- 1Graduate School of Agriculture, Tokyo University of Agriculture and Technology, Tokyo, Japan
- 2Institute for Agriculture, Medicine and the Environment, Shizuoka, Japan
A nature farming system is an ecological farming practice that entails cultivating crops without using chemical fertilizers and pesticides. To understand the diversity and functions of root microbiomes associated with nature farming systems, we compared the root microbial community of rice under nature farming conditions with those under conventional farming conditions. High-throughput amplicon analysis demonstrated a higher abundance and greater diversity of the root microbiome under unfertilized nature farming conditions than under conventional conditions. The application of chemical fertilizers reduced the microbial diversity and abundance of some beneficial taxa important for plant growth and health. Subsequently, we isolated and identified 46 endo- and epiphytic bacteria from rice roots grown under nature farming conditions and examined their plant growth-promoting activity. Six potential isolates were selected for plant growth assessment in insoluble P- and K-containing media. Most of the isolates promoted rice growth, and Pseudomonas koreensis AEPR1 was able to enhance rice growth significantly in both insoluble P- and K-containing media. Our data indicated that nature farming systems create a distinct root microbiome that is comparatively more diverse and supports plant growth under low-input cultivation practices than under conventional practices. The potential isolates could be exploited as sources with potential applications in sustainable agriculture.
Introduction
The root microbiome plays a key role in plant growth and productivity, making it an important factor in agroecosystems (Edwards et al., 2015; de Vries and Wallenstein, 2017; Wasai and Minamisawa, 2018). Notably, plant root-associated microbiomes have attracted attention in recent years owing to their roles in maintaining plant productivity by contributing to conferring plant biotic and abiotic stress and resilience through various mechanisms (Tkacz and Poole, 2015; Ding et al., 2019). These include plant growth promotion (PGP) through fixation or solubilization of growth-limiting nutrients, growth promotion by the production of plant growth hormones and antagonism of plant pathogens through the production of antibiotics and resource competition. The utilization of plant growth-promoting microbes may serve as a sustainable alternative to the use of chemical fertilizers (Vejan et al., 2016; Backer et al., 2018). Therefore, understanding how root-associated microbial communities respond to farming management practices is of great agronomic interest.
Conventional farming heavily relies on the use of agrochemicals such as synthetic fertilizer and pesticide to boost agricultural productivity to meet the global crop production demand. The intensification of agrochemical application, however, has led to adverse impacts. These include increases in greenhouse gas emissions, nutrient leaching, soil degradation, loss of plant and soil microbes, increased susceptibility of crops to pests and diseases and loss of biodiversity (Xu et al., 2018; Guo et al., 2019). Concern over these problems has led to the search for alternative practices to ensure more sustainable food production and environmental conservation practices. Of these practices, organic farming is the most adopted in many countries worldwide as demand for sustainability increases (Eyhorn et al., 2019), and this includes “nature farming,” which is the major organic farming practice in Japan. The cultivation practices of nature farming systems are more or less similar or comparable with other organic farming practices conducted in other countries to improve soil biological and chemical properties as well as crop production (Li et al., 2012; Hartmann et al., 2015; Wang et al., 2016). “Nature farming,” proposed by Mokichi Okada (1882–1955) in Japan in 1935, is an ecological farming system based on sound agronomic principles (Xu, 2006). One of the concepts of nature farming systems is to advocate a lack of reliance on the application of agrochemicals in cultivating crops (Okada, 1991, 1993). The farming strategies in nature farming systems to improve plant and soil health include not relying on the application of agrochemicals such as fertilizer and pesticides, reducing soil tillage and plowing, using mulches with crop residue, crop rotation, intercropping and incorporating biofertilizer. These farming systems may enhance the abundance and diversity of plant and soil microbes, maintain soil physiochemical properties and improve the quality and safety of food production (Lenc et al., 2015; Gu et al., 2017; Liao et al., 2019).
Under nature farming systems, plants mostly rely on the root microbiome for plant health and productivity. However, we still have limited knowledge on the effects of nature farming systems on the root microbial community in crop plants. Little is known about the root microbial community associated with nature farming and its plant growth-promoting potential under low-input conditions, and no study has been performed on the root microbial community of naturally farmed rice plants in Japan. A previous study reported that rice fields managed with organic farming practices have a distinct bacterial community composition compared with that of conventionally farmed fields (Suzuki et al., 2019). The root microbial abundance was significantly higher in organically managed crops than in conventionally managed crops (Lenc et al., 2015), and a higher abundance of nutrition-related bacteria was observed (Wang et al., 2016). Organic farming harbors a much more complex root microbial structure than that of conventional farming with highly significant connectivity (Banerjee et al., 2019). The culturable bacterial and fungal endophytes associated with plants grown under organic farming were reported to be able to promote tomato growth and yield (Xia et al., 2015, 2019).
In the current study, we used high-throughput sequencing of 16S rRNA gene markers to evaluate the root microbial community composition of naturally farmed rice plants. We also aimed to identify culturable bacterial isolates associated with nature farming practices that are likely to have plant growth-promoting traits that might be beneficial to plant growth, health, and yield. This study determined the root microbial community and its plant growth-promoting function in rice plants growing under nature farming conditions in Japan.
Materials and Methods
Soil Preparation
A nature farming soil sample was collected on 26th November 2019 from the field of Ohito Farm (Shizuoka Prefecture, 35°02′ N, 139°00′ E, 357 AMSL), which has continuously practiced non-chemical fertilizer and non-pesticide application for 17 years. A conventional farming soil sample was collected on 3rd December 2019 from Fuchu Honmachi Paddy Field (Tokyo, 35°41′ N, 139°29′ E, 46 AMSL). Soil samples were collected at a depth of 0–15 cm. Subsequently, each of the soil samples (40 L) was divided into two parts to be used as unfertilized and fertilized soil-treated samples. A 1/5000 Wagner pots were filled with 2.5 L of soil, and fertilized soils were mixed with 2.6 g pot−1 potassium oxide (60% K2O), 8.9 g pot−1 superphosphate (17% P2O5), and 4.16 g pot−1 ammonium sulfate (21% NH4SO4).
Physicochemical Properties of Soil
Soil pH and electric conductivity (EC) were determined in deionized water (H2O) and 1 M KCl at a soil-to-solution ratio of 1:5. Total carbon (TC) and total nitrogen (TN) were quantified using the dry combustion method with an NC analyzer (SUMIGRAPH NC TR22, Sumika Chemical Analysis Service, Ltd., Osaka, Japan). Inorganic N in the soil was promptly extracted from 6 g of soil with 2 M KCl for 30 min. Then, the suspension was centrifuged and filtered through filter paper (Advantec No. 5C; Advantec, Tokyo, Japan). Several drops of 1 mg kg-1 copper(II) bromide solution were added to the extract and kept at 4°C. Then, N-NH4 in the extract was analyzed by the modified indophenol blue method (Rhine et al., 1998) using a spectrophotometer (UV-1200; Shimadzu Co., Ltd., Kyoto, Japan), and N-NO3 in the extract was analyzed by flow injection analysis using a flow-through visible spectrophotometer (S/3250; Soma-Kogaku Co., Ltd., Tokyo, Japan) connected to a double-plunger pump (PE-230; Aqua-lab Co., Ltd., Tokyo, Japan) equipped with a copperized cadmium column to reduce nitrate to nitrite in the sample solutions. Available phosphate (Available-P) was evaluated by the Bray 2 method (Nanzyo, 1997).
Rice Pot Experiment
We used two rice (Oryza sativa L.) cultivars, namely, the Koshihikari and Asahi cultivars. Koshihikari is one of the main cultivars of paddy rice in Japan, and we used this cultivar as a model of a modern variety. The Asahi cultivar is one of the native varieties separated from the old cultivar Hinode since 1909, and it is optimized under low-nutrient soil conditions (Takahashi and Yoshida, 2008). The rice seeds were surface-sterilized by immersion in 70% ethanol for 30 s, followed by soaking in 5% sodium hypochlorite solution for 20 min and washing five times with sterilized distilled water. Subsequently, the sterilized seeds were soaked in PPM solution for 48 h at 4°C and washed three times with sterilized distilled water. The seeds were incubated for 48 h at 28°C under dark conditions for germination. The germinated seeds were sown directly into prepared pots, with four plants per pot and a total of four replications per experiment. Plants were grown in a controlled-environmental chamber (Koito-tron Koito-industries, Japan) at Tokyo University of Agriculture and Technology and kept at 28°C for 16 h and 25°C for 8 h with 60% humidity for 30 days, and plants were watered with sterilized distilled water.
DNA Extraction and Amplicon Analysis
DNA extraction was carried out according to Ikeda et al. (2004) with slight modification. Root samples were washed thoroughly under running tap water to remove soil debris. Ten-gram root samples were first lyophilized and ground to a fine powder with liquid nitrogen. Then, 0.3 g of ground root sample was suspended in 350 μl of DNA extraction buffer [0.5 M Tris (pH 8.0), 0.1 M EDTA, 0.1 M NaCl, 2% SDS] and 350 μl of 0.3 M sodium phosphate buffer (pH 8.0) in 2 ml screw-capped tubes. After adding 0.5 g of ϕ0.1 mm zirconia/silica beads (BioSpec product, Inc., Bartlesville, USA), the tubes were processed for 20 s four times using a FastPrep-24TM instrument (MP Biomedicals, Santa Ana, California, USA). The tubes were centrifuged for 5 min at 13,000 rpm and room temperature. Then, 600 μl of supernatant was collected into a new tube, and 120 μl of 8 M potassium acetate was added. After 5 min of incubation, the tubes were centrifuged for 5 min at 13,000 rpm. The supernatants were transferred into new tubes and mixed with 0.1 volumes of 3 M sodium acetate (pH 5.2) and 0.5 volumes of isopropanol. After incubation for 5 min at room temperature, the tubes were centrifuged at 13,000 rpm at room temperature. The pellets were washed with 500 μl of 70% ethanol twice and suspended in 100 μl of sterilized Milli-Q water. DNA was assessed for quantity and quality by using a Nanodrop 2000 (Thermo Fisher and Scientific, USA) and stored at −20°C until sequencing.
