- 1Department of Pharmacy, Faculty of Science, University of Rajshahi, Rajshahi, Bangladesh
- 2Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 3Laboratory for Pharmacogenomics, Somdech Phra Debaratana Medical Center (SDMC), Ramathibodi Hospital, Bangkok, Thailand
- 4Department of Pharmacy, Daffodil International University, Dhaka, Bangladesh
- 5Department of Pharmaceutical Life Sciences, Faculty of Pharmacy, Universiti Malaya, Kuala Lumpur, Malaysia
- 6Department of Clinical Pharmacy and Pharmacy Practice, Faculty of Pharmacy, Universiti Malaya, Kuala Lumpur, Malaysia
- 7Pharmacogenomics and Precision Medicine, The Preventive Genomics & Family Check-up Services Center, Bumrungrad International Hospital, Bangkok, Thailand
- 8Faculty of Pharmaceutical Sciences, Burapha University, Saensuk, Chonburi, Thailand
Background: The safety or efficacy of drugs may be affected by the genetic variability of CYP2C9 or VKORC1. Patients may be at increased risk of drug-related toxicities, for example, bleeding events, if they carry CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910), or VKORC1 c.-1639G>A (rs9923231) genetic variants.
Methods: The allele frequencies of CYP2C9*2, *3, and VKORC1 c.-1639G>A were obtained from the 1000 Genomes Project Phase III in line with Fort Lauderdale principles. Predictive risk phenotypes and correlations were assigned based on the carrying of characteristic allele carriers following international pharmacogenomics (PGx)-based dosing guidelines.
Results: Intermediate and poor metabolizers were predicted to be risk phenotypes (17.8%; 95% CI 16.3%–19.3%) due to inheriting CYP2C9*2 and *3 genetic variants. These risk phenotypes were highest in European (35%; 95% CI 30.8%–39.2%), followed by South Asian (26.8%; 95% CI 22.9%–30.7%), American (25.9%; 95% CI 21.3%–30.5%), East Asian (6.7%; 95% CI 4.5%–8.9%), and African populations (2.1%; 95% CI 1%–3.2%). In addition, sensitive and highly sensitive responders were considered risk phenotypes (33.1%; 95% CI 31.3%–35%) when combining CYP2C9*2 and *3 variants with VKORC1c.-1639G>A. These risk phenotypes were most prevalent in East Asian (79.6%; 95% CI 76%–83.1%), followed by European (38.6%; 95% CI 34.3%–42.8%), American (30%; 95% CI 25.2%–34.8%), South Asian (25.2%; 95% CI 21.3%–29%), and African populations (1.2%; 95% CI 0.4%–2%), respectively. The prevalence of risk phenotypes in different ethnic groups was statistically significant (p < 0.05; 1.94 × 10−175, χ2 test). According to clinical annotations in the PharmGKB, the safety or efficacy of at least 29 commonly prescribed drugs is impacted by the genetic polymorphisms of CYP2C9/VKORC1 c.-1639G>A to varying degrees. The PGx label information is available for at least 23 drugs, and the Clinical Pharmacogenetics Implementation Consortium (CPIC) has already recommended PGx-based dosing guidelines for at least 11 of these medications, based on the genetic variability of CYP2C9/VKORC1 c.-1639G>A.
Conclusion: To enhance the safety of at least 11 clinically significant drugs, the current study found that approximately one-fifth of the global population is at risk based on the CYP2C9*2 and *3 genotypes. Additionally, approximately one-third of the population is at risk when considering the combination of CYP2C9 and VKORC1 c.-1639G>A genotypes. Further studies are warranted to evaluate the clinical benefits of integrating PGx-based therapies in routine practice.
Introduction
Genetic polymorphism refers to gene variations occurring at a frequency of 1% or higher in the human population (Brookes, 1999; Chiarella et al., 2023). Individual drug response is chiefly determined by the drug’s pharmacodynamic and pharmacokinetic features, which are directly or indirectly linked to the polymorphism in genes encoding the enzymes responsible for drug metabolism (Ahmed et al., 2016). To optimize both effectiveness and safety in treatment, it is advisable to employ precision medicine, which tailors patient care based on individual genetic traits. In this approach, particular attention is given to the polymorphisms of genes encoding cytochrome P450 (CYP) enzymes, as approximately 80% of phase-1 drug biotransformation is facilitated by a diverse array of CYP enzymes (Ashley, 2016; Collins and Varmus, 2015; Tornio and Backman, 2018; Zanger and Schwab, 2013; Zhou et al., 2017; Biswas, 2021). Of these, CYP2C9 is the predominantly expressed CYP2 isoform in the liver, making up ∼20% of the hepatic CYP proteins measured by mass spectrometry (Zhang et al., 2016). It is involved in the metabolism of a range of widely prescribed drugs such as coumarin anticoagulants, statins, non-steroidal anti-inflammatory drugs (NSAIDs), phenytoin, and sulfonylureas (Cooper-Dehoff et al., 2022; Daly et al., 2017; Johnson et al., 2017; Karnes et al., 2021; Relling and Klein, 2011; Yasar et al., 2003). CYP2C9 is a highly polymorphic gene with at least 85 known variant alleles identified to date (Babaev et al., 2025). CYP2C9*2 (p.R144C; rs1799853) and CYP2C9*3 (p.I359L; rs1057910) are two well-studied genetic variants known to alter the safety and effectiveness of various medications. For instance, individuals carrying the CYP2C9*2 allele exhibit a 30%–40% reduction in CYP2C9 function, which leads to increased systemic exposure to fluvastatin. Those with the CYP2C9*3 allele experience a significantly more pronounced impact, with an approximately 80% reduction in CYP2C9 function (Cooper-Dehoff et al., 2022). Similarly, individuals inheriting one or two copies of CYP2C9*2 or *3 exhibited a greater risk of bleeding toxicity with warfarin therapy, as CYP2C9*2 and *3, respectively, impair 30%–40% and 80%–90% of the metabolism of S-warfarin (Lee et al., 2002; Mega et al., 2015). Due to the substantial impact of CYP2C9 variants, the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) recommended genotyping for CYP2C9 in the product characteristics summary or drug labels of 19 drugs (Zhou et al., 2023). The Clinical Pharmacogenetics Implementation Consortium (CPIC) also made similar recommendations, emphasizing the testing and adjusting dosages for several commonly prescribed drugs (Karnes et al., 2021; Johnson et al., 2017; Theken et al., 2020; Cooper-Dehoff et al., 2022).
On the other hand, VKORC1 is situated on chromosome 16 and encodes the vitamin K epoxide reductase protein, which is associated with the metabolism of coumarin anticoagulants like warfarin (Gaikwad et al., 2014; Johnson et al., 2017). Among the several identified single-nucleotide polymorphisms (SNPs) in the VKORC1 gene, VKORC1 c.-1639G>A (rs9923231) is specifically notable because of its impact on warfarin dosing by causing changes in the enzymatic activities of VKORC that significantly affects warfarin’s anticoagulant activity (Liu S. et al., 2021; Yang et al., 2010; Eriksson and Wadelius, 2012). VKORC1 c.-1639G>A alters the gene promoter activity, leading to lower warfarin doses being required for individuals inheriting allele A compared to people with the GG wild type (Panchenko et al., 2020; Ghafoor et al., 2022; Altawil and Youssef, 2023). Consequently, a reduction of warfarin dose by 70% in patients carrying the AA genotype was suggested by the Food and Drug Administration (FDA) in the 2007 drug label (Eriksson and Wadelius, 2012). Combinations of CYP2C9 variants and VKORC1 c.-1639G>A have been used as the indication for choosing effective warfarin doses and managing the incidents of bleeding toxicity and have been extensively studied in many clinical studies (Yee et al., 2019; Vuorinen et al., 2021; Sridharan et al., 2021; Shukla et al., 2019; Liu J. et al., 2021; Dietz et al., 2020; Wang et al., 2024; Fahmi et al., 2024).
Previous studies exhibited substantial differences in the allele frequencies of CYP2C9 and VKORC1 c.-1639G>A among various populations and ethnic groups (Sangkuhl et al., 2021; Nizamuddin et al., 2021; Gaikwad et al., 2013; Lund et al., 2012; Ma et al., 2012; Pourgholi et al., 2016; Vuorinen et al., 2021). Given the critical role in clinical outcomes, studies of the prevalence of CYP2C9 and VKORC1 c.-1639G>A variants in global populations are therefore warranted. Only a few such studies were identified, but those did not include VKORC1 c.-1639G>A, and individuals within the same geographic group most commonly conformed to subpopulations (Céspedes-Garro et al., 2015; Sangkuhl et al., 2021; Sistonen et al., 2009; Zhou et al., 2023). No study has been identified to date that correlates the risk phenotypes with the number of clinically prescribed medications.
