEXN-AS1 nexilin F-actin binding protein antisense RNA 1
PDCD4 programmed cell death 4
PPARA proliferator-activated receptor alpha
RAPIA associated with the progression and intervention of atherosclerosis
SOCS2 cytokine signalling 2
SQLE squalene epoxidase
TNRC6A trinucleotide repeat-containing gene 6a
BC bladder cancer
CYTOR cytoskeleton regulator RNA
FAM83H-AS1 FAM83H antisense RNA 1
FOXD2-AS1 FOXD2 adjacent opposite strand RNA 1
GBM glioblastoma
GC gastric cancer
HCC hepatocellular carcinoma
KIRC kidney renal clear cell carcinoma
LC laryngeal cancer
LncRNA-ATB lncRNA activated by TGF β
CRC colorectal cancer
LPM lncRNA prognostic model
LUAD lung adenocarcinoma
MSC-AS1 MSC antisense RNA 1
OSCC oral squamous cell carcinoma
PANDAR promoter of CDKN1A antisense DNA damage activated RNA
PCa prostate cancer
PTC papillary thyroid cancer
PTENP1 phosphatase and tensin homolog pseudogene 1
SAMMSON survival associated mitochondrial melanoma-specific oncogenic non-coding RNA
SMARCC2 SWI/SNF related, matrix associated, actin-dependent regulator of chromatin subfamily c member 2
SNHG18 small nucleolar RNA host gene 18
Keywords: lncRNAs, therapeutic targets, lipid metabolism, lipid-related diseases, biomarkers
Citation: Huang S-F, Peng X-F, Jiang L, Hu CY and Ye W-C (2021) LncRNAs as Therapeutic Targets and Potential Biomarkers for Lipid-Related Diseases. Front. Pharmacol. 12:729745. doi: 10.3389/fphar.2021.729745
Received: 23 June 2021; Accepted: 26 July 2021;
Published: 04 August 2021.
Edited by:
Yi-Chao Zheng, Zhengzhou University, ChinaReviewed by:
Shobana Sugumar, SRM Institute of Science and Technology, IndiaSiti Aishah Sulaiman, National University of Malaysia, Malaysia
Jens Claus Hahne, Institute of Cancer Research, United Kingdom
Copyright © 2021 Huang, Peng, Jiang, Hu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Chu Ye, eWV3ZW5jaHVAZ3pobXUuZWR1LmNu
†These authors have contributed equally to this work
 Shi-Feng Huang
Shi-Feng Huang Xiao-Fei Peng1†
Xiao-Fei Peng1† Ching Yuan Hu
Ching Yuan Hu Wen-Chu Ye
Wen-Chu Ye