- 1Department of Critical Care Medicine, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Sir Run Run Shaw Hospital, Hangzhou, China
- 3Department of Rehabilitation, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 4Department of Infectious Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 5Department of Emergency Medicine, Lanxi People’s Hospital, Lanxi, China
- 6Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is highly prevalent and poses a significant threat to public health. In critically ill patients, gut colonization is considered to be the reservoir of recurrent CRKP infection. Therefore, eliminating CRKP carriage in the intestine is critical for preventing subsequent CRKP infection. In the present study, Lactobacillus plantarum LP1812, a probiotic that can inhibit CRKP in vitro, was used as a candidate probiotic to investigate its efficacy for CRKP anticolonization. Compared with the control, mice fed with 1×10 8 CFU L. plantarum LP1812 exhibited significant CRKP clearance from 1×10 4 CFU/mg to less than 10 CFU/mg in mice feces. Furthermore, 16S RNA gene sequencing revealed that L. plantarum LP1812 modulated mice microbiota by increasing the relative abundance of the genus Halomanas, Blautia, and Holdemania. Further KEGG pathway enrichment analysis revealed that fatty acid-utilizing bacteria, such as acetate-producing Bacteroidetes and Blautia flourished in mice fed with L. plantarum LP1812. Moreover, we found that the concentration of acetic acid was higher in L. plantarum LP1812, which inhibited the growth of K. pneumoniae strains in vitro. Meanwhile, mice intragastrically administered with acetic acid exhibited significantly increased CRKP elimination in vivo. In conclusion, L. plantarum LP1812 is a potential candidate for intestinal CRKP anticolonization by regulating the intestinal microbiota and inhibiting CRKP via increased acetic acid in the intestinal lumen.
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is an increasingly common nosocomial pathogen that causes infection in a variety of locations, including the lower respiratory tract, urinary tract, and the bloodstream (Pendleton et al., 2013), posing a significant threat to public health (Michalopoulos et al., 2011; Musicha et al., 2017). Among other infections, CRKP bacteremia is hazardous for having a high all-cause mortality rate of up to 40% (Kontopoulou et al., 2019; Wang et al., 2019). Notably, patients with CRKP gut colonization have a greater risk of bloodstream infection and recurrent infection (Kontopoulou et al., 2019; Wang et al., 2019). Therefore, eliminating intestinal carriage of CRKP and thus reducing secondary infection is an intuitively appealing concept.
Numerous decolonization studies have been undertaken, including selective digestive decontamination with colistin or aminoglycosides (Machuca et al., 2016; Stoma et al., 2018), custom-made bacteriophage (Hua et al., 2017; Corbellino et al., 2020; Liu et al., 2021) and fecal microbiota transplantation (FMT) (Alagna et al., 2020; Gouveia et al., 2020; Ueckermann, et al., 2020). Although those methods demonstrated some decolonization effects, the possibility of drug resistance induced by gentamycin or colistin, the high cost and prolonged waiting time for phage, and the possibility of infectious agent transmission accompanied by FMT rendered them impractical for generalization. Recent advances in pathogen decolonization have included the use of novel treatments that are microbiome-based or microbiome-targeted (Mullineaux-Sanders et al., 2018; Kontopoulou et al., 2019). Using these methods, two patients with Clostridioides difficile infection were successfully treated with 33 commensal isolates (Petrof et al., 2013). Araos et al. (2016) reported that patients with a higher abundance of Lactobacillus spp. were less likely to be colonized with antibiotic-resistant bacteria during hospitalization. Therefore, probiotics are promising candidates for eradicating CRKP in the intestine with fewer adverse effects and expenses.
As a common probiotic, Lactobacillus spp. has been largely recognized as safe for the human body, and its anti-pathogen effect has been confirmed (Ripamonti et al., 2011). However, the exact mechanisms of action of Lactobacillus spp. are equivocal. In this study, we established that Lactobacillus plantarum LP1812 can improve mammalian gut health by increasing intestinal fatty acid-utilizing bacteria such as Bacteroidetes and Blautia. In addition to the strong CRKP elimination and killing effect of Bacteroidetes and L. plantarum per se, abundant acetic acid secreted by L. plantarum can acidify the CRKP intracellular environment and inhibit its growth. These synergistic effects result in efficient intestinal CRKP clearance.
Materials and Methods
Bacteria Strains and Culture Conditions
The four Klebsiella pneumoniae strains: K. pneumoniae ATCC1705, NTUH-K2044, ZKP25 (carbapenem-resistant) and ZKP16 (carbapenem sensitive) were obtained from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. MRS (de Man, Rogosa & Sharpe), and LB were used as basic growth media for Lactobacilli spp. and K. pneumoniae, respectively. All strains were frozen at -80°C in 20% glycerin bouillon. Pathogen resuscitation was performed by inoculating them on MH (Mueller-Hinton) agar at 37°C overnight.
Co-Culture Assays
K. pneumoniae and Lactobacilli spp. strains were revived on MH agar, then cultured in their basic broth with a single colon and shook overnight. Subsequently, 50 μL K. pneumoniae culture liquid was incubated in MRS broth as a blank control, while the test groups were incubated in modified MRS broth containing 10% overnight Lactobacilli spp. suspension obtained by filtering Lactobacilli spp. culture liquid using a 0.45 μM membrane. After that, the mixtures were shaken anaerobically for 12 hours at 37°C. Every 2 hours, sterile phosphate-buffered saline (PBS) was added to the co-culture system, and the serial dilutions were plated on ampicillin 0.1 mg/mL MH screening agar. The exact number of K. pneumoniae colonies at each time point was determined the next day. All experiments were repeated three times.
Growth Curve
After reviving and filtering the four Lactobacilli spp. strains, the supernatant was collected and mixed with LB broth into different concentrations (5%, 10%, 15%, and 20%). Additionally, modified LB broth was prepared by combining it with glacial acetic acid and HCl to ensure that each liter LB broth contained 50 mmol acetates and had a final pH of 3. The modified LB was added to normal LB at 5%, 10%, 20%, and 50% concentrations. Subsequently, 10μL of overnight K. pneumoniae was inoculated into 1 mL of a separate culture broth. The blank controls were MRS and 0% LB in different batch assays. Bioscreen C was used to detect the OD600 value of samples at different time points (every five minutes) and BioScreener was used to collect and store data.
Murine CRKP Colonization Model
We initially prepared the cell concentration of ZKP25 and LP1812 overnight bacterial culture fluid, which was subsequently used for dilutions to obtain the final concentration. Mice were purchased from Zhejiang University’s Institutional Animal Ethics Committee with the number ZJU20160154. All BALC/c mice were female and weighed 20-25 g, when purchased from Ziyuan, Hangzhou, China.
Figure 1A shows the procedure for establishing the CRKP intestinal colonization model. In our set-up, we first cleared the animal gut from day -10 to day -4 using a broad-spectrum antibiotic combination, KGCVM, containing kanamycin 0.4 mg/mL, gentamycin 0.035 mg/mL, polymyxin B 0.085 mg/mL, vancomycin 0.045 mg/mL, and metronidazole 0.215 mg/mL (Chen et al., 2008). Subsequently, mice in each group were gavaged with different treatments for three days (day-3 to day-1). Mice in the L.P. group were treated with 200 μL 5×108 colony-forming units (CFU) per milliliter bacterial suspension, while mice in the FMT group were treated with 200 μL fecal suspension. Suspensions of L. plantarum and feces were combined in a 1:1 ratio, and mice in the MIX group were gavaged with 200 μL. Additionally, all mice in the Blank group were given 200 μL PBS, as a negative control. Animals in four groups were all orally inoculated with about 200 μL ZKP25 (5×107 CFU/mL) on day 0. The CFU of K. pneumoniae strains in mice intestines was reflected by culturing mouse feces samples on-screen plants MEM 8 (meropenem 8 μg/mL). The mouse feces were collected on days 1, 2, 3, 5, and 7, then weighted and mixed with 1 mL sterile PBS, diluted ten times, and inoculated on MH screening agar containing 8 μg/mL meropenem. Parts of the collected feces were stored at -80°C for subsequent microbiome sequencing.
