- 1Department of Agriculture and Animal Health, University of South Africa, Florida, South Africa
- 2Agricultural Research Council - Grain Crops Institute, Potchefstroom, South Africa
- 3Department of Plant and Soil Sciences, Faculty of Science, Engineering and Agriculture, University of Venda, Thohoyandou, South Africa
Finger millet is a climate-resilient cereal notable for its exceptional nutritional properties, yet it remains underutilized mainly because of its narrow genetic diversity and slow breeding progress. The loss of genetic variation from prolonged directional selection further hampers the development of improved cultivars suited to changing climatic conditions. Mutation breeding, through the use of physical and chemical mutagens has demonstrated efficacy in generating novel genetic diversity and enhancing desirable traits in finger millet. This review highlights (i) the role of mutation breeding in expanding genetic variability with emphasis on improving grain yield and nutrient composition; (ii) mutagenesis techniques and their application in developing elite mutant lines; and (iii) integration of mutation breeding with advanced omics technologies for efficient screening and target trait selection. Combining mutation breeding with advanced molecular approaches provides a strategic pathway to accelerate the development of high-yielding and nutrient-rich finger millet mutants.
1 Introduction
Finger millet (Eleusine coracana (L.) Gaertn, 2n = 4x = 36, AABB) is an allotetraploid cereal crop with two distinct subspecies: subsp. africana (wild finger millet) and subsp. coracana (cultivated finger millet) (Hilu, 1994; Admasu and Belete, 2020). The A genome originated from a diploid progenitor related to wild Eleusine indica (genome donor), while the B genome progenitor remains unknown or extinct species resembling Eleusine floccifolia (Hatakeyama et al., 2018; Kamenya et al., 2021). The modern finger millet (E. coracana subsp. coracana) is believed to be domesticated from wild finger millet (E. coracaca subsp. africana) populations about 5,000 years ago in western Uganda and Ethiopian highlands (Hittalmani et al., 2017). This long domestication history, combined with finger millet's polyploid genome, has contributed to its high adaptability to drought-prone and marginal environments. Finger millet grains are rich in essential nutrients, including calcium, iron, and dietary fiber, and the crop is also known for its long shelf life (Ramesh et al., 2019); (Rathore et al., 2019). In sub-Saharan Africa and Asia, finger millet is cultivated by smallholder farmers for household consumption, with surplus grain sold in local and regional markets (Gebreyohannes et al., 2024). The crop holds strong niche market potential due to its increasing popularity among both rural and urban populations, driven by its nutritional benefits and perceived food quality (Gebreyohannes et al., 2021; Ramashia et al., 2018).
Despite its significance, finger millet remains underutilized when compared to major cereals like wheat (Triticum aestivum) and maize (Zea mays), mainly due to limited genetic research and narrow genetic diversity (Rebecca et al., 2018; Wright and Devos, 2024). Over the years, the genetic diversity of the crop has declined as traditional landraces have been progressively replaced by a narrow range of high-yielding cultivars. This shift, coupled with repeated directional selection, has significantly reduced the genetic variability within cultivated gene pools, leading to a lower mean yield of < 1.0 t/ha (Govindaraj et al., 2015). Therefore, broadening the genetic base of finger millet is essential to enhance its breeding potential and achieve sustainable productivity through the development of improved varieties.
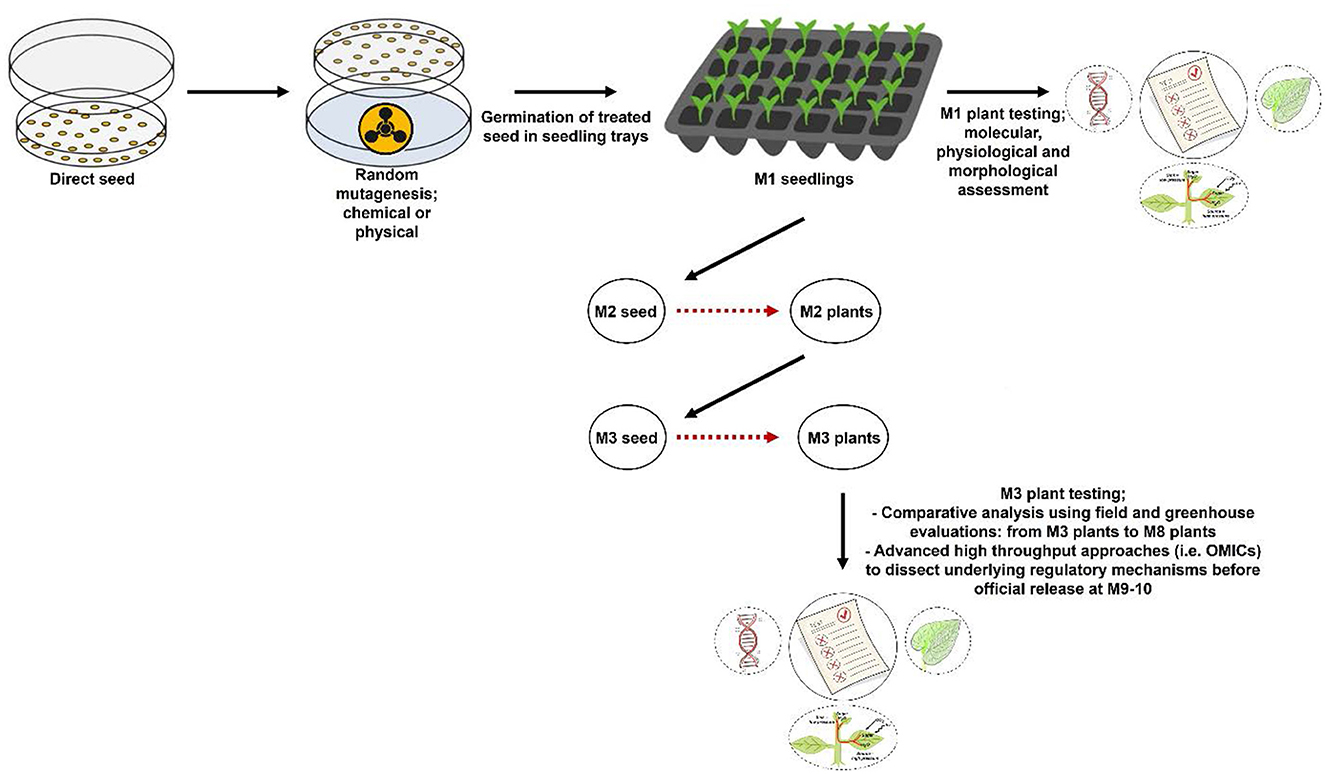
Various strategies have been employed to generate and harness genetic variation. Conventional approaches and biotechnological techniques have played a significant role in this regard (Kumar et al., 2017). In conventional breeding, genetic variation is harnessed through crosses with divergent and complementary genetic backgrounds (Martínez-Fortún et al., 2022). However, conventional breeding takes a long time before genetically distinct, uniform and stable varieties are developed and released (Lamichhane and Thapa, 2022). This creates a critical bottleneck for cultivar development under a rapidly changing environment. Therefore, mutation breeding could revolutionize finger millet breeding by creating genetic variation and enabling the development of superior finger millet varieties in a short period using the workflow presented in Figure 1.
Mutation breeding is a plant improvement technique that involves the artificial induction of genetic mutations using physical or chemical mutagens to generate novel and beneficial traits (Oladosu et al., 2015; Thangwana et al., 2021). Physical mutagens (such as gamma rays and X-rays) induce DNA damage through chromosomal breaks or rearrangements, while chemical mutagens [including ethyl methanesulfonate (EMS)] cause point mutations by alkylating DNA bases (Türkoglu et al., 2022; Sawarkar and Mansoori, 2023). These mutations can lead to changes in gene function, protein activity and downstream metabolic pathways. This approach has been successfully applied in other crops such as wheat, sorghum and rice to improve yield potential and other desirable traits (Kalpande et al., 2021; Shamshad et al., 2023). Therefore, induced mutations can enhance attributes in finger millet such as grain yield, grain size, tillering ability and grain nutrient content (Waghmode et al., 2020; Ganapathy et al., 2021; Sellapillai et al., 2022). These traits contribute to the crop's productivity and adoption in different agro-climatic conditions.
The genotypes developed through mutation breeding require efficient screening to identify superior mutant lines (Supplementary Figure 1). Various phenotyping approaches, such as conducting field, greenhouse and laboratory experiments, have been utilized to identify high-yielding finger millet mutant lines (Ganapathy et al., 2021; Sellapillai et al., 2022; Nair and Devi, 2024). In recent years, the high-throughput phenotyping tools, including liquid chromatography-mass spectrometry, satellite-imaging and unmanned aerial vehicles, have been widely used to identify key agro-morphological traits, proteins and metabolites (Chawade et al., 2019; Hall et al., 2022), associated with improved grain yield and grain nutrient content.
Building on these advancements, the integration of phenotyping with omics technologies has emerged as a powerful strategy to dissect the complex genetic and biochemical basis of key traits. The traits such as grain yield and grain nutrient composition in finger millet are complex traits controlled by multiple genes and environmental conditions (Sharma et al., 2018; Singhal et al., 2021). Recent studies suggest that advanced molecular approaches, including genomics, proteomics and metabolomics, can help to evaluate the molecular mechanisms underlying these traits in cereal crop cultivars (Broyart et al., 2009; Khakimov et al., 2017; Bernardi et al., 2019). Hence, the link between grain yield, grain nutrient content, genomic, proteomic and metabolic background are vital to develop superior finger millet mutant lines. Protein and metabolite profiling provide insights into how mutations influence grain composition and plant physiology. Moreover, correlation and path coefficient analysis are commonly utilized to identify direct and indirect relationships between agronomic traits and grain nutrient content, providing insights into trait contribution toward improving yield and nutritional quality. This association will help finger millet breeders pinpoint key traits contributing to high grain yield and grain nutrient content.
Mutation breeding holds significant promise as an underutilized strategy to enhance both productivity and nutritional quality in finger millet by creating novel genetic diversity and enabling selection of superior lines (Zuge et al., 2017; Waghmode et al., 2020). This breeding strategy has a long history in crop improvement starting from 300BC, with the first natural mutant plant in cereals found over 2,317 years ago in China (Kharkwal, 2023) and its modern use expanded significantly in the mid-twentieth century through induced mutagenesis (Hamdan and Tan, 2024). Nonetheless, the prominence of mutation breeding declined after the 1990s, due to the emergence of more targeted genetic technologies and a shift in focus to adopt these technologies (Mohan Jain and Suprasanna, 2011) such as transgenic approaches, which offered greater precision and control. Despite these advances, finger millet remains an under-researched crop, with limited genomic resources and little systematic application of mutation breeding compared to major cereals. This gap highlights the need for a focused synthesis that highlights how mutation breeding, when integrated with modern omics technologies, can accelerate the development of nutrient-dense and high-yielding finger millet varieties. Therefore, this review aimed to present the highlights on (i) the role of mutation breeding in enhancing genetic variability in finger millet, with a focus on improving grain yield and nutrient composition; (ii) the techniques employed in mutagenesis, their application in developing elite mutant lines; and (iii) the integration of mutation breeding with advanced omics technologies and approaches for efficient screening and trait selection.
2 Genetic variation in finger millet
Genetic variation refers to heritable differences in DNA sequences among individuals of the same species and forms the basis for phenotypic, proteomic and metabolic diversity in crop improvement (Fernie et al., 2006; Dwiningsih and Alkahtani, 2022). It supports the expression of key agronomic and grain nutrient traits (Harrigan et al., 2007; Dwiningsih and Alkahtani, 2022). However, the genetic variation in finger millet is relatively narrow due to its predominantly self-pollinating nature (Nagaraja et al., 2022) and limited exploitation of wild relatives in breeding programs. Although global germplasm collections are maintained in several countries (such as 10 507 accessions at ICAR-National Bureau of Plant Genetic Resources in India, 6,804 at International Crops Research Institute for the Semi-Arid Tropics in India, 6,257 at All India Coordinated Small Millets Improvement Project in Bengaluru, 2,156 at Ethiopian Biodiversity Institute in Ethiopia, 2,875 at the Kenya Agricultural and Livestock Research Organization in Kenya, and smaller collections in the United State of America, Uganda, Zambia, Nepal, and Japan), they are mainly landraces and indigenous materials with modest genetic diversity (Joshi et al., 2021). This limited variability constrains breeding progress for yield stability and nutritional traits. Mutation breeding, among other breeding approaches, has been applied to induce new variability. For instance, (Zuge et al. 2017) and (Waghmode et al. 2020), demonstrated that mutation induction can create genetic variation, enhance grain calcium and iron concentrations in selected mutant lines, which directly contribute to nutritional improvement in finger millet. Recent advances in molecular markers and genomics can further facilitate the characterization of induced variants, thereby broadening the genetic pool and supporting crop improvement efforts (Broyart et al., 2009; Khalil et al., 2018; Francis et al., 2023).
2.1 Sources of genetic variation in finger millet
Genetic variation in cereals arises mainly from genetic recombination, gene transfer, and both natural and induced mutations (Yali and Mitiku, 2022; Salgotra and Chauhan, 2023). Spontaneous mutations occur at low frequencies and often fail to produce the level of diversity needed for improving desirable traits (Yali and Mitiku, 2022). However, induced mutations offer a faster and more effective way to expand the genetic pool. These mutations can be achieved through chemical or physical agents. The physical and chemical agents alter the DNA and introduce new genetic variations (Sharma et al., 2024). The genetic recombination and crossing over during sexual reproduction contribute to genetic diversity by shuffling alleles between parental lines (Danguy des Déserts et al., 2021). Recombination plays a crucial role in finger millet breeding where wide genetic variation is essential for enhancing traits such as yield and grain quality. Furthermore, genetic variation influences both qualitative and quantitative traits. Qualitative traits are usually controlled by one or a few genes (Serpico, 2020) and exhibit clear phenotypic categories that are inherited in Mendelian ratios. In contrast, quantitative traits such as plant height and grain yield, are influenced by multiple genes, proteins and metabolites (Li et al., 2025; Zhang et al., 2021), resulting in a continuous range of phenotypes. Mutation breeding has become a useful method for introducing new genetic variation that does not occur naturally in populations (Hamdan and Tan, 2024). It is especially helpful in improving the agronomic and nutritional qualities of finger millet (Pandit et al., 2021; Tripathi et al., 2025). Therefore, combining genetic recombination with mutation breeding provides a strong approach for developing mutants with superior traits.
