- 1Orient Science & Technology College of Hunan Agricultural University, Changsha, China
- 2Yuelushan Laboratory, Changsha, China
- 3Hunan Crop Research Institute, Hunan Academy of Agricultural Sciences, Changsha, Hunan, China
- 4College of Agronomy, Hunan Agricultural University, Changsha, China
Introduction: Agricultural land degradation threatens food security and agricultural ecosystem sustainability, necessitating phytoremediation to address this problem. Ramie (Boehmeria nivea L.), a cash crop known for its resilience in marginal environments and multiple ecological benefits, represents a promising candidate for this purpose. However, lack of varieties tolerant to poor soil limits this potential, necessitating genetic improvement.
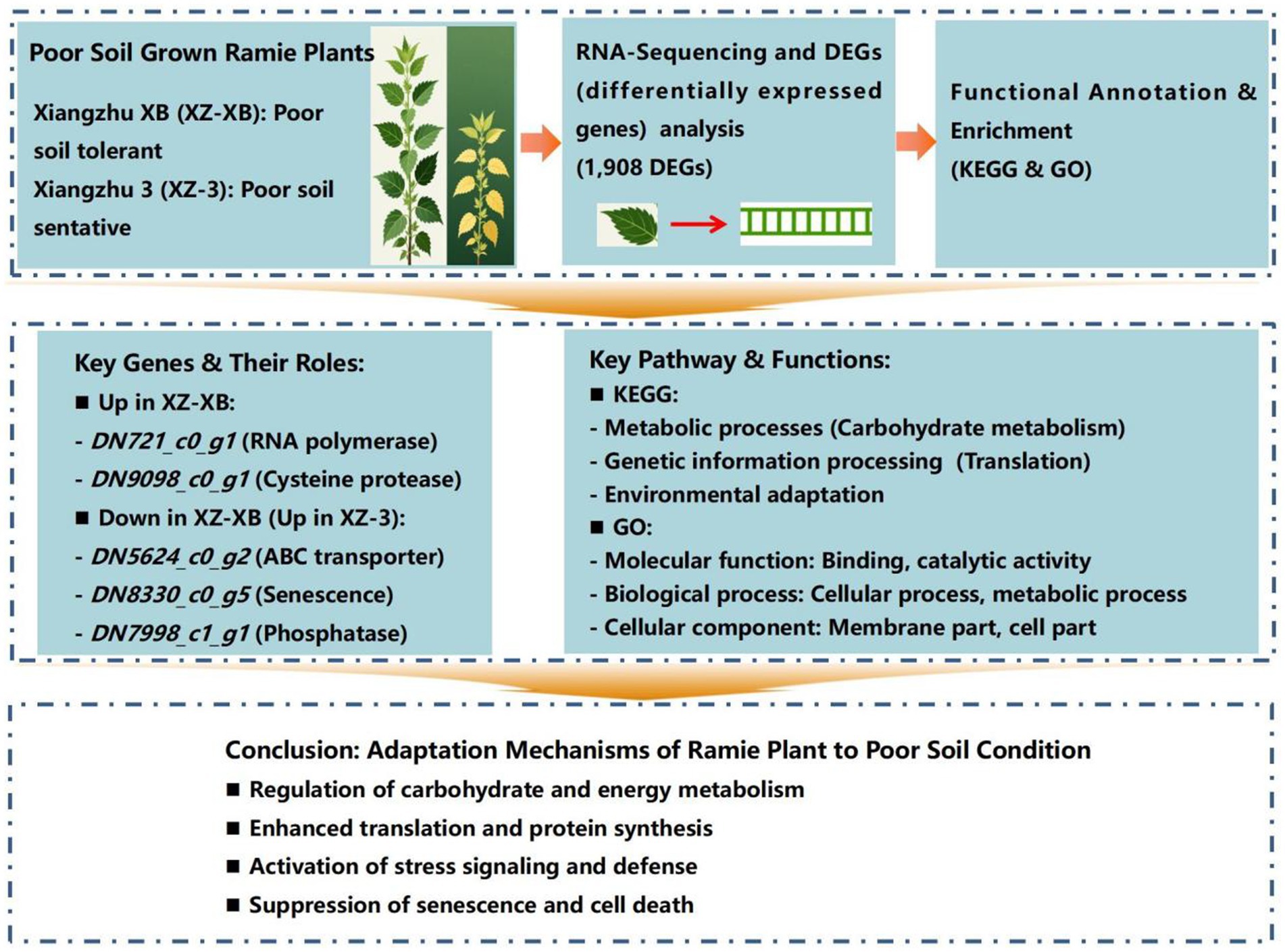
Methods: This study was therefore designed to identify key genes involved in ramie’s adaptation to poor soil conditions and to further explore the underlying molecular mechanisms. Leaf RNA from two ramie varieties, the tolerant Xiangzhu XB (XZ-XB) and the sensitive Xiangzhu 3 (XZ-3), was analyzed using high-throughput sequencing.
Results: After processing high-quality clean data, comparative transcriptome analysis revealed 1,908 differentially expressed genes (DEGs) between XZ-XB and XZ-3, among which 1,116 were up-regulated and 792 were down-regulated in XZ-XB relative to XZ-3. Notably, four up-regulated DEGs displayed fold changes greater than 9,500, while four down-regulated DEGs showed fold changes exceeding 1,000. Functional annotation linked the DEGs to critical processes as transporter activity, proteases regulation, purple acid phosphatase activity, etc. The findings also revealed that tolerant genotype likely enhance survival under poor soil condition by down-regulating senescence-promoting genes and up-regulating stress-signaling pathways.
Discussion: Results of this study provide valuable genetic resources and candidate targets for molecular breeding of ramie varieties with enhanced resilience to nutrient-poor soils. By the help of molecular breeding, the process will be speed up to develop nutrients-deficiency resilient ramie varieties as a sustainable, plant-based strategy for restoring degraded agricultural ecosystems and enhancing land productivity.
1 Introduction
Agricultural land degradation poses a significant threat to global food security and ecosystem sustainability. More than 60% of the world’s agricultural land is already degraded (Pravalie et al., 2021), a situation exacerbated by intensive farming, climate change, and industrial pollution (Talukder et al., 2021; Chaudhuri et al., 2023). Degraded soils are typically characterized by nutrient depletion, heavy metal contamination, structural deterioration (e.g., soil compaction), and loss of organic matter (Hossain et al., 2020). These characteristics, acting as environmental stress factors, ultimately undermine both crop productivity and environmental health. As the contradiction between population growth and reduced grain yield caused by soil degradation intensifies, along with the growing conflict between people’s pursuit of high-quality food and the decline in food quality due to soil degradation, the restoration and fertility improvement of degraded soils have become increasingly urgent (Mosier et al., 2021).
Currently, a comprehensive set of techniques has been developed to manage degraded cropland, targeting various degradation situations. For instance, application of some amendments has been confirmed effective in treating saline-alkali degraded land (Xu et al., 2023; Li et al., 2025). Similarly, the application of soil conditioners and lime to adjust pH has proven beneficial in addressing degradation in red soil cropland (Li et al., 2024; He et al., 2025). Furthermore, for commonly encountered degraded croplands, degradation can be halted through strategies such as rational crop rotation and intercropping, the cultivation of green manure and cover crops, and the promotion of conservation tillage (Hussain et al., 2021; Perveen et al., 2025). These techniques have proven effective in ameliorating lightly to moderately degraded soils. However, their efficacy is limited in severely degraded soils. In such settings, phytoremediation has emerged as a preferred strategy due to its cost-effectiveness, ease of implementation, and multifunctional ecological benefits (Phang et al., 2024). However, the widespread adoption of phytoremediation is constrained by the lack of plant species that combine strong adaptability, high remediation efficiency, and particularly the ability to generate economic returns (Lee et al., 2025).
Ramie (Boehmeria nivea L.), a perennial fiber crop known for its resilience in marginal environments and multiple ecological benefits, is a promising candidate for the phytoremediation and sustainable utilization of degraded lands (Wu et al., 2022; Chen et al., 2023). The well-developed underground system (deep root, extensive rhizomes/radish roots) allows it to improve soil structure after planting, and decomposition of these biomass can contribute to an increase of soil organic carbon content (Zhu et al., 2021; Liu, 2022). Furthermore, the soil stabilization provided by these extensive underground systems, coupled with the rainwater interception by the high-coverage leaves, gives ramie cultivation excellent potential for preventing soil erosion (Wang et al., 2021). Additionally, ramie exhibits remarkable tolerance to multiple heavy metals as Cd, Mn, and Cr (Wu et al., 2022; An et al., 2023). Coupled with its well-established processing technologies for non-food applications (such as in textiles), this makes it a key plant species for the utilization and remediation of heavy metal-contaminated soils. Despite these advantages, most existing ramie varieties have been bred for multiple annual harvests, generally exhibiting high soil nutrient requirements. When applied to the remediation of degraded soils, these varieties tend to accelerate the decline of soil fertility. Moreover, these high nutrient-demand varieties limit the remediation potential of ramie in poor, degraded soil scenarios. Therefore, breeding nutrient-efficient ramie varieties is essential to fully realize the crop’s potential in marginal environments.
