- 1Cawthron Institute, Nelson, New Zealand
- 2Institute of Marine Science, University of Auckland, Warkworth, New Zealand
- 3National Institute of Water and Atmospheric Research, Hamilton, New Zealand
- 4Waikato Regional Council, Hamilton, New Zealand
- 5National Institute of Water and Atmospheric Research, Christchurch, New Zealand
The Mediterranean fanworm, Sabella spallanzanii, is listed as an introduced and established “unwanted species” in New Zealand, subject to nationwide targeted surveillance in port, marina, urban and natural environments. Sabella spallanzanii has the potential to change soft-sediment benthic habitats due to the physical presence of the fanworm's tube and associated biological activities, particularly suspension feeding and bio-deposition. A 6-month field experiment was conducted to investigate the impacts of S. spallanzanii on the structure and diversity of existing communities within invaded soft-sediment habitats. Macrofaunal communities were assessed using traditional sampling and identification via microscopy, while microbial and eukaryotic communities were characterized using metabarcoding of 16S and 18S ribosomal genes, respectively. Live and mimic S. spallanzanii were transplanted at different densities (10–50 individuals per m2) into experimental plots with existing assemblages, to test for potential biological and/or physical effects on benthic communities. Analyses revealed significant differences in macrofaunal, eukaryote, and bacterial assemblages in the presence of live S. spallanzanii and mimics, underpinned by changes in the relative abundance of several taxa, indicating that these effects are brought about by biological and physical functions. The presence of S. spallanzanii did not alter total abundance and taxa richness of benthic assemblages but resulted in compositional differences. We found no effect of live or mimic worm density on the structure and diversity of the studied communities. Changes in the structure of native benthic communities, as indicated by this study, can potentially impact functioning of soft-sediment habitats, through alterations to nutrient cycling, bioturbation and benthic-pelagic coupling. Quantitative measurements of impacts are crucial to understand the trajectory of marine invasions, their roles in re-structuring communities, and to guide management efforts.
Introduction
Non-indigenous species (NIS) can have profound impacts on marine coastal ecosystem functioning, by altering community structure, native species richness and ecological processes (Ruiz et al., 1999; Molnar et al., 2008). The magnitude and extent of these impacts vary across temporal and spatial scales (Carlsson et al., 2009), but can be extensive and irreversible (Edelist et al., 2013). Some NIS can regulate the availability of resources to other species by changing the physical state of the ecosystems they invade. Such species are termed “ecosystem engineers” (Jones et al., 1994) or “transformier species” (Richardson et al., 2000), which have the potential to disproportionately affect the functioning of coastal habitats (Cuddington and Hastings, 2004). Ecosystem engineers modulate invaded environments through physical and biological mechanisms. Physical mechanisms are associated with the creation of habitats and modification of the local environment by the presence of body structures of the engineers themselves. For example, introduced mussels or oysters are well-recognized ecosystem engineers that form dense beds or reefs, which can increase local diversity by providing novel hard habitat in soft-sediment environments for many species to colonize (Sousa et al., 2009). On the other hand, biological mechanisms alter the flux of resources through mechanical or chemical functions, such as filtration and bio-deposition that can shift the flow of energy between pelagic and benthic habitats. For example, the proliferation of an invasive filter-feeding clam in San Francisco Bay (USA) caused persistent changes in planktonic food webs and benthic macrofaunal community structure through alteration of benthic-pelagic coupling (Peterson, 2002; Cloern and Jassby, 2012).
The Mediterranean fan worm, Sabella spallanzanii (Gmelin, 1791), a large sabellid tube worm, is considered an ecosystem engineer owing to its capacity to form dense canopies of tubes extending to 50 cm above the sediment. The physical structure provided by the leathery tubes and spiral feeding fans of S. spallanzanii canopies can alter near-seabed hydrodynamics and reduce light penetration below. In Australia, changes in hydrodynamics caused by dense aggregations of invasive S. spallanzanii have been linked to alterations in understory invertebrate larval abundance and recruitment patterns of epifaunal assemblages (Holloway and Keough, 2002a,b). Sabella spallanzanii canopies can also provide structurally complex habitat for other species to inhabit and seek refuge from predators, thus potentially changing the structure of benthic assemblages. Additionally, S. spallanzanii is a suspension feeder that can filter large quantities of water (ca. 12 m3 per day, Stabili et al., 2006). Its efficient removal of organic matter from the water column, and conversion to fecal material and other waste products, may alter bentho-pelagic coupling in invaded habitats. Previous studies conducted in south-eastern Australia revealed changes in benthic community structure and composition associated with S. spallanzanii (O'Brien et al., 2006; Ross et al., 2007). The impact of NIS is frequently context dependent (Didham et al., 2007; Thomsen et al., 2011), making it difficult to extrapolate findings from one location to another. This merits further efforts to understand the generality of their effects or to add context across regions that may attempt to manage their incursion.
Sabella spallanzanii is native to the Mediterranean and east Atlantic coast of Europe and has established introduced populations along the southern coast of Australia and in New Zealand, and has also been recorded in Brazil (Currie et al., 2000). Sabella spallanzanii was first detected in New Zealand in 2008 (Read et al., 2011) and has since become established in several regions around the country. It occurs subtidally at a range of densities in multiple geographic locations (100s of km apart) including on man-made structures and in natural reef and soft-sediment habitats. Although S. spallanzanii is an epifaunal species that requires a solid substrate on which to attach, it can successfully invade soft sediments by using hard substrate within the sediment as anchoring points, such as a fragment of cobble, shell or debris. The density of an introduced species often drives the magnitude of impact. Worm density is generally higher in artificial than natural habitats, and in subtidal soft sediment environments it generally increases with depth, up to a maximum of 30 m. In these natural soft sediment habitats worms typically grow in clumps (50–200 worms) attached to dead shells or live mussels and ascidians (Currie et al., 2000). Its reproductive biology, including high fecundity rates, “sperm-casting” fertilization strategy, and extended reproductive season (Giangrande et al., 2000), contribute to it being found in dense aggregations as a numerically dominant, habitat-forming species (Currie et al., 2000). It has been designated an Unwanted Organism under New Zealand's Biosecurity Act 1993 and is subject to targeted surveillance in harbors, ports, marinas, and high-value natural environments around the country. However, there is still relatively scarce knowledge about the associated adverse effects of established populations. Given S. spallanzanii's wide environmental tolerance and its rapid spread potential into natural habitats, there is a need for a better understanding of its impacts on ecosystem diversity and functioning. This information is critical to understanding resource allocation and benefits of management.
This study aimed to elucidate the ecological impacts of S. spallanzanii on soft-sediment ecosystems in New Zealand. We used a manipulative field experiment to quantify density dependent effects of S. spallanzanii on the structure of soft sediment macrofaunal, eukaryote, and microbial assemblages. We manipulated the density of live and mimic S. spallanzanii to separate biological and physical effects of the worms on macrofaunal and microbial assemblages.
Methods
Experimental Design, Sample Collection, and Processing
A field experiment was conducted from September 2017 until March 2018 in subtidal soft-sediment habitats along the Rangitoto Channel, Waitemata Harbor, Auckland, New Zealand (−36.813, 174.8396). The site had a water depth of 8 m and consisted of relatively flat soft-sediment habitat. The site is situated just outside Waitemata Harbor and is located approximately 1 km outside the commercial shipping lane to the Port of Auckland. An earlier survey revealed the presence of S. spallanzanii in this area, at varying densities (0–20 individuals per m2).
