- Universidad UTE, Facultad de Ciencias de la Salud Eugenio Espejo, Centro de Investigación Genética y Genómica, Quito, Ecuador
Statins have been primarily used for the management of low-density lipoprotein cholesterol and cardiovascular diseases However, in recent years, research has identified potential applications beyond cholesterol regulation. Statins exhibit pleiotropic effects, due to their ability to modulate gene expression via epigenetic mechanisms, including DNA methylation, histone acetylation, and microRNA regulation. Clinical studies have correlated these epigenetic changes with various pathological conditions, such as inflammation, atherosclerosis, cancer, diabetes, and autoimmune disorders. Despite encouraging findings, further research is required to fully understand the molecular pathways associated with the epigenetic actions of statins and disease pathogenesis. This review describes the potential role of statins as epigenetic modulators and their relevance in human disease management.
Introduction
Statins are the primary pharmacological approach for reducing elevated levels of low-density lipoprotein (LDL) cholesterol (Guadamuz et al., 2022). Their clinical importance is highlighted by their inclusion in the World Health Organization (WHO) Model List of Essential Medicines (EML) for the management of cardiovascular diseases (CVD) (Kishore et al., 2018).
The therapeutic effect of statins involves the inhibition of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase, a key regulatory enzyme in the cholesterol synthesis pathway. This enzyme catalyzes the conversion of HMG-CoA into L-mevalonate, a crucial precursor in endogenous cholesterol production (Sizar et al., 2024; Zaky et al., 2023). As a result of this, there is an increase in the upregulation of LDL receptors on cell surfaces, enhancing the uptake of circulating LDL cholesterol (Zaky et al., 2023). Given the strong association between elevated LDL cholesterol and CVDs (Zambrano et al., 2023; Jung et al., 2022), statins are widely recommended for the management and prevention of these diseases (Guadamuz et al., 2022).
Beyond the lipid-lowering properties of statins, they exert a range of pleiotropic effects by modulating the mevalonate pathway, thereby influencing various cellular processes (Kim et al., 2019). One key effect is their anti-inflammatory potential, which acts by reducing LDL cholesterol, an established contributor to systemic inflammation, statins indirectly reduce inflammatory responses (Yan et al., 2024). Additionally, statins can directly interfere with the production of pro-inflammatory cytokines such as interferon-gamma and tumor necrosis factor-alpha (TNF-α), thereby reducing immune system activation and inflammation. Statins have also been shown to reduce of C-reactive protein levels in human hepatocytes, further supporting its anti-inflammatory properties and suggesting liver-specific interactions (Kim et al., 2019).
Emerging evidence has also correlated statins with epigenetic modulation, including changes in DNA methylation, histone acetylation, and microRNA expression (Awosika et al., 2023). For instance, statins have been shown to inhibit the expression of histone deacetylases (HDAC) while enhancing histones H3 and H4 acetylation, promoting a transcriptionally active chromatin state (Awosika et al., 2023; Karlic et al., 2015; Feig et al., 2011; Singh et al., 2016; Tikoo et al., 2015). In addition, they may influence gene regulation by enhancing the expression of DNA methyltransferases (DNMTs) at promoter regions (Awosika et al., 2023; Karlic et al., 2015; Kodach et al., 2011).
The present review aims to describe the intricate interaction between statins and epigenetic mechanisms, emphasizing their broader implications in various disorders beyond cardiovascular disease.
Epigenetics and statins: molecular mechanisms
Statins act as epigenetic regulators through four main mechanisms: DNA methylation, histone modifications, microRNA expression, and long non-coding RNA regulation (Figure 1). These mechanisms contribute to their therapeutic impact in cardiovascular, metabolic, and inflammatory diseases.
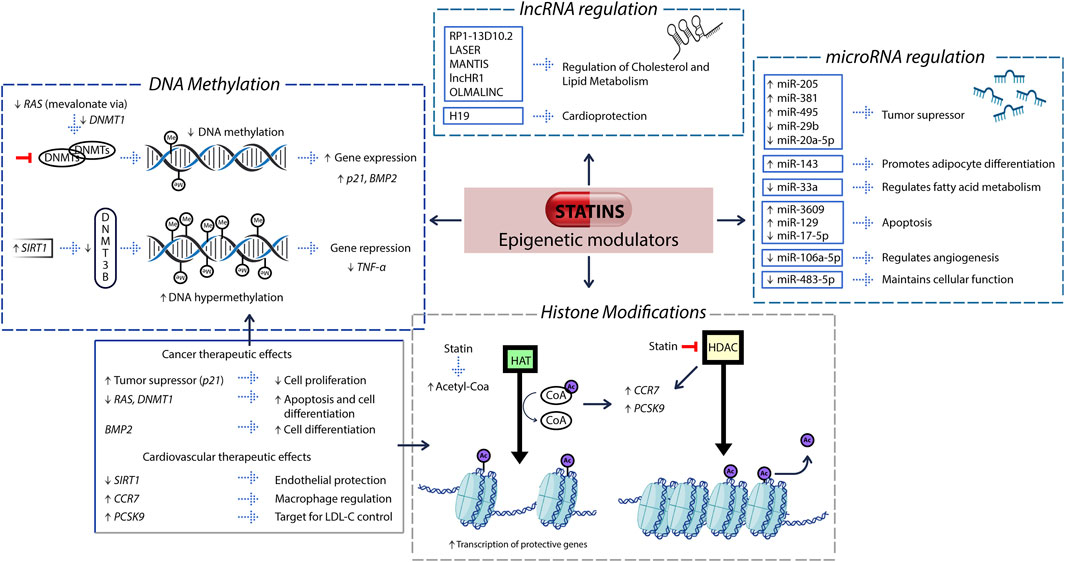
Figure 1. Epigenetic pathways modulated by statins and their functional implications. Statins modulate gene expression through four epigenetic mechanisms: (i) DNA methylation, (ii) histone modifications, (iii) microRNA regulation, and (iv) long non-coding RNA (lncRNA) regulation. (i) By inhibiting the mevalonate–RAS signaling pathway, statins downregulate DNA methyltransferase 1 (DNMT1), resulting in hypomethylation of promoter regions in tumor suppressor genes such as p21 and BMP2. Concurrently, they upregulate SIRT1, which enhances the recruitment of DNMT3B, leading to hypermethylation and transcriptional silencing of pro-inflammatory genes such as TNF-α. These DNA methylation changes are represented in the figure as gray spheres labeled “Me.” (ii) Statins inhibit the activity of histone deacetylases (HDACs), increasing the intracellular availability of acetyl-CoA, a key substrate for histone acetyltransferases (HATs). HATs catalyze the addition of acetyl groups to histone tails, shown in the figure as purple spheres labeled “Ac,” resulting in chromatin relaxation and enhanced accessibility for transcription factors. This mechanism promotes the expression of genes such as PCSK9 and CCR7, involved in cholesterol metabolism and immune cell trafficking, respectively. (iii) Statins modulate the expression of specific microRNAs, which regulate diverse cellular processes such as tumor suppression, lipid metabolism, apoptosis, and angiogenesis. (iv) Statins also influence the expression of long non-coding RNAs such as LASER, MANTIS, and H19, which play key roles in lipid homeostasis, vascular function, and cardiometabolic protection. These interconnected epigenetic pathways underline the multifaceted therapeutic potential of statins across oncology and cardiovascular health.
