Abstract
Estimating the heritability and genotype by environment (GxE) interactions of performance-related traits (e.g., growth, survival, reproduction) under future ocean conditions is necessary for inferring the adaptive potential of marine species to climate change. To date, no studies have used quantitative genetics techniques to test the adaptive potential of large pelagic fishes to the combined effects of elevated water temperature and ocean acidification. We used an experimental approach to test for heritability and GxE interactions in morphological traits of juvenile yellowtail kingfish, Seriola lalandi, under current-day and predicted future ocean conditions. We also tracked the fate of genetic diversity among treatments over the experimental period to test for selection favoring some genotypes over others under elevated temperature and CO2. Specifically, we reared kingfish to 21 days post hatching (dph) in a fully crossed 2 × 2 experimental design comprising current-day average summer temperature (21°C) and seawater pCO2 (500 μatm CO2) and elevated temperature (25°C) and seawater pCO2 (1,000 μatm CO2). We sampled larvae and juveniles at 1, 11, and 21 dph and identified family of origin of each fish (1,942 in total) by DNA parentage analysis. The animal model was used to estimate heritability of morphological traits and test for GxE interactions among the experimental treatments at 21 dph. Elevated temperature, but not elevated CO2 affected all morphological traits. Weight, length and other morphological traits in juvenile yellowtail kingfish exhibited low but significant heritability under current day and elevated temperature. However, there were no measurable GxE interactions in morphological traits between the two temperature treatments at 21 dph. Similarly, there was no detectable change in any of the measures of genetic diversity over the duration of the experiment. Nonetheless, one family exhibited differential survivorship between temperatures, declining in relative abundance between 1 and 21 dph at 21°C, but increasing in relative abundance between 1 and 21 dph at 25°C. This suggests that this family line could perform better under future warming than in current-day conditions. Our results provide the first preliminary evidence of the adaptive potential of a large pelagic fisheries species to future ocean conditions.
Introduction
Ocean warming and acidification are predicted to have far-reaching impacts on marine biodiversity and fisheries before the end of this century (Doney et al., 2012; Hoegh-Guldberg et al., 2014; Pörtner et al., 2014; Phillips and Pérez-Ramírez, 2017). Indeed, changes in geographical distributions and phenology that are consistent with climate change predictions have already been observed in many marine species (Simpson et al., 2011; Poloczanska et al., 2013). Marine heatwaves are also increasing in frequency and duration, with devastating effects in some ecosystems (Hughes et al., 2018; Richardson et al., 2018; Stuart-Smith et al., 2018). Significant biological effects of contemporary ocean acidification are more difficult to ascertain, but there is evidence that anthropogenic carbon dioxide is already exacerbating the impacts of corrosive seawater incursions that occur during periodic upwelling events in some coastal ecosystems (Bednaršek et al., 2014; Ekstrom et al., 2015; Chan et al., 2017). Predictions about the impacts of future ocean warming and ocean acidification on marine organisms are primarily derived from laboratory experiments that rear animals at projected future temperatures, ocean acidification conditions, or both, and assess the performance of the animals in these conditions compared with a current-day control (Kroeker et al., 2013; Boyd et al., 2018). Such experiments usually last for several weeks, or maybe months at the most. Thus, while these experiments are informative about the potential effects of climate change conditions on the performance of marine organisms, they do not account for adaptation that might occur over the timeframe that the environmental changes are actually occurring (Riebesell and Gattuso, 2015). Therefore, to provide robust predictions about the likely impacts of ocean warming and acidification, it is also necessary to assess the adaptive potential of marine species to these environmental changes (Munday et al., 2013; Sunday et al., 2014).
The larval and early juvenile stages of marine species are generally more susceptible to higher temperatures and ocean acidification conditions than are older juveniles and adults (Pörtner and Farrell, 2008; Melzner et al., 2009; Pörtner and Peck, 2010). In marine fish, temperatures above the summer average can affect a range of performance-related traits in larval stages, including developmental rate, growth, survivorship, aerobic scope and swimming ability (reviewed in Pankhurst and Munday, 2011; Llopiz et al., 2014), with implications for population recruitment and fisheries productivity (Hollowed et al., 2013; Llopiz et al., 2014). Elevated CO2 levels have also been observed to affect developmental rate, growth, survivorship, aerobic scope and swimming ability in larval stages of some marine fishes (reviewed in Heuer and Grosell, 2014; Cattano et al., 2018; Hannan and Rummer, 2018). As observed with temperature, negative effects of elevated CO2 on larval development and survivorship could have serious implications for population recruitment and fisheries productivity in some species, such as Atlantic cod (Stiasny et al., 2016; Koenigstein et al., 2018). Therefore, determining the potential for fish populations to adapt to warming, ocean acidification, and their combined effects, is crucial for making predictions about the future impacts of these environmental stressors on fish population, and the fisheries productivity they support (Heenan et al., 2015).
Adaptation to climate change depends on the presence of heritable genetic variation in fitness related traits that respond to the changing environment (Lynch and Walsh, 1998). Therefore, a first step in assessing evolutionary potential to climate change is to estimate the heritability of thermally sensitive traits that could limit individual performance in the future (Munday et al., 2013; Reusch, 2014). Genotype by environment interactions (GxE) are another important component of assessing adaptive potential to climate change, because they describe the response of different genotypes under current-day and projected future conditions. In particular, they indicate if all genotypes might exhibit the same change in performance, or if there are some genotypes that outperform others under altered environmental conditions (Falconer and Mackay, 1996). Finally, it is necessary to consider genetic correlations between traits measured in different environments (e.g., warming vs. acidification) because they influence the potential for a trait to evolve under the combined effects of multiple environmental stressors (Munday et al., 2013; Sunday et al., 2014). In particular, if a negative genetic correlation is present, selection will be acting orthogonally to the direction of maximum genetic variation in the population, thereby reducing the response to selection.
To date, only a handful of studies have estimated trait heritability and adaptive potential of fishes under elevated temperature (Muñoz et al., 2015; Munday et al., 2017) or ocean acidification conditions (Malvezzi et al., 2015; Welch and Munday, 2017; Tasoff and Johnson, 2018), and none have estimated heritability under the combined effect of both environmental stressors. For example, Munday et al. (2017) showed that morphometric traits and aerobic performance measured during early life were highly heritable in the spiny damselfish, Acanthochromis polyacanthus, under current-day and projected future temperatures. Furthermore, there were significant GxE interactions, indicating that some genetic lineages had greater performance than others at higher temperatures. Similarly, Malvezzi et al. (2015) found that survivorship of larval Atlantic silverside, Menidia menidia, had a significant heritable genetic component under elevated CO2. These estimates of heritability are valuable, because they indicate that evolutionary responses to warming and ocean acidification are likely in these species. Moreover, they can potentially be used to estimate the rate of evolutionary change that might be expected under different selection intensities (Falconer and Mackay, 1996). However, many more estimates of trait heritability and GxE interactions, in other species, for other traits, and under combined effects of elevated temperature and ocean acidification conditions, are needed to determine if adaptive potential to these environmental stressors is widespread in marine fishes.
