- 1Department of Biology, Case Western Reserve University, Cleveland, OH, United States
- 2The Holden Arboretum, Kirtland, OH, United States
Plant-soil feedbacks have important implications for community composition and restoration. However, relatively few field trials test the influence of plant-soil feedbacks, especially on longer-lived species, such as trees. Here we present a field restoration experiment with 10 ectomycorrhizal fungal tree species native to eastern North America. Trees were inoculated with soils collected from conspecifics in the field or from a heterospecific: Quercus rubra. Following 16 months of growth in the field, Carya ovata diameter increase was significantly greater in trees receiving the heterospecific inoculant. This plant-soil feedback is consistent with C. ovata's natural co-occurrence with Q. rubra. Conversely, Quercus macrocarpa diameter increase and Carya cordiformis height increase were significantly greater when inoculated with conspecific soils, and this positive plant-soil feedback is consistent with their numerical dominance in natural communities. We found no evidence for phylogenetic Janzen-Connell effects or conservation of soil mutualists across tree species. We also quantified differences in soil fungal community structure with next generation sequencing methods (Illumina MiSeq) following 16 months in the field. Shannon's diversity of fungal taxa was greater in heterospecific soils of seven of our nine experimental species, consistent with a diversifying influence of Quercus rubra soil inocula. However, only one genus, Ulmus, exhibited differences in fungal community composition derived from conspecific and heterospecific sources, suggesting a stronger effect of focal tree species than of soil inocula source. The relatedness among focal tree species also influenced fungal community composition, with tree families and genera displaying different fungal communities. We suggest that future experiments should determine whether more diverse tree and fungal communities might have enhanced ecosystem functioning in tree restoration sites.
Introduction
Understanding the influence of plant-soil feedbacks on plant growth, community composition, and restoration ecology has become a growing area of focus in plant ecology (Kulmatiski et al., 2008; Brinkman et al., 2010; Bever et al., 2012), in part because plant-soil feedbacks have the potential to influence species coexistence (Mangan et al., 2010; Anacker et al., 2014; Bennett et al., 2017). For example, reciprocal negative feedbacks predict frequency-dependent coexistence and could be a diversity-enhancing mechanism in plant communities (Bever et al., 1997). Conversely, positive individual plant-soil feedback predict the numerical dominance of species that benefit from conspecific soils (Klironomos, 2002). Plant-soil feedbacks occur because plants influence the soils where they grow (Ehrenfeld et al., 2005) including soil microbial community composition and soil nutrient availability (reviewed in Ehrenfeld et al., 2005; e.g., Burns et al., 2017). A large body of greenhouse experiments suggest that individual plant-soil feedbacks can have a strong influence on plant growth (Kulmatiski et al., 2008). However, far fewer studies have determined whether such short-term greenhouse results translate into longer-term effects in the field (but see e.g., Schittko et al., 2016). Further, while plant-soil feedbacks are often quantified by comparing conspecific vs. heterospecific conditioned soils, the role of plant relatedness in plant-soil feedbacks is still unclear (Liu et al., 2012; Anacker et al., 2014; Mehrabi and Tuck, 2015; Crawford et al., 2019). Here, we ask whether individual plant-soil feedbacks influence soil microbial community composition and tree growth for nine temperate ectomycorrhizal fungal tree species in a field restoration experiment.
A common goal of ecological restoration is to facilitate the return of ecosystem function to disturbed environments, simultaneously providing substantial socio-economic and ecological benefits (BenDor et al., 2015). In eastern North America, the restoration of temperate deciduous forests is a common objective of many restoration projects, including those targeting abandoned agricultural land and anthropogenically disturbed urban habitats (Cernasky, 2018). While a multitude of factors influence the success or failure of these projects, soil microbial communities may have an important influence on restoration outcomes (Harris, 2009; Kardol and Wardle, 2010). Manipulation of soil microbial community structure may influence the result of temperate forest restoration and has become a commonly employed method intended to improve tree survival and/or growth.
One efficient method of manipulating soil microbial community structure is to inoculate trees with forest collected soils prior to outplanting in a restoration site (Maltz and Tresder, 2015; St-Denis et al., 2017). This method transfers potentially beneficial microbes, including mycorrhizal fungi, as well as potentially antagonistic microbes including pathogenic fungi and/or bacteria. Practitioners looking to collect soils for use as an inoculant could either avoid collecting from mature conspecific individuals if pathogens are found to have a primary influence on tree survivorship and growth (e.g., Packer and Clay, 2000), or conversely, target mature conspecific individuals if specific mutualisms elicit improved tree performance (e.g., den Bakker et al., 2004; Ishida et al., 2007). Plant-soil feedbacks that result from the conditioning of soil communities by different focal tree species have a profound influence on the microbial composition of forest soil transfers and the subsequent response of plants to inoculation during restoration (Wubs et al., 2016; Lance et al., 2019). Understanding the factors that influence the development of plant-soil feedbacks is essential to developing best practices for soil microbial community manipulation and is critical for ecological restoration of temperate forest communities. For example, the Janzen-Connell hypothesis (Janzen, 1970; Connell, 1971), in which adult individuals inhibit the growth of conspecific or closely related recruits (Liu et al., 2012), may prove to be an important predictor of tree response to inoculation with forest soils.
Knowledge of the phylogenetic relationships between individuals that condition soil inocula and those receiving the inocula may be of value to both restoration practitioners and ecologists looking to further understand the connection between soil microbial communities, plant performance, and plant community composition (e.g., Reinhart et al., 2012a). Phylogenetic relationships amongst plants are known to influence the soil microbial communities with which they associate (Ishida et al., 2007; Burns et al., 2015). Phylogenetic conservatism in plant-soil feedbacks would result in closely related species responding in a similar fashion to inoculation with forest soil transfers. For example, pathogenic soil microbes are known to negatively influence closely related sub-tropical tree species (Liu et al., 2012), a pattern called “phylogenetic Janzen-Connell” effects. When pathogens effect close relatives similarly, practitioners should avoid collecting soils from close relatives. Alternatively, if closely related species response similarly to soil mutualists like mycorrhizal fungi (Reinhart et al., 2012b), then forest restoration might be enhanced by soils collected from close relatives. A lack of phylogenetic effect in plant-soil feedbacks might limit local soil collections to conspecifics for access to mutualists or heterospecifics for avoidance of pathogens (Fitzpatrick et al., 2017), at least where soil communities are species-specific (Ehrenfeld et al., 2005).
In this study we determine whether plant-soil feedbacks and plant relatedness influence tree growth and soil microbial community composition and diversity in a 16 month field experiment. We estimated a phylogeny for ten ectomycorrhizal tree species native to eastern North America, then performed a field experiment in which trees were inoculated with soil conditioned by a conspecific or heterospecific source. Our heterospecific source was Quercus rubra, a widely distributed ectomycorrhizal tree. We asked three primary questions: (1) Did individual plant-soil feedbacks (conspecific/heterospecific) influence tree growth and survival for ectomycorrhizal tree species? (2) Are plant-soil feedbacks consistent with a phylogenetic Janzen-Connell effect of conserved pathogens or do conserved mutualisms improve tree performance? (3) Were rhizosphere soil fungal diversity and community composition influenced by conditioning soil source (conspecific/heterospecific), focal tree species, and plant relatedness after 16 months in the field?