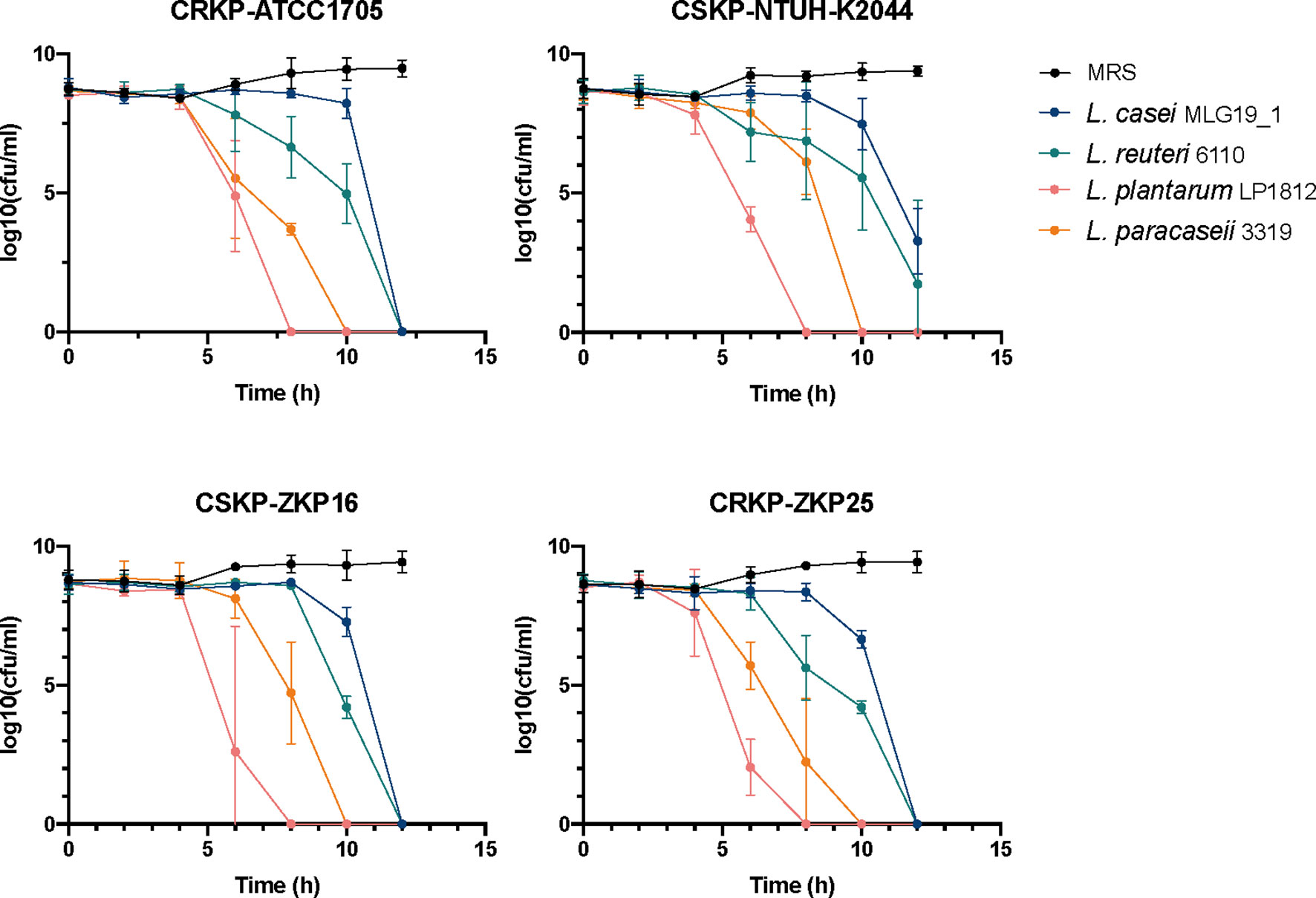
Figure 1 The effects of co-culture trails of K. pneumoniae. and Lactobacillus spp. NTUH-K2044, a classical hypervirulent K. pneumoniae isolate, ATCC1705, a standard carbapenem-resistant K. pneumoniae isolate, ZKP16, a carbapenem-sensitive clinical isolate, ZKP25, a carbapenem-resistant clinical isolate.
Blank group mice were given normal LB broth instead of PBS in subsequent CRKP anti-colonization assays (Figures 5 and S3D). In addition, mice in the acetic acid group were gavaged with a modified LB broth that we had previously prepared. The acetic acid + FMT group was treated with a combination of modified LB and FMT suspension (1:1 ratio).
16S rRNA Sequencing and Analysis
DNA extraction was performed on 136 feces samples to obtain information on intestinal microbiota changes using the PowerSoil® DNA Isolation Kit (MO BIO, Cat.No.12888). To amplify 16S rRNA genes from distinct regions, specific primers with barcodes were used, and PCR reactions were performed using Phusion® High-Fidelity PCR Master Mix (New England Biolabs). TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) was used to generate the sequencing libraries, which were subsequently sequenced on an Ion S5TM platform to yield 400 bp/600 bp single-end reads. After analyzing sequences using the Uparse software (Uparse V8.1.1861), OTUs (Operational Taxonomic Units) were obtained. To obtain species information, the SILVA database was used as a reference to annotate representative sequences in OTUs. Multiple sequence alignments were performed using the MUSCLE software (Version 3.8.31) to determine the phylogenetic relationship of different OTUs and the differences in the dominant species in different samples (groups).
The LDA score was calculated using LEfSe after dividing all feces samples into two categories based on the OTUs table: KP free and KP high loads (the ratio of K. pneumoniae > 0.01). The aim was to identify specific enriched bacteria species in KP-free feces. Principal coordinate analyses (PCoA) in R software were used to determine the microbiota structure of different samples. Tax4Fun was used to estimate KEGG function and calculate group differences based on functional abundance among samples, and analyze bacterial function in various fecal samples.
Evaluation of Intracellular pH
Initial preparation of the acetic acid modified LB solutions were required for the CRKP intracellular pH assays. The pH-matched LB broth was then prepared by combining varying amounts of HCl to the pH of 3, 4, 5, 6, 7, 8 as determined by pH strips. The ratio-metric dye BCECF-AM was added to the culture conditions, which were detected using an enzyme-labeled instrument. The ratio of fluorescence emission from 485 and 444 spectra was used to determine intracellular pH. To draw a standard curve for subsequent intracellular pH detection, 10 μL of overnight cultured CRKP strains were inoculated into 1 mL of the pH-matched LB mixed with 1 μM BCECF-AM and 50 μM CCCP (a cell membrane depolarizer). The emission ratio of BCECF-AM was monitored throughout a 2 h period at 37°C to allow the dye to equilibrate. The ratio of λem 485 to λem 444 was calculated and used to generate a standard curve. The ratio-metric measurements of BCECF-AM at different concentrations of acetic acid and extracellular pH were converted to absolute intracellular pH values using linear interpolation to the previously obtained standard curve measurements.
