- 1Institute of Molecular Infection Biology, University of Würzburg, Würzburg, Germany
- 2Centre for Infection Medicine, Institute of Microbiology and Epizootics, Free University of Berlin, Berlin, Germany
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) of clonal complex CC398 typically carry various antimicrobial resistance genes, many of them located on plasmids. In the bovine LA-MRSA isolate Rd11, we previously identified plasmid pAFS11 in which resistance genes are co-localized with a novel ica-like gene cluster, harboring genes required for polysaccharide intercellular adhesin (PIA)-mediated biofilm formation. The ica genes on pAFS11 were acquired in addition to a pre-existing ica locus on the S. aureus Rd11 chromosomal DNA. Both loci consist of an icaADBC operon and icaR, encoding a corresponding icaADBC repressor. Despite carrying two biofilm gene copies, strain Rd11 did not produce PIA and transformation of pAFS11 into another S. aureus strain even slightly diminished PIA-mediated biofilm formation. By focusing on the molecular background of the biofilm-negative phenotype of pAFS11-carrying S. aureus, we identified the pAFS11-borne ica locus copy as functionally fully active. However, transcription of both plasmid- and core genome-derived icaADBC operons were efficiently suppressed involving IcaR. Surprisingly, although being different on the amino acid sequence level, the two IcaR repressor proteins are mutually replaceable and are able to interact with the icaA promoter region of the other copy. We speculate that this regulatory crosstalk causes the biofilm-negative phenotype in S. aureus Rd11. The data shed light on an unexpected regulatory interplay between pre-existing and newly acquired DNA traits in S. aureus. This also raises interesting general questions regarding functional consequences of gene transfer events and their putative implications for the adaptation and evolution of bacterial pathogens.
Introduction
Staphylococcus aureus is a common human and animal pathogen, causing a wide range of clinical manifestations (Tong et al., 2015; Ballhausen et al., 2017). Due to the capability to readily acquire many different resistance genes, S. aureus and other staphylococcal species are regarded as pathogens of concern for public health (Foster, 2017; Lakhundi and Zhang, 2018). Thus, methicillin-resistant S. aureus (MRSA) and coagulase-negative staphylococci (MR-CoNS) are among the most common causes of healthcare-associated infections (Lee et al., 2018a; Becker et al., 2020). In this respect, the ability to form biofilms on the inert surfaces of medical devices is considered as important pathomechanism that contributed to the establishment of staphylococci as notorious nosocomial pathogens (Heilmann et al., 2019; Becker et al., 2020; Schilcher and Horswill, 2020). Biofilms are understood as bacterial communities that adhere to surfaces by encasing into a self-produced extracellular polymeric matrix (Costerton et al., 1999). The staphylococcal biofilm matrix may contain exopolysaccharides (Heilmann et al., 1996) and proteins (Rohde et al., 2005) as well as extracellular (e)DNA (Qin et al., 2007) [for a recent review see reference (Schilcher and Horswill, 2020)]. The key exopolysaccharide component of staphylococcal biofilms is PIA (polysaccharide intercellular adhesin), a beta-1,6 linked N-acetyl glucosaminoglycan, whose synthesis enzymes are encoded by the ica (intercellular adhesin) locus [recently reviewed in (Nguyen et al., 2020)]. PIA/ica was originally discovered in Staphylococcus epidermidis and was later also detected in S. aureus and other staphylococcal species (Heilmann et al., 1996; Mack et al., 1996; Cramton et al., 1999). Interestingly, ica locus homologs also exist in phylogenetically unrelated bacteria such as Escherichia coli (Wang et al., 2004), suggesting an eminent role of the factor in the evolution of bacterial biofilm functions. In these organisms, PIA is also often referred to as PNAG (poly-1,6-N-acetylglucosamine). The staphylococcal ica locus consists of two divergently oriented transcription units, one comprising the icaADBC operon (encoding the enzymes required for PIA synthesis) and the other harboring icaR which codes for a transcription factor of the TetR family (Figure 1A). IcaR, for which the crystal structure was solved, binds to a region upstream of icaA and represents a potent repressor of icaADBC operon transcription (Conlon et al., 2002a; Jefferson et al., 2004; Jeng et al., 2008). Regulation of the ica locus is highly complex and a plethora of environmental cues are known to influence PIA production many of which either directly or indirectly influencing icaR transcription (Conlon et al., 2002b; Cerca et al., 2008; Fey and Olson, 2010; Cue et al., 2012; Hoang et al., 2019; Nguyen et al., 2020). Expression of icaR is further controlled post-transcriptionally through RNA-mediated mechanisms that influence stability and translation of the icaR mRNA, with direct consequences for PIA production and biofilm formation (Ruiz de los Mozos et al., 2013; Rochat et al., 2018; Bronesky et al., 2019; Lerch et al., 2019; Schoenfelder et al., 2019). While nearly all S. aureus genomes carry the ica locus, distribution of the gene cluster among S. epidermidis and other CoNS species is more diverse and often associated with distinct clonal lineages (Kozitskaya et al., 2005; Conlan et al., 2012; Thomas et al., 2014; Méric et al., 2015; Méric et al., 2018; Lee et al., 2018b; Espadinha et al., 2019). The ica locus is usually located in the bacterial chromosomal DNA in all staphylococcal species. Previously, however, we detected an ica gene cluster of unknown genetic origin on plasmid pAFS11 in the bovine MRSA isolate S. aureus Rd11 (Feßler et al., 2017). S. aureus Rd11 is a livestock-associated (LA)-MRSA strain of sequence type ST398, a clonal lineage known for its potential to carry a broad range of both common and novel antibiotic resistance genes, many of which located on plasmids (Kadlec et al., 2012; Feßler et al., 2018). On pAFS11, antimicrobial and heavy metal resistance genes were found to be co-localized with a novel ica gene cluster. The ica locus on pAFS11 differed both on nucleotide and protein levels from the copy in the S. aureus Rd11 chromosome, and initial analyses (i.e. BLAST searches against the entire non-redundant sequence collections at NCBI) suggested that the plasmid-borne ica locus might have its origin in the CoNS species Staphylococcus sciuri [recently re-classified as Mammaliicoccus sciuri (Madhaiyan et al., 2020)] (Feßler et al., 2017). The mosaic structure of pAFS11 further suggests that the plasmid arose by a series of recombination events and was acquired by S. aureus Rd11 through horizontal gene transfer (HGT). As a result, S. aureus Rd11 carries two ica loci. Surprisingly, however, the strain did not produce biofilm when tested in standard tissue culture plate assays. Also, transformation of the pAFS11 plasmid into another S. aureus strain did not prompt biofilm formation, but even slightly reduced it (Feßler et al., 2017). In this study, we address the molecular mechanism underlying the biofilm-negative phenotype of pAFS11-bearing S. aureus. We identified an unexpected IcaR-mediated regulatory crosstalk between the plasmid-borne and chromosomally encoded ica loci, resulting in downregulation of biofilm formation. We discuss these findings in the context of co-evolution of virulence and resistance traits and raise the question of how genes newly acquired by HGT might become integrated into the regulatory network of host bacteria.