For question (1), we hypothesized that trees receiving the heterospecific inoculant would display greater growth and survival than those receiving the conspecific inoculant, if pathogens exhibit more host specificity than mutualists (but see e.g., Burgess et al., 2017). Host specificity is displayed in a small proportion of ectomycorrhizal fungal relationships and large numbers of ectomycorrhizal fungi can be found colonizing one tree (Bruns, 1995; Palmer et al., 2008). In addition, most cases of ectomycorrhizal fungal specificity occur in the genera Pinus and Alnus, neither of which was included in our study design (Bruns et al., 2002; Tedersoo et al., 2009). Thus we expect pathogen escape in heterospecific soils (i.e., negative plant-soil feedbacks). Alternatively, because our tree species are ectomycorrhizal, and because some prior work has observed positive plant-soil feedbacks in most ectomycorrhizal trees (Bennett et al., 2017), we might expect positive plant-soil feedbacks. To answer question (2), we explored how inoculation with distant relative's heterospecific conditioned soil might enable trees to form beneficial ectomycorrhizal fungal relationships while avoiding pathogens, consistent with a phylogenetic Janzen-Connell effect (Liu et al., 2012). Thus, we predicted more neutral plant-soil feedbacks in the close relatives of Q. rubra (e.g., Q. palustris) and more positive effects in distant relatives (e.g., pathogen escape). Alternatively, plant performance could be greater in conspecific and close relatives' soils, consistent with the positive plant-soil feedbacks observed in some ectomycorrhizal fungal trees (Bennett et al., 2017). In this case, we would predict more neutral plant-soil feedbacks in the close relatives of Q. rubra and more negative in distant relatives (e.g., loss of mutualist services). Conversely, if pathogens and mutualists are not influenced by plant relatedness, plant-soil feedbacks might not scale with phylogenetic distance (Mehrabi and Tuck, 2015). Finally, to answer question (3), we used next generation sequencing following 16 months in the field to explore the roles of soil treatment (conspecific, heterospecific), focal species, and plant relatedness in determining fungal community composition on roots and in rhizosphere soils. We predicted that fungal communities derived from heterospecific soils would be more diverse than those derived from conspecific soils, because heterospecific soils have both the fungal community in the inocula as well as any fungal species cultivated by the focal tree. We also predicted that rhizosphere fungal community structure would significantly differ between the heterospecific and conspecific treatments within each species. We predicted effects of focal tree species on fungal community composition, if focal trees influence soil microbial communities as they grow in our restoration site. Finally, we predicted that closely related focal tree species might have similar fungal community composition (e.g., Burns et al., 2015), if such effects are conserved (e.g., Liu et al., 2012; but see Mehrabi and Tuck, 2015; Sweet and Burns, 2017).
Methods
Experimental Overview and Plant Material
Our experiment focused on the restoration of 10 ectomycorrhizal fungal tree species (see Figure 1), which are native to the northeastern United States. Our design included two experimental treatments: conspecific or heterospecific soil inocula, each with 10 replicates (except for Q. rubra, which received only conspecific soil). Thus, the total design was 9 species × 2 soils (conspecific/heterospecific) × 10 replicates (180 trees) + 20 Q. rubra in conspecific soil for a total of 200 trees. Quercus rubra served as the heterospecific soil source for all other tree species (see details below), because it grows in a number of forest types and can be found in association with all of our other experimental tree species (Lance, personal observation). We manipulated the soil microbial community associated with each tree by adding field collected soils to pots prior to planting in a former community garden at Squire Valleevue Farm (Hunting Valley, OH, USA). The region in which our site is located is characterized by a humid continental climate, with a mean annual precipitation of approximately 990 mm. Soils at our research site were classified as Ellsworth silt loams (USDA, 2019). We monitored tree growth and soil fungal community structure after two growing seasons to understand how plant-soil feedbacks may influence restoration efforts.
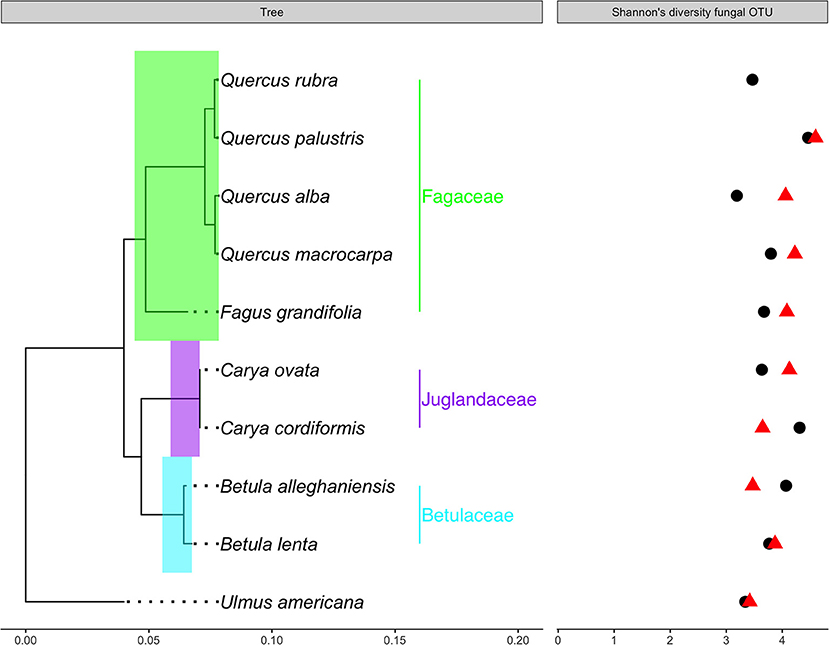
Figure 1. A maximum likelihood phylogeny for 10 tree species, which associate with ectomycorrhizal fungi. Two DNA regions, matK and rbcL were used to estimate this phylogeny (Supplementary Material S2). Branch lengths are in substitutions per site. Shannon's diversity of fungal communities following inoculation with conspecific soil (black dots) or heterospecific soils (red triangles) are represented to the right of the phylogeny.
Trees for use as experimental phytometers were sourced from a native plant nursery in Indiana, USA in April 2017. We obtained 20 trees of each of our 10 focal species. Trees arrived in 4 to 12-liter pots, except for Fagus grandifolia, which arrived in 18-liter pots. Variation in initial size was accounted for by calculating relative growth rates (see Statistical approach below). Trees were watered during the period between arrival from the nursery and planting into the field site.
Inoculation and Planting
We collected soils to use as soil inoculum in April 2017. We collected soil from 3 separate mature (diameter at breast height > 20 cm) trees of each of the 10 species in our field restoration experiment [as suggested in (Reinhart and Rinella, 2016)]. Replicate soil collections were not mixed following collection. To obtain soils from all 10 species, soils were collected at two locations (Squire Vallevue Farm and Holden Arboretum) located approximately 15 km apart. The three dominant soil types at both locations are Mahoning silt loams, Ellsworth silt loams, and Haskins loams (USDA, 2019). Collection implements were sterilized with 80% ethanol and air dried between replicate soil collections. Trees targeted for soil collection were growing in natural temperate forests except for Quercus macrocarpa and Quercus palustrus where two of the three replicate collections were made from Holden Arboretum's planted collection. Approximately 0.5 kg of soil was collected within 1 m of the bole of each tree; collection locations were void of herbaceous vegetation and measured approximately 2 m × 2 m. Only the top 10 cm of soil was collected, and large root fragments were removed in the field. Following collection, soils were dried at ambient room temperature for 1 week then sieved with a 0.5 cm sieve. Each of the 3 replicate collections per species were kept separate throughout the inoculum preparation process; we avoid mixing soils to avoid potential problems with pseudoreplication, such as mixing a rare pathogen into all samples (Smith-Ramesh and Reynolds, 2017; Rinella and Reinhart, 2018).
Ten trees of each species were randomly selected for inoculation with either conspecific or heterospecific soil, resulting in a total of 20 trees per species being planted in the restoration site (except for 20 Q.rubra, which received only conspecific inoculation). We equally divided the three replicate collections per species during inoculation. Therefore, three trees received soil from one of two replicate collections while four trees received the randomly selected third replicate collection. We thus replicated both soil collections within species and phytometer/tree genotype within soil replicate. We abraded the surface of each pot with a sterile gloved hand, then applied 50 g of dried soil to the surface. Trees were immediately watered with 0.5 L water. Trees remained in their pots following inoculation for 2 weeks before being planted in May, 2017.
