- 1Key Laboratory of Green Chemical Engineering Process of Ministry of Education, Hubei Key Laboratory of Novel Reactor and Green Chemical Technology, School of Chemical Engineering and Pharmacy, Wuhan Institute of Technology, Wuhan, China
- 2School of Medicine, Jiujiang University, Jiujiang, China
- 3Medical University of South Carolina, Charleston, SC, United States
Objective: Traditional Chinese medicine (TCM) has garnered attention for its potential in cancer therapy. Yinchen Wuling San (YWLS), a classical herbal formula, has been traditionally used for liver-related conditions, but its bioactive components and molecular mechanisms relevant to hematologic malignancies such as acute myeloid leukemia (AML) remain unclear. This study aims to identify the active compounds and potential molecular targets of Yinchen Wuling San in the context of AML through network pharmacology analysis, and to experimentally validate the effects of selected candidate compounds in AML models.
Methods: Active ingredients from six YWLS herbs were screened via the TCMSP database using oral bioavailability ≥30% and DL ≥0.18 thresholds. Targets were predicted using SwissTargetPrediction, and AML-related genes were obtained from DisGeNET and GeneCards. Key overlapping targets were analyzed via STRING PPI networks and GO/KEGG enrichment. Molecular docking was performed between three core compounds (genkwanin, isorhamnetin, quercetin) and hub proteins (e.g., SRC) using Sybyl-X. ADME profiles were predicted using SwissADME, and molecular dynamics simulations (GROMACS) assessed complex stability. These compounds were further evaluated in vitro (viability, apoptosis, cell cycle, RT-qPCR, flow cytometry) and in vivo using an AML xenograft mouse model.
Results: Of 621 YWLS targets, 113 overlapped with 1,247 AML-related genes. PPI analysis identified hub genes, including AKT1, SRC, and EGFR. Enrichment analysis highlighted PI3K-AKT, MAPK, and JAK-STAT pathways. Genkwanin, isorhamnetin, and quercetin were predicted to target SRC, with strong molecular docking affinities. ADME analysis suggested favorable pharmacokinetics, and molecular dynamics simulations confirmed structural stability. In vitro, these compounds exhibited dose-dependent cytotoxicity, induced apoptosis, modulated the cell cycle, and downregulated SRC expression. Notably, Genkwanin promoted CD8+ T cell proliferation and inhibited leukemia growth, improving survival in a leukemia xenograft model.
Conclusion: YWLS compounds, particularly Genkwanin, exhibit significant anti-leukemic activity via apoptosis induction, cell cycle modulation, and promote T cells proliferation. Genkwanin emerges as a promising therapeutic candidate for AML, warranting further clinical investigation.
Introduction
Acute myeloid leukemia (AML) is one of the most common and severe types of malignant leukemia in adults and children worldwide (Cai and Levine, 2019). It is an aggressively hematopoietic malignancy that characterized with high heterogeneity, an increased number of accumulative immature clonal myeloid cells in the bone marrow and blood, inhibiting the hematopoiesis of normal blood cells, leading high mortality rate (Bruzzese et al., 2025; Juliusson et al., 2009). Despite advances in oncology, AML remains an aggressive hematopoietic malignancy characterized by rapid progression, marked heterogeneity, and poor clinical outcomes. The 5-year survival of patients with AML has barely improved over 40 years. The severity of AML is reflected in its persistently high mortality rate; the 5-year survival rate has shown only modest improvement over the past 4 decades. According to the Statistics of the American Cancer Society the overall survival rate (OS) of AML patients under 60 years old in recent 5 years is 26% (Miller et al., 2019). The 5-year OS of elderly patients over 60 years old is only 3%–8%, and the median OS is less than 1 year (Pandolfi et al., 2015; Zhang B. et al., 2021; Jing et al., 2024). Currently, the main treatments for AML are chemotherapy, molecular targeted therapy, immune-based therapy, and hematopoietic stem cell transplantation (Yang and Wang, 2018; Chen T. et al., 2024; Fu et al., 2021a). Although these advanced treatments offer available therapeutic options. However, these approaches are limited by issues such as drug resistance, disease relapse, high toxicity, and unsatisfactory long-term outcomes. More than half of young patients face the risk of relapse or refractory treatment, and the elderly or debilitated patients are unable to tolerate the serious side effects of conventional chemotherapy, 40% of patients with traditional regimen still fail to achieve complete response and have low OS (O'Donnell et al., 2017). High relapse was observed in patients with chemotherapy with the reformulation of standard therapy, such as Vyxeo™, cytarabine and daunorubicin liposome administration (Kantarjian et al., 2021). Novel FLT3 inhibitor is restricted to be applied for mutation targeted therapy (Sanz et al., 2009). Antibody-toxin Gemtuzumab ozogamicin conjugates that targeting CD33+ AML with high toxicity (Venugopal et al., 2021). Although chimeric antigen receptor (CAR) T-cell therapy offers a novel immunotherapy approach, its clinical application is hampered by high costs and risks of serious adverse effects, such as cytokine release syndrome (Tabata et al., 2021). These significant limitations underline the urgent need for novel, safer, and more effective therapeutic strategies for AML.
Given these challenges, increasing attention has turned to alternative and complementary medical approaches. Traditional Chinese Medicine (TCM), with its holistic treatment philosophy and multi-targeted therapeutic properties, has shown promise in supporting cancer therapy, including hematologic malignancies. Several TCM-derived phytochemicals, such as curcumin, berberine, and resveratrol, have been found to enhance chemotherapy efficacy and sensitize AML cells to treatment (Jin et al., 2013; Xiang et al., 2019). However, these effects are attributed to individual compounds rather than TCM as a comprehensive therapeutic system. Some clinical studies have reported that certain Chinese herbal medicines or integrative approaches combining herbal medicine with conventional chemotherapy may improve complete response rates and help mitigate chemotherapy-induced adverse effects such as diarrhea, vomiting, nausea, and bleeding or infection resulting from bone marrow suppression (Fleischer et al., 2017; Chen and Zhuang, 2024).
Yinchen Wuling San Prescription (YWLS), a classical TCM prescription originating from Synopsis of Golden Chamber, consists of six herbals: Artemisiae Scopariae Herba (ASH, Chinese name: Yinchen), Poria Cocos (PC, Chinese name: Fuling), Alisma Orientale (AO, Chinese name: Zexie), Atractylodes Macrocephala Koidz (AMK, Chinese name: Baizhu), Polyporus Umbellatus (PU, Chinese name: Zhuling), and Cinnamomi Ramulus (CR, Chinese name: Guizhi) (Haiqiang et al., 2016). Traditionally, YWLS is prepared by grinding the herbs into fine powder and taken orally with rice soup on an empty stomach, three times daily, as recorded in the ancient medical text Jin Gui Yao Lue. In modern clinical settings, YWLS is commonly administered as a decoction. The raw herbs are soaked and boiled twice (first for 2 h, then for 1 h), with the filtrates combined and concentrated into a specific volume. The extract can then be dried into granules or processed into capsules for ease of use (Leong et al., 2024). For experimental use, the decoction is typically concentrated and administered orally in animal models or reconstituted in suitable solvents for in vitro studies (Wu et al., 2022). YWLS has been widely used in the treatment of liver-related disorders such as icteric hepatitis (Zhang Y. et al., 2021), hepatic fibrosis, and non-alcoholic fatty liver disease (Liu and Zhao, 2024). Several individual herbal components of YWLS have shown potential anti-leukemic activities. For instance, Alisol B 23-acetate (ABA), a bioactive compound isolated from Alismatis Rhizoma (AO), exhibited P-glycoprotein (P-gp) reversal activity in a multidrug-resistant (MDR) leukemia cell line (K562-DR) (Wang et al., 2004), and demonstrated synergistic effects when combined with chemotherapeutic agents such as cisplatin and fluorouracil (5-FU) (Fong et al., 2007). In a study by Willimott et al., the water extract of Alismatis Rhizoma (AO), one of the herbal components of the traditional formula Long Dan Xie Gan Wan, was found to exhibit cytotoxic activity against human promyelocytic leukemia HL-60 cells, suggesting that AO extract may possess anti-leukemic properties when used as part of multi-herbal preparations (Willimott et al., 2009). Triterpenes separated from PC were identified to play a key role in anti-leukemia (Xu et al., 2020; Choi, 2015; Ukiya et al., 2002). AMK can induce apoptosis of human leukemia cells through increased production of reactive oxygen species (Huang et al., 2005; Huang et al., 2016). Polyporusterone derived from PU exhibited cytotoxic action on leukemia cell proliferation (Ohsawa et al., 1992). CR can alleviate chemoradiotherapy-induced gastrointestinal reaction, such as nausea, vomiting and constipation (Bu et al., 2024). Extracts isolated from ASH have anti-inflammation and anti-cancer activities (Cai et al., 2020). For example, Capillin potently inhibited human leukemia HL-60 cells growth through inducing apoptosis (Masuda et al., 2015). These indicated that active ingredients from YWLS may exert synergistic anti-leukemia effects. Despite these promising observations, the bioactive components of YWLS and their anti-leukemic mechanisms remain largely unexplored. Comprehensive elucidation of its molecular mechanisms is crucial for potential development into evidence-based, adjunctive AML therapies.
Network pharmacology acting as an emerging analytical approach has been widely employed to predict potential molecular mechanisms of pharmacological action for TCM through integrating drugs, diseases, and their corresponding targets into biomolecular network (Li and Zhang, 2013; Sun et al., 2021). In this study, we utilized a network pharmacology approach to systematically predict the active compounds, targets, and pathways associated with YWLS against AML (Supplementary Figure S1). Molecular docking and ADME (Absorption, Distribution, Metabolism, and Excretion) analysis were employed to evaluate drug-target interactions and drug-likeness. Molecular dynamics simulations were conducted to assess the stability of compound-protein complexes. Finally, the anti-leukemic potential of selected active ingredients was validated through in vitro assays. This study aims to uncover the underlying pharmacological mechanisms of YWLS against AML and to provide a scientific basis for its potential application as a novel anti-AML therapeutic strategy. Further in vivo investigations will be warranted to strengthen these findings.
Methods and materials
Identification of active ingredients of YWLS and its pharmacological targets
YWLS, a common traditional Chinese prescription, contains six herbs ASH, PC, AO, AMK, CR, and PU. A variety of compounds identified from these herbs were found to have anti-inflammation/oxidant/tumor activities and immune regulatory functions (Huang et al., 2016). The active ingredients of these herbs were retrieved from TCMSP database (https://tcmsp-e.com/tcmsp.php) (Ru et al., 2014) with parameters of drug-like properties (DL) ≥0.18 and oral bioavailability (OB) ≥30%.
The pharmacological targets of the ingredients of YWLS were screened from SwissTargetPrediction database (http://www.swisstargetprediction.ch/) (Daina et al., 2019). Additional unpredicted targets for these ingredients were included through literature review. The UniProt database (https://www.drugbank.ca/) (Toth et al., 2020) was employed to standardize gene names of targets. While this approach is widely adopted in network pharmacology studies, it may be biased toward well-characterized and frequently studied targets due to the nature of the underlying training data in these databases.
Screening disease targets
Pathological targets related to acute myeloid leukemia (AML) were obtained from the DisGeNET database (https://www.disgenet.org/) with a gene-disease association (GDA) score ≥0.1 and the GeneCards database (https://www.genecards.org/) with a relevance score ≥10. After removing duplicate genes, the intersecting targets of YWLS active ingredients and AML-related genes were identified using the “VennDiagram” package (version 1.7.3) in R software (version 4.2.0).
Protein-protein interaction (PPI) network and gene enrichment analysis
The STRING database (https://string-db.org/, version 11.5) was used to analyze interactions among the intersected genes, with a minimum interaction score threshold set at 0.4 (Szklarczyk et al., 2021). The resulting PPI network was visualized using Cytoscape (version 3.8.0) (Shannon et al., 2003). Densely connected modules were identified using the MCODE plugin (Bader and Hogue, 2003) with default parameters (Degree Cutoff = 2, Node Score Cutoff = 0.2, K-Core = 2, Max Depth = 100).
Network centrality was assessed using the CytoNCA plugin (Tang et al., 2015) (based on betweenness, closeness, and degree centralities), and hub genes were further screened with the cytoHubba plugin (Chin et al., 2014) using the Maximal Clique Centrality (MCC) method.
Gene Ontology (GO) functional enrichment (including biological process [BP], molecular function [MF], and cellular component [CC]) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted using the “clusterProfiler” R package (version 4.2.2) with a cutoff of p. adjust <0.05. Key AML-related signaling pathways were visualized using the Pathview R package (Luo and Brouwer, 2013) (version 1.38.0).
Construction of drug-target-disease network
Relationships between active ingredients, targets, and disease were organized using Perl scripts (Perl version 5.32.1). The integrated Drug-Target-Disease network was visualized by Cytoscape (version 3.8.0) (Shannon et al., 2003).
Gene expression profiling and prognostic evaluation of identified targets in AML
Gene expression data (log2(FPKM+1) format) and clinical information for AML patients were downloaded from the UCSC Xena database (https://xenabrowser.net/datapages/). Wilcoxon rank-sum test was used to compare gene expression levels between normal and AML samples. Univariate Cox regression analysis was performed using the “survival” R package (version 3.4.0) to identify targets associated with overall survival (OS). A p-value <0.05 was considered statistically significant.
Molecular docking
Structures of active compounds were downloaded from the TCMSP database (https://tcmsp-e.com/tcmsp.php) in.mol2 format, and the crystal structures of target proteins were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/). Molecular docking was performed using Sybyl-X 2.0 software. Ligand Preparation: Small molecules underwent energy minimization using the Tripos force field, applying 1,000 iterations and a convergence criterion of 0.005 kcal/(mol·Å). Protein Preparation: Proteins were preprocessed by removing water molecules, adding hydrogens, and optimizing side chains. Docking scores were calculated, and binding poses were evaluated. Docking interactions were visualized using PyMOL software (version 2.5.4).