A two-factor experimental design was used to examine the influence of fanworms on soft sediments at the study location. A first factor, “Treatment,” had three levels: “Sabella,” corresponding to the addition (via transplants, see below) of live S. spallanzanii worms to experimental plots; “Mimic” corresponding to the addition of mimic S. spallanzanii worms; and “Control” corresponding to plots without worms. Live S. spallanzanii were obtained by divers from adjacent soft sediment invaded areas and transplanted into the plots using garden pegs. During a 2017 pilot study conducted at a nearby marina, the use of U-shaped steel garden pegs had been identified as a reliable method to anchoring and ensuring the viability of transplanted live and mimic worms (Atalah, unpubl. data). Only healthy undamaged worms with intact tubes were transplanted for this treatment. Many of the live worms collected at the study site were attached to shell fragments, sometimes with several individuals per shell, and thus treatments were created by transplanting naturally occurring clusters of worms into experimental plots to achieve the target densities. The average tube length of transplanted worms was 32.1 ± 0.66 cm (mean ± standard error), with a range of 10–57 cm. Sabella spallanzanii mimics was made from elastic bungee cord (of a length and diameter similar to real S. spallanzanii) with frayed ends that resembles the worms' crown structure. These were threaded onto pegs in clumps of two, three or five individuals, similar to the natural aggregations observed at that site (Supplementary Video 1).
A second factor, “Density,” had five levels: zero (control), 10, 20, 35, and 50 worms (or mimics) per plot. Twenty-four 1 m2 plots, separated by approximately 2 m, were arranged along two perpendicular 30 m transect with a common vertex (Figure 1, n = 12 plots per transect). Plots were positioned on each side of the transects, separated by a corridor to allow divers access. Four replicate plots were assigned to Controls, two replicate plots were assigned to 10, 20, and 35 density level plots (both Sabella and Mimic), and four replicates to the 50 individual density level (Figure 1). Extra replicates in the end member treatments (i.e., four replicates each in the control and 50 density treatments, rather than two each) limited the influence of potential outliers and provided greater statistical power. To ensure interspersion, experimental plots were randomly assigned to treatment combinations. Control plots were treated in the same manner as Sabella and Mimic plots (i.e., garden pegs inserted into the sediment), but no worms were transplanted.
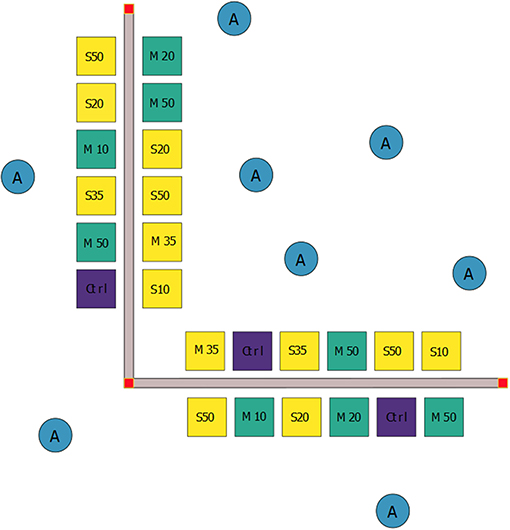
Figure 1. Schematic diagram of the design and set-up of the field experiment showing the random allocation of treatments to 1 m2 experimental plots: live Sabella spallanzanii worms (S, yellow squares), mimic S. spallanzanii worms (M, green squares) and controls (Ctrl, purple squares) without worms. Live and mimic worms were transplanted at four different densities (i.e., 10, 20, 35, and 50 worms (or mimics) per plot). Additionally, eight ambient samples (A, blue circles) were collected from adjacent uninvaded areas as procedural controls.
Experimental plots were established on 19th September 2018 and left undisturbed until 21st March 2019, when destructive sampling of macrofaunal assemblages and sediment characteristics was carried out to conclude the experiment. Two small sediment cores (3 cm internal diameter, 2 cm deep) were collected from each experimental plot: one for sediment grain size and organic matter content analysis (Gatehouse, 1971; Mook and Hoskin, 1982), the other for sediment chlorophyll-a and phaeopigment content (Sartory and Grobbelaar, 1984). One core (13 cm internal diameter, 15 cm deep) was collected from a randomly selected area within each of the 24 plots and the material retained on a 500 μm mesh sieve preserved in 70% isopropyl alcohol for macrofaunal community analysis. Eight additional set of samples were collected from non-invaded adjacent “Ambient” areas (i.e., without S. spallanzanii) to serve as procedural controls. Macrofauna were separated from sediment and shell hash after staining with Rose Bengal and identified to the lowest feasible taxonomic level and their abundance quantified using a binocular dissecting microscope. A random subset of samples was quality checked by an independent taxonomy expert. All samples and vouchers specimens of the identifications were kept for later verification. The macrofauna dataset was dominated by infaunal taxa, but by nature of the sampling method also encompassed surface-dwelling organisms (e.g., epifauna growing on shell hash or coarser sediment fractions). All organisms were included in the analyses, as we were interested in changes in all components of soft-sediment communities. Molecular samples were collected in parallel with macrofauna samples to enable direct comparison of the metabarcoding results (i.e., microbial and eukaryotic communities) with the traditional macrofaunal methods. Specifically, triplicate 50 g sediment samples were obtained by scraping the top 2 cm layer from haphazardly selected intact areas within each of the 24 plots using sterilized plastic vials. Molecular samples were frozen and transported to the laboratory and immediately stored at −80°C until further processing.
High-Throughput Sequencing and Bioinformatics Analyses
The microbial and eukaryotic community compositions of the triplicate sediment samples collected at the 24 plots, represented by 10 Mimic plots (n = 30 samples), 10 Sabella plots (n = 30 samples), 4 control plots (n = 12 samples), and 7 ambient plots (n = 21 samples), were assessed using DNA metabarcoding and following a similar high-throughput sequencing library preparation method as outlined in Keeley et al. (2018). Briefly, environmental DNA was extracted from pre-homogenized 0.2 g of surface sediment using the DNeasy PowerSoilTM DNA extraction kit (Qiagen, Hilden, Germany), including DNA extraction blanks (i.e., extraction controls), followed by DNA quality and purity verification using a spectrophotometer (Eppendorf, Leipzig, Germany). A two-step tailed Polymerase Chain Reaction (PCR) amplicon procedure (Kozich et al., 2013) was applied to all DNA samples (n = 95; including pooled extraction and pooled PCR blanks), targeting ca. 400 base-pair (bp) fragments of a) the nuclear 16S rRNA bacterial gene (V3-4 region) and b) the nuclear 18S rRNA eukaryotic gene (V4 region), and using the primers, reagents and PCR thermocycling conditions described in von Ammon et al. (2018). Detailed information on sequence library preparation, demultiplexing and follow-up bioinformatics analyses are provided in Supplemental Material 2.
Data Analyses
All three datasets (i.e., macrofauna, eukaryotes and bacteria) were analyzed in a similar manner using R software (R Core Team, 2018). Amplicon sequence variants (ASVs) of bacteria and eukaryotes were rarefied at a sequencing depth of 5,000 reads per sample using the “rarefy_even_depth” function in the phyloseq package (McMurdie and Holmes, 2013). This resulted in two and one triplicate samples removed for bacteria and eukaryotes, respectively, because they contained fewer reads than the sample size. Additionally, ASVs represented with a mean <1−4 and present <3 times in at least 20% of the samples in each dataset were removed. Rarefaction curves were calculated for each sample using the vegan package (Oksanen et al., 2018). Differences in assemblage structure between treatments were tested using permutational analysis of variance (PERMANOVA, Anderson et al., 2002) based on Bray-Curtis similarities of the log (x + 1) transformed abundance data using the “Adonis” function in the vegan package. “Treatment” was considered a fixed factor and “Density” included as a continuous covariate representing the final density of fanworms per plot. Significant terms were then investigated using pair-wise comparisons with the PERMANOVA t statistic and 999 permutations. Assemblage differences among treatment levels were visualized using canonical analysis of principal coordinates (CAP, Anderson and Willis, 2003). Indicator taxa for each treatment were determined using the R package “indicspecies” (Cáceres and Legendre, 2009). Indicator species were selected if they were significantly associated with each treatment and their relative abundance visually compared among treatments. Univariate analyses of covariances (ANCOVA) were used to test changes of taxa richness of all three datasets and macrofaunal total abundance using the same experimental design described above for the multivariate data. Tukey tests with Bonferroni adjustment for multiple comparisons were used to compare means when factor “Treatment” was found to be statistically significant in the ANCOVA analyses. In all cases the “Treatment × Density” interaction was not significant (P > 0.3), thus it was removed from the ANCOVA models. Model assumptions were checked by inspecting the model residuals. Type III sums of squares (and its direct multivariate analog) were used to analyse the unbalanced designs.