Our experimental site measured ~50 m × 50 m; the site had previously been used as a community garden but was idle for 1 year prior to the start of our experiment. We tilled the site twice: once in the fall of 2016 and again in spring of 2017 to remove herbaceous vegetation and facilitate the planting process. Tilling has also been shown to increase ectomycorrhizal colonization in temperate tree restoration (Bauman et al., 2013). Trees were planted in 15 rows in completely randomized positions. Approximately 3 m separated each tree and row. We irrigated the site during planting and throughout the summer of 2017. Irrigation was not provided during 2018. Trees also received an ~1 m diameter ring of fresh wood chips (not composted) immediately following planting. Trees were wrapped with plastic protective wraps (ArborGuard, Gempler's Supply, Janesville, WI, USA) to prevent small mammal herbivory. Areas surrounding trees and wood chips were seeded with a mixture of annual and perennial grasses.
Measurements and Soil Collection
We took a baseline measurement of tree size immediately following planting in May 2017. Subsequent measurements were collected in September 2018. Height was determined by measuring to the apical bud of each tree using a meter stick (height < 140 cm) or Sokkia telescoping height pole (Senshin Industry, Osaka, Japan). We measured diameter 10 cm above the root collar of each tree using a Mitutoyo digital caliper (Mitutoyo Corporation, Kanagawa, Japan).
Soils for molecular analysis were collected in September 2018. We sampled 6 trees per species: 3 inoculated with conspecific soils (one per replicate collection location) and three inoculated with heterospecific soils (one per replicate collection location) for a total of 57 samples [9 species × 2 soil origins (conspecific, heterospecific) × 3 collection locations + 3 conspecific inoculated Q. rubra (one per each of the 3 collection locations)]. Two separate cores of the top 15 cm of soil were taken for each sampled tree and immediately homogenized in the field. Samples consisted of fine root fragments and soil that adhered to the roots (rhizosphere soil). Cores were sterilized with 80% ethanol between collections. Samples were immediately placed on ice in the field, then frozen at −70°C before processing.
Next Generation Sequencing With Illumina Mi-Seq
We utilized next generation sequencing methods to examine general fungal communities on roots and in rhizosphere soils of all 57 soil samples. DNA was extracted from samples following a phenol-chloroform protocol (Burke, 2008). We made amplicons of the fungal ITS-2 gene region using the primers 58A2F and ITS4 with Illumina overhang adapters. The primer sequences are included in the Supplementary Material S1. The “16S Metagenomic Sequence Library Preparation” (Illumina technology protocol) was utilized as a guide during primer selection. Each reaction included 2 units of FastStart Taq DNA polymerase (Sigma-Aldritch, Inc., St. Louis, MO, USA), 2 mM MgCl2, 0.2 uM of both primers 58A2F and ITS4, 0.5 ug/ul bovine serum albumin, and 0.8 mM dNTP mix. Our thermocycling conditions were an initial denaturation at 95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 5 min. Amplicons were then purified, indexed, and sequenced as 2 × 250 bp reads on the Illumina MiSeq V3 sequencer at the Case Western Reserve University Genomics Core facility.
The Blaxter lab's metabarcoding processing pipeline (version 1.0.1) was used as a guide for our sample processing (Blaxter, 2016) with the UPARSE pipeline (Edgar, 2013). We merged forward and reverse reads with the fastq_mergepairs command in USEARCH, version 11.0 (Edgar, 2010). We removed control phiX prior to merging reads using the filter_phiX command. Primers were removed with Cut Adapt (v1.10) (Martin, 2011). We implemented the UCLUST algorithm (Edgar, 2010) for OTU clustering at 97% similarity and removal of chimeras using the UCHIME algorithm (Edgar et al., 2011). Singletons OTUs were removed. We made taxonomic assignments for each OTU by utilizing the SINTAX algorithm (Edgar, 2016) and comparing against the UNITE database (v 8.0, release date 2018-11-18) (Aberenkov et al., 2010; Koljalg et al., 2013).
Phylogeny Estimation
A molecular phylogeny based on matK and rbcL gene sequences was estimated for 10 ectomycorrhizal fungal tree species (Figure 1). Sequences for both gene regions were available for all 10 species (see Supplementary Material S2). The program MUSCLE (Edgar, 2004a,b) in the MEGA platform (version 7.0.14) was used to align each gene sequence separately. Alignments were checked by eye and preliminary phylogenies for each gene region were used to diagnose outliers. Because both regions generated consistent preliminary phylogenies, they were concatenated to construct a “total evidence” or “super matrix” phylogeny. Garli (version.951) was utilized to conduct a maximum likelihood tree search with 100 bootstrap replicates. We rooted the tree using Ulmus americana (Hinchliff et al., 2015), which is in the Rosales (Rees and Cranston, 2017). All other taxa in our sample are in the Fagales (Rees and Cranston, 2017).
Statistical Analyses
We conducted all statistical analyses in R version 3.5.1 unless otherwise specified.
Did Plant-Soil Feedbacks Influence Tree Growth and Survival for Ectomycorrhizal Tree Species?
To compare growth rates across species, we first standardized by initial size by calculating relative growth rates (RGR), then compared responses to conspecific and heterospecific soil treatments using log response ratios (Brinkman et al., 2010). Betula lenta was not included in these analyses due to high mortality and Quercus rubra had only the conspecific soil treatment, leaving 8 tree species in these analyses. Relative growth rate (RGR) in each treatment was calculated as (ln X-ln Y)/16, where X was a size measurement (height or diameter) at the conclusion of the experiment and Y was the corresponding size measurement at the beginning of the experiment. Our experiment lasted for 16 months as indicated by the denominator in our relative growth rate equation. We examined differences in tree growth response (stem elongation and diameter increase) to conspecific and heterospecific inoculants by calculating pairwise natural-log response ratios (lnRR) (Brinkman et al., 2010; Larios and Suding, 2015). We averaged growth data by replicate soil collection location in order to avoid pseudoreplication. Ratios were calculated as ln(RGR in conspecific/RGR in heterospecific) (Brinkman et al., 2010). This procedure resulted in n = 3 lnRR per species. Ninety five percentage confidence intervals were calculated for each ratio in order to determine statistical significance. If plant-soil feedbacks are primarily positive (greater plant performance in conspecific soils), we would see positive log response ratios. If plant-soil feedbacks are primarily negative (greater performance in heterospecific soils), we would see negative log response ratios. Note that this metric is an individual plant-soil feedback metric, and measures the absolute difference in response between plant performance in conspecific and heterospecific soils. This is an important measure of plant-soil feedback effects, and corresponds to differences in abundance in the field in some cases (e.g., Klironomos, 2002). However, it does not make a coexistence prediction, as do net-pairwise plant-soil feedback metrics (Bever et al., 1997).
Tree survival was examined using a generalized linear model with a binomial error distribution in which survival was analyzed as a function of treatment (conspecific or heterospecific), soil replicate, species, row, and an interaction of treatment and species. We predicted significant differences in survival across species; furthermore, if tree species respond to conspecific and heterospecific soils in a species-specific manner, we predict a treatment × species interaction. We also analyzed the within species patterns to test for effects of treatment, again including row as a blocking effect. If trees had higher survival in the conspecific compared with heterospecific treatment, this is a positive plant-soil feedback. If tree had higher survival in the heterospecific treatment, this is a negative plant-soil feedback.
Were Plant-Soil Feedbacks Consistent With a Phylogenetic Janzen-Connell Effect or Conserved Mutualists?