ADME analysis
The pharmacokinetic properties (absorption, distribution, metabolism, and excretion), drug-likeness, and medicinal chemistry profiles of the selected compounds were evaluated using the SwissADME database (http://www.swissadme.ch/) (Daina et al., 2017).
Molecular dynamics simulation
Molecular dynamics simulations were conducted using GROMACS software (version 2020.3) (Abraham et al., 2015; Van Der Spoel et al., 2005). Ligand Topology: Ligand parameters were generated via SwissParam (https://www.swissparam.ch/) (Zoete et al., 2011). Force Field and Solvation: CHARMM36 force field and the TIP3P explicit water model were applied (Kutzner et al., 2015; Ul Haq et al., 2017). Complexes were solvated in a cubic box with at least 1.0 nm distance from the box edges, neutralized with Na+/Cl− ions. Energy Minimization: Conducted using 50,000 steps each of steepest descent and conjugate gradient methods. System Equilibration: NVT ensemble (constant number of particles, volume, and temperature) for 100 ps at 300 K using the V-rescale thermostat. NPT ensemble (constant number of particles, pressure, and temperature) for 100 ps at 1 atm using the Parrinello-Rahman barostat (Fatima et al., 2014). Production Run: 50 ns MD simulation with a 2 fs timestep using the leap-frog integration algorithm. Trajectory analyses included: Root-Mean-Square Deviation (RMSD) to monitor structural stability. Root-Mean-Square Fluctuation (RMSF) to assess residue flexibility. Radius of Gyration (Rg) to evaluate protein compactness. Solvent Accessible Surface Area (SASA) to explore solvation effects. Hydrogen Bond (Hbond) Analysis to characterize protein-ligand interactions. All analyses were conducted using GROMACS built-in modules and visualized with OriginPro 2022b. Binding Free Energy Calculation: The Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method was applied to estimate binding free energies of the protein-ligand complexes using the “g_mmpbsa” package. The total binding free energy was calculated based on the last 10 ns of the simulation trajectory (Monteiro et al., 2018; Patidar et al., 2019).
Cell culture
MOLM14 cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/strptomycin and grown at 37°C with 5% CO2.
Quercetin, Isorhamnetin, and Genkwanin (Sigma-Aldrich) were dissolved in Dimethylsulfoxide (DMSO) to prepare the stock solutions of 100 mmol/L. Different Genkwanin, Quercetin, and Isorhamnetin working solutions of 10, 20, 40, and 80 μmol/L were diluted from the stock solutions and used to treat MOLM14 cells at different assays.
Cell viability analysis
MOLM14 cells (1 × 105) were plated in 96-well plates. Cells per well were treated for 72 h with various concentrations of different compounds followed by Viability Dye eFluor™ 506 (Invitrogen, catalog: 65-0866-14) to each well for 30 min incubation. The cell viability was then assessed by flow cytometry. The cell viability was determined by normalizing to vehicle group.
Dose-response modeling and interpolation
To estimate tumor cell growth inhibition at additional concentrations, a four-parameter logistic (4 PL) model was fitted to the experimental dose-response data for Quercetin, Isorhamnetin, and Genkwanin. The fitted model was used to interpolate or extrapolate inhibition rates at untested concentrations of 0.1, 1, 10, and 100 μM. The 4 PL model is defined as:
where Y is the predicted inhibition rate, X is the drug concentration, and H is the Hill slope. Curve fitting and prediction were performed using the scipy.optimize.curve_fit function in Python.
Cell cycle and apoptosis assay
For cell cycle analysis of MOLM14 cells treated with Genkwanin, Quercetin, and Isorhamnetin (10, 20, 40, and 80 μmol/L) for 72-h. Cells in 96-well plate were harvested and washed with 1x phosphate buffered saline (PBS). The washed cells were fixed using Foxp3/Transcription Factor Staining Buffer Set and the Fixation/Permeabilization Kit (Cat.No: 00-5523-00, Invitrogen) for 30 min at room temperature. Ki-67 at indicated concentration was added to each well and incubated at 4°C for 30 min. DAPI was added at 2 mM/L for each well and analyzed on a BD LSRFortessa in 30 min.
Apoptosis assay was performed in MOLM14 cells treated with Genkwanin, Quercetin, and Isorhamnetin (10, 20, 40, and 80 μM) at 72-h following the PE-Annexin V Apoptosis Detection Kit (BD Pharmingen™, catalog: 559763) instructions, and analyzed on a BD LSRFortessa.
Mitochondria function assay
MOLM14 cells were treated with genkwanin, quercetin, or isorhamnetin (10, 20, 40, and 80 μM) in a 96-well plate for 72 h. After treatment, cells were centrifuged to collect a cell pellet and the supernatant was aspirated. Cells were gently resuspended in pre-warmed (37°C) RPMI 1640 medium containing MitoTracker® Deep Red FM (Thermo Fisher, Cat. No. M22426) and MitoTracker® Red CMXRos (Thermo Fisher, Cat. No. M7512) and incubated for 30 min at 37°C. After staining, cells were washed, resuspended in pre-warmed 1xPBS buffer, and analyzed by flow cytometry. Debris and apoptotic bodies were excluded based on FSC/SSC gating to focus the analysis on intact cells. All samples were prepared and stained under identical conditions, and comparisons were made relative to control-treated cells.
Quantitative RT-PCR (qRT-PCR)
Total RNA from MOLM14 cells was extracted using the RNeasy Mini Kit (QIAGEN, Cat. No. 74104) according to the manufacturer’s protocol. RNA concentration and purity (A260/A280 ratio) were assessed using a NanoDrop spectrophotometer, and samples with acceptable purity (A260/A280 between 1.8 and 2.0) were used for downstream analysis. One microgram of RNA was reverse transcribed to cDNA using the iScript™ cDNA synthesis kit (Bio-Rad, Cat. No. 1708891). Fifty nanograms of cDNA were used for qPCR on a CFX96 Real-Time PCR Detection System (Bio-Rad). qPCR reactions were performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Cat. No. 172-5270) with the following program: 95°C for 30 s; 40 cycles of 95°C for 15 s and 60°C for 1 min.
Relative gene expression was calculated using the 2−ΔΔCT method and normalized to GAPDH expression. Primers for SRC gene expression were described previously (Zhang et al., 2020). The 72-h treatment time point was selected to capture late-stage transcriptional changes, including those involved in apoptotic and stress response pathways. Despite partial loss of viability at this time point, sufficient viable cells remained for reliable RNA extraction and analysis.
Flow cytometry
All antibodies used for intracellular, or surface staining used in this study were purchased from eBioscience (San Diego, CA) or BD Bioscience (San Jose, CA) or Biolegend (San Diego, CA) unless otherwise stated. For cell surface antibody staining, each antibody was added at recommended concentration and incubated at 4°C for 30 min by blocking with 20% FBS for 15 min at 4°C, which prevents non-specific binding. For intracellular and cytokines staining, cells were fixed with Foxp3/Transcription Factor Staining Buffer Set and the Fixation/Permeabilization Kit (Cat. No: 00-5523-00, Invitrogen) for 30 min at room temperature followed by permeabilization with 1x permeabilization buffer. For the cytokine staining, samples were stimulated with phorbol 12-myristate 13-acetate (PMA) (50 ng/mL; Sigma-Aldrich), ionomycin (1 μg/mL; Sigma-Aldrich), and in the presence of GolgiStop monensin (1 μg/mL) for 4–5 h at 37°C. Cells were washed twice with 1X permeabilization buffer. Finally, samples were run on BD LSRII, BD LSR Fortessa (X-20) and analyzed using FlowJo 10.7.0 software (TreeStar).
T cell proliferation assay
Splenocytes were harvested from C57BL/6 mice and purified using a Pan T Cell Isolation Kit (Catalog No. 130-095-130). T cells were labeled with CFSE (5 μM) for 5 min at room temperature. A total of 1 × 105 cells per well were seeded into a 96-well round-bottom plate containing 200 μL of RPMI 1640 medium supplemented with 10% FBS, 2.5% 1M HEPES, 1% non-essential amino acids, 1% penicillin/streptomycin, and 1% sodium pyruvate.
Quercetin, Isorhamnetin, and Genkwanin were added to the respective wells at final concentrations of 0, 10, 20, 40, and 80 μM, with 0.1% DMSO serving as the vehicle control. After 72 h of culture, cells were harvested and analyzed by flow cytometry. Staining included CD4 (PE), CD8 (PE-Tex-Red), and viability dye (eFluor™ 506) to assess cell proliferation.
In vivo study of Genkwanin using a leukemia xenograft model
To evaluate the anti-leukemic efficacy of Genkwanin in vivo, we developed a human leukemia xenograft model using 8–10-week-old NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice, both male and female. Each mouse was injected intravenously with 1 × 105 MOLM14gfp cells. Four days after leukemia cell engraftment, mice were randomized into treatment and control groups. The treatment group received intraperitoneal injections of Genkwanin (5 mg/kg) twice weekly for 3 weeks, while the control group received vehicle only.
To simulate and evaluate the potential immunomodulatory effects of Genkwanin within a limited human immune context, mice received weekly intravenous adoptive transfers of 1 × 106 human peripheral blood mononuclear cells (PBMCs) isolated from healthy donors, starting on the first day of treatment. This approach enabled partial and transient reconstitution of human immune cells—primarily T lymphocytes—allowing for exploratory assessment of T cell-mediated responses, particularly CD8+ T cell activity, in the presence of Genkwanin.
Several studies have adopted similar strategies, using NSG mice humanized with PBMCs with or without leukemic cell co-engraftment to explore immunomodulatory functions of therapeutic agents or gene targets in leukemia and lymphoma models (Fu et al., 2021b; Adom et al., 2020; Wellhausen et al., 2023; Pyzer et al., 2017). These models provide valuable insight into human immune cell dynamics and treatment responses, despite not fully recapitulating the patient-matched immune milieu of autologous PDX systems.
Survival was monitored daily, and on Day 21 post-transfer, tumor burden and frequencies of CD8+ T cells in the bone marrow were quantified by flow cytometry. All animal experiments were conducted in accordance with protocols approved by the Animal Ethics Committee at Jiujiang University (Approval No. JJU20240076).
Results
Uncovering active ingredients in YWLS and their target genes
A total of 58 active ingredients in YWLS were identified through the TCMSP database and supplemented with findings from literature review (Table 1). These include 13 ingredients from ASH, seven from CR, 10 from AO, 11 from PU, 15 from PC, and seven from AMK. Notably, several compounds are shared between different sources: ASH and CR both contain beta-sitosterol; PC and PU share ergosta-7,22E-dien-3beta-ol and cerevisterol; AO and CR both include sitosterol; and PU and CR share peroxyergosterol. To identify the targets of these active ingredients, 621 targets were retrieved from the Swiss Target Prediction database (Supplementary Table S1).
Active ingredient-target-disease network analysis
To identify genes that are potentially related to AML pathogenesis, we retrieved 1,018 and 407 AML disease related genes (ADRGs) from DisGeNET and GeneCards, respectively (Figure 1A). A total of 1,247 ADRGs were obtained after deduplication (Supplementary Table S2). Targets of YWLS and AML were intersected using Venny, 113 targets of the active ingredients from YWLS overlapped with ADRGs (Figure 1B; Supplementary Table S3). Furthermore, we developed YWLS-ingredients-targets-AML network which includes 156 nodes and 841 edges using Perl language and visualized through Cytoscape (Figure 1C). The 14-acetyl-12-senecioyl-2E,8E,10E-atractylentriol, Cerevisterol and 12-senecioyl-2E,8E,10E-atractylentriolareare are active ingredients that have the most targets than other ingredients.

Figure 1. Construction of a compound-target-disease network based on the targets of active ingredients from YWLS and AML-associated genes. (A) Screening of acute myeloid leukemia related genes from DisGeNET and GeneCards databases. (B) The intersection of targets of active ingredients of YWLS and AML-related genes. (C) The active component-target-disease network. The red diamond represented the disease; The light green rectangle represented intersecting targets; The light purple diamond represented YWLS; The blue triangle represented active compounds. The edges represented the connection among active components, targets, and disease. (D) Two modules (AKT1 and PIK3CA) identified from the whole PPI network of 113 targets of active ingredients in YWLS and AML-associated genes. (E) 20 core targets determined by the degrees. Color represented the target degree.
Protein-protein interaction and gene enrichment analysis of YWLS-AML targets
The PPI network analysis was conducted to explore potential interactions of 113 intersection targets using the STRING database. Two modules were identified through the MCODE plugin via the Cytohubba plugin (Figure 1D). The first module, named AKT1, contains 39 nodes and 1,072 edges, while the second module includes 21 nodes and 168 edges. Among the hub genes, 17 out of 20 genes are part of the first module, including AKT1, SRC, EGFR, MTOR, CCND1, and HSP90AA1. The second module retains PIK3CA, CYN, and KIT as its top genes. Numerous reports have highlighted the association of these genes with AML pathogenesis and inflammation.
Constitutive activation of the PI3K/Akt/mTOR pathway occurs in 50%–80% of AML patients and is linked to reduced OS (Bertacchini et al., 2014; Darici et al., 2020). Downregulation of GLUT3 via the EGFR signaling pathway promotes apoptosis of AML cells (Zhuang et al., 2018). The deregulated activity of Src tyrosine kinases induces malignant transformation, and patients with leukemia have benefited from Src tyrosine kinase inhibitors (Rivera-Torres and San José, 2019). SRC is expressed and activated in most AML primary samples and cell lines, and its inhibitors can suppress the proliferation of AML cell lines and promote apoptosis (MacDonald et al., 2018). Additionally, mutations in CCND1 have been identified in adult AML patients, suggesting its potential as a therapeutic target (Eisfeld et al., 2017).
