- National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bengaluru, India
Pigmentation is involved in a wide array of biological functions across insect orders, including body patterning, thermoregulation, and immunity. The melanin pathway, in particular, has been characterized in several species. However, molecular evolution of the genes involved in this pathway is poorly explored. We traced the molecular evolution of six melanin pathway genes in 53 species of Lepidoptera covering butterflies and moths, and representing over 100 million years of diversification. We compared the rates of synonymous and non-synonymous substitutions within and between these genes to study the signatures of selection at the level of individual sites, genes, and branches of the gene tree. We found that molecular evolution of all six genes was governed by strong purifying selection. Yet, a number of sites showed signs of being under positive selection, including in the highly conserved domain regions of three genes. Further, we traced the expression of these genes across developmental stages, tissues, and sexes in the Papilio polytes butterfly using a developmental transcriptome dataset. We observed that the expression patterns of the genes in P. polytes largely reflected their known tissue-specific function in other species. The expression of sequentially acting genes in the melanin pathway was correlated. Interestingly, melanin pathway genes also showed a sexually dimorphic pattern of developmental heterochrony, i.e., females showed prominent upregulation of melanin pathway genes in pre-pupal stage compared to males, while males showed prominent upregulation in 9-day pupal wings compared to females. Our evolutionary and developmental analyses suggest that the vast diversity of wing patterning and pigmentation in Lepidoptera may have evolved despite largely constrained sequence evolution, with potential contribution from differential developmental expression of genes in a highly conserved pathway.
Introduction
The evolution of color patterns contributes to the striking diversity of lifeforms. Coloration is a critical adaptation that impacts an organism’s intra- and inter-specific communication and interactions. In several cases, the emergence of sexual dimorphism and polymorphism is also linked to the evolution and differential expression of pigmentation genes (Wittkopp et al., 2009; Miyazaki et al., 2014; Yassin et al., 2016). Among Lepidoptera, wing coloration and patterning are remarkably diverse, which have shaped several adaptations such as aposematism, crypsis, mimicry, and thermoregulation (True et al., 2005; Hegna et al., 2013; Olofsson et al., 2013; Kronforst and Papa, 2015; Nadeau, 2016; Deshmukh et al., 2018; van’t Hof et al., 2019). In insects, melanins, ommochromes, and pterins are three major biosynthesized pigments deposited in developing wing scales (Wittkopp and Beldade, 2009). Melanin pathway and its components—being involved in a wide range of physiological processes—are conserved across insects. For instance, increased melanization in high-altitude moths aids in thermoregulation, resulting in a trade-off between warning coloration and hindwing melanization (Hegna et al., 2013). Melanization is also associated with immunity, wound healing, and protection from both ultraviolet light and parasitoids (Yassine et al., 2012; Bilandžija et al., 2017). Melanin and related pigments—such as DOPA-melanin, dopamine-melanin, NBAD sclerotin, and NADA sclerotin—contribute to the production of black, brown, and yellow coloration (Wright, 1987; Koch et al., 2000a; Zhang et al., 2017a). The availability of genetic manipulation techniques in non-model systems has allowed elucidation of the role of melanin pathway genes in color adaptations (Zhang and Reed, 2017; Zhang et al., 2017a; Matsuoka and Monteiro, 2018). While the developmental mechanisms underlying pigmentation and patterning are being extensively studied, the evolutionary trajectories of these pigmentation genes remain poorly explored.
We chose six melanin pathway genes—tan, black, ebony, pale (tyrosine hydroxylase), arylalkylamine N-acetyltransferase 1 (aaNAT), and DOPA decarboxylase (DDC)—whose developmental function in Lepidoptera coloration is well characterized (Zhang et al., 2017a; Matsuoka and Monteiro, 2018). The functions and phenotypic effects of these genes are summarized in Figure 1 and Supplementary Table S1. Melanin synthesis occurs by a branched pathway with tyrosine and uracil as precursors which are shunted into the pathway by pale and DDC (Zhang et al., 2017a). black and ebony are involved in the synthesis of N-β-alanyl dopamine (NBAD), which is responsible for yellow coloration, while tan breaks down NBAD to dopamine, thus countering the synthesis of NBAD and the activity of ebony. aaNAT facilitates the formation of colorless cuticles essential for wing pigmentation (Figure 1 and Supplementary Table S1). These genes are thus crucial for melanin production and its deposition on Lepidoptera wings. The pigments observed in adults start appearing on the wings and body tissues during late pupal stages, and black melanin is usually the last one to be deposited (Koch et al., 1998; Wittkopp and Beldade, 2009; Matsuoka and Monteiro, 2018). In this study, we trace the molecular evolution of these six genes in 53 species across nine superfamilies of Lepidoptera (total over 100 million years of divergence) and additionally characterize their expression patterns across various developmental stages and tissues of Papilio polytes to address the following questions: (i) What selection pressures have shaped the evolution of these genes in the Lepidoptera? (ii) Do these genes show differential activity in sexually dimorphic species? and (iii) Do they have similar expression patterns during larval pigmentation and adult wing patterning and pigmentation?