Amplicon sequencing of the V3/V4 region was carried out with the MiSeq platform at Bioengineering Lab. Co. (Sagamihara, Japan). The primers V3/V4f_MIX and V3/V4r_MIX were used to amplify the bacterial V3/V4 region (341f-805r). The first PCR was prepared in a final volume of 20 μl comprised of DNA template (100 ng) using an Amplitaq Gold 360 polymerase kit (Thermo Fisher Scientific, USA), and the PCR conditions were set according to the manufacturer's instructions. The PCR products were purified using AMPure XP (Beckman Coulter) and quantified using Synergy H1 (Bio Tek) and the QuantiFlour dsDNA system. A quality check of the libraries was performed using the fragment analyzer and sdDNA 915 reagent kit (Advanced Analytical Technologies). The libraries were pooled together and loaded into an Illumina MiSeq instrument following the manufacturer's instructions (Illumina, San Diego, CA, USA).
The Quantitative Insights into Microbial Ecology (QIIME 2.0) toolkit was used to process the raw high-throughput sequencing data. Then, the high-throughput sequencing data obtained from the 16S amplicon were analyzed using MicrobiomeAnalyst (Dhariwal et al., 2017), and the OTUs were annotated as SILVA labels. The amplicon data were subjected to rarefaction curve, microbial alpha- and beta-diversity and relative abundance analyses using the ranacapa and phyloseq packages, respectively. Preferential microbial taxa were compared across treatments using linear discriminate analysis (LDA) effect size (LEfSe, LDA score > 4.0). The raw sequencing reads were submitted to DRA/DDBJ under accession no. DRA010459.
Isolation and Characterization of Bacteria From Rice Roots Grown in Nature Farming Soil
For isolation of endophytic and epiphytic bacteria, roots were collected from plants grown on unfertilized nature farming soils. The isolation of epiphytic bacteria was carried out by immersion of 0.5 g root samples into 10 ml of 0.05% Tween 20 solution, and 100 μl of the solution was plated onto NB agar medium (Eiken Chemical Co., Ltd. Tochigi, Japan) for bacteria and incubated at 28°C for 7 days. Next, the roots were surface-sterilized with 70% ethanol for 30 s and 1% sodium hypochlorite for 1 min and washed five times using sterilized distilled water for the isolation of endophytic bacteria. The root tissue was macerated in a sterilized mortar in the presence of 1 ml of 0.85% sodium chloride solution. The extract was serially diluted, and 100 μl of diluted suspension was spread onto NB and PDA media, followed by incubation at 28°C for 7 days. Colonies with different morphologies based on size, shape and color were selected and subcultured on fresh NB agar media to obtain pure cultures. Stock cultures of each isolated strain were preserved in 50% glycerol and kept at −80°C. These strains were routinely grown in NB agar medium as necessary.
The bacterial isolates were identified by targeting 16S rDNA. Colony PCR was conducted by using the primer sets V3V4f 5′-CCTACGGGNGGCWGCAG-3′ and V3V4r 5′-GACTACHVGGGTATCTAATCC-3′ using GoTaq (Promega). PCR was conducted at 95°C for 3 min and 30 cycles of denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and 30 s extension at 72°C using the Veriti 96-well thermal cycler (Applied Biosystems; California, USA). After electrophoresis, the bands were cut and purified using a FastGeneTM Gel/PCR Extraction kit following the manufacturer's protocol. The DNA concentration and purity were examined using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, USA) and sent for sequencing. The 16S rRNA gene sequences of the bacterial isolates associated with nature farming practices were compared with bacterial sequences in the GenBank database through the BLAST program, and MEGA X version 10.1 was used to estimate the phylogenetic relationship with the obtained sequences. The sequences obtained in this study have also been deposited at the DDBJ under accession number.
Plant Growth Promotion (PGP) Assays
Indole-3-Acetic Acid (IAA) Production
For the IAA evaluation, bacterial isolates (adjusted OD600 = 0.01) cultured in NB medium supplemented with 100 mg L−1 L-tryptophan were inoculated with bacterial isolates and incubated for 48 h in the dark at 28°C. Bacterial cell suspensions were centrifuged at 10,000 rpm for 15 min, and the concentration of IAA in the supernatant was measured using the Salkowski colorimetric technique (Glickmann and Dessaux, 1995) by measuring absorbance at 530 nm with a microplate reader (Spectra MaxParadigm, molecular device Japan Co., Ltd, Osaka, Japan).
P- and K-Solubilizing Activities
Five microliters of a bacterial suspension (adjusted OD600 = 0.01) was spotted onto National Botanical Research Institute's Phosphate (NBRIP) medium (Nautiyal, 1999) containing 0.5% tricalcium phosphate [Ca3(PO4)2] and slightly modified Aleksandrov medium (Hu et al., 2006) containing 0.2% sericite mica for insoluble phosphate and potassium solubilization screening, respectively. Plates were incubated at 28°C for 7 d in the dark. The P- and K-solubilizing activities of each isolate were evaluated by measuring the size of the clear zone around the colony and comparing it with previously isolated P- and K-solubilizing isolates, Paenibacillus rhizosphaerae (NBRC109635) and Micrococcus yunnanensis, respectively.
Siderophore Production
Five microliters of the bacterial suspension (adjusted OD600 = 0.01) was spotted onto slightly modified Chrome-azurol S (CAS) medium (Hu, 2011). Plates were incubated at 28°C for 2 d in the dark. The siderophore production activity of each isolate was evaluated by measuring the size of the orange or yellow zone around the colony.
Effects of Bacterial Isolates on Rice Growth
Seed coats were removed from rice seeds (cv. Nipponbare). The dehusked seeds were surface-sterilized with 70% ethanol for 30 sec, shaken in 5% (w/v) sodium hypochlorite for 10 min and washed five times with sterilized distilled water. The sterilized seeds were incubated for 2 d at 28°C under dark conditions for germination.
Bacterial isolates were grown on NB medium at 28°C for 48 h and washed twice with sterilized distilled water. Bacterial cells were resuspended in sterilized distilled water to a final density of 109 CFU mL−1. Bacterial suspensions were diluted 10-fold to a final density of 2 × 107 CFU mL−1. Pregerminated sterilized seeds were soaked in the prepared bacterial suspension for 2 d at 28°C in the dark. P. rhizosphere and M. yunnanensis isolates were used as positive strains, and isolate KENR28 was used as a negative strain in our study.
Inoculated seeds were transferred into plant boxes (CUL-JAR300; Iwaki, Tokyo, Japan) containing 100 mL of 25% semisolid modified phosphate-deficient Hoagland and 25% semisolid modified potassium-deficient Hoagland medium supplemented with 2 g l−1 Ca3(PO4)2 as the sole P source and 2 g l−1 sericite mica as the sole K source. Non-inoculated seeds planted on modified P-deficient and K-deficient Hoagland media acted as negative controls. Five replicates of all controls and inoculated treatments were conducted. The plants were incubated in a plant growth chamber (LPH-240SP; NK system, Osaka, Japan) under 16-h light: 8-h dark conditions at 25°C for 7 days. Growth parameters were analyzed to assess the response of bacteria to inoculation, including plant height, root length, shoot biomass and root biomass.
Statistical Analysis
Statistical analyses were performed using one-way analysis of variance (ANOVA) followed by Duncan's multiple range test. Different lowercase letters represent significantly different values (p < 0.05).
Results
Soil Physicochemical Properties
Soil samples were collected from naturally and conventionally farmed fields and subjected to soil nutrient analysis before the cultivation of crops began. The soil pH of nature farming soil was slightly lower than that of conventional farming soil (Supplementary Table 1). The total carbon and nitrogen contents were observed to be slightly higher in the nature farming soil. Moreover, the nature farming soil had lower soil nitrate nitrogen (NO3-N), ammonium nitrogen (NH4-N) and available P contents than the conventional farming soil.
Diversity of the Root Microbial Community Under Nature Farming and Conventional Farming Conditions With Fertilization Treatments
A total of 1,340,485 16S rRNA sequences were obtained with a median read count per sample of 56,446 (range: 39,586–69,191) obtained from the 24 root samples (two farming systems, two fertilization treatments and two rice cultivars; three replicates each). The reads were clustered into 1,225 microbial OTUs. Rarefaction curves increased steadily as the number of sequences increased (Supplementary Figure 1), indicating that the sample size was sufficient for subsequent data analysis.
Root Microbial Diversity as a Function of Farming Practice, Fertilization Treatment and Rice Cultivar
Univariate analysis showed that microbial abundance (Chao1) and diversity (Shannon index) were significantly different (p < 0.005) within the fertilization treatments irrespective of farming system and rice cultivar (Figure 1). The fertilized groups had lower microbial abundances and diversity than the unfertilized groups (Figure 1B). However, there was no significant difference in root microbial richness or diversity between the farming systems or rice cultivars (Figures 1A,C).