3 Finger millet mutation breeding
Mutation breeding is an effective method for creating genetic variation in finger millet within a short period (Bolbhat and Thikekar Chaitali, 2020). It is beneficial when continuous hybridization through conventional breeding is used to reduce the risks of genetic drift. Induced mutagenesis enables the development of novel traits by altering the plant's genetic makeup, leading to enhanced agronomic performance. Mutations may result in chromosomal changes such as inversions, translocations, duplications, deletions, or gene substitutions (Ramesh et al., 2019). These mutations are generally classified into two categories: micro-mutations, which involve subtle genetic changes that are not visible phenotypically and macro-mutations, which cause clear morphological alterations. Both have contributed to the development of improved cultivars in several cereal crops, including finger millet (Animasaun and Oguntoye, 2024; Mohanta et al., 2025).
Artificial mutagenesis enhances genetic variation that would otherwise occur in nature at very low frequencies to be fully exploited for breeding purposes (Jain, 2010). The methods utilized include physical and chemical mutagenesis to increase the frequency of mutations, which depends on the nature of the plant part mutated (Alemu, 2016). Each method has been used in numerous instances with relative success. There is variable information on mutation treatment conditions for many cereal crops such as wheat, maize, finger millet and rice, which have been widely investigated (Bado et al., 2023; Chaudhary and Kumar, 2023; Animasaun and Oguntoye, 2024; Mohanta et al., 2025). However, the treatment conditions still need to be optimized to increase mutation frequency and reduce losses. The success rates and treatment conditions reported by different researchers show that the resultant mutations are unpredictable and are specific to environmental conditions (Elena and de Visser, 2003). Therefore, there is a need to determine the most effective method between physical and chemical mutagenesis in line with the objectives of the crop breeding program. Both physical and chemical mutagens have been used successfully to create variation and develop new finger millet cultivars with improved yield potential (Bolbhat and Thikekar Chaitali, 2020; Sellapillai et al., 2022).
3.1 Physical mutagens
Physical mutagenesis involves exposing plant materials to ionizing radiation, such as gamma rays, X-rays, neutrons and charged particles, which causes direct DNA damage and induces heritable mutations (Bado et al., 2023). Among these, gamma rays are the most commonly used due to their strong tissue penetration and ability to cause a broad spectrum of mutations, including point mutations, chromosomal rearrangements, and gene deletions (Bharat et al., 2024). Accurate determination of the lethal dose is crucial for repeatable, large-scale trials (Oladosu et al., 2015). Gamma irradiation accounts for over 60% of registered mutant cereal varieties globally (Ghanim, 2023), highlighting its significance in breeding programs. Several studies have demonstrated the effectiveness of gamma irradiation in generating useful mutations. For instance, (Ambavane et al. 2015) and (Ganapathy et al. 2021) reported that gamma radiation induced mutants with enhanced yield and yield-related traits in finger millet under field conditions. Other physical mutagens include x-rays, neutrons, beta and alpha particles, protons and ion beams; they vary in source, penetration and hazard level (Animasaun and Oguntoye, 2024). For example, while X-rays are electromagnetic like gamma rays, they have shallower penetration. Neutrons and ion beams are more hazardous and require advanced safety infrastructure, while alpha and beta particles are less penetrating but highly ionizing. Such diversity in physical mutagens allows breeders to select specific tools based on crop type, desired mutation spectrum, and facility availability. Despite their potential, the use of physical mutagens is limited in sub-Saharan Africa due to the high operation costs, radiation safety regulations and the lack of specialized laboratory facilities (Olaolorun, 2020). Therefore, there is a pressing need to invest in accessible and safe technologies for physical mutation breeding to support crop improvement programs in developing regions.
3.2 Chemical mutagens
Initially, mutation breeding was predominantly based on physical mutagens. However, advancements in the field led to the discovery and increasing use of chemical mutagens (Supplementary Figure 2), which significantly expanded the scope and efficiency of induced mutagenesis. (Auerbach and Robson 1946) were among the first to provide a detailed account of chemical mutagens, reporting that dichloro-diethyl-sulphide or mustard gas could induce mutations. Since then, several chemical agents summarized by (Animasaun and Oguntoye 2024), have demonstrated mutagenic potency comparable to that of physical mutagens. Among these, EMS and Methyl Methanesulfonate (MMS) are widely employed due to their high mutagenic efficiency, ability to induce point mutations and ease of use. These agents function primarily by alkylating DNA bases, leading to base-pair substitutions that can disrupt protein function and modify metabolic pathways (Ramesh et al., 2019; Hu et al., 2024). Alkylation typically involves the replacement of hydrogen atoms in nitrogenous bases with alkyl groups from the mutagen (Larrañaga et al., 2017), a process that can cause miscoding and replication errors. The EMS is widely used due to its high mutation frequency, environmental safety and ease of disposal (Chen et al., 2023; Ji and Li, 2025). However, the EMS dose must be precisely optimized because high doses may reduce seed viability, while low doses may not produce sufficient genetic variability.
These properties make chemical mutagens, especially EMS, particularly suitable for mutation breeding applications in diverse agroecological conditions. For example, their accessibility, low equipment requirements and high efficiency have made them more applicable in sub-Saharan Africa as compared to physical methods (Mishra et al., 2024). Furthermore, chemical mutagenesis has facilitated the identification of key genes, proteins and metabolites linked to desirable traits. Nonetheless, because chemically induced mutations are random and often unpredictable, rigorous screening under controlled and multi-environmental field conditions is essential to identify stable and superior lines. Additionally, maintaining accurate dosimetry is challenging due to environmental influences. Integrating chemical mutagenesis with advanced omics tools can help uncover underlying biochemical pathways associated with favorable traits.
4 Screening mutagenized plant population
4.1 Methods of screening
Several screening methods based on physiological and agronomic traits have been widely reported in cereals (Jael et al., 2022; Mude et al., 2022). These techniques have facilitated the development of finger millet lines by enabling the identification of superior lines for crop production and hybrid breeding. The effectiveness of a screening method largely depends on the use of highly heritable traits that are strongly associated with key yield-related traits such as grain yield and grain size. Phenotyping of these superior lines can be conducted under diverse environmental conditions in screen houses and field settings.
4.1.1 Screen house conditions
A screen house is a controlled environment structure typically covered with glass, plastic or other transparent materials that permits the cultivation of mutants under regulated conditions (Jáquez-Gutiérrez et al., 2019; Rotasperti et al., 2022). These controlled parameters include air humidity, light intensity, temperature, and soil moisture, which can be precisely adjusted to simulate specific environmental conditions. This setup facilitates consistent and efficient evaluation of agronomic traits, including grain yield and shoot biomass, whilst minimizing environmental variability (Peirone et al., 2018; Rotasperti et al., 2022). Screenhouses provide an ideal setting to detect subtle phenotypic differences in mutants that might be masked under open-field conditions. This is especially important when screening for traits related to yield and nutritional quality. However, translation between results obtained under field and controlled conditions is difficult (Poorter et al., 2016). Moreover, screenhouses can be equipped with the Internet of Things (IoT) which includes advanced imaging technologies and sensor-based systems to enhance the accuracy of physiological trait measurements such as canopy temperature and chlorophyll content (Gao et al., 2024). The uniformity of growth conditions in a screenhouse makes it easy to assess genotypic variation among mutants. The detailed phenotyping under greenhouse conditions enhance precise monitoring of plants and faster release of drought-tolerant cultivars (Peirone et al., 2018). However, care must be taken in experimental design because pot size and root restriction can influence trait expression. Therefore, ensuring that these limitations are minimized is essential for obtaining reliable and translatable phenotypic data.
4.1.2 Field conditions
Field phenotyping is essential for evaluating the yield and yield-related traits of new mutants under real-world conditions, enabling the selection of superior mutants for crop production (Johnson et al., 2017). However, field evaluations are influenced by environmental factors which affect genotype performance (de Leon et al., 2016). These factors obscure the expression of genes, proteins and metabolites (Kosová et al., 2021; Sahil et al., 2021), which complicate the detection of quantitative trait loci associated with beneficial traits. In addition, the presence of uncontrolled biotic stresses such as diseases and pests may further confound phenotypic data (Alonso et al., 2018). These complexities make it challenging to estimate trait heritability, genetic variance and selection efficiency. However, standardized field management practices, including uniform fertilization and irrigation scheduling can improve the reliability and repeatability of phenotypic assessments. Furthermore, conducting multi-location trials across diverse environments enables the identification of high-yielding and stable genotypes across environments (Valenzuela-Antelo et al., 2023; Aboye and Edo, 2024). Incorporating genotype performance data from different optimal locations provides a more realistic assessment of adaptability and resilience. However, genotype-by-environment interactions remain a key challenge due to differential genotype responses across locations (Temesgen, 2021; Goa et al., 2022) and the reduction of the correlation between phenotypic and genotypic values (Kang, 2002). Therefore, the use of correlation analysis, path analysis, multivariate analysis and mixed linear models, along with high-throughput phenotyping tools, enhances the accuracy of selection under field conditions.
4.2 Target traits in phenotyping for finger millet mutants
4.2.1 Seedling phenotypic traits
Mutants exhibiting high germination rates, shoot length, root length and strong seedling vigor often demonstrate superior early growth and development (Fauzi et al., 2022). During this critical phase, these lines efficiently mobilize stored metabolites (such as sugars and amino acids) that support active cell division and elongation in both shoots and roots (Canarini et al., 2019; Seerat et al., 2025). This metabolic activity contributes to increased root biomass and a higher root-to-shoot ratio, thereby improving the plant's capacity to access water and nutrients (Gargallo-Garriga et al., 2014). The rapid establishment of roots and shoots enhances survival under field conditions and positively influences yield potential (Comas et al., 2013; Grossnickle and Ivetić, 2022). Early vigor traits are routinely targeted in mutation breeding programs to evaluate the effects of different mutagenic treatments (Jency et al., 2016). The key parameters include germination rate, shoot length and root length, which help to identify promising mutants at an early stage (Dimaano et al., 2020). This enables breeders to eliminate poorly performing mutants before advancing to resource-intensive field evaluations.
Furthermore, recent technological advances, such as Iot, including high-throughput image-based phenotyping and machine learning algorithms, are used in early trait assessments (Zhang et al., 2018, 2025). These tools allow for accurate and rapid measurement of seedling traits across large populations. However, it is important to note that seedling-stage traits do not always correlate with yield-related traits, which are influenced by additional genetic and environmental factors (Tsago et al., 2014). Despite these challenges, early seedling vigor remains a key component of crop resilience under marginal conditions. Therefore, integrating early-stage vigor traits with later-stage agronomic and physiological traits is essential to maximize breeding efficiency.
4.2.2 Physiological traits
Physiological traits such as stomatal conductance and photosynthetic rate are essential for optimizing growth and improving grain yield in finger millet (Reddy, 2020). Stomatal conductance regulates carbon dioxide uptake for photosynthesis and controls water loss through transpiration, thereby influencing overall plant productivity (Urban et al., 2017; Lv et al., 2024). The optimal stomatal conductance promotes high carbon assimilation and supports biomass accumulation and grain filling. However, disruption in stomatal regulation, either excessive or insufficient, can limit productivity or lead to unnecessary water loss (Davies et al., 2002; Hasanuzzaman et al., 2023; Qiao et al., 2024). High photosynthetic activity is closely associated with increased dry matter accumulation (Xie et al., 2012). The differences in these physiological responses across developmental stages and environments make it essential to identify genotypes with consistent performance. The lines that demonstrate stable photosynthetic efficiency and gas exchange offer a valuable resource for improving grain yield (Furbank et al., 2015; Nazari et al., 2024).
Furthermore, the internal anatomical features must be considered to fully optimize photosynthetic efficiency (Mathan et al., 2021). These features, including mesophyll cell density and chloroplast organization, contribute to light interception and carbon fixation (Tholen et al., 2012; Mathan et al., 2021). The advanced phenotyping tools, including infrared thermography, gas exchange analyzers and chlorophyll fluorescence imaging, allow accurate measurement of these traits across large populations (Estrada et al., 2025; He et al., 2025). Therefore, utilizing physiological data in mutation breeding programs improves the early identification of elite mutants with enhanced yield potential.
4.2.3 Agronomic traits
Agronomic traits are essential indicators of plant performance and are used in the evaluation and selection of high-yielding mutant lines in crop breeding programs (Gallegos-Cedillo et al., 2021; Ganapathy et al., 2021). They provide direct insights into the adaptability and productivity of the crop under various environmental conditions (Mochida et al., 2020). For instance, early flowering and maturity are advantageous in drought-prone environments, allowing the plants to complete their life cycle before severe water stress negatively impacts yield (Shavrukov et al., 2017). The tiller number and panicle size are crucial yield components that contribute directly to grain production (Huang et al., 2020). Higher tillering capacity increases the number of productive stems and biomass accumulation which support grain yield production (Kalaitzidis et al., 2025). The short finger millet mutants are typically preferred in areas prone to wind or heavy rainfall, as they are less likely to lodge. However, taller varieties may produce higher biomass (Nascimento et al., 2024), which can improve forage yield and biomass utilization in certain cropping systems.
Moreover, agronomic traits such as harvest index and shoot biomass offer valuable information about the efficiency of assimilate partitioning to the grain. High harvest index values are key in plant breeding as they indicate better allocation of carbon assimilates to grain (Unkovich et al., 2010), rather than vegetative growth. Both genetic and environmental factors highly influence harvest index and shoot biomass (Porker et al., 2020), making their evaluation under different environments crucial for selecting stable and high-performing lines. Genotype by environment interactions significantly affect the expression of these traits, highlighting the importance of multi-location trials in breeding programs. Therefore, agronomic traits must be evaluated across a range of environmental conditions to ascertain yield stability and adaptation of mutant lines (OlaOlorun et al., 2021).
