- 1College of Agriculture, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Utilization and Conservation of Food and Medicinal Resources in Northern Region, Shaoguan University, Shaoguan, China
- 3Department of Biology, College of science, University of Basrah, Basrah, Iraq
- 4Plant Protection, College of Agriculture, University of Misan, AL-amarah, Iraq
- 5Al-Manara College for Medical Sciences, Maysan, AL-Amarah, Iraq
- 6Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
Eggplant production in China is significantly impacted by Verticillium wilt caused by Verticillium dahliae, leading to substantial yield losses. This study was designed to investigate the potential of rhizobacterial species for the biocontrol of Verticillium wilt in eggplant. Among the 42 bacterial isolates tested, strain ARF4 demonstrated the strongest antagonistic effect by inhibiting V. dahliae growth by 84.49%, in addition to showing antifungal properties against four other plant pathogens. We found the strain ARF4 is closely related to Bacillus velezensis with high bootstrap values (100) through a phylogenetic tree based on 16S, rpoB, and gyrA gene sequences. The ARF4 produces important antifungal compounds such as chitinase, protease, β-glucosidase, and lipopeptide bacillomycin D, which contribute to its antifungal properties. The extracted lipopeptide of the ARF4 exhibited strong inhibition of conidial germination in V. dahliae. Scanning Electron Microscopy (SEM) showed that hyphae treated with the extracted lipopeptide exhibited considerable deformation. Transmission electron microscopy results revealed lysis of the cell walls and plasma membrane, a decreased inner cytoplasmic matrix and a number of mitochondria, and disintegration of internal organelles. Greenhouse trials demonstrated that eggplants treated with strain ARF4 experienced a significant disease severity reduction of 68.45%. This study offers B. velezensis ARF4 biological approach to Verticillium wilt control in eggplants as an alternative to chemical fungicides and contributes to sustainable agriculture practices.
1 Introduction
Eggplant (Solanum melongena) is a vital agricultural crop in China due to its significant contribution to both the economy and diet. China is the largest producer of eggplant globally, accounting for over 60% of global production. In 2023, China produced approximately 38 million tons of eggplant, showcasing its dominance in the market (Canton, 2021). Zhejiang province stands out as a major hub for producing eggplants in China, with its cultivated area expanding to 26,666 ha by 2018, “according to data from the Agriculture Department, Zhejiang Province.” Eggplant is not only a dietary staple rich in vitamins, minerals, and antioxidants but also a crucial export commodity, enhancing China’s agricultural income (Lyu et al., 2024). Its usefulness in cooking makes it an important component of many traditional Chinese dishes. Despite high production, eggplant farming in China meets major challenges due to various diseases. One of the common diseases impacting eggplant crops includes Verticillium wilt caused by Verticillium dahliae (V. dahliae), which has main symptoms such as yellowing leaves, wilting, and yield loss (Ibanga et al., 2023; Scholz et al., 2018; Yang et al., 2019; Zhang et al., 2024).
Verticillium dahliae, the causative agent of Verticillium wilt, is a soil-borne fungus that is considered a major threat to eggplant production in China, leading to considerable economic losses (Tomah et al., 2023). The occurrence of Verticillium wilt in eggplant cultivation areas in China is responsible for yield losses ranging from 34.1 to 42.5%, which is a major blow to the agricultural economy and causes significant economy impact (Zhu et al., 2021). Urgent research is required to produce resilient, sustainable, and eco-friendly approaches for handling soil-borne diseases affecting eggplant, such as Verticillium wilt. Biological control is considered being widely recognized eco-friendly alternative to traditional chemical methods. It plays a crucial role in integrated pest management by employing natural predators to regulate pest populations. This approach offers several significant economic and ecological advantages, making it an important approach to sustainable agriculture (Barzman et al., 2015; Lahlali et al., 2022). Biocontrol offers a sustainable and economical pest management solution, reducing the risk of pesticide resistance while maintaining long-term effectiveness at minimal cost (Pandit et al., 2022; Shang et al., 2024). This method may decrease our dependency on chemical pesticides, which can harm non-target species such as birds, beneficial insects, and aquatic life (Wan et al., 2025). Implementing biological control to minimize reliance on chemical pesticides can offer substantial environmental and health benefits, as chemical pesticides enhance agricultural productivity, while their improper or excessive use can lead to severe environmental damage, polluting soil, air, and water. While also threatening human health with long-term effects like cancer, endocrine disorders, and poisoning. By minimizing the need for these chemicals, biocontrol endorses safer food production and supports healthier ecosystems (Farah et al., 2024; Munir et al., 2024). Rhizobacteria are important biocontrol agents and play a crucial rule, extensively used against Verticillium wilt diseases, which infect several plants, such as tomatoes and olives as well. Additionally, they are also effective in the management of other plant diseases such as pepper gray mold, apple bitter rot, and tobacco bacterial wilt (Chen and Wang, 2022; Dhouib et al., 2019; Jiang et al., 2018; Kim et al., 2021; Triki et al., 2006; Wang et al., 2023). Studies have shown that some rhizobacteria species produce a variety of antimicrobial compounds, including lipopeptides and polyketides, which prevent the growth of pathogens and induce systemic resistance in plants (Fazle Rabbee and Baek, 2020). Furthermore, the application of rhizobacteria as a biocontrol agent not only reduces the occurrence of diseases but also increases plant health. Rhizobacteria with antifungal properties can provide promising and sustainable approach to the control of Verticillium wilt diseases. The current study was carried out with the objectives to isolate and screen antifungal rhizobacteria strains against V. dahliae and to assess their in vivo effectiveness in reducing Verticillium wilt in eggplant.
2 Materials and methods
2.1 Fungus pathogen and bacterial strain isolation
The highly pathogenic fungus V. dahliae H6 was obtained from the Plant Pathology Laboratory, Agriculture and Biotechnology College, Zhejiang University, Hangzhou, China (Tomah et al., 2023). The fields infestated with Verticillium wilt prolonged in Yongle Township, Youjiang District, Guangxi, China, in June 2023 were selected. Rhizosphere soil samples were collected from healthy eggplants. Totally 10 samples, each from a distinct field, were collected and analyzed in the laboratory. Soil samples were serially diluted in order to extract bacterial strains and cultured on Lysogeny broth agar (comprising of 5-g yeast extract, 10-g tryptone, 10-g NaCl, and 15-g agar, with a pH range of 7.0–7.5). Bacterial colonies were selected based on morphological characteristics, including color, size, shape, and surface texture. Each colony was then subcultured three times for further isolation and stored in 30% glycerol. The isolated strains were subsequently preserved in the Culture Collection facility at South China Agricultural University, China.
2.2 Screening of rhizobacterial isolates for antifungal activity against the Verticillium dahliae
The antifungal activity of all the isolated rhizobacterial strains was evaluated against the V. dahliae growth via direct confrontation through the dual-culture plate method, as outlined by Meena et al. (2024). Petri plates of 90-mm diameter containing solidified and sterile potato dextrose agar (PDA) medium were prepared in a fume hood. The center of the PDA plate was inoculated with the disc of a pathogenic fungus (0.5 cm in diameter) which was taken from a 15-day-old colony. A 30-μL aliquot of each bacterial suspension previously grown in Luria Bertani broth (LB broth) in a ZWY-211B rotary shaker at 200 rpm at 30°C for 8 h was put on four of the 5-mm diameter paper discs put around the V. dahliae inocula at a distance of 25 mm. The plates inoculated with a pathogenic fungus and without bacteria were regarded as the control treatment. All the plates were incubated at 25°C. The V. dahliae colonies’ growth diameters (mm) in the antagonistic and control treatments were measured after 8 days of incubation. The percentage inhibition of radial growth (PIRG) was calculated through the formula: PIRG = [(Ct − Vt)/Ct] × 100, where Ct is the growth in the control treatment, while Vt is the growth with cell suspensions of the rhizobacteria strain’s treatment. The values were determined as the mean of four replicates, while every experiment was repeated at least three times. The rhizobacteria strain demonstrating the strongest antagonistic activity was selected for further investigations.
2.3 Evaluation of the broad-spectrum antifungal activity of strain ARF4
A highly effective strain ARF4 was assayed for its impact toward radial growth of highly aggressive plant pathogenic fungi, viz. V. dahliae H6, Fusarium oxysporum f. sp. niveum ZJ1 (Huang et al., 2023) Phytophthora capsici HZ07 (Tomah et al., 2020a), and Sclerotinia sclerotiorum YY01 (Tomah et al., 2020b), obtained from the plant pathology lab, previously identified and placed in the Culture Collection facility of Institute of Biotechnology, Zhejiang University, Zhejiang, China. The strain ARF4 was routinely reactivated at 37°C in LB broth, while the plant pathogens were reactivated on PDA, at 25°C, in dark. To detect the direct activity of an antifungal strain ARF4 in vitro, the disc diffusion technique was applied as described by Meena et al. (2024). In brief, the sterile PDA media were poured into 90 mm a diameter Petri dish plates and left to solidify in a fume hood. The 5-mm diameter mycelial plugs were places at the center of the plates (taken from the edge of colonies of 5-day-old plant-pathogen), and then the four of the 5-mm diameter paper discs were put around the fungal inocula at a distance of 25 mm. The paper discs were inoculated with 30 μL of the bacterial cell suspension which had been cultured on LB broth using a ZWY-211B rotary shaker at 200 rpm at 30°C for 12 h.
To evaluate the indirect antagonistic activity of strain ARF4, the poisoned food method was followed as described by Swati Verma et al. (2020). The bacteria were cultured in LB broth in a ZWY-211B rotary shaker at 200 rpm at 25°C for 48 h. Cell-free culture supernatant (CFCS) was obtained after centrifugation at 6000 rpm for 10 min and filtration using a Millipore filter paper of 0.22-μm diameters. The CFCS was incorporated into the molten PDA at 50%, mixed well, and immediately, 10 mL were poured into the Petri plates. After the medium was solidified in the plates inside a fume hood, the mycelial plugs measuring 5 mm in diameter, carefully excised from the 5-day-old plant-pathogen culture, were positioned centrally on plates. Parafilm strips were used to seal the plates. V. dahlias plates were incubated for 14 days in the dark at 25°C, while other plant pathogens were incubated for 5 days only. The plant pathogen colonies’ growth diameters (mm) in both direct and indirect antagonistic tests were measured, and the PIRG was determined by applying the formula: PIRG = [(Ct − Cf)/Ct] × 100; where Ct; the growth in control treatments while the Cf; the growth in CFCS treatments. The recorded values represent the average of three repeats, and the entire experiment was repeated at least three times.
2.4 Identification of bacterial strains
The ARF4 bacterial strain was selected for further experiments, grown on LB agar for 20–24 h at 30°C and characterized following the procedures outlined by Zhang et al. (2018). For 16S ribosomal RNA (rRNA) genes analysis, universal primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3′) were used. The Polymerase Chain Reaction (PCR) products were analyzed by separating them on a 1% agarose gel and visualizing them under ultraviolet light, using a transilluminator (GenoSens 1850, Clinx Science Instruments Co., Ltd., China) after staining with ethidium bromide, in order to verify the presence and size of the amplified 16S rRNA gene fragments (da Silva et al., 2013; Hayward, 2000). The PCR products were subsequently forwarded for sequencing to Sangon Biotech Company Limited (Shanghai, China) via the Sanger method on an automated DNA sequencer (ABI 3730xl, Applied Biosystems, USA). To identify bacterial species, rpoB gene was amplified using primers rpoB-f (5′-AGGTCAACTAGTTCAGTATGGAC-3′) and rpoB-r (5′-AAGAACCGTAACCGGCAACTT-3′) (Qiu et al., 2018), whereas the gyrA gene was amplified with primer pair gyrA-f (5′-CAGTCAGGAAATGCGTACGTCCTT-3′) and gyrA-r (5′-CAAGGTAATGCTCCAGGCATTGCT-′3) (Chun and Bae, 2000). The resulted amplified sequences were edited by BioEdit 7.19, aligned with ClustalX 1.83, and Basic Local Alignment Search Tools (BLAST) against the GenBank database to identify the closely related sequences. Representative strain sequences were deposited in the GenBank in order to acquire accession numbers. These representative sequences were then used to build a phylogenetic tree, with a neighbor-joining/maximum-likelihood (NJ/ML) approach using the MEGA 7.0 (Kumar et al., 2016), with bootstrap analysis (1,000 replication) conducted to evaluate statistical support of the tree nodes in the phylogenetic trees. Bayesian inference of posterior probability (BIPP) analyses are performed with MrBayes v. 3.2.6 (Ronquist et al., 2012). This method was used to analyze evolutionary connections between the identified bacterial strains and other known species.
