- 1Department of Plant Sciences, University of the Free State, Bloemfontein, South Africa
- 2Maize Improvement Program, CSRI-Savanna Agricultural Research Institute, Tamale, Ghana
- 3Plant Breeding Program, ARC-Grain Crops, Potchefstroom, South Africa
- 4Zambia Seed Company Limited (Zamseed), Lusaka, Zambia
Introduction: Biofortified maize varieties could contribute to the fight against hunger and malnutrition of the increasing human population and help meet the high demand of maize for human consumption, industrial use and feed for animal and poultry. The understanding of the genetic mechanisms conditioning the inheritance of grain yield and other agronomic and quality traits is essential in the development of superior maize genotypes. The main objective of this study was to determine the combining ability for grain yield and other agronomic traits of QPM and non-QPM inbred lines crossed with two QPM and two non-QPM testers.
Materials and methods: A total of 130 hybrids were obtained by crossing 10 non-QPM and 23 QPM lines with four elite testers (two QPM and two non-QPM). The 130 single cross hybrids were evaluated at 13 sites in 2018 and 2019.
Results and discussion: The results showed significant general combining ability (GCA) for lines and testers as well as significant specific combining ability (SCA) for hybrids for most of the measured traits. Non-additive gene action controlled the inheritance of grain yield while agronomic and quality traits were controlled by additive gene action. QPM lines 11, 14 and 28 and non-QPM tester CML444 showed desirable GCA effects for grain yield, indicating that these lines in combination with tester CML444 should be considered when targeting development of superior maize genotypes with QPM traits.
Conclusions: Based on the SCA values, crosses 120, 108, 105, 99, 85, and 41 were identified as the best hybrids across the locations. It is recommended that maize breeding programs targeting the development of high yielding QPM hybrids should exploit QPM lines with high GCA values for heterosis to be realized. In addition, the identified superior hybrids may be further evaluated and consequently promoted for commercial release which could increase incomes of farmers and help to alleviate poverty, hunger and malnutrition in southern Africa and sub-Saharan Africa at large.
Introduction
Maize (Zea mays L.) ranks among the top three most important cereal crops worldwide based on its yield potential and wide adaptability. In maize breeding programs, inbred lines are used to generate hybrids for commercial production. Because of their increased yield potential, hybrid maize varieties are used worldwide to attain higher grain yield. In breeding, it is essential to understand how inbred lines or parents combine for the development of superior hybrids (Amegbor et al., 2017; de Abreu et al., 2019). Vacaro et al. (2002) and Wegary et al. (2013) noted that the performance of any genetic material is dependent on its potential per se performance as well as the combining ability of the lines in crosses. However, since heterosis for superior performance is controlled primarily by specific combining ability (SCA), lines should be selected based on their SCA with testers. Combining ability studies generate useful information on the genetic mechanisms governing how quantitative traits are inherited. This serves as a guide for plant breeders in selecting superior parental lines for further crop improvement or use in hybrid production for commercial purposes (Abuali et al., 2012; Ali et al., 2012; Saif-ul-Malook et al., 2016).
Two types of combining abilities are commonly used in breeding and biometrical genetics, and these are general combining ability (GCA) and SCA (Abuali et al., 2012; Haydar and Paul, 2014). The GCA is the average contribution an inbred makes to hybrid performance in a series of hybrid combinations in comparison to other inbreds in the same series of hybrid combinations and this is due to additive gene action. The SCA, on the other hand, is the contribution of an inbred to hybrid performance in a cross with a specific inbred in relation to its contributions in crosses with an array of other inbreds (Haydar and Paul, 2014). In developing hybrids, one must choose an appropriate mating design to generate the hybrids. Even though there are several mating designs used in plant breeding, they all come with advantages and disadvantages. Several researchers use the diallel and North Carolina mating designs. However, it is very expensive to use a diallel when there is a high number of lines used in generating the hybrids. Therefore, in that case it is better to use the line x tester mating design which estimates GCA and SCA and has been extensively used in studies (Nduwumuremyi et al., 2013; Wegary et al., 2014; Tilahun et al., 2019).
Developing maize hybrids with improved nutritional quality through biofortification can help improve nutritional deficiencies in sub-Sahara Africa (SSA) and other developing regions. This is attainable if biofortified crop varieties are produced and readily accessible by the end users (De Groote et al., 2010, 2014). Globally, two billion individuals have been reported to be adversely affected by micronutrient deficiencies and there are about 800 million malnourished people in the world (Global Nutrition Report, 2017; Hossain et al., 2019).
There are several studies on QPM under stress and non-stress conditions in Southern Africa and this has led to the release of about 40 QPM varieties and hybrids in South Africa, Zambia, Lesotho, Zimbabwe, Malawi and Mozambique (Twumasi-Afriye et al., 2016). In spite of this large number varieties and hybrids that were released, only two varieties are being grown, thus ZS261 and Q623 in Zimbabwe and South Africa, respectively (Goredema-Matongera et al., 2021). Consumption of maize products with enhanced lysine and tryptophan content could help alleviate the growing challenge posed by malnutrition in several parts of the world (Menkir et al., 2008; Mpofu et al., 2012). Several authors have reported on the nutritional advantages of QPM in feeding experiments involving livestock, especially monogastric animals; and children (Mpofu et al., 2012). Children who were fed with meals made from QPM were healthier than those fed with food products from normal maize (Gunaratna et al., 2010; Tandzi et al., 2017).
Research on daily intake of QPM food products of primary school children in countries such as Malawi, Lesotho and Zambia have shown its importance in providing sufficient amounts of essential amino acids in human diets (Nuss and Tanumihardjo, 2011; Tandzi et al., 2017). Several studies of QPM combining ability under varying environmental conditions, with the aim to identify QPM donors and testers using several mating designs, have been reported (Mbuya et al., 2011; Amin et al., 2014; Sarika et al., 2018). QPM genotypes generally had lower grain yield, therefore, the major aim of QPM breeding programs currently is to improve both grain yield and nutritional value instead of focusing only on quality traits (Li and Vasal, 2016). Even though there are variations between QPM and non-QPM genotypes for insect attacks, kernel rots and moisture content, there are no differences in physico-chemical properties such as starch and oil contents (Li and Vasal, 2016; Synrem et al., 2017).
Even though several studies have been conducted on QPM for grain yield and nutritional characteristics to address hidden hunger in resource limited populations, important information on the combining ability of genotypes for these quality traits is lacking for the QPM inbred lines recently developed by the International Maize and Wheat Improvement Center (CIMMYT) Zimbabwe maize breeding program. Information on combining ability of lines used in crop development will identify superior lines to be crossed to develop hybrids with improved yield and quality traits. Genetic enhancement of grain yield and its associated traits depends on the nature and the degree of genetic variation in available germplasm. Grain yield is the most important trait of interest to the breeder and farmer. However, grain yield is a polygenically inherited trait determined by several genes. Amegbor et al. (2022a) observed that the grain yield of QPM crosses with non-QPM was higher than non-QPM crossed with non-QPM and QPM crossed with QPM. It is therefore necessary to understand the mode of inheritance of yield in QPM and non-QPM inbred lines. The aim of this study was to determine the combining ability of QPM and non-QPM lines with QPM and non-QPM testers for grain yield and quality trait performance.
Materials and methods
Germplasm, field evaluation, and data collection
A total of 33 inbred lines (23 QPM and 10 non-QPM) and four inbred testers were obtained from CIMMYT, Zimbabwe. The inbred lines were developed from germplasm with varied genetic backgrounds by conversion of early and medium maturing elite maize with tolerance to drought, low N and maize streak virus (MSV) with introgression genes from temperate germplasm (Supplementary Table 1). The four testers were crossed with the 33 inbred lines to create 132 single cross hybrids. Two of the crosses were not included in the evaluation due to insufficient seed (Supplementary Table 2).
Experimental sites, design, agronomic practices and obtaining seed samples for laboratory analysis
The trial was planted at 13 sites in the 2017/2018 and 2018/2019 seasons in two countries in Southern Africa. At Zimbabwe, the trial was planted at eight sites during 2017/2018 cropping season. In South Africa, the study was conducted at five sites. These included two sites at Potchefstroom and one site at Cedara during the 2017/2018 cropping season. During the 2018/2019 cropping season, the trial was repeated at one location each at Potchefstroom and Cedera (Supplementary Table 3).
