- 1Department of Biotechnology, Guru Ghasidas Vishwavidyalaya (A Central University), Bilaspur, Chhattisgarh, India
- 2Department of Botany, Shaheed Mahendra Karma Vishwavidyalaya, Bastar, Chhattisgarh, India
- 3Department of Chemistry, National Institute of Technology, Jamshedpur, Jharkhand, India
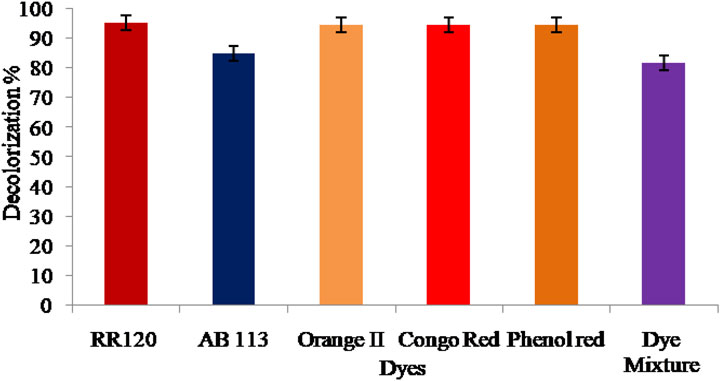
Currently, the textile industry is among the most rapidly expanding areas of the economy and is an important source of water pollution. There are many efficient chemical and physical methods for treating textile effluents, but they produce secondary pollutants. Therefore, there is a need to manage textile wastewater. The potential Lysinibacillus capsici bacterial strain has been isolated from the bark borer insect tunnel of the Peltophorum pterocarpum plant and has been determined to be effective in >95% decolorization of Reactive Red 120 (RR120) and other azo dyes, such as AB-113 (85%), orange II (94.62%), Congo red (94.62%), phenol red (94.54%), and mixtures of azo dyes (81.66%). Physico-chemical factors were optimized manually, including Taguchi design. Fabric discolorations by L. capsici was qualitatively studied. FT-IR, GC–MS, and UV absorbance studies also confirmed that the dye had been broken down into its amines. Research findings using enzyme assays have shown that the bacteria Lysinibacillus capsici can utilize laccase and manganese peroxidase and are capable of degrading dyes significantly. According to this work, immobilized L. capsici cells and the studied four bacterial consortia, namely, Lysinibacillus capsici, Alcaligenes faecalis subsp. phenolicus, Acinetobacter baumannii, and Pseudomonas aeruginosa, may be used to degrade RR120 effectively, and it is concluded that L. capsici is significantly efficient in textile effluent treatment.
Introduction
Color helps make the world more beautiful and colorful (Ayele et al., 2021). Dye is an organic chemical substance that is bright, ionizing, and aromatic (Samykannu, 2025). Synthetic dyes cause serious environmental pollution (Carrascal-Hernández et al., 2025). In the textile sector, mostly azo dyes are used to dye clothes, and due to their capacity for oxidative breakdown, synthetic dyes persist in the environment for a long duration. Even in low quantities, the manufacturing of synthetic dyes has the potential to be dangerous, carcinogenic, and mutagenic. Common uses for dyes are in paper, textiles, leather, rubber, plastics, cosmetics, tannery, printing and dyeing, and food. Humans and nature have been exposed to synthetic toxic dyes for a few million years. Every year, an estimated 700,000 tons of synthetic dyes are produced in approximately 100,000 types (Rehaman et al., 2021). Azo dyes are the most commonly used category of dyes. Azo dyes are mainly utilized in the textile industry (Florencio et al., 2021). An azo bond (–N=N–) is present in azo dyes and is related to the class of heterocyclic aromatic compounds (Carolin et al., 2021). Recently, more than 2,000 azo dyes have been used in various industries, among which the textile coloring industry is the largest.
Earlier studies have reported that, only 10% of dyes bind to the fabrics, while 90% are released into water resources, which is the root cause of various skin problems (Kour et al., 2023; Lata et al., 2024a). Water pollution adversely affects aquatic life and ecological balance, increases chemical and biological oxygen demand (BOD), and alters the pH and chemical composition of the water body; these are just a few of the detrimental effects of releasing textile effluent into the aqueous ecosystem (Lata et al., 2024a; Sreedharan et al., 2021). The entry of dye-contaminated wastewater into the soil harms the soil nutrients and microbial communities (Ayele et al., 2021; Lalnunhlimi and Krishnaswamy, 2016), which are directly and indirectly associated with soil health and crop productivity. In addition, synthetic dyes decrease dissolved oxygen (DO) levels, leading to many harmful effects on marine flora and fauna.
The most commonly used physicochemical techniques for treating textile wastewater in the world are flocculation and coagulation. However, many problems, including the use of chemicals, secondary contamination, large sludge production, low efficiency, and high operating costs, deter the use of these techniques. As an alternative, the more advanced technique known as bioremediation, which uses the capabilities of fungi, bacteria, or their combination, has shown potential in the treatment of textile wastewater (Sonal and Mishra, 2021).
Sometimes, the compounds formed during aerobic degradation of azo dyes are more toxic than the parent azo dyes. These are usually recognized as aromatic amines. As a result, their degradation requires an additional aerobic stage. It has been observed that oxidative enzymes function better under aerobic conditions and allow potential decolorization and additional detoxification of produced amines, whereas reductive enzymes break down the azo link of azo dyes under aerobic conditions. Consequently, azo dye degradation by successive aerobic processes emerged as a sustainable approach for textile effluent treatment (Lade et al., 2015).
An alternate technology that has recently gained popularity among these methods is the immobilization of microbes in a polymeric matrix, such as sodium alginate (SA). SA is the sodium equivalent of alginic acid, a polymer that is widely distributed along its main chain with free hydroxyl and carboxyl groups. Its non-toxicity, biodegradability, and environmental friendliness are its key benefits. Alginate beads have been shown in many studies to significantly improve pollutant removal in treatment plants (Bustos-Terrones et al., 2022). According to Martinez et al. (2009), residential wastewater contains nutrients and organic matter that can be broken down by SA-immobilized microorganisms while producing less sludge. Other benefits of immobilized beads included a shorter hydraulic retention time, a reduction in the loss of suspended bacteria, and a greater ability to withstand environmental stress. Microorganisms can enter a resting phase in which their cells remain viable but are not cultivable when exposed to harmful contaminants (Martinez et al., 2009).
Different microorganisms in bioreactors are used for the decolorization of textile dyes (Chaturvedi et al., 2021). The most cost-effective and environmentally friendly technique for managing dye-containing wastewater is microbial degradation. The bacterial aerobic degradation pathway breaks down the aromatic ring of azo dyes via monooxygenase or dioxygenase, which is then further degraded and mineralized (Chen et al., 2021). According to Dixit and Garg (2021), Klebsiella pneumoniae has the potential to degrade toxic azo dyes. Previous studies showed that methyl red and MG can be degraded by the Klebsiella pneumoniae strain WA-1 (Dixit and Garg, 2021). Recently, it has been reported that various Bacillus species have the capability to decolorize textile wastewater. B. Cohnii RAPT1 significantly decolorizes Reactive Red 120 (RR120) (Guembri et al., 2021). However, to date, only a limited number of aerobic and anaerobic bacterial species have been reported to degrade azo dyes.
Wastewater from textile dyes usually contains a large number of heavy metals. Human beings, plants, and microorganisms are all extremely sensitive to a variety of heavy metals. Among the several ions of heavy metals and hexavalent compounds [Cr (VI)], some are commonly present with azo dyes in the effluents of the leather and textile industries (Hussain et al., 2020). Shewanella decolorization is caused by MBTD16, Paenibacillus glucanolyticus SLM1, and Bacillus amyloliquefaciens, which are examples of ligninolytic bacteria that remove dyes from textile wastewater. These microorganisms represent oxidative enzymes such as lignin peroxidases and laccases. Oxidative enzymes such as laccases, tyrosinases, veratryl alcohol oxidases, peroxidases, and azoreductases biodegrade dyes in wastewater. Mixed bacterial cultures decolorize dye molecules faster and more completely than single cultures because they contain several enzymes that attack the dye molecule at many sites (Kamal et al., 2021).
The main research objectives were to (a) isolate plant growth-promoting rhizobacteria (PGPR) from bark borer insect tunnels of the Peltophorum pterocarpum plant for dye degradation, (b) optimize the effects of different nutritional and physio-chemical factors, (c) develop a consortium for dye decolorization, (d) decolorize dyes using immobilized bacteria cells, (e) quantitatively observe azo dye decolorization, (f) qualitatively investigate fabric decolorization and dye decolorization in the presence of metal ions, and (g) analyze metabolites using FT-IR and GC–MS.
Materials and methods
Sample collection and bacterial culture
The standard sample RR120 dye was procured from Sigma-Aldrich, Begaluru, Karnataka, India. All the chemicals, salts, acids, buffers, sugars (carbon source), and nitrogen source were procured from HiMedia, Wagle Industrial Estate, Thane (West) Maharashtra, India. Bacteria were isolated from the bark borer insect tunnel of the Peltophorum pterocarpum plant grown at Guru Ghasidas Vishwavidyalaya, Bilaspur, Chhattisgarh, India.
To study the bio-decolorization and biodegradation of RR120 dyes, the Bushnell Hass medium (BHM) was prepared with the following composition of BHM broth: (gl−1) K2HPO4 1.0, MgSO4 0.2, FeCl3 0.05, CaCl2 0.02, and NH4NO3 1.0, supplemented with yeast extract (0.2% w/v), glucose (0.4% w/v), and 1% RR120 dye. To isolate microorganisms that decolorize dyes, 1 g of the sample was placed in tubes, and 3 g of the sample was placed individually in a flask containing Reactive Red 120 dye–BHM broth and incubated for 3–5 days under static and shaking conditions at 35 °C. Three consecutive transfers were made in a fresh dye-containing medium until the medium became decolorized. The decolorized samples were serially diluted (10-fold), and 100 µL aliquots of 10−5 to 10−7 dilution were spread and poured onto BHM agar plates containing 100 mg L−1 Reactive Red dye and subsequently incubated for 24–48 h at 35 °C. After incubation, the colonies emerged and were repeatedly streaked to obtain pure cultures. Colony and culture morphology were used as the basis for the primary screening of the isolated bacteria. In secondary screening, the culture was incubated in BHM broth at 35 °C for 24 h. One milliliter of bacterial suspension was taken from the incubated medium and transferred to 30 mL of BHM broth containing the dye; it was incubated under static conditions at 35 °C for 72 h, and absorbance was measured at 12-h intervals. The culture having >95% decolorization with RR120 dye within 12 h was finally represented by four bacterial strains: WR-1 and WR-2 (isolated from the bark borer sample) and TS-1 and TS-3 (isolated from the termite mound soil sample). Secondary and final screening of these cultures was carried out following primary screening, based on their rapid and high degradation efficiency of RR120 dye.