The relationship between species data and sediment environmental variables was analyzed using multivariate multiple regression (McArdle and Anderson, 2001), more specifically using the adonis routine in the vegan package. A conditional test was used where individual variables were fitted separately to test their relationship with the assemblage data (ignoring other variables) based on Bray-Curtis dissimilarities of the log (x + 1) transformed abundance data. Models were visualized using redundancy analysis ordination biplots (Borcard et al., 2011).
Results
Macrofaunal, Eukaryote, and Bacterial Diversity
Overall, the macrofaunal data comprised 75 different taxa, classified into 10 different phyla, 13 classes, 22 orders, 45 families, and 68 genera. Species level identifications were made for 27 of the 75 unique taxa. Macrofaunal assemblages were largely dominated by annelids, molluscs and crustaceans (52, 19, and 16% of all specimens, respectively, Figure 2A), with the non-indigenous bivalve Theora lubrica numerically dominant, followed by polychaetes from the Paraonidae and Cirratulidae families.
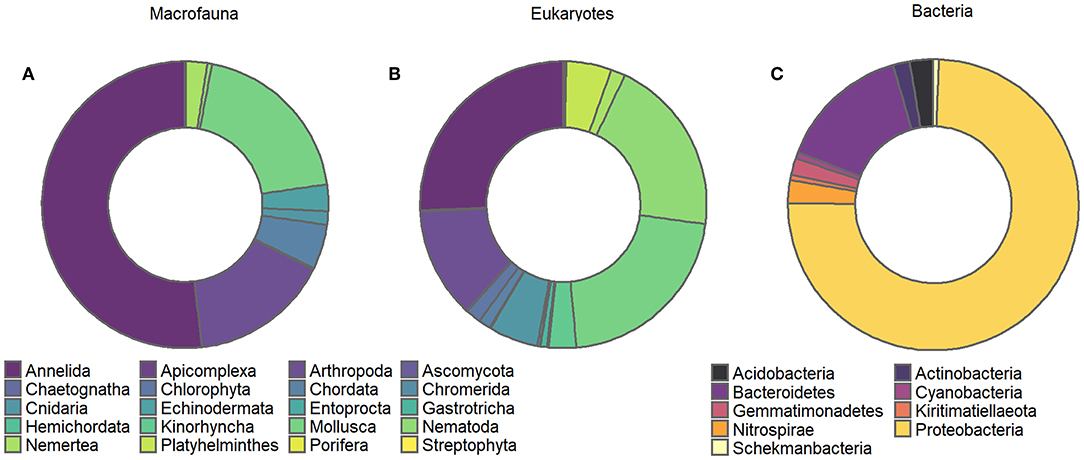
Figure 2. Chart of proportional abundance of different: (A) macrofauna, (B) eukaryote, and (C) bacterial phyla identified from soft sediment assemblages inhabiting experimental plots.
The illuminaTM MiSeq sequencing run generated a total of 12,717,386 sequences, consisting of 5,592,988 bacterial 16S rRNA gene sequences and 7,124,398 eukaryotic 18S rRNA gene sequences. Following filtering (denoising, merging, and chimera removal), the number of high-quality 16S and 18S sequences retained for taxonomic assignments of Amplicon Sequence Variants (ASVs) were 1,848,310 and 3,111,615, respectively. Following reads rarefaction and the removal of ASVs represented with a mean <1−4 and present <3 times in at least 20% of the samples, the number of ASVs retained for downstream statistical analyses for the eukaryote and bacterial datasets were 1,210 and 6,947, respectively; with rarefaction of samples indicating that all samples reached saturation (Supplemental Material 3). Eukaryotic ASVs were assigned to 29 different phyla, 64 classes, 137 orders, 232 families, 304 genera, and 339 distinct species. The output of the metabarcoding identification resulted in a somewhat similar abundance-based partitioning of higher taxa (Figure 2B) with phyla Annelida, Mollusca and Nematoda dominating the eukaryote dataset (26, 21, 20% of all detected specimens, respectively, Figure 2B). Bacterial ASVs were assigned to 44 different phyla, 92 classes, 206 subclasses, 281 families. Bacterial assemblages were largely dominated by Protobacteria, in particular from the family Woeseiaceae and Desulfobulbaceae, followed by bacteria from the phylum Bacteroidetes (Figure 2C). All sequence data are deposited in GenBank's Short Reads Archive, BioProject PRJNA552692, Accession number SRR9644676 to SRR9644588.
Treatment Effects on Assemblage
We found no differences between control plots and ambient (procedural) sediment samples (i.e., the effects of garden pegs and manipulation) on macrofaunal total abundance (t-test, P > 35), richness (t-test, P > 0.25) or assemblage structure (PERMANOVA, P > 0.05) for all three communities (i.e., macrofauna, eukaryotes, and bacteria, Supplemental Material 4). Therefore, results measured from ambient plots were excluded from all further analyses. There was a significant effect of treatment on assemblage structure consistent across macrofaunal, eukaryote and bacterial datasets (P < 0.05, Table 1), although there was no main or interactive effect of the factor Density for all three datasets (P > 0.1, Table 1). The CAP plots illustrate this pattern, showing a clear separation between assemblages from the three treatments in all three datasets (Figure 3). For the macrofaunal data, pair-wise comparisons distinguished between Sabella and Control (P < 0.05); and Sabella and Mimic (P < 0.05), but not between Control and Mimic (P > 0.1). The most significant indicators for Control plots included the brushworm Phylo novazealandiae, amphipods, the burrowing crab Pilumnus novaezealandiae, the burrowing holothurian Taeniogyrus dendyi, and the polychaete Aglaophamus sp. Significant indicator taxa for Mimic plots included the bivalves Monia zelandica and Limaria africana, the polychaetes Armandia maculata, unidentified worms from the family Spionidae and Heteromastus filiformis (Figure 4A). Sabella plots were associated with polychaete family Paraonidae, nemerteans and the polychaete Prionospio sp. (Figure 4A).
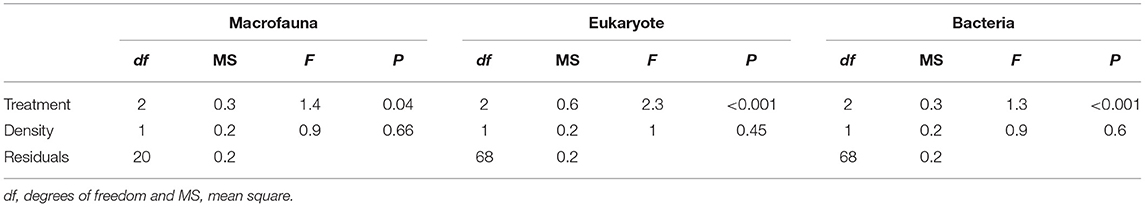
Table 1. Results of PERMANOVA analyses to test the effect of Treatment (“Sabella,” “Mimic,” and “Control”) and Sabella spallanzanii Density on macrofaunal, eukaryote and bacterial assemblages.
