- Beijing Key Laboratory for Source Control Technology of Water Pollution, Engineering Research Center for Water Pollution Source Control and Ecoremediation, Beijing Forestry University, Beijing, China
The release and fate of estrogens have attracted more and more public attention. Biodegradation is an important method for estrogen removal from the environment. However, few comparative studies concentrated on the degradation of 17β-estradiol (E2) by fungi and bacteria. In this study, the removal efficiencies of E2 by fungi (Candida utilis CU-2) and bacteria (Lactobacillus casei LC-1) were investigated through influencing factors, kinetics, and biodegradation pathways. The results demonstrated that both C. utilis CU-2 and L. casei LC-1 have the same degradation efficiency, and they can effectively degrade E2 (10 μM) with nearly 97% degradation efficiency. However, the biodegradation efficiency of the two strains only reached 20% when E2 was used as a sole carbon source, while it increased to 97% with 1.2 g/L sucrose, glucose, or sodium acetate supply, indicating the occurrence of co-metabolism. In addition, the results indicated that 35°C and 0.6 g/L sucrose favored the degradation. However, the addition of excessive carbon sucrose (10 g/L) significantly inhibited the biodegradation of E2. Besides, the degradation of E2 with ~0–10 g/L sucrose as co-substrate followed the first-order kinetics well. Through intermediate products analysis, 12 degradation products were identified, and they were mainly produced via hydroxylation and methylation, among others, among which C14H22O4 (m/z:[M + H]+ = 255) was detected as the product with the smallest amount of carbon in this study. Based on the detected products and previous studies, five biodegradation pathways were proposed. To our knowledge, there are few reports about the comparisons of E2 removal between fungi and bacteria. Moreover, the results confirmed that the strain CU-2 and the strain LC-1 may have similar degradation characteristics and metabolic mechanisms in the degradation of E2. This study may provide a promising bio-treatment method with low energy consumption for E2 removal from aqueous environments and help in understanding their biodegradation mechanisms.
Introduction
Natural estrogens, including 17β-estradiol (E2), estrone (E1), and estriol (E3), mainly come from human activity and animal excretion (Ying et al., 2002; Zhao et al., 2019). With the growth of population and the demand for livestock products, the release of estrogens to the environment significantly increased in recent years (Wise et al., 2011; Whitman, 2017). Besides, the reports on the exposure of estrogens in the environment increased gradually. Lange et al. (Lange et al., 2002) reported that the annual estrogen contents coming from the excretion of farm animals were ~33–49 tons in the European Union and the US. In addition, estrogens could cause long-term toxicity to organism, especially to the endocrine and reproductive organs (Cevasco et al., 2008). Previous studies reported that E2, as a typical type of estrogens (Li et al., 2019), was of great significance for the growth, maintenance, and contraception of female reproductive tissues (Sacdal et al., 2020), and it could induce a male to produce female characteristics even at very low concentrations (≥1 ng/L) (Peiris et al., 2020), showing its great harm to human and animal physiology and behavior (Young et al., 2002; Adeel et al., 2017). In Europe, intersex fish had been found near sewage treatment plants (Jobling et al., 2005). Besides, the phenomenon of intersex was also found in black basses (Micropterus spp.) in the US (Hinck et al., 2009). Moreover, Lambert et al. (2015) found that the offspring sex ratios of wild frogs in suburban ponds were affected by E2 exposure. Owing to its chemical stability, it was very hard to completely remove E2 in sewage treatment plants (Ting and Praveena, 2017). Therefore, it is of great significance to develop an effective method for E2 removal.
Until now, various treatment processes, such as physical methods, chemical oxidation, and microbial degradation, have been applied in the removal of E2. Physical methods such as activated carbon adsorption and membrane process have high operation cost and high energy consumption (Snyder et al., 2007). Chemical oxidation methods such as chemical treatment and electrochemistry also have the problem of high energy consumption (Klavarioti et al., 2009). While, microbial degradation technology was considered as a promising and effective method because of its advantages, including no secondary pollution, low energy consumption, high efficiency, low cost, etc (Yu et al., 2013). It was reported that many bacteria (Rhodococcus spp., Sphingomonas spp., Pseudomonas spp., etc.) and some fungi showed a high capacity to degrade E2 and convert it to harmless products (Hyungkeun and Kung-Hui, 2010). Fujii et al. (2002) found that Novosphingobium ARI-1 could completely degrade 5 mg/L of E2 in 20 days. Yu et al. (2016) reported that 1 mg/L of E2 could be completely degraded by Rhodococcus spp. DS201 with 1% inoculation quantity in 3 days at 30°C. Xiong et al. also claimed that a 90% removal efficiency of 10 mg/L of E2 could be achieved by the strain SJTH1 with the initial OD600 of 0.05 (Xiong et al., 2020). All these above strains could effectively degrade E2, but it was difficult to compare their degradation efficiencies due to different experimental conditions. Furthermore, co-substrate or co-metabolism also had important effects on the degradation of pollutants. The degradation ratio of ciprofloxacin could be promoted with the presence of glucose and sodium acetate (Xiong et al., 2017). Stenotrophomonas maltophilia DT1 exhibited the highest degradation efficiency for tetracycline when glucose was added compared with when glycerol, lactose, or sucrose was added (Leng et al., 2016). Therefore, it can be inferred that the removal efficiency of E2 was significantly related to the species of strains and experimental conditions.
Khattab et al. (2018) investigated the removal of E2 by two kinds of bacteria, namely Klebsiella and Bacillus, and the degradation efficiencies of the strains of Klebsiella and Bacillus to E2 were 100 and 91.56%, respectively. Li et al. (2018) also investigated the removal efficiencies of E2 by two kinds of bacteria and found that the E2 removal efficiency by Acinetobacter LM1 was higher than that by Pseudomonas LY1. Except for the bacterial strains, three kinds of fungi (Pycnoporus spp. SYBC-L3, Trametes versicolor TV, and Hymenochaete spreta PL-1) were investigated, and the degradation ratios of E2 were 78.4, 62.7, and 14.5%, respectively (Liu et al., 2016). Although these studies have enhanced the comparability of E2 degradation efficiency, most of them only focus on single fungi or bacteria, and there are few studies on the comparison of E2 removal by fungi and bacteria. Thus, it is desirable to explore the difference and mechanism of E2 removal by fungi and bacteria.
Aside from these research studies, the degradation products and pathways of E2 have also been concerned. In the degradation process of E2, E1 was identified as the first step in previous studies (Shi et al., 2004). While other E2 conversion products have also been found in recent years, Kurisu et al. (2010) observed a new intermediate metabolite, named 4-hydroxyestradiol, and they affirmed that it was converted from the hydroxylation of the C-4 site on E2 rather than first converting E2 to E1. Besides, the formation of estratetranol (E0) also confirmed the fact that it was formed by the dehydration of the C-17 site on E2 (Nakai et al., 2011). Furthermore, another new degradation product X1 contained lactones in the D ring, which was proposed in a previous study (Yu et al., 2013), and X1 was considered to be further degraded by entering the tricarboxylic acid cycle. The results provided a new idea for the degradation pathway of E2. According to the previous studies, although some biodegradation intermediate metabolites of E2 have been reported, the degradation mechanism of E2 is not clear.
In this study, Lactobacillus casei LC-1 and Candida utilis CU-2 were selected to be the representative strains of bacteria and fungi for E2 removal. The degradation efficiency and mechanism of E2 by the two strains were explored. Furthermore, the differences in the following aspects were mainly compared: (i) the effects of carbon source substrate, sucrose concentration, and temperature on the biodegradation of E2; (ii) the biodegradation kinetics of E2 under different sucrose concentrations; and (iii) the intermediate products analysis and the degradation pathways of E2. This research may provide a new idea for E2 degradation through the comparative study of fungi and bacteria and elaborate their degradation mechanisms via the degradation pathways of E2.
Materials and Methods
Chemicals and Materials
17β-estradiol (purity ≥ 98%) was purchased from Beijing InnoChem Science & Technology Co., Ltd. (Beijing, China). The concentration of E2 stock solution was 2 MM. It was prepared by dissolving 54.4 mg of E2 in 150 ml of methanol solution. Sucrose (purity 99%), glucose (purity 99%), and sodium acetate (purity 99%) were obtained from Sinopharm Chemical Reagent Beijing Co., Ltd. (Beijing, China). Methanol (chromatographic grade) and acetonitrile (chromatographic grade) were purchased from J.T. Baker, Inc. (Phillipsburg, NJ, USA). The Milli-Q ultrapure water (18.2 MΩ) used in this study was provided by Beijing Wahaha Company (Beijing, China).
A mineral medium (MM) containing 1.5 g/L K2HPO4, 0.5 g/L KH2PO4, 0.2 g/L MgSO4, and 1.5 g/L NH4Cl was used for the enrichment of estrogen-degrading strains (Cao et al., 2019). A Luria–Bertani medium (LB) containing 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl was used for the cultivation of the strain CU-2, and strain LC-1 was cultivated with a minimal salt medium (MSM), which was composed of 10 g/L casein peptone, 10 g/L beef extract, 5 g/L yeast extract, 5 g/L glucose, 5 g/L sodium acetate, 2 g/L diamine citrate, 1 g/L Tween 80, 2 g/L K2HPO4, 0.2 g/L MgSO4·7H2O, 0.05 g/L MnSO4·H2O, and 20 g/L CaCO3. A solid medium was prepared by adding 15 g/L agar to the medium above.
Strain Sources and Cultivation Conditions
The strains were purchased from the China General Microbiological Culture Collection Center (CGMCC), and they were L. casei CGMCC 1.29 (LC-1) and C. utilis CGMCC 2.2878 (CU-2), respectively.
The strains were activated by the solid medium and cultured at 25°C. After about 2 days of cultivation, colonies were obtained, and then, a single colony was picked and inoculated into liquid medium at 30°C for 36 h to prepare for the next experiment. The strain CU-2 and strain LC-1 were enriched in LB and MSM, respectively.
Morphological Observation of Strains
The activated strains were observed by biochemical methods. The cells cultured for 24 h were treated with Gram staining, and the Gram reaction was observed with an optical microscope (Shanghai Yongheng Optical Instrument Manufacturing Co., Ltd., XSP-63XD, Shanghai, China).
Biodegradation Experiments
The biodegradation experiments were performed with strain CU-2 and strain LC-1 in 150-ml Erlenmeyer flasks containing 75 ml MM. All experiments were performed in a shaker with a speed of 150 rpm, and three parallel experiments were conducted for each group. C. utilis CU-2 and L. casei LC-1 reached an exponential growth phase in liquid LB medium and liquid MSM for about 24 h, and then, cells were harvested by centrifugation at 4,500 rpm for 5 min under ambient temperature. In this step, the strain was washed twice by sterilized 0.9% NaCl solution (pH = 7.0) and then resuspended in 0.9% NaCl solution, and the concentration of cells in the strain were adjusted to optical density (OD) 600 = 0.8 (Pan et al., 2017). Finally, the treated bacterial solution was added to 75 ml MM (2%, v/v) in a 150-ml Erlenmeyer flask. Based on preliminary experimental results, three influencing factors, namely, carbon substrate (glucose, sucrose, and sodium acetate, all at 1.2 g/L), sucrose concentration (0.05–10 g/L) and temperature (15, 25, 35, and 40°C), were designed to explore their effects on E2 degradation by the two strains. For the earlier experiments, the initial E2 concentration of each group was 10 μM, and the blank control group without strain was added in all series of experiments except the temperature series. In addition, liquid samples were collected daily for 6 days and filtered through a 0.22-μm nylon filter before further analysis.
The first-order kinetics (Lu et al., 2019) was used in this study to evaluate the biodegradation rate of E2 under different sucrose concentrations.
Analytical Methods
Bacterial Growth
Cell density was measured by an UV-visible spectrophotometer (Thermo evolution 201, Thermo Technology Co., Ltd., Chachoengsao, Thailand) at an OD of 600 nm to assess bacterial growth (Pan et al., 2017; Moreira et al., 2018).
Quantitative Analysis of E2
To measure E2 concentration, 1.5 ml liquid sample was drawn with a 2-ml syringe and passed through a 0.22-μm filter to eliminate impurities, and it was finally preserved in brown flaskets at 4°C. Stored samples were analyzed by high-performance liquid chromatography (HPLC, Agilent Technologies, Inc., Santa Clara, CA, USA). A column C18 (4.6 × 75 mm, 5 μm, Agilent Technologies, Inc., Santa Clara, CA, USA) was operated with the mobile phase consisting of 40% water and 60% acetonitrile at a flow rate of 0.3 ml/min. The total analysis time of each sample was 5 min, and the injection volume was 90 μl. In addition, the instrument was operated at a column temperature of 35°C, and the detection wavelength was 279 nm.
Identification of Biodegradation Products of E2
The detection and identification of the biodegradation products of E2 were carried out by liquid chromatography tandem mass spectrometry (LC-MS/MS) equipped with an electrospray ionization mode (ESI). The ESI was used in a positive ion source (ESI+), and the mass spectrums were operated in a full-scan mode (100–1,500 m/z). The samples were pretreated before determination. Using a solid phase extraction cartridge, 50-ml sample solutions were extracted and eluted with 5 ml of methanol for concentration. Chromatography was operated using a Waters Acquity UPLC BEH C18 (2.1 × 100 mm, 1.7 μm), and the flow rate of the mobile phase was 0.3 ml/min. Additional details are shown in Supplementary Text 1.
Results and Discussion
Morphological Characteristics of Strains
As shown in Figure 1, both L. casei LC-1 and C. utilis CU-2 were gram-positive. The strain LC-1 was rod-shaped and the strain CU-2 was elliptical.
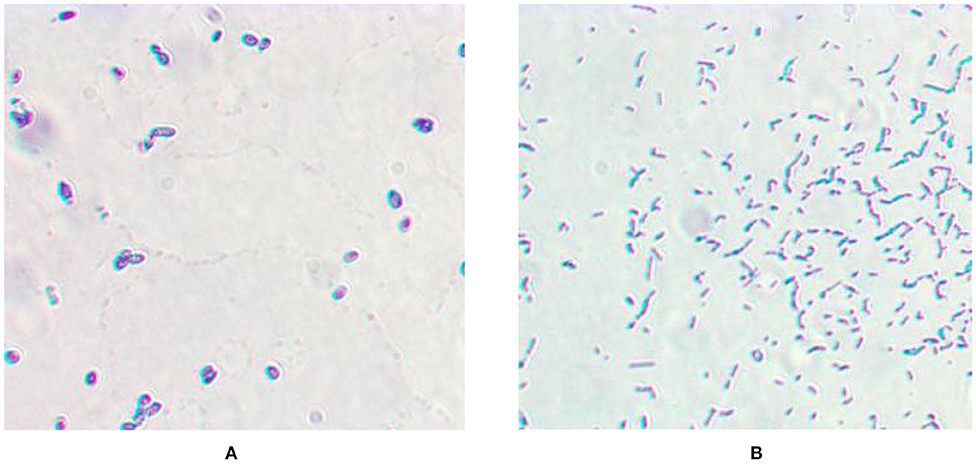
Figure 1. Microscopic morphology. (A) Gram staining of Candida utilis CU-2; (B) Gram staining of Lactobacillus casei LC-1.
Biodegradation of E2 by C. utilis CU-2 and L. casei LC-1
Effects of Different Carbon Substrates on E2 Biodegradation
The effects of the addition of substrates on E2 biodegradation by L. casei LC-1 and C. utilis CU-2 were shown in Figure 1, and the experiments were carried out with three single carbon sources, namely, sucrose, glucose, or sodium acetate, with the concentration of 1.2 g/L. The experiments were conducted in MM with 10 μM of E2. The results showed that all groups with the supply of carbon sources exhibited higher degradation efficiency than the control group (without the addition of carbon sources) (Figures 2A,C). Consistent with E2 degradation, the cell growth of the two strains was also more rapid than the control group (Figures 2B,D). The addition of carbon sources promoted cell growth and E2 degradation. This phenomenon could be explained by the occurrence of biological activity and co-metabolism (Dawas-Massalha et al., 2014). C. utilis CU-2, as shown in Figure 1A, could eventually remove about 97% of E2 in 6 days with supplemented substrates, indicating that it was a highly effective strain for E2 degradation. Besides, 92% of E2 could be removed in 2 days by C. utilis CU-2 with sucrose addition. However, the removal of E2 under glucose and sodium acetate supplement could only reach 84 and 28% in 2 days. The results indicated that sucrose addition resulted in higher removal efficiency of E2 than the other two carbon sources. This could be explained by the fact that sucrose was more easily utilized by the strain than the other carbon sources (Chen et al., 2013). For L. casei LC-1, as shown in Figures 2C,D, the degradation efficiencies of E2 were 94, 92, and 39%, respectively, after adding sucrose, glucose, or sodium acetate in 2 days, which were higher than those of C. utilis CU-2. This difference in E2 degradation efficiencies may be related to the difference in the growth of the two strains, that is, the cell concentration of L. casei LC-1 was higher than that of C. utilis CU-2 in the early stage. This could be accounted to that high biological activity was beneficial to the degradation of E2. While, over time, C. utilis CU-2 and L. casei LC-1 could basically achieve equal biomass at the later stage in Figures 2B,D, and the final removal efficiency of E2 by L. casei LC-1 could also reach about 97% with the increase of reaction time under three different carbon sources.
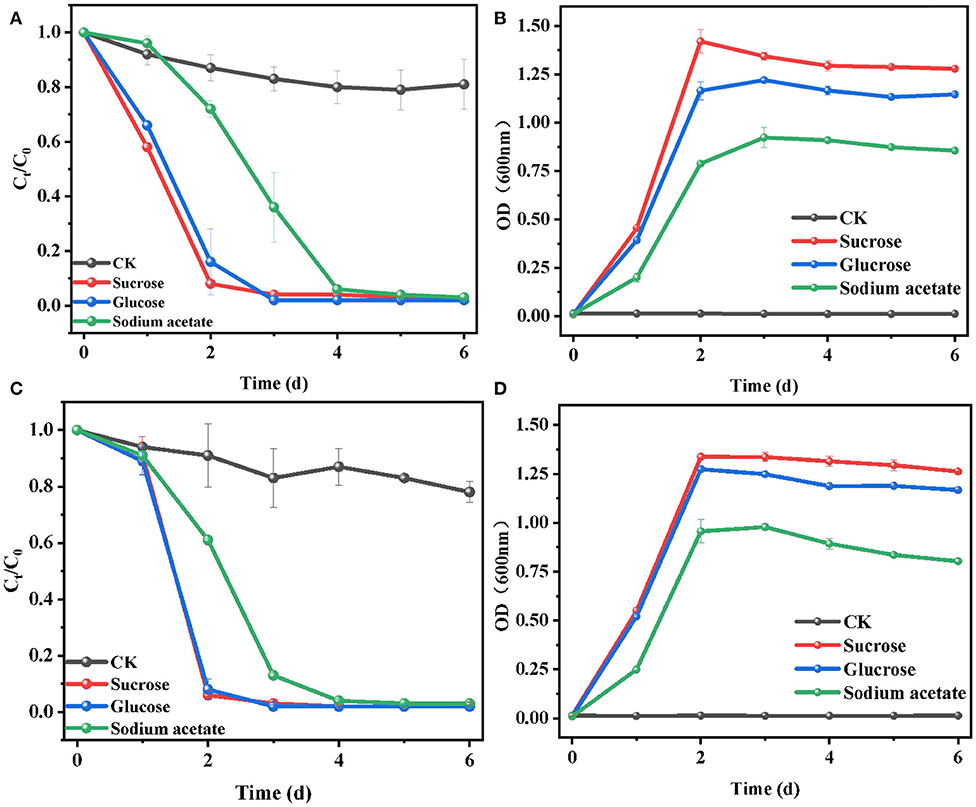
Figure 2. Effects of sucrose, glucose, or sodium acetate on the biodegradation of E2. (A) Degradation efficiency of E2 by C. utilis CU-2; (B) cell growth of C. utilis CU-2, measured with optical density (OD) at λ = 600 nm; (C) degradation efficiency of E2 by L. casei LC-1; (D) cell growth of L. casei LC-1, measured with OD at λ = 600 nm. The concentrations of all carbon sources were 1.2 g/L, and CK represented the control group without carbon source added ([E2]0 = 10 μM, the initial pH = 7.0 ± 0.2).
In summary, these results indicated that the addition of carbon substrate (sucrose, glucose, or sodium acetate) could accelerate cell growth and promote E2 removal compared with the control group, which was due to co-metabolism. Moreover, when sucrose was used as carbon source, the cell growth rate and the E2 biodegradation efficiency were faster than that with glucose or sodium acetate as a carbon source. Thus, sucrose was selected as the optimal carbon source and was used for the following experiments.
Effects of Different Sucrose Concentration on E2 Biodegradation
The effect of sucrose concentration (~0.05–10 g/L) on E2 removal was investigated, and the results are shown in Figure 3. It can be seen that the biodegradation efficiency of E2 improved with the increase of sucrose concentration (~0.05–1.2 g/L), while it decreased when the sucrose concentration increased to 10 g/L. For cell growth (Figures 3B,D), all sucrose added at different concentrations improved the growth of the two strains to different extents. Moreover, cell growth was accelerated with the increase of sucrose concentration (~0.05–10 g/L). To some extent, cell growth may explain the increasing removal efficiency of E2, V:that is, the enhancement of biological activity in high cell concentration accelerated the biodegradation to a certain range (Ziagova and Liakopoulou-Kyriakides, 2007). Meanwhile, the degradation of E2 (Figures 3A,C) was slowly removed with the addition of 0.05 g/L sucrose. When the sucrose concentration ranged from 0.6 to 1.2 g/L, over 95% of E2 was rapidly degraded in 2 days. However, when the initial concentration of sucrose was increased to 10 g/L, the removal efficiency of E2 significantly decreased to about 40% in 2 days, and the maximum removal efficiency of E2 appeared until the sixth day. The results suggested that increasing sucrose concentration promoted cell growth, and the suitable concentration of sucrose was beneficial to the degradation of E2. Too high or too low carbon source concentration would affect the carbon-to-nitrogen ratio, leading to weak biodegradation (Dawas-Massalha et al., 2014). Similar phenomena have been reported in the degradation of some other contaminants by microorganisms. Min et al. (2018) reported that 5 g/L succinate supplement had a lower removal efficiency compared with 0.5 g/L succinate in the degradation of 2,6-dibromo-4-nitrophenol by Cupriavidus sp. strain CNP-8. Pan et al. (2017) also proposed that suitable sodium acetate concentration accelerated the biodegradation by Thermus sp. in the degradation of ciprofloxacin. Therefore, it is necessary to select the appropriate concentration of carbon source for the removal of pollutants.
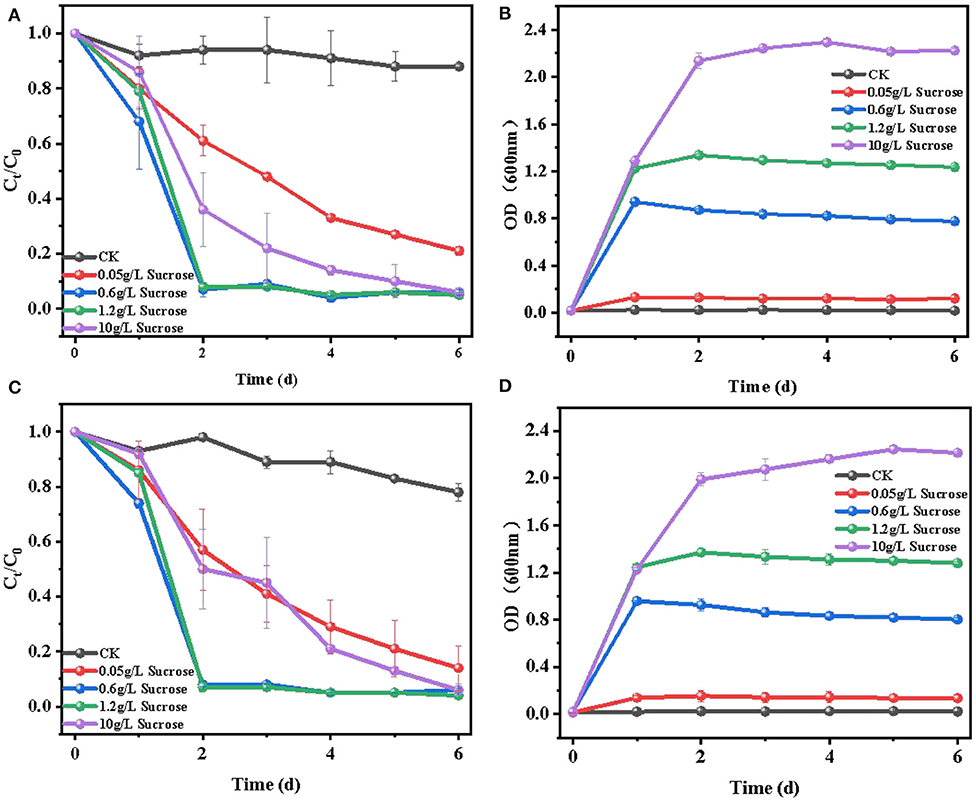
Figure 3. Effects of 0.05–10 g/L sucrose concentration on the degradation of E2: (A) degradation efficiency of E2 by C. utilis CU-2; (B) cell growth of C. utilis CU-2, measured with OD at λ = 600 nm; (C) degradation efficiency of E2 by L. casei LC-1; (D) cell growth of L. casei LC-1, measured with OD at λ = 600 nm. CK represented the control group without carbon source added ([E2]0 = 10 μM, the initial pH = 7.0 ± 0.2).
In conclusion, there was no significant difference in E2 degradation between C. utilis CU-2 and L. casei LC-1 at different sucrose concentrations. In addition, the results also revealed that the suitable sucrose concentration could result in a rapid degradation of E2. Simultaneously, considering the degradation efficiency of E2 and cell growth, the optimal concentration of sucrose was 0.6 g/L for further experiments.
Effects of Temperature on E2 Biodegradation
In the biodegradation process, temperature had a great influence on the growth of microorganisms and the activity of some important enzymes. Choosing an optimal culture temperature was a key environmental factor for E2 removal. To investigate the influence of temperature on E2 biodegradation and the growth of C. utilis CU-2 or L. casei LC-1, a series of experiments were carried out at 15, 25, 35, and 40°C. The results, as shown in Figure 3, demonstrated that E2 degradation was positively influenced by the increase of temperature in the range of 15–35°C, while the biodegradation efficiency of E2 significantly decreased at 40°C. For C. utilis CU-2 (Figures 4A,B), when the temperature ranged from 15 to 35°C, the biodegradation of E2 and the bacterial growth rate presented an upward trend with the increasing temperature. Furthermore, the fastest degradation efficiency within 1 day and the most rapid cell growth within 12 h was observed at 35°C. Meanwhile, as the temperature was raised to 40°C, about 90% removal efficiency of E2 was not achieved until the fourth day, and the maximum cell concentration was 0.34 (OD600), which was about half of that (OD600 = 0.81) at other temperatures. It was well-known that each microorganism had an optimal temperature range. On the one hand, too high or too low temperatures could reduce the growth and reproduction rates of microorganisms, and it also reduced the number of effective microorganisms. On the other hand, although denaturation of proteins was not caused at 40°C, the proteins in microbial cells could not maintain structural integrity and catalytic function. Meanwhile, when the temperature was too low, the growth of bacteria was slow and the enzyme activity was also inhibited (Jagdale and Gordon, 1997). Overall, it is generally stated that each microorganism had an appropriate temperature range, and the results also indicated that the optimum temperature of C. utilis CU-2 for E2 degradation was 35°C. As shown in Figures 4C,D, the optimum temperature of L. casei LC-1 was also 35°C. However, unlike C. utilis CU-2, L. casei LC-1 was more sensitive at 40°C. The cell growth of L. casei LC-1 was lower than that of C. utilis CU-2 at 40°C. At the same time, the maximum degradation efficiency of E2 by C. utilis CU-2 reached 92% within 4 days, while that of L. casei LC-1 was only 42%. Overall, L. casei LC-1 was similar to C. utilis CU-2 in cell growth and E2 biodegradation, and the optimal temperature for E2 biodegradation of the two strains was 35°C.
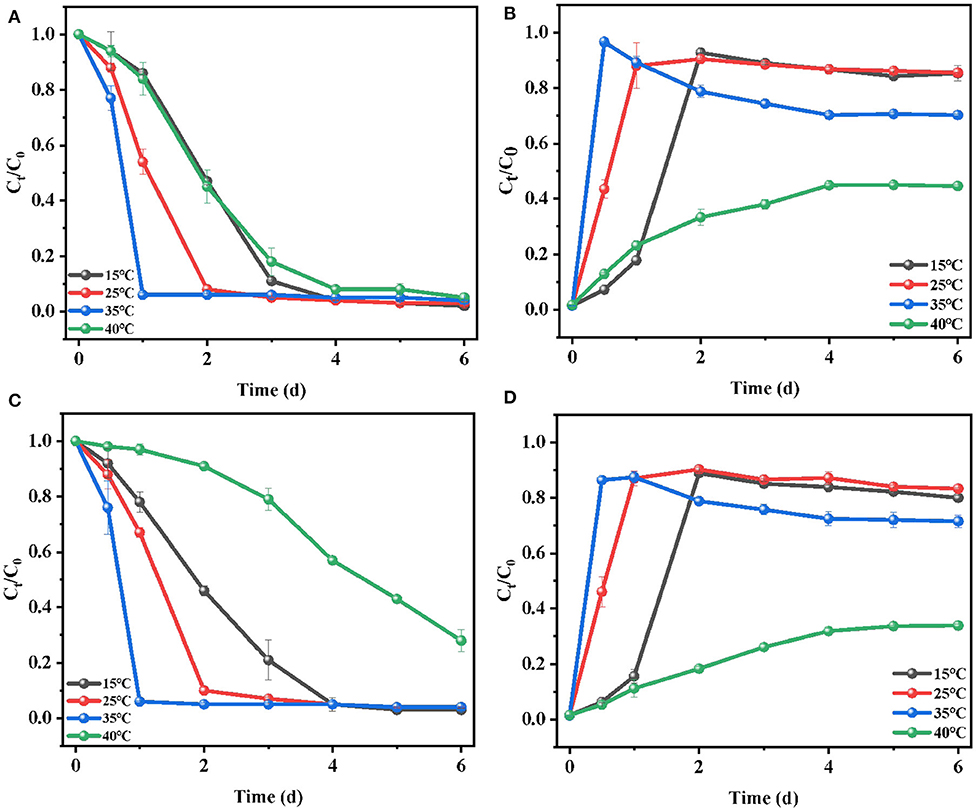
Figure 4. Effects of temperature on the biodegradation of E2: (A) degradation efficiency of E2 by C. utilis CU-2; (B) cell growth of C. utilis CU-2, measured with OD at λ = 600 nm; (C) degradation efficiency of E2 by L. casei LC-1; (D) cell growth of L. casei LC-1, measured with OD at λ = 600 nm. The concentration of sucrose addition was at 0.6 g/L, ([E2]0 = 10 μM, the initial pH = 7.0 ± 0.2).
Kinetic Modeling of E2 Biodegradation at Different Sucrose Concentrations
The first-order kinetics was usually used to fit the biodegradation of E2 (10 mM) at different sucrose concentrations. As shown in Table 1, the kinetics of E2 biodegradation at five concentration sucrose gradients was evaluated. The R2 values of C. utilis CU-2 and L. casei LC-1 were ~0.77–0.99 and 0.85–0.96, respectively, indicating that the removal of E2 at different sucrose concentrations could fit the first-order kinetic models well. During the biodegradation process of E2 by C. utilis CU-2, the k-values calculated at different concentrations of sucrose (0–0.6 g/L) were 0.03 d−1, 0.27 d−1, and 0.82 d−1, showing a gradual increasing degradation trend. However, the removal rate constants of 1.2 and 10 g/L of sucrose supplement decreased, which was consistent with the previous statement (Figure 2). Similarly, for L. casei LC-1, the removal rate constants at the sucrose concentrations ranging from 0 to 0.6 g/L were 0.06 d−1, 0.28 d−1, and 0.78 d−1, respectively, and the k-value of 10 g/L sucrose decreased to 0.37 d−1, also confirming the above results. That is, when the concentrations of sucrose exceeded a certain range, it would inhibit the degradation of E2. Besides, the maximum rate constants of the two strains were 0.82 d−1 and 0.74 d−1, respectively. They all occurred at the condition of 0.6 g/L sucrose addition. Thus, it was also reconfirmed that 0.6 g/L was the optimal concentration of sucrose. On the other hand, the k-value of sucrose group was higher than that of control group, which reflected that sucrose as co-substrate accelerated cell growth and promoted the biodegradation of E2.
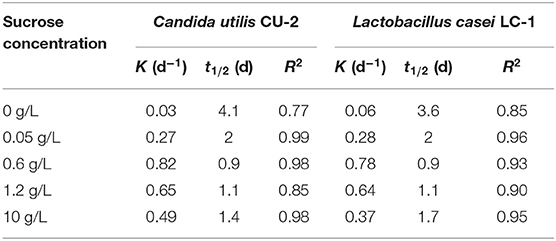
Table 1. First-order kinetic rate constant (k) and half-life (t1/2) for E2 degradation by Candida utilis CU-2 or Lactobacillus casei LC-1.
Biodegradation Products and Metabolic Pathways
The degradation intermediates of E2 by C. utilis CU-2 and L. casei LC-1 were detected by LC-MS/MS, and the degradation pathways were speculated based on accurate MS. In the E2 biodegradation, 12 intermediate products were identified (Supplementary Table 1). According to the detected products and the previously reported degradation pathways, five degradation pathways of E2 were proposed. For E2 degradation intermediates, P1 (C18H24O3, m/z:[M + H]+ = 289), P2 (E0:C18H22O, m/z:[M + H]+ = 255), and P3 (E1:C18H22O, m/z:[M + H]+ = 271) were primary products, and P4 (C18H22O3, m/z:[M + H]+ = 287)—P12 (C19H26O3, m/z:[M + H]+ = 304) were secondary products. It was indicated that P4–P11 might be generated from the further degradation of P3. The formation of these products was mainly through hydroxylation, oxidation, and ring cleavage. All intermediate products were detected in the metabolic fluids of C. utilis CU-2 and L. casei LC-1, indicating that the functions of degrading enzymes produced by C. utilis CU-2 and L. casei LC-1 may be similar. This needs further analysis and verification.
On the basis of the LC-MS/MS results, the biodegradation pathway of E2 was proposed (Figure 5). Specifically, P3, emerging at the time of 16.25 min, contained a mass ion of m/z 271. Its molecular formula was calculated as C18H22O2, which was determined to be E1 (estrone). In addition, E1 was formed by the dehydrogenation process at the C-17 position of the D ring of E2, and it was suggested that the formation of E1 was caused by 17β-hydroxysteroid dehydrogenases (Xiong et al., 2020). In previous studies, E1 was generally regarded as the initial step of estrogen degradation (Shi et al., 2004). However, new degradation pathways have been found in recent years, which was not degraded to E1 first. This study also reconfirmed this discovery, and the new pathways were illustrated in pathway I and pathway II. In pathway I, P2 (E0 m/z:[M + H]+ = 255) was formed by dehydration at the C-17 position of the D ring of E2, which was also reported by Nakai et al. (2011). In pathway II, P1 (C18H24O3, m/z:[M + H]+ = 289) was inferred to be formed by the hydroxylation of E2 (Wang et al., 2019). In addition, the hydroxyl in A ring of P1 was further methylated to form P12 (m/z:[M + H]+ = 303). However, in other three pathways, the products were transformed on the basis of E1. In pathway III, the chemical composition of P4 was calculated to be C18H22O3, and its structural formula was determined by MS (Supplementary Table 1). P4 contained an ether bond, which could be inferred to be formed by the oxidation of E1 saturated ring. This product was also reported in the E2 degradation by microalgaes (Wang et al., 2019). In addition, P4 was further transformed to P5 (m/z:[M + H]+ = 305) by the cleavage of D-ring, and P5 (m/z:[M + H]+ = 305) was further hydroxylated to form P6 (m/z:[M + H]+ = 321) (Wang et al., 2019). In pathway IV, P7 (m/z:[M + H]+ = 288) was identified as 4OH-E1, which was formed by hydroxylation of E1 at C-4 (Yu et al., 2013). Besides, further oxidation between C-13 and C-17 of D-ring on P7 resulted in the formation of degradation intermediate of P8 (m/z:[M + H]+ = 303) (Wang et al., 2019). Another degradation product, P9 (m/z:[M + H]+ = 237), was detected in MS, but its intensity was very low. P9 (C14H20O3) was further transformed to P10 (C14H22O4) through ring cleavage. Both of these intermediate products were proposed by Wang et al. (2019). Furthermore, in pathway IV, P10 was the degradation product with the smallest amount of carbon in molecular mass. Some studies had also found that E2 was degraded into small molecules (Ma and Yates, 2018), but such findings were rare. The detection of P10 also indicated the possibility of further biodegradation or even the possibility of E2 being completely degraded into carbon dioxide and water (Yu et al., 2013), but this needs to be confirmed by further research. In pathway V, P11 (m/z:[M + H]+ = 237) was calculated as C18H20O2, and a double bond was formed by dehydrogenation on the C-ring of E1 (C18H22O2).
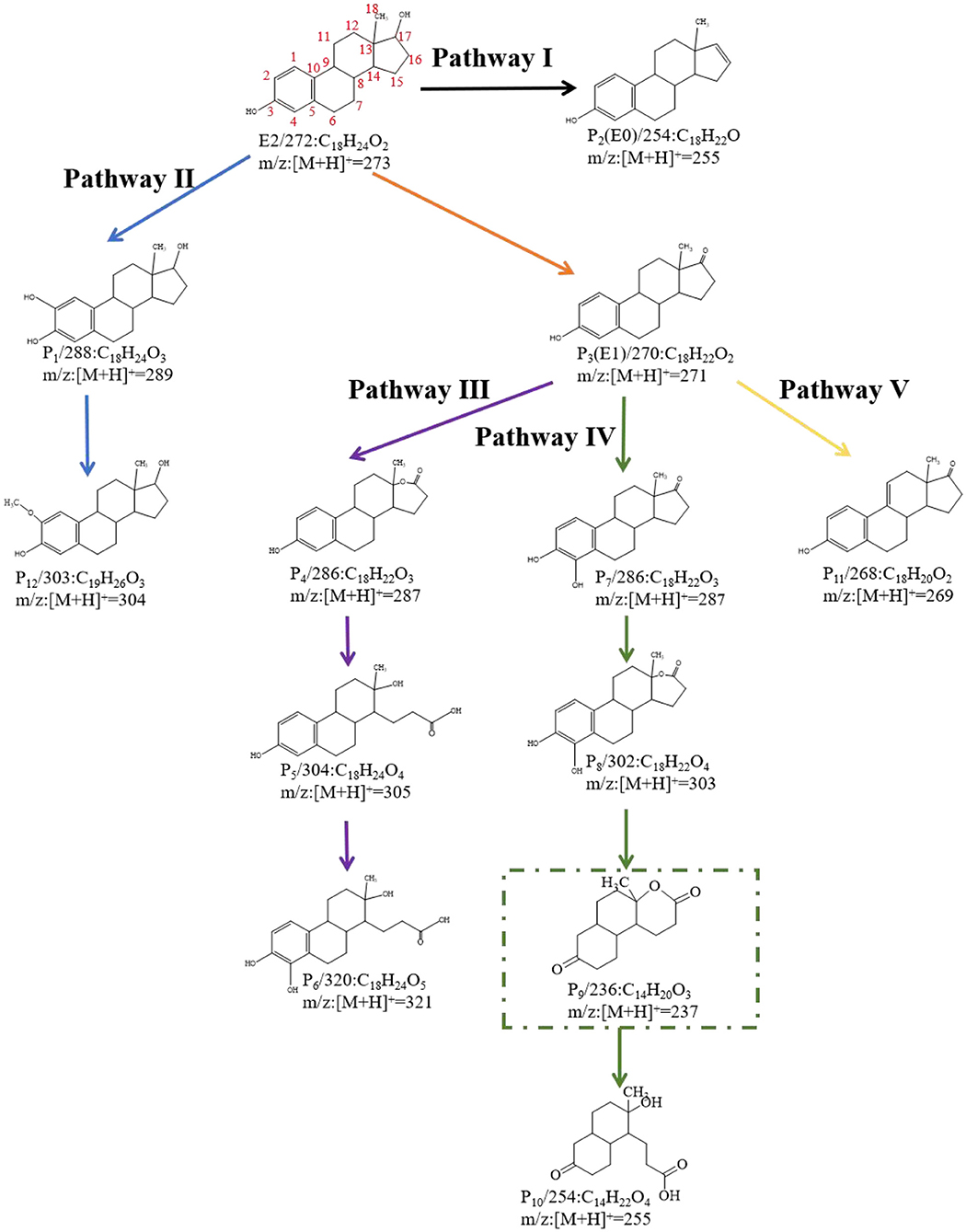
Figure 5. E2 degradation products and predicted biodegradation pathways. The green rectangle indicated the metabolite that was not detected by liquid chromatography tandem mass spectrometry (LC-MS/MS) (the black, blue, purple, green, and yellow lines represent Pathway I, Pathway II, Pathway III, Pathway IV, and Pathway V, respectively).
Based on the intermediate degradation products detected by LC-MS/MS, some possible degradation pathways of E2 were proposed. All degradation products were detected in the reaction system of L. casei LC-1 and C. utilis CU-2. Therefore, it was speculated that L. casei LC-1 and C. utilis CU-2 may produce the same or similar functional enzymes, while more research and analysis were needed.
Conclusions
In this paper, the contrastive study of E2 biodegradation by fungi and bacteria was investigated. Taking C. utilis CU-2 and L. casei LC-1 as functional microorganisms, the environmental impact factors, kinetics, and degradation pathways were analyzed. The results showed that both strains could achieve an optimal degradation efficiency at 35°C with 0.6 g/L sucrose addition. The addition of suitable concentration of sucrose (0.05–0.6 g/L) could significantly promote cell growth and the degradation of E2 with over 97% after reacting for 5 days. However, a high concentration of 10 g/L sucrose as co-substrate would inhibit the biodegradation of E2 compared with the low concentration. C. utilis CU-2 or L. casei LC-1 could degrade E2 into 12 intermediate products through 5 degradation pathways, among which C14H22O4 as the minimum-carbon-content degradation product was detected, and other smaller degradation products were not detected due to equipment intensity or low levels. In general, there was no significant difference between C. utilis CU-2 and L. casei LC-1 in E2 biodegradation characteristics and degradation products, but both of them were E2-efficient degradation strains. It was helpful to understand the degradation mechanism of C. utilis CU-2 and L. casei LC-1 and provide a new idea for the removal of E2.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HG: writing-original draft, conceptualization, methodology, data curation, and formal analysis. LZ: conceptualization, formal analysis, writing-review—editing, and supervision. ZD, YL, and LF: validation and investigation. YZ, SW, and YF: methodology and investigation. BL and LY: writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (41977317), the Natural Science Foundation of Beijing Municipality (8182037), and the Fundamental Research Funds for the Central Universities (2015ZCQ-HJ-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2021.661850/full#supplementary-material
The method of LC/MS/MS, basic information, and the secondary MS of E2-degrading products can be found in the Electronic Supplementary Information.
References
Adeel, M., Song, X., Wang, Y., Francis, D., and Yang, Y. (2017). Environmental impact of estrogens on human, animal and plant life: a critical review. Environ. Int. 99, 107–119. doi: 10.1016/j.envint.2016.12.010
Cao, L., Zhang, J., Zhao, R., Deng, Y., Liu, J., Fu, W., et al. (2019). Genomic characterization, kinetics, and pathways of sulfamethazine biodegradation by Paenarthrobacter sp. A01. Environ. Int. 131, 104961–104961. doi: 10.1016/j.envint.2019.104961
Cevasco, A., Urbatzka, R., Bottero, S., Massari, A., Pedemonte, F., Kloas, W., et al. (2008). Endocrine disrupting chemicals (EDC) with (anti)estrogenic and (anti)androgenic modes of action affecting reproductive biology of Xenopus laevis: II. Effects on gonad histomorphology. Comp. Biochem. Phys. C 147, 241–251. doi: 10.1016/j.cbpc.2007.10.001
Chen, S., Yin, H., Ye, J., Peng, H., Zhang, N., and He, B. (2013). Effect of copper(II) on biodegradation of benzo[a]pyrene by Stenotrophomonas maltophilia. Chemosphere 90, 1811–1820. doi: 10.1016/j.chemosphere.2012.09.009
Dawas-Massalha, A., Gur-Reznik, S., Lerman, S., Sabbah, I., and Dosoretz, C. G. (2014). Co-metabolic oxidation of pharmaceutical compounds by a nitrifying bacterial enrichment. Bioresour. Technol 167, 336–342. doi: 10.1016/j.biortech.2014.06.003
Fujii, K., Kikuchi, S., Satomi, M., Ushio-Sata, N., and Morita, N. (2002). Degradation of 17β-estradiol by a gram-negative bacterium isolated from activated sludge in a sewage treatment plant in Tokyo, Japan. Appl. Environ. Microbiol. 68, 2057–2060. doi: 10.1128/AEM.68.4.2057-2060.2002
Hinck, J. E., Blazer, V. S., Schmitt, C. J., Papoulias, D. M., and Tillitt, D. E. (2009). Widespread occurrence of intersex in black basses (Micropterus spp.) from U.S.rivers, 1995–2004. Aquatic. Toxicol. 95, 60–70. doi: 10.1016/j.aquatox.2009.08.001
Hyungkeun, R., and Kung-Hui, C. (2010). A 17β-estradiol-utilizing bacterium, sphingomonas strain KC8: part I - characterization and abundance in wastewater treatment plants. Environ. Sci. Technol. 44, 4943–4950. doi: 10.1021/es1001902
Jagdale, G. B., and Gordon, R. (1997). Effect of temperature on the activities of glucose-6-phosphate dehydrogenase and hexokinase in entomopathogenic nematodes (Nematoda: Steinernematidae). Comp. Biochem. Physiol. A Physiol. 118, 1151–1156. doi: 10.1016/S0300-9629(97)00034-0
Jobling, S., Williams, R. J., Johnson, A. C., Taylor, A., Grosssorokin, M., Nolan, M., et al. (2005). Predicted exposures to steroid estrogens in U.K. Rivers correlate with widespread sexual disruption in wild fish populations. Environ. Health Persp. 114, 32–39. doi: 10.1289/ehp.8050
Khattab, R. A., Elnwishy, N., Hannora, A., Mattiasson, B., Omran, H. O., Alharbi, O. M. L., et al. (2018). Biodegradation of 17β-estradiol in water. Int. J. Environ. Sci. Technol. 16, 4935–4944. doi: 10.1007/s13762-018-1929-y
Klavarioti, M., Mantzavinos, D., and Kassinos, D. (2009). Removal of residual pharmaceuticals from aqueous systems by advanced oxidation processes. Environ Int. 35, 402–417.
Kurisu, F., Ogura, M., Saitoh, S., Yamazoe, A., and Yagi, O. (2010). Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J. Biosci. Bioeng. 109, 576–582. doi: 10.1016/j.jbiosc.2009.11.006
Lambert, M. R., Giller, G. S. J., Barber, L. B., Fitzgerald, K. C., and Skelly, D. K. (2015). Suburbanization, estrogen contamination, and sex ratio in wild amphibian populations. Proc. Natl. Acad. Sci. U.S.A. 112, 11881–11886. doi: 10.1073/pnas.1501065112
Lange, I. G., Daxenberger, A., Schiffer, B., Witters, H., Ibarreta, D. H., and Meyer, H. H. D. (2002). Sex hormones originating from different livestock production systems: fate and potential disrupting activity in the environment. Anal. Chim. Acta 473, 27–37. doi: 10.1016/S0003-2670(02)00748-1
Leng, Y., Bao, J., Chang, G., Zheng, H., Li, X., Du, J., et al. (2016). Biotransformation of tetracycline by a novel bacterial strain Stenotrophomonas maltophilia DT1. J. Hazard Mater. 318, 125–133. doi: 10.1016/j.jhazmat.2016.06.053
Li, M., Zhao, X., Zhang, X., Wu, D., and Leng, S. (2018). Biodegradation of 17β-estradiol by Bacterial Co-culture Isolated from Manure. Sci. Rep. 8:3787. doi: 10.1038/s41598-018-22169-0
Li, Y., Sun, Y., Zhang, H., Wang, L., Zhang, W., Niu, L., et al. (2019). The responses of bacterial community and N2O emission to nitrogen input in lake sediment: estrogen as a co-pollutant. Environ. Res. 179:108769. doi: 10.1016/j.envres.2019.108769
Liu, J., Luo, Q., and Huang, Q. (2016). Removal of 17β-estradiol from poultry litter via solid state cultivation of lignolytic fungi. J. Clean Prod. 139, 1400–1407. doi: 10.1016/j.jclepro.2016.09.020
Lu, Z., Sun, W., Li, C., Ao, X., Yang, C., and Li, S. (2019). Bioremoval of non-steroidal anti-inflammatory drugs by Pseudoxanthomonas sp. DIN-3 isolated from biological activated carbon process. Water Res. 161, 459–472. doi: 10.1016/j.watres.2019.05.065
Ma, L., and Yates, S. R. (2018). Degradation and metabolite formation of 17β-estradiol-3-glucuronide and 17β-estradiol-3-sulphate in river water and sediment. Water Res. 139, 1–9. doi: 10.1016/j.watres.2018.03.071
Min, J., Chen, W., and Hu, X. (2018). Biodegradation of 2,6-dibromo-4-nitrophenol by Cupriavidus sp. strain CNP-8: Kinetics, pathway, genetic and biochemical characterization. J. Hazard Mater. 361, 10–18. doi: 10.1016/j.jhazmat.2018.08.063
Moreira, I. S., Bessa, V. S., Murgolo, S., Piccirillo, C., Mascolo, G. P., et al. (2018). Biodegradation of Diclofenac by the bacterial strain Labrys portucalensis F11. Ecotoxicol. Environ. Saf. 152, 104–113. doi: 10.1016/j.ecoenv.2018.01.040
Nakai, S., Yamamura, A., Tanaka, S., Shi, J., Nishikawa, M., Nakashimada, Y., et al. (2011). Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environ. Chem. Lett. 9, 1–6. doi: 10.1007/s10311-010-0308-9
Pan, L. J., Li, J., Li, C. X., Tang, X. D., Yu, G. W., and Wang, Y. (2017). Study of ciprofloxacin biodegradation by a Thermus sp. isolated from pharmaceutical sludge. J. Hazard Mater. 343, 59–67. doi: 10.1016/j.jhazmat.2017.09.009
Peiris, C., Nawalage, S., Wewalwela, J. J., Gunatilake, S. R., and Vithanage, M. (2020). Biochar based sorptive remediation of steroidal estrogen contaminated aqueous systems: a critical review. Environ. Res. 191:110183. doi: 10.1016/j.envres.2020.110183
Sacdal, R., Madriaga, J., and Espino, M. P. (2020). Overview of the analysis, occurrence and ecological effects of hormones in lake waters in Asia. Environ Res. 182:109091. doi: 10.1016/j.envres.2019.109091
Shi, J., Fujisawa, S., Nakai, S., and Hosomi, M. (2004). Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea. Water Res. 38, 2322–2329. doi: 10.1016/j.watres.2004.02.022
Snyder, S. A., Adham, S., Redding, A. M., Cannon, F. S., DeCarolis, J., Oppenheimer, J., et al. (2007). Role of membranes and activated carbon in the removal of endocrine disruptors and pharmaceuticals. Desalination, 202, 181–453.
Ting, Y. F., and Praveena, S. M. (2017). Sources, mechanisms, and fate of steroid estrogens in wastewater treatment plants: a mini review. Environ. Monit. Assess. 189:178. doi: 10.1007/s10661-017-5890-x
Wang, Y., Sun, Q., Li, Y., Wang, H., Wu, K., and Yu, C. (2019). Biotransformation of estrone, 17β-estradiol and 17α-ethynylestradiol by four species of microalgae. Ecotox. Environ. Saf. 180, 723–732. doi: 10.1016/j.ecoenv.2019.05.061
Whitman, W. B. (2017). Bacteria and the fate of estrogen in the environment. Chem. Biol. 24, 652–653. doi: 10.1016/j.chembiol.2017.05.028
Wise, A., Obrien, K., and Woodruff, T. J. (2011). Are oral contraceptives a significant contributor to the estrogenicity of drinking water. Environ. Sci. Technol. 45, 51–60. doi: 10.1021/es1014482
Xiong, J. Q., Kurade, M. B., Kim, J. R., Roh, H. S., and Jeon, B. H. (2017). Ciprofloxacin toxicity and its co-metabolic removal by a freshwater microalga Chlamydomonas mexicana. J. Hazard Mater. 323, 212–219. doi: 10.1016/j.jhazmat.2016.04.073
Xiong, W., Yin, C., Wang, Y., Lin, S., Deng, Z., and Liang, R. (2020). Characterization of an efficient estrogen-degrading bacterium Stenotrophomonas maltophilia SJTH1 in saline-, alkaline-, heavy metal-contained environments or solid soil and identification of four 17β-estradiol-oxidizing dehydrogenases. J. Hazard Mater. 385, 121611–121615. doi: 10.1016/j.jhazmat.2019.121616
Ying, G., Kookana, R. S., and Ru, Y. (2002). Occurrence and fate of hormone steroids in the environment. Environ. Int. 28, 545–551. doi: 10.1016/S0160-4120(02)00075-2
Young, W. F., Whitehouse, P., Johnson, I., and Sorokin, N. (2002). Proposed predicted-no-effect-concentrations (PNES) for natural and synthetic steroid oestrogens in surface water. Environment Agency R&D Technical report P2-T04/1, 93–95.
Yu, C., Deeb, R. A., and Chu, K. (2013). Microbial degradation of steroidal estrogens. Chemosphere 91, 1225–1235. doi: 10.1016/j.chemosphere.2013.01.112
Yu, Q., Wang, P., Liu, D., and Gao, R. (2016). Degradation characteristics and metabolic pathway of 17β-estradiol (E2) by Rhodococcus sp. DS201. Biotechnol. Bioproc. E 21, 804–813. doi: 10.1007/s12257-016-0283-5
Zhao, X., Grimes, K. L., Colosi, L. M., and Lung, W. (2019). Attenuation, transport, and management of estrogens: a review. Chemosphere 230, 462–478. doi: 10.1016/j.chemosphere.2019.05.086
Keywords: biodegradation, 17β-estradiol, co-metabolism, degradation products, pathways
Citation: Ge H, Yang L, Li B, Feng Y, Wang S, Zheng Y, Feng L, Liu Y, Du Z and Zhang L (2021) A Comparative Study on the Biodegradation of 17β-Estradiol by Candida utilis CU-2 and Lactobacillus casei LC-1. Front. Energy Res. 9:661850. doi: 10.3389/fenrg.2021.661850
Received: 31 January 2021; Accepted: 15 March 2021;
Published: 29 April 2021.
Edited by:
Yong Jiang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Weiwe Cai, Beijing Jiaotong University, ChinaShengbing He, Shanghai Jiao Tong University, China
Copyright © 2021 Ge, Yang, Li, Feng, Wang, Zheng, Feng, Liu, Du and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqiu Zhang, emhhbmdsaXFpdUAxNjMuY29t
 Huiru Ge
Huiru Ge Shurui Wang
Shurui Wang Liqiu Zhang
Liqiu Zhang