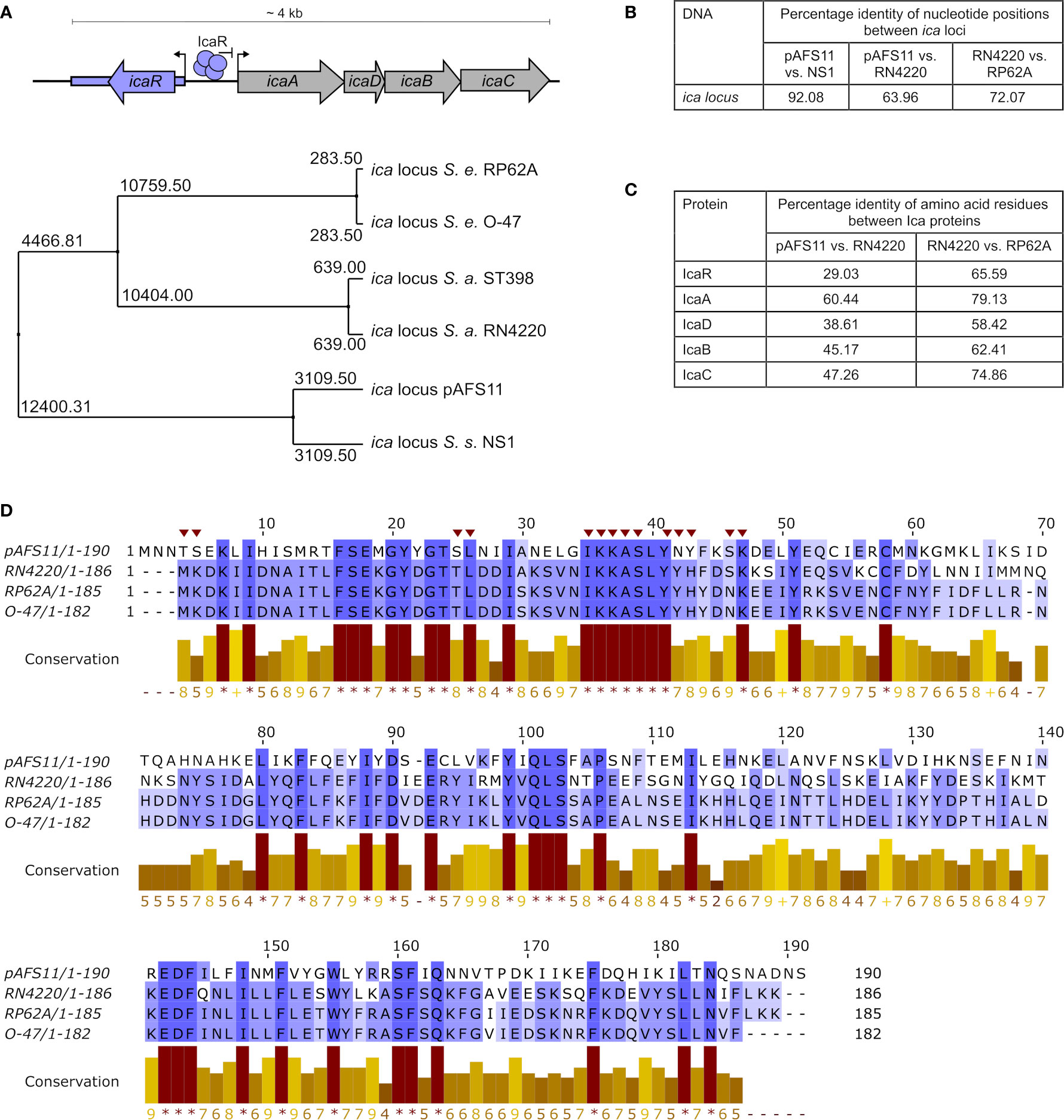
Figure 1 Conservation of pAFS11 ica locus. (A) TOP: schematic view of the organization of the ica locus including the IcaR tetramer and its function as repressor of transcription. BOTTOM: average distances between the ica loci of S. epidermidis O-47, S. epidermidis RP62A, S. aureus RN4220, S. aureus lineage ST398, S. sciuri NS1 and plasmid pAFS11. (B) Pairwise Alignment shown as percentage identity of nucleotide positions between the ica loci of pAFS11 vs. NS1, pAFS11 vs. RN4220 and RN4220 vs. RP62A. (C) Pairwise Alignment shown as percentage identity of amino acid residues between the Ica proteins of RN4220 vs. pAFS11 and RN4220 vs. RP62A. (D) Multiple sequence alignment of IcaR protein from pAFS11, RN4220, RP62A and O-47. Conservation is visualized as a histogram and a score is given for each column: conserved residues are indicated by ‘*’, and columns with residues, where all properties are conserved are marked with ‘+’. Putative icaA operon- interacting residues on IcaR in S. epidermidis are marked with a red triangle on top of the sequence (Jeng et al., 2008). All comparisons shown in (A–D) were calculated with the aid of Jalview (Waterhouse et al., 2009) from CLUSTAL Omega multiple sequence alignments (Madeira et al., 2019).
Materials and Methods
Sequence Alignments and Data Base Searches
Alignments of the nucleotide sequences of ica loci as well as that of amino acid sequences of Ica proteins from different species were performed with CLUSTAL Omega multiple sequence alignments (https://www.ebi.ac.uk/Tools/msa/clustalo) (Madeira et al., 2019). Average distance between the ica loci and pairwise alignments were calculated with the aid of Jalview (Waterhouse et al., 2009). Strains and sequences included into the analyses comprised S. epidermidis RP62A (accession no. NC_002976), S. epidermidis O-47 (accession no. CP040883), S. aureus ST398 (accession no. AM990992.1), S. aureus RN4220 (accession no. AFGU01000118.1), S. sciuri NS1 (accession no. LDTK01000031.1) and plasmid pAFS11 (accession no. FN806789.3). Data base queries with nucleotide and protein sequences were performed using the Basic Local Alignment Search Tool available at the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Plasmid and Strain Construction
Strains, plasmids and oligonucleotides used for this work are listed in Tables 1 and 2, respectively.
Construction of a Markerless icaADBC Mutant
The markerless ica mutant was obtained via allelic replacement with inducible counter-selection using the pBASE6 shuttle vector (Bae and Schneewind, 2006; Geiger et al., 2012). pBASE6 vector was linearized using primers SLIC_pBASE_R and SLIC_pBASE_F. Total deletion was achieved by overlapping PCR using as template gDNA from RN4220 with primers Flank_A_SLIC together with Flank_A_rev and Flank_B_rev together with Flank_B_SLIC. The amplicon was introduced into the linearized pBASE6 vector using the in vivo E. coli cloning (iVEC) method (Nozaki and Niki, 2019). The resulting plasmid (pAK17) was transformed into the restriction-deficient strain RN4220. Mutagenesis was performed as described elsewhere (Bae and Schneewind, 2006). The deletion was verified by PCR with oligonucleotides spanning the deletion region.
Construction of Complementation Plasmids
All plasmids were created following the iVEC method (Nozaki and Niki, 2019). As iVEC turned out to be more efficient in the presence of buffer, the ligation buffer from the QuickLigation™ Kit (NEB, #M2200S) was added to the reactions. Sanger sequencing was used to verify accuracy of all plasmids. To create plasmids pGM10 and pGM12, the icapAF and icaRN operons were amplified from pAFS11 and RN4220, respectively, and introduced in the linearized pRB473. Deletion of icaR coding regions from plasmids pGM10 and pGM12 (resulting in plasmids pGM11 and pGM13, respectively) was achieved by overlapping PCRs which amplified the respective vectors in two fragments that overlapped in the icaR deletion and multiple cloning site regions. Of note, this approach left putative icaR 5’ and 3’ untranslated regions (UTRs) intact which might be involved in post-transcriptional regulation of icaR. icaRpAF was amplified from pAFS11 and introduced in the linearized pGM12, resulting in plasmid pGM14. icaRRN was amplified from RN4220 and introduced in the linearized pGM10, resulting in plasmid pGM15. The vectors containing mutated sequences which alter the palindromes (pGM16 to pGM21) were generated by PCR site-directed mutagenesis amplifying the original vectors (see Table 2 for details) in two fragments that overlapped in the region containing the mutated palindrome and in the multiple cloning site region. In Table 2, the mutated palindrome sequences are shown as lower-case characters in the primer sequences.
Preparation of Total RNA and qRT-PCR
Total RNA of bacteria was isolated as described previously (Lerch et al., 2019). Briefly, RNA was precipitated with 1x volume isopropanol (Sigma-Aldrich, #I9516) for 10 minutes at room temperature. Pelleted RNA was washed with 70 % ethanol and solved in RNase‐free double-distilled water (ddH2O). The transcript abundance of icaApAF, icaRpAF and icaARN from three independent experiments was determined by real-time qRT-PCR. Thus, 5 µg of each RNA sample was treated with DNaseI (Thermofisher, #AM2235) for 45 min at 37°C and the reaction was stopped by phenol/chloroform/isoamylalcohol extraction (25:24:1, Carl Roth GmbH, #X985.2) with the aid of PLG heavy tubes (5 Prime, #2302830). RNA was precipitated overnight at -20°C with 4.67x volume ethanol/3M sodium acetate pH 6.5 (Thermofisher, #AM9740) (30:1 mix). Pelleted RNA was washed with 70% ethanol, dissolved in 30 µl RNase‐free ddH2O and diluted 1:10. To check for efficiency of DNA digestion, a PCR was set up with the same amount of RNA (1 µl of 1:10) and same primers used for qRT-PCR. One-step qRT–PCR was performed using an amplification kit with SYBR Green (Power SYBR™, Green RNA-to-CT™ 1-Step Kit; Thermofisher, # 4389986) with the primers listed in Table 2 and run on Biorad CFX according to the manufacturer’s instructions. Transcript abundance was calculated using a logarithmic dilution series of one sample to generate a standard curve for each gene. Relative quantification of the genes of interest was expressed in relation to the expression of the constitutive reference gene gyrase B (gyrB). The means were calculated from three biological replicates run in technical duplicates. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package.
Biofilm Assay
Biofilm formation was tested on 96-well, polystyrene tissue culture plates (Greiner Bio-One, # 655180) as described previously (Christensen et al., 1985), using Trypticase™ Soy Broth (BD BBL™, #211768) supplemented with 4% NaCl as growth medium. S. epidermidis RP62A and S. carnosus TM300 were used as positive and negative controls, respectively. For strains carrying resistance genes, antimicrobial agents were used at the following concentrations: 25 µg ml−1 erythromycin (for both overnight and day culture) and 30 µg ml-1 (for overnight culture) or 10 µg ml-1 (for day culture) chloramphenicol. Bacterial overnight cultures were freshly diluted to OD600 of 0.05 and 200 µl filled in each well (two technical replicates per strain). To distinguish between total, protein and PIA matrix-mediated biofilm production, three tissue culture plates were set up in parallel and incubated at 30°C for 18 h. Cultures were then discarded and adherent cells washed twice with 1x PBS buffer. The control plate for measuring the total biofilm was dried and heat-fixed at 65°C for 1 h. To discern between PIA- and protein- mediated biofilm, biofilms were either treated with 1 mg ml−1 proteinase K (Merck, #1245680500) for 4 h at 37°C or 40 mM NaIO4 (Carl Roth GmbH, #2603.1) for 24 h at 4°C. Afterwards the plates were washed with 1x PBS, dried and heat-fixed. All three plates were stained with 10 mg ml−1 crystal violet (Merck, #115940) for 2 min, washed twice with double-distilled water before measuring the absorbance at 492 nm (ELISA plate reader, Multiskan Ascent). The means were calculated from three biological replicates. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package.