Identification of RR120 dye-decolorizing bacteria
Genomic DNA of four bacterial isolates, WR-1, WR-2, TS-1, and TS-3, was extracted using the phenol/chloroform extraction method. After that, the 16S rRNA gene was amplified by PCR using the universal primers 16F27 [5′-CCA GAG TTT GAT CMT GGC TCA G-3'] and 16R1492 [5′-TAC GGY TAC CTT GTT ACG ACT T-3']. The 16S rRNA gene PCR product was amplified and purified using PEG-NaCl precipitation and then sequenced immediately on an ABI® 3730XL automated DNA sequencer (Applied Biosystems, Inc., Foster City, CA, United States) at the NCMR sequencing laboratory of the National Centre for Cell Science in Pune, Maharashtra, India, according to the manufacturer’s instructions. The Lasergene package was used for assembly, while the EzBioCloud database was used for identification. The neighbor-joining tree method was used in BLAST and MEGA 11 software to generate a phylogenetic tree (Figure 1).
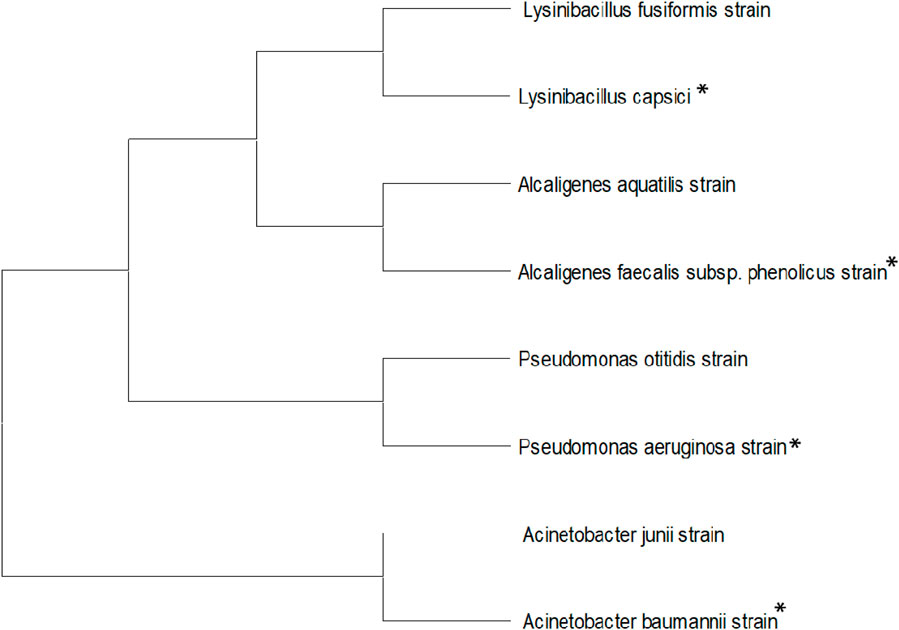
Figure 1. Neighbor-joining tree for identified bacterial isolates, including L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa.
Decolorization studies
Four identified bacterial isolates, TS-1, BOS-24, WR-1, and WR-2, were individually inoculated in BHM broth (gl−1) supplemented with yeast extract (0.2% w/v), glucose (0.4% w/v), and 1% RR120 dye; furthermore, the culture tubes were kept under static conditions at 37 °C for 72 h for decolorization. A similar procedure was applied to the control sample (with dye but without inoculation) and the blank sample (without dye and inoculation). To study the effects of aeration, inoculum size, and incubation time, 5 mL aliquots of the decolorized and control samples were drawn at regular 12-h intervals, and the pellets were removed through centrifugation at 4 °C for 10 min at 10,000 rpm. RR120 decolorization was studied using the supernatant and was determined by measuring absorbance at 524 nm using a UV–VS spectrophotometer. Decolorization % was determined using the formula given below (Ayed et al., 2022; Lata et al., 2024b).
Aeration, inoculum size, and time for dye decolorization
Several physical and chemical variables directly impact the decolorization ability of microorganisms during biodegradation processes (Sreedharan et al., 2021). Therefore, to improve the rate of dye decolorization, decrease the time required for decolorization, and increase the process affordability, various physicochemical parameters were optimized.
Fresh bacterial cultures were cultivated in BHM broth and incubated at 35 °C–37 °C for 24 h under anaerobic conditions. One milliliter of bacterial inoculum was cultivated in 30 mL screw-cap tubes containing BHM broth supplemented with 1% RR120 dye and incubated at 35 °C–37 °C for 72 h under shaking (aerobic) conditions. Then, 1 mL of culture was added to a 100-mL Erlenmeyer flask containing 40 mL of BHM broth and incubated for 72 h in an incubator shaker at 120 rpm and 35 °C–37 °C. The absorbance of the culture-inoculated sample and control was measured every 12 h. The decolorization potential of the WR-1 strain was tested under shaking and static conditions at the optimum pH of 7.0. Cell growth was measured as the optical density of the culture at 600 nm (Ayed et al., 2022; Telke et al., 2008). The impact of different inoculum percentages—1% (0.3 mL), 2% (0.6 mL), 4% (1.5 mL), 6% (1.6 mL), 8% (2.4 mL), and 10% (3 mL)—for decolorization of RR 120 dye was studied under anoxic static conditions at 35 °C. Various concentrations of inoculum were added to the 30 mL screw-cap tubes, and BHM broth was supplemented with 1% RR120 dye. The effect of various incubation times on the decolorization of Reactive Red 120 was studied under anaerobic static conditions at 35 °C and pH 7. One milliliter inoculum was added to the BHM medium supplemented with 1% RR120 dye and incubated at 35 °C for 72 h under static conditions.
Optimization using Taguchi methods
The Taguchi approach for optimization of the pH, temperature, carbon source, nitrogen source, and dye concentration parameters was followed according to Pourbabaee et al. (2013). The factors were optimized using the Taguchi method. The criteria were selected according to their effect on the biodegradation of RR120 dye. The experiments were designed using the following five variables: carbon sources (glucose, fructose, sucrose, lactose, and starch) and nitrogen sources (yeast, peptone, ammonium sulfate, ammonium nitrate, and urea). Nitrogen sources were added to the BHM culture media. The BHM media with different ranges of pH 4, 5, 6, 7, and 8 and different incubation temperatures of 25 °C–45 °C were used. Dye concentrations of 0.1, 0.2, 0.3, 0.4, and 0.5 mg/L were added to the BHM culture media (Pourbabaee et al., 2013). The experiments were set up using the L25 orthogonal array after selecting the factors in the mixed-level design. Minitab 19 statistical software was used for data analysis (Marvi-Mashhadi et al., 2021).
Development of consortium
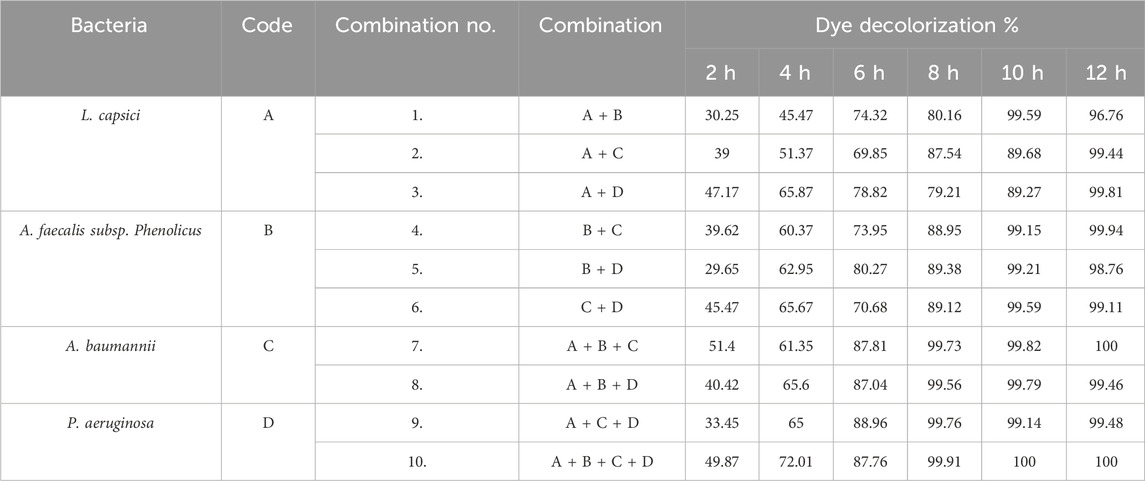
Compatibility tests of L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa were carried out using the agar well diffusion method. All four bacterial cultures were individually cultivated aerobically in BHM broth and incubated at 35 °C–37 °C for 24 h under static conditions. These bacterial cultures were prepared by mixing them into 10 combinations. These cultures were cultivated anaerobically in 30 mL of BHM broth with 3% RR120 dye-containing medium inoculated according to culture combination and incubated for 12 h at 35 °C–37 °C under static conditions (Thiruppathi et al., 2021). The decolorization percentage of RR120 dye was analyzed using a UV spectrophotometer by calculating the absorbance of the supernatant at 524 nm. Finally, comparative decolorization percentages by L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa bacterial monocultures and the consortium were recorded. The bacterial combination that showed the highest dye degradation percentage in the shortest time was used as the best combination for further analysis.
Immobilization of dye-degrading bacteria
L. capsici was inoculated in an LB liquid medium and incubated for 24 h at 35 °C. After incubation, 5 mL of medium from well-grown bacterial culture media was used as the bacterial source. SA was used as a bio-carrier in this study. As the stability of SA in water is very low, a well-known synthetic polymer polyvinyl alcohol (PVA) was used in immobilization (Hameed and Ismail, 2018; Ibrahim et al., 2024). Sterile 3% SA and 3% PVA were added. A 5 mL alginate–alcohol solution was added to 5 mL of bacterial solution, and this SA–PVA–bacteria mixture was then dropped, drop by drop using a syringe, into a cold 3% CaCl2 solution. The beads–CaCl2 solution was kept on a shaker for 1 h and then stored in a freezer at 4 °C for 24 h. Beads (0.5 mm-sized 10 beads) were used for dye decolorization; however, first, the beads were washed using double-distilled water after 24 h (Kathiravan et al., 2014). The immobilized bacterial beads were washed several times using a phosphate buffer (pH 7) and then added to 30 mL BHM dye-containing broth and incubated for 12 h at 35 °C under static conditions. Cell-free immobilized beads were added to the control (BHM broth with dye) and the blank sample (BHM broth without dye) and incubated under the same conditions. After 12 h of incubation, the decolorized sample absorbance was measured, and the beads were collected. The collected beads were washed three times with phosphate buffer, placed in fresh BHM broth, and incubated under the same conditions. The same procedures were repeated six times.
Decolorization of various azo dyes and mixture of dyes
The optimized parameters were used for the decolorization of various azo dyes, viz., RR120, AB-113, Orange II, CR, PR, and mixed azo dyes. These dyes were mixed at a concentration of 20 mgl−1 each in BHM broth. The BHM broth was transferred to 30 mL tubes, and bacterial cultures were inoculated and then incubated under the above-optimized conditions. After incubation, the decolorization percentage was analyzed using a UV spectrophotometer by calculating the absorbance of the supernatant at λmax of the respective azo dyes and a mixture of dyes.