Statins and DNA methylation
Statins have been implicated in the regulation of epigenetic mechanisms, particularly by inhibiting DNMTs, which leads to reduced DNA methylation at gene promoter regions and subsequent activation of gene expression (Kayzuka et al., 2025). Statins can inhibit DNMT1 through two primary pathways. First, by blocking the mevalonate pathway, statins inhibit the isoprenylation of GTP-binding proteins, leading to the suppression of downstream signaling and reducing DNMT1 expression. Second, statins lower the production of interleukin-6 (IL-6), thereby interfering with the IL-6/JAK2/STAT3 signaling pathway, an established inducer of DNMT1 expression (Dongoran et al., 2020).
For instance, statins have been shown to downregulate the RAS/PI3K/mTOR signaling cascade by the inhibiting the mevalonate pathway through DNA demethylation and the downregulation of the histone deacetylase HDAC2. This process begins with the inhibition of GTPase isoprenylation, leading to the reduced activity of RAS proteins and the MAPK pathway (Karlic et al., 2015). Another study found that statins were associated with the downregulation of DNMT 1, which may contribute to the overexpression of the cyclin-dependent kinase inhibitor p21, possibly reversing aberrant promoter p21 hypermethylation. However, this hypothesis has not been solved and remains to be conclusively demonstrated (Kodach et al., 2011; Dongoran et al., 2020).
In the context of CVDs, statins have also been associated with the upregulation of endothelial nitric oxide synthase (eNOS), which promotes vasodilatation, prevents thrombosis, and improves endothelial cell function in patients with hypertension and atherosclerosis (Chen et al., 2024). This effect may be partially mediated through epigenetic modulation.
Notably, statins have also been reported to modulate DNA hypermethylation in specific contexts. For instance, they can promote the overexpression of sirtuin 1 (SIRT1), which recruits DNMT 3B to CpG islands, leading to transcription repression of target genes (Zhang and Kraus, 2010; Allen and Mamotte, 2017). Furthermore, under high simvastatin doses, reduced acetylation of NF-κB has been observed, which suppresses its transcriptional activity and downregulates expression of pro-inflammatory genes such as TNF-α (Du et al., 2014).
Taken together, these findings highlight the regulatory capacity of statins on DNA methylation depending on the cellular context and target pathways. Further research on DNMT regulation by statins could provide further comprehension into novel epigenetic therapies aimed at modulating gene expression in pathological processes such as cancer or CVDs.
Statins and histone modifications
Statins have been also involved in the regulation of epigenetic histone modifications, contributing to their broad spectrum of biological effects (Allen and Mamotte, 2017). Various studies have described that statins can influence gene expression through increased histone acetylation of histones H3 (Tikoo et al., 2015) and H4 (Singh et al., 2016). One proposed mechanism involves the inhibition of the mevalonate pathway, which leads to the intracellular accumulation of acetyl-CoA. This excess of acetyl-CoA may serve as a substrate for histone acetyltransferases, enhancing acetylation of the gene promoter regions and thereby promoting transcriptional activation (Allen and Mamotte, 2017; Cooney, 2010).
In addition to increasing acetyl-CoA availability, statins may also directly inhibit HDACs by binding to their active sites, suppressing their deacetylase activity (Singh et al., 2016; Kayzuka et al., 2025; Lin et al., 2008). This inhibition promotes histone acetylation, which neutralizes the positive charge on histones, allowing the loosening of chromatin structure. As a result, DNA becomes more accessible for the binding of transcription factors to promoter regions (Kayzuka et al., 2025; Allen and Mamotte, 2017; Bannister and Kouzarides, 2011).
Furthermore, statins have been correlated with the inhibition of histone methyltransferases (HMTs), potentially leading to the hypomethylation of histones and enhanced transcriptional activity (Kayzuka et al., 2025). These combined effects on histone acetylation and methylation suggest a significant role for statins in chromatin remodeling and gene regulation. Further molecular studies exploring how different types of statins influence these epigenetic processes could elucidate the underlying mechanisms of their protective roles in chronic diseases.
Several specific pathways have been described to illustrate these mechanisms. For example, statins downregulate the histone methyltransferase enhancer of zeste homolog 2 (EZH2), which in turn promotes the upregulation of HDAC5 and overexpression of the cyclin-dependent kinase inhibitor p27KIP1 (Ishikawa et al., 2014). Statins also downregulate HDAC activity, leading to increased histone-H3 acetylation at Sp1 binding sites within the p21 promoter (Lin et al., 2008). Moreover, inhibition of geranylgeranyl pyrophosphate (GGPP) synthesis by statins has been linked to the overexpression of p21, reinforcing their role in cell cycle regulation (Fuchs et al., 2008). Additionally, statins inhibit the GGTase–RhoA–YAP–SOX9 signaling axis, contributing to chromatin remodeling and further supporting their involvement in epigenetic regulation (Chen et al., 2024; Liu et al., 2023).
Statins and microRNA regulation
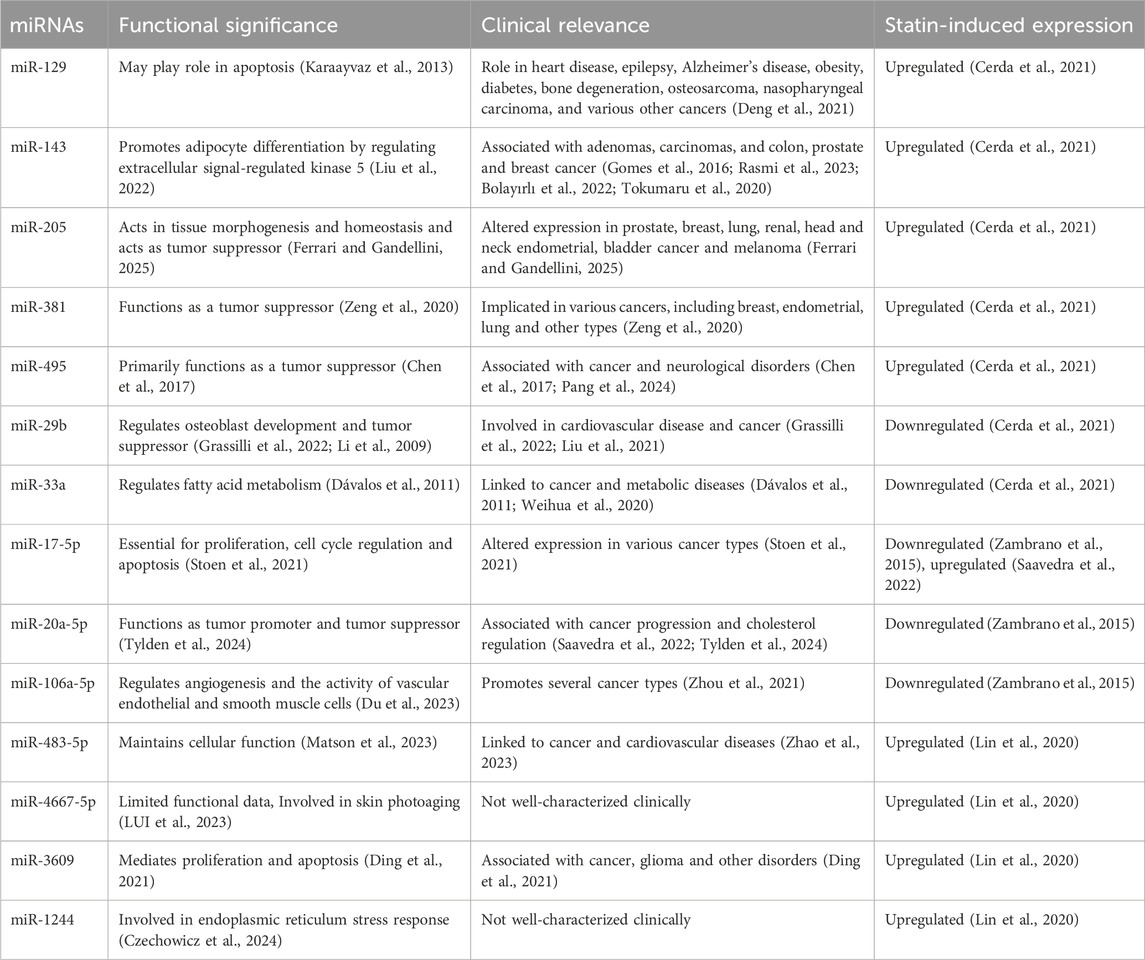
MicroRNAs (miRNAs) are small non-coding RNAs, typically 18–25 nucleotides in length, that regulate gene expression at the post-transcriptional level (Yao et al., 2019). miRNAs can act as epigenetic modulators by targeting enzymes involved in chromatin remodeling and epigenetic modifications, such as DNMTs, HDACs, and HMTs (Yao et al., 2019). Conversely, miRNA expression is itself subject to regulation by epigenetic mechanisms. DNA methylation and histone modifications can modulate the expression of miRNAs, indicating a complex bidirectional interaction between miRNAs and epigenetic processes (Yao et al., 2019). Table 1 describes the miRNAs associated with statins role in functional significance and clinical relevance in various human diseases.
