- 1The First Clinical Medical College, Shandong University of Traditional Chinese Medicine, Jinan, China
- 2Affiliated Hospital of Shandong University of Chinese Medicine, Jinan, China
Objective: This study aimed to predict and verify the mechanism of curculigoside in treating osteoporosis using network pharmacology, molecular docking technology, and micro-CT technology.
Methods: Herb databases were searched to identify and screen potential targets of curculigoside. The GeneCards platform was utilized to mine osteoporosis-related targets. Cytoscape 3.6.0 software was employed to construct a compound-target-disease network. A protein–protein interaction (PPI) network for curculigoside in osteoporosis treatment was established, and core targets were screened. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and GO biological process analyses were performed using the Metascape database. Finally, molecular docking and micro-CT were used to validate core targets relevant to osteoporosis.
Results: A total of 166 potential curculigoside targets and 4,313 osteoporosis-related targets were identified, with 91 common targets. Ten key targets, including matrix metalloproteinase (MMP)3, MMP9, interleukin (IL)-6, and caspase-3, were screened. KEGG pathway enrichment analysis indicated involvement in 10 pathways, such as the Rap1 signaling pathway and tumor necrosis factor (TNF) signaling pathway. Molecular docking results demonstrated strong binding affinity between curculigoside and the core targets. Micro-CT analysis revealed that curculigoside not only improved BMD, BV/TV, BS/BV, and Tb.Th but also reduced Tb.Sp in osteoporotic bone.
Conclusions: Curculigoside is likely to treat osteoporosis through targets such as MMP3, MMP9, IL-6, and caspase-3, acting on signaling pathways including Rap1 and TNF. These results indicate that curculigoside exhibits multitarget and multipathway characteristics in osteoporosis treatment, providing a theoretical basis for further clinical investigation.
1 Introduction
Bone is a special type of connective tissue that is dynamically mineralized and has a variety of physiological functions (1, 2). Recognized for its mechanical properties, it serves as an attachment point for muscles, providing a structural basis and aiding movement (3, 4). Bone undergoes lifelong remodeling, including bone formation by osteoblasts and bone resorption by osteoclasts, which are essential for maintaining healthy bone mass (5–7). The entire process of bone remodeling is strictly controlled and coordinated by a variety of cells, including osteoblasts, which mediate bone formation, and osteoclasts, which mediate bone resorption (8, 9). Osteoblasts originate from mesenchymal stem cells in the bone marrow stroma and are responsible for the synthesis of the bone matrix and its subsequent mineralization (10, 11). Osteoclasts are large multinucleated giant cells formed by the fusion of mononuclear progenitors of monocytes/macrophages, and they are responsible for bone resorption (12, 13). The formation, proliferation, differentiation, and activity of these cells are controlled by local and systemic factors (14, 15).
Osteoporosis results from an imbalance between bone formation and resorption. It represents a common group of skeletal disorders characterized by destruction of bone microstructure, reduced bone mass, and increased bone fragility (16–18). The dynamic equilibrium between new bone formation by osteoblasts and old bone resorption by osteoclasts is crucial for maintaining bone tissue metabolism (19, 20). Osteoblasts, as the principal bone-forming cells, play a major role in the metabolic balance, growth, development, and repair of bone tissue (21, 22). During bone formation, osteoblasts undergo three stages: proliferation, differentiation, and apoptosis. These three stages involve: (1) the proliferation stage, where osteoblast precursor cells multiply; (2) the differentiation stage, where cells mature to secrete bone matrix; and (3) the apoptosis stage, where excess cells undergo programmed death to maintain tissue balance (23). Abnormalities in osteoblast proliferation, differentiation, or apoptosis play a critical role in the development of osteoporosis (24, 25).
Curculigoside, a naturally occurring phenolic compound, has been traditionally employed in many Asian countries for the treatment of osteoporosis (26). Its documented effects include antioxidant, anti-aging, immunomodulatory, and anti-inflammatory activities, along with the prevention of bone loss. Curculigoside demonstrates osteoprotective properties (27). Studies indicate that it improves bone microstructure and biomechanical properties while enhancing antioxidant enzyme activity in serum and bone tissue through the regulation of bone metabolic homeostasis (28, 29). Furthermore, curculigoside reduces bone loss, promotes osteogenesis, and inhibits adipogenesis in ovariectomized rats by upregulating endoplasmic reticulum-dependent bone morphogenetic protein-2 (BMP) (30). During aging, declining estrogen levels and excessive accumulation of reactive oxygen species (ROS) in bone tissue activate the nuclear factor kappa B (NF-κB) and MAPK pathways, inducing apoptosis and osteoclastogenesis (31). These findings suggest that curculigoside exerts bone-protective effects, though its precise mechanism of action remains unclear.
Traditional Chinese medicine is characterized by multiple chemical components, multiple targets, and multiple effects. Network pharmacology explores the complex relationships among drugs, targets, diseases, and pathways, enabling the identification of multiple components, targets, and signaling pathways. In this way, network pharmacology helps elucidate the therapeutic mechanisms of traditional Chinese medicine (32, 33). In this study, we applied a network pharmacological approach to identify potential targets of curculigoside and to elucidate its mechanism of action in the treatment of osteoporosis. Effective targets for curculigoside and osteoporosis were mined from multiple databases. Pathway enrichment and protein–protein interaction (PPI) network analyses were then performed on the overlapping curculigoside and osteoporosis to identify potential therapeutic pathways for osteoporosis treatment. To verify the protective effect of curculigoside on osteoporosis, in vitro validation was conducted by detecting the expression of cross-targets (e.g., IL-6, tumor necrosis factor alpha [TNF-α], matrix metalloproteinase (MMP)3, MMP9, caspase-3) in rat bone tissue using reverse transcription quantitative polymerase chain reaction (RT-qPCR) and enzyme-linked immunosorbent assay (ELISA). The results were consistent with the network pharmacology predictions.
2 Materials and methods
2.1 Animals
Thirty female SD rats of specific pathogen-free (SPF) grade, 8 weeks old, were purchased from Shandong Jinan Panyue Experimental Animal Co. Ltd. (Animal Production License No. SCXK [Lu] 20190003). All animals were housed under controlled, identical SPF standard environmental conditions (23°C ± 2°C, 12-h light/dark cycle), had free access to food, and were allowed free movement. The study protocol was approved by the Experimental Animal Ethics Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine (approval number: SDSZYYAWE20241105005).
2.2 Screening potential gene targets of curculigoside
Target gene data for curculigoside were collected from the Encyclopaedia of Traditional Chinese Medicine, SwissTargetPrediction (http://swisstargetprediction.ch), and the Similarity Ensemble Approach (https://sea.bkslab.org). The obtained gene targets were summarized and de-duplicated by searching the database using “cynarin” as the keyword. Target names were normalized using the UniProt database (https://www.uniprot.org/) (34).
2.3 Osteoporosis disease-target collection
The DisGeNET (https://www.disgenet.org/home/) and GeneCards (https://www.genecards.org/) (35) databases were used to search for relevant targets using the keyword “osteoporosis”. A network target map was constructed to identify overlapping targets between osteoporosis and the active ingredient. The identified overlapping targets are considered to play key roles in the antiosteoporosis activity of curculigoside.
2.4 Network construction
To elucidate the relationship among curculigoside, gene targets, and osteoporosis, Cytoscape 3.7.2 software was used to construct the curculigoside-target and osteoporosis-target networks (36). The drug-target network and disease-target network were established. In the network, nodes of different colors represent various drug and disease targets. Edges represent the relationships between two nodes, and their number, defined as the “degree,” determines the size of each node. A Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was then generated to identify the intersection of targets between curculigoside and osteoporosis, thereby revealing the potential targets of curculigoside against osteoporosis.
2.5 KEGG pathway enrichment and Gene Ontology analyses
The intersecting targets were used as inputs for the functional annotation tool DAVID 6.8 (https://david.ncifcrf.gov/). The identifier was set to OFFICIAL_GENE SYMBOL, and Homo sapiens and Gene List were selected. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and Gene Ontology (GO) enrichment analyses were then performed. p < 0.01 and FDR < 0.05 were set as the screening criteria to identify the biological processes and signaling pathways associated with Traditional Chinese Medicine (TCM) disease interventions.
2.6 Protein–protein interaction network construction
The potential targets of curculigoside in osteoporosis treatment were imported into the STRING 11.5 platform (https://cn.string-db.org/) (37). The species was set to Homo sapiens before conducting the PPI analysis. The PPI analysis output was saved in TSV file format, with the minimum interaction threshold set to 0.9. Cytoscape 3.7.2 was used to construct the PPI network and evaluate the node and edge degrees.
2.7 Molecular docking
Molecular docking was used to clarify the relationships among the potential curculigoside targets versus osteoporosis and the corresponding active ingredients. The 3D structure of the core target protein was retrieved from the RSCB PDB database (https://www.rcsb.org/) (38). The chemical structure of curculigoside was acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (39). AutoDock (https://vina.scripps.edu) was used to perform the molecular docking, and PYMOL (https://pymol.org/2/) was used to visualize the results.