Results
The Two ica Locus Copies in S. aureus Rd11 Are of Different Genetic Origin
We previously reported that database searches against the entire non-redundant nucleotide collection at NCBI (including whole-genome shotgun contigs) returned similarities of pAFS11 to an ica-like gene cluster present in some S. sciuri isolates [now M. sciuri (Madhaiyan et al., 2020)] (Feßler et al., 2017). Of note, the putative ica locus on pAFS11 was found to differ on nucleotide level from ica sequences present in S. aureus and S. epidermidis (Feßler et al., 2017). For further phylogenetic analysis, we therefore performed multiple sequence alignments of icaR/icaADBC nucleotide sequences from two S. aureus (i.e. RN4220 and ST398, to which Rd11 belongs) and two S. epidermidis (i.e. RP62A and O-47) strains as well as from the pAFS11 ica locus (referred to as icapAF hereafter). Finally, based on the nucleotide BLAST query results, an ica-like locus from the CoNS species S. sciuri was included into the analysis as well. Average distances were calculated from the alignment data and the tree displayed in Figure 1A illustrates that icapAF is most distantly related to the two S. aureus-derived ica loci. In addition, the two ica loci from S. epidermidis are highly divergent from icapAF, while they are closer related to ica from S. aureus. Interestingly, however, the icapAF nucleotide sequence is closely related to the ica-like locus from S. sciuri strain NS1 (Figures 1A, B). The data suggest that the two ica loci present in Rd11 are of different genetic origin, with icapAF most likely being derived from another species for which S. sciuri (M. sciuri) is a putative candidate. Although, as expected, interspecies conservation on the nucleotide level was found to be low (Figure 1B), the icapAF genes translate into amino acid sequences that are identified by the BLASTP algorithm as Ica-associated proteins with (again) some sequence differences between species. Thus, Figure 1C shows the percentage of identical amino acid positions upon pairwise alignments of Ica proteins from icapAF, S. aureus RN4220 and S. epidermidis RP62A. While comparisons between RN4220 and RP62A revealed high conservation of S. aureus and S. epidermidis Ica proteins, identical amino acid positions were much lower between icapAF and S. aureus-derived Ica proteins. In this respect, IcaR was the protein with the lowest conservation (29%), indicating that the two IcaR repressor proteins harbored by S. aureus Rd11 differ significantly on the protein sequence level (Figure 1C). Despite this apparent divergence, IcaRpAF exhibits a number of amino acid residues (marked by asterisks in the conservation histogram in Figure 1D) that are highly conserved in IcaR proteins of S. aureus and S. epidermidis as well (Figure 1D). These include the putative icaA operator-interacting residues shown for IcaR S. epidermidis (Jeng et al., 2008) (marked with red triangles on top of the sequence in Figure 1D).
Plasmid pAFS11 Has a Negative Effect on S. aureus PIA-Mediated Biofilm Formation
As mentioned above, we previously reported that S. aureus Rd11 carrying pAFS11 does not produce biofilm, nor does S. aureus RN4220 into which the plasmid was transformed, suggesting that icapAF on the plasmid might be inactive (Feßler et al., 2017). To challenge this hypothesis, we cloned the entire icapAF from pAFS11 onto the shuttle vector pRB473 (resulting in plasmid picapAF) to enable ready genetic manipulation of the locus. As a first step, we deleted the icaRpAF coding region from the vector, yielding plasmid picapAF_ΔicaR. Both plasmids (with or without icaRpAF) were transformed into S. aureus RN4220 as recipient strain and biofilm assays were performed with the constructs and corresponding wild types (Figure 2). The biofilm assays allow to detect total biofilm formation as well as to differentiate between PIA and protein matrix-mediated biofilm production (see material and methods for details). We display here (and in the following figures) solely the data for PIA-mediated biofilm formation (Figures 2–5). The entire data sets on total, protein and PIA biofilm formation can be found in Supplementary Figure S1. The assays confirmed the PIA biofilm-negative phenotype of S. aureus Rd11 and revealed that the S. aureus RN4220 wild type is a weak, but detectable PIA biofilm producer. Upon acquisition of pAFS11, PIA biofilm formation of RN4220 did not increase, but on the contrary was even slightly reduced, although this reduction was statistically not significant (+pAFS11, Figure 2). The same phenomenon occurred when S. aureus RN4220 was transformed with plasmid picapAF, carrying the entire icapAF locus from pAFS11 (+picapAF, Figure 2). In contrast, however, PIA biofilm formation of RN4220 massively increased when the icaRpAF gene was deleted from the icapAF copy on the vector (+picapAF_ΔicaR, Figure 2). In the S. aureus RN4220 +picapAF_ΔicaR strain, PIA biofilm levels even exceeded that of the S. epidermidis RP62A positive control and were much higher than in the RN4220 wild type, suggesting that the icapAF copy on the vector contributes to PIA production, but only when the IcaRpAF repressor is absent. We conclude from this that pAFS11 may exert its negative effect on S. aureus PIA biofilm formation most likely via IcaRpAF which also seems to negatively influence the ica locus on the RN4220 chromosome (hereinafter referred to as icaRN).
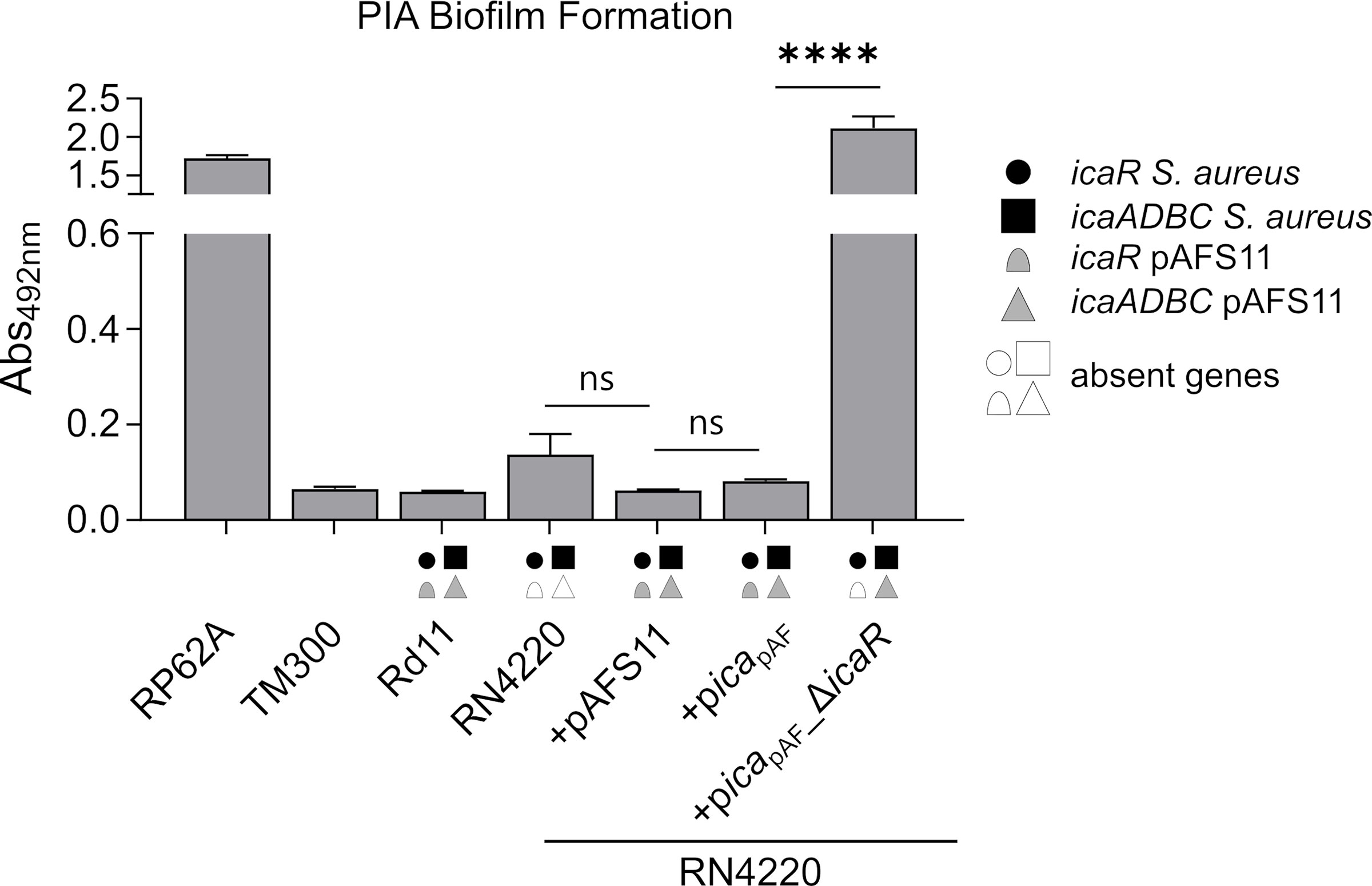
Figure 2 Effect of pAFS11 on PIA biofilm formation. Analysis of PIA biofilm production by static 96-well microtiter plate biofilm assays of Rd11, RN4220 and RN4220 transformed with plasmid pAFS11 or with a plasmid carrying the whole ica operon from pAFS11 (+picapAF) or with icaR deletion (+picapAF_ΔicaR). RP62A served as positive control, TM300 as negative control. The means were calculated from three biological replicates run in duplicates. The ica genes distinctive for each strain are depicted as symbols, with filled symbols indicating presence and empty symbols indicating absence of a given gene (as indicated in the legend). The entire data sets on total, protein and PIA biofilm formation can be found in Supplementary Figure S1. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package. ns: P = 0.1234; ****P < 0.0001.