2.5 Detection and production of β-glucosidase, chitinase, and protease enzymes
To explore the production of some extracellular hydrolytic enzymes by bacterium strain ARF4 which may be related to antagonism toward pathogen growth, the minimal salt medium (MSM) was used with contents following KH2PO4 0.2 g, (NH4)2SO4 1 g, MgSO4•7H2O 0.2 g, K2HPO4 1.6 g, FeSO4•7H2O 0.01 g, CaCl2•2H2O 0.02, g, and NaCl 0.1 g. For glucanase activity, the MSM medium was supplemented with 0.5% β-1,3-glucan (Howard et al., 2003; Renwick et al., 1991). The MSM medium was prepared by adding 1% (w/v) colloidal chitin and 1.5% (w/v) agar, with the final pH adjusted to 7.0 for the chitinase activity test (Alsalman et al., 2022), while for protease activity, the medium supplemented with 1% skim milk was used. The 10 μL of suspension of bacteria (standardized to 1 × 108 CFU/mL) and 10 μL of LB broth as control were dropped in the center of the plate and incubated for 2–3 days at 30°C. The chitin plates were treated with 0.1% (w/v) Congo red stain and washed with 1% NaCl. The presence of bacterial hydrolytic enzyme activities was revealed by clear zones of hydrolysis around the bacterial colonies.
2.6 Characterization of lipopeptides of strain ARF4
The inhibition zone in the direct antifungal trial was targeted to collect and identify lipopeptides involved in V. dahliae inhibition. The inhibition zone located between the ARF4 strain colony and the V. dahliae colony was identified on fifty of the LBA media plates. The sections of the LBA media were carefully cut out from the inhibition zone using a sterile scalpel, ensuring to avoid contamination from other areas. The parts of the media around strain ARF4 that grew alone without V. dahliae were collected as a control. An amount of 100 g of cut sections of the medium was added to a 500-ml flask, followed by the addition of 200 mL of ethyl acetate. The mixture was mechanically shaken overnight at 25°C. The ethyl acetate extract obtained from LBA medium sections was filtered using the Whatman filter paper. Subsequently, the ethyl acetate was evaporated under reduced pressure using an evaporator vacuum shaker at 40°C to obtain a dry extract, which was dissolved in a methanol solvent of 40% for analysis.
The ethyl acetate extract was rinsed through a silica gel column using different ratios of methanol and methylene chloride (1:2, 3:1, and 5:1, respectively).
After drying, the extract was purified further with the reversed-phase high-performance liquid chromatography (RP-HPLC) device. The dry extract was dissolved in 40% methanol and filtered using a 0.22-μm pore. A 10-μl aliquot was put into an Inert Sustain C18 column (5 μm, 250 mm × 4.6 mm) in the HPLC system. Mobile phase A was acetonitrile with 0.1% (vol/vol) trifluoroacetic acid (TFA), and mobile phase B was Milli-Q water with 0.1% (vol/vol) TFA. Purification was performed using a solvent containing 45% mobile phase A and 55% mobile phase B at a flow rate of 8 mL/min. The UV absorption at 220 nm was recorded at running times between 0 min and 35 min for detection. The extract of RP-HPLC-purified and bacillomycin D (CAS:76012–17-4, Guangzhou Yaoguang Technology Co., Ltd., China), as a standard, was subjected to a preparative silica gel thin-layer chromatography (TLC) plate (GF254) using n-butanol–methanol–water (39: 10: 20, v/v/v) as the mobile phase. The TLC plate was visualized under the wavelength of 254 nm.
The extract of RP-HPLC-purified was analyzed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Bruker, Bremen, Germany) equipped with a 337-nm smart laser beam for desorption and ionization. The study focused on detecting and characterizing lipopeptides by analyzing their mass-to-charge (m/z) ratios using MALDI-TOF mass spectrometry. The analysis was conducted using a Bruker instrument (Bremen, Germany) equipped with a 337-nm smart laser beam for desorption and ionization (Ayed et al., 2014).
2.7 Effect of the extracted lipopeptide of strain ARF4 on conidia germination of Verticillium dahliae
The extracted lipopeptide of strain ARF4 was tested for its inhibitory effect on the conidial germination of V. dahliae using the agar well diffusion technique. A 0.5 mL of conidia suspension of V. dahliae (106 spores/mL) was evenly spread onto pre-prepared PDA plates (90-mm diameter). Wells (5-mm diameter) were created in the center of the plate by a sterilized cork borer. The extracted lipopeptide (50 μg/mL prepared in 50% w/v DMSO) was added to wells, while 50 μL of DMSO was added to wells as control. The plates were kept at 25°C for two 48 h, and antifungal activity was assessed by measuring the inhibition zone around the wells.
2.8 Structural change assays in SEM and TEM
SEM was used to observe the morphological changes in the fungal hyphae treated with extracted lipopeptide of strain ARF4. The fungal hyphae were treated with 50 μg/mL of extracted lipopeptide using the agar well diffusion technique for 2 days. The 10-mm diameters of mycelial plugs were taken from the inhibition zone and control (50 μL of DMSO). Both samples were fixed in a 2.5% glutaraldehyde solution for 12 h, following the method outlined by Pretorius et al. (2015) and Xiao et al. (2021). After fixation, the samples were washed three times with 100-mM phosphate buffer, each lasting 10 min. Subsequently, the samples were post-fixed in 1% osmium tetroxide for 3 h. Dehydration was then carried out using a graded ethanol series, ranging from 50 to 100%. After mounting onto an aluminium stub, the samples were sputter-coated with a layer of gold particles and examined using a scanning electron microscope (SEM, SU8010, HITACHI, Japan), followed by imaging. For ultrastructural analysis of extracted lipopeptide effect on hyphae, transmission electron microscopy (TEM) was used. Prior to TEM, mycelial plugs were fixed for 12 h in 2.5% glutaraldehyde solution at 4°C. The 100-mM phosphate buffer was used to rinse for 10 min, and the 1% osmium tetroxide solution for 3 h was used to fix the cells and the gradient of ethanol to dehydrate. With the help of LKB-V Ultratome, the gold-coated samples were cut into ultra-thin sections and stained with uranyl acetate for examination under TEM (H-600, Hitachi, Japan) at 15-kV voltage.
2.9 Management of Verticillium wilt disease by ARF4 strain
The potential of the selected rhizobacteria strain ARF4 as a biocontrol agent against Verticillium wilt disease was evaluated in susceptible eggplant plants (cv. Zheqie 3, an eggplant F1 hybrid) grown in potted soil under greenhouse conditions. Plastic pots were filled with sterilized soil composed of a mixture of peat, vermiculite, and farmyard soil in a 2:1:1 ratio. The conidial suspension of V. dahliae was prepared from a liquid culture grown in Potato Dextrose broth for 7 days at a temperature of 24°C and 130 rpm. The conidia were gently scraped from a sporulating fungal culture using a sterile loop. Then, the spores were suspended in a sterile diluent (distilled water, 0.01% Tween 80) to prevent clumping. Conidia were then filtered through sterile cheesecloth (40 μm) to remove hyphal debris, and its concentration was adjusted to (107 conidia/mL). A 5 μL of the conidial suspension was loaded onto a hemocytometer. Spore counts were conducted on the central grid under a microscope (40 × magnification). To calculate the conidia concentration, the following formula was used: Conidia/mL = Average count per square × Dilution factor × 104 (Gayoso et al., 2010). The cell suspension of bacterium strain ARF4 was cultured in LB medium at a concentration of 107 CFU/mL and incubated on a rotary shaker at 130 rpm and 30 ± 2°C for 24 h, then adjusted to a final concentration of 107 conidia/mL. The experiment was done using a completely randomized design with three replicates, 10 plants per replicate. The experimental treatments were carried out as follows: the soil of a group of pots was inoculated with 20 mL of conidia suspension of Verticillium and 20 mL of a suspension of bacterial cells. The soil of a group of pots was inoculated with 20 mL of conidia suspension of Verticillium only, as negative controls, while the soil of a group of pots was left without inoculation with suspensions of bacterial cells and conidia of Verticillium, as positive controls. The three-leaf-old eggplant seedlings previously prepared were transplanted into the inoculated pots. The pots were placed in the greenhouse under controlled conditions, maintaining the humidity level to 70–80% and temperatures of 25 ± 3°C with a light, dark cycle of 16/8 h and light intensity of 500 μmol/m2 was used (Yang et al., 2023). To assess the potential of a bacterial biocontrol agent, the symptoms of the development of Verticillium wilt disease on seedlings were monitored after 20-day post-pathogen inoculation. Assessment of disease severity was applied using a 0–5 scale based on foliar symptoms: 0, no diseased leaf; 1, <10%; 2, 11–25%; 3, 26–50%; 4, >50%; and 5, plant killed (Li et al., 2024). The disease severity (%) and the biological control efficacy (%) were evaluated.
2.10 Statistical analysis
The data analysis was performed using IBM SPSS statistics 21 (Georgia, USA). The data presented in the results represent the average value along with the standard error, which is calculated from a minimum of three values obtained in each independent experiment. The significant differences between groups are determined by the least significant difference test, one-way analysis of variance (ANOVA), and Duncan’s test.
3 Results
3.1 Bacterial isolations and antifungal test
In the current study, 42 bacterial isolates were isolated from the rhizosphere of eggplant plants. The bacterial isolates were purified and designated sequentially as ARF1 to ARF42 (Table 1). The antifungal effect of these bacterial strains was evaluated against the radial growth of V. dahliae using the dual-culture plate method. One-way ANOVA and Duncan’s test were used to show the differences among tested bacterial strains in the rates of radial growth inhibition of V. dahliae (Table 1). Among these bacterial strains, ARF4 was found to be the most proficient antagonistic strain, showing the maximum in vitro inhibition rate of 84.49% against radial growth of V. dahliae. Furthermore, 10 of the bacterial strains isolated had caused growth inhibition of V. dahliae, ranging from 18.60 to 18.87%. With a lower inhibition rate, 11 of the bacterial strains isolated caused growth inhibition of V. dahliae, ranging from 16.07 to 16.92%. Also, eight of the bacterially produced inhibition zones were between 15.07 and 15.99%. While three of the bacterial strains had recorded the zone of inhibition, ranging from 12.03 to 12.66%, five of the bacterial strains had recorded the zone of inhibition, ranging from 11.17 to 11.96%, and four bacterial strains recorded the zone of inhibition from 10.16 to 10.86%. Overall, although most isolates demonstrated only slight antifungal activity, strain ARF4 outperformed other isolates against V. dahliae.
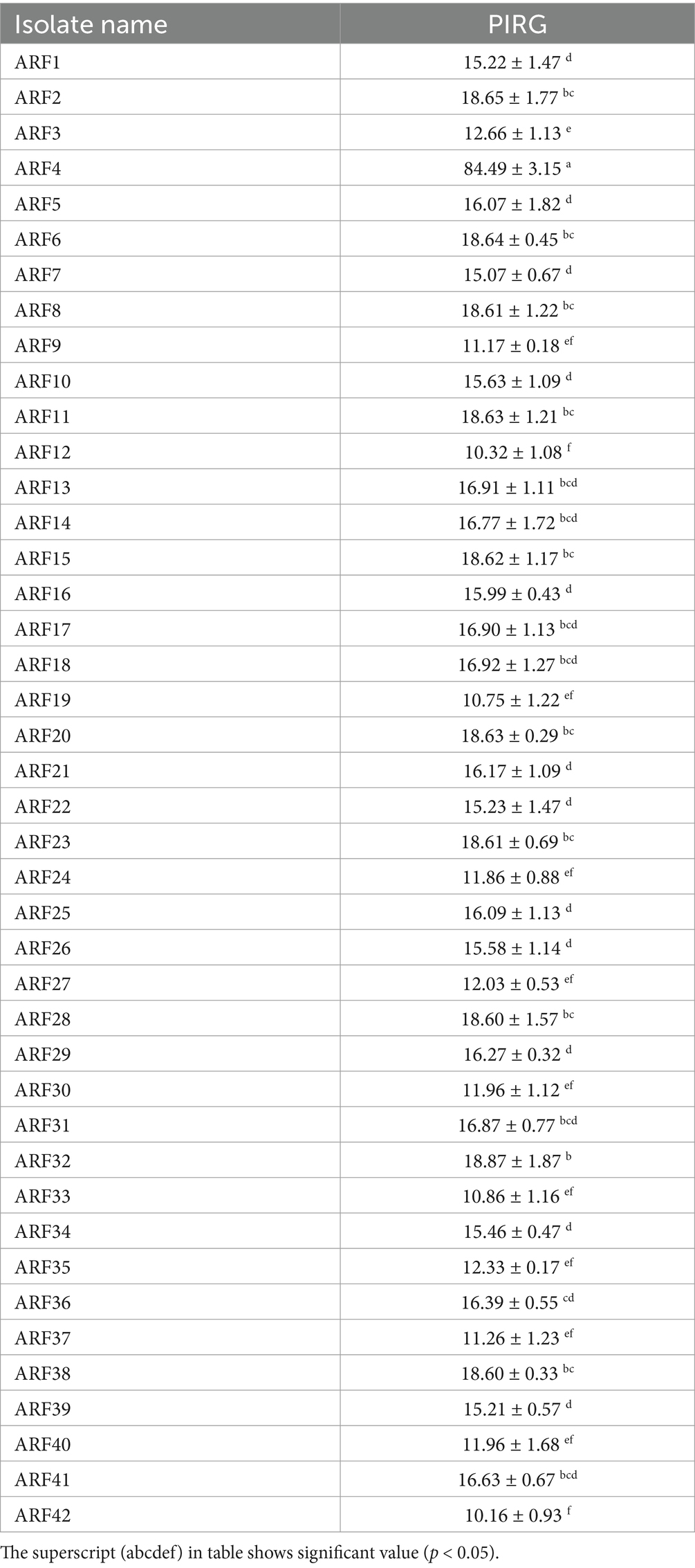
Table 1. Antifungal activity on Verticillium dahliae by rhizobacteria strains isolated from rhizosphere zone of eggplant plants.