2.8 Establishment of animal model
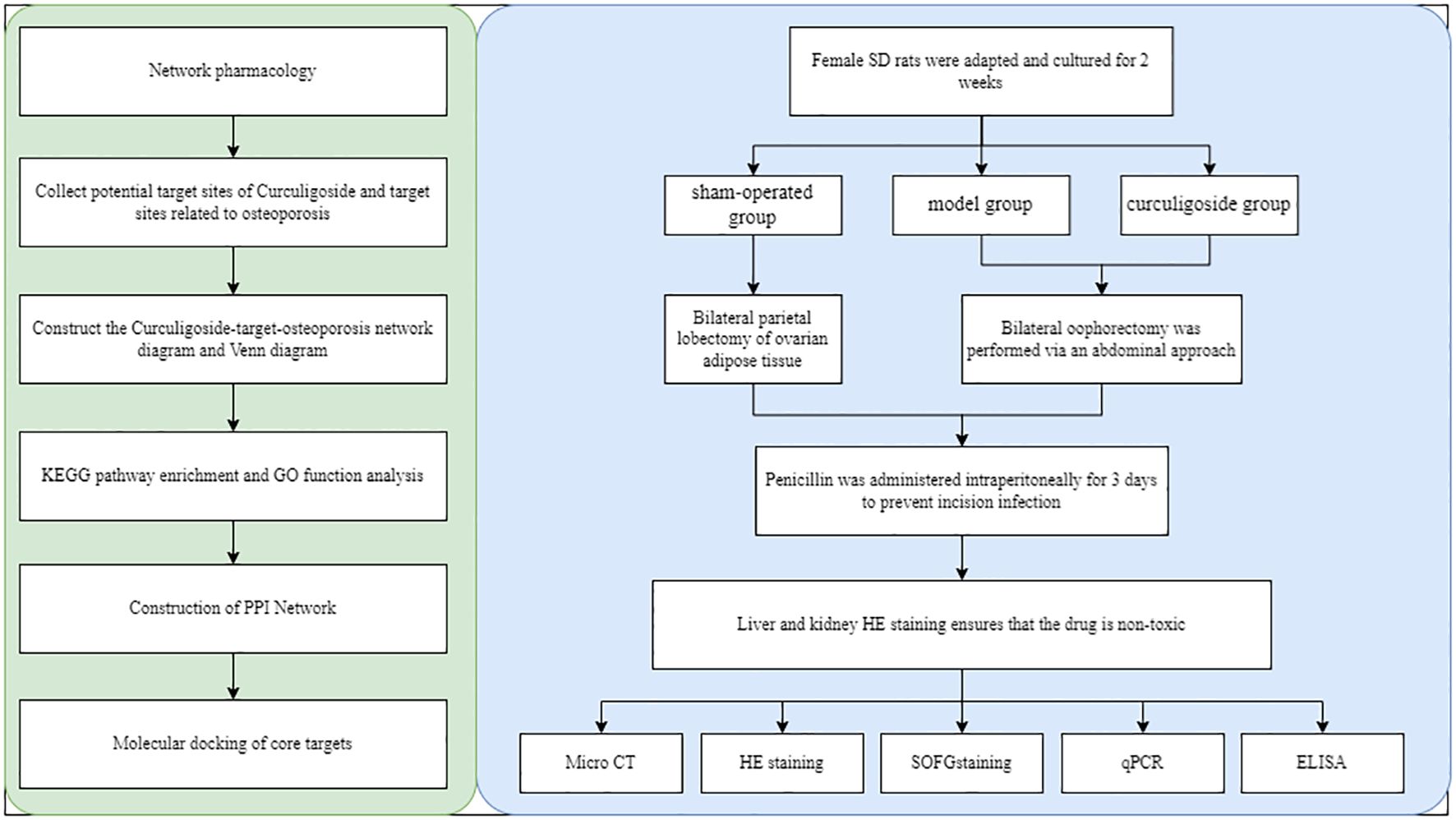
After 2 weeks of acclimatization, all female SD rats were randomly divided into three groups: sham-operated (sham) group, model (OVX) group, and curculigoside (curculigoside) group, with 10 rats in each group. Both the model group and the curculigoside group underwent bilateral ovariectomy via the abdominal approach, while the sham-operated group underwent excision of bilateral parietal ovarian adipose tissue. Penicillin was administered intraperitoneally to the rats daily for 3 days to prevent incisional infection. Penicillin at a concentration of 40,000 IU/mL was injected into rats at a volume of 1 mL/kg for 3 consecutive days to prevent infection (40). The rats in the curculigoside group were injected intraperitoneally at 7.5 mg/kg. In this study, three doses of Curculigoside were set: 5, 7.5, and 10 mg/kg. Based on hematoxylin–eosin (HE) staining and TRAP staining to observe the expression of bone trabeculae and osteoclasts, no significant difference was found between the 7.5- and 10-mg/kg intervention groups. Therefore, 7.5 mg/kg was selected as the optimal dose. The experimental data are provided in Supplementary File 1. All rats were treated continuously for 12 weeks, and bone mineral density testing was performed.
2.9 HE staining and SOFG staining
After fixation, decalcification, and paraffin embedding, HE staining was performed, and the pathological structure of the bone tissue was observed following dehydration and clearing. Safranine O and Fast Green (SOFG) staining was carried out after dewaxing and hydration of the sections. The sections were sequentially stained with hematoxylin, solid green, and mesene O, followed by dehydration and clearing, to more precisely display structural changes such as trabeculae and bone marrow cavities. For the liver and kidney tissues of rats, the HE staining procedure was as follows: the tissues were fixed and embedded in paraffin, then sectioned. After dewaxing and hydration, the sections were stained with hematoxylin–eosin. Following dehydration and clearing, the tissue structures were observed to evaluate whether any of the treatment groups caused toxic damage to the liver and kidneys.
2.10 Microcomputed tomography
The removed femur was placed in 4% paraformaldehyde (4°C, 24 h) for fixation. The microstructure of the femur was scanned and analyzed using the SkyScan imaging system. Each sample was carefully placed so that the femoral stem was oriented as vertically as possible. Bone morphometric parameters of the femur were obtained, including bone mineral density (BMD) on total volume, bone surface area to bone volume ratio (BS/BV), bone surface area to tissue volume ratio (BS/TV), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp).
2.11 Measurement of inflammatory cytokines
The plasma of mice was centrifuged, and the supernatant was collected to determine the levels of inflammatory factors. The expression levels of IL-6, TNF-α, IL-1β, osteocalcin (OCN), and insulin-like growth factor-1 (IGF-1) were measured using ELISA kits according to the manufacturer’s instructions.
2.12 RNA isolation and real-time PCR
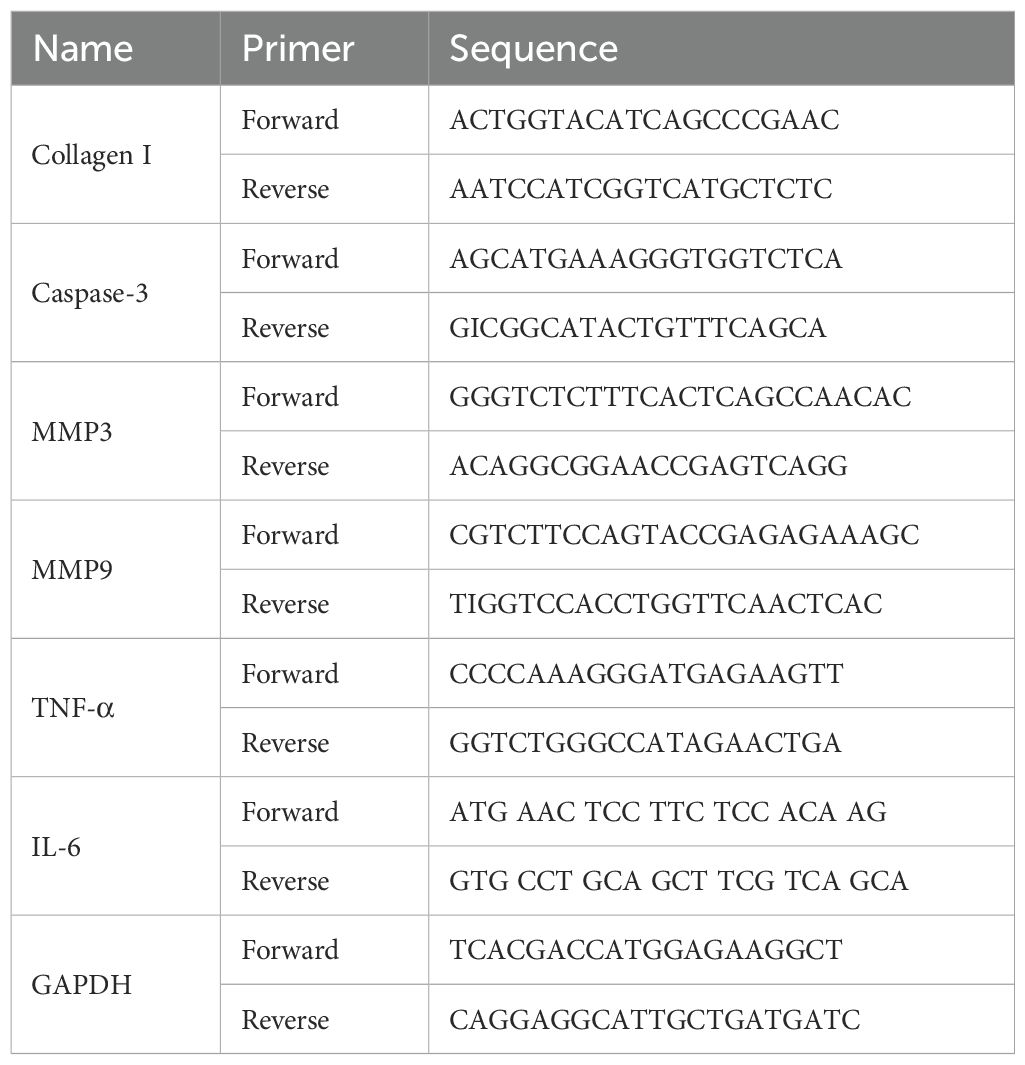
Total RNA was extracted from rat bone tissue using the Spark Jade Science & Technology Co., Ltd. (SPARK) easy Improved Tissue/Cell RNA Kit (Spark Jade, AC0202). RNA reverse transcription was performed using the SPARK script IIRT Plus Kit (Spark Jade, AG0304) according to the manufacturer’s instructions. qPCR reactions, prepared with 2 × SYBR Green qPCR Mix (Spark Jade, AH0104), were run at 94°C for 3 min. Primers for collagen I, IL-6, TNF-α, MMP3, MMP9, and caspase-3 were used for the RT-qPCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the internal reference. The relative expression levels of the target and reference genes were quantified using the 2−ΔΔCT method (Table 1).