Aims
Most genetic frequency studies commonly use subpopulations living within the same geographic area, which greatly limits the generalizability of the findings. We aimed to determine the predictive prevalence of CYP2C9*2, *3, and VKORC1 c.-1639G>A genetic polymorphisms on a global scale by utilizing the genetic data from the 1000 Genomes Project, comprising 2,504 participants from 26 different populations, to determine the predictive risk phenotypes. We also aimed to establish a correlation between the identified risk phenotypes and important commonly prescribed clinical drugs, whose safety and effectiveness may be affected by the risk phenotypes due to carrying genetic variants.
Methods
Study participants
This study analyzed genetic data for CYP2C9*2, *3, and VKORC1 c.-1639G>A from five different continental groups consisting of America (AMR), Africa (AFR), Europe (EUR), East Asia (EAS), and South Asia (SAS) (Auton et al., 2015; Sudmant et al., 2015). A total of 347 healthy individuals from four distinct ethnic groups (MXL = Mexican ancestry in Los Angeles, California; CLM = Colombian in Medellin, Colombia; PUR = Puerto Rican in Puerto Rico; and PEL = Peruvian in Lima, Peru) participated in the 1000 Genomes project for the continent of America. Similarly, across Europe, 503 healthy volunteers from five different ethnic groups (FIN = Finnish in Finland; CEU = Utah residents with Northern and Western European ancestry; GBR = British in England and Scotland; TSI = Tuscany in Italy; and IBS = Iberian populations in Spain) participated in the 1,000 Genome project. For the continent of Africa, seven ethnic groups (ASW = African ancestry in Southwest US; ACB = African Caribbean in Barbados; ESN = Esan in Nigeria; LWK = Luhya in Webuye, GWD = Gambian in Western Division, The Gambia; Kenya; YRI = Yoruba in Ibadan, Nigeria; and MSL = Mende in Sierra Leone) comprising 661 healthy volunteers were included in the 1,000 Genome project. Five different ethnic groups (GIH = Gujarati Indian in Houston; BEB, Bengali in Bangladesh; ITU = Indian Telugu in the United Kingdom; STU = Sri Lankan Tamil in the United Kingdom; and PJL = Punjabi in Lahore, Pakistan) containing 489 healthy individuals were included in the 1000 Genomes project for the continent of South Asia. For East Asia, 504 healthy volunteers from five ethnic groups (CHB = Han Chinese in Beijing; CDX = Chinese Dai in Xishuangbanna, China; CHS = Southern Han Chinese, China; KHV = Kinh in Ho Chi Minh City, Vietnam; and JPT = Japanese in Tokyo, Japan) participated in the 1000 Genomes project.
Genetic data
Adhering to the Fort Lauderdale principles, the frequency of the genotypes and alleles of 2,504 participants of 26 distinct populations from five continental groups across the world carrying CYP2C9*1, CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910), or VKORC1 c.-1639G>A (rs9923231) were extracted from 1000 Genomes Project Phase III. We then classified them into genotype groups based on carriage of different combinations of the aforementioned alleles. For instance, a participant carrying two copies of the CYP2C9*3 allele was assigned as a genotype of CYP2C9*3/*3. Likewise, other genotypes were assigned based on the characteristic alleles carried by the participants.
Determination of predicted phenotypes and risk phenotypes
Three phenotype groups were assigned based on the genotypes as detailed in the Clinical Pharmacogenetics Implementation Consortium Guideline. Individuals with CYP2C9*1/*1 were classified as the normal metabolizer (NM) group. Similarly, individuals carrying CYP2C9*1/*2, CYP2C9*1/*3, and CYP2C9*2/*2 conformed to the intermediate metabolizer (IM) group, while individuals inheriting CYP2C9*2/*3 and CYP2C9*3/*3 were assigned as poor metabolizers (PM) (Karnes et al., 2021; Theken et al., 2020; Cooper-Dehoff et al., 2022).
For warfarin, three functional genotype groups were then assigned based on combinations of CYP2C9 and VKORC1 c.-1639G>A genotypes that correspond to the categories of updated warfarin labels by the FDA. Individuals carrying CYP2C9*1/*1 + VKORC1 c.-1639G/G, CYP2C9*1/*1 + VKORC1 c.-1639A/G, or CYP2C9*1/*2 + VKORC1 c.-1639G/G were grouped as normal responders (NR). In contrast, the carriers of CYP2C9*1/*1 + VKORC1 c.-1639A/A, CYP2C9*1/*2 + VKORC1 c.-1639A/G, CYP2C9*1/*2 + VKORC1 c.-1639A/A, CYP2C9*1/*3 + VKORC1 c.-1639G/G, CYP2C9*1/*3 + VKORC1 c.-1639A/G, or CYP2C9*2/*2 + VKORC1 c.-1639G/G were grouped as sensitive responders (SR). Individuals with CYP2C9*1/*3 + VKORC1 c.-1639A/A, CYP2C9*2/*2 + VKORC1 c.-1639A/G, CYP2C9*2/*2 + VKORC1 c.-1639A/A, CYP2C9*2/*3 + VKORC1 c.-1639G/G, CYP2C9*2/*3 + VKORC1 c.-1639A/G, CYP2C9*2/*3 + VKORC1 c.-1639A/A, CYP2C9*3/*3 + VKORC1 c.-1639G/G, CYP2C9*3/*3 + VKORC1 c.-1639A/G, or CYP2C9*3/*3 + VKORC1 c.-1639A/A formed the highly sensitive responder (HSR) group (Mega et al., 2015). Table 1 sums up the categories.
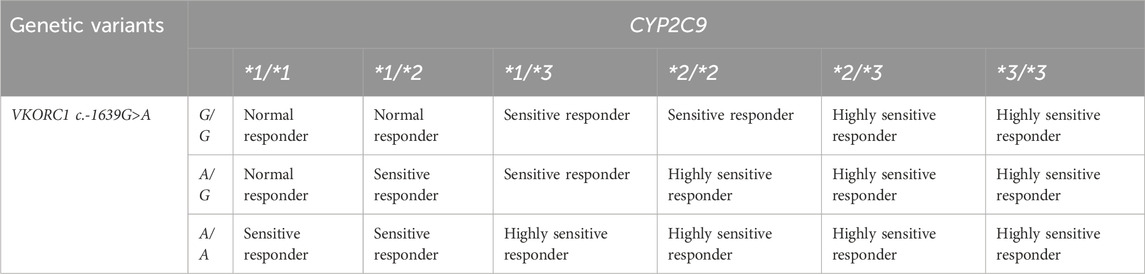
Table 1. Predictive phenotypes based on the combined genotypes of CYP2C9*2, CYP2C9*3, and VKORC1 c.-1639G>A genetic variants.
According to the CPIC dosing guidelines for several clinically significant drugs, the IM and PM groups of predicted phenotypes based on the CYP2C9 genotype pose a safety risk. Therefore, these phenotypes (IM and PM) are considered risk phenotypes (Cooper-Dehoff et al., 2022; Karnes et al., 2021; Theken et al., 2020). The association between the presence of genetic variants CYP2C9*2, *3, and VKORC1 c.-1639G>A and the bleeding risk with warfarin therapy has been explored and established in several previous studies (Bazan et al., 2014; Bedewy et al., 2018; Biswas et al., 2018; Eriksson et al., 2016; Ferder et al., 2010; Gaikwad et al., 2013; Higashi et al., 2002; Limdi et al., 2008; Shukla et al., 2019; Vuorinen et al., 2021). A higher risk of major hemorrhages (up to 2–5-fold) has been observed among genetically sensitive and highly sensitive patients (Mega et al., 2015; Limdi et al., 2008; Yang et al., 2019). Therefore, they were the risk population in this study.
Linking risk CYP2C9*2, *3, and VKORC1 c.-1639G>A phenotypes with the safety and effectiveness of medications
The clinical annotations and PGx label information of CYP2C9*2, *3, and VKORC1 c.-1639G>A for various clinically important drugs were sourced from internationally renowned pharmacogenomics working bodies, including the FDA-approved drug label, PharmGKB, the EMA-approved drug label, and the Health Canada Santé Canada (HCSC) approved drug label. All of the information was sourced from the PharmGKB website (Barbarino et al., 2018; Relling and Klein, 2011). The correlation between the safety or efficacy and the predicted risk phenotype was established using freely available pharmacogenomic-based CPIC dosing guidelines for various drugs (Karnes et al., 2021; Johnson et al., 2017; Cooper-Dehoff et al., 2022; Theken et al., 2020).
