- Department of Entomology and Plant Pathology, NC State University, Raleigh, NC, United States
Mycological herbaria contain important records of plant biodiversity and past outbreaks of plant disease. Mycological herbaria have been used to (1) understand the life history of plant pathogens; (2) identify outbreak strains and track their source; (3) understand the landscape ecology, biodiversity, and distribution of native plants and their diseases; and (4) examine the impact of climate change on pests and plant diseases. In this review, recent ecological and evolutionary questions being addressed using mycological herbarium specimens will be discussed followed by a case study on the evolution and migration of the historic outbreak strain of the Irish famine pathogen, Phytophthora infestans. Herbarium specimens are providing new information on the population biology and source of one of oldest plant diseases.
Introduction
Some of the biodiversity on the earth including plant diversity is maintained in dried reference specimens in the world’s herbarium collections (Thiers, 2018). Originally preserved by European physicians for medicinal purposes, specimens bound into books were used as sources for herbal medicine (Fertig, 2016). Over time, their uses became more diverse. There are more than 300 active herbaria in the world today that contain an estimated 387 million specimens documented from earth’s vegetation for the past 400 years (Thiers, 2018). These collections can be used to study the ecology, evolution, and biodiversity of not only the earth’s plant species but also their associated microbes, including plant pathogens that cause disease (Ristaino et al., 2001; Soltis, 2017).
Mycological herbaria provide a glimpse into past genetic information of extinct and extant fungal and oomycete plant pathogens and have been used to address unsolved and intriguing questions in plant biology (Ristaino et al., 2001; Martin et al., 2013). Traditionally, specimens in mycological herbaria were used to understand the life history of plant pathogens. Disputes about taxonomy or nomenclature of organisms, and studies of systematics, phylogenies, function, and evolution of genes can be resolved. Herbarium specimens have been used to identify outbreak strains of plant pathogens and track their source. Examples of these studies will follow. Origins of pathogen populations and causal agents of historical epidemics have been discerned. Changes in genetic structure of “old” versus “new” populations have also been compared. Specimens can be used to understand the landscape ecology and distribution of native plant and their plant diseases. Increasingly, herbarium records are being used to provide baseline data on the impact of climate change on pest and plant disease distribution.
In this review, examples of the use of mycological herbarium specimens to address these questions will be given. Today, questions on the ecology and evolution of pathogens are being addressed using herbarium specimens containing a range of plant pathogens. We make the case for the value and necessity of these collections, which need continued curation. This is followed by a detailed case study of the use of mycological herbarium specimens to address several important research questions related to the evolution, source, and migration of Phytophthora infestans, the famed Irish famine pathogen.
Genetic Structure of “Old” Versus “New” Populations
Citrus bacterial canker caused by Xanthomonas axonopodis pv. citri (Xac) was first documented in India and Java in the mid-19th century and causes severe disease on leaves, stems, and fruit of citrus that affect marketability of fruit. DNA sequencing was used to sequence insertion sequences from 90 herbarium samples from Japan and Florida and study DNA diversity present within the pathogen populations in herbarium specimens over the past century (Li et al., 2007). Four different genotypes of the pathogen were identified in early citrus samples from Florida. All of these genotypes were preserved in herbarium specimens from Japan, thus suggesting that Japan was the source of the original outbreak of citrus canker in Florida in 1911 according to Li et al. (2007). Multiple introductions of the pathogen into citrus in Florida were inferred based on Florida populations that contained all four global genotypes and had greater genetic diversity than genotypes from other regions of the world.
A larger-scale global study of citrus bacterial canker lineages using multilocus minisatellites revealed four clusters of genotypes of the citrus canker pathogen and that a single lineage caused the original outbreaks in Asia (Pruvost et al., 2014). Work is underway in France to sequence the whole genomes from early outbreak strains of the bacterial canker pathogen found in citrus specimens in plant herbaria that were not originally collected for that purpose (Gagnevin, 2017). Unidentified leaf spots are common in plant herbaria and provide a time stamp of disease outbreaks from the past. The study documented the usefulness of searching outside traditional mycological herbaria for plant specimens not labeled as diseased for plant disease records. Interestingly, positive samples of the citrus canker pathogen were found in citrus samples from the 1820s in Japan, many years before it was even documented that bacteria could cause disease and the citrus canker bacterium was identified.
Taxonomic Disputes
Disputes about the taxonomy and classification of plant pathogens can be resolved with herbarium samples (Table 1). For example, the taxonomic status of the spinach downy mildew pathogen was resolved using herbarium specimens from four continents (Choi et al., 2007). These workers constructed phylogenetic trees using the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) within Peronospora accessions (Choi et al., 2007). The work led to reinstatement of Peronospora effusa as the name of the pathogen on spinach (Spinacea oleracea) (Choi et al., 2007).
Similarly, authenticated herbarium specimens were used to change the nomenclature of an anthracnose pathogen of safflower, garland chrysanthemum, and pot marigold in the Asteraceae from Gloeosporium carthami to Colletotrichum carthami comb. nov. (Uematsu et al., 2012). The cause of Peronospora blight on opium poppy was also determined to be Peronospora arborescens and not Paraschistura cristata using type specimens from the early 19th century (Montes-Borrego et al., 2008). Support for the name of the postharvest rot of apple as Botrytis mali was shown after conducting molecular phylogenies from DNA extracted from the earliest described diseased apples in herbarium samples (O’Gorman et al., 2008). A real-time PCR diagnostic was developed to distinguish common corn rust, Puccinia sorghi, from southern corn rust, Puccinia polysora. Rust spores from samples from herbaria were used to develop the primers and validate the specimen labels (Crouch and Szabo, 2011). These examples document the usefulness of mycological herbaria in designating fungal species names, revising names, giving naming priority, and developing pathogen diagnostic tools.
Landscape Ecology and Distribution of Pathogens of Native Plants
The spatial distribution of anther smut fungi in the eastern United States was studied using plant herbarium specimens. Herbaria containing Silene virginica, Silene caroliniana, Smilax rotundifolia, and Silene latifolia were used to survey the incidence of anther-smut disease caused by Microbotryum violaceum sensu lato, in the eastern United States (Antonovics et al., 2003). Landscape level maps of plant species occurrence and the occurrence of the anther-smut pathogen were made by visual confirmation of symptoms on anthers stored in plant herbaria specimens. Disease was found in two of the four species, S. virginica and S. caroliniana but not S. rotundifolia, and S. latifolia, in the eastern United States. Relative abundance of disease was related to species and latitude with more disease occurring further north in S. virginica than S. caroliniana. Comparisons with modern native populations and disease outbreaks on these Silene species revealed different results and indicated a recent introduction of the disease on S. latifolia that was not identified previously in plant herbaria records.