3.2 Broad spectrum antifungal activity
The inhibitory activity of rhizobacteria strain ARF4 was assessed against four plant pathogens in vitro. In the disc diffusion assay, bacterial strain ARF4 exhibited a strong direct antagonistic effect and inhibited all tested plant pathogen radial growth compared to the control (Figure 1). Similarly, in the poisoned food method, indirect antagonism using the cell-free culture supernatant (CFCS) of strain ARF4 (55%) also exhibited significant inhibition of radial growth in all pathogens (Figure 1). Visual assessments indicated that the highest inhibition was recorded against V. dahliae in both direct and indirect assays. Based on these results, strain ARF4 is considered an effective antagonist in inhibiting the growth of tested plant pathogens.
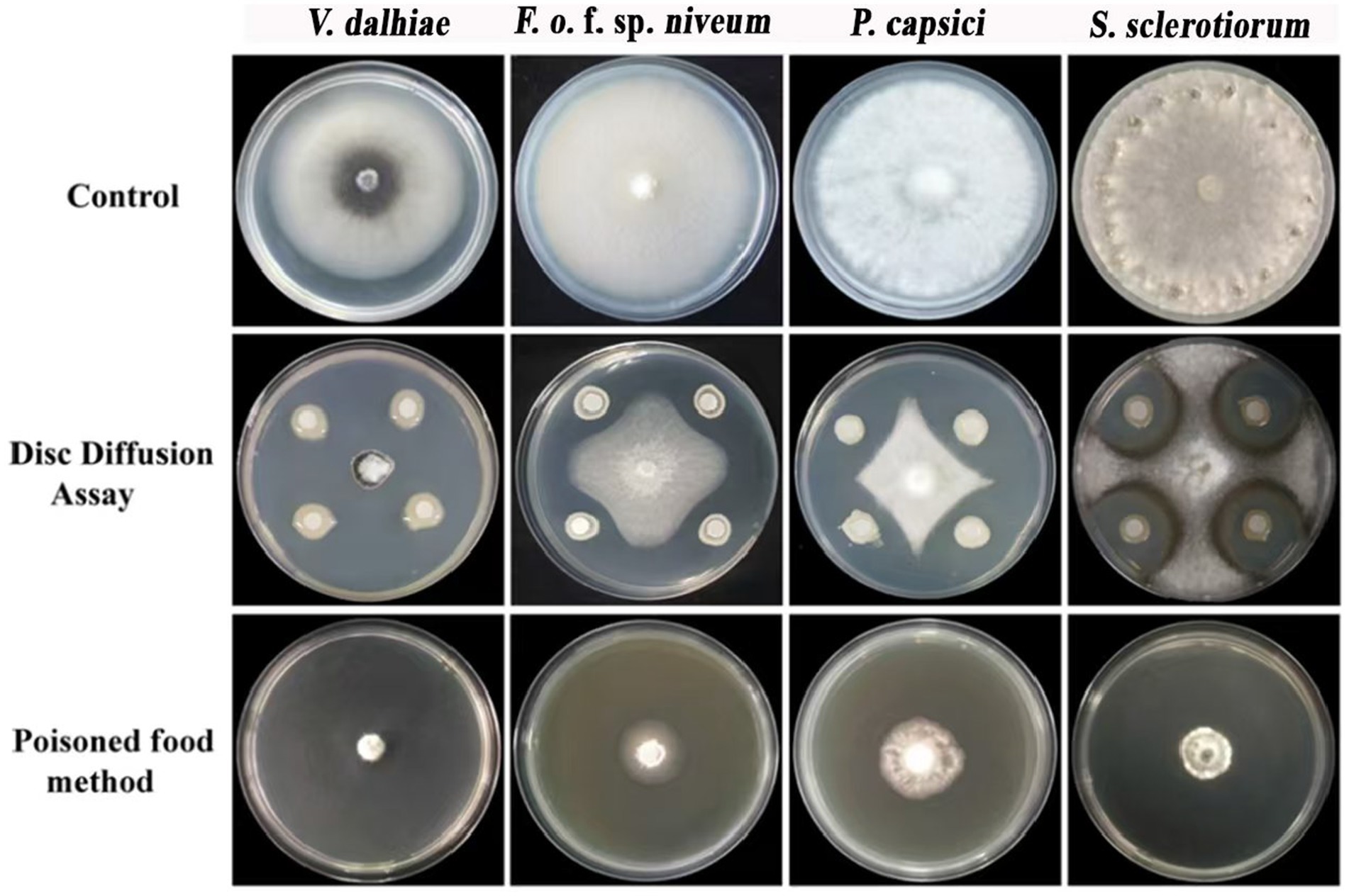
Figure 1. In vitro antagonistic activity of strain ARF4 against four plant pathogens. Disc diffusion and poisoned food assays show inhibition of radial growth with maximal suppression against V. dahliae (vs. control).
Statistical analysis of PIRG revealed that strain ARF4 exhibited strong inhibitory potential against plant pathogens with significant difference (p < 0.05) observed among them (Table 2). In the disc diffusion assay, the strain ARF4 strain showed the highest PIRG against V. dahliae (80.21 ± 0.58%) while the least recorded was against F. o. f. sp. niveum was (48.88 ± 0.95%). The PIRG values were found to be (64.67 ± 0.88%) and (57.49 ± 0.97%) against P. capsici and S. sclerotiorum, respectively. In the poisoned food method, the CFCS of the bacterial strain ARF4 recorded significant inhibition differences in the radial growth among the plant pathogens. The highest PIRG was again recorded against V. dahliae (86.57 ± 0.60%), while the lowest inhibition was recorded against P. capsici (67.29 ± 0.92%). However, the high values of PIRG (79.60 ± 0.96%) and (69.19 ± 0.64%) were also recorded against S. sclerotiorum and F. o. f. sp. niveum, respectively.
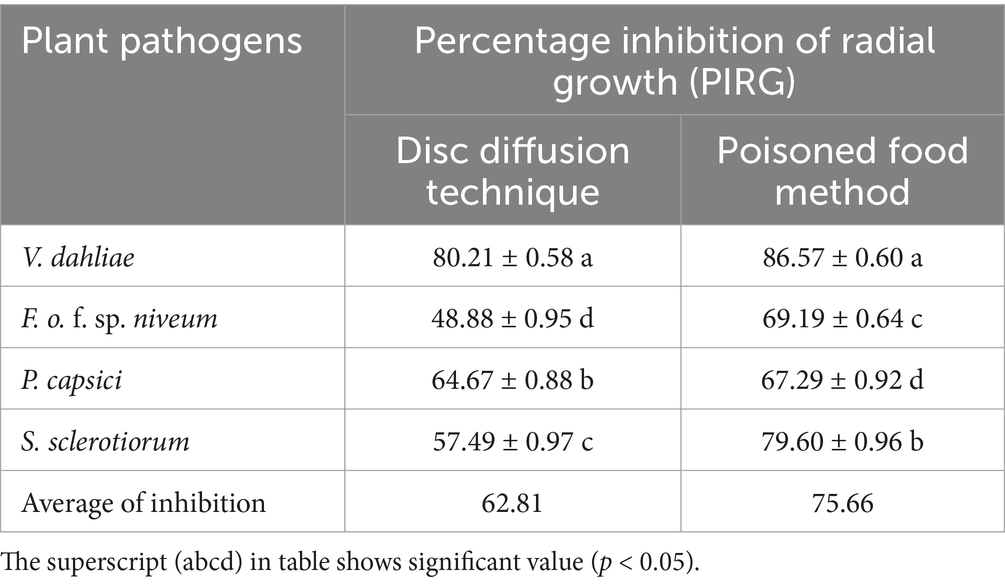
Table 2. Antifungal activity of strain ARF4 against four of the plant pathogens using disc diffusion assay and poisoned food method.
3.3 Identification of bacterial strains
The DNA of strain ARF4 was amplified by the 16S gene, and a sequence of 1,420 bp was obtained. The BLAST results showed that the strain belongs to the genus Bacillus spp. The rpoB and gyrA genes were amplified to determine a species, and 959 and 546 bp of sequences were obtained, respectively. The BLAST results showed that the species identity of the two genes was aligned 100% with the species of B. velezensis of strain BY6 and strain LPL061, respectively. In a phylogenetic tree, the topology of the scoring NJ/ML bootstrap tree analysis was congruent with the BIPP tree for the concatenated three-locus dataset. Strain ARF4 is grouped with B. velezensis BY6 and B. velezensis LPL061 with a bootstrap support of NJ/ML = 98 and BIPP = 0.99 (Figure 2). This high bootstrap value indicates strong support for the grouping, suggesting that strain ARF4 is closely related to these strains of B. velezensis. Strain ARF4 was submitted to GenBank under accession numbers OQ918063.1, OQ920283.1, and OQ920284.1 for 16S, rpoB, and gyrA genes.
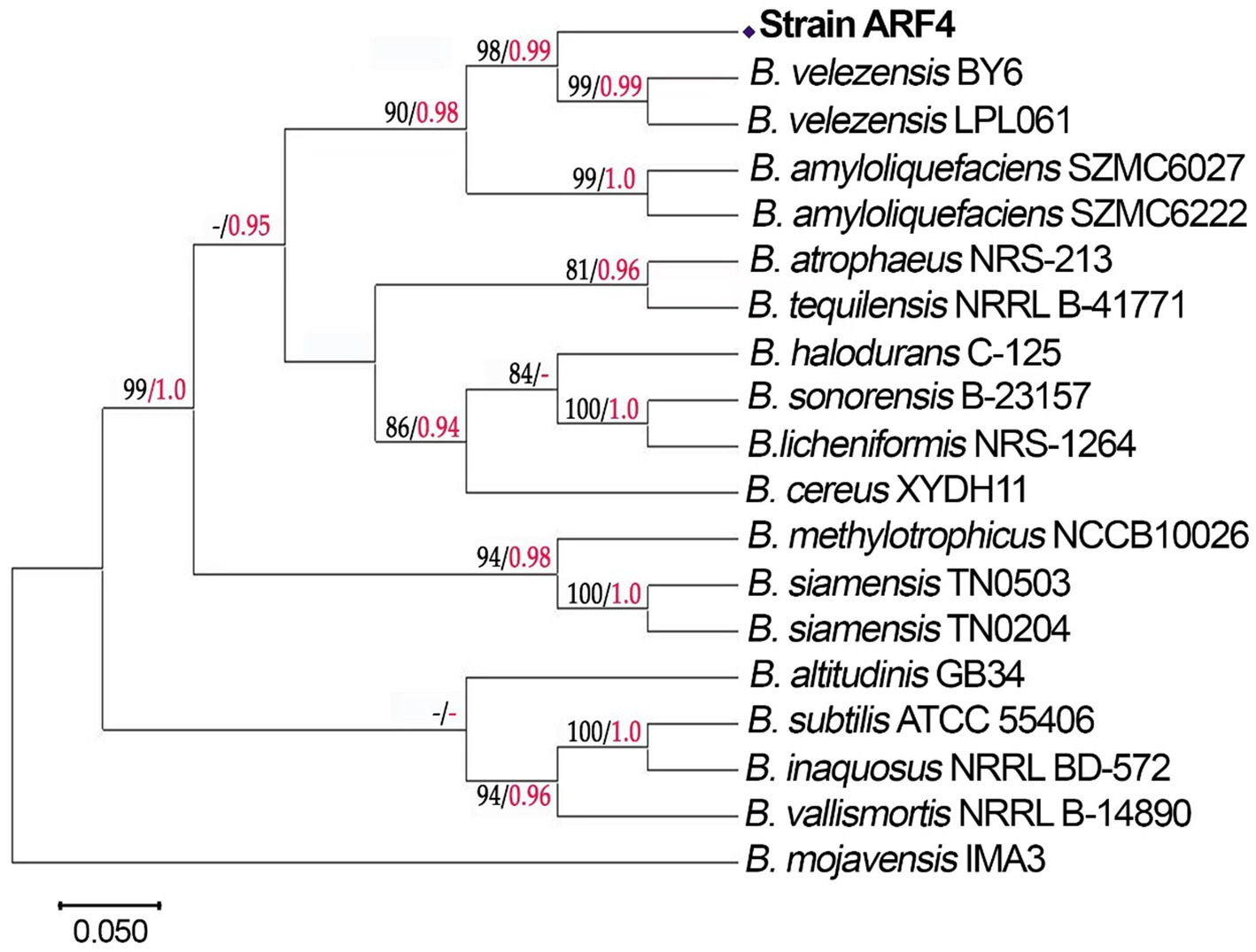
Figure 2. The phylogenetic tree of the Strain ARF4 was built based on 16S rRNA, rpoB, and gyrA gene sequences using MEGA version X with neighbor-joining and maximum-likelihood and Bayesian inference (BI) methods. NJ/ML bootstrap values are indicated by black numbers, while BIPP values are indicated by red. Dashes indicate support values lower than 80% NJ/ML and 0.95 BIPP.