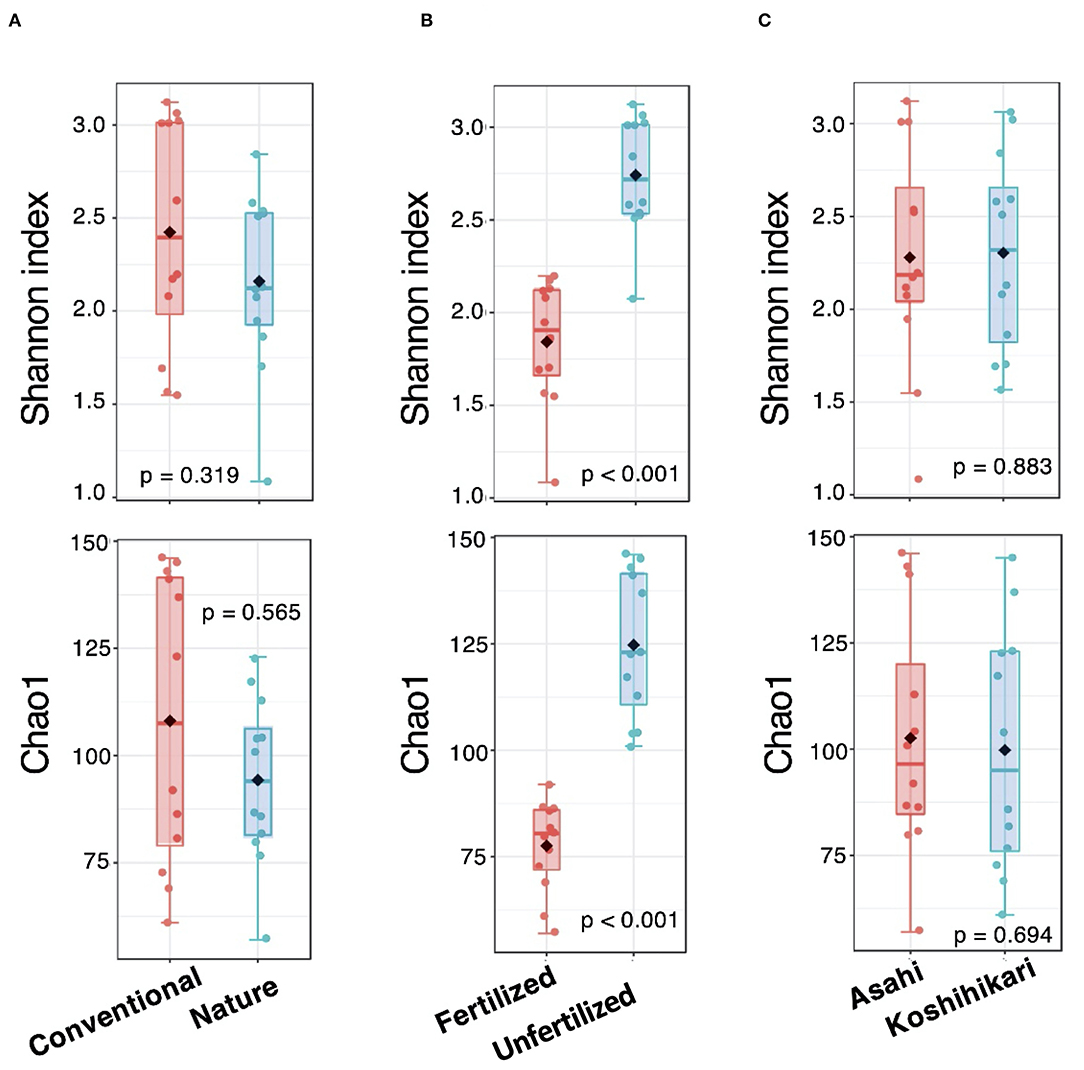
Figure 1. Richness estimators (Chao1) and the diversity index (Shannon) for (A) farming systems, (B) fertilization treatments, and (C) rice cultivar factors.
Unconstrained principal coordinate analysis (PCoA) indicated that the bacterial community clearly clustered based on fertilization treatments, and PERMANOVA revealed that fertilization treatments (p < 0.001) led to significant shifts in the bacterial communities in both farming systems (p < 0.004) (Supplementary Table 2). There was a significant effect of farming practice (nature faming vs. conventional farming) on the root microbial community structure of rice (Figure 2A). Unfertilized nature farming has a distinct root microbial community composition from fertilized conventional farming (Figure 2B). However, the rice cultivars Asahi and Koshihikari showed no significant difference in root microbial community structure (Figure 2C). Our results suggested that the application of chemical fertilizer influenced the root microbial community across the farming systems.
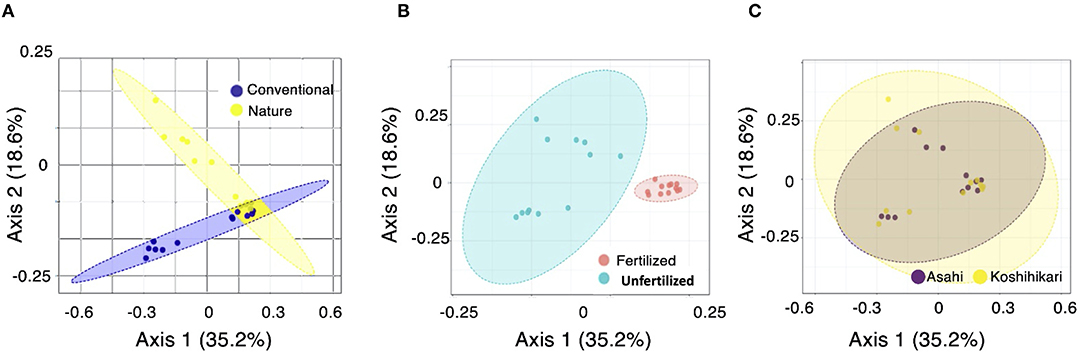
Figure 2. Unconstrained principal coordinate analysis (PCoA) based on Bray-Curtis distance for the effects of (A) farming system, (B) fertilization treatments, and (C) rice cultivar factors on root microbial community structure.
To clarify the root microbial community composition within the two farming systems, the phylum-level dataset was analyzed first. The four most dominant phyla across the treatments were V5, Proteobacteria, Firmicutes and Bacteroidetes (Figure 3A). The application of chemical fertilizer reduced the abundances of Proteobacteria, Firmicutes, Chloroflexi, Spirochaetes and certain other phyla, irrespective of farming system or rice cultivar (Figure 3B). The root microbial community composition within the farming systems (Figure 3C) and rice cultivars (Figure 3D) showed no significant difference in their relative abundance of phyla.
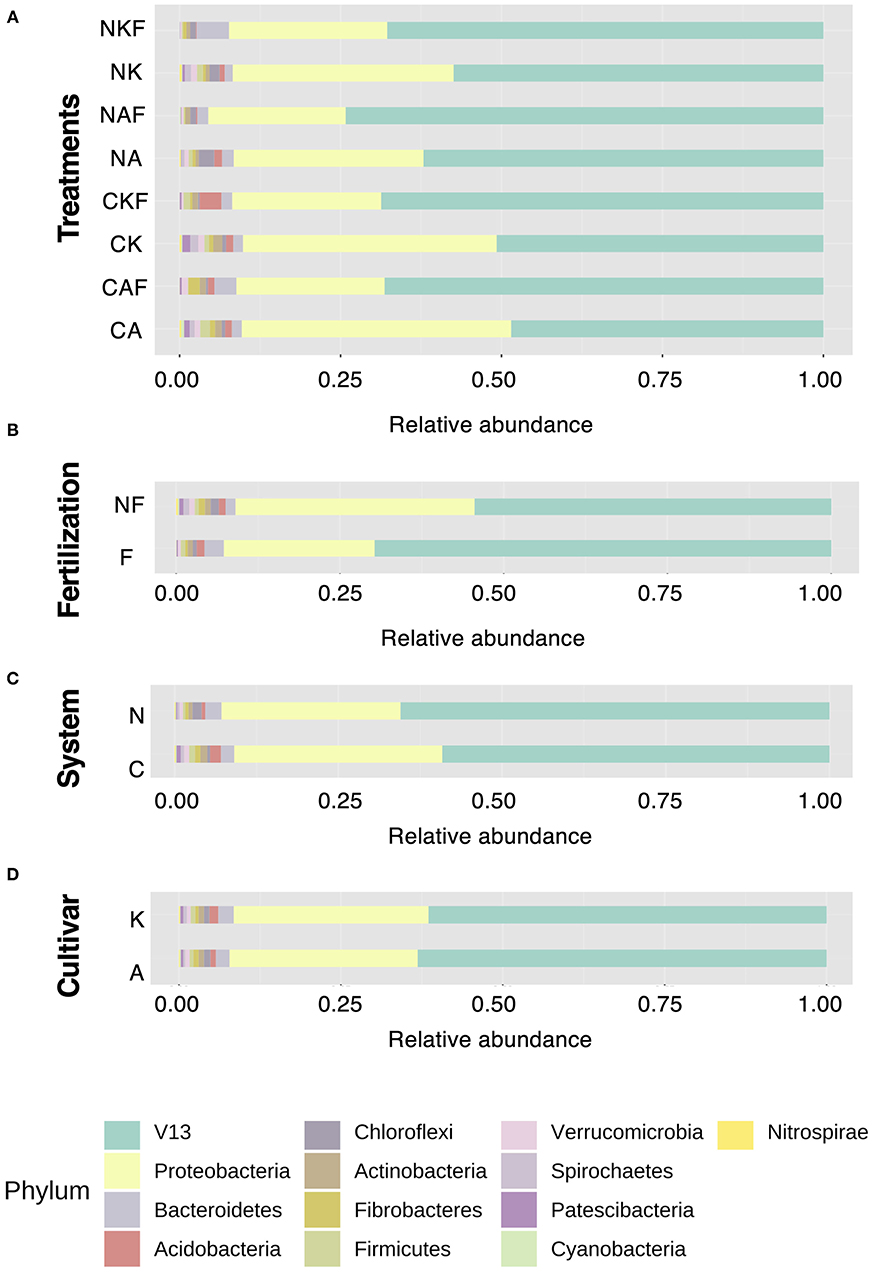
Figure 3. Relative abundance of dominant phyla (A) across the samples, (B) fertilization, (C) farming systems, and (D) rice cultivars. (A) NKF, Koshihikari grown in nature farming soil treated with chemical fertilizer; NK, Koshihikari grown in nature farming soil without chemical fertilizer; NAF, Asahi grown in nature farming soil treated with chemical fertilizer; NA, Asahi grown in nature farming soil without chemical fertilizer; CKF, Koshihikari grown in conventional farming soil treated with chemical fertilizer; CK, Koshihikari grown in conventional farming soil without chemical fertilizer; CAF, Asahi grown in conventional farming soil treated with chemical fertilizer; and CA, Asahi grown in conventional farming soil without chemical fertilizer. (B) NF, no application of chemical fertilizers; F, application of chemical fertilizer. (C) N, Nature farming soil; C, conventional farming soil. (D) K, Koshihikari cultivar; A, Asahi cultivar. V13 refer to taxonomy labels which not assigned or other category cannot be projected to lower taxonomy.