4.2.4 Grain nutritional quality traits
Beyond yield and yield-related traits, the grain nutritional quality traits are crucial targets in finger millet mutant screening because the crop is recognized as a rich source of calcium, iron and protein (Zuge et al., 2017; Waghmode et al., 2020). However, despite its reputation as a “nutri-cereal” (Saini et al., 2021), the variation for many of these traits within available germplasm collections remains limited, and identifying mutants with enhanced nutritional profiles has become a priority in mutation breeding programs. Screening for grain nutritional quality heavily relies on a combination of destructive and non-destructive approaches (Duduzile Buthelezi et al., 2019; Liu et al., 2022). The destructive methods include bicinchoninic acid assay and high-performance liquid chromatography-mass spectrometry for protein analysis and nutritional profiling (Burnouf and Bietz, 1984; Langyan et al., 2022; Navami et al., 2023). These approaches provide precise measurements but are resource-intensive and require the grinding of grain samples. Furthermore, the non-destructive approaches can be used for large-scale mutant screening. The near-infrared reflectance spectroscopy enables the rapid estimation of grain protein, starch, and mineral content without destroying seed samples (Johnson, 2020), and soft x-ray technology has also been applied to measure grain quality efficiently in intact grains (Shao et al., 2020). More recently, hyperspectral imaging has been explored for simultaneous detection of multiple nutritional attributes (Liang et al., 2025). These high-throughput tools can be valuable in mutation breeding programs where large mutant populations must be screened efficiently. Therefore, integrating destructive and non-destructive techniques ensures both accuracy and scalability, which helps to identify mutants with superior nutritional profiles early in the selection pipeline.
5 Progress in finger millet improvement using mutation breeding techniques
Mutation breeding has contributed meaningfully to the cereal crop improvement by generating novel genetic variation and enhancing traits related to yield and nutritional quality (Ganapathy et al., 2021; OlaOlorun et al., 2021; Mawcha et al., 2024; Xiao et al., 2025). Several mutant lines exhibiting superior characteristics such as early maturity, increased grain size, and higher micronutrient content have been developed and evaluated, supporting the effectiveness of this approach in addressing key breeding targets (Table 1). The release of high-yielding mutant varieties in various regions underscores the breeding value of induced mutants. For example, single-trait selection has led to significant improvements in traits such as flowering time and grain morphology, which are particularly important for adaptation to local agroecological conditions. These results demonstrate how targeted mutagenesis can enhance specific traits without the need for extensive hybridization cycles. Despite these advances, the adoption of mutation breeding in finger millet improvement programs in developing countries remains limited. One major constraint is the underutilization of mutants as foundational breeding material. Optimizing their use in hybrid development could significantly accelerate genetic gains. (Nazarenko et al. 2018) emphasized the potential of mutant lines in generating elite progeny. This agrees with (Githinji and Birithia 2015), who revealed that the development of high-performing F1 progenies from wheat mutant lines, highlights the breeding value of these genetic resources. In addition, finger millet mutation breeding has shown considerable promise in improving key agronomic traits (Ganapathy et al., 2021; Sellapillai et al., 2022) due to its wide genetic base. The wide genetic diversity could also facilitate the selection of genotypes with enhanced grain yield, improved grain size, and increased iron and calcium content. Therefore, these improvements are critical for addressing food security and micronutrient deficiencies in regions where finger millet is a dietary staple.
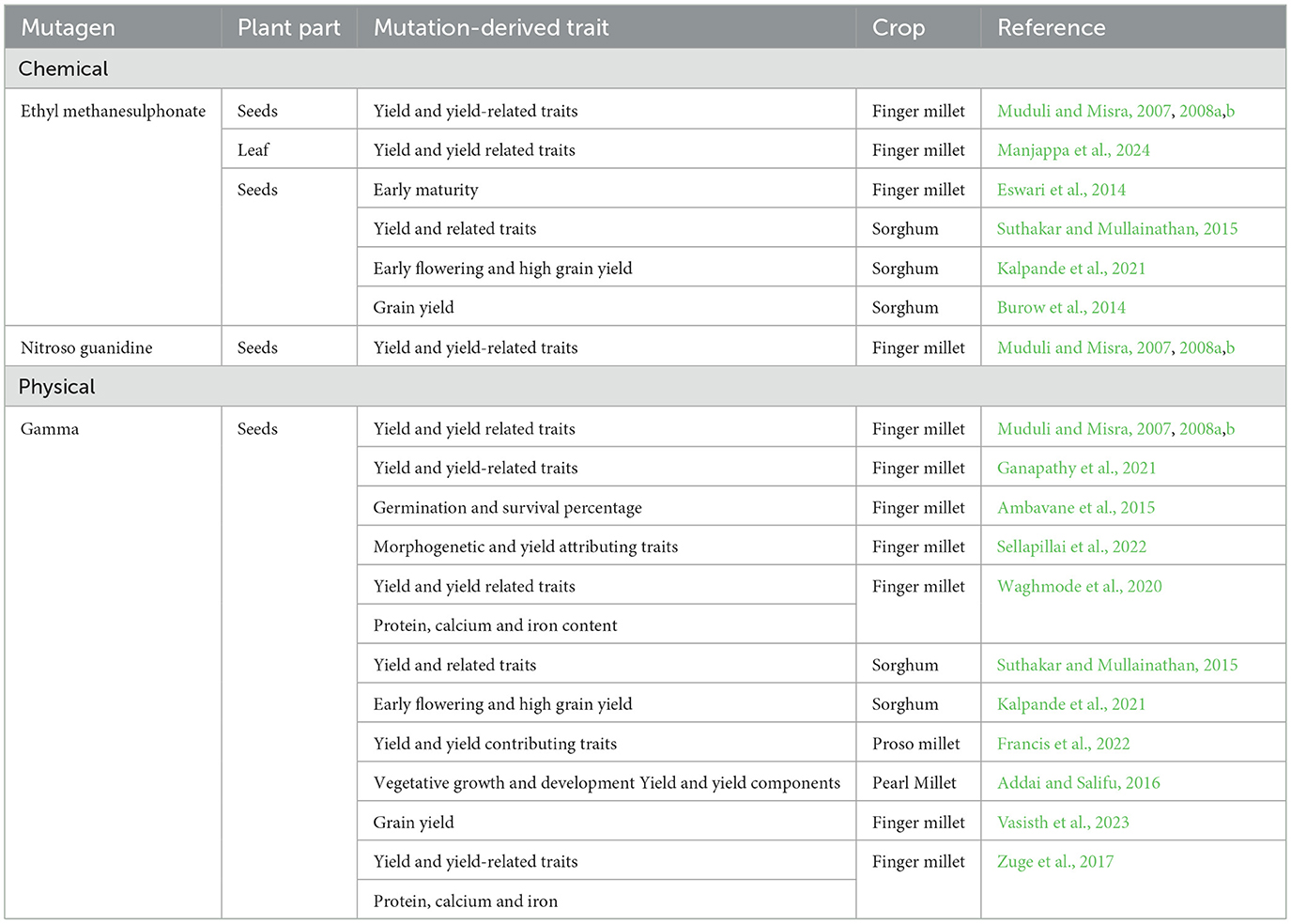
Table 1. Summary of mutagen-induced traits in millets across different countries in the past two decades.
6 Integrating mutation breeding with omics-assisted breeding approaches
6.1 Genomics
Mutation breeding induces genetic variation by generating novel alleles that can lead to desirable traits in crop genomes (Mba, 2013; Ayan et al., 2022). Despite its effectiveness, this approach has traditionally relied on phenotypic selection, which can overlook beneficial mutations not expressed through visible traits (Jankowicz-Cieslak et al., 2017a). This limitation can be addressed by integrating mutation breeding with molecular and genomic tools, which enable deeper exploration of induced genetic variation at a deeper level. Over the past 3 decades, the application of mutagenesis has expanded into genomic studies (Li et al., 2001), facilitating mutant characterization and accelerating cereal crop improvement (Prasanna and Jain, 2017).
This integration has led to the emergence of muta-genomics, a modern approach that combines conventional mutagenesis with functional genomics to enhance the detection and exploitation of genetic variation. Muta-genomics supports rapid mutation screening and accurate selection of mutant lines with improved phenotypes (Olaolorun, 2020). High-throughput techniques such as microarrays, differential display, high-resolution melt analysis and Targeting Induced Local Lesions in Genomes (TILLING) have been widely adopted in various crops (Mohan Jain and Suprasanna, 2011). Among these, TILLING has emerged as a key method, allowing mutation detection at the DNA level without requiring prior knowledge of gene function (Sestili et al., 2009; Uauy et al., 2009; Mishra et al., 2024). The method involves isolating chromosomal DNA from mutant populations and screening for nucleotide changes using molecular tools, which is further enhanced by high-throughput DNA sequencing (King et al., 2015). However, the success of TILLING depends on well-organized logistical operations. Critical procedures, including proper harvesting, cleaning, labeling, and storage of seeds from individual mutant lines, are vital to avoid contamination and maintain genetic integrity over generations (Sikora et al., 2011). Additionally, managing large mutant populations requires the development of databases and barcoding systems to track lines and their associated DNA samples. These systems are essential for long-term research and selection but may present technical and financial challenges in developing countries.
Furthermore, the advances in genomics have transformed mutation breeding by enabling the precise identification of induced mutations at the DNA level. The technologies such as whole-genome sequencing and genotyping-by-sequencing facilitate the detection of single-nucleotide polymorphisms, insertions, deletions and structural variants (Hittalmani et al., 2017; Francis et al., 2022; Xiong et al., 2023; Geethanjali et al., 2024; Zulfiqar et al., 2024). For example, whole-genome sequencing in cereals (such as sorghum and foxtail millets) revealed that all the genes involved in C4 carbon fixation pathways were also present in C3 plants, but these functional changes in these genes in the C3 cycle can lead to the C4 pathway's evolution (Banshidhar et al., 2023). These technologies accelerate the screening process and assist in identifying lines carrying favorable alleles and key traits (Bibi et al., 2010; Dhillon et al., 2014; Xiong et al., 2023). Mapping mutations to specific genomic regions facilitates the discovery of candidate genes controlling key traits (Luo et al., 2019), while the integration of genotypic and phenotypic data improves selection accuracy and breeding efficiency (Cobb et al., 2013). Furthermore, the finger millet genomic data have been instrumental in understanding traits such as grain yield and micronutrient accumulation, which are influenced by environmental factors. Therefore, researchers can dissect the genetic architecture of these traits and identify key functional variants by integrating genomic information with mutation breeding.
6.2 Transcriptomics
Transcriptomics provides a powerful platform for evaluating the molecular basis of key traits targeted in mutation breeding programs. It can identify candidate genes associated with yield enhancement and nutritional quality improvement in cereal crops and enables comparison of expression profiles between the lines (Zhu et al., 2023). The high-throughput RNA sequencing interrogates thousands of transcripts simultaneously to reveal regulatory networks (Ozsolak and Milos, 2011), which directly influence grain yield and grain nutrient composition, but are undetectable through phenotypic assessment alone. According to (Bhat et al. 2024) and (Zhang et al. 2024), plants show that transcriptional activation of genes involved in osmolyte accumulation and metabolite biosynthesis enhances yield potential. Differential expression analysis across diverse mutants further reveals transcriptional signatures associated with yield performance, whilst the co-expression analyses refine these insights by identifying the genes that regulate physio-biochemical processes and key traits (Tan et al., 2017; Xiong et al., 2020). Therefore, the integration of transcriptomic data with RNA-sequencing variants facilitates the detection of induced Single-Nucleotide Polymorphisms (SNPs) that alter transcript structure, thereby linking mutagenesis to functional gene variation. Furthermore, transcript profiling reveals gene modules correlated with key traits and yield quality (Li et al., 2022; Verma et al., 2022). These candidate genes can be mined for induced SNPs (Morgil et al., 2020) and converted into molecular markers, which accelerate the fixation of beneficial alleles. Transcriptomic data also inform expression-based assays for the early screening of plants that can be affected by stress (Gong et al., 2020; Zhou et al., 2020), while reducing reliance on late-stage phenotyping. Therefore, integrating transcriptomics with other omic tools provide a view of regulatory mechanisms and metabolic adjustments that allow the identification of key genes and pathways causing trait variation in mutant lines.
6.3 Proteomics
Proteomics is the large-scale quantitative analysis of the entire set of proteins produced or modified by a plant under certain conditions (Hu et al., 2015). It offers a powerful approach for understanding the molecular effects of induced mutations in crop improvement (Tachioka et al., 2016). Unlike the genome, which remains relatively static, the proteome is highly dynamic and reflects the plant's physiological state at different growth stages and environmental conditions (Kumar et al., 2025). In mutation breeding, proteomic analysis enables researchers to detect changes in protein abundance, post-translational modifications (PTMs) and protein—protein interactions that result from mutagenic treatments, especially EMS and γ-rays treatments (Asif et al., 2019). These molecular shifts often underlie phenotypic changes in traits such as yield and nutrient content.
Furthermore, the advanced mass spectrometry (MS)-based platforms, including Orbitrap and time-of-flight analyzers coupled with tandem MS (MS/MS), allow high-throughput identification and quantification of proteins in crop cultivars (Kumar et al., 2025). Both label-free and label-based techniques such as isobaric tags for relative and absolute quantification (iTRAQ) and Tandem Mass Tags (TMT) can be used to compare mutant and wild-type proteomes. Although gel-free methods dominate modern workflows, two-dimensional gel electrophoresis (2DGE) remains useful for detecting isoforms and PTM-affected proteins, especially when combined with LC–MS/MS (Görg et al., 2000).