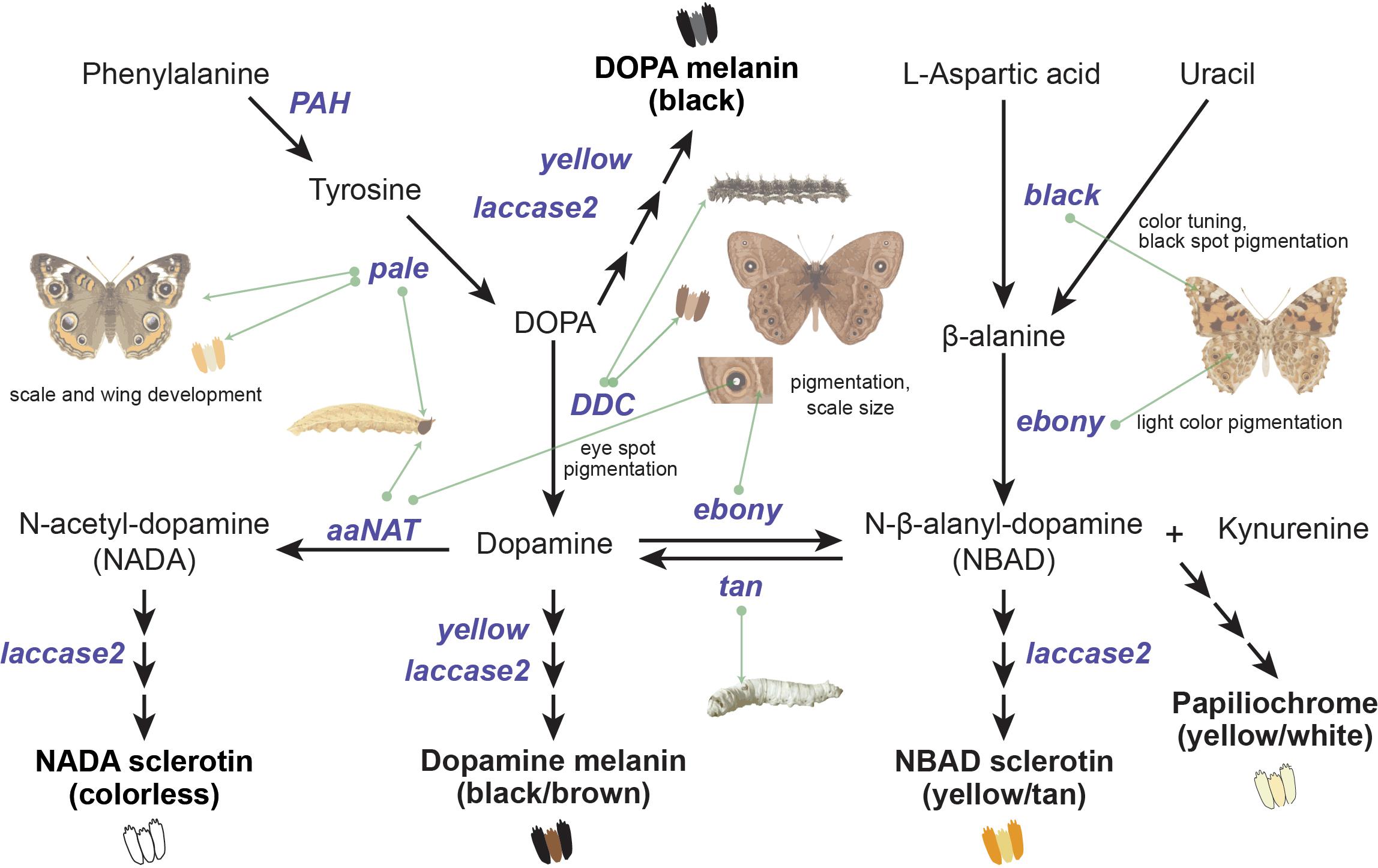
Figure 1. Melanin pathway genes and their phenotypic effects in Lepidoptera. The proposed melanin pathway (Wright, 1987; Zhang et al., 2017a) illustrates the genes/enzymes (colored in purple) that catalyze different steps in the production of four major pigments. The end products of the pathway conjugate with sclerotin to produce differently colored scales (Vavricka et al., 2010), as illustrated. Each gene also regulates individual wing elements in different species of butterflies (green arrows), as deciphered from their knockout phenotypes (summarized in Supplementary Table S1).
Materials and Methods
Gene Sequences and Multiple Sequence Alignment
We downloaded whole genome sequences of 53 species of Lepidoptera from GenBank1, LepBase2, and GigaDB3, which best represented different families. We selected six genes which are central to pigmentation and patterning pathways, namely tan, black, ebony, pale (tyrosine hydroxylase), aaNAT (Arylalkylamine N-acetyltransferase 1), and DDC (dopa decarboxylase) (Wright, 1987; Zhang et al., 2017a; Matsuoka and Monteiro, 2018). We performed exon-wise NCBI local tBLASTn to locate these genes within genomes of the selected species (Supplementary Table S2). For all the genes, we chose the longest isoform for performing tBLASTn as it incorporates maximum sequence information for the given gene. Using these gene coordinates, we extracted respective genes from the genome using an in-house python script. We chose the hits with the highest similarity scores to avoid misidentification. The downloaded genome sequences showed no evidence of duplication for any of the six melanin pathway genes. Several genome files (such as those of Bicyclus anynana, P. polytes, Papilio xuthus, and Bombyx mori) had annotated two copies of aaNAT and black, but they shared low similarity scores (∼40%) and were therefore not considered as duplicates in our analysis. We performed multiple sequence alignment of each gene with MEGA X MUSCLE aligner with codon alignment option (Kumar et al., 2018). We included homologous gene sequences from Diptera as outgroups for respective genes.
Gene Tree Construction and Phylogenetic Analysis
We constructed a species-level phylogeny with two mitochondrial (cytochrome c oxidase I, acetyl-CoA acetyltransferase) and ten nuclear genes (elongation factor 1-alpha, wingless, ribosomal protein S5, ribosomal protein S2, isocitrate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, malate dehydrogenase, catalase, CAD, ribosomal protein S27a/hairy cell leukemia) that are used commonly in phylogenetic reconstruction (Wahlberg and Wheat, 2008). We performed multiple sequence alignment on these 13 markers using the codon aligner PRANK v150803 (Löytynoja and Goldman, 2005). For construction of species and gene trees, we calculated the best partition scheme and corresponding sequence evolution model using Partition Finder 2.1.1 (Lanfear et al., 2016). We chose greedy algorithm and Mr. Bayes model of evolution. We selected Bayesian information criterion to compare and choose from the best-fit models. We used a split frequency below 0.01 to assess stationarity and to set the burn-in in Mr. Bayes and then built a consensus tree using the remaining trees (Ronquist et al., 2012).