Human ethics approval
For the current study, we gathered all the presented human genetic data from Phase III of the 1000 Genomes Project that adhered to Fort Lauderdale principles. The dictates for publishing any result utilizing data from the 1000 Genomes Project require no additional human ethics approval, as it has been previously published in another study (Auton et al., 2015; Sudmant et al., 2015).
Statistical analysis
Data analyses were performed using descriptive statistics, and MS Excel was applied for both analysis and graphical representations. We utilized descriptive statistics to determine the frequency and risk profiles, and a chi-square test was employed to establish the statistical significance across the population. The findings are presented in a line chart.
Validation of data analysis
All the genetic data included in the present study were sourced as raw data from the 1000 Genomes Project, which were then grouped into genotype groups by utilizing the COUNTIFS function in MS Excel, followed by descriptive analyses. Two researchers independently carried out all the analyses, and the corresponding author then double-checked all the analyzed data collected from the two researchers and corrected any anomalies accordingly.
Results
Prevalence of CYP2C9*2, *3, and VKORC1 c.-1639G>A alleles and associated genotypes in 26 populations
The allele frequency of CYP2C9*2 was 4.8% (95% CI 4%–5.6%) in the global populations that participated in the 1000 Genomes Project and varied among all the populations studied. The European population had the highest prevalence (12.4%; 95% CI 11.1%–13.7%), and the American population ranked second with a frequency of 9.9% (95% CI 8.8%–11.1%). The allele frequency was found to be progressively lower in the South Asian (3.5%; 95% CI 2.8%–4.2%), African (0.8%; 95% CI 0.5%–1.2%), and East Asian populations (0.1%; 95% CI 0%–0.2%). The allele frequency of CYP2C9*3 was 4.9% (95% CI 4%–5.7%) in the global populations that participated in the 1000 Genomes Project, where the South Asian population had the highest prevalence of CYP2C9*3 (10.9%; 95% CI 9.7%–12.2%). The European population had the second highest prevalency group with a frequency of 7.3% (95% CI 6.2%–8.3%), followed by the American (3.7%; 95% CI 3%–4.5%), East Asian (3.4%; 95% CI 2.7%–4.1%), and African (0.2%; 95% CI 0%–0.4%) populations, respectively.
Similar variability was observed for the allele frequency of VKORC1 c.-1639G>A. The prevalence of this variant allele was 35.6% (95% CI 33.7%–37.4%) in the global populations that participated in the 1000 Genomes Project. The highest prevalence was in the East Asian population with a frequency of 88.5% (95% CI 87.2%–89.7%), followed by American (41.1%; 95% CI 39.1%–43%), European (38.8%; 95% CI 36.9%–40.7%), South Asian (14.5%; 95% CI 13.1%–15.9%), and African (5.4%; 95% CI 4.6%–6.3%) populations. For the A/G type genotype, however, the American population exhibited the highest frequency of 47.6% (95% CI 42.3%–52.8%), closely followed by the European group with 46.1% (95% CI 41.8%–50.5%). The lowest frequency (10% 95% CI 7.7%–12.3%) was observed for the African population. South Asian and East Asian populations showed frequencies of 24.1% (95% CI 20.3%–27.9%) and 19.4% (95% CI 16.0%–22.9%), respectively. The A/A genotype was predominantly observed in the East Asian population, where its prevalence reached 78.8% (95% CI: 75.2%–82.3%). In the American population, it was found in 17.3% (95% CI: 13.3%–21.3%), making it the second-most common group, followed by the European population with a prevalence of 15.7% (95% CI: 12.5%–18.9%). South Asian and African populations had a low prevalence of 2.5% (95% CI 1.1%–3.8%) and 0.5% (95% CI 0%–1%), respectively.
The predictive genotypes were established using different combinations of the different variants of CYP2C9, as shown in Table 2. Participants inheriting two normal copies, that is, CYP2C9*1/*1, were the most prevalent (82.2%; 95% CI 80.7%–83.7%) in all populations. Individuals with a normal copy and a *3 allele (CYP2C9*1/*3) were the second most prevalent, with a global frequency of 8.5% (95% CI 7.4%–9.6%), followed by CYP2C9*1/*2 (7.7%; 95% CI 6.7%–8.7%), CYP2C9*2/*3 (0.8%; 95% CI 0.5%–1.1%), CYP2C9*2/*2 (0.6%; 95% CI 0.3%–0.9%), and CYP2C9*3/*3 (0.2%; 95% CI 0%–0.4%). Likewise, for VKORC1 c.-1639G>A, the average prevalence in all populations was the highest for G/G (50.9%; 95% CI 48.9%–52.9%), followed by A/G (27.1%; 95% CI 25.4%–28.8%) and A/A (22%; 95% CI 20.4%–23.6%).
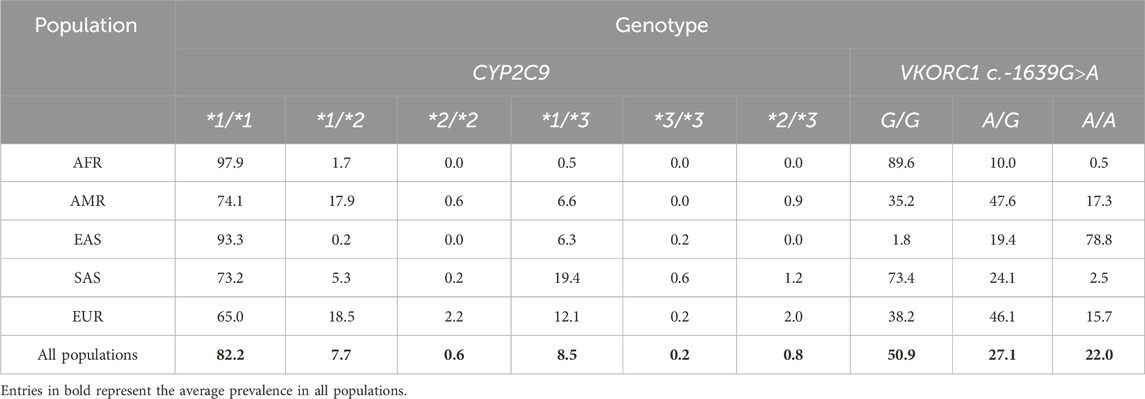
Table 2. Frequency of genotypes associated with carrying either CYP2C9*2, CYP2C9*3, or VKORC1 c.-1639G>A genetic variants in the world populations participating in the 1000 Genomes Project.
Prevalence of predicted phenotypes in 26 populations
As detailed in the methods section, predictive phenotype groups, such as NMs, IMs, and PMs, were assigned according to genotype, as shown in Figure 1. The NMs had an average prevalence of 82.2% (95% CI 80.7%–83.7%) for all 26 populations, and NM had the highest prevalence in the African population (97.9%; 95% CI 96.8%–99%), followed by East Asian (93.3%; 95% CI 91.1%–95.4%), American (74.1%; 95% CI 69.5%–78.7%), South Asian (73.2%; 95% CI 69.3%–77.1%), and European populations (65%; 95% CI 60.8%–69.2%). In contrast, the average prevalence of IMs in all populations was 16.8% (95% CI 15.3%–18.3%). Geographical group analysis showed the prevalence of IMs in European (32.8%; 95% CI 28.7%–36.9%), South Asian (24.9%; 95% CI 21.1%–28.8%), American (25.1%; 95% CI 20.5%–29.6%), East Asian (6.5% 95% CI 4.4%–8.7%) and African populations (2.1% 95% CI 1%–3.2%). Finally, PMs had an average prevalence of 1% (95% CI 0.6%–1.3%) for all populations, and geographical group analysis showed the prevalence of PMs in European (2.2%; 95% CI 0.9%–3.5%), South Asian (1.8%; 95% CI 0.6%–3%), American (0.9%; 95% CI 0%–1.8%) and East Asian populations (0.2%; 95% CI 0%–0.5%). PMs were not observed in the African population.
