- 1Naiman Desertification Research Station, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, China
- 2Key Laboratory of Stress Physiology and Ecology in Cold and Arid Regions, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, China
- 3University of Chinese Academy of Sciences, Beijing, China
Long-term ecological restoration plays an important role in the sustainable development of degraded grassland ecosystem. In this study, the levels of species diversity, genetic diversity and soil microbial diversity in restored grassland were measured by vegetation survey, DNA barcoding and soil microbial high-throughput sequencing technology, so as to explore the relationship between above- and belowground biodiversity and its driving factors in Horqin sandy grassland. In this study, the results found that herb are dominated in restoration grassland types. Plant species richness (SR) from post-non-grazing restoration plot (NGR) communities was significantly higher than other restoration communities (10 ± 1.1, p = 0.004). Genetic diversity indices of dominant plant species in chloroplast DNA (cpDNA), were remarkable greater than nuclear DNA (nrDNA) in each recovering sandy grassland plots (amplitude of difference was 44.8%–70.5% in allelic richness (AR), 81.9%–128.1% in expected heterozygosity (HE)). The soil bacterial and fungal richness from natural mobile dune grassland (NM) communities was notably lower than that from recovering grassland types (1641.9 ± 100.4, p < 0.001; 533 ± 16.6, p < 0.001). In this study, heterogeneous levels of genetic variability among different recovering sandy grassland types were detected. Correlation analyses revealed that there were positive correlations between species diversity and genetic diversity (SR & AR: r = 0.56, R2 = 0.31, p < 0.001; SR & HE: r = 0.33, R2 = 0.11, p = 0.045) and a negative correlation between soil microbial diversity and genetic diversity (r = -0.44, R2 = 0.19, p = 0.005). The final structural equation model explained 38% of the variance in SR, 57% in AR, 52% in soil microbial diversity (SD), 49% in aboveground biomass (AGB), 87% in soil organic carbon (SOC), 47% in soil alkali-hydrolyzable nitrogen (SAN) and 69% in soil available phosphorus (SOP). Long-term ecological restoration had significant direct positive effects on AGB, SOC, SAN, SOP, AR, SR and SD. There was a negative correlation between above- and belowground biodiversity and biological and abiotic factors. The results of this study have clarified the above- and underground biodiversity levels of sandy grassland and the relationship with driving factors under long-term ecological restoration measures, and will provide effective support for the management and sustainable development of sandy grassland.
Introduction
Land degradation is one of the most serious environmental and socio-economic problems at global scale, which reduces the resilience of natural ecosystems (Bai et al., 2008), involving serious soil erosion, degradation of vegetation, loss of biodiversity, and pollution of water bodies and so on (García-Díaz et al., 2017). In recent years, natural grassland degradation or desertification has become a universal ecological environment obstacle in the world and a major threat for the sustainability of agriculture and economic development (Zhao et al., 2003; Xue et al., 2021). Corresponding to that, some large long-term ecological restoration projects have been implemented in China, such as the “Three-North Shelter Forest Program” and the “Grain for Green Project” (Stokes et al., 2010). Numerous studies have found that grassland fence can effectively accelerate vegetation succession and is one of the cost-effective strategies used to restore plant and soil communities (Lenka et al., 2012; Deng et al., 2018; Zhao et al., 2019). The process of vegetation restoration affects biodiversity by regulating the interaction between aboveground plant diversity and subsurface soil microbial diversity (Suding, 2011). Vegetation restoration of degraded grassland ecosystems has become increasingly important to conserve biodiversity (Porto et al., 2009).
A long-term ecological restoration measure will change plant-soil community composition and directly or indirectly impact on biodiversity. At the same time, biodiversity also affects corresponding ecosystem processes, for instance, primary production, nutrient and water cycling, and soil formation and retention (Liang and Wei, 2020). Interest in studying the interaction between plants and soil microbial communities is increasing over recent years. However, examples of (cor)relations between above and below-ground biodiversity are confusing, demonstrating that they can be positive, negative or non-existent (Hooper et al., 2000). This phenomenon may be due to the variety of their interaction mechanisms. Vegetation could straight affect the composition and diversity of the soil community (Yuan J. et al., 2020; Yuan Z. Q. et al., 2020). In the meantime, most of the biodiversity in soil will be indirectly linked to vegetation characteristics such as litter type and root exudates (Korthals et al., 2001). For grassland ecosystems dominated by perennial herbaceous plants, lasting ecological management policies will change both vegetation and soil characteristics and composition of communities, resulting in changes in aboveground and subsurface biodiversity.
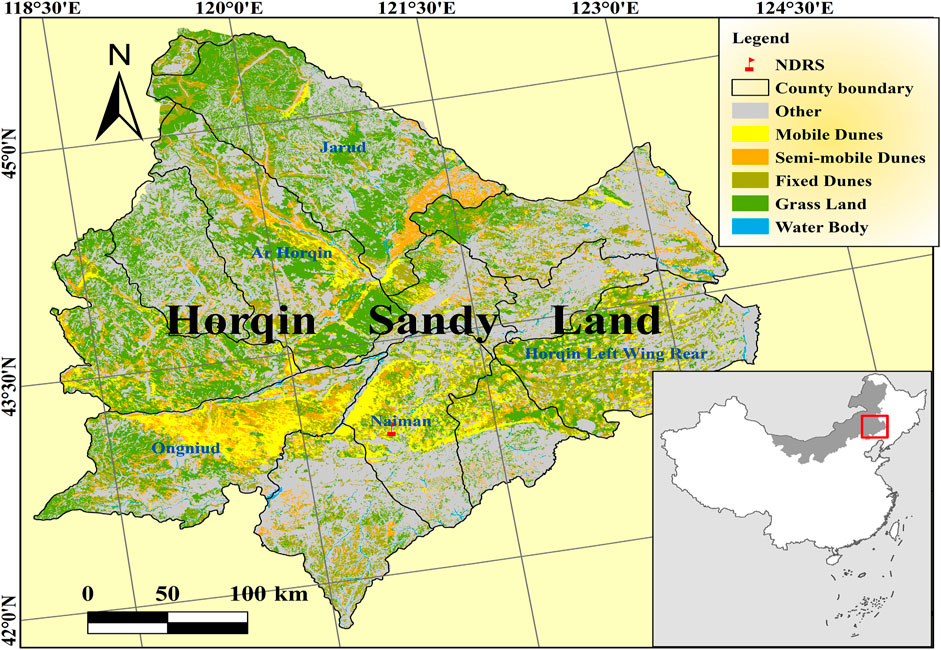
Horqin Sandy Land, as a representative sensitive ecological region, is located in the agro-pastoral transitional zone between the Inner Mongolian Plateau and the Northeast Plains (42°41′–45°45′N, 118°35′–123°30′E), and is one of the four largest sandy areas in northern China; it covers an area of approximately 139,300 km2, which had been degraded sandy land area up to 71,884 km2 (Wang, 2003; Zhao et al., 2003). Landscape in this area is characterized by sand dunes that alternate with gently undulating lowland areas (Li et al., 2005). This area belongs to the continental semi-arid monsoon climate and is in the temperate zone, with a mean annual temperature (AMT) of 3°C–7°C and mean annual rainfall (AP) of 350–500 mm (Zhao et al., 2003). Over recent decades, this region has undergone severe degradation due to climate change and human activities (Li et al., 2000; Li et al., 2004), and has displayed the northern moving phenomenon of the interlocked agro-pasturing area of north China in the most recent hundred years (Zhao et al., 2000; Zhao et al., 2002). These altered environmental conditions may caused strong effects on above-and belowground biodiversity. Therefore, the protection of above-and belowground biodiversities plays an important role in maintaining the community stability and controlling degradation.
The relationship between above-and underground biodiversity has always been a hot topic in ecological research. Long-term ecological restoration, especially fencing policy, is the main measure to protect biodiversity and maintain ecosystem function of degraded grassland communities. Here, this study examined how biodiversity aboveground (plant species richness and genetic diversity) and belowground (soil bacterial operational taxonomic units (OTUs) and fungal richness) and biological and abiotic factors interacted along restoration gradients in the Horqin sandy grassland. This study assessed 1) the biodiversity levels and differences in plants, soil bacteria and soil fauna along restoration gradients; 2) the correlations between plant and soil biodiversity along different restoration gradients; and 3) the combined effects of biological and abiotic factors on above- and belowground biodiversity in the Horqin sandy grassland. The results of this study will provide a scientific basis for biodiversity conservation in sandy grassland ecosystems, so as to provide effective support for the management and sustainable development of sandy grassland.
Materials and methods
Site description
Field study was done at the observation field in Naiman Desertification Research Station of the Chinese Academy of Sciences in the eastern part of Inner Mongolia, China (42°55′-42°56′N, 120°41′-120°42′E) (Figure 1). Naiman located in the Northern China’s agro-pastoral ecotone in the southwestern part of the Horqin Sandy Land, which is roughly 400 × 400 km in size and represents the most desertification threatened area in north China (Li et al., 2005). This area has a continental semi-arid monsoon climate in the temperate zone. The mean annual precipitation is 377 mm, and nearly 75% is concentrated in the growing season (May to August) (Zuo et al., 2012). The average annual temperature is 6.8°C. Vegetation cover in the study area ranged from 50% to 70%.
Experimental design
The experimental field consisted of four plots along a restoration gradient, i.e., Plot A, natural mobile dune grassland plot (NM); Plot B, post-non-grazing restoration plot (NGR); Plots C and D, post-light-grazing restoration plot (LGR) and post-heavy-grazing restoration plot (HGR). NM, NGR, LGR and HGR were long-term observation fields, and many studies have been carried out (Su et al., 2006; Zhao et al., 2009; Zhang et al., 2016). The NM Plot has been monitored since 2005 and has an area of 0.60 ha (200 × 30 m). The vegetation cover was less than 5%. The NM Plot was dominated by the annual herb Agriophyllum squarrosum. The NGR Plot was established and fenced in 1985 to assist with the natural recovery of the sandy grassland and covers 0.66 ha (200 × 33 m). The vegetation cover in the NGR Plot ranged from 30% to 50%. The NGR Plot was dominated by the perennial grasses Artemisia halodendron, Melissitus ruthenicus, and Euphorbia humifusa. The LGR and HGR Plot were established in 1992 to allow natural restoration of the 5-years lightly and heavily grazed lands, has 1.5 ha (200 × 75 m) each. The grazing intensities were 2 sheep units/hm2 and 6 sheep units/hm2, respectively. Vegetation cover in LGR Plot was more than 70%. The dominant plant species were annual herbs, including Artemisia scoparia, Setaria viridis, Salsola collina, and Corispermum hyssopifolium (Niu et al., 2018). Vegetation cover in the HGR Plot was about 60%. The HGR Plot is dominated by the perennial grass, Pennisetum centrasiaticum, S. viridis, Digitaria cilliaris, and Phragmites communis (Zhao et al., 2007).
Community survey and sampling
The number of plants was determined in mid-August 2019, at which time coverage, height and frequency were measured and used to calculate their importance value. In each plot, 9 random quadrats (1 × 1 m) were used to measure vegetation characteristics. Young healthy leaves from 3 dominant plant species in each quadrat were arbitrarily sampled from plants and immediately stored with silica gel in zip lock plastic bags for later DNA extraction. Aboveground biomass (AGB) in each quadrat was harvested in August, standing biomass and litter separated, oven-dried at 65°C for 48 h and weighed (Zhao et al., 2016).
About 36 soil samples (0–10 cm; 9 samples per plot) were collected from the 4 recovering grassland plots. Soil samples were air-dried and hand-sieved through a 2-mm screen to remove roots and other debris. A portion of each air-dried soil sample was ground to pass a 0.1 mm mesh for soil nutrient analyses. Soil organic carbon (SOC) concentration was determined using the Walkley-Black dichromate oxidation procedure (Nelson and Sommers, 1982). Soil available nitrogen (SAN) was determined using the alkaline diffusion method, and soil phosphorus (SOP) was determined using the Olsen method (Olsen et al., 1954). Soil samples for molecular analysis were packed in polyethylene bags, immediately stored in portable refrigerator powered with a car battery and then stored in the lab at −80°C until processing.
DNA extraction and sequencing
Plant and soil total DNA was extracted using an AxyPrep genomic DNA mini kit (Axygen, Beijing, China) and Qiagen Dneasy Powersoil kit (Qiagen, Beijing, China), respectively. DNA was quantified spectrophotometrically; samples that yielded high quantities of good-quality DNA were used in consecutive experiments.
Soil bacterial and fungal communities were sequenced separately in 16S rDNA (amplified by primers 515F/806R) (Walters et al., 2016) and ITS rDNA (amplified by primers ITS5-1737F/ITS2- 2043R) (Zhou et al., 2020) (Appendix S1). Polymerase chain reactions (PCR) was performed in a 20 µL reaction volume, containing 10 ng of template DNA, 0.4 µL Pfu DNA polymerase (TransGen Biotech, Beijing, China), dNTPs (4 µL of 2.5 mM), 5 × reaction buffer (4 µL), each primer (0.8 µL of 5 µM). The amplification condition for bacteria are as follow: 95 °C for 3 min, by 27 cycles of 30 s at 95°C, 30 s at 55°C, 45 s at 72°C, and a final 10 min extension step at 72°C. The amplification condition for fungal are as: 94 °C for 1 min, followed by 30 cycles of 20 s at 94°C, 30 s at 56°C, 30 s at 72°C, and a final 5 min extension step at 72°C. High-throughput sequencing was performed on an Illumina NovaSeq platform (Illumina, San Diego, CA, United States) in Novogene Co., Ltd. (Beijing, China).
In this study, DNA barcoding markers were used to assess genetic diversity and variation in dominant plants. Several pairs of plant cpDNA and nrDNA primers designed by Hamilton (1999), Taberlet et al. (1991) and Sang et al. (1997), were used in the initial screening (Appendix S1). Marks trnL-F, matK and psbA-trnH were chose as primers for plants from Chenopodiaceae and Leguminosae amplification; ITS, rps16 and matK as primers for plants from Gramineae amplification; trnL-F, ITS and psbA-trnH as primers for plants from other families amplification. PCR was performed in a 25 µL reaction volume, containing 40 ng of genomic DNA, 1.0 U of Taq polymerase (Axygen Inc., Beijing, China), 3 mM MgCl2, 500 µM each dNTP, 20 mM Tris-HCl (pH 8.3), 100 mM KCl, 0.3 µM each primer. The amplification condition was an initial denaturation step of 94°C for 3 min, followed by 30 cycles of 30 s at 94°C, 30 s at an appropriate annealing temperature, 1 min at 72°C, and a final 5 min extension step at 72°C. The PCR products were determined by 1.0% agarose gel electrophoresis. The amplification products were purified using AxyPrep PCR purification kit following the manufacturer’s protocol (Axygen Inc., Beijing, China). Purified plant DNA was sequenced by the MEIJI sequencing company in Shanghai, China, applying the PCR-primers as sequencing primers.
Statistical analyses
Two measures of plant species diversity from 4 recovering grassland plots were used: species richness (SR) and Simpson’s diversity index (D). D was calculated from the formula,
where Ni = important value, which is the average of relative density, relative height, and relative biomass.
The number of species was taken as a measure of species richness. The measures of plant genetic diversity from recovering gradients plots were calculated with Arlequin 3.0 (Excoffier et al., 2005), including allelic richness (AR) and expected heterozygosity (HE), as they are analogous to SR and D, respectively (Etienne, 2005; Evanno et al., 2009).
The raw reads of soil DNA sequencing were demultiplexed, quality-filtered by FASTQ, and merged by FLASH (Magoč and Salzberg, 2011). Quality-filtered sequences were aligned with SILVA alignment (Quast et al., 2013), and clustered into the operational taxonomic units (OTUs) with 97% sequence similarity using UPARSE (Edgar, 2013). Two measures of soil microbial diversity from recovering gradients plots were used: soil bacterial operational taxonomic unit (OTUs) and soil fungal richness.
The relationships between plant species diversity (SR) and plant genetic diversity (AR and HE) and between plant diversity (PD) and soil microbial diversity (SD) were examined. Before analysis of variance (ANOVA) and correlation analysis, data were tested for normality to ensure normal distribution. Statistical significance was determined by one-way (plant species and soil microbial diversity index) or two-way (plant genetic diversity index) ANOVA followed by Tukey’s post hoc test. All statistical analyses were performed using SPSS 19.0 for windows (SPSS, Inc. Chicago, IL). Confirmatory analysis of structural equation model (SEM) was applied to the data to determine direct and indirect impacts of long-term ecological restoration on above-and belowground biodiversities. SEM analysis used lavaan packages in R version 3.5.2 (https://www.r-project.org/).
Results
Plant species diversity
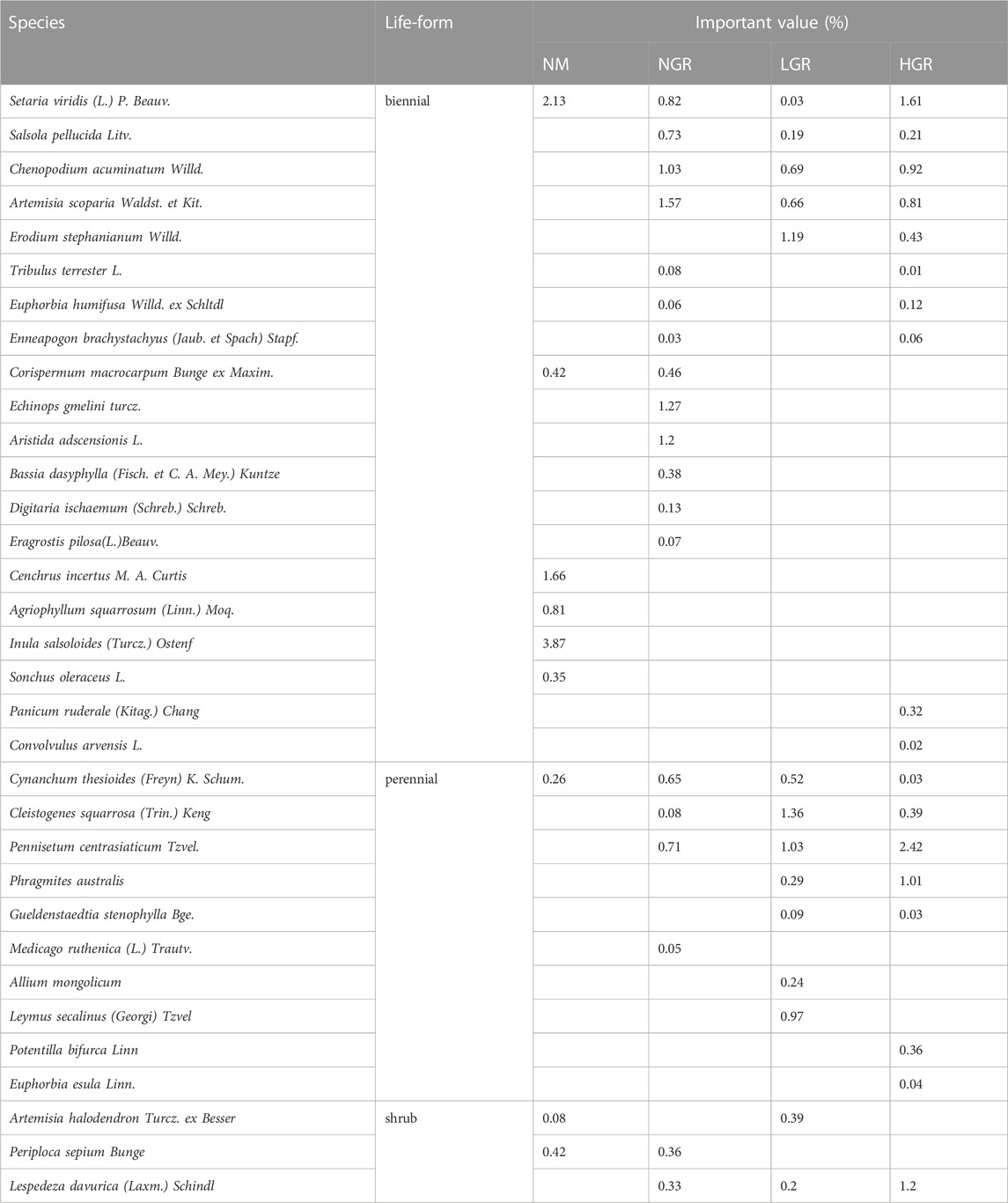
Thirty-three species were recorded (Table 1). Specifically, there are 9 species in natural mobile dune grassland plot (NM), 19 species in post-non-grazing restoration plot (NGR), 14 species in post-light-grazing restoration plot (LGR) and 18 species in post-heavy-grazing restoration plot (HGR). The results showed that herbaceous are dominated in all grassland plots. Biennials were dominant in NM and NGR, accounting for 92.40% and 78.22%, respectively. Perennials were dominant in LGR and HGR, accounting for 57.32% and 44.35% of the total importance value. According to the species importance value, the results showed that S. viridis and C. incertus were the dominant plant species in NM, A. scoparia and E. gmelini were the dominant plant species in NGR, E. stephanianum and C. squarrosa were the dominant plant species in LGR, S. viridis and P. centrasiaticum were the dominant plant species in HGR. One-way ANOVAs explained that grassland restoration were notable impacted on plant species richness (SR) and Simpson’s diversity index (D) (p < 0.001) (Figure 2). The results indicated that the plant species richness from the NGR communities was significantly higher than that from the other recovering grassland plots (10 ± 1.1, p = 0.004), and the plant species richness from the NM communities was significantly lower than that from the other recovering grassland plots (4 ± 1.2, p < 0.05). The plant Simpson’s diversity index in the NM communities was remarkably higher than that in other recovering grassland plots (0.417 ± 0.036, p < 0.001) and exhibited no significant change in the NGR, LGR, and HGR communities.
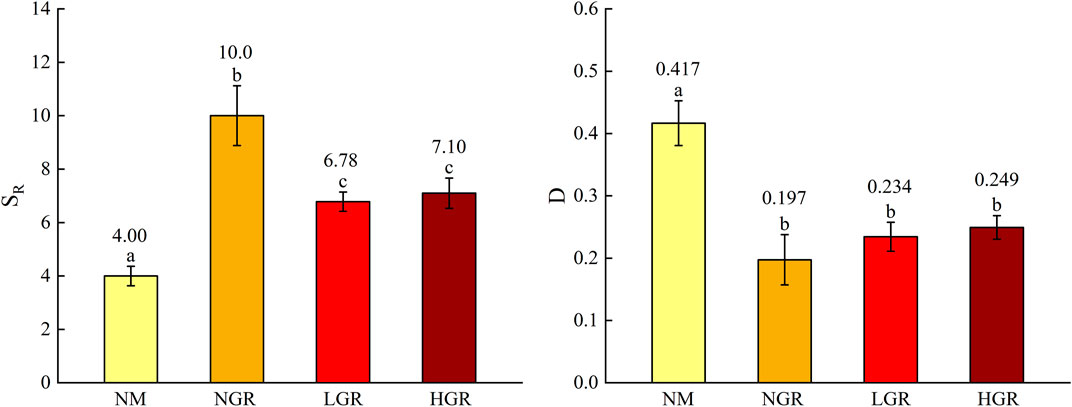
FIGURE 2. The plant species diversity (SR, species richness; D, Simpson’s diversity index) along long-term ecological restoration gradients. Values are mean ± SE. Different lowercase letters indicate significant differences (p < 0.05) among ecological restoration gradients treatments. NM, natural mobile dune grassland plot; NGR, post-natural-assisted recovering plot; LGR, post-lightly-grazed recovering plot; HGR, post-heavily-grazed recovering plot.
Plant genetic diversity
Genetic diversity of dominant plant species from different recovering grassland plots is given in Figure 3. Two-way ANOVAs revealed that genetic diversity indices were affected significantly by different grassland plots, and the allelic richness (AR) and expected heterozygosity (HE) of dominant plant species in cpDNA was outstanding greater than those in nrDNA in each recovering sandy grassland plots. Among the restoration plots, the AR of dominant plant species in nrDNA and cpDNA was varied from 1.931 to 2.590 and from 3.844 to 3.930, respectively. The AR values of nrDNA and cpDNA in NGR were significantly higher than those in NM. The HE of the dominant plant species in nrDNA and cpDNA varied from 0.2989 to 0.4822, and from 0.6924 to 0.7007, respectively, and the values of each recovering grassland plot were not significant (amplitude of difference up to 44.8%–70.5% in AR, 81.9%–128.1% in HE).
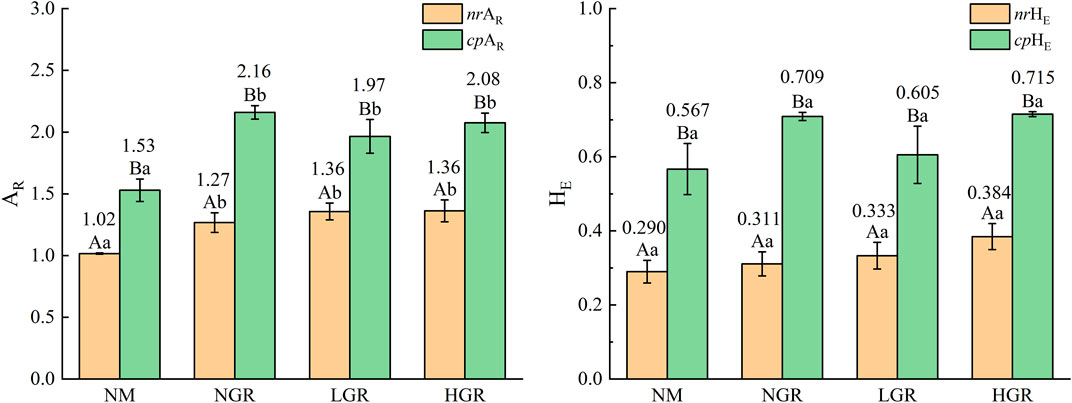
FIGURE 3. The plant genetic diversity (AR, allelic richness; HE, expected heterozygosity) along long-term ecological restoration gradients. Values are mean ± SE. Bars labelled with different lowercase letters represent significant differences among ecological restoration gradients treatments (p < 0.05). Bars labelled with different capital letters differ significantly between nuclear and chloroplast DNA genetic diversity treatments (p < 0.05). nrAR and cpAR, nuclear and chloroplast DNA allelic richness; nrHE and cpHE, nuclear and chloroplast DNA expected heterozygosity; NM, natural mobile dune grassland plot; NGR, post-natural-assisted recovering plot; LGR, post-lightly-grazed recovering plot; HGR, post-heavily-grazed recovering plot.
Soil microbial diversity
The bacterial operational taxonomic units (OTUs) were assigned into 37 phyla, 216 families and 487 genera. According to the annotated OTUs, Actinobacteria, Proteobacteria and Acidobacteria represented the most abundant phyla in all samples, comprising 21.87%–53.15%, 27.62%–45.37%, and 3.31%–18.60%, respectively, of the total OTUs in each treatment. These three phyla occupied more than 80% of each plot. The fungal OTUs were assigned into 16 phyla, 220 families and 388 genera. According to the annotated OTUs, Mortierellomycota and Ascomycota represented the most abundant phyla in all samples, comprising 23.59%–44.90% and 21.47%–44.94%, respectively, of the total OTUs in each treatment. These two phyla occupied more than 50% of each plot (Figure 4).
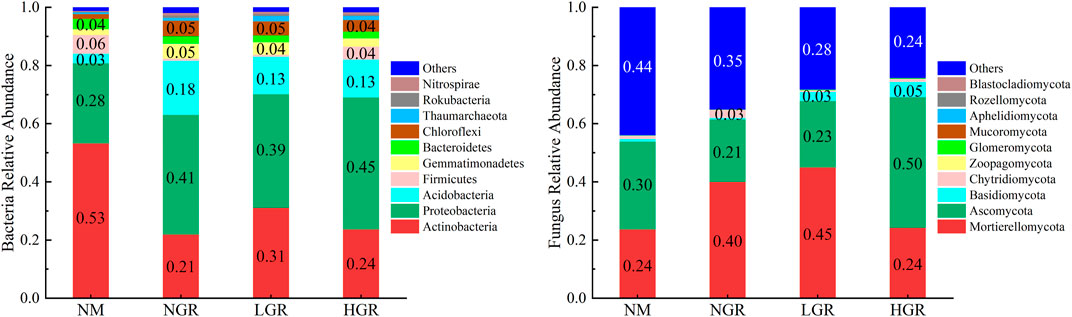
FIGURE 4. Soil bacteria and fungus relative abundance along long-term ecological restoration gradients. NM, natural mobile dune grassland plot; NGR, post-natural-assisted recovering plot; LGR, post-lightly-grazed recovering plot; HGR, post-heavily-grazed recovering plot.
In different restoration grassland plots, one-way ANOVAs identified that community types had sharp influence on soil microbial richness (p < 0.001) (Figure 5). The results found that soil bacterial and fungal richness from NM communities were obvious fewer than recovering grassland plots (1641.9 ± 100.4, p < 0.001; 533.2 ± 16.6, p < 0.001). The soil bacterial richness from the HGR communities was greater than that from the NM and NGR communities (2714.1 ± 38.8, p < 0.05). However, the soil fungal richness demonstrated no clear change in the NGR, LGR and HGR communities (Figure 5).
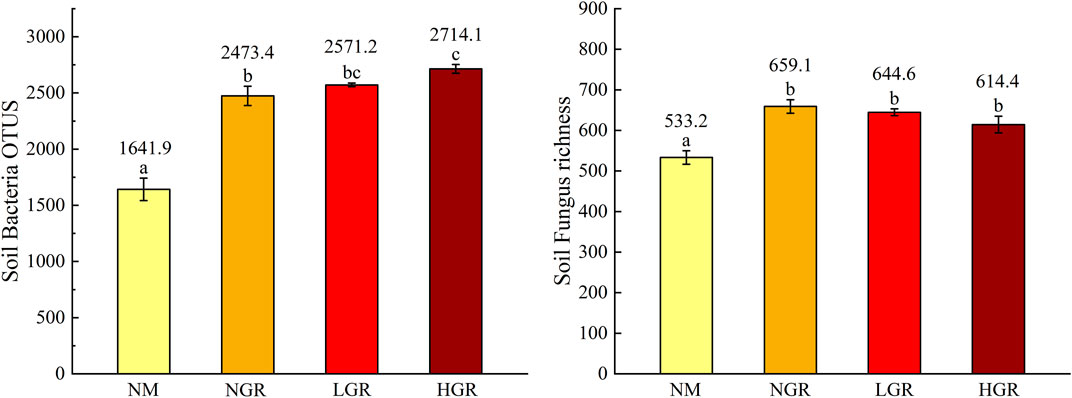
FIGURE 5. Soil bacteria and fungus richness along long-term ecological restoration gradients. Values are mean ± SE. Different lowercase letters indicate significant differences (p < 0.05) among ecological restoration gradients treatments. OTUS, operational taxonomic unit; NM, natural mobile dune grassland plot; NGR, post-natural-assisted recovering plot; LGR, post-lightly-grazed recovering plot; HGR, post-heavily-grazed recovering plot.
Correlations in plant and soil biodiversity
The heterogeneous levels of genetic variability among different restoration grassland plots were detected. Composite allelic richness of cpDNA and nrDNA in NM, NGR, LGR and HGR communities varied from 1.02 to 1.78, 2.25 to 2.37, 1.72 to 2.32, 1.74 to 2.36, respectively. Composite expected heterozygosity of cpDNA and nrDNA varied from 0.34 to 0.78, 0.72 to 0.77, 0.72 to 0.76, 0.71 to 0.77, respectively. The heterogeneous levels of soil microbial diversity among different recovering grassland plots were revealed. Composite soil microbial shannon diversity of bacterial and fungal in NM, NGR, LGR and HGR communities varied from 4.728 to 6.253, 6.210 to 7.173, 6.616 to 7.090, 6.138 to 7.349, respectively. Composite plant diversity of species diversity and genetic diversity varied from 0.164 to 0.368, 0.099 to 0.496, 0.217 to 0.579, 0.147 to 0.355, respectively (Figure 6).
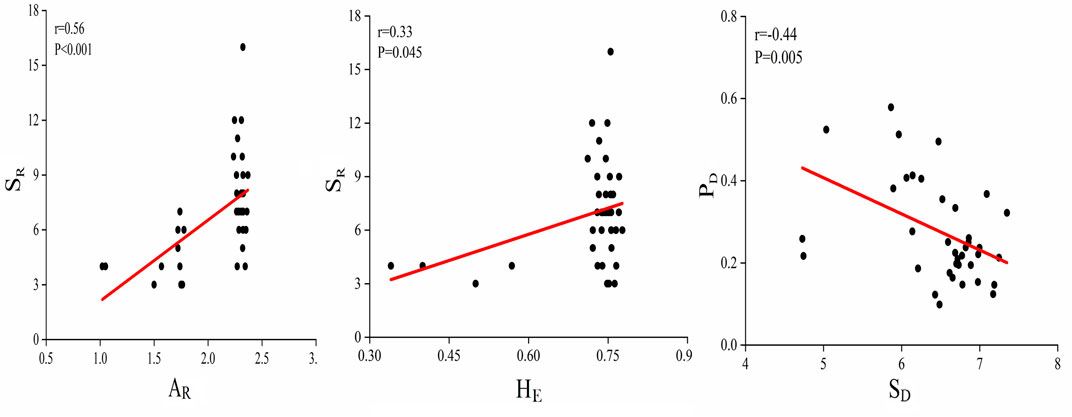
FIGURE 6. Correlations between above-and belowground biodiversity. SR, species richness; AR, allelic richness; HE, expected heterozygosity; PD, plant diversity; SD, soil microbial diversity.
Correlation analyses disclosed that there were positive correlations in species diversity and genetic diversity (Figure 6). Data illustrated that there was a prominent correlation between SR and AR (r = 0.56, R2 = 0.31, p < 0.001), and a striking correlation between SR and HE (r = 0.33, R2 = 0.11, p = 0.045). The results also showed that there was a striking negative correlation between plant diversity (PD) of different grassland types and soil microbial diversity (SD) (r = -0.44, R2 = 0.19, p = 0.005) (Figure 6).
Effects of long-term ecological restoration on above-and belowground biodiversity
The final structural equation model (SEM) for projecting the direct and indirect effects of long-term restoration on SR, AR and SD emerged when aboveground biomass (AGB), soil organic carbon (SOC), soil alkali-hydrolyzable nitrogen (SAN) and soil phosphorus (SOP) were the variables (PChi-squre = 0.914, PRMSEA = 0.979, df = 11). The final model explained 38% of the variance of SR, 57% of AR, 52% of SD, 49% of AGB, 87% of SOC, 47% of SAN and 69% of SOP (Figure 7). Long-term ecological restoration had outstanding direct active effects on AGB, SOC, SAN, SOP, AR, SR and SD. There was an inactive correlation between aboveground and belowground biodiversity and biological and abiotic factors; that is, SR was significantly negatively correlated with AGB, AR was significantly negatively correlated with AGB, SAN and SOP, and SD was negatively correlated with SAN but not significantly. The effect of SOC on aboveground plant diversity was mediated by AGB and SAN.
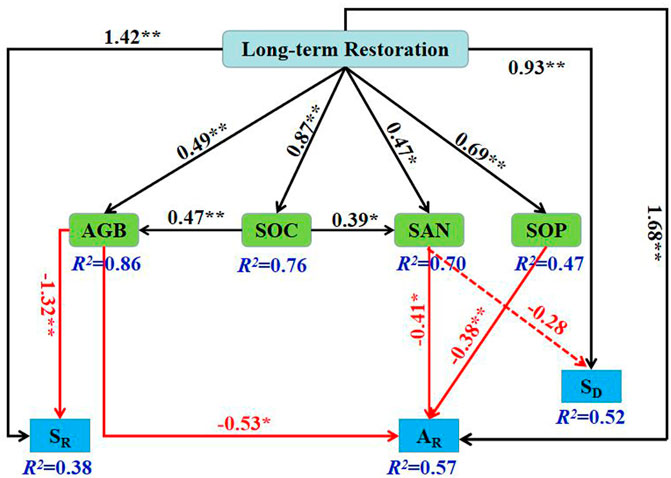
FIGURE 7. Structural equation model of abiotic and biotic factors as predictors of above-and belowground biodiversity. Solid black arrows represent positive paths, solid red arrows represent negative paths. The values on the arrows are standardized path coefficients, asterisks following the numbers denote significant relationships (**p < 0.01 and *p < 0.05), and the R2 indicates the extent to which this variable is explained. SR, species richness; AR, allelic richness; SD, soil microbial diversity; AGB, aboveground biomass; SOC, soil organic carbon; SAN, soil available nitrogen; SOP, soil phosphorus.
Discussion
The relationship between above- and belowground biodiversity is the key to biodiversity research. It is mainly affected by environmental conditions, especially the impact of climate change and human activities on plants composition, structure of aboveground and soil microbial communities of belowground. In this study, above- and belowground biodiversity of plant communities in sandy grassland was studied for long-term ecological restoration. The purpose was to explore the relationship between plant and soil microbial diversity and the main influencing factors, and to provide scientific basis for the multi-function research of sandy grassland ecosystem.
Plant-soil microbial community characters
Long-term ecological restoration greatly affects the structure and function of grassland ecosystem, especially the vegetation characteristics. Different forms of restoration will have different influences. The results showed that the number of species in NGR, LGR and HGR was more than NM (Table 1), which might be due to ecological restoration had improved the original habitat conditions and increased the number of species suitable for living. The data pointed out that the proportion of herbaceous was higher than that of shrubs in all treatments, and perennial plants were the dominant species in LGR and HGR, and biennial plants were the dominant species in NM and NGR (Table 1). These results indicated that long-term ecological restoration affected the composition of life forms of grazing and natural grassland vegetation and changed the community structure. It is consistent with the research results of Lv et al. (2016) and Zhao et al. (2008).
In this study, long-term ecological restoration altered soil bacterial and fungal communities and different microbial taxa responded unequally. The results stated that in all restoration gradients, the dominant phyla of bacteria and fungi were Actinobacteria, Proteobacteria and Acidobacteria, Mortierellomycota and Ascomycota, respectively, which was consistent with relevant research datas in Horqin Sandy Land (Wang et al., 2018; Zhang et al., 2020). This indicated that soil microbial community composition was relatively stable in sandy grassland with different restoration modes. The abundance of Proteobacteria and Acidobacteria in the restoration plot (NGR, LGR, HGR) was higher than that in the NM. These results indicated that ecological restoration promoted the bacterial transformation from slow growth type to fast growth type (Xun et al., 2018; Zheng et al., 2022). The reason was that vegetation regrew after enclosed-fencing, soil nutrient content increased, and nutrient-loving bacteria were promoted to grow stably and multiply rapidly. The relative abundance of Mortierellomycota in NGR and LGR were much greater than that in NM and HGR, while that value of Ascomycota in HGR were much greater than that in other plots. This is the same as the research results of Wang et al. (2019) on the short-term grazing ban (14 and 19 years) in semi-arid steppe, that is, the diversity of soil fungi becomes richer with the increase of the years of enclosure. The reason may be that the increase of vegetation biomass and coverage in the process of ecological restoration changes the rate of soil nutrient conversion and migration, and accelerates the demand for saprophytic nutrient fungi in grassland ecosystem (Zhang et al., 2013) (Figure 4).
Plant-soil microbial diversity
Ecological restoration measures are of great significance to the sustainable development of sandy grassland ecosystem. In this study, the results discovered that enclosure had a certain effect on aboveground plant species richness, allele diversity and belowground soil fungi and bacteria richness in sandy grassland (Figures 2, 3, 5). The species richness (SR) from NGR was significantly higher than that of other restoration plots (Figure 2), because the original habitat conditions of NGR are more superior than other plots, which is more conducive to plant growth and community construction. The overall trend was consistent with that the aboveground plant diversity (SR and AR) and belowground soil microbiology diversity (bacterial OTUS and fungal richness) with NM were significantly lower than those of other treatments (Figures 2, 3, 5). This is due to the poor original habitat conditions in NM lead to low species richness and soil microbiology diversity, and habitat fragmentation leads to reduced allelic richness. The soil bacterial OTUS in HGR was outstanding more than that of other treatments, possibly because severe grazing had positive and lasting effects on soil bacterial community. There were no significant differences in Simpson’s diversity index (D), expected heterozygosity (HE) and soil microbiology diversity values from NGR, LGR and HGR indicating that after years of restoration and conservation, the grazing grassland of different intensities in the early stage has been repaired well, and the habitat conditions are similar to the natural grassland in the same period.
Correlations in above-and belowground biodiversity
The relationship between above- and belowground biodiversity in Horqin sandy grassland community was significantly affected by long-term ecological restoration, but the trend was not consistent. The data detected a positive correlation between plant species diversity and genetic diversity parameters, which was supported by many similar studies (He et al., 2008; Crawford and Rudgers, 2013). In this study, SR was extremely significant vigorous correlated with AR, and was significantly vigorous correlated with HE, indicating that plant species richness determines the genetic differences of dominant species and plays a key role in the level of genetic diversity of a sandy grassland community. The results showed that there was a significant negative correlation between above- and belowground biodiversity in sandy grassland community, which suggested that plant species richness and genetic diversity index inhibited soil bacteria and fungi diversity in enclosure conditions (Figure 6).
Driving factors of above-and belowground biodiversity
Previous studies have shown that different management measures responded differently to biodiversity conservation (Lindenmayer et al., 2006; Kernecker et al., 2022). According to the SEM in the present study, long-term ecological restoration did active affect above- and belowground biodiversity directly in sandy grassland (Figure 7). However, biological and abiotic factors did inactive affect them directly. AGB was the main direct driving factor for SR. SAN and SOP were the main direct driving factor for AR. SAN was the main direct driving factor for SD. SOC was affected plant diversity through AGB and SAN, and was an indirect driver of plant diversity.
Conclusion
Understanding the relationships between above- and belowground biodiversity to long-term ecological restoration is crucial to developing biodiversity conservation measures in Horqin Sandy Land. The results emerged that: long-term ecological restoration had changed the above- and belowground community structure and composition of sandy grassland vegetation, had effected on plant species richness, allele diversity and soil fungi and bacteria richness in sandy grassland; it was a energetic correlation between plant species diversity and genetic diversity parameters, a significant passive correlation between above- and belowground biodiversity; long-term ecological restoration measure did positive affect above- and belowground biodiversity directly, biological and abiotic factors did negative affect them directly. In this study, the finding provide scientific basis for biodiversity conservation of sandy grassland ecosystem. In the future, the effects of above- and belowground biodiversity on ecosystem function of sandy land should be further analyzed, so as to provide effective support for the management and sustainable development of sandy grassland.
Data availability statement
The datasets presented in the study are deposited in the NCBI repository, accession number PRJNA985896, OR141062-OR141064, OR177791-OR177831, OR182494-OR182532, OR194037-OR194068.
Author contributions
WH designed the experiment, performed the laboratory analysis and wrote the manuscript. YH and HW conducted the field trial and analyzed the data. XZ performed the laboratory analysis. YZ analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (41971144) and China National Key Research and Development Plan (2017FY100205).
Acknowledgments
The authors thank all the members of Naiman Desertification Research Station, and Key Laboratory of Stress physiology and Ecology, Gansu Province, Northwest Institute of Eco-Environment and Resources, China Academy of Sciences (CAS), for their help in field work and laboratory studies. We also wish to express our appreciation to the reviewers for valuable comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai, Z. G., Dent, D. L., Olsson, L., and Schaepman, M. E. (2008). Proxy global assessment of land degradation. Soil Use Manag. 24, 223–234. doi:10.1111/j.1475-2743.2008.00169.x
Crawford, K. M., and Rudgers, J. A. (2013). Genetic diversity within a dominant plant outweighs plant species diversity in structuring an arthropod community. J. Ecol. 100, 1025–1035. doi:10.1890/12-1468.1
Deng, L., Kim, D. G., Peng, C. H., and Shangguan, Z. (2018). Controls of soil and aggregate associated organic carbon variations following natural vegetation restoration on the Loess Plateau in China. Land Degrad. Dev. 29, 3974–3984. doi:10.1002/ldr.3142
Edgar, R. C. (2013). Uparse: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 10, 996–998. doi:10.1038/nmeth.2604
Etienne, R. S. (2005). A new sampling formula for neutral biodiversity. Ecol. Lett. 8, 253–260. doi:10.1111/j.1461-0248.2004.00717.x
Evanno, G., Castella, E., Antoine, C., Paillat, G., and Goudet, J. (2009). Parallel changes in genetic diversity and species diversity following a natural disturbance. Mol. Ecol. 18, 1137–1144. doi:10.1111/j.1365-294X.2009.04102.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform 1, 117693430500100–117693430500150. doi:10.1177/117693430500100003
García-Díaz, A., Bienes, R., Sastre, B., Novara, N., Gristina, L., and Cerdà, A. (2017). Nitrogen losses in vineyards under different types of soil groundcover. A field runoff simulator approach in central Spain. Agric. Ecosyst. Environ. 236, 256–267. doi:10.1016/j.agee.2016.12.013
Hamilton, M. B. (1999). Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol. Ecol. 8, 521–523. doi:10.1046/j.1365-294X.1999.00510.x
He, T. H., Lamont, B. B., Krauss, S. L., Enright, N. J., and Miller, B. P. (2008). Covariation between intraspecific genetic diversity and species diversity within a plant functional group. J. Ecol. 96, 956–961. doi:10.1111/j.1365-2745.2008.01402.x
Hooper, D. U., Bignell, D. E., Brown, V. K., Brussaard, L., Dangerfield, M. J., Wall, D. H., et al. (2000). Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: Patterns, mechanisms, and feedbacks. Bioscience 50, 1049–1061. doi:10.1641/0006-3568(2000)050[1049:IBAABB]2.0.CO;2
Kernecker, M., Fienitz, M., Nendel, C., Ptzig, M., Walzl, K. P., Raatz, L., et al. (2022). Transition zones across agricultural field boundaries for integrated landscape research and management of biodiversity and yields. Ecol. Solutions Evid. 3 (1), e12122. doi:10.1002/2688-8319.12122
Korthals, G. W., Smilauer, P., Dijk, C., and Van Der Putten, W. H. (2001). Linking above- and below-ground biodiversity: Abundance and trophic complexity in soil as a response to experimental plant communities on abandoned arable land. Funct. Ecol. 15, 506–514. doi:10.1046/j.0269-8463.2001.00551.x
Lenka, N. K., Dass, A., Sudhishri, S., and Patnaik, U. S. (2012). Soil carbon sequestration and erosion control potential of hedgerows and grass filter strips in sloping agricultural lands of eastern India. Agr. Ecosyst. Environ. 158, 31–40. doi:10.1016/j.agee.2012.05.017
Li, F. R., Zhang, A. S., Duan, S. S., and Kang, L. F. (2005). Patterns of reproductive allocation in Artemisia halodendron inhabiting two contrasting habitats. Acta Oecol 28, 57–64. doi:10.1016/j.actao.2005.02.005
Li, F. R., Zhao, L. Y., Zhang, H., and Shirato, Y. (2004). Wind erosion and airborne dust deposition in farmland during spring in the Horqin Sandy Land of eastern Inner Mongolia, China. Soil Till Res. 75, 121–130. doi:10.1016/j.still.2003.08.001
Li, S. G., Harazono, Y., Oikawa, T., Zhao, H. L., He, Z. Y., and Xue, L. C. (2000). Grassland desertification by grazing and the resulting micrometeorological changes in Inner Mongolia. Agric For Meteorol 102, 125–137. doi:10.1016/S0168-1923(00)00101-5
Liang, W. J., and Wei, X. (2020). Relationships between ecosystems above and below ground including forest structure, herb diversity and soil properties in the mountainous area of Northern China. Glob. Ecol. Conserv. 24, e01228. doi:10.1016/j.gecco.2020.e01228
Lindenmayer, D. B., Franklin, J. F., and Fischer, J. (2006). General management principles and a checklist of strategies to guide forest biodiversity conservation. Biol. Conserv. 131 (3), 433–445. doi:10.1016/j.biocon.2006.02.019
Lv, P., Zuo, X. A., Zhang, J., Zhou, X., Lian, J., and Liu, L. X. (2016). Effects of grazing intensity on vegetation in sandy grassland of Horqin. J. Desert Res. 36 (1), 34–39. (in Chinese with English summary). doi:10.7522/j.issn.1000-694X.2015.00197
Magoč, T., and Salzberg, S. L. (2011). Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi:10.1093/bioinformatics/btr507
Nelson, D. W., and Sommers, L. E. (1982). “Total carbon, organic carbon and organic matter,” in Methods of soil analysis, Part 2 Editors A. L. Page, H. Miller, and D. R. Keeney second ed. (Madison: John Wiley).
Niu, Y. Y., Li, Y. Q., Wang, X. Y., Gong, X. W., Luo, Y. Q., and Tian, D. Y. (2018). Characteristics of annual variation in net carbon dioxide flux in a sandy grassland ecosystem during dry years. Acta Prataculturae Sin. 27, 215–221. (in Chinese with English summary). doi:10.11686/cyxb2017231
Olsen, S. R., Cole, C. W., Watanabe, F. S., and Dean, L. A. C. (1954). Estimation of available phosphorus in soil by extraction with sodium bicarbonate. Misc. Pap. Inst. Agicultural 939.
Porto, P., Walling, D. E., and Callegari, G. (2009). Investigating the effects of afforestation on soil erosion and sediment mobilisation in two small catchments in Southern Italy. Catena 79, 181–188. doi:10.1016/j.catena.2009.01.007
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi:10.1093/nar/gks1219
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi:10.2307/2446155
Stokes, A., Sotir, R., Chen, W., and Ghestem, M. (2010). Soil bio- and eco-engineering in China: Past experience and future priorities. Ecol. Eng. 36 (3), 247–257. doi:10.1016/j.ecoleng.2009.07.008
Su, Y. Z., Li, Y. L., and Zhao, H. L. (2006). Soil properties and their spatial pattern in a degraded sandy grassland under post-grazing restoration, Inner Mongolia, northern China. Biogeochemistry 79, 297–314. doi:10.1007/s10533-005-5273-1
Suding, K. N. (2011). Toward an era of restoration in ecology: Successes, failures, and opportunities ahead. Annu. Rev. Ecol. Evol. Syst. 42, 465–487. doi:10.1146/annurev-ecolsys-102710-145115
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi:10.1007/BF00037152
Walters, W., Hyde, E. R., Berglyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. Msystems 1 (1), e00009–e00015. doi:10.1128/mSystems.00009-15
Wang, S. K., Zuo, X. A., Zhao, X. Y., Awada, T., Luo, Y. Q., Li, Y. Q., et al. (2018). Dominant plant species shape soil bacterial community in semiarid sandy land of northern China. Ecol. Evol. 8, 1693–1704. doi:10.1002/ece3.3746
Wang, T. (2003). Desert and desertification in China. Shijiazhuang, China: Hebei Science and Technology Press.
Wang, Z., Zhang, Q., Christopher, S., Gao, H. L., Satoshi, I., Wei, X. R., et al. (2019). Impact of long-term grazing exclusion on soil microbial community composition and nutrient availability. Biol. Fert. Soils 55 (2), 121–134. doi:10.1007/s00374-018-01336-5
Xue, Z. J., Zhou, Z. C., and An, S. S. (2021). Changes in the soil microbial communities of different soil aggregations after vegetation restoration in a semiarid grassland, China. Soil Ecol. Lett. 3 (1), 6–21. doi:10.1007/s42832-020-0055-1
Xun, W. B., Yan, R. R., Ren, Y., Xiong, W., Zhang, G. S., Cui, Z. L., et al. (2018). Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome 6, 170. doi:10.1186/s40168-018-0544-y
Yuan, J., Wen, T., Zhang, H., Zhao, M. L., Pento, R., Thomashow, L. S., et al. (2020a). Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 14, 2936–2950. doi:10.1038/s41396-020-0720-5
Yuan, Z. Q., Ali, A., Ruiz-Benito, P., Jucker, T., Mori, A. S., Wang, S. P., et al. (2020b). Above- and below-ground biodiversity jointly regulate temperate forest multifunctionality along a local-scale environmental gradient. J. Ecol. 108, 2012–2024. doi:10.1111/1365-2745.13378
Zhang, J. Y., Gu, P. F., Li, L. Y., Zong, L. Y., and Zhao, W. J. (2016). Changes of soil particle size fraction along a chronosequence in sandy desertified land: A fundamental process for ecosystem succession and ecological restoration. J. Soil Sediments 16, 2651–2656. doi:10.1007/s11368-016-1454-x
Zhang, S. N., Yan, D. R., Huang, H. G., and Hu, X. L. (2020). Effects of short-term fencing on soil microbial community structure in Ulmus pumila scattered woodland of Horqin Sandy Land. Chin. J. Ecol. 39 (9), 2860–2867. doi:10.13292/j.1000-4890.202009.003
Zhang, X. F., Zhao, L., Xu, S. J., Liu, Y. Z., Liu, H. Y., and Cheng, G. D. (2013). Soil moisture effect on bacterial and fungal community in Beilu River (Tibetan Plateau) permafrost soils with different vegetation types. J. Appl. Microbiol. 114 (4), 1054–1065. doi:10.1111/jam.12106
Zhao, F. Z., Bai, L., Wang, J. Y., Deng, J., Ren, C. J., Han, X. H., et al. (2019). Change in soil bacterial community during secondary succession depend on plant and soil characteristics. Catena 173, 246–252. doi:10.1016/j.catena.2018.10.024
Zhao, H. L., Li, Y. Q., and Zhou, R. L. (2007). Effects of desertification on C and N storages in grassland ecosystem on Horqin sandy land. Chin. J. Appl. Ecol. 18, 2412–2417. doi:10.1360/yc-007-1324
Zhao, H. L., Okuro, T., Li, Y. L., Zuo, X. A., Huang, G., and Zhou, R. L. (2009). Changes of plant community in grazing and restoration processes in Horqin Sand Land, Inner Mongolia. J. Desert Res. 29 (2), 229–235.
Zhao, H. L., Okuro, T., Li, Y. L., Zuo, X. A., Huang, G., and Zhou, R. L. (2008). Effects of human activities and climate changes on plant diversity in Horqin sandy grassland, Inner Mongolia. Acta Prataculturae Sin. 17 (5), 1–8. doi:10.1007/s10499-007-9164-4
Zhao, H. L., Zhao, X. Y., and Zhang, T. H. (2003). Beijing. Desertification process and its recovery mechanism in Horqin sandy land. Beijing, China: Ocean Press.
Zhao, H. L., Zhao, X. Y., and Zhang, T. H. (2000). Causes, processes and countermeasures of desertification in the interlocked agro-pasturing area of North China. J. Desert Res. 20, 22–28. doi:10.7522/j.issn.1000-694X.2000.00227
Zhao, H. L., Zhao, X. Y., Zhang, T. H., and Zhou, R. L. (2002). Boundary line on agro-pasture zigzag zone in North China and its problems on eco-environment. Adv. Earth Sci. 17, 739–747. doi:10.1007/s11769-002-0041-9
Zhao, J. X., Luo, T. X., Li, R. C., Li, X., Tian, L. H., and Luo, T. X. (2016). Grazing effect on growing season ecosystem respiration and its temperature sensitivity in alpine grasslands along a large altitudinal gradient on the central Tibetan Plateau. Agric For Meteorol 218-219, 114–121. doi:10.1016/j.agrformet.2015.12.005
Zheng, J. H., Zhao, M. L., Wang, Q., Zhang, F., Zhang, B., and Zhang, J. (2022). Effects of management regime on soilmicrobial community structure and diversity of Stipa grandis grassland. Acta Ecol. Sin. 42 (12), 4998–5008. doi:10.5846/stxb202103290818
Zhou, H., Gao, Y., Jia, X. H., Wang, M. M., Ding, J. J., Cheng, L., et al. (2020). Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol. Biochem. 144, 107782. doi:10.1016/j.soilbio.2020.107782
Appendix S1: Primers sequence in selected plant and soil.
Keywords: sandy grassland, long-term ecological restoration, aboveground diversity, belowground diversity, structural equation model
Citation: Huang W, He Y, Wang H, Zhao X and Zhu Y (2023) Long-term effects of restoration on the links between above-and belowground biodiversity in degraded Horqin sandy grassland, Northern China. Front. Environ. Sci. 11:1128187. doi: 10.3389/fenvs.2023.1128187
Received: 20 December 2022; Accepted: 05 June 2023;
Published: 20 July 2023.
Edited by:
Stephen M. Mureithi, University of Nairobi, KenyaReviewed by:
Vincent Kathumo, University of Nairobi, KenyaXiaojing Qin, Henan Polytechnic University, China
Copyright © 2023 Huang, He, Wang, Zhao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenda Huang, aHVhbmd3ZW5kYTIwMDhAMTYzLmNvbQ==
 Wenda Huang
Wenda Huang Yuanzheng He1,3
Yuanzheng He1,3