Herbarium samples of the forest sedge Carex blanda were examined for the presence of Anthracoidea blanda (smut), Puccinia caricina (rust), and a chalcid insect on forest sedge in Kansas (Alexander et al., 2007). The latitude and longitude of the plant host were not related to the presence of the insect seed predator or the two pathogens. However, the smut fungus was dispersed more locally than the rust and both pathogens were found less frequently at the western borders of the plant host range. There was also a significant increase in occurrence of both plant pathogens over time revealed in the herbarium sample records.
Both of these examples demonstrated the spatial dynamics of plant diseases in native plant populations. Understanding disease spread in native plant populations is important as native plants may provide reservoirs for pathogens to migrate into agricultural crops. In addition, the work on anther smut has led to elegant models on the transmission dynamics of plant pathogens in native plant populations that are relevant for understanding disease management in agroecosystems (Antonovics, 2017).
The associated metadata from specimen labels can be used to study and map pathogen and host species distributions over time and could provide important baseline data for regulatory agencies interested in new pathogen introductions. The DNA sequence data generated from specimens can be linked to reference specimens in herbaria using data aggregating portals such as the Global Biodiversity Information Facility (GBIF), the United States Geological Survey’s portal (Biodiversity Information Serving Our Nation; BISON), iDigBio (Integrated Digitized Biocollections), and the MyCoPortal1. The data can then be archived and used by groups of scientists on biodiversity projects such as the microbial tree of life to provide baseline data for pathogen phylogenetic trees. The Atlas of Living Australian Life is an example of a project that uses mycological herbarium data to create spatial maps of disease outbreaks. These data are useful to provide early records of plant disease or new reports of diseases and can be very useful to regulatory and plant quarantine personnel trying to control an outbreak.
Impacts of Climate Change on the Distribution of Pests or Pathogen Outbreaks
Historical plant collections have been used in global change research to investigate how plant species interactions shift with time (Meineke et al., 2018). The impact of climate on herbivory caused by plant feeding insects on hickory, oak, trefoil, and blueberry in the northeastern United States was examined using a large number of plant herbarium specimens that were not originally collected for this purpose (Meineke et al., 2018). There was increased herbivory in more recent than older specimens of the plant species investigated. In addition, higher winter temperatures were associated with increased herbivory, and there was less herbivory in samples from urban than rural areas (Meineke et al., 2019).
A bias in the collection of historical plant collections can impact interpretation of results, as herbarium samples are not typically collected randomly. In addition, there tends to be greater numbers of collections made in the developed than developing world so data gaps can be evident. However, there is the advantage that many plant specimens present in herbaria were collected before wide-scale human changes to climate occurred, so the specimens provide useful baseline data for plant, pest, or pathogen distribution in climate change studies.
An example of the impact of air pollutants on historic pathogen occurrence was done using the Rothamsted long-term wheat trials. The temporal occurrence of two major wheat pathogens was investigated from specimens collected in the long-term trials on wheat at Rothamsted Research in the United Kingdom over a 160-year period (Bearchell et al., 2005). The abundance of Phaeosphaeria nodorum and Mycosphaerella graminicola were monitored in wheat from samples from the Broadbalk experiments where systematic sampling from the same sentinel plots at Rothamsted has occurred since 1844 (Table 1). DNA was extracted from samples and a beta tubulin gene was amplified using real-time PCR to identify each species. Strong correlations between changes in the ratio of the two pathogens over the 160-year period with changes in atmospheric SO2 emissions were observed. The abundance of P. nodorum was positively correlated with SO2 emissions, while the abundance of Mycosphaerella graminícola was negatively correlated with SO2. The study was one of the first to use long-term data from archival collections for understanding and investigating anthropogenic factors such as air pollutants on the dynamics of pathogens on wheat.
Climate change is expected to affect not only the abundance but also the distribution of plant species. The distribution of plant diseases is also likely to change as climates warm with some plant diseases expanding into new ranges and at higher latitudes that were previously inhospitable for disease (Bebber, 2015). Knowledge of baseline pathogen distributions can be discerned from data-mining and mapping plant pathogen occurrence records of mycological herbaria. Much of the data records in the world’s mycological herbaria are now being digitized and uses of specimens have changed from traditional taxonomy and systematics to DNA analyses, global change biology, and biodiversity informatics (Heberling et al., 2019). These examples suggest the importance of historic plant and mycological herbarium collections and the need for not only curation but deposits of both diseased and healthy plant specimens into collections.
A Case Study Using Mycological Herbarium Specimens: the Irish Famine Pathogen P. infestans
Phytophthora infestans, an oomycete plant pathogen, caused the Irish potato famine. The disease first occurred in the United States in 1843 and subsequently spread into Europe and Ireland in 1845 (Bourke, 1964). My laboratory has studied both modern and historic outbreaks of the disease for many years. Mycological herbaria have been used to address several important research questions including (1) understanding the life history of the pathogen, (2) identification of the strain that caused famine-era outbreaks and its source, (3) the spatial scale of the outbreak strain, (4) the center of origin of the disease, (5) the genetic structure and evolutionary history of P. infestans and sister lineages, and (6) changes in pathogen virulence through time.
Life History of the Pathogen and Mycological Collections
Herbarium specimens played an early role in identifying and understanding the pathogen that caused the Irish potato famine (Figure 1). There was no consensus on the cause of the potato disease and weather, insects, “the wrath of God,” the “temperament of the Irish,” and a minute “fungus” were suggested (Berkeley, 1846; Bourke, 1993). Teschemacher, a member of Boston’s Natural History Society, associated a fungus with the lesions on the potato plant (Teschemacher, 1845). In 1846, Montagne in France and Miles J. Berkeley in the United Kingdom (Berkeley, 1846; Montagne, 1846) gave the name Botrytis infestans for the pathogen. These scientists collected and exchanged specimens of infected potato leaves and wrote letters to colleagues. M. J. Berkeley’s letter to W. G. Farlow of Harvard University is preserved in the Farlow archives (Figure 2) and includes early descriptions of the pathogen including the asexual fruiting bodies called sporangia and discussion of putative sexual spores or oospores (Ristaino, 1998).

