- 1Institute of Natural and Environmental Sciences, University of Hyogo, Tamba, Japan
- 2Laboratory of Geographical Ecology, Department of Biology, Faculty of Science, Toho University, Chiba, Japan
- 3Kaiyo High School, Gamagori, Japan
- 4School of Biological Sciences, The University of Sydney, Sydney, NSW, Australia
In order to understand molecular and genetic mechanism of color pattern formation, not only adult phenotypes but also processes and mechanisms of color production and pattern formation during embryonic and postembryonic stages should be described. The pigment cell based color production and pattern formation during embryogenesis were reviewed for the recent studies on lizards and snakes, by focusing on different color production mechanisms in terms of epidermal and dermal pigment cell architectures, and then discuss the genetic determinants of pattern formation considering both biologically relevant theoretical models which consider pigment cell specification, migration, and architecture differentiation. Clarifying the contributions of pigment cells and genetic factors improves our general understanding of reptilian color pattern evolution.
Introduction
Why do we study color pattern production mechanisms in squamate reptiles. Basically, animal color pattern is a longstanding and important topic in evolutionary biology in general, and squamates in particular, which exhibit a wide variety of color patterns such as warning signal of venomous coral snakes and its mimicry by the distantly related non-venomous snakes (Figure 1A), and convergent evolution of stripes and vivid blue tails by small lizards (Figure 1B), are important target of several agents of natural selection for effective visual signals to conspecifics for reproduction, to predators for avoidance, and to sometimes prey for feeding (e.g., Cooper and Greenberg, 1992; Pianka and Vitt, 2003; Stuart-Fox et al., 2008; McKinnon and Pierotti, 2010; Kronforst et al., 2012; Allen et al., 2013; Olsson et al., 2013). The rich diversity of reptiles, as a stem lineage of terrestrial vertebrates living in various ecosystems, provides an invaluable opportunity to study coloration by mainly focusing on functional mechanisms of adaptation for avoiding predation (Ruxton et al., 2018) such as aposematic coloration and coral snake mimicry (Wallace, 1867; Jackson et al., 1976; Pfennig et al., 2001), back ground matching for cryptic coloration (Poulton, 1890; Cott, 1940; Rosenblum et al., 2004), and predator avoidance by motion dazzle coloration (Thayer, 1909; Stevens et al., 2008; Murali et al., 2018; Kodandaramaiah et al., 2020). Yet, we could point out that here have been no recent reviews on the proximate mechanisms underlying various color patterns in reptiles, though several important studies in this topic have been published to date.

Figure 1. Diverse reptile coloration. (A) An example of well known aposematic coloration by the venomous coral snake Micrurus alleni (photo by R. Fukuyama) and the non-venomous Colubrid mimic Oxyrhopus petolarius (photo by K. Fujishima). (B) An example of convergent evolution of the motile dazzle coloration of striped and blue tail of the small skinks of Emoia caeruleocauda of Micronesia (left) and Plestiodon latiscutatus of Japan (right).
Our brief review attempt to fill this gap of proximate mechanisms of color pattern formation by extending the conceptual frame-work presented by Arnold (1983) into color pattern evolution (Figure 2). According to the Arnold’s conceptual frame-work or rationale linking traits, performance and fitness of organisms, function of prey’s color pattern and its performance should be measured by considering the interactions between sender and receiver of visual signals under a variety of light environment (Endler, 1978, 1992, 1993). Therefore, when we hope to understand function and performance of prey’s color pattern from a predator’s view (Endler, 1978), we have to know how prey produce color signal and how predator perceive prey coloration. Under the classic mathematical theory of communication by Shannon and Weaver (1948), we first simply ask how reptile skin is capable of producing a wide array of color signals (Kuriyama et al., 2006; Kikuchi et al., 2014), no matter how it is perceived by other organisms (Figure 2).
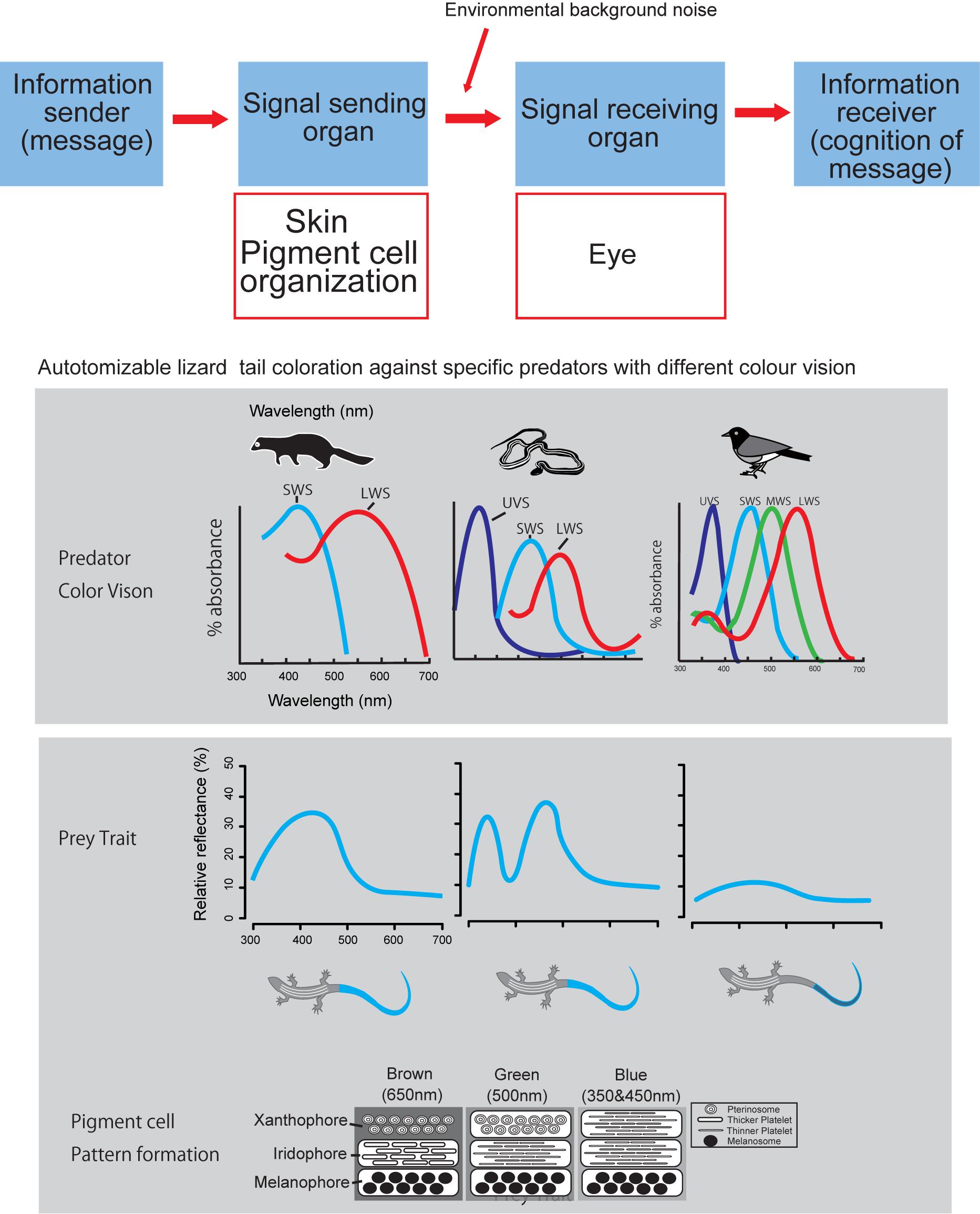
Figure 2. Logical relationship between color producing pigment cell organization and visual signal perception by eye, according to the Shannon – Weaver model of communication (Shannon and Weaver, 1948). Layered organization of pigment cells act as signal transmitter, and eye of predator act as signal receiver.
Admitting that predator’s visual perception is strictly important for understanding predator avoidance function of prey’s coloration (Endler, 1978), we would like to present a case study of lizard tail color which functions to deflect predator attacks from body to tail (Cooper and Vitt, 1985; Murali et al., 2018; Kodandaramaiah et al., 2020). Kuriyama et al. (2016a) showed that vividness of blue tail color was associated with the differences in color vision capabilities of lizard predator species, i.e., lizard tails with vivid blue reflectance evolved in communities with either weasel or snake predators, whereas, cryptic brown tail evolved independently on the islands where birds are the primary predator (Brandley et al., 2014). This review comes from our past effort of understanding mechanism of color pattern formation in reptiles, under Arnold’s conceptual framework of evolutionary biology (Arnold, 1983), with intent to obtain broader perspective of understanding function and performance of color pattern in the complex prey-predator interactions (Endler, 1978; Hämäläinen et al., 2015).
Elucidating the spatial and vertical architecture of pigment cells within skin tissues is a fundamental step to understand color pattern formation (Kuriyama et al., 2006; Saenko et al., 2013; Mallarino et al., 2016). It is necessary to not just describe the composition and spatial arrangement of pigment cells in adults, but also to characterize the processes of pigment cell specification, migration, and spatial organization during embryonic and postembryonic development (Kelsh et al., 2009; Kuriyama et al., 2013; Murakami et al., 2016, 2017). These analyses can provide insight into gene regulatory networks (GRNs) involved in color pattern formation (Mallarino et al., 2016). However, our current understanding of the color pattern formation in vertebrates is primarily based on the studies of mouse for a simple single pigment cell type (Mills and Patterson, 2009) and zebrafish for multiple pigment cell types (e.g., Ziegler, 2003; Kelsh, 2004; Parichy, 2006; Kelsh et al., 2009; Greenhill et al., 2011; Parichy and Spiewak, 2015; Petratou et al., 2018), and little is known for the developmental mechanisms of color pattern in reptiles.