At all sites, the trials were planted in a 5 x 26 alpha lattice design with two replications. Each plot consisted of a single 4 m row, with a 0.75 and 0.25 m inter and intra-row spacing, respectively. Seeding rates were two kernels per sowing hill, with one seedling maintained per hill at 28 days after sowing. The final plant population was 53,333 plants per hectare. At 4 weeks after sowing (4 WAS), compound fertilizer (NPK) 3:2:1 (25) + Zn was used for basal application at Potchefstroom at a rate of 200 kg ha−1. For top-dressing, limestone ammonium nitrate (LAN) containing 28% N was applied at 8 WAS at a rate of 100 kg ha−1. The compound fertilizer used at Cedara was monoammonium phosphate (MAP) applied at a rate of 250 kg ha−1. A total of 150 kg ha−1 of LAN was top-dressed two times at 4 WAS and 8 WAS. Recommended fertilizer for optimal maize production was adopted for the trials established in Zimbabwe with 250 kg ha−1 N, 83 kg ha−1 P and 111 kg ha−1 K. NPK was applied at 4 WAS and supplementary N was top-dressed at 8 WAS. Weeds were controlled using chemical and cultural methods at all sites.
Collection of samples for biochemical analyses
Seeds for laboratory analysis were generated in the field by self-pollinating two ears in each experimental unit for four sites in South Africa (Potchefstroom 2017/2018 and 2018/2019; and Cedera 2017/2018 and 2018/2019) and two sites from Zimbabwe (Harare and Glendale 2017/2018). Because the fields were planted with QPM and non-QPM, the main purpose for the self-pollination was to prevent xenia effect. Cobs from each experimental unit were dried after harvest and the kernels were bulked after shelling for determination of tryptophan, starch, moisture, protein and oil. In this study, lysine was not determined because of the strong positive correlation reported between lysine and tryptophan in several studies (Vivek et al., 2008; Sarika et al., 2017).
Determination of tryptophan
For tryptophan determination, 80 mg of defatted flour was weighed and digested with 3 ml of 4 mg ml−1 papain solution for 16 h. The hydrolysate was mixed with 0.1 M glyoxylic acid, 1.8 mM ferric chloride and 7N H2SO4 as described by Nurit et al. (2009). Absorbance of tryptophan was detected at 560 nm via a mass spectrophotometer (Jenway Spectrophotometer Model 7315). The concentration of tryptophan (%) was estimated using a standard regression as described by Nurit et al. (2009). Absorbance of tryptophan was detected at 560 nm via a mass spectrophotometer (Jenway Spectrophotometer Model 7315). The concentration of tryptophan (%) was estimated using a standard regression as described by Nurit et al. (2009).
Determination of protein, starch, oil, and moisture content
A 500 g sample of maize seeds was taken from each experimental unit after harvesting and drying of the seeds. Relative contents of grain protein, starch, oil and moisture contents were estimated (percentage weight basis, %wt) using a Grain Analyzer, Model DA 7250 (Perten Instruments, Sweden). The measurement of the characteristics was based on the calibration of this near-infrared spectrometer (NIR) by Agri-Environ Solutions (www.aelab.co.za).
Estimation of quality index
The protein QI was estimated as the concentration of tryptophan over protein content expressed as:
Trait measurement
Data was obtained for days to 50% pollen shed (DA) and days to 50% silk emergence (DS). Anthesis-silking interval (ASI) was estimated as the interval between DS and DA. Other traits recorded were plant height, ear height, root lodging, stalk lodging, plant aspect and ear aspect. At harvest, ears per plant (EPP) was estimated by dividing the total number of ears harvested per experimental unit by the plant stand recorded at harvest. In addition, ear rot per plot was scored and grain yield (t ha−1) was estimated based on 80% shelling percentage following Amegbor et al. (2022b).
Data analysis
The data obtained from the 13 sites were combined to estimate GCA and SCA for the field experiment while the laboratory data was obtained from six sites. Line x tester analysis was performed for both field and laboratory experiments. The GCA and SCA and their standard errors were computed for grain yield and other measured traits using the PROC Mixed Procedure in SAS version 9.4 (SAS, 2011). The variation among hybrids was partitioned into sources due to lines, testers and line x testers. GCA of testers (male) was obtained based on their performance in the F1 hybrid combinations with all possible lines (females). Similarly, GCA of each line was also determined based on the performance of F1 hybrids with all possible testers. GCA and SCA effects were determined for each agronomic trait for each location and across locations following the procedure described by Kaur et al. (2019).
Results
Analysis of variance for general and specific combining ability at Potchefstroom, Cedara, and Zimbabwe
The ANOVA for combining ability for trials conducted at Potchefstroom during 2018 and 2019 cropping seasons showed significant effects of line and tester and for line x tester for all the traits examined except for stalk lodging and husk cover (Table 1). At Cedara, line x tester SCA effect was also significant for most of the traits except stalk lodging and husk cover. Apart from grain yield, ear aspects and root lodging, other traits measured were not significant for site x line x tester.
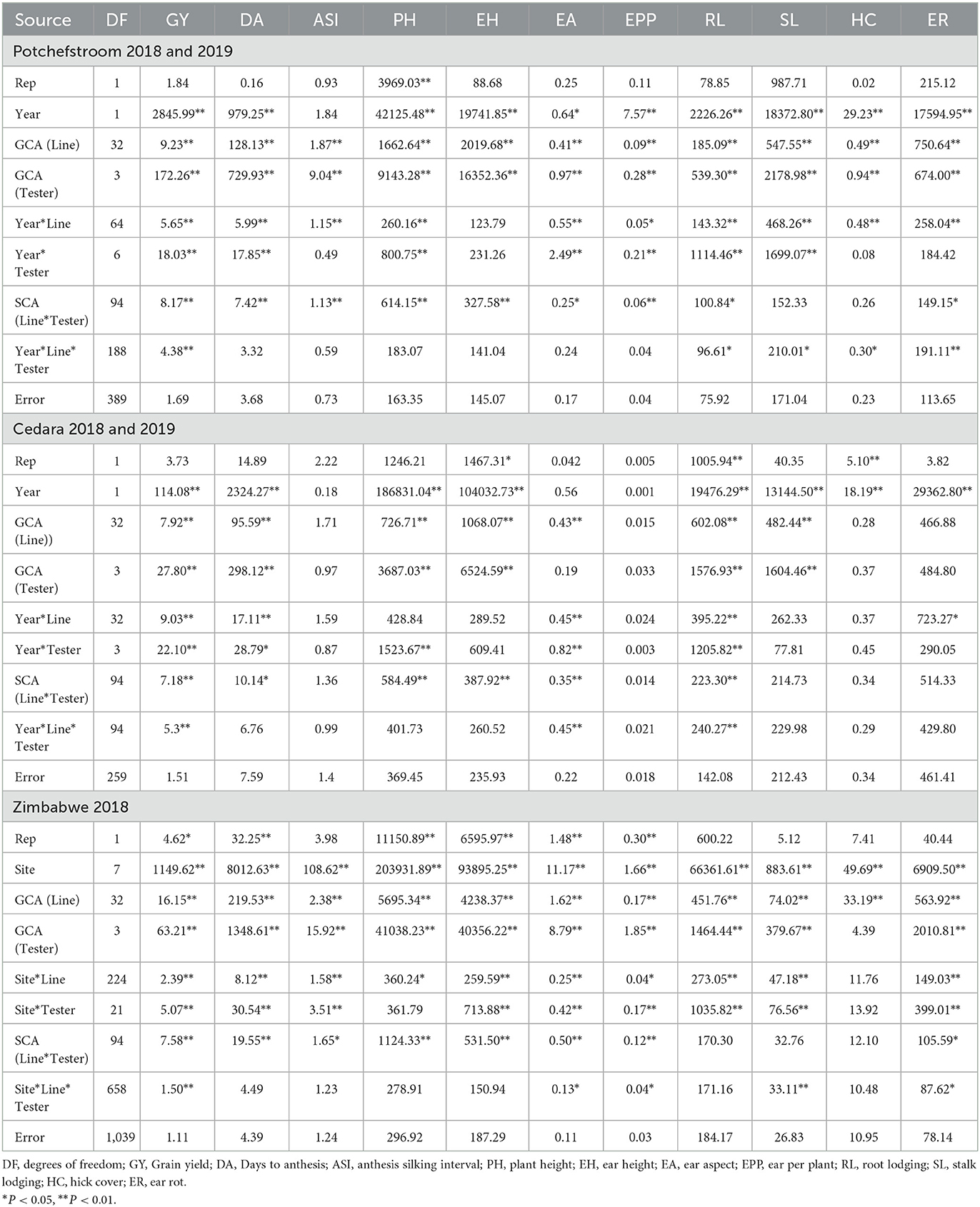
Table 1. Mean squares from analysis of variance for general and specific combining ability for grain yield and other agronomic traits of QPM lines at Potchefstroom, Cedara and Zimbabwe during the 2018 and 2019 cropping seasons.