To test for an effect of phylogenetic distance on the strength of plant-soil feedbacks, we conducted a phylogenetic “meta-analysis” on the plant-soil feedback log response ratio. Because log response ratios have an associated variance, we used this analysis method to take that variance into account. Plant soil feedbacks were again measured as the log response ratios for diameter and height, and we conducted two separate models, one for each log response ratio. As in standard meta-analysis, the log response ratios were weighted by the inverse of their variance (Koricheva and Gurevitch, 2013). We used the rma.mv function in the metafor (Viechtbauer, 2010) package with species treated as a random effect and phylogeny incorporated into the error structure as a variance-covariance matrix. This test takes non-independence of the branch lengths in the phylogeny into account and is thus preferable to a linear model. We asked whether the strength of plant-soil feedbacks were a function of phylogenetic distance to the heterospecific (Quercus rubra). If a phylogenetic Janzen-Connell effect is present, we predict that Quercus will have the most neutral plant-soil feedbacks, followed by Fagus grandifolia, then the Caryas and Betula, and Ulmus americana will have the most positive plant-soil feedback (i.e., a positive slope). In other words, we predict a positive slope for the phylogenetic distance effect in these models (Liu et al., 2012). Alternatively, if mutualist effects are phylogenetically conserved (Reinhart et al., 2012b), we predict the most neutral plant soil feedbacks for Quercus, followed Fagus grandifolia, then the Caryas and Betula, and finally Ulmus americana will have the most negative plant-soil feedback (i.e., loss of mutualist benefits). In other words, we predict a negative slope for the phylogenetic distance effect [as found in (Crawford et al., 2019)].
Were Soil Fungal Diversity and Community Composition Influenced by Plant-Soil Feedbacks, Focal Tree Species, and Plant Relatedness After 16 Months in the Field?
To characterize the soil fungal community, we utilized next generation sequencing on rhizosphere soils. We were able to generate OTU matrix tables for 52 of the 57 collected samples. Five samples did not produce useable reads (one B. allegheniensis, one B. lenta, one C. cordiformis, and two Q. palustrus samples). We normalized the matrix of sequence counts generated by our next generation sequencing effort with the RLE normalization using the edgeR package (version 3.22.5) in R prior to statistical analysis. Variance stabilizing normalizations such as the RLE normalization are superior to rarifying microbiome data during statistical analysis due to a higher retention of data (McMurdie and Holmes, 2014).
We compared general fungal community composition on roots and in rhizosphere soils with non-metric multidimensional scaling (NMDS) procedures in the “vegan” package of R (Oksanen et al., 2017). Our NMDS procedures utilized the Sorenson (Bray-Curtis) distance metric, a random starting configuration, and three dimensions. Three dimensions were selected in order to minimize ordination stress. All permutations consisted of 4,999 iterations and were stratified by row (position in the experimental design). Non-parametric permutation procedures (PERMANOVA) were utilized to test for significant differences in fungal communities between genera, families, and inoculant source (conspecific or heterospecific). We analyzed all species collectively.
To determine how focal tree species and soil treatment (conspecific/heterospecific) influenced ectomycorrhizal fungal abundance, we extracted normalized abundances for six focal ectomycorrhizal fungal taxa (Entoloma, Laccaria, Russula, Scleroderma, Tomentella, and Tuber) from the sequencing data set. Some of these ectomycorrhizal fungal genera contained several taxa. To summarize these data, we summed abundances for each of these six genera across the RLE transformed matrix counts for each species/fungal genus combination. These values represent normalized abundances of these fungal genera and were then plotted as a heatmap against our tree phylogeny.
To determine whether fungal community diversity differed across treatments (conspecific/heterospecific), we calculated Shannon's diversity on the total fungal OTU matrix. We used a paired t-test and a paired sign test to test the prediction that heterospecific soils will be more diverse than conspecific soils. The paired t-test tests the prediction that the diversity values for heterospecific soil are greater than conspecific soils within species. The paired sign test tests the prediction that the direction of the effect (a priori prediction: heterospecific > conspecific) is consistent across species, for the 9 trees with both heterospecific and conspecific treatments (note that Quercus rubra only had a conspecific treatment).
Results
Plant-Soil Feedbacks Influenced Tree Growth or Survival for Three Ectomycorrhizal Tree Species After 16 Months in the Field
We found three significant trends in tree growth in response to inoculation with either heterospecific or conspecific soils. Following two growing seasons, C. cordiformis stem elongation was significantly greater when trees received the conspecific inoculant (Figure 2). Inoculation with conspecific soils also facilitated diameter increase in Q. macrocarpa (Figure 2). Conversely, C. ovata trees receiving the heterospecific inoculant displayed greater diameter increase following two growing seasons when compared to trees receiving the conspecific inoculant. We did not find additional significant growth trends in the remaining species (Figure 2).
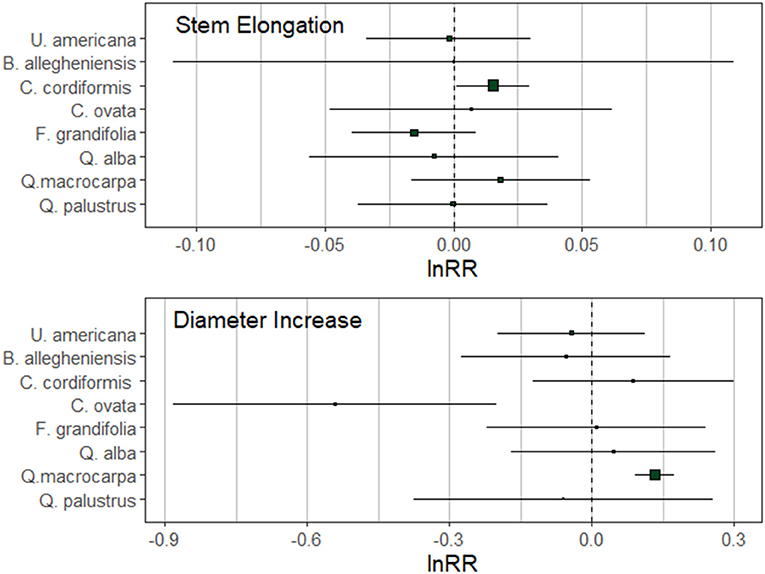
Figure 2. Natural log-response ratios (lnRR) for stem elongation and diameter increase at the conclusion of the second growing season. Mean lnRR and boundaries of the 95% confidence intervals are represented on the figure. Positive plant-soil feedbacks have positive values and negative plant-soil feedbacks have negative values of lnRR.
We found significant differences in the survival of the 10 experimental species (Table 1). Only 33% of the Betula lenta trees survived through two growing seasons. Three other species, C. ovata, C. cordiformis, and B. allegheniensis exhibited more than more one fatality throughout the course of the experiment; 80% of the planted trees survived for each of the above three species. Carya cordiformis survival was significantly influenced by soil treatment (p = 0.03), with significantly greater survival following inoculation with heterospecific soil. The remaining species had 100% survival throughout the experiment, with the exception of Q. rubra, in which one tree (5%) died. We did not find a treatment × species interaction on tree survival over 16 months in the field (Table 1).
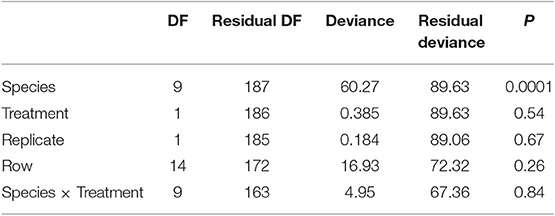
Table 1. Results of the survival analyses for tree survival across 9 species in a tree restoration experiment with two soil treatments (conspecific soil, heterospecific soil from Quercus rubra).
Plant-Soil Feedbacks Were Not Consistent With a Phylogenetic Janzen-Connell Effect or Conserved Mutualists
There was not an effect of phylogenetic distance on the strength of plant-soil feedbacks as measured by either growth in tree height or diameter (Table 2, Figure 3). In general, both diameter and height growth data suggested that the three closest relatives to heterospecific Quercus rubra (other Quercus) responded similarly to conspecific soil and heterospecific soil, performing weakly better in conspecific soil in most cases (Figure 3). Focal species at intermediate phylogenetic distances to Q. rubra were highly variable in their plant-soil feedbacks (Figure 3). The Caryas were especially diverse in their plant-soil feedbacks, with the largest and smallest effect sizes in diameter in our data. The most distant relative to heterospecific Quercus rubra, Ulmus americana, had a neutral feedback (responded similarly to conspecific and heterospecific soils (Figures 2, 3).