Dye decolorization in the presence of metal ions
Decolorization of RR120 (50 mg/L) and the heavy metals Cu, Pd, Zn, Ni, and Cd (1 mg/L to 6 mg/L) was simultaneously examined. The absorbance of the test and control media samples was measured after incubation (24 h) to determine the percentage of dye decolorization when heavy metals were present at varying concentrations. Using the previously mentioned technique, the azo dye’s decolorization efficacy was determined. Every experiment was conducted in triplicate.
Decolorization of textile fabric
Commercial 100% cotton two-set fabric (red and blue fabric), purchased from the local textile market in Bilaspur, Chhattisgarh, India, was cut into small, equal-sized pieces. The fabric pieces were packed well and sterilized in an autoclave. Sterilized red and blue fabric pieces were placed in separate tubes containing sterilized BHM broth under aseptic conditions. A 1% inoculum of the bacterial culture was added to the test sample tubes, while the control media contained no culture. All tubes were incubated at 35 °C–37 °C under anaerobic and static conditions for 5 days. The fabric was removed from every tube, washed three times with phosphate buffer, and then dried at room temperature (Kaur et al., 2023).
Decolorization and biodegradation analysis
The decolorization and biodegradation of RR120 were studied under static conditions using FT-IR, UV–visible, and GC–MS. During decolorization, the azo bond of the chromophore conjugated in RR120 was degraded due to azoreductase activity, and the peak gradually reduced and fully disappeared within 12 h (Patel and Bhatt, 2015). For this, 30 mL of BHM broth with dye was sampled after 12 h of decolorization and then centrifuged at 10,000 × g for 10 min at 4 °C to remove pellets. After centrifugation and dye extraction, metabolites were recovered from the supernatant using an equal amount of ethyl acetate. The extracted metabolite was dried over anhydrous Na2SO4 in an oven. During UV–Vis spectral analysis, changes in the absorption spectrum in the decolorized medium (400–800 nm) were recorded compared with the decolorization percentage outcomes from the control sample. The biodegraded RR120 was evaluated and compared with the control dye using Fourier-transform infrared spectroscopy (Lata et al., 2024c).
UV–Vis spectroscopic study
Decolorization was confirmed via UV–Vis spectroscopy. UV–Vis analysis was carried out in dye decolorization conformation, in which 5 mL of incubated decolorized media, control media, and blank media were taken in centrifuge tubes and centrifuged for 10 min at 10,000 rpm and 4 °C. After removing the cell debris, the absorbance of the supernatant in the centrifuged medium was measured at 524 nm (Vinayak and Singh, 2022).
Analysis of dye metabolites using FT-IR and GC–MS
Dried extracted dye metabolites were mixed with KBr in a ratio of 5:95. Mixed samples were pressed using a hydraulic press to obtain transparent pellets. Pellets were used to obtain FT-IR spectra in the region between 4,000 and 400 cm−1. The acquired sample spectra were compared between the range of dye metabolites treated with bacteria and the untreated dye control (Singh and Dwivedi, 2020). Spectral library software NIST1.10 (Shimadzu, Nakagyo-ku, Kyoto, Japan) was used to confirm and match the molecular weight, retention times, molecular formula, and peak area and height of the metabolites. A GC–MS study of dye-degraded metabolites was carried out using a GCMS-TQ8050 NX (Shimadzu). The initial column temperature was 80 °C with a 2-min hold and then increased linearly at 10 °C per minute to 100 °C, with a 2-min hold. The temperature of the injection port was 230 °C, and the GC–MS interface was maintained at 280 °C. The injector temperature was maintained at 280 °C. The compounds were identified based on mass spectra using the NIST library (Abd El-Rahim et al., 2021).
Enzyme assay
Bacterial cultures were grown in LB broth for 24 h, and after growth, the culture solution was centrifuged at 10,000 rpm for 10 min at 4 °C. The pellets were washed three times and suspended in 5 mL of 50 mM potassium phosphate buffer at pH 7.0, and the bacterial cells were disrupted by centrifugation at 10,000 ×g for 5 min at 4 °C. Cell-free supernatants (crude cell extract) were used as a reaction mixture for screening laccase and manganese peroxidase (Thanavel et al., 2020). To screen for laccase enzyme activity, 0.5 mL of guaiacol was added to 0.5 mL of the reaction mixture as a substrate and then incubated for 5 min; the enzyme activity was assessed based on the color change in the control and test suspensions. To evaluate the presence of the manganese peroxidase enzyme, 0.5 mL of 50 mM sodium tartrate buffer (pH 5) and 0.5 mL of 20 mM guaiacol were added to the 0.5 mL reaction mixture and incubated for 5 min. Finally, 0.5 mL of 2 mM H2O2 was added, and the brownish-red color changed. The presence of manganese peroxidase enzyme was confirmed based on the color change in the control and test suspensions.
Result
Identification of RR120 dye-decolorizing bacteria
RR120 dye-decolorizing potent bacterial strains were identified according to biochemical and morphological characteristics and the 16S rRNA gene sequence. Four bacterial strains, namely, Lysinibacillus capsici PB300 (T), Alcaligenes faecalis subsp. Phenolicus DSM 16503 (T), Acinetobacter baumannii ATCC 19606 (T), and Pseudomonas aeruginosa JCM 5962 (T), were identified using the 16S rRNA gene sequence. L. capsici, A. faecalis subsp. Phenolicus, strains isolated from bark borer samples, A. baumannii, and P. aeruginosa were isolated from termite-mound soil. L. capsici (accession no 13606), A. faecalis subsp. Phenolicus (accession no 13436), A. baumannii (accession no 13438), and P. aeruginosa (accession no 13437) bacterial strains were deposited in the Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh, Panjab, India.
Decolorization studies
Among four bacterial isolates, Lysinibacillus capsici showed the highest decolorization percentage (Figure 2). It showed >95% decolorization within 12 h at a dye concentration of 100 mg L−1, pH 7–8, and temperature of 35 °C–37 °C under static conditions.
Optimization of various parameters for the decolorization of RR120 dye
Effects of aeration, incubation time, and inoculum size
Under the anoxic static and shaking conditions, between 12 and 72 h, 90% and 83.33% decolorization and cell growth were observed; however, under the shaking condition, cell growth (Figure 3a) was much greater than under the static condition, but decolorization was lower (less than 80%). In the growth stage of the cell, oxygen has a major impact on its physiological properties. Oxygen can affect the rate at which microbes degrade azo dyes. The findings demonstrated that RR120 required more than 12 h under static conditions and 72 h under shaking conditions to fully decolorize (Figure 3b). Incubation time has a significant impact on the growth and enzymatic activity of microorganisms, which, in turn, affects their optimal decolorization performance. Following a 12-h incubation period, a significant amount of decolorization was observed over the temperature range of 35 °C–37 °C. After achieving more than 99 percent decolorization in 12 h, the experiment was repeated to study the effect of time, with decolorization measured every 2 h. Figure 3b shows >95% decolorization between 12 and 72 h. An increase in the inoculum size was correlated with an increased rate of decolorization in a limited period. Over 95% of the decolorization occurred in 12 h at a concentration of 1%–10% inoculum, which yielded the highest decolorization. Figure 3c shows that inoculum size significantly affected decolorization. Observation showed that a 1% inoculum size was effective for full decolorization in 12 h. Faster and greater maximum decolorization was observed with an increase in inoculum size.
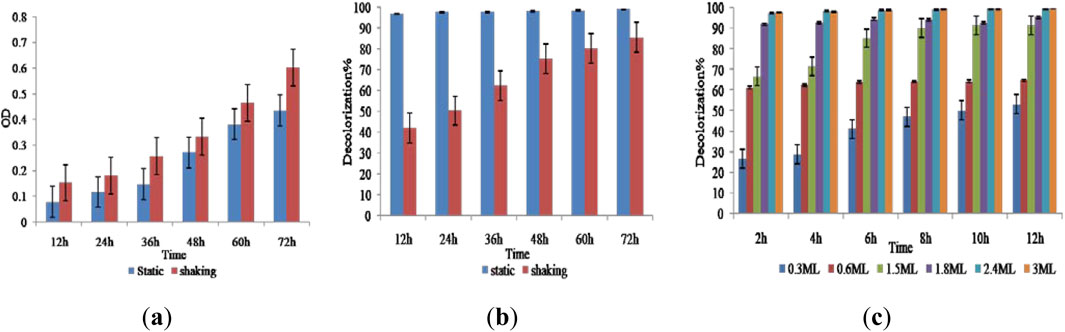
Figure 3. (a) Effect of cell growth, (b) aeration and incubation time (static and shaking), and (c) inoculum size on the decolorization of RR120 dye.
Taguchi statistical analysis
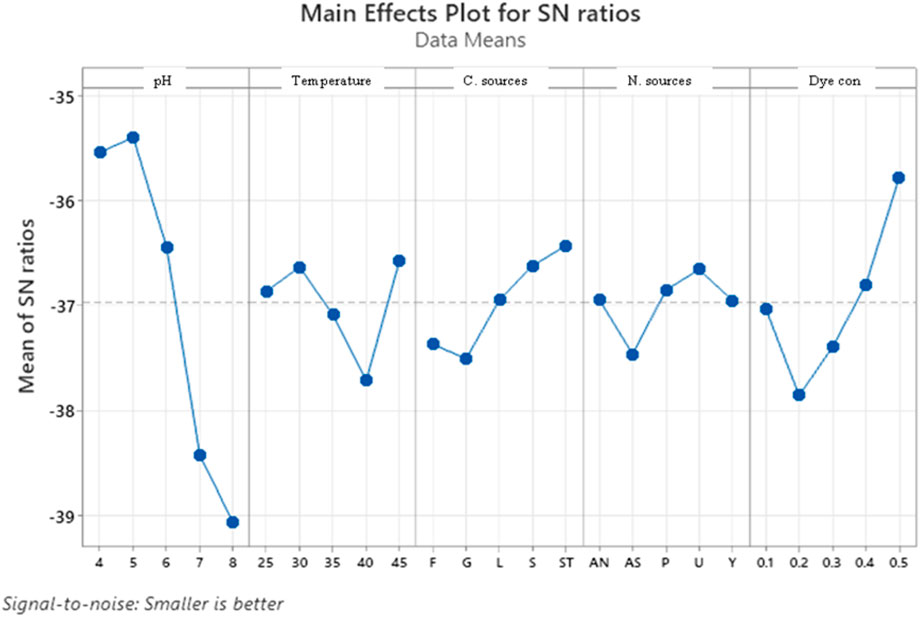
The Taguchi orthogonal array was used to determine the best operating settings, having a beneficial impact on the best combination of several parameter sets (Table 1). Since the average mean response for each level of each factor was estimated first, the delta value for each control factor was also calculated. After that, estimates of the control factors were made using their delta values. In the Taguchi technique, the terms “signal” and “noise” refer to the desired and unwanted values, respectively, for the output characteristic. The Taguchi technique uses the S/N ratio to quantify the quality characteristic that is different from the desired value. According to the type of characteristic, the S/N ratios change. The factor’s importance and the delta have a positive correlation with RR120 biodegradation. Hence, the results showed (Table 2) that pH (delta 30.5, rank 1) has the highest level of biodegradation of RR120 dye, followed by dye concentration (delta 15.55, rank 2), temperature (delta 8.4, rank 3), carbon source (delta 7.95, rank 4), and nitrogen source (delta 5.41, rank 5). It was easy to find the best levels of the five control parameters now in levels because Taguchi is adaptable, which was then varied, and the mean response for each run in the Taguchi array was calculated. As a result, process variability was minimized by reducing the influence of noise or uncontrollable factors during operation. A high S/N ratio indicates that the control factors perform as efficiently as possible and that the noise factors have minimal impact. Hence, the results showed (Table 2 S/N Ratios) that pH (delta 3.68, rank 1) has the highest level of biodegradation of RR120 dye, followed by dye concentration (delta 2.08, rank 2), temperature (delta 1.15, rank 3), carbon source (delta 1.08, rank 4), and nitrogen source (delta 0.81, rank 5). Additionally, the Taguchi technique identified the optimal level for each of the five factors within the study, which changed accordingly. The biodegradation level of RR120 was determined to be 98.67%, and run No. L18 in the Taguchi array was reported to have recovered (Marvi-Mashhadi et al., 2021). Based on the findings presented in Figures 4, 5, the optimal effective parameters were pH 8, temperature 40 °C, dye concentration 0.2 mg/L, ammonium nitrate as the nitrogen source, and glucose as the carbon source.
