- 1School of Breeding and Multiplication (Sanya Institute of Breeding and Multiplication), Hainan University, Sanya, China
- 2School of Tropical Agriculture and Forestry, Hainan University, Haikou, China
- 3Institute of Tropical Crops Genetic Resources, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China
- 4Key Laboratory of Plant Disease and Pest Control of Hainan Province/Institute of Plant Protection, Hainan Academy of Agricultural Sciences, Haikou, China
Introduction: Macrotyloma uniflorum is an important legume fodder crop and green fertilizer. Salinity impedes plant growth and productivity of legume crops by disrupting the ionic and osmotic balance and hormonal regulation. Plant growth-promoting rhizobacteria (PGPR) are rhizosphere bacteria that contribute to the improvement of plant growth through diverse physiological mechanisms.
Methods: In this study, the growth promoting characteristics of the isolated strain Bacillus amyloliquefaciens PG-4 were analyzed, and to further investigated the possible mechanism of PG-4 in mitigating the damage caused by salt stress in M. uniflorum plants through pot experiments.
Results: In presence of different salt levels, PG-4 showed a high potentiality to produce several plant growth promoting metabolites such as NH3, siderophore, 1-aminocyclopropane-1-carboxylic acid deaminase (ACC-deaminase), and hydrolytic enzymes. Inoculation of the PG-4 significantly enhanced plant tolerance to salt stress, as demonstrated by promotion of plant growth (shoot and root biomass) under salt stress condition. Furthermore, PG-4 improved salt tolerance of Macrotyloma uniflorum seedlings by affecting the antioxidant enzymes including peroxidase (POD) and superoxide dismutase (SOD), by increasing the levels of proline, soluble sugars and chlorophyll. Treatment with PG-4 increased the K+ content while decreased the Na+ concentration level under salt stress. Transcriptomic analysis revealed 5525 genes were differentially expressed (PG-4-inoculated versus non-inoculated samples) at 0 mM NaCl, of which 3277 were upregulated and 2248 downregulated, while 1298 genes were differentially expressed at 100 mM NaCl, of which 819 were upregulated and 479 were downregulated. GO and KEGG enrichment analyses showed that these DEGs were significantly enriched in several terms and pathways mainly involved in the regulation of the cellular redox state, cell wall modification, metabolic adjustments, hemoglobin, biosynthesis of secondary metabolites and plant hormone signal transduction.
Discussion: These data showed that Bacillus amyloliquefaciens PG-4 significantly enhance salt stress tolerance in Macrotyloma uniflorum plants during salt stress conditions. Therefore, the results may be useful for explaining the mechanism by which PGPR inoculation regulates the salt tolerance of crops.
Introduction
Soil salinity is one of the most serious problems associated with soil and is considered a major abiotic threat to agricultural production worldwide. Currently, on a global scale, 20% of the land used for agriculture is affected, and annually, 2,500–5,000 km2 of crop production is lost annually due to salinity (Singh et al., 2022). Moreover, it has been estimated that by 2050, nearly 50% of arable land will be affected by salinity (Butcher et al., 2016). Salinity can have various effects on all plants, reducing crop growth and output by altering morphological, biochemical and physiological processes (Ali et al., 2022). An excessive accumulation of sodium (Na+) and chloride (Cl−) ions during salt stress causes ionic toxicity and leads to the generation of reactive oxygen species (ROS) in plant cells (Munns and Tester, 2008; Farhangi-Abriz et al., 2020). These ROS include superoxide, hydrogen peroxide, hydroxyl radicals, singlet oxygen, etc., which in excess amounts threaten plant cell integrity by initiating lipid peroxidation, damaging nucleic acids and activating programmed cell death and enzyme inhibition (Farhangi-Abriz et al., 2020). In addition, higher salt concentrations in the soil cause lower water availability, a decrease in plant nutrient uptake, and disturbed soil properties such as porosity, aeration and water conductance (Ibarra-Villarreal et al., 2021). However, in response to salt-stressed environments, plants activate various self-defense mechanisms, including osmotic adjustment, antioxidant metabolism and plant hormones (Liu et al., 2022; Wang Y. et al., 2022; Wang Q. et al., 2022). Specifically, osmotic adjustment occurs through the accumulation of organic solutes and inorganic ions in plants (Wang Y. et al., 2022; Wang Q. et al., 2022; Pistelli et al., 2023). Plant antioxidant metabolism involves nonenzymatic components such as ascorbic acid (AsA), glutathione (GSH), carotenoids and phenolic compounds, as well as enzymes such as dehydroascorbate reductase (DHAR); ascorbate peroxidase (APX); glutathione reductase; catalase (CAT); superoxide dismutase (SOD); glutathione peroxidase (GSH-Px); and glutathione S-transferase (GST), which clear excess ROS (Garcia-Caparros et al., 2021). Furthermore, plant hormones, such as ethylene, abscisic acid (ABA) and 3-indoleacetic acid (IAA), are known to endogenously regulate the homeostasis of plants and lead plants to perceive and respond to salt stress (Singh et al., 2022). However, these mechanisms are insufficient for alleviating salt stress beyond a certain limit.
To boost crop yield in salinized land, a large amount of chemical fertilizer is applied to the soil, but the increase in chemical input is not directly proportional to the increase in crop yield (Wang et al., 2018). This behavior has also led to serious soil ecological and environmental problems, including secondary salinization, soil consolidation, soil acidification and microecological imbalance (Ma et al., 2021). Furthermore, several molecular breeding and genetic engineering methods have been developed for improving crop yield under salt stress based on plant defense mechanisms. Nonetheless, these methods are expensive and time-consuming (Haque et al., 2021; Ashraf and Munns, 2022). To overcome the above issues, a new biocontrol approach has been developed to protect plants from salt stress in soil by utilizing beneficial microorganisms (Etesami and Maheshwari, 2018; Etesami and Glick, 2020). Plant growth-promoting rhizobacteria (PGPR) are an important cluster of beneficial, root-colonizing bacteria thriving in the plant rhizosphere, and positive effects on plant growth, development and metabolism (Li et al., 2020). When combined with roots and other tissues, PGPR improve the nutritional supply of crop plants through several mechanisms. PGPR have direct effects, including nitrogen fixation, phosphorus solubilization, NH3 production, IAA, and siderophores, and indirect effects, including antioxidant defense, volatile organic compound (VOC) production, exopolysaccharide (EPS) production, and osmotic balance mechanisms for improving plant growth and enhancing tolerance against salt stress (Etesami and Maheshwari, 2018; Mohanty et al., 2021). Therefore, an increasing number of scholars are studying how soil microorganisms play a role in plant growth to achieve eco-friendly and sustainable agriculture.
Diverse members of the Bacillus genus, which is one of the most extensively studied genera with beneficial microorganisms in the rhizosphere, have also been shown to be involved in promoting plant growth and tolerance against environmental stresses (Egamberdieva et al., 2019; Ibarra-Villarreal et al., 2021; Patani et al., 2023). Several mechanisms underlying the interactions between plants and Bacillus strains under salt stress have been revealed. B. atrophaeus WZYH01 protected maize from salt stress by regulating the levels of plant hormones (IAA and ABA) and increasing nutrient acquisition (Hou et al., 2022). Similarly, B. pumilus NCT4, B. firmus NCT1, B. licheniformis LCT4, B. cereus LAT3, and B. safensis LBM4 enhanced the tolerance of tomato plants to salt stress through increased total soluble sugar, proline and chlorophyll contents (Patani et al., 2023). B. subtilis helps mitigate salt-induced oxidative stress in soybean plants by modulating antioxidant defense and glyoxalase systems while maintaining ion homeostasis and osmotic adjustment (Hasanuzzaman et al., 2022). Woo et al. (2020) analyzed the ameliorative effects of the B. subtilis strain GOT9 on Arabidopsis thaliana and Brassica campestris under salt stress. Collectively, these results show that a variety of Bacillus strains interact with their specific plant hosts and contribute to enhanced tolerance against salt stress. The acclimation of plants to environmental stress after PGPR induction is a complex and coordinated response involving multiple physiological, biochemical, metabolic, and molecular mechanisms. Hence, a thorough understanding of the mechanisms induced by the strain in plants is imperative for enhancing plant yield and quality, as well as sustainable agricultural development.
Legumes are susceptible to a range of abiotic threats, with salinity being one of the most critical barriers to crop yield (Yilmaz and Kulaz, 2019). Horsegram [Macrotyloma uniflorum (Lam.) Verdc] is a dicotyledonous plant that belongs to the Leguminosae family, extensively distributed across South Asia, Australia, West Indies and Africa. This species is cultivated mainly for providing nutritional security in the form of food, livestock supplements and green manure because of its high nitrogen availability, nutritious composition, indomitable pest resistance, and drought tolerance (Bolbhat and Dhumal, 2009), and it has been identified as a potential, future food source by the U.S. National Academy of Sciences (Kadam and Salunkhe, 1985). Like several other legume crops, M. uniflorum is susceptible to salinity stress. Therefore, it is necessary to improve the tolerance of M. uniflorum to the deleterious effects of salinity, which could be achieved by using PGPR. However, knowledge of the potential of PGPR isolates and their effects on the growth and physiological characteristics of M. uniflorum under salt stress remains quite limited. The present study was carried out to determine the behavior of the selected PGPR strains under salinity stress conditions as well as their role in enhancing M. uniflorum growth. Thus, the present study was undertaken. (a) Study of the growth-promoting traits of isolated strains, (b) identification and characterization of the isolated bacteria based on biochemical characteristics and confirmation of the bacterial genera through 16S rRNA sequence analysis, and (c) evaluation of the effects of the bacteria on the growth, physiology, and expression level of stress tolerance genes in M. uniflorum seedlings under salt stress. Our results not only provide valuable information for exploring the mechanism by which PGPR alleviate the osmotic and oxidative stress of salt-stressed plants but also provide an eco-friendly method for cultivating high-quality and high-yield M. uniflorum in salt environments, and provide theoretical reference for the biological improvement of salt soil.
Materials and methods
Materials
M. uniflorum seeds of the native cultivar Yazhou with a viability >98% were obtained from the Institute of Tropical Crop Genetic Resources, Chinese Academy of Tropical Agricultural Sciences. Healthy seeds were selected and stored in a Kraft paper bag at 4°C until use. The PGPR strain of B. amyloliquefaciens PG-4 (16S rRNA GenBank: PP320457) was isolated from M. uniflorum rhizosphere soil in Danzhou, Hainan Province, China (19°31’N, 109°34′E). The isolation procedure was the same as that described in Hmaeid et al. (2019) and Patani et al. (2023). The strain was stored in 25% glycerol solution at −80°C until use. The strain was preserved in the China Center for Type Culture Collection, with preservation numbers of CCTCC AB 2024049.
Morphological observation of strains and homology analysis of 16S rDNA sequences
After preparing a 24-h culture, the strain was smeared on slides and subsequently fixed with heat. The samples were treated with crystal violet for 1 min following rinsing with running water. The samples were again flooded with gram iodine solution for 1 min following washing with alcohol. Finally, the samples were flooded with safranin, washed with water, dried, and observed under oil immersion conditions (Masi et al., 2021).
Genomic DNA was extracted using an OMEGA Genomic DNA Extraction Kit. Next, 16S rDNA fragments were PCR-amplified with the universal primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). The PCR mixture (30 μL) consisted of 15 μL of 2 × PCR mix, 1 μL of DNA, 1 μL of the upstream primer (10 mM), 1 μL of the downstream primer (10 mM), and 12 μL of ddH2O. Reaction conditions: 95°C 5 min, 94°C 45 s, 55°C 45 s, 72°C 1 min 15 s, 32 cycles, 72°C 10 min. After the reaction was completed, the size and specificity of the amplified fragments were verified by 1% agarose gel electrophoresis and photographed using a gel imaging system. The PCR amplification products were purified and sequenced by Nanshan Biotech (China) Co., Ltd. The 16S rDNA sequencing results of the obtained strains were analyzed using BLAST, and the related sequences with high homology were selected for relationship analysis. A phylogenetic tree was constructed via the neighbor method in MEGA6.0 software.
Plant growth promotion assays
The plant growth-promoting properties of PG-4, including nitrogen fixation, siderophore production, NH3 production, ACC deaminase production, cellulase production, pectinase production and amylase production, were evaluated in the presence of 0 mM, 50 mM, 100 mM and 200 mM NaCl. The determination method is described below.
Nitrogen fixation
A nitrogen (N2) fixation test was performed in malate nitrogen-free mineral media with modifications (Baldani and Döbereiner, 1980). The inoculated media were incubated for 72 h at 30 ± 2°C. A color change from pale green to blue was considered to indicate a positive effect of N2-fixing activity.
Siderophore production
Siderophore secretion by the isolated bacteria was assessed using blue agar plates containing chrome azurol S (CAS) (Sigma–Aldrich) (Schwyn and Neilands, 1987). A positive reaction was indicated by the formation of an orange zone around the colony, indicating siderophore excretion.
NH3 production
NH3 production was checked on peptone water according to Cappuccino and Sherman (1992). Fresh cultures grown in LB broth were inoculated into 10 mL of peptone water and incubated at 30 ± 2°C. After 48 h, 500 μL of Nessler’s reagent was added to each tube. Ammonia production is indicated by the development of a color change from brown to yellow.
ACC deaminase production
The 1-aminocyclopropane-1-carboxylic acid deaminase activity of PG-4 was detected by using DF medium and ADF medium (Shanghai Yuanye Bio-Technology Co., Ltd., China). These media were inoculated with 1% (v/v) fresh PG-4 cultures and incubated in a shaker (180 rpm, 28°C) for 5 days. The optical density was measured at 600 nm by a spectrophotometer from three replicates; the optical density result was considered to indicate a positive result for ACC deaminase production for PG-4 that was grown in ADF media and that showed no growth on DF media.
Assay for enzyme production
Cellulase production was tested using carboxymethylcellulose (CMC) agar medium (Gupta et al., 2012). After 24 h of incubation at 30°C, the CMC plates were flooded twice: first, with an aqueous solution of Congo red (1% w/v) for 15 min, followed by 1 M NaCl for 15 min. A clear halo was considered to indicate cellulase production. Pectinase production was tested as described by Plazinski and Rolfe (1985) using ammonium mineral agar medium, with some modifications. After 3 days at 30 ± 2°C, the plates were flooded with 2% hexadecyltrimethylammonium bromide. Pectinase production was detected by clear halos around the colonies. Amylase production was determined by soluble starch agar medium. After 2 days of incubation at 28°C, the ability of PG-4 to hydrolyze amylase was determined by the appearance of a halo zone around the colonies, which was confirmed by immersion in Lugol’s iodine solution for 15 min and 70% ethanol (Zhou et al., 2021). The noninoculated plates were used as controls.