Figure 1. Phytophthora infestans-infected potato collected in (A) Sweden in 1882 by Jacob Eriksson from the herbarium in Uppsala, Sweden. (B) Sample collected in Bogata, Colombia by J. M. Vargas Vergana in 1913 (BPI186968). Disease in both samples was caused by the FAM-1 lineage of P. infestans.

Figure 2. Letter and drawing by Berkeley sent to W. G. Farlow at Harvard University on July 10, 1875 showing the sporangiophore, sporangia, zoospores and putative oogonia (female), and antheridia (male) of P. infestans. Letters deposited in the Farlow Library Archives, Harvard University.
The work on late blight by M. J. Berkeley and others predated work by Louis Pasteur on the germ theory and clearly documented that a microbe could cause disease (Berkeley, 1846). The work was a major contribution to both the development of the science of plant pathology and the germ theory of disease and is often unrecognized by microbiologists. In 1876, Anton de Bary elucidated the full life cycle of the pathogen and based on morphology of sporangia and sporangiophore characteristics changed the pathogen name from Peronospora to P. infestans (De Bary, 1876). The word Phytophthora means “plant destroyer.”
Mycological specimens linked with archival documents have been used to understand the life history of P. infestans (Ristaino, 1998). The occurrence of the sexual oospores of the pathogen in field samples was investigated and documented in herbarium sample records. Herbarium specimen labels from the early 20th century collected by G.P. Clinton in Connecticut indicated that oospores (survival spores) of P. infestans were observed in infected potatoes and PCR and microscopic evidence was published that corroborate his work and that of others who also observed oospores of the pathogen in potato in the early 20th century (Ristaino, 1998).
We searched the Index Herbariorum and the MyCoPortal for plant herbarium specimens infected with P. infestans. Over 1280 specimens of P. infestans-infected plant leaves are stored in global mycological herbaria (Figure 3). Most samples were collected between 1843 and 1950, while far fewer samples were collected more recently (Figure 3A). The distribution of historic samples is biased toward the developed world and temperate climates (Figure 3). The greatest number of samples were from potato (n = 771) followed by tomato (n = 134), other wild Solanum species (n = 48), and petunia (n = 6) (Figure 3B). The largest numbers of specimens were collected from the United States, followed by Germany, the United Kingdom, Sweden, Canada, France, New Zealand, India, and Italy (Supplementary Figure 1). Samples from many other countries were also found in the global database and span most continents where potatoes are currently grown (Figure 3A). The greatest numbers of P. infestans-infected plant specimens are housed at the United States National Fungus Collections (BPI) in Maryland followed by the Kew Mycological Collections (Kew) in the United Kingdom and Harvard’s Farlow Herbarium (FH) in Cambridge, MA, United States (Supplementary Figure 1B).
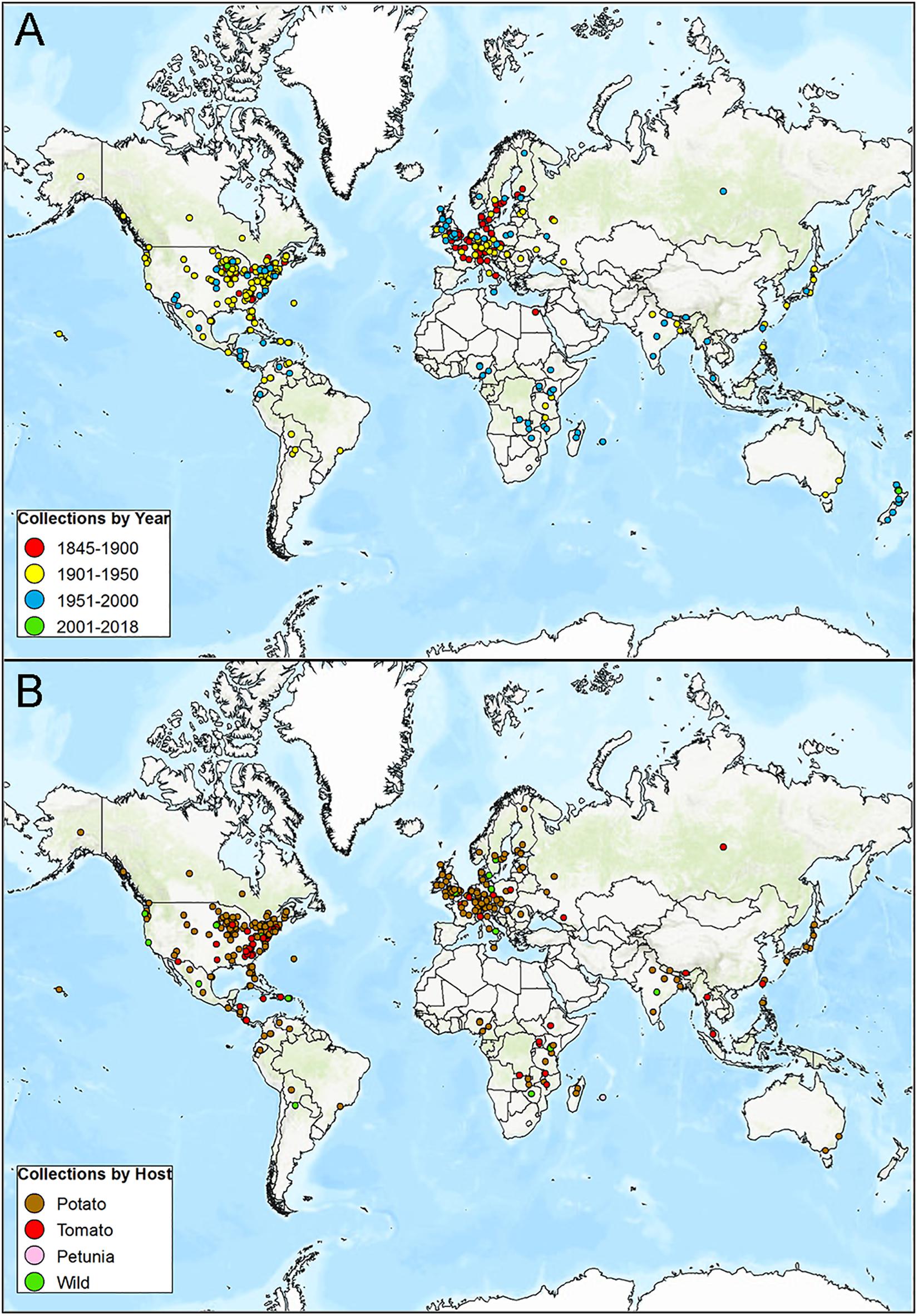
Figure 3. Distribution of late blight caused by P. infestans herbarium records. (A) Sorted by year from 1845 to 2018. (B) Collections sorted by host including potato, tomato, wild Solanum species, or petunia. Herbarium records mined from a search of MyCoPortal.