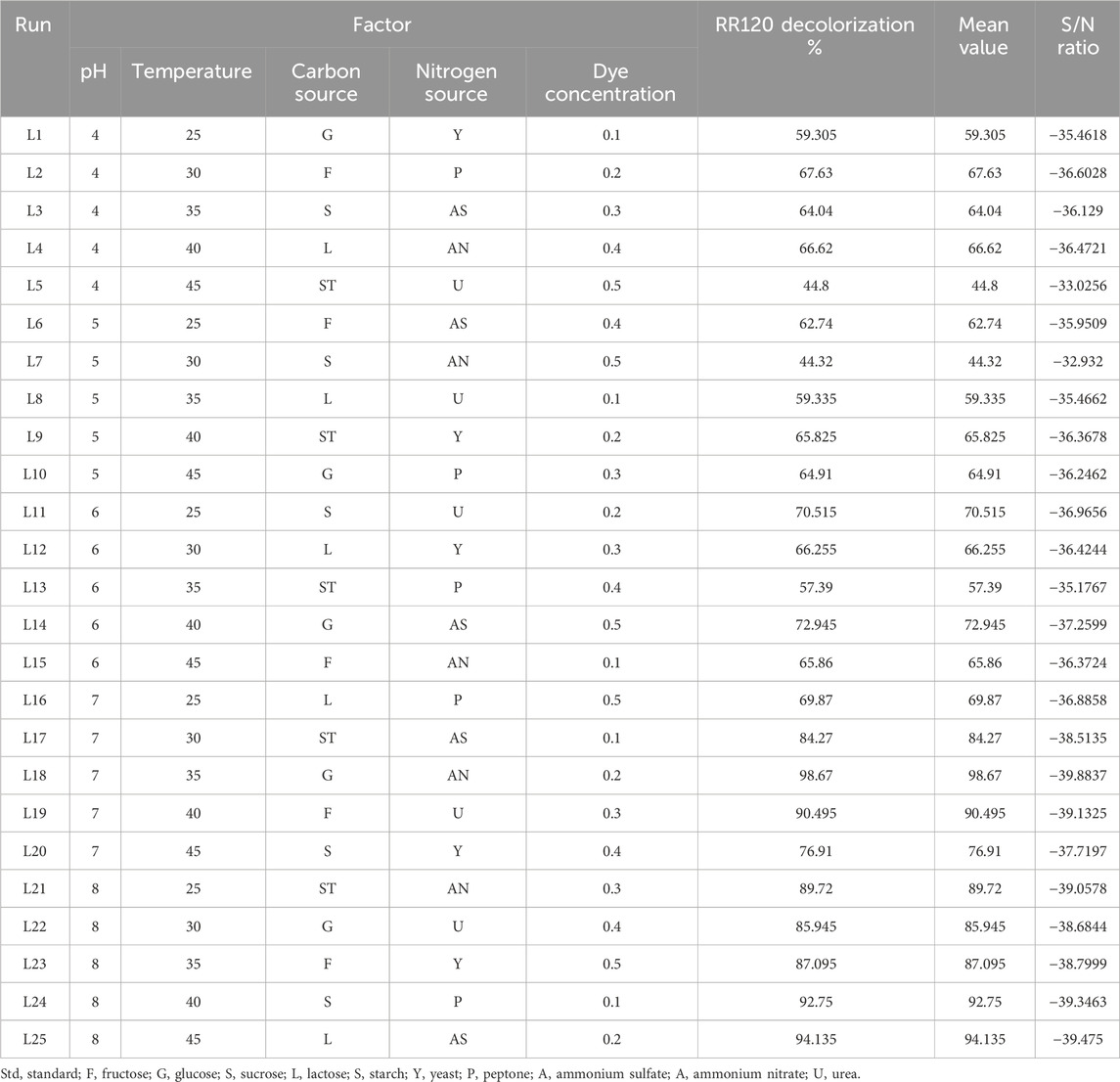
Table 1. Taguchi’s L25 orthogonal array, decolorization percentage, mean value, and S/N ratio for the optimization of RR120 decolorization using the L. capsici bacterial strain.
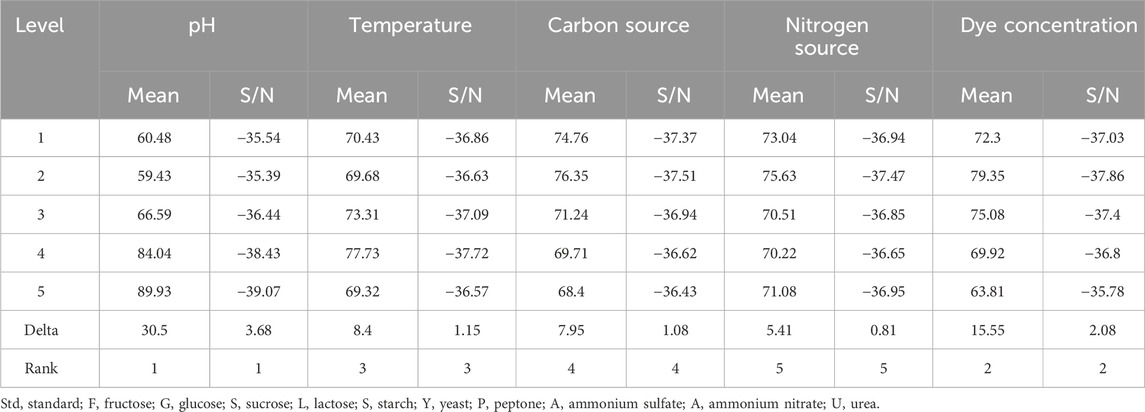
Table 2. Mean response and signal-to-noise ratio calculation of Taguchi data for the optimization of the physical and nutritional factors and their highest levels of RR120 dye decolorization by the L. capsici.
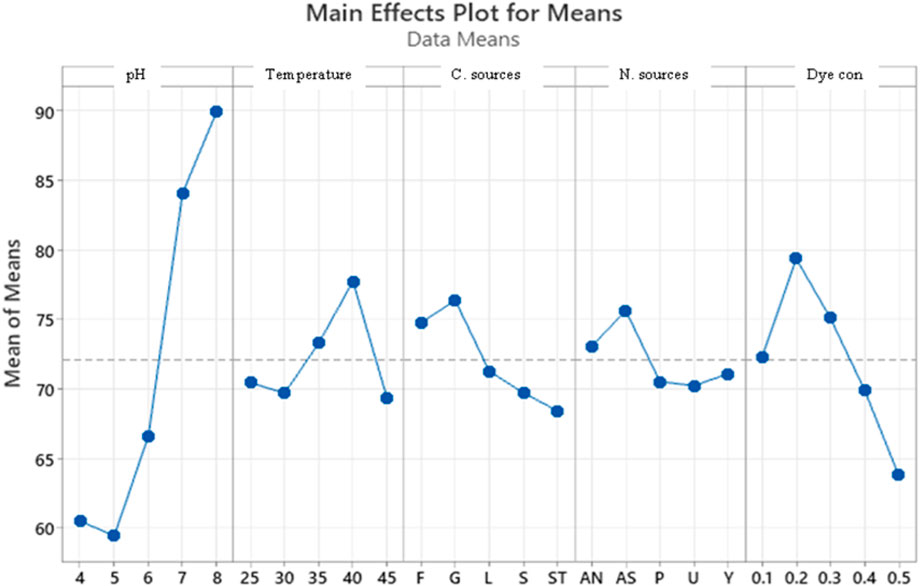
Figure 4. Main effects plot for mean response. Std, standard; F, fructose; G, glucose; S, sucrose; L, lactose; S, starch; Y, yeast; P, peptone; A, ammonium sulfate; A, ammonium nitrate; U, urea.
Dye decolorization by the consortium
For decolorization of dyes, the selected consortia consisted of four bacterial strains, namely, L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa. All four bacterial strains survived in the consortium. These four identified bacteria were selected individually, which can degrade >95% of the RR120 dye at a pH of 7–8 and 35 °C–40 °C within 12 h of incubation. Selected bacterial strains were mixed in 10 combinations, and their degradation ability in the RR120 azo dye was studied (Figure 6). According to the consortium results, complete degradation of the dye by the consortium occurred in less than 12 h, compared to the monoculture. The details of the developed consortia and the results for degradation by a consortium of dyes are provided in Table 3. The RR120 dye bacterial degradation was confirmed using UV–visible spectroscopy of samples inserted with bacterial consortia and untreated control samples. Methyl red and carbon fuchsin dyes showed decolorization percentages of 100% and 96%, respectively, indicating a greater decolorization capacity over their isolates in a consortium of three bacteria (Acinetobacter spp., Proteus spp., and Pseudomonas spp.). At pH 7 and a temperature of 40 °C, a two-bacterial consortium (Galactomyces geotrichum and B. laterosporus) suggested a decolorization percentage of 92% for Remazole Red 198 dye; in contrast, the individual bacteria demonstrated only 58% and 42% decolorization, respectively (Si et al., 2013).
