- 1Jiangsu Key Lab of Zoonosis/Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety, Yangzhou University, Yangzhou, China
- 3Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agri-food Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
Introduction: The CRISPR-Cas system serves as a defense mechanism in bacteria and archaea, protecting them against the invasion of mobile genetic elements. Staphylococcus argenteus, a Gram-positive bacterium that diverged from Staphylococcus aureus, is characterized by the rare presence of the CRISPR-Cas system in only a few isolates.
Methods: In this study, we analyzed the prevalence of the type III-A CRISPR-Cas system in 368 S. argenteus genome sequences from animals, food sources, and humans across 26 countries, available in public database.
Results: Our findings revealed that 44.0% of these strains carry this immune system, with 98.1% of them belonging to the sequence type 2250 (ST2250). Genomic localization analysis indicated that the CRISPR-Cas is closely associated with SCCmec (mecA-ΔmecR1-IS1272-ccrB2-ccrA2) or Insertion sequence 1272 (IS1272) transposase. Further analysis identified a common IS1272 target inverted repeats (IR) sequence in ST2250 strains, providing insights into why these strains are more likely to acquire the CRISPR-Cas system. CRISPR typing identified 41 sequences types, classifying these strains into two clusters, with Cluster II being the predominant one. Homology analysis of spacers revealed that all the identified 15 spacers exhibited homology to sequences from plasmids, lytic phages, or prophages.
Conclusion: This study suggests that the acquisition of the CRISPR-Cas system in S. argenteus enhances its resistance to phage attacks and plasmid invasions in environmental settings, potentially posing significant challenges for clinical treatment of infections caused by these strains and hindering efforts to control their spread in food products using phage-based interventions.
1 Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins are considered an adaptive immune system (CRISPR-Cas) distributed in approximately 40% of bacteria and 90% of archaea, protecting them against foreign genetic elements, including phages or plasmids (Barrangou et al., 2007; Kunin et al., 2007; Grissa et al., 2007). A small foreign DNA fragment, ranging mainly from 26 bp to 72 bp, can be inserted into the CRISPR array as a spacer (Grissa et al., 2007). The spacer can be transcribed and processed into mature crRNA, which guides Cas protein complexes to homologous foreign DNA sequences, enabling the digestion of the invading DNA through the activity of Cas nucleases (Hille et al., 2018).
Until now, two classes of CRISPR-Cas systems consist of six types and almost 33 subtypes have been identified (Makarova et al., 2020). The distribution of CRISPR-Cas systems varies across different bacterial species. For example, most Salmonella isolates carry the type I-E CRISPR-Cas system, while only a few strains of S. aureus and S. epidermidis have been reported to contain CRISPR-Cas system (Zhang et al., 2024; Mikkelsen et al., 2023). In staphylococci, the CRISPR-Cas system belongs to type III-A and demonstrates strong activity against phages or plasmids that are targeted by its spacers (Marraffini and Sontheimer, 2008; Li et al., 2021). Besides, the type III-A CRISPR-Cas system doesn’t require a Protospacer Adjacent Motif (PAM) sequence to recognize targeted sequence, distinguishing it from other CRISPR systems like type II CRISPR-Cas9 systems, which relies on a PAM sequence (Pyenson et al., 2017; Gleditzsch et al., 2019). The type III-A CRISPR-Cas system has additional notable characteristics: it can cleave both DNA and RNA targets and induce non-specific immune responses mediated by the production of cyclic oligoadenylate (cOA) by Cas10, which can accumulate nucleases to degrade both foreign and host RNA, leading to an antiviral defense state (Niewoehner et al., 2017; Kazlauskiene et al., 2017). Although the type III-A CRISPR-Cas system show strong immune response against foreign DNAs or RNAs, it is not prevalent in strains of different staphylococci species. Cruz-López et al., found that only 0.83% (6/716) of the analyzed 716 S. aureus genomes from GENOMES-NCBI harbored the CRISPR-Cas system (Cruz-López et al., 2021); while our previous study revealed that 2.9% of MRSA isolates in Denmark carried this system (Mikkelsen et al., 2023). A recent study showed that the CRISPR-Cas system existed in all the 40 MDR S. aureus isolated from poultry meat in Pakistan (Shabbir et al., 2024). Therefore, it is essential to elucidate the prevalence of type III-A CRISPR-Cas system in staphylococci strains and the genomic characteristics of these strains.
S. argenteus was reported as a distinct staphylococcal species diverged from S. aureus in 2015, and have been found globally (Tong et al., 2015). It can cause various infections, including bloodstream infection, skin and soft tissue infections, osteomyelitis, and brain abscess like S. aureus (Chantratita et al., 2016; Imam et al., 2025; Lee et al., 2024). The major phenotypic difference between the two species is the pigmentation: S. aureus typically produces a golden pigment, while S. argenteus exhibits a silvery-white appearance (Holt et al., 2011). rpoB sequencing is another reliable method to differentiate S. argenteus from S. aureus, as it reveals species-specific genetic variations in the RNA polymerase β subunit gene (Argudín et al., 2016; Mellmann et al., 2006). Previous studies have analyzed the genomic characteristics of 132 global S. argenteus strains from published databases between 2005 and 2008, revealing that ST2550 S. argenteus strains exhibit a tendency to carry the type III-A CRISPR-Cas system (Goswami et al., 2021). Since 2008, an increasing number of S. argenteus genomic sequences have been submitted to the NCBI GenBank database, and the sources of these strains have expanded. This study further analyzed the prevalence, genetic characteristics, genomic location, and spacer content of the CRISPR-Cas system in 368 S. argenteus isolates from across the globe.
2 Materials and methods
2.1 Data collection of S. argenteus strains
A total of 370 genome sequences of S. argenteus were obtained from the NCBI GenBank database as of December 31, 2024. Since the S. argenteus strains SH3 and DSM 28299 were sequenced twice, 368 unique S. argenteus strains and their genomic sequences were included in the analysis for this study. Strain information, including the name, assembly name, accession number, submission data, bioproject number, host, and country of origin, was collected from the database. Additional information was corrected or added based on published papers for the respective strains. The complete information for all 368 strains is provided in the Supplementary Table 1.
2.2 MLST and SCCmec analysis
Multi-locus sequence typing of all S. argenteus strains was performed using the PubMLST platform (https://pubmlst.org/organisms/staphylococcus-aureus) designed for S. aureus. Although the MLST sequence types (STs) of some strains were known when the genome sequences were submitted to the NCBI GenBank database, all submitted STs were subsequently verified using the PubMLST platform. To identify the presence of mecA or mecC in the chromosome of S. argenteus, we used the SCCmecFinder 1.2 platform (https://cge.food.dtu.dk/services/SCCmecFinder/), which also provides information on the type of SCCmec elements and the presence of IS1272 in the chromosome.