Determination of Synonymous (dS) and Non-synonymous (dN) Substitution Rates
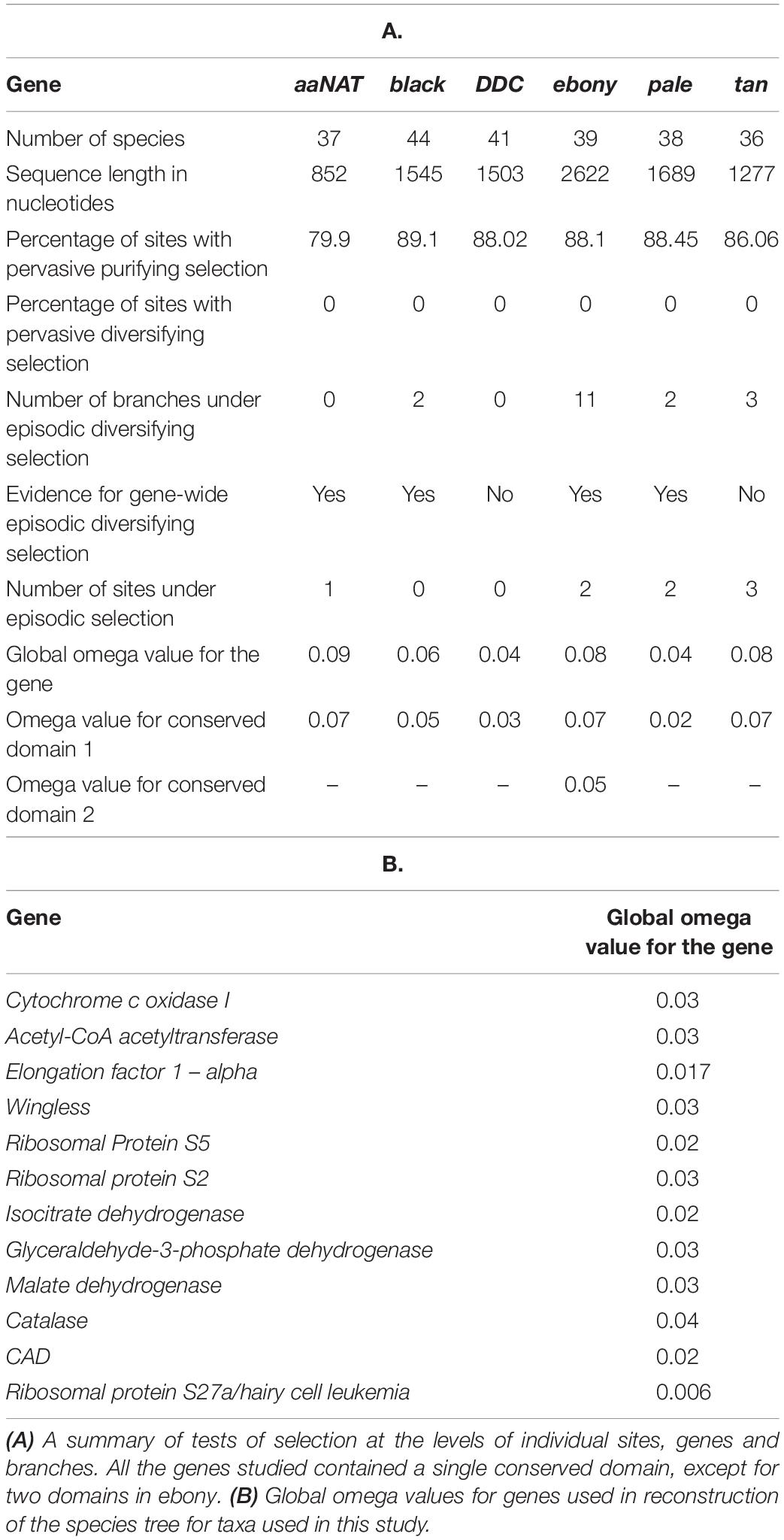
To calculate site-wise synonymous and non-synonymous substitution rates of each gene, we used their respective MUSCLE alignments and the fixed effect likelihood (FEL) method with a maximum-likelihood (ML) approach (Kosakovsky and Frost, 2005). We plotted and compared codon-wise dN, dS, and dN/dS values for each gene (Figure 2) and performed a Kruskal–Wallis test followed by Dunn’s test with Bonferroni correction in R (Derek et al., 2020). We also identified sites that have been subjected to pervasive diversification (dN/dS > 1) or purifying selection (dN/dS < 1) within every gene using FEL. We calculated global omega (R value, which represents dN/dS for the entire sequence) for respective alignments using AnalyzeCodonData function of HyPhy 2.3.14 (Pond et al., 2005; Kosakovsky et al., 2020; Table 1). For comparison, we also calculated R for genes used in species-level phylogenetic reconstruction. To identify conserved domains in the given proteins, we used the Conserved Domain Database (CDD) and the CD-Search Tool (Marchler-Bauer et al., 2012). We separately performed FEL analysis and estimated global omega for these domains to compare gene-wide and domain-wide signatures of selection (Figure 2 and Table 1).
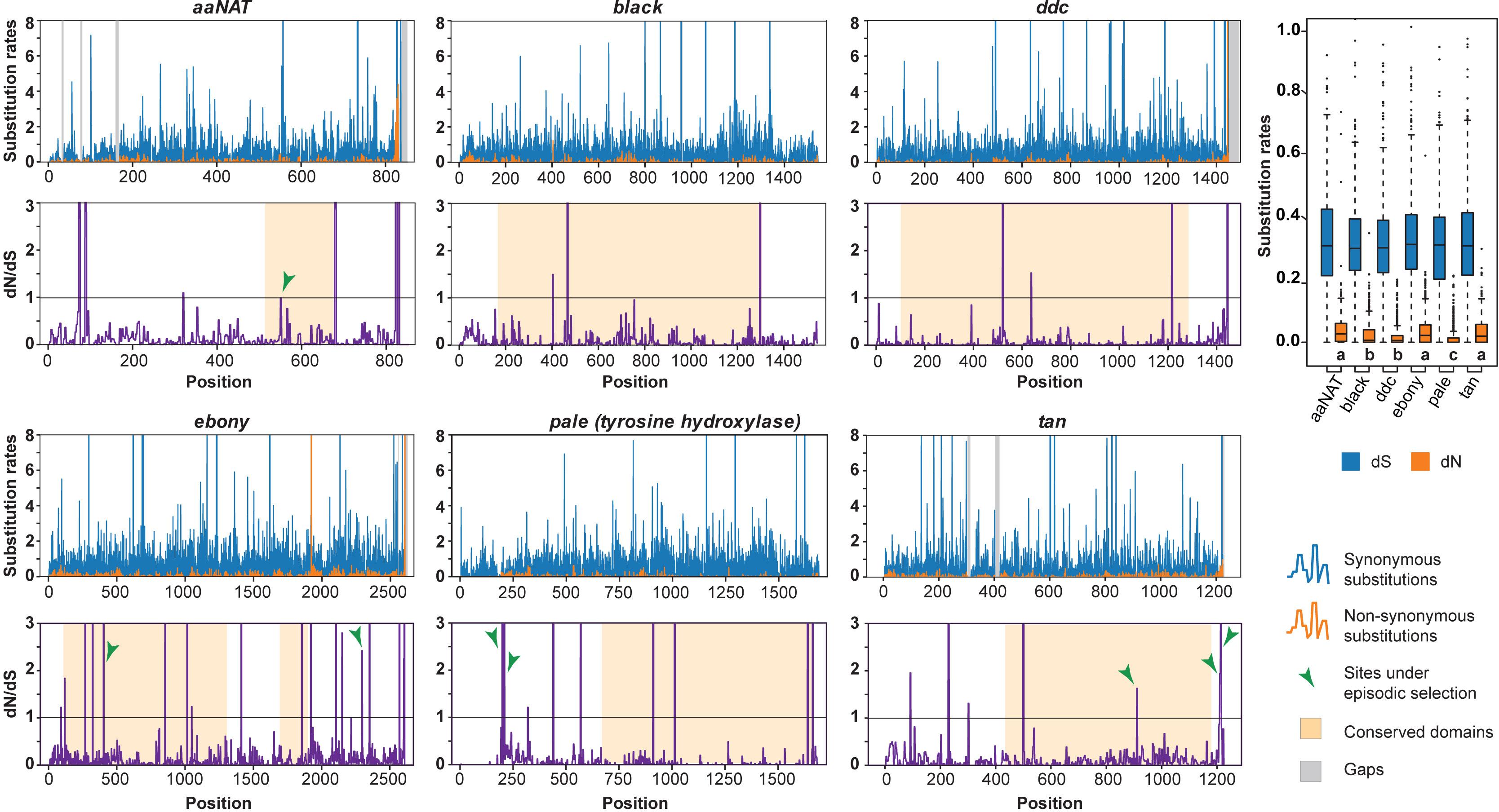
Figure 2. Molecular evolution of selected genes in the melanin pathway. Molecular evolution is shown with individual rates of synonymous (blue) and non-synonymous substitutions (orange) for each codon (top panels) and dN/dS ratios for individual codons (lower panels). Functional domains for each gene are highlighted along with sites under episodic diversifying selection (green arrows, see Supplementary Table S4). Rates of substitutions are compared between genes in the boxplot (right) where dN values significantly differ across genes (p < 0.001, see Supplementary Table S3). Groups that do not differ statistically are represented by the same letter.