The Identification of the Strain That Caused Famine-Era Outbreaks and Its Source
A single clonal lineage named US-1 [Ib mitochondrial DNA (mtDNA) haplotype] was globally distributed in the late 20th century and was proposed as the “famine-era” lineage of P. infestans (Goodwin et al., 1994). The lineage was identified at that time by RFLP fingerprinting and mitochondrial haplotying. However, my laboratory conducted the first direct PCR amplification of mitochondrial genes from actual 19th-century herbarium samples of blight-afflicted potato leaves, and our work led to a different conclusion (Ristaino et al., 2001). Historical P. infestans from famine-era outbreaks were the Ia and not the Ib mtDNA haplotype and thus US-1 was not the culprit (May and Ristaino, 2004). In addition, evidence of other mtDNA haplotypes in Nicaragua (type IIb) in the mid-20th century were found, suggesting that there was more than one clonal lineage circulating by the 1950s in the Americas (May and Ristaino, 2004).
My laboratory in collaboration with Tom Gilbert and Mike Martin (Martin et al., 2013) and another group simultaneously (Yoshida et al., 2013) conducted Illumina sequencing of whole genomes from historical P. infestans derived from infected potato samples in Europe. Both studies corroborated our previous work (Ristaino et al., 2001), concluding that the historical lineage was not the US-1. Yoshida called the mitochondrial lineage “HERB-1” and suggested that it was more closely related to the US-1 lineage (Ib mtDNA haplotype) than the Ia mtDNA haplotype and that the lineage was rare or possibly extinct (Birch and Cooke, 2013; Yoshida et al., 2013). Subsequently, we sequenced 45 additional mitochondrial genomes from historic and modern lineages (Martin et al., 2014) and results indicated that the famine lineage was not extinct but present in both Mexico and Ecuador, thus refuting the Yoshida claim (Yoshida et al., 2013). The divergence time of the HERB-1 and other mitochondrial lineages was also dated to 75 years prior to the Irish famine outbreaks (Martin et al., 2014). Highly supported structure within the HERB-1 lineage indicated that multiple distinct mitogenome haplotypes were present in 19th-century Europe, a situation compatible with either multiple introductions of different haplotypes of the pathogen or a single introduction containing multiple haplotypes into Europe. The HERB-1 mitochondrial lineages radiated within Europe over time and formed a sister lineage that was most closely related to the type Ia mitochondrial lineage.
The Spatial Scale of the Outbreak Strain
PCR primers were developed based on full mitogenome sequences to differentiate HERB-1/Ia mitogenomes from other reported modern mtDNA lineages (Ib, IIa, and IIb) (Avila-Adame et al., 2006; Gomez-Alpizar et al., 2007; Martin et al., 2014; Lassiter et al., 2015). The HERB-1 mitogenome shared all the SNPs in the P2, P3, and P4 regions of the mitochondrial genome that were reported in previous studies of historic mtDNA lineages and was thus a type Ia mitochondrial DNA haplotype (Ristaino et al., 2001; Martin et al., 2014; Saville et al., 2016). A survey of an even larger set of global herbarium specimens documented the frequent occurrence of the HERB-1 mitochondrial haplotype in archival collections from 1866 to 1875 and 1906 to 1915 and the later rise of the Ib mitochondrial lineage (US-1) between 1936 and 1955 (Saville et al., 2016). The HERB-1 mtDNA lineage was identified in the Andean hybrid species Phytophthora andina, suggesting that this mtDNA lineage was not exclusive to P. infestans and that it still exists in South America (Martin et al., 2014, 2016; Saville et al., 2016). For that reason, we renamed the actual famine lineage found in P. infestans as FAM-1 (Saville et al., 2016).
A large set of historic United States and European P. infestans from specimens collected from 1846 to 1970 and more recent isolates from 1992 to 2014 were genotyped with 12 plex SSR multilocus genotyping (Saville et al., 2016; Ristaino et al., 2020). The same, unique SSR multilocus genotype, FAM-1 was found. This lineage caused outbreaks in both the United States and Europe and was widely distributed. Interestingly, the FAM-1 lineage shared allelic diversity and grouped with the oldest known historic specimens infected with P. infestans collected in Colombia (Figure 1B) and FAM-1 was also found in Costa Rica, thus confirming the presence of the famine lineage in Central and South America in the early 20th century. The FAM-1 lineage of P. infestans formed a genetic group that was distinct from more recent aggressive lineages of the pathogen (US-8, US-22, US-23) found in the United States. The US-1 lineage formed a second, mid-20th-century group and shared allelic diversity with lineages from Peru suggesting a South American source of US-1. The US-1 lineage was first found in the United States in herbarium records in samples from Texas in 1931. In contrast, many recent modern United States lineages with the exception of the US-23 lineage, grouped with Mexican lineages, suggesting a Mexican source of recent aggressive United States lineages of P. infestans (Saville and Ristaino, 2019). Tomatoes and potatoes grown in Mexico are imported into the United States frequently, and are likely sources of introductions of recent clonal lineages of the pathogen.
Evidence from historic herbarium collections from the United States show that the FAM-1 lineage first emerged on potato as all the 19th-century samples genotyped on potato were FAM-1 (Figure 4A). In contrast, FAM-1 was not confirmed on tomato until after 1900 (Figure 4B). A host jump to tomato may have occurred later after initial introductions of FAM-1 on potato. We are continuing to sequence more Solanum species from global populations to provide further information on host jumps. It is clear that the FAM-1 lineage survived for almost 100 years in the United States, and was geographically more widespread than originally assumed (Saville et al., 2016). In contrast, the US-1 lineage was not identified on potato between 1845 and 1900 but was more common on potato in later samples from 1901 to 1950 (Figure 4A). In addition, the US-1 lineage was found in a greater percentage of the samples genotyped from tomato than potato from 1901 to 1950 (Figure 4B), suggesting tomato may have been the initial host of introduction of the US-1 lineage.