2.13 Statistical analysis
All data were analyzed with GraphPad Prism v.8 (GraphPad Software, La Jolla, CA, USA). One-way analysis of variance (ANOVA) was used to detect significant differences between groups. p < 0.05 indicated statistical significance. Figure 1 shows the overall flowchart of the research.
3 Results
3.1 Potential targets of curculigoside against osteoporosis
The chemical molecular formula of curculigoside is C22H26O11, and its molecular structure is shown in Figure 2A. A total of 188 potential targets were obtained by searching the Encyclopaedia of Chinese Medicine database, SwissTargetPrediction, and the Similarity Ensemble Approach database. A total of 4,313 osteoporosis-related targets were retrieved from the DisGeNET and GeneCards databases. By linking the two groups of targets, 91 overlapping targets were identified as valid targets for the treatment of osteoporosis with curculigoside. Finally, a network target map was constructed using Cytoscape (3.7.1) software and Venny (2.1.0) for the 91 valid targets (Figures 2B, C).

Figure 2. Network target diagram and targets common to curculigoside and osteoporosis. (A) Molecular structure of curculigoside. (B) Network target diagram. (C) Venn diagram of shared targets.
3.2 Construction of a protein–protein interaction network
The validated targets of cenobacterial glycosides for osteoporosis were imported into the STRING (11.5) database, set to “Homo sapiens”, and subjected to PPI analysis. The PPI data were imported into Cytoscape software, and topological and clustering analyses were performed using the Cytoscape plug-in cytoHubba and MCODE. The top 10 central genes obtained from the topological analysis were Vascular Endothelial Growth Factor A (VEGFA), GAPDH, Epidermal Growth Factor Receptor (EGFR), CASP3, IL6, Fibroblast Growth Factor 2 (FGF2), MMP9, Kinase Insert Domain Receptor (KDR), SRC, and MMP3. Among these genes, Vascular Endothelial Growth Factor A (VEGFA) acts as an active growth factor in angiogenesis and endothelial cell growth; it induces endothelial cell proliferation, promotes cell migration, and inhibits apoptosis. GAPDH possesses 3-phosphoglyceraldehyde dehydrogenase and nitrosylase activities, which play roles in glycolysis and nuclear function, respectively. FGF2 plays an important role in the regulation of cell survival, cell division, cell differentiation, and cell migration. KDR and SRC play an important role in the regulation of angiogenesis and vascular permeability. MMP9 and MMP3 play an important role in local protein hydrolysis and leukocyte migration in the extracellular matrix, as well as in osteoclast-mediated bone resorption (Figures 3A, B). The results of the clustering analysis are shown in Figure 3C.
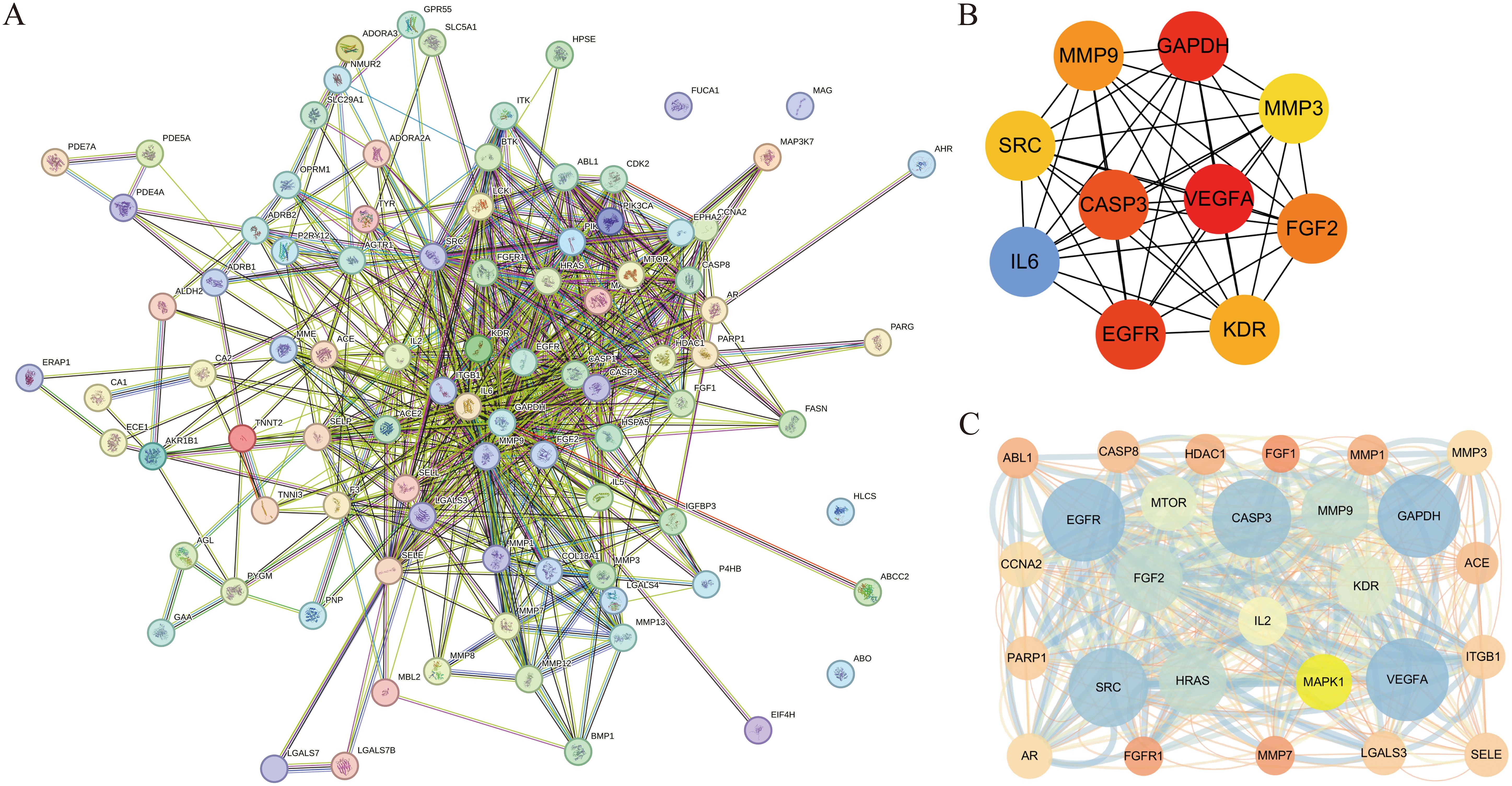
Figure 3. Construction of protein–protein interaction (PPI) network and disease topology analysis. (A) PPI network of overlapping targets. (B) Intersecting genes from the STRING database were constructed using Cytoscape. (C) Clustering analysis.
3.3 KEGG enrichment and GO functional analyses
A total of 69 signaling pathways were identified by GO and KEGG functional enrichment analyses of the validated targets using Metascape software. Using a threshold of p < 0.01 and sorting by ascending p-value, 47 KEGG pathways were obtained, with the top 10 shown in Figure 4. Further analysis of GO functional enrichment results, using the same screening criteria (p < 0.01 and ascending p-value), yielded a total of 514 BPs, 5 CCs, and 31 MFs, as shown in Figure 4. Supplementary File 2 provides the detailed GO analysis results, while Supplementary File 3 presents the specific KEGG analysis outcomes.