Branch-Level, Gene-Wide, and Site-Wise Assessment of Molecular Evolution
We used aBSREL (adaptive Branch-Site Random Effects Likelihood) to test for positive selection along each gene tree (Kosakovsky et al., 2011; Smith et al., 2015). We performed aBSREL analysis in exploratory mode which tests for positive selection along a branch of the gene tree and corrects for multiple testing. aBSREL models site and branch-level omega values but does not test for site-level selection. We used BUSTED to investigate gene-level positive selection. It calculates site-specific evidence ratios and combines this information from sites with weak positive selection to infer selection at the gene level. We carried out MEME (Mixed Effects Model of Evolution) which takes ML approach to test whether individual sites have been subjected to episodic positive or diversifying selection (dN/dS > 1). We performed FEL, MEME, BUSTED, and aBSREL on the Datamonkey Adaptive Evolution Server (Weaver et al., 2018), processing the data using in-house scripts.
Sample Collection, RNA Extraction, and Transcriptome Sequencing for Quantification of Expression
We bred greenhouse populations of P. polytes from mated wild-caught females. We maintained larvae at 28 ± 4°C on lemon (Citrus sp.) and curry plants (Murraya koenigii) and adults on Birds ChoiceRM butterfly nectar. We preserved P. polytes at different stages of metamorphosis in TRIzolRM. We collected eggs at 2, 10, and 24 h, and at 3 days after oviposition, and pooled five eggs for each sample to get sufficient amount of RNA. We sampled larvae at 1st, 3rd, and 5th instars. For RNA extraction, we used gutted bodies of 1st and 3rd instar larvae and dissected wing disks from 5th instar larvae and pre-pupae to get three tissue types—forewings, hindwings, and gutted body. We collected pupae at 3, 6, and 9 days after pupation and dissected them to separate the forewings, hindwings, abdomen, thorax, and head. Several pigmentation pathway genes participate in transport and phototransduction in insect eyes (True et al., 2005; Ziegler et al., 2013). To account for altered sensitivity and visual perception due to the mating status of the butterflies, if any, we collected unmated and mated adults separately and further dissected them to separate abdomen, head, eyes, and thorax. We used pure-breeding mimetic lines for this study. We sampled eggs, 1st and 3rd instar larvae in triplicates, 5th instar larvae, pre-pupae, and pupae in duplicates for males and females each, and adults in quadruplicates for each sex.
We extracted RNA from preserved tissues using the chloroform–isopropanol-based extraction method. We prepared libraries using TruSeq® RNA Sample Preparation Kit v2 and used Qubit fluorometric quantification. We also used Bioanalyzer to check library profiles. For transcriptome sequencing, we used 2 × 100 PE runs on Illumina HiSeq 2500 and obtained ∼20 million reads for each sample. We performed quality check on these reads using FastQC, trimmed them using Trimmomatic, and aligned them to P. polytes reference genome (Nishikawa et al., 2015) using STAR and HISAT2 aligners. We chose the alignments showing higher mapping percentages. We used HTSeq for obtaining raw counts and edgeR pipeline for analyzing and plotting gene expression for all the genes. To determine the effect of sex on expression, we performed a principal component analysis (PCA) in R using the normalized counts for five melanin pathway genes. We excluded tan as it did not show sexual dimorphism in its expression profile. We also performed pairwise correlation tests in R (Pearson correlation) to test similarities of expression profiles for all six melanin pathway genes.
Results
Melanin Pathway Genes Are Single-Copy Conserved Genes Under Strong Purifying Selection in Lepidoptera
We reconstructed gene trees for each of the six genes (Supplementary Figure S1). Each gene tree was well supported, and it essentially mirrored the species tree in terms of species relationships and divergence, except aaNAT, which had poor resolution with a high degree of polytomy. Absence of long branches and broad similarity with the species tree suggest that these genes have evolved with species divergence and do not seem to have undergone rapid evolution that would be incongruent with the species tree in any lineage.
We plotted dN/dS values for each site in each gene (Figure 2). The molecular evolution of these genes was characterized by predominantly synonymous substitutions, suggesting constrained evolution (dN/dS values for most sites were well below 1, with mean dN/dS value for each gene ranging from 0.04 to 0.10). Rates for gene-wide synonymous substitutions were similar, but non-synonymous rates were significantly different between three groups of genes: black and DDC; ebony, tan and aaNAT; and pale (Dunn’s test, p < 0.001) (Supplementary Table S3). Overall, >80% sites showed signatures of pervasive purifying selection (p < 0.05), but no sites showed signatures of pervasive diversifying selection in any melanin pathway genes (Table 1).
All the genes had a single functional domain except for ebony, which contained two (Figure 2). R values for the six genes ranged between 0.04 and 0.09 and were found to be consistently lower for functional domains compared to the gene-wide values (Table 1A). Similarly, R values for genes used in species-level phylogenetic reconstruction were considerably lower and ranged between 0.006 and 0.04. We observed that the non-synonymous substitution rate was lowest in pale, followed by black and DDC (Figure 2, p < 0.05). Since DDC, black, and pale are essential for the synthesis of dopamine—a precursor for multiple pathways including neuromodulation, immune functions, and the circadian cycle (Basu and Dasgupta, 2000; Allen et al., 2011; Shang et al., 2011)—the low dN/dS as well as global R values were expected for these genes. On the other hand, tan, aaNAT, and ebony yielded high R values, indicating that they may have evolved under relatively relaxed selection (Figure 2 and Table 1).
Melanin Pathway Genes Show Signs of Episodic Positive Selection
In spite of the overall constrained molecular evolution of the six genes, eight sites showed signatures of episodic diversifying selection (p < 0.05), including some in the otherwise highly conserved domain regions (Figure 2, Table 1, and Supplementary Table S4). Interestingly, although pale generally shows very constrained molecular evolution similar to DDC and black, it also contained two of the eight sites under episodic positive selection. While BUSTED could not detect gene-wide episodic diversifying selection in DDC and tan, aBSREL found evidence for it in all the genes except DDC.