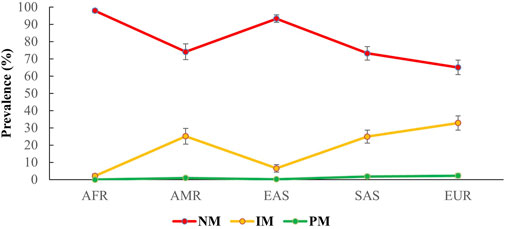
Figure 1. Predicted prevalence of different phenotypes for CYP2C9 among different populations participating in the 1000 Genomes Project. The phenotypes were grouped on the basis of being a carrier of CYP2C9*1, *2, and *3 alleles following international guidelines. Here, AFR: Africans, AMR: Ad.Mix Americans, EAS: East Asians, SAS: South Asians, EUR: Europeans. NM, normal metabolizers; IM, intermediate metabolizers; PM, poor metabolizers.
For the combination of CYP2C9 and VKORC1 c.-1639G>A, we classified the predictive phenotypes into three distinguished groups, that is, NR, SR, and HSR, as described in the methods section. After grouping the combined genotypes into subsequent predictive phenotype groups, based on the geographical analysis, we observed the highest prevalence of NR in the African population (98.8%; 95% CI 98%–99.6%), sequentially followed by the South Asian (74.8%; 95% CI 71%–78.7%), American (70%; 95% CI 65.2%–74.8%), European (61.4%; 95% CI 57.2%–65.7%), and, finally, the East Asian population (20.4%; 95% CI 16.9%–24%). Interestingly, the East Asian population marked the highest prevalence (73.8%; 95% CI 70%–77.6%) of SR, while the lowest prevalence of SR was observed in the African population (1.2%; 95% CI 0.4%–2%). The prevalence of SR in European, American, and South Asian populations was 33.2% (95% CI 29.1%–37.3%), 27.1% (95% CI 22.4%–31.8%), and 22.7% (95% CI 19%–26.4%), respectively. Finally, for HSR, the highest frequency was observed in the East Asian population (5.8%; 95% CI 3.7%–7.8%), followed by the European (5.4%; 95% CI 3.4%–7.3%), American population (2.9%; 95% CI 1.1%–4.6%), and South Asian populations (2.5%; 95% CI 1.1%–3.8%). HSRs were not identified in the African population. The frequency of the phenotype groups for different populations is shown in Figure 2.
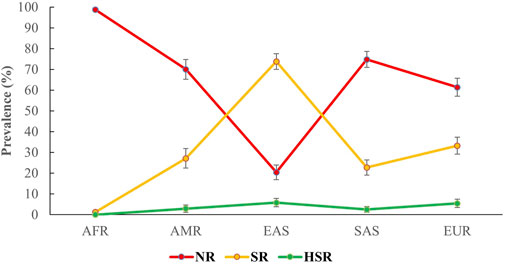
Figure 2. Predicted prevalence of different phenotypes of the combinations of CYP2C9 and VKORC1 c.-1639G>A among different populations participating in the 1000 Genomes Project. The phenotypes were grouped on the basis of being a carrier of the variants of CYP2C9 and VKORC1 c.-1639G>A alleles, following international guidelines. Here, AFR: Africans, AMR: Americans, EAS: East Asians, SAS: South Asians, EUR: Europeans. NR, normal responders; SR, sensitive responders; HRS, highly sensitive responders.
Prevalence of risk phenotypes in 26 populations
As outlined in the method portion, the risk phenotypes of CYP2C9 were determined by the frequency of IM and PM. Among the 2,504 participants of the 1000 Genomes Project, we identified approximately one-fifth (17.8%; 95% CI 16.3%–19.3%) of the CYP2C9 risk phenotypes. Geographical analysis revealed variability in the frequency of risk phenotypes among different populations. The European group had the highest prevalence, with a frequency of 35% (95% CI 30.8%–39.2%). The South Asian population followed with a frequency of 26.8% (95% CI 22.9%–30.7%), while the American population had a prevalence of 25.9% (95% CI 21.3%–30.5%). The East Asian population showed a frequency of 6.7% (95% CI 4.5%–8.9%), and the African population had the lowest prevalence at 2.1% (95% CI 1.1%–3.2%). For the combined CYP2C9 and VKORC1 c.-1639G>A genotypes, SR and HSR comprised the risk phenotypes, which we found to be approximately one-third (33.1%; 95% CI 31.3%–35%) of the 2,504 individuals partaking in the 1000 Genomes Project. Geographical group analysis revealed that the East Asian population is in the lead with a frequency of 79.6% (95% CI 76%–83.1%) of the risk phenotypes, and the African population, with only 1.2% (95% CI 0.4%–2%) frequency, was the group with the lowest prevalence. The European, American, and South Asian populations had frequencies of 38.6% (95% CI 34.3%–42.8%), 30% (95% CI 25.2%–34.8%), and 25.2% (95% CI 21.3%–29%), respectively. The prevalence of risk phenotypes in different ethnic groups was statistically significant (p < 0.05; 1.94 × 10−175, χ2 test).
Linking CYP2C9 and VKORC1 c.-1639G>A risk phenotypes with the safety or effectiveness of drugs
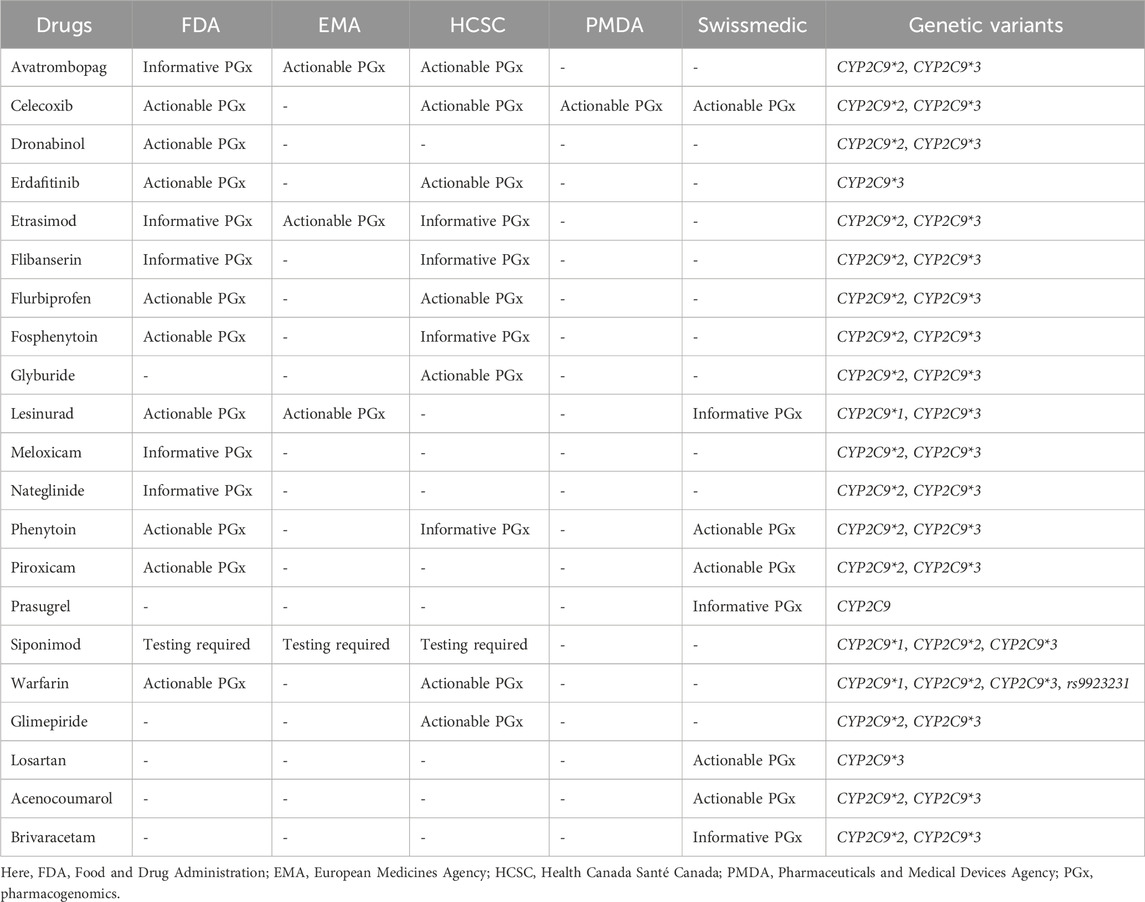
According to clinical annotations of the PharmGKB, the safety or efficacy of at least 29 commonly prescribed clinical drugs is impacted by the genetic polymorphism of CYP2C9 in varying degrees, as depicted through the evidence levels ranging from Level 4 (preliminary) to Level 1A (Pharmgkb, 2025a). In total, 11 of them, that is, warfarin, celecoxib, flurbiprofen, fluvastatin, ibuprofen, lornoxicam, meloxicam, phenytoin, piroxicam, siponimod, and tenoxicam, were said to have strong evidence (Level 1A). One drug (acenocoumarol) had Level 1B evidence; at least 14 drugs had a low level of evidence (Level 3) and at least three drugs had Level 4 (unsupported) evidence for interfering with the safety or effectiveness due to the presence of a CYP2C9*2 or *3 allele. In contrast, VKORC1 c.-1639G>A was associated with the safety of at least three drugs, that is, warfarin, acenocoumarol, and phenprocoumon, all having Level 1A evidence, as described in clinical annotations of the PharmGKB (Pharmgkb, 2025b).