Evaluation of the plant growth-promoting ability of B. amyloliquefaciens PG-4
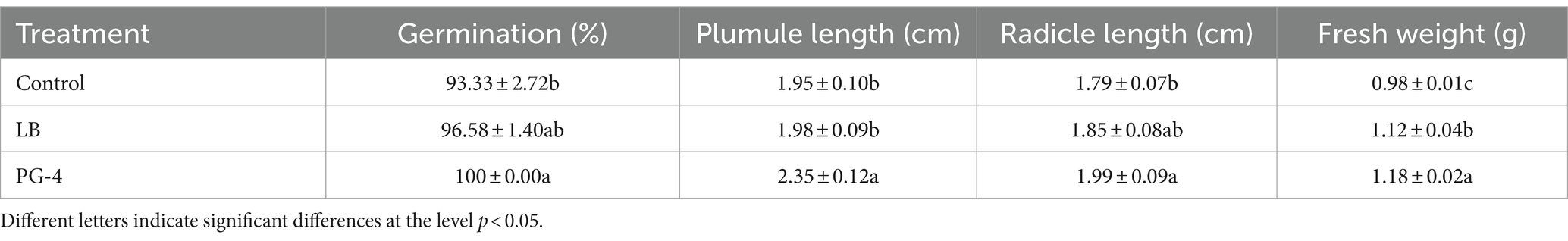
The healthy seeds were surface-sterilized by immersion in 70% ethanol for 50 s, followed by rinsing three times with sterile distilled water, submerging in 10% (v/v) sodium hypochlorite for 15 min and subsequent washing 5 times with sterile distilled water. The seed germination experiment was carried out as described by Li et al. (2021) with minor modifications. The sterilized plants were completely soaked in 20 mL of PG-4 suspension (OD600 = 0.6) for 2 h to allow PG-4 to colonize the M. uniflorum seeds, which served as the inoculation treatment. Seeds soaked in LB media or distilled water served as uninoculated controls. Next, seeds subjected to different treatments were air-dried at room temperature and subsequently transferred to Petri dishes (10 × 10 cm) containing 0.7% water agar at 25°C under a 12 h light/12 h dark cycle for germination. Each treatment had five replicates, and each replicate contained 18 seeds. The germinated seeds were counted every day until they reached a constant number (approximately 2 days). The germination percentage, seedling plumule length, radicle length, and fresh weight were measured at 3 days after germination.
Pot experimental design
The experimental treatments included (i) no bacterial inoculum or salt stress (CN), (ii) bacterial inoculum or salt stress (BN), (iii) no bacterial inoculum with salt stress (CS), and (iv) bacterial inoculum and salt stress (BS). Surface-sterilized M. uniflorum seeds with normal and healthy appearances were selected. Before sowing, the seeds were inoculated by immersion for 2 h in the respective bacterial inoculum (OD600 = 0.6) or in sterile distilled water (bacterial inoculum treatment and uninoculated controls). The seeds were subsequently transferred to a separate pot (10 cm × 8 cm × 10 cm) filled with a mixture of autoclaved vermiculite and nutrient soil (1:1, v:v). After 7 and 14 days, the seedlings were thinned to one per pot and then reinoculated with 1.0 mL of the corresponding bacterial inoculum (OD600 = 0.6) (BN and BS) or sterile distilled water (CN and CS) was applied to the stem base. Salt stress was imposed beginning on day 12 by supplementing the watering solution (water or nutritive solution) with 100 mM NaCl. The plants were grown at 25°C under a 16 h light/8 h dark cycle at Hainan University (Haikou, Hainan, China). Pots were arranged in a randomized block design, and each group consisted of 8 pots. Each treatment was kept in independent trays to avoid contamination, and the trays were rotated weekly to randomize possible position effects. After 14 days of stress, the SPAD values; plant height; plant diameter; and shoot and root fresh weights (FWs) were recorded. Then, all plants were removed from the pots, immediately cleaned with distilled water, and then stored at −80°C to measure physiological characteristics. In the remaining three pots of each treatment, all the plants in the pot were removed from the pot and cleaned immediately with distilled water. Then, the samples were oven dried at 60°C for 72 h to measure their dry weight and Na+ and K+ concentrations.
Physiological characterization and determination of ion contents
Prior to harvest, a SPAD LD-YD chlorophyll meter (LANENDE, Shandong, China) was used to directly determine the relative chlorophyll contents of the plant leaves via soil and plant analyzer development (SPAD). The mature leaves of 10 plants were tested 3 times, and the average values were obtained (Li et al., 2020). The contents of proline (Pro), total soluble sugar (TSS) and malondialdehyde (MDA) were measured by a PRO BC0295 assay kit (Beijing Solarbio Science & Technology Co., Ltd), a BC0035 Plant Soluble Sugar Content Assay Kit (Beijing Solarbio Science & Technology Co., Ltd), and a BC0025 MDA assay kit (Beijing Solarbio Science & Technology Co., Ltd.) following the manufacturer’s instructions, respectively. The activities of SOD and POD were measured by a BC0175 SOD assay kit (Beijing Solarbio Science & Technology Co., Ltd.) and a POD BC0095 assay kit (Beijing Solarbio Science & Technology Co., Ltd.) following the manufacturer’s instructions, respectively. All leaves used in testing were collected from mature plants at a single location.
A fine powder of dry plant leaves was mixed with acetic acid (0.1 N) (w/v) at 90°C for 2 h, and flame emission spectroscopy was used to measure the Na+ concentrations (Rus et al., 2001), the contents of K+ were measured by a Potassium Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
RNA sequencing procedures
RNA was extracted from twelve samples (four treatments × three biological replicates) using a TRIzol reagent kit (Invitrogen, Carlsbad, CA, United States). Total RNA was reverse transcribed to cDNA using an NEBNext Ultra RNA Library Prep Kit for Illumina (NEB #7530, New England Biolabs, Ipswich, MA, USA). Library construction, quality detection and Illumina sequencing were carried out by Gene Denovo Biotechnology Co., Guangzhou, China. An Agilent Bioanalyzer 2,100 system was used to evaluate RNA quality, and library sequencing was performed through an Illumina NovaSeq 6,000 platform. After removing adaptors, the unknown nucleotides and those of low quality were filtered from the raw reads to obtain clean reads. All the downstream analyses were based on these clean reads. De novo assembly was carried out using the de novo assembly software Trinity.1
Differential expression analysis
To compare the read count values of every gene to the original expression of the gene, HTSeq (0.9.1) statistics were used to standardize the expression by using fragments for exon kilobase per million fragments mapped (FPKM) values. Genes with a false discovery rate (FDR) < 0.05 and an absolute fold change ≥2 were defined as differentially expressed genes (DEGs). The software package R language heatmap (1.0.8) was used for bidirectional clustering analysis of all genes for all samples. For various samples, a heatmap was generated using the Euclidean method to measure the distance to and complete relation method to the cluster according to the expression level of the same and different genes in the same sample.
Gene annotation and pathway analysis
For annotations, all unigenes that were proven to be longer than 200 bp were subjected to a BLAST search (E-value <1e−5) against the NCBI nonredundant (NR) protein database (Pruitt et al., 2007), manually annotated and reviewed protein sequence database Swiss-Prot (Apweiler et al., 2004), Gene Ontology (GO) (Ashburner et al., 2000), Clusters of Orthologous Groups of proteins (KOG/COG) (Tatusov et al., 2000; Koonin et al., 2004), protein family (Pfam) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Finn et al., 2008; Kanehisa et al., 2008). The Gene Ontology database2 was used to map all the DEGs; however, Blast2GO software was used to calculate the differentially enriched genes. KEGG analysis of all the DEGs was performed by using KOBAS 2.0 software. For further analysis, significant pathways were selected.
Quantitative real-time PCR
The expression patterns of ten genes were analyzed via qRT-PCR. A pair of primers for each gene was designed using Primer 3.0 and are listed in Supplementary Table S1. qRT-PCR was subsequently performed using a Real-Time PCR System with a total reaction volume of 10 μL, which consisted of 1 μL of cDNA template, 0.5 μL of forward and reverse primers, 5 μL TB Green Premix Ex Taq II (2×) and 3 μL of sterilized ddH2O. The PCR procedure was as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 59 s, and 60°C for 15 s. The expression levels of ten genes were determined based on the 2−ΔΔCT method (Schmittgen and Livak, 2008), and relative changes in gene expression were calculated from the qRT-PCR data using the TCTP gene as a reference gene.
Statistical analysis
The collected data are presented as the means of the three replicates ± SE (standard error). One-way analysis of variance (ANOVA) was used to determine statistically significant differences between the means of treatments according to Duncan’s multiple range test (p = 0.05). All statistical analyses of data were performed using IBM SPSS Statistics 26.0. The calculation formula for Z-Score in the heat map is as follows: Z = (FPKM - mean)/standard deviation.
Results
Plant growth-promoting activity of the PG-4 strain
The PG-4 strain showed the potential to secrete several PGP substances in the presence of different salt concentrations (Control, 50, 100 and 200 mM), as reported in Supplementary Table S2. The PG-4 strain has the potential to fix nitrogen and produce siderophores. Spectrophotometric analysis revealed that strain PG-4 grew better on ADF media than on control DF media under different salt stress conditions, suggesting that PG-4 has the ability to produce ACC deaminase. The analysis also showed that the PG-4 strain has a widespread ability to secrete hydrolytic enzymes, including cellulase, pectinase, and amylase, under different salt stress conditions. These results suggest that the PG-4 strain may be an effective agent for supporting plant resistance to salt stress.
Growth-promoting efficacy of the PG-4 strain
The growth-promoting efficacy of PG-4 on M. uniflorum seeds was examined, and the results are shown in Table 1. PG-4 had a growth-promoting effect on seed germination and seedling growth, significantly increasing the germination percentage, plumule and radicle length, and fresh weight (p < 0.05). The average germination percentage, plumule length, radicle length, and fresh weight were 100 ± 0.00%, 2.35 ± 0.12 cm, 1.99 ± 0.09 cm, and 1.18 ± 0.02 g, respectively, which were equivalent to increases of 7.15, 20.51, 11.17, and 20.4%, respectively, relative to those of the control after PG-4 was inoculated for 3 days. Overall, these results revealed that, compared with that of the noninoculated control seedlings, PG-4 promoted the growth of inoculated M. uniflorum seedlings.
Identification of the PG-4 strain
The colony morphology of the PG-4 strain is shown in Figures 1A,B, and the 16S rDNA gene sequences were obtained and compared with GenBank sequences. The strain was identified as gram-positive and facultative anaerobic following the protocol described in the “Common Bacterial System Identification Manual” and “Bergey’s Manual of Determinative Bacteriology” with slight modifications (Dong and Cai, 1999). The 16S rRNA amplicon sequencing results revealed a total of 1,404 bases with homology between PG-4 and Bacillus amyloliquefaciens NBRC 15535 (NCBI accession number: NR041455.1) of 99.3%. Their close relationship was further supported by a phylogenetic tree (Figure 1C) (Nautiyal et al., 2013; Kim et al., 2017; Liu et al., 2017; Tanaka et al., 2017; Kazerooni et al., 2021). Accordingly, the PG-4 strain was identified as B. amyloliquefaciens.
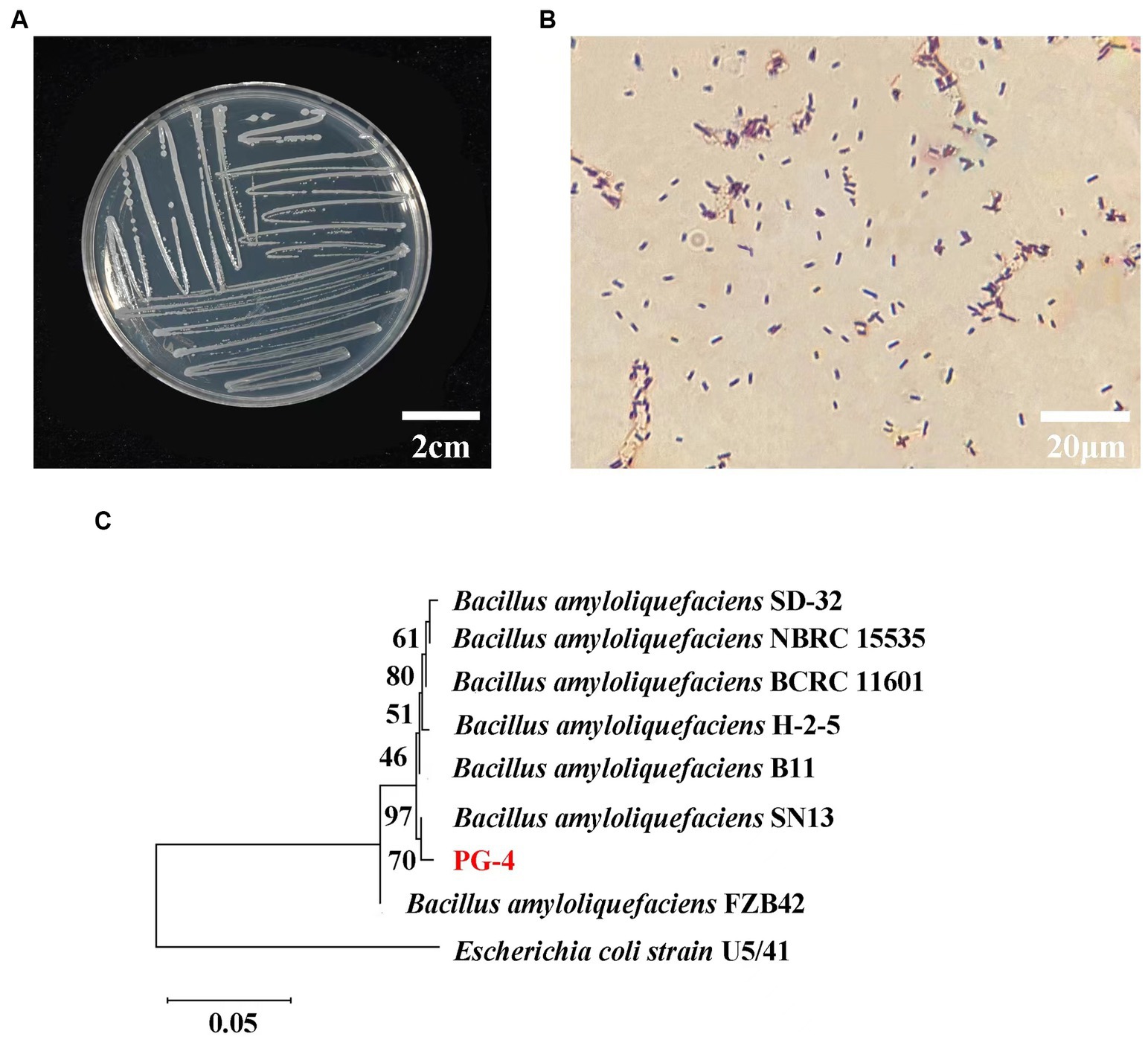
Figure 1. Molecular identification and biological characteristics analysis of strain PG-4. (A) Morphological characteristics of colony of strain PG-4. (B) Gram staining of strain PG-4. (C) A phylogenetic tree among eight Bacillus strains was derived from alignment of the 16S rRNA gene sequence. Scale bar, 0.05 substitutions per nucleotide position. Information on their isolation sources was obtained from the references and NCBI (BioSample).
Effects of B. amyloliquefaciens PG-4 on M. uniflorum seedlings under salt stress
After the interaction of B. amyloliquefaciens PG-4 with M. uniflorum under control conditions (without salt stress) and stress conditions (100 mM NaCl), the growth patterns of the plants were evaluated for 14 days. The growth of the plant seedlings under all treatments (CN, BN, CS, and BS) is shown in Figure 2. Greater plant growth was observed in the plants treated with PG-4 under salt stress (Figure 2A). The shoot length of the treated plants (BS) under salt stress was significantly (p < 0.05) different from that of the untreated control (CS) plants. The shoot length of the plants treated with CS was approximately 9.65 cm, whereas the shoot length of the BS-treated plants was approximately 10.05 cm (Figure 2B). Moreover, there was no significant (p > 0.05) difference in the roots of the untreated and treated plants under salt stress conditions (Figure 2C). In addition, under salt stress, compared with those in the control group without inoculation (CS), the shoot and root fresh weights of the plants treated with BS increased by 2.7 and 1.8 times, respectively (Figures 2D,E), and the shoot and root dry weights increased by 2.9 and 1.5 times, respectively (Figures 2F,G).
