- 1Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland
- 2Swiss Institute of Bioinformatics, Lausanne, Switzerland
Most animals use olfaction to obtain important information from the environment, including the presence of food or mates. Insects detect odorants through receptors that are expressed in the sensory neurons of the olfactory sensilla, which cover the surface of the antennae. The olfactory capacities of an insect thus depend largely on the repertoire of the odorant receptors. Here, we study the repertoire of olfactory proteins in the stick insect Timema cristinae. We first generate transcriptomes from the antennae of adult males and females and identify, via homology searches, putative olfactory proteins of three different families: odorant binding proteins, odorant receptors, and chemosensory proteins (CSPs). We then attempt to categorize olfactory proteins likely involved in sexual communication by comparing gene expression between adults and juveniles, as well as between males and females. Notably, the olfactory proteins involved in the perception of food or abiotic environmental components, should be expressed in both adults and juveniles. By contrast, the olfactory proteins involved in sexual communication, such as the detection of sex pheromones, should be expressed in adults and often comprise different repertoires in males and females. Finally, we also tested whether olfactory proteins in general and the subset, with putative roles in sexual communication in particular, are under relaxed selection in the asexual species T. monikensis, a close relative of T. cristinae. We found that olfactory proteins are typically differentially expressed between juveniles and adults, but there is little overlap of differential expression between developmental stages and the level of sex bias in adults. Furthermore, while we find evidence that olfactory proteins are indeed under relaxed selection in the asexual species, there is no evidence that this is necessarily the case for olfactory genes with a putative role in sexual communication. Nevertheless, the list of olfactory genes generated in our study provides a useful tool for future studies on olfaction in Timema and other stick insects.
Introduction
Olfaction, the sense of smell, is of critical importance for survival and reproduction in insects. For the majority of species studied, it is the main sense that guides the location of resources, mate recognition, selection of oviposition sites and predator avoidance (Pickett and Glinwood, 2007; Leal, 2013). In stick insects, experiments on different species have demonstrated the role of olfactory cues for the selection of suitable host plant species, as well as for sexual communication and intra-and interspecific mate discrimination (Nosil et al., 2007; Schwander et al., 2013a; Burke et al., 2015; Myers et al., 2015; Riesch et al., 2017). In contrast, the molecular mechanisms underlying the detection and processing of olfactory cues for host plant selection or mate discrimination remain unexplored. This constrains studies of how olfaction contributes to adaptive divergence between stick insect populations and species.
The two organs insects primarily used to detect odors are the antennae and maxillary palps (specialized mouthparts). These organs comprise large numbers of olfactory receptor neurons which house receptors for scent molecules in their cell membrane (reviewed in Pelosi et al., 2006). Specifically, the detection of odors depends on different protein families, with odorant-binding proteins (OBPs), chemosensory proteins (CSPs), and odorant receptors, playing a key role (Hallem et al., 2006; Touhara and Vosshall, 2009; Leal, 2013). Odorant-binding and CSPs are involved in the first step of the recognition of chemical signals, by binding to hydrophobic molecules from the environment and delivering them to the receptors (Hallem et al., 2006; Touhara and Vosshall, 2009; Leal, 2013). However, members of these protein families are also involved in functions not linked to the detection of odors. Members of the chemosensory protein family in particular have more diverse functions in chemoreception, growth, and development (Pelosi et al., 2018).
Here, we provide a first step toward developing a better understanding of the mechanisms underlying olfaction in stick insects, by generating a catalog of olfactory proteins that are expressed in the antennae of the stick insect Timema cristinae. Timema, the sister group of all remaining stick and leaf insects (Euphasmatodea) (Whiting et al., 2003; Tilgner, 2009), are a small group of stick insects native to the western part of the United States of America and Mexico (Vickery and Sandoval, 2001). Previous work on Timema has shown that olfaction plays an important role in mate discrimination, both within and between species (Nosil et al., 2007; Arbuthnott and Crespi, 2009; Schwander et al., 2013a; Riesch et al., 2017).