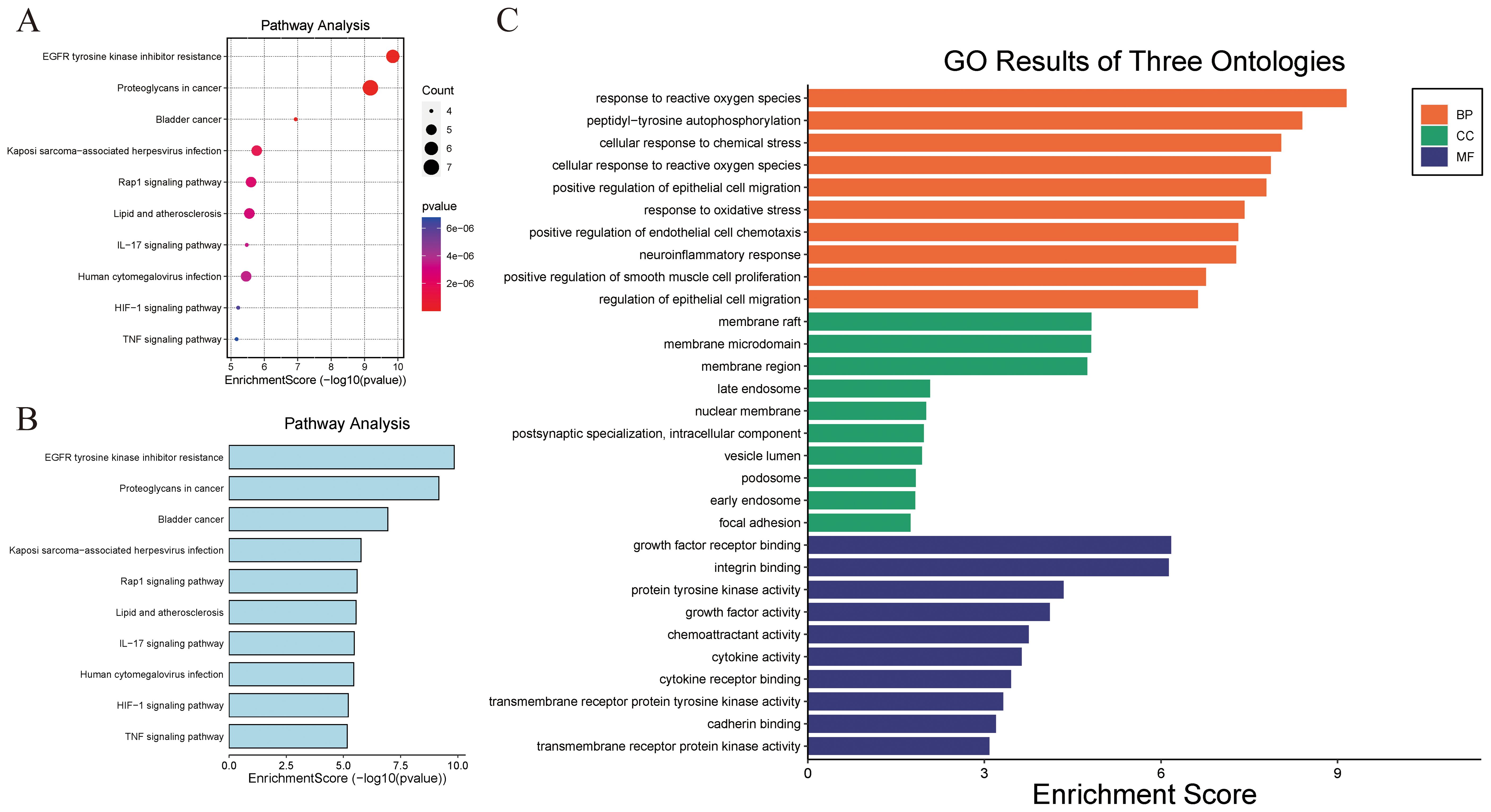
Figure 4. KEGG and GO functional enrichment analyses. (A, B) KEGG pathway enrichment analysis bubble map. (B) GO Ontology analysis.
3.4 Molecular docking
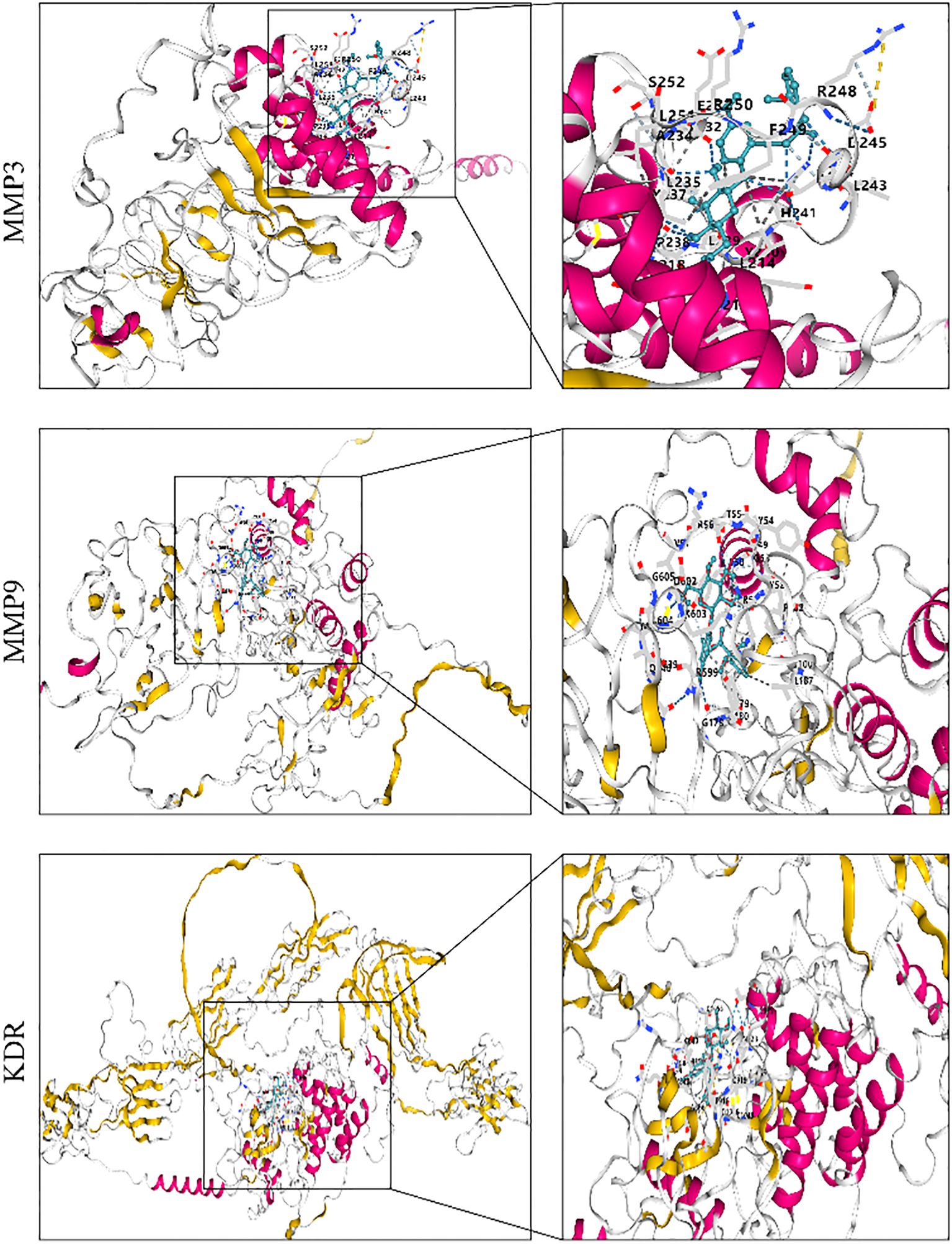
Molecular docking analysis was used to assess the binding ability of curculigoside to key proteins. To evaluate its potential antiosteoporotic effect, molecular docking was performed between curculigoside and the key proteins VEGFA, GAPDH, EGFR, CASP3, IL-6, FGF2, MMP9, KDR, SRC, and MMP3. A lower binding energy indicates higher docking affinity and stronger binding ability. The molecular docking results showed binding energies ranging from − 5.8 to − 8.2. Targets with binding energies ≤ − 6 were considered potentially active, while those ≤ − 8 indicated strong stability and activity. Among them, curculigoside exhibited strong binding affinity to MMP3, MMP9, and KDR with binding energies ≤ − 8, suggesting high binding stability and potential biological activity (41) (as shown in Figure 5).