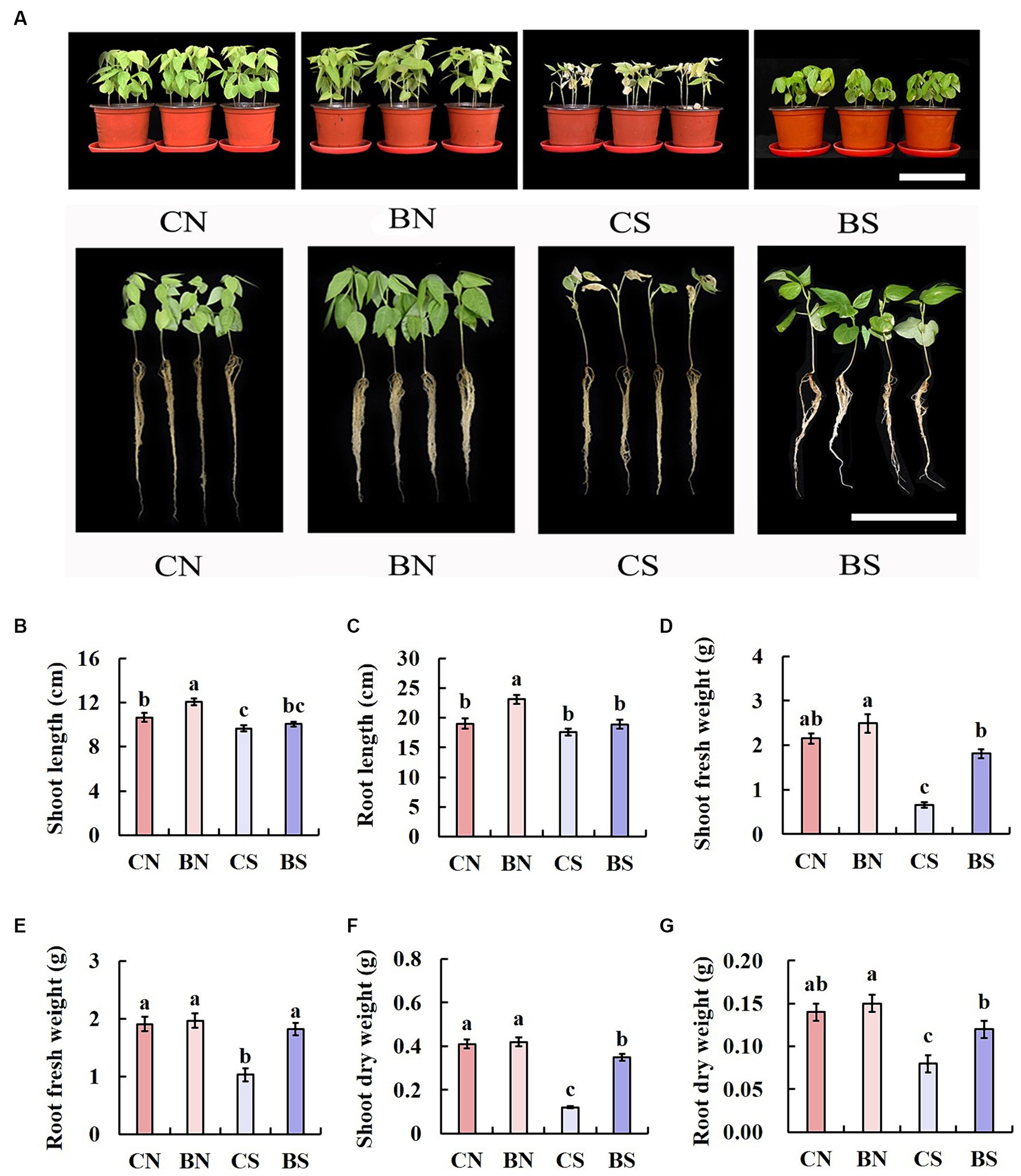
Figure 2. Effect of PG-4 inoculation on the growth of Macrotyloma uniflorum seedlings under salt stress in a pot experiment. (A) Growth status of M. uniflorum seedlings under the four treatments. Scale bar, 10 cm. (B) Shoot length. (C) Root length. (D) Shoot fresh weight. (E) Root fresh weights. (F) Shoot dry weight. (G) Root dry weight. CN, CS, BN, and BS represented the treatments of control and non-salt, control and salt, PG-4 inoculated and non-salt, and PG-4 inoculated and salt, respectively.
The chlorophyll content is an important index for predicting the health and photosynthetic capacity of plants under salt stress. Therefore, we measured the leaf chlorophyll content to examine the impact of PG-4 on the photosynthetic capacity of plants under salt stress. The results showed that salinity significantly (p < 0.05) reduced the chlorophyll content in the leaves of M. uniflorum. However, our findings also showed that, compared with that of the uninoculated control plant, PG-4 inoculation significantly increased the chlorophyll content in the plants under salt stress or those under no-salt conditions (Figure 3A).
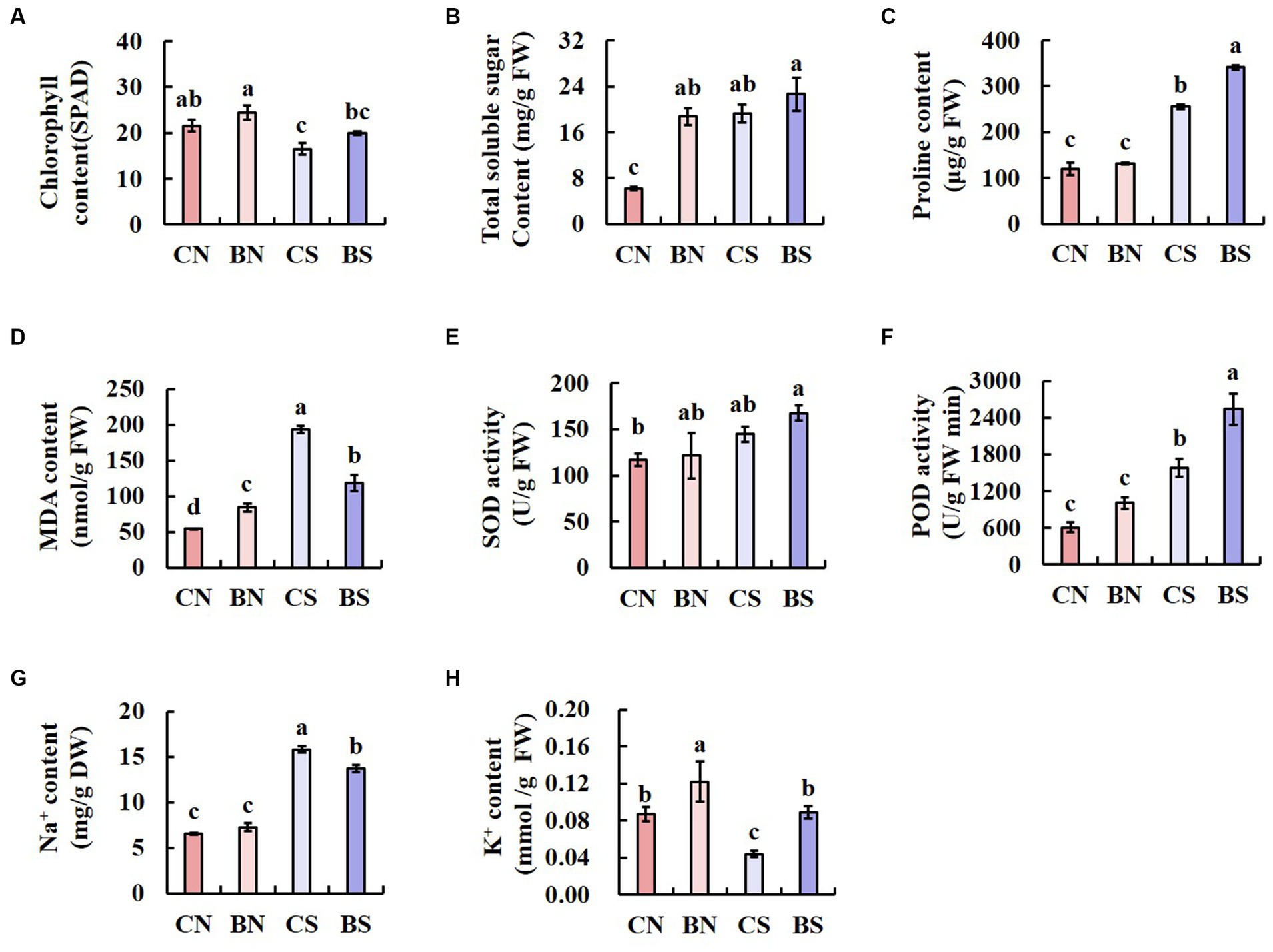
Figure 3. The physiological parameters of M. uniflorum seedlings under different treatments. Bars represent means ± SE. Different letters indicate significant differences at the level p < 0.05 according to Duncan’s multiple range test test (n = 3 independent replicates). (A) ChlorophyII content. (B) Total soluble sugar content. (C) Proline content. (D) MDA content. (E) SOD activity. (F) POD activity. (G) Na+ content. (H) k+ content.
Salinity significantly (p < 0.05) increased the TSS content in the leaves of M. uniflorum. However, whether the plants were under salt stress or control conditions, the TSS and Pro content in the leaves of M. uniflorum inoculated with PG-4 was significantly higher than that in the leaves of the uninoculated control plants (Figures 3B,C). Salt stress significantly (p < 0.05) increased the MDA content. However, under salt stress, inoculation with PG-4 significantly reduced the MDA content in M. uniflorum leaves compared with that in uninoculated control plants (Figure 3D). Overall, these results indicated that inoculation with PG-4 can significantly (p < 0.05) reduce oxidative stress-related MDA levels and improve the salt tolerance more effectively in plants under salt stress. Furthermore, the data analysis showed that SOD and POD activity was increased by PG-4 inoculation and salinity stress, with the highest activity in BS-treated M. uniflorum leaves (Figures 3E,F). The analysis of Na+ and K+ content showed that salt stress strongly increased the absorption of Na+ and decreased K+ uptake. However, under salt stress, inoculation with PG-4 significantly (p < 0.05) increased the accumulation of potassium in plant leaves, while reduced the accumulation of sodium (Figures 3G,H). These data suggest the positive effect of B. amyloliquefaciens PG-4 on K+ absorption under salt conditions.
Overview of the transcriptome analysis and read assembly
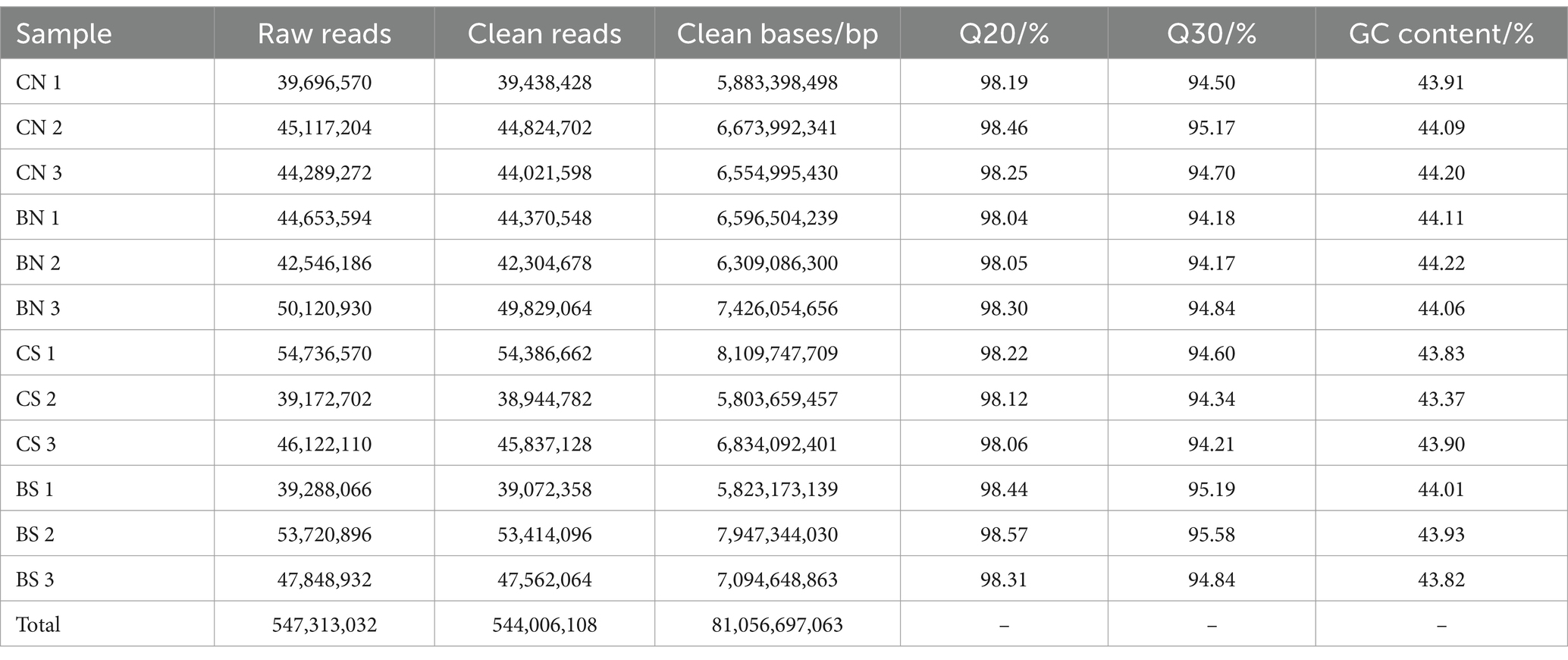
To unravel the mechanism of B. amyloliquefaciens PG-4-mediated salt tolerance in M. uniflorum plants, transcriptome profiling was carried out using an RNA-Seq approach. In this study, RNA libraries were constructed and sequenced for a total of 12 samples from four groups of M. uniflorum samples, namely, CN, BN, CS, and BS. Among them, there were 547,313,032 original fragments. Adapter sequences and low-quality reads, at least 544,006,108 clean reads remained. Q30 (sequences with a sequencing error rate of less than 0.1%) was ≥94%, and the average GC content was 43.95% (Table 2). Trinity software was used to generate 42,179 unigenes via de novo transcriptome assembly. The average length of unigene is 1,314 bp (Supplementary Table S3; Figure 4A).