Molecular breeding offers a powerful approach to accelerate the development of such improved varieties. Although preliminary studies (Tan, 2015; Zheng, 2014; Hou, 2018) have identified genes (such as BnNRT1.1 and the BnGS gene family) involved in ramie’s tolerance to nitrogen, phosphorus, and potassium deficiencies, these findings remain fragmented. A systematic understanding of the molecular mechanisms underlying ramie’s adaptation to poor soil is still lacking, hindering the development of targeted breeding strategies.
As a highly heterozygous species with an unannotated genome, ramie presents challenges for genomic studies. For such species, transcriptome sequencing technology is an efficient and cost-effective means for functional gene discovery, and it has been widely used, particularly for screening key genes involved in stress responses. Therefore, this study employed a comparative transcriptomic analysis of two ramie varieties with contrasting adaptability to poor soil. Our objectives were to identify critical genes and regulatory pathways associated with nutrient stress tolerance, (1) uncover the molecular basis of ramie’s adaptation to poor soil, and (2) provide genetic resources and candidate genes for molecular breeding aimed at enhancing ramie’s resilience in degraded environments.
2 Materials and methods
2.1 Experimental materials and sampling methods
Based on long-term screening results for poor soil tolerance germplasm, Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3) were identified as tolerant and sensitive materials, respectively, under nutrient-deficient soil conditions (Wu et al., 2021a; Wu et al., 2021b). XZ-XB’s tolerance stems from its strong root system, efficient nutrient retranslocation to roots, conservative aboveground nutrient use, and supportive rhizosphere microbiome. In contrast, XZ-3’s sensitivity is due to its weaker root system, inefficient nutrient cycling, higher nutrient loss during harvest, and less favorable microbial community. These traits collectively explain their contrasting adaptability to nutrient-deficient soils. Utilizing these two genotypes, we conducted a comprehensive transcriptomic analysis to compare their gene expression profiles, with the aim of identifying key regulatory genes associated with ramie’s tolerance to poor soil. Sampling was carried out in early December 2020 from the long-term impoverished soil experimental base of the Ramie Research Institute of Hunan Agricultural University in Huarong (29°32ʹ46ʹʹN, 112°39ʹ57ʹʹE, 73 m a.s.l.). This experiment was established at 2010 on a poor sandy red soil, which is characterized to have a total nitrogen of 0.69 g kg−1, available phosphorus of 9.62 mg kg−1 and exchangeable potassium of 56.53 mg kg−1. From each of the three biological replicate plots per variety, three normal and disease-free plants per replicate were selected and pooled to form one biological replicate. Young leaves were collected, washed, surface-sterilized with 75% ethanol, and approximately 100 g of tissue was immediately frozen in liquid nitrogen for subsequent RNA extraction.
2.2 High-throughput sequencing workflow
Total RNA was extracted from the collected samples using the TRIzol Plant RNA Extraction Kit, followed by concentration, purity, and integrity detection. After passing quality checks, the mRNA was enriched using Oligo dT, fragmented, reverse-transcribed into cDNA, and adaptors were ligated. Finally, sequencing was performed on the Illumina Novaseq 6,000 platform provided by Majorbio Bio-pharm Technology Co., Ltd.
2.3 Analysis of the sequencing data
The raw sequencing reads were subjected to quality control inspection and filtering to generate clean data. Subsequent analyses were then performed using the Majorbio Cloud Platform.1 On this platform, the clean data from each sample were assembled using the Trinity software. Differential expression analysis was conducted with the DESeq2 software, with screening thresholds set at |log2FC| ≥ 1 and padj < 0.05, where FC represents the fold change. Functional annotation of the differentially expressed genes was conducted by aligning the assembled transcript sequences against six major databases: NR (Non-Redundant Protein Database), Swiss-Prot (Annotated Protein Sequence Database), Pfam (Protein Family Database), COG (Clusters of Orthologous Groups), GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes). Annotation results from each database were compiled and statistically summarized.
2.4 Real-time fluorescence quantitative PCR (qRT-PCR) detection
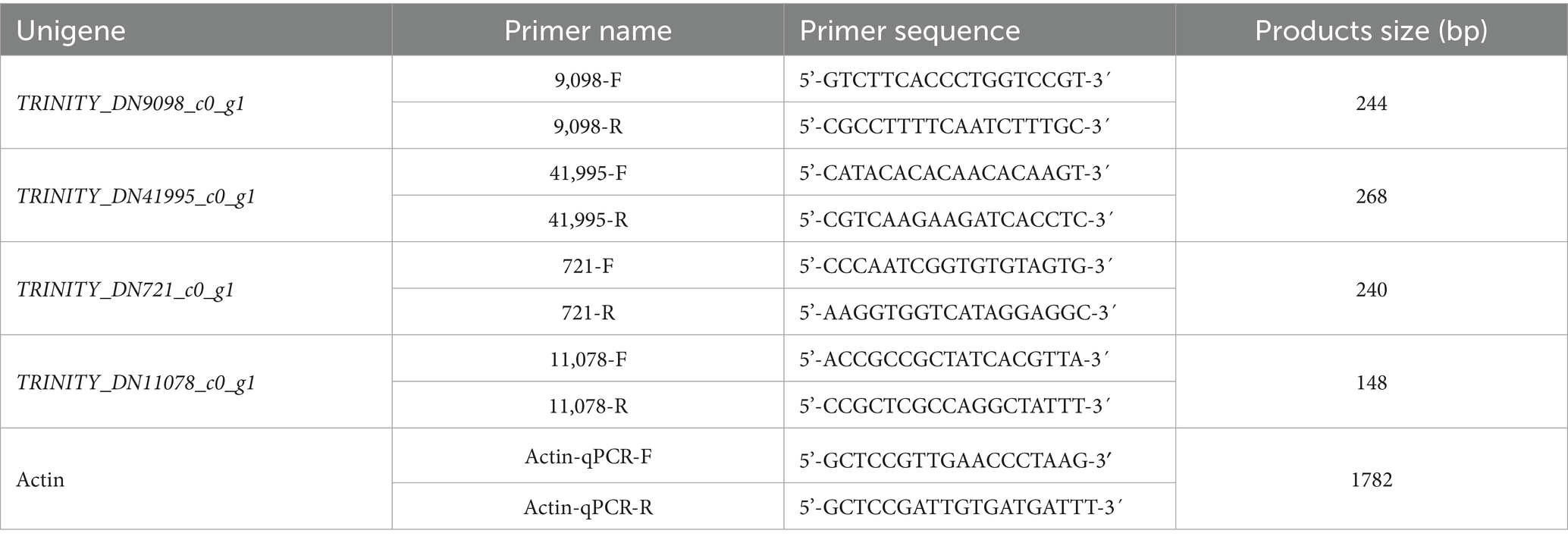
To verify the reliability of the transcriptome sequencing results, four genes with significantly differential expression levels were selected for qRT-PCR detection to analyze their expression patterns. The same batch of RNA used for transcriptome sequencing was employed for qRT-PCR. The ramie actin gene (Actin) was used as the internal reference gene, and the Applied Biosystems™ PowerUp™ SYBR™ Green Master Mix (Thermo Fisher, Shanghai, China) was used to prepare the reaction system according to the manufacturer’s instructions. The reaction system was processed using an ABI 7300 Real-Time PCR System (Applied Biosystems, USA). Primers for each target gene were designed using Primer 3 software (as shown in Table 1), while the internal reference gene primers were referenced from the sequence provided in Ma’s study (Ma et al., 2017). The PCR amplification program was as follows: pre-denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30s, annealing at 55 °C for 30s, and extension at 72 °C for 30s. After completion, the relative expression levels of the target genes were calculated using the 2-ΔΔCt method.
3 Results
3.1 Transcriptome sequencing quality analysis and unigene functional annotation
This study obtained a total of 38.08 Gb clean data from the six samples of XZ-XB and XZ-3, with each sample yielding over 6.18 Gb of clean data. All samples exhibited Q20 values of >97% and Q30 values of >93%, indicating that the data quality met the requirements for subsequent analyses. After assembly and quality evaluation of all high-quality reads using Trinity, 88,861 transcripts and 52,759 unigenes were obtained, with N50 values of 1,827 and 1,824, respectively. The length of the unigenes ranged from 201 bp to 13,439 bp, with an average length of 934.23 bp.