2.3 CRISPR-Cas identification
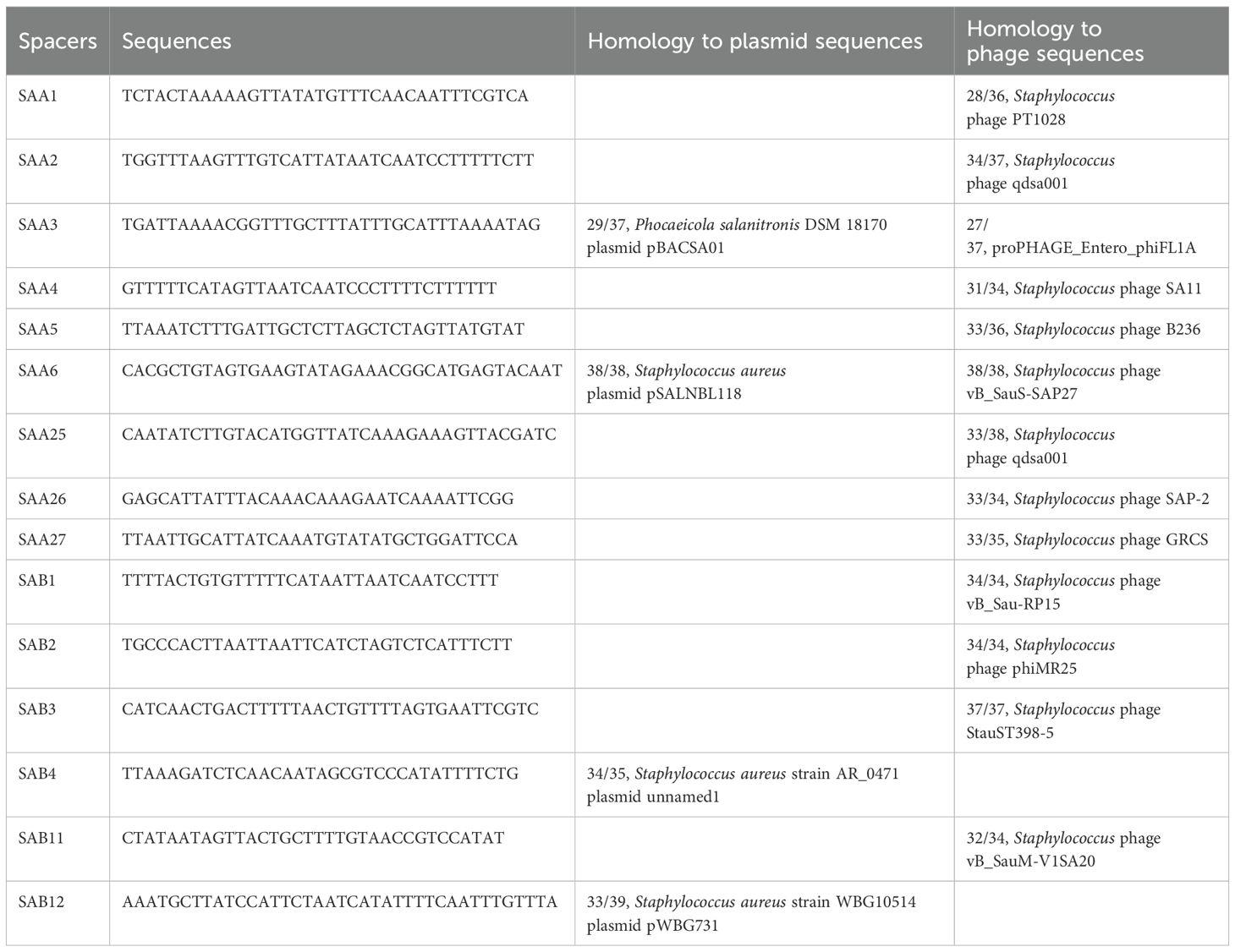
The presence of type III-A CRISPR-Cas system in S. argenteus is determined by the cas genes (cas1, cas2, cas10, csm2, csm3, csm4, csm5, csm6, cas6) and CRISPR arrays (CRISPR1 and CRISPR2). To assess whether this system is present in S. argenteus strains, the CRISPRCasFinder platform (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder) were used to analyze complete genome sequences or assembled contigs. The size of flanking regions for each analyzed CRISPR arrays were set to 100bp, and the repeat length threshold was defined with a minimum of 23 and a maximum of 55. According to the characteristics of CRISPR arrays upstream and downstream of the cas gene clusters (Li et al., 2018), small CRISPR-like elements were excluded from the analysis. All the repeats identified in the type III-A CRISPR-Cas system of S. argenteus share over 90% homology with the sequences: GATCGATACCCACCCCGAAGAAAAGGGGACGAGAAC. The spacers were extracted from the CRISPR arrays and designed as previously reported in S. aureus (Li et al., 2016). All the 15 identified spacers have been previously documented in S. aureus (Li et al., 2016).
2.4 CRISPR-Cas typing and phylogenic analysis
After extracting the CRISPR arrays from each strain, the spacer arrangements of CRISPR1 and CRISPR2 was analyzed using the spacer names as previously described (Li et al., 2016). Each unique arrangement of spacers in CRISPR1 were designated as “SgCTA + NO.” to represent the CRISPR1 type, while “SgCTB + NO.” was used for the CRISPR2 type. The combination of CRISPR1 and CRISPR2 types was represented as “SgCT + NO.” to indicate the overall CRISPR type for each strain. To perform the genomic analysis of S. argenteus strains using CRISPR type, a binary file was constructed based on the presence or absence of spacers. The presence of a spacer was represented by “1”, while the absence was denoted by “0” in the binary file. A phylogenetic tree based on the CRISPR types was then generated using Bionumericus 7.5 software (Applied Maths, Belgium).
2.5 Homology analysis of spacers
Previous studies have demonstrated that obtaining homologous sequences of spacers using BLSATn in the NCBI GenBank database is challenging (Li et al., 2016). The advancement of genome sequencing technology and the expansion of phage and plasmid sequences in various databases have significantly improved homology analysis. In this study, the CRISPRTarget (http://crispr.otago.ac.nz/CRISPRTarget/) was used to analyze the homologous sequences of each spacer, employing the GenBank-Phage, RefSeq-Plasmid, and ACLAME databases to represent phage, plasmid, and mobile genetic elements, respectively. The cutoff score was set at 20.
2.6 Location of CRISPR-Cas system in chromosome
The CRISPR-Cas system is not present in all the Staphylococcus strains; it is considered the genetic elements frequently located within the SCCmec region in S. aureus (Mikkelsen et al., 2023). Interestingly, most CRISPR-Cas-positive S. argenteus strains are methicillin-sensitive. To investigate it further, the contigs or sequences containing the CRISPR-Cas system from these S. argenteus strains were extracted and analyzed for the upstream and downstream gene clusters using the SnapGene software (Dotimatics, Boston, USA). Additionally, we searched for IS1272-targeted inverted repeats (IR) in the sequences surrounding the CRISPR-Cas system as previously described (Wan et al., 2017).
3 Results
3.1 Global distribution of S. argenteus published in the database
Since S. argenteus was classified as a separate species distinct from S. aureus in 2015, the number of S. argenteus isolates has reached 368 based on the NCBI GenBank database. From 2015 to 2018, 132 publicly available sequences were recorded, while since 2019, more than 230 bacterial genomic sequences have been submitted to the database (Supplementary Table 1). The 368 isolates were collected from 26 countries, including Thailand (21.2%, 78), Netherlands (17.1%, 63), China (13.3%, 49), Japan (11.4%, 42), Denmark (7.9%, 29), USA (7.3%, 27), Canada (4.3%, 16), Malaysia (2.7%, 10) and other countries (<10 isolates), such as Australia, Brazil, Colombia, Fiji, France, Gabon, Germany, Israel, Italy, Samoa, Saudi Arabia, Singapore, South Korea, Sri Lanka, Sweden, United Arab Emirates, United Kingdom, and Viet Nam (Figure 1A). The geographic distribution of these isolates indicates that S. argenteus is now found in Asia, Europe, North America, South America, Oceania, and Africa (Figure 1A). The majority of these isolates are sourced from humans (78.3%, 288), including patients and healthy people; however, animals and food products also serve as reservoirs, including poultry (12.0%, 44), fish and shrimp (3.5%, 13), and other livestock (Figure 1B). Some isolates have been obtained from vegetables and environmental samples, such as Capra aegagrus hircus, chilled water in slaughterhouse, and surface swabs in dental clinics. These findings suggest that S. argenteus may be involved in cross-contamination or transmission between foods, animals, and humans.