PGx label information from the FDA, EMA, HCSC, Pharmaceuticals and Medical Devices Agency (PMDA), and Swissmedic for at least 23 drugs mentioned the genetic variability of CYP2C9. The FDA’s table of Pharmacogenomic Biomarkers in Drug Label was the source for most of the PGx label information. However, the PGx label information of at least five drugs, that is, avatrombopag, etrasimod, prasugrel, rimegepant, and Siponimod, was identified from the FDA, EMA, and HCSC. The label emphasizes the requirement of PGx testing for siponimod alongside the label of “Actionable PGx” for 15 drugs, stating the need for dose adjustment or alternative therapy for individuals with certain problematic genotypes or risk phenotypes if known, although it does not recommend genotype testing prior to the use of these drugs (Pharmgkb, 2025). On the other hand, “No Clinical PGx” offers specific genetic information and its impact on drug concentration, metabolism, and side effects, but no specific clinical guideline is provided. Three drugs, that is, abrocitinib, rimegepant, and prasugrel, were identified as being in the “No Clinical PGx” category due to CYP2C9 variants. As for VKORC1 c.-1639G>A, warfarin is the only drug with drug-level annotation. Both the FDA and HCSC deemed the association as actionable PGx. Detailed lists of the drugs can be found in Table 3.
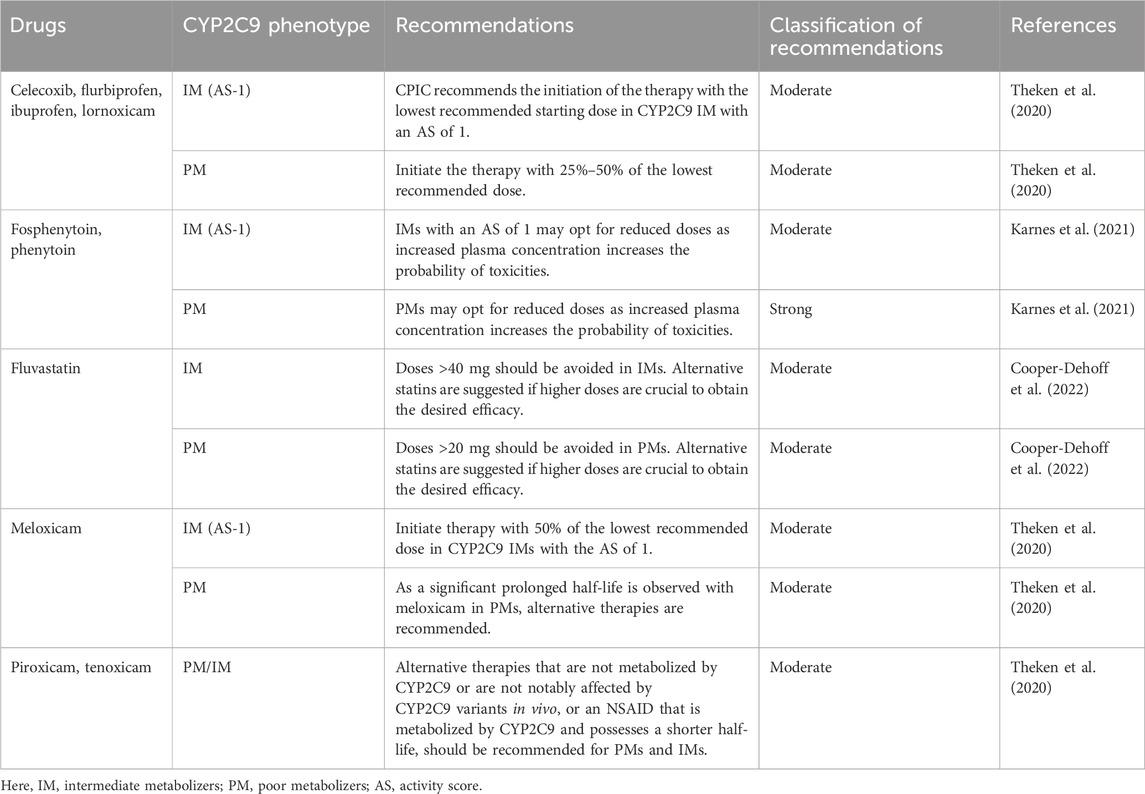
To date, the CPIC has provided PGx-based dosing guidelines for 11 drugs based on CYP2C9 variability (Cooper-Dehoff et al., 2022; Johnson et al., 2017; Karnes et al., 2021; Theken et al., 2020). In contrast, for warfarin, a detailed guideline based on the ancestry and the presence of genetic variants in the CYP2C9, VKORC1, and CYP4F2 genes, as well as rs12777823, has been recommended by the CPIC (Johnson et al., 2017). The guidelines have been summarized in Tables 4, 5 which strongly advise the careful handling of the IM and PM phenotypes for the optimization of the safety or effectiveness of many clinically significant drugs. For example, alternative therapies that are not impacted by the genetic variability of CYP2C9, are not metabolized by CYP2C9, or are metabolized by CYP2C9 but have a short half-life should be considered instead of piroxicam and tenoxicam, as the phenotypes correspond to CYP2C9 IM and PM.
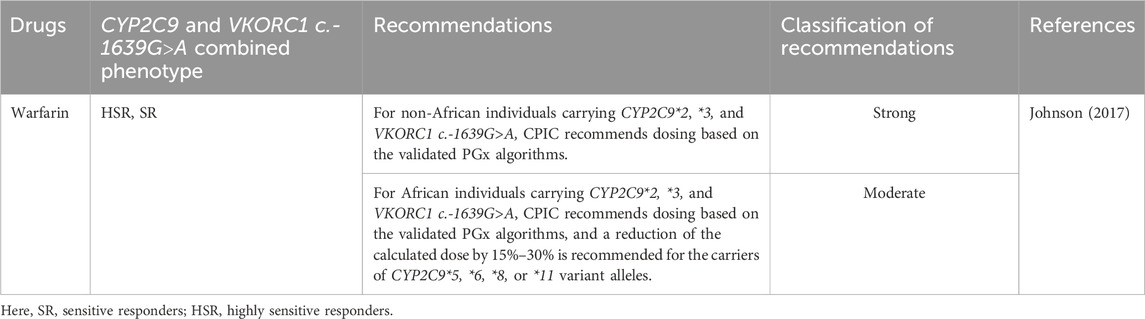
Table 5. The CPIC dosing guidelines for warfarin based on the CYP2C9 and VKORC1 c.-1639G>A genetic polymorphisms.
Discussion
We found that almost one-fifth (17.8%; 95% CI 16.3%–19.3%) of the participants of the 1000 Genomes Project are at an increased risk of drug-related toxicities of several clinically significant drugs that are affected by the genetic variability of CYP2C9. Similarly, the combination of CYP2C9 and VKORC1 c.-1639G>A genetic polymorphisms imposes risk on 33.1% (95% CI 31.3%–35%) of all the individuals who participated in the 1000 Genomes Project if they are taking warfarin therapy. This study also observed that South Asian and European populations are particularly at high risk for CYP2C9 variability, and East Asian and European populations are the most affected by the combined genotype of CYP2C9 and VKORC1 c.-1639G>A. The insightful findings can be viewed as new information, and further studies may be conducted, keeping these populations in mind, to design therapies to optimize the safety of the drugs of concern.
The global prevalence data observed in the current study for CYP2C9*2 and *3 from 26 different ethnic groups conforming to five geographical populations were in line with the previously published literature and provide supporting evidence for them (Céspedes-Garro et al., 2015; Sangkuhl et al., 2021; Sistonen et al., 2009). Likewise, the frequency of VKORC1 c.-1639G>A variants among different world populations also complied with the frequency found in several studies focusing on specific populations (Dandara et al., 2011; Buzoianu et al., 2012; Gaikwad et al., 2014; Meckley et al., 2008).