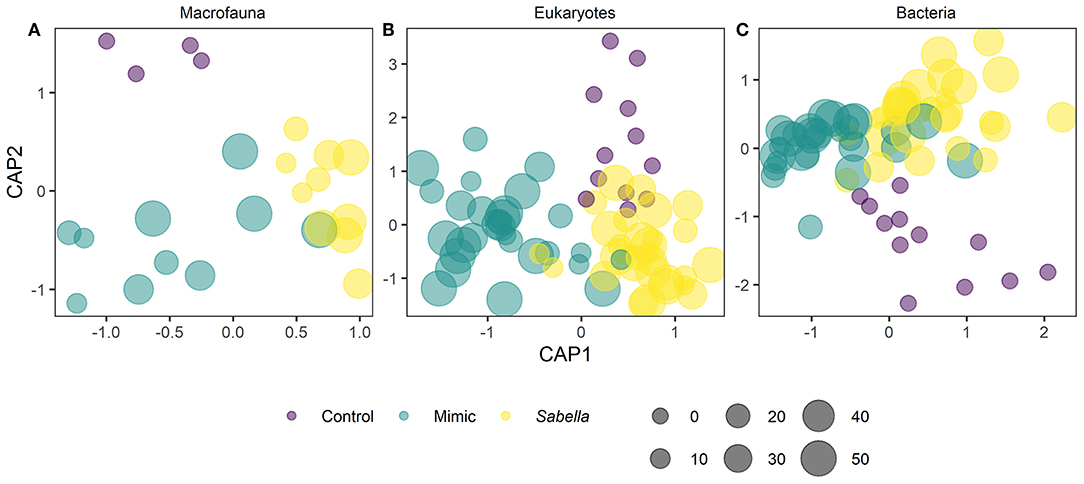
Figure 3. Principal coordinate ordination (PCO) and Canonical analysis of principal coordinates of assemblages in “Control” (purple circles), “Mimic” (green circles) and “Sabella” (yellow circles) experimental plots on the basis of Bray–Curtis similarities of the log (x +1) transformed abundance of macrofauna (A), eukaryotes (B), and bacteria (C).
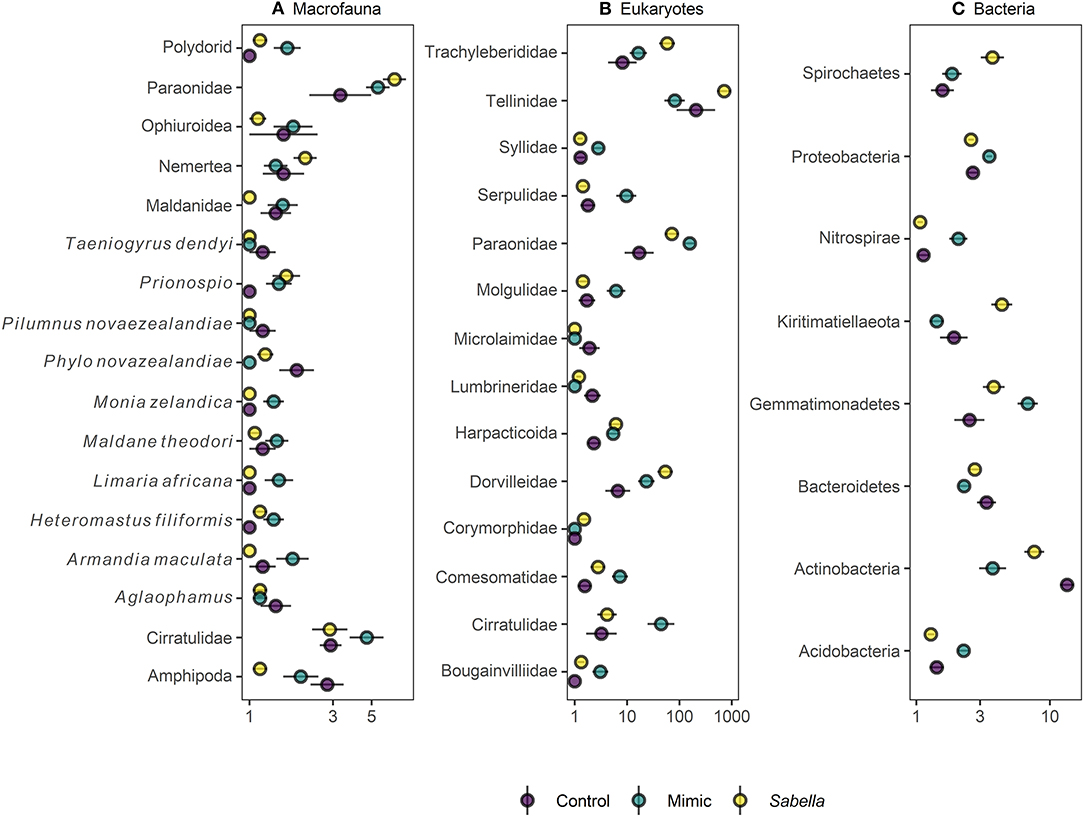
Figure 4. Mean (± SE) relative abundance (%) of indicator taxa for (A) macrofaunal, (B) eukaryote, and (C) bacterial communities in “Control” (purple circles), “Mimic” (green circles) and “Sabella” (yellow circles) experimental plots.
For the eukaryote communities, pair-wise comparisons found significant differences among all three treatments (P < 0.05). Controls were significantly associated with higher abundances of polychaetes from the family Lumbrineridae and nematodes from the family Microlaimidae (Figure 4B). Most significant indicator eukaryote taxa for the Mimic treatment included polychaetes from the families Paraonidae and the non-indigenous Serpulidae tubeworm Hydroides elegans and ascidian Molgula manhattensis, whereas tellinid bivalves, harpacticoid copepods and dorvilleid polychaetes were more abundant on Sabella plots (Figure 4B).
For bacterial assemblages, pair-wise comparisons also revealed significant differences among all three treatments (P < 0.01 for Control vs. Mimic and Mimic vs. Sabella, and P < 0.05 for Control vs. Sabella) and 55 indicator taxa were identified as being significantly associated with these differences. Most significant indicator taxa for the Control treatment included families in the phyla Actinobacteria, Bacteroidetes and Proteobacteria (Figure 4C), whereas for the Mimic treatment they included bacteria in the phyla Acidobacteria, Gemmatimonadetes, Nitrospirae, and Proteobacteria (Figure 4C). Significant indicator bacterial taxa for the Sabella treatment most notably included families in the phyla Kiritimatiellaeota (e.g., Kiritimatiellaceae), Spirochaetes (e.g., Spirochaetaceae) and Proteobacteria (Figure 4C), including significant higher abundance of Desulfobacteraceae.
Total abundance and richness of macrofaunal and bacterial assemblages did not vary significantly among Treatment or Density (Figure 5 and Table 2), although there was a non-significant trend for higher numbers of macrofauna taxa richness in the Mimic compared to the Control plots. Treatment had a significant effect on eukaryote taxa richness, with Mimic having higher number of taxa compared to Control plots (P < 0.05, Figure 5 and Table 2).