In this paper, we review recent advances in our understanding of the roles of pigment cells in the production of various skin colors and patterns in reptilian sauropsids (Kuriyama et al., 2006, 2013, 2016a,b; Murakami et al., 2017; Alibardi, 2011, 2012, 2013, 2015; Kikuchi and Pfennig, 2012; Saenko et al., 2013; Kikuchi et al., 2014; Murakami et al., 2016, 2017; Szydłowski et al., 2016; Kuriyama and Hasegawa, 2017; Jindřich et al., 2019). We approach this topic from the viewpoint of theoretical studies of pattern formation (Murray and Myerscough, 1991; Chang et al., 2009; Allen et al., 2013) and molecular studies of gene systems involved in generating color patterns (Rosenblum et al., 2004; Manceau et al., 2010; Saenko et al., 2015; Irizarry and Bryden, 2016; Iwanishi et al., 2018). Owing to the lack of extensive genomic resources, early studies relied on sequence analyses of a few candidate genes, such as MC1R (Rosenblum et al., 2004; Cox et al., 2013). Positional cloning to identify mutations or genomic intervals harboring causal mutations requires both controlled pedigrees to generate recombinant mapping populations and a dense and large set of genetic markers that co-segregate with the phenotype of interest. Therefore, Saenko et al. (2015) and Tzika et al. (2015) adopted an unbiased next-generation sequencing and exome assembly approach, with extensive genotyping and candidate gene sequencing.
Pigment Cell–Based Understanding of Color and Pattern Formation
In reptiles, four basic types of pigment cells have been identified in the dermal skin: xanthophores, erythrophores, iridophores, and melanophores (Taylor and Hadley, 1970; Cooper and Greenberg, 1992; Morrison, 1995; Morrison et al., 1995; Bagnara and Matsumoto, 2006). The spatial arrangement and architecture of the pigment cells produce a variety of skin colors in reptiles (e.g., Kuriyama et al., 2013, 2017; Saenko et al., 2013; Murakami et al., 2016), tuatara (Alibardi, 2012), crocodiles (Alibardi, 2011), and chelonians (Alibardi, 2013; Brejcha et al., 2019). These layered organization of pigment cells (xanthophores on top, iridophores in the middle, and melanophores on the bottom) is conserved in zebrafish (Hirata et al., 2003) but not in mammals and birds (Olsson et al., 2013). An evolutionarily independent transition from the conserved core layering of multiple pigment cells to single melanophore layer is worth emphasizing, because it is well known but least understood phenomena in terms of genetic and molecular control (Bagnara et al., 1968; Grether et al., 2004; Hofreiter and Schöneberg, 2010).
For lizards and snakes, the color of a given patch of skin is determined not only by the static structural combination of pigment cells (Kuriyama et al., 2006), but also by dynamic interactions between chemical and physical parameters (Saenko et al., 2013; Teyssier et al., 2015). Xanthophores and erythrophores produce yellow to red coloration through the selective absorption of short wave length of light by pterinosomes containing pteridine and carotenoid vesicles in cells. Saenko et al. (2013) revealed that the pH or redox states reversibly change yellow xanthophores to red erythrophores and vice versa. Melanophores are light-absorbing pigment cells that produce black or brown colors. The melanophore density controls the lightness or darkness of skin. Iridophores are important light-reflecting cells containing light-reflecting platelets made of crystalline purines and pteridines, which generate structural colors by thin-layer interference and the scatter or diffraction of light from stacks of reflecting platelets (Huxley, 1968; Fujii, 1993; Bagnara and Matsumoto, 2006). The wavelength of light reflected by iridophores is determined by size, shape, orientation, number, and conformation of reflecting platelets and their cytoplasmic spacing (e.g., Land, 1972; Rohrlich and Porter, 1972; Morrison, 1995; Morrison et al., 1995; Kuriyama et al., 2006, 2016a, 2017; Saenko et al., 2013). Rohrlich (1974) noted that extensive filament networks in passive iridophores of Anolis lizards play a cytoskeletal function to maintain crystal sheets in their strict parallel array, while the motile filament system may mediate cellular changes by altering the array, spacing, or tilt of cellular crystals for rapidly changing colors due to dispersion/aggregation of pigment-containing organelles within dermal pigment cells. Recently, Teyssier et al. (2015) showed that chameleons can rapidly change their coloration through active tuning of a lattice of guanine nanocrystals within a superficial thick layer of dermal iridophores. Additionally, a group of iridophores with larger crystals layered in deeper dermis reflected the near-infrared range of light to produce red color. Therefore, the iridophores organized into two superposed layers is an evolutionarily novel mechanism functioning both for efficient camouflage and potentially avoiding passive thermal stress. Collectively, these complex interactions amongst pigment cells can not only produce various colors in the visible spectrum (Figure 3) but also provide various physiological functions.
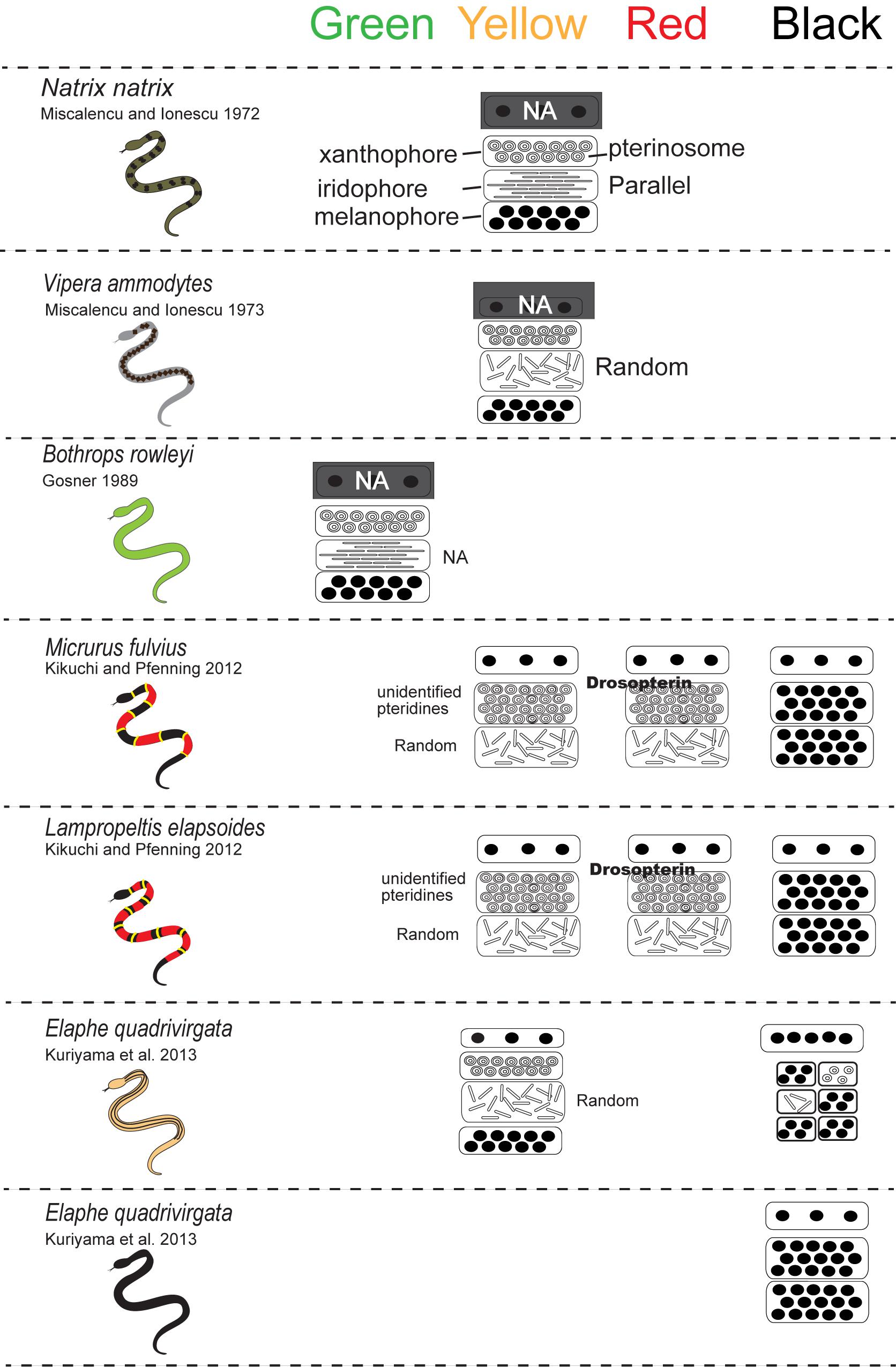
Figure 3. A schematic drawing of the vertical combination and relative thickness of epidermal and dermal pigment cells producing different skin coloration in snakes.
Recently, studies of lizard body color pattern formation revealed that, a quasi-hexagonal lattice of skin scales, rather than individual chromatophore cells, established a green and black labyrinthine pattern of skin color in ocellated lizards, and that this pattern was produced a cellular automaton that dynamically computed the color states of individual mesoscopic skin scales to produce the corresponding macroscopic color pattern (Manukyan et al., 2017), indicating that cellular automata can directly correspond to a continuous Turing reaction-diffusion system’s processes generated by biological evolution. However, due to our little understandings of physiological mechanisms controlling environmentally derived pigments such as carotenoids (Kikuchi et al., 2014), this review did little consideration of phenotypic plasticity in color patterns of squamates, admitting that there exist important topic of behavioral ecology and physiology in squamate reptiles (Pérez i de Lanuzaa et al., 2014).
Blue Tail Coloration and Stripe Formation in Lizards
Blue coloration of the lizard is produced by the vertical organization of iridophores with thin platelets in the uppermost layer and melanophores in the basal layer (Figure 4; Kuriyama et al., 2006; Bagnara et al., 2007). In the lizards with blue colored tail, iridophores with thin platelets are found only in skin tissues in the tail (Kuriyama et al., 2006, 2016a). Although the precise border between iridophores with thin or thick platelets have not yet been specified, it is possible that iridophore types radically shift along the longitudinal axis of the trunk and tail. During the embryonic development, iridophores with thin platelets appear in a specific region of the tail, and this position well coincides with the boundary between the blue coloration and brown coloration after hatching (Figure 5). The factors controlling the locations of iridophores with thinner or ticker platelets differ among populations of the same species and among species within the genus (Kuriyama et al., 2016a). After hatching, juveniles with a vivid blue tail gradually shifted to the adult coloration with a uniform brown tail, and the loss of blue tail during ontogeny is due to the change in iridophores with thin platelets into iridophores with thicker platelets and the appearance of xanthophores (Kuriyama et al., unpublished).