We first annotated OBPs, CSPs, and odorant receptors in a previously published reference transcriptome of T. cristinae and screened for their expression in the antennae of juvenile and adult individuals of both sexes. We then combined two approaches in an attempt to distinguish between olfactory proteins likely involved in sexual communication vs. proteins likely involved in odor detection linked to host plants and other environmental cues (or in functions not related to olfaction). For the first approach, we identified olfactory proteins with a higher expression in adults than in juveniles, as only adults react to pheromone and hydrocarbon stimuli of the opposite sex (Schwander et al., 2013b). Functions such as host plant selection, predator avoidance and the detection of other cues from the environment are not expected to differ extensively between juveniles and adults given that different developmental stages live on the same individual host plant and are exposed to similar predator guilds (Sandoval, 1993). For the second approach, we took advantage of the fact that several functionally apomictic asexual species are known in the genus Timema (Schwander and Crespi, 2009).
A previous study showed that females in asexual species feature reduced pheromone production and altered surface smells, likely because selection favored the reduction of costly sexual traits in asexual species where such traits are not required for reproduction (Schwander et al., 2013b). Thus, olfactory proteins involved sexual communication are expected to be under relaxed (or even negative) selection in asexual species, while olfactory proteins involved in vital functions are expected to be maintained. By combining these two approaches, we generated a list of olfactory genes which will be a useful tool for future studies on olfaction in Timema and other stick insects.
Methods
Identifying Odorant-Related Genes
We identified OBPs, CSPs, and odorant receptors (ORs) in a previously published reference transcriptome of T. cristinae (Bast et al., 2018) using a blast approach. Protein reference sequences were downloaded from the protein database at NCBI, found by searching the titles for the following terms: “Odorant binding,” “Chemosensory protein,” or “Odorant receptor” for the following insect taxa: Drosophila melanogaster, Tribolium castaneum, Orthoptera, Hemiptera, and Blattodea (for a full list of accession numbers, see Supplementary Table 2). We also used the putative OBP, CSP, and OR sequences of Clitarchus hookeri provided in Wu et al. (2016), and OR sequences of Phyllium siccifolium provided in Missbach et al. (2014).
The protein reference sequences were blasted (tblastn) to the whole T. cristinae transcriptome, keeping only hits with an e-value of < 1 × 10−5. From these, sequences with significant hits were classed as putative OBPs, CSPs, or ORs. If a sequence had hits from different sequence groups (OBPs, CSPs, or ORs), the sequence was then classified to the group which provided the greatest number of significant blast for that sequence. Note identical Protein reference sequences were discarded before blasting.
Antennal Samples
Antenna transcriptomes were generated from individuals collected as second or third instars in the field (spring 2018, field site coordinates 34.514781, −119.779256) and then reared in the laboratory for ~4 weeks in net cages on Ceanothus thyrsiflorus. Fourth instar juveniles and virgin adults of both sexes (collected 5 days after their final molt) were collected simultaneously and CO2 anesthetized before dissections.
Total RNA was extracted from the whole antennae of four individuals per developmental stage and sex. This was done by first freezing the tissue in liquid nitrogen, followed by the addition of Trizol (Life Technologies) and crushing with beads (Sigmund Lindner). The homogenized tissue was then treated with chloroform and ethanol and the aqueous layer transferred to RNeasy MinElute Columns (Qiagen). Following the RNeasy Mini Kit protocol, potential DNA in the sample was digested, RNA was eluted into water and stored at 80°C. RNA quantity and quality was measured using NanoDrop (Thermo Scientific) and Bioanalyzer (Agilent). RNA extracts were pooled and fragmented to 120 nt for strand-specific library preparation. Paired-end sequencing with a read length of 100 bp was performed on a HiSeq 2500 platform at the CIG (Center of Integrative Genomics, Lausanne, Switzerland).
Expression Analyses
To quantify gene expression of olfactory genes in the antennae, we mapped reads to the reference transcriptome using Kallisto (v. 0.43.1) (Bray et al., 2016) with the following options: –bias –rf-stranded. Before mapping, adapter sequences were trimmed from raw reads with CutAdapt (Martin, 2011). Reads were then quality trimmed using Trimmomatic v 0.36 (Bolger et al., 2014), clipping leading or trailing bases with a phred score of <10 from the read, before using a sliding window from the 5′ end to clip the read if 4 consecutive bases had an average phred score of <20. Any reads with a sequence length of <80 after trimming were discarded. Any unpaired reads after this trimming were then also discarded.