Here, we used an experimental approach to test for heritability and GxE interactions in early life history traits of the yellowtail kingfish, Seriola lalandi, under elevated temperature and CO2 conditions. Furthermore, we compared genetic diversity among the treatments at key developmental stages to test for selection favoring some genotypes over others under elevated temperature and CO2. Specifically, we reared juvenile kingfish from a mass spawning to 21 days post hatching (dph) in a fully crossed 2 × 2 experimental design that consisted of: (1) current-day average summer temperature (21°C) and seawater pCO2 (500 μatm CO2) for the study location, (2) current-day average summer temperature (21°C) and elevated seawater pCO2 (1,000 μatm CO2), (3) elevated temperature (25°C) and current-day seawater pCO2 (500 μatm CO2) and (4) elevated temperature (25°C) and elevated seawater pCO2 (1,000 μatm CO2). The elevated temperature and CO2 levels were consistent with projections for the ocean by the year 2100 under Representative Carbon Pathway (RCP) 8.5 (Collins et al., 2013). We sampled fish from all the treatments at 1, 11, and 21 dph and identified the family of origin of each sampled fish by DNA parentage analysis. This allowed us to track the fate of genetic diversity among the experimental treatments through time and test for evidence of selection on specific genotypes under elevated temperature, elevated CO2 or their combination. For fish sampled at 21 dph we also coupled the parentage information for each fish with data on weight, length and other morphological traits measured in the same fish (Watson et al., 2018) to estimate the heritability of these traits and test for GxE interactions among the experimental treatments. Our aim was to investigate the adaptive potential to ocean conditions that could occur by the end of the twenty first century in the yellowtail kingfish, a high-value and commercially harvested fish species for human food consumption.
Materials and Methods
Study Location and Species
This study focused on the New Zealand population of the yellowtail kingfish, Seriola lalandi, and was conducted at the National Institute of Water and Atmospheric Research (NIWA) Northland Marine Research Centre, Ruakaka, New Zealand. The yellowtail kingfish is a large coastal pelagic fish with a circumglobal distribution in subtropical waters. In New Zealand waters it reaches up to 1.7 m in length and over 50 kg in weight (Taylor and Willis, 1998; Walsh et al., 2003; McKenzie et al., 2014). It is a powerful swimmer adapted to a pelagic lifestyle and supports an important recreational and commercial fishery in New Zealand, Australia, Japan and other subtropical regions. The yellowtail kingfish is emerging as an important species for testing the effects of environmental change on large pelagic fishes (e.g., Watson et al., 2018), because it is one of the few species that can be bred and reared in captivity (Symonds et al., 2014; Sicuro and Luzzana, 2016). The ability to breed and rear yellowtail kingfish in captivity also makes is amenable to quantitative genetic studies, such as those conducted here, because these approaches require the ability to identify and compare traits among individuals of known relatedness (e.g., full sibs, half sibs).
Broodstock, Eggs, and Larval Culture
Spawning stocks of yellowtail kingfish were maintained outdoors in 5 × 20 m3 circular tanks. The broodstock consisted of 26 locally sourced, wild-caught adult fish (four to six fish per tank with approximately equal sex ratio in each tank) that had been domesticated in tanks for up to 9 years. In the current study, experiments were conducted in conjunction with a natural spawning event that occurred on the night of 23/01/2017. Four broodstock tanks containing a total of 9 females, 10 males and one fish of unknown sex contributed to spawning. Time of spawning was consistent across tanks, occurring within the last 2 h of daylight. Long-term mean summer ocean temperature for the region is 21°C (Shears and Bowen, 2017), however, local conditions vary naturally and ambient water temperature was 19–20°C in the 5 days immediately prior to spawning and then dropped to 18.2°C on the night of spawning. Ambient pHT and pCO2 were 7.91 and 589 μatm, respectively, in the spawning tanks (Table S1). A representative proportion of floating eggs from the four contributing tanks were collected the morning after spawning, mixed, rinsed with oxygenated seawater for 5 min, and disinfected with Tosylchloramide (chloramine-T) at 50 ppm for 15 min. Eggs were then rinsed with seawater and distributed into 24 conical 400 L incubation tanks at a density of approximately 100,000 eggs per tank at 12:45 h on 24/01/2017. The 400 L incubation tanks were at ambient ocean temperature (18.2°C) at stocking. Heating was turned on at 15:30 h and allowed to slowly rise to the treatment set points of 21°C and 25°C overnight. Tanks received flow-through seawater at a flow rate of 3 L min−1 with a photoperiod of 14 h light and 10 h dark. Eggs hatched 2 days after stocking at 25°C and 3 days after stocking at 21°C and larvae were reared for a further 1 day in the incubation tanks before transfer to grow-out tanks.
At 1 dph, larvae were transferred from their rearing tanks into 24 reciprocal grow-out tanks at a density of approximately 45,000 larvae per tank (44,227 ± 2,152). Grow-out tanks were 1,500 L circular tanks with slightly sloping bottoms with a black internal surface. Each grow-out tank received flow-through seawater at either 21 or 25°C with a photoperiod of 14 h light and 10 h dark and at a flow rate of 3 L min−1 and gentle aeration. Larvae were fed with rotifers enriched with S.presso (INVE Aquaculture, Belgium) up to 4 times per day until 14 dph, transitioning to S.presso enriched Artemia up to 4 times per day from 11 dph until the end of the experiment.
Experimental System and Water Chemistry
Seawater pumped from the ocean was filtered through mixed-media (sand), bag filtered to 5 μm, UV light treated to 150 mW.cm−2 and delivered to large header tanks. Oxygen diffusers in the header tanks maintained baseline minimum dissolved oxygen (100% saturation) and foam fractionators removed any additional organics. Seawater from each header tank was gravity-fed into eight separate 100 L sump tanks where temperature was maintained at ambient control 21°C or elevated to 25°C and CO2 was maintained at ambient control (~500 μatm) or elevated (~1,000 μatm) CO2 in a fully-crossed 2 × 2 experimental design with 2 replicate sumps for each treatment. Seawater from each of the eight treatment sumps was pumped into three of the 400 L incubation tanks during the egg incubation stage and three of the 1,500 L rearing tanks during the grow-out stage, so that there were six replicate experimental tanks at each temperature and CO2 level throughout the experiment.
The pHtotal and temperature of each rearing tank were measured daily (SG8 SevenGo Pro, Mettler Toledo, Switzerland). The pH electrode was calibrated with Tris buffers obtained from Prof. A.G. Dickson (Scripps Institution of Oceanography, batch number 26). Water samples for total alkalinity (TA) analysis were taken from all rearing tanks at the start, middle and end of the experiment to match the fish sampling at 1, 11, and 21 dph. TA and salinity determination was conducted by the University of Otago Research Centre for Oceanography, Dunedin, New Zealand. See Watson et al. (2018) for further details of CO2 treatment and water sampling methods.
Carbonate chemistry parameters in each tank were calculated in CO2SYS using the measured values of pHtotal, salinity, temperature and TA and the constants K1, K2 from Mehrbach et al. (1973) refit by Dickson and Millero (1987) and Dickson et al. (2007) for KHSO4. Seawater carbonate chemistry parameters are shown in Table S1.