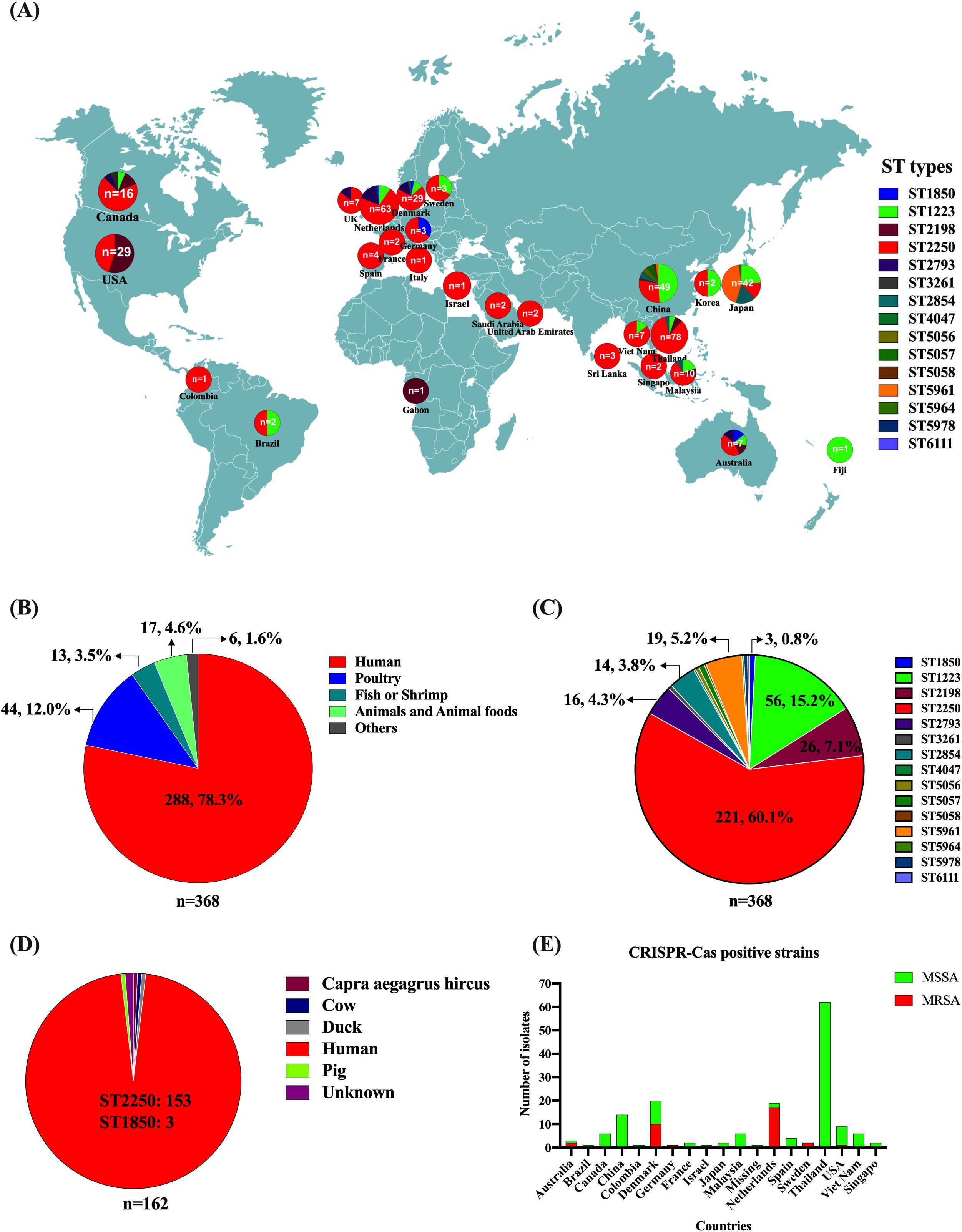
Figure 1. Distribution, Sources, and Characteristics of Global S. argenteus isolates. (A) Geographic locations and MLST types of S. argenteus isolates. (B) Number of S. argenteus isolates obtained from different hosts. (C). Number of S. argenteus with different MLST types. (D) Distribution of CRISPR-Cas-positive S. argenteus isolates from different hosts. (E) Distribution of methicillin-sensitive S. argenteus (MSSA) and methicillin-sensitive S. argenteus (MRSA) isolates carrying the CRISPR-Cas system across different countries.
3.2 MLST analysis of S. argenteus
For the isolates available prior to 2019, the reported MLST types of S. argenteus include ST1223, ST1850, ST2198, ST2250, ST2793, ST2854, and ST3261. Since 2019, however, several new sequence types have emerged and increased in number, such as ST5961, ST4067, ST5056, ST5057, ST5058, ST5978, ST5964, and ST6111 (Figure 1C; Supplementary Table 1). The most prevalent sequence type is ST2250 (60.1%, 221), followed by ST1223 (15.2%, 56), ST2198 (7.1%, 26), ST5961 (5.2%, 19), ST2793 (4.3%, 16), and ST2854 (3.8%, 14). Although the strain MSHR1132 was considered the first S. argenteus stain isolated from an indigenous woman with necrotizing fasciitis in 2006, its sequence type is ST1850 (Holt et al., 2011; Li et al., 2016), which has only been reported in three strains (Figure 1C; Supplementary Table 1). The predominant sequence type remains ST2250, accounting for 60.1% of the global isolates (Figure 1C).
3.3 CRISPR-Cas positive S. argenteus
Staphylococcal species possess a type III-A CRISPR-Cas system located on the chromosome, though not all isolates carry this system. The type III-A CRISPR-Cas system in S. epidermidis and S. aureus has been shown to protect bacteria against phage attacks and plasmid invasion. The presence of the CRISPR-Cas system is more common in Staphylococcal isolates with specific MLST sequence types; for example, 50% of S. aureus ST630 isolates carry the system. In S. argenteus, the CRISPR-Cas system is present in all ST2250 isolates, except for three ST1850 isolates that also carry the system. It is not found in any other ST isolates (Figure 1D). The three ST1850 isolates were collected from human infections in Australia, Germany, and Denmark (Figure 1D). Among the 221 ST2250 isolates, 159 (71.9%) carry the CRISPR-Cas system, while 62 are negative of the system. The 159 CRISPR-Cas-positive ST2250 isolates were collected from 17 out of 24 countries (Figure 1E). The three ST1850 isolates were methicillin-resistant due to the presence of mecA gene. In contrast among the 159 CRISPR-Cas-positive ST2250 isolates, only 30 (18.9%) were MRSA, with the majority of these strains collected from Netherlands and Denmark (Figure 1E).
3.4 CRISPR types of S. argenteus
Although only 15 spacers were identified in all the CRISPR-Cas-positive isolates, 41 S. argenteus CRISPR types (SgCTs) were detected according to the arrangement of spacers (Figure 2A). As shown in Figure 2A, the most prevalent CRISPR type was SgCT1, accounting for 42.6% (69) of the 162 CRISPR-Cas-positive isolates. The spacers arrangement of SgCT1 is SAA26-SAA27-SAA25-SAA5-SAA6 (SgCTA1) for CRISPR1 locus and SAAB11-SAB12-SAB2-SAB3-SAB4 (SgCTB1) for CRISPR2 locus (Figures 2A, B). Analysis of the CRISPR types for each CRISPR locus showed that 90 isolates share the SgCTA1 for CRISPR1 locus, and 107 isolates share the SgCTB1 for CRISPR2 locus (Figure 2B). Ten isolates shared the SgCT2, which has SAB12 deleted in the CRISPR2 locus compared to that of SgCT1 (Figure 2A). Eight isolates belong to the SgCT3, which has SAA27 and SAA25 deleted in the CRISPR1 locus compared to that of SgCT1 (Figure 2A). Deletion or addition of spacers led to the emergence of new CRISPR types in S. argenteus. Cluster analysis grouped the 41 SgCTs into two clusters, Cluster I and Cluster II (Figure 2A). Seventeen SgCTs, covering 49 (30.2%) strains, belong to Cluster I, while 24 SgCTs, covering 113 (69.8%) strains, belong to Cluster II (Figure 2A). SgCT1 and SgCT2, located in Cluster II, account for 48.8% of the strains, indicating that Cluster II is the predominant group among CRISPR-Cas-positive S. argenteus strains (Figure 2A).