Statistical Analysis
All statistical analyses were performed in R software (Version R 4.1.0), while ggplot2 was used to draw the graphs. In addition, we introduced python 2.7 to calculate LDA score (Segata et al., 2011).
Results
Lactobacillus spp. Can Kill Klebsiella spps. In Vitro
To determine the inhibitory effect of Lactobacilli spp. against K. pneumoniae, we respectively co-cultured four lactobacilli strains, Lactobacillus casei MLG19_1, Lactobacillus reuteri 6110, Lactobacillus paracaseii 3319 and Lactobacillus plantarum LP1812 with four different K. pneumoniae strains, which were classical hypervirulent K. pneumoniae NTUH-K2044, Carbapenem-sensitive K. pneumoniae (CSKP) ZKP16, and CRKP ATCC1705, clinical CRKP ZKP25 isolates (Figure 1). After 12 hours of co-incubation, the four probiotics had completely eradicated ZKP16 and two CRKP strains. In addition, L. plantarum LP1812 was able to eradicate K. pneumoniae strains in 8 hours.
To determine whether the above-mentioned pathogen-killing effects were attributable to the probiotics per se or certain elements in the supernatant, we performed growth curve assays between K. pneumoniae strains and the supernatant of four distinct Lactobacillus spp. at different concentrations. As shown in Figure 2, the supernatant of all four Lactobacillus spp. strains significantly inhibited K. pneumoniae growth at a 10% concentration, and completely inhibited its growth at 20%, except for hypervirulent K. pneumoniae NTUH-K2044 (Figure 2). Similar to the co-culture results, L. plantarum LP1812 had the highest inhibitory effect against K. pneumoniae, whereas L. casei MLG19_1 had the least.
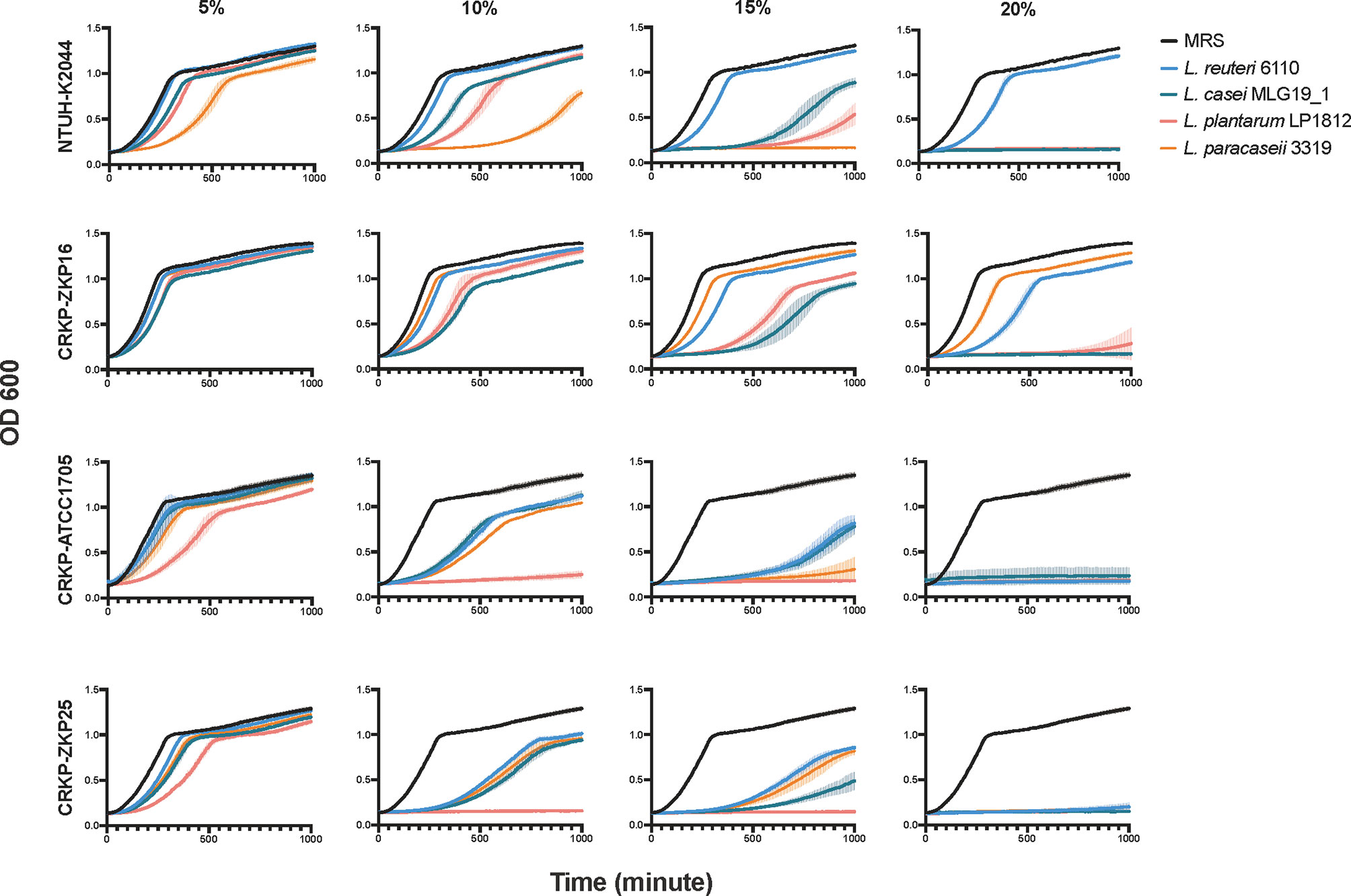
Figure 2 The growth curves of K. pneumoniae in Lactobacillus spp. supernatants of different species and concentrations. The amount of K. pneumoniae was exhibited by variations in absorption values at OD600.
L. plantarum LP1812 Exhibits Anti-CRKP Colonization Ability In Vivo
Because Lactobacillus spp. showed a significant anti-CRKP effect in vitro, we wondered if pre-treatment with Lactobacillus spp. strains could interfere with K. pneumoniae colonization. Therefore, we depleted the microbiota in adult mice using a broad spectrum KGCVM antibiotic combination and then pre-treated with different interventions before CRKP ZKP25 inoculation (Figure 3A). Given that FMT is commonly used in clinical practice to restore intestinal microbiota and consequently clear intestinal colonized pathogens (Liptak et al., 2021), it was considered as a positive control. Before ZKP25 inoculation, four interventions were administered, including Blank (control), FMT, L. plantarum LP1812 (L.P. group), and MIX (combined L. plantarum LP1812 with FMT). Mice pretreated with L. plantarum LP1812 showed comparable intestinal protectivity with FMT (Figure 3B), and the CRKP strains were below the limit of detection by day 5. Furthermore, when the L. plantarum LP1812 and FMT were combined, this obvious impact was amplified (MIX group).