3.4 Detection production of enzymes of β-glucosidase, chitinase, and protease
The lytic enzymes produced by bacteria play crucial role in the biological transformation of nitrogen, hydrogen, and carbon. These enzymes deform pathogenic fungi’s cell walls and act as critical mechanisms of eco-friendly soil-borne pathogen control. ARF4 strain produced a halo zone of 1.6 cm around its colony on MSM with 1% skim milk, indicating its capability to produce protease enzyme (Figure 3A). Similarly, ARF4 also exhibited the potential to produce chitinase and β-glucosidase hydrolytic enzymes as indicated by clear halo zones of 1.7- and 1.5-cm diameters on chitin agar and MSM amended with glucan media, respectively (Figures 3B,C).
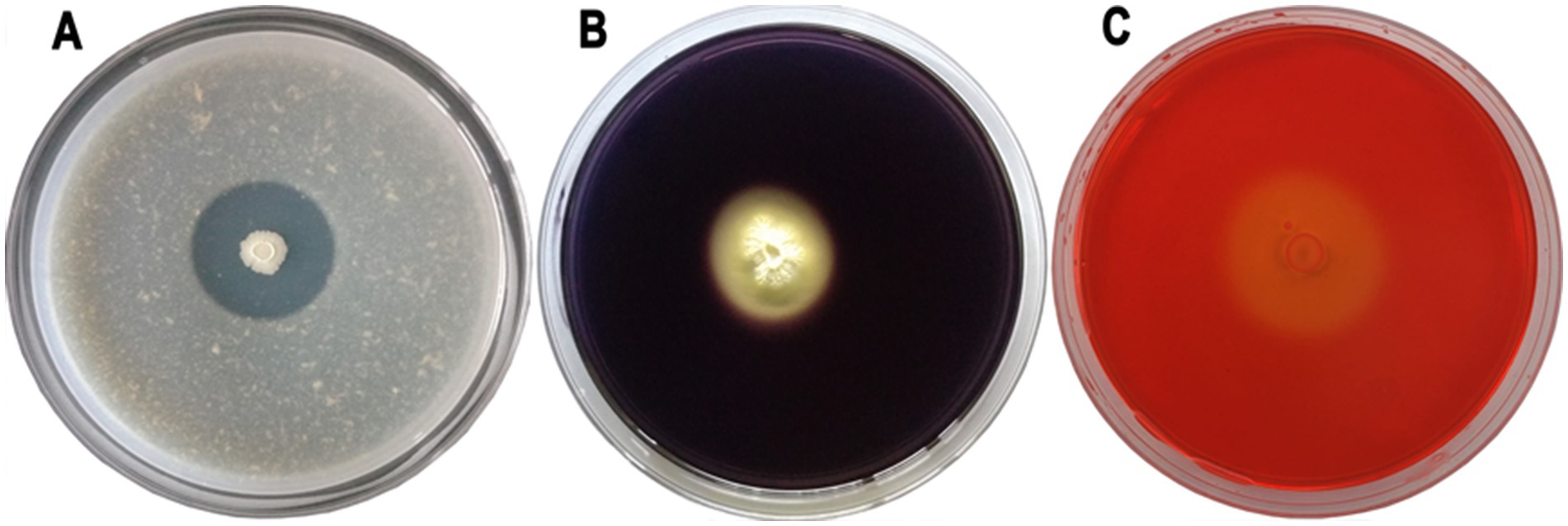
Figure 3. The image displays enzyme activity on agar plates incubated with the ARF4 strain. These results highlight the ability of the ARF4 strain to produce enzymes, (A) protease, (B) β-glucosidase, and (C) chitinase, as observed through distinct color changes and clear zones on the agar plates.
3.5 Identification of lipopeptides from strain ARF4 extract
The production of specialized lipopeptide by the B. velezensis ARF4 cultured on a solid LB medium was analyzed. RP-HPLC analysis showed only one large peak appeared at a retention time19.023 min (Figure 4A). Characterization of the extract produced by strain ARF4 was carried out by TLC. Results obtained by TLC indicate the lipopeptide nature of the bacillomycin D produced, which was observed against standard bacillomycin D (Figure 4B).
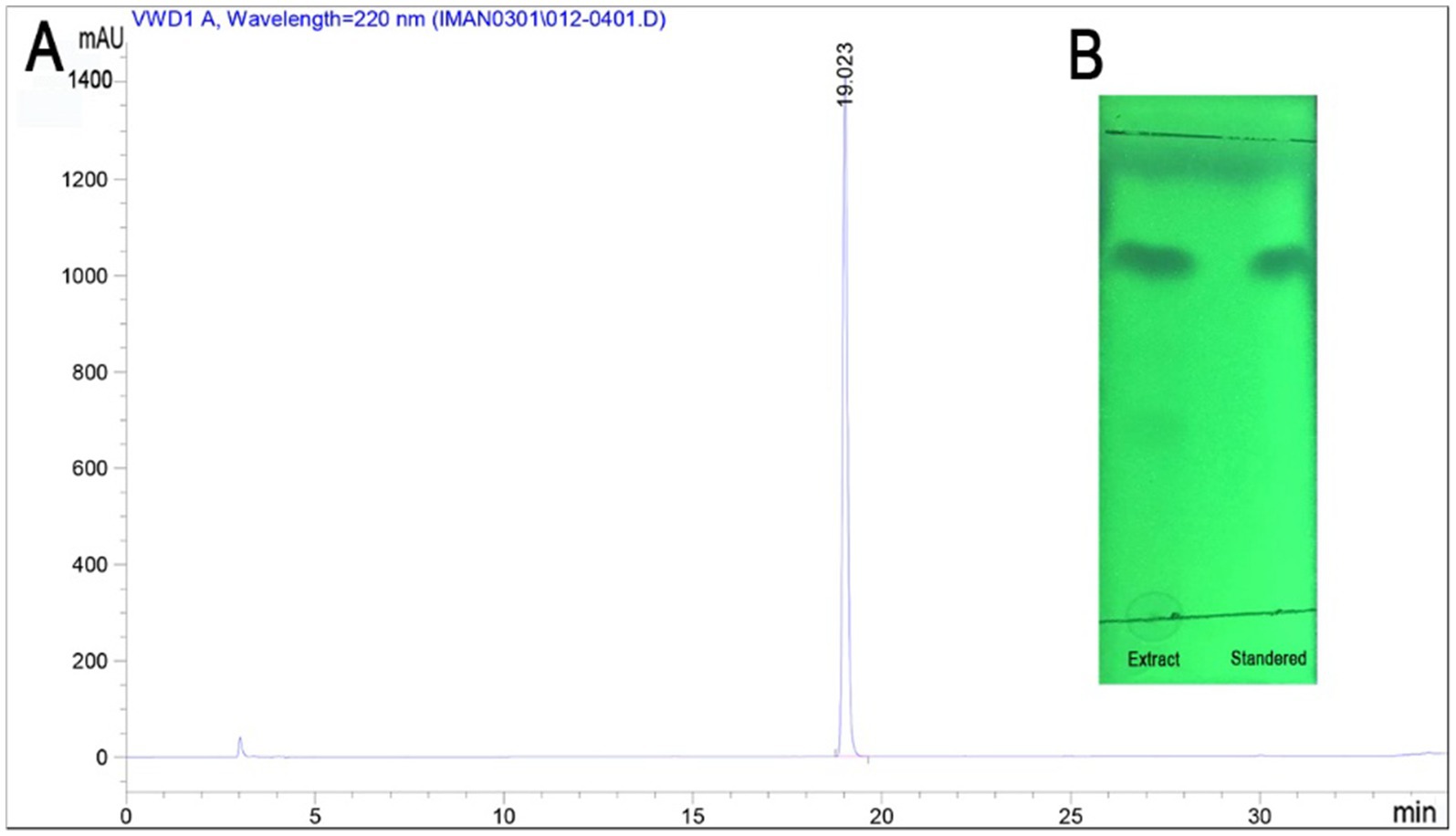
Figure 4. Reversed-phase high-performance liquid chromatography (RP-HPLC) and preparative silica gel TLC plate (GF254) analyses. (A) RP-HPLC analysis of extract-product by B. velezensis strain ARF4. A single peak was observed at 220 nm with a retention time of 19.02 min. (B) TLC profile of extract-product in comparison to standard bacillomycin D.
Using MALDI-TOF, the resulted mass spectrometry data were compared with the known Bacillus metabolite, confirming the presence of a cyclic lipopeptides compound of the iturin family. This compound was homologous to bacillomycin D (m/z 1081.336 [M + Na]+), which includes a heptapeptide molecule attached to a β-amino fatty acid chain consisting of 15 carbon atoms (Table 3; Figures 5A,B). The results of MALDI-TOF mass spectra of strain ARF4, which was grown alone (without V. dahliae) as a control, also showed the same lipopeptide indicating polymyxin, which was separated but with shorter peaks (Figure 5C).
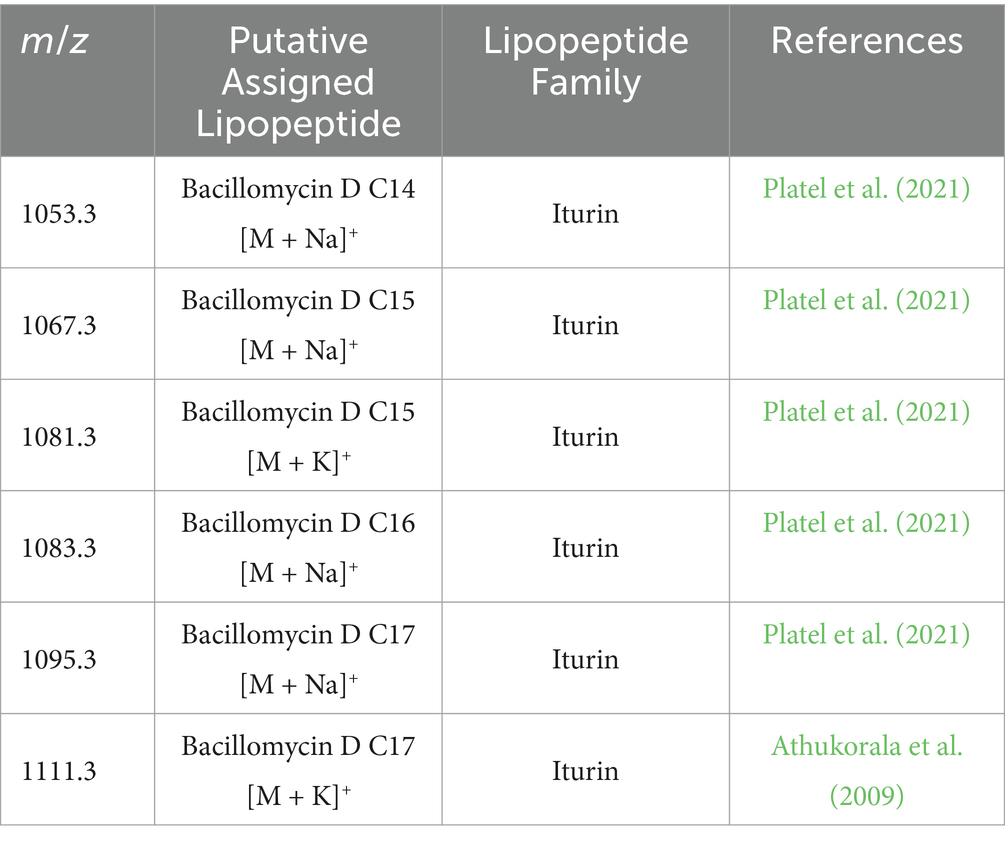
Table 3. Specialized extracted lipopeptide from the B. velezensis ARF4, extracted from the inhibition zone on the PDA medium, detected by MALDI-TOF.