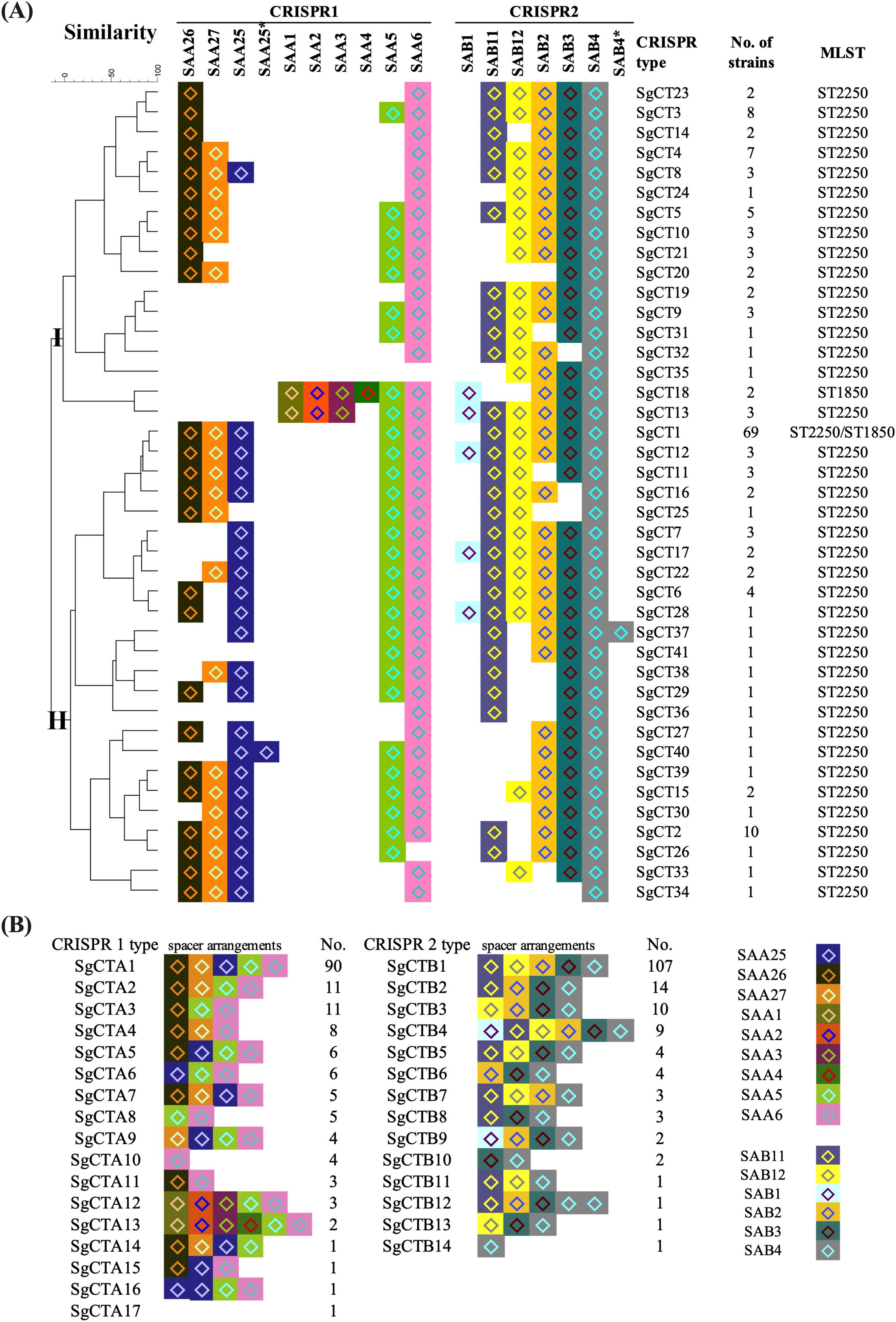
Figure 2. CRISPR typing of CRISPR-Cas-positive S. argenteus isolates. (A) Phylogenetic tree of CRISPR-Cas-positive S. argenteus isolates based on 41 distinct CRISPR types. The spacer arrangements in the CRISPR 1 and CRISPR 2 loci were used to define CRISPR types. The number of strains corresponding to each CRISPR type and their associated MLST types are indicated in the right columns. (B). Distribution of strains with CRISPR 1 or CRISPR 2 types, with spacer arrangements shown for each type.
Among the nine spacers in the CRISPR1 locus, the most prevalent spacer is SAA6 (160), followed by SAA5 (150), SAA26 (136), SAA25 (125), and SAA27 (119) (Figure 2B). However, the spacers SAA1, SAA2, and SAA3 in CRISPR1 locus were found only in three ST2250 and two ST1850 isolates; while the spacer SAA4 was detected in the two ST1850 isolates (Figure 2A). Among the six spacers in the CRISPR2 locus, SAB4 is present in all 162 CRISPR-Cas-positive isolates, followed by SAB3 (157), SAB2 (150), SAB11 (141), and SAB12 (115). The spacer SAB1 was found only in two ST1850 and nine ST2250 isolates (Figure 2A).
The minimum spanning tree graph (MST) graph, generated using the BioNumerics v7.5 advanced cluster analysis tool, revealed the relationships between each SgCT (Figure 3). Interestingly, all 41 CgSTs include isolates with either IS1272 or mecA in their genomes; while seven SgCTs (SgCT1, 2, 4, 6, 8, 10, and 11) contain 14 isolates that lack both IS1272 and mecA, indicating that 91.3% (146/160) of the isolates harbor IS1272 in the chromosome (Figure 3; Supplementary Table S3). Among the methicillin-resistant S. argenteus isolates, two types of SCCmec were identified: type_IVa(2B) and type_IVc(2B) (Supplementary Table S3).
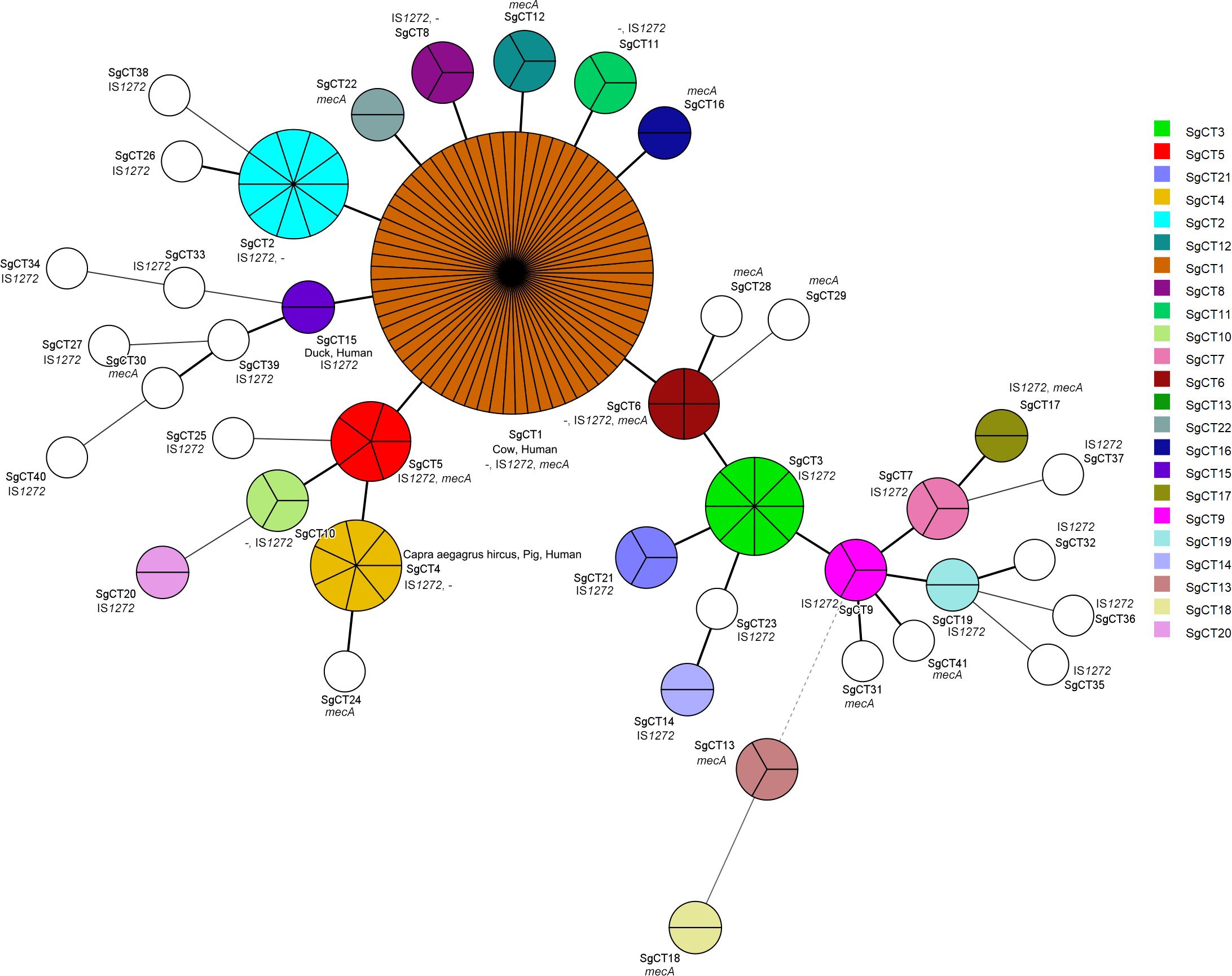
Figure 3. Genetic relationship and characteristics of CRISPR-Cas-positive S. argenteus isolates. The Minimum spanning tree of CRISPR-Cas-positive S. argenteus isolates were constructed using BioNumerics 7.5 software based on CRISPR types. Each circle represents the strains sharing a single CRISPR type. The presence of mecA and IS1272 is indicated in each circle, while “-” indicates strains lacking both mecA and IS1272.