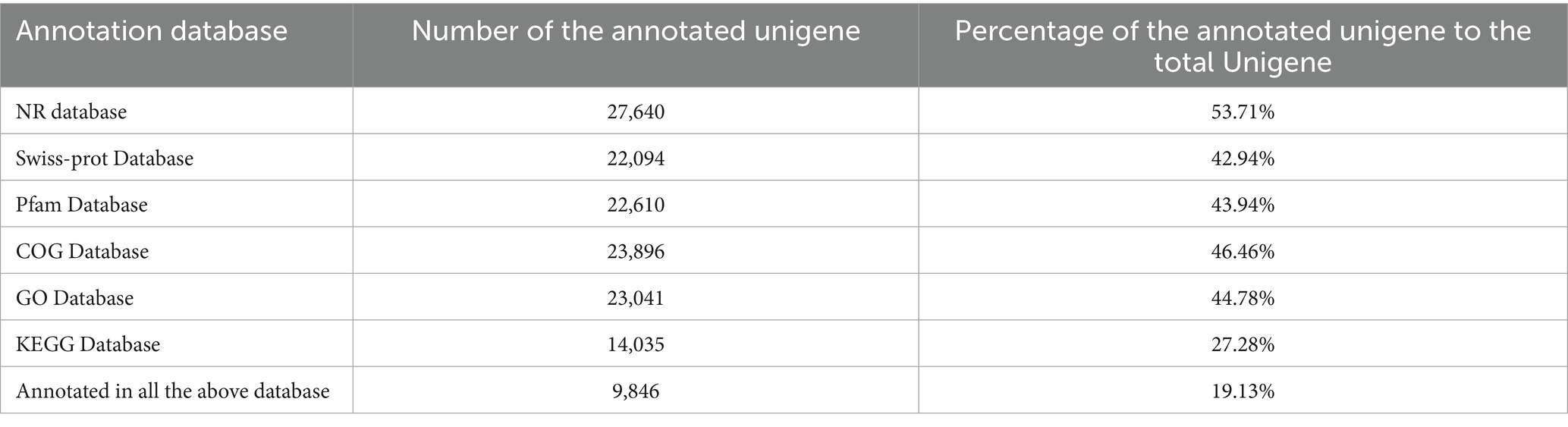
The assembled unigenes were aligned against six major databases (NR, Swiss-Prot, Pfam, COG, GO, and KEGG) using BLAST software to retrieve functional annotations. As shown in Table 2, among these databases, the NR database had the highest annotation rate, with 27,640 unigenes annotated, accounting for 53.71% of the total. In contrast, the KEGG database had the lowest annotation rate of 27.28%, with only 14,035 unigenes annotated. The annotation rates in the other four databases were relatively consistent (approximately 45%), which correspond to about 23,000 annotated unigenes each.
Integrative analysis of the annotation results from all six databases revealed that 9,846 unigenes were co-annotated across all databases, representing 19.13% of the total unigenes. These results indicate that the functional annotation rate of ramie unigenes remains relatively low.
3.2 Analysis of differential gene expression patterns
Differential gene expression between XZ-XB and XZ-3 under poor soil conditions was analyzed using DESeq2 (results shown in Figure 1). Using the criteria of FC ≥ 2 and FDR (False Discovery Rate) ≤ 0.01, a total of 1,908 differentially expressed genes (DEGs) with significant (p < 0.05) expression differences were identified. Among these, 1,116 genes were up-regulated and 792 were down-regulated in XZ-XB compared to XZ-3. In the up-regulated DEGs, DN721_c0_g1, DN9098_c0_g1, DN7103_c0_g1, and DN11078_c0_g1 exhibited expression levels in XZ-XB approximately 10,000 times higher than those in Xiangzhu 3, specifically 17,508-fold, 12,396-fold, 11,266-fold, and 9,735-fold, respectively. Among the down-regulated DEGs, DN41995_c0_g1, DN5624_c0_g2, DN8330_c0_g5, and DN7998_c1_g1 showed expression levels in XZ-3 only about 1/1000 of those in XZ-XB, specifically 1/2793, 1/1630, 1/1218, and 1/1006-fold, respectively (Table 3).
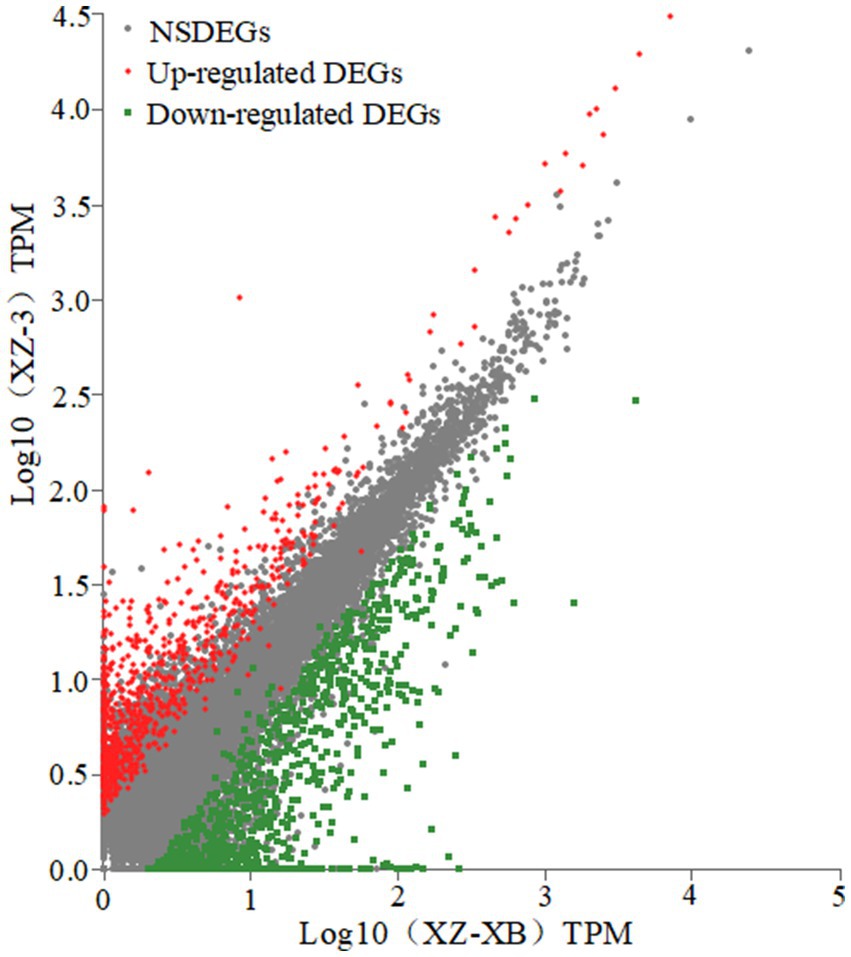
Figure 1. Overview of the differentially expressed unigenes (DEGs) in poor soil grown Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3), which is characterized to have the strongest and weakest adaptability to poor soil, respectively. Gray dots represent the non-significantly differentially expressed genes (NSDEGs).
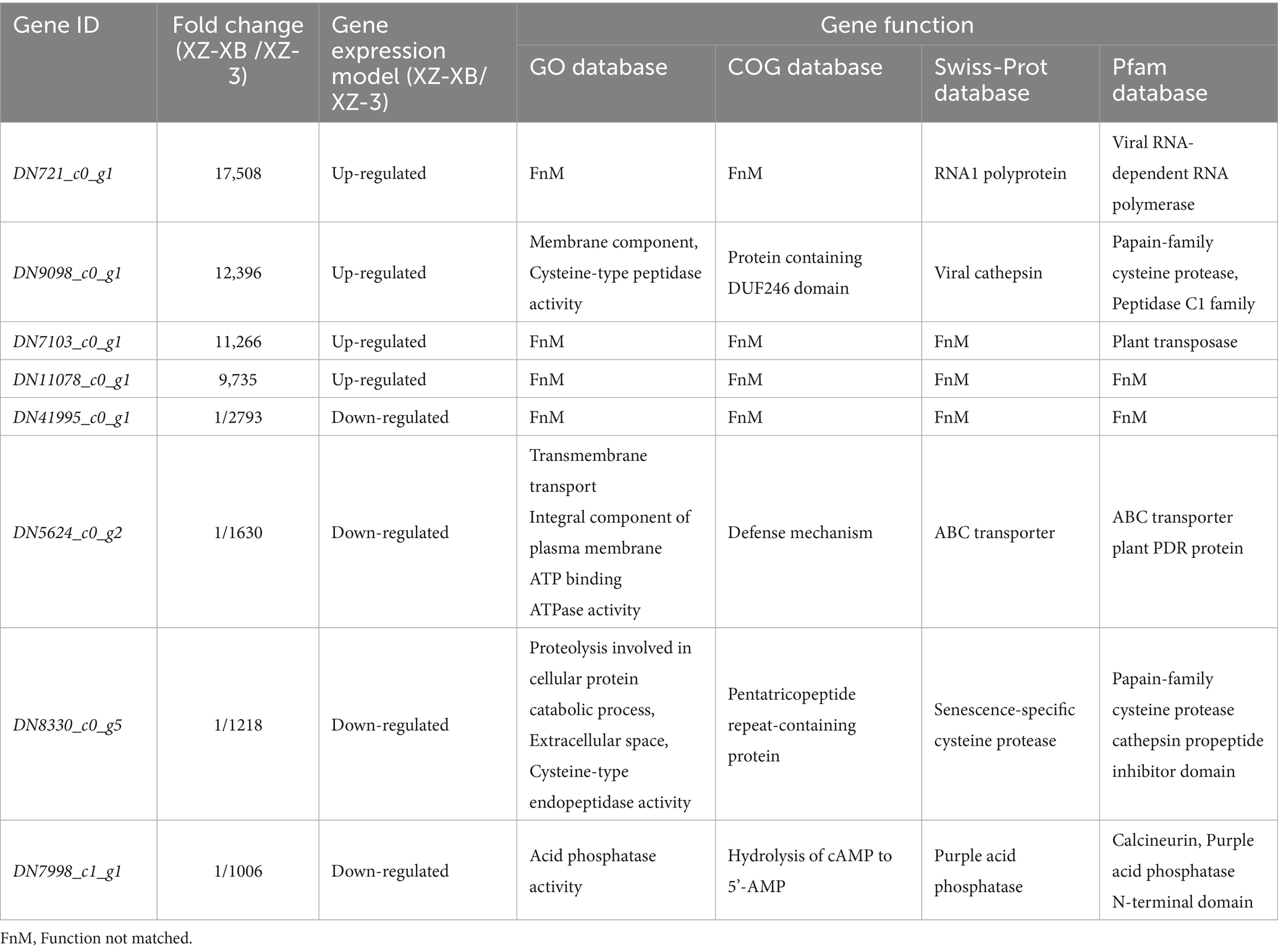
Table 3. The first and last top four different expressed genes between Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3) under poor field soil condition.