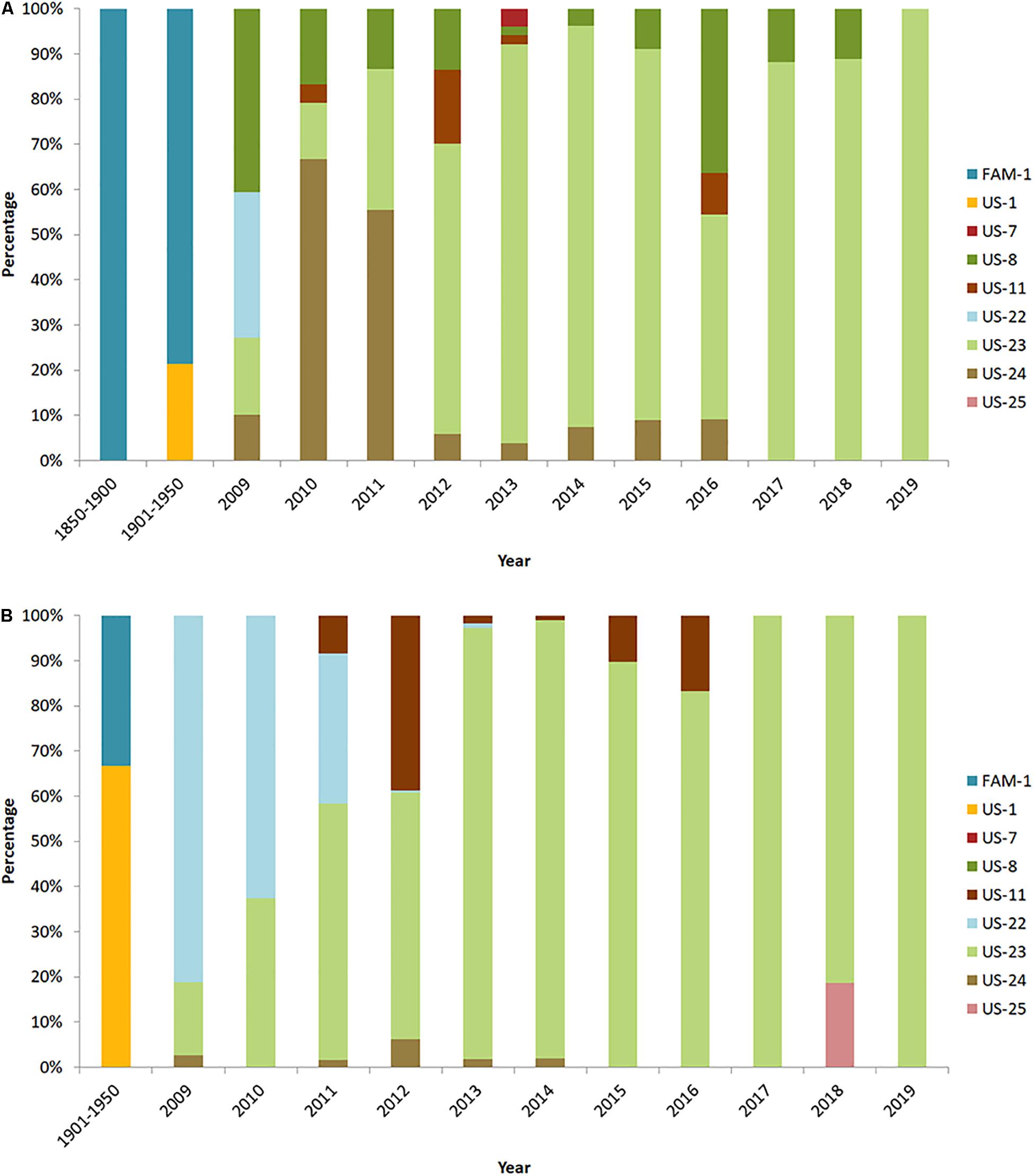
Figure 4. Distribution of United States lineages of P. infestans from herbaria (1850–1950) and modern United States collections (2009–2019) on (A) potato and (B) tomato. Data from 2011 to 2019 are from USABlight.org.
Center of Origin of Disease
The first researchers who studied late blight said the pathogen originated in the Andes of South America (Berkeley, 1846; De Bary, 1863). These workers cited reports by Spanish explorers and Jesuit priests that stated the pathogen was widely known in the Andean region of Colombia (Abad and Abad, 1997). In addition, in Europe, potato seed recently imported from Andean sources on fast-moving steam ships was reported to harbor the pathogen. The higher temperatures in the ship holds of earlier vessels would have likely killed P. infestans (Jones et al., 1912).
In the 20th century, Reddick and others suggest that Mexico was the center of origin of the pathogen and the source of 19th-century outbreaks (Reddick, 1939; Fry et al., 2015). Evidence includes the presence of sexual oospores that were described in field populations in Mexico (Gallegly and Galindo, 1958) and highly diverse sexual lineages in modern populations there (Fry et al., 2015). In addition, resistance to the pathogen in Solanum demissum, which was widely used as a source of R genes, came from Mexico.
Gomez-Alpizar (Gomez-Alpizar et al., 2007), using a global set of isolates and more lineages from South America than previous work, conducted multilocus genotyping of nuclear and mitochondrial genes and presented gene genealogies that provided evidence of a South American origin of the pathogen. Coalescent-based genealogies of all loci were congruent and demonstrate the existence of two lineages leading to present-day haplotypes of P. infestans on potatoes. The oldest lineage associated with isolates from the section Anarrhichomenun including Solanum tetrapetalum from Ecuador (later identified as P. andina) evolved from a common ancestor of P. infestans. Mexican haplotypes were derived from only one of the two lineages.
Gomez-Alpizar’s work stimulated research by others to do further multilocus genotyping with a wider set of Mexican samples, and in that work, a Mexican center of origin was proposed (Goss et al., 2014). However, the P. infestans lineages from South America studied in that work were most likely reintroductions from Europe, and their migration analysis supports this idea. The disparate results stimulated further work by our team using the population genomics approaches described in the next section that have shed more light on the historic controversy “Whence came P. infestans” (Martin et al., 2016).