Fish Sampling and Morphology
A random subset of approximately 30 fish per tank was sampled at 1, 11, and 21 dph. At 1 and 21 dph, fish were sampled from all 24 tanks (6 replicate tanks per treatment). At 11 dph, fish were sampled from the first 16 grow-out tanks only (4 tanks per treatment). Each fish was weighed and then photographed with a Leica DFC 420 camera fitted to a Leica MZ7.5 stereo microscope or, for larger individuals, a Canon G16 series camera fitted to a stand. Fish were then fixed in 90% ethanol for DNA extraction and genotyping of microsatellite markers. Fish collected at 21 dph were stored individually so that morphological traits measured from the photographs could be matched to the genotype of each fish. From the photographs we measured a range of standard morphometric traits that are indicators of growth and development in larval fishes: standard length (SL), total length (TL), total depth at vent including fins (fin depth at vent) (FDV), eye diameter (ED), mandible length (M), head length (HL), head depth (HD) (see Watson et al., 2018 for an illustration showing each trait). Morphometric traits were extracted from the photographs using Image J software with the image displayed on a high-resolution computer screen. The observer was blinded to the treatments when extracting morphological data from the photographs.
DNA, Microsatellites and Genetic Diversity Analyses
DNA was extracted from caudal fin tissue of all sampled fish using DNeasy 96 Blood & Tissue Kits (Qiagen) according to the manufacturer instructions. Eleven microsatellite loci were amplified in one multiplex reaction. Primer sequences and exact primer amount can be found in the Supplementary Materials (Table S2). Each of the primers was color labeled and amplified using a PCR cycle of 95°C for 15 min followed by a denaturing at 95°C for 38 cycles of 30 s. Annealing was performed at 60°C for 90s with a 60s extension at 72°C and a final extension of 60°C for 30 min. Each reaction of 15ul consisted of 7.5 μl of Qiagen Taq PCR Master Mix, 0.84 μl of primer mix. For the 1 dph and 11 dph samples 5 μl of DNA was added (approximately 100–200 ng) and mixed with 1.66 μl H20. For the larger 21 dph samples, 2 μl of DNA for an equal amount of DNA was added and mixed with 4.66 μl H20. The amplified PCR products were then purified with ExoSAP-IT (Affymetrix, Inc). Screening was performed on an ABI 3700 sequencer in the core lab facilities of the King Abdullah University of Science and Technology. The internal size standard genescan-500LIZ (Applied Biosystems, Inc.) was used to size and score alleles with Geneious R8 microsatellite plugin (Kearse et al., 2012). Alele scoring was performed while blinded to the sample treatments.
Genotypes for 1942 individual offspring sampled at 1 (n = 735), 11 (n = 488), and 21 (n = 719) dph were used to estimate genetic diversity in the sample population and to test for any effect of treatments on genetic diversity between days 1, 11, and 21 dph. The measures of genetic diversity used were allelic richness, allele frequencies, number of private alleles, Shannon's diversity index and heterozygosity. Each diversity measure was calculated for each of the 11 microsatellite loci for each of the four CO2 × temperature treatments and three sampling times (1, 11, and 21 dph). Allele frequencies and genetic diversity metrics were calculated using GenePop (Rousset, 2008) and GenAlEx version 6.5 (Peakall and Smouse, 2012) was used to calculate observed and expected heterozygosity. Comparisons of genetic diversity measures between treatment groups were performed with a PERMANOVA with the particular diversity index (separately as well as combined) as the dependent variable and treatment (4 levels) and sampling time (3 levels) as fixed effects. Data transformation was done using square root, distance was then measured with the Bray-Curtis index and tests were performed with 999 permutations in the package vegan v2.5.5 (Oksanen et al., 2019) in R (R Core Team, 2018). Tests were performed to evaluate the differences among sample points and treatments in heterozygosity, allelic richness and frequency for all family lines included as well as for the five dominant family lines only. Fisher's Exact tests were used to compare the relative changes in abundance between family lines and treatments.
Parental Assignment
Parentage of the 1942 sampled offspring was determined by matching the 11 microsatellite markers in the offspring with the same ll microsatellite markers in the 26 genotyped broodstock fish (i.e., putative parents). We included parental genotypes from the non-spawning tank as an additional control for false assingments. Two programs (Colony v2.0.6.4 and Cervus v 3.0.7) were run to establish parent-offspring trios and only consensus trios were used in the further analysis. All potential parents were known as well as the exact number of potential parents per tank (2–3 females and 2–4 males). In Colony, parameters were set to allow for female and male polygamy and the length of the run was chosen to be long (Jones and Wang, 2010). In Cervus, the parental pairs (sexes known) option was selected (Kalinowski et al., 2007).
Heritability and GxE Interactions
Heritabilities of yellowtail kingfish morphometric traits at 21 dph were estimated by fitting an animal model (Henderson, 1984) using residual maximum likelihood approaches in ASRmel 4.1 (VNSI, UK):
where y is the vector of the (Log10 transformed) phenotypic observations of each trait (weight, SL, TL, FDV, ED, M, HL, HD), β is the vector of the fixed effects (water temperature, two levels and CO2 concentration, two levels), a and t are respective vectors of random animal additive genetic and common environment tank effects, e is a random error associated with the individual record, i.e., residual effects, X, Z, and E are incidence matrices that relate observations to the respective effects. Conditional Wald F-tests determined that the effects of water temperature (P < 0.05), but not CO2 levels (P > 0.05) were statistically significant within models. As such, CO2 was removed as an environmental effect from heritability and GxE analyses. Heritability (h2) of each trait was estimated as /( + + ), where , , and were the variances attributed to additive genetic, random tank and residual error effects, respectively. Similarly, common environmental tank effects (c2) were estimated as /( + + ).
Due to constraints in the pedigree structure, specifically the limited number of half siblings and the presence of many full-siblings in the dataset, it was not possible to isolate potential maternal environmental effects, such as differential yolk provisioning, from family effects. This was due to the inherent nature of breeding this large pelagic fish in captivity, specifically the need to distribute adults among four separate spawning tanks to avoid overcrowding (see broodstock section above), which prevented potential crosses among fish allocated in different tanks. Furthermore, not all adults within each tank contributed to breeding due to the inherent biology of natural spawning, whereby the spawning synchronicity among breeders is uncontrollable and individual breeder contributions to a cohort are variable, resulting in certain families dominating the cohort (e.g., Domingos et al., 2014a). Despite such limitations, we proceeded with genetic analysis without factoring for maternal environmental effects as, in our experience with larvae of large and prolific natural spawners, in which females produce millions of small eggs (~1 mm in diameter) and yolk reserves are exhausted within the first week, environmental maternal effects are non-significant past metamorphosis (Domingos et al., 2014b).
To investigate for potential GxE interactions, bivariate models were used to estimate additive genetic covariances of the same morphometric trait measured in the different (water temperature) environments, modeled as different traits (Falconer, 1952). As only one observation per trait was taken for each individual, the residual covariance was set to zero and the covariance matrix was constrained to be positive definite. GxE interaction was determined as the genetic correlation (rg) between the same trait, calculated as rg = σA1A2/[√() √()], where A1A2 is the additive genetic covariance between the same trait in the different environments. Statistical significance of heritabilities and GxE interactions were further evaluated with likelihood ratio tests (Wilson et al., 2010; Munday et al., 2017).
