- Department of Environmental Engineering Science, Graduate School of Science and Technology, Gunma University, Kiryu, Japan
The diamondback moth (Plutella xylostella L.), a major global pest of cruciferous vegetables, has resisted conventional insecticides, necessitating novel pest control strategies. In this study, we evaluated four non-Bt bacterial strains—Mesobacillus thioparans CC8, Bacillus mobilis CC13, Bacillus subtilis CC18, Chromobacterium rhizoryzae 4C2, and the effectiveness of bacterial consortia against P. xylostella L. larvae. The extracellular hemolysins and insecticidal activity utilizing the leaf-dip method were investigated. The effect of cell concentration, larval size, and exposure methods on insecticidal efficacy were examined. The fecal pellet examination was used to evaluate the presence of microbial communities, while scanning electron microscopy assessed gut damage. The findings demonstrated 100% larval mortality within 48 h of exposure, with the artificial selection, comprising four non-Bt bacterial strains, exhibiting enhanced efficacy compared to individual applications. Utilizing second-instar larvae with precisely dose-dependent cell densities increased mortality. Both leaf-dip and direct-spray application methods showed comparable efficacy, offering flexibility for practical applications. The target insecticidal bacteria were detected in the larval fecal microbiota, while the larva’s external features showed damage after exposure. This study highlights the potential of non-Bt insecticidal bacteria as an alternative strategy for managing P. xylostella L., contributing to the development of sustainable pest management solutions.
1 Introduction
The diamondback moth, Plutella xylostella L. (Lepidoptera: Plutellidae), is a significant pest of cruciferous vegetables globally. Managing P. xylostella L. has become increasingly challenging, making it one of the most difficult pests to control worldwide (Shehzad et al., 2023). The larvae of P. xylostella L. feed on the leaves of Brassica crops, causing substantial yield losses, particularly in broccoli, cabbage, and cauliflower. An estimated US$ 1.4 billion is spent annually on pesticide costs for Brassicaceae vegetable crops (Farias et al., 2020). Outbreaks can make controlling larvae and adults difficult, often necessitating multiple insecticide applications (Calvin and Palumbo, 2024). This pest contributes to global annual losses and control costs of US$ 4–5 billion (Zalucki et al., 2012) and has developed resistance to 104 active ingredients and pesticides, presenting considerable control challenges (Arthropod Pesticide Resistance Database, 2025). Although synthetic pesticides have been widely used to mitigate crop losses, they pose risks to both human health and the environment, such as water contamination, soil microbial community disruption, and biodiversity loss (European Environment Agency, 2023; Fenibo et al., 2022).
Microbial insecticides have been significantly reported for pest management, effectively targeting and killing insect pests (da Silva et al., 2020; Mnif and Ghribi, 2015; Thakur et al., 2020). The most widely utilized biological pesticide to control insect pests is the product from Bacillus thuringiensis (Bt). These insecticidal proteins can withstand long-term storage. Recent studies highlight the potential of certain bacterial species, other than B. thuringiensis (non-Bt), to produce toxic proteins that target insect larvae (Barbieri et al., 2021; Beltrán Pineda and Castellanos-Rozo, 2025). Over the past decade, there has been a notable shift toward using new insecticidal properties, particularly those derived from non-Bt bacteria (Barry et al., 2023). Identifying novel non-Bt insecticidal bacteria is crucial for developing new insecticides targeting coleopteran pests (Mi et al., 2023). Previous studies have identified non-Bt insecticidal bacteria effective against P. xylostella L., including Bacillus popilliae, B. lentimorbus, B. sphaericus, Pseudomonas taiwanesis, P. entomophila, P. cedrina, P. paralactis, P. aeruginosa, Klebsiella pneumoniae (Beltrán Pineda and Castellanos-Rozo, 2025), Photorhabdus luminescens (Guo et al., 1999), Yersinia entomophaga (Hurst et al., 2019), and P. cedrina (Liu et al., 2019).
Utilizing non-Bt bacteria with insecticidal properties introduces alternative modes of action against insects (Beltrán Pineda and Castellanos-Rozo, 2025). A recent study by Srujana et al. (2022) showed that bioactive secondary metabolites produced by microbial biopesticides accelerated metabolic processes and induced toxicity in P. xylostella L. larvae. Hemolytic activity was used to assess insecticidal effectiveness (Brillard et al., 2001; Bravo et al., 2018). B. thuringiensis (Bt), a well-known biological insecticide, has been extensively studied in insecticidal bioassays and is recognized for its hemolytic properties (Wu et al., 2008; Nair et al., 2018). However, non-Bt insecticidal bacteria may also exhibit hemolytic properties, offering additional avenues for pest control.
This study aimed to identify non-Bt insecticidal bacteria and to evaluate the effectiveness of a bacterial consortium derived from wastewater treatment sludge and dam sediment in combating the larvae of the diamondback moth, P. xylostella L., the most destructive pest of cruciferous plants. We demonstrated 100% larval mortality within 48 h of exposure, with the artificial selection of four non-Bt bacterial strains exhibiting enhanced efficacy compared to individual applications. Our goal is not only to conduct research but also to go beyond purely laboratory-based research, emphasizing the practical application of our findings in real-world settings.
2 Materials and methods
2.1 Sample collection
The dam sediment sample was collected at the Shimokubo Dam, located on the border between Gunma and Saitama prefectures (N36°71′57″, E139°1′21″). At the time of sampling, the water depth of the dam was approximately 53 meters. Sampling was conducted on June 5, 2024, using a submersible pump (Smashing cutter pump, Model 80CA43.7, Tsurumi Manufacturing Co., Ltd., Japan) from a small barge. The pump was placed at the bottom of the dam to collect the bottom mud. The near-surface sediment, approximately 30 cm from the surface of the sediment, was pumped up at a rate of 900 kg/h and immediately dewatered. The dewatered sediment primarily consisted of clay and silt, with 88% of the sampled sediment being fine particles. The total solids (TS) and volatile solids (VS) of the sediment were 4.9 and 7.6%, respectively. The water at the bottom of the dam had a temperature of 6–7°C, pH of 7.0–7.2, dissolved oxygen (DO) of 5.9 mg/L, and electrical conductivity (EC) of 21 mS/m. The surface water temperature was 24–25°C. On the other hand, the activated sludge sample was obtained from the wastewater treatment plant of Tomioka Foods (Sano City, Japan). For dam sediment and activated sludge, approximately 1 kg of samples were collected in sterile 50 mL tubes and plastic bags, placed in a cool box, and transported to the laboratory within 2 h. Each sample was then stored in a refrigerator at 3°C and used within 24 h.
2.2 Isolation and identification
The bacteria were isolated using dam-dredged sediment and activated sludge samples. For the dam sediment sample, one gram of the sample was suspended in 9.0 mL of sterile distilled water using a Vortex Mixer, and a serial dilution was conducted. Aliquots of 100 μL were plated on nutrient agar (NA, pH 7.0), which served as the enriched medium. The nutrient agar contained 15.0 g/L of agar, 5.0 g/L of peptone, 5.0 g/L of sodium chloride, 2.0 g/L of yeast extract, and 1.0 g/L of meat extract. The plates were incubated at 30°C for 48 h. Afterward, the isolated colonies were selected for purification based on their morphological characteristics. Pure cultures were obtained by repeatedly picking and streaking the colonies onto nutrient agar plates. For the activated sludge sample, after homogenizing 20 mL of activated sludge using a tissue homogenizer (DREMEL 300) equipped with a φ7 mm probe, a 0.2 mL aliquot of the homogenized activated sludge was mixed with 1.8 mL of 10% phosphate-buffered saline (pH 7.2). A serial dilution was performed, and aliquots were plated on nutrient agar. The plates were incubated at 37°C for 48 h. The following isolation process was the same as that conducted for the dam sediment. The purified colonies were preserved in nutrient broth (NB) containing 50% glycerol at −80°C for future use (Note: 50% glycerol was used without a specific reason and is higher than the typical 10–30% used). The full-length 16S rRNA gene sequences of the colonies were analyzed using a colony polymerase chain reaction, followed by Sanger sequencing, which was performed at GENEWIZ Japan. The sequencing data are deposited in the GenBank/DDBJ/ENA database under the accession numbers LC868238 to LC868265.
2.3 Hemolytic activity assay
The extracellular hemolysins produced by isolated strains were assessed using Sheep Blood Agar plates (Nippon Becton Dickinson Company Ltd., Japan). Each fresh colony on the LB plate culture was pin-spotted on the Sheep Blood Agar plates and then incubated for 96 h at 30°C. The B. thuringiensis (NBRC 101235) and Escherichia coli strain K-12 (NBRC 3301) served as positive and negative controls, respectively, purchased from the National Biological Resource Center, NITE (NBRC, Japan). The formation of a clear zone around the bacterial colonies indicated extracellular hemolysin production. The diameter of the clear zone was measured with a caliper at intervals of 0, 24, 48, 72, and 96 h. The experiments were performed in triplicate.
2.4 Insecticidal activity assay using leaf-dip method
The P. xylostella L. larvae were purchased from Sumika Technoservice Corp (Takarazuka, Japan). A mixture of larvae at both the 3rd and 4th instar was used unless otherwise stated. The larvae were deprived of diet for 6 h before the experiments were conducted. Luria-Bertani (LB) medium, consisting of 15.0 g/L of agar, 10.0 g/L of tryptone, 10.0 g/L of sodium chloride, and 5.0 g/L of yeast extract, was used to culture all isolated strains at 30°C. A bacterial suspension was prepared from 24-h colonies grown on LB plates. The cells were collected and resuspended in sterile distilled water.
The bacterial strains that exhibited positive hemolytic activity were selected to assess their insecticidal properties. The screening for the insecticidal bioassay was conducted using an optimal bacterial suspension of 1010 CFU/mL, the measured concentration was 3 × 1010 CFU/mL. Fresh leaves of cabbage (Brassica oleracea var. capitata L.) were used due to their significant susceptibility to P. xylostella L. Freshly clipped, cleaned leaves of B. oleracea var. capitata L. were dipped in a bacterial suspension for 30 s before being transferred to a plastic box (17.5 cm × 10 cm × 14 cm) containing ten starved P. xylostella L. larvae. Leaves dipped in sterile distilled water were used as the blank control. The B. thuringiensis (Bt) and E. coli K-12 were served as positive and negative controls, respectively. Larval mortality was recorded at 0, 6, 24, 36, and 48 h post-exposure. Larval movement and mortality were observed using an Illuminated Magnifier lamp (Otsuka Optics Co., Ltd., Tokyo, Japan). Deceased larvae were photographed using an OLYMPUS Tough TG-3 digital camera (12 MP CMOS sensor, f/2.0; Olympus Corporation, Japan). All experiments were carried out in triplicate.
2.5 Effect of cell concentration on insecticidal efficacy
This study examined the impact of dose-dependent cell density on insecticidal efficiency. The bacteria exhibiting insecticidal activity were further selected to evaluate the cell concentration on insecticidal efficacy. The consortium of non-Bt bacteria was created using isolated bacterial strains with insecticidal potential. The insecticidal effectiveness of each strain and the consortium was assessed using the leaf-dip method. Concentrations of bacterial suspensions were set in 10-fold dilutions between 106 and 1010. The measured results, with the bacterial suspension concentrations of 5 × 106, 5 × 107, 4 × 108, 3 × 109, and 4 × 1010 CFU/mL, were utilized to assess the effect of cell concentration on insecticidal efficacy. Larval mortality was recorded at 48 h post-exposure. Probit regression analysis was conducted to assess the dose–response mortality of P. xylostella L. to the bacterial consortium using IBM SPSS Statistics version 25. The number of dead larvae was used as the response frequency, with log10-transformed CFU/mL as the covariate. The lethal concentration (LC50) and its 95% confidence interval were estimated from the regression output. Model fit was evaluated using the Pearson chi-square (χ2) goodness-of-fit test. Dose–response curves and residual analyses were generated to assess model adequacy and predictive performance. Controlled studies were conducted with sterile distilled water without bacterial suspension, while B. thuringiensis (Bt) was utilized as a positive control. The insecticidal bioassay for each strain and consortium was conducted in triplicate.
2.6 Effect of larval size on insecticidal efficacy
Two sizes of P. xylostella L. (2nd and 4th instar larvae) were examined in this study. We aimed for bacterial suspensions with a concentration of 1010 CFU/mL for all strains. The measured concentrations were 7 × 1010 for CC8, 6 × 1010 for CC13, 8 × 1010 for CC18, 6 × 1010 for 4C2, 8 × 1010 for Bt, and 8 × 1010 CFU/mL for the bacteria consortium. These suspensions, obtained after dilution, were employed to evaluate larval mortality using a leaf-dip method. The bacteria exhibiting insecticidal activity were constructed as a consortium and assessed for its insecticidal efficacy compared with individual strains. The second and fourth instar larvae mortality rates were compared at 0, 24, 36, and 48 h post-exposure. Sterile distilled water without bacterial suspension was utilized as a blank control, and B. thuringiensis (Bt) was utilized as the positive control, respectively. Each insecticidal bioassay was conducted in triplicate.
2.7 Microbial community of larval feces
Fecal pellets were collected from larvae during the insecticidal activity assay and stored in sterile microcentrifuge tubes at −80°C. The fecal samples were then sent to the Bioengineering Lab. Co., Ltd. (Atsugi, Japan) for DNA extraction, library preparation, sequencing, and phylogenetic analysis. To extract DNA from the fecal samples, they were first freeze-dried and ground using a Multi Beads Shocker (Yasuikikai, Japan) at 1,500 rpm for 2 min. Lysis Solution F (Nippon Gene, Japan) was then added to the homogenized samples, followed by incubation at 65°C for 10 min. The samples were subsequently centrifuged at 12,000 × g for 2 min, and the supernatant was collected. DNA purification was performed using the Lab-Aid824s DNA Extraction Kit (ZEESAN, China). For library preparation, the 2-step tailed PCR method was employed. PCR amplification of the 16S rRNA gene was performed using the bacterial primer pairs 341f and 805r (Takahashi et al., 2014). The quality of the prepared library was assessed using the Fragment Analyzer and dsDNA 915 Reagent Kit (Agilent Technologies, USA). Sequencing was conducted using the MiSeq system and MiSeq Reagent Kit v3 (Illumina, USA) with a 2 × 300 bp configuration. Sequence data processing was performed using the fastx_barcode_splitter tool from FASTX-Toolkit (ver. 0.0.14) to organize reads according to barcode information. Primer sequences were subsequently removed using the fastx_trimmer tool from the same toolkit. Next, low-quality sequences with a quality score below 20 were filtered out using sickle (ver. 1.33), and sequences shorter than 130 bases, along with their paired sequences, were discarded. Read merging was conducted using the paired-end read assembly tool FLASH (ver. 1.2.11). Finally, chimera and noise filtering were performed using the dada2 plugin in QIIME2 (ver. 2024.10), yielding representative sequences and an Amplicon Sequence Variant (ASV) table. Taxonomic classification was carried out using the feature-classifier plugin, comparing representative sequences against the Greengene (ver. 13_8) 97% OTU database (DeSantis et al., 2006).
2.8 SEM analysis of larval midgut
SEM samples were prepared following the protocol as described by Bozzola and Russell (1999). Freshly deceased larvae were rinsed with phosphate-buffered saline (PBS, pH 7.2) before carefully dissection of their midguts. The gut tissues were fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.2) at 4°C for 24 h. Afterward, the samples were rinsed thrice for 10 min each with 0.1 M PBS (pH 7.2). The gut tissues were processed with a two-step dehydration process utilizing an ethanol series of 30, 50, 70, 80, 90, 95, and 100% ethanol. Each dehydration step lasts 15 min. The specimens were affixed to SEM stubs using carbon adhesive tape and air-dried overnight. Subsequently, the samples were coated with a conductive layer of osmium metal using a plasma osmium coater (OSMIUM COATER HPC-1SW, Japan). The surface damage, including disruptions, smoothness changes, epithelial lining detachment, pore formation, erosion, and the collapse of gut microstructures, were analyzed by SEM (SSX-550, Shimazu Co. Ltd., Japan).
2.9 Effect of exposure method on insecticidal efficacy
The effect of bioassay methods on insecticidal efficacy was examined. Two practically distinct methods of exposure, leaf dip and direct spray, were employed to investigate insecticidal efficacy. The leaf-dip method is to administer an oral dose to the larvae. Directly spraying onto the larval body is to administer a dermal dose. The leaf-dip method was described above. The direct-spray method involved the use of bacterial suspensions at a concentration of 9 × 1010 CFU/mL. Ten starved P. xylostella L. larvae were placed in a sterile Petri dish. A 200 μL suspension of bacterial strains exhibiting insecticidal activity was sprayed onto the larvae and cleaned B. oleracea var. capitata L. leaves, with each Petri dish receiving 3 sprays. The effects of different exposure methods on larval mortality were studied at 0, 24, 36, and 48 h. Leaves dipped in sterile distilled water were used as the blank and B. thuringiensis (Bt) was used as the positive control.
2.10 Statistical analysis
The data were analyzed using a One-way ANOVA (IBM SPSS Statistics 25) to identify any statistical differences (p = 0.05). A Tukey HSD Post-Hoc Test was conducted to evaluate pairwise group mean differences, where means sharing the same letter indicate no significant difference (p > 0.05). The mean difference is considered significant at the 0.05 level. A compact letter display (CLD) was generated using R version 4.4.2. Results are presented as triplicate samples ± standard deviation (SD).
2.11 Molecular analysis
We submitted colony samples of our strains to Azenta Life Sciences Inc. (Tokyo, Japan) for 16S rRNA gene sequencing. The submitted colony samples underwent crude NaOH lysis to be directly used in PCR. PCR was performed according to the company’s standard operating procedures, using primers designed to amplify regions V1-V9 of the 16S rRNA gene, resulting in an amplicon approximately 1.4 kb in size. Following PCR amplification, enzymatic cleanup was carried out using Exonuclease I and Shrimp Alkaline Phosphatase (ExoSAP). Dye-terminator sequencing was performed with Applied Biosystems BigDye version 3.1, and sequencing-specific primers were utilized to generate bidirectional reads. The reactions were analyzed on an Applied Biosystems 3730xl DNA Analyzer. We received de novo assemblies of each strain and compared the resulting sequences to the NCBI BLASTn database (Altschul et al., 1990), specifically the RefSeq 16S database. The sequence with the highest alignment score and an E-value of 0.0 was identified as the most closely related species.
3 Results and discussion
3.1 Isolation and identification of potential insecticidal bacteria
Different species of bacteria were obtained based on their morphological characteristics. Through 16S rRNA gene sequencing analysis, a total of 16 bacterial species from 10 distinct genera (Acinetobacter, Alkalihalobacillus, Bacillus, Brevundimonas, Enterobacter, Exiguobacterium, Klebsiella, Mesobacillus, Metabacillus, and Stenotrophomonas) were identified from sediment samples. Furthermore, 11 bacterial species from 10 different genera (Bacillus, Brevibacillus, Chromobacterium, Chryseobacterium, Corynebacterium, Empedobacter, Microbacterium, Neobacillus, Ralstonia, and Staphylococcus) were isolated from the wastewater treatment sludge. As a result, 27 bacterial species belonging to 19 different genera were successfully isolated from both sampled sites (Table 1). Both activated sludge from the wastewater treatment plant and dam-dredged sediment have been reported to exhibit a high microbial diversity that is vital for biotechnological applications (Röske et al., 2012; Ito et al., 2021). The successful isolation and identification of bacterial species from sediment and wastewater treatment sludge samples emphasized the rich microbial diversity in these environments. The current study consists of previous findings highlighting bacterial strains from sediment and wastewater treatment plants that have been reported to affect Lepidoptera species (Mohammedi et al., 2006; Fathy et al., 2024). The presence of Bacillus in both environments demonstrates its ecological versatility. Moreover, Bacillus bacteria have been reported to control various Lepidoptera species effectively (Nelly et al., 2024) and to disrupt the midgut membrane of these insect larvae (Kobisi et al., 2024).
3.2 Hemolytic activity of bacterial isolates
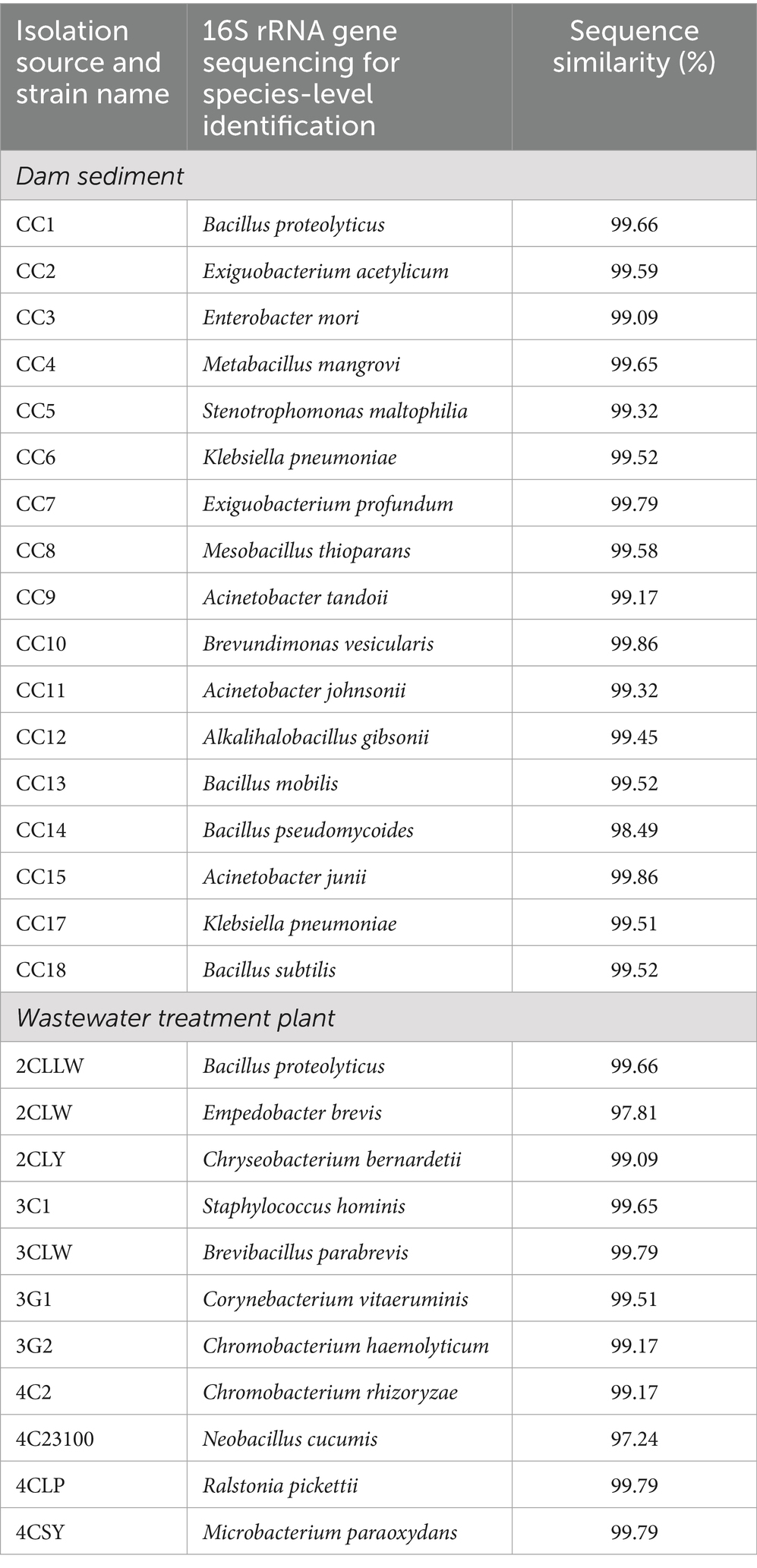
The hemolytic activity was evaluated to determine the potential of insecticidal toxins as microbial biopesticides that disrupt midgut epithelial cells. An isolated strain was further evaluated for its hemolytic activity. The Bacillus subtilis strain CC18 showed the greatest activity among the isolated strains, with halo diameters increasing from 4 cm at 24 h to 7.3 cm at 96 h. The positive control, Bt, exhibited a halo diameter of 3.5 cm at 24 h, increasing to 7.9 cm at 96 h, with no significant difference (p < 0.05) from the B. subtilis strain CC18. The Acinetobacter junii strain CC15 and Bacillus mobilis strain CC13 showed moderate activity. CC15 exhibited a halo diameter of 1.1 cm at 24 h, increasing to 2.5 cm at 96 h. CC13 started at 1.9 cm at 24 h, rising to 3.9 cm by 96 h. Chromobacterium rhizoryzae strain 4C2 had a halo diameter of 1.1 cm at 24 h, reaching 2.7 cm at 96 h, as shown in Figure 1A. Previous studies found that B. subtilis produces secondary metabolites with hemolytic activity and insecticidal effects against lepidopteran pest larvae (Osouli and Afsharmanesh, 2016). This bacterium shows strong larvicidal activity against lepidopterans and produces bioactive lipopeptides (Ghribi et al., 2012; Susetyo et al., 2023). Acinetobacter has identified β-hemolysis as the most common in this genus (Boone et al., 2021). Similarly, Chromobacterium species are strongly hemolytic and toxic to various lepidopteran insects (Han et al., 2008; Blackburn et al., 2017).
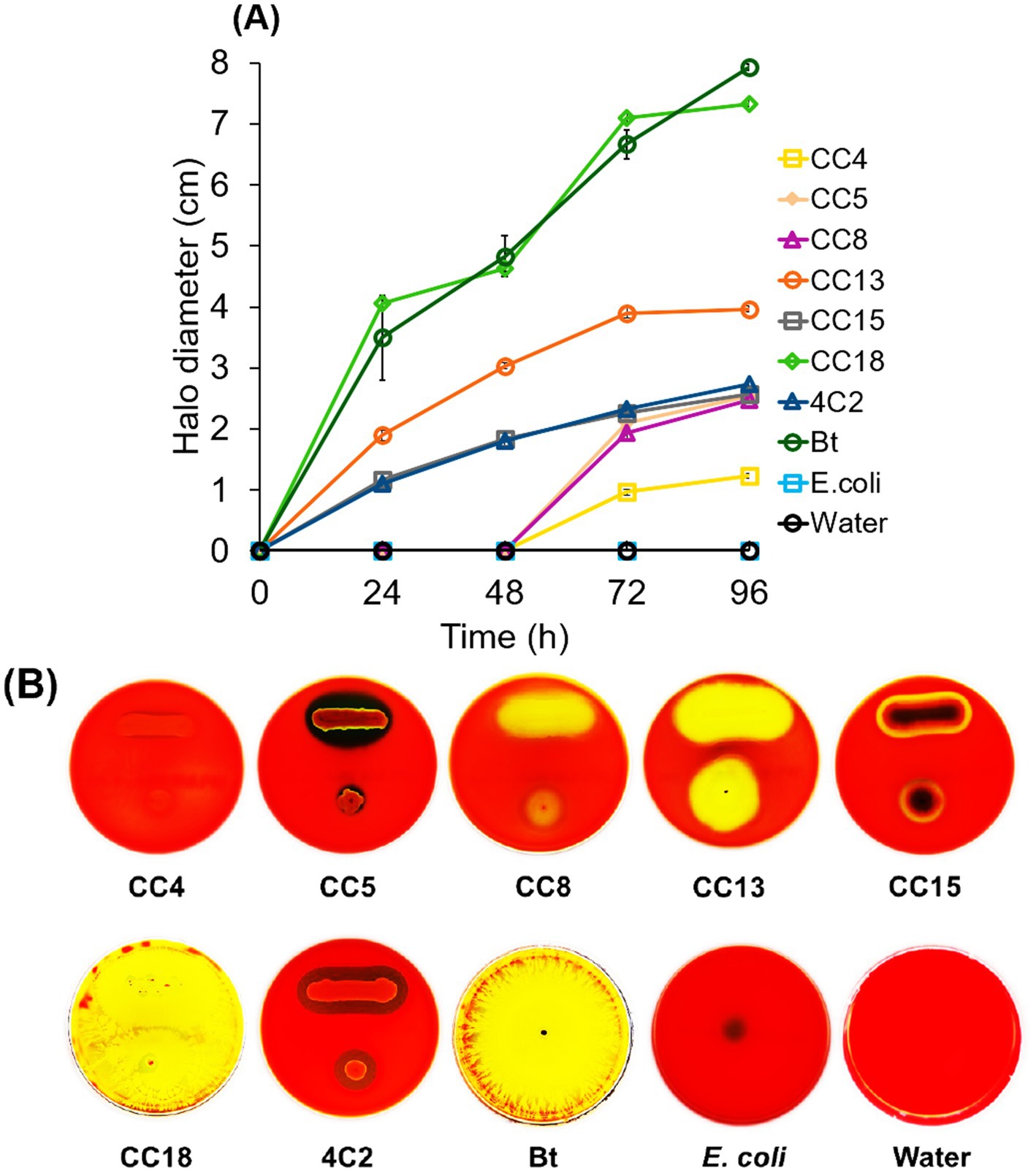
Figure 1. Hemolytic activity of bacterial isolates. (A) Time course monitoring of halo diameter of the clear zones appearing on blood agar plates incubated with the isolated strains CC4, CC5, CC8, CC13, CC15, CC18, and 4C2. Bt served as a positive control. E. coli K-12 served as a negative control. Sterile distilled water was used as a blank control. (B) Images showing the breakdown of red blood cells on the blood agar plates were observed over 96 h at 30°C. The hemolytic activity assay was performed in triplicate for each strain. Each plot is presented with the standard deviation (SD).
In contrast, Mesobacillus thioparans strain CC8, Stenotrophomonas maltophilia strain CC5, and Metabacillus mangrovi strain CC4 revealed limited activity, with only minor halo increases over 96 h. E. coli K-12 and sterile distilled water showed no halo diameter. Figure 1B displays the halo formation among the bacterial strains, revealing variability in activity. The hemolysis patterns demonstrated B. subtilis strain CC18, A. junii strain CC15, M. thioparans strain CC8, B. thuringiensis (Bt), S. maltophilia strain CC5, B. mobilis strain CC13, and C. rhizoryzae strain 4C2 exhibit β-hemolysis, characterized by a clear and transparent zone surrounding the bacterial colonies, indicating destruction of red blood cells. In contrast, M. mangrovi strain CC4 displays α-hemolysis, identified by partially destroying red blood cells and oxidizing hemoglobin. E. coli K-12 is classified as γ-hemolysis, indicated by no visible change to the agar around the bacterial colonies, which signifies no hemolysis.
Insecticidal bacteria produce hemolysin, which is evident as a lysis zone on blood agar plates. Isolated strains showed positive hemolysis around their colonies, indicating biochemical and insecticidal properties against lepidopteran pests. Previous studies indicated that bacteria with hemolytic activities may possess potential insecticidal properties (Naimov et al., 2008; Nair et al., 2018; Mollah et al., 2020). While hemolytic activity may indicate possible insecticidal properties, it also raises concerns about its effects on non-target organisms (Belousova et al., 2021). Therefore, using hemolytic strains to identify insecticidal toxins should be carefully ensured to avoid non-target species. As a result, promising isolates with significant hemolytic activity, particularly strains CC4, CC5, CC8, CC13, CC15, CC18, and 4C2, have been chosen for an insecticidal activity assay using the leaf-dip method.
3.3 Insecticidal activity assay
The leaf-dip method was utilized to assess insecticidal activity. Isolated bacterial strains with hemolytic activity were selected for insecticidal potential through the leaf-dip method, with mortality rates recorded after specific exposure intervals. The larvae were observed to be sluggish and stopped feeding, while the dead larvae turned black (Figure 2), as described by Devi et al. (2022). As illustrated in Figure 3, B. subtilis strain CC18 showed an increasing mortality rate, reaching 33% at 24 h, 50% at 36 h, and 100% at 48 h. Similarly, B. mobilis strain CC13 and C. rhizoryzae strain 4C2 demonstrated a significant increase in mortality, starting at 36 and 50% at 24 h, respectively, and reaching 100% by 48 h. Bt displayed a rapid biopesticidal effect, achieving 100% mortality within 24 h, showcasing its well-established entomopathogenic properties as a biopesticide. The A. junii strain CC15, S. maltophilia strain CC5, and M. mangrovi strain CC4 showed low mortality rates of around 26 and 23% after 48 h, indicating limited pesticidal potential.
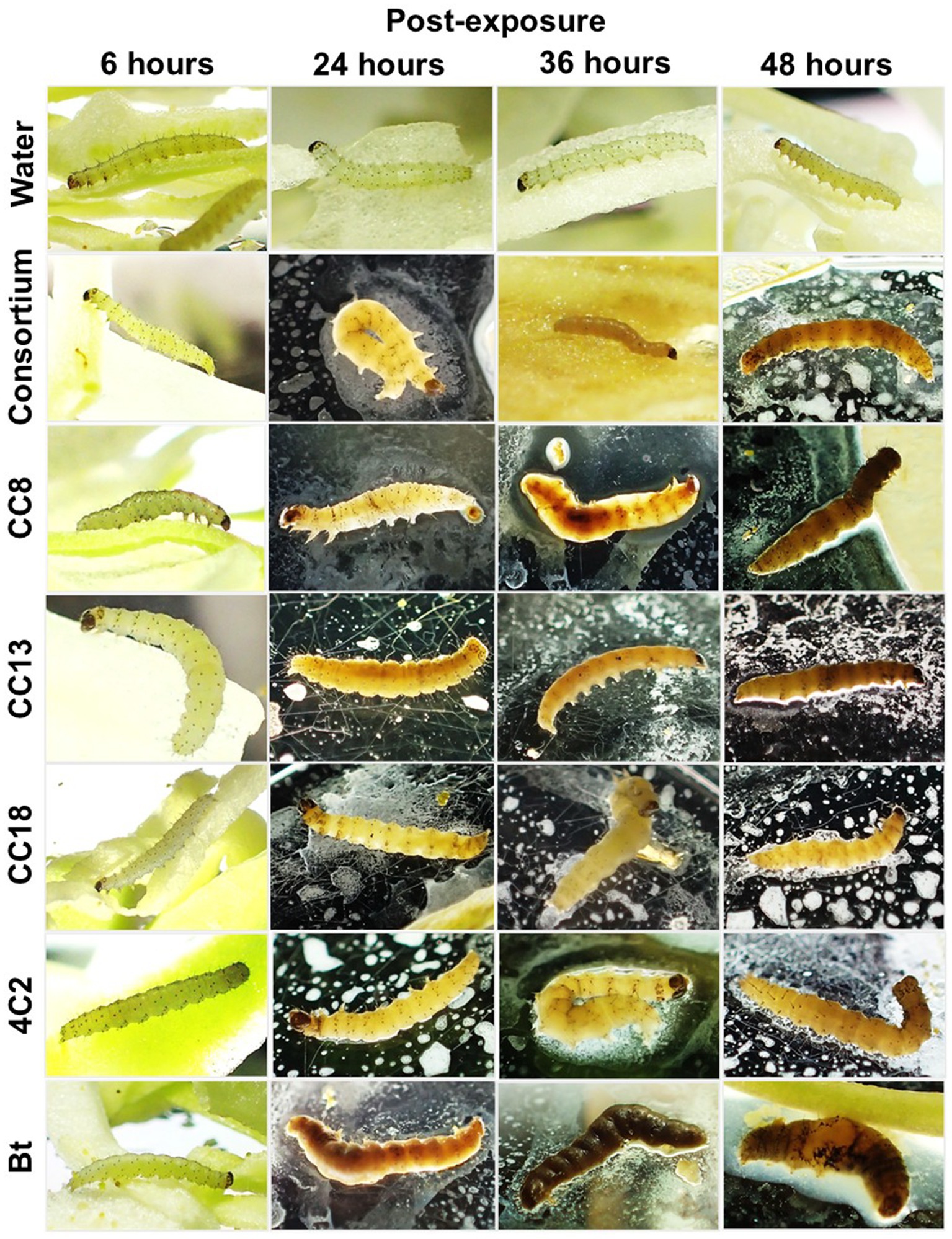
Figure 2. Plutella xylostella L. larvae at 6, 24, 36, and 48 h post-exposure to the selected isolated strains CC8, CC13, CC18, 4C2, and a consortium (a mixture of CC8, CC13, CC18, and 4C2). Bt served as a positive control. Sterile distilled water was used as a blank control.
Figure 3. Screening and evaluation of insecticidal activity of isolated strains against Plutella xylostella L. The isolated strains CC4, CC5, CC8, CC13, CC15, CC18, and 4C2 were tested. Bt served as a positive control. Escherichia col K-12 served as a negative control. Sterile distilled water was used as a blank control. The insecticidal activity was evaluated in triplicate for each strain, with standard deviations (SD) presented for each plot.
In contrast, M. thioparans strain CC8 had a slower onset, reaching 43% mortality at 36 h and 100% at 48 h, suggesting it may still be a viable biopesticidal agent. As a negative control, E. coli K-12 showed minimal mortality at 13% after 48 h, confirming the effectiveness of other tested strains. Sterile distilled water maintained 0% mortality, indicating no external pesticidal effects. Among our strains, B. subtilis CC18, M. thioparans CC8, B. mobilis CC13, and C. rhizoryzae 4C2 achieved complete mortality within 48 h. Strains such as A. junii CC15, S. maltophilia CC5, and M. mangrovi CC4 showed limited efficacy. Although CC5 and CC15 demonstrated hemolysis comparable to CC8 and 4C2, their insecticidal activity efficacy indicated the characteristics of the species. Further investigation is needed to explore specific metabolites and characterize the bioactive compounds responsible for the insecticidal effects. Future studies focusing on these aspects will enhance our mechanistic understanding of these non-Bt strains.
Strains CC18, CC8, CC13, and 4C2 demonstrated β-hemolysis (Figure 1B), indicating a link between hemolytic activity and the efficacy observed in the insecticidal bioassay (Figure 3). B. subtilis species has been reported to produce insecticidal metabolites (Abd El-Salam et al., 2011; Yang et al., 2017; Rocha et al., 2023), and the genus Bacillus is widely distributed in sustainable agriculture, making it one of the most commonly used insecticides (Villarreal-Delgado et al., 2018). Mesobacillus has been reclassified from Bacillus (Patel and Gupta, 2020), and the Bacillus species is most well-known for its insecticidal properties. The insecticidal potential of Mesobacillus is not as well established. However, our findings indicate that Mesobacillus species might have insecticidal properties. Chromobacterium is known for its production of insecticidal compounds (Blackburn et al., 2017; Tikhe et al., 2024).
3.4 Effect of cell concentration on insecticidal efficacy
Cell concentration significantly affected insecticidal efficacy, demonstrating a dose-dependent relationship with mortality rates. The findings highlight the importance of sufficient cell density for optimal effectiveness. The selected individual strains CC8, CC13, CC18, and 4C2 showed low mortality rates (less than 20%) 5 × 106 CFU/mL and 5 × 107 CFU/mL. However, they showed significantly higher mortality rates (50–57%) at 4 × 108 CFU/mL and achieved 100% mortality at 4 × 1010 CFU/mL. Interestingly, the consortium, mixed use of the single strains (CC8, CC13, CC18, and 4C2), caused 50% mortality at 5 × 106 CFU/mL, and increased mortality as the cell concentration increased, achieving 100% at 3 × 109 CFU/mL (Figure 4A). The insecticidal efficacy of the bacterial consortium was evaluated through a dose–response bioassay using cell concentrations ranging from 6.7 to 10.6 log10 (Dose) (5 × 106 to 4 × 1010 CFU/mL). Probit regression analysis revealed a significant dose-dependent increase in larval mortality, and the model demonstrated an adequate goodness-of-fit (χ2 = 1.182, df = 3, p = 0.757), indicating no significant deviation between observed and expected mortality rates. The estimated LC50 was 6.91 log10 (Dose) (approximately 8 × 106 CFU/mL) (Figure 4B), confirming the effectiveness of the bacterial consortium against P. xylostella L. larvae. The individual bacterial strains of the consortium exhibited β-hemolysis. The higher insecticidal performance of the bacterial consortium might indicate a synergistic hemolysis phenomenon (Gubash, 1978). Although the preliminary test on bacterial competition using nutrient broth (NB) agar plates indicated that the combination of strains CC8, CC13, CC18, and 4C2 did not inhibit one another, combining this with a CAMP-like test would provide further insight for constructing an insecticidal consortium. The recent study conducted by Devi et al. (2022) found that the mortality rate increased significantly in a dose-dependent manner. Furthermore, Gebremariam et al. (2021) conducted a similar study using various concentrations of spore-crystal Bt. The highest concentrations resulted in the highest mortality rates. Elevated concentrations of bacteria lead to increased insecticidal toxins that disrupt the larval midgut’s protective membrane, causing rapid damage, paralysis, and death (Zhu, 2008; Dahmana et al., 2020). High bacterial loads can compromise gut integrity, potentially causing leakage and septicemia, while lower concentrations might extend immune responses. The recent findings indicated that dose dependency is essential for non-Bt insecticidal activity, as higher bacterial concentrations increase larval mortality.
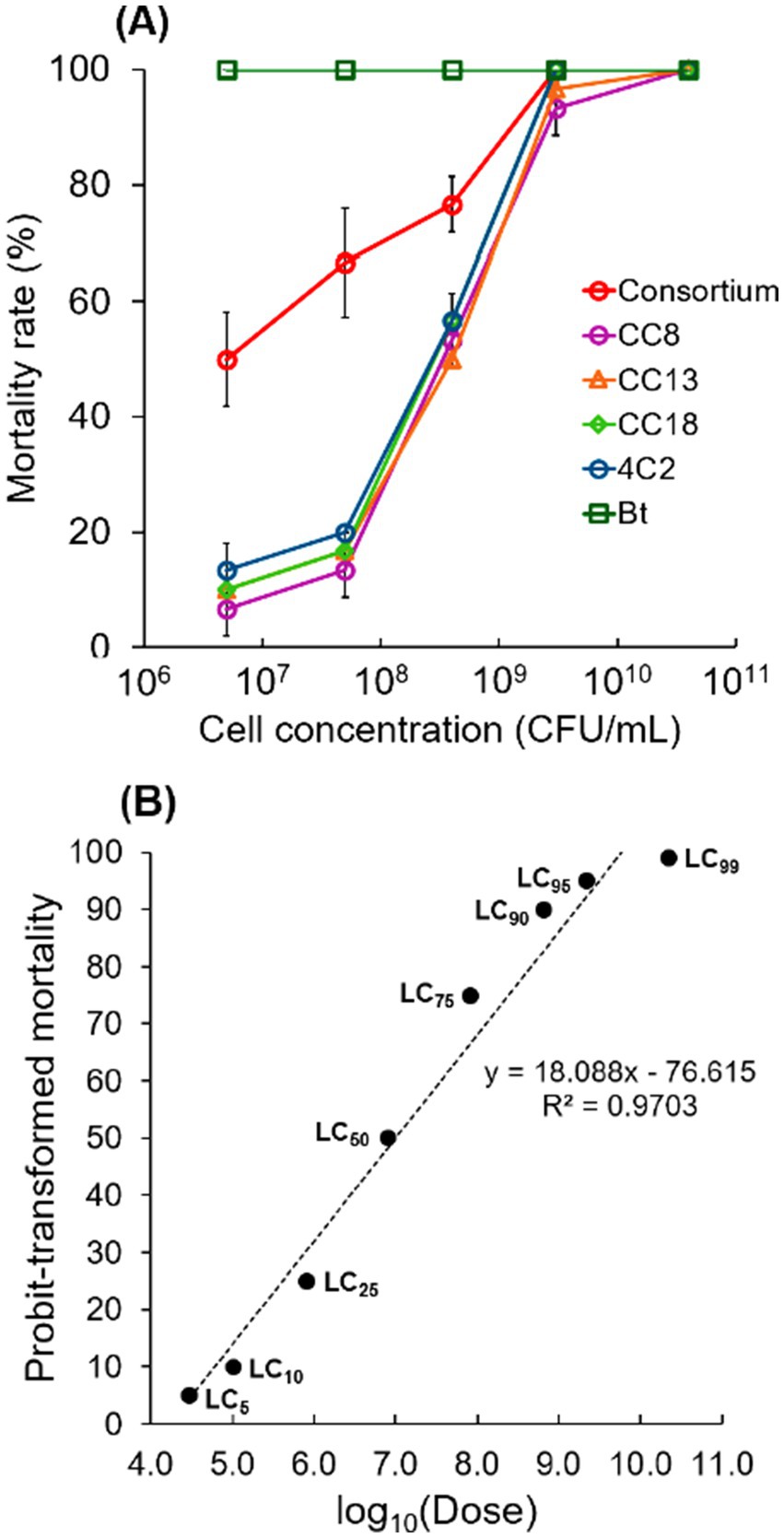
Figure 4. Effect of cell concentration on insecticidal efficacy. (A) The mortality rate was recorded after a 48-h exposure period. The selected isolated strains CC8, CC13, CC18, and 4C2, a consortium (a mixture of CC8, CC13, CC18, and 4C2), and Bt were tested. The effect of cell concentration on insecticidal efficacy was analyzed in triplicate for each strain. Each plot shows the standard deviation (SD). (B) Probit regression showing the relationship between log10-transformed bacterial dose (log10Dose) and probit-transformed larval mortality of Plutella xylostella L.
Furthermore, utilizing mixed insecticidal bacterial strains exposes larvae to multiple toxins, creating a synergistic effect that enhances mortality rates (Chang et al., 2024). Moreover, strain combinations can exhibit significant toxicity, resulting in larval death through their diverse mechanisms of action (Ma et al., 2023). These mechanisms include metabolic inhibition (Maharana et al., 2022), which involves interrupting or slowing cellular metabolic activities, and immune suppression (Xiao et al., 2023), which weakens the immune system, making it less capable of combating infections. Additionally, enzyme production has insecticidal properties (Choi et al., 2023), and bacteria produce insecticidal toxin diversity (Das et al., 2025). This study showed that cell concentration impacted insecticidal efficacy, while the combined use of individual strains in a consortium significantly enhanced insecticidal effectiveness compared to using the strains alone.
3.5 Effect of larval size on insecticidal efficacy
Larval size is critical in pest management, influencing feeding damage, resistance, and the timing of interventions. Larval size significantly impacted insecticidal efficacy. The mortality rates of second and fourth-instar P. xylostella L. larvae exposed to individual strains and the consortium were evaluated at 24, 36, and 48 h (Figure 5). After being exposed for 24 h, second-instar larvae demonstrated significantly higher mortality rates (37% for CC8, 43% for CC13, CC18 for 46, 46% for 4C2, and 60% for consortium) compared to fourth-instar larvae (3% for CC8, 3% for CC13, 7% for CC18, 10% for 4C2, and 23% for consortium). The mean differences were statistically significant at the 0.05 level. Bt caused 100% mortality in second-instar larvae at 24 h and 76% mortality in fourth-instar larvae. Second-instar larvae still continuously have maximum efficacy over 36 h of exposure. Interestingly, at 48 h of exposure, the bacterial consortium showed a significant difference from individual strains when fourth-instar larvae were used (Figure 5). Moreover, when the consortium was used to study the effect of cell concentration on insecticidal efficacy, the mortality rate was higher than that of individual strains when used at lower cell concentrations (Figure 4). These findings strongly suggest that the bacterial consortium exhibits enhanced efficacy compared to individual applications. After 48 h, all strains and the consortium achieved 100% mortality in second-instar larvae, with slight increases in fourth-instar larvae mortality rates (33% for CC8, 30% for CC13, CC18 for 23, 43% for 4C2, and 66% for consortium). Bt exhibited the highest efficacy, while consortium outperformed the individual strains. Second-instar larvae showed faster and higher mortality rates than fourth-instar larvae when exposed to the tested strains and consortium. Second-instar larvae, as early instar larvae, were reported to be more sensitive than fourth-instar larvae, highlighting their potential for pest management by targeting these early developmental stages of P. xylostella L. larvae.
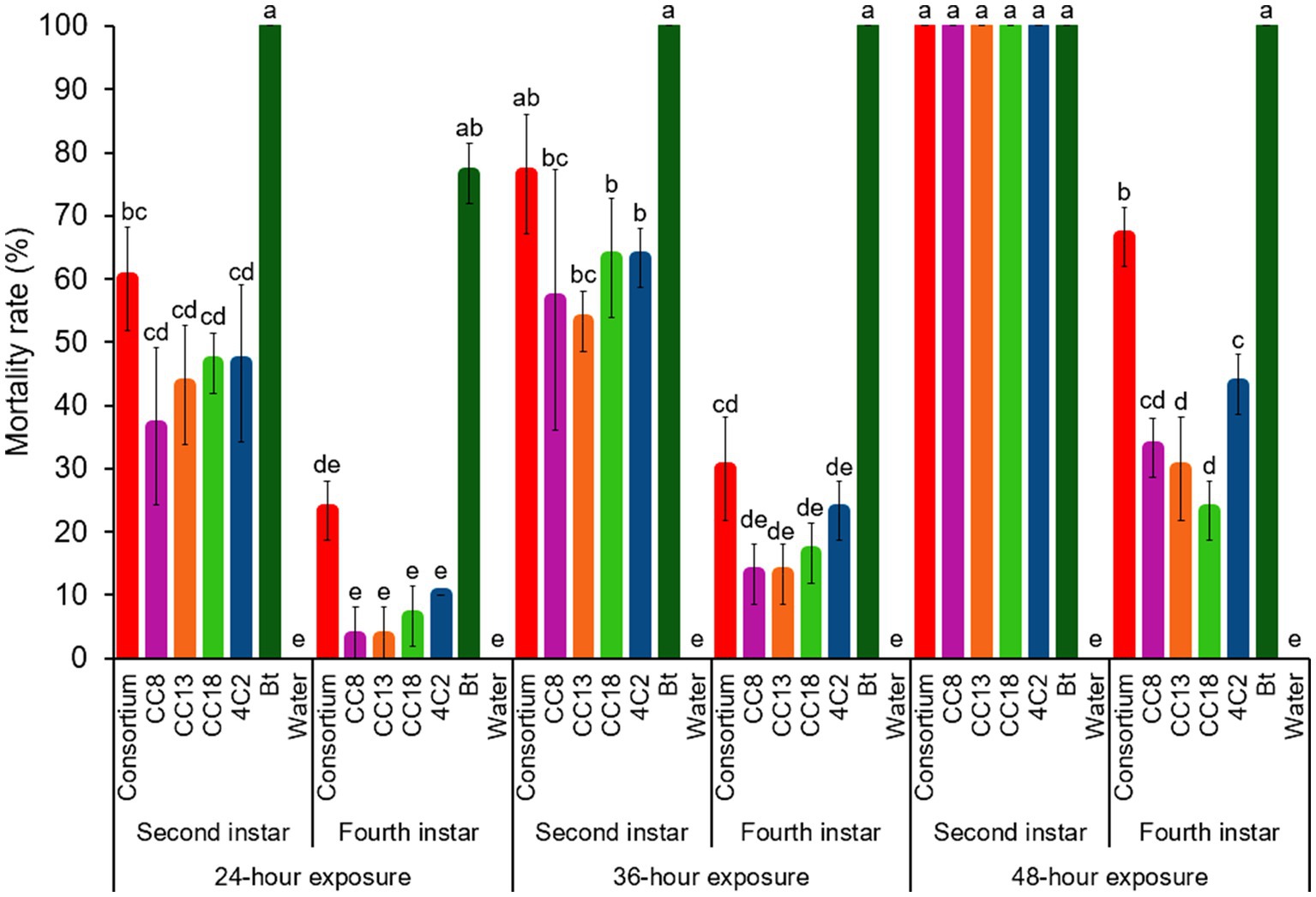
Figure 5. Insecticidal activity of isolated strains and a consortium against second instar and fourth instar larvae. The selected isolated strains CC8, CC13, CC18, and 4C2, a consortium (a mixture of CC8, CC13, CC18, and 4C2), and Bt were tested. Sterile distilled water was used as a blank control. Error bars represent standard deviations (SD) of triplicate experiments.
The findings suggested that second-instar larvae exhibit more rapid and higher mortality rates during the early stages of their development compared to fourth-instar larvae when exposed to the bacteria exhibiting insecticidal activity. This is likely attributed to the underdeveloped immune responses of the early-instar larvae, which are less effective in combating bacterial infections. Li et al. (2018) demonstrated that insecticidal bacteria can effectively control P. xylostella L. by suppressing its immune system. Xia et al. (2018) confirmed that gut bacteria play a critical role in the insecticide resistance of P. xylostella L. and may also affect the immune system. Previous studies have shown that the immune response in lepidopteran larvae is induced after they feed on insecticidal bacteria. This immune induction may result from localized cellular damage to the gut lining (Rahman et al., 2007). Furthermore, Rahman et al. (2007) remarked that the larvae’s tolerance to insecticidal properties, such as crystal endotoxins, correlates to an enhanced immune status in the larvae.
3.6 Microbial community of larvae feces
Analyzing microbial communities in larval feces offers valuable insights into gut health and the impact of insecticidal treatments. The microbial community of the feces of P. xylostella L. larvae revealed distinct composition and dominance patterns, as illustrated in Figure 6. In the feces of the larvae exposed to B. mobilis strain CC13, 33 microbial species were identified. The target strain B. mobilis was identified with a relative frequency of 42%. The result indicates that B. mobilis was dominated among 33 microbial species in the feces. In the feces of the larvae exposed to the B. subtilis strain CC18, B. subtilis was the most dominant species with a relative frequency of 43% among 30 microbial species. In the feces of larvae exposed to the M. thioparans strain CC8, M. thioparans was the most dominant species with a relative frequency of 45% among 31 microbial species. C. rhizoryzae showed a dominant relative frequency of 89% of the 20 identified species in the fecal microbiota of larvae exposed to the C. rhizoryzae strain 4C2. In the feces of the larvae exposed to B. thuringiensis (Bt), Bt was the most dominant species with a relative frequency of 80% among 24 microbial species. The larvae exposed to E. coli K-12, 32 microbial species were identified. The E. coli was identified with a relative frequency of 60%. The feces from the larvae exposed to distilled water contained 28 different species. The most prevalent was Enterobacteriaceae at 41%, followed by P. viridiflava at 16%, Carnobacterium at 14%, Weissella at 9%, Enterococcus at 6%, and Janthinobacterium at 3%. Enterobacteriaceae, P. viridiflava, Carnobacterium, Enterococcus, and Janthinobacterium were found in all feces from the larvae exposed to the individual bacterial strains.
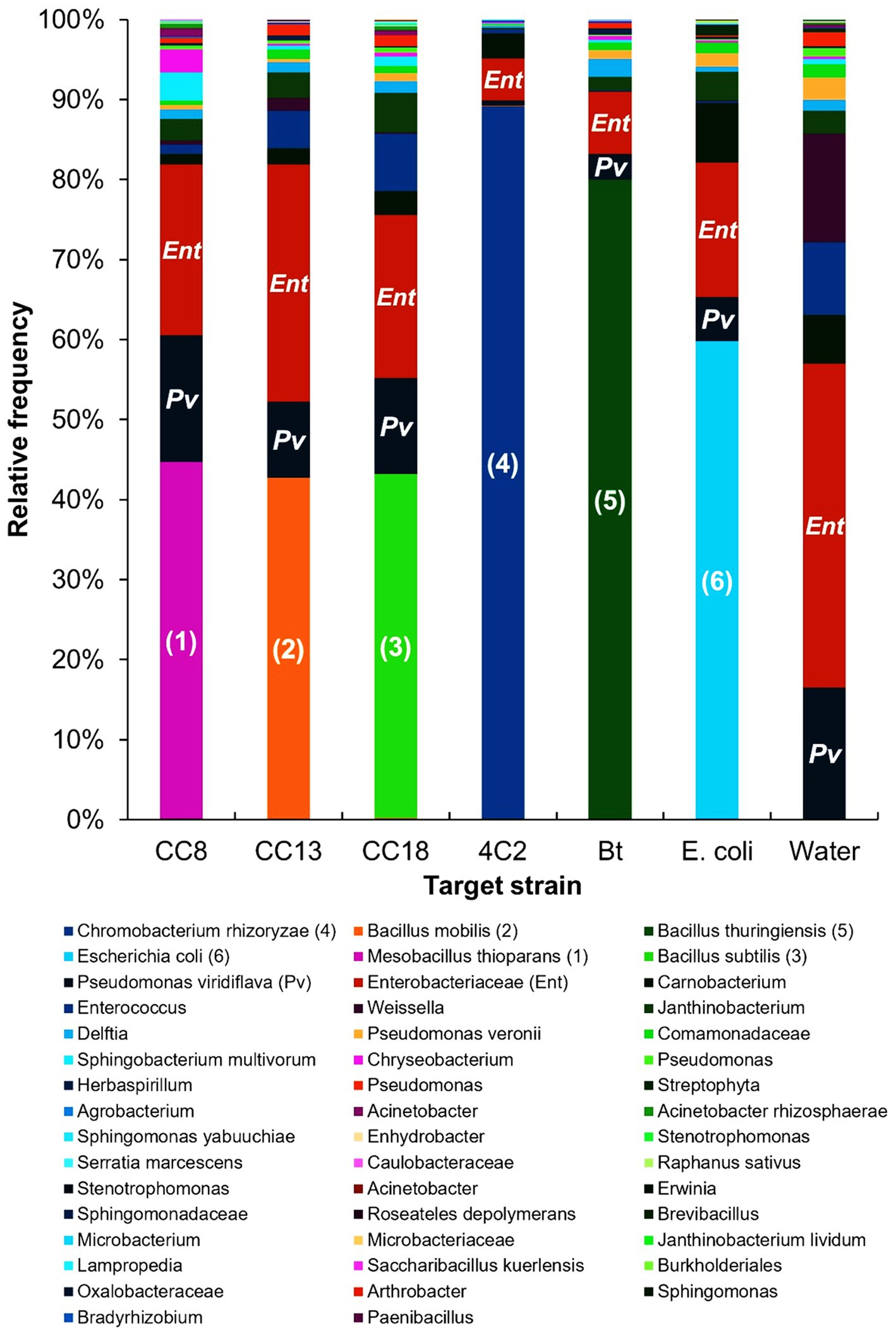
Figure 6. Microbial communities present in the feces of Plutella xylostella L. larvae, collected during the insecticidal activity assay. The target strains in the larval feces are indicated as follows: (1) Mesobacillus thioparans, (2) Bacillus mobilis, (3) Bacillus subtilis, (4) Chromobacterium rhizoryzae, (5) Bacillus thuringiensis, (6) Escherichia coli, (Pv) Pseudomonas viridiflava, and (Ent) Enterobacteriaceae.
Hammer et al. (2017) reported that resident gut microbiomes were very low in caterpillars, and microbes in the gut were leaf-derived, with the leaf-associated microbes reflected in the feces of caterpillars. The microbiomes in the gut and feces of larvae might be more strongly affected by leaf-associated microbes than those of caterpillars. Our analysis of larvae feces showed that the bacterial strains used in the leaf-dip method were predominant in the feces of P. xylostella L., indicating their abundance within the larvae’s gut. These bacterial strains were likely temporally abundant and not integrated with the resident microbiota. The fecal microbial community analysis indicates that the larvae consumed the leaf-dipped bacteria, which can affect the bacterial composition of the larvae gut. However, the presence of leaf-dipped bacteria in the feces does not necessarily indicate their activity in the larvae gut. Future research should focus on the correlation between larval death and the insecticidal activity of microbes in the larvae gut.
The gut’s functional bacteria and microflora in larval P. xylostella L. play important roles in food digestion, development, and environmental suitability. Analyzing the microbial community in feces can provide insights into the status of insecticide bacteria in the larvae’s gut. The study by Lu et al. (2023) indicated that feeding preferences can affect the gut microbiota of lepidopteran larvae. The analysis shows that target strains prevail in the gut of P. xylostella L. after larval exposure. Fecal microbiota analysis shows insecticidal bacteria that affect the larval gut microbiome, contributing valuable information for pest management.
3.7 SEM examination of deceased larval midgut
SEM analysis of the damaged midgut in deceased P. xylostella L. larvae revealed significant morphological differences between healthy and bacteria-exposed midguts. Larvae treated with sterile distilled water displayed smooth epithelial surfaces with intact microvilli (Figure 7A). In contrast, larvae exposed to the strains exhibiting insecticidal activity showed considerable epithelial damage, including cell detachment and degradation of the brush border microvilli (Figure 7B). Additionally, midgut tissue exhibited signs of cell collapse and membrane rupture (Figure 7C), structural erosion and pore formation (Figure 7D), tissue sloughing (Figures 7E,F), and irregular tissue morphology (Figure 7G), indicating cytotoxic activity.
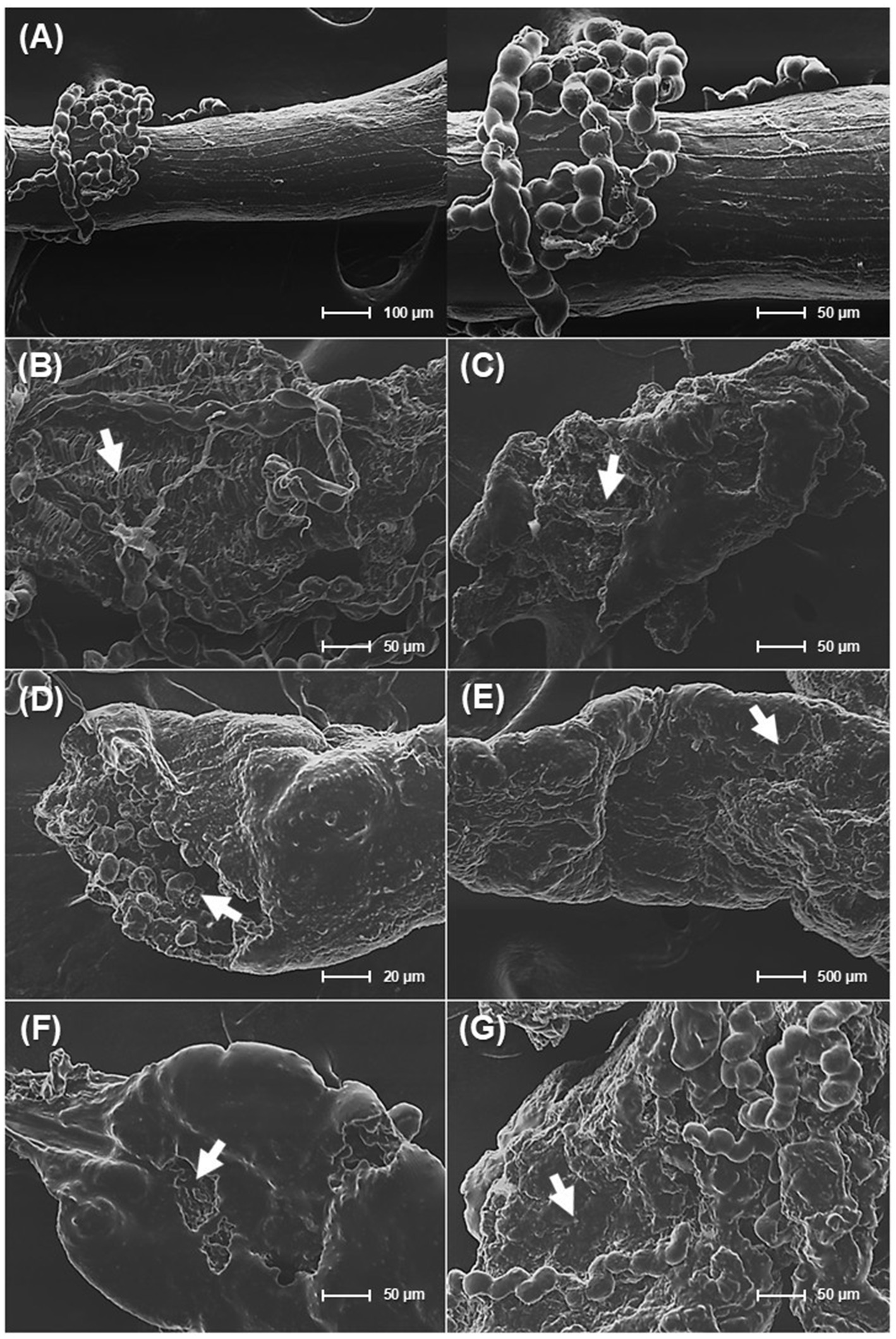
Figure 7. Scanning electron microscopy (SEM) analysis of the midgut epithelium in Plutella xylostella L. larvae. (A) healthy midgut tissue from larvae treated with sterile distilled water. (B–G) midgut tissues from larvae exposed to the selected isolated strains: (B) midgut tissues from larvae exposed to microbial consortium (a mixture of CC8, CC13, CC18, and 4C2), (C) midgut tissues from larvae exposed to Mesobacillus thioparans CC8, (D) midgut tissues from larvae exposed to Bacillus mobilis CC13, (E) midgut tissues from larvae exposed to Bacillus subtilis CC18, (F) midgut tissues from larvae exposed to Chromobacterium rhizoryzae 4C2, and (G) midgut tissues from larvae exposed to Bacillus thuringiensis (Bt).
The analysis of deceased P. xylostella L. larvae using SEM revealed gut epithelial damage compared to healthy larvae. This damage, characterized by membrane rupture and pore formation, suggests bacterial virulence factors like hydrolytic enzymes and secondary metabolites play a crucial role. Hydrolytic enzymes can dismantle midgut proteins and damage cell membranes, leading to cell lysis (Lopes et al., 2021). Additionally, secondary metabolites like biosurfactants can disrupt cell membranes, causing further disorganization of gut tissue (Khedher et al., 2015; Wang et al., 2024). These findings demonstrate that M. thioparans CC8, B. mobilis CC13, B. subtilis CC18, and C. rhizoryzae 4C2, along with a bacterial consortium, can damage midgut integrity, ultimately leading to larval death due to physical destruction and systemic toxicity.
3.8 Effect of exposure method on insecticidal efficacy
Two exposure methods, leaf dip and direct spraying, were evaluated for their insecticidal efficacy. Tukey’s HSD analysis indicated no significant differences in mortality rates (Figure 8). Using the dipping method, the strains M. thioparans CC8, B. mobilis CC13, B. subtilis CC18, and C. rhizoryzae 4C2 exhibited mortality rates from 16 to 50% after 24 h of exposure. In comparison, the direct spray method resulted in mortality rates between 13 and 53% after the same 24-h period. The mortality rates for the dipping method increased from 43 to 60% at 36 h, with complete mortality (100%) after 48 h of exposure. In contrast, the direct spray method increased mortality rates from 23 to 63% after 36 h, reaching 100% mortality by the 48-h exposure. After 48 h of exposure, both methods resulted in complete mortality (100%), with no significant differences observed. These results indicate that leaf dip and direct spray methods are interchangeable regarding insecticidal effectiveness, providing flexibility in method selection based on experimental needs.
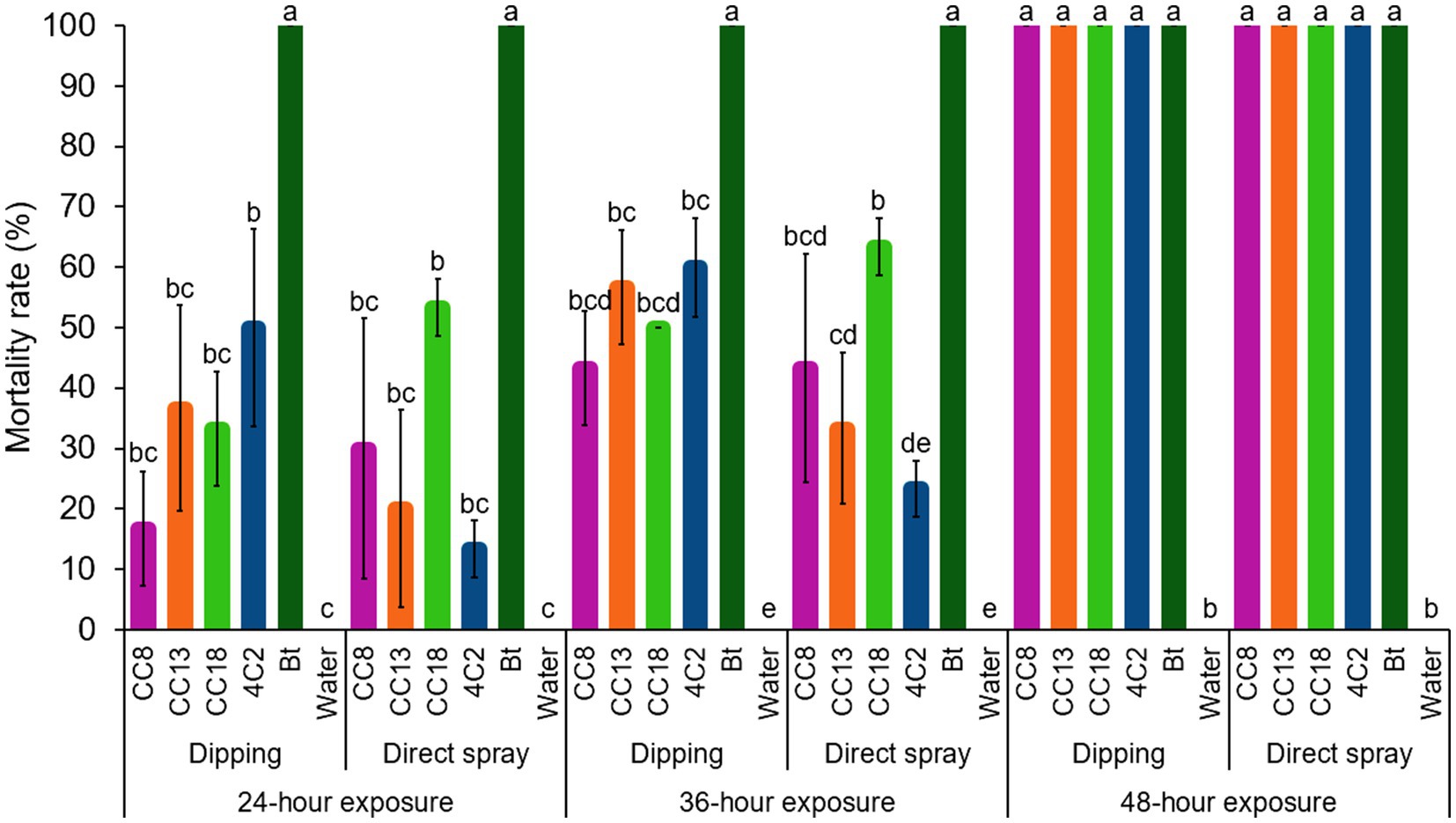
Figure 8. Effect of two bioassay methods on insecticidal efficacy, comparing the dipping method and the direct spraying method. The selected isolated strains CC8, CC13, CC18, 4C2, and Bt were tested. Sterile distilled water was used as a blank control. Error bars represent standard deviations (SD) of triplicate experiments.
Insecticide toxicity may be influenced by feeding behavior, insecticidal properties, exposure methods, the age of the larvae at the time of exposure, larval weight, detoxification enzymes in larvae, or even insecticide penetration (Abro et al., 2013). Similarly, different exposure methods of leaf dip and direct spraying can also affect the results (Zhao and Grafius, 1993; Abro et al., 2013). While both approaches evaluate pesticide contact toxicity, our understanding suggests that leaf-dip bioassays are one of the most common exposure methods used under laboratory conditions due to realistic feeding exposure, controlled application, simplicity, and repeatability. In comparison, direct spraying better represents practical application. Hence, leaf dipping is valuable in controlled lab studies. In contrast, spraying provides a more comprehensive evaluation of an insecticide’s efficacy and can be used to guide application strategies in integrated pest management.
4 Conclusion
The diamondback moth (P. xylostella L.) is a significant global pest of cruciferous vegetables and has resisted conventional insecticides, highlighting the need for new pest control strategies. Our four non-Bt bacterial strains, including M. thioparans strain CC8, B. mobilis strain CC13, B. subtilis strain CC18, and C. rhizoryzae strain 4C2, achieved 100% larval mortality within 48 h of exposure, while the created consortium of four non-Bt bacterial strains enhanced efficacy compared to individual strains. Managing second-instar larvae with precisely dose-dependent cell densities influenced insecticidal efficacy. After exposure, the target bacterial strains became dominant in the larvae’s fecal microbiota, and scanning electron microscopy revealed damage to the larvae’s gut. Utilizing leaf dip and direct spraying demonstrated comparable effectiveness, supporting the viability of various application strategies. This study enhances our understanding of non-Bt insecticidal bacteria and demonstrates the effectiveness of creating bacterial consortia for managing P. xylostella L., providing an alternative approach to improving pest management strategies.
Data availability statement
The nucleotide sequencing data presented in the study are deposited in the DNA Data Bank of Japan (DDBJ) repository, accession numbers LC868238 to LC868265. The data supporting the findings of this study are available from the authors upon reasonable request.
Author contributions
CC: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. GE: Investigation, Writing – review & editing. TI: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research project received support from the Ministry of Land, Infrastructure, Transport and Tourism (MLIT, Japan) through the initiative on “Research and Development of River and Erosion Control Technology.” Chanchao Chem was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan through the Japanese Government (MEXT) Scholarship Program (Scholarship Number: 232027).
Acknowledgments
The authors would like to thank Chen Wei for technical assistance with bacterial isolation. The surface damage of larval midgut was analyzed using a SEM at the Center for Instrumental Analysis of Gunma University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El-Salam, A. M. E., Nemat, A. M., and Magdy, A. (2011). Potency of Bacillus thuringiensis and Bacillus subtilis against the cotton leafworm, Spodoptera littoralis (Bosid.) larvae. Arch. Phytopathol. Plant Protect. 44, 204–215. doi: 10.1080/03235400902952129
Abro, G. H., Syed, T. S., Kalhoro, A. N., Sheikh, G. H., Awan, M. S., Jessar, R. D., et al. (2013). Insecticides for control of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae) in Pakistan and factors that affect their toxicity. Crop Prot. 52, 91–96. doi: 10.1016/j.cropro.2013.05.017
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arthropod Pesticide Resistance Database, Michigan State University. (2025). Available online at: http://www.pesticideresistance.org (Accessed on May 5, 2025).
Barbieri, G., Ferrari, C., Mamberti, S., Gabrieli, P., Castelli, M., Sassera, D., et al. (2021). Identification of a novel Brevibacillus laterosporus strain with insecticidal activity against Aedes albopictus larvae. Front. Microbiol. 12:624014. doi: 10.3389/fmicb.2021.624014
Barry, J. K., Simmons, C. R., and Nelson, M. E. (2023). Beyond Bacillus thuringiensis: new insecticidal proteins with potential applications in agriculture. Adv. Insect Physiol. 65, 185–233. doi: 10.1016/bs.aiip.2023.09.004
Belousova, M. E., Malovichko, Y. V., Shikov, A. E., Nizhnikov, A. A., and Antonets, K. S. (2021). Dissecting the environmental consequences of Bacillus thuringiensis application for natural ecosystems. Toxins 13:355. doi: 10.3390/toxins13050355597
Beltrán Pineda, M. E., and Castellanos-Rozo, J. (2025). Bacterial insecticides beyond Bacillus thuringiensis. Phytopathol. Res. 7:19. doi: 10.1186/s42483-024-00306-0
Blackburn, M. B., Farrar, R. R. Jr., Sparks, M. E., Kuhar, D., Mitchell, A., and Gundersen-Rindal, D. E. (2017). Chromobacterium sphagni sp. nov., an insecticidal bacterium isolated from Sphagnum bogs. Int. J. Syst. Evol. Microbiol. 67, 3417–3422. doi: 10.1099/ijsem.0.002127
Boone, R. L., Whitehead, B., Avery, T. M., Lu, J., Francis, J. D., Guevara, M. A., et al. (2021). Analysis of virulence phenotypes and antibiotic resistance in clinical strains of Acinetobacter baumannii isolated in Nashville, Tennessee. BMC Microbiol. 21, 1–12. doi: 10.1186/s12866-020-02082-1
Bozzola, J. J., and Russell, L. D. (1999). “Electron microscopy: principles and techniques for biologists” in Jones and Bartlett series in biology (Sudbury, Massachusetts: Jones & Bartlett Learning, United States), 48–71.
Bravo, A., López-Diaz, J. A., Yamamoto, T., Harding, K., Zhao, J. Z., Mendoza, G., et al. (2018). Susceptible and mCry3A resistant corn rootworm larvae killed by a non-hemolytic Bacillus thuringiensis Cyt1Aa mutant. Sci. Rep. 8:17805. doi: 10.1038/s41598-018-36205-6
Brillard, J., Ribeiro, C., Boemare, N., Brehélin, M., and Givaudan, A. (2001). Two distinct hemolytic activities in Xenorhabdus nematophila are active against immunocompetent insect cells. Appl. Environ. Microbiol. 67, 2515–2525. doi: 10.1128/AEM.67.6.2515-2525.2001
Calvin, W., and Palumbo, J. C. (2024). Chlorantraniliprole resistance associated with diamondback moth (Lepidoptera: Plutellidae) outbreaks in Arizona Brassica crops. J. Econ. Entomol. 117, 2608–2617. doi: 10.1093/jee/toae212
Chang, T. Y., Hsieh, C., and Wu, L. H. (2024). Synergistic insecticidal effect of Photorhabdus luminescens and Bacillus thuringiensis against fall armyworm (Spodoptera frugiperda). Agriculture 14:864. doi: 10.3390/agriculture14060864
Choi, S. I., Ajuna, H. B., Won, S. J., Choub, V., Kim, C. W., Moon, J. H., et al. (2023). The insecticidal potential of Bacillus velezensis CE 100 against Dasineura jujubifolia Jiao & Bu (Diptera: Cecidomyiidae) larvae infestation and its role in the enhancement of yield and fruit quality of jujube (Zizyphus jujuba Miller var. inermis Rehder). Crop Prot. 163:106098. doi: 10.1016/j.cropro.2022.106098
da Silva, W. J., Pilz-Júnior, H. L., Heermann, R., and da Silva, O. S. (2020). The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: a review. Parasit. Vectors 13, 376–314. doi: 10.1186/s13071-020-04236-6
Dahmana, H., Raoult, D., Fenollar, F., and Mediannikov, O. (2020). Insecticidal activity of bacteria from larvae breeding site with natural larvae mortality: screening of separated supernatant and pellet fractions. Pathogens. 9:486. doi: 10.3390/pathogens9060486
Das, K., Das, P., Nath, R. K., and Sharma, P. (2025). “Microbial insecticides and their potential use in insect pest management” in Advances in organic farming. eds. B. Sanchita, U. Vinod, K. N. Rupak, S. Ranjit, and P. D. Elangbam (Apple Academic Press), 565–580.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Devi, S., Saini, H. S., and Kaur, S. (2022). Insecticidal and growth inhibitory activity of gut microbes isolated from adults of Spodoptera litura (fab.). BMC Microbiol. 22:71. doi: 10.1186/s12866-022-02476-3
European Environment Agency (2023). How pesticides impact human health and ecosystems in Europe. Briefing no. 06/2023. doi: 10.2800/760240
Farias, E. S., Santos, A. A., Ribeiro, A. V., Carmo, D. G., Paes, J. S., and Picanço, M. C. (2020). Climate and host plants mediating seasonal dynamics and within-plant distribution of the diamondback moth (Plutella xylostella). Crop Prot. 134:105172. doi: 10.1016/j.cropro.2020.105172
Fathy, H. M., Awad, M., Alfuhaid, N. A., Ibrahim, E. D. S., Moustafa, M. A., and El-Zayat, A. S. (2024). Isolation and characterization of Bacillus strains from Egyptian mangroves: exploring their Endophytic potential in maize for biological control of Spodoptera frugiperda. Biology 13:1057. doi: 10.3390/biology13121057
Fenibo, E. O., Ijoma, G. N., and Matambo, T. (2022) in Biopesticides in sustainable agriculture: Current status and future prospects. New and future development in biopesticide research: Biotechnological exploration. eds. S. D. Mandal, G. Ramkumar, S. Karthi, and F. Jin (Singapore: Springer), 1–53.
Gebremariam, A., Chekol, Y., and Assefa, F. (2021). Isolation, characterization, and bio-insecticidal efficiency of Ethiopian isolates of Bacillus thuringiensis against galleria mellonella L. (Lepidoptera: Pyralidae) and tomato whitefly, Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae). Egypt. J. Biol. Pest Control. 31, 1–12. doi: 10.1186/s41938-021-00375-9
Ghribi, D., Elleuch, M., Abdelkefi, L., and Ellouze-Chaabouni, S. (2012). Evaluation of larvicidal potency of Bacillus subtilis SPB1 biosurfactant against Ephestia kuehniella (Lepidoptera: Pyralidae) larvae and influence of abiotic factors on its insecticidal activity. J. Stored Prod. Res. 48, 68–72. doi: 10.1016/j.jspr.2011.10.002
Gubash, S. M. (1978). Synergistic hemolysis phenomenon shown by an alpha-toxin-producing Clostridium perfingens and streptococcal CAMP factor in presumptive streptococcal grouping. J. Clin. Microbiol. 8, 480–488. doi: 10.1128/jcm.8.5.480-488.1978
Guo, L., Fatig, R. O., Gregory, L. O., Schafer, B. W., Strickland, J. A., Sukhapinda, K., et al. (1999). Photorhabdus luminescens W-14 insecticidal activity consists of at least two similar but distinct proteins: PURIFICATION AND CHARACTERIZATION OF TOXIN a AND TOXIN B. J. Biol. Chem. 274, 9836–9842. doi: 10.1074/jbc.274.14.9836
Hammer, T. J., Janzen, D. H., Hallwachs, W., Jaffe, S. P., and Fierer, N. (2017). Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA 114, 9641–9646. doi: 10.1073/pnas.1707186114
Han, X. Y., Han, F. S., and Segal, J. (2008). Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. Int. J. Syst. Evol. Microbiol. 58, 1398–1403. doi: 10.1099/ijs.0.64681-0
Hurst, M. R. H., Jones, S. A., Beattie, A., van Koten, C., Shelton, A. M., Collins, H. L., et al. (2019). Assessment of Yersinia entomophaga as a control agent of the diamondback moth Plutella xylostella. J. Invertebr. Pathol. 162, 19–25. doi: 10.1016/j.jip.2019.02.002
Ito, T., Yamanashi, Y., Noguchi, N., Miyazato, N., and Aoi, T. (2021). Microbial communities and nitrogen-utilizing bacteria of rotating biological contactors and activated sludge treating public sewage and night soil/johkasou sludge. J. Water Environ. Technol. 19, 109–119. doi: 10.2965/jwet.20-188
Khedher, S. B., Boukedi, H., Kilani-Feki, O., Chaib, I., Laarif, A., Abdelkefi-Mesrati, L., et al. (2015). Bacillus amyloliquefaciens AG1 biosurfactant: putative receptor diversity and histopathological effects on Tuta absoluta midgut. J. Invertebr. Pathol. 132, 42–47. doi: 10.1016/j.jip.2015.08.010
Kobisi, A. A., Imam, A. I., and Mansour, A. N. (2024). Biocontrol efficacy of native protease-producing bacterial isolates against the olive leaf moth, Palpita unionalis (Hübner) (Lepidoptera: Pyralidae). Egypt. J. Biol. Pest Control. 34:20. doi: 10.1186/s41938-024-00783-7
Li, S., Xu, X., Shakeel, M., Xu, J., Zheng, Z., Zheng, J., et al. (2018). Bacillus thuringiensis suppresses the humoral immune system to overcome defense mechanism of Plutella xylostella. Front. Physiol. 9:1478. doi: 10.3389/fphys.2018.01478
Liu, F. H., Lin, X. L., Kang, Z. W., Tian, H. G., and Liu, T. X. (2019). Isolation and characterization of Pseudomonas cedrina infecting Plutella xylostella (Lepidoptera: Plutellidae). Arch. Insect Biochem. Physiol. 102:e21593. doi: 10.1002/arch.21593
Lopes, F. C., Martinelli, A. H. S., John, E. B. O., and Ligabue-Braun, R. (2021). “Microbial hydrolytic enzymes: powerful weapons against insect pests” in Microbes for sustainable insect Pest management. eds. K. Aslam and W. Ahmad (Springer Cham), 1–31.
Lu, J., Su, X., Yang, Z., and Hu, P. (2023). The correlation between the gut microbiota of Endoclita signifer (Lepidoptera, Hepialidae) larvae and their host preferences. Insects. 14:919. doi: 10.3390/insects14120919
Ma, X., Hu, J., Ding, C., Portieles, R., Xu, H., Gao, J., et al. (2023). New native Bacillus thuringiensis strains induce high insecticidal action against Culex pipiens pallens larvae and adults. BMC Microbiol. 23:100. doi: 10.1186/s12866-023-02842-9
Maharana, C., Padala, V. K., Hubballi, A. B., Raj, M. N., Paschapur, A., Bhat, C., et al. (2022). “Secondary metabolites of Microbials as potential pesticides” in Sustainable Management of Potato Pests and Diseases. eds. S. K. Chakrabarti, S. Sharma, and M. A. Shah (Singapore: Springer), 111–142.
Mi, L., Gu, Z., Li, Y., Xu, W., Shu, C., Zhang, J., et al. (2023). Enterobacter strain IPPBiotE33 displays a synergistic effect with Bacillus thuringiensis Bt185. Int. J. Mol. Sci. 24:14193. doi: 10.3390/ijms241814193
Mnif, I., and Ghribi, D. (2015). Potential of bacterial derived biopesticides in pest management. Crop Prot. 77, 52–64. doi: 10.1016/j.cropro.2015.07.017
Mohammedi, S., Subramanian, S. B., Yan, S., Tyagi, R. D., and Valéro, J. R. (2006). Molecular screening of Bacillus thuringiensis strains from wastewater sludge for biopesticide production. Process Biochem. 41, 829–835. doi: 10.1016/j.procbio.2005.10.023
Mollah, M. M. I., Yeasmin, F., and Kim, Y. (2020). Benzylideneacetone and other phenylethylamide bacterial metabolites induce apoptosis to kill insects. J. Asia Pac. Entomol. 23, 449–457. doi: 10.1016/j.aspen.2020.03.008
Naimov, S., Boncheva, R., Karlova, R., Dukiandjiev, S., Minkov, I., and de Maagd, R. A. (2008). Solubilization, activation, and insecticidal activity of Bacillus thuringiensis serovar thompsoni HD542 crystal proteins. Appl. Environ. Microbiol. 74, 7145–7151. doi: 10.1128/AEM.00752-08
Nair, K., Al-Thani, R., Al-Thani, D., Al-Yafei, F., Ahmed, T., and Jaoua, S. (2018). Diversity of Bacillus thuringiensis strains from Qatar as shown by crystal morphology, δ-endotoxins and cry gene content. Front. Microbiol. 9:708. doi: 10.3389/fmicb.2018.00708
Nelly, N., Hamid, H., Lina, E. C., Yunisman, Y., Rusli, R., Yanti, Y., et al. (2024). Effectiveness of Bacillus spp. from West Sumatra, Indonesia in controlling Spodoptera frugiperda (Lepidoptera: Noctuidae). Biodiversitas 25, 1472–1478. doi: 10.13057/biodiv/d250415
Osouli, S., and Afsharmanesh, H. (2016). To evaluate the effects of secondary metabolites produced by Bacillus subtilis mutant M419 against Papilio demoleus L. and aspergillus flavus. Acta Ecol. Sin. 36, 492–496. doi: 10.1016/j.chnaes.2016.08.002
Patel, S., and Gupta, R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. Nov., Cytobacillus gen. Nov., Mesobacillus gen. Nov., Neobacillus gen. Nov., Metabacillus gen. Nov. and Alkalihalobacillus gen. Nov. Int. J. Syst. Evol. Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Rahman, M. M., Roberts, H. L., and Schmidt, O. (2007). Tolerance to Bacillus thuringiensis endotoxin in immune-suppressed larvae of the flour moth Ephestia kuehniella. J. Invertebr. Pathol. 96, 125–132. doi: 10.1016/j.jip.2007.03.018
Rocha, G. T., Queiroz, P. R. M., Grynberg, P., Togawa, R. C., de Lima Ferreira, A. D. C., Do Nascimento, I. N., et al. (2023). Biocontrol potential of bacteria belonging to the Bacillus subtilis group against pests and diseases of agricultural interest through genome exploration. Antonie Van Leeuwenhoek 116, 599–614. doi: 10.1007/s10482-023-01822-3
Röske, K., Sachse, R., Scheerer, C., and Röske, I. (2012). Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany). Syst. Appl. Microbiol. 35, 35–44. doi: 10.1016/j.syapm.2011.09.002
Shehzad, M., Bodlah, I., Siddiqui, J. A., Bodlah, M. A., Fareen, A. G. E., and Islam, W. (2023). Recent insights into pesticide resistance mechanisms in Plutella xylostella and possible management strategies. Environ. Sci. Pollut. Res. 30, 95296–95311. doi: 10.1007/s11356-023-29271-5
Srujana, Y., Hugar, P. S., and Krishnaraj, P. U. (2022). Exploring the entomotoxic ability of actinomycetes against diamondback moth, Plutella xylostella L. (Lepidoptera: Plutellidae). Int. J. Trop. Insect Sci. 42, 2559–2566. doi: 10.1007/s42690-022-00783-w
Susetyo, R. D., Geraldi, A., Supriyanto, A., Nurhariyat, T., and Nafidiastri, F. A. (2023). Biosurfactant production of entomopathogenic Bacillus subtilis BK7. 1, as potential biocontrol bacteria, isolated from Baluran National Park, East Java, Indonesia. Biodiversitas 24, 1785–1792. doi: 10.13057/biodiv/d240342
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T., and Nishijima, M. (2014). Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592
Thakur, N., Kaur, S., Tomar, P., Thakur, S., and Yadav, A. N. (2020). “Microbial biopesticides: current status and advancement for sustainable agriculture and environment” in New and future developments in microbial biotechnology and bioengineering. eds. A. A. Rastegari, et al. (Elsevier), 243–282.
Tikhe, C. V., Issiaka, S., Dong, Y., Kefi, M., Tavadia, M., Bilgo, E., et al. (2024). Chromobacterium biopesticide overcomes insecticide resistance in malaria vector mosquitoes. Sci. Adv. 10:eads3658. doi: 10.1126/sciadv.ads3658
Villarreal-Delgado, M. F., Villa-Rodríguez, E. D., Cira-Chávez, L. A., Estrada-Alvarado, M. I., Parra-Cota, F. I., and Santos-Villalobos, S. D. L. (2018). The genus Bacillus as a biological control agent and its implications in the agricultural biosecurity. Rev. Mex. Fitopatol. 36, 95–130. doi: 10.18781/r.mex.fit.1706-5
Wang, X., An, J., Cao, T., Guo, M., and Han, F. (2024). Application of biosurfactants in medical sciences. Molecules 29:2606. doi: 10.3390/molecules29112606
Wu, Y., Gao, M., Dai, S., Yi, D., and Fan, H. (2008). Investigation of the cyt gene in Bacillus thuringiensis and the biological activities of Bt isolates from the soil of China. Biol. Control 47, 335–339. doi: 10.1016/j.biocontrol.2008.08.020
Xia, X., Sun, B., Gurr, G. M., Vasseur, L., Xue, M., and You, M. (2018). Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L.). Front. Microbiol. 9:25. doi: 10.3389/fmicb.2018.00025
Xiao, Z., Yao, X., Bai, S., Wei, J., and An, S. (2023). Involvement of an enhanced immunity mechanism in the resistance to Bacillus thuringiensis in lepidopteran pests. Insects. 14:151. doi: 10.3390/insects14020151
Yang, S. Y., Lim, D. J., Noh, M. Y., Kim, J. C., Kim, Y. C., and Kim, I. S. (2017). Characterization of biosurfactants as insecticidal metabolites produced by Bacillus subtilis Y9. Entomol. Res. 47, 55–59. doi: 10.1111/1748-5967.12200
Zalucki, M. P., Shabbir, A., Silva, R., Adamson, D., Shu-Sheng, L., and Furlong, M. J. (2012). Estimating the economic cost of one of the world's major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): just how long is a piece of string? J. Econ. Entomol. 105, 1115–1129. doi: 10.1603/EC12107
Zhao, J. Z., and Grafius, E. (1993). Assessment of different bioassay techniques for resistance monitoring in the diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 86, 995–1000. doi: 10.1093/jee/86.4.995
Keywords: bacterial consortium, biopesticides, dam sediment, insecticidal bacteria, wastewater treatment sludge
Citation: Chem C, Eslamloo G and Ito T (2025) Insecticidal efficacy of non-Bt bacterial strains against diamondback moth, Plutella xylostella (L.). Front. Sustain. Food Syst. 9:1591114. doi: 10.3389/fsufs.2025.1591114
Edited by:
Emmanuel Oliver Fenibo, Hensard University, NigeriaReviewed by:
Li-Hsin Wu, National Pingtung University of Science and Technology, TaiwanPravara S. Rupawate, Sangamner Nagarpalika Arts, D. J. Malpani Commerce and B. N. Sarda Science Autonomous College, India
Ciro Pedro Guidotti Pinto, São Paulo State University, Brazil
Jianyang Bai, Anhui Agricultural University, China
Copyright © 2025 Chem, Eslamloo and Ito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsukasa Ito, dC5pdG9AZ3VubWEtdS5hYy5qcA==
 Chanchao Chem
Chanchao Chem Ghazaleh Eslamloo
Ghazaleh Eslamloo Tsukasa Ito
Tsukasa Ito