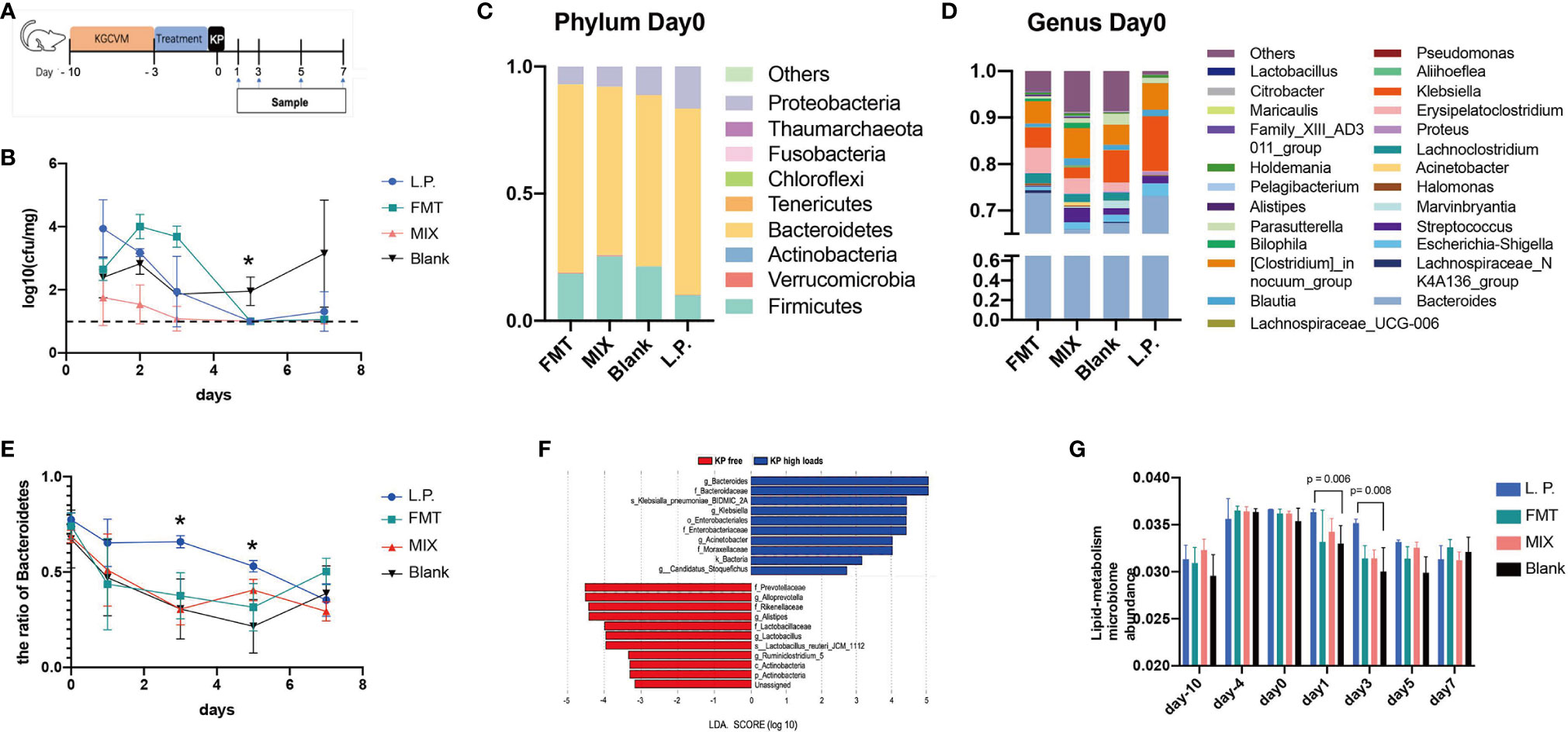
Figure 3 CRKP anticolonization experiments and 16S analysis. (A) Schedule design for animal trails. (B) The anti-CRKP assay findings. The Y-axis represents loads of CRKP strains in mouse feces (n = 4). The limit of detection is represented by the imaginary line. Blank represents the negative control, whereas FMT represents the positive control. L.P. means inoculating mice with L. plantarum LP1812, FMT stands for fecal microbiota transplantation, and MIX refers to pretreatment mice with mixtures of LP1812 and FMT suspensions. The percentages of intestinal microbiota community abundance at the Phylum (C) and Genus (D) levels. The relative abundances of various bacteria were calculated by averaging the data from five/four replicates within each group (FMT and Blank have four mice, L.P. and MIX have five mice) obtained from 16S OTU tables. (E) The ratio of Bacteroidetes in mouse intestinal microbiome calculated from the OTU tables. (F) LEfSe analysis between two groups that were re-classified in OTU tables based on loads of CRKP ZKP25. (G) The lipid metabolism relative microbiome ratios in feces microbiomes. The ratios were obtained from KEGG analysis. *denotes p-value < 0.05.
L. plantarum LP1812 Affects CRKP Colonization by Modulating Intestinal Microbiota in Mice
To investigate the changes in the microbiota of mouse feces following the aforementioned pretreatments, we sequenced the 16S rRNA of the collected feces and determined the microbial community composition in different groups. By day 5, the intestinal microbiota of the four groups was restored to normal (Figure S1). Pre-treatment with L. plantarum LP1812 was beneficial to mouse gut microbiota since both L.P. and MIX exhibited significantly higher α-diversity when compared with the Blank and FMT, respectively. Additionally, at phylum levels, Bacteroidetes dominated the mouse intestinal microbiota on day 0 (after three days of treatment), followed by Firmicutes and Proteobacteria (Figure 3C). Between day 0 and day 5, the L. P. group exhibited the highest Bacteroidetes abundance among the four groups (Figure 3E). After comparing the composition of the intestinal microbiota at the genus level (Figure 3D), we found that the abundance of beneficial bacteria such as Halomanas, Blautia, and Holdemania was increased in all three treatment groups, with the first being significantly higher when comparing L.P. and Blank (p-value = 0.010). In the Blank group, in contrast, Erysipelatoclostridium and Citrobacter were significantly increased (compared with the L.P. group, the p-value was 0.018, 0.002, and 0.030, respectively).
To investigate which bacteria in mice gut were highly correlated with CRKP colonization, we calculated LDA scores for each bacteria strain in the microbiome after dividing the microbiomes of the four groups (day5, day7) according to the K. pneumoniae level (KP free; KP high loads) obtained from 16S OTU tables. The LEfSe with LDA score findings indicated that Lactobacillus was significantly enriched in KP free feces (LDA > 3.0) (Figure 3F), while probiotics such as Bacteroidetes and Actinobacteria were also increased. Therefore, numerous probiotics such as Lactobacillus, Bacteroidetes, and Actinobacteria were associated with decreased K. pneumoniae levels.
KEGG Pathway Analysis Reveals That L. plantarum LP1812 Treatment Increases the Abundance of Fatty Acid Metabolizing Bacteria
To investigate the impact of L. plantarum LP1812 on the functions of the mice intestinal microbiome, we annotated our 16S sequences using the KEGG database and identified 274 genes distributed across 42 different pathways (including unclassified pathway, data not shown). There were 13 distinct metabolic pathways (Figure S2A), and the abundance changes of metabolism genes indicated that the mice intestinal microbiome of the L. P. group had a significant variation in metabolism from day 0 to day 3 when compared to the other three groups (Figure S2B). Additionally, the changes in group clustering demonstrated that the MIX group was considerably closer to L. P. on day 5 and that Blank clustered away from the other three trail groups on the last day. Further analysis of different metabolic pathways indicated that bacteria involved in lipid metabolism were more abundant in the L. P. group which exhibited a greater abundance of lipid metabolism genes than the other three groups from day 1 to day 5, particularly when compared with the Blank (Figure 3G). All of these findings suggest that genes involved in lipid metabolism may contribute to the CRKP-anticolonization process.