3.5 Liver and kidney tests show no toxicity
Figure 6 shows that the liver and kidney tissue structures in the sham group, OVX group, and curculigoside group were all normal. No lesions such as steatosis, necrosis, inflammatory cell infiltration, or renal tubular epithelial cell injury were observed, indicating that the treatments did not cause liver or kidney toxicity. In bone tissue, the results of HE staining and SOFG staining confirmed the successful establishment of the osteoporosis model in the OVX group, which exhibited pathological features such as sparse and fractured trabeculae and an enlarged bone marrow cavity. The trabecular bone structure in the curculigoside group was improved compared to that in the OVX group and was closer to the normal state, indicating that curculigoside has a therapeutic effect on osteoporosis.
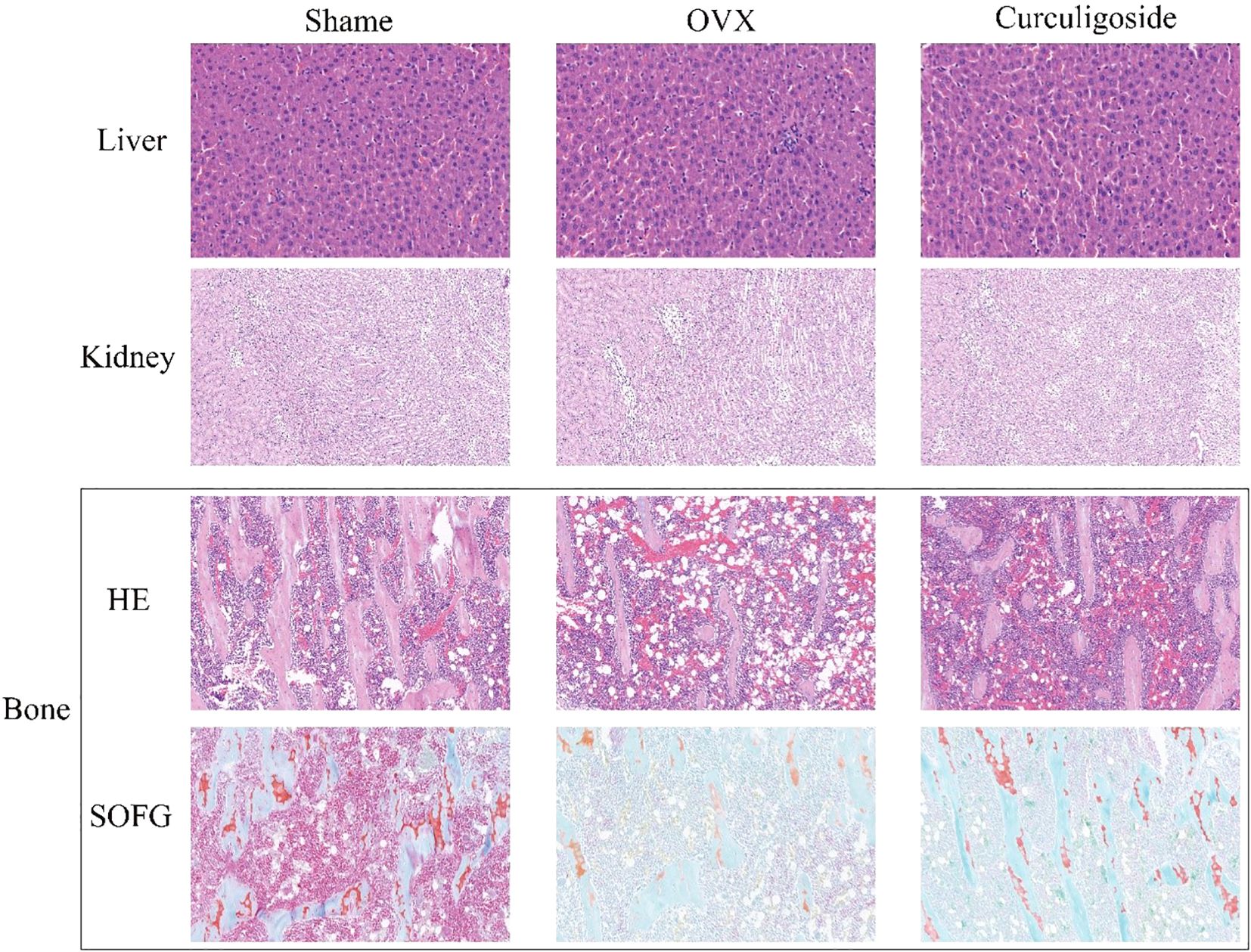
Figure 6. Histopathological sections of the liver, kidney, and bone tissues of rats in each group (H&E and SOFG staining).
3.6 Bone density and CT evaluation
Results from the OVX and curculigoside groups showed a significant decrease in femoral bone mineral density compared to the sham group. The curculigoside group showed a significant increase in BMD compared to the OVX group (p < 0.05). Daily administration of cenicaloside further increased femoral bone density. 2D micro-CT scans revealed the trabecular bone microstructure at the femoral epiphysis after 12 weeks of curculigoside treatment. Quantitative parameters included BMD, BV/TV, BS/BV, Tb.Sp, and Tb.Th. Cenchoside treatment positively affected all CT parameters (Figure 7).
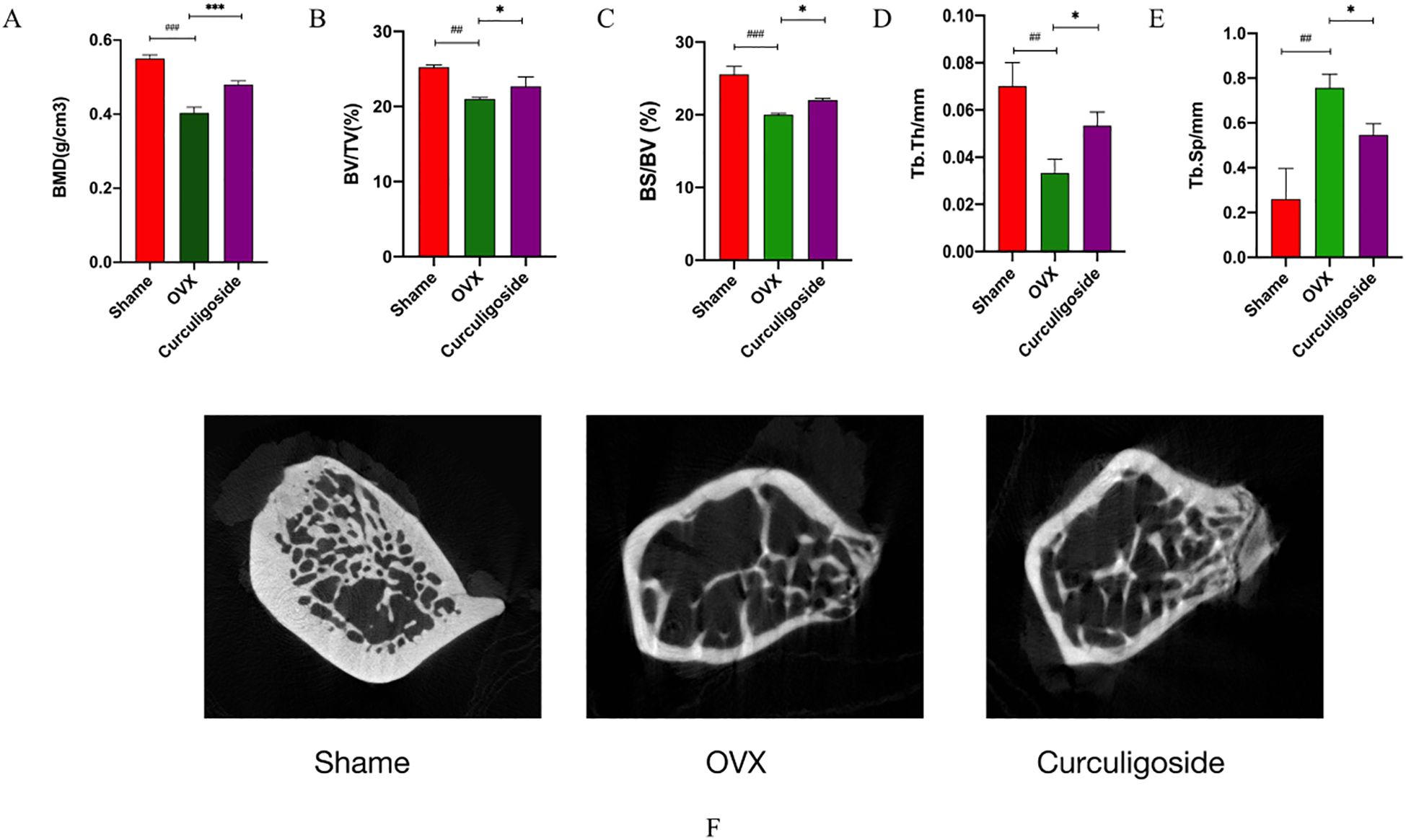
Figure 7. Detection of bone tissue by micro-CT. (A–E) Bone tissue-related index measurements. (F) CT images. n = 3, *p < 0.05; **p < 0.01. #p < 0.05; ##p < 0.01; ###p < 0.01.