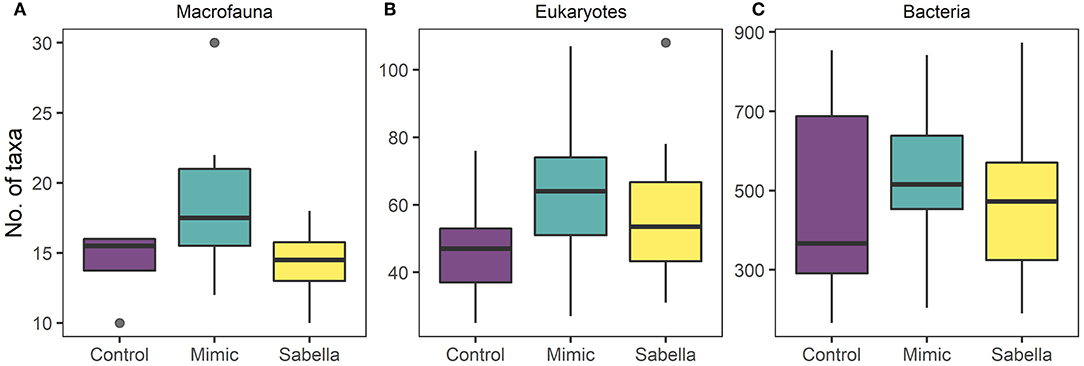
Figure 5. Boxplots of taxa richness for (A), macrofauna (B), eukaryote (C), and bacterial communities in “Control” (purple), “Mimic” (green) and “Sabella” (yellow) experimental plots.
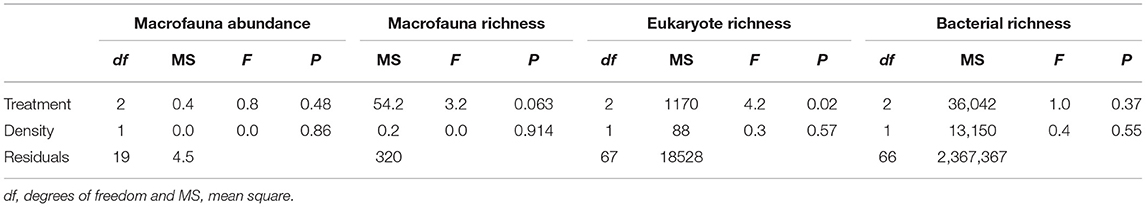
Table 2. Results of univariate analyses of covariance (ANCOVAs) to test the effect of Treatment (“Sabella,” “Mimic” and “Control”) and Sabella spallanzanii Density on macrofaunal, eukaryote, and bacterial univariate indices.
Relationship Between Soft-Sediment Assemblages and Environmental Variables
The sediment at the site was relatively poor in terms of organic content (mean 2.5% LOI ± 0.1 S.E.) and was dominated by very fine sand (63–500 μm, 46.6% ± 0.9), silt particles (< 63 μm, 29.3% ± 1.1) and clay (11% ± 1.1) grain size fractions. Mean chlorophyll-a and phaeopigments contents were on average 1.92 μg g−1 (± 0.1) and 4.5 μg g−1 (± 0.3), respectively. Sabella treated plots had higher fine sand, chlorophyll-a, organic and phaeopigments compared to Mimic and Control plots (Figure 6). On the other hand, Mimic and Sabella plots had higher content of medium and coarse sediment in relation to the controls (Figure 6). The multivariate multiple regression analysis showed that gravel, medium and fine sediment had a significant relationship with macrofaunal assemblage structure, explaining 8, 7, and 9% of the variation in the data, respectively. Eukaryote assemblage structure was significantly related to sediment chlorophyll-a concentration and marginally to the fine sediment fraction and total organic content; whereas bacterial assemblage structure was not significantly related to sediment characteristics. Although no significant relationships were detected between bacterial assemblages and sediment characteristics, there was a trend of bacterial associations with the fine sediment fraction and phaeopigment concentration (P < 0.1). The dbRDA plots (Figure 7) illustrate that all three benthic assemblages were distinguishable among treatments constrained to the variation in sediment characteristics. In all three cases Sabella plots assemblages were generally well separated from the other treatments and correlated with higher phaeopigment, chlorophyll-a, organics and the fine sediment fraction, whereas Mimic plots were related to larger sediment fractions (i.e., medium and gravel).
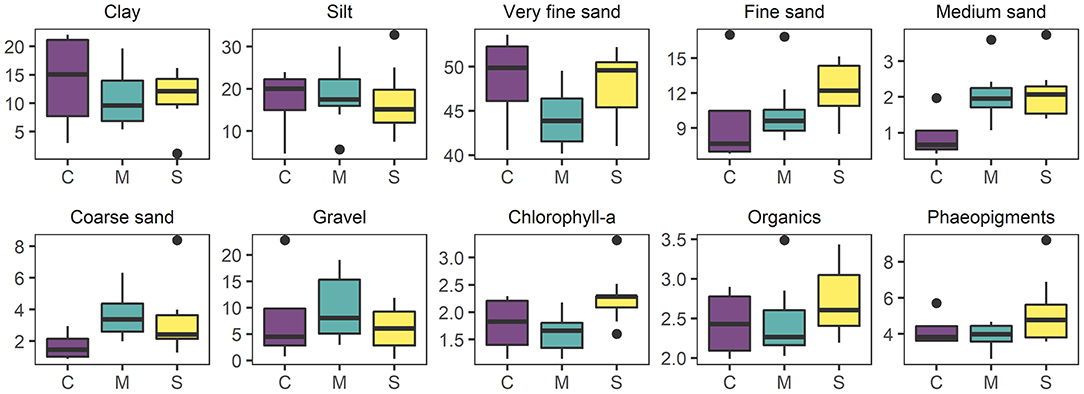
Figure 6. Boxplots of soft sediment physico-chemical environmental variables: clay (< 0.98 μm), Silt (3.9–62.5 μm), very fine sand (62.5–125 μm), fine sand (12–250 μm), medium sand (0.28–0.5 mm), coarse sand (0.5–1.0 mm), gravel (>1 mm), chlorophyll-a (μg g−1), organic content (% LOI), and phaeopigments (μg g−1) in relation to experimental treatments: C, “Control”; M, “Mimic”; S, “Sabella”.
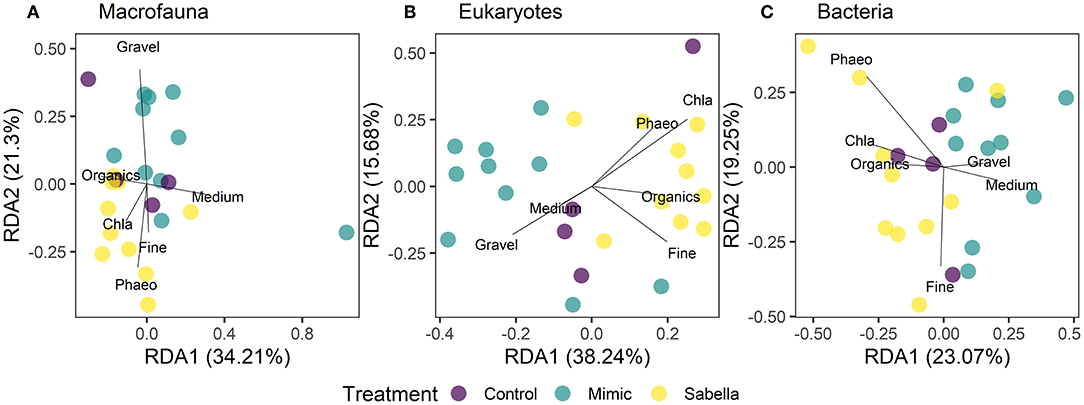
Figure 7. Distance-based redundancy analysis (dbRDA) ordination of first and second fitted axes relating the environmental variables to the faunal data (A) macrofaunal, (B) eukaryotes, and (C) bacterial communities in “Control” (purple circles), “Mimic” (green circles) and “Sabella” (yellow circles) experimental plots. Biplot projections are given for the sediment environmental variables. Vectors are plotted showing the correlations of environmental variables with dbRDA axis 1 and dbRDA axis 2, with the length and direction of the arrow representing the strength and direction of the relationship.