At Cedara, ANOVA for the two seasons revealed highly significant (P < 0.01) line GCA for grain yield, days to pollen shed, plant and ear heights, ear aspect, root and stalk lodging. GCA for tester was not significant for anthesis-silking interval, ear aspect, ears per plant, husk cover and number of ear rots (Table 1). Site significantly interacted with line for grain yield, days to pollen shed, ear aspect, root lodging and ear rot, while the site x tester effect was significant for grain yield, days to pollen shed, plant height, ear aspect and root lodging.
The ANOVA of the trials conducted at eight sites in Zimbabwe showed significant line GCA for all the traits measured (Table 1). Similarly, there was significant tester GCA for all the traits examined with the exception of husk cover. Site x line effect was also significant for all the traits measured, except for husk cover, while, apart from plant height and husk cover, site x tester effect was significant for all measured traits. The SCA for line x tester was not significant for root lodging, stalk lodging and husk cover. Site x line x tester effect in Zimbabwe trials was significant for grain yield, ear aspect, ears per plant, stalk lodging and ear rot.
Analysis of variance for GCA and SCA of 130 hybrids evaluated across 13 locations
Site mean squares were significant (P < 0.05) in the combined ANOVA for all measured traits (Table 2). The GCA of lines was significant (P < 0.05) for all the measured traits while the GCA of the testers was also significant (P < 0.01) for all the measured traits, except husk cover. Site x line effect was only significant (P < 0.05) for anthesis-silking interval, ear aspect, husk cover and root lodging, while site x tester effect was significant for the majority of the traits measured, except days to pollen shed, plant height, husk cover and ear rot. The line x tester SCA was significant for grain yield, anthesis-silking interval, plant and ear heights, ear aspect, husk cover, root lodging and ears per plant. However, root lodging was the only trait with significant site x line x tester effects. Across the 13 locations, percentage sum of squares due to SCA was higher than that due to GCA for grain yield, anthesis-silking interval, ears per plant and husk cover. On the other hand, GCA sum of squares primarily explained variation for days to pollen shed, plant and ear height, ear aspect, root and stacking lodging and ear rot.
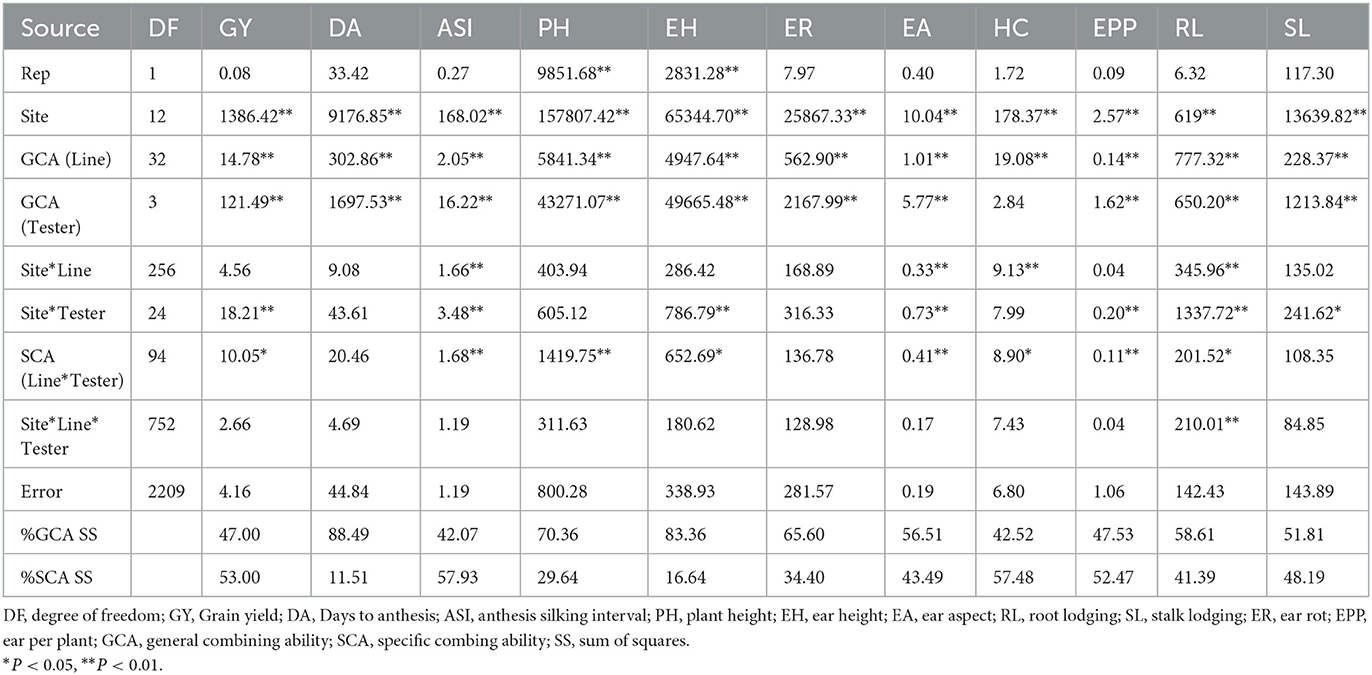
Table 2. Mean squares from analysis of variance for general and specific combining ability for grain yield and other agronomic traits across 13 locations in South Africa and Zimbabwe during the 2018 and 2019 cropping seasons.
GCA effects of lines for grain yield and traits measured from the field experiments across 13 locations
For grain yield, lines 11, 13, 14, and 28 had positive and significant GCA while lines 16, 18, and 32 had significant (P < 0.05) negative GCA for grain yield (Table 3). A total of 15 lines, representing 45.45% of the total lines, had significant and negative GCA for days to pollen shed. For anthesis-silking interval, only lines 20 and 32 GCA were negative and significant (P < 0.05). Lines 11, 26, 28, and 29 showed significant and negative GCA effects for ear aspect. Five lines representing 15.15% of the total, recorded positive and significant GCA for ears per plant. None of the lines had negative and significant GCA effects for root lodging while lines 10, 11, and 21 showed positive and significant GCA for root lodging. Lines 8 and 32 had negative and significant (P < 0.05) GCA for stalk lodging while lines 5, 6, and 18 recorded positive and significant GCA. For ear rot, lines 14 and 20 had positive and significant GCA while line VL0510130 exhibited negative and significant GCA effects.
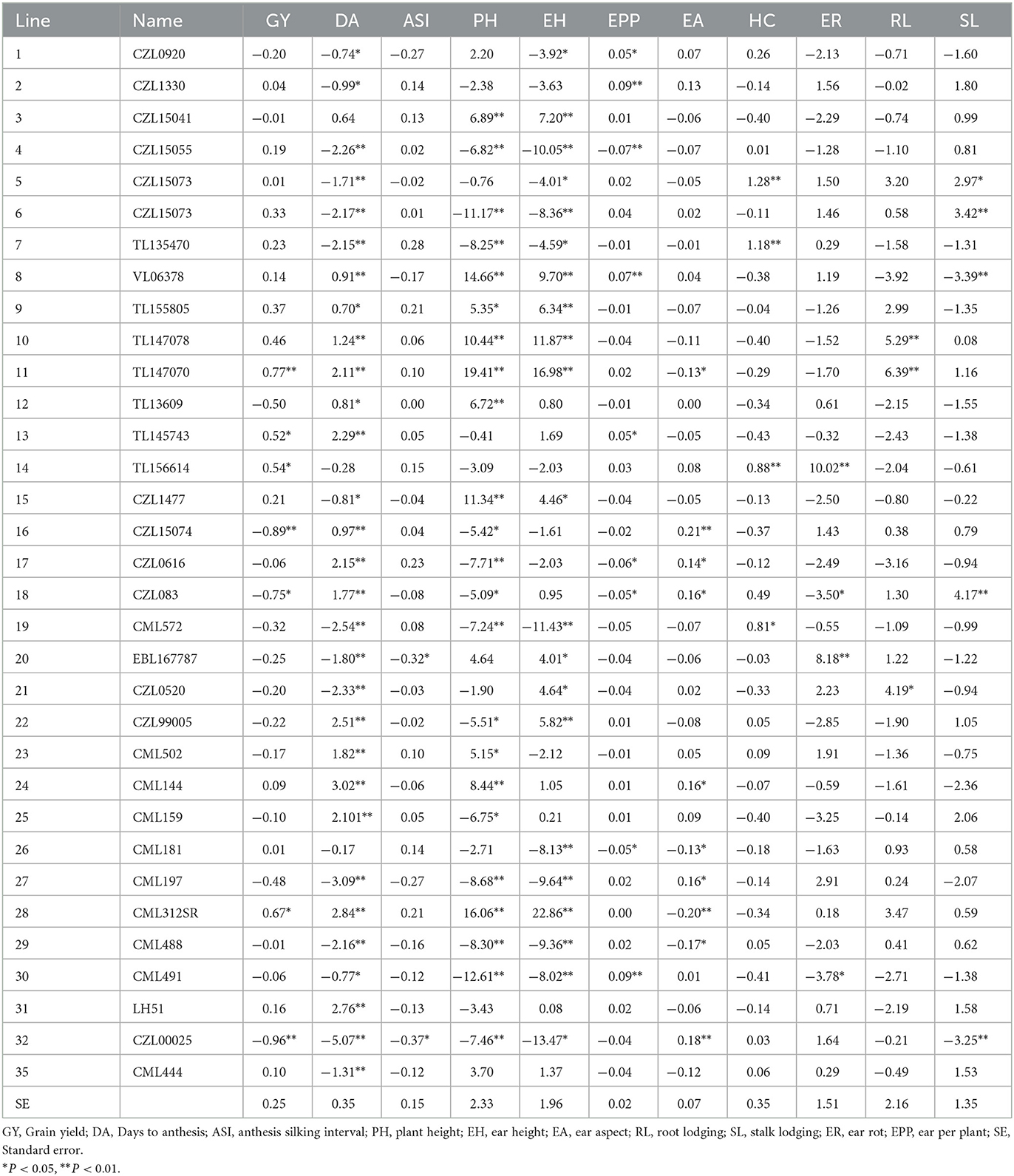
Table 3. General combining ability effects of 33 lines for grain yield and other traits evaluated at 13 locations during the 2018 and 2019 seasons in South Africa and Zimbabwe.