Expression analyses were performed using the Bioconductor package EdgeR (v. 3.18.1) (Robinson et al., 2010) in R (v. 3.4.1) (R Core Team, 2017). Firstly, gene-level counts were obtained from the transcript-level estimates using the tximport package (Soneson et al., 2015). Genes with a low expression in the antennae samples (transcripts per million (TPM) <0.5 in 2 or more libraries) were excluded from the expression analyses. Normalization factors for each library were computed using the TMM method in EdgeR. To estimate dispersion, we then fit a generalized linear model (GLM) with negative binomial distribution with developmental stage, sex, and their interaction as explanatory variables. To identify genes that differed in expression between developmental stages and between the sexes, we used a quasi-F test to determine the significance for each gene by comparing appropriate model contrasts. P-values were then corrected for multiple tests using Benjamini and Hochberg's algorithm (Benjamini and Hochberg, 1995), with statistical significance set to 5%.
Sequence Evolution of Olfactory Genes in the Asexual Species T. monikensis
To test for differences in the rate of evolutionary divergence between olfactory genes and other genes in the asexual species T. monikensis, we calculated dN/dS ratios for each of the one-to-one orthologs between T cristinae, T. monikensis, and T. californicum (outgroup species) using the ortholog matrix from Bast et al. (2018). All of the one-to-one orthologs were aligned with prank (v.100802, codon-mode) (Löytynoja and Goldman, 2005) and then curated with Gblocks (v. 0.91b, type = codons, minimum block length = 4) (Talavera and Castresana, 2007). These alignments were then used as input for codeml of the PAML package (Yang, 2007) to generate maximum likelihood estimates of dN/dS for the T. monikensis terminal branch in the phylogeny (using the “free model”). Note we did not use a more complex model for examining differences in evolutionary rate (e.g., branch-site models) as we are interested in identifying genes which have increased accumulation of deleterious changes due to the reduced purifying selection (indicated by an increase in dN/dS), rather than genes showing evidence of positive selection. dN/dS estimates where dS = 0 or dN/dS > 1 were discarded. Differences of dN/dS between gene categories were assessed using a Wilcoxon test in R.
Results
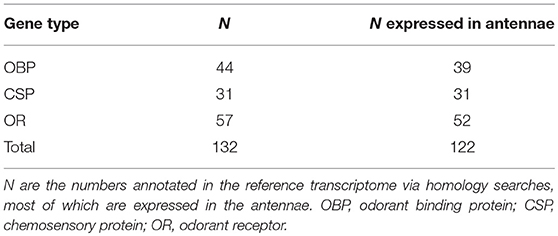
We identified a total of 132 putative olfactory genes in T. cristinae, 122 of which were expressed at least at a low level in the antennae (Table 1, Supplementary Table 1). Factors influencing antennal expression pattern of olfactory genes varied by olfactory gene type, with sex inferencing OBP expression more than the developmental stage, and vice versa for CSPs and ORs (Figure 1). The majority of olfactory genes (68%) showed a significant change in expression between developmental stages in males and/or females (OBPs: 28/39, CSPs: 23/31, ORs: 32/52) while only a minority of them were sex-biased in adults (OBPs: 18/39, CSPs: 7/31, ORs: 16/52).
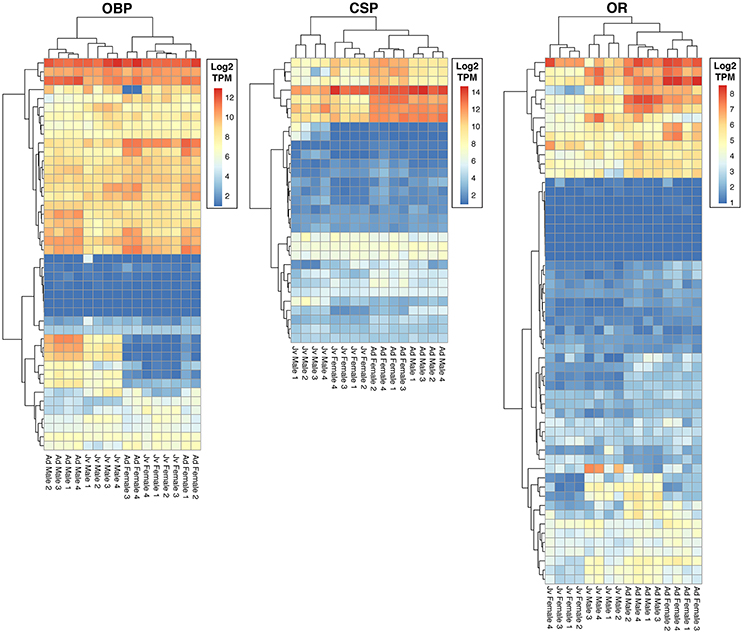
Figure 1. Heatmaps of expression of olfactory genes in antennae of juvenile and adult males and females of Timema cristinae. OBP, odorant binding proteins; CSP, chemosensory proteins; OR, odorant receptors. Jv, Juvenile; Ad, Adult.