The Genetic Structure and Evolutionary History of P. infestans and Sister Lineages
Several sister lineages of P. infestans have been described in association with wild Solanaceous species in South America, including Phytophthora betacei (Mideros et al., 2018) and the hybrid species P. andina (Adler et al., 2004; Oliva et al., 2010; Goss et al., 2011). P. andina (Adler et al., 2004; Oliva et al., 2010) is a close relative of P. infestans and hypothesized to have arisen through multiple hybridizations between P. infestans and an as yet undescribed species within Phytophthora clade 1c (Gomez-Alpizar et al., 2008; Goss et al., 2011; Blair et al., 2012; Lassiter et al., 2015). P. andina is a pathogen on various Solanum species in the Andean highlands and, in some South American locales, occurs sympatrically with P. infestans on Solanum betaceum (tree tomato) and Solanum muricatum (pear melón) (Perez et al., 2001; Adler et al., 2004; Oliva et al., 2010; Forbes et al., 2013, 2016).
Historic genomes of P. infestans were sequenced using Illumina sequencing and have been compared with more recent genomes from collections of modern aggressive lineages of P infestans and P. andina, suggesting an Andean origin of the outbreak strain of P. infestans (Martin et al., 2016). The famine lineage from historical Europe is one of the parents of the hybrid P. andina that is endemic to the Andean highlands of South America on wild Solanum species (Gomez-Alpizar et al., 2008; Martin et al., 2016). As P. andina lineages are the most basal in a phylogeny that includes much of the known diversity of P. infestans, this indicates that the sister hybrid species P. andina, although described only recently (Oliva et al., 2010), actually diverged early on in the history of P. infestans in South America. Thus, the closest relative of P. infestans is likely on a wild host and is likely endemic in South America. This is evidence that FAM-1-like genotypes responsible for the Irish famine once existed in Ecuador, Colombia, and/or Peru, the known range of P. andina (Martin et al., 2016). FAM-1 lineages were indeed present in the oldest herbarium samples tested to date from Colombia (Figure 1B) (Saville et al., 2016). The work illustrates the need for further sampling of P. infestans and related sister species on wild Solanum species in South America. Further sequencing of P. infestans from additional wild species present in mycological herbaria (Figure 3) is also needed to further understand the ancestry of the entire 1c clade of Phytophthora.
Changes in Pathogen Virulence Over Time
Next-gen sequencing of genomes from historic P. infestans has also enabled us to ask questions about the virulence of the FAM-1 lineage (Martin et al., 2013; Yoshida et al., 2013). Computational analysis of pathogen genomes from plant herbaria can have many challenges, and these challenges have been reviewed and analyzed elsewhere (Yoshida et al., 2015). P. infestans produces proteins known as effectors present in gene sparse regions of the genome that are recognized by host resistance genes (R) or not to trigger host immunity or susceptibility (Gilroy et al., 2011). The first genome sequence of P. infestans revealed over 500 effectors present in gene sparse regions of the pathogen genome (Haas et al., 2009). Many Avr genes have been cloned and identified (Vleeshouwers et al., 2011). Many of the Avr genes known to be essential for virulence in modern P. infestans were absent in the historic lineages (Vleeshouwers et al., 2011; Martin et al., 2013; Yoshida et al., 2013). Results showing the absence of Avr genes in historic outbreak strains known to be essential for virulence in modern P. infestans suggest the likely role of plant breeding in accelerating development of new lineages with expanded effector repertoires.
Conclusion
Our scientific ancestors could never have imagined the diverse uses and questions now being addressed with plant pathogen herbaria. Mycological herbaria form a window into epidemics of the past. As genome sequencing comes down in price and specimen digitization occurs, further sequencing of historic outbreak samples are expected from diverse plant pathosystems. Unfortunately, funding to maintain, preserve, and study these historic collections has been reduced and some collections are threatened. Linking DNA sequence data back to voucher specimens in herbaria is important, as presently much of the sequence data are in separate databases from the collections. Further work to mine specimen labels and link sequence data with larger data portals and geospatial mapping is needed. In addition, further deposits of modern-day pathogen-infected plants into mycological herbaria are needed to preserve the temporal scale of archived samples. Going back to archival samples to predict future emerging disease outbreaks can yield valuable information on changes in pathogen virulence, abundance, and evolution of important plant diseases.
Author Contributions
JR conceived the ideas, analyzed the data, and wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Funding was provided by the North Carolina Agricultural Research Service and the National Institute of Food and Agriculture, U.S. Department of Agriculture (https://nifa.usda.gov/) under sub award number 2011-68004-30154 to JR.
Acknowledgments
The collaboration of Dr. Michael Martin, Norwegian University of Science and Technology, Trondheim, Norway on organizing this special issue and Mike Martin and Tom Gilbert, University of Copenhagen, Denmark on the great collaborative research published previously are acknowledged. Amanda Saville, research assistant Department of Entomology and Plant Pathology, NC State collected statistics on herbarium specimens and prepared figures.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00521/full#supplementary-material
Footnotes
References
Abad, Z. G., and Abad, J. A. (1997). Another look at the origin of late blight of potatoes, tomatoes, and pear melon in the andes of South America. Plant Dis. 81, 682–688. doi: 10.1094/pdis.1997.81.6.682
Adler, N. E., Erselius, L. J., Chacon, M., Flier, W. G., Ordoñez, M. E., Kroon, L. P., et al. (2004). Genetic diversity of Phytophthora infestans sensu lato in Ecuador provides new insight into the origin of this important plant pathogen. Phytopathology 94:154. doi: 10.1094/PHYTO.2004.94.2.154
Alexander, H. M., Price, S., Houser, R., Finch, D., and Tourtellot, M. (2007). Is there reduction in disease and pre-dispersal seed predation at the border of a host plant’s range? Field and herbarium studies of Carex blanda. J. Ecol. 95, 446–457. doi: 10.1111/j.1365-2745.2007.01228.x
Antonovics, J. (2017). Transmission dynamics: critical questions and challenges. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372:20160087. doi: 10.1098/rstb.2016.0087
Antonovics, J., Hood, M. E., Thrall, P. H., Abrams, J. Y., and Duthie, G. M. (2003). Herbarium studies on the distribution of anther-smut fungus (Microbotryum violaceum) and Silene species (Caryophyllaceae) in the eastern United States America. J. Bot. 90, 1522–1531. doi: 10.3732/ajb.90.10.1522
Avila-Adame, C., Gómez-Alpizar, L., Zismann, V., Jones, K. M., Buell, C. R., and Ristaino, J. B. (2006). Mitochondrial genome sequences and molecular evolution of the Irish potato famine pathogen, Phytophthora infestans. Curr. Genet. 49:39. doi: 10.1007/s00294-005-0016-3
Bearchell, S. J., Fraaije, B. A., Shaw, M. W., and Fitt, B. D. (2005). Wheat archive links long-term fungal pathogen population dynamics to air pollution. Proc. Natl. Acad. Sci. U.S.A. 102, 5438–5442. doi: 10.1073/pnas.0501596102
Bebber, D. P. (2015). Range-expanding pests and pathogens in a warming world. Annu. Rev. Plant Pathol. 53, 335–356. doi: 10.1146/annurev-phyto-080614-120207
Berkeley, M. J. (1846). Observations, botanical and physiological, on the potato murain. J. Hortic. Soc. Lond. 1:9.