Root Microbial Community Composition Between Unfertilized Nature Farming and Fertilized Conventional Farming Soils
The unfertilized nature farming soil showed a distinctly different root microbial community structure than that of fertilized conventional farming soil, which was significantly influenced by the application of chemical fertilizer. The microbial community composition analysis between the unfertilized nature farming and fertilized conventional farming soils, irrespective of rice cultivar, revealed that the abundances of Proteobacteria, Firmicutes, Chloroflexi and Spirochaetes were lower under fertilized conventional farming conditions (Figure 4A). This was supported by LEfSe analysis (LDA score>4.0), which showed that the unfertilized nature farming soil had higher numbers of differential taxa than the fertilized conventional farming soil (Figure 4B). The relative abundances of the OTUs of uncultured bacteria (LDA score = 5.32) were observed to be the highest under unfertilized nature farming conditions. This was followed by the genera Anaromyxobacter, Haliangium, Sideroxydans, Methylomonas, Candidatus Methylospira, Bradyrhizobium, Piscinibacter, Curvibacter, and Acidibacter. Notably, the preferential taxa under fertilized conventional farming conditions were Allorhizobium, Asticcacaulis, Dyella, Thermomonas, Massilia, Devosia, Azospirillum, and Magnetospirillum.
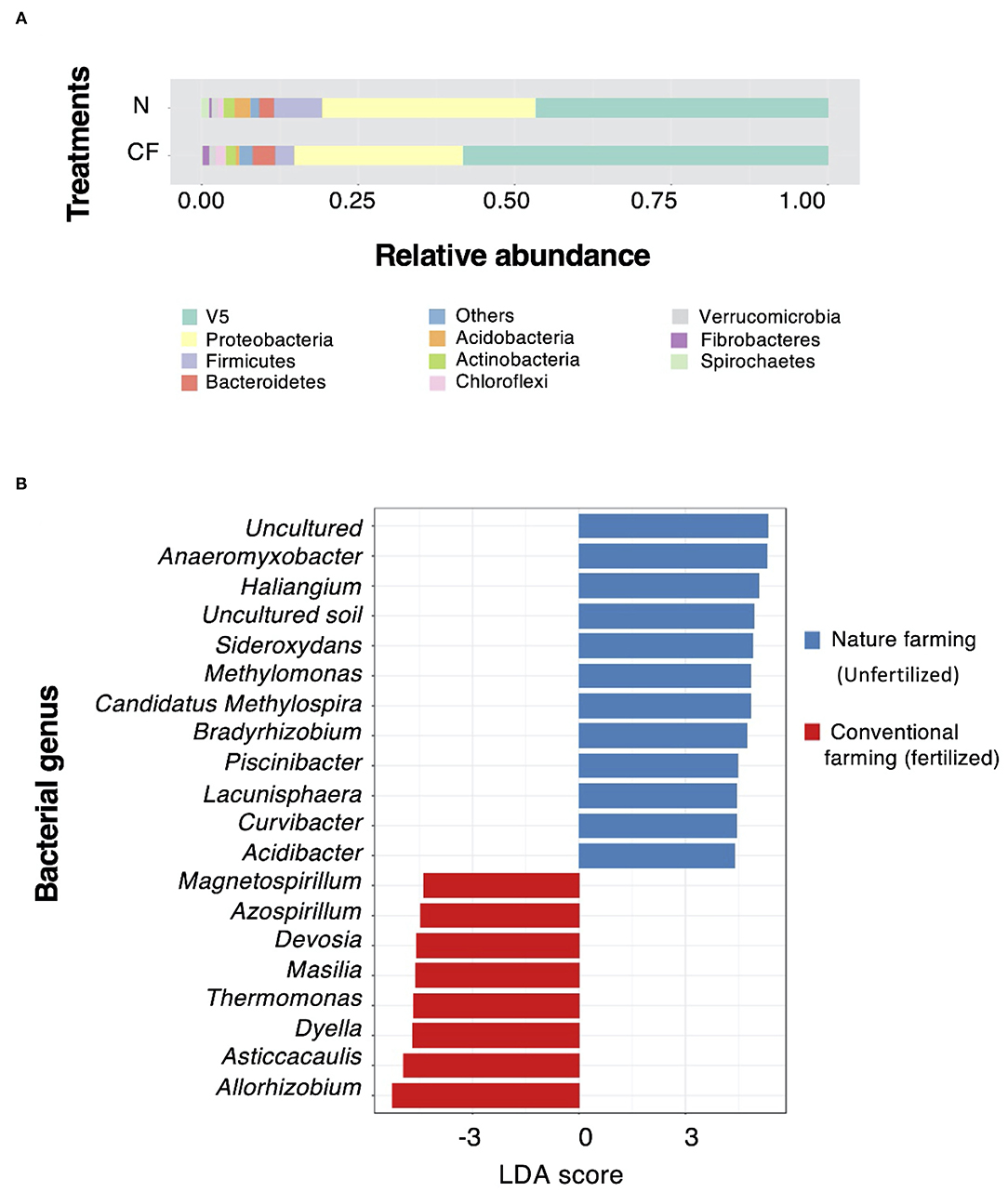
Figure 4. Root microbial community composition between unfertilized nature farming and fertilized conventional farming. (A) Relative abundance of dominant phyla under unfertilized nature farming (N) and fertilized conventional farming (CF) conditions. (B) Linear discriminate analysis (LDA) of the effect size (LEfSe) of the preferential taxa at the genus level under unfertilized nature farming and fertilized nature farming conditions. Only taxa with LDA > 0.4 are shown. V5 refer to taxonomy labels which not assigned or other category cannot be projected to lower taxonomy.
Identification of Bacterial Isolates Associated With Rice Roots Grown Under Nature Farming Conditions
A total of 46 bacteria were isolated from rice roots grown in the nature farming soil. Based on 16S rRNA identification, the isolates were categorized into 16 different genera and grouped into six groups according to the phylogenetic tree (Figure 5): G1, Pseudomonas; G2, Enterobacter, Kosakonia, Flavobacterium, Klebsiella, Aeromonas and Leclercia; G3, Achromobacter; G4, Sinorhizobium and Brevundimonas, G5, Microbacterium; and G6, Bacillus, Paenibacillus, Lysinibacillus, Ureibacillus and Brevibacillus. Of the 46 isolates, 27 isolates were endophytic bacteria and 19 isolates were epiphytic bacteria. The dominant genera were Pseudomonas, Bacillus and Paenibacillus at 35, 20, and 11%, respectively.
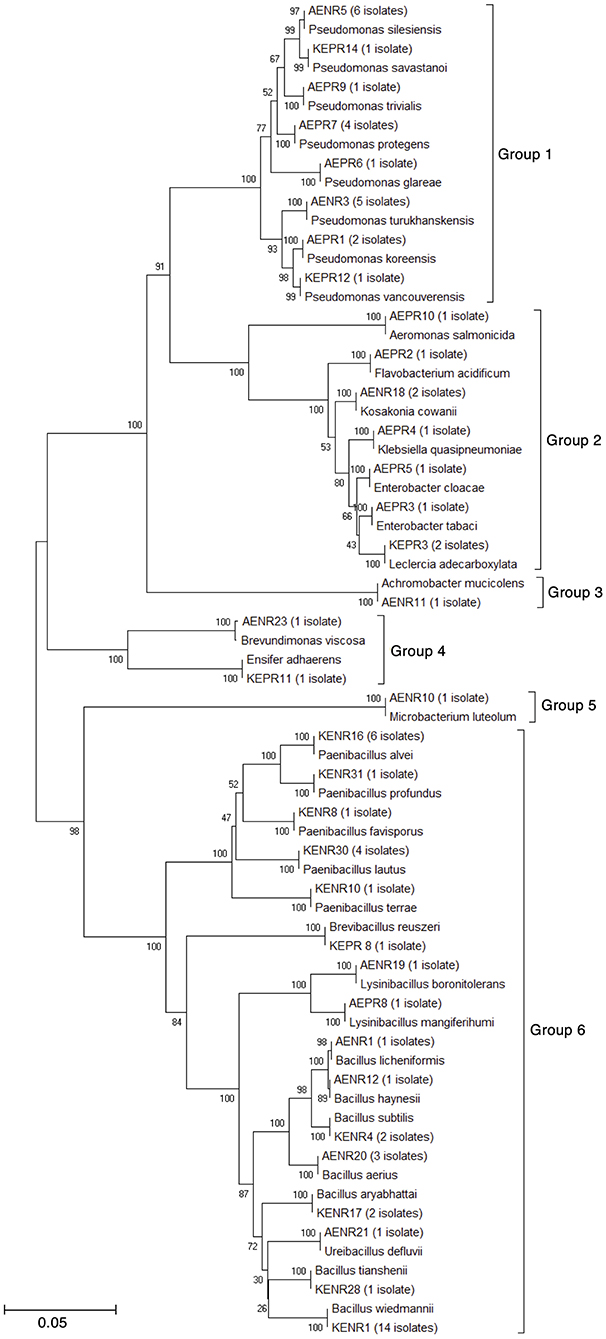
Figure 5. Phylogenetic tree of bacterial isolates associated with rice roots grown under unfertilized nature farming.
Evaluation of Plant Growth-Promoting Traits of Bacterial Isolates Associated With Nature Farming Rice Roots
We evaluated plant growth-promoting (PGP) traits, such as IAA production, P solubilization, K solubilization and siderophore production, of the isolates (Table 1). From the IAA production assays, 13 of the isolates exhibited IAA production traits through the observation of pink or red color development. AENR23 and KEPR3, which were closely identified as B. viscosa and L. adecarboxylata, respectively, showed notable ability to produce IAA according to color change observations.