Olfactory genes evolve significantly faster than genes with other functions in the asexual species T. monikensis (Figure 2A). However, the comparison is only significant if the three gene categories are pooled; only ORs show significantly elevated rates of evolution if the three gene categories are analyzed separately (Figure 2B). Note, a similar result was found when using dN/dS values for T. cristinae (Supplementary Figure 1). We were only able to calculate a dN/dS ratio for 58 of the 122 expressed olfactory genes (47%), in large part due to a lack of orthologous sequences. This percentage is not specific to olfactory genes as we obtained a similar percentage for non-olfactory genes (45%) (Supplementary Table 1).
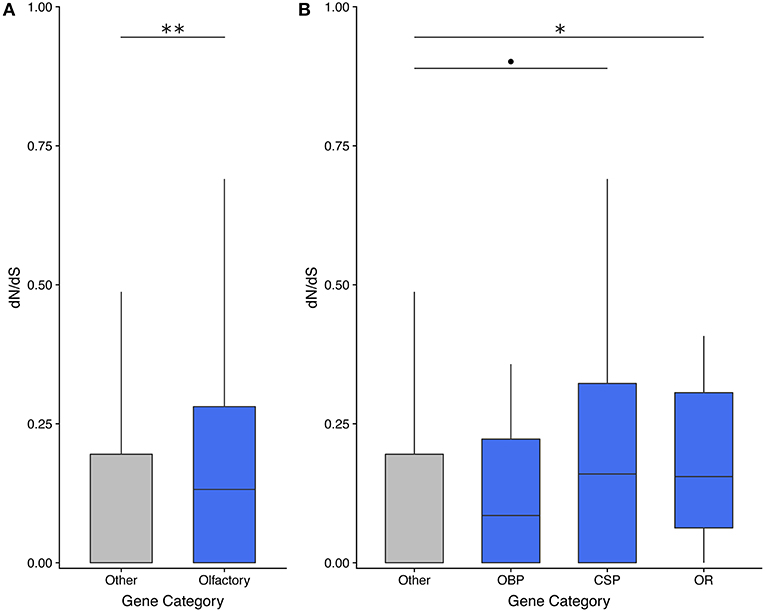
Figure 2. dN/dS analyses reveal that olfactory genes evolve faster than genes with other functions in the asexual species T. monikensis. OBP, odorant binding proteins; CSP, chemosensory proteins; OR, odorant receptors. Codes for levels of significance are 0.01 “**,” 0.05 “*,”0.1 “•” from a Wilcoxon test.
We then assessed whether any genes featured patterns consistent with expectations for olfactory genes involved in sexual communication. This would be the case for genes featuring increased expression in adults, sex-bias and an elevated rate of evolution. However, when we combined expression shifts between developmental stages, sex bias and evolutionary rates of olfactory genes (Figure 3) we found no genes that fit these predictions of olfactory genes.