Immobilization of dye-degrading L. capsici bacteria
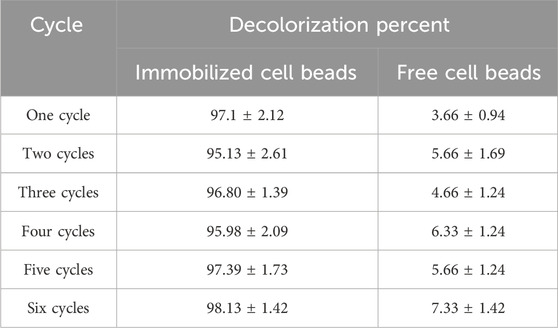
The dye decolorization and reusability capacity of immobilized beads were analyzed using a repeated-batch reactor. SA–PVA-immobilized bacterial cells showed >96% decolorization of RR 120 aliquots (300 mg/L) within six cycles at a 12-h incubation time. After six cycles, there was a 10% reduction in decolorization efficiency. As a result, the immobilized bacteria-mediator beads showed good reusability for the decolorization of RR120 (Figure 7; Table 4).
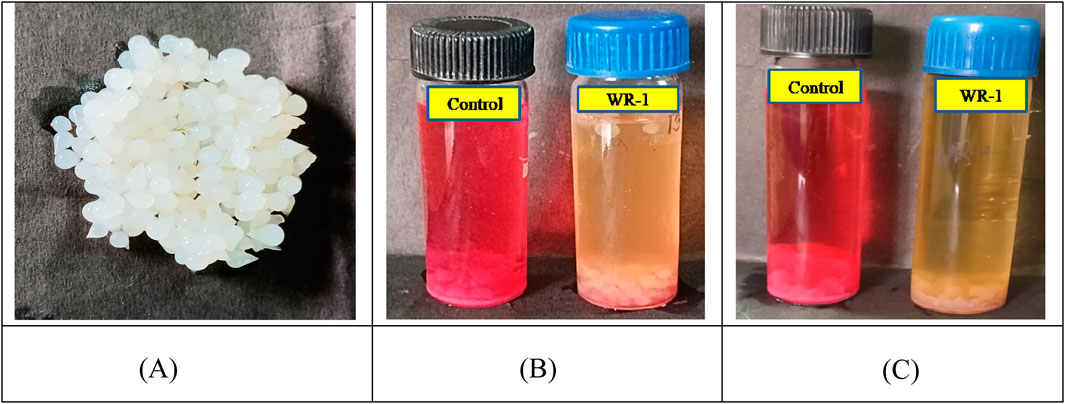
Figure 7. Immobilized bacterial cells and decolorized samples using immobilized beads. (A) Immobilized beads. (B) Batch decolorization. (C) Decolorization by transferred beads.
Decolorization of reactive dye and dye mixtures
Since synthetic dyes with a variety of structures are frequently used in the textile processing industry, the composition of effluents from various industries varies significantly. The decolorization efficacy of various azo dyes, used alone and in combination with BHM broth, was examined (Figure 8). According to our findings, L. capsici decolorized azo dyes with efficiencies of 95.15% for RR120, 85% for AB-113, 94.62% for Orange II, 94.62% for CR, 94.54% for PR, and 81.66% for mixed azo dyes, reaching 90.2% after 96 h under static conditions at 40 °C and pH 8.
Effect of heavy metal ions on the decolorization of RR120
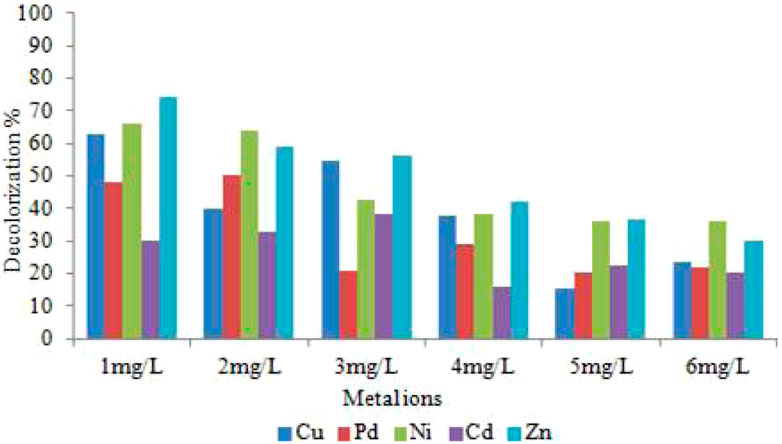
By supplementing Cu, Pd, Zn, Ni, and Cd heavy metal concentrations (1 mg/L to 6 mg/L) in the decolorization assay medium, we were able to determine the effects of heavy metal ions on L. capsici bacterial decolorization and the WR-1 strain’s tolerance to heavy metals. The dye decolorization activity result (Figure 9) did not show the same results when all heavy metal ions were utilized. The presence and concentration of heavy metals such as Cu, Pd, Zn, Ni, and Cd had a significant impact on RR120 dye decolorization; a significant difference was found in the decolorization %. Decolorization was not found in the control. Increasing concentrations of Cu, Pd, Zn, Ni, and Cd heavy metals showed a significant decrease in decolorization. A significant enhancement of decolorization was observed with Cu2+. When 1, 5, and 10 mm Cu2+ were implemented, decolorization increased by 39%, 33%, and 6.5%, respectively, compared with the control. According to this finding, the decolorization efficiency reduces as the concentrations of the heavy metals Cu, Ni, Pd, Zn, and Cd increase. Previous works have also reported on the decolorization of textile dyes in the presence of heavy metal ions. When compared to other heavy metals used, the rate of decolorization was reduced in the presence of Cd. At different concentrations of Zn metal ions (1 mg/L–6 mg/L), decolorization levels were 74%, 59%, 56.01%, 41.96%, 36.55%, and 29.91%, respectively, within 12 h.
Decolorization of textile fabric
To assess the effectiveness of bacteria in colored fabric, we used red and blue colored cotton fabric. The fabric pieces were dipped in bacteria-inoculated and uninoculated broth, and the activity of the colored fabric toward dye was analyzed. The change in fabric color was completely observed during the incubation period, resulting in complete fading, as shown in Figure 10. The fabric of the infected tubes was significantly lighter in color than that of the uninoculated tubes after washing and drying. From this study, the presence of dye-degrading bacterial enzymes and their enzymatic activity were determined. According to the observation of the result, these bacteria caused the red-colored fabric to fade completely, whereas the blue-colored fabric could not fade completely under the same conditions.
Decolorization and biodegradation analysis
UV–Vis spectroscopic analysis
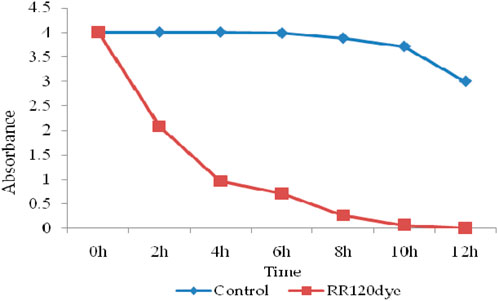
For dye decolorization and degradation analysis, control and test samples were centrifuged every 2 h, and the UV–Vis absorption of the supernatant was measured. RR120 dye absorbance optical density decreased in the UV–Vis absorption spectrum (524 nm) and was confirmed for decolorization compared to the control samples. The calculated OD and time graph (Figure 11) confirmed that, in the test sample compared to the control, the OD of the samples decreased with increasing time. The peak at 524 nm, which represents the optical absorbance of RR120, decreased compared to the initial absorbance as biodegradation was completed (12 h) (Guembri et al., 2021).
FT-IR analysis of dye RR120 metabolites
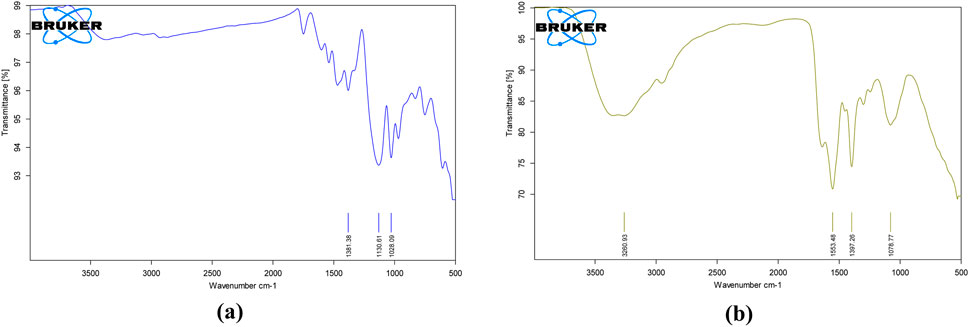
Essential bands such as aromatic rings, C–N stretching, and the azo chromophore (–N=N–) are considered when evaluating dye degradation. The decolorized compounds’ FT-IR spectra revealed significant differences in peak positions compared to the control dye spectrum. The dyes underwent biodegradation, as shown by the variation in their FT-IR spectra and those of their metabolites obtained during decolorization. The results are displayed in Figures 12a,b. After combining the samples with spectroscopically pure KBr, analyses were performed. FT-IR spectra of the compounds extracted with RR120 azo dye showed a peak for O–H bending at 1,381.38 cm−1, along with a supporting medium peak for the C–N stretching group and amine class at 1,130.61 cm−1 and another medium peak for the C–N stretching functional group and amine class at 1,028.09 cm−1, which indicates the formation of amines. FT-IR spectra of the degraded RR120 dye metabolites indicated the presence of O–H stretching, characteristic of alcohols, with a broad and strong peak observed at 3,260.93 cm-1. The strong peaks at 1,553.48 cm−1 showed the N–O stretching and nitro class compound. However, additional medium peaks at 1,397.26 cm−1 and 1,078.77 cm−1 confirmed the presence of functional groups, specifically O–H bending (carboxylic acid) and C–N stretching (amine compounds), respectively.
GC–MS analysis of metabolites produced by L. capsici
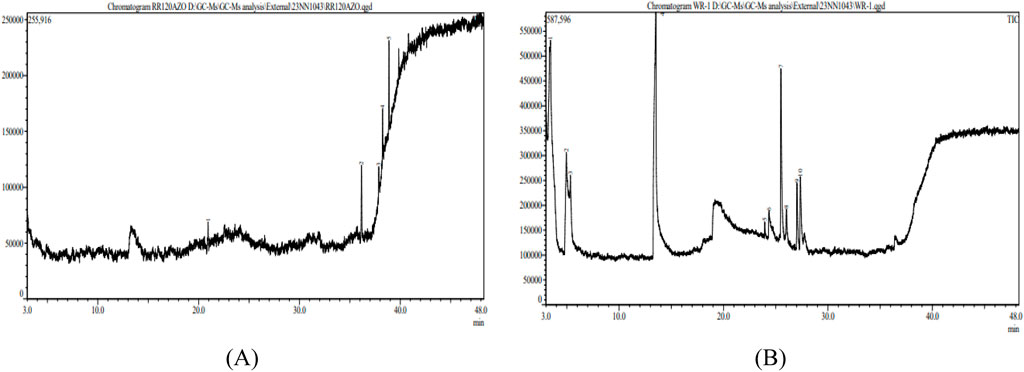
In RR120 dye, five peaks were found during GC–MS analysis (Figure 13A). RR120 dye components were identified as dodecane, 2,6,11-trimethyldodecane (molecular mass 212 g/mol, area % 7.96, and retention time 20.935 min), tetrapentacontane (molecular mass 758 g/mol, area % 27.77, and retention time 36.147 min), tetrapentacontane (molecular mass 758 g/mol, area % 26.78, and retention time 37.866 min), bis(2-ethylhexyl) phthalate (phthalic acid derivative; molecular mass 390 g/mol, area % 13.41, and retention time 38.244 min), and hexatriacontane (n-hexatriacontane; molecular mass 506 g/mol, area % 24.09, and retention time 38.874 min).