GCA effects of testers for grain yield and traits measured from the field experiments across 13 locations
QPM tester 2 had negative and significant GCA for grain yield while non-QPM tester 4 had positive and significant (P < 0.01) GCA for grain yield (Table 4). For days to pollen shed, testers 1 and 4 recorded negative effects while testers 2 and 3 had significant positive GCA effects. Tester 3 had significant positive effects for anthesis-silking interval, while tester 4 had a significant negative effect. QPM tester 2 recorded significant positive GCA effects for ear aspect and number of ears per plant. Non-QPM tester 3 had significant negative GCA effects for ear aspect and number of ears per plant. None of the testers displayed neither positive and significant nor significant negative GCA effects for root lodging. QPM testers 1 and 2 had positive and significant GCA values for husk cover, while the non-QPM tester had negative and significant GCA effects for husk cover. Tester 1 recorded positive and significant GCA effects for ear rot, while tester 2 had negative and significant effects for ear rot.
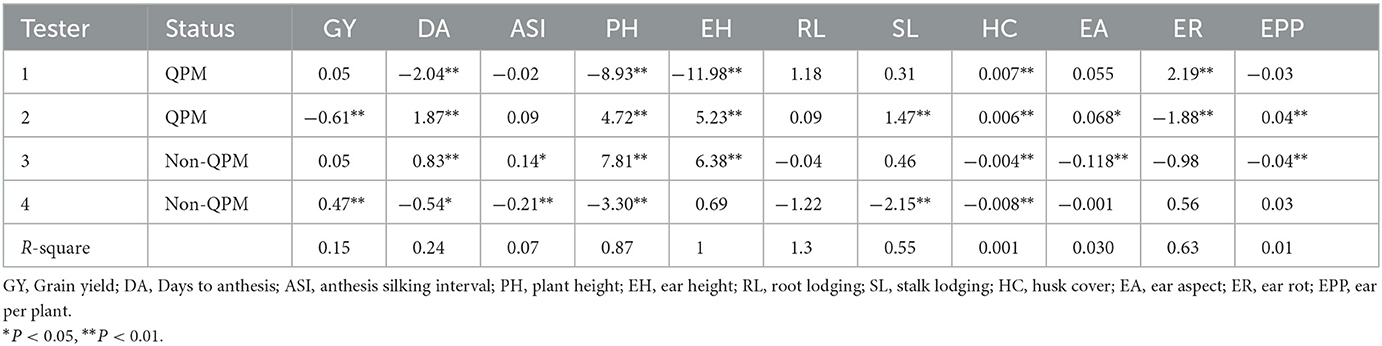
Table 4. General combining ability effects of four testers for grain yield and other traits evaluated at 13 locations during the 2018 and 2019 seasons.
SCA effects for the crosses for grain yield and traits measured from the field experiments across 13 locations
The SCA values for the crosses ranged from −3.00 to 1.17 for yield. Thirty crosses, representing 23.08% of the total, showed both positive and negative SCA effects (Supplementary Table 4). Of these, 17 crosses had positive and significant effects while 13 crosses showed negative and significant SCA effects. Hybrid 22 x 2 (entry 85) showed the highest SCA effects for grain yield. Twelve hybrids showed significant and negative SCA effects for days to pollen shed. For anthesis-silking interval, only four crosses showed significant negative SCA effects while for ear aspect, only nine crosses of the 130, had negative and significant SCA effects (Supplementary Table 4). Only eight hybrids recorded significant positive SCA effects for number of ears per plant with hybrid 18 x 1 having the highest value. Across the 13 locations, only hybrid 10 x 1 out of the 130 crosses recorded significant and negative SCA effects for root lodging, while for stalk lodging, three crosses had positive and significant SCA effects and 6 x 4 had negative and significant SCA effects for stalk lodging. The SCA of ear rot, ranged from −5.33 for 21 x 1 to 6.82 for 8 x 4. Of all the hybrids, only three had significant and negative SCA effects and three also recorded positive and significant SCA values for ear rot.
Analysis of variance for general and specific combining ability of lines and testers evaluated for quality characteristics at Potchefstroom, Cedara and Zimbabwe
The ANOVA for combining ability for the seed samples from Cedara during the 2017/2018 and 2018/2019 seasons showed significant line GCA and tester GCA for all the traits measured, except tester GCA for moisture content (Table 5). Apart from oil content, there were significant line x tester (SCA) effects for all the quality traits. With the exception of protein and starch content, all the quality traits showed significant site x line x tester effects (Table 5).
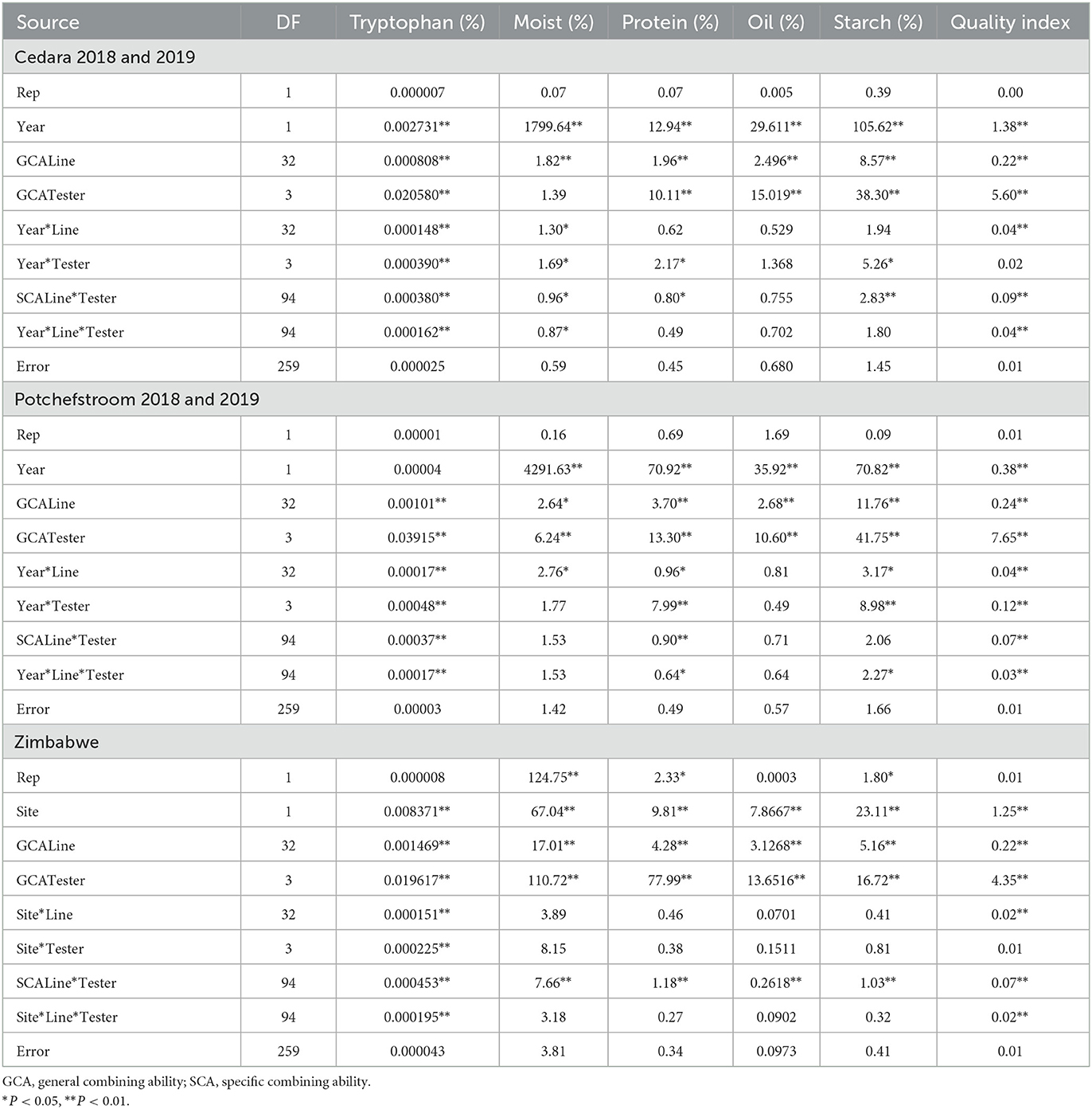
Table 5. Mean squares from analysis of variance for general and specific combining ability for six quality traits of 33 inbred lines and four testers at Potchefstroom, Cedara and Zimbabwe during the 2017/2018 and 2018/2019 cropping seasons.