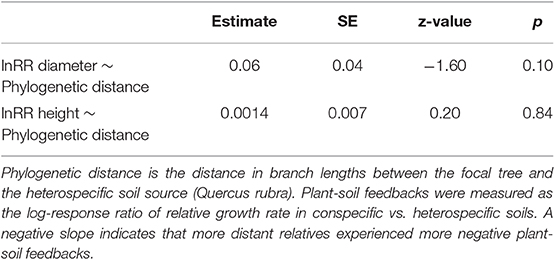
Table 2. Test for phylogenetic Janzen-Connell effects on plant-soil feedbacks (lnRR) (Figure 3).
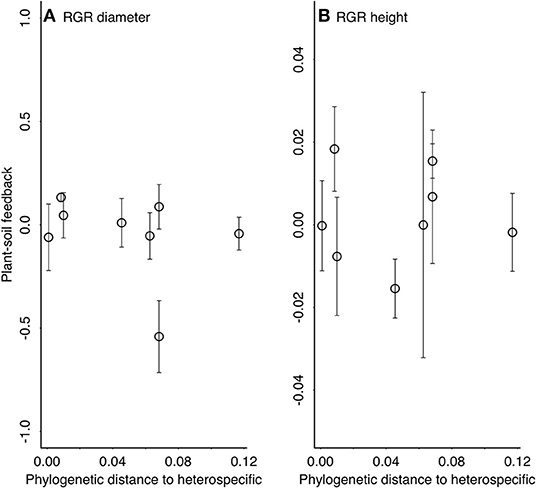
Figure 3. The strength of plant-soil feedbacks [lnRR = ln(RGR conspecific/RGR heterospecific)] across the phylogenetic distance from the focal tree to the heterospecific soil inocula source (Quercus rubra) (Table 2). Relative growth rates were used to calculate lnRR for (A) plant diameter and (B) plant height across 16 months in the field. Positive plant-soil feedbacks have positive values and negative plant-soil feedbacks have negative values of lnRR. Means ± 1 SE.
Were Soil Fungal Diversity and Community Composition Influenced by Plant-Soil Feedbacks, Focal Tree Species, and Plant Relatedness After 16 Months in the Field?
Our next generation sequencing methods generated over 14.1 million reads, which were mapped to 3,360 OTUs in the UPARSE pipeline. 780 OTUs (~23%) could be assigned to either the genus or species level. 833 OTUs (~25%) could not be assigned to a taxonomic level below “fungi.” Richness ranged from 229 OTUs per sample to 1,186 OTUs per sample. A diversity of fungal functional groups was represented in our OTU database. OTUs included saprotrophic fungi (e.g., Mortierella, Coprinellus, and Pleurotheciella), ectomycorrhizal fungi (e.g., Scleroderma, Tomentella, and Tuber), and arbuscular mycorrhizal fungi including the genera Glomus and Funneliformis.
Plant-soil feedbacks influenced fungal diversity (Figure 1) but did not influence fungal community composition after 16 months in the field (Figure 4). We found marginally significantly greater (p = 0.07) Shannon's diversity of general fungal communities in heterospecific than conspecific soils (Figure 1). Trees receiving the heterospecific inoculant had greater diversity than those receiving the conspecific inoculant in seven of the nine species. Our paired t-tests, however, yielded insignificant results, suggesting that Shannon's diversity was not greater in heterospecific soils (t = −0.64, p = 0.53). Across all species, soil treatment (conspecific and heterospecific) did not have a significant influence on fungal community structure [F(1, 50) = 0.87, p = 0.80]. Our limited number of replicates precluded the use of PERMANOVA tests on single species; however, we implemented NMDS ordination plots to provide a visual representation of fungal communities derived from conspecific and heterospecific inoculants. We noted few visual differences in community structure between communities derived from conspecific and heterospecific inoculants, except in the genus Ulmus in which conspecific and heterospecific communities were visually distinct.
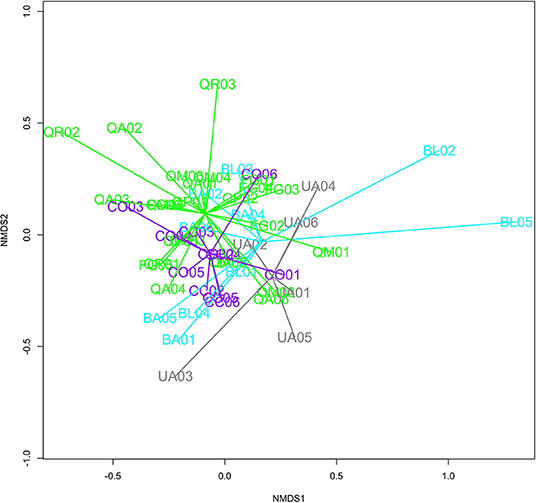
Figure 4. Radar graph for general fungi communities in rhizosphere soils of each of the four experimental plant families at the end of the second growing season. Both conspecific and heterospecific treatments are represented. Families include Fagaceae (Green), Juglandaceae (Purple), Ulmaceae (Gray), and Betulaceae (Teal).
Focal tree species had different normalized abundances of ectomycorrhizal fungi and there was a great deal of variation in normalized abundances, with some ectomycorrhizal fungi being much more abundant than others (Figure 5). The most abundant ectomycorrhizal fungal genus in our experiment was Tuber. The genera Entoloma and Russula had low abundances in association with all of the tree species. Tuber associated strongly with distantly related tree species, suggesting limited host specificity, at least in this fungal genus. Additionally, the genus Quercus associated with more ectomycorrhizal fungi than other tree genera (Figure 5).
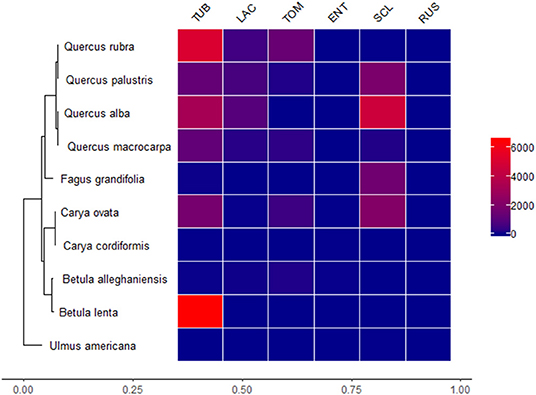
Figure 5. Heatmap representing relationships between ectomycorrhizal fungi genera and the experimental tree species. Heatmap values indicate cumulative edge R normalized scores for each fungal genus in our OTU matrix. Ectomycorrhizal fungi taxa are TUB (Tuber), LAC (Laccaria), TOM (Tomentella), ENT (Entoloma), SCL (Scleroderma), and RUS (Russula). Red values are greater normalized abundances and blue colors are lower normalized abundances.
Our soil fungal community analysis indicated significant taxonomic conservatism amongst our experimental tree species. In other words, closely related focal tree species had similar fungal community composition after 16 months in the field. Our PERMANOVA analysis indicated that both tree genera [F(4, 46) = 1.39, p = 0.0002] and family [F(4, 47) = 1.41, p = 0.001] had significant influences on fungal community composition, indicating that closely related trees shared similar soil fungal community structure (Figure 4). Row also had a significant effect in our genera model [F(1, 47) = 1.56, p = 0.03].
Discussion
Our experiment demonstrates that individual plant-soil feedbacks can persist for at least two growing seasons in temperate tree restoration. Tree responses to conspecific and heterospecific inoculation varied by species following two growing seasons. We found negative plant-soil feedbacks in growth for one species and positive for another, in addition to a negative plant-soil feedback in survival for a third species. These effects of soil inocula on tree growth were not consistent with a phylogenetic Janzen-Connell effect or conserved soil mutualists. Though greater fungal community diversity in heterospecific Quercus rubra soils persisted over 16 months in the field, our analysis of community composition suggests that trees planted into the restoration experiment might be “conditioning” the soils in their root zones. In other words, we likely observed convergence between soil general fungal communities derived from heterospecific and conspecific soil inoculants over time, though a baseline characterization of the fungal community would have been necessary to confirm this pattern. Further, we found a tree taxonomic influence on the structure of soil general fungal communities, with tree genera and families exhibiting different communities. Thus, plant-soil feedbacks are likely to interact with a species receiving an inoculant in a way that depends upon the plant species identity and evolutionary history.