3.3 Functional annotation of differentially expressed unigenes
Table 3 presents the functional annotations of the top four unigenes with the most significant differential expression between XZ-XB and XZ-3. Among the significantly up-regulated genes in XZ-XB compared to XZ-3, only DN9098_c0_g1 has a relatively well-defined function, primarily associated with cysteine protease activity in plants. The functions of the other three genes remain largely uncharacterized. The gene of DN721_c0_g1 was annotated in the Swiss-Prot and Pfam databases as involved in the formation and function of RNA polymerase. Among the significantly down-regulated genes, the function of DN41995_c0_g1 remains unknown. DN5624_c0_g2 is primarily involved in the synthesis of transmembrane transport proteins. DN8330_c0_g5 mainly affects the synthesis and function of plant cysteine proteases, and DN7998_c1_g1 is associated with the regulation of phosphatase activity.
KEGG functional annotation analysis of the differentially expressed unigenes (Figure 2) revealed that 185 up-regulated and 116 down-regulated unigenes were mapped. They were assigned to five major functional categories: metabolic processes, cellular processes, organismal systems, genetic information processing, and environmental information processing. In terms of metabolic processes, the down-regulated genes were associated with: amino acid metabolism (11 unigenes), biosynthesis of other secondary metabolites (10 unigenes), carbohydrate metabolism (17 unigenes), energy metabolism (7 unigenes), terpenoid and polyketide metabolism (9 unigenes), lipid metabolism (4 unigenes), metabolism of cofactors and vitamins (4 unigenes), metabolism of other amino acids (4 unigenes), and nucleotide metabolism (1 unigene). The up-regulated genes were involved in the same nine subcategories as above, with unigene counts of 12, 11, 23, 15, 7, 8, 10, 12, and 2, respectively, and were additionally associated with glycan biosynthesis and metabolism (1 unigene). Among all metabolic process-related genes, those affecting carbohydrate metabolism were the most abundant, with 23 up-regulated and 17 down-regulated unigenes. For genetic information processing, the DEGs were annotated to the following functional subcategories: folding, sorting, and degradation (11 vs. 6 unigenes), replication and repair (6 vs. 1 unigene), transcription (4 vs. 1 unigene), translation (18 vs. 3 unigenes). Notably, translation-related genes were the most abundant among up-regulated unigenes (18), while folding, sorting, and degradation were most represented among down-regulated unigenes (6). Within the remaining three functional categories, only one specific function was annotated in each: environmental information processing (signal transduction), cellular processes (transport and catabolism), organismal systems (environmental adaptation). Overall, environmental adaptation contained the highest number of unigenes, including 40 up-regulated and 20 down-regulated genes.
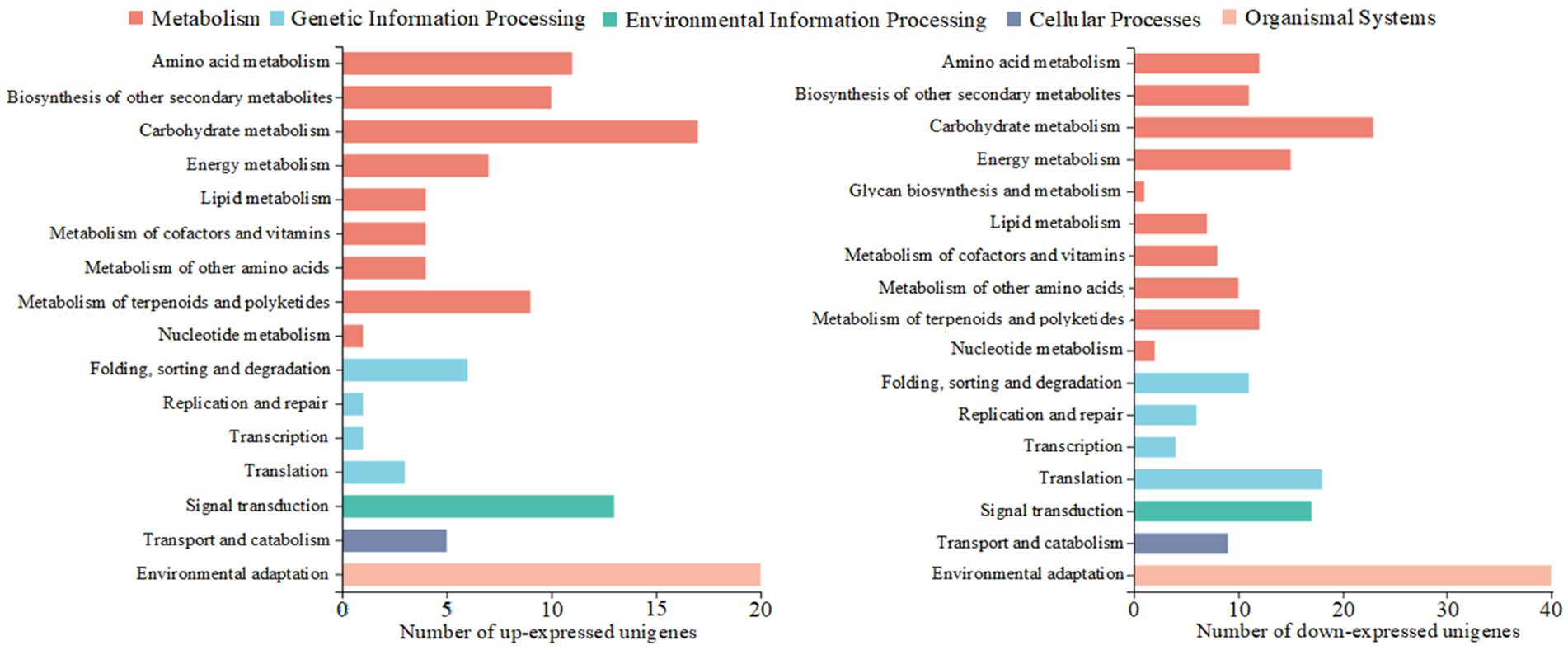
Figure 2. KEGG annotation results of the different expressed unigenes (DEGs) in Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3) under poor soil condition.
Further in-depth analysis of the differentially expressed genes related to environmental adaptation is presented in Table 4. The results indicate that the highest number of DEGs were associated with the synthesis of proteins containing NB-ARC and LRR domains, including 15 up-regulated and 12 down-regulated genes. This was followed by genes related to calcium-binding proteins and disease resistance proteins, with 7 and 5 up-regulated DEGs, respectively, and only one down-regulated gene in each category. In addition to the above genes that were both up- and down-regulated, three genes were exclusively up-regulated. These are involved in the functions of calcium-dependent protein kinase (DN13170_c0_g1), transcription activator of pathogenesis-related genes (DN1117_c0_g1), calcineurin (DN38256_c0_g1). Furthermore, five genes were exclusively down-regulated, functioning in death-associated protein kinase (DN23138_c0_g1), cyclic nucleotide-gated ion channels (DN20183_c0_g3, DN712_c0_g1), pathogenesis-related protein (DN13478_c0_g2), and cysteine protease (DN37443_c0_g1).