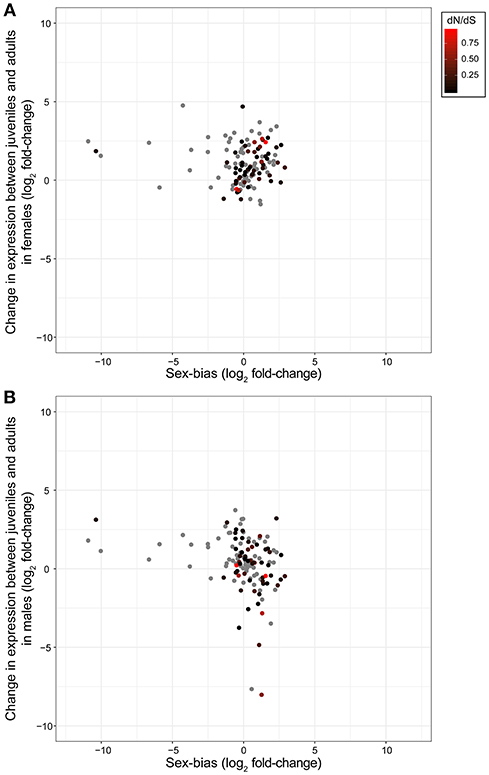
Figure 3. Expression changes of olfactory genes between T. cristinae developmental stages in females (A) and males (B), and between adult males and females. Olfactory genes involved in sexual communication are expected to mostly be expressed in adults and feature sex-biased expression. They are further expected to be under relaxed selection in asexual species (red points, see legend) where sexual communication no longer occurs. (No dN/dS rates could be determined for the gray points because of lack of polymorphism or orthology). None of the candidate olfactory genes identified in T. cristinae clearly fit with these expectations. Positive values for sex-bias indicate higher expression in females, and positive values for expression between developmental stages indicate higher expression in adults.
Discussion
We identified putative genes for olfactory proteins in Timema cristinae and studied their expression in antennae. These proteins include 44 odorant binding proteins, 31 CSPs, and 57 odorant receptors. The number of olfactory proteins identified in T. cristinae is higher than in other phasmids whose olfactory repertoire has been studied [Clitarchus hookeri: OBP = 10, CSP = 12, and OR = 16 (Wu et al., 2016), and in Phyllium siccifolium: OR = 30 (Missbach et al., 2014)], for unknown reasons. Such differences in olfactory protein numbers are unlikely to be driven primarily by differences in study design as all studies used an RNA-seq approach which included antennal tissue. Despite this, the number of genes in these different gene families are known to vary extensively among insects (Brito et al., 2016; Pelosi et al., 2018), and in the vast majority of species, there are no studies designed to identify the subset of these genes that are directly involved in odorant reception vs. those are involved in other functions. As a consequence, the diversification of genes in different olfactory gene families does not inform the complexity of the chemical communication system in a given species.
We expected that olfactory proteins involved in sexual functions such as the detection of pheromones should be expressed mostly in adults and not in juveniles, and generally feature sex biased expression. However, there was no set of olfactory proteins that fit this description and we found no relation between expression shifts in juveniles and adults and the level of sex bias. There was also no group of olfactory genes featuring particularly fast evolutionary rates in the asexual species, as could be expected for olfactory genes involved in sexual communication. A possible explanation for the lack of a pattern in our data is that we used homology of known insect olfactory proteins to identify olfactory proteins in Timema. This approach constrains the set of analyzed proteins to those that are relatively conserved among distantly related species. However, proteins involved in sexual communication often diverge very rapidly between species (e.g., Picimbon and Gadenne, 2002; Guo and Kim, 2007; Sánchez-Gracia et al., 2009). Therefore, the lack of pattern in our data may stem from the fact that our set of proteins may largely lack the set involved in sexual functions. An additional complication may be that our study only examined gene expression levels, but not protein expression levels. It is therefore possible that our study could have missed important differences in expression levels if there was extensive post-translational regulation. Future work examining both protein and gene expression levels are needed to investigate this.
Olfactory proteins overall evolve faster in the asexual species T. monikensis than proteins with other functions. We were able to detect such accelerated evolution in spite of the fact that there is little power to detect rate variations among genes in asexuals. Gene level selection is relatively inefficient in asexuals given all alleles co-segregate as a single linkage block in the absence of meiosis and outcrossing. The implications are that the differences in selection pressures, acting on olfactory proteins vs. other proteins in T. monikensis, must be quite extensive. Because we could not identify subsets of olfactory proteins likely associated with sexual functions, we could not test whether rapid evolution of olfactory proteins overall is a consequence of relaxed selection for sexual functions in asexuals, or whether it reflects the fact that olfactory proteins are relatively “dispensable” overall. The fact that we also observed an elevated rate of sequence evolution for olfactory proteins in the sexual species T. cristinae suggests the latter, however, it is also likely that sexual selection may increase the dN/dS of olfactory proteins in sexual species (e.g., Guo and Kim, 2007; Sánchez-Gracia et al., 2009). There is extensive evidence for relaxed and/or negative selection on sexual phenotypes in asexual species (van der Kooi and Schwander, 2014). By contrast, it has thus far been difficult to detect corresponding signatures in genes underlying sexual traits, perhaps because sexual trait decay proceeds largely via an altered expression of certain gene networks, rather than the pseudogenization of specific genes. Only a single study, on an asexual wasp species, detected a disproportionate accumulation of deleterious mutations in genes likely involved in male functions (Kraaijeveld et al., 2016). However, this wasp produces haploid eggs via meiosis and diploidy in the eggs is restored secondarily via fusion of the daughter cells of the first mitotic division (so-called gamete duplication). This allows for both, gene-specific selection (because normal meiosis occurs) as well as the rapid fixation of beneficial mutations (because they are immediately homozygous).