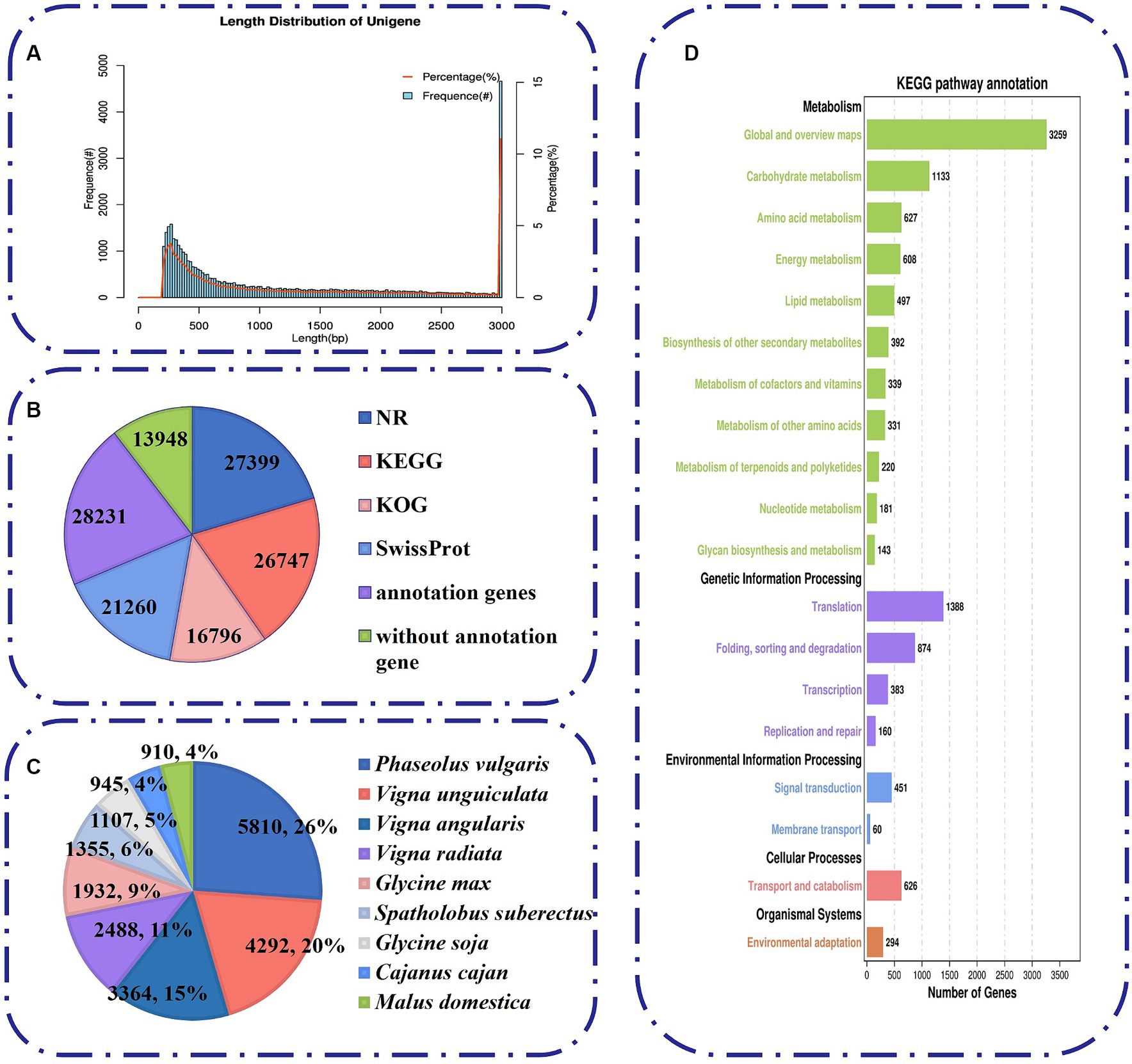
Figure 4. Characteristics of unigenes generated by Illumina sequencing. (A) The length distribution of all assembled unigenes. (B) The number of unigenes annotated by different databases, including NR, KEGG, KOG and Swiss-Prot. (C) Species distribution of the top BLAST hits for all homologous sequences. (D) A total of 6,938 unigenes were assigned to different KEGG terms.
Functional annotation
To analyze the comprehensive functional annotation of the 42,179 unigenes, we used BLAST against four databases, namely, the NR, KEGG, KOG, and SwissProt databases. A total of 28,231 unigenes were annotated in at least one database (Figure 4B). Among them, 16,796 unigenes were assigned to 25 functional clusters according to the KOG database. “General function prediction only” (17.4%, 3,611) was the largest category (Figure 4B; Supplementary Figure S1A). In addition, we obtained functional information on the similarity between the sequences of M. uniflorum and related species by blasting against other plant species via the NR database. The M. uniflorum isoforms had the highest number of hits to Phaseolus vulgaris (5,810 hits), followed by Vigna unguiculata (4,292 hits) and Vigna angularis (3,364 hits) (Figure 4C).
To study the function of these genes in biological pathways, the unigenes were identified via the KEGG pathway database. A total of 6,938 unigenes were grouped into five main KEGG functional categories and 138 KEGG pathways (Figure 4D; Supplementary Table S4). A high proportion of unigenes were distributed in “Metabolism” (7,730) (Figure 4D), such as “Global and overview maps” (3,259), “Carbohydrate metabolism” (1,133), and “Amino acid metabolism” (627). To further classify the M. uniflorum unigenes, GO annotation was performed based on three major categories, namely, biological process, cellular component, and molecular function (Supplementary Figure S1B; Supplementary Table S5). For biological process classification, “cellular process” (14,775), “metabolic process” (12,742), and “biological regulation” (4,309) were the three major categories. Unigenes involved in “binding” (12,266), “catalytic activity” (11,136) and “transporter activity” (1,807) were highly represented in the molecular function subgroups. The major categories of cellular component were “cellular anatomical entity (10,809),” “protein-containing complex (3,876)” and “virion component (101).”
Differential expression gene (DEG) and functional enrichment analysis among different treatments
The expression of these unigenes was subjected to principal component analysis (PCA) (Supplementary Figure S2A). The first two components, which explained 49.6 and 22.8% of the variation, respectively, were able to distinguish the four different treatment groups, with three biological replicates clustering together, and each group had certain differences in their gene expression profiles. A heatmap of Pearson’s correlation coefficients between all sample pairs also showed that each sample was reliable with good reproducibility (Supplementary Figure S2B). Based on the read counts, DEGs were assessed with DESeq2 software. In this study, DEGs between treatments were defined on the basis of an absolute fold change ≥2 and a false discovery rate (FDR) < 0.05. Hierarchical clustering analysis was used to determine the expression patterns of DEGs under different experimental conditions (Figure 5A). Four different comparison groups were obtained by comparing the DEGs between different treatments: BN vs. CN, BS vs. BN, CS vs. CN, and BS vs. CS. The expression trends and numbers of DEGs identified in the different comparisons are shown in Figure 5B. There were 5,525 DEGs between the BN vs. CN groups, 3,277 upregulated genes and 2,248 downregulated genes. A total of 1,473 DEGs were found between BS vs. BN, which included 859 upregulated genes and 614 downregulated genes (Figure 5C). A total of 3,783 DEGs were found between CS vs. CN; these included 2,280 upregulated genes and 1,503 downregulated genes (Figure 5C). A total of 1,298 DEGs were found between BS vs. CS groups, which included 819 upregulated genes and 479 downregulated genes (Figure 5C). The total number of unique downregulated DEGs in the BS vs. BN comparison was 443, with 1,332 in the CS vs. CN comparison, while 171 DEGs were common between the treatments; 819 unique upregulated DEGs were found in the BS vs. BN comparison; and 1,827 were found in the CS vs. CN comparison, while 40 DEGs were common between the two treatments (Figure 5D).
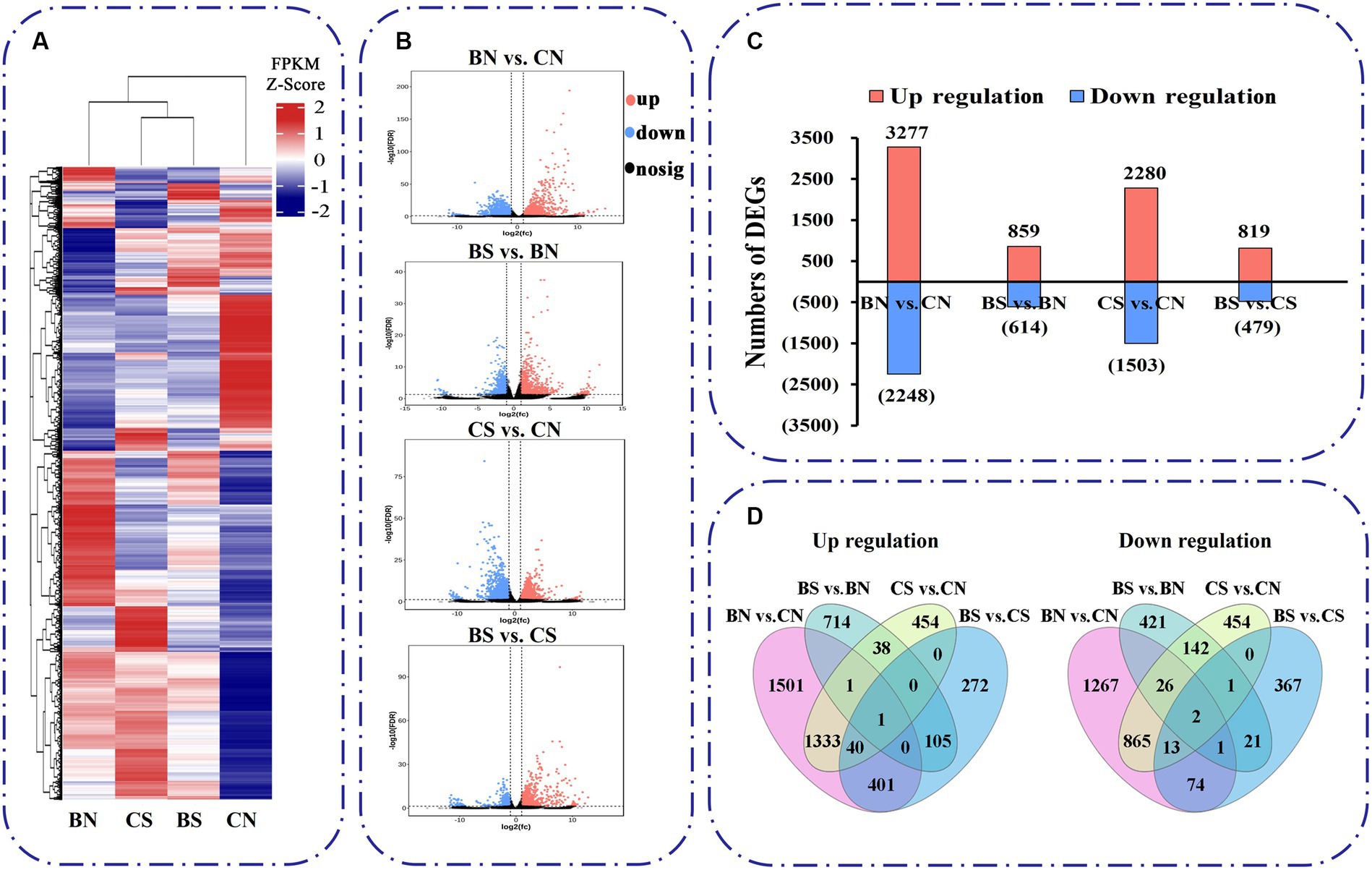
Figure 5. Transcriptional variations in M. uniflorum roots under different treatments. (A) Hierarchical clustrering analysis of the DEGs under different treatments. The color scale represents FPKM values in each treatment, normalized using z-score. (B) Significance analysis of the DEGs in different treatments by Volcano plots. (C) The number of up- and down-regulated genes in different treatments. (D) Venn diagrams showed the proportions of the up-regulated and down-regulated genes in four treatments.
Functional annotation and classification of differentially expressed genes
To further predict the DEGs induced by PG-4 inoculation and salt stress, functional annotation of the DEGs in the four groups was performed based on GO and KEGG analyses. Among the four comparative groups, BN vs. CN, BS vs. BN, CS vs. CN, and BS vs. CS, 3,904, 1,038, 2,763 and 905 DEGs, respectively, were subdivided into at least one GO term to describe their biological processes, molecular functions, and cellular components (Supplementary Figure S3). Under no-salt stress, a higher percentage of the PG-4-induced DEGs involved in plant growth regulation were found; these DEGs were associated mainly with “Cellular process,” “metabolic process” and “response to stimulus” in the biological process category (Supplementary Figure S3A). According to the “response to stimulus” classification, 832 genes were changed by PG-4 inoculation under no-salt stress, whereas only 203 DEGs were regulated by salt stress in PG-4-inoculated seedlings (Supplementary Figures S3A,B). In addition, all DEGs induced by salt stress and PG-4 inoculation were mapped via KEGG to reference canonical pathways to identify the associated biological pathways (Supplementary Figure S4). A total of 347 salt stress-related DEGs in PG-4-inoculated plants were assigned to 109 pathways, which were classified into 19 KEGG pathway subcategories, of which the most common DEGs were “global and overview maps” followed by “carbohydrate metabolism,” “biosynthesis of other secondary metabolites,” and “translation” (Supplementary Figure S4A; Supplementary Table S6). The DEGs induced by PG-4 in salt-treated plants were assigned to 104 pathways, mainly “global and overview maps,” “carbohydrate metabolism,” “biosynthesis of other secondary metabolites” and “amino acid metabolism” (Supplementary Figure S4B; Supplementary Table S7).
Go and KEGG pathway enrichment analysis of differentially expressed genes
To elucidate the biological functions of these DEGs, the enriched GO terms were analyzed. The top 20 enriched GO terms are shown in Supplementary Figure S5. The DEGs of the different comparison groups were investigated through GO classification and KEGG enrichment. According to the GO enrichment analysis, 2,763 DEGs in the CS treatment group compared with those in the CN treatment group were significantly enriched (q-value <0.05) in 246 terms, and the most enriched GO terms that involved DEGs between CS vs. CN were related to “catalytic activity,” “oxidoreductase activity,” and “Small molecule metabolic process” (Supplementary Figure S5A). Moreover, 1,038 DEGs in the BS group compared with those in the BN group were significantly (q value <0.05) enriched in 39 terms, and the DEGs between the BS vs. BN groups were associated mainly with “heme binding,” “calcium ion binding,” “carbohydrate metabolic process” and the activity of various enzymes, such as oxidoreductase, peroxidase and glycosyltransferase (Supplementary Figure S5B). The lists of all identified significantly (q-value <0.05) enriched GO pathways are shown in Supplementary Tables S8, S9. KEGG pathway analysis was performed to explore the biological pathways represented by DEGs in the four comparison treatments. Several significant pathways were identified in the different comparisons (Figure 6). KEGG pathway enrichment revealed that the DEGs between CS vs. CN were involved primarily in “metabolic pathways,” “biosynthesis of secondary metabolites,” “phenylpropanoid biosynthesis,” “glycosylphosphatidylinositol (GPI)-anchor biosynthesis,” “circadian rhythm-plant,” “isoflavonoid biosynthesis,” “cyanoamino acid metabolism,” “alpha-linolenic acid metabolism” and “glycerolipid metabolism” (Figure 6A). The DEGs in the BS vs. BN comparison were assigned to “phenylpropanoid biosynthesis,” “plant-pathogen interaction,” “metabolic pathways,” “biosynthesis of secondary metabolites,” “pentose and glucuronate interconversions,” “starch and sucrose metabolism” and “plant hormone signal transduction” (Figure 6B). The lists of all identified KEGG pathways are shown in Supplementary Tables S10, S11.