Statins and lncRNA regulation
Long non-coding RNAs (lncRNAs) are transcripts of approximately 200 nucleotides that do not encode proteins. Despite being non protein-coding regions, lncRNAs play diverse and essential biological roles, including participation in chromosomal organization, telomere maintenance, and the structural organization of subcellular compartments. Notably, lncRNAs can also mediate epigenetic regulation by modulating chromatin structure, transcription, and post-transcriptional processes (Mercer et al., 2009).
Recent studies suggest that, in addition to their effect on miRNAs, statins also regulate lncRNAs, contributing to their pleiotropic actions (Tsilimigras et al., 2021). For example, the lncRNA RP1-13D10.2, has been shown to regulate LDLR expression and modulate the individual response to statin therapy (Mitchel et al., 2016). Similarly, another study identified LASER, a lncRNA involved in cholesterol homeostasis, may serve as a therapeutic target to enhance statins efficacy (Li et al., 2019). The lncRNA MANTIS has also been associated with statin-mediated vascular protection (Leisegang et al., 2019). Additionally, the lncRNA H19 has been implicated in the statin-mediated therapeutic response in patients with acute myocardial infarction (Huang et al., 2020).
In the context of atherosclerosis, statin were found to regulate pyroptosis-associated lncRNAs such as NEX-AS1 and NEXN, exerting protective effects that are independent of lipid-lowering activity (Wu et al., 2020). Furthermore OLMALINC, an oligodendrocyte maturation-associated long intergenic noncoding RNA, has been linked to the epigenetic regulation of genes involved in cholesterol biosynthesis, such as stearoyl-coenzyme A desaturase and shows strong associations with both statins use and serum triglycerides levels (Benhammou et al., 2019).
A more recent study reported that RP1-13D10.2, MANTIS, and lncHR1 were overexpressed in individuals with hypercholesterolemia, and that atorvastatin treatment significantly suppressed lncHR1 expression (Paez et al., 2023).
Collectively, these findings underscore the important role of lncRNAs in the epigenetic regulation mediated by statins. The identification of statin-responsive lncRNAs opens new avenues for personalized medicine and suggests novel molecular targets for improving the therapeutic efficacy of statins across a range of lipid-related and inflammatory diseases.
Clinical evidence on statins regulating epigenetic modifications
Cardiovascular diseases and atherosclerosis
Clinical evidence supports the protective role of statins in reducing cardiovascular risk, extending beyond their lipid-lowering effects. Emerging research highlights that epigenetic mechanisms contribute significantly to the pleiotropic benefits of statins. These compounds influence gene expression through modulation of DNA methylation, histone post-translational modifications, and non-coding RNAs, particularly in vascular endothelial cells (Kayzuka et al., 2025).
For instance, simvastatin has been shown to suppress the epigenetic activation of the YAP-SOX9 axis, thereby inhibiting endothelial-to-mesenchymal transition (EndMT)—a process implicated in vascular dysfunction and atherosclerosis progression (Liu et al., 2023). Similarly, atorvastatin has been reported to upregulate SIRT1 expression at both the transcriptional and protein levels in patients with coronary artery disease, linking statin therapy to pathways associated with endothelial protection and cellular longevity (Tabuchi et al., 2012).
In experimental models of atherosclerosis, rosuvastatin enhances histone H3 and H4 acetylation by inhibiting HDAC6 and HDAC7, leading to increased expression of CCR7, a chemokine receptor involved in macrophage migration and plaque remodeling (Feig et al., 2011). This effect is mediated through SREBP-2-dependent displacement of HDAC6/7 from the CCR7 promoter, allowing recruitment of histone acetyltransferases (HATs) such as p300, therey promoting transcriptional activation via histone acetylation (Feig et al., 2011).
Statins also epigenetically regulate PCSK9, a key gene in cholesterol homeostasis and fatty acid metabolism. They increase PCSK9 expression through SREBP2 activation, which recruits cofactors like NPAT and TRRAP to facilitate histone H4 acetylation (Dong et al., 2010; Li and Liu, 2012). This chromatin remodeling enables the recruitment of HATs such as p300 and CBP, promoting active transcription via H3K9 acetylation and H3K4 trimethylation (Duddu et al., 2025). Although this upregulation of PCSK9 may reduce the lipid-lowering efficacy of statins, it uncovers a precise epigenetic mechanism that could be pharmacologically targeted.
In addition to chromatin remodeling, statins modulate miRNA expression, contributing to both lipid regulation and inflammation control (Ruiz-Pozo et al., 2023). In HepG2 cells, atorvastatin upregulates miR-129, miR-143, miR-205, miR-381, and miR-495, while downregulating miR-29b and miR-33a—miRNAs involved in lipogenesis and lipid metabolism (Cerda et al., 2021). Other studies have reported a decrease in hsa-miR-17-5p, hsa-miR-20a-5p, and hsa-miR-106a-5p with atorvastatin treatment (Zambrano et al., 2015), although contrasting findings suggest that miR-17-5p may also be upregulated and associated with LDLR suppression (Saavedra et al., 2022).
Furthermore statin treatment has been associated with the upregulation of miR-483-5p, miR-4667-5p, miR-3609, and miR-1244, all of which are implicated in the regulation of inflammatory responses. Notably, miR-483-5p may inhibit RhoA-mediated pathways, which are critical for monocyte migration and cytoskeletal dynamics. These miRNAs also appear to interact with the TGF-β signaling pathway, known for its dual role in immune modulation within atherosclerotic plaques (Lin et al., 2020).
Collectively, these findings underscore the role of statins as epidrugs—agents capable of modulating the epigenome—offering new avenues for therapeutic optimization in cardiovascular disease management.
Cancer
Although statins are primarily recognized for their cholesterol-lowering effects, increasing evidence supports their potential as anticancer agents, particularly through the modulation of epigenetic mechanisms involved in tumorigenesis. These effects include alterations in DNA methylation, histone modifications, and non-coding RNA expression, which collectively influence gene regulation, cell cycle progression, and tumor cell differentiation (Awosika et al., 2023; Kayzuka et al., 2025; Mohammadzadeh et al., 2020).