Melanin Pathway Genes Show Stage-Specific, Often Sexually Dimorphic Expression
We traced the expression of the six genes across different developmental stages, tissues, and sexes in the developmental transcriptome of P. polytes. Broadly, we identified two groups with similar expression patterns across stages and tissues: aaNAT, DDC, and pale formed one group, while black and ebony formed another group (Figure 3). Since pale, DDC, and aaNAT convert tyrosine to N-acetyl-dopamine in successive reactions with no intermediates (Figure 1), we expected similar activity patterns of these genes. aaNAT, DDC, and pale showed similar activity across stages with a peak in the pre-pupal stage (Figure 3 and Supplementary Figure S2), suggesting a more general function than melanization-specific activity. Although ebony combines the products of black and DDC to produce NBAD (Figure 1), its expression pattern was similar to that of black, not DDC. This suggested that ebony activity was more dependent on black than on DDC. We tested this possibility with an analysis of co-expression between these genes, which indeed showed that the expression profiles of ebony and black were most strongly correlated compared to any other pairs among these six genes (Figure 3C). Interestingly, DDC showed a strong positive correlation with all the other genes except tan, which can be attributed to the central function of DDC in the melanin pathway (Figure 3C), whose products feed into several branches. These correlations need to be experimentally verified.
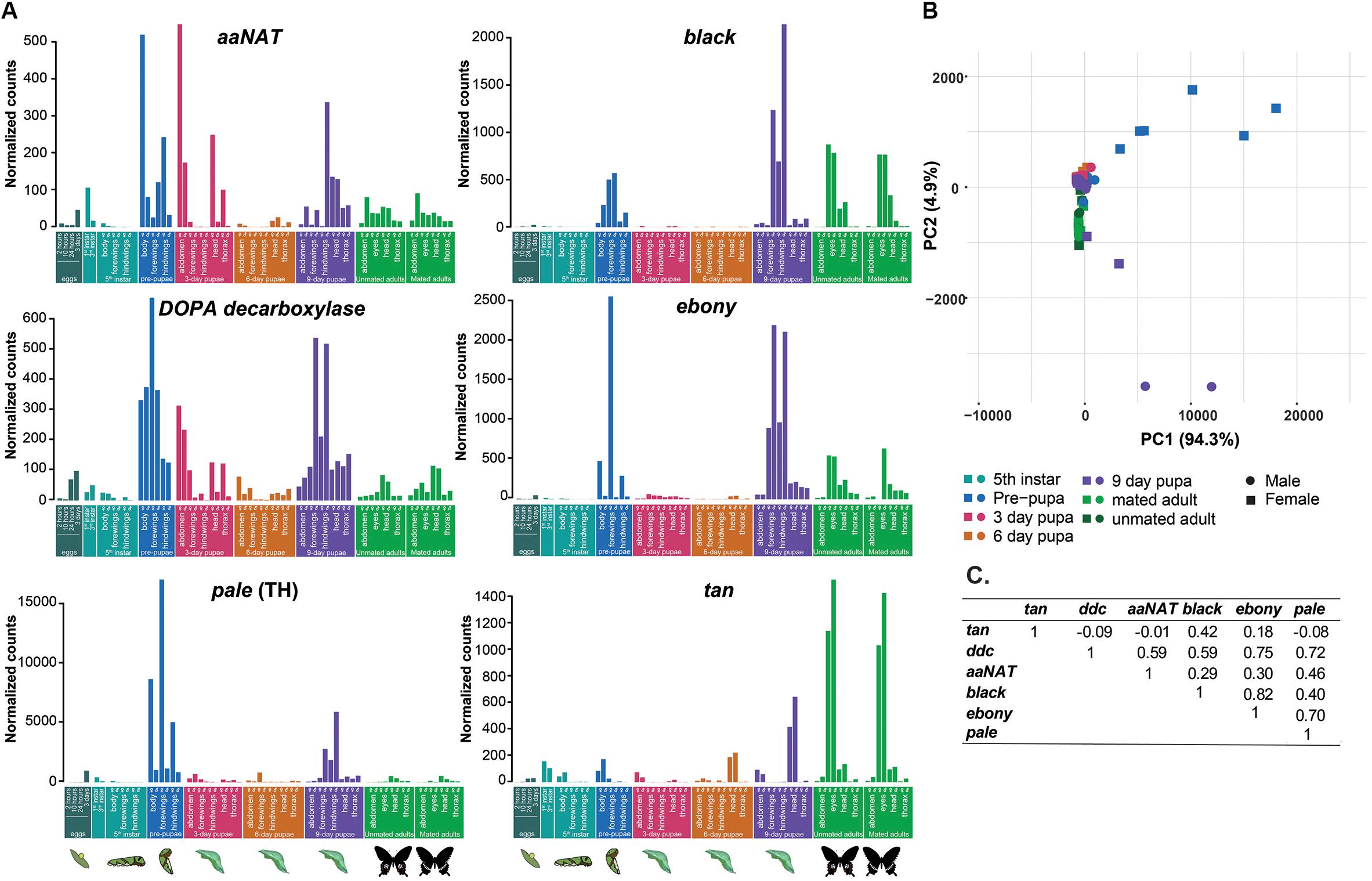
Figure 3. (A) Expression of the six melanin pathway genes across developmental stages of Papilio polytes. Means of normalized counts for each stage, tissue and sex (beyond 5th instar larvae) obtained from transcriptome data are plotted. The developmental stages are color-coded and pictorially represented at the bottom (instar indicates larval instars). (B) Principal component analysis (PCA) of normalized expression data across developmental stages, tissues and sexes for five genes. (C) Pairwise Pearson correlation coefficients for co-expression profiles of the six genes.
Unlike any of the other genes, tan had a remarkably distinct pattern of expression across development. It showed little to no expression in wings but it was expressed in the 1st and 3rd instar larvae, suggesting possible involvement in larval melanization. Expression of tan, black, and ebony in adult eyes and head may have resulted from their role in photo-transduction. For example, tan is involved in metabolism of neurotransmitters in photoreceptors, while ebony and black play a role in signal transduction in the optic lobe (True et al., 2005; Ziegler et al., 2013). Because of this, we further tested whether mated and unmated females show differential expression since female visual systems may switch from mate choice to host plant search after mating. However, expression of these genes was similar in unmated and mated females as well as in males (Figure 3B and Supplementary Table S5).