3.7 The effect of curculigoside on inflammatory and proapoptotic factors
Collagen I, a major constituent of the extracellular matrix, is sensitive to external stimulation. RT-qPCR was used to detect the messenger RNA (mRNA) expression of collagen I, IL-6, TNF-α, MMP3, MMP9, and caspase-3, while ELISA kits were employed to measure the levels of IL-6, TNF-α, IL-1β, OCN, and IGF-1. The results demonstrated that, in the OVX group, collagen I mRNA expression was significantly downregulated and OCN and IGF-1 levels were reduced. Conversely, the mRNA levels of IL-6, TNF-α, MMP3, MMP9, and caspase-3, as well as the secretion levels of IL-6, TNF-α, and IL-1β, were significantly increased. Treatment with curculigoside reversed these changes by upregulating collagen I mRNA and increasing OCN and IGF-1 levels, while downregulating the mRNA and protein levels of IL-6, TNF-α, MMP3, MMP9, and caspase-3 (Figure 8).
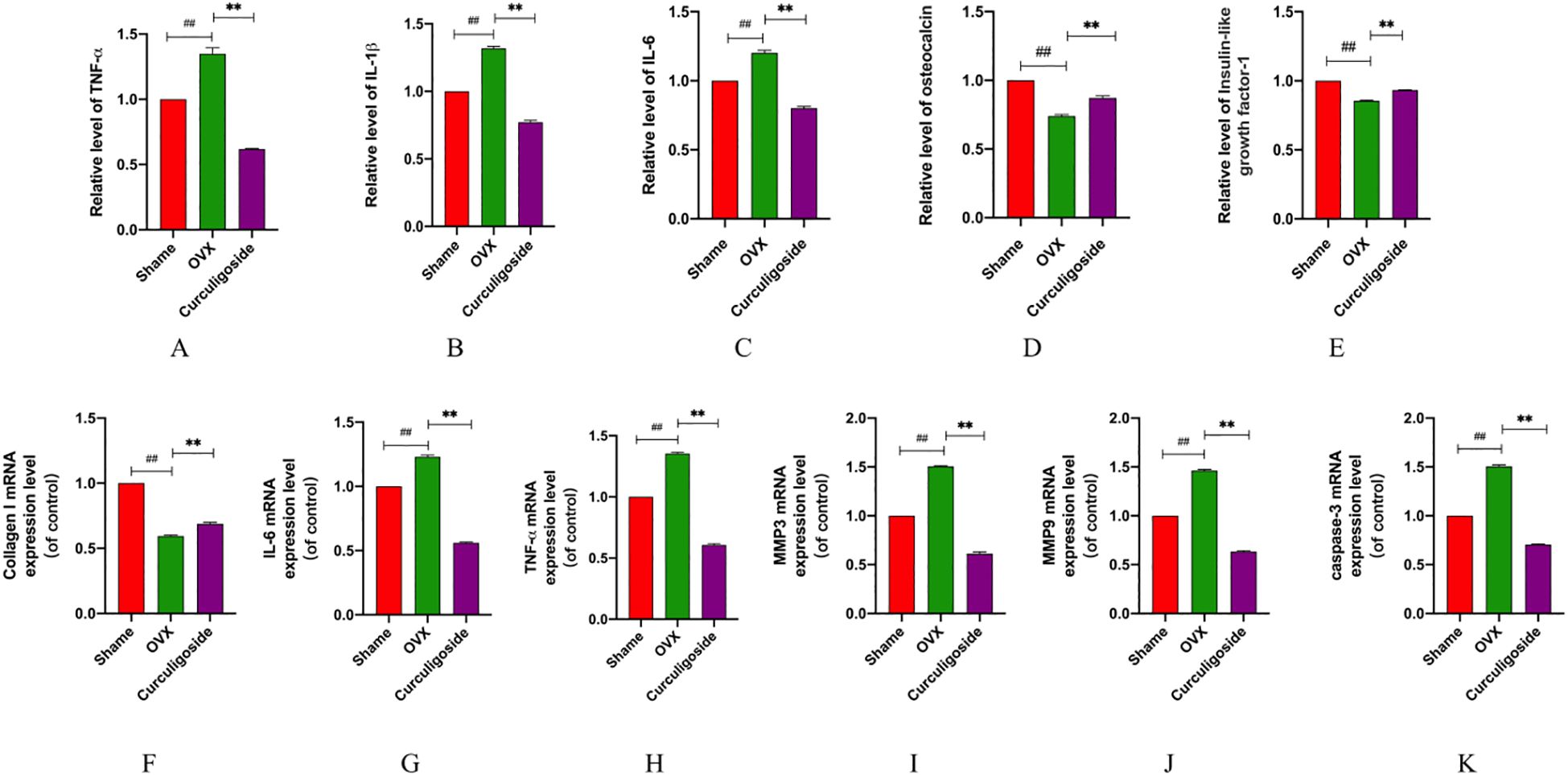
Figure 8. Detection of inflammatory and proapoptotic factors. (A–E) The expression levels of IL-6, IL-1β, TNF-α, OCN, and IGF-1 were measured by ELISA. (F–K) mRNA levels of IL-6, TNF-α, MMP3, MMP9, caspase-3, and collagen (I) n = 3, *p < 0.05; **p < 0.01; ***p < 0.01. #p < 0.05; ##p < 0.01; ###p < 0.01.
4 Discussion
Osteoporosis is a common skeletal metabolic disease characterized primarily by reduced bone mass. In recent years, its prevalence has risen significantly and has increasingly affected younger populations. Consequently, identifying improved treatments has become a major focus of current research. Cyberpharmacology enables comprehensive visualization and analysis of drug chemistry, disease targets, and pathways of action, facilitating the exploration of curculigoside’s potential mechanisms for treating osteoporosis (42, 43). Employing network pharmacology and molecular docking techniques, this study enabled the initial identification of curculigoside’s potential targets and complex molecular mechanisms in osteoporosis.
A Venn diagram of the “curculigoside-target-disease” network was constructed, identifying 166 cross-targets and 10 related signaling pathways. These pathways are primarily involved in the negative regulation of reactive oxygen species, oxidative stress, and cell proliferation and differentiation, through the regulation of the Rap1 signaling pathway, interleukin (IL)-17 signaling pathway, and Hypoxia-Inducible Factor 1 (HIF-1) signaling pathway. The IL-17, HIF-1, and TNF signaling pathways may contribute to curculigoside’s therapeutic effects in osteoporosis. The protein–protein interaction network was analyzed using Cytoscape 3.6.0, identifying 10 core targets. Curculigoside may exert its therapeutic effect on osteoporosis by binding to these key targets. The results showed that curculigoside reduced the mRNA expression of IL-6, TNF-α, and IL-1β; decreased the damage caused by inflammatory factors to osteoblasts; downregulated the expression of MMP3, MMP9, and caspase-3; and upregulated the expression of the collagen I gene. It also reduced the secretion levels of IL-6, IL-1β, and TNF-α; increased the secretion levels of OCN and IGF-1; improved the status of type I collagen; promoted the proliferation of osteoblasts; and slowed the progression of osteoporosis. In this study, the downregulation of IL-6, TNF-α, IL-1β, MMP3, MMP9, and caspase-3 by curculigoside aligns with previous findings that these factors mediate osteoblast injury and extracellular matrix degradation in osteoporosis (44–46). For example, Min et al. demonstrated that IL-17-driven inflammation promotes osteoclastogenesis and osteoblast apoptosis, which is consistent with our observation of reduced proinflammatory cytokine levels. Additionally, the upregulation of collagen I, OCN, and IGF-1 by curculigoside supports its role in enhancing osteoblast function, paralleling the reported mechanisms of VEGFA and FGF2 in promoting bone formation (46). These results collectively suggest that curculigoside may inhibit osteoclast differentiation and promote osteoblast proliferation through multitarget regulation of inflammatory and oxidative stress pathways. We suggest that cynarin may be involved in the regulation of ROS-related biological processes, estrogen signaling, osteoblast apoptosis, and osteoclast differentiation pathways in the treatment of osteoporosis. Both osteoclast and osteoblast differentiation, as well as their proliferation, are closely related to reactive oxygen species (47). ROS can cause severe damage to bone tissue by accelerating bone resorption, which is strongly associated with the upregulation of osteoclast differentiation via the NF-κB and calcium-regulated neurophosphatase pathways (48). Cynarin processes multiple biological activities, such as antioxidant and anti-inflammatory properties, which have been demonstrated in both animal and cellular experiments (26, 27). Liu et al. (49) suggested that cynarin could attenuate oxidative stress and inhibit osteoclastogenesis and MMP9-specific gene expression by modulating the Nrf2/NF-κB signaling pathway in RAW264.7 cells. This is consistent with our KEGG and GO enrichment results, which highlighted the involvement of NF-κB and ROS-related pathways in curculigoside’s mechanism, further validating its antiosteoporotic effects through the regulation of oxidative stress and inflammation. Mitochondrial impairment exacerbates the accumulation of reactive oxygen species, leading to oxidative stress and activation of osteoclast activity, as well as increased expression of MMPs and secretion of inflammatory factors through NF-κB activation. The results of KEGG and GO functional enrichment suggest that curculigoside may treat osteoporosis by inhibiting the overproduction of osteoclastic cytokines and their induction of oxidative damage, primarily through modulation of the Rap1, IL-17, HIF-1, and TNF signaling pathways. This modulation may reduce oxidative damage, restore mitochondrial dysfunction, decrease osteoblast apoptosis, and help maintain the dynamic balance between bone resorption and bone formation.