In conclusion, we found that putative olfactory proteins are typically differentially expressed between juveniles and adults in T. cristinae, but there is little overlap of differential expression between developmental stages and the level of sex bias in adults. Furthermore, while we found evidence that olfactory proteins are indeed under relaxed selection in the asexual species T. monikensis, there is no evidence that this is necessarily the case for olfactory genes with a putative role in sexual communication. Nevertheless, the list of olfactory genes generated in our study provides a useful tool for future studies on olfaction in Timema and other stick insects.
Data Availability
Raw reads have been deposited in the SRA with the following accession numbers: SRR8182935-SRR8182950.
Author Contributions
TS and DP designed the study. TS collected samples. DP and JD analyzed the data with input TS. DP and TS wrote the manuscript with input from all authors.
Funding
This study was supported by Swiss FNS grants PP00P3_170627 and CRSII3_160723.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00101/full#supplementary-material
References
Arbuthnott, D., and Crespi, B. J. (2009). Courtship and mate discrimination within and between species of Timema walking-sticks. Anim. Behav. 78, 53–59. doi: 10.1016/j.anbehav.2009.02.028
Bast, J., Parker, D. J., Dumas, Z., Jalvingh, K. M., Tran Van, P., Jaron, K. S., et al. (2018). Consequences of asexuality in natural populations: insights from stick insects. Mol. Biol. Evol. 35, 1668–1677. doi: 10.1093/molbev/msy058
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Brito, N. F., Moreira, M. F., and Melo, A. C. A. (2016). A look inside odorant-binding proteins in insect chemoreception. J. Insect. Physiol. 95, 51–65. doi: 10.1016/j.jinsphys.2016.09.008
Burke, N. W., Crean, A. J., and Bonduriansky, R. (2015). The role of sexual conflict in the evolution of facultative parthenogenesis: a study on the spiny leaf stick insect. Anim. Behav. 101, 117–127. doi: 10.1016/j.anbehav.2014.12.017
Guo, S., and Kim, J. (2007). Molecular evolution of Drosophila odorant receptor genes. Mol. Biol. Evol. 24, 1198–1207. doi: 10.1093/molbev/msm038
Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1146/annurev.ento.51.051705.113646
Kraaijeveld, K., Anvar, S. Y., Frank, J., Schmitz, A., Bast, J., Wilbrandt, J., et al. (2016). Decay of sexual trait genes in an asexual parasitoid wasp. Genome Biol. Evol. 8, 3685–3695. doi: 10.1093/gbe/evw273
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Löytynoja, A., and Goldman, N. (2005). An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U.S.A. 102, 10557–10562. doi: 10.1073/pnas.0409137102
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. Elife 3:e02115. doi: 10.7554/eLife.02115
Myers, S. S., Buckley, T. R., and Holwell, G. I. (2015). Mate detection and seasonal variation in stick insect mating behaviour (Phamatodea: Clitarchus hookeri). Behaviour 152, 1325–1348. doi: 10.1163/1568539X-00003281
Nosil, P., Crespi, B. J., Gries, R., and Gries, G. (2007). Natural selection and divergence in mate preference during speciation. Genetica 129, 309–327. doi: 10.1007/s10709-006-0013-6
Pelosi, P., Iovinella, I., Zhu, J., Wang, G., and Dani, F. R. (2018). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 93, 184–200. doi: 10.1111/brv.12339
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Picimbon, J. F., and Gadenne, C. (2002). Evolution of noctuid pheromone binding proteins: identification of PBP in the black cutworm moth, Agrotis ipsilon. Insect Biochem. Mol. Biol. 32, 839–846. doi: 10.1016/S0965-1748(01)00172-2
Pickett, J. A., and Glinwood, R. T. (2007). “Chemical ecology,” in Aphids as Crop Pests, eds H. F. van Emden and R. Harrington (Wallingford: CAB International Press), 235–260.