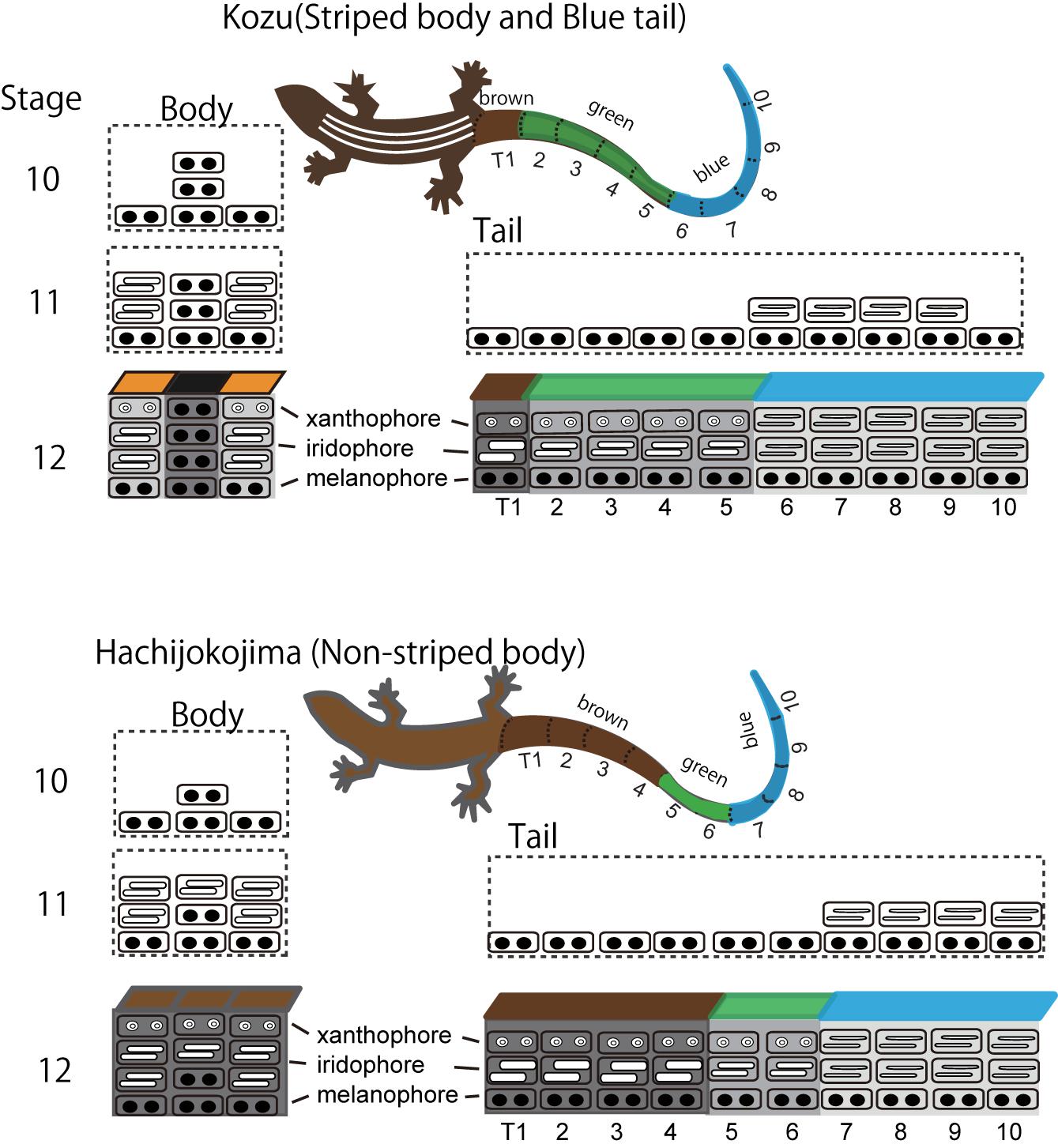
Figure 4. A schematic drawing of stripe pattern and tail color formation during embryogenesis in Plestiodon latiscutatus. The melanophore density in the black background region was higher in lizards with vivid stripes in Kozu island of Japan than in lizards with drab stripes in Hachijokojima island. Iridophores started to fill the dermal space that was not yet occupied by melanophores, which resulted in a higher iridophore density in stripes than in the inter-stripe regions. Iridophores with thin platelets reflecting blue structural coloration appeared at a specific region in the tail at stage 11. The position of the tail where iridophores emerged coincided with the boundary area separating anterior brown and green colored tail from posterior blue colored tail after hatching. Figure from Kuriyama and Hasegawa (2017) with permissions.

Figure 5. Dorso-lateral views of Plestiodon latiscutatus juveniles and full adult males from Kozu and Hachijokojima. (A) A juvenile in Kozu island of Japan has five yellowish-white longitudinal lines in the trunk and a green tail in the anterior half that transitions to blue in the posterior region. (B) A full adult male in Kozu is almost uniformly brown. (C) A juvenile in Hachijokojima island of Japan is almost uniformly brown, except for a blue tail tip. (D) A full adult male in Hachijokojima is almost uniformly brown.
For stripe pattern formation during embryogenesis of P. latiscutatus, melanophores initially appear before the appearance of iridophores and xanthophores to form stripe pattern (Kuriyama and Hasegawa, 2017), and iridophores subsequently occupy the area above the melanophore layers. Iridophores start to fill the dermal space that is not yet occupied by melanophores, resulting in a higher iridophore density in yellowish-white stripes than in the inter-stripe region. This observation suggests that mechanisms controlling the density gradient of melanophores during embryogenesis are crucial to understand GRNs involved in naturally occurring variation in stripe pattern formation.
The stripe pattern formation is a dynamic process also in the zebrafish (Singh et al., 2014), but the role of irridophores in pattern formation seems to differ between the zebrafish and the lizard. In the zebrafish, iridophore instead of melanophore proliferate, disperse and aggregate to organize the interstripe framework along the dorsoventral axis. However, it seems premature to conclude there there is a fundamental difference between fish and reptiles, even though fish and lizard irridophores shared same iridophore types with thin and thick platelets.
Iridophores differentiate from the NC cells during embryonic development (Bagnara et al., 1979), and the endoplasmic reticulum and Golgi apparatus, which produce platelets in iridophores, possibly play a certain role (Bagnara et al., 1979). Although the mechanisms controlling platelet thickness and orientation remain unknown, it seems possible to hypothesize that the initial volume of vesicles derived via budding from the endoplasmic reticulum and Golgi apparatus are responsible for the size of reflecting platelets (Figure 6). The process by which platelets are arranged is poorly understood at present, and additional observations of microtubule and/or microfilament formation in iridophores are needed; Rohrlich (1974) suggested that extensive filament networks in iridophores have cytoskeletal functions to maintain crystal sheets in a strict parallel array, while a motile filament system may mediate cellular changes by altering the array, spacing, or tilt of cellular crystals during embryonic development. Further comparative studies of various iridophore types with parallel or random and thick or thin platelets will be indispensable for inferring the genetic basis of reflecting platelet formation.
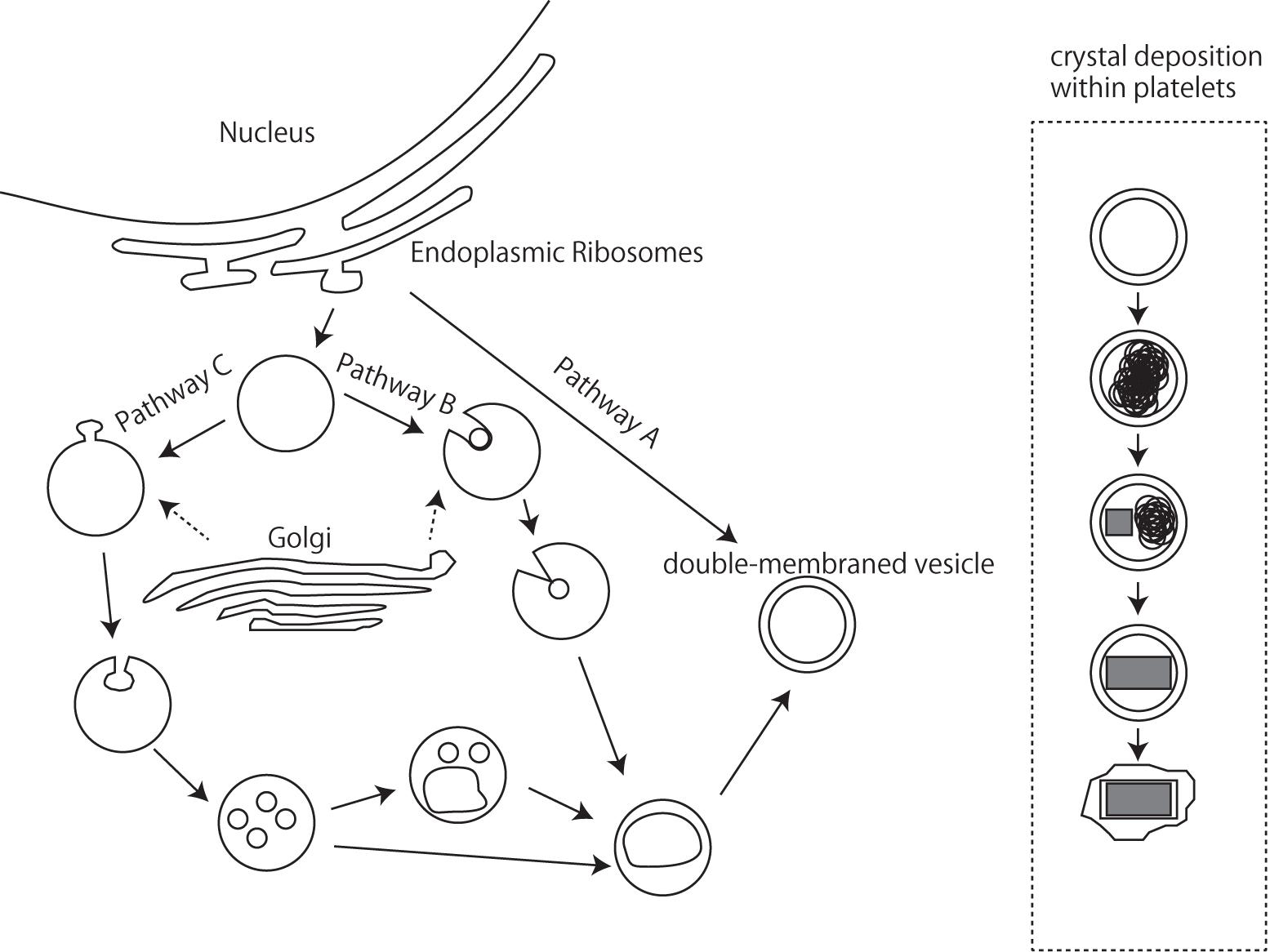
Figure 6. A schematic drawing of iridophore organellogenesis by Morrison and Frost-Mason, 1991. Three pathways of origin of the primordial double membraned vesicles and crystal deposition.