In this study, micro-CT imaging was employed to assess histopathological changes in the bone tissues of ovariectomized rats, providing both quantitative and qualitative evidence for curculigoside’s effects on bone microarchitecture. The technique helped confirm that curculigoside improved bone density and microstructural parameters (e.g., trabecular thickness and connectivity) (50–52), supporting the study’s overall conclusion on the drug’s antiosteoporotic efficacy. However, it should be noted that micro-CT alone cannot fully characterize cellular and molecular mechanisms. Future studies could combine histomorphological analysis and immunohistochemistry to further validate these findings.
5 Conclusion
This study combined network pharmacology, molecular docking, and in vivo experiments to investigate curculigoside’s mechanism against osteoporosis. Network analysis identified 91 shared targets (e.g., MMP3, MMP9, IL-6, caspase-3) enriched in the Rap1 and TNF signaling pathways. Molecular docking showed strong binding affinity (≤ − 8 kcal/mol) to core targets. In ovariectomized rats, curculigoside improved bone parameters (BMD, BV/TV, Tb.Th↑; Tb.Sp↓) and regulated inflammatory (IL-6, TNF-α↓) and osteogenic (collagen I, IGF-1↑) markers. These results indicate that curculigoside exerts multitarget, multipathway effects on bone remodeling, inflammation, and apoptosis, providing a basis for its clinical use.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by The Experimental Animal Ethics Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CW: Conceptualization, Writing – original draft. WZ: Data curation, Writing – original draft. CL: Formal Analysis, Methodology, Visualization, Writing – original draft. NL: Writing – review & editing. HC: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Shandong Provincial Natural Science Foundation Project (ZR202211240011); the Key Project of the Shandong Gerontological Society (LKJGG2021Z009); the Construction Project of the Inheritance Studio of Famous Old TCM Experts in Shandong Province (Lu Weihan (2019) No. 92, Shandong Provincial Health Commission); and the Shandong Provincial Traditional Chinese Medicine Science and Technology Project Task Book (M20241843).
Acknowledgments
The author sincerely thanks the Experimental Center of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine for its support and guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fendo.2025.1690095.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1549471/full#supplementary-material
References
1. Wang CJ and McCauley LK. Osteoporosis and periodontitis. Curr Osteoporos Rep. (2016) 14:284–91. doi: 10.1007/s11914-016-0330-3, PMID: 27696284
2. Ma J, Lin X, Chen C, Li S, Zhang S, Chen Z, et al. Circulating miR-181c-5p and miR-497-5p Are Potential Biomarkers for Prognosis and Diagnosis of Osteoporosis. J Clin Endocrinol Metab. (2020) 105:dgz300. doi: 10.1210/clinem/dgz300, PMID: 31872255
4. Kobayakawa T, Miyazaki A, Saito M, Suzuki T, Takahashi J, and Nakamura Y. Denosumab versus romosozumab for postmenopausal osteoporosis treatment. Sci Rep. (2021) 11:11801. doi: 10.1038/s41598-021-91248-6, PMID: 34083636
5. Xu X, Jia X, Mo L, Liu C, Zheng L, Yuan Q, et al. Intestinal microbiota: a potential target for the treatment of postmenopausal osteoporosis. Bone Res. (2017) 5:17046. doi: 10.1038/boneres.2017.46, PMID: 28983411
6. Noh JY, Yang Y, and Jung H. Molecular mechanisms and emerging therapeutics for osteoporosis. Int J Mol Sci. (2020) 21:7623. doi: 10.3390/ijms21207623, PMID: 33076329
7. Johnston CB and Dagar M. Osteoporosis in older adults. Med Clin North Am. (2020) 104:873–84. doi: 10.1016/j.mcna.2020.06.004, PMID: 32773051
8. Rossi LMM, Copes RM, Dal Osto LC, Flores C, Comim FV, and Premaor MO. Factors related with osteoporosis treatment in postmenopausal women. Med (Baltimore). (2018) 97:e11524. doi: 10.1097/MD.0000000000011524, PMID: 29995822
9. Choi YJ, Shin HB, Park B, Kim DJ, and Chung YS. Temporal change in the diagnosis and treatment rates of osteoporosis: results from the Korea National Health and Nutrition Examination Survey. Osteoporos Int. (2021) 32:1777–84. doi: 10.1007/s00198-021-05864-z, PMID: 33630130
10. Park E, Kim J, Kim MC, Yeo S, Kim J, Park S, et al. Anti-osteoporotic effects of kukoamine B isolated from lycii radicis cortex extract on osteoblast and osteoclast cells and ovariectomized osteoporosis model mice. Int J Mol Sci. (2019) 20:2784. doi: 10.3390/ijms20112784, PMID: 31174394
11. Yang L, Liu S, Mu S, Guo R, Zhou L, and Fu Q. Paeoniflorin attenuates dexamethasone-induced apoptosis of osteoblast cells and promotes bone formation via regulating AKT/mTOR/autophagy signaling pathway. Evid Based Complement Alternat Med. (2021) 2021:6623464. doi: 10.1155/2021/6623464, PMID: 33880124
12. Ouyang X, Li S, Ding Y, Xin F, and Liu M. Foxf1 gene increases the risk of osteoporosis in rats by inhibiting osteoblast formation and promoting osteoclast differentiation through the upregulation of NF-κB pathway. J Musculoskelet Neuronal Interact. (2022) 22:242–50., PMID: 35642703
13. Chen L, Shi X, Weng SJ, Xie J, Tang JH, Yan DY, et al. Vitamin K2 can rescue the dexamethasone-induced downregulation of osteoblast autophagy and mitophagy thereby restoring osteoblast function in vitro and in vivo. Front Pharmacol. (2020) 11:1209. doi: 10.3389/fphar.2020.01209, PMID: 32848799
14. Yue C, Jin H, Zhang X, Li W, Wang D, Tong P, et al. Aucubin prevents steroid-induced osteoblast apoptosis by enhancing autophagy via AMPK activation. J Cell Mol Med. (2021) 25:10175–84. doi: 10.1111/jcmm.16954, PMID: 34612603
15. Vakili S, Zal F, Mostafavi-Pour Z, Savardashtaki A, and Koohpeyma F. Quercetin and vitamin E alleviate ovariectomy-induced osteoporosis by modulating autophagy and apoptosis in rat bone cells. J Cell Physiol. (2021) 236:3495–509. doi: 10.1002/jcp.30087, PMID: 33030247
16. Rachner TD, Khosla S, and Hofbauer LC. Osteoporosis: now and the future. Lancet. (2011) 377:1276–87. doi: 10.1016/S0140-6736(10)62349-5, PMID: 21450337
17. Kerschan-Schindl K. Prevention and rehabilitation of osteoporosis. Wien Med Wochenschr. (2016) 166:22–7. doi: 10.1007/s10354-015-0417-y, PMID: 26769298
18. Armas LA and Recker RR. Pathophysiology of osteoporosis: new mechanistic insights. Endocrinol Metab Clin North Am. (2012) 41:475–86. doi: 10.1016/j.ecl.2012.04.006, PMID: 22877425
19. Oreira LD, Oliveira ML, Lirani-Galvão AP, Marin-Mio RV, Santos RN, and Lazaretti-Castro M. Physical exercise and osteoporosis: effects of different types of exercises on bone and physical function of postmenopausal women. Arq Bras Endocrinol Metabol. (2014) 58:514–22. doi: 10.1590/0004-2730000003374, PMID: 25166042
20. Fischer V and Haffner-Luntzer M. Interaction between bone and immune cells: Implications for postmenopausal osteoporosis. Semin Cell Dev Biol. (2022) 123:14–21. doi: 10.1016/j.semcdb.2021.05.014, PMID: 34024716
21. Bonjour JP. Protein intake and bone health. Int J Vitam Nutr Res. (2011) 81:134–42. doi: 10.1024/0300-9831/a000063, PMID: 22139564
22. Marozik P, Alekna V, Rudenko E, Tamulaitiene M, Rudenka A, Mastaviciute A, et al. Bone metabolism genes variation and response to bisphosphonate treatment in women with postmenopausal osteoporosis. PloS One. (2019) 14:e0221511. doi: 10.1371/journal.pone.0221511, PMID: 31437227
23. Chung SI, Ryu SN, and Kang MY. Changes in bone metabolism and antioxidant defense systems in menopause-induced rats fed bran extract from dark purple rice (Oryza sativa L. Cv. Superjami). Nutrients. (2021) 13:2926. doi: 10.3390/nu13092926, PMID: 34578804