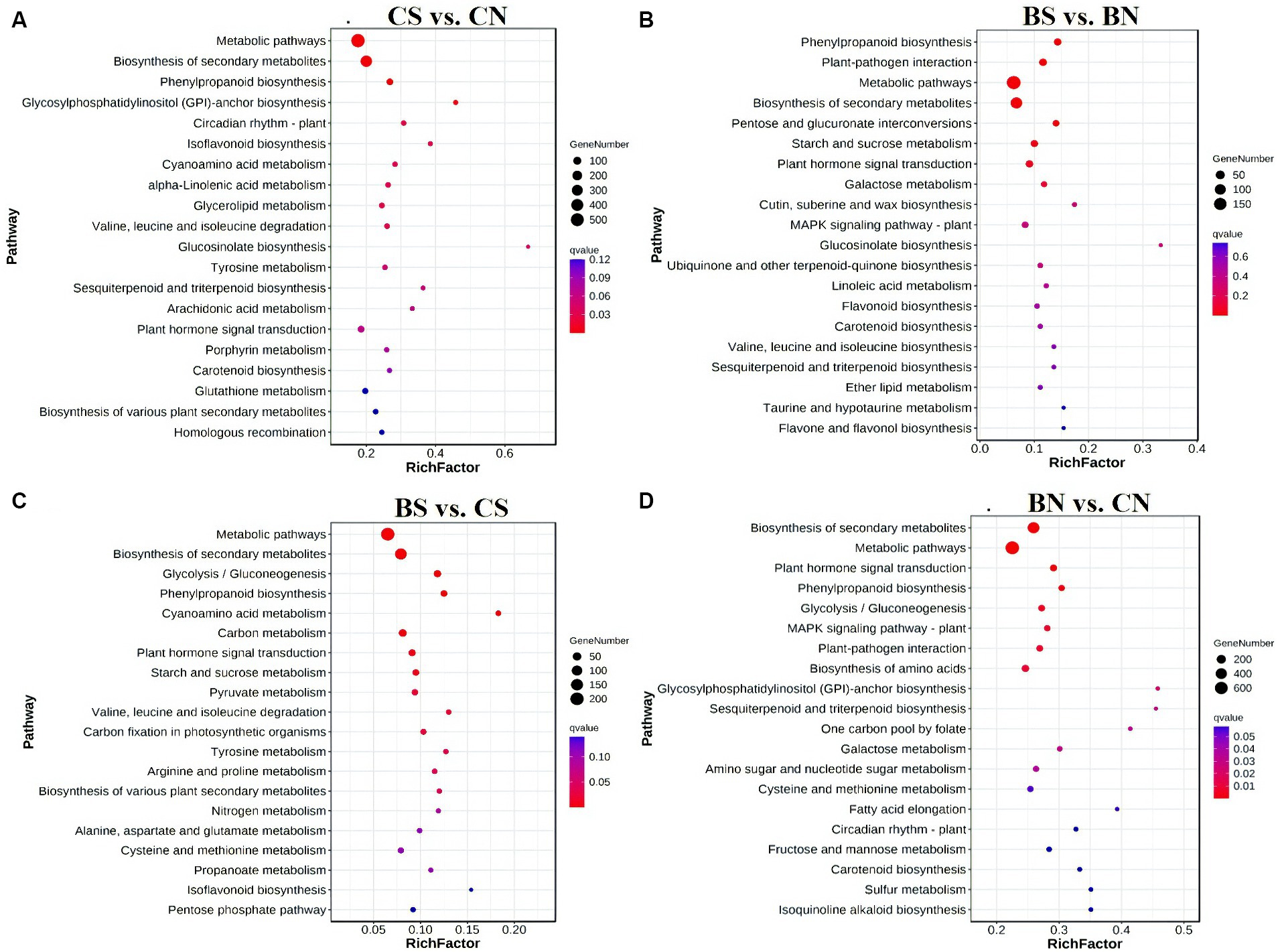
Figure 6. Enriched KEGG pathways analysis on of genes with significant differences between groups. (A) Enriched KEGG pathways of differential genes between CS and CN. (B) Enriched KEGG pathways of differential genes between BS and BN. (C) Enriched KEGG pathways of differential genes between BS and CS. (D) Enriched KEGG pathways of differential genes between BN and CN. False discovery rate (FDR) is the corrected p-value.
Response of key unigenes to hormone signal transduction
Phytohormones, such as ethylene, IAA, cytokinin and ABA, play critical roles in eliciting a salinity stress adaptation response in plants (Singh et al., 2022). The three protein components related to auxin signal transduction are the auxin receptor-associated SCF complex (SKP1, Cullin and F-box complex), the auxin protein (Aux/IAA), which has regulatory functions, and the auxin response factor (ARF). Early auxin response genes can be divided into three categories: Aux/IAAs, GH3s, and small auxin-up RNAs (SAURs) (Hagen, 2015). The AUX/IAA, ARF, SAUR, GH3, and SAUR genes in M. uniflorum under salt stress were significantly downregulated to varying degrees, while these genes were upregulated by PG-4 in M. uniflorum under salt stress (Figure 7A; Supplementary Table S12). Three AUX/IAA genes (Unigene0023452, Unigene0023453 and Unigene0035539), one ARF gene (Unigene0030592), one SAUR genes (Unigene0028233) and one GH3 genes (Unigene0015319) were downregulated under salt stress, while they were significantly upregulated by PG-4 under salt stress. The genes Unigene0023453, Unigene0023452, and Unigene0015319 were down regulated by 2.9-, 2.5- and 2.8-fold, respectively, by salt stress, while they were significantly downregulated 2.4-, 2.4- and 1.8-fold, respectively, by PG-4 under salt stress conditions. In the ABA signaling pathway, only one PYL gene (Unigene0002166), three protein phosphatase 2C (PP2C) genes (Unigene0030091, Unigene0031036 and Unigene0031406) and one ABF gene (Unigene0003719) were differentially expressed in the BS vs. CS comparison (Figure 7B; Supplementary Table S12). The protein phosphatase 2C 51 isoform X2 (Unigene0031406) were significantly downregulated by salt stress but upregulated by PG-4 inoculation. In the BR signaling pathway, three TCH4 genes (Unigene0036355, Unigene0022458, and Unigene0027059) were significantly downregulated by 1.3-, 1.2- and 1.2-fold under salt stress, respectively, while they were significantly upregulated by 1.6-, 2.6- and 2.3-fold after PG-4 inoculation under salt stress (Figure 7C; Supplementary Table S12). A differentially expressed JAZ genes (in JA signaling pathway) were present in BS vs. CS (Figure 7D; Supplementary Table S12), and two differentially expressed TGA genes (in SA signaling pathway) were present in BN vs. CN (Figure 7E; Supplementary Table S12). Overall, these results indicated that PG-4 can affect the signal transduction process of plants in response to salt stress, which can help plants prepare for future salt stress.
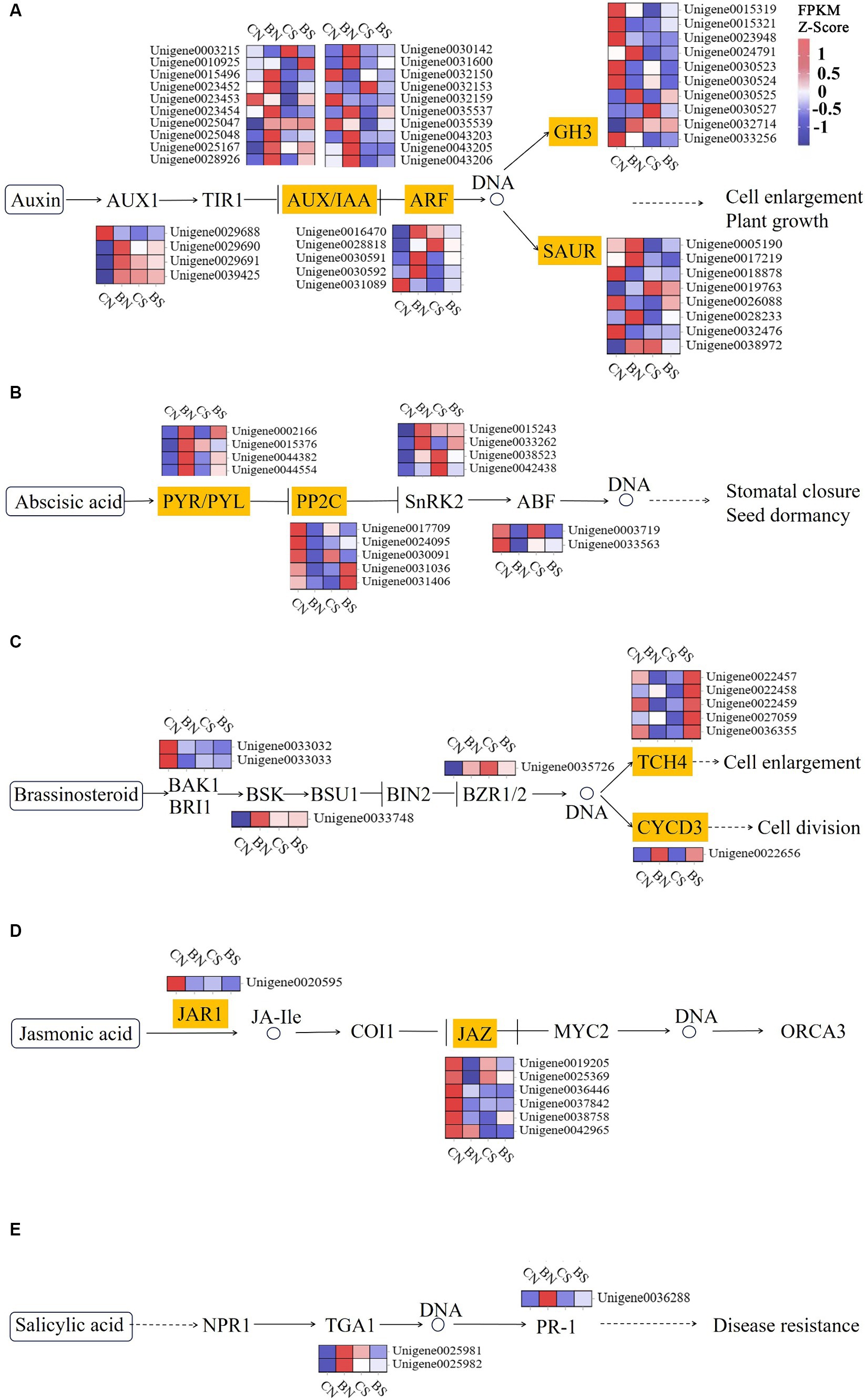
Figure 7. PG-4 activated plant hormone signal transduction pathway under salt stress. (A) Auxin signaling pathway. (B) ABA signaling pathway. (C) BR signaling pathway. (D) JA signaling pathway. (E) SA signaling pathway. The color scale represents FPKM values in each treatment, normalized using z-score.
Response of key unigenes to transporter proteins and key cellular metabolic processes
Among the thirteen DEGs related to ABC transporters expressed in seedlings, seven ABC transporter DEGs (Unigene0003435, Unigene0006341, Unigene0019507, Unigene0022111, Unigene0022624, Unigene0027432, and Unigene0029184) were significantly downregulated, and three (Unigene0000140, Unigene0033713, and Unigene0037140) were significantly upregulated in the CS vs. CN comparison (Supplementary Figure S6A; Supplementary Table S13). In the BS vs. CS comparison, three ABC transporter DEGs (Unigene0003435, Unigene0006339, and Unigene0022513) were significantly upregulated, while only one (Unigene0020052) was significantly downregulated. In addition, the expression of the ABC transporter B family member 11 (Unigene0003435) was significantly downregulated by 1.94-fold in response to salt but significantly upregulated by 6.50-fold in response to PG-4 inoculation under salt stress (Supplementary Figure S6A; Supplementary Table S13). In soluble sugar metabolism, Unigene0038428 and Unigene0038427, which encode invertase, were significantly downregulated, and Unigene0014202, Unigene0010635, and Unigene0019276, which encode sucrose synthase, were significantly upregulated in the CS vs. CN comparison. In the BS vs. CS comparison, the granule-Unigene0003715, glucan endo-1,3-beta-glucosidase 1 (Unigene0042124) and 12 (Unigene0016553) genes were significantly upregulated (Supplementary Figure S6B; Supplementary Table S14).
Pectinesterase (EC 3.1.1.11) is an enzyme that catalyzes the hydrolysis of pectin to produce pectinic acid and methanol and can regulate cell wall strength, cell growth and stress resistance. The expression of DEGs related to pectin metabolism was significantly upregulated by PG-4 inoculation under salt conditions. The pectinesterase/pectinesterase inhibitor 47 (Unigene0031614) and pectinesterase (Unigene0035002) were significantly downregulated (1.50-fold and 1.48-fold, respectively) by salt stress, while they were significantly upregulated (1.79-fold and 1.74-fold, respectively) by PG-4 under salt stress (Supplementary Figure S6C; Supplementary Table S15). In addition to the above transcripts encoding carbohydrate-active enzymes, the xyloglucan endotransglucosylase/hydrolase protein (XTH) transcript required for cell wall synthesis and modification was also shown to be induced by PG-4 inoculation under salinity stress (Supplementary Figure S6D; Supplementary Table S16). In the BS vs. CS comparison, seven XTH DEGs (Unigene0007670, Unigene0036223, Unigene0022458, Unigene00036355, Unigene0022457, Unigene0022459 and Unigene0027059) were significantly upregulated, and only two XTH DEGs (Unigene0033881 and Unigene44199) were significantly downregulated. Overall, these results suggested that the changes in the cell wall structure of M. uniflorum in response to salt stress could be adjusted by the inoculation of PG-4 (Supplementary Figure S6D; Supplementary Table S16).
Classification of transcription factors (TFs) under PG-4 inoculation and salt stress
In this study, 74 and 166 differentially expressed TFs were identified in the CS vs. BS comparison and the CN vs. CS comparison, respectively, and mainly belonged to the TF families APETALA2/ethylene responsive factor (AP2/ERF), WRKY, myeloblastosis protein (MYB) superfamily, NAC domain protein (NAC), basic helix–loop–helix (bHLH), and basic leucine zipper (bZIP), which are known to be associated with plant abiotic and biotic stress response and regulation (Figure 8; Supplementary Table S17). As shown in Figure 8A, these TFs were extensively upregulated by PG-4 under salinity stress, suggesting the special role of PG-4 in the transcriptional regulation of stress responses. In contrast, 60.64% of the TFs were downregulated by salinity stress, which may reflect the compromised defense response in M. uniflorum plants. AP2/ERF is one of the largest TF families involved in the response to salt stress (Xie et al., 2019). Among these TFs, ERF106 (Unigene0018746), PTI5 (Unigene0023663) and ERF095 (Unigene0025360) were significantly downregulated by salt treatment but significantly upregulated upon PG-4 inoculation (Figure 8B; Supplementary Table S17).
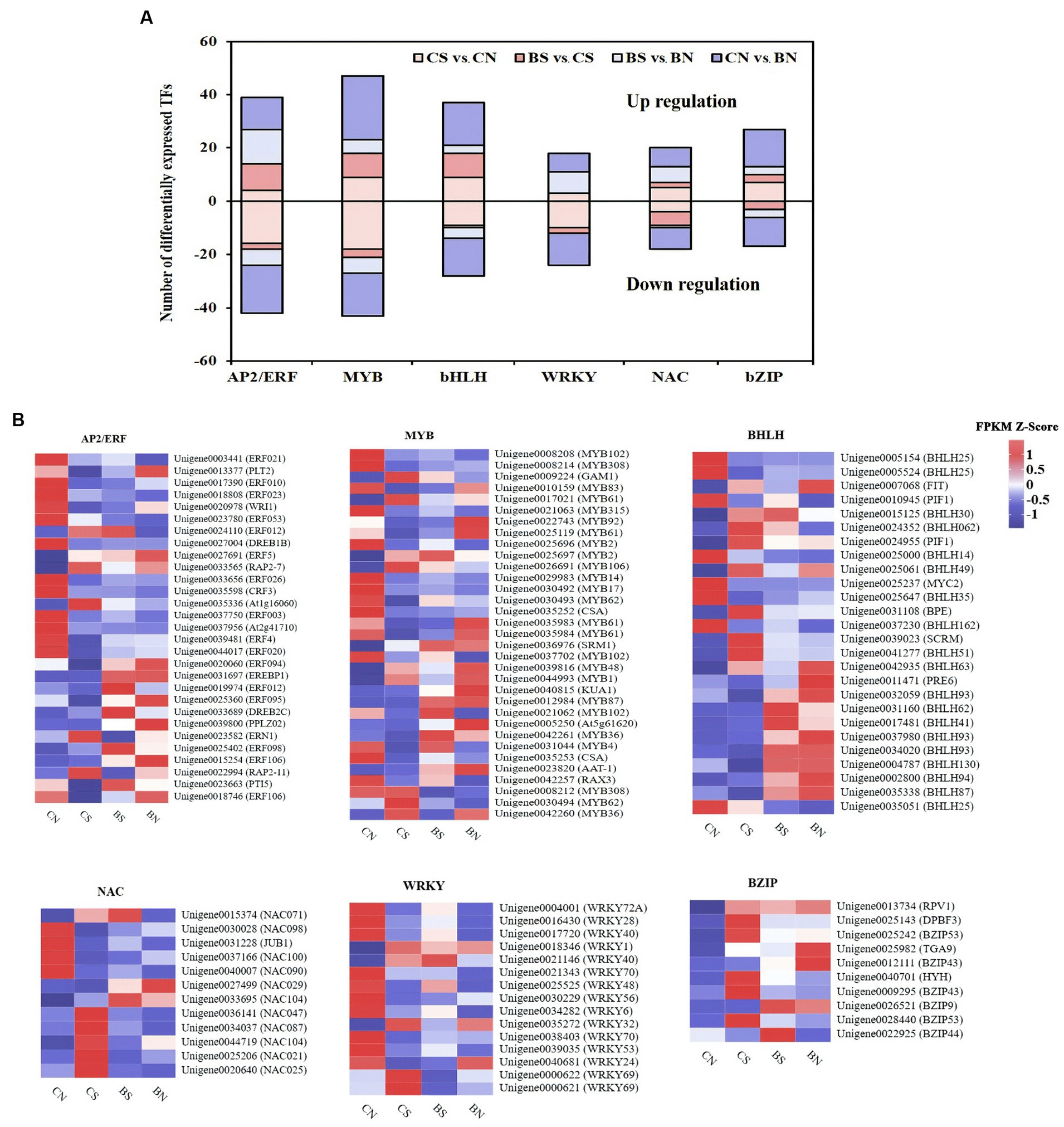
Figure 8. The differential expression pattern of the six TF families. (A) The number and expression pattern of differentially expressed TFs. The positive numbers indicate the up-regulated TFs and the negative numbers indicate the down-regulated TFs. (B) The differential expression pattern of the six TF families were depicted by a heatmap. The color scale represents FPKM values in each treatment, normalized using z-score.