In finger millet, where genomic resources are limited, proteomics offers a practical tool for characterizing functional variation induced by mutagenesis. Differential protein expression analysis in EMS-treated lines can reveal candidate proteins linked to stress tolerance and biomass accumulation (Khalil et al., 2018). These proteins may serve as early biomarkers for phenotypic selection, reducing breeding cycle time. Integrating proteomic insights with field-based phenotyping and metabolomics enhances the ability to dissect complex traits, especially those related to yield potential and yield quality. Although such integrated omics approaches have been successfully applied in cereals like rice and wheat (Liu et al., 2019; Naaz et al., 2024), their application in finger millet remains underexplored and presents a promising direction for future mutation breeding efforts.
6.4 Metabolomics
The application of mutation breeding in crop improvement has led to the development of new genotypes with desirable traits, including early maturity and improved yield potential (Table 1). However, the reliance of this approach on conventional phenotypic screening can limit the detection of subtle or non-visible traits. Therefore, metabolomics has emerged as a complementary tool that offers a more precise and efficient approach for trait identification and mutant selection. Furthermore, metabolomics provides valuable insights into the physiological and adaptive capacity of plants under various environmental conditions (Samsami and Maali-Amiri, 2024). The identified metabolite traits are used to complement morphological and physiological selection because they offer a deeper understanding of the molecular and cellular responses of plants (Zhang et al., 2024; Ali et al., 2025). The metabolites that are regulated in plants are grouped into primary and secondary metabolites (Nezafatian et al., 2024). The primary metabolites such as sugars (sucrose and fructose), amino acids (proline and glutamate) and organic acids (malate and citrate) are crucial for energy supply and osmotic regulation (le Roux et al., 2021; Saeidi et al., 2024). These compounds often accumulate in response to abiotic stresses such as drought and salinity stress helping to stabilize cellular functions and maintain growth (Bao et al., 2025; Pompelli et al., 2025). In addition, secondary metabolites including phenolics and flavonoids play critical roles in plant defense, signaling and oxidative stress regulation (Upadhyay et al., 2024; Rao et al., 2025), thereby protecting cellular structures from damage. The elevated levels of polyphenols and flavonoids have been associated with improved yield and enhanced nutritional quality (Turfan, 2025). Therefore, harnessing the potential of metabolite-based selection increases the selection accuracy of superior mutants. For instance, mutants with increased accumulation of proline and sugars are often linked to superior performance, making these metabolites reliable biomarkers (le Roux et al., 2021; Sen et al., 2023).
However, metabolomics research heavily depends on different analytical platforms with differences in resolution, sensitivity and metabolite coverage to samples and diverse small molecules (Liao et al., 2025). The nuclear Magnetic Resonance (NMR) spectroscopy is used for its reproducibility and capacity to provide structural information across a broad spectrum of metabolites, but it has low sensitivity affect the detection of metabolites present in each sample (Nagana Gowda and Raftery, 2021). However, gas chromatography mass spectrometry and liquid chromatograph-mass spectrometry offers high sensitivity and resolution which allow the detection of low-abundance metabolites with greater accuracy (Lee et al., 2010). The mass spectrometry imaging extends these capabilities by adding spatial information, allowing visualization of metabolite distribution within tissues (Bjarnholt et al., 2014). Furthermore, the recently established approaches, including capillary electrophoresis—mass spectrometry and fourier transform infrared spectroscopy, are gaining prominence due to their accuracy in identifying metabolite biomarkers (Maia et al., 2023; Soga, 2023). These biomarkers can serve as selection indices in mutation breeding and accelerate the identification of desirable traits. Despite their potential, the application and adoption of metabolomics in mutation breeding programs remain limited due to high costs and a lack of skilled personnel. Therefore, capacity building, collaborative research networks and investments are encouraged in shared metabolomics facilities to translate this technology into mutation breeding.
6.5 Gene editing
Mutation breeding has been widely applied to generate genetic diversity in crops such as finger millet, yet its random nature complicates the establishment of direct links between mutations and desirable traits. However, gene editing technologies (CRISPR-Cas9, base editing, and prime editing) offer precise and predictable modification of specific genomic changes and sequences (Kantor et al., 2020; Chen and Liu, 2023). These tools enable targeted changes in coding regions, promoters or regulatory elements associated with yield and nutritional composition (Chen et al., 2024; Hamdan and Tan, 2024). The high specificity of gene editing assists in random mutagenesis, where large populations must be screened to recover useful variants (Zimmermann et al., 2024). For example, CRISPR-mediated knockouts of negative regulators create genotypes with improved yield and other traits (Sun et al., 2025), base editing enables single-nucleotide changes without double-strand breaks to reduce unintended genomic alterations (Li et al., 2023), and prime editing facilitates precise insertions, deletions and substitutions with reduced off-target activity (Chen and Liu, 2023). These technologies provide breeders with tools for precision crop improvement, supporting mutation breeding procedures. Therefore, the complementary strengths of mutation breeding and gene editing highlight the potential of integrated approaches. Furthermore, the broad spectrum of genetic variability generated during mutagenesis makes it crucial for the discovery of new alleles and pathways in underutilized crops. These novel variants can be characterized through transcriptomic and metabolomic analyses, however gene editing allows a precise modification of validated targets without introducing foreign genes (Hamdan and Tan, 2024). When candidate genes are identified through mutagenesis and functional analysis, CRISPR-Cas9 or base editing is used to reproduce beneficial alleles and traits (Ansori et al., 2023) in preferred varieties. According to (Wolter et al. 2019), the CRISPR/Cas technologies enable rapid and directed generation of genetic diversity at multiple genomic sites, thereby accelerating plant breeding compared to traditional mutation breeding methods.
7 Mutation breeding in finger millet for yield gain and nutritional quality
In the last few decades, induced mutagenesis has played a pivotal role in generating mutant lines with improved grain yield and related agronomic traits (Table 1). The primary objective has been to enhance the commercial value of cultivars by modifying key characteristics such as plant height, maturity period and grain size. According to (Ambavane et al. 2015), the economic success of a new mutant variety is largely dependent on its yield potential and adaptability to specific agro-ecological conditions. Although mutation breeding has been effective in developing short-statured lines and reducing days to maturity in cereals, its targeted application in improving biomass allocation and enhancing grain nutritional quality in finger millet remains limited. Most efforts have concentrated on aboveground traits, particularly height reduction, which may indirectly influence total shoot biomass. However, traits associated with root architecture are critical for drought adaptation but have received minimal attention, despite their importance in enhancing water uptake from deeper soil layers (Eziz et al., 2017).
Phenotyping root traits remains a significant challenge due to the complexity and labor-intensive nature of belowground measurements (Keerthi et al., 2025). As a result, root improvement has often been overlooked, and many modern cultivars possess underdeveloped root systems that reduce resilience under water-limited conditions (White et al., 2015). The long-term selection for plant height and grain yield has inadvertently narrowed the genetic variation in rooting traits, making it difficult to breed for both high yield and robust root systems. Furthermore, the trade-off often exists between grain yield and root biomass, complicating efforts to simultaneously improve both traits. This highlights the need for integrated breeding strategies that combine mutation breeding with advanced phenotyping and molecular tools to develop balanced genotypes with improved yield and grain nutrient content. Recent advances, such as high-throughput root imaging platforms and non-invasive phenotyping tools, help to accelerate selection for hidden traits like root depth, length, density and branching (Clark et al., 2012; Atkinson et al., 2019; Angidi et al., 2025). Moreover, modern mutation breeding tools (including TILLING), combined with downstream analytical tools including proteomic and metabolomic profiling, enable the identification of functional mutations affecting biomass partitioning and grain filling. For example, EMS-induced mutations in other cereals (rice and wheat) have led to altered expression of key proteins and enzymes (Yu and Tian, 2018; Irshad et al., 2022) that can be associated with improved source—sink balance and grain nutrient. These approaches, though underutilized in finger millet, hold significant promise for trait dissection and breeding efficiency. Therefore, future mutation breeding in finger millet should prioritize multi-trait selection frameworks that target yield potential and root traits, especially across sub-Saharan Africa, where finger millet remains an underexploited.
Furthermore, mutation breeding has made notable contributions to the nutritional enhancement of millets (Table 1). Gamma irradiation has been effective in producing mutant lines with significantly higher grain calcium and iron contents compared with the original varieties. For instance, (Zuge et al. 2017) reported that the four mutant lines had more than a 3.43% increase in protein content compared to their mother variety, whereas calcium content increased by more than 1.30% in the four mutant lines. Additionally, three mutant lines (DML-4, DML-7, and DML-10) showed higher iron content compared to the mother variety. That agrees with (Waghmode et al., 2020) who revealed that mutant lines have a significantly higher protein, calcium and iron content as compared to their mother variety. These nutritional gains are relevant because finger millet is traditionally consumed in semi-arid and marginal regions where mineral deficiencies are common. However, published evidence of EMS directly increasing mineral concentrations in finger millet remains limited, although EMS is applied to broaden genetic variability (Chen et al., 2023). Furthermore, since finger millet already possesses essential nutrients, including proteins, calcium and iron in higher levels compared to other cereals (Ramesh et al., 2019; Rathore et al., 2019). Therefore, mutation breeding can complement this natural advantage by expanding the range of genotypes with enhanced nutritional composition. However, more detailed studies are needed to evaluate the bioavailability of these nutrients in mutant lines, considering the influence of antinutritional compounds such as tannins, phytate and oxalate (Kumar et al., 2016). Therefore, integration of mutation breeding with advanced molecular approaches will help to better understand the biochemical regulation of nutrient accumulation.
8 Challenges and limitations of mutation breeding in finger millet
Despite the considerable potential of mutation breeding to broaden the genetic base and improve key agronomic and nutritional traits in finger millet (Table 1), several challenges negatively affect its widespread application and efficiency. One major limitation is the random and unpredictable nature of induced mutations, which often generate a high proportion of deleterious or neutral variants alongside beneficial ones. This random nature of mutagenesis necessitates the screening of large mutant populations, which is labor-intensive, time-consuming and resource-demanding in environments with limited research infrastructure (Shahwar et al., 2023). Low mutation frequencies of desirable traits require efficient population management to select superior mutants. The interactions between genotype and environment affects the accuracy needed during mutant lines evaluation (Dyulgerova and Dyulgerov, 2019), resulting in extended breeding cycles. In addition, the lack of integrated mutant resource databases and standardized protocols for finger millet mutant line maintenance often results in genetic contamination and loss of valuable alleles.
The emerging biotechnologies and integrative breeding approaches offer viable strategies to overcome these challenges and enhance mutation breeding efficiency (Jankowicz-Cieslak et al., 2017b). The integration of molecular marker-assisted selection and high-throughput phenotyping platforms accelerate the detection and validation of beneficial mutations while reducing labor and cost. Moreover, the omics-assisted breeding (including genomics, transcriptomics, proteomics, and metabolomics) evaluates the molecular and biochemical bases of beneficial traits, which supports targeted mutant selection. However, financial and technical constraints in finger millet-producing regions challenge the adoption of these advanced tools. Therefore, strengthening institutional support, capacity building, policy and collaborative frameworks that prioritize capacity building, resource allocation and technology access are essential to unleash the full potential of mutation breeding for finger millet improvement on a broad scale.
9 Conclusion and outlooks
Mutation breeding in finger millet has proven to be a valuable tool in enhancing traits that are critical for improving productivity and nutritional value. Through the use of physical and chemical mutagens, a wide array of genetic variability can be generated, offering significant opportunities for selecting superior mutant lines. The application of advanced screening methods, including genomics, proteomics, metabolomics, and phenomics through the use of IoT applications can enhance the precision and efficiency of identifying desirable mutants. This approach allows for a deeper understanding of the molecular changes associated with beneficial traits, which facilitate the selection of superior lines for future breeding. As mutation breeding continues to advance, integrating these techniques with other modern plant breeding tools will expedite the development of improved finger millet varieties that can contribute to achieve global food and nutritional security. The ability to optimize genetic diversity and harness beneficial mutations will enhance crop productivity and help address climate change challenges and nutritional deficiencies. However, the progress is slowed by limited genomic resources and the lack of high-throughput phenotyping platforms. Stronger integration of multi-omics with field-based validation is needed to translate laboratory findings into practical outcomes. Future research should focus on combining mutation breeding with omics-driven selection and IoT applications to enhance productivity and nutritional quality, supported by policies that ensure rapid development and release of improved varieties to farmers.
Author contributions
MM: Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Resources. SF: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. NS: Conceptualization, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. EG: Conceptualization, Investigation, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing, Data curation, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was based on generous financial support from the National Research Foundation of the Republic of South Africa (PMDS240601223016).
Acknowledgments
The University of South Africa, the University of Venda and the Agricultural Research Council of South Africa are thanked for their overall research support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2025.1679819/full#supplementary-material
References
Aboye, B. M., and Edo, M. A. (2024). Exploring genotype by environment interaction in sunflower using genotype plus genotype by environment interaction (GGE) and best linear unbiased prediction (BLUP) approaches. Dis. Appl. Sci. 6:431. doi: 10.1007/s42452-024-06136-1
Addai, I. K., and Salifu, B. (2016). Selection of mutants with improved growth and total grain yield in the M2 generation of pearl millet (Pennicetum glaceum L.) in the Northern Region of Ghana. J. Agron. 15, 88–93. doi: 10.3923/ja.2016.88.93
Admasu, S., and Belete, T. (2020). Finger millet (Eleusine coracana (L.) Gaertn) breeding, major production challenges and future prospects. J. Agri. Res. Adv. 2, 33–39.