Discussion
Soft-sediment communities in Waitemata differed significantly in the presence and absence of Mediterranean fanworms and their mimics. This effect was consistent across benthic macrofaunal, eukaryote and bacterial assemblages, and indicates that S. spallanzanii may affect benthic community structure through both biological and physical processes and mechanisms. The measured differences among treatments were based on relative abundances of several taxa between plots with transplanted live and mimic Sabella and control plots rather than differences in overall abundance or richness. These differences in community structure can have important consequences for ecosystem functioning of soft-sediment habitats (Thrush et al., 2017) based on shifts in feeding, burrowing and sediment irrigation intensity that can drive crucial functions, such as nutrient and organic matter processing.
Small mobile taxa (mainly amphipods) and larger bioturbator taxa such as ophiuroids, a burrowing crab (Pilumnus novaezealandiae), and a burrowing holothurian (Taeniogyrus dendyi) were less abundant, or absent, in treatment plots compared to controls. This may be a direct or indirect consequence of both physical presence and biological activities of fanworms. Resident communities may be affected by small-scale changes in hydrodynamics linked to the physical structures of S. spallanzanii canopies, as changes in water velocities can alter sediment stability and concentrations of key solutes (e.g., oxygen, sulfide; O'Brien et al., 2006). Mobile bioturbator taxa can substantially change the physical and chemical properties of soft sediment habitats and thus are crucial for the functioning of these ecosystems (Lohrer et al., 2004). On the other hand, S. spallanzanii plots in our study were also associated with significantly higher abundance (compared to controls) of direct deposit-feeders, such as polychaetes from the family Paraonidae, Dorvilleidae and Spionidae, and Nematoda. The biodeposition activity of the fanworm may has the potential to change nutrient cycling and the microbial community, with knock-on effects on deposit feeders. Suspension feeders, such as S. spallanzanii can increase sedimentation of organic material and produce abundant feces and pseudofeces rich in organic matter, which may serve as food sources for deposit feeding organisms (Commito and Boncavage, 1989). This biological activity may explain the observed patterns of higher concentration of chlorophyll-a, organics and phaeopigments in Sabella treated plots. As such, there has been some concern about the effect of S. spallanzanii on nitrogen cycling including changes in nutrient availability and its consequences for community structure (Harris et al., 1996).
Changes in bacterial communities in response to experimental treatments were consistent with those observed for macrofaunal and eukaryote communities. Soft-sediment dwelling bacteria play a crucial role in the functioning of these habitats and are recognized for their sensitivity to environmental change. Recent studies have highlighted the ability of S. spallanzanii to filter, accumulate and concentrate bacteria from the surrounding environment (Stabili et al., 2006; Licciano et al., 2007). Sabella spallanzanii have also been shown to exude mucus with antibacterial properties, which may influence the composition of microbial biofilms on the worm's tube and surrounding sediments (Stabili et al., 2011). In contrast, nutrient enrichment and increased primary production observed in Sabella treated plots may decrease the redox potential in sediments and favor sulfide-oxidizing and sulfate-reducing bacteria for example, as illustrated by the observed increase in the abundance of Desulfobacteraceae. Regardless of the mechanisms that may be responsible for these effects, the observed changes in bacterial communities may have important consequences for the functioning of soft-sediment habitats, most notably on nutrient and organic matter cycling.
It is often assumed that the impacts of invasive species are density-dependent, although few studies have assessed this contention (Thomsen et al., 2011) and this may not always be the case. We found no effect of density in our simulated invasion of such an engineer throughout our study, which may indicate that no real effect occurred, that the range of experimental densities was too small to have an effect, or that the duration of the study was insufficient for density-based differences to emerge. Non-indigenous filter-feeder species, such as S. spallanzanii, are more likely to affect communities over longer time scales than provide instantaneous shifts or impacts (Byrnes and Stachowicz, 2009). Nonetheless, the simple presence of fanworms or mimics (≥10 m−2) appeared to prompt community differences compared to plots without them. One mechanism is through secondary colonization of S. spallanzanii tubes that can provide additional habitat for a range of epifauna typically associated with hard substrates (O'Brien et al., 2006). Although no significant changes in total macrofaunal abundance were detected, the presence of S. spallanzanii and mimics was associated with higher eukaryote taxa richness and higher abundance of encrusting and non-indigenous organisms such as tube worms (Hydroides elegans), ascidians and hydroids, evidenced by the macrofaunal and eukaryote datasets. These changes were reflected in a non-significant trend for an increase in macrofauna taxa richness in mimic plots compared to the controls. Although metabarcoding identification resulted in similar taxonomic partitioning, estimated taxa richness was an order of magnitude higher than that using morphological identification. Metabarcoding provides a more holistic view of the metazoan taxonomic diversity, regardless of the size and developmental stage (e.g., including eggs or larvae). The eukaryote data not only included macrofaunal taxa that dominated morphological samples, but also small-sized meiofaunal species (<1 mm), extending the scope of the analysis. Novel epifaunal taxa facilitated by the presence of S. spallanzanii may have important implications in the functioning of invaded soft-sediment habitats, as the combined filtering capacity of these organisms is considerably enhanced compared to uninvaded habitats (Lemmens et al., 1996). Furthermore, positive and potentially synergistic interactions among invasive species may lead to accelerated impacts on native ecosystems (Simberloff and Von Holle, 1999).
The results reported here are generally consistent with previous studies looking at the impacts of S. spallanzanii in soft sediments assemblages (O'Brien et al., 2006; Ross et al., 2007). However, the effects recorded in these Australian studies from Port Phillip Bay were dependent on spatial distribution and density of S. spallanzanii, with negligible effects recorded at low densities (<1 individual per m2). However, similarly to this study, localized dense clumps (15 individuals per m2), caused changes in benthic infauna structure characterized by lower abundances of cumaceans, ostracods and harpacticoid copepods (O'Brien et al., 2006). Ross et al. (2007) found no effect of S. spallanzanii on the resident macrofauna, with the exception of lumbrinerid polychaetes and gammarid amphipods; however, these taxa only represented a small proportion of those present. Ross et al. (2013) found no difference in the total abundance of macrofauna between S. spallanzanii and control plots. However, the composition of assemblages did change significantly in the presence of S. spallanzanii, with a significant increase in the abundance of echinoderms (largely attributable to brittle stars) in the presence of S. spallanzanii at all three sites investigated (Ross et al., 2013).
Our study site experiences relatively high velocity tidal currents (average 1.25 m s−1, Greig and Proctor, 1988), which may pose physical stress on organisms and are unfavorable for the accumulation of organic matter. It is likely that these prevailing hydrographic conditions and resulting sedimentary physical conditions may exert a stronger meso-scale effect on biological communities compared to the relatively subtle observed response to the experimental treatments. In this context, the generality of observed effects to a regional scale or other locations is difficult to gauge. On the other hand, conditions that promote a mixed particle size benthic surface (larger fragments within a sandy-silt matrix) and regular water flow may be ideal for S. spallanzanii settlement and growth. Sabella spallanzanii is also among the only or very few organisms in this area's benthic community that can extend as high as 50 cm above the seafloor into the water column to take advantage of passing food-laden currents. A key factor for future work is to monitor population density, especially clusters of high density, because the magnitude of effects would undoubtedly increase if such novel canopies of this species proliferated.