We also identified 20 hub genes in the network by ranking degree using the Cytohubba plugin in Cytoscape (Figure 1E; Supplementary Table S4). These findings prompted us to conduct a comprehensive analysis of their biological processes through gene function enrichment. Our analysis revealed that these genes are primarily involved in processes such as peptidyl-tyrosine phosphorylation, protein serine/threonine kinase activity regulation, and phosphatidylinositol 3-kinase signaling. The cellular components were predominantly enriched in processes related to transferring phosphorus-containing groups and membrane rafts. The primary molecular functions included protein tyrosine/serine/threonine kinase activity, phosphatase and growth factor binding, and transmembrane receptor protein kinase activity (Supplementary Figure S2A). This suggests that the active ingredients of YWLS may inhibit leukemia growth by regulating kinase activity, protein and growth factor binding, and membrane raft dynamics.
KEGG analysis revealed notably enriched pathways, including PI3K-AKT, MAPK, Ras, JAK-STAT, acute myeloid leukemia, EGFR tyrosine kinase inhibitors, and endocrine resistance (Supplementary Figure S2B). These leukemia-promoting signaling pathways further emphasize the crucial role of these targets in AML pathogenesis and treatment response. Additionally, we observed the involvement of PD-L1 expression, and the PD-1 checkpoint pathway in cancer, indicating that the identified targets may play a role in immunotherapy-based treatment responses by modulating the tumor immune microenvironment.
Correlation of intersected targets expression with patients’ overall survival
We explored the relationship between 113 intersection targets and AML prognosis using clinical data from 132 AML patients in the TCGA database. Kaplan-Meier analysis and the log-rank test were applied for survival analysis. Univariate Cox regression identified 20 targets significantly associated with AML prognosis (Figure 2A). Overexpression of PIK3CA, STAT5B, MDM4, ITGA4, PI3CB, CASP3, EPHX1, CDK6, CCND2, and MPO were linked to a favorable prognosis, while higher expression of PTGS1, PIM1, ITGAL, IDH1, SRC, SYK, PARP1, G6PD, CCND3, and GRK6 indicated poor prognosis. Validation using the Beat-AML dataset confirmed the prognostic relevance of CCND2, CCND3, and SRC, aligning with previous studies (Figure 2B). As shown in Figure 2C, low CCND2 and high SRC expression were associated with worse survival outcomes, consistent with earlier reports (Zhang X. et al., 2022; Patel et al., 2019).
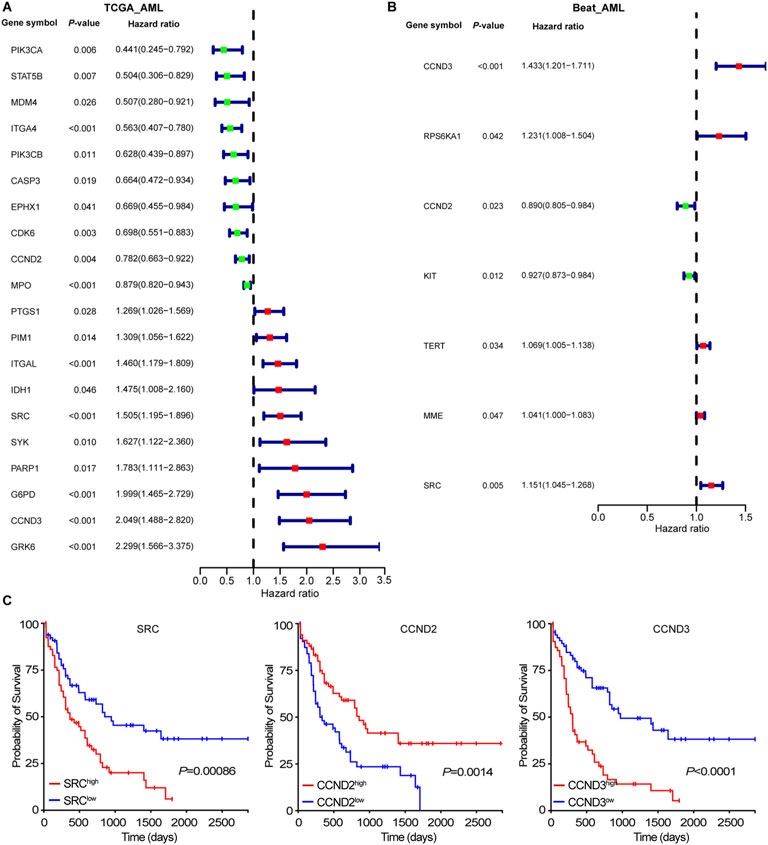
Figure 2. Associations of intersecting targets with patients’ outcome. (A) Forest plot represented the associations of intersecting targets with patients’ outcome in TCGA-AML cohort. (B) Forest plot represented the associations of intersecting targets with patients’ outcome in Beat-AML cohort. (C) The Kaplan-Meier curve of SRC, CCND2, and CCND3 in TCGA-AML cohort.
Molecular docking
To explore the potential interactions between bioactive compounds in YWLS and key molecular targets associated with AML, molecular docking studies were performed using SybylX-2.0. We selected three core AML-related proteins—PIK3CA, SRC, and CASP3—based on their relevance in leukemogenesis and their identification as key nodes in our network pharmacology analysis.
Docking simulations revealed that multiple compounds exhibited favorable binding affinities with these proteins. Specifically, five compounds demonstrated excellent affinity (docking score >7), 10 compounds showed good affinity (score 5–7), and six compounds exhibited moderate affinity (score 4–5) (Table 2). These findings may suggest the potential for YWLS-derived compounds to modulate AML-related signaling pathways through direct interaction with critical molecular targets.
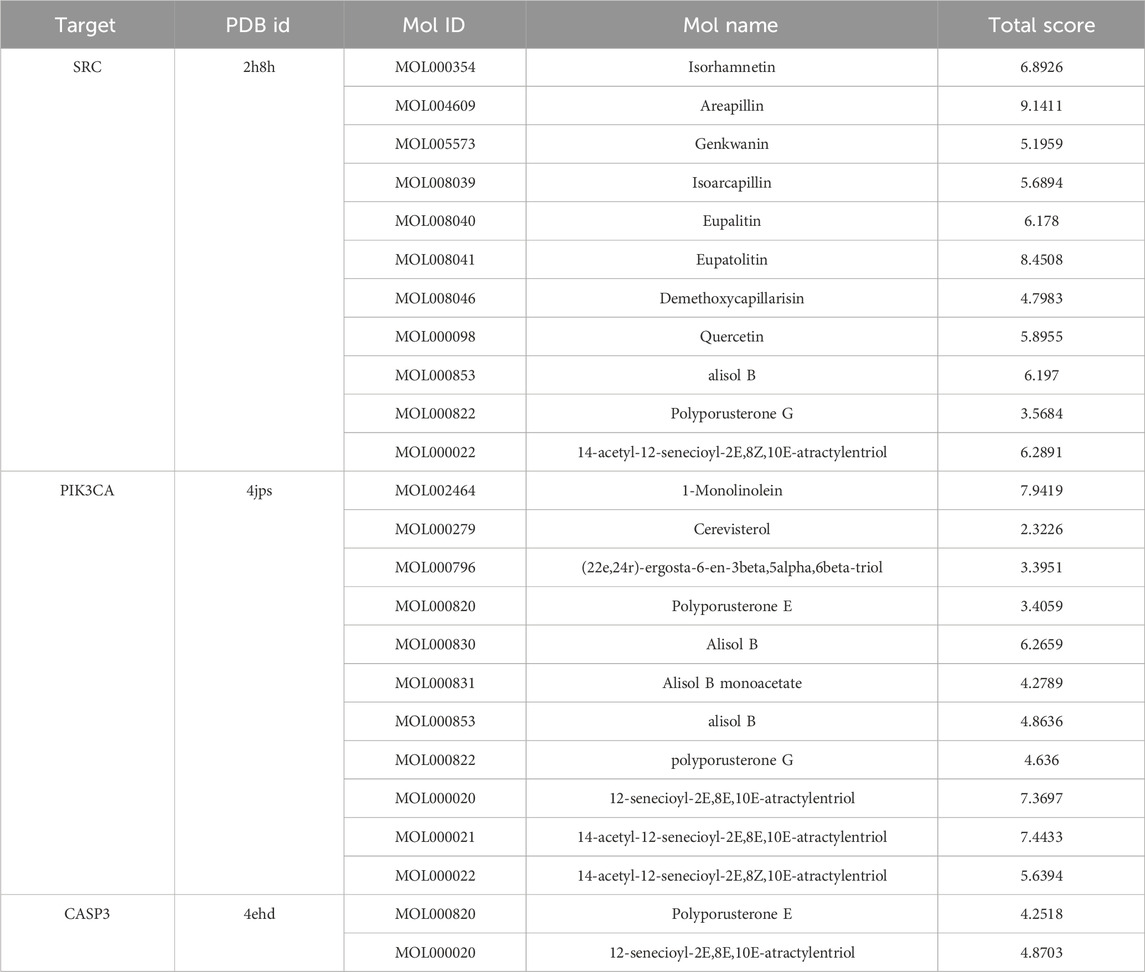
Table 2. Binding affinities between key target genes (SRC, PIK3CA, CASP3) and active YWLS compounds by molecular docking.
To further illustrate these interactions, Figure 3 presents the docking conformations of three representative YWLS compounds—Isorhamnetin, Genkwanin, and Quercetin—with SRC, a non-receptor tyrosine kinase implicated in AML cell proliferation and survival. All three compounds were predicted to stably occupy the active binding pocket of SRC, engaging in a combination of hydrogen bonding and hydrophobic interactions that support high-affinity binding. These interactions may underlie the potential of YWLS components to inhibit SRC-mediated oncogenic signaling, reinforcing their candidacy as therapeutic agents in AML (Lohning et al., 2017).

Figure 3. Molecular docking of selected active compounds with the core AML-related target protein SRC. (A) Isorhamnetin-SRC. (B) Genkwanin-SRC. (C) Quercetin-SRC.
ADME prediction analysis
ADME prediction using the SwissADME database assessed the pharmacokinetic properties of selected YWLS compounds. Fifteen active ingredients met key druggability indicators (Table 3), demonstrating favorable gastrointestinal absorption, bioavailability, and druglikeness. Flavonoids like Genkwanin, Isorhamnetin, and Quercetin were particularly promising, while compounds such as Eupalitin could cross the blood-brain barrier, suggesting broader therapeutic potential. These findings support further evaluation of YWLS compounds for AML treatment, including their pharmacokinetic profiles and safety.
Molecular dynamics simulation
The RMSD values of four systems (unloaded SRC, SRC-isorhamnetin, SRC-genkwanin, and SRC-quercetin) reached equilibrium after a simulation period (Figure 4A). The RMSD values were 1.97 Å, 2.38 Å, 4.07 Å, and 1.34 Å, respectively, indicating that the complexes were stable. As shown in Figure 4B, the RMSF values varied among the systems due to different interactions between the active molecules and SRC. Despite these differences, the RMSF trends were similar, with notable increases in the regions between residues 113–117 and 519–529, where the SRC loop structure exhibits flexible space. Figure 4C shows that the radius of gyration (RG) of unloaded SRC (2.46 nm), SRC-isorhamnetin (2.45 nm), SRC-genkwanin (2.47 nm), and SRC-quercetin (2.46 nm) remained stable during the simulation, confirming that the complex structures remained stable throughout binding, consistent with the RMSD analysis.
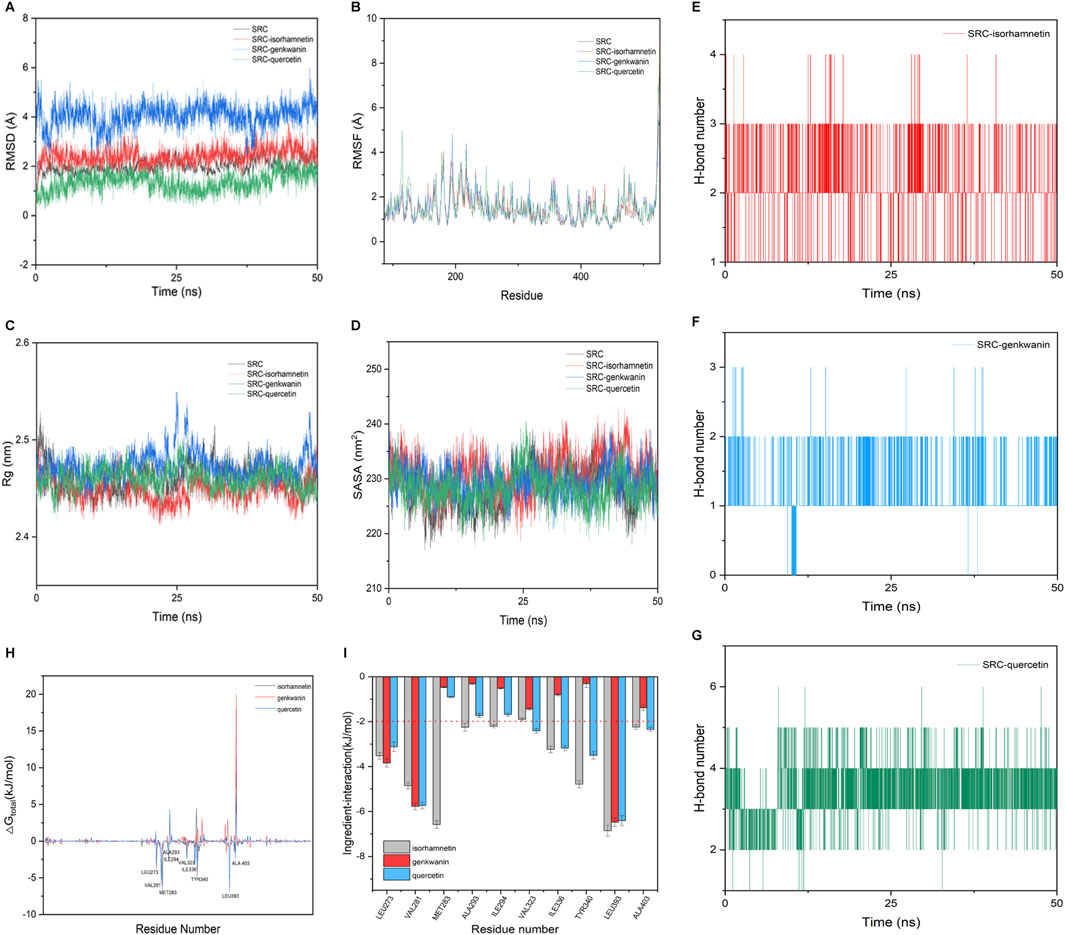
Figure 4. Molecular dynamics simulation of Genkwanin, isorhamnetin, and quercetin and SRC. (A) RMSD of Genkwanin, isorhamnetin, and quercetin in complex with SRC, respectively. (B) RMSF of amino acid residues of Genkwanin, isorhamnetin, and quercetin in complex with SRC, respectively. (C) Rg of Genkwanin, isorhamnetin, and quercetin in complex with SRC, respectively. (D) SASA of Genkwanin, isorhamnetin, and quercetin in complex with SRC, respectively. (E–G) H-bond number of isorhamnetin, Genkwanin, and quercetin in complex with SRC, respectively. (H) Gibbs binding free energy of isorhamnetin, Genkwanin, and quercetin in complex with SRC, respectively. (I) Interaction energy between the isorhamnetin, Genkwanin, and quercetin and key amino acid residues of SRC, respectively.