24. Capozzi A, Scambia G, Migliaccio S, and Lello S. Role of vitamin K2 in bone metabolism: a point of view and a short reappraisal of the literature. Gynecol Endocrinol. (2020) 36:285–8. doi: 10.1080/09513590.2019.1689554, PMID: 31711322
25. Trevisan C, Alessi A, Girotti G, Zanforlini BM, Bertocco A, Mazzochin M, et al. The impact of smoking on bone metabolism, bone mineral density and vertebral fractures in postmenopausal women. J Clin Densitom. (2020) 23:381–9., PMID: 31350204
26. Zhang Q, Zhao L, Shen Y, He Y, Cheng G, Yin M, et al. Curculigoside protects against excess-iron-induced bone loss by attenuating akt-foxO1-dependent oxidative damage to mice and osteoblastic MC3T3-E1 cells. Oxid Med Cell Longev. (2019) 2019:9281481. doi: 10.1155/2019/9281481, PMID: 31949885
27. You WL and Xu ZL. Curculigoside promotes osteogenic differentiation of ADSCs to prevent ovariectomized-induced osteoporosis. J Orthop Surg Res. (2021) 16:279. doi: 10.1186/s13018-021-02389-3, PMID: 33902663
28. Han J, Wan M, Ma Z, Hu C, and Yi H. Prediction of targets of curculigoside A in osteoporosis and rheumatoid arthritis using network pharmacology and experimental verification. Drug Des Devel Ther. (2020) 14:5235–50. doi: 10.2147/DDDT.S282112, PMID: 33273808
29. Gong W, Liu M, Zhang Q, Zhang Q, Wang Y, Zhao Q, et al. Orcinol Glucoside Improves Senile Osteoporosis through Attenuating Oxidative Stress and Autophagy of Osteoclast via Activating Nrf2/Keap1 and mTOR Signaling Pathway. Oxid Med Cell Longev. (2022) 2022:5410377. doi: 10.1155/2022/5410377, PMID: 35585885
30. Wang X, Li Z, Wang Z, Liu H, Cui Y, Liu Y, et al. Incorporation of bone morphogenetic protein-2 and osteoprotegerin in 3D-printed ti6Al4V scaffolds enhances osseointegration under osteoporotic conditions. Front Bioeng Biotechnol. (2021) 9:754205. doi: 10.3389/fbioe.2021.754205, PMID: 34805113
31. An Y, Zhang H, Wang C, Jiao F, Xu H, Wang X, et al. Activation of ROS/MAPKs/NF-κB/NLRP3 and inhibition of efferocytosis in osteoclast-mediated diabetic osteoporosis. FASEB J. (2019) 33:12515–27. doi: 10.1096/fj.201802805RR, PMID: 31461386
32. Qin L, Chen H, Ding X, Guo M, Lang H, Liu J, et al. Utilizing network pharmacology to explore potential mechanisms of YiSui NongJian formula in treating myelodysplastic syndrome. Bioengineered. (2021) 12:2238–52. doi: 10.1080/21655979.2021.1933867, PMID: 34098848
33. Xu L, Zhang J, Wang Y, Zhang Z, Wang F, and Tang X. Uncovering the mechanism of Ge-Gen-Qin-Lian decoction for treating ulcerative colitis based on network pharmacology and molecular docking verification. Biosci Rep. (2021) 41:BSR20203565. doi: 10.1042/BSR20203565, PMID: 33409535
34. UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. (2021) 49:D480–9. doi: 10.1093/nar/gkaa1100, PMID: 33237286
35. Rebhan M, Chalifa-Caspi V, Prilusky J, and Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. (1998) 14:656–64. doi: 10.1093/bioinformatics/14.8.656, PMID: 9789091
36. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303, PMID: 14597658
37. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. (2017) 45:D362–8. doi: 10.1093/nar/gkw937, PMID: 27924014
38. Westbrook J, Feng Z, Jain S, Bhat TN, Thanki N, Ravichandran V, et al. The Protein Data Bank: unifying the archive. Nucleic Acids Res. (2002) 30:245–8. doi: 10.1093/nar/30.1.245, PMID: 11752306
39. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. (2021) 49:D1388–95. doi: 10.1093/nar/gkaa971, PMID: 33151290
40. Wang QJ, Zhang P, Zhang RX, and Li BB. Effect of curculigoside on bone metabolism in osteoporotic rats by regulating lncRNA KCNQ1OT1/miR-214-5p axis. Chin J Osteoporosis. (2024) 30:1261–7. doi: 10.3969/j.issn.1006-7108.2024.09.003
41. Alwan SM. Computational calculations of molecular properties and molecular docking of new and reference cephalosporins on penicillin binding proteins and various β-lactamases. J Pharm Pharmacol. (2016) 4:212–24.
42. Fang T, Liu L, and Liu W. Network pharmacology-based strategy for predicting therapy targets of Tripterygium wilfordii on acute myeloid leukemia. Med (Baltimore). (2020) 99:e23546. doi: 10.1097/MD.0000000000023546, PMID: 33327305
43. Luo P, Wang Y, Zhao C, Guo J, Shi W, Ma H, et al. Bazedoxifene exhibits anti-inflammation and anti-atherosclerotic effects via inhibition of IL-6/IL-6R/STAT3 signaling. Eur J Pharmacol. (2021) 893:173822. doi: 10.1016/j.ejphar.2020.173822, PMID: 33347820
44. Min HK, Choi J, Lee SY, Lee AR, Min BM, Cho ML, et al. Vitronectin-derived bioactive peptide prevents spondyloarthritis by modulating Th17/Treg imbalance in mice with curdlan-induced spondyloarthritis. PloS One. (2022) 17:e0262183. doi: 10.1371/journal.pone.0262183, PMID: 34986165
45. Yamamoto H, Saito M, Goto T, Ueshima K, Ishida M, Hayashi S, et al. Heme oxygenase-1 prevents glucocorticoid and hypoxia-induced apoptosis and necrosis of osteocyte-like cells. Med Mol Morphol. (2019) 52:173–80. doi: 10.1007/s00795-018-00215-0, PMID: 30706191
46. Geng Q, Gao H, Yang R, Guo K, and Miao D. Pyrroloquinoline quinone prevents estrogen deficiency-induced osteoporosis by inhibiting oxidative stress and osteocyte senescence. Int J Biol Sci. (2019) 15:58–68. doi: 10.7150/ijbs.25783, PMID: 30662347
47. Zhu FB, Wang JY, Zhang YL, Quan RF, Yue ZS, Zeng LR, et al. Curculigoside regulates proliferation, differentiation, and pro-inflammatory cytokines levels in dexamethasone-induced rat calvarial osteoblasts. Int J Clin Exp Med. (2015) 8:12337–46., PMID: 26550143
48. Zhu FB, Wang JY, Zhang YL, Quan RF, Yue ZS, Zeng LR, et al. Preventive effects of evodiamine on dexamethasone-induced osteoporosis in zebrafish. BioMed Res Int. (2019) 2019:5859641. doi: 10.1155/2019/5859641, PMID: 30805367
49. Liu M, Liu S, Zhang Q, Fang Y, Yu Y, Zhu L, et al. Curculigoside attenuates oxidative stress and osteoclastogenesis via modulating Nrf2/NF-κB signaling pathway in RAW264.7 cells. J Ethnopharmacol. (2021) 275:114129. doi: 10.1016/j.jep.2021.114129, PMID: 33878416
50. Boutin RD and Lenchik L. Value-added opportunistic CT: insights into osteoporosis and sarcopenia. AJR Am J Roentgenol. (2020) 215:582–94. doi: 10.2214/AJR.20.22874, PMID: 32755187
51. Zhang V, Koa B, Borja AJ, Padmanhabhan S, Bhattaru A, Raynor WY, et al. Diagnosis and monitoring of osteoporosis with total-body 18F-sodium fluoride-PET/CT. PET Clin. (2020) 15:487–96. doi: 10.1016/j.cpet.2020.06.011, PMID: 32768370
Keywords: curculigoside, osteoporosis, network pharmacology, molecular docking, micro CT technology
Citation: Wei C, Zhang W, Lou C, Li N and Cao H (2025) Mechanism of action of curculigoside ameliorating osteoporosis: an analysis based on network pharmacology and experimental validation. Front. Endocrinol. 16:1549471. doi: 10.3389/fendo.2025.1549471
Received: 08 February 2025; Accepted: 24 July 2025;
Published: 21 August 2025; Corrected: 09 September 2025.
Edited by:
Yuankun Zhai, Henan University, ChinaReviewed by:
Jiawen Han, The Central Laboratory of the First Hospital of Jilin University, ChinaPeng Zhang, Guangzhou University of Chinese Medicine, China
Viney Kumar, Albert Einstein College of Medicine, United States
Copyright © 2025 Wei, Zhang, Lou, Li and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Cao, Y2FvaHVpNjNAMTYzLmNvbQ==; Nianhu Li, dGlnZXJsZWUwNzNAMTI2LmNvbQ==
†These authors have contributed equally to this work
‡These authors share first authorship
 Chuanfu Wei1,2‡
Chuanfu Wei1,2‡ Nianhu Li
Nianhu Li Hui Cao
Hui Cao