The ica Locus on pAFS11 Is Inactive Due to Efficient IcaR Repression
The data obtained so far strongly suggest that the icapAF locus is functional and capable to enable PIA synthesis. However, by our initial experimental set-up (i.e. by employing the S. aureus RN4220 wild type with an intact chromosomal icaRN locus) it was difficult to distinguish between icapAF- and icaRN-derived PIA production. Therefore, we constructed a markerless icaRN deletion mutant in RN4220 via allelic replacement, and transformed the resulting RN4220 Δica strain with plasmids pAFS11, picapAF and picapAF_ΔicaR. Biofilm assays with the constructs confirmed loss of PIA production in the RN4220 Δica deletion mutant (Figure 3A). Providing the mutant with an entire icapAF locus either on plasmid pAFS11 or picapAF did not result in biofilm formation (Figure 3A). However, biofilm formation was triggered and highly significantly increased, when the icaRpAF repressor-encoding gene was deleted from the icapAF locus (+picapAF_ΔicaR, Figure 3A), indicating that the icaADBCpAF genes of icapAF are indeed able to mediate PIA biofilm formation, once IcaRpAF-dependent repression is alleviated. To further corroborate this assumption, we monitored transcription of icaApAF and icaRpAF by qRT-PCR in the various constructs. In strain RN4220 Δica, transformed with either pAFS11 or picapAF, weak icaApAF transcription was detectable (Figure 3B). Upon deletion of icaRpAF from the plasmid (+picapAF_ΔicaR, Figure 3B), icaRpAF transcription was no longer detectable (as expected) and icaApAF transcription levels massively increased (i.e. 200-fold) compared to the intact icapAF copy (+picapAF, Figure 3B). These findings are in agreement with the biofilm test results. From the combined data we conclude that (i) the icapAF locus on pAFS11 is functionally fully intact and (ii) icaADBCpAF operon transcription is efficiently repressed by its cognate IcaRpAF repressor.
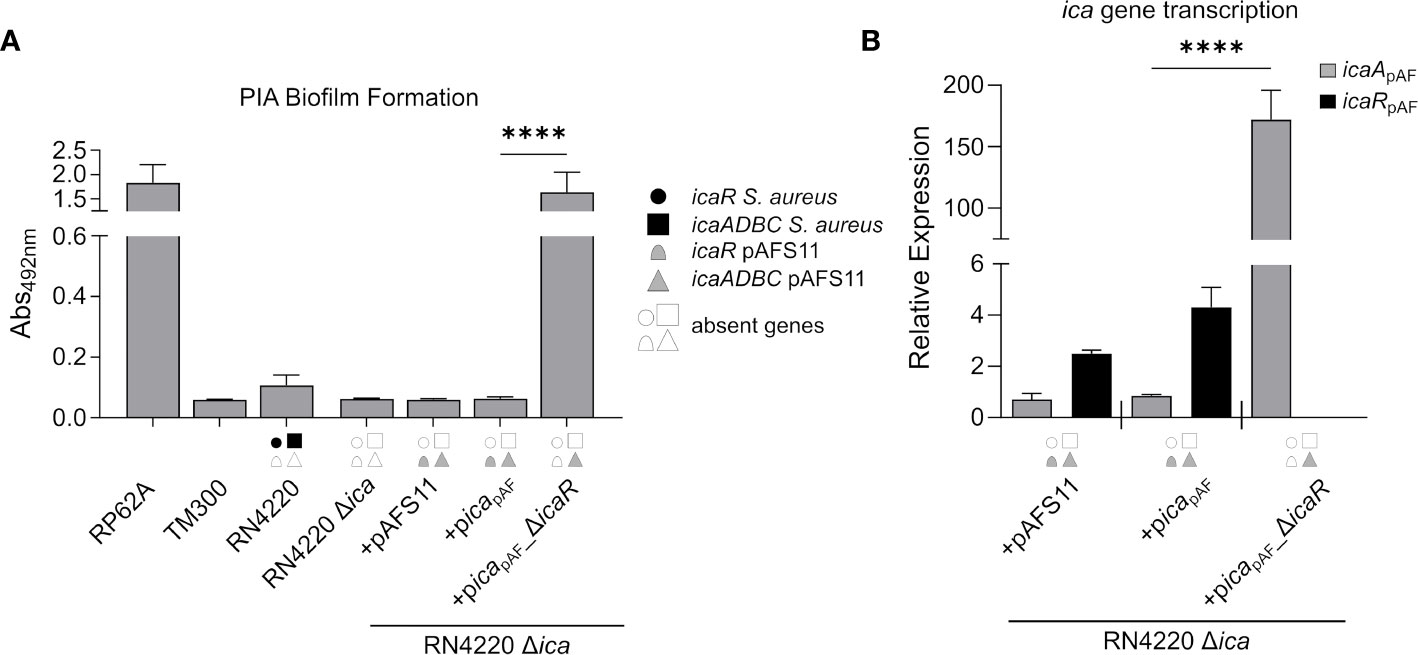
Figure 3 The ica genes of pAFS11 lead to biofilm formation. (A) Analysis of PIA biofilm production by static 96-well microtiter plate biofilm assays of strain RN4220 wild type and Δica alone or complemented with plasmid pAFS11 or with a plasmid carrying the whole ica operon from pAFS11 (+picapAF) or with icaR deletion (+picapAF_ΔicaR). RP62A served as positive control, TM300 as negative control. The entire data sets on total, protein and PIA biofilm formation can be found in Supplementary Figure S1. (B) Quantification of icaApAF and icaRpAF transcripts by qRT-PCR of strains from (A). The graph displays relative mRNA amounts using gyrB expression as reference. (A, B) The ica genes distinctive for each strain are depicted as symbols, with filled symbols indicating presence and empty symbols indicating absence of a given gene (as indicated in the legend). The means were calculated from three biological replicates run in duplicates. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package. ns: P = 0.1234; ****P < 0.0001.
IcaR From pAFS11 Represses the ica Locus in the S. aureus Chromosomal DNA and vice versa
Our initial experiments with wild type S. aureus RN4220 indicated that IcaRpAF may also inhibit icaADBCRN expression on the RN4220 chromosome (Figure 2). To substantiate this hypothesis, we constructed another set of plasmids carrying (i) the ica locus from RN4220 (picaRN), (ii) the ica locus from RN4220 lacking icaRRN (picaRN_ΔicaRRN) and (iii) the ica locus from RN4220 where we exchanged icaRRN from RN4220 with icaRpAF from pAFS11 (picaRN_ΔicaRRN_icaRpAF). All plasmids were transformed into the RN4220 Δica mutant background and the resulting strains were analyzed for their ability to form PIA biofilm (Figure 4A). Complementation of RN4220 Δica with its own icaRN locus restored PIA-mediated biofilm formation, and upon icaRRN repressor gene deletion, PIA biofilm production significantly increased, demonstrating functionality of the vector-borne icaRN locus, including icaRRN-mediated regulation (Figure 4A). Accordingly, qRT-PCR analysis confirmed that IcaRRN efficiently represses transcription of its cognate icaADBCRN operon (Figure 4B). We then asked the question whether or not expression of the chromosomal icaADBCRN operon can undergo control by the foreign IcaRpAF repressor from pAFS11. Thus, we performed biofilm tests and quantitative transcription analyses with vector picaRN_ΔicaR_icaRpAF, in which icaADBCRN was combined with the icaRpAF gene from pAFS11. As shown in Figure 4, presence of icaRpAF significantly diminished PIA production and transcription of the icaADBCRN operon, suggesting the capability of IcaRpAF to control the icaADBCRN copy from RN4220 (Figure 4). Vice versa, we next investigated, if IcaRRN from RN4220 can influence icaADBCpAF from pAFS11. For this purpose, we additionally constructed vector picapAF_ΔicaRpAF_icaRRN which was transformed into the RN4220 Δica mutant background. Biofilm testing revealed a highly significant reduction of PIA production when icaADBCpAF was combined with icaRRN, suggesting that IcaR from S. aureus RN4220 is indeed able to repress the icaADBCpAF genes from pAFS11 (Figure 4C).