Acetic Acid Inhibits K. pneumoniae In Vitro by Acidifying Its Intracellular Environment
Because the aforementioned results showed a difference in intestinal lipid metabolism between the trail and blank groups, and given short-chain fatty acids (SCFAs) can help to protect the intestines from pathogenic bacteria colonization (LeBlanc et al., 2017), we hypothesized that SCFAs might play a role in the L. plantarum anti-CRKP colonization process. Consequently, the levels of SCFAs in Klebsiella spp. and Lactobacillus spp. were determined. Acetic acid was the most abundant SCFA in all investigated bacterial strains, with an estimated concentration of 50 mmol/L in Lactobacillus spp. (Table 1). Hence, we further modified LB broth with 50 mmol/L glacial acetic acid which was equivalent to that in Lactobacillus spp. supernatant. The modified LB broth was utilized to replace probiotic supernatant and mixed with normal LB broth as we performed in the assays of Figure 2. Those acetic mixtures significantly inhibited K. pneumoniae at 20% concentration and weakly at 5% and 10% concentration (Figure 4A), which was consistent with probiotic supernatants assays. Acetic acid was shown to be a significant bacteriostat in Lactobacillus spp. supernatants.
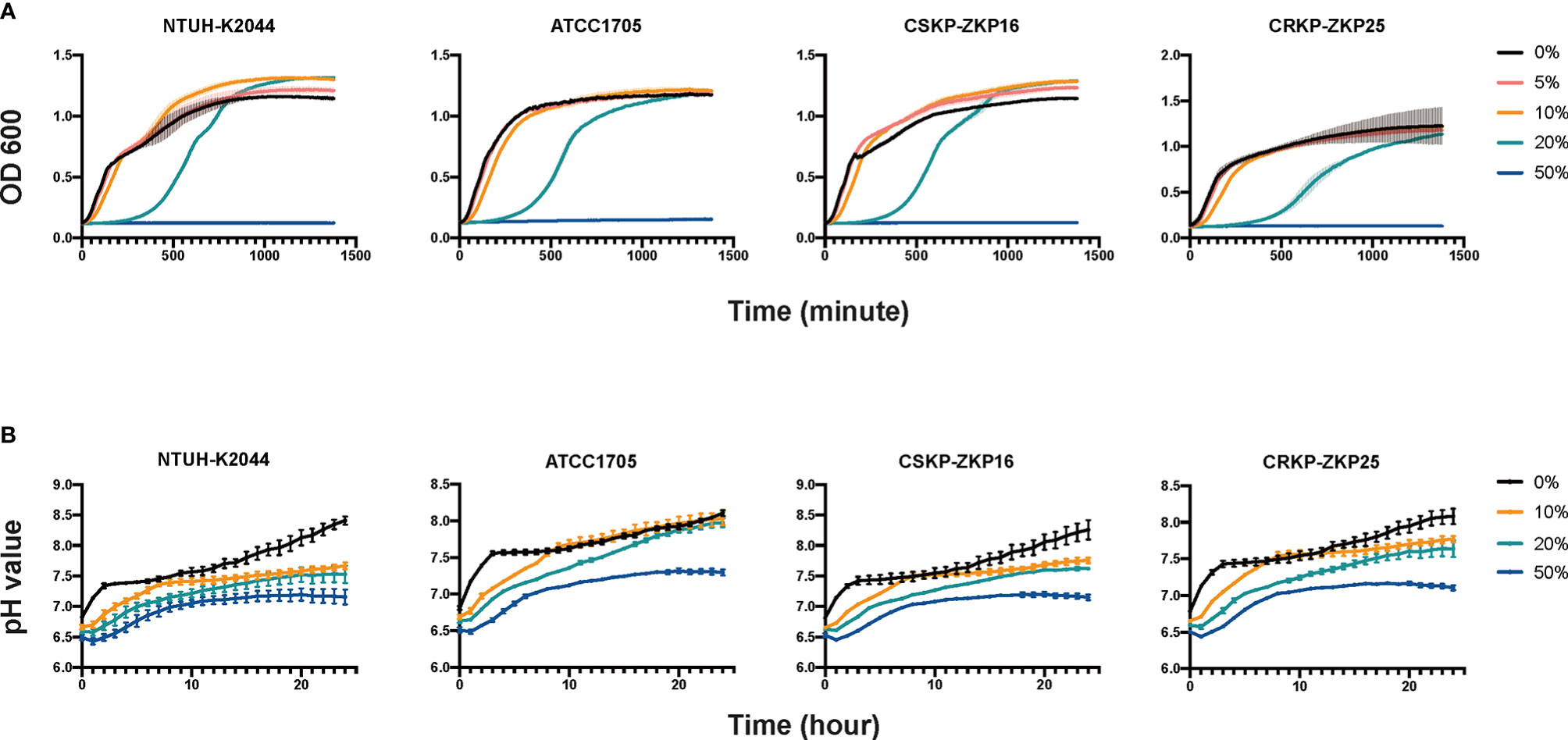
Figure 4 K. pneumoniae growth curves and intracellular pH measurements. (A) Culturing K. pneumoniae in acetic acid-containing LB broth. LB with 50 mmol/L acetic acids (pH=3) is considered as modified LB that is combined with normal LB broth to achieve various concentrations of 5%, 10%, 20%, and 50%. (B) Intracellular pH values of K. pneumoniae. The aforementioned modified LB broth was combined with normal LB at 10%, 20%, and 50% concentrations.
Amanda reported that propionic acid may acidify the intracellular environment of Salmonella to delay its growth (Jacobson et al., 2018). Therefore, we hypothesized that acetic acid could inhibit K. pneumoniae through a mechanism similar to propionic acid. Interestingly, the pH testing results (Figure 4B) were consistent with the growth assays results as shown in Figure 4A. As 10% and 20% modified LB broth increased the intra-cellular H+ ions of K. pneumoniae, whist the pH value of K. pneumoniae that added with 50% acetic modified LB remained less than 7.5 (Figure 4B) which made the bacteria could not proliferate all the time (Figure 4A). In summary, the CRKP inhibitory effect of Lactobacillus spp. was mediated by acetic acid through acidifying the pathogen’s intracellular environment.
Anticolonization Effect of Acetic Acid on CRKP Colonization In Vivo
To further elucidate the role of acetic acid in the anticolonization of CRKP, we repeated the CRKP anti-colonization experiment and replaced L. plantarum LP1812 with acetic acid. As shown in Figure 5A, acetic acid alone exhibited a significant anti-CRKP colonization effect. More importantly, when combined with FMT, the intestinal protection was significantly enhanced, and the CRKP anticolonization capacity was comparable between acetic acid and L. plantarum LP1812 (Figure 5B). Therefore, the anticolonization of CRKP by L. plantarum LP1812 was mainly mediated by acetic acid.