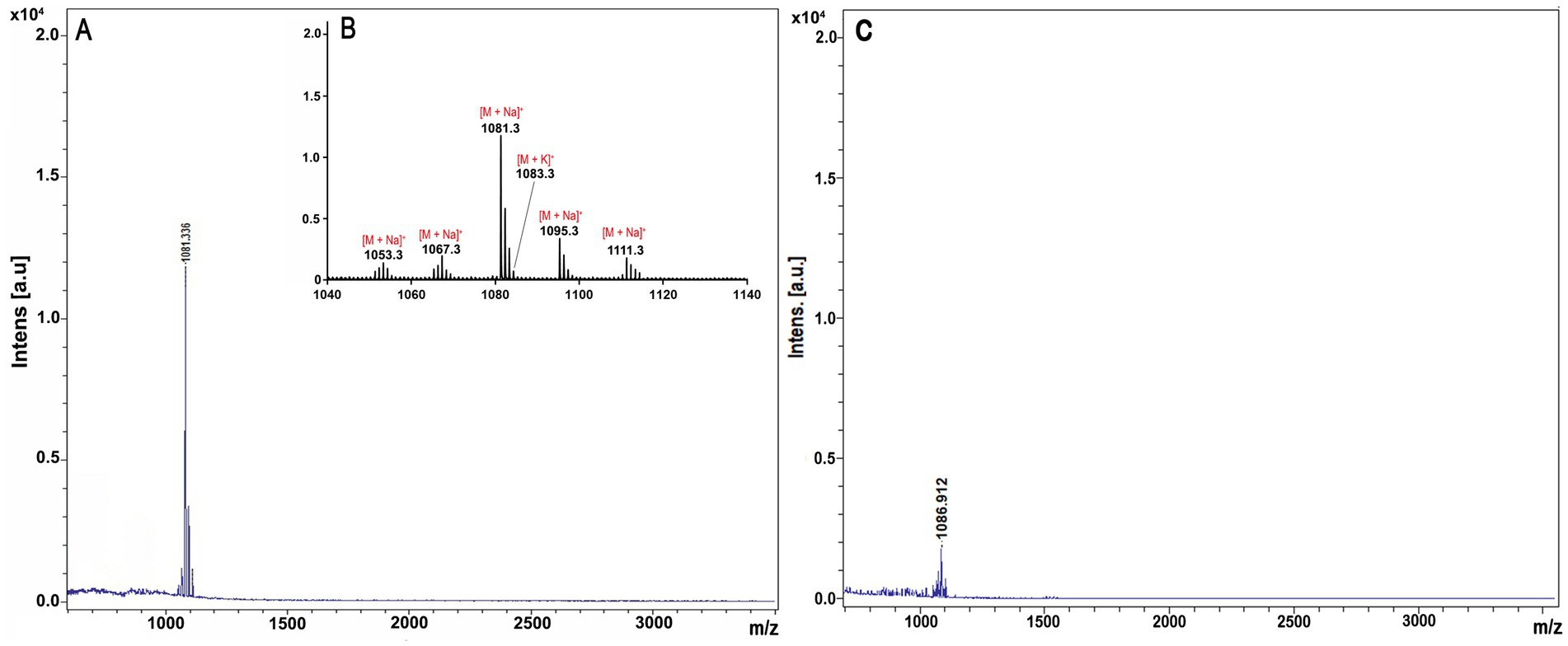
Figure 5. MALDI-TOF MS analysis of the extracted lipopeptides produced by the ARF4 strain. (A) High mass intensity in the m/z range 500–3,500 is proposed to contain bacillomycin D. (B) High mass intensity at m/z 1,040–1,140 is proposed to contain bacillomycin D. (C) Lipopeptide was detected in a sample of strain ARF4 without fungi as a control.
3.6 Effect of extracted lipopeptides of the ARF4 on germination Verticillium dahliae spores and hyphae structure
The antifungal activity of extracted lipopeptides of strain ARF4 was assessed using the agar well diffusion technique. The extract exhibited strong inhibition of conidial germination in V. dahliae, as evidenced by the presence of inhibition zones around the wells, compared to the DMSO control (Figures 6A,B).
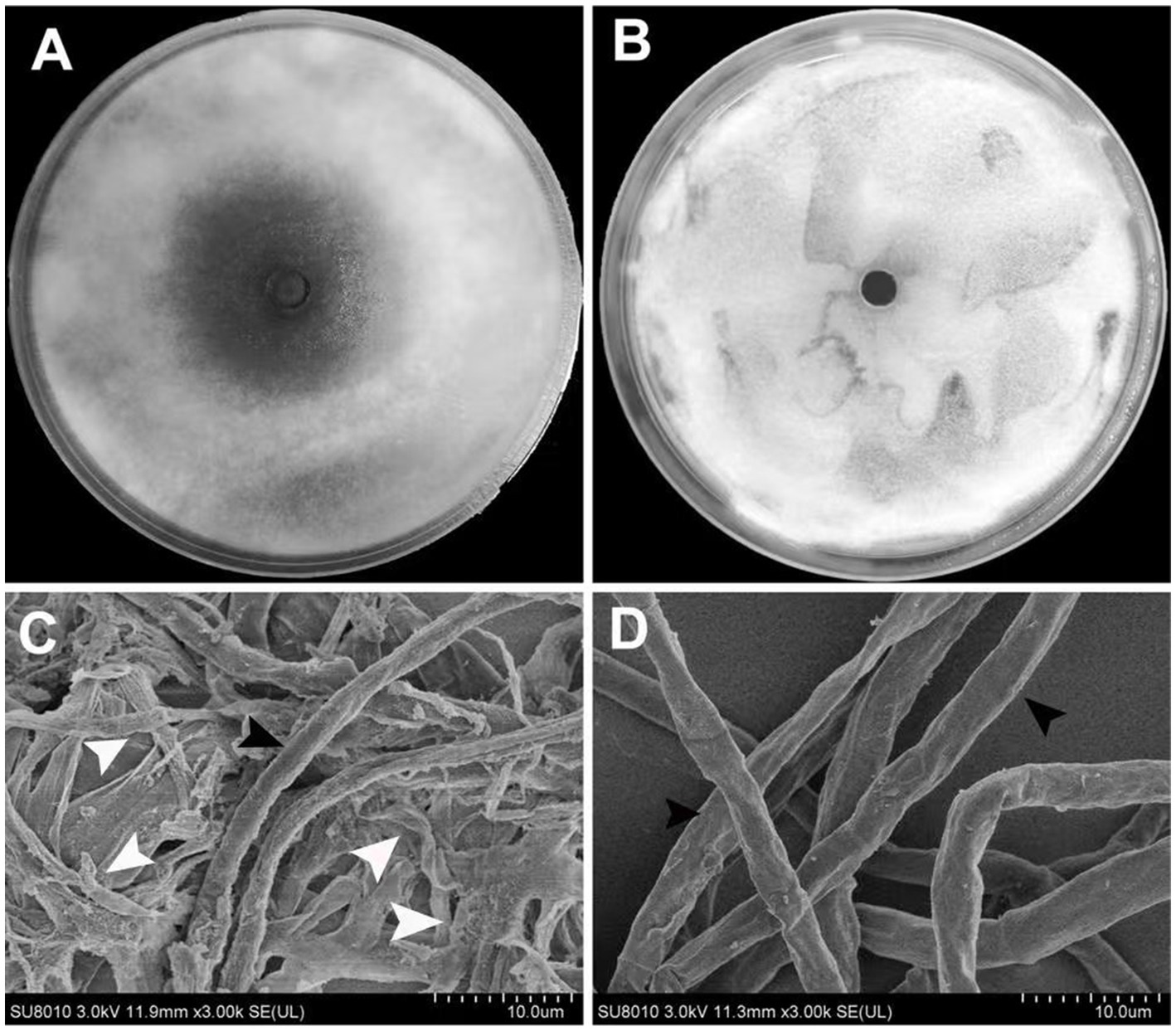
Figure 6. Antifungal activity of extracted lipopeptide of the strain ARF4 against V. dahliae. (A) Antifungal activity of extracted lipopeptide on conidia germination and (B) Control treatment using DMSO. (C) Scanning electron microscopy images of V. dahliae hyphae were treated with extracted lipopeptide of ARF4 and (D) were hyphae treated with DMSO (control). White arrows point to hyphae deterioration. Black arrowheads point to plump and intact hyphae.
In order to study the effects of the extracted lipopeptide on the morphology of V. dahliae hyphae, scanning electron microscopy (SEM) was conducted. Agar slices of 0.5 cm2 from hyphal zones treated with the extract and from regions treated with Dimethylsulfoxide (DMSO) were prepared, and the resulted morphological variations were examined by SEM. The morphology of V. dahliae hyphae treated with extracted lipopeptide was significantly different from that of DMSO-treated control. The hyphae treated with extracted lipopeptide exhibited considerable deformation, appearing irregular with extensive collapse, and the normal plump cylindrical shape of the hyphae was lost, likely due to the outflow of cytoplasm, which may have led to cell death (Figure 6C). In contrast, the hyphae from the DMSO-treated control group displayed regular, plump, intact cylindrical structures (Figure 6D).
The cell wall and plasma membrane treated with DMSO showed uniform and typical cellular morphology, with a clear and intact cell wall. The cytoplasmic matrix was dense, and essential organelles, such as the nucleus and mitochondria, exhibited normal and well-defined structures (Figures 7A,C). In contrast, the hyphae treated with the extracted lipopeptide displayed a reduced cytoplasmic matrix and unclear ultrastructure, with a reduced number of mitochondria, and the internal organelles appeared disintegrated (Figure 7B). Additionally, the cell walls and plasma membrane of the treated hyphae were rough and blurred, with visible signs of cell wall lysis (Figure 7D).
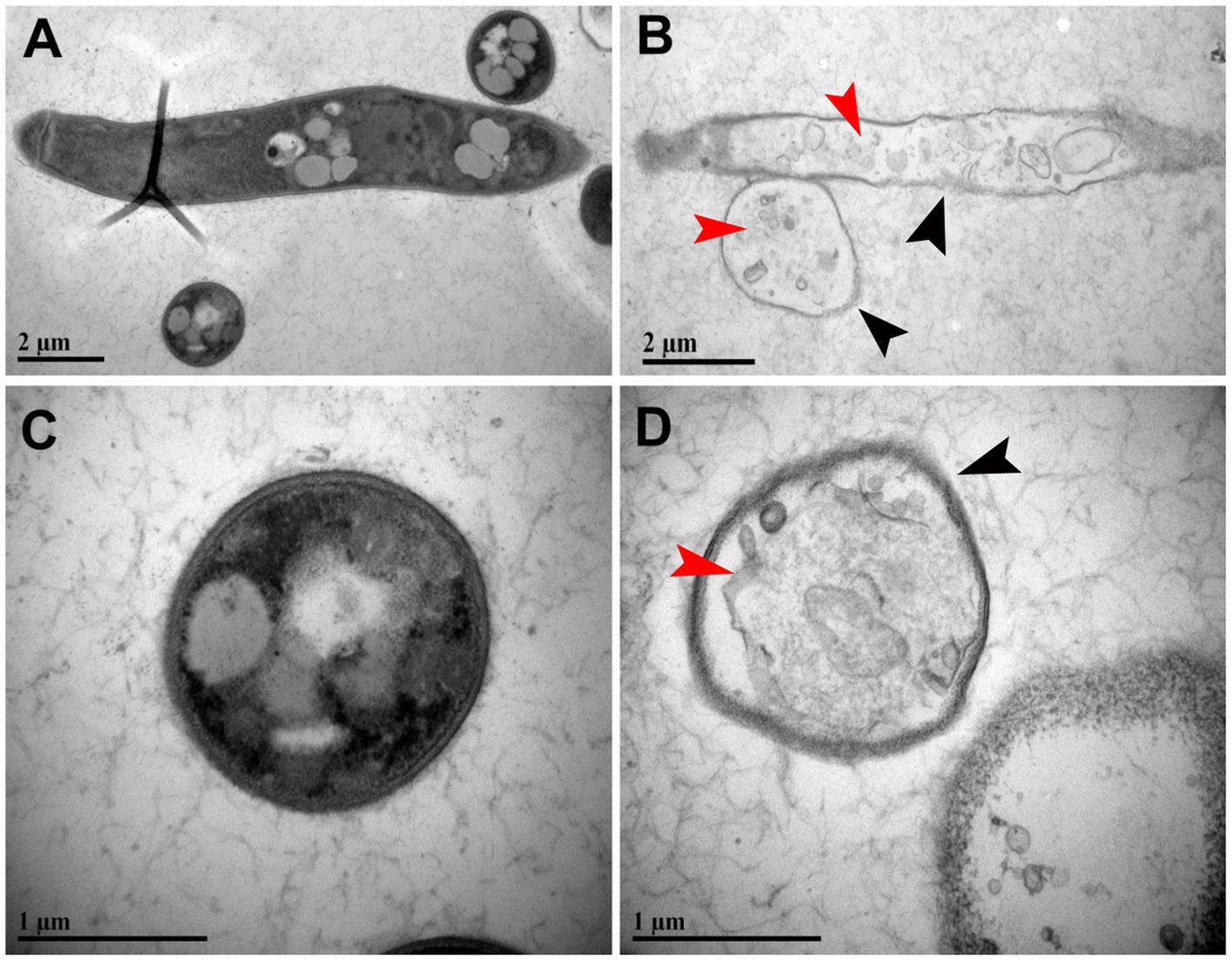
Figure 7. Transmission electron microscopy (TEM) analysis showing the effect of lipopeptide produced by the ARF4 strain on the ultrastructure of V. dahliae hyphae. (A,C) Hyphal cells were treated with 40 μg/mL of DMSO (control). (B,D) Hyphal cells were treated with 40 μg/mL of the extracted lipopeptide. Black arrowheads point to the cell wall and plasma membrane lysis of hyphae. Red arrowheads point to a reduced cytoplasmic matrix and unclear ultrastructure, as well as a reduced number of mitochondria, with the internal organelles appearing disintegrated.