Figure 4D shows that the overall solvent accessible surface area (SASA) of these complexes showed minimal changes from the unloaded SRC, suggesting that the binding of isorhamnetin, genkwanin, and quercetin had little effect on the hydrophilicity and hydrophobicity of SRC. The role of hydrogen bonds in maintaining the stability of these complexes is shown in Figures 4E–G. All three compounds formed hydrogen bonds with SRC, with quercetin forming the most frequent bonds, followed by isorhamnetin and genkwanin, highlighting quercetin’s significant role in binding to SRC.
To calculate the binding free energy, we selected the RMSD stationary phase in the last 5 ns of the simulation and extracted one conformation every 10 ps, resulting in 500 conformations. The binding free energies and components for the SRC-isorhamnetin, SRC-genkwanin, and SRC-quercetin complexes are presented in Table 4. The binding free energies were −60.42 ± 1.48 kJ/mol for SRC-isorhamnetin, −48.78 ± 1.59 kJ/mol for SRC-genkwanin, and −61.04 ± 1.49 kJ/mol for SRC-quercetin. The van der Waals energies were −148.66 ± 1.86 kJ/mol for SRC-isorhamnetin, −117.71 ± 1.36 kJ/mol for SRC-genkwanin, and −132.77 ± 1.72 kJ/mol for SRC-quercetin. The electrostatic energies were −70.20 ± 1.07 kJ/mol, −44.96 ± 0.91 kJ/mol, and −77.18 ± 1.64 kJ/mol, respectively. Additionally, the polar solvation energies were 175.18 ± 1.40 kJ/mol, 129.92 ± 0.92 kJ/mol, and 164.73 ± 1.06 kJ/mol. These findings indicate that van der Waals and electrostatic interactions significantly contribute to binding, while polar solvation inhibits interaction.
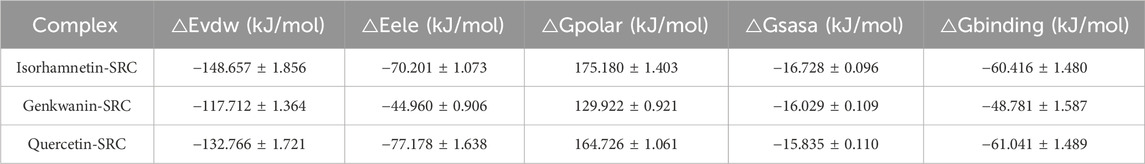
Table 4. Binding free energy and energy components of (isorhamnetin, genkwanin, quercetin)-SRC complex.
To further investigate the binding mechanism, we used residue-free energy decomposition to identify key residues contributing to the binding of isorhamnetin, genkwanin, and quercetin to SRC. Figure 4H compares the binding free energy of each compound with SRC, while Figure 4I highlights the key residues contributing to the binding process. The residues LEU273, VAL281, VAL323, LEU393, and ALA403 were identified as significant contributors to binding across all three complexes. The binding energies of these residues were as follows: −3.5, −4.85, −1.90, −6.86, −2.24 kJ/mol for isorhamnetin; −3.84, −5.78, −1.44, −6.48, −1.39 kJ/mol for genkwanin; and −3.84, −5.78, −1.44, −6.48, −1.39 kJ/mol for quercetin. Additionally, in the isorhamnetin-SRC complex, nine SRC residues (LEU273, VAL281, MET283, ALA293, ILE294, ILE336, TYR340, LEU393, and ALA403) interacted with isorhamnetin with binding energies greater than −2 kJ/mol.
Cytotoxic effects of Quercetin, Isorhamnetin, and Genkwanin on AML cells
To assess the cytotoxic effects of selected YWLS compounds on AML cells, MOLM14, a human acute myeloid leukemia cell line, was treated with varying concentrations of Quercetin, Isorhamnetin, and Genkwanin for 72 h. Cell viability was evaluated using eFluor™ 506 Viability Dye staining followed by flow cytometry. The results demonstrated a clear dose-dependent reduction in cell viability for all three compounds, compared to the vehicle-treated control. Among them, Genkwanin exhibited the strongest cytotoxic effect, followed by Isorhamnetin and Quercetin, suggesting that these compounds may exert anti-leukemic activity through direct effects on AML cell survival (Figure 5A).
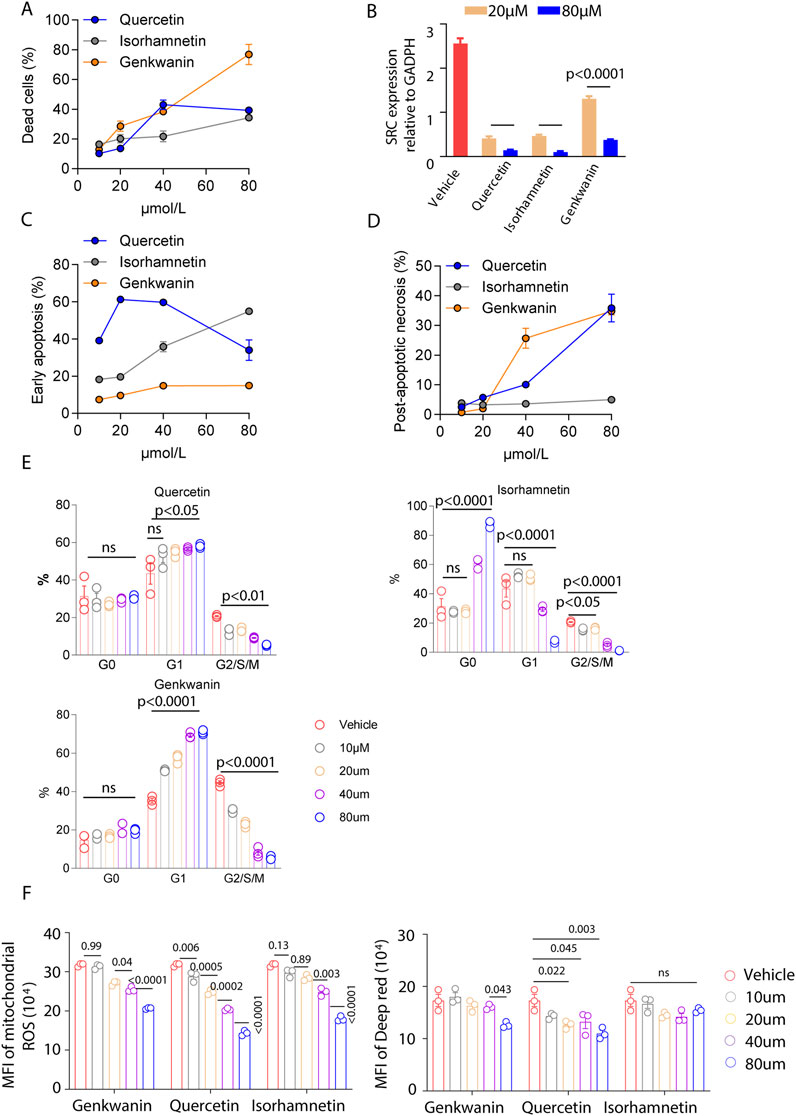
Figure 5. Effects of Genkwanin, Isorhamnetin, and Quercetin on leukemia cell apoptosis, cell cycle, and mitochondrial function. (A) Cytotoxic effects of Genkwanin, Isorhamnetin, and Quercetin on leukemic cells, presented relative to the vehicle control. (B) SRC expression in leukemic cells at 72 h following treatment with different concentrations of Genkwanin, Isorhamnetin, and Quercetin. (C) Early apoptosis of leukemic cells at 72 h after treatment with different concentrations of Genkwanin, Isorhamnetin, and Quercetin. (D) Late apoptosis of leukemic cells at 72 h after treatment with different concentrations of Genkwanin, Isorhamnetin, and Quercetin. (E) Cell cycle distribution of leukemic cells at 72 h after treatment with different concentrations of Genkwanin, Isorhamnetin, and Quercetin. (F) Reactive oxygen species levels and mitochondrial membrane potential in leukemic cells at 72 h following treatment with different concentrations of Genkwanin, Isorhamnetin, and Quercetin.
Based on the fitted dose-response models, predicted inhibition rates at 0.1, 1, 10, and 100 μM were obtained for each compound. IC50 analysis for Quercetin, Isorhamnetin, and Genkwanin derived from four-parameter logistic curve fitting were as follows: Quercetin: 24.49 μM, Isorhamnetin: 162.41 μM, and Genkwanin: 55.71 μM. As shown in Supplementary Table S5, inhibition was minimal at lower concentrations (≤10 μM), consistent with experimental observations in Figure 5A. Genkwanin exhibited a marked increase in predicted inhibition at 100 μM, consistent with its steep dose-response profile.
Since previous analyses suggested that these compounds target SRC, qRT-PCR was performed to evaluate SRC expression in MOLM14 cells post-treatment. A significant downregulation of SRC was observed at concentrations of 20 µM and 80 μM, compared to the vehicle group (Figure 5B), indicating that SRC is a viable target for these compounds. These findings suggest that these compounds may be promising pharmacological agents for AML treatment.
To further investigate the mechanisms underlying the observed cytotoxic effects, we performed an apoptosis assay using Annexin V staining. Isorhamnetin treatment induced early apoptosis in MOLM14 cells in a dose-dependent manner, with lower concentrations of Quercetin also promoting early apoptosis (Figure 5C; Supplementary Figure S4). Genkwanin demonstrated a mild enhancement of early apoptosis. For late apoptosis (Figure 5D; Supplementary Figure S4), both Quercetin and Genkwanin significantly increased post-apoptotic necrosis, whereas Isorhamnetin had a minimal effect. Overall, these compounds appear to induce tumor cell death primarily by promoting apoptosis.
Additionally, cell cycle analysis revealed distinct effects of the compounds on the cell cycle. Quercetin promoted G1/S/M phase arrest, even at low concentrations (Figure 5E). High concentrations of Isorhamnetin caused an increase in the G0 phase, keeping tumor cells in a quiescent state while reducing progression through the G1/S/M phases. Genkwanin exhibited a dose-dependent increase in the G1 phase and G2/S/M phase arrest, with these effects being more pronounced at higher concentrations.
Mitochondrial function is critical for tumor cell proliferation, as mitochondria are central to cellular energy metabolism and survival. Previous studies have emphasized the importance of mitochondrial alterations in cancer progression and immunity (Li et al., 2024; Wang et al., 2023). In our study, we assessed mitochondrial reactive oxygen species (ROS) levels and membrane potential, finding that all three compounds significantly reduced ROS levels and mitochondrial membrane potential (Figure 5F). These results suggest that the compounds may disrupt the cellular energy balance, impairing leukemia cell metabolism and inhibiting cell proliferation.
In conclusion, Quercetin, Isorhamnetin, and Genkwanin exhibit cytotoxic effects against AML cells through the induction of apoptosis, modulation of the cell cycle, and disruption of mitochondrial function, highlighting their potential as therapeutic agents.
Genkwanin as a therapeutic pharmacological target for AML
The three compounds demonstrated significant cytotoxicity against AML cells, prompting us to investigate their potential impact on immune cells, which play a critical role in modulating the tumor microenvironment. A T cell proliferation assay revealed that quercetin and genkwanin did not significantly affect CD4+ T cell proliferation, as measured by CFSE, compared to the vehicle group (Supplementary Figure S3A). However, isorhamnetin exhibited a dose-independent proliferative effect. For CD8+ T cells, quercetin and isorhamnetin had minimal effects on proliferation, whereas genkwanin showed a pronounced proliferative effect at 10 μM, comparable to effects observed at higher concentrations (20 μM and 80 µM) (Supplementary Figure S3B). These in vitro findings suggest that the tested compounds are not toxic to healthy splenic T cells and may modulate T cell activity. Given the importance of CD8+ T cells in anti-leukemia immunity, these observations provided the rationale for subsequent in vivo studies to evaluate whether genkwanin could enhance anti-leukemia immune responses within the tumor microenvironment.