Stripe Pattern Formation in Snakes
The Japanese four-lined snake Elaphe quadrivirgata is polymorphic with striped, pale-striped, non-striped, banded, and melanistic morphs (Mori et al., 2005; Kuriyama et al., 2011, 2013). Vivid striped and non-striped morphs possess the same set of epidermal melanophores and dermal pigment cells (Figure 7, xanthophores at the top, iridophores in the middle, and melanophores at the bottom), but the spatial aggregation and concentrations of epidermal and dermal melanophores differ between the morphs in terms of the sharpness of the boundary between dark- and light-colored scales (Kuriyama et al., 2013). Even at hatching, stripe and non-stripe patterns are detectable (Murakami et al., 2014, 2016), indicating that the basic stripe pattern might be established during embryonic development (Murakami et al., 2016).
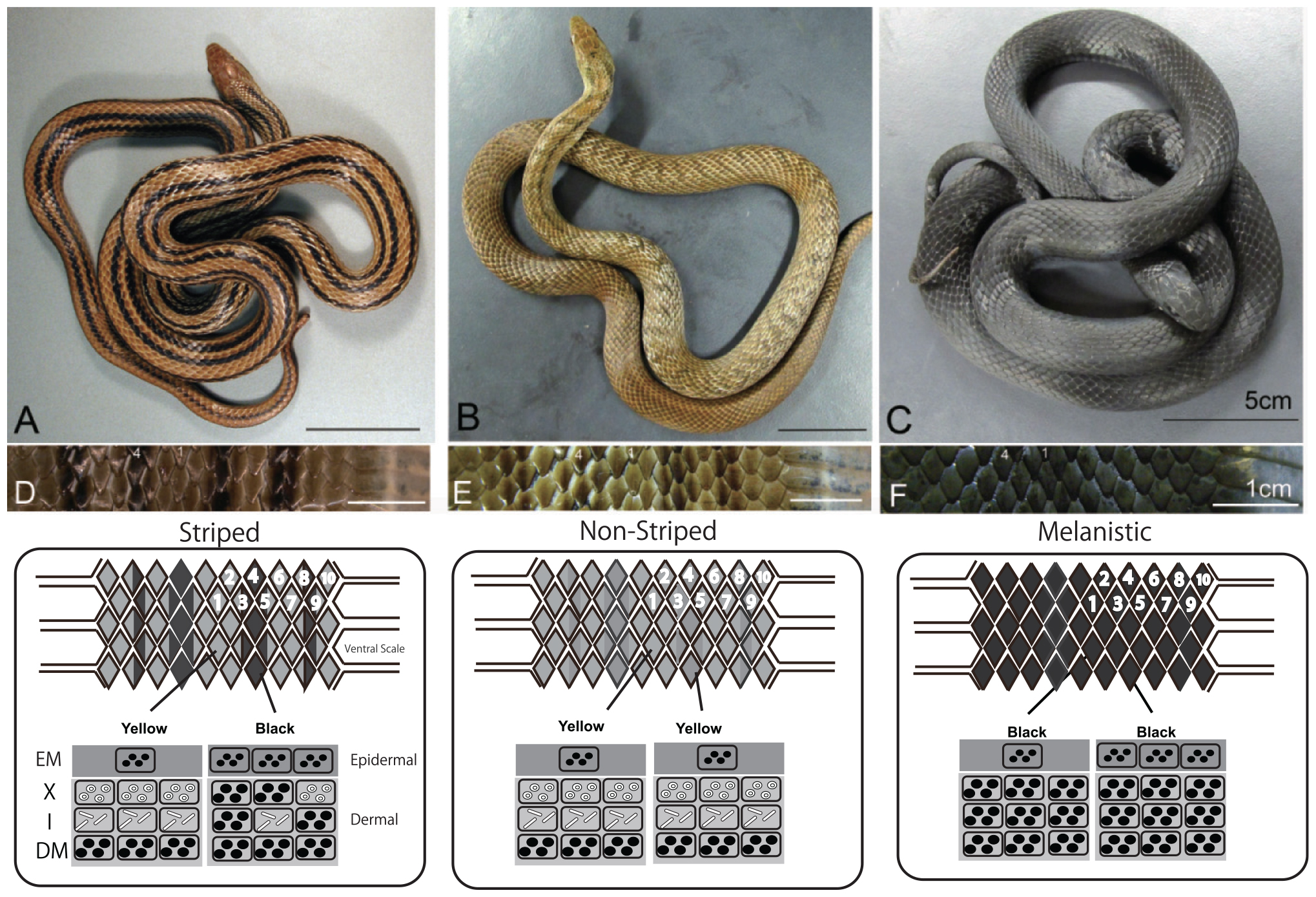
Figure 7. Dorso-lateral views and vertical combination of pigment cells of three-color morphs of Elaphe quadrivirgata. (A) The striped morph with four vivid dark-brown longitudinal stripes over yellowish-brown back ground. (B) The non-striped morph with a uniform brown or light-brown ground color without a trace of the stripe pattern except the region just behind head. (C) The uniformly jet black melanistic morph with no sign of stripe pattern. (D) Magnified view of the dorsal surface of a striped morph. Two dorsal scales along the lateral direction were selected as standard landmark scales to describe stripedness (No. 1) and background coloration (No. 4). (E) Magnified view of the dorsal surface of a non-striped morph. (F) Magnified view of the dorsal surface of a melanistic morph. A schematic drawing of the vertical combination and relative thickness of epidermal and dermal pigment cells producing different skin coloration and color pattern in Elaphe quadrivirgata. Number, shape, and size of pigment cells and organelles are simplified and exaggerated. EM, epidermal melanophore; X, xanthophore; I, iridophore; DM, dermal melanophore. Figure from Kuriyama et al. (2013) with permissions.
During embryonic development, the initial positions of melanophores do not correspond exactly to the positions of stripes (Murakami et al., 2017). Melanophores first appearing in the epidermis may form a precursor stripe pattern, and a region of highly dense melanophores in hatchlings follows these precursor stripes. An increase in the density gradient of epidermal melanophores proceeds the development of the stripe pattern due to dermal melanophores. A distinct stripe pattern is observed during late embryonic stages in association with the appearance of dermal melanophores. However, the higher density and the double stratum of dermal melanophores in the stripe region, which are characteristics of adult snakes (Kuriyama et al., 2013), are not developed by the time of hatching. Unlike the stripe pattern of Plestiodon lizards, which is fully developed at the time of hatching but decays during ontogeny (Kuriyama and Hasegawa, 2017), the snake stripe pattern continues to develop during ontogeny after hatching (Murakami et al., 2016). The density of dermal melanophores presumably increases to replace xanthophores and to form the double stratum in dark-brown scales during postembryonic growth.
Regarding the developmental timing of pigment cells in forming stripe pattern in snakes, there is a lesson from zebrafish. It was thought, for a long time, that melanophores are the first cell type to appear in the metamorphic skin from juvenile to adult zebrafish. Our observation did not pose question if melanophores are not the first cell type canalizing pattern formation in reptiles. However, advent of pigment-independent cell labeling techniques showed that xanthophores are the first cell type that cover the skin, followed by iridophores and then melanophores (Mahalwar et al., 2014). In order to determine that developmental timing of pattern formation in snakes, one has to utilize pigment-independent markers of cell types, if earlier appearance of xanthophores prior to melanophores was controlled pattern formation in the snakes too.
Color Pattern Polymorphisms
Studies of the mode of inheritance of particular color patterns in polymorphic species are important for exploring standing genetic variation contributing to amelanistic or melanistic coloration and stripe pattern polymorphism (Blanchard and Blanchard, 1940; Bechtel, 1978; Zweifel, 1981; Bechtel and Whitecar, 1983; Betchtel and Betchtel, 1989; King, 2003). A pigment cell–based understanding of color polymorphism facilitates the identification of types of pigment cell architectures corresponding to gains or losses of function to produce color and pattern polymorphisms. Higher order mutations, such as insertions of transposable elements, translocations, inversions, and recombination events, may contribute substantially to phenotypic variation, depending on their location in protein-coding regions or cis-regulatory DNA sequences (Wray, 2007; Wittkopp and Kalay, 2012). The precise roles of these events in the variation of color patterns in reptiles now have been revealing.