In oral squamous cell carcinoma (OSCC), cerivastatin and simvastatin have been shown to significantly suppress DNMT1, a key enzyme responsible for maintaining promoter hypermethylation of tumor suppressor genes. This suppression leads to reactivation of genes such as p21, resulting in G0/G1 cell cycle arrest and reduced tumor proliferation (Dongoran et al., 2020). Given the frequent overexpression of DNMT1 in various malignancies, these findings highlight a promising epigenetic mechanism for statin-mediated tumor suppression.
In a broader oncological context, simvastatin and ibandronate have been shown to modulate the mevalonate pathway in breast, prostate, and osteosarcoma cell lines. This inhibition reduces the isoprenylation of small GTPases like RAS, leading to downregulation of DNMT1, HDACs, and specific miRNAs. These epigenetic changes promote the demethylation and activation of pro-apoptotic and differentiation-related genes. Notably, simvastatin significantly upregulates miR-612, a miRNA associated with reduced tumor cell pluripotency and enhanced sensitivity to 5-fluorouracil, suggesting a potential suggesting a potential chemosensitizing role for statins (Karlic et al., 2015).
In colorectal cancer (CRC), statins such as simvastatin, fluvastatin, and atorvastatin exert epigenetic effects that are independent of the mevalonate pathway. These include inhibition of EZH2, a HMT that represses tumor suppressor genes. EZH2 inhibition leads to upregulation of p27KIP1, promoting cellular differentiation and improved patient survival. Furthermore, combining statins with class II HDAC inhibitors has been shown to enhance these anticancer effects synergistically (Ishikawa et al., 2014).
Another mechanism involves lovastatin, which promotes demethylation of the BMP2 gene, encouraging tumor differentiation and reducing aggressiveness. This protein is part of the Bone Morphogenetic Proteins (BMPs) family involved in intestinal epithelial cell differentiation, inhibition of stem cell activity, and maintenance of adult tissue homeostasis. DNMT inhibition facilitates BMP2 demethylation and upregulation in CRC cells, sensitizing tumors to chemotherapeutic agents. While additional studies are required to validate its efficacy and define its clinical application, these findings underscore the potential of statins as adjuvant epigenetic agents (Wang et al., 2014).
Despite these promising findings, some studies have reported inconsistent results, with no significant changes in histone acetylation and, in some cases, increased DNMT activity following statin treatment (Bridgeman et al., 2019). These discrepancies underscore the complexity of statin-epigenome interactions and the need for further mechanistic and translational research.
In summary, statins are emerging as multifunctional agents with potential applications in oncology, particularly as adjuvant modulators of the epigenome. Their ability to influence chromatin remodeling, gene expression, and non-coding RNA networks supports their integration into personalized cancer therapies, pending further validation in preclinical and clinical settings.
Diabetes and insulin resistance
Growing evidence suggests a paradoxical association between statin therapy and an increased risk of type 2 diabetes mellitus (T2DM), primarily through mechanisms that promote insulin resistance (Paseban et al., 2019). T2DM is a multifactorial disease influenced by genetic predisposition, environmental exposures, and pharmacological interventions (Beulens et al., 2021). Among these, epigenetic mechanisms have emerged as critical contributors to the pathogenesis of insulin resistance and impaired glucose metabolism (Awosika et al., 2023).
An important study involving approximately 4,760 participants from the Framingham Heart Study Offspring cohort (FHS) and the Women’s Health Initiative (WHI) identified a specific epigenetic marker associated with statin use. DNA methylation at CpG site cg06500161 within the ABCG1 gene was positively correlated with statin therapy, elevated fasting glucose, increased insulin levels, and a higher risk of T2DM (Qie et al., 2021). Given ABCG1’s dual role in cholesterol efflux and glucose homeostasis, this finding underscores the gene’s central role in the metabolic interplay between lipid and glucose regulation (Kotlyarov and Kotlyarova, 2025; Liu et al., 2020).
Further supporting this, a comparative epigenome-wide association study in statin-treated versus non-treated T2DM patients identified 79 differentially methylated CpG sites, with three—cg17901584 (DHCR24), cg27243685 (ABCG1), and cg05119988 (SC4MOL)—showing strong associations with statin exposure (Schrader et al., 2021). While DHCR24 and SC4MOL are primarily involved in cholesterol biosynthesis, methylation at DHCR24 was also linked to glucose metabolism, suggesting a shared epigenetic axis between lipid and glycemic pathways (Peeples et al., 2024).
In addition to DNA methylation, miRNA dysregulation has also been implicated in statin-induced metabolic changes. For instance, rosuvastatin has been shown to deregulate miR-27a and miR-221, both of which are involved in insulin signaling and glucose uptake (Serik et al., 2021). Simvastatin dose-dependently increases miR-27a expression in hepatic cells, which indirectly reduces LDL receptor (LDLR) levels by upregulating PCSK9, a protein that promotes LDLR degradation. Under hyperglycemic conditions, this dysregulation may impair lipid clearance and exacerbate insulin resistance (Galicia-Garcia et al., 2020).
Moreover, miR-33a and miR-33b, which regulate ABCA1 and ABCG1, are reportedly overexpressed in statin users. These genes are essential for pancreatic beta-cell function, and their suppression may impair insulin secretion and glucose regulation (Awosika et al., 2023). Clinical studies have corroborated these molecular findings. A 10-week trial of high-intensity atorvastatin therapy demonstrated increased insulin resistance and compensatory insulin secretion, suggesting a shift toward glucose intolerance in susceptible individuals (Abbasi et al., 2021). A systematic review further confirmed that statin use is associated with reduced insulin sensitivity and increased insulin resistance, raising concerns for patients at risk of developing diabetes (Dabhi et al., 2023).
Collectively, these findings highlight the epigenetic complexity underlying statin-induced metabolic effects. Through DNA methylation and miRNA modulation, statins may inadvertently disrupt glucose homeostasis. These results emphasize the need for personalized risk assessment and epigenetic monitoring in patients undergoing long-term statin therapy.
Other diseases
Conversely, statins exhibit notable immunomodulatory and anti-inflammatory properties, making them promising candidates for the treatment of autoimmune diseases. Statins can regulate immune responses through both mevalonate pathway-dependent and -independent mechanisms, affecting antigen-presenting cells and T-cell functions (Dehnavi et al., 2020). Evidence has demonstrated improvements in conditions such as rheumatoid arthritis, lupus, and multiple sclerosis, including enhancements in cytokine profiles and clinical markers like C-reactive protein and erythrocyte sedimentation rate (ESR). However, the precise mechanisms and optimal doses required for these immunomodulatory effects remain unclear (Dehnavi et al., 2020).
Furthermore, statins have demonstrated significant neuroprotective effects, potentially reducing the incidence of neurodegenerative diseases. For instance, a large retrospective cohort study involving 288,515 participants found that statin use is associated with a substantial reduction in the risk of various neurodegenerative diseases, including Alzheimer’s disease, dementia, multiple sclerosis, Parkinson’s disease, and amyotrophic lateral sclerosis (Torrandell-Haro et al., 2020). These findings have fueled interest in drug repurposing, as the anti-inflammatory and antioxidant properties of statins may help reduce amyloid plaque formation and protein aggregation, both central to the pathogenesis of Alzheimer’s and Parkinson’s diseases (Bhat et al., 2020). Moreover, a meta-analysis of 55 observational studies encompassing over seven million patients revealed that prolonged statin exposure (more than 3 years) significantly enhances dementia risk reduction, with rosuvastatin displaying the most pronounced protective effects (Westphal Filho et al., 2025).