RR120 dye was treated with L. capsici, and 10 peaks were found during GC–MS analysis (Figure 13B). Degraded metabolites were identified using the NIST1.10 library (Si et al., 2013) and included butanoic acid (retention time 3.433 min, molecular mass 102 g/mol, and area % 49.38), pentanoic acid (molecular mass 116 g/mol, retention time 4.953 min, and area % 3.22), pentanoic acid (molecular mass 116 g/mol, area % 0.90, and retention time 5.345 min), 2-piperidinone (molecular mass 99 g/mol, area % 26.72, and retention time 13.527 min), tridecanoic acid (molecular mass 242 g/mol, area % 0.37, and retention time 23.945 min), 1,4-diazabicyclo [4.3.0] nonan-2,5-dione (molecular mass 168 g/mol, area % 2.27, and retention time 24.362 min), cyclo (L-prolyl-L-valine) (molecular mass 196 g/mol, area % 8.77, and retention time 25.498 min), pyrrolo [1,2-a]pyrazine-1,4-dione (molecular mass 210 g/mol, area % 1.18, and retention time 26.020 min), L-leucine (molecular mass 409 g/mol, area % 3.18, and retention time 27.036 min), and pyrrolo [1,2-a]pyrazine-1,4-dione (molecular mass 210 g/mol, area % 4.01, and retention time 27.356 min), as detailed in Table 5.
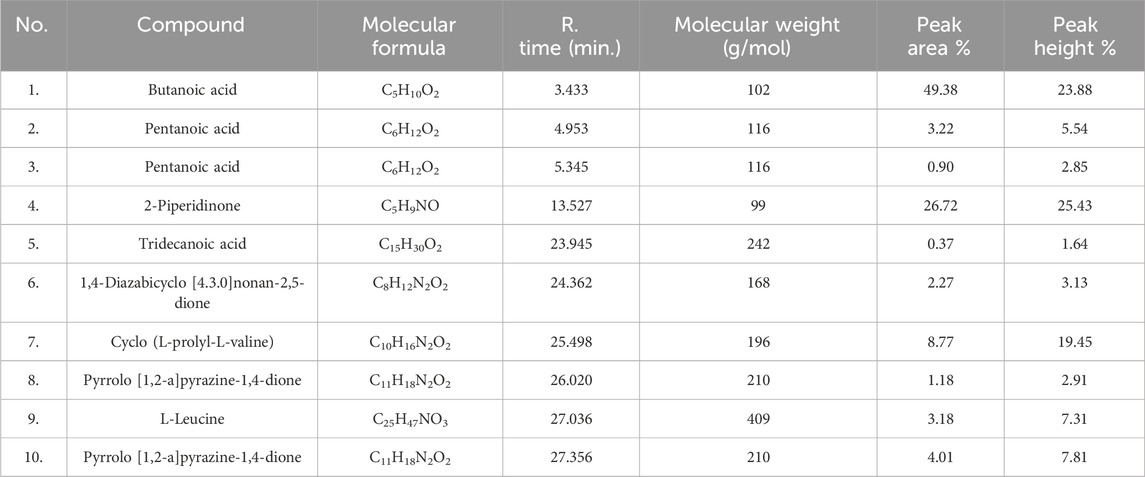
Table 5. Compound, molecular formula, R. time, and molecular weight of dye RR120-degraded metabolites studied using GC–MS.
Enzyme assay
Laccase and MnP enzymes were screened during the dye decolorization test. Laccase and manganese peroxidase enzyme screening tests finally showed reddish-brown color and brownish-red color in the test sample. The L. capsici bacterial strain showed positive activity in laccase and manganese peroxidase enzymes. These enzymes convert toxins into less hazardous or non-toxic metabolites, which is beneficial for detoxifying several types of industrial substances. In the decolorization experiment, laccase was found to be the protein that is most prevalent and least frequently produced. Our research showed that enzymes were essential in promoting the dye’s decolorization (Henagamage and Peries, 2023).
Discussion
Isolation of dye-decolorizing bacterial strains
We found that three isolates showed significant RR120 decolorization, with efficiency higher than 90%, even though the bacteria were isolated from the Indarbela tetraonis bark borer insect tunnels at Guru Ghasidas Vishwavidyalaya, Bilaspur, Chhattisgarh, India. In the present study, WR-1 strains showed 99.9% decolorization of BHM containing glucose as the carbon source, ammonium nitrate as the nitrogen source, and 0.2 mg/L RR120 at 28 °C and pH 7 after 12 h of incubation under anaerobic static conditions. Likewise, a few PGPR bacteria can also remove the color of azo dyes, including Rhizobium meliloti NCIM 2757, Bacillus sp. SR 2-1/1, B. megaterium NCIM 2054, Pseudomonas japonica I-15, and Azotobacter vinelandii MTCC 1241 (Haque et al., 2024). In the MS media containing 150 mg L−1 of the respective dye at 30 °C and pH 6 after 48 h of incubation, Bacillus sp. MR-1/2, a PGPR strain isolated from the root zone of maize crop, was shown to decolorize 78, 83, 63, and 65% of Reactive Black 5, Reactive Red 120, Direct blue 1, and Congo red, respectively, under microaerophilic conditions (Shahid et al., 2018). Moreover, it was found that Bacillus sp. SR-2-1/1, isolated from the root zone of sorghum plants grown in soil contaminated with textile wastewater, could remove 83, 40, 71, and 55% of Congo red, Reactive Red 120, Reactive black 5, and Direct blue 1, respectively, from the MS media after 72 h of incubation under static conditions (Mahmood et al., 2017).
Effects of various parameters
In the present investigation, the isolated bacterium L. capsici strain was able to decolorize RR120 in BHM under anaerobic conditions. In the presence or absence of oxygen, dyes are susceptible to biological degradation. Although the first step of azo-linkage cleavage primarily involves the breakdown of azo dyes under anaerobic conditions, bacteria may mainly destroy the aromatic amines generated in the second step in an aerobic environment. As a result, combining the anaerobic and aerobic phases of a treatment process may accelerate and enhance it (Pham et al., 2023). Under static conditions, Kocuria rosea MTCC 1532, a Gram-positive bacterium, has been reported to totally (100%) decolorize MO (50 mg L−1). However, under shaking conditions, this strain is unable to decolorize it. pH is one of the environmental factors that control the synthesis of enzymes and the biodegradation of dye molecules into microbial cells (Parshetti et al., 2010). Different organic nitrogen sources, such as BE, Pep, and YE, are usually used to enhance the color degradation of azo dyes by different bacterial strains (Guo et al., 2020). The MO was properly decolored by ESR7, ESR13, ESR20, ESR21, ESD8, and ESB18 at all pH tests conducted under microaerophilic conditions. However, ESD8 was able to decolorize 33.7, 57.0, 32.9, and 31.9% at initial pH 5, pH 6, pH 8, and pH 9, respectively, under aerobic conditions (Haque et al., 2024). Micrococcus glutamicus NCIM-2168 was used to break down the sulfonated diazo dye C.I. Reactive Green 19A. Under static conditions, this process was faster than that under shaking conditions (Sharma et al., 2016). Similar results using Acinetobacter calcoaceticus showed that azo-dyed amaranth had maximal decolorization in a static environment. When comparing static culture versus shaking (aerobic) conditions, several studies found that static culture results in more effective dye decolorization. Rhizobium radiobacter was shown to decolorize Direct Black 38 by 90% under static conditions and only 6% under shaking conditions (Telke et al., 2008). Direct Black 38 was decolored in 5 days by Pseudomonas sp. under anaerobic conditions, and C. I. Acid Red 88 was decolored in 60 h by Stenotrophomonas acidaminiphila (BN-3). Under static situations, the Staphylococcus hominis RMLRT03 strain decolored Acid Orange at 85.52%, while under shaking conditions, decolorization was only 32.47% (Singh et al., 2014). The bacterial isolate Acinetobacter baumannii JC359 could decolorize RR120 and showed 96% decolorization at 37 °C and pH 9 within 48 h (Ameenudeen et al., 2021). S. haliotis, A. hydrophila, and S. putrefaciens were reported to decolorize RR120 dye azo at pH 8.0 and 35 °C by 27.71%, 10.39%, and 27.28%, respectively (Radhika and Aruna, 2022). The Acinetobacter junii strain FA10 showed maximum a 80% Reactive Red 120 degradation efficiency at 30 °C within 72 h of incubation (Anwar et al., 2014). The best inoculum size and time conditions for RR120 decolorization in this study were 1% (0.3 mL) inoculums during 12 h of incubation under both microaerophilic situations. It was found that variations in pH, temperature, carbon–nitrogen sources, dye concentration, inoculum size, and aeration conditions had a significant impact on the RR120 breakdown percentage by L. capsici. The growth rate of L. capsici was significantly reduced under a static state compared to that under shaking conditions.
RR120 decolorization by immobilized L. capsici
The A. jandaei strain SCS5-immobilized cells were able to decolorize MR, and compared to immobilized cells alone, the decolorization rate was greatly increased by immobilized cells combined with mediators such as anthraquinone-2, 6-disulfonate and magnetic Fe3O4 nanoparticles (Sharma et al., 2016). The maximum decolorization was observed in the study during the biodegradation of RR120 in a batch continuous bioreactor using an immobilized L. capsici PB300 (T) bacterial cell.
RR120 decolorization by consortium L. capsici
The four bacterial consortia, L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa, showed the highest dye degradation percentage in <8 h. Previous research indicated that the rate of dye degradation increases when multiple cultures are combined in a consortium. In wastewater treatment and dye bioremediation, microbial consortia are more effective than pure isolates. Multiple microbial compounds within the consortium co-metabolize, contributing to the expected complete and rapid destruction of the target pollutants (Agrawal et al., 2023). S. paucimobilis, Rhizobium radiobacter, and B. subtilis were the three bacterial species that revealed high decolorization of azo dye in the bacterial consortium throughout a single culture system. Compared to their isolates, which took 72 h, a group of six bacterial strains demonstrated an 84% decolorization percentage for a dye mixture in only 24 h (Si et al., 2013).