Albinism or amelanistic color variation was explained by a mutation that cause loss-of-function in Phelsuma guttata (Saenko et al., 2015). These mutations occur in the coding region of a cell surface protein receptor regulating inner cell pH. In particular, a retrotransposon insertion in an exon of OCA, similar to genetic variation in MC1R (responsible for the dysfunction of melanin synthesis), explains the whitish coloration in lizards living in white sand dunes (Rosenblum et al., 2004). A complete lack of tyrosinase gene due to a nonsense mutation in the coding region of the tyrosinase gene (TYR) in Elaphe climacophora (Iwanishi et al., 2018) explains another albinistic color variant. In these cases, color variation results from dysfunctions in melanin bio-synthesis. Interestingly, Saenko et al. (2015) hypothesized that even though melanin bio-synthesis is suppressed, melanophores without melanin can still play crucial roles in pattern formation.
Melanism is a typical variant of animal pigmentation (Kettlewell, 1973; Majerus, 1998; True, 2003), and reptilian melanism could be realized by one of two processes involving distinct pigment cell mechanisms: an increase in the relative amount of melanophores and a lack of pigment cells other than melanophores (Kuriyama et al., 2016b). As an example of an increase in the melanophore density in the skin layer, Morrison et al. (1995) studied the melanism of the iguanid lizard Sceloporus undulatus erythrocheilus, and revealed that melanophores are four times more abundant, producing darker skin than that of typical individuals. The complete dominance inheritance system with two alleles at one locus explain normal and melanistic morphs in T. sirtalis (King, 2003) and E. quadrivirgata (Tanaka, 2009). Because the melanistic morph has only epidermal and dermal melanophores and lacks dermal xanthophores and iridophores in E. quadrivirgata, it is reasonable to assume that the melanistic morph can be explained by the loss of function to differentiate xanthophores and iridophores. The underlying genetic alterations seem to occur in cis-regulatory elements within GRNs for pigment cell differentiation from chromatoblasts or melanophores to xanthophores and iridophores.
In the studies of striped and non-striped polymorphism, striped pattern is a dominant trait in the gopher snake Pituophis melanoleucus (Bechtel and Whitecar, 1983), the California king snake Lampropeltis getula californiae (Zweifel, 1981) and in the Japanese four-lined snake E. quadrivirgata. Because vivid striped and non-striped morphs possess the same set of epidermal and dermal pigment cells, Murakami et al. (2014) hypothesized that the alleles producing the striped morph were dominant under a co-dominance model of one locus with two alleles. Murakami et al. (2016, 2017) later proposed that the alleles of genes controlling the distribution and density of melanophores would additively influence individual variation in the vividness of stripes, and that the vividness of dorsal stripes is controlled by genes involved in the density gradient of melanophores at particular stages during embryonic development. This inheritance mode of the stripe and non-stripe polymorphism is compatible with the simulation result for the cell-chemotaxis model showing that a stronger chemotactic response generates a vivid longitudinal stripe pattern (Murray and Myerscough, 1991).
Pigment Cell Specification Gene Network and Evolution of Color Pattern Formation
To understand color pattern formation in reptiles with four types of melanocytes (i.e., epidermal melanophores, dermal melanophores, xanthophores, and iridophores), it seems instructive to consider GRNs controlling pattern formation and color production step by step. Pigment cells are differentiated from the NC, a multipotent and migratory cell population, along with other diverse derivatives, such as the facial skeleton and peripheral nervous system (Bagnara et al., 1979). Candidate GRNs for color pattern formation would include genes responsible for cell differentiation, proliferation, death, and localization (Chang et al., 2009; Olsson et al., 2013; Parichy and Spiewak, 2015). Therefore, in this section, we first summarize potential genetic mechanisms orchestrating pigment cell formation from NC cells, from the viewpoint of recognizing evolutionarily important GRNs and mutations in both coding and cis-regulatory DNA elements (Wray, 2007; Stern and Orgogozo, 2008; Wittkopp and Kalay, 2012). Later in this section, we consider theoretical analysis of pattern-generating mechanisms, such as cell-chemotaxis mechanisms (Oster and Murray, 1989; Murray and Myerscough, 1991), because it is promising to identify molecular components of the biological system by mathematical modeling to provide a basis for experimental studies and to refine and improve our knowledge of the mechanisms underlying pattern formation (Chang et al., 2009). Later in this section, potential roles of mutations in upstream GRNs for generating novel color phenotypes under domestication (Wilkins et al., 2014) will be reviewed for speculating evolutionary mechanism of color pattern evolution.
During embryonic development, Neural Crest cells undergo cytoskeletal rearrangements and morphological changes to lose cell–cell adhesion, allowing them to delaminate and emigrate from the neuroepithelium, with the concomitant acquisition of migratory ability, cell–surface receptors, metalloproteases, and adhesion molecules that allow them to respond properly to cell–cell interactions and environmental cues influencing migration pathways (reviewed in Sauka-Spengler and Bronner-Fraser, 2008). Although many kinds of GRNs have the potential to contribute to cell motility, cell–cell interactions, and cell population growth rates before the terminal differentiation of pigment cells, the core genetic mechanisms underlying the formation of NC cell derivatives are relatively uniform and conserved among vertebrate lineages. This is instructive for understanding characteristics of intermediate and terminally differentiated pigment cells essential for pattern formation. Based on zebrafish studies, the genes responsible for differentiation from NC cells into pigment and neural cells have been identified (Pavan and Raible, 2012). The subsequent specification of pigment cells and development of pigment cells occur in temporal order from melanophores, xanthophores, and iridophores (Ziegler, 2003). The regulation of pigment cell differentiation involves protein synthesis; transcription factor (TF) binding to cis-regulatory DNA sequences; the promotion or suppression of target gene expression, subsequent expression of TFs, signaling molecules, and cell surface receptor proteins; regulation of standing or activated signal transduction pathways; and the sequential production of pigment synthesis enzymes and other essential products for specific pigment cells. In zebrafish, the core GRNs governing melanocyte specification have been established (Greenhill et al., 2011; Pavan and Raible, 2012), and iridophore GRNs have recently been established by a systems biology approach (Higdon et al., 2013; Petratou et al., 2018). GRNs for xanthophores have not yet been fully established, but the development and patterning of pteridine synthesis and the regulation of the pteridine pathway and of its patterning are largely known in zebrafish (Ziegler, 2003). Consequently, we have established components and logical maps for GRNs of melanophore, iridophore, and xanthophore differentiation in zebrafish and in reptiles, given the high conservation of core GRNs for the same set of pigment cells.
A theoretical model was developed that explicitly considers actual cell movement, chemically mediated cell–cell interactions, and growth of the integumental domain during the patterning process (Murray and Myerscough, 1991). Pattern-generating mechanisms such as reaction-diffusion models (Turing, 1952), however, are unable to generate many common snake patterns, and other models are similarly insufficient to mimic snake pattern. A cell chemotaxis model has been developed considering chromatoblasts, which are NC progenitors; chemotaxis may be a factor in the migration of chromatoblasts to the dermis. According to the model, chromatoblasts both respond to and produce their own chemoattractant to promote the localization of differentiated cells in certain regions of skin. The model thus formalizes cell density in a given region as the product of chemically mediated interactions among dispersive and aggregative effects on cell motility and independent cell population growth rates. The chemoattractant produced by chromatoblasts diffuse and decay. Murray and Myerscough (1991) showed through numerical simulations that longitudinal stripes are most likely to form when both the chemotactic response (α) and cell population growth rate (r) are extremely high (Figure 8). This corresponds to the rapid production or slow diffusion or decay of the chemoattractant in a confined integumental domain. The lateral stripe is likely to form when the chemotactic response is weak. Chemically mediated cell motility (chemotaxis) and cell–cell interactions considered in the model thus provide a hypothetical basis for recognizing candidate GRNs for pattern formation. Both cell–cell and cell–environment interactions are thought to have important roles in directing immature pigment cell to appropriate sites. Assuming that the final transformation to mature pigment cells occurs after NC progenitors migrate to certain regions, GRNs responsible for color pattern formation will be those involved in the timing and speed of motility, sensitivity to chemical signals, cell population growth and death, and the strength of cell–cell and cell–environment interactions during migration from the NC to correct sites.
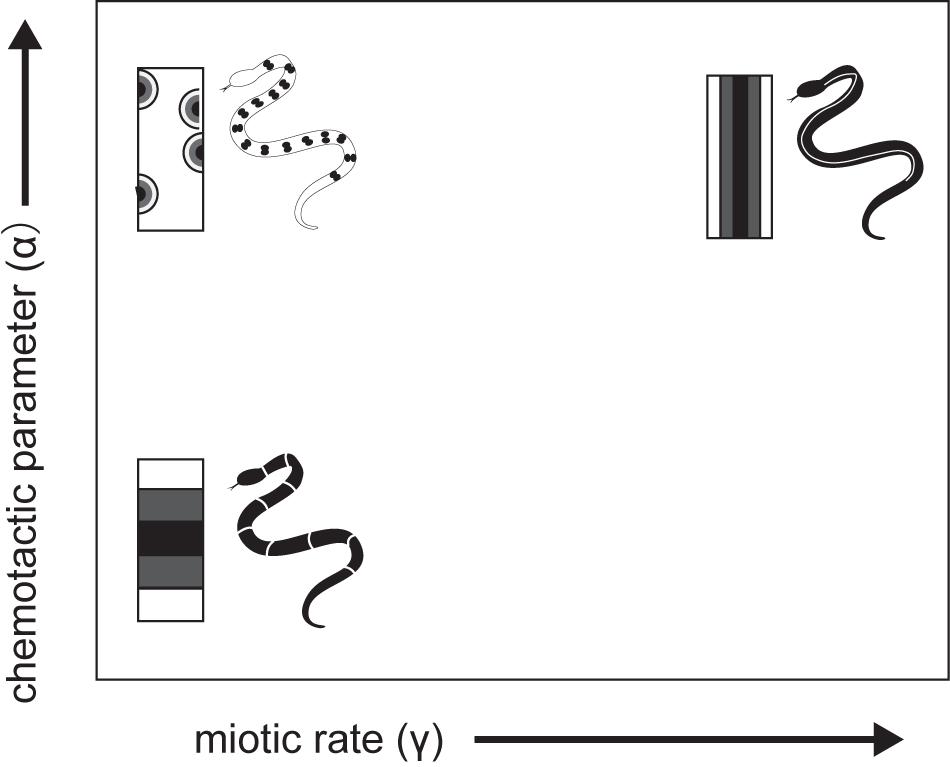
Figure 8. A schematic drawing of cell -chemotaxis model mechanism of snake dorsal color pattern by Murray and Myerscough (1991).