Emerging evidence also suggests that statins may delay cellular aging and combat senescence. These agents have been shown to improve cellular function, mitigate telomere shortening, reduce apoptosis, and counteract the senescence-associated secretory phenotype (SASP) (Bahrami et al., 2020; Guaraldi et al., 2023; Strazhesko et al., 2016). Together, these findings illustrate the broader physiological impacts of statins, highlighting their potential benefits beyond lipid regulation while also underscoring the need for careful evaluation of long-term safety.
Limitations and future perspectives
Together, these findings illustrate the broader physiological impacts of statins, highlighting their potential benefits beyond lipid regulation while also underscoring the need for careful evaluation of long-term safety (Awosika et al., 2023). Advancing the understanding of statin-induced epigenetic modifications could expand their therapeutic applications beyond cardiovascular disease, potentially informing the management of cancer, neurodegenerative disorders, autoimmune diseases, and metabolic dysfunctions. To fully harness this potential, healthcare providers and researchers must recognize the epigenetic dimensions of statin action and support research strategies that prioritize both mechanistic depth and clinical relevance.
Conclusion
In conclusion, while statins are primarily prescribed for the management of LDL cholesterol, increasing evidence supports their role as modulators of the epigenome. Their capacity to influence DNA methylation, histone modifications, and miRNA expression implicates statins in the regulation of key biological processes, including inflammation, endothelial function, and tumor suppression.
Preclinical and clinical studies have demonstrated that these epigenetic mechanisms may underlie the beneficial effects of statins across a range of conditions, including cardiovascular disease, cancer, and diabetes. However, statins have also been linked to adverse metabolic effects, such as increased insulin resistance and heightened risk of T2DM. Therefore, it is essential to further investigate the molecular pathways involved in statin-mediated disease modulation.
Large-scale, longitudinal studies incorporating epigenomic profiling and integrative molecular analyses are needed to more precisely define the benefits and risks of statin therapy. Such efforts will be critical to developing personalized therapeutic strategies that optimize statin efficacy while minimizing unintended effects.
Author contributions
RT-T: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. PG-R: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. SC-U: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. VRP: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. EP-C: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. AKZ: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The experimentation and publication fee of this article are funded by Universidad UTE.
Acknowledgments
We are grateful to Universidad UTE for supporting the researchers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi, F., Lamendola, C., Harris, C. S., Harris, V., Tsai, M. S., Tripathi, P., et al. (2021). Statins are associated with increased insulin resistance and secretion. Arterioscler. Thromb. Vasc. Biol. 41 (11), 2786–2797. doi:10.1161/ATVBAHA.121.316159
Allen, S. C., and Mamotte, C. D. S. (2017). Pleiotropic and adverse effects of statins—do epigenetics play a role? J. Pharmacol. Exp. Ther. 362 (2), 319–326. doi:10.1124/jpet.117.242081
Awosika, A., Omole, A. E., Adabanya, U., Anand, N., and Millis, R. M. (2023). “Statins and epigenetics: a putative mechanism for explaining pleiotropic effects,” in Statins - from lipid-lowering benefits to pleiotropic effects (United Kingdom: IntechOpen). Available online at: www.intechopen.com.
Bahrami, A., Bo, S., Jamialahmadi, T., and Sahebkar, A. (2020). Effects of 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors on ageing: molecular mechanisms. Ageing Res. Rev. 58, 101024. doi:10.1016/j.arr.2020.101024
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21 (3), 381–395. doi:10.1038/cr.2011.22
Benhammou, J. N., Ko, A., Alvarez, M., Kaikkonen, M. U., Rankin, C., Garske, K. M., et al. (2019). Novel lipid long intervening noncoding RNA, oligodendrocyte maturation-associated long intergenic noncoding RNA, regulates the liver steatosis gene stearoyl-coenzyme A desaturase as an enhancer RNA. Hepatol. Commun. 3 (10), 1356–1372. doi:10.1002/hep4.1413
Beulens, J. W. J., Pinho, M. G. M., Abreu, T. C., den Braver, N. R., Lam, T. M., Huss, A., et al. (2021). Environmental risk factors of type 2 diabetes—an exposome approach. Diabetologia 65, 263–274. doi:10.1007/s00125-021-05618-w
Bhat, A., Dalvi, H., Jain, H., Rangaraj, N., Singh, S. B., and Srivastava, S. (2020). Perspective insights of repurposing the pleiotropic efficacy of statins in neurodegenerative disorders: an expository appraisal. Curr. Res. Pharmacol. Drug Discov. 2, 100012. doi:10.1016/j.crphar.2020.100012
Bolayırlı, I. M., Önal, B., Adıgüzel, M., Konukoğlu, D., Demirdağ, Ç., Kurtuluş, E. M., et al. (2022). The clinical significance of circulating miR-21, miR-142, miR-143, and miR-146a in patients with prostate cancer. J. Med. Biochem. 41 (2), 191–198. doi:10.5937/jomb0-32046
Bridgeman, S., Northrop, W., Ellison, G., Sabapathy, T., Melton, P. E., Newsholme, P., et al. (2019). Statins do not directly inhibit the activity of major epigenetic modifying enzymes. Cancers 11, 516. doi:10.3390/cancers11040516
Cerda, A., Bortolin, R. H., Manriquez, V., Salazar, L., Zambrano, T., Fajardo, C. M., et al. (2021). Effect of statins on lipid metabolism-related microRNA expression in HepG2 cells. Pharmacol. Rep. 73 (3), 868–880. doi:10.1007/s43440-021-00241-3
Chen, H., Wang, X., Bai, J., and He, A. (2017). Expression, regulation and function of miR-495 in healthy and tumor tissues. Oncol. Lett. 13 (4), 2021–2026. doi:10.3892/ol.2017.5727
Chen, W. H., Chen, C. H., Hsu, M. C., Chang, R. W., Wang, C. H., and Lee, T. S. (2024). Advances in the molecular mechanisms of statins in regulating endothelial nitric oxide bioavailability: interlocking biology between eNOS activity and L-arginine metabolism. Biomed. and Pharmacother. 171, 116192. doi:10.1016/j.biopha.2024.116192
Cooney, C. A. (2010). Drugs and supplements that may slow aging of the epigenome. Drug Discov. Today Ther. Strateg. 7 (3–4), 57–64. doi:10.1016/j.ddstr.2011.03.001