R Core Team (2017). R: A Language and Environment for Statistical Computing. Available online at: https://www.R-project.org/ (accessed March 23, 2019).
Riesch, R., Muschick, M., Lindtke, D., Villoutreix, R., Comeault, A. A., Farkas, T. E., et al. (2017). Transitions between phases of genomic differentiation during stick-insect speciation. Nat. Ecol. Evol. 1:0082. doi: 10.1038/s41559-017-0082
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sánchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Sandoval, C. P. (1993). Geographic, Ecological and Behavioral Factors Affecting Spatial Variation in Color Morph Frequency in the Walking-Stick, Timema Cristinae. Ph.D. Thesis, University of California, Santa Barbara, CA.
Schwander, T., Arbuthnott, D., Gries, R., Gries, G., Nosil, P., and Crespi, B. J. (2013a). Hydrocarbon divergence and reproductive isolation in Timema stick insects. BMC Evol. Biol. 13:151. doi: 10.1186/1471-2148-13-151
Schwander, T., and Crespi, B. J. (2009). Multiple direct transitions from sexual reproduction to apomictic parthenogenesis in Timema stick insects. Evolution 63, 84–103. doi: 10.1111/j.1558-5646.2008.00524.x
Schwander, T., Crespi, B. J., Gries, R., and Gries, G. (2013b). Neutral and selection-driven decay of sexual traits in asexual stick insects. Proc. Biol. Sci. 280:20130823. doi: 10.1098/rspb.2013.0823
Soneson, C., Love, M. I., and Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4:1521. doi: 10.12688/f1000research.7563.1
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Tilgner, E. H. (2009). “Phasmida: Stick and Leaf Insects,” in Encyclopedia of Insects, 2nd Edn., eds V. H. Resh and R. T. Cardé (London: Elsevier), 765–766.
Touhara, K., and Vosshall, L. B. (2009). Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332. doi: 10.1146/annurev.physiol.010908.163209
van der Kooi, C. J., and Schwander, T. (2014). On the fate of sexual traits under asexuality. Biol. Rev. Camb. Philos. Soc. 89, 805–819. doi: 10.1111/brv.12078
Vickery, V. R., and Sandoval, C. P. (2001). Descriptions of three new species of Timema (Phasmatoptera: Timematodea: Timematidae) and notes on three other species. J. Orthoptera Res. 10, 53–61. doi: 10.1665/1082-6467(2001)010[0053:DOTNSO]2.0.CO;2
Whiting, M. F., Bradler, S., and Maxwell, T. (2003). Loss and recovery of wings in stick insects. Nature 421, 264–267. doi: 10.1038/nature01313
Wu, C., Crowhurst, R. N., Dennis, A. B., Twort, V. G., Liu, S., Newcomb, R. D., et al. (2016). De novo transcriptome analysis of the common New Zealandstick insect Clitarchus hookeri (Phasmatodea) reveals genes involved in olfaction, digestion and sexual reproduction. PLoS ONE 11:e0157783. doi: 10.1371/journal.pone.0157783
Keywords: timema, gene expression, olfacation, selection, antennae
Citation: Parker DJ, Djordjevic J and Schwander T (2019) Olfactory Proteins in Timema Stick Insects. Front. Ecol. Evol. 7:101. doi: 10.3389/fevo.2019.00101
Received: 30 November 2018; Accepted: 13 March 2019;
Published: 05 April 2019.
Edited by:
Sven Bradler, University of Göttingen, GermanyReviewed by:
Francesca Romana Dani, University of Florence, ItalyDmitry Yurievich Sherbakov, Limnological Institute (RAS), Russia
Copyright © 2019 Parker, Djordjevic and Schwander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren J. Parker, ZGFycmVuSmFtZXMuUGFya2VyQHVuaWwuY2g=
 Darren J. Parker
Darren J. Parker Jelisaveta Djordjevic1
Jelisaveta Djordjevic1 Tanja Schwander
Tanja Schwander