Overall, the estimated risk profile reported provides a strong base for the initial assessment of the global population. However, genetic data from the Middle East and North Africa (MENA) region and the Oceania continents are absent in the 1000 Genomes Project, and this may impact the overall estimation of the global risk population. In addition, phenoconversion (the mismatch between the genotype-oriented drug metabolism prediction and the true individual phenotype due to the comorbidity, comedication, or other non-genetic factors) should also be considered. Phenoconversion may affect genotypes in different ways (Klomp et al., 2020). For example, investigation of concomitant treatment with a CYP2C9 substrate (flurbiprofen) and a CYP2C9 inhibitor (fluconazole) revealed that with a 200 mg dose of fluconazole, CYP2C9 NMs are converted to IMs, while the IMs convert to PMs. With the 400 mg dose of fluconazole, both NMs and IMs convert to PMs (Kumar et al., 2008). Because these factors may limit the generalizability of the result presented, more specific and large-scale assessments are required to reach a comprehensive understanding of the risk population.
Several international PGx working groups are providing clinical guidelines to translate PGx data into clinical practice. By compiling the approved PGx information together and categorizing it into various evidence levels, the PharmGKB is now emerging as a significant source of PGx information. For example, the PharmGKB offered varying degrees of evidence of clinical association for at least 29 drugs for CYP2C9*2 and *3 and at least three drugs for VKORC1 c.-1639G>A variants. These associations are then translated into clinical practice as “Testing Required,” “Testing Recommended,” “Actionable PGx,” “Informative PGx,” “No Clinical PGx,” and “Criteria Not Met.” The FDA is at the forefront in offering PGx information compared to other global regulatory authorities. Around three-fourths of the PGx information of CYP2C9 (73.9%) was provided by the FDA alone. However, this information also existed in different combinations from other PGx working bodies like the EMA, HCSC, PMDA, and Swissmedic.
The CPIC has thus far provided dosing guidelines for at least 10 drugs based on the genetic polymorphism of CYP2C9, particularly considering the presence of CYP2C9*2 and CYP2C9*3. The CPIC also provided a detailed dosing guideline for warfarin, taking the combination of CYP2C9 and VKORC1 c.-1639G>A into account. Adhering to the consensus standardization for characterizing phenotypes based on carriers of certain genotypes, as detailed in the CPIC, this study is uniquely constructed to identify phenotypes based on the CPIC dosing guidelines. No study thus far has explored the prevalence of the variants of CYP2C9 and VKORC1 c.-1639G>A together on a global scale to categorize them in predictive phenotype groups, thereby associating them with the safety of several clinical drugs and available dosing guidelines.
The findings of the frequency of predictive risk phenotype for CYP2C9 variants (17.8%; 95% CI 16.3%–19.3%) and the combination of CYP2C9 and VKORC1 c.-1639G>A variants (33.1%; 95% CI 31.3%–35%) among 26 different world populations represented by 2,504 participants of the 1000 Genomes Project thus give uniqueness to the study. This further reinforces the need for the implementation of PGx-based therapy for the drugs in concern among the risk phenotypes. For example, meloxicam, with a longer half-life (15–20 h), imposes the risk of increased exposure in impaired meloxicam metabolizers. Therefore, the CPIC recommended alternative therapy for CYP2C9 poor metabolizers and in IM with an AS of 1, initiating the therapy with 50% of the lowest recommended dose to avoid toxicities (Theken et al., 2020).
It should also be emphasized that the genetic variability of CYP2C9 may not always cause all of the pharmacokinetic variation. Alongside CYP2C9, other genetic factors may also contribute to the safety or effectiveness of the same drug. For example, fluvastatin is impacted by SLCO1B1 and CYP2C9 variants, phenytoin metabolism is affected by the variants of CYP2C19, CYP1A1, and EPHX1, and, on the other hand, the safety of warfarin therapy is dependent on the genetic variabilities of CYP2C9, VKORC1, CYP4F2, and rs12777823 (Cooper-Dehoff et al., 2022; Johnson et al., 2017; Karnes et al., 2021; Hung et al., 2012; Wilke et al., 2005). Therefore, developing pharmacogenomic polygenic response scores (PGxRS) can serve as a valuable source for predicting the occurrence of side effects associated with certain drugs, especially when multiple genetic polymorphisms are expected to alter the safety or effectiveness profile of the drug.
Although several international PGx working bodies are working to translate the PGx information into clinical practice, insufficient evidence and incomplete correlation of genetic variation and phenotypic outcomes remain the most significant setbacks for the widespread application of PGx-driven precision medicine into clinical practice. These authorities continue to provide updates on PGx-based dosing; however, the application is still limited. The present study aimed to bridge the existing knowledge gap by providing robust scientific evidence for the authorities and stakeholders to make more informed decisions, with an emphasis on PGx integration for the drugs with a high association with genetic variabilities to optimize the safety of certain drugs, as reported in this study.
Limitations
Because it is a predictive analysis, variation in the prevalence of the estimated risk phenotype may arise because the CYP2C9 and VKORC1 enzyme function was not directly measured; rather, various combinations of genotype were employed. The absence of genetic data from the Middle East and North Africa (MENA) region and the Oceania continents in the 1000 Genomes Project limited the global population coverage of the study.
Future directions
As the present study established the association of the safety of at least 11 drugs with CYP2C9*2, *3, and VKORC1 c.-1639G>A genetic polymorphisms, future studies should emphasize large-scale longitudinal PGx-based therapy to assess toxicities in real-world clinical settings. In addition, the drugs categorized into different evidence levels on the PharmGKB should be further explored to gather sufficient evidence for conclusive decisions.
Conclusion
For optimizing the safety of at least 11 clinically significant drugs, the present study identified that approximately one-fifth and one-third of the global population are at risk based on the CYP2C9 and the combination of CYP2C9 and VKORC1 c.-1639G>A genotypes, respectively. Further large-scale longitudinal studies on different ethnicities are warranted to evaluate the clinical benefits of integrating PGx-based therapies in routine practice and achieve precision medicine.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
For the current study, we gathered all the presented human genetic data from Phase III of the 1000 Genomes Project that adheres to Fort Lauderdale principles, that dictates for publishing any result utilizing data from 1000 Genomes Project requires no additional human ethics approval as it has been previously published in another study.