Plant-Soil Feedbacks Influenced Tree Growth and Survival for Some Ectomycorrhizal Tree Species After 16 Months in the Field
Tree growth responses in our experiment varied by species, supporting the findings of a previous pot experiment, which utilized temperate trees (McCarthy-Neumann and Kobe, 2010). Prior studies have indicated that many ectomycorrhizal trees native to eastern North America facilitate the recruitment of their own seedlings/saplings (Bennett et al., 2017). Field experiments utilizing ectomycorrhizal fungal Tsuga canadensis provides support for these findings, as saplings of this species have shown improved performance in the soils of conspecifics compared to heterospecifics (O'Brien et al., 2011, but see Reinhart et al., 2012b). We identified conspecific facilitation in only one of our ten experimental species (Q. macrocarpa). The conspecific facilitation we observed in Q. macrocarpa is consistent with dominance patterns in natural plant communities, as this species commonly grows in open grassland communities where it alone can dominate the tree community.
Interestingly, Q. rubra and C. ovata are common associates in natural forests throughout the ecoregion in which our study took place (the glaciated Allegheny plateau). The facilitation of C. ovata growth by inoculation with Q. rubra conditioned soils may represent a diversity enhancing mechanism in these forests. C. ovata may have enhanced growth in tree fall gaps creating by Q. rubra, a pattern which would suggest that plant-soil feedbacks can persist for long periods of time and have important implications for structuring tree communities in natural forests (Bennett et al., 2017).
Plant-Soil Feedbacks Were Not Consistent With a Phylogenetic Janzen-Connell Effect or Conserved Mutualists
Our study differs from some previous work, which found evidence consistent with a phylogenetic Janzen-Connell effect (Liu et al., 2012; Sweet and Burns, 2017; Crawford et al., 2019). Some of these studies had comparable sample sizes to ours, including Liu et al. (2012) with 8 species of tree and Sweet and Burns (2017) with 7 species of herbaceous plants. There could be several, non-mutually exclusive, reasons for this apparent discrepancy. First, we have only a single species, Ulmus americana, that is highly phylogenetically distant from heterospecific Quercus rubra, and more distant relatives, might be needed to detect larger trends. Second, these trees have only grown for 16 months in the field, and increases in growth, especially height, were relatively modest. Longer time periods may be needed to detect significant effects of soil inocula on the growth of these tree species. Third, variance among species in tree sources could add variance to our data, potentially making detecting phylogenetic patterns more difficult. Fourth, meta-analyses using an alternative measure of plant-soil feedbacks, reciprocal pairwise plant-soil feedbacks (Bever et al., 1997), find for plants that share a mycorrhizal guild, more distantly related species have more negative plant-soil feedbacks (Crawford et al., 2019). Alternatively, we used the individual plant-soil feedback metric, perhaps hinting that the type of metric used to measure plant-soil feedbacks influences the results (i.e., plants might perform relatively, but not absolutely, better in the soils of distant relatives, compared with close relatives). Finally, these analyses do not consider predictions about the variance in plant responses over evolutionary time, again because of the single sampled species at greater phylogenetic distances. However, Brownian motion evolution suggests that the variance in plant responses should be greater in the soils of more distant relatives (Cadotte et al., 2017). Thus we need more experimental tests with replication both across and, critically, within, phylogenetic distances (Burns et al., 2019). The greater variance among the Betula and Carya species than among Quercus (Figure 3) hints that this prediction might hold true with increased sampling at greater phylogenetic distances.
Root and Rhizosphere Fungal Diversity or Community Composition Were Influenced by Plant-Soil Feedbacks, Focal Tree Species, and Plant Relatedness After 16 Months in the Field
Our next generation sequencing methods returned an incredible diversity of fungi in our early successional restoration site. Comparable hyperdiversity has previously been reported in general fungi communities of boreal forest communities (Taylor et al., 2014); however, we know of no similar estimations of fungal diversity from temperate forested systems or temperate restoration sites. While experimental treatments may have driven the diversity observed in rhizosphere soils, other sources of fungi such as nursery soils or the addition of wood chips could also have contributed to overall fungal diversity. The maintenance of such diversity through the establishment of a diverse array of tree species may result in increased fine-scale niche partitioning (Taylor et al., 2014) and increased microbial function (Carnovale et al., 2019). The marginally greater Shannon's diversity observed in trees receiving the heterospecific Quercus rubra inoculant suggests that heterospecific inoculation may introduce fungal taxa not typically associated with certain tree genera, and that these fungal taxa can persist for more than two seasons in field conditions.
Prior work in agricultural areas has also shown tree genus to be an important determinant of soil microbial community structure (Carnovale et al., 2019), although the mechanisms driving this pattern were unidentified in the cited study. Most studies connecting tree species or genera with soil microbial community structure investigate relationships between leaf litter traits and microbial community composition (e.g., Bardgett and Shine, 1999; Thoms and Gleixner, 2013). Our finding, however, could not be explained by interspecies variability in litter traits, as our soil samples were taken prior to leaf fall and excluded any leaf material which may have been present. Therefore, our finding is more likely the result of species having specific differences in belowground traits such as root carbon exudation (Broeckling et al., 2008). Above ground traits, however, can also influence belowground mechanisms. Growth rate, a trait in which our experimental species differed, has been shown to interact with certain microbial functional groups (Pei et al., 2016). The complexity of individual species responses to inoculation and the influence of tree relatedness on subsequent fungal community formation makes the development of broadly applicable methods for inoculation challenging.
We observed similar fungal community composition in the soils under congeneric and confamilial trees, consistent with some other studies (e.g., Burns et al., 2015). Such phylogenetic effects on soil fungal communities has the potential to lead to “phylogenetic” Janzen-Connell effects (Liu et al., 2012), where plants perform better in soils influenced by distant relatives and less well in soils from close relatives (see also Sweet and Burns, 2017). Escape from pathogens in the soil could potentially help explain such patterns. However, in this tree restoration experiment, tree growth over the first 16 months in the field did not suggest such phylogenetic Janzen-Connell effects. Rather, plant-soil feedback effects were generally species specific (McCarthy-Neumann and Kobe, 2010; St-Denis et al., 2017). We also noted that inoculation with heterospecific soils had the greatest influence on fungal community assembly within Ulmus, the genus most distantly related to Quercus. This suggests that increased phylogenetic distance between the soil conditioning species and the species receiving the soil transfer can result in more profound changes to fungal community structure than soil transfers between close relatives, though greater sampling of distant relatives would be needed to confirm this hypothesis.
In conclusion, we found plant-soil feedbacks after 16 months in the field for three tree species, with 2 out of 8 species showing effects on growth and 1 out of 9 for survival. Thus our results reinforce the hypothesis that the influence of plant-soil feedbacks may be exaggerated in glasshouse studies when compared to studies under natural conditions (Schittko et al., 2016; Heinze and Joshi, 2018). Our next generation sequencing approach found an influence of inocula treatment, tree species, and tree relatedness on rhizosphere fungal diversity or community structure. Therefore, planting a phylogenetically diverse tree restoration site could result in a more diverse fungal community. Future studies should measure ecosystem function (Lance et al., 2019), especially across a phylogenetic diversity gradient in tree restoration.
Data Availability Statement
The datasets generated for this study can be found in the https://osf.io/cv9kf/.
Author Contributions
AL and JB designed the study. AL, SC-K, and DB conducted the lab work and analyzed the fungal sequencing data. JB conducted the phylogenetic analyses. AL and JB conducted the statistical analyses and wrote the first version of the manuscript. SC-K and DB contributed to manuscript revisions.