Czechowicz, P., Gebert, M., Bartoszewska, S., Kalinowski, L., Collawn, J. F., and Bartoszewski, R. (2024). The Yin and Yang of hsa-miR-1244 expression levels during activation of the UPR control cell fate. Cell Commun. Signal. 22 (1), 577. doi:10.1186/s12964-024-01967-2
Dabhi, K. N., Gohil, N. V., Tanveer, N., Hussein, S., Pingili, S., Makkena, V. K., et al. (2023). Assessing the link between statins and insulin intolerance: a systematic review. Cureus 15 (7), e42029. doi:10.7759/cureus.42029
Dávalos, A., Goedeke, L., Smibert, P., Ramírez, C. M., Warrier, N. P., Andreo, U., et al. (2011). miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U. S. A. 108 (22), 9232–9237. doi:10.1073/pnas.1102281108
Dehnavi, S., Sohrabi, N., Sadeghi, M., Lansberg, P., Banach, M., Al-Rasadi, K., et al. (2020). Statins and autoimmunity: state-of-the-art. Pharmacol. Ther. 214, 107614. doi:10.1016/j.pharmthera.2020.107614
Deng, B., Tang, X., and Wang, Y. (2021). Role of microRNA-129 in cancer and non-cancerous diseases (Review). Exp. Ther. Med. 22 (3), 918. doi:10.3892/etm.2021.10350
Ding, R., Duan, Z., Yang, M., Wang, X., Li, D., and Kan, Q. (2021). High miR-3609 expression is associated with better prognosis in TNBC based on mining using systematic integrated public sequencing data. Exp. Ther. Med. 23 (1), 54. doi:10.3892/etm.2021.10976
Dong, B., Wu, M., Li, H., Kraemer, F. B., Adeli, K., Seidah, N. G., et al. (2010). Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 51 (6), 1486–1495. doi:10.1194/jlr.M003566
Dongoran, R. A., Wang, K. H., Lin, T. J., Yuan, T. C., and Liu, C. H. (2020). Anti-proliferative effect of statins is mediated by DNMT1 inhibition and p21 expression in OSCC cells. Cancers (Basel) 12 (8), 2084. doi:10.3390/cancers12082084
Du, G., Song, Y., Zhang, T., Ma, L., Bian, N., Chen, X., et al. (2014). Simvastatin attenuates TNF-α-induced apoptosis in endothelial progenitor cells via the upregulation of SIRT1. Int. J. Mol. Med. 34 (1), 177–182. doi:10.3892/ijmm.2014.1740
Du, W., Fan, L., and Du, J. (2023). Neuroinflammation-associated miR-106a-5p serves as a biomarker for the diagnosis and prognosis of acute cerebral infarction. BMC Neurol. 23 (1), 248–249. doi:10.1186/s12883-023-03241-3
Duddu, S., Katakia, Y. T., Chakrabarti, R., Sharma, P., and Shukla, P. C. (2025). New epigenome players in the regulation of PCSK9-H3K4me3 and H3K9ac alterations by statin in hypercholesterolemia. J. Lipid Res. 66 (1), 100699. doi:10.1016/j.jlr.2024.100699
Feig, J. E., Shang, Y., Rotllan, N., Vengrenyuk, Y., Wu, C., Shamir, R., et al. (2011). Statins promote the regression of atherosclerosis via activation of the CCR7-Dependent emigration pathway in macrophages. PLoS One 6 (12), e28534. doi:10.1371/journal.pone.0028534
Ferrari, E., and Gandellini, P. (2025). Unveiling the ups and downs of miR-205 in physiology and cancer: transcriptional and post-transcriptional mechanisms. Cell Death Dis. 11, 980. doi:10.1038/s41419-020-03192-4
Fuchs, D., Berges, C., Opelz, G., Daniel, V., and Naujokat, C. (2008). HMG-CoA reductase inhibitor simvastatin overcomes bortezomib-induced apoptosis resistance by disrupting a geranylgeranyl pyrophosphate-dependent survival pathway. Biochem. Biophys. Res. Commun. 374 (2), 309–314. doi:10.1016/j.bbrc.2008.07.012
Galicia-Garcia, U., Jebari, S., Larrea-Sebal, A., Uribe, K. B., Siddiqi, H., Ostolaza, H., et al. (2020). Statin treatment-induced development of type 2 diabetes: from clinical evidence to mechanistic insights. Int. J. Mol. Sci. 21 (13), 4725. doi:10.3390/ijms21134725
Gomes, S. E., Simões, A. E. S., Pereira, D. M., Castro, R. E., Rodrigues, C. M. P., Borralho, P. M., et al. (2016). miR-143 or miR-145 overexpression increases cetuximab-mediated antibody-dependent cellular cytotoxicity in human colon cancer cells. Oncotarget 7 (8), 9368–9387. doi:10.18632/oncotarget.7010
Grassilli, S., Bertagnolo, V., and Brugnoli, F. (2022). Mir-29b in breast cancer: a promising target for therapeutic approaches. Diagnostics 12, 2139. doi:10.3390/diagnostics12092139
Guadamuz, J. S., Shooshtari, A., and Qato, D. M. (2022). Global, regional and national trends in statin utilisation in high-income and low/middle-income countries, 2015-2020. BMJ Open 12 (9), e061350. doi:10.1136/bmjopen-2022-061350
Guaraldi, G., Erlandson, K. M., Milic, J., Landay, A. L., and Montano, M. A. (2023). Can statin preventative treatment inform geroscience-guided therapeutics? Aging Cell 22 (12), e13998. Available online at. doi:10.1111/acel.13998
Huang, P., Wang, L., Li, Q., Tian, X., Xu, J., Xu, J., et al. (2020). Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res. 116 (2), 353–367. doi:10.1093/cvr/cvz139
Ishikawa, S., Hayashi, H., Kinoshita, K., Abe, M., Kuroki, H., Tokunaga, R., et al. (2014). Statins inhibit tumor progression via an enhancer of zeste homolog 2-mediated epigenetic alteration in colorectal cancer. Int. J. Cancer 135 (11), 2528–2536. doi:10.1002/ijc.28672
Jung, E., Kong, S. Y., Ro, Y. S., Ryu, H. H., and Do, S. S. (2022). Effects of integrative cognitive function improvement program on cognitive function, oral health, and mental health in older people: a randomized clinical trial. Int. J. Environ. Res. Public Health 19, 14339. doi:10.3390/ijerph192114339
Karaayvaz, M., Zhai, H., and Ju, J. (2013). miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death and Dis. 4 (6), e659. doi:10.1038/cddis.2013.193
Karlic, H., Thaler, R., Gerner, C., Grunt, T., Proestling, K., Haider, F., et al. (2015). Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells. Cancer Genet. 208 (5), 241–252. doi:10.1016/j.cancergen.2015.03.008
Kayzuka, C., Rondon-Pereira, V. C., Nogueira Tavares, C., Pacheco Pachado, M., Monica, F. Z., Tanus-Santos, J. E., et al. (2025). “Epigenetics is involved in the pleiotropic effects of statins,” in Expert opinion on drug metabolism and toxicology. United Kingdom: Taylor & Francis.
Kim, S. W., Kang, H. J., Jhon, M., Kim, J. W., Lee, J. Y., Walker, A. J., et al. (2019). Statins and inflammation: new therapeutic opportunities in psychiatry. Front. Psychiatry 10 (MAR), 103. doi:10.3389/fpsyt.2019.00103
Kishore, S. P., Blank, E., Heller, D. J., Patel, A., Peters, A., Price, M., et al. (2018). “Modernizing the world health organization list of essential medicines for preventing and controlling cardiovascular diseases. Vol. 71,” in Journal of the American College of cardiology (USA: Elsevier), 564–574.