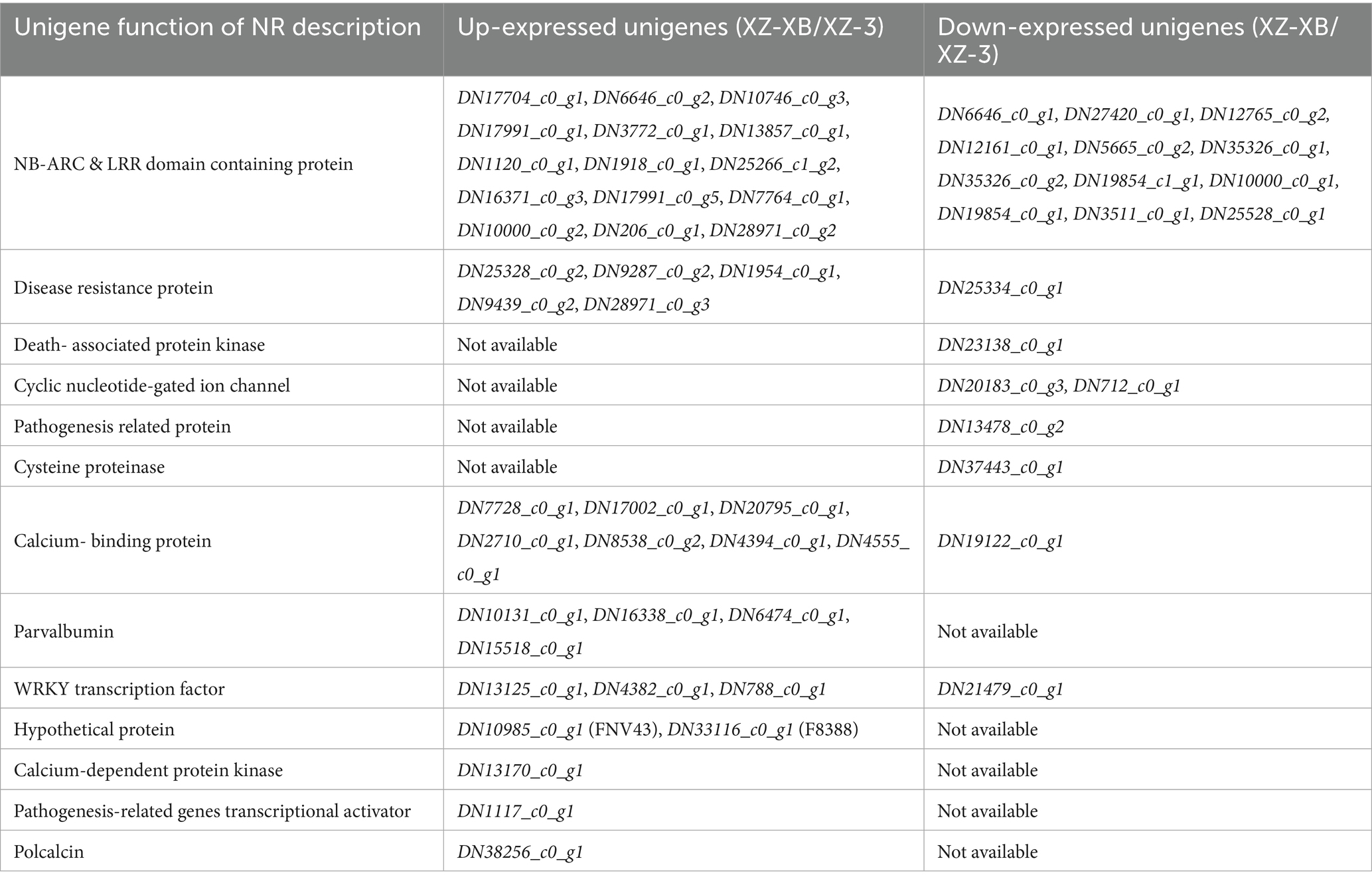
Table 4. Function of environment adaption related unigenes between Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3).
GO functional annotation analysis of the DEGs (Figure 3) revealed that among the 1,908 differentially expressed genes, multiple unigenes were co-annotated in the categories of molecular function, biological process, and cellular component. Overall, the largest number of DEGs were annotated in molecular function (1,356), followed by cellular component (1,196) and biological process (1,006). Within the molecular function category, the most annotated terms were “binding” and “catalytic activity,” with 618 and 565 unigenes, respectively. The other four functions (translation regulator activity, structural molecule activity, transporter activity, and transcription regulator activity) had relatively fewer annotated unigenes, ranging between 15 and 50. In the biological process category, the most annotated term was “cellular process” (323 unigenes), followed by “metabolic process” (291 unigenes). The number of unigenes annotated as “response to stimulus” and “biological regulation” were nearly identical, with 114 and 117, respectively. Terms such as “reproduction,” “cellular localization,” and “cellular component organization or biogenesis” had relatively fewer unigenes, each numbering around 40. For the cellular component category, the term “membrane part” contained the highest number of unigenes (441), followed by “cell part” (315). The remaining terms included “organelle” (173), “membrane” (90), “organelle part” (64), “protein-containing complex” (54), and “extracellular region” (21).
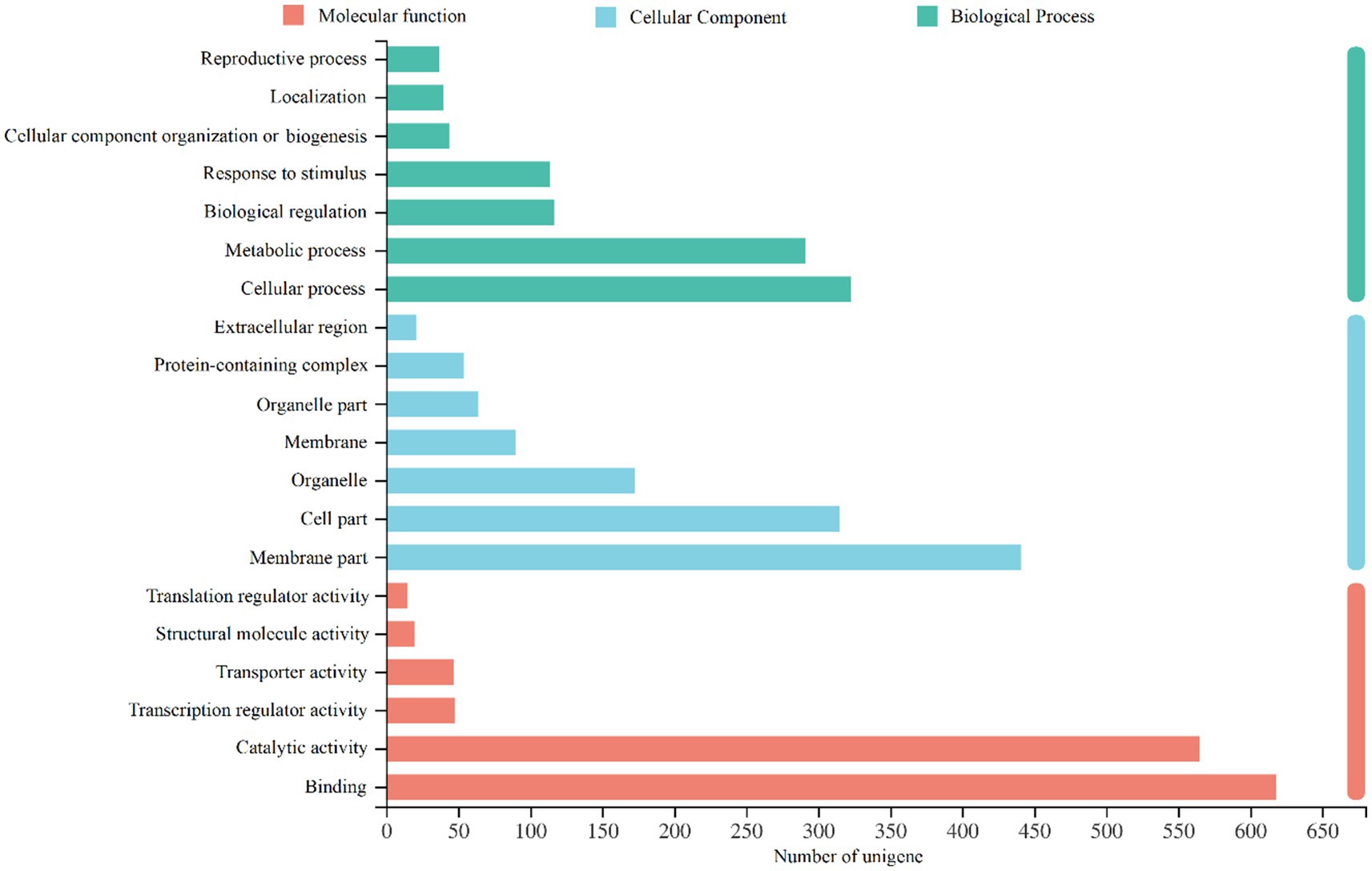
Figure 3. GO annotation results of the different expressed unigenes (DEGs) in Xiangzhu XB (XZ-XB) and Xiangzhu 3 (XZ-3) under poor soil condition.