We utilized an established leukemia xenograft model by injecting 1 × 105 MOLM14gfp cells into NSG mice (Figure 6A). Genkwanin was administered intraperitoneally at 5 mg/kg starting on day 4 post-leukemia cell injection, with a total of six doses given twice weekly. Additional treatment groups included mice receiving genkwanin in combination with weekly adoptive transfer of 1 × 106 human PBMCs, as well as mice treated with PBMCs alone. The results showed that genkwanin monotherapy significantly prolonged survival compared to the vehicle-treated group (Figure 6B). However, there was no significant difference between the genkwanin-only group and the PBMC-only group. Notably, mice treated with the combination of genkwanin and PBMCs exhibited further improved survival and a marked reduction in leukemia burden by Day 21 post-challenge, compared to genkwanin treatment alone (Figure 6C).
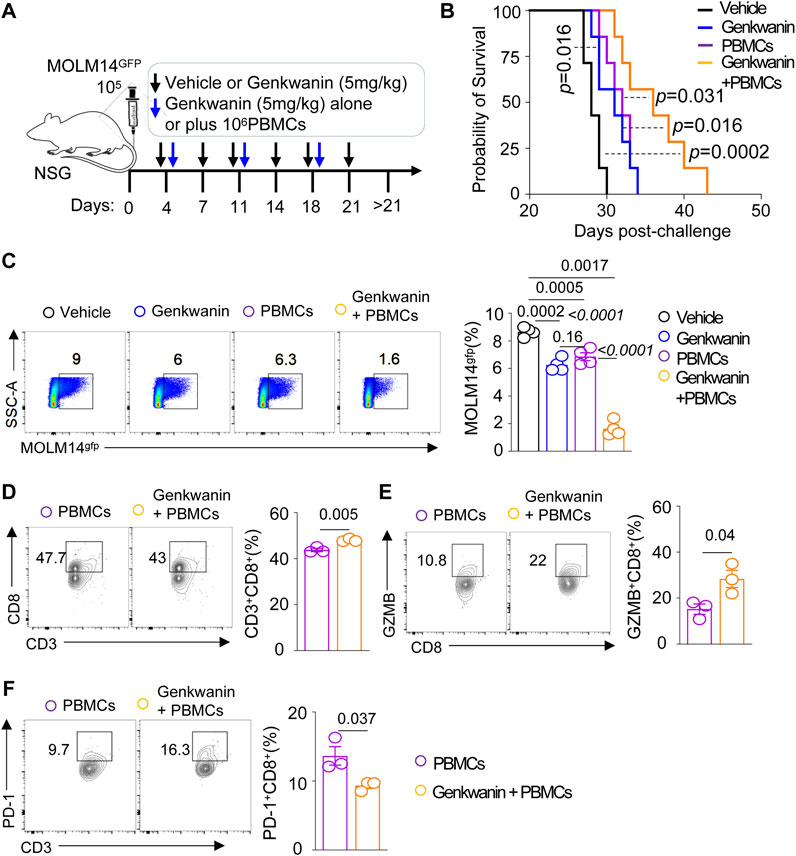
Figure 6. Genkwanin as a pharmacological agent in a leukemia model. (A) Schematic representation of Genkwanin treatment alone or in combination with adoptive PBMC transfer. (B) Kaplan-Meier survival curve of mice treated with Genkwanin alone or in combination with adoptive PBMC transfer. (C) Frequency of leukemia cells in the bone marrow on Day 21 post-challenge. (D) Frequency of CD3+CD8+T cells in the bone marrow in mice transferred with PBMCs alone or Genkwanin + PBMCs on Day 21 post-challenge. (E) Frequency of GZMB+CD8+T cells in the bone marrow in mice transferred with PBMCs alone or Genkwanin + PBMCs on Day 21 post-challenge. (F) Frequency of PD-1+CD8+T cells in the bone marrow in mice transferred with PBMCs alone or Genkwanin + PBMCs on Day 21 post-challenge.
To determine whether genkwanin modulates human T cell responses in vivo, we analyzed the bone marrow of NSG mice co-engrafted with MOLM14 cells and human PBMCs. Flow cytometric analysis revealed an increased frequency of human CD3+CD8+ T cells in mice receiving genkwanin plus PBMCs compared to those treated with PBMCs alone (Figure 6D). A similar trend was observed for granzyme B (GZMB)+CD8+ T cells, suggesting enhanced cytotoxic potential (Figure 6E). Moreover, the frequency of PD-1+CD8+ T cells was reduced in the genkwanin-treated group (Figure 6F), indicating a possible alleviation of T cell exhaustion. Collectively, these data suggest that genkwanin may promote the expansion and functional activation of CD8+ T cells in vivo, and may hold promise as a potential pharmacological target for AML therapy.
Discussion
AML is a highly aggressive and prevalent hematological malignancies, characterized by the accumulation of immature myeloid progenitor in the bone marrow, resulting in impaired hematopoiesis and poor clinical outcomes (Garcia-Manero, 2023; Fu et al., 2023; Huang et al., 2019). Despite advancements in targeted therapies—such as FLT3, IDH, and BCL-2 inhibitors—treatment resistance and relapse rates remain high, and the 5-year survival rate for AML patients remains disappointingly low (Zhang B. et al., 2021; Forsberg and Konopleva, 2024; Mohamed Jiffry et al., 2023). Immunotherapies, including monoclonal antibodies (e.g., gemtuzumab ozogamicin targeting CD33), bispecific T-cell engagers (BiTEs), and CAR T-cell therapies targeting antigens like CD123, represent promising modalities but are limited by antigen heterogeneity, toxicity, off-target effects, and the immuosuppressive bone marrow microenvironment (Restelli et al., 2024; Abaza et al., 2024).
In this context, traditional Chinese medicine (TCM) offers a holistic approach, focusing on restoring physiological balance and supporting immune function (Xiang et al., 2019; Zhang et al., 2024; Kim et al., 2024; Yang et al., 2020). In the context of AML, specific herbal compounds such as curcumin, berberine, and astragalus polysaccharides exhibit anti-inflammatory, antioxidant, and pro-apoptotic effects, targeting critical pathways involved in disease progression, including apoptosis, angiogenesis, and metastasis (de Waure et al., 2023; Huang et al., 2023; Yang et al., 2024). In addition to these direct anti-leukemic agents, broader traditional Chinese medicine (TCM) therapies have been employed to alleviate treatment-related side effects—such as fatigue, nausea, and immune suppression—commonly experienced by AML patients undergoing chemotherapy or stem cell transplantation (Ling, 2013). The integration of TCM with contemporary AML treatments holds considerable promise. For instance, TCM may complement targeted therapies like FLT3 and IDH inhibitors or immune-based approaches, such as CAR T-cell therapy, by enhancing immune responses, mitigating side effects, and potentially improving therapeutic outcomes (Ruglioni et al., 2024; Ge et al., 2022; Marvin-Peek et al., 2022). Ongoing research and clinical trials are actively exploring the synergistic effects of combining TCM with chemotherapy, targeted therapies, and immunotherapies, aiming to offer more effective and holistic treatment strategies for AML patients (Liu et al., 2020; Wu et al., 2024; Shi et al., 2021).
TCM bridges the gap between traditional and modern approaches, offering a promising avenue for integrated cancer care in AML. YWLS is believed to regulate Qi and improve blood flow, alleviating cancer-related fatigue and improving patients’ overall wellbeing (Su and Tai, 2020). Studies suggest that its anti-inflammatory properties play a key role in inhibiting tumor progression and metastasis (Mou et al., 2022). Additionally, its detoxifying and liver-protecting effects may reduce liver damage caused by cancer treatments, enhancing chemotherapy efficacy (Zhu et al., 2016). Flavonoids in YWLS, known for their anti-cancer properties, inhibit tumor proliferation and induce apoptosis by downregulating the PI3K/AKT/mTOR signaling pathway (Kopustinskiene et al., 2020; Mir et al., 2024; Zhang et al., 2018). Given the complex composition of TCM formulations, preclinical studies have focused on identifying the molecular targets and mechanisms of individual components within YWLS. For example, Genkwanin—a flavonoid present in several medicinal plants—has demonstrated potential anticancer activity, particularly in head and neck squamous cell carcinoma, and emerging evidence suggests that Genkwanin may exert its effects by inducing apoptosis and inhibiting cancer cell proliferation (Grattan, 2013; Zhang B. et al., 2022). While these findings are encouraging, they relate to specific active compounds rather than the entire YWLS formulation. Therefore, further investigation into the individual constituents of YWLS and their role in tumorigenesis is warranted. Such studies could provide a scientific foundation for the potential integration of these herbal components into broader cancer treatment strategies.
Our study highlights the potential of YWLS, a classical TCM formula, as a novel source of anti-leukemia agents. In this study, we identified 58 active ingredients of YWLS from the TCMSP database. Some ingredients have been implicated in tumorigenesis, suggesting their potential role in treating leukemia. Among these, cerevisterol and (22E, 24R)-ergosta-6-en-3β, 5α, 6β-triol are steroids known for their anti-inflammatory, immunomodulatory, and anti-cancer activities (Calpe-Berdiel et al., 2007). Steroids have shown efficacy in cancers such as breast (Grattan, 2013), gastric (De Stefani et al., 2000), and lung (Mendilaharsu et al., 1998). Polyporusterone E has a dose-independent inhibitory effect in the cell proliferation of leukemia L-1210 (Ohsawa et al., 1992). Additionally, Genkwanin, a major non-glycosylated flavonoid, exhibits anti-inflammatory effects by modulating the miR-101/MKP-1/MAPK pathway (Gao et al., 2014). Isorhamnetin has demonstrated anti-tumor activity across cancers, including colorectal, skin, lung, and breast, through apoptosis induction and signaling pathway inhibition (Ruan et al., 2015; Hu et al., 2015). A total of 113 target genes were identified for YWLS active ingredients, intersecting with AML-related genes. Pathway analysis revealed enrichment in PI3K-AKT, MAPK, JAK-STAT, PD-1/PD-L1, and leukemia-specific signaling pathways, which are closely linked to AML carcinogenesis and drug resistance (Park et al., 2010). Among these, 10 genes correlated with favorable survival (e.g., PIK3CA/B, ITGA4, CCND2, MPO, and CASP3), while others, such as PIM1 (Cheng et al., 2017), IDH1 (Messina et al., 2022), SRC (Patel et al., 2019), and CCND3 (Matsuo et al., 2018), were associated with poor survival, consistent with previous evidence. High CCND3 and SRC expression, validated in an independent dataset, correlated with unfavorable survival. Dysregulated D-type cyclins (e.g., CCND1, CCND2, and CCND3) drive the G1-to-S phase cell cycle transition via CDK4/6, contributing to AML pathogenesis (Matsuo et al., 2018; Choi et al., 2012). Src, a tyrosine kinase, regulates cell proliferation, motility, and survival and is implicated in AML drug resistance (Ozawa et al., 2008). While inhibitors like A-419259 and TL02-59 target Src family kinases such as Hck and Fgr, acquired resistance remains a significant challenge (Yang et al., 2022; Negi et al., 2021), underscoring the need for novel inhibitors to improve AML outcomes.
Using molecular docking, we identified several compounds from the active ingredients of YWLS that are predicted to target SRC, including quercetin, isorhamnetin, alisol B, eupalitin, eupatolitin, areapillin, and genkwanin. To evaluate the drug potential of these compounds for AML patients, ADME analysis revealed that most SRC-targeting compounds possess favorable pharmacological properties, including good drug-likeness and bioavailability scores comparable to current clinical agents. Flavonoids such as genkwanin, isorhamnetin, and quercetin exhibited excellent gastrointestinal absorption, druggability, and bioavailability. Notably, eupalitin, 12-senecioyl-2E, 8E, 10E-atractylentriol, and 14-acetyl-12-senecioyl-2E, 8E, 10E-atractylentriol demonstrated blood-brain barrier penetration but may accumulate in the body due to inhibition of liver drug-metabolizing enzymes. Flavonoids are known to suppress cancer cell growth by inhibiting proliferation and inducing apoptosis through the downregulation of the PI3K/AKT/mTOR/ULK signaling pathway (Mir et al., 2024; Zhang et al., 2018; Slika et al., 2022). In in vitro killing assays, the compounds inhibited AML cell growth to varying degrees and reduced SRC expression in a dose-dependent manner, suggesting their potential as therapeutic agents for AML.
We further focused on quercetin, isorhamnetin, and genkwanin due to their commercial availability and supporting evidence from previous studies. Molecular dynamics simulations indicated that the binding stability of quercetin and isorhamnetin to the SRC protein skeleton was greater than that of genkwanin. Despite this, the flexibility and intensity of protein-ligand movements during binding were comparable across the three compounds. Hydrogen bonding played a crucial role in stabilizing the interactions between SRC and these compounds, as reported in prior studies (Huang et al., 2014; Chen et al., 2016). The binding stability correlated with the comparable number of hydrogen bonds formed. Free energy analysis identified key amino acid residues critical to binding stability (Chen J. et al., 2024). For example, MET283 and ILE336 were crucial for isorhamnetin binding, while LEU273 and LEU393 were essential for all three compounds. These findings provide a molecular-level foundation for targeting SRC with these compounds. However, further validation through site-directed mutagenesis is necessary to confirm these results (Liao et al., 2001).