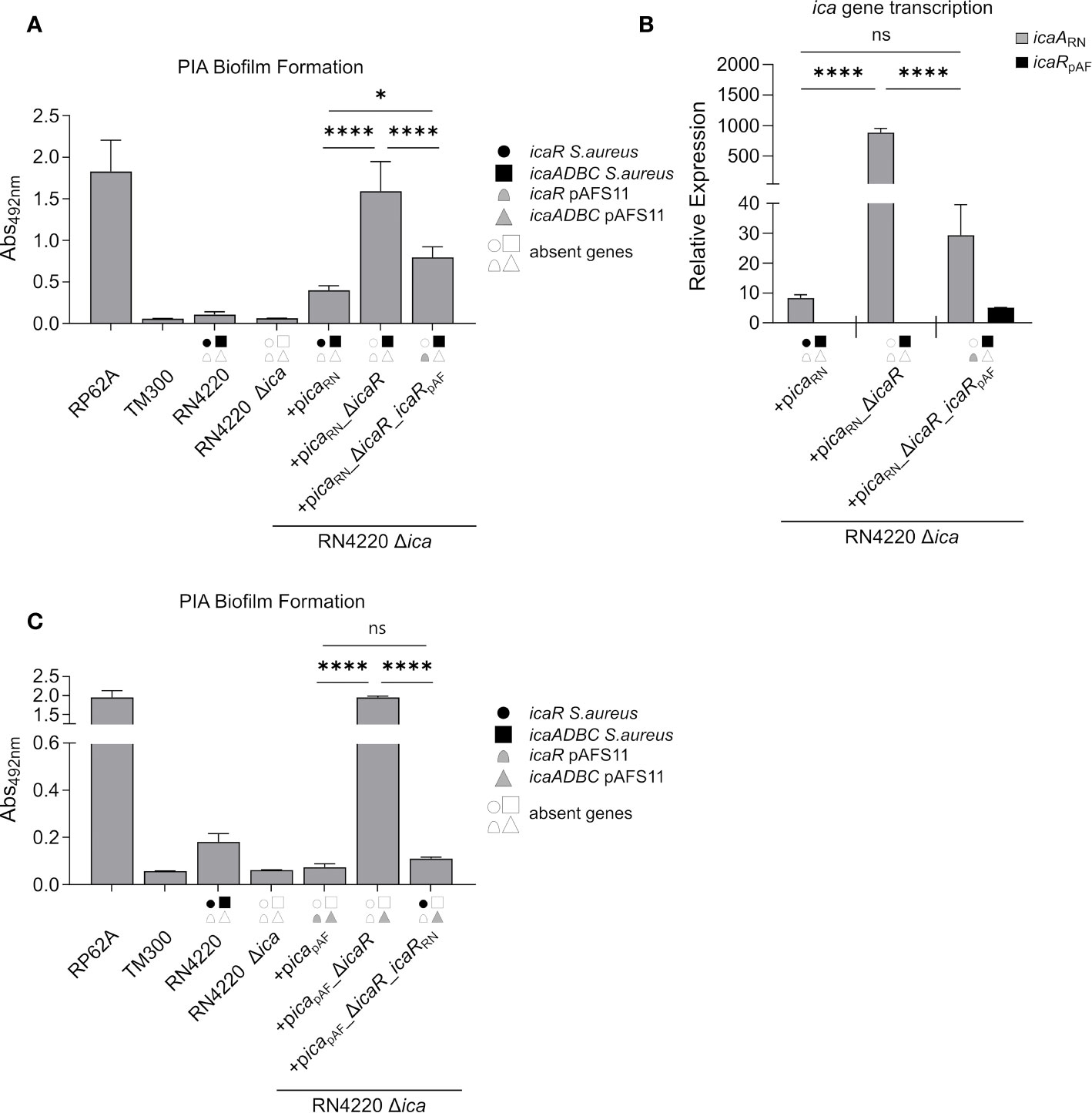
Figure 4 IcaRpAF controls expression of ica genes from RN4220 and vice versa. (A) Analysis of PIA biofilm production by static 96-well microtiter plate biofilm assays of strain RN4220 wild type and Δica alone or complemented with a plasmid carrying the ica operon of RN4220 (+picaRN) or with icaR deletion (+picaRN_ΔicaR) or with icaRpAF instead of icaRRN(+picaRN_ΔicaR_icaRpAF). The entire data sets on total, protein and PIA biofilm formation can be found in Supplementary Figure S1. (B) Quantification of icaARN and icaRpAF transcripts by qRT-PCR of complemented strains from (A). The graph displays relative mRNA amounts using gyrB expression as reference. (C) Analysis of PIA biofilm production by static 96-well microtiter plate biofilm assays of RN4220 wild type and Δica alone or complemented with the ica operon of plasmid pAFS11 on a working plasmid (+picapAF) or the ica operon of plasmid pAFS11 with icaR deletion (+picapAF_ΔicaR) or with icaRRN instead of icaRpAF (+picapAF_ΔicaR_icaRRN). (A, C) RP62A served as positive control, TM300 as negative control. (A–C) The ica genes distinctive for each strain are depicted as symbols, with filled symbols indicating presence and empty symbols indicating absence of a given gene (as indicated in the legend). The means were calculated from three biological replicates run in duplicates. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package. ns: P = 0.1234; *P = 0.0332; ****P < 0.0001.
A Palindrome Sequence in the icaApAF Upstream Region Is Required for IcaR-Mediated Biofilm Repression
The data obtained so far demonstrate that the pAFS11- and RN4220-derived IcaR repressors, which differ on amino acid level (Figure 1D), are able to control the icaADBC operon of the respective other ica locus copy. To understand the molecular prerequisites for the IcaR interactions with their DNA targets, we focused on the nucleotide sequence constraints known to be involved in IcaR binding. IcaR was previously shown to bind as a dimer to a specific palindrome sequence (ACCTANCTNNC/GNNAGNTAGGT) present in the icaA operator of S. epidermidis (Jeng et al., 2008). This palindrome is 22 nucleotides long and contains six mismatches (22,6) (Figure 5A, top). Of note, the sequence is highly conserved and is also present in the S. aureus icaA promoter region (Figure 5A, top). Surprisingly, although IcaR from S. aureus is clearly able to control PIA production from pAFS11, the S. aureus-like palindrome sequence stretch lacks in icapAF (Figure 5A, bottom). Instead, the icaApAF upstream region displays two other palindromes which differ at the nucleotide level from that of the known S. aureus/S. epidermidis recognition site. Thus, palindrome A (TNAAAATNNTA/TANNATTTTNA) is 22 nucleotides long and harbors six mismatches (22,6), while palindrome B (CNAACNANC/GNTNGTTNG) consists of 18 nucleotides with six mismatches (18,6) (Figure 5A, bottom). To investigate the putative involvement of these palindromes in IcaR function, we mutated either palindrome A or B in (i) a plasmid carrying the whole icapAF, (ii) in an icapAF plasmid with an icaRpAF deletion as well as (iii) in an icaRpAF deletion vector complemented with icaRRN. The plasmids were again transformed into the RN4220 Δica mutant background and analyzed for PIA-mediated biofilm formation. Figure 5B demonstrates that an altered palindrome A sequence did not influence icaR-mediated biofilm control. Thus, upon palindrome A mutation, PIA-mediated biofilm production remained repressed in picapAF(A***), indicating that IcaRpAF does not require this nucleotide sequence stretch for action. As expected, PIA production was derepressed when icaRpAF was lacking in the palindrome A mutant (+picapAF(A***)_ΔicaR, Figure 5B). Most importantly, however, icaRRN was still able to completely downregulate PIA-mediated biofilm production in this construct, confirming that palindrome A is not an interaction site for IcaR, neither for IcaR proteins derived from pAFS11 nor from RN4220 (Figure 5B). In contrast, mutation of palindrome B had a profound impact on biofilm regulation by IcaR. Firstly, PIA production was found to be deregulated and increased in an icapAF construct carrying an altered palindrome B nucleotide sequence (+picapAF(B***)), suggesting that control by the cognate IcaRpAF is significantly impaired when integrity of this sequence stretch is disturbed (Figure 5B). Moreover, PIA-mediated biofilm production further increased in a palindrome B mutant in which icaRpAF was deleted (+picapAF(B***)_ΔicaR, Figure 5B), which speaks in favor of residual IcaRpAF repressor activity in the palindrome B mutant. Finally, when providing the mutant with IcaRRN from RN4220, biofilm repression was partially, but not fully restored which (again) is in good agreement with residual IcaR repression in the palindrome B mutant. Together, the combined data strongly suggest that IcaR from both pAFS11 and RN4220 require an intact palindrome B (but not palindrome A) for unfolding their repressor activity on the icapAF locus.