Author contributions
MB: Writing – original draft, Formal Analysis, Methodology, Data curation, Visualization, Investigation, Writing – review and editing, Conceptualization, Validation, Supervision. MM: Formal Analysis, Writing – original draft, Investigation. ME: Writing – review and editing. MK: Writing – review and editing. MS: Writing – review and editing. BI: Writing – review and editing. CS: Conceptualization, Funding acquisition, Visualization, Supervision, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, S., Zhou, Z., Zhou, J., and Chen, S. Q. (2016). Pharmacogenomics of drug metabolizing enzymes and transporters: relevance to precision medicine. Genomics Proteomics Bioinforma. 14, 298–313. doi:10.1016/j.gpb.2016.03.008
Altawil, Y., and Youssef, L. A. (2023). Frequencies of VKORC1-1639G>A and rs397509427 in patients on warfarin and healthy Syrian subjects. Cardiovasc Ther. 2023, 8898922. doi:10.1155/2023/8898922
Ashley, E. A. (2016). Towards precision medicine. Nat. Rev. Genet. 17, 507–522. doi:10.1038/nrg.2016.86
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi:10.1038/nature15393
Babaev, D., Kutumova, E., and Kolpakov, F. (2025). Mathematical modeling of pharmacokinetics and pharmacodynamics of losartan in relation to CYP2C9 allele variants. Front. Syst. Biol. 5, 1504077. doi:10.3389/fsysb.2025.1504077
Barbarino, J. M., Whirl-Carrillo, M., Altman, R. B., and Klein, T. E. (2018). PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 10, e1417. doi:10.1002/wsbm.1417
Bazan, N. S., Sabry, N. A., Rizk, A., Mokhtar, S., and Badary, O. A. (2014). Factors affecting warfarin dose requirements and quality of anticoagulation in adult Egyptian patients: role of gene polymorphism. Ir. J. Med. Sci. 183, 161–172. doi:10.1007/s11845-013-0978-y
Bedewy, A. M. L., Sheweita, S. A., Mostafa, M. H., and Kandil, L. S. (2018). The influence of CYP2C9 and VKORC1 gene polymorphisms on the response to warfarin in Egyptians. Indian J. Hematol. Blood Transfus. 34, 328–336. doi:10.1007/s12288-016-0725-4
Biswas, M. (2021). Global distribution of CYP2C19 risk phenotypes affecting safety and effectiveness of medications. Pharmacogenomics J. 21, 190–199. doi:10.1038/s41397-020-00196-3
Biswas, M., Bendkhale, S. R., Deshpande, S. P., Thaker, S. J., Kulkarni, D. V., Bhatia, S. J., et al. (2018). Association between genetic polymorphisms of CYP2C9 and VKORC1 and safety and efficacy of warfarin: results of a 5 years audit. Indian Heart J. 70 (Suppl. 3), S13-S19–s19. doi:10.1016/j.ihj.2018.02.005
Buzoianu, A. D., Trifa, A. P., Mureşanu, D. F., and Crişan, S. (2012). Analysis of CYP2C9*2, CYP2C9*3 and VKORC1 -1639 G>A polymorphisms in a population from South-Eastern Europe. J. Cell Mol. Med. 16, 2919–2924. doi:10.1111/j.1582-4934.2012.01606.x
Céspedes-Garro, C., Fricke-Galindo, I., Naranjo, M. E., Rodrigues-Soares, F., Fariñas, H., De Andrés, F., et al. (2015). Worldwide interethnic variability and geographical distribution of CYP2C9 genotypes and phenotypes. Expert Opin. Drug Metab. Toxicol. 11, 1893–1905. doi:10.1517/17425255.2015.1111871
Chiarella, P., Capone, P., and Sisto, R. (2023). Contribution of genetic polymorphisms in human health. Int. J. Environ. Res. Public Health 20, 912. doi:10.3390/ijerph20020912
Collins, F. S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795. doi:10.1056/NEJMp1500523
Cooper-Dehoff, R. M., Niemi, M., Ramsey, L. B., Luzum, J. A., Tarkiainen, E. K., Straka, R. J., et al. (2022). The Clinical pharmacogenetics implementation consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated musculoskeletal symptoms. Clin. Pharmacol. Ther. 111, 1007–1021. doi:10.1002/cpt.2557
Daly, A. K., Rettie, A. E., Fowler, D. M., and Miners, J. O. (2017). Pharmacogenomics of CYP2C9: functional and clinical considerations. J. Pers. Med. 8, 1. doi:10.3390/jpm8010001
Dandara, C., Lombard, Z., Du Plooy, I., Mclellan, T., Norris, S. A., and Ramsay, M. (2011). Genetic variants in CYP (-1A2, -2C9, -2C19, -3A4 and -3A5), VKORC1 and ABCB1 genes in a Black South African population: a window into diversity. Pharmacogenomics 12, 1663–1670. doi:10.2217/pgs.11.106
Dietz, N., Ruff, C., Giugliano, R. P., Mercuri, M. F., and Antman, E. M. (2020). Pharmacogenetic-guided and clinical warfarin dosing algorithm assessments with bleeding outcomes risk-stratified by genetic and covariate subgroups. Int. J. Cardiol. 317, 159–166. doi:10.1016/j.ijcard.2020.03.055
Eriksson, N., and Wadelius, M. (2012). Prediction of warfarin dose: why, when and how? Pharmacogenomics 13, 429–440. doi:10.2217/pgs.11.184
Eriksson, N., Wallentin, L., Berglund, L., Axelsson, T., Connolly, S., Eikelboom, J., et al. (2016). Genetic determinants of warfarin maintenance dose and time in therapeutic treatment range: a RE-LY genomics substudy. Pharmacogenomics 17, 1425–1439. doi:10.2217/pgs-2016-0061
Fahmi, A. M., Bardissy, A. E., Saad, M. O., Fares, A., Sadek, A., Elshafei, M. N., et al. (2024). Accuracy of an internationally validated genetic-guided warfarin dosing algorithm compared to a clinical algorithm in an Arab population. Curr. Probl. Cardiol. 49, 102865. doi:10.1016/j.cpcardiol.2024.102865
Ferder, N. S., Eby, C. S., Deych, E., Harris, J. K., Ridker, P. M., Milligan, P. E., et al. (2010). Ability of VKORC1 and CYP2C9 to predict therapeutic warfarin dose during the initial weeks of therapy. J. Thromb. Haemost. 8, 95–100. doi:10.1111/j.1538-7836.2009.03677.x
Gaikwad, T., Ghosh, K., Kulkarni, B., Kulkarni, V., Ross, C., and Shetty, S. (2013). Influence of CYP2C9 and VKORC1 gene polymorphisms on warfarin dosage, over anticoagulation and other adverse outcomes in Indian population. Eur. J. Pharmacol. 710, 80–84. doi:10.1016/j.ejphar.2013.04.006
Gaikwad, T., Ghosh, K., and Shetty, S. (2014). VKORC1 and CYP2C9 genotype distribution in Asian countries. Thromb. Res. 134, 537–544. doi:10.1016/j.thromres.2014.05.028
Ghafoor, M. B., Yasmeen, F., Khalid, A. W., Mustafa, G., Khaliq, S., and Mohsin, S. (2022). VKORC1 gene polymorphism (-1639G>A) in warfarin therapy patients of Pakistani population. J. Pak Med. Assoc. 72, 418–423. doi:10.47391/jpma.01042
Higashi, M. K., Veenstra, D. L., Kondo, L. M., Wittkowsky, A. K., Srinouanprachanh, S. L., Farin, F. M., et al. (2002). Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. Jama 287, 1690–1698. doi:10.1001/jama.287.13.1690
Hung, C. C., Huang, H. C., Gao, Y. H., Chang, W. L., Ho, J. L., Chiou, M. H., et al. (2012). Effects of polymorphisms in six candidate genes on phenytoin maintenance therapy in Han Chinese patients. Pharmacogenomics 13, 1339–1349. doi:10.2217/pgs.12.117
Johnson, J. A., Caudle, K. E., Gong, L., Whirl-Carrillo, M., Stein, C. M., Scott, S. A., et al. (2017). Clinical pharmacogenetics implementation consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404. doi:10.1002/cpt.668
Karnes, J. H., Rettie, A. E., Somogyi, A. A., Huddart, R., Fohner, A. E., Formea, C. M., et al. (2021). Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2C9 and HLA-B genotypes and phenytoin dosing: 2020 update. Clin. Pharmacol. Ther. 109, 302–309. doi:10.1002/cpt.2008
Klomp, S. D., Manson, M. L., Guchelaar, H. J., and Swen, J. J. (2020). Phenoconversion of cytochrome P450 metabolism: a systematic review. J. Clin. Med. 9, 2890. doi:10.3390/jcm9092890
Kumar, V., Brundage, R. C., Oetting, W. S., Leppik, I. E., and Tracy, T. S. (2008). Differential genotype dependent inhibition of CYP2C9 in humans. Drug Metab. Dispos. 36, 1242–1248. doi:10.1124/dmd.108.020396
Lee, C. R., Goldstein, J. A., and Pieper, J. A. (2002). Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics 12, 251–263. doi:10.1097/00008571-200204000-00010
Limdi, N. A., Mcgwin, G., Goldstein, J. A., Beasley, T. M., Arnett, D. K., Adler, B. K., et al. (2008). Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin. Pharmacol. Ther. 83, 312–321. doi:10.1038/sj.clpt.6100290
Liu, J., Wang, G., Qin, L., Wu, Y., Zou, Y., Wang, X., et al. (2021a). Incremental value of genotype bins over the HAS-BLED score for the prediction of bleeding risk in warfarin-treated patients with atrial fibrillation. Cardiol. Res. Pract. 2021, 9030005. doi:10.1155/2021/9030005
Liu, S., Li, S., Shen, G., Sukumar, N., Krezel, A. M., and Li, W. (2021b). Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science 371, eabc5667. doi:10.1126/science.abc5667
Lund, K., Gaffney, D., Spooner, R., Etherington, A. M., Tansey, P., and Tait, R. C. (2012). Polymorphisms in VKORC1 have more impact than CYP2C9 polymorphisms on early warfarin international normalized ratio control and bleeding rates. Br. J. Haematol. 158, 256–261. doi:10.1111/j.1365-2141.2012.09150.x
Ma, C., Zhang, Y., Xu, Q., Yang, J., Zhang, Y., Gao, L., et al. (2012). Influence of warfarin dose-associated genotypes on the risk of hemorrhagic complications in Chinese patients on warfarin. Int. J. Hematol. 96, 719–728. doi:10.1007/s12185-012-1205-8
Meckley, L. M., Wittkowsky, A. K., Rieder, M. J., Rettie, A. E., and Veenstra, D. L. (2008). An analysis of the relative effects of VKORC1 and CYP2C9 variants on anticoagulation related outcomes in warfarin-treated patients. Thromb. Haemost. 100, 229–239. doi:10.1160/th07-09-0552
Mega, J. L., Walker, J. R., Ruff, C. T., Vandell, A. G., Nordio, F., Deenadayalu, N., et al. (2015). Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet 385, 2280–2287. doi:10.1016/s0140-6736(14)61994-2
Nizamuddin, S., Dubey, S., Singh, S., Sharma, S., Machha, P., and Thangaraj, K. (2021). CYP2C9 variations and their pharmacogenetic implications among diverse South Asian populations. Pharmgenomics Pers. Med. 14, 135–147. doi:10.2147/pgpm.s272015
Panchenko, E., Kropacheva, E., Dobrovolsky, A., Titaeva, E., Zemlyanskaya, O., Trofimov, D., et al. (2020). CYP2C9 and VKORC1 genotyping for the quality of long-standing warfarin treatment in Russian patients. Pharmacogenomics J. 20, 687–694. doi:10.1038/s41397-020-0157-2
Pharmgkb (2025). Drug label information and legend. Available online at: https://www.pharmgkb.org/page/drugLabelLegend (Accessed February 21, 2025).