Deficiencies in upstream GRNs for melanophore specification are often related to skin cancer, neural diseases, or behavioral abnormalities (e.g., Jackson, 1994; Asher et al., 1996), implying that the evolution of color pattern would involve a balance between the removal of lethal mutations and neutral evolution or positive selection on non-lethal mutations in GRNs for the divergence or generation of novel color patterns. Along this line of thoughts, Wilkins et al. (2014) have hypothesized that the domestication syndrome in mammals such as a combination of behavioral tameness, pigmentation changes, and reduced facial skeleton and tooth size results predominantly from mild neural crest cell failure during embryonic development. Throat and ventral color polymorphism in lizards is often associated with behavioral, immune functional, or life history variation, which is advantageous or disadvantageous in an intraspecific social context, and color polymorphisms can evolve under the varieties of social and ecological selection regimes (Sinervo et al., 2001; Sinervo and Svensson, 2002), presumably due to mutations and subsequent correlational selection on GRNs affecting differentiation from NC cells to peripheral neurons and pigment cell progenitors upstream of GRNs for pigment cell differentiation. Similarly, non-lethal but likely adaptive mutations responsible for pattern formation may exist before the final transformation to mature pigment cells (Wilkins et al., 2014). The GRNs responsible for color pattern formation are expected to function in the timing and speed of motility, sensitivity to chemical signaling, cell population growth and death, and the strength of cell–cell and cell–environment interactions during migration from NC to correct sites (Sauka-Spengler and Bronner-Fraser, 2008), as discussed on the model of cell-chemotaxis (Murray and Myerscough, 1991).
Conclusion
According to the new perspective of the molecular mechanisms that regulate gene expression, we are increasingly appreciating that mutations affecting cis-regulatory sequences (as opposed to trans-regulatory sequences encoding TFs that bind to cis-regulatory sequences) are the most frequent cause of phenotypic divergence (Wray, 2007; Carroll, 2008; Stern and Orgogozo, 2008; Jones et al., 2012; Wittkopp and Kalay, 2012). Despite the significance in evolutionary and developmental biology, few studies have successfully connected changes in cis-regulatory and coding DNA with changes in pigment cells and color pattern variation. Advances in population genetic approaches for detecting either positive or balancing selection will facilitate the identification of genetic mechanisms underlying the fascinating adaptive color pattern divergence in reptiles.
Comparative studies for elucidating proximate mechanisms of color pattern convergence amongst the distantly related coral snake model and mimics (Kikuchi et al., 2014) and blue tail coloration amongst the closely related and distantly related groups of lizard species (Richmond and Reeder, 2002; Richmond, 2006) are the promising study projects to uncover gene regulatory networks (GRNs) for convergent phenotypic evolution. Under the extended conceptual framework linking GRN, developmental biology, morphology, performance and fitness, fruitful crops of evolutionary biology will be harvested from a sort of classic but long-term mark-release-recapture study of lizard and snake, associated with individually obtained genetic information, because lizards and snakes are particularly affordable animals for both field and laboratory studies (e.g., Sinervo and Svensson, 2002) for detecting natural selection played by the prey lizard and predator snakes.
Author Contributions
TK and MH wrote the manuscript and prepared the figures. AM and MB reviewed the manuscript before submission. TK, MH, and AM discussed the scope of review before stating the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review work was financially supported by the Japan Society for the Promotion of Science (JSPS) to MH (19H03307), and original field and laboratory studies were supported by the JSPS grants to MH (19570026, 21570024, 24570031, and 15H04426) and G. Morimoto (20770018), and the Sasakawa Scientific Research Grant from the Japan Science Society and the Fujiwara Natural History Foundation to TK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank H. Abe, S. Fukasawa, K. Furuse, H. Misawa, T. Mizuta, K. Osawa, T. Okamoto, H. Takahashi, H. Takeda, K. Tomizawa, H. Sanayama, H. Usuda, the members of the Laboratory of Geographical Ecology of Toho University, and T. Okamoto of Kyoto University for their field assistances, A. Mori, R. Fukuyama, and K. Fujishima for providing us the photos of coral snake and its mimics, and two reviewers for constructive criticism.
References
Alibardi, L. (2011). Histology, ultrastructure, and pigmentation in the horny scales of growing crocodilians. Acta Zool. 92, 187–200. doi: 10.1111/j.1463-6395.2010.00469.x
Alibardi, L. (2012). Cytology and localization of chromatophores in the skin of the Tuatara (Sphenodon punctaus). Acta Zool. 93, 330–337. doi: 10.1111/j.1463-6395.2011.00506.x
Alibardi, L. (2013). Observations on the ultrastructure and distribution of chromatophores in the skin of chelonians. Acta Zool. 94, 222–232. doi: 10.1111/j.1463-6395.2011.00546.x
Alibardi, L. (2015). Ultrastructural features of skin pigmentation in the lizard Heloderma suspectum with emphasis on xanto-melanophores. Acta Zool. 96, 154–159. doi: 10.1111/azo.12062
Allen, W. L., Baddeley, R., Scott-Samuel, N. E., and Cuthill, I. C. (2013). The evolution and function of pattern diversity in snakes. Behav. Ecol. 24, 1237–1250. doi: 10.1093/beheco/art058
Arnold, S. J. (1983). Morphology, performance and fitness. Am. Zool. 23, 347–361. doi: 10.1093/icb/23.2.347
Asher, J. H., Harrison, R. W., Morell, R., Carey, M. L., and Fridman, T. B. (1996). Effects of Pax3 modifier genes on craniofacial morphology, pigmentation, and viability: a murine model of Waardenburg syndrome variation. Genomics 34, 285–298. doi: 10.1006/geno.1996.0289
Bagnara, J. T., Fernandez, P. J., and Fujii, R. (2007). On the blue coloration of vertebrates. Pigm. Cell Res. 20, 14–26. doi: 10.1111/j.1600-0749.2006.00360.x
Bagnara, J. T., and Matsumoto, J. (2006). “Comparative anatomy and physiology of pigment cells in nonmammalian tissues,” in The Pigmentary System, Physiology and Pathophysiology, 2nd Edn, eds J. J. Nordlund, R. E. Boissy, V. J. Hearing, R. A. King, and J. Ortonne (New York, NY: Oxford University Press), 9–40.
Bagnara, J. T., Matsumoto, J., Ferris, W., Frost, S., Turner, W., Tchen, T., et al. (1979). Common origin of pigment cells. Science 203, 410–415. doi: 10.1126/science.760198
Bagnara, J. T., Taylor, J. D., and Hadley, M. E. (1968). The dermal chromatophore unit. J. Cell Biol. 38, 67–79. doi: 10.1083/jcb.38.1.67
Bechtel, E. R., and Whitecar, T. (1983). Genetics of striping in the gopher snake, Pituophis melanoleucus. J. Herpetol. 17, 362–370.
Betchtel, H. B., and Betchtel, E. R. (1989). Color mutations in the corn Snake (Elaphe guttata guttata): review and additional breeding data. J. Hered. 80, 272–276. doi: 10.1093/oxfordjournals.jhered.a110853
Blanchard, F., and Blanchard, F. (1940). The inheritance of melanism in the garter snake Thamnophis sirtalis sirtalis (Linnaeus) and some further evidence of effective autumn mating. Michigan Acad. Sci. Arts Lett. 26, 177–193.
Brandley, M. C., Kuriyama, T., and Hasegawa, M. (2014). Snake and bird predation drive the repeated convergent evolution of correlated life history traits and phenotype in the Izu Island scincid lizard (Plestiodon latiscutatus). PLoS One 9:e92233. doi: 10.1371/journal.pone.0092233
Brejcha, J., Bataller, J. V., Bosáková, Z., Geryk, J., Havlíková, M., Kleisner, K., et al. (2019). Body coloration and mechanisms of colour production in Archelosauria: the case of deirocheline turtles. R. Soc. open sci. 6:190319. doi: 10.1101/556670
Carroll, S. B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. doi: 10.1016/j.cell.2008.06.030
Chang, C., Wu, P., Baker, R. E., Maini, P. K., Alibardi, L., and Chuong, C. M. (2009). Reptile scale paradigm: Evo-Devo, pattern formation and regeneration. Int. J. Dev. Biol. 53, 813–826. doi: 10.1387/ijdb.072556cc
Cooper, W. E., and Greenberg, N. (1992). “Reptilian coloration and behavior,” in Biology of the Reptilia. Physiology E, Hormones, Brain, and Behavior, Vol. 18, eds C. Gans and D. Crews (Chicago, IL: University of Chicago Press), 298–422.
Cooper, W. E., and Vitt, L. J. (1985). Blue tails and autotomy: enhancement of predation avoidance in juvenile skinks. Z. Tierpsychol. 70, 265–276. doi: 10.1111/j.1439-0310.1985.tb00518.x
Cox, C. L., Rabosky, A. R., and Chippindale, R. T. (2013). Sequence variation in the Mc1r gene for a group of polymorphic snakes. Gene 513, 282–286. doi: 10.1016/j.gene.2012.10.065
Endler, J. A. (1978). A predator’s view of animal color patterns. Evol. Biol. 11, 319–364. doi: 10.1007/978-1-4615-6956-5_5
Endler, J. A. (1992). Signals, signal conditions, and the direction of evolution. Am. Nat. 139, s125–s153. doi: 10.1086/285308
Endler, J. A. (1993). The color of light in forests and its implications. Ecol. Monogr. 63, 1–27. doi: 10.2307/2937121
Fujii, R. (1993). Cytophysiology of fish chromatophores. Int. Rev. Cytol. 143, 191–255. doi: 10.1016/s0074-7696(08)61876-8
Gosner, K. L. (1989). Histological notes on the green coloration of arboreal pit vipers: genus Bothrops. J. Herpetol. 23, 318–320.