Finally, to test for sexual dimorphism in the expression patterns of the melanin pathway genes, we performed a PCA using normalized counts of five genes excluding tan. Most stages and tissues had low expression of melanin pathway genes, which caused most of the points to cluster around zero. Two stages that stood out—pre-pupal tissues and 9-day pupal wings—showed sexual dimorphism. Melanin pathway genes were upregulated in female pre-pupae and male wings of 9-day pupae, indicating a sex-specific developmental heterochrony in activity of these genes in developing pupae of P. polytes.
Discussion
The melanin pathway is complex and versatile, with products of intermediately placed genes feeding into cross-pathways, performing diverse biological functions (Figure 1). The feasibility of genetic manipulation in non-model organisms has made it possible to understand the extent of role played by each gene in pigment production in a wide range of organisms (Mazo-Vargas et al., 2017; Zhang et al., 2017a, b; Connahs et al., 2019). We supplement this understanding by studying the molecular evolution of six genes (tan, black, ebony, pale, aaNAT, and DDC) in the melanin pathway that have prominent roles in color production in butterflies and moths (Koch, 1995; Koch et al., 2000a, b; Futahashi and Fujiwara, 2005; Futahashi et al., 2010; Zhang et al., 2017a; Matsuoka and Monteiro, 2018). With each gene governing a generalized or specialized step, the pathway experiences varied constraints at different levels. We estimated synonymous and non-synonymous substitution rates for each gene using sequence data and tested for signatures of selection. Our study shows that these six genes are highly conserved, despite 100 million years of evolutionary divergence across the sampled Lepidopteran superfamilies (Misof et al., 2014). However, the degree of conservation varies across genes, and most of them even show some signatures of episodic and positive selection.
Our selection analysis revealed that all the melanin pathway genes studied here show signatures of strong purifying selection. The dearth of non-synonymous substitutions across these genes indicates a lack of pervasive diversifying selection at a sequence level, although the developmental regulation of these genes produces dissimilar color patterns across species (Wittkopp et al., 2009; Miyazaki et al., 2014; Yassin et al., 2016). While testing for episodic diversifying selection at the levels of individuals sites, branches and genes, aaNAT, black, ebony, pale, and tan, but not DDC, showed evidence of diversifying selection. Similarly, we did not find any site-level evidence for diversifying selection in black. These differential signatures of selection across the genes are consistent with their estimated non-synonymous substitutions, which shows that black and DDC have followed more constrained evolutionary trajectories compared to the other four genes. The slight difference between domain-wide and gene-wide R values suggests that these six genes are nearly as conserved as their functional domains, and in some cases, this may be a result of the domain spanning a large portion of the gene. The melanin pathway genes showed slightly higher R values compared to the genes used for phylogenetic reconstruction, reiterating their high degree of conservation. Our study does not offer insights into the functional roles of these sites under diversifying selection. However, it does point out potential developmental genetic targets for future manipulative experiments. The functional roles of these sites may also become clear using protein structural analysis.
We obtained a few contradictory results in our selection analysis with the three methods used. For example, gene-level analysis using BUSTED inferred the presence of episodic diversifying selection in black, but MEME could not detect individual sites under selection. This may be because BUSTED is capable of combining information from sites with weak signals of positive selection to infer presence of selection at the whole-gene level, unlike MEME, which detects individual sites and might therefore underestimate sites under selection at the gene level. Similarly, we performed branch-level tests of diversifying selection with aBSREL, whose exploratory module is less likely to detect selection compared to targeted testing (Kosakovsky et al., 2011; Smith et al., 2015). This may have resulted in aaNAT not showing branch-level selection despite showing site-level and gene-level diversifying selection using other methods.
We explored the spatio-temporal activity of melanin pathway genes in P. polytes using a developmental transcriptome dataset. The first four instars of P. polytes larvae mimic bird droppings with black, brown, and white coloration on their bodies. We expected to detect expression of melanin pathway genes in the eggs and 1st and 3rd larval instars, similar to what we see in 9-day old pupae. However, we did not find substantial larval expression of any of the six genes (except tan) even though knockout phenotypes in previous studies have reported a lack of larval pigmentation (Zhang et al., 2017a; Matsuoka and Monteiro, 2018). It is possible that the low expression that we detected suffices in triggering melanization, or we may have missed the transient stage where melanin pathway genes are upregulated in early development. Exploring the expression patterns of other genes in the pathway, such as laccase2 and yellow, may help corroborate either hypothesis. The expression patterns of genes also showed signs of sex-specific developmental heterochrony in melanin production, with expression during pupal development peaking early in females (pre-pupae) and late in males (9-day pupae), as corroborated by PCA. While this could be an artifact of sampling, in some cases (especially between male and female pre-pupae and 9-day old pupae), the difference is quite stark and observed across five of the six genes to be attributed to sampling error. It is possible that males and females differentially invest in melanin production and immunity (or other functions that require melanin pathway intermediates), which would also explain the heterochrony we observed. However, this needs further investigation. With the additional complexity of mimicry in this species, and the use of only mimetic females in this study, the effect of mimicry-related wing-pattern reorganization on melanin pathway genes remains to be explored and could perhaps be linked to the developmental heterochrony.
Melanin pathway genes such as tan, ebony, and yellow are involved in the evolution of color pattern-related sexual dimorphism and polymorphism (Wittkopp et al., 2009; Miyazaki et al., 2014; Yassin et al., 2016), suggesting that these genes have the potential to evolve novel functions despite evolutionarily constrained sequences. Many butterflies and moths exhibit sexual dimorphism and/or female polymorphism, so they are good system to study signatures of selection in such clades. This would also require information regarding functional allelic variation and duplicated copies of these genes, which we did not encounter in the genes and species that we studied. Alternatively, production of certain color patterns is associated with changes in proximal and/or distal cis regulatory regions which govern gene expression. For instance, gain of novel transcription factor binding sites in the cis-regulatory region of yellow has influenced wing pattern evolution in Drosophila biarmipes (Gompel et al., 2005). Examining the regulatory regions of such highly conserved genes and tracing the molecular evolution of upstream regulators might help detect sites that influence color pattern-related sexual dimorphism or polymorphism. Furthermore, selective manipulation of protein-coding versus cis-regulatory regions of melanin pathway genes might also help dissect the relative contributions of these genetic elements in pigment production. We have excluded yellow gene family from our study even though substantial work has been done on this in flies and butterflies (Arnoult et al., 2013; Miyazaki et al., 2014; Camino et al., 2015; Zhang et al., 2017a; Matsuoka and Monteiro, 2018). We were unable to identify sequences of this gene family from the downloaded genomes with sufficient confidence due to issues with sequence similarity scores. However, the evolution of this gene family has been explored previously (Ferguson et al., 2011).