Shallow subtidal soft sediment habitats are an extensive component of bays and estuaries and provide an important environment for S. spallanzanii establishment and spread. This species represents a relatively novel introduction as an epibenthic (rather than infaunal) invader that is not (almost) exclusively associated with hard-bottom habitats, like a majority of marine introduced species in New Zealand (Cranfield et al., 1998) and elsewhere (Byrnes et al., 2007; Ruiz et al., 2011). Given that these habitats are a dominant and important feature in the study area and wider region, it would be valuable to replicate this study in other areas to better understand the generality of effects of this invasion. As it is a listed “unwanted” organism under New Zealand legislation, there are active campaigns to (a) prevent its anthropogenic spread (mainly via shipping and boating) and (b) locally extirpate any nascent populations that are detected in discrete new areas using targeted removals (Read et al., 2011). These efforts, combined with monitoring hotspot populations that are not being managed, provide a precautionary approach to S. spallanzanii in New Zealand (Champion, 2018). An eradication attempt in the Auckland region is considered too uncertain and potentially expensive to initiate at present using existing tools and strategies. This view may change as marine eradication matures and if impacts of NIS on benthic communities, functioning, and maritime industries (e.g., aquaculture) proliferate (Edwards and Leung, 2009; Soliman and Inglis, 2018).
NIS are generally considered among one of the greatest threats to marine ecosystems, however our understanding about their overall impacts is often insufficient (Thomsen et al., 2011), partly because of the lack of adequate studies and context-dependency of their effects. Elucidating and predicting context-dependent impacts of NIS are crucial for assessing the associated risks to biodiversity and ecosystem functioning, and to underpin management efforts. At a global scale it has been shown that NIS have significant, although modest in magnitude, ecological effects (Anton et al., 2019). Using a combination of morphological and molecular taxonomical techniques, our study provides insights into the nature and magnitude of local-scale effects on different communities inhabiting soft-sediment habitats, which may inform the implementation of effective knowledge-based management and mitigation measures that reduce the impacts of NIS.
Data Availability Statement
All data and R scripts used for the analyses are available at https://github.com/jatalah/sabella_impacts_experiment.git.
Author Contributions
JA, OF, MT, LT, and AL: conceived and designed the experiments. JA, OF, MT, LT, and AL: performed the experiments. JA: analyzed the data. XP: analyzed molecular samples. JA, XP, and OF: wrote the paper.
Funding
This study was funded by the New Zealand Ministry of Business, Innovation and Employment, under the Programme What's at stake?—Enabling decision-making through better measurement, forecasting, and evaluation of the impacts of non-native organisms in NZ's changing ocean (C01X1511).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, GR, declared a shared affiliation, with no collaboration, with several of the authors, MT, LT, and AL, to the handling editor at the time of review.
Acknowledgments
We thank Isaac Eckert and Fiona Gower (Cawthron Institute) for helping with sample processing, Samantha Happy from Auckland Council for helping with pilot study, Westhaven Marina for access to the study sites and Dr. Ian Davidson for his valuable insights comments on this manuscript. We also acknowledge MPI—New Zealand Biosecurity (Kathy Walls) for processing permits for the transplanting of Sabella spallanzanii.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00481/full#supplementary-material
Supplementary Video 1. Video footage of the field set-up of the experiment in conducted in Waitemata Harbour, Auckland, New Zealand showing the treatments to 1 m2 experimental plots: live Sabella spallanzanii worms, mimic S. spallanzanii worms and controls without worms. Live and mimic worms were transplanted at four different densities [i.e. 10, 20, 35 and 50 worms (or mimics) per plot].
References
Anderson, M., Hewitt, J., and Thrush, S. (2002). The Development of Criteria for Assessing Community Health of Urban Intertidal Flats. NIWA report prepared for Auckland Regional Council Technical Report No. 184. National Institute of Water and Atmospheric Research.
Anderson, M. J., and Willis, T. J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2
Anton, A., Geraldi, N. R., Lovelock, C. E., Apostolaki, E. T., Bennett, S., Cebrian, J., et al. (2019). Global ecological impacts of marine exotic species. Nat. Ecol. Evol. 3, 787–800. doi: 10.1038/s41559-019-0851-0
Borcard, D., Gillet, F., and Legendre, P. (2011). “Canonical ordination,” in Numerical Ecology with R (New York, NY: Springer), 153–225. doi: 10.1007/978-1-4419-7976-6_6
Byrnes, J., and Stachowicz, J. J. (2009). Short and long term consequences of increases in exotic species richness on water filtration by marine invertebrates. Ecol. Lett. 12, 830–841. doi: 10.1111/j.1461-0248.2009.01339.x
Byrnes, J. E., Reynolds, P. L., and Stachowicz, J. J. (2007). Invasions and extinctions reshape coastal marine food webs. PLoS ONE 2:e295. doi: 10.1371/journal.pone.0000295
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Carlsson, N. O. L., Sarnelle, O., and Strayer, D. L. (2009). Native predators and exotic prey –an acquired taste? Front. Ecol. Environ. 7, 525–532. doi: 10.1890/080093
Champion, P. D. (2018). Knowledge to action on aquatic invasive species: island biosecurity–the New Zealand and South Pacific story. Manag. Biol. Invasions 9, 383–394. doi: 10.3391/mbi.2018.9.4.02
Cloern, J. E., and Jassby, A. D. (2012). Drivers of change in estuarine-coastal ecosystems: discoveries from four decades of study in San Francisco Bay. Rev. Geophys. 50, 1–33. doi: 10.1029/2012RG000397
Commito, J. A., and Boncavage, E. M. (1989). Suspension-feeders and coexisting infauna: an enhancement counterexample. J. Exp. Mar. Biol. Ecol. 125, 33–42. doi: 10.1016/0022-0981(89)90214-1
Cranfield, H. J., Gordon, D., Willan, R., Marshall, B., Battershill, C., Francis, M., et al. (1998). Adventive Marine Species in New Zealand. Technical Report 34. National Institute of Water and Atmospheric Research, Wellington.
Cuddington, K., and Hastings, A. (2004). Invasive engineers. Ecol. Modell. 178, 335–347. doi: 10.1016/j.ecolmodel.2004.03.010
Currie, D. R., McArthur, M. A., and Cohen, B. F. (2000). Reproduction and distribution of the invasive European fanworm Sabella spallanzanii (Polychaeta: Sabellidae) in Port Phillip Bay, Victoria, Australia. Mar. Biol. 136, 645–56. doi: 10.1007/s002270050724
Didham, R. K., Tylianakis, J. M., Gemmell, N. J., Rand, T. A., and Ewers, R. M. (2007). Interactive effects of habitat modification and species invasion on native species decline. Trends Ecol. Evol. 22, 489–496. doi: 10.1016/j.tree.2007.07.001
Edelist, D., Rilov, G., Golani, D., Carlton, J. T., and Spanier, E. (2013). Restructuring the sea: profound shifts in the world's most invaded marine ecosystem. Divers. Distrib. 19, 69–77. doi: 10.1111/ddi.12002
Edwards, P. K., and Leung, B. (2009). Re-evaluating eradication of nuisance species: invasion of the tunicate, Ciona intestinalis. Front. Ecol. Environ. 7, 326–332. doi: 10.1890/070218
Gatehouse, J. (1971). “Sedimentary analysis,” in Procedures in Sedimentary Petrology, ed R. E. Carver (New York, NY: John Wiley & Sons Incorporated), 59–94.