Analysis of the Potchefstroom trials showed significant (P < 0.01) GCA for lines and testers for all the quality traits (Table 5). The SCA for line x tester was significant only for tryptophan, protein, oil and fiber. Site x line x tester effect was only significant for tryptophan, protein and starch (Table 5).
The ANOVA for combining ability from two sites in Zimbabwe showed significant (P < 0.01) line GCA and significant tester GCA for all the traits that were analyzed. The site x line x tester effect was significant only for tryptophan, while there was a significant site effect for all the quality traits (Table 5). There was significant SCA for line x tester for all quality traits (Table 5).
Analysis of variance of GCA and SCA of 33 lines and four testers for six quality traits across six environments
The combined ANOVA showed significant genotype effects for all the quality traits and QI (P < 0.05). GCA of lines as well as testers were significant for all the traits (P < 0.01). The SCA of line x tester was also significant for all the quality traits and QI (Table 6). Similar observations were recorded for site, site x line and site x tester for all the traits analyzed. Apart from oil and fiber contents, all quality traits showed significant site x line x tester effects across the six environments. The percentage sum of squares for GCA was higher than for SCA for all the quality traits analyzed and calculated QI, indicating dominance of additive effects over non-additive gene effects for the traits (Table 6).
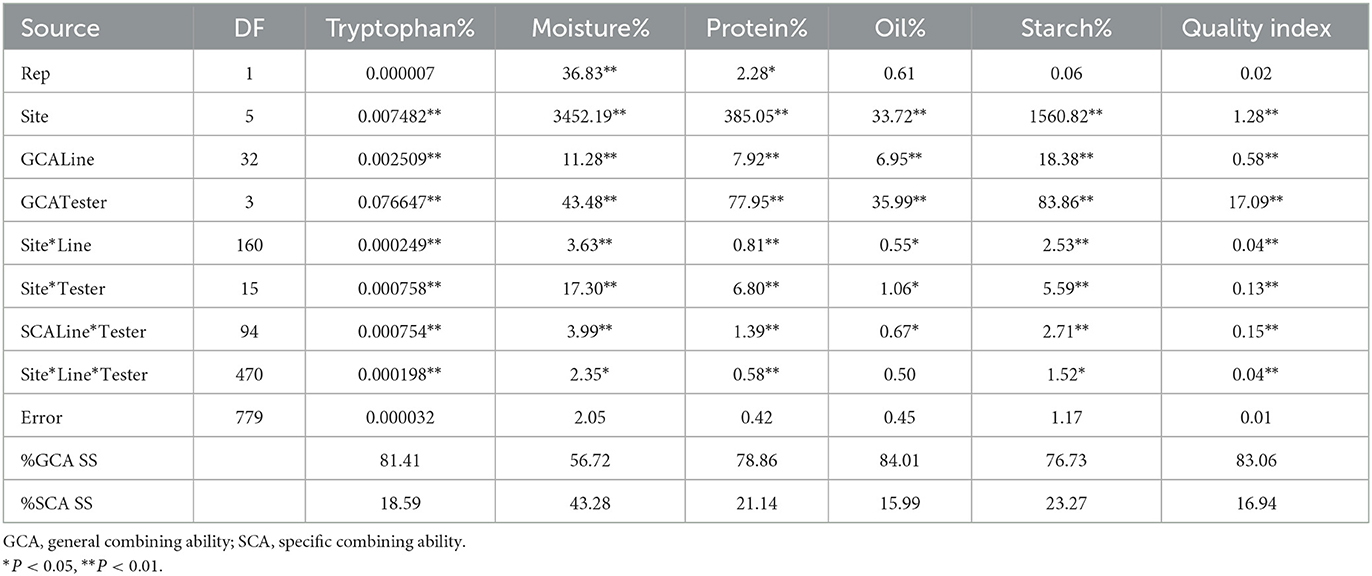
Table 6. Mean squares from combined analysis of variance for general and specific combining ability for six quality traits of 33 inbred lines and four testers across six sites during the 2017/2018 and 2018/2019 cropping seasons.
GCA effects of 33 lines for six quality traits across six locations
The GCA effects for the lines varied for the quality traits analyzed (Table 7). For tryptophan, only lines 1, 2, 6, 7, 8, 12, 13, 15, 17, and 18 recorded positive and significant GCA, while lines 19, 20, 21, 22, 28, 29, 30 32, and 35 had significant and negative GCA. Lines 4, 7, 10 and 14 had significant and positive GCA for moisture content (Table 6). For protein content, 14 lines (42.42% of entries) had positive and significant GCA effects while 13 lines (33.39% of entries) had negative but significant GCA effects (Table 7). Eight lines (24.24% of entries) recorded positive and significant GCA effects and nine lines (33.39% of entries) had negative and significant GCA for oil content. For fiber content, nine lines had positive and significant GCA effects while 10 lines recorded significant but negative GCA effects. Lines 5, 13, 16, 19, 26, 27, 29 and 31 showed positive and significant GCA effects for starch while nine lines also recorded negative and significant GCA effects for starch (Table 7). Inbred lines 1, 3, 6, 7, 12, 13, 15, 16, 17, 18, and 31 showed positive and significant GCA effects for QI.
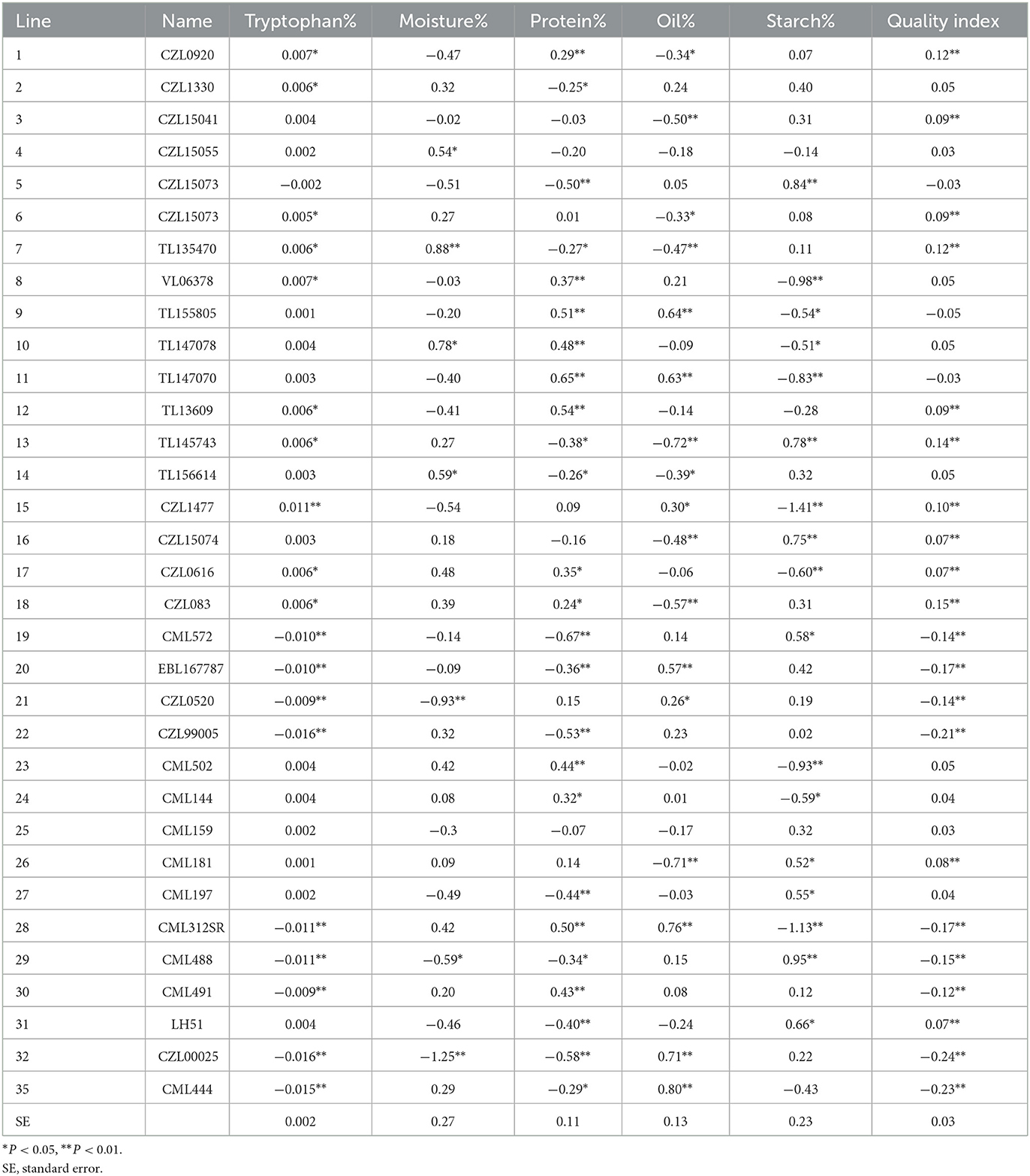
Table 7. General combining ability effects of 33 lines for quality traits across six sites during the 2017/2018 and 2018/2019 cropping seasons in South Africa and Zimbabwe.