3.4 Results of qRT-PCR detection
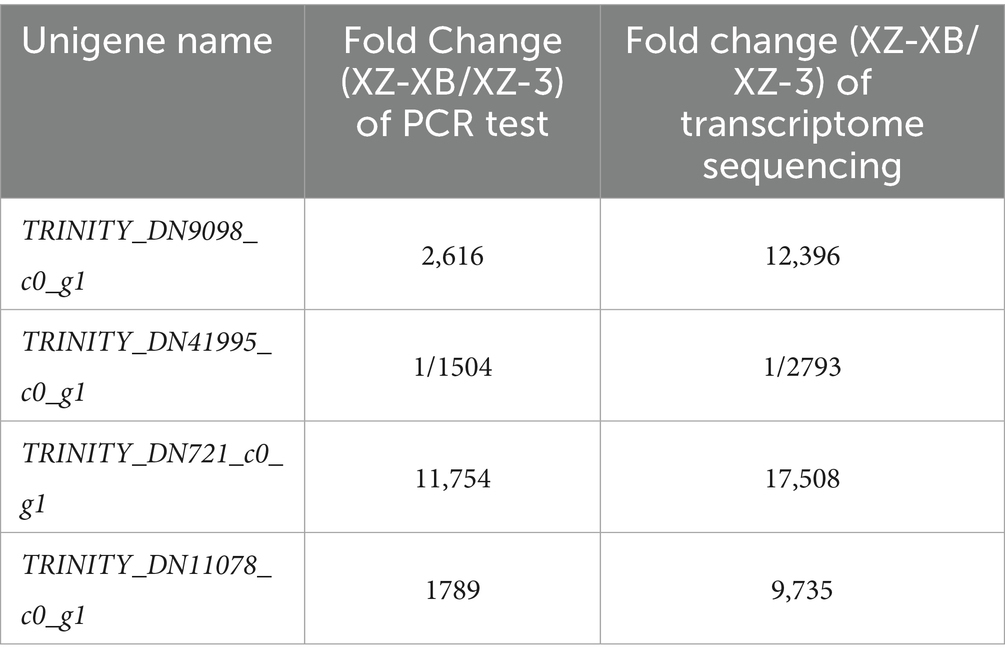
While transcriptome sequencing reflects the overall trend in gene expression, it may not perfectly capture the expression dynamics of every single gene. In contrast, qRT-PCR measures the actual expression levels of specific genes. To validate the accuracy of transcriptome sequencing results, it is common practice to select a subset of genes for verification using qRT-PCR. Consistency in expression trends between the two methods is generally considered successful validation. To assess the reliability of the transcriptome sequencing results in this study, four genes with substantial expression differences were selected for qRT-PCR analysis using the Actin gene as the internal reference. The results are presented in Table 5. Although the fold changes in gene expression detected by qRT-PCR differed somewhat from those obtained in the transcriptome sequencing analysis, the overall expression trends were consistent. The strong correlation between RNA-seq and qRT-PCR results (Table 5) validates the reliability of our transcriptomic data for identifying DEGs.
4 Discussion
4.1 The ramie transcriptome characteristics under poor soil conditions
The number of unigenes assembled in this study was significantly higher than the number of expressed genes obtained in the transcriptome study of ramie by Liu et al. (2013). This discrepancy may be attributed to the induction of gene expression under poor soil stress that is not activated under normal growth conditions (as in the experimental setup of Liu’s study). However, compared to the 77.7% unigene functional annotation rate reported in Liu’s study, the highest annotation rate achieved in this study was only 53.71%. As a non-model plant without a publicly available reference genome, functional annotation of unigenes relies solely on information from related species in public databases (Chen et al., 2014; Ma et al., 2017). Previous studies on plant responses to abiotic stress have largely focused on drought, salinity, alkali, and heavy metal tolerance, while mechanisms underlying adaptation to nutrient-poor conditions remain poorly explored (Wu et al., 2016). This has resulted in a scarcity of genetic information related to poor soil tolerance in existing databases, ultimately leading to the limited functional annotation of many expressed genes in this study.
4.2 The contribution of DEGs in helping ramie adapting to poor soil conditions
Among the annotated differentially expressed genes identified in this study, DN5624_c0_g2 is associated with ABC transporters. As a superfamily of proteins involved in the transmembrane transport of nutrients, photosynthetic products, and hormones, ABC transporters play a positive role in promoting root absorption of nutritional elements (Rentsch et al., 2007; Courty et al., 2015). For example, the orthologs of the ABC transports G subfamily in white lupin (Lupinus albus), such as L.albABCG29, are crucial for enhancing phosphorus accumulation under low-phosphorus conditions and have been linked to improved plant growth through mechanisms like promoted root development and rhizosheath formation (Aslam et al., 2021). Similarly, in maize (Zea mays L.), a set of Zm ABC genes such as ZmABC012, 013, 031, contribute to lead stress tolerance (Guo et al., 2022). However, the up-regulation of DN5624_c0_g2 in the poor soil-sensitive variety XZ-3 suggests that it may act as a suppressor of ABC transporter synthesis or its function might be specific to transporting compounds that are detrimental under nutrient-deficient stress. This highlights the complex and sometimes contradictory roles of transporter families, necessitating functional validation in ramie.
Another highly up-regulated gene in XZ-3, DN8330_c0_g5, is annotated with functions related to pentatricopeptide repeat (PPR) proteins and senescence-specific cysteine proteases. The PPR proteins influence RNA metabolism through editing, splicing, and other mechanisms, primarily promoting RNA translation (Huang and Huang, 2018). Cysteine proteases, widely involved in various physiological processes, induce or participate in both natural and stress-induced senescence (Liu et al., 2018). The co-upregulation of these functional domains strongly suggests that under poor soil condition, DN8330_c0_g5 in the sensitive variety promotes the translation of RNA associated with senescence-specific cysteine proteases, leading to increased production of senescence-promoting enzymes and accelerating plant aging and death. This mechanism is conserved across species. For example, in creeping bentgrass (Agrostis stolonifera), heat stress induces general protease activities that accelerate leaf senescence (Rossi and Huang, 2024). This indicates that DN8330_c0_g5 represents a promising target for gene silencing or knockout to delay stress-induced senescence and enhance poor soil tolerance in ramie.
Additionally, DN7998_c1_g1, which is highly expressed in XZ-3, may function as an inhibitor of purple acid phosphatases (PAPs). The PAPs enhance the activation of organic phosphorus, thereby helping plants adapt to low-phosphorus environments (Liu et al., 2019). Their function has been validated in several plants, including Arabidopsis (Wang et al., 2011), rice (Lu et al., 2016), and common bean (Wu et al., 2018). As a member of the cysteine protease family, papain-like cysteine proteases are primarily associated with programmed cell death in plants (Liu et al., 2018; Ueda et al., 2000). Previous studies have shown that under environmental stresses such as drought and salinity, plants accumulate papain-like cysteine proteases to promote senescence (Cilliers et al., 2018; Havé et al., 2018). Therefore, the highly expressed DN8330_c0_g5 in XZ-3 may be a key gene regulating the synthesis of papain-like cysteine proteases in ramie, while the highly expressed DN9098_c0_g1 in XZ-XB may act as a suppressor. This presents a direct breeding application: selecting for alleles of DN7998_c1_g1 that confer low expression could be a strategy to enhance phosphorus scavenging capacity in new ramie varieties.
4.3 The potential mechanisms underlying ramie tolerance to poor soil stress
Through functional enrichment analysis of differentially expressed genes, this study found that the largest number of genes were associated with carbohydrate metabolism and environmental adaptation. This suggests that ramie varieties with strong adaptability to poor soil conditions may mitigate the impact of nutrient deficiency on photosynthetic product accumulation (i.e., carbohydrates) by regulating specific genes, while also modulating environment adaptation-related genes to reduce the effects of nutrient scarcity on plant growth.
Among the differentially expressed genes involved in environmental adaptation, annotated functions were primarily associated with the synthesis and activity of death-associated protein kinases, pathogenesis-related proteins, and cysteine proteases (which induce plant senescence). Thus, it can be inferred that ramie germplasms with high tolerance to poor soil reduce the expression of genes that promote senescence and cell death, thereby alleviating the damage caused by nutrient stress and enhancing adaptation to poor environments.
Among the genes positively correlated with ramie’s adaptability to poor soil, functions were related to critical proteins such as calcium-dependent protein kinases and calcineurin. Calcium-dependent protein kinases transduce environmental stress signals and defense responses (Ni and Wei, 2002) but rely on calcium signaling. Calcineurin maintains high calcium ion concentrations to ensure the activity of calcium-dependent protein kinases, thereby facilitating signal transduction. Therefore, the ability to rapidly and sensitively recognize poor soil signals and transmit them to the plant for swift defensive responses represents a key mechanism enabling ramie germplasms with high poor soil tolerance to adapt to nutrient-deficient environments.