Our work demonstrates that molecular sequences of the six melanin pathway genes are highly conserved across the vastly diverse Lepidoptera, yet they show some signatures of positive selection. The pattern of conservation and divergence of these genes may be a direct outcome of their functions. Such studies on molecular evolution of genes associated with ecological adaptations (Ferguson et al., 2011; Hughes, 2011; Baral et al., 2019) can help establish baselines against which evolutionary patterns of other genes could be compared. We also explored the developmental expression of these genes across tissues and sexes, which recapitulates what we observe in adult stages, but does not provide insights into larval pigmentation. Our work provides a framework in which broad comparisons can be made across genes and species to understand the genetics and evo-devo of complex pathways that produce the remarkable color patterns displayed by insects.
Data Availability Statement
The raw RNA seq data are deposited in the NCBI SRA database (BioProject PRJNA634605, Accession nos. SAMN15001929 to SAMN15002046; https://dataview.ncbi.nlm.nih.gov/object/PRJNA634605?reviewer=mf4q72fcfvq67spjf5gl1sduop). The accession numbers for genomes used in the molecular evolutionary analysis are provided in Supplementary Table S2.
Author Contributions
RD and KK designed the study. MK and AP downloaded the sequence data and performed the molecular evolution analysis. RD performed the developmental transcriptome sequencing and analysis. MK and RD prepared the figures and wrote the manuscript. KK conceived and directed the project. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by an NCBS Research Grant to KK, support of the Department of Atomic Energy, Government of India, under project nos. 12-R&D-TFR-5.04-0800 and 12-R&D-TFR-5.04-0900, and a CSIR Shyama Prasad Mukherjee Fellowship to RD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Atharva Khotpal, Bhavanarayeni R., Sai Guha, and Sarvesh Menon for assistance in downloading and analyzing genome sequences, Saurav Baral and Vinod Shankar for help with molecular evolution analysis, Sergei L. Kosakovsky Pond for valuable inputs on HyPhy, NCBS Sequencing Facility for transcriptome sequencing, and NCBS Greenhouse Facility for butterfly breeding facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00226/full#supplementary-material
Footnotes
References
Allen, J. M., Van Kummer, B. H., and Cohen, R. W. (2011). Dopamine as an anorectic neuromodulator in the cockroach Rhyparobia maderae. J. Exp. Biol. 214, 3843–3849. doi: 10.1242/jeb.062430
Arnoult, L., Su, K. F. Y., Manoel, D., Minervino, C., Magriña, J., Gompel, N., et al. (2013). Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339, 1423–1426. doi: 10.1126/science.1233749
Baral, S., Gandhimathi, A., Deshmukh, R., and Kunte, K. (2019). Genetic architecture and sex-specific selection govern modular, male-biased evolution of doublesex. Sci. Adv. 5:eaau3753. doi: 10.1126/sciadv.aau3753
Basu, S., and Dasgupta, P. S. (2000). Dopamine, a neurotransmitter, influences the immune system. J. Neuroimmunol. 102, 113–124. doi: 10.1016/S0165-5728(99)00176-9
Bilandžija, H., Laslo, M., Porter, M. L., and Fong, D. W. (2017). Melanization in response to wounding is ancestral in arthropods and conserved in albino cave species. Sci. Rep. 7:17148. doi: 10.1038/s41598-017-17471-2
Camino, E. M., Butts, J. C., Ordway, A., Vellky, J. E., Rebeiz, M., and Williams, T. M. (2015). The evolutionary origination and diversification of a dimorphic gene regulatory network through parallel innovations in cis and trans. PLoS Genet. 11:e1005136. doi: 10.1371/journal.pgen.1005136
Connahs, H., Tlili, S., van Creij, J., Loo, T. Y. J., Banerjee, T., and Das et al. (2019). Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process. Development 146:dev169367. doi: 10.1242/dev.169367
Derek, O., Powell, W., and Dinno, A. (2020). FSA: Fisheries Stock Analysis. Available online at: https://github.com/droglenc/FSA (accessed February 2020).
Deshmukh, R., Baral, S., Gandhimathi, A., Kuwalekar, M., and Kunte, K. (2018). Mimicry in butterflies: co-option and a bag of magnificent developmental genetic tricks. WIREs Dev. Biol. 7:e291. doi: 10.1002/wdev.291
Ferguson, L. C., Green, J., Surridge, A., and Jiggins, C. D. (2011). Evolution of the insect yellow gene family. Mol. Biol. Evol. 28, 257–272. doi: 10.1093/molbev/msq192
Futahashi, R., Banno, Y., and Fujiwara, H. (2010). Caterpillar color patterns are determined by a two-phase melanin gene prepatterning process: new evidence from tan and laccase2. Evol. Dev. 12, 157–167. doi: 10.1111/j.1525-142X.2010.00401.x
Futahashi, R., and Fujiwara, H. (2005). Melanin-synthesis enzymes coregulate stage-specific larval cuticular markings in the swallowtail butterfly, Papilio xuthus. Dev. Genes Evol. 215, 519–529. doi: 10.1007/s00427-005-0014-y
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A., and Carroll, S. B. (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487. doi: 10.1038/nature03235
Hegna, R. H., Nokelainen, O., Hegna, J. R., and Mappes, J. (2013). To quiver or to shiver: increased melanization benefits thermoregulation, but reduces warning signal efficacy in the wood tiger moth. Proc. R. Soc. B 280:20122812. doi: 10.1098/rspb.2012.2812
Hughes, A. L. (2011). Runaway evolution of the male-specific exon of the doublesex gene in Diptera. Gene 472, 1–6. doi: 10.1016/j.gene.2010.10.015
Koch, B. P., Behnecke, B., Weigmann-Lenz, M., and Ffrench-Constant, R. H. (2000a). Insect pigmentation: activities of beta-alanyldopamine synthase in wing color patterns of wild-type and melanic mutant swallowtail butterfly Papilio glaucus. Pigment cell Res. 13(Suppl. 8) 54–58. doi: 10.1111/j.0893-5785.2000.130811.x
Koch, P. B., Behnecke, B., and Ffrench-Constant, R. H. (2000b). The molecular basis of melanism and mimicry in a swallowtail butterfly. Curr. Biol. 10, 591–594. doi: 10.1016/s0960-9822(00)00494-2
Koch, B. P., Keys, D. N., Rocheleau, T., Aronstein, K., Blackburn, M., Carroll, S. B., et al. (1998). Regulation of dopa decarboxylase expression during colour pattern formation in wild-type and melanic tiger swallowtail butterflies. Development 125, 2303–2313.
Koch, P. B. (1995). Colour pattern specific melanin synthesis is controlled by ecdysteroids via dopa decarboxylase in wings of Precis coenia (Lepidoptera: Nymphalidae). Eur. J. Entomol. 92, 161–167.