Results
Microsatellite analysis revealed that a total of 5 adult females and 10 adult males contributed to the parentage of the offspring (Table S3). No assignments were made to adults in the non-spawning tank, supporting the robustness of assignments. A total of 1903 (1 dph = 703, 11 dph = 461, and 21 dph = 709) well-supported parental assignments were made for the 1942 fish sampled. These 1903 fish comprised 12 family genotypes (Table S3). Fish from one spawning tank (JA001) consisted of just one family genotype (i.e., all were full sibs). Fish from two spawning tanks, JA003 and JA004, comprised three and four maternal half sibs (i.e., shared the same mother, but with different fathers) with three and four different fathers, respectively. Genotypes from spawning tank JA006 comprised two pairs of maternal half sibs and one pair of paternal half sibs (i.e., shared the same father, but with different mothers). Five families dominated the sample, comprising 93.5% of the total fish sampled across the three time points (Table S3).
There was no detectable change in genetic diversity over the course of the experiment. Genetic heterozygosity did not differ among treatment or sampling time points [Table 1; F(6, 120) = 0.2627, P = 1.00]. Similarly, allelic richness and frequency exhibited close similarity among treatments and sampling time points [Table 1; all F(6, 120) < 0.0641, P = 1.00]. Combined diversity measures also did not show any differences [all F(6, 120) < 0.0504, P = 1.00]. Because 5 families dominated the samples, we also assessed combined diversity measures among treatments and time points for just these families and there were no significant differences [all F(6, 120) = 0.3278, P = 0.922], as observed in the complete data set. Nevertheless, one family had improved performance at 25°C compared with 21°C, irrespective of CO2 treatment. Specifically, Family 288_359, which had the greatest proportional contribution overall, declined in relative abundance between 1 and 21 dph at 21°C, whereas it increased in relative abundance between 1 and 21 dph at 25°C (Figure 1). This change in relative abundance was statistically significant when comparing between 21 and 25°C in both ambient CO2 (Fisher's exact test P = 0.006) and elevated CO2 (Fisher's exact test P = 0.028). No other family exhibited a statistically significant difference in relative abundance between temperature or CO2 treatments.
Table 1
| Days post hatching | 1 | 11 | 21 | 1 | 11 | 21 | 1 | 11 | 21 | 1 | 11 | 21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO2 treatment | Ambient | Ambient | Ambient | Ambient | Ambient | Ambient | High | High | High | High | High | High |
| Temperature | 21°C | 21°C | 21°C | 25°C | 25°C | 25°C | 21°C | 21°C | 21°C | 25°C | 25°C | 25°C |
| Na | 8.273 | 7.545 | 8.091 | 7.455 | 8.182 | 7.818 | 8.364 | 8.364 | 6.818 | 9.091 | 7.000 | 7.455 |
| Na Freq. ≥ 5% | 4.818 | 4.727 | 4.545 | 4.727 | 4.818 | 4.273 | 5.000 | 4.909 | 4.364 | 4.273 | 3.909 | 4.545 |
| Ne | 3.935 | 3.937 | 3.639 | 3.966 | 3.991 | 3.806 | 4.164 | 4.059 | 3.573 | 3.864 | 3.316 | 3.680 |
| SII | 1.468 | 1.453 | 1.400 | 1.453 | 1.464 | 1.439 | 1.528 | 1.507 | 1.392 | 1.485 | 1.325 | 1.389 |
| No. Private Alleles | 0.273 | 0.182 | 0.182 | 0.000 | 0.182 | 0.182 | 0.182 | 0.000 | 0.091 | 0.273 | 0.091 | 0.000 |
| He | 0.688 | 0.687 | 0.667 | 0.686 | 0.689 | 0.683 | 0.705 | 0.695 | 0.664 | 0.687 | 0.643 | 0.671 |
| Ho | 0.759 | 0.765 | 0.752 | 0.749 | 0.761 | 0.763 | 0.767 | 0.766 | 0.749 | 0.748 | 0.755 | 0.761 |
Genetic diversity of juvenile yellowtail kingfish (Seriola lalandi) reared at 21°C and 25°C in current-day ambient and elevated CO2 and sampled at 1, 11, and 21 days post hatching.
Na, No. of Different Alleles; Na (Freq ≥ 5%), No. of Different Alleles with a Frequency ≥ 5%; Ne, No. of Effective Alleles = 1 (Sum pi2); SII, Shannon's Information Index = −1* Sum [pi * Ln (pi)]; No. Private Alleles, No. of Alleles Unique to a Single Population; He, Expected Heterozygosity = 1 − Sum pi2; Ho, Observed Heterozygosity.
Figure 1
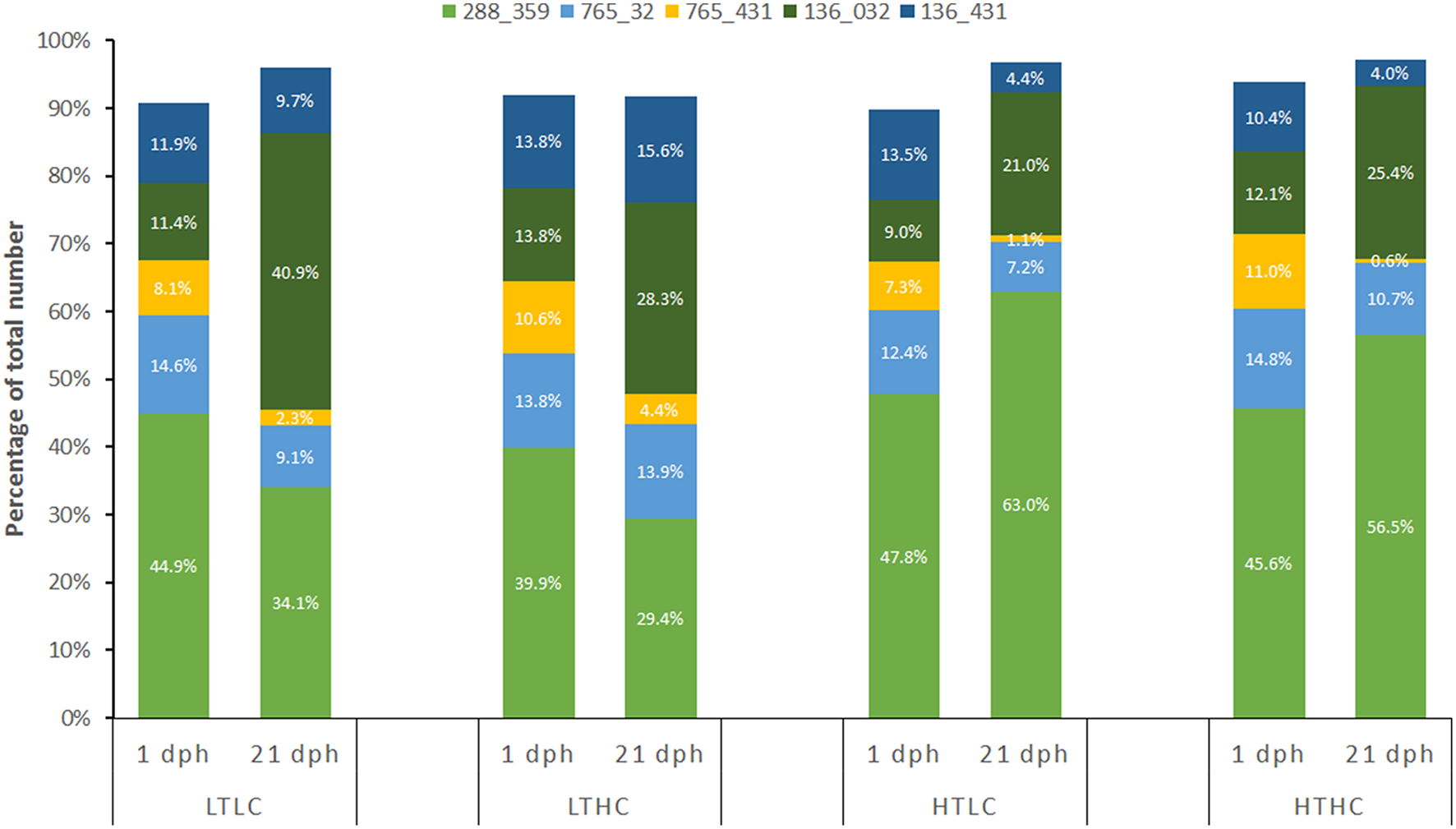
Relative proportions of the five most abundant families at 1 and 21 dph in each of the four experimental treatments. LTLC = 21°C and 500 μatm CO2; LTHC = 21°C and 1,000 μatm CO2; HTLC = 25°C and 500 μatm CO2; HTHC = 25°C and 1,000 μatm CO2. Family code is FatherID_MotherID as per Table S3.