Funding
Squire Valluevue and Valley Ridge Farms provided financial support for the implementation of this project. Additional funding came from the Native Plant Society of Northeastern Ohio. Funding was also provided by Case Western Reserve University Department of Biology Oglebay Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Squire Vallevue and Valley Ridge Farms, particularly Shane Brown and Ana Locci, for logistical support of our experiment. We also thank the CWRU Ecology and Evolution reading group and the Burns lab for assistance with planting and manuscript review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00500/full#supplementary-material
References
Aberenkov, K., Niellsen, H. R., Larsson, K. H., Aleksander, R. J., Eberhart, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi- recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Anacker, B. L., Klironomos, J. N., Maherali, H., Reinhart, K. O., and Strauss, S. Y. (2014). Phylogenetic conservatism in plant-soil feedback and its implications for plant abundance. Ecol. Lett. 17, 1613–1621. doi: 10.1111/ele.12378
Bardgett, R. D., and Shine, A. (1999). Linkages between plant litter diversity, soil microbial biomass and ecosystem function in temperate grasslands. Soil Biol. Biochem. 31, 317–321. doi: 10.1016/S0038-0717(98)00121-7
Bauman, J. M., Keiffer, C. H., Hiremath, S., and McCarthy, B. C. (2013). Soil preparation methods promoting ectomycorrhizal colonization and American chestnut Castanea dentata establishment in coal mine restoration. J. Appl. Ecol. 50, 721–729. doi: 10.1111/1365-2664.12070
BenDor, T., Lester, T. W., Livengood, A., Davis, A., and Yonavjak, L. (2015). Estimating the size and impact of the ecological restoration economy. PLoS ONE 10:e0128339. doi: 10.1371/journal.pone.0128339
Bennett, J. A., Maherali, H., Reinhart, K. O., Lekburg, Y., Hart, M. M., and Klironomos, J. (2017). Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184. doi: 10.1126/science.aai8212
Bever, J. D., Platt, T. G., and Morton, E. R. (2012). Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 66, 265–283. doi: 10.1146/annurev-micro-092611-150107
Bever, J. D., Westover, K. M., and Antonovics, J. (1997). Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85, 561–573. doi: 10.2307/2960528
Blaxter, M. (2016). blaxterlab/metabarcoding_pipeline: v1.0.1 (Version v1.0.1). Zenodo. doi: 10.5281/zenodo.154423
Brinkman, E. P., Van der Putten, W. H., Bakker, E. J., and Verhoeven, K. J. F. (2010). Plant-soil feedback: experimental approaches, statistical analyses and ecological interpretations. J. Ecol. 98:10631073. doi: 10.1111/j.1365-2745.2010.01695.x
Broeckling, C. D., Broz, A. K., Bergelson, J., Manter, D. K., and Vivanco, J. M. (2008). Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 74, 738–744. doi: 10.1128/AEM.02188-07
Bruns, T. D. (1995). Thoughts on the processes that maintain local species diversity of ectomycorrhizal fungi. Plant Soil 170, 66–73. doi: 10.1007/BF02183055
Bruns, T. D., Bidortondo, M. I., and Taylor, D. L. (2002). Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integ. Comp. Biol. 42, 352–359. doi: 10.1093/icb/42.2.352
Burgess, T. I., Scott, J. K., McDougall, K. L., Stukely, M. J., Crane, C., Dunstan, W. A., et al. (2017). Current and projected global distribution of Phytophthora cinnamomi, one of the world's worst plant pathogens. Glob. Chang. Biol. 23, 1661–1674. doi: 10.1111/gcb.13492
Burke, D. J. (2008). Effects of Alliaria petiolata (garlic mustard: Brassicaceae) on mycorrhizal colonization and community structure in three herbaceous plants in a mixed deciduous forest. Am. J. Bot. 95, 1416–1425. doi: 10.3732/ajb.0800184
Burns, J. H., Anacker, B. L., Strauss, S. Y., and Burke, D. J. (2015). Soil microbial community variation correlates most strongly with plant species identity, followed by soil chemistry, spatial location and plant genus. AoB Plants 7:plv030. doi: 10.1093/aobpla/plv030
Burns, J. H., Brandt, A. J., Murphy, J. E., Kaczowka, A. M., and Burke, D. J. (2017). Spatial heterogeneity of plant-soil feedbacks increases per capita reproductive biomass of species at an establishment disadvantage. Oecologia 183, 1077–1086. doi: 10.1007/s00442-017-3828-1
Burns, J. H., Murphy, J. E., and Zheng, Y. L. (2019). Tests of alternative evolutionary models are needed to enhance our understanding of biological invasions. New Phytol. 222, 701–707. doi: 10.1111/nph.15584
Cadotte, M. W., Davies, J., and Peres-Neto, P. R. (2017). Why phylogenies do not always predict ecological differences. Ecol. Monogr. 87, 535–551. doi: 10.1002/ecm.1267
Carnovale, D., Bissett, A., Thrall, P. H., and Baker, G. (2019). Plant genus (Acacia and Eucalyptus) alters soil microbial community structure and relative abundance within revegetated shelterbelts. Appl. Soil Ecol. 133, 1–11. doi: 10.1016/j.apsoil.2018.09.001
Connell, J. H. (1971). “On the role of natural enemies in preventing competitive exclusion in some marine animals and in rainforest trees,” in Dynamics of Populations, eds P. J. den Boer and G. R. Gradwell (Wageningen: Center for Agricultural Publishing and Documentation), 298–312.
Crawford, K. M., Bauer, J. T., Comita, L. S., Eppinga, M. B., Johnson, D. J., Mangan, S. A., et al. (2019). When and where plant-soil feedback may promote plant coexistence: a meta-analysis. Ecol. Lett. 22, 1274–1284. doi: 10.1111/ele.13278
den Bakker, H. C., Zuccarello, G. C., Kuyper, T. W., and Noordeloos, M. E. (2004). Evolution and host specificity in the ectomycorrhizal genus Leccinum. New Phytol. 163, 201–215. doi: 10.1111/j.1469-8137.2004.01090.x
Edgar, R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences form microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C. (2016). SINTAX: a simple non-Bayesian taxonomy classifier for 16s and ITS sequences. bioRxiv. doi: 10.1101/074161
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Ehrenfeld, J. G., Ravit, B., and Elgersma, K. (2005). Feedback in the plant-soil system. Annu. Rev. Environ. Resour. 30, 75–115. doi: 10.1146/annurev.energy.30.050504.144212
Fitzpatrick, C. R., Gehant, L., Kotanen, P. M., and Johnson, M. T. (2017). Phylogenetic relatedness, phenotypic similarity and plant-soil feedbacks. J. Ecol. 105, 786–800. doi: 10.1111/1365-2745.12709
Harris, J. (2009). Soil microbial communities and restoration ecology: facilitators or followers? Science 325, 573–574. doi: 10.1126/science.1172975
Heinze, J., and Joshi, J. (2018). Plant–soil feedback effects can be masked by aboveground herbivory under natural field conditions. Oecologia 186, 235–246. doi: 10.1007/s00442-017-3997-y
Hinchliff, C. E., Smith, S. A., Allman, J. F., Burleigh, J. G., Chaudhary, R., Coghill, L. M., et al. (2015). Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. U.S.A. 112, 12764–12769. doi: 10.1073/pnas.1423041112
Ishida, T. A., Nara, K., and Hogetsu, T. (2007). Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer–broadleaf forests. New Phytol. 174, 430–440. doi: 10.1111/j.1469-8137.2007.02016.x
Janzen, D. H. (1970). Herbivores and the number of tree species in tropical forests. Am. Nat. 104, 501–508. doi: 10.1086/282687
Kardol, P., and Wardle, D. A. (2010). How understanding aboveground-belowground linkages can assist restoration ecology. Trends Ecol. Evol. 25, 670–679. doi: 10.1016/j.tree.2010.09.001
Klironomos, J. N. (2002). Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70. doi: 10.1038/417067a
Koljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Koricheva, J., and Gurevitch, J. (2013). “Place of meta-analysis among other methods of research synthesis,” in Handbook of Meta-analysis in Ecology and Evolution, eds J. Koricheva, J. Gurevitch, and K. Mengersen (Princeton, NJ: Princeton University Press), 3–13.