SCA effects among the crosses for quality traits across six locations
The SCA values for tryptophan of the crosses ranged from −0.013 to 0.018 for entries 113 and 78, respectively (Supplementary Table 5). A total of 21 hybrids (6 QPM hybrids and 15 non-QPM hybrids) representing 16.15% of the total crosses had significant and positive SCA effects for tryptophan. For moisture content, 11 crosses had significant and positive GCA effects and 11 crosses again showed negative and significant SCA effects. Ten crosses had positive and significant SCA effects for oil content, while four crosses showed negative and significant SCA effects. Hybrid L7 x CML444 (non-QPM) (27) showed the highest SCA effects for oil content. Seventeen hybrids (13.08%) showed significant and positive SCA effects for protein content with that of hybrid L35 x CZL15049 (non-QPM) (127) being the highest. For fiber content, a total of 20 crosses showed significant negative SCA effects while 22 crosses also had positive and significant SCA effects. Hybrids L10 x CML444 (non-QPM) (39) and (L10 x CZL15049 (QPM) (37) recorded the least and the highest SCA values for fiber content, respectively. Only eight hybrids had positive and significant SCA effects for starch content with L15 x CZL059 (QPM) (58) being the highest, while 13 crosses recorded significant and negative SCA effects with L10 x CZL15049 (QPM) (37) recording the lowest value (Supplementary Table 5). The genetic analysis for QI indicated that, 25 hybrids recorded positive and significant SCA. Hybrid 78 (L20 x CML444 non-QPM) recorded the highest SCA value of 0.24 for QI (Supplementary Table 5).
Discussion
The results from this study, involving QPM and non-QPM lines crossed with QPM and non-QPM testers, showed variation and significant differences for GCA for lines and testers and SCA effects for the single cross hybrids. This indicates the presence of additive and non-additive genetic effects conditioning the inheritance of grain yield and most of the characters examined. The variance due to GCA sum of squares and SCA sum of squares as percentage of total sum of squares was estimated to quantify gene effects that determined the inheritance of the traits measured. Non-additive gene effects primarily controlled grain yield. These findings differ from earlier reports by Abbas et al. (2016) and Chiuta and Mutengwa (2020), who found additive gene action controlling the inheritance of grain yield under non-stress conditions. However, the findings from our experiments are consistent with the earlier results reported by Asefa et al. (2008) who found SCA to be a more important contributor to grain yield than GCA. These differences might be due to the varied germplasm used with diverse genetic makeup, as well as the conditions under which the study was carried out, since environment is an important determinant of genotype performance.
Positive and significant GCA is important for grain yield improvement of inbreds, but SCA is most important for superior hybrid performance. Lines or testers with positive GCA values suggest that they will contribute positively to grain yield, while lines and testers with negative GCA are undesirable as they may not contribute positively to the traits of interest.
The significant effects of site, site × GCALine and site × GCATester on grain yield and most of the characters measured suggested that site had a large effect on GCA and that there was a site with GCA interaction for both lines and testers; and GCA effects ranking was different between sites. However, the absence of significant site x line x tester effect on grain yield and some of the other traits examined suggested that the yield performance of the set of hybrids evaluated was constant across the 13 locations. The substantial GCALine, GCATester, and SCALine × Tester mean squares for ears per plant and ear aspect emphasized the crucial roles of additive and non-additive genetic effects in the expression of these secondary traits.
The positive and significant GCA effects recorded for lines 11, 13, 14, and 28 (all QPM) for grain yield. Among the testers, only non-QPM tester 4 showed positive significant GCA for GCA. This suggests that these QPM lines have the prospects to be utilized in breeding superior hybrids when crossed with non-QPM lines. Using these parents should lead to progenies with high grain yield because of being sourced from genetically diverse backgrounds (Fasahat et al., 2016; Amegbor et al., 2022b). Since ear rot is an important trait in breeding for QPM genotypes, lines with negative and significant GCA effects need to be selected and used in making crosses to overcome the issues surrounding soft kernels of QPM genotypes. Lines 18 and 30 have the potential to pass kernel hardness to their progeny, thereby preventing kernel rots (Vancetovic et al., 2015).
In the current study, there was larger contribution of GCA sum of squares for all the quality traits analyzed than the SCA sum of squares. This indicated that the quality traits measured were determined predominantly by additive genetic effects. This is in line with results reported in several studies involving QPM and non-QPM genetic materials for quality traits, particularly tryptophan, protein, starch and oil contents (Njeri et al., 2017; Darshan and Marker, 2019). With the predominance of additive genetic effects and the large genetic variability for the quality traits, selection methods such as recurrent selection using half-sibs can be used to improve this germplasm.
Inbred line CZL1477 (15), with significant positive GCA effects for tryptophan across the six sites, is the only inbred line having the favorable alleles to be used by breeding programs with the aim of developing hybrids with high tryptophan. On the other hand, the two QPM testers (CZL15049 and CZL059) were superior contributors with positive and significant GCA effects for tryptophan content. The significant GCA effects of the 14 parental lines for protein content across the six sites denotes their ability to transfer this trait to their progeny. Therefore, lines with high and stable protein content with significant positive GCA effects are good genetic materials to be used in QPM inbred line recycling programs (Njeri et al., 2017). Similarly, lines and testers having positive and significant GCA effects for oil content are desirable as contributors in breeding for high oil contents. This agrees with the studies by Laude and Carena (2014), Kostadinović et al. (2016) and Synrem et al. (2017). For nutritional benefits, it is desirable to select lines with negative and significant GCA effects for starch and fiber. However, industries with the aim of producing genotypes with high starch content could select lines with significant positive GCA effects for starch and lines with low GCA for fiber contents (Cuevas-Rodríguez et al., 2006; Mamun et al., 2015; Ai and Jane, 2016).
The crosses showing positive and significant specific combinations for tryptophan, protein, and oil contents are desirable for commercial hybrid production and vice versa for the hybrids with negative and significant SCA effects for starch and fiber contents. Even though only 10 inbred lines recorded significant GCA for tryptophan, several hybrids were found to have positive and significant SCA, with hybrids L20 x CML444 (78) and L21 x CML444 (82) being the superior genotypes. Similarly, non-QPM hybrid L35 x CZL15049 (127) was the best genotype for protein content. These findings were similar with the results reported by Wegary et al. (2013), Williams and Kucheryavskiy (2016), Synrem et al. (2017), and Tilahun et al. (2019) on significant SCA effects for quality traits.
The genetic analysis for QI indicated that the majority of the QPM lines recorded positive and significant GCA, while the non-QPM lines recorded negative and significant GCA. Some of the QPM lines such as 5, 9, and 11 recorded non-significant negative GCA. Lines with significant GCA values could be selected for hybrid development with enhanced quality traits. These lines could also be useful in the development of populations. Interestingly, some of the hybrids that are classified as non-QPM for example 78 (L20 x CML444; non-QPM) and 82 (L21 x ML444; non-QPM) showed significant and positive SCA effects for QI, more than the QPM hybrids. This may be linked to one of the parents being QPM for those hybrids, implying that heterosis for QI could be achieved when QPM lines are crossed with non-QPM lines because of the genetic distance the lines and testers. Inheritance of QI traits for the genetic materials studied was controlled by additive genetic effects (Amegbor et al., 2022a). The observed results are consistent with the study reported by Machida et al. (2010) and Ignjatovic-Micic et al. (2013) who also detected the predominance of GCA over SCA.