Pharmgkb (2025a). CYP2C9 clinical annotations. Available online at: https://www.pharmgkb.org/gene/PA126/clinicalAnnotation (Accessed February 21, 2025).
Pharmgkb (2025b). CYP2C9 clinical annotations. Available online at: https://www.pharmgkb.org/gene/PA133787052/clinicalAnnotation (Accessed February 21, 2025).
Pourgholi, L., Goodarzynejad, H., Mandegary, A., Ziaee, S., Talasaz, A. H., Jalali, A., et al. (2016). Gene polymorphisms and the risk of warfarin-induced bleeding complications at therapeutic international normalized ratio (INR). Toxicol. Appl. Pharmacol. 309, 37–43. doi:10.1016/j.taap.2016.08.026
Relling, M. V., and Klein, T. E. (2011). CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89, 464–467. doi:10.1038/clpt.2010.279
Sangkuhl, K., Claudio-Campos, K., Cavallari, L. H., Agundez, J. A. G., Whirl-Carrillo, M., Duconge, J., et al. (2021). PharmVar GeneFocus: CYP2C9. Clin. Pharmacol. Ther. 110, 662–676. doi:10.1002/cpt.2333
Shukla, A., Jain, A., Kahalekar, V., Bendkhale, S., Gogtay, N., Thatte, U., et al. (2019). Mutations in CYP2C9 and/or VKORC1 haplotype are associated with higher bleeding complications in patients with Budd-Chiari syndrome on warfarin. Hepatol. Int. 13, 214–221. doi:10.1007/s12072-018-9922-6
Sistonen, J., Fuselli, S., Palo, J. U., Chauhan, N., Padh, H., and Sajantila, A. (2009). Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 19, 170–179. doi:10.1097/FPC.0b013e32831ebb30
Sridharan, K., Al Banna, R., Malalla, Z., Husain, A., Sater, M., Jassim, G., et al. (2021). Influence of CYP2C9, VKORC1, and CYP4F2 polymorphisms on the pharmacodynamic parameters of warfarin: a cross-sectional study. Pharmacol. Rep. 73, 1405–1417. doi:10.1007/s43440-021-00256-w
Sudmant, P. H., Rausch, T., Gardner, E. J., Handsaker, R. E., Abyzov, A., Huddleston, J., et al. (2015). An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81. doi:10.1038/nature15394
Theken, K. N., Lee, C. R., Gong, L., Caudle, K. E., Formea, C. M., Gaedigk, A., et al. (2020). Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin. Pharmacol. Ther. 108, 191–200. doi:10.1002/cpt.1830
Tornio, A., and Backman, J. T. (2018). Cytochrome P450 in pharmacogenetics: an update. Adv. Pharmacol. 83, 3–32. doi:10.1016/bs.apha.2018.04.007
Vuorinen, A. L., Lehto, M., Niemi, M., Harno, K., Pajula, J., Van Gils, M., et al. (2021). Pharmacogenetics of anticoagulation and clinical events in warfarin-treated patients: a register-based cohort study with biobank data and national health registries in Finland. Clin. Epidemiol. 13, 183–195. doi:10.2147/clep.s289031
Wang, X., Zhao, D., Ma, J., Wang, X., and Liu, J. (2024). Correlation between metabolic parameters and warfarin dose in patients with heart valve replacement of different genotypes. Rev. Cardiovasc Med. 25, 128. doi:10.31083/j.rcm2504128
Wilke, R. A., Reif, D. M., and Moore, J. H. (2005). Combinatorial pharmacogenetics. Nat. Rev. Drug Discov. 4, 911–918. doi:10.1038/nrd1874
Yang, L., Ge, W., Yu, F., and Zhu, H. (2010). Impact of VKORC1 gene polymorphism on interindividual and interethnic warfarin dosage requirement--a systematic review and Meta analysis. Thromb. Res. 125, e159–e166. doi:10.1016/j.thromres.2009.10.017
Yang, T., Zhou, Y., Chen, C., Lu, M., Ma, L., and Cui, Y. (2019). Genotype-guided dosing versus conventional dosing of warfarin: a meta-analysis of 15 randomized controlled trials. J. Clin. Pharm. Ther. 44, 197–208. doi:10.1111/jcpt.12782
Yasar, U., Bennet, A. M., Eliasson, E., Lundgren, S., Wiman, B., De Faire, U., et al. (2003). Allelic variants of cytochromes P450 2C modify the risk for acute myocardial infarction. Pharmacogenetics 13, 715–720. doi:10.1097/00008571-200312000-00002
Yee, J., Kim, W., Chang, B. C., Chung, J. E., Lee, K. E., and Gwak, H. S. (2019). Effects of polymorphisms in myc-related genes on bleeding complications in patients with stable warfarin responses. Cardiovasc Ther. 2019, 1813747. doi:10.1155/2019/1813747
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141. doi:10.1016/j.pharmthera.2012.12.007
Zhang, H. F., Wang, H. H., Gao, N., Wei, J. Y., Tian, X., Zhao, Y., et al. (2016). Physiological content and intrinsic activities of 10 cytochrome P450 isoforms in human normal liver microsomes. J. Pharmacol. Exp. Ther. 358, 83–93. doi:10.1124/jpet.116.233635
Zhou, Y., Ingelman-Sundberg, M., and Lauschke, V. M. (2017). Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 102, 688–700. doi:10.1002/cpt.690
Keywords: genetic polymorphisms, CYP2C9, VKORC1, safety, efficacy, pharmacogenomics, precision medicine, clinical practice
Citation: Biswas M, Murad MA, Ershadian M, Kali MSK, Shin Sim M, Ibrahim B and Sukasem C (2025) Prevalence of risk phenotypes associated with CYP2C9*2, *3, and VKORC1 c.-1639G>A genetic polymorphisms in world populations: implications in clinical practice. Front. Pharmacol. 16:1597379. doi: 10.3389/fphar.2025.1597379
Received: 21 March 2025; Accepted: 05 August 2025;
Published: 19 September 2025.
Edited by:
Rossana Roncato, University of Udine, ItalyReviewed by:
Varun Sharma, NMC Healthcare (NMC Genetics), IndiaNela Maksimovic, University of Belgrade, Serbia
Copyright © 2025 Biswas, Murad, Ershadian, Kali, Shin Sim, Ibrahim and Sukasem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chonlaphat Sukasem, Y2hvbmxhcGhhdC5zdWtAbWFoaWRvbC5hYy50aA==
†ORCID: Mohitosh Biswas, https://orcid.org/0000-0003-1432-7701; Chonlaphat Sukasem, https://orcid.org/0000-0003-0033-5321
‡These authors have contributed equally to this work
 Mohitosh Biswas
Mohitosh Biswas Murshadul Alam Murad
Murshadul Alam Murad Maliheh Ershadian
Maliheh Ershadian Most. Sumaiya Khatun Kali
Most. Sumaiya Khatun Kali Maw Shin Sim
Maw Shin Sim Baharudin Ibrahim6
Baharudin Ibrahim6 Chonlaphat Sukasem
Chonlaphat Sukasem