Kodach, L. L., Jacobs, R. J., Voorneveld, P. W., Wildenberg, M. E., Verspaget, H. W., Van Wezel, T., et al. (2011). Statins augment the chemosensitivity of colorectal cancer cells inducing epigenetic reprogramming and reducing colorectal cancer cell ‘stemness’ via the bone morphogenetic protein pathway. Gut 60 (11), 1544–1553. doi:10.1136/gut.2011.237495
Kotlyarov, S., and Kotlyarova, A. (2025). Biological functions and clinical significance of the ABCG1 transporter. Biology 14, 8. doi:10.3390/biology14010008
Leisegang, M. S., Bibli, S. I., Günther, S., Pflüger-Müller, B., Oo, J. A., Höper, C., et al. (2019). Pleiotropic effects of laminar flow and statins depend on the Krüppel-like factor-induced lncRNA MANTIS. Eur. Heart J. 40 (30), 2523–2533. doi:10.1093/eurheartj/ehz393
Li, C., Hu, Z., Zhang, W., Yu, J., Yang, Y., Xu, Z., et al. (2019). Regulation of cholesterol homeostasis by a novel long non-coding RNA LASER. Sci. Rep. 9, 7693. doi:10.1038/s41598-019-44195-2
Li, H., and Liu, J. (2012). The novel function of HINFP as a co-activator in sterol-regulated transcription of PCSK9 in HepG2 cells. Biochem. J. 443 (3), 757–768. doi:10.1042/BJ20111645
Li, Z., Hassan, M. Q., Jafferji, M., Aqeilan, R. I., Garzon, R., Croce, C. M., et al. (2009). Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 284 (23), 15676–15684. doi:10.1074/jbc.M809787200
Lin, H. J., Yu, S. L., Su, T. C., Hsu, H. C., Chen, M. F., Lee, Y. T., et al. (2020). Statin-induced microRNAome alterations modulating inflammation pathways of peripheral blood mononuclear cells in patients with hypercholesterolemia. Biosci. Rep. 40 (9), BSR20201885. doi:10.1042/BSR20201885
Lin, Y. C., Lin, J. H., Chou, C. W., Chang, Y. F., Yeh, S. H., and Chen, C. C. (2008). Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 68 (7), 2375–2383. doi:10.1158/0008-5472.CAN-07-5807
Liu, C., Shen, M., Tan, W. L. W., Chen, I. Y., Liu, Y., Yu, X., et al. (2023). Statins improve endothelial function via suppression of epigenetic-driven EndMT. Nat. Cardiovasc. Res. 2 (5), 467–485. doi:10.1038/s44161-023-00267-1
Liu, J., Wang, H., Zeng, D., Xiong, J., Luo, J., Chen, X., et al. (2022). The novel importance of miR-143 in obesity regulation. Int. J. Obes. 47 (2), 100–108. doi:10.1038/s41366-022-01245-6
Liu, M. N., Luo, G., Gao, W. J., Yang, S. J., and Zhou, H. (2021). miR-29 family: a potential therapeutic target for cardiovascular disease. Pharmacol. Res. 166, 105510. doi:10.1016/j.phrs.2021.105510
Liu, Y., Shen, Y., Guo, T., Parnell, L. D., Westerman, K. E., Smith, C. E., et al. (2020). Statin use associates with risk of type 2 diabetes via epigenetic patterns at ABCG1. Front. Genet. 11, 622. doi:10.3389/fgene.2020.00622
Lui, K. H., Zhao, H., Sun, J., Shen, Z., and Xu, J. (2023). Analysis of the expression profile of miRNAs related to skin photoaging in the GEO database. Chin. J. Plastic Reconstr. Surg. 5 (2), 53–59. doi:10.1016/j.cjprs.2023.05.003
Matson, K., Macleod, A., Mehta, N., Sempek, E., Tang, X., Matson, K., et al. (2023). Impacts of MicroRNA-483 on human diseases. Non-Coding RNA 9 (4), 37. doi:10.3390/ncrna9040037
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10 (3), 155–159. doi:10.1038/nrg2521
Mitchel, K., Theusch, E., Cubitt, C., Dosé, A. C., Stevens, K., Naidoo, D., et al. (2016). RP1-13D10.2 is a novel modulator of statin-induced changes in cholesterol. Circ. Cardiovasc Genet. 9 (3), 223–230. doi:10.1161/CIRCGENETICS.115.001274
Mohammadzadeh, N., Montecucco, F., Carbone, F., Xu, S., Al-Rasadi, K., and Sahebkar, A. (2020). Statins: epidrugs with effects on endothelial health? Eur. J. Clin. Invest 50 (12), e13388. Available online at:. doi:10.1111/eci.13388
Paez, I., Prado, Y., Loren, P., Ubilla, C. G., Rodríguez, N., and Salazar, L. A. (2023). Cholesterol-related lncRNAs as response predictors of atorvastatin treatment in Chilean hypercholesterolemic patients: a pilot study. Biomedicines 11 (3), 742. doi:10.3390/biomedicines11030742
Pang, Y., Ruan, X., Liu, W., Hou, L., Yin, B., Shu, P., et al. (2024). MicroRNA-495 modulates neuronal layer fate determination by targeting Tcf4. Int. J. Biol. Sci. 20 (15), 6207–6221. doi:10.7150/ijbs.94739
Paseban, M., Butler, A. E., and Sahebkar, A. (2019). Mechanisms of statin-induced new-onset diabetes. J. Cell Physiol. 234 (8), 12551–12561. doi:10.1002/jcp.28123
Peeples, E. S., Mirnics, K., and Korade, Z. (2024). Chemical inhibition of sterol biosynthesis. Biomolecules 14 (4), 410. doi:10.3390/biom14040410
Qie, R., Chen, Q., Wang, T., Chen, X., Wang, J., Cheng, R., et al. (2021). Association of ABCG1 gene methylation and its dynamic change status with incident type 2 diabetes mellitus: the rural Chinese cohort study. J. Hum. Genet. 66 (4), 347–357. doi:10.1038/s10038-020-00848-z
Rasmi, Y., Mohamed, Y. A., Alipour, S., Ahmed, S., and Abdelmajed, S. S. (2023). The role of miR-143/miR-145 in the development, diagnosis, and treatment of diabetes. J. Diabetes Metab. Disord. 23 (1), 39–47. doi:10.1007/s40200-023-01317-y
Ruiz-Pozo, V. A., Cadena-Ullauri, S., Guevara-Ramírez, P., Paz-Cruz, E., Tamayo-Trujillo, R., and Zambrano, A. K. (2023). Differential microRNA expression for diagnosis and prognosis of papillary thyroid cancer. Front. Med. (Lausanne) 10, 1139362. doi:10.3389/fmed.2023.1139362
Saavedra, K., Leal, K., Saavedra, N., Prado, Y., Paez, I., Ubilla, C. G., et al. (2022). MicroRNA-20a-5p downregulation by atorvastatin: a potential mechanism involved in lipid-lowering therapy. Int. J. Mol. Sci. 23 (9), 5022. doi:10.3390/ijms23095022
Schrader, S., Perfilyev, A., Martinell, M., García-Calzón, S., and Ling, C. (2021). Statin therapy is associated with epigenetic modifications in individuals with type 2 diabetes. Epigenomics 13 (12), 919–925. doi:10.2217/epi-2020-0442
Serik, S., Riabukha, V., Serdobinska-Kanivets, E., Bondar, T., and Ovrakh, T. (2021). Dose-dependent effects of rosuvastatin on circulating microRNAs-27a and 221 levels in patients with coronary artery disease with type 2 diabetes mellitus. Eur. Heart J. 42 (Suppl. ment_1). Available online at:. doi:10.1093/eurheartj/ehab724.2942
Singh, R. S., Chaudhary, D. K., Mohan, A., Kumar, P., Chaturvedi, C. P., Ecelbarger, C. M., et al. (2016). Greater efficacy of atorvastatin versus a Non-statin lipid-lowering agent against renal injury: potential role as a histone deacetylase inhibitor. Sci. Rep. 6, 38034. doi:10.1038/srep38034
Sizar, O., Khare, S., Raja, P., and Affiliations, T. (2024). Statin medications. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK430940/?report=printable.