A total of 23 bacterial isolates were able to solubilize insoluble Ca3(PO4)2 based on the visibility of the halo zone formed around the colony. The size of the clear zone varied between 0.7 and 1.4 cm among the isolates (Table 1, Supplementary Figure 2). Most of the isolates showed better phosphate solubilization activity than that of the previously reported phosphate solubilizer bacterium P. rhizosphere (0.6 cm). AENR18, AENR22 (K. cowanii) and AEPR2 (F. acidificum) isolates formed the largest halo zone, with a diameter of 1.4 cm.
From the potassium solubilization assays, 25 isolates exhibited solubilization activity of insoluble sericite mica and showed better activity than that of the previously reported potassium solubilizer bacterium M. yunnanensis (0.7 cm) (Table 1, Supplementary Figure 3). The size of the halo zone formed varied between 0.7 and 2.3 cm among the isolates. AEPR2 and AEPR3 isolates, which were closely identified as F. acidificum and E. tabaci, formed the largest halo zone, with a diameter of 2.3 cm.
For the siderophore production assay, 22 isolates exhibited siderophore production activity by the formation of a yellow zone around the colony. The size of the yellow zone formed varied between 1.0 and 2.9 cm among the isolates (Table 1, Supplementary Figure 4). AEPR1 (P. koreensis) and AEPR3 (E. tabaci) isolates recorded the largest yellow zone diameters of 2.9 and 2.7 cm, respectively.
Among the 46 isolates, 35 isolates showed IAA production, P- and K-solubilization and siderophore production abilities in qualitative assays. Twenty isolates exhibited three PGP traits, and the remaining isolates exhibited two or fewer PGP traits. Most of the isolates belonging to G1 (Pseudomonas sp.) exhibited more PGP traits than isolates from the other groups. However, only 2 isolates, AEPR1 and AEPR2, which were closely identified as P. koreensis and Flavobacterium acidificum, respectively, were capable of carrying out all four assays.
Effects of Bacterial Inoculation on Rice Growth in Low-Nutrient Media
Six potential isolates (AENR22, AEPR1, AEPR2, AEPR6, AEPR7, and KEPR14) were selected for the plant inoculation test based on their PGP activity. The inoculation effects of the selected isolates were evaluated on rice plants grown in low-nutrient media for 7 days.
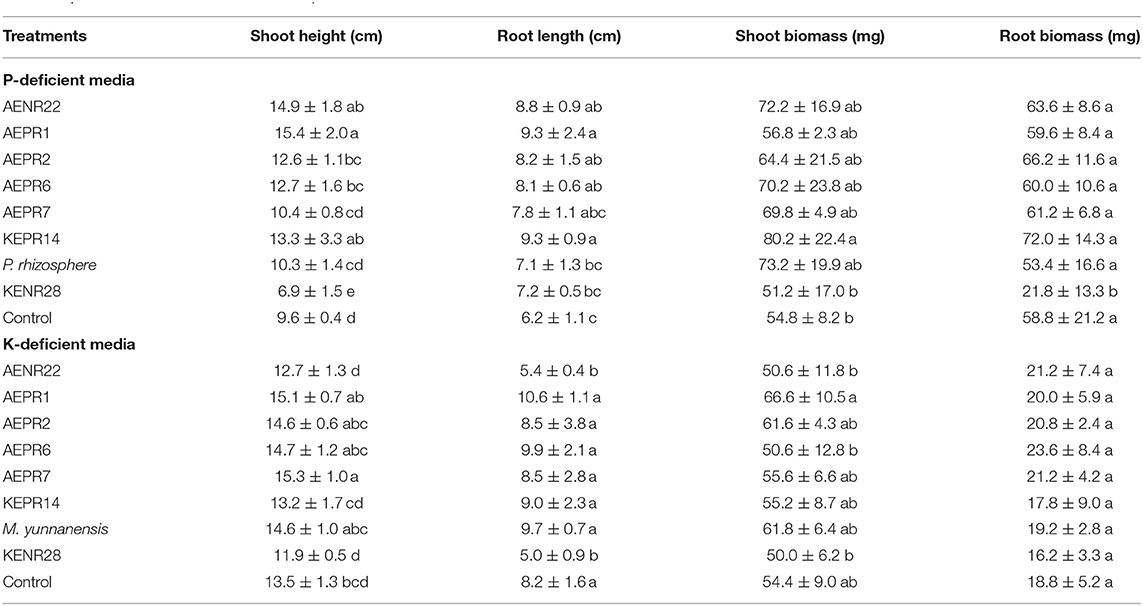
Under phosphate-deficient conditions, most of the isolates significantly promoted shoot height and root length (Table 2). Among the isolates, AEPR1 (P. koreensis) significantly increased the shoot height and root length by 15.4 and 9.3 cm, respectively. This was followed by KEPR14 (P. savastanoi), which also significantly increased the root length (9.3 cm) and shoot biomass (80.2 mg). There was no significant difference in the root biomass of the inoculated and non-inoculated plants. However, most of the inoculated plants showed an increase in root biomass compared with that of the control treatment. Most of the isolates associated with nature farming were able to promote rice growth under p-deficient conditions.
For the potassium-deficient media (Table 2), among the treatments, only AEPR7 (P. protegens) significantly increased the shoot height. AEPR7 increased the shoot height by 15.3 cm compared to the control at 13.5 cm. AENR22 significantly decreased the root length to 5.4 cm compared to the control at 8.2 cm. The inoculated plants showed no significant difference in shoot and root biomass compared with the control treatment. Among all the tested isolates, AEPR1, which was closely identified as P. koreensis, was able to promote rice seedling growth on both phosphorus- and potassium-limited media. Plants inoculated with isolate KENR28 showed the least activity in promoting rice growth compared to other tested isolates associated with nature farming because of its lower phosphorus and potassium solubilization activity.
Discussion
Root microbiomes host a large variety of bacteria, many of which are associated with the host plant and enhance plant nutrition, stress tolerance or health. Previous studies have focused on soil microbial communities under organically or naturally managed farms (Li F. et al., 2017; Liao et al., 2018, 2019), but a limited number of studies have reported on plant-associated microbes. Elucidating whether farming practices, such as nature farming vs. conventional practices that employ chemical fertilizers, can influence root microbial communities is therefore of considerable interest. The present study targeted the root microbial community associated with rice plants cultivated under nature farming practices.
Overall, our results showed that the diversity and structures of the root microbial community were significantly influenced by fertilization practices, in which unfertilized nature farming had distinctly more diverse and abundant root microbial community structures than fertilized conventional farming. We further explored the differential microbial taxa that may possess important ecological functions in corresponding farming systems. Moreover, we identified bacterial isolates associated with nature farming systems that have PGP traits and are able to promote rice growth under low nutrient conditions. Our results will contribute to the augmentation of the available knowledge on root-associated microbes of rice plants, especially those of nature farming.
Root Microbial Community Composition Under Different Farming Practices
Root microbial abundance and diversity are fundamental to plant health and productivity. Our results revealed lower microbial richness and diversity under chemical fertilizer application. Huang et al. (2019) reported that long-term application of chemical fertilizers reduced the richness and diversity of soil microbes. On the other hand, organically managed fields had more positive effects on alpha-diversity parameters than conventionally managed fields (Hartmann et al., 2015; Gu et al., 2017; Liao et al., 2019). Xia et al. (2015) reported that the diversity of the endophytic community associated with plants was higher under organic practices than under conventional practices. The microbial diversity difference between these two systems might have differences in the rhizosphere microbiome that were influenced by the presence of chemical fertilizers, which could presumably result in differential bacterial colonization into the plant roots. We suggest that the application of chemical inputs may be the main factor that alters root microbial alpha-diversity.
Our findings demonstrated that farming type led to a significant variation in the root microbiome structure, although the microbial alpha-diversity exhibited relatively low variability, and this effect could be influenced by the application of inorganic fertilizers. This was in line with previous findings in which long-term fertilization greatly influenced the root microbial community structure, although their microbial alpha-diversity was less impacted by different cropping systems (Hartman et al., 2018; Bai et al., 2020). The variation in microbial community structures may not necessarily influence microbial diversity or richness, as the changes in some taxonomic groups may be compensated by changes in other groups (Hartmann and Widmer, 2006). Edwards et al. (2015) also reported that organic cultivation had a significant separation from eco-farming or conventional cultivation practices and that this effect was exhibited across the rhizosphere compartment consisting of the rhizosphere, rhizoplane and endosphere. Taken together with previous findings, our results showed that avoidance of chemical fertilization was favorable in shaping the root microbial community.
Root Microbial Community Composition Between Unfertilized Nature Farming and Fertilized Conventional Farming
Our results demonstrated that the application of chemical fertilizer significantly influenced the bacterial community structure. Notably, the unfertilized nature farming soil was enriched for Proteobacteria, Firmicutes, Chloroflexi and Spirochaetes, whereas the fertilized conventional farming soil was enriched in Actinobacteria. Bai et al. (2020) also reported that the phylum Proteobacteria was less diverse in the rhizosphere and root endosphere of walnut with the long-term use of chemical fertilizer, while Actinobacteria was enriched in these regions with fertilization. However, a study by Dai et al. (2018) reported that the abundance of Proteobacteria was significantly increased in response to the addition of chemical fertilizer, which differs from our results.