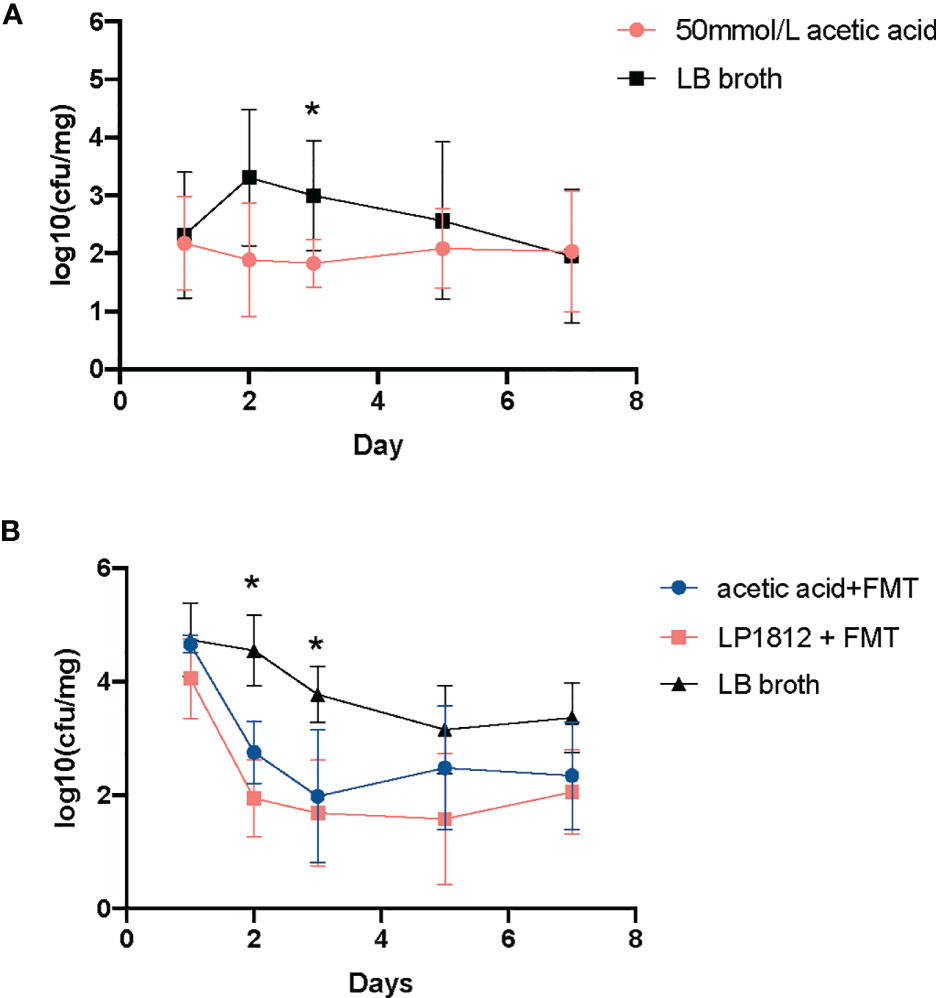
Figure 5 The results of anti-CRKP assays. (A) The mice (n = 5) treated with LB broth containing 50 mmol/L acetic acid. (B) The mice treated with acetic acid + FMT (n = 5) or L. plantarum LP1812 +FMT (n = 5). *denotes the p-value < 0.05.
Discussion
CRKP belongs to Enterobacterales, is regarded as a main nosocomial pathogen that can cause severe infection in critically ill patients (Medrzycka-Dabrowska et al., 2021), and its intestinal colonization is a risk factor for recurrent bloodstream infection (Kontopoulou et al., 2019). Therefore, intestinal clearance of CRKP is important for public health. Lactobacillus spp. is generally regarded as safe and has strong pathogen-killing abilities, making them a potential CRKP elimination agent. In the present study, we demonstrated the bactericidal activity of L. plantarum LP1812 against CRKP and revealed the critical function of acetic acid in the process of LP1812 clearing CRKP from mice intestines.
The Lactobacillus spp. strains used were L. casei MLG19_1, L. reuteri 6110, L. paracesei 3319, as well as L. plantarum LP1812. All four strains exhibited a prominent CRKP-killing ability, with L. plantarum LP1812 being the most potent. These findings were consistent with the findings of Chi-Chung Chen et al. that L. plantarum exhibited a strong pathogen inhibition effect when compared to other Lactobacillus spp. (Chen et al., 2019) Mohamed et al. (El-Mokhtar et al., 2020) discovered that this inhibitory effect was mainly achieved by Lactobacillus spp. supernatant as we proved in Figure 2 and L. plantarum LP1812 exhibit the strongest CRKP inhibition effect as well. These findings suggested L. plantarum LP1812 may be able to alter CRKP intestinal colonization statues.
Although Lactobacillus spp. is a potential candidate to eliminate CRKP, the decolonization effect of Lactobacillus spp. towards CRKP is not-ideal in clinical trials (Ekmekciu et al., 2017; Ljungquist et al., 2020), which might be due to diverse Lactobacillus spp. isolates exert different pathogen-killing and intestinal colonization abilities. We have validated the powerful capacity of L. plantarum LP1812 to kill CRKP strains, and the results in Figure S3B revealed L. plantarum LP1812 can exist in mice intestinal and suffer from pepsin and bile salt, which has been proved by Q Zhou (Zhou et al., 2021) and Mathara (Mathara et al., 2008). In other words, L. plantarum LP1812 can normally reproduce and secrete CRKP-killing substances in mice intestinal. More importantly, L. plantarum LP1812 indeed decreased the load of CRKP strains in mice feces (Figure 3B). Those findings hinted LP1812 might be a potential candidate for utilize in human digestive tract to prevent CRKP colonization.
Additionally, a single inoculation of L. plantarum LP1812 can increase the proportions of intestinal probiotics, such as Bacteroidetes (Sequeira et al., 2020), Blautia (Xia et al., 2021), Halomanas (Yu et al., 2021), and Holdemania (Raimondi et al., 2021), and the former two can generate acetic acid, which can improve the quantity of SCFAs in the gut (Aoki et al., 2021). Chi-Chung Chen et al. (Chen et al., 2020) reported that acetic acid in the Lactobacillus spp. supernatant is a critical factor in mediating CRKP inhibition. Our assays also demonstrated acetic acid’s ability to suppress K. pneumoniae growth (Figure 4A) and further illustrated the exact CRKP-inhibition mechanism of acetic acid in vitro, which was acidifying K. pneumoniae intracellular environment (Figure 4B). That will damage bacterial vigor and fecundity like Shuichi Nakamura et al. (Nakamura et al., 2009) and Amanda et al. (Jacobson et al., 2018) reported. Although 50 mM acetate used in the vivo study is quite a high concentration, the concentration of acetic acid in the lumen of the colon ranges from 10 to 100 mM (Koh et al., 2016), thus acetate in mice intestinal could reach up to the CRKP inhibition concentration (50 mM). Furthermore, acetic acid in the colon can enhance poly-reactive IgA and alter the IgA pool’s ability to bind to Enterobacterales (Takeuchi et al., 2021). All of these mechanisms mediated the in vivo CRKP inhibition and elimination effect of acetic acid, indicating that acetic acid may contribute to CRKP anticolonization. Our in vivo experiments shown in Figure 5A confirmed our assumption that acetic acid enhances anti-CRKP intestinal eradication.
Given that, when combined with FMT, LP1812 and acetic acid had equivalent CRKP anti-colonization efficacy (Figure 5B), we speculated that acetic acid is a key component in LP1812 that promotes the CRKP elimination process. In other words, the acetic acid in LP1812 that we added to FMT suspensions contributed to alterations in the intestinal microbiota and subsequently achieved CRKP anti-colonization. Unfortunately, we did not perform 16S sequences in acetic acid animal experiments. Further tests are needed to validate this hypothesis.