Conclusions
Significant variations for grain yield, agronomic and quality traits were found for lines, testers, and line x tester. Inbred lines and testers having positive and significant GCA effects for grain yield and quality traits have the potential to transfer traits related to high yield and enhanced nutritional value to their first filial generation. For the field experiments, non-additive genetic effects controlled more than 50% of inheritance of grain yield and the inheritance of anthesis-silking interval, husk cover and ears per plant while additive genetic effects controlled the inheritance of ear rot, plant and ear heights, days to pollen shed, ear aspect as well as root and stalk lodging. Inbred lines 11 and 14 and non-QPM tester CML444 showed desirable GCA effects for grain yield. Inbred lines 1, 2, 6, 7, 8, 12, 13, 15, 17, and 18 and QPM testers CZL15049 and CZL059 showed desirable GCA effects for tryptophan, suggesting these lines and testers should be considered for QPM inbred development for high tryptophan content. The superior hybrids identified for various quality traits will be important for the food and animal feed industries to enhance human and animal nutrition. It is also recommended that these hybrids should be further evaluated across several locations due to the environment's interaction with these quality traits. Inbred lines and testers with good GCA effects for quality traits could be utilized by other maize improvement programs for development of superior QPM hybrids. Promotion and adoption of these superior QPM hybrids for grain yield and quality traits from the present study would contribute significantly to fight against malnutrition in SSA.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
Conceptualization, formal analysis, and writing—original draft: IA and ML. Data curation: IA, NS, ML, and AB. Investigation, methodology, and writing—review and editing: IA, AB, ML, NS, and AT. Project administration: ML and AT. Resources: ML, NS, and AT. Supervision: AB, NS, ML, and AT. Validation: ML, AB, and IA. All authors have read and approved the final version of the manuscript.
Funding
This study was funded by the National Research Foundation of South Africa through the SARChI chair initiative (UID 84647).
Acknowledgments
The authors acknowledge CIMMYT-Zimbabwe for providing us the genetic and assisting in the data collection for the field trials conducted in Zimbabwe. The authors would also like to thank ARC Grain Crops Institute for the technical support given to us at Potchefstroom and Cedera. This is an extract from IA's thesis submitted to the University of the Free State.
Conflict of interest
AT was employed the International Maize and Wheat Improvement Center at the time the study was conducted and is currently employed the Zambia Seed Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1123224/full#supplementary-material
References
Abbas, G., Ahsan, M., Saleem, M., and Ahmad., R. (2016). Inheritance study of different agronomic traits in mung × mash interspecific recombinant inbred lines. J. Animal Plant Sci. 26, 149–155. doi: 10.1016/j.cj.2016.10.005
Abuali, A. I., Abdelmulla, A. A., Khalafalla., M. M., Idris, A. E., and Osman, A. M. (2012). Combining ability and heterosis for yield and yield components in maize. Aus. J. Basic Appl. Sci. 6, 36–41.
Ai, Y., and Jane, J. L. (2016). Macronutrients in corn and human nutrition. Compr. Rev. Food Sci. Food Saf. 15, 581–598. doi: 10.1111/1541-4337.12192
Ali, F., Irfan, A. S., Rahman, H., Mohammad, N., Durrishahwar Muhammad, Y. K., et al. (2012). Heterosis for yield and agronomic attributes in diverse maize germplasm. Aust. J. Crop Sci. 6, 455–462.
Amegbor, I. K., Badu-Apraku, B., and Annor, B. (2017). Combining ability and heterotic patterns of extra-early maturing white maize inbreds with genes from Zea diploperennis under multiple environments. Euphytica 213, 1–16. doi: 10.1007/s10681-016-1823-y
Amegbor, I. K., Van Biljon, A., Shargie, N., Tarekegne, A., and Labuschagne, M. T. (2022a). Grain quality and yield potential of hybrids from quality and non-quality protein maize inbred lines. J. Cereal Sci. 107, 1–6. doi: 10.1016/j.jcs.2022.103544
Amegbor, I. K., Van Biljon, A., Shargie, N., Tarekegne, A., and Labuschagne, M. T. (2022b). Does the quality protein maize trait cause hybrid yield losses? A case study in Southern Africa. Euphytica 218, 1–13. doi: 10.1007/s10681-022-03041-9
Amin, M. N., Amiruzzaman, M., Ahmed, A., and Ali, M. R. (2014). Evaluation of inbred lines of maize (Zea mays L.) through line × tester method. Bang. J. Agri. Res. 39, 675–683. doi: 10.3329/bjar.v39i4.22547
Asefa, B., Mohammed, H., and Zelleke, H. (2008). Combining ability of transitional highland maize inbred lines. East Af. J. Sci. 2, 19–24. doi: 10.4314/eajsci.v2i1.40359
Chiuta, N. E., and Mutengwa, C. S. (2020). Combining ability of quality protein maize inbred lines for yield and morpho-agronomic traits under optimum as well as combined drought and heat-stressed conditions. Agronomy 10, 1–14. doi: 10.3390/agronomy10020184
Cuevas-Rodríguez, E. O., Verdugo-Montoya, N. M., Angulo-Bejarano, P. I., Milán-Carrillo, J., Mora-Escobedo, R., Bello-Pérez, L. A., et al. (2006). Nutritional properties of tempeh flour from quality protein maize (Zea mays L.). LWT 39, 1072–1079. doi: 10.1016/j.lwt.2005.07.003
Darshan, S. S., and Marker, S. (2019). Heterosis and combining ability for grain yield and its component characters in quality protein maize (Zea mays L.) hybrids. Elect. J. Plant Breed. 10, 111–118. doi: 10.5958/0975-928X.2019.00013.9
de Abreu Von Pinho, V. M., Mendes-Resende, É. V., Balestre, M. P., Lima, M. A. C., Santos, H. O., et al. (2019). Combining ability and heterosis of maize genotypes under water stress during seed germination and seedling emergence. Crop Sci. 59, 33–43. doi: 10.2135/cropsci2018.03.0161
De Groote Gunaratna, H., Ergano, N. K., and Friesen, D. (2010). Extension and adoption of biofortified crops: Quality protein maize in East Africa. Paper presented at the Joint 3rd African Association of Agricultural Economists (AAAE) and 48th Agricultural Economists Association of South Africa (AEASA) Conference, Cape Town, South Africa.
De Groote Gunaratna, H., Okuro, N., Wondimu, J., Chege, A. C., and Tomlins, K. (2014). Consumer acceptance of quality protein maize (QPM) in East Africa. J. Sci. Food Agric. 94, 3201–3212. doi: 10.1002/jsfa.6672
Fasahat, P., Rajabi, A., Rad, J. M., and Derera, J. (2016). Principles and utilization of combining ability in plant breeding. Biomet. Biostat. Int. J.4, 1–24. doi: 10.15406/bbij.2016.04.00085
Global Nutrition Report (2017). Available online at: https://globalnutritionreport.org/documents/822/Global_Nutrition_Report_2017.pdf (accessed October 2, 2022).
Goredema-Matongera, N., Ndhlela, T., Magorokosho, C., Kamutando, C. N., van Biljon, A., Labuschagne, M., et al. (2021). Multinutrient Biofortification of Maize (Zea mays L.) in Africa: current Status, Opportunities and Limitations. Nutrients. 13, 1039. doi: 10.3390/nu13031039
Gunaratna, N. S., Groote De Nestel, H., Pixley, P. K. V., and McCabe, G. (2010). Evaluating the impact of biofortification: A meta-analysis of community-based studies on quality protein maize (QPM). Food Policy 35, 202–210. doi: 10.1016/j.foodpol.2009.11.003
Haydar, F. M. A., and Paul, N. K. (2014). Combining ability analysis for different yield trails in maize. Bang. J. Plant Breed. Gen.27, 1–5. doi: 10.3329/bjpbg.v27i1.23973
Hossain, F., Sarika, K., Muthusamy, V., Zunjare, R. U., and Gupta, H. S. (2019). Quality protein maize for nutritional security. In: Quality Breeding in Field Crops. eds A. M. I. Qureshi, Z. A. Dar and S. H Wani (Switzerland:Springer), pp. 217–237. doi: 10.1007/978-3-030-04609-5_11
Ignjatovic-Micic, D., Kostadinovic, M., Stankovic, G., Markovic, K., Vancetovic, J., Bozinovic, S., et al. (2013). Biochemical and agronomic performance of quality protein maize hybrids adapted to temperate regions. Maydica 58, 311–317.