Furthermore, at the genus level, we observed that the bacterial community structure in the rice roots differed between unfertilized nature farming and fertilized conventional farming soils. LEfSe analysis revealed several taxa to be keystone taxa in unfertilized nature farming and fertilized conventional farming systems, which were linked to special functions. Some of the genera that were more abundant when associated under unfertilized nature farming management play an important role in plant health and development. For example, Haliangium produces haliangicin as an antifungal compound that can inhibit the growth of a wide spectrum of fungi (Fudou et al., 2001), and Bradyrhizobum is known as PGPR and is able to fix atmospheric nitrogen (Greetatorn et al., 2019). A greater abundance of iron-reducing bacteria, including the Sideroxydans, Acidibacter and Anaeromyxobacter genera, was found under unfertilized nature farming conditions than under conventional farming conditions (Hori et al., 2010; Falagán and Johnson, 2014; Jin et al., 2017). Unfertilized nature farming also enriched the genera belonging to Betaproteobacteria, including Curvibacter and Piscinibacter. Most Betaproteobacteria have been reported as PGPR (Vacheron et al., 2013; Bruto et al., 2014). It has been reported that the relative abundances of Curvibacter and Anaeromyxobacter were greater in unfertilized soils than in fertilized soils (Kumar et al., 2018; Hui et al., 2019). Chen et al. (2020) also reported that the relative abundance of the Piscinibacter decreased over a long-term monocropping practice of peanut crops. Our results demonstrated the enrichment of the beneficial bacterial community with unfertilized nature farming. Another interesting result that emerged from this study was that the uncultured bacterial genera (LEfSe score > 4.0) were enriched under unfertilized nature farming, revealing the complexity of the root microbiome under unfertilized nature farming.
Some of the enriched genera under fertilized conventional farming, including Dyella, Thermomonas, Massilia, and Devosia, have been previously reported to be found under long-term fertilization conditions (Zhao et al., 2013; Zheng et al., 2017; O'Brien et al., 2018). The genus Dyella was reported to play an important role in mineral weathering and the bioremediation of hydrocarbon-contaminated soil (Kong et al., 2013; Zhao et al., 2013). The results reveal that unfertilized nature farming might enrich beneficial microbes responsible for plant growth-promoting activity. Low input management was more favorable than fertilization in shaping the beneficial bacterial community structure to improve plant health and growth, highlighting the vital importance of suitable management strategies.
Culturable Bacteria Associated With Nature Farming Systems and Their PGP Characteristics
In the present study, we isolated bacterial strains from rice roots grown under a nature farming system and examined their potential as plant growth-promoting rhizobacteria. The present study revealed that the genera Pseudomonas, Bacillus, and Paenibacillus were the most predominant groups isolated from nature farming systems. These genera were previously reported to be the most dominant and most commonly found in various plant studies (Zahid et al., 2015). Previous studies also reported Pseudomonas and Bacillus as the dominant genera isolated from organic farming (Xia et al., 2015; Armalyte et al., 2019). Rodrigues et al. (2018) reported Burkholderia, Bacillus and Rhizobium as the most dominant genera of diazotrophic bacterial isolated from organically grown sugarcane crops. Xia et al. (2015) also managed to identify Stenotrophomonas sp., Micrococcus sp., Denococcus sp. and Burkholderia sp. to be associated with organic farming systems that are not isolated in our study.
Our results also showed that most of the isolates belonging to the Pseudomonas genus were able to yield positive results in all the PGP tests conducted. This was in line with a previous study that reported that the production of all PGP traits was higher among Pseudomonas genera than among other bacterial groups isolated from organic farming (Sagar et al., 2017). They reported that Pseudomonas isolates possessed IAA production; phosphorus solubilization; siderophore, ammonia and hydrogen cyanide production; and ACC deaminase activity. Many studies have reported the potential of Pseudomonas as a PGP in other crops (Islam et al., 2014; Sharma et al., 2014; Vacheron et al., 2016; Qessaoui et al., 2019). In our study, Pseudomonas isolates showed higher siderophore production activity than those of other genera. Pseudomonas has been previously reported to exhibit siderophore production and antifungal activity against fungal phytopathogens (Sasirekha and Srividya, 2016; Abo-Zaid et al., 2020). Costa et al. (2006) reported that Pseudomonas strains isolated from organically managed maize possessed siderophore production activity and antifungal activity against Ralstonia solanacearum.
Most of the isolates belonged to the Bacilli group in our study, especially Bacillus, Paenibacillus and Lysinibacillus, which exhibited IAA production activity. The production of IAA by bacteria may aid in plant growth, such as cell elongation and rooting. Our results are in accordance with a previous study that reported that Bacilli possessed the ability to produce IAA (Wagi and Ahmed, 2019). In phosphorus and potassium solubilization assays, the isolate identified as F. acidificum exhibited the highest P- and K-solubilizing activity among other isolates. Phosphorus- and potassium-solubilizing bacteria play an important role in converting insoluble phosphate or potassium into their soluble form for plant uptake. Similar findings on the phosphorus solubilizing ability of the Flavobacterium group have been reported (Walitang et al., 2017); however, studies on the roles of Flavobacterium as a potassium solubilizer are limited. Isolates belonging to the Enterobacteriaceae family, such as Enterobacter, Leclercia, Kosakonia, and Klebsiella, were also reported to exhibit phosphorus and potassium solubilization and siderophore production (Chakdar et al., 2018; Pramanik et al., 2018; Bendaha and Belaouni, 2019; Khani et al., 2019). Klebsiella sp. isolated from the organic farming system were reported to be efficient at solubilizing phosphate, producing phytohormones and siderophores and inhibiting the mycelial growth of various phytopathogenic fungi (Melo et al., 2016).
Plant Growth Promotion Effects of Bacterial Isolates Associated With Nature Farming Systems
Based on the PGP assays, we selected six isolates for the plant growth test and evaluated their growth potential on rice plants under low-nutrient media. These isolates positively promoted different growth parameters of rice seedlings. In the plant growth test, AEPR1, which is closely identified as P. koreensis, showed a greater increase in rice seedling growth on both deficient media than the other isolates. P. koreensis was previously reported to enhance the growth of various crops, including rice, sugarcane and vegetable crops (Li H. B. et al., 2017; Hafez et al., 2019; Kang et al., 2019). Moreover, the production of biosurfactants by P. koreensis suppressed Pythium ultimum in tomato and Phytophthora infestans in potato (Hultberg et al., 2010a,b). Kaur and Reddy (2013) reported the potential of other Pseudomonas sp. isolated from an organic field, including Pseudomonas plecoglossicida, which possesses phosphate solubilization abilities and significantly increases plant growth, organic carbon and available P in soils as well as P uptake in plants. The present study suggested the strong potential of these selected bacterial isolates to benefit agriculture, not only through the enhancement of crop growth and yield but also through a reduction in chemical inputs. Further works are required to determine the antagonistic activity and the effectiveness of these bacteria to promote plant growth under field conditions.
The present study provides information on the impacts of unfertilized nature farming on the diversity, composition and PGP role of the root microbial community, which has the potential to improve crop production in the face of increasing chemical fertilizer use. However, it should be noted that our study has limitations, as we only used one source of nature farming soil and focused on comparing the effects of chemical fertilizer application in nature and conventional farming systems. In this regard, our study might not represent the root microbiomes of other nature farming fields and only provides insight into the root microbiomes influenced by chemical fertilizer but not the effects of other farming practices. Previous studies have shown that geographical location and different nature farming management strategies may have distinct effects on plants and soil microbes (Gardner et al., 2011; Lenc et al., 2015; Liao et al., 2019; Zhang et al., 2019; Mohammad Golam Dastogeer et al., 2020). Our study may provide information on root microbial communities, which could serve as a reference for assessing root microbiomes under different farming practices and geographical locations of nature farming systems. We also did not investigate the fungal communities associated with the roots, but future studies might consider examining the effects of nature farming practices on the fungal communities along with their function as PGPs to better understand the root microbiomes and their characteristics and roles under nature farming. The current study also did not investigate the relationship between soil physiochemical properties and root-associated microbes; thus, future studies might need to examine the roles of root microbes in enhancing soil fertility, which is essential for plant health.
In this study, we highlighted the impacts of agricultural systems and chemical fertilizer application on the root microbial communities. On the basis of root microbial richness and diversity, unfertilized nature farming appeared to have a significant advantage over the fertilized conventional farming and also present a distinct root microbial community structure. We demonstrated that different farming practices significantly changed the relative abundance of Proteobacteria, Firmicutes, Chloroflexi, Spirochaetes, and Actinobacteria. Fertilization appeared to have strong impacts on shifting the richness, diversity and microbial community structure of rice root microbiome, which could be important in plant growth and health. In addition, differential taxa were enriched under unfertilized nature farming, suggesting unfertilized management provides a conducive environment for the enrichment of various bacterial taxa. The enriched taxa in unfertilized nature farming, such as Haliangium, Bradyrhizobium and iron-reducing bacteria, have previously been reported to improve plant health and development. In contrast, fertilized conventional farming enriched the abundance of genera related to mineral weathering and bioremediation of hydrocarbon contaminated soil. Moreover, we identified 46 end- and epiphytic bacteria associated with rice roots grown under unfertilized nature farming, with 35 of it possessing PGP traits for such as IAA production, phosphorus- and potassium solubilization, and siderophore production. Among all the tested isolates, P. koreensis AEPR1 appeared to have better activity in enhancing the rice seedling growth on both phosphorus- and potassium-limited media.
The current study will help in understanding the diversity, community structure and functions of root microbial communities in nature farming crops. This study further suggests that PGP bacterial isolates associated with nature farming systems can be promising components of integrated plant health and disease management. The development of such beneficial microbial inocula can serve as agro-inputs in nature or organic farming systems of various crops. Taken together, this study indicates that unfertilized nature farming practices beneficially alter the root microbiome and could be regarded as appropriate management for sustainable agricultural production.
Data Availability Statement
The raw sequencing reads were submitted to DRA/DDBJ under accession no. DRA010459.