Giangrande, A., Licciano, M., Pagliara, P., and Gambi, M. (2000). Gametogenesis and larval development in Sabella spallanzanii (Polychaeta: Sabellidae) from the Mediterranean Sea. Mar. Biol. 136, 847–861. doi: 10.1007/s002279900251
Greig, M. J., and Proctor, R. (1988). A numerical model of the Hauraki Gulf, New Zealand. N. Z. J. Mar. Freshw. Res. 22, 379–390. doi: 10.1080/00288330.1988.9516309
Harris, G., Batley, G., Fox, D., Hall, D., Jernakoff, P., Molloy, R., et al. (1996). Port Phillip Bay Environmental Study Final Report. CSIRO Publishing, Collingwood, VIC. doi: 10.4225/08/5856cf3221739
Holloway, M. G., and Keough, M. J. (2002a). Effects of an introduced polychaete, Sabella spallanzanii, on the development of epifaunal assemblages. Mar. Ecol. Prog. Ser. 236, 137–154. doi: 10.3354/meps236137
Holloway, M. G., and Keough, M. J. (2002b). An introduced polychaete affects recruitment and larval abundance of sessile invertebrates. Ecol. Appl. 12, 1803–1823. doi: 10.1890/1051-0761(2002)012[1803:AIPARA]2.0.CO;2
Jones, C. G., Lawton, J. H., and Shachak, M. (1994). Organisms as ecosystem engineers. Oikos 69, 373–386. doi: 10.2307/3545850
Keeley, N., Wood, S. A., and Pochon, X. (2018). Development and preliminary validation of a multi-trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecol. Indic. 85, 1044–1057. doi: 10.1016/j.ecolind.2017.11.014
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lemmens, J., Clapin, G., Lavery, P., and Cary, J. (1996). Filtering capacity of seagrass meadows and other habitats of Cockburn Sound, Western Australia. Mar. Ecol. Prog. Ser. 143, 187–200. doi: 10.3354/meps143187
Licciano, M., Terlizzi, A., Giangrande, A., Cavallo, R. A., and Stabili, L. (2007). Filter-feeder macroinvertebrates as key players in culturable bacteria biodiversity control: a case of study with Sabella spallanzanii (Polychaeta: Sabellidae). Mar. Environ. Res. 64, 504–513. doi: 10.1016/j.marenvres.2007.04.004
Lohrer, A. M., Thrush, S. F., and Gibbs, M. M. (2004). Bioturbators enhance ecosystem function through complex biogeochemical interactions. Nature 431, 1092–1095. doi: 10.1038/nature03042
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Molnar, J. L., Gamboa, R. L., Revenga, C., and Spalding, M. D. (2008). Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 6, 485–492. doi: 10.1890/070064
Mook, D. H., and Hoskin, C. M. (1982). Organic determinations by ignition: caution advised. Estuar. Coast. Shelf Sci. 15, 697–699. doi: 10.1016/0272-7714(82)90080-4
O'Brien, A. L., Ross, D. J., and Keough, M. J. (2006). Effects of Sabella spallanzanii physical structure on soft sediment macrofaunal assemblages. Mar. Freshw. Res. 57, 363–371. doi: 10.1071/MF05141
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2018). vegan: Community Ecology Package. R package version 2.4-6. Available online at: https://CRAN.R-project.org/package=vegan (accessed July 15, 2018).
Peterson, H. A. (2002). Long-term benthic community change in a highly invaded estuary (Ph.D. thesis). San Francisco State University, San Francisco, CA.
R Core Team (2018). A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Read, G. B., Inglis, G., Stratford, P., and Ahyong, S. T. (2011). Arrival of the alien fanworm Sabella spallanzanii (Gmelin, 1791) (Polychaeta: Sabellidae) in two New Zealand harbours. Aquat. Invasions 6, 273–279. doi: 10.3391/ai.2011.6.3.04
Richardson, D. M., Pysek, P., Rejmanek, M., Barbour, M. G., Panetta, F. D., and West, C. J. (2000). Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib. 6, 93–107. doi: 10.1046/j.1472-4642.2000.00083.x
Ross, D. J., Keough, M. J., Longmore, A. R., and Knott, N. A. (2007). Impacts of two introduced suspension feeders in Port Phillip Bay, Australia. Mar. Ecol. Prog. Ser. 340, 41–53. doi: 10.3354/meps340041
Ross, D. J., Longmore, A. R., and Keough, M. J. (2013). Spatially variable effects of a marine pest on ecosystem function. Oecologia 172, 525–538. doi: 10.1007/s00442-012-2497-3
Ruiz, G. M., Fofonoff, P., Hines, A. H., and Grosholz, E. D. (1999). Non-indigenous species as stressors in estuarine and marine communities: assessing invasion impacts and interactions. Limnol. Oceanogr. 44, 950–972. doi: 10.4319/lo.1999.44.3_part_2.0950
Ruiz, G. M., Fofonoff, P. W., Steves, B., Foss, S. F., and Shiba, S. N. (2011). Marine invasion history and vector analysis of California: a hotspot for western North America. Divers. Distrib. 17, 362–373. doi: 10.1111/j.1472-4642.2011.00742.x
Sartory, D., and Grobbelaar, J. (1984). Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 114, 177–187. doi: 10.1007/BF00031869
Simberloff, D., and Von Holle, B. (1999). Positive Interactions of nonindigenous species: invasional meltdown? Biol. Invasions 1, 21–32. doi: 10.1023/A:1010086329619
Soliman, T., and Inglis, G. J. (2018). Forecasting the economic impacts of two biofouling invaders on aquaculture production of green-lipped mussels Perna canaliculus in New Zealand. Aquacult. Environ. Interac. 10, 1–12. doi: 10.3354/aei00249
Sousa, R., Gutierrez, J. L., and Aldridge, D. C. (2009). Non-indigenous invasive bivalves as ecosystem engineers. Biol. Invasions 11, 2367–2385. doi: 10.1007/s10530-009-9422-7
Stabili, L., Licciano, M., Giangrande, A., Fanelli, G., and Cavallo, R. A. (2006). Sabella spallanzanii filter-feeding on bacterial community: ecological implications and applications. Mar. Environ. Res. 61, 74–92. doi: 10.1016/j.marenvres.2005.06.001
Stabili, L., Schirosi, R., Di Benedetto, A., Merendino, A., Villanova, L., and Giangrande, A. (2011). First insights into the biochemistry of Sabella spallanzanii (Annelida: Polychaeta) mucus: a potentially unexplored resource for applicative purposes. J. Mar. Biol. Assoc. U.K. 91, 199–208. doi: 10.1017/S0025315410001013
Thomsen, M. S., Wernberg, T., Olden, J. D., Griffin, J. N., and Silliman, B. R. (2011). A framework to study the context-dependent impacts of marine invasions. J. Exp. Mar. Biol. Ecol. 400, 322–327. doi: 10.1016/j.jembe.2011.02.033
Thrush, S. F., Hewitt, J. E., Kraan, C., Lohrer, A. M., Pilditch, C. A., and Douglas, E. (2017). Changes in the location of biodiversity-ecosystem function hot spots across the seafloor landscape with increasing sediment nutrient loading. Proc. Biol. Sci. 284:20162861. doi: 10.1098/rspb.2016.2861
Keywords: introduced species, soft sediment, invasive species, impact study, field experiment, environmental DNA, Mediterranean fanworm
Citation: Atalah J, Floerl O, Pochon X, Townsend M, Tait L and Lohrer AM (2019) The Introduced Fanworm, Sabella spallanzanii, Alters Soft Sediment Macrofauna and Bacterial Communities. Front. Ecol. Evol. 7:481. doi: 10.3389/fevo.2019.00481
Received: 07 October 2019; Accepted: 25 November 2019;
Published: 10 December 2019.
Edited by:
Elena Tricarico, University of Florence, ItalyReviewed by:
Agnese Marchini, University of Pavia, ItalyGeoff Read, National Institute of Water and Atmospheric Research (NIWA), New Zealand
Copyright © 2019 Atalah, Floerl, Pochon, Townsend, Tait and Lohrer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Javier Atalah, amF2aWVyLmF0YWxhaEBjYXd0aHJvbi5vcmcubno=
 Javier Atalah
Javier Atalah Oliver Floerl1
Oliver Floerl1 Xavier Pochon
Xavier Pochon Michael Townsend
Michael Townsend Leigh Tait
Leigh Tait Andrew M. Lohrer
Andrew M. Lohrer