In parallel, dose-response modeling using the four-parameter logistic (4 PL) model provided valuable insights into the tumor cell growth inhibition rates of Quercetin, Isorhamnetin, and Genkwanin at various concentrations. The model predicted minimal inhibition at concentrations ≤10 μM, consistent with experimental observations, indicating that higher concentrations are needed for significant anti-tumor effects. Genkwanin showed a notably steep dose-response curve, as evidenced by the sharp increase in inhibition at 100 μM. This behavior highlights its potential for greater efficacy at higher doses, supporting its consideration for further investigation in therapeutic applications. In contrast, Quercetin and Isorhamnetin showed more gradual increases in tumor growth inhibition, achieving approximately 40% inhibition at 100 μM. These findings suggest that while Quercetin and Isorhamnetin may have moderate anti-tumor effects, their potency may be limited at concentrations typically used in clinical or experimental settings.
Beyond tumor cell inhibition, CD8+ T cells are central to anti-tumor immunity, but their exhaustion often renders them incapable of effective tumor killing (Desai et al., 2023). Among the three compounds, genkwanin demonstrated a superior ability to promote T-cell proliferation, particularly of CD8+ T cells, even at low concentrations. Genkwanin also reduced mitochondrial ROS levels, suggesting it may restore T-cell function while inhibiting tumor growth. These observations align with previous studies showing genkwanin induces apoptosis, mitigates oxidative stress, and scavenges free radicals (El Menyiy et al., 2023). In addition, genkwanin induced G2/S/M cell cycle arrest in AML cells. In a leukemia xenograft model, genkwanin significantly inhibited leukemic cell growth, improved survival, and did not cause observable toxicity, such as colitis. These data suggest that genkwanin’s dual-targeting strategy—enhancing CD8+ T-cell function and inhibiting leukemia cell growth—holds promise as a therapeutic strategy for AML by SRC targeting.
Compared to current targeted therapies, which often address single genetic mutations (e.g., FLT3, IDH1/2), the multi-targeted nature of YWLS-derived compounds offers broader anti-leukemia effects and may overcome the limitation of clonal heterogeneity. Additionally, unlike antibody-drug conjugates or CAR-T cells that carry risks of severe cytokine storms, YWLS compounds demonstrated a safer profile with immunomodulatory benefits. These findings suggest that YWLS active ingredients could be developed as either standalone therapies or adjuncts to existing regimens.
Despite these promising results, our study has several limitations that warrant further investigation. One key limitation lies in the target identification process. The use of public databases such as TCMSP and SwissTargetPrediction, although valuable for hypothesis generation, is inherently biased toward well-studied proteins and targets. As a result, potential interactions with less-characterized or novel targets may be underrepresented. This limitation may affect the comprehensiveness of the predicted compound–target interactions and should be considered when interpreting the results. Further studies using unbiased proteomic and transcriptomic approaches are needed to comprehensively map compound-target interactions. The precise mechanism by which these compounds modulate SRC signaling and ROS levels warrants deeper exploration, potentially involving site-directed mutagenesis and metabolic flux analysis. Then, while downregulation of SRC was observed, the precise mechanism remains unclear. Given the complexity of the SRC family, identifying the specific family member targeted by these compounds in AML progression could provide deeper molecular insights. Moreover, combination strategies integrating YWLS ingredients with standard chemotherapies, targeted agents (e.g., FLT3 inhibitors), or immunotherapies (e.g., checkpoint blockade) should be investigated to evaluate potential synergistic effects. Finally, clinical trials assessing the safety, pharmacokinetics, and efficacy of YWLS-derived compounds in AML patients are critical next steps toward translation. In addition, we note that a viability dye was not used prior to mitochondrial staining, which could represent a limitation. Future studies incorporating viability-based gating will further refine analysis of mitochondrial function specifically in surviving cells. Finally, the effect of these compounds on T-cell activation needs to be clarified to strengthen the case for a dual-targeting strategy. Addressing these aspects in future studies will provide a more comprehensive understanding of the therapeutic potential of genkwanin and related compounds in AML.
Conclusion
In summary, through network pharmacology analysis, molecular docking, and molecular dynamics simulations, we predicted and identified potential therapeutic targets of YWLS active ingredients in the treatment of AML. Three key compounds—genkwanin, quercetin, and isorhamnetin—were validated for their ability to inhibit AML cell growth in vitro, with genkwanin demonstrating additional benefits in promoting T-cell proliferation and enhancing survival. These findings highlight the potential of YWLS active ingredients, particularly flavonoids targeting SRC, as promising pharmacological candidates for AML therapy.
Future research should focus on several key areas: (1) further mechanistic exploration to elucidate how individual YWLS compounds modulate SRC signaling, mitochondrial metabolism, and T-cell function; (2) validation of the predicted compound-target interactions using genetic and proteomic approaches; (3) evaluation of combination strategies integrating YWLS compounds with conventional chemotherapies or immunotherapies to assess potential synergistic effects; and (4) advancement toward clinical translation through pharmacokinetic studies, toxicity assessments, and early-phase clinical trials. These efforts will be crucial to fully realize the therapeutic potential of YWLS and its active components in AML treatment.
Data availability statement
The data analyzed in this study are available in the following repositories: 1. TCGA: https://portal.gdc.cancer.gov/; 2. UCSC xena: https://xena.ucsc.edu/; 3. TCMSP: https://old.tcmsp-e.com/tcmsp.php; 4. DisGeNET: https://www.disgenet.org/; 5. GeneCards: https://www.genecards.org/; 6. STRING: https://string-db.org/; 7. RCSB PDB: https://www.rcsb.org; 8. PubChem: https://pubchem.ncbi.nlm.nih.gov/; 9. SwissTargetPrediction: http://www.swisstargetprediction.ch/; 10. SwissADME: http://www.swissadme.ch/.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by The Animal Ethics Committee at Jinjiang University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Resources, Writing – original draft, Writing – review and editing. DF: Formal Analysis, Methodology, Software, Visualization, Writing – original draft, Writing – review and editing. XW: Data curation, Methodology, Resources, Supervision, Writing – original draft, Writing – review and editing. XH: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Science and Technology Research Project of the Jiangxi Province Department of Education (GJJ201837).
Acknowledgments
We are grateful to all contributors to the public databases used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1591164/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Workflow of the study.
SUPPLEMENTARY FIGURE S2 | GO and KEGG enrichment analysis of intersecting targets. (A) The top 10 enriched GO items of BP, CC, and MF. (B) The top 20 enriched KEGG pathways. (C) The important target genes were mainly distributed in the MAPK signaling pathway. (D) The important target genes were mainly distributed in the PI3K-AKT pathway.
SUPPLEMENTARY FIGURE S3 | Effects of Genkwanin, Isorhamnetin, and Quercetin on T cells proliferation. (A) Effects of Genkwanin, Isorhamnetin, and Quercetin on CD4+T cells proliferation. (B) Effects of Genkwanin, Isorhamnetin, and Quercetin on murine CD8+T cell proliferation.
SUPPLEMENTARY FIGURE S4 | Representative flow cytometry plots showing apoptosis induced by Genkwanin, Isorhamnetin, and Quercetin in MOLM14 cells.
SUPPLEMENTARY TABLE S1 | Potential targets of active ingredients in YWLS.
SUPPLEMENTARY TABLE S2 | Acute myeloid leukemia-related genes from DisGeNET and GeneCards databases.
SUPPLEMENTARY TABLE S3 | 113 targets were identified from 621 targets of 58 active ingredients in YWLS overlapped with 1,247 genes associated with AML.
SUPPLEMENTARY TABLE S4 | Degree value of 20 top targets of YWLS active components analyzed by network analysis.
SUPPLEMENTARY TABLE S5 | Predicted tumor growth inhibition (%) at selected concentrations based on model fitting.
Abbreviations
AML, acute myeloid leukemia; AMK, Atractylodes Macrocephala Koidz; ASH, Artemisiae Scopariae Herba; ADME, absorption, distribution, metabolism, and excretion; AO, Alisma Orientale; BP, Biological Process; CC, Cellular Component; CR, Cinnamomi Ramulus; KEGG, Kyoto Encyclopedia of Genes and Genomes; MCODE. Molecular Complex Detection; MF, Molecular Function; OS, Overall Survival; PU, Polyporus Umbellatus; PPI, Protein-Protein Interaction; PC, Poria Cocos; TCGA, The Cancer Genome Atlas; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; YWLS, Yinchen wuling san.
References
Abaza, Y., McMahon, C., and Garcia, J. S. (2024). Advancements and challenges in the treatment of AML. Am. Soc. Clin. Oncol. Educ. Book 44 (3), e438662. doi:10.1200/EDBK_438662
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. doi:10.1016/j.softx.2015.06.001
Adom, D., Dillon, S. R., Yang, J., Liu, H., Ramadan, A., Kushekhar, K., et al. (2020). ICOSL+ plasmacytoid dendritic cells as inducer of graft-versus-host disease, responsive to a dual ICOS/CD28 antagonist. Sci. Transl. Med. 12 (564), eaay4799. doi:10.1126/scitranslmed.aay4799
Bader, G. D., and Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4, 2. doi:10.1186/1471-2105-4-2
Bertacchini, J., Guida, M., Accordi, B., Mediani, L., Martelli, A. M., Barozzi, P., et al. (2014). Feedbacks and adaptive capabilities of the PI3K/Akt/mTOR axis in acute myeloid leukemia revealed by pathway selective inhibition and phosphoproteome analysis. Leukemia 28 (11), 2197–2205. doi:10.1038/leu.2014.123
Bruzzese, A., Martino, E. A., Labanca, C., Mendicino, F., Lucia, E., Olivito, V., et al. (2025). Advances and challenges in Quizartinib-based FLT3 inhibition for acute myeloid leukemia: mechanisms of resistance and prospective combination therapies. Eur. J. Haematol. 114 (4), 584–595. doi:10.1111/ejh.14383
Bu, Z., Xu, Y., Zhou, X., Wang, X., Liu, S., Wang, L., et al. (2024). Exploring the therapeutic potential of “Xiaochaihu Decoction”: a systematic review and meta-analysis on the clinical effectiveness and safety in managing cancer-related fever. Front. Pharmacol. 15, 1359866. doi:10.3389/fphar.2024.1359866
Cai, S. F., and Levine, R. L. (2019). Genetic and epigenetic determinants of AML pathogenesis. Semin. Hematol. 56 (2), 84–89. doi:10.1053/j.seminhematol.2018.08.001
Cai, Y., Zheng, Q., Sun, R., Wu, J., Li, X., and Liu, R. (2020). Recent progress in the study of Artemisiae Scopariae Herba (Yin Chen), a promising medicinal herb for liver diseases. Biomed. Pharmacother. 130, 110513. doi:10.1016/j.biopha.2020.110513
Calpe-Berdiel, L., Escolà-Gil, J. C., Benítez, S., Bancells, C., González-Sastre, F., Palomer, X., et al. (2007). Dietary phytosterols modulate T-helper immune response but do not induce apparent anti-inflammatory effects in a mouse model of acute, aseptic inflammation. Life Sci. 80 (21), 1951–1956. doi:10.1016/j.lfs.2007.02.032
Chen, D., Oezguen, N., Urvil, P., Ferguson, C., Dann, S. M., and Savidge, T. C. (2016). Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2 (3), e1501240. doi:10.1126/sciadv.1501240
Chen, J., Chen, C., Zhang, Z., Zeng, F., and Zhang, S. (2024b). Exploring the key amino acid residues surrounding the active center of lactate dehydrogenase A for the development of ideal inhibitors. Molecules 29 (9), 2029. doi:10.3390/molecules29092029
Chen, T., Zhang, Y., Zhang, D., and Zhou, H. (2024a). Immune-based subgroups uncover diverse tumor immunogenicity and implications for prognosis and precision therapy in acute myeloid leukemia. Front. Immunol. 15, 1451486. doi:10.3389/fimmu.2024.1451486