Author Contributions
GS, MY, and SO designed research. GS, MY, and YN performed research. CL, KD, HT, HN, and SD contributed new reagents and analytic tools. GS, MY, CL, and KD analyzed data. GS, MY, KD, and SO wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Yanmar Environmental Sustainability Support Association and JSPS KAKENHI Grant Number 19F19102.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Suzuki for her kind help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.629942/full#supplementary-material
References
Abo-Zaid, G. A., Soliman, N. A. M., Abdullah, A. S., El-Sharouny, E. E., Matar, S. M., and Sabry, S. A. F. (2020). Maximization of siderophores production from biocontrol agents, Pseudomonas aeruginosa F2 and Pseudomonas fluorescens JY3 using batch and exponential fed-batch fermentation. Processes 8:455. doi: 10.3390/pr8040455
Armalyte, J., Skerniškyte, J., Bakiene, E., Krasauskas, R., ŠiugŽdiniene, R., Kareiviene, V., et al. (2019). Microbial diversity and antimicrobial resistance profile in microbiota from soils of conventional and organic farming systems. Front. Microbiol. 10:892. doi: 10.3389/fmicb.2019.00892
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Bai, Y. C., Chang, Y. Y., Hussain, M., Lu, B., Zhang, J. P., Song, X. B., et al. (2020). Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8:694. doi: 10.3390/microorganisms8050694
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Bendaha, M. E. A., and Belaouni, H. A. (2019). Tomato growth and resistance promotion by Enterobacter hormaechei subsp. steigerwaltii EB8D. Arch. Phytopathol. Plant Prot. 52, 318–332. doi: 10.1080/03235408.2019.1620511
Bruto, M., Prigent-Combaret, C., Muller, D., and Moënne-Loccoz, Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related proteobacteria. Sci. Rep. 4:6261. doi: 10.1038/srep06261
Chakdar, H., Dastager, S. G., Khire, J. M., Rane, D., and Dharne, M. S. (2018). Characterization of mineral phosphate solubilizing and plant growth promoting bacteria from termite soil of arid region. 3 Biotech 8:463. doi: 10.1007/s13205-018-1488-4
Chen, M., Liu, H., Yu, S., Wang, M., Pan, L., Chen, N., et al. (2020). Long-term continuously monocropped peanut significantly changed the abundance and composition of soil bacterial communities. PeerJ 8:e9024. doi: 10.7717/peerj.9024
Costa, R., Gomes, N. C. M., Peixoto, R. S., Rumjanek, N., Berg, G., Mendonça-Hagler, L. C. S., et al. (2006). Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biol. Biochem. 38, 2434–2447. doi: 10.1016/j.soilbio.2006.03.003
Dai, Z., Su, W., Chen, H., Barberán, A., Zhao, H., Yu, M., et al. (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of actinobacteria and proteobacteria in agro-ecosystems across the globe. Glob. Change Biol. 24, 3452–3461. doi: 10.1111/gcb.14163
de Vries, F. T., and Wallenstein, M. D. (2017). Below-ground connections underlying above-ground food production: a framework for optimising ecological connections in the rhizosphere. J. Ecol. 105, 913–920. doi: 10.1111/1365-2745.12783
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., and Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. doi: 10.1093/nar/gkx295
Ding, L. J., Cui, H. L., Nie, S. A., Long, X. E., Duan, G. L., and Zhu, Y. G. (2019). Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol. Ecol. 95:fiz040. doi: 10.1093/femsec/fiz040
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Eyhorn, F., Muller, A., Reganold, J. P., Frison, E., Herren, H. R., Luttikholt, L., et al. (2019). Sustainability in global agriculture driven by organic farming. Nat. Sustain. 2, 253–255. doi: 10.1038/s41893-019-0266-6
Falagán, C., and Johnson, D. B. (2014). Acidibacter ferrireducens gen. nov., sp. nov.: an acidophilic ferric iron-reducing gammaproteobacterium. Extremophiles 18, 1067–1073. doi: 10.1007/s00792-014-0684-3
Fudou, R., Iizuka, T., and Yamanaka, S. (2001). Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium. 1. Fermentation and biological characteristics. J. Antibiot. 54, 149–152. doi: 10.7164/antibiotics.54.149
Gardner, T., Acosta-Martinez, V., Senwo, Z., and Dowd, S. E. (2011). Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity 3, 308–328. doi: 10.3390/d3030308
Glickmann, E., and Dessaux, Y. (1995). A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. doi: 10.1128/AEM.61.2.793-796.1995
Greetatorn, T., Hashimoto, S., Sarapat, S., Tittabutr, P., Boonkerd, N., Uchiumi, T., et al. (2019). Empowering rice seedling growth by endophytic Bradyrhizobium sp. SUTN9-2. Lett. Appl. Microbiol. 68, 258–266. doi: 10.1111/lam.13114
Gu, Y., Wang, Y., Lu, S., Xiang, Q., Yu, X., Zhao, K., et al. (2017). Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil. Front. Microbiol. 8:1516. doi: 10.3389/fmicb.2017.01516
Guo, Z., Han, J., Li, J., Xu, Y., and Wang, X. (2019). Effects of long-term fertilization on soil organic carbon mineralization and microbial community structure. PLoS ONE 14:e0211163. doi: 10.1371/journal.pone.0211163
Hafez, E. M., Alsohim, A. S., Farig, M., Omara, A. E. D., Rashwan, E., and Kamara, M. M. (2019). Synergistic effect of biochar and plant growth promoting Rhizobacteria on alleviation of water deficit in rice plants under salt-affected soil. Agronomy 9:847. doi: 10.3390/agronomy9120847
Hartman, K., van der Heijden, M. G. A., Wittwer, R. A., Banerjee, S., Walser, J. C., and Schlaeppi, K. (2018). Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14. doi: 10.1186/s40168-018-0456-x
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hartmann, M., and Widmer, F. (2006). Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 72, 7804–7812. doi: 10.1128/AEM.01464-06
Hori, T., Müller, A., Igarashi, Y., Conrad, R., and Friedrich, M. W. (2010). Identification of iron-reducing microorganisms in anoxic rice paddy soil by 13C-acetate probing. ISME J. 4, 267–278. doi: 10.1038/ismej.2009.100
Hu, Q. P. (2011). A simple double-layered chrome azurol S agar (SD-CASA) plate assay to optimize the production of siderophores by a potential biocontrol agent Bacillus. Afr. J. Microbiol. Res. 5, 4321–4327. doi: 10.5897/AJMR11.238
Hu, X., Chen, J., and Guo, J. (2006). Two phosphate- and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J. Microbiol. Biotechnol. 22, 983–990. doi: 10.1007/s11274-006-9144-2
Huang, R., McGrath, S. P., Hirsch, P. R., Clark, I. M., Storkey, J., Wu, L., et al. (2019). Plant-microbe networks in soil are weakened by century-long use of inorganic fertilizers. Microb. Biotechnol. 12, 1464–1475. doi: 10.1111/1751-7915.13487
Hui, C., Liu, B., Wei, R., Jiang, H., Zhao, Y., Liang, Y., et al. (2019). Dynamics, biodegradability, and microbial community shift of water-extractable organic matter in rice–wheat cropping soil under different fertilization treatments. Environ. Pollut. 249, 686–695. doi: 10.1016/j.envpol.2019.03.091
Hultberg, M., Alsberg, T., Khalil, S., and Alsanius, B. (2010a). Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis. BioControl 55, 435–444. doi: 10.1007/s10526-009-9261-6
Hultberg, M., Bengtsson, T., and Liljeroth, E. (2010b). Late blight on potato is suppressed by the biosurfactant-producing strain Pseudomonas koreensis 2.74 and its biosurfactant. BioControl 55, 543–550. doi: 10.1007/s10526-010-9289-7
Ikeda, S., Watanabe, K. N., Minamisawa, K., and Ytow, N. (2004). Evaluation of soil DNA from arable land in Japan using a modified direct-extraction method. Microbes Environ. 19, 301–309. doi: 10.1264/jsme2.19.301
Islam, F., Yasmeen, T., Ali, Q., Ali, S., Arif, M. S., Hussain, S., et al. (2014). Influence of Pseudomonas aeruginosa as PGPR on oxidative stress tolerance in wheat under Zn stress. Ecotoxicol. Environ. Saf. 104, 285–293. doi: 10.1016/j.ecoenv.2014.03.008
Jin, L., Lee, C. S., Ahn, C. Y., Lee, H. G., Lee, S., Shin, H. H., et al. (2017). Abundant iron and sulfur oxidizers in the stratified sediment of a eutrophic freshwater reservoir with annual cyanobacterial blooms. Sci. Rep. 7:43814. doi: 10.1038/srep43814
Kang, S. M., Adhikari, A., Lee, K. E., Park, Y. G., Shahzad, R., and Lee, I. J. (2019). Gibberellin producing rhizobacteria Pseudomonas koreensis MU2 enhance growth of lettuce (Lactuca sativa) and Chinese cabbage (Brassica rapa, chinensis). J. Microbiol. Biotechnol. Food Sci. 9, 166–170. doi: 10.15414/jmbfs.2019.9.2.166-170
Kaur, G., and Reddy, M. S. (2013). Phosphate solubilizing rhizobacteria from an organic farm and their influence on the growth and yield of maize (Zea mays L.). J. Gen. Appl. Microbiol. 59, 295–303. doi: 10.2323/jgam.59.295
Khani, A. G., Enayatizamir, N., and Masir, M. N. (2019). Impact of plant growth promoting rhizobacteria on different forms of soil potassium under wheat cultivation. Lett. Appl. Microbiol. 68, 514–521. doi: 10.1111/lam.13132
Kong, C., Wang, L., Li, P., Qu, Y., Tang, H., Wang, J., et al. (2013). Genome sequence of Dyella ginsengisoli strain LA-4, an efficient degrader of aromatic compounds. Genome Announc. 1, e00961–e00913. doi: 10.1128/genomeA.00961-13
Kumar, U., Kumar Nayak, A., Shahid, M., Gupta, V. V. S. R., Panneerselvam, P., Mohanty, S., et al. (2018). Continuous application of inorganic and organic fertilizers over 47 years in paddy soil alters the bacterial community structure and its influence on rice production. Agric. Ecosyst. Environ. 262, 65–75. doi: 10.1016/j.agee.2018.04.016
Lenc, L., Kwaśna, H., Sadowski, C., and Grabowski, A. (2015). Microbiota in wheat roots, rhizosphere and soil in crops grown in organic and other production systems. J. Phytopathol. 163, 245–263. doi: 10.1111/jph.12313
Li, F., Chen, L., Zhang, J., Yin, J., and Huang, S. (2017). Bacterial community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Front. Microbiol. 8:187. doi: 10.3389/fmicb.2017.00187
Li, H. B., Singh, R. K., Singh, P., Song, Q. Q., Xing, Y. X., Yang, L. T., et al. (2017). Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 8, 1268. doi: 10.3389/fmicb.2017.01268
Li, R., Khafipour, E., Krause, D. O., Entz, M. H., de Kievit, T. R., and Fernando, W. G. D. (2012). Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PLoS ONE 7:e51897. doi: 10.1371/journal.pone.0051897
Liao, J., Liang, Y., and Huang, D. (2018). Organic farming improves soil microbial abundance and diversity under greenhouse condition: a case study in Shanghai (Eastern China). Sustainability 10:3825. doi: 10.3390/su10103825
Liao, J., Xu, Q., Xu, H., and Huang, D. (2019). Natural farming improves soil quality and alters microbial diversity in a cabbage field in Japan. Sustainability 11:3131. doi: 10.3390/su11113131
Melo, J., Carolino, M., Carvalho, L., Correia, P., Tenreiro, R., Chaves, S., et al. (2016). Crop management as a driving force of plant growth promoting rhizobacteria physiology. Springerplus 5, 1574–1574. doi: 10.1186/s40064-016-3232-z
Mohammad Golam Dastogeer, K., Oshita, Y., Yasuda, M., Kanasugi, M., Matsuura, E., Xu, Q., et al. (2020). Host specificity of endophytic fungi from stem tissue of nature farming tomato (Solanum lycopersicum Mill.) in Japan. Agronomy 10:1019. doi: 10.3390/agronomy10071019
Nanzyo, M. (1997). “Available phosphate,” in Methods of Soil Environment Analysis (Dojyo-Kankyo-Bunsekihou), ed Editorial Committee for Methods of Soil Environment Analysis (Dojyo-Kankyo-Bunsekihou-Henshu-Iinkai) (Tokyo: Hakuyu-sha), 267–273.