3.5 Homology analysis of CRISPR spacers
Although no novel spacers were identified in S. argenteus, the homology analysis of these spacers was significantly enhanced due to the increased availability of genomic sequences of phages, plasmids, prophages in public databases. Consequently, we conducted homology analysis of these spacers in the CRISPRTarget platform (Table 1). Among the 15 spacers, 13 show similarities to phage sequences, with12 spacers specifically homologous to Staphylococcus phage sequences. Additionally, four spacers exhibited homology to sequence in plasmids, three of which were identified as S. aureus plasmids. In details, the spacer SAA2 and SAA25 showed homology to different sequences in the same Staphylococcus phage qdsa001, a lytic phage isolated from urban sewage and used to inactivate S. aureus in ready-to-eat milk in China. The spacer SAA6 showed 100% homology to sequences located in a gene encoding a DUF1270 family protein, which is present in both the S. aureus plasmid pSALNBL118 and the lytic phage vB_SauS-SAP27. Although SAA3 showed similarity to sequences in both the Phocaeicola salanitronis plasmid pBACSA01 and the proPHAGE_Entero_phiFL1A, its homology rate is much lower compared to other spacers. The spacers SAA4, SAA26, SAA27, and SAB1 exhibited homology to sequences in the lytic phages SA11, SAP-2, GRCS, and vB_Sau-RP15, respectively. Meanwhile, SAA1, SAA5, SAB2, and SAB3 showed homology to sequences in the prophages PT1028, B236, phiMR25, and StauST398-5, respectively. Additionally, SAB4 and SAB12 were homologous to sequences located in a plasmid of S. aureus strain AR_0471 and the plasmid pWBG731, respectively.
3.6 Genetic location of CRISPR-Cas system in S. argenteus
To analyze the genomic location of the type III-A CRISPR-Cas system in S. argenteus, the contigs containing the CRISPR-Cas system were collected for identification of the sequences upstream of CRISPR1 array and downstream of CRISPR2 array. According to the difference of MLST sequence types, the presence of mecA or IS1272, we determined the location sites of the CRISPR-Cas system for four different types of strains (Figure 4). The three ST1850 strains were identified as methicillin-resistant S. argenteus, with the CRISPR-Cas system located downstream of hsdR and adjacent to the SCCmec type Iva(2B) cassette (Figure 4). Among the 30 ST2250 methicillin-resistant S. argenteus strains, the CRISPR-Cas system exhibited a similar genomic location to that of the ST1850 strains, but these strains carried two types of SCCmec cassettes: type IVa(2B) and type IVc(2B) (Figure 4). Among the 129 CRISPR-Cas-positive ST2250 methicillin-sensitive S. argenteus strains, 115 (89.1%) strains carry IS1272 elements in their chromosomes (Figure 4). Interestingly, at the downstream location of the CRISPR2 locus, a common IS1272 target inverted repeat (IR) site, GGAGGAAACTAAAATTCCTCC, was identified, which is located near the gene encoding tRNA dihydrouridine synthase (Figure 4). Additionally, all ST2250 strains carry the target IR site, suggesting that ST2250 strains are more likely to acquire the CRISPR-Cas system compared to strains of other sequence types. The presence of IS1272 within the SCCmec cassette further explains why the acquisition of the CRISPR-Cas system is closely associated with the presence of mecA.
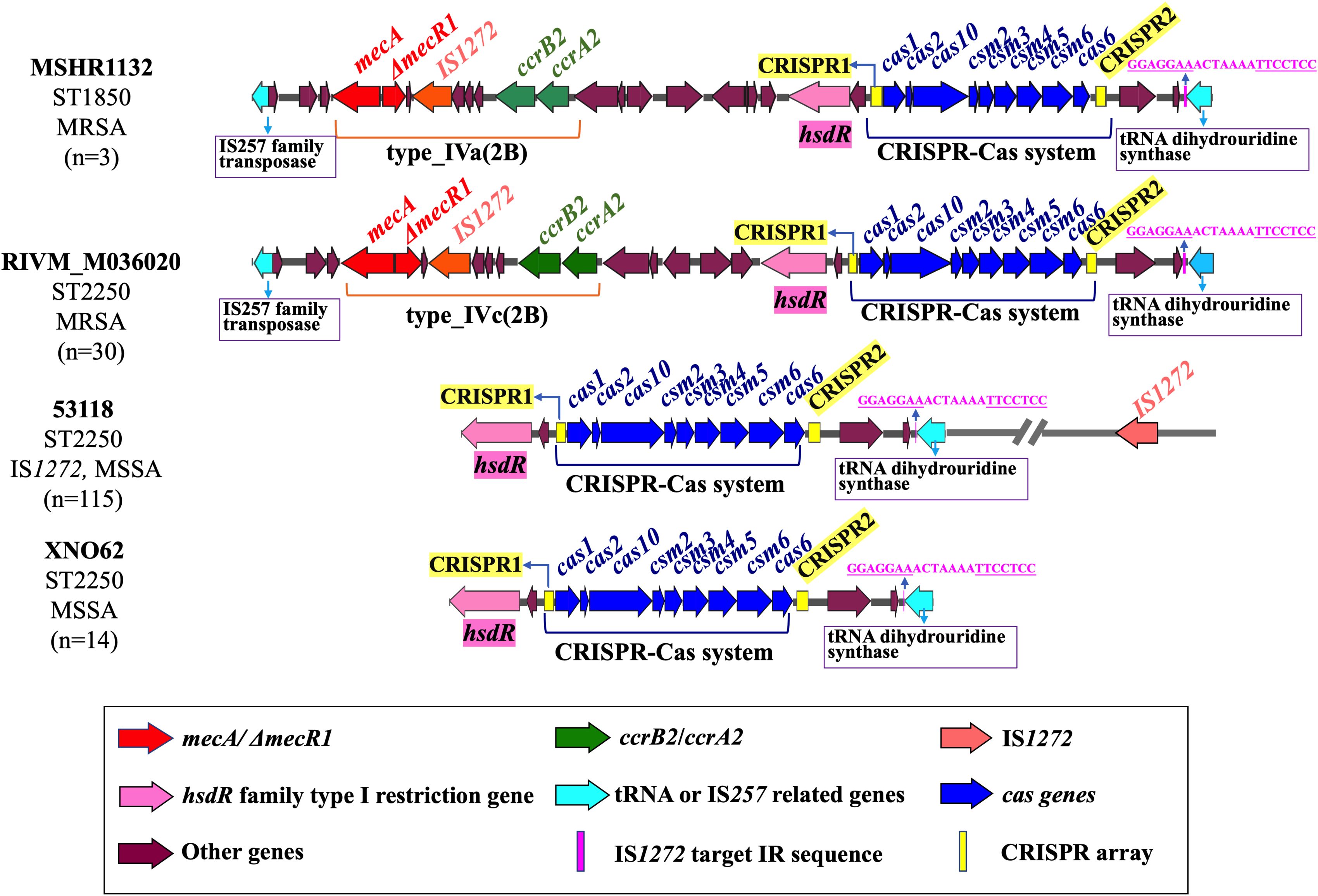
Figure 4. Genetic location of CRISPR-Cas system in the S. argenteus chromosome. The arrangement and chromosomal location of the CRISPR-Cas system are illustrated in MSSA and MRSA strains with two distinct MLST types. The conserved IS1272 target IR sequences are also highlighted in these strains.