Weight and all morphological traits at 21 dph exhibited low, but statistically significant heritabilities, ranging between 0.062 and 0.113 (Table 2). In the genetic models, temperature had a significant effect on phenotypic variances for all the measured traits (Table 2). By contrast, CO2 treatment did not affect any of the phenotypic variances. Rearing tank explained a large proportion of phenotypic variation, ranging from 0.254 to 0.398 among traits (Table 2).
Table 2
| Trait | Source of variation | Mean ± S.E | P-value | Fixed effect (P-value) | |
|---|---|---|---|---|---|
| Temp | CO2 | ||||
| W | h2 | 0.0853 ± 0.0868 | 0.004161 | <0.001 | 0.299 |
| c2 | 0.2958 ± 0.0719 | <0.0001 | |||
| SL | h2 | 0.0619 ± 0.0648 | 0.003217 | <0.001 | 0.384 |
| c2 | 0.3632 ± 0.0779 | <0.0001 | |||
| TL | h2 | 0.0641 ± 0.0670 | 0.002852 | <0.001 | 0.348 |
| c2 | 0.3530 ± 0.0771 | <0.0001 | |||
| FDV | h2 | 0.0687 ± 0.0723 | 0.013216 | <0.001 | 0.361 |
| c2 | 0.3976 ± 0.0803 | <0.0001 | |||
| ED | h2 | 0.0710 ± 0.0727 | 0.000279 | <0.001 | 0.408 |
| c2 | 0.2795 ± 0.0695 | <0.0001 | |||
| ML | h2 | 0.0902 ± 0.0902 | 0.000453 | <0.001 | 0.418 |
| c2 | 0.2539 ± 0.0664 | <0.0001 | |||
| HL | h2 | 0.1128 ± 0.1054 | <0.0001 | <0.001 | 0.354 |
| c2 | 0.2881 ± 0.0714 | <0.0001 | |||
| HD | h2 | 0.0965 ± 0.0924 | 0.000116 | <0.001 | 0.504 |
| c2 | 0.3255 ± 0.0753 | <0.0001 | |||
Heritability (h2), common environmental tank effects (c2) effects, and direct effects of temperature and CO2 treatment for 21 dph yellowtail kingfish (Seriola lalandi) weight and morphometric traits.
W, weight; SL, standard length; TL, total length; FDV, fin depth at vent; ED, eye diameter; ML, mandible length; HL, head length; HD, head depth.
As temperature, but not CO2 affected phenotypic variance, GxE interactions were only tested for the two temperature treatments. High genetic correlations (close to unity) between the same trait measured in different thermal environments indicate an absence of GxE interactions (Table 3). However, this result should be interpreted with caution, because the standard errors are large and the estimated genetic correlations where either very close to, or did not reach, statistical significance.
Table 3
| Trait | GxE (mean ± S.E) | P-value |
|---|---|---|
| Weight | 0.9990 ± 0.3033 | 0.058 |
| SL | 0.9908 ± 0.4776 | 0.040 |
| TL | 0.9786 ± 0.5210 | 0.048 |
| FDV | 0.9521 ± 0.3544 | 0.074 |
| ML | 0.9791 ± 0.2832 | 0.098 |
| ED | 0.9887 ± 0.1921 | 0.040 |
| HL | 0.9953 ± 0.4701 | 0.023 |
| HD | 0.9885 ± 0.2991 | 0.036 |
Genotype by environment (GxE) interactions for 21 dph yellowtail kingfish (Seriola lalandi) weight and morphometric traits measured at 21 and 25°C.
W, weight; SL, standard length; TL, total length; FDV, fin depth at vent; ED, eye diameter; ML, mandible length; HL, head length; HD, head depth.
Discussion
Estimating the heritability of performance and fitness related traits under predicted future ocean conditions is important for determining the adaptive potential of marine species to climate change, but has rarely been attempted for a large pelagic fish. Here we show that weight, length and other morphological traits in juvenile yellowtail kingfish at 21 dph exhibit low, but statistically significant heritabilities under current day and elevated temperature and CO2 conditions. Elevated temperature, but not elevated CO2 had an experiment-wide effect on morphological traits, as has been reported previously (Watson et al., 2018). However, there were no measurable GxE interactions between the temperature treatments at 21 dph, suggesting that different genotypes exhibit similar responses to elevated temperature as they do at current day temperatures, although this result must be treated with caution as we had low power to test for these interactions. Similarly, there was no change in any of the allelic measures of genetic diversity in the sample population between 1 and 21 dph, indicating that most families responded similarly to temperature through time. Nevertheless, one dominant family in the experiment did exhibit significant effects of temperature on relative abundance. Family 288_359 declined in relative abundance between 1 and 21 dph at 21°C, whereas it increased in relative abundance between 1 and 21 dph at 25°C. This suggests that this family could perform better under future warming than it does in current-day conditions. By contrast, no other families exhibited a significant difference in relative abundance through time between temperature treatments. The smaller number of fish in other families may have reduced the statistical power to detect significant changes in relative abundance through time, but importantly, none of the other families exhibited even a trend toward increased relative abundance at 25°C compared with 21°C.
Early life stages are typically more sensitive to environmental stressors than the adults. Our results confirm earlier observations that elevated temperature has a greater effect than elevated CO2 on growth and development of juvenile yellowtail kingfish. Importantly, all the traits we measured exhibited significant additive genetic variance (i.e., they were heritable), which could enable them to adapt under warmer conditions. However, it is important to note that the low heritability values also indicate that their ability to adapt could be constrained by the intensity and speed of ocean warming, as the response to selection is inversely related to the generation interval of this large pelagic fish. Yet, the fact that one genetic family had higher relative survivorship through time at the elevated temperature provides encouraging evidence that this population of kingfish has some capacity to adapt to ocean warming. To date, only five other studies have estimated trait heritabilities and adaptive potential of juveniles fishes under elevated temperature (Muñoz et al., 2015; Munday et al., 2017) or ocean acidification conditions (Malvezzi et al., 2015; Welch and Munday, 2017; Tasoff and Johnson, 2018), and none have estimated trait heritabilities under the combined effect of both environmental stressors, or in a large pelagic species. Consequently, this is the first evidence that the early life stages of a large pelagic fish has some capacity to adapt to rising temperatures and ocean acidification.