Alemu, H. (2016). Review paper on mutation breeding as applied in groundnut (Arachis hypogea L.) improvement. Gene Cell Ther. 1, 35–40. doi: 10.11648/j.gct.20160105.11
Ali, S., Mir, R. A., Haque, M. A., Danishuddin, A.lmalki, M. A., Alfredan, M., et al. (2025). Exploring physiological and molecular dynamics of drought stress responses in plants: challenges and future directions. Front Plant Sci. 16:1565635. doi: 10.3389/fpls.2025.1565635
Alonso, C., Ramos-Cruz, D., and Becker, C. (2018). The role of plant epigenetics in biotic interactions. New Phytol. 221, 731–737. doi: 10.1111/nph.15408
Ambavane, A. R., Sawardekar, S. V., Sawantdesai, S. A., and Gokhale, N. B. (2015). Studies on mutagenic effectiveness and efficiency of gamma rays and its effect on quantitative traits in finger millet (Eleusine coracana L. Gaertn). J. Radiat. Res. Appl. Sci. 8, 120–125. doi: 10.1016/j.jrras.2014.12.004
Angidi, S., Madankar, K., Tehseen, M. M., and Bhatla, A. (2025). Advanced high-throughput phenotyping techniques for managing abiotic stress in agricultural crops—a comprehensive review. Crops 5:8. doi: 10.3390/crops5020008
Animasaun, D. A., and Oguntoye, E. O. (2024). Mutagenesis in crop improvement: methods and applications. J. Crop. Improv. 38, 156–178. doi: 10.1080/15427528.2024.2336257
Ansori, A. N. M., Antonius, Y., Susilo, R. J. K., Hayaza, S., Kharisma, V. D., Parikesit, A. A., et al. (2023). Application of CRISPR-Cas9 genome editing technology in various fields: a review. Narrat. J. 3:e184. doi: 10.52225/narra.v3i2.184
Asif, A. K., Ansari, M. Y., Hashem, A., Tabassum, B., Abd Allah, E. F., et al. (2019). Proteome profiling of the mutagen-induced morphological and yield macro-mutant lines of Nigella sativa L. Plants 8:321. doi: 10.3390/plants8090321
Atkinson, J. A., Pound, M. P., Bennett, M. J., and Wells, D. M. (2019). Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 55, 1–8. doi: 10.1016/j.copbio.2018.06.002
Auerbach, C., and Robson, J. M. (1946). Chemical production of mutations. Nature 157, 302–302. doi: 10.1038/157302a0
Ayan, A., Meriç, S., Gümüş, T., and Atak, Ç. (2022). Current strategies and future of mutation breeding in soybean improvement. doi: 10.5772/intechopen.104796
Bado, S., Forster, B. P., and Maghuly, F. (2023). “Physical and chemicals mutagenesis in plant breeding,” in Mutation Breeding for Sustainable Food Production and Climate Resilience, eds S. Penna and S. M. Jain (Singapore: Springer Nature), 57–97. doi: 10.1007/978-981-16-9720-3_3
Banshidhar, P. S., Singh, A., Jaiswal, P., Singh, M. K., Meena, K. R., et al. (2023). The potentialities of omics resources for millet improvement. Funct. Integr. Genomics 23:210. doi: 10.1007/s10142-023-01149-2
Bao, Q., Wu, Y., Du, H., Wang, Y., and Zhang, Y. (2025). Phenotypic physiological and metabolomic analyses reveal crucial metabolic pathways in quinoa (Chenopodium quinoa Willd.) in response to PEG-6000 induced drought stress. Int. J. Mol. Sci. 26:2599. doi: 10.3390/ijms26062599
Bernardi, J., Battaglia, R., Bagnaresi, P., Lucini, L., and Marocco, A. (2019). Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize. Plant Sci. 281, 133–145. doi: 10.1016/j.plantsci.2019.01.027
Bharat, R. A., Prathmesh, S. P., Sarsu, F., and Suprasanna, P. (2024). Induced mutagenesis using gamma rays: biological features and applications in crop improvement. OBM Genet. 8, 1–27. doi: 10.21926/obm.genet.2402233
Bhat, B. A., Mir, R. A., Mir, W. R., Hamdani, S. S., and Mir, M. A. (2024). Transcription factors-golden keys to modulate the plant metabolism to develop salinity tolerance. Plant Stress 11:100409. doi: 10.1016/j.stress.2024.100409
Bibi, S., Dahot, M. U., Nizamani, G. S., Khan, I. A., Khatri, A., Naqvi, M. H., et al. (2010). Molecular marker assisted selection for drought tolerant wheat genotypes. Pak. J. Bot. 42, 2443–2452.
Bjarnholt, N., Li, B., D'Alvise, J., and Janfelt, C. (2014). Mass spectrometry imaging of plant metabolites–principles and possibilities. Nat. Prod. Rep. 31, 818–837. doi: 10.1039/C3NP70100J
Bolbhat, S. N., and Thikekar Chaitali, S. (2020). Studies on effect of induced mutagenesis on finger millet [Eleusine Coracana (L.) Gaertn.] in M1 Generation. Int. J. Creat. Res.Thought. 8.
Broyart, C., Fontaine, J., Molini,é, R., Cailleu, D., Tercé-Laforgue, T., Dubois, F., et al. (2009). Metabolic profiling of maize mutants deficient for two glutamine synthetase isoenzymes using 1H-NMR-based metabolomics. Phytochem. Anal. 21, 102–109. doi: 10.1002/pca.1177
Burnouf, T., and Bietz, J. A. (1984). Reversed-phase high-performance liquid chromatography of durum wheat gliadins: relationships to durum wheat quality. J. Cereal Sci. 2, 3–14. doi: 10.1016/S0733-5210(84)80002-8
Burow, G., Xin, Z., Hayes, C., and Burke, J. (2014). Characterization of a multiseeded (msd1) mutant of sorghum for increasing grain yield. Crop Sci. 54, 2030–2037. doi: 10.2135/cropsci2013.08.0566
Canarini, A., Kaiser, C., Merchant, A., Richter, A., and Wanek, W. (2019). Corrigendum: root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10:00420. doi: 10.3389/fpls.2019.00420
Chaudhary, N., and Kumar, G. (2023). “Mutagenic radiations: X-rays, ionizing particles, and ultraviolet,” in Biotechnologies and Genetics in Plant Mutation Breeding (Apple Academic Press), 45–67. doi: 10.1201/9781003305064-3
Chawade, A., van Ham, J., Blomquist, H., Bagge, O., Alexandersson, E., and Ortiz, R. (2019). High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 9:258. doi: 10.3390/agronomy9050258
Chen, F., Chen, L., Yan, Z., Xu, J., Feng, L., He, N., et al. (2024). Recent advances of CRISPR-based genome editing for enhancing staple crops. Front. Plant Sci. 15:1478398. doi: 10.3389/fpls.2024.1478398
Chen, L., Duan, L., Sun, M., Yang, Z., Li, H., Hu, K., et al. (2023). Current trends and insights on EMS mutagenesis application to studies on plant abiotic stress tolerance and development. Front Plant Sci. 13:1052569. doi: 10.3389/fpls.2022.1052569
Chen, P. J., and Liu, D. R. (2023). Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 24, 161–177. doi: 10.1038/s41576-022-00541-1
Clark, R. T., Famoso, A. N., Zhao, K., Shaff, J. O. N. E., Craft, E. J., Bustamante, C. D., et al. (2012). High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 36, 454–466. doi: 10.1111/j.1365-3040.2012.02587.x
Cobb, J. N., DeClerck, G., Greenberg, A., Clark, R., and McCouch, S. (2013). Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 126, 867–887. doi: 10.1007/s00122-013-2066-0
Comas, L. H., Becker, S. R., Cruz, V. M. V., Byrne, P. F., and Dierig, D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:00442. doi: 10.3389/fpls.2013.00442
Danguy des Déserts, A., Bouchet, S., Sourdille, P., and Servin, B. (2021). Evolution of recombination landscapes in diverging populations of bread wheat. Gen. Biol. Evol. 13:evab152. doi: 10.1093/gbe/evab152
Davies, W. J., Wilkinson, S., and Loveys, B. (2002). Stomatal control by chemical signalling and the exploitation of this mechanism to increase water use efficiency in agriculture. New Phytol. 153, 449–460. doi: 10.1046/j.0028-646X.2001.00345.x
de Leon, N., Jannink, J., Edwards, J. W., and Kaeppler, S. M. (2016). Introduction to a special issue on genotype by environment interaction. Crop Sci. 56, 2081–2089. doi: 10.2135/cropsci2016.07.0002in
Dhillon, R. S., Saharan, R. P., Jattan, M., Rani, T., Sheokand, R. N., Dalal, V., et al. (2014). Molecular characterization of induced mutagenesis through gamma radiation using RAPD markers in Jatropha curcas L. Afr. J. Biotechnol. 13, 806–813. doi: 10.5897/AJB12.2934
Dimaano, N. G. B., Ali, J., Mahender, A., Sta. Cruz, P. C., Baltazar, A. M., Diaz, M. G. Q., et al. (2020). Identification of quantitative trait loci governing early germination and seedling vigor traits related to weed competitive ability in rice. Euphytica 216:026948. doi: 10.1007/s10681-020-02694-8
Duduzile Buthelezi, N. M., Tesfay, S. Z., Ncama, K., and Magwaza, L. S. (2019). Destructive and non-destructive techniques used for quality evaluation of nuts: a review. Sci. Horticult. 247, 138–146. doi: 10.1016/j.scienta.2018.12.008
Dwiningsih, Y., and Alkahtani, J. (2022). Phenotypic variations, environmental effects and genetic basis analysis of grain elemental concentrations in rice (Oryza sativa L.) for improving human nutrition PsyAxiv preprint. doi: 10.20944/preprints202209.0263.v1
Dyulgerova, B., and Dyulgerov, N. (2019). Genotype by environment interaction for grain yield of barley mutant lines. Agriculture 65, 51–58. doi: 10.2478/agri-2019-0006
Elena, S. F., and de Visser, J. A. G. (2003). Environmental stress and the effects of mutation. J. Biol. 2, 1–4. doi: 10.1186/1475-4924-2-12
Estrada, F., Gonzàlez-Meler, M. A., Dias de Oliveira, E. A., del Pozo, A., and Lobos, G. A. (2025). Morphophysiological plant phenotyping for the development of plant breeding under drought and heat conditions: a practical approach. Food Energy Secur. 14:70030. doi: 10.1002/fes3.70030
Eswari, K., Gogulan, G., Hari Prathab, K. A., and Sakila, M. (2014). Development of early maturing mutants in finger millet. Res. J. Agric. For. Sci. 2, 1–9.
Eziz, A., Yan, Z., Tian, D., Han, W., Tang, Z., and Fang, J. (2017). Drought effect on plant biomass allocation: a meta-analysis. Ecol. Evol. 7, 11002–11010. doi: 10.1002/ece3.3630
Fauzi, A. R., Junaedi, A., Lubis, I., Ghulamahdi, M., Aswidinnoor, H., and Sakagami, J.-I. (2022). Evaluation of rice genotypes on seed attributes and agronomic performance for developing direct-seeded cultivar. AIMS Agricult. Food 7, 1–21. doi: 10.3934/agrfood.2022001
Fernie, A. R., Tadmor, Y., and Zamir, D. (2006). Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9, 196–202. doi: 10.1016/j.pbi.2006.01.010
Francis, N., Rajasekaran, R., Krishnamoorthy, I., Muthurajan, R., Thiyagarajan, C., and Alagarswamy, S. (2022). Gamma irradiation to induce beneficial mutants in proso millet (Panicum miliaceum L.): an underutilized food crop. Int. J. Radiat. Biol. 98, 1277–1288. doi: 10.1080/09553002.2022.2024292
Francis, N., Rajasekaran, R., Rajagopalan, V. R., Bakya, S. V., Muthurajan, R., Kumar, A. G., et al. (2023). Molecular characterization and SNP identification using genotyping-by-sequencing in high-yielding mutants of proso millet. Front. Plant Sci. 14:1108203. doi: 10.3389/fpls.2023.1108203
Furbank, R. T., Quick, W. P., and Sirault, X. R. R. (2015). Improving photosynthesis and yield potential in cereal crops by targeted genetic manipulation: prospects, progress and challenges. Field Crops Res. 182, 19–29. doi: 10.1016/j.fcr.2015.04.009
Gallegos-Cedillo, V. M., Diánez, F., Nájera, C., and Santos, M. (2021). Plant agronomic features can predict quality and field performance: a bibliometric analysis. Agronomy 11:2305. doi: 10.3390/agronomy11112305
Ganapathy, K. N., Patro, T. S. S. K., Palanna, K. B., Das, I. K., Elangovan, M., Prashant, B., et al. (2021). Development of improved mutants for grain yield and related traits in finger millet (Eleusine coracana L Gaertn.) through gamma mutagenesis. Int. J. Plant Soil Sci. 74, 225–235. doi: 10.9734/ijpss/2021/v33i1830592
Gao, T., Sheng, W., Zhang, Z., Li, H., and Zhang, M. (2024). Greenhouse Phenotyping Measurement Techniques and Systems: A Review,” 43–59. doi: 10.1007/978-981-97-6441-9_3
Gargallo-Garriga, A., Sardans, J., Pérez-Trujillo, M., Rivas-Ubach, A., Oravec, M., Vecerova, K., et al. (2014). Opposite metabolic responses of shoots and roots to drought. Sci. Rep. 4:06829. doi: 10.1038/srep06829
Gebreyohannes, A., Shimelis, H., Laing, M., Mathew, I., Odeny, D. A., and Ojulong, H. (2021). Finger millet production in ethiopia: opportunities, problem diagnosis, key challenges and recommendations for breeding. Sustainability 13:13463. doi: 10.3390/su132313463
Gebreyohannes, A., Shimelis, H., Mashilo, J., Odeny, D. A., Tadesse, T., and Ojiewo, C. O. (2024). Finger millet (Eleusine coracana) improvement: challenges and prospects—a review. Plant Breed. 143, 350–374. doi: 10.1111/pbr.13169
Geethanjali, S., Kadirvel, P., and Periyannan, S. (2024). Wheat improvement through advances in single nucleotide polymorphism (SNP) detection and genotyping with a special emphasis on rust resistance. Theor. Appl. Genet. 137:224. doi: 10.1007/s00122-024-04730-w
Ghanim, A. M. (2023). “Physical mutagenesis in cereal crops,” in Mutation Breeding and Efficiency Enhancing Technologies for Resistance to Striga in Cereals, eds A. M. A. Ghanim, S. Sivasankar, and P. J. Rich (Berlin, Heidelberg: Springer Berlin Heidelberg), 13–27. doi: 10.1007/978-3-662-68181-7_2
Githinji, G. G., and Birithia, R. K. (2015). Effects of induced mutagenesis and single crossing on agronomic traits of wheat (Triticum Aestivum L.). J. Agricult. Life Sci. 2, 31–37.