Decolorization of many azo dyes and dye mixtures
In this study, we found that L. capsici is effective in decolorization of a mixture of azo dyes, viz., Reactive Red 120 and other azo dyes (e.g., 85% AB-113, 94.62% orange II, 94.62% Congo red, 94.54% phenol red, and 81.66%). Similarly, Pseudomonas sp. WS-D/183 is efficient in decolorization of dyes (100 mg/L concentration.) RB5 (53.5% ± 1.1%), followed by RR120 (18.3% ± 1.3%), RY2 (17.7% ± 3.2%), Blue Direct (14.7 ± 1.7), Congo Red direct (7.5 ± 2.7), and Orange Direct (3.4 ± 1.1) at 30 °C and 24 h (Chen et al., 2021). Reactive purple, Reactive Red M5B, malachite green, and reactive pink MB mixed dyes (200 mg/L) were 70.8% decolorized by Bacillus subtilis and B. cereus under aerobic conditions at pH 7 and 37 °C after 8 days (Maheswari and Sivagami, 2016). Pseudomonas aeruginosa SVM16 decolorizes mixed dyes (50 mg/L) such as RR 21 and RO16 at 90.2% after 96 h under static conditions of 40 °C at pH 8.2, when it was supplemented with jack fruit seed powder (Mishra and Maiti, 2019). Likewise, Clostridium bifermentans SL186 can decolorize mixed dyes (100 mg/L) reactive black-5, reactive red 3B-A, and reactive yellow 3G-P in anaerobic medium at pH 10 and 35 °C after 36 h (Joe et al., 2008). Brevibacillus laterosporus MTCC 2298 decolored the mixture (70 mg/L) of structurally distinct dyes, including Remazol red, methyl red, golden yellow HER, Rubine GFL, brown 3-REL, brilliant blue GL, scarlet RR, and yeast extract in a static micro-aerobic medium at pH 6–8 and 40 °C after 24 h (Kurade et al., 2011).
Heavy metal ion tolerance activity
Many of the strains linked to P. australis also showed strong resistance to metals such as zinc, nickel, and copper, when added either singly or in combination in the growth medium. Since it may improve plant metal uptake and transport to above-ground tissues, metal tolerance is a beneficial characteristic in CW macrophyte plant-related bacteria (Zhuang et al., 2020). When heavy metal ions are present in large concentrations, PGPR Bacillus sp. SR-2-1/1 exhibits excellent potential for decolorizing various azo dyes (Mahmood et al., 2017).
Enzyme assay
Reactive Red 120 dye is 95%–99% decolorized by laccase and manganese peroxidase producing L. capsici, according to our findings. The presence of manganese peroxidase and laccase in the L. capsici bacterial strain has been proven by the enzymes and laccase. These enzymes contribute to the detoxification of numerous chemicals used in the industry by converting poisons into metabolites that are less dangerous or non-toxic. Laccase is the most prevalent enzyme but accumulates the least during the decolorization experiment. Our findings demonstrated the critical role enzymes played in expediting the process of dye decolorization. Likewise, a strain of Serratia liquefaciens that produced over 90% of the lipase was responsible for decolorizing AB dye, whereas laccase and a strain of Streptomyces microflavus that produced lipase were responsible for decolorizing textile industrial wastewater. The lipase- and laccase-producing Aeromonas hydrophila strain decolorized CV dye by 99% (Kishor et al., 2021).
Conclusion
One of the most impactful types of industrial effluent is thought to originate from the textile industry. The purpose of this study was to decolorize Reactive Red 120 dye using strains of L. capsici bacteria that were obtained from the bark borer sample. Under optimized conditions using manual and Taguchi strategies, more than 99% dye decolorization was achieved with glucose, ammonium nitrate, 1% inoculum size, and dye concentrations of 0.2 mg/L under static conditions at pH 8 and 40 °C within 12 h. Four bacterial strains, namely, L. capsici, A. faecalis subsp. Phenolicus, A. baumannii, and P. aeruginosa, were used for consortium development, and the consortium results showed that the time for complete decolorization decreased. As a result, microbial consortia and cell immobilization bioremediation are emerging as one of the most promising methods because of their ability to break down various types of environmental pollutants, strong resistance to complex environments, affordable cost, and environmental friendliness. When heavy metal ions such as Zn, Pd, Cu, Ni, and Cd are present, L. capsici can degrade the dye. It was reported that the L. capsici strain could decolorize colored fabric. FT-IR, UV–Vis, and GC–MS analyses confirmed that the dye has been converted to its amines by bacterial metabolism, which is less toxic than the parent dye.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, L. capsici (accession no 13606), A. faecalissubsp. Phenolicus (accession no 13436), A. baumanni (accession no 13438), and P. aeruginosa (accession no 13437).
Author contributions
Kusumlata: Writing – original draft. RS: Writing – review and editing. BA: Formal analysis, Visualization, Writing – review and editing. AK: Writing – review and editing, Formal analysis, Investigation, Supervision, Visualization.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors are thankful to the Central Instrumentation Lab, Central University of Punjab, Bhatinda for GC-MS analysis and National Centre for Microbial Resource, Pune, Maharashtra for Molecular identification of bacterial isolates.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El-Rahim, W. M., Moawad, H., Azeiz, A. Z., and Sadowsky, M. J. (2021). Biodegradation of azo dyes by bacterial or fungal consortium and identification of the biodegradation products. Egypt. J. Aquatic Res. 47 (3), 269–276. doi:10.1016/j.ejar.2021.06.002
Agrawal, S., Tipre, D., and Dave, S. R. (2023). Biotreatment of azo dye containing textile industry effluent by a developed bacterial consortium immobilised on brick pieces in an indigenously designed packed bed biofilm reactor. World J. Microbiol. Biotechnol. 39 (3), 83. doi:10.1007/s11274-023-03521-7
Ameenudeen, S., Unnikrishnan, S., and Ramalingam, K. (2021). Statistical optimization for the efficacious degradation of reactive azo dyes using Acinetobacter baumannii JC359. J. Environ. Manag. 279, 111512. doi:10.1016/j.jenvman.2020.111512
Anwar, F., Hussain, S., Ramzan, S., Hafeez, F., Arshad, M., Imran, M., et al. (2014). Characterization of reactive red-120 decolorizing bacterial strain Acinetobacter junii FA10 capable of simultaneous removal of azo dyes and hexavalent chromium. Water, Air, Soil Pollut. 225, 2017–16. doi:10.1007/s11270-014-2017-7
Ayed, L., Bekir, K., and Jabeur, C. (2022). Modelling and optimization of biodegradation of methylene blue by Staphylococcus aureus through a statistical optimization process: a sustainable approach for waste management. Water Sci. Technol. 86 (2), 380–394. doi:10.2166/wst.2022.211
Ayele, A., Getachew, D., Kamaraj, M., and Suresh, A. (2021). Phycoremediation of synthetic dyes: an effective and eco-friendly algal technology for the dye abatement. J. Chem. 2021, 1–14. doi:10.1155/2021/9923643
Bustos-Terrones, Y. A., Bandala, E. R., Moeller-Chavez, G. E., and Bustos-Terrones, V. (2022). Enhanced biological wastewater treatment using sodium alginate-immobilized microorganisms in a fluidized bed reactor. Water Sci. Eng. 15 (2), 125–133. doi:10.1016/j.wse.2022.02.002
Carolin, C. F., Kumar, P. S., and Joshiba, G. J. (2021). Sustainable approach to decolourize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technol. Environ. Policy 23, 173–181. doi:10.1007/s10098-020-01934-8
Carrascal-Hernández, D. C., Orozco-Beltrán, E. J., Insuasty, D., Márquez, E., and Grande-Tovar, C. D. (2025). Systematic evaluation of biodegradation of azo dyes by microorganisms: efficient species, physicochemical factors, and enzymatic systems. Int. J. Mol. Sci. 26 (16), 7973. doi:10.3390/ijms26167973
Chaturvedi, A., Rai, B. N., Singh, R. S., and Jaiswal, R. P. (2021). A computational approach to incorporate metabolite inhibition in the growth kinetics of indigenous bacterial strain Bacillus subtilis MN372379 in the treatment of wastewater containing Congo red dye. Appl. Biochem. Biotechnol. 193, 2128–2144. doi:10.1007/s12010-021-03538-4
Chen, G., An, X., Li, H., Lai, F., Yuan, E., Xia, X., et al. (2021). Detoxification of azo dye direct black G by thermophilic Anoxybacillus sp. PDR2 and its application potential in bioremediation. Ecotoxicol. Environ. Saf. 214, 112084. doi:10.1016/j.ecoenv.2021.112084
Dixit, S., and Garg, S. (2021). Enzymatic degradation of sulphonated azo dye using purified azoreductase from facultative Klebsiella pneumoniae. Folia Microbiol. 66, 79–85. doi:10.1007/s12223-020-00824-2
Florencio, T. D. M., de Godoi, L. A., Rocha, V. C., Oliveira, J. M. S., Motteran, F., Gavazza, S., et al. (2021). Anaerobic structured-bed reactor for azo dye decolorization in the presence of sulfate ions. J. Chem. Technol. Biotechnol. 96 (6), 1700–1708. doi:10.1002/jctb.6695
Guembri, M., Neifar, M., Saidi, M., Ferjani, R., Chouchane, H., Mosbah, A., et al. (2021). Decolorization of textile azo dye novacron red using bacterial monoculture and consortium: response surface methodology optimization. Water Environ. Res. 93 (8), 1346–1360. doi:10.1002/wer.1521
Guo, G., Hao, J., Tian, F., Liu, C., Ding, K., Xu, J., et al. (2020). Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: systematic investigations of performance, pathway and metagenome. Ecotoxicol. Environ. Saf. 204, 111073. doi:10.1016/j.ecoenv.2020.111073
Hameed, B. B., and Ismail, Z. Z. (2018). Decolorization, biodegradation and detoxification of reactive red azo dye using non-adapted immobilized mixed cells. Biochem. Eng. J. 137, 71–77. doi:10.1016/j.bej.2018.05.018
Haque, M. M., Hossen, M. N., Rahman, A., Roy, J., Talukder, M. R., Ahmed, M., et al. (2024). Decolorization, degradation and detoxification of mutagenic dye methyl orange by novel biofilm-producing plant growth-promoting rhizobacteria. Chemosphere 346, 140568. doi:10.1016/j.chemosphere.2023.140568
Henagamage, A. P., and Peries, C. M. (2023). Degradation and decolorization of textile azo dyes by effective fungal-bacterial consortium. Mol. Biol. Rep. 50 (11), 8901–8914. doi:10.1007/s11033-023-08741-6