Kaur, N., Singh, B., and Sharma, S. (2019). Comparison of quality protein maize (QPM) and normal maize with respect to properties of instant porridge. LWT-Food Sci. Tech. 99, 291–298. doi: 10.1016/j.lwt.2018.09.070
Kostadinović, M., Ignjatović-Micić, D., Vancetović, J., Ristić, D., Bozinović, S., Stanković, G., et al. (2016). Development of high tryptophan maize near isogenic lines adapted to temperate regions through marker assisted selection—Impediments and benefits. PLoS ONE 11, 1–17. doi: 10.1371/journal.pone.0167635
Laude, T. P., and Carena, M. J. (2014). Diallel analysis among 16 maize populations adapted to the northern U.S. Corn Belt for grain yield and grain quality traits. Euphytica 200, 29–44. doi: 10.1007/s10681-014-1125-1
Li, J. S., and Vasal, S. K. (2016). “Maize: quality protein maize.” in Encyclopedia of Food Grains: Second Edition. eds C. Wrigley, H. Corke, K. Seetharaman and J. Faubion (Elsevier) pp. 420–424. doi: 10.1016/B978-0-12-394437-5.00223-0
Machida, L., Derera, J., Tongoona, P., and MacRobert, J. (2010). Combining ability and reciprocal cross effects of elite quality protein maize inbred lines in subtropical environments. Crop Sci. 50, 1708–1717. doi: 10.2135/cropsci2009.09.0538
Mamun, A. A., Heim, H. P., and Bledzki, A. K. (2015). “The use of maize, oat, barley and rye fibres as reinforcements in composites,” in Biofiber Reinforcements in Composite Materials. Woodhead Publishing, eds O. Faruk and M. Sain (pp. 454–487). doi: 10.1533/9781782421276.4.454
Mbuya, K., Nkongolo, K. K., and Kalonji-Mbuyi, A. (2011). Nutritional analysis of quality protein maize varieties selected for agronomic characteristics in a breeding program. Int. J. Plant Breed. Gen. 5, 317–327. doi: 10.3923/ijpbg.2011.317.327
Menkir, A., White, W. S., Maziya-Dixon, B., Rocheford, T., and Weiping, L. (2008). Carotenoid diversity in tropical adapted yellow maize inbred lines. Food Chem. 109, 521–529. doi: 10.1016/j.foodchem.2008.01.002
Mpofu, I. D. T., Sibanda, S., Shonihwa, A., and Pixley, K. (2012). The nutritional value of quality protein maize for weaner pigs. J. Petrol. Eviron. Biotechnol. 3, 1–4. doi: 10.4172/2157-7463.1000129
Nduwumuremyi, A., Tongoona, P., Habimana, S., and Husbandry, A. (2013). Mating designs: helpful tool for quantitative plant breeding analysis. J. Plant Breed. Genetics 1, 117–129.
Njeri, S. G., Makumbi, D., Warburton, M. L., Diallo, A., Jumbo, M. D. B., and Chemining'wa, G. (2017). Genetic analysis of tropical quality protein maize (Zea mays L.) germplasm. Euphytica 213: 1–19. doi: 10.1007/s10681-017-2048-4
Nurit, E., Tiessen, A., Pixley, K., and Palacios-Rojas, N. (2009). Reliable and inexpensive colorimetric method for deter- mining protein-bound tryptophan in maize kernels. J. Agric. Food Chem. 57, 7233–7238. doi: 10.1021/jf901315x
Nuss, E. T., and Tanumihardjo, S. A. (2011). Quality protein maize for Africa: closing the protein inadequacy gap in vulnerable populations. Adv. Nutri. 2, 217–224. doi: 10.3945/an.110.000182
Saif-ul-Malook, A. Q., Ahsan, M., Shabaz, M. K., Waseem, M., and Aamir, A. (2016). Combining ability analysis for evaluation of maize hybrids under drought stress. J. Nat. Sci. Found. Sri Lanka 44, 223–230. doi: 10.4038/jnsfsr.v44i2.8003
Sarika, K., Hossain, F., Muthusamy, V., Baveja, A., Zunjare, R., Goswami, R., et al. (2017). Exploration of novel opaque16 mutation as a source for high lysine and tryptophan in maize endosperm. Indian J. Gen. Plant Breed. 77:59–64. doi: 10.5958/0975-6906.2017.00008.6
Sarika, K., Hossain, F., Muthusamy, V., Zunjare, R. U., Baveja, A., Goswami, R., et al. (2018). Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci. 272, 142–152. doi: 10.1016/j.plantsci.2018.04.014
Synrem, G. J., Marker, S., and Ramteke, P. W. (2017). Gene action and combining ability analysis for grain yield and quality parameters in sub-tropical maize (Zea mays L.). Vegetos 30, 139–147. doi: 10.5958/2229-4473.2017.00048.9
Tandzi, L. N., Mutengwa, C. S., Ngonkew, E. L. M., Woïn, N., and Gracen, V. (2017). Breeding for quality protein maize (QPM) varieties: a review. Agronomy 7, 1–16. doi: 10.3390/agronomy7040080
Tilahun, B., Azimach, G., Keno, T., Chibsa, T., Garoma, B., Abebe, B., et al. (2019). Test cross performance and combining ability of newly introduced quality protein maize (Zea mays) inbred lines for grain yield and agronomic traits evaluated in mid-altitude agro-ecological zones of Ethiopia. South Af. J. Plant Soil 36, 173–180. doi: 10.1080/02571862.2018.1537009
Twumasi-Afriye, S., Palacios Rojas, N., Friesen, D., Teklewold, A., Gissa, D. W., Groote De, et al. (2016). Guidelines for the Quality Control of Quality Protein Maize (QPM) Seed and Grain: Technical Bulletin. 38 p. Addis Ababa, Ethiopia: CGIAR. CIMMYT 38pp.
Vacaro, E., Fernandes, J., Neto, B., Pegoraro, D. G., Nuss, C. N., Conccicao, L. H., et al. (2002). Combining ability of twelve maize populations. Pesquisa Agropecuária Brasileira 37, 67–72. doi: 10.1590/S0100-204X2002000100009
Vancetovic, J., Bozinovic, S., Ignjatovic-Micic, D., Delic, N., Kravic, N., Nikolic, N., et al. (2015). A diallel cross among drought tolerant maize populations. Euphytica 205, 1–16. doi: 10.1007/s10681-015-1372-9
Vivek, B. S., Krivanek, A. F., Palacios-Rojas, N., and Twumasi-Afiriye, S. Diallo, A. O. (2008). Breeding Quality Protein Maize (QPM) Cultivars: Protocols for Developing QPM Cultivars. Mexico: CIMMYT.
Wegary, D., Vivek, B., and Labuschagne, M. (2013). Association of parental genetic distance with heterosis and specific combining ability in quality protein maize. Euphytica 191, 205–216. doi: 10.1007/s10681-012-0757-2
Wegary, D., Vivek, B. S., and Labuschagne, M. T. (2014). Combining ability of certain agronomic traits in quality protein maize under stress and non-stress environments in Eastern and Southern Africa. Crop Sci. 54, 1004–1014. doi: 10.2135/cropsci2013.09.0585
Keywords: combining ability, biofortification, quality traits, grain yield, inbred lines
Citation: Amegbor IK, van Biljon A, Shargie NG, Tarekegne A and Labuschagne MT (2023) Combining ability estimates for quality and non-quality protein maize inbred lines for grain yield, agronomic, and quality traits. Front. Sustain. Food Syst. 7:1123224. doi: 10.3389/fsufs.2023.1123224
Received: 13 December 2022; Accepted: 10 February 2023;
Published: 09 March 2023.
Edited by:
Silvia Tavarini, University of Pisa, ItalyReviewed by:
S. K. Singh, National Bureau of Plant Genetic Resources (ICAR), IndiaNuan Wen, Washington State University, United States
Aleck Kondwakwenda, University of Fort Hare, South Africa
Copyright © 2023 Amegbor, van Biljon, Shargie, Tarekegne and Labuschagne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isaac Kodzo Amegbor, aXNhYWNhbWVnYm9yQGdtYWlsLmNvbQ==; Maryke T. Labuschagne, bGFidXNjbUB1ZnMuYWMuemE=
 Isaac Kodzo Amegbor
Isaac Kodzo Amegbor Angeline van Biljon
Angeline van Biljon Nemera Geleta Shargie
Nemera Geleta Shargie Amsal Tarekegne
Amsal Tarekegne Maryke T. Labuschagne
Maryke T. Labuschagne