Goa, Y., Mohammed, H., Worku, W., and Urage, E. (2022). Genotype by environment interaction and yield stability of cowpea (Vigna unguiculata (L.) Walp.) genotypes in moisture limited areas of Southern Ethiopia. Heliyon 8:e09013. doi: 10.1016/j.heliyon.2022.e09013
Gong, L., Han, S., Yuan, M., Ma, X., Hagan, A., and He, G. (2020). Transcriptomic analyses reveal the expression and regulation of genes associated with resistance to early leaf spot in peanut. BMC Res. Notes 13:381. doi: 10.1186/s13104-020-05225-9
Görg, A., Obermaier, C., Boguth, G., Harder, A., Scheibe, B., Wildgruber, R., et al. (2000). The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V
Govindaraj, M., Vetriventhan, M., and Srinivasan, M. (2015). Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet. Res. Int. 2015, 1–14. doi: 10.1155/2015/431487
Grossnickle, S. C., and Ivetić, V. (2022). Root system development and field establishment: effect of seedling quality. New For. 53, 1021–1067. doi: 10.1007/s11056-022-09916-y
Hall, R. D., D'Auria, J. C., Silva Ferreira, A. C., Gibon, Y., Kruszka, D., Mishra, P., et al. (2022). High-throughput plant phenotyping: a role for metabolomics? Trends Plant Sci. 27, 549–563. doi: 10.1016/j.tplants.2022.02.001
Hamdan, M. F., and Tan, B. C. (2024). Genetic modification techniques in plant breeding: a comparative review of CRISPR/Cas and GM technologies. Horticult. Plant J. 11, 1807–1829. doi: 10.1016/j.hpj.2024.02.012
Harrigan, G. G., Martino-Catt, S., and Glenn, K. C. (2007). Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 3, 259–272. doi: 10.1007/s11306-007-0076-0
Hasanuzzaman, M.d., Zhou, M., and Shabala, S. (2023). How does stomatal density and residual transpiration contribute to osmotic stress tolerance? Plants 12:494. doi: 10.3390/plants12030494
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2018). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25, 39–47. doi: 10.1093/dnares/dsx036
He, J., Ning, K., Naznin, A., Wang, Y., Chen, C., Zuo, Y., et al. (2025). Technological advances in imaging and modelling of leaf structural traits: a review of heat stress in wheat. J. Exp. Bot. 4:eraf070. doi: 10.1093/jxb/eraf070
Hilu, K. (1994). Validation of the combination Eleusine coracana subspecies africana (Kennedy-O'Byrne) Hilu et Dewet. Phytologia 76, 410–411. doi: 10.5962/bhl.part.9964
Hittalmani, S., Mahesh, H. B., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y. R., et al. (2017). Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom. 18:465. doi: 10.1186/s12864-017-3850-z
Hu, J., Liu, M., Wang, D., Liang, Y., Zong, Y., Li, Y., et al. (2024). Transcriptional and genetic characteristic of chimera pea generation via double ethyl methanesulfonate-induced mutation revealed by transcription analysis. Front. Plant Sci. 15:1439547. doi: 10.3389/fpls.2024.1439547
Hu, J., Rampitsch, C., and Bykova, N. V. (2015). Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 6:00209. doi: 10.3389/fpls.2015.00209
Huang, M., Shan, S., Cao, J., Fang, S., Tian, A., Liu, Y., et al. (2020). Primary-tiller panicle number is critical to achieving high grain yields in machine-transplanted hybrid rice. Sci. Rep. 10:2811. doi: 10.1038/s41598-020-59751-4
Irshad, A., Guo, H., Ur Rehman, S., Gu, J., Wang, C., Xiong, H., et al. (2022). Screening of induced mutants led to the identification of starch biosynthetic genes associated with improved resistant starch in wheat. Int. J. Mol. Sci. 23:10741. doi: 10.3390/ijms231810741
Jael, M., Paul, K. K., and Pascal, P. O. O. (2022). Identification of drought tolerant finger millet (Eleusine coracana) lines based on morpho-physiological characteristics and grain yield. Afr. J. Plant Sci. 16, 47–60. doi: 10.5897/AJPS2022.2225
Jain, S. M. (2010). Mutagenesis in crop improvement under the climate change. Rom Biotech. Lett. 15, 88–106.
Jankowicz-Cieslak, J., Mba, C., and Till, B. J. (2017a). “Mutagenesis for crop breeding and functional genomics,” in Biotechnologies for Plant Mutation Breeding, eds J. Jankowicz-Cieslak, T. H. Tai, J. Kumlehn, and B. J. Till (Cham: Springer International Publishing), 3–18. doi: 10.1007/978-3-319-45021-6_1
Jankowicz-Cieslak, J., Tai, T. H., Kumlehn, J., and Till, B. J. (2017b). Biotechnologies for Plant Mutation Breeding: Protocols, eds B. J. Till, T. H. Tai, and J. Kumlehn (Springer Nature). doi: 10.1007/978-3-319-45021-6
Jáquez-Gutiérrez, M., Atarés, A., Pineda, B., Angarita, P., Ribelles, C., García-Sogo, B., et al. (2019). Phenotypic and genetic characterization of tomato mutants provides new insights into leaf development and its relationship to agronomic traits. BMC Plant Biol. 19:141. doi: 10.1186/s12870-019-1735-9
Jency, J. P., Ravikesavan, R., Sumathi, P., and Raveendran, M. (2016). Determination of lethal dose and effect of physical mutagen on germination percentage and seedling parameters in kodomillet variety CO 3. Electron. J. Plant Breed. 7:1122. doi: 10.5958/0975-928X.2016.00155.1
Ji, Z., and Li, J. (2025). Ethyl methanesulfonate mutagenesis conditions of walnut seeds and preliminary construction of mutant library. Int. J. Agricult. Environ. Inform. Syst. 16, 1–13. doi: 10.4018/IJAEIS.373124
Johnson, J. B. (2020). An overview of Near-Infrared Spectroscopy (NIRS) for the detection of insect pests in stored grains. J. Stored Prod. Res. 86:101558. doi: 10.1016/j.jspr.2019.101558
Johnson, S. D., Taylor, D. R., and Tai, T. H. (2017). Field evaluation of mutagenized rice material. Biotechnol. Plant Mut. Breed. 144-156. doi: 10.1007/978-3-319-45021-6_9
Joshi, D. C., Meena, R. P., and Chandora, R. (2021). “Genetic resources: collection, characterization, conservation, and documentation,” in Millets and Pseudo Cereals. (Elsevier), 19–31. doi: 10.1016/B978-0-12-820089-6.00003-3
Kalaitzidis, A., Kadoglidou, K., Mylonas, I., Ghoghoberidze, S., Ninou, E., and Katsantonis, D. (2025). Investigating the impact of tillering on yield and yield-related traits in European rice cultivars. Agriculture 15:616. doi: 10.3390/agriculture15060616
Kalpande, H. V., Surashe, S. M., Badigannavar, A., More, A., and Ganapathi, T. R. (2021). Induced variability and assessment of mutagenic effectiveness and efficiency in sorghum genotypes [Sorghum bicolor (L.) Moench]. Int. J. Radiat. Biol. 98, 230–243. doi: 10.1080/09553002.2022.2003466
Kamenya, S. N., Mikwa, E. O., Song, B., and Odeny, D. A. (2021). Genetics and breeding for climate change in orphan crops. Theor. Appl. Genet. 134, 1787–1815. doi: 10.1007/s00122-020-03755-1
Kang, M. S. (2002). “Genotype-environment interaction: progress and prospects.,” in Quantitative genetics, genomics and plant breeding (UK: CABI Publishing), 221–243. doi: 10.1079/9780851996011.0221
Kantor, A., McClements, M. E., and MacLaren, R. E. (2020). CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21:6240. doi: 10.3390/ijms21176240
Keerthi, G. M., Mallikarjuna, M. G., Jha, S. K., Pandey, R., Veeraya, P., Lohithaswa, H. C., et al. (2025). Unravelling root system architecture plasticity in response to abiotic stresses in maize. Sci. Rep. 15:20433. doi: 10.1038/s41598-025-04123-z
Khakimov, B., Rasmussen, M. A., Kannangara, R. M., Jespersen, B. M., Munck, L., and Engelsen, S. B. (2017). From metabolome to phenotype: GC-MS metabolomics of developing mutant barley seeds reveals effects of growth, temperature and genotype. Sci. Rep. 7:8195. doi: 10.1038/s41598-017-08129-0
Khalil, F., Naiyan, X., Tayyab, M., and Pinghua, C. (2018). Screening of EMS-induced drought-tolerant sugarcane mutants employing physiological, molecular and enzymatic approaches. Agronomy 8:226. doi: 10.3390/agronomy8100226
Kharkwal, M. C. (2023). “History of plant mutation breeding and global impact of mutant varieties,” in Mutation Breeding for Sustainable Food Production and Climate Resilience, eds S. Penna and S. M. Jain (Singapore: Springer Nature), 25–55. doi: 10.1007/978-981-16-9720-3_2
King, R., Bird, N., Ramirez-Gonzalez, R., Coghill, J. A., Patil, A., Hassani-Pak, K., et al. (2015). Mutation scanning in wheat by exon capture and next-generation sequencing. PLoS ONE 10:e0137549. doi: 10.1371/journal.pone.0137549
Kosová, K., Vítámvás, P., Prášil, I. T., Klíma, M., and Renaut, J. (2021). Plant proteoforms under environmental stress: functional proteins arising from a single gene. Front. Plant Sci. 12:793113. doi: 10.3389/fpls.2021.793113
Kumar, M., Ahmad, A., Singh, M. K., Chouhan, S. K., Adhimoolam, P., Das, R., et al. (2025). The power of omics: genomics and proteomics approaches for crop improvement. Int. J. Adv. Biochem. Res. 9, 27–37. doi: 10.33545/26174693.2025.v9.i5Sa.4300
Kumar, S., Babu, C., Reddy, V., and Swathi, B. (2016). Anti-nutritional factors in finger millet. J. Nutr. Food Sci. 6:1000491. doi: 10.4172/2155-9600.1000491
Kumar, S., Modi, A. R., Parekh, M. J., Mahla, H. R., Sharma, R., Fougat, R. S., et al. (2017). Role of conventional and biotechnological approaches for genetic improvement of cluster bean. Ind. Crops Prod. 97, 639–648. doi: 10.1016/j.indcrop.2017.01.008
Lamichhane, S., and Thapa, S. (2022). Advances from conventional to modern plant breeding methodologies. Plant Breed. Biotechnol. 10, 1–14. doi: 10.9787/PBB.2022.10.1.1
Langyan, S., Bhardwaj, R., Radhamani, J., Yadav, R., Gautam, R. K., Kalia, S., et al. (2022). A quick analysis method for protein quantification in oilseed crops: a comparison with standard protocol. Front. Nutr. 9:892695. doi: 10.3389/fnut.2022.892695
Larrañaga, O., de Cózar, A., and Cossío, F. P. (2017). Mono-and di-alkylation processes of DNA bases by nitrogen mustard mechlorethamine. Chem. Phys. Chem. 18, 3390–3401. doi: 10.1002/cphc.201700937
le Roux, M.-S. L., Burger, N. F. V., Vlok, M., Kunert, K. J., Cullis, C. A., and Botha, A.-M. (2021). EMS derived wheat mutant BIG8-1 (Triticum aestivum L.)—a new drought tolerant mutant wheat line. Int. J. Mol. Sci. 22:5314. doi: 10.3390/ijms22105314
Lee, D. Y., Bowen, B. P., and Northen, T. R. (2010). Mass spectrometry—based metabolomics, analysis of metabolite-protein interactions, and imaging. Biotechniques 49, 557–565. doi: 10.2144/000113451
Li, F., Liu, Y., Zhang, X., Liu, L., Yan, Y., Ji, X., et al. (2022). Transcriptome and metabolome analyses reveals the pathway and metabolites of grain quality under phytochrome B in rice (Oryza sativa L.). Rice 15:52. doi: 10.1186/s12284-022-00600-5
Li, X., Song, Y., Century, K., Straight, S., Ronald, P., Dong, X., et al. (2001). A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 27, 235–242. doi: 10.1046/j.1365-313x.2001.01084.x
Li, X., Wan, Y., Wang, D., Li, X., Wu, J., Xiao, J., et al. (2025). Spatiotemporal transcriptomics reveals key gene regulation for grain yield and quality in wheat. Genome Biol. 26:93. doi: 10.1186/s13059-025-03569-8
Li, Y., Liang, J., Deng, B., Jiang, Y., Zhu, J., Chen, L., et al. (2023). Applications and prospects of CRISPR/Cas9-mediated base editing in plant breeding. Curr. Issues Mol. Biol. 45, 918–935. doi: 10.3390/cimb45020059
Liang, Y., Li, Z., Shi, J., Zhang, N., Qin, Z., Du, L., et al. (2025). Advances in hyperspectral imaging technology for grain quality and safety detection: a review. Foods 14:2977. doi: 10.3390/foods14172977
Liao, G.-Q., Tang, H.-M., Yu, Y.-D., Fu, L.-Z., Li, S.-J., and Zhu, M.-X. (2025). Mass spectrometry-based metabolomic as a powerful tool to unravel the component and mechanism in TCM. Chin. Med. 20:62. doi: 10.1186/s13020-025-01112-2
Liu, Y., Lu, S., Liu, K., Wang, S., Huang, L., and Guo, L. (2019). Proteomics: a powerful tool to study plant responses to biotic stress. Plant Methods 15:135. doi: 10.1186/s13007-019-0515-8
Liu, Y., Zhang, J., Yuan, H., Song, M., Zhu, Y., Cao, W., et al. (2022). Non-destructive quality-detection techniques for cereal grains: a systematic review. Agronomy 12:3187. doi: 10.3390/agronomy12123187
Luo, H., Pandey, M. K., Khan, A. W., Guo, J., Wu, B., Cai, Y., et al. (2019). Discovery of genomic regions and candidate genes controlling shelling percentage using seq approach in cultivated peanut (Arachis hypogaea L.). Plant Biotechnol. J. 17, 1248–1260. doi: 10.1111/pbi.13050
Lv, Y., Gu, L., Man, R., Liu, X., and Xu, J. (2024). Response of stomatal conductance, transpiration, and photosynthesis to light and CO2 for rice leaves with different appearance days. Front. Plant Sci. 15:1397948. doi: 10.3389/fpls.2024.1397948
Maia, M., Figueiredo, A., Cordeiro, C., and Sousa Silva, M. (2023). FT-ICR-MS-based metabolomics: a deep dive into plant metabolism. Mass Spectr. Rev. 42, 1535–1556. doi: 10.1002/mas.21731
Manjappa, G. M. V. C., Rangaiah, S., Sachin, S. J., and Sujay, V. (2024). A novel virescence mutant simplifies hybridization in finger millet breeding. Genet. Res. Crop Evol. 72, 2653–2664. doi: 10.1007/s10722-024-02112-1
Martínez-Fortún, J., Phillips, D. W., and Jones, H. D. (2022). Natural and artificial sources of genetic variation used in crop breeding: a baseline comparator for genome editing. Front. Genome Edit. 4:937853. doi: 10.3389/fgeed.2022.937853
Mathan, J., Singh, A., Jathar, V., and Ranjan, A. (2021). High photosynthesis rate in two wild rice species is driven by leaf anatomy mediating high Rubisco activity and electron transport rate. J. Exp. Bot. 72, 7119–7135. doi: 10.1093/jxb/erab313
Mawcha, K. T., Ndolo, D., Yang, W., and Babalola, O. O. (2024). Development of EMS mutagenized wheat mutant lines resistant to fusarium crown rot and fusarium head blight. Plant Breed Biotechnol. 12, 98–121. doi: 10.9787/PBB.2024.12.98
Mba, C. (2013). Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 3, 200–231. doi: 10.3390/agronomy3010200
Mishra, S., Kumar, S., and Srivastava, R. C. (2024). Genetic Improvement of Small Millets. Springer Nature. doi: 10.1007/978-981-99-7232-6
Mochida, K., Nishii, R., and Hirayama, T. (2020). Decoding plant–environment interactions that influence crop agronomic traits. Plant Cell Physiol. 61, 1408–1418. doi: 10.1093/pcp/pcaa064
Mohan Jain, S., and Suprasanna, P. (2011). Induced mutations for enhancing nutrition and food production. Gene Conserv. 10, 201-215.