4 Discussion
As an adaptive immune system of bacteria, the CRISPR-Cas system provides protection against foreign phages and plasmids. However, this system is not widespread among staphylococci species, as only certain isolates with specific characteristics tend to carry it (Rossi et al., 2017). For instance, more than 50% of ST630 MRSA strains have been found to carry the CRISPR-Cas system (Mikkelsen et al., 2023). S. argenteus is a recently identified species capable of causing human infections and is frequently detected in food, particularly in poultry products (Wakabayashi et al., 2022; Li et al., 2019). S. argenteus was first identified in Australia and has since been increasingly reported worldwide, particularly in tropical regions (McDonald et al., 2006). In Thailand, it has been associated with community-acquired invasive infections since 2006 (Thaipadungpanit et al., 2015). As a result, numerous strains have been collected and sequenced for comparative analysis with S. aureus (Chantratita et al., 2016). Additionally, an increasing number of S. argenteus-related cases have been reported in other countries, including the Netherlands, Japan, China, etc (Aung et al., 2025; Chen et al., 2023; Bank et al., 2021). To date, 15 STs have been identified in S. argenteus, with ST2250 (60.1%) and ST1223 (15.2%) being the most prevalent STs globally. In Japan, ST2250 took up 49% of clinical S. argenteus isolates collected from 2020 to 2023 (Aung et al., 2025). ST2250 has been reported as the predominant ST in both food sources and clinical isolates from various regions, including Indonesia (Supriadi et al., 2024), Hong Kong (China) (Chen et al., 2023), Guangdong (China) (Rong et al., 2023), Myanmar (Kyaw et al., 2023), North America (Eshaghi et al., 2021), Hokkaido (Japan) (Aung et al., 2021). In our study, 71.9% of the ST2250 strains were found to carry the CRISPR-Cas system, suggesting that these strains may have enhanced resistance against phage infections. This is supported by the previous findings that the CRISPR-Cas system in S. aureus and S. epidermidis are functionally active in providing immunity against phage and plasmid infections (Marraffini and Sontheimer, 2008; Li et al., 2021). This may also explain the higher prevalence of CRISPR-Cas-positive S. argenteus in Thailand compared to other countries, as nearly 84.6% (66/78) of the clinical isolates belonged to ST2250. Although the genetic location of the CRISPR-Cas system in S. aureus and S. argenteus has been reported to be closely associated with SCCmec (Mikkelsen et al., 2023; Goswami et al., 2021), the characteristics of the specific insertion sites for the CRISPR-Cas system have not yet been analyzed in detail. Here, we found that the CRIPSR-Cas system in S. argenteus is most closely related to the IS1272 transposase, which is also detected in SCCmec elements of methicillin-resistance S. argenteus, suggesting that IS1272 may play a role in the acquisition of the CRISPR-Cas system in S. argenteus. Additionally, in all the CRISPR-Cas-positive S. argenteus, a conserved recognition site for IS1272 inverted repeat (IR) elements is consistently observed at the right end of the CRISPR2 locus. CRISPR typing, developed as a molecular typing method, has been widely used to reveal the genetic difference and evolutionary relationships among different bacterial isolates, showing strong correspondence with cgMLST (core genome multilocus sequence typing) and cgSNP (core genome single nucleotide polymorphism) typing methods (Li et al., 2016; Yassine et al., 2022). In this study, CRISPR typing classified the CRISPR-Cas-positive S. argenteus isolates into two clusters, and the Cluster II emerging as the predominant group among these strains.
The spacers within CRISPR array play a crucial role in providing adaptive immunity against foreign genetic elements. The 15 spacers identified in S. argenteus have also been reported in S. aureus, suggesting that both species encounter and survive in environments with similar phages and plasmids. However, obtaining the homologous sequences for these spacers has been challenging due to the limited availability of comprehensive databases in earlier studies (Mikkelsen et al., 2023; Li et al., 2019). Here, the homologous sequences for all 15 spacers were successfully identified using multiple databases, providing new insights into the targets of these spacers and their role in adaptive immunity. Among the identified spacers, four showed homology to sequences in plasmids. pWBG731 is a multidrug resistance plasmid frequently found in community-associated methicillin-resistant S. aureus (CA-MRSA) (Yui Eto et al., 2019). The plasmid carries genes conferring resistance to mupirocin, trimethoprim, cadmium, and penicillin, as well as mobile genetic elements related to the horizontal dissemination of multidrug resistance in CA-MRSA (Yui et al., 2019). pSALNBL118 is a phage like plasmid originating from S. aureus strain B3–4A, isolated from beef liver (Karki et al., 2020). The plasmid is thought to play a significant role in horizontal gene transfer (Goerke et al., 2009) and transmission of virulence factors.
Among the 13 spacers with homology to phage sequences, seven correspond to lytic phages, while six were associated with lysogenic phages. SA11 is a lytic phage isolated from a wastewater treatment facility in Gwa-Chon, South Korea (Kim and Myung, 2012). It has been successfully used in combination with antibiotics to effectively inhibit the growth of antibiotic-resistant S. aureus under simulated intestinal conditions (Zhou et al., 2023). SAP-2 is a podoviridae lytic bacteriophage that encodes a cell-wall degrading enzyme, SAL-2, which can disrupt biofilm formation of S. aureus, including MRSA (Son et al., 2010). GRCS is a podoviridae lytic phage isolated from sewage in India, and it showed more efficient in treating both diabetic and non-diabetic septicemic mice than oxacillin antibiotic alone (Plumet et al., 2022). The vB_Sau-RP15 phage was isolated from raw milk and developed as a promising agent against S. aureus contamination in pasteurized milk (Imklin et al., 2023). Phage vB_SauM-V1SA20, isolated from wastewater, exhibits a broad host activity against S. aureus, including CC80 strains (Kolenda et al., 2022). Phage vB_SauS-SAP27 (ϕSAP27) is a Siphovirdiae phage that infects S. aureus and was isolated from sewage (Park et al., 2021).
The homology of spacers to temperate phages was not given much consideration during the CRISPR-Cas system analysis. Among the six temperate phages, phage B236 has been identified as an eta (Exfoliative toxin A, ETA) phage, contributing to the toxic phenotype of the S. aureus SA236 strain (Botka et al., 2015). This kind of phages are able to mediate the transfer of eta gene to prophage-free S. aureus strains. ETA is potentially a major toxin responsible for staphylococcal skin blistering infections (Botka et al., 2015). Phage phiMR25 was a lysogenic phage isolated from an MRSA strain MR25 by mitomycin C induction (Hoshiba et al., 2010). Although it is a lysogenic phage, is showed a broad host range and can protect mice against S. aureus infection (Hoshiba et al., 2010). The prophage StauST398–5 was specifically identified in non-LA CC398 isolates, where it protects bacteria from horizontal genetic transfer to its host and carries genes related to bacterial virulence and adaptation (van der Mee-Marquet et al., 2013).