3.7 Management of Verticillium wilt disease by ARF4 strain
The potential of the ARF4 strain in controlling Verticillium wilt disease was rigorously evaluated through a series of pot experiments designed to simulate real-world conditions. The results of these assays were highly promising, indicating that inoculation with the ARF4 strain significantly reduced both the incidence and severity of Verticillium wilt in eggplants compared to negative controls (Figure 8). Specifically, the seedling’s inoculation with conidia of V. dahliae led to the appearance of a major infection, represented by yellowing and wilting of most leaves (Figure 8A). While the yellowing and wilting were observed to be reduced on the leaves of plants co-inoculated with ARF4 strain and conidia of V. dahliae (Figure 8B). While no symptoms of yellowing and wilt were observed in the positive control (Figure 8C). Statistically, the disease severity observed in eggplants inoculated solely with V. dahliae reached 73.28 ± 1.87 after 20 days, reflecting substantial infection. In contrast, eggplants treated with both V. dahliae and the ARF4 strain showed a markedly reduced disease severity of 23.05 ± 0.72 (Figure 8D), corresponding to a control efficacy of 68.45 ± 0.85% (Figure 8E).
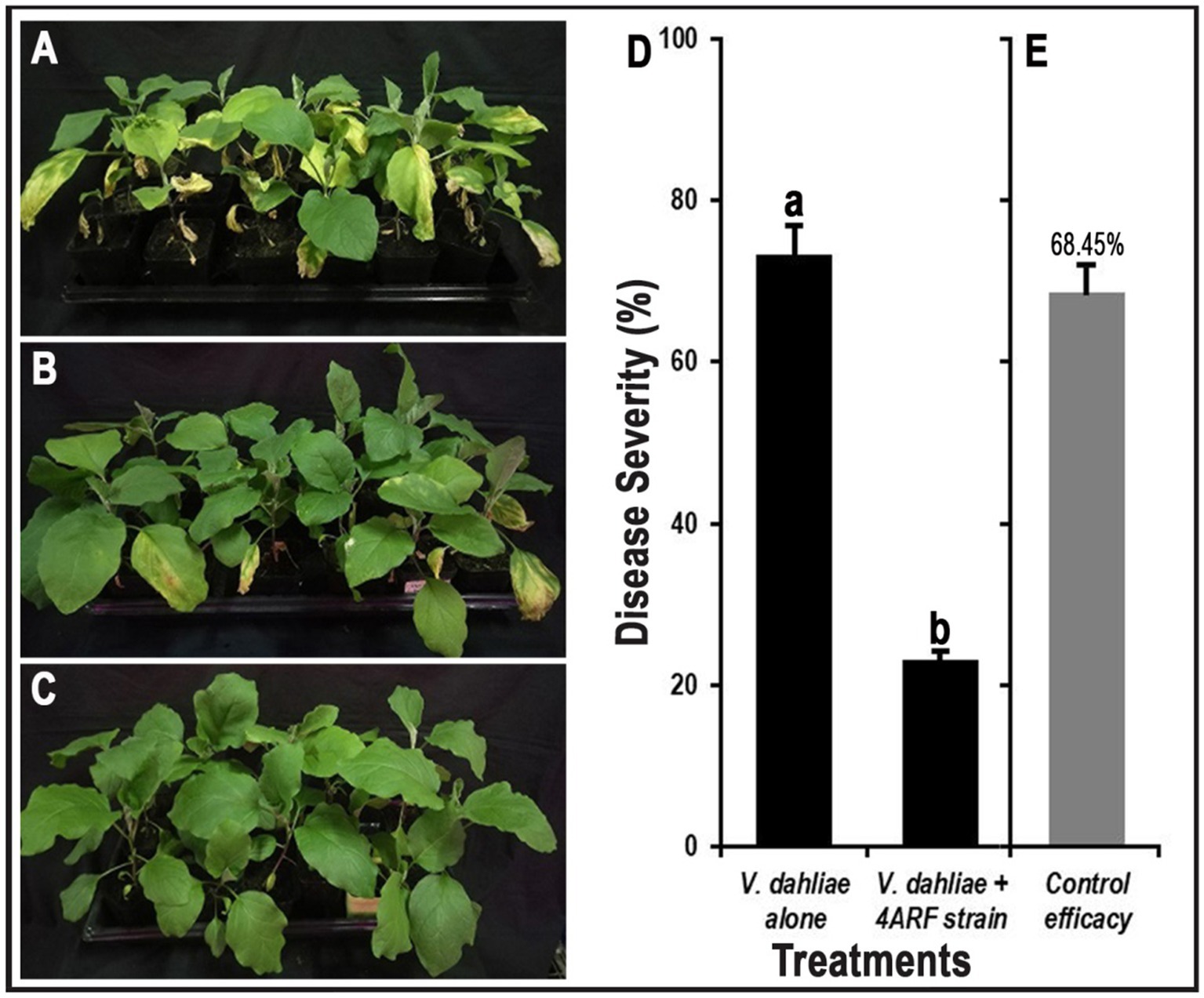
Figure 8. Inhibitory efficacy of rhizobacteria strain ARF4 against eggplant Verticillium wilt. (A) Inoculation with conidia suspension of V. dahliae. (B) Coinoculation with strain ARF4 and conidia suspension of V. dahliae. (C) Without inoculation with V. dahliae and strain ARF4 (control). (D) The disease severity of a Verticillium wilt after 20 days. (E) Control efficacy to the strain ARF4.
4 Discussion
In the present study, overall, 42 rhizospheric isolates were extracted from healthy eggplant rhizosphere soil samples, and their antagonistic potential was evaluated using the dual-culture plate method as described by Meena et al. (2024) and Soylu et al. (2021). The PIRG was estimated, and the bacterial strain with strong antagonistic activity was chosen for further experiments. The strain ARF4 had the highest PIRG percentage, which was 84.49%. These results align with other studies where antagonists from the Bacillus genus exhibit comparable inhibition against different pathogens (Fira et al., 2018; Soylu et al., 2021). Phylogenetic analysis revealed that strain ARF4 is closely related to these strains of Bacillus velezensis (Figure 2). B. velezensis (ARF4) was further tested for broad-spectrum antifungal activity against V. dahliae, F. oxysporum f. sp. niveum, P. capsici, and S. sclerotiorum YY0 through disc diffusion assay, as well as poisoned food method and the results showed that it is considered being an effective antagonist in inhibiting the growth of tested plant pathogens (Figure 1). The dual-culture assay used in this study is in line with the previous studies conducted by Prom and Shi, who effectively demonstrated the antagonistic potential of B. velezensis against a range of pathogens like F. o. f. sp. niveum and V. dahliae (Prom et al., 2023; Shi et al., 2024). In our study, the identified strain B. velezensis ARF4 has potent biocontrol activity, particularly in managing Verticillium wilt, a significant disease affecting eggplant (Solanum melongena). This bacterium is highly effective against a broader range of plant pathogens, especially V. dahliae, due to its broad-spectrum antifungal activity. These bacteria produce various enzymes such as β-glucosidase, chitinase, and protease, as well as the antifungal lipopeptide bacillomycin D (Figures 3, 4). Bacillomycin D is produced by several Bacillus strains using fermentative production in liquid media, such as B. amyloliquefaciens (Gu et al., 2017; Lv et al., 2024), B. subtilis (Gong et al., 2014), and B. velezensis HN-2 (Jin et al., 2020). Primary extraction for bacillomycin D produced by B. velezensis ARF4 was carried out by using solid LBA media, purification by RH-HPLC, and characterization for bacillomycin D was carried out by using TLC using bacillomycin D as a standard.
While TLC and MALDI-TOF MS analyses provided useful insights, it is important to acknowledge that these techniques yield only preliminary evidence for the identification of bacillomycin D. The lack of structural validation via advanced techniques such as nuclear magnetic resonance (NMR) spectroscopy or tandem mass spectrometry (MS–MS) represents a limitation of the current study. Therefore, we explicitly recognize that the identification of bacillomycin D in this study remains tentative. Future studies should incorporate NMR-based structural elucidation and comparative antifungal bioassays using purified standards to confirm the identity and bioactivity of bacillomycin D and its homologs with higher precision.
Bioactive metabolites and lipopeptides are typically extracted from cell-free culture filtrates. However, Zihalirwa Kulimushi et al. (2017) demonstrated, they can also be extracted directly from inhibition zones. While this method may offer practical advantages, such as ease of isolation from actively antagonistic regions, it presents the drawback of potential co-extraction of contaminants. These contaminants may obscure the true efficacy of the antimicrobial agents by introducing other compounds that could interfere with the inhibition zone, leading to an inaccurate assessment of bioactivity. We include this caveat upon the reviewer’s request to clarify limitations inherent in our current extraction approach.
Chitinases are key enzymatic players in the antifungal arsenal of Bacillus species, targeting chitin found in fungal cell walls. Their antifungal efficacy is attributed to hydrolyzing chitin, leading to morphological distortions and stunted growth of fungi (Nazeer, 2023; Toufiq et al., 2018). This hydrolytic action is crucial in biocontrol strategies, as chitinase production has been correlated with notable antifungal activity (Abdelmoteleb et al., 2017; Zhang et al., 2021). Similarly, proteases contribute to this antifungal action by degrading proteins within the fungal cell wall, further facilitating cell lysis (Allioui et al., 2021). Together, these enzymes disrupt the structural integrity of fungal cells, enhancing the overall antifungal potential of the Bacillus species. On the other hand, lipopeptides such as iturin, fengycin, and surfactin emerge as versatile antimicrobial agents that exhibit broad-spectrum antifungal activity. These compounds can disrupt fungal cell membranes, leading to cell lysis and death through mechanisms distinct from those of chitinases and proteases (Guillén-Navarro et al., 2023; Liu et al., 2020). The evidence suggesting that viable Bacillus cultures showcase superior antifungal effects compared to individually applied lipopeptides suggests that additional metabolites or a collaborative interaction may be at play (Deng et al., 2023; Jumpathong et al., 2022). The interplay between these components points toward a synergistic mechanism. The simultaneous action of chitinases, proteases, and lipopeptides could create a multipronged approach to fungal inhibition. For example, while lipopeptides compromise the fungal membrane, chitinases and proteases could facilitate further degradation of cellular architecture. This was illustrated in experiments where the combined action of hydrolytic enzymes and lipopeptides led to enhanced antifungal effects compared to either component alone (Abdelmoteleb et al., 2017; Al-Mutar et al., 2023; Soylu et al., 2022). The evidence suggests that these factors do not act in isolation but rather synergistically enhance the overall antifungal potential, allowing Bacillus species to effectively combat a spectrum of phytopathogenic fungi as well as in the enhancement of plant health.