Birch, P. R. J., and Cooke, D. E. L. (2013). The early days of late blight. eLife 2:e00954. doi: 10.7554/eLife.00954
Blair, J. E., Coffey, M. D., and Martin, F. N. (2012). Species tree estimation for the late blight pathogen, Phytophthora infestans, and close relatives. PLoS One 7:e37003. doi: 10.1371/journal.pone.0037003
Bourke, P. M. A. (1964). Emergence of Potato blight, 1843-46. Nature 203, 805–808. doi: 10.1038/203805a0
Bourke, P. M. A. (1993). The Visitation of God: The Potato and the Great Irish Famine. Dublin: Lilliut Press.
Choi, Y.-J., Hong, S.-B., and Shin, H.-D. (2007). Re-consideration of Peronospora farinosa infecting Spinacia oleracea as distinct species, Peronospora effusa. Mycol. Res. 111, 381–391. doi: 10.1016/j.mycres.2007.02.003
Crouch, J. A., and Szabo, L. J. (2011). Real-time PCR detection and discrimination of the southern and common corn rust pathogens Puccinia polysora and Puccinia sorghi. Plant Dis. 95, 624–632. doi: 10.1094/PDIS-10-10-0745
De Bary, A. (1876). Researches into the nature of the potato-fungus - Phytophthora infestans. J. R. Agr. Soc. 12, 239–268.
Fertig, W. (2016). Are herbaria still relevant in the 21st century. Biodivers. Knowl. Inform. Center 39, 1–6.
Forbes, G. A., Gamboa, S., Lindqvist-Kreuze, H., Oliva, R. F., and Perez, W. (2016). Identification of an A2 population of Phythophthora andina attacking tree tomato in Peru indicates a risk of sexual reproduction in this pathosystem. Plant Pathol. 65, 1109–1117. doi: 10.1111/ppa.12531
Forbes, G. A., Morales, J. G., Restrepo, S., Pérez, W., Gamboa, S., Ruiz, R., et al. (2013). “Phytophthora infestans and Phytophthora andina on solanaceous hosts in South America,” in Phytophthora: A Global Perspective, ed. K. Lamour, (Oxfordshire England: CABI International), 48–58. doi: 10.1079/9781780640938.0048
Fry, W. E., Birch, P. R. J., Judelson, H. S., Grünwald, N. J., Danies, G., Everts, K. L., et al. (2015). Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105, 966–981. doi: 10.1094/PHYTO-01-15-0005-FI
Gagnevin, L. (2017). Exploring the Naturalis Biodiversity Center Herbarium for Xanthomonas symptoms on Rutaceae and Euphorbiaceae in Order to Reconstruct the Evolution and Emergence of Diseases on Citrus and Cassava. Short Term Scientific Mission Report No. 16107. Brussels: European Cooperation in Science and Technology.
Gallegly, M. E., and Galindo, J. (1958). Mating types and oospores of Phytophthora infestans in nature in Mexico. Phytopathology 48:274.
Gilroy, E. M., Breen, S., Whisson, S., Squire, J., Hein, I., Lokossou, A., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191:763. doi: 10.1111/j.1469-8137.2011.03736.x
Gomez-Alpizar, L., Carbone, I., and Ristaino, J. B. (2007). An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 104, 3306–3311. doi: 10.1073/pnas.0611479104
Gomez-Alpizar, L., Hu, C. H., Olivia, R., Forbes, G., and Ristaino, J. B. (2008). Phylogenetic relationships of a new species, Phytophthora andina, from the highlands of Ecuador that is closely related to the Irish potato famine pathogen Phytophthora infestans. Mycologia 100, 590–602. doi: 10.3852/07-074r1
Goodwin, S. B., Cohen, B. A., and Fry, W. E. (1994). Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc. Natl. Acad. Sci. U.S.A. 91:11591. doi: 10.1073/pnas.91.24.11591
Goss, E. M., Cardenas, M. E., Myers, K., Forbes, G. A., Fry, W. E., Restrepo, S., et al. (2011). The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans. PLoS One 6:e24543. doi: 10.1371/journal.pone.0024543
Goss, E. M., Tabima, J. F., Cooke, D. E., Restrepo, S., Fry, W. E., Forbes, G. A., et al. (2014). The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. Proc. Nat. Acad. Sci. U.S.A. 111:8791. doi: 10.1073/pnas.1401884111
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393. doi: 10.1038/nature08358
Heberling, J. M., Prather, L. A., and Tonsor, S. J. (2019). The changing uses of herbarium data in an era of global change: an overview using automated content analysis. Bioscience 69, 812–822. doi: 10.1093/biosci/biz094
Jones, L. R., Giddings, N. J., and Lutman, B. F. (1912). Investigations of the potato fungus, Phytophthora infestans. Exp. Stn. Bull. 168, 1–100.