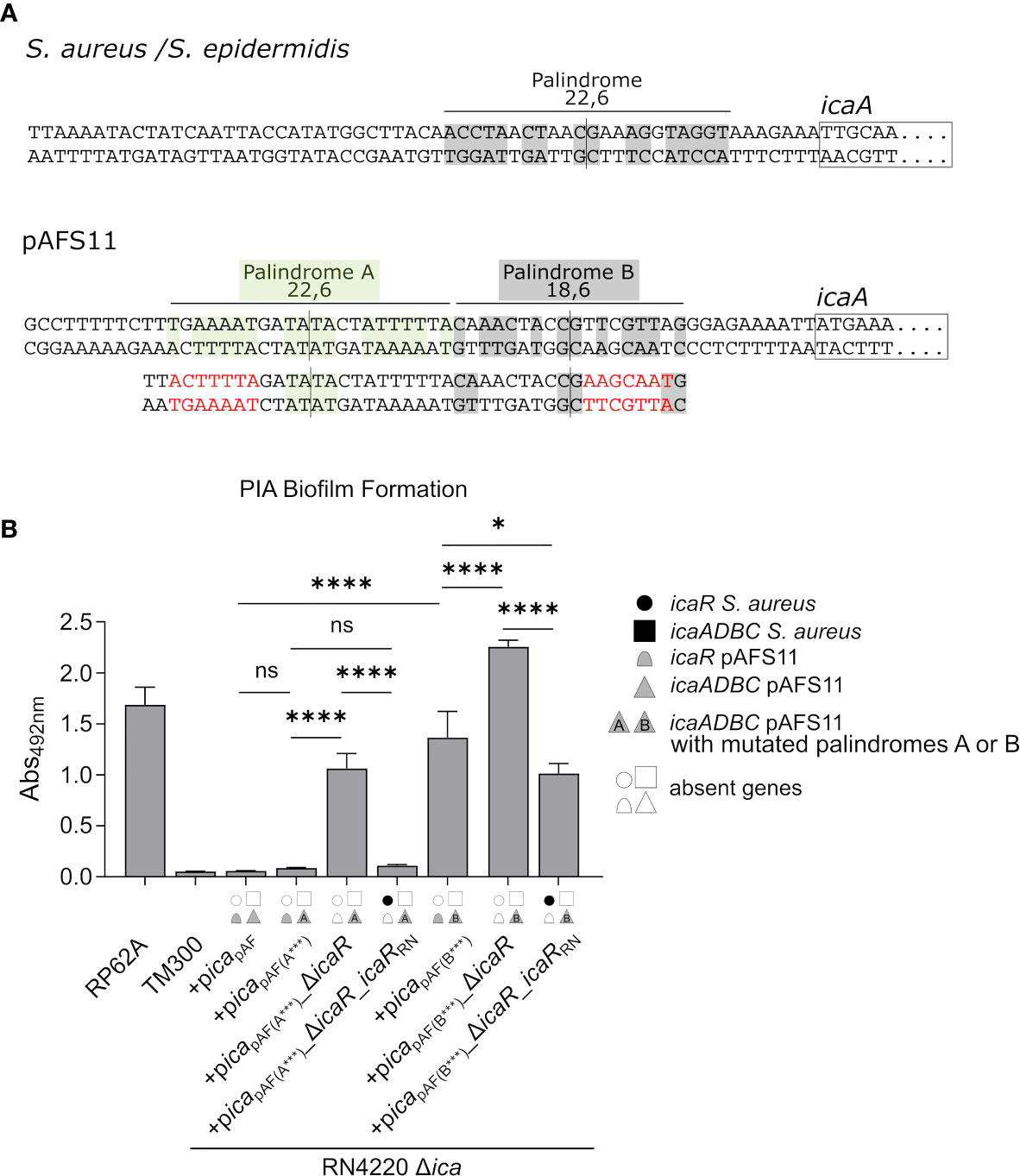
Figure 5 IcaR proteins from S. aureus and from pAFS11 require a palindrome on the icaApAF operator for action. (A) TOP: Known palindrome sequence on S. aureus/S. epidermidis is shown at the top of the panel (Jeng et al., 2008). 22,6 stands for a palindrome which is 22 nt long and carries six mismatches. BOTTOM: Putative palindromes A and B on icaApAF operator are shown and their characteristics are reported. The nucleotide mutations introduced are shown in red below the wild type sequence alongside with the resulting palindrome perturbations. (B) Analysis of PIA biofilm production by static 96-well microtiter plate biofilm assays of strain RN4220 Δica complemented with the ica operon from pAFS11 wild type (+picapAF) or with deletion of icaRpAF (+picapAF_ΔicaR) or with icaRRN instead of icaRpAF (+picapAF_ΔicaR_icaRRN), carrying a mutated palindrome A (icapAF(A***)) or B (icapAF(B***)). The ica genes distinctive for each strains are depicted as symbols, with filled symbols indicating presence and empty symbols indicating absence of a given gene (as indicated in the legend). The entire data sets on total, protein and PIA biofilm formation can be found in Supplementary Figure S1. The means were calculated from three biological replicates run in duplicates. Statistical analysis was performed using one-way ANOVA by employing the GraphPad Prism software package. ns: P = 0.1234; *P = 0.0332; ****P < 0.0001.
Discussion
Acquisition of mobile genetic elements (MGEs) is often beneficial for bacteria by providing novel metabolic and resistance traits. However, MGE carriage may also come at considerable cost for the recipient bacterial cell (Slater et al., 2008; San Millan and MacLean, 2017). Thus, resources will be required to replicate and maintain MGEs (e.g. plasmids) on which beneficial genes are located and their (inappropriate) expression may impose a metabolic burden, resulting in reduced fitness and competitiveness of MGE-bearing bacterial cells (Baltrus, 2013). Accordingly, bacteria have evolved sophisticated mechanisms to control both MGE uptake and maintenance as well as expression of horizontally acquired genes (Brantl, 2014; Kwong et al., 2017; Firth et al., 2018). In case of plasmid-mediated HGT, this often involves a regulatory crosstalk between chromosomal factors and the newly acquired plasmid (Huang et al., 1990; Charles and Nester, 1993; Baños et al., 2009). Interestingly, these control networks are not unidirectional and there is growing evidence to suggest that plasmids are able to influence chromosomal gene expression as well in a wide range of bacterial species [recently reviewed in (Vial and Hommais, 2020)]. In the study presented here, we extend these examples by the Gram-positive pathogen S. aureus and reveal that horizontally acquired and core genome genes have the capacity to mutually influence each other in this organism. Thus, we demonstrate that a transcription factor (i.e. IcaR), located on a large multi-resistance plasmid, is able to target a pathogenicity factor (i.e. icaADBC-mediated PIA biofilm formation) from the S. aureus core genome. Vice versa, the IcaR homolog from the S. aureus core genome was found to be able to silence transcription of plasmid-borne icaADBC genes, creating a bi-directional regulatory crosstalk between plasmid- and chromosomally encoded factors that eventually hindered metabolically costly PIA-mediated biofilm formation.
PIA consists of N-acetyl-glucosamines (GlcNac) molecules, and ample sugar and energy supplies are fuelled into GlcNac synthesis to provide the building blocks of the exopolysaccharide. Accordingly, ica gene expression is intimately linked to central carbon flux control and energy balance (Vuong et al., 2005; Seidl et al., 2008; Zhu et al., 2009; Sadykov et al., 2011; Lindgren et al., 2014) which also involves the action of non-coding RNAs to appropriately adjust metabolic patterns (Rochat et al., 2018; Bronesky et al., 2019; Marincola et al., 2019; Schoenfelder et al., 2019). Presence of two fully functional ica gene clusters in strain S. aureus Rd11 is likely to represent a major metabolic challenge and the observed downregulation of PIA production in this strain makes sense to prevent metabolic overload. Paradoxically, it is just the additionally acquired icapAF locus copy on plasmid pAFS11 that mediates a biofilm-negative phenotype. Indeed, acquisition of pAFS11 or the icapAF locus alone abolished PIA biofilm formation in S. aureus (Figure 2). This effect is accomplished through the IcaRpAF repressor which can also target the chromosomal ica locus copy (Figure 4). Moreover, the tight self-control of the icapAF copy on pAFS11 by its cognate IcaRpAF repressor further contributes to a biofilm-negative phenotype (Figures 2, 3). Remarkably, icaADBCpAF expression can be additionally repressed by IcaRN from the core genome (Figure 4C). Thus, although being different from the canonical IcaR recognition site known from S. epidermidis and S. aureus (Jeng et al., 2008), IcaRN interacts with a palindrome sequence present in the upstream region of icaADBCpAF (palindrome B in Figure 5A), revealing a certain flexibility of IcaR-like proteins in DNA target selection (Figure 5).
Phylogenetic analyses revealed that the two ica loci in Rd11 differ both on nucleotide and protein sequence levels and are only distantly related to each other (Figure 1). Thus, the horizontally acquired icapAF copy on pAFS11 is likely to originate from an unknown bacterium for which the soil and animal dwelling species S. sciuri [recently re-classified as M. sciuri (Madhaiyan et al., 2020)] might represent a putative candidate (Feßler et al., 2017). But clearly more detailed investigations will be required to substantiate this hypothesis. Interestingly, inhibition of core genome-encoded exopolysaccharide production by plasmids seems to be a common theme in the bacterial world. Thus, in nitrogen-fixing Rhizobium tropici, exopolysaccharide production was found to be downregulated by the NrcR transcription factor, encoded on an acquired plasmid (Del Cerro et al., 2016), and in the nosocomial pathogen Acinetobacter baumanii, PNAG (PIA) production was described to be diminished upon acquisition of a multi-resistance plasmid (Venanzio et al., 2019). In the latter case, this large conjugative multiresistance plasmid facilitates its own transmission by downregulating chromosomally encoded type-6-secretion systems (T6SS) that usually hamper HGT (Venanzio et al., 2019). Together with the effect on PNAG production, the A. baumanii plasmids represent interesting examples for a plasmid-chromosome regulatory crosstalk that influences simultaneously both virulence and resistance traits.