Kulmatiski, A., Beard, K. H., Stevens, J. R., and Cobbold, S. M. (2008). Plant-soil feedbacks: a meta analytical review. Ecol. Lett. 1, 980–992. doi: 10.1111/j.1461-0248.2008.01209.x
Lance, A. C., Burke, D. J., Hausman, C. E., and Burns, J. H. (2019). Microbial inoculation influences arbuscular mycorrhizal fungi community structure and nutrient dynamics in temperate tree restoration. Restor. Ecol. 27, 1084–1093. doi: 10.1111/rec.12962
Larios, L., and Suding, K. M. (2015). Competition and soil resource environment alter plant–soil feedbacks for native and exotic grasses. AoB Plants 7:plu077. doi: 10.1093/aobpla/plu077
Liu, X., Liang, M., Etienne, R. S., Wang, Y., Staehelin, C., and Yu, S. (2012). Experimental evidence for a phylogenetic Janzen-Connell effect in a subtropical forest. Ecol. Lett. 15, 111–118. doi: 10.1111/j.1461-0248.2011.01715.x
Maltz, M. R., and Tresder, K. K. (2015). Sources of inocula influence mycorrhizal colonization of plants in restoration projects: a meta-analysis. Restor. Ecol. 23, 625–634. doi: 10.1111/rec.12231
Mangan, S. A., Schnitzer, S. A., Herre, E. A., Mack, K. M., Valencia, M. C., Sanchez, E. I., et al. (2010). Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–756. doi: 10.1038/nature09273
Martin, M. (2011). Cutadapt removes adapter dequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McCarthy-Neumann, S., and Kobe, R. K. (2010). Conspecific and heterospecific plant-soil feedbacks influence survivorship and growth of temperate tree seedlings. J. Ecol. 98, 408–418. doi: 10.1111/j.1365-2745.2009.01620.x
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: why rareifying microbiome data is inadmissible. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Mehrabi, Z., and Tuck, S. L. (2015). Relatedness is a poor predictor of negative plant–soil feedbacks. New Phytol. 205, 1071–1075. doi: 10.1111/nph.13238
O'Brien, M. J., Gomala, C. E., and Horton, T. R. (2011). The effect of forest soil and community composition on ectomycorrhizal colonization and seedling growth. Plant Soil 341, 321–331. doi: 10.1007/s11104-010-0646-1
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). Vegan: Community Ecology Package. R package version 2.4-3. Available online at: http://CRAN.R-project.org/package=vegan
Packer, A., and Clay, C. (2000). Soil pathogens and spatial patterns of seedling mortality in a temperate tree. Nature 404, 278–281. doi: 10.1038/35005072
Palmer, J. M., Lindner, D. L., and Volk, T. J. (2008). Ectomycorrhizal characterization of an American chestnut (Castanea dentata) -dominated community in western Wisconsin. Mycorrhiza 19, 27–36. doi: 10.1007/s00572-008-0200-7
Pei, Z., Eichenberg, D., Bruelheide, H., Krober, W., Kuhn, P., Li, Y., et al. (2016). Soil and tree species traits both shape soil microbial communities during early growth of Chinese subtropical forests. Soil Biol. Biochem. 96, 180–190. doi: 10.1016/j.soilbio.2016.02.004
Rees, J., and Cranston, K. (2017). Automated assembly of a reference taxonomy for phylogenetic data synthesis. Biodivers. Data J. 5:e12581. doi: 10.3897/BDJ.5.e12581
Reinhart, K. O., Johnson, D., and Clay, K. (2012b). Effects of trees on their recruits in the southern Appalachians, USA. For. Ecol. Manage. 263, 268–274. doi: 10.1016/j.foreco.2011.09.038
Reinhart, K. O., and Rinella, M. J. (2016). A common soil handling technique can generate incorrect estimates of soil biota on plants. New Phytol. 210, 786–789. doi: 10.1111/nph.13822
Reinhart, K. O., Wilson, G. W. T., and Rinella, M. J. (2012a). Predicting plant responses to mycorrhizae: integrating evolutionary history and plant traits. Ecol. Lett. 15, 689–695. doi: 10.1111/j.1461-0248.2012.01786.x
Rinella, M. J., and Reinhart, K. O. (2018). Toward more robust plant-soil feedback research. Ecology 99, 550–556. doi: 10.1002/ecy.2146
Schittko, C., Runge, C., Strupp, M., Wolff, S., and Wurst, S. (2016). No evidence that plant–soil feedback effects of native and invasive plant species under glasshouse conditions are reflected in the field. J. Ecol. 104, 1243–1249. doi: 10.1111/1365-2745.12603
Smith-Ramesh, L. M., and Reynolds, H. L. (2017). The next frontier of plant–soil feedback research: unraveling context dependence across biotic and abiotic gradients. J. Veg. Sci. 28, 484–494. doi: 10.1111/jvs.12519
St-Denis, A., Kneeshaw, D., Belanger, N., Simard, S., Laforest-Lapointe, I., and Messier, C. (2017). Species-specific responses to forest soil inoculum in planted saplings in an abandoned agricultural field. Appl. Soil Ecol. 112, 1–10. doi: 10.1016/j.apsoil.2016.12.008
Sweet, D. D., and Burns, J. H. (2017). Plant performance was greater in the soils of more distantly related plants for an herbaceous understory species. AoB Plants 9:plx005. doi: 10.1093/aobpla/plx005
Taylor, D. L., Hollingsworth, T. N., McFarland, J. W., Lennon, N. J., Nusbaum, C., and Ruess, R. W. (2014). A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecol. Monogr. 84, 3–20. doi: 10.1890/12-1693.1
Tedersoo, L., Suvi, T., Jairus, T., Ostonen, I., and Põlme, S. (2009). Revisiting ectomycorrhizal fungi of the genus Alnus: differential host specificity, diversity and determinants of the fungal community. New Phytol. 182, 727–735. doi: 10.1111/j.1469-8137.2009.02792.x
Thoms, C., and Gleixner, G. (2013). Seasonal differences in tree species' influence on soil microbial communities. Soil Biol. Biochem. 66, 239–248. doi: 10.1016/j.soilbio.2013.05.018
USDA (2019). Web Soil Survey. Natural Resources Conservation Service, United States Department of Agriculture, Washington, DC. Available online at: https://websoilsurvey.sc.egov.usda.gov/ (accessed February 07, 2019).
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48. doi: 10.18637/jss.v036.i03
Keywords: plant-soil feedbacks, ectomycorrhizae, temperate trees, restoration, Illumina Mi-Seq
Citation: Lance AC, Carrino-Kyker SR, Burke DJ and Burns JH (2020) Individual Plant-Soil Feedback Effects Influence Tree Growth and Rhizosphere Fungal Communities in a Temperate Forest Restoration Experiment. Front. Ecol. Evol. 7:500. doi: 10.3389/fevo.2019.00500
Received: 29 May 2019; Accepted: 05 December 2019;
Published: 09 January 2020.
Edited by:
Martijn Bezemer, Netherlands Institute of Ecology (NIOO-KNAW), NetherlandsReviewed by:
Scott Mangan, Washington University in St. Louis, United StatesKurt Reinhart, Livestock and Range Research Laboratory, Agricultural Research Service, USDA, United States
Copyright © 2020 Lance, Carrino-Kyker, Burke and Burns. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew C. Lance, YWNsOTFAY2FzZS5lZHU=
 Andrew C. Lance
Andrew C. Lance Sarah R. Carrino-Kyker
Sarah R. Carrino-Kyker David J. Burke1,2
David J. Burke1,2 Jean H. Burns
Jean H. Burns