Lassiter, E. S., Russ, C., Nusbaum, C., Zeng, Q., Saville, A., Olarte, R., et al. (2015). Mitochondrial genome sequences reveal evolutionary relationships of the Phytophthora Ic clade species. Curr. Gen. 61, 567–577. doi: 10.1007/s00294-015-0480-3
Li, W., Song, Q., Brlansky, R. H., and Hartung, J. S. (2007). Genetic diversity of citrus bacterial canker pathogens preserved in herbarium specimens. Proc. Nat. Acad. Sci. U.S.A. 104, 18427–18432. doi: 10.1073/pnas.0705590104
Martin, M. D., Cappellini, E., Samaniego, J. A., Zepeda, M. L., Campos, P. F., Seguin-Orlando, A., et al. (2013). Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 4:2172. doi: 10.1038/ncomms3172
Martin, M. D., Ho, S. Y. W., Wales, N., Ristaino, J. B., and Gilbert, M. T. P. (2014). Persistence of the mitochondrial lineage responsible for the Irish potato famine in extant New World Phytophthora infestans. Mol. Biol. Evol. 31, 1414–1420. doi: 10.1093/molbev/msu086
Martin, M. D., Vieira, U., Filipe, G., Ho, I., Simon, Y. W., Jean, B., et al. (2016). Genomic characterization of a South American Phytophthora hybrid mandates reassessment of the geographic origins of Phytophthora infestans. Mol. Biol. Evol. 33, 478–491. doi: 10.1093/molbev/msv241
May, K. J., and Ristaino, J. B. (2004). Identity of the mtDNA haplotype(s) of Phytophthora infestans in historical specimens from the Irish potato famine. Myco. Res. 108:471. doi: 10.1017/s0953756204009876
Meineke, E., Davis, C. C., and Davies, T. J. (2018). The unrealized potential of herbaria for global change biology. Ecol. Mon. 88, 1–21. doi: 10.1002/ecm.1307
Meineke, E., Nathan, A. T. C., Sanders, J. Jonathan, and Davies, T. (2019). Herbarium specimens reveal increasing herbivory over the past century. J. Ecol. 107, 105–117. doi: 10.1111/1365-2745.13057
Mideros, M. F., Turissini, D. A., Guayazán, N., Ibarra-Avila, H., Danies, G., Cárdenas, M., et al. (2018). Phytophthora betacei, a new species within Phytophthora clade 1c causing late blight on Solanum betaceum in Colombia. Persoonia 41, 39–55. doi: 10.3767/persoonia.2018.41.03
Montagne, J. F. C. (1846). Note sur la maladie qui ravage les pommes de terre et caractères du Botrytis infestans. Mem. Institut. France 609, 989–101.
Montes-Borrego, M., Muñoz Ledesma, F. J., Muñoz Ledesma, B. B., and Landa, B. B. (2008). A nested-polymerase chain reaction protocol for detection and population biology studies of Peronospora arborescens, the downy mildew pathogen of opium poppy, using herbarium specimens and asymptomatic, fresh plant tissues. Phytopathology 99, 73–81. doi: 10.1094/PHYTO-99-1-0073
O’Gorman, D. T., Sholberg, P. L., Stokes, S. C., and Ginns, J. (2008). DNA sequence analysis of herbarium specimens facilitates the revival of Botrytis mali, a postharvest pathogen of apple. Mycologia 100, 227–235. doi: 10.3852/mycologia.100.2.227
Oliva, R. F., Kroon, L., Chacón, G., Flier, W. G., Ristaino, J. B., and Forbes, G. A. (2010). Phytophthora andina sp. nov., a newly identified heterothallic pathogen of solanaceous hosts in the Andean highlands. Plant Pathol. 59, 613–625. doi: 10.1111/j.1365-3059.2010.02287.x
Perez, W. G., Gamboa, J. S., Falcon, Y. V., Coca, M., Raymundo, R. M., and Nelson, R. J. (2001). Genetic structure of Peruvian populations of Phytophthora infestans. Phytopathology 91, 956–965. doi: 10.1094/PHYTO.2001.91.10.956
Pruvost, O., Magne, M., Boyer, M., Leduc, A., Tourterel, C., Drevet, C., et al. (2014). A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage. PLoS One 9:e98129. doi: 10.1371/journal.pone.0098129
Ristaino, J. B. (1998). The importance of archival and herbarium materials in understanding the role of oospores in Late blight epidemics of the past. Phytopathology 88, 1120–1130. doi: 10.1094/PHYTO.1998.88.11.1120
Ristaino, J. B., Cooke, D. E. L., Acuña, I., and Muñoz, M. (2020). “The threat of late blight to global food security,” in Emerging Plant Disease and Global Food Security, Chap. 6, eds A. Records, and J. B. Ristaino (St. Paul: American Phytopathological Society Press).
Ristaino, J. B., Groves, C. T., and Parra, G. R. (2001). PCR amplification of the Irish potato famine pathogen from historic specimens. Nature 411, 695–697. doi: 10.1038/35079606
Saville, A., and Ristaino, J. B. (2019). Genetic structure and subclonal variation of extant and recent US lineages of Phytophthora infestans. Phytopathology 109, 614–627. doi: 10.1094/PHYTO-09-18-0357-R
Saville, A. C., Martin, M. D., and Ristaino, J. B. (2016). Historic late blight outbreaks caused by a widespread dominant lineage of Phytophthora infestans (Mont.) de Bary. PLoS One 11:e0168381. doi: 10.1371/journal.pone.0168381
Soltis, P. S. (2017). Digitization of herbaria enables novel research. Am. J. Bot. 104, 1281–1284. doi: 10.3732/ajb.1700281
Teschemacher, J. E. (1845). Observations on the Potato Disease. London: Gardeners’ Chronicle and Agricultural Gazette.
Thiers, B. (2018). Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York, NY: Botanical Garden’s Virtual Herbarium.
Uematsu, S., Kageyama, K., Moriwaki, J., and Sato, T. (2012). Colletotrichum carthami comb. nov., an anthracnose pathogen of safflower, garland chrysanthemum and pot marigold, revived by molecular phylogeny with authentic herbarium specimens. J. Gen. Plant Pathol. 78, 316–330. doi: 10.1007/s10327-012-0397-3
Vleeshouwers, V. G., Raffaele, S., Vossen, J. H., Champouret, N., Oliva, R., Segretin, M. E., et al. (2011). Understanding and exploiting late blight resistance in the age of effectors. Ann. Rev. Phytopathol. 49, 507–531. doi: 10.1146/annurev-phyto-072910-095326
Yoshida, K., Sasaki, E., and Kamoun, S. (2015). Computational analyses of ancient pathogen DNA from herbarium samples: challenges and prospects. Front. Plant Sci. 6:771. doi: 10.3389/fpls.2015.00771
Keywords: plant disease, mycological herbaria, Phytophthora infestans, climate change, potato famine
Citation: Ristaino JB (2020) The Importance of Mycological and Plant Herbaria in Tracking Plant Killers. Front. Ecol. Evol. 7:521. doi: 10.3389/fevo.2019.00521
Received: 31 October 2018; Accepted: 23 December 2019;
Published: 04 February 2020.
Edited by:
Freek T. Bakker, Wageningen University & Research, NetherlandsReviewed by:
Waldir M. Berbel-Filho, Swansea University, United KingdomNathan Wales, University of York, United Kingdom
Copyright © 2020 Ristaino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean B. Ristaino, amVhbl9yaXN0YWlub0BuY3N1LmVkdQ==
 Jean B. Ristaino
Jean B. Ristaino