LA-MRSA lineages of the clonal complex CC398 (to which strain Rd11 belongs to) can thrive in very different habitats (e.g. animals, humans, environment etc.) where they are exposed to numerous stress conditions. It is tempting to speculate that PIA biofilm formation, which is an important factor in S. aureus pathogenesis and survival (Fluckiger et al., 2005; Kropec et al., 2005), might become a selection advantage at some stage. In this respect, it is an interesting question by which mechanism(s) biofilm formation could be restored in the Rd11 isolate. Plasmid pAFS11 displays a striking mosaic structure and has most likely arisen by recombination of genes from various origins. Assuming that icapAF locus integration into pAFS11 and acquisition of the plasmid by S. aureus is an evolutionarily recent event, it is conceivable that control of pAFS11-encoded gene expression is not fully integrated into the regulatory network of the S. aureus Rd11 recipient (yet). One obvious possibility to (re)gain a biofilm positive phenotype would be (spontaneous) mutation and inactivation of icaRpAF on pAFS11. At first sight, this idea seems to be contradictory to our experimental findings showing that IcaRRN has the potential to take over and replace the lacking IcaRpAF activity. This mutual IcaR replacement seems to work particularly well, when both factors (icaR and icaADBC) are on the same replicon and are probably in an appropriate stoichiometric repressor/DNA equilibrium (Figure 4C). In agreement with this assumption, the IcaRRN repressor effect was found to be less efficient when a single icaRRN copy resides on the chromosome and icaADBCpAF is located on a (multi-copy) plasmid (Figure 2). Here, icaRpAF deletion on the vector enabled PIA production, and it is reasonable to suggest that the insufficient repressor activity of IcaRN in this experimental set-up might be associated with the copy number of the icapAF-bearing vector which increased the number of DNA targets for the IcaRRN repressor. At the present stage of experimental work this is mere speculation. However, recent research demonstrates that available IcaR protein amounts are critical for the appropriate control of icaADBC transcription, and multiple cis- and trans-acting factors have been identified that target icaR mRNA molecules to fine-tune their translation (Ruiz de los Mozos et al., 2013; Rochat et al., 2018; Bronesky et al., 2019; Lerch et al., 2019; Schoenfelder et al., 2019). Plasmid copy number effects may have the potential to interfere with this delicate repressor/DNA target balance. In this respect, the data obtained with icapAF-bearing plasmids in the icaRN locus proficient S. aureus RN4220 background may reflect very well the natural situation in pAFS11 carrying isolates (Figure 2). Thus, from an evolutionary point of view, mutational inactivation of icaRpAF on the pAFS11 plasmid would make sense to readily enable PIA production. The same effect could be achieved by acquiring mutations in the icapAF palindrome B sequence, which represents the target site of both the IcaRpAF and IcaRRN proteins. Indeed, at least for S. aureus core genome ica loci, mutations in icaA upstream regions were previously described. The mutations had direct consequences for PIA production and occurred both in vitro and in vivo, indicating that the icaA promoter region undergoes selection and is a suitable target to modulate PIA production (Jefferson et al., 2003; Schwartbeck et al., 2016). It will be interesting to explore if the icapAF locus on pAFS11 might become subject to mutational variation. Long-term in vitro passage experiments together with surveillance of the evolution of pAFS11-like plasmids in field studies will be suitable approaches to give answers to how control of plasmid-borne ica locus expression will be integrated into the regulatory network of S. aureus.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
GM, AF, SS, and WZ: conceived and designed the experiments. GM, GJ and A-KK performed the experiments. GM, GJ, A-KK, FW and WZ analyzed the data. GM and WZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the German Research Council (DFG) through grant ZI665/3-1 as well as by the German Federal Ministry of Education and Research (BMBF), grant numbers 01KI1727E, 01KI1727D, 01KI2009D as part of the Research Network Zoonotic Infectious Diseases as well as grant number 16GW0297 within the program ‘Target validation for pharmaceutical drug development’. This publication was supported by the Open Access Publication Fund of the University of Wuerzburg.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Sonja Schoenfelder for critical discussions and her continuous interest in this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.660702/full#supplementary-material
References
Bae, T., Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Ballhausen, B., Kriegeskorte, A., van Alen, S., Jung, P., Köck, R., Peters, G., et al. (2017). The pathogenicity and host adaptation of livestock-associated MRSA CC398. Vet. Microbiol. 200, 39–45. doi: 10.1016/j.vetmic.2016.05.006
Baltrus, D. A. (2013). Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495. doi: 10.1016/j.tree.2013.04.002
Baños, R. C., Vivero, A., Aznar, S., García, J., Pons, M., Madrid, C., et al. (2009). Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 5, e1000513. doi: 10.1371/journal.pgen.1000513
Becker, K., Both, A., Weißelberg, S., Heilmann, C., Rohde, H. (2020). Emergence of coagulase-negative staphylococci. Expert Rev. Anti Infect. Ther. 18, 349–366. doi: 10.1080/14787210.2020.1730813
Brantl, S. (2014). Plasmid Replication Control by Antisense RNAs. Microbiol. Spectr. 2, PLAS-0001-2013. doi: 10.1128/microbiolspec.PLAS-0001-2013
Bronesky, D., Desgranges, E., Corvaglia, A., François, P., Caballero, C. J., Prado, L., et al. (2019). A multifaceted small RNA modulates gene expression upon glucose limitation in Staphylococcus aureus. EMBO J. 38, e99363. doi: 10.15252/embj.201899363
Brückner, R., Wagner, E., Götz, F. (1993). Characterization of a sucrase gene from Staphylococcus xylosus. J. Bacteriol. 175, 851–857. doi: 10.1128/jb.175.3.851-857.1993
Cerca, N., Brooks, J. L., Jefferson, K. K. (2008). Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190, 6530–6533. doi: 10.1128/JB.00482-08
Charles, T. C., Nester, E. W. (1993). A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175, 6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993
Christensen, G. D., Simpson, W. A., Younger, J. J., Baddour, L. M., Barrett, F. F., Melton, D. M., et al. (1985). Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006. doi: 10.1128/JCM.22.6.996-1006.1985
Conlan, S., Mijares, L. A., NISC Comparative Sequencing Program, B, Becker, J., Blakesley, R. W., Bouffard, G. G., et al. (2012). Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64. doi: 10.1186/gb-2012-13-7-r64
Conlon, K. M., Humphreys, H., O’Gara, J. P. (2002a). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184, 4400–4408. doi: 10.1128/jb.184.16.4400-4408.2002
Conlon, K. M., Humphreys, H., O’Gara, J. P. (2002b). Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216, 171–177. doi: 10.1111/j.1574-6968.2002.tb11432.x
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., Götz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433. doi: 10.1128/IAI.67.10.5427-5433.1999
Cue, D., Lei, M. G., Lee, C. Y. (2012). Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell. Infect. Microbiol. 2:38. doi: 10.3389/fcimb.2012.00038
Del Cerro, P., Rolla-Santos, A. A. P., Valderrama-Fernández, R., Gil-Serrano, A., Bellogín, R. A., Gomes, D. F., et al. (2016). NrcR, a New Transcriptional Regulator of Rhizobium tropici CIAT 899 Involved in the Legume Root-Nodule Symbiosis. PLoS One 11, e0154029. doi: 10.1371/journal.pone.0154029
Espadinha, D., Sobral, R. G., Mendes, C. I., Méric, G., Sheppard, S. K., Carriço, J. A., et al. (2019). Distinct Phenotypic and Genomic Signatures Underlie Contrasting Pathogenic Potential of Staphylococcus epidermidis Clonal Lineages. Front. Microbiol. 10:1971. doi: 10.3389/fmicb.2019.01971
Feßler, A. T., Zhao, Q., Schoenfelder, S., Kadlec, K., Brenner Michael, G., Wang, Y., et al. (2017). Complete sequence of a plasmid from a bovine methicillin-resistant Staphylococcus aureus harbouring a novel ica-like gene cluster in addition to antimicrobial and heavy metal resistance genes. Vet. Microbiol. 200, 95–100. doi: 10.1016/j.vetmic.2016.07.010
Feßler, A., Kadlec, K., Wang, Y., Zhang, W.-J., Wu, C., Shen, J., et al. (2018). Small Antimicrobial Resistance Plasmids in Livestock-Associated Methicillin-Resistant Staphylococcus aureus CC398. Front. Microbiol. 9:2063. doi: 10.3389/fmicb.2018.02063
Fey, P. D., Olson, M. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. doi: 10.2217/fmb.10.56
Firth, N., Jensen, S. O., Kwong, S. M., Skurray, R. A., Ramsay, J. P. (2018). Staphylococcal Plasmids, Transposable and Integrative Elements. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.GPP3-0030-2018
Fluckiger, U., Ulrich, M., Steinhuber, A., Döring, G., Mack, D., Landmann, R., et al. (2005). Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73, 1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. doi: 10.1093/femsre/fux007
Geiger, T., Francois, P., Liebeke, M., Fraunholz, M., Goerke, C., Krismer, B., et al. (2012). The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8, e1003016. doi: 10.1371/journal.ppat.1003016