5 Conclusion
In this study, the presence of the type III-A CRISPR-Cas system in 368 S. argenteus strains obtained from public database were analyzed to reveal the genetic characteristics of CRISPR-Cas-positive strains. The CRISPR-Cas system is present in 44.0% (162) of S. argenteus strains, but only in ST2250 and ST1850 strains. Notably, ST2250 strains, which are the predominant sequence type of S. argenteus, show that 71.9% of these strains carry the CRISPR-Cas system. Additionally, the presence of IS1272 and its target IR site is likely the reason for the acquisition of the CRISPR-Cas system in ST2250 strains. Homology analysis confirmed that all 15 identified spacers in the CRISPR array showed homology to sequences in plasmids, phages, or prophages, indicating that the acquisition of the CRISPR-Cas system may provide protection against phage attacks and plasmid invasion. These findings highlight the potential role of the CRISPR-Cas system in enhancing the adaptive immunity of S. argenteus in environments rich in mobile genetic elements.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
XC: Writing – original draft, Methodology, Data curation. LX: Writing – original draft, Formal analysis, Data curation, Investigation. ZL: Writing – original draft, Visualization, Resources. LW: Writing – original draft, Validation. ZW: Writing – review & editing, Data curation, Software. YL: Validation, Writing – review & editing. XJ: Supervision, Conceptualization, Writing – review & editing. QL: Supervision, Writing – review & editing, Conceptualization, Software, Project administration, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Key Research and Development Program of China (2024YFE0198800; 2023YFD1800503); Jiangsu Key Laboratory of Zoonosis Major Independent Research Project (RZZ202302); Postgraduate Research & Practice Innovation Program of Jiangsu Province (Yangzhou University) (no. KYCX24_3850); the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Acknowledgments
The authors declare our appreciation to all the students and teachers who participated in our study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1644286/full#supplementary-material
References
Argudín, M. A., Dodémont, M., Vandendriessche, S., Rottiers, S., Tribes, C., Roisin, S., et al. (2016). Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1017–1022. doi: 10.1007/s10096-016-2632-x
Aung, M. S., Osada, M., Urushibara, N., Kawaguchiya, M., Ohashi, N., Hirose, M., et al. (2025). Molecular characterization of methicillin-susceptible/resistant Staphylococcus aureus from bloodstream infections in northern Japan: The dominance of CC1-MRSA-IV, the emergence of human-associated ST398 and livestock-associated CC20 and CC97 MSSA. J. Global antimicrobial resistance 41, 77–87. doi: 10.1016/j.jgar.2024.12.010
Aung, M. S., Urushibara, N., Kawaguchiya, M., Hirose, M., Ike, M., Ito, M., et al. (2021). Distribution of virulence factors and resistance determinants in three genotypes of Staphylococcus argenteus clinical isolates in Japan. Pathogens 10, 163. doi: 10.3390/pathogens10020163
Bank, L. E. A., Bosch, T., Schouls, L. M., Weersink, A. J. L., Witteveen, S., Wolffs, P. F. G., et al. (2021). Methicillin-resistant Staphylococcus argenteus in the Netherlands: not a new arrival. Eur. J. Clin. Microbiol. Infect. Dis. 40, 1583–1585. doi: 10.1007/s10096-021-04204-7
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Botka, T., Růžičková, V., Konečná, H., Pantůček, R., Rychlík, I., Zdráhal, Z., et al. (2015). Complete genome analysis of two new bacteriophages isolated from impetigo strains of Staphylococcus aureus. Virus Genes 51, 122–131. doi: 10.1007/s11262-015-1223-8
Chantratita, N., Wikraiphat, C., Tandhavanant, S., Wongsuvan, G., Ariyaprasert, P., Suntornsut, P., et al. (2016). Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin. Microbiol. infection 22, 458.e11–9. doi: 10.1016/j.cmi.2016.01.008
Chen, J. H. K., Leung, H. Y., Wong, C. M. C., Yuen, K. Y., and Cheng, V. C. C. (2023). Prevalence and characteristics of invasive Staphylococcus argenteus among patients with bacteremia in Hong Kong. Microorganisms 11, 2435. doi: 10.3390/microorganisms11102435
Cruz-López, E. A., Rivera, G., Cruz-Hernández, M. A., Martínez-Vázquez, A. V., Castro-Escarpulli, G., Flores-Magallón, R., et al. (2021). Identification and characterization of the CRISPR/Cas system in Staphylococcus aureus strains from diverse sources. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.656996
Eshaghi, A., Bommersbach, C., Zittermann, S., Burnham, C. A., Patel, R., Schuetz, A. N., et al. (2021). Phenotypic and genomic profiling of Staphylococcus argenteus in Canada and the United States and recommendations for clinical result reporting. J. Clin. Microbiol. 59, e02470–e02420. doi: 10.1128/JCM.02470-20
Gleditzsch, D., Pausch, P., Müller-Esparza, H., Özcan, A., Guo, X., Bange, G., et al. (2019). PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517. doi: 10.1080/15476286.2018.1504546
Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. bacteriology 191, 3462–3468. doi: 10.1128/JB.01804-08
Goswami, C., Fox, S., Holden, M., Leanord, A., and Evans, T. J. (2021). Genomic analysis of global Staphylococcus argenteus strains reveals distinct lineages with differing virulence and antibiotic resistance gene content. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.795173
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinf. 8, 172. doi: 10.1186/1471-2105-8-172
Hille, F., Richter, H., Wong, S. P., Bratovič, M., Ressel, S., and Charpentier, E. (2018). The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259. doi: 10.1016/j.cell.2017.11.032
Holt, D. C., Holden, M. T., Tong, S. Y., Castillo-Ramirez, S., Clarke, L., Quail, M. A., et al. (2011). A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 3, 881–895. doi: 10.1093/gbe/evr078
Hoshiba, H., Uchiyama, J., Kato, S., Ujihara, T., Muraoka, A., Daibata, M., et al. (2010). Isolation and characterization of a novel Staphylococcus aureus bacteriophage, phiMR25, and its therapeutic potential. Arch. Virol. 155, 545–552. doi: 10.1007/s00705-010-0623-2
Imam, O., Tang, P., Almaslamani, E., Sawahreh, M., Suleiman, M., Sharma, A., et al. (2025). First reported case of brain abscess in an infant caused by Staphylococcus argenteus. Pediatr. Infect. Dis. J. 44, e107. doi: 10.1097/INF.0000000000004600
Imklin, N., Chaengphaniad, P., Šimoliūnas, E., and Nasanit, R. (2023). A novel Staphylococcus phage, vB_Sau-RP15, and its application in contaminated milk. Lett. Appl. Microbiol. 76, ovac003. doi: 10.1093/lambio/ovac003
Karki, A. B., Neyaz, L., and Fakhr, M. K. (2020). Comparative genomics of plasmid-bearing Staphylococcus aureus strains isolated from various retail meats. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.574923
Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G., and Siksnys, V. (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. doi: 10.1126/science.aao0100
Kim, M. S. and Myung, H. (2012). Complete genome of Staphylococcus aureus phage SA11. J. Virol. 86, 10232. doi: 10.1128/JVI.01574-12
Kolenda, C., Medina, M., Bonhomme, M., Laumay, F., Roussel-Gaillard, T., Martins-Simoes, P., et al. (2022). Phage therapy against Staphylococcus aureus: selection and optimization of production protocols of novel broad-spectrum Silviavirus phages. Pharmaceutics 14, 1885. doi: 10.3390/pharmaceutics14091885
Kunin, V., Sorek, R., and Hugenholtz, P. (2007). Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8, R61. doi: 10.1186/gb-2007-8-4-r61
Kyaw, W. K., Aung, M. S., San, T., Maw, W. W., Mu, K. K., Mon, W. L. Y., et al. (2023). Molecular epidemiological characterization of Staphylococcus aureus and Staphylococcus argenteus clinical isolates from a national tertiary care hospital in Myanmar: co-isolation of multiple clones and identification of novel Staphylocoagulase genotype. Microbial Drug resistance 29, 127–137. doi: 10.1089/mdr.2022.0191