The primary reasons for a lack of evolutionary studies on climate change effects in large pelagic fishes is the inherent difficulty in rearing these animals in captivity, especially in the type of structured breeding designs needed to disentangle genetic and environmental components of phenotypic variation among individuals (Munday et al., 2013). Most large pelagic fishes do not spawn in captivity and/or there is insufficient knowledge and capacity to rear the larvae and juveniles with even moderate success. The yellowtail kingfish is one of the few exceptions, along with dolphinfish (Mahi-Mahi) and cobia, and is now successfully cultured in New Zealand, Australia, Japan and several other countries (Symonds et al., 2014; Sicuro and Luzzana, 2016). However, even for the few species of large pelagic fishes that can be reared in captivity, an additional problem for experimental studies is that adults must be housed in very large tanks in order to accommodate their size and active swimming lifestyle. Here, adult yellowtail kingfish were maintained in replicate 20 m3 circular tanks, each containing between four and six adult fish weighing in excess of 17 kg each. Financial and logistical constraints on the number of broodstock tanks that can be maintained in optimal breeding conditions therefore limits the total number of adults that could contribute to genetic diversity in a quantitative genetics experiment. A further constraint is that the 3–4 males and females in each tank cannot breed with fish in other tanks, thus limiting the total number of crosses and half-sib families that can be produced. Furthermore, not all adults within each tank will contribute to a particular spawning event due to the inherent variability of reproductive synchrony. Finally, individual breeder contributions are highly variable, resulting in certain families dominating the experimental population. All of these factors reduce the power of quantitative genetics models (e.g., the animal model) and indices of genetic diversity (e.g., genetic heterozygosity) to detect important genetic variance. Despite, these substantial limitations, we were still able to detect significant additive genetic variance in morphological traits of juvenile yellowtail kingfish. By contrast we did not detect GxE interactions at 21 dph, or any changes in overall genetic structure of the population between 1 and 21 dph. However, the absence of GxE interactions or changes in genetic structure through time should be treated with caution due to the low genetic diversity in the experiment (12 families total, but overwhelmingly dominated by five abundant families). The large standard error around the estimated GxE interactions and borderline significance values are indicative of both low trait heritability and reduced pedigree structure to provide stronger statistical power to detect significant effects in this analysis. One option to improve the power of future studies to detect genetic variance and GxE interactions would be to conduct the experiment multiple times with the same parents; however, the logistical and financial resources to do this do not currently exist.
Despite the challenges in conducting quantitative genetics studies with large pelagic fishes, these investigations are necessary for evaluating adaptive potential when making predictions about the impacts of warming and ocean acidification on fish populations (Munday et al., 2013). Growth and development of larval and juvenile kingfish in our experiment was substantially faster at 25°C compared with 21°C (Watson et al., 2018). For example, juvenile kingfish were approximately 5 × heavier and 40% longer at 21 dph in 25°C compared with 21°C. By contrast, survivorship to 21 dph was lower at 25°C compared with 21°C, and elevated CO2 had no effect on growth, morphological development or survivorship (Watson et al., 2018). Here, we found that between 6 and 11% of the variation in growth and morphological development between individuals could be attributed to additive genetic variance. A far greater proportion of variance (25–40%) in growth and morphological development was due to common tank effects. Some of this common environmental variation could also have been due to maternal effects, because we were not able to separate maternal effects from other environmental effects. Taken together, the results of the current analysis and the earlier study by Watson et al. (2018) on mean changes in growth and development indicate that phenotypic plasticity is the primary driver of phenotypic changes under elevated temperature. This comparison indicates that growth and development will respond flexibly to rising water temperature, but nevertheless there is also some capacity for genetic adaptation. Ultimately, whether kingfish will adapt will depend on the pace of ocean warmining relative to the generation time of this fish. Importantly, one of the five abundant families in the experiment exhibited a relative increase in abundance at 25°C, suggesting that survivorship of some genotypes will be favored over others at higher temperatures, leading to changes in population genetic structure.
New studies are showing that some freshwater and marine fishes exhibit heritable phenotypic variation that could assist them adapt to warming (Muñoz et al., 2015; Munday et al., 2017) and ocean acidification (Malvezzi et al., 2015; Tasoff and Johnson, 2018). To date, most quantitative genetics studies have been conducted on species of smaller sizes and that are amenable to structured breeding designs in the laboratory, either by artificially crossing gametes from a large number of males and females, or by tracking performance in a multigenerational pedigree. Ours is one of the first studies to attempt to estimate genetic traits important to adaptive potential to climate change in a large pelagic fish. While we confirmed that projected CO2 levels have not impacted early life history traits of juvenile yellowtail kingfish, we are still able to demonstrate that growth, morphological development and survivorship have components of genetic variance that will likely aid them in adapting to a warming ocean.
Statements
Ethics statement
This study was carried out in accordance with the recommendations of the James Cook University Animal Ethics Committee. The protocol was approved by the James Cook University Animal Ethics Committee # A2357.
Author contributions
PM, SN, DP, SMJP, SP, and NS designed the study. SMJP, SP, AS, BA, and PM reared the fish and managed the experiment. BA, PM, AS, and DP collected the samples. CS and TR analyzed the parentage and genetic data. JD did the quantitative genetics analysis. PM, JD, and CS wrote the first draft of the paper. All authors commented on the article and approved the final version.
Funding
This project was supported by Tommy Moore, project manager of the South Pacific Regional Environment Programme (SPREP) and The Pacific Community (SPC) Pacific Islands Ocean Acidification Partnership (PIOAP). Funding for the project was provided by the Government of New Zealand and the Principality of Monaco (PIOAP), the Australian Research Council (FT130100505), King Abdullah University of Science and Technology (BAS/1/1016-01-01), the ARC Centre of Excellence for Coral Reef Studies, and New Zealand's National Institute of Water and Atmospheric Research (NIWA).
Acknowledgments
We thank all the staff at the Northland Marine Research Centre, Shannon McMahon and Megan Welch for assistance with the experiments and data compilation, and Rita Bartossek for help with DNA extractions and microsatellite loci amplification.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00253/full#supplementary-material
References
1
BednaršekN.FeelyR. A.ReumJ. C. P.PetersonB.MenkelJ.AlinS. R.et al. (2014). Limacina helicina shell dissolution as an indicator of declining habitat suitability owing to ocean acidification in the California Current Ecosystem. Proc. R. Soc. B Biol. Sci.281:20140123. 10.1098/rspb.2014.0123
2
BoydP. W.CollinsS.DupontS.FabriciusK.GattusoJ. P.HavenhandJ.et al. (2018). Experimental strategies to assess the biological ramifications of multiple drivers of global ocean change—A review. Glob. Chang. Biol.24, 2239–2261. 10.1111/gcb.14102
3
CattanoC.ClaudetJ.DomeniciP.MilazzoM. (2018). Living in a high CO2 world: a global meta-analysis shows multiple trait-mediated fish responses to ocean acidification. Ecol. Monogr.88, 320–335. 10.1002/ecm.1297
4
ChanF.BarthJ. A.BlanchetteC. A.ByrneR. H.ChavezF.CheritonO.et al. (2017). Persistent spatial structuring of coastal ocean acidification in the California Current System. Sci. Rep.7, 1–7. 10.1038/s41598-017-02777-y
5
CollinsM.KnuttiR.ArblasterJ.DufresneJ.-L.FichefetT.FriedlingsteinP.et al. (2013). Long-term climate change: projections, commitments and irreversibility, in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds StockerT. F.QinD.PlattnerG.-K.TignorM.AllenS. K.BoschungJ.et al. (Cambridge; New York, NY: Cambridge University Press), 1029–1136.