RNA-seq results validated by qRT–PCR
To verify the reliability of the RNA-seq data, we randomly selected 10 unigenes (Unigene0023113, Unigene0029497, Unigene0027804, Unigene0022458, Unigene0022457, Unigene0036457, Unigene0004787, Unigene0030152, Unigene0037512, and Unigene0027059) for further investigation via qRT-PCR (Supplementary Table S1; Supplementary Figure S7A). The results showed that the expression patterns determined via qRT-PCR were highly consistent with the RNA–seq data, with a relative R2 of 0.94 (Supplementary Figure S7B).
Discussion
To address the increasing the demand for food due to the growing world population under various abiotic stresses, a more sustainable and environmentally friendly approach is required. The use of PGPR has recently become a new option for improving salt stress tolerance in plants due to its beneficial interactions with plants (Bhat et al., 2020; Chen et al., 2021). Bacillus is the most common genus found in PGPR and represent the most widely used and well-studied bacteria in agricultural production (Shultana et al., 2022; Singh et al., 2022; Ditta and Ullah, 2023; Pistelli et al., 2023). Many Bacillus spp. have been reported to enhance plant tolerance to salinity and mitigate the salt stress-induced inhibition of plant growth. For example, Akbar et al. (2022) reported that B. subtilis and B. pumilus significantly enhanced salt stress tolerance in cotton plants under salt stress conditions. Ansari et al. (2019) suggested that the B. pumilus strain FAB10 contributes to salt stress alleviation in wheat plants through multiple modes of action. Similarly, B. amyloliquefaciens has been demonstrated to increase salt tolerance in a variety of host plants, such as Arabidopsis (Liu et al., 2020), maize (Chen et al., 2016), rice (Nautiyal et al., 2013) and Codonopsis pilosula (Han et al., 2017). In the present study, the PGP activity of B. amyloliquefaciens PG-4 was evaluated under detached and greenhouse conditions (Supplementary Table S2). The results indicated that the plant-beneficial effects were supported by the successful colonization of M. uniflorum by PG-4 under both mock and salt conditions. The ability of PGPR to alleviate salt stress damage and improve plant growth could result from the production of bioactive secondary metabolites or compounds. PG-4 produced ACC deaminase, siderophores, nitrogen-fixing enzymes and major enzymes, such as protease, amylase, and cellulase. ACC is a precursor for ethylene, one of the most important regulatory hormones for plant growth (Glick, 2014). The production of siderophores is one of the most vital mechanisms for preventing plant pathogens (Etesami and Maheshwari, 2018), suggesting the potential biocontrol function of PG-4.
Salt stress usually triggers ROS accumulation by reducing CO2 assimilation, interfering with plant metabolism and causing ion toxicity (Chen et al., 2022; Wang Y. et al., 2022; Wang Q. et al., 2022). The rapid and excessive accumulation of ROS causes lipid peroxidation, protein denaturation, membrane damage and nucleic acid dysfunction in plants (Gupta and Pandey, 2020; Wang Y. et al., 2022; Wang Q. et al., 2022). The level of malondialdehyde (MDA), produced during oxidative damage caused by lipids (lipid peroxidation), is regarded as the main indicator of salinity-induced oxidative stress injury in plant tissues (Gupta and Pandey, 2020; Wang et al., 2021). Compared with that of control plants, the PG-4-inoculated plants presented a lower content of MDA under salt stress (Figure 3D), which agrees with the findings of previous studies (Hmaeid et al., 2019; Gupta and Pandey, 2020; Masmoudi et al., 2021). Wang Y. et al. (2022) and Wang Q. et al. (2022) also showed that B. cereus G2 alleviated the toxic effect of oxidative bursts in Glycyrrhiza uralensis Fisch. resulting from salt stress by inhibiting the production rate of O2− and correspondingly decreasing the concentration of MDA. Therefore, we speculate that the inoculation of plants with PG-4 reduces the MDA content and reduces ROS production and accumulation at the initial stage of salt stress, which might be an important mechanism for enhancing the salt tolerance of plants. Antioxidant enzymes play an important role in maintaining the normal functioning of plants by repairing and detoxifying the oxidative damage caused by stress-generated ROS (Akhter et al., 2022; Malea et al., 2022). In the antioxidant enzyme system, the conversion of superoxide anions (O2−) into H2O and H2O2 by SOD is the “first line of defense” for plants to cope with oxidative stress. Peroxidase (POD) is an H2O2 scavenging enzyme (Akhter et al., 2022; Wang Y. et al., 2022; Wang Q. et al., 2022). In the present study, compared to that in non-salt -stressed plants, the POD activity in plant leaves significantly increased after 2 weeks of salt stress (Figure 3F), indicating that salinity increases the activities of antioxidants involved in scavenging ROS (Abd_Allah et al., 2018; Wang et al., 2020). Most notably, inoculation with PG-4 significantly increased SOD and POD activity in salt-stressed M. uniflorum plants, enabling them to clear excess ROS and alleviate oxidative stress in plants. Similar studies by Hegazi et al. (2017) and Masmoudi et al. (2021) have shown that inoculation with beneficial microorganisms can trigger antioxidant defense mechanisms in plants to induce the synthesis of antioxidant enzymes such as SOD, POD, and CAT. Moreover, osmotic adjustment occurs by reducing osmotic potential to maintain cell volume and turgor, thereby reducing the adverse effects of salt stress by balancing solute potential, which subsequently promotes cell growth (Zhang et al., 2019). In the present study, the soluble sugar and proline contents in M. uniflorum leaves also significantly increased under salt stress (Figures 3B,C), which was consistent with previous findings in soybean (Soliman et al., 2020), maize (Wang Y. et al., 2022; Wang Q. et al., 2022) and Pisum sativum (Gupta et al., 2022). In particular, PG-4 further increased the proline and soluble sugar contents in salt-stressed M. uniflorum plants, suggesting that PG-4 could lower the osmotic potential and maintain cell turgor by promoting proline and soluble sugar accumulation, eventually helping M. uniflorum respond to the osmotic stress caused by salt stress. Soluble sugars and proline are involved not only in osmotic adjustment but also in scavenging ROS and protecting cells from oxidative damage (Zouari et al., 2019).
Salt stress results in the accumulation of Na+ ions within plants (Joshi et al., 2022). Excessive Na+ in plants competes with other ions to bind proteins, which leads to enzyme inactivation and affects the uptake of K+ and other ions through the impact of ion balance between the two sides of the cytoplasmic membrane, causing severe ion toxicity (Zhu, 2016; Liu et al., 2022). Therefore, reducing the accumulation of Na+ in the cytoplasm to maintain ion balance in plants is necessary for plants to reduce ion toxicity and enhance salt tolerance. We observed that salt stress led to a decrease in the K+ concentration in the M. uniflorum seedlings, while inoculation with PG-4 significantly increased the K+ concentration in the seedlings (Figures 3G,H). Our findings indicated that PG-4 reconstructs the ion balance of host plants by increasing K+ and/or reducing Na+ in the cycloplasm, thereby reducing the harmful effects of Na+ on plants (Chen et al., 2016, 2017; Yilmaz et al., 2020; Shultana et al., 2022). Tank and Saraf (2010) suggested that PGPR inoculation increases the production of exopolysaccharides, which retain Na+ in salt soil and prevent absorption by plant roots. Moreover, after inoculation with PG-4, the chlorophyll content of the M. uniflorum plants was significantly higher than control plants under salt stress (Figure 3A). These results may be due to the increased absorption and concentration of K+ and reduced absorption of Na+ by inoculation of B. amyloliquefaciens under salt stress, which increase the content of antioxidant compounds and improve photosynthesis (Ruíz-Sánchez et al., 2011; ALKahtani et al., 2021). PGPR can generate exopolysaccharides, which enhance soil structure, increase the availability of soil water, improving plant physiological characteristics, such as chlorophyll content, particularly under stress conditions (Tank and Saraf, 2010; ALKahtani et al., 2020).
In recent years, researchers have gradually begun to study the effect of PGPR on plant molecular responses through the use of the transcriptome method, with the final objective of improving plant salt tolerance (Wang Y. et al., 2022; Wang Q. et al., 2022; Xiao et al., 2022). The investigation of the transcriptome-wide response of M. uniflorum leaves revealed that PG-4-initiated salt tolerance is likely attributed to the regulation of several key biological processes, such as stress signaling components and regulators, plant hormone signal transduction and putative regulators of cell wall organization. Previous studies have shown that PGPR can enhance antioxidant enzyme activity and enhance host plant antioxidant capacity by inducing the upregulation of antioxidant enzyme-encoding genes (Khanna et al., 2021; Nivetha et al., 2021; Maleki et al., 2023). In the present study, GO terms related to the cellular redox state, such as “oxidoreductase activity,” “response to oxidative stress,” “antioxidant activity” and “catalytic activity,” were significantly enriched with DEGs in the BS vs. CS comparison, which was in line with the effects of PG-4 on the enhancement of antioxidant enzyme activity in Macrotyloma uniflorum under salt stress. Moreover, GO terms related to hemoglobin, such as “iron ion binding,” “heme binding” and “tetrapyrrole binding,” were significantly enriched (Supplementary Figure S5). Hemoglobin and its catalyzed products can act as strong antioxidant enzymes against various abiotic stresses, as well as inducers that function in different developmental processes, including adventitious and lateral root development (Zhao et al., 2008; Xu et al., 2011). These results suggested that PG-4 may ameliorate the negative effects of salinity stress on Macrotyloma uniflorum by enhancing its ROS scavenging capacity.
The plant cell wall is the first site at which plants perceive and respond to salt stress (Kaashyap et al., 2018). The stress signal triggers cell wall remodeling to maintain flexibility and protect the cell against ionic imbalance (Kaashyap et al., 2018; Colin et al., 2023). Cell wall modification under abiotic stress conditions is accomplished by modulating the cross-linking of cellulose, pectins and lignin. Xyloglucans are involved in modulating these cross-links and regulating cell wall metabolism processes (Kaashyap et al., 2018; Ganie and Ahammed, 2021). Xyloglucans are linked to at least two cellulose molecules and are reported to remodel stomatal cell walls to prevent excess water loss during osmotic stress (Gall et al., 2015; Byrt et al., 2018). Recently, studies have indicated that XTH participates in plant cell wall construction and modification and that the overexpression of XTH confers increased plant tolerance under high salinity conditions (Zhang et al., 2021). Xyloglucan-modifying enzymes reportedly impart salt and drought tolerance when they are overexpressed in pepper and wheat plants in response to osmotic stress and salt stress (Abuqamar et al., 2013). Kaashyap et al. (2018) reported that the upregulation of xyloglucan genes in the tolerant genotype clearly suggested that their activity in ‘cell wall loosening’ may impart salt stress tolerance to chickpea plants. In the present study, GO terms related to the cell wall, such as “pectinesterase activity,” “cell wall organization,” “cell wall modification” and “cell wall organization or biogenesis,” were also significantly enriched with DEGs in the CS vs. BS comparison, “xyloglucan: xyloglucosyl transferase activity” and “xyloglucan metabolic process” were significantly enriched with DEGs in the CS vs. BS comparison (Supplementary Figure S5). Under salinity stress, the expression levels of DEGs encoding pectinesterase and xyloglucan endotransglucosylase/hydrolase proteins were mainly downregulated. However, the addition of PG-4 facilitated the upregulation of these transcripts under salinity stress (Supplementary Figures S6C,D). This result indicated that there was a connection between the cell wall synthesis and modification induced by PG-4 inoculation and plant tolerance of salt stress.
PGPR play a key role in regulating plant signal transduction under salt stress. Liu et al. (2017) reported that FZB42 might use the ethylene and jasmonic acid transduction pathways to induce systemic salt tolerance in Arabidopsis plants. Lastochkina et al. (2017) reported that B. subtilis enhances salt stress tolerance in wheat by enhancing JA signaling. B. pumilus enhances the salt tolerance of cotton plants and is related to SA signaling (Akbar et al., 2022). In this study, the auxin, cytokinin, gibberellin, abscisic acid, ethylene, brassinosteroid, JA and salicylic acid signaling pathways were triggered, as reflected in the DEGs (Figure 7). These genes might be involved in the plant mechanism underlying the salt stress adaptation conferred by PG-4.
TFs play important roles in regulating gene expression, biotic and abiotic stress responses, and signal transduction by acting as gene activators, repressors, or both (Zhang et al., 2021; Wang Y. et al., 2022; Wang Q. et al., 2022). The roles of TFs in salt stress tolerance have been clearly demonstrated in important plants, such as pepper (Capsicum frutescens L.) (Wang Y. et al., 2022; Wang Q. et al., 2022), asparagus (Asparagus officinalis) (Zhang et al., 2021), rice (Tang et al., 2019), and tomato (Solanum lycopersicum) (Devkar et al., 2020). For example, Gruber et al. (2009) identified 46 TFs as salt-inducible genes in Medicago truncatula roots, including members of the AP2/ERF, WRKY, zinc-finger bZIP, zinc finger, GRAS, ZIM, Bhlh, MYB and bZIP families. In this study, several of the TFs belonging to the AP2/ERF, MYB, bHLH, WRKY, NAC, and bZIP families were downregulated by salt stress and upregulated by PG-4 under salt stress (Figure 8; Supplementary Figure S17), indicating the critical involvement of these TFs in the salt stress response and the important roles of PG-4 in the development of salt tolerance in plants.
Conclusion
The application of PGPR plays a crucial role in advancing agricultural resilience and productivity. PGPRs markedly enhance soil health, nutrient assimilation, and plant development, culminating in increased crop yields and a diminished dependency on chemical fertilizers, and its application is of great significance for sustainable agricultural. Furthermore, PGPRs are promising tools for increasing crop salt stress resistance. In the present study, the beneficial effects of B. amyloliquefaciens PG-4 on M. uniflorum plants under salt stress were evaluated. As shown in Figure 9, PG-4 significantly mitigated the deleterious effects of salt stress on M. uniflorum plants by improving biomass growth, increasing chlorophyll content, limiting the cellular absorption of Na+ and export of K+,” and maintaining membrane integrity by reducing lipid peroxidation and increasing antioxidant production. Additionally, our transcriptome analysis revealed several key mediators of PG-4-induced transcriptional regulation in M. uniflorum, including plant hormone signal transduction, cell wall synthesis and modification and transporters. Moreover, transcription factors may also play important roles in mitigating the detrimental effects of salt stress on M. uniflorum, such as MYB, AP2/ERF, and bHLH. Our findings are helpful for providing important insights into the salt tolerance mechanism induced by PGPR and for providing a solid basis for future studies on the enhancement of legume forage tolerance to salinity stress, and providing theoretical references for bio-mediated soil improvement.