In this research, the analysis of MALDI-TOF MS showed that antimicrobial lipopeptides produced by rhizobacteria strain ARF4 against V. dahliae were of itrurin family having cyclic structure and homologs to bacillomycin D. These lipopeptides suppress the germination of fungal spores and destroy the fungal cell membrane. Bacillomycin D has been reported to effectively destroy the hyphal structure of V. dahliae and, thereby, its pathogenicity (Figure 6; Hammad et al., 2023). In addition, bacillomycin D extracted from B. velezensis ARF4 strain has also been shown to inhibit the germination of the V. dahliae spores, which further highlights its biocontrol potential (Prom et al., 2023). The biological role of these lipopeptides in biocontrol is well documented, but comprehensive structure–function studies are needed to delineate their specific mechanisms and interactions. The activity of the β-glucosidase and chitinase enzymes also add to the antagonism of this bacterium as they are essential in the breakdown of cell walls of fungal pathogens (Bayisa et al., 2021). Moreover, these enzymes, in addition to antifungal properties, increase nutrient bioavailability in the rhizosphere and promote plant growth and disease resistance (Khan et al., 2020; Songwattana et al., 2023).
Furthermore, a genomic study on B. velezensis has revealed that it contains multiple gene clusters related to the biosynthesis of antimicrobial compounds, including non-ribosomal peptide synthetases responsible for lipopeptide production (Hammad et al., 2023; Nakkeeran et al., 2021). This genetic potential highlights the adaptability of B. velezensis as a biocontrol agent, capable of combating multiple pathogens effectively in a range of environmental conditions. Also, the targeting of specific strains with improved antifungal activities is a step toward the efficient implementation of B. velezensis in agriculture (Grady et al., 2019; Vahidinasab et al., 2022). Field and greenhouse studies have demonstrated that inoculation with B. velezensis significantly reduces both the incidence and severity of Verticillium wilt in eggplants compared to untreated controls. For example, research indicates that B. velezensis strains can effectively lower disease severity by inhibiting the mycelial growth of V. dahliae and enhancing plant defense mechanisms (Hasan et al., 2020; Prom et al., 2023). The presence of B. velezensis in the rhizosphere stimulates the plant’s immune response, leading to increased production of defense-related compounds and improved overall plant health (Ying et al., 2024). This biocontrol strategy not only alleviates Verticillium wilt impact but also maintains sustainable agricultural practices by reducing the reliance on chemical fungicides.
5 Conclusion
Bacillus velezensis, with its multiple mechanisms of action, such as the production of antifungal compounds and hydrolytic enzymes, might be considered an important biocontrol agent against Verticillium wilt in eggplant. The disease incidence and severity were significantly reduced with the inoculation B. velezensis, proving its potential as a sustainable alternative to chemical fungicides in crop protection strategies. Continued research into the specific mechanisms and optimal application methods will further enhance the efficacy and increase its effectiveness in environmentally friendly disease management strategies of B. velezensis in agricultural practices.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: GenBank accession numbers OQ918063.1, OQ920283.1, and OQ920284.1.
Author contributions
AK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. XC: Writing – review & editing, Supervision, Funding acquisition, Resources. IA: Methodology, Writing – review & editing. AT: Software, Writing – review & editing. RH: Writing – review & editing, Software. DA: Writing – review & editing, Supervision, Project administration, Funding acquisition. ME: Writing – review & editing, Funding acquisition. MA: Writing – review & editing, Data curation, Formal analysis, Project administration, Software, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by King Saud University, Riyadh, Saudi Arabia (RSP2025R190).
Acknowledgments
The authors extend their appreciation to the Researchers supporting project number (RSP2025R190), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. ChatGPT4o and DeepSeek were used for rephrasing of the article.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelmoteleb, A., Troncoso-Rojas, R., Gonzalez-Soto, T., and González-Mendoza, D. (2017). Antifungical activity of autochthonous Bacillus subtilis isolated from Prosopis juliflora against phytopathogenic fungi. Mycobiology 45, 385–391. doi: 10.5941/MYCO.2017.45.4.385
Allioui, N., Driss, F., Dhouib, H., Jlail, L., Tounsi, S., and Frikha-Gargouri, O. (2021). Two novel Bacillus strains (subtilis and simplex species) with promising potential for the biocontrol of Zymoseptoria tritici, the causal agent of Septoria tritici blotch of wheat. Biomed. Res. Int. 2021:6611657. doi: 10.1155/2021/6611657
Al-Mutar, D. M. K., Alzawar, N. S. A., Noman, M., Azizullah Li, D., and Song, F. (2023). Suppression of Fusarium wilt in watermelon by Bacillus amyloliquefaciens DHA55 through extracellular production of antifungal Lipopeptides. J. Fungi 9:336. doi: 10.3390/jof9030336
Alsalman, A. J., Farid, A., Al Mohaini, M., Muzammal, M., Khan, M. H., Dadrasnia, A., et al. (2022). Chitinase activity by chitin degrading strain (Bacillus Salmalaya) in shrimp waste. Int. J. Curr. Res. Rev. 14, 11–17. doi: 10.31782/IJCRR.2022.141107
Athukorala, S. N., Fernando, W. D., and Rashid, K. Y. (2009). Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can. J. Microbiol. 55, 1021–1032. doi: 10.1139/W09-067
Ayed, H. B., Hmidet, N., Béchet, M., Chollet, M., Chataigné, G., Leclère, V., et al. (2014). Identification and biochemical characteristics of lipopeptides from Bacillus mojavensis A21. Process Biochem. 49, 1699–1707. doi: 10.1016/j.procbio.2014.07.001
Barzman, M., Bàrberi, P., Birch, A. N. E., Boonekamp, P., Dachbrodt-Saaydeh, S., Graf, B., et al. (2015). Eight principles of integrated pest management. Agron. Sustain. Dev. 35, 1199–1215. doi: 10.1007/s13593-015-0327-9
Bayisa, R. A., Cho, J. Y., and Kim, K. Y. (2021). Purification and identification of a new antifungal dipeptide from Bacillus velezensis AR1 culture supernatant. Pest Manag. Sci. 77, 775–779. doi: 10.1002/ps.6078
Canton, H. (2021). “Food and agriculture organization of the United Nations—FAO” in The Europa directory of international organizations 2021, 297–305.
Chen, Q., and Wang, H. (2022). Biocontrol activity and action mechanism of Bacillus velezensis strain SDTB038 against Fusarium crown and root rot of tomato. Front. Microbiol. 13:994716. doi: 10.3389/fmicb.2022.994716
Chun, J., and Bae, K. S. (2000). Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 78, 123–127. doi: 10.1023/A:1026555830014
da Silva, M. A. C., Cavalett, A., Spinner, A., Rosa, D. C., Jasper, R. B., Quecine, M. C., et al. (2013). Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. Springerplus 2, 1–10. doi: 10.1186/2193-1801-2-127
Deng, Y. J., Chen, Z., Ruan, C. Q., Xiao, R. F., Lian, H. P., Liu, B., et al. (2023). Antifungal activities of Bacillus velezensis FJAT-52631 and its lipopeptides against anthracnose pathogen Colletotrichum acutatum. J. Basic Microbiol. 63, 594–603. doi: 10.1002/jobm.202200489
Dhouib, H., Zouari, I., Abdallah, D. B., Belbahri, L., Taktak, W., Triki, M. A., et al. (2019). Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol. Control 139:104092. doi: 10.1016/j.biocontrol.2019.104092
Farah, I. F., dos Santos, C. R., Pinto, M. C. F., Araújo, C. R., and Amaral, M. C. S. (2024). Pesticides in aquatic environment: occurrence, ecological implications and legal framework. J. Environ. Chem. Eng. 12:114072. doi: 10.1016/j.jece.2024.114072
Fazle Rabbee, M., and Baek, K.-H. (2020). Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 25:4973. doi: 10.3390/molecules25214973
Fira, D., Dimkić, I., Lozo, J., and Stanković, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55.
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., and Martínez de Ilárduya, Ó. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PALgene expression. BMC Plant Biol. 10:232. doi: 10.1186/1471-2229-10-232
Gong, Q., Zhang, C., Lu, F., Zhao, H., Bie, X., and Lu, Z. (2014). Identification of bacillomycin D from Bacillus subtilis fmbJ and its inhibition effects against Aspergillus flavus. Food Control 36, 8–14. doi: 10.1016/j.foodcont.2013.07.034
Grady, E. N., MacDonald, J., Ho, M. T., Weselowski, B., McDowell, T., Solomon, O., et al. (2019). Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 19, 1–14. doi: 10.1186/s12866-018-1380-8
Gu, Q., Yang, Y., Yuan, Q., Shi, G., Wu, L., Lou, Z., et al. (2017). Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 83, e01075–e01017. doi: 10.1128/AEM.01075-17
Guillén-Navarro, K., López-Gutiérrez, T., García-Fajardo, V., Gómez-Cornelio, S., Zarza, E., De la Rosa-García, S., et al. (2023). Broad-spectrum antifungal, biosurfactants and bioemulsifier activity of Bacillus subtilis subsp. spizizenii—a potential biocontrol and bioremediation agent in agriculture. Plan. Theory 12:1374. doi: 10.3390/plants12061374
Hammad, M., Ali, H., Hassan, N., Tawab, A., Salman, M., Jawad, I., et al. (2023). Food safety and biological control; genomic insights and antimicrobial potential of Bacillus velezensis FB2 against agricultural fungal pathogens. PLoS One 18:e0291975. doi: 10.1371/journal.pone.0291975
Hasan, N., Farzand, A., Heng, Z., Khan, I. U., Moosa, A., Zubair, M., et al. (2020). Antagonistic potential of novel endophytic Bacillus strains and mediation of plant defense against Verticillium wilt in upland cotton. Plan. Theory 9:1438. doi: 10.3390/plants9111438
Hayward, A. (2000). Ralstonia solanacearum. Encyclopedia of microbiology, vol. 4. Cambridge, MA: Academic Press.