6
DicksonA. G.MilleroF. J. (1987). A comparison of the equilibrium constants for the dissociation of carbonic acid in seawater media. Deep Sea Res. A Oceanogr. Res. Pap.34, 1733–1743. 10.1016/0198-0149(87)90021-5
7
DicksonA. G.SabineC. L.ChristianJ. R. (2007). Guide to best practices for ocean CO2 measurements. PICES Spec. Publ. 3:191. Available online at: https://www.oceanbestpractices.net/handle/11329/249
8
DomingosJ. A.Smith-KeuneC.HarrisonP.JerryD. R. (2014b). Early prediction of long-term family growth performance based on cellular processes - A tool to expedite the establishment of superior foundation broodstock in breeding programs. Aquaculture 428–429, 88–96. 10.1016/j.aquaculture.2014.02.037
9
DomingosJ. A.Smith-KeuneC.JerryD. R. (2014a). Fate of genetic diversity within and between generations and implications for DNA parentage analysis in selective breeding of mass spawners: a case study of commercially farmed barramundi, Lates calcarifer. Aquaculture 424–425, 174–182. 10.1016/j.aquaculture.2014.01.004
10
DoneyS. C.RuckelshausM.Emmett DuffyJ.BarryJ. P.ChanF.EnglishC. A.et al. (2012). Climate change impacts on marine ecosystems. Ann. Rev. Mar. Sci.4, 11–37. 10.1146/annurev-marine-041911-111611
11
EkstromJ. A.SuatoniL.CooleyS. R.PendletonL. H.WaldbusserG. G.CinnerJ. E.et al. (2015). Vulnerability and adaptation of US shellfisheries to ocean acidification. Nat. Clim. Chang.5, 207–214. 10.1038/nclimate2508
12
FalconerD. S. (1952). The problem of environment and selection. Am. Nat.86, 293–298. 10.1086/281736
13
FalconerD. S.MackayT. F. C. (1996). Introduction to Quantitative Genetics.Harlow: Pearson Education Limited.
14
HannanK. D.RummerJ. L. (2018). Aquatic acidification: a mechanism underpinning maintained oxygen transport and performance in fish experiencing elevated carbon dioxide conditions. J. Exp. Biol.221:jeb154559. 10.1242/jeb.154559
15
HeenanA.PomeroyR.BellJ.MundayP. L.CheungW.LoganC.et al. (2015). A climate-informed, ecosystem approach to fisheries management. Mar. Policy57, 182–192. 10.1016/j.marpol.2015.03.018
16
HendersonC. R. (1984). Applications of Linear Models in Animal Breeding.Guelph, ON: University of Guelph.
17
HeuerR. M.GrosellM. (2014). Physiological impacts of elevated carbon dioxide and ocean acidification on fish. AJP Regul. Integr. Comp. Physiol.307, R1061–R1084. 10.1152/ajpregu.00064.2014
18
Hoegh-GuldbergO.CaiR.PoloczanskaE. S.BrewerP. G.SundbyS.HilmiK.et al. (2014). The Ocean,in Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds BarrosV. R.FieldC. B.DokkenD. J.MastrandreaM. D.MachK. J.BilirT. E.et al. (Cambridge; New York, NY: Cambridge University Press), 688.
19
HollowedA. B.BarangeM.BeamishR. J.BranderK.CochraneK.DrinkwaterK.et al. (2013). Projected impacts of climate change on marine fish and fisheries. ICES J. Mar. Sci.70, 1023–1037. 10.1093/icesjms/fst081
20
HughesT. P.KerryJ. T.BairdA. H.ConnollyS. R.DietzelA.EakinC. M.et al. (2018). Global warming transforms coral reef assemblages. Nature556, 492–496. 10.1038/s41586-018-0041-2
21
JonesO. R.WangJ. (2010). COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour.10, 551–555. 10.1111/j.1755-0998.2009.02787.x
22
KalinowskiS. T.TaperM. L.MarshallT. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol.16, 1099–1106. 10.1111/j.1365-294X.2007.03089.x
23
KearseM.MoirR.WilsonA.Stones-HavasS.CheungM.SturrockS.et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics28, 1647–1649. 10.1093/bioinformatics/bts199
24
KoenigsteinS.DahlkeF. T.StiasnyM. H.StorchD.ClemmesenC.PörtnerH. O. (2018). Forecasting future recruitment success for Atlantic cod in the warming and acidifying Barents Sea. Glob. Chang. Biol.24, 526–535. 10.1111/gcb.13848
25
KroekerK. J.KordasR. L.CrimR.HendriksI. E.RamajoL.SinghG. S.et al. (2013). Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Chang. Biol.19, 1884–1896. 10.1111/gcb.12179
26
LlopizJ. K.CowenR. K.HauffM. J.JiR.MundayP. L.MuhlingB. A.et al. (2014). Early life history and fisheries oceanography: new questions in a changing world. Oceanography27, 26–41. 10.5670/oceanog.2014.84
27
LynchM.WalshB. (1998). Genetics and Analysis of Quantitative Triats.Sunderland: Sinauer Associates, Inc.
28
MalvezziA. J.MurrayC. S.FeldheimK. A.DibattistaJ. D.GarantD.GoblerC. J.et al. (2015). A quantitative genetic approach to assess the evolutionary potential of a coastal marine fish to ocean acidification. Evol. Appl.8, 352–362. 10.1111/eva.12248
29
McKenzieJ. R.SmithM.WatsonT.FrancisM. Ó.MaolagáinC.PoortenaarC.et al. (2014). Age, Growth, Maturity and Natural Mortality of New Zealand Kingfish (Seriola lalandi lalandi). Wellington: Ministry for Primary Industries.