Lee, M., Choi, Y., Choi, S. J., Moon, S. M., Kim, E. S., Kim, H. B., et al. (2024). Staphylococcus argenteus bacteremia in the Republic of Korea. Microbiol. Spectr. 12, e0279823. doi: 10.1128/spectrum.02798-23
Li, Q., Li, Y., Tan, Y., Meng, C., Ingmer, H., and Jiao, X. (2019). Prevalence and characterisation of Staphylococcus aureus and Staphylococcus argenteus in chicken from retail markets in China. Food Control 96, 158–164. doi: 10.1016/j.foodcont.2018.08.030
Li, Y., Mikkelsen, K., Lluch I Grané, O., Wang, Z., Tang, Y., Jiao, X., et al. (2021). Functional characterization of type III-A CRISPR-Cas in a clinical human methicillin-R Staphylococcus aureus strain. CRISPR J. 4, 686–698. doi: 10.1089/crispr.2021.0046
Li, Q., Wang, X., Yin, K., Hu, Y., Xu, H., Xie, X., et al. (2018). Genetic analysis and CRISPR typing of Salmonella enterica serovar Enteritidis from different sources revealed potential transmission from poultry and pig to human. Int. J. Food Microbiol. 266, 119–125. doi: 10.1016/j.ijfoodmicro.2017.11.025
Li, Q., Xie, X., Yin, K., Tang, Y., Zhou, X., Chen, Y., et al. (2016). Characterization of CRISPR-Cas system in clinical Staphylococcus epidermidis strains revealed its potential association with bacterial infection sites. Microbiological Res. 193, 103–110. doi: 10.1016/j.micres.2016.09.003
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Marraffini, L. A. and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
McDonald, M., Dougall, A., Holt, D., Huygens, F., Oppedisano, F., Giffard, P. M., et al. (2006). Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J. Clin. Microbiol. 44, 3720–3727. doi: 10.1128/JCM.00836-06
Mellmann, A., Becker, K., von Eiff, C., Keckevoet, U., Schumann, P., and Harmsen, D. (2006). Sequencing and staphylococci identification. Emerging Infect. Dis. 12, 333–336. doi: 10.3201/eid1202.050962
Mikkelsen, K., Bowring, J. Z., Ng, Y. K., Svanberg Frisinger, F., Maglegaard, J. K., Li, Q., et al. (2023). An endogenous Staphylococcus aureus CRISPR-Cas system limits phage proliferation and is efficiently excised from the genome as part of the SCCmec cassette. Microbiol. Spectr. 11, e0127723. doi: 10.1128/spectrum.01277-23
Niewoehner, O., Garcia-Doval, C., Rostøl, J. T., Berk, C., Schwede, F., Bigler, L., et al. (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. doi: 10.1038/nature23467
Park, D. W., Lee, Y. D., and Park, J. H. (2021). Characteristics for phage-encoded cell wall hydrolase of LysSAP27 to reduce staphylococcal food poisoning. Food Sci. Biotechnol. 30, 745–753. doi: 10.1007/s10068-021-00910-2
Plumet, L., Ahmad-Mansour, N., Dunyach-Remy, C., Kissa, K., Sotto, A., Lavigne, J. P., et al. (2022). Bacteriophage therapy for Staphylococcus aureus infections: a review of animal models, treatments, and clinical trials. Front. Cell. infection Microbiol. 12. doi: 10.3389/fcimb.2022.907314
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O., and Marraffini, L. A. (2017). Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22, 343–353.e3. doi: 10.1016/j.chom.2017.07.016
Rong, D., Liu, Z., Huang, J., Zhang, F., Wu, Q., Dai, J., et al. (2023). Prevalence and characterization of Staphylococcus aureus and Staphylococcus argenteus isolated from rice and flour products in Guangdong, China. Int. J. Food Microbiol. 406, 110348. doi: 10.1016/j.ijfoodmicro.2023.110348
Rossi, C. C., Souza-Silva, T., Araújo-Alves, A. V., and Giambiagi-deMarval, M. (2017). CRISPR-Cas systems features and the gene-reservoir role of coagulase-negative Staphylococci. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01545
Shabbir, M. A. B., Ul-Rahman, A., Iftikhar, M. R., Rasheed, M., Maan, M. K., Sattar, A., et al. (2024). Exploring the interplay of the CRISPR-CAS system with antibiotic resistance in Staphylococcus aureus: a poultry meat study from Lahore, Pakistan. Medicina (Kaunas Lithuania) 60, 130. doi: 10.3390/medicina60010130
Son, J. S., Lee, S. J., Jun, S. Y., Yoon, S. J., Kang, S. H., Paik, H. R., et al. (2010). Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl. Microbiol. Biotechnol. 86, 1439–1449. doi: 10.1007/s00253-009-2386-9
Supriadi, I. R., Santosaningsih, D., Budayanti, N. S., Zandijk, W. H. A., Rijfkogel, A., Klaassen, C. H. W., et al. (2024). Identification and characterization of Staphylococcus argenteus from Indonesia. Int. J. Med. Microbiol. 316, 151629. doi: 10.1016/j.ijmm.2024.151629
Thaipadungpanit, J., Amornchai, P., Nickerson, E. K., Wongsuvan, G., Wuthiekanun, V., Limmathurotsakul, D., et al. (2015). Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J. Clin. Microbiol. 53, 1005–1008. doi: 10.1128/JCM.03049-14
Tong, S. Y. C., Schaumburg, F., Ellington, M. J., Corander, J., Pichon, B., Leendertz, F., et al. (2015). Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int. J. systematic evolutionary Microbiol. 65, 15–22. doi: 10.1099/ijs.0.062752-0
van der Mee-Marquet, N., Corvaglia, A. R., Valentin, A. S., Hernandez, D., Bertrand, X., Girard, M., et al. (2013). Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infection Genet. Evol. 18, 299–308. doi: 10.1016/j.meegid.2013.06.009
Wakabayashi, Y., Takemoto, K., Iwasaki, S., Yajima, T., Kido, A., Yamauchi, A., et al. (2022). Isolation and characterization of Staphylococcus argenteus strains from retail foods and slaughterhouses in Japan. Int. J. Food Microbiol. 363, 109503. doi: 10.1016/j.ijfoodmicro.2021.109503
Wan, T. W., Higuchi, W., Khokhlova, O. E., Hung, W. C., Iwao, Y., Wakayama, M., et al. (2017). Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS1272 transposition through a novel inverted repeat-replacing mechanism. PloS One 12, e0187288. doi: 10.1371/journal.pone.0187288
Yassine, I., Lefèvre, S., Hansen, E. E., Ruckly, C., Carle, I., Lejay-Collin, M., et al. (2022). Population structure analysis and laboratory monitoring of Shigella by core-genome multilocus sequence typing. Nat. Commun. 13, 551. doi: 10.1038/s41467-022-28121-1
Yui Eto, K., Firth, N., Davis, A. M., Kwong, S. M., Krysiak, M., Lee, Y. T., et al. (2019). Evolution of a 72-Kilobase Cointegrant, Conjugative multiresistance plasmid in community-associated methicillin-resistant Staphylococcus aureus isolates from the early 1990s. Antimicrobial Agents chemotherapy 63, e01560–e01519. doi: 10.1128/AAC.01560-19
Zhang, K., Wang, P., Li, S., Xie, X., Wang, Z., Li, Y., et al. (2024). Type I-E CRISPR-Cas system regulates fimZY and T3SS1 genes expression in Salmonella enterica serovar Pullorum. Veterinary Microbiol. 299, 110301. doi: 10.1016/j.vetmic.2024.110301
Zhou, W. Y., Wen, H., Li, Y. J., Gao, Y. J., Zheng, X. F., Li, H. X., et al. (2023). WGS analysis of two Staphylococcus aureus bacteriophages from sewage in China provides insights into the genetic feature of highly efficient lytic phages. Microbiological Res. 271, 127369. doi: 10.1016/j.micres.2023.127369
Keywords: Staphylococcus argenteus, CRISPR-Cas, poultry, SCCmec, IS1272
Citation: Chen X, Xu L, Luo Z, Wang L, Wang Z, Li Y, Jiao X and Li Q (2025) Prevalence and genomic insights into type III-A CRISPR-Cas system acquisition in global Staphylococcus argenteus strains. Front. Cell. Infect. Microbiol. 15:1644286. doi: 10.3389/fcimb.2025.1644286
Received: 10 June 2025; Accepted: 08 July 2025;
Published: 28 July 2025.
Edited by:
Xiancai Rao, Army Medical University, ChinaReviewed by:
Yujie Li, University of Science and Technology of China, ChinaRasha Othman, University of Basrah, Iraq
Copyright © 2025 Chen, Xu, Luo, Wang, Wang, Li, Jiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Li, eWFuZ19saUB5enUuZWR1LmNu; Qiuchun Li, cWNsaUB5enUuZWR1LmNu
 Xinhai Chen1,2
Xinhai Chen1,2 Yang Li
Yang Li Xinan Jiao
Xinan Jiao Qiuchun Li
Qiuchun Li