Gill, S. R., Fouts, D. E., Archer, G. L., Mongodin, E. F., Deboy, R. T., Ravel, J., et al. (2005). Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., Götz, F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Heilmann, C., Ziebuhr, W., Becker, K. (2019). Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 25, 1071–1080. doi: 10.1016/j.cmi.2018.11.012
Hoang, T.-M., Zhou, C., Lindgren, J. K., Galac, M. R., Corey, B., Endres, J. E., et al. (2019). Transcriptional Regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J. Bacteriol. 201, 1–17. doi: 10.1128/jb.00524-18
Huang, M. L., Cangelosi, G. A., Halperin, W., Nester, E. W. (1990). A chromosomal Agrobacterium tumefaciens gene required for effective plant signal transduction. J. Bacteriol. 172, 1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990
Jefferson, K. K., Cramton, S. E., Götz, F., Pier, G. B. (2003). Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48, 889–899. doi: 10.1046/j.1365-2958.2003.03482.x
Jefferson, K. K., Pier, D. B., Goldmann, D. A., Pier, G. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. doi: 10.1128/jb.186.8.2449-2456.2004
Jeng, W. Y., Ko, T. P., Liu, C. I., Guo, R. T., Liu, C. L., Shr, H. L., et al. (2008). Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 36, 1567–1577. doi: 10.1093/nar/gkm1176
Kadlec, K., Feßler, A. T., Hauschild, T., Schwarz, S. (2012). Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 745–755. doi: 10.1111/j.1469-0691.2012.03842.x
Kozitskaya, S., Olson, M. E., Fey, P. D., Witte, W., Ohlsen, K., Ziebuhr, W. (2005). Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43, 4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005
Kropec, A., Maira-Litran, T., Jefferson, K. K., Grout, M., Cramton, S. E., Götz, F., et al. (2005). Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 73, 6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005
Kwong, S. M., Ramsay, J. P., Jensen, S. O., Firth, N. (2017). Replication of Staphylococcal Resistance Plasmids. Front. Microbiol. 8:2279. doi: 10.3389/fmicb.2017.02279
Lakhundi, S., Zhang, K. (2018). Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 31. doi: 10.1128/CMR.00020-18
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018a). Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 4, 18033. doi: 10.1038/nrdp.2018.33
Lee, J. Y. H., Monk, I. R., Gonçalves da Silva, A., Seemann, T., Chua, K. Y. L., Kearns, A., et al. (2018b). Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 3, 1175–1185. doi: 10.1038/s41564-018-0230-7
Lerch, M. F., Schoenfelder, S. M. K., Marincola, G., Wencker, F. D. R., Eckart, M., Förstner, K. U., et al. (2019). A non-coding RNA from the intercellular adhesion (ica) locus of Staphylococcus epidermidis controls polysaccharide intercellular adhesion (PIA)-mediated biofilm formation. Mol. Microbiol. 111, 1571–1591. doi: 10.1111/mmi.14238
Lindgren, J. K., Thomas, V. C., Olson, M. E., Chaudhari, S. S., Nuxoll, A. S., Schaeffer, C. R., et al. (2014). Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J. Bacteriol. 196, 2277–2289. doi: 10.1128/JB.00051-14
Mack, D., Fischer, W., Krokotsch, A., Leopold, K., Hartmann, R., Egge, H., et al. (1996). The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183. doi: 10.1128/jb.178.1.175-183.1996
Madeira, F., Park, Y., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Madhaiyan, M., Wirth, J. S., Saravanan, V. S. (2020). Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 70, 5926–5936. doi: 10.1099/ijsem.0.004498
Marincola, G., Wencker, F. D. R., Ziebuhr, W. (2019). The Many Facets of the Small Non-coding RNA RsaE (RoxS) in Metabolic Niche Adaptation of Gram-Positive Bacteria. J. Mol. Biol. 431, 4684–4698. doi: 10.1016/j.jmb.2019.03.016
Méric, G., Miragaia, M., de Been, M., Yahara, K., Pascoe, B., Mageiros, L., et al. (2015). Ecological Overlap and Horizontal Gene Transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol. 7, 1313–1328. doi: 10.1093/gbe/evv066
Méric, G., Mageiros, L., Pensar, J., Laabei, M., Yahara, K., Pascoe, B., et al. (2018). Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat. Commun. 9, 5034. doi: 10.1038/s41467-018-07368-7
Monk, I. R., Shah, I. M., Xu, M., Tan, M.-W., Foster, T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3. doi: 10.1128/mBio.00277-11
Nair, D., Memmi, G., Hernandez, D., Bard, J., Beaume, M., Gill, S., et al. (2011). Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193, 2332–2335. doi: 10.1128/JB.00027-11
Nguyen, H. T. T., Nguyen, T. H., Otto, M. (2020). The staphylococcal exopolysaccharide PIA - Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 18, 3324–3334. doi: 10.1016/j.csbj.2020.10.027
Nozaki, S., Niki, H. (2019). Exonuclease III (XthA) Enforces In Vivo DNA Cloning of Escherichia coli To Create Cohesive Ends. J. Bacteriol. 201, 1–13. doi: 10.1128/JB.00660-18
Qin, Z., Ou, Y., Yang, L., Zhu, Y., Tolker-Nielsen, T., Molin, S., et al. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153, 2083–2092. doi: 10.1099/mic.0.2007/006031-0
Que, Y.-A., Haefliger, J.-A., Piroth, L., François, P., Widmer, E., Entenza, J. M., et al. (2005). Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201, 1627–1635. doi: 10.1084/jem.20050125
Rochat, T., Bohn, C., Morvan, C., Le Lam, T. N., Razvi, F., Pain, A., et al. (2018). The conserved regulatory RNA RsaE down-regulates the arginine degradation pathway in Staphylococcus aureus. Nucleic Acids Res. 46, 8803–8816. doi: 10.1093/nar/gky584
Rohde, H., Burdelski, C., Bartscht, K., Hussain, M., Buck, F., Horstkotte, M. A., et al. (2005). Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x
Ruiz de los Mozos, I., Vergara-Irigaray, M., Segura, V., Villanueva, M., Bitarte, N., Saramago, M., et al. (2013). Base pairing interaction between 5’- and 3’-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 9, e1004001. doi: 10.1371/journal.pgen.1004001
Sadykov, M. R., Hartmann, T., Mattes, T. A., Hiatt, M., Jann, N. J., Zhu, Y., et al. (2011). CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157, 3458–3468. doi: 10.1099/mic.0.051243-0
San Millan, A., MacLean, R. C. (2017). Fitness Costs of Plasmids: a Limit to Plasmid Transmission. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.MTBP-0016-2017
Schilcher, K., Horswill, A. R. (2020). Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 84. doi: 10.1128/MMBR.00026-19
Schoenfelder, S. M. K., Lange, C., Prakash, S. A., Marincola, G., Lerch, M. F., Wencker, F. D. R., et al. (2019). The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog. 15, e1007618. doi: 10.1371/journal.ppat.1007618
Schwartbeck, B., Birtel, J., Treffon, J., Langhanki, L., Mellmann, A., Kale, D., et al. (2016). Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. 12, e1006024. doi: 10.1371/journal.ppat.1006024
Seidl, K., Goerke, C., Wolz, C., Mack, D., Berger-Bächi, B., Bischoff, M. (2008). Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76, 2044–2050. doi: 10.1128/IAI.00035-08
Slater, F. R., Bailey, M. J., Tett, A. J., Turner, S. L. (2008). Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 66, 3–13. doi: 10.1111/j.1574-6941.2008.00505.x
Thomas, J. C., Zhang, L., Robinson, D. A. (2014). Differing lifestyles of Staphylococcus epidermidis as revealed through Bayesian clustering of multilocus sequence types. Infect. Genet. Evol. 22, 257–264. doi: 10.1016/j.meegid.2013.06.020
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Venanzio, G., Moon, K. H., Weber, B. S., Lopez, J., Ly, P. M., Potter, R. F., et al. (2019). Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc. Natl. Acad. Sci. U. S. A. 116, 1378–1383. doi: 10.1073/pnas.1812557116
Vial, L., Hommais, F. (2020). Plasmid-chromosome cross-talks. Environ. Microbiol. 22, 540–556. doi: 10.1111/1462-2920.14880
Vuong, C., Kidder, J. B., Jacobson, E. R., Otto, M., Proctor, R. A., Somerville, G. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187, 2967–2973. doi: 10.1128/JB.187.9.2967-2973.2005
Wang, X., Preston, J. F., Romeo, T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734. doi: 10.1128/jb.186.9.2724-2734.2004
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Keywords: Staphylococcus aureus, biofilm regulation, PIA/ica, IcaR, horizontal gene transfer, plasmid-chromosome crosstalk
Citation: Marincola G, Jaschkowitz G, Kieninger A-K, Wencker FDR, Feßler AT, Schwarz S and Ziebuhr W (2021) Plasmid-Chromosome Crosstalk in Staphylococcus aureus: A Horizontally Acquired Transcription Regulator Controls Polysaccharide Intercellular Adhesin-Mediated Biofilm Formation. Front. Cell. Infect. Microbiol. 11:660702. doi: 10.3389/fcimb.2021.660702
Received: 29 January 2021; Accepted: 02 March 2021;
Published: 22 March 2021.
Edited by:
Yinduo Ji, University of Minnesota Twin Cities, United StatesReviewed by:
Hanne Ingmer, University of Copenhagen, DenmarkChia Y. Lee, University of Arkansas for Medical Sciences, United States
Iñigo Lasa, Public University of Navarre, Spain
Copyright © 2021 Marincola, Jaschkowitz, Kieninger, Wencker, Feßler, Schwarz and Ziebuhr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella Marincola, Z2FicmllbGxhLm1hcmluY29sYUB1bmktd3VlcnpidXJnLmRl, Z2FicmllbGxhQG1hcmluY29sYS5ldQ==; Wilma Ziebuhr, dy56aWVidWhyQG1haWwudW5pLXd1ZXJ6YnVyZy5kZQ==
†Present address: Greta Jaschkowitz, Department of Urology and Pediatric Urology, Saarland University, Homburg, Germany
Ann-Katrin Kieninger, Interfaculty Institute of Microbiology and Infection Medicine Tübingen, University of Tübingen, Tübingen, Germany
 Gabriella Marincola
Gabriella Marincola Greta Jaschkowitz
Greta Jaschkowitz Ann-Katrin Kieninger
Ann-Katrin Kieninger Freya D.R. Wencker
Freya D.R. Wencker Andrea T. Feßler
Andrea T. Feßler Stefan Schwarz
Stefan Schwarz Wilma Ziebuhr
Wilma Ziebuhr