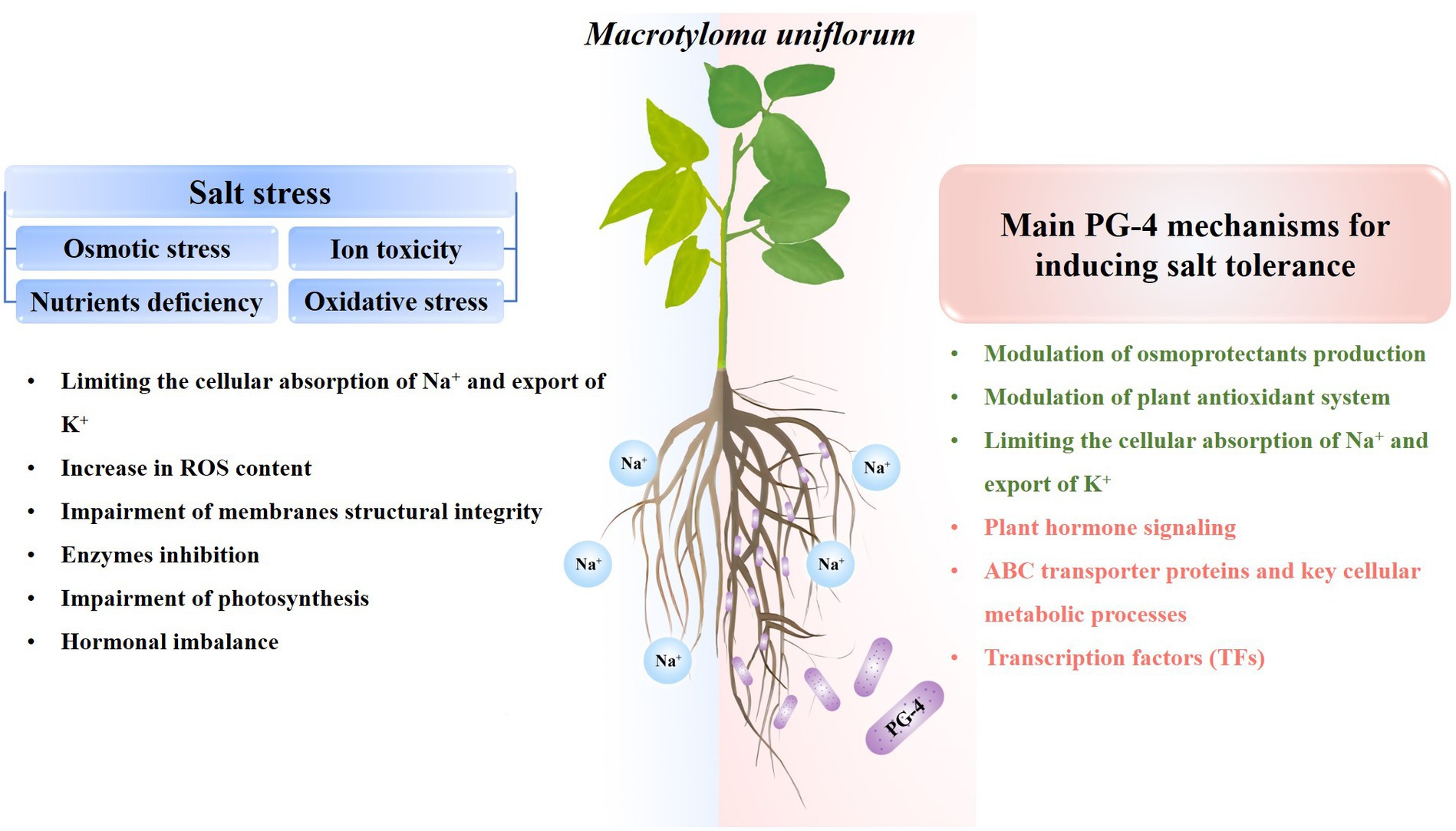
Figure 9. General overview of salt stress effects on plants (left) and PG-4 mechanisms involved in salt-stress tolerance (right) in Macrotyloma uniflorum seedlings. Green texts represent the physiological mechanism, while red texts represent the molecular mechanism by which PG-4 enhances the salt tolerance of M. uniflorum seedlings.
Data availability statement
The data presented in the study are deposited in the SRA: https://www.ncbi.nlm.nih.gov/sra/PRJNA1075426, accession number PRJNA1075426.
Author contributions
YW: Writing – original draft, Writing – review & editing. CG: Writing – review & editing. YX: Writing – review & editing. XL: Resources, Supervision, Writing – review & editing. DY: Resources, Supervision, Writing – review & editing. QW: Writing – review & editing. HL: Writing – review & editing. YZ: Conceptualization, Supervision, Writing – review & editing. RZ: Conceptualization, Supervision, Writing – review & editing. KL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Finance Science and Technology Project of Hainan Province (SQKY2022-0005), the National Natural Science Foundation of China (32160324), Innovative Team Project of Safety Prevention and Control of Major Tropical Fruit and Vegetable Pests of the Hainan Academy of Agricultural Sciences (HAAS2023TDYD12), and Collaborative Innovation Center of Nanfan and High-Efficiency Tropical Agriculture, Hainan University (XTCX2022NYC10).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1386079/full#supplementary-material
Footnotes
References
Abd_Allah, E. F., Alqarawi, A. A., Hashem, A., Radhakrishnan, R., al-Huqail, A. A., al-Otibi, F. O. N., et al. (2018). Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 13, 37–44. doi: 10.1080/17429145.2017.1414321
Abuqamar, S., Ajeb, S., Sham, A., Enan, M. R., and Iratni, R. (2013). A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol. Plant Pathol. 14, 813–827. doi: 10.1111/mpp.12049
Akbar, A., Han, B., Khan, A. H., Feng, C., Ullah, A., Khan, A. S., et al. (2022). A transcriptomic study reveals salt stress alleviation in cotton plants upon salt tolerant PGPR inoculation. Environ. Exp. Bot. 200:104928. doi: 10.1016/j.envexpbot.2022.104928
Akhter, M. S., Noreen, S., Ummara, U., Aqeel, M., Saleem, N., Ahmed, M. M., et al. (2022). Silicon-induced mitigation of NaCl stress in barley (Hordeum vulgare L.), associated with enhanced enzymatic and non-enzymatic antioxidant activities. Plan. Theory 11:2379. doi: 10.3390/plants11182379
Ali, B., Wang, X., Saleem, M. H., Sumaira, H., Hafeez, A., Afridi, M. S., et al. (2022). PGPR-mediated salt tolerance in maize by modulating plant physiology, antioxidant defense, compatible solutes accumulation and bio-surfactant producing genes. Plan. Theory 11:345. doi: 10.3390/plants11030345
ALKahtani, M. D. F., Fouda, A., Attia, K. A., al-Otaibi, F., Eid, A. M., Ewais, E. E.-D., et al. (2020). Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 10:1325. doi: 10.3390/agronomy10091325
ALKahtani, M., Hafez, Y., Attia, K., al-Ateeq, T., Ali, M. A. M., Hasanuzzaman, M., et al. (2021). Bacillus thuringiensis and silicon modulate antioxidant metabolism and improve the physiological traits to confer salt tolerance in lettuce. Plan. Theory 10:1025. doi: 10.3390/plants10051025
Ansari, F. A., Ahmad, I., and Pichtel, J. (2019). Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10. Appl. Soil Ecol. 143, 45–54. doi: 10.1016/j.apsoil.2019.05.023
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., and Ferro, S. (2004). Uni Prot: the universal protein knowledgebase. Nucleic Acids Res. 32, 115D–1119D. doi: 10.1093/nar/gkh131
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Ashraf, M., and Munns, R. (2022). Evolution of approaches to increase the salt tolerance of crops. Crit. Rev. Plant Sci. 41, 128–160. doi: 10.1080/07352689.2022.2065136
Baldani, V. L. D., and Döbereiner, J. (1980). Host-plant specificity in the infection of cereals with Azospirillum spp. Soil. Biolo. Biochem. 12, 433–439. doi: 10.1016/0038-0717(80)90021-8
Bhat, M. A., Kumar, V., Bhat, M. A., Wani, I. A., Dar, F. L., Farooq, I., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 11:1952. doi: 10.3389/fmicb.2020.01952
Bolbhat, S. N., and Dhumal, K. N. (2009). Induced macromutations in horsegram [Macrotyloma uniflorum (lam.) Verdc]. Legum. Res. 32, 278–281.
Butcher, K., Wick, A. F., DeSutter, T., Chatterjee, A., and Harmon, J. (2016). Soil Salinity: A Threat to Global Food Security. Agron J, 108, 2189–2200. doi: 10.2134/agronj2016.06.0368
Byrt, C. S., Munns, R., Burton, R. A., Gilliham, M., and Wege, S. (2018). Root cell wall solutions for crop plants in saline soils. Plant Sci. 269, 47–55. doi: 10.1016/j.plantsci.2017.12.012
Cappuccino, J. C., and Sherman, N. (1992). Ammonia Production. Microbiol. A Lab. Manual, 3rd edn. Redw, City, CA, USA: Benjamin/Cummings Pub Co., pp. 125–179.
Chen, J. T., Aroca, R., and Romano, D. (2021). Molecular aspects of plant salinity stress and tolerance. Int. J. Mol. Sci. 22:4918. doi: 10.3390/ijms22094918
Chen, G., Han, H., Yang, X., Du, R., and Wang, X. (2022). Salt tolerance of rice is enhanced by the SS3 gene, which regulates ascorbic acid synthesis and ROS scavenging. Int. J. Mol. Sci. 23:10338. doi: 10.3390/ijms231810338
Chen, L., Liu, Y., Wu, G., Veronican Njeri, K., Shen, Q., Zhang, N., et al. (2016). Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plantarum 158, 34–44. doi: 10.1111/ppl.12441
Chen, L., Liu, Y., Wu, G., Zhang, N., Shen, Q., and Zhang, R. (2017). Beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 induces plant salt tolerance through spermidine production. Mol. Plant Microbe In. 30, 423–432. doi: 10.1094/MPMI-02-17-0027-R
Colin, L., Ruhnow, F., Zhu, J. K., Zhao, C., Zhao, Y., and Persson, S. (2023). The cell biology of primary cell walls during salt stress. Plant Cell 35, 201–217. doi: 10.1093/plcell/koac292
Devkar, V., Thirumalaikumar, V. P., Xue, G. P., Vallarino, J. G., Turečková, V., Strnad, M., et al. (2020). Multifaceted regulatory function of tomato SlTAF1 in the response to salinity stress. New Phytol. 225, 1681–1698. doi: 10.1111/nph.16247
Ditta, A., and Ullah, N. (2023). “Perspectives of using plant growth-promoting rhizobacteria under salinity stress for sustainable crop production” in Plant Stress Mitigators. eds. M. Ghorbanpour and M. A. Shahid (Cambridge, MA: Academic Press), 231–247.
Dong, X., and Cai, M. (1999). Handbook for the identification of common bacteria. Beijing: Science Press 353–365
Egamberdieva, D., Wirth, S., Bellingrath-Kimura, S. D., Mishra, J., and Arora, N. K. (2019). Salt-tolerant plant growth promoting rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 10:2791. doi: 10.3389/fmicb.2019.02791
Etesami, H., and Glick, B. R. (2020). Halotolerant plant growth–promoting bacteria: prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 178:104124. doi: 10.1016/j.envexpbot.2020.104124
Etesami, H., and Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotox. Environ. Safe. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Farhangi-Abriz, S., Tavasolee, A., Ghassemi-Golezani, K., Torabian, S., Monirifar, H., and Rahmani, H. A. (2020). Growth-promoting bacteria and natural regulators mitigate salt toxicity and improve rapeseed plant performance. Protoplasma 257, 1035–1047. doi: 10.1007/s00709-020-01493-1
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H. R., et al. (2008). The Pfam protein families database. Nucleic Acids Res. 32, 138D–1141D. doi: 10.1093/nar/gkh121
Gall, H. L., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plan. Theory 4, 112–166. doi: 10.3390/plants4010112
Ganie, S. A., and Ahammed, G. J. (2021). Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 40, 437–459. doi: 10.1007/s00299-020-02649-2
Garcia-Caparros, P., De Filippis, L., Gul, A., Hasanuzzaman, M., Ozturk, M., Altay, V., et al. (2021). Oxidative stress and antioxidant metabolism under adverse environmental conditions: a review. The Bot. Rev. 87, 421–466. doi: 10.1007/s12229-020-09231-1
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. FEMS Microbiol. Rev. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Gruber, V., Blanchet, S., Diet, A., Zahaf, O., Boualem, A., Kakar, K., et al. (2009). Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol. Gen. Genomics. 281, 55–66. doi: 10.1007/s00438-008-0392-8
Gupta, S., and Pandey, S. (2020). Enhanced salinity tolerance in the common bean (Phaseolus vulgaris) plants using twin ACC deaminase producing rhizobacterial inoculation. Rhizosphere 16:100241. doi: 10.1016/j.rhisph.2020.100241
Gupta, A., Rai, S., Bano, A., Sharma, S., Kumar, M., Binsuwaidan, R., et al. (2022). ACC deaminase produced by pgpr mitigates the adverse effect of osmotic and salinity stresses in Pisum sativum through modulating the antioxidants activities. Plan. Theory 11:3419. doi: 10.3390/plants11243419
Gupta, P., Samant, K., and Sahu, A. (2012). Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. microbiol. 2012:578925. doi: 10.1155/2012/578925
Han, Q. Q., Wu, Y. N., Gao, H. J., Xu, R., Paré, P. W., Shi, H., et al. (2017). Improved salt tolerance of medicinal plant Codonopsis pilosula by Bacillus amyloliquefaciens GB03. Acta Physiol. Plant. 39, 1–7. doi: 10.1007/s11738-016-2325-1
Haque, M. A., Rafii, M. Y., Yusoff, M. M., Ali, N. S., Yusuff, O., Datta, D. R., et al. (2021). Advanced breeding strategies and future perspectives of salinity tolerance in rice. Agronomy 11:1631. doi: 10.3390/agronomy11081631
Hasanuzzaman, M., Raihan, M. R. H., Nowroz, F., and Fujita, M. (2022). Insight into the mechanism of salt-induced oxidative stress tolerance in soybean by the application of Bacillus subtilis: coordinated actions of osmoregulation, ion homeostasis, antioxidant defense, and methylglyoxal detoxification. Antioxidants 11:1856. doi: 10.3390/antiox11101856
Hegazi, A. M., El-Shraiy, A. M., and Ghoname, A. A. (2017). Mitigation of salt stress negative effects on sweet pepper using arbuscular mycorrhizal fungi (AMF), bacillus megaterium and brassinosteroids (BRs). Gesunde Pflanz. 69, 91–102. doi: 10.1007/s10343-017-0393-9
Hmaeid, N., Wali, M., Mahmoud, O. M. B., Pueyo, J. J., Ghnaya, T., and Abdelly, C. (2019). Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl. Soil Ecol. 133, 104–113. doi: 10.1016/j.apsoil.2018.09.011
Hou, Y., Zeng, W., Ao, C., Luo, Y., Wang, Z., Hou, M., et al. (2022). Bacillus atrophaeus WZYH01 and planococcus soli WZYH02 improve salt tolerance of maize (Zea mays l.) in saline soil. Front. Plant Sci. 13:891372. doi: 10.3389/fpls.2022.891372
Ibarra-Villarreal, A. L., Gándara-Ledezma, A., Godoy-Flores, A. D., Herrera-Sepúlveda, A., Díaz-Rodríguez, A. M., Parra-Cota, F. I., et al. (2021). Salt-tolerant bacillus species as a promising strategy to mitigate the salinity stress in wheat (Triticum turgidum subsp. durum). J. Arid Environ. 186:104399. doi: 10.1016/j.jaridenv.2020.104399
Joshi, S., Nath, J., Singh, A. K., Pareek, A., and Joshi, R. (2022). Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants. Physiol. Plantarum 174:e13702. doi: 10.1111/ppl.13702
Kaashyap, M., Ford, R., Kudapa, H., Jain, M., Edwards, D., Varshney, R., et al. (2018). Differential regulation of genes involved in root morphogenesis and cell wall modification is associated with salinity tolerance in chickpea. Sci. Rep. 8:4855. doi: 10.1038/s41598-018-23116-9
Kadam, S., and Salunkhe, D. K. (1985). Nutritional composition, processing, and utilization of horse gram and moth bean. Crit. Rev. Food Sci. Nutr. 22, 1–26. doi: 10.1080/10408398509527416
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kazerooni, E. A., Maharachchikumbura, S. S., Adhikari, A., Al-Sadi, A. M., Kang, S. M., Kim, L. R., et al. (2021). Rhizospheric Bacillus amyloliquefaciens protects Capsicum annuum cv. Geumsugangsan from multiple abiotic stresses via multifarious plant growth-promoting attributes. Front. Plant Sci. 12:669693. doi: 10.3389/fpls.2021.669693
Khanna, K., Kohli, S. K., Sharma, P., Kour, J., Singh, A. D., Sharma, N., et al. (2021). “Antioxidant potential of plant growth-promoting rhizobacteria (PGPR) in agricultural crops infected with root-knot nematodes” in Antioxidants in plant-microbe interaction. eds. A. K. Singh, S. Kumar, and T. Sinha (Berlin: Springer)
Kim, M. J., Radhakrishnan, R., Kang, S. M., You, Y. H., Jeong, E. J., Kim, J. G., et al. (2017). Plant growth promoting effect of Bacillus amyloliquefaciens H-2-5 on crop plants and influence on physiological changes in soybean under soil salinity. Physiol. Mol. Biol. Pla. 23, 571–580. doi: 10.1007/s12298-017-0449-4
Koonin, E. V., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Krylov, D. M., Makarova, K. S., et al. (2004). A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7–R28. doi: 10.1186/gb-2004-5-2-r7
Lastochkina, O., Pusenkova, L., Yuldashev, R., Babaev, M., Garipova, S., Blagova, D. Y., et al. (2017). Effects of Bacillus subtilis on some physiological and biochemical parameters of Triticum aestivum L. (wheat) under salinity. Plant Physiol. Bioch. 121, 80–88. doi: 10.1016/j.plaphy.2017.10.020
Li, G. E., Kong, W. L., Wu, X. Q., and Ma, S. B. (2021). Phytase-producing Rahnella aquatilis JZ-GX1 promotes seed germination and growth in corn (Zea mays L.). Microorganisms 9:1647. doi: 10.3390/microorganisms9081647
Li, Y., Li, W., Hu, D., Shen, P., Zhang, G., and Zhu, Y. (2020). Comparative analysis of the metabolome and transcriptome between green and albino zones of variegated leaves from Hydrangea macrophylla ‘Maculata’ infected by hydrangea ringspot virus. Plant Physiol. Bioch. 157, 195–210. doi: 10.1016/J.Plaphy.2020.10.012
Liu, S., Hao, H., Lu, X., Zhao, X., Wang, Y., Zhang, Y., et al. (2017). Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana. Sci. Rep. 7:10795. doi: 10.1038/s41598-017-11308-8
Liu, S., Tian, Y., Jia, M., Lu, X., Yue, L., Zhao, X., et al. (2020). Induction of salt tolerance in Arabidopsis thaliana by volatiles from Bacillus amyloliquefaciens FZB42 via the jasmonic acid signaling pathway. Front. Microbiol. 11:562934. doi: 10.3389/fmicb.2020.562934
Liu, Y., Xun, W., Chen, L., Xu, Z., Zhang, N., Feng, H., et al. (2022). Rhizosphere microbes enhance plant salt tolerance: toward crop production in saline soil. Comput. Struct. Biotec. 20, 6543–6551. doi: 10.1016/j.csbj.2022.11.046(30)
Ma, T., Zeng, W., Lei, G., Wu, J., and Huang, J. (2021). Predicting the rooting depth, dynamic root distribution and the yield of sunflower under different soil salinity and nitrogen applications. Ind. Crop. Prod. 170:113749. doi: 10.1016/j.indcrop.2021.113749
Malea, P., Kokkinidi, D., Kevrekidou, A., and Adamakis, I. S. (2022). The enzymatic and non-enzymatic antioxidant system response of the seagrass Cymodocea nodosa to bisphenol-a toxicity. Int. J. Mol. Sci. 23:1348. doi: 10.3390/ijms23031348
Maleki, M., Fatehi, F., and Ghorbanpour, M. (2023). “PGPR reduces the adverse effects of abiotic stresses by modulating morphological and biochemical properties in plants” in Plant Stress Mitigators. eds. A. Vaishnav, S. S. Arya, and D. K. Choudhary (Singapore: Springer)
Masi, C., Gemechu, G., and Tafesse, M. (2021). Isolation, screening, characterization, and identification of alkaline protease-producing bacteria from leather industry effluent. Ann. Microbiol. 71, 1–11. doi: 10.1186/S13213-021-01631-X
Masmoudi, F., Tounsi, S., Dunlap, C. A., and Trigui, M. (2021). Endophytic halotolerant Bacillus velezensis FMH2 alleviates salt stress on tomato plants by improving plant growth and altering physiological and antioxidant responses. Plant Physiol. Bioch. 165, 217–227. doi: 10.1016/j.plaphy.2021.05.025
Mohanty, P., Singh, P. K., Chakraborty, D., Mishra, S., and Pattnaik, R. (2021). Insight into the role of PGPR in sustainable agriculture and environment. Front. Sustain. Food Syst. 5:667150. doi: 10.3389/fsufs.2021.667150
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nautiyal, C. S., Srivastava, S., Chauhan, P. S., Seem, K., Mishra, A., and Sopory, S. K. (2013). Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Bioch. 66, 1–9. doi: 10.1016/j.plaphy.2013.01.020
Nivetha, N., Lavanya, A. K., Vikram, K. V., Asha, A. D., Sruthi, K. S., Bandeppa, S., et al. (2021). “PGPR-mediated regulation of antioxidants: prospects for abiotic stress management in plants” in Antioxidants in plant-microbe interaction. eds. A. K. Singh, S. Kumar, and T. Sinha (Berlin: Springer)
Patani, A., Prajapati, D., Ali, D., Kalasariya, H., Yadav, V. K., Tank, J., et al. (2023). Evaluation of the growth-inducing efficacy of various bacillus species on the salt-stressed tomato (Lycopersicon esculentum mill.). Front. Plant Sci. 14:1168155. doi: 10.3389/fpls.2023.1168155
Pistelli, L., Libik-Konieczny, M., and Hasanuzzaman, M. (2023). Plant-microbe interactions and their role in salinity tolerance. Front. Plant Sci. 14:1142563. doi: 10.3389/fpls.2023.1142563
Plazinski, J., and Rolfe, B. G. (1985). Analysis of the pectolytic activity of Rhizobium and Azospirillum strains isolated from Trifolium repens. J. Plant Physiol. 120, 181–187. doi: 10.1016/S0176-1617(85)80021-3
Pruitt, K. D., Tatusova, T., and Maglott, D. R. (2007). NCBI reference sequence (ref Seq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65. doi: 10.1093/nar/gkl842
Ruíz-Sánchez, M., Armada, E., Muñoz, Y., García de Salamone, I. E., Aroca, R., Ruíz-Lozano, J. M., et al. (2011). Azospirillum and arbuscular mycorrhizal colonization enhance rice growth and physiological traits under well-watered and drought conditions. J. Plant Physiol. 168, 1031–1037. doi: 10.1016/j.jplph.2010.12.019
Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. P. Natl. Acad. Sci. USA. 98, 14150–14155. doi: 10.1073/pnas.241501798
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shultana, R., Zuan, A. T. K., Naher, U. A., Islam, A. M., Rana, M. M., Rashid, M. H., et al. (2022). The PGPR mechanisms of salt stress adaptation and plant growth promotion. Agronomy 12:2266. doi: 10.3390/Agronomy12102266
Singh, P., Choudhary, K. K., Chaudhary, N., Gupta, S., Sahu, M., Tejaswini, B., et al. (2022). Salt stress resilience in plants mediated through osmolyte accumulation and its crosstalk mechanism with phytohormones. Front. Plant Sci. 13:1006617. doi: 10.3389/fpls.2022.1006617
Soliman, M., Elkelish, A., Souad, T., Alhaithloul, H., and Farooq, M. (2020). Brassinosteroid seed priming with nitrogen supplementation improves salt tolerance in soybean. Physiol. Mol. Biol. Plants 26, 501–511. doi: 10.1007/s12298-020-00765-7
Tanaka, K., Fukuda, M., Amaki, Y., Sakaguchi, T., Inai, K., Ishihara, A., et al. (2017). Importance of prumycin produced by Bacillus amyloliquefaciens SD-32 in biocontrol against cucumber powdery mildew disease. Pest Manag. Sci. 73, 2419–2428. doi: 10.1002/ps.4630
Tang, Y., Bao, X., Zhi, Y., Wu, Q., Guo, Y., Yin, X., et al. (2019). Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 10:168. doi: 10.3389/fpls.2019.00168
Tank, N., and Saraf, M. (2010). Salinity-resistant plant growth promoting rhizobacteria ameliorates sodium chloride stress on tomato plants. J. Plant Interact. 5, 51–58. doi: 10.1080/17429140903125848
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Wang, Y., Huang, L., Du, F., Wang, J., Zhao, X., Li, Z., et al. (2021). Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci. Rep. 11:5166. doi: 10.1038/s41598-021-84638-3
Wang, D., Lu, X., Chen, X., Wang, S., Wang, J., Guo, L., et al. (2020). Temporal salt stress-induced transcriptome alterations and regulatory mechanisms revealed by Pac bio long-reads RNA sequencing in Gossypium hirsutum. BMC Genomics 21, 1–15. doi: 10.1186/s12864-020-07260-z
Wang, Q., Peng, X., Lang, D., Ma, X., and Zhang, X. (2022). Physio-biochemical and transcriptomic analysis reveals that the mechanism of Bacillus cereus G2 alleviated oxidative stress of salt-stressed Glycyrrhiza uralensis Fisch. Seedlings. Ecotox. Environ. Safe 247:114264. doi: 10.1016/j.ecoenv.2022.114264
Wang, Y., Sun, Q., Liu, J., Wang, L., Wu, X., Zhao, Z., et al. (2022). Suaeda salsa root-associated microorganisms could effectively improve maize growth and resistance under salt stress. Microbiol. Spectr. 10, e01349–e01322. doi: 10.1128/spectrum.01349-22
Wang, W., Wu, Z., He, Y., Huang, Y., Li, X., and Ye, B. C. (2018). Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by bacillus isolated from saline soil in Xinjiang. Ecotox. Environ. Safe 164, 520–529. doi: 10.1016/j.ecoenv.2018.08.070
Woo, O. G., Kim, H., Kim, J. S., Keum, H. L., Lee, K. C., Sul, W. J., et al. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 148, 359–367. doi: 10.1016/j.plaphy.2020.01.032
Xiao, X., Wang, Q., Ma, X., Lang, D., Guo, Z., and Zhang, X. (2022). Physiological biochemistry-combined transcriptomic analysis reveals mechanism of Bacillus cereus G2 improved salt-stress tolerance of Glycyrrhiza uralensis Fisch. Seedlings by balancing carbohydrate metabolism. Front. Plant Sci. 12:712363. doi: 10.3389/fpls.2021.712363
Xie, Z., Nolan, T. M., Jiang, H., and Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10:228. doi: 10.3389/fpls.2019.00228
Xu, S., Hu, B., He, Z., Ma, F., Feng, J., Shen, W., et al. (2011). Enhancement of salinity tolerance during rice seed germination by presoaking with hemoglobin. Int. J. Mol. Sci. 12, 2488–2501. doi: 10.3390/ijms12042488
Yilmaz, Y., Erdinc, C., Akköprü, A., and Kıpçak, S. (2020). Use of plant growth promoting rhizobacteria against salt stress for tomato (Solanum lycopersicum L.) seedling growth. Acta Sci. Pol. Hortorum. Cultus. 19, 15–29. doi: 10.24326/asphc.2020.6.2
Yilmaz, H., and Kulaz, H. (2019). The effects of plant growth promoting rhizobacteria on antioxidant activity in chickpea (Cicer arietinum L.) under salt stress. Legum. Res. 42, 72–76. doi: 10.18805/LR-435
Zhang, X., Gao, H., Liang, Y., and Cao, Y. (2021). Full-length transcriptome analysis of asparagus roots reveals the molecular mechanism of salt tolerance induced by arbuscular mycorrhizal fungi. Environ. Exp. Bot. 185:104402. doi: 10.1016/j.envexpbot.2021.104402
Zhang, F., Wang, Y., Liu, C., Chen, F., Ge, H., Tian, F., et al. (2019). Trichoderma harzianum mitigates salt stress in cucumber via multiple responses. Ecotox. Environ. Safe. 170, 436–445. doi: 10.1016/j.ecoenv.2018.11.084
Zhao, L., Gu, R., Gao, P., and Wang, G. (2008). A nonsymbiotic hemoglobin gene from maize, ZmHb, is involved in response to submergence, high-salt and osmotic stresses. Plant Cell Tiss. Org. 95, 227–237. doi: 10.1007/s11240-008-9436-3
Zhou, H., Ren, Z. H., Zu, X., Yu, X. Y., Zhu, H. J., Li, X. J., et al. (2021). Efficacy of plant growth-promoting bacteria Bacillus cereus YN917 for biocontrol of rice blast. Front. Microbiol. 12:684888. doi: 10.3389/fmicb.2021.684888
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zouari, M., Hassena, A. B., Trabelsi, L., Rouina, B. B., Decou, R., and Labrousse, P. (2019). “Exogenous proline-mediated abiotic stress tolerance in plants: possible mechanisms” in Osmoprotectant-mediated abiotic stress tolerance in plants: Recent advances and future perspectives. eds. M. A. Hossain, V. Kumar, D. J. Burritt, M. Fujita, and P. S. A. Mäkelä (Cham: Springer)
Keywords: plant growth-promoting rhizobacteria, Bacillus amyloliquefaciens, Macrotyloma uniflorum, salt stress tolerance, transcriptome
Citation: Wu Y, Guo C, Xiao Y, Li X, Yu D, Wan Q, Liu H, Zhang Y, Zhang R and Luo K (2024) Physio-biochemical and transcriptomic analysis of Bacillus amyloliquefaciens PG-4-induced salt stress tolerance in Macrotyloma uniflorum. Front. Sustain. Food Syst. 8:1386079. doi: 10.3389/fsufs.2024.1386079
Edited by:
Zhou Li, Guizhou University, ChinaReviewed by:
Yuguo Wu, Guangxi University, ChinaRamadoss Dhanuskodi, Massey University, New Zealand
Copyright © 2024 Wu, Guo, Xiao, Li, Yu, Wan, Liu, Zhang, Zhang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhang, emhhbmdydWlAaGFpbmFudS5lZHUuY24=; Kai Luo, bHVva0BoYWluYW51LmVkdS5jbg==
 Yanna Wu1,2
Yanna Wu1,2 Daogeng Yu
Daogeng Yu Qi Wan
Qi Wan Rui Zhang
Rui Zhang Kai Luo
Kai Luo