Greenhill, E. R., Rocco, A., Vibert, L., Nikaido, M., and Kelsh, R. N. (2011). An iterative genetic and dynamical modelling approach identifies novel features of the gene regulatory network underlying melanocyte development. PLoS Genet. 7:e1002265. doi: 10.1371/journal.pgen.1002265
Grether, G. F., Kolluru, G. R., and Nersissian, K. (2004). Individual color patches as multicomponent signals. Biol. Rev. 79, 583–610. doi: 10.1017/s1464793103006390
Hämäläinen, L., Valkonen, J., Mappes, J., and Rojas, B. (2015). Visual illusions in predator-prey interactions: birds find moving patterned prey harder to catch. Anim. Cogn. 18, 1059–1068. doi: 10.1007/s10071-015-0874-0
Higdon, C. W., Mitra, R. D., and Johnson, S. L. (2013). Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One 8:e67801. doi: 10.1371/journal.pone.0067801
Hirata, M., Nakamura, K., Kanemaru, T., Shibata, Y., and Kondo, S. (2003). Pigment cell organization in the hypodermis of zebrafish. Dev. Dyn. 227, 497–503. doi: 10.1002/dvdy.10334
Hofreiter, M., and Schöneberg, T. (2010). The genetic and evolutionary basis of colour variation in vertebrates. Cell. Mol. Life Sci. 67, 2591–2603. doi: 10.1007/s00018-010-0333-7
Huxley, A. F. (1968). A theoretical treatment of the reflection of light by multilayer structures. J. Exp. Biol. 48, 227–245.
Irizarry, K. J. L., and Bryden, R. L. (2016). In silico analysis of gene expression network components underlying pigmentation phenotypes in the Python identified evolutionarily conserved clusters of transcription factor binding sites. Adv. Bioinformatics 2016:1286510.
Iwanishi, S., Zaitsu, S., Shibata, H., and Nitasaka, E. (2018). An albino mutant of the Japanese rat snake (Elaphe climacophora) carries a nonsense mutation in the tyrosinase gene. Genes Genet. Syst. 93, 163–167. doi: 10.1266/ggs.18-00021
Jackson, I. J. (1994). Molecular and developmental genetics of mouse coat color. Annu. Rev. Genet. 28, 189–217. doi: 10.1146/annurev.ge.28.120194.001201
Jackson, J. F., Ingram, I. I. I. W., and Campbell, H. W. (1976). The dorsal pigmentation pattern of snakes as an antipredator strategy: a multivariate approach. Am. Nat. 110, 1029–1053. doi: 10.1086/283125
Jindřich, B., Jose Vicente, B., Bosáková, Z., Geryk, J., Havlíková, M., Kleisner, K., et al. (2019). Body coloration and mechanisms of colour production in Archelosauria: the case of deirocheline turtles6R. R. Soc. Open Sci. 6:190319. doi: 10.1098/rsos.190319
Jones, F. C., Grabherr, M. G., Chan, Y. F., Russell, P., Mauceli, E., Johnson, J., et al. (2012). The genomic basis of adaptive evolution in threespine stickleback. Nature 484, 55–61.
Kelsh, R. N. (2004). Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 17, 326–336. doi: 10.1111/j.1600-0749.2004.00174.x
Kelsh, R. N., Harris, M. L., Colanesi, S., and Erickson, C. A. (2009). Stripes and belly-spots—A review of pigment cell morphogenesis in vertebrates. Semin. Cell Dev. Biol. 20, 90–104. doi: 10.1016/j.semcdb.2008.10.001
Kettlewell, B. (1973). The Evolution of Melanism: The Study of a Recurring Necessity, with Special Reference to Industrial Melanism in the Lepidoptera. Oxford: Clarendon Press.
Kikuchi, D. W., and Pfennig, D. W. (2012). A Batesian mimic and its model share color production mechanisms. Curr. Zool. 58, 658–667. doi: 10.1093/czoolo/58.4.658
Kikuchi, D. W., Seymoure, B. M., and Pfenning, D. W. (2014). Mimicry’s palette:widespread use of conserved pigments in the aposematic signals of snakes. Evol. Dev. 16, 61–67. doi: 10.1111/ede.12064
King, R. B. (2003). Mendelian inheritance of melanism in the garter snake Thamnophis sirtalis. Herpetologica 59, 484–489. doi: 10.1655/02-93
Kodandaramaiah, U., Palathingal, S., Kurup, G. P., and Murali, G. (2020). What makes motion dazzle markings effective against predation? Behav. Ecol. 31, 43–53.
Kronforst, M. R., Barsh, G. S., Kopp, A., Mallet, J., Monteiro, A., Mullen, S. P., et al. (2012). Unraveling the thread of nature’s tapestry: the genetics of diversity and convergence in animal pigmentation. Pigment Cell Melanoma Res. 25, 411–433. doi: 10.1111/j.1755-148x.2012.01014.x
Kuriyama, T., Brandley, M. C., Katayama, A., Mori, A., Honda, M., and Hasegawa, M. (2011). A time-calibrated phylogenetic approach to assessing the phylogeography, colonization history and phenotypic evolution of snakes in the Japanese Izu Islands. J. Biogeogr. 38, 259–271. doi: 10.1111/j.1365-2699.2010.02403.x
Kuriyama, T., Esashi, J., and Hasegawa, M. (2017). Light reflection from crystal platelets in iridophores determines green or brown skin coloration in Takydromus lizards. Zoology 121, 83–90. doi: 10.1016/j.zool.2016.11.003
Kuriyama, T., and Hasegawa, M. (2017). Embryonic developmental process governing the conspicuousness of body stripes and blue tail coloration in the lizard Plestiodon latiscutatus. Evol. Dev. 19, 29–39. doi: 10.1111/ede.12214
Kuriyama, T., Misawa, H., Miyaji, K., Sugimoto, M., and Hasegawa, M. (2013). Pigment cell mechanisms underlying dorsal color-pattern polymorphism in the Japanese four-lined snake. J. Morphol. 274, 1353–1364. doi: 10.1002/jmor.20182
Kuriyama, T., Miyaji, K., Sugimoto, M., and Hasegawa, M. (2006). Ultrastructure of the dermal chromatophores in a lizard (Scincidae: Plestiodon latiscutatus) with conspicuous body and tail coloration. Zool. Sci. 23, 793–799. doi: 10.2108/zsj.23.793
Kuriyama, T., Morimoto, G., Miyaji, K., and Hasegawa, M. (2016a). Cellular basis of anti-predator adaptation in a lizard with autotomizable blue tail against specific predators with different colour vision. J. Zool. 300, 89–98. doi: 10.1111/jzo.12361
Kuriyama, T., Okamoto, T., Miyaji, K., and Hasegawa, M. (2016b). Iridophore- and xanthophore-deficient melanistic color variant of the lizard Plestiodon latiscutatus. Herpetologica 72, 189–195. doi: 10.1655/herpetologica-d-15-00022
Land, M. F. (1972). The physics and biology of animal reflectors. Prog. Biophys. Mol. Biol. 24, 75–106. doi: 10.1016/0079-6107(72)90004-1
Mahalwar, P., Walderich, B., Singh, A. P., and Nusslei-Volhard, C. (2014). Local recorganizationof xanthophores fine-tunes and colors the striped pattern of zebrafish. Science 345, 1362–1364. doi: 10.1126/science.1254837
Mallarino, R., Henegar, C., Mirasierra, M., Manceau, M., Schradin, C., Vallejo, M., et al. (2016). Developmental mechanisms of stripe patterns in rodents. Nature 539, 518–523. doi: 10.1038/nature20109
Manceau, M., Domingues, V. S., Linnen, C. R., Rosenblum, E. B., and Hoekstra, H. E. (2010). Convergence in pigmentation at multiple levels: mutations, genes and function. Philos. Trans. R. Soc. B Biol. Sci. 365, 2439–2450. doi: 10.1098/rstb.2010.0104
Manukyan, L., Montandon, S. A., Fofonjka, A., Smirnov, S., and Milinkovitch, M. C. (2017). A living mesoscopic cellular automaton made of skin scales. Nature 544, 173–179. doi: 10.1038/nature22031
McKinnon, J. S., and Pierotti, M. E. R. (2010). Colour polymorphism and correlated characters: genetic mechanisms and evolution. Mol. Ecol. 19, 5101–5125. doi: 10.1111/j.1365-294x.2010.04846.x
Mills, M. G., and Patterson, L. B. (2009). Not just black and white: pigment pattern development and evolution in vertebrates. Semin. Cell Dev. Biol. 20, 72–81. doi: 10.1016/j.semcdb.2008.11.012
Miscalencu, D., and Ionescu, M. D. (1972). Fine structure of dermal chromatophores in the Natrix natrix (L.) snake. Anat. Anz. 131, 470–475.
Miscalencu, D., and Ionescu, M. D. (1973). Fine-structure of epidermis and dermal chromatophores in Vipera ammodytes (L.). Acta Anat. 86, 111–122. doi: 10.1159/000144113
Mori, A., Tanaka, K., Moriguchi, H., and Hasegawa, M. (2005). Color variations in Elaphe quadrivirgata throughout Japan. Bull. Herpetol. Soc. Jpn. 2005, 22–38. doi: 10.14880/hrghsj1999.2005.22
Morrison, R. L. (1995). A transmission electron microscopic (TEM) method for determining structural colors reflected by lizard iridophores. Pigment Cell Res. 8, 28–36. doi: 10.1111/j.1600-0749.1995.tb00771.x
Morrison, R. L., and Frost-Mason, S. K. (1991). Ultrastructure analysis of iridophore organellogenesis in a lizard, Sceloporus graciosus (Reptilia, Phynosomatidae). J. Morphol. 209, 229–239. doi: 10.1002/jmor.1052090209
Morrison, R. L., Rand, M. S., and Frost-Mason, S. K. (1995). Cellular basis of color differences in three morphs of the lizard Sceloporus undulatus erythrocheilus. Copeia 1995, 397–408.