30
MehrbachC.CulbersonC. H.HawleyJ. E.PytkowiczR. N. (1973). Measurement of the apparent dissociation constants of carbonic acid in seawater at atmospheric pressure. Limnol. Oceanogr.18, 897–907. 10.4319/lo.1973.18.6.0897
31
MelznerF.GutowskaM. A.LangenbuchM.DupontS.LucassenM.ThorndykeM. C.et al. (2009). Physiological basis for high CO2 tolerance in marine ectothermic animals: pre-adaptation through lifestyle and ontogeny?Biogeosciences6, 2313–2331. 10.5194/bg-6-2313-2009
32
MundayP. L.DonelsonJ. M.DomingosJ. A. (2017). Potential for adaptation to climate change in a coral reef fish. Glob. Chang. Biol.23, 307–317. 10.1111/gcb.13419
33
MundayP. L.WarnerR. R.MonroK.PandolfiJ. M.MarshallD. J. (2013). Predicting evolutionary responses to climate change in the sea. Ecol. Lett.16, 1488–1500. 10.1111/ele.12185
34
MuñozN. J.FarrellA. P.HeathJ. W.NeffB. D. (2015). Adaptive potential of a Pacific salmon challenged by climate change. Nat. Clim. Chang.5, 163–166. 10.1038/nclimate2473
35
OksanenJ.BlanchetF. G.FriendlyM.KindtR.LegendreP.McGlinnD.et al. (2019). Vegan: Community Ecology Package. R package version 2.5-5. Available online at: https://CRAN.R-project.org/package=vegan
36
PankhurstN. W.MundayP. L. (2011). Effects of climate change on fish reproduction and early life history stages. Mar. Freshw. Res.62, 1015–1026. 10.1071/MF10269
37
PeakallR.SmouseP. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics28, 2537–2539. 10.1093/bioinformatics/bts460
38
PhillipsB. F.Pérez-RamírezM. (2017). Climate Change Impacts on Fisheries and Aquaculture: A Global Analysis.Vol. 1.Hoboken: John Wiley & Sons. 10.1002/9781119154051
39
PoloczanskaE. S.BrownC. J.SydemanW. J.KiesslingW.SchoemanD. S.MooreP. J.et al. (2013). Global imprint of climate change on marine life. Nat. Clim. Chang.3, 919–925. 10.1038/nclimate1958
40
PörtnerH.-O.KarlD. M.BoydP. W.CheungW. W. L.Lluch-CotaS. E.NojiriY.et al. (2014). Ocean systems, in Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds FieldC. B.BarrosV. R.DokkenD. J.MachK. J.MastrandreaM. D.BilirT. E.et al. (Cambridge; New York, NY: Cambridge University Press), 411–484.
41
PörtnerH. O.FarrellA. P. (2008). Physiology and climate change. Science322, 690–692. 10.1126/science.1163156
42
PörtnerH. O.PeckM. A. (2010). Climate change effects on fishes and fisheries: towards a cause-and-effect understanding. J. Fish Biol.77, 1745–1779. 10.1111/j.1095-8649.2010.02783.x
43
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
44
ReuschT. B. H. (2014). Climate change in the oceans: Evolutionary versus phenotypically plastic responses of marine animals and plants. Evol. Appl.7, 104–122. 10.1111/eva.12109
45
RichardsonL. E.GrahamN. A. J.PratchettM. S.EurichJ. G.HoeyA. S. (2018). Mass coral bleaching causes biotic homogenization of reef fish assemblages. Glob. Chang. Biol.24, 3117–3129. 10.1111/gcb.14119
46
RiebesellU.GattusoJ. P. (2015). Lessons learned from ocean acidification research. Nat. Clim. Chang.5, 12–14. 10.1038/nclimate2456
47
RoussetF. (2008). GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour.8, 103–106. 10.1111/j.1471-8286.2007.01931.x
48
ShearsN. T.BowenM. M. (2017). Half a century of coastal temperature records reveal complex warming trends in western boundary currents. Sci. Rep.7, 1–9. 10.1038/s41598-017-14944-2
49
SicuroB.LuzzanaU. (2016). The state of Seriola spp. other than yellowtail (S. quinqueradiata) farming in the world. Rev. Fish. Sci. Aquac.24, 314–325. 10.1080/23308249.2016.1187583
50
SimpsonS. D.JenningsS.JohnsonM. P.BlanchardJ. L.SchönP. J.SimsD. W.et al. (2011). Continental shelf-wide response of a fish assemblage to rapid warming of the sea. Curr. Biol.21, 1565–1570. 10.1016/j.cub.2011.08.016
51
StiasnyM. H.MittermayerF. H.SswatM.VossR.JutfeltF.ChiericiM.et al. (2016). Ocean acidification effects on Atlantic cod larval survival and recruitment to the fished population. PLoS ONE11:e0155448. 10.1371/journal.pone.0155448
52
Stuart-SmithR. D.BrownC. J.CeccarelliD. M.EdgarG. J. (2018). Ecosystem restructuring along the Great Barrier Reef following mass coral bleaching. Nature560, 92–96. 10.1038/s41586-018-0359-9
53
SundayJ. M.CalosiP.DupontS.MundayP. L.StillmanJ. H.ReuschT. B. H. (2014). Evolution in an acidifying ocean. Trends Ecol. Evol.29, 117–125. 10.1016/j.tree.2013.11.001
54
SymondsJ. E.WalkerS. P.PetherS.GublinY.McQueenD.KingA.et al. (2014). Developing yellowtail kingfish (Seriola lalandi) and hāpuku (Polyprion oxygeneios) for New Zealand aquaculture. New Zeal. J. Mar. Freshw. Res.48, 371–384. 10.1080/00288330.2014.930050
55
TasoffA. J.JohnsonD. W. (2018). Can larvae of a marine fish adapt to ocean acidification? Evaluating the evolutionary potential of California Grunion (Leuresthes tenuis). Evol. Appl.12, 560–571. 10.1111/eva.12739
56
TaylorR. B.WillisT. J. (1998). Relationships amongst length, weight and growth of north-eastern New Zealand reef fishes. Mar. Freshw. Res.49, 255–260. 10.1071/MF97016
57
WalshC.McKenzieJ.McGregorG.PoortenaarC.HartillB.SmithM. (2003). Information Available for the Management of New Zealand Kingfish (Seriola lalandi lalandi) Stocks. Wellington: New Zealand Fisheries Assessment Report 2003/25. Available online at: https://fs.fish.govt.nz/Doc/17351/2003~FARs/03_25_FAR.pdf.ashx
58
WatsonS.-A.AllanB. J. M.McQueenD. E.NicolS.ParsonsD. M.PetherS. M. J.et al. (2018). Ocean warming has a greater effect than acidification on the early life history development and swimming performance of a large circumglobal pelagic fish. Glob. Chang. Biol.24, 4368–4385. 10.1111/gcb.14290
59
WelchM. J.MundayP. L. (2017). Heritability of behavioural tolerance to high CO2 in a coral reef fish is masked by nonadaptive phenotypic plasticity. Evol. Appl.10, 682–693. 10.1111/eva.12483
60
WilsonA. J.RéaleD.ClementsM. N.MorrisseyM. M.PostmaE.WallingC. A.et al. (2010). An ecologist's guide to the animal model. J. Anim. Ecol.79, 13–26. 10.1111/j.1365-2656.2009.01639.x
Summary
Keywords
climate change, adaptation, heritability, early life-history, morphology, the animal model, pelagic fish, Seriola lalandi
Citation
Munday PL, Schunter C, Allan BJM, Nicol S, Parsons DM, Pether SMJ, Pope S, Ravasi T, Setiawan AN, Smith N and Domingos JA (2019) Testing the Adaptive Potential of Yellowtail Kingfish to Ocean Warming and Acidification. Front. Ecol. Evol. 7:253. doi: 10.3389/fevo.2019.00253
Received
23 January 2019
Accepted
17 June 2019
Published
09 July 2019
Volume
7 - 2019
Edited by
Dennis Murray, Trent University, Canada
Reviewed by
Ajith Premachandra, University of the Sunshine Coast, Australia; Davi Castro Tavares, Leibniz Centre for Tropical Marine Research (LG), Germany
Updates
Copyright
© 2019 Munday, Schunter, Allan, Nicol, Parsons, Pether, Pope, Ravasi, Setiawan, Smith and Domingos.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip L. Munday philip.munday@jcu.edu.au
This article was submitted to Population and Evolutionary Dynamics, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.