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
O'Brien, F. J. M., Dumont, M. G., Webb, J. S., and Poppy, G. M. (2018). Rhizosphere bacterial communities differ according to fertilizer regimes and cabbage (Brassica oleracea var. capitata L.) harvest time, but not aphid herbivory. Front. Microbiol. 9:1620. doi: 10.3389/fmicb.2018.01620
Pramanik, K., Mitra, S., Sarkar, A., Soren, T., and Maiti, T. K. (2018). Characterization of a Cd2+-resistant plant growth promoting rhizobacterium (Enterobacter sp.) and its effects on rice seedling growth promotion under Cd2+-stress in vitro. Agric. Nat. Resour. 52, 215–221. doi: 10.1016/j.anres.2018.09.007
Qessaoui, R., Bouharroud, R., Furze, J. N., El Aalaoui, M., Akroud, H., Amarraque, A., et al. (2019). Applications of new rhizobacteria Pseudomonas Isolates in agroecology via fundamental processes complementing plant growth. Sci. Rep. 9:12832. doi: 10.1038/s41598-019-49216-8
Rhine, E. D., Mulvaney, R. L., Pratt, E. J., and Sims, G. K. (1998). Improving the berthelot reaction for determining ammonium in soil extracts and water. Soil Sci. Soc. Am. J. 62, 473–480. doi: 10.2136/sssaj1998.03615995006200020026x
Rodrigues, A. A., AraÚJo, M. V. F., Soares, R. S., De Oliveira, B. F. R., Ribeiro, I. D. A., Sibov, S. T., et al. (2018). Isolation and prospection of diazotrophic rhizobacteria associated with sugarcane under organic management. An. Acad. Bras. Ciênc. 90, 3813–3829. doi: 10.1590/0001-3765201820180319
Sagar, A., Debbarma, V., Abraham, T. K, and Shukla, P.W.. (2017). Functional diversity of soil bacteria from organic agro ecosystem. Int. J. Curr. Microbiol. Appl. Sci. 6, 3500–3518. doi: 10.20546/ijcmas.2017.612.408
Sasirekha, B., and Srividya, S. (2016). Siderophore production by Pseudomonas aeruginosa FP6, a biocontrol strain for Rhizoctonia solani and Colletotrichum gloeosporioides causing diseases in chilli. Agric. Nat. Resour. 50, 250–256. doi: 10.1016/j.anres.2016.02.003
Sharma, A., Shankhdhar, D., Sharma, A., and Shankhdhar, S. C. (2014). Growth promotion of the rice genotypes by pgprs isolated from rice rhizosphere. J. Soil Sci. Plant Nutr. 14, 505–517. doi: 10.4067/S0718-95162014005000040
Suzuki, K., Takemura, M., Miki, T., Nonaka, M., and Harada, N. (2019). Differences in soil bacterial community compositions in paddy fields under organic and conventional farming conditions. Microbes Environ. 34, 108–111. doi: 10.1264/jsme2.ME18101
Takahashi, Y., and Yoshida, T. (2008). Studies on the paddy rice cultivar “Ashi-no-yume” method of fertilizer application in tomo area of gunma prefecture. Jpn. J. Crop. Sci. 77, 84–93. doi: 10.1626/jcs.77.84
Tkacz, A., and Poole, P. (2015). Role of root microbiota in plant productivity. J. Exp. Bot. 66, 2167–2175. doi: 10.1093/jxb/erv157
Vacheron, J., Desbrosses, G., Bouffaud, M. L., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. doi: 10.3389/fpls.2013.00356
Vacheron, J., Moënne-Loccoz, Y., Dubost, A., Gonçalves-Martins, M., Muller, D., and Prigent-Combaret, C. (2016). Fluorescent pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front. Plant Sci. 7:1212. doi: 10.3389/fpls.2016.01212
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., and Nasrulhaq Boyce, A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21:573. doi: 10.3390/molecules21050573
Wagi, S., and Ahmed, A. (2019). Bacillus spp.: potent microfactories of bacterial IAA. PeerJ 7:e7258. doi: 10.7717/peerj.7258
Walitang, D. I., Kim, K., Madhaiyan, M., Kim, Y. K., Kang, Y., and Sa, T. (2017). Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of Rice. BMC Microbiol. 17:209. doi: 10.1186/s12866-017-1117-0
Wang, W., Wang, H., Feng, Y., Wang, L., Xiao, X., Xi, Y., et al. (2016). Consistent responses of the microbial community structure to organic farming along the middle and lower reaches of the Yangtze river. Sci. Rep. 6:35046. doi: 10.1038/srep35046
Wasai, S., and Minamisawa, K. (2018). Plant-associated microbes: from rhizobia to plant microbiomes. Microbes Environ. 33, 1–3. doi: 10.1264/jsme2.ME3301rh
Xia, Y., DeBolt, S., Dreyer, J., Scott, D., and Williams, M. A. (2015). Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 6:490. doi: 10.3389/fpls.2015.00490
Xia, Y., Sahib, M. R., Amna, A., Opiyo, S. O., Zhao, Z., and Gao, Y. G. (2019). Culturable endophytic fungal communities associated with plants in organic and conventional farming systems and their effects on plant growth. Sci. Rep. 9:1669. doi: 10.1038/s41598-018-38230-x
Xu, Y., Tang, H., Xiao, X., Li, W., Li, C., Sun, G., et al. (2018). Effects of long-term fertilization management practices on soil microbial carbon and microbial biomass in paddy soil at various stages of rice growth. Rev. Bras. Ciênc. Solo 42:e0170111. doi: 10.1590/18069657rbcs20170111
Zahid, M., Abbasi, M. K., Hameed, S., and Rahim, N. (2015). Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Front. Microbiol. 6:207. doi: 10.3389/fmicb.2015.00207
Zhang, H., Zheng, X., Bai, N., Li, S., Zhang, J., and Lv, W. (2019). Responses of soil bacterial and fungal communities to organic and conventional farming systems in East China. J. Microbiol. Biotechnol. 29, 441–453. doi: 10.4014/jmb.1809.09007
Zhao, F., Guo, X. Q., Wang, P., He, L. Y., Huang, Z., and Sheng, X. F. (2013). Dyella jiangningensis sp. nov., a γ-proteobacterium isolated from the surface of potassium-bearing rock. Int. J. Syst. Evol. Microbiol. 63, 3154–3157. doi: 10.1099/ijs.0.048470-0
Keywords: nature farming, root microbiome, plant growth-promoting bacteria, rice, sustainable agriculture
Citation: Sinong GF, Yasuda M, Nara Y, Lee CG, Dastogeer KMG, Tabuchi H, Nakai H, Djedidi S and Okazaki S (2021) Distinct Root Microbial Communities in Nature Farming Rice Harbor Bacterial Strains With Plant Growth-Promoting Traits. Front. Sustain. Food Syst. 4:629942. doi: 10.3389/fsufs.2020.629942
Received: 16 November 2020; Accepted: 31 December 2020;
Published: 28 January 2021.
Edited by:
Saveetha Kandasamy, A&L Canada Laboratories, CanadaReviewed by:
Wu Xiong, Utrecht University, NetherlandsAgnieszka Barbara Najda, University of Life Sciences of Lublin, Poland
Copyright © 2021 Sinong, Yasuda, Nara, Lee, Dastogeer, Tabuchi, Nakai, Djedidi and Okazaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shin Okazaki, c29rYXpha2lAY2MudHVhdC5hYy5qcA==
 Grace Flavyeliz Sinong
Grace Flavyeliz Sinong Michiko Yasuda
Michiko Yasuda Yoshiyuki Nara2
Yoshiyuki Nara2 Chol Gyu Lee
Chol Gyu Lee Khondoker Mohammad Golam Dastogeer
Khondoker Mohammad Golam Dastogeer Shin Okazaki
Shin Okazaki