4.4 Agroecological benefits derived by the ramie cultivation on degraded lands
Soil degradation, particularly of arable land, is mainly driven by some interconnected factors as soil erosion, structural deterioration from intensive tillage, and ecological imbalance caused by the excessive use of fertilizers, pesticides, and plastic mulch (Oliveira et al., 2022; Dadzie et al., 2023). The development of stress-tolerant ramie varieties offers a dual-benefit strategy: enabling productive utilization of degraded land while concurrently restoring soil health through agroecological mechanisms.
As a perennial, ramie requires only a single establishment, but provides repeated harvests over 5–10 years or more. This long-term growth habit minimizes soil disturbance, thereby reducing structural damage and erosion risk. Its well-developed root and rhizome system enhances soil stabilization, while the dense canopy effectively intercepts rainfall (Wang et al., 2021). Together, these traits enable ramie to offer soil and water conservation benefits comparable to those of 5–6 year-old regenerating forests or perennial grasslands (Liao et al., 2011). Breeding ramie for improved adaptation to poor soils can further reduce reliance on synthetic fertilizer. This not only improves nutrient use efficiency within ramie systems, but also helps mitigate the acidification and compaction often associated with the chemical fertilizer application in conventional crop production (Pahalvi et al., 2021). Moreover, the practice of leaf stripping during harvesting, coupled with natural root turnover, continuously returns organic matter to the soil (Bene et al., 2011). This, together with root exudates supply, builds stable organic carbon, which is vital energy to soil microbiota, significantly enhancing microbial diversity and activity (Zhu et al., 2018).
In summary, ramie serves as a multifunctional crop for sustainable land management. Its cultivation on degraded or marginal lands can simultaneously improve soil structure, enhance fertility, and reactivate soil biological communities—all while maintaining economic returns through sustainable fiber production.
4.5 Implications of the screened genes for molecular breeding
The DEGs identified in this study provide a valuable resource for molecular breeding of ramie with enhanced adaptation to nutrients-decifient soils. The highly expressed genes in XZ-XB (the poor soil tolerant germplasm), such as DN721_c0_g1, DN9098_c0_g1, can be used as molecular markers for marker-assisted selection. Breeders can screen germplasm collections for these favorable alleles to rapidly introgress the tolerance trait. Conversely, the genes highly expressed in the sensitive XZ-3 (e.g., DN8330_c0_g5 and DN7998_c1_g1) are targets for gene editing. Using gene editing tools such as CRISPR-Cas9 to knock out these genes in elite but sensitive varieties could directly convert them into nutrient-efficient cultivars. Furthermore, the suite of DEGs related to carbohydrate metabolism, calcium signaling, and defense responses provides a foundation for developing multi-gene selection models, enabling the pyramiding of several advantageous transcriptional alleles for robust and durable tolerance to poor soil.
5 Conclusion
This study identified key genes involved in ramie’s adaptation to poor soil condition through transcriptome analysis. A total of 1,908 differentially expressed genes (DEGs) were detected, among which seven unigenes showed large expression differences (>1,000-fold change). Functional annotation linked these DEGs to the functions of nutrient transport, senescence, and stress signaling. Results uncovered here provide valuable genetic resources and candidate targets to breed advanced ramie varieties that are genetically equipped to thrive in poor soil condition. The development of such varieties could transform ramie cultivation from conventional cropping system into an integrated agroecological solutions. This ramie production system simultaneously contributes to fiber production, soil restoration, and alleviates competition between food and fiber crops for arable land.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SW: Data curation, Formal analysis, Visualization, Writing – original draft. HJ: Visualization, Writing – review & editing. HX: Conceptualization, Writing – review & editing. YJ: Conceptualization, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the Yuelushan Laboratory Breeding Project (YLS-2025-ZY02056), Natural Science Foundation of China (32071940 and 31872877), and the National Science and Technology Resource Sharing Service Platform Project (NCGRC-2025-48).
Acknowledgments
We acknowledge all those who contributed to this work but are not named as authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
An, X., Wei, J. Q., Liu, Q., Ying, J. Y., Zhou, H. P., Luo, X. H., et al. (2023). Progress in the study of the absorption, accumulation, and tolerance of ramie to the heavy metal cadmium. Mol. Plant Breed. 14, 1–8. doi: 10.5376/mpb.2023.14.0003
Aslam, M. M., Waseem, M., Zhang, Q., Ke, W., Zhang, J., and Xu, W. (2021). Identification of ABC transporter G subfamily in white lupin and functional characterization of L.albABGC29 in phosphorus use. BMC Genomics 22:723. doi: 10.1186/s12864-021-08015-0
Bene, C. D., Tavarini, S., Mazzoncini, M., and Angelini, L. G. (2011). Changes in soil chemical parameters and organic matter balance after 13 years of ramie [Boehmeria nivea (L.) gaud.] cultivation in the Mediterranean region. Eur. J. Agron. 35, 154–163. doi: 10.1016/j.eja.2011.05.007
Chaudhuri, S., Roy, M., McDonald, L. M., and Emendack, Y. (2023). Land degradation–desertification in relation to farming practices in India: an overview of current practices and agro-policy perspectives. Sustainability 15:6383. doi: 10.3390/su15086383
Chen, K., Li, Y., Yu, C., Chen, P., Chen, J., Gao, G., et al. (2023). Systematic evaluation of ramie (Boehmeria nivea L.) for phytoremediation of cadmium contaminated soil and the mechanism of microbial regulation. Chemosphere 337:139298. doi: 10.1016/j.chemosphere.2023.139298
Chen, J., Pei, Z. H., Dai, L. J., Wang, B., Liu, L. J., An, X., et al. (2014). Transcriptome profiling using pyrosequencing shows genes associated with bast fiber development in ramie (Boehmeria nivea L.). BMC Genomics 15:919. doi: 10.1186/1471-2164-15-919
Cilliers, M., Wyk, S. G., Heerden, P. D. R., Kunert, K. J., and Vorster, B. J. (2018). Identification and changes of the drought-induced cysteine protease transcriptome in soybean (Glycine max) root nodules. Environ. Exp. Bot. 148, 59–69. doi: 10.1016/j.envexpbot.2017.12.005
Courty, P. E., Smith, P., Koegel, S., Redecker, D., and Wipf, D. (2015). Inorganic nitrogen uptake and transport in beneficial plant root-microbe interactions. Crit. Rev. Plant Sci. 34, 4–16. doi: 10.1080/07352689.2014.897897
Dadzie, F. A., Egidi, E., Stewart, J., Eldridge, D. J., Molesworth, A., Singh, B. K., et al. (2023). “Agricultural soil degradation in Australia” in Impact of agriculture on soil degradation I. The handbook of environmental chemistry. eds. P. Pereira, M. Muñoz-Rojas, I. Bogunovic, and W. Zhao (Cham: Springer). doi: 10.1007/698_2023_966
Guo, Z., Yuan, X., Li, L., Zeng, M., Yang, J., Tang, H., et al. (2022). Genome-wide analysis of the ATP-binding cassette (ABC) transporter family in Zea mays L. and its response to heavy metal stresses. Int. J. Mol. Sci. 23:2109. doi: 10.3390/ijms23042109
Havé, M., Balliau, T., Cottyn-Boitte, B., Dérond, E., Cueff, G., Soulay, F., et al. (2018). Increases in activity of proteasome and papain-like cysteine protease in Arabidopsis autophagy mutants: back-up compensatory effect or cell-death promoting effect? J. Exp. Bot. 69, 1369–1385. doi: 10.1093/jxb/erx482
He, X., Wu, Y., Liu, K., Ji, J., Wu, C., Li, J., et al. (2025). The combined application of inorganic and organic materials over two years improves soil pH, slightly increases soil organic carbon, and enhances crop yields in severely acidic red soil. Agronomy 15:498. doi: 10.3390/agronomy15020498
Hossain, A., Krupnik, T. J., Timsina, J., Mahboob, M. G., Chaki, A. K., Farooq, M., et al. (2020). “Agricultural land degradation: processes and problems undermining future food security” in Environment, climate, plant and vegetation growth. ed. S. Fahad (Cham: Springer).
Hou, M. (2018). Screening of high nitrogen use efficiency varieties of ramie (Boehmeria nivea L.) and analysis of expression on BnNRT1.1. Beijing: Chinese Academy of Agricultural Sciences.