In conclusion, we found that L. plantarum LP1812 can completely eradicate CRKP strains in vitro and improve CRKP clearance in vivo via altering mice intestinal microbiota. The abundance of bacteria producing acetic acid increased in the colon, ensuring a longer and stronger CRKP-free status. These findings show that L. plantarum LP1812 is one of the potential candidates for anticolonization strategies against CRKP infection, which has to be confirmed in clinical practice.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA776100.
Ethics Statement
The animal study was reviewed and approved by The Institutional Animal Ethics Committee of Zhejiang University with the number of ZJU20160154.
Author Contributions
WL provided four Lactobacillus spp. strains and YY isolated strains ZKP25 and ZKP16. WL and JZ designed the study and all experiments. RY and YL carried out the assays. XiW, PY, and PL analyzed the data. YR and XuW drafted this manuscript. JY and XP revised the manuscript. All authors have read and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China (No. 81672067, No. 81830069), Project of Zhejiang Bureau of Traditional Chinese Medicine (No. 2022ZB375), Hangzhou Agricultural and Society Development Project (grant No. 202004A20), and Zhejiang Public welfare project (LGN19C010003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.804253/full#supplementary-material
References
Alagna, L., Palomba, E., Mangioni, D., Bozzi, G., Lombardi, A., Ungaro, R., et al. (2020). Multidrug-Resistant Gram-Negative Bacteria Decolonization in Immunocompromised Patients: A Focus on Fecal Microbiota Transplantation. Int. J. Mol. Sci. 21 (16), 5619. doi: 10.3390/ijms21165619
Aoki, R., Onuki, M., Hattori, K., Ito, M., Yamada, T., Kamikado, K., et al. (2021). Commensal Microbe-Derived Acetate Suppresses NAFLD/NASH Development via Hepatic FFAR2 Signalling in Mice. Microbiome 9 (1), 188. doi: 10.1186/s40168-021-01125-7
Araos, R., Tai, A. K., Snyder, G. M., Blaser, M. J., D’Agata, E. M. C. (2016). Predominance of Lactobacillus Spp. Among Patients Who Do Not Acquire Multidrug-Resistant Organisms. Clin. Infect. Dis. 63 (7), 937–943. doi: 10.1093/cid/ciw426
Chen, X., Katchar, K., Goldsmith, J. D., Nanthakumar, N., Cheknis, A., Gerding, D. N., et al. (2008). A Mouse Model of Clostridium Difficile-Associated Disease. Gastroenterology 135 (6), 1984–1992. doi: 10.1053/j.gastro.2008.09.002
Chen, C. C., Lai, C. C., Huang, H. L., Huang, W. Y., Toh, H. S., Weng, T. C., et al. (2019). Antimicrobial Activity of Lactobacillus Species Against Carbapenem-Resistant Enterobacteriaceae. Front. Microbiol. 10, 789. doi: 10.3389/fmicb.2019.00789
Chen, C. C., Lai, C. C., Huang, H. L., Su, Y. T., Chiu, Y. H., Toh, H. S., et al. (2020). Antimicrobial Ability and Mechanism Analysis of Lactobacillus Species Against Carbapenemase-Producing Enterobacteriaceae. J. Microbiol. Immunol. Infect 54 (3), 447–456. doi: 10.1016/j.jmii.2020.01.005
Corbellino, M., Kieffer, N., Kutateladze, M., Balarjishvili, N., Leshkasheli, L., Askilashvili, L., et al. (2020). Eradication of a Multidrug-Resistant, Carbapenemase-Producing Klebsiella Pneumoniae Isolate Following Oral and Intra-Rectal Therapy With a Custom Made, Lytic Bacteriophage Preparation. Clin. Infect. Dis. 70 (9), 1998–2001. doi: 10.1093/cid/ciz782
Ekmekciu, I., Fiebiger, U., Stingl, K., Bereswill, S., Heimesaat, M. M. (2017). Amelioration of Intestinal and Systemic Sequelae of Murine Campylobacter Jejuni Infection by Probiotic VSL3 Treatment. Gut. Pathog. 9, 17. doi: 10.1186/s13099-017-0168-y
El-Mokhtar, M. A., Hassanein, K. M., Ahmed, A. S., Gad, G. F., Amin, M. M., Hassanein, O. F. (2020). Antagonistic Activities of Cell-Free Supernatants of Lactobacilli Against Extended-Spectrum Beta-Lactamase Producing Klebsiella Pneumoniae and Pseudomonas Aeruginosa. Infect. Drug Resist. 13, 543–552. doi: 10.2147/IDR.S235603
Gouveia, C., Palos, C., Pereira, P., Roque Ramos, L., Cravo, M. (2020). Fecal Microbiota Transplant in a Patient Infected With Multidrug-Resistant Bacteria: A Case Report. GE. Port. J. Gastroenterol. 28 (1), 56–61. doi: 10.1159/000507263
Hua, Y., Luo, T., Yang, Y., Dong, D., Wang, R., Wang, Y., et al. (2017). Phage Therapy as a Promising New Treatment for Lung Infection Caused by Carbapenem-Resistant Acinetobacter Baumannii in Mice. Front. Microbiol. 8, 2659. doi: 10.3389/fmicb.2017.02659
Jacobson, A., Lam, L., Rajendram, M., Tamburini, F., Honeycutt, J., Pham, T., et al. (2018). A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 24 (2), 296–307 e297. doi: 10.1016/j.chom.2018.07.002
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., Backhed, F. (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165 (6), 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kontopoulou, K., Iosifidis, E., Antoniadou, E., Tasioudis, P., Petinaki, E., Malli, E., et al. (2019). The Clinical Significance of Carbapenem-Resistant Klebsiella Pneumoniae Rectal Colonization in Critically Ill Patients: From Colonization to Bloodstream Infection. J. Med. Microbiol. 68 (3), 326–335. doi: 10.1099/jmm.0.000921
LeBlanc, J. G., Chain, F., Martin, R., Bermudez-Humaran, L. G., Courau, S., Langella, P. (2017). Beneficial Effects on Host Energy Metabolism of Short-Chain Fatty Acids and Vitamins Produced by Commensal and Probiotic Bacteria. Microb. Cell Fact. 16 (1), 79. doi: 10.1186/s12934-017-0691-z
Liptak, R., Gromova, B., Gardlik, R. (2021). Fecal Microbiota Transplantation as a Tool for Therapeutic Modulation of Non-Gastrointestinal Disorders. Front. Med. (Lausanne) 8, 665520. doi: 10.3389/fmed.2021.665520
Liu, D., Van Belleghem, J. D., de Vries, C. R., Burgener, E., Chen, Q., Manasherob, R., et al. (2021). The Safety and Toxicity of Phage Therapy: A Review of Animal and Clinical Studies. Viruses 13 (7), 1268. doi: 10.3390/v13071268
Ljungquist, O., Kampmann, C., Resman, F., Riesbeck, K., Tham, J. (2020). Probiotics for Intestinal Decolonization of ESBL-Producing Enterobacteriaceae: A Randomized, Placebo-Controlled Clinical Trial. Clin. Microbiol. Infect. 26 (4), 456–462. doi: 10.1016/j.cmi.2019.08.019