Murakami, A., Hasegawa, M., and Kuriyama, T. (2014). Identification of Juvenile Color Morphs for Evaluating Heredity Model of Stripe/Non-stripe Pattern Polymorphism in Japanese four-lined snake Elaphe quadrivirgata. Curr. Herpetol. 33, 68–74. doi: 10.5358/hsj.33.68
Murakami, A., Hasegawa, M., and Kuriyama, T. (2016). Pigment cell mechanism of postembryonic stripe pattern formation in the Japanese four-lined snake. J. Morphol. 277, 196–203. doi: 10.1002/jmor.20489
Murakami, A., Hasegawa, M., and Kuriyama, T. (2017). Developmental mechanisms of longitudinal stripes in the Japanese four-lined snake. J. Morphol. 279, 27–36. doi: 10.1002/jmor.20750
Murali, G., Merilait, S., and Kodandaramaiah, U. (2018). Grab my tail: evolution of dazzle stripes and colourful tails in lizards. J. Evol. Biol. 31, 1675–1688. doi: 10.1111/jeb.13364
Murray, J. D., and Myerscough, M. R. (1991). Pigmentation pattern formation on snakes. J. Theor. Biol. 149, 339–360. doi: 10.1016/s0022-5193(05)80310-8
Olsson, M., Stuart-Fox, D., and Ballen, C. (2013). Genetics and evolution of colour patterns in reptiles. Semin. Cell Dev. Biol. 24, 529–541. doi: 10.1016/j.semcdb.2013.04.001
Oster, G. F., and Murray, J. D. (1989). Pattern formation models and developmental constraints. J. Exp. Zool. 251, 186–202. doi: 10.1002/jez.1402510207
Parichy, D. M. (2006). Evolution of danio pigment pattern development. Heredity 97, 200–210. doi: 10.1038/sj.hdy.6800867
Parichy, D. M., and Spiewak, J. E. (2015). Origins of adult pigmentation: diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell Melanoma Res. 28, 31–50. doi: 10.1111/pcmr.12332
Pavan, W. J., and Raible, D. W. (2012). Specification of neural crest into sensory neuron and melanocyte lineages. Dev. Biol. 366, 55–63. doi: 10.1016/j.ydbio.2012.02.038
Pérez i de Lanuzaa, G., Carazoa, P., and Font, E. (2014). Colours of quality: structural (but not pigment) coloration informs about male quality in a polychromatic lizard. Anim. Behav. 90, 73–81. doi: 10.1016/j.anbehav.2014.01.017
Petratou, K., Subkhankulova, T., Lister, J. A., Rocco, A., Schwetlick, H., Kelsh, R. N., et al. (2018). A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genet. 14:e1007402. doi: 10.1371/journal.pgen.1007402
Pfennig, D. W., Harcombe, W. R., and Pfennig, K. S. (2001). Frequency-dependent Batesian mimicry. Nature 410:323. doi: 10.1038/35066628
Pianka, E. R., and Vitt, L. J. (2003). Lizards: Windows to the Evolution of Diversity. Berkeley, CA: University of California Press.
Richmond, J. Q. (2006). Evolutionary basis of parallelism in North American scincid lizards. Evol. Dev. 8, 477–490. doi: 10.1111/j.1525-142x.2006.00121.x
Richmond, J. Q., and Reeder, T. W. (2002). Evidence for pallalel ecological speciation in scinced lizards of the Eumeces skiltonianus species group (Squamata: Sincidae). Evolution 56, 1498–1513. doi: 10.1111/j.0014-3820.2002.tb01461.x
Rohrlich, S. T. (1974). Fine structural demonstration of ordered arrays of cytoplasmic filaments in vertebrate iridophores, a comparative survey. J. Cell Biol. 62, 295–304. doi: 10.1083/jcb.62.2.295
Rohrlich, S. T., and Porter, K. R. (1972). Fine structural observations relating to the production of color by the iridophores of a lizard, Anolis carolinensis. J. Cell Biol. 53, 38–52. doi: 10.1083/jcb.53.1.38
Rosenblum, E. B., Hoekstra, H. E., and Nachman, M. W. (2004). Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution 58, 1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x
Ruxton, G. D., Allen, W. L., Speed, M. P., and Sherratt, T. N. (2018). Avoiding Attack: the Evolutionary Ecology of Crypsis, Aposematism, and Mimicry. Oxford: Oxford University Press.
Saenko, S. V., Lamichhaney, S., Barrio, A. M., Rafati, N., Andersson, L., and Milinkovitch, M. C. (2015). Amelanism in the corn snake is associated with the insertion of an LTR-retrotransposon in the OCA2 gene. Sci. Rep. 5:17118.
Saenko, S. V., Teyssier, J., van der Marel, D., and Milinkovitch, M. (2013). Precise co-localization of interacting structural and pigmentary elements generates extensive color pattern variation in Phelsuma lizards. BMC Biol. 11:105. doi: 10.1186/1741-7007-11-105
Sauka-Spengler, T., and Bronner-Fraser, M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568. doi: 10.1038/nrm2428
Shannon, C. E. W., and Weaver, W. (1948). The Mathematical Theory of Communication. Champaign, IL: University of Illinois press.
Sinervo, B., Bleay, C., and Adamopoulou, C. (2001). Social causes of correlational selection and the resolution of a heritable throat color polymorphism in a lizard. Evolution 55, 2040–2052. doi: 10.1111/j.0014-3820.2001.tb01320.x
Sinervo, B., and Svensson, E. (2002). Correlational selection and the evolution of genomic architecture. Heredity 89, 329–338. doi: 10.1038/sj.hdy.6800148
Singh, A. P., Schach, U., and Nüsslein-Volhard, C. (2014). Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat. Cell Biol. 16, 604–611. doi: 10.1038/ncb2955
Stern, D. L., and Orgogozo, V. (2008). The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x
Stevens, M., Yule, D. H., and Ruxton, G. D. (2008). Dazzle coloration and prey movement. Proc. Biol. Sci. 275, 2639–2643. doi: 10.1098/rspb.2008.0877
Stuart-Fox, D., Moussalli, A., and Whiting, M. J. (2008). Predator-specific camouflage in chameleons. Biol. Lett. 4, 326–329. doi: 10.1098/rsbl.2008.0173
Szydłowski, P., Madej, J. P., and Mazurkiewicz-Kania, M. (2016). Ultrastructure and distribution of chromatophores in the skin of the leopard gecko (Eublepharis macularius). Acta Zool. 97, 370–375. doi: 10.1111/azo.12132
Tanaka, K. (2009). Function of color pattern and adaptive significance of color polymorphism in snakes. Jpn. J. Herpetol. 2009, 175–187.
Taylor, J. D., and Hadley, M. E. Z. (1970). Chromatophores and color change in the lizard, Anolis carolinensis. Zellforsch. Mikrosk. Anat. 104, 282–294. doi: 10.1007/bf00309737
Teyssier, J., Saenko, S. V., Van der Marel, D., and Milinkovitch, M. C. (2015). Photonic crystals cause active colour change in chameleons. Nat. Commun. 6:6368.
Thayer, G. H. (1909). Concealing-Coloration in the Animal Kingdom; An Exposition of the Laws of Disguise Through Color and Pattern: Being a Summary of Abbott H. Thayer’s Discoveries. New York, NY: Macmillan.
True, J. R. (2003). Insect melanism: the molecules matter. Trends Ecol. Evol. 18, 640–647. doi: 10.1016/j.tree.2003.09.006
Turing, A. M. (1952). The chemical basis for morphogenesis. Philos. Trans. R. Soc. Lond. B 237, 37–72.
Tzika, A. C., Ullate-Agote, A., Grbic, D., and Milinkovitch, M. C. (2015). Reptilian transcriptome v2.0: an extensive resource for sauropsida genomics and transcriptomics. Genome Biol. Evol. 7, 1827–1841. doi: 10.1093/gbe/evv106
Wallace, A. R. (1867). Mimicry, and Other Protective Resemblances Among Animals. (Vancouver: Read Books), 1–27.
Wilkins, A. S., Wrangham, R. W., and Fitch, W. T. (2014). The “domestic syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197, 795–808. doi: 10.1534/genetics.114.165423
Wittkopp, P. J., and Kalay, G. (2012). Cis-regulatory elements molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wray, G. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216. doi: 10.1038/nrg2063
Ziegler, I. (2003). The pteridine pathway in Zebrafish: regulation and Specification during the Determination of Neural Crest Cell-Fate. Pigment Cell Res. 16, 172–182. doi: 10.1034/j.1600-0749.2003.00044.x
Keywords: pigment cells, embryogenesis, polymorphism, melanism, reptile
Citation: Kuriyama T, Murakami A, Brandley M and Hasegawa M (2020) Blue, Black, and Stripes: Evolution and Development of Color Production and Pattern Formation in Lizards and Snakes. Front. Ecol. Evol. 8:232. doi: 10.3389/fevo.2020.00232
Received: 13 December 2019; Accepted: 25 June 2020;
Published: 21 August 2020.
Edited by:
Marcus Kronforst, The University of Chicago, United StatesReviewed by:
David Pfennig, The University of North Carolina at Chapel Hill, United StatesAjeet Pratap Singh, Novartis Institutes for BioMedical Research, United States
Copyright © 2020 Kuriyama, Murakami, Brandley and Hasegawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masami Hasegawa, bWhhc2VAYmlvLnNjaS50b2hvLXUuYWMuanA=
 Takeo Kuriyama
Takeo Kuriyama Arata Murakami2,3
Arata Murakami2,3 Masami Hasegawa
Masami Hasegawa