Chen, Z., and Zhuang, X. (2024). Meta-analysis of the efficacy and safety of integrated Chinese and Western medicine in the treatment of acute myeloid leukaemia in elderly people. Heliyon 10 (1), e23398. doi:10.1016/j.heliyon.2023.e23398
Cheng, H., Huang, C., Xu, X., Hu, X., Gong, S., Tang, G., et al. (2017). PIM-1 mRNA expression is a potential prognostic biomarker in acute myeloid leukemia. J. Transl. Med. 15 (1), 179. doi:10.1186/s12967-017-1287-4
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 (Suppl. 4), S11. doi:10.1186/1752-0509-8-S4-S11
Choi, Y. H. (2015). Induction of apoptosis by an ethanol extract of Poria cocos Wolf. in human leukemia U937 cells. Oncol. Rep. 34 (5), 2533–2540. doi:10.3892/or.2015.4256
Choi, Y. J., Li, X., Hydbring, P., Sanda, T., Stefano, J., Christie, A. L., et al. (2012). The requirement for cyclin D function in tumor maintenance. Cancer Cell 22 (4), 438–451. doi:10.1016/j.ccr.2012.09.015
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. doi:10.1038/srep42717
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47 (W1), W357–W364. doi:10.1093/nar/gkz382
Darici, S., Alkhaldi, H., Horne, G., Jørgensen, H. G., Marmiroli, S., and Huang, X. (2020). Targeting PI3K/Akt/mTOR in AML: rationale and clinical evidence. J. Clin. Med. 9 (9), 2934. doi:10.3390/jcm9092934
Desai, P. N., Wang, B., Fonseca, A., Borges, P., Jelloul, F. Z., Reville, P. K., et al. (2023). Single-cell profiling of CD8+ T cells in acute myeloid leukemia reveals a continuous spectrum of differentiation and clonal hyperexpansion. Cancer Immunol. Res., OF1–OF18. doi:10.1158/2326-6066.CIR-22-0961
De Stefani, E., Boffetta, P., Ronco, A. L., Brennan, P., Deneo-Pellegrini, H., Carzoglio, J. C., et al. (2000). Plant sterols and risk of stomach cancer: a case-control study in Uruguay. Nutr. Cancer 37 (2), 140–144. doi:10.1207/S15327914NC372_4
de Waure, C., Bertola, C., Baccarini, G., Chiavarini, M., and Mancuso, C. (2023). Exploring the contribution of curcumin to cancer therapy: a systematic review of randomized controlled trials. Pharmaceutics 15 (4), 1275. doi:10.3390/pharmaceutics15041275
Eisfeld, A. K., Kohlschmidt, J., Schwind, S., Nicolet, D., Blachly, J. S., Orwick, S., et al. (2017). Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia 31 (6), 1278–1285. doi:10.1038/leu.2016.332
El Menyiy, N., Aboulaghras, S., Bakrim, S., Moubachir, R., Taha, D., Khalid, A., et al. (2023). Genkwanin: an emerging natural compound with multifaceted pharmacological effects. Biomed. Pharmacother. 165, 115159. doi:10.1016/j.biopha.2023.115159
Fatima, N., Kalsoom, A., Mumtaz, A., and Muhammad, S. A. (2014). Computational drug designing of fungal pigments as potential aromatase inhibitors. Bangladesh J. Pharmacol. 9 (4), 575–579. doi:10.3329/bjp.v9i4.20435
Fleischer, T., Chang, T. T., Chiang, J. H., Sun, M. F., and Yen, H. R. (2017). Improved survival with integration of Chinese herbal medicine therapy in patients with acute myeloid leukemia: a nationwide population-based cohort study. Integr. Cancer Ther. 16 (2), 156–164. doi:10.1177/1534735416664171
Fong, W. F., Wang, C., Zhu, G. Y., Leung, C. H., Yang, M. S., and Cheung, H. Y. (2007). Reversal of multidrug resistance in cancer cells by Rhizoma Alismatis extract. Phytomedicine 14 (2-3), 160–165. doi:10.1016/j.phymed.2006.03.004
Forsberg, M., and Konopleva, M. (2024). AML treatment: conventional chemotherapy and emerging novel agents. Trends Pharmacol. Sci. 45 (5), 430–448. doi:10.1016/j.tips.2024.03.005
Fu, D., Jiang, H., Long, A., Guo, H. f., Capitano, M. L., Ramdas, B., et al. (2021b). Immunotherapy targeting ST2/IL-33 signaling in myeloid leukemia stem cells. Blood 138, 23. doi:10.1182/blood-2021-149672
Fu, D., Zhang, B., Wu, S., Feng, J., and Jiang, H. (2023). Molecular subtyping of acute myeloid leukemia through ferroptosis signatures predicts prognosis and deciphers the immune microenvironment. Front. Cell Dev. Biol. 11, 1207642. doi:10.3389/fcell.2023.1207642
Fu, D., Zhang, B., Wu, S., Zhang, Y., Xie, J., Ning, W., et al. (2021a). Prognosis and characterization of immune microenvironment in acute myeloid leukemia through identification of an autophagy-related signature. Front. Immunol. 12, 695865. doi:10.3389/fimmu.2021.695865
Gao, Y., Liu, F., Fang, L., Cai, R., Zong, C., and Qi, Y. (2014). Genkwanin inhibits proinflammatory mediators mainly through the regulation of miR-101/MKP-1/MAPK pathway in LPS-activated macrophages. PLoS One 9 (5), e96741. doi:10.1371/journal.pone.0096741
Garcia-Manero, G. (2023). Myelodysplastic syndromes: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 98 (8), 1307–1325. doi:10.1002/ajh.26984
Ge, S. S., Liu, S. B., and Xue, S. L. (2022). Developments and challenges of FLT3 inhibitors in acute myeloid leukemia. Front. Oncol. 12, 996438. doi:10.3389/fonc.2022.996438
Grattan, B. J. (2013). Plant sterols as anticancer nutrients: evidence for their role in breast cancer. Nutrients 5 (2), 359–387. doi:10.3390/nu5020359
Haiqiang, Y., Zengliang, Z., Ji, W., Yu, C., Libing, Z., Xiaoke, L., et al. (2016). Efficacy and safety of Yinchenwuling powder for hyperlipidemia: a systematic review and Meta-analysis. J. Tradit. Chin. Med. 36 (2), 135–143. doi:10.1016/s0254-6272(16)30019-x
Hu, S., Huang, L., Meng, L., Sun, H., Zhang, W., and Xu, Y. (2015). Isorhamnetin inhibits cell proliferation and induces apoptosis in breast cancer via Akt and mitogen-activated protein kinase kinase signaling pathways. Mol. Med. Rep. 12 (5), 6745–6751. doi:10.3892/mmr.2015.4269
Huang, H. L., Chen, C. C., Yeh, C. Y., and Huang, R. L. (2005). Reactive oxygen species mediation of baizhu-induced apoptosis in human leukemia cells. J. Ethnopharmacol. 97 (1), 21–29. doi:10.1016/j.jep.2004.09.058
Huang, H. L., Lin, T. W., Huang, Y. L., and Huang, R. L. (2016). Induction of apoptosis and differentiation by atractylenolide-1 isolated from Atractylodes macrocephala in human leukemia cells. Bioorg Med. Chem. Lett. 26 (8), 1905–1909. doi:10.1016/j.bmcl.2016.03.021
Huang, S., Zhang, B., Fan, W., Zhao, Q., Yang, L., Xin, W., et al. (2019). Identification of prognostic genes in the acute myeloid leukemia microenvironment. Aging (Albany NY) 11 (22), 10557–10580. doi:10.18632/aging.102477
Huang, Y. M., Kang, M., and Chang, C. E. (2014). Switches of hydrogen bonds during ligand-protein association processes determine binding kinetics. J. Mol. Recognit. 27 (9), 537–548. doi:10.1002/jmr.2377
Huang, Z., Shen, Y., Liu, W., Yang, Y., Guo, L., Yan, Q., et al. (2023). Berberine targets the electron transport chain complex I and reveals the landscape of OXPHOS dependency in acute myeloid leukemia with IDH1 mutation. Chin. J. Nat. Med. 21 (2), 136–145. doi:10.1016/S1875-5364(23)60391-7
Jin, J., Wang, J. X., Chen, F. F., Wu, D. P., Hu, J., Zhou, J. F., et al. (2013). Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 14 (7), 599–608. doi:10.1016/S1470-2045(13)70152-9
Jing, L., Zhang, B., Sun, J., Feng, J., and Fu, D. (2024). Prognostic insights and immune microenvironment delineation in acute myeloid leukemia by ferroptosis-derived signature. Heliyon 10 (6), e28237. doi:10.1016/j.heliyon.2024.e28237
Juliusson, G., Antunovic, P., Derolf, A., Lehmann, S., Möllgård, L., Stockelberg, D., et al. (2009). Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood 113 (18), 4179–4187. doi:10.1182/blood-2008-07-172007
Kantarjian, H., Kadia, T., DiNardo, C., Daver, N., Borthakur, G., Jabbour, E., et al. (2021). Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 11 (2), 41. doi:10.1038/s41408-021-00425-3
Kim, S. D., Kim, J. H., Kim, D. H., Park, J. H., Gong, Y., Sun, C., et al. (2024). Comprehensive evaluation of traditional herbal medicine combined with adjuvant chemotherapy on post-surgical gastric cancer: a systematic review and meta-analysis. Integr. Cancer Ther. 23, 15347354231226256. doi:10.1177/15347354231226256
Kopustinskiene, D. M., Jakstas, V., Savickas, A., and Bernatoniene, J. (2020). Flavonoids as anticancer agents. Nutrients 12 (2), 457. doi:10.3390/nu12020457
Kutzner, C., Páll, S., Fechner, M., Esztermann, A., de Groot, B. L., and Grubmüller, H. (2015). Best bang for your buck: GPU nodes for GROMACS biomolecular simulations. J. Comput. Chem. 36 (26), 1990–2008. doi:10.1002/jcc.24030
Leong, P. Y., Chen, H. H., Gau, S. Y., Chen, C. Y., Su, Y. C., and Wei, J. C. C. (2024). Traditional Chinese medicine in the treatment of patients with hyperuricemia: a randomized placebo-controlled double-blinded clinical trial. Int. J. Rheum. Dis. 27 (1), e14986. doi:10.1111/1756-185X.14986
Li, S., and Zhang, B. (2013). Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 11 (2), 110–120. doi:10.1016/S1875-5364(13)60037-0
Li, Z., Zhang, W., Guo, S., Qi, G., Huang, J., Gao, J., et al. (2024). A review of advances in mitochondrial research in cancer. Cancer Control. 31, 10732748241299072. doi:10.1177/10732748241299072
Liao, D., Basarab, G. S., Gatenby, A. A., Valent, B., and Jordan, D. B. (2001). Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis. Structure 9 (1), 19–27. doi:10.1016/s0969-2126(00)00548-7
Ling, Y. (2013). Traditional Chinese medicine in the treatment of symptoms in patients with advanced cancer. Ann. Palliat. Med. 2 (3), 141–152. doi:10.3978/j.issn.2224-5820.2013.04.05
Liu, N., Wu, C., Jia, R., Cai, G., Wang, Y., Zhou, L., et al. (2020). Traditional Chinese medicine combined with chemotherapy and cetuximab or bevacizumab for metastatic colorectal cancer: a randomized, double-blind, placebo-controlled clinical trial. Front. Pharmacol. 11, 478. doi:10.3389/fphar.2020.00478
Liu, X., and Zhao, L. (2024). Cases of nonalcoholic fatty liver treated with Yinchen Wuling powder. Shanxi J. Tradit. Chin. Med. 32 (05), 520–521.
Lohning, A. E., Levonis, S. M., Williams-Noonan, B., and Schweiker, S. S. (2017). A practical guide to molecular docking and homology modelling for medicinal chemists. Curr. Top. Med. Chem. 17 (18), 2023–2040. doi:10.2174/1568026617666170130110827
Luo, W., and Brouwer, C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29 (14), 1830–1831. doi:10.1093/bioinformatics/btt285
MacDonald, R. J., Bunaciu, R. P., Ip, V., Dai, D., Tran, D., Varner, J. D., et al. (2018). Src family kinase inhibitor bosutinib enhances retinoic acid-induced differentiation of HL-60 leukemia cells. Leuk. Lymphoma 59 (12), 2941–2951. doi:10.1080/10428194.2018.1452213
Marvin-Peek, J., Savani, B. N., Olalekan, O. O., and Dholaria, B. (2022). Challenges and advances in chimeric antigen receptor therapy for acute myeloid leukemia. Cancers (Basel) 14 (3), 497. doi:10.3390/cancers14030497
Masuda, Y., Asada, K., Satoh, R., Takada, K., and Kitajima, J. (2015). Capillin, a major constituent of Artemisia capillaris Thunb. flower essential oil, induces apoptosis through the mitochondrial pathway in human leukemia HL-60 cells. Phytomedicine 22 (5), 545–552. doi:10.1016/j.phymed.2015.03.008
Matsuo, H., Yoshida, K., Fukumura, K., Nakatani, K., Noguchi, Y., Takasaki, S., et al. (2018). Recurrent CCND3 mutations in MLL-rearranged acute myeloid leukemia. Blood Adv. 2 (21), 2879–2889. doi:10.1182/bloodadvances.2018019398
Mendilaharsu, M., De Stefani, E., Deneo-Pellegrini, H., Carzoglio, J., and Ronco, A. (1998). Phytosterols and risk of lung cancer: a case-control study in Uruguay. Lung Cancer 21 (1), 37–45. doi:10.1016/s0169-5002(98)00044-0
Messina, M., Piciocchi, A., Ottone, T., Paolini, S., Papayannidis, C., Lessi, F., et al. (2022). Prevalence and prognostic role of IDH mutations in acute myeloid leukemia: results of the GIMEMA AML1516 protocol. Cancers (Basel) 14 (12), 3012. doi:10.3390/cancers14123012
Miller, K. D., Nogueira, L., Mariotto, A. B., Rowland, J. H., Yabroff, K. R., Alfano, C. M., et al. (2019). Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69 (5), 363–385. doi:10.3322/caac.21565
Mir, S. A., Dar, A., Hamid, L., Nisar, N., Malik, J. A., Ali, T., et al. (2024). Flavonoids as promising molecules in the cancer therapy: an insight. Curr. Res. Pharmacol. Drug Discov. 6, 100167. doi:10.1016/j.crphar.2023.100167
Mohamed Jiffry, M. Z., Kloss, R., Ahmed-Khan, M., Carmona-Pires, F., Okam, N., Weeraddana, P., et al. (2023). A review of treatment options employed in relapsed/refractory AML. Hematology 28 (1), 2196482. doi:10.1080/16078454.2023.2196482