Stoen, M. J., Andersen, S., Rakaee, M., Pedersen, M. I., Ingebriktsen, L. M., Bremnes, R. M., et al. (2021). High expression of miR-17-5p in tumor epithelium is a predictor for poor prognosis for prostate cancer patients. Sci. Rep. 11, 13864. doi:10.1038/s41598-021-93208-6
Strazhesko, I. D., Tkacheva, O. N., Akasheva, D. U., Dudinskaya, E. N., Plokhova, E. V., Pykhtina, V. S., et al. (2016). Atorvastatin therapy modulates telomerase activity in patients free of atherosclerotic cardiovascular diseases. Front. Pharmacol. 7 (SEP), 347. doi:10.3389/fphar.2016.00347
Tabuchi, T., Satoh, M., Itoh, T., and Nakamura, M. (2012). MicroRNA-34a regulates the longevity-associated protein SIRT1 in coronary artery disease: effect of statins on SIRT1 and microRNA-34a expression. Clin. Sci. 123 (3), 161–171. doi:10.1042/CS20110563
Tikoo, K., Patel, G., Kumar, S., Karpe, P. A., Sanghavi, M., Malek, V., et al. (2015). Tissue specific up regulation of ACE2 in rabbit model of atherosclerosis by atorvastatin: role of epigenetic histone modifications. Biochem. Pharmacol. 93 (3), 343–351. doi:10.1016/j.bcp.2014.11.013
Tokumaru, Y., Oshi, M., Katsuta, E., Matsuhashi, N., Futamura, M., Yoshida, K., et al. (2020). Clinical relevance of miR-143 expression in ER-positive breast cancer patients. J. Clin. Oncol. 38 (15_Suppl. l), e12574. doi:10.1200/JCO.2020.38.15_suppl.e12574
Torrandell-Haro, G., Branigan, G. L., Vitali, F., Geifman, N., Zissimopoulos, J. M., and Brinton, R. D. (2020). Statin therapy and risk of Alzheimer’s and age-related neurodegenerative diseases. Alzheimer’s and Dementia Transl. Res. and Clin. Interventions 6 (1), e12108. Available online at:. doi:10.1002/trc2.12108
Tsilimigras, D. I., Bibli, S. I., Siasos, G., Oikonomou, E., Perrea, D. N., Filis, K., et al. (2021). Regulation of long non-coding RNAs by statins in atherosclerosis. Biomolecules 11 (5), 623. doi:10.3390/biom11050623
Tylden, E. S., Delgado, A. B., Lukic, M., Moi, L., Busund, L. T. R., Pedersen, M. I., et al. (2024). Roles of miR-20a-5p in breast cancer based on the clinical and multi-omic (CAMO) cohort and in vitro studies. Sci. Rep. 14, 25022. doi:10.1038/s41598-024-75557-0
Wang, R. N., Green, J., Wang, Z., Deng, Y., Qiao, M., Peabody, M., et al. (2014). Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1 (1), 87–105. doi:10.1016/j.gendis.2014.07.005
Weihua, Z., Guorong, Z., Xiaolong, C., and Weizhan, L. (2020). MiR-33a functions as a tumor suppressor in triple-negative breast cancer by targeting EZH2. Cancer Cell Int. 20 (1), 85–12. doi:10.1186/s12935-020-1160-z
Westphal Filho, F. L., Moss Lopes, P. R., Menegaz de Almeida, A., Sano, V. K. T., Tamashiro, F. M., Gonçalves, O. R., et al. (2025). Statin use and dementia risk: a systematic review and updated meta-analysis. Alzheimer’s and Dementia Transl. Res. and Clin. Interventions 11 (1), e70039. Available online at. doi:10.1002/trc2.70039
Wu, L. M., Wu, S. G., Chen, F., Wu, Q., Wu, C. M., Kang, C. M., et al. (2020). Atorvastatin inhibits pyroptosis through the lncRNA NEXN-AS1/NEXN pathway in human vascular endothelial cells. Atherosclerosis 293, 26–34. doi:10.1016/j.atherosclerosis.2019.11.033
Yan, C., Bao, J., and Jin, J. (2024). Exploring the interplay of gut microbiota, inflammation, and LDL-cholesterol: a multiomics Mendelian randomization analysis of their causal relationship in acute pancreatitis and non-alcoholic fatty liver disease. J. Transl. Med. 22 (1), 179. doi:10.1186/s12967-024-04996-0
Yao, Q., Chen, Y., and Zhou, X. (2019). The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 51, 11–17. doi:10.1016/j.cbpa.2019.01.024
Zaky, M. Y., Fan, C., Zhang, H., and Sun, X. F. (2023). Unraveling the anticancer potential of statins: mechanisms and clinical significance. Cancers 15, 4787. doi:10.3390/cancers15194787
Zambrano, A. K., Cadena-Ullauri, S., Guevara-Ramírez, P., Paz-Cruz, E., Tamayo-Trujillo, R., Ruiz-Pozo, V. A., et al. (2023). The autosomal short tandem repeat polymorphisms are potentially associated with cardiovascular disease predisposition in the Latin American population: a mini review. Biomed. Res. Int. 2023, 1–9. doi:10.1155/2023/6152905
Zambrano, T., Hirata, R. D. C., Hirata, M. H., Cerda, Á., and Salazar, L. A. (2015). Altered microRNome profiling in statin-induced HepG2 cells: a pilot study identifying potential new biomarkers involved in lipid-lowering treatment. Cardiovasc Drugs Ther. 29 (6), 509–518. doi:10.1007/s10557-015-6627-0
Zeng, X., Cao, Z., Luo, W., Zheng, L., and Zhang, T. (2020). MicroRNA-381—A key transcriptional regulator: its biological function and clinical application prospects in cancer. Front. Oncol. 10, 535665. doi:10.3389/fonc.2020.535665
Zhang, T., and Kraus, W. L. (2010). SIRT1-dependent regulation of chromatin and transcription: linking NAD+ metabolism and signaling to the control of cellular functions. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1804 (8), 1666–1675. doi:10.1016/j.bbapap.2009.10.022
Zhao, Y., Song, X., Ma, Y., Liu, X., and Peng, Y. (2023). Circulating mir-483-5p as a novel diagnostic biomarker for acute coronary syndrome and its predictive value for the clinical outcome after PCI. BMC Cardiovasc Disord. 23 (1), 360. doi:10.1186/s12872-023-03387-5
Keywords: pharmacology, pharmacoepigenetics, epigenetics, statins, molecular biology, healthcare and well-being
Citation: Tamayo-Trujillo R, Guevara-Ramírez P, Cadena-Ullauri S, Ruiz Pozo VA, Paz-Cruz E and Zambrano AK (2025) Statins and their impact on epigenetic regulation: insights into disease. Front. Pharmacol. 16:1621163. doi: 10.3389/fphar.2025.1621163
Received: 30 April 2025; Accepted: 04 July 2025;
Published: 17 July 2025.
Edited by:
Luis Abel Quiñones, University of Chile, ChileReviewed by:
Tzong-Shyuan Lee, National Taiwan University, TaiwanCopyright © 2025 Tamayo-Trujillo, Guevara-Ramírez, Cadena-Ullauri, Ruiz Pozo, Paz-Cruz and Zambrano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Karina Zambrano, YW5hemFtYnJhbm8xN0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Rafael Tamayo-Trujillo
Rafael Tamayo-Trujillo Patricia Guevara-Ramírez
Patricia Guevara-Ramírez Santiago Cadena-Ullauri
Santiago Cadena-Ullauri Viviana A. Ruiz Pozo
Viviana A. Ruiz Pozo Elius Paz-Cruz
Elius Paz-Cruz Ana Karina Zambrano
Ana Karina Zambrano