Huang, J. R., and Huang, W. C. (2018). Research progress on the pentatricopeptide-repeat protein. Hubei Agric. Sci. 57, 19–31. doi: 10.14088/j.cnki.issn0439-8114.2018.23.004
Hussain, S., Hussain, S., Guo, R., Sarwar, M., Ren, X., Krstic, D., et al. (2021). Carbon sequestration to avoid soil degradation: a review on the role of conservation tillage. Plants 10:2001. doi: 10.3390/plants10102001
Lee, Y.-Y., Cho, K.-S., and Yun, J. (2025). Phytoremediaton strategies for co-contaminated soils: overcoming challenges, enhancing efficiency, and exploring future advancements and innovations. PRO 13:132. doi: 10.3390/pr13010132
Li, Q., Jiang, W., and Lyu, J. (2024). Soil health assessment of an acidic red soil agricultural area and its restoration with biochar soil conditioners. Soil Use Manag. 40:e13002. doi: 10.1111/sum.13002
Li, Z., Kekeli, M. A., Jiang, Y., and Rui, Y. (2025). Progress and prospect of saline-alkaline soil management technology: a review. Appl. Sci. 15:4567. doi: 10.3390/app15084567
Liao, M. Q., Li, J., Huang, Q. R., Ye, C., Yu, X. C., Tang, X. J., et al. (2011). A comparative study on soil and water conservation of ramie and peanut fields in red soil sloping land. Soil 43, 657–661. doi: 10.13758/j.cnki.tr.2011.04.025
Liu, C. (2022). Phytostabilization of ion-adsorption rare earth element mine tailings using the fiber plant ramie (Boehmeria nivea L.). Guangzhou: Sun Yat-sen University.
Liu, H., Hu, M., Wang, Q., Cheng, L., and Zhang, Z. (2018). Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 9:1717. doi: 10.3389/fpls.2018.01717
Liu, P. D., Huang, R., Xu, W. R., Luo, J. J., Chen, Z. J., and Liu, G. D. (2019). Research progress of purple acid phosphatase in plants. Chin. J. Trop. Crops 40, 410–416. doi: 10.3969/j.issn.1000-2561.2019.02.028
Liu, T., Zhu, S., Tang, Q., Chen, P., Yu, Y., and Tang, S. (2013). De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. gaud). BMC Genomics 14:125. doi: 10.1186/1471-2164-14-125
Lu, L., Qiu, W., Gao, W., Tyerman, S. D., Shou, H., and Wang, C. (2016). OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ. 39, 2247–2259. doi: 10.1111/pce.12794
Ma, X., Tan, S. L., Xing, H. C., and Liao, S. B. (2017). Establishment of real-time quantitative PCR system for ramie. Plant Fiber Sci. China 39, 111–119.
Mosier, S., Córdova, S. C., and Robertson, G. P. (2021). Restoring soil fertility on degraded lands to meet food, fuel, and climate security needs via perennialization. Front. Sustain. Food Syst. 5:706142. doi: 10.3389/fsufs.2021.706142
Ni, T. H., and Wei, Y. Z. (2002). The physiological function of calcium-dependent protein kinases in plants. J. Northwest Sci. Tech. Univ. Agric. For. 30, 241–246. doi: 10.13207/j.cnki.jnwafu.2002.06.059
Oliveira, P. T. S., de Faria Godoi, R., Colman, C. B., Motta, J. S., Sone, J. S., and Almagro, A. (2022). “Agricultural land degradation in Brazil” in Impact of agriculture on soil degradation I. The Handbook of Environmental Chemistry. eds. P. Pereira, M. Muñoz-Rojas, I. Bogunovic, and W. Zhao (Cham: Springer).
Pahalvi, H. N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A. N. (2021). “Chemical fertilizers and their impact on soil health” in Microbiota and biofertilizers, Vol 2. eds. G. H. Dar, R. A. Bhat, M. A. Mehmood, and K. R. Hakeem (Cham: Springer).
Perveen, I., Javed, M. M., Ahmad, I., Arif, N., and Kausar, S. (2025). “Advances in agronomy for soil conservation and plant health” in Soils and sustainable agriculture. ed. M. Shaaban (Cham: Frontier Studies in Soil Science, Springer).
Phang, L. Y., Mingyuan, L., Mohammadi, M., and Hosseini, S. (2024). Phytoremediation as a viable ecological and socioeconomic management strategy. Environ. Sci. Pollut. Res. 31, 50126–50141. doi: 10.1007/s11356-024-34585-z
Pravalie, R., Patriche, C., Borrelli, P., Panagos, P., Roșca, B., Dumitrascu, M., et al. (2021). Arable lands under the pressure of multiple land degradation processes. A global perspective. Environ. Res. 194:110697. doi: 10.1016/j.envres.2020.110697
Rentsch, D., Schmidt, S., and Tegeder, M. (2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 581, 2281–2289. doi: 10.1016/j.febslet.2007.04.013
Rossi, S., and Huang, B. (2024). Protease inhibitors suppressed leaf senescence in creeping bentgrass exposed to heat stress in association with inhibition of protein degradation into free amino acids. Plant Growth Regul. 102, 65–75. doi: 10.1007/s10725-023-00977-3
Talukder, B., Ganguli, N., Matthew, R., vanLoon, G. W., Hipel, K. W., and Orbinski, J. (2021). Climate change-triggered land degradation and planetary health: a review. Land Degrad. Dev. 32, 4509–4522. doi: 10.1002/ldr.4056
Tan, L. T. (2015). Screening and expression analysis of ramie (Boehmeria nivea L.) with nitrogen efficiency. Beijing: Chinese Academy of Agricultural Sciences.
Ueda, T., Seo, S., Ohashi, Y., and Hashimoto, J. (2000). Circadian and senescence-enhanced expression of a tobacco cysteine protease gene. Plant Mol. Biol. 44, 649–657. doi: 10.1023/A:1026546004942
Wang, L., Li, Z., Qian, W., Guo, W., Gao, X., Huang, L., et al. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 157, 1283–1299. doi: 10.1104/pp.111.183723
Wang, D., Zeng, R., Hu, Z., Li, X., Guo, Y., Yang, P., et al. (2021). Soil conservation effects and economic benefit of different crops cultivated on gentle slopes in the south mining area, China. Bangladesh J. Bot. 50, 755–761. doi: 10.3329/bjb.v50i5.56426
Wu, S., Jie, H., and Jie, Y. (2021b). Role of rhizosphere soil microbes in adapting ramie (Boehmeria nivea L.) plants to poor soil conditions through N-fixing and P-solubilization. Agronomy 11:2096. doi: 10.3390/agronomy11112096
Wu, W., Lin, Y., Liu, P., Chen, Q., Tian, J., and Liang, C. (2018). Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots. J. Exp. Bot. 69, 603–617. doi: 10.1093/jxb/erx441
Wu, Z., Tang, Q., Wang, Y., Qiu, C., Long, S., Zhao, X., et al. (2022). Ramie (Boehmeria nivea) as phytoremediation crop for heavy metal-contaminated paddy soil in southern China: variety comparison, cd accumulation, and assessment of fiber recycling. J. Nat. Fibers 19, 11078–11091. doi: 10.1080/15440478.2021.2009400
Wu, S., Xue, S., Iqbal, Y., Xing, H., and Jie, Y. (2021a). Seasonal nutrient cycling and enrichment of nutrient-related soil microbes aid in the adaptation of ramie (Boehmeria nivea L.) to nutrient-deficient conditions. Front. Plant Sci. 12:644904. doi: 10.3389/fpls.2021.644904
Wu, Y. R., Zhong, K., and Xie, Q. (2016). Research progress and prospective of plant response to abiotic stresses. China Basic Sci. 1, 35–40. doi: 10.3969/j.issn.1009-2412.2016.01.006
Xu, X., Guo, L., Wang, S., Wang, X., Ren, M., Zhao, P., et al. (2023). Effective strategies for reclamation of saline-alkali soil and response mechanisms of the soil-plant system. Sci. Total Environ. 905:167179. doi: 10.1016/j.scitotenv.2023.167179
Zheng, J. S. (2014). Study on glutamine synthetase genes cloning and over-expression of ramie (Boehmeria nivea L.). Beijing: Chinese Academy of Agricultural Sciences.
Zhu, S., Wang, Y., Xu, X., Liu, T., Wu, D., Zheng, X., et al. (2018). Potential use of high-throughput sequencing of soil microbial communities for estimating the adverse effects of continuous cropping on ramie (Boehmeria nivea L. gaud). PLoS One 13:e0197095. doi: 10.1371/journal.pone.0197095
Keywords: fiber crop, marginal land, infertile soil, molecular breeding, gene screening
Citation: Wu S, Jie H, Xing H and Jie Y (2025) Mining key genes for ramie (Boehmeria nivea L.) adaptation to poor soil condition using transcriptome analysis. Front. Sustain. Food Syst. 9:1716412. doi: 10.3389/fsufs.2025.1716412
Edited by:
Chaochen Tang, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaReviewed by:
Wen Yin, Gansu Agricultural University, ChinaYuanyuan Chen, Guangdong Academy of Agricultural Sciences (GDAAS), China
Copyright © 2025 Wu, Jie, Xing and Jie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yucheng Jie, ZmJmY2p5Y0BodW5hdS5lZHUuY24=
 Shenglan Wu
Shenglan Wu Hongdong Jie
Hongdong Jie Hucheng Xing2,4
Hucheng Xing2,4