Kosakovsky, P. S. L., and Frost, S. D. W. (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222. doi: 10.1093/molbev/msi105
Kosakovsky, P. S. L., Murrell, B., Fourment, M., Frost, S. D. W., Delport, W., and Scheffler, K. (2011). A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 28, 3033–3043. doi: 10.1093/molbev/msr125
Kosakovsky, P. P. L., Poon, A. F. Y., Velazquez, R., Weaver, S., Hepler, N. L., Murrell, B., et al. (2020). HyPhy 2.5—A customizable platform for evolutionary hypothesis testing using phylogenies. Mol. Biol. Evol. 37, 295–299. doi: 10.1093/molbev/msz197
Kronforst, M. R., and Papa, R. (2015). The functional basis of wing patterning in Heliconius butterflies: the molecules behind mimicry. Genetics 200, 1–19. doi: 10.1534/genetics.114.172387
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Löytynoja, A., and Goldman, N. (2005). An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U.S.A. 102, 10557–10562. doi: 10.1073/pnas.0409137102
Marchler-Bauer, A., Zheng, C., Chitsaz, F., Derbyshire, M. K., Geer, L. Y., Geer, R. C., et al. (2012). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352. doi: 10.1093/nar/gks1243
Matsuoka, Y., and Monteiro, A. (2018). Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Rep. 24, 56–65. doi: 10.1016/j.celrep.2018.05.092
Mazo-Vargas, A., Concha, C., Livraghi, L., Massardo, D., Wallbank, R. W. R., Zhang, L., et al. (2017). Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl. Acad. Sci. U.S.A. 114, 10701–10706. doi: 10.1073/pnas.1708149114
Misof, B., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767.
Miyazaki, S., Okada, Y., Miyakawa, H., Tokuda, G., Cornette, R., Koshikawa, S., et al. (2014). Sexually dimorphic body color is regulated by sex-specific expression of yellow gene in ponerine ant Diacamma sp. PLoS One 9:e92875. doi: 10.1371/journal.pone.0092875
Nadeau, N. J. (2016). Genes controlling mimetic colour pattern variation in butterflies. Curr. Opin. Insect Sci. 17, 24–31. doi: 10.1016/j.cois.2016.05.013
Nishikawa, H., Iijima, T., Kajitani, R., Yamaguchi, J., Ando, T., Suzuki, Y., et al. (2015). A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat. Genet. 47, 1–7. doi: 10.1038/ng.3241
Olofsson, M., Løvlie, H., Tibblin, J., Jakobsson, S., and Wiklund, C. (2013). Eyespot display in the peacock butterfly triggers antipredator behaviors in naïve adult fowl. Behav. Ecol. 24, 305–310. doi: 10.1093/beheco/ars167
Pond, S. L. K., Frost, S. D. W., and Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. doi: 10.1093/bioinformatics/bti079
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shang, Y., Haynes, P., Pírez, N., Harrington, K. I., Guo, F., Pollack, J., et al. (2011). Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat. Neurosci. 14, 889–895. doi: 10.1038/nn.2860
Smith, M. D., Wertheim, J. O., Weaver, S., Murrell, B., Scheffler, K., and Kosakovsk, P. S. L. (2015). Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 32, 1342–1353. doi: 10.1093/molbev/msv022
True, J. R., Yeh, S.-D., Hovemann, B. T., Kemme, T., Meinertzhagen, I. A., Edwards, T. N., et al. (2005). Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 1:e63. doi: 10.1371/journal.pgen.0010063
van’t Hof, A. E., Reynolds, L. A., Yung, C. J., Cook, L. M., and Saccheri, I. J. (2019). Genetic convergence of industrial melanism in three geometrid moths. Biol. Lett. 15:20190582. doi: 10.1098/rsbl.2019.0582
Vavricka, C. J., Christensen, B. M., and Li, J. (2010). Melanization in living organisms: a perspective of species evolution. Protein Cell 1, 830–841. doi: 10.1007/s13238-010-0109-8
Wahlberg, N., and Wheat, C. W. (2008). Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 57, 231–242. doi: 10.1080/10635150802033006
Weaver, S., Shank, S. D., Spielman, S. J., Li, M., Muse, S. V., and Kosakovsky, P. S. L. (2018). Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777. doi: 10.1093/molbev/msx335
Wittkopp, P. J., and Beldade, P. (2009). Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin. Cell Dev. Biol. 20, 65–71. doi: 10.1016/j.semcdb.2008.10.002
Wittkopp, P. J., Stewart, E. E., Arnold, L. L., Neidert, A. H., Haerum, B. K., Thompson, E. M., et al. (2009). Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 326, 540–544. doi: 10.1126/science.1176980
Wright, T. R. F. (1987). The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 24, 127–222. doi: 10.1016/S0065-2660(08)60008-5
Yassin, A., Bastide, H., Chung, H., Veuille, M., David, J. R., and Pool, J. E. (2016). Ancient balancing selection at tan underlies female colour dimorphism in Drosophila erecta. Nat. Commun. 7:10400. doi: 10.1038/ncomms10400
Yassine, H., Kamareddine, L., and Osta, M. A. (2012). The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 8:e1003029. doi: 10.1371/journal.ppat.1003029
Zhang, L., Martin, A., Perry, M. W., van der Burg, K. R. L., Matsuoka, Y., Monteiro, A., et al. (2017a). Genetic basis of melanin pigmentation in butterfly wings. Genetics 205, 1537–1550. doi: 10.1534/genetics.116.196451
Zhang, L., Mazo-Vargas, A., and Reed, R. D. (2017b). Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc. Natl. Acad. Sci. U.S.A. 114, 10707–10712. doi: 10.1073/pnas.1709058114
Zhang, L., and Reed, R. D. (2017). “A practical guide to CRISPR/Cas9 genome editing in Lepidoptera,” in Diversity and Evolution of Butterfly Wing Patterns, eds T. Sekimura and H. F. Nijhout (Singapore: Springer), 155–172. doi: 10.1007/978-981-10-4956-9_8
Keywords: melanization, pigmentation, developmental heterochrony, butterfly wing coloration, wing patterns
Citation: Kuwalekar M, Deshmukh R, Padvi A and Kunte K (2020) Molecular Evolution and Developmental Expression of Melanin Pathway Genes in Lepidoptera. Front. Ecol. Evol. 8:226. doi: 10.3389/fevo.2020.00226
Received: 12 March 2020; Accepted: 19 June 2020;
Published: 14 July 2020.
Edited by:
Marcus Kronforst, The University of Chicago, United StatesReviewed by:
Heather Hines, Pennsylvania State University (PSU), United StatesSimon Martin, The University of Edinburgh, United Kingdom
Copyright © 2020 Kuwalekar, Deshmukh, Padvi and Kunte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krushnamegh Kunte, a3J1c2huYW1lZ2hAbmNicy5yZXMuaW4=
†These authors have contributed equally to this work
 Muktai Kuwalekar
Muktai Kuwalekar Riddhi Deshmukh
Riddhi Deshmukh Ajay Padvi
Ajay Padvi Krushnamegh Kunte
Krushnamegh Kunte