Monteiro, A. F. M., Viana, J. D. O., Nayarisseri, A., Zondegoumba, E. N., Mendonça Junior, F. J. B., Scotti, M. T., et al. (2018). Computational studies applied to flavonoids against alzheimer's and Parkinson's diseases. Oxid. Med. Cell Longev. 2018, 7912765. doi:10.1155/2018/7912765
Mou, Y., Wang, X., Wang, T., Wang, Y., Wang, H., Zhao, H., et al. (2022). Clinical application and pharmacological mechanism of Wuling powder in the treatment of ascites: a systematic review and network pharmacological analysis. Biomed. Pharmacother. 146, 112506. doi:10.1016/j.biopha.2021.112506
Negi, P., Cheke, R. S., and Patil, V. M. (2021). Recent advances in pharmacological diversification of Src family kinase inhibitors. Egypt. J. Med. Hum. Genet. 22, 52–16. doi:10.1186/s43042-021-00172-x
O'Donnell, M. R., Tallman, M. S., Abboud, C. N., Altman, J. K., Appelbaum, F. R., Arber, D. A., et al. (2017). Acute myeloid leukemia, version 3.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 15 (7), 926–957. doi:10.6004/jnccn.2017.0116
Ohsawa, T., Yukawa, M., Takao, C., Murayama, M., and Bando, H. (1992). Studies on constituents of fruit body of Polyporus umbellatus and their cytotoxic activity. Chem. Pharm. Bull. 40 (1), 143–147. doi:10.1248/cpb.40.143
Ozawa, Y., Williams, A. H., Estes, M. L., Matsushita, N., Boschelli, F., Jove, R., et al. (2008). Src family kinases promote AML cell survival through activation of signal transducers and activators of transcription (STAT). Leuk. Res. 32 (6), 893–903. doi:10.1016/j.leukres.2007.11.032
Pandolfi, A., Stanley, R. F., Yu, Y., Bartholdy, B., Pendurti, G., Gritsman, K., et al. (2015). PAK1 is a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood 126 (9), 1118–1127. doi:10.1182/blood-2014-12-618801
Park, S., Chapuis, N., Tamburini, J., Bardet, V., Cornillet-Lefebvre, P., Willems, L., et al. (2010). Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica 95 (5), 819–828. doi:10.3324/haematol.2009.013797
Patel, R. K., Weir, M. C., Shen, K., Snyder, D., Cooper, V. S., and Smithgall, T. E. (2019). Expression of myeloid Src-family kinases is associated with poor prognosis in AML and influences Flt3-ITD kinase inhibitor acquired resistance. PLoS One 14 (12), e0225887. doi:10.1371/journal.pone.0225887
Patidar, K., Panwar, U., Vuree, S., Sweta, J., Sandhu, M. K., Nayarisseri, A., et al. (2019). An in silico approach to identify high affinity small molecule targeting m-TOR inhibitors for the clinical treatment of breast cancer. Asian Pac J. Cancer Prev. 20 (4), 1229–1241. doi:10.31557/APJCP.2019.20.4.1229
Pyzer, A. R., Stroopinsky, D., Rosenblatt, J., Anastasiadou, E., Rajabi, H., Washington, A., et al. (2017). MUC1 inhibition leads to decrease in PD-L1 levels via upregulation of miRNAs. Leukemia 31 (12), 2780–2790. doi:10.1038/leu.2017.163
Restelli, C., Ruella, M., Paruzzo, L., Tarella, C., Pelicci, P. G., and Colombo, E. (2024). Recent advances in immune-based therapies for acute myeloid leukemia. Blood Cancer Discov. 5 (4), 234–248. doi:10.1158/2643-3230.BCD-23-0202
Rivera-Torres, J., and San José, E. (2019). Src tyrosine kinase inhibitors: new perspectives on their immune, antiviral, and senotherapeutic potential. Front. Pharmacol. 10, 1011. doi:10.3389/fphar.2019.01011
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform 6, 13. doi:10.1186/1758-2946-6-13
Ruan, Y., Hu, K., and Chen, H. (2015). Autophagy inhibition enhances isorhamnetin-induced mitochondria-dependent apoptosis in non-small cell lung cancer cells. Mol. Med. Rep. 12 (4), 5796–5806. doi:10.3892/mmr.2015.4148
Ruglioni, M., Crucitta, S., Luculli, G. I., Tancredi, G., Del Giudice, M. L., Mechelli, S., et al. (2024). Understanding mechanisms of resistance to FLT3 inhibitors in adult FLT3-mutated acute myeloid leukemia to guide treatment strategy. Crit. Rev. Oncol. Hematol. 201, 104424. doi:10.1016/j.critrevonc.2024.104424
Sanz, M., Burnett, A., Lo-Coco, F., and Löwenberg, B. (2009). FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr. Opin. Oncol. 21 (6), 594–600. doi:10.1097/CCO.0b013e32833118fd
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shi, G., Yu, D., Wu, J., Liu, Y., Huang, R., and Zhang, C. S. (2021). A systematic review and meta-analysis of traditional Chinese medicine with chemotherapy in breast cancer. Gland. Surg. 10 (5), 1744–1755. doi:10.21037/gs-21-284
Slika, H., Mansour, H., Wehbe, N., Nasser, S. A., Iratni, R., Nasrallah, G., et al. (2022). Therapeutic potential of flavonoids in cancer: ROS-mediated mechanisms. Biomed. Pharmacother. 146, 112442. doi:10.1016/j.biopha.2021.112442
Su, P. H., and Tai, C. J. (2020). Current development in integrative therapy of traditional Chinese medicine for cancer treatment: a mini-review. J. Tradit. Complement. Med. 10 (5), 429–433. doi:10.1016/j.jtcme.2019.07.001
Sun, J., Han, T., Yang, T., and Huang, J. (2021). Interpreting the molecular mechanisms of yinchenhao decoction on hepatocellular carcinoma through absorbed components based on network pharmacology. Biomed. Res. Int. 2021, 6616908. doi:10.1155/2021/6616908
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49 (D1), D605–D612. doi:10.1093/nar/gkaa1074
Tabata, R., Chi, S., Yuda, J., and Minami, Y. (2021). Emerging immunotherapy for acute myeloid leukemia. Int. J. Mol. Sci. 22 (4), 1944. doi:10.3390/ijms22041944
Tang, Y., Li, M., Wang, J., Pan, Y., and Wu, F. X. (2015). CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 127, 67–72. doi:10.1016/j.biosystems.2014.11.005
Toth, E., Varga, É., Kulcsár, P. I., Kocsis-Jutka, V., Krausz, S. L., Nyeste, A., et al. (2020). Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res. 48 (7), 3722–3733. doi:10.1093/nar/gkaa110
Ukiya, M., Akihisa, T., Tokuda, H., Hirano, M., Oshikubo, M., Nobukuni, Y., et al. (2002). Inhibition of tumor-promoting effects by poricoic acids G and H and other lanostane-type triterpenes and cytotoxic activity of poricoic acids A and G from Poria cocos. J. Nat. Prod. 65 (4), 462–465. doi:10.1021/np0103721
Ul Haq, F., Abro, A., Raza, S., Liedl, K. R., and Azam, S. S. (2017). Molecular dynamics simulation studies of novel beta-lactamase inhibitor. J. Mol. Graph Model 74, 143–152. doi:10.1016/j.jmgm.2017.03.002
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. C. (2005). GROMACS: fast, flexible, and free. J. Comput. Chem. 26 (16), 1701–1718. doi:10.1002/jcc.20291
Venugopal, S., Daver, N., and Ravandi, F. (2021). An update on the clinical evaluation of antibody-based therapeutics in acute myeloid leukemia. Curr. Hematol. Malig. Rep. 16 (1), 89–96. doi:10.1007/s11899-021-00612-w
Wang, C., Zhang, J. X., Shen, X. L., Wan, C. K., Tse, A. K. W., and Fong, W. F. (2004). Reversal of P-glycoprotein-mediated multidrug resistance by Alisol B 23-acetate. Biochem. Pharmacol. 68 (5), 843–855. doi:10.1016/j.bcp.2004.05.021
Wang, S. F., Tseng, L. M., and Lee, H. C. (2023). Role of mitochondrial alterations in human cancer progression and cancer immunity. J. Biomed. Sci. 30 (1), 61. doi:10.1186/s12929-023-00956-w
Wellhausen, N., O'Connell, R. P., Lesch, S., Engel, N. W., Rennels, A. K., Gonzales, D., et al. (2023). Epitope base editing CD45 in hematopoietic cells enables universal blood cancer immune therapy. Sci. Transl. Med. 15 (714), eadi1145. doi:10.1126/scitranslmed.adi1145
Willimott, S., Barker, J., Jones, L. A., and Opara, E. I. (2009). An in vitro based investigation of the cytotoxic effect of water extracts of the Chinese herbal remedy LD on cancer cells. Chem. Cent. J. 3, 12. doi:10.1186/1752-153X-3-12
Wu, J., Tang, G., Cheng, C. S., Yeerken, R., Chan, Y. T., Fu, Z., et al. (2024). Traditional Chinese medicine for the treatment of cancers of hepatobiliary system: from clinical evidence to drug discovery. Mol. Cancer 23 (1), 218. doi:10.1186/s12943-024-02136-2
Wu, P., Zhang, J., Xiao, J., Huang, G., Li, J., and Zhou, Z. (2022). Efficacy and safety of Yinchenwuling Powder for nonalcoholic fatty liver disease: a protocol for systematic review and meta-analysis of randomized controlled trials. Med. Baltim. 101 (48), e32088. doi:10.1097/MD.0000000000032088
Xiang, Y., Guo, Z., Zhu, P., Chen, J., and Huang, Y. (2019). Traditional Chinese medicine as a cancer treatment: modern perspectives of ancient but advanced science. Cancer Med. 8 (5), 1958–1975. doi:10.1002/cam4.2108
Xu, H., Wang, Y., Zhao, J., Jurutka, P. W., Huang, D., Liu, L., et al. (2020). Triterpenes from Poria cocos are revealed as potential retinoid X receptor selective agonists based on cell and in silico evidence. Chem. Biol. Drug Des. 95 (5), 493–502. doi:10.1111/cbdd.13610
Yang, J., Zhu, X., Yuan, P., Liu, J., Wang, B., and Wang, G. (2020). Efficacy of traditional Chinese Medicine combined with chemotherapy in patients with non-small cell lung cancer (NSCLC): a meta-analysis of randomized clinical trials. Support Care Cancer 28 (8), 3571–3579. doi:10.1007/s00520-020-05433-w
Yang, Q., Meng, D., Zhang, Q., and Wang, J. (2024). Advances in research on the anti-tumor mechanism of Astragalus polysaccharides. Front. Oncol. 14, 1334915. doi:10.3389/fonc.2024.1334915
Yang, X., and Wang, J. (2018). Precision therapy for acute myeloid leukemia. J. Hematol. Oncol. 11 (1), 3. doi:10.1186/s13045-017-0543-7
Yang, Y., Li, S., Wang, Y., Zhao, Y., and Li, Q. (2022). Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective. Signal Transduct. Target Ther. 7 (1), 329. doi:10.1038/s41392-022-01168-8
Zhang, B., Liu, G., Wang, X., and Hu, X. (2022b). Identification of molecular targets and potential mechanisms of yinchen wuling san against head and neck squamous cell carcinoma by network pharmacology and molecular docking. Front. Genet. 13, 914646. doi:10.3389/fgene.2022.914646
Zhang, B., Yang, L., Wang, X., and Fu, D. (2021a). Identification of survival-related alternative splicing signatures in acute myeloid leukemia. Biosci. Rep. 41 (7). doi:10.1042/BSR20204037
Zhang, H. W., Hu, J. J., Fu, R. Q., Liu, X., Zhang, Y. H., Li, J., et al. (2018). Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells. Sci. Rep. 8 (1), 11255. doi:10.1038/s41598-018-29308-7
Zhang, J., Wu, Y., Tian, Y., Xu, H., Lin, Z. X., and Xian, Y. F. (2024). Chinese herbal medicine for the treatment of intestinal cancer: preclinical studies and potential clinical applications. Mol. Cancer 23 (1), 217. doi:10.1186/s12943-024-02135-3
Zhang, S., Yang, Z., Bao, W., Liu, L., You, Y., Wang, X., et al. (2020). SNX10 (sorting nexin 10) inhibits colorectal cancer initiation and progression by controlling autophagic degradation of SRC. Autophagy 16 (4), 735–749. doi:10.1080/15548627.2019.1632122
Zhang, X., Xiong, H., Duan, J., Chen, X., Liu, Y., and Huang, C. (2022a). The prognostic significance of the BIN1 and CCND2 gene in adult patients with acute myeloid leukemia. Indian J. Hematol. Blood Transfus. 38 (3), 481–491. doi:10.1007/s12288-021-01479-w
Zhang, Y., Zhao, M., Liu, Y., Liu, T., Zhao, C., and Wang, M. (2021b). Investigation of the therapeutic effect of Yinchen Wuling Powder on CCl(4)-induced hepatic fibrosis in rats by (1)H NMR and MS-based metabolomics analysis. J. Pharm. Biomed. Anal. 200, 114073. doi:10.1016/j.jpba.2021.114073
Zhu, H., Peng, Z., Dai, M., Zou, Y., Qin, F., Chen, J., et al. (2016). Efficacy and safety of Wuling San for treatment of breast-cancer-related upper extremity lymphoedema: study protocol for a pilot trial. BMJ Open 6 (12), e012515. doi:10.1136/bmjopen-2016-012515
Zhuang, Y., Zhao, J., Xu, X., and Bi, L. (2018). Downregulation of GLUT3 promotes apoptosis and chemosensitivity of acute myeloid leukemia cells via EGFR signaling. Arch. Iran. Med. 21 (2), 73–78.
Keywords: acute myeloid leukemia, Yinchen Wuling San, network pharmacology, molecular docking, molecular dynamics simulation, pharmacological target
Citation: Zhang B, Fu D, Wang X and Hu X (2025) Network-based analysis and experimental validation of identified natural compounds from Yinchen Wuling San for acute myeloid leukemia. Front. Pharmacol. 16:1591164. doi: 10.3389/fphar.2025.1591164
Received: 10 March 2025; Accepted: 09 May 2025;
Published: 30 May 2025.
Edited by:
Syed Shams ul Hassan, Shanghai Jiao Tong University, ChinaReviewed by:
Diwakar Bastihalli Tukaramrao, The Pennsylvania State University, United StatesChengqian Pan, Jiangsu University, China
Copyright © 2025 Zhang, Fu, Wang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuelei Hu, MDQwMDIxNDhAd2l0LmVkdS5jbg==; Xin Wang, d2FuZ3hpbjAwNzJAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Biyu Zhang
Biyu Zhang Denggang Fu
Denggang Fu Xin Wang
Xin Wang Xuelei Hu1*
Xuelei Hu1*