Machuca, I., Gutierrez-Gutierrez, B., Perez Cortes, S., Gracia-Ahufinger, I., Serrano, J., Madrigal, M. D., et al. (2016). Oral Decontamination With Aminoglycosides Is Associated With Lower Risk of Mortality and Infections in High-Risk Patients Colonized With Colistin-Resistant, KPC-Producing Klebsiella Pneumoniae. J. Antimicrob. Chemother. 71 (11), 3242–3249. doi: 10.1093/jac/dkw272
Mathara, J. M., Schillinger, U., Kutima, P. M., Mbugua, S. K., Guigas, C., Franz, C., et al. (2008). Functional Properties of Lactobacillus Plantarum Strains Isolated From Maasai Traditional Fermented Milk Products in Kenya. Curr. Microbiol. 56 (4), 315–321. doi: 10.1007/s00284-007-9084-6
Medrzycka-Dabrowska, W., Lange, S., Zorena, K., Dabrowski, S., Ozga, D., Tomaszek, L. (2021). Carbapenem-Resistant Klebsiella Pneumoniae Infections in ICU COVID-19 Patients-A Scoping Review. J. Clin. Med. 10 (10), 2067. doi: 10.3390/jcm10102067
Michalopoulos, A., Falagas, M. E., Karatza, D. C., Alexandropoulou, P., Papadakis, E., Gregorakos, L., et al. (2011). Epidemiologic, Clinical Characteristics, and Risk Factors for Adverse Outcome in Multiresistant Gram-Negative Primary Bacteremia of Critically Ill Patients. Am. J. Infect. Control. 39 (5), 396–400. doi: 10.1016/j.ajic.2010.06.017
Mullineaux-Sanders, C., Suez, J., Elinav, E., Frankel, G. (2018). Sieving Through Gut Models of Colonization Resistance. Nat. Microbiol. 3 (2), 132–140. doi: 10.1038/s41564-017-0095-1
Musicha, P., Cornick, J. E., Bar-Zeev, N., French, N., Masesa, C., Denis, B., et al. (2017). Trends in Antimicrobial Resistance in Bloodstream Infection Isolates at a Large Urban Hospital in Malaw–2016): A Surveillance Study. Lancet Infect. Dis. 17 (10), 1042–1052. doi: 10.1016/s1473-3099(17)30394-8
Nakamura, S., Kami-ike, N., Yokota, J. P., Kudo, S., Minamino, T., Namba, K. (2009). Effect of Intracellular pH on the Torque-Speed Relationship of Bacterial Proton-Driven Flagellar Motor. J. Mol. Biol. 386 (2), 332–338. doi: 10.1016/j.jmb.2008.12.034
Pendleton, J. N., Gorman, S. P., Gilmore, B. F. (2013). Clinical Relevance of the ESKAPE Pathogens. Expert Rev. Anti Infect. Ther. 11 (3), 297–308. doi: 10.1586/eri.13.12
Petrof, E. O., Gloor, G. B., Vanner, S. J., Weese, S. J., Carter, D., Daigneault, M. C., et al. (2013). Stool Substitute Transplant Therapy for the Eradication of Clostridium Difficile Infection: ‘Repoopulating’ the Gut. Microbiome 1 (1), 3. doi: 10.1186/2049-2618-1-3
Raimondi, S., Musmeci, E., Candeliere, F., Amaretti, A., Rossi, M. (2021). Identification of Mucin Degraders of the Human Gut Microbiota. Sci. Rep. 11 (1), 11094. doi: 10.1038/s41598-021-90553-4
Ripamonti, B., Agazzi, A., Bersani, C., De Dea, P., Pecorini, C., Pirani, S., et al. (2011). Screening of Species-Specific Lactic Acid Bacteria for Veal Calves Multi-Strain Probiotic Adjuncts. Anaerobe 17 (3), 97–105. doi: 10.1016/j.anaerobe.2011.05.001
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic Biomarker Discovery and Explanation. Genome Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60
Sequeira, R. P., McDonald, J. A. K., Marchesi, J. R., Clarke, T. B. (2020). Commensal Bacteroidetes Protect Against Klebsiella Pneumoniae Colonization and Transmission Through IL-36 Signalling. Nat. Microbiol. 5 (2), 304–313. doi: 10.1038/s41564-019-0640-1
Stoma, I., Karpov, I., Iskrov, I., Krivenko, S., Uss, A., Vlasenkova, S., et al. (2018). Decolonization of Intestinal Carriage of MDR/XDR Gram-Negative Bacteria With Oral Colistin in Patients With Hematological Malignancies: Results of a Randomized Controlled Trial. Mediterr. J. Hematol. Infect. Dis. 10 (1), e2018030. doi: 10.4084/MJHID.2018.030
Takeuchi, T., Miyauchi, E., Kanaya, T., Kato, T., Nakanishi, Y., Watanabe, T., et al. (2021). Acetate Differentially Regulates IgA Reactivity to Commensal Bacteria. Nature 595 (7868), 560–564. doi: 10.1038/s41586-021-03727-5
Ueckermann, V., Hoosien, E., De Villiers, N., Geldenhuys, J. (2020). Fecal Microbial Transplantation for the Treatment of Persistent Multidrug-Resistant Klebsiella Pneumoniae Infection in a Critically Ill Patient. Case Rep. Infect. Dis. 2020, 8462659. doi: 10.1155/2020/8462659
Wang, X., Wang, Q., Cao, B., Sun, S., Zhang, Y., Gu, B., et al. (2019). Retrospective Observational Study From a Chinese Network of the Impact of Combination Therapy Versus Monotherapy on Mortality From Carbapenem-Resistant Enterobacteriaceae Bacteremia. Antimicrob. Agents Chemother. 63 (1), e01511–18. doi: 10.1128/AAC.01511-18
Xia, X., Ni, J., Yin, S., Yang, Z., Jiang, H., Wang, C., et al. (2021). Elevated Systemic and Intestinal Inflammatory Response Are Associated With Gut Microbiome Disorder After Cardiovascular Surgery. Front. Microbiol. 12, 686648. doi: 10.3389/fmicb.2021.686648
Yu, D., Xia, Y., Ge, L., Tan, B., Chen, S. (2021). Effects of Lactococcus Lactis on the Intestinal Functions in Weaning Piglets. Front. Nutr. 8, 713256. doi: 10.3389/fnut.2021.713256
Keywords: carbapenem-resistant, Klebsiella pneumoniae, Lactobacillus plantarum, anticolonization, Acetic acid
Citation: Yan R, Lu Y, Wu X, Yu P, Lan P, Wu X, Jiang Y, Li Q, Pi X, Liu W, Zhou J and Yu Y (2021) Anticolonization of Carbapenem-Resistant Klebsiella pneumoniae by Lactobacillus plantarum LP1812 Through Accumulated Acetic Acid in Mice Intestinal. Front. Cell. Infect. Microbiol. 11:804253. doi: 10.3389/fcimb.2021.804253
Received: 02 November 2021; Accepted: 29 November 2021;
Published: 15 December 2021.
Edited by:
Yang Zhang, University of Pennsylvania, United StatesReviewed by:
Yu Zhou, Institut Pasteur of Shanghai (CAS), ChinaHao Zhang, University of Pennsylvania, United States
Copyright © 2021 Yan, Lu, Wu, Yu, Lan, Wu, Jiang, Li, Pi, Liu, Zhou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiancang Zhou, amlhbmNhbmd6aG91QHpqdS5lZHUuY24=; Wei Liu, YmlvbHdlaUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Rushuang Yan1,2†
Rushuang Yan1,2† Peng Lan
Peng Lan Xueqing Wu
Xueqing Wu Yan Jiang
Yan Jiang Wei Liu
Wei Liu Jiancang Zhou
Jiancang Zhou Yunsong Yu
Yunsong Yu