Mohanta, R., Maiti, P., Sharangi, A. B., Roy, S., Hazra, S., Chakraborty, S., et al. (2025). Directed mutagenesis in fruit crops. Biotech. 3:15. doi: 10.1007/s13205-025-04268-8
Morgil, H., Gercek, Y. C., and Tulum, I. (2020). Single Nucleotide Polymorphisms (SNPs) in Plant Genetics and the Recent Topics Genetic Polymorphisms. doi: 10.5772/intechopen.91886
Mude, L. N., Mondam, M., Gujjula, V., Jinka, S., Pinjari, O. B., Panditi, V., et al. (2022). Morpho-physiological responses of finger millet genotypes to PEG-induced osmotic stress at an early seedling stage. Proceed. Nat. Acad. Sci. India Sec B Biol. Sci. 93, 337–350. doi: 10.1007/s40011-022-01421-8
Muduli, K. C., and Misra, R. C. (2007). Efficacy of mutagenic treatments in producing useful mutants in finger millet (Eleusine coracana Gaertn.). Indian J. Genet. Plant Breed. 67, 232–237.
Muduli, K. C., and Misra, R. C. (2008a). Genetic divergence analysis among micromutant lines in finger millet (Eleusine coracana G.). J. Crop Sci. Biotechnol. 11, 63–68.
Muduli, K. C., and Misra, R. C. (2008b). Induced polygenic variability in M2 generation and its relationship with production of high-yielding mutants in finger millet. Indian J. Genet. Plant Breed. 68, 419–425.
Naaz, N., Choudhary, S., and Hasan, N. (2024). “Proteomics application in plant breeding,” in Plant Molecular Breeding in Genomics Era: Concepts and Tools, eds J. M. Al-Khayri, K. P. Ingle, S. M. Jain, and S. Penna (Cham: Springer Nature Switzerland), 243–282. doi: 10.1007/978-3-031-68586-6_10
Nagana Gowda, G. A., and Raftery, D. (2021). “NMR-based metabolomics,” in Cancer Metabolomics: Methods and Applications, ed S. Hu (Springer), 19–37. doi: 10.1007/978-3-030-51652-9_2
Nagaraja, T. E., Bhat, S., and Nandini, C. (2022). Current scenario of crop improvement of finger millet [Eleusine coracana (L.)] in India: a review. Agricult. Rev. doi: 10.18805/ag.R-2545
Nair, V. D., and Devi, R. (2024). “Breeding finger millet for abiotic stress tolerance: strategies and challenges,” in Genetic improvement of Small Millets (Singapore: Springer Nature Singapore), 225–277. doi: 10.1007/978-981-99-7232-6_11
Nascimento, L. A., do, Simões, W. L., Oliveira, A. R., de, Salviano, A. M., Barros, J. R. A., Silva, W. O., et al. (2024). Productive performance of biomass sorghum (Sorghum bicolor (L.) Moench) and Cowpea (Vigna unguiculata (L.) Walp) cultivars in different cropping systems and planting times. Agronomy 14:1970. doi: 10.3390/agronomy14091970
Navami, M. M., Abraham, B., Archana, H., and Nisha, P. (2023). Nutritional profiling and quantitative analysis of amino acids and vitamins using in selected raw and germinated ancient grains. JSFA Rep. 3, 377–386. doi: 10.1002/jsf2.141
Nazarenko, M., Lykholat, Y., Grygoryuk, I., and Khromikh, N. (2018). Optimal doses and concentrations of mutagens for winter wheat breeding purposes. Part I. grain productivity. J. Cent. Eur. Agricult. 19, 194–205. doi: 10.5513/JCEA01/19.1.2037
Nazari, M., Kordrostami, M., Ghasemi-Soloklui, A. A., Eaton-Rye, J. J., Pashkovskiy, P., Kuznetsov, V., et al. (2024). Enhancing photosynthesis and plant productivity through genetic modification. Cells 13:1319. doi: 10.3390/cells13161319
Nezafatian, E., Farhadian, O., Daneshvar, E., and Bhatnagar, A. (2024). Investigating the effects of salinity and light stresses on primary and secondary metabolites of Tetraselmis tetrathele: total phenolic compounds, fatty acid profile, and biodiesel properties. Biomass Bioenergy 181:107050. doi: 10.1016/j.biombioe.2024.107050
Oladosu, Y., Rafii, M. Y., Abdullah, N., Hussin, G., Ramli, A., Rahim, H. A., et al. (2015). Principle and application of plant mutagenesis in crop improvement: a review. Biotechnol. Biotechnol. Equip. 30, 1–16. doi: 10.1080/13102818.2015.1087333
Olaolorun, B. M. (2020). Breeding wheat (Triticum Aestivum L.) for drought tolerance, improved yield and biomass allocation through chemical mutagenesis. (PhD Thesis). Pietermaritzburg: University of KwaZulu-Natal.
OlaOlorun, B. M., Shimelis, H., Laing, M., and Mathew, I. (2021). Development of Wheat (Triticum aestivum L.) Populations for Drought Tolerance and Improved Biomass Allocation Through Ethyl Methanesulphonate Mutagenesis. Frontiers in Agronomy 3. doi: 10.3389/fagro.2021.655820
Ozsolak, F., and Milos, P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12, 87–98. doi: 10.1038/nrg2934
Pandit, R., Bhusal, B., Regmi, R., Neupane, P., Bhattarai, K., Maharjan, B., et al. (2021). mutation breeding for crop improvement: a review. Rev. Food Agricult. 2, 31–35. doi: 10.26480/rfna.01.2021.31.35
Peirone, L. S., Pereyra Irujo, G. A., Bolton, A., Erreguerena, I., and Aguirrezábal, L. A. N. (2018). Assessing the efficiency of phenotyping early traits in a greenhouse automated platform for predicting drought tolerance of soybean in the field. Front. Plant Sci. 9:00587. doi: 10.3389/fpls.2018.00587
Pompelli, M. F., Seleiman, M. F., Dey, P., Suarez-Padrón, I. E., Zumaqu,é, L. E. O., Jarma-Orozco, A., et al. (2025). MALDI-ToF/ToF-MS detection of differential protein expression and metabolomic profiles in Jatropha curcas under salinity: advancing the understanding of salt stress mechanisms. J. Agron. Crop Sci. 211:70058. doi: 10.1111/jac.70058
Poorter, H., Fiorani, F., Pieruschka, R., Wojciechowski, T., van der Putten, W. H., Kleyer, M., et al. (2016). Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol. 212, 838–855. doi: 10.1111/nph.14243
Porker, K., Straight, M., and Hunt, J. R. (2020). Evaluation of G × E × M interactions to increase harvest index and yield of early sown wheat. Front. Plant Sci. 11:994. doi: 10.3389/fpls.2020.00994
Prasanna, S., and Jain, S. M. (2017). Mutant resources and mutagenomics in crop plants. Emir. J. Food Agric. 29:651. doi: 10.9755/ejfa.2017.v29.i9.86
Qiao, M., Hong, C., Jiao, Y., Hou, S., and Gao, H. (2024). Impacts of drought on photosynthesis in major food crops and the related mechanisms of plant responses to drought. Plants 13:1808. doi: 10.3390/plants13131808
Ramashia, S. E., Gwata, E. T., Meddows-Taylor, S., Anyasi, T. A., and Jideani, A. I. O. (2018). Some physical and functional properties of finger millet (Eleusine coracana) obtained in sub-Saharan Africa. Food Res. Int. 104, 110–118. doi: 10.1016/j.foodres.2017.09.065
Ramesh, M., Vanniarajan, C., Ravikesavan, A. K. E. A., and Mahendran, P. P. (2019). Mutagenic effectiveness and efficiency in barnyard millet (Echinochloa frumentacea) using physical, chemical and combination of mutagens. Electron. J. Plant Breed. 10:949. doi: 10.5958/0975-928X.2019.00122.4
Rao, M. J., Duan, M., Zhou, C., Jiao, J., Cheng, P., Yang, L., et al. (2025). Antioxidant defense system in plants: reactive oxygen species production, signaling, and scavenging during abiotic stress-induced oxidative damage. Horticulturae 11:477. doi: 10.3390/horticulturae11050477
Rathore, T., Singh, R., Kamble, D. B., Upadhyay, A., and Thangalakshmi, S. (2019). Review on finger millet: processing and value addition. Pharma Innov. J. 8, 283–291.