Howard, M. B., Ekborg, N. A., Weiner, R. M., and Hutcheson, S. W. (2003). Detection and characterization of chitinases and other chitin-modifying enzymes. J. Ind. Microbiol. Biotechnol. 30, 627–635. doi: 10.1007/s10295-003-0096-3
Huang, Z., Lou, J., Gao, Y., Noman, M., Li, D., and Song, F. (2023). FonTup1 functions in growth, conidiogenesis and pathogenicity of Fusarium oxysporum f.sp. niveum through modulating the expression of the tricarboxylic acid cycle genes. Microbiol. Res. 272:127389. doi: 10.1016/j.micres.2023.127389
Ibanga, U. P., Otobong, U. F., Anthony, E. O., Effiong, U. E., and Udo, N. N. (2023). Effect of application of palm bunch ash on the growth and yield of eggplant (Solanum melongena L.) in a pineapple orchard. Am. J. Life Sci. 12, 50–55. doi: 10.11648/j.ajls.20231104.11
Jiang, C.-H., Liao, M.-J., Wang, H.-K., Zheng, M.-Z., Xu, J.-J., and Guo, J.-H. (2018). Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 126, 147–157. doi: 10.1016/j.biocontrol.2018.07.017
Jin, P., Wang, H., Tan, Z., Xuan, Z., Dahar, G. Y., Li, Q. X., et al. (2020). Antifungal mechanism of bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides Penz. Pestic. Biochem. Physiol. 163, 102–107. doi: 10.1016/j.pestbp.2019.11.004
Jumpathong, W., Intra, B., Euanorasetr, J., and Wanapaisan, P. (2022). Biosurfactant-producing Bacillus velezensis PW192 as an anti-fungal biocontrol agent against Colletotrichum gloeosporioides and Colletotrichum musae. Microorganisms 10:1017. doi: 10.3390/microorganisms10051017
Khan, M. S., Gao, J., Chen, X., Zhang, M., Yang, F., Du, Y., et al. (2020). The endophytic bacteria Bacillus velezensis Lle-9, isolated from Lilium leucanthum, harbors antifungal activity and plant growth-promoting effects. J. Microbiol. Biotechnol. 30, 668–680. doi: 10.4014/jmb.1910.10021
Kim, Y. S., Lee, Y., Cheon, W., Park, J., Kwon, H.-T., Balaraju, K., et al. (2021). Characterization of Bacillus velezensis AK-0 as a biocontrol agent against apple bitter rot caused by Colletotrichum gloeosporioides. Sci. Rep. 11:626. doi: 10.1038/s41598-020-80231-2
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lahlali, R., Ezrari, S., Radouane, N., Kenfaoui, J., Esmaeel, Q., El Hamss, H., et al. (2022). Biological control of plant pathogens: a global perspective. Microorganisms 10:596. doi: 10.3390/microorganisms10030596
Li, X., Liang, Z., Yang, G., Lin, T., and Liu, B. (2024). Assessing the severity of Verticillium wilt in cotton fields and constructing pesticide application prescription maps using unmanned aerial vehicle (UAV) multispectral images. Drones 8:176. doi: 10.3390/drones8050176
Liu, Y., Teng, K., Wang, T., Dong, E., Zhang, M., Tao, Y., et al. (2020). Antimicrobial Bacillus velezensis HC6: production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 128, 242–254. doi: 10.1111/jam.14459
Lv, Z., Li, R., Shi, C., Lu, Z., Meng, F., and Bie, X. (2024). The efficient synthesis strategy of bacillomycin D in Bacillus amyloliquefaciens fmbJ. Process Biochem. 145, 131–138. doi: 10.1016/j.procbio.2024.06.026
Lyu, J., Jin, N., Ma, X., Yin, X., Jin, L., Wang, S., et al. (2024). A comprehensive evaluation of nutritional quality and antioxidant capacity of different Chinese eggplant varieties based on multivariate statistical analysis. Antioxidants 14:10. doi: 10.3390/antiox14010010
Meena, P. N., Meena, A. K., Tiwari, R. K., Lal, M. K., and Kumar, R. (2024). Biological control of stem rot of groundnut induced by Sclerotium rolfsii sacc. Pathogens 13:632. doi: 10.3390/pathogens13080632
Munir, S., Azeem, A., Zaman, M. S., and Haq, M. Z. U. (2024). From field to table: ensuring food safety by reducing pesticide residues in food. Sci. Total Environ. 922:171382. doi: 10.1016/j.scitotenv.2024.171382
Nakkeeran, S., Saranya, N., Senthilraja, C., Renukadevi, P., Krishnamoorthy, A., El Enshasy, H. A., et al. (2021). Mining the genome of Bacillus velezensis VB7 (CP047587) for MAMP genes and non-ribosomal peptide synthetase gene clusters conferring antiviral and antifungal activity. Microorganisms 9:2511. doi: 10.3390/microorganisms9122511
Nazeer, W. W. (2023). Isolation, purification and characterization of antifungal Chitinase from Phaseolus vulgaris Var. paulista. Egyptian Acad. J. Biol. Sci. 14, 69–79. doi: 10.21608/eajbsh.2023.297612
Pandit, M. A., Kumar, J., Gulati, S., Bhandari, N., Mehta, P., Katyal, R., et al. (2022). Major biological control strategies for plant pathogens. Pathogens 11:273. doi: 10.3390/pathogens11020273
Platel, R., Sawicki, M., Esmaeel, Q., Randoux, B., Trapet, P., El Guilli, M., et al. (2021). Isolation and identification of lipopeptide-producing Bacillus velezensis strains from wheat phyllosphere with antifungal activity against the wheat pathogen Zymoseptoria tritici. Agronomy 12:95. doi: 10.3390/agronomy12010095
Pretorius, D., Van Rooyen, J., and Clarke, K. (2015). Enhanced production of antifungal lipopeptides by Bacillus amyloliquefaciens for biocontrol of postharvest disease. New Biotechnol. 32, 243–252. doi: 10.1016/j.nbt.2014.12.003
Prom, L. K., Medrano, E. G., and Liu, J. (2023). In vitro antagonistic action by Bacillus velezensis strain LP16S against cotton wilt pathogens. J. Agric. Crops 9, 372–375. doi: 10.32861/jac.93.372.375
Qiu, Z., Lu, X., Li, N., Zhang, M., and Qiao, X. (2018). Characterization of garlic endophytes isolated from the black garlic processing. Microbiol. Open 7:e00547. doi: 10.1002/mbo3.547
Renwick, A., Campbell, R., and Coe, S. (1991). Assessment of in vivo screening systems for potential biocontrol agents of Gaeumannomyces graminis. Plant Pathol. 40, 524–532. doi: 10.1111/j.1365-3059.1991.tb02415.x
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Scholz, S. S., Schmidt-Heck, W., Guthke, R., Furch, A. C., Reichelt, M., Gershenzon, J., et al. (2018). Verticillium dahliae-Arabidopsis interaction causes changes in gene expression profiles and jasmonate levels on different time scales. Front. Microbiol. 9:217. doi: 10.3389/fmicb.2018.00217
Shang, H., He, D., Li, B., Chen, X., Luo, K., and Li, G. (2024). Environmentally friendly and effective alternative approaches to pest management: recent advances and challenges. Agronomy 14:1807. doi: 10.3390/agronomy14081807
Shi, Y., Niu, X., Yang, H., Chu, M., Wang, N., Bao, H., et al. (2024). Optimization of the fermentation media and growth conditions of Bacillus velezensis BHZ-29 using a Plackett–Burman design experiment combined with response surface methodology. Front. Microbiol. 15:1355369. doi: 10.3389/fmicb.2024.1355369
Songwattana, P., Boonchuen, P., Piromyou, P., Wongdee, J., Greetatorn, T., Inthaisong, S., et al. (2023). Insights into antifungal mechanisms of Bacillus velezensis S141 against Cercospora leaf spot in mungbean (V. radiata). Microbes Environ. 38:ME22079. doi: 10.1264/jsme2.ME22079
Soylu, S., Kara, M., Soylu, E. M., Uysal, A., and Kurt, Ş. (2022). Determination of biocontrol potentials of Endophytic Bacteria in biological control of Citrus sour rot disease caused by Geotrichum citri-aurantii. Tekirdağ Ziraat Fakültesi Dergisi 19, 177–191. doi: 10.33462/jotaf.944704
Soylu, S., Kara, M., Uysal, A., Kurt, Ş., and Soylu, E. (2021). Determination of antagonistic potential of endophytic bacteria isolated from lettuce against lettuce white mould disease caused by Sclerotinia sclerotiorum. Zemdirbyste-Agriculture 108, 303–312. doi: 10.13080/z-a.2021.108.039
Swati Verma, R., Chauhan, A., Shandilya, M., Li, X., Kumar, R., and Kulshrestha, S. (2020). Antimicrobial potential of ag-doped ZnO nanostructure synthesized by the green method using Moringa oleifera extract. J. Environ. Chem. Eng. 8:103730. doi: 10.1016/j.jece.2020.103730
Tomah, A. A., Abd Alamer, I. S., Li, B., and Zhang, J.-Z. (2020a). A new species of Trichoderma and gliotoxin role: a new observation in enhancing biocontrol potential of T. virens against Phytophthora capsici on chili pepper. Biol. Control 145:104261. doi: 10.1016/j.biocontrol.2020.104261
Tomah, A. A., Alamer, I. S. A., Khattak, A. A., Ahmed, T., Hatamleh, A. A., Al-Dosary, M. A., et al. (2023). Potential of Trichoderma virens HZA14 in controlling Verticillium wilt disease of eggplant and analysis of its genes responsible for Microsclerotial degradation. Plan. Theory 12:3761. doi: 10.3390/plants12213761
Tomah, A. A., Alamer, I. S. A., Li, B., and Zhang, J.-Z. (2020b). Mycosynthesis of silver nanoparticles using screened Trichoderma isolates and their antifungal activity against Sclerotinia sclerotiorum. Nano 10:1955. doi: 10.3390/nano10101955
Toufiq, N., Tabassum, B., Bhatti, M. U., Khan, A., Tariq, M., Shahid, N., et al. (2018). Improved antifungal activity of barley derived chitinase I gene that overexpress a 32 kDa recombinant chitinase in Escherichia coli host. Braz. J. Microbiol. 49, 414–421. doi: 10.1016/j.bjm.2017.05.007
Triki, M., Hassaïri, A., and Mahjoub, M. (2006). Premières observations de Verticillium dahliae sur olivier en Tunisie. EPPO Bulletin 36, 69–71. doi: 10.1111/j.1365-2338.2006.00941.x
Vahidinasab, M., Adiek, I., Hosseini, B., Akintayo, S. O., Abrishamchi, B., Pfannstiel, J., et al. (2022). Characterization of Bacillus velezensis UTB96, demonstrating improved lipopeptide production compared to the strain B. velezensis FZB42. Microorganisms 10:2225. doi: 10.3390/microorganisms10112225
Wan, N.-F., Fu, L., Dainese, M., Kiær, L. P., Hu, Y.-Q., Xin, F., et al. (2025). Pesticides have negative effects on non-target organisms. Nat. Commun. 16:1360. doi: 10.1038/s41467-025-56732-x
Wang, J., Peng, Y., Xie, S., Yu, X., Bian, C., Wu, H., et al. (2023). Biocontrol and molecular characterization of Bacillus velezensis D against tobacco bacterial wilt. Phytopathol. Res. 5:50. doi: 10.1186/s42483-023-00204-x
Xiao, J., Guo, X., Qiao, X., Zhang, X., Chen, X., and Zhang, D. (2021). Activity of fengycin and iturin A isolated from Bacillus subtilis Z-14 on Gaeumannomyces graminis Var. tritici and soil microbial diversity. Front. Microbiol. 12:682437. doi: 10.3389/fmicb.2021.682437
Yang, H., Wang, T., Ji, F., Zhou, Q., and Wang, J. (2023). Effects of LED light spectrum on the growth and energy use efficiency of eggplant transplants. Int. J. Agric. Biol. Eng. 16, 23–29. doi: 10.25165/j.ijabe.20231603.7260
Yang, X., Zhang, Y., Cheng, Y., and Chen, X. (2019). Transcriptome analysis reveals multiple signal network contributing to the Verticillium wilt resistance in eggplant. Sci. Hortic. 256:108576. doi: 10.1016/j.scienta.2019.108576
Ying, Y., Zhang, N., Li, B., Liu, D., Zhang, Q., Yang, Z., et al. (2024). Biocontrol potential of Bacillus velezensis ZF-10 inhibiting potato virus Y and promoting growth of tobacco plant. J. Phytopathol. 172:e70002. doi: 10.1111/jph.70002
Zhang, F., Li, X.-L., Zhu, S.-J., Ojaghian, M. R., and Zhang, J.-Z. (2018). Biocontrol potential of Paenibacillus polymyxa against Verticillium dahliae infecting cotton plants. Biol. Control 127, 70–77. doi: 10.1016/j.biocontrol.2018.08.021
Zhang, D., Xiong, X., Wang, Y., Gao, Y., Ren, Y., Wang, Q., et al. (2021). Bacillus velezensis WLYS23 strain possesses antagonistic activity against hybrid snakehead bacterial pathogens. J. Appl. Microbiol. 131, 3056–3068. doi: 10.1111/jam.15162
Zhang, Y., Zhang, D., Zhang, Y., Cheng, F., Zhao, X., Wang, M., et al. (2024). Early detection of verticillium wilt in eggplant leaves by fusing five image channels: a deep learning approach. Plant Methods 20:173. doi: 10.1186/s13007-024-01291-3
Zhu, W., Liu, X., Chen, M., Tao, N., Tendu, A., and Yang, Q. (2021). A new miRNA miRm0002 in eggplant participates in the regulation of defense responses to Verticillium wilt. Plan. Theory 10:2274. doi: 10.3390/plants10112274
Keywords: biocontrol, phytopathogenic, bacteria, green technology, sustainable agriculture
Citation: Khattak AA, Chen X, AbdAlamer IS, Tomah AA, Hussein RA, Al Farraj DA, Elshikh MS and Afzal M (2025) Isolation and characterization of Bacillus velezensis: investigating its mechanisms as an effective biocontrol agent against Verticillium wilt in eggplants. Front. Sustain. Food Syst. 9:1581883. doi: 10.3389/fsufs.2025.1581883
Edited by:
Mohamed Ait-El-Mokhtar, University of Hassan II Casablanca, MoroccoReviewed by:
Soner Soylu, Mustafa Kemal University, TürkiyeTalwinder Kaur, Guru Nanak Dev University, India
Jubair Al-Rashid, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Khattak, Chen, AbdAlamer, Tomah, Hussein, Al Farraj, Elshikh and Afzal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Afzal, bWFmemFsLmt1c3RAZ21haWwuY29t
†ORCID: Muhammad Afzal, orcid.org/0000-0001-9530-1356
 Arif Ali Khattak
Arif Ali Khattak Xiaoyuan Chen2
Xiaoyuan Chen2 Iman Sabah AbdAlamer
Iman Sabah AbdAlamer Ali Athafah Tomah
Ali Athafah Tomah Mohamed Soliman Elshikh
Mohamed Soliman Elshikh Muhammad Afzal
Muhammad Afzal