Hussain, S., Maqbool, Z., Shahid, M., Shahzad, T., Muzammil, S., Zubair, M., et al. (2020). Simultaneous removal of reactive dyes and hexavalent chromium by a metal-tolerant Pseudomonas sp. WS-D/183 harbouring plant growth-promoting traits. Int. J. Agric. Biol. 23, 241–252. doi:10.17957/IJAB/15.1282
Ibrahim, A. H. H., Cihangir, N., Idil, N., and Aracagök, Y. D. (2024). Adsorption of azo dye by biomass and immobilized Yarrowia lipolytica; equilibrium, kinetic and thermodynamic studies. World J. Microbiol. Biotechnol. 40 (5), 140. doi:10.1007/s11274-024-03949-5
Joe, M. H., Lim, S. Y., Kim, D. H., and Lee, I. S. (2008). Decolorization of reactive dyes by Clostridium bifermentans SL186 isolated from contaminated soil. World J. Microbiol. Biotechnol. 24, 2221–2226. doi:10.1007/s11274-008-9733-3
Kamal, I. M., Abdeltawab, N. F., Ragab, Y. M., Farag, M. A., and Ramadan, M. A. (2021). Biodegradation, decolorization, and detoxification of di-azo dye direct red 81 by halotolerant, alkali-thermo-tolerant bacterial mixed cultures. Microorganisms 10 (5), 994. doi:10.3390/microorganisms10050994
Kathiravan, M. N., Praveen, S. A., Gim, G. H., Han, G. H., and Kim, S. W. (2014). Biodegradation of methyl Orange by alginate-immobilized Aeromonas sp. in a packed bed reactor: external mass transfer modelling. Bioprocess Biosyst. Eng. 37, 2149–2162. doi:10.1007/s00449-014-1192-7
Kaur, J., Mudgal, G., Negi, A., Tamang, J., Singh, S., Singh, G. B., et al. (2023). Reactive Black-5, Congo red and methyl Orange: chemical degradation of azo-dyes by agrobacterium. Water 15 (9), 1664. doi:10.3390/w15091664
Kishor, R., Purchase, D., Saratale, G. D., Ferreira, L. F., Bilal, M., Iqbal, H. M., et al. (2021). Environment friendly degradation and detoxification of Congo red dye and textile industry wastewater by a newly isolated Bacillus cohnni (RKS9). Environ. Technol. and Innovation. 22, 101425. doi:10.1016/j.eti.2021.101425
Kour, D., Khan, S. S., Kour, H., Kaur, T., Devi, R., Rai, P. K., et al. (2023). Microbe-mediated bioremediation: current research and future challenges. J. Appl. Biol. Biotechnol. 10 (2), 6–24. doi:10.7324/jabb.2022.10s202
Kurade, M. B., Waghmode, T. R., and Govindwar, S. P. (2011). Preferential biodegradation of structurally dissimilar dyes from a mixture by Brevibacillus laterosporus. J. Hazard. Mater. 192 (3), 1746–1755. doi:10.1016/j.jhazmat.2011.07.004
Lade, H., Govindwar, S., and Paul, D. (2015). Low-cost biodegradation and detoxification of textile azo dye CI reactive blue 172 by Providencia rettgeri strain HSL1. J. Chem. 2015, 1–10. doi:10.1155/2015/894109
Lalnunhlimi, S., and Krishnaswamy, V. (2016). Decolorization of azo dyes (direct blue 151 and direct red 31) by moderately alkaliphilic bacterial consortium. Braz. J. Microbiol. 47, 39–46. doi:10.1016/j.bjm.2015.11.013
Lata, S., Ambade, B., Kumar, A., and Gautam, S. (2024a). Sustainable solutions: reviewing the future of textile dye contaminant removal with emerging biological treatments. Limnol. Rev. 24 (2), 126–149. doi:10.3390/limnolrev24020007
Lata, S., Singh, R. P., and Kumar, A. (2024b). Biodecolorization of azo dye by bacteria Alcaligenes faecalis sub sp. phenolicus isolated from a bark-beetle tunnel developed in Peltophorum pterocarpum plant. Curr. World Environ. 19 (2), 824–840. doi:10.12944/cwe.19.2.25
Lata, S., Singh, R. P., and Kumar, A. (2024c). Biodecolorization and degradation of textile wastewater and RR120 dye using Acinetobacter baumanni bacteria isolated from textile industry. Afr. J. Bio.Sc. 14 (6). doi:10.12944/CWE.19.2.25
Maheswari, N. U., and Sivagami, S. (2016). Biological degradation of textile dyes using marine Bacillus species. Int. J. Pure Appl. Biosci. 4 (4), 123–128. doi:10.18782/2320-7051.2326
Mahmood, F., Shahid, M., Hussain, S., Shahzad, T., Tahir, M., Ijaz, M., et al. (2017). Potential plant growth-promoting strain Bacillus sp. SR-2-1/1 decolorized azo dyes through NADH-ubiquinone: oxidoreductase activity. Bioresour. Technol. 235, 176–184. doi:10.1016/j.biortech.2017.03.098
Martinez, A., Sergio, D., and Bustos, T. Y. (2009). Biodegradation of wastewater pollutants by activated sludge encapsulated inside calcium-alginate beads in a tubular packed bed reactor. Biodegradation 20 (5), 709–715. doi:10.1007/s10532-009-9258-y
Marvi-Mashhadi, A., Sharifmoghadam, M. R., Goharimanesh, M., Marvi-Mashhadi, M., Dehghan, H., and Bahreini, M. (2021). Methyl red biodegradation based on Taguchi method by two novel bacteria. Int. J. Environ. Sci. Technol. 19, 1357–1368. doi:10.1007/s13762-021-03264-8
Mishra, S., and Maiti, A. (2019). Optimization of process parameters to enhance the bio-decolorization of reactive red 21 by Pseudomonas aeruginosa 23N1. Int. J. Environ. Sci. Technol. 16, 6685–6698. doi:10.1007/s13762-018-2023-1
Parshetti, G. K., Telke, A. A., Kalyani, D. C., and Govindwar, S. P. (2010). Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J. Hazard Mater. 176 (1–3), 503–509. doi:10.1016/j.jhazmat.2009.11.058
Patel, V. R., and Bhatt, N. (2015). Isolation, development and identification of salt-tolerant bacterial consortium from crude-oil-contaminated soil for degradation of di-azo dye reactive blue 220. Water Sci. Technol. 72 (2), 311–321. doi:10.2166/wst.2015.208
Pham, V. H., Kim, J., Chang, S., and Bang, D. (2023). Investigating bio-inspired degradation of toxic dyes using potential multi-enzyme producing extremophiles. Microorganisms 11 (5), 1273. doi:10.3390/microorganisms11051273
Pourbabaee, A. A., Ramezani, S., and Javaheri Daneshmand, H. (2013). Biodegradation of malachite green by Klebsiella terrigenaptcc 1650: the critical parameters were optimized using Taguchi optimization method. J. Bioremed. Biodeg. 4, 175. doi:10.4172/2155-6199.1000175
Radhika, B., and Aruna, K. (2022). Elucidating a pathway for degradation of azo dye reactive red 120 BY bacterial consortium. J. Appl. Biol. Sci. 16 (3), 396–417. doi:10.71336/jabs.1042
Rehaman, S., Aravindan, G., and Karthick, G. (2021). Spectral studies of azo dye degradation using selected biofertilizer of Pseudomonas fluorescens. Int. J. Life Sci. Pharma Res. 11 (1), L73–L79. doi:10.22376/ijpbs/lpr.2021.11.1.L73-79
Samykannu, M. (2025). Eco-friendly approaches to azo dye removal: the role of microbial azo-reductases. Appl. Biochem. Biotechnol., 1–19. doi:10.1007/s12010-025-05343-9
Shahid, M., Mahmood, F., Hussain, S., Shahzad, T., Haider, M. Z., Noman, M., et al. (2018). Enzymatic detoxification of azo dyes by a multifarious Bacillus sp. strain MR-1/2-bearing plant growth-promoting characteristics. 3 Biotech. 8, 425–12. doi:10.1007/s13205-018-1442-5
Sharma, S. C. D., Sun, Q., Li, J., Wang, Y., Suanon, F., Yang, J., et al. (2016). Decolorization of azo dye methyl red by suspended and co-immobilized bacterial cells with mediators anthraquinone-2, 6-disulfonate and Fe3O4 nanoparticles. Int. Biodeterior. Biodegrad. 112, 88–97. doi:10.1016/j.ibiod.2016.04.035
Si, J., Peng, F., and Cui, B. (2013). Purification, biochemical characterization and dye decolorization capacity of an alkali-resistant and metal-tolerant laccase from Trametes pubescent. Bioresour. Technol. 128, 49–57. doi:10.1016/j.biortech.2012.10.085
Singh, G., and Dwivedi, S. K. (2020). Decolorization and degradation of direct Blue-1 (azo dye) by newly isolated fungus Aspergillus terreus GS28, from sludge of carpet industry. Environ. Technol. and innovation 18, 100751. doi:10.1016/j.eti.2020.100751
Singh, R. P., Singh, P. K., and Singh, R. L. (2014). Bacterial decolorization of textile azo dye acid orange by Staphylococcus hominis RMLRT03. Toxicol. Int. 21 (2), 160. doi:10.4103/0971-6580.139797
Sonal, S., and Mishra, B. K. (2021). “Role of coagulation/flocculation technology for the treatment of dye wastewater: trend and future aspects,” in Water pollution and management practices (Springer), 303–331.
Sreedharan, V., Saha, P., and Rao, K. V. B. (2021). Dye degradation potential of Acinetobacter baumannii strain VITVB against commercial azo dyes. Bioremediation J. 25 (4), 347–368. doi:10.1080/10889868.2020.1871317
Telke, A., Kalyani, D., Jadhav, J., and Govindwar, S. (2008). Kinetics and mechanism of reactive red 141 degradation by a bacterial isolate Rhizobium radiobacter MTCC 8161. Acta Chim. 55 (2).
Thanavel, M., Bankole, P. O., Selvam, R., Govindwar, S. P., and Sadasivam, S. K. (2020). Synergistic effect of biological and advanced oxidation process treatment in the biodegradation of remazol yellow RR dye. Sci. Rep. 10 (1), 20234. doi:10.1038/s41598-020-77376-5
Thiruppathi, K., Rangasamy, K., Ramasamy, M., and Muthu, D. (2021). Evaluation of textile dye degrading potential of ligninolytic bacterial consortia. Environ. Challenges 4, 100078. doi:10.1016/j.envc.2021.100078
Vinayak, A., and Singh, G. B. (2022). Synthetic azo dye bio-decolorization by Priestia sp. RA1: process optimization and phytotoxicity assessment. Archives Microbiol. 204 (6), 318. doi:10.1007/s00203-022-02931-9
Keywords: reactive red 120, dye degradation, biodegradation, bacterial consortia, metal-ion-tolerant, immobilized cell
Citation: Kusumlata , Singh RP, Ambade B and Kumar A (2025) Biodecolorization of Reactive Red 120 azo dye by metal-ion-tolerant Lysinibacillus capsici and its potential application in textile effluent treatment. Front. Environ. Sci. 13:1688655. doi: 10.3389/fenvs.2025.1688655
Received: 19 August 2025; Accepted: 29 September 2025;
Published: 04 November 2025.
Edited by:
Antonio Albuquerque, University of Beira Interior, PortugalReviewed by:
Oscar Manuel Rodriguez Narvaez, Centro de Innovación Aplicada en Tecnologías Competitivas (CIATEC), MexicoShantkriti S., SRM Institute of Science and Technology, Tiruchirappalli, India
Copyright © 2025 Kusumlata, Singh, Ambade and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashish Kumar, cHJvZmJhbmphcmFhc2hpc2hAZ21haWwuY29t
 Kusumlata
Kusumlata Rajat Pratap Singh1
Rajat Pratap Singh1 Balram Ambade
Balram Ambade Ashish Kumar
Ashish Kumar