Rebecca, J., Peter, D., George, O., Patience, M., Paul, K., Maximilian, W., et al. (2018). Factors affecting the adoption of agricultural innovations on underutilized cereals: the case of finger millet among smallholder farmers in Kenya. Afr. J. Agric. Res. 13, 1888–1900. doi: 10.5897/AJAR2018.13357
Reddy, Y. A. N. (2020). Studies on photosynthetic rate, anatomical characters, and grain yield in finger millet genotypes. Curr. J. Appl. Sci. Technol. 31–39. doi: 10.9734/cjast/2020/v39i2330854
Rotasperti, L., Tadini, L., Chiara, M., Crosatti, C., Guerra, D., Tagliani, A., et al. (2022). The barley mutant happy under the sun 1 (hus1): an additional contribution to pale green crops. Environ. Exp. Bot. 196:104795. doi: 10.1016/j.envexpbot.2022.104795
Saeidi, M., Ahmadi, A., Moradi, F., and Hajirezaei, M.-R. (2024). Comparative metabolome profiling of two contrasting wheat cultivars in late-season water deficit. Front. Plant Physiol. 2:1386473. doi: 10.3389/fphgy.2024.1386473
Sahil, K. R., Patra, A., Mehta, S., Abdelmotelb, K. F., Lavale, S. A., et al. (2021). “Expression and regulation of stress-responsive genes in plants under harsh environmental conditions,” in Harsh Environment and Plant Resilience, ed A. Husen (Cham: Springer International Publishing), 25–44. doi: 10.1007/978-3-030-65912-7_2
Saini, S., Saxena, S., Samtiya, M., Puniya, M., and Dhewa, T. (2021). Potential of underutilized millets as nutri-cereal: an overview. J. Food Sci. Technol. 58, 4465–4477. doi: 10.1007/s13197-021-04985-x
Salgotra, R. K., and Chauhan, B. S. (2023). Genetic diversity, conservation, and utilization of plant genetic resources. Genes 14:174. doi: 10.3390/genes14010174
Samsami, H., and Maali-Amiri, R. (2024). Global insights into intermediate metabolites: signaling, metabolic divergence and stress response modulation in plants. Plant Physiol. Biochem. 213:108862. doi: 10.1016/j.plaphy.2024.108862
Sawarkar, H. A., and Mansoori, M. H. (2023). “Methodology for physical and chemical mutagenic treatments,” in Biotechnologies and Genetics in Plant Mutation Breeding (Apple Academic Press), 15–44. doi: 10.1201/9781003305064-2
Seerat, A., Aslam, M. A., Rafique, M. T., Chen, L., and Zheng, Y. (2025). Interplay between phytohormones and sugar metabolism in dendrocalamus latiflorus. Plants 14:305. doi: 10.3390/plants14030305
Sellapillai, L., Dhanarajan, A., Raina, A., and Ganesan, A. (2022). Gamma ray induced positive alterations in morphogenetic and yield attributing traits of finger millet (Eleusine coracana (L.) Gaertn.) in M2 generation. Plant Sci. Today doi: 10.14719/pst
Sen, A., Kecoglu, I., Ahmed, M., Parlatan, U., and Unlu, M. B. (2023). Differentiation of advanced generation mutant wheat lines: conventional techniques versus Raman spectroscopy. Front. Plant Sci. 14:1116876. doi: 10.3389/fpls.2023.1116876
Serpico, D. (2020). Beyond quantitative and qualitative traits: three telling cases in the life sciences. Biol. Philos.35. doi: 10.1007/s10539-020-09750-6
Sestili, F., Botticella, E., Bedo, Z., Phillips, A., and Lafiandra, D. (2009). Production of novel allelic variation for genes involved in starch biosynthesis through mutagenesis. Mol. Breed. 25, 145–154. doi: 10.1007/s11032-009-9314-7
Shahwar, D., Ahn, N., Kim, D., Ahn, W., and Park, Y. (2023). Mutagenesis-based plant breeding approaches and genome engineering: a review focused on tomato. Mut. Res. Rev. Mut. Res. 792:108473. doi: 10.1016/j.mrrev.2023.108473
Shamshad, A., Rashid, M., Jankuloski, L., Ashraf, K., Sultan, K., Alamri, S., et al. (2023). Effect of ethyl methanesulfonate mediated mutation for enhancing morpho-physio-biochemical and yield contributing traits of fragrant rice. Peer J. 11:e15821. doi: 10.7717/peerj.15821
Shao, X., Yang, X., Xu, S., Li, H., and Jitendra, P. (2020). Detection of the growth stage of rice weevil as a stored-grain pest based on soft X-ray imaging. Trans. Chin. Soc. Agric. Eng. 36, 309–314. doi: 10.3390/foods12132484
Sharma, D., Tiwari, A., Sood, S., Jamra, G., Singh, N. K., Meher, P. K., et al. (2018). Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS ONE 13:e0199444. doi: 10.1371/journal.pone.0199444
Sharma, V., Thakur, M., Maan, S. S., Verma, K., Thakur, A., and Penna, S. (2024). In Vitro mutagenesis: a non-invasive technology for effective crop improvement to assure food and nutritional security—current trends, advancements and future perspectives. J. Plant Growth Regul. 44, 484–507. doi: 10.1007/s00344-024-11484-8
Shavrukov, Y., Kurishbayev, A., Jatayev, S., Shvidchenko, V., Zotova, L., Koekemoer, F., et al. (2017). Early flowering as a drought escape mechanism in plants: how can it aid wheat production? Front. Plant Sci. 8:01950. doi: 10.3389/fpls.2017.01950
Sikora, P., Chawade, A., Larsson, M., Olsson, J., and Olsson, O. (2011). Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genomics 113:314829. doi: 10.1155/2011/314829
Singhal, T., Satyavathi, C. T., Singh, S. P., Kumar, A., Sankar, S. M., Bhardwaj, C., et al. (2021). Multi-environment quantitative trait loci mapping for grain iron and zinc content using bi-parental recombinant inbred line mapping population in pearl millet. Front. Plant Sci. 12:659789. doi: 10.3389/fpls.2021.659789
Soga, T. (2023). Advances in capillary electrophoresis mass spectrometry for metabolomics. TrAC Trends Anal. Chem. 158:116883. doi: 10.1016/j.trac.2022.116883
Sun, Y., Yang, R., Liu, M., Liu, Y., Yuan, X., Chen, L., et al. (2025). CRISPR/Cas9-mediated knockout of PvTCP19/22 enhances tiller number and biomass yield in switchgrass. Ind Crops Prod. 226:120689. doi: 10.1016/j.indcrop.2025.120689
Suthakar, V., and Mullainathan, L. (2015). Studies on effect of physical and chemical mutagens in Sorghum (Sorghum bicolor (L.) Moench) in M2 Generation. Int. Lett. Nat. Sci. 37, 46–50. doi: 10.56431/p-fsymt9
Tachioka, M., Sugimoto, N., Nakamura, A., Sunagawa, N., Ishida, T., Uchiyama, T., et al. (2016). Development of simple random mutagenesis protocol for the protein expression system in Pichia pastoris. Biotechnol. Biofuels 9:199. doi: 10.1186/s13068-016-0613-z
Tan, M., Cheng, D., Yang, Y., Zhang, G., Qin, M., Chen, J., et al. (2017). Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 17:194. doi: 10.1186/s12870-017-1143-y
Temesgen, B. (2021). Role and economic importance of crop genetic diversity in food security. Int. J. Agricult. Sci. Food Technol.7, 164–169. doi: 10.17352/2455-815X.000104
Thangwana, A., Gwata, E. T., and Zhou, M. M. (2021). Impact of chemical mutagenesis using ethyl methane sulphonate on tepary bean seedling vigour and adult plant performance. Heliyon 7:e06103. doi: 10.1016/j.heliyon.2021.e06103
Tholen, D., Ethier, G., Genty, B., Pepin, S., and Zhu, X. (2012). Variable mesophyll conductance revisited: theoretical background and experimental implications. Plant Cell Environ. 35, 2087–2103. doi: 10.1111/j.1365-3040.2012.02538.x
Tripathi, T., Tanwar, H., Pareek, S., and Pheirim, R. (2025). Harnessing breeding and genomic advances for nutritional and climate-resilient minor millets. J. Adv. Biol. Biotechnol. 28, 622–632. doi: 10.9734/jabb/2025/v28i42222
Tsago, Y., Andargie, M., and Takele, A. (2014). In vitro selection of sorghum (Sorghum bicolor (L) Moench) for polyethylene glycol (PEG) induced drought stress. Plant Sci. Today 1, 62–68. doi: 10.14719/pst.2014.1.2.14
Turfan, N. (2025). Revealing the impact of L-cysteine and L-phenylalanine supplements on the yield, nutritional quality and storage of garlic. J. Plant Nutr. 48, 1742–1763. doi: 10.1080/01904167.2025.2459255
Türkoglu, A., Tosun, M., and Haliloglu, K. (2022). Evaluation of ethyl methanesulfonate-induced in vitro mutagenesis, polymorphism and genomic instability in wheat (Triticum aestivum L.). J. Crop Sci. Biotechnol. 26, 199–213. doi: 10.1007/s12892-022-00172-2
Uauy, C., Paraiso, F., Colasuonno, P., Tran, R. K., Tsai, H., Berardi, S., et al. (2009). A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9:115. doi: 10.1186/1471-2229-9-115
Unkovich, M., Baldock, J., and Forbes, M. (2010). “Variability in Harvest Index of Grain Crops and Potential Significance for Carbon Accounting,” Advances in Agronomy (Elsevier),173–219. doi: 10.1016/S0065-2113(10)05005-4
Upadhyay, R., Saini, R., Shukla, P. K., and Tiwari, K. N. (2024). Role of secondary metabolites in plant defense mechanisms: a molecular and biotechnological insights. Phytochem. Rev. 24, 953–983. doi: 10.1007/s11101-024-09976-2
Urban, J., Ingwers, M., McGuire, M. A., and Teskey, R. O. (2017). Stomatal conductance increases with rising temperature. Plant Sign. Behav. 12:e1356534. doi: 10.1080/15592324.2017.1356534
Valenzuela-Antelo J. L. Benitez-Riquelme I. Vargas-Hernandez M. Huerta-Espino J. Bentley A. R. Villaseñor-Mir H. E. . (2023). Multi-location trials identify stable high-yielding spring bread and durum wheat cultivars in Mexico. Crop Sci. 63, 2103–2114. doi: 10.1002/csc2.20993
Vasisth, P., Rangaiah, S., Sharma, M., Balar, V., and Chittora, V. (2023). Induced polygenic variability for identification of high yielding mutants in M 3-M 4 generations of finger millet (Eleusine coracana L. Gaertn). Electron. J. Plant Breed. 14, 511–520. doi: 10.37992/2023.1402.057
Verma, S. K., Mittal, S., Gayacharan, W.ankhede, D. P., Parida, S. K., Chattopadhyay, D., et al. (2022). Transcriptome analysis reveals key pathways and candidate genes controlling seed development and size in ricebean (Vigna umbellata). Front. Genet. 12:791355. doi: 10.3389/fgene.2021.791355
Waghmode, B. D., Sable, P. S., Sonone, N. G., and Burondkar, M. M. (2020). Genetical studies of mutant lines in M3 generation of finger millet (Eleusine coracana (L.) Gaertn). Int. J. Curr. Microbiol. Appl. Sci. 9, 1833–1844. doi: 10.20546/ijcmas.2020.903.212
White, C. A., Sylvester-Bradley, R., and Berry, P. M. (2015). Root length densities of UK wheat and oilseed rape crops with implications for water capture and yield. J. Exp. Bot. 66, 2293–2303. doi: 10.1093/jxb/erv077
Wolter, F., Schindele, P., and Puchta, H. (2019). Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19:176. doi: 10.1186/s12870-019-1775-1
Wright, H., and Devos, K. M. (2024). Finger millet: a hero in the making to combat food insecurity. Theor. Appl. Genet. 137. doi: 10.1007/s00122-024-04637-6
Xiao, Y., Khangura, R. S., Wang, Z., Dilkes, B. P., and Eveland, A. L. (2025). Targeted seed EMS mutagenesis reveals a basic helix–loop–helix transcription factor underlying male sterility in sorghum. Genetics 230:iyaf017. doi: 10.1093/genetics/iyaf017
Xie, X. J. I. N., LI, B. I. N. B. A. I., and Shen, S. H. E. (2012). Impact of high temperature stress on photosynthetic characteristic and yield of rice (Oryza sativa) at heading. Indian J. Agricult. Sci. 82, 516–522. doi: 10.56093/ijas.v82i6.19014
Xiong, H., Guo, H., Fu, M., Xie, Y., Zhao, L., Gu, J., et al. (2023). A large-scale whole-exome sequencing mutant resource for functional genomics in wheat. Plant Biotechnol. J. 21, 2047–2056. doi: 10.1111/pbi.14111
Xiong, H., Zhou, C., Guo, H., Xie, Y., Zhao, L., Gu, J., et al. (2020). Transcriptome sequencing reveals hotspot mutation regions and dwarfing mechanisms in wheat mutants induced by γ-ray irradiation and EMS. J. Radiat. Res. 61, 44–57. doi: 10.1093/jrr/rrz075
Yali, W., and Mitiku, T. (2022). Mutation breeding and its importance in modern plant breeding. J. Plant Sci. 10:64. doi: 10.11648/j.jps.20221002.13
Yu, S., and Tian, L. (2018). Breeding major cereal grains through the lens of nutrition sensitivity. Mol. Plant 11, 23–30. doi: 10.1016/j.molp.2017.08.006
Zhang, C., Si, Y., Lamkey, J., Boydston, R. A., Garland-Campbell, K. A., and Sankaran, S. (2018). High-throughput phenotyping of seed/seedling evaluation using digital image analysis. Agronomy 8:63. doi: 10.3390/agronomy8050063
Zhang, F., Rosental, L., Ji, B., Brotman, Y., and Dai, M. (2024). Metabolite-mediated adaptation of crops to drought and the acquisition of tolerance. Plant J. 118, 626–644. doi: 10.1111/tpj.16634
Zhang, L., Zhang, F., Du, W., Hu, M., Hao, Y., Ding, S., et al. (2025). Machine learning analysis of maize seedling traits under drought stress. Biology 14:787. doi: 10.3390/biology14070787
Zhang, S., Ghatak, A., Bazargani, M. M., Bajaj, P., Varshney, R. K., Chaturvedi, P., et al. (2021). Spatial distribution of proteins and metabolites in developing wheat grain and their differential regulatory response during the grain filling process. Plant J. 107, 669–687. doi: 10.1111/tpj.15410
Zhou, X., Liu, L., Li, Y., Li, K., Liu, X., Zhou, J., et al. (2020). Integrative metabolomic and transcriptomic analyses reveal metabolic changes and its molecular basis in rice mutants of the strigolactone pathway. Metabolites 10:425. doi: 10.3390/metabo10110425
Zhu, M., He, Q., Lyu, M., Shi, T., Gao, Q., Zhi, H., et al. (2023). Integrated genomic and transcriptomic analysis reveals genes associated with plant height of foxtail millet. Crop J. 11, 593–604. doi: 10.1016/j.cj.2022.09.003
Zimmermann, A., Prieto-Vivas, J. E., Voordeckers, K., Bi, C., and Verstrepen, K. J. (2024). Mutagenesis techniques for evolutionary engineering of microbes-exploiting CRISPR-Cas, oligonucleotides, recombinases, and polymerases. Trends Microbiol. 32, 884–901. doi: 10.1016/j.tim.2024.02.006
Zuge, S. S., Sawardekar, S. V., Bhave, S. G., Gokhale, N. B., Dhekale, J. S., Mahadik, S. G., et al. (2017). Study of M4 and M5 generations of finger millet (Eleusine coracana L. Gaertn) and quality analysis of promising mutants. Environ. Ecol. 35, 2266–2270.
Keywords: finger millet, genetic diversity, physical mutagens, chemical mutagens, grain yield
Citation: Mutanda M, Figlan S, Shargie NG and Gwata ET (2025) Mutation breeding: an underutilized strategy for improving finger millet productivity and nutritional quality. Front. Sustain. Food Syst. 9:1679819. doi: 10.3389/fsufs.2025.1679819
Received: 05 August 2025; Accepted: 10 September 2025;
Published: 01 October 2025.
Edited by:
Afiya John, The University of the West Indies St. Augustine, Trinidad and TobagoReviewed by:
Muhammad Zayed, Menoufia University, EgyptNeethu Francis, Kansas State University, United States
Faizo Kasule, Iowa State University, United States
Copyright © 2025 Mutanda, Figlan, Shargie and Gwata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maltase Mutanda, MTkyODM4NDlAbXlsaWZlLnVuaXNhLmFjLnph
†ORCID: Maltase Mutanda orcid.org/0009-0008-3881-5834
Sandiswa Figlan orcid.org/0000-0003-0395-8612
Nemera G. Shargie orcid.org/0000-0003-2590-0999
Eastonce T. Gwata orcid.org/0000-0001-6282-7650
 Maltase Mutanda
Maltase Mutanda Sandiswa Figlan
Sandiswa Figlan Nemera G. Shargie
Nemera G. Shargie